- 1Department of Pediatric Hematology/Oncology and Cell and Gene Therapy, IRCCS, Ospedale Pediatrico Bambino Gesù, Rome, Italy
- 2Core Facilities-Clinical Proteomics and Metabolomics, IRCCS, Istituto Giannina Gaslini, Genoa, Italy
- 3Ophthalmology Unit, IRCCS, Ospedale Pediatrico Bambino Gesù, Rome, Italy
- 4Department of Laboratories, Pathology Unit, IRCCS, Ospedale Pediatrico Bambino Gesù, Rome, Italy
- 5Neuroradiology Unit, Imaging Department, Bambino Gesù Children’s Hospital, IRCCS, Rome, Italy
- 6Catholic University of the Sacred Heart, Rome, Italy
This study focuses on the proteomic analysis of cerebrospinal fluid (CSF) in a patient with stage III retinoblastoma (RB) with the aim to identify molecular changes associated with central nervous system (CNS) relapse. The child received systemic chemotherapy and intrathecal topotecan as CNS prophylaxis, along with enucleation of the left eye. After two chemotherapy cycles, CNS relapse occurred, evidenced by positive CSF findings and magnetic resonance imaging (MRI) showing leptomeningeal involvement at the anterior skull base. The child’s condition deteriorated, and two months later, he died due to progressive CNS disease. The aim of the study was to analyze serial CSF samples collected at different stages of treatment, as well as a control sample, to identify differences in CSF protein expression profiles during CNS RB relapse. Using mass spectrometry, a total of 1,029 proteins were identified across all CSF samples, samples were analyzed in duplicate ensuring technical replication. An unsupervised heatmap revealed 46 differentially expressed proteins. Over-regulated proteins in CSF-RB samples were primarily involved in inflammation, extracellular matrix remodeling, epithelial mesenchymal transition initiation, migration, invasion, and cellular metabolism (PON1, RNPEP, MCAM, NEGR1, NID1, SERPINA1, FAT2, RELN, NEGR1, and SEZ6). These processes are key drivers of cancer progression and metastasis. Proteomic analysis could be valuable in identifying proteins modulated in CSF during disease progression in RB patients, offering potential for new prognostic biomarkers.
Background
Retinoblastoma (RB) represents the most common intraocular malignancy of infancy and childhood (1). Although the survival rates are over 95% for intraocular forms, in low-income countries the prognosis remains poor, ranging from 20% to 60% (2, 3). Among the dissemination routes, RB may spread to the central nervous system (CNS) through the optic nerve. This occurrence correlates with poor prognosis despite intensive multimodal treatment (4). In particular, in patients with advanced RB, CNS remains the predominant site of relapse. Thus, an early detection of CNS involvement is crucial to set up timely and tailored therapeutic approaches. The conventional diagnosis of CNS involvement relies on magnetic resonance imaging (MRI) and on the detection of tumor cells in CSF. Detection of biomarkers in the CSF could be useful in identifying patients at risk of developing CNS disease, even before the evidence of CNS involvement. In recent years, mass spectrometry has emerged as a sensitive tool in characterizing proteomic profiles of different biofluids, such as CSF, in neurological diseases and brain tumors (5–8). Importantly, the proteomic analysis of CSF may detect the presence of brain tumors before the onset of symptoms or imaging in animal models (9), reveling that CSF may be altered even at the earliest stages of malignancy. In this report, we perform a proteomic study of CSF in a patient affected by stage III (IRSS) RB in order to detect proteins changes occurring during CNS recurrence.
Case presentation
Clinical data
An 8-month-old male child from Africa was admitted to the emergency department of the Bambino Gesù Children Hospital. He had a 6-months history of strabismus of the left eye associated with leukocoria diagnosed as cataract in his country and treated with eye drops. Due to the onset of exophthalmos, the child underwent MRI, which led to the diagnosis of RB (Figures 1A, a), and received 2 cycles of chemotherapy with etoposide and carboplatin. After his arrival at our hospital, the diagnostic workup documented a sporadic unilateral extraocular RB of the left eye, stage IIIa for orbit invasion, without CNS and bone-marrow involvement (Figures 1B, b). The patient underwent left eye debulking surgery (macroscopically incomplete enucleation) and received 3 cycles according to ICE schedule associated with intrathecal Topotecan at 0.3 mg per cycle. After 2 ICE cycles, the MRI and bone scintigraphy showed no local or CNS disease recurrence (Figures 1 C, c). However, the cytologic evaluation of CSF, performed during the third intrathecal injection of Topotecan, resulted positive for neoplastic cells. A new MRI showed impregnation of the leptomeninges of the anterior cranial bases confirming a disease recurrence at the CNS level. In light of these findings, the child received chemotherapy with Etoposide and Carboplatin for 5 days, in combination with intrathecal topotecan and thiotepa. CSF evaluation after chemotherapy was still positive for neoplastic cells. Due to incoercible vomiting and generalized hypotonia, the child was admitted to the emergency department where a Computerized Tomography (CT) scan showed massive CNS disease progression. The patient progressively worsened with cardiorespiratory arrest occurring 9 days after admission.
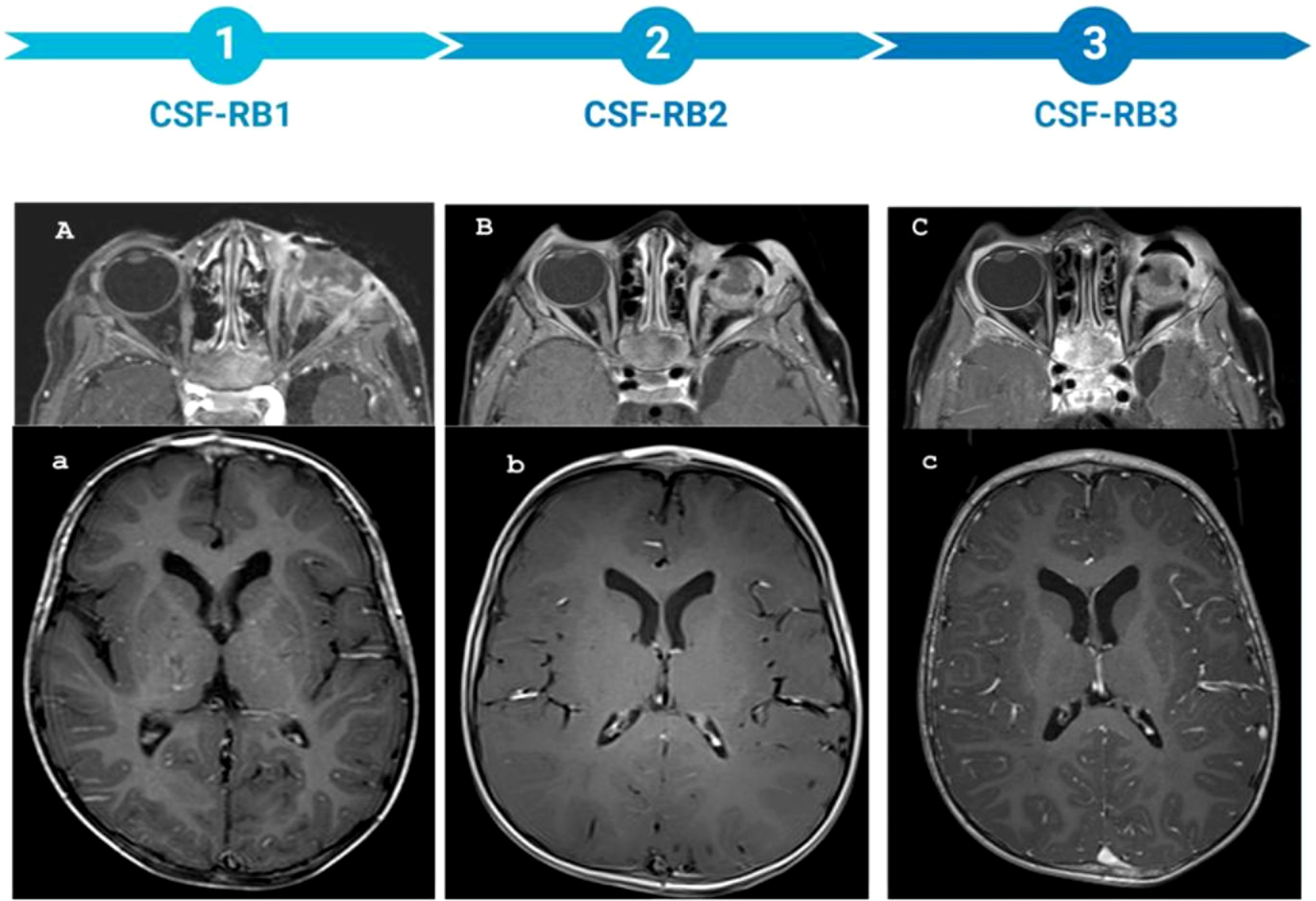
Figure 1. MRI of the patient throughout the entire treatment, along with the corresponding sample time points. (A–C) axial DIXON T1 post-CM: the timeline of optic nerve involvement. (A) presence of pathological tissue localized in the left retro bulbar site. This tissue takes the contrast medium (MDC) which localizes in the left retro bulbar region, is characterized by irregular profiles and extends to partially involve the extrinsic muscles, making their distal portion difficult to identify. The retro bulbar segment of the optic nerve is also partially affected, showing pathological enhancement. Additionally, there is pathological enhancement of the periorbital soft tissues. (B) Extensive involvement of the left optic nerve is evident following enucleation (C) Further progression of optic nerve involvement is observed, with irregular profiles of optic nerve. (a–c) axial T1w pst-CM: timeline of meningeal involvement. (a) no meningeal enhancement, (b) no meningeal enhancement, (c) intense meningeal enhancement.
Proteomic analysis
CSF samples were collected at the Bambino Gesù Children Hospital, Rome Italy. Parental informed consent was obtained for each patient. Three CSF samples were taken from a patient with RB (CSFRB) by lumbar puncture just before the injection of intrathecal Topotecan at the following time points: before starting chemotherapy (CSF-RB1), negative for neoplastic cells; after the 3th cycle of chemotherapy with Carboplatin, Etoposide, Ifosfamide (ICE), (CSF-RB2), positive for neoplastic cells; and just before starting chemotherapy with Etoposide and Carboplatin, 12 days after the previous sampling (CSF-RB3), still positive for neoplastic cells. We used one sample as normal control (CSFCTRL), collected from a 2-year-old boy with a history of post hemorrhagic tetra-ventricular hydrocephalus, during the peritoneal ventricular shunt procedure. All these samples were stored at −80°C until their use. For proteomic analysis, CSF was processed by in-StageTip (iST) method, samples were analyzed in duplicate ensuring technical replication (10). The tryp- column, thermostated at 55°C. The peptides were separated with an organic solvent at a flow rate of 250 nL/min, using a non-linear gradient of 5–45% solution B (80% CAN and 20% H2O, 0.1% FA) in 140 min, and analyzed using an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific Instruments, Bremen, Germany). Orbitrap detection was used for both MS1 and MS2 measurements at re-solving powers of 120 K and 30 K (at m/z 200), respectively. Data dependent MS/MS analysis was performed in top speed mode with a 2 sec cycle-time, during which pre-cursors detected within the range of m/z 375−1500 were selected for activation in or-der of charge state, using CHarge Ordered Parallel Ion aNalysis (CHOPIN) (11). MaxQuant software was used to process the raw data, setting a false discovery rate (FDR) of 0.01 for the identification of proteins, peptides and PSM (peptide-spectrum match), a minimum length of 6 amino acids for peptide identification was required. Andromeda engine, incorporated into MaxQuant software, was used to search MS/MS spectra against Uniprot human database. All bioinformatics analyses were done with the Perseus software of the MaxQuant computational platform (12). Protein groups were filtered to require 70% valid values in total. The label-free intensities were ex-pressed as base log2, and empty values were imputed with random numbers from a normal distribution for each column, to best simulate low abundance values close to noise level. The Venn diagram of identified proteins was calculated using an online tool (13). For each comparison, a B significance test (Benjamini–Hochberg FDR <0.05) was used (14). To visualize the profile of the experiment, a hierarchical clustering of resulting proteins was performed on log2 intensities after z-score normalization of the data for each sample, using Euclidean distances.
Results
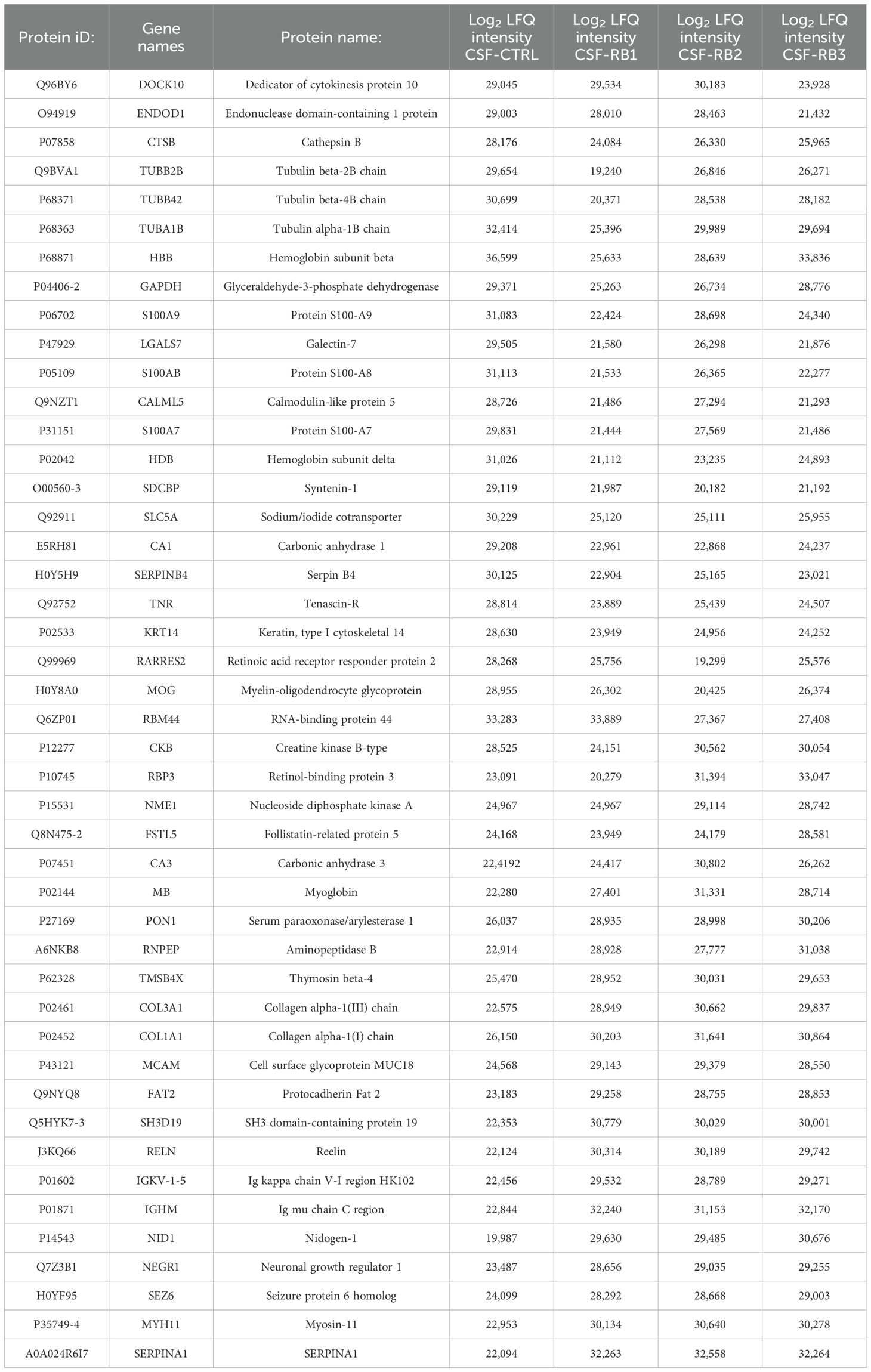
A total of 1029 proteins were identified in all CSF samples, of these 500 proteins were in common in all samples (Figure 2A). The Heatmap, resulting from the comparisons among the samples, showed 46 differentially expressed proteins (Figure 2B, Table 1). In all 3 CSF-RB samples we identified seven downregulated and seventeen upregulated proteins compared with the CSF-CTRL (Figure 2B, Table 1). By gene enrichment analysis performed by FunRich software (15), the downregulated proteins were mainly involved in cell growth, metabolism and cell communication, whereas the upregulated ones in cell growth, signal transduction and metabolism (Figure 2C). Four proteins were commonly and differentially expressed in CSF-RB2 and CSF-RB3 samples (positive for neoplastic cells). Specifically, 3 proteins were up-regulated [Creatine kinase B-type (CKB), Retinolbinding protein 3 (RBP3), Nucleoside diphosphate kinase A (NME1)] and one down-regulated [RNAbinding protein 44 (RBM44)]. Four proteins were down-regulated in CSF-RB1: Tubulin beta proteins (TUBB4B, TUBB2B, TUBA1B) and Cathepsin B (CTSB). Interestingly, FSTL5 was upregulated only in CSF-RB3, whereas 2 proteins (DOCK10 and ENDOD1) were down regulated in the same sample.

Figure 2. (A) Venn diagram showing a total of 1029 proteins detected in all CSF samples (CSF-CTRL, CSF-RB1, CSF-RB2, CSf-RB3), 500 proteins were in common in all samples. 733, 713, 809 and 789 proteins were found in CSF-CTRL, CSF-RB1, CSF-RB2 and CSF-RB3 respectively; 131, 18, 27 and 31 were exclusively present in CSF-CTRL, CSF-RB1, CSF-RB2 and CSF-RB3 respectively (B) Unsupervised hierarchical-clustered heatmap of proteins identified by significance B test. Heatmap showing the distinctive proteomic signature of the CSF-CTRL vs CSF-RB1, CSF-RB2, CSF-RB3. A red-blue scale corresponds to high and low protein amount, respectively. C-D) Graphical representation of biological processes gene enrichment obtained by FunRich software for low expressed (C) and high expressed (D) protein in CSF-RB (CSF-RB1; CSF-RB2; CSF- RB3) vs CSF-CTRL.
Discussion
Although RB primarily affects the eyes, advanced disease mostly associated with delayed diagnosis, can lead to CNS involvement, including the brain and CSF, associated with extremely poor prognosis. Therefore, early detection of CNS involvement is crucial for allowing early therapeutic intervention. In RB, different biofluids, such as plasma, aqueous humor or CSF have been studied to detect tumor related biomarkers as prognostic predictors of disease evolution or treatment response (16). CSF samples from patients with non-metastatic RB and high-risk pathologic features showed that, among four cases with CSF relapse, two patients had GD2 synthase positivity in CSF as marker of minimal dissemination disease (MD) (17). In view of these findings, Laurent et al. suggested early and intensified intrathecal treatment for children with MD in CSF in order to improve the outcomes of these patients. Dimaras et al. also demonstrated that MD could be detected in patients with metastatic RB by using RB1 mutational analysis in CSF (18). Proteomic analysis involves the comprehensive study of proteins present in a biological sample. Since CSF is considered a primary route for metastatic dissemination (19), its proteomic analysis has been useful in improving the understanding of CNS tumors (20). Furthermore, proteins that are overexpressed in the CSF may serve as targets for developing new tailored treatment. For the afore mentioned reasons in this study, we have performed a proteomic evaluation of serial CSF samples in a patient affected by stage III RB with the aim to identify molecular changes occurring during treatment and CNS relapse. The differential analysis revealed a specific proteomics signature in CSF-RB samples. Among the upregulated proteins in CSF-RB, several have structural and physiological functions in the eye (COL1A1, COL3A1, TMSB4X, NID1) (21). NID1 has been described to promote tumor cell migration, invasion, epithelial-to-mesenchymal transition (EMT) and chemo-resistance in different tumors (22–24) similarly to MCAM (25). Other upregulated proteins are involved in neuronal/retinal development and cell migration (Fat2, RELN, NEGR1, SEZ6) (26–29). RELN is an extracellular matrix protein expressed during normal retinogenesis and seems to play an important role in regulating stem cell trafficking in neuronal and non-neuronal tissues (30). SEZ6 has been found be elevated in the CSF from patients with multiple sclerosis and inflammatory conditions (31), furthermore, has been shown to promote metastatic spread by affecting cellular adhesion, cytoskeletal and extracellular matrix remodeling and inducing EMT in cancer cells (32). Of note, RNPEP and PON1 progressively increased during treatment. RNPEP is a secreted enzyme belonging to M1 metallopeptidase family (33) and its circulating mRNA levels have been demonstrated to be an independent prognostic factor in different tumors (34). PON1 is an enzyme associated with high-density lipoprotein (HDL) and plays a role in detoxification processes (35); this protein has potential implications in ocular disorders associated with oxidative stress and inflammation (36–38). Similarly, SERPINA1, is an acute inflammatory protein involved in corneal defense and neuroinflammation (39). Among the down-regulated proteins in CSF-RB, we identified proteins involved in glucose transport (SLC5A) (40), synthesis of glycogen and lipids (CA1) (41), and exosomes biogenesis (SDCBP) (42); those proteins have been described to contribute to aggressive behavior of tumor via metabolic reprogramming and cell signaling. Other downregulated proteins were: protease inhibitor (SERPINB4), which may regulate the immune response against tumor cells (43), protein involved in brain development and synaptic plasticity (TNR) (44) and keratin (KRT14), which provides structural support and integrity to epithelial cells and is abnormal expressed in several tumors (45). The CSF samples that were positive for neoplastic cells, CSF-RB2 and -RB3, presented two commonly upregulated proteins involved in visual pathways: RBP3, which is involved in retinol transport (46), and CK-B, an enzyme that provides energy to photoreceptors for the visual cycle particularly expressed in retinal diseases (47, 48). Another protein, NME1, is involved in oxidative stress response (49) and its increased expression correlates with tumor aggressiveness in specific cancer types (melanoma, liver and breast) (50). CSF-RB3 showed two down-regulated proteins (DOCK10 and ENDOD1) and the upregulation of FSTL5. FSTL5 is a secretory glycoprotein expressed in restricted areas of CNS and few studies indicate its role on neo-plastic transformation and metastasis development (51, 52). DOCK 10 is also expressed in the brain and seems to have a role in neuroinflammation and cancer development by regulating cell adhesion and migration (53). ENDOD1 is a member of nucleases, which participate in DNA repair with abnormal expression in different tumors (54). Lastly, in the CSF-RB1 sample, which was negative for neoplastic cells, we found down-regulated three tubulin beta proteins (TUBB4B, TUBB2B, TUBA1B). Tubulins are microtubules subunits involved in cell shape maintenance (55). In particular, TUBB4B expression is correlated whit EMT initiation and affection of cell polarity (56) and TUBB2B is involved in visual development and function (57); mutations of TUBB4B and TUBB2B have been linked to several ocular disorders (57, 58).
Conclusion
Overall, the proteomic analysis of CSF showed in our patient deregulation of proteins mainly involved in inflammation, extracellular matrix remodeling, EMT initiation, migration, invasion and cell metabolism. The CSF samples positive for neoplastic cells revealed proteins with a prominent role in visual pathways, including transport of retinol, photoreceptor’s function and oxidative stress response. The timepoint negative for tumor infiltration revealed several TUBB proteins that are related with cellular shape maintenance, cell polarity and EMT initiation. These results demonstrate how proteomic analysis of CSF can help to improve our understanding in RB. Further studies including large cohorts of patients with RB should be conducted to identify new prognostic biomarkers and potential therapeutic targets in this tumor.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://www.proteomexchange.org/, PXD056783.
Ethics statement
The study was conducted to the guidelines of the Declaration of Helsinki and approved by the Ethics committee of Ospedale Pediatrico Bambino Gesù IRCCS (protocol code 1189_OPBG_2016; date of approval: 12/08/2016. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author contributions
AG: Conceptualization, Data curation, Formal Analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. VD: Data curation, Investigation, Resources, Writing – review & editing. CL: Conceptualization, Data curation, Formal Analysis, Methodology, Writing – review & editing. IR: Investigation, Resources, Writing – review & editing. AR: Investigation, Resources, Writing – review & editing. EM: Visualization, Writing – review & editing. RD: Investigation, Resources, Writing – review & editing. DL: Data curation, Investigation, Writing – review & editing. AP: Conceptualization, Data curation, Formal Analysis, Supervision, Writing – review & editing. FL: Supervision, Writing – review & editing. AD: Conceptualization, Data curation, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Italian Ministry of Health with Current Research funds.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Abramson DH, Schefler AC. Update on retinoblastoma. Retina;. (2004) 24:828–48. doi: 10.1097/00006982-200412000-00002
2. Canturk S, Qaddoumi I, Khetan V, Ma Z, Furmanchuk A, Antoneli CB, et al. Survival of retinoblastoma in less-developed countries impact of socioeconomic and health-related indicators. Br J Ophthalmol. (2010) 94:1432–6. doi: 10.1136/bjo.2009.168062
3. Chantada GL, Qaddoumi I, Canturk S, Khetan V, Ma Z, Kimani K, et al. Strategies to manage retinoblastoma in developing countries. Pediatr Blood Cancer. (2011) 56:341–8. doi: 10.1002/pbc.22843
4. Dunkel IJ, Khakoo Y, Kernan NA, Gershon T, Gilheeney S, Lyden DC, et al. Intensive multimodality therapy for patients with stage 4a metastatic retinoblastoma. Pediatr Blood Cancer. (2010) 55:55–9. doi: 10.1002/pbc.22504
5. Parnetti L, Castrioto A, Chiasserini D, Persichetti E, Tambasco N, El-Agnaf O, et al. Cerebrospinal fluid biomarkers in Parkinson disease. Nat Rev Neurol. (2013) 9:131–40. doi: 10.1038/nrneurol.2013.10
6. Rajagopal MU, Hathout Y, MacDonald TJ, Kieran MW, Gururangan S, Blaney SM, et al. Proteomic profiling of cerebrospinal fluid identifies prostaglandin D2 synthase as a putative biomarker for pediatric medulloblastoma: A pediatric brain tumor consortium study. Proteomics. (2011) 11:935–43. doi: 10.1002/pmic.201000198
7. Saratsis AM, Yadavilli S, Magge S, Rood BR, Perez J, Hill DA, et al. Insights into pediatric diffuse intrinsic pontine glioma through proteomic analysis of cerebrospinal fluid. Neuro Oncol. (2012) 14:547–60. doi: 10.1093/neuonc/nos067
8. Khwaja FW, Reed MS, Olson JJ, Schmotzer BJ, Gillespie GY, Guha A, et al. Proteomic identification of biomarkers in the cerebrospinal fluid (CSF) of astrocytoma patients. J Proteome Res. (2007) 6:559–70. doi: 10.1021/pr060240z
9. Whitin JC, Jang T, Merchant M, Yu TT, Lau K, Recht B, et al. Alterations in cerebrospinal fluid proteins in a presymptomatic primary glioma model. PloS One. (2012) 7:e49724. doi: 10.1371/journal.pone.0049724
10. Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. (2014) 11:319–24. doi: 10.1038/nmeth.2834
11. Davis S, Charles PD, He L, Mowlds P, Kessler BM, Fischer R. Expanding proteome coverage with CHarge ordered parallel ion aNalysis (CHOPIN) combined with broad specificity proteolysis. J Proteome Res. (2017) 16:1288–99. doi: 10.1021/acs.jproteome.6b00915
12. Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. (2016) 13:731–40. doi: 10.1038/nmeth.3901
13. Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinf. (2014) 15:293. doi: 10.1186/1471-2105-15-293
14. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. (2008) 26:1367–72. doi: 10.1038/nbt.1511
15. Mathivanan S, Ji H, Simpson RJ. Exosomes: Extracellular organelles important in intercellular communication. J Proteom. (2010) 73:1907–20. doi: 10.1016/j.jprot.2010.06.006
16. Ghose N, Kaliki S. Liquid biopsy in Retinoblastoma: A review. Semin Ophthalmol. (2022) 37:813–9. doi: 10.1080/08820538.2022.2078165
17. Laurent VE, Torbidoni AV, Sampor C, Ottaviani D, Vazquez V, Gabri MR, et al. Minimal disseminated disease in nonmetastatic retinoblastoma with high-risk pathologic features and association with disease-free survival. JAMA Ophthalmol. (2016), 1374–9. doi: 10.1001/jamaophthalmol.2016.4158
18. Dimaras H, Rushlow D, Halliday W, Doyle JJ, Babyn P, Abella EM, et al. Using RB1 mutations to assess minimal residual disease in metastatic retinoblastoma. Transl Res. (2010) 156:91–7. doi: 10.1016/j.trsl.2010.05.009
19. Spreafico F, Bongarzone I, Pizzamiglio S, Magni R, Taverna E, De Bortoli M, et al. Proteomic analysis of cerebrospinal fluid from children with central nervous system tumors identifies candidate proteins relating to tumor metastatic spread. Oncotarget. (2017) 8:46177–90. doi: 10.18632/oncotarget.17579
20. Blennow K, Hampel H, Weiner M, Zetterberg H. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat Rev Neurol. (2010) 6:131–44. doi: 10.1038/nrneurol.2010.4
21. Yuan M, He Q, Long Z, Zhu X, Xiang W, Wu Y, et al. Exploring the pharmacological mechanism of liuwei dihuang decoction for diabetic retinopathy: A systematic biological strategy-based research. Evid Based Complement Alternat Med. (2022). doi: 10.1155/2022/9790813
22. Gaggero S, Bruschi M, Petretto A, Parodi M, Del Zotto G, Lavarello C, et al. Nidogen-1 is a novel extracellular ligand for the NKp44 activating receptor. Oncoimmunology. (2018) 7:e1470730. doi: 10.1080/2162402X.2018.1470730
23. Zhou Y, Zhu Y, Fan X, Zhang C, Wang Y, Zhang L, et al. NID1, a new regulator of EMT required for metastasis and chemoresistance of ovarian cancer cells. Oncotarget. (2017) 8:33110–21. doi: 10.18632/oncotarget.16145
24. Pedrola N, Devis L, Llauradó M, Campoy I, Martinez-Garcia E, Garcia M, et al. Nidogen 1 and Nuclear Protein 1: novel targets of ETV5 transcription factor involved in endometrial cancer invasion. Clin Exp Metastasis. (2015) 32:467–78. doi: 10.1007/s10585-015-9720-7
25. Chen Y, Sumardika IW, Tomonobu N, Winarsa Ruma IM, Kinoshita R, Kondo E, et al. Melanoma cell adhesion molecule is the driving force behind the dissemination of melanoma upon S100A8/A9 binding in the original skin lesion. Cancer Lett. (2019) 28:452:178–190. doi: 10.1016/j.canlet.2019.03.023
26. Wu Q, Maniatis T. Large exons encoding multiple ectodomains are a characteristic feature of protocadherin genes. Proc Natl Acad Sci U S A. (2000) 97:3124–9. doi: 10.1073/pnas.97.7.3124
27. Schulze M, Violonchi C, Swoboda S, Welz T, Kerkhoff E, Hoja S, et al. RELN signaling modulates glioblastoma growth and substrate-dependent migration. Brain Pathol. (2018) 28:695–709. doi: 10.1111/bpa.12584
28. Kim H, Hwang JS, Lee B, Hong J, Lee S. Newly identified cancer-associated role of human neuronal growth regulator 1 (NEGR1). J Cancer. (2014) 5:598–608. doi: 10.7150/jca.8052
29. Yu ZL, Jiang JM, Wu DH, Xie HJ, Jiang JJ, Zhou L, et al. Febrile seizures are associated with mutation of seizure-related (SEZ) 6, a brain-specific gene. J Neurosci Res. (2007) 85:166–72. doi: 10.1002/jnr.21103
30. Pulido JS, Sugaya I, Comstock J, Sugaya K. Reelin expression is upregulated following ocular tissue injury. Graefes Arch Clin Exp Ophthalmol. (2007) 245:889–93. doi: 10.1007/s00417-006-0458-4
31. Roitman M, Edgington-Mitchell LE, Mangum J, Ziogas J, Adamides AA, Myles P, et al. Sez6 levels are elevated in cerebrospinal fluid of patients with inflammatory pain-associated conditions. Pain Rep. (2019) 4:e719. doi: 10.1097/PR9.0000000000000719
32. Wang L, Ling X, Zhu C, Zhang Z, Wang Z, Huang S, et al. Upregulated seizure-related 6 homolog-like 2 is a prognostic predictor of hepatocellular carcinoma. Dis Markers. (2020) 13:2020:7318703. doi: 10.1155/2020/7318703
33. Pham VL, Cadel MS, Gouzy-Darmon C, Hanquez C, Beinfeld MC, Nicolas P, et al. Aminopeptidase B, a glucagon-processing enzyme: site directed mutagenesis of the Zn2+-binding motif and molecular modelling. BMC Biochem. (2007) 31:21. doi: 10.1186/1471-2091-8-21
34. Miettinen JJ, Kumari R, Traustadottir GA, Huppunen ME, Sergeev P, Majumder MM, et al. Aminopeptidase expression in multiple myeloma associates with disease progression and sensitivity to melflufen. Cancers (Basel). (2021) 13:1527. doi: 10.3390/cancers13071527
35. Kar S, Patel MA, Tripathy RK, Bajaj P, Pande AH. Oxidized-phospholipids in reconstituted high density lipoprotein particles affect structure and function of recombinant paraoxonase 1. Biochim Biophys Acta. (2013) 1831:1714–20. doi: 10.1016/j.bbalip.2013.08.008
36. Oczos J, Grimm C, Barthelmes D, Sutter F, Menghini M, Kloeckener-Gruissem B, et al. Regulatory regions of the paraoxonase 1 (PON1) gene are associated with neovascular age-related macular degeneration (AMD). Age (Dordr). (2013) 35:1651–62. doi: 10.1007/s11357-012-9467-x
37. Yilmaz N, Coban DT, Bayindir A, Erol MK, Ellidag HY, Giray O, et al. Higher serum lipids and oxidative stress in patients with normal tension glaucoma, but not pseudoexfoliative glaucoma. Bosn J Basic Med Sci. (2016) 16:21–7. doi: 10.17305/bjbms.2016.830
38. Chen Y, Meng J, Li H, Wei H, Bi F, Liu S, et al. Resveratrol exhibits an effect on attenuating retina inflammatory condition and damage of diabetic retinopathy via PON1. Exp Eye Res. (2019) 181:356–66. doi: 10.1016/j.exer.2018.11.023
39. Funding M, Vorum H, Nexø E, Ehlers N. Alpha-1–antitrypsin in aqueous humor from patients with corneal allograft rejection. Acta Ophthalmol Scand. (2005) 83:379–84. doi: 10.1111/j.1600-0420.2005.00457.x
40. Petersen AM, Small CM, Yan YL, Wilson C, Batzel P, Bremiller RA, et al. Evolution and developmental expression of the sodium-iodide symporter (NIS, slc5a5) gene family: Implications for perchlorate toxicology. Evol Appl. (2022) 15:1079–98. doi: 10.1111/eva.13424
41. Swenson ER. Carbonic anhydrase inhibitors and hypoxic pulmonary vasoconstriction. Respir Physiol Neurobiol. (2006) 151:209–16. doi: 10.1016/j.resp.2005.10.011
42. Qian XL, Li YQ, Yu B, Gu F, Liu FF, Li WD, et al. Syndecan binding protein (SDCBP) is overexpressed in estrogen receptor negative breast cancers, and is a potential promoter for tumor proliferation. PloS One. (2013) 8:e60046. doi: 10.1371/journal.pone.0060046
43. Riaz N, Havel JJ, Kendall SM, Makarov V, Walsh LA, Desrichard A, et al. Recurrent SERPINB3 and SERPINB4 mutations in patients who respond to anti-CTLA4 immunotherapy. Nat Genet. (2016) 48:1327–9. doi: 10.1038/ng.3677
44. Anlar B, Gunel-Ozcan A. Tenascin-R: role in the central nervous system. Int J Biochem Cell Biol. (2012) 44:1385–9. doi: 10.1016/j.biocel.2012.05.009
45. Hanley CJ, Henriet E, Sirka OK, Thomas GJ, Ewald AJ. Tumor-resident stromal cells promote breast cancer invasion through regulation of the basal phenotype. Mol Cancer Res. (2020) 18:1615–22. doi: 10.1158/1541-7786.MCR-20-0334
46. Ramesh S, Bonshek RE, Bishop PN. Immunolocalization of opticin in the human eye. Br J Ophthalmol. (2004) 88:697–702. doi: 10.1136/bjo.2003.031989
47. Takagi Y, Yasuhara T, Gomi K. Creatine kinase and its isozymes. Rinsho Byori. (2001) Suppl 116:52–61.
48. Zhu L, Shen W, Zhu M, Coorey NJ, Nguyen AP, Barthelmes D, et al. Anti-retinal antibodies in patients with macular telangiectasia type 2. Invest Ophthalmol Vis Sci. (2013) 54:5675–83. doi: 10.1167/iovs.13-12050
49. Boissan M, Dabernat S, Peuchant E, Schlattner U, Lascu I, Lacombe ML. The mammalian Nm23/NDPK family: from metastasis control to cilia movement. Mol Cell Biochem. (2009) 329:51–62. doi: 10.1007/s11010-009-0120-7
50. Puts GS, Leonard MK, Pamidimukkala NV, Snyder DE, Kaetzel DM. Nuclear functions of NME proteins. Lab Invest. (2018) 98:211–8. doi: 10.1038/labinvest.2017.109
51. Remke M, Hielscher T, Korshunov A, Northcott PA, Bender S, Kool M, et al. FSTL5 is a marker of poor prognosis in non-WNT/non-SHH medulloblastoma. J Clin Oncol. (2011) 29:3852–61. doi: 10.1200/JCO.2011.36.2798
52. Schmit SL, Schumacher FR, Edlund CK, Conti DV, Raskin L, Lejbkowicz F, et al. A novel colorectal cancer risk locus at 4q32.2 identified from an international genome-wide association study. Carcinogenesis. (2014) 35:2512–9. doi: 10.1093/carcin/bgu148
53. Gadea G, Blangy A. Dock-family exchange factors in cell migration and disease. Eur J Cell Biol. (2014) 93:466–77. doi: 10.1016/j.ejcb.2014.06.003
54. Qiu J, Peng S, Si-Tu J, Hu C, Huang W, Mao Y, et al. Identification of endonuclease domain-containing 1 as a novel tumor suppressor in prostate cancer. BMC Cancer. (2017) 17:360. doi: 10.1186/s12885-017-3330-5
55. Sallee MD, Feldman JL. Microtubule organization across cell types and states. Curr Biol. (2021) 31:R506–11. doi: 10.1016/j.cub.2021.01.042
56. Sobierajska K, Ciszewski WM, Wawro ME, Wieczorek-Szukała K, Boncela J, Papiewska-Pajak I, et al. TUBB4B downregulation is critical for increasing migration of metastatic colon cancer cells. Cells. (2019) 8:810. doi: 10.3390/cells8080810
57. Breuss M, Morandell J, Nimpf S, Gstrein T, Lauwers M, Hochstoeger T, et al. The expression of tubb2b undergoes a developmental transition in murine cortical neurons. J Comp Neurol. (2015) 523:2161–86. doi: 10.1002/cne.23836
Keywords: cerebrospinal fluid (CSF), retinoblastoma, proteomic analysis, liquid biopsy, case report
Citation: Galardi A, Di Paolo V, Lavarello C, Russo I, Romanzo A, Miele E, Vito RD, Longo D, Petretto A, Locatelli F and Di Giannatale A (2025) Case Report: Proteomic analysis of cerebrospinal fluid in a retinoblastoma patient. Front. Oncol. 15:1511594. doi: 10.3389/fonc.2025.1511594
Received: 15 October 2024; Accepted: 01 April 2025;
Published: 24 April 2025.
Edited by:
Andrea Di Cataldo, University of Catania, ItalyReviewed by:
Paula Schaiquevich, Garrahan Hospital, ArgentinaDemirkan Gursel, Northwestern University, United States
Peiwei Chai, Shanghai Jiao Tong University, China
Copyright © 2025 Galardi, Di Paolo, Lavarello, Russo, Romanzo, Miele, Vito, Longo, Petretto, Locatelli and Di Giannatale. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angela Galardi, YW5nZWxhLmdhbGFyZGlAaXN0aXR1dG90dW1vcmkubWkuaXQ=
 Angela Galardi
Angela Galardi Virginia Di Paolo
Virginia Di Paolo Chiara Lavarello2
Chiara Lavarello2 Ida Russo
Ida Russo Evelina Miele
Evelina Miele Rita De Vito
Rita De Vito Angela Di Giannatale
Angela Di Giannatale