- 1Department of Pathology, Changde Hospital, Xiangya School of Medicine, Central South University, Changde, China
- 2Department of Clinical Laboratory, Changde Hospital, Xiangya School of Medicine, Central South University, Changde, China
Gut microbes are emerging as critical regulators in cancer therapy, influencing the efficacy and toxicity of radiotherapy, chemotherapy, immunotherapy, targeted therapy, Traditional Chinese Medicine, and rehabilitation interventions. Acting through metabolic reprogramming, immune modulation, DNA damage, and tumor microenvironment remodeling, specific microbial taxa and their metabolites can either enhance or hinder treatment outcomes. However, these interactions are highly context-dependent and shaped by individual factors such as diet, geography, and host immunity. While microbial interventions such as probiotics, fecal microbiota transplantation, and engineered bacteria show promise, their translation into precise and safe clinical applications remains limited by interindividual variability, regulatory hurdles, and incomplete mechanistic understanding. Future efforts should focus on defining high-evidence microbial signatures, clarifying causal mechanisms, and developing personalized microbiome-based therapeutic strategies, potentially integrated with nanotechnology. This review underscores the need for interdisciplinary approaches to harness gut microbiota as co-targets in cancer treatment.
1 Introduction
The gut, as the body’s largest immune organ, plays a central role in immune surveillance and tolerance. Constantly exposed to dietary and microbial antigens, it also serves as a potential entry point for pathogens. Approximately 30–40 dominant bacterial species shape the adult gut microbiota, whose composition is dynamic and influenced by diet, smoking, medications (e.g., antibiotics), probiotics, and host physiology (1, 2). Along the gastrointestinal tract, bacterial density increases distally, with anaerobes dominating the colon. Microbial colonization begins at birth and stabilizes by age two, facilitating mucosal immune maturation and the balance between inflammation and immune tolerance (3, 4).
Gut microbiota contributes to host health by regulating nutrient metabolism, xenobiotic detoxification, epithelial development, immune modulation, and resistance to pathogen colonization (5–10). Conversely, dysbiosis can promote malignancies, notably colorectal and hepatocellular carcinomas (11). In rodent models, germ-free or antibiotic-treated conditions have revealed microbiota-driven tumorigenesis, independent of inflammation (12). Microbial biofilms may also reshape the tumor microenvironment (TME) by promoting metabolic cross-talk and immune evasion (13–15).
Beyond tumorigenesis, gut microbes profoundly influence the efficacy and toxicity of anticancer therapies, especially immunotherapy (15, 16). The TME has emerged as a critical determinant of therapy response and is shaped by microbial-derived metabolites and immune signaling (17–20). Specific microbes enhance immune cell infiltration and antigen presentation, while others hinder treatment through immune suppression. Novel strategies such as probiotic supplementation, fecal microbiota transplantation (FMT), bacterial engineering, and phage therapy are under investigation for enhancing therapeutic outcomes (21–24).
Despite rapid progress, challenges remain. Interindividual variability in microbiota composition, limited mechanistic understanding, and regulatory constraints hinder clinical translation. Moreover, the interplay between gut microbes, host immunity, and cancer remains complex and context-dependent. This review provides an updated synthesis of how gut microbiota modulate responses to multiple cancer therapies, including radiotherapy, chemotherapy, targeted therapy, immunotherapy, Traditional chinese medicine (TCM), and rehabilitation interventions. We highlight key mechanisms—ranging from metabolic reprogramming and immune modulation to TME remodeling—and discuss their translational potential. Looking forward, personalized microbiota-based interventions and interdisciplinary innovations such as AI-driven microbial profiling may pave the way for safer and more effective cancer treatment strategies.
2 Influence of gut microbes on multiple antitumor therapies
2.1 Radiation therapy and chemotherapy
Radiotherapy and chemotherapy remain cornerstone treatments for various malignancies. However, increasing evidence highlights the gut microbiota as a critical modulator of both their therapeutic efficacy and associated toxicities.
Radiation-induced damage not only alters tumor tissues but also disrupts intestinal microbial homeostasis. Certain microbial populations can exacerbate the toxicity of radiation therapy. Animal studies have demonstrated that radiation-induced alterations in the gut microbiota promote the secretion of interleukin-1β (IL-1β) and the generation of reactive oxygen species (ROS), which in turn disrupt intestinal tight junctions and amplify inflammatory processes, thereby further aggravating mucosal inflammation in mice (25, 26). These effects compromise quality of life and may necessitate dose reductions or treatment suspension. Conversely, specific commensal strains—particularly those with anti-inflammatory or mucosal barrier-protective properties—can mitigate such adverse effects (27). Certain intestinal microorganisms—such as Lactobacillus rhamnosus, Lactobacillus acidophilus, Bifidobacterium, members of the families Lachnospiraceae and Enterococcaceae, as well as Akkermansia—have been reported to mitigate the side effects of radiation therapy (27, 28).
Clinical studies reinforce these findings. In a large double-blind, placebo-controlled trial, Delia et al. demonstrated that probiotic supplementation significantly reduced radiation-induced diarrhea in postoperative cancer patients (29). Similarly, Sharma et al.found that probiotics lowered the incidence of grade III-IV oral mucositis in patients receiving radiotherapy for head and neck cancers, improving treatment completion rates (30). These results support the integration of targeted probiotic interventions into radiotherapy regimens to reduce complications and improve therapeutic adherence.
Gut microbes can influence chemotherapy through multiple mechanisms, including drug metabolism, immune modulation, and barrier integrity maintenance. Numerous studies have demonstrated that gut microbiota can modulate the efficacy of chemotherapeutic agents (31). Specific bacterial species, such as Bacteroides fragilis and Lactobacillus acidophilus, can influence the bioavailability and antitumor activity of chemotherapeutic drugs through metabolic interactions. For example, Bacteroides fragilis has been shown to metabolize agents like 5-fluorouracil (5-FU), thereby affecting its therapeutic impact (32). Additionally, oxaliplatin (OXP) chemotherapy has been reported to enhance local immune responses by modulating the ileal microbiota, ultimately improving its clinical antitumor efficacy. In contrast, cisplatin can induce alterations in commensal gut bacteria, exacerbating mucosal injury, increasing tumor burden, and triggering systemic inflammation. Notably, these adverse effects can be reversed by the administration of Lactobacillus acidophilus (33).
Microbial metabolites also play crucial roles. Butyrate enhances gemcitabine-induced apoptosis, while microbial β-glucuronidase can reactivate irinotecan’s active metabolite SN-38, leading to toxicity—an effect reversible by co-administering β-glucuronidase inhibitors. Conversely, microbial enzymes like cytidine deaminase may inactivate gemcitabine, promoting drug resistance in pancreatic and colorectal cancers.
Gut microbiota act as both mediators and modulators of chemo-radiotherapeutic outcomes (Figure 1). Their dual role in enhancing efficacy and limiting toxicity opens promising avenues for microbiota-informed oncologic strategies. However, interpatient variability, context-dependent responses, and incomplete mechanistic understanding remain significant challenges. Future research should aim to identify predictive microbial signatures, explore metabolite-host-drug interactions in depth, and design microbiome-based adjuvant therapies tailored to individual tumor types and treatment protocols.
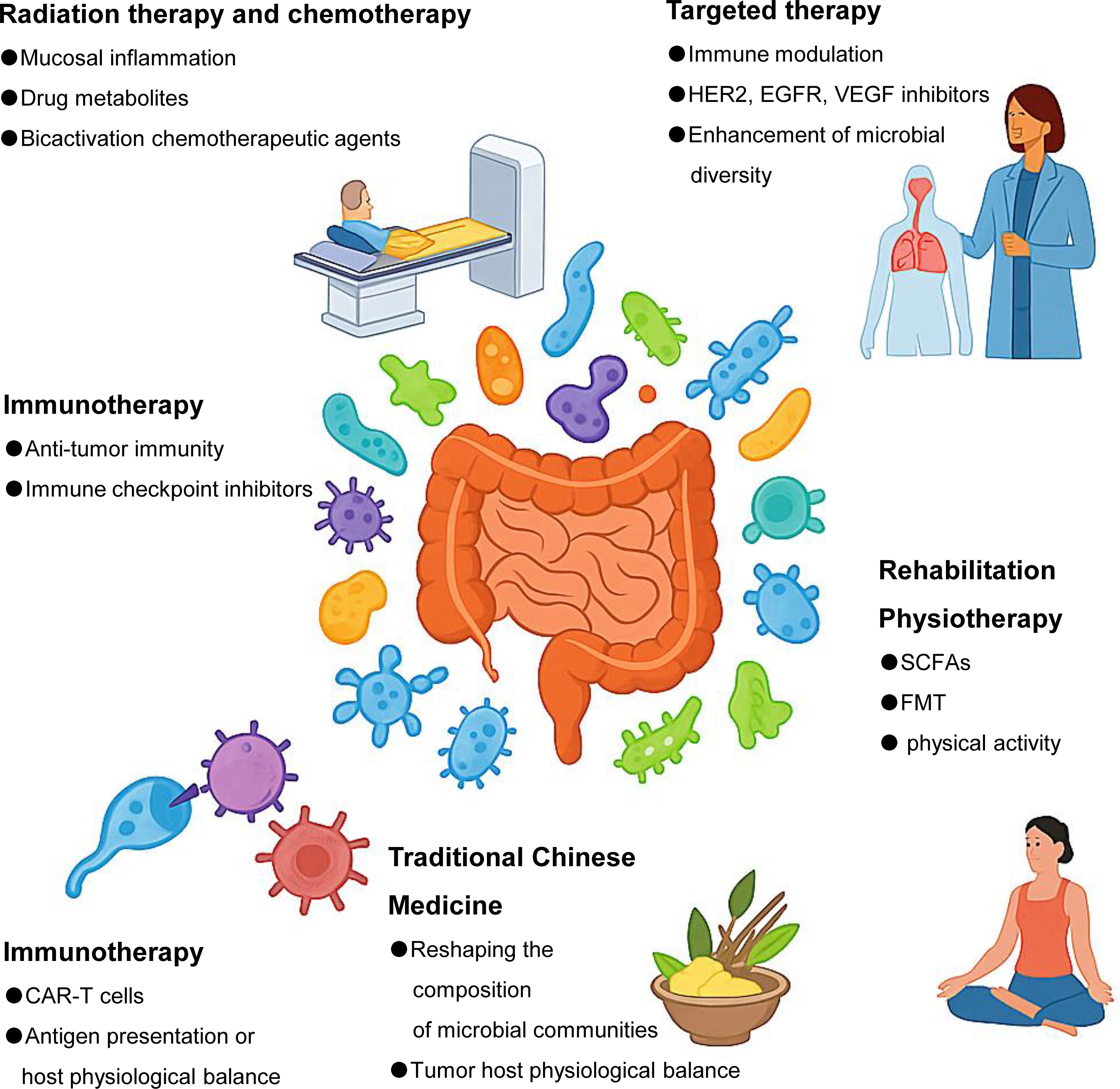
Figure 1. Gut microbes are associated with a wide range of tumor therapies. Gut microbes influence multiple aspects of cancer treatment, including radiotherapy, chemotherapy, immunotherapy, targeted therapy, TCM, and rehabilitation strategies such as dietary intervention and FMT. Specific microbial taxa can modulate treatment efficacy and toxicity through diverse mechanisms, such as regulating immune responses, altering drug metabolism, maintaining intestinal barrier integrity, or shaping the tumor microenvironment.
2.2 Targeted therapy
Molecular targeted therapies, designed to interfere with specific oncogenic signaling pathways, have significantly improved cancer treatment (34). However, recent studies reveal that gut microbiota can modulate the efficacy of several commonly used targeted agents, including trastuzumab (HER2 inhibitor), cetuximab (EGFR inhibitor), and bevacizumab (VEGF inhibitor) (35). A phase I trial (NCT03772899) evaluated healthy donor fecal microbiota transplantation (FMT) combined with PD-1 inhibitors in 20 treatment-naïve advanced melanoma patients. FMT alone was safe, with no grade 3 events; combined therapy led to a 65% response rate (including 20% complete responses) but 25% grade 3 immune-related adverse events. Microbiome analysis showed donor strain engraftment and increased similarity over time only in responders, alongside enrichment of immunogenic and reduction of harmful bacteria. Mouse models confirmed enhanced anti-PD-1 efficacy, supporting further investigation of FMT as an adjunct to immunotherapy (36).
In HER2-positive breast cancer, low intestinal abundances of Trichoderma, Zygomycetes, Bifidobacterium, and Prevotella were found in trastuzumab-nonresponsive patients. In murine models, antibiotic-induced microbiota depletion reduced trastuzumab efficacy by impairing CD4+ T cell and granzyme B+ cell infiltration, dendritic cell activation, and IL-12 secretion within tumors (37). These findings suggest that gut microbes may enhance trastuzumab’s therapeutic effect through immune modulation.
Similarly, gut microbiota diversity has been associated with improved outcomes in colorectal cancer (CRC) patients treated with cetuximab or bevacizumab. In contrast, high levels of Klebsiella pneumoniae, Lactobacillus, Bifidobacterium, and Clostridium perfringens correlated with disease progression and poorer prognosis, indicating that not all bacteria exert beneficial effects (38).
In prostate cancer, the microbiome’s influence extends to hormonal therapy. Patients receiving androgen axis-targeted therapies (e.g., bicalutamide, enzalutamide, abiraterone) exhibited enriched bacterial taxa capable of steroid biosynthesis (39). Notably, Coccidioides and Mycobacterium species were more abundant in castration-resistant patients and were shown to convert androgen precursors into active forms, thereby compromising the efficacy of androgen deprivation therapy (40).
These studies underscore the role of gut microbiota not only as passive biomarkers but also as active modulators of targeted therapy response. Their immunoregulatory capacity and metabolic flexibility—such as influencing cytokine signaling or steroid metabolism—can either enhance or undermine treatment efficacy. Future directions should include identifying microbial signatures predictive of response to targeted agents and exploring microbiome modulation as a strategy to overcome resistance in precision oncology (Figure 1).
2.3 Immunotherapy
Immunotherapy has revolutionized cancer treatment, offering durable responses across various malignancies through approaches such as immune checkpoint inhibitors (ICIs), CAR-T cells, oncolytic viruses, cancer vaccines, and cytokine therapies (41–43). However, increasing evidence reveals that gut microbiota critically shape both the efficacy and toxicity of these immune-based therapies (44, 45).
Clinical and preclinical studies have demonstrated that gut microbes modulate antitumor immunity, particularly in T-cell-mediated therapies (17, 33). Disruption of the gut microbiota, such as through antibiotics, has been shown to impair responses to CAR-T therapy. For instance, patients treated with antibiotics exhibited increased tumor burden and systemic inflammation, while those colonized with Bifidobacterium longum and peptidoglycan-producing microbes prior to CAR-T therapy showed improved 6-month survival and reduced tumor progression (46).
Pioneering studies by Sivan et al. (47) first identified the microbiota-dependency of ICI efficacy. Bifidobacterium intestinalis and Lactobacillus casei paracasei enhanced CD8+ T-cell infiltration and dendritic cell (DC) activation, boosting anti-PD-L1 activity in colorectal cancer models (38,39). Likewise, B. longum improved anti-PD-L1 responses in melanoma by promoting tumor-specific CD8+ T-cell effector functions (48).
Subsequent studies confirmed that Bifidobacterium pseudomallei and B. bifidum similarly enhanced ICI efficacy across multiple tumor models through oral administration or modulation of DC function (49). In addition, Clostridium perfringens activated the STING pathway, promoting PD-L1 expression and IFN-γ+ CD8+ TIL accumulation, further sensitizing tumors to PD-L1 blockade (50). Interestingly, the efficacy of ICIs may be influenced not only by microbial composition but also by tumor type, as different malignancies induce distinct microbiota alterations that can either enhance or impair immunotherapeutic response (51).
Gut microbiota plays a dual role in immunotherapy: they can augment antitumor immunity or contribute to resistance. Key taxa such as Bifidobacterium spp. and Clostridium spp. exert their effects through modulation of antigen presentation, cytokine signaling, and T-cell recruitment. Given the complex and context-dependent interactions, future work should focus on identifying microbial biomarkers predictive of ICI responsiveness and developing microbial adjuvants or preconditioning strategies to optimize immune-based therapies (Figure 1).
2.4 Traditional Chinese medicine
In recent years, cancer treatment strategies have become increasingly diversified, ranging from modern immunotherapies to TCM (52). While these approaches differ significantly in their theoretical foundations, mechanisms of action, and clinical application, they also exhibit potential complementarities. ICIs, a hallmark of modern immunotherapy, offer targeted interventions against tumor immune evasion with robust clinical efficacy and scientific reproducibility. However, they are often associated with variable patient responses and immune-related adverse events. In contrast, TCM adopts a holistic and multi-targeted approach, emphasizing the principles of “reinforcing the body’s vital energy and eliminating pathogenic factors” and “harmonizing organ function.” It has shown promise in enhancing immune function, mitigating treatment-related toxicity, and improving patients’ quality of life. Emerging evidence suggests that TCM may enhance immunotherapy outcomes by modulating the gut microbiota and reducing systemic inflammation, indicating its potential as a valuable adjunct to modern treatments. Future research exploring the synergistic mechanisms of microbiota regulation and immune modulation between TCM and immunotherapy may pave the way for integrated cancer treatment paradigms.
Traditional chinese medicine has demonstrated notable efficacy in complex diseases, including malignancies, where it contributes to symptom relief, immune regulation, and prevention of metastasis and recurrence (53). Recent studies highlight a bidirectional interaction between TCM and gut microbiota, positioning this interplay as a potential mediator of TCM’s therapeutic effects in oncology.
On one hand, TCM can reshape gut microbial composition and metabolism, thereby restoring host physiological balance and alleviating tumor-promoting conditions. Herbal compounds may promote beneficial taxa and suppress pathogenic ones, contributing to anti-inflammatory and antitumor effects. On the other hand, gut microbes are involved in the biotransformation of TCM components, enhancing the bioavailability and activity of pharmacologically relevant metabolites. However, some microbial species may antagonize TCM efficacy by degrading active compounds or interfering with their absorption.
Mechanistically, TCM exerts its antitumor effects through modulation of host–microbiota axes, including the gut–liver, gut–brain, and gut–immune pathways, thereby influencing endocrine and immune networks (54). This modulation helps to disrupt the tumor-favorable microenvironment and restore systemic immune homeostasis.
Specific herbal formulations further exemplify this mechanism. For instance, the Paeonia lactiflora Soft Liver Combination significantly reduced Mycobacterium avium abundance, while Jiawei Yuxuan decoction altered gut microbial profiles and regulated key metabolites—such as primary bile acids and IFN-γ—in a hepatocellular carcinoma model, thereby enhancing antitumor immunity (17). These findings support the microbiota-dependent therapeutic potential of TCM in liver and colorectal cancers.
The gut microbiota–TCM axis represents a promising frontier in integrative oncology. However, the dualistic nature of this interaction—where gut microbes can both enhance and hinder TCM efficacy—necessitates careful characterization of host-microbe-drug dynamics. Future research should aim to identify microbial biomarkers predictive of TCM responsiveness, optimize herbal compound formulation for microbiota compatibility, and avoid unintended microbial interference. Particularly in palliative care, where TCM offers symptomatic relief with minimal toxicity, microbiota-informed TCM strategies may become valuable adjuncts to mainstream cancer therapies (Figure 1).
2.5 Rehabilitation physiotherapy
Rehabilitation therapies for cancer patients increasingly recognize the role of gut microbiota as a modifiable factor influencing prognosis, treatment tolerance, and quality of life. Modulating the gut microbial ecosystem—through dietary interventions, probiotics, FMT, physical activity, or circadian rhythm regulation—has shown potential in supporting CRC management and general oncologic recovery.
Dietary fiber is a key determinant of gut microbiota composition. Fermentation of fiber in the colon produces Short-Chain Fatty Acids (SCFAs)—such as acetate, propionate, and butyrate—which enhance mucosal integrity, suppress inflammation, and inhibit tumor proliferation (55). High-fiber diets also reduce carcinogenic secondary bile acids and support beneficial microbial populations.
Personalized nutrition, tailored to the unique microbial profile of CRC patients, has been proposed as a preventive and therapeutic strategy (56). Specially formulated diets rich in vegetables, fruits, oilseeds, low-sugar complex carbohydrates, and unsaturated fatty acids offer antioxidant and anti-inflammatory benefits (57, 58). Microecological formulas incorporating short peptides, Lactobacillus, Bifidobacterium, and herbal extracts have demonstrated immune-enhancing and anticancer potential, though further validation is needed to identify microbial or metabolic predictors of dietary response (59, 60).
Probiotics play a restorative role in gut barrier function, immune modulation, and malnutrition correction. Butyrate-producing strains, for example, alleviate intestinal wall atrophy in malnourished tumor patients (61). FMT from healthy donors has shown efficacy in resolving therapy-induced complications, such as Clostridium difficile infections, by restoring microbial diversity (62). Notably, FMT from immune checkpoint inhibitor-responsive donors enhanced PD-1 blockade efficacy in germ-free mice, associated with elevated Akkermansia muciniphila levels (63).
Moderate exercise (e.g., yoga, swimming, walking) has been linked to increased microbial diversity and SCFA production, enhancing gut barrier integrity and immune responsiveness (64–66). Disruption of circadian rhythms negatively impacts microbiota structure and immune balance. Taurocholic acid metabolism, epigenetically regulated by microbial activity, can promote MDSC accumulation and lung metastasis in CRC models when circadian patterns are disturbed (67).
Microbiota-targeted rehabilitation represents a promising adjunct in cancer care, extending beyond tumor suppression to systemic recovery. Future research should prioritize stratified approaches based on microbiome profiling, define optimal combinations of diet, probiotics, and lifestyle interventions, and explore their synergy with frontline oncologic treatments. These strategies not only support tumor rehabilitation but may contribute broadly to patient resilience and survivorship (Figure 1).
3 Mechanisms by which gut microbes affect tumor therapy
3.1 Gut microbes intervene in tumor cell metabolism and metastasis through flora metabolites
A growing body of evidence suggests that gut microbial metabolites play a pivotal role in modulating tumor cell behavior, influencing both therapeutic response and metastatic potential (45, 68–71)(Table 1).
One representative example is indole-3-acetic acid (3-IAA), a tryptophan-derived metabolite produced by Bacteroides fragilis and B. polymorphicus, which was enriched in pancreatic ductal adenocarcinoma (PDAC) patients who responded to chemotherapy. Exogenous 3-IAA supplementation or a high-tryptophan diet enhanced therapeutic efficacy, highlighting its potential as a microbial co-adjuvant in treatment. Similarly, reuterin, secreted by Lactobacillus reuteri, has shown anti-cancer properties in colon cancer models by inducing protein oxidation and suppressing ribosome biogenesis, thereby inhibiting tumor progression (72).
In contrast, some metabolites may promote tumor development. For example, indole-3-acrylic acid (IDA), mainly produced by Streptococcus species enriched in CRC patients, was shown to accelerate CRC progression in mice by inhibiting ferroptosis via the AHR–ALDH1A3 pathway, a mechanism associated with poor prognosis (73).
In addition to soluble metabolites, microbial extracellular vesicles (EVs) can directly modulate tumor behavior. Fn-OMVs, derived from clostridium nucleatum, promote lung metastasis by activating autophagic flux in tumor cells. Inhibiting autophagy with chloroquine significantly reduced metastases in murine models, confirming the pro-metastatic role of microbial vesicles in CRC (74).
These findings underscore a dualistic role of microbial metabolites in cancer: some exert therapeutic potential, while others facilitate tumor progression. The regulatory landscape is shaped by metabolite structure, producing species, and host context. Understanding these mechanisms provides opportunities to: Identify metabolite biomarkers predictive of treatment response; Design diet or microbiota-based interventions to modulate metabolite production; Target pathogenic metabolite pathways, such as ferroptosis suppression or autophagy activation.
Future therapeutic strategies may involve precision modulation of microbiota-derived metabolites, either by microbial engineering or metabolite-mimicking drugs, to enhance antitumor efficacy while mitigating metastatic risk (Figure 2).
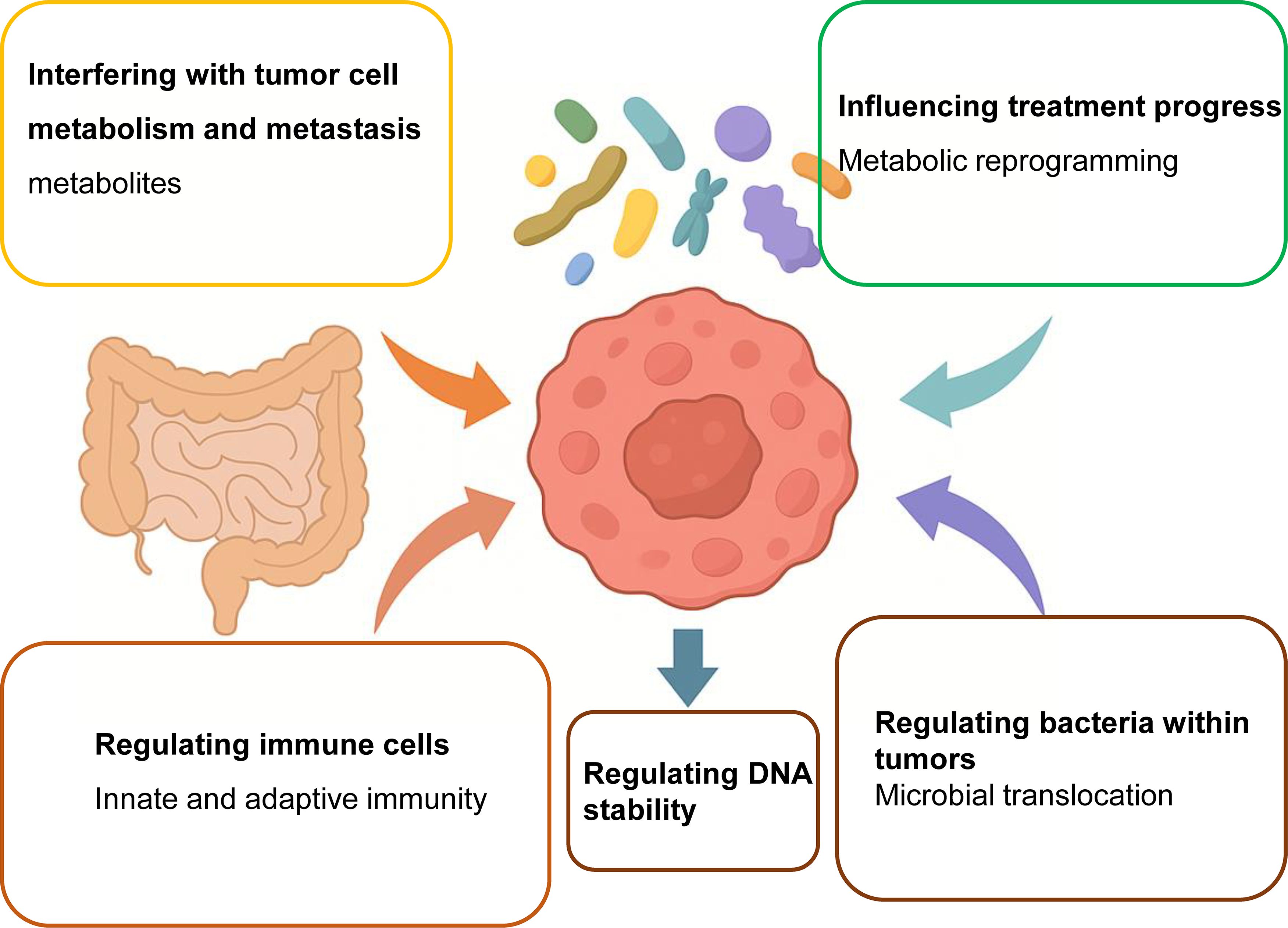
Figure 2. Gut microbes modulate the efficacy of cancer therapies through multiple mechanisms. Gut microbiota exerts a profound influence on cancer therapy outcomes by orchestrating a range of biological processes within the host and tumor microenvironment. These mechanisms include: (1) metabolic reprogramming of tumor cells via microbial metabolites such as SCFAs and tryptophan derivatives, which can either enhance or impair therapeutic efficacy; (2) modulation of the tumor immune microenvironment by influencing T-cell function, antigen presentation, and immune checkpoint expression; (3) remodeling of tumor biomechanical properties such as extracellular matrix stiffness and intercellular adhesion, thereby affecting tumor invasion and immune infiltration; (4) translocation of bacteria from the gut to tumor tissue, contributing to the formation of intratumoral microbiota that can interact with local immune cells; and (5) induction of DNA damage and genomic instability through genotoxic bacterial products like colibactin. Together, these pathways illustrate the multifaceted regulatory roles of gut microbes in shaping therapeutic response, toxicity, and resistance, suggesting new opportunities for microbiota-targeted adjuvant interventions.
3.2 Gut microbes influence tumor cell therapeutic progression by regulating the metabolic reprogramming of tumor cells
Metabolic reprogramming is a hallmark of cancer, underpinning genomic instability, inflammation, and immune escape (75). As a “second genome,” the gut microbiota play a crucial role in modulating host metabolic pathways, including glucose, lipid, and amino acid metabolism, thereby indirectly reshaping tumor cell behavior and response to therapy (76, 77).
Enhanced glycolysis (the Warburg effect) in CRC leads to lactate accumulation and acidification of the tumor microenvironment (TME), fostering tumor progression (78, 79). Recent studies have linked this metabolic shift to specific gut microbes. Clostridium nucleatum and Clostridium perfringens are enriched in CRC tissues with elevated glucose metabolism, as confirmed via PET/CT imaging and qPCR. Mechanistically, C. perfringens promotes glycolysis by epigenetically modulating histone acetylation through lncRNA ENO1-IT1-mediated regulation of ENO1, a glycolytic enzyme, revealing a microbe-epigenetic-metabolism axis in CRC (80, 81).
Lipid biosynthesis is often upregulated in tumor cells. Circulating tumor-derived lipids can alter intestinal microbiota by damaging bacterial membranes and shifting microbial composition, leading to dysbiosis and inflammation (82). Conversely, gut microbial metabolites such as butyrate and propionate activate PPARγ signaling, which promotes lipid catabolism, reduces hepatic fat accumulation, and may counteract tumor-driven metabolic shifts (83).
Although the role of microbial fatty acids in tumor lipid anabolism remains unclear, they represent promising candidates for therapeutic exploitation (84).Tumor cells exhibit glutamine addiction under glycolytic stress, relying on glutaminolysis for ATP, nucleotide, and redox homeostasis. While direct microbial regulation of glutamine metabolism in tumors is poorly defined, studies suggest that glutamine supplementation can reshape gut microbial communities—reducing the Firmicutes/Bacteroidetes ratio—and elevate protective secretory IgA (SIgA), which maintains intestinal immune integrity (85, 86).Disruption of this axis may promote the leakage of harmful microbial metabolites into circulation, fueling tumor progression.
The gut microbiota influence tumor metabolism through bidirectional nutrient and metabolite exchange. Key microbial products—including SCFAs and lncRNA-regulated epigenetic modifiers—affect tumor energy metabolism, redox balance, and epigenetic landscape. Future research should focus on: Identifying microbe-metabolite-target networks driving therapeutic resistance; Modulating microbial composition to reverse oncogenic metabolic states; Integrating microbiota data into metabolic precision oncology platforms. These insights may unlock novel microbial adjuvants or diet-microbiome interventions that synergize with metabolic-targeted cancer therapies (Figure 2).
3.3 Gut microbes regulate immune cells in the immune microenvironment
The TME often displays strong immunosuppressive features that hinder the effectiveness of cancer immunotherapies. Emerging evidence indicates that gut microbiota can reshape the immune landscape of the TME by influencing both innate and adaptive immunity, thereby modulating therapeutic response (33, 49, 87–89).
Gut microbes release microbe-associated molecular patterns (MAMPs)—such as lipopolysaccharides, flagellin, and peptidoglycans—that are sensed by host pattern recognition receptors (e.g., TLRs, NOD-like receptors), influencing both systemic and intratumoral immune responses (21, 82, 90). For instance, dietary fiber enhances cyclic di-adenosine secretion from gut microbes, which activates the STING pathway and promotes type I interferon production in the TME, facilitating antigen presentation and boosting the efficacy of immune checkpoint blockade (ICB) therapies (91, 92).
Additionally, microbial sensing impacts therapy-induced immunotoxicity. For example, TLR9 or MYD88 deficiency reduces graft-versus-host disease (GVHD) in mice, while TLR2 signaling attenuates methotrexate toxicity by inducing compensatory metabolic pathways (17, 93).
Increased gut permeability during tumor progression or therapy allows translocation of live bacteria and metabolites into secondary lymphoid tissues or tumors, influencing immune activation (84). Fusobacterium nucleatum (Fn), a CRC-associated microbe, has been shown to localize preferentially in tumors and interfere with ICB efficacy. Fn-derived succinic acid suppresses IFN-I signaling and CD8+ T-cell infiltration, while its surface protein FAP2 binds TIGIT on T/NK cells to inhibit antitumor immunity (94–96).
SCFAs such as butyrate, acetate, and propionate, as well as tryptophan metabolites, play key roles in modulating immune responses (97, 98). Butyrate enhances CD8+ T-cell activation by inhibiting histone deacetylases and inducing ID2 expression, thereby improving responses to ICB and radiotherapy (99). However, it may also suppress type I IFN production in dendritic cells, reducing radiotherapy efficacy in certain contexts (100).
In contrast, microbial SCFAs also exhibit cytoprotective effects, alleviating treatment-related toxicity. For example, butyrate from Prevotella loescheii mitigates cardiotoxicity associated with PD-1/PD-L1 blockade (101), while propionate and indole-derived metabolites reduce hematologic and gastrointestinal toxicity during radiotherapy (102).
Gut microbiota also influences immune checkpoint expression. Coprobacillus cateniformis, for example, downregulates PD-L2 in dendritic cells, enhancing CD8+ T-cell–mediated antitumor immunity. In germ-free mice, blockade of PD-L2 or its receptor RGMb restores responsiveness to anti-PD-1/PD-L1 therapy, in a MYD88-dependent manner (103).
The gut-tumor immune axis is a dynamic interface with both therapeutic potential and challenges. While some bacteria promote immune activation, others contribute to therapy resistance or immune suppression. Conflicting findings across studies highlight the influence of host genetic background, tumor type, and microbiota composition.
Identifying key microbial species and metabolites that predict or modulate immunotherapy outcomes; Targeting microbial pathways (e.g., SCFA production, MAMP sensing) to enhance immune responses; Designing microbiome-informed immunotherapy protocols, including prebiotic/probiotic combinations or microbial-derived adjuvants (Figure 2).
3.4 Regulation of the biophysical properties of the tumor microenvironment
The biophysical landscape of the TME—including matrix stiffness, cellular adhesion, and mechanical stress—plays a crucial role in cancer progression and immune regulation. Recent studies reveal that gut microbiota, particularly specific bacterial species and their bioactive factors, can significantly influence these mechanical properties, thereby modulating tumor invasiveness and therapeutic response.
In CRC, Fusobacterium nucleatum (Fn) has been shown to enhance tumor cell adhesion to endothelial cells via upregulation of ICAM1, a key adhesion molecule, facilitating extravasation and metastasis in in vivo models. This process is mediated by the ALPK1-NF-κB signaling axis (104).Furthermore, circulating tumor cells experience fluid shear stress and other mechanical forces that can cause cytoskeletal damage and limit metastatic potential (105, 106). However, cancer cells invaded by certain bacteria—termed intratumoral microbes (InTM)—exhibit enhanced survival under these conditions. In murine models, bacterial invasion induced RhoA-ROCK-actin remodeling, which strengthens cytoskeletal integrity and increases resistance to mechanical stress, promoting distant metastasis (107).
The exact microbial trigger for this phenotype remains under investigation, but bacterial factors such as C3 ribosyl transferase from Clostridium botulinum, known to alter actin dynamics, are potential candidates (108). Microbial dysbiosis also influences extracellular matrix (ECM) dynamics within tumors. For instance, reduced abundance of Faecalibacterium prausnitzii in CRC patients correlates with enhanced MMP2 activation, leading to chronic inflammation, fibrosis, and excessive ECM deposition (109). These changes result in increased TME stiffness, which has been observed across multiple cancers (e.g., breast, liver, pancreatic, prostate) and is associated with poor prognosis, immune evasion, and therapeutic resistance (105).The stiffened ECM promotes tumor progression through mechanotransduction pathways, enabling cancer cells to sense and respond to physical cues via integrins and YAP/TAZ transcriptional regulation, further reinforcing invasive phenotypes.
Gut microbes not only influence biochemical signaling but also remodel the physical architecture of the TME through: Modulation of adhesion molecules (e.g., ICAM1); Cytoskeletal reprogramming under shear stress; Fibrosis-induced ECM stiffening. These biomechanical changes contribute to metastasis, immune suppression, and therapy resistance. Understanding how specific microbial species interact with tumor biophysics offers a novel therapeutic avenue. Targeting bacteria-ECM-cytoskeleton crosstalk may allow us to “soften” the TME and enhance antitumor immunity or drug penetration (Figure 2).
3.5 Gut microbes influence intracellular bacteria in tumors to regulate tumor development
Studies have increasingly demonstrated that microorganisms can negatively impact antitumor immunity, particularly in pancreatic cancer. In a genetically engineered mouse model of PDAC, Pushalkar et al. identified a high abundance of bacteria within tumor tissues, with up to 20% originating from the gut microbiota—an observation further supported by human surgical specimens in which these bacteria were absent from adjacent non-tumor areas (110, 111). Notably, pancreatic tumorigenesis was found to induce time-dependent disruptions in gut microbial composition, which correlated with Kras activation, highlighting a dynamic gut-tumor microbiome interaction during disease progression (112, 113).
Beyond direct colonization, the intratumoral microbiome can also be shaped indirectly by gut microbiota modulation. FMT from short-term survivors (STS), long-term survivors (LTS), and healthy individuals into tumor-bearing mice led to distinct differences in tumor microbiome composition, immune infiltration, and tumor growth, underscoring the profound influence of gut-derived microbial signals on the tumor immune microenvironment (17, 114).
The origins of the intratumoral microbiota are believed to include mucosal surfaces (e.g., gastrointestinal tract, lungs), nearby normal tissues, and the circulatory system (115). Tumors at mucosal sites are particularly susceptible to microbial infiltration due to barrier dysfunction. For example, PDAC-associated bacteria have been shown to translocate from the gut to the pancreas via the pancreatic duct, facilitated by the unique inflammatory and immunosuppressive microenvironment of adenocarcinoma (116, 117). However, even tumors in non-mucosal sites such as the breast harbor microorganisms, suggesting alternative routes of microbial entry, possibly via blood or immune cells (118).
A 2020 study further revealed that tumor-resident microbiota closely resembles the bacterial communities in adjacent normal tissues, suggesting NATs as a potential seeding source for intratumoral bacteria (119). Given the multisource and tissue-specific nature of tumor microbiota, systematic comparisons across tumor types and anatomical regions may help uncover tumor-specific microbial signatures. These findings open new avenues for cancer prevention and precision therapeutics by targeting microbial components of the tumor microenvironment (Figure 2).
3.6 Regulating DNA stability
Gut microbes have emerged as key players in CRC pathogenesis through their capacity to induce genotoxic stress, compromise genome integrity, and drive malignant transformation (120–122). One of the most well-characterized mechanisms involves colibactin, a genotoxin synthesized by pks+ strains of Escherichia coli, which causes DNA double-strand breaks, triggers DNA damage response cascades, and increases chromosomal instability and mutation frequency. In vivo experiments have shown that inhibiting colibactin production can suppress tumor development, underscoring its critical role in carcinogenesis.
Furthermore, colibactin-producing E. coli can act synergistically with enterotoxin-producing Bacteroides fragilis-like species. These bacteria degrade the protective mucus barrier, facilitating the colonization of the colonic mucosa by pks+ E. coli, thereby amplifying DNA damage within epithelial cells. This coordinated microbial assault accelerates neoplastic initiation in the colon.
To explain these dynamics, researchers have proposed the “driver–passenger” model: early “driver” bacteria possess oncogenic traits that initiate DNA damage and tumor formation. As tumor progression remodels the local microenvironment—through inflammation, nutrient shifts, and immune suppression—it becomes more permissive to “passenger” or opportunistic bacteria. These later colonizers, although not directly oncogenic, benefit from the altered niche and further exacerbate tumor development through immune modulation or metabolic interactions (31, 123).
In summary, gut microbes contribute to CRC not only by initiating genotoxic events but also by participating in a dynamic ecological succession that sustains and promotes tumor progression. This interplay highlights the potential of targeting specific microbial signatures or their genotoxins as a strategy for CRC prevention and intervention (Figure 2).
Recent studies have further elucidated the diverse mechanisms by which gut microbes influence cancer progression and therapeutic responses. Notably, the STING signaling pathway has emerged as a critical mediator linking microbial-derived signals to innate immune activation, thereby shaping antitumor immunity (124). Additionally, autophagy regulation has been shown to intersect with microbial cues, affecting tumor cell survival and responsiveness to treatment (125). Microbial metabolites also play pivotal roles in driving epigenetic reprogramming within the tumor microenvironment, altering gene expression patterns that can either suppress or promote tumorigenesis (15). Moreover, Bifidobacterium species have been reported to enhance dendritic cell maturation and T-cell activation, strengthening host antitumor immune responses (126). Together, these insights underscore the multifaceted ways in which gut microbes and their metabolites modulate cancer biology and highlight promising avenues for therapeutic intervention.
4 Microbiota-targeted therapeutics: clinical perspectives and challenges
4.1 Current clinical applications of FMT, probiotics, and next-generation probiotics
In recent years, microbiota-targeted interventions have been increasingly explored in cancer therapy, with FMT emerging as a promising strategy for microbiota reconstruction. FMT has shown preliminary clinical value in modulating immunity and enhancing therapeutic efficacy (127–131). Particularly in patients with poor responses to ICIs, FMT has been demonstrated to restore microbial diversity and metabolic function, thereby improving the immune microenvironment and therapeutic outcomes (Table 2). For instance, FMT from ICI-responsive donors has been applied to patients with melanoma and non-small cell lung cancer, leading to significant alterations in gut microbial composition and immune cell infiltration (132). Additionally, probiotics and next-generation probiotics have exhibited potential in supporting radiotherapy, chemotherapy, or ICI therapy in several phase I/II clinical trials (133, 134). However, current applications are largely empirical, lacking standardized and personalized clinical protocols. The substantial functional heterogeneity among strains, challenges in maintaining formulation viability, and the complex clinical backgrounds of recipients all obscure the translational pathway. Therefore, future efforts should focus on identifying functional microbial biomarkers and elucidating underlying mechanisms to advance gut microbial interventions from empirical approaches toward mechanism-driven precision strategies.
4.2 The potential of synthetic biology-engineered bacteria and phage therapy in cancer treatment
With advances in synthetic biology and microbial engineering, engineered bacteria and phage therapy have opened unprecedented avenues in cancer treatment (135, 136). By genetically modifying gut-colonizing bacteria such as Escherichia coli, it is now possible to endow them with the ability to selectively release cytokines, immune-activating molecules, or anti-tumor metabolites within the tumor microenvironment—effectively functioning as an “in vivo micro-factory” for targeted therapy (137, 138). For instance, engineered E. coli Nissle strains have been developed to express PD-L1 nanobodies, with the potential to enhance the penetration and efficacy of immune checkpoint therapies (137). In addition, synthetic phages can be designed to selectively eliminate oncogenic bacterial populations (e.g., Fusobacterium nucleatum), thereby mitigating their roles in promoting cancer cell adhesion and immune suppression (139, 140).
However, several technical challenges remain in the clinical translation of these strategies, including biosafety concerns, microbial co-adaptation, and the risk of genetic drift. Achieving precise control over in vivo activity—such as spatiotemporal release, modulation of immune tolerance, and avoiding unintended disruption of the gut microbiota—remains a critical bottleneck. Moving forward, integrating dynamic simulation modeling, nanocarrier delivery platforms, and CRISPR-based regulatory systems is expected to enhance the precision, controllability, and clinical viability of these synthetic microbiome-based interventions.
4.3 Controllability, safety, and interindividual variability in microbiota-based interventions
Although microbiota-targeted interventions have demonstrated promising therapeutic effects, significant interindividual variability remains one of the primary barriers to clinical translation (33, 127). The gut microbiota exhibits highly personalized characteristics influenced by factors such as genetic background, dietary patterns, antibiotic usage history, and baseline immune status, leading to substantial differences in response to the same microbial intervention across individuals (141–143). Furthermore, the diffusion, retention time, and interactions of FMT and engineered microbial preparations during intervention are complex and can lead to unpredictable efficacy, immune responses, or dysbiosis (144).
In terms of safety, there is currently a lack of systematic assessment regarding the potential pathogenicity and cumulative toxicity associated with the long-term use of engineered bacteria or FMT (143, 145, 146). Serious infection events reported in some clinical cases of FMT have also exposed shortcomings in donor screening and risk management protocols. Therefore, establishing standardized recipient/donor matching criteria, predictive models for pre-intervention microbiota structure and function, and incorporating dynamic monitoring alongside pharmacokinetic profiling are crucial strategies to enhance the reliability and safety of microbiota-based cancer interventions.
4.4 Challenges in standardized modeling and multi-omics integration
The lack of standardization and integration of multi-omics data significantly hampers the clinical advancement of microbiota-based therapies. In clinical settings, microbiota sequencing and functional prediction often suffer from inconsistencies in data dimensions, non-uniform analytical methods, and poor reproducibility, making cross-study comparisons and longitudinal accumulation of evidence difficult (147). Moreover, current intervention strategies lack mechanisms for synergistic interpretation with other omics data (e.g., metabolomics, transcriptomics, proteomics), which hinders the establishment of clear causal links between microbiota changes and therapeutic outcomes (148).
Although some studies have identified specific microbial taxa associated with treatment responses, elucidating their signaling pathways or the role of microbial metabolites remains a challenge (20, 133, 149–151). Therefore, future efforts should focus on developing standardized microbiota intervention models, unifying protocols for sampling, sequencing, and analysis, and integrating clinical cohort data with mechanistic studies. Leveraging big data platforms to build a cross-omics analytical framework will be crucial to strengthen the evidence base and provide traceable mechanistic insights for microbiota-targeted therapies.
4.5 AI and microbiome integration for therapeutic prediction and precision intervention design
Encouragingly, the integration of artificial intelligence (AI) technologies offers a novel approach to achieving precision microbiome interventions. AI models can extract features and uncover associations within the complex tripartite relationship between the microbiota, host, and therapy, enabling predictive modeling of key microbial taxa linked to therapeutic responses (152, 153). On this basis, the incorporation of multi-omics datasets—such as scRNA-seq, metabolomics, and 16S rRNA sequencing—allows AI to optimize personalized intervention strategies and enhance both therapeutic efficacy and safety margins (154, 155).
Recent studies have demonstrated that deep learning can identify characteristic microbial signatures in responders to ICIs, thereby supporting donor selection and improving the predictive accuracy of FMT outcomes (156). Furthermore, reinforcement learning algorithms can simulate the impact of different intervention pathways on microbial succession, assisting in the selection of intervention targets and the timing of therapeutic decisions (157).
However, current AI systems remain limited by the scale of training data, the precision of microbial taxonomic annotation, and insufficient causal inference capabilities. Moving forward, it is essential to leverage multicenter clinical cohorts and construct high-quality, well-annotated datasets. The development of interpretable and generalizable AI models will be critical to achieving an intelligent leap from population-level microbial “common pattern recognition” to truly individualized microbiome regulation.
5 Conclusion and perspectives
In recent years, gut microbiota has emerged as a crucial player in host immune modulation and metabolic regulation, gaining increasing prominence in the field of cancer therapy. A growing body of evidence indicates that the composition of the gut microbiota, its metabolic products, and its interactions with host cells significantly influence the outcomes of various antitumor treatments, including immunotherapy, chemotherapy, and radiotherapy. From enhancing treatment response rates to alleviating adverse effects and reshaping the tumor microenvironment, gut microbes exhibit a “triple role” of response prediction, therapeutic potentiation, and toxicity mitigation. As such, targeting the gut microbiome has become a new research frontier with the potential to transform next-generation cancer intervention strategies.
Despite impressive progress, the complex mechanisms through which gut microbes affect cancer therapy remain incompletely understood. High interindividual variability—driven by host genetics, immune status, and metabolic profiles—poses a challenge in decoding causative microbial-host interactions from vast multi-omics datasets. Moreover, the phenomenon of microbial “co-morbidity-coexistence-co-therapy” complicates efforts in precise clinical targeting. Current studies largely remain at the level of association analysis, with limited functional validation or mechanistic elucidation, which restricts the clinical application of gut microbiota as reliable biomarkers or therapeutic targets.
The controllability and safety of microbial interventions also represent major bottlenecks in translational applications. Existing strategies such as FMT, probiotic/next-generation microbial formulations, and synthetic engineered bacteria have shown therapeutic promise to some extent. However, issues such as poor stability, undefined side effect profiles, and potential interference with host immunity and metabolism persist. This is especially critical in the context of cancer, where patients often exhibit compromised immune systems, narrowing the “therapeutic safety window.” Therefore, intervention strategies must be accompanied by enhanced precision and controllability under dynamic immunological conditions.
Future research should emphasize the development of an integrated “tumor × microbiome × immunity × metabolism” framework, leveraging single-cell sequencing, spatial multi-omics, and metabolomics to enable in-depth analysis from population-wide to single-cell resolution (Figure 3). The incorporation of AI will be pivotal in overcoming existing limitations. AI-driven tools for microbiome prediction modeling, immune response forecasting, and individualized intervention optimization will significantly improve the clinical interpretability of microbiome data, fostering the evolution from “experience-based” to “mechanism-driven precision microbiome therapy.”
Figure 3. Future directions for gut microbes in cancer therapy. Harnessing the potential of gut microbes represents a promising frontier in precision oncology. Future research should focus on identifying key microbial taxa that influence specific anti-tumor therapeutic modalities—including chemotherapy, radiotherapy, immunotherapy, and targeted therapies—through detailed microbiome profiling. Mechanistic studies are needed to elucidate how gut microbes and their metabolites modulate treatment response, therapeutic resistance, and host immunity. Moreover, robust clinical trials must be conducted to validate the efficacy and safety of microbiota-based interventions such as probiotics, FMT, and microbial metabolite supplementation in cancer patients. With the advancement of synthetic biology and nanotechnology, the design of “artificial anti-tumor bacteria” engineered to deliver drugs, modulate the tumor microenvironment, or boost host immune responses could enable precise and controllable microbial therapy. Ultimately, integrating microbiome science with multi-omics and artificial intelligence tools will drive the development of individualized, microbiota-informed strategies for improving cancer treatment outcomes.
Ultimately, personalized microbiome-based therapy is poised to become a central component of future anticancer strategies. Given the regional, dietary, lifestyle, and genetic diversity of microbiomes, constructing high-resolution population microbiota maps and dynamic evaluation systems will be a key research priority. By establishing a three-dimensional interactive network of “microbiota-host-tumor,” it may become feasible to design early screening strategies, personalized interventions, and synergistic treatment pathways based on microbiota status—achieving truly symbiotic and microbiota-empowered anticancer approaches.
Author contributions
DL: Writing – original draft, Writing – review & editing. YH: Conceptualization, Investigation, Writing – original draft. YC: Formal Analysis, Funding acquisition, Software, Supervision, Writing – original draft, Writing – review & editing. LL: Conceptualization, Funding acquisition, Writing – review & editing. MZ: Funding acquisition, Visualization, Writing – review & editing. WD: Supervision, Project administration, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was funded by the science and technology innovation Program of Changde city (No. 2023ZD69).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Malczewski AB, Navarro S, Coward JI, and Ketheesan N. Microbiome-derived metabolome as a potential predictor of response to cancer immunotherapy. J Immunother Cancer. (2020) 8:e001383. doi: 10.1136/jitc-2020-001383
2. Weersma RK, Zhernakova A, and Fu J. Interaction between drugs and the gut microbiome. Gut. (2020) 69:1510–9. doi: 10.1136/gutjnl-2019-320204
3. Sawhney SS, Thanert R, Thanert A, Hall-Moore C, Ndao IM, Mahmud B, et al. Gut microbiome evolution from infancy to 8 years of age. Nat Med. (2025) 31:2004–15. doi: 10.1038/s41591-025-03610-0
4. Gensollen T, Iyer SS, Kasper DL, and Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. (2016) 352:539–44. doi: 10.1126/science.aad9378
5. Clemente JC, Ursell LK, Parfrey LW, and Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. (2012) 148:1258–70. doi: 10.1016/j.cell.2012.01.035
6. Qin Y, Tong X, Mei WJ, Cheng Y, Zou Y, Han K, et al. Consistent signatures in the human gut microbiome of old- and young-onset colorectal cancer. Nat Commun. (2024) 15:3396. doi: 10.1038/s41467-024-47523-x
7. Kumari S, Srilatha M, and Nagaraju GP. Effect of gut dysbiosis on onset of GI cancers. Cancers (Basel). (2024) 17:90. doi: 10.3390/cancers17010090
8. Singh V, Yeoh BS, Chassaing B, Xiao X, Saha P, Aguilera Olvera R, et al. Dysregulated microbial fermentation of soluble fiber induces cholestatic liver cancer. Cell. (2018) 175:679–94 e22. doi: 10.1016/j.cell.2018.09.004
9. Xia C, Su J, Liu C, Mai Z, Yin S, Yang C, et al. Human microbiomes in cancer development and therapy. MedComm (2020). (2023) 4:e221. doi: 10.1002/mco2.221
10. Wu J, Xia C, Liu C, Zhang Q, and Xia C. The role of gut microbiota and drug interactions in the development of colorectal cancer. Front Pharmacol. (2023) 14:1265136. doi: 10.3389/fphar.2023.1265136
11. Heymann CJF, Bard JM, Heymann MF, Heymann D, and Bobin-Dubigeon C. The intratumoral microbiome: Characterization methods and functional impact. Cancer Lett. (2021) 522:63–79. doi: 10.1016/j.canlet.2021.09.009
12. Su J, Lin X, Li D, Yang C, Lv S, Chen X, et al. Prevotella copri exhausts intrinsic indole-3-pyruvic acid in the host to promote breast cancer progression: inactivation of AMPK via UHRF1-mediated negative regulation. Gut Microbes. (2024) 16:2347757. doi: 10.1080/19490976.2024.2347757
13. Simpson RC, Shanahan ER, Scolyer RA, and Long GV. Towards modulating the gut microbiota to enhance the efficacy of immune-checkpoint inhibitors. Nat Rev Clin Oncol. (2023) 20:697–715. doi: 10.1038/s41571-023-00803-9
14. Sun J, Chen F, and Wu G. Potential effects of gut microbiota on host cancers: focus on immunity, DNA damage, cellular pathways, and anticancer therapy. ISME J. (2023) 17:1535–51. doi: 10.1038/s41396-023-01483-0
15. Yang Q, Wang B, Zheng Q, Li H, Meng X, Zhou F, et al. A review of gut microbiota-derived metabolites in tumor progression and cancer therapy. Adv Sci (Weinh). (2023) 10:e2207366. doi: 10.1002/advs.202207366
16. Zhang Q, Zhao Q, Li T, Lu L, Wang F, Zhang H, et al. Lactobacillus plantarum-derived indole-3-lactic acid ameliorates colorectal tumorigenesis via epigenetic regulation of CD8(+) T cell immunity. Cell Metab. (2023) 35:943–60 e9. doi: 10.1016/j.cmet.2023.04.015
17. Park EM, Chelvanambi M, Bhutiani N, Kroemer G, Zitvogel L, and Wargo JA. Targeting the gut and tumor microbiota in cancer. Nat Med. (2022) 28:690–703. doi: 10.1038/s41591-022-01779-2
18. de Visser KE and Joyce JA. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell. (2023) 41:374–403. doi: 10.1016/j.ccell.2023.02.016
19. Wang H, Bo W, Feng X, Zhang J, Li G, and Chen Y. Strategies and recent advances on improving efficient antitumor of lenvatinib based on nanoparticle delivery system. Int J Nanomed. (2024) 19:5581–603. doi: 10.2147/IJN.S460844
20. Tang Y, Cai Q, Tian Z, Chen W, and Tang H. Crosstalk between gut microbiota and cancer immunotherapy: present investigations and future perspective. Res (Wash D C). (2025) 8:600. doi: 10.34133/research.0600
21. Blake SJ, Wolf Y, Boursi B, and Lynn DJ. Role of the microbiota in response to and recovery from cancer therapy. Nat Rev Immunol. (2024) 24:308–25. doi: 10.1038/s41577-023-00951-0
22. Drobner JC, Lichtbroun BJ, Singer EA, and Ghodoussipour S. Examining the role of microbiota-centered interventions in cancer therapeutics: applications for urothelial carcinoma. Technol Cancer Res Treat. (2023) 22:15330338231164196. doi: 10.1177/15330338231164196
23. Zhang H, Wu J, Li N, Wu R, and Chen W. Microbial influence on triggering and treatment of host cancer: An intestinal barrier perspective. Biochim Biophys Acta Rev Cancer. (2023) 1878:188989. doi: 10.1016/j.bbcan.2023.188989
24. El Tekle G, Andreeva N, and Garrett WS. The role of the microbiome in the etiopathogenesis of colon cancer. Annu Rev Physiol. (2024) 86:453–78. doi: 10.1146/annurev-physiol-042022-025619
25. Yao J, Ning B, and Ding J. The gut microbiota: an emerging modulator of drug resistance in hepatocellular carcinoma. Gut Microbes. (2025) 17:2473504. doi: 10.1080/19490976.2025.2473504
26. Ge Z, Chen C, Chen J, Jiang Z, Chen L, Wei Y, et al. Gut microbiota-derived 3-hydroxybutyrate blocks GPR43-mediated IL6 signaling to ameliorate radiation proctopathy. Adv Sci (Weinh). (2024) 11:e2306217. doi: 10.1002/advs.202306217
27. Guo H, Chou WC, Lai Y, Liang K, Tam JW, Brickey WJ, et al. Multi-omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science. (2020) 370:eaay9097. doi: 10.1126/science.aay9097
28. Riehl TE, Alvarado D, Ee X, Zuckerman A, Foster L, Kapoor V, et al. Lactobacillus rhamnosus GG protects the intestinal epithelium from radiation injury through release of lipoteichoic acid, macrophage activation and the migration of mesenchymal stem cells. Gut. (2019) 68:1003–13. doi: 10.1136/gutjnl-2018-316226
29. Blanarova C, Galovicova A, and Petrasova D. Use of probiotics for prevention of radiation-induced diarrhea. Bratisl Lek Listy. (2009) 110:98–104.
30. Sharma A, Rath GK, Chaudhary SP, Thakar A, Mohanti BK, and Bahadur S. Lactobacillus brevis CD2 lozenges reduce radiation- and chemotherapy-induced mucositis in patients with head and neck cancer: a randomized double-blind placebo-controlled study. Eur J Cancer. (2012) 48:875–81. doi: 10.1016/j.ejca.2011.06.010
31. Liu L and Shah K. The potential of the gut microbiome to reshape the cancer therapy paradigm: A review. JAMA Oncol. (2022) 8:1059–67. doi: 10.1001/jamaoncol.2022.0494
32. Ding X, Ting NL, Wong CC, Huang P, Jiang L, Liu C, et al. Bacteroides fragilis promotes chemoresistance in colorectal cancer, and its elimination by phage VA7 restores chemosensitivity. Cell Host Microbe. (2025) 33:941–56 e10. doi: 10.1016/j.chom.2025.05.004
33. Chrysostomou D, Roberts LA, Marchesi JR, and Kinross JM. Gut microbiota modulation of efficacy and toxicity of cancer chemotherapy and immunotherapy. Gastroenterology. (2023) 164:198–213. doi: 10.1053/j.gastro.2022.10.018
34. Nagini S, Kallamadi PR, Tanagala KKK, and Reddy GB. Aldo-keto reductases: Role in cancer development and theranostics. Oncol Res. (2024) 32:1287–308. doi: 10.32604/or.2024.049918
35. Li BT, Smit EF, Goto Y, Nakagawa K, Udagawa H, Mazieres J, et al. Trastuzumab deruxtecan in HER2-mutant non-small-cell lung cancer. N Engl J Med. (2022) 386:241–51. doi: 10.1056/NEJMoa2112431
36. Routy B, Lenehan JG, Miller WH Jr., Jamal R, Messaoudene M, Daisley BA, et al. Fecal microbiota transplantation plus anti-PD-1 immunotherapy in advanced melanoma: a phase I trial. Nat Med. (2023) 29:2121–32. doi: 10.1038/s41591-023-02453-x
37. Di Modica M, Gargari G, Regondi V, Bonizzi A, Arioli S, Belmonte B, et al. Gut microbiota condition the therapeutic efficacy of trastuzumab in HER2-positive breast cancer. Cancer Res. (2021) 81:2195–206. doi: 10.1158/0008-5472.CAN-20-1659
38. Chen YC, Chuang CH, Miao ZF, Yip KL, Liu CJ, Li LH, et al. Gut microbiota composition in chemotherapy and targeted therapy of patients with metastatic colorectal cancer. Front Oncol. (2022) 12:955313. doi: 10.3389/fonc.2022.955313
39. Sfanos KS, Markowski MC, Peiffer LB, Ernst SE, White JR, Pienta KJ, et al. Compositional differences in gastrointestinal microbiota in prostate cancer patients treated with androgen axis-targeted therapies. Prostate Cancer Prostatic Dis. (2018) 21:539–48. doi: 10.1038/s41391-018-0061-x
40. Pernigoni N, Zagato E, Calcinotto A, Troiani M, Mestre RP, Cali B, et al. Commensal bacteria promote endocrine resistance in prostate cancer through androgen biosynthesis. Science. (2021) 374:216–24. doi: 10.1126/science.abf8403
41. Waldman AD, Fritz JM, and Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. (2020) 20:651–68. doi: 10.1038/s41577-020-0306-5
42. Liu L, Liang L, Luo Y, Han J, Lu D, Cai R, et al. Unveiling the power of gut microbiome in predicting neoadjuvant immunochemotherapy responses in esophageal squamous cell carcinoma. Res (Wash D C). (2024) 7:529. doi: 10.34133/research.0529
43. Chen Q, Guo X, and Ma W. Opportunities and challenges of CD47-targeted therapy in cancer immunotherapy. Oncol Res. (2023) 32:49–60. doi: 10.32604/or.2023.042383
44. Tito RY, Verbandt S, Aguirre Vazquez M, Lahti L, Verspecht C, Llorens-Rico V, et al. Microbiome confounders and quantitative profiling challenge predicted microbial targets in colorectal cancer development. Nat Med. (2024) 30:1339–48. doi: 10.1038/s41591-024-02963-2
45. Xie J, Liu M, Deng X, Tang Y, Zheng S, Ou X, et al. Gut microbiota reshapes cancer immunotherapy efficacy: Mechanisms and therapeutic strategies. Imeta. (2024) 3:e156. doi: 10.1002/imt2.156
46. Stein-Thoeringer CK, Saini NY, Zamir E, Blumenberg V, Schubert ML, Mor U, et al. A non-antibiotic-disrupted gut microbiome is associated with clinical responses to CD19-CAR-T cell cancer immunotherapy. Nat Med. (2023) 29:906–16. doi: 10.1038/s41591-023-02234-6
47. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. (2015) 350:1084–9. doi: 10.1126/science.aac4255
48. Bender MJ, McPherson AC, Phelps CM, Pandey SP, Laughlin CR, Shapira JH, et al. Dietary tryptophan metabolite released by intratumoral Lactobacillus reuteri facilitates immune checkpoint inhibitor treatment. Cell. (2023) 186:1846–62 e26. doi: 10.1016/j.cell.2023.03.011
49. McCulloch JA, Davar D, Rodrigues RR, Badger JH, Fang JR, Cole AM, et al. Intestinal microbiota signatures of clinical response and immune-related adverse events in melanoma patients treated with anti-PD-1. Nat Med. (2022) 28:545–56. doi: 10.1038/s41591-022-01698-2
50. Gao Y, Bi D, Xie R, Li M, Guo J, Liu H, et al. Fusobacterium nucleatum enhances the efficacy of PD-L1 blockade in colorectal cancer. Signal Transduct Target Ther. (2021) 6:398. doi: 10.1038/s41392-021-00795-x
51. Galeano Nino JL, Wu H, LaCourse KD, Kempchinsky AG, Baryiames A, Barber B, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. (2022) 611:810–7. doi: 10.1038/s41586-022-05435-0
52. Wahi A, Bishnoi M, Raina N, Singh MA, Verma P, Gupta PK, et al. Recent updates on nano-phyto-formulations based therapeutic intervention for cancer treatment. Oncol Res. (2023) 32:19–47. doi: 10.32604/or.2023.042228
53. Tian J, Bai B, Gao Z, Yang Y, Wu H, Wang X, et al. Alleviation effects of GQD, a traditional chinese medicine formula, on diabetes rats linked to modulation of the gut microbiome. Front Cell Infect Microbiol. (2021) 11:740236. doi: 10.3389/fcimb.2021.740236
54. He J, Jin Y, He C, Li Z, Yu W, Zhou J, et al. Danggui Shaoyao San: comprehensive modulation of the microbiota-gut-brain axis for attenuating Alzheimer’s disease-related pathology. Front Pharmacol. (2023) 14:1338804. doi: 10.3389/fphar.2023.1338804
55. Yang J, Wei H, Lin Y, Chu ESH, Zhou Y, Gou H, et al. High soluble fiber promotes colorectal tumorigenesis through modulating gut microbiota and metabolites in mice. Gastroenterology. (2024) 166:323–37 e7. doi: 10.1053/j.gastro.2023.10.012
56. Dai R, Kelly BN, Ike A, Berger D, Chan A, Drew DA, et al. The impact of the gut microbiome, environment, and diet in early-onset colorectal cancer development. Cancers (Basel). (2024) 16:676. doi: 10.3390/cancers16030676
57. Tayyem RF, Shehadah I, Abu-Mweis SS, Bawadi HA, Bani-Hani KE, Al-Jaberi T, et al. Fruit and vegetable intake among Jordanians: results from a case-control study of colorectal cancer. Cancer Control. (2014) 21:350–60. doi: 10.1177/107327481402100412
58. Martin-Perez M, Urdiroz-Urricelqui U, Bigas C, and Benitah SA. The role of lipids in cancer progression and metastasis. Cell Metab. (2022) 34:1675–99. doi: 10.1016/j.cmet.2022.09.023
59. Iacucci M, Santacroce G, Majumder S, Morael J, Zammarchi I, Maeda Y, et al. Opening the doors of precision medicine: novel tools to assess intestinal barrier in inflammatory bowel disease and colitis-associated neoplasia. Gut. (2024) 73:1749–62. doi: 10.1136/gutjnl-2023-331579
60. AtChade AM, Williams JL, Mermelstein L, and Nemesure B. Unraveling the complexities of early-onset colorectal cancer: a perspective on dietary and microbial influences. Front Public Health. (2024) 12:1370108. doi: 10.3389/fpubh.2024.1370108
61. Panebianco C, Villani A, Pisati F, Orsenigo F, Ulaszewska M, Latiano TP, et al. Butyrate, a postbiotic of intestinal bacteria, affects pancreatic cancer and gemcitabine response in in vitro and in vivo models. BioMed Pharmacother. (2022) 151:113163. doi: 10.1016/j.biopha.2022.113163
62. Andrlova H, Miltiadous O, Kousa AI, Dai A, DeWolf S, Violante S, et al. MAIT and Vdelta2 unconventional T cells are supported by a diverse intestinal microbiome and correlate with favorable patient outcome after allogeneic HCT. Sci Transl Med. (2022) 14:eabj2829. doi: 10.1126/scitranslmed.abj2829
63. Xu H, Cao C, Ren Y, Weng S, Liu L, Guo C, et al. Antitumor effects of fecal microbiota transplantation: Implications for microbiome modulation in cancer treatment. Front Immunol. (2022) 13:949490. doi: 10.3389/fimmu.2022.949490
64. Lensu S and Pekkala S. Gut microbiota, microbial metabolites and human physical performance. Metabolites. (2021) 11:716. doi: 10.3390/metabo11110716
65. Hughes RL and Holscher HD. Fueling gut microbes: A review of the interaction between diet, exercise, and the gut microbiota in athletes. Adv Nutr. (2021) 12:2190–215. doi: 10.1093/advances/nmab077
66. Clauss M, Gerard P, Mosca A, and Leclerc M. Interplay between exercise and gut microbiome in the context of human health and performance. Front Nutr. (2021) 8:637010. doi: 10.3389/fnut.2021.637010
67. Liu JL, Xu X, Rixiati Y, Wang CY, Ni HL, Chen WS, et al. Dysfunctional circadian clock accelerates cancer metastasis by intestinal microbiota triggering accumulation of myeloid-derived suppressor cells. Cell Metab. (2024) 36:1320–34 e9. doi: 10.1016/j.cmet.2024.04.019
68. Zhou Y, Han W, Feng Y, Wang Y, Sun T, and Xu J. Microbial metabolites affect tumor progression, immunity and therapy prediction by reshaping the tumor microenvironment (Review). Int J Oncol. (2024) 65:73. doi: 10.3892/ijo.2024.5661
69. Park PH, Keith K, Calendo G, Jelinek J, Madzo J, Gharaibeh RZ, et al. Association between gut microbiota and CpG island methylator phenotype in colorectal cancer. Gut Microbes. (2024) 16:2363012. doi: 10.1080/19490976.2024.2363012
70. Li Y, He P, Chen Y, Hu J, Deng B, Liu C, et al. Microbial metabolite sodium butyrate enhances the anti-tumor efficacy of 5-fluorouracil against colorectal cancer by modulating PINK1/Parkin signaling and intestinal flora. Sci Rep. (2024) 14:13063. doi: 10.1038/s41598-024-63993-x
71. Tintelnot J, Xu Y, Lesker TR, Schonlein M, Konczalla L, Giannou AD, et al. Microbiota-derived 3-IAA influences chemotherapy efficacy in pancreatic cancer. Nature. (2023) 615:168–74. doi: 10.1038/s41586-023-05728-y
72. Bell HN, Rebernick RJ, Goyert J, Singhal R, Kuljanin M, Kerk SA, et al. Reuterin in the healthy gut microbiome suppresses colorectal cancer growth through altering redox balance. Cancer Cell. (2022) 40:185–200 e6. doi: 10.1016/j.ccell.2021.12.001
73. Cui W, Guo M, Liu D, Xiao P, Yang C, Huang H, et al. Gut microbial metabolite facilitates colorectal cancer development via ferroptosis inhibition. Nat Cell Biol. (2024) 26:124–37. doi: 10.1038/s41556-023-01314-6
74. Zhou P, Yang D, Sun D, and Zhou Y. Gut microbiome: New biomarkers in early screening of colorectal cancer. J Clin Lab Anal. (2022) 36:e24359. doi: 10.1002/jcla.24359
75. Chen Z, Zhao M, Liang J, Hu Z, Huang Y, Li M, et al. Dissecting the single-cell transcriptome network underlying esophagus non-malignant tissues and esophageal squamous cell carcinoma. EBioMedicine. (2021) 69:103459. doi: 10.1016/j.ebiom.2021.103459
76. Van Rossum T, Ferretti P, Maistrenko OM, and Bork P. Diversity within species: interpreting strains in microbiomes. Nat Rev Microbiol. (2020) 18:491–506. doi: 10.1038/s41579-020-0368-1
77. Zhu Z, Cai J, Hou W, Xu K, Wu X, Song Y, et al. Microbiome and spatially resolved metabolomics analysis reveal the anticancer role of gut Akkermansia muciniphila by crosstalk with intratumoral microbiota and reprogramming tumoral metabolism in mice. Gut Microbes. (2023) 15:2166700. doi: 10.1080/19490976.2023.2166700
78. Zhang L, Jiang C, Zhong Y, Sun K, Jing H, Song J, et al. STING is a cell-intrinsic metabolic checkpoint restricting aerobic glycolysis by targeting HK2. Nat Cell Biol. (2023) 25:1208–22. doi: 10.1038/s41556-023-01185-x
79. San-Millan I, Sparagna GC, Chapman HL, Warkins VL, Chatfield KC, Shuff SR, et al. Chronic lactate exposure decreases mitochondrial function by inhibition of fatty acid uptake and cardiolipin alterations in neonatal rat cardiomyocytes. Front Nutr. (2022) 9:809485. doi: 10.3389/fnut.2022.809485
80. Hong J, Guo F, Lu SY, Shen C, Ma D, Zhang X, et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut. (2021) 70:2123–37. doi: 10.1136/gutjnl-2020-322780
81. Wang N and Fang JY. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. (2023) 31:159–72. doi: 10.1016/j.tim.2022.08.010
82. Coker OO, Liu C, Wu WKK, Wong SH, Jia W, Sung JJY, et al. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome. (2022) 10:35. doi: 10.1186/s40168-021-01208-5
83. Bian X, Liu R, Meng Y, Xing D, Xu D, and Lu Z. Lipid metabolism and cancer. J Exp Med. (2021) 218:e20201606. doi: 10.1084/jem.20201606
84. Liu HH, Xu Y, Li CJ, Hsu SJ, Lin XH, Zhang R, et al. An SCD1-dependent mechanoresponsive pathway promotes HCC invasion and metastasis through lipid metabolic reprogramming. Mol Ther. (2022) 30:2554–67. doi: 10.1016/j.ymthe.2022.03.015
85. Leone RD, Zhao L, Englert JM, Sun IM, Oh MH, Sun IH, et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science. (2019) 366:1013–21. doi: 10.1126/science.aav2588
86. Li TT, Chen X, Huo D, Arifuzzaman M, Qiao S, Jin WB, et al. Microbiota metabolism of intestinal amino acids impacts host nutrient homeostasis and physiology. Cell Host Microbe. (2024) 32:661–75 e10. doi: 10.1016/j.chom.2024.04.004
87. Mao J, Wang D, Long J, Yang X, Lin J, Song Y, et al. Gut microbiome is associated with the clinical response to anti-PD-1 based immunotherapy in hepatobiliary cancers. J Immunother Cancer. (2021) 9:e003334. doi: 10.1136/jitc-2021-003334
88. Coutzac C, Jouniaux JM, Paci A, Schmidt J, Mallardo D, Seck A, et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat Commun. (2020) 11:2168. doi: 10.1038/s41467-020-16079-x
89. Silveira MAD, Bilodeau S, Greten TF, Wang XW, and Trinchieri G. The gut-liver axis: host microbiota interactions shape hepatocarcinogenesis. Trends Cancer. (2022) 8:583–97. doi: 10.1016/j.trecan.2022.02.009
90. Bruning EE, Coller JK, Wardill HR, and Bowen JM. Site-specific contribution of Toll-like receptor 4 to intestinal homeostasis and inflammatory disease. J Cell Physiol. (2021) 236:877–88. doi: 10.1002/jcp.29976
91. Lam KC, Araya RE, Huang A, Chen Q, Di Modica M, Rodrigues RR, et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell. (2021) 184:5338–56 e21. doi: 10.1016/j.cell.2021.09.019
92. Spencer CN, McQuade JL, Gopalakrishnan V, McCulloch JA, Vetizou M, Cogdill AP, et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science. (2021) 374:1632–40. doi: 10.1126/science.aaz7015
93. Lu H, Xu X, Fu D, Gu Y, Fan R, Yi H, et al. Butyrate-producing Eubacterium rectale suppresses lymphomagenesis by alleviating the TNF-induced TLR4/MyD88/NF-kappaB axis. Cell Host Microbe. (2022) 30:1139–50 e7. doi: 10.1016/j.chom.2022.07.003
94. Zhang X, Yu D, Wu D, Gao X, Shao F, Zhao M, et al. Tissue-resident Lachnospiraceae family bacteria protect against colorectal carcinogenesis by promoting tumor immune surveillance. Cell Host Microbe. (2023) 31:418–32 e8. doi: 10.1016/j.chom.2023.01.013
95. Luo B, Zhang S, Yu X, Tan D, Wang Y, and Wang M. Gasdermin E benefits CD8(+)T cell mediated anti-immunity through mitochondrial damage to activate cGAS-STING-interferonbeta axis in colorectal cancer. biomark Res. (2024) 12:59. doi: 10.1186/s40364-024-00606-9
96. Jiang SS, Xie YL, Xiao XY, Kang ZR, Lin XL, Zhang L, et al. Fusobacterium nucleatum-derived succinic acid induces tumor resistance to immunotherapy in colorectal cancer. Cell Host Microbe. (2023) 31:781–97 e9. doi: 10.1016/j.chom.2023.04.010
97. Mann ER, Lam YK, and Uhlig HH. Short-chain fatty acids: linking diet, the microbiome and immunity. Nat Rev Immunol. (2024) 24:577–95. doi: 10.1038/s41577-024-01014-8
98. Liu Y, Wong CC, Ding Y, Gao M, Wen J, Lau HC, et al. Peptostreptococcus anaerobius mediates anti-PD1 therapy resistance and exacerbates colorectal cancer via myeloid-derived suppressor cells in mice. Nat Microbiol. (2024) 9:1467–82. doi: 10.1038/s41564-024-01695-w
99. Yang K, Hou Y, Zhang Y, Liang H, Sharma A, Zheng W, et al. Suppression of local type I interferon by gut microbiota-derived butyrate impairs antitumor effects of ionizing radiation. J Exp Med. (2021) 218:e20201915. doi: 10.1084/jem.20201915
100. Uribe-Herranz M, Rafail S, Beghi S, Gil-de-Gomez L, Verginadis I, Bittinger K, et al. Gut microbiota modulate dendritic cell antigen presentation and radiotherapy-induced antitumor immune response. J Clin Invest. (2020) 130:466–79. doi: 10.1172/JCI124332
101. Chen Y, Liu Y, Wang Y, Chen X, Wang C, Chen X, et al. Prevotellaceae produces butyrate to alleviate PD-1/PD-L1 inhibitor-related cardiotoxicity via PPARalpha-CYP4X1 axis in colonic macrophages. J Exp Clin Cancer Res. (2022) 41:1. doi: 10.1186/s13046-021-02201-4
102. Yang Q, Qin B, Hou W, Qin H, and Yin F. Pathogenesis and therapy of radiation enteritis with gut microbiota. Front Pharmacol. (2023) 14:1116558. doi: 10.3389/fphar.2023.1116558
103. Park JS, Gazzaniga FS, Wu M, Luthens AK, Gillis J, Zheng W, et al. Targeting PD-L2-RGMb overcomes microbiome-related immunotherapy resistance. Nature. (2023) 617:377–85. doi: 10.1038/s41586-023-06026-3
104. Zhang Y, Zhang L, Zheng S, Li M, Xu C, Jia D, et al. Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF-kappaB/ICAM1 axis. Gut Microbes. (2022) 14:2038852. doi: 10.1080/19490976.2022.2038852
105. Gensbittel V, Krater M, Harlepp S, Busnelli I, Guck J, and Goetz JG. Mechanical adaptability of tumor cells in metastasis. Dev Cell. (2021) 56:164–79. doi: 10.1016/j.devcel.2020.10.011
106. Follain G, Herrmann D, Harlepp S, Hyenne V, Osmani N, Warren SC, et al. Fluids and their mechanics in tumour transit: shaping metastasis. Nat Rev Cancer. (2020) 20:107–24. doi: 10.1038/s41568-019-0221-x
107. Dasgupta I and McCollum D. Control of cellular responses to mechanical cues through YAP/TAZ regulation. J Biol Chem. (2019) 294:17693–706. doi: 10.1074/jbc.REV119.007963
108. Rohrbeck A and Just I. Cell entry of C3 exoenzyme from clostridium botulinum. Curr Top Microbiol Immunol. (2017) 406:97–118. doi: 10.1007/82_2016_44
109. Cambria E, Coughlin MF, Floryan MA, Offeddu GS, Shelton SE, and Kamm RD. Linking cell mechanical memory and cancer metastasis. Nat Rev Cancer. (2024) 24:216–28. doi: 10.1038/s41568-023-00656-5
110. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. (2019) 178:795–806 e12. doi: 10.1016/j.cell.2019.07.008
111. Ghaddar B, Biswas A, Harris C, Omary MB, Carpizo DR, Blaser MJ, et al. Tumor microbiome links cellular programs and immunity in pancreatic cancer. Cancer Cell. (2022) 40:1240–53 e5. doi: 10.1016/j.ccell.2022.09.009
112. Burdziak C, Alonso-Curbelo D, Walle T, Reyes J, Barriga FM, Haviv D, et al. Epigenetic plasticity cooperates with cell-cell interactions to direct pancreatic tumorigenesis. Science. (2023) 380:eadd5327. doi: 10.1126/science.add5327
113. Huang Z, Huang X, Huang Y, Liang K, Chen L, Zhong C, et al. Identification of KRAS mutation-associated gut microbiota in colorectal cancer and construction of predictive machine learning model. Microbiol Spectr. (2024) 12:e0272023. doi: 10.1128/spectrum.02720-23
114. Yang L, Li A, Wang Y, and Zhang Y. Intratumoral microbiota: roles in cancer initiation, development and therapeutic efficacy. Signal Transduct Target Ther. (2023) 8:35. doi: 10.1038/s41392-022-01304-4
115. Jiang Z, Zhang W, Zhang Z, Sha G, Wang D, and Tang D. Intratumoral microbiota: A new force in diagnosing and treating pancreatic cancer. Cancer Lett. (2023) 554:216031. doi: 10.1016/j.canlet.2022.216031
116. Raghavan S, Winter PS, Navia AW, Williams HL, DenAdel A, Lowder KE, et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell. (2021) 184:6119–37 e26. doi: 10.1016/j.cell.2021.11.017
117. Yousuf S, Qiu M, Voith von Voithenberg L, Hulkkonen J, Macinkovic I, Schulz AR, et al. Spatially resolved multi-omics single-cell analyses inform mechanisms of immune dysfunction in pancreatic cancer. Gastroenterology. (2023) 165:891–908 e14. doi: 10.1053/j.gastro.2023.05.036
118. Wong-Rolle A, Wei HK, Zhao C, and Jin C. Unexpected guests in the tumor microenvironment: microbiome in cancer. Protein Cell. (2021) 12:426–35. doi: 10.1007/s13238-020-00813-8
119. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. (2020) 368:973–80. doi: 10.1126/science.aay9189
120. Cao Y, Oh J, Xue M, Huh WJ, Wang J, Gonzalez-Hernandez JA, et al. Commensal microbiota from patients with inflammatory bowel disease produce genotoxic metabolites. Science. (2022) 378:eabm3233. doi: 10.1126/science.abm3233
121. Dougherty MW, Valdes-Mas R, Wernke KM, Gharaibeh RZ, Yang Y, Brant JO, et al. The microbial genotoxin colibactin exacerbates mismatch repair mutations in colorectal tumors. Neoplasia. (2023) 43:100918. doi: 10.1016/j.neo.2023.100918
122. Lin C, Cai X, Zhang J, Wang W, Sheng Q, Hua H, et al. Role of gut microbiota in the development and treatment of colorectal cancer. Digestion. (2019) 100:72–8. doi: 10.1159/000494052
123. Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, et al. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature. (2020) 580:269–73. doi: 10.1038/s41586-020-2080-8
124. Chen W, Zhu Y, Chen J, Jing X, Xiong Y, Zou L, et al. Potentiating the systemic immunity by bacteria-delivered sting activation in a tumor microenvironment. Advanced Funct Materials. (2023) 33. doi: 10.1002/adfm.202307001
125. Wang Y, Du J, Wu X, Abdelrehem A, Ren Y, Liu C, et al. Crosstalk between autophagy and microbiota in cancer progression. Mol Cancer. (2021) 20:163. doi: 10.1186/s12943-021-01461-0
126. Chen Z, Qin YT, Li QR, He JL, Deng XC, Zhang Y, et al. Layer-by-layer deposition of antigen peptides on bifidobacterium for subintestinal lymphatic system-guided personalized tumor immunotherapy. Adv Mater. (2025) 37:e2503571. doi: 10.1002/adma.202503571
127. Fernandes MR, Aggarwal P, Costa RGF, Cole AM, and Trinchieri G. Targeting the gut microbiota for cancer therapy. Nat Rev Cancer. (2022) 22:703–22. doi: 10.1038/s41568-022-00513-x
128. Zhao LY, Mei JX, Yu G, Lei L, Zhang WH, Liu K, et al. Role of the gut microbiota in anticancer therapy: from molecular mechanisms to clinical applications. Signal Transduct Target Ther. (2023) 8:201. doi: 10.1038/s41392-023-01406-7
129. Jia D, Wang Q, Qi Y, Jiang Y, He J, Lin Y, et al. Microbial metabolite enhances immunotherapy efficacy by modulating T cell stemness in pan-cancer. Cell. (2024) 187:1651–65 e21. doi: 10.1016/j.cell.2024.02.022
130. Kim Y, Kim G, Kim S, Cho B, Kim SY, Do EJ, et al. Fecal microbiota transplantation improves anti-PD-1 inhibitor efficacy in unresectable or metastatic solid cancers refractory to anti-PD-1 inhibitor. Cell Host Microbe. (2024) 32:1380–93 e9. doi: 10.1016/j.chom.2024.06.010
131. Kelly CR, Yen EF, Grinspan AM, Kahn SA, Atreja A, Lewis JD, et al. Fecal microbiota transplantation is highly effective in real-world practice: initial results from the FMT national registry. Gastroenterology. (2021) 160:183–92 e3. doi: 10.1053/j.gastro.2020.09.038
132. Lin A, Jiang A, Huang L, Li Y, Zhang C, Zhu L, et al. From chaos to order: optimizing fecal microbiota transplantation for enhanced immune checkpoint inhibitors efficacy. Gut Microbes. (2025) 17:2452277. doi: 10.1080/19490976.2025.2452277
133. Nobels A, van Marcke C, Jordan BF, Van Hul M, and Cani PD. The gut microbiome and cancer: from tumorigenesis to therapy. Nat Metab. (2025) 7:895–917. doi: 10.1038/s42255-025-01287-w
134. Wang L, Yu KC, Hou YQ, Guo M, Yao F, and Chen ZX. Gut microbiome in tumorigenesis and therapy of colorectal cancer. J Cell Physiol. (2023) 238:94–108. doi: 10.1002/jcp.30917
135. Baker ZR, Zhang Y, Zhang H, Franklin HC, Serpa PBS, Southard T, et al. Sustained in situ protein production and release in the mammalian gut by an engineered bacteriophage. Nat Biotechnol. (2025). doi: 10.1038/s41587-025-02570-7
136. Chang Z, Guo X, Li X, Wang Y, Zang Z, Pei S, et al. Bacterial immunotherapy leveraging IL-10R hysteresis for both phagocytosis evasion and tumor immunity revitalization. Cell. (2025) 188:1842–57 e20. doi: 10.1016/j.cell.2025.02.002
137. Gurbatri CR, Radford GA, Vrbanac L, Im J, Thomas EM, Coker C, et al. Engineering tumor-colonizing E. coli Nissle 1917 for detection and treatment of colorectal neoplasia. Nat Commun. (2024) 15:646. doi: 10.1038/s41467-024-44776-4
138. Yang S, Sheffer M, Kaplan IE, Wang Z, Tarannum M, Dinh K, et al. Non-pathogenic E. coli displaying decoy-resistant IL18 mutein boosts anti-tumor and CAR NK cell responses. . Nat Biotechnol. (2024) 43:1311–23. doi: 10.1038/s41587-024-02418-6
139. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. (2017) 170:548–63 e16. doi: 10.1016/j.cell.2017.07.008
140. Dong X, Pan P, Zheng DW, Bao P, Zeng X, and Zhang XZ. Bioinorganic hybrid bacteriophage for modulation of intestinal microbiota to remodel tumor-immune microenvironment against colorectal cancer. Sci Adv. (2020) 6:eaba1590. doi: 10.1126/sciadv.aba1590
141. Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, and Knight R. Current understanding of the human microbiome. Nat Med. (2018) 24:392–400. doi: 10.1038/nm.4517
142. Liang J, Li T, Zhao J, Wang C, and Sun H. Current understanding of the human microbiome in glioma. Front Oncol. (2022) 12:781741. doi: 10.3389/fonc.2022.781741
143. Aggarwal N, Kitano S, Puah GRY, Kittelmann S, Hwang IY, and Chang MW. Microbiome and human health: current understanding, engineering, and enabling technologies. Chem Rev. (2023) 123:31–72. doi: 10.1021/acs.chemrev.2c00431
144. Kamath S, Bryant RV, Costello SP, Day AS, Forbes B, Haifer C, et al. Translational strategies for oral delivery of faecal microbiota transplantation. Gut. (2025). doi: 10.1136/gutjnl-2025-335077
145. Puurunen MK, Vockley J, Searle SL, Sacharow SJ, Phillips JA 3rd, Denney WS, et al. Safety and pharmacodynamics of an engineered E. coli Nissle for the treatment of phenylketonuria: a first-in-human phase 1/2a study. Nat Metab. (2021) 3:1125–32. doi: 10.1038/s42255-021-00430-7
146. Hu M, Zhu X, Huang X, Hua L, Lin X, Zhang H, et al. Optimizing anti-PD-1/PD-L1 therapy efficacy and fecal microbiota transplantation donor selection through gut mycobiome-based enterotype. Cell Rep. (2025) 44:115589. doi: 10.1016/j.celrep.2025.115589
147. Bindels LB, Watts JEM, Theis KR, Carrion VJ, Ossowicki A, Seifert J, et al. A blueprint for contemporary studies of microbiomes. Microbiome. (2025) 13:95. doi: 10.1186/s40168-025-02091-0
148. Turjeman S, Rozera T, Elinav E, Ianiro G, and Koren O. From big data and experimental models to clinical trials: Iterative strategies in microbiome research. Cell. (2025) 188:1178–97. doi: 10.1016/j.cell.2025.01.038
149. Battaglia TW, Mimpen IL, Traets JJH, van Hoeck A, Zeverijn LJ, Geurts BS, et al. A pan-cancer analysis of the microbiome in metastatic cancer. Cell. (2024) 187:2324–35 e19. doi: 10.1016/j.cell.2024.03.021
150. Lu Y, Yuan X, Wang M, He Z, Li H, Wang J, et al. Gut microbiota influence immunotherapy responses: mechanisms and therapeutic strategies. J Hematol Oncol. (2022) 15:47. doi: 10.1186/s13045-022-01273-9
151. Elkrief A, Montesion M, Sivakumar S, Hale C, Bowman AS, Begum Bektas A, et al. Intratumoral escherichia is associated with improved survival to single-agent immune checkpoint inhibition in patients with advanced non-small-cell lung cancer. J Clin Oncol. (2024) 42:3339–49. doi: 10.1200/JCO.23.01488
152. Xiao L and Zhao F. Exploring the frontier of microbiome biomarker discovery with artificial intelligence. Natl Sci Rev. (2024) 11:nwae325. doi: 10.1093/nsr/nwae325
153. Li M, Liu J, Zhu J, Wang H, Sun C, Gao NL, et al. Performance of gut microbiome as an independent diagnostic tool for 20 diseases: cross-cohort validation of machine-learning classifiers. Gut Microbes. (2023) 15:2205386. doi: 10.1080/19490976.2023.2205386
154. Zhang SL, Cheng LS, Zhang ZY, Sun HT, and Li JJ. Untangling determinants of gut microbiota and tumor immunologic status through a multi-omics approach in colorectal cancer. Pharmacol Res. (2023) 188:106633. doi: 10.1016/j.phrs.2022.106633
155. Gu F, Zhu S, Tang Y, Liu X, Jia M, Malmuthuge N, et al. Gut microbiome is linked to functions of peripheral immune cells in transition cows during excessive lipolysis. Microbiome. (2023) 11:40. doi: 10.1186/s40168-023-01492-3
156. Bhutiani N and Wargo JA. Gut microbes as biomarkers of ICI response - sharpening the focus. Nat Rev Clin Oncol. (2022) 19:495–6. doi: 10.1038/s41571-022-00634-0
157. Amit G and Bashan A. Top-down identification of keystone taxa in the microbiome. Nat Commun. (2023) 14:3951. doi: 10.1038/s41467-023-39459-5
158. Colbert LE, El Alam MB, Wang R, Karpinets T, Lo D, Lynn EJ, et al. Tumor-resident Lactobacillus iners confer chemoradiation resistance through lactate-induced metabolic rewiring. Cancer Cell. (2023) 41:1945–62 e11. doi: 10.1016/j.ccell.2023.09.012
159. Vivarelli S, Salemi R, Candido S, Falzone L, Santagati M, Stefani S, et al. Gut microbiota and cancer: from pathogenesis to therapy. Cancers (Basel). (2019) 11:38. doi: 10.3390/cancers11010038
160. Ma W, Zhang L, Chen W, Chang Z, Tu J, Qin Y, et al. Microbiota enterotoxigenic Bacteroides fragilis-secreted BFT-1 promotes breast cancer cell stemness and chemoresistance through its functional receptor NOD1. Protein Cell. (2024) 15:419–40. doi: 10.1093/procel/pwae005
161. Lee J, McClure S, Weichselbaum RR, and Mimee M. Designing live bacterial therapeutics for cancer. Adv Drug Delivery Rev. (2025) 221:115579. doi: 10.1016/j.addr.2025.115579
162. Zheng Y, Fang Z, Xue Y, Zhang J, Zhu J, Gao R, et al. Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes. (2020) 11:1030–42. doi: 10.1080/19490976.2020.1737487
163. Wang X, Fang Y, Liang W, Wong CC, Qin H, Gao Y, et al. Fusobacterium nucleatum facilitates anti-PD-1 therapy in microsatellite stable colorectal cancer. Cancer Cell. (2025) 43:564–74. doi: 10.1016/j.ccell.2025.02.023
164. Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. (2018) 360:eaan5931. doi: 10.1126/science.aan5931
165. Lee MH, Nuccio SP, Mohanty I, Hagey LR, Dorrestein PC, Chu H, et al. How bile acids and the microbiota interact to shape host immunity. Nat Rev Immunol. (2024) 24:798–809. doi: 10.1038/s41577-024-01057-x
166. Nan K, Zhong Z, Yue Y, Shen Y, Zhang H, Wang Z, et al. Fasting-mimicking diet-enriched Bifidobacterium pseudolongum suppresses colorectal cancer by inducing memory CD8(+) T cells. Gut. (2025) 74:775–86. doi: 10.1136/gutjnl-2024-333020
167. Preet R, Islam MA, Shim J, Rajendran G, Mitra A, Vishwakarma V, et al. Gut commensal Bifidobacterium-derived extracellular vesicles modulate the therapeutic effects of anti-PD-1 in lung cancer. Nat Commun. (2025) 16:3500. doi: 10.1038/s41467-025-58553-4
168. Li H, Dong T, Tao M, Zhao H, Lan T, Yan S, et al. Fucoidan enhances the anti-tumor effect of anti-PD-1 immunotherapy by regulating gut microbiota. Food Funct. (2024) 15:3463–78. doi: 10.1039/d3fo04807a
169. Wang X, Fang Y, Liang W, Wong CC, Qin H, Gao Y, et al. Fusobacterium nucleatum facilitates anti-PD-1 therapy in microsatellite stable colorectal cancer. Cancer Cell. (2024) 42:1729–46 e8. doi: 10.1016/j.ccell.2024.08.019
170. Huang J, Liu D, Wang Y, Liu L, Li J, Yuan J, et al. Ginseng polysaccharides alter the gut microbiota and kynurenine/tryptophan ratio, potentiating the antitumour effect of antiprogrammed cell death 1/programmed cell death ligand 1 (anti-PD-1/PD-L1) immunotherapy. Gut. (2022) 71:734–45. doi: 10.1136/gutjnl-2020-321031
Keywords: gut microbes, tumor therapy, tumor microenvironment, drug resistance, immunotherapy
Citation: Liao L, Zeng M, Liu D, He Y, Du W and Cao Y (2025) Focus on gut microbes: new direction in cancer treatment. Front. Oncol. 15:1505656. doi: 10.3389/fonc.2025.1505656
Received: 03 October 2024; Accepted: 19 August 2025;
Published: 29 August 2025.
Edited by:
Abbas Yadegar, Shahid Beheshti University of Medical Sciences, IranReviewed by:
Li Zhang, Brown University, United StatesChenglai Xia, Foshan Women and Children Hospital, China
Sameer Ullah Khan, Oregon Health and Science University, United States
Copyright © 2025 Liao, Zeng, Liu, He, Du and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanlin Cao, MTU5MDczNjYxNDhAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Lingshan Liao1†
Lingshan Liao1† Yanlin Cao
Yanlin Cao