- 1Division of Hematology-Medical Oncology, Upstate Cancer Center, SUNY Upstate Medical University, Syracuse, NY, United States
- 2Foundation Medicine, Cambridge, MA, United States
- 3Department of Pathology, SUNY Upstate Medical University, Syracuse, NY, United States
Background: Fusion of the RET gene resulting in clinically significant Genomic Alteration (GA) occur in 1-2% of NSCLC in the United States and has emerged as a major target for RET inhibitors which are first line treatment options in the Stage 4 setting. RET fusions have also been well-described as acquired resistance mutations in cases of EGFR-driven NSCLC treated with anti-EGFR tyrosine kinase inhibitors including erlotinib and osimertinib. The aim of this study was to determine whether RET fusion positive (RETfus+) NSCLC represents a unique histologic subtype of the disease with a unique genomic profile.
Methods: We selected 503 of 72,596 (0.7%) total NSCLC that were reported as RETfus+ from the Foundation One database. The cases were centrally evaluated for predominant histology and underwent hybrid capture based CGP to evaluate diverse GA. Cases with EGFR mutations were excluded. PD-L1 expression was determined by Immunohistochemistry (IHC) (Dako 22C3) with Tumor Proportion Score (TPS) ≥50% = high expression. For statistical comparisons, the false discovery rate was corrected using Benjamini/Hochberg adjustment.
Results: Potentially targetable GAs found less frequently in the RETfus+ group included BRCA1, BRAF, FGF12, FGFR1, KEAP1, KMT2D, KRAS, MDM2, MET, NF1, NSD3, PIK3CA, RB1, AND TP53. The presence of HRD, APOBEC and Tobacco gene signatures were also lower in frequencies in the RETFus+ NSCLC cases. SETD2 was the only GA found to be higher in the RETfus+ group. While markers predictive of checkpoint therapy response including TMB high level was more frequent in the RETfus- cases, PD-L1 high expression was more in RETfus+ samples. Surgical pathology analysis revealed that the high grade solid non-acinar pattern at 32% was the most frequent histologic subtype.
Conclusions: RETfus+ NSCLC features a unique genomic signature which can further impact therapy selection. With recent expanded approval of more specific RET kinase targeting inhibitors (selpercatinib and Pralsetinib) in the pan-cancer treatment setting, further study of RETfusion+ NSCLC histology and genomic/biomarker status appears warranted.
Introduction
Precision medicine has revolutionized the management of clinically advanced and metastatic Non-Small Cell Lung Cancer (mNSCLC), to the extent that it has become standard practice to perform Next Generation Sequencing (NGS) for formulating a treatment strategy (1, 2). NGS has expanded to limited stage disease and there is an evolving spectrum for Targeted agents for various settings (3). Currently, 350 individuals per day die from lung cancer, making it the leading cause of cancer death (4), prompting the need for continued research that will enable better understanding of the molecular drivers of the disease.
RET fusion (RETfus+) are drivers in the oncogenesis of several malignancies including mNSCLC and thyroid cancers. Gene fusions in the RET kinase domain result in the production of constitutively active chimeric RET homodimers that result in unchecked and aberrant cellular proliferation. This is usually seen in mNSCLC. Whereas in medullary thyroid cancer, germline, or somatic point mutations of kinase domain result in RET kinase activation (5). RETfus+ is seen in 1-2% of mNSCLCs, Today, patients have oral therapeutic options like selpercatinib and pralisetinib for treatment of this type of NSCLC (6, 7). These agents are relatively well-tolerated and have also demonstrated intracranial activity particularly with selpercatinib (6, 7). Although traditionally believed to be mutually exclusive, the hypothesis that RETfus+ may be a unique pathologic entity originated with reports showing its association with other alterations and biomarkers. Co-occurrence with EGFR mutations, MET amplification, low Tumor Mutational Burden (TMB) and PD-L1 expression have also been documented (8). RETfus+ NSCLC demonstrates increased responsiveness to pemetrexed based chemotherapy (9) and may also have a role in resistance to other targets like EGFR (10). Therefore, understanding the unique distribution of genomic alterations (GA) that occurs in RETfus+ mNSCLC is essential (8, 11).
Methods
The study was approved, and a waiver of consent was obtained from the Western Institutional Review Board (Protocol No. 20152817) and SUNY Upstate IRB (Project No. 2144270). FoundationOne is a CLIA-certified and CAP-accredited reference molecular laboratory. A database query was done for all mNSCLC cases submitted to Foundation Medicine. All cases were clinically advanced, either inoperable or metastatic. Basic details such as patient age and gender, routine histology, immunohistochemical staining results and confirmation of the diagnosis, were extracted from the pathology reports. All cases were sequenced between January 2018 and July 2022. Cases needed have had adequate tissue sample, DNA amount of 50 ng or greater and minimum of 20% tumor nuclear area versus benign nuclear area either before or after pathologist-guided macro-enrichment. Cases with low tumor purity on sequencing or inadequate sequencing coverage depth as described in the Foundation Medicine US FDA approval were excluded from the analysis (12).
Sample sequencing was done for Comprehensive genomic profiling (CGP) on 72,506 mNSCLC tissue samples. DNA was extracted using hybridization-capture- adaptor ligation–based libraries (FoundationOneCDx, Foundation Medicine, Inc.). Assay was done using all coding exons from 324 cancer related genes, plus select introns from at least 31 genes frequently rearranged in various malignancies. Specimens were evaluated for all classes of GAs including base substitutions, insertions, deletions, copy number alterations (amplifications and homozygous deletions), and for select gene fusions/rearrangements. Bioinformatics processes included Bayesian algorithms to detect base substitutions, local assembly algorithms to detect short insertions and deletions, a comparison with process-matched normal control samples to detect gene copy number alterations and an analysis of chimeric read pairs to identify gene fusions. An oncoprint plot was generated with the online tools of the cbio portal as described by Gao et al (13) and Cerami et al (14). TMB scores were defined by mutation/Mb on 0.83–1.14 Mb of sequence. Assessment of microsatellite instability was performed from DNA sequencing at least 95 loci. Each microsatellite locus had repeat length of 7–39 bp. The next-generation sequencing based “microsatellite instability score” was translated into categorical “microsatellite instability high”, “microsatellite instability intermediate”, or “microsatellite stable” by unsupervised clustering of specimens for which microsatellite instability status was assessed via gold standard methods (15). PD-L1 expression was measured using the DAKO 22C3 assay with low-positive tumor cell scoring defined as 1%-49% staining and high-positive tumor cell scoring defined as >50% staining (16). Anti-PD-L1 staining was done using the Dako 22C3 IHC kit, following the instructions provided in the kit protocol. Results were scored using the widely used tumor proportional score system (TPS) (17). Chi-square test and Mann Whitney U test were used in the statistical comparisons of the 2 groups. Statistical significance was defined as p < 0.05. Odds Ratio (OR) estimates were utilized to study the difference in the likelihood of a GA occurring between RETfus+ and RETfus-. The technique has been described in our previous publications, in which similar templates were followed (18).
Results
A total of 72,506 mNSCLC patients were identified of which 503 (0.69%) were RETfus+ and 72,003 (99.31%) were RET-fusion-. The age, sex distribution and descriptive analysis of various GAs are depicted in Table 1. The mean age was 66 years in RETfus+ and 69 years in the RETfus- group. Sex distribution was similar with 243 (48.31%) males in the RETfus+ cohort and 35,971 (49.96%) in the RETfus- group. The average GA/tumor was 4.6 for RETfus+ and 6 for RETfus-.
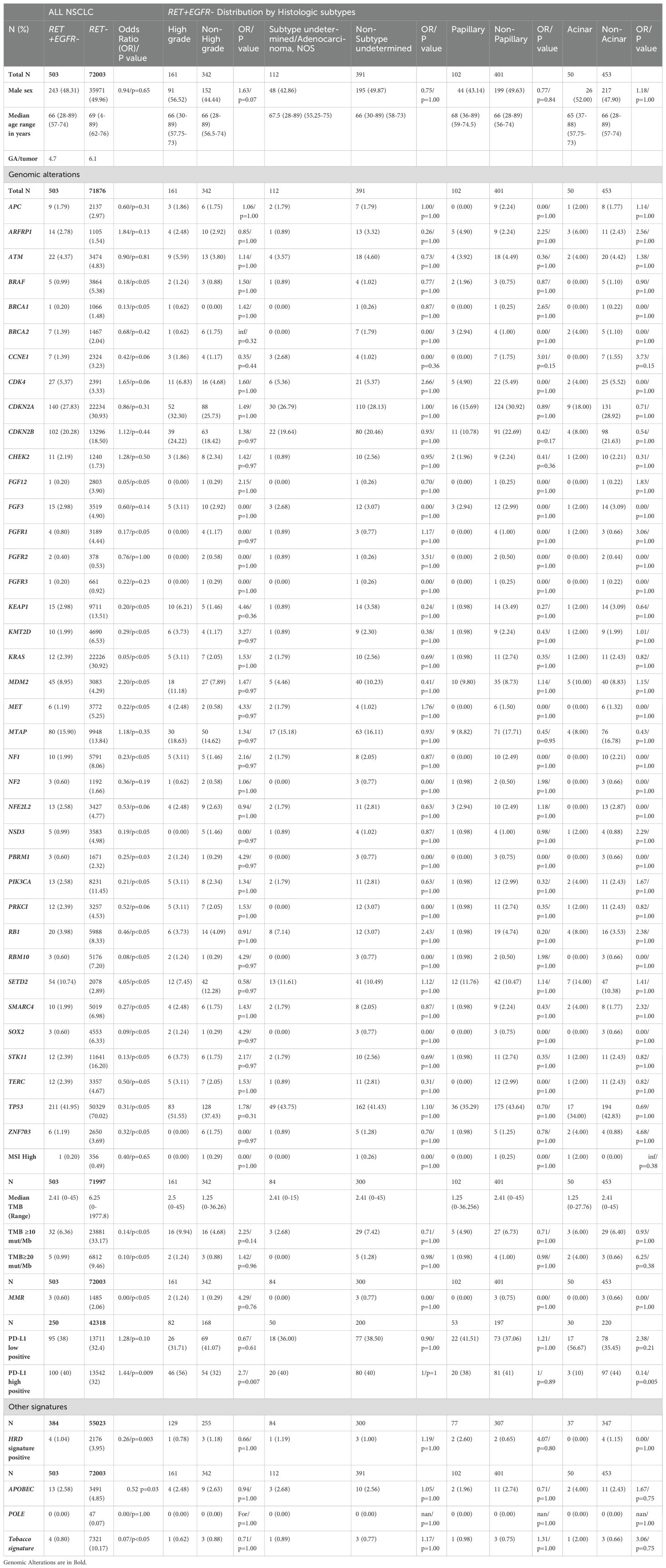
Table 1. Distribution of various biomarkers among RETfus+ and RETfus- NSCLC overall and based on histology.
This study identified more than 25 different RET fusions in the NSCLC cohort. The most frequent alterations accounting for more than 90% of the RET fusions included RET - KIF5B (60.3%), RET-CCDC6 (17.2%), RET–RET (9.6%) and RET-NCOA4 (3.2%).
We further divided the cohort into histologic subtypes and provided a descriptive analysis of the distribution of the various GAs. Table 1 shows the distribution of various biomarkers and GA in our cohort along with their distribution based on histologic subtypes of adenocarcinoma. The distribution of the histology revealed that high grade (HG) solid non-acinar 161 (32%) was the most frequent subtype, followed by Undetermined subtype 112 (22.27%), papillary (PAP) 102 (20.28%), and acinar (AC) 50 (9.94%). Other subtypes seen were mucinous/sigmoid (MUCSIG) 49 (9.74%), SCC (2.78%), sarcomatoid (SAR) 8 (1.59%), Lepidic 5 (1%), and SEC 2 (0.4%).
On analyzing potentially targetable GAs, most alterations were found in lower frequency in the RETfus+ vs RETfus- cohort. These included BRCA1 1(0.20%) vs 1066 (1.48%) OR 0.13/p<0.05], and BRAF [5 (0.99) vs 3864(5.38) OR 0.18/p<0.05]. Others included FGFR1 [4 (0.80) vs 3189 (4.44) OR 0.17/p<0.05], KEAP1 [15 (2.98) vs 9711 (13.51) OR 0.20/p<0.05], KMT2D [10 (1.99) vs 4690 (6.53) OR 0.29/p<0.05], KRAS [12 (2.39) vs 22226 (30.92) OR 0.05/p<0.05], MDM2 [45 (8.95) vs 3083 (4.29) OR 2.20/p<0.05], MET [6 (1.19) vs 3772 (5.25) OR 0.22/p<0.05], NF1 [10 (1.99) vs 5791 (8.06) OR 0.23/p<0.05], NSD3 [5 (0.99) vs 3583 (4.98) OR 0.19/p<0.05], PBRM1 [3 (0.60) vs 1671 (2.32) OR 0.25/p=0.03], PIK3CA [13 (2.58) vs 8231 (11.45) OR 0.21/p<0.05], RB1 [20(3.98) vs 5988 (8.33) OR 0.46/p<0.05], SMARC4 [10 (1.99) vs 5019 (6.98) OR 0.27/p<0.05], SOX2 [3 (0.60) vs 4553 (6.33) OR 0.09/p<0.05], STK11 [12 (2.39) vs 11641 (16.20) OR 0.13/p<0.05], TERC [12 (2.39) vs 3357 (4.67) OR 0.50/p=0.05], TP53 [211 (41.95) vs 50329 (70.02) OR 0.31/p<0.05], and ZNF703 [6 (1.19) vs 2650 (3.69) OR 0.32/p<0.05]. SETD2 [54 (10.74) vs 2078 (2.89) OR 4.05/p<0.05] was the only marker that was elevated in RETfus+ NSCLC.
Signatures were analyzed from a sub-cohort of 384 RETfus+ and 55023 RETfus- NSCLC. All signatures such as HRD [4(1.04) vs 2176(3.95) OR 0.26/p<0.05], APOBEC [13 (2.58) vs 3491 (4.85) OR 0.52 p=0.03], and Tobacco [4 (0.80) vs 7321 (10.17) OR 0.07/p<0.05] were lower in RETfus+ NSCLC.
The sub cohort studying TMB involved 503 RETfus+ and 71997 RETfus- NSCLC. The median TMB was 2.41 and 6.25 respectively. Both high [32 (6.36) vs 23881 (33.17) OR 0.14/p<0.05] and ultra-high [5 (0.99) vs 6812 (9.46) OR 0.10/p<0.05] were lower in RETfus+. However, from a cohort of 250 RETfus+ and 42318 RETfus- NSCLC, High positive PD-L1 [100 (40) vs 13542 (32) OR 1.44/p=0.009] was significantly more in RETfus+ NSCLC.
On reviewing the distribution of the various GAs RETfus+ and RETfus- NSCLC stratified by histologic subgroups, none of them revealed a statistically significant difference. This data is shown in Table 1.
For the RETfus+ and EGFR short variant mutation positive NSCLC cases (28 total cases), the top 7 co-altered genes with both RET and EGFR were TP53 (75.9%), CDKN2A (37.9%), MTAP (27.8%), NFKBIA (27.6%), CDKN2B (24.1%), NKX2-1 (24.1%) and CTNNB1 (20.7%).
Discussion
From a very large database, we show that advanced NSCLC with RETfus+ has a unique genomic profile and may represent and unique subtype within NSCLC, both clinically and pathologically. RETfus+ occurs in less than 2% of all NSCLC. Most studies have found RETfus+ to occur between 1-2%, which is close to what we found in our analysis (0.69%) (6, 19). From the COSMIC database, RET alterations were noted in 4.06% (239/5882) in all lung cancer specimens (20). Since RET alterations can occur as acquired resistance to EGFR therapy (21), the above analysis excluded patients with EGFR alteration, which improves the validity of our analysis.
RETfus+ NSCLC in our analysis was found to have significantly lesser frequency of most targetable and un-targetable GAs. This is likely because RET fusion is the primary driver in these cases. However, understanding the molecular spectrum is important to understand treatment strategies for when the disease progresses on RET inhibitors, as well as for developing further targeted agents (22). Understanding the mutational signatures is also important to analyzing the mechanism and development of resistance to RET directed therapy (23). There are around 45 RET fusions with various partners, with KIF5B-RET being the most common. This may have treatment implications as RET inhibitors like Vandetanib and Pralsetinib may show a difference in response based on the type of fusion partner (24–26). Several mechanisms of resistance, both intrinsic and acquired have been described. MET amplification is one of the most reported mechanisms of resistance to RET inhibitors (27). MET amplification can occur even after modest treatment periods with RET inhibitors like selpercatinib, showing that mutational signatures can change with the clinical course. RET alterations can disappear and re-appear as the clinical course fluxes between response and progressive disease (28). Acquired resistance to RET via MET amplification can be overcome by combining RET inhibitors with MET directed therapy like crizotinib. Limited anecdotal evidence for this is available from preclinical studies and small case series. MET alteration may also occur as an intrinsic mutation causing primary resistance (27). Only 6 patients with MET GA in our relatively large RETfus+ cohort were found, highlighting the challenge faced in studying this phenomenon. Other MAPK-activating changes have been reported as a cause of acquired resistance. These include KRAS, BRAF and FGFR1, where were observed at a frequency of 2.39%, 1% and 0.8% in our RETfus+ cohort respectively. It can be hypothesized that RET inhibitors like Selpercatinib may eliminate the RET positive cells and create a selective pressure enabling cells with other MAPK alteration to proliferate (5). Primary resistance may be caused by alterations in PI3K pathway such as PIK3CA (2.58% in our cohort) or PTEN mutations. GA in MAPK pathway like KRAS alleles can also cause primary resistance (5). SMARCA4 (2% in our cohort) is another GA associated with primary RET resistance (29). Besides these, several gatekeeper mutations of RET itself result in both primary and secondary resistance (22). RET alteration is an important mechanism of acquired resistance to EGFR TKIs including osimertinib (30). When these co-exist, studies have hypothesized and demonstrated re-sensitization of malignant cells to EGFR inhibition when combined with RET inhibition. A similar strategy could potentially be evaluated when resistance develops to RET inhibitors, but very little data is available on the same (31). SETD2 (SET domain containing 2, histone lysine methyltransferase) occurs in about 7% of all NSCLC (32). In our RETfus+ cohort, this occurred at around 11%. SETD2 is involved in DNA methylation and DNA repair through depletion of its product, histone H3 lysine 36 (H3K36) trimethylase. These sensitize them to Hypomethylating agents which has been demonstrated in studies utilizing cell lines. Combining hypomethylating agents and PARP inhibitors may be a strategy that can be explored in the future (33).
Several advancements have been made in the past decade in the therapeutic spectrum of RETfus+ NSCLC (34). Initial trials investigated multikinase inhibitors like Cabozantinib, Lenvatinib and Vandetinib. Cabozantinib had response rates ranging between 28-50%, Vandetinib 18-47% and Lenvatinib at 16% (34). Other agents like sunitinib, sorafenib, alectinib, nintedanib, ponatinib, and regorafenib had modest responses, and were not extensively studied (34). It was only a matter of time until new selective RET inhibitors made its way, which started with the LIBRETTO-001 trial (35). 105 patients with platinum refractory or relapsed RETfus+ NSCLC were enrolled and treated with Selpercatinib. The objective response rate (ORR) was 64%. In the untreated setting, the ORR was 85% among 39 patients. Among 11 patients with CNS disease the ORR was 91% (6). Similar results, but with smaller numbers were noted with Pralisetinib (7). The NCCN guidelines recommend selpercatinib, Pralesertinib and Cabozantinib as first line options for RETfus+ NSCLC (36). Pemetrexed based chemotherapy remains the standard second line option but results with immune checkpoint therapy have yielded underwhelming results. Results on checkpoint therapy use in this setting from real-word and retrospective analysis have shown a varying result, but the consensus remains that it does not work, due to the “biologically cold” nature of RETfus+ tumors (22). Checkpoint markers in our analysis have shown conflicting results with PD-L1 high seen more and high/ultra-high TMB seen in lesser frequency in the RETfus+ cohort. STK11 was also less often seen in the RETfus+ group. These data support the hypothesis that checkpoint therapy is not efficacious in mutationally driven NSCLC and may even be detrimental if started early by augmenting toxicities like pneumonitis (37).
Lung adenocarcinoma has distinct histologic subtypes and each subtype carry unique genomic and prognostic significance (38). From our study, HG and PAP were noted more frequently in RETfus+ NSCLC. Both are high-grade variants and may be representative of aggressive disease with poor prognosis (39, 40).
Other studies have analyzed the genomic characteristics of RETfus+ NSCLC. A study on Chinese NSCLC patients showed that RETfus+ was noted 1.43% of 174 and KIF5B-RET was the most common fusion partner (41). Another study reported low frequencies of co-existing alterations like EGFR, KRAS and ALK (42). But our analysis gives a descriptive overview of other GAs that co-occur in RETfus+ NSCLC, from a relatively large cohort, which may help understand the disease biology and support future studies. Our study does carry limitations due to the lack of correlating clinical outcome data. Regardless, understanding the GA of distinct molecular subtypes of NSCLC is important as evidenced by our prior work along the same lines (11).
In summary, RETfus+ NSCLC represents a relatively rare genomic category of NSCLC that is usually devoid of other alterations. Although targetable with oral agents (6), more research is needed to determine the best beyond first line treatment options. In addition, these findings are important as a guide to increasing accuracy in genomic testing especially for small sample sizes such as fine-needle aspiration biopsies and, in the future, for patient who have had liquid biopsies that feature a low tumor fraction. The combination of histologic and genomic features can help determine if RETfus+ has been falsely negative in specimens constrained by tumor cell availability.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by SUNY Upstate Institutional Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin because No identifiable patient information has been used. Written informed consent was not obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article because No identifiable patient information has been used.
Author contributions
PA: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. MC: Data curation, Methodology, Validation, Writing – original draft, Writing – review & editing. AB: Data curation, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. DP: Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. RH: Data curation, Formal Analysis, Investigation, Methodology, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. SG: Formal Analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. JR: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
All Foundation Medicine co-authors disclose that they are employees of Foundation Medicine and own shares in Roche Holdings.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman JR, Bharat A, et al. Non–small cell lung cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Network. (2022) 20:497–530. doi: 10.6004/JNCCN.2022.0025
2. Kumar PA, Karimi M, Basnet A, Seymour L, Kratzke R, Brambilla E, et al. Association of molecular profiles and mutational status with distinct histological lung adenocarcinoma subtypes. An analysis of the LACE-bio data. Clin Lung Cancer. (2023) 24(6):528–40. doi: 10.1016/J.CLLC.2023.06.002
3. Lu S, Kato T, Dong X, Ahn M-J, Quang L-V, Soparattanapaisarn N, et al. Osimertinib after chemoradiotherapy in stage III EGFR-mutated NSCLC. New Engl J Med. (2024) 391(7):585–97. doi: 10.1056/NEJMOA2402614
4. Siegel RL, Miller KD, Fuchs HE, and Jemal A. Cancer statistics, 2022. CA Cancer J Clin. (2022) 72:7–33. doi: 10.3322/CAAC.21708
5. Rosen EY, Won HH, Zheng Y, Cocco E, Selcuklu D, Gong Y, et al. The evolution of RET inhibitor resistance in RET-driven lung and thyroid cancers. Nat Commun. (2022) 13:1–9. doi: 10.1038/s41467-022-28848-x
6. Drilon A, Oxnard GR, Tan DSW, Loong HHF, Johnson M, Gainor J, et al. Efficacy of selpercatinib in RET fusion–positive non–small-cell lung cancer. New Engl J Med. (2020) 383:813–24. doi: 10.1056/NEJMOA2005653/SUPPL_FILE/NEJMOA2005653_DATA-SHARING.PDF
7. Gainor JF, Curigliano G, Kim DW, Lee DH, Besse B, Baik CS, et al. Pralsetinib for RET fusion-positive non-small-cell lung cancer (ARROW): a multi-cohort, open-label, phase 1/2 study. Lancet Oncol. (2021) 22:959–69. doi: 10.1016/S1470-2045(21)00247-3
8. Andrini E, Mosca M, Galvani L, Sperandi F, Ricciuti B, Metro G, et al. Non-small-cell lung cancer: how to manage RET-positive disease. Drugs Context. (2022) 11:2022–1-5. doi: 10.7573/DIC.2022-1-5
9. Song Z, Yu X, and Zhang Y. Clinicopathologic characteristics, genetic variability and therapeutic options of RET rearrangements patients in lung adenocarcinoma. Lung Cancer. (2016) 101:16–21. doi: 10.1016/j.lungcan.2016.09.002
10. Klempner SJ, Bazhenova LA, Braiteh FS, Nikolinakos PG, Gowen K, Cervantes CM, et al. Emergence of RET rearrangement co-existing with activated EGFR mutation in EGFR-mutated NSCLC patients who had progressed on first- or second-generation EGFR TKI. Lung Cancer. (2015) 89:357–9. doi: 10.1016/j.lungcan.2015.06.021
11. Ashok Kumar P, Graziano SL, Danziger N, Pavlick D, Severson EA, Ramkissoon SH, et al. Genomic landscape of non-small-cell lung cancer with methylthioadenosine phosphorylase (MTAP) deficiency. Cancer Med. (2023) 12(2):1157–66. doi: 10.1002/CAM4.4971
12. Milbury CA, Creeden J, Yip WK, Smith DL, Pattani V, Maxwell K, et al. Clinical and analytical validation of FoundationOne®CDx, a comprehensive genomic profiling assay for solid tumors. PLoS One. (2022) 17:e0264138. doi: 10.1371/JOURNAL.PONE.0264138
13. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. (2013) 6(269):pl1. doi: 10.1126/SCISIGNAL.2004088
14. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. (2012) 2:401–4. doi: 10.1158/2159-8290.CD-12-0095
15. Trabucco SE, Gowen K, Maund SL, Sanford E, Fabrizio DA, Hall MJ, et al. A novel next-generation sequencing approach to detecting microsatellite instability and pan-tumor characterization of 1000 microsatellite instability–high cases in 67,000 patient samples. J Mol Diagn. (2019) 21:1053. doi: 10.1016/J.JMOLDX.2019.06.011
16. Roach C, Zhang N, Corigliano E, Jansson M, Toland G, Ponto G, et al. Development of a companion diagnostic PD-L1 immunohistochemistry assay for pembrolizumab therapy in non–small-cell lung cancer. Appl Immunohistochemistry Mol Morphology. (2016) 24:392. doi: 10.1097/PAI.0000000000000408
17. Ilie M, Khambata-Ford S, Copie-Bergman C, Huang L, Juco J, Hofman V, et al. Use of the 22C3 anti–PD-L1 antibody to determine PD-L1 expression in multiple automated immunohistochemistry platforms. PLoS One. (2017) 12(8):e0183023. doi: 10.1371/JOURNAL.PONE.0183023
18. Ashok Kumar P, Serinelli S, Zaccarini DJ, Huang R, Danziger N, Janovitz T, et al. Genomic landscape of clinically advanced KRAS wild-type pancreatic ductal adenocarcinoma. Front Oncol. (2023) 13:1169586/BIBTEX. doi: 10.3389/FONC.2023.1169586/BIBTEX
19. Wang R, Hu H, Pan Y, Li Y, Ye T, Li C, et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol. (2012) 30:4352–9. doi: 10.1200/JCO.2012.44.1477
20. Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. (2017) 45:D777–83. doi: 10.1093/NAR/GKW1121
21. Wang C, Zhang Z, Sun Y, Wang S, Wu M, Ou Q, et al. RET fusions as primary oncogenic drivers and secondary acquired resistance to EGFR tyrosine kinase inhibitors in patients with non-small-cell lung cancer. J Transl Med. (2022) 20:1–13. doi: 10.1186/S12967-022-03593-3/FIGURES/4
22. Novello S, Califano R, Reinmuth N, Tamma A, and Puri T. RET fusion-positive non-small cell lung cancer: the evolving treatment landscape. Oncologist. (2023) 28:402. doi: 10.1093/ONCOLO/OYAC264
23. Lin JJ, Liu SV, McCoach CE, Zhu VW, Tan AC, Yoda S, et al. Mechanisms of resistance to selective RET tyrosine kinase inhibitors in RET fusion-positive non-small cell lung cancer. Ann Oncol. (2020) 31:1725. doi: 10.1016/J.ANNONC.2020.09.015
24. 984P Relationship between RET fusion partner and treatment outcomes in patients (pts) with non-small cell lung cancer (NSCLC) from the phase I/II ARROW study and real-world data (RWD) - Annals of Oncology. Available online at: https://www.annalsofoncology.org/article/S0923-7534(22)02962-3/fulltext (Accessed November 12, 2023).
25. Regua AT, Najjar M, and Lo HW. RET signaling pathway and RET inhibitors in human cancer. Front Oncol. (2022) 12:932353. doi: 10.3389/FONC.2022.932353
26. Tan AC, Seet AOL, Lai GGY, Lim TH, Lim AST, Tan GS, et al. Molecular characterization and clinical outcomes in RET-rearranged NSCLC. J Thorac Oncol. (2020) 15:1928–34. doi: 10.1016/J.JTHO.2020.08.011
27. Rosen EY, Johnson ML, Clifford SE, Somwar R, Kherani JF, Son J, et al. Overcoming MET-dependent resistance to selective RET inhibition in patients with RET fusion⇓positive lung cancer by combining selpercatinib with crizotinib. Clin Cancer Res. (2021) 27:34–42. doi: 10.1158/1078-0432.CCR-20-2278/77512/AM/OVERCOMING-MET-DEPENDENT-RESISTANCE-TO-SELECTIVE
28. Zhu VW, Madison R, Schrock AB, and Ignatius Ou SH. Emergence of high level of MET amplification as off-target resistance to selpercatinib treatment in KIF5B-RET NSCLC. J Thorac Oncol. (2020) 15:e124–7. doi: 10.1016/j.jtho.2020.03.020
29. Marinello A, Vasseur D, Conci N, Fallet V, Audigier-Valette C, Cousin S, et al. 1007P Mechanisms of primary and secondary resistance to RET inhibitors in patients with RET-positive advanced NSCLC. Ann Oncol. (2022) 33:S1013–4. doi: 10.1016/j.annonc.2022.07.1133
30. Wang C, Zhang Z, Sun Y, Wang S, Wu M, Ou Q, et al. RET fusions as primary oncogenic drivers and secondary acquired resistance to EGFR tyrosine kinase inhibitors in patients with non-small-cell lung cancer. J Transl Med. (2022) 20:390. doi: 10.1186/S12967-022-03593-3
31. Piotrowska Z, Isozaki H, Lennerz JK, Gainor JF, Lennes IT, Zhu VW, et al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discovery. (2018) 8:1529. doi: 10.1158/2159-8290.CD-18-1022
32. Walter DM, Gladstein AC, Doerig KR, Natesan R, Baskaran SG, Gudiel AA, et al. Setd2 inactivation sensitizes lung adenocarcinoma to inhibitors of oxidative respiration and mTORC1 signaling. Commun Biol. (2023) 6(1):255. doi: 10.1038/S42003-023-04618-3
33. Zhou X, Sekino Y, Li HT, Fu G, Yang Z, Zhao S, et al. SETD2 deficiency confers sensitivity to dual inhibition of DNA methylation and PARP in kidney cancer. Cancer Res. (2023) 83:3813. doi: 10.1158/0008-5472.CAN-23-0401
34. Rocco D, Sapio L, Della Gravara L, Naviglio S, and Gridelli C. Treatment of advanced non-small cell lung cancer with RET fusions: reality and hopes. Int J Mol Sci. (2023) 24(3):2433. doi: 10.3390/IJMS24032433
35. Drilon A, Subbiah V, Gautschi O, Tomasini P, De Braud F, Solomon BJ, et al. Selpercatinib in patients with RET fusion-positive non-small-cell lung cancer: updated safety and efficacy from the registrational LIBRETTO-001 phase I/II trial. J Clin Oncol. (2023) 41:385–94. doi: 10.1200/JCO.22.00393
36. Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman JR, Bharat A, et al. NCCN guidelines® Insights: non-small cell lung cancer, version 2.2023. J Natl Compr Canc Netw. (2023) 21:340–50. doi: 10.6004/JNCCN.2023.0020
37. Calles A, Riess JW, and Brahmer JR. Checkpoint Blockade in Lung Cancer With Driver Mutation: Choose the Road Wisely. 2318 Mill Road, Suite 800, Alexandria, VA 22314: American Society of Clinical Oncology Educational Book ASCO (2020). pp. 372–84. pp. 372–84. doi: 10.1200/EDBK_280795.
38. Caso R, Sanchez-Vega F, Tan KS, Mastrogiacomo B, Zhou J, Jones GD, et al. The underlying tumor genomics of predominant histologic subtypes in lung adenocarcinoma. J Thorac Oncol. (2020) 15:1844. doi: 10.1016/J.JTHO.2020.08.005
39. Moreira AL, Ocampo PSS, Xia Y, Zhong H, Russell PA, Minami Y, et al. A grading system for invasive pulmonary adenocarcinoma: A proposal from the international association for the study of lung cancer pathology committee. J Thorac Oncol. (2020) 15:1599. doi: 10.1016/J.JTHO.2020.06.001
40. Yaldiz D, Acar A, Kaya Şörs, Aydoğdu Z, Gürsoy S, and Yaldiz S. Papillary predominant histological subtype predicts poor survival in lung adenocarcinoma. Turkish J Thorac Cardiovasc Surg. (2019) 27:360. doi: 10.5606/TGKDC.DERGISI.2019.17284
41. Wu G, Guo L, Gu Y, Huang T, Liu M, Zou X, et al. The genomic characteristics of RET fusion positive tumors in Chinese non-small cell lung cancer (NSCLC) patients. J Cancer Res Clin Oncol. (2023) 149:1019–28. doi: 10.1007/S00432-022-03959-6/FIGURES/6
Keywords: non-small cell lung cancer, RET fusion, targeted therapy, genomic profiling, lung cancer
Citation: Ashok Kumar P, Connolly M, Basnet A, Pavlick D, Huang R, Graziano S and Ross J (2025) RET fusion driven (RETfus+) non-small cell lung cancer: a comprehensive genomic profiling study with histologic correlation. Front. Oncol. 15:1477910. doi: 10.3389/fonc.2025.1477910
Received: 08 August 2024; Accepted: 16 April 2025;
Published: 16 May 2025.
Edited by:
Francesco Facchinetti, Dana–Farber Cancer Institute, United StatesReviewed by:
Akshita Bhatt, American Association For Cancer Research, United StatesXiangliang Liu, The First Hospital of Jilin University, China
Luis Mas, Auna Oncosalud, Peru
Copyright © 2025 Ashok Kumar, Connolly, Basnet, Pavlick, Huang, Graziano and Ross. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prashanth Ashok Kumar, YXNob2trdXBAdXBzdGF0ZS5lZHU=
†ORCID: Prashanth Ashok Kumar, orcid.org/0000-0001-6236-2822
 Prashanth Ashok Kumar
Prashanth Ashok Kumar Michael Connolly
Michael Connolly Alina Basnet
Alina Basnet Dean Pavlick
Dean Pavlick Richard Huang
Richard Huang Steven Graziano
Steven Graziano Jeffrey Ross1,2,3
Jeffrey Ross1,2,3