- 1Department of Pathology and Laboratory Medicine, Weill Cornell Medicine, New York, NY, United States
- 2Englander Institute for Precision Medicine, Weill Cornell Medicine, New York, NY, United States
- 3BostonGene Corporation, Waltham, MA, United States
- 4Department of Radiology, Weill Cornell Medicine, New York, NY, United States
- 5Department of Medicine, Weill Cornell Medicine, New York, NY, United States
Cancer of unknown primary (CUP) represents a significant diagnostic and therapeutic challenge, being the third to fourth leading cause of cancer death, despite advances in diagnostic tools. This article presents a successful approach using a novel genomic analysis in the evaluation and treatment of a CUP patient, leveraging whole-exome sequencing (WES) and RNA sequencing (RNA-seq). The patient, with a history of multiple primary tumors including urothelial cancer, exhibited a history of rapid progression on empirical chemotherapy. The application of our approach identified a molecular target, characterized the tumor expression profile and the tumor microenvironment, and analyzed the origin of the tumor, leading to a tailored treatment. This resulted in a substantial radiological response across all metastatic sites and the predicted primary site of the tumor. We argue that a comprehensive genomic and molecular profiling approach, like the BostonGene© Tumor Portrait, can provide a more definitive, personalized treatment strategy, overcoming the limitations of current predictive assays. This approach offers a potential solution to an unmet clinical need for a standardized approach in identifying the tumor origin for the effective management of CUP.
Introduction
Cancer of unknown primary (CUP) is a diagnosis of a heterogeneous group of metastatic tumors for which no tissue of origin has been identified after extensive pathological, radiological, and clinical assessment (1, 2). The characterization of CUP to identify effective treatments that may improve patient survival continues to be challenging despite the increased accessibility to imaging, immunohistochemistry, and molecular tools. The treatment of CUP does not only require the identification of molecular targets for treatment but also the identification of the tumor of origin because the response to targeted treatment is often also dependent on cancer subtyping. An example of this is the different response to BRAF-targeted treatment in different tumors harboring the BRAF V600E mutation (3).
Currently, 3% to 5% of all cancer diagnosis is CUP, and it is the third to fourth most common cause of cancer death (4, 5). In the past five decades, there has been an observable biphasic trend in the diagnosis of CUP. Initially, there was an increase, largely attributable to the increased sensitivity of diagnostic tools, including imaging modalities. Following this, a decline was observed, which can be primarily credited to enhancements in the capabilities to detect the primary neoplastic site, including immunohistochemistry, molecular testing, and improved imaging (6). Nevertheless, survival of patients with CUP is poor with a prognosis typically less than a year, most often with empirical treatment (7). This poor prognosis is caused primarily by the advanced stage at diagnosis and resistance to empirical treatment (8, 9).
For patients with a diagnosis of CUP, molecular profiling is being developed to better characterize the tumor of origin (10). A better characterization of those tumors gives a better chance for the patients to respond to approved systemic or targeted therapies or to be enrolled in clinical trials for specific cancer types (11–13).
Several predictive assays have been developed using various techniques, including a genome-wide expression assay for predicting tumor origin, a 2000-gene classification RNA-based model (CancerTYPE ID; Biotheranostics, San Diego, CA, USA), and a 64-tissue-specific microRNA assay (Tissue of Origin; Cancer Genetics, Rutherford, NJ, USA), which are all commercially available (14–16). Further approaches in development, such as miRNA expression profiling and an assay using methylation signatures, aim to predict the tissue of origin (17, 18). However, while these developments mark progress, the clinical landscape of managing CUP is yet to be fully shaped by a standardized testing modality. The practical advantages of these methods need to be thoroughly evaluated in clinical trials, as their true impact on patient treatment is still unclear. This highlights an unmet need for a definitive, standardized approach in identifying the tumor origin for the effective management of CUP.
There is a strong belief that knowing the site of origin could improve therapeutic strategies and increase the survival of patients with responsive CUP tumor types (19). However, one clinical trial reported that there was no benefit from site-specific chemotherapy compared with empirical chemotherapy (10). Thus, more complex and molecular profiling for the determination of treatment modalities is needed for the management of patients with CUP. In addition to determining the origin of the tumor, molecular evaluation is required to identify therapeutic targets, such as hotspot mutations and mutational signatures (20, 21).
In this article, we present a successful approach using a novel genomic analysis for the evaluation and treatment of a patient with CUP. The patient had a history of multiple primary tumors, including urothelial cancer and metastatic disease without an origin after radiological and pathological evaluation. He was being treated for metastatic urothelial cancer in the lung. Whole-exome sequencing (WES) and RNA sequencing (RNA-seq) integrated analysis was done after rapid progression on empirical chemotherapy. Our approach identified a molecular target, characterized the tumor expression profile and the tumor microenvironment, and analyzed the origin of the tumor. This led to tailored treatment that resulted in substantial radiological response in all the metastatic sites and the predicted primary site of the tumor.
Case history
An 81-year-old gentleman presented to the emergency department with an ulnar pathologic fracture and leg pain after falling at home (Figure 1). The patient, currently a non-smoker, has a history of 12.5 pack-years of smoking. He also had previous diagnoses of anxiety, aortic stenosis (treated with balloon valvuloplasty), hypersensitivity lung disease, hyperlipidemia, ischemic stroke, zoster, and excised melanoma (15 years prior). In addition to the balloon valvuloplasty and the melanoma excision, his surgical history included the removal of a back epidermoid cyst and umbilical and inguinal hernia repair. His family history included colorectal cancer in his mother, sudden death in his father, Raynaud’s disease in his son, Crohn’s disease in his daughter, and heart disease in his brother. He was being followed for urothelial carcinoma that was diagnosed 2 years prior to presentation. Abdomino-pelvic computed tomography (CT) scan at the time of the diagnosis of the urothelial carcinoma showed a 2.5-cm bladder lesion and a 1.5-cm left iliac lymph node, and the chest CT scan showed an irregular cystic focus of the right lung with peripheral fibrosis. A transurethral biopsy of the bladder lesion showed invasive high-grade urothelial carcinoma with undifferentiated sarcomatous features. PD-L1 immunohistochemistry (IHC) was positive with a combined positive score (CPS) of more than 50. The lung cystic lesion could not be biopsied due to the small size of the solid component and his severe emphysema. The urothelial carcinoma was managed with six cycles of neoadjuvant carboplatin and gemcitabine that resulted in interval resolution of the bladder lesion and the prominent lymph node. Radical cystoprostatectomy showed no residual invasive urothelial carcinoma (pTis pN0) with therapy-related changes and an incidental organ-confined Gleason grade 6 prostatic adenocarcinoma (pT2 pN0).
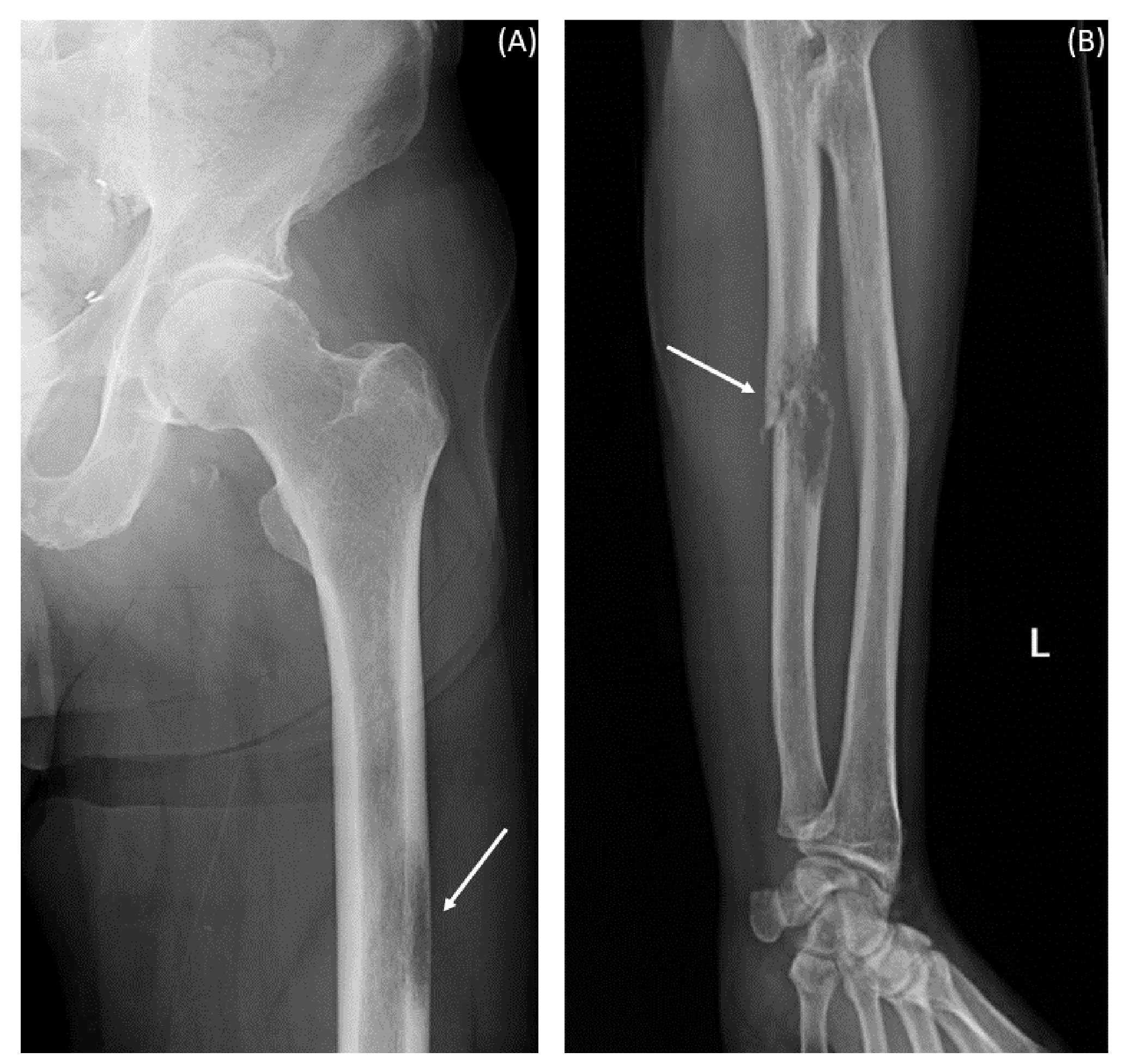
Figure 1 X-ray images at presentation with pain after fall. The arrows indicate hypodensities suggestive of metastatic lesions. (A) Left femur. (B) Left forearm.
After a year and a half of loss to follow-up to oncology, the patient presented after a fall with left forearm and leg pain. Physical exam was positive for swelling and tenderness in his left forearm. An X-ray showed a permeative osseous lesion with cortical erosion at the diaphysis of the ulna and a cortical lucency in the lateral aspect of the left femur, both concerning metastasis (Figure 1). A positron emission tomography (PET) CT scan showed a fluorodeoxyglucose (FDG) avid retrocaval lymph node, hypermetabolic abdominopelvic lymphadenopathy, left femoral and ulnar lesions, posterior elements of the T11 vertebral body, and a hypermetabolic lung lesion at the location of the previously seen cystic lesion (Figure 2A). The ulnar and femoral lesions were excised for diagnostic and treatment purposes. Histopathology showed a poorly differentiated metastatic carcinoma with sarcomatoid elements. By immunohistochemistry, the tumor was positive for pan-cytokeratin (CK) and CK7 and negative for CK20, p63, GATA3, TTF1, PAX8, NKX3.1, and CDX2 (Figure 3). PD-L1 immunohistochemical staining was negative (CPS = 0). This immunoprofile was diagnostic of carcinoma but did not support the origin of either colon (CDX2 negative), lung (TTF1 negative), renal (PAX-8 negative), or bladder (GATA-3 negative) primary sites. Along with tumor histomorphology, the immunoprofile also excluded metastatic malignant melanoma. While the histological and immunohistochemical profile and comparison to prior urothelial carcinoma biopsy could suggest metastatic sarcomatoid carcinoma from prior urothelial carcinoma, the site of origin could not be certainly determined. The patient was started on treatment with carboplatin/gemcitabine for what was presumed to be a metastatic urothelial carcinoma and showed an overall progression on FDG PET scan of widespread FDG avid disease (Figure 2B).
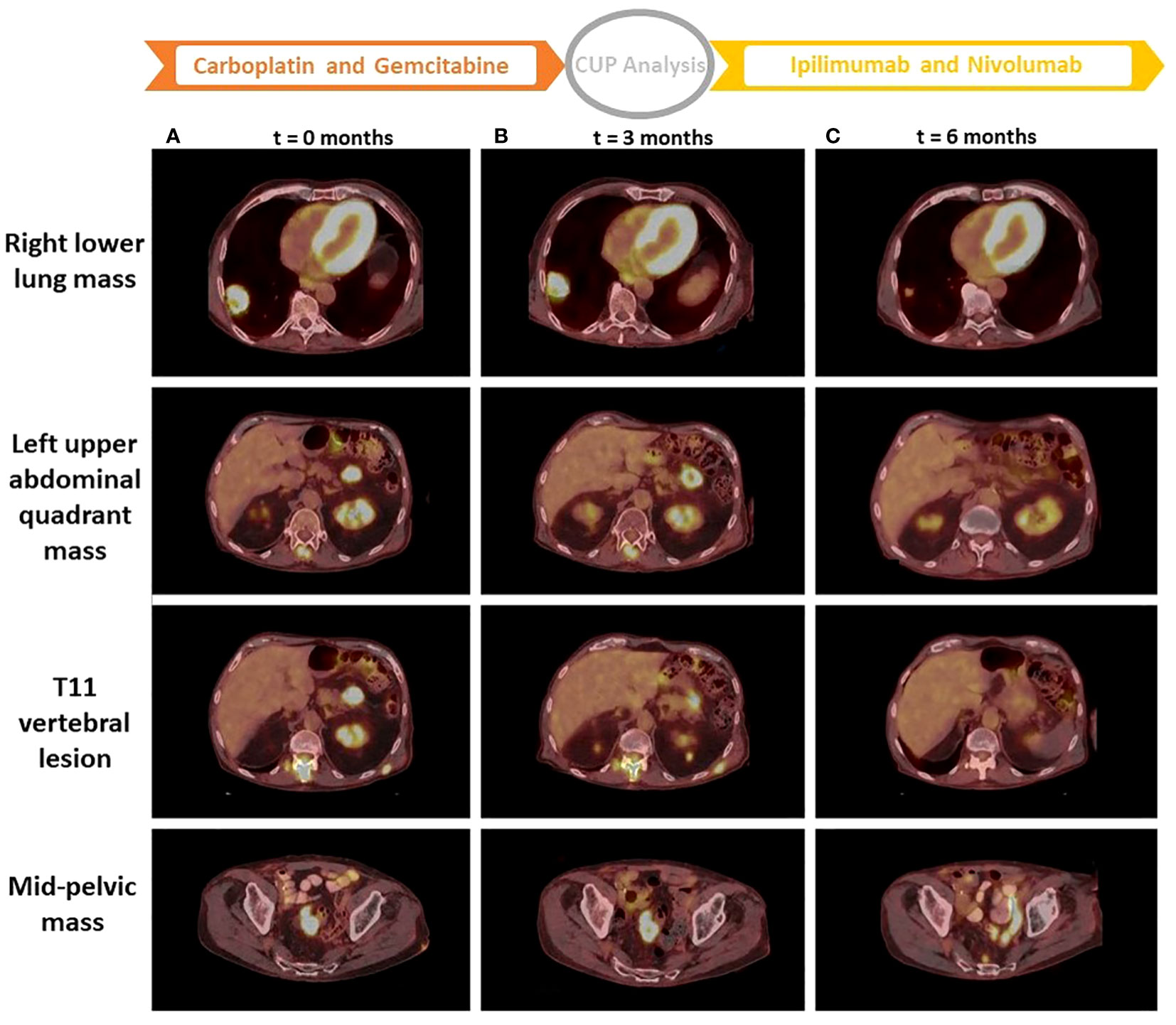
Figure 2 PET CT scan images from three time points. Images of column (A) are from a PET CT scan at the start of treatment with carboplatin and gemcitabine (t = 0 months). Images of column (B) are from a PET CT scan at the start of treatment with ipilimumab + nivolumab after performing the CUP analysis (t = 3 months). Images of column (C) are from a PET CT scan 3 months after treatment with ipilimumab + nivolumab (t = 6 months). t, time; CUP, cancer of unknown primary.
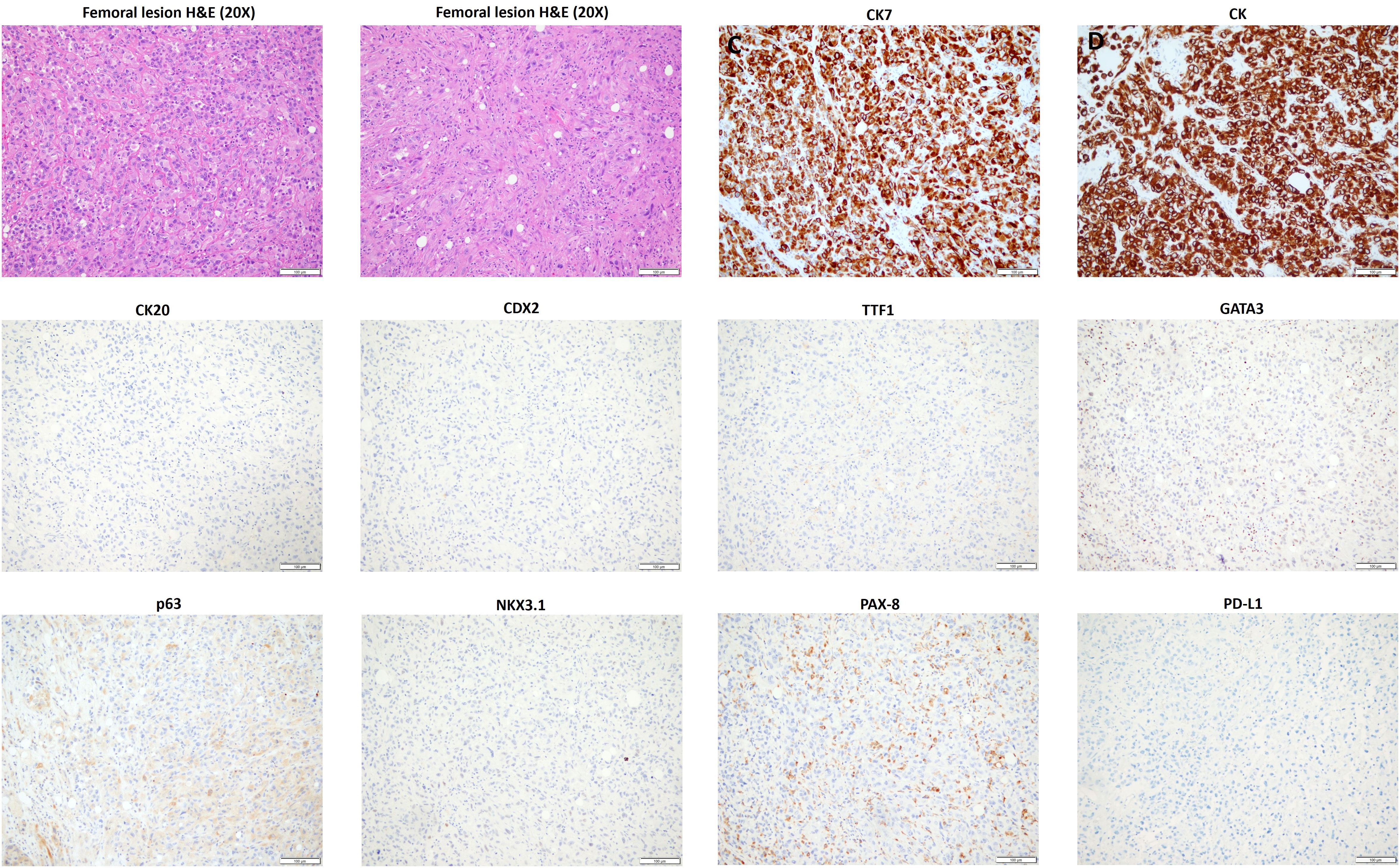
Figure 3 Histopathology and immunoprofile of the excised femoral lesion. H&E, hematoxylin and eosin; CK, cytokeratin.
While the patient was on chemotherapy, further attempts were made to characterize the metastatic tumor. Thus, TruSight Oncology 500 (TSO500), a panel-based next-generation sequencing assay of the formalin-fixed paraffin-embedded (FFPE) tissue from his femoral lesion, showed KRAS (c.34G>T) and BRAF (c.1780G>A) mutations with high tumor mutational burden (TMB) and no microsatellite instability (MSI). TSO500 of the urothelial carcinoma showed BRCA1 (c.3430_3458del29), BRCA2 (c.7963C>T and c.8084C>G), TP53 (c.273G>A), and CDKN2A (c.355G>T) mutations with high TMB and microsatellite-stable (MSS) status. Based on those molecular tests, the primary urothelial carcinoma and the metastatic tumor had different molecular profiles. Due to the molecular differences between the bladder and femoral tumors, combined with the uncertainty of pathological and clinical assessments, a diagnosis of CUP was made.
To better characterize the metastatic disease and to find the best management, the patient was enrolled in a prospective study for CUP patients (IRB protocol 20-05022007). Frozen tissue specimen from the femoral metastatic lesion was extracted for DNA and RNA that was used for WES and RNA-seq. BostonGene© Tumor Portrait was established from the analysis of WES and RNA-seq. To produce an unbiased molecular profile, the analysis was blinded to the clinical, pathological, IHC, and radiological data. The results of the tumor profiling were then analyzed in the context of the clinical testing done. The tumor profiling confirmed the BRAF and KRAS mutations and it showed high TMB. BostonGene Tissue of origin classifier trained and validated on both genomic and transcriptomic molecular patterns was applied to predict the tumor origin of the sample and clearly favored a lung adenocarcinoma with the highest probability of 72.8% (Figure 4A). The top second prediction with a much lower probability was gastrointestinal (6.9%), consisting of colorectal (4.5%) and stomach (1.2%) (Figure 4A). The molecular events that played a major role in lung adenocarcinoma prediction were the presence of tobacco smoking mutational signature performed independently (22), KRAS G12 somatic mutation, TMB, and several characteristic copy-number alterations (CNAs) (Figure 4B), whereas for GI adenocarcinoma, only CNA features were in favor of diagnosis (Figure 4C). Indeed, when compared with the distributions of TMB (Figure 4D) and smoking signature rank (Figure 4E) and frequencies of KRAS G12 (Figure 4F) among The Cancer Genome Atlas (TCGA) cohorts, the sample showed the highest similarity with the TCGA-LUAD (lung adenocarcinoma) cohort rather than the TCGA-COREAD (colorectal adenocarcinoma) and TCGA-BLCA (bladder carcinoma). RNA-seq showed a high expression of PD-L1. Although a gastrointestinal tract primary could not be ruled out by the blinded Tumor Portrait, it was low on the list of clinical differentials because IHC of the metastatic tumor was CK20 and CDX2 negative, and on radiology, there was a low suspicion of a gastrointestinal tract primary tumor.
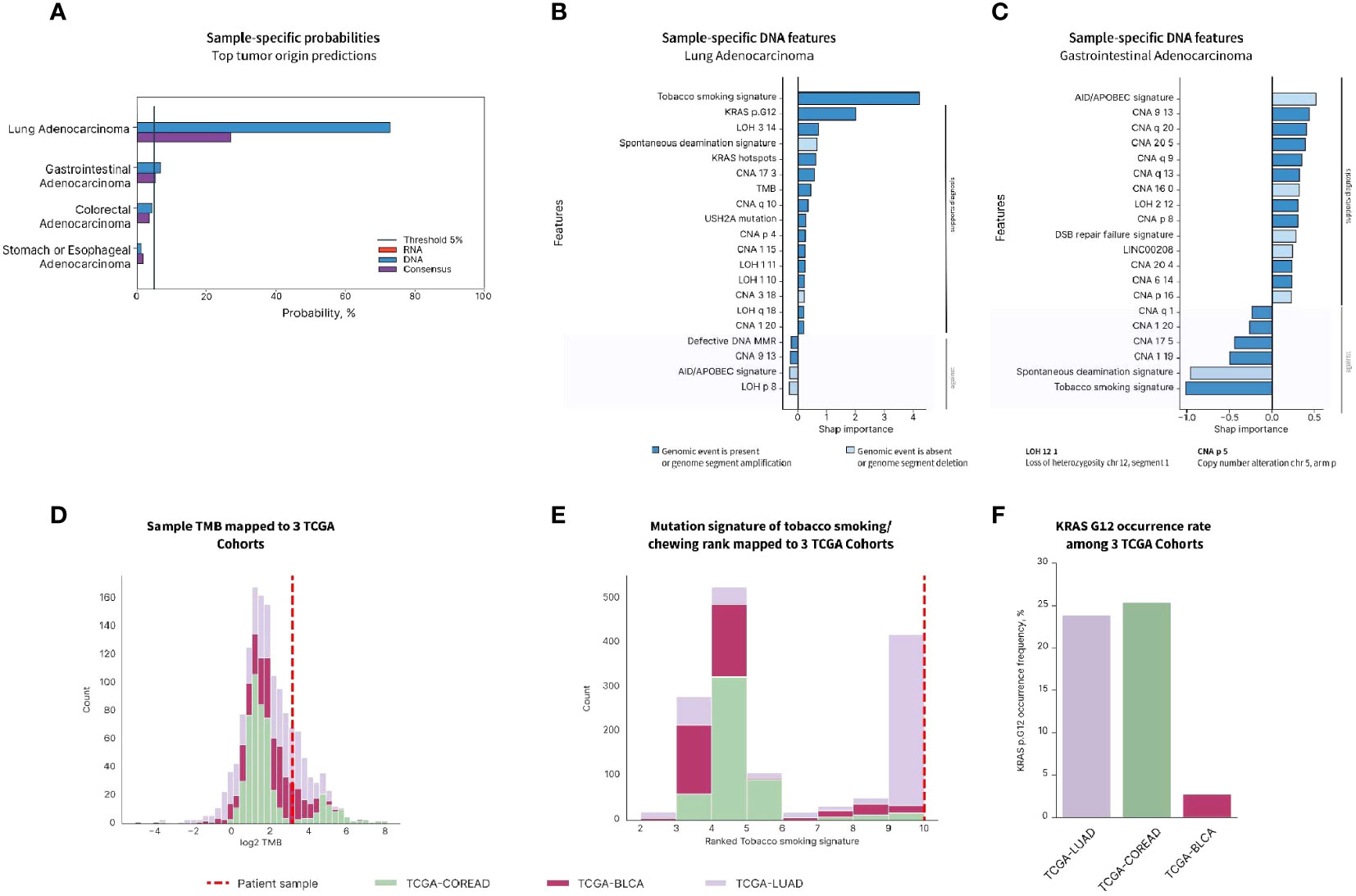
Figure 4 Tumor origin prediction using BostonGene Tumor of origin classifier. (A) Probabilities of top tumor origins predicted by DNA, RNA, and consensus classifiers. Feature importance plots provided by the DNA classifier for lung adenocarcinoma (B) and gastrointestinal adenocarcinoma (C) show molecular events with major roles in tumor origin prediction. Distributions of TMB (D), tobacco smoking mutation signature rank (E), and KRAS G12 mutation rate (F) among the three TCGA cohorts: colorectal (TCGA-COREAD), urothelial (TCGA-BLCA), and lung adenocarcinoma (TCGA-LUAD). The patient’s value is represented with a dashed line. The data presented has been elaborated based on the commercially available service.
By accounting for all the results of clinical and molecular profiling of the tumor, a multidisciplinary discussion led to stopping the treatment with carboplatin and gemcitabine. The patient was started on treatment with nivolumab plus ipilimumab for a highly probable primary non-small cell lung cancer (NSCLC) with high TMB and negative PD-L1 (23). Other observations such as high RNA expression of PD-L1 (24) and a high degree of tumor infiltration by immune cells suggested a more favorable prognosis for the patient undergoing this treatment (25, 26). The patient was treated with three cycles of ipilimumab and nivolumab and experienced no adverse effects. A follow-up PET CT scan showed an overall partial response to treatment based on the Response Evaluation Criteria in Solid Tumours (RECIST) 1.1 criteria. The abdominal and mid-pelvic masses have demonstrated a decrease in size with a resolution of FDG uptake (Figure 2C). Also, increased sclerosis and resolved FDG uptake of osseous metastases including T11 vertebral metastasis (Figure 2C) further indicated treatment response.
Discussion
Cancer of unknown primary is a diagnosis of exclusion. It is a metastatic disease with poor differentiation and an unspecific immunohistochemical profile (2, 9). Multiple assay-based panels that used real-time PCR, RNA-seq, and methylation studies have been developed to identify the tumor of origin. Both the FDA-approved panels and the experimental panels for tumor of origin identification have shown good accuracy but present questionable clinical implications. Finding the tumor of origin does not imply a better management strategy or better response to treatment (3, 10). Also, the reported accuracy of the developed panels has two major flaws. First, cancers of known primary used as control cases for these panels have better differentiation and more specific molecular profiles compared with CUP cases (8, 9). Second, there is no available standard assay to determine the accuracy of the panels in testing CUP cases (13). The BostonGene© Tumor Portrait is a comprehensive genomic and molecular profiling report. It integrates WES and RNA-seq and offers several key analyses: determining the tumor of origin, identifying potential treatment targets, characterizing the tumor microenvironment, and determining the tumor composition and mutational signatures. This approach covers the precision medicine aspects that have the potential of directing the clinical management of patients with CUP.
In this article, we report a case of CUP diagnosis in a patient with a history of multiple cancer primaries. Despite the histologic similarities with the previously diagnosed urothelial carcinoma, the distinct mutational profile of the metastatic tumor led to a diagnosis of CUP. The lack of response to empirical chemotherapy necessitated further attempts at detailed tumor characterization. Thus, the patient was enrolled in a prospective study to identify the tumor of origin in an attempt to offer a treatment alternative. The BostonGene© Tumor Portrait test was performed on deidentified samples as part of a research collaboration. An integrated tissue of origin WES/RNA-seq analysis ruled out urothelial carcinoma and suggested a lung primary with a high probability. The Tumor Portrait confirmed the high TMB and microenvironment characterized by infiltration by immune cells and a high RNA expression of PD-L1 and PD-1 and a smoking mutational signature. This characterization was needed because even with the identification of the lung primary, empirical treatment could not be offered to this patient with stage IV lung cancer with widespread progressing metastasis and worsening performance status. The patient was treated with ipilimumab + nivolumab because this treatment has been shown to extend survival duration in patients with high TMB, regardless of their PD-L1 status (27). In addition to that, studies have shown that immune infiltration of the tumor is associated with a better response to immune checkpoint inhibitors in general (25). Although it has been reported that RNA-seq PD-L1 and IHC PD-L1 results could be concordant, our case showed a high RNA-seq expression with negative IHC (24). This could be due to unknown technical or biological reasons. However, the high level of expression could be correlated to the response to immune checkpoint inhibitors (28). After three cycles of treatment with ipilimumab + nivolumab, a PET CT scan showed a dramatic radiological response of the primary tumor in the lung and all metastatic tumor lesions in the bone and soft tissue. This report demonstrated that tumor of origin identification using NGS-based tests, like the BostonGene CUP algorithm, could help with improved diagnosis and effective treatment selection. The tumor regression provides evidence that treating CUP patients should go beyond identifying the tumor’s molecular target and origin.
Despite being effective, this method has limitations when managing CUP patients. The availability and quality of the tissue affect the algorithm’s performance and accuracy, similar to other molecular panels. Utilizing FFPE may reduce the DNA and RNA extraction’s quality, which would then make analysis more challenging. Additionally, it may be difficult to biopsy metastatic locations in CUP patients for accurate characterization, as was the case in our patient when the lung lesion could not be biopsied due to extensive emphysema. Despite all the developments, CUP continues to be one of the most challenging cancer types to diagnose and treat. To demonstrate the clinical utility of Tumor Portrait, it must be performed on a large number of CUP patients.
Data availability statement
The raw data supporting the conclusions of this article is available at (https://www.ncbi.nlm.nih.gov/sra/docs/) via BioProject accession number: PRJNA1063915.
Ethics statement
The studies involving humans were approved by The Weill Cornell Medicine Institutional Review Board (WCM-IRB). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
MA: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. NS: Conceptualization, Data curation, Project administration, Writing – review & editing. MS: Data curation, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. JM: Data curation, Investigation, Project administration, Resources, Supervision, Writing – review & editing. ZA: Data curation, Formal analysis, Writing – review & editing. NK: Data curation, Formal analysis, Validation, Writing – review & editing. DK: Data curation, Formal analysis, Writing – review & editing. AN: Data curation, Formal analysis, Writing – review & editing. MK: Data curation, Formal analysis, Writing – review & editing. AT: Data curation, Formal analysis, Writing – review & editing. AM: Data curation, Formal analysis, Writing – review & editing. VF: Data curation, Formal analysis, Writing – review & editing. JD: Data curation, Formal analysis, Writing – review & editing. AB: Data curation, Formal analysis, Writing – review & editing. NA: Data curation, Formal analysis, Project administration, Resources, Supervision, Writing – review & editing. DW: Conceptualization, Investigation, Methodology, Visualization, Writing – review & editing. JE: Formal analysis, Investigation, Validation, Visualization, Writing – review & editing. AS: Data curation, Methodology, Supervision, Validation, Writing – review & editing. OE: Conceptualization, Project administration, Supervision, Writing – review & editing. CS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Writing – review & editing. DN: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Writing – review & editing. JM: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Englander Institute for Precision Medicine at Weill Cornell Medicine and NewYork-Presbyterian.
Acknowledgments
We appreciate Jessica H. Brown from BostonGene for her kind review and edits.
Conflict of interest
NS, ZA, NK, DK, AN, MK, AT, AM, VF, JD, AB, and NA are employees of Bostongene.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wagland R, Bracher M, Drosdowsky A, Richardson A, Symons J, Mileshkin L, et al. Differences in experiences of care between patients diagnosed with metastatic cancer of known and unknown primaries: mixed-method findings from the 2013 cancer patient experience survey in England. BMJ Open (2017) 7(9):e017881. doi: 10.1136/bmjopen-2017-017881
2. Krämer A, Bochtler T, Pauli C, Baciarello G, Delorme S, Hemminki K, et al. Cancer of unknown primary: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up☆. Ann Oncol (2023) 34(3):228–46. doi: 10.1016/j.annonc.2022.11.013
3. Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay J-Y, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. New Engl J Med (2015) 373(8):726–36. doi: 10.1056/NEJMoa1502309
4. Qaseem A, Usman N, Jayaraj JS, Janapala RN, Kashif T. Cancer of unknown primary: a review on clinical guidelines in the development and targeted management of patients with the unknown primary site. Cureus. (2019) 11(9). doi: 10.7759/cureus.5552
5. Pavlidis N, Pentheroudakis G. Cancer of unknown primary site. Lancet (2012) 379(9824):1428–35. doi: 10.1016/S0140-6736(11)61178-1
6. Jones W, Allardice G, Scott I, Oien K, Brewster D, Morrison DS. Cancers of unknown primary diagnosed during hospitalization: a population-based study. BMC canc (2017) 17(1):1–7. doi: 10.1186/s12885-017-3083-1
7. Kolling S, Ventre F, Geuna E, Milan M, Pisacane A, Boccaccio C, et al. “Metastatic cancer of unknown primary” or “primary metastatic cancer”? Front Oncol (2020) 9:1546. doi: 10.3389/fonc.2019.01546
8. Economopoulou P, Mountzios G, Pavlidis N, Pentheroudakis G. Cancer of unknown primary origin in the genomic era: elucidating the dark box of cancer. Cancer Treat Rev (2015) 41(7):598–604. doi: 10.1016/j.ctrv.2015.05.010
9. Alshareeda AT, Al-Sowayan BS, Alkharji RR, Aldosari SM, Alghuwainem A. Cancer of unknown primary site: real entity or misdiagnosed disease? J Cancer (2020) 11(13):3919. doi: 10.7150/jca.42880
10. Hayashi H, Kurata T, Takiguchi Y, Arai M, Takeda K, Akiyoshi K, et al. Randomized phase II trial comparing site-specific treatment based on gene expression profiling with carboplatin and paclitaxel for patients with cancer of unknown primary site. J Clin Oncol (2019) 37(7):570–9. doi: 10.1200/JCO.18.00771
11. Kato S, Alsafar A, Walavalkar V, Hainsworth J, Kurzrock R. Cancer of unknown primary in the molecular era. Trends canc (2021) 7(5):465–77. doi: 10.1016/j.trecan.2020.11.002
12. Gu H, Zhang S, He S, Zhu X, Wang Y, Xie Q, et al. DNA methylation profiling to determine the primary sites of metastatic cancers using formalin-fixed paraffin-embedded tissues. JCO (2022) 40:3079–3079. doi: 10.1200/JCO.2022.40.16_suppl.3079
13. Zhang Y, Xia L, Ma D, Wu J, Xu X, Xu Y. 90-gene expression profiling for tissue origin diagnosis of cancer of unknown primary. Front Oncol (2021) 11:722808. doi: 10.3389/fonc.2021.722808
14. Ojala KA, Kilpinen SK, Kallioniemi OP. Classification of unknown primary tumors with a data-driven method based on a large microarray reference database. Genome Med (2011) 3(9):1–12. doi: 10.1186/gm279
15. Erlander MG, Ma X-J, Kesty NC, Bao L, Salunga R, Schnabel CA. Performance and clinical evaluation of the 92-gene real-time PCR assay for tumor classification. J Mol Diagnostics (2011) 13(5):493–503. doi: 10.1016/j.jmoldx.2011.04.004
16. Meiri E, Mueller WC, Rosenwald S, Zepeniuk M, Klinke E, Edmonston TB, et al. A second-generation microRNA-based assay for diagnosing tumor tissue origin. oncologist. (2012) 17(6):801–12. doi: 10.1634/theoncologist.2011-0466
17. Søkilde R, Vincent M, Møller AK, Hansen A, Høiby PE, Blondal T, et al. Efficient identification of miRNAs for classification of tumor origin. J Mol Diagnostics (2014) 16(1):106–15. doi: 10.1016/j.jmoldx.2013.10.001
18. Moran S, Martínez-Cardús A, Sayols S, Musulén E, Balañá C, Estival-Gonzalez A, et al. Epigenetic profiling to classify cancer of unknown primary: a multicentre, retrospective analysis. Lancet Oncol (2016) 17(10):1386–95. doi: 10.1016/S1470-2045(16)30297-2
19. Ding Y, Jiang J, Xu J, Chen Y, Zheng Y, Jiang W, et al. Site-specific therapy in cancers of unknown primary site: a systematic review and meta-analysis. ESMO Open (2022) 7(2):100407. doi: 10.1016/j.esmoop.2022.100407
20. Selves J, Long-Mira E, Mathieu M-C, Rochaix P, Ilié M. Immunohistochemistry for diagnosis of metastatic carcinomas of unknown primary site. Cancers. (2018) 10(4):108. doi: 10.3390/cancers10040108
21. Cacho-Díaz B, García-Botello DR, Wegman-Ostrosky T, Reyes-Soto G, Ortiz-Sánchez E, Herrera-Montalvo LA. Tumor microenvironment differences between primary tumor and brain metastases. J Trans Med (2020) 18:1–12. doi: 10.1186/s12967-019-02189-8
22. Chevalier A, Yang S, Khurshid Z, Sahelijo N, Tong T, Huggins JH, et al. The Mutational Signature Comprehensive Analysis Toolkit (musicatk) for the discovery, prediction, and exploration of mutational signatures. Cancer Res (2021) 81(23):5813–7. doi: 10.1158/0008-5472.CAN-21-0899
23. Hellmann MD, Ciuleanu T-E, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. New Engl J Med (2018) 378(22):2093–104. doi: 10.1056/NEJMoa1801946
24. Vannitamby A, Hendry S, Makadia T, Danks J, Slavin J, Irving L, et al. A novel approach to detect programed death ligand 1 (PD-L1) status and multiple tumor mutations using a single non–small-cell lung cancer (NSCLC) bronchoscopy specimen. J Mol Diagnostics (2019) 21(2):186–97. doi: 10.1016/j.jmoldx.2018.10.001
25. Bai R, Lv Z, Xu D, Cui J. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. biomark Res (2020) 8:1–17. doi: 10.1186/s40364-020-00209-0
26. Kümpers C, Jokic M, Haase O, Offermann A, Vogel W, Grätz V, et al. Immune cell infiltration of the primary tumor, not PD-L1 status, is associated with improved response to checkpoint inhibition in metastatic melanoma. Front Med (2019) 6:27. doi: 10.3389/fmed.2019.00027
27. Hellmann MD, Paz-Ares L, Bernabe Caro R, Zurawski B, Kim S-W, Carcereny Costa E, et al. Nivolumab plus ipilimumab in advanced non–small-cell lung cancer. New Engl J Med (2019) 381(21):2020–31. doi: 10.1056/NEJMoa1910231
Keywords: cancer of unknown primary, molecular targets, genomic testing, tumor of origin, diagnostic tools, personalized treatment, biomarkers
Citation: Al Assaad M, Shin N, Sigouros M, Manohar J, Antysheva Z, Kotlov N, Kiriy D, Nikitina A, Kleimenov M, Tsareva A, Makarova A, Fomchenkova V, Dubinina J, Boyko A, Almog N, Wilkes D, Escalon JG, Saxena A, Elemento O, Sternberg CN, Nanus DM and Mosquera JM (2024) Deciphering the origin and therapeutic targets of cancer of unknown primary: a case report that illustrates the power of integrative whole-exome and transcriptome sequencing analysis. Front. Oncol. 13:1274163. doi: 10.3389/fonc.2023.1274163
Received: 07 August 2023; Accepted: 18 December 2023;
Published: 22 January 2024.
Edited by:
Giulia Maria Stella, University of Pavia, ItalyReviewed by:
Xiaowei Zhang, Fudan University Shanghai Cancer Center, ChinaCarla Boccaccio, IRCCS Candiolo Cancer Institute, Italy
Copyright © 2024 Al Assaad, Shin, Sigouros, Manohar, Antysheva, Kotlov, Kiriy, Nikitina, Kleimenov, Tsareva, Makarova, Fomchenkova, Dubinina, Boyko, Almog, Wilkes, Escalon, Saxena, Elemento, Sternberg, Nanus and Mosquera. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Miguel Mosquera, am1tOTAxOEBtZWQuY29ybmVsbC5lZHU=
†These authors share senior authorship
 Majd Al Assaad
Majd Al Assaad Nara Shin3
Nara Shin3 Michael Sigouros
Michael Sigouros Nikita Kotlov
Nikita Kotlov Daria Kiriy
Daria Kiriy Mikhail Kleimenov
Mikhail Kleimenov Anastasiya Makarova
Anastasiya Makarova Alexandra Boyko
Alexandra Boyko Ashish Saxena
Ashish Saxena Olivier Elemento
Olivier Elemento Cora N. Sternberg
Cora N. Sternberg David M. Nanus
David M. Nanus Juan Miguel Mosquera
Juan Miguel Mosquera