- 1Jiangsu Province Hospital on Integration of Chinese and Western Medicine, Nanjing, China
- 2Nanjing University of Chinese Medicine, Nanjing, China
Background: Renal cancer (RC) is one of the most common malignant tumors of the urinary system, and molecular targets for the specific diagnosis and treatment of RC have been widely explored. The purpose of this study was to systematically analyze circular RNAs (circRNAs), which may serve as novel tumor markers in terms of the diagnosis, prognosis and clinicopathological characteristics of RC.
Methods: PubMed and Web of Science were systematically searched for literature as up to July 30, 2021. All included studies were evaluated by the evaluation system, and the results were satisfactory. Hazard ratios (HRs) and odds ratios (ORs) were used to assess the association of circRNAs with diagnostic and clinicopathological indicators. The sensitivity (SEN), specificity (SPE), positive likelihood ratio, negative likelihood ratio, diagnostic odds ratio and area under the summary receiver operating characteristic curve (AUC) were combined to evaluate the diagnostic performance of circRNAs in RC.
Results: We included 22 studies that met the criteria, including 18 that were prognostic, 4 that were diagnostic, and 12 that were clinicopathologically relevant. In terms of prognosis, we found that upregulated circRNAs were positively associated with poor overall survival in patients with RC (HR=1.63, 95% CI=1.43–1.85). In terms of diagnosis, the combined SEN, SPE and AUC of circRNAs in the diagnosis of RC were 0.82, 0.84 and 0.89 (0.86–0.91), respectively. In terms of clinicopathological features, upregulated circRNAs were associated with the Fuhrman grade (OR=0.641, 95% CI=0.471–0.873), T stage (OR=0.236, 95% CI=0.141–0.396), TNM stage (OR=0.225, 95% CI=0.158–0.321) and lymphatic metastasis (OR=0.329, 95% CI=0.193–0.560).
Conclusion: Our meta-analysis confirms that circRNAs may be candidate biomarkers for the diagnosis, prognosis, and clinicopathological indicators of RC.
1 Introduction
Among tumors of the urinary system, renal cancer (RC) is characterized by a high incidence, high degree of malignancy, and poor prognosis (1, 2). According to the 2004 World Health Organization classification, RC can be divided into 10 types, including renal cell carcinoma (RCC), the most common type (3, 4). Distant metastases are found in approximately 1/3 of patients with RCC at the first visit, and the 5-year survival rate for patients with advanced RC is only approximately 20% (5). At present, the diagnosis of RC mainly depends on imaging, but it is difficult to distinguish it from a renal cyst or renal hamartoma in the early stage. In addition, the lack of specific serum markers makes early diagnosis more difficult (6). In terms of treatment, although targeted drugs, including sunitinib, have shown some success in treating RC, resistance remains a key issue to be addressed (7). Therefore, exploring the molecular basis and mechanism of the occurrence and development of RC, actively studying high-quality diagnostic markers of RC, and providing accurate therapeutic targets and prognostic indicators have become the primary tasks of current RC research.
Circular RNAs (circRNAs)—a particular type of endogenous noncoding RNA—were first identified in viruses in the 1990s (8). Initially, circRNAs were considered to be the product of missplicing. With the continuous progress of sequencing technology and molecular purification technology, an increasing number of studies have proven that circRNAs play different roles in the regulation of a variety of cell activities (9–12). Especially in recent years, a large number of experiments have proven that circRNAs play an important role in many fields, especially in tumor development. Many studies have demonstrated that different subtypes of circRNAs expressed in different tumors can play different inhibitory or promotive roles. In pancreatic cancer (PC), Yang et al. found that circRHOBTB3 was highly expressed in PC tissues and positively correlated with clinicopathological data, such as the clinical prognosis of patients. In terms of mechanism, circRNAs can promote the autophagy of PC cells and regulate the proliferation of PC cells by competitively binding miR-600 to upregulate NACC1. Similarly, a large number of circRNAs have been studied in RC (13, 14). For example, Cen et al. used bioinformatics to screen out the high expression of circSDHC in RC tissue. In subsequent clinical sample validation, it was further found that high circSDHC expression was closely associated with a poor patient prognosis and the TNM stage. Mechanistically, circSDHC can promote the proliferation and metastasis of RC cells in vivo and in vitro by downregulating miR-127-3p to promote the CDKN3/E2F1 signaling pathway (15). In addition, Frey et al. investigated the clinical value of 7 circRNAs in patients with RC. The results showed that circEGLN3, circEHD2, and circNETO2 may be used for the diagnosis of RC. Interestingly, high and low circEHD2 expression were also found to be independent predictors of the postoperative prognosis in patients with RC (16).
To determine whether circRNAs promote or inhibit RC, we summarized the results of different circRNA subtypes in RC based on current studies. Different software programs were used to meta-analyze the correlation of RC-related circRNAs with prognosis, diagnosis, and clinicopathology to explore the possible role and value of circRNAs in RC and to provide a reliable basis for the early diagnosis and precise treatment of RC in the future.
2 Materials and Methods
2.1 Literature Search Strategy
The literature search used in this paper was carried out in accordance with the preferred reporting items of the PRISMA statement standard. The article ran a search of related English language articles through PubMed and Web of Science until 30 July 2021 using the following related terms: (a) “renal carcinoma” or “kidney cancer” or “renal cancer” or “kidney neoplasm” or “renal neoplasm”; and (b) “circular RNA” or “circ RNA”. The literature search is performed independently by two researchers (WW and SX), who make appropriate assessments and data extraction. If there is any disagreement, discuss and decide with the third researcher (FG).
2.2 Inclusion Criteria and Exclusion Criteria
Studies concerning the value of circRNA expression in terms of the prognosis, diagnosis or clinicopathological characteristics of RC were eligible for quantitative synthesis. The inclusion criteria were as follows: (a) case-control study design; (b) diagnosis with RC by histopathology; and (c) confirmation of the relationship between circRNAs and the prognosis, diagnosis and clinicopathological characteristics of RC. The exclusion criteria were as follows: (a) ambiguous data or insufficient data leading to a study being defined as having unsuitable statistics; (b) reviews, comments, letters, or case reports; and (c) work unrelated to RC or circRNAs.
2.3 Data Extraction and Quality Assessment
The two researchers (WW and SX) separately extracted data from the included studies based on uniform criteria. Data extracted in terms of prognosis included the following: name of the first author; name of the circRNA; year of publication; country of origin; expression level in RC; cut-off value; sample size; detected sample; detection method; follow-up time; survival outcome; and survival analysis and hazard ratio (HR) with 95% confidence interval (CI) for overall survival (OS), progression-free survival (PFS), cancer-specific survival (CCS), metastasis-free survival (MFS) or disease-free survival (DFS). Data extracted in terms of diagnosis included the following: odds ratio (OR) and 95% CI used to summarize the above information and evaluate the accuracy of the diagnosis by the true positive (TP), false positive (FP), false negative (FN), true negative (TN) and area under the receiver operating characteristic (ROC) curve (AUC) in each study to synthesize the sensitivity (SEN), specificity (SPE), positive likelihood ratio (PLR), negative likelihood ratio (NLR) and diagnostic odds ratio (DOR). Data extracted in terms of clinicopathological characteristics included the following: gender; age; tumor size; Fuhrman grade; T stage; lymphatic metastasis; M stage; and TNM stage.
2.4 Statistical Analysis
The full-text data were statistically analyzed using Stata software (version 15.1). Each article was graded by Review Manager software (version 5.4), and only those meeting the criteria were included. In relation to the prognosis of patients, we mainly referred to the OS, PFS, CCS, MFS, and DFS curves in the study and extracted the relevant HRs and 95% CIs. The HRs and 95% CIs can usually be directly accessed from the study. When the prognosis was described as a Kaplan-Meier curve, we extracted the corresponding data using Engauge Digitizer version 4.1 and calculated the corresponding HRs and 95% CIs (17). In relation to the diagnosis of patients, the TP, TN, FP, and FN values were calculated from the known SEN and SPE values of the ROC curves using GetData Graph Digitizer software, and the NLR, PLR and DOR required for the results were further calculated (18). In relation to clinicopathological correlations, we used ORs and 95% CIs to analyze the clinical value of circRNAs associated with RC. An OR and 95% CI both > 1 suggested a positive correlation between the clinicopathological indicator and circRNA expression; otherwise, a negative correlation was indicated. In the heterogeneity test, I2 and Q tests were used to analyze the existence of heterogeneity. If I2>50% or P <0.1, heterogeneity was indicated. When the heterogeneity was obvious, a random-effects model was used; otherwise, a fixed-effects model was used (19). At the same time, we conducted a sensitivity analysis of all the included studies and excluded each individual study to compare its influence on the overall effect of the results of the meta-analysis. All tests were two-sided, and P < 0.05 was considered statistically significant. Publication bias was quantified by Egger’s test and Begg’s test and reflected by funnel plot analysis.
3 Results
3.1 Literature Retrieval and Screening
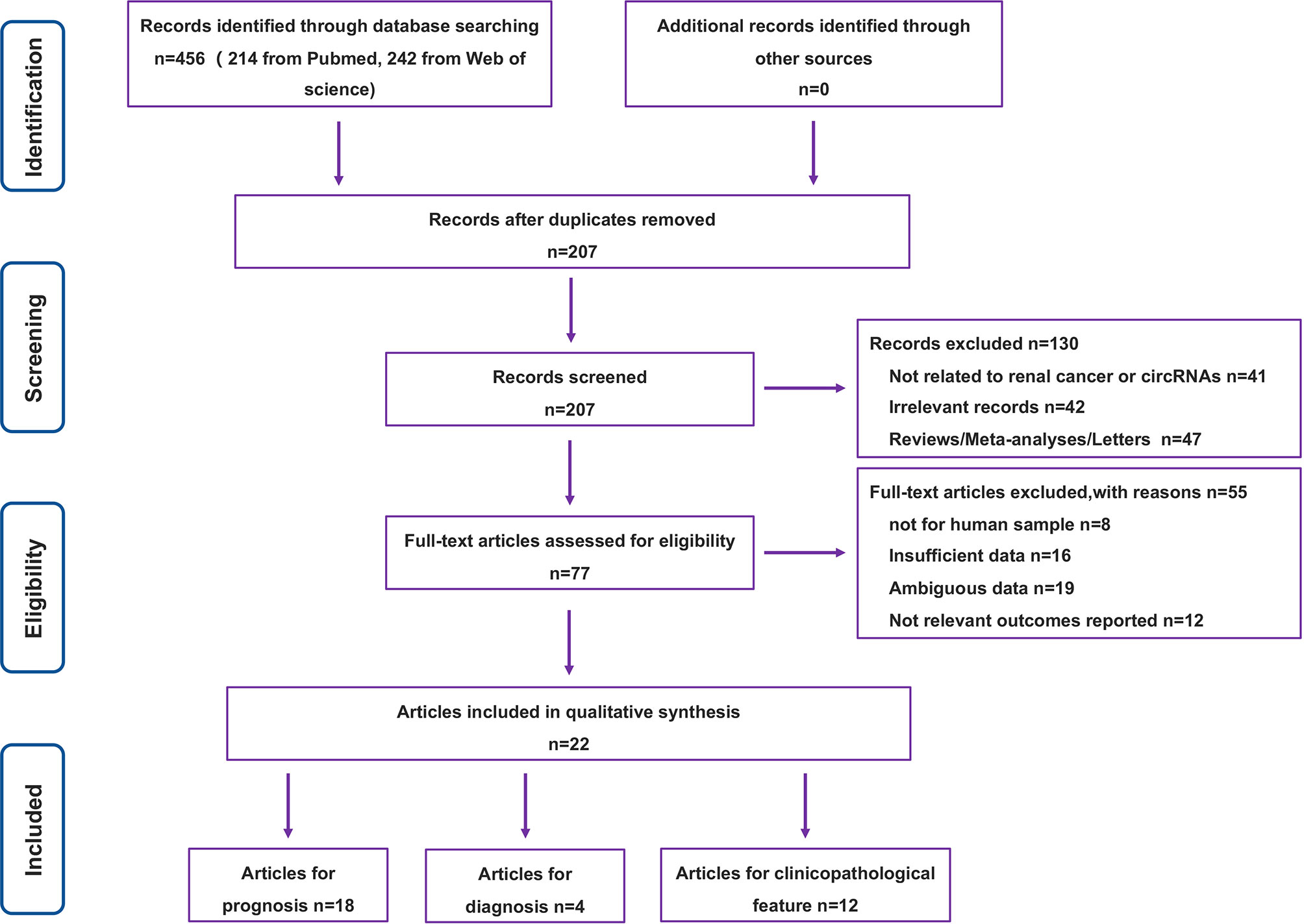
The literature search involved in this paper and the process of filtering through relevant conditions is represented in Figure 1. A total of 456 studies were retrieved from the above database, and the remaining 207 studies were extracted after preliminary screening. Of the 207 studies, we also excluded studies unrelated to RC or circRNAs, irrelevant records, reviews and other studies that did not meet the requirements of format and content, leaving 77 studies. After the analysis and evaluation of the remaining 77 studies, we found that some studies did not meet the inclusion criteria, and a total of 22 studies were actually available for reference after elimination (15, 16, 20–39). Among them, 18 were related to prognosis, 4 were related to diagnosis, and 12 were related to clinicopathology.
3.2 Analysis and Evaluation of Research Results
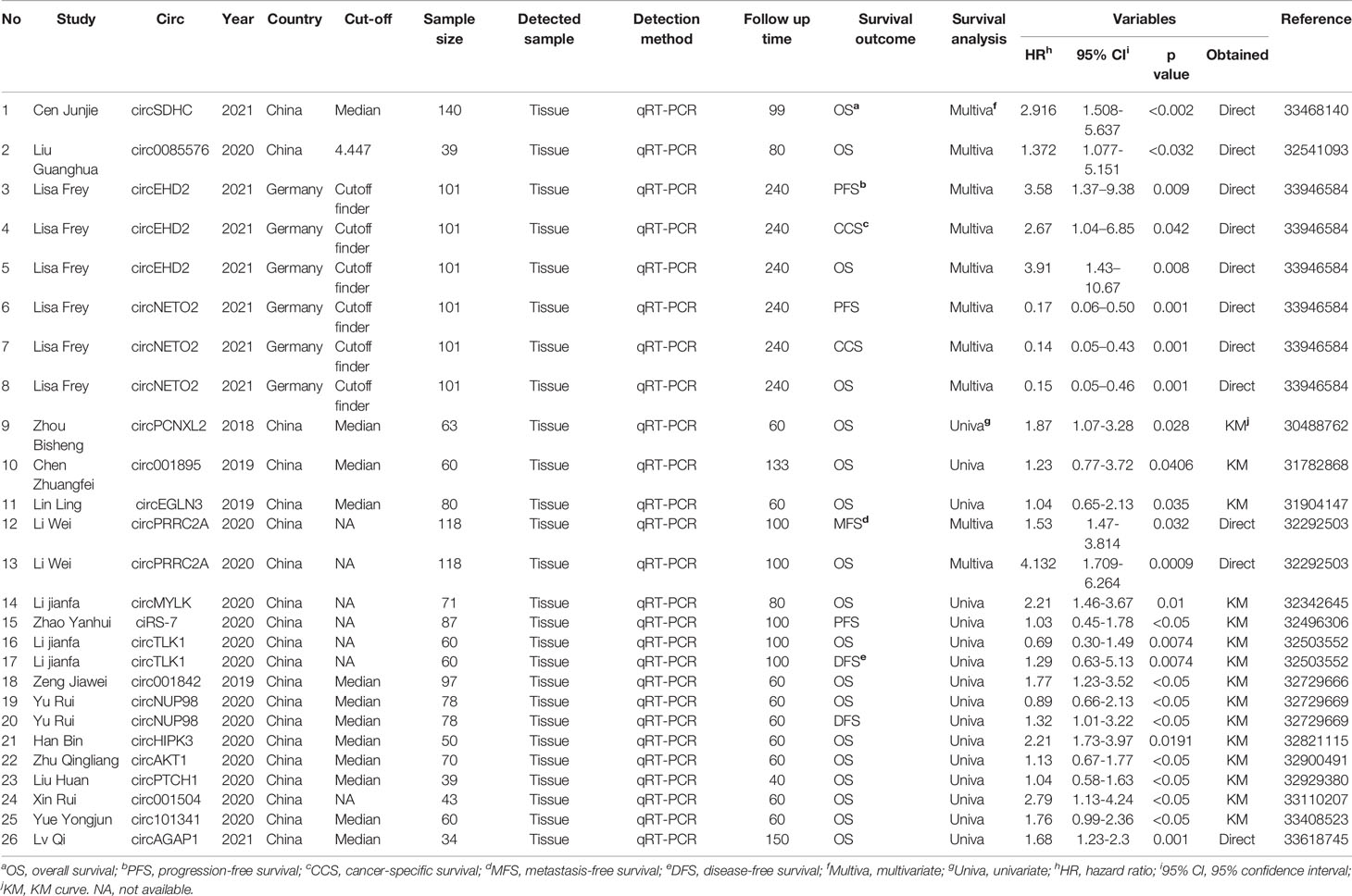
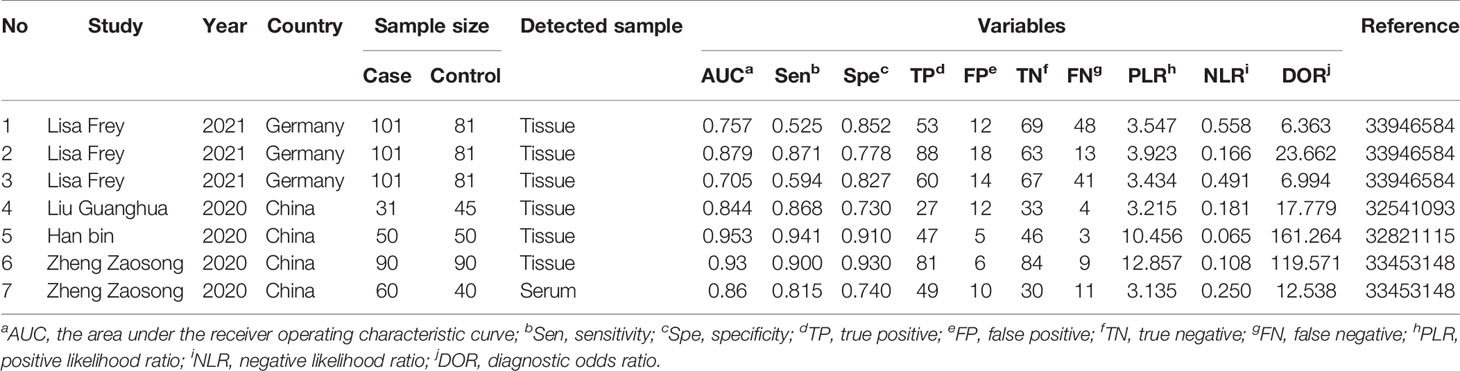
The above studies were all published between 2019 and 2021, most of them were from China, and only one was from Germany. The minimum follow-up time was 40 months, and the longest was 240 months. A total of 22 circRNAs were mentioned, and the expression of circRNAs in RC was detected by quantitative real-time polymerase chain reaction (qRT-PCR). We found that the expression of all circRNAs in these articles was increased. The above research features are summarized in Table 1. We scored the prognostic and diagnostic components of the included studies by the Newcastle-Ottawa score (NOS) and the Quality Assessment for Studies of Diagnostic Accuracy II (QUADAS II) checklist, suggesting that the prognostic studies had scores ≥7 or the diagnostic studies had scores ≥4, which means that the methods and articles used in the studies were of high quality and could be used for reference (40, 41). The specific scores can be seen in Figures 2 and 3.
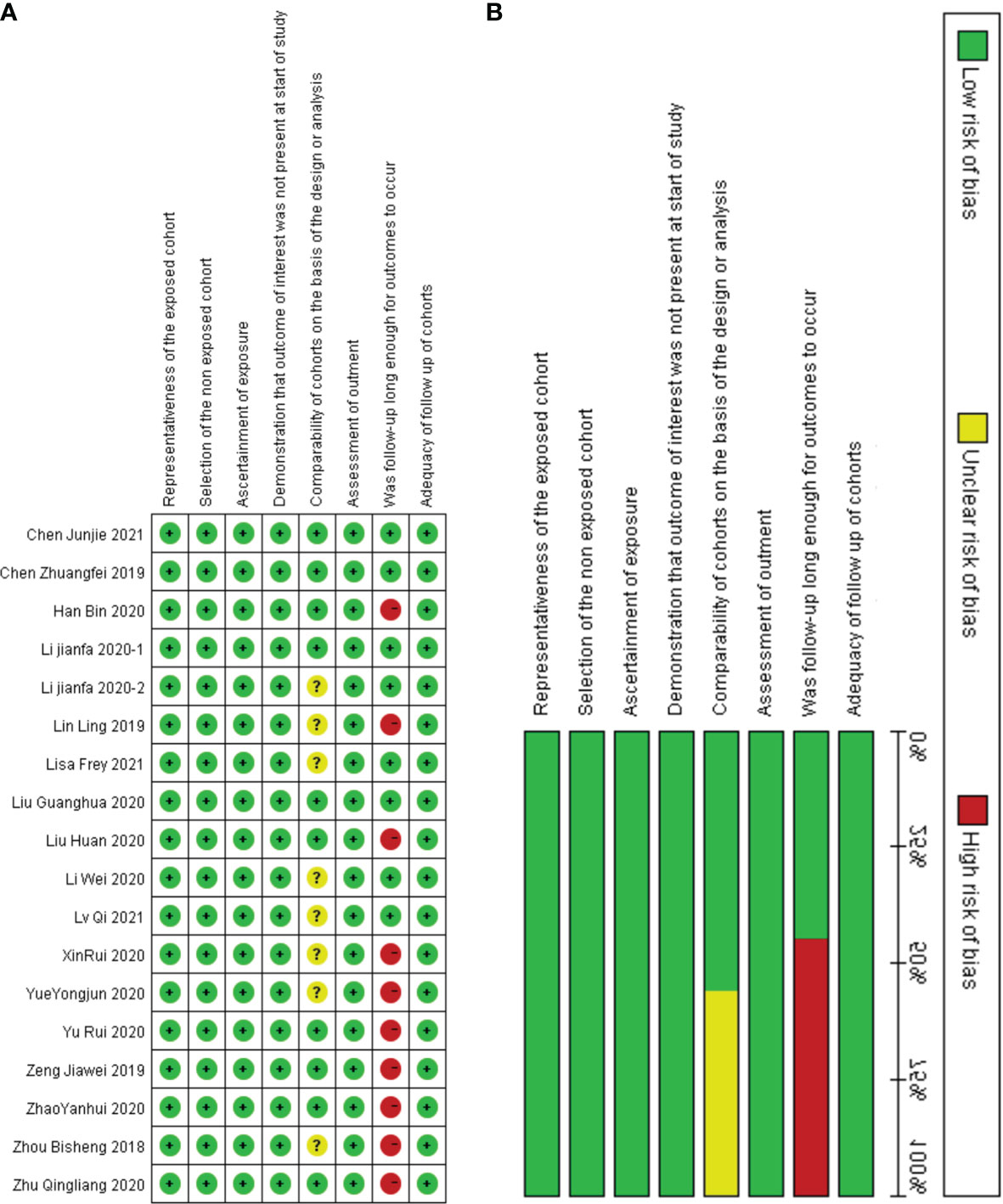
Figure 2 Quality assessment by the NOS. (A) each bias risk item for each included study; (B) each bias risk item is presented as a percentage for all included studies.
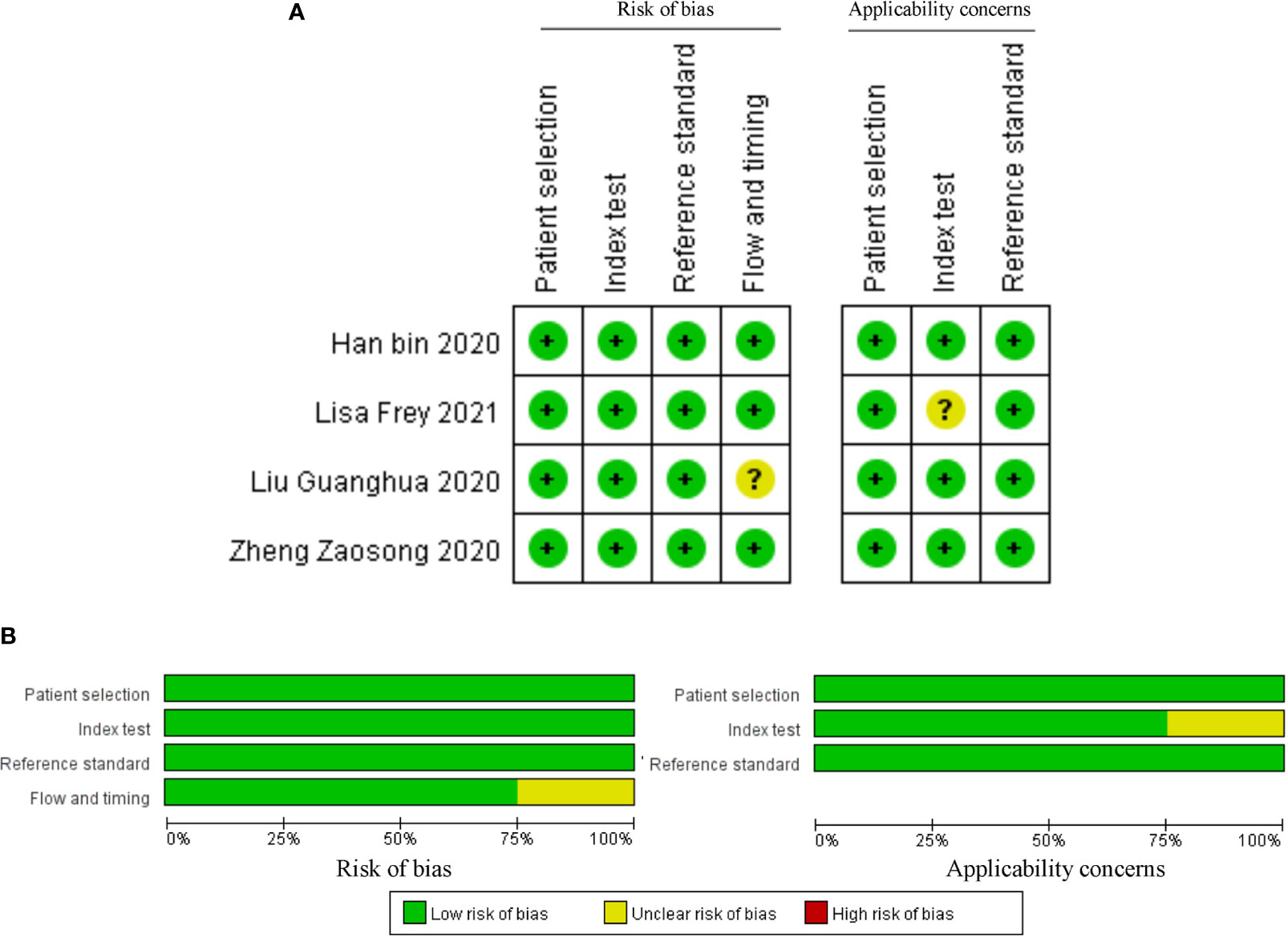
Figure 3 Quality assessment by the QUADAS II. (A) each bias risk item for each included study; (B) each bias risk item is presented as a percentage for all included studies.
3.2.1 Role of circRNAs in the Prognosis of RC
The characteristics of circRNAs related to the prognosis of RC are summarized in Table 1. In the 18 articles on prognosis we identified, we further found that all 18 circRNAs involved in the above studies were overexpressed in RC, and comprehensive evaluation of the OS was performed (HR=1.63, 95% CI =1.43–1.85, p<0.001, I2 = 69.0%). These results suggest reduced OS in RC patients with elevated circRNA expression. In Lisa Frey’s study, although circNETO2 was highly expressed in RC tissues, it was negatively correlated with the risk of death (HR=0.15, 95% CI =0.05–0.46), suggesting that circNETO2 had an inhibitory effect on cancer. In addition, we summarized and analyzed the relationship between circRNAs and the PFS, CCS, MFS and DFS of patients with RC and extracted the corresponding data (HR=1.17, 95% CI=0.90–1.52, p<0.001, I2 = 81.3%). The specific values are shown in Figure 4.
On the basis of the above contents, we found that there was heterogeneity in the factors affecting the prognosis of patients with RC. Therefore, we conducted a subgroup analysis of the contents summarized in Table 1 to further understand the specific factors affecting the OS of patients, as shown in Figure 5. We separately evaluated and analyzed factors such as the country of origin (China or Germany), cutoff value (median or nonmedian), sample size (>60 or ≤60), follow-up time (>60 months or ≤60 months), and survival analysis (multivariate or univariate). Similarly, we also performed a subgroup analysis of PFS, CCS, MFS and DFS to obtain the relevant data. The results are shown in Figure 6.
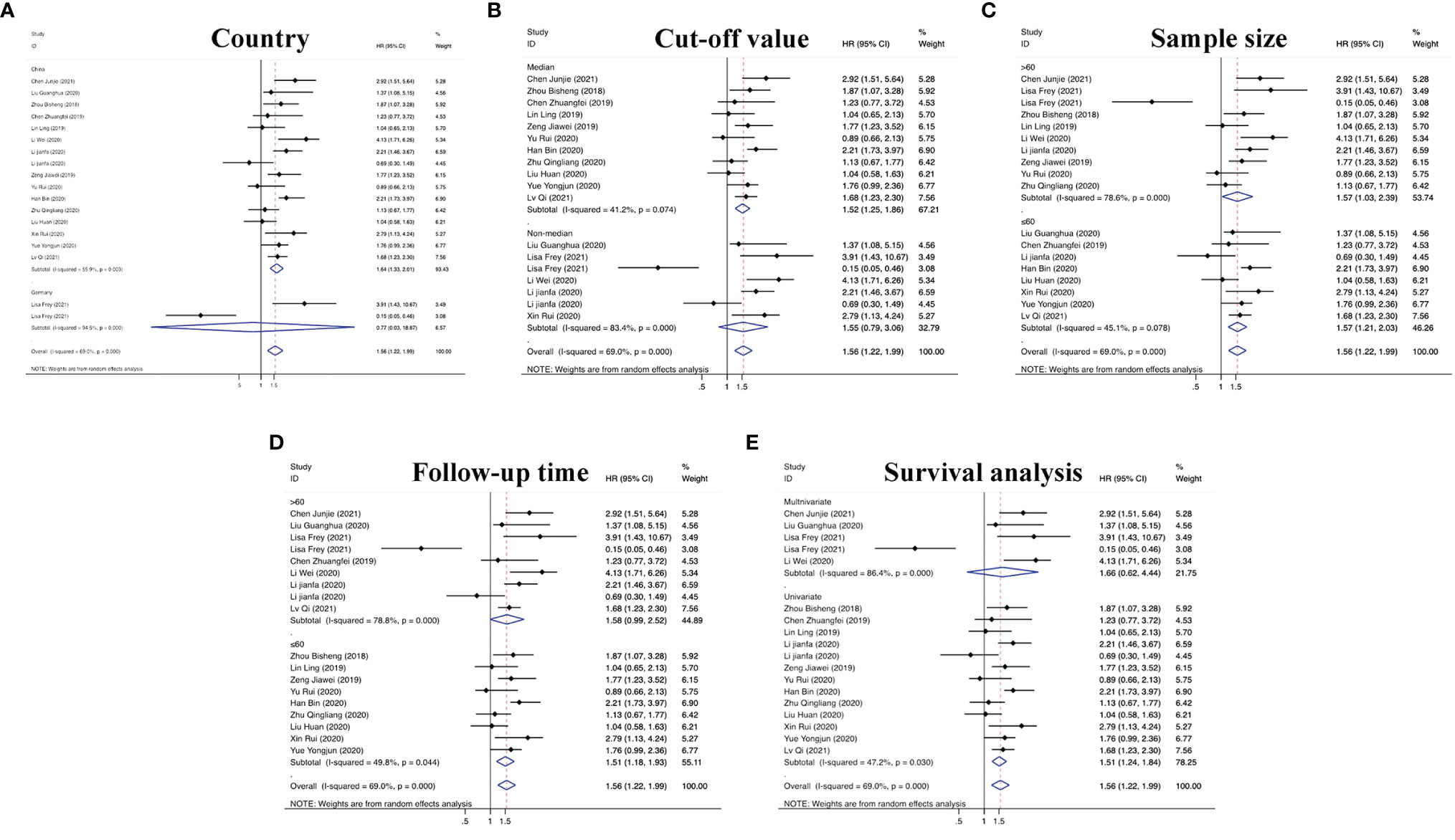
Figure 5 Subgroup analyses of OS for circRNAs, stratified by (A) country, (B) cut-off value, (C) sample size, (D) follow-up time, and (E) survival analysis.
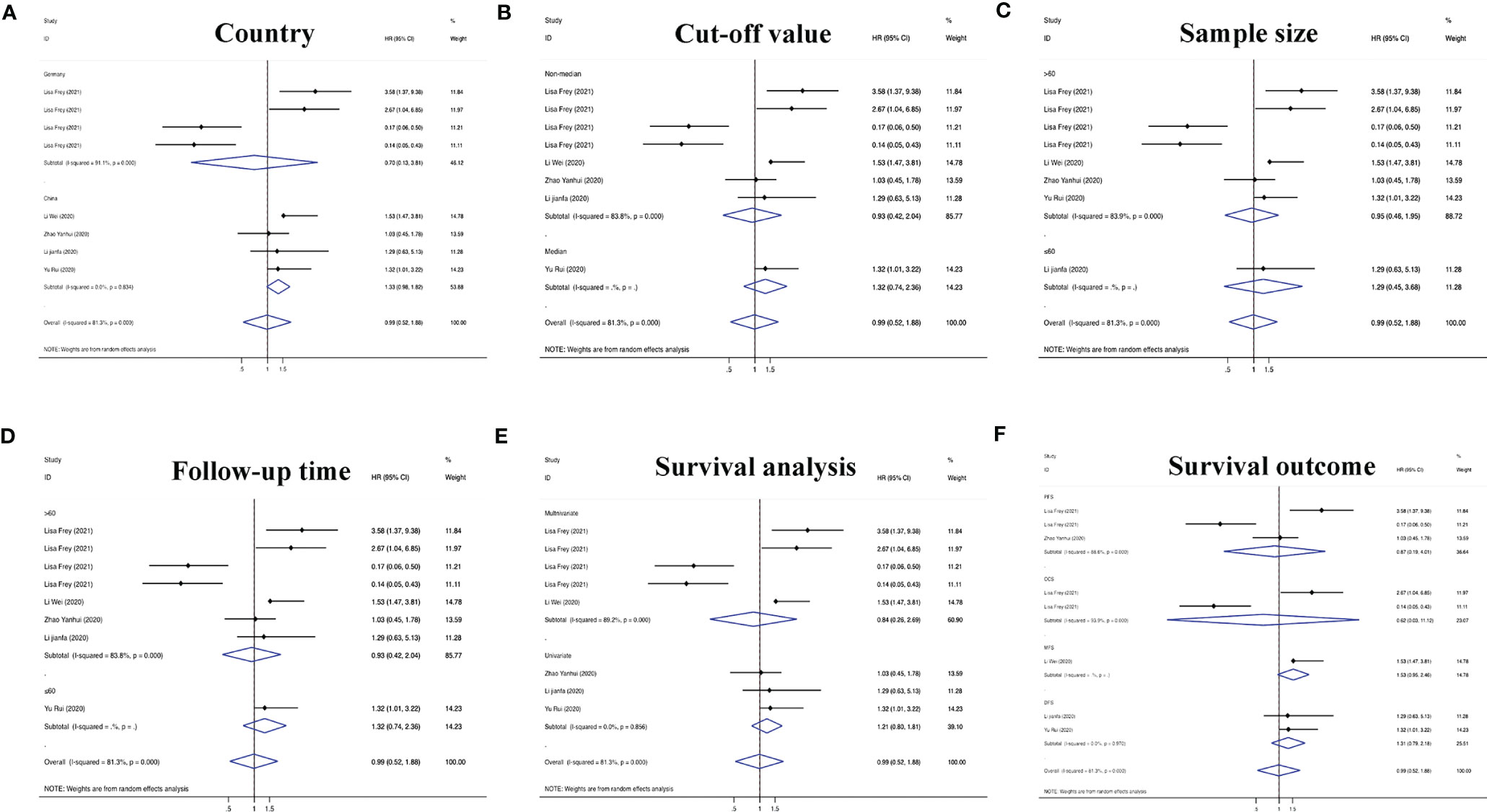
Figure 6 Subgroup analyses of PFS/CCS/MFS/DFS for circRNAs, stratified by (A) country, (B) cut-off value, (C) sample size, (D) follow-up time, (E) survival analysis, and (F) survival outcome.
3.2.2 Role of circRNAs in the Diagnosis of RC
After accurate screening of the retrieved literature, we finally sorted out four articles related to diagnosis and scored the four articles using QUADAS II software, resulting in scores greater than 6 for all studies, which indicated high quality. The above articles were summarized, and corresponding data were extracted, including the SEN, SPE, PLR, NLR and DOR, as shown in Table 2. First, the SROC (summary ROC) curve was drawn, and the AUC was calculated to be 0.89 (0.86–0.91), as shown in Figure 7G. The results suggested that circRNAs had diagnostic significance in RC, and some known specific circRNAs could be detected for the diagnosis of RC. Moreover, we drew forest plots (Figures 7A–E) of the SEN (HR=0.82, 95% CI=0.69–0.90), SPE (HR=0.84, 95% CI=0.78–0.88), DLR (HR=5.03, 95% CI=3.47–7.31), NLR (HR=0.22, 95% CI=0.12–0.39) and DOR (HR=22.97, 95% CI=9.72–54.30). The ROC curve was drawn to analyze whether there was a threshold effect, and it was found that there was no shoulder arm shape, so it was considered that there was no threshold diagnostic effect (Figure 7F). To further analyze the clinical value of circRNAs associated with RC, Fagan’s nomograms and scatter plots of the PLR and NLR (Figures 8C, D) were plotted. When the probability was set to 20% before validation, the PLR of circRNAs as a diagnostic indicator increased to 56% after model correction, while the NLR decreased to 5%. In summary, circRNAs can play a very sensitive and accurate role in the clinical diagnosis of RC.
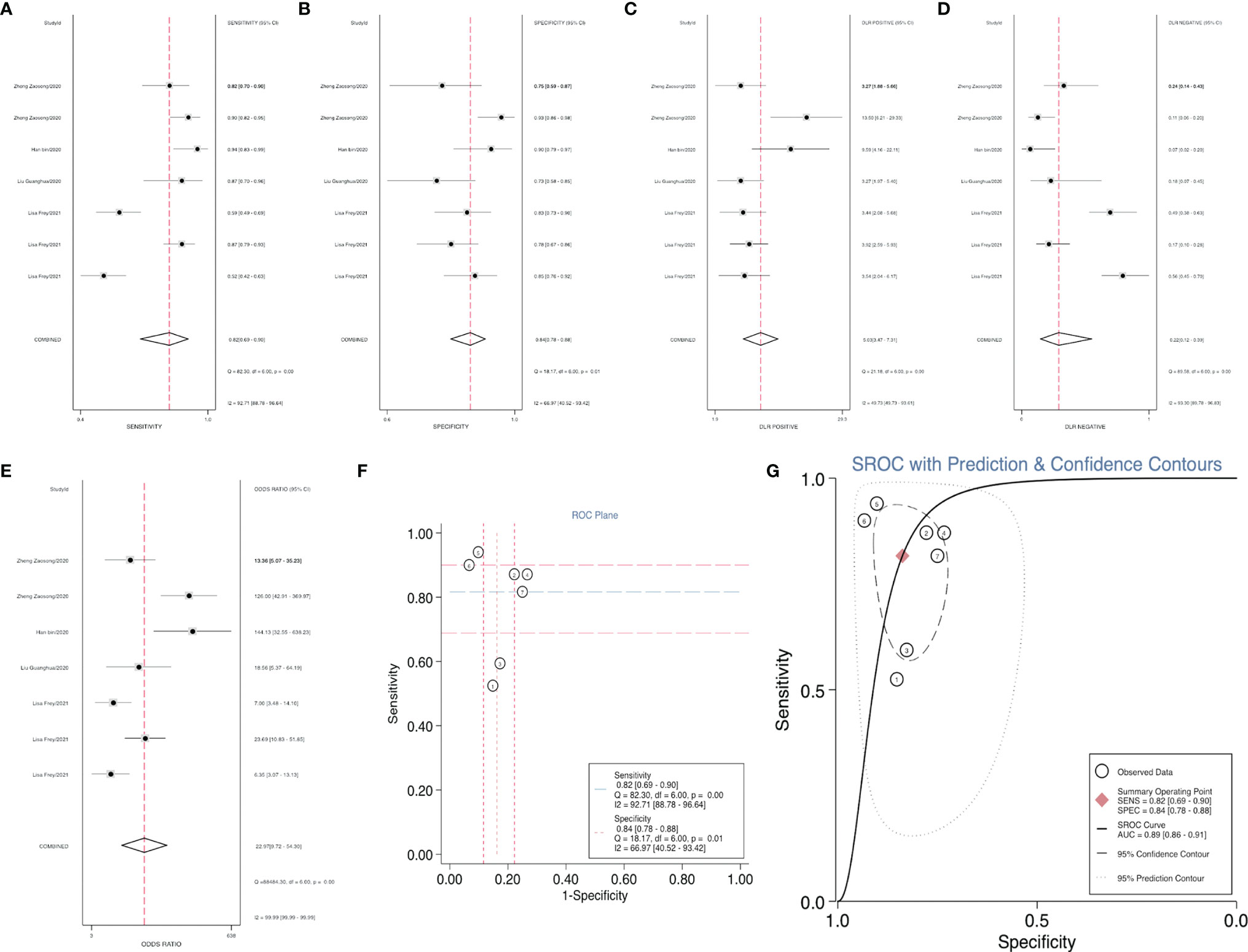
Figure 7 Forest plots of the combined (A) SEN, (B) SPE, (C) PLR, (D) NLR, (E) DOR, (F) ROC curve, and (G) SROC curve to illustrate the diagnosis of RC.
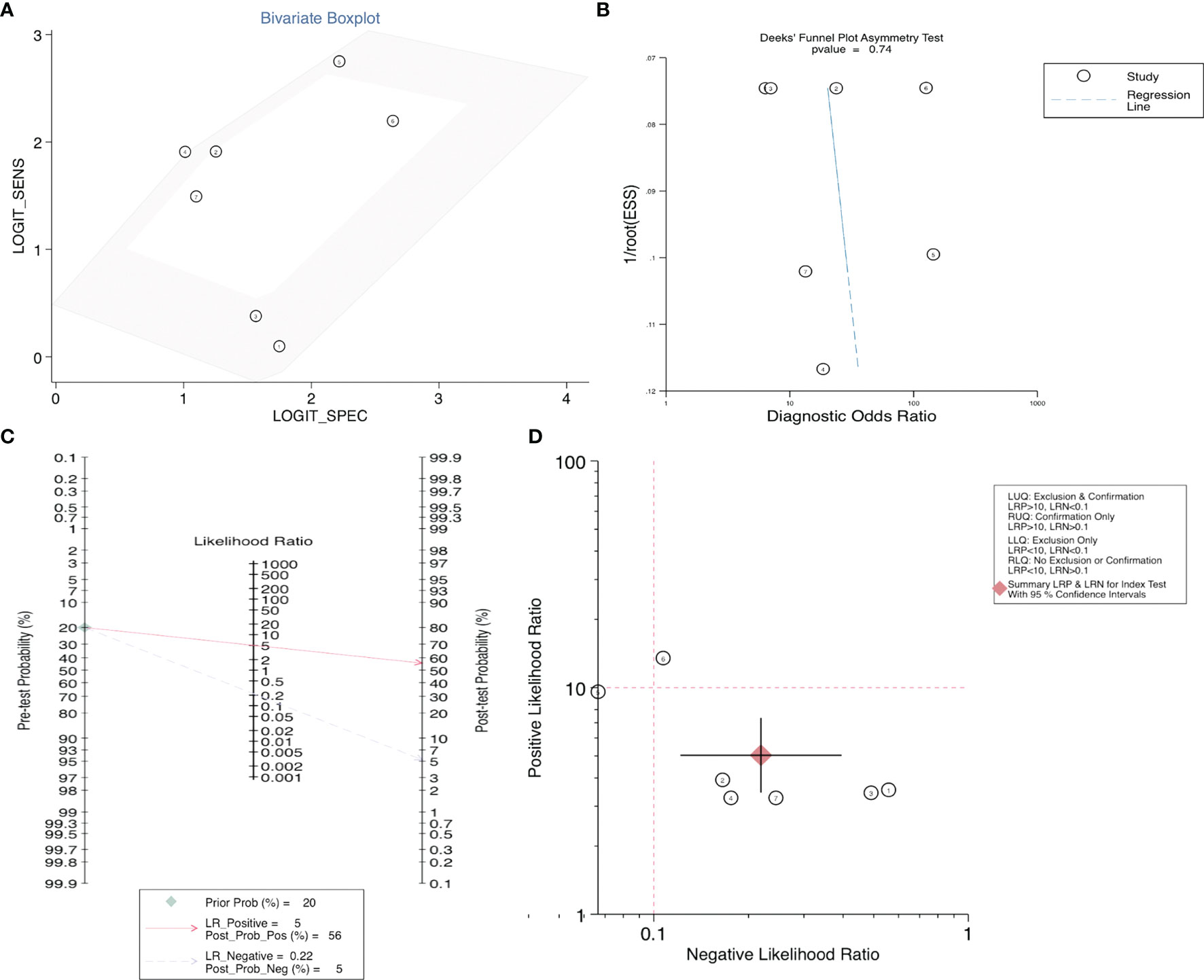
Figure 8 (A) Bivariate boxplot, (B) Deeks’ funnel plot, (C) Scatter plot, and (D) Fagan’s nomogram to illustrate the effect of circRNAs on the diagnosis of RC.
3.2.3 Role of circRNAs in the Clinicopathologic Characteristics of RC
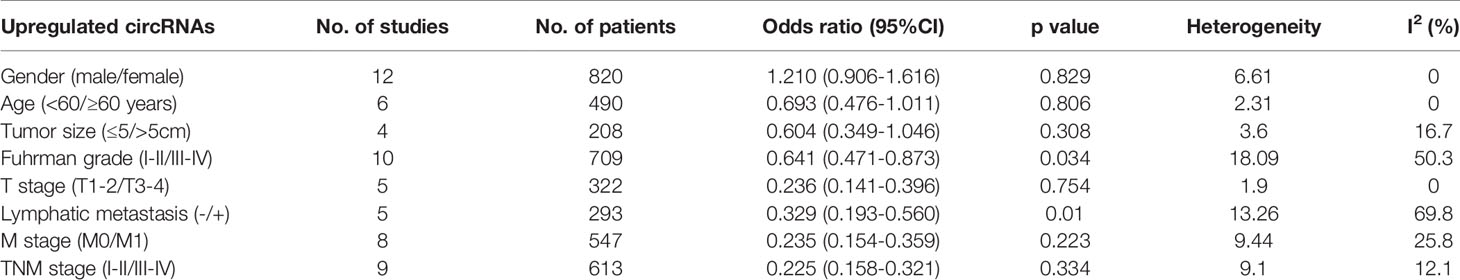
In addition to the role of circRNAs in evaluating the prognosis of RC patients and assisting in the diagnosis of RC, circRNAs are also closely related to many clinicopathological indicators of RC patients. In our meta-analysis, to quantify the relationship between circRNAs and the clinicopathologic characteristics of RC, we aggregated 12 studies with a total of 820 people (Table 3) and summarized the contents of the same influencing factor in different articles in the above 12 studies. The ORs (including 95% CIs) of different influencing factors were calculated and summarized. As shown in Figure 9, we found that high circRNA expression was correlated with a poor Fuhrman grade (OR=0.641, 95% CI=0.471–0.873), high T stage (OR=0.236, 95% CI=0.141–0.396) and TNM stage (OR=0.225, 95% CI=0.158–0.321), and the existence of lymphatic (OR=0.329, 95% CI=0.193–0.560) and distant metastasis (OR=0.235, 95% CI=0.154–0.359). In addition, we found no association of gender (OR=1.210, 95% CI=0.906–1.616), age (OR=0.693, 95% CI=0.476–1.011), or tumor size (OR=0.604, 95% CI=0.349–1.046) with high circRNA expression (Figure 10). Due to insufficient statistical data, some hematology-related influencing factors were not included in the table, such as the estimated glomerular filtration rate (eGFR), creatinine (Cr) level, and urine albumin creatine ratio (UACR), among others.
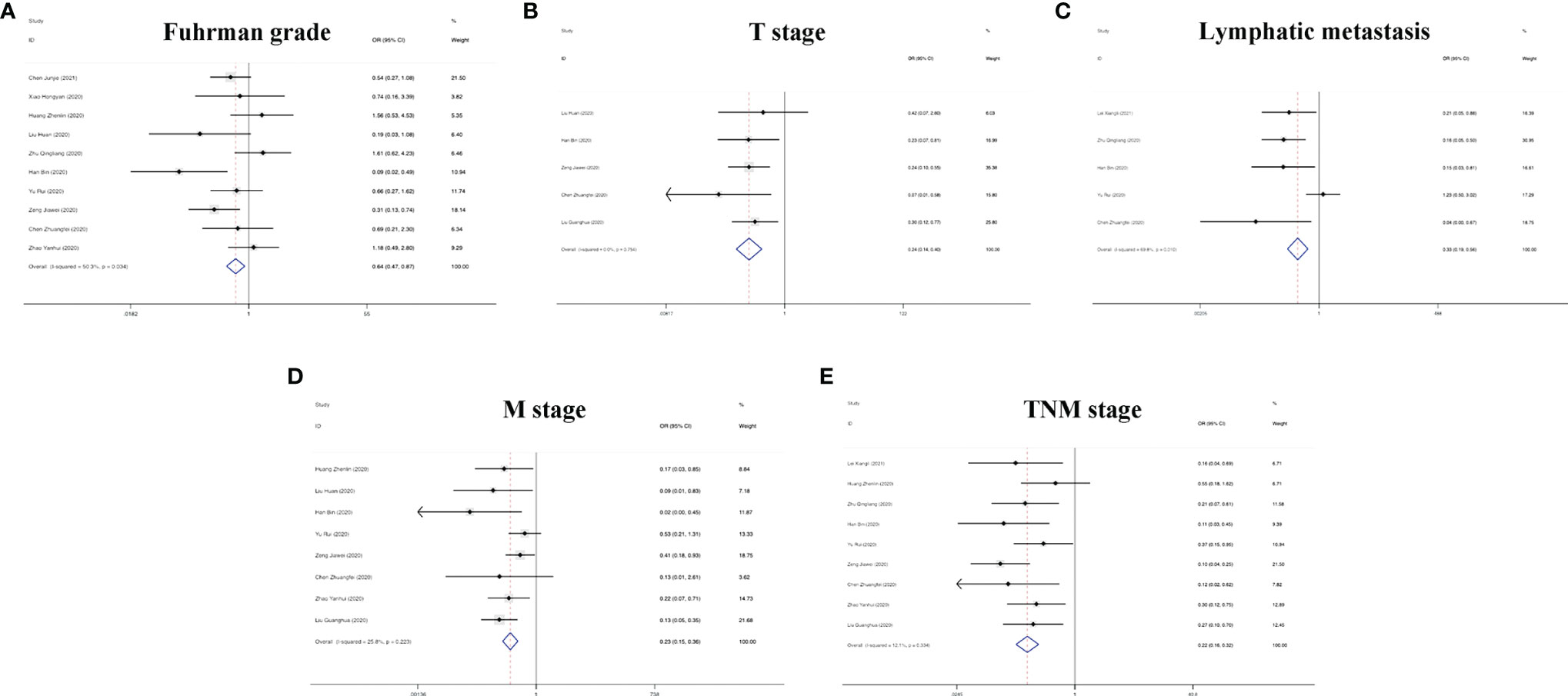
Figure 9 Forest plots of the clinicopathological characteristics, (A) Fuhrman grade, (B) T stage, (C) lymphatic metastasis, (D) M stage, and (E) TNM stage for circRNAs in RC.

Figure 10 Forest plots of the clinicopathological characteristics, (A) gender, (B) age, and (C) tumor size for circRNAs in RC.
3.3 Sensitivity Analysis
To evaluate the stability of our results, we performed a sensitivity analysis of the included studies by category, including prognosis (Figures S1A, D) and clinicopathologically related indicators (Figure S2). The results showed that excluding each study did not change the overall effect of circRNAs on the combination of HRs and ORs, suggesting that the conclusions of our meta-analysis were reliable.
3.4 Publication Bias
Egger’s and Begg’s tests were used to analyze the prognostic indicators of RC with high circRNA expression and evaluate whether there was obvious publication bias in the operating system. As shown in Figure S1, the results showed p > 0.05 for Egger’s and Begg’s tests of OS, as well as for the PFS, CCS, MFS, and DFS, indicating no significant publication bias. At the same time, Egger’s and Begg’s tests were carried out for the corresponding influencing factors, such as gender, age, tumor size, Fuhrman grade, T stage, lymphatic metastasis, M stage and TNM stage, and funnel plots were drawn. According to the obtained p values, there was no significant publication bias. Finally, in the diagnostic study of renal cancer-related circRNAs, bivariate and Deeks models (Figures 8A, B) were used to analyze the sensitivity, and no significant publication bias was found.
4 Discussion
As the third leading cause of death related to the urinary system, RC is difficult to detect in early stages, with a high metastasis rate and poor prognosis (42, 43). The advent of molecularly targeted drugs offers a novel approach for the treatment of patients with RC, with tumor-related signaling pathways being currently targeted, including VEGFR and mTOR inhibitors (44). Nevertheless, more than 30% of patients will likely become resistant to targeted drugs. Therefore, it is particularly important to comprehensively study the mechanism of the occurrence and development of RC as well as the molecules involved to generate precise treatment plans for different patients (45).
Many biomarkers have been found to be useful for the early diagnosis or long-term prognosis of patients with RC, such as mutation rates of VHL and PBRM1 in somatic cells, urine proteomics detection, and immune detection points (46–48). As a recent hot topic in tumor development, the role of non-coding RNA (ncRNA) in RC has been gradually explored (49). MiR-196a affects progression of RC by targeting BRAM1 to regulate the SMAD and MAPK signaling pathways, and miR-196 can be used as a prognostic marker for RC (50). Jasmine et al. also found 11 lncRNAs able to guide diagnosis of RC (51). As ncRNAs, circRNAs are formed via covalent bonding. With the rapid development of high-throughput sequencing and bioinformatics, the nature of circRNAs has been gradually revealed. Compared with mRNA, circRNA is more stable due to its closed ring structure, and it is not easily hydrolyzed by RNA enzymes. In addition, circRNAs formation is mostly driven by the exon lariat structure, and they are highly conserved and specific. Finally, circRNAs are widely present in exosomes and plasma, conferring distinct advantages as novel diagnostic markers (52–54).
According to defined inclusion and exclusion criteria, 22 studies of a total of 26 circRNAs associated with RC were analyzed. Regarding prognostic risk factors, 18 circRNAs were found to be associated with OS in RC patients, 3 circRNAs were associated with PFS, 2 circRNAs were associated with CCS, 2 circRNAs were associated with DFS, and 1 circRNA was associated with MFS (15, 16, 20–33, 36, 38). Overall, the risk of death in patients with high circRNA expression was 1.63 times higher than that in patients without high circRNA expression (95% CI =1.43–1.85). Interestingly, only Frey et al.’s study found that circNETO2 was highly expressed in RC, but prognostic analysis showed that circNETO2 may play a protective role in RC (16). No detailed experimental study has been conducted thus far, and determination of its mechanism may be a direction for the future development of targeted therapies. In the subsequent subgroup analysis, we further found that China as the country (HR=1.64, 95% CI=1.33–2.01), the median as the cutoff value (HR=1.52, 95% CI=1.25–1.86), a follow-up time ≤60 months (HR=1.51, 95% CI=1.18–1.93), univariate analysis (HR=1.51, 95% CI=1.24–1.84) and the sample size (HR=1.56, 95% CI=1.03–2.39) were risk factors affecting OS. However, there was no strong correlation between circRNAs and non-OS indicators in patients with RC (HR=1.17, 95% CI=0.90–1.52), which may be related to an insufficient sample size.
For RC diagnosis, although there were only 4 studies addressed this topic, 7 indicators were revealed, providing a valid basis for the diagnosis of RC patients (16, 27, 30, 37). Using SROC curve analysis, we calculated an AUC of 0.89 for circRNAs, indicating that 89% of our randomized RC patients had higher circRNA levels than the controls. The SEN was 82%, and the SPE was 84%. In addition, Fagan’s nomogram analysis of the clinical value of circRNAs can be used to indicate the sensitivity and specificity of a biomarker for the diagnosis of RC, and it showed positive predictive value. In addition, in our study, we listed some serum indicators, such as UACR, eGFR, Cr and cystatin C (CysC), to explore their relationship with the diagnosis of RC. However, due to the small number of studies involved and the lack of universality, they were not included in our study. In future studies, a composite evaluation index, rather than a single evaluation index, may be more valuable for the diagnosis of RC.
Finally, we performed a meta-analysis of clinicopathologic data from patients with RC, including a total of 840 participants in 12 studies (15, 21, 25, 27–32, 34, 35, 39). The results showed that high circRNA expression was consistent with a poor Fuhrman grade, lymph node metastasis, and poor clinicopathological features in patients with RC. In addition, other factors, such as age and gender. In conclusion, circRNA overexpression in patients with RC can indicate poor clinicopathological characteristics and can be used as a reference for clinical diagnosis and evaluation of therapeutic efficacy.
Of course, there are still many limitations to our research. On the one hand, although most studies have suggested that circRNAs have a cancer-promoting effect in RC, there are also a few contrary results, suggesting that there can be multiple effects of circRNAs. Second, the sample size of the diagnostic studies is relatively small, and most of them rely on the postoperative analysis of patient tissues. More circRNAs in preoperative samples of serum, urine and other body fluids from patients are expected to be studied. Finally, our study only examined overexpressed circRNAs, and the other side of the role of differentially expressed circRNAs in the occurrence and development of RC remains to be explored. Therefore, future efforts should be made to increase the quantity and quality of included studies to improve the accuracy and stability of the results.
5 Conclusion
In summary, our study found that the abnormal expression of circRNAs in patients with RC was closely related to its diagnosis, prognosis and clinicopathological features; that is, it was associated with the growth, progression and differentiation of RC tumors. Therefore, achieving an in-depth understanding of the biological and clinical effects of circRNAs, making efforts to screen potential targets for the treatment of RC, and developing new ideas for the diagnosis and treatment of RC will be the top priorities of future research.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
WW and SX designed and carried out this study. WW and DY participated in collecting the literature on circ related to renal cancer. SX and DH participated in the analysis of clinicopathological features. WW and SX participated in the assessment of prognosis and diagnostic value. LF and FG were critically revised it critically for important intellectual content. All the authors approved the final version to be published.
Funding
This study was supported by the Natural Science Foundation for Nanjing University of Chinese Medicine (XZR2020029), the Hospital Pharmaceutical Research Foundation for Nanjing Pharmaceutical Association and Changzhou Siyao Pharma (2019YX001), and the Second Batch of Science and Technology Projects for Traditional Chinese Medicine in Jiangsu Province (FY201805). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.773236/full#supplementary-material
Supplementary Figure 1 | Sensitivity analysis (A, D) and Publication bias judged by Egger’s (B, E) and Begg’s (C, F) funnel plots of circRNAs for the OS and PFS/CCS/MFS/DFS in RC.
Supplementary Figure 2 | Sensitivity analysis of the circRNAs for (A) Fuhrman grade, (B) T stage, (C) lymphatic metastasis, (D) M stage, (E) TNM stage, (F) gender, (G) age, and (H) tumor size in RC.
Supplementary Figure 3 | Publication bias judged by Egger’s funnel plots of the circRNAs for (A) Fuhrman grade, (B) T stage, (C) lymphatic metastasis, (D) M stage, (E) TNM stage, (F) gender, (G) age, and (H) tumor size in RC.
Supplementary Figure 4 | Publication bias judged by Begg’s funnel plots of the circRNAs for (A) Fuhrman grade, (B) T stage, (C) lymphatic metastasis, (D) M stage, (E) TNM stage, (F) gender, (G) age, and (H) tumor size in RC.
Abbreviations
RC, renal cancer; CircRNA, circular RNA; HR, hazard ratio; OR, odds ratio; SEN, sensitivity; SPE, specificity; PLR, positive likelihood ratio; NLR, negative likelihood ratio; DOR, diagnostic odds ratio; SROC, summary receiver operator characteristic; AUC, area under the SROC; OS, overall survival; RCC, renal cell carcinoma; PC, pancreatic cancer; CI, confidence interval; PFS, progression-free survival; CCS, cancer-specific survival; MFS: metastasis-free survival; DFS, disease-free survival; TP, true positive; FP, false positive; TN, true negative; FN: false negative; NOS, newcastle-ottawa score; QUADAS II, quality assessment for studies of diagnostic accuracy II; eGFR, estimated glomerular filtration rate; Cr, creatinine; UACR, urine albumin creatine ratio; CysC, cystatin C; ncRNA, non-coding RNA.
References
1. Mir M, Derweesh I, Porpiglia F, Zargar H, Mottrie A, Autorino R. Partial Nephrectomy Versus Radical Nephrectomy for Clinical T1b and T2 Renal Tumors: A Systematic Review and Meta-Analysis of Comparative Studies. Eur Urol (2017) 71(4):606–17. doi: 10.1016/j.eururo.2016.08.060
2. Linehan W, Ricketts C. The Cancer Genome Atlas of Renal Cell Carcinoma: Findings and Clinical Implications. Nat Rev Urol (2019) 16(9):539–52. doi: 10.1038/s41585-019-0211-5
3. Lopez-Beltran A, Scarpelli M, Montironi R, Kirkali Z. WHO Classification of the Renal Tumors of the Adults. Eur Urol (2006) 49(5):798–805. doi: 10.1016/j.eururo.2005.11.035
4. Skolarus T, Serrano M, Berger D, Bullock T, Yan Y, Humphrey P, et al. The Distribution of Histological Subtypes of Renal Tumors by Decade of Life Using the 2004 WHO Classification. J Urol (2008) 179(2):439–43; discussion 43-4. doi: 10.1016/j.juro.2007.09.076
5. Chung J, Son N, Lee S, Hong S, Lee S, Kwak C, et al. Overall Survival and Renal Function After Partial and Radical Nephrectomy Among Older Patients With Localised Renal Cell Carcinoma: A Propensity-Matched Multicentre Study. Eur J Cancer (Oxford Engl 1990) (2015) 51(4):489–97. doi: 10.1016/j.ejca.2014.12.012
6. Steffens S, Junker K, Roos F, Janssen M, Becker F, Henn D, et al. Small Renal Cell Carcinomas–How Dangerous Are They Really? Results of a Large Multicenter Study. Eur J Cancer (Oxford Engl 1990) (2014) 50(4):739–45. doi: 10.1016/j.ejca.2013.11.020
7. Haas N, Manola J, Dutcher J, Flaherty K, Uzzo R, Atkins M, et al. Adjuvant Treatment for High-Risk Clear Cell Renal Cancer: Updated Results of a High-Risk Subset of the ASSURE Randomized Trial. JAMA Oncol (2017) 3(9):1249–52. doi: 10.1001/jamaoncol.2017.0076
8. Huang Y, Zhu Q. Mechanisms Regulating Abnormal Circular RNA Biogenesis in Cancer. Cancers (2021) 13(16):4185. doi: 10.3390/cancers13164185
9. Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: Functions and Properties of a Novel Potential Biomarker for Cancer. Mol Cancer (2017) 16(1):94. doi: 10.1186/s12943-017-0663-2
10. Shang Q, Yang Z, Jia R, Ge S. The Novel Roles of circRNAs in Human Cancer. Mol Cancer (2019) 18(1):6. doi: 10.1186/s12943-018-0934-6
11. Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H, et al. Circular RNAs in Body Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Mol Cancer (2021) 20(1):13. doi: 10.1186/s12943-020-01298-z
12. Xu T, Wang M, Jiang L, Ma L, Wan L, Chen Q, et al. CircRNAs in Anticancer Drug Resistance: Recent Advances and Future Potential. Mol Cancer (2020) 19(1):127. doi: 10.1186/s12943-020-01240-3
13. Yang T, Shen P, Chen Q, Wu P, Yuan H, Ge W, et al. FUS-Induced Circrhobtb3 Facilitates Cell Proliferation via miR-600/NACC1 Mediated Autophagy Response in Pancreatic Ductal Adenocarcinoma. J Exp Clin Cancer Res CR (2021) 40(1):261. doi: 10.1186/s13046-021-02063-w
14. Chen Q, Shen P, Ge W, Yang T, Wang W, Meng L, et al. Roundabout Homolog 1 Inhibits Proliferation via the YY1-ROBO1-CCNA2-CDK2 Axis in Human Pancreatic Cancer. Oncogene (2021) 40(15):2772–84. doi: 10.1038/s41388-021-01741-5
15. Cen J, Liang Y, Huang Y, Pan Y, Shu G, Zheng Z, et al. Circular RNA circSDHC Serves as a Sponge for miR-127-3p to Promote the Proliferation and Metastasis of Renal Cell Carcinoma via the CDKN3/E2F1 Axis. Mol Cancer (2021) 20(1):19. doi: 10.1186/s12943-021-01314-w
16. Frey L, Klümper N, Schmidt D, Kristiansen G, Toma M, Ritter M, et al. CircEHD2, CircNETO2 and CircEGLN3 as Diagnostic and Prognostic Biomarkers for Patients With Renal Cell Carcinoma. Cancers (2021) 13(9):2177. doi: 10.3390/cancers13092177
17. Shi X, Chen Q, Wang F. Mesenchymal Stem Cells for the Treatment of Ulcerative Colitis: A Systematic Review and Meta-Analysis of Experimental and Clinical Studies. Stem Cell Res Ther (2019) 10(1):266. doi: 10.1186/s13287-019-1336-4
18. Wang R, Yao Q, Chen W, Gao F, Li P, Wu J, et al. Stem Cell Therapy for Crohn's Disease: Systematic Review and Meta-Analysis of Preclinical and Clinical Studies. Stem Cell Res Ther (2021) 12(1):463. doi: 10.1186/s13287-021-02533-0
19. DerSimonian R, Laird N. Meta-Analysis in Clinical Trials Revisited. Contemp Clin Trials (2015) 45:139–45. doi: 10.1016/j.cct.2015.09.002
20. Zhou B, Zheng P, Li Z, Li H, Wang X, Shi Z, et al. CircPCNXL2 Sponges miR-153 to Promote the Proliferation and Invasion of Renal Cancer Cells Through Upregulating ZEB2. Cell Cycle (Georgetown Tex) (2018) 17(23):2644–54. doi: 10.1080/15384101.2018.1553354
21. Chen Z, Xiao K, Chen S, Huang Z, Ye Y, Chen T. Circular RNA Hsa_Circ_001895 Serves as a Sponge of microRNA-296-5p to Promote Clear Cell Renal Cell Carcinoma Progression by Regulating SOX12. Cancer Sci (2020) 111(2):713–26. doi: 10.1111/cas.14261
22. Lin L, Cai J. Circular RNA Circ-EGLN3 Promotes Renal Cell Carcinoma Proliferation and Aggressiveness via miR-1299-Mediated IRF7 Activation. J Cell Biochem (2020) 121(11):4377–85. doi: 10.1002/jcb.29620
23. Li W, Yang F, Sun C, Huang J, Zhang H, Li X, et al. Circprrc2a Promotes Angiogenesis and Metastasis Through Epithelial-Mesenchymal Transition and Upregulates TRPM3 in Renal Cell Carcinoma. Theranostics (2020) 10(10):4395–409. doi: 10.7150/thno.43239
24. Li J, Huang C, Zou Y, Yu J, Gui Y. Circular RNA MYLK Promotes Tumour Growth and Metastasis via Modulating miR-513a-5p/VEGFC Signalling in Renal Cell Carcinoma. J Cell Mol Med (2020) 24(12):6609–21. doi: 10.1111/jcmm.15308
25. Zhao Y, Wang Z, Zhang N, Cui T, Zhang Y. Effect of ciRS-7 Expression on Clear Cell Renal Cell Carcinoma Progression. Chin Med J (2020) 133(17):2084–9. doi: 10.1097/CM9.0000000000000867
26. Li J, Huang C, Zou Y, Ye J, Yu J, Gui Y. CircTLK1 Promotes the Proliferation and Metastasis of Renal Cell Carcinoma by Sponging miR-136-5p. Mol Cancer (2020) 19(1):103. doi: 10.1186/s12943-020-01225-2
27. Liu G, Zhou J, Piao Y, Zhao X, Zuo Y, Ji Z. Hsa_circ_0085576 Promotes Clear Cell Renal Cell Carcinoma Tumorigenesis and Metastasis Through the miR-498/YAP1 Axis. Aging (2020) 12(12):11530–49. doi: 10.18632/aging.103300
28. Zeng J, Feng Q, Wang Y, Xie G, Li Y, Yang Y, et al. Circular RNA Circ_001842 Plays an Oncogenic Role in Renal Cell Carcinoma by Disrupting microRNA-502-5p-Mediated Inhibition of SLC39A14. J Cell Mol Med (2020) 24(17):9712–25. doi: 10.1111/jcmm.15529
29. Yu R, Yao J, Ren Y. A Novel circRNA, Circnup98, a Potential Biomarker, Acted as an Oncogene via the miR-567/ PRDX3 Axis in Renal Cell Carcinoma. J Cell Mol Med (2020) 24(17):10177–88. doi: 10.1111/jcmm.15629
30. Han B, Shaolong E, Luan L, Li N, Liu X. CircHIPK3 Promotes Clear Cell Renal Cell Carcinoma (ccRCC) Cells Proliferation and Metastasis via Altering of miR-508-3p/CXCL13 Signal. OncoTargets Ther (2020) 13:6051–62. doi: 10.2147/OTT.S251436
31. Zhu Q, Zhan D, Zhu P, Chong Y, Yang Y. CircAKT1 Acts as a Sponge of miR-338-3p to Facilitate Clear Cell Renal Cell Carcinoma Progression by Up-Regulating CAV1. Biochem Biophys Res Commun (2020) 532(4):584–90. doi: 10.1016/j.bbrc.2020.08.081
32. Liu H, Hu G, Wang Z, Liu Q, Zhang J, Chen Y, et al. Circptch1 Promotes Invasion and Metastasis in Renal Cell Carcinoma via Regulating miR-485-5p/MMP14 Axis. Theranostics (2020) 10(23):10791–807. doi: 10.7150/thno.47239
33. Xin R, Qu D, Xu H, Chen D. Circ_001504 Promotes the Development of Renal Cell Carcinoma by Sponging microRNA-149 to Increase NUCB2. Cancer Gene Ther (2021) 28(6):667–78. doi: 10.1038/s41417-020-00247-8
34. Huang Z, Ding Y, Zhang L, He S, Jia Z, Gu C, et al. Circpdk1upregulated Promotes RCC Cell Migration and Invasion by Regulating the Axis in Renal Cell Carcinoma. OncoTargets Ther (2020) 13:11237–52. doi: 10.2147/OTT.S280434
35. Xiao H, Shi J. Exosomal Circular RNA_400068 Promotes the Development of Renal Cell Carcinoma via the Mir−210−5p/SOCS1 Axis. Mol Med Rep (2020) 22(6):4810–20. doi: 10.3892/mmr.2020.11541
36. Yue Y, Cui J, Zhao Y, Liu S, Niu W. Circ_101341 Deteriorates the Progression of Clear Cell Renal Cell Carcinoma Through the miR- 411/EGLN3 Axis. Cancer Manage Res (2020) 12:13513–25. doi: 10.2147/CMAR.S272287
37. Zheng Z, Chen Z, Zhong Q, Zhu D, Xie Y, Shangguan W, et al. CircPVT1 Promotes Progression in Clear Cell Renal Cell Carcinoma by Sponging miR-145-5p and Regulating TBX15 Expression. Cancer Sci (2021) 112(4):1443–56. doi: 10.1111/cas.14814
38. Lv Q, Wang G, Zhang Y, Shen A, Tang J, Sun Y, et al. CircAGAP1 Promotes Tumor Progression by Sponging miR-15-5p in Clear Cell Renal Cell Carcinoma. J Exp Clin Cancer Res CR (2021) 40(1):76. doi: 10.1186/s13046-021-01864-3
39. Lei X, Yang M, Xiao Z, Zhang H, Tan S. Circtlk1 Facilitates the Proliferation and Metastasis of Renal Cell Carcinoma by Regulating miR-495-3p/CBL Axis. Open Life Sci (2021) 16(1):362–74. doi: 10.1515/biol-2021-0041
40. Lo C, Mertz D, Loeb M. Newcastle-Ottawa Scale: Comparing Reviewers' to Authors' Assessments. BMC Med Res Method (2014) 14:45. doi: 10.1186/1471-2288-14-45
41. Kim G, Shim K, Baik S. Diagnostic Accuracy of Hepatic Vein Arrival Time Performed With Contrast-Enhanced Ultrasonography for Cirrhosis: A Systematic Review and Meta-Analysis. Gut Liver (2017) 11(1):93–101. doi: 10.5009/gnl16031
42. Blick C, Ritchie A, Eisen T, Stewart G. Improving Outcomes in High-Risk, Nonmetastatic Renal Cancer: New Data and Ongoing Trials. Nat Rev Urol (2017) 14(12):753–9. doi: 10.1038/nrurol.2017.123
43. Riazalhosseini Y, Lathrop M. Precision Medicine From the Renal Cancer Genome. Nat Rev Nephrol (2016) 12(11):655–66. doi: 10.1038/nrneph.2016.133
44. Tuma R. Three Molecularly Targeted Drugs Tested in Kidney Cancer Clinical Trials. J Natl Cancer Inst (2004) 96(17):1270–1. doi: 10.1093/jnci/96.17.1270
45. Groenland S, Verheijen R, Joerger M, Mathijssen R, Sparreboom A, Beijnen J, et al. Precision Dosing of Targeted Therapies Is Ready for Prime Time. Clin Cancer Res an Off J Am Assoc Cancer Res (2021) 27(24):6644–52. doi: 10.1158/1078-0432.CCR-20-4555
46. Moore A, Batavia A, Kuipers J, Singer J, Burcklen E, Schraml P, et al. Spatial Distribution of Private Gene Mutations in Clear Cell Renal Cell Carcinoma. Cancers (2021) 13(9):2163. doi: 10.3390/cancers13092163
47. Senturk A, Sahin A, Armutlu A, Kiremit M, Acar O, Erdem S, et al. Quantitative Proteomics Identifies Secreted Diagnostic Biomarkers as Well as Tumor-Dependent Prognostic Targets for Clear Cell Renal Cell Carcinoma. Mol Cancer Res MCR (2021) 19(8):1322–37. doi: 10.1101/2021.02.08.430238
48. Zhang X, Yin X, Zhang H, Sun G, Yang Y, Chen J, et al. Differential Expressions of PD-1, PD-L1 and PD-L2 Between Primary and Metastatic Sites in Renal Cell Carcinoma. BMC Cancer (2019) 19(1):360. doi: 10.1186/s12885-019-5578-4
49. Jan van Zonneveld A, Kölling M, Bijkerk R, Lorenzen J. Circular RNAs in Kidney Disease and Cancer. Nat Rev Nephrol (2021) 17(12):814–26. doi: 10.1038/s41581-021-00465-9
50. Cui J, Yuan Y, Shanmugam M, Anbalagan D, Tan T, Sethi G, et al. MicroRNA-196a Promotes Renal Cancer Cell Migration and Invasion by Targeting BRAM1 to Regulate SMAD and MAPK Signaling Pathways. Int J Biol Sci (2021) 17(15):4254–70. doi: 10.7150/ijbs.60805
51. Blondeau J, Deng M, Syring I, Schrödter S, Schmidt D, Perner S, et al. Identification of Novel Long Non-Coding RNAs in Clear Cell Renal Cell Carcinoma. Clin Epigenet (2015) 7:10. doi: 10.1186/s13148-015-0047-7
52. Dong Y, He D, Peng Z, Peng W, Shi W, Wang J, et al. Circular RNAs in Cancer: An Emerging Key Player. J Hematol Oncol (2017) 10(1):2. doi: 10.1186/s13045-016-0370-2
53. Wang Y, Zhang Y, Wang P, Fu X, Lin W. Circular RNAs in Renal Cell Carcinoma: Implications for Tumorigenesis, Diagnosis, and Therapy. Mol Cancer (2020) 19(1):149. doi: 10.1186/s12943-020-01266-7
Keywords: circRNA, renal cancer, diagnostic, prognostic, clinicopathological, meta-analysis
Citation: Wang W, Xie S, Yuan D, He D, Fang L and Ge F (2022) Systematic Review With Meta-Analysis: Diagnostic, Prognostic and Clinicopathological Significance of CircRNA Expression in Renal Cancer. Front. Oncol. 11:773236. doi: 10.3389/fonc.2021.773236
Received: 16 September 2021; Accepted: 31 December 2021;
Published: 28 January 2022.
Edited by:
Weiqiang Lin, Zhejiang University, ChinaReviewed by:
Ziheng Wang, Affiliated Hospital of Nantong University, ChinaJuan Ren, The First Affiliated Hospital of Xi’an Jiaotong University, China
Linhui Wang, Second Military Medical University, China
Gonghui Li, Sir Run Run Shaw Hospital, China
Copyright © 2022 Wang, Xie, Yuan, He, Fang and Ge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liming Fang, ZmFuZ2xpbWluZzcyQDEyNi5jb20=; Fengfeng Ge, Z2dnZWZlbmdmZW5nQDEyNi5jb20=
†These authors have contributed equally to this work
 Wujun Wang
Wujun Wang Shengfang Xie
Shengfang Xie Dongping Yuan
Dongping Yuan Dandan He
Dandan He Liming Fang
Liming Fang Fengfeng Ge
Fengfeng Ge