- 1Department of Clinical Laboratory, Affiliated Hospital of Jiangxi University of Chinese Medicine, Nanchang, China
- 2School of Medicine, Tongji University, Shanghai, China
- 3Department of Pediatric Surgery, the First Affiliate Hospital of Gannan Medical University, Ganzhou, China
- 4Jiangxi Provincial Clinical Research Center for Vascular Anomalies, Gannan Medical University, Ganzhou, China
- 5Jiangxi Province Key Research Laboratory of Chinese Medicine for the Prevention and Treatment of Hemangioma, Jiangxi University of Chinese Medicine, Nanchang, China
Objective: To evaluate the role of POGLUT2 in pan-cancer through bioinformatics analysis and experimental verification.
Methods: Expression, gene mutation and amplification, methylation, and copy number alteration (CNA) of POGLUT2 were evaluated using The Cancer Genome Atlas (TCGA), Cancer Cell Line Encyclopedia (CCLE), and Genotype-Tissue Expression (GTEx) databases. Moreover, POGLUT2 on survival and disease progression in pan-cancer was performed using TCGA data. Immune infiltration and tumor microenvironment evaluations were assessed by ImmuneScore, ImmuCellAI, and TIMER databases. POGLUT2 correlated drug resistance analysis was performed using the GDSC2 database. Furthermore, POGLUT2 knockdown of breast cancer cells was established, followed by in vitro biological function assays and in vivo tumor growth study. The mechanisms of POGLUT2 in breast cancer were briefly evaluated via its connection with Notch signaling.
Results: Increased levels of POGLUT2 were found in multiple types of cancer tissues and cell lines. Moreover, increased gene mutation and amplification, methylation, and CNA of POGLUT2 were found in several types of cancers. POGLUT2 was mainly expressed in stromal cells as verified by StromalScore, ESTIMATEScore, ImmuneScore, and Tumor purity, and POGLUT2 was positively correlated with cancer-associated fibroblasts, macrophages, monocytes, and neutrophils in the tumor microenvironment. In vitro and in vivo results showed that POGLUT2 knockdown could delay tumor growth and progression. Notch signaling components were related to the function of POGLUT2.
Conclusions: Increased levels of POGLUT2 could result in the dysregulated immune cell infiltration and tumor microenvironment and showed a significant regulatory effect on the progression of breast cancer through Notch-related signaling.
1 Introduction
Cancer and its related mortality are one of the most serious public health problems worldwide (1). With the advancement of science and technology, novel diagnostic therapies (such as chimeric antigen receptor T cell (CAR-T) and immune checkpoint monoclonal antibody treatment (2)) tools are emerging and bringing the hope to cure the disease; however, the objective to achieve an idea survival time has not been fulfilled (3). Moreover, increased financial burden (4) and life quality improvement requests remain the main troubles for the patients and their families (5). Therefore, it is necessary to study the molecular mechanisms involved in cancer in detail (6), thereby helping to solve the above problems.
Protein O-glucosyltransferase 2 (POGLUT2), a gene located on chromosome 5, is a protein product located at the lumen of the endoplasmic reticulum. As a member of the endoplasmic reticulum protein family, POGLUT2 functions to prevent the secretion of all endoplasmic reticulum resident proteins via receptor–ligand interaction with a protein in the Golgi apparatus by a Lys-Asp-Glu-Leu or KDEL motif located at its C-terminus (7, 8). On the biochemistry function, POGLUT2 can catalyze to transfer the glucose from UDP-glucose to a serine residue and specifically target extracellular epidermal growth factor (EGF) repeats of proteins such as Notch signaling members (NOTCH1 and NOTCH3) (9, 10), thereby affecting Notch signaling pathway (11). However, its role in cancer has not been determined yet.
In the present study, we aimed to evaluate the role of POGLUT2 in pan-cancer through bioinformatics analysis and experimental verification.
2 Materials and methods
2.1 Collection and processing of data
2.1.1 Differential expression analysis
The Cancer Genome Atlas (TCGA) pan-cancer, Cancer Cell Line Encyclopedia (CCLE), the Genotype-Tissue Expression (GTEx), and corresponding clinical data of the patients were downloaded from the University of California Santa Cruz (UCSC) Xena browser (https://xenabrowser.net/datapages/). Differential expression analysis of the genes in pan-caner tissue and normal tissues was performed using the combined data from TCGA and GTEx and processed with the ggplot2 package in R, whereas the mean expression of POGLUT2 in tumor tissue from TCGA, normal tissue from GTEx, and cell lines from CCLE were processed using the ggradar package in R. The protein expression of POGLUT2 was evaluated using the data from the UALCAN database (12) (http://ualcan.path.uab.edu).
2.1.2 Gene mutation and amplification, methylation, and copy number alteration analysis
The data on gene mutation, amplification, and methylation was obtained from the cBioportal database (https://www.cbioportal.org/) (13). The association analysis was calculated using Pearson’s correlation method and plotted using the ggplot2 package in R.
2.1.3 Enrichment analysis
Gene Set Variation Analysis (GSVA) (14) was carried out using the GSVA package, and the results were plotted using the ggplot2 and ggpubr packages in R. Gene Set Enrichment Analysis (GSEA) (15) was performed using the clusterpofiler package, and corresponding plots were fulfilled with the pheatmap package in R.
2.2 Kaplan–Meier curve construction for survival and disease progression analyses
The effects of POGLUT2 on four survival and disease progression-related parameters, including overall survival (OS), progression-free interval (PFI), disease-free survival (DFI), and disease-specific survival (DSS), were calculated using the expression data and clinical data obtained above and plotted using the survminer and survival packages in R. Moreover, optimal cutoff values were calculated according to Youden’s index and using the survminer and survival packages in R.
2.3 Association of POGLUT2 with immune infiltration and tumor microenvironment
Stromalcore, ImmuneScore, and ESTIMAScore (sum of Stromalcore and ImmuneScore) (16) were calculated using the ESTIMATE package in R. The Tumor Immune Estimation Resource (TIMER) database (17) was employed for the characterization of the cells and pathways involved in the effects of POGLUT2. Immune infiltration analysis was carried out using the CIBERSORT database (https://cibersort.stanford.edu) (18). Immune Cell Abundance Identifier (19) (ImmuCellAI; http://bioinfo.life.hust.edu.cn/ImmuCellAI#!/) was also employed for immune infiltration score calculation. The plots were achieved by either the ggplot2 or circlize package in R.
2.4 Drug resistance analysis
Drug resistance analysis was fulfilled using data from the Genomics of Drug Sensitivity in Cancer 2 (GDSC2) database (https://www.cancerrxgene.org/) and processed using Spearman’s correlation method.
2.5 Cell line culture and establishment of POGLUT2 knockdown cells
Breast cancer cell lines MDA-MB-468, MDA-MB-231, MCF-7, and BT474 were purchased from American Type Culture Collection (ATCC, Gaithersburg, MD, USA). Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) or 1640 medium supplemented with 10% fetal bovine serum (FBS; Corning, CA, USA) and 1% penicillin–streptomycin solution (Beyotime, Nantong, China) and maintained at 37°C, 5% CO2 condition. POGLUT2 knockdown MCF-7 and MDA-MB-231 cell line establishment was achieved by GenePharma Gene Co., Ltd. (Shanghai, China) and verified through quantitative PCR and Western blotting experiment described below.
2.6 Immunohistochemistry staining of POGLUT2 in breast cancer tissues
Three paired breast cancer tissue and corresponding para-tumor normal tissues were obtained and processed for series alcohol fixation, paraffin embedding, and sections. Then the sections were further handled for antigen retrieval, blocking, primary and secondary antibody staining, and detection. The antibody used for POGLUT2 staining was from NOVUS (1:200, NBP1-97469, CA, USA), whereas the two-step detection kit was from ebiogo Co., Ltd. (B001, Shanghai, China).
2.7 In vitro cell proliferation, apoptosis, cell cycle, clone formation, wound healing assay, cell migration, and invasion assay
Cell proliferation was assayed using the Cell Counting Kit-8 (CCK-8) reagent from Biomiky (BL001B, China). Briefly, 10 µl of CCK-8 was added into 100 µl of cell suspension in a 96-well plate, and the assay was performed at different time points.
Cell apoptosis was assayed using the Annexin V-PI staining kit Beyotime (22837, China). Briefly, cells were seeded in a six-well plate and grown for 48 h, followed by trypsinization, cell suspension preparation, phosphate-buffered saline (PBS) wash, binding buffer suspension, and Annexin V-PI staining for 30 min before undergoing analysis by a FACS analyzer (BD FACSCanto, San Jose, CA, USA).
Cell cycle assay was also performed using the propidium iodide (PI) staining kit from Beyotime (C1052, China). Briefly, cells were seeded in a 6-cm dish and grown into 80% confluency, followed by trypsinization, cell suspension preparation, PBS wash, ethanol fixation, and PI staining for 30 min before undergoing analysis by a FACS analyzer.
Clone formation was carried out as follows: (0.4–1) × 103 cells/well were seeded in six-well plates and cultured for 14 days for clone formation. Then the wells were fixed with 4% paraformaldehyde, washed with PBS, stained with crystal violet for 20 min at room temperature, and photographed.
The wound healing assay was carried out as follows: cells were seeded into a 12-well plate and grown into 95% confluency, followed by a vertical scape made by 200 µl pipette tips and 24- or 48-h incubation in 37°C, 5% CO2 condition before being photographed under a microscope. Cell migration ability quantification was processed via measurement of the width of the wound area in the images.
Transwell cell migration and invasion were assayed in a similar manner except that a Matrigel was added in the upper chamber in the setting of invasion before cell seeding. The upper chamber was seeded with 5 × 104 cells/well, and the lower chamber was added with 600 µl of complete medium with 20% FBS. Then the chamber was subjected to 37°C, 5% CO2 condition for a specific time period before undergoing fixation, washing, staining with crystal violet, and imaging. Three random fields (×100) were selected in each upper chamber for data processing.
2.8 In vivo tumor formation assay
Control siRNA and POGLUT2 siRNA MCF-7 cancer cells (2 × 107) were subcutaneously administrated into the back of the nude mice aged 6 weeks (each cell type with six mice). The tumor growth was monitored for a period of 28 days by measuring the length (L) and width (W) using the following equation: volume = (W2 × L)/2. Animals were maintained under a cycle of 12/12 h (light/dark) at 24°C with free access to water and food. At the end time point, all the mice were euthanized with an overdose of pentobarbital sodium, and the tumors were isolated, photographed, and weighed for data collection.
2.9 Quantitative PCR
RNA from the Control siRNA and POGLUT2 siRNA-treated cells or different types of breast cancer cells were isolated using TRIzol® Plus RNA Purification Kit (12183-555; Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Reverse transcription and quantitative PCR were respectively carried out using PrimeScript™ RT Master Mix (RR036A; Takara, Dalian, China) and TB Green Premix Ex Taq II (RR820A; Takara). Quantification of the relative expression levels was performed using the 2−ΔΔCT method. The sequences of used primers were as follows: POGLUT2 forward 5′-TGGCAGGAGCAACTCAAGTT-3′, reverse 5′-CCCTGGCGTTCTGCTGTC-3′; GAPHD forward 5′-GATGAGATTGGCCCGATG-3′; reverse 5′-GACTGAGATTGGCCCGATG-3′.
2.10 Western blotting
Cell proteins were prepared from the above-indicated cells using T-PER Tissue Protein Extraction Reagent containing Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher, Pleasanton, CA, USA). Then cell protein was measured using BCA kit from Beyotime (P0010) before being subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) electrophoresis, polyvinylidene difluoride (PVDF) membrane transferring, 5% milk blocking, and primary and secondary antibody incubation. The antibody used for POGLUT2 detection was from SANTA CRUZ (1:1,000, sc-390065, Santa Cruz, CA, USA); simultaneously, the GAPDH antibody from Proteintech Co., Ltd. (Chicago, IL, USA; 10494-1-AP) was employed as the internal control. For the experiments on Notch signaling evaluation, antibodies were from Abcam (Cambridge, UK; ab234987 for CSL) and Proteintech (55114-1-AP for Notch3; 28580-1-AP for Notch2; 66890-1-Ig for Jagged1; 20687-1-AP for Notch1). Quantification of the intensity of the protein was determined using ImageJ software (NIH, MA, USA).
3 Results
3.1 Expression analysis of POGLUT2 in pan-cancer and its correlation with clinical pathology
Compared to normal tissue, elevated and decreased levels of POGLUT2 mRNA were found in BLCA, BRCA, CHOL, COAD, DLBC, ESCA, GBM, HNS, KIRC, KIRP, LAML, LGG, LIHC, LUAD, LUSC, OV, PAAD, PCPG, PRAD, READ, STAD, TGCT, THCA, THYM, UCS, UVM, KICH, OV, THCA, and UCEC, whereas no difference was found in ACC, CESC, MESO, and SARC (Figure 1A). Moreover, the elevated mean expression of POGLUT2 was confirmed in tumor tissue and cancer cell lines (Figures 1B–D). In the results of pair tissue comparison, elevated POGLUT2 gene levels were found in most tumor tissue compared to normal tissue except in PRAD and KICH (Figures 1E–R). Moreover, elevated POGLUT2 in tumor tissue compared to normal tissue was found in breast cancer, colon cancer, and renal clear cell carcinoma, whereas no difference was found in ovarian cancer and UCEC according to the data from the UALCAN database (Figure 1S). If the clinical data were combined, then an elevated POGLUT2 mRNA level was found in tumors derived from high stages compared to lower stages (Figure 1T).
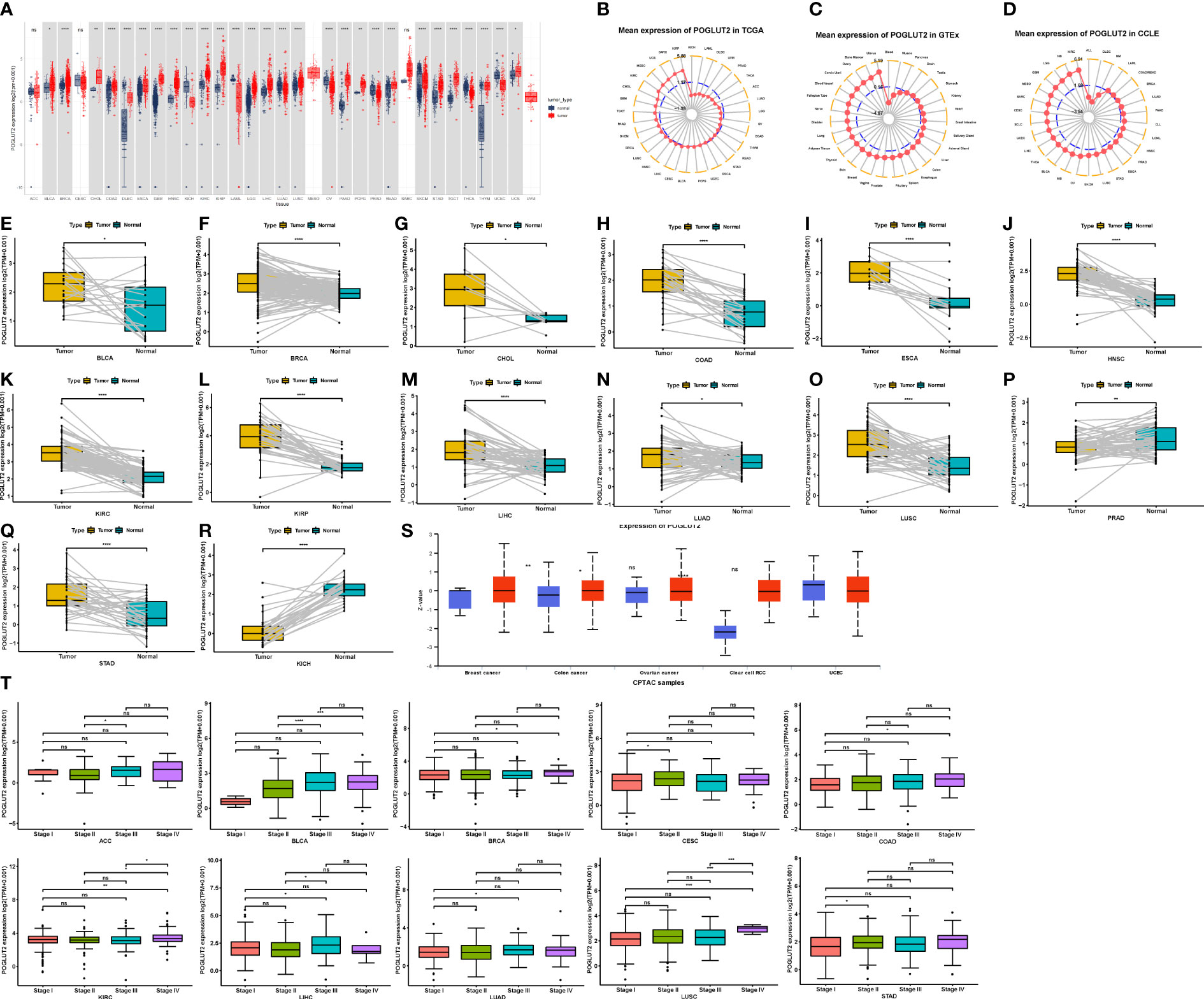
Figure 1 Expression pattern of POGLUT2 mRNA in different types of tumors and paired tumor and para-tumor normal tissues. (A) Compared to normal tissue, elevated and decreased levels of POGLUT2 mRNA were respectively found in BLCA, BRCA, CHOL, COAD, DLBC, ESCA, GBM, HNS, KIRC, KIRP, LAML, LGG, LIHC, LUAD, LUSC, OV, PAAD, PCPG, PRAD, READ, STAD, TGCT, THCA, THYM, UCS, UVM, and KICH, OV, THCA, and UCEC, whereas no difference was found in ACC, CESC, MESO, and SARC. (B) Mean expression of POGLUT2 mRNA in different types of tumors from TCGA. (C) Mean expression of POGLUT2 mRNA in different types of tumors from GTEx. (D) Mean expression of POGLUT2 mRNA in different types of tumors from CCLE. Elevated (E–O) and decreased (P–R) levels of POGLUT2 mRNA in paired tumor and para-tumor normal tissues. (S) Expression of POGLUT2 protein in different types of tumors. Elevated POGLUT2 in tumor tissue compared to normal tissue was found in breast cancer, colon cancer, and renal clear cell carcinoma, whereas no difference was found in ovarian cancer and UCEC (data are from UALCAN database). (T) The relationship between POGLUT2 mRNA level and pathology stages in different types of tumors. Elevated POGLUT2 mRNA level was found in tumors derived from high stages compared to lower stages. TCGA, The Cancer Genome Atlas; GTEx, Genotype-Tissue Expression; CCLE, Cancer Cell Line Encyclopedia. ns, no significant. *P < 0.05, **P < 0.01, ***P < 0.001.
3.2 The role of POGLUT2 in gene mutation and amplification, methylation, and copy number alteration
Considering the critical role of gene mutation and amplification, methylation, and copy number alteration (CNA) in tumor development and progression, we also plotted figures that showed the ratio variation of POGLUT2 in different types of cancers (Figure 2A). Moreover, the correlation parameter calculation results indicated that on CNA and POGLUT2 expression, significant positive correlations were found in LIHC, UCEC, UCS, LUSC, SARC, OV, GBM, BLCA, KIRC, STAD, SKCM, KICH, CESC, UVM, BRCA, ESCA, LUAD, HNSC, COAD, MESO, and PCPG, whereas no correlation was found in LGG, DLBC, THYM, ACC, PRAD, THCA, READ, LAML, CHOL, PAAD, KIRP, and TGCT (Figure 2B). On the relationship between methylation and POGLUT2 mRNA expression, significant negative correlations between methylation and POGLUT2 mRNA expression were found in HNSC, STAD, CESC, ESCA, LIHC, PRAD, LUAD, LUSC, THYM, UCEC, BRCA, LAML, KIRP, SARC, UVM, DLBC, ACC, TGCT, UCS, and SKCM but not in other types of cancers (Figure 2C).
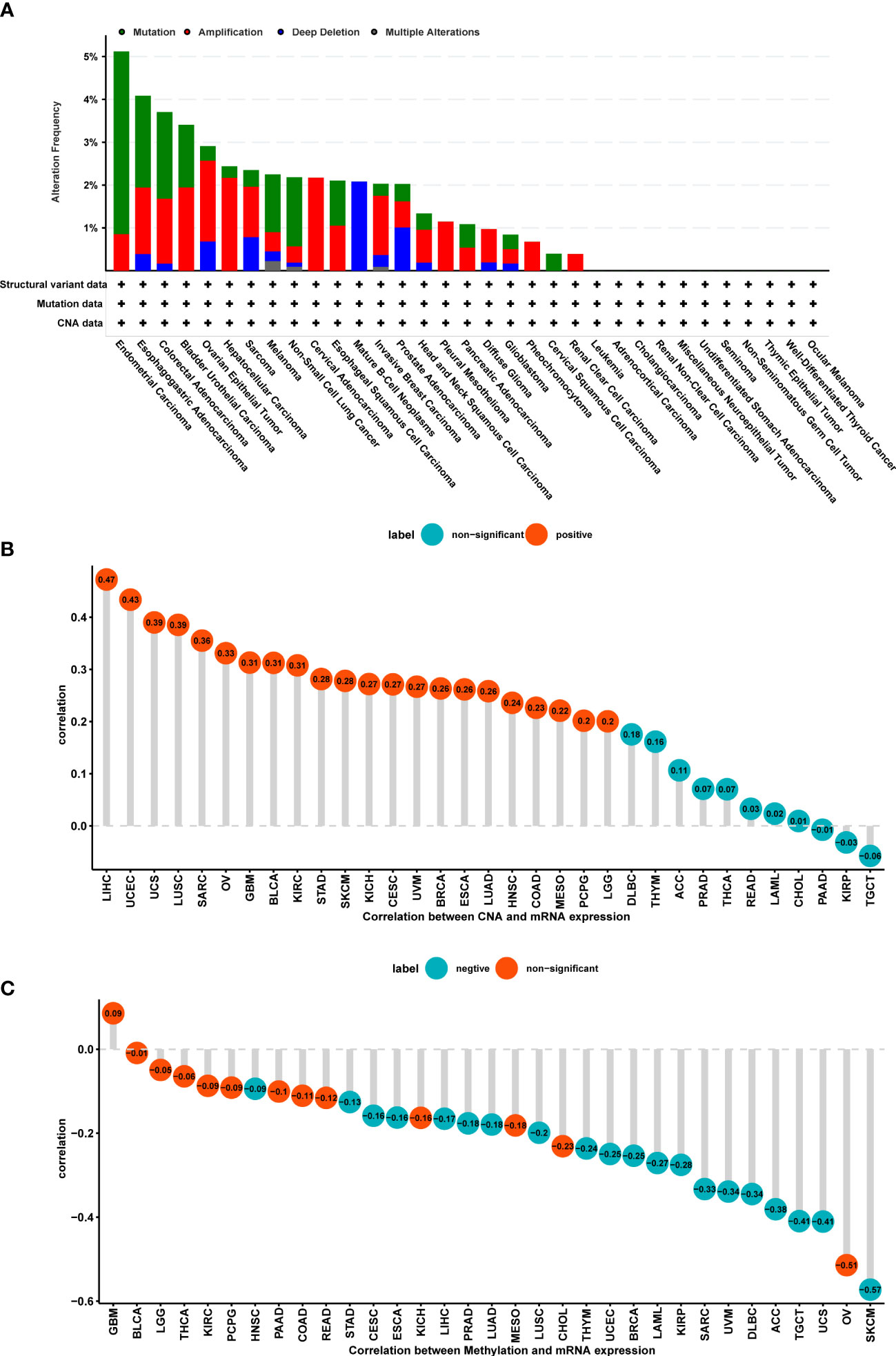
Figure 2 The role of POGLUT2 in gene mutation and amplification, methylation, and copy number alterations (CNAs). (A). Ratio of gene mutation (green), amplification (red), deep deletion (blue), and multiple alterations (gray) in different types of tumors. (B) Correlation between CAN and POGLUT2 expression. Significant positive correlations were found in LIHC, UCEC, UCS, LUSC, SARC, OV, GBM, BLCA, KIRC, STAD, SKCM, KICH, CESC, UVM, BRCA, ESCA, LUAD, HNSC, COAD, MESO, and PCPG, whereas no correlation was found in LGG, DLBC, THYM, ACC, PRAD, THCA, READ, LAML, CHOL, PAAD, KIRP, and TGCT. (C) Correlation between methylation and POGLUT2 mRNA expression. Significant negative correlations between methylation and POGLUT2 mRNA expression were found in HNSC, STAD, CESC, ESCA, LIHC, PRAD, LUAD, LUSC, THYM, UCEC, BRCA, LAML, KIRP, SARC, UVM, DLBC, ACC, TGCT, UCS, and SKCM, but not in other types of cancers.
3.3 The impact of POGLUT2 on survival and disease progression in pan-cancer
We further analyze the impact of POGLUT2 on survival and disease progression in pan-cancer; the results showed that POGLUT2 could affect the OS in ACC, BLCA, BRCA, HNSC, KICH, KIRC, LGG, LIHC, LUAD, MESO, PCPG, PRAD, and STAD; DSS in ACC, BLCA, BRCA, COAD, HNSC, KICH, LGG, LIHC, LUSC, MESO, PCPG, and PRAD, but not in other types of cancers; DFI in ACC, CHOL, PAAD, and PCPG; and PFI in ACC, BLCA, BRCA, HNSC, KICH, KIRC, LGG, LIHC, LUAD, MESO, and PRAD, but not in other types of cancers (Figures 3A–D). Moreover, the high- and low-expression grouping also confirmed that significantly increased survival in POGLUT2 low-expression patients in most types of cancers (Figure 3E).
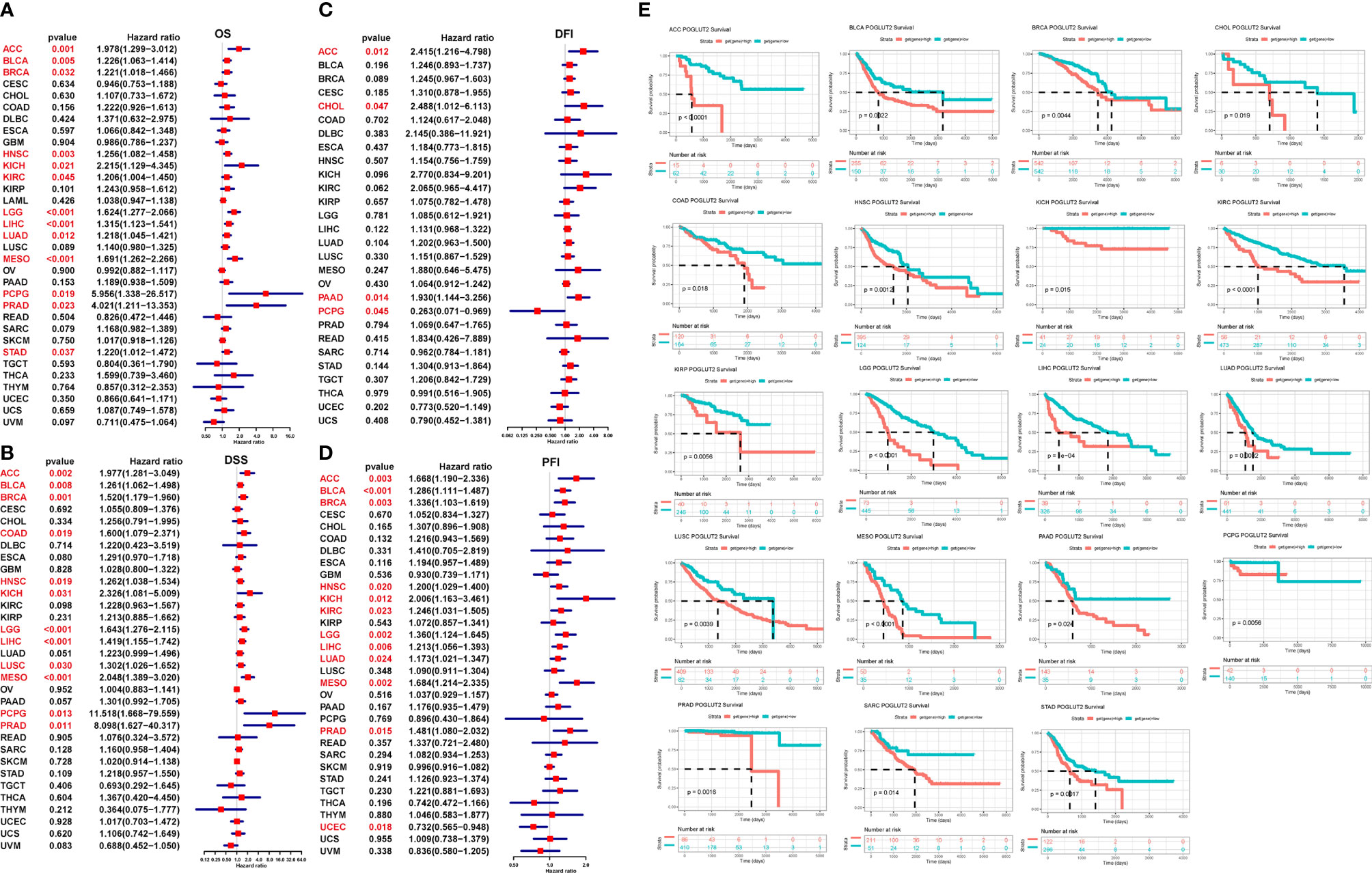
Figure 3 The impact of POGLUT2 on survival of pan-cancer by univariate logistic regression and receiver operating characteristic (ROC) curves. Red characters represent the data of significance. (A) Overall survival (OS). (B) Disease-specific survival (DSS). (C) Disease-free interval (DFI). (D) Progression-free interval (PFI). (E) ROC curve for survival in pan-cancer classified by high and low expression of POGLUT2.
3.4 Enrichment analysis of POGLUT2 in breast cancer
We also performed GSVA and GSEA. In GSVA, epithelial–mesenchymal transition, apical junction, and angiogenesis are the top 3 upregulated pathways that correlated with POGLUT2, whereas oxidative phosphorylation, peroxisome, and bile acid metabolism are the top 3 downregulated pathways that correlated with POGLUT2 (Figures 4A, B) In GSEA, we listed the heatmap of the top 50 positive and negative correlated genes of POGLUT2, and the top 3 were respectively RECK, DZIP1, and DPYSL3 and MCRIP2, NECAB3, and SIGIRR (Figures 4C, D). Moreover, the GSEA-GO, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome results were also listed, and the top 3 terms were skeletal system development, formation of the primary germ layer, and blood vessel development in GO; extracellular matrix (ECM)–receptor interaction, focal adhesion, TGF-beta signaling in KEGG; and disease of glycosylation, L1CAM interaction, and regulation of insulin-like growth factor in Reactome (Figures 4E–G). Moreover, the univariate (Figure 4H) and multivariate (Figure 4I) Cox regression analyses demonstrated that POGLUT2 (p < 0.05) acted as an independent prognostic indicator of breast cancer together with age and stage (p < 0.001).
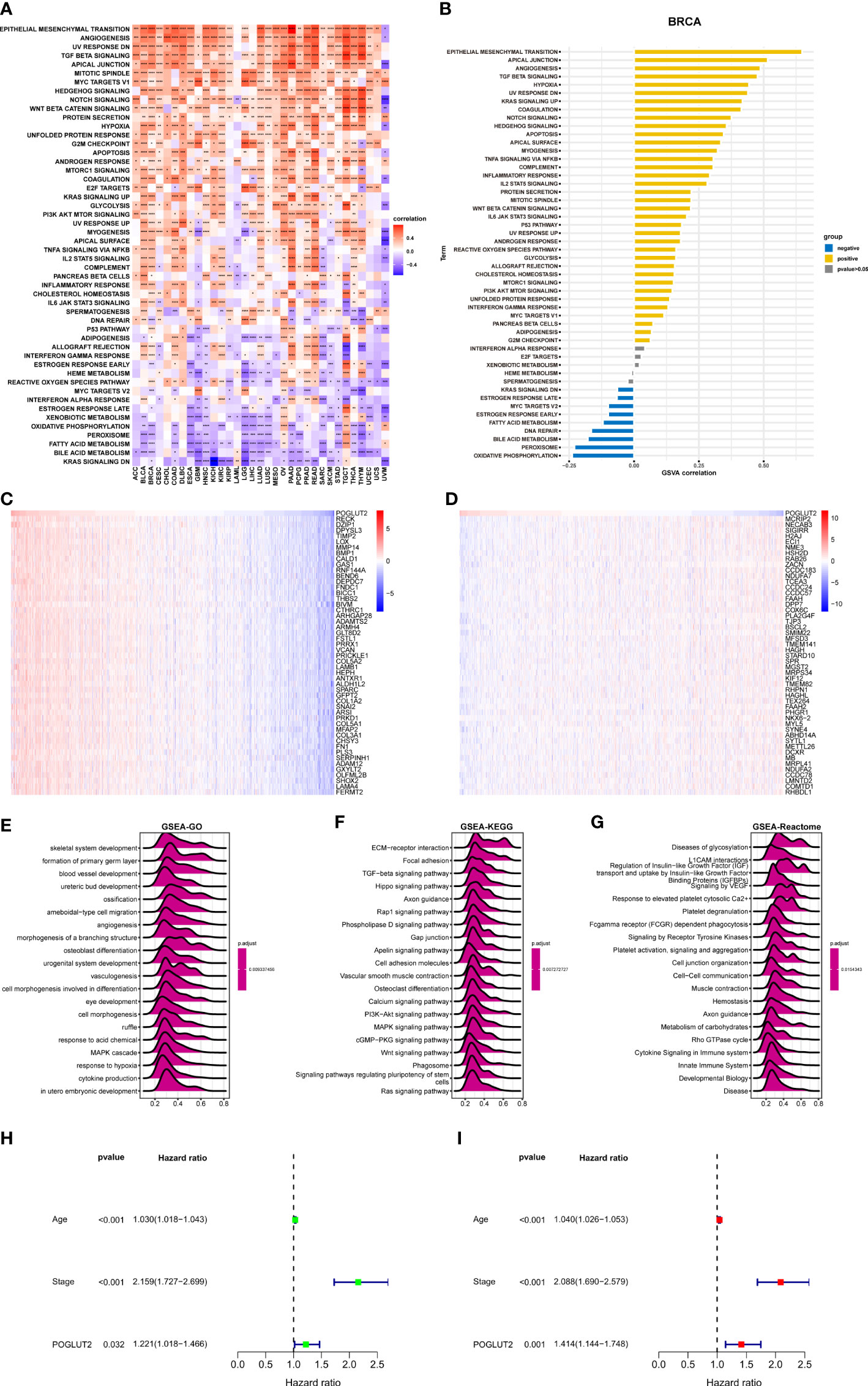
Figure 4 GSVA enrichment analysis. (A). Top 50 POGLUT2 correlated hallmark pathways in pan-cancer. (B) Top 50 POGLUT2 correlated hallmark pathways in breast cancer. (C) Heatmap of top 50 positively correlated genes. (D) Heatmap of top 50 negative correlated genes. (E) GSEA-GO. (F) GSEA KEGG. (G) GSEA Reactome enrichment analysis using the correlation genes. Top 20 results were illustrated. (H, I) Univariate (H) and multivariate (I) Cox regression analyses assessing the prognostic value of POGLUT2 in breast cancer. GSVA, Gene Set Variation Analysis; GSEA, Gene Set Enrichment Analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes.
3.5 The effects of POGLUT2 on immune cell infiltration and tumor microenvironment
We also calculated the effects of POGLUT2 on immune cell infiltration. Firstly, a significant correlation could be found between POGLUT2 and StromalScore, ESTIMATEScore, ImmuneScore, and Tumor purity (Figure 5A). In specific breast cancer, POGLUT2 was positively correlated with StromalScore, ESTIMATEScore, and ImmuneScore and negatively correlated with Tumor purity (Figure 5B). The role of POGLUT2 on immune infiltration was also figured using the data from the TIMER2 dataset (Figure 5C). The results indicated that POGLUT2 was positively correlated with cancer-associated fibroblasts, macrophages, monocytes, and neutrophils and negatively correlated with B cells, T cells, and NK cells in most of the cancers. In specific breast cancer using the data from CIBERSORT, POGLUT2 was found positively correlated with monocyte, macrophage, and infiltration scores (Figure 5D) and negatively correlated with B cells, CD8 T cells, and conventional T cells. In a specific BRCA setting, POGLUT2 was found positively correlated with iTreg, macrophage, DC, and Th2 cells and negatively correlated with neutrophils (Figure 5E).
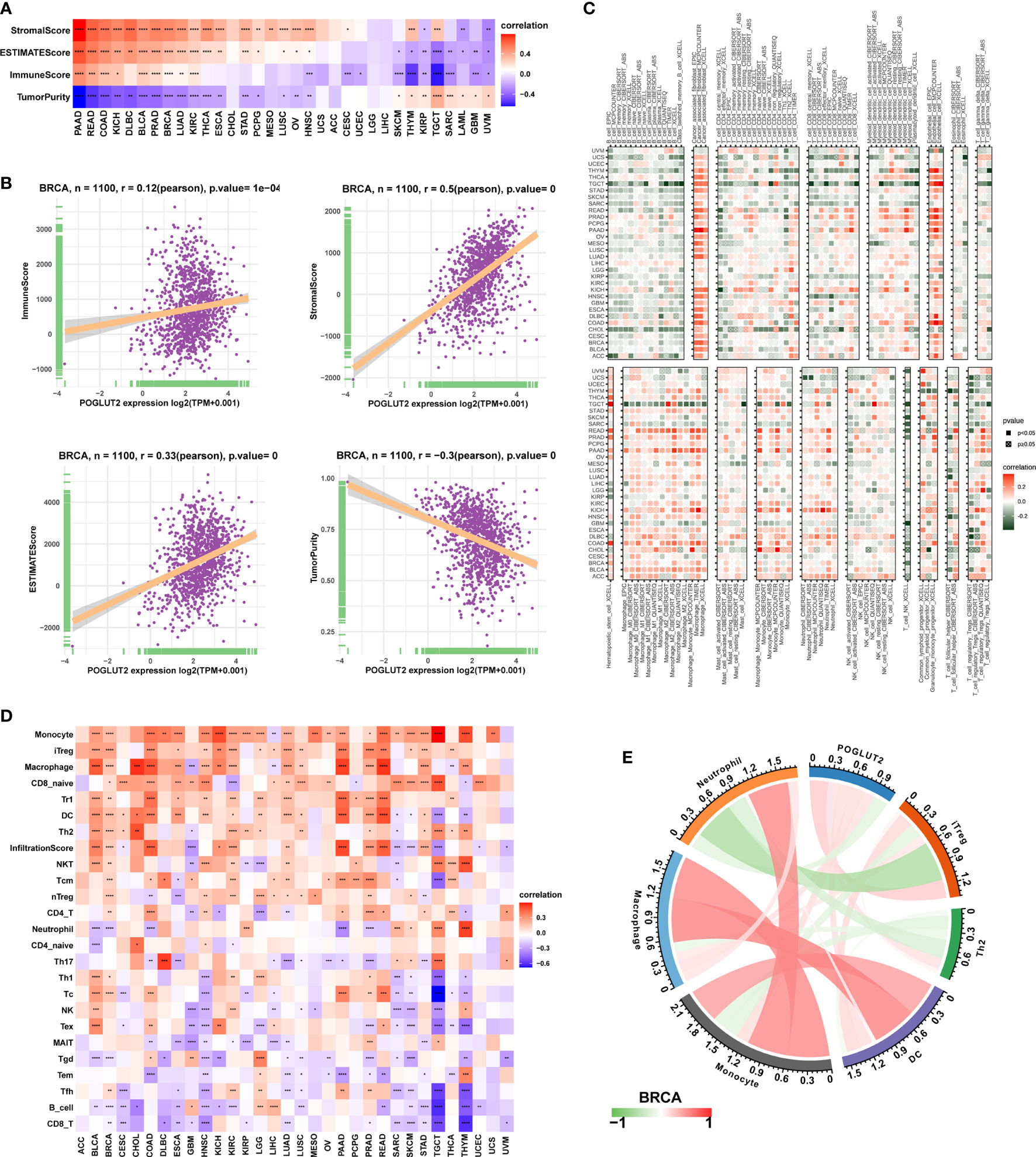
Figure 5 The role of POGLUT2 in tumor microenvironment. (A) POGLUT2 correlated Stromal Score, ESTIMATEScore, ImmuneScore, and Tumor purity in different types of cancers. (B) The relationship between POGLUT2 mRNA level and ImmuneScore, Stromal Score, ESTIMATEScore, and Tumor purity in BRCA dataset. (C) The role of POGLUT2 on immune infiltration using the data from the TIMER2 database. (D) The role of POGLUT2 on immune cell infiltration using data from ImmuCellAI database (http://bioinfo.life.hust.edu.cn/ImmuCellAI#!/). (E) The role of POGLUT2 on immune cell infiltration in the setting of breast cancer.
3.6 The association between POGLUT2 and immune-related genes
On immune inhibitory genes, the top 3 POGLUT2 positively correlated genes were TGFBR1, KDR, and IL10RB, whereas the top 3 negatively correlated genes were IDO1, KIR2DL1, and LAG3 (Figure 6A). On immune checkpoint genes in breast cancers, POGLUT2 was positively correlated with CTLA4, CD274, TIGIT, LAG3, and PDCD1 (Figure 6B). On MHC genes, the top 3 POGLUT2 positively correlated genes were TAP2, TAP1, and B2M, whereas the top 3 negatively correlated genes were HLA-DOB, HLA-DMA, and HLA-DQB1 (Figure 6C). On chemokines, the top 3 POGLUT2 positively correlated genes were CXCL12, CCL2, and CXCL5, whereas the top 3 negatively correlated genes were CXCL17, CCL-15, and CCL-25 (Figure 6D). On chemokine receptors, the top 3 POGLUT2 positively correlated genes were CCR1, CCR10, and CCR8, whereas the top 3 negatively correlated genes were CXCR5, CCR9, and CCR6 (Figure 6E). Moreover, the association between POGLUT2 and immune inhibitory genes in pan-cancer in specific TGF-beta1 and Wnt-beta-catenin signaling pathways was also illustrated. In TGF-beta1 pathways, the top 3 POGLUT2 positively correlated genes were SMURF2, ACVR1, and WWTR1, whereas the top 3 negatively correlated genes were CDH1, ID1, and JUNB (Figure 6F). In Wnt-beta-catenin pathways, the top 3 POGLUT2 positively correlated genes were FZD1, ADAM17, and HDAC2, whereas the top 3 negatively correlated genes were DKK4, WNT1, and FRAT1 (Figure 6G).
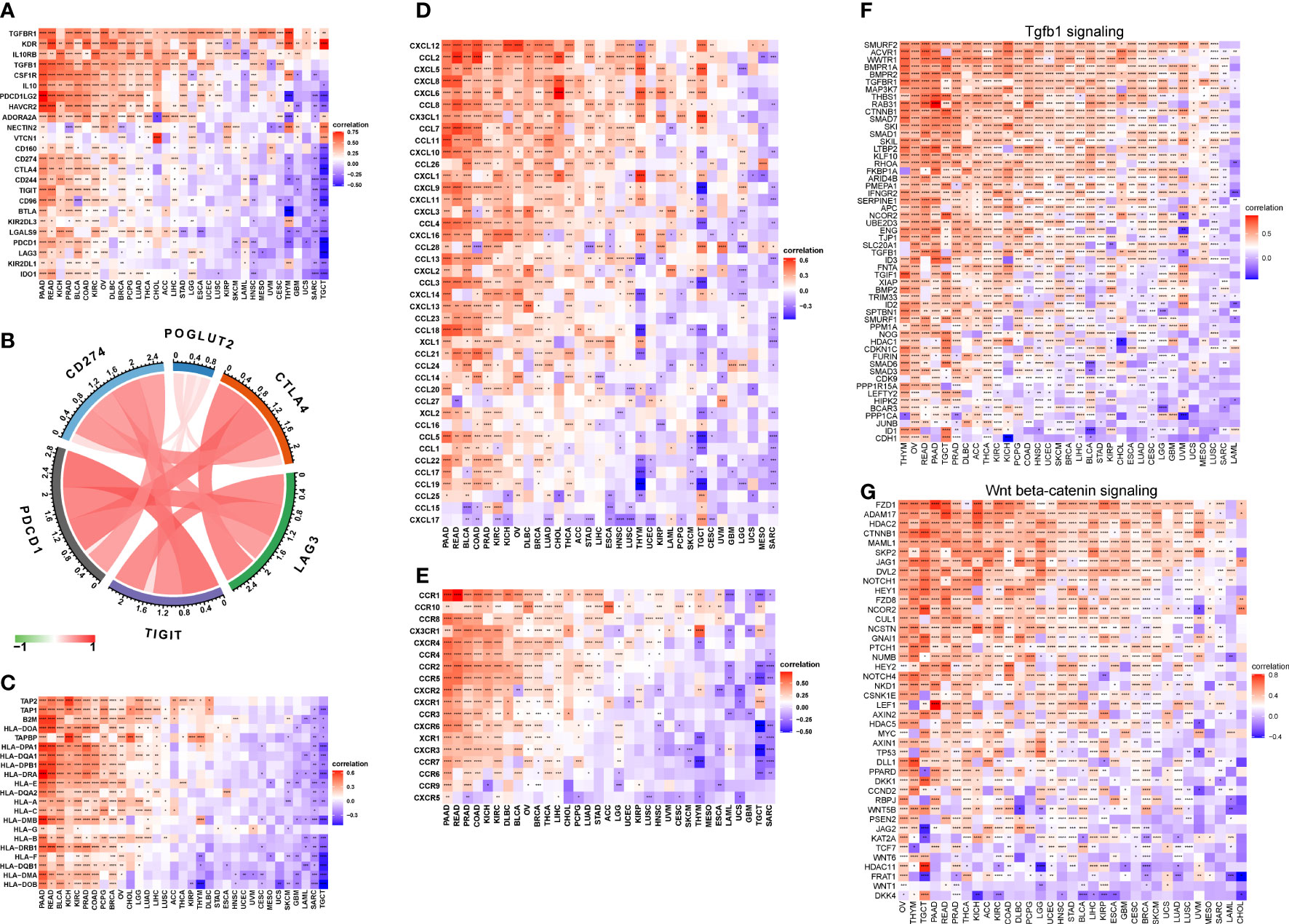
Figure 6 The association between POGLUT2 and immune-related genes. (A) The association between POGLUT2 and immune inhibitory genes in pan-cancer. (B) The association between POGLUT2 and immune checkpoint genes in breast cancers. (C) The association between POGLUT2 and MHC genes. (D) The association between POGLUT2 and chemokines. (E) The association between POGLUT2 and chemokine receptors. (F) The association between POGLUT2 and immune inhibitory genes in pan-cancer in specific signaling pathways. Left, TGF-beta1 signaling. Right, Wnt-beta-catenin signaling. (G) the top 3 POGLUT2 positively correlated genes were assessed.
3.7 POGLUT2 correlated drug resistance analysis
The association between POGLUT2 and IC50 of 198 drugs was calculated through Spearman’s correlation, and the top 6 POGLUT2 positively correlated drugs from the GDSC2 database (Figure 7) were doramapimod, acetalax, afatinib, sapitinib, LGK974, and selumetinib.
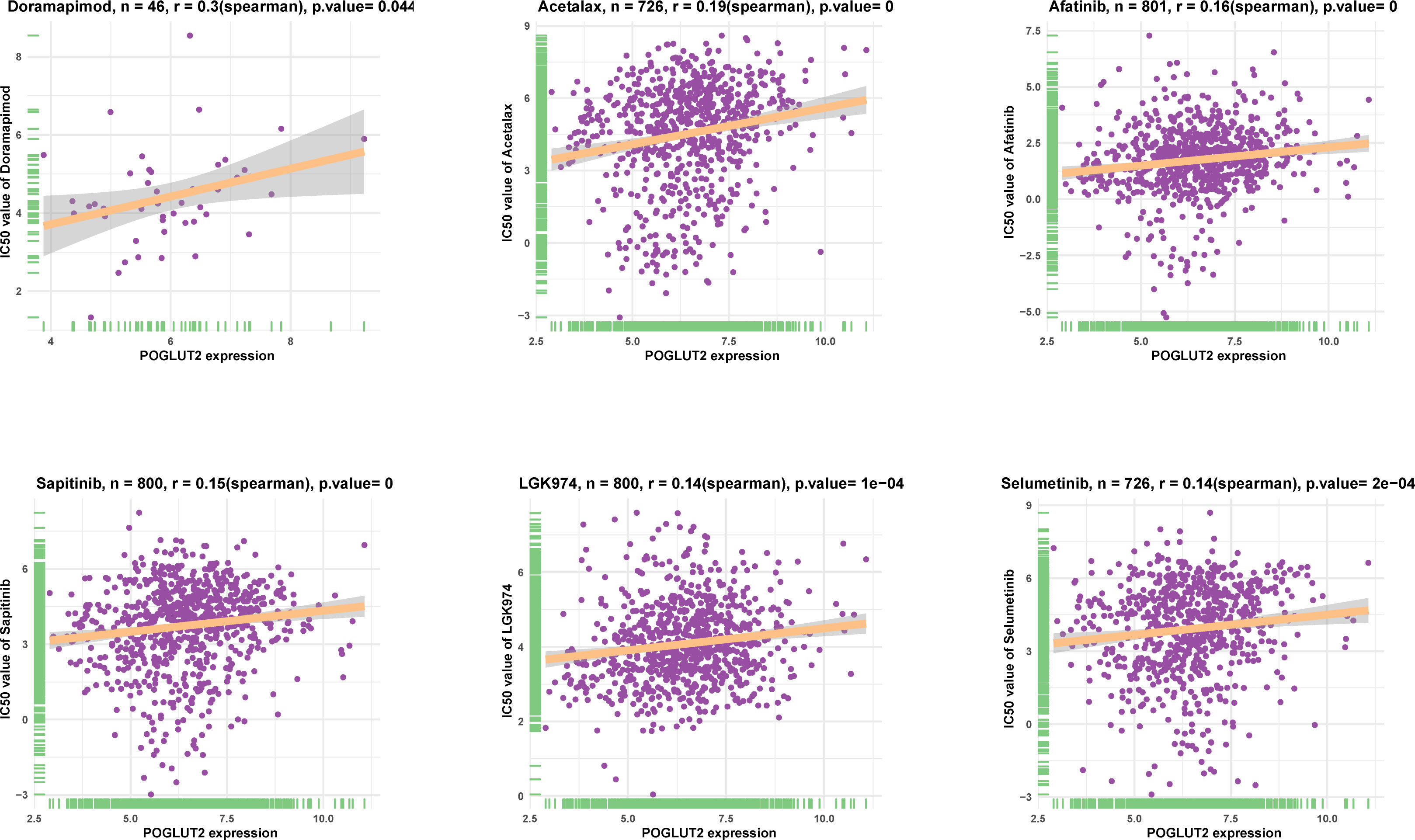
Figure 7 The top 6 POGLUT2 positively correlated drugs from GDSC2 database. The association between POGLUT2 and IC50 of 198 drugs was calculated through Spearman’s correlation.
3.8 Expression evaluation of POGLUT2 in breast cancer cells and tissues
Expression of POGLUT2 was determined in four breast cancer cell lines, including MDA-MB-468, MDA-MB-231, MCF-7, and BT474, and the highest gene and protein expression were respectively found in MCF-7 and MDA-MB-231 cells (Figures 8A, B). Meanwhile, immunohistochemistry analysis of POGLUT2 in breast cancer tissues revealed increased levels of POGLUT2 compared to control tissues (Figure 8C). Moreover, we also established the POGLUT2 knockdown MCF-7 and MDA-MB-231 cells, and significantly decreased levels of gene and protein expression of POGLUT2 were found in these cells after siRNA transfection (Figures 8D–G).
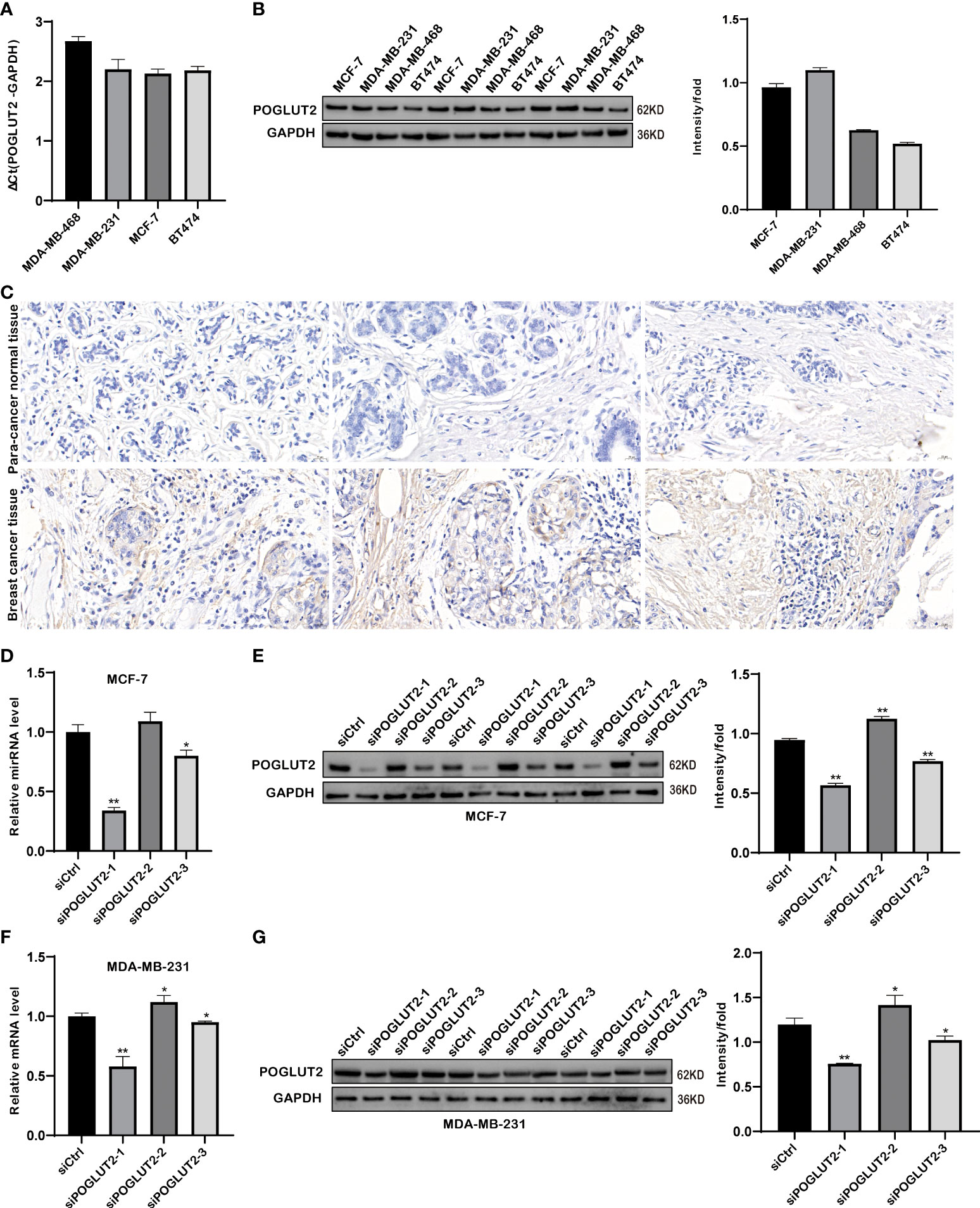
Figure 8 Gene and protein expression evaluation of POGLUT2 in breast cancer cells and tissues. (A) Gene expression of POGLUT2 was determined in four breast cancer cell lines, including MDA-MB-468, MDA-MB-231, MCF-7, and BT474, and highest expression was found in MCF-7 cells. (B) Protein expression of POGLUT2 was determined in four breast cancer cell lines, including MDA-MB-468, MDA-MB-231, MCF-7, and BT474, and highest expression was found in MDA-MB-231 cells. (C) Immunohistochemistry analysis of POGLUT2 in breast cancer tissues compared to control tissues. (D) Significant POGLUT2 expression downregulation was found in MCF-7 cells after siRNA transfection. (E) Significantly POGLUT2 expression downregulation was found in MDA-MB-231 cells after siRNA transfection. (F) Significantly POGLUT2 protein level downregulation was found in MCF-7 cells after siRNA transfection. (G) Significantly POGLUT2 protein level downregulation was found in MDA-MB-231 cells after siRNA transfection. *p < 0.05; **p < 0.01.
3.9 POGLUT2 regulates the cell proliferation, apoptosis, cell cycle, clone formation, migration, and invasion in breast cancer cells in vitro
We then established POGLUT2 knockdown cell lines to examine their biological behavior; the results showed that decreased cell proliferation, increased cell apoptosis, increased G1 phase and decreased S and G2 phase, decreased cell clone formation, decreased cell migration in 2-D and 3-D systems, and decreased cell invasion in a 3-D system were found in POGLUT2 knockdown MCF-7 (Figures 9A–F) and MDA-MB-231 cells (Figures 10A–F).
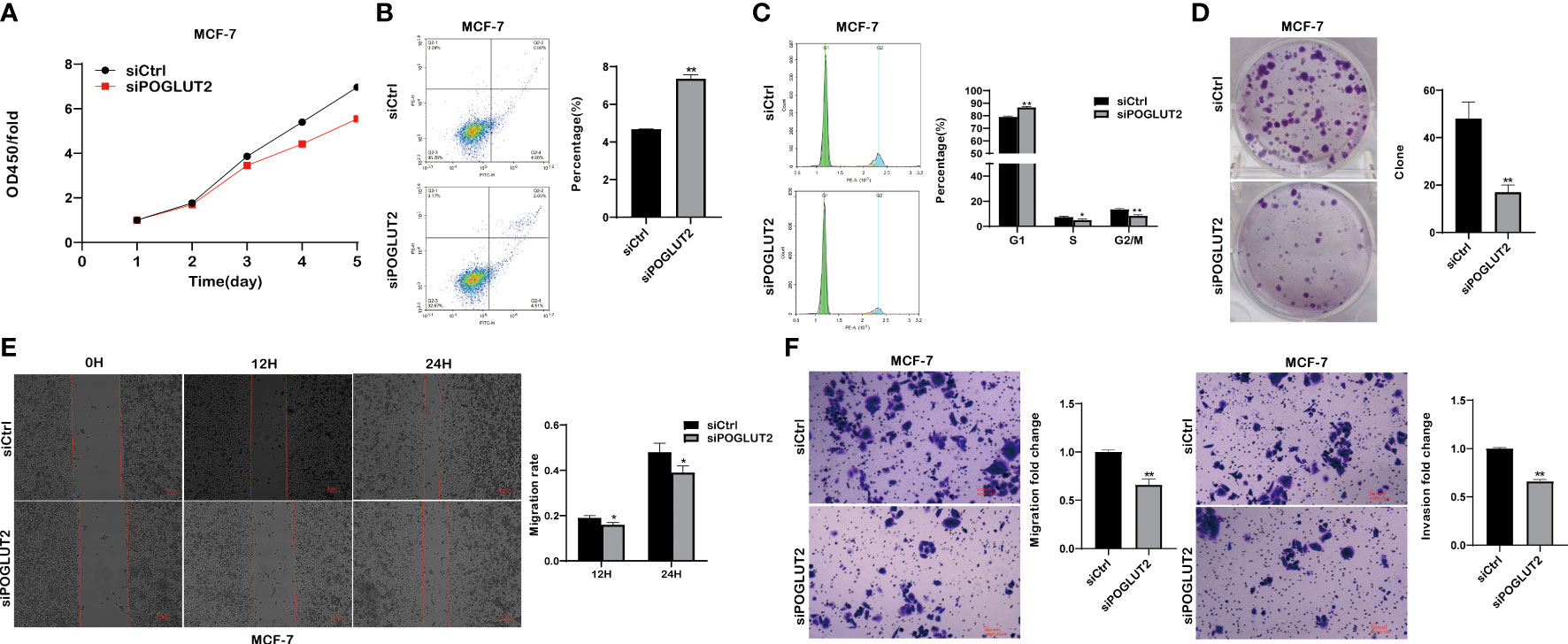
Figure 9 POGLUT2 regulates cell proliferation, apoptosis, cell cycle, clone formation, migration, and invasion in breast cancer cell line MCF-7 in vitro. (A) Decreased cell proliferation was found in POGLUT2 knockdown MCF-7 cells. (B) Increased cell apoptosis was found in POGLUT2 knockdown MCF-7 cells. (C) Increased G1 phase and decreased S and G2 phase in POGLUT2 knockdown MCF-7 cells. (D) Decreased cell clone formation in POGLUT2 knockdown MCF-7 cells. (E) Decreased cell migration of POGLUT2 knockdown MCF-7 cells in 2-D wound healing assay. (F) Decreased cell migration and invasion of MCF-7 cells in 3-D transwell assay. *p < 0.05; **p < 0.01.
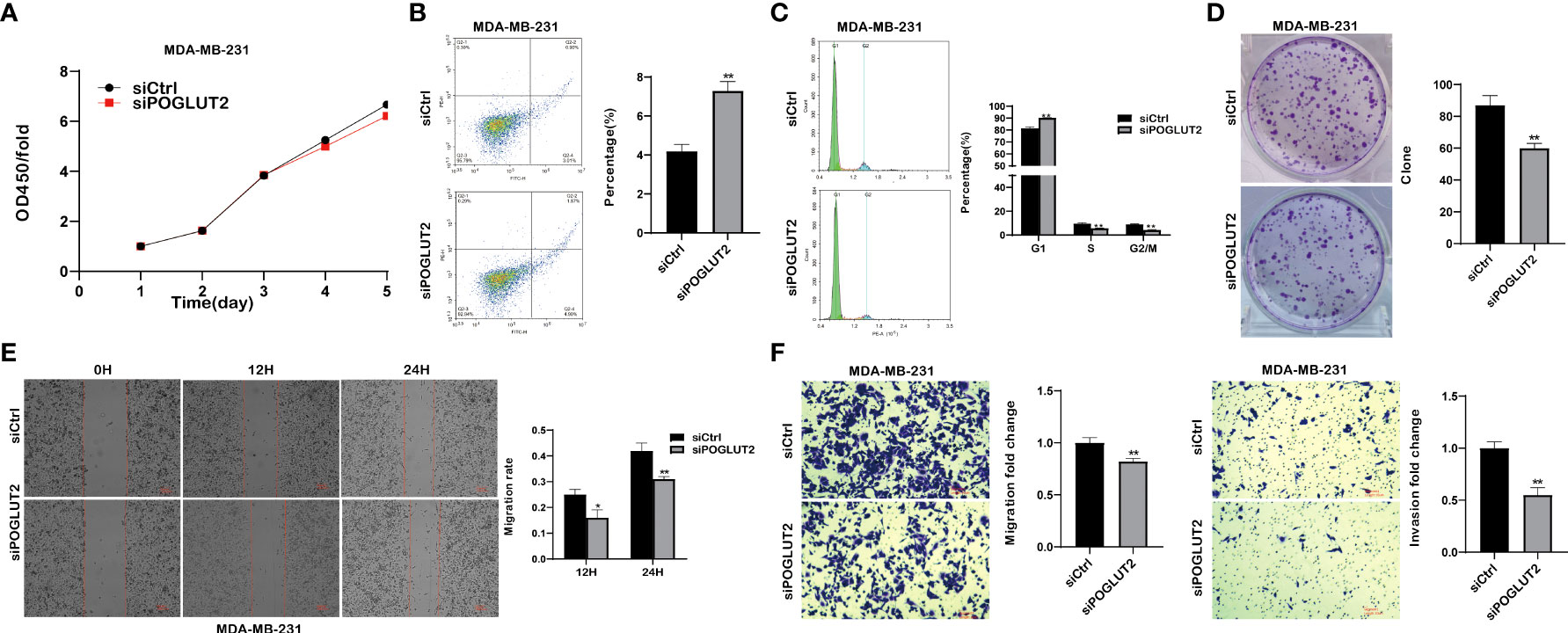
Figure 10 POGLUT2 regulates cell proliferation, apoptosis, cell cycle, clone formation, migration, and invasion in breast cancer cell line MDA-MB-231 in vitro. (A) Decreased cell proliferation was found in POGLUT2 knockdown MDA-MB-231. (B) Increased cell apoptosis was found in POGLUT2 knockdown MDA-MB-231. (C) Increased G1 phase and decreased S and G2 phase in POGLUT2 knockdown MDA-MB-231. (D) Decreased cell clone formation in POGLUT2 knockdown MDA-MB-231. (E) Decreased cell migration of POGLUT2 knockdown MDA-MB-231 in 2-D wound healing assay. (F) Decreased cell migration and invasion of MDA-MB-231 in 3-D transwell assay. *p < 0.05; **p < 0.01.
3.10 POGLUT2 knockdown decreased the tumor growth in vivo
In vivo results indicated that POGLUT2 knockdown decreased the tumor growth as verified by tumor size, tumor volume, and tumor weight (Figures 11A–C).
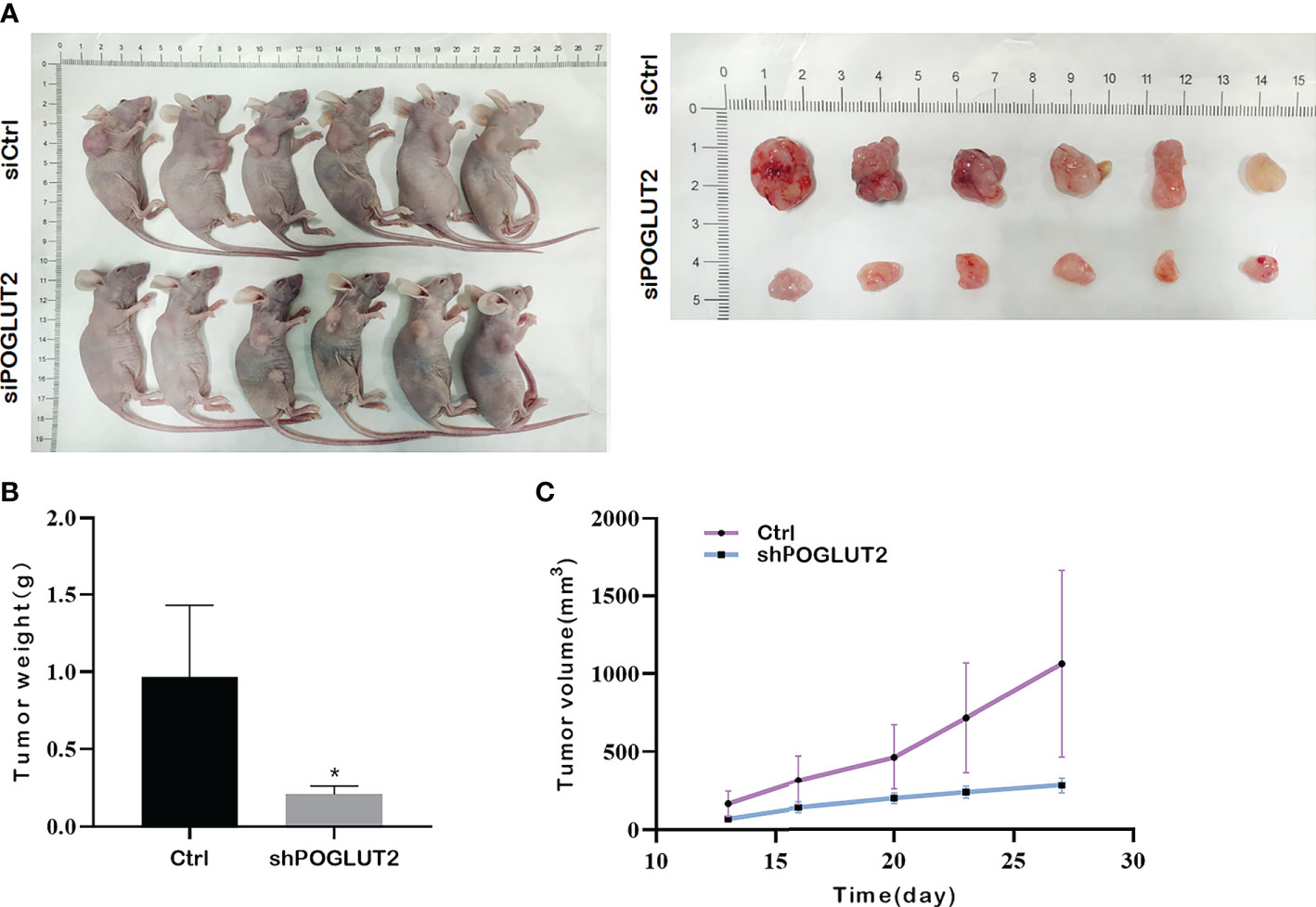
Figure 11 POGLUT2 knockdown decreased the tumor growth in a xenograft transplantation model. (A) Photographs of tumor-bearing nude mice and corresponding tumor tissues. (B) Line chart quantification indicated that deceased tumor volume was found in mice injected with POGLUT2 knockdown MCF-7 cells. (C) Bar graph quantification indicated decreased tumor weight in mice with POGLUT2 knockdown MCF-7 cells. *p < 0.05.
3.11 Notch pathway is involved in the effects of POGLUT2 in breast cancer cells
To further test the possible molecular mechanisms, we performed Western blotting to check the effects of POGLUT2 knockdown in two breast cancer cell lines. The results indicated that increased levels of CSL and decreased levels of Jagged1, Notch1, Notch2, and Notch3 were found in POGLUT2 knockdown MCF-7 and MDA-MB-231 cells (Figures 12A, B).
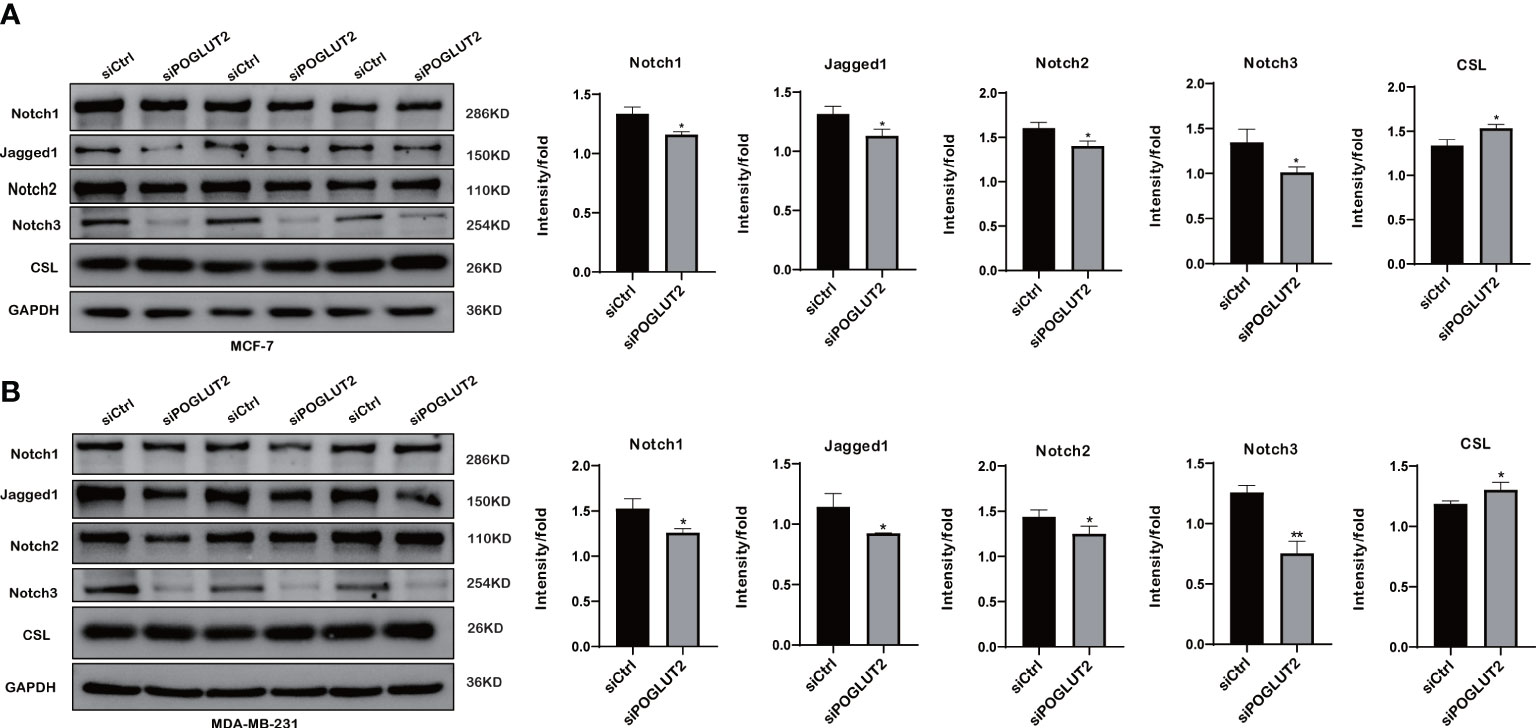
Figure 12 Notch pathway is involved in the effects of POGLUT2 in breast cancer cells. (A) Increased levels of CSL and decreased levels of Jagged1, Notch1, Notch2, and Notch3 were found in POGLUT2 knockdown MCF-7 cells. (B) Increased levels of CSL and decreased levels of Jagged1, Notch1, Notch2, and Notch3 were found in POGLUT2 knockdown MDA-MB-231 cells. *p < 0.05.
4 Discussion
Glycosylation is a kind of widespread and important protein post-translational modification process, which is catalyzed by glycosyltransferases (20). Specific glycosylation modifications are essential for protein activity, and they are presented in various proteins with clinical therapeutic purposes, such as antibodies and vaccines (21). The chemical enzymatic synthesis method based on glycosyltransferase is one of the effective ways to obtain uniform glycoproteins (22). Therefore, understanding the role and effects of glycosyltransferases in the biological process could help us to discover novel therapeutic targets regarding their biochemical properties.
According to previous reports, the Notch signaling pathway, as one of the most conserved cellular signaling pathways, plays a critical role in animal development and tissue homeostasis (23, 24). Recent visualization of Notch receptor structures in complex with Notch ligands and those of Notch-modifying glycosyltransferases has promoted the understanding of the significance of glycosylation in Notch signaling component regulation (25–27). In the present study, we evaluated the expression, gene mutation and amplification, methylation, and CNA of a novel O-glucosyltransferase POGLUT2 using TCGA, CCLE, and GTEx databases. Moreover, we also evaluated the role of POGLUT2 on survival and disease progression in pan-cancer based on TCGA data. Moreover, we also found that immune infiltration and tumor microenvironment evaluation using the ImmuneScore, ImmuCellAI, and TIMER databases and POGLUT2 correlated with drug resistance analysis using the GDSC2 database. Furthermore, POGLUT2 knockdown of breast cancer cells was established, followed by in vitro biological function assays and in vivo tumor growth study. Finally, we explored the mechanisms of POGLUT2 in breast cancer via a brief evaluation of its connection with Notch signaling.
Our results showed that increased levels of POGLUT2 were found in multiple types of cancer tissues and cell lines. Moreover, increased gene mutation and amplification, methylation, and CNA of POGLUT2 were found in several types of cancers. In recent studies by Williamson et al., POGLUT2 was found to modify not only a small fraction of Notch molecules with EGF repeats but also over half of fibrillin proteins with the EGF repeats (FBN1, FBN2, and LTBP1), which are the major components of extracellular matrix microfibrils and hold an important role in elastic tissue rational development, including the major organs (heart and lungs). Williamson et al. concluded that POGLUT2 together with O-glycosylated protein substrates is involved in the secretion of substrate proteins. In our cell knockdown experiment, we also found increased levels of CSL and decreased levels of Jagged1, Notch1, Notch2, and Notch3, which is consistent with Williamson’s study.
Here, we found that POGLUT2 was mainly expressed in stromal cells, which is verified by StromalScore, ESTIMATEScore, ImmuneScore, and Tumor purity, and POGLUT2 was positively correlated with cancer-associated fibroblasts, macrophages, monocytes, and neutrophils in the tumor microenvironment. These results indicated that POGLUT2 could regulate the immune status in the tumor microenvironment, thereby facilitating the development and progression of cancer. However, further studies are still required for obtaining detailed information about POGLUT2.
To confirm the specific function of POGLUT2 in human cancer, in vitro and in vivo validations were carried out. The results showed that POGLUT2 knockdown could delay the tumor growth and progression of breast cancer. Notch signaling components were related to the function of POGLUT2. According to the previous studies about POGLUT1, a member from the same family of POGLUT2, elevated POGLUT1 expression was found in hematopoietic malignancies, including primary acute myelogenous leukemia and T-ALL (28). Moreover, POGLUT1 amplification and overexpression were found in NSCLC (29). RNAi-mediated POGLUT1 knockdown in NSCLC cell lines A549 could result in a significant reduction in the expression of HEY1 and HES2, both of which are Notch downstream target genes that inhibit cell proliferation, migration, and survival. Furthermore, POGLUT1 was confirmed as a protein O-glucosyltransferase that transfers glucose and xylose to the EGF-like domains on Notch (30). In addition, based on the expression and clinical data, POGLUT1 was identified as a novel negative prognostic factor and could serve as a potential therapeutic target for NSCLC.
There are some limitations in our present study. The detailed cell identities related to POGLUT2 are not determined. Considering the prosperity of single-cell analysis technology (31), sorted POGLUT2-expressed cells might be collected for further analysis. Therefore, it is possible that novel mechanisms could be expected on POGLUT2. Additionally, the enhanced effect of POGLUT2 on breast cancer development has been confirmed in the present study. Due to the heterogeneity of gene expression and function, the specific role and mechanism of POGLUT2 need further investigations in various human cancers.
In summary, our results here demonstrated that increased levels of POGLUT2 in cancer could result in dysregulated immune cell infiltration and tumor microenvironment. Validated in breast cancer, the knockdown of POGLUT2 exerted a significant inhibitory effect on cell growth, apoptosis, and metastasis and suppressed tumor progression in rats by regulating Notch-related signaling.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was reviewed and approved by The Biomedical Ethics Committee of Gannan medical University.Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
Conceived and designed the study: XX and QL. Performed the literature search and data extraction: GX. Analyzed the data: MX. Drafted the manuscript: XX and GX. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Key Projects of Nature Science Foundation of Jiangxi (grant no. 20192ABCL20005), the Nature Science Foundation of Jiangxi (grant no. 20181BAB205052), and The Technology Plan Fund of Jiangxi Province Department of Education Project (grant no. GJJ211534).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.962540/full#supplementary-material
Glossary

References
1. Lippman SM, Abate-Shen C, Colbert Maresso KL, Colditz GA, Dannenberg AJ, Davidson NE, et al. AACR white paper: shaping the future of cancer prevention–a roadmap for advancing science and public health. Cancer Prev Res (2018) 11(12):735–78. doi: 10.1158/1940-6207.CAPR-18-0421
2. Hosseinkhani N, Derakhshani A, Kooshkaki O, Abdoli Shadbad M, Hajiasgharzadeh K, Baghbanzadeh A, et al. Immune checkpoints and CAR-T cells: the pioneers in future cancer therapies? Int J Mol Sci (2020) 21(21):8305. doi: 10.3390/ijms21218305
3. Crosby D, Lyons N, Greenwood E, Harrison S, Hiom S, Moffat J, et al. A roadmap for the early detection and diagnosis of cancer. Lancet Oncol (2020) 21(11):1397–9. doi: 10.1016/S1470-2045(20)30593-3
4. de Oliveira C, Weir S, Rangrej J, Krahn MD, Mittmann N, Hoch JS, et al. The economic burden of cancer care in Canada: a population-based cost study. Can Med Assoc Open Access J (2018) 6(1):E1–E10. doi: 10.9778/cmajo.20170144
5. Coens C, Pe M, Dueck AC, Sloan J, Basch E, Calvert M, et al. International standards for the analysis of quality-of-life and patient-reported outcome endpoints in cancer randomised controlled trials: recommendations of the SISAQOL consortium. Lancet Oncol (2020) 21(2):e83–96. doi: 10.1016/S1470-2045(19)30790-9
6. Mikubo M, Inoue Y, Liu G, Tsao MS. Mechanism of drug tolerant persister cancer cells: The landscape and clinical implication for therapy. J Thorac Oncol (2021) 16(11):1798–809. doi: 10.1016/j.jtho.2021.07.017
7. Yu H, Takeuchi H, Takeuchi M, Liu Q, Kantharia J, Haltiwanger RS, et al. Structural analysis of notch-regulating rumi reveals basis for pathogenic mutations. Nat Chem Biol (2016) 12(9):735–40. doi: 10.1038/nchembio.2135
8. Li Z, Han K, Pak JE, Satkunarajah M, Zhou D, Rini JM. Recognition of EGF-like domains by the notch-modifying O-fucosyltransferase POFUT1. Nat Chem Biol (2017) 13(7):757–63. doi: 10.1038/nchembio.2381
9. Takeuchi H, Fernández-Valdivia RC, Caswell DS, Nita-Lazar A, Rana NA, Garner TP, et al. Rumi functions as both a protein O-glucosyltransferase and a protein O-xylosyltransferase. Proc Natl Acad Sci USA (2011) 108(40):16600–5. doi: 10.1073/pnas.1109696108
10. Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, et al. Rumi is a CAP10 domain glycosyltransferase that modifies notch and is required for notch signaling. Cell (2008) 132(2):247–58. doi: 10.1016/j.cell.2007.12.016
11. Takeuchi H, Schneider M, Williamson DB, Ito A, Takeuchi M, Handford PA, et al. Two novel protein O-glucosyltransferases that modify sites distinct from POGLUT1 and affect notch trafficking and signaling. Proc Natl Acad Sci USA (2018) 115(36):E8395–402. doi: 10.1073/pnas.1804005115
12. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia (2017) 19(8):649–58. doi: 10.1016/j.neo.2017.05.002
13. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery (2012) 2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095
14. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf (2013) 14:7. doi: 10.1186/1471-2105-14-7
15. Canzler S, Hackermuller J. multiGSEA: a GSEA-based pathway enrichment analysis for multi-omics data. BMC Bioinf (2020) 21(1):561. doi: 10.1186/s12859-020-03910-x
16. Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun (2013) 4:2612. doi: 10.1038/ncomms3612
17. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res (2017) 77(21):e108–10. doi: 10.1158/1538-7445.AM2017-108
18. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol (2018) 1711:243–59. doi: 10.1007/978-1-4939-7493-1_12
19. Miao YR, Zhang Q, Lei Q, Luo M, Xie GY, Wang H, et al. ImmuCellAI: A unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci (Weinh) (2020) 7(7):1902880. doi: 10.1002/advs.201902880
20. Wandall HH, Nielsen MAI, King-Smith S, de Haan N, Bagdonaite I. Global functions of O-glycosylation: promises and challenges in O-glycobiology. FEBS J (2021) 288(24):7183–212. doi: 10.1111/febs.16148
21. Shi J, Ku X, Zou X, Hou J, Yan W, Zhang Y. Comprehensive analysis of O-glycosylation of amyloid precursor protein (APP) using targeted and multi-fragmentation MS strategy. Biochim Biophys Acta (BBA)-General Subj (2021) 1865(10):129954. doi: 10.1016/j.bbagen.2021.129954
22. Kightlinger W, Warfel KF, DeLisa MP, Jewett MC. Synthetic glycobiology: parts, systems, and applications. ACS synthetic Biol (2020) 9(7):1534–62. doi: 10.1021/acssynbio.0c00210
23. Kovall RA, Gebelein B, Sprinzak D, Kopan R. The canonical notch signaling pathway: Structural and biochemical insights into shape, sugar, and force. Dev Cell (2017) 41(3):228–41. doi: 10.1016/j.devcel.2017.04.001
24. Artavanis-Tsakonas S, Muskavitch MA. Notch: the past, the present, and the future. Curr Top Dev Biol (2010) 92:1–29. doi: 10.1016/S0070-2153(10)92001-2
25. Li Z, Fischer M, Satkunarajah M, Zhou D, Withers SG, Rini JM. Structural basis of notch O-glucosylation and O-xylosylation by mammalian protein-o-glucosyltransferase 1 (POGLUT1). Nat Commun (2017) 8(1):185. doi: 10.1038/s41467-017-00255-7
26. Luca VC, Kim BC, Ge C, Kakuda S, Wu D, Roein-Peikar M, et al. Notch-jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science (2017) 355(6331):1320–4. doi: 10.1126/science.aaf9739
27. Taylor P, Takeuchi H, Sheppard D, Chillakuri C, Lea SM, Haltiwanger RS, et al. Fringe-mediated extension of O-linked fucose in the ligand-binding region of Notch1 increases binding to mammalian notch ligands. Proc Natl Acad Sci USA (2014) 111(20):7290–5. doi: 10.1073/pnas.1319683111
28. Wang Y, Chang N, Zhang T, Liu H, Ma W, Chu Q, et al. Overexpression of human CAP10-like protein 46 KD in T-acute lymphoblastic leukemia and acute myelogenous leukemia. Genet Test Mol Biomarkers (2010) 14(1):127–33. doi: 10.1089/gtmb.2009.0145
29. Chammaa M, Malysa A, Redondo C, Jang H, Chen W, Bepler G, et al. RUMI is a novel negative prognostic marker and therapeutic target in non-small-cell lung cancer. J Cell Physiol (2018) 233(12):9548–62. doi: 10.1002/jcp.26858
30. Takeuchi H, Yu H, Hao H, Takeuchi M, Ito A, Li H, et al. O-Glycosylation modulates the stability of epidermal growth factor-like repeats and thereby regulates notch trafficking. J Biol Chem (2017) 292(38):15964–73. doi: 10.1074/jbc.M117.800102
Keywords: POGLUT2, pan cancer, breast cancer, bioinformatic analysis, immune infiltration
Citation: Xu X, Xie G, Xie M and Liu Q (2022) A comprehensive role evaluation and mechanism exploration of POGLUT2 in pan-cancer. Front. Oncol. 12:962540. doi: 10.3389/fonc.2022.962540
Received: 06 June 2022; Accepted: 16 August 2022;
Published: 08 September 2022.
Edited by:
Zhanzhan Li, Xiangya Hospital, Central South University, ChinaReviewed by:
Mingchi Luo, Second Affiliated Hospital of Tianjin University of Traditional Chinese Medicine, ChinaYan Zhang, Second Affiliated Hospital of Zhengzhou University, China
Copyright © 2022 Xu, Xie, Xie and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Liu, bGl1cWlhbmdtdTIwMTdAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Xianyun Xu
Xianyun Xu Guangming Xie
Guangming Xie Mingfeng Xie
Mingfeng Xie Qian Liu
Qian Liu