- 1Department of Gynecology, The First People’s Hospital of Changzhou and the Third Affiliated Hospital of Soochow University, Changzhou, China
- 2Clinical Medical Research Center, The First People’s Hospital of Changzhou and the Third Affiliated Hospital of Soochow University, Changzhou, China
- 3Changzhou Medical Center, Nanjing Medical University, Changzhou, China
Background: Human papillomavirus (HPV) infection, especially high-risk types like HPV16 and HPV18, is a primary cause of cervical cancer. The p53 gene influences cellular response to DNA damage and has a functional polymorphism (rs1042522, p.Arg72Pro) that affects susceptibility to degradation by HPV E6 protein. This study aims to analyze the relationship among p53 genotypes, high-risk HPV infection, and hematological parameters in cervical cancer development and to develop a predictive model.
Methods: This retrospective cross-sectional study collected cervical cancer specimens and brush samples from patients at the First People’s Hospital of Changzhou between January 2020 and August 2024. HPV types and p53 genotyping were performed using PCR. Inflammatory markers like neutrophil-to-lymphocyte ratio (NLR), systemic immune-inflammation index (SII), and platelet-to-lymphocyte ratio (PLR) were calculated. Statistical analyses including logistic regression and LASSO were used to construct a predictive model.
Results: The study included 147 female patients with cervical cancer and controls. HPV16 and HPV18 had high infection rates. In the log-additive model, each additional p53 C allele reduced the risk by 48% (OR = 0.52, 95% CI: 0.27-0.98, P = 0.038). Significant interactions were found between p53 genotypes and HPV18 infection on cervical cancer risk (P = 0.026). Cervical cancer patients showed reduced red blood cell count and hemoglobin. The predictive model, including p53 genotype, HPV16, HPV18, and hematological parameters, had an AUC of 0.920 (95% CI: 0.875–0.965).
Conclusion: The study identified significant differences in p53 genotypes, HPV infection, and hematological parameters between cervical cancer patients and controls. The predictive model demonstrated high discriminatory ability for cervical cancer risk assessment. The interaction between HPV18 and p53 genotypes suggests a potential protective effect of the p53 C allele. Larger studies are needed to validate these findings.
1 Introduction
Cervical cancer is one of the most common malignant tumors among women worldwide, especially in developing countries, where the incidence and mortality rates remain high (1). Human papillomavirus (HPV) infection is considered the main cause of cervical cancer, particularly high-risk HPV types (such as HPV16 and HPV18), which are closely associated with the development of the disease (2–5). However, not all women infected with high-risk HPV will develop cervical cancer, suggesting that individual genetic susceptibility may play an important role in this process (6, 7). The p53 gene, a critical tumor suppressor, may influence the cellular response to DNA damage when mutated or polymorphic. Studies have shown that the p53 gene has a functional polymorphism (rs1042522, p.Arg72Pro), where the p.72Arg variant of p53 is more susceptible to degradation mediated by the HPV E6 protein compared to the p.72Pro variant. Therefore, individuals carrying the p.72Arg variant of p53 have a significantly higher risk of developing cervical cancer (8, 9). However, no consistent conclusions have been reached across different ethnic populations (10, 11). This study will further analyze the relationship between p53 genotypes and the risk of cervical cancer.
In recent years, researchers have gradually recognized the potential value of hematological indicators in the early diagnosis and prognosis of tumors. Inflammatory markers such as the neutrophil to lymphocyte ratio (NLR), the systemic immune-inflammation index (SII), and the platelet to lymphocyte ratio (PLR) are considered to be closely related to the occurrence, progression, and prognosis of tumors (12–14). Thus, the integration of HPV infection status, p53 genotype, and routine blood parameters may offer a new approach for the preliminary screening and risk assessment of cervical cancer. This study aims to investigate the interrelationships between high-risk HPV infection, p53 genotypes, and routine blood parameters in the development of cervical cancer, identify factors associated with its onset, and develop an effective nomogram predictive model to support clinical decision-making.
2 Materials and methods
2.1 Study design and participants
This study is a retrospective cross-sectional study that collected cervical cancer specimens from patients who underwent surgical treatment at the First People’s Hospital of Changzhou between January 2020 and August 2024 (cervical cancer group, 55 cases). Additionally, cervical brush samples were collected from women with normal or benign lesions (cervicitis or cervical intraepithelial neoplasia grade I) diagnosed by pathology following cervical biopsy at the same Hospital during the same period (control group, 92 cases).
Inclusion criteria: 1) Age between 18–75 years; 2) No use of medications that could affect study outcomes within the past two weeks; 3) Availability of complete clinical data.
Exclusion criteria: 1) History of cervical surgery; 2) History of pelvic radiotherapy; 3) History of chemotherapy; 4) Presence of severe heart, liver, or kidney disease; 5) Presence of autoimmune diseases; 6) Co-existing with other malignancies.
2.2 PCR analysis system components and reagents
The fully automated medical PCR analysis system SLAN-96S (Shanghai Hongshi Medical Technology Co., Ltd., China) was used. PCR reaction system included: 10× buffer, 50 mM MgCl2, IMMOLASE™ DNA polymerase (Midian Biotechnology Inc., USA), and dNTPs (Takara, Japan). The quantitative PCR reaction system included: 10× buffer, 25 mM MgCl2, Taq DNA polymerase, dNTPs (Shanghai Bocai Biotechnology Co., Ltd., China), and the SYSMEX XN-9000 fully automated blood analyzer (Sysmex Corporation, Japan).
2.3 Typing method for high-risk HPV and p53 and RB1
The method established by Zhang Jun et al. (15), was used to detect 16 types of high-risk HPV and related tumor suppressor genes, p53 and RB1. This method is based on high-throughput two-dimensional PCR technology (2D-PCR) (16). Briefly, specific primers are designed according to the DNA sequences of 16 different types of high-risk HPV, as well as the p53 and RB1 genes. The upstream primers for the different types of high-risk HPV, p53, and RB1 are labeled with corresponding tags. After the PCR reaction is completed, a melting curve analysis is performed. In three fluorescence detection channels, the probes and the complementary sequences of corresponding tags bind together and dissociate as the temperature increases, resulting in clearly distinguishable melting valleys, which allows for accurate determination of the genotypes.
2.4 Blood routine parameters
A fully automated blood analyzer was utilized to analyze the routine indicators of peripheral venous blood from the subjects included in this study. These indicators included white blood cell count (WBC), red blood cell count (RBC), hemoglobin (HGB), platelet count (PLT), neutrophil count (NEUT), neutrophil percentage (NEUT%), eosinophil count (EO), eosinophil percentage (EO%), basophil count (BASO), basophil percentage (BASO%), lymphocyte count (LY), lymphocyte percentage (LY%), monocyte count (MONO), monocyte percentage (MONO%), hematocrit (HCT), red cell distribution width-coefficient of variation (RDW-CV), mean corpuscular hemoglobin (MCH), and mean corpuscular hemoglobin concentration (MCHC).
The inflammatory indicators calculated based on the above complete blood count parameters included the systemic inflammation response index (SIRI), NLR, SII, PLR, lymphocyte to monocyte ratio (LMR), and neutrophil to platelet ratio (NPR). The formulas for calculation were as follows:
● SIRI = NEUT × MONO/LY
● NLR = NEUT/LY
● SII = PLT × NEUT/LY
● PLR = PLT/LY
● LMR = LY/MONO
● NPR = NEUT/PLT
2.5 Statistical analysis
The SNPStats online analysis tool (https://www.snpstats.net/start.htm) was employed to examine the allele and genotype frequencies of the p53 gene, a cervical cancer susceptibility gene, and its interaction with high-risk HPV infection. Continuous variables that followed a normal distribution were presented as means with standard deviations (SD), while non-normally distributed continuous variables were described using medians and interquartile ranges (IQR). Categorical variables were summarized using counts and percentages (%).
The process for constructing the optimal clinical predictive model for cervical cancer diagnosis involved several steps. First, a univariate logistic regression analysis was performed on all variables to identify those with a P-value below 0.20, which were selected as candidate variables for further modeling. Second, the least absolute shrinkage and selection operator (LASSO) regression was applied to refine these candidate variables, using 10-fold cross-validation to determine the optimal regularization parameter (λ). Next, the variables selected through LASSO regression were incorporated into a multivariate logistic regression analysis, with the final variables being selected via a backward stepwise method (BACKWARD) to construct the nomogram prediction model.
Subsequently, the model’s discriminative ability was assessed using a receiver operating characteristic (ROC) curve, calibration was evaluated through a calibration curve, and the clinical applicability of the model was analyzed using decision curve analysis (DCA). To further assess and validate the model’s performance, 1000 bootstrap resampling iterations were conducted.
2.6 Ethics statement
This study was approved by the Ethics Committee of the First People’s Hospital of Changzhou on December 17, 2023 (Approval No (2023):195). The Ethics Committee granted a waiver of written informed consent due to the retrospective nature of the study and the anonymization of all samples prior to analysis.
3 Results
3.1 Study population characteristics and HPV infection rates
As shown in Supplementary Table S1, a total of 147 female patients were included in this study. The mean age was 48.76 ± 11.26 years (range: 20–74 years). The mean age of the cervical cancer group was significantly higher than that of the control group (P = 0.023). Among the total participants, HPV16 and HPV18 had relatively high infection rates, accounting for 22% and 12%, respectively. Significant differences in high-risk HPV infection rates between the cervical cancer and control groups were observed for HPV16 (P < 0.001), HPV18 (P = 0.027), and HPV58 (P = 0.032).
All 147 patients in this study had the A/A genotype for the RB1 gene, and thus, this gene was excluded from further analysis. The allele and genotype frequencies of the p53 gene are shown in Supplementary Table S2. The Hardy-Weinberg equilibrium exact test results indicated P > 0.05 (Supplementary Table S3), suggesting that the study population was genetically stable and representative, providing a solid foundation for subsequent association analyses and interaction studies with high-risk HPV.
3.2 Adjusted association analysis of p53 gene variants with cervical cancer risk
After adjusting for age and multiple high-risk HPV types, including HPV16, HPV18, HPV31, HPV33, HPV39, HPV45, HPV51, HPV52, HPV56, HPV58, HPV59, HPV66, HPV68, and HPV82, the association analysis between the p53 gene and cervical cancer is presented in Table 1. In the codominant model, using the G/G genotype as the reference group, the C/C genotype showed a 76% reduction in risk (OR = 0.24, 95% CI: 0.06-0.95), although this was not statistically significant (P > 0.05). In the log-additive model, each additional C allele was associated with a 48% reduction in cervical cancer risk, which was statistically significant (OR = 0.52, 95% CI: 0.27-0.98, P = 0.038).
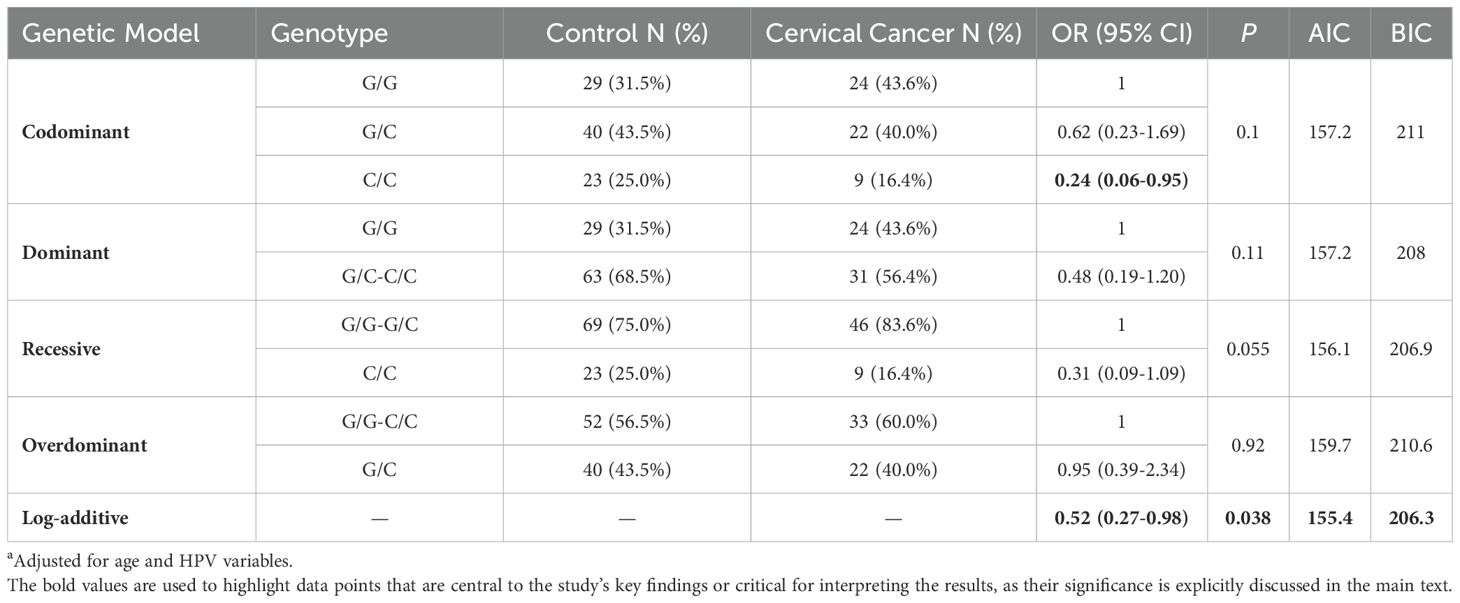
Table 1. Association analysis between p53 gene and cervical cancera.
3.3 Interaction analysis of p53 gene variants with HPV16 and HPV18 infections on cervical cancer risk
This study examined the interaction between the p53 gene and the two most prevalent high-risk HPV types, HPV16 and HPV18. Supplementary Table S4 presents the interaction analysis between p53 genotypes and HPV16 status, with data adjusted for age and other HPV types (including HPV18, HPV31, HPV33, HPV39, HPV45, HPV51, HPV52, HPV56, HPV58, HPV59, HPV66, HPV68, and HPV82). Individuals with the G/G genotype and HPV16 infection exhibited a significantly increased risk of cervical cancer, with an odds ratio (OR) of 16.63. This finding suggests that HPV16 infection exerts a strong carcinogenic effect in individuals with the G/G genotype. For those with the G/C or C/C genotypes, the risk of cervical cancer increased 6.38 times when infected with HPV16. While HPV16 infection increases cervical cancer risk in both genotypes, the magnitude of risk is lower for individuals with the G/C or C/C genotype compared to those with the G/G genotype. Despite this difference, the interaction P-value was 0.84, indicating no statistically significant interaction between p53 genotypes and HPV16 infection, and suggesting that the two factors independently influence cervical cancer risk.
In contrast, as shown in Supplementary Table S5, the interaction between HPV18 infection and the p53 genotype on cervical cancer risk was statistically significant (P = 0.026). This finding indicates that the p53 genotype modulates cervical cancer risk in individuals infected with HPV18. Table 2 demonstrates that, among HPV18-negative individuals, those with the G/C or C/C genotype had a 31% lower risk of cervical cancer compared to the G/G genotype, although this difference did not reach statistical significance. However, among HPV18-positive individuals, using the G/G genotype as the reference (OR = 1), the OR for cervical cancer risk in individuals with the G/C or C/C genotype was 0. This suggests that the p53 genotype may influence cervical cancer risk in the context of HPV18 infection. Specifically, individuals carrying the C allele may have some protective effect against cervical cancer in the presence of HPV18, while those with the G/G genotype may experience a significantly increased risk following HPV18 infection.
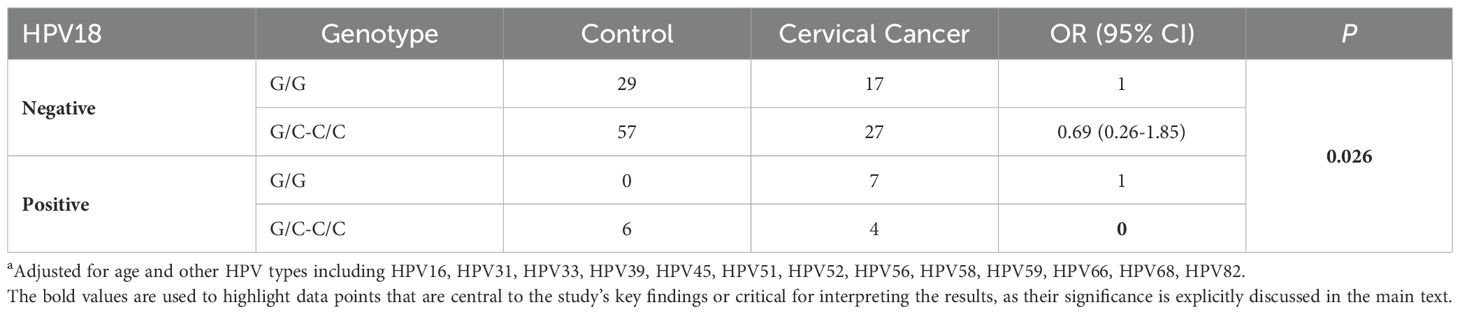
Table 2. Impact of different p53 genotypes on cervical cancer risk in HPV18 infectiona.
3.4 Hematological parameters and inflammatory biomarkers in cervical cancer patients
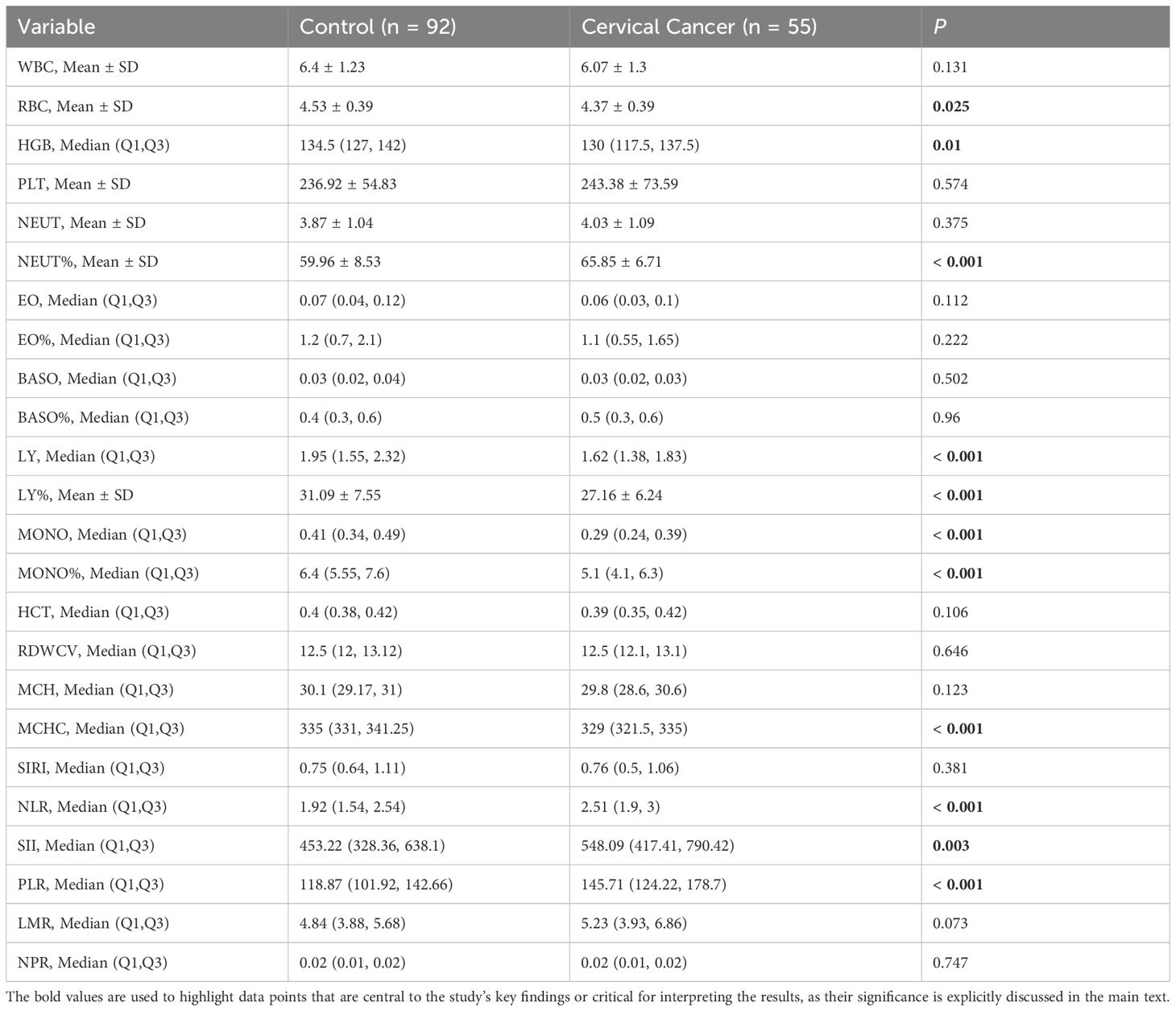
As presented in Table 3, significant differences were observed between the cervical cancer group and the control group across several hematological parameters, particularly in RBC, HGB, NEUT%, LY, MONO, and MONO%. These findings indicate that cervical cancer patients may exhibit distinct hematological alterations and inflammatory responses. Such results provide valuable insights for the early diagnosis and clinical monitoring of cervical cancer. In particular, potential clinical biomarkers, such as the NLR, SII and PLR, may hold significant promise in the evaluation and assessment of cervical cancer.
3.5 Logistic regression and LASSO analysis for cervical cancer risk prediction
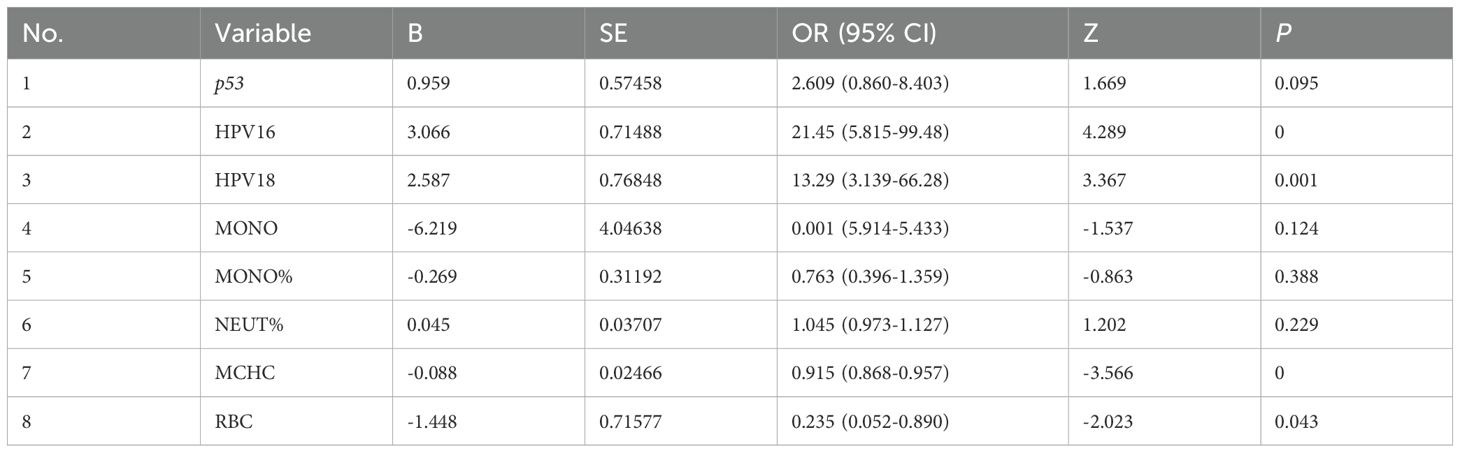
A univariate logistic regression analysis was conducted on variables including high-risk HPV, p53 genotypes (G/G and G/C-C/C), blood routine parameters, and inflammatory markers. Using a significance threshold of P < 0.20, a total of 23 variables were selected for further analysis (Supplementary Table S6). Subsequently, a LASSO regression analysis with 10-fold cross-validation was applied to these 23 variables (Supplementary Figure S1), and the optimal regularization parameter λ was identified (lambda.1SE = 0.05218946). Eight key variables were found to be significantly associated with cervical cancer risk: p53 genotype, HPV16, HPV18, MONO, MONO%, NEUT%, MCH, and RBC. These variables exhibited strong predictive value for cervical cancer within the model, laying a solid foundation for subsequent multivariate logistic regression analysis and clinical applications.
3.6 Development and validation of a nomogram prediction model for cervical cancer risk
A multivariate logistic regression analysis was performed using the 8 key variables identified through LASSO regression. BACKWARD was applied to select the final variables, as presented in Table 4. Based on these selected variables, a nomogram prediction model was constructed (Figure 1). This model calculates an individual score for each predictor and generates a total score to estimate the probability of cervical cancer occurrence, thus assisting in the diagnosis of cervical cancer.

Figure 1. Clinical nomogram for predicting cervical cancer diagnosis. p53: 0 represents GC or CC genotype, 1 represents GG genotype. HPV16 and HPV18: 0 represents negative, 1 represents positive.
The model’s discriminative ability was evaluated using a ROC curve (Figure 2A) and validated with 1000 bootstrap resampling iterations (Figure 2B). The results demonstrated an AUC of 0.920 (95% CI: 0.875–0.965), indicating strong performance in distinguishing cervical cancer cases from non-cancer cases. Moreover, the ROC rationality analysis (Figure 2C) showed that the nomogram model’s curve was the closest to the upper left corner, further confirming its superior performance compared to individual variable models in differentiating between cervical cancer and non-cervical cancer cases.
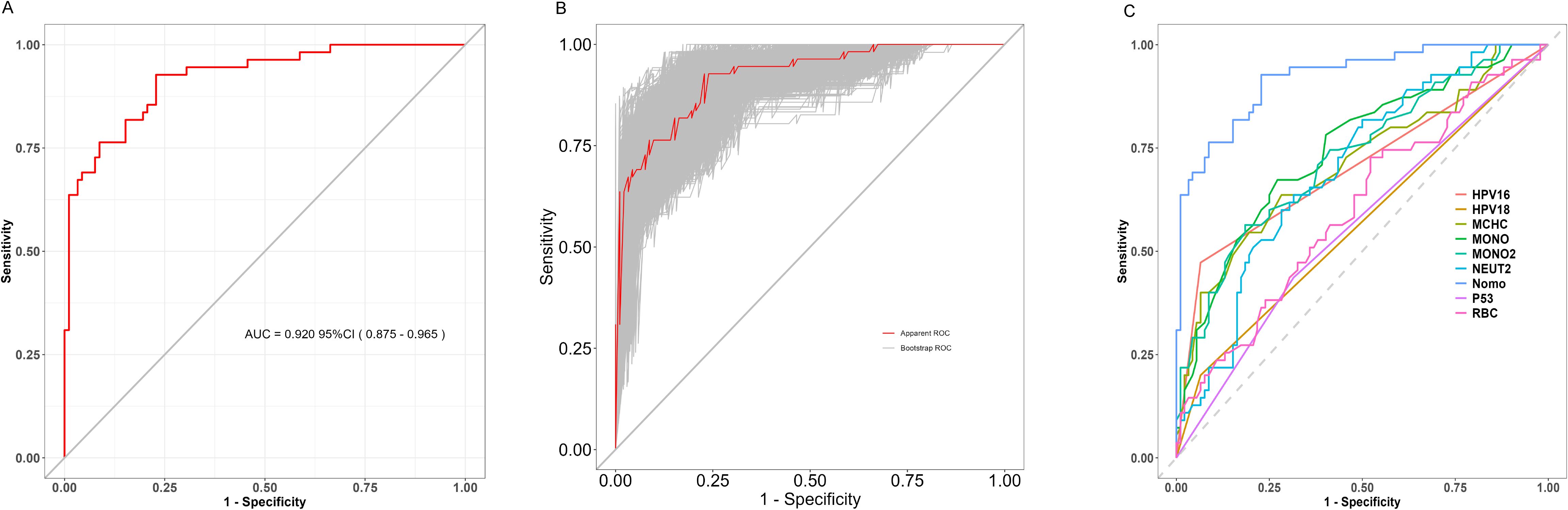
Figure 2. (A) ROC curve; (B) Bootstrap Validation of Model Stability (1000 Resampling Iterations); (C) ROC Curve Analysis of Diagnostic Model Performance.
The calibration of the model was further assessed using the Hosmer-Lemeshow goodness-of-fit test and a calibration curve. The Hosmer-Lemeshow test yielded a χ² value of 5.1654, P = 0.7398 (P > 0.05), indicating no statistically significant difference between the predicted and observed values. Figure 3 shows the proximity between the prediction curve, drawn from 1000 bootstrap resamples, and the reference line, demonstrating good agreement between the model’s predictions and actual outcomes.
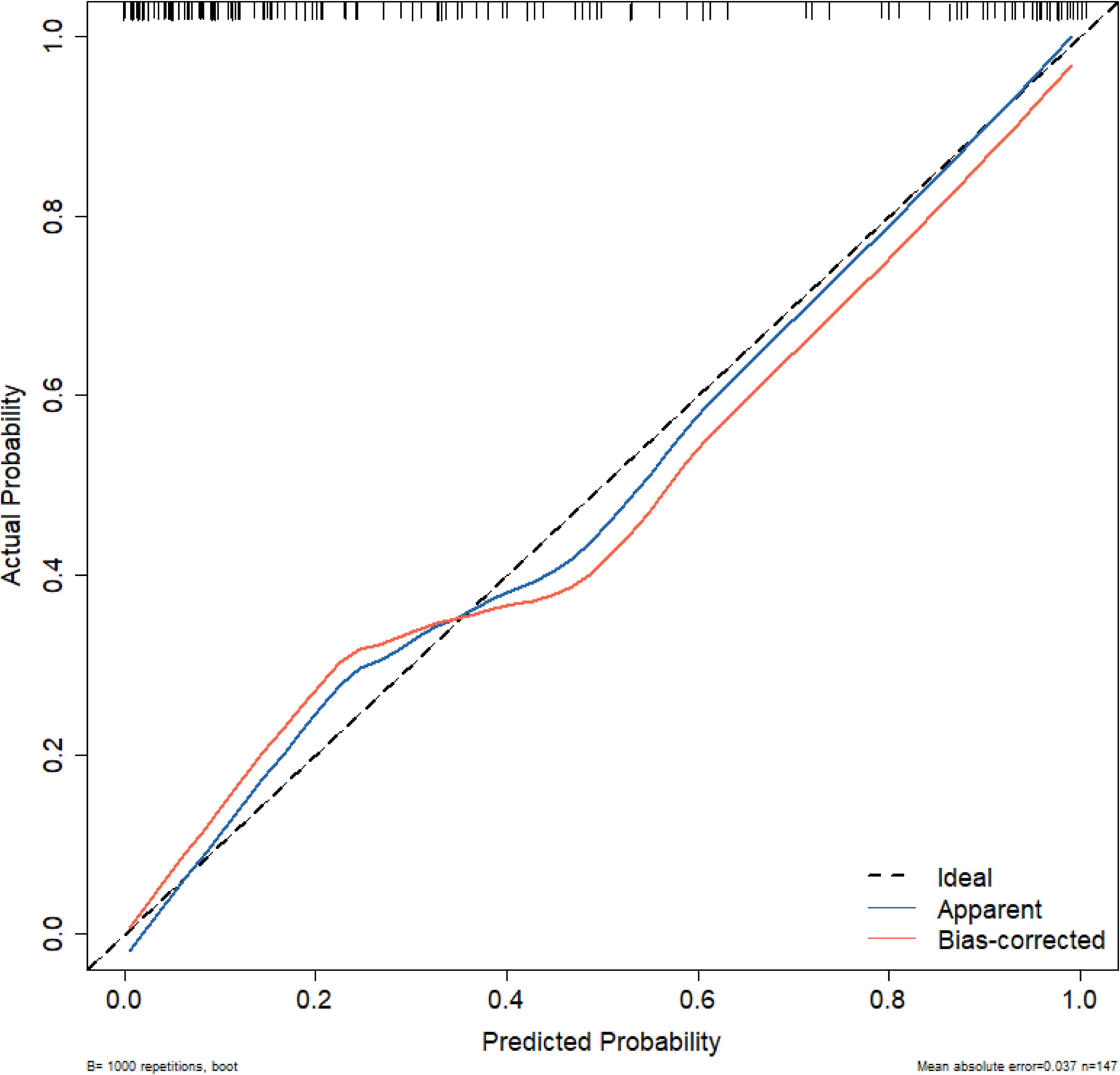
Figure 3. Calibration curve for the nomogram model. Ideal: The ideal curve; Apparent: The original data model; Bias-corrected: The model prediction after 1000 bootstrap resampling corrections.
Lastly, Figure 4 presents the DCA, generated from 1000 bootstrap resamples. The results suggest that the model provides substantial clinical benefit and can be a valuable tool for guiding clinical decision-making.
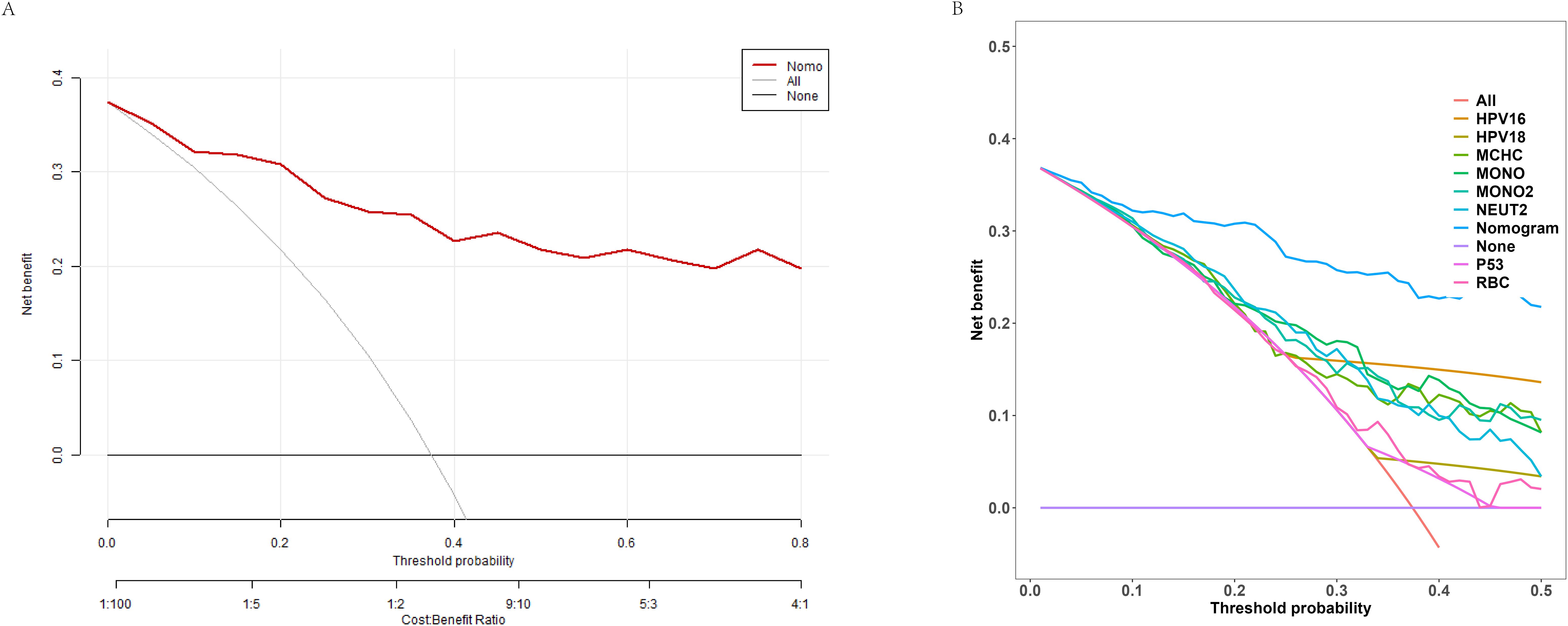
Figure 4. Decision curve analysis (DCA) of the nomogram model. (A) Net benefit curve of the nomogram predictive model at different decision probability thresholds. (B) Comparison of net benefits between different variables and the predictive model at various probability thresholds.
4 Discussion
The association analysis between the p53 gene and cervical cancer (Table 1) revealed that the C/C genotype significantly reduced the risk of cervical cancer, suggesting a potential protective effect of the C allele, which aligns with findings from previous studies (8, 17). Although statistical significance was not achieved in other models, the results indicated that the C allele might be associated with a lower risk of cervical cancer. The log-additive model results further support the idea that the C allele may exert a dose-dependent protective effect. These findings suggest that the p53 genotype may influence cervical cancer risk through a complex genetic interplay. Overall, these results provide valuable insights for further investigation into the role of the p53 genotype in the development of cervical cancer.
Infection with HPV16 significantly increases the risk of cervical cancer, with a more pronounced risk observed in individuals with the G/G genotype. This finding may suggest that the C allele offers some degree of protection; however, no significant interaction between the two was identified. This insight enhances our understanding of the complex relationship between genotype and HPV infection in the development of cervical cancer, which should be further validated in larger-scale studies in the future.
In individuals infected with HPV18, those with the G/C or C/C genotype exhibited an OR of 0 when compared to the G/G genotype (Table 2). This finding is particularly intriguing, as an OR of 0 suggests that individuals carrying at least one C allele (G/C or C/C) appear to have no risk, or an extremely low risk, of developing cervical cancer in the context of HPV18 infection. The p53 genotype significantly influences cervical cancer risk among HPV18-infected individuals, indicating that different p53 variants may have varying effects in response to HPV18.
The G/C and C/C genotypes appear to confer a strong protective effect against cervical cancer in those infected with HPV18, suggesting that the C allele could play a crucial role in resisting cancer progression induced by HPV18, potentially through more effective suppression of HPV18-driven oncogenesis. If validated, these results could have significant implications for risk assessment and management strategies in HPV18-infected individuals, with those carrying the G allele possibly requiring tailored follow-up and intervention protocols. However, it is essential to note that the sample size of this study (n = 147) is relatively small, which may affect the stability and generalizability of the findings. Furthermore, an OR of 0 is an extreme outcome, highlighting the need for larger studies to validate this result and explore the underlying mechanisms further.
Significant differences were observed in several blood routine parameters between the cervical cancer group and the control group. The reduction in RBC and HGB may indicate an anemic state in cervical cancer patients, potentially linked to the biological characteristics of the tumor, malnutrition, or chronic bleeding. The NEUT% and decrease in LY suggest the presence of a systemic inflammatory response, which may be associated with the tumor’s immune evasion mechanisms (18). Additionally, the decrease in MONO and its percentage may reflect immune suppression (19, 20). It is noteworthy that age may also influence hematologic parameters (21). For instance, increasing patient age may lead to decreased hemoglobin levels and affect systemic inflammatory indices. In this study, we implemented an age-matched design between the two groups to mitigate such confounding effects. These findings provide critical insights for the early diagnosis and clinical monitoring of cervical cancer. Specifically, indicators such as the NLR, SII, and PLR may possess significant clinical value in assessing cervical cancer. This underscores the rationale for including these parameters in the predictive model for cervical cancer developed in this study.
Following the univariate logistic regression and LASSO regression analyses of variables such as high-risk HPV, p53 genotypes, and blood routine indicators, eight variables were identified as being significantly associated with cervical cancer risk: p53 genotype, HPV16, HPV18, MONO, MONO%, NEUT%, MCH, and RBC. The significance of HPV16 and HPV18 underscores their critical roles in the development of cervical cancer, aligning with findings in the existing literature (22). Furthermore, the significance of MONO and MONO% indicates that variations in monocyte counts may be linked to immune responses within the tumor microenvironment (23). Additionally, the significance of MCH and RBC suggests that red blood cells and related parameters may also play an important role in the pathogenesis of cervical cancer.
The AUC was 0.920, demonstrating that the model developed in this study has a remarkably high discriminatory ability, effectively distinguishing cervical cancer patients from non-cancer cases. Furthermore, the results of the Hosmer-Lemeshow goodness-of-fit test and calibration curve (χ² = 5.1654, P = 0.7398) indicate that the model’s predictions closely align with actual outcomes. The DCA further shows that in the moderate to high probability threshold range, the model provides a substantial net benefit, reinforcing its practical value in clinical applications.
In conclusion, this study analyzed the p53 genotype, high-risk HPV types, and routine blood parameters in patients with cervical cancer and a control group. We identified significant differences between the two groups and highlighted the potential clinical value of these variables. Key variables associated with cervical cancer risk were identified, and an effective predictive model was constructed. This model demonstrated good discrimination and calibration, and showed substantial benefits in clinical decision-making, providing new insights for preliminary screening and risk prediction of cervical cancer. The study also emphasized the interaction between HPV infection and p53 tumor suppressor gene mutations, highlighting their importance in cancer development. In particular, the interaction between HPV16, HPV18, and p53 genotypes, and its impact on cervical cancer onset, was explored—a topic that has not been thoroughly addressed in existing literature. Future studies should further validate the applicability of this model in larger sample sizes and investigate its potential use in other cancer types. By optimizing detection methods and risk assessment tools, we hope to provide more effective strategies for clinical practice to reduce the incidence and mortality of cervical cancer.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by the Ethics Committee of the First People’s Hospital of Changzhou on December 17, 2023 (Approval No (2023):195). The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’ legal guardians/next of kin because the retrospective nature of the study and the anonymization of all samples prior to analysis.
Author contributions
CS: Data curation, Investigation, Visualization, Writing – original draft, Writing – review & editing. JZ: Formal analysis, Investigation, Methodology, Writing – review & editing. LP: Formal analysis, Writing – review & editing. SY: Funding acquisition, Methodology, Writing – review & editing. FZ: Validation, Writing – review & editing. LJ: Validation, Writing – review & editing. MY: Funding acquisition, Methodology, Writing – review & editing. GL: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Visualization, Writing – review & editing. XJ: Conceptualization, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by Natural Science Foundation of Jiangsu Province (BK20211063), Jiangsu Maternal and Child Health Association Project (FYX202015), Changzhou Applied Basic Research Project (CJ20210113), and Leading Talent of Changzhou “The 14th Five-Year Plan” High-Level Health Talents Training Project (grant no. 2022260) of China.
Acknowledgments
We extend our heartfelt gratitude to all the participants who contributed to this study, as well as to the dedicated staff at our hospital who offered invaluable support throughout the program.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1541928/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Yu YQ, Hao JQ, Mendez MJG, Mohamed SB, Fu SL, Zhao FH, et al. The prevalence of cervical HPV infection and genotype distribution in 856,535 chinese women with normal and abnormal cervical lesions: A systemic review. J Cytol. (2022) 39:137–47. doi: 10.4103/joc.joc_42_22
3. Li T, Yang Z, Luo P, Yang Y, Lin Z, and Mei B. Genetic variability of human papillomavirus type 18 based on E6, E7 and L1 genes in central China. Virol J. (2024) 21:152. doi: 10.1186/s12985-024-02424-9
4. Holthaus D, Rogmans C, Gursinski I, Quevedo-Olmos A, Ehsani M, Mangler M, et al. Inhibition of ADAM17 increases the cytotoxic effect of cisplatin in cervical spheroids and organoids. Front Oncol. (2024) 14:1432239. doi: 10.3389/fonc.2024.1432239
5. Mai Q, Yang X, Cheng H, Wu G, and Wu Z. Prevalence and genotype distribution of human papillomavirus among women with cervical lesions in Shenzhen city, China. Hum Vaccin Immunother. (2021) 17:965–71. doi: 10.1080/21645515.2020.1805993
6. Pu X, Gu Z, and Wang X. Polymorphisms of the interleukin 6 gene and additional gene-gene interaction contribute to cervical cancer susceptibility in Eastern Chinese women. Arch Gynecol Obstet. (2016) 294:1305–10. doi: 10.1007/s00404-016-4175-x
7. Yi K, Yang L, Lan Z, and Xi M. The association between MTHFR polymorphisms and cervical cancer risk: a system review and meta analysis. Arch Gynecol Obstet. (2016) 294:579–88. doi: 10.1007/s00404-016-4037-6
8. SHSAMPDe. Is there a biological plausability for p53 codon 72 polymorphism influence on cervical cancer development? Acta Med Portuguesa. (2011) 24:127–34. doi: 10.20344/amp.335
9. Alsbeih GA, Al-Harbi NM, Bin Judia SS, Khoja HA, Shoukri MM, and Tulbah AM. Reduced rate of human papillomavirus infection and genetic overtransmission of TP53 72C polymorphic variant lower cervical cancer incidence. Cancer. (2017) 123:2459–66. doi: 10.1002/cncr.v123.13
10. Mostaid MS, Mumu SB, Haque MA, Sharmin S, Jamiruddin MR, Sayedur Rahman GM, et al. Elevated serum expression of p53 and association of TP53 codon 72 polymorphisms with risk of cervical cancer in Bangladeshi women. PloS One. (2021) 16:e0261984. doi: 10.1371/journal.pone.0261984
11. Apu MNH, Rashed AZM, Bashar T, Rahman MM, and Mostaid MS. TP53 genetic polymorphisms and susceptibility to cervical cancer in Bangladeshi women: a case-control study. Mol Biol Rep. (2020) 47:4357–64. doi: 10.1007/s11033-020-05523-2
12. Nost TH, Alcala K, Urbarova I, Byrne KS, Guida F, Sandanger TM, et al. Systemic inflammation markers and cancer incidence in the UK Biobank. Eur J Epidemiol. (2021) 36:841–8. doi: 10.1007/s10654-021-00752-6
13. Nakamoto S, Ohtani Y, Sakamoto I, Hosoda A, Ihara A, and Naitoh T. Systemic immune-inflammation index predicts tumor recurrence after radical resection for colorectal cancer. Tohoku J Exp Med. (2023) 261:229–38. doi: 10.1620/tjem.2023.J074
14. Zhao H, Wu L, Yan G, Chen Y, Zhou M, Wu Y, et al. Inflammation and tumor progression: signaling pathways and targeted intervention. Signal Transduct Target Ther. (2021) 6:263. doi: 10.1038/s41392-021-00658-5
15. Zhang J, Yao S, Yu Y, Yu M, and Luo G. Development of a typing detection method for high−risk human papillomavirus and related tumor suppressor genes p53 and RB1 based on two−dimensional PCR technology. Zhonghua Jian Yan Yi Xue Za Zhi. (2024) 47:391–400. doi: 10.3760/cma.j.cn114452-20240204-00069
16. Zhan Y, Zhang J, Yao S, and Luo G. High-throughput two-dimensional polymerase chain reaction technology. Anal Chem. (2020) 92:674–82. doi: 10.1021/acs.analchem.9b02030
17. Guo H, Wen Z, Yang S, and Qi H. Association of p73 G4C14-A4T14 and p53 codon 72 polymorphism with cervical cancer in Chinese population. Indian J Cancer. (2022) 59:33–8. doi: 10.4103/ijc.IJC_538_19
18. Kao KC, Vilbois S, Tsai CH, and Ho PC. Metabolic communication in the tumour-immune microenvironment. Nat Cell Biol. (2022) 24:1574–83. doi: 10.1038/s41556-022-01002-x
19. Wang M, Chen S, He X, Yuan Y, and Wei X. Targeting inflammation as cancer therapy. J Hematol Oncol. (2024) 17:13. doi: 10.1186/s13045-024-01528-7
20. Coussens LM and Werb Z. Inflammation and cancer. Nature. (2002) 420:860–7. doi: 10.4103/aam.aam_56_18
21. Abukanna AMA, AlAnazi FM, AlAnazi ZM, AlAnazi FAL, AlAnaz AHO, and AlAnazi RML. Aging and changes in white blood cells count and immunity: A systematic review. Clin Cancer Invest J. (2022) 11:25–30. doi: 10.51847/kcAWdH6o97
22. Bansal A, Singh MP, and Rai B. Human papillomavirus-associated cancers: A growing global problem. Int J Appl Basic Med Res. (2016) 6:84–9. doi: 10.4103/2229-516X.179027
Keywords: cervical cancer, blood routine parameters, p53, high-risk HPV, nomogram
Citation: Sun C, Zhang J, Pan L, Yao S, Zhang F, Ji L, Yu M, Luo G and Jiang X (2025) Innovative nomogram for cervical cancer prediction: integrating high-risk HPV infection, p53 genotype, and blood routine parameters. Front. Oncol. 15:1541928. doi: 10.3389/fonc.2025.1541928
Received: 09 December 2024; Accepted: 28 April 2025;
Published: 20 May 2025.
Edited by:
Ajaz Bhat, Sidra Medicine, QatarReviewed by:
Katerina Kubelka-Sabit, Department od histopathology and cytology, North MacedoniaYara Lucia Furtado, Federal University of Rio de Janeiro, Brazil
Copyright © 2025 Sun, Zhang, Pan, Yao, Zhang, Ji, Yu, Luo and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guanghua Luo, c2hpbmVyb2FyQDE2My5jb20=; Xiping Jiang, MTY5NTE3MTQ2MkBxcS5jb20=
†These authors have contributed equally to this work
‡ORCID: Cheng Sun, orcid.org/0009-0001-8705-7186
Jun Zhang, orcid.org/0000-0002-1826-6099
Lili Pan, orcid.org/0000-0001-8164-7890
Shuang Yao, orcid.org/0000-0003-3686-5404
Fenghua Zhang, orcid.org/0009-0009-6088-1309
Linjuan Ji, orcid.org/0009-0002-7655-5777
Miaomei Yu, orcid.org/0000-0002-8287-0725
Guanghua Luo, orcid.org/0000-0001-8339-2828
Xiping Jiang, orcid.org/0009-0007-5429-7902
 Cheng Sun1†‡
Cheng Sun1†‡ Miaomei Yu
Miaomei Yu Guanghua Luo
Guanghua Luo