- 1Department of Pathology, University of California San Francisco, San Francisco, CA, United States
- 2Syneos Health, Morrisville, NC, United States
- 3Department of Pathology, Stanford University, Stanford, CA, United States
- 4Department of Pathology and Laboratory Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, United States
- 5Department of Pathology, Kaiser Permanente, Los Angeles, CA, United States
- 6Department of Pathology, University of Utah and ARUP Laboratories, Salt Lake City, UT, United States
Anaplastic lymphoma kinase (ALK) positive large B-cell lymphoma (ALK+ LBCL) is an aggressive and rare subtype of B-cell lymphoma. Patients typically present with advanced clinical stage disease and do not respond to conventional chemotherapy; the median overall survival is 1.8 years. The genetic landscape of this entity remains poorly understood. Here we report a unique case of ALK+ LBCL harbouring a rare TFG::ALK fusion. Targeted next-generation sequencing showed no significant single nucleotide variants, insertions/deletions, or other structural variants beyond the TFG::ALK fusion; deep deletions of FOXO1, PRKCA, and the MYB locus were also detected. Our case report draws attention to this rare disease, highlights a need for larger genetic profiling studies, and focuses on the pathogenesis and potential therapeutic targets of this aggressive disease. To our knowledge, this is the first report of a TFG::ALK fusion in ALK+ LBCL.
Introduction
Anaplastic lymphoma kinase (ALK) positive large B cell lymphoma (ALK+ LBCL) is a rare aggressive subtype of large B-cell lymphoma (LBCL), accounting for less than 1% of all diffuse large B-cell lymphoma (DLBCL) (1–3). It was first reported in 1997 by Delsol and colleagues (4) but has since been described in only small or individual case reports; to this date fewer than 200 cases have been reported in the literature. This disease occurs in both adult and pediatric patients with the median age at diagnosis of 38 years [range of 9-90 years (5)] but most often is seen in immunocompetent male patients with a male:female ratio of 4.2 (5). Advanced disease with cervical lymph node and/or extranodal involvement is a common presentation of this disease. Patients often do not respond to treatment with CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisolone), and median survival is 1.8 years (5).
The diagnosis of ALK+ LBCL is challenging because of its rarity and its morphologic and immunophenotypic resemblance to other hematopoietic and non-hematopoietic neoplasms (6). Histologically, it can show sinusoidal or a diffuse growth pattern consisting of sheets of large monomorphic immunoblastic or plasmablastic cells. By immunophenotype, the tumor cells importantly express ALK protein, and commonly co-express plasma cell markers such as CD38, CD138, VS38c, multiple myeloma oncogene 1 (MUM1); are negative for T-cell markers, and most cases are negative or only partially express B-cell markers (CD19, CD20, CD79a, PAX5) (1, 7, 8).
ALK overexpression in ALK+ LBCL is due to its translocation which generates a fusion protein. The most common translocation is t(2;17)(p23;q23) involving clathrin (CLTC) on chromosome 17q23 and ALK on chromosome 2p23 resulting in a CLTC::ALK fusion translocation (1, 9). ALK fusion proteins lead to ligand independent dimerization of the intracellular tyrosine kinase domain of ALK (10) and result in constitutive activation which promotes oncogenesis (11). Here we report the first case of an ALK+ LBCL with TFG::ALK fusion and describe the unique clinical, morphologic, immunophenotypic, and molecular features.
Materials and methods
This study was approved by the University of California, San Francisco (UCSF) institutional review board. Clinical information was obtained from Kaiser Permanente, Los Angeles. All hematoxylin and eosin (H&E) and immunohistochemistry slides were reviewed for this study. Immunohistochemistry was performed on whole slide sections to confirm the diagnosis of ALK+ LBCL, including ALK-1, Oct2, P53, c-MYC, BOB1, CD45RB (LCA), CD79a, CD10, Bcl-6, CD138, MUM-1, IgG, INI-1. Ki-67, Pancytokeratin, CK7, CK20, CK5/6, EMA, P63, P40, CK903, TTF-1, PAX5, CD20, CD30, CD2, CD3, CD4, CD5, CD7, CD8, CD43, CD21, CD23, Bcl-2, Cyclin D1, CD56, CD57, CD1a, CD68 (PGM1), SOX10, Melan-A, S100, HMB-45, SALL4, synaptophysin, chromogranin, INSM1, TdT, myeloperoxidase, CD34, CD31, ERG, desmin, myogenin, SF-1, GFAP, HHV-8, EBV by in situ hybdridization (ISH), IgM, IgA, IgD, and Kappa and Lambda ISH.
Capture-based next-generation sequencing (NGS) was performed at the UCSF Clinical Cancer Genomics Laboratory with UCSF500, a targeted sequencing panel consisting of all coding regions of 529 cancer-related genes and selected introns from 47 genes. Analysis of single nucleotide variants, insertion/deletions, structural variants including gene fusions, genome-wide copy number, and zygosity analysis, with a total sequencing footprint of 2.8 Mb was performed and bioinformatic analysis was performed using custom pipelines as previously described (12, 13).
Case report
A 23-year-old previously healthy male developed 7 months of nasal congestion and obstruction. At the time of presentation, he admitted to fatigue, diaphoresis, abdominal pain, and nausea. A physical exam revealed right cervical lymphadenopathy. A PET scan demonstrated a 2.3 cm left nasopharyngeal mass, bilateral cervical lymphadenopathy, and right retropharyngeal lymphadenopathy. A subsequent brain MRI scan was negative and endoscopy demonstrated a friable nasopharyngeal mass which was biopsied.
H&E stained slides of the nasopharyngeal mass showed sheets of large cells with round to oval nuclei, dispersed chromatin and prominent, often single, centrally located nucleoli (Figure 1A). These cells had moderate to abundant amounts of lightly eosinophilic cytoplasm. Mitotic figures were easily identified and there was abundant necrosis.

Figure 1 Histologic features and immunohistochemical stains in this case of ALK+ LBCL. Histology demonstrated sheets of large monomorphic immunoblastic or plasmablastic cells with moderate to abundant amounts of lightly eosinophilic cytoplasm and abundant necrosis (A). The tumor cells were negative for CD3 (B), CD30 (C), and pancytokeratin (D); the tumor cells were positive for ALK (E), Oct2 (F), BOB1 (G), dim CD79a (H), CD45RB (I), IgG (J), TP53 (K), and c-MYC (L).
A broad panel of immunostains was performed and the neoplastic cells were positive for ALK, Oct2, c-MYC, BOB1, CD45RB (LCA), CD79a (dim), CD10 (dim), Bcl-6 (subset), CD138, MUM-1, IgG. (Figures 1B–L). TP53 was positive in 30-40% of tumor cells. INI-1 staining was intact. Kappa and Lambda ISH show kappa light chain restriction on cells. Ki-67 demonstrated 70-80% proliferation in the tumor cells. The following immunohistochemical stains were negative in the neoplastic cells: pancytokeratin, CK7, CK20, CK5/6, EMA, P63, P40, CK903, TTF-1, PAX5, CD20, CD30, CD3, CD4, CD5, CD7, CD8, CD43, CD21, CD23, Bcl-2, Cyclin D1, CD56, CD57, CD1a, CD68 (PGM1), SOX10, Melan-A, S100, HMB-45, SALL4, synaptophysin, chromogranin, INSM1, TdT, myeloperoxidase, CD34, CD31, ERG, desmin, myogenin, SF-1, GFAP, HHV-8, EBV-ISH, IgM, IgA, IgD. A staging bone marrow biopsy showed no evidence of involvement by LBCL.
Molecular results
NGS using a DNA targeted sequencing panel identified a TFG::ALK1 fusion involving exons 1-4 of TFG and exons 20-29 of ALK (Figure 2A). No Tier 1 single nucleotide variants (SNV) were identified. One PRDM1 p.N421S variant of uncertain significance (VUS) was identified at a variant allele frequency (VAF) of 48%, likely representing a germline variant given the VAF. Deep deletions at FOXO1, MYB and PRKCA were identified. A custom microbial analysis pipeline did not identify any microbial organisms.

Figure 2 Next-generation sequencing using a DNA targeted sequencing panel identified a TFG::ALK1 fusion involving exons 1-4 in TFG and exons 20-29 in ALK (A). The TFG-ALK fusion protein results in downstream up-regulation of the ALK pathway implicated in many oncogenic cellular pathways, including cell survival, transformation, invasion and proliferation (B). PB1, Phox and Bem1p domain; C, coiled coil region; PQ rich, proline and glutamine rich region; SP, signal peptide, MAM, Meprin, A5 protein and protein tyrosine phosphatase Mu domain; LBD, low density lipoprotein receptor domain; G-rich, glycrine-rich region, TM, transmembrane domain; Kinase domain, tyrosine kinase domain; ER, endoplasmic reticulum.
Treatment and follow-up
The patient was treated with EPOCH (etoposide phosphate, prednisone, oncovin/vincristine sulfate, cyclophophamide, hydroxydaunomycin/doxorubicin hydrocholoride) and provided pembrolizumab (Keytruda) for maintenance after testing positive for PD-L1. Six months after diagnosis, the patient was refractory to treatment with no significant change upon PET scan.
Discussion
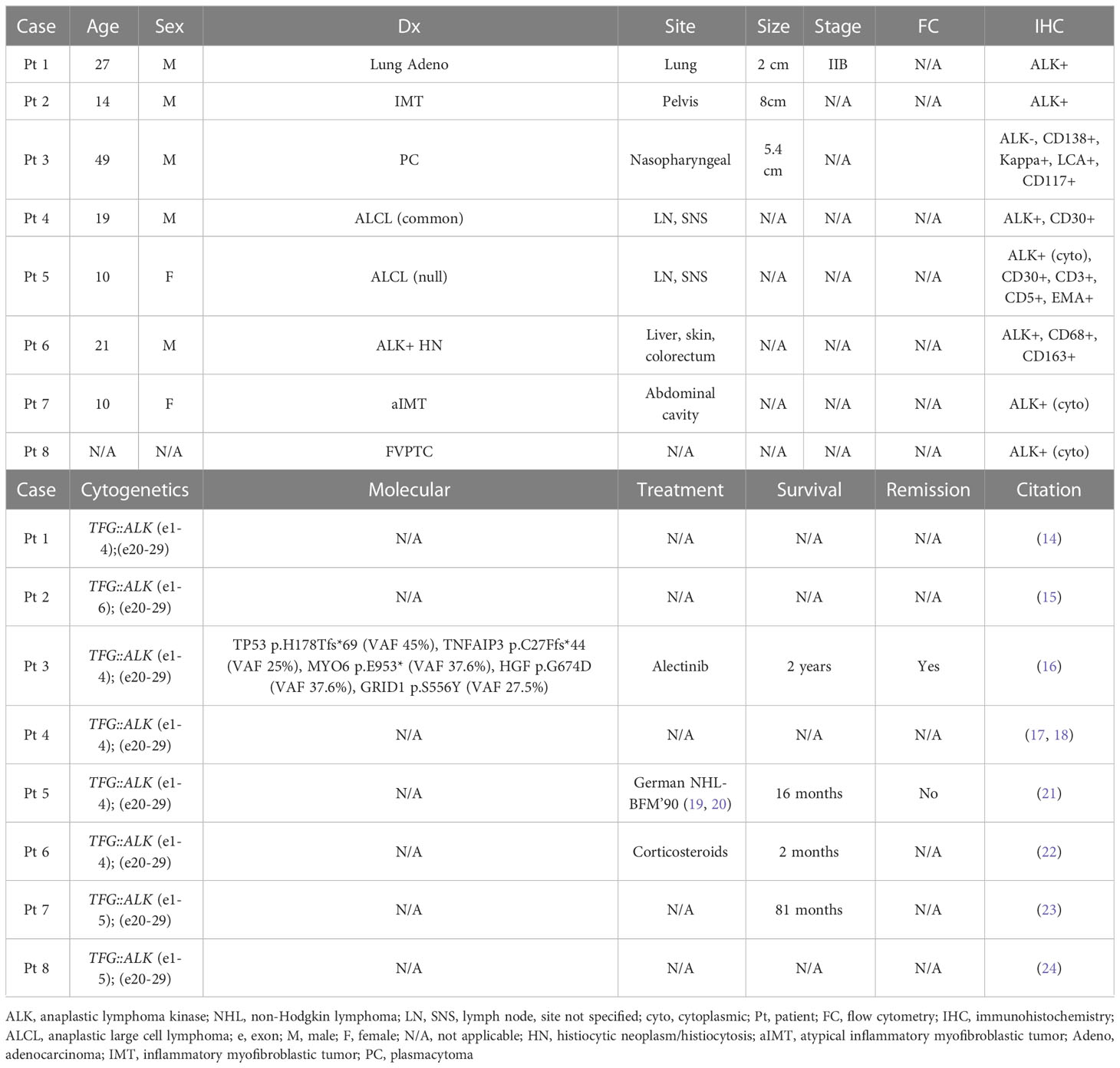
We report the clinicopathologic features of a rare case of ALK+ LBCL with a TFG::ALK fusion analyzed by NGS. TFG::ALK fusions have been reported in a case of lung adenocarcinoma, inflammatory myofibroblastic tumor, anaplastic large cell lymphomas, a histiocytic neoplasm, a plasma cell neoplasm, and papillary thyroid carcinomas (Table 1) (14–18, 21–26). To our knowledge, this is the first report of a TFG::ALK fusion in an ALK+ LBCL.
Trk-fused gene (TFG) was originally identified as the fusion partner of the NRTK1 gene (27). Subsequent studies identified TFG as a partner to other genes such as ALK (17). Since its early discovery, functional studies have shown that TFG, in octamer form, plays an important role in protein trafficking from the endoplasmic reticulum to the Golgi apparatus (28). Multimerization of TFG is possible due to its N-terminal Phox and Bem1p (PB1) and coiled-coil (CC) regions, interaction domains that allow TFG to self-oligomerize (Figure 2B).
In the case of the TFG-ALK fusion protein, presumably the fusion of the TFG PB1 and CC domains to the ALK kinase domain allows for the ALK fusion protein to dimerize, and become constitutively activated (Figure 2B). This mechanism was illustrated by in vitro experiments demonstrating that expression of the TFG-ALK protein resulted in 1) increased proliferation of cells 2) invasiveness of cells, and 3) transformation in NIH3T3 cells (Figure 2B) (11).
It is interesting to note that this particular t(2;3)(p23;q21);ALK::TFG fusion is represented so broadly in diverse tumor types originating from various cells (i.e. B-cell, T-cell, plasma cell, myofibroblast, histocyte and lung epithelium). Perhaps this translocation is not responsible for cell fate and other somatic genetic abnormalities drive the mutated cancer stem cell towards B-cell, T-cell or other cell type differentiation. Another possibility is that the translocation may occur in a very early pluripotent stem cell which is local to or migrates to different tissues and then the cancer cells differentiate according to anatomic site. Further in vivo large-scale studies could help understand the mechanism of disease here.
ALK+ LBCL is a rare neoplasm and most cases show an ALK fusion partner t(2;17)(p23;q23) involving clathrin (CLTC), which results in a CTCL::ALK rearrangement. Other identified fusion proteins include nucleophosmin (NPM1)-ALK, SEC31A-ALK, SQSTM1-ALK, RANBP2-ALK, EML4-ALK, and GORASP2-ALK (1, 7, 8).
Studies exploring the genomic landscape of ALK+ LBCL are limited and, to our knowledge, no previous NGS study on ALK+ LBCL has been performed. Our sequencing analysis only demonstrated a TFG::ALK fusion and no other pathogenic somatic mutations were identified. In contrast, mutational analysis of 20 diffuse large B-cell lymphoma, not otherwise specified (DLBCL, NOS), using this same panel showed a median single nucleotide variant mutational burden of approximately 7.3 (range 0 to 23). The relatively silent cancer genome of this TFG::ALK ALK+ LBCL, save the translocation, is consistent with oncogenically defining translocations seen in many neoplasms where that characteristic driver fusion mutation determines the cellular behavior of a tumor, and further mutations are not critical or necessary for the propagation of that tumor. This driver effect is important to consider as it indicates that inhibition of this central fusion gene and associated pathways could potentially reduce tumor growth. Importantly, studies have shown that ALK translocated tumors are highly sensitive to targeted ALK inhibitor therapies (10, 16, 29). Presently, crizotinib is the most well studied ALK inhibitor in ALK+ LBCL and it induces a transient improvement in lymphadenopathy and serum LDH levels, though this has been followed by rapid progression and a survival time of less than 6 months (5). Soumerai et al. evaluated the therapeutic potential of higher potency ALK inhibitors alectinib and lorlatinib in patient-derived xenograft models. They hypothesize that crizotinib resistance in ALK+LBCL may be overcome by these higher potency ALK inhibitors via upregulation of bypass signalling pathways possibly engaged by the tumor microenvironment (29).
In conclusion, we present the first report of ALK+ LBL harboring a TFG::ALK translocation identified by NGS. Our findings provide novel insight into the pathogenesis of this unique disease process. Further large cohort genetic studies are necessary to expand upon our initial molecular profiling, which may also inform therapeutic management.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics statement
Written informed consent was obtained from the participant/patient(s) for the publication of this case report.
Author contributions
AX, PL and RO wrote the manuscript, conceived of the idea, and analyzed data. NS, JK, and AN contributed to writing the manuscript and analyzing data. JZ, PD, LL and FH edited the manuscript and analyzed data. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Pan Z, Hu S, Li M, Zhou Y, Kim YS, Reddy V, et al. ALK-positive Large b-cell lymphoma: a clinicopathologic study of 26 cases with review of additional 108 cases in the literature. Am J Surg Pathol Jan (2017) 41(1):25–38. doi: 10.1097/PAS.0000000000000753
2. Campo E, Jaffe ES, Cook JR, Quintanilla-Martinez L, Swerdlow SH, Anderson KC, et al. The international consensus classification of mature lymphoid neoplasms: a report from the clinical advisory committee. Blood (2022) 140(11):1229–53. doi: 10.1182/blood.2022015851
3. Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, Araujo IBO, Berti E, et al. The 5th edition of the world health organization classification of haematolymphoid tumours: lymphoid neoplasms. Leukemia (2022) 36(7):1720–48. doi: 10.1038/s41375-022-01620-2
4. Delsol G, Lamant L, Mariame B, Pulford K, Dastugue N, Brousset P, et al. A new subtype of large b-cell lymphoma expressing the ALK kinase and lacking the 2; 5 translocation. Blood (1997) 89(5):1483–90.
5. Castillo JJ, Beltran BE, Malpica L, Marques-Piubelli ML, Miranda RN. Anaplastic lymphoma kinase-positive large b-cell lymphoma (ALK + LBCL): a systematic review of clinicopathological features and management. Leuk Lymphoma (2021) 62(12):2845–53. doi: 10.1080/10428194.2021.1941929
6. Morgan EA, Nascimento AF. Anaplastic lymphoma kinase-positive large b-cell lymphoma: an underrecognized aggressive lymphoma. Adv Hematol (2012) 2012:529572. doi: 10.1155/2012/529572
7. Jiang XN, Yu BH, Wang WG, Zhou XY, Li XQ. Anaplastic lymphoma kinase-positive large b-cell lymphoma: clinico-pathological study of 17 cases with review of literature. PloS One (2017) 12(6):e0178416. doi: 10.1371/journal.pone.0178416
8. Salat H, Din NU, Moatter T, Kayani N, Ahmed A. Anaplastic lymphoma kinase protein positive diffuse large b cell lymphoma; a developing world experience. Pathol Res Pract (2017) 213(6):649–53. doi: 10.1016/j.prp.2017.02.017
9. Gascoyne RD, Lamant L, Martin-Subero JI, Lestou VS, Harris NL, Müller-Hermelink HK, et al. ALK-positive diffuse large b-cell lymphoma is associated with clathrin-ALK rearrangements: report of 6 cases. Blood (2003) 102(7):2568–73. doi: 10.1182/blood-2003-03-0786
10. Hallberg B, Palmer RH. The role of the ALK receptor in cancer biology. Ann Oncol (2016) 27(Suppl 3):iii4–iii15. doi: 10.1093/annonc/mdw301
11. Armstrong F, Duplantier MM, Trempat P, Hieblot C, Lamant L, Espinos E, et al. Differential effects of X-ALK fusion proteins on proliferation, transformation, and invasion properties of NIH3T3 cells. Oncogene (2004) 23(36):6071–82. doi: 10.1038/sj.onc.1207813
12. Butzmann A, Sridhar K, Jangam D, Kumar J, Sahoo MK, Shahmarvand N, et al. A comprehensive analysis of RHOA mutation positive and negative angioimmunoblastic T-cell lymphomas by targeted deep sequencing, expression profiling and single cell digital image analysis. Int J Mol Med (2020) 46(4):1466–76. doi: 10.3892/ijmm.2020.4686
13. Guney E, Lucas CG, Qi Z, Yu J, Zhang R, Ohgami RS, et al. A genetically distinct pediatric subtype of primary CNS large b-cell lymphoma is associated with favorable clinical outcome. Blood Adv (2022) 6(10):3189–93. doi: 10.1182/bloodadvances.2021006018
14. Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell (2007) 131(6):1190–203. doi: 10.1016/j.cell.2007.11.025
15. Lovly CM, Gupta A, Lipson D, Otto G, Brennan T, Chung CT, et al. Inflammatory myofibroblastic tumors harbor multiple potentially actionable kinase fusions. Cancer Discov (2014) 4(8):889–95. doi: 10.1158/2159-8290.CD-14-0377
16. Masood A, Christ T, Asif S, Rajakumar P, Gustafson BA, Shune LO, et al. Non-secretory multiple myeloma with unusual TFG-ALK fusion showed dramatic response to ALK inhibition. NPJ Genom Med (2021) 6(1):23. doi: 10.1038/s41525-021-00186-9
17. Hernandez L, Pinyol M, Hernandez S, Beà S, Pulford K, Rosenwald A, et al. TRK-fused gene (TFG) is a new partner of ALK in anaplastic large cell lymphoma producing two structurally different TFG-ALK translocations. Blood (1999) 94(9):3265–8.
18. Hernandez L, Bea S, Bellosillo B, Pinyol M, Falini B, Carbone A, et al. Diversity of genomic breakpoints in TFG-ALK translocations in anaplastic large cell lymphomas: identification of a new TFG-ALK(XL) chimeric gene with transforming activity. Am J Pathol (2002) 160(4):1487–94. doi: 10.1016/S0002-9440(10)62574-6
19. Reiter A, Schrappe M, Tiemann M, Ludwig WD, Yakisan E, Zimmermann M, et al. Improved treatment results in childhood b-cell neoplasms with tailored intensification of therapy: a report of the Berlin-Frankfurt-Munster group trial NHL-BFM 90. Blood (1999) 94(10):3294–306.
20. Reiter A, Schrappe M, Ludwig WD, Tiemann M, Parwaresch R, Zimmermann M, et al. Intensive ALL-type therapy without local radiotherapy provides a 90% event-free survival for children with T-cell lymphoblastic lymphoma: a BFM group report. Blood (2000) 95(2):416–21.
21. Rosenwald A, Ott G, Pulford K, Katzenberger T, Kühl J, Kalla J, et al. t(1;2)(q21;p23) and t(2;3)(p23;q21): two novel variant translocations of the t(2;5)(p23;q35) in anaplastic large cell lymphoma. Blood (1999) 94(1):362–4.
22. Kemps PG, Picarsic J, Durham BH, Hélias-Rodzewicz Z, Hiemcke-Jiwa L, van den Bos C, et al. ALK-positive histiocytosis: a new clinicopathologic spectrum highlighting neurologic involvement and responses to ALK inhibition. Blood (2022) 139(2):256–80. doi: 10.1182/blood.2021013338
23. Lee JC, Li CF, Huang HY, Zhu MJ, Mariño-Enríquez A, Lee CT, et al. ALK oncoproteins in atypical inflammatory myofibroblastic tumours: novel RRBP1-ALK fusions in epithelioid inflammatory myofibroblastic sarcoma. J Pathol (2017) 241(3):316–23. doi: 10.1002/path.4836
24. McFadden DG, Dias-Santagata D, Sadow PM, Lynch KD, Lubitz C, Donovan SE, et al. Identification of oncogenic mutations and gene fusions in the follicular variant of papillary thyroid carcinoma. J Clin Endocrinol Metab (2014) 99(11):E2457–62. doi: 10.1210/jc.2014-2611
25. Evangelista AF, Zanon MF, Carloni AC, de Paula FE, Morini MA, Ferreira-Neto M, et al. Detection of ALK fusion transcripts in FFPE lung cancer samples by NanoString technology. BMC Pulm Med (2017) 17(1):86. doi: 10.1186/s12890-017-0428-0
26. Drexler HG, Gignac SM, von Wasielewski R, Werner M, Dirks WG. Pathobiology of NPM-ALK and variant fusion genes in anaplastic large cell lymphoma and other lymphomas. Leukemia (2000) 14(9):1533–59. doi: 10.1038/sj.leu.2401878
27. Greco A, Mariani C, Miranda C, Lupas A, Pagliardini S, Pomati M, et al. The DNA rearrangement that generates the TRK-T3 oncogene involves a novel gene on chromosome 3 whose product has a potential coiled-coil domain. Mol Cell Biol (1995) 15(11):6118–27. doi: 10.1128/MCB.15.11.6118
28. Witte K, Schuh AL, Hegermann J, Sarkeshik A, Mayers JR, Schwarze K, et al. TFG-1 function in protein secretion and oncogenesis. Nat Cell Biol (2011) 13(5):550–8. doi: 10.1038/ncb2225
Keywords: ALK+ large B-cell lymphoma, TFG, ALK, ALK translocation, ALK fusion
Citation: Xiao A, Shahmarvand N, Nagy A, Kumar J, Van Ziffle J, Devine P, Huang F, Lezama L, Li P and Ohgami RS (2023) TFG::ALK fusion in ALK positive large B-cell lymphoma: a case report and review of literature. Front. Oncol. 13:1174606. doi: 10.3389/fonc.2023.1174606
Received: 26 February 2023; Accepted: 24 April 2023;
Published: 25 May 2023.
Edited by:
Lorenzo Leoncini, University of Siena, ItalyReviewed by:
Konnie Hebeda, Radboud University Medical Centre, NetherlandsGiorgio Alberto Croci, University of Milan, Italy
Copyright © 2023 Xiao, Shahmarvand, Nagy, Kumar, Van Ziffle, Devine, Huang, Lezama, Li and Ohgami. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert S. Ohgami, cm9iZXJ0Lm9oZ2FtaUBhcnVwbGFiLmNvbQ==
†These authors have contributed equally to this work
 Andrew Xiao1
Andrew Xiao1 Jyoti Kumar
Jyoti Kumar Jessica Van Ziffle
Jessica Van Ziffle Peng Li
Peng Li Robert S. Ohgami
Robert S. Ohgami