- 1Department of Stomatology Center, The First People’s Hospital of Foshan, Foshan, China
- 2Department of Stomatology, Foshan Hospital of Traditional Chinese Medicine, Foshan, China
Background: Oral leukoplakia (OL) is the most common potentially malignant disease of the oral cavity. In recent years, studies have identified a correlation between the gut microbiota (GM) and oral cancer, in addition, inflammation-related proteins have been reported to play an important role in the development of OL. However, the causal relationship between gut microbiota and OL, as well as whether cytokines play a mediating role, remain unclear.
Methods: In this Mendelian randomization (MR) study, the genome-wide association studies (GWAS) (n=18340) of the MiBioGen consortium microbiome was used as exposure data. Genetic variation data related to OL were extracted from the Finngen R9 project (513 cases of OL and 411668 controls). The 91 inflammation-related proteins obtained in the literature serve as potential mediators. Two-sample Mendelian randomization analysis was applied to infer causality using Inverse Variance Weighted (IVW), MR Egger, weighted media, simple mode and weighted mode method. Subsequently, sensitivity analysis was conducted to ensure the robustness of the MR results. In addition, we conducted reverse MR analysis to alleviate reverse causality. Finally, we used mediation analysis to determine the pathway mediated by inflammation-related proteins from the gut microbiota to OL.
Results: The five bacterial taxa in the gut microbiota indicate a potential causal relationship with the development of OL. Notably, family Clostridiaceae1 was negatively correlated with the risk of OL development, while genus Dorea, genus Ruminococcus1, genus Senegalimasilia and genus Veillonella were positively associated with the risk of OL development. In addition, this study identified a potential causal relationship between interleukin-10 receptor subunit alpha (IL-10RA), interleukin-18 receptor 1(IL18-R1) and the occurrence of OL. In addition, intermediary analysis indicates that IL18-R1 mediated the pathway between the gut microbiota genus Senegalimasilia and OL.
Conclusions: In summary, our research emphasize the complex relationship between gut microbiota, inflammation-related proteins and OL. The identified associations and mediating effects provide new insights into potential therapeutic approaches for targeting the gut microbiota in the management of OL, and contribute to its prevention, diagnosis and treatment.
1 Introduction
Oral potentially malignant disease (OPMD) is characterized by tissue changes that are more likely to transform into oral cancer than normal tissue (1). Oral squamous cell carcinoma (OSCC) is the most common type of oral cancer, accounting for more than 90% of oral malignancies (2). According to the World Health Organization, oral leukoplakia (OL), defined as “a suspected risky white plaque that excludes other known diseases but does not increase the risk of cancer,” is the most common OPMD (1, 3, 4),with an estimated global prevalence of between 1.5 and 4.1% (5). Current research indicates that the pooled proportion of malignant transformation (MT) from OL to OSCC is 9.8% (6). Until now, there is no effective treatment to prevent MT in OL (7). Regular follow-up may help in the early detection of potentially malignant transformation, but its cost-effectiveness and best practices still need to be further investigated.
The gut microbiota is a vast microbial community that lives in the human gut and is considered the largest microbial reservoir in the human body, including bacteria, archaea, viruses, fungi, protozoa, and parasites (8, 9). It is especially noteworthy that the community contains over 10 trillion different microorganisms, accounting for approximately 90% of the total human microbial population (10). The gut microbiota plays an important role in many physiological and health-related processes in humans, including immune system development, vitamin production and nutrient absorption, through interactions with the host (11). The oral microbiota is the second largest microbiota in the human body after the intestinal microbiota. The oral microbiota is composed of over 700 types of bacteria, as well as fungi, viruses, and protozoa (12). Current research has found that OL is associated with a decrease in Firmicutes levels in the oral microbiota, including an increase in the abundance of anaerobic bacteria such as Fusobacterium nucleatum, Prevotella intermedia and Porphyromonas gingivalis, which are considered periodontal pathogens (13). The consumption of Firmicutes is often reported as precancerous and malignant lesions (14, 15). Considering that the oral mucosa and gastrointestinal mucosa are physically connected, and existing research has demonstrated the interrelationship between the oral and intestinal microbiota (16), it is reasonable to hypothesize that gut flora may influence the development of OL in certain form.
Inflammation is a defensive response of the body to external stimuli and an important component of the immune system, playing an indispensable role in resisting damage and infection. However, when inflammation loses control or persists for a long time and becomes chronic, it can lead to cell damage, tissue destruction, cancer, and even death (17, 18). Previous studies have supported the role of inflammatory mediators in the pathogenesis of OL to OSCC (19). The results of the study showed that salivary concentrations of interleukin-6 (IL-6) were significantly higher in patients with clinically proven OL compared to healthy individuals. In addition, elevated levels of tumor necrosis factor alpha (TNF-α) and IL-8 were found in OL samples (20, 21). However, the exact role of these cytokines in OL is still unclear. The current research has established a connection between gut microbiota and inflammatory mediators. The changes in gut microbiota are usually related to abnormal immune responses and dysregulation of inflammatory cytokine production (22, 23). Therefore, it is reasonable to speculate that there may be a relationship between gut microbiota, inflammatory mediators and OL. Our research aims to elucidate these potential correlations and identify specific inflammatory cytokines that can serve as valuable tools for early diagnosis and potential clinical treatment.
Mendelian randomization (MR) is a genetic epidemiological method for determining causal relationships between exposures and outcomes by using genetic variation as an instrumental variable (24, 25). Since genetic variants detected in fertilized eggs are identified at the time of conception, MR is less subject to traditional environmental confounding variables and reverse causation than observational studies (26, 27). MR has become a key epidemiological method for inferring causal relationships (28). Therefore, based on the significant reliability of previous MR studies in determining the etiology, in this study, we aim to explore the correlation between gut microbiota, inflammatory mediators and OL through MR analysis of the aggregated data from publicly available genome-wide association studies (GWAS). These analyses aim to dissect the intricate correlations between these variables and provide valuable insights into their causal relationships.
2 Materials and methods
2.1 Study design
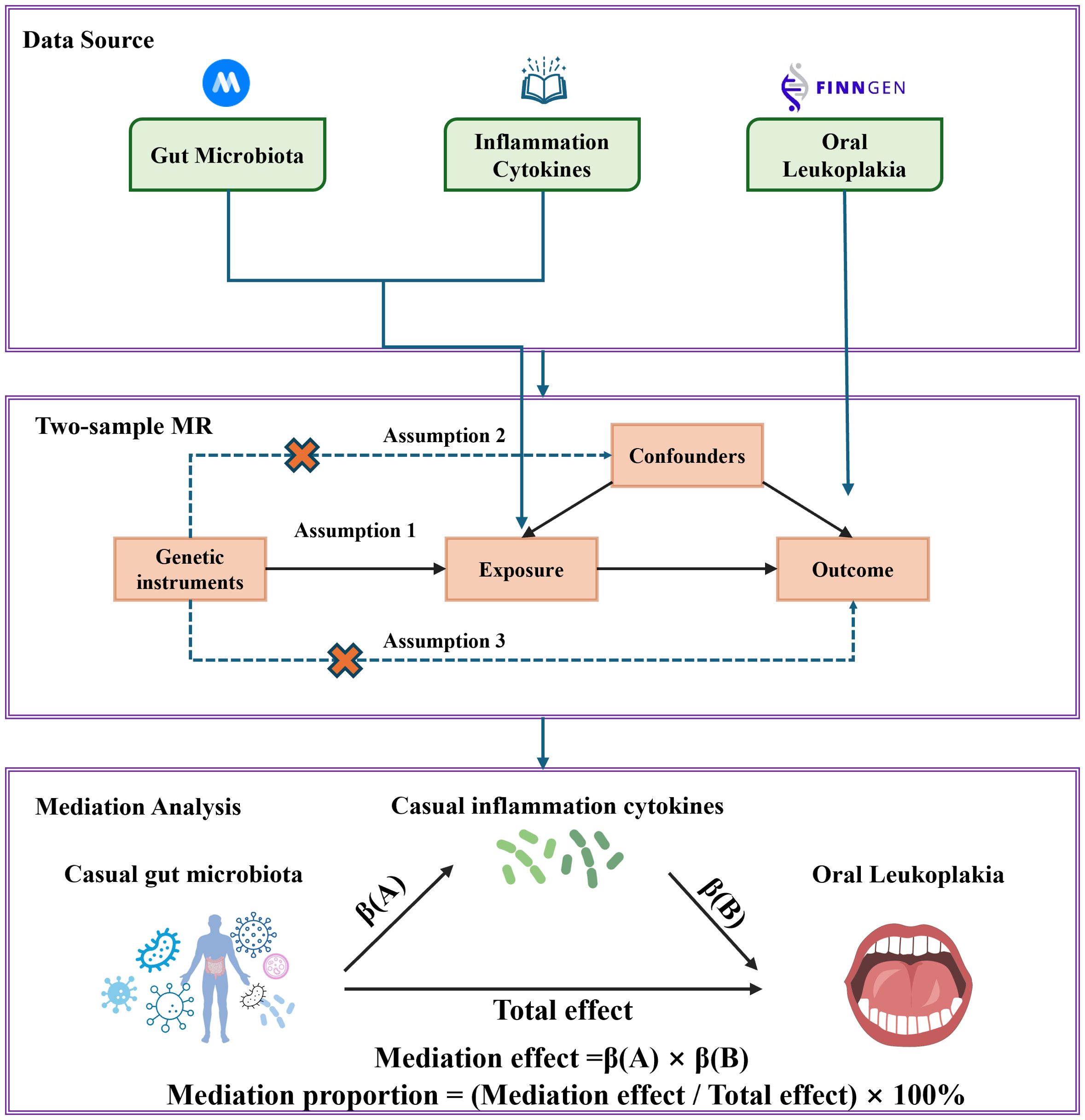
The research flowchart is shown in Figure 1. Firstly, we obtained summary statistics from published GWAS that included characteristics of the gut microbiota, inflammation-related proteins and OL. Secondly, two-sample MR analysis was used to assess causal relationships between gut microbiota, inflammation-related proteins and OL. Finally, two-step and multivariate MR (MVMR) analyses were used to determine the mediating role of inflammation-related proteins on the relationship between gut microbiota and OL. In order to obtain unbiased causal effects, MR analyses should adhere to the following three hypotheses: (1) Correlation hypothesis: instrumental variables (IVs) are strongly associated with exposure. (2) Independence hypothesis: IVs must be independent of potential confounders. (3) Exclusion limitation hypothesis: IVs affect the outcome of interest only through association with the exposure of interest, and there is no other way to play a role (29). This study followed the reporting guidelines of STROBE-MR (25). As this study is based on publicly available data, no additional ethical approval or consent is required.
2.2 Data source
Gut microbiota GWAS data were obtained from the MiBioGen study (30), which includes genome-wide genotypes of 18340 individuals (24 cohorts) from 11 countries, as well as fecal microbiota data obtained through 16S rRNA gene sequencing. The GWAS summary data includes 211 taxa of gut microbiota (131 genera, 35 families, 20 orders, 16 classes, 9 phyla), which is the most extensive multi group study on gut microbiota to date. The consortium has carefully adjusted for variables such as sex, age, and genetic principal components (PCs). They also included alpha diversity indices and technical covariates like DNA isolation methods and genotyping platforms. Quality control procedures, including minor allele frequency (MAF) thresholds and outlier removal, were also applied (31).
The GWAS summary data for oral leukoplakia is sourced from the 10th version of FinnGen Consortium(https://r10.risteys.finngen.fi/endpoints/K11_ORAL_LEUCOPLACIA) (32), which included 513 individuals diagnosed with oral leukoplakia and 411668 control subjects (FinnGen data field: K11_ORAL_LEUCOPLACIA). To maintain uniformity, all study participants are of European descent.
We obtained updated GWAS summary statistics for 91 inflammation-related proteins (the GWAS catalog ranges from GCST0001391 to GCST0002121) from the study by Zhao et al. (33), which recruited 14,824 participants mainly from Europe. Detailed information on data processing and complete data sources are available in the original literature.
2.3 Instrumental variables selection
Firstly, we utilized single nucleotide polymorphisms (SNPs) as IVs to investigate the causal relationship between exposure and outcomes at the genetic level. To ensure the accuracy and effectiveness of the causal relationship between gut microbiota and oral leukoplakia, we selected SNPs (P<1 × 10-5) that are significantly correlated with gut microbiota (34). In order to maximize the available instrumental variables for each inflammation-related proteins, we selected SNPs with a P-value of 5 × 10-6 as the threshold to obtain more comprehensive results and explore more potential relationships between OL and inflammation-related proteins (35). Then we excluded SNPs with linkage disequilibrium (LD) in the analysis. The selected SNPs with LD should meet the conditions of r2<0.001 and distance>10,000 kb with gut microbiota to minimize the linkage imbalance effect that violates random allele allocation. In addition, SNPs exhibiting palindromic or fuzzy features are systematically excluded from MR analysis. (Palindrome SNPs are SNPs with A/T or G/C alleles.) The strength of IVs is evaluated by calculating the F-statistic (Equation 1). Among them, N represents the sample size, K represents the number of instruments, R2 is used to represent the exposure variance explained by the selected SNP. If the corresponding F-statistic is less than 10, IVs are considered weak IVs and then excluded; On the contrary, F-statistics above 10 indicate that IVs have sufficient strength and are considered to have no significant weak instrument bias.
2.4 MR analysis and Mediation analysis
Causal relationships between gut microbiota, inflammation-related proteins, and oral leukoplakia were assessed using MR methods. The Wald ratio (WR) method was used to estimate the effect of individual IVs on causal estimates (36). For exposures that included multiple IVs, causality was inferred using the inverse variance weighted (IVW), MR Egger, weighted median, simple mode, and weighted mode methods. Of these, the IVW, which weights the inverse variance of the causal effects of different genetic variants on a trait and then combines the weighted effect estimates, was used as the primary method, while the other methods were used as supplements. We aimed to minimize bias and obtain reliable estimates that could alter the causal relationship between exposure and the outcome of interest.
Mediation analysis aims to evaluate the pathways from exposure to outcomes through mediation, which helps to explore the potential mechanisms by which exposure affects outcomes. Firstly, two sample MR methods were used to evaluate the causal relationship between gut microbiota and inflammation-related proteins to obtain β(A). Next, we determined the direct impact of gut microbiota on oral leukoplakia through MVMR to obtain β(B). We calculate the mediating effect by multiplying β (A) by β (B). To quantify the degree of mediation, we obtained the total impact of gut microbiota on obesity in the previous two-sample MR. We divided the mediating effect by the total effect of gut microbiota in OL. Two steps of MR were significant (P<0.05) and overlapping gut microbiota were considered as mediators.
2.5 Sensitivity analysis
To evaluate the robustness of causal relationships, we conducted sensitivity analysis. Sensitivity analysis includes multiple validity analysis, heterogeneity analysis and leave-one-out analysis. MR Egger regression analysis is used to evaluate the potential pleiotropic effects of SNPs used as IVs, The MR Egger intercept is used to indicate horizontal pleiotropy. Excessive horizontal pleiotropy indicates that the analysis violates the basic assumptions of MR analysis, making the results less reliable. The Cochran Q-test is used to measure the heterogeneity of IVs (37). In addition, a “leave-one-out” analysis was conducted to determine the potential bias effect of a single SNP on MR analysis, and this impact was re estimated by sequentially removing one SNP at a time. In order to clearly and concisely visualize the results of the MR method, the “TwoSampleMR” software package was used (38). All statistical analyses in this study were conducted using the R package in R (v4.2.1) statistical software.
2.6 Reverse MR analysis
To investigate whether oral leukoplakia has a causal effect on the identified significant gut microbiota (PIVW<0.05), we performed reverse MR analysis using SNPs associated with OL as IVs, and identified gut pathogens as the outcomes. The reverse MR analysis program is similar to the program used for MR analysis to elucidate the directional causal relationship between exposure and outcome.
3 Results
3.1 Causal effects of gut microbiota on OL
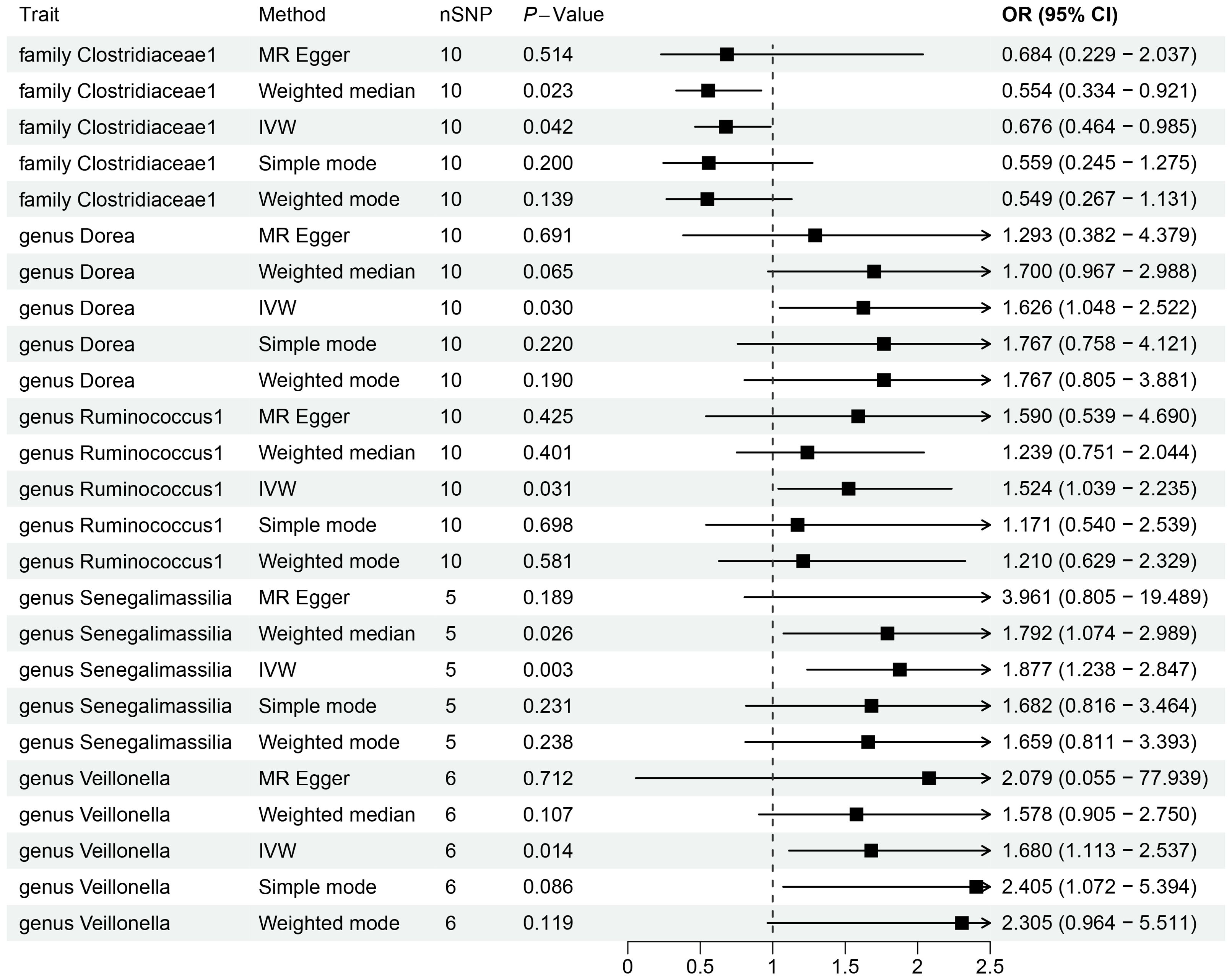
By utilizing Mendelian randomization, we delved into the relationship between specific gut microbiota and OL (Figure 1). The preliminary findings indicate that 5 out of 211 gut bacterial taxa are causally associated with oral leukoplakia (Figures 2, 3). The results of IVW analysis of these 5 bacterial taxa are shown below: family Clostridiaceae1(P= 0.042;odds ratio [OR] 95% confidence interval [CI]= 0.676 [0.464 − 0.985]), genus Dorea (P= 0.030; OR[95%CI]= 1.626 [1.048 − 2.522]), genus Ruminococcus1(P= 0.031; OR=95%[CI]= 1.524 [1.039 − 2.235]), genus Senegalimassilia(P= 0.003; OR[95%CI]= 1.877 [1.238 − 2.847]), genus Veillonella(P= 0.014; OR[95%CI]= 1.680 [1.113 − 2.537]). The family Clostridiaceae1 was negatively associated with the risk of oral leukoplakia development, whereas the other taxa were positively associated with the risk of oral leukoplakia development, suggesting that their abundance in the gut microbiota may contribute to the development of OL. The Figure 4; Supplementary Materials provide detailed information on MR analysis of gut microbiota and oral leukoplakia (Supplementary Table S1). Heterogeneity test of gut microbiota and oral microbiota showed that all p-values exceeded 0.05, and no evidence of horizontal pleiotropy was found using the MR Egger method (Supplementary Table S4). The sensitive analysis further validated these findings (Supplementary Tables S2–S4).
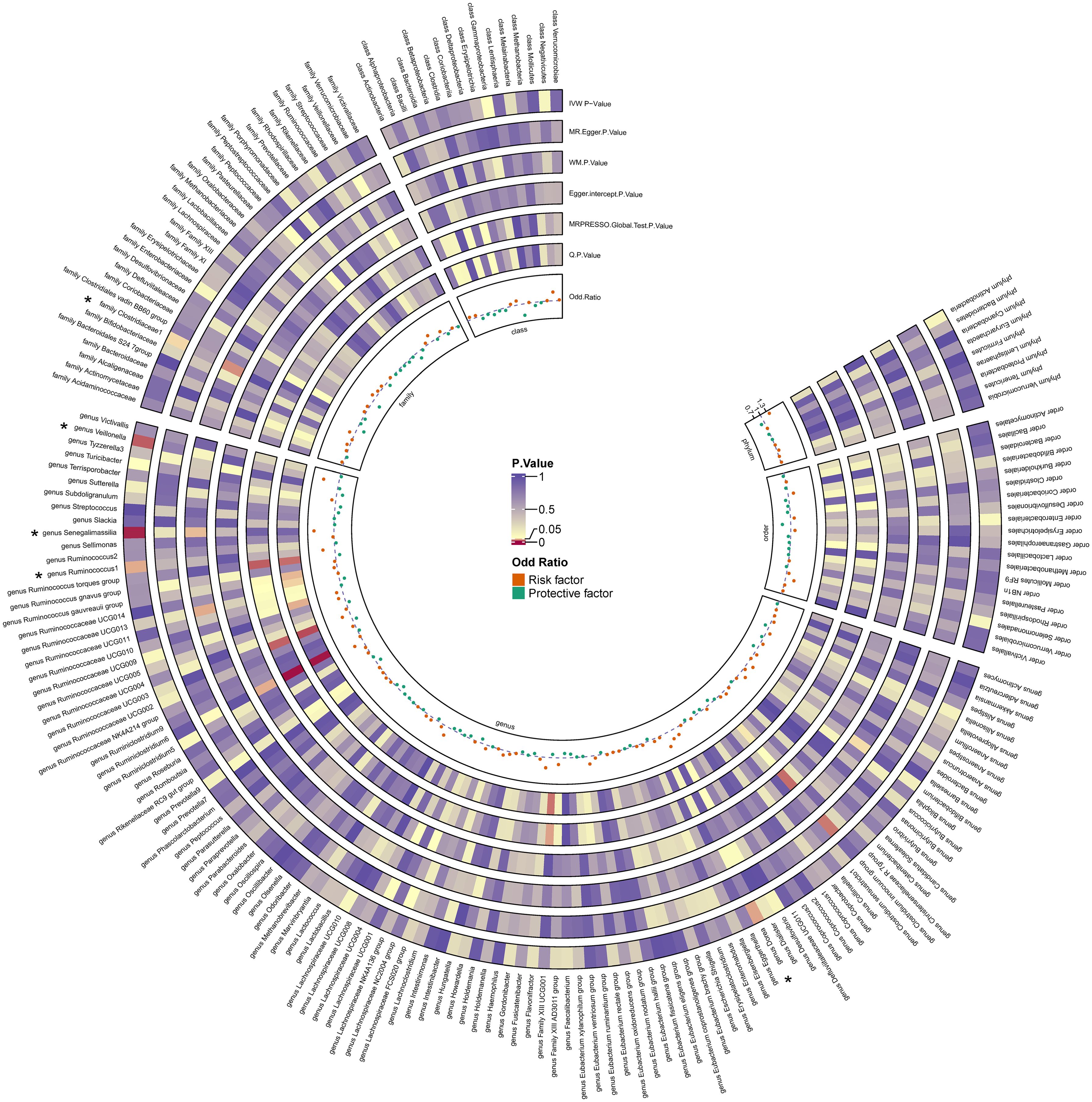
Figure 3. Heatmap for causality of gut microbiota on OL. Causality of 211 gut bacterial taxa on OL. Results that reached statistical significance in the IVW analysis are marked with an asterisk (*) and represent a P.value of less than 0.05.
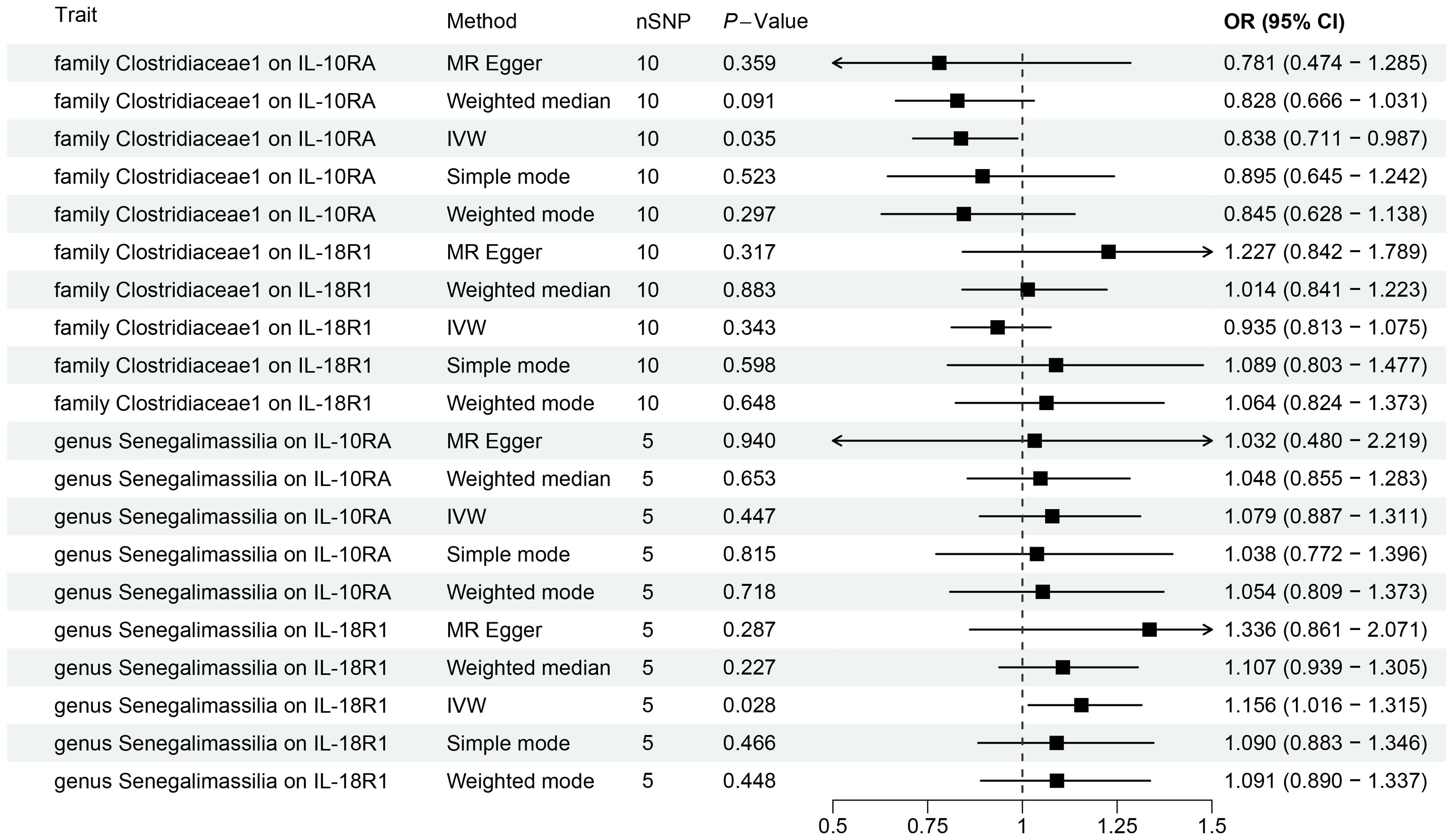
Figure 4. Forest plots for Mendelian randomization results of causal effects between gut microbiotas and 2 inflammation-related proteins (IL-10RA and IL18-R1).
For the causal relationship between the gut microbiota and oral leukoplakia mentioned above, we conducted reverse MR (Supplementary Table S5), and found a causal relationship between oral leukoplakia and family Bacteroidales S24 7group(P=0.0048), family Christensenellacea (P=0.0414), genus Intestinibacte (P=0.0479) and genus Lachnospiraceae (P=0.0298).
3.2 The causal effect of inflammation-related proteins on OL
This study revealed a causal relationship between 91 inflammation-related proteins and oral leukoplakia, with only 2 inflammation-related proteins exhibiting a potential causal relationship with oral leukoplakia (Supplementary Table S6; Figure 5). IVW analysis shows that, IL-10RA (OR[95%CI]= 1.327[1.022-1.722], P= 0.034) and IL18-R1 (OR[95%CI]=1.201[1.037-1.392], P= 0.015) are risk factors for oral leukoplakia. Sensitivity analysis was used to verify that these results have no heterogeneity or level pleiotropy (Supplementary Tables S7–S9).
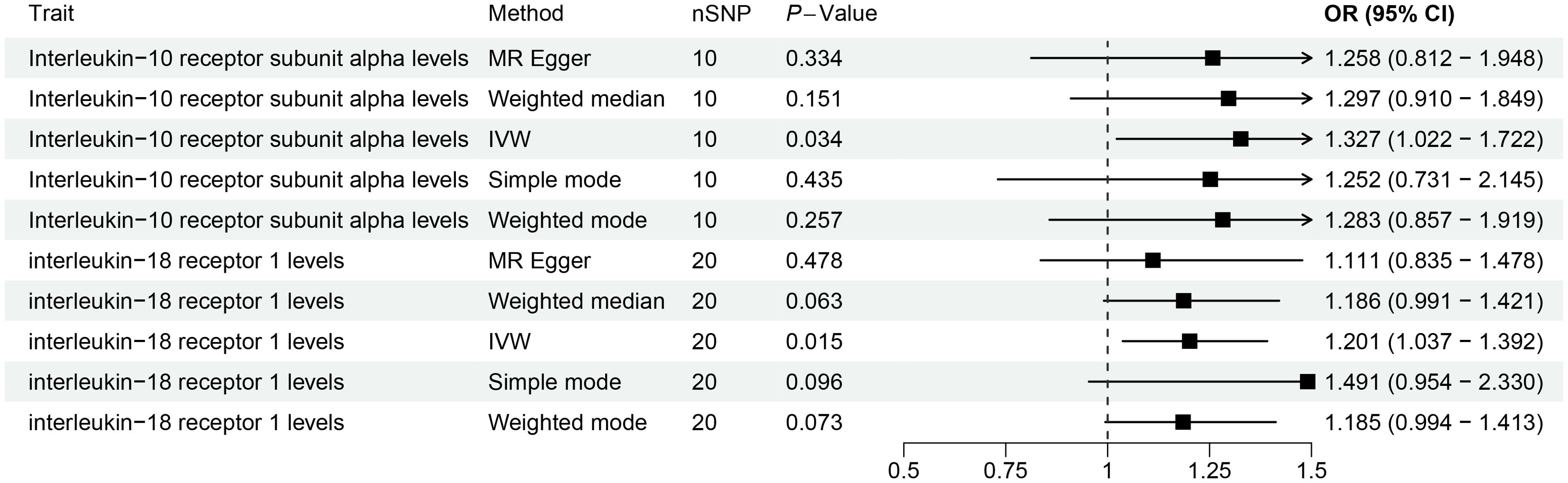
Figure 5. The causal association results of causal effects between 2 inflammation-related proteins (IL-10RA and IL18-R1) and OL.
3.3 Mendelian randomization analysis of gut microbiota and inflammation-related proteins
We conducted MR analysis on the gut microbiota and 91 inflammation-related proteins to further elucidate the role of these inflammation-related proteins in the relationship between gut microbiota and oral leukoplakia (Supplementary Table S11; Figure 4). The IVW analysis results showed a causal relationship between family Clostridiaceae1 and IL-10RA (OR[95%CI]=0.84[0.71, 0.99], P=0.035). There is a causal relationship between genus Senegalimassilia and IL18-R1 (OR[95%CI]=1.16 [1.02, 1.31], P=0.028), and no significant association was observed between any other bacterial taxa and inflammation-related proteins.
3.4 Mediation analysis results of gut microbiota, inflammation-related proteins and OL
To explore the potential mechanisms underlying the occurrence and development of oral leukoplakia, we conducted a mediation analysis to determine the causal pathway mediated by inflammation-related proteins from the gut microbiota to oral leukoplakia. This analysis focuses on the previously identified gut microbiota and inflammation-related proteins associated with oral leukoplakia in two-sample MR. In this study, both gut microbiota and inflammation-related proteins have a causal effect on OL inflammation-related proteins seem to play a mediating role in the pathways of gut microbiota and oral leukoplakia. Mediation analysis shows that the indirect effect of gens Senegalimassilia on oral leukoplakia through IL18-R1 is β= 0.026 (95%CI: 0.001-0.063), with a mediation ratio of 4.21%. Additionally, we also conducted a leave-one-out analysis, which confirmed the stability of our findings. The leave-one-out plot is shown in Figure 6, with heterogeneity results and pleiotropy analysis results provided in Supplementary Tables S2–S4. The leave-one-out analysis helps assess the stability and reliability of the results in Mendelian randomization study by showing how sensitive the overall findings are to the inclusion or exclusion of individual genetic variants. This indicates that IL18-R1 play a mediating role in the pathway between the gut microbiota genus Senegalimasilia and oral leukoplakia.
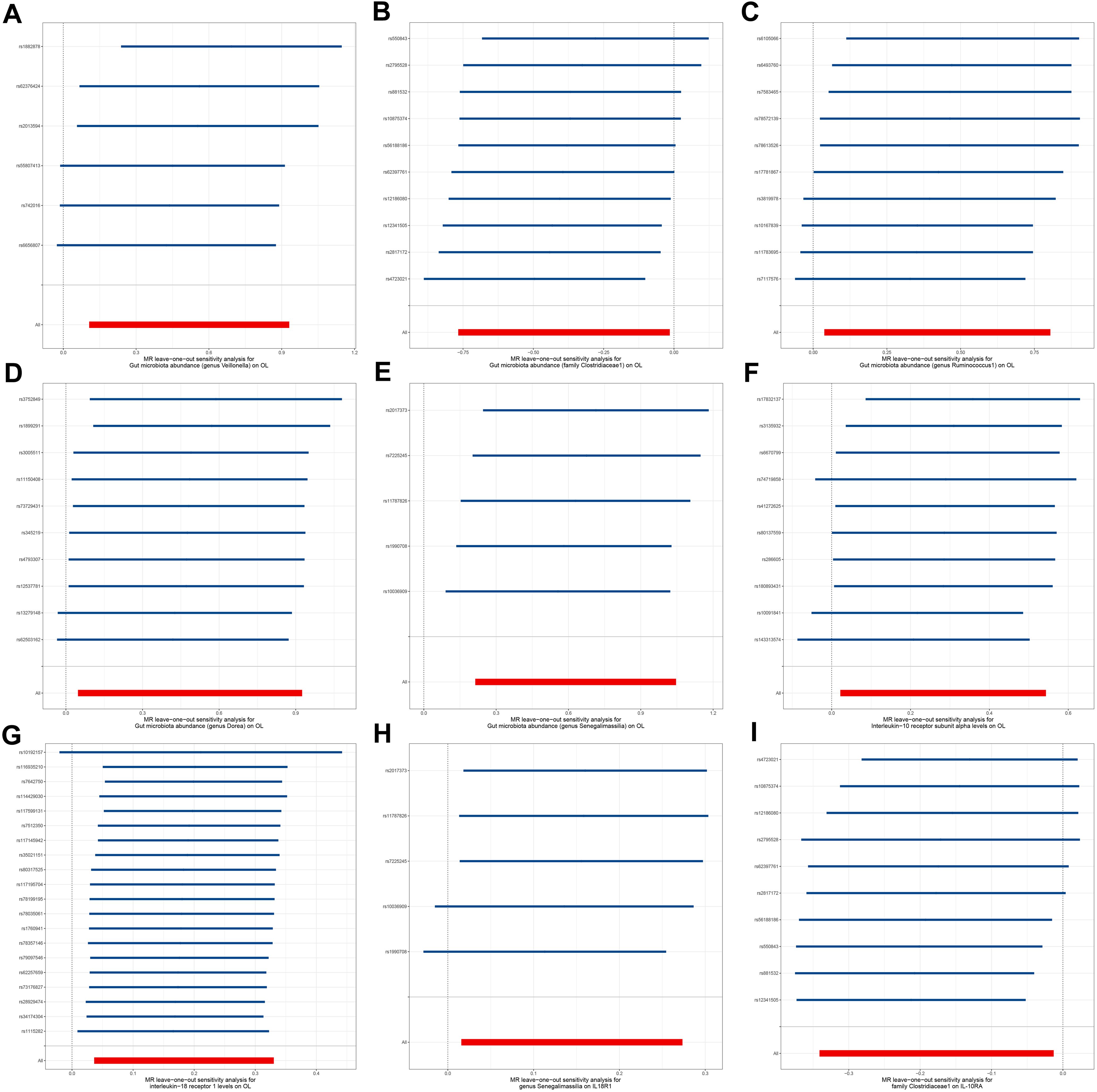
Figure 6. Leave-one-out plot. (A) Leave-one-out plot of genus Veillonella and OL, (B) Leave-one-out plot of family Clostridiaceae1 and OL; (C) Leave-one-out plot of genus Ruminococcus1 and OL; (D) Leave-one-out plot of genus Dorea and OL; (E) Leave-one-out plot of genus Senegalimassilia and OL; (F) Leave-one-out plot of IL-10RA and OL; (G) Leave-one-out plot of IL18-R1 and OL; (H) Leave-one-out plot of genus Senegalimassilia and IL18-R1; (I) Leave-one-out plot of family Clostridiaceae1 and IL-10RA.
4 Discussion
Few studies have explored the relationship between gut microbiota and oral leukoplakia. In this study, we used MR studies to explore the potential causal relationship between gut microbiota and oral leukoplakia. We analyzed the relationship between the abundance of 211 common gut microbiota and oral leukoplakia. The results indicate that certain specific gut microbiota are risk factors, while others are protective factors for oral leukoplakia. High abundance of family Clostridiaceae1 can reduce the risk of oral leukoplakia, while high abundance of genus Dorea, genus Ruminococcus1, genus Senegalimasilia, and genus Veillonella may increase the risk of oral leukoplakia. The available studies usually consider genus Dorea as a harmful microorganism. Chiu et al. showed a significant correlation between genus Dorea and allergic rhinitis (39). Nitsan et al. found that luminal proportions of genus Dorea were significantly higher in fecal samples from patients with irritable bowel syndrome (IBS) than in healthy controls (40). Genus Ruminococcus1 is an anaerobic bacterium that, as a gram-positive bacterium, plays different roles in various diseases. Although genus Ruminococcus1 can ferment complex sugars and produce short chain fatty acids (SCFAs), SCFAs have been shown to be beneficial in reducing the risk of certain cardiovascular metabolic diseases by reducing visceral fat. The research results of Zhang et al. indicate that genus Ruminococcus1 has a protective effect on blood vessels and can reduce the risk of Intracranial aneurysm and aneurysm subarachnoid hemorrage (41). While in some digestive system diseases, spondylitis and asthma, the abundance of genus Ruminococcus1 significantly increases (42). The results of Li et al. found that genus Ruminococcus1 may act synergistically with other bacteria to promote an increase in lipopolysaccharide and exacerbate intestinal barrier damage (43).Genus Senegalimassilia are nonmotile, Gram-stain-positive, anaerobic and coccobacilli. The genus Senegalimassilia is positively associated with potential cardiovascular disease risk biomarkers (44), Wang et al. showed that genus Senegalmassilia was positively associated with blood pressure (45), Hypertension can damage endothelial cells, affect vascular dilation and blood flow, and thus affect the health status of the oral mucosa. Previous studies have reported an increase in the abundance of genus Veillonella in patients with oral leukoplakia, which is consistent with our observations (46). This indicates a potential connection between specific gut microbiota and the development of oral leukoplakia.
The human gut microbiota contains over 10 trillion microbes that reside in the human intestine. Similarly, the oral microbiota, the second-largest microbiota in the body, consists of over 700 bacterial species (12). Given the physical connection between the oral and gastrointestinal mucosa, and the ingestion of saliva daily, there is a significant interplay between oral and gut microbiota (47). Oral bacteria can influence the gut microbiota through various routes, including the enteral route via saliva (48), the hematogenous route via bloodstream spread (49), and immune cell migration (50). These interactions underscore the close relationship between oral and gut microbiota, with the former potentially impacting gut health and systemic diseases (16). Recent studies have highlighted a complex relationship between OL and alterations in the oral microbiota. While results vary across investigations, they generally indicate a shift in microbial communities associated with OL. Alfa-diversity studies have reported inconsistent findings, ranging from increased bacterial richness to depletion or no difference between OL and healthy controls. However, beta-diversity has consistently shown differences between these groups, though some results are complicated by variations in subsites or subgroups (51, 52). Several studies have identified specific bacteria associated with OL. Ganly et al. found an enrichment of periodontal pathogens like Fusobacterium, Prevotella, and Alloprevotella in OL, along with a depletion of commensal Streptococcus (46). Amer et al. reported decreased levels of Firmicutes and an increase in Fusobacterium nucleatum, Campylobacter spp., and Rothia mucilaginosa in OL, while healthy mucosa was enriched in Streptococcus mitis and Gemella haemolysans (51). Similarly, Decsi et al. observed increased Fusobacterium nucleatum and decreased Streptococcus mitis in OL (14). In addition to bacterial shifts, fungal colonization, particularly by Candida species, has been noted in OL. Amer et al. found Candida in 35% of OL samples, significantly higher than in healthy controls, with specific clusters showing a strong association with Candida (51). The role of fungi in oral premalignant disorders is ambivalent, with some species like Malassezia and Schizophyllum showing anti-carcinogenic potential, while Candida species have been linked to carcinogenesis. The potential interaction between bacteria, fungi, and viruses in OL pathogenesis remains largely unexplored and warrants further investigation. In conclusion, OL is associated with significant changes in the oral microbiota, characterized by a decrease in commensals like Streptococci and an increase in anaerobes such as Fusobacterium nucleatum and Porphyromonas gingivalis. These microbial shifts may serve as markers for the progression of OL to oral squamous cell carcinoma (OSCC) (53). While there is indeed a substantial body of literature on the oral microbiome’s role in oral cancer and OL, the gut microbiome’s potential influence remains underexplored. Our study aims to contribute novel insights by investigating whether gut microbiota can have a systemic effect on the pathogenesis of OL, thus broadening the understanding of microbial contributions beyond the oral cavity. This approach may reveal new therapeutic targets or preventive strategies by modulating the gut microbiome, which could complement existing oral microbiome-focused interventions.
This study determined the impact of gut microbiota on oral leukoplakia by their relative abundance expression. However, the exact mechanism by which gut microbiota leads to oral leukoplakia has not been determined. We assume that inflammation-related proteins may be a mediating factor between the gut microbiota and oral leukoplakia. According to MR analysis, we found that IL-10RA and IL18-R1 significantly increased the risk of oral leukoplakia. Our study provides genetic evidence that IL18-R1 plays a mediating role in the effect of genus Senegalimasilia on OL.
Our MR study provides compelling genetic evidence that 2 inflammation-related proteins (IL-10RA and IL18-R1) are associated with the etiology of oral leukoplakia. Notably, IL18 was identified as a risk factor for patients with OL. In a previous report, Zhang et al. used an enzyme-linked immunosorbent assay to screen serum and salivary concentrations of IL-18 and showed that elevated serum and salivary IL-18 correlated with disease severity in patients with OL, and these findings could be considered the predictive or prognostic value of elevated IL-18 for OL (21). The key pro-inflammatory cytokine IL-18 plays an important role in the control of innate and adaptive immunity. The receptor IL18-R1 belongs to the interleukin-1 receptor family and is expressed in Th1 cells. After binding with IL18, IL-18 triggers the recruitment of IL18RAP, thereby initiating signal transduction (54, 55). IL-18 is secreted by antigen-presenting cells, stimulates IFN - γ synthesis, and participates in the synergistic activation of interleukin-12 to promote Th1 mediated immune responses. IL18-R1 plays an important role in cancer progression. For example, IL18R1 was over-expressed in breast cancer tissues compared with nearby non-cancerous tissues (56). In addition, the expression level of IL18-R1 is positively correlated with disease-free survival (DFS) in patients with triple negative breast cancer (57), This indicates that it is useful as a prognostic marker for this malignant tumor. Human IL-10R is a heterotetramer composed of receptor chains IL-10RA and IL-10RB, belonging to class II cytokine receptors (58), expressed on innate immune cells and adaptive immune cells. The IL-10RA chain plays a dominant role in mediating high affinity ligand binding and signal transduction. The interaction between IL-10 and IL-10R complex stabilizes the dimerization of two IL-10R subunits, activating Janus kinase 1 and tyrosine kinase 2, and catalyzing phosphorylation of themselves, and induction of intercellular signal transduction. Usually, The expression level of IL-10RA is related to the intensity of IL-10’s effect on immune cells (59). Research has shown that due to its immunosuppressive function, the presence of Tregs in the subcutaneous matrix of vitiligo is a potential indicator of cancer progression (60). The expression of higher quantities of Treg related proteins (including IL-10) may have potential predictive effects in malignant transformation (61). Previous studies have shown that mutations in the first splicing site of IL10RA lead to inflammatory bowel disease (IBD) in infants. In addition, the phenotypes of IL10RA polymorphism include severe arthritis and Very early onset Ulcerative Colitis (62). Our research findings indicate that IL-10RA and IL-18R1 are risk factors for precancerous lesions associated with oral leukoplakia, thereby strengthening their association with oral leukoplakia and providing new insights into their role in the pathogenesis of oral leukoplakia.
This is the first large-scale MR analysis of the causal relationship between gut microbiota, inflammation-related proteins and OL. The exploration of the relationship between gut microbiota, inflammation-related proteins and OL not only elucidates potential prevention and treatment strategies, but also helps to deepen understanding of the pathogenesis of OL. In addition, the identification of specific gut microbiota associated with the risk of OL provides promising targets for future interventions and treatment methods aimed at regulating gut microbiota to reduce the risk of OL. But our research has some limitations. Firstly, since the samples come from European ancestry, the applicability of our research results is limited, which may not accurately reflect the impact of genetic and lifestyle diversity on the gut microbiota of different populations. Secondly, the association between certain microbial communities we have observed and the risk of oral leukoplakia is preliminary. Considering the complexity of the interactions between gut microbiota, further research should be conducted on their mechanisms of action. Thirdly, we highlight MR method’s ability to infer causal relationships using genetic variants as instrumental variables, thereby reducing confounding factors. However, we also acknowledge the limitations, such as the potential for pleiotropy and the assumption that the genetic variants only influence the outcome through the exposure of interest. By addressing these points, we aim to provide a balanced perspective on the use of MR in our study. Finally, although we investigated the mediating role of 91 inflammation-related proteins between different gut microbiota abundance and oral leukoplakia, the mechanism by which gut microbiota affects the occurrence and development of oral leukoplakia through inflammation-related proteins remains to be studied.
5 Conclusion
This study provides new evidence for the causal relationship between gut microbiota, inflammation and OL. Our MR study suggests that inflammation-related proteins mediate the pathway of gut microbiota leading to OL. These findings provide new insights into the pathogenesis of OL, indicating their potential significance in preventing and treating OL.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
JD: Conceptualization, Data curation, Formal analysis, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. YR: Writing – original draft, Formal analysis, Investigation, Methodology. SL: Writing – original draft, Conceptualization, Project administration, Validation. NW: Writing – review & editing, Data curation, Methodology, Software. LW: Writing – original draft, Data curation, Formal analysis, Investigation. ZF: Writing – original draft, Conceptualization, Methodology, Resources. YL: Writing – original draft, Methodology, Project administration, Visualization. YP: Writing – original draft, Methodology, Software, Validation. JL: Supervision, Writing – original draft, Writing – review & editing, Investigation, Resources. JC: Supervision, Writing – original draft, Writing – review & editing, Formal analysis, Investigation.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We appreciate the strong support of our colleagues and the constructive comments of the team members for this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1443123/full#supplementary-material
References
1. Warnakulasuriya S, Kujan O, Aguirre-Urizar JM, Bagan JV, González-Moles MÁ, Kerr AR, et al. Oral potentially Malignant disorders: A consensus report from an international seminar on nomenclature and classification, convened by the WHO Collaborating Centre for Oral Cancer. Oral Dis. (2021) 27(8):1862–80. doi: 10.1111/odi.13704
2. Evren I, Najim AM, Poell JB, Brouns ER, Wils LJ, Peferoen LA, et al. The value of regular follow-up of oral leukoplakia for early detection of Malignant transformation. Oral Dis. (2024) 30(5):2991–3003. doi: 10.1111/odi.14797
3. Warnakulasuriya S, Johnson NW, van der Waal I. Nomenclature and classification of potentially Malignant disorders of the oral mucosa. J Oral Pathol Med. (2007) 36(10):575–80. doi: 10.1111/j.1600-0714.2007.00582.x
4. Wils LJ, Poell JB, Brink A, Evren I, Brouns ER, de Visscher JG, et al. Elucidating the genetic landscape of oral leukoplakia to predict Malignant transformation. Clin Cancer Res. (2023) 29(3):602–13. doi: 10.1158/1078-0432.CCR-22-2210
5. Mello FW, Miguel AFP, Dutra KL, Porporatti AL, Warnakulasuriya S, Guerra ENS, et al. Prevalence of oral potentially Malignant disorders: a systematic review and meta-analysis. J Oral Pathol Med. (2018) 47(7):633–40. doi: 10.1111/jop.2018.47.issue-7
6. Aguirre-Urizar JM, Lafuente-Ibáñez de Mendoza I, Warnakulasuriya S. Malignant transformation of oral leukoplakia: systematic review and meta-analysis of the last 5 years. Oral Dis. (2021) 27(8):1881–95. doi: 10.1111/odi.13810
7. Lodi G, Franchini R, Warnakulasuriya S, Varoni EM, Sardella A, Kerr AR, et al. Interventions for treating oral leukoplakia to prevent oral cancer. Cochrane Database System Rev. (2016) 7. doi: 10.1002/14651858.CD001829.pub4
8. Alkasir R, Li J, Li X, Jin M, Zhu B. Human gut microbiota: the links with dementia development. Protein Cell. (2017) 8(2):90–102. doi: 10.1007/s13238-016-0338-6
9. Matijašić M, Meštrović T, Čipčić Paljetak H, Perić M, Barešić A, Verbanac D. Gut microbiota beyond bacteria—mycobiome, virome, archaeome, and eukaryotic parasites in IBD. Int J Mol Sci. (2020) 21(8):2668.
10. Enaud R, Vandenborght L-E, Coron N, Bazin T, Prevel R, Schaeverbeke T, et al. The mycobiome: a neglected component in the microbiota-gut-brain axis. Microorganisms. (2018) 6(1):22. doi: 10.3390/microorganisms6010022
11. Ursell LK, Haiser HJ, Van Treuren W, Garg N, Reddivari L, Vanamala J, et al. The intestinal metabolome: an intersection between microbiota and host. Gastroenterology. (2014) 146(6):1470–6. doi: 10.1053/j.gastro.2014.03.001
12. Kilian M, Chapple I, Hannig M, Marsh P, Meuric V, Pedersen A, et al. The oral microbiome–an update for oral healthcare professionals. British Dental J. (2016) 221(10):657–66. doi: 10.1038/sj.bdj.2016.865
13. Pietrobon G, Tagliabue M, Stringa LM, De Berardinis R, Chu F, Zocchi J, et al. Leukoplakia in the oral cavity and oral microbiota: a comprehensive review. Cancers. (2021) 13(17):4439. doi: 10.3390/cancers13174439
14. Decsi G, Soki J, Pap B, Dobra G, Harmati M, Kormondi S, et al. Chicken or the egg: microbial alterations in biopsy samples of patients with oral potentially Malignant disorder. Pathol Oncol Res. (2019) 25:1023–33. doi: 10.1007/s12253-018-0457-x
15. Hashimoto K, Shimizu D, Hirabayashi S, Ueda S, Miyabe S, Oh-iwa I, et al. Changes in oral microbial profiles associated with oral squamous cell carcinoma vs leukoplakia. J Investig Clin Dentist. (2019) 10(4):. doi: 10.1111/jicd.12445
16. Tan X, Wang Y, Gong T. The interplay between oral microbiota, gut microbiota and systematic diseases. J Oral Microbiol. (2023) 15(1):2213112. doi: 10.1080/20002297.2023.2213112
17. Greten FR, Grivennikov SI. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity. (2019) 51(1):27–41. doi: 10.1016/j.immuni.2019.06.025
18. Liu Y, Zhu Q, Guo G, Xie Z, Li S, Lai C, et al. Causal associations of genetically predicted gut microbiota and blood metabolites with inflammatory states and risk of infections: a Mendelian randomization analysis. Front Microbiol. (2024) 15:1342653. doi: 10.3389/fmicb.2024.1342653
19. Babiuch K, Kuśnierz-Cabala B, Kęsek B, Okoń K, Darczuk D, Chomyszyn-Gajewska M. Evaluation of proinflammatory, NF-kappaB dependent cytokines: IL-1α, IL-6, IL-8, and TNF-α in tissue specimens and saliva of patients with oral squamous cell carcinoma and oral potentially Malignant disorders. J Clin Med. (2020) 9(3):867. doi: 10.3390/jcm9030867
20. Brailo V, Vucicevic-Boras V, Lukac J, Biocina-Lukenda D, Zilic-Alajbeg I, Milenovic A, et al. Salivary and serum interleukin 1 beta, interleukin 6 and tumor necrosis factor alpha in patients with leukoplakia and oral cancer. Med Oral Patol Oral Cir Bucal. (2012) 17(1):e10. doi: 10.4317/medoral.17323
21. Kaur J, Jacobs. Proinflammatory cytokine levels in oral lichen planus, oral leukoplakia, and oral submucous fibrosis. J Korean Assoc Oral Maxillofac Surg. (2015) 41(4):171. doi: 10.5125/jkaoms.2015.41.4.171
22. Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. (2016) 167(4):1125–36. e8. doi: 10.1016/j.cell.2016.10.020
23. Nigam M, Devi K, Coutinho HD, Mishra AP. Exploration of gut microbiome and inflammation: A review on key signalling pathways. Cell Signal. (2024) 111140:. doi: 10.1016/j.cellsig.2024.111140
24. Birney E. Mendelian randomization. Cold Spring Harb Perspect Med. (2021) a041302. doi: 10.1101/cshperspect.a041302
25. Skrivankova VW, Richmond RC, Woolf BA, Yarmolinsky J, Davies NM, Swanson SA, et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the STROBE-MR statement. Jama. (2021) 326(16):1614–21. doi: 10.1001/jama.2021.18236
26. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. (2008) 27(8):1133–63. doi: 10.1002/sim.v27:8
27. Carter AR, Sanderson E, Hammerton G, Richmond RC, Davey Smith G, Heron J, et al. Mendelian randomisation for mediation analysis: current methods and challenges for implementation. Eur J Epidemiol. (2021) 36(5):465–78. doi: 10.1007/s10654-021-00757-1
28. Zhou S, Sun Q, Gao N, Hu Z, Jia J, Song J, et al. The role of inflammatory biomarkers in mediating the effect of inflammatory bowel disease on nonmalignant digestive system diseases: A multivariable mendelian randomized study. Can J Gastroenterol Hepatol. (2024) 2024(1):1266139. doi: 10.1155/2024/1266139
29. Bowden J, Holmes MV. Meta-analysis and Mendelian randomization: a review. Res Synthesis Methods. (2019) 10(4):486–96. doi: 10.1002/jrsm.v10.4
30. Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Gen. (2021) 53(2):156–65.
31. Chen Y, Tang S. Gut microbiota and immune mediation: a Mendelian randomization study on granulomatosis with polyangiitis. Front Immunol. (2023) 14:1296016. doi: 10.3389/fimmu.2023.1296016
32. Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. (2023) 613(7944):508–18.
33. Zhao JH, Stacey D, Eriksson N, Macdonald-Dunlop E, Hedman ÅK, Kalnapenkis A, et al. Genetics of circulating inflammatory proteins identifies drivers of immune-mediated disease risk and therapeutic targets. Nat Imunol. (2023) 24(9):1540–51. doi: 10.1038/s41590-023-01588-w
34. Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Gen. (2017) 13(11):e1007081. doi: 10.1371/journal.pgen.1007081
35. Ji D, Chen W-Z, Zhang L, Zhang Z-H, Chen L-J. Gut microbiota, circulating cytokines and dementia: a Mendelian randomization study. J Neuroinflammation. (2024) 21(1):2. doi: 10.1186/s12974-023-02999-0
36. Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. (2017) 26(5):2333–55. doi: 10.1177/0962280215597579
37. Cohen JF, Chalumeau M, Cohen R, Korevaar DA, Khoshnood B, Bossuyt PM. Cochran’s Q test was useful to assess heterogeneity in likelihood ratios in studies of diagnostic accuracy. J Clin Epidemiol. (2015) 68(3):299–306. doi: 10.1016/j.jclinepi.2014.09.005
38. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. elife. (2018) 7:e34408. doi: 10.7554/eLife.34408
39. Chiu C-Y, Chan Y-L, Tsai M-H, Wang C-J, Chiang M-H, Chiu C-C. Gut microbial dysbiosis is associated with allergen-specific IgE responses in young children with airway allergies. World Allergy Organ J. (2019) 12(3):100021. doi: 10.1016/j.waojou.2019.100021
40. Maharshak N, Ringel Y, Katibian D, Lundqvist A, Sartor RB, Carroll IM, et al. Fecal and mucosa-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Dig Dis Sci. (2018) 63:1890–9. doi: 10.1007/s10620-018-5086-4
41. Zhang H, Jiang X, Li A, Wang X. Causal associations between gut microbiota and cerebrovascular diseases. World Neurosur. (2024) 183:e587–97. doi: 10.1016/j.wneu.2023.12.150
42. Henke MT, Kenny DJ, Cassilly CD, Vlamakis H, Xavier RJ, Clardy J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc Natl Acad Sci U S A. (2019) 116(26):12672–7. doi: 10.1073/pnas.1904099116
43. Li N, Wang H, Pei H, Wu Y, Li L, Ren Y, et al. Genus_Ruminococcus and order_Burkholderiales affect osteoporosis by regulating the microbiota-gut-bone axis. Front Microbiol. (2024) 15:1373013. doi: 10.3389/fmicb.2024.1373013
44. Romo-Vaquero M, Cortés-Martín A, Loria-Kohen V, Ramírez-de-Molina A, García-Mantrana I, Collado MC, et al. Deciphering the human gut microbiome of urolithin metabotypes: association with enterotypes and potential cardiometabolic health implications. Mol Nutr Food Res. (2019) 63(4):1800958. doi: 10.1002/mnfr.201800958
45. Wang B, Liu J, Lei R, Xue B, Li Y, Tian X, et al. Cold exposure, gut microbiota, and hypertension: A mechanistic study. Sci Total Environ. (2022) 833:155199. doi: 10.1016/j.scitotenv.2022.155199
46. Ganly I, Yang L, Giese RA, Hao Y, Nossa CW, Morris LG, et al. Periodontal pathogens are a risk factor of oral cavity squamous cell carcinoma, independent of tobacco and alcohol and human papillomavirus. Int J Cancer. (2019) 145(3):775–84. doi: 10.1002/ijc.v145.3
47. Kitamoto S, Nagao-Kitamoto H, Hein R, Schmidt T, Kamada N. The bacterial connection between the oral cavity and the gut diseases. J Dent Res. (2020) 99(9):1021–9. doi: 10.1177/0022034520924633
48. Bao J, Li L, Zhang Y, Wang M, Chen F, Ge S, et al. Periodontitis may induce gut microbiota dysbiosis via salivary microbiota. Int J Oral Sci. (2022) 14(1):32. doi: 10.1038/s41368-022-00183-3
49. Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. (2015) 15(1):30–44. doi: 10.1038/nri3785
50. Kitamoto S, Nagao-Kitamoto H, Jiao Y, Gillilland MG, Hayashi A, Imai J, et al. The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell. (2020) 182(2):447–62.e14.
51. Amer A, Galvin S, Healy CM, Moran GP. The microbiome of potentially Malignant oral leukoplakia exhibits enrichment for Fusobacterium, Leptotrichia, Campylobacter, and Rothia species. Front Microbiol. (2017) 8:2391. doi: 10.3389/fmicb.2017.02391
52. Gopinath D, Kunnath Menon R, Chun Wie C, Banerjee M, Panda S, Mandal D, et al. Salivary bacterial shifts in oral leukoplakia resemble the dysbiotic oral cancer bacteriome. J Oral Microbiol. (2021) 13(1):1857998. doi: 10.1080/20002297.2020.1857998
53. Lee W-H, Chen H-M, Yang S-F, Liang C, Peng C-Y, Lin F-M, et al. Bacterial alterations in salivary microbiota and their association in oral cancer. Sci Rep. (2017) 7(1):1–11. doi: 10.1038/s41598-017-16418-x
54. Ovsyannikova IG, Haralambieva IH, Kennedy RB, O’Byrne MM, Pankratz VS, Poland GA. Genetic variation in IL18R1 and IL18 genes and inteferon γ ELISPOT response to smallpox vaccination: an unexpected relationship. J Infect Dis. (2013) 208:1422–30. doi: 10.1093/infdis/jit341
55. Haralambieva IH, Ovsyannikova IG, Dhiman N, Kennedy RB, O’Byrne M, Pankratz VS, et al. Common SNPs/haplotypes in IL18R1 and IL18 genes are associated with variations in humoral immunity to smallpox vaccination in caucasians and african americans. J Infect Dis. (2011) 204:433–41. doi: 10.1093/infdis/jir268
56. Sattari A, Hussen BM, Ghafouri-Fard S, Alihashemi A, Omrani MD, Zekri A, et al. Expression of T helper 1-associated lncRNAs in breast cancer. Exp Mol Pathol. (2021) 119:104619. doi: 10.1016/j.yexmp.2021.104619
57. Marchetti P, Antonov A, Anemona L, Vangapandou C, Montanaro M, Botticelli A, et al. New immunological potential markers for triple negative breast cancer: IL18R1, CD53, TRIM, Jaw1, LTB, PTPRCAP. Discov Oncol. (2021) 12:6. doi: 10.1007/s12672-021-00401-0
58. Hussein P, Zahran F, Ashour Wahba A, Ahmad A, Ibrahiem M, Shalaby S, et al. Interleukin 10 receptor alpha subunit (IL-10RA) gene polymorphism and IL-10 serum levels in Egyptian atopic patients. J Investig Allergol Clin Immunol. (2010) 20(1):20–6.
59. Yang X-K, Li P, Li S, Zhang C, Li B-Z, Leng R-X, et al. Genetic variant of IL-10RA and susceptibility to rheumatoid arthritis in a Chinese population. Clin Rheumatol. (2017) 36:825–30. doi: 10.1007/s10067-016-3449-9
60. Sakata J, Yoshida R, Matsuoka Y, Kawahara K, Arita H, Nakashima H, et al. FOXP3 lymphocyte status may predict the risk of Malignant transformation in oral leukoplakia. J Oral Maxillofac Surg Med Pathol. (2020) 32(1):33–9. doi: 10.1016/j.ajoms.2019.06.005
61. González-Arriagada WA, Canedo-Marroquin G, Adorno-Farías D, Fernández-Ramires R. New insights into the role of the oral leukoplakia microenvironment in Malignant transformation. Front Oral Health. (2024) 5:1363052. doi: 10.3389/froh.2024.1363052
Keywords: gut microbiota, oral leukoplakia, inflammation-related proteins, Mendelian randomization, mediation analysis
Citation: Da J, Ren Y, Liu S, Wang N, Wang L, Fu Z, Liang Y, Pan Y, Li J and Chen J (2024) Effect of the gut microbiome and inflammation-related proteins on oral leukoplakia: a Mendelian randomization study and mediation analysis. Front. Oncol. 14:1443123. doi: 10.3389/fonc.2024.1443123
Received: 03 June 2024; Accepted: 02 September 2024;
Published: 25 September 2024.
Edited by:
Manuel Weber, University of Erlangen Nuremberg, GermanyReviewed by:
Bettina Grötsch, University of Erlangen Nuremberg, GermanyVinay Jain, Bhabha Atomic Research Centre (BARC), India
Copyright © 2024 Da, Ren, Liu, Wang, Wang, Fu, Liang, Pan, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin Li, bGlqaW5rcTk5NjU1QDE2My5jb20=; Jufeng Chen, Y2pmZW5nQGZzeXl5LmNvbQ==
†These authors have contributed equally to this work
 Junlong Da
Junlong Da Yinting Ren
Yinting Ren Shiwei Liu1
Shiwei Liu1