- 1Department of General Surgery, the First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, China
- 2Department of Gynecologic Oncology, the Shaanxi Provincial Cancer Hospital, Xi’an, China
Background: The Naples prognostic score (NPS) determined by the nutritional and inflammatory condition of an individual is attracting growing attention for predicting postoperative outcomes in a variety of malignancies. The study aimed to assess the clinical significance of a modified NPS (M-NPS) and establish and validate nomograms incorporating M-NPS in curative stage II-III colon cancer patients.
Methods: We retrospectively analyzed 328 stage II-III colon cancer patients receiving radical surgical resection at our hospital from January 2011 to December 2016. Kaplan–Meier (KM) survival analysis and Cox regression analysis were executed for overall survival (OS) and cancer-specific survival (CSS). Independent predictive indicators were applied to develop nomograms. The model’s performance was evaluated using many different methods.
Results: Of a total of 328 cases, 153 cases were in group 0, 145 in group 1, and 30 in group 2. In terms of OS or CSS, there were obvious differences between groups 0 and 1, and between groups 0 and 2. Age, obstruction, N stage, gross tumor type, and M-NPS group were independent prognostic indicators for OS, while obstruction, gross tumor type, M-NPS group, and N stage were independent predictive parameters for CSS. Furthermore, the training and validation sets were randomly allocated among a cohort of 328 patients. OS and CSS prediction nomograms were developed. In the training and validation cohort, the C-index and ROC analysis showed good discrimination, calibration curves exhibited an excellent level of consistency between model-predicted survival and actual survival outcomes, and DCA curves demonstrated good clinical performance.
Conclusion: M-NPS is a reliable survival predictor in patients with curative stage II-III colon cancer. Nomograms incorporating M-NPS for OS and CSS have good predictive performance and clinical utility.
Introduction
Colorectal cancer (CRC) ranks third in prevalence and is the second leading cause of cancer deaths globally, according to recent statistics (1). Smoking, alcohol, obesity, diabetes, poor diets and low physical activity increase CRC risk (2, 3). Approximately 50% of colon cancer patients are stage II or III at the time of diagnosis (4). Radical surgery is the primary therapeutic approach for II-III colon cancer (4, 5). Although surgical resection significantly improves the survival of stage II-III patients, postoperative prognostic monitoring of these individuals continues to be a great challenge as a result of the disease’s highly heterogeneous nature. The current widely accepted prognostic assessment of colon cancer is based on postoperative pathological indicators (6). These histological factors can be obtained for assessment only after surgery. Therefore, the identification of a preoperatively available prognostic marker is extremely important for physicians’ clinical decision-making and prognostic risk stratification.
It is generally accepted that the prognosis of cancer individuals is influenced not only by the biology of the cancer but also by host-related conditions (7, 8). Inflammation, a leading hallmark of cancer, has a significant impact on tumorigenesis and progression (9). The cytokine release mediated by the NLRP3 inflammasome in various cell types contributes to the formation of an inflammatory tumor microenvironment through autocrine and paracrine signaling mechanisms (10). In colon cancer, activation of NLRP3 functions as a tumor suppressor by mediating the production of IL-18, which reinforces the killing activity of natural killer cells against metastatic tumor cells or down-regulates IL-22BP to suppress tumorigenesis (10, 11). The nutritional, inflammatory, and immune conditions of individuals are thought to be tightly associated with the outcome of CRC (12–18). Prior research has indicated that a variety of serum inflammation-related biomarkers can be beneficial for assessing the outcome in CRC (17), such as neutrophil-to-lymphocyte ratio (NLR) (19, 20), platelet-to-lymphocyte ratio (PLR) (21, 22) and lymphocyte-to-monocyte ratio (LMR) (23, 24). Additionally, several scoring systems, including the controlling nutritional status score (25, 26), the systemic inflammation score (27), the prognostic nutritional index (12, 28), and the Glasgow prognostic score (27, 29), had been confirmed as promising predictive markers for CRC outcome. NPS, a new scoring system to assess cancer outcomes, has attracted widespread attention. NPS based on inflammatory and nutritional status, first defined by Gennaro et al., is constructed from total cholesterol (CHOL), serum albumin (ALB), LMR, and NLR (30). Currently, the prognostic significance of NPS in CRC, obstructive CRC, metastatic CRC (mCRC) and rectal cancer has been reported by related studies (28, 30–32). The clinical significance of NPS in stage II-III patients, particularly in those receiving radical resection, has yet to be documented.
This study aims to construct a M-NPS for stage II-III patients, assess its prognostic value, and create a nomogram to forecast survival outcomes.
Patients and methods
Patient cohort
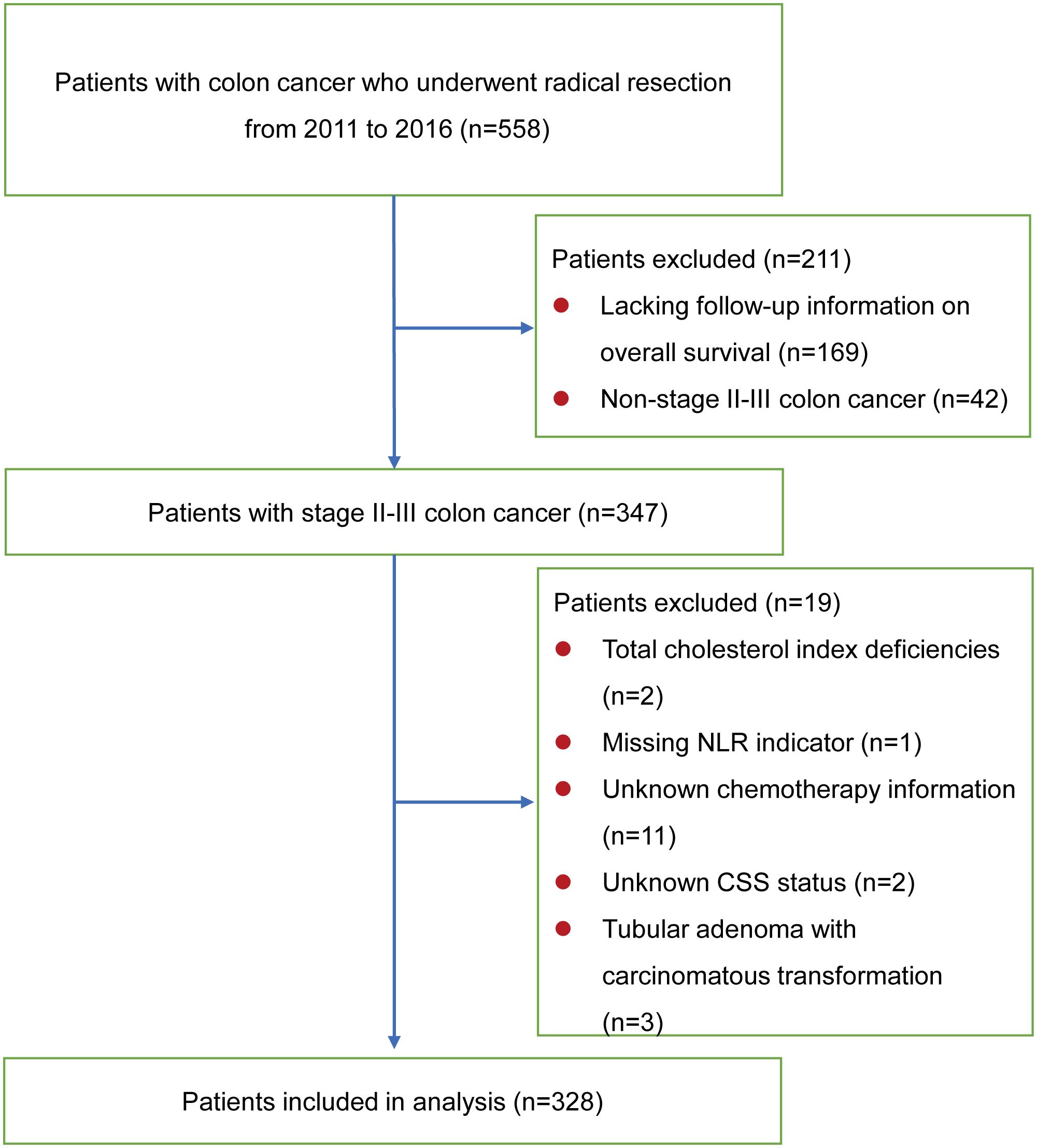
This study ultimately enrolled 328 stage II-III colon cancer patients, who underwent curative surgical resection at the Department of General Surgery, the First Affiliated Hospital of Xi’an Jiaotong University from January 2011 to December 2016 (Figure 1). All subjects were diagnosed with preoperative biopsy or postoperative pathological examination. All patients were diagnosed with colon cancer and not in combination with other cancers, and none of them underwent neoadjuvant chemotherapy before surgery. The following were the exclusion criteria: definite inflammatory or hematologic diseases affecting immune or nutritional status; unknown prognostic information due to lost to follow-up; missing blood test results within one week before surgery. Patients were followed up via outpatient services or telephone interviews to obtain their health condition. The follow-up concluded in December 2022. The outcome was specified as either OS or CSS. OS was described as the interval between the surgical procedure and mortality resulting from any cause. The period from surgery to mortality as a result of recurrence or metastasis of the primary malignancy was referred to as CSS. For CSS, a patient’s death was defined as a non-endpoint event if the cause of death was not due to cancer. Cases lacking an outcome event were censored in the analysis.
Data gathering
The clinicopathological indicators extracted from the patient’s electronic health data include age, sex, underlying disease, anemia, tumor location, American Society of Anesthesiologists (ASA) classification, surgical approach, ascites, intestinal perforation, obstruction, tumor size, gross tumor type, pathology type, differentiation, N stage, T stage, TNM stage, lymph node dissection, perineural invasion, vessel invasion and chemotherapy. Laboratory examinations within one week before surgery include ALB, CHOL, total counts of neutrophils, lymphocytes, and monocytes.
Construction of M-NPS
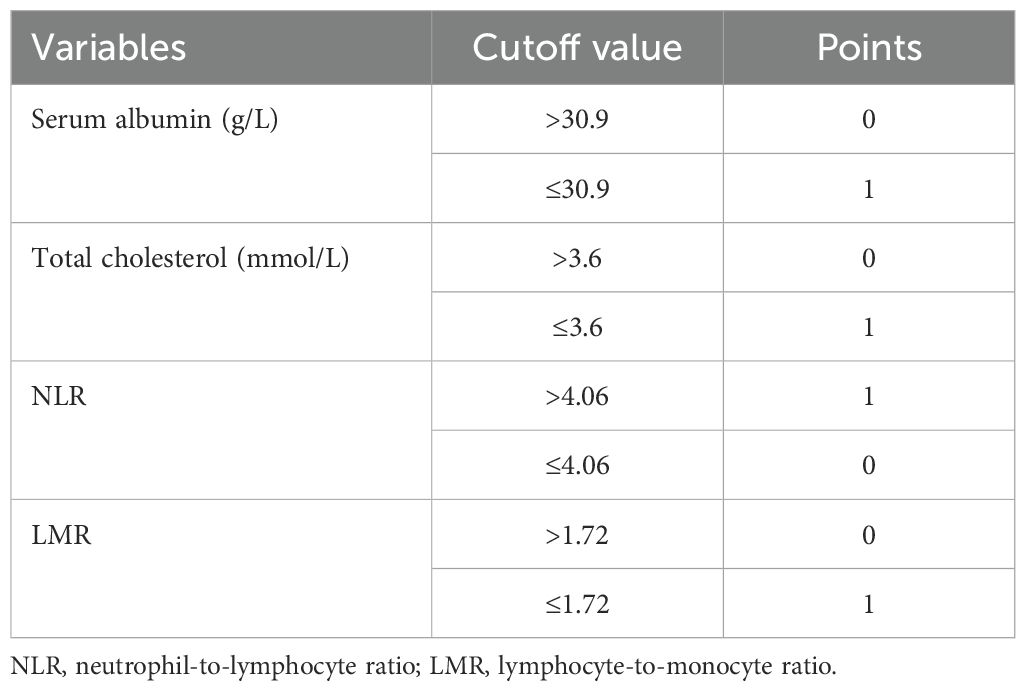
M-NPS is a scoring system calculated from four factors: ALB, CHOL, LMR and NLR. Given that traditional NPS failed to exhibit substantial clinical utility in the study population, X-Tile 3.6.1 software (Yale University, New Haven, USA) (33) was employed to identify the most suitable threshold value for the four indicators according to the maximum chi-square and the lowest P values of log-rank tests (Figure 2). Table 1 provides the comprehensive scoring criteria for the M-NPS. Patients were classified into three groups according to their total M-NPS scores: group 1 for scores of 0, group 2 for scores of 1-2, and group 3 for scores of 3-4.
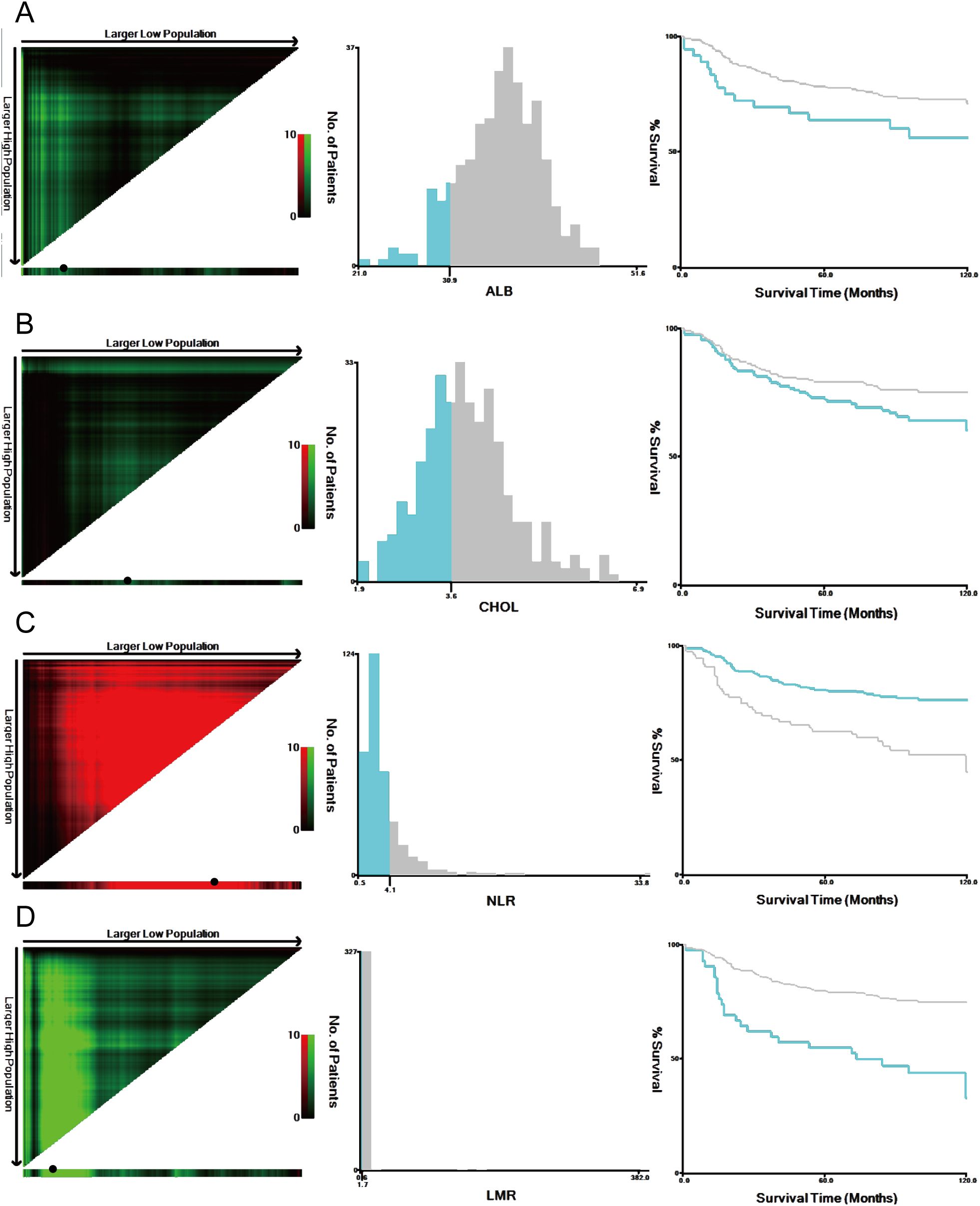
Figure 2. Optimal cutoff values were identified by the X-Tile analysis. The plots showed the χ 2 log‐rank values produced when dividing the cohort into two groups with optimal cut-points. Red coloration of cut points indicates an inverse correlation with survival, while green ones positively correlate. The optimal cutoff values highlighted by the black circles in the left panels are displayed in histograms of the entire cohort (middle panels), and Kaplan–Meier plots are shown in the right panels. For overall survival, the optimal thresholds for ALB (A), CHOL (B), NLR (C), and LMR (D) were identified as 30.9 g/L, 3.6 mmol/L, 4.06, and 1.72, respectively.
Procedures of treatment
All curative surgeries for patients enrolled in the study were conducted by a skilled surgical team. They underwent open resection or laparoscopic operation, subject to the location of the disease and the surgical team’s preference. Adjuvant chemotherapy was implemented within two months postoperatively according to pathologic TNM staging results and the patient’s condition. Some patients discontinue chemotherapy midway because they cannot tolerate the side effects.
Statistical analysis
The medians and interquartile ranges (IQRs) of continuous variables are shown, while categorical variables are described as percentages and numbers. Continuous variables were compared by the Kruskal-Waills test or the Mann-Whitney U test. For comparison of categorical variables between two or more groups, chi-square or Fisher’s exact tests were implemented. The survival and survminer R packages were employed to plot KM survival curves and compared their survival rates with a log-rank test. Independent prognostic analysis for OS or CSS was determined by multivariate Cox regression. A multivariate analysis was performed only on parameters with P<0.05 in a univariate analysis. The dataset was randomly divided in 6.5:3.5 into training and validation sets using the sample function. Independent prognostic variables were employed to develop nomograms in the training set using the rms R package. The model discrimination can be assessed with the C-index and the ROC curve. The calibration curve was generated with the rms R package to evaluate the prediction ability of the model. Additionally, the DCA was employed to evaluate the clinical efficacy of the model using the ggDCA R package. Version 4.3.2 of the R software was used to conduct all statistical analyses. Statistical tests with P< 0.05 are significantly difference.
Results
Patients’ baseline features and connection between M-NPS and clinicopathological parameters of patients
Figure 1 displays the inclusion of 328 patients in the ultimate statistical analysis. Of the 328 patients, the age at diagnosis had a median value of 62 years. The research comprised 165 male and 163 female participants. The study subjects consisted of 113 patients with underlying diseases and 215 patients without underlying diseases. There were 148 patients with left hemicolon cancers, while 180 patients had right hemicolon cancers. 199 patients received open resection and 129 underwent laparoscopic surgery (Table 2). The median OS and CSS were both 86 months. Based on M-NPS scores, the cohort was stratified into three groups: 153 patients in group 0, 145 in group 1, and 30 in group 2. Of all clinicopathologic features, male, no underlying disease, open surgery and larger tumor size were associated with higher M-NPS scores. Lower ASA classification, ascites, no intestinal perforation, no obstruction and chemotherapy were related to lower M-NPS scores (Table 2). In addition, ALB, CHOL and LMR were all negatively correlated with M-NPS scores, whereas NLR was positively related to M-NPS scores (Table 2).
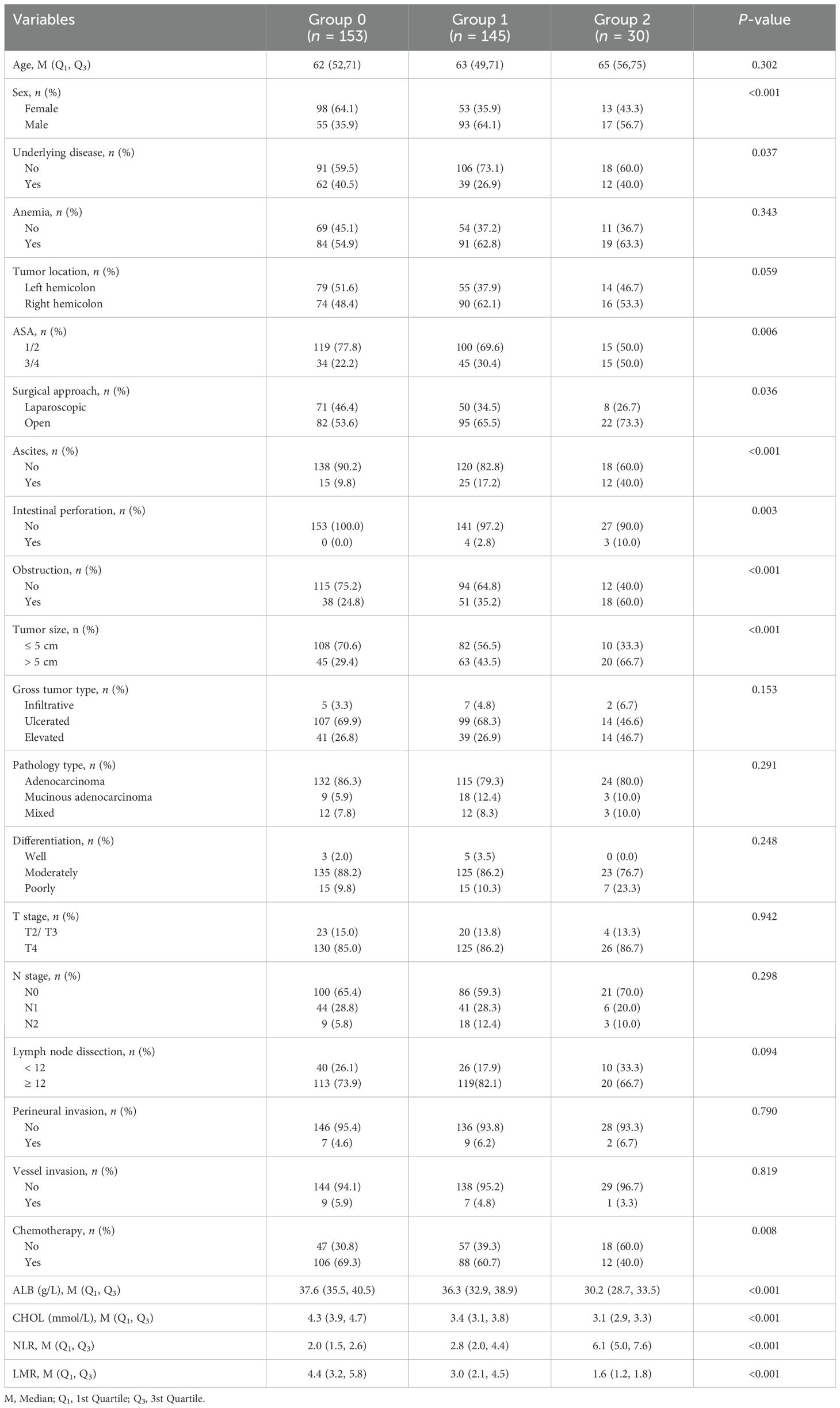
Table 2. Association of modified Naples prognostic score and clinicopathological characteristics in 328 stage II-III colon cancer patients.
Patient’s prognosis (OS and CSS) according to M-NPS
Survival analysis of OS and CSS based on M-NPS grouping was performed and survival curves were plotted in Figure 3. For OS in stage II-III patients, the median survival for group 2 was 95 months. Group 0 had more favorable survival than both group 1 and group 2 (Figure 3A). Further stratified survival analysis using staging showed that in comparison to group 0 and group 1, group 2 had the poorest prognosis for stage II patients (Figure 3B). In stage III patients, group 0 had superior survival than group 1 (Figure 3C), and a significant survival difference was not observed between group 1 and 2, nor between group 0 and 2. In terms of CSS for stage II-III patients, the median survival time in group 2 was 119 months. Group 0 had the best survival compared to group 1 and group 2 (Figure 3D). Similarly, among stage II patients, group 2 had poorer CSS compared to those in group 1 and 0 (Figure 3E). For stage III patients, group 0 had the most favorable CSS, and the remaining two groups did not appear to differ markedly in survival (Figure 3F).
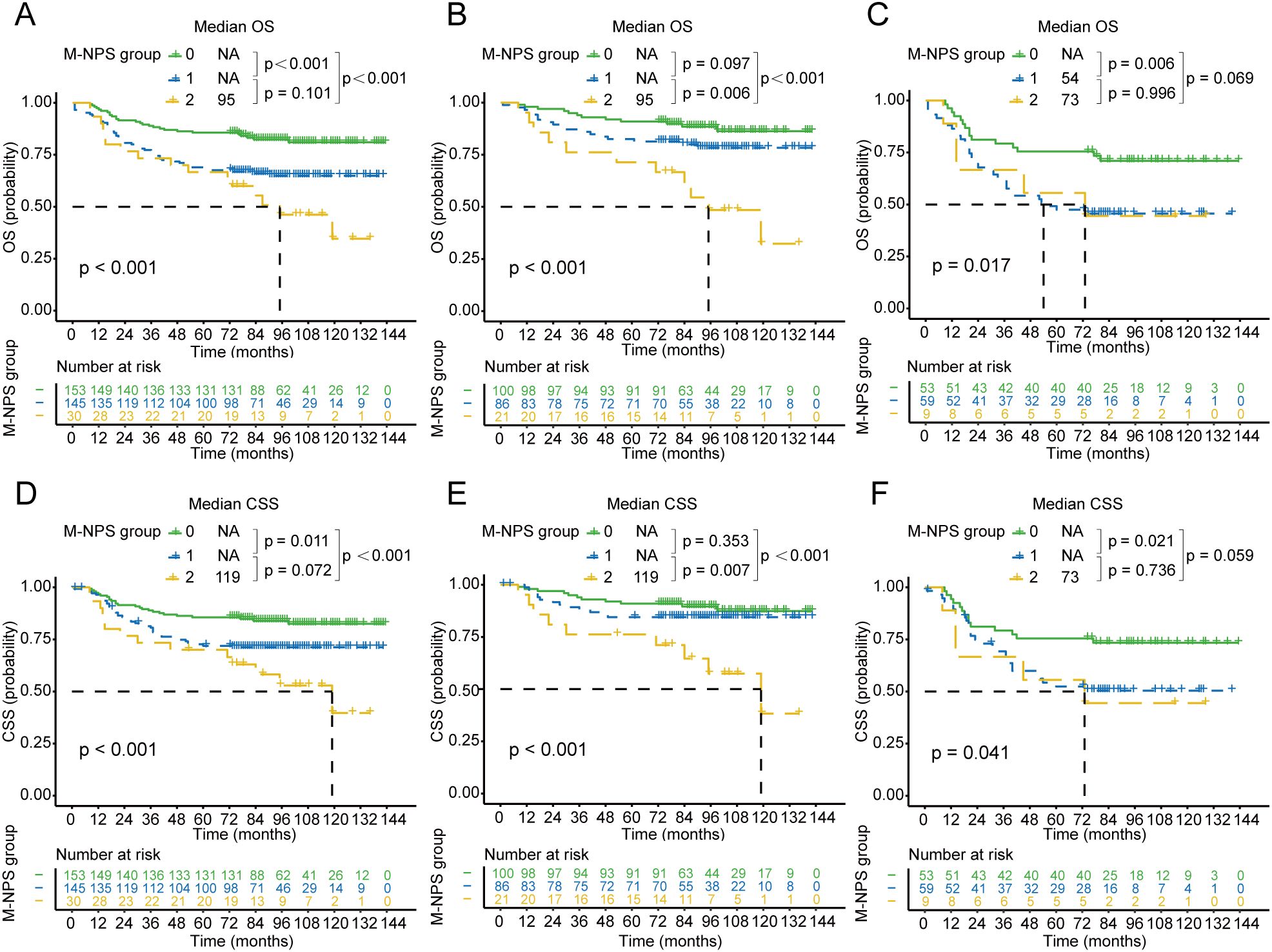
Figure 3. Kaplan–Meier survival curves of OS and CSS for each M-NPS group. Differences in OS among the three groups in patients with stage II-III (A), II (B), and III (C) colon cancer. Differences in CSS among the three groups in patients with stage II-III (D), II (E), and III (F) colon cancer. OS, overall survival; CSS, cancer-specific survival.
Univariate and multivariate analyses of prognostic factors for OS and CSS
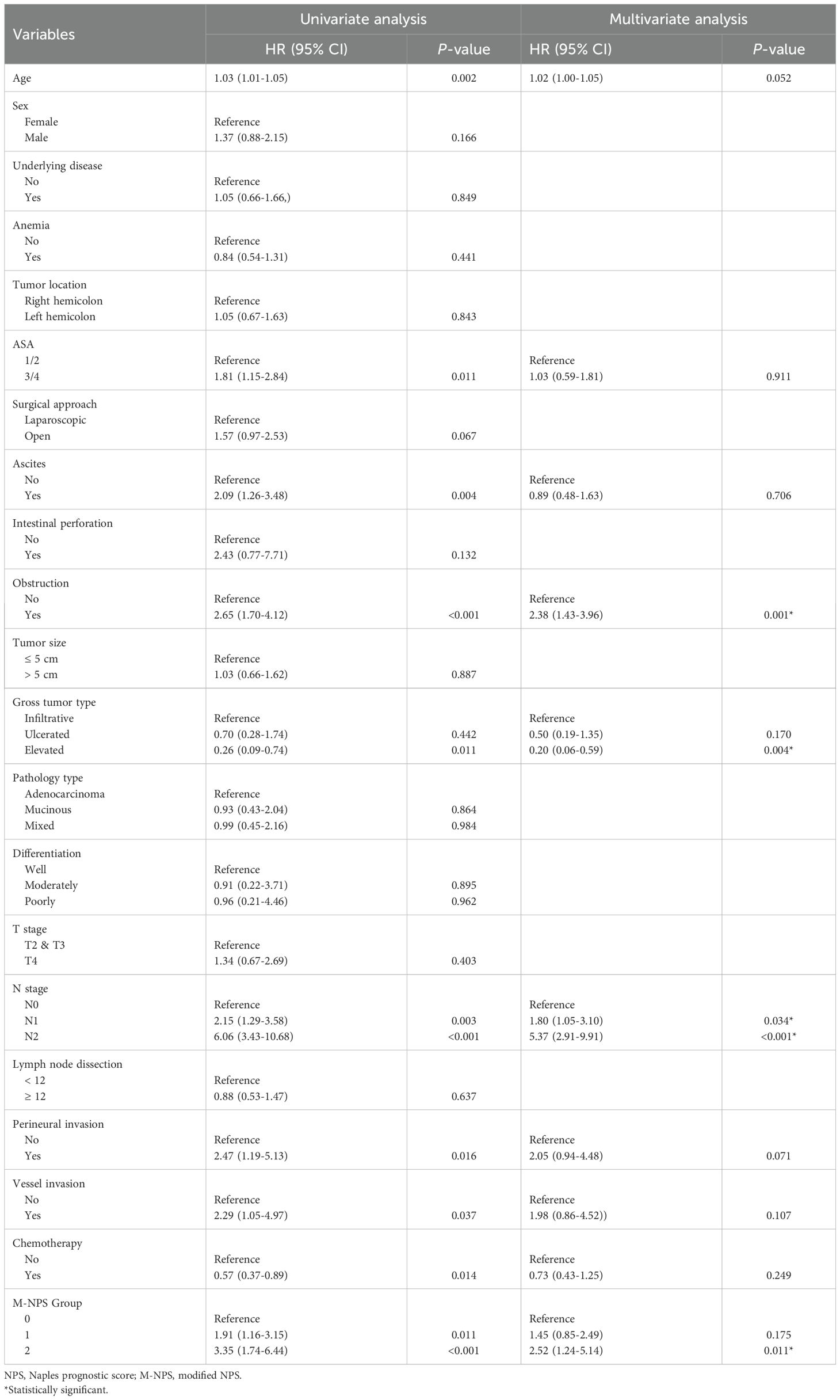
We conducted univariate and multivariate Cox regression analysis on OS for stage II-III patients. The results are displayed in Table 3. OS had a strong association with the M-NPS group, as indicated by the univariate analysis. Additional multivariate analysis revealed that the M-NPS group was an independent risk indicator for OS. Group 1 and group 2 had worse OS compared to group 0 [G1 vs G0: 1.73 (1.05–2.86); G2 vs G0: 2.55 (1.29–5.03)]. Other independent prognostic risk indicators included age, obstruction, gross tumor type, and N stage.
In addition, the same analytical methods as for OS were executed for CSS (Table 4). CSS was notably correlated with the M-NPS group in the univariate analysis. CSS was independently influenced by the M-NPS group in multivariate analysis. Group 2 had worse CSS compared to group 0 [G2 vs G0: 2.52 (1.24–5.14)]. Obstruction, gross tumor type, and N stage were also independent prognostic risk parameters of CSS.
Division of the dataset and construction of the nomogram
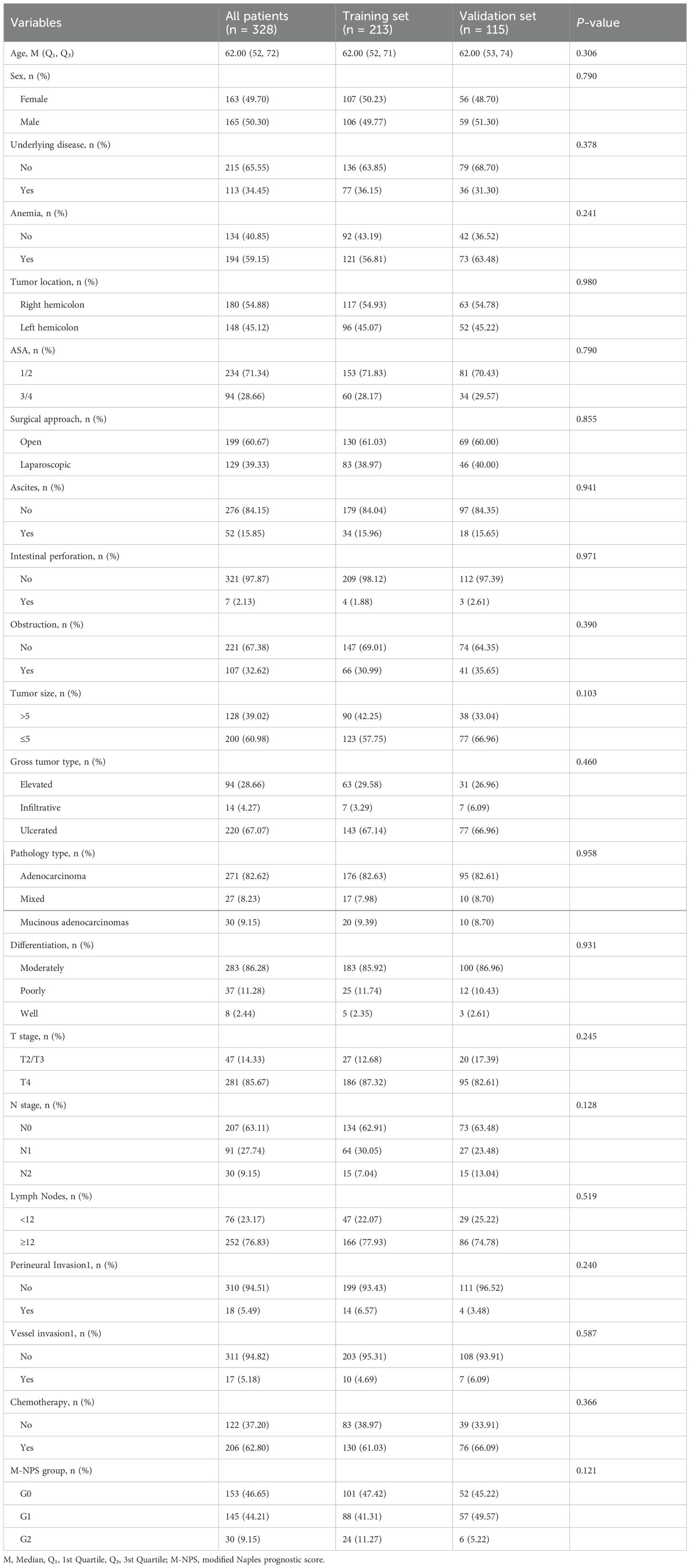
The total of 328 patients finally included were stochastically allocated into two cohorts: 213 in the training set and 115 in the validation set. All clinicopathological characteristics in two sets are listed in Table 5. No statistical difference in clinicopathological characteristics between the two cohorts proved that the allocation was completely randomized (Table 5).
OS and CSS nomograms were created to forecast the survival rates at 3, 5, and 10 years, using independent parameters that influence OS and CSS, respectively (Figure 4). Nomograms can personalize the forecast of OS and CSS for each patient by calculating the sum of the corresponding score for each variable in the models.
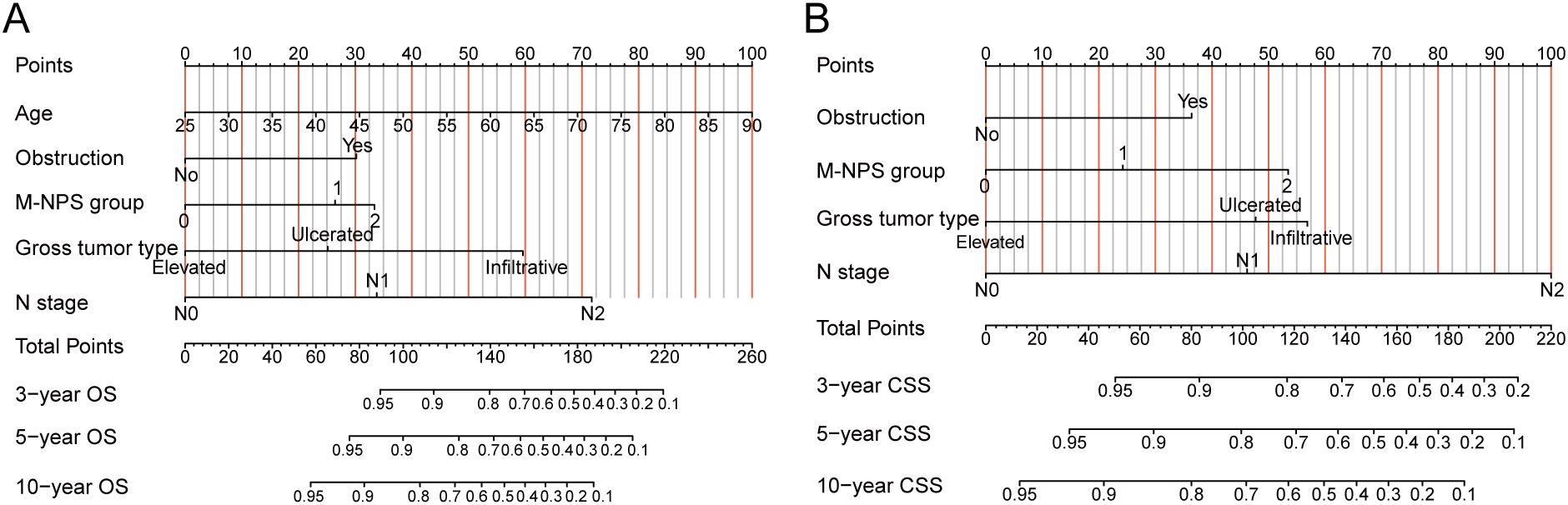
Figure 4. Nomograms for OS and CSS. (A) Nomogram for predicting 3-, 5- and 10-year OS. (B) Nomogram for predicting 3-, 5- and 10-year CSS. OS, overall survival; CSS, cancer-specific survival.
Evaluation of nomogram prediction models
The C-indexes for OS and CSS nomograms in the training cohort were 0.802 (0.747–0.857) and 0.783 (0.722–0.844), respectively. In the validation cohort, the C-indexes for OS and CSS nomograms were 0.742 (0.656–0.828) and 0.745 (0.649–0.841), respectively.
ROC analysis indicated that the nomogram for OS in the training cohort achieved AUC values of 0.82, 0.82, and 0.87 for the 3-, 5-, and 10-year predictions, respectively (Figure 5A). In the validation cohort, the predictive model for OS reached AUC values of 0.75, 0.77, and 0.82, respectively (Figure 5B). The nomogram’s calibration curves demonstrated that excellent alignment between predicted and actual 3-, 5-, and 10-year OS in both the training (Figure 5C) and validation sets (Figure 5D). In addition, DCA curves showed that the predictive model of OS had excellent clinical efficacy at 3, 5, and 10 years in clinical practice (Figures 5E, F).
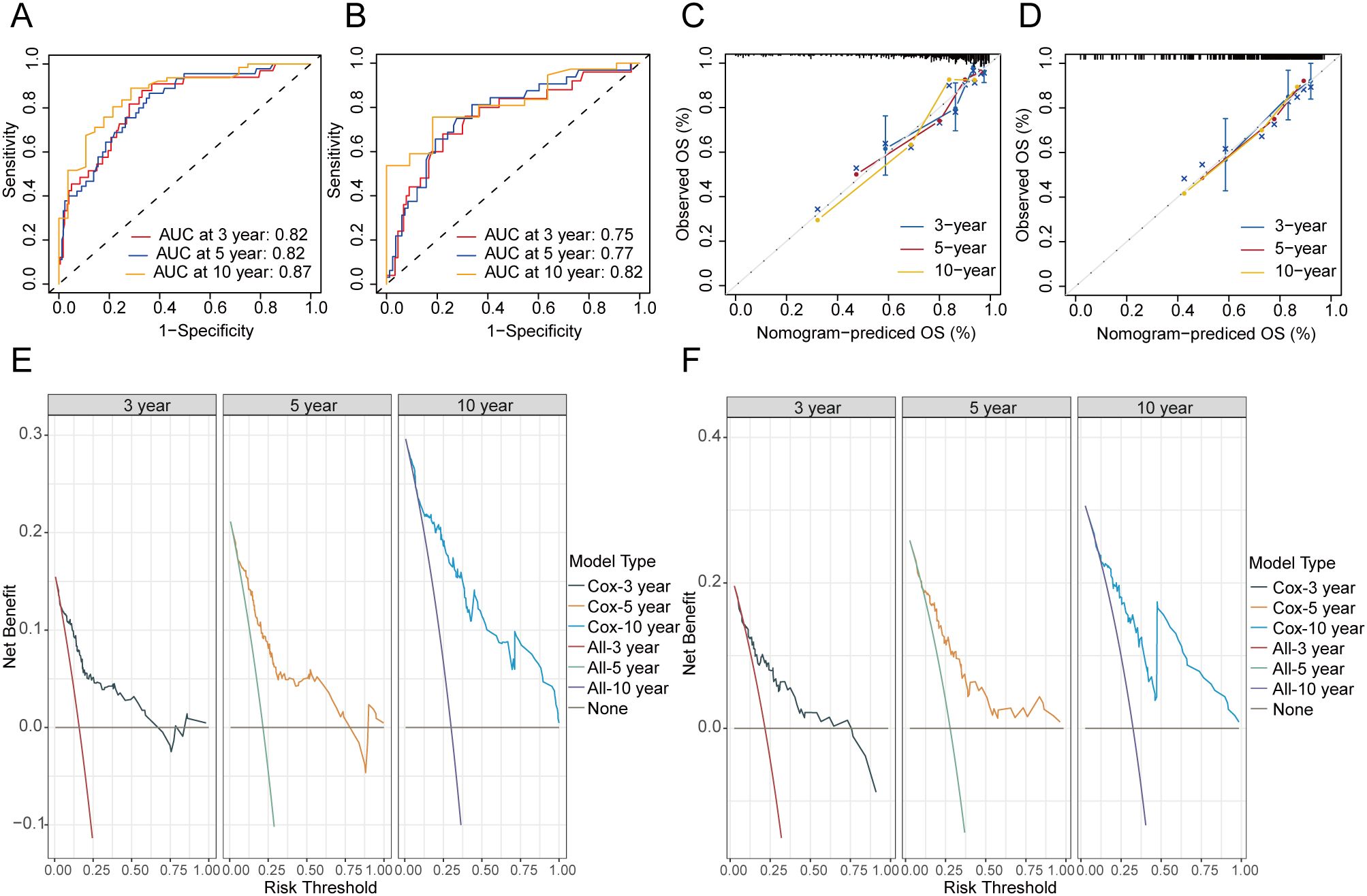
Figure 5. Evaluation of the nomogram of OS. ROC curves of nomograms for 3-, 5- and 10-year OS in the training (A) and validation cohort (B). Calibration curves of the nomogram for the probability of 3-, 5- and 10-year OS in the training (C) and validation cohort (D). DCA curves of the nomogram for 3-, 5- and 10-year OS in the training (E) and validation cohort (F). OS, overall survival.
At the same time, the CSS nomogram in the training set attained AUC values of 0.81, 0.80, and 0.80 for the 3-, 5-, and 10-year predictions, respectively (Figure 6A). In the validation set, the CSS nomogram got AUC values of 0.73, 0.75, and 0.80 for the 3-, 5-, and 10-year predictions, respectively (Figure 6B). In the training (Figure 6C) and validation cohort (Figure 6D), the nomogram’s calibration curves exhibited an excellent level of consistency between the model-forecasted CSS and the actual survival outcomes. Additionally, DCA curves for the nomogram of CSS indicated that the model had favorable clinical efficacy in both the training (Figure 6E) and validation cohort (Figure 6F).
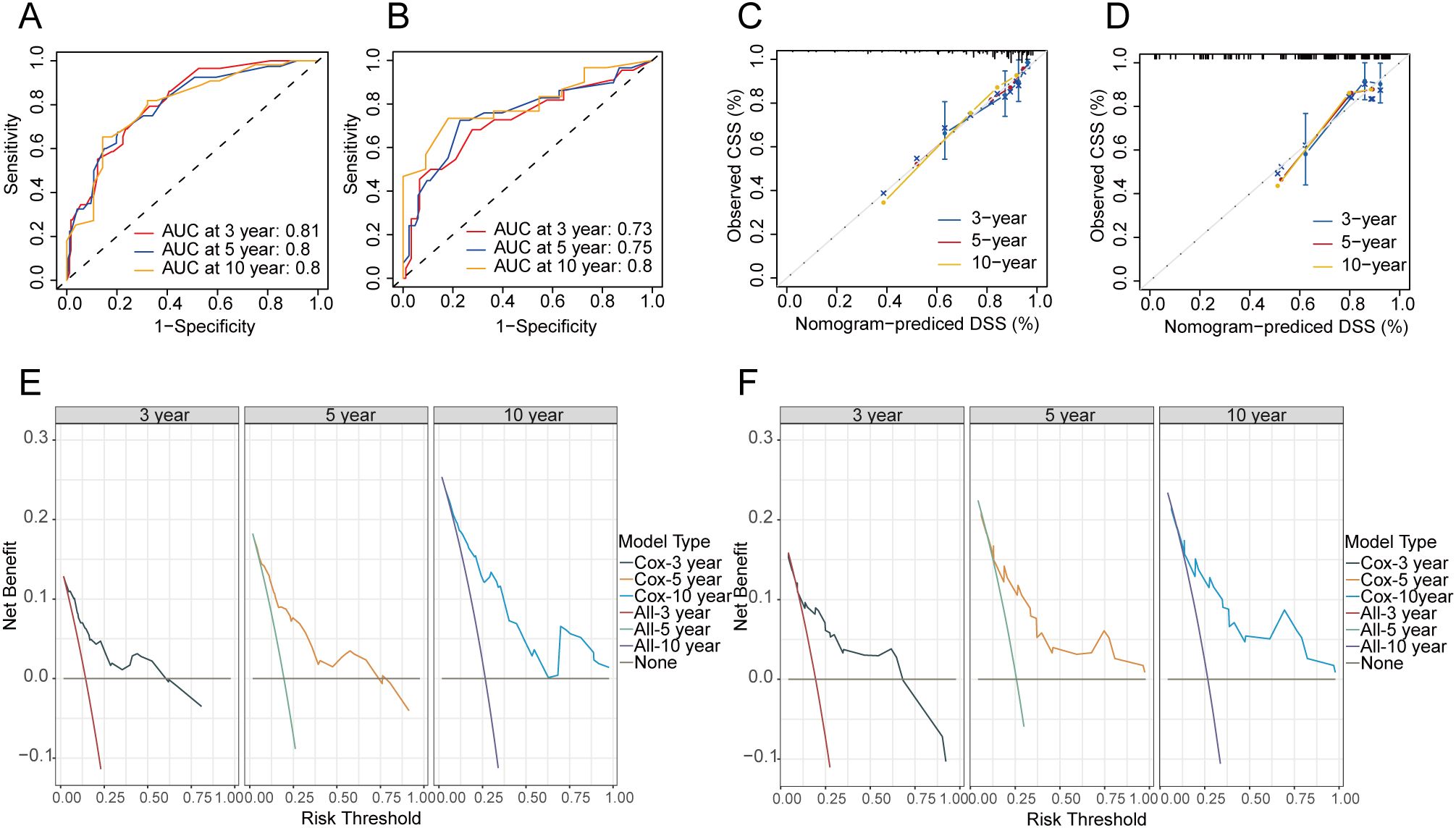
Figure 6. Evaluation of the nomogram of CSS. ROC curves of nomograms for 3-, 5- and 10-year CSS in the training (A) and validation cohort (B). Calibration curves of the nomogram for the probability of 3-, 5- and 10-year CSS in the training (C) and validation cohort (D). DCA curves for training (E) and validation cohort (F) at 3-, 5-, and 10-year CSS. CSS, cancer-specific survival.
Discussion
Recently, research has been conducted extensively on the application of NPS to predict the outcome of numerous types of cancer due to the integration of multiple indicators of nutritional, inflammatory, and immune status (28, 30, 34–36). NPS thresholds in different cancer types and treatment contexts remain controversial. In our research, the X-Tile was employed to construct M-NPS based on stage II-III patients in the research center. Furthermore, we examined the connection between M-NPS and patients’ clinicopathological parameters and assessed the predictive significance of M-NPS for OS and CSS. Finally, OS and CSS nomograms containing M-NPS are established and validated.
NLR is a highly reliable indicator of systemic inflammation. The threshold value for NLR in NPS was originally defined by Gennaro et al. (30) as 4.44, and the cutoff value identified in this study was very close to 4.04. LMR is used as a marker of the host’s immunological response. We determined a cutoff value of 1.72 for LMR, however, the cutoff value for LMR in NPS is 2.96. In addition, the cutoff values for ALB and CHOL levels, which represent nutritional status, also differed from those defined by Gennaro et al. (30). The above differences may originate from the race, lifestyle, and dietary structure of Eastern and Western populations.
Accumulating evidence suggests that patients with CRC in a hyperinflammatory state portend an unfavorable prognosis, whereas patients in a good nutritional state show longer survival (15). Our study showed that colon cancer patients with low NLR, high ALB and high CHOL had a more favorable survival compared to those with high NLR, low ALB and low CHOL. Therefore, as recommended by the guidelines (37), an appropriate anti-inflammatory and nutritious diet is crucial in preventing the occurrence of colon cancer and reducing postoperative colon cancer deaths.
Our study revealed that obstruction was a significant predictive indicator for OS and CSS in stage II-III patients. Relevant studies have pointed out that obstruction as a marker of malignancy portends an unfavorable prognosis in CRC patients even after radical surgery (38–40). Obstruction causes dysbiosis and increased toxins, resulting in local and systemic inflammatory responses to remodel the tumor microenvironment (41, 42). Inflammation associated with cancer not only facilitates the growth of malignant tumors but can also reduce response to chemotherapeutic agents by inducing genetic instability (43). We have also found in the study that the gross tumor type was also an independent prognostic factor for the prognosis of patients with stage II-III colon cancer. As we all know, the gross tumor type reflects the biology of the cancer itself. Infiltrative colon cancers are the most malignant and elevated colon cancers are the least malignant. Previous studies have also revealed that gross tumor type could serve as a marker of survival and peritoneal metastasis in CRC (44, 45), which is consistent with our results.
NPS, as a novel and powerful scoring system, has been widely applied in the construction of prognostic nomograms for a variety of cancers, including gastric cancer (46), small cell lung cancer (47), esophageal squamous cell carcinoma (48, 49), cholangiocarcinoma (50), gallbladder cancer (51) and osteosarcoma (52). Currently, there are no reports related to the application of NPS to the prognostic nomogram of colon cancer. For the first time, we incorporated M-NPS into the predictive nomogram for colon cancer patients in stages II–III. Nomograms integrating M-NPS exhibited excellent predictive ability for 3, 5, and 10-year OS or CSS.
This study relies on a national clinical key specialty platform with a superb surgical team, sufficient surgical patients and advanced instrumentation. In addition, the professional follow-up process provides a strong warrant for the reliability of the study’s conclusions. However, there exist some constraints in our research. Initially, the retrospective study is susceptible to selection bias. Besides, the sample size of a single center is relatively limited and the number of outcome events is relatively few. Hence, the clinical significance of M-NPS has to be validated through prospective multicenter trials involving a substantial number of participants.
Conclusions
Taken together, M-NPS is a robust survival predictor in curative stage II-III patients. Nomograms incorporating M-NPS for OS and CSS have good predictive performance and clinical utility.
Data availability statement
The data used in the study are not publicly available due to privacy restrictions but are available from the corresponding author on reasonable request.
Ethics statement
The studies involving humans were approved by the Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. This study is a clinical study and did not involve the ethics of animal and cellular experiments.
Author contributions
XL: Writing – original draft, Formal analysis. CC: Data curation, Writing – review & editing. XH: Project administration, Writing – review & editing. CZ: Data curation, Formal analysis, Writing – review & editing. HY: Formal analysis, Visualization, Writing – review & editing. GC: Formal analysis, Visualization, Writing – review & editing. JY: Formal analysis, Visualization, Writing – review & editing. MM: Project administration, Supervision, Writing – review & editing. XS: Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the National Natural Science Foundation of China (Grant No. 81972720). This study was funded by the IIT Project of the First Affiliated Hospital of Xi'an Jiaotong University (Grant No: XJTU1AF-CRF-2024-012).
Acknowledgments
We would like to thank CC for his great contribution to the prognostic follow-up of colon cancer cases.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J For Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. (2019) 16:713–32. doi: 10.1038/s41575-019-0189-8
3. Murphy N, Ward HA, Jenab M, Rothwell JA, Boutron-Ruault M-C, Carbonnel F, et al. Heterogeneity of colorectal cancer risk factors by anatomical subsite in 10 european countries: A multinational cohort study. Clin Gastroenterol Hepatology: Off Clin Pract J Am Gastroenterological Assoc. (2019) 17:1323 1331.e6. doi: 10.1016/j.cgh.2018.07.030
4. Miller KD, Nogueira L, Devasia T, Mariotto AB, Yabroff KR, Jemal A, et al. Cancer treatment and survivorship statistics, 2022. CA: Cancer J For Clin. (2022) 72:409–36. doi: 10.3322/caac.21731
5. Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, et al. Colorectal cancer. Nat Rev Dis Primers. (2015) 1:15065. doi: 10.1038/nrdp.2015.65
6. Weiser MR. AJCC 8th edition: colorectal cancer. Ann Surg Oncol. (2018) 25:1454–5. doi: 10.1245/s10434-018-6462-1
7. Ikeda M, Natsugoe S, Ueno S, Baba M, Aikou T. Significant host- and tumor-related factors for predicting prognosis in patients with esophageal carcinoma. Ann Surg. (2003) 238:197–202. doi: 10.1097/01.sla.0000080822.22415.cb
8. Mohri Y, Tanaka K, Ohi M, Yokoe T, Miki C, Kusunoki M. Prognostic significance of host- and tumor-related factors in patients with gastric cancer. World J Surg. (2010) 34:285–90. doi: 10.1007/s00268-009-0302-1
9. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discovery. (2022) 12:31–46. doi: 10.1158/2159-8290.CD-21-1059
10. Sharma BR, Kanneganti T-D. NLRP3 inflammasome in cancer and metabolic diseases. Nat Immunol. (2021) 22:550–9. doi: 10.1038/s41590-021-00886-5
11. Huber S, Gagliani N, Zenewicz LA, Huber FJ, Bosurgi L, Hu B, et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. (2012) 491:259–63. doi: 10.1038/nature11535
12. Tokunaga R, Sakamoto Y, Nakagawa S, Miyamoto Y, Yoshida N, Oki E, et al. Prognostic nutritional index predicts severe complications, recurrence, and poor prognosis in patients with colorectal cancer undergoing primary tumor resection. Dis Colon Rectum. (2015) 58:1048–57. doi: 10.1097/DCR.0000000000000458
13. Tamai M, Kiuchi J, Kuriu Y, Arita T, Shimizu H, Ohashi T, et al. Clinical impact of postoperative prognostic nutritional index in colorectal cancer patients undergoing adjuvant chemotherapy. Am J Cancer Res. (2021) 11:4947–55.
14. Chen J-H, Zhai E-T, Yuan Y-J, Wu K-M, Xu J-B, Peng J-J, et al. Systemic immune-inflammation index for predicting prognosis of colorectal cancer. World J Gastroenterol. (2017) 23:6261–72. doi: 10.3748/wjg.v23.i34.6261
15. Fu J, Zhu J, Du F, Zhang L, Li D, Huang H, et al. Prognostic inflammatory index based on preoperative peripheral blood for predicting the prognosis of colorectal cancer patients. Cancers. (2020) 13:3–23. doi: 10.3390/cancers13010003
16. Ruan G-T, Xie H-L, Yuan K-T, Lin S-Q, Zhang H-Y, Liu C-A, et al. Prognostic value of systemic inflammation and for patients with colorectal cancer cachexia. J Cachexia Sarcopenia Muscle. (2023) 14:2813–23. doi: 10.1002/jcsm.13358
17. Yamamoto T, Kawada K, Obama K. Inflammation-related biomarkers for the prediction of prognosis in colorectal cancer patients. Int J Mol Sci. (2021) 22:8002–15. doi: 10.3390/ijms22158002
18. Malka D, Lièvre A, André T, Taïeb J, Ducreux M, Bibeau F. Immune scores in colorectal cancer: Where are we? Eur J Cancer (Oxford England: 1990). (2020) 140:105–18. doi: 10.1016/j.ejca.2020.08.024
19. Li Y, Jia H, Yu W, Xu Y, Li X, Li Q, et al. Nomograms for predicting prognostic value of inflammatory biomarkers in colorectal cancer patients after radical resection. Int J Cancer. (2016) 139:220–31. doi: 10.1002/ijc.30071
20. Proctor MJ, Morrison DS, Talwar D, Balmer SM, Fletcher CD, O'Reilly DSJ, et al. A comparison of inflammation-based prognostic scores in patients with cancer. A Glasgow Inflammation Outcome Study. Eur J Cancer (Oxford England: 1990). (2011) 47:2633–41. doi: 10.1016/j.ejca.2011.03.028
21. Kim JH, Lee JY, Kim HK, Lee JW, Jung SG, Jung K, et al. Prognostic significance of the neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio in patients with stage III and IV colorectal cancer. World J Gastroenterol. (2017) 23:505–15. doi: 10.3748/wjg.v23.i3.505
22. Dudani S, Marginean H, Tang PA, Monzon JG, Raissouni S, Asmis TR, et al. Neutrophil-to-lymphocyte and platelet-to-lymphocyte ratios as predictive and prognostic markers in patients with locally advanced rectal cancer treated with neoadjuvant chemoradiation. BMC Cancer. (2019) 19:664. doi: 10.1186/s12885-019-5892-x
23. Chan JCY, Diakos CI, Chan DLH, Engel A, Pavlakis N, Gill A, et al. A longitudinal investigation of inflammatory markers in colorectal cancer patients perioperatively demonstrates benefit in serial remeasurement. Ann Surg. (2018) 267:1119–25. doi: 10.1097/SLA.0000000000002251
24. Chan JCY, Chan DL, Diakos CI, Engel A, Pavlakis N, Gill A, et al. The lymphocyte-to-monocyte ratio is a superior predictor of overall survival in comparison to established biomarkers of resectable colorectal cancer. Ann Surg. (2017) 265:539–46. doi: 10.1097/SLA.0000000000001743
25. Iseki Y, Shibutani M, Maeda K, Nagahara H, Ohtani H, Sugano K, et al. Impact of the preoperative controlling nutritional status (CONUT) score on the survival after curative surgery for colorectal cancer. PloS One. (2015) 10:e0132488. doi: 10.1371/journal.pone.0132488
26. Tokunaga R, Sakamoto Y, Nakagawa S, Ohuchi M, Izumi D, Kosumi K, et al. CONUT: a novel independent predictive score for colorectal cancer patients undergoing potentially curative resection. Int J Colorectal Dis. (2017) 32:99 106. doi: 10.1007/s00384-016-2668-5
27. Suzuki Y, Okabayashi K, Hasegawa H, Tsuruta M, Shigeta K, Kondo T, et al. Comparison of preoperative inflammation-based prognostic scores in patients with colorectal cancer. Ann Surg. (2018) 267:527–31. doi: 10.1097/SLA.0000000000002115
28. Miyamoto Y, Hiyoshi Y, Daitoku N, Okadome K, Sakamoto Y, Yamashita K, et al. Naples prognostic score is a useful prognostic marker in patients with metastatic colorectal cancer. Dis Colon Rectum. (2019) 62:1485–93. doi: 10.1097/DCR.0000000000001484
29. Lu X, Guo W, Xu W, Zhang X, Shi Z, Zheng L, et al. Prognostic value of the Glasgow prognostic score in colorectal cancer: a meta-analysis of 9,839 patients. Cancer Manage Res. (2019) 11:229–49. doi: 10.2147/CMAR.S185350
30. Galizia G, Lieto E, Auricchio A, Cardella F, Mabilia A, Podzemny V, et al. Naples prognostic score, based on nutritional and inflammatory status, is an independent predictor of long-term outcome in patients undergoing surgery for colorectal cancer. Dis Colon Rectum. (2017) 60:1273–84. doi: 10.1097/DCR.0000000000000961
31. Gu J, Deng S, Jiang Z, Mao F, Xue Y, Qin L, et al. Modified Naples prognostic score for evaluating the prognosis of patients with obstructive colorectal cancer. BMC Cancer. (2023) 23:941. doi: 10.1186/s12885-023-11435-8
32. Sugimoto A, Fukuoka T, Nagahara H, Shibutani M, Iseki Y, Kasashima H, et al. Predictive value of the Naples prognostic score on postoperative outcomes in patients with rectal cancer. Langenbeck's Arch Surg. (2023) 408:113. doi: 10.1007/s00423-023-02851-2
33. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Research: an Off J Am Assoc For Cancer Res. (2004) 10:7252–9. doi: 10.1158/1078-0432.CCR-04-0713
34. Guo D, Liu J, Li Y, Li C, Liu Q, Ji S, et al. Evaluation of predictive values of naples prognostic score in patients with unresectable stage III non-small cell lung cancer. J Inflammation Res. (2021) 14:6129–41. doi: 10.2147/JIR.S341399
35. Lieto E, Auricchio A, Tirino G, Pompella L, Panarese I, Del Sorbo G, et al. Naples prognostic score predicts tumor regression grade in resectable gastric cancer treated with preoperative chemotherapy. Cancers. (2021) 13:4676–90. doi: 10.3390/cancers13184676
36. Nakagawa N, Yamada S, Sonohara F, Takami H, Hayashi M, Kanda M, et al. Clinical implications of naples prognostic score in patients with resected pancreatic cancer. Ann Surg Oncol. (2020) 27:887–95. doi: 10.1245/s10434-019-08047-7
37. Quagliariello V, D'Aiuto G, Iaffaioli RV, Berretta M, Buccolo S, Iovine M, et al. Reasons why COVID-19 survivors should follow dietary World Cancer Research Fund/American Institute for Cancer Research (WCRF/AICR) recommendations: from hyper-inflammation to cardiac dysfunctions. Eur Rev For Med Pharmacol Sci. (2021) 25:3898–907. doi: 10.26355/eurrev_202105_25957
38. Dahdaleh FS, Sherman SK, Poli EC, Vigneswaran J, Polite BN, Sharma MR, et al. Obstruction predicts worse long-term outcomes in stage III colon cancer: A secondary analysis of the N0147 trial. Surgery. (2018) 164:1223–9. doi: 10.1016/j.surg.2018.06.044
39. Okuda Y, Shimura T, Yamada T, Hirata Y, Yamaguchi R, Sakamoto E, et al. Colorectal obstruction is a potential prognostic factor for stage II colorectal cancer. Int J Clin Oncol. (2018) 23:1101–11. doi: 10.1007/s10147-018-1307-2
40. Yang Z, Wang L, Kang L, Xiang J, Peng J, Cui J, et al. Clinicopathologic characteristics and outcomes of patients with obstructive colorectal cancer. J Gastrointestinal Surgery: Off J Soc For Surg Alimentary Tract. (2011) 15:1213–22. doi: 10.1007/s11605-011-1563-1
41. Brennan CA, Garrett WS. Gut microbiota, inflammation, and colorectal cancer. Annu Rev Microbiol. (2016) 70:395–411. doi: 10.1146/annurev-micro-102215-095513
42. Diakos CI, Charles KA, McMillan DC, Clarke SJ. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. (2014) 15:e493–503. doi: 10.1016/S1470-2045(14)70263-3
43. Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. (2009) 30:1073–81. doi: 10.1093/carcin/bgp127
44. Vasile L, Olaru A, Munteanu M, Pleşea IE, Surlin V, Tudoraşcu C. Prognosis of colorectal cancer: clinical, pathological and therapeutic correlation. Romanian J Morphology Embryology = Rev Roumaine Morphologie Et Embryologie. (2012) 53:383–91.
45. Song X-Q, Liu Z-X, Kong Q-Y, He Z-H, Zhang S. Nomogram for prediction of peritoneal metastasis risk in colorectal cancer. Front In Oncol. (2022) 12:928894. doi: 10.3389/fonc.2022.928894
46. Wang H, Fang T, Yin X, Lou S, Han B, Gao J, et al. Prognostic importance of the preoperative New-Naples prognostic score for patients with gastric cancer. Cancer Med. (2023) 12:1358–75. doi: 10.1002/cam4.5017
47. Chen S, Liu S, Xu S, Cao S, Han Z, Kong L, et al. Naples prognostic score is an independent prognostic factor in patients with small cell lung cancer and nomogram predictive model established. J Inflammation Res. (2022) 15:3719–31. doi: 10.2147/JIR.S371545
48. Feng J-F, Zhao J-M, Chen S, Chen Q-X. Naples prognostic score: A novel prognostic score in predicting cancer-specific survival in patients with resected esophageal squamous cell carcinoma. Front In Oncol. (2021) 11:652537. doi: 10.3389/fonc.2021.652537
49. Xu S-J, Wang P-L, Chen C, You C-X, Chen R-Q, Wu W-W, et al. Inflammatory and nutritional status influences outcomes of minimally invasive esophagectomy. World J Surg. (2023) 47:1003–17. doi: 10.1007/s00268-023-06890-0
50. Xu B, Zhu J, Wang R, Pang X, Wang X, Lian J, et al. Clinical implications of naples prognostic score for patients with resected cholangiocarcinoma: A real-world experience. J Inflammation Res. (2024) 17:655–67. doi: 10.2147/JIR.S446735
51. Yang J, Lv L, Zhao F, Mei X, Zhou H, Yu F. The value of the preoperative Naples prognostic score in predicting prognosis in gallbladder cancer surgery patients. World J Surg Oncol. (2023) 21:303. doi: 10.1186/s12957-023-03198-0
Keywords: colon cancer, modified Naples prognostic score, prognostic factors, curative resection, nomogram
Citation: Li X, Cheng C, Huo X, Zhao C, Yuan H, Chen G, Yu J, Mu M and Sun X (2024) Clinical significance of the modified Naples prognostic score in patients with stage II-III colon cancer undergoing curative resection: a retrospective study from the real world. Front. Oncol. 14:1403666. doi: 10.3389/fonc.2024.1403666
Received: 03 April 2024; Accepted: 27 August 2024;
Published: 16 September 2024.
Edited by:
Sharon R. Pine, University of Colorado Anschutz Medical Campus, United StatesReviewed by:
Vincenzo Quagliariello, Istituto Nazionale Tumori-IRCCS-Fondazione G. Pascale, ItalyJohannes Doerner, Helios Klinikum Wuppertal, Germany
Copyright © 2024 Li, Cheng, Huo, Zhao, Yuan, Chen, Yu, Mu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingchao Mu, bXVtaW5nY2hhb0BtYWlsLnhqdHUuZWR1LmNu; Xuejun Sun, c3VueHlAbWFpbC54anR1LmVkdS5jbg==
 Xiaopeng Li
Xiaopeng Li Chen Cheng2
Chen Cheng2 Junhui Yu
Junhui Yu Mingchao Mu
Mingchao Mu Xuejun Sun
Xuejun Sun