- 1Department of Urology, Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Department of Burn Surgery, The First Affiliated Hospital of Naval Medical University, Shanghai, China
- 3Department of Nursing, Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, China
Background: The vital role metabolism plays in RCC, a global disease with huge disease burden, has been widely acknowledged. However, bibliometric analysis remains underexplored in the context of metabolism in RCC.
Methods: The Web of Science database was adopted to obtain relevant publications for further bibliometric analysis of countries, institutions, authors, journals, publications, references and keywords. Literature reading and keyword co-occurrence analysis were employed to figure out major points and hotspots in this field. The analysis was conducted by biblioshiny based on Bibliometrix package in R version 4.3.2.
Results: From 15 May 2015 to 15 May 2025, 3010 relevant publications were retrieved. China was the most productive country and USA was the country with the highest total citations. The most productive institution was “Harvard University”. WANG Y published 46 publications with an H-index of 16. Core journals were identified with Bradford’s law. Additionally, three major points and hotspots were identified and discussed through bibliometric analysis and targeted literature reading.
Conclusion: Our research provided a reference for future basic and clinical research through summarizing past research findings, analyzing current research hotspots, and prospecting the future development of research. “Metabolic alterations in RCC”, “Metabolic syndrome and RCC”, “Microbiome and RCC” were major points and hot spots in this field. In the future, a broader metabolic map could be made and more researches concerning metabolism during RCC treatment and drug resistance might bring more clinical significance.
1 Introduction
In 2022, kidney cancer ranked the 14th globally, with about 434,419 new cases, causing 155,702 deaths (1). Renal cell carcinoma (RCC), accounting for over 90% of the malignancies in the kidney (2), refers to malignant neoplasm derived from the epithelium of renal tubule (3). The vital role that metabolism plays in RCC has been widely acknowledged (4, 5), and researches concerning metabolism in RCC has progressed quite fast in the past few years due to the advancement of understanding towards metabolism and related technologies.
People’s understanding towards “metabolism” has been continuously evolving and progressing. Around the 1960s, metabolism was generally viewed as a set of biochemical pathways. Krebs and colleagues identified the tricarboxylic acid cycle (6), laying the foundation for succeeding metabolic researches. During this period, most researches concerning metabolism in RCC focused on enzyme activity (7) and the levels of metabolites themselves (8). From the 1970s to the 1990s, breakthroughs in molecular biology and genetics, such as gas chromatography (9) and gene sequencing technology by Sanger (10) enabled scientists to connect metabolic pathways with gene regulation. This revealed how metabolism is precisely controlled at the genetic level. For example, researchers identified that abnormal CpG islands methylation in RCC might be involved in aberrant inactivation of VHL gene (11) and thus initiating metabolic alterations including upregulated glycolysis. Since 2000s, systems biology and high-throughput technologies, such as the next-generation sequencing (12), RNA sequencing (13) and single-cell RNA sequencing (14), have revolutionized metabolism research. Researchers began to view metabolism as an interconnected network intricately linked to cell fate determination, epigenetic regulation, and disease pathogenesis. The Warburg effect was reinterpreted in this context, revealing that cancer cells could gain survival advantages through metabolic reprogramming, such as the overexpression of the LDHA gene (15). Today, metabolism is widely recognized as a central regulator of cell fate, immunity, aging, and environmental sensing.
The advancement of metabolomics-related technologies and the evolving understanding of metabolism have evolved into the concept of metabolomics. In 1998, the term “metabolomics” was first put forward. It means the comprehensive study of all the small molecules in a biological sample, representing biochemistry on a large scale (16). The focus of metabolomic studies, as the advancement of technologies and people’s understanding, is transitioning from merely profiling chemicals or molecules to uncovering the biological narratives.
Metabolism in RCC is a fast-paced field, especially in recent years with the development of advanced concepts and technologies. Hence, it is of necessity to timely describe and summarize the knowledge structure and trend topics of this field in recent years. Bibliometric could provide us with a comprehensive landscape of a specific field in a statistical and quantitative manner, discovering hot spots and trend topics in a field. This effective analytical method has been applied in various filed, from breast cancer (17), prostate cancer (18) to lung cancer (19), etc. However, bibliometric analysis remains a void in the field of metabolism in RCC. Here, we tried to summarize past research findings, analyze current research hotspots, and prospect the future development of this field from 2015 to 2025. As a bibliometric study, we hope our work can serve as a foundational reference for future basic and clinical research in this field.
2 Materials and methods
2.1 Data sources and retrieval methods
This bibliometric study was mainly based on the Web of Science (WoS) database. Search term as follows was applied to collect data comprehensively from 15 May 2015 to 15 May 2025: ((TS = ((renal OR kidney) NEAR/2 (cancer* OR tumor* OR tumor* OR neoplasm* OR carcinoma* OR oncology))) AND ((TS = metabolism) OR (TS = metabolic) OR (TS = metabolome) OR (TS = metabolite) OR (TS = metabonomics) OR (TS = metabolomics))). A total of 3173 publications in Web of Science (Core Collection) were obtained and 3010 of them, comprising articles and reviews, were ultimately included in our study.
2.2 Bibliometric strategies
All data were integrated in one TXT file, and were uploaded to Biblioshiny, a web application based on Bibliometrix package (20) in R version 4.3.2 for further analysis. After systematically analyzing the data from the perspective of authors, countries, institutions, journals, publications, keywords, etc., a relatively comprehensive understanding of this field could be obtained. Here, we adopted number of publications and citations to provide a brief overview of this field. And H-index (21), which means H of one’s publications were cited at least H times, were used as a criterion for assessing both the quantity and quality of an author or a journal. The M-index of an author equals the H-index/the years since first publication (22). Publications of an author is ranked in decreased order of citations, and G-index is the largest sequence number (denoted as G) of a publication that has at least G2 citations (23). Bradford’s law (24) divides journals into several zones in a ratio of 1: k: k2: … (k > 1), with each of the zones accounting for equal publications in a certain field. The first zone with the least journals is perceived as core collections. Lotka’s law (25) states that the number of authors with one publication is n2 times as many as those with n publications. In co-citation network, the weight of lines between two articles indicates the frequency of which they are cited simultaneously. Similarly, keyword co-occurrence measures the correlation between keywords occurred in the identical publication. The topics could be classified into four groups by the thematic map with the “Density” axis representing the development degree and the “Centrality” axis signifying the relevance degree of these themes or fields: “Niche Themes” might be some small, specialized but well-developed territories with high “Density” but low “Centrality”. “Motor Themes” might be some core areas that received continuous attention in a field with both high “Density” and “Centrality”. “Basic Themes” might not be the hot spots at present, but served as corner stones in a field with high “Centrality” but low “Density”. “Emerging or Declining Themes” might refer to new ideas that existed for a relatively short period of time.
The parameters used during the analytical process were as follows:
The collaboration network between institutions adopted Walktrap algorithm with 30 nodes. The collaboration network between authors adopted Walktrap algorithm with 30 nodes. The co-citation network between publications adopted Walktrap algorithm with 50 nodes. The keywords co-occurrence network adopted Louvein algorithm with 50 nodes. The thematic evolution plot adopted Walktrap algorithm with 3 cutting points, namely 2019, 2021 and 2023. The remaining settings utilized custom settings, or could be intuitively seen from the plot.
3 Result
3.1 Annual publications
The complete retrieval and analytic process was shown in Figure 1. From 15 May 2015 to 15 May 2025, 3010 publications (reviews or articles) in Web of Science (core collection) were written by 16884 authors with an annual growth rate of 8.86%, published on 1002 journals. Annual scientific production could be utilized to partially measure the popularity of a specific field. As depicted in Supplementary Figure S1, the number of publications and citations manifested steady growth from 2015 to 2024, indicating that the field of “metabolism in RCC” is gaining more and more attention. Although the number of publications and citations seemed to drop in 2025, this might be attributed to the statistics ended at 15 May 2025, which only accounted for less than a half of the whole year.
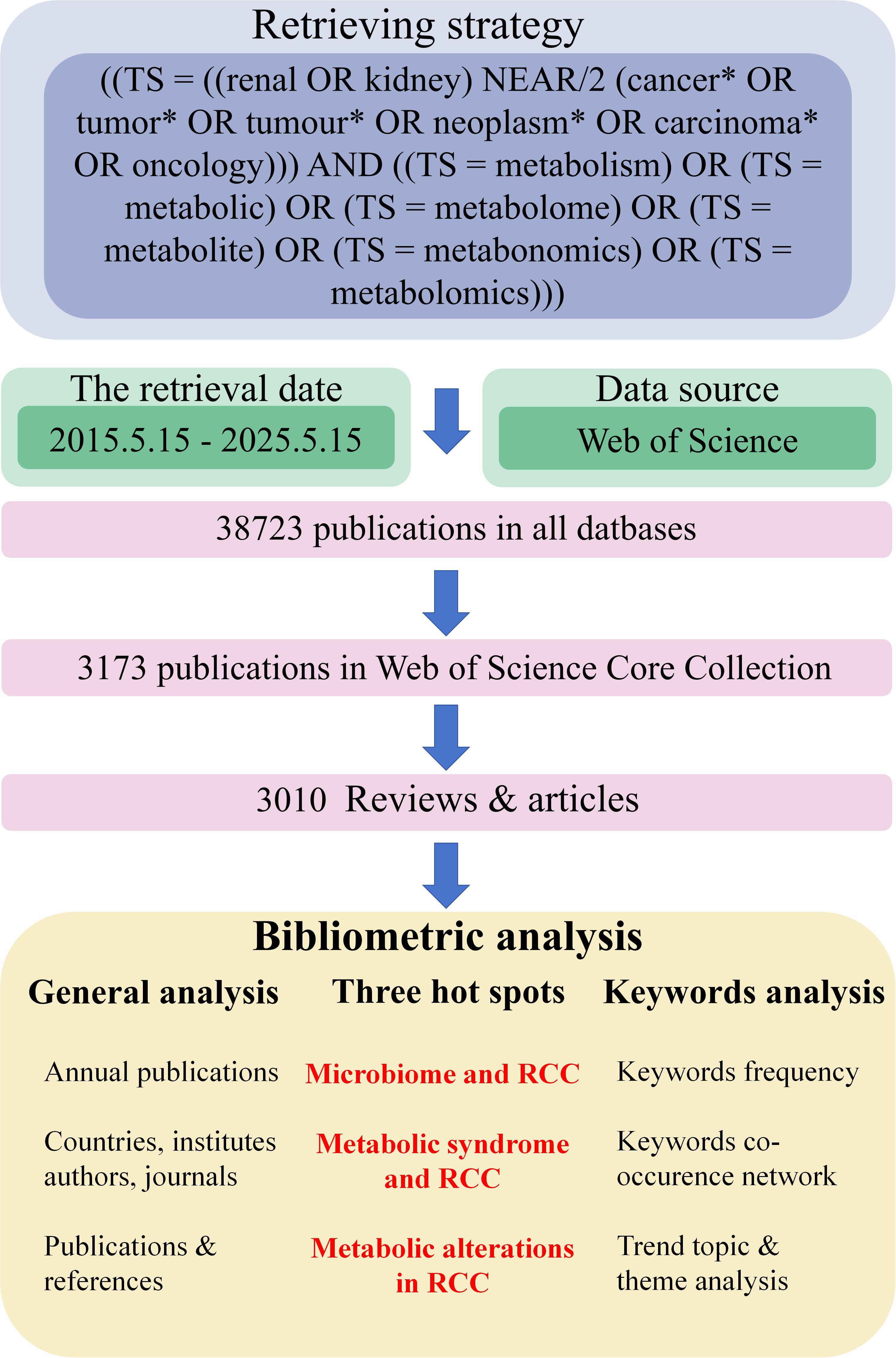
Figure 1. A simple flowchart of our study. Data source, researching process and bibliometrics analysis were demonstrated in this figure. RCC, renal cell carcinoma.
3.2 Countries and institutions
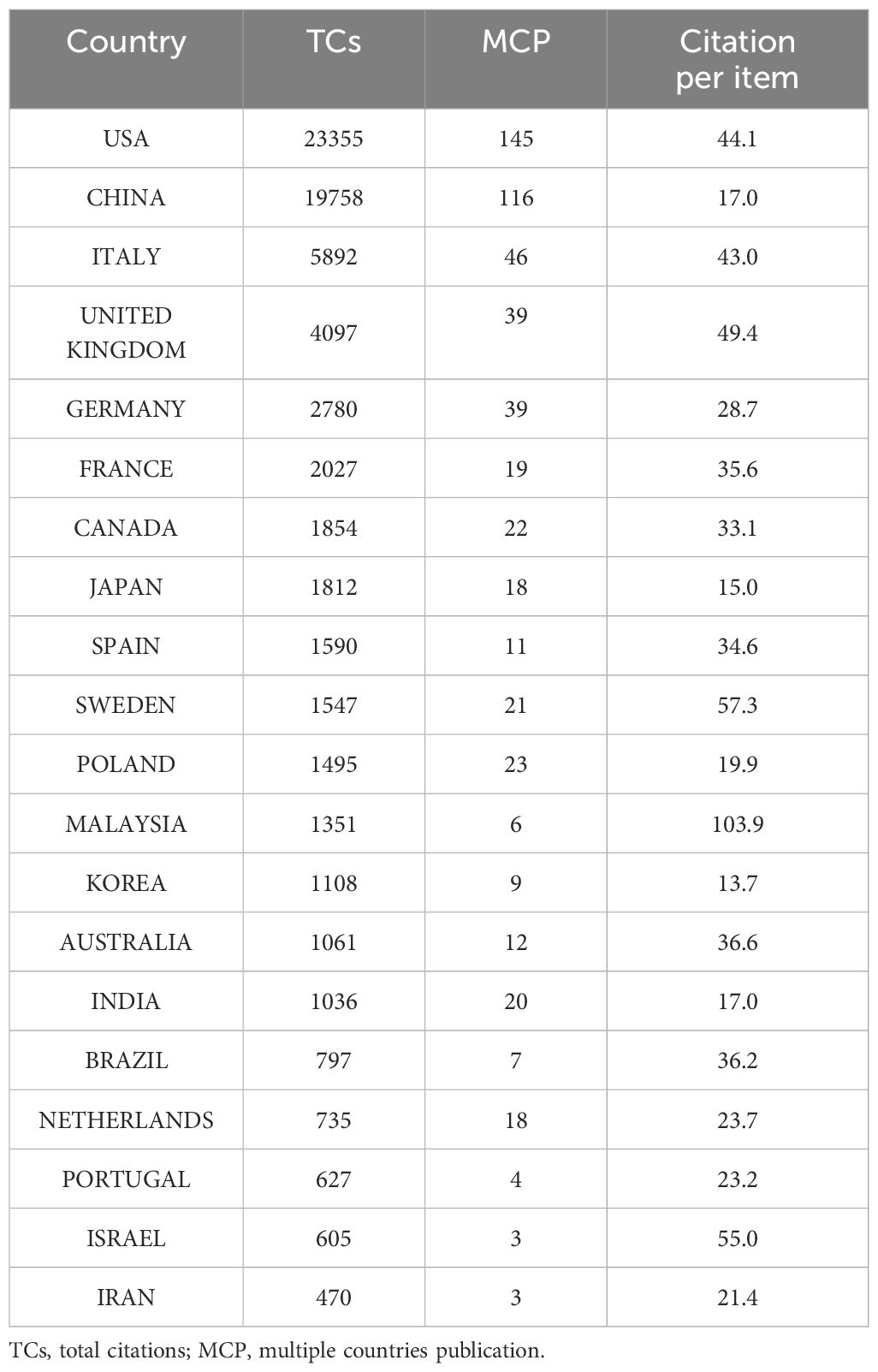
A total of 66 countries contributed to this field. USA, China, ITALY were the top 3 among these countries according to total citations. The USA dominated the list with a total citation per item of 44.1, while China ranked second with a total citation per item of 17.0 (Table 1). Countries’ number of publications could be intuitively observed in Figure 2A, where the darkness of the country indicated the quantity of publications. China was the most productive country and USA, China and ITALY were also the top 3 countries with highest MCP (multiple countries publication), indicating that they were the countries with the most intention to cooperate (Figure 2B).
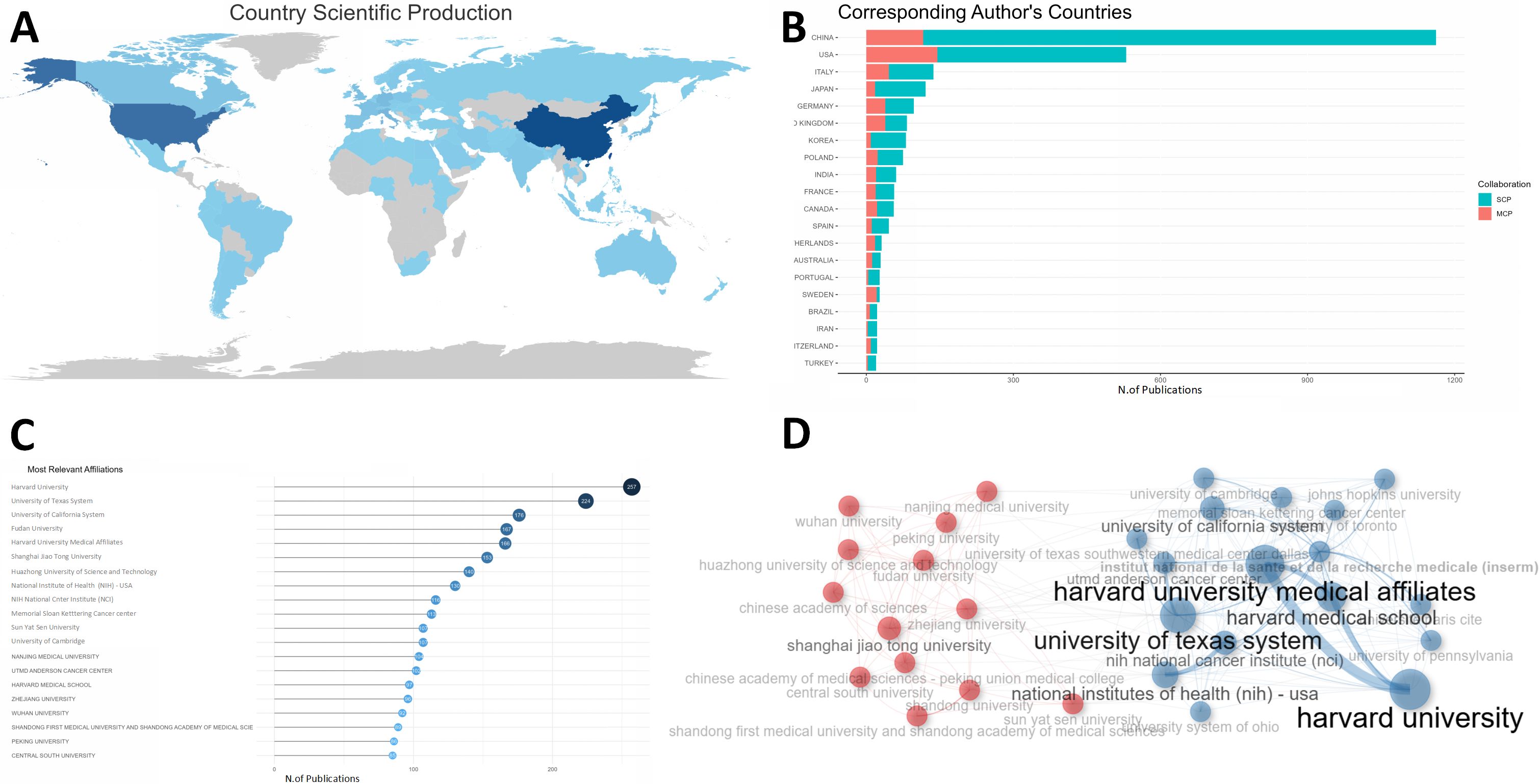
Figure 2. Country and institute analysis. (A) This world map shows the collaborations between different countries. The darkness of the country indicated the quantity of publications and the number of lines manifested cooperation between countries. (B) This plot shows the top 20 most productive countries measured by number of publications. SCP indicates researches conducted independently by scientists from the same country. MCP refers to studies completed collaboratively by scientists with different nationalities, indicating the degree of cooperation between countries. The USA, China, and ITALY were the top 3 countries with the highest MCP. (C) The top 20 most prolific institutions were listed. (D) The collaboration network manifested the relationships among different institutes in this field. SCP, single country publication; MCP, multiple countries publication; RCC, renal cell carcinoma.
As of institutions, Figure 2C showed the top 20 most prolific institutes, among which Harvard University in USA dominated the list with a production of 257 publications. Fudan University, Shanghai Jiao Tong University and Huazhong University of Science and Technology in China ranked fourth, sixth and seventh, respectively. In Figure 2D, the institutions could be classified into different groups, and each group had close relationships or common features, like publications they have contributed to, etc.
3.3 Authors
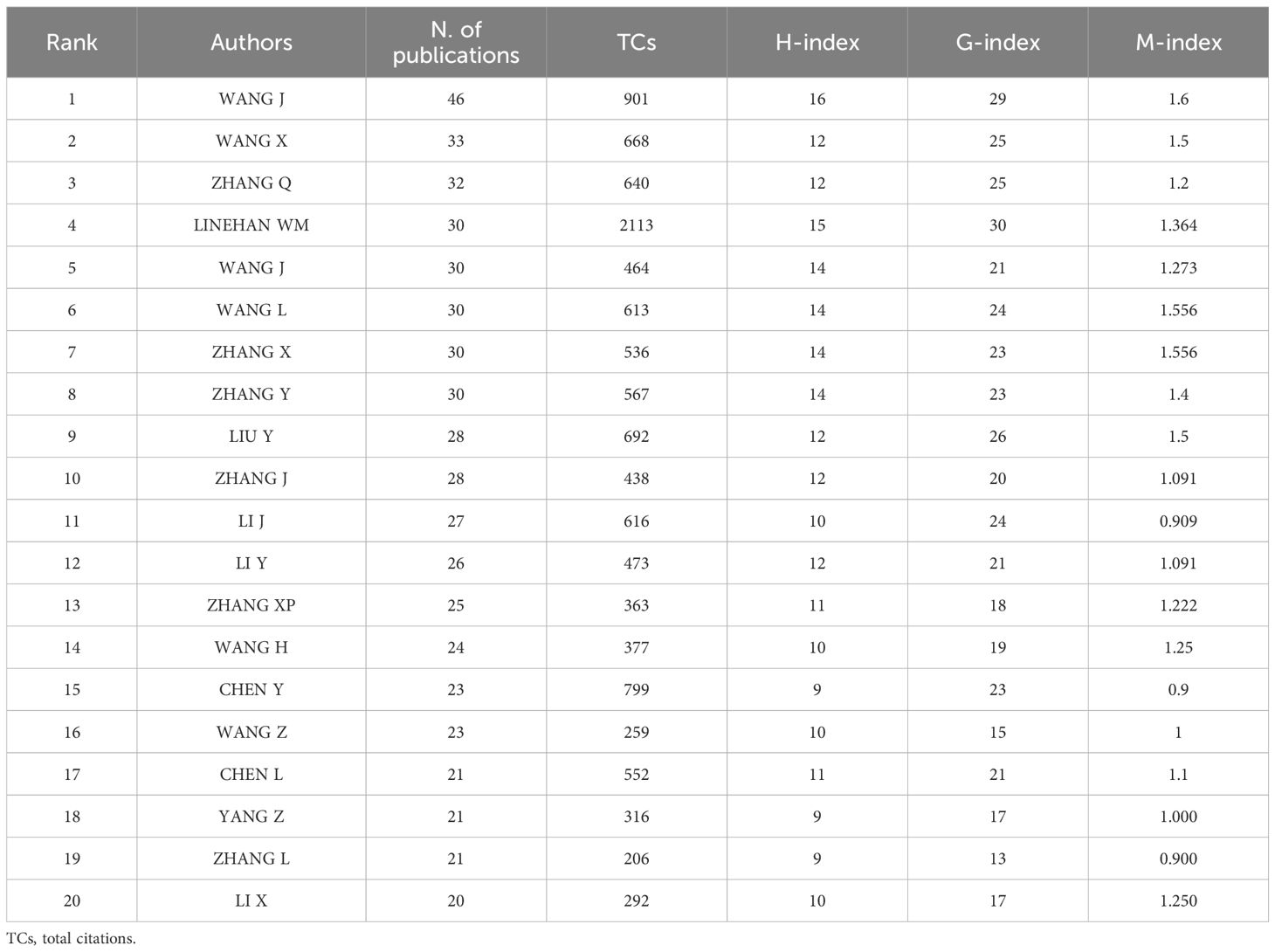
Up to 16884 authors contributed to this field. The top 20 most productive authors were listed in Figure 3A, where WANG Y and WANG X dominated the first and second place with 46 and 33 publications, surpassing other authors. As was shown in Figure 3B, the H-index of the top 20 authors varied from 12 to 16, with WANG Y still ranked first and LINEHAN WM ranked second (H-index = 15). In terms of local citations, HAKIMI AA topped the list with 648 local citations, closely followed by HSIEH JJ (Supplementary Figure S2A). LINEHAN WM dominated the list of G-index (Supplementary Figure S2B), but the list of M-index was dominated by “LAIMON YN” (Supplementary Figure S2C). Figure 3C presented the timelines of authors’ related researches in this field. “WANG Y” published 46 publications with an H-index of 16, indicating that “WANG Y” was the most productive and prominent author. The distribution of different authors with different number of publications was roughly in accordance with Lotka’s law (Supplementary Figure S2D). The authors could also be divided into different groups (Figure 3D). The top 20 most productive authors were also listed in Table 2 with their number of publications, total citations, H-index, G-index and M-index.
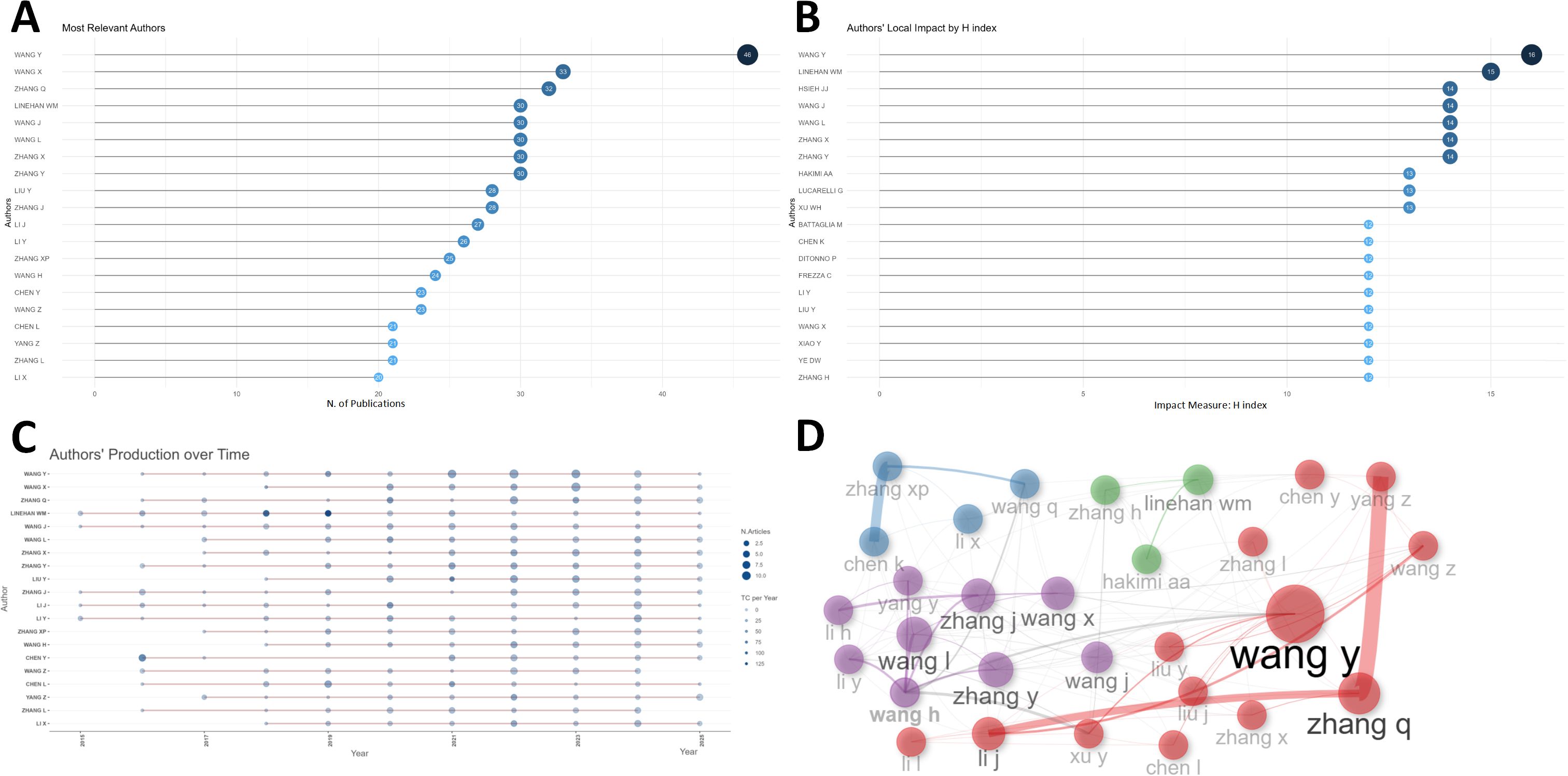
Figure 3. Author analysis. (A) The top 20 authors with most articles were listed in this plot. WANG Y and WANG X dominated the first and second place with 46 and 33 publications. (B) The top 20 authors with highest h-index were listed. WANG Y ranked first and LINEHAN WM ranked second. (C) The dynamic plot shows the timelines of the top 20 authors’ related researches of this field. The size and color-density of the nodes were positively correlated with the quantity of publication and total citation per year, respectively. (D) The collaboration network manifested the relationships among different authors in this field.
3.4 Journals
Supplementary Figure S3A demonstrated the top 20 journals extracted from 1002 journals according to number of publications, Local citations, total citations and H-index were also adopted to evaluate journals’ impact, as is shown in Supplementary Figures S3B–D. Supplementary Figure S3E displayed 41 core journals according to the Bradford’s law in the field of metabolism in RCC.
Detailed information of the top 20 journals with highest number of publications, including their impact factor, Quartile in category, H-index and total citations, was shown in Supplementary Table S1.
3.5 Publications and references
Figure 4A demonstrated the top 20 publications based on total citations, Obesity and inflammation: the linking mechanism and the complications (total citations = 1240), Epidemiology of Renal Cell Carcinoma (total citations = 1061), Shared Risk Factors in Cardiovascular Disease and Cancer (total citations = 1036) had the most total citations, surpassing other publications whose total citations were all below 1000. Figure 4B presented the top 20 publications with the largest local citations, An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma with local citations of 244 topped the list, while local citations of other publications were all lower than 200. Co-cited publications were put in the same cluster with identical color, and different clusters of publications might indicate different hotspots in this field (Figure 4C).
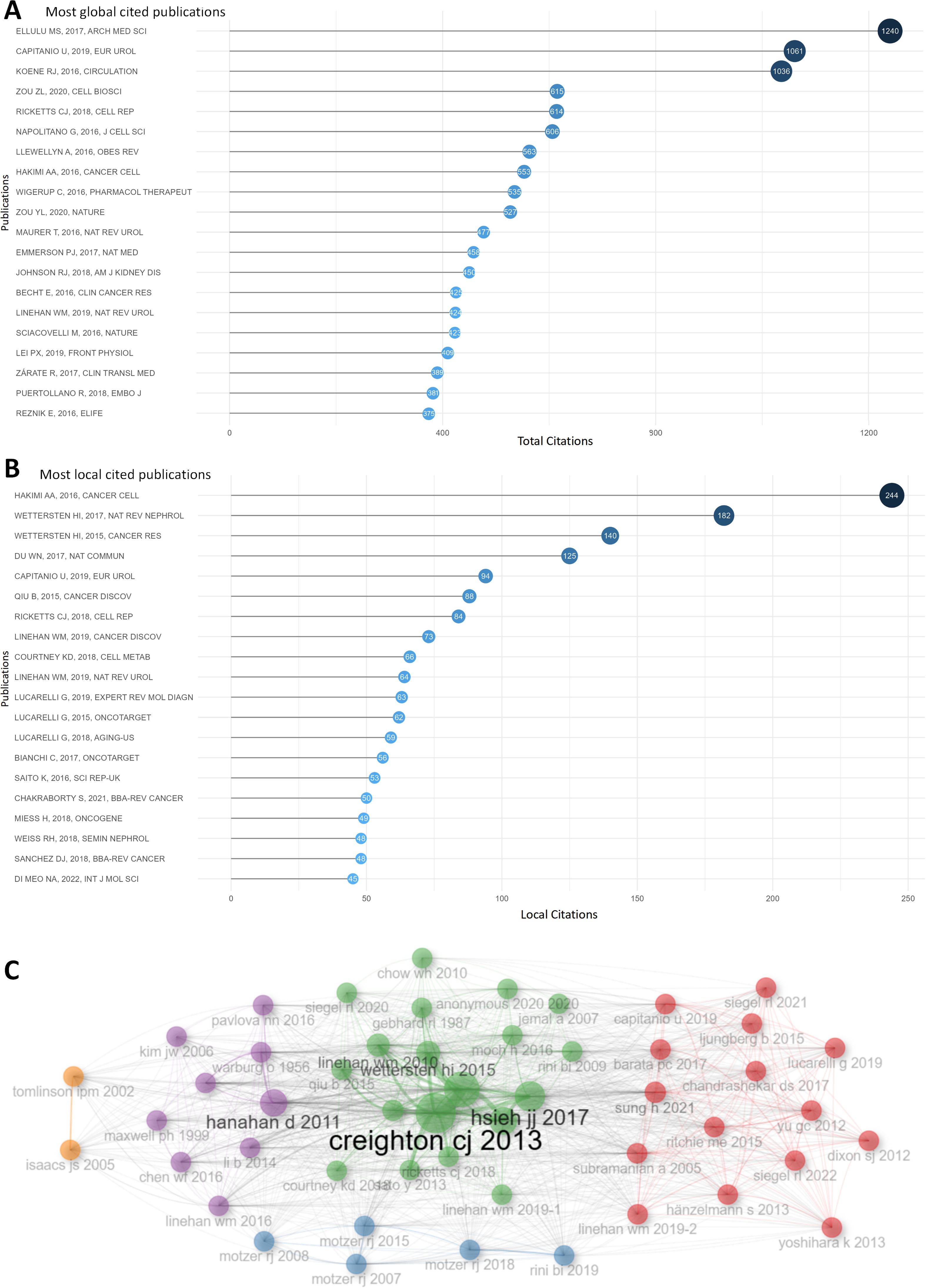
Figure 4. Publication analysis. (A) The top 20 publications were ranked according to the total citations. (B) The top 20 publications were listed according to local citations. (C) This plot demonstrates the publications clusters. Co-cited publications were put in the same cluster with identical color, and different clusters of publications might indicate different hot spots in this fields.
As for references, Supplementary Table S2 illustrated the top 20 references out of more than 144,000 references in total according to local citations. Comprehensive molecular characterization of clear cell renal cell carcinoma, An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma, Hallmarks of cancer: the next generation were the top 3 in the list, indicating their fundamental impact in this fields. These publications could probably be regarded as the cornerstone of this fields.
3.6 Keywords, trend topic and themes analysis
Keywords could be regarded as the essence of publications, which concisely summarized major topics and main information of a publications. A total of more than 6000 keywords were extracted from our retrieval data. “Cancer”, “expression”, “metabolism”, “renal cell carcinoma” were the top 4 keywords with frequencies over 400, while those of other keywords were lower than 200, which could also be intuitively observed in Supplementary Figure S4A.
In keyword co-occurrence network, as is shown in Figure 5A, the red cluster was characterized by terms “metabolism”, “expression”, “cancer”, “activation”, “hypoxia”, “mutations”, indicating metabolic alterations in RCC and their influences on RCC cells. The purple cluster focused possibly on the correlations between metabolic syndrome and RCC, as terms like “metabolic syndrome”, “body-mass index”, “obesity”, “kidney cancer” could be seen in this cluster. The green cluster mainly reflected therapy techniques and potential mechanisms in this field, as terms like “sunitinib”, “therapy” could be observed. The blue cluster might focus on some shared mechanisms among RCC and other cancer types. We would make further explorations in the discussion part later about the red and purple clusters.
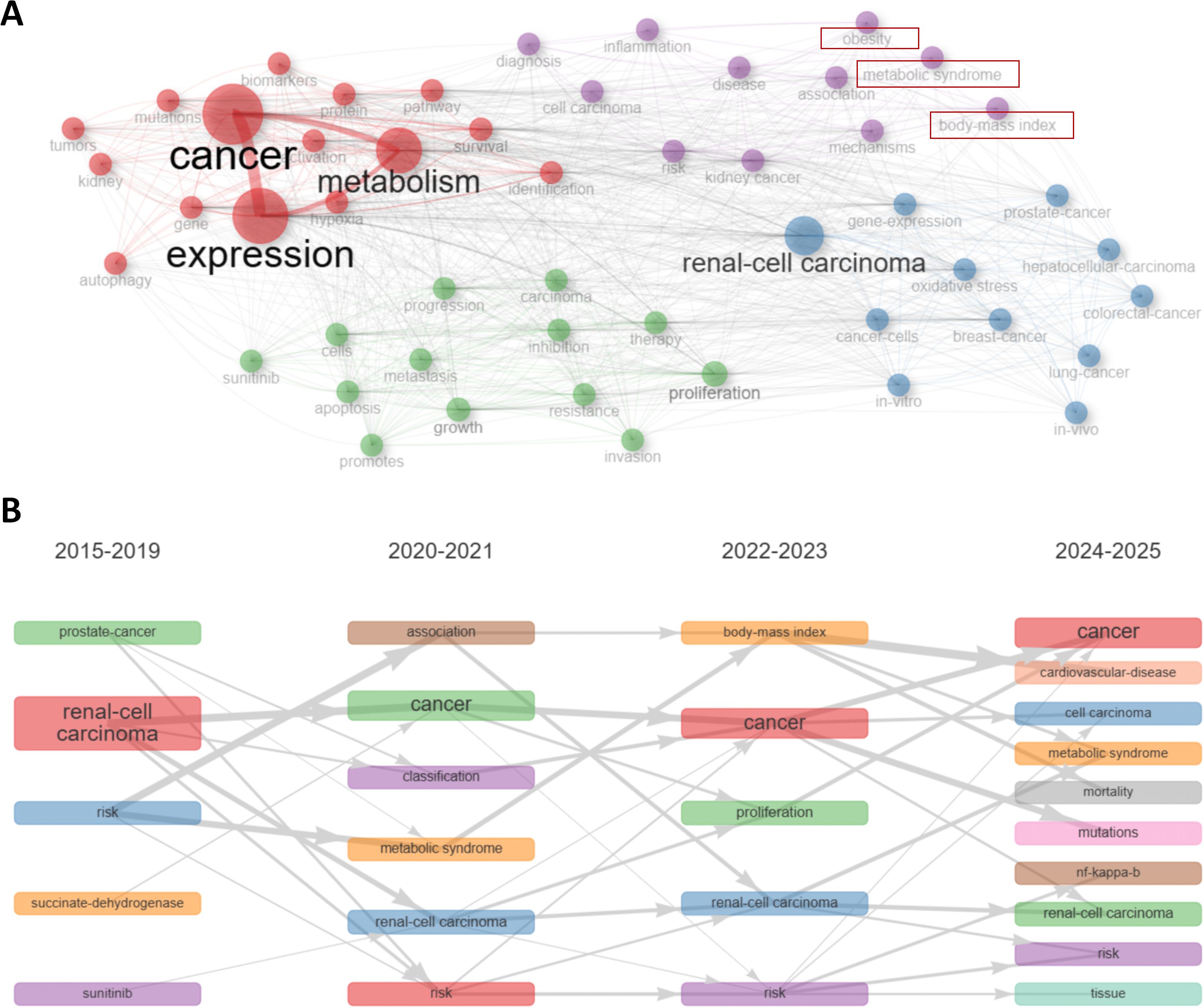
Figure 5. Keyword and hot spot analysis. (A) Keywords were distributed in different clusters in the keyword co-occurrence network. The red cluster indicated metabolic alterations in RCC. The purple cluster focused possibly on the correlations between metabolic syndrome and RCC. The green cluster mainly reflected therapy techniques and potential mechanisms. The blue cluster might focus on some shared mechanisms among RCC and other cancer types. (B) This dynamic plot demonstrated the thematic evolution. “metabolic syndrome”, as well as its related term “body-mass index”, were emerging keywords recent years from 2020, indicating metabolic syndrome and RCC might be a hotspot of this field recent years.
Trend shifts in a particular field could be succinctly presented by keywords dynamics. As can be observed in Figure 5B, “metabolic syndrome”, as well as its related term “body-mass index”, were emerging keywords recent years from 2020, indicating metabolic syndrome and RCC might be a hotspot of this field recent years. Development of trend topics overtime was shown in Supplementary Figure S4B. The classified topics were displayed in Supplementary Figure S4C by a thematic map.
4 Discussion
RCC is a global disease with heavy burden, and metabolism is considered to play vital roles in RCC. The concept of metabolism and related technologies have undergone rapid development, which has led to significant progress in the field of “metabolism in RCC”. However, bibliometric analysis in this field remains a void. Therefore, we conducted this study, hoping to provide a foundational reference for future basic and clinical research in this field.
In this study, we tried to summarize past research findings, analyze current research hotspots, and prospect the future development of this field by reading and analyzing crucial articles and reviews from 2015 to 2025 in the Web of Science (Core Collection) with the help of bibliometric methods.
4.1 General information
The annual publications and citation in this field increased steadily from 2015 to 2024 with a drop in 2025, possibly for the statistic end point of 15 May 2025. As for countries, USA, China and UK were in the top 3 list according to total citations, and they were also the countries with most cooperation as indicated by their high MCP. For institutions’ productivity, Harvard University in USA dominated the list. Fudan University, Shanghai Jiao Tong University and Huazhong University of Science and Technology in China ranked fourth, sixth and seventh, respectively. As for authors, WANG Y dominated the list of productivity and H-index, while HAKIMI AA, LINEHAN WM, LAIMON YN ranked the first in terms of local citations, G-index, M-index, respectively. Journals were analyzed based on number of publications, total citations, local citations and h-index. Core journals were identified with Bradford’s law. Publications on these journals would be conductive to keep pace with the latest development in this field, as well as obtain basic knowledge in this field. After analyzing the keyword co-occurrence network, combined with targeted literature reading, we identified several major points and hotspots over the past decade in the field of metabolism in RCC, namely the metabolic alterations in RCC, the metabolic syndrome and RCC, the microbiome and RCC.
4.2 Metabolic alterations in RCC
4.2.1 Metabolic gene reprogramming in RCC
A series of genes were involved in metabolic reprogramming in RCC. Generally, RCC could be majorly classified into three subtypes: clear cell RCC (ccRCC), which was the most prevalent (26), papillary RCC (pRCC), and chromophobe RCC (chRCC). Apart from these, there were some rare subtypes, such as medullary RCC, collecting duct RCC, and hereditary leiomyomatosis renal cell cancer (HLRCC) (2, 26–28).
VHL gene mutation and subsequent HIF accumulation (4, 11) are widely recognized as the most common pathogenic events in ccRCC. HIF act as a transcriptional regulator that promote the expression of a range of glycolysis-related genes, including GLUT1 (glucose transporter-1), PGK (phosphoglycerate kinase), LDHA (lactate dehydrogenase), PDK1 (pyruvate dehydrogenase kinase), and HK (hexokinase) (29, 30). Besides, HIFs could also contribute to the downregulation of TCA cycle and oxidative phosphorylation (31). Genes involved in PI3K/AKT/mTOR signaling pathway, including PTEN, TSC1/2, and PIK3CA (5, 32), were also commonly mutated in ccRCC based on TCGA database (33). Notably, mTORC1 could augment the expression of HIF (34). In ccRCC cells, MYC was another commonly overexpressed gene, which might function during metabolic reprogramming of glutamine and fatty acid synthesis (35–37).
As for other cancer types, FH was regarded as the gene responsible for HLRCC (38), SDH was found to be associated with familial renal cancer (4), and MET was thought to be correlate with hereditary pRCC (4).
4.2.2 Carbohydrate metabolism
Glycolysis was upregulated in ccRCC cells. GLUT-1 was upregulated (29), which subsequently led to increased glucose uptake. After entering the cancer cell, glucose was further catalyzed into G-6-P by upregulated HK (15). Other enzymes involved in glycolysis or subsequent lactate fermentation, including GPI, PGK, LDH-A, etc. were also upregulated (15), further substantiated the upregulation of glycolysis. By upregulated enzyme G6PD (39), G-6-P could then enter Pentose Phosphate Pathway (PPP), where NADPH and ribose sugars were produced. TCA and ETC was downregulated in RCC cells. Two critical enzymes, PDH and PC, which help pyruvate to enter TCA cycle, were downregulated in RCC (40, 41), implicating a decreased shunt flux into TCA cycle (40). However, citrate, an intermediate in TCA, was upregulated (42, 43). Downregulation of oxidative phosphorylation complexes (II, III, IV) of the respiratory chain and ATP synthase was associated with RCC (44), implicating downregulation of ETC.
Metabolic alterations in carbohydrate indicate that RCC tends to use glycolysis rather than oxidative phosphorylation to survive, which was in consistent with the definition of Warburg effect (45). The Warburg effect enabled cancer cells to thrive in a nutrient-deficient condition (46), while the PPP was utilized to antagonize oxidative stress (47) as well as providing nucleotides (46).
4.2.3 Lipid metabolism
Lipogenesis was upregulated in ccRCC. Actually, lipids storage was not only a morphological manifestation of ccRCC, but also an indicator of malignancy. Recently, mesoderm induction early response 2 (MIER2) was identified as a new biomarker for RCC, which could promote malignancy and sunitinib resistance by inducing lipids accumulation in RCC (48). This further indicated the vital role of lipid metabolism.
4.2.3.1 Fatty acid metabolism
Various alterations contributed to fatty acid synthesis in ccRCC. Stearoyl-CoA desaturase 1 (SCD1), an enzyme that elongates and desaturates fatty acids to produce unsaturated fatty acids like triglycerides and phospholipids (49), was upregulated in ccRCC tissues (50). Compared with non-malignant renal tissue, the mRNA level of ACC, the rate-limiting enzyme during FA synthesis (51), was upregulated in ccRCC and had a positive correlation with unfavorable overall survival (52). Besides, the IHC staining intensity of FASN, the terminal enzyme in de novo FA synthesis, was shown to be positively correlated with tumor staging and distant metastasis in 120 RCC patients (53). Besides, the expression and activity of Carnitine palmitoyl transferase 1A (CPT1A), which functions to transport lipids into mitochondria for further oxidative degradation (54), was down regulated in ccRCC (55). Moreover, after lipids were produced and protected from degradation, another protein called Perilipin 2 (PLIN2) would promote lipids storage. PLIN2, a protein on the surface of lipids droplets (56), was proved to be over-expressed in ccRCC patients’ samples and was shown to promote lipids storage, tumor progression and tumor proliferation in ccRCC xenografts (57).
Apart from alterations in lipids metabolism, many altered metabolites in carbohydrates and amino acids metabolism could influence lipids synthesis. Citrate, which was upregulated in TCA cycle, could generate intracellular acetyl-CoA via the action of ATP citrate lyase (ACL) (58) for further fatty acid synthesis. Interestingly, ACL was observed to be upregulated in RCC cells (59), and knockdown of ACL could inhibit proliferation and induce apoptosis in RCC cells. In fact, citrate could also be obtained from reductive carboxylation of glutamine (60, 61), where amino acid metabolism showed its importance during lipids synthesis.
4.2.3.2 Upregulated cholesteryl ester metabolism
CE metabolism was upregulated in ccRCC (62–64), and inhibition of 3 beta-hydroxy steroid dehydrogenase type 7 (HSD3B7), an enzyme concerning CE metabolism, might induce apoptosis in ccRCC cells (65). Mechanically, CE could be oxidized to 7α-hydroxycholesterol (7α-OHC), an intermediate product triggering apoptosis, and HSD3B7 could catalyze 7α-OHC into cholic acid or chenodeoxycholic acid (CDCA), and thus prevent apoptosis (65).
4.2.3.3 Clinical significance of lipids metabolism in ccRCC
Metabolic reprogramming of lipids made it a promising endeavor to adopt lipids as biomarkers for ccRCC diagnosis or prognosis. In fact, researchers have found by lipidomics and machine learning that a 26-lipids panel could have excellent performance in distinguishing stage I and II with stage III and IV ccRCC (66). In another study focusing on using metabolites to predict ICI response, researchers found that most of the selected marker metabolites (9 in 10) were very-long-chain FAs (67). Furthermore, in 2025, another group of researchers developed a fatty acid metabolism signature (68) based on TCGA and GEO database, which could effectively predict the survival and response to targeted therapy and immunotherapy among ccRCC patients. This further validated the vital role lipids metabolism played in ccRCC.
4.2.4 Amino acid metabolism
Upregulated glutamine metabolism (42) might prevent RCC from cell death. GSH generated in this pathway could act to hinder ROS, hence protecting the cancer cells from oxidative stress (40). Interestingly, inhibition of GSH synthesis could induce ferroptosis in ccRCC cells (VHL- deficient) (69), and ferroptosis is characterized by lipid-peroxidation (70).
Increased downstream metabolites in Kynurenine pathways, including kynurenine and quinolinate, was identified with decreased tryptophan level in RCC (40), and IDO, which functions during tryptophan catabolism, was upregulated in endothelial cells of RCC compared with normal tissues (71). These implied an upregulated Kynurenine pathway, whose downstream metabolites might have immunosuppressive effects (40). Furthermore, the combined administration of IDO inhibitor and IFN-α was shown to inhibit RENCA cell (renal cancer cell line in mice) while IFN-α or IDO inhibitor along couldn’t inhibit tumor growth (71). Besides, in vitro experiment also indicated that DCs treated by IDO inhibitor, namely the epacadostat, could possibly enhance the oncolysis function of T cells (72).
ASS1, the key enzyme during arginine synthesis, was downregulated in ccRCC patients’ biopsy samples (73), indicating an arginine dependency in cancer cells. Arginine deprivation was thought to be a promising therapy for ccRCC (5).
4.3 Metabolic syndrome and RCC
Metabolic syndrome was defined by WHO as a pathological condition consisting abdominal obesity, insulin resistance, hypertension, and hyperlipidemia (74). A growing body of evidence suggested a close association between metabolic syndrome and RCC. Metabolic syndrome has already been identified as an independent prognostic factor in localized ccRCC patients (75) back to 2019, and a meta-analysis (76) composed of six studies in 2025 reconfirmed its independent correlation with RCC. In addition, some specific diseases related to metabolic syndrome, including diabetes, obesity and hypertension was discovered to be correlated with RCC as well. Diabetes was identified as an independent risk factor for TNM staging in ccRCC patients (77). Obesity was a well-recognized risk factor for RCC, and BMI was positively correlated with the incidence (78, 79) and pathological upstaging (80) of RCC. Hypertension was identified as an independent prognostic factor for RCC survival (81), and a positive link between both diastolic blood pressure and systolic blood pressure and RCC risk (82) was discovered.
Mechanically, hyperinsulinemia was thought to be the shared potential mechanism between diabetes, obesity and RCC. Notably, hyperinsulinemia was not only seen in diabetes, but also existed in in obesity state (83). Insulin can enhance IGF-1 synthesis and activation. Both insulin and IGF-1 have the effects of fostering cell proliferation and restraining apoptosis (84). Upon activation of insulin receptors (INSR) and IGF1 receptors (IGF1R), a series of signaling pathways is triggered. These include pathways such as PI3K/AKT, cyclin D1, HIF-1α and VEGF. The activation of these pathways could contribute to key characteristics of cancer, including enhancing cell proliferation, stimulating angiogenesis, and diminishing apoptosis (85–87). However, some researchers observed a positive correlation between better prognosis and increased IGF-1 levels (88), which might suggest a more complicated mechanisms behind IGF-1 and RCC.
High level of glucose might play a role between diabetes and RCC by stimulating the metabolism of RCC cells (84). It has been proposed that hyperglycemia can enhance cancer cell proliferation via elevating the levels of protein kinase C (PKC) and peroxisome proliferator-activated receptors (PPARs), which can in turn accelerate cellular metabolism and thereby foster cell proliferation (89).
Pro-inflammatory factors and leptin might play roles between obesity and RCC. It was believed that chronic inflammation existed in adipose tissue under obesity state (90). Obesity-related chronic inflammation might promote tumor by releasing pro-inflammatory factors, such as IL-6 with anti-apoptotic and cell proliferation effect via JAK2 and PI3K/AKT (91, 92), as well as TNF-α with anti-apoptotic effect via NF-kB (87). Notably, glutamine, which was correlated with decreasing inflammation and proinflammatory cytokines, was downregulated in obesity (93), which further implied the role of inflammation in mediating the link between obesity and RCC. Moreover, leptin, an adipocyte-specific protein functions to regulate satiety and bodyweight (94), might also promote RCC by promoting cell proliferation, upregulating VEGF and inhibiting apoptosis, possibly through HIF-1α and NF-κB (95). Leptin was also shown to be correlated with worse OS and migration of ccRCC cells (96).
Though the precise and detailed mechanism between RCC and hypertension has not been fully understood, some researchers (97) hypothesized that renin-angiotensin-system might play a potential role. Ang- (1-7) (generated from Ang I and Ang II) could promote xenograft tumor growth in nude mice and migration of caki-1 and caki-2 cell lines in vitro (98), and combined administration of sunitinib and telmisartan (a renin-angiotensin-system antagonist) was observed to induce more necrosis and less neo-angiogenesis in 786-O cell line xenograft in mice, indicating the potential role and therapeutic value of renin-angiotensin-system in RCC (99).
4.4 Microbiome and RCC
Studies have demonstrated that there were significant differences in the relative abundance of 20 gut bacteria species between RCC patients and control group (100). Besides, antibiotic use may reduce the efficacy of immunotherapy in metastatic renal cell carcinoma (101), yet using antibiotics to target Bacteroides spp. in stool can improve progression-free survival (PFS) in metastatic RCC patients receiving first-line VEGF-TKI therapy (102). Additionally, CBM588, a live bacterial product, enhance clinical outcomes in patients with metastatic renal cell carcinoma treated with nivolumab-ipilimumab (103). These findings suggest that microbiome may play significant roles in RCC.
The precise and comprehensive mechanism behind microbiome and RCC were not fully understood yet. However, there seemed to be some indirect evidences revealing potential mechanisms.
The microbiome may impact RCC through tryptophan metabolism. Some endogenous tryptophan metabolites can act as ligands for aryl hydrocarbon receptor (AhR) signaling (104), and AhR has been linked to the invasion of RCC (105). Researchers investigated the relationship between gut bacterial abundance and the expression levels of certain tryptophan metabolites (106). Kynurenic acid showed a negative correlation with the gut bacteria Prevotella-9 and Akkermansia, and kynurenine (Kyn) was found to enhance the migration and invasion of 786-O cells via AhR. These findings suggest that gut microbiota may activate AhR through its tryptophan metabolite kynurenine, thereby mediating RCC metastasis. The microbiome might also influence RCC by modulating immunity through lipid metabolism. In 2025, a research team (107) discovered that low intra-tumoral mycobiome abundance was associated with suppressed lipid catabolism, CD8+ T cell depletion, and poor prognosis. This implies that the intra-tumoral mycobiome might affect RCC via immune regulation mediated by lipid metabolism. Carbohydrate metabolism and related molecules may represent another target through which the microbiome affect RCC. SUCNR1, the receptor of succinate, an important molecule during TCA cycle, was linked to a variety of microbes, including beneficial bacteria possibly contributing to a better disease-specific survival rate in ccRCC (108). Additionally, taurine might also play a intermediate role between microbiome and RCC. Upregulated bacteria desulfovibrionaceae downregulated Lactobacillus was observed in ccRCC (100) while the serum level of taurine, which was considered to be consumed by bacteria desulfovibrionaceae (109) and promoted by Lactobacillus (110), also decreased in the ccRCC (100).
In the future, with the aid of continuously advancing new technologies, it may be possible for researchers to further map the spatial and temporal landscapes of metabolism in RCC, thereby more comprehensively elucidating the role of metabolism throughout the entire process of cancer initiation. Additionally, leveraging sophisticated omics technologies and the progress in microbiome-related research, it may be feasible in the future to construct a holistic metabolic map of the human-microbiome ecosystem, providing a more comprehensive understanding of the role of metabolism in renal cancer. Lastly, it is anticipated that more studies will focus on the role of metabolism in renal cancer treatment and drug resistance, revealing the close connection between metabolism and renal cancer from a more clinically significant perspective.
5 Conclusion
Through bibliometric analysis over 3010 articles and reviews, together with critical literature reading, our research summarized past research findings, analyzed current research hotspots, and prospected the future development in this field. Specifically, three major points were proposed. Firstly, the metabolic alterations in RCC. Metabolic reprogramming and the biological significance of metabolic changes were discussed from the perspective of carbohydrate, lipids and amino acid metabolism. Secondly, the metabolic syndrome and RCC. The correlation and potential mechanism between RCC and metabolic diseases, including obesity, diabetes, and hypertension were explored. Thirdly, the microbiome and RCC. Microorganisms may influence the development of RCC through specific metabolites, revealing a close connection between RCC and metabolism from a novel viewpoint. In the future, a broader metabolic map with spatial, temporal and human-microbiome interaction perspective could be made and more researches concerning metabolism’s role during RCC treatment and drug resistance might bring more clinical significance.
6 Limitations
In our study, publications from 2015 to 2025 in the field of metabolism in RCC were summarized by bibliometric analysis. Major points and hotspots over the past decade were explored and discussed. However, some flaws might still exist because of data acquisition method. All the publications were from Web of Science (core collection), and hence data from other sources were not included in our study. Besides, data after our retrieving date could not be included in our study as well. Data from multiple sources could be collected in future studies to make our understandings about this field more comprehensive and accurate. Moreover, this study aimed at providing a fundamental reference for future by summarizing major points and hotspots in this filed. Therefore, this study may not be able to provide conclusions with high level of evidence-based hierarchy like a meta-analysis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
YFL: Conceptualization, Formal analysis, Writing – original draft. JH: Formal analysis, Writing – original draft. DL: Formal analysis, Writing – original draft. XY: Formal analysis, Writing – original draft. HZ: Formal analysis, Writing – original draft. ST: Formal analysis, Writing – original draft. YY: Formal analysis, Writing – original draft. MW: Formal analysis, Writing – original draft. YAL: Formal analysis, Writing – original draft. ZZ: Formal analysis, Writing – original draft. RH: Conceptualization, Formal analysis, Writing – review & editing. BL: Conceptualization, Data curation, Formal analysis, Writing – review & editing. XX: Conceptualization, Data curation, Formal analysis, Writing – review & editing. XP: Conceptualization, Data curation, Formal analysis, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was sponsored by the National Natural Science Foundation of China (No. 82072806); Shanghai Rising-Star Program (23QC1401400); Shanghai Rising-Star Program (Sailing Special Program) (23YF1458400). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
We thank the Web of Science™ (WOS) team for allowing us to use their data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1537805/full#supplementary-material
References
1. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2024) 74:229–63. doi: 10.3322/caac.21834
2. Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, et al. Renal cell carcinoma. Nat Rev Dis Primers. (2017) 3:17009. doi: 10.1038/nrdp.2017.9
3. Moch H. An overview of renal cell cancer: pathology and genetics. Semin Cancer Biol. (2013) 23:3–9. doi: 10.1016/j.semcancer.2012.06.006
4. Linehan WM, Srinivasan R, and Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol. (2010) 7:277–85. doi: 10.1038/nrurol.2010.47
5. Chakraborty S, Balan M, Sabarwal A, Choueiri TK, and Pal S. Metabolic reprogramming in renal cancer: Events of a metabolic disease. Biochim Biophys Acta Rev Cancer. (2021) 1876:188559. doi: 10.1016/j.bbcan.2021.188559
7. Braunstein H and Adelman JU. Histochemical study of the enzymatic activity of human neoplasms. II. Histogenesis of renal cell carcinoma. Cancer. (1966) 19:935–8. doi: 10.1002/1097-0142(196607)19:7<935::AID-CNCR2820190706>3.0.CO;2-D
8. Lindlar F. Hypernephroid carcinoma and kidney carcinoma. Lipid chemical analysis of 24 kidney tumors. Verh Dtsch Ges Pathol. (1961) 45:144–9.
9. Gates SC and Sweeley CC. Quantitative metabolic profiling based on gas chromatography. Clin Chem. (1978) 24:1663–73. doi: 10.1093/clinchem/24.10.1663
10. Sanger F, Nicklen S, and Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U.S.A. (1977) 74:5463–7. doi: 10.1073/pnas.74.12.5463
11. Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U.S.A. (1994) 91:9700–4. doi: 10.1073/pnas.91.21.9700
12. Kumar KR, Cowley MJ, and Davis RL. Next-generation sequencing and emerging technologies. Semin Thromb Hemost. (2024) 50:1026–38. doi: 10.1055/s-0044-1786397
13. Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. (2008) 320:1344–9. doi: 10.1126/science.1158441
14. Shapiro E, Biezuner T, and Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. (2013) 14:618–30. doi: 10.1038/nrg3542
15. Wettersten HI, Aboud OA, Lara P, and Weiss RH. Metabolic reprogramming in clear cell renal cell carcinoma. Nat Rev Nephrol. (2017) 13:410–9. doi: 10.1038/nrneph.2017.59
16. Baker M. Metabolomics: from small molecules to big ideas. Nat Methods. (2011) 8:117–21. doi: 10.1038/nmeth0211-117
17. Shi Y, Wei W, Li L, Wei Q, Jiang F, Xia G, et al. The global status of research in breast cancer liver metastasis: a bibliometric and visualized analysis. Bioengineered. (2021) 12:12246–62. doi: 10.1080/21655979.2021.2006552
18. Lu B, Liu Y, Yao Y, Yang T, Zhang H, Yang X, et al. Advances in sequencing and omics studies in prostate cancer: unveiling molecular pathogenesis and clinical applications. Front Oncol. (2024) 14:1355551. doi: 10.3389/fonc.2024.1355551
19. Yang Q, Zhai X, and Lv Y. A bibliometric analysis of triptolide and the recent advances in treating non-small cell lung cancer. Front Pharmacol. (2022) 13:878726. doi: 10.3389/fphar.2022.878726
20. Aria M and Cuccurullo C. bibliometrix: An R-tool for comprehensive science mapping analysis. J Informetrics. (2017) 11:959–75. doi: 10.1016/j.joi.2017.08.007
21. Hirsch JE. An index to quantify an individual’s scientific research output. Proc Natl Acad Sci U.S.A. (2005) 102:16569–72. doi: 10.1073/pnas.0507655102
22. Musbahi A, Rao CB, and Immanuel A. A bibliometric analysis of robotic surgery from 2001 to 2021. World J Surg. (2022) 46:1314–24. doi: 10.1007/s00268-022-06492-2
23. LEO E. Theory and practise of the g-index. Scientometrics. (2006) 69:131–52. doi: 10.1007/s11192-006-0144-7
24. Venable GT, Shepherd BA, Loftis CM, McClatchy SG, Roberts ML, Fillinger ME, et al. Bradford’s law: identification of the core journals for neurosurgery and its subspecialties. J Neurosurg. (2016) 124:569–79. doi: 10.3171/2015.3.JNS15149
25. Kushairi N and Ahmi A. Flipped classroom in the second decade of the Millenia: a Bibliometrics analysis with Lotka’s law. Educ Inf Technol (Dordr). (2021) 26:4401–31. doi: 10.1007/s10639-021-10457-8
26. Linehan WM and Ricketts CJ. The Cancer Genome Atlas of renal cell carcinoma: findings and clinical implications. Nat Rev Urol. (2019) 16:539–52. doi: 10.1038/s41585-019-0211-5
27. Rathmell WK, Rathmell JC, and Linehan WM. Metabolic pathways in kidney cancer: current therapies and future directions. J Clin Oncol. (2018), Jco2018792309. doi: 10.1200/JCO.2018.79.2309
28. Moch H, Cubilla AL, Humphrey PA, Reuter VE, and Ulbright TM. The 2016 WHO classification of tumours of the urinary system and male genital organs-part A: renal, penile, and testicular tumours. Eur Urol. (2016) 70:93–105. doi: 10.1016/j.eururo.2016.02.029
29. Ozcan A, Shen SS, Zhai QJ, and Truong LD. Expression of GLUT1 in primary renal tumors: morphologic and biologic implications. Am J Clin Pathol. (2007) 128:245–54. doi: 10.1309/HV6NJVRQKK4QHM9F
30. Schönenberger D, Harlander S, Rajski M, Jacobs RA, Lundby AK, Adlesic M, et al. Formation of renal cysts and tumors in vhl/trp53-deficient mice requires HIF1α and HIF2α. Cancer Res. (2016) 76:2025–36. doi: 10.1158/0008-5472.Can-15-1859
31. LaGory EL, Wu C, Taniguchi CM, Ding CC, Chi JT, von Eyben R, et al. Suppression of PGC-1α Is critical for reprogramming oxidative metabolism in renal cell carcinoma. Cell Rep. (2015) 12:116–27. doi: 10.1016/j.celrep.2015.06.006
32. Dibble CC and Cantley LC. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. (2015) 25:545–55. doi: 10.1016/j.tcb.2015.06.002
33. Haake SM, Weyandt JD, and Rathmell WK. Insights into the genetic basis of the renal cell carcinomas from the cancer genome atlas. Mol Cancer Res. (2016) 14:589–98. doi: 10.1158/1541-7786.MCR-16-0115
34. Toschi A, Lee E, Gadir N, Ohh M, and Foster DA. Differential dependence of hypoxia-inducible factors 1 alpha and 2 alpha on mTORC1 and mTORC2. J Biol Chem. (2008) 283:34495–9. doi: 10.1074/jbc.C800170200
35. Shroff EH, Eberlin LS, Dang VM, Gouw AM, Gabay M, Adam SJ, et al. MYC oncogene overexpression drives renal cell carcinoma in a mouse model through glutamine metabolism. Proc Natl Acad Sci U.S.A. (2015) 112:6539–44. doi: 10.1073/pnas.1507228112
36. Tang SW, Chang WH, Su YC, Chen YC, Lai YH, Wu PT, et al. MYC pathway is activated in clear cell renal cell carcinoma and essential for proliferation of clear cell renal cell carcinoma cells. Cancer Lett. (2009) 273:35–43. doi: 10.1016/j.canlet.2008.07.038
37. Shi W, Xu X, Yan F, Wang B, Zhao H, Chan A, et al. N-Myc downstream-regulated gene 2 restrains glycolysis and glutaminolysis in clear cell renal cell carcinoma. Oncol Lett. (2017) 14:6881–7. doi: 10.3892/ol.2017.7024
38. Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. (2002) 30:406–10. doi: 10.1038/ng849
39. Zhang Q, Ni Y, Wang S, Agbana YL, Han Q, Liu W, et al. G6PD upregulates Cyclin E1 and MMP9 to promote clear cell renal cell carcinoma progression. Int J Med Sci. (2022) 19:47–64. doi: 10.7150/ijms.58902
40. Wettersten HI, Hakimi AA, Morin D, Bianchi C, Johnstone ME, Donohoe DR, et al. Grade-dependent metabolic reprogramming in kidney cancer revealed by combined proteomics and metabolomics analysis. Cancer Res. (2015) 75:2541–52. doi: 10.1158/0008-5472.CAN-14-1703
41. Kaelin WG Jr. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. (2008) 8:865–73. doi: 10.1038/nrc2502
42. Hakimi AA, Reznik E, Lee CH, Creighton CJ, Brannon AR, Luna A, et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell. (2016) 29:104–16. doi: 10.1016/j.ccell.2015.12.004
43. Li B, Qiu B, Lee DS, Walton ZE, Ochocki JD, Mathew LK, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. (2014) 513:251–5. doi: 10.1038/nature13557
44. Simonnet H, Alazard N, Pfeiffer K, Gallou C, Béroud C, Demont J, et al. Low mitochondrial respiratory chain content correlates with tumor aggressiveness in renal cell carcinoma. Carcinogenesis. (2002) 23:759–68. doi: 10.1093/carcin/23.5.759
45. Vander Heiden MG, Cantley LC, and Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. (2009) 324:1029–33. doi: 10.1126/science.1160809
46. Pavlova NN and Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. (2016) 23:27–47. doi: 10.1016/j.cmet.2015.12.006
47. Zhang Q, Yang Z, Han Q, Bai H, Wang Y, Yi X, et al. G6PD promotes renal cell carcinoma proliferation through positive feedback regulation of p-STAT3. Oncotarget. (2017) 8:109043–60. doi: 10.18632/oncotarget.22566
48. Wei Z, Ye Y, Liu C, Wang Q, Zhang Y, Chen K, et al. MIER2/PGC1A elicits sunitinib resistance via lipid metabolism in renal cell carcinoma. J Adv Res. (2025) 70:287–305. doi: 10.1016/j.jare.2024.04.032
49. Enoch HG, Catalá A, and Strittmatter P. Mechanism of rat liver microsomal stearyl-CoA desaturase. Studies of the substrate specificity, enzyme-substrate interactions, and the function of lipid. J Biol Chem. (1976) 251:5095–103. doi: 10.1016/S0021-9258(17)33223-4
50. von Roemeling CA, Marlow LA, Wei JJ, Cooper SJ, Caulfield TR, Wu K, et al. Stearoyl-CoA desaturase 1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Clin Cancer Res. (2013) 19:2368–80. doi: 10.1158/1078-0432.CCR-12-3249
51. Tan SK, Hougen HY, Merchan JR, Gonzalgo ML, and Welford SM. Fatty acid metabolism reprogramming in ccRCC: mechanisms and potential targets. Nat Rev Urol. (2023) 20:48–60. doi: 10.1038/s41585-022-00654-6
52. Zhao Z, Liu Y, Liu Q, Wu F, Liu X, Qu H, et al. The mRNA expression signature and prognostic analysis of multiple fatty acid metabolic enzymes in clear cell renal cell carcinoma. J Cancer. (2019) 10:6599–607. doi: 10.7150/jca.33024
53. Horiguchi A, Asano T, Asano T, Ito K, Sumitomo M, and Hayakawa M. Fatty acid synthase over expression is an indicator of tumor aggressiveness and poor prognosis in renal cell carcinoma. J Urol. (2008) 180:1137–40. doi: 10.1016/j.juro.2008.04.135
54. Moustacchi E and Williamson DH. Physiological variations in satellite components of yeast DNA detected by density gradient centrifugation. Biochem Biophys Res Commun. (1966) 23:56–61. doi: 10.1016/0006-291X(66)90268-3
55. Du W, Zhang L, Brett-Morris A, Aguila B, Kerner J, Hoppel CL, et al. HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. Nat Commun. (2017) 8:1769. doi: 10.1038/s41467-017-01965-8
56. Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, et al. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. (2011) 121:2102–10. doi: 10.1172/JCI46069
57. Qiu B, Ackerman D, Sanchez DJ, Li B, Ochocki JD, Grazioli A, et al. HIF2α-dependent lipid storage promotes endoplasmic reticulum homeostasis in clear-cell renal cell carcinoma. Cancer Discov. (2015) 5:652–67. doi: 10.1158/2159-8290.CD-14-1507
58. Icard P, Wu Z, Fournel L, Coquerel A, Lincet H, Alifano M, et al. ATP citrate lyase: A central metabolic enzyme in cancer. Cancer Lett. (2020) 471:125–34. doi: 10.1016/j.canlet.2019.12.010
59. Teng L, Chen Y, Cao Y, Wang W, Xu Y, Wang Y, et al. Overexpression of ATP citrate lyase in renal cell carcinoma tissues and its effect on the human renal carcinoma cells in vitro. Oncol Lett. (2018) 15:6967–74. doi: 10.3892/ol.2018.8211
60. Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. (2011) 481:385–8. doi: 10.1038/nature10642
61. Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. (2011) 481:380–4. doi: 10.1038/nature10602
62. Zhang S, Fang T, He Y, Feng W, Yu Z, Zheng Y, et al. VHL mutation drives human clear cell renal cell carcinoma progression through PI3K/AKT-dependent cholesteryl ester accumulation. EBioMedicine. (2024) 103:105070. doi: 10.1016/j.ebiom.2024.105070
63. Mohtar MA, Hasanah Mohd Yusuf SN, and Syafruddin SE. Cholesterol accumulation in ccRCC: the role of ccRCC-initiating VHL-HIFα pathway. EBioMedicine. (2024) 103:105112. doi: 10.1016/j.ebiom.2024.105112
64. Drabkin HA and Gemmill RM. Cholesterol and the development of clear-cell renal carcinoma. Curr Opin Pharmacol. (2012) 12:742–50. doi: 10.1016/j.coph.2012.08.002
65. Riscal R, Gardner SM, Coffey NJ, Carens M, Mesaros C, Xu JP, et al. Bile acid metabolism mediates cholesterol homeostasis and promotes tumorigenesis in clear cell renal cell carcinoma. Cancer Res. (2024) 84:1570–82. doi: 10.1158/0008-5472.CAN-23-0821
66. Manzi M, Palazzo M, Knott ME, Beauseroy P, Yankilevich P, Giménez MI, et al. Coupled mass-spectrometry-based lipidomics machine learning approach for early detection of clear cell renal cell carcinoma. J Proteome Res. (2021) 20:841–57. doi: 10.1021/acs.jproteome.0c00663
67. Mock A, Zschäbitz S, Kirsten R, Scheffler M, Wolf B, Herold-Mende C, et al. Serum very long-chain fatty acid-containing lipids predict response to immune checkpoint inhibitors in urological cancers. Cancer Immunol Immunother. (2019) 68:2005–14. doi: 10.1007/s00262-019-02428-3
68. Wei Z, Cheng G, Ye Y, Le C, Miao Q, Chen J, et al. A fatty acid metabolism signature associated with clinical therapy in clear cell renal cell carcinoma. Front Genet. (2022) 13:894736. doi: 10.3389/fgene.2022.894736
69. Miess H, Dankworth B, Gouw AM, Rosenfeldt M, Schmitz W, Jiang M, et al. The glutathione redox system is essential to prevent ferroptosis caused by impaired lipid metabolism in clear cell renal cell carcinoma. Oncogene. (2018) 37:5435–50. doi: 10.1038/s41388-018-0315-z
70. Chen J, Li X, Ge C, Min J, and Wang F. The multifaceted role of ferroptosis in liver disease. Cell Death Differ. (2022) 29:467–80. doi: 10.1038/s41418-022-00941-0
71. Trott JF, Kim J, Abu Aboud O, Wettersten H, Stewart B, Berryhill G, et al. Inhibiting tryptophan metabolism enhances interferon therapy in kidney cancer. Oncotarget. (2016) 7:66540–57. doi: 10.18632/oncotarget.11658
72. Jochems C, Fantini M, Fernando RI, Kwilas AR, Donahue RN, Lepone LM, et al. The IDO1 selective inhibitor epacadostat enhances dendritic cell immunogenicity and lytic ability of tumor antigen-specific T cells. Oncotarget. (2016) 7:37762–72. doi: 10.18632/oncotarget.9326
73. Yoon CY, Shim YJ, Kim EH, Lee JH, Won NH, Kim JH, et al. Renal cell carcinoma does not express argininosuccinate synthetase and is highly sensitive to arginine deprivation via arginine deiminase. Int J Cancer. (2007) 120:897–905. doi: 10.1002/ijc.22322
74. Saklayen MG. The global epidemic of the metabolic syndrome. Curr Hypertens Rep. (2018) 20:12. doi: 10.1007/s11906-018-0812-z
75. Liu Z, Wang H, Zhang L, Li S, Fan Y, Meng Y, et al. Metabolic syndrome is associated with improved cancer-specific survival in patients with localized clear cell renal cell carcinoma. Transl Androl Urol. (2019) 8:507–18. doi: 10.21037/tau.2019.10.04
76. Zhou Y, Chen Y, Yang H, Xu Z, Zhuang J, Bian Q, et al. Metabolic syndrome and increased susceptibility to renal cell carcinoma - a meta-analysis. BMC Nephrol. (2025) 26:102. doi: 10.1186/s12882-025-04013-6
77. Zhang Q, Chen P, Tian R, He J, Han Q, and Fan L. Metabolic syndrome is an independent risk factor for fuhrman grade and TNM stage of renal clear cell carcinoma. Int J Gen Med. (2022) 15:143–50. doi: 10.2147/IJGM.S346972
78. Liu X, Sun Q, Hou H, Zhu K, Wang Q, Liu H, et al. The association between BMI and kidney cancer risk: An updated dose-response meta-analysis in accordance with PRISMA guideline. Med (Baltimore). (2018) 97:e12860. doi: 10.1097/MD.0000000000012860
79. Graff RE, Wilson KM, Sanchez A, Chang SL, McDermott DF, Choueiri TK, et al. Obesity in relation to renal cell carcinoma incidence and survival in three prospective studies. Eur Urol. (2022) 82:247–51. doi: 10.1016/j.eururo.2022.04.032
80. Pruthi DK, Miller G, Ankerst DP, Neumair M, Capitanio U, Correa AF, et al. Diabetes, obesity, and pathological upstaging in renal cell carcinoma: results from a large multi-institutional consortium. J Urol. (2023) 210:750–62. doi: 10.1097/JU.0000000000003650
81. Eskelinen TJ, Kotsar A, Tammela TLJ, and Murtola TJ. Components of metabolic syndrome and prognosis of renal cell cancer. Scand J Urol. (2017) 51:435–41. doi: 10.1080/21681805.2017.1352616
82. Alcala K, Mariosa D, Smith-Byrne K, Nasrollahzadeh Nesheli DN, Carreras-Torres R, Ardanaz Aicua E, et al. The relationship between blood pressure and risk of renal cell carcinoma. Int J Epidemiol. (2022) 51:1317–27. doi: 10.1093/ije/dyac042
83. Renehan AG, Roberts DL, and Dive C. Obesity and cancer: pathophysiological and biological mechanisms. Arch Physiol Biochem. (2008) 114:71–83. doi: 10.1080/13813450801954303
84. Labochka D, Moszczuk B, Kukwa W, Szczylik C, and Czarnecka AM. Mechanisms through which diabetes mellitus influences renal cell carcinoma development and treatment: A review of the literature. Int J Mol Med. (2016) 38:1887–94. doi: 10.3892/ijmm.2016.2776
85. Calle EE and Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. (2004) 4:579–91. doi: 10.1038/nrc1408
86. Aurilio G, Piva F, Santoni M, Cimadamore A, Sorgentoni G, Lopez-Beltran A, et al. The role of obesity in renal cell carcinoma patients: clinical-pathological implications. Int J Mol Sci. (2019) 20. doi: 10.3390/ijms20225683
87. Hursting SD and Dunlap SM. Obesity, metabolic dysregulation, and cancer: a growing concern and an inflammatory (and microenvironmental) issue. Ann N Y Acad Sci. (2012) 1271:82–7. doi: 10.1111/j.1749-6632.2012.06737.x
88. Rasmuson T, Grankvist K, Jacobsen J, Olsson T, and Ljungberg B. Serum insulin-like growth factor-1 is an independent predictor of prognosis in patients with renal cell carcinoma. Acta Oncol. (2004) 43:744–8. doi: 10.1080/02841860410017260
89. Ryu TY, Park J, and Scherer PE. Hyperglycemia as a risk factor for cancer progression. Diabetes Metab J. (2014) 38:330–6. doi: 10.4093/dmj.2014.38.5.330
90. Nieman KM, Romero IL, Van Houten B, and Lengyel E. Adipose tissue and adipocytes support tumorigenesis and metastasis. Biochim Biophys Acta. (2013) 1831:1533–41. doi: 10.1016/j.bbalip.2013.02.010
91. Huang B, Lang X, and Li X. The role of IL-6/JAK2/STAT3 signaling pathway in cancers. Front Oncol. (2022) 12:1023177. doi: 10.3389/fonc.2022.1023177
92. Smith DA, Kiba A, Zong Y, and Witte ON. Interleukin-6 and oncostatin-M synergize with the PI3K/AKT pathway to promote aggressive prostate Malignancy in mouse and human tissues. Mol Cancer Res. (2013) 11:1159–65. doi: 10.1158/1541-7786.MCR-13-0238
93. Petrus P, Lecoutre S, Dollet L, Wiel C, Sulen A, Gao H, et al. Glutamine links obesity to inflammation in human white adipose tissue. Cell Metab. (2020) 31:375–390.e11. doi: 10.1016/j.cmet.2019.11.019
94. Margetic S, Gazzola C, Pegg GG, and Hill RA. Leptin: a review of its peripheral actions and interactions. Int J Obes Relat Metab Disord. (2002) 26:1407–33. doi: 10.1038/sj.ijo.0802142
95. Gonzalez-Perez RR, Xu Y, Guo S, Watters A, Zhou W, and Leibovich SJ. Leptin upregulates VEGF in breast cancer via canonic and non-canonical signalling pathways and NFkappaB/HIF-1alpha activation. Cell Signal. (2010) 22:1350–62. doi: 10.1016/j.cellsig.2010.05.003
96. Fan WL, Yeh YM, Liu TT, Lin WM, Yang TY, Lee CW, et al. Leptin is associated with poor clinical outcomes and promotes clear cell renal cell carcinoma progression. Biomolecules. (2021) 11. doi: 10.3390/biom11030431
97. Guang Y, Yan W, Zhi-Wei L, Hua Z, Yue Z, and Fei S. Renin-angiotensin-system and clear cell renal carcinoma: research advances and future perspectives. J Cancer Metastasis Treat. (2024) 10:26. doi: 10.20517/2394-4722.2024.40
98. Sobczuk P, Trzcinska-Danielewicz J, Koperski L, Girstun A, and Cudnoch-Jedrzejewska A. Angiotensin-(1-7) can promote cell migration and tumor growth of clear cell renal cell carcinoma. J Physiol Pharmacol. (2022) 73. doi: 10.26402/jpp.2022.6.04
99. Verhoest G, Dolley-Hitze T, Jouan F, Belaud-Rotureau MA, Oger E, Lavenu A, et al. Sunitinib combined with angiotensin-2 type-1 receptor antagonists induces more necrosis: a murine xenograft model of renal cell carcinoma. BioMed Res Int. (2014) 2014:901371. doi: 10.1155/2014/901371
100. Yang BY, Zhao FZ, Li XH, Zhao MS, Lv JC, Shi MJ, et al. Alteration of pro-carcinogenic gut microbiota is associated with clear cell renal cell carcinoma tumorigenesis. Front Microbiol. (2023) 14:1133782. doi: 10.3389/fmicb.2023.1133782
101. Lalani AA, Xie W, Braun DA, Kaymakcalan M, Bossé D, Steinharter JA, et al. Effect of antibiotic use on outcomes with systemic therapies in metastatic renal cell carcinoma. Eur Urol Oncol. (2020) 3:372–81. doi: 10.1016/j.euo.2019.09.001
102. Hahn AW, Froerer C, VanAlstine S, Rathi N, Bailey EB, Stenehjem DD, et al. Targeting bacteroides in stool microbiome and response to treatment with first-line VEGF tyrosine kinase inhibitors in metastatic renal-cell carcinoma. Clin Genitourin Cancer. (2018) 16:365–8. doi: 10.1016/j.clgc.2018.05.001
103. Dizman N, Meza L, Bergerot P, Alcantara M, Dorff T, Lyou Y, et al. Nivolumab plus ipilimumab with or without live bacterial supplementation in metastatic renal cell carcinoma: a randomized phase 1 trial. Nat Med. (2022) 28:704–12. doi: 10.1038/s41591-022-01694-6
104. Liu JR, Miao H, Deng DQ, Vaziri ND, Li P, and Zhao YY. Gut microbiota-derived tryptophan metabolism mediates renal fibrosis by aryl hydrocarbon receptor signaling activation. Cell Mol Life Sci. (2021) 78:909–22. doi: 10.1007/s00018-020-03645-1
105. Zhao H, Chen L, Yang T, Feng YL, Vaziri ND, Liu BL, et al. Aryl hydrocarbon receptor activation mediates kidney disease and renal cell carcinoma. J Transl Med. (2019) 17:302. doi: 10.1186/s12967-019-2054-5
106. Dai G, Chen X, and He Y. The gut microbiota activates ahR through the tryptophan metabolite kyn to mediate renal cell carcinoma metastasis. Front Nutr. (2021) 8:712327. doi: 10.3389/fnut.2021.712327
107. Mou W, Deng Z, Zhu L, Jiang A, Lin A, Xu L, et al. Intratumoral mycobiome heterogeneity influences the tumor microenvironment and immunotherapy outcomes in renal cell carcinoma. Sci Adv. (2025) 11:eadu1727. doi: 10.1126/sciadv.adu1727
108. Najm R, Hachim MY, and Kandasamy RK. Divulging a pleiotropic role of succinate receptor SUCNR1 in renal cell carcinoma microenvironment. Cancers (Basel). (2022) 14. doi: 10.3390/cancers14246064
109. Hu H, Shao W, Liu Q, Liu N, Wang Q, Xu J, et al. Gut microbiota promotes cholesterol gallstone formation by modulating bile acid composition and biliary cholesterol secretion. Nat Commun. (2022) 13:252. doi: 10.1038/s41467-021-27758-8
Keywords: RCC, bibliometric analysis, metabolism, metabolic syndrome, microbiome, metabolic reprogramming
Citation: Liu Y, He J, Lyu D, Yang X, Zhang H, Tu S, Yao Y, Wei M, Li Y, Zhao Z, Huang R, Lu B, Xu X and Pan X (2025) Uncovering the research evolution and hotspots of metabolism in renal cell carcinoma over the last decade. Front. Oncol. 15:1537805. doi: 10.3389/fonc.2025.1537805
Received: 06 January 2025; Accepted: 28 July 2025;
Published: 26 August 2025.
Edited by:
Liangyou Gu, People’s Liberation Army General Hospital, ChinaReviewed by:
Alessandra di Masi, Roma Tre University, ItalyZhihao Wei, Huazhong University of Science and Technology, China
Copyright © 2025 Liu, He, Lyu, Yang, Zhang, Tu, Yao, Wei, Li, Zhao, Huang, Lu, Xu and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiuwu Pan, cGFueGl1d3VAMTI2LmNvbQ==; Xiao Xu, eHV4aWFvbGlvbkBob3RtYWlsLmNvbQ==; Bingnan Lu, OTcyMDgxMDYwQHFxLmNvbQ==; Runzhi Huang, cnVuemhpaHVhbmcyMDIyQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Yifan Liu
Yifan Liu Junzhe He
Junzhe He Donghao Lyu1†
Donghao Lyu1† Siqi Tu
Siqi Tu Yuntao Yao
Yuntao Yao Maodong Wei
Maodong Wei Yuanan Li
Yuanan Li Runzhi Huang
Runzhi Huang Bingnan Lu
Bingnan Lu Xiuwu Pan
Xiuwu Pan