- 1Department of Medical Oncology, The First Affiliated Hospital of University of Science and Technology of China (USTC), Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
- 2Geneseeq Research Institute, Nanjing Geneseeq Technology Inc., Nanjing, China
- 3Department of Thyroid and Breast Surgery, The First Affiliated Hospital of University of Science and Technology of China (USTC), Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
Background: Neoadjuvant chemotherapy (NAC) has been expanded to hormone receptor (HR) positive breast cancer (BC) patients with operable disease, to increase the likelihood of breast-conserving surgery. Genomic profiling at baseline would reveal NAC response relevant genomic features and signaling pathways, guiding clinical NAC utilization based on patients’ genomic characteristics.
Methods: We prospectively studied stage II/III BC patients who were eligible for breast-conserving surgery. Patients received epirubicin and cyclophosphamide for 4 cycles, followed by another 4-cycle docetaxel, and human epidermal growth factor receptor (HER2) positive patients were additionally treated with herceptin when using docetaxel (EC-T(H)). NAC responses were evaluated as pathologic complete response (pCR) or non-pathologic complete response (non-pCR). Genomic features related to NAC responses were identified by profiling baseline tumor tissues sampled one day before NAC, using whole-exome sequencing. Differentially expressed genes and up-/down-regulated pathways were investigated by performing RNA-sequencing.
Results: A total of 25 stage II/III BC patients were enrolled, including 5 patients ultimately evaluated as pCR and 20 patients evaluated as non-pCR. PIK3CA (48%) and TP53 (40%) mutations were enriched in patients not achieving pCR. Mutated phosphatidylinositol-3-kinase-AKT (PI3K-AKT) pathway and homologous recombinational repair pathway were also more frequently observed in patients evaluated as non-pCR. Significant arm-level amplifications (8q24.23 and 17q12) and deletions (1p32.2, 4p14, 7q11.23, 10q21.3, 11q23.3, etc.) were identified among patients not achieving pCR, while patients achieving pCR displayed no significant copy number alterations. Significantly up-regulated expression of PI3K-AKT pathway genes was also detected among patients failed to achieve pCR, compared to patients achieving pCR.
Conclusion: Compared to BC patients achieving pCR to NAC, aberrant activation of PI3K-AKT pathway genes were more frequently observed in patients not achieving pCR, consistent with the significant up-regulation of PI3K-AKT pathway gene expression in the non-pCR subgroup. Together, these findings indicate that upregulated PI3K-AKT pathway serves as a potential indicator of lack of response to NAC in stage II/III BC patients, and other effective therapeutic options are urgently needed for those resistant patients.
Introduction
Breast cancer (BC) is the most common cancer among females, and its mortality rate is also the highest for female cancer patients worldwide (1). Neoadjuvant chemotherapy (NAC) has become a widely used systemic therapy before definitive surgery, as it could reduce the extent of surgery, control the tumor size, and increase the probability of curability in early-stage BC (2, 3). For triple-negative breast cancer (TNBC) patients, it was suggested that anthracycline/taxane-based NAC should be offered if patients were node-positive and/or at least T1c. Similarly, anthracycline/taxane-based regimen in combination with trastuzumab were recommended to human epidermal growth factor receptor (HER2) positive BC patients with node-positive or high-risk node-negative (4). Although no evidence directly indicated the optimal NAC regimen in hormone receptor (HR)-positive/HER2-negative BC patients, NAC was suggested to be used instead of adjuvant chemotherapy, as patients treated with NAC had similar prognosis as those with adjuvant chemotherapy (5).
The most common endpoint of NAC is pathologic complete response (pCR), usually defined as “no invasive cancer and in situ cancer in the breast and axillary nodes” or “no invasive cancer in the breast and axillary nodes, irrespective of ductal carcinoma in situ” or “absence of invasive cancer in the breast irrespective of ductal carcinoma in situ or nodal involvement”. pCR can serve as a surrogate endpoint for the prognosis of disease-free survival and overall survival (6). Compared to the patients with non-pathologic complete response (non-pCR), patients achieving pCR had significantly superior disease-free survival, especially for HER2-enriched and TNBC patients (7).
Previous studies indicated that clinical characteristics and some biomarkers were related with pCR. Higher pCR percentages were observed among HR–/HER2+ (38.9%, 95% CI 33.2-44.9%) and TNBC (38.9%, 95% CI 33.2-44.9%) subtypes, compared to HR+/HER2– and HR+/HER2+ (8). Rates of pCR were also higher among patients with family BC history or carrying BRCA1/2 mutations (9, 10). Additionally, higher ratios of CD8+ to forkhead box protein 3+ tumor infiltrating lymphocytes were detected in non-pCR TNBC patients with lower residual cancer burden (RCB) after NAC, representing that anti-tumor immune response was related with the response to NAC (11). However, genomic profiles do not act as a routine guideline for NAC regimen in BC, as the association of BC-related genes or signaling pathways with the response to NAC has not been comprehensively investigated (4).
Whole-exome sequencing (WES), covering almost all protein coding regions in the human genome, has previously been used to identify BC susceptibility genes and biomarkers of treatment response (12, 13). RNA sequencing (RNA-Seq), using next-generation sequencing (NGS) technology, can reveal the quantity of RNA, and detect differentially expressed genes (DEGs) by performing statistical analyses (14). DEGs in specific subgroup will be classified as up- or down-regulated genes, compared to a reference subgroup. Biological processes or signaling pathways associated with DEGs can further be identified (15).
In this study, we aimed to explore genomic features associated with NAC response among stage II/III BC patients who were eligible for breast-conserving surgery. Patients’ tumor tissue samples were performed with WES profiling before receiving epirubicin, cyclophosphamide, followed by docetaxel with or without herceptin (EC-T(H)). We then investigated DEGs between patients achieving and not achieving pCR, and identified up-/down-regulated pathways by RNA-Seq. Our results provided insights into baseline genomic indicators and key signaling pathways of NAC responses, and our findings could potentially help guide clinical NAC utilization.
Methods
Participants and Neoadjuvant Chemotherapy
Female patients with stage II/III primary BC, eligible for breast-conserving surgery, and suitable for receiving NAC were included in this study. All patients received EC-T (H): epirubicin (100 mg/m2), and cyclophosphamide (600 mg/m2) intravenous drip every 3 weeks for 4 cycles, followed by docetaxel (100 mg/m2) intravenously every 3 weeks for 4 cycles with or without herceptin. HER2+ patients were additionally treated with herceptin (8 mg/kg at first infusion, and then 6 mg/kg) every 3 weeks for 4 cycles when receiving docetaxel. Key exclusion criteria included receiving other BC treatment at the same time (e.g., neoadjuvant endocrine therapy and traditional Chinese medicine treatment), with NAC regimen changed depending on physicians’ decisions, no response records of NAC, or unavailable baseline tumor biopsies sampled one day before the beginning of NAC. Each participant provided informed written consent, and this study was approved by the ethics committees of Anhui Provincial Hospital (ethics number 2019KY086).
Receptor Status Determination
The patient’s estrogen receptor (ER) and progesterone receptor (PR) status were determined by immunohistochemistry (IHC). The expression of ER and PR were quantified by the percent of stained cell nuclei. If the expression of each receptor was 1% or greater, the status of ER or PR was considered positive; otherwise, it was identified as negative status. Patients’ HER2 status were primarily analyzed by IHC, with scores of 0, 1+, 2+, or 3+. If the score was 0 or 1+, the status of HER2 was considered to be negative. HER2 positive samples included tumor samples with cores of 3+. Fluorescence in situ hybridization (FISH) was further performed on tumor tissues whose scores were 2+. In FISH analyses, a HER2 positive sample was identified if the ratio of HER2 to centromere 17 was over 2.0 (16).
NAC Evaluation and Pathological Review
The response to NAC was primarily evaluated as pCR or non-pCR. After completing NAC, patients underwent surgical resection, and then specimens were sent to the pathology department for ascertaining surgical margin status. If residual invasive cancer was absent in the complete resected specimen and sampled regional lymph nodes, it was identified as pCR; otherwise, it was identified as non-pCR. NAC responses were further classified into 4 RCB categories: RCB-0 (pCR), RCB-1 (minimal residual disease), RCB-2 (moderate residual disease) and RCB-3 (extensive residual disease) (17).
Baseline Sample Collection and Sequencing
The baseline tumor tissue, sampled one day before receiving NAC, underwent the complete procedure of formalin-fixed paraffin-embedded (FFPE) sample preparation before sequencing.
For WES, genomic DNA from the baseline FFPE sample was extracted with QIAamp DNA FFPE Tissue Kit (QIAGEN). Whole blood samples were used as normal controls, and genomic DNA from control samples was extracted with DNeasy Blood and Tissue Kit (QIAGEN). Extracted genomic DNAs were quantified by Qubit 3.0 using dsDNA HS Assay Kit (ThermoFisher Scientific). All samples were analyzed at a centralized clinical testing center (Nanjing Geneseeq Technology Inc.). Sequencing libraries were prepared using KAPA Hyper Prep Kit (KAPA Biosystems), and targeted enrichment was performed with the xGen Exome Research Panel and Hybridization and Wash Reagents Kit (Integrated DNA Technology). Enriched libraries were sequenced on Illumina Hiseq4000 platform using PE150 sequencing chemistry (Illumina), with a mean coverage depth of 150x for tumor tissue samples and 50x for matched normal blood control samples.
For RNA-seq, total RNA from FFPE samples was extracted with RNeasy FFPE Kit (QIAGEN) and quantified using Bioanalyzer 2100 (Agilent Technologies). Ribosomal RNA and residual genomic DNA were depleted by the KAPA Standard RNA-Seq Kit with RiboErase (HMR) and DNase digestion, followed by purification using the Agencourt RNA Clean XP Beads. Illumina compatible sequencing libraries were prepared using KAPA Stranded RNA-Seq Library Preparation Kit, including RNA fragmentation and priming, double-stranded cDNA synthesis, adaptor ligation and polymerase chain reaction amplification. Libraries were sequenced on Illumina Hiseq NGS platforms (Illumina).
Mutation Calling and Sequencing Data Processing
For WES, FASTQ file quality was controlled by Trimmomatic, removing leading/trailing low quality (reading < 15) or N bases (18). High-quality reads were aligned to the reference human genome (hg19) using the Burrows-Wheeler Aligner Maximal Exact Match (BWA-MEM) with the default parameters (19). Local realignment around indels and base-quality score recalibration were applied using the Genome Analysis Toolkit (GATK) (20). Somatic single-nucleotide variants and small indels were called using MuTect and SCALPEL, respectively (21, 22). Somatic mutation and indel calls were further filtered by excluding: (1) alternate reads < 3 or variant allele frequency < 2%, (2) located in a non-protein-encoding region, or (3) the presence of one variant in public databases (Exome Variant Server, 1000 Genomes Project, and Exome Aggregation Consortium) at a population frequency > 1%. single-nucleotide variant and indel annotation was performed using ANNOVAR. Next, Somatic copy number alteration (CNA) analysis was performed by Genomic Identification of Significant Targets in Cancer (GISTIC), with the reference human genome of hg19. Aberrant regions with False Discovery Rates (q-values) below 0.2 were considered significant, and the most likely gene targets were identified within each significant region.
For RNA-seq, base calling was performed on bal2fastq (Illumina) to generate sequence reads in the format of FASTQ, followed by FASTQ file quality control using Trimmomatic (18). RNA-seq reads passing the quality control were then aligned to the reference human genome (hg19) using STAR algorithm (23).
Differential Expression and Pathway Analysis
Differential expression analysis of RNA-seq data was performed between patients achieving and not achieving pCR, taking normalized read count data and discovering quantitative differences in expression levels, by DESeq2 based on negative binomial distribution. The significant DEG was considered if the adjusted p-value was less than 0.05 and absolute fold-change was greater than 2.
Statistical Analysis
Fisher’s exact test was performed to compare the frequencies between subgroups. All quoted p-values of Fisher’s exact tests were two-tailed, and values less than 0.05 were considered to be statistically significant. Data were analyzed using R software (version 4.0.3).
Results
Patient Overview
Between January 2019 and January 2021, 60 stage II/III primary BC patients planning to receive NAC before breast-conserving surgery in Anhui Provincial Hospital were eligible for participation. Thirty-five of them were excluded due to unavailable baseline tumor biopsy (n=32) or no response records of NAC (n=3). The remaining 25 patients were enrolled in this study, and their baseline tumor tissue samples were profiled using WES. However, 3 of 25 patients did not have enough tumor tissues for further RNA-seq. Therefore, 25 patients were included in the WES analyses, and 22 of them were included in the RNA-seq analyses (Figure 1).
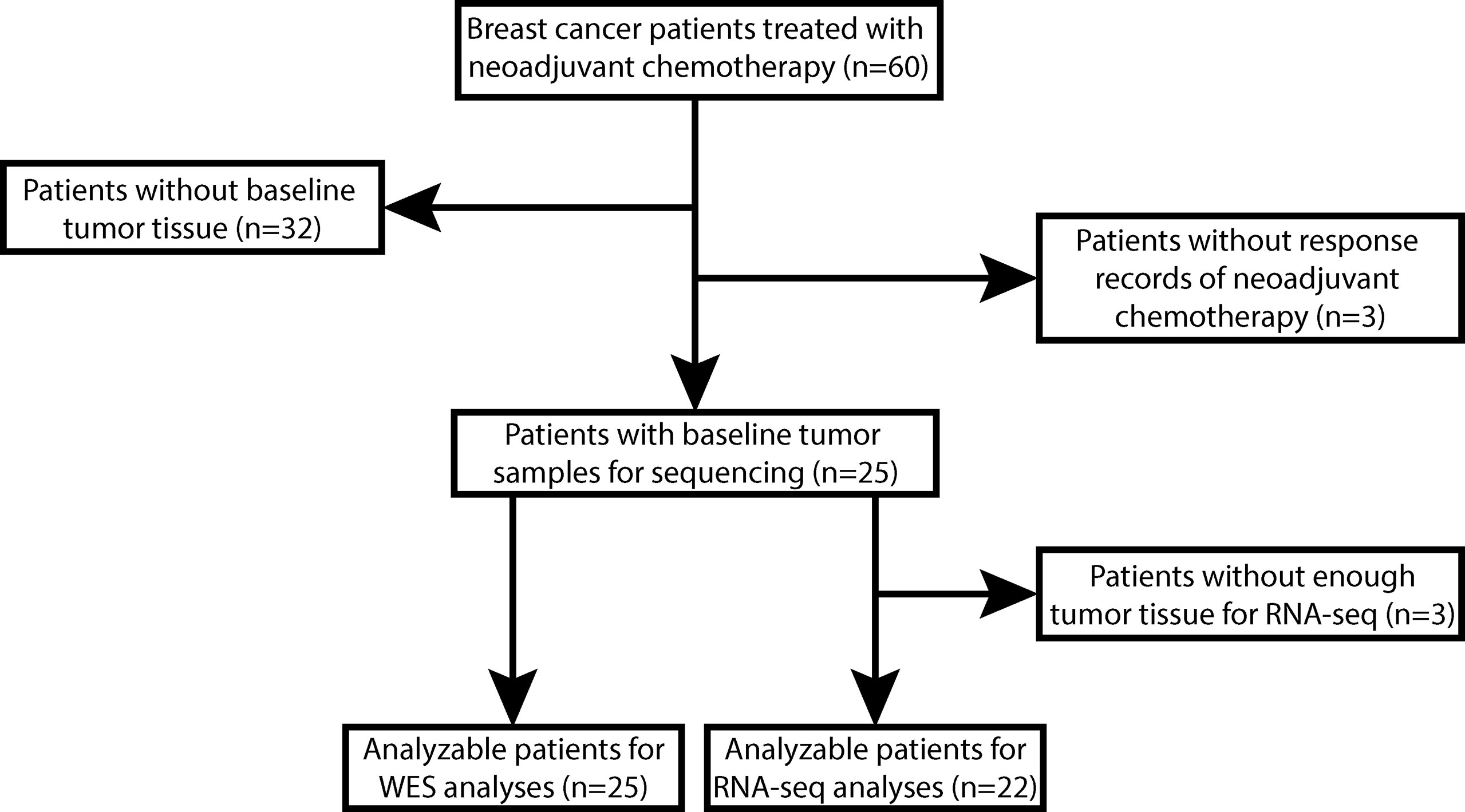
Figure 1 The flowchart of enrollment and analyzable patients. A total of 60 stage II/III breast cancer (BC) patients planned to receive breast-conserving surgery and neoadjuvant chemotherapy (NAC) of epirubicin, cyclophosphamide plus docetaxel with or without herceptin (EC-T(H)) were eligible for participation. We excluded 32 patients without available baseline tumor tissues and 3 patients without NAC response records, and the remaining 25 patients were enrolled in this study. All these 25 patients were analyzable for whole-exome sequencing (WES) data analyses, but 3 patients without enough tumor tissue for RNA sequencing (RNA-Seq) were excluded from RNA-Seq data analyses.
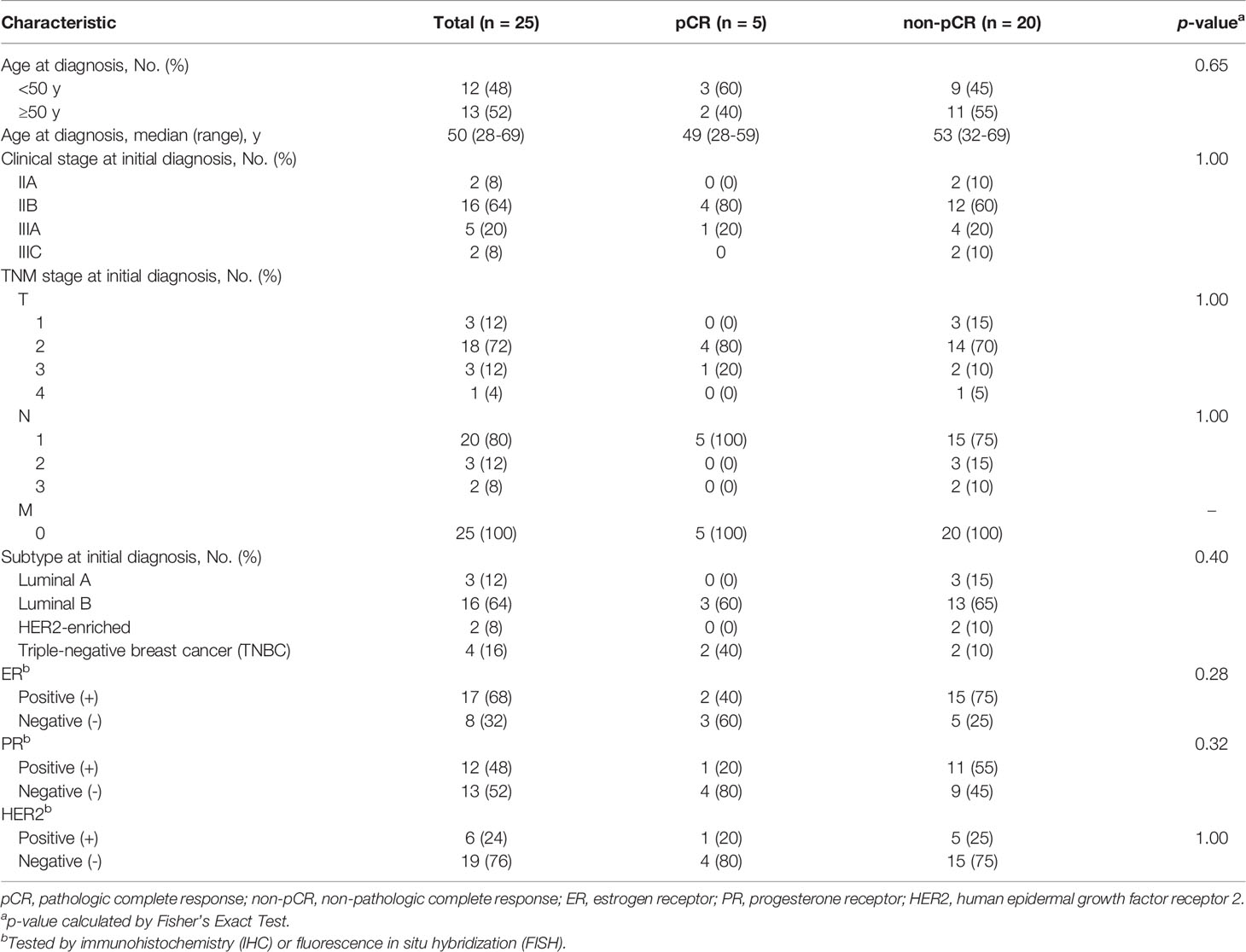
The clinical characteristics of the 25 enrolled patients were summarized in Table 1. Five patients were evaluated as pCR after completing NAC, while the other 20 patients achieved non-pCR responses. Neither Luminal A nor HER2-enriched patients were evaluated as pCR. The median age at diagnosis of these 25 patients was 50 years (range, 28 to 69), and 13 (52%) patients were 50 years or older. At initial diagnosis, 18 of 25 (72%) patients were in clinical stage II, and 16 of 25 (64%) patients were classified as Liminal B subtype. No significant differences in clinical characteristics were detected between patients achieving and not achieving pCR, which was probably due to the limited sample size (Table 1).
Baseline Somatic Mutations and CNAs Associated With Responses to NAC
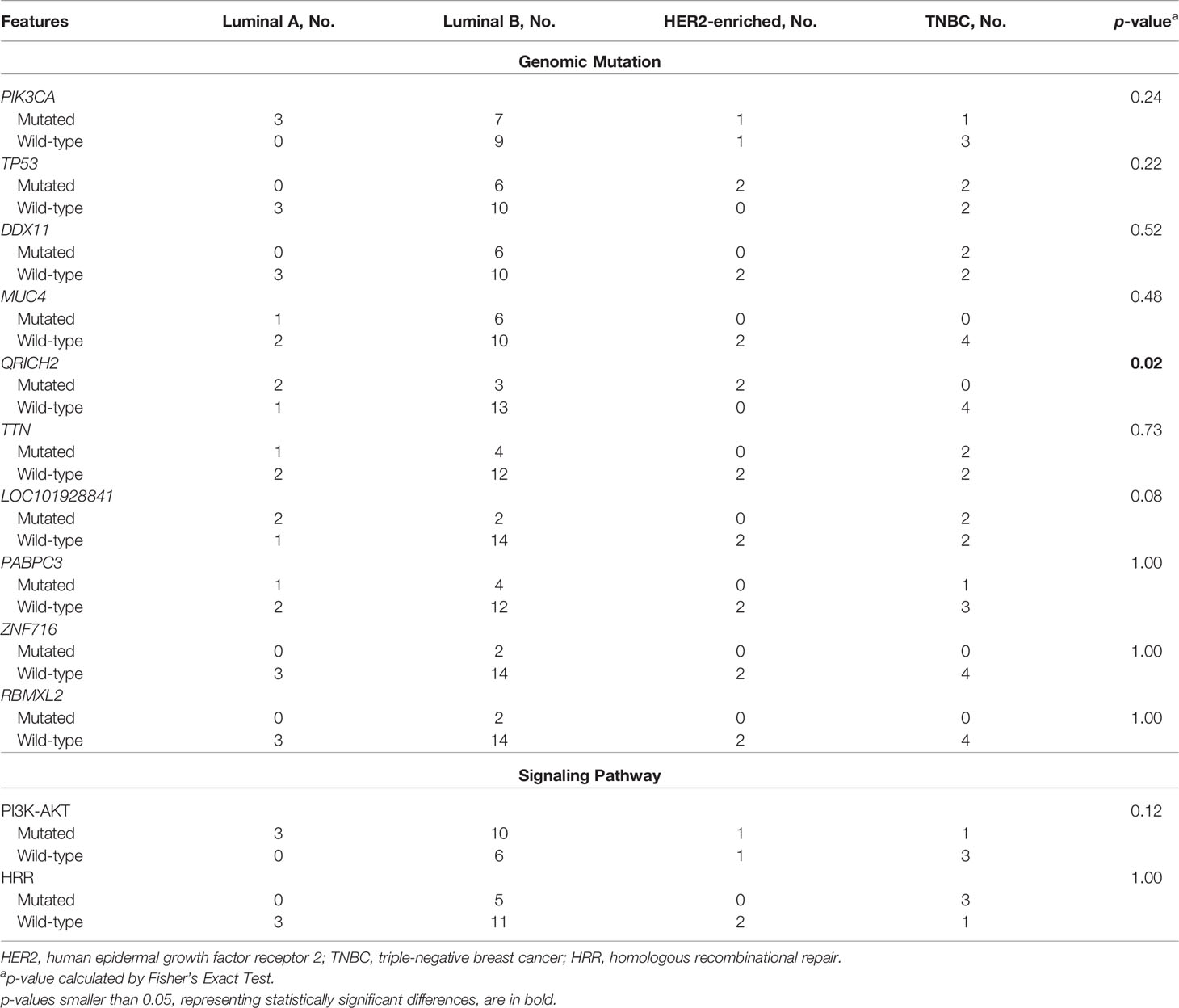
We performed WES on baseline tumor tissue samples of 25 enrolled patients. The top-three mutated genes were PIK3CA (48%), TP53 (40%) and DDX11 (32%). Only 1 of 12 PIK3CA mutations was observed in the 5 patients evaluated as pCR, and no TP53 mutations were identified in these 5 patients. Although some mutated genes were exclusively detected in the non-pCR subgroup (e.g., AK9, PRR12, PTPRD, SLITRK2, etc.), their frequencies were relatively low. Additionally, baseline somatic mutations in 2 key BC-related signaling pathways, the phosphatidylinositol-3-kinase-AKT (PI3K-AKT) pathway and the homologous recombinational repair (HRR) pathway, were more likely to be observed in patients not achieving pCR (Figure 2).
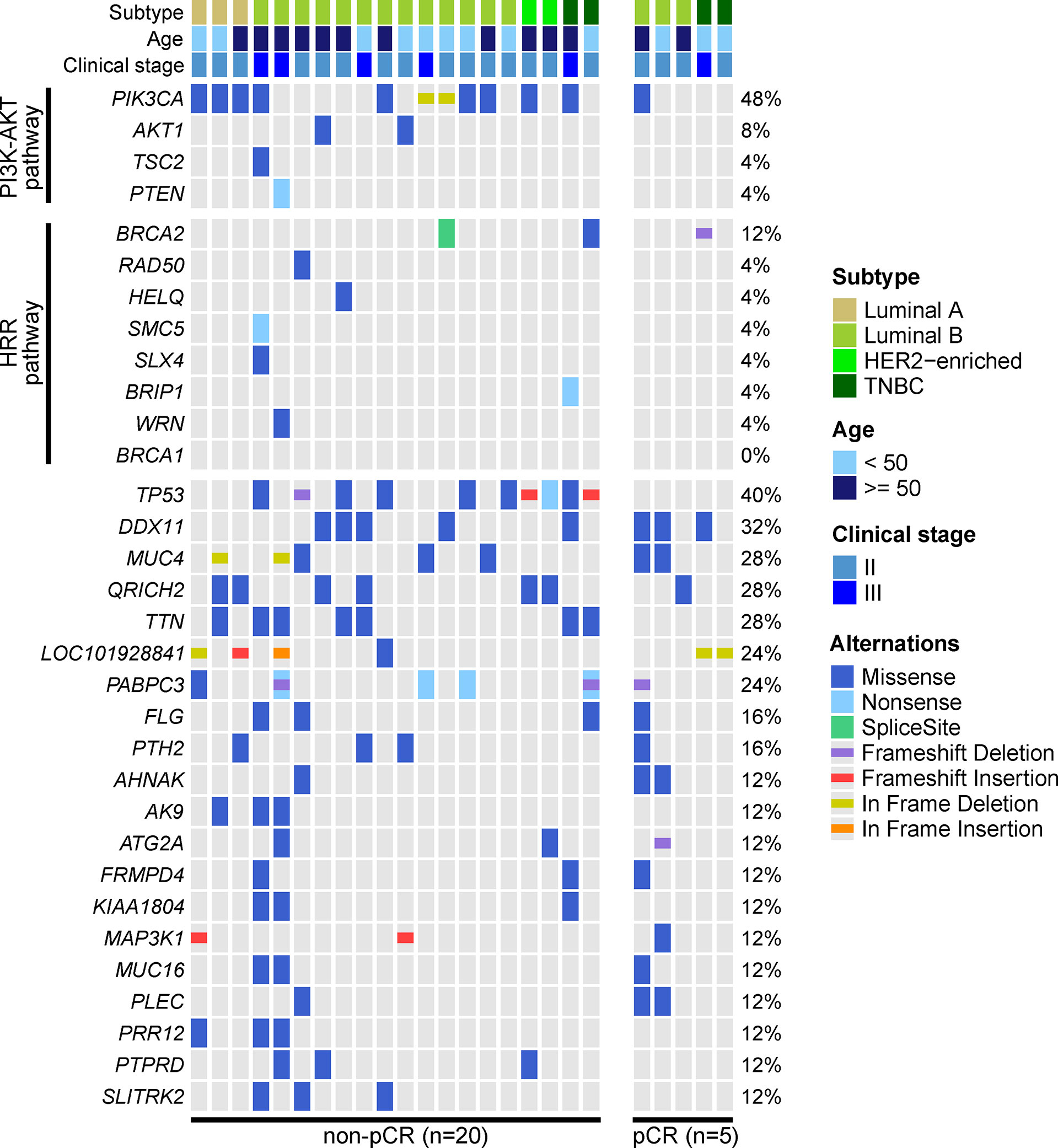
Figure 2 Genetic profile of baseline tumor tissue samples. The baseline tumor tissue samples of 25 patients were performed by WES. Altered genes of the phosphatidylinositol-3-kinase-AKT (PI3K-AKT) pathway and homologous recombinational repair (HRR) pathway and altered genes over 10% frequency were displayed. The most frequently altered genes were PIK3CA, TP53, DDX11, MUC4, QRICH2, and TTN. PIK3CA and TP53 mutations were enriched in patients not achieving pCR, and mutated PI3K-AKT pathway and HRR pathway were also more frequently observed in patients not achieving pCR.
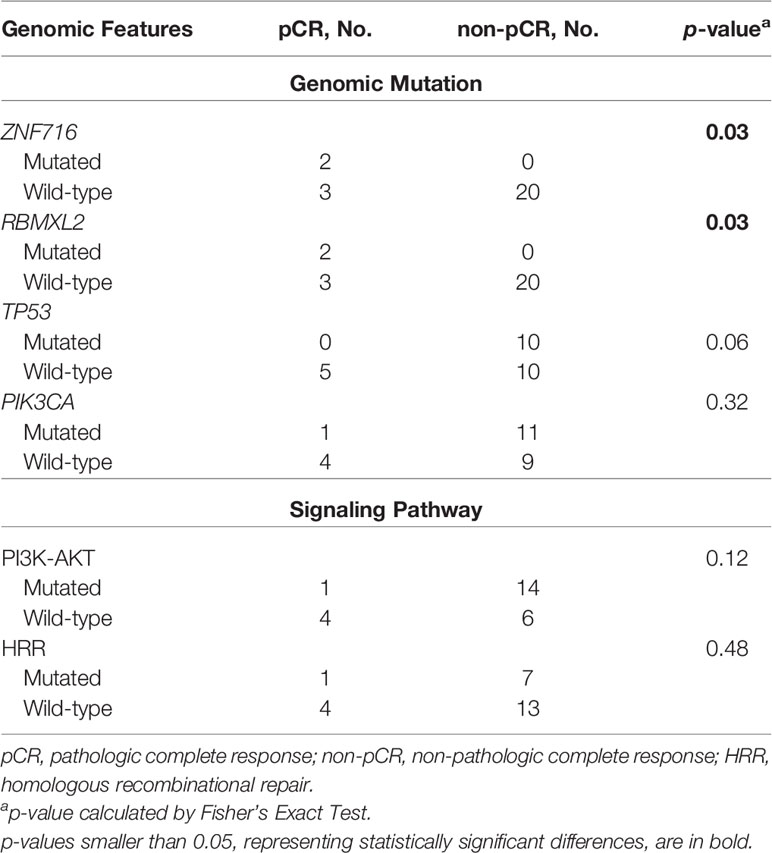
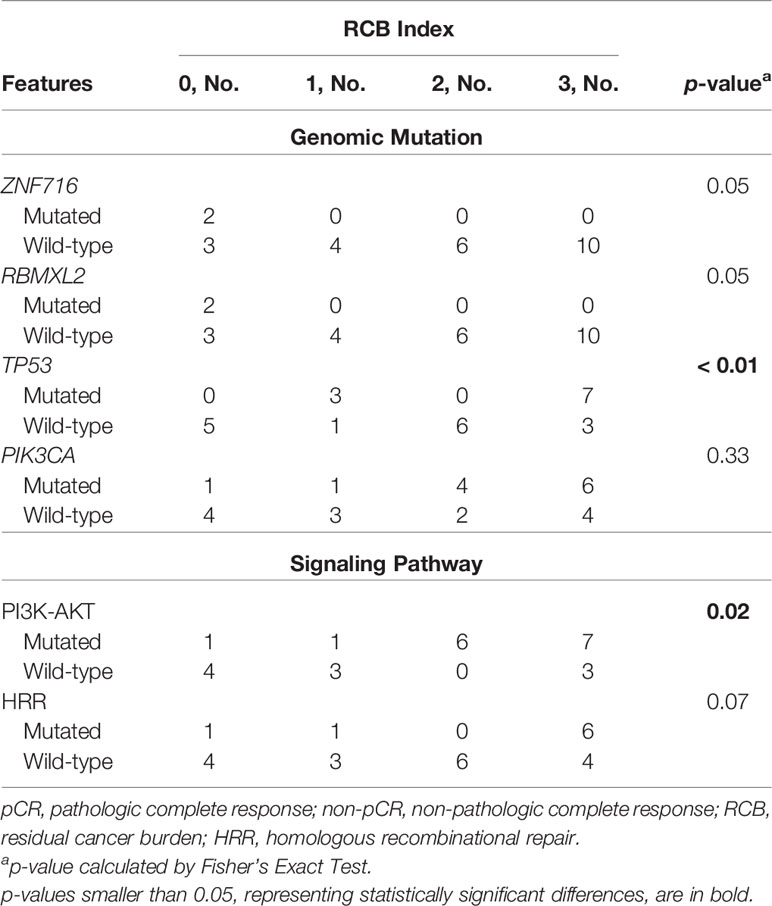
The association between baseline mutations and responses to NAC was explored. ZNF716 (p=0.03) and RBMXL2 (p=0.03) were statistically associated with NAC responses, but extremely low frequencies (n=2) of these 2 mutated genes were observed in patients achieving pCR. We also observed a trend of TP53 and PIK3CA mutations being enriched in patients not achieving pCR, while the associations were not statistically significant (TP53, p=0.06; PIK3CA, p=0.32). Similarly, neither PI3K-AKT (p=0.12) nor HRR pathway (p=0.48) showed statistically significant relationship with NAC responses (Table 2). The baseline somatic mutation of each RCB category and BC subtype were summarized in Tables 3 and 4, respectively. Although TP53 mutations appeared to be enriched in patients not achieving pCR, no patients in RCB Index 2 were detected as TP53 mutation positive. Mutated PI3K-AKT pathway were more frequently observed in RCB Index 2 or 3 patients than patients in the RCB Index 0 or 1 subgroup (Table 3).
We further studied the association of NAC responses with baseline CNAs at the chromosomal arm level. Many BC-related genes, such as ERBB2, GRB7, MIEN1 and STARD3, were located in the region of chromosome 17q12, where significant amplification was observed among patients with non-pCR. Significant amplifications were also detected on chromosome 8q24.23, in the non-pCR subgroup (Figure 3A). Compared to amplifications, deletions were more common in patients not achieving pCR, such as 1p32.3, 4p14, 7q11.23, 10q21.3,11q23.3, etc., and BC relevant genes were frequently observed in the wide peak of each deletion region (Figure 3B). All baseline arm-level CNAs, with wide peak boundaries and genes in peaks, were shown in the Supplementary Table.
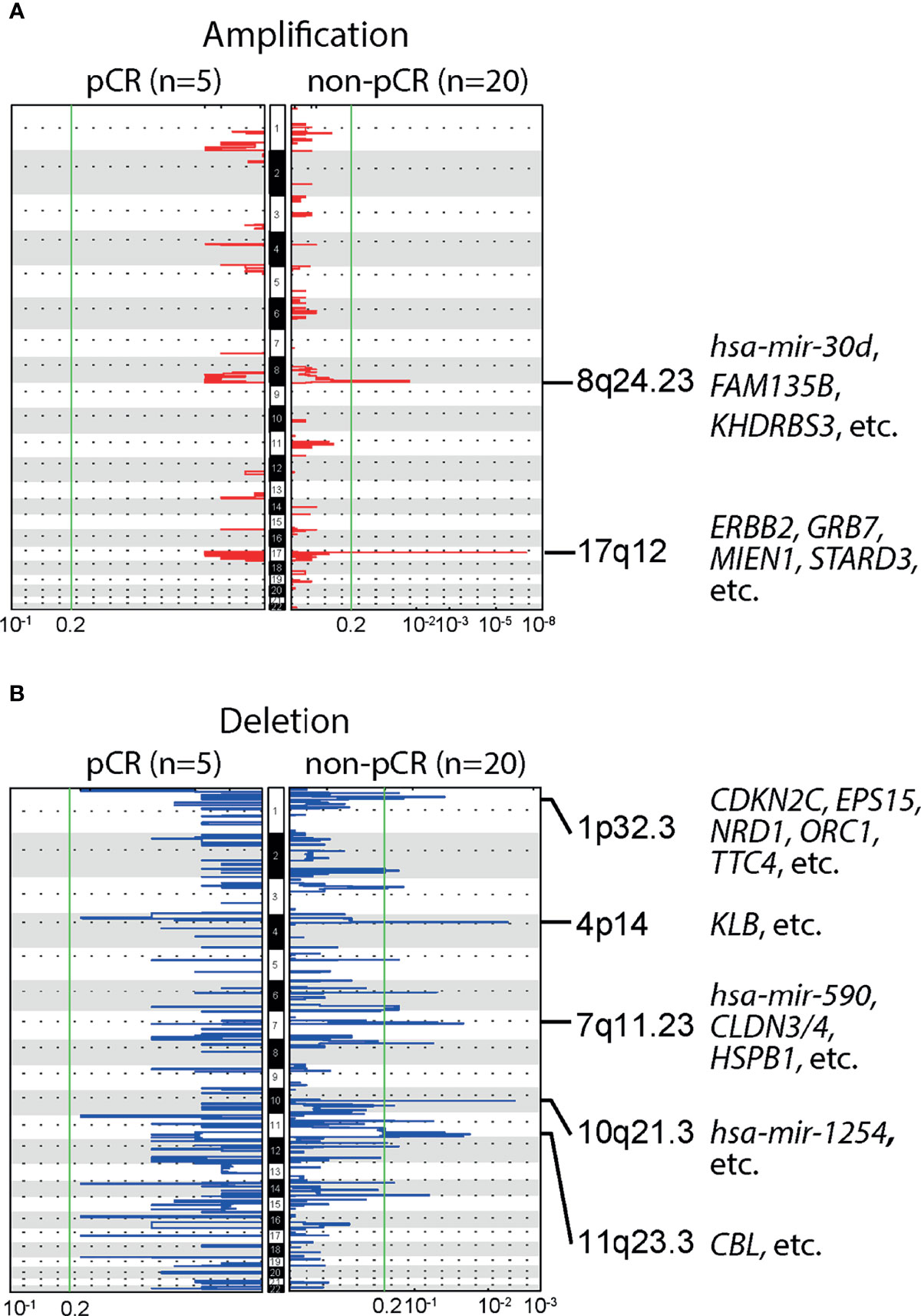
Figure 3 Baseline arm-level copy number alterations (CNAs) and responses to neoadjuvant chemotherapy (NAC). (A) Significant amplifications were detected in the chromosomal region 8q24.23 and 17q12, in patients not achieving pCR, but no significant amplifications were observed in patients achieving pCR. Of note, ERBB2, a gene closely related to breast cancer (BC) was located in 17q12. (B) Multiple significant deletions were detected in patients not achieving pCR, and five regions with the highest q-values were 1p32.3, 4p14, 7q11.23, 10q21.3,11q23.3.
Differential Expression and Up/Down-Regulated Pathways
Our WES data showed that most observed mutations of PIK3CA gene in patients who failed to achieve pCR were gain-of-function mutations, including PIK3CA-H1047R, PIK3CA-E545K, PIK3CA-C420R, etc. We thus compared 3 patients achieving pCR with 19 patients not achieving pCR, investigating whether gain-of-function mutations of PIK3CA gene could influence the gene expression of PI3K-AKT signaling pathway (Figure 4). Compared to the patients who failed to achieve pCR after NAC, 27 DEGs involved in several down-regulated pathways were expressed at significantly lower levels among patients achieving pCR, including COL1A1, COL1A2, COMP, and CREB3L1 genes belonging to PI3K-AKT pathway. In other words, patients not achieving pCR displayed up-regulated gene expression of PI3K-AKT pathway, among whom gain-of-function mutations of PI3K-AKT pathway were identified in previous WES analysis. On the other hand, 34 DEGs were expressed significantly higher among pCR patients, and the most frequently up-regulated pathways, such as metabolic pathways, neuroactive ligand-receptor interaction, glutamatergic synapse, etc., did not have strong relationship with BC development or lack of response of NAC.
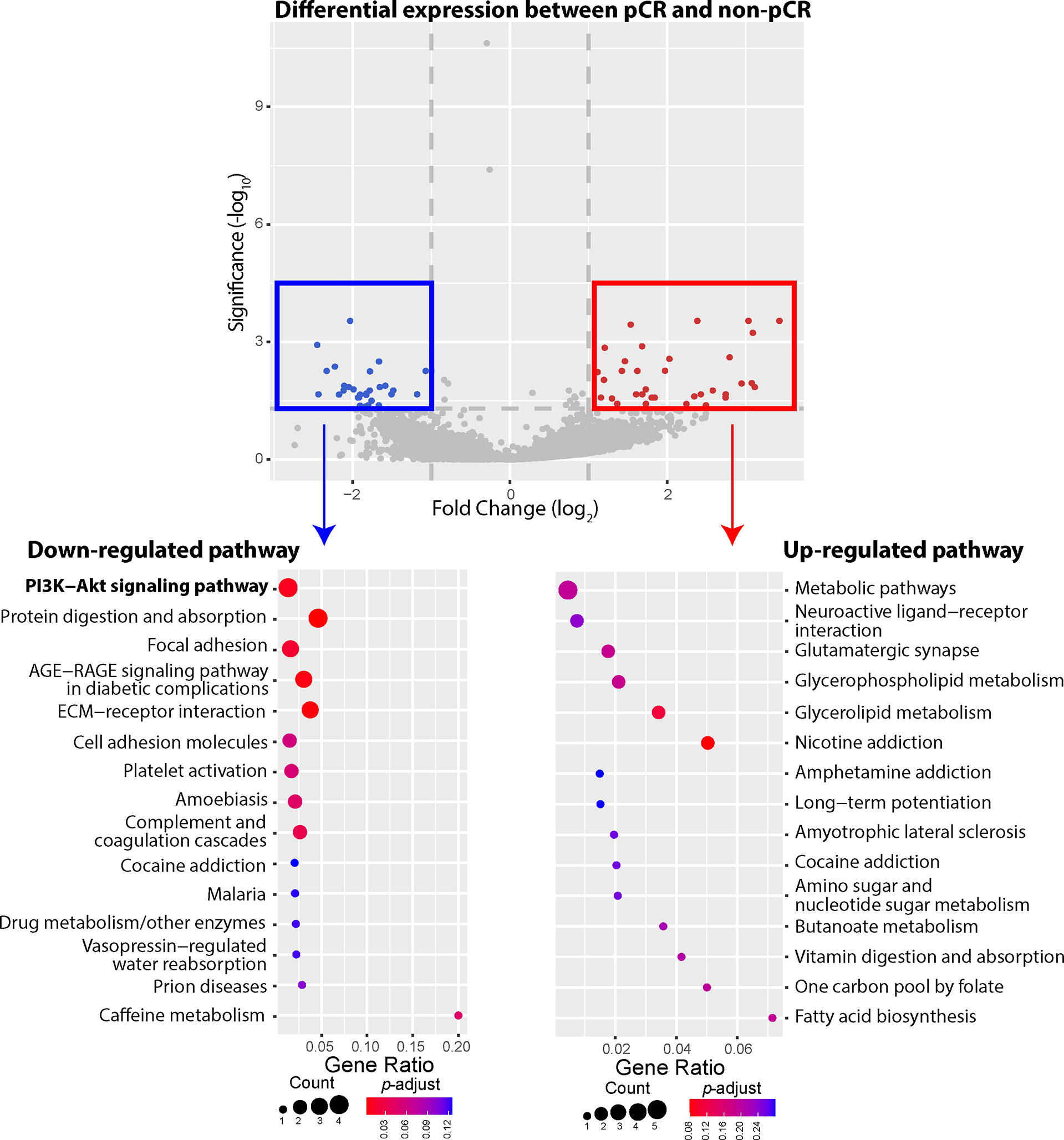
Figure 4 Differentially expressed genes (DEGs) and up-/down-regulated signaling pathways between pathologic complete response (pCR) patients or non-pathologic complete response (non-pCR) patients. A total of 61 DEGs were identified between the pCR subgroup and the non-pCR subgroup, including 34 genes expressed at higher levels and 27 genes expressed at lower levels, in the pCR subgroup, with absolute fold-change greater than 2. The phosphatidylinositol-3-kinase-AKT (PI3K-AKT) pathway was significantly down regulated among patients achieving pCR, with 4 lower expressed genes. Other down-regulated pathways included protein digestion and absorption, focal adhesion, AGE-RAGE, ECM-receptor, etc. On the other hand, higher expressed genes belonged to the pathways of metabolic pathways, neuroactive ligand-receptor interaction, glutamatergic synapse, glycerophospholipid metabolism, glycerolipid metabolism, nicotine addiction pathways, etc.
Discussion
This study reported WES and RNA-Seq data on baseline tumor tissue samples of patients who were treated with NAC of EC-T(H). We compared the genomic features of mutated genes and signaling pathways, DEGs and up-/down-regulated signaling pathways between patients achieving and not achieving pCR.
PIK3CA and TP53 mutations were more frequently observed in patients not achieving pCR than patients achieving pCR, while this trend was not statistically significant due to the relatively small sample size. However, our findings of mutated PIK3CA genes aligned to a previous study including 851 BC patients. In that study, Loibi et al. reported a significantly higher pCR rate among non-carriers of PIK3CA mutation, especially HER2+ patients. An reduced response rate of nab-paclitaxel was also observed in BC patients harboring PIK3CA mutations (24). In another study of Loibi et al. after receiving taxane-based chemotherapy plus herceptin and/or lapatinib, a significantly lower pCR rate and worse disease-free survival were observed in HR+ patients with PIK3CA mutations (25). Additionally, in our study, of 11 patients with altered PIK3CA gene in the non-pCR subgroup, 5 patients harbored PIK3CA-H1047R mutation. GeparSixto, a prospective randomized phase II clinical trial including 92 TNBC patients treated with anthracycline-based neoadjuvant chemotherapy, reported a strong association between PIK3CA-H1047R mutation and a lower pCR rate (26). On the other hand, the association between TP53 mutations and NAC response rate was not completely consistent across different studies. For instance, one study including 450 BC patients receiving anthracycline/taxane-based NAC indicated that TP53 mutations were not statistically associated with pCR (27); however, a meta-analysis suggested that altered TP53 gene was related to better response rates, and this association remained significant when BC patients were stratified according to their NAC regimens (28). Hence, further larger and well-designed prospective studies are required to evaluate the predictive role of TP53 mutation in NAC of EC-T(H).
In our study, altered RBMXL2 and ZNF716 genes were significantly enriched in patients achieving pCR to the EC-T(H) regimen, whereas the frequencies of these two gene were extremely low. The association of RBMXL2 gene, controlling for splicing patterns during meiosis and being essential for male fertility (29), with BC progression or NAC resistance was not well investigated by previous studies. Additionally, few studies have revealed the relationship between ZNF716 and BC, even though another type of zinc finger protein could inhibit the proliferation of BC cells by regulating P53 stability (30). Thus, our findings on these two genes should be interpreted with caution, and further studies with large sample size are needed.
The 8q24.23 chromosomal region of patients not achieving pCR was amplified, in which several genes related to treatment outcomes of BC patients, such as ST3GAL1 and miR-30b. Fan et al. reported that silencing ST3GAL1 gene could reduce GDNF-mediated cell proliferation of BC cells in vitro, and ST3GAL1 overexpression was associated with poor clinical outcomes in BC patients with late-stage disease (31). miR-30b was identified as a herceptin response regulator in another in vitro study on HER2+ BC cell lines, as the target gene of miR-30b, CCNE2, could probably contribute to herceptin resistance (32). Amplification was also detected in the 17q12 chromosomal region of patients with non-pCR responses, including the ERBB2 gene which is related to BC development and progression. It was demonstrated that the amplification of ERBB2 could affect the sensitivity of doxorubicin, another type of anthracyclines, to BC, which was consistent with our findings (33). GRB7, another gene playing an important role in BC development, was also located in this region. Previous study suggested that GRB7 overexpression was an independent factor of poor prognosis (34). Furthermore, we observed several arm-level deletions of BC tumor suppressor genes and favorable BC prognostic genes in patients who failed to achieve pCR, including CDKN2C (35), EPS15 (36), etc. Of note, there were a considerable number of genes related to BC development and progression in the regions where deletions were identified, including CDKN2C, EPS15, CBL, etc. Their associations with pCR rate of NAC should be further investigated separately.
Our WES data also demonstrated that PIK3CA mutations were the main mutations in PI3K-AKT signaling pathway, which comprises a group of intracellular signal transducer enzymes. This pathway was very likely to be associated to the response to NAC for BC patients. PIK3CA mutations and AKT activation by phosphorylation (pAKT) are frequently detected in BC, and pAKT regulates growth, proliferation, differentiation, tumorigenesis and other critical cellular activities (37, 38). BC tumor cells with PIK3CA mutations had lower probability of early apoptosis after being treated with epirubicin, and abnormal protein expression in PI3K-AKT signaling pathway, suggesting the association between chemotherapy resistance and activated PI3K-AKT pathway in TNBC (39). In one BC study, mutated PI3K-AKT pathways were detected among 29.8% TNBC patients, and over 70% of these patients carried PIK3CA mutations, which was consistent with our findings in this study including 4 BC subtypes (40). We further observed that the PI3K-AKT pathway was up-regulated among patients not achieving pCR, compared to those with pCR to NAC. Although there is no sufficient evidence supporting that mutated PIK3CA genes or mutated PI3K-AKT pathways are associated with treatment benefit of anthracyclines for BC, the interaction between mutated PIK3CA gene/PI3K-AKT pathway and chemotherapy agents, like anthracyclines and taxanes, has been indicated in vitro (41). For example, doxorubicin could induce AKT activation in BC cells, and overexpression of AKT in BC cells transfected with HER2 could result in the resistance to doxorubicin or other chemotherapeutic agents (42, 43). Additionally, lower pCR rates were observed in HER2+ BC patients with PIK3CA alterations than without PIK3CA alterations, even though the association between PI3K-AKT pathway biomarkers and docetaxel benefit was not identified (44–46). In our study, both epirubicin and docetaxel were included in the NAC regimen, and arm-level ERBB2 amplification was of note detected within patients not achieving pCR. Thus, the combination of up-regulated PI3K-AKT pathway and ERBB2 amplification could partially explain why some patients achieved relatively poor responses to NAC, in addition to other altered genes that we discussed before.
According to the National Comprehensive Cancer Network guidelines, the Oncotype DX breast recurrence score tests could help personalize systemic adjuvant treatment of HR+/HER2- and node-negative BC patients with tumor size greater than 0.5 cm. This genomic test is able to predict BC patients’ benefits of chemotherapy, by analyzing 21 cancer related genes and assigning a recurrence score (47). However, currently, genomic profiles are not strongly recommended as a routine guideline for clinical decisions on NAC, even though many studies have investigated whether genomic profiles could help BC patients select the optimal NAC regimen. A previous study suggested that chemoresistant genes existed prior to NAC, and clonal dynamics of these genes were involved in chemoresistance development (48). Another study demonstrated that BC patients with deficient BRCA mutations displayed higher pCR rates than BRCA-proficient or wild type patients, and the relationship remained significant in Luminal BC subtypes. Of note, no differences in survival were observed between patients with and without BRCA mutations (49). However, another research on taxane- and/or anthracycline-based NAC revealed that the BRCA alternation was associated with reduced progression-free survival in BC patients, in addition to poorer clinical response rates (50). Other BC related genetic aberrations predicting chemosensitivity included TP53 mutation, PIK3CA mutation, ERBB2 amplification, and CCND1/2 amplification (51). Moreover, conducting genomic testing before NAC could also help define mutations predicting sensitivity of chemotherapy and survival in poor-prognosis BC patients, such as MUC17 and PCNX1 genes (52). Hence, we proposed that BC patients could take genomic sequencing for selecting optimal options of NAC and detecting potential NAC drug resistance.
It is of great clinical importance to explore more effective therapeutic options for patients who are resistant to the EC-T(H) NAC regimen. In general, extended adjuvant therapy, including chemotherapy, target therapy, and radiotherapy, may be an option for patients who fail to achieve pCR to EC-T(H). Similar to distinctive strength of associations between pCR and prognosis in each BC subtype (6), the development of subsequent treatment plans for BC patients with different subtypes varies considerably. The CREATE-X study suggested that patients not achieving pCR could consider the addition of capecitabine to improve the prognosis, especially TNBC patients, with 13.7% increase in 5-year disease-free survival rate (53). Endocrine therapy (ET) plus CDK4/6 inhibitors might be an option for HR+/HER2- patients, among whom significant improvement in prognosis was not observed in the CREATE-X study. In the monarchE trial, HR+/HER2- early BC patients receiving Abemaciclib for 2 years combined with ET had significantly superior invasive disease-free survival than those using ET alone (54). Hence, U.S. Food and Drug Administration has approved Abemaciclib with ET in the treatment of early BC. However, neither the PENELOPE-B trial nor the PALLAS trial showed HR+/HER2- patients could have improved invasive disease-free survival by receiving Palbociclib for 1 or 2 years combined with ET (55, 56). For HER2+ BC patients not achieving pCR, they could potentially receive T-DM1 in adjuvant therapy, with reduced free of invasive disease rate and distant recurrence rate by 11.3% and 6.7% in 3-year follow-up, respectively (57). Postmastectomy radiotherapy might be another option for patients with residual disease after NAC. Fowble et al. suggested that patients in clinical stage III could benefit from radiotherapy, regardless of their responses to previous NAC (58). Furthermore, BC patients with potential resistance to EC-T(H) NAC regimen might consider receiving other neoadjuvant therapy regimens to improve pCR rates. The GeparSixto trial suggested that the pCR rate increased by 16.3% when carboplatin was added into the anthracycline-based NAC regimen for TNBC patients, whereas the improvement of pCR rate was not significant in HER2+ BC patients (59). A meta-analysis including 9 randomized clinical trials also suggested higher a pCR rate in patients receiving platinum-based NAC, compared to those treated with platinum-free NAC; however, BC patients with BRCA mutations did not show significantly increased pCR rates when receiving platinum-based NAC (60). For patients harboring PIK3CA mutations, the addition of letrozole to neoadjuvant ET might help improve patients’ disease-free survival and overall survival (61). However, the NeoALTTO trial demonstrated that HER2+ patients with PIK3CA mutations did not had a pCR rate to neoadjuvant HER2-targeted therapy as high as patients with wide-type counterparts, and suggested the possibility of using anti-HER2 treatment combined with PI3K inhibitors (62). Compared to pan-PI3K inhibitors [such as Buparlisib and Pictilisib (63)], frequently added in ET or paclitaxel treatment for advanced BC patients, isoform-specific PI3K inhibitors [such as Alpelisib and Taselisib (63)] were generally used according to specific mutated genes in PI3K-AKT pathway. The addition of Taselisib to neoadjuvant ET was associated with higher proportion of objective response among HR+/HER2- stage I-III BC patients, but the improvement in pCR rate was not significant (64). Another study focused on HR+/HER2- BC patients with T1c-T3 disease showed that the combination of Alpelisib with 24-week neoadjuvant letrozole treatment could not significantly improve patients’ responses (65). Although our study revealed that mutated PI3K-AKT pathway could potentially indicate the lack of response to EC-T(H) regimen, further prospective studies or clinical trials should be launched to investigate how to personalize optimal treatment to BC patients with NAC resistance, according to patients’ receptor status and genomic profiles.
The main strength of this study was using WES and RNA-Seq simultaneously to explore genomic characteristics related to the NAC response for BC patients. The association of PI3K-AKT pathway with NAC responses was primarily investigated by somatic mutations in baseline tumor tissues and then confirmed by pathway analyses based on RNA-Seq data. Higher-expressed genes and up-regulated PI3K-AKT pathway in patients not achieving pCR could probably explain how mutated PI3K-AKT pathway genes, such as PIK3CA, AKT1 and PTEN, influence the response to the NAC of EC-T(H). Another strength was the time point of sampling baseline tumor tissues. Compared to other retrospective studies where baseline tumor tissues were sampled at initial diagnoses or couples of weeks before NAC, our baseline tumor tissue biopsies were sampled one day before the beginning of NAC. This means we did not need to consider the confounding effects of mutation changes, induced by unknown treatments between diagnosis and NAC.
On the other hand, the primary limitation of our study was the relatively small sample size. Although the EC-T(H) was one of top recommended NAC regimens, we had to exclude patients who could and did receive other potentially beneficial and feasible BC treatment concurrently with NAC of EC-T(H). In addition, our patients were counselled to avoid using non-evidence-based treatment or other treatment without healthcare professionals’ authorization, whereas patients’ compliance to the EC-T(H) regimen was not as good as expected, leading to a part of potentially eligible participants being further excluded. As a result, we observed a series of trends without statistical significance, such as the enrichment of mutated PIK3CA and TP53 genes in patients not achieving pCR compared to patients with non-pCR, and the potential relationship between mutated PI3K-AKT pathway and non-pCR. Therefore, in the next phase, multi-center research with a larger sample size is needed. Another limitation of our study was that both HER2+ patients with herceptin treatment and HER2- patients not receiving herceptin were included in this study. However, due to the limited sample size, we were not able to perform subgroup analyses in HER2+ or HER2- patients separately, to further explore the potential association between mutated PI3K-AKT pathway and lack of response to NAC in each subpopulation. In our data, a total of 6 patients with positive HER2 status, including 4 Luminal B and 2 HER2-enriched patients, were treated with herceptin, and 1 of 6 successfully achieved pCR to EC-T(H). Hence, further studies focused on HER2+ BC patients receiving NAC of EC-T(H) could be launched in the future. For instance, comparing the strength of association of mutated PI3K-AKT pathway with lacking response to the EC-T(H) NAC regimen, between Luminal B patients with and without positive HER2 status. Finally, the tumor tissue biopsies were collected for sequencing in our study, but tissue samples might not comprehensively capture the entire genomic profile of the tumor due to the heterogenicity of BC (54).
Conclusion
In summary, our data suggest that patient’s baseline genomic profile could be taken into consideration when making NAC regimen decisions for stage II/III BC patients. Higher aberrancy of PI3K-AKT pathway genes and up-regulated gene expression were identified in baseline tumor biopsies of patients not achieving pCR to EC-T(H) regimen, as compared to patients achieving pCR. Our WES data also revealed that patients not achieving pCR had significant arm-level CNAs of breast cancer-relevant genes, including ERBB2 amplification. Together, these findings indicate that upregulated PI3K-AKT signaling pathway serves as potential indicator of lack of response to NAC in stage II/III BC patients, and other effective therapeutic options are urgently needed for those resistant patients.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://ngdc.cncb.ac.cn/gsa-human/, HRA002168. Further inquiries can be directed to the corresponding author/s.
Ethics Statement
The studies involving human participants were reviewed and approved by The Ethics Committees of Anhui Provincial Hospital (Ethics Number 2019KY086). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
MD and BS contributed equally to the work. YP designed the study. MD was responsible for patient recruitment and registration. BS and XH collected samples and data. XM and YP provided administrative support. XZ, FW, LZ, and QO analyzed data and interpreted results. All authors wrote and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This investigation was supported by University of Science and Technology of China (New Medicine Joint Foundation of USTC, grant No. WK9110000058), Anhui Provincial Department of Science and Technology (Anhui Science and Technology Research Project, 1704a0802148; Anhui Key Research and Development Projects, grant No. 1804h08020259 & 202104j07020040), and Natural Science Foundation of Anhui Province (grant No. 1908805MH260).
Conflict of Interest
XZ, FW, LZ, and QO are employees of Nanjing Geneseeq Technology Inc., China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank all the patients who participated in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.784985/full#supplementary-material
Abbreviations
BC, breast cancer; DEG, differentially expressed gene; ER, estrogen receptor; ET, endocrine therapy; FFPE, formalin-fixed paraffin-embedded; FISH, Fluorescence in situ hybridization; HER2, human epidermal growth factor receptor 2; HR, hormone receptor; IHC, immunohistochemistry; NAC, neoadjuvant chemotherapy; NGS, next-generation sequencing; non-pCR, non-pathologic complete response; pCR, pathologic complete response; PR, progesterone receptor; RNA-Seq, RNA sequencing; TNBC, triple-negative breast cancer; WES, whole-exome sequencing.
References
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int J Cancer (2015) 136(5):E359–E86. doi: 10.1002/ijc.29210
2. Charfare H, Limongelli S, Purushotham A. Neoadjuvant Chemotherapy in Breast Cancer. J Br Surg (2005) 92(1):14–23. doi: 10.1002/bjs.4840
3. Schott AF, Hayes DF. Defining the Benefits of Neoadjuvant Chemotherapy for Breast Cancer. J Clin Oncol (2012) 30(15):1747–9. doi: 10.1200/JCO.2011.41.3161
4. Korde LA, Somerfield MR, Carey LA, Crews JR, Denduluri N, Hwang ES, et al. Neoadjuvant Chemotherapy, Endocrine Therapy, and Targeted Therapy for Breast Cancer: ASCO Guideline. J Clin Oncol (2021) 39(13):1485–505. doi: 10.1200/JCO.20.03399
5. Fisher B, Brown A, Mamounas E, Wieand S, Robidoux A, Margolese RG, et al. Effect of Preoperative Chemotherapy on Local-Regional Disease in Women With Operable Breast Cancer: Findings From National Surgical Adjuvant Breast and Bowel Project B-18. J Clin Oncol (1997) 15(7):2483–93. doi: 10.1200/JCO.1997.15.7.2483
6. Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, et al. Pathological Complete Response and Long-Term Clinical Benefit in Breast Cancer: The CTNeoBC Pooled Analysis. Lancet (2014) 384(9938):164–72. doi: 10.1016/S0140-6736(13)62422-8
7. Von Minckwitz G, Untch M, Blohmer J-U, Costa SD, Eidtmann H, Fasching PA, et al. Definition and Impact of Pathologic Complete Response on Prognosis After Neoadjuvant Chemotherapy in Various Intrinsic Breast Cancer Subtypes. J Clin Oncol (2012) 30(15):1796–804. doi: 10.1200/JCO.2011.38.8595
8. Houssami N, Macaskill P, von Minckwitz G, Marinovich ML, Mamounas E. Meta-Analysis of the Association of Breast Cancer Subtype and Pathologic Complete Response to Neoadjuvant Chemotherapy. Eur J Cancer (2012) 48(18):3342–54. doi: 10.1016/j.ejca.2012.05.023
9. Paluch-Shimon S, Friedman E, Berger R, Papa MZ, Dadiani M, Friedman N, et al. Does Pathologic Complete Response Predict for Outcome in BRCA Mutation Carriers With Triple-Negative Breast Cancer?, Chicago, Illinois:American Society of Clinical Oncology (2014).
10. Von Minckwitz G, Hahnen E, Fasching PA, Hauke J, Schneeweiss A, Salat C, et al. Pathological Complete Response (pCR) Rates After Carboplatin-Containing Neoadjuvant Chemotherapy in Patients With Germline BRCA (G BRCA) Mutation and Triple-Negative Breast Cancer (TNBC): Results From GeparSixto. Am Soc Clin Oncol (2014) 32(15_supp):1005–1005. doi: 10.1200/jco.2014.32.15_suppl.1005
11. Miyashita M, Sasano H, Tamaki K, Chan M, Hirakawa H, Suzuki A, et al. Tumor-Infiltrating CD8+ and FOXP3+ Lymphocytes in Triple-Negative Breast Cancer: Its Correlation With Pathological Complete Response to Neoadjuvant Chemotherapy. Breast Cancer Res Treat (2014) 148(3):525–34. doi: 10.1007/s10549-014-3197-y
12. Beltran H, Eng K, Mosquera JM, Sigaras A, Romanel A, Rennert H, et al. Whole-Exome Sequencing of Metastatic Cancer and Biomarkers of Treatment Response. JAMA Oncol (2015) 1(4):466–74. doi: 10.1001/jamaoncol.2015.1313
13. Chandler MR, Bilgili EP, Merner ND. A Review of Whole-Exome Sequencing Efforts Toward Hereditary Breast Cancer Susceptibility Gene Discovery. Hum Mutat (2016) 37(9):835–46. doi: 10.1002/humu.23017
14. Chu Y, Corey DR. RNA Sequencing: Platform Selection, Experimental Design, and Data Interpretation. Nucleic Acid Ther (2012) 22(4):271–4. doi: 10.1089/nat.2012.0367
15. Calura E, Martini P. Summarizing RNA-Seq Data or Differentially Expressed Genes Using Gene Set, Network, or Pathway Analysis. In: RNA Bioinformatics. New York, NY: Springer (2021). p. 147–79.
16. Hammond MEH, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Immunohistochemical Testing of Estrogen and Progesterone Receptors in Breast Cancer. J Clin Oncol (2010) 28(16):2784–95. doi: 10.1200/jco.2009.25.6529
17. Symmans WF, Peintinger F, Hatzis C, Rajan R, Kuerer H, Valero V, et al. Measurement of Residual Breast Cancer Burden to Predict Survival After Neoadjuvant Chemotherapy. J Clin Oncol (2007) 25(28):4414–22. doi: 10.1200/JCO.2007.10.6823
18. Bolger AM, Lohse M, Usadel B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics (2014) 30(15):2114–20. doi: 10.1093/bioinformatics/btu170
19. Li H, Durbin R. Fast and Accurate Short Read Alignment With Burrows–Wheeler Transform. Bioinformatics (2009) 25(14):1754–60. doi: 10.1093/bioinformatics/btp324
20. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A Framework for Variation Discovery and Genotyping Using Next-Generation DNA Sequencing Data. Nat Genet (2011) 43(5):491–8. doi: 10.1038/ng.806
21. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive Detection of Somatic Point Mutations in Impure and Heterogeneous Cancer Samples. Nat Biotechnol (2013) 31(3):213–9. doi: 10.1038/nbt.2514
22. Fang H, Bergmann EA, Arora K, Vacic V, Zody MC, Iossifov I, et al. Indel Variant Analysis of Short-Read Sequencing Data With Scalpel. Nat Protoc (2016) 11(12):2529–48. doi: 10.1038/nprot.2016.150
23. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics (2013) 29(1):15–21. doi: 10.1093/bioinformatics/bts635
24. Loibl S, Treue D, Budczies J, Weber K, Stenzinger A, Schmitt WD, et al. Mutational Diversity and Therapy Response in Breast Cancer: A Sequencing Analysis in the Neoadjuvant GeparSepto Trial. Clin Cancer Res (2019) 25(13):3986–95. doi: 10.1158/1078-0432.CCR-18-3258
25. Loibl S, Majewski I, Guarneri V, Nekljudova V, Holmes E, Bria E, et al. PIK3CA Mutations Are Associated With Reduced Pathological Complete Response Rates in Primary HER2-Positive Breast Cancer: Pooled Analysis of 967 Patients From Five Prospective Trials Investigating Lapatinib and Trastuzumab. Ann Oncol (2016) 27(8):1519–25. doi: 10.1093/annonc/mdw197
26. Guo S, Loibl S, von Minckwitz G, Darb-Esfahani S, Lederer B, Denkert C. PIK3CA H1047R Mutation Associated With a Lower Pathological Complete Response Rate in Triple-Negative Breast Cancer Patients Treated With Anthracycline-Taxane–Based Neoadjuvant Chemotherapy. Cancer Res Treat (2020) 52(3):689. doi: 10.4143/crt.2019.497
27. Darb-Esfahani S, Denkert C, Stenzinger A, Salat C, Sinn B, Schem C, et al. Role of TP53 Mutations in Triple Negative and HER2-Positive Breast Cancer Treated With Neoadjuvant Anthracycline/Taxane-Based Chemotherapy. Oncotarget (2016) 7(42):67686. doi: 10.18632/oncotarget.11891
28. Chen Q, Sjölander A, Långström N, Rodriguez A, Serlachius E, D’Onofrio BM, et al. Maternal Pre-Pregnancy Body Mass Index and Offspring Attention Deficit Hyperactivity Disorder: A Population-Based Cohort Study Using a Sibling-Comparison Design. Int J Epidemiol (2014) 43(1):83–90. doi: 10.1093/ije/dyt152
29. Ehrmann I, Crichton JH, Gazzara MR, James K, Liu Y, Grellscheid SN, et al. An Ancient Germ Cell-Specific RNA-Binding Protein Protects the Germline From Cryptic Splice Site Poisoning. Elife (2019) 8:e39304. doi: 10.7554/eLife.39304
30. Hu R, Peng G, Dai H, Breuer E-K, Stemke-Hale K, Li K, et al. ZNF668 Functions as a Tumor Suppressor by Regulating P53 Stability and Function in Breast Cancer. Cancer Res (2011) 71(20):6524–34. doi: 10.1158/0008-5472.CAN-11-0853
31. Fan T-C, Yeo HL, Hsu H-M, Yu J-C, Ho M-Y, Lin W-D, et al. Reciprocal Feedback Regulation of ST3GAL1 and GFRA1 Signaling in Breast Cancer Cells. Cancer Lett (2018) 434:184–95. doi: 10.1016/j.canlet.2018.07.026
32. Tormo E, Adam-Artigues A, Ballester S, Pineda B, Zazo S, González-Alonso P, et al. The Role of miR-26a and miR-30b in HER2+ Breast Cancer Trastuzumab Resistance and Regulation of the CCNE2 Gene. Sci Rep (2017) 7(1):1–9. doi: 10.1038/srep41309
33. Järvinen TA, Tanner M, Rantanen V, Bärlund M, Borg Å, Grénman S, et al. Amplification and Deletion of Topoisomerase Iiα Associate With ErbB-2 Amplification and Affect Sensitivity to Topoisomerase II Inhibitor Doxorubicin in Breast Cancer. Am J Pathol (2000) 156(3):839–47. doi: 10.1016/S0002-9440(10)64952-8
34. Ramsey B, Bai T, Newell AH, Troxell M, Park B, Olson S, et al. GRB7 Protein Over-Expression and Clinical Outcome in Breast Cancer. Breast Cancer Res Treat (2011) 127(3):659–69. doi: 10.1007/s10549-010-1010-0
35. Wang W-J, Wang H, M-s W, Huang Y-Q, Ma Y-Y, Qi J, et al. Assessing the Prognostic Value of Stemness-Related Genes in Breast Cancer Patients. Sci Rep (2020) 10(1):1–10. doi: 10.1038/s41598-020-73164-3
36. Dai X, Liu Z, Zhang S. Over-Expression of EPS15 Is a Favorable Prognostic Factor in Breast Cancer. Mol Biosyst (2015) 11(11):2978–85. doi: 10.1039/C5MB00219B
37. Paplomata E, O’Regan R. The PI3K/AKT/mTOR Pathway in Breast Cancer: Targets, Trials and Biomarkers. Ther Adv Med Oncol (2014) 6(4):154–66. doi: 10.1177/1758834014530023
38. Yang SX, Polley E, Lipkowitz S. New Insights on PI3K/AKT Pathway Alterations and Clinical Outcomes in Breast Cancer. Cancer Treat Rev (2016) 45:87–96. doi: 10.1016/j.ctrv.2016.03.004
39. Hu H, Zhu J, Zhong Y, Geng R, Ji Y, Guan Q, et al. PIK3CA Mutation Confers Resistance to Chemotherapy in Triple-Negative Breast Cancer by Inhibiting Apoptosis and Activating the PI3K/AKT/mTOR Signaling Pathway. Ann Trans Med (2021) 9(5):410. doi: 10.21037/atm-21-698
40. Kriegsmann M, Endris V, Wolf T, Pfarr N, Stenzinger A, Loibl S, et al. Mutational Profiles in Triple-Negative Breast Cancer Defined by Ultradeep Multigene Sequencing Show High Rates of PI3K Pathway Alterations and Clinically Relevant Entity Subgroup Specific Differences. Oncotarget (2014) 5(20):9952. doi: 10.18632/oncotarget.2481
41. Liedtke C, Cardone L, Tordai A, Yan K, Gomez HL, Figureoa LJB, et al. PIK3CA-Activating Mutations and Chemotherapy Sensitivity in Stage II–III Breast Cancer. Breast Cancer Res (2008) 10(2):1–10. doi: 10.1186/bcr1984
42. Clark AS, West K, Streicher S, Dennis PA. Constitutive and Inducible Akt Activity Promotes Resistance to Chemotherapy, Trastuzumab, or Tamoxifen in Breast Cancer Cells. Mol Cancer Ther (2002) 1(9):707–17.
43. Knuefermann C, Lu Y, Liu B, Jin W, Liang K, Wu L, et al. HER2/PI-3k/Akt Activation Leads to a Multidrug Resistance in Human Breast Adenocarcinoma Cells. Oncogene (2003) 22(21):3205–12. doi: 10.1038/sj.onc.1206394
44. Ellis P, Barrett-Lee P, Johnson L, Cameron D, Wardley A, O'Reilly S, et al. Sequential Docetaxel as Adjuvant Chemotherapy for Early Breast Cancer (TACT): An Open-Label, Phase III, Randomised Controlled Trial. Lancet (2009) 373(9676):1681–92. doi: 10.1016/S0140-6736(09)60740-6
45. Guarneri V, Dieci MV, Frassoldati A, Maiorana A, Ficarra G, Bettelli S, et al. Prospective Biomarker Analysis of the Randomized CHER-LOB Study Evaluating the Dual Anti-HER2 Treatment With Trastuzumab and Lapatinib Plus Chemotherapy as Neoadjuvant Therapy for HER2-Positive Breast Cancer. Oncologist (2015) 20(9):1001. doi: 10.1634/theoncologist.2015-0138
46. Loibl S, Von Minckwitz G, Schneeweiss A, Paepke S, Lehmann A, Rezai M, et al. PIK3CA Mutations Are Associated With Lower Rates of Pathologic Complete Response to Anti-Human Epidermal Growth Factor Receptor 2 (Her2) Therapy in Primary HER2-Overexpressing Breast Cancer. J Clin Oncol (2014) 32(29):3212–20. doi: 10.1200/JCO.2014.55.7876
47. Giordano SH, Elias AD, Gradishar WJ. NCCN Guidelines Updates: Breast Cancer. J Natl Compr Cancer Netw (2018) 16(5S):605–10. doi: 10.6004/jnccn.2018.0043
48. Becher I, Andrés-Pons A, Romanov N, Stein F, Schramm M, Baudin F, et al. Pervasive Protein Thermal Stability Variation During the Cell Cycle. Cell (2018) 173(6):1495–507. e18. doi: 10.1016/j.cell.2018.03.053
49. Grandal B, Evrevin C, Laas E, Jardin I, Rozette S, Laot L, et al. Impact of BRCA Mutation Status on Tumor Infiltrating Lymphocytes (TILs), Response to Treatment, and Prognosis in Breast Cancer Patients Treated With Neoadjuvant Chemotherapy. Cancers (2020) 12(12):3681. doi: 10.3390/cancers12123681
50. Akashi-Tanaka S, Watanabe C, Takamaru T, Kuwayama T, Ikeda M, Ohyama H, et al. BRCAness Predicts Resistance to Taxane-Containing Regimens in Triple Negative Breast Cancer During Neoadjuvant Chemotherapy. Clin Breast Cancer (2015) 15(1):80–5. doi: 10.1016/j.clbc.2014.08.003
51. Yang L, Ye F, Bao L, Zhou X, Wang Z, Hu P, et al. Somatic Alterations of TP53, ERBB2, PIK3CA and CCND1 Are Associated With Chemosensitivity for Breast Cancers. Cancer Sci (2019) 110(4):1389–400. doi: 10.1111/cas.13976
52. Al Amri WS, Allinson LM, Baxter DE, Bell SM, Hanby AM, Jones SJ, et al. Genomic and Expression Analyses Define MUC17 and PCNX1 as Predictors of Chemotherapy Response in Breast Cancer. Mol Cancer Ther (2020) 19(3):945–55. doi: 10.1158/1535-7163.MCT-19-0940
53. Masuda N, Lee S-J, Ohtani S, Im Y-H, Lee E-S, Yokota I, et al. Adjuvant Capecitabine for Breast Cancer After Preoperative Chemotherapy. N Engl J Med (2017) 376(22):2147–59. doi: 10.1056/NEJMoa1612645
54. Johnston SR, Harbeck N, Hegg R, Toi M, Martin M, Shao ZM, et al. Abemaciclib Combined With Endocrine Therapy for the Adjuvant Treatment of HR+, HER2–, Node-Positive, High-Risk, Early Breast Cancer (Monarche). J Clin Oncol (2020) 38(34):3987–98. doi: 10.1200/JCO.20.02514.
55. Loibl S, Marmé F, Martin M, Untch M, Bonnefoi H, Kim S-B, et al. Palbociclib for Residual High-Risk Invasive HR-Positive and HER2-Negative Early Breast Cancer—The Penelope-B Trial. J Clin Oncol (2021) 39(14):1518–30. doi: 10.1200/JCO.20.03639
56. Gnant M, Dueck AC, Frantal S, Martin M, Burstein HJ, Greil R, et al. Adjuvant Palbociclib for Early Breast Cancer: The PALLAS Trial Results (ABCSG-42/AFT-05/BIG-14-03). J Clin Oncol (2021), JCO. 21.02554. doi: 10.1200/JCO.21.02554
57. Von Minckwitz G, Huang C-S, Mano MS, Loibl S, Mamounas EP, Untch M, et al. Trastuzumab Emtansine for Residual Invasive HER2-Positive Breast Cancer. N Engl J Med (2019) 380(7):617–28. doi: 10.1056/NEJMoa1814017
58. Fowble BL, Einck JP, Kim DN, McCloskey S, Mayadev J, Yashar C, et al. Role of Postmastectomy Radiation After Neoadjuvant Chemotherapy in Stage II-III Breast Cancer. Int J Radiat Oncol Biol Phys (2012) 83(2):494–503. doi: 10.1016/j.ijrobp.2012.01.068
59. Von Minckwitz G, Schneeweiss A, Loibl S, Salat C, Denkert C, Rezai M, et al. Neoadjuvant Carboplatin in Patients With Triple-Negative and HER2-Positive Early Breast Cancer (GeparSixto; GBG 66): A Randomised Phase 2 Trial. Lancet Oncol (2014) 15(7):747–56. doi: 10.1016/S1470-2045(14)70160-3
60. Poggio F, Bruzzone M, Ceppi M, Pondé N, La Valle G, Del Mastro L, et al. Platinum-Based Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer: A Systematic Review and Meta-Analysis. Ann Oncol (2018) 29(7):1497–508. doi: 10.1093/annonc/mdy127
61. Skriver SK, Jensen M-B, Eriksen J-O, Ahlborn LB, Knoop AS, Rossing M, et al. Induction of PIK3CA Alterations During Neoadjuvant Letrozole may Improve Outcome in Postmenopausal Breast Cancer Patients. Breast Cancer Res Treat (2020) 184(1):123–33. doi: 10.1007/s10549-020-05833-w
62. Majewski IJ, Nuciforo P, Mittempergher L, Bosma AJ, Eidtmann H, Holmes E, et al. PIK3CA Mutations Are Associated With Decreased Benefit to Neoadjuvant Human Epidermal Growth Factor Receptor 2–Targeted Therapies in Breast Cancer. J Clin Oncol (2015) 33(12):1334. doi: 10.1200/JCO.2014.55.2158
63. Ellis H, Ma CX. PI3K Inhibitors in Breast Cancer Therapy. Curr Oncol Rep (2019) 21(12):1–9. doi: 10.1007/s11912-019-0846-7
64. Saura C, Hlauschek D, Oliveira M, Zardavas D, Jallitsch-Halper A, de la Peña L, et al. Neoadjuvant Letrozole Plus Taselisib Versus Letrozole Plus Placebo in Postmenopausal Women With Oestrogen Receptor-Positive, HER2-Negative, Early-Stage Breast Cancer (LORELEI): A Multicentre, Randomised, Double-Blind, Placebo-Controlled, Phase 2 Trial. Lancet Oncol (2019) 20(9):1226–38. doi: 10.1016/S1470-2045(19)30334-1
65. Mayer IA, Prat A, Egle D, Blau S, Fidalgo JAP, Gnant M, et al. A Phase II Randomized Study of Neoadjuvant Letrozole Plus Alpelisib for Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Breast Cancer (NEO-ORB). Clin Cancer Res (2019) 25(10):2975–87. doi: 10.1158/1078-0432.CCR-18-3160
Keywords: neoadjuvant chemotherapy (NAC), pathologic complete response (pCR), PIK3CA mutations, PI3K-AKT pathway, stage II/III breast cancer (BC)
Citation: Dong M, Shan B, Han X, Zhao X, Wang F, Zhu L, Ou Q, Ma X and Pan Y (2022) Baseline Mutations and Up-Regulation of PI3K-AKT Pathway Serve as Potential Indicators of Lack of Response to Neoadjuvant Chemotherapy in Stage II/III Breast Cancer. Front. Oncol. 11:784985. doi: 10.3389/fonc.2021.784985
Received: 28 September 2021; Accepted: 28 December 2021;
Published: 04 April 2022.
Edited by:
Adam Brufsky, University of Pittsburgh Medical Center, United StatesReviewed by:
Gilda Alves Brown, Rio de Janeiro State University, BrazilAzadeh Nasrazadani, University of Pittsburgh Medical Center, United States
Copyright © 2022 Dong, Shan, Han, Zhao, Wang, Zhu, Ou, Ma and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yueyin Pan, cGFueXVleWluQHVzdGMuZWR1LmNu; Xiaopeng Ma, eGlhb3BlbmdtYUBmc3l5LnVzdGMuZWR1LmNu
†These authors have contributed equally to this work
 Menghao Dong
Menghao Dong Benjie Shan1†
Benjie Shan1† Xinghua Han
Xinghua Han Xiaotian Zhao
Xiaotian Zhao Qiuxiang Ou
Qiuxiang Ou Yueyin Pan
Yueyin Pan