- 1Department of Urology, Mianyang Central Hospital, School of Medicine, University of Electronic Science and Technology of China, Mianyang, China
- 2National Health Commission (NHC) Key Laboratory of Nuclear Technology Medical Transformation, Mianyang Central Hospital, School of Medicine, University of Electronic Science and Technology of China, Mianyang, China
- 3Department of Hepatobiliary Surgery, Mianyang Central Hospital, School of Medicine, University of Electronic Science and Technology of China, Mianyang, China
Prostate cancer (PCa) is one of the most common urological malignancies in older male patients. Castration-resistant prostate cancer (CRPC) is an aggressive and refractory stage of PCa and is the leading cause of PCa-related deaths. Exosomes are small spherical vesicles with a lipid bilayer membrane structure, secreted by cells, which carry large amounts of nucleic acids, proteins, lipids, and various important reactive small molecules. Numerous studies have demonstrated that exosomes are involved in the development of CRPC by delivering various biomolecules that regulate biological processes in recipient cells. Despite the advancement in treatments, CRPC remains poorly managed, underscoring the urgent need for novel treatment strategies.As research into exosomes continues, they have shown significant potential in the diagnosis and treatment of CRPC.Unlike previous reviews,this review not only provides an overview of exosomes but also comprehensively explores their role in the CRPC tumor microenvironment, angiogenesis, immune escape, metastasis, and drug resistance, with a focus on the potential value of exosomes in the diagnosis and treatment of CRPC.The literature review includes studies published up to June 2024, and the search strategy involved exosomes, CRPC, diagnosis,and treatment using Pubmed.
1 Introduction
Prostate cancer (PCa) ranks as one of the most prevalent forms of malignant tumors affecting the male genitourinary system globally. In 2023, PCa had the highest incidence rate among cancers in the United States, accounting for 29% of cases, and it is the second leading cause of death among men in the United States (1). Although the incidence of PCa is lower in Asia than in Europe or the United States, it continues to rise (2). Early PCa lacks apparent symptoms, leading to most patients being diagnosed at the middle to late stages of the disease when the opportunity for radical surgery is lost (3, 4). The survival rate of patients with PCa depends on early diagnosis and treatment (5). PCa screening (6). Despite improving the early diagnostic rate of PCa, prostate-specific antigen (PSA) has drawbacks, such as low specificity and susceptibility to false-positive results (7). PCa is androgen-dependent, and androgen deprivation therapy (ADT) is currently the mainstay of treatment for PCa, with most patients achieving desirable results early in endocrine therapy. However, as the disease progresses, most patients progress to the more aggressive form known as castration-resistant prostate cancer (CRPC) after an average of 2 years of treatment (8).
CRPC occurs when the original ADT loses its therapeutic effect on the tumor, and the tumor tissue continues to grow normally in a substantially low androgen environment (9). CRPC can be divided into non-metastatic and metastatic, with metastatic CRPC often progressing from non-metastatic CRPC and being prone to distant metastasis, such as to the bone, lymph nodes, lungs, liver, and other tissues, which portends worse prognosis and survival (10–12). The current major mechanisms of CRPC development include persistent activation of the androgen receptor (AR) signaling pathway, genetic and epigenetic alterations, and changes in the tumor microenvironment (13–15). However, the exact mechanism by which CRPC develops remains unclear. The diagnosis of CRPC is based on two main conditions: serum testosterone reaching desmoplastic levels and disease progression continuing after testosterone reaches desmoplastic conditions (16). Therapeutic approaches for CRPC include novel endocrine therapeutic agents, radiopharmaceuticals, immunotherapeutic agents, and therapeutically relevant drugs for bone metastases (16). Nonetheless, no cure exists for CRPC, and the main goals of treatment are to control symptoms and prolong life. Currently, there are no widely recognized biomarkers that can guide the diagnosis and treatment of CRPC. Therefore, identifying new diagnostic and therapeutic targets is imperative for the effective treatment of this class of malignant tumors.
Exosomes are minuscule vesicles, ranging from 30 to 150 nanometers in diameter, released by various types of cells. These vesicles are generated through the fusion of multivesicular bodies (MVBs) with the cell membrane (17, 18). They are rich in a diverse array of biological macromolecules, including proteins, lipids, and nucleic acids, all of which contribute significantly to processes such as tumor formation, progression, and diagnosis (19). Exosomes play an important role in intercellular information transfer by releasing active substances that bind to cell membranes or cell surface receptors (20). Additionally, exosomes released by tumor cells can form a premetastatic microenvironment in distal organs and regulate immune cells and factors in the tumor microenvironment, thus promoting immune escape from the tumor (21). They also participate in the metabolic exchange between tumor cells and other cells in the microenvironment, affecting the energy and material metabolism of tumors (22). Exosomes can promote tumor metastasis by promoting tumor neovascularization, increasing vascular permeability, and promoting the epithelial-mesenchymal transition (EMT) of tumor cells (23). Moreover, exosomes contain various genetic materials through which genetic information can be transmitted between cells to influence tumor progression and drug resistance (24). Exosomes are ideal drug delivery systems for oncological therapeutics because of their natural biocompatibility and targeting properties (25). Overall, exosomes play an important role in tumorigenesis, tumor development, and therapy, affecting the biological behavior of tumors through various mechanisms and providing new ideas and targets for tumor diagnosis and treatment. Therefore, this study reviews the value of exosomes in the diagnosis of CRPC and their potential therapeutic applications.
2 Overview of exosomes
Based on their origin, size, and biogenesis, extracellular vesicles are primarily classified into exosomes, microvesicles, and apoptotic bodies (17). Exosomes represent an important subset of extracellular vesicles. Exosomes are nanoscale vesicles secreted around cells by phospholipid bilayer teato-like structures (26). They were discovered in 1983 in sheep reticulocytes and named “exosomes” by Johnstone in 1987 (27, 28). Originally, exosomes were thought to serve as a means for cells to excrete waste products; however, in 2007, researchers began to discover that exosomes have diverse functions (29). Almost all cells secrete exosomes, which are found in various body fluids, including blood, lymph fluid, semen, urine, saliva, cerebrospinal fluid, and breast milk (30–36). Exosome production is a multi-step, multifactorial process. It begins with the formation of MVBs inside the cell, some of which fuse with the cytoplasmic membrane and secrete intraluminal vesicles (ILVs) outside the cell through the outgrowth process to form exosomes (with a diameter of approximately 30–150 nm) (37). Microvesicles, predominantly formed through outward “budding” of the plasma membrane, exhibit a larger diameter ranging approximately from 100 to 1000 nm (17). Apoptotic bodies, primarily generated through plasma membrane fragmentation during apoptosis, represent the largest extracellular vesicles with diameters ranging from approximately 50 to 5000 nm (17). The biogenesis of exosomes is regulated by various biomolecules and organelles. Exosomes carry substances such as proteins, lipids, and nucleic acids that act directly on receptor cells to regulate their biological activities (18). Exosomes are heterogeneous and their function depends on component-selective sorting; even exosomes secreted by the same cells are functionally distinct (38). Moreover, as natural nano-biological drug carriers, exosomes can achieve precise targeted drug delivery, improve bioavailability, and reduce toxic side effects (39). Additionally, analyzing the composition of exosome contents allows for the identification of the cell type from which they originate, a property that can be developed as a biomarker for the early diagnosis of clinical diseases and therapeutic monitoring (40).
2.1 Formation and secretion of exosomes
The mechanism of exosome formation is complex and consists of several steps in the endocytosis pathway (Figure 1). Initially, the cell membrane undergoes invagination, leading to the formation of early endosomes that contain cell-surface proteins and soluble proteins linked to the extracellular space (41). These early endosomes then develop into late endosomes via a process of double invagination of the plasma membrane, eventually maturing into MVBs (42). The diameter of MVBs is approximately 250–1000 nm; hence, MVBs can carry multiple ILVs with diameters of 30–150 nm (43). MVBs have two possible pathways: they can either merge with lysosomes or autophagosomes, leading to their degradation, or they can fuse with the plasma membrane. In the latter case, the ILVs within the MVBs are expelled as exosomes (44). The formation and transport of ILVs and MVBs require the involvement of multiple proteins. The classical protein-dependent pathway are endosomal sorting complexes required for transport (ESCRT) (45). ESCRT is a protein complex localized on the cytoplasmic side of endosomes that regulates the formation of ILVs and access to their components (18). Small GTPases in the Rab family play important roles in the intracellular transport chain (46). Syndecan and syntenin affect the targeting of transmembrane proteins to ILVs in MVBs through protein interactions (47). Notably, there is another non-dependent ESCRT exosome biodiscovery pathway in which four transmembrane proteins directly regulate the transport, membrane partitioning, and function of heat shock proteins that act as molecular chaperones (48).
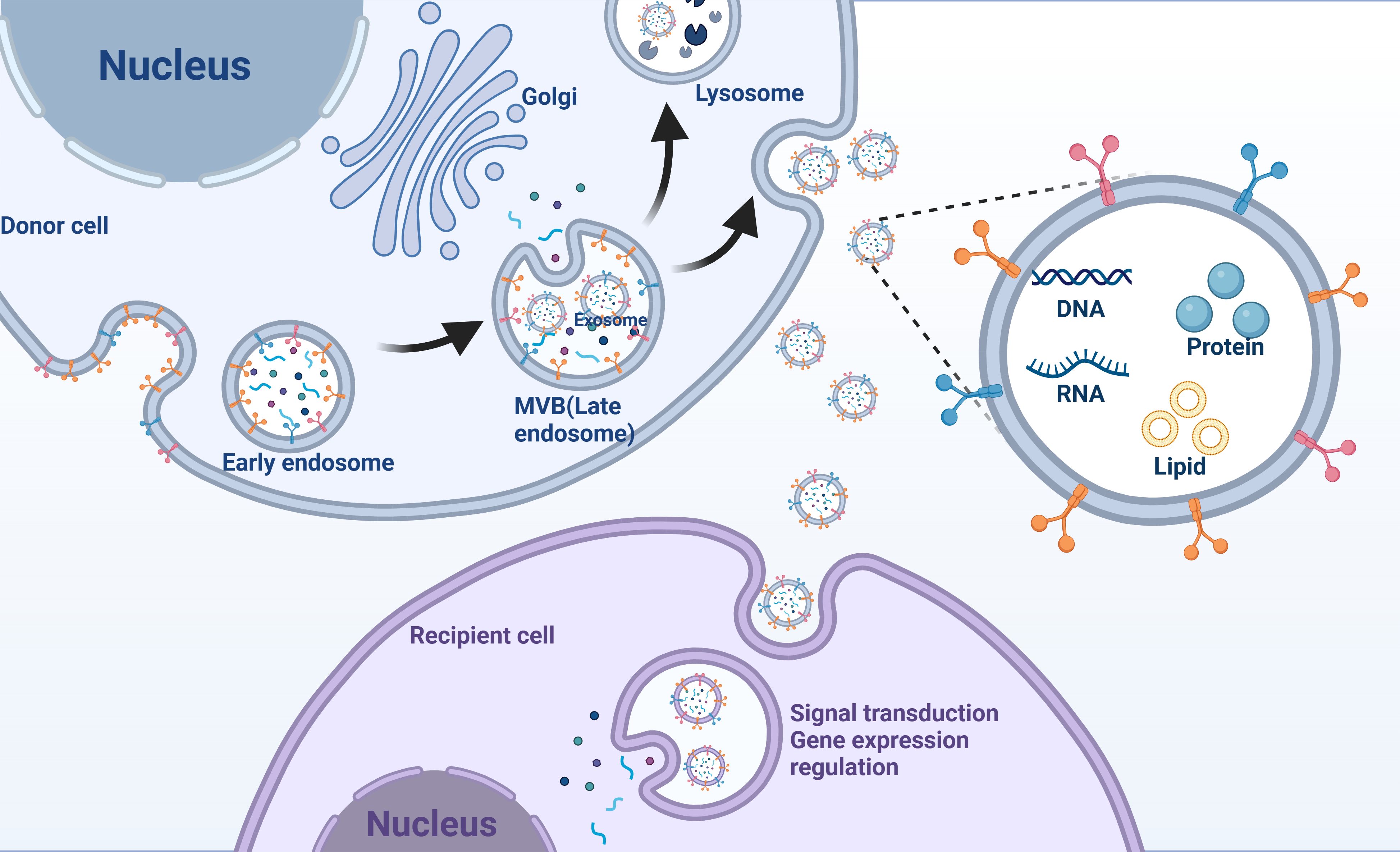
Figure 1. Exosomes mediate intercellular communication. Exosomes originate from early endosomes and subsequently form MVBs (multivesicular bodies). MVBs fuse with the cell membrane, releasing exosomes that enter recipient cells, carrying nucleic acids, proteins, and lipids involved in signal transduction or gene expression regulation.
2.2 Contents of exosomes
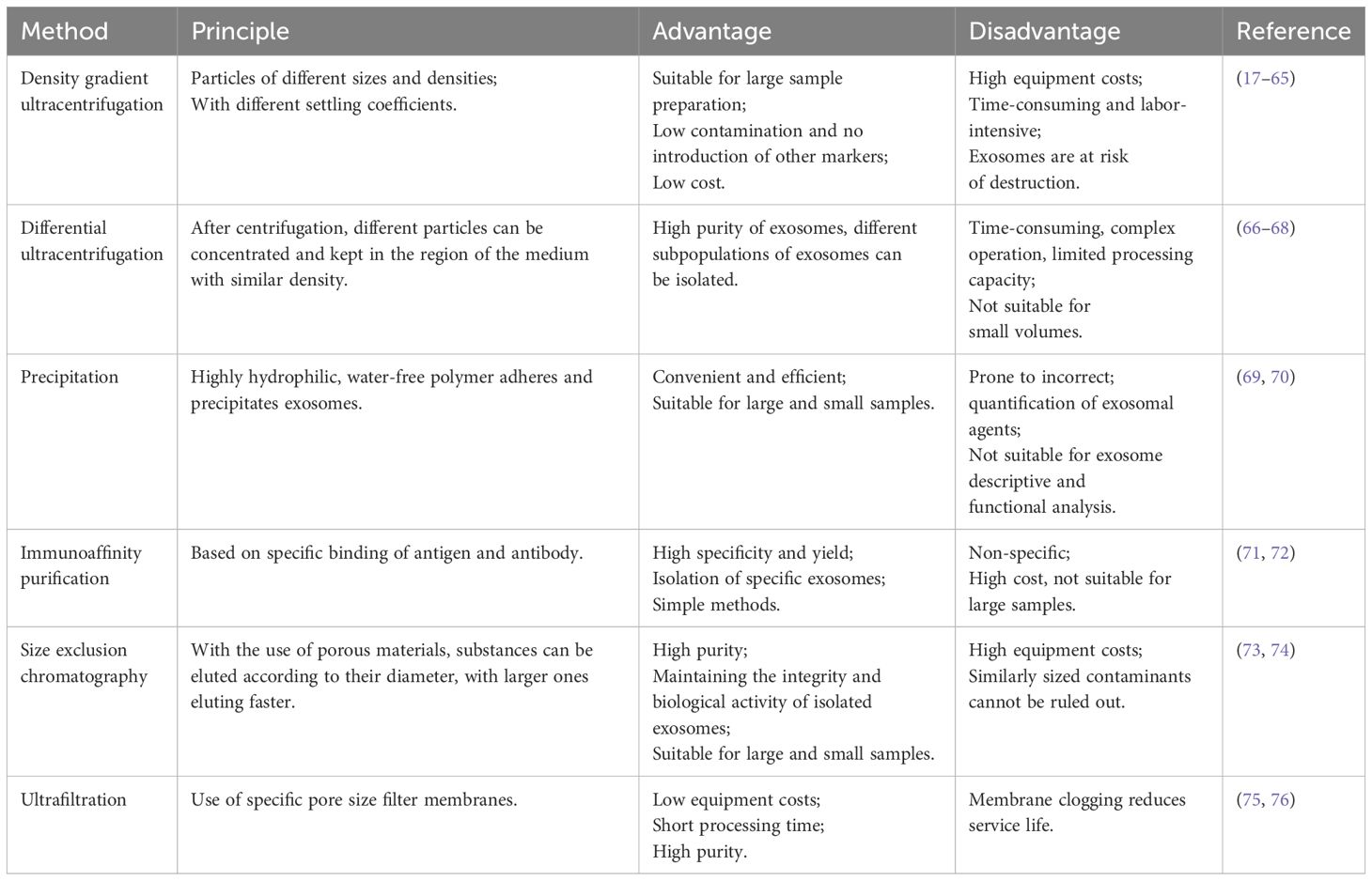
When viewed under an electron microscope, the exosome membrane, similar to that of cells, is a phospholipid bilayer structure containing various components, including nucleic acids, proteins, lipids, and small-molecule metabolites (ATP, amino acids, sugars, and amides) (Table 1) (59, 60). Nucleic acids in exosomes mainly include miRNA, DNA, long-stranded non-coding RNA (lncRNA), circular RNA (circRNA), mRNA, and interfering RNA (siRNA), which are involved in the remote transmission of genetic material and the diagnosis of diseases (49–54). The proteins mainly include tetraspanins (including CD9, CD63, CD81, and CD82), heat shock proteins (such as Hsp60, Hsp70, and Hsp90), membrane transporters and fusion proteins (Annexins (I, II, IV, V, VI), Rab proteins (GTPases), proteins incorporated during MVB formation (Alix, Tsg101, Clatherin), signaling proteins (EGFR, HIF-1a, ARF1, PI3K), lipid raft (Flotillins, Ceramide, Cholesterol), antigen presentation (MHC I, II, IV, VI), and antigen presentation (MHC I, MHC II), cytoskeletal proteins (actin, myocin), which are involved in the process of exosome formation and release (55, 56). Exosomal lipids are one of the least studied but most important compoents of exosomes (61). Exosomes are rich in lipids, primarily sphingolipids, sphingomyelins, phosphatidylserine, phosphatidylinositol, cholesterol, ceramides, diacylglycerides, and diphosphatidic acid (57, 58). Lipids not only play an important role in the structure of exosome membranes but also facilitate exosome formation and release into the extracellular environment (57). Similarly, small metabolic molecules in exosomes play important roles. Although less diverse compared to nucleic acids and proteins, these small metabolic molecules are involved in cell-to-cell proximity communication and are suitable biomarkers for disease diagnosis (18).
2.3 Isolation and identification of exosomes
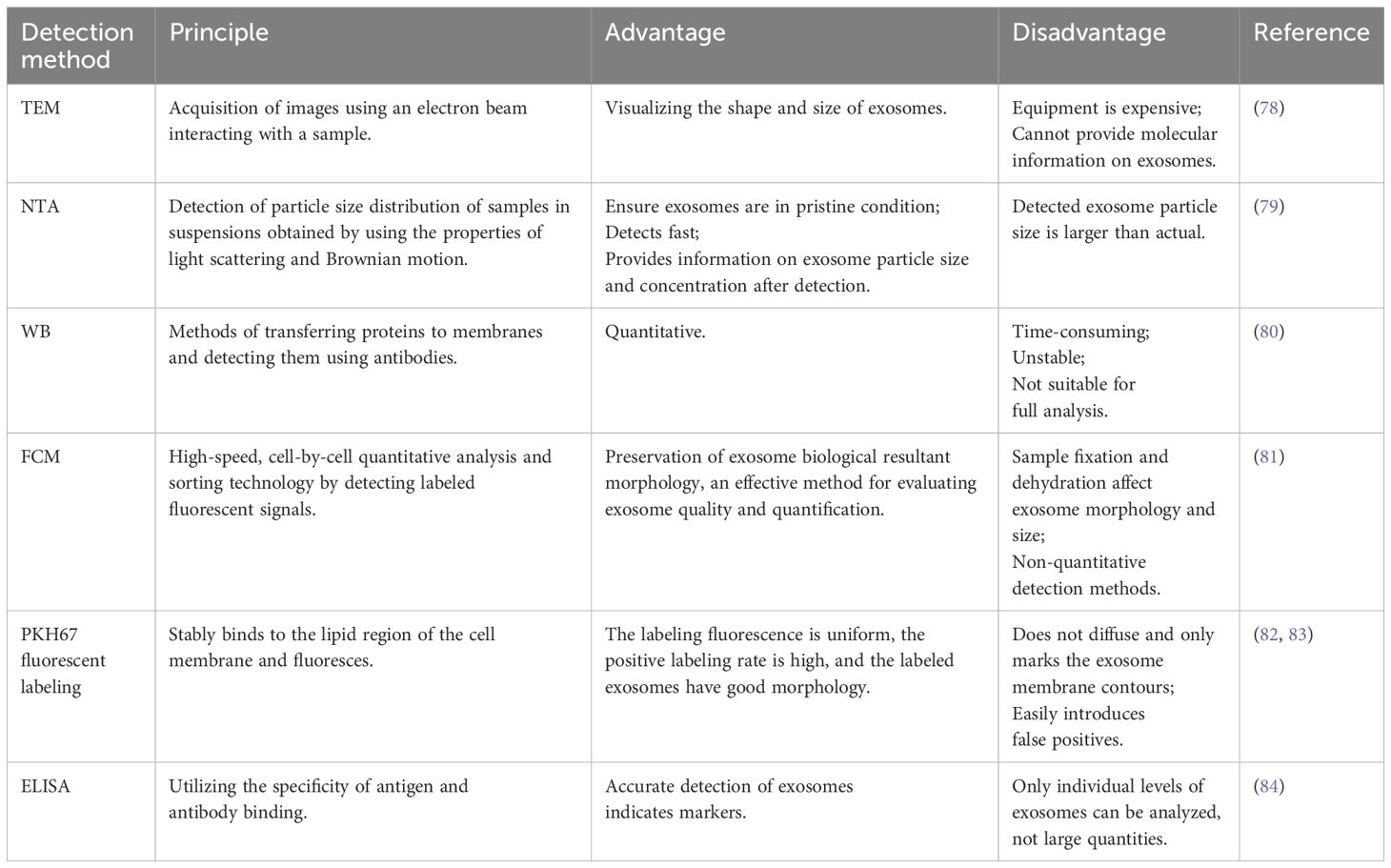
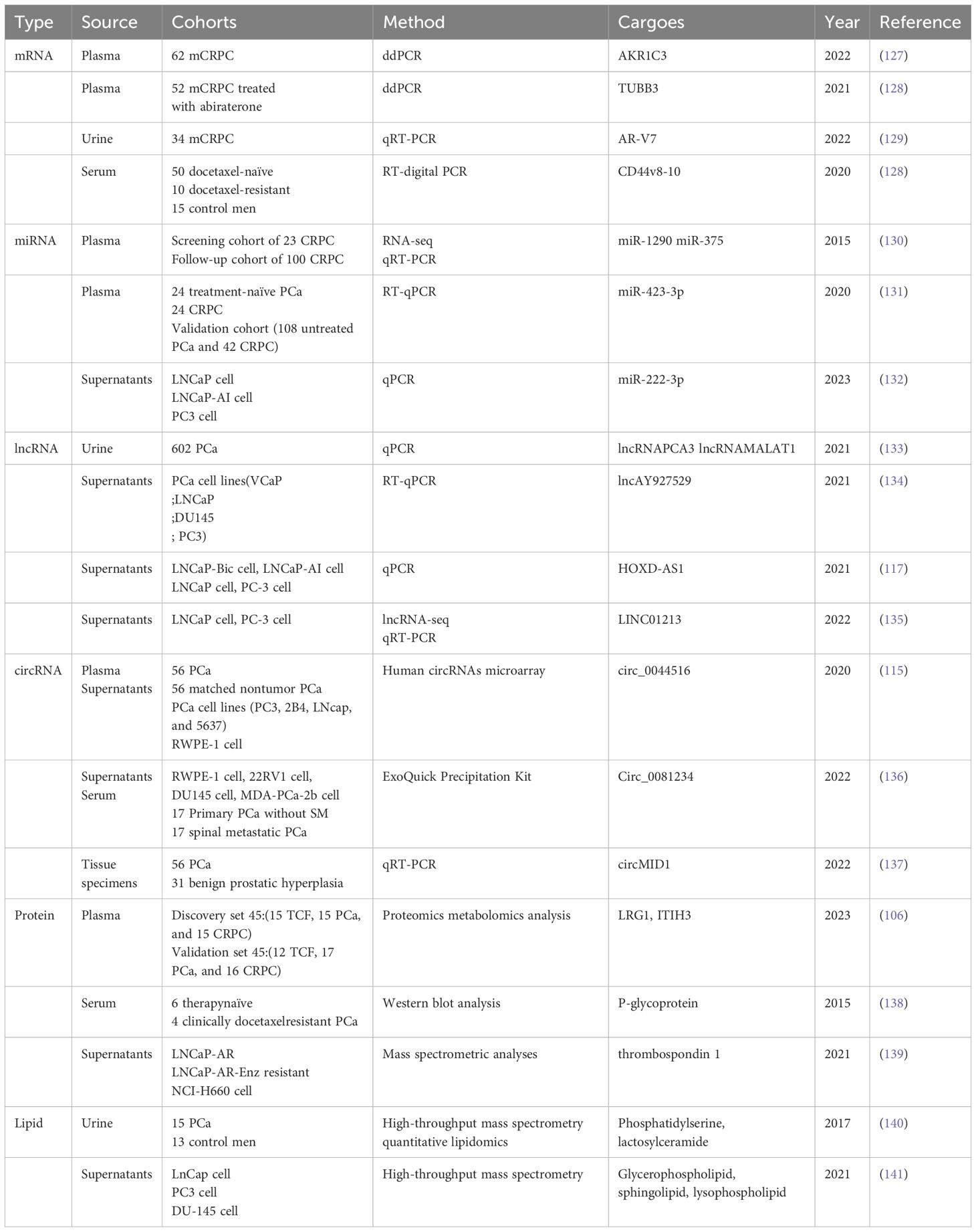
The isolation of exosomes from biological fluids is mainly based on their size, density, and immunological properties (62). Currently, exosomes are separated and purified using ultracentrifugation, precipitation, immunoaffinity purification, size exclusion chromatography, and ultrafiltration (63). Each method has its advantages and disadvantages (64). For example, ultracentrifugation is recognized as the “gold standard” in exosome isolation methods. It is also the most widely used method for exosome isolation and purification, with advantages such as suitability for preparing large samples, small contamination, no introduction of other markers, and low cost. However, it has shortcomings, including the high cost of equipment, being time-consuming, labor-intensive, and carrying the risk of destroying exosomes (65). Ultracentrifugation can be classified into density-gradient centrifugation and ordinary differential ultracentrifugation (63). Further details are presented in Table 2. Exosomes are identified mainly based on factors such as particle size, concentration, morphology, and marker-carrying capacity (77). Currently, the common methods of identification include transmission electron microscopy (78), nanoparticle tracking analysis (79), western blot (80), flow cytometry (81), PKH67 fluorescent labeling (82, 83), and enzyme-linked immunosorbent assay (84). Transmission electron microscopy can visualize the shape, size, and partial structure of exosomes, among other advantages. However, its disadvantages include the expensive equipment and an inability to provide molecular information on exosomes (78). For further details, please refer to Table 3. Different isolation and identification methods have their advantages and disadvantages, and suitable combinations of methods must be selected according to the specific research objectives and sample characteristics. With continuous technological progress, exosome isolation and identification methods are also being updated and improved, providing more possibilities for exosome research and utilization.
2.4 Function of exosomes
Exosomes are packed with numerous biologically active substances, including proteins, lipids, nucleic acids, and metabolites. These components are engaged in a variety of intricate physiological and pathological processes, such as cancer development, immune responses, pregnancy, cardiovascular conditions, viral infections, and neurodegenerative disorders (29, 85–89). Exosomes influence biological processes through two primary mechanisms. The first involves direct interaction, where surface proteins or lipid ligands on the exosome engage directly with receptors on the target cell’s membrane. This leads to the formation of signaling complexes and the activation of intracellular signaling pathways. The second mechanism is the direct fusion of exosomes with the target cell’s lipid bilayer, allowing the transfer of their own proteins, nucleic acids, lipids, and other active molecules into the cell. This process affects cellular functions and biological behaviors. In the context of physiological and pathological mechanisms, exosomes derived from tumor cells, tumor-associated immune cells (such as myeloid-derived suppressor cells (MDSCs)), and tumor-associated stromal cells (like tumor-associated fibroblasts) play a role in modifying the tumor microenvironment. They contribute to angiogenesis, immune evasion, tumor progression and metastasis, and are involved in the development of drug resistance (90). For example, exosomes with circPACRGL released by colorectal cancer cells can promote colorectal cancer proliferation, migration, and invasion by regulating the miR-142-3p/miR-506-3p-TGF-β1 axis (91). By releasing exosomes, tumor cells can regulate the immunosuppressive function of MDSCs, thereby affecting tumorigenesis and progression. For example, activation of MDSCs by HSP70-containing exosomes from renal cancer cell sources can exert antigen-specific immunosuppressive effects on cytotoxic T lymphocytes, which specifically promote renal tumor growth and immune escape (92). Exosomes secreted by resistant tumors can induce resistance to chemotherapy. The secretion of lincROR-rich exosomes by PCa cells can propagate the chemoresistant phenotype to recipient cells and promote PCa resistance to doxorubicin (93).
3 Exosomes and castration-resistant prostate cancer
Various roles for exosomes in CRPC have been elucidated (Figure 2).
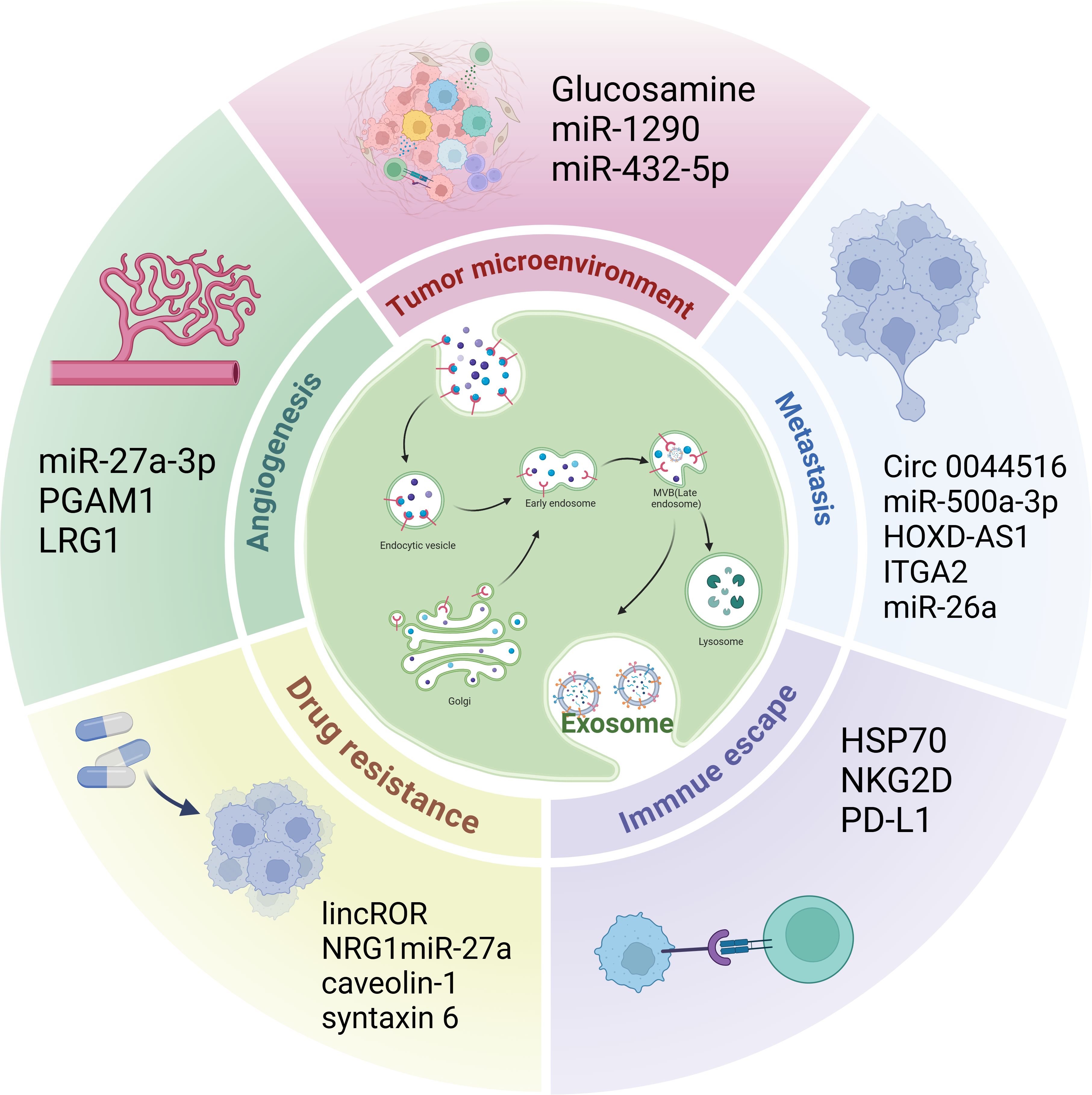
Figure 2. The role of exosomes in CRPC. Exosomes play a role in the CRPC tumor microenvironment, angiogenesis, immune escape, metastasis, and drug resistance.
3.1 Tumor microenvironment
Tumor progression is intricately linked to the tumor microenvironment. Key elements of this environment comprise stromal cells (including cancer-associated fibroblasts (CAFs) and mesenchymal stromal cells), immune cells (such as tumor-associated macrophages, neutrophils, dendritic cells, and regulatory T-cells), vascular cells (like pericytes), and the extracellular matrix. These components interact with each other, facilitating both tumor initiation and advancement (94). CAFs are among the most important cellular components of the tumor microenvironment (95). Several studies have shown that CAF-derived exosomes play important roles in CRPC progression. For example, Cui et al. (96) found that, in PCa cells, glucosamine secreted by CAFs promotes the elevation of O-GlcNAcylation, a post-translational modification that attaches O-conjugated N-acetylglucosamine to proteins to orchestrate nutritional and stress responses (97). This modification further promotes EIK1-induced transcription of HSD3B1, thereby inducing androgen synthesis, activating ARs, and promoting the progression of CRPC. Wang et al. (98) identified ADT-induced changes in CAF subtypes, such as SPP1+ myofibroblasts, as key stromal components driving the development of desmoplasia-resistant PCa. Wang et al. (99) discovered that elevated levels of exosomal miR-1290, originating from CAFs in PCa, markedly increased PCa cell migration, invasion, and EMT. Subsequent research has shown that exosomal miR-1290 facilitates PCa cell proliferation and metastasis by targeting and inhibiting GSK3β mRNA, thereby disrupting the GSK3β/β-catenin signaling pathway. Additionally, exosomal miR-432-5p secreted by CAFs promotes PCa resistance to docetaxel (100). CAF reprogramming is considered a key factor driving the development of CRPC. CAFs play a multifaceted role in the tumor microenvironment through their secreted exosomes, including the promotion of tumor growth, metastasis, and chemotherapy resistance. Therefore, these studies highlight the importance of therapeutic strategies targeting the tumor microenvironment, especially CAFs and their exosomes, to improve the therapeutic efficacy of CRPC.
3.2 Angiogenesis
During tumorigenesis and development, tumor tissues require neovascularization to raise sufficient nutrients and oxygen for infinitely proliferating tumor cells (101). Angiogenesis is a crucial factor in the development and advancement of CRPC (102). Numerous studies have demonstrated that exosomes originating from tumors significantly enhance angiogenesis (103). Prigol et al. (104) found that the human PCa cell line PC-3-derived exosome miR-27a-3p, treated with a vascular endothelial cell line derived from the human umbilical vein, showed enhanced invasive and angiogenic capacities. Luo et al. (105) reported that Phosphoglycerate mutase 1 PCa-derived exosomes bind to γ-actin and promote peduncle formation and neovascularization sprouting in human umbilical vein endothelial cells, thereby enhancing lung metastasis in vivo. Additionally, Liu et al. (106) found that leucine-rich alpha2-glycoprotein 1 (LRG1) derived from exosomes in LNCaP cells promotes tube formation in human umbilical vein endothelial cells, suggesting that exosomal LRG1 protein may be involved in CRPC angiogenesis. Despite the crucial role of angiogenesis in CRPC, clinical studies on CRPC have demonstrated that anti-angiogenic therapy has not resulted in the expected clinical benefit but rather increased toxicity (107). Current therapies targeting anti-angiogenesis in CRPC are poorly developed, and the above studies have revealed the key role of exosomes in CRPC progression, especially in promoting neovascularization, suggesting that exosomes have the potential for anti-angiogenic therapy in CRPC.
3.3 Immune escape
Several studies have demonstrated that tumor-derived exosomes contribute to immune escape and promote tumor cell proliferation and metastasis (108). The immune escape mechanism in CRPC is complex. First, the tumor immune microenvironment in CRPC is highly immunosuppressive, hindering the function of the immune system and allowing tumors to escape immune surveillance and attacks. Second, recognition by the immune system can be avoided by inhibiting immune cell activity or selectively expressing hypoimmunogenic antigens, making monoimmunotherapy a poor option in patients with CRPC (109). The accumulation of MDSCs at high concentrations in the CRPC tumor microenvironment is an important reason for CRPC immune escape (110). Li et al. (111) showed that HSP70-enriched exosomes derived from the mouse PCa cell line, RM-1, enhanced the activation of the TLR2/NF-κB signaling pathway, which resulted in the upregulation of CXCR4 expression in MDSCs and the accumulation of MDSCs in the tumor microenvironment, accelerating immune escape. Natural killer cells are the first line of defense against cancer and play crucial roles in anti-tumor activity and immune surveillance (112). Lundholm et al. (113) found that plasma-derived exosomes from patients with CRPC downregulate the expression of NKG2D on natural killer cells, which may be one of the mechanisms of immune escape from CRPC cells. Li et al. found that higher levels of programmed death ligand 1 (PD-L1) were detected in exosomes derived from the highly malignant PC3 and DU145 cell lines and that PD-L1 can inhibit CD8+ T cell function to help PCa tumor cells evade the immune system. Therefore, inhibiting exosome secretion or preventing PD-L1 sorting into exosomes may improve the response of CRPC cells to anti-PD-L1 therapy (114). These findings provide important biological information for immunotherapy using exosomes in CRPC, which will help develop more precise therapeutic strategies and improve efficacy.
3.4 Cancer cell and metastasis
Metastasis is the main cause of poor tumor prognosis and survival. Numerous investigations have indicated that exosomes serve as crucial regulators in the metastasis of PCa (105). For instance, circ_0044516, found in exosomes, is markedly upregulated in both PCa patients and cell lines, and its functional downregulation has been shown to impede PCa cell metastasis (115). Additionally, recent research discovered that exosomal miR-500a-3p, which is secreted by CAFs, is significantly increased in hypoxic conditions. This upregulation promotes PCa metastasis in both in vivo and in vitro studies, with further analysis revealing that CAF-derived exosomes facilitate PCa metastasis through the miR-500a-3p/FBXW7/HSF1 signaling pathway (116). Furthermore, in a bone metastasis mouse model, Jiang et al. demonstrated that the exosome HOXD-AS1 derived from CRPC cells accelerates distant PCa metastasis in vivo (117). The EMT is well-established as a principal mechanism driving cancer cell migration and invasion (118). Gaballa et al. found that CRPC cell-derived exosomal ITGA2 promoted the invasive behavior of AR-positive PCa cells through the EMT pathway (119). Wang et al. (120) reported that exosomal miR-26a overexpression inhibited the migration and invasion ability of the mCRPC cell line (PC-3). By assessing the levels of proteins associated with the EMT, they proposed that exosomal miR-26a might play a role in suppressing the EMT process in PCa. These studies suggest that exosomes play an important role in the metastasis of CRPC, providing new directions and methods of treatment to inhibit CRPC metastasis.
3.5 Tumor drug resistance
Drug resistance in tumors is a major challenge affecting the efficacy of current oncology drugs. The emergence of drug resistance in CRPC is mainly caused by alterations in the AR signaling pathway and the influence of the microenvironment (121, 122). Exosomes play an important role in the mechanism of CRPC resistance and can promote chemotherapy resistance in CRPC (123). For example, lincROR is highly expressed in doxorubicin-resistant PCa cell lines. LincROR is packaged into exosomes in an hnRNPA1-dependent manner, which then propagates the chemoresistant phenotype to recipient cells to promote PCa resistance to doxorubicin (93). Zhang et al. (15) demonstrated that neuregulin 1 (NRG1) can promote PCa resistance to anti-androgen therapy. Further studies revealed that CAFs activate human epidermal growth factor receptor 3 (HER3) in tumor cells by secreting NRG1, and pharmacological blockade of the NRG1-HER3 axis can inhibit the development of PCa hormone resistance. Additionally, Cao et al. (124) found that exosomal miR-27a produced by primary prostate fibroblasts (PSC-27) ameliorates PCa resistance to chemical drugs by negatively regulating p53 expression. Lin et al. (125) showed that mCRPC-derived exosome caveolin-1 induces radiation and chemoresistance in recipient cells. Further, Peak et al. found that exosome syntaxin 6 secreted by mCRPC increased the survival of PCa cells resistant to enzalutamide (126). The study of exosome-mediated resistance in CRPC not only helps to elucidate the resistance mechanism of CRPC but also provides a basis for the diagnosis and treatment of CRPC.
4 Exosomes in the diagnosis of castration-resistant prostate cancer
Compared to traditional CRPC biomarkers, exosomes and their contents (including proteins, nucleic acids, and lipids) are rich in biological information and can provide more relevant specific biomarker information. Monitoring CRPC blood and urine for specific biomarkers derived from exosomes would be a convenient, non-invasive diagnostic method to track disease progression (Table 4).
4.1 mRNA
Exosomes transport large amounts of mRNA to act on receptor cells (142). Various exosomal mRNAs are highly expressed in patients with CRPC. Zhu et al. measured plasma samples from 62 patients with mCRPC using digital droplet polymerase chain reaction (ddPCR) and showed positive expression of exosomal AKR1C3 mRNA. Further studies revealed that exosomal AKR1C3 mRNA was associated with poor overall survival and progression-free survival under first-line abiraterone use (ABI-PFS) prognosis in patients (overall survival: 16.2 vs. 32.5 months), P < 0.001; ABI-PFS: 3.9 vs. 10.1 months, P < 0.001, suggesting that it can be used as a biomarker for mCRPC (127). Zhu et al. also used ddPCR to detect plasma exosomal TUBB3 mRNA expression in 52 abiraterone-treated patients with mCRPC and found that strong positive exosomal TUBB3 mRNA correlated with poor prostate-specific antigen-PFS prognosis in mCRPC patients treated with abiraterone (7.9 months for positive vs. 11.0 months for no TUBB3, P = 0.014) (128). Similarly, Wang et al. examined the urinary exosomal AR-V7 mRNA status of 34 patients with mCRPC using quantitative real-time polymerase chain reaction assay (qRT-PCR). They found that 32.4% of the urine samples were positive for urinary exosomal AR-V7 mRNA, suggesting that exosomal AR-V7 mRNA can be used as a diagnostic marker for CRPC, especially in abiraterone-resistant (ABI-Res) patients with CRPC (129). Kato et al. utilized RT digital PCR to evaluate the levels of cluster of differentiation 44 variant 8-10 (CD44v8-10) mRNA in the blood of 50 patients with PCa initially treated with docetaxel, 10 patients with CRPC who were resistant to docetaxel, and 15 individuals undergoing prostate biopsies, while excluding males in the PCa control group. Their results revealed that serum exosomal CD44v8–10 mRNA was significantly elevated in docetaxel-resistant patients with CRPC compared to docetaxel-naïve and control groups (median values: 46, 12, and 17 copies/mL serum, respectively; P = 0.032, Kruskal–Wallis test), indicating that serum extracellular vesicle CD44v8–10 mRNA may be a diagnostic biomarker for docetaxel-resistant CRPC (143). These studies suggest that exosomal mRNA levels can be used as molecular markers for the diagnosis of CRPC.
4.2 MiRNA
MiRNAs can act as oncogenic and carcinogenic agents involved in tumor development (144). Exosomes regulate gene expression and distal cell functions by transporting miRNAs that mediate paracrine and endocrine communication between tissues (145). Huang et al. analyzed miRNA sequencing in 23 patients with CRPC to identify and assess the prognostic impact of plasma exosomal miRNAs. They found that high expression of exosomal miR-1290 and miR-375 was significantly associated with worse overall survival in patients with CRPC and that these miRNAs can be used as biomarkers for the diagnosis of advanced CRPC (130). Similarly, Guo et al. conducted RNA sequencing and RT-qPCR validation of plasma exosomes from 24 patients with primary PCa and 24 patients with CRPC, as well as RT-qPCR validation of a validation cohort (108 patients with untreated PCa and 42 patients with CRPC). Their findings revealed that exosomal miR-423-3p exhibited the most significant differential expression. This finding was also validated in a separate study center of 30 patients with untreated PCa and 30 patients with CRPC, suggesting that miR-423-3p has the potential to serve as a biomarker for the early detection and prediction of CRPC (131). Wang et al. (132) used qPCR to detect the expression level of exosomal miR-222-3p in androgen‐dependent PCa and androgen‐independent prostate cancer (AIPC) cells. They found that exosomal miR-222-3p was highly expressed in AIPC cells and that exosomal miR-222-3p could promote the transformation of androgen‐dependent PCa cells to AIPC cells. They further analyzed the expression level of AIPC cells using bioinformatic databases and whole transcriptome sequencing analysis and found that exosomal miR-222-3p promoted CRPC progression by targeting midnolin to activate mTOR signaling, suggesting that exosomal miR-222-3p is a promising biomarker for the early diagnosis of CRPC. These studies not only reveal the mechanism of action of exosomal miRNAs in CRPC progression but also provide directions for the development of novel diagnostic biomarkers.
4.3 LncRNA
Owing to the genome-wide expression patterns of lncRNAs in various tissues and their tissue-specific expression characteristics, they hold great promise as novel tumor markers (146). LncRNAs can enter the human circulatory system via exosomes and are stable in body fluids such as blood and urine, making them potential tumor diagnostic biomarkers (147). Li et al. examined the expression levels of urinary exosomes lncRNA PCa antigen 3 and lncRNA metastasis-associated lung adenocarcinoma transcript 1 using qPCR in 602 patients with PCa clarified by prostate puncture biopsy. They found that these urinary exosomes were overexpressed in PCa and the more aggressive PCa (p < 0.001), suggesting that they have potential as early diagnostic biomarkers for PCa and high-grade PCa (133). Qi et al. found that exosomal lncAY927529 was upregulated in PCa cells and PCa cells derived from PCa patient serum and human PCa cells. They additionally discovered that exosomal lncAY927529 from PCa cells can enhance both the proliferation and invasion of PCa cells by modulating CXCL14 levels in bone marrow stromal cell lines within the bone microenvironment. This indicates that exosomal lncAY927529 could potentially serve as a molecular diagnostic marker for PCa (134). Jiang et al. observed that the levels of exosomal lncRNA HOXD-AS1 were elevated in exosomes from CRPC cell lines compared to those from PCa cell lines. Moreover, they discovered that exosomal lncRNA HOXD-AS1 might be taken up directly by PCa cells and function as a competing endogenous RNA, thereby influencing the miR-361-5p/FOXM1 pathway and facilitating distant metastasis of PCa (117). Additionally, Guo et al. demonstrated that LINC01213 was increased in exosomes from the AIPC cell line, and these AIPC-derived exosomes were found to induce the transition of androgen-dependent PCa cells to AIPC cells, both in vitro and in vivo (135). Exosomal lncRNAs have important applications as biomarkers in the diagnosis of CRPC, as they can help to improve diagnostic accuracy and enable early detection. With in-depth research, these findings are expected to result in new breakthroughs in the diagnosis of CRPC.
4.4 CircRNA
CircRNA has been found to be enriched in exosomes (148). However, the role of exosomal circRNAs in CRPC has not yet been elucidated. Li et al. found that exosomal circ _0044516 was significantly upregulated in patients with PCa and cell lines, negatively correlated with the expression level of miR-29a-3p. Additionally, they found that the downregulation of exosome circ_0044516 inhibited PCa cell proliferation and metastasis (115). Zhang et al. found that circ_0081234 was significantly elevated in PCa with spinal metastases. Further analysis showed that exosomal circ_0081234 facilitates the migration, invasion, and EMT of PCa cells by modulating the miR-1/MAP3K1 signaling pathway (136). Gao et al. used qRT-PCR to clarify differences in circMID1 expression in PCa and observed that circMID1 expression was higher in patients with CRPC than in those with hormone-sensitive PCa, making it a promising biomarker for CRPC (137). The role of exosomal circRNAs in CRPC is multifaceted, including the regulation of biological behaviors and serving as potential biomarkers. These are important for an in-depth understanding of the pathogenesis of CRPC and the development of new biomarkers; however, few studies have been conducted, and further research is needed.
4.5 Proteins
Exosomal proteins reflect the characteristics of the cells from which they originate. These proteins are characterized by high stability and long half-life and can act directly on target cells to provide abundant, stable, sensitive, and unique diagnostic information. Exosomal proteins are important biomarkers for early tumor diagnosis. Liu et al. (106) analyzed plasma exosomes collected from 17 patients with PCa and 16 patients with CRPC using comprehensive proteomics and metabolomics analyses. They found that the levels of exosome-derived LRG1 and inter-alpha-trypsin inhibitor heavy chain H3 (ITIH3) were 1.7-fold and 2.04-fold higher in the CRPC group than those in the PCa group, respectively, suggesting that LRG1 and ITIH3 could serve as potential molecular diagnostic markers for CRPC. Docetaxel is the first-line chemotherapeutic agent for CRPC, and timely determination of docetaxel resistance in CRPC is important for subsequent treatment. Kato et al. found that P-glycoprotein was highly expressed in the serum of patients with docetaxel-resistant CRPC, suggesting that P-glycoprotein molecular markers have the potential to facilitate the diagnosis of docetaxel resistance in CRPC (138). Neuroendocrine PCa is highly malignant, and Bhagirath et al. found that thrombospondin 1 can be used to detect neuroendocrine differentiation in advanced CRPC (139). Exosome-derived proteins are highly stable and specific for CRPC diagnosis, providing new possibilities for the early detection, diagnosis, and treatment of CRPC. With further development of related technologies, the prospects of exosome protein diagnostic applications in CRPC are expected to expand.
4.6 Lipids
Lipids are important components of biofilms and are involved in tumor cell proliferation, motility, adhesion, apoptosis, signal transduction, and cell cycle regulation, among other activities (149). Lipids are among the most important components of exosomes and play a role in cell signaling, membrane structure composition, and function (150). Skotland et al. analyzed the differences in lipids in urinary exosomes of 15 patients with PCa and 13 healthy controls using high-throughput mass spectrometry quantitative lipidomics. Their findings revealed that phosphatidylserine and lactosylceramide were the most differentiated, demonstrating the potential utility of urinary exosomal lipids as biomarkers of PCa (140). Changes in lipid composition occur at different PCa stages. For example, Yi et al. used mass spectrometry to comprehensively examine three LnCap, PC3, and DU-145 cell lines with different degrees of malignancy. They found that glycerophospholipids were the most abundant lipids in exosomes originating from LnCap cells, whereas sphingolipids were the most abundant lipids in exosomes originating from PC3 and DU-145 cells. The glycerophospholipid and lysophospholipid contents were similarly differentiated among LnCap, PC3, and DU-145 cell-derived exosomal lipids (141). Lipids of exosomal origin are currently a hot topic in tumor diagnosis and treatment; however, they are less studied in CRPC. Further research is necessary to improve our understanding of the role of exosomal lipids in CRPC and provide new insights into the diagnosis and treatment of CRPC.
5 Exosomes in the treatment of castration-resistant prostate cancer
With the gradual unraveling of the pathological and physiological mechanisms of exosomes, like for the traditional delivery systems mainly including liposomes, polymer nanoparticles, etc., exosomes have the core advantages of information carriers, low immunogenicity, natural targeting, and biocompatibility. Important progress has been made in the research of CRPC therapy, mainly in the three major directions of drug carriers, Therapeutic target and immunotherapy (Figure 3).
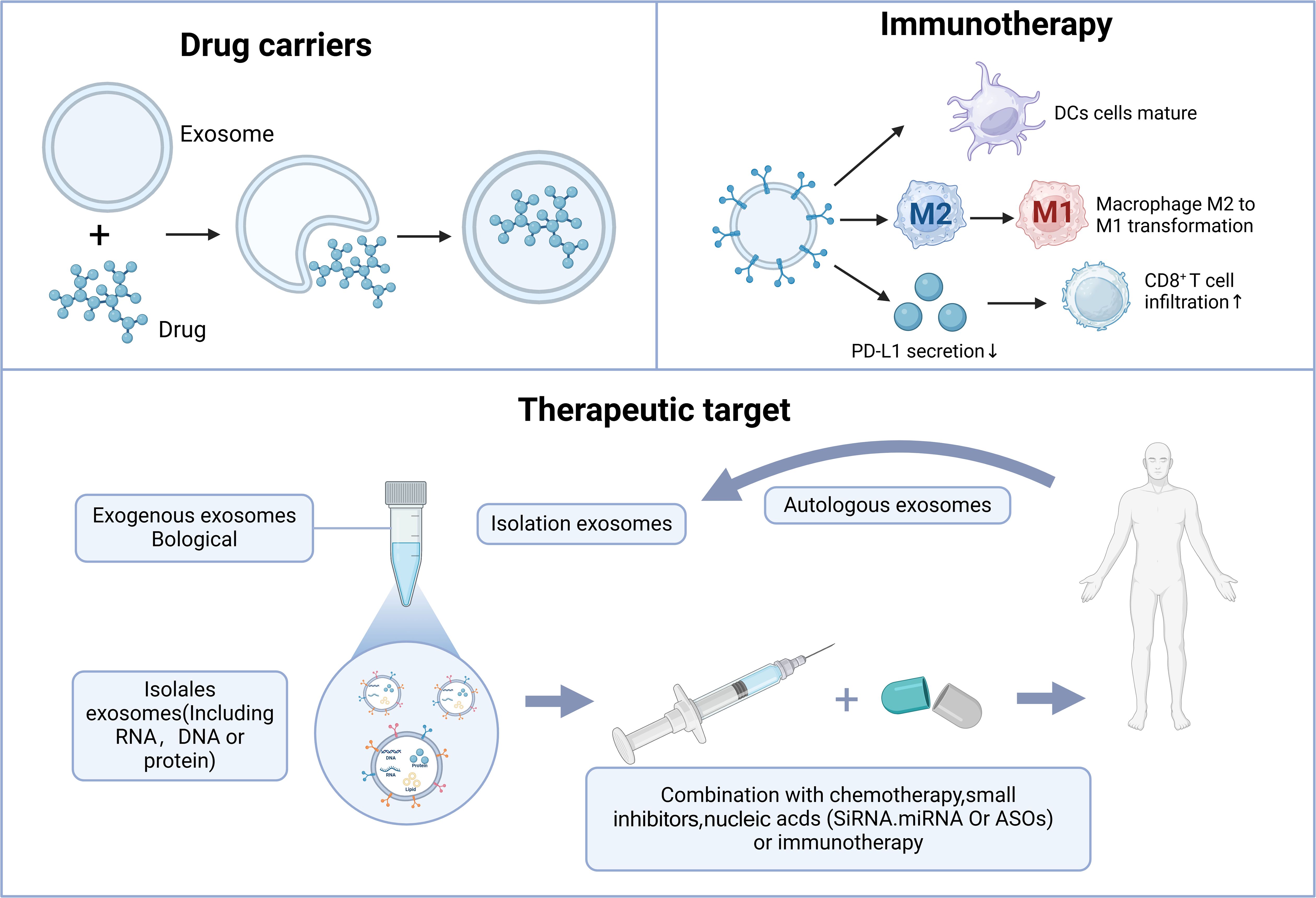
Figure 3. The role of exosomes in CRPC treatment. Exosomes contribute to CRPC treatment primarily through drug delivery, therapeutic targeting, and immune therapy.
5.1 Drug carriers
Exosomes are nano-sized double-layered membrane vesicle structures used as natural nano-biological drug carriers with the ability to transport different cargoes and target specific cells or tissues, which can achieve precise, targeted delivery of drugs. Exosomes also offer advantages such as biocompatibility, low immunogenicity, high physicochemical stability, and high tissue penetration ability (151). In recent years, several studies have used exosomes as drug-delivery systems for different diseases (152). Han et al. (153) identified SIRT6 as a promising new therapeutic target in CRPC and significantly inhibited tumorigenesis by intravenously injecting mice with therapeutic SIRT6 siRNA aptamer-modified exosomes twice weekly, 2 weeks after implantation of DU145 tumor cells in a subcutaneous xenograft tumor model. Kurniawati et al. (154) found that exogenous miR-lethal 7c can be successfully packaged into mesenchymal stem cell exosomes and that these exosomes can be targeted as a therapeutic lethal 7c delivery system, which can lead to a significant reduction in PC3 and CWR22Rv1 cells in terms of cell proliferation and migration. Gan et al. (155) reported that plasma levels of exosomal miR-375 were markedly higher in patients with CRPC compared to denudation-sensitive patients with PCa. Additionally, they found that exosomes derived from human umbilical cord mesenchymal stem cells could carry miR-375 antisense oligonucleotides (e-375i), which are effectively taken up by PCa cells. This suggests that targeting miR-375 could provide a novel therapeutic approach for PCa, particularly in cases of CRPC with elevated AR levels. Overall, exosomes show immense potential as drug delivery vehicles, particularly for improving drug targeting and reducing side effects. However, the clinical use of exosomes still faces challenges, such as production scale, purification technology, and quality control, which require further exploration.
5.2 Therapeutic target
Exosomes can transfer oncogenic proteins and nucleic acids to participate in tumor development, thus allowing them to be used as targets in tumor therapy (19). Lshizuya et al. found that actinin-4 (ACTN4) was significantly elevated in patients with CRPC via serum exosomal proteomic analysis of patients with CRPC versus ADT-treated patients with metastatic PCa. Experimental studies showed that RNA interference-based reduction of ACTN4 markedly decreased the proliferation and invasion of DU145 cells. This indicates that ACTN4 present in exosomes could represent a promising novel therapeutic target for CRPC (156). Based on the lethality of neuroendocrine PCa, Lin et al. (157) found that adipocyte differentiation-related protein can be released from PCa cells into exosomes and can induce neuroendocrine differentiation in neighboring cells in a paracrine manner. Further in-depth studies on the role of adipocyte differentiation-related protein in neuroendocrine differentiation will help identify new therapeutic targets for the treatment of advanced CRPC. The androgen receptor splice variant 7 (AR-V7) is associated with hormone therapy resistance in CRPC. Plasma exosomal AR-V7 was detected via digital droplet polymerase chain reaction in 26 patients with CRPC treated with abiraterone and 10 patients with enzalutamide. Overall, 39% of patients were found to be AR-V7 positive. AR-V7 negative patients had a significantly longer median progression-free survival than that of AR-V7 positive patients (20 months vs. 3 months; P < 0.001), suggesting that targeting exosomal AR-V7 may be a potential treatment modality for CRPC (158). Gan et al. observed an increased expression of miR-375 in exosomes derived from PCa and proposed that targeting miR-375 could offer a potential alternative treatment for PCa, particularly in cases of CRPC with elevated AR levels (155). The value and potential of exosomes as therapeutic targets for CRPC have been widely recognized, and their application deserves further research and exploration.
5.3 Immunotherapy
Immunotherapy for PCa is one of the hotspots of current research. Exosomes have a great potential for immunotherapy in PCa. Wang et al. (159) found that exosomes encapsulated with the acoustic sensitizer chlorin e6 (Ce6) and the immune adjuvant R848 (i.e., ExoCe6+R848) enhanced R848-mediated maturation of dendritic cells under ultrasound irradiation and simultaneously reprogrammed macrophages from an immunosuppressive M2-like phenotype to an anti-tumor M1-like phenotype, thereby activating effector T cells and restoring the immunosuppressive microenvironment. Further animal experiments revealed that engineering the ExoCe6+R848 type under ultrasound irradiation inhibited PCa progression in a hormonal mouse model. Tumor-associated macrophages are among the most prevalent immune cell types within the tumor microenvironment (160). Peng et al. (161) found that the exosome biosynthesis inhibitor GW4869 inhibited the release of exosomes from PCa cells, which suppressed the M2 differentiation of macrophages and their pro-tumorigenic effects in the tumor microenvironment. Additionally, Liu et al. (162) revealed that the novel P300/CBP inhibitor A485 can block the transcription of CD274, cutting off the secretion of PCa exosomal PD-L1 and increasing the infiltration of CD8T cells. This can convert “cold” tumors into “hot” tumors, thus promoting the efficacy of immunotherapy. These findings provide new ideas and a clinical rationale for PCa immunotherapy. Exosomes play important roles in immunotherapy for PCa, functioning as drug carriers, influencing the tumor immune microenvironment, and being part of novel immunotherapeutic strategies. With a deeper understanding of exosomes and immunotherapy, additional exosome-based immunotherapeutic approaches may emerge for CRPC.
6 Discussion
Early diagnosis and treatment are crucial for improving tumor survival rates, and exosomes play a significant role in both. Tumor cell exosomes contain various proteins, lipids, nucleic acids, and other components involved in regulating tumor progression. In CRPC, exosomes are secreted in large quantities into the blood, urine, and other body fluids, making them easily accessible, non-invasive, and specific. Exosomes are rich in bioactive molecules that provide potential biomarkers and help in diagnosing CRPC. They play crucial roles in intercellular communication and have been shown to be involved in various aspects of CRPC, including genesis, metastasis, angiogenesis, immune escape, and drug resistance. This underscores their importance in CRPC therapy. However, most studies have focused on early PCa, with limited research on advanced CRPC. Future research should focus on exosomes that are active in the development of CRPC. Questions such as “How different are the exosomal contents of early PCa and CRPC?” and “Do the exosomes secreted by early PCa cells support CRPC formation?” remain largely unexplored. Moreover, most existing studies are retrospective and single-center studies. To promote the clinical application of exosomes, more rigorous prospective correlative studies are needed, which must be validated by multicenter repetitive studies to generate reproducible biomarkers. Further development of exosomal drug carriers is necessary to facilitate precise therapy for CRPC and clarify the causes and mechanisms of drug resistance in targeted therapy.
Author contributions
YC: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. JW: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Writing – original draft, Writing – review & editing. HL: Conceptualization, Data curation, Investigation, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing. YW: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Validation, Writing – original draft, Writing – review & editing. XC: Conceptualization, Formal analysis, Investigation, Methodology, Resources, Validation, Writing – original draft, Writing – review & editing. TZ: Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Visualization, Writing – original draft, Writing – review & editing. YL: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. DW: Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by Incubation Project of Mianyang Central Hospital (Grant No. 2023FH009, 2023FH005), and the National Natural Science Foundation of China (NSFC) (Grant No.82400758).
Acknowledgments
We appreciate all the reviewers who participated in the review, as well as Editage for providing English editing services during the preparation of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ACTN4, Actinin-4; ADRP, Adipocyte Differentiation-Related Protein; ADT, Androgen Deprivation Therapy; AIPC, Androgen-Independent Prostate Cancer; AR, Androgen Receptor; AR-V7, Androgen Receptor Splice Variant 7;CAF, Cancer-Associated Fibroblast; CRPC, Castration-Resistant Prostate Cancer; EMT, Epithelial-to-Mesenchymal Transition; LRG1, Leucine-Rich Alpha-2 Glycoprotein 1; PCa, Prostate Cancer; PD-L1, Programmed Death-Ligand 1; PSC-27, Primary Prostate Fibroblasts; qRT-PCR, Quantitative Real-Time Polymerase Chain Reaction.
References
1. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. (2023) 73:17–48. doi: 10.3322/caac.21763
2. Zhu Y, Mo M, Wei Y, Wu J, Pan J, Freedland SJ, et al. Epidemiology and genomics of prostate cancer in Asian men. Nat Rev Urol. (2021) 18:282–301. doi: 10.1038/s41585-021-00442-8
3. Hugosson J, Roobol MJ, Månsson M, Tammela TLJ, Zappa M, Nelen V, et al. A 16-yr follow-up of the european randomized study of screening for prostate cancer. Eur Urol. (2019) 76:43–51. doi: 10.1016/j.eururo.2019.02.009
4. Dejous C, Krishnan UM. Sensors for diagnosis of prostate cancer: Looking beyond the prostate specific antigen. Biosens Bioelectron. (2020) 173:112790. doi: 10.1016/j.bios.2020.112790
5. Gandaglia G, Leni R, Bray F, Fleshner N, Freedland SJ, Kibel A, et al. Epidemiology and prevention of prostate cancer. Eur Urol Oncol. (2021) 4:877–92. doi: 10.1016/j.euo.2021.09.006
6. Kohaar I, Petrovics G, Srivastava S. A rich array of prostate cancer molecular biomarkers: opportunities and challenges. Int J Mol Sci. (2019) 20:1813. doi: 10.3390/ijms20081813
7. Liu F, Shi X, Wang F, Han S, Chen D, Gao X, et al. Evaluation and multi-institutional validation of a novel urine biomarker lncRNA546 to improve the diagnostic specificity of prostate cancer in PSA gray-zone. Front Oncol. (2022) 12:946060. doi: 10.3389/fonc.2022.946060
8. Achard V, Putora PM, Omlin A, Zilli T, Fischer S. Metastatic prostate cancer: treatment options. Oncology. (2022) 100:48–59. doi: 10.1159/000519861
9. Chen M, Feuerstein MA, Levina E, Baghel PS, Carkner RD, Tanner MJ, et al. Hedgehog/Gli supports androgen signaling in androgen deprived and androgen independent prostate cancer cells. Mol Cancer. (2010) 9:89. doi: 10.1186/1476-4598-9-89
10. Meng F, Han X, Min Z, He X, Zhu S. Prognostic signatures associated with high infiltration of Tregs in bone metastatic prostate cancer. Aging (Albany NY). (2021) 13:17442–61. doi: 10.18632/aging.203234
11. Chen H, Qu M, Shi H, Dong Z, Wang Y, Gao X. Adjuvant radiotherapy in patients with node-positive prostate cancer after radical prostatectomy. J Cancer Res Clin Oncol. (2023) 149:4925–32. doi: 10.1007/s00432-022-04409-z
12. van den Bergh GPA, Kuppen MCP, Westgeest HM, Mehra N, Gerritsen WR, Aben KKH, et al. Incidence and survival of castration-resistant prostate cancer patients with visceral metastases: results from the Dutch CAPRI-registry. Prostate Cancer Prostatic Dis. (2023) 26:162–9. doi: 10.1038/s41391-022-00605-7
13. Dai C, Dehm SM, Sharifi N. Targeting the androgen signaling axis in prostate cancer. J Clin Oncol. (2023) 41:4267–78. doi: 10.1200/JCO.23.00433
14. Imamura J, Ganguly S, Muskara A, Liao RS, Nguyen JK, Weight C, et al. Lineage plasticity and treatment resistance in prostate cancer: the intersection of genetics, epigenetics, and evolution. Front Endocrinol (Lausanne). (2023) 14:1191311. doi: 10.3389/fendo.2023.1191311
15. Zhang Z, Karthaus WR, Lee YS, Gao VR, Wu C, Russo JW, et al. Tumor microenvironment-derived NRG1 promotes antiandrogen resistance in prostate cancer. Cancer Cell. (2020) 38:279–296.e9. doi: 10.1016/j.ccell.2020.06.005
16. Schaeffer EM, Srinivas S, Adra N, An Y, Barocas D, Bitting R, et al. Prostate cancer, version 4.2023, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. (2023) 21:1067–96. doi: 10.6004/jnccn.2023.0050
17. Doyle LM, Wang MZ. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. (2019) 8:727. doi: 10.3390/cells8070727
18. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. (2020) 367:eaau6977. doi: 10.1126/science.aau6977
19. Zhou Y, Zhang Y, Gong H, Luo S, Cui Y. The role of exosomes and their applications in cancer. Int J Mol Sci. (2021) 22:12204. doi: 10.3390/ijms222212204
20. Yu J, Ostowari A, Gonda A, Mashayekhi K, Dayyani F, Hughes CCW, et al. Exosomes as a source of biomarkers for gastrointestinal cancers. Cancers (Basel). (2023) 15:1263. doi: 10.3390/cancers15041263
21. Paskeh MDA, Entezari M, Mirzaei S, Zabolian A, Saleki H, Naghdi MJ, et al. Emerging role of exosomes in cancer progression and tumor microenvironment remodeling. J Hematol Oncol. (2022) 15:83. doi: 10.1186/s13045-022-01305-4
22. Yuan Y, Li H, Pu W, Chen L, Guo D, Jiang H, et al. Cancer metabolism and tumor microenvironment: fostering each other? Sci China Life Sci. (2022) 65:236–79. doi: 10.1007/s11427-021-1999-2
23. Aslan C, Maralbashi S, Salari F, Kahroba H, Sigaroodi F, Kazemi T, et al. Tumor-derived exosomes: Implication in angiogenesis and antiangiogenesis cancer therapy. J Cell Physiol. (2019) 234:16885–903. doi: 10.1002/jcp.28374
24. Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. (2019) 18:75. doi: 10.1186/s12943-019-0991-5
25. Familtseva A, Jeremic N, Tyagi SC. Exosomes: cell-created drug delivery systems. Mol Cell Biochem. (2019) 459:1–6. doi: 10.1007/s11010-019-03545-4
26. Yu M, Liu W, Li J, Lu J, Lu H, Jia W, et al. Exosomes derived from atorvastatin-pretreated MSC accelerate diabetic wound repair by enhancing angiogenesis via AKT/eNOS pathway. Stem Cell Res Ther. (2020) 11:350. doi: 10.1186/s13287-020-01824-2
27. Harding C, Stahl P. Transferrin recycling in reticulocytes: pH and iron are important determinants of ligand binding and processing. Biochem Biophys Res Commun. (1983) 113:650–8. doi: 10.1016/0006-291x(83)91776-x
28. Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. (1987) 262:9412–20. doi: 10.1016/S0021-9258(18)48095-7
29. Liu Q, Piao H, Wang Y, Zheng D, Wang W. Circulating exosomes in cardiovascular disease: Novel carriers of biological information. BioMed Pharmacother. (2021) 135:111148. doi: 10.1016/j.biopha.2020.111148
30. Muller L, Hong CS, Stolz DB, Watkins SC, Whiteside TL. Isolation of biologically-active exosomes from human plasma. J Immunol Methods. (2014) 411:55–65. doi: 10.1016/j.jim.2014.06.007
31. Wang S, Li F, Ye T, Wang J, Lyu C, Qing S, et al. Macrophage-tumor chimeric exosomes accumulate in lymph node and tumor to activate the immune response and the tumor microenvironment. Sci Transl Med. (2021) 13:eabb6981. doi: 10.1126/scitranslmed.abb6981
32. Barceló M, Castells M, Bassas L, Vigués F, Larriba S. Semen miRNAs contained in exosomes as non-invasive biomarkers for prostate cancer diagnosis. Sci Rep. (2019) 9:13772. doi: 10.1038/s41598-019-50172-6
33. Street JM, Koritzinsky EH, Glispie DM, Star RA, Yuen PS. Urine exosomes: an emerging trove of biomarkers. Adv Clin Chem. (2017) 78:103–22. doi: 10.1016/bs.acc.2016.07.003
34. Song M, Bai H, Zhang P, Zhou X, Ying B. Promising applications of human-derived saliva biomarker testing in clinical diagnostics. Int J Sci. (2023) 15:2. doi: 10.1038/s41368-022-00209-w
35. Tsakonas G, Tadigotla V, Chakrabortty SK, Stragliotto G, Chan D, Lewensohn R, et al. Cerebrospinal fluid as a liquid biopsy for molecular characterization of brain metastasis in patients with non-small cell lung cancer. Lung Cancer. (2023) 182:107292. doi: 10.1016/j.lungcan.2023.107292
36. Yi DY, Kim SY. Human breast milk composition and function in human health: from nutritional components to microbiome and microRNAs. Nutrients. (2021) 13:3094. doi: 10.3390/nu13093094
37. Han QF, Li WJ, Hu KS, Gao J, Zhai WL, Yang JH, et al. Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer. Mol Cancer. (2022) 21:207. doi: 10.1186/s12943-022-01671-0
38. Ferguson SW, Nguyen J. Exosomes as therapeutics: The implications of molecular composition and exosomal heterogeneity. J Control Release. (2016) 228:179–90. doi: 10.1016/j.jconrel.2016.02.037
39. Liang Y, Duan L, Lu J, Xia J. Engineering exosomes for targeted drug delivery. Theranostics. (2021) 11:3183–95. doi: 10.7150/thno.52570
40. Barile L, Vassalli G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol Ther. (2017) 174:63–78. doi: 10.1016/j.pharmthera.2017.02.020
41. Delenclos M, Trendafilova T, Mahesh D, Baine AM, Moussaud S, Yan IK, et al. Investigation of endocytic pathways for the internalization of exosome-associated oligomeric alpha-synuclein. Front Neurosci. (2017) 11:172. doi: 10.3389/fnins.2017.00172
42. Buratta S, Tancini B, Sagini K, Delo F, Chiaradia E, Urbanelli L, et al. Lysosomal exocytosis, exosome release and secretory autophagy: the autophagic- and endo-lysosomal systems go extracellular. Int J Mol Sci. (2020) 21:2576. doi: 10.3390/ijms21072576
43. Huotari J, Helenius A. Endosome maturation. EMBO J. (2011) 30:3481–500. doi: 10.1038/emboj.2011.286
44. Liu J, Ren L, Li S, Li W, Zheng X, Yang Y, et al. The biology, function, and applications of exosomes in cancer. Acta Pharm Sin B. (2021) 11:2783–97. doi: 10.1016/j.apsb.2021.01.001
45. Vietri M, Radulovic M, Stenmark H. The many functions of ESCRTs. Nat Rev Mol Cell Biol. (2020) 21:25–42. doi: 10.1038/s41580-019-0177-4
46. Xu L, Nagai Y, Kajihara Y, Ito G, Tomita T. The regulation of rab GTPases by phosphorylation. Biomolecules. (2021) 11:1340. doi: 10.3390/biom11091340
47. Addi C, Presle A, Frémont S, Cuvelier F, Rocancourt M, Milin F, et al. The Flemmingsome reveals an ESCRT-to-membrane coupling via ALIX/syntenin/syndecan-4 required for completion of cytokinesis. Nat Commun. (2020) 11:1941. doi: 10.1038/s41467-020-15205-z
48. Fan Y, Pionneau C, Cocozza F, Boëlle PY, Chardonnet S, Charrin S, et al. Differential proteomics argues against a general role for CD9, CD81 or CD63 in the sorting of proteins into extracellular vesicles. J Extracell Vesicles. (2023) 12:e12352. doi: 10.1002/jev2.12352
49. Yu X, Odenthal M, Fries JW. Exosomes as miRNA carriers: formation-function-future. Int J Mol Sci. (2016) 17:2028. doi: 10.3390/ijms17122028
50. Sharma A, Johnson A. Exosome DNA: Critical regulator of tumor immunity and a diagnostic biomarker. J Cell Physiol. (2020) 235:1921–32. doi: 10.1002/jcp.29153
51. Huang X, Wu W, Jing D, Yang L, Guo H, Wang L, et al. Engineered exosome as targeted lncRNA MEG3 delivery vehicles for osteosarcoma therapy. J Control Release. (2022) 343:107–17. doi: 10.1016/j.jconrel.2022.01.026
52. Zhang X, Xu Y, Ma L, Yu K, Niu Y, Xu X, et al. Essential roles of exosome and circRNA_101093 on ferroptosis desensitization in lung adenocarcinoma. Cancer Commun (Lond). (2022) 42:287–313. doi: 10.1002/cac2.12275
53. Yan X, Liu L, Yao S, Chen Y, Yu Q, Jiang C, et al. LncRNA and mRNA profiles of human milk-derived exosomes and their possible roles in protecting against necrotizing enterocolitis. Food Funct. (2022) 13:12953–65. doi: 10.1039/d2fo01866g
54. Zhao L, Gu C, Gan Y, Shao L, Chen H, Zhu H. Exosome-mediated siRNA delivery to suppress postoperative breast cancer metastasis. J Control Release. (2020) 318:1–15. doi: 10.1016/j.jconrel.2019.12.005
55. Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. (2019) 9:19. doi: 10.1186/s13578-019-0282-2
56. Wei D, Zhan W, Gao Y, Huang L, Gong R, Wang W, et al. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. (2021) 31:157–77. doi: 10.1038/s41422-020-00409-1
57. Skotland T, Hessvik NP, Sandvig K, Llorente A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J Lipid Res. (2019) 60:9–18. doi: 10.1194/jlr.R084343
58. Tenchov R, Sasso JM, Wang X, Liaw WS, Chen CA, Zhou QA. Exosomes─Nature’s lipid nanoparticles, a rising star in drug delivery and diagnostics. ACS Nano. (2022) 16:17802–46. doi: 10.1021/acsnano.2c08774
59. Hade MD, Suire CN, Suo Z. Mesenchymal stem cell-derived exosomes: applications in regenerative medicine. Cells. (2021) 10:1959. doi: 10.3390/cells10081959
60. Liu Z, Yan J, Tong L, Liu S, Zhang Y. The role of exosomes from BALF in lung disease. J Cell Physiol. (2022) 237:161–8. doi: 10.1002/jcp.30553
61. Skotland T, Sandvig K, Llorente A. Lipids in exosomes: Current knowledge and the way forward. Prog Lipid Res. (2017) 66:30–41. doi: 10.1016/j.plipres.2017.03.001
62. Willms E, Cabañas C, Mäger I, Wood MJA, Vader P. Extracellular vesicle heterogeneity: subpopulations, isolation techniques, and diverse functions in cancer progression. Front Immunol. (2018) 9:738. doi: 10.3389/fimmu.2018.00738
63. Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in exosome isolation techniques. Theranostics. (2017) 7:789–804. doi: 10.7150/thno.18133
64. Wang W, Sun H, Duan H, Sheng G, Tian N, Liu D, et al. Isolation and usage of exosomes in central nervous system diseases. CNS Neurosci Ther. (2024) 30:e14677. doi: 10.1111/cns.14677
65. Lai JJ, Chau ZL, Chen SY, Hill JJ, Korpany KV, Liang NW, et al. Exosome processing and characterization approaches for research and technology development. Adv Sci (Weinh). (2022) 9:e2103222. doi: 10.1002/advs.202103222
66. Lischnig A, Bergqvist M, Ochiya T, Lässer C. Quantitative proteomics identifies proteins enriched in large and small extracellular vesicles. Mol Cell Proteomics. (2022) 21:100273. doi: 10.1016/j.mcpro.2022.100273
67. Raj DA, Fiume I, Capasso G, Pocsfalvi G. A multiplex quantitative proteomics strategy for protein biomarker studies in urinary exosomes. Kidney Int. (2012) 81:1263–72. doi: 10.1038/ki.2012.25
68. Langevin SM, Kuhnell D, Orr-Asman MA, Biesiada J, Zhang X, Medvedovic M, et al. Balancing yield, purity and practicality: a modified differential ultracentrifugation protocol for efficient isolation of small extracellular vesicles from human serum. RNA Biol. (2019) 16:5–12. doi: 10.1080/15476286.2018.1564465
69. Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, Leimgruber A, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles. (2015) 4:27031. doi: 10.3402/jev.v4.27031
70. He L, Zhu D, Wang J, Wu X. A highly efficient method for isolating urinary exosomes. Int J Mol Med. (2019) 43:83–90. doi: 10.3892/ijmm.2018.3944
71. Lima Moura S, Martì M, Pividori MI. Matrix effect in the isolation of breast cancer-derived nanovesicles by immunomagnetic separation and electrochemical immunosensing-A comparative study. Sensors (Basel). (2020) 20:965. doi: 10.3390/s20040965
72. Yousif G, Qadri S, Parray A, Akhthar N, Shuaib A, Haik Y. Exosomes derived neuronal markers: immunoaffinity isolation and characterization. Neuromolecular Med. (2022) 24:339–51. doi: 10.1007/s12017-021-08696-6
73. Ludwig N, Razzo BM, Yerneni SS, Whiteside TL. Optimization of cell culture conditions for exosome isolation using mini-size exclusion chromatography (mini-SEC). Exp Cell Res. (2019) 378:149–57. doi: 10.1016/j.yexcr.2019.03.014
74. Sidhom K, Obi PO, Saleem A. A review of exosomal isolation methods: is size exclusion chromatography the best option? Int J Mol Sci. (2020) 21:6466. doi: 10.3390/ijms21186466
75. Kurian TK, Banik S, Gopal D, Chakrabarti S, Mazumder N. Elucidating methods for isolation and quantification of exosomes: A review. Mol Biotechnol. (2021) 63:249–66. doi: 10.1007/s12033-021-00300-3
76. Diaz G, Bridges C, Lucas M, Cheng Y, Schorey JS, Dobos KM, et al. Protein digestion, ultrafiltration, and size exclusion chromatography to optimize the isolation of exosomes from human blood plasma and serum. J Vis Exp. (2018) 134:57467. doi: 10.3791/57467
77. Omrani M, Beyrampour-Basmenj H, Jahanban-Esfahlan R, Talebi M, Raeisi M, Serej ZA, et al. Global trend in exosome isolation and application: an update concept in management of diseases. Mol Cell Biochem. (2024) 479:679–91. doi: 10.1007/s11010-023-04756-6
78. Dong B, Wang C, Zhang J, Zhang J, Gu Y, Guo X, et al. Exosomes from human umbilical cord mesenchymal stem cells attenuate the inflammation of severe steroid-resistant asthma by reshaping macrophage polarization. Stem Cell Res Ther. (2021) 12:204. doi: 10.1186/s13287-021-02244-6
79. Ramos T L, Sánchez-Abarca LI, Muntión S, Preciado S, Puig N, López-Ruano G, et al. MSC surface markers (CD44, CD73, and CD90) can identify human MSC-derived extracellular vesicles by conventional flow cytometry. Cell Commun Signal. (2016) 14:2. doi: 10.1186/s12964-015-0124-8
80. Siwaponanan P, Kaewkumdee P, Phromawan W, Udompunturak S, Chomanee N, Udol K, et al. Increased expression of six-large extracellular vesicle-derived miRNAs signature for nonvalvular atrial fibrillation. J Transl Med. (2022) 20:4. doi: 10.1186/s12967-021-03213-6
81. Lozano-Andrés E, Libregts SF, Toribio V, Royo F, Morales S, López-Martín S, et al. Tetraspanin-decorated extracellular vesicle-mimetics as a novel adaptable reference material. J Extracell Vesicles. (2019) 8:1573052. doi: 10.1080/20013078.2019.1573052
82. Hu J, Jiang Y, Wu X, Wu Z, Qin J, Zhao Z, et al. Exosomal miR-17-5p from adipose-derived mesenchymal stem cells inhibits abdominal aortic aneurysm by suppressing TXNIP-NLRP3 inflammasome. Stem Cell Res Ther. (2022) 13:349. doi: 10.1186/s13287-022-03037-1
83. Kamei N, Nishimura H, Matsumoto A, Asano R, Muranaka K, Fujita M, et al. Comparative study of commercial protocols for high recovery of high-purity mesenchymal stem cell-derived extracellular vesicle isolation and their efficient labeling with fluorescent dyes. Nanomedicine. (2021) 35:102396. doi: 10.1016/j.nano.2021.102396
84. Hu Z, Chen G, Zhao Y, Gao H, Li L, Yin Y, et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol Cancer. (2023) 22:55. doi: 10.1186/s12943-023-01759-1
85. Yu D, Li Y, Wang M, Gu J, Xu W, Cai H, et al. Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer. (2022) 21:56. doi: 10.1186/s12943-022-01509-9
86. Feng Z, Zhou J, Liu Y, Xia R, Li Q, Yan L, et al. Epithelium- and endothelium-derived exosomes regulate the alveolar macrophages by targeting RGS1 mediated calcium signaling-dependent immune response. Cell Death Differ. (2021) 28:2238–56. doi: 10.1038/s41418-021-00750-x
87. Ghafourian M, Mahdavi R, Akbari Jonoush Z, Sadeghi M, Ghadiri N, Farzaneh M, et al. The implications of exosomes in pregnancy: emerging as new diagnostic markers and therapeutics targets. Cell Commun Signal. (2022) 20:51. doi: 10.1186/s12964-022-00853-z
88. Li J, Zhang Y, Luo B. Effects of exosomal viral components on the tumor microenvironment. Cancers (Basel). (2022) 14. doi: 10.3390/cancers14143552
89. Tian Y, Fu C, Wu Y, Lu Y, Liu X, Zhang Y. Central nervous system cell-derived exosomes in neurodegenerative diseases. Oxid Med Cell Longev. (2021) 2021:9965564. doi: 10.1155/2021/9965564
90. van der Pol E, Böing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. (2012) 64:676–705. doi: 10.1124/pr.112.005983
91. Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. (2020) 19:117. doi: 10.1186/s12943-020-01235-0
92. Gao Y, Xu H, Li N, Wang H, Ma L, Chen S, et al. Renal cancer-derived exosomes induce tumor immune tolerance by MDSCs-mediated antigen-specific immunosuppression. Cell Commun Signal. (2020) 18:106. doi: 10.1186/s12964-020-00611-z
93. Jiang X, Xu Y, Liu R, Guo S. Exosomal lincROR Promotes Docetaxel Resistance in Prostate Cancer through a β-catenin/HIF1α Positive Feedback Loop. Mol Cancer Res. (2023) 21:472–82. doi: 10.1158/1541-7786.MCR-22-0458
94. Tiwari A, Trivedi R, Lin SY. Tumor microenvironment: barrier or opportunity towards effective cancer therapy. J BioMed Sci. (2022) 29:83. doi: 10.1186/s12929-022-00866-3
95. Mao X, Xu J, Wang W, Liang C, Hua J, Liu J, et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer. (2021) 20:131. doi: 10.1186/s12943-021-01428-1
96. Cui D, Li J, Zhu Z, Berk M, Hardaway A, McManus J, et al. Cancer-associated fibroblast-secreted glucosamine alters the androgen biosynthesis program in prostate cancer via HSD3B1 upregulation. J Clin Invest. (2023) 133:e161913. doi: 10.1172/JCI161913
97. Yang X, Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol. (2017) 18:452–65. doi: 10.1038/nrm.2017.22
98. Wang H, Li N, Liu Q, Guo J, Pan Q, Cheng B, et al. Antiandrogen treatment induces stromal cell reprogramming to promote castration resistance in prostate cancer. Cancer Cell. (2023) 41:1345–1362.e9. doi: 10.1016/j.ccell.2023.05.016
99. Wang S, Du P, Cao Y, Ma J, Yang X, Yu Z, et al. Cancer associated fibroblasts secreted exosomal miR-1290 contributes to prostate cancer cell growth and metastasis via targeting GSK3β. Cell Death Discov. (2022) 8:371. doi: 10.1038/s41420-022-01163-6
100. Zhao J, Shen J, Mao L, Yang T, Liu J, Hongbin S. Cancer associated fibroblast secreted miR-432-5p targets CHAC1 to inhibit ferroptosis and promote acquired chemoresistance in prostate cancer. Oncogene. (2024) 43:2104–14. doi: 10.1038/s41388-024-03057-6
101. Lugano R, Ramachandran M, Dimberg A. Tumor angiogenesis: causes, consequences, challenges and opportunities. Cell Mol Life Sci. (2020) 77:1745–70. doi: 10.1007/s00018-019-03351-7
102. Eisermann K, Fraizer G. The androgen receptor and VEGF: mechanisms of androgen-regulated angiogenesis in prostate cancer. Cancers (Basel). (2017) 9:32. doi: 10.3390/cancers9040032
103. Ludwig N, Whiteside TL. Potential roles of tumor-derived exosomes in angiogenesis. Expert Opin Ther Targets. (2018) 22:409–17. doi: 10.1080/14728222.2018.1464141
104. Prigol AN, Rode MP, Silva AH, Cisilotto J, Creczynski-Pasa TB. Pro-angiogenic effect of PC-3 exosomes in endothelial cells in vitro. Cell Signal. (2021) 87:110126. doi: 10.1016/j.cellsig.2021.110126
105. Luo JQ, Yang TW, Wu J, Lai HH, Zou LB, Chen WB, et al. Exosomal PGAM1 promotes prostate cancer angiogenesis and metastasis by interacting with ACTG1. Cell Death Dis. (2023) 14:502. doi: 10.1038/s41419-023-06007-4
106. Liu P, Wang W, Wang F, Fan J, Guo J, Wu T, et al. Alterations of plasma exosomal proteins and motabolies are associated with the progression of castration-resistant prostate cancer. J Transl Med. (2023) 21:40. doi: 10.1186/s12967-022-03860-3
107. Ioannidou E, Moschetta M, Shah S, Parker JS, Ozturk MA, Pappas-Gogos G, et al. Angiogenesis and anti-angiogenic treatment in prostate cancer: mechanisms of action and molecular targets. Int J Mol Sci. (2021) 22:9926. doi: 10.3390/ijms22189926
108. Li J, Wang K, Yang C, Zhu K, Jiang C, Wang M, et al. Tumor-associated macrophage-derived exosomal LINC01232 induces the immune escape in glioma by decreasing surface MHC-I expression. Adv Sci (Weinh). (2023) 10:e2207067. doi: 10.1002/advs.202207067
109. Bou-Dargham MJ, Sha L, Sang QA, Zhang J. Immune landscape of human prostate cancer: immune evasion mechanisms and biomarkers for personalized immunotherapy. BMC Cancer. (2020) 20:572. doi: 10.1186/s12885-020-07058-y
110. Jeong JH, Zhong S, Li F, Huang C, Chen X, Liu Q, et al. Tumor-derived OBP2A promotes prostate cancer castration resistance. J Exp Med. (2023) 220:e20211546. doi: 10.1084/jem.20211546
111. Li N, Wang Y, Xu H, Wang H, Gao Y, Zhang Y. Exosomes Derived from RM-1 Cells Promote the Recruitment of MDSCs into Tumor Microenvironment by Upregulating CXCR4 via TLR2/NF-κB Pathway. J Oncol. (2021) 2021:5584406. doi: 10.1155/2021/5584406
112. Hodgins JJ, Khan ST, Park MM, Auer RC, Ardolino M. Killers 2.0: NK cell therapies at the forefront of cancer control. J Clin Invest. (2019) 129:3499–510. doi: 10.1172/JCI129338
113. Lundholm M, Schröder M, Nagaeva O, Baranov V, Widmark A, Mincheva-Nilsson L, et al. Prostate tumor-derived exosomes down-regulate NKG2D expression on natural killer cells and CD8+ T cells: mechanism of immune evasion. PLoS One. (2014) 9:e108925. doi: 10.1371/journal.pone.0108925
114. Li D, Zhou X, Xu W, Chen Y, Mu C, Zhao X, et al. Prostate cancer cells synergistically defend against CD8+ T cells by secreting exosomal PD-L1. Cancer Med. (2023) 12:16405–15. doi: 10.1002/cam4.6275
115. Li T, Sun X, Chen L. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J Cell Biochem. (2020) 121:2118–26. doi: 10.1002/jcb.28239
116. Liu Z, Lin Z, Jiang M, Zhu G, Xiong T, Cao F, et al. Cancer-associated fibroblast exosomes promote prostate cancer metastasis through miR-500a-3p/FBXW7/HSF1 axis under hypoxic microenvironment. Cancer Gene Ther. (2024) 31:698–709. doi: 10.1038/s41417-024-00742-2
117. Jiang Y, Zhao H, Chen Y, Li K, Li T, Chen J, et al. Exosomal long noncoding RNA HOXD-AS1 promotes prostate cancer metastasis via miR-361-5p/FOXM1 axis. Cell Death Dis. (2021) 12:1129. doi: 10.1038/s41419-021-04421-0
118. Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu Rev Pathol. (2018) 13:395–412. doi: 10.1146/annurev-pathol-020117-043854
119. Gaballa R, Ali HEA, Mahmoud MO, Rhim JS, Ali HI, Salem HF, et al. Exosomes-mediated transfer of itga2 promotes migration and invasion of prostate cancer cells by inducing epithelial-mesenchymal transition. Cancers (Basel). (2020) 12:2300. doi: 10.3390/cancers12082300
120. Wang X, Wang X, Zhu Z, Li W, Yu G, Jia Z, et al. Prostate carcinoma cell-derived exosomal MicroRNA-26a modulates the metastasis and tumor growth of prostate carcinoma. BioMed Pharmacother. (2019) 117:109109. doi: 10.1016/j.biopha.2019.109109
121. Dai C, Heemers H, Sharifi N. Androgen signaling in prostate cancer. Cold Spring Harb Perspect Med. (2017) 7:a030452. doi: 10.1101/cshperspect.a030452
122. El-Kenawi A, Dominguez-Viqueira W, Liu M, Awasthi S, Abraham-Miranda J, Keske A, et al. Macrophage-derived cholesterol contributes to therapeutic resistance in prostate cancer. Cancer Res. (2021) 81:5477–90. doi: 10.1158/0008-5472.CAN-20-4028
123. Zhang H, Li M, Zhang J, Shen Y, Gui Q. Exosomal circ-XIAP promotes docetaxel resistance in prostate cancer by regulating miR-1182/TPD52 axis. Drug Des Devel Ther. (2021) 15:1835–49. doi: 10.2147/DDDT.S300376
124. Cao Z, Xu L, Zhao S. Exosome-derived miR-27a produced by PSC-27 cells contributes to prostate cancer chemoresistance through p53. Biochem Biophys Res Commun. (2019) 515:345–51. doi: 10.1016/j.bbrc.2019.05.120
125. Lin CJ, Yun EJ, Lo UG, Tai YL, Deng S, Hernandez E, et al. The paracrine induction of prostate cancer progression by caveolin-1. Cell Death Dis. (2019) 10:834. doi: 10.1038/s41419-019-2066-3
126. Peak TC, Panigrahi GK, Praharaj PP, Su Y, Shi L, Chyr J, et al. Syntaxin 6-mediated exosome secretion regulates enzalutamide resistance in prostate cancer. Mol Carcinog. (2020) 59:62–72. doi: 10.1002/mc.23129
127. Zhu S, Ni Y, Wang Z, Zhang X, Zhang Y, Zhao F, et al. Plasma exosomal AKR1C3 mRNA expression is a predictive and prognostic biomarker in patients with metastatic castration-resistant prostate cancer. Oncologist. (2022) 27:e870–7. doi: 10.1093/oncolo/oyac177
128. Zhu S, Ni Y, Sun G, Wang Z, Chen J, Zhang X, et al. Exosomal TUBB3 mRNA expression of metastatic castration-resistant prostate cancer patients: Association with patient outcome under abiraterone. Cancer Med. (2021) 10:6282–90. doi: 10.1002/cam4.4168
129. Wang C, Liu X, Li H, Zhao L, Kong G, Chen J, et al. Urinary exosome-based androgen receptor-variant 7 detection in metastatic castration-resistant prostate cancer patients. Transl Androl Urol. (2022) 11:202–12. doi: 10.21037/tau-21-1136
130. Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. (2015) 67:33–41. doi: 10.1016/j.eururo.2014.07.035
131. Guo T, Wang Y, Jia J, Mao X, Stankiewicz E, Scandura G, et al. The Identification of Plasma Exosomal miR-423-3p as a Potential Predictive Biomarker for Prostate Cancer Castration-Resistance Development by Plasma Exosomal miRNA Sequencing. Front Cell Dev Biol. (2020) 8:602493. doi: 10.3389/fcell.2020.602493
132. Wang W, Kong P, Feng K, Liu C, Gong X, Sun T, et al. Exosomal miR-222-3p contributes to castration-resistant prostate cancer by activating mTOR signaling. Cancer Sci. (2023) 114:4252–69. doi: 10.1111/cas.15948
133. Li Y, Ji J, Lyu J, Jin X, He X, Mo S, et al. A Novel Urine Exosomal lncRNA Assay to Improve the Detection of Prostate Cancer at Initial Biopsy: A Retrospective Multicenter Diagnostic Feasibility Study. Cancers (Basel). (2021) 13:4075. doi: 10.3390/cancers13164075
134. Li Q, Hu J, Shi Y, Xiao M, Bi T, Wang C, et al. Exosomal lncAY927529 enhances prostate cancer cell proliferation and invasion through regulating bone microenvironment. Cell Cycle. (2021) 20:2531–46. doi: 10.1080/15384101.2021.1992853
135. Guo Z, Lu X, Yang F, He C, Qin L, Yang N, et al. Exosomal LINC01213 plays a role in the transition of androgen-dependent prostate cancer cells into androgen-independent manners. J Oncol. (2022) 2022:8058770. doi: 10.1155/2022/8058770
136. Zhang G, Liu Y, Yang J, Wang H, Xing Z. Inhibition of circ_0081234 reduces prostate cancer tumor growth and metastasis via the miR-1/MAP 3 K1 axis. J Gene Med. (2022) 24:e3376. doi: 10.1002/jgm.3376
137. Gao F, Xu Q, Tang Z, Zhang N, Huang Y, Li Z, et al. Exosomes derived from myeloid-derived suppressor cells facilitate castration-resistant prostate cancer progression via S100A9/circMID1/miR-506-3p/MID1. J Transl Med. (2022) 20:346. doi: 10.1186/s12967-022-03494-5
138. Kato T, Mizutani K, Kameyama K, Kawakami K, Fujita Y, Nakane K, et al. Serum exosomal P-glycoprotein is a potential marker to diagnose docetaxel resistance and select a taxoid for patients with prostate cancer. Urol Oncol. (2015) 33:385.e15–20. doi: 10.1016/j.urolonc.2015.04.019
139. Bhagirath D, Liston M, Akoto T, Lui B, Bensing BA, Sharma A, et al. Novel, non-invasive markers for detecting therapy induced neuroendocrine differentiation in castration-resistant prostate cancer patients. Sci Rep. (2021) 11:8279. doi: 10.1038/s41598-021-87441-2
140. Skotland T, Ekroos K, Kauhanen D, Simolin H, Seierstad T, Berge V, et al. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur J Cancer. (2017) 70:122–32. doi: 10.1016/j.ejca.2016.10.011
141. Yi X, Li Y, Hu X, Wang F, Liu T. Changes in phospholipid metabolism in exosomes of hormone-sensitive and hormone-resistant prostate cancer cells. J Cancer. (2021) 12:2893–902. doi: 10.7150/jca.48906
142. Qin W, Dallas SL. Exosomes and extracellular RNA in muscle and bone aging and crosstalk. Curr Osteoporos Rep. (2019) 17:548–59. doi: 10.1007/s11914-019-00537-7
143. Kato T, Mizutani K, Kawakami K, Fujita Y, Ehara H, Ito M. CD44v8–10 mRNA contained in serum exosomes as a diagnostic marker for docetaxel resistance in prostate cancer patients. Heliyon. (2020) 6:e04138. doi: 10.1016/j.heliyon.2020.e04138
144. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. (2017) 16:203–22. doi: 10.1038/nrd.2016.246
145. Mori MA, Ludwig RG, Garcia-Martin R, Brandão BB, Kahn CR. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. (2019) 30:656–73. doi: 10.1016/j.cmet.2019.07.011
146. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. (2017) 77:3965–81. doi: 10.1158/0008-5472.CAN-16-2634
147. Chen F, Wang N, Tan HY, Guo W, Zhang C, Feng Y. The functional roles of exosomes-derived long non-coding RNA in human cancer. Cancer Biol Ther. (2019) 20:583–92. doi: 10.1080/15384047.2018.1564562
148. Song J, Chen ZH, Zheng CJ, Song KH, Xu GY, Xu S, et al. Exosome-transported circRNA_0000253 competitively adsorbs microRNA-141-5p and increases IDD. Mol Ther Nucleic Acids. (2020) 21:1087–99. doi: 10.1016/j.omtn.2020.07.039
149. Bian X, Liu R, Meng Y, Xing D, Xu D, Lu Z. Lipid metabolism and cancer. J Exp Med. (2021) 218:e20201606. doi: 10.1084/jem.20201606
150. Donoso-Quezada J, Ayala-Mar S, González-Valdez J. The role of lipids in exosome biology and intercellular communication: Function, analytics and applications. Traffic. (2021) 22:204–20. doi: 10.1111/tra.12803
151. Lorenc T, Klimczyk K, Michalczewska I, Słomka M, Kubiak-Tomaszewska G, Olejarz W. Exosomes in prostate cancer diagnosis, prognosis and therapy. Int J Mol Sci. (2020) 21:2118. doi: 10.3390/ijms21062118
152. Batrakova EV, Kim MS. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J Control Release. (2015) 219:396–405. doi: 10.1016/j.jconrel.2015.07.030
153. Han Q, Xie QR, Li F, Cheng Y, Wu T, Zhang Y, et al. Targeted inhibition of SIRT6 via engineered exosomes impairs tumorigenesis and metastasis in prostate cancer. Theranostics. (2021) 11:6526–41. doi: 10.7150/thno.53886
154. Kurniawati I, Liu MC, Hsieh CL, Do AD, Sung SY. Targeting castration-resistant prostate cancer using mesenchymal stem cell exosomes for therapeutic microRNA-let-7c delivery. Front Biosci (Landmark Ed). (2022) 27:256. doi: 10.31083/j.fbl2709256
155. Gan J, Liu S, Zhang Y, He L, Bai L, Liao R, et al. MicroRNA-375 is a therapeutic target for castration-resistant prostate cancer through the PTPN4/STAT3 axis. Exp Mol Med. (2022) 54:1290–305. doi: 10.1038/s12276-022-00837-6
156. Ishizuya Y, Uemura M, Narumi R, Tomiyama E, Koh Y, Matsushita M, et al. The role of actinin-4 (ACTN4) in exosomes as a potential novel therapeutic target in castration-resistant prostate cancer. Biochem Biophys Res Commun. (2020) 523:588–94. doi: 10.1016/j.bbrc.2019.12.084
157. Lin LC, Gao AC, Lai CH, Hsieh JT, Lin H. Induction of neuroendocrine differentiation in castration resistant prostate cancer cells by adipocyte differentiation-related protein (ADRP) delivered by exosomes. Cancer Lett. (2017) 391:74–82. doi: 10.1016/j.canlet.2017.01.018
158. Del Re M, Biasco E, Crucitta S, Derosa L, Rofi E, Orlandini C, et al. The detection of androgen receptor splice variant 7 in plasma-derived exosomal RNA strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients. Eur Urol. (2017) 71:680–7. doi: 10.1016/j.eururo.2016.08.012
159. Wang D, Wan Z, Yang Q, Chen J, Liu Y, Lu F, et al. Sonodynamical reversion of immunosuppressive microenvironment in prostate cancer via engineered exosomes. Drug Deliv. (2022) 29:702–13. doi: 10.1080/10717544.2022.2044937
160. Wang N, Wang S, Wang X, Zheng Y, Yang B, Zhang J, et al. Research trends in pharmacological modulation of tumor-associated macrophages. Clin Transl Med. (2021) 11:e288. doi: 10.1002/ctm2.288
161. Peng Y, Zhao M, Hu Y, Guo H, Zhang Y, Huang Y, et al. Blockade of exosome generation by GW4869 inhibits the education of M2 macrophages in prostate cancer. BMC Immunol. (2022) 23:37. doi: 10.1186/s12865-022-00514-3
Keywords: biomarker, exosomes, castration-resistant prostate cancer, diagnosis, treatment
Citation: Cao Y, Wang J, Luo H, Wang Y, Cai X, Zhang T, Liao Y and Wang D (2025) Role of exosomes in castration-resistant prostate cancer. Front. Oncol. 15:1498733. doi: 10.3389/fonc.2025.1498733
Received: 26 September 2024; Accepted: 18 April 2025;
Published: 14 May 2025.
Edited by:
Shiv K Gupta, Mayo Clinic, United StatesReviewed by:
Zakaria Y. Abd Elmageed, Edward Via College of Osteopathic Medicine (VCOM), United StatesHojat Dehghanbanadaki, Tehran University of Medical Sciences, Iran
Copyright © 2025 Cao, Wang, Luo, Wang, Cai, Zhang, Liao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yougang Liao, TGlhb3lvdWdhbmc4OTUzQDE2My5jb20=; Decai Wang, ZGVjYWl3YW5nXzIwMjBAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Yuan Cao1,2†
Yuan Cao1,2† Jianjun Wang
Jianjun Wang Xianfu Cai
Xianfu Cai Decai Wang
Decai Wang