- 1Department of Pathology, Hunan Cancer Hospital, The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, Hunan, China
- 2Department of Physiology, School of Basic Medical Science, Central South University, Changsha, Hunan, China
- 3Postdoctoral Research Station of Biology, School of Basic Medical Science, Central South University, Changsha, Hunan, China
Introduction: Non-small cell lung cancer (NSCLC) remains a principal cause of cancer-related mortality. The discovery of novel biomarkers is pivotal for enhancing early diagnosis, refining prognostic evaluations, and optimizing targeted therapeutic strategies. Proteasome 26S subunit ATPase 4 (PSMC4), a proteasome subunit essential for protein degradation, influences tumor progression regulatory mechanisms. Despite its recognized importance, the specific contributions of PSMC4 to NSCLC progression are not well defined.
Methods: This investigation employs a combination of bioinformatics and histological methods to delineate the expression profile of PSMC4 in NSCLC and its correlations with clinicopathological characteristics, diagnostic efficacy, prognostic value, and tumor microenvironment.
Results: We reveal an elevated level of PSMC4 in various malignancies, notably lung adenocarcinoma. Elevated levels of PSMC4 are strongly associated with higher pathological T stages, N stages, and pathological stages. Analysis using receiver operating characteristic curves demonstrates the high diagnostic sensitivity and specificity of PSMC4. Furthermore, patients with elevated PSMC4 levels experience markedly reduced overall survival, disease-specific survival, and progression-free intervals. Both univariate and multivariate Cox regression analyses substantiate that PSMC4 serves as an independent prognostic marker. Analysis of differentially expressed genes and functional annotation demonstrate that genes related to PSMC4 are crucial across a spectrum of biological processes, including DNA replication, chromatin assembly, and mitotic prophase. Gene set enrichment analysis reveals significant correlations between PSMC4 and essential signaling pathways such as the G2/M DNA damage checkpoint, WNT signaling pathway, and cellular senescence. Moreover, immunohistochemical evaluations confirm the increased expression of PSMC4 in NSCLC tissues. Functional assays reveal that PSMC4 accelerates the proliferation of lung cancer cells and tumor growth in xenograft models.
Discussion: These results highlight the potential of PSMC4 as a diagnostic and prognostic marker in NSCLC and elucidate its integral role within the tumor immune microenvironment. Consequently, targeting PSMC4 emerges as a viable therapeutic approach for NSCLC.
1 Introduction
Lung adenocarcinoma (LUAD) is the most common histological subtype of non-small cell lung cancer (NSCLC), which accounts for approximately 85% of all lung cancer cases (1, 2). Lung cancer remains the leading cause of cancer-related mortality worldwide, with an estimated 1.8 million deaths annually (3). Despite advances in diagnostic and therapeutic strategies, the five-year survival rate for lung adenocarcinoma patients remains dismally low, primarily due to late-stage diagnosis and the development of resistance to conventional therapies (4–6). Current treatment modalities, including surgery, chemotherapy, radiotherapy, and targeted therapy, have shown limited efficacy in improving long-term survival, underscoring the urgent need for novel biomarkers and therapeutic targets to enhance early diagnosis and treatment outcomes (7, 8).
The proteasome 26S subunit, ATPase 4 (PSMC4), is a crucial component of the 26S proteasome complex involved in the degradation of ubiquitinated proteins, thereby regulating various cellular processes such as cell cycle progression, apoptosis, and DNA repair (9, 10). Recent studies have implicated PSMC4 in various cancers, including breast cancer (10), endometrial cancer (11, 12), prostate carcinoma (13, 14), and oral squamous cell carcinoma (15), where its dysregulation contributes to tumorigenesis and cancer progression. Research has revealed that elevated expression of PSMC4 correlates positively with reduced survival rates in breast cancer patients (10). Additionally, evidence suggests that PSMC4 may influence the progression of prostate cancer by modulating the CBX3-EGFR-PI3K-AKT-mTOR signaling pathway (14). These findings suggest that PSMC4 may serve as a potential biomarker and therapeutic target in cancer. However, the role of PSMC4 in lung cancer remains largely unexplored.
In this study, we investigated the expression levels of PSMC4 in lung adenocarcinoma and assessed its associations with clinicopathological features, immune cell infiltration, and functional enrichment. Utilizing data from public repositories, including The Cancer Genome Atlas (TCGA), the Genotype-Tissue Expression project (GTEx), and the Gene Expression Omnibus (GEO), we analyzed the expression of PSMC4 and its correlation with clinical outcomes. Considering the pivotal role of immune status in the survival of lung cancer patients (16, 17), we also explored the impact of PSMC4 expression on immune cell infiltration. Furthermore, we conducted comprehensive in vitro and in vivo experiments to delineate the functional role of PSMC4 in the progression of lung cancer.
2 Materials and methods
2.1 Data processing and ethical statement
For this investigation, comprehensive gene expression data were sourced from prominent public databases, including The Cancer Genome Atlas (TCGA) from the National Cancer Institute, the Genotype-Tissue Expression (GTEx) project, and the Gene Expression Omnibus (GEO). These repositories provided a robust foundation for analyzing the expression patterns of the PSMC4 gene in lung adenocarcinoma as well as a spectrum of other cancers. We extracted mRNA expression profiles for 33 cancer types and their corresponding normal tissues, primarily from the TCGA database. Additionally, the GEO dataset GSE19804, which comprises expression data from 60 lung cancer specimens alongside 60 control samples, was utilized for comparative analyses.
Data from TCGA were retrieved directly from its official portal (https://portal.gdc.cancer.gov/), encompassing extensive gene expression data and relevant clinical information across 33 cancer types. GTEx data were accessed via its dedicated portal (https://gtexportal.org/), providing a comprehensive reference for normal tissue gene expression. The dataset GSE19804 from GEO was downloaded from its platform (https://www.ncbi.nlm.nih.gov/geo/), offering balanced gene expression data from both lung cancer and normal samples.
Following data acquisition, rigorous procedures—including quality control, normalization, and statistical analyses—were implemented to ensure the integrity and consistency of the data. Given that all samples were derived from publicly accessible databases with the necessary ethical approvals, this study was exempt from further ethical review. Throughout the research, stringent measures were taken to protect participant privacy and rights.
2.2 Correlation analysis between PSMC4 gene expression and clinicopathological features in lung adenocarcinoma
To elucidate the association between PSMC4 gene expression and the clinicopathological characteristics of lung adenocarcinoma, we conducted a series of statistical analyses. These included chi-square tests for categorical variables, logistic regression for binary outcomes, and Wilcoxon rank-sum tests for continuous variables. The diagnostic potential of the PSMC4 gene was assessed through Receiver Operating Characteristic (ROC) analysis. Survival probabilities were estimated using Kaplan-Meier curve analysis, while the impact of variables on survival was quantified using both univariate and multivariate Cox proportional hazards regression models. Visualization of prognostic indicators was achieved through the construction of nomograms, utilizing the “rms” package in R for both nomogram creation and calibration plot generation.
2.3 Differential gene expression and functional enrichment analysis related to PSMC4 expression levels in lung adenocarcinoma patients
Patients in the TCGA lung adenocarcinoma cohort were stratified into low and high PSMC4 expression groups based on the median expression level of PSMC4 mRNA. Differential gene expression between these groups was analyzed using the “limma” package in R, with results visually represented in volcano plots and ranked difference plots to highlight the significant contrasts. For further insight into the biological implications of these expression differences, functional enrichment analyses were performed. This included assessing Gene Ontology (GO) categories, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and Gene Set Enrichment Analysis (GSEA), using the “clusterProfiler” (18) package in R to identify enriched pathways and biological processes associated with the dichotomized expression of PSMC4.
2.4 Exploration of the association between PSMC4 and immune cell infiltration in lung adenocarcinoma
To elucidate the link between PSMC4 gene expression and immune cell infiltration in lung adenocarcinoma, we utilized the single-sample Gene Set Enrichment Analysis (ssGSEA) algorithm, as implemented in the “GSVA” (19) package in R. This approach allowed for the quantification of enrichment across 24 prevalent immune cell types. The relationship between PSMC4 expression levels and the respective infiltration scores of these immune cell types was assessed using Spearman correlation analysis.
2.5 Patient samples and tissue collection
Patient samples and tissues for this study were obtained from individuals diagnosed with lung cancer at the Hunan Cancer Hospital over the period from 2021 to 2023. All procedural aspects of sample collection were conducted in compliance with ethical standards set forth by the Ethics Committee of Hunan Cancer Hospital (2024101). Informed consent was secured from each participant prior to sample collection.
2.6 Immunohistochemical staining
Immunohistochemical analysis was performed in accordance with established protocols and evaluation criteria (20). For the detection of PSMC4, tissues were stained using a rabbit anti-PSMC4 antibody (1:100; Proteintech, Wuhan, China; 11389-1-AP).
2.7 Cell culture and gene transfection
Human lung adenocarcinoma cell lines, A549 and H1299, were cultured in RPMI-1640 medium enriched with 10% fetal bovine serum. These cells were maintained at 37°C in a 5% CO2 humidified incubator. For studies on gene function, cells underwent transfection with plasmids containing either PSMC4 short hairpin RNA or PSMC4 complementary DNA using Lipofectamine 3000 (Invitrogen), adhering strictly to the manufacturer’s guidelines. Cells were collected 48 hours after transfection for further analysis.
2.8 Western blot
Proteins were harvested using RIPA buffer from the cells and quantified with a BCA Protein Assay Kit. Following separation by SDS-PAGE, proteins were transferred to PVDF membranes. These membranes were then probed with a rabbit anti-PSMC4 antibody (1:1000; Proteintech, Wuhan, China; 11389-1-AP) and a mouse anti-GAPDH antibody (1:7000; Proteintech, Wuhan, China; 60004-1-Ig). Detection of protein bands was facilitated by enhanced chemiluminescence detection.
2.9 Cell proliferation and colony formation assays
Cell proliferation was evaluated using the CCK-8 assay. After transfection, cells were plated at a density of 5,000 cells per well in 96-well plates. CCK-8 solution (Yeasen Biotechnology, Shanghai, China) was administered every 24 hours, and absorbance was measured with a microplate reader to monitor cell growth. For colony formation assays, cells were seeded in 6-well plates and cultured for two weeks, followed by fixation with methanol and staining with crystal violet to facilitate colony enumeration.
2.10 Establishment of mouse tumor models
For the establishment of mouse tumor models, six-week-old nude mice were randomly assigned into groups of two. Human lung adenocarcinoma H1299 cells, transfected with either PSMC4 overexpression plasmids or control plasmids (1×10^6 cells per mouse), were subcutaneously injected into the right flank of each mouse. Tumor growth was monitored every week, with tumor volume calculated using the formula (V = 0.5 × length × width²). After the experiment, mice were euthanized, and tumors were excised, weighed, and subjected to further analysis. All experimental procedures were conducted in accordance with ethical guidelines and approved by the Ethics Committee of Hunan Cancer Hospital (D2021-142).
2.11 Statistical analysis
Statistical analyses were performed using SPSS version 20.0 (IBM, Armonk, NY, USA) and GraphPad Prism version 8.0.2 (San Diego, California, USA). We designated a p-value of less than 0.05 as statistically significant. To assess differences in quantitative and categorical data between groups, we applied one-way analysis of variance (ANOVA) and Student’s t-tests for continuous variables, and chi-square tests or Fisher’s exact tests for categorical variables, as appropriate.
3 Results
3.1 Elevated expression of PSMC4 across diverse cancer types, including lung cancer
Utilizing TCGA dataset and the GTEx database, our comprehensive analysis of PSMC4 mRNA expression across 33 cancer types and their corresponding normal tissues revealed significant upregulation of PSMC4 in 26 out of these 33 types, most notably in hepatocellular carcinoma, lung adenocarcinoma, squamous cell carcinoma of the lung, and gastric cancer (Figure 1A). Paired sample analysis robustly confirmed PSMC4’s elevated expression in these malignancies (Figure 1B). A detailed examination of 539 lung adenocarcinoma samples and 59 normal lung samples from TCGA highlighted a pronounced increase in PSMC4 expression in lung adenocarcinoma (Figure 1C). This elevation was corroborated through differential analysis of paired samples, where lung adenocarcinoma exhibited significantly higher levels of PSMC4 compared to the normal lung tissues (Figure 1D). Further integration of data from TCGA and GTEx, encompassing an expanded cohort of 347 normal tissues, also demonstrated a marked upregulation of PSMC4 in LUAD (Figure 1E). Moreover, analysis of the GEO dataset GSE19804, which includes 60 lung cancer samples and 60 matched normal samples, consistently supported the upregulation of PSMC4 in lung cancer (Figure 1F). These observations collectively underscore the widespread upregulation of PSMC4 across multiple cancer types, emphasizing its potential role in tumorigenesis.
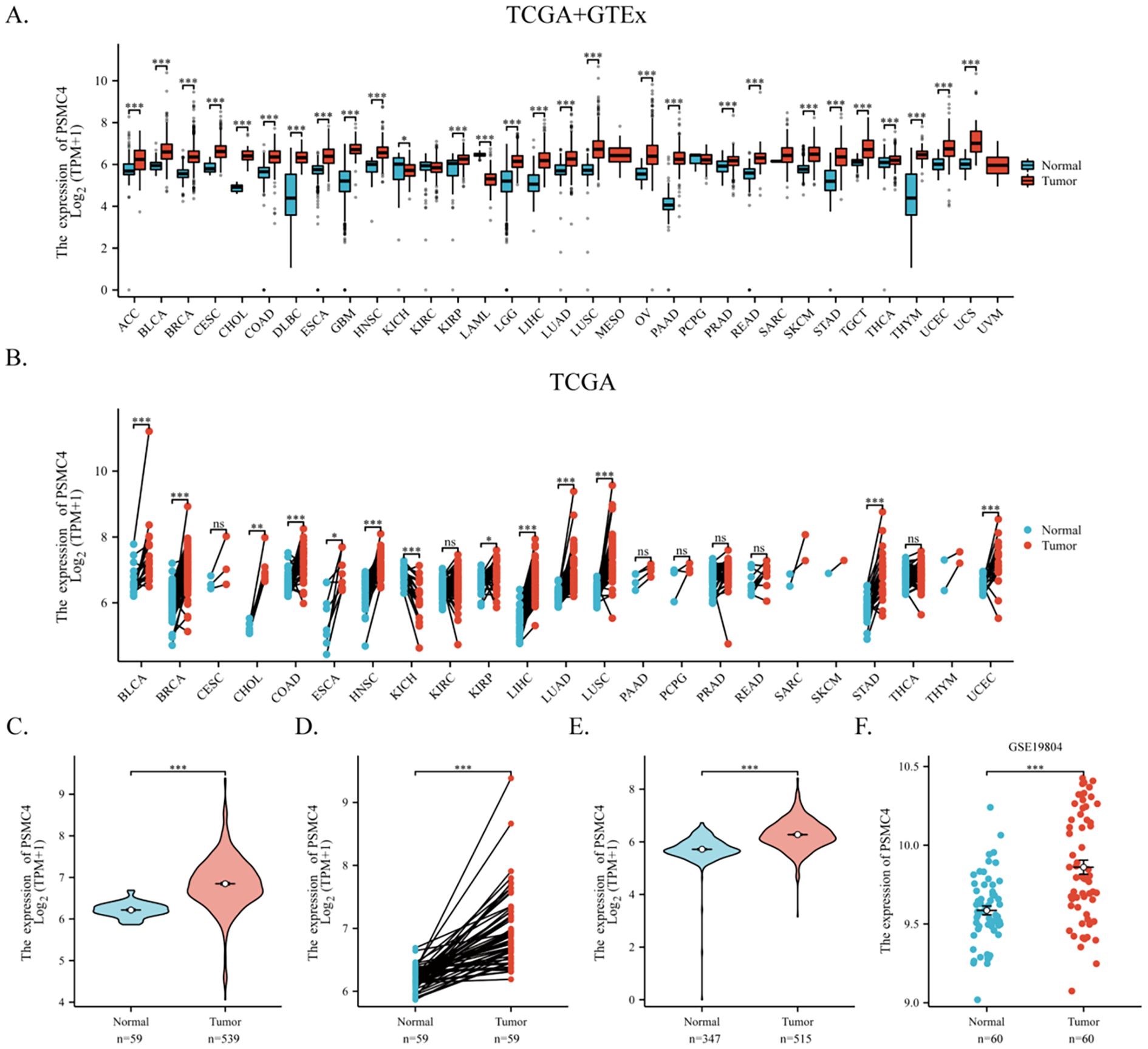
Figure 1. Upregulation of PSMC4 expression in lung cancer. (A, B) Expression levels of PSMC4 in various cancer tissues and their adjacent normal tissues from TCGA and GTEx databases. (C, D) Comparison of PSMC4 expression levels between normal tissues and lung adenocarcinoma tissues from the TCGA database. (E) Comparison of PSMC4 expression levels between lung adenocarcinoma and adjacent normal tissues from TCGA and GTEx databases. (F) Analysis of PSMC4 expression levels in the GEO dataset GSE19804 shows significant differences (ns: p>0.05, *p<0.05, **p<0.01, ***p<0.001) (ACC, Adrenocortical carcinoma; BLCA, Bladder urothelial carcinoma; BRCA, Breast invasive carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, Cholangiocarcinoma; COAD, Colon adenocarcinoma; DLBC, Diffuse large B-cell lymphoma; ESCA, Esophageal carcinoma; GBM, Glioblastoma multiforme; HNSC, Head and neck squamous cell carcinoma; KICH, Kidney chromophobe; KIRC, Kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LAML, Acute myeloid leukemia; LGG, Low-grade glioma; LIHC, Liver hepatocellular carcinoma; LUAD, Lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; MESO, Mesothelioma; OV, Ovarian serous cystadenocarcinoma; PAAD, Pancreatic adenocarcinoma; PCPG, Pheochromocytoma and paraganglioma; PRAD, Prostate adenocarcinoma; READ, Rectum adenocarcinoma; SARC, Sarcoma; SKCM, Skin cutaneous melanoma; STAD, Stomach adenocarcinoma; TGCT, Testicular germ cell tumors; THCA, Thyroid carcinoma; THYM, Thymoma; UCEC, Uterine corpus endometrial carcinoma; UCS, Uterine carcinosarcoma; UVM, Uveal melanoma).
3.2 Correlation between PSMC4 expression and clinical pathologic features in lung adenocarcinoma
Our investigation elucidates the relationship between the expression of PSMC4 and various clinical-pathological characteristics in lung adenocarcinoma. Table 1 illustrates that patients with elevated PSMC4 expression tend to present with more advanced pathological T stages, N stages, and overall pathologic stages compared to those exhibiting lower expression levels. However, no notable differences were observed in other clinical-pathological parameters. Further stratification of PSMC4 expression against different pathological indices demonstrated a significant positive correlation with the severity of T stage, N stage, and overall pathologic stage (Figures 2A–C). Specifically, increased PSMC4 levels were significantly associated with advanced stages in these categories. Logistic regression analysis substantiated this positive association, affirming that higher PSMC4 expression correlates with more advanced T stage, N stage, and pathologic stage in patients with lung adenocarcinoma (Table 2). This correlation underscores the potential of PSMC4 as a prognostic marker for tumor progression in lung adenocarcinoma.
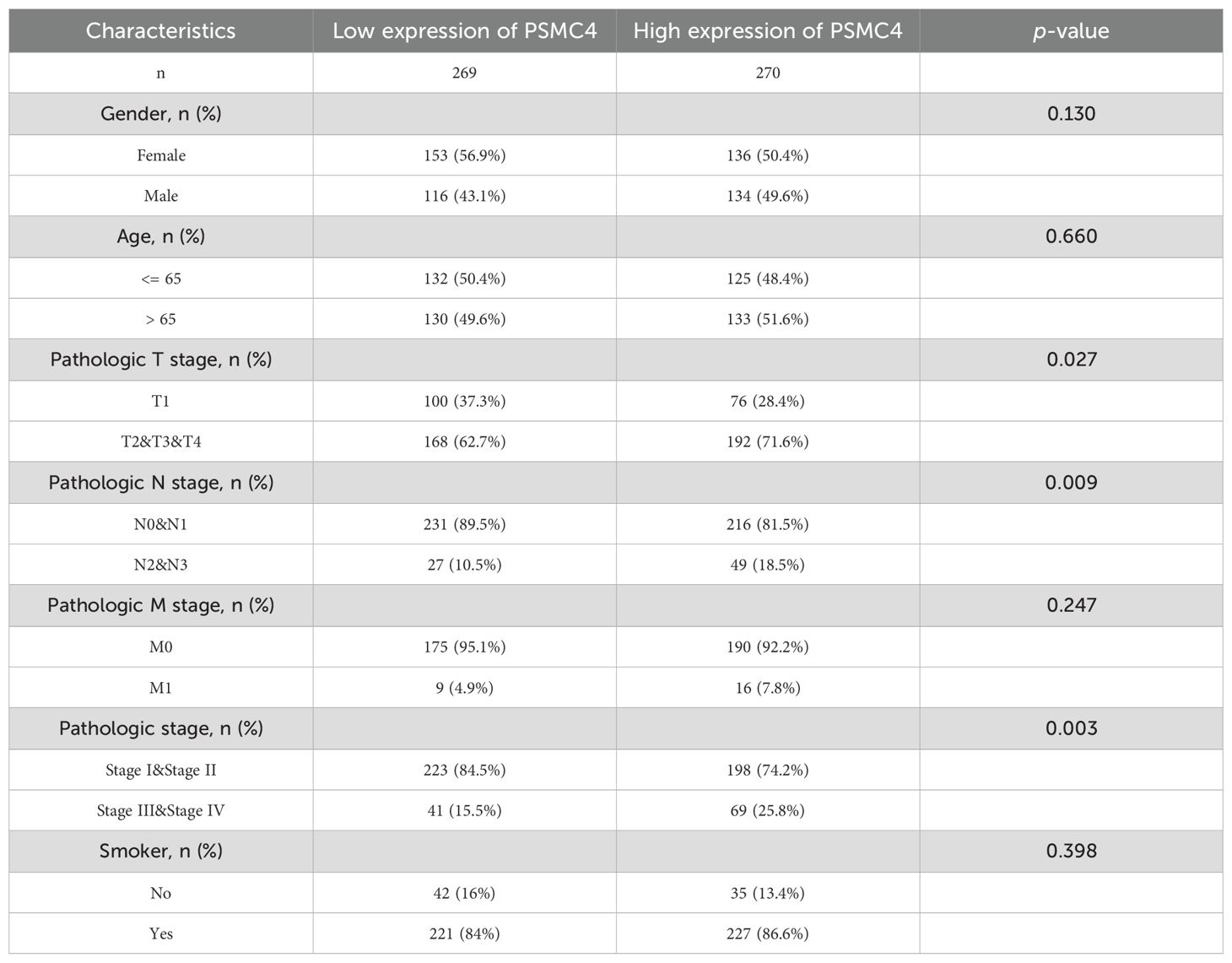
Table 1. Correlation analysis of PSMC4 expression with clinicopathological features in lung adenocarcinoma patients.
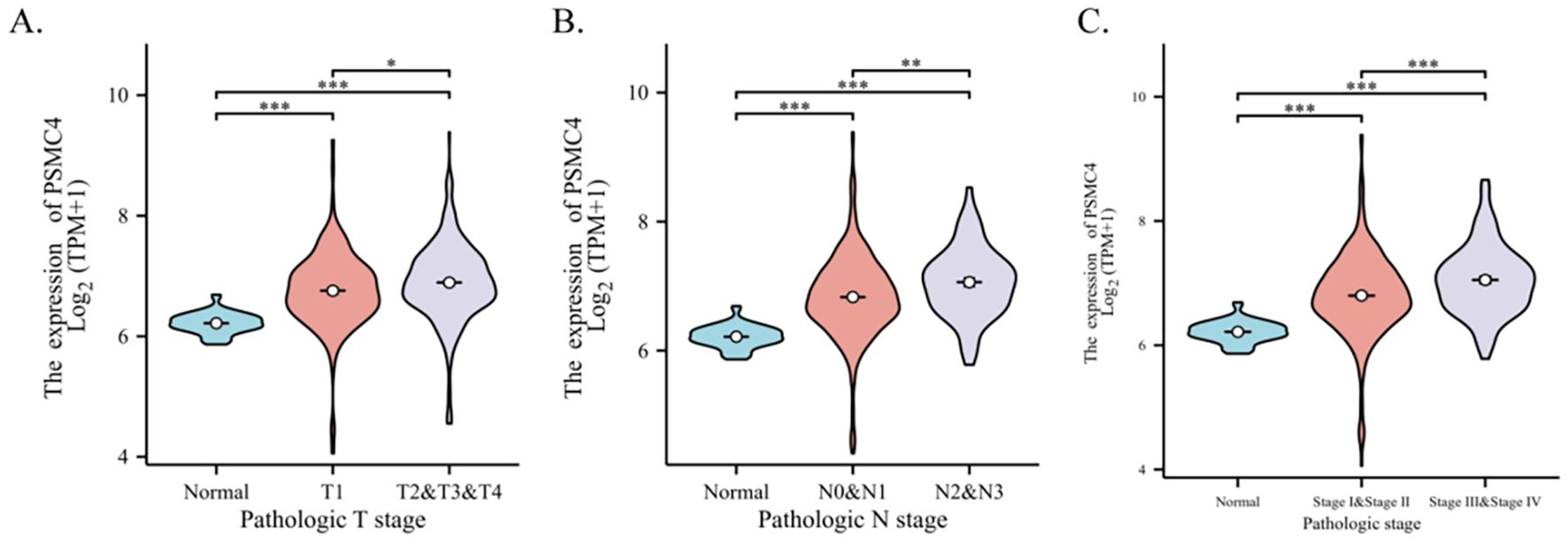
Figure 2. Correlation between PSMC4 expression and clinical pathological features in lung adenocarcinoma patients. The association of PSMC4 expression with T stage (A), N stage (B), and pathologic stage (C) in lung adenocarcinoma patients were analyzed using the Wilcoxon rank-sum test (*p<0.05, **p<0.01, ***p<0.001).
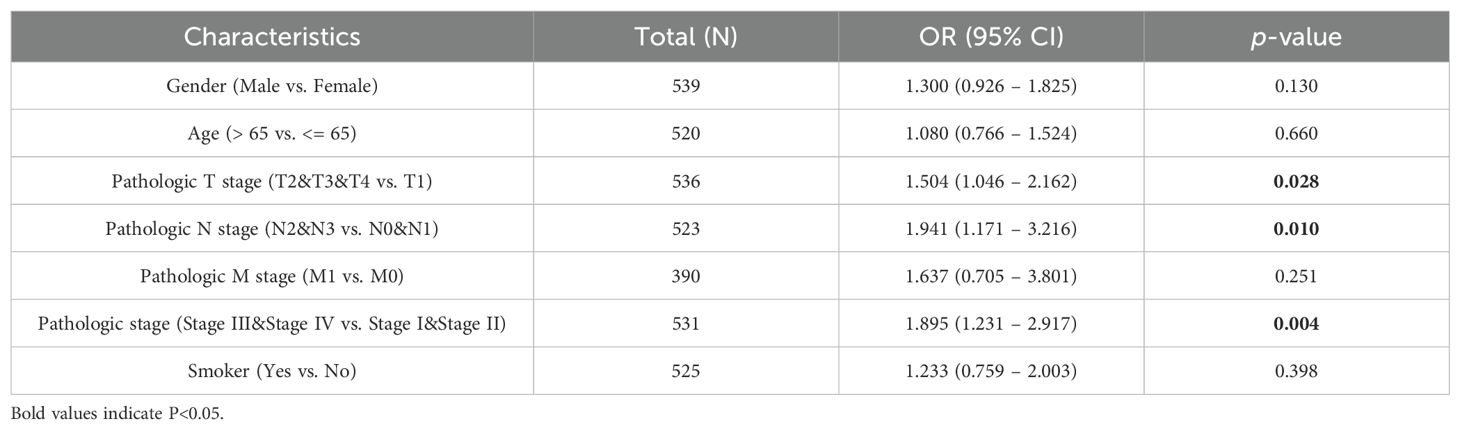
Table 2. Logistic regression model assessing the association between PSMC4 expression and clinicopathological characteristics in lung adenocarcinoma patients.
3.3 PSMC4 as a potential diagnostic and prognostic biomarker for lung adenocarcinoma
The diagnostic capabilities of PSMC4 in lung adenocarcinoma were determined through ROC curve analysis, revealing an area under the curve (AUC) of 0.876, which reflects high diagnostic sensitivity and specificity (Figure 3A). The diagnostic efficacy of PSMC4 varied with pathological stages, manifesting AUC values of 0.861 for stages I-II and 0.939 for stages III-IV, underscoring its stage-dependent utility (Figures 3B,C). Kaplan-Meier survival analysis was employed to ascertain the prognostic significance of PSMC4 expression in clinical outcomes. Notably, elevated PSMC4 expression was associated with significantly reduced overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI), highlighting its adverse impact on survival metrics (p<0.01) (Figures 4A–C). In subgroup analyses, high PSMC4 expression particularly compromised prognoses in patients with early-stage lung adenocarcinoma (N0/N1 and pathological stages I-II), indicating its potential as an early adverse prognostic marker (Figures 4D–I).
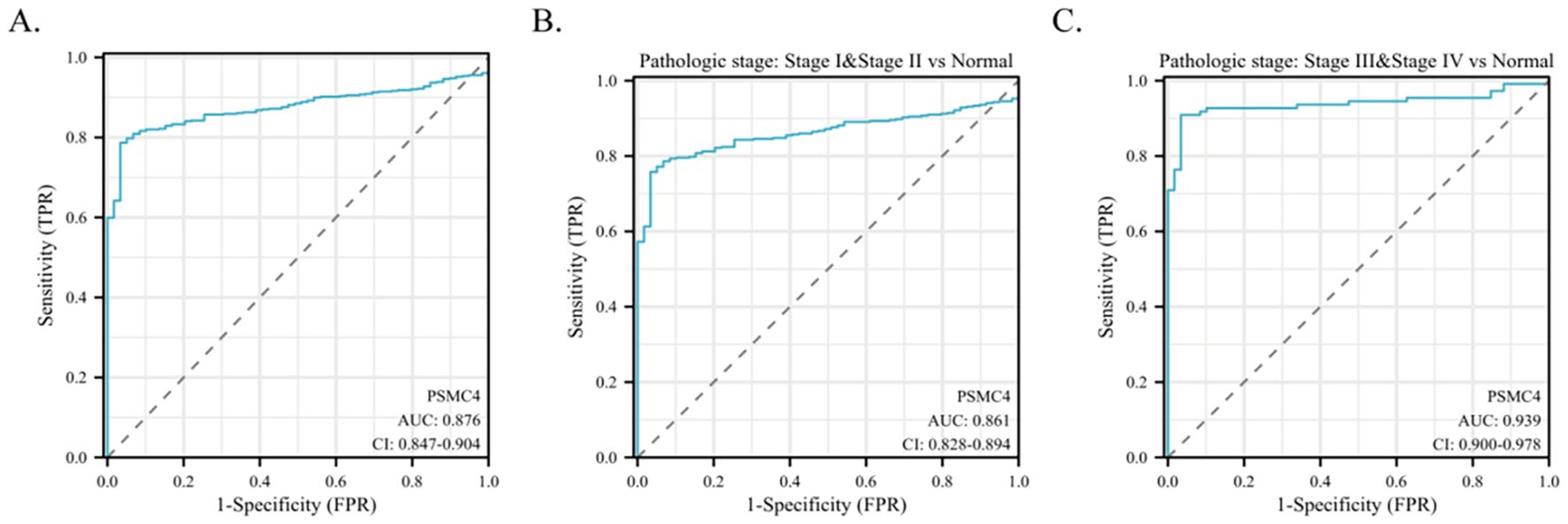
Figure 3. Predictive potential of PSMC4 in lung adenocarcinoma diagnosis. (A) ROC curve evaluation of the diagnostic value of PSMC4 in lung adenocarcinoma. (B, C) Subgroup analysis based on pathological stages I-II and III-IV.
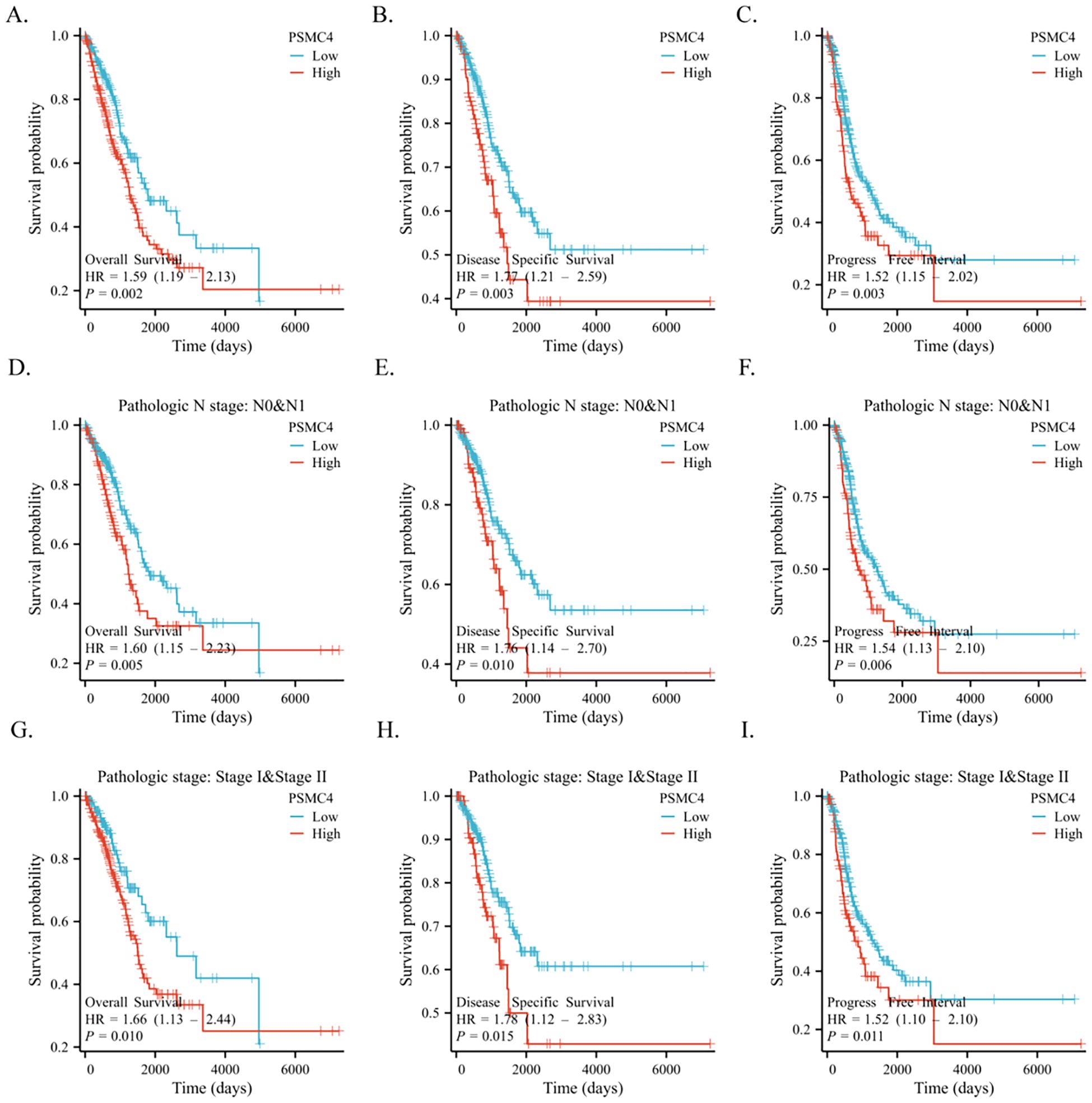
Figure 4. Application of PSMC4 in the clinical prognosis of lung adenocarcinoma. Kaplan-Meier survival analysis comparing high and low PSMC4 expression levels in lung adenocarcinoma patients in terms of overall survival (A), disease-specific survival (B), and progression-free interval (C). Additionally, the same analysis was performed for N0/N1 stage (D-F) and pathological stages I-II (G-I) lung adenocarcinoma patients.
To further delineate risk factors influencing overall survival, both univariate and multivariate Cox regression analyses were executed. Univariate analysis underscored the association of T stage, N stage, pathologic stage, and PSMC4 expression with poor prognosis (Table 3, p<0.05). The multivariate analysis, adjusting for confounders, identified the T stage, pathologic stage, and PSMC4 expression as independent prognostic indicators (p<0.05). Additionally, the prognostic model we developed, which integrates these significant independent factors, demonstrated robust predictive accuracy with a concordance index (c-index) of 0.744, affirming its utility in clinical prognostication (Figures 5A, B).
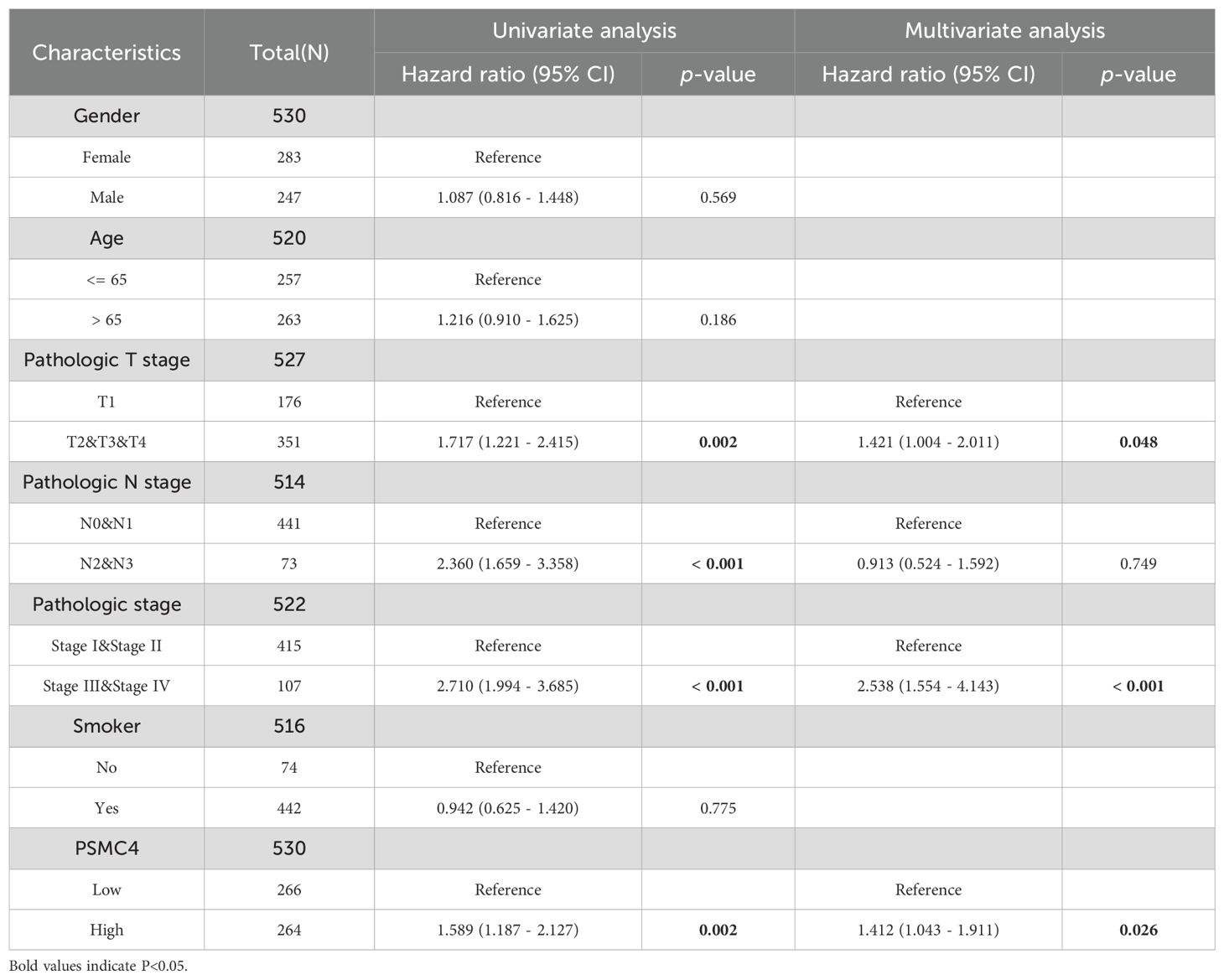
Table 3. Cox regression analysis of overall survival in lung adenocarcinoma patients based on clinical outcomes.
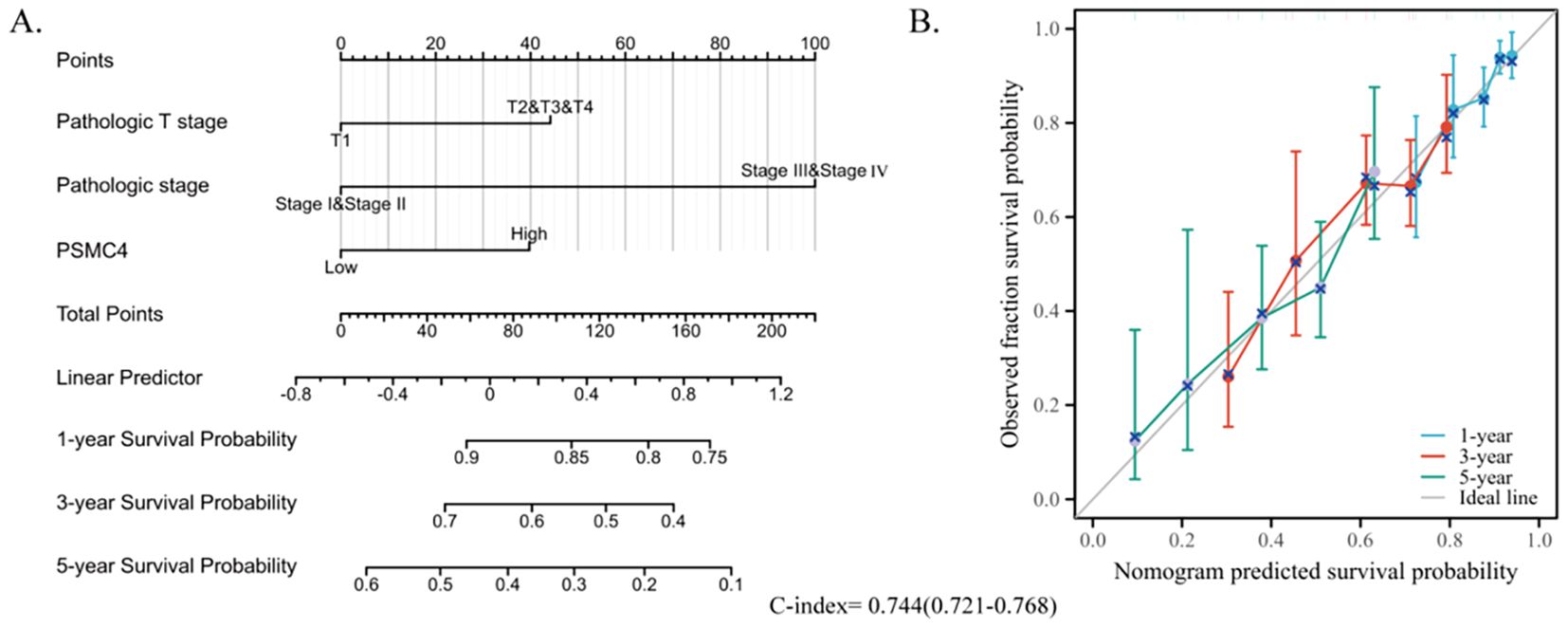
Figure 5. Development of a prognostic nomogram based on PSMC4 expression. (A) Nomogram depicting the 1-year, 3-year, and 5-year overall survival risks based on PSMC4 expression. (B) Calibration plot validating the accuracy of the nomogram in predicting overall survival.
3.4 Comparative analysis of differentially expressed genes in lung adenocarcinoma patients with varying PSMC4 expression levels
We stratified 539 lung adenocarcinoma patients into groups exhibiting high and low expression of PSMC4 based on the median PSMC4 expression. This stratification enabled a comprehensive analysis of differentially expressed genes (DEGs) between these cohorts. In the high PSMC4 expression group, 86 genes were identified as significantly differentially expressed (p<0.05, |log2 fold change (FC)| ≥ 1), including 26 upregulated and 60 downregulated genes (Figures 6A, B). A functional analysis of these genes will be detailed subsequently.
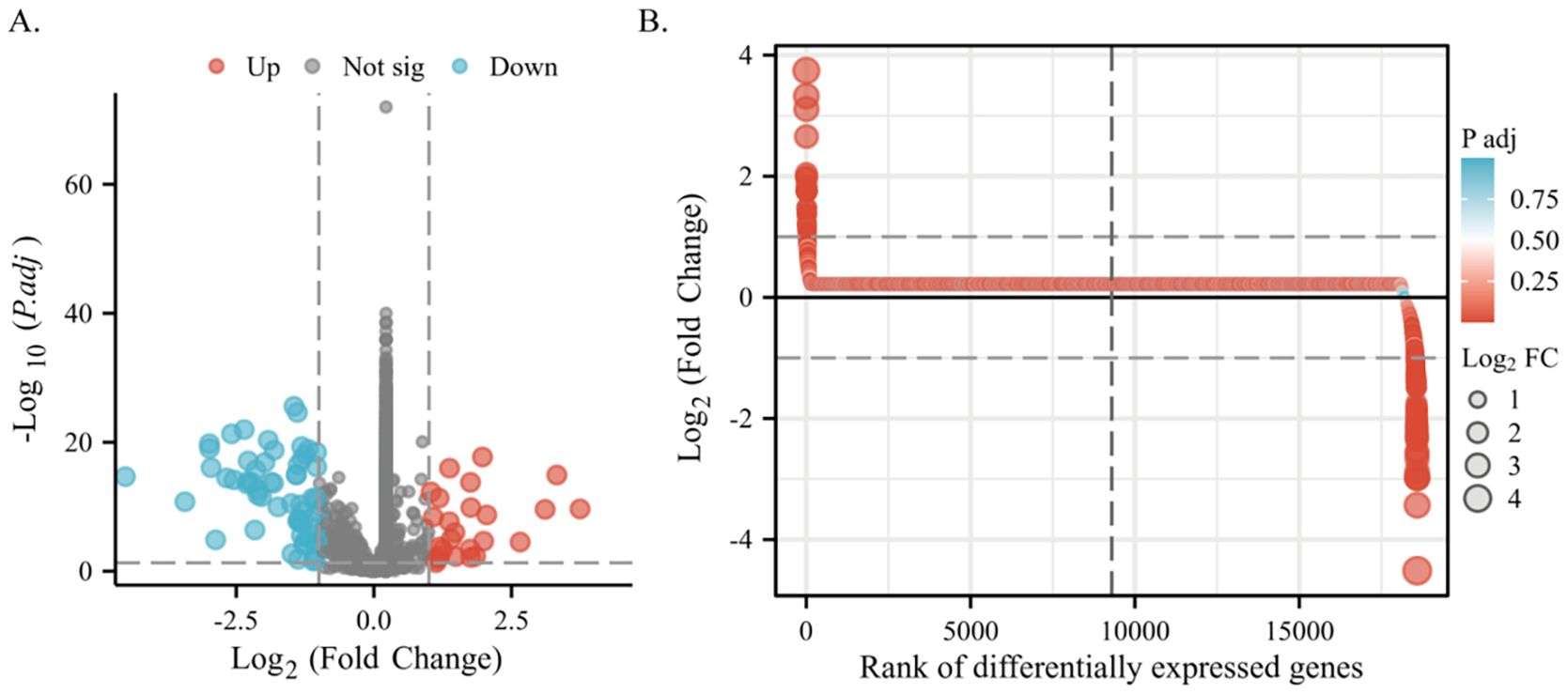
Figure 6. Differential gene expression associated with PSMC4 in lung adenocarcinoma. (A) Volcano plot displaying detailed DEGs between high and low PSMC4 expression lung adenocarcinoma groups. (B) Differential heatmap further illustrates the expression differences of these genes.
3.5 Functional annotation of differentially expressed genes associated with PSMC4
A thorough examination using the “ClusterProfiler” R package uncovered the functional roles of 86 differentially expressed genes across various biological processes (BP). These encompass crucial activities such as DNA replication-dependent chromatin assembly, nucleosome organization, negative regulation of megakaryocyte differentiation, and protein-DNA complex assembly, among others (Figure 7A). Moreover, these genes demonstrate molecular functions primarily characterized by protein heterodimerization activity (Figure 7A). Regarding their cellular localization, they are prominently situated within structures like the nucleosome, DNA packaging complex, and nuclear chromosome (Figure 7B). Notably, KEGG pathway analysis shed light on their involvement in diverse pathways including neutrophil extracellular trap formation, viral carcinogenesis, transcriptional misregulation in cancer, shigellosis, and necroptosis, underlining their significance in disease processes (Figure 7B).
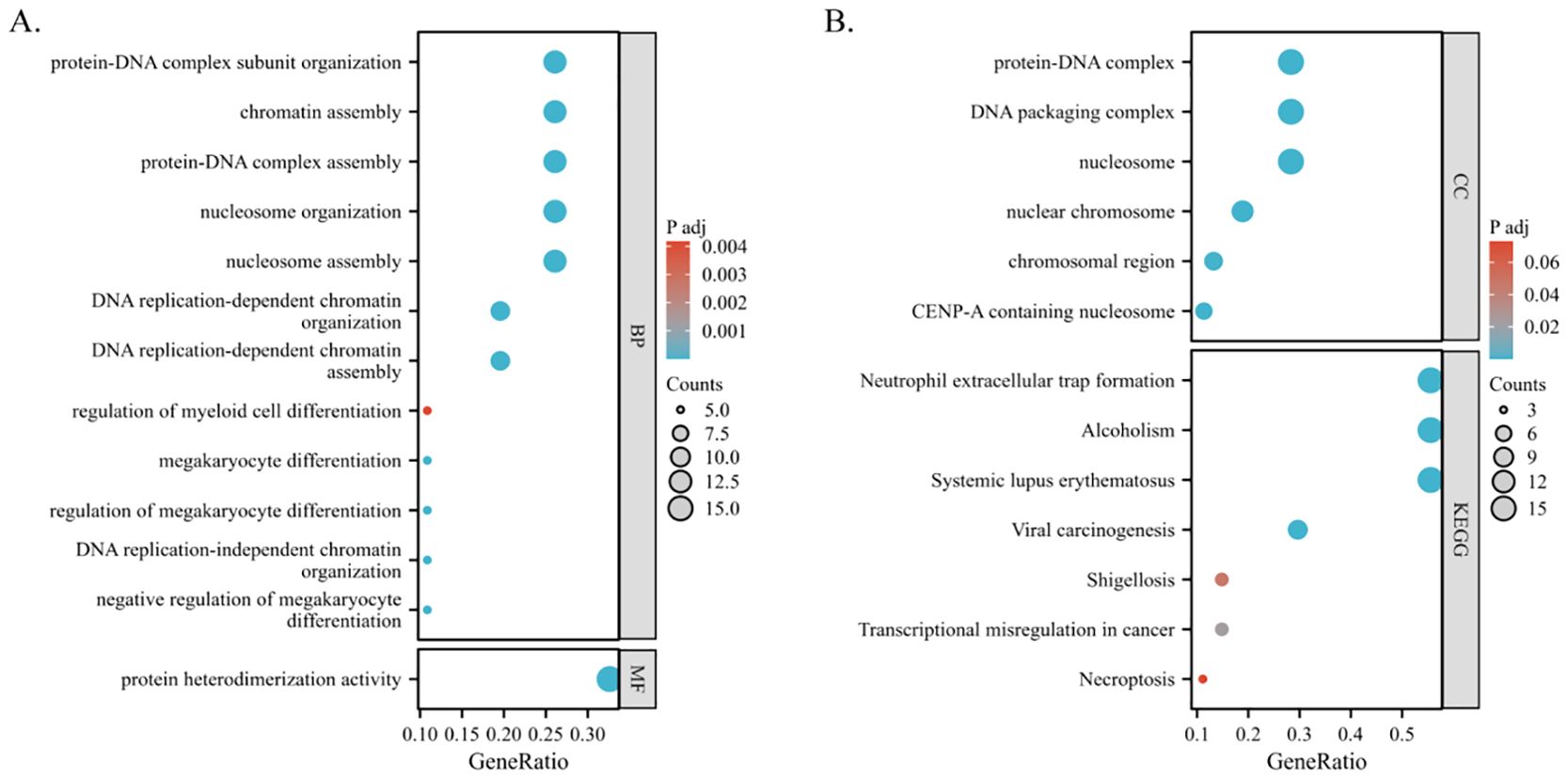
Figure 7. Functional enrichment analysis of PSMC4-related differentially expressed genes in lung adenocarcinoma. (A, B) Biological functions and biological pathways associated with PSMC4 expression-related genes were studied using GO and KEGG pathway analysis.
Furthermore, GSEA revealed that PSMC4 is intricately linked to key biological pathways. These include the G2/M DNA Damage Checkpoint, G2/M Checkpoints, Mitotic Prophase, Epigenetic Regulation of Gene Expression, DNA Replication, Signaling by Notch, Cellular Senescence, Signaling by WNT, Formation of the Beta-Catenin TCF Transactivating Complex, Diseases of Programmed Cell Death, DNA Replication Pre-Initiation, and TCF Dependent Signaling in Response to WNT (Figures 8A–L). These insights are critical for advancing our understanding of the molecular dynamics associated with PSMC4 in the progression of lung adenocarcinoma.
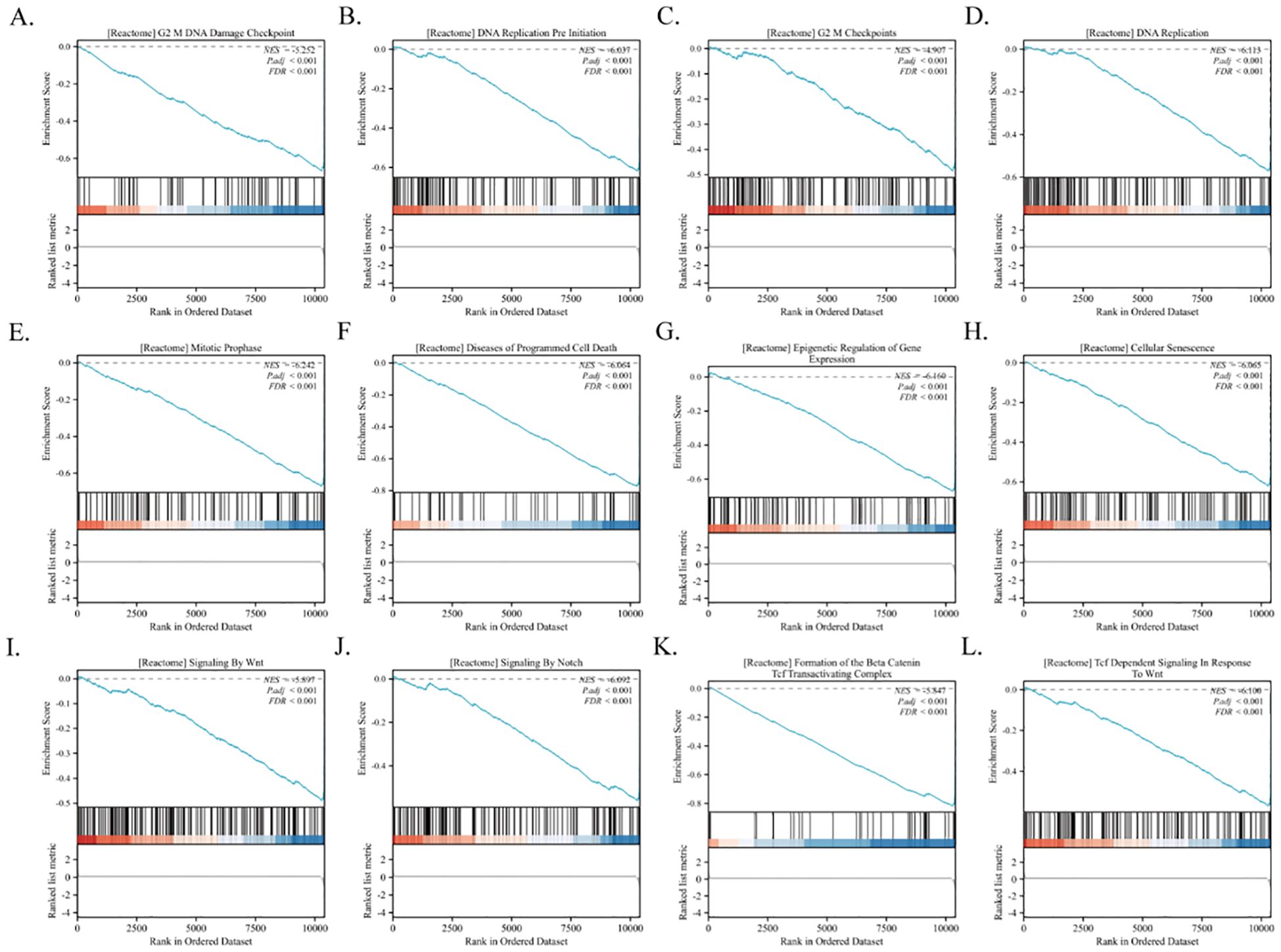
Figure 8. Identification of PSMC4-related signaling pathways in lung adenocarcinoma. (A-L) GSEA was conducted to explore the biological functions and potential signaling pathways of differentially expressed genes related to PSMC4 in lung adenocarcinoma.
3.6 Correlation between PSMC4 expression and immune cell infiltration in lung adenocarcinoma
Utilizing ssGSEA, our study assessed the presence of 24 immune cell types within lung adenocarcinoma tissues, concurrently exploring the relationship between PSMC4 expression and immune cell infiltration using Spearman correlation analysis. Our findings underscore a marked linkage of PSMC4 expression with various immune cell subtypes. Notably, PSMC4 expression correlated positively with Th2 cells (R=0.396, p<0.001) and gamma delta T cells (R=0.175, p<0.001), but negatively with central memory T cells (Tcm) (R=-0.281, p<0.001), mast cells (R=-0.235, p<0.001), effector memory T cells (Tem) (R=-0.232, p<0.001), and B cells (R=-0.203, p<0.001). Subsequently, we verified the infiltration levels of the six most correlated immune cell types, affirming the consistency of our initial analysis (Figures 9A–G).
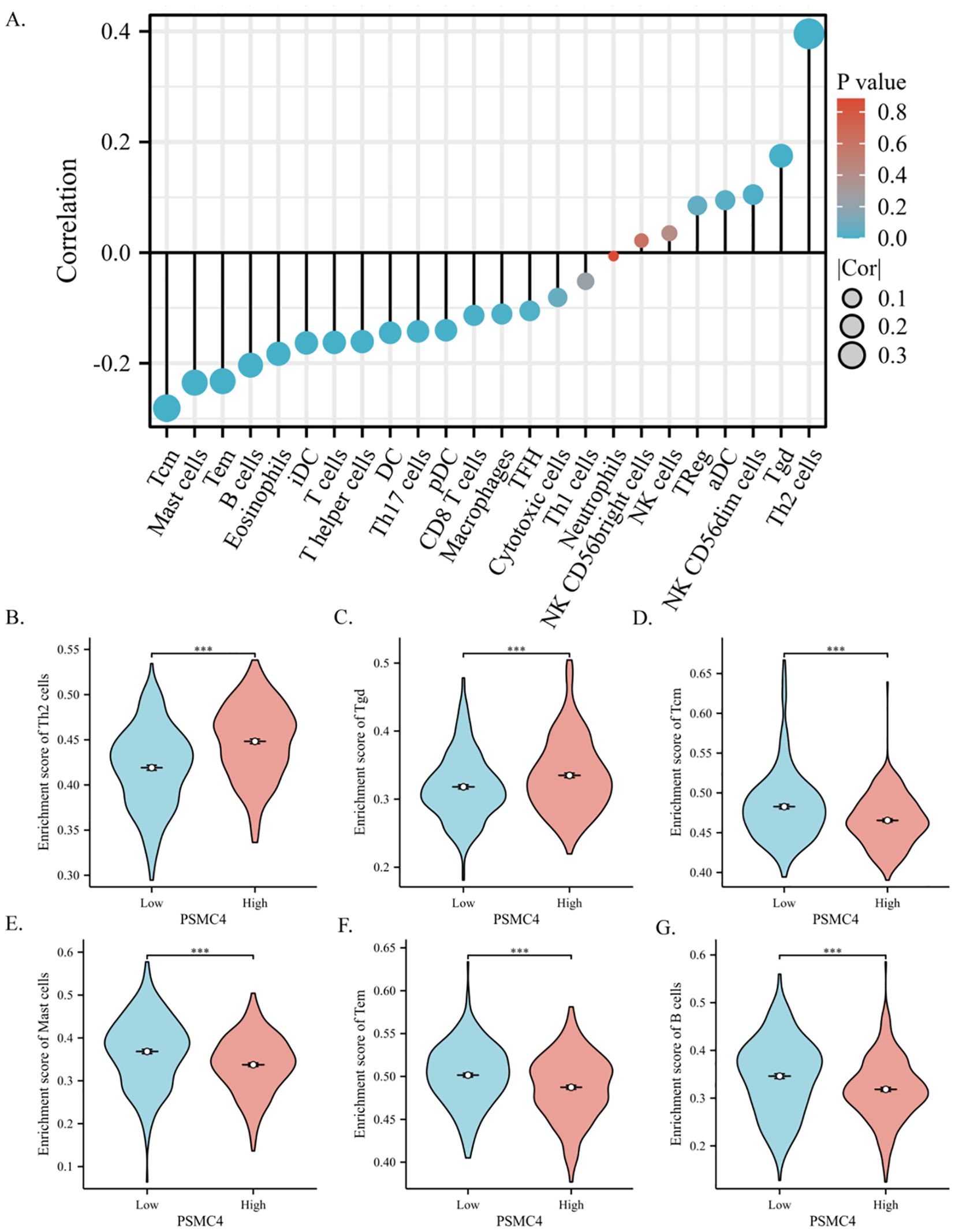
Figure 9. Correlation of PSMC4 with the immune microenvironment in lung adenocarcinoma. (A) Spearman correlation analysis of PSMC4 expression levels with the infiltration levels of 24 immune cells in lung adenocarcinoma. (B–G) Comparison of immune cell infiltration levels between PSMC4 high expression group and low expression group in Th2 cells, Tgd, Tcm, Mast cells, Tem, and B cells. (***p<0.001).
3.7 Validation of PSMC4 expression in non-small cell lung cancer tissues
We procured tumor and adjacent normal tissue samples from 88 individuals diagnosed with NSCLC and scrutinized PSMC4 expression and its subcellular distribution using immunohistochemistry. The analysis demonstrated a predominant cytoplasmic localization of PSMC4 (Figure 10A) and a significant upregulation in NSCLC tissues compared to adjacent normal tissues (p<0.05) (Figure 10B). Stratification of patients into high and low PSMC4 expression groups unveiled significant associations of elevated PSMC4 expression with advanced clinical T stage (p=0.031) and N stage (p=0.027) (Table 4). Notably, patients with NSCLC at T1–T2 stages exhibited lower PSMC4 expression compared to those at T3–T4 stages (Figure 10C), and a similar trend was observed for N0–N1 stage patients, who showed reduced PSMC4 expression relative to those at N2–N3 stages (Figure 10D). Furthermore, the diagnostic efficacy of PSMC4 in non-small cell lung cancer was evaluated through ROC curve analysis. The results indicated an AUC of 0.757, reflecting a high level of diagnostic sensitivity and specificity (Figure 10E).
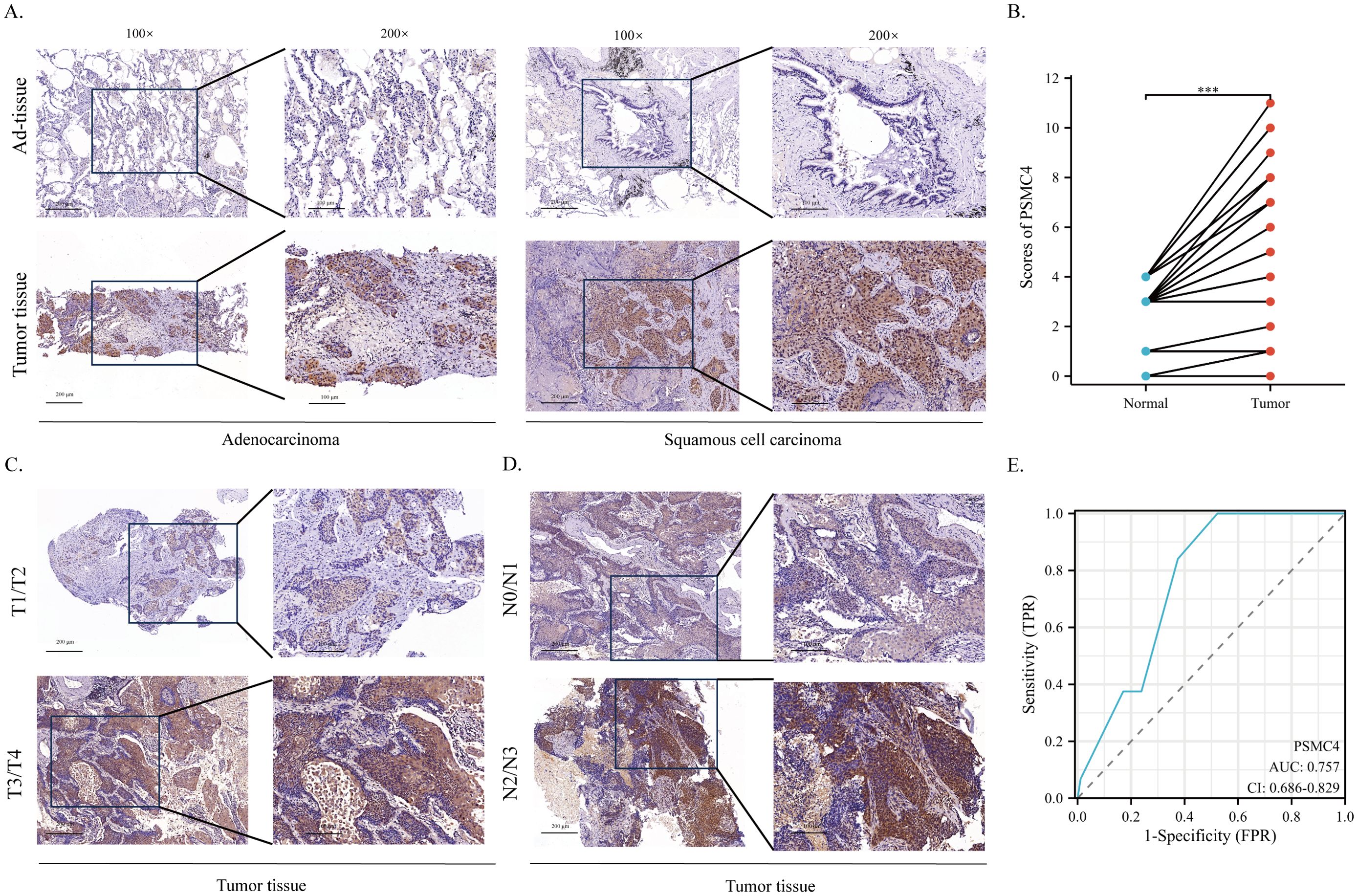
Figure 10. Expression of PSMC4 in non-small cell lung cancer tissues. (A) Representative immunohistochemical staining images illustrating PSMC4 expression in adenocarcinoma, squamous cell carcinoma, and normal lung tissues. The enlarged regions of interest are shown in the right panels (scale bars: 100 μm, 200 μm). (B) Quantitative analysis revealed significantly elevated PSMC4 protein levels in non-small cell lung cancer tissues (***p < 0.001). (C) Representative immunohistochemical staining images showing PSMC4 expression in NSCLC at T1–T2 and T3–T4 stages, with the magnified views of the rectangular regions shown on the right (scale bars: 100 μm, 200 μm). (D) Representative immunohistochemical staining images showing PSMC4 expression in NSCLC at N0–N1 and N2–N3 stages, with the magnified views of the rectangular regions shown on the right (scale bars: 100 μm, 200 μm). (E) ROC curve evaluation of the diagnostic value of PSMC4 in NSCLC.
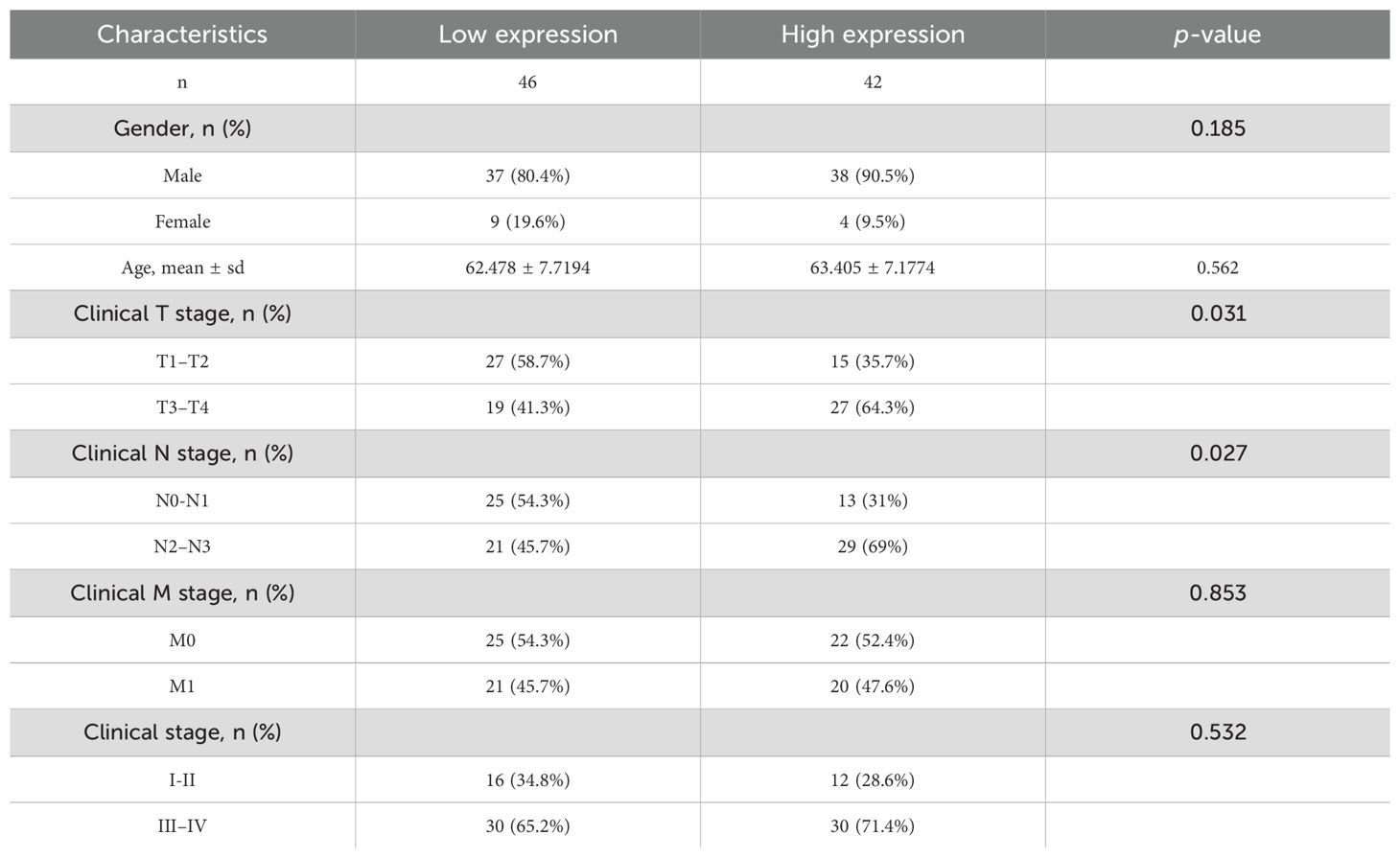
Table 4. Immunohistochemical analysis of PSMC4 expression and its correlation with clinicopathological features in non-small cell lung cancer.
3.8 Impact of PSMC4 on lung cancer cell proliferation
Functional assays were initiated by silencing PSMC4 in A549 cells and enhancing its expression in H1299 cells (Figures 11A, B). Relative to control groups, suppression of PSMC4 substantially reduced the viability of A549 cells (p<0.0001) (Figure 11C), while its overexpression significantly increased the viability of H1299 cells (p<0.0001) (Figure 11D). Moreover, colony formation assays aligned with these findings, further validating PSMC4’s pivotal role in modulating lung cancer cell proliferation (Figures 11E, F).
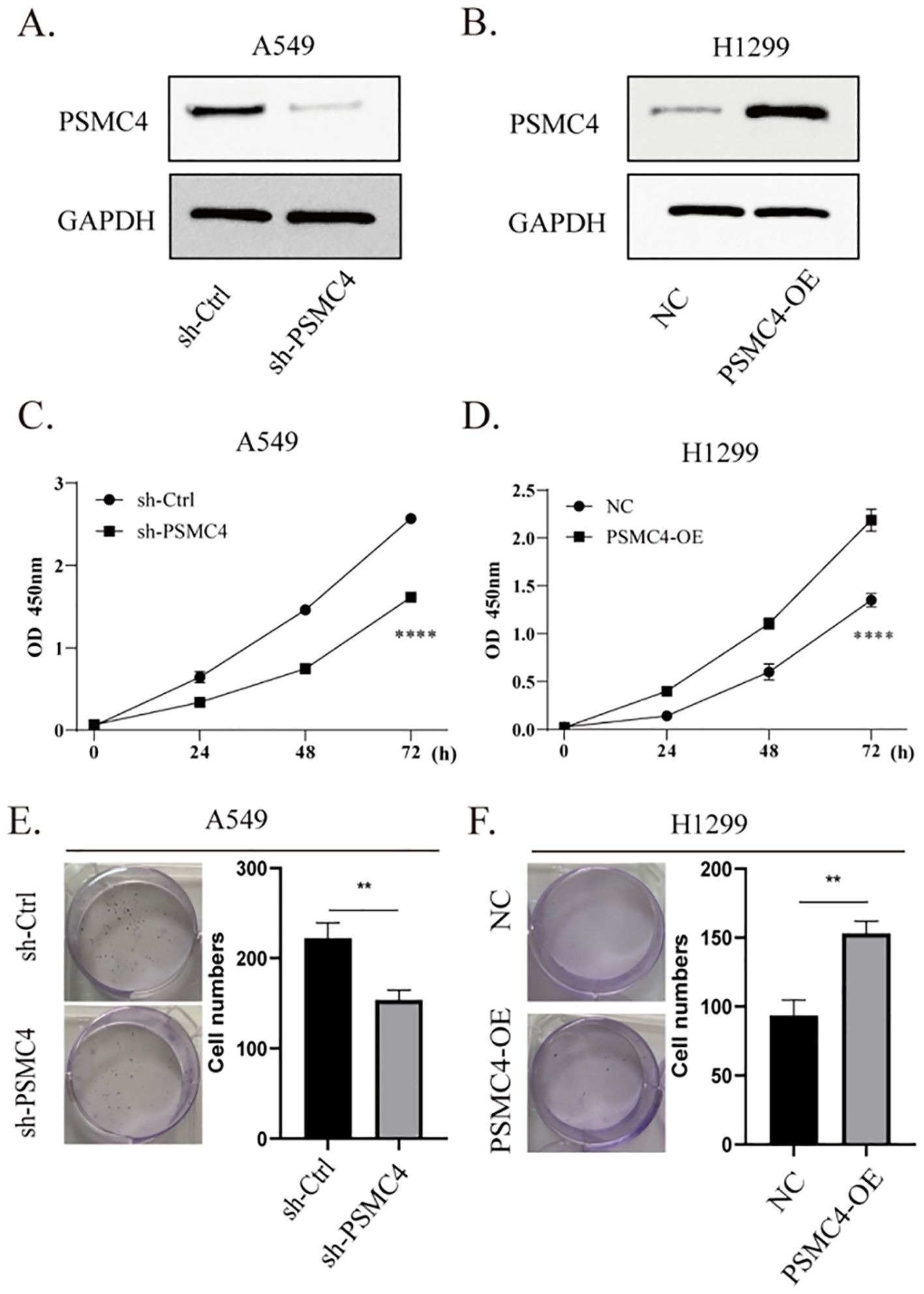
Figure 11. Promoting effect of PSMC4 on lung cancer cell proliferation. (A) Downregulation of PSMC4 expression in A549 cells. (B) Overexpression of PSMC4 in H1299 cells. (C, D) Evaluation of the impact of PSMC4 on cell proliferation by CCK-8 analysis. (E, F) Further investigation of the promoting effect of PSMC4 on cell growth through colony formation assay (**p<0.01, ****p<0.0001).
3.9 Effect of PSMC4 overexpression on tumor growth in mouse xenograft models
Using a subcutaneous xenograft model in mice, H1299 cells with upregulated PSMC4 exhibited tumors of significantly greater volume and weight than those in the control group (Figures 12A–C). These results underscore the potential of PSMC4 to facilitate tumor growth in vivo.
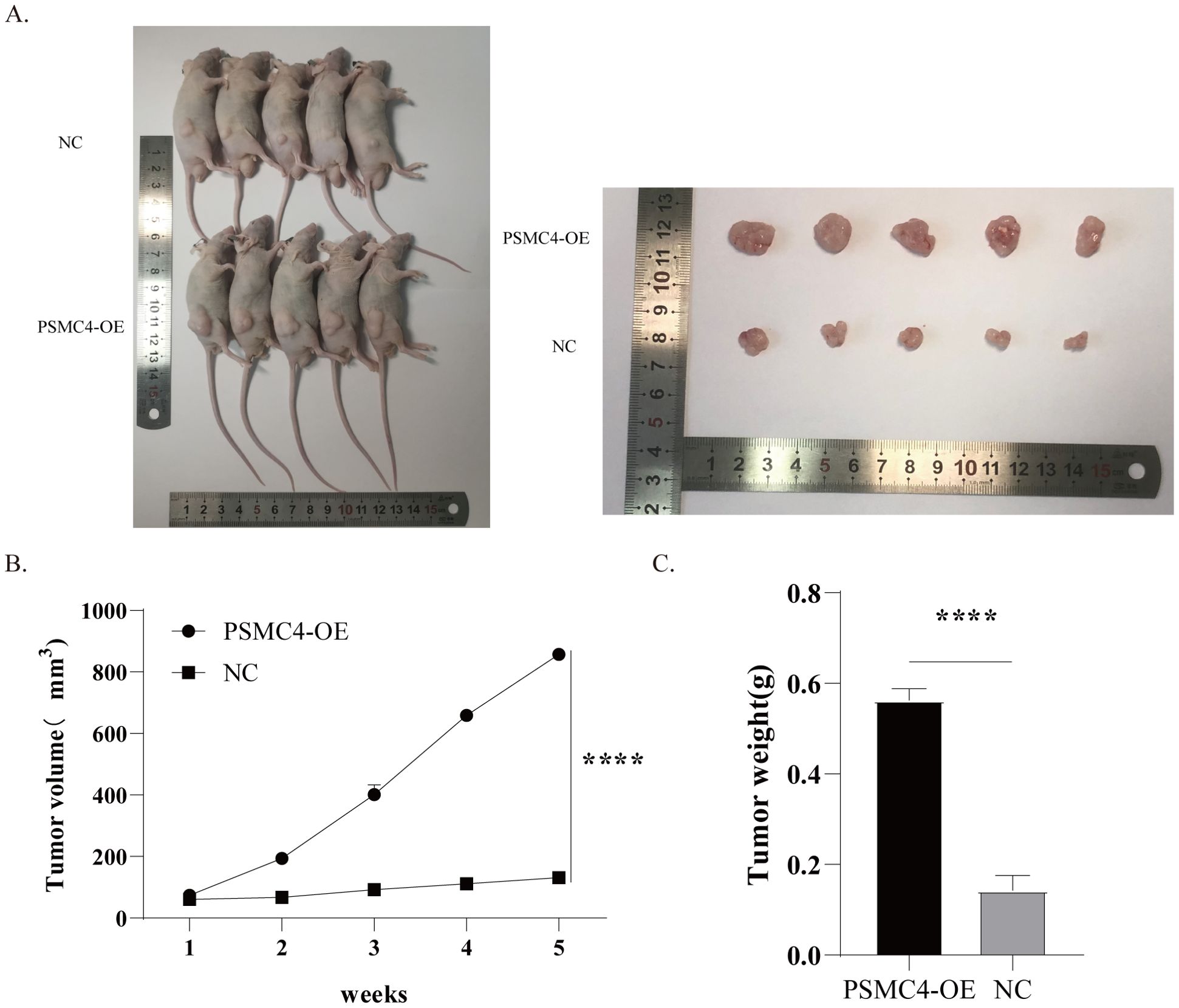
Figure 12. Enhanced tumor growth by overexpression of PSMC4 in mice xenografts. (A) Tumor samples after subcutaneous implantation of transfected H1299 cells in SCID mice for 5 weeks. (B) Tumor growth curve. (C) Comparison of tumor weights between the NC group and PSMC4 overexpression group. (****p<0.0001).
4 Discussion
In this investigation, we explored the role of the PSMC4 gene, a critical player in protein degradation and cell cycle regulation (21, 22), in LUAD. Our comprehensive analysis encompassed PSMC4 expression profiling in LUAD, alongside its associations with clinical-pathological features, immune cell infiltration, and functional pathways. Our findings reveal that elevated PSMC4 expression correlates significantly with more advanced T stages, N stages, and overall pathological stages. It also shows specific patterns of immune cell infiltration, underscoring its potential as both a diagnostic and prognostic biomarker. Moreover, our data highlight PSMC4’s involvement in crucial biological pathways that facilitate tumor progression and immune escape, such as the G2/M DNA damage checkpoint, Notch signaling, neutrophil extracellular trap formation, and the negative regulation of megakaryocyte differentiation. We further delineated PSMC4’s functional roles through in vitro and in vivo studies on lung cancer cells, suggesting its utility in crafting targeted and personalized treatments that could enhance the prognosis of LUAD patients. These insights not only deepen our understanding of LUAD pathogenesis but also suggest new avenues for therapeutic intervention, potentially transforming patient management and outcomes.
PSMC4, a subunit of the 26S proteasome, is integral to protein degradation and the maintenance of cellular homeostasis (23). Through meticulous analysis of the TCGA and GTEx databases, we noted the ubiquitous overexpression of PSMC4 across various cancers, particularly LUAD. This consistent overexpression highlights PSMC4’s potential as a robust biomarker for oncological assessments. Previous research across breast and prostate cancers suggests that elevated PSMC4 levels are closely linked with tumor progression (10, 14). We extend these findings to LUAD, showing that high PSMC4 expression correlates significantly with increased T stage, N stage, and overall pathological stage, indicative of its role in promoting tumor aggressiveness and metastatic capacity. Our statistical analyses confirm the relationship between PSMC4 overexpression and advanced pathological stages, suggesting its involvement in facilitating local invasion and lymph node metastasis. Additionally, we utilized ROC curve analysis to assess PSMC4’s diagnostic capabilities in LUAD. The AUC demonstrates high diagnostic sensitivity and specificity, affirming PSMC4’s utility as an effective diagnostic biomarker. Survival analyses further reveal that LUAD patients with elevated PSMC4 expression experience significantly reduced OS, DSS, and PFS relative to those with lower expression levels. These results align with and extend previous findings (14), underscoring PSMC4’s potential not just as a diagnostic tool but also as a predictor of clinical outcomes in cancer therapy.
The enriched analysis of PSMC4-associated differentially expressed genes, as detailed through comprehensive GO and KEGG pathways, significantly enhances our understanding of the molecular mechanisms by which PSMC4 influences lung adenocarcinoma progression. Notably, the GO analysis highlights the pivotal role of these genes in fundamental biological processes such as DNA replication-dependent and independent chromatin organization, protein-DNA complex assembly, and regulation of megakaryocyte differentiation. Such involvement underscores the influence of PSMC4 on chromatin dynamics, which is critical for cellular proliferation and genome stability-key elements in tumorigenesis (24, 25). Furthermore, the KEGG pathway analysis elucidates the association of these genes with several cancer-related pathways, including transcriptional misregulation in cancer and viral carcinogenesis, pointing to a broader impact of PSMC4 on oncogenic processes. Importantly, the GSEA reveals that PSMC4 is significantly linked with pathways like the G2/M DNA damage checkpoint and mitotic prophase, which are crucial for cell cycle control and the maintenance of chromosomal integrity during cell division (26, 27). The association with pathways such as signaling by NOTCH and WNT, as well as the formation of the beta-catenin TCF transactivating complex, further suggests a role for PSMC4 in signal transduction mechanisms that govern cellular differentiation, proliferation, and apoptosis. These insights are consistent with findings from previous studies, which have established the involvement of these pathways in the regulation of apoptosis and cell cycle progression in various cancers. For instance, disruptions in the NOTCH signaling pathway have been linked to a variety of cancers, including lung adenocarcinoma, by influencing cell fate decisions and maintaining the balance between cell proliferation and death (28, 29). Additionally, aberrant WNT signaling has been implicated in tumorigenesis through its effects on cellular senescence and differentiation (30, 31) By correlating these pathways with PSMC4 activity, our study not only confirms the gene’s pivotal role in cellular regulatory mechanisms but also highlights potential therapeutic targets. Modulating PSMC4 expression or its downstream pathways could offer new strategies for the treatment of lung adenocarcinoma, a prospect that warrants further investigation.
Within the immune environment of LUAD, the expression of PSMC4 is closely linked with the infiltration of specific immune cell subsets, underscoring its potential influence on tumor immune dynamics. Employing ssGSEA, we quantified the presence of 24 distinct immune cell types in LUAD tissues. Our Spearman correlation analysis demonstrated a significant association between PSMC4 expression and various immune cell subtypes, indicative of PSMC4’s crucial role in the immune evasion strategies characteristic of LUAD. Notably, increased PSMC4 expression correlates positively with the presence of Th2 cells and γδ T cells (Tgd). This relationship may skew the immune milieu of LUAD towards either a pro-inflammatory or immunosuppressive state through the enhanced recruitment of these cells. Typically, Th2 cell activity is linked with a diminished anti-tumor response and heightened inflammatory conditions (32, 33), consistent with the pro-tumorigenic influence of PSMC4. Conversely, although γδ T cells can exert anti-tumor effects under certain conditions, their presence may also promote tumor growth and facilitate immune escape in other scenarios (34, 35). Conversely, PSMC4 expression inversely correlates with Tcm, mast cells, Tem, and B cells. These cells are generally pivotal to robust anti-tumor immune responses. Thus, lower levels of PSMC4 could favor the accumulation of these beneficial immune cell types, potentially curtailing tumor growth. Particularly, a decrease in central memory T cells and effector memory T cells might compromise sustained and long-term anti-tumor immunity, exacerbating tumor immune evasion (36–39).
Our immunohistochemical analysis has revealed a significant upregulation of PSMC4 expression in NSCLC tissues compared to adjacent normal tissues. The cytoplasmic localization of PSMC4 in cancer cells indicates its involvement in proteasomal degradation pathways, which are commonly utilized by cancer cells to promote survival and proliferation (40, 41). The correlation between high PSMC4 expression and advanced clinical T and N stages further emphasizes its role in driving cancer aggressiveness. These findings are in line with previous studies (42), underscoring the crucial role of proteasome subunits in supporting cancer cell growth and survival. Therefore, targeting PSMC4 and the proteasome pathway may present novel therapeutic opportunities for NSCLC patients. Functional analysis has demonstrated that PSMC4 significantly impacts the proliferation and growth of lung cancer cells. Knockdown of PSMC4 in A549 cells resulted in reduced cell viability and colony formation, while its overexpression in H1299 cells enhanced these properties. These results indicate that PSMC4 functions as an oncogene in lung adenocarcinoma, promoting cell proliferation and tumorigenesis. This is consistent with previous research showing that PSMC4 knockout inhibits proliferation, cell cycle progression, and in vivo migration of prostate cancer cells while inducing apoptosis (14). In vivo xenograft models have further validated these findings, showing that PSMC4 overexpression leads to increased tumor volume and weight. In our experiments, although PSMC4-knockout A549 cells and PSMC4-overexpressing H1299 cells showed only a 1.5-fold proliferation difference in vitro, this discrepancy increased 3- to 8-fold in immunodeficient mouse models. Several factors may explain this: PSMC4 could enhance tumor growth by modulating tumor microenvironmental cells, such as immune and endothelial cells, promoting angiogenesis, and recruiting fibroblasts. Additionally, the longer duration in xenograft models allows for metastasis and angiogenesis, and PSMC4 might aid immune evasion, further promoting tumor growth in the absence of functional immune surveillance. These functional insights highlight the potential of PSMC4 as a therapeutic target, suggesting that inhibiting its expression or function could impede the progression of lung cancer.
Despite the comprehensive approach and robust methodologies employed in this study, several limitations warrant acknowledgment. First, the research predominantly relies on bioinformatics analyses and lacks extensive wet lab validation, which could provide more definitive evidence regarding the role of PSMC4 in lung adenocarcinoma. Second, the sample size—particularly for clinical validation—is relatively small, potentially constraining the generalizability of the findings. Furthermore, the absence of a thorough clinical validation analysis is a significant limitation, as such analysis is crucial for translating these findings into clinical practice.
In conclusion, this study offers valuable insights into the expression of the PSMC4 gene in lung adenocarcinoma and its association with clinical pathological features, immune cell infiltration, and functional enrichment. The findings suggest that PSMC4 may serve as a promising diagnostic biomarker and therapeutic target for lung adenocarcinoma. Future research should prioritize larger, more diverse cohorts and incorporate extensive wet lab validation to corroborate these findings. Additionally, integrating clinical validation analyses will be essential for translating these discoveries into practical clinical applications. The results of this study lay the groundwork for further exploration of PSMC4’s role in cancer biology and its potential to enhance the diagnosis and treatment of lung adenocarcinoma.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by the Ethics Committee of Hunan Cancer Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal study was approved by the Ethics Committee of Hunan Cancer Hospital. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LZ: Investigation, Writing – original draft. YL: Investigation, Writing – original draft. YX: Project administration, Writing – review & editing. JL: Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Fundamental Research Funds for the Central Universities of Central South University (2020zzts222), the Major special project of the Research Climbing Plan for the Department of Hunan Cancer Hospital (ZX2021006), and the Fundamental Research Funds for the Health Commission of Hunan Province (D202301047860).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Barta JA, Powell CA, Wisnivesky JP. Global epidemiology of lung cancer. Ann Glob Health. (2019) 85:532–46. doi: 10.5334/aogh.2419
3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
4. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. (2023) 73:17–48. doi: 10.3322/caac.21763
5. Duma N, Santana-Davila R, Molina JR. Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc. (2019) 94:1623–40. doi: 10.1016/j.mayocp.2019.01.013
6. Alduais Y, Zhang H, Fan F, Chen J, Chen B. Non-small cell lung cancer (NSCLC): A review of risk factors, diagnosis, and treatment. Med (Baltimore). (2023) 102:e32899. doi: 10.1097/MD.0000000000032899
7. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. (2018) 553:446–54. doi: 10.1038/nature25183
8. Li Y, Yan B, He S. Advances and challenges in the treatment of lung cancer. BioMed Pharmacother. (2023) 169:115891. doi: 10.1016/j.biopha.2023.115891
9. Finley D, Chen X, Walters KJ. Gates, channels, and switches: elements of the proteasome machine. Trends Biochem Sci. (2016) 41:77–93. doi: 10.1016/j.tibs.2015.10.009
10. Kao TJ, Wu CC, Phan NN, Liu YH, Ta HDK, Anuraga G, et al. Prognoses and genomic analyses of proteasome 26S subunit, ATPase (PSMC) family genes in clinical breast cancer. Aging (Albany NY). (2021) 13:17970. doi: 10.18632/aging.203345
11. Ayakannu T, Taylor AH, Konje JC. Selection of endogenous control reference genes for studies on type 1 or type 2 endometrial cancer. Sci Rep. (2020) 10:8468. doi: 10.1038/s41598-020-64663-4
12. Zhou H, Zhang C, Li H, Chen L, Cheng X. A novel risk score system of immune genes associated with prognosis in endometrial cancer. Cancer Cell Int. (2020) 20:240. doi: 10.1186/s12935-020-01317-5
13. Hellwinkel OJ, Asong LE, Rogmann JP, Sültmann H, Wagner C, Schlomm T, et al. Transcription alterations of members of the ubiquitin-proteasome network in prostate carcinoma. Prostate Cancer Prostatic Dis. (2011) 14:38–45. doi: 10.1038/pcan.2010.48
14. Liu K, Zhang S, Gong Y, Zhu P, Shen W, Zhang Q. PSMC4 promotes prostate carcinoma progression by regulating the CBX3-EGFR-PI3K-AKT-mTOR pathway. J Cell Mol Med. (2023) 27:2437–47. doi: 10.1111/jcmm.v27.16
15. Fan Y, Zhang J, Shi J, Chen L, Long J, Zhang S, et al. Genetic cross-talk between oral squamous cell carcinoma and type 2 diabetes: the potential role of immunity. Dis Markers. (2022) 2022:6389906. doi: 10.1155/2022/6389906
16. Reck M, Remon J, Hellmann MD. First-line immunotherapy for non-small-cell lung cancer. J Clin Oncol. (2022) 40:586–97. doi: 10.1200/JCO.21.01497
17. Mezquita L, Auclin E, Ferrara R, Charrier M, Remon J, Planchard D, et al. Association of the lung immune prognostic index with immune checkpoint inhibitor outcomes in patients with advanced non-small cell lung cancer. JAMA Oncol. (2018) 4:351–7. doi: 10.1001/jamaoncol.2017.4771
18. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. (2012) 16:284–7. doi: 10.1089/omi.2011.0118
19. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. (2013) 14:7. doi: 10.1186/1471-2105-14-7
20. Lv SL, Guo P, Zou JR, Chen RS, Luo LY, Huang DQ. Prognostic significance and relationship of SMAD3 phospho-isoforms and VEGFR-1 in gastric cancer: A clinicopathological study. World J Gastrointest Oncol. (2024) 16:118–32. doi: 10.4251/wjgo.v16.i1.118
21. Sakao Y, Kawai T, Takeuchi O, Copeland NG, Gilbert DJ, Jenkins NA, et al. Mouse proteasomal ATPases Psmc3 and Psmc4: genomic organization and gene targeting. Genomics. (2000) 67:1–7. doi: 10.1006/geno.2000.6231
22. Zavodszky E, Peak-Chew SY, Juszkiewicz S, Narvaez AJ, Hegde RS. Identification of a quality-control factor that monitors failures during proteasome assembly. Science. (2021) 373:998–1004. doi: 10.1126/science.abc6500
23. Zhang W, Hu Y, Qian M, Mao L, Yuan Y, Xu H, et al. A novel APA-based prognostic signature may predict the prognosis of lung adenocarcinoma in an East Asian population. iScience. (2023) 26:108068. doi: 10.1016/j.isci.2023.108068
24. Zhang B, Liu Q, Wen W, Gao H, Wei W, Tang A, et al. The chromatin remodeler CHD6 promotes colorectal cancer development by regulating TMEM65-mediated mitochondrial dynamics via EGF and Wnt signaling. Cell Discov. (2022) 8:130. doi: 10.1038/s41421-022-00478-z
25. Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. (2013) 502:462–71. doi: 10.1038/nature12749
26. Smith HL, Southgate H, Tweddle DA, Curtin NJ. DNA damage checkpoint kinases in cancer. Expert Rev Mol Med. (2020) 22:e2. doi: 10.1017/erm.2020.3
27. Pedroza-Garcia JA, Xiang Y, De Veylder L. Cell cycle checkpoint control in response to DNA damage by environmental stresses. Plant J. (2022) 109:490–507. doi: 10.1111/tpj.v109.3
28. Zhou B, Lin W, Long Y, Yang Y, Zhang H, Wu K, et al. Notch signaling pathway: architecture, disease, and therapeutics. Signal Transduct Target Ther. (2022) 7:95. doi: 10.1038/s41392-022-00934-y
29. Zhang H, Liang J, Lu T, Li M, Shan G, Bi G, et al. AGRN promotes lung adenocarcinoma progression by activating Notch signaling pathway and acts as a therapeutic target. Pharmacol Res. (2023) 194:106819. doi: 10.1016/j.phrs.2023.106819
30. Chen D, Gao S, Gao F, Liu A, Li J, Li J, et al. SMEK1 promotes lung adenocarcinoma proliferation and invasion by activating Wnt/β-catenin signaling pathway. Clin Transl Oncol. (2023) 25:976–86. doi: 10.1007/s12094-022-03001-8
31. Zhang HJ, Chang WJ, Jia CY, Qiao L, Zhou J, Chen Q, et al. Destrin contributes to lung adenocarcinoma progression by activating wnt/β-catenin signaling pathway. Mol Cancer Res. (2020) 18:1789–802. doi: 10.1158/1541-7786.MCR-20-0187
32. Su H, Jin Y, Tao C, Yang H, Yang E, Zhang WG, et al. Th2 cells infiltrating high-grade serous ovarian cancer: a feature that may account for the poor prognosis. J Gynecol Oncol. (2023) 34:e48. doi: 10.3802/jgo.2023.34.e48
33. Gao S, Hsu TW, Li MO. Immunity beyond cancer cells: perspective from tumor tissue. Trends Cancer. (2021) 7:1010–9. doi: 10.1016/j.trecan.2021.06.007
34. Mensurado S, Blanco-Domínguez R, Silva-Santos B. The emerging roles of γδ T cells in cancer immunotherapy. Nat Rev Clin Oncol. (2023) 20:178–91. doi: 10.1038/s41571-022-00722-1
35. Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell. (2019) 176:998–1013.e16. doi: 10.1016/j.cell.2018.12.040
36. Zheng L, Qin S, Si W, Wang A, Xing B, Gao R, et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science. (2021) 374:abe6474. doi: 10.1126/science.abe6474
37. Wang F, Cheng F, Zheng F. Stem cell like memory T cells: A new paradigm in cancer immunotherapy. Clin Immunol. (2022) 241:109078. doi: 10.1016/j.clim.2022.109078
38. Damei I, Trickovic T, Mami-Chouaib F, Corgnac S. Tumor-resident memory T cells as a biomarker of the response to cancer immunotherapy. Front Immunol. (2023) 14:1205984. doi: 10.3389/fimmu.2023.1205984
39. Yenyuwadee S, Sanchez-Trincado Lopez JL, Shah R, Rosato PC, Boussiotis VA. The evolving role of tissue-resident memory T cells in infections and cancer. Sci Adv. (2022) 8:eabo5871. doi: 10.1126/sciadv.abo5871
40. Gou Q, Dong C, Xu H, Khan B, Jin J, Liu Q, et al. PD-L1 degradation pathway and immunotherapy for cancer. Cell Death Dis. (2020) 11:955. doi: 10.1038/s41419-020-03140-2
41. Rousseau A, Bertolotti A. Regulation of proteasome assembly and activity in health and disease. Nat Rev Mol Cell Biol. (2018) 19:697–712. doi: 10.1038/s41580-018-0040-z
Keywords: PSMC4, non-small cell lung cancer, prognosis, immune infiltration, cell proliferation
Citation: Zhu L, Li Y, Xu Y and Lei J (2025) The role of PSMC4 in non-small cell lung cancer: implications for prognosis, diagnosis, and immune microenvironment modulation. Front. Oncol. 15:1503466. doi: 10.3389/fonc.2025.1503466
Received: 29 September 2024; Accepted: 09 April 2025;
Published: 02 May 2025.
Edited by:
Kentaro Jingushi, Osaka University, JapanCopyright © 2025 Zhu, Li, Xu and Lei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunfei Xu, MjIwMjIxODlAY3N1LmVkdS5jbg==; Jian Lei, bGVpamlhbkBobmNhLm9yZy5jbg==
†These authors have contributed equally to this work and share first authorship
 Lili Zhu1†
Lili Zhu1† Yunfei Xu
Yunfei Xu