- 1Department of Nuclear Medicine, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Hubei Province Key Laboratory of Molecular Imaging, Wuhan, China
- 3Key Laboratory of Biological Targeted Therapy, the Ministry of Education, Wuhan, China
- 4Cancer Center, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei, China
- 5Department of Breast and Thyroid Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei, China
The incidence of thyroid cancer has been rising in recent years. While tumorigenesis has traditionally been attributed to the accumulation of genetic mutations in oncogenes and tumor suppressor genes, increasing attention has been directed toward the role of epigenetic regulation in cancer development. Since the 1980s, however, it has been acknowledged that the role of another key regulatory system in carcinogenesis: epigenetics, shedding light on the regulation of gene expression without altering the DNA sequence.This review synthesizes current literature on epigenetic alterations in follicular cell-derived thyroid cancers, focusing on DNA methylation, histone modifications, chromatin remodeling, and RNA regulation. Evidence indicates that dysregulation of these epigenetic processes is prevalent in thyroid cancer, influencing tumor initiation, progression, and resistance to therapy. Several epigenetic inhibitors are under development, some demonstrating synergy with existing chemotherapies and immunotherapies. Understanding these mechanisms may facilitate the development of novel, more effective strategies for early detection and treatment.
1 Introduction
Classical genetics has posited that genetic information is primarily determined by the DNA sequence (1). However, as scientific exploration advanced, it became evident that classical genetics alone could not fully explain certain biological phenomena. For instance, Morgan et al. demonstrated that the expression of the white gene in fruit flies is influenced by its location in either heterochromatin or euchromatin, a phenomenon linked to the local environment of the nucleus (2). Building on such findings, developmental biologist Waddington introduced the term “epigenetics” to describe the interaction between genes and environmental factors in shaping phenotypes, establishing it as a distinct field in biology (3). Epigenetics now refers to heritable modifications in gene expression that do not involve changes to the DNA sequence itself (4). These modifications can be stably transmitted through cell division, and serve as a crucial link between environmental cues and gene expression. They are essential for various biological processes including development, cell proliferation, aging and others. On the other hand, the dysregulation of epigenetic mechanisms is fundamentally involved in numerous pathophysiological processes like cancer, psychiatric diseases, cardiovascular diseases, metabolic diseases, autoimmune diseases, and so on (5–7). In the 1980s, Feinberg and Vogelstein first correlated epigenetic changes with cancer, marking a turning point in understanding cancer biology. Emerging evidence has highlighted the prevalence of epigenetic modifications in most tumors, underscoring their critical role in tumor initiation and progression (8). For instance, recent research from the Van Andel Institute revealed that the loss of two key protective proteins induces abnormal DNA methylation, converting healthy lung cells into cancer cells (9). This finding demonstrated that cancer cells can be exclusively engendered by epigenetic modifications, highlighting the significance of epigenetic changes in cancer. Thus, understanding the underlying epigenetic mechanisms driving cancer occurrence and progression is imperative.
Thyroid cancer stands as the most common malignant tumor of the endocrine system, with a global incidence that continues to rise (10, 11). It was the ninth most common cancer worldwide in 2020 and is projected to become the fourth most diagnosed cancer in the United States by 2030, imposing a substantial socioeconomic burden (12). Follicular cell-derived thyroid cancers include papillary thyroid cancer (PTC), follicular thyroid cancer (FTC), and anaplastic thyroid cancer (ATC). Differentiated thyroid cancer (DTC), which include PTC and FTC, account for over 90% of cases and typically have an excellent prognosis, with a 5-year survival rate exceeding 90% (13, 14). ATC is a rare form of thyroid cancer, comprising less than 5% of all thyroid cancers, but displays a highly aggressive phenotype (15, 16). ATC has a dismal prognosis and high lethality and accounts for 14–39% of thyroid cancer related death with a median survival of just 5–6 months (17). The diverse molecular, histological, and clinical profiles of these subtypes necessitate the identification of reliable biomarkers for precise diagnosis and effective management.
Early detection and accurate diagnosis are paramount for improving thyroid cancer outcomes. Clinically, the diagnosis of thyroid nodules is mainly based on physical examination by clinicians and thyroid ultrasound examination. However, differentiating between benign and malignant thyroid nodules remains challenging. Fine-needle aspiration biopsy (FNAC) provides high sensitivity (62–89%) and specificity (71–100%), approximately 15–30% of thyroid nodules yield indeterminate results (18–21). This diagnostic uncertainty complicates clinical decision-making, as benign nodules generally require observation, while malignant ones necessitate surgical intervention (22). Therefore, there is a pressing need for non-invasive diagnostic tools to differentiate benign from malignant nodules preoperatively.
Surgical resection remains the cornerstone of thyroid cancer management, with radioactive iodine (RAI) therapy serving as a standard adjuvant treatment for DTC following thyroidectomy (23). Although the prognosis for most DTC patients is favorable, approximately two-thirds of metastatic DTC cases exhibit RAI resistance, limiting therapeutic options (24). For ATC, a multidisciplinary approach combining surgery, radiotherapy, and chemotherapy is employed, yet the prognosis remains grim (25). These thyroid cancers currently account for the majority of thyroid cancer-related deaths, highlighting the necessity of exploring molecular mechanisms driving thyroid cancer progression and developing more effective therapeutic strategies.
Epigenetic modifications are heritable but also differ considerably based on tissue types, different developmental stages, and environmental factors (26). Epigenetics holds substantial implications for health, disease, and genetic disorders, as environmental and lifestyle factors can influence epigenetic changes, affecting an individual’s health and disease risk. While the precise etiology and pathogenesis of thyroid cancer remain incompletely understood, some risk factors for thyroid cancer have been identified, most of which can be detected and prevented early. These risk factors primarily include genetic factors (e.g., gene mutations in B-raf proto-oncogene serine/threonine kinase (BRAF), rat sarcoma (RAS) gene family, family history of thyroid cancer), demographic factors (e.g., higher incidence in females, white and Asian populations), and lifestyle habits (e.g., obesity, abnormal iodine intake, history of radiation exposure, stress) (27, 28). The phenotype is shaped by a complex interplay of environmental factors, genetic elements, and epigenetic mechanisms that interact with each other (Figure 1).
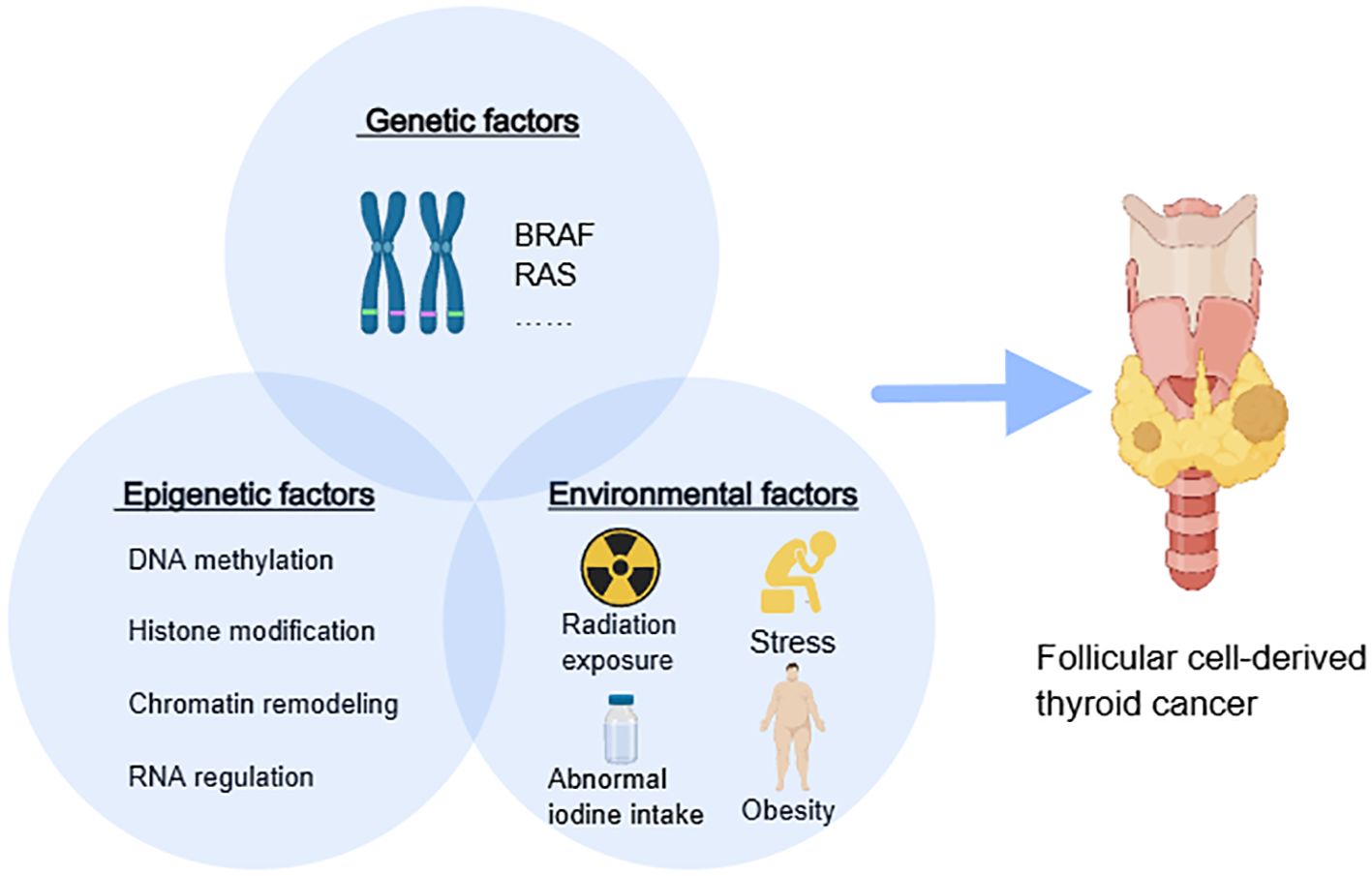
Figure 1. The interaction among epigenetics, genetics, and the environment. BRAF, B-Raf proto-oncogene serine/threonine kinase; RAS, rat sarcoma gene family.
Mounting evidence indicates that epigenetic modifications play a significant role in the initiation and progression of follicular cell-derived thyroid cancers (29). However, on the one hand, current literature predominantly focuses on isolated events such as DNA methylation or histone modifications, neglecting their dynamic interplay with chromatin architecture and signaling pathways. For instance, recent findings reveal that SETMAR-mediated activation of SMARCA2, a core component of the switching defective/sucrose nonfermenting (SWI/SNF) chromatin remodeling complex, drives thyroid cancer differentiation by altering chromatin accessibility of key differentiation genes (30). On the other hand, the progress in translating epigenetic insights into precise treatments in clinical practice is lagging behind (31). Therefore, this review provides an overview of the prevailing mechanisms of epigenetic modifications in follicular cell-derived thyroid cancer including PTC, FTC ATC, prospects the future development of epigenetics and postulates opportunities for applications in thyroid diseases.
2 Epigenetic alterations in thyroid cancer
Thyroid cancer, especially follicular cell-derived thyroid cancer, has some unique epigenetic patterns, such as relatively low whole genome methylation levels, specific non-coding RNA expression patterns, and specific histone modification patterns, which are closely related to the occurrence and development of thyroid cancer (32, 33). Compared with other types of cancer, the epigenetic features of thyroid cancer may be more stable and have specific molecular markers, providing potential research directions for early diagnosis, prognosis evaluation, and targeted therapy of thyroid cancer. Epigenetic modifications primarily regulate chromatin structure and gene expression through four mechanisms, including DNA methylation, histone modifications, chromatin remodeling, and RNA regulation (Figure 2) (34). These aberrant epigenetic modifications are associated with all stages of tumor development, covering tumor genesis, invasion, metastasis, as well as therapy resistance (35).
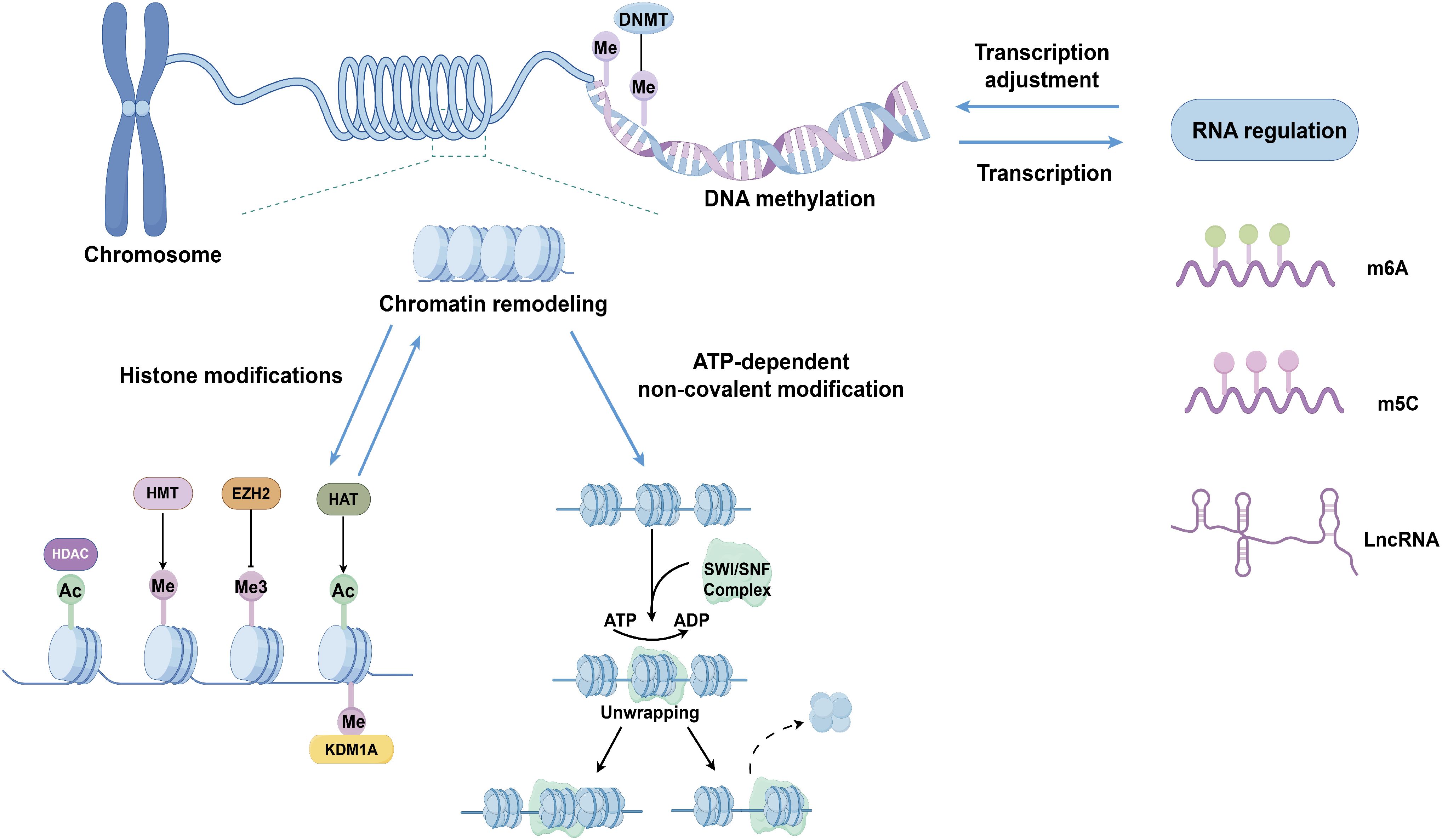
Figure 2. Epigenetic mechanisms involved in follicular cell-derived thyroid cancer: DNA methylation, histone modifications, chromatin remodeling, and RNA regulation. DNMT, DNA methyltransferase; Me, methylation; Me3 trimethylation Ac, acetylation; HDAC, histone deacetylase; KDM1A, lysine demethylase 1A; HMT Histone methyltransferase; EZH2, enhancer of zeste homolog 2; HAT, histone acetyltransferase; SWI/SNF, switching defective/sucrose nonfermenting; lncRNA, long non-coding RNA.
2.1 DNA methylation
DNA methylation, one of the earliest discovered epigenetic mechanisms, plays a pivotal role in regulating diverse cellular processes, including differentiation, proliferation, chromosomal stability, genomic imprinting, and X-chromosome inactivation (36). Aberrant DNA methylation is frequently implicated in various diseases, including cancers. DNA methylation refers to the covalent addition of a methyl group to the 5-position carbon of cytosine in cytosine-phosphate-guanine (CpG) island (regions in DNA with a high density of cytosine-guanine dinucleotides), forming 5-methylcytosine (5mC) (37). This process is mediated by DNA methyltransferases (DNMTs), which play essential roles in establishing and maintaining DNA methylation patterns (38). In cancer cells, DNA methylation alterations are characterized by hypermethylation of promoter CpG islands and global genomic hypomethylation, leading to the activation of oncogenes and inactivation of tumor suppressor genes (39, 40). Promoter hypermethylation of tumor suppressor genes silences their expression, facilitating tumorigenesis. Tumor suppressor genes such as RAS association domain family 1 isoform A, phosphatase and tensin homolog, retinoic acid receptor 2, and tissue inhibitor of metalloproteinase 3 (TIMP3) exhibit hypermethylation in thyroid cancer, leading to their inactivation (41–43). For example, TIMP3 promoter methylation in papillary thyroid cancer (PTC) correlates with tumor aggressiveness and the presence of the BRAFV600E mutation, underscoring its potential role as an oncosuppressor (44). Thyroid-specific genes such as thyrotropin-stimulating hormone receptor (TSHR) and sodium-iodide symporter (NIS) also undergo abnormal methylation in thyroid cancer. This results in reduced iodine uptake and resistance to RAI therapy (45, 46). DNA hypomethylation, one of the characteristics of tumor cells, is considered to be associated with chromosomal instability, causing aberrant cellular proliferation and ultimately inducing tumors (47). DNA hypomethylation in repetitive sequences, such as Alu elements, contributes to genomic instability and the activation of oncogenes. In healthy cells, the pericentric heterochromatin is highly methylated, which effectively silences repetitive genomic sequences, thereby ensuring the integrity and stability of the genome. However, in numerous tumor cells, this regulatory mechanism becomes disrupted, resulting in the loss of DNA methylation in regions that are typically inactive. Consequently, transposons are reactivated and can integrate into any genomic site, thereby inducing mutations and genomic instability. These alterations may potentially contribute to tumor progression or various disease stages. Alu hypomethylation has been observed in distant metastatic DTC and ATC, suggesting a role in tumor progression and dedifferentiation (48).
DNA methylation alterations often occur early in tumorigenesis, making them promising biomarkers for early cancer detection (49). Cell-free DNA (cfDNA) carrying DNA methylation information has emerged as promising non-invasive targets for cancer detection (50). Hong et al. developed and validated ThyMet classifier, a cfDNA-based methylation marker panel that non-invasively and accurately differentiates PTC from benign thyroid nodule (BTN) (51). The ThyMet demonstrated enhanced performance in differentiating PTC from BTN compared to thyroid ultrasonography grades, while maintaining a comparable sensitivity. The integration of ThyMet and thyroid ultrasonography grades further improved the accuracy. In a word, ThyMet combined ultrasonography has complementary advantages and can provide an effective way to optimize the management of thyroid nodule population. DNA hydroxymethylation, another type of DNA modification, serves as a sign of active DNA demethylation processes (52). Reduced 5-hydroxymethylcytosine (5-hmC) levels have been documented in PTC and ATC, particularly in cases with telomerase reverse transcriptase (TERT) promoter mutations, highlighting its potential as a biomarker for distinguishing cancer patients from healthy individuals (53, 54).
Emerging evidence also links DNA methylation to drug resistance (55, 56). Downregulation of tumor suppressor expression by DNA methylation promotes tumor growth and enhances drug tolerance. For instance, promoter hypermethylation of insulin-like growth factor binding protein-3 (IGFBP-3), a tumor suppressor gene, has been associated with enhanced tumor growth and drug resistance (57).
From a therapeutic perspective, DNMT inhibitors such as decitabine and 5-azacytidine, approved for treating myelodysplastic syndromes, have shown promise in thyroid cancer (Table 1). They are nucleoside analogues which bind to DNA and form irreversible complexes with DNMTs during the S phase of the cell cycle, leading to the degradation of DNMTs. Studies have demonstrated that decitabine treatment effectively inhibits the proliferation of both undifferentiated and differentiated thyroid cancer cells (58, 59). Notably, a phase II clinical trial revealed that patients with metastatic PTC or FTC, who were unresponsive to RAI, exhibited recovered RAI uptake in metastatic lesions subsequent to decitabine therapy. This suggests the combination of DNMT inhibitors and radioactive iodine therapy may provide a new option for cancer patients who do not respond well to current treatment options. Iodide serves as an essential constituent of thyroid hormones and is actively transported into thyroid follicular cells via the basilar membrane-bound sodium-iodide symporter (NIS) protein. The functional integrity of NIS is critical for the effective concentration of radioactive iodine within the thyroid gland, where it becomes incorporated into colloid and decomposes into cytotoxic beta and gamma rays, inducing DNA damage, apoptosis, and cell death. Consequently, the absence or dysfunction of NIS expression is postulated to play a significant role in the development of iodine-resistant thyroid tumors (60). Hypermethylation of the NIS gene promoter and global epigenetic dysregulation of differentiation related genes may lead to the development of RAI resistance and affect clinical efficacy (61).
DNA methylation holds significant promise in thyroid cancer pathogenesis, offering valuable diagnostic and therapeutic potential. Molecular diagnostics targeting methylation alterations can complement existing diagnostic methods, while DNMT inhibitors hold promise for overcoming treatment resistance and enhancing patient outcomes. Continued exploration of DNA methylation biomarkers and therapies will likely transform thyroid cancer management.
2.2 Histone modifications
Histones, the most abundant proteins bound to DNA, are key components of chromatin and play a vital role in gene regulation. Histones have five main types: H1, H2A, H2B, H3 and H4. They have similar structures, but differ slightly in some ways to accommodate different chromosomal structures and functions. Post-translational modifications to the N-terminal tails of histones, including acetylation, methylation, ubiquitination, and phosphorylation, regulate chromatin accessibility and transcriptional activity (62). Relaxed chromatin states facilitate promoter region accessibility, promoting gene expression, while condensed chromatin restricts access, silencing genes (63, 64). Among these, acetylation and methylation are the most extensively studied and are recognized as critical epigenetic markers (65).
2.2.1 Histone acetylation
Histone acetylation is dynamically regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). Histone acetylation occurs mostly on the N-terminal lysine residues of histones H3 and H4. Acetylation neutralizes the positive charge on lysine (K) residues, weakening histone-DNA interactions and resulting in an open chromatin configuration that promotes transcription (66, 67). Studies have revealed a strong correlation between histone acetylation levels and thyroid cancer progression. Pupppin et al. reported that acetylation lysine residues at positions 4 and 9 of histone H3 were elevated in follicular adenomas, PTC, FTC, and ATC compared with normal tissue, with different characteristics in different subtypes (68). Specifically, acetylation at H3K18 was observed in differentiated thyroid cancers but remained unaltered in ATC, suggesting its potential as a marker for disease progression. Further research is warranted to explore whether these histone acetylation profiles can improve the diagnostic accuracy of fine-needle aspiration cytology (FNAC) in thyroid nodules.
Histone deacetylase inhibitors (HDACis) are promising agents for reprogramming thyroid cancer cells by restoring histone acetylation (69–71). HDACis, such as valproic acid (VA), panobinostat, vorinostat (SAHA), and trichostatin A (TSA), have demonstrated the ability to reinduce NIS expression, thereby enhancing iodine uptake in thyroid cells (72–74). However, clinical efficacy has been limited. For instance, a phase II clinical trial using VA in patients with radioiodine-refractory follicular thyroid cancer revealed only modest reductions in serum thyroglobulin levels without significant tumor shrinkage (75). Despite these limitations, HDACis have shown potential in combination therapies. SAHA, when combined with chemotherapeutic agents such as doxorubicin, carboplatin, and paclitaxel, exhibited synergistic anti-tumor effects (76). Moreover, panobinostat combined with MAP kinase inhibitors (MAPKi) demonstrated enhanced BRAFV600E-dependent redifferentiation effects, highlighting the potential for combination strategies to improve therapeutic outcomes (72).
2.2.2 Histone methylation
Histone methylation occurs primarily at arginine and lysine residues of histones H3 and H4 and is mediated by histone methyltransferases (HMTs) and histone demethylases (HDMs) (77). Histone lysine residues can be monomethylated, dimethylated or trimethylated (m1, m2 and m3, respectively) (78). Histone methylation can activate or repress gene transcription, depending on which residue is methylated. For example, methylation at H3K4 is associated with transcriptional activation, while methylation at H3K9 is linked to gene repression (79). Lysine-specific demethylase 1A (LSD1/KDM1A), the first identified histone lysine demethylase, plays a critical role in thyroid cancer progression by regulating stemness and activating the Wnt/β-catenin signaling pathway (80, 81). KDM1A demethylates H3K4me1/2 of the adenomatous polyposis coli 2 (APC2) promoter region and the nonhistone substrate HIF-1α (hypoxia-inducible factor 1α), inhibiting APC2 transcription and activating HIF-1α/microRNA-146a/Dickkopf-1 axis, resulting in activating Wnt signaling pathway (79). GSK-LSD1, a KDM1A inhibitor, has demonstrated significant efficacy in inhibiting thyroid cancer progression and increasing chemotherapy sensitivity, suggesting potential clinical applications for advanced thyroid cancers (79).
Enhancer of zeste homolog 2 (EZH2), a key component of polycomb repressive complex 2 (PRC2), mediates H3K27 trimethylation, repressing transcription (82). The combination of the EZH2 inhibitor tazemetostat with MAPKi has shown promising results in PTC cells harboring BRAFV600E mutations. Tazemetostat alone modestly increased iodine-metabolizing gene expression and radioiodine uptake, effects significantly enhanced when combined with MAPKi (83). This combination synergistically reduced H3K27 trimethylation and improved PTC differentiation, suggesting its potential for differentiation therapy in radioiodine-refractory thyroid cancers.
Histone modifications, particularly acetylation and methylation, are emerging as critical regulators of thyroid cancer progression. The integration of histone modification profiling into diagnostic workflows, such as FNAC, may enhance preoperative diagnostic precision. Additionally, HDACis and HMT inhibitors hold promise in therapeutic strategies (Table 1), especially in combination with targeted therapies. Future research should focus on translating these findings into clinical applications to improve the management and outcomes of thyroid cancer patients.
2.3 Chromatin remodeling
DNA wraps around histones to form nucleosomes, the fundamental repeating subunits of chromatin (84). Chromatin remodeling refers to the dynamic modification of chromatin structure, which regulates gene expression by altering DNA accessibility for transcription factors and RNA polymerases (85, 86). Chromatin remodeling exists in two forms: highly compressed and silent heterochromatin, and relatively open and active euchromatin. In euchromatin, the DNA is in an “open” state, allowing transcription factors and RNA polymerases to bind to the DNA, thus activating or promoting gene expression. On the other hand, heterochromatin will hinder this process, causing gene expression to be blocked. Chromatin remodeling involves two main mechanisms (1): chemical modifications of histones, as previously discussed, and (2) ATP-dependent chromatin remodeling driven by specialized protein complexes that use ATP hydrolysis to reposition nucleosomes and alter DNA-histone interactions (87, 88). Four primary families of chromatin remodeling complexes have been identified based on structural and functional domains: SWI/SNF, imitation SWI (ISWI), chromodomain helicase DNA binding (CHD), and inositol requiring 80 (INO80) (89).
The SWI/SNF complex, also known as the BRG1/BRM-associated (BAF) complex, uses ATP hydrolysis to remodel nucleosomes, thereby enhancing DNA accessibility for transcriptional machinery (90). The complex binds to promoters, enhancers, and DNA replication initiation sites, and interacts with proteins involved in various cellular processes like the cell cycle (91). A study by Saqcena et al. demonstrated the essential role of SWI/SNF complexes in maintaining thyroid cell differentiation and suppressing thyroid cancer progression. Loss of SWI/SNF subunits in BRAFV600E-mutant thyroid cells reduced transcription factor expression and chromatin accessibility, impairing differentiation and radioiodine uptake (92). Compared with normal thyroid cells, BRAFV600E-mutant mouse thyroid cell lines showed lower expression of transcription factors and the accessibility to specific DNA binding sites, resulting in obstacles to thyroid differentiation gene expression and impaired RAI uptake. Blocking the MAPK pathway can reverse these effects in PTC without BRAF mutation, but not in BRAF-mutant thyroid cancer due to loss of SWI/SNF subunits, rendering them insensitive to its redifferentiation effects. These findings highlight SWI/SNF complexes as pivotal for thyroid cancer differentiation and potential therapeutic targets for RAI-refractory thyroid cancers.
2.4 RNA regulation
2.4.1 RNA modifications
It is well known that more than 170 types of RNA modifications have been discovered thus far that regulate gene expression, among which methylation modification is one of the most dominant forms (93). RNA methylation participates in the post-transcriptional gene expression regulation processes like RNA degradation, alternative, splicing and translational efficiency regulation (94).
N6-methyladenosine (m6A) methylation refers to the addition of a methyl group to the 6th nitrogen atom of the adenine base in RNA, facilitated by methyltransferases (95). m6A regulates RNA stability, splicing, translation, and degradation. Its role in cancer progression is context-dependent, with methyltransferase-like protein 3 (METTL3), a core methyltransferase, exerting both oncogenic and tumor-suppressive effects in cancers (96). METTL3 stabilizes the mRNA of six-transmembrane epithelial antigen of prostate 2 (STEAP2) through m6A modification, which is recognized by the m6A reader protein YTH N6-methyladenosine RNA-binding protein F1 (YTHDF1). STEAP2 acts as a tumor suppressor by inhibiting the Hedgehog signaling pathway and epithelial-to-mesenchymal transition (EMT), thereby reducing PTC cell proliferation, invasion, and migration. Rescue experiments further demonstrated that silencing STEAP2 reverses the tumor-suppressive effects induced by METTL3 overexpression, highlighting the METTL3-STEAP2 axis as a therapeutic target in PTC (97). METTL3 also plays a crucial role in maintaining cell viability and proliferation in ATC. Silencing METTL3 reduces overall m6A levels and significantly inhibits cell viability in ATC cell lines, demonstrating its importance in this aggressive thyroid cancer subtype (98). High-throughput sequencing revealed METTL3-dependent m6A modification on mRNAs associated with key pathways such as apoptosis and angiogenesis, suggesting potential pathways for therapeutic intervention. METTL3 promotes the stability of the long non-coding RNA LINC00894 through m6A modification in a YTHDC2-dependent manner. LINC00894 exerts a tumor-suppressive effect in PTC by inhibiting lymphangiogenesis and cancer cell proliferation via the Hippo signaling pathway (99). Downregulation of LINC00894 is associated with PTC progression, lymph node metastasis, and poor patient prognosis.
5-methylcytosine (m5C) modification refers to the addition of an active methyl group from the donor to the 5th carbon of the cytosine base in RNA (100). The RNA methyltransferase NOP2/Sun RNA Methyltransferase 2 (NSUN2) is significantly upregulated in ATC and promotes codon-dependent oncogenic translation by stabilizing tRNA (101). The study reveals that NSUN2 is significantly upregulated in ATC and is associated with tumor dedifferentiation and poor prognosis. NSUN2 enhances the stability of tRNA by catalyzing tRNA m5C modification, thereby supporting selective codon-dependent oncogenic protein synthesis. NSUN2 also promotes the maturation of its upstream transcription factor c-Myc and exhibits broad anti-cancer effects in both in vitro and in vivo experiments. Moreover, NSUN2 maintains the drug resistance of cancer cells to chemotherapy drugs. The research results indicate that NSUN2-mediated m5C tRNA modification is closely related to the progression of ATC, providing a new molecular basis for the development of potential therapeutic strategies targeting ATC.
m6A and m5C RNA modifications play crucial roles in thyroid cancer by regulating gene expression and translation. m6A, driven by METTL3, modulates key oncogenic and tumor-suppressor pathways, while m5C, mediated by NSUN2, supports codon-dependent translation for cancer progression. Both modifications show potential as biomarkers and therapeutic targets, offering new strategies for diagnosis, prognosis, and treatment of thyroid cancer (102, 103).
2.4.2 Non-coding RNAs
Only a small fraction of the transcriptional products of the human genome are translated into proteins to participate in biological activities, while the majority are non-coding RNAs (ncRNAs), constituting more than 90% of the genome. Recent studies have shown that ncRNAs, especially long non-coding RNAs (lncRNAs), play an important role in the occurrence and progression of thyroid cancer (104).
LncRNAs, longer than 200 nucleotides, can be located in the nucleus or cytoplasm and serve as key regulators of tumor pathogenesis, playing critical roles in tumor cell invasion, apoptosis and metastasis (105–107). LncRNAs can mediate the recruitment of chromatin-regulatory complexes to specific genomic loci and regulate gene expression through chromatin modifications (108–110). Common lncRNAs implicated in thyroid cancer include HOX antisense intergenic RNA (HOTAIR), papillary thyroid carcinoma susceptibility candidate 3, plasmacytoma variant translocation 1, metastasis associated lung adenocarcinoma transcript 1, growth arrest specific 5 (GAS5) and BRAF-activated non-protein coding RNA (111–114). Up-regulation of HOTAIR has been found in thyroid cancer and is associated with metastasis and poor prognosis (114). Thyroid cancer cells exhibit upregulated expression of HOTAIR compared to normal thyroid cells. Additionally, knockdown of HOTAIR inhibited thyroid cancer cell growth and invasion. Thus, lncRNAs hold promise as noninvasive and readily accessible biomarkers for the diagnosis and prognosis of thyroid cancer. In addition, lncRNAs in the cytoplasm are involved in the regulation of mRNA stability and function as miRNA sponges, thereby regulating gene expression (115). A study conducted by Li et al. found that GAS5 acts as a sponge for miR-362-5p, promoting sensitivity of thyroid cancer cells to iodine 131 by upregulating suppressor of morphogenesis in genitalia 1 (SMG1) and deactivating the Akt/mammalian target of rapamycin signaling pathway (112). The overexpression of GAS5 enhanced sensitivity to iodine 131 and inhibited the growth of thyroid cancer cells, while the upregulation of miR-362-5p had the opposite effect. The upregulation of miR-362-5p counteracted the effects of GAS5, and the overexpression of SMG1 negated the impact of miR-362-5p upregulation on iodine 131-resistant thyroid cancer cells. GAS5 competitively binds to miR-362-5p, and SMG1 is targeted by miR-362-5p.
Well-known functional ncRNAs are basically involved in epigenetic modification. They mainly function on the mRNA of target genes, and down-regulate the expression of target genes by degrading mRNA or inhibiting their translation. As research on ncRNAs in thyroid cancer progresses, lncRNAs are poised to offer significant potential for application in the diagnosis and prognosis assessment of thyroid cancer. LncRNAs are associated with treatment efficacy and prognosis, influencing radiotherapy resistance/sensitivity. Although there are currently no drugs targeting for the treatment of thyroid cancer that target non-coding RNAs, non-coding RNAs appear to be promising therapeutic targets that merit further research. Nevertheless, further investigation is required to elucidate the underlying molecular mechanisms and address challenges related to the translation of these findings into clinical practice.
Epigenetic mechanisms, including DNA methylation, histone modifications, chromatin remodeling, and RNA regulation, are interconnected in regulating gene expression during tumorigenesis. For instance, the methylation state of histone proteins in chromatin changes with different DNA modification modes and sequence changes, and the methylation state of histone lysine in chromatin may affect the modification of DNA itself (116, 117). These interconnected pathways shape tumor progression, metastasis, and therapeutic responses.
3 Future perspectives
In this review, we discuss epigenetic modifications that play a pivotal role in thyroid cancer. Firstly, we briefly introduced the concept and the characteristics of epigenetics. Then, we reviewed the mechanisms of epigenetic modifications, encompassing DNA methylation, histone modifications, chromatin remodeling and RNA regulation, and their roles in thyroid cancer. From a clinical point of view, most of the early clinical manifestations of thyroid cancer are thyroid nodules. Therefore, early detection and identification of patients with malignant thyroid nodules are an important preventive measure to reduce the risk of thyroid cancer death. The current study found that in the earliest stage of tumor development, the epigenetic changes may precede the gene mutations, thus epigenome detection is of great significance for the early diagnosis of tumors. DNA methylation and ncRNAs are promising biomarkers of thyroid cancer. Many candidate biomarkers have been identified in thyroid cancer and many epigenetic markers are under investigation (Table 2). The ideal biomarker set should be able to distinguish malignant and benign thyroid lesions with high diagnostic accuracy, and future studies should focus on identifying combinations of biomarkers to assist in the diagnosis of uncertain cytological cases. Different from genetic mutations, epigenetic modifications are mostly reversible. Therefore, from a theoretical point of view, it is easier to target epigenetics than genetics in cancer treatment, opening up new possibilities for cancer treatment development (118). Researchers are developing a variety of small molecule epigenetic inhibitors for tumor treatment. However, it has been found through available data that only a very small number of patients have a complete recovery from thyroid cancer (119, 120), which may be related to the significant differences in epigenetic abnormalities among different subtypes of thyroid cancer and the abnormal activation of the immune system caused by epigenetic drugs. In addition, it is worth noting that iodine levels or radiation exposure can greatly affect epigenetic changes and weaken drug sensitivity, which may be related to DNA methylation abnormalities and histone modification dysregulation caused by DNA damage and free radical generation (121, 122)
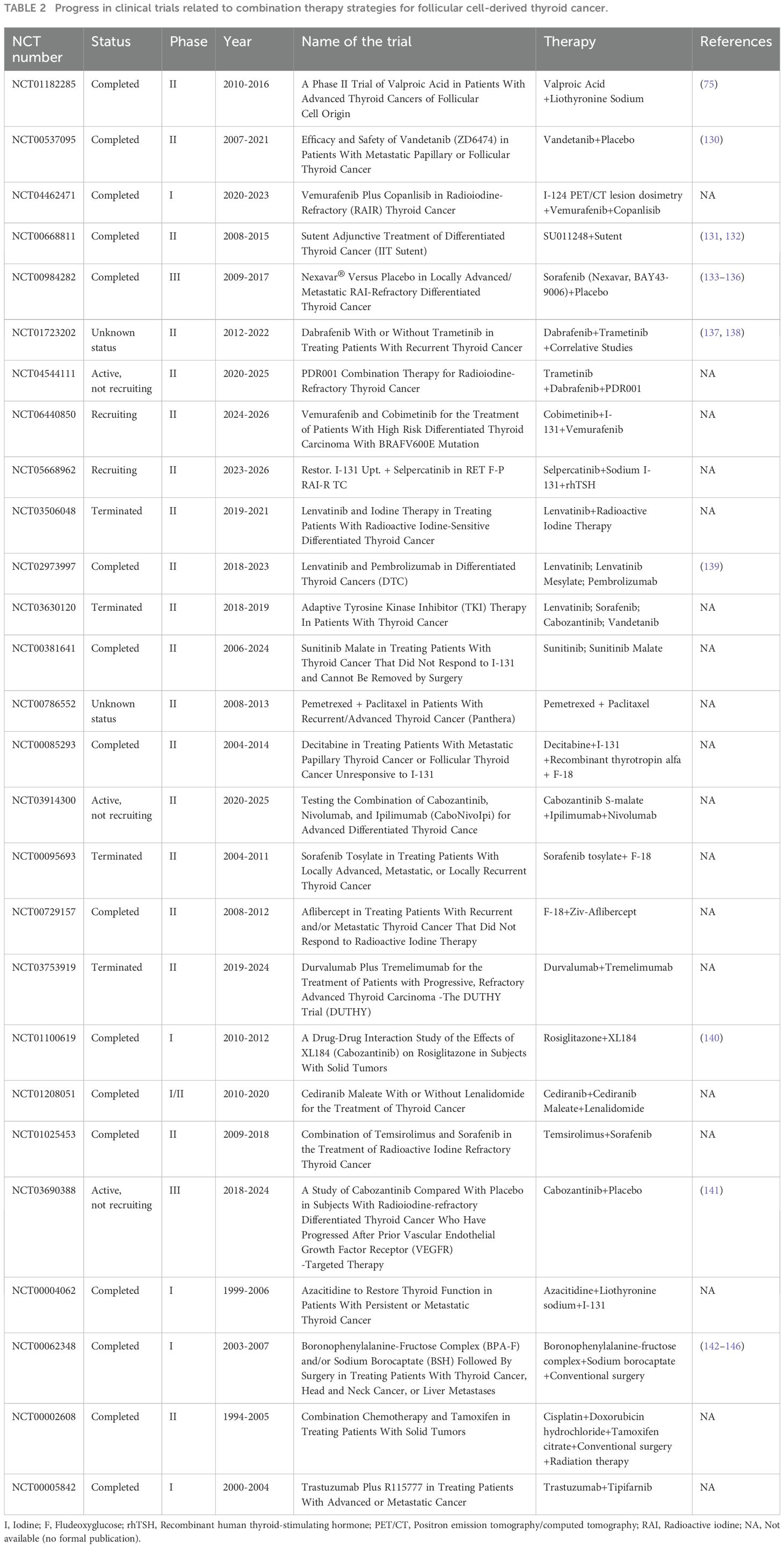
Table 2. Progress in clinical trials related to combination therapy strategies for follicular cell-derived thyroid cancer.
There is still much work to be done to determine the real effectiveness of epigenetic drugs alone for the treatment of thyroid cancer. Although there are many problems to be solved, epigenetic therapy has shown clear new advantages over classical genetic theory. The core point is that epigenetic therapy is used to reprogram tumor cells, rather than induce cytotoxicity, which is a completely different mechanism from chemotherapy targeted therapy. Resistance to traditional anti-tumor drugs has always been a difficult problem to solve, but epigenetic drugs can inhibit the generation of resistance, enhance the efficacy, and even directly act on the tumor cells that have developed resistance. Consequently, the concurrent administration of two or more epigenetic drugs may represent a promising strategy for enhancing the therapeutic effectiveness against tumors. For instance, HDACis TSA or VA in combination with demethylating agent 5-azacytidine inhibit cell growth in FTC and PTC cell lines (123). In addition, epigenetic therapy in combination with other therapies may significantly enhance treatment response rates in thyroid cancer, including chemotherapy, targeted drugs, and immunotherapy. Future trials should prioritize combined therapies to achieve more effective results in the treatment of thyroid cancer. In the last, more studies linking environmental exposure to epigenetic and genetic changes in thyroid cancer are needed. In summary, despite the clear evidence of epigenetic modifications in thyroid cancer, further research is needed to better understand the pathogenesis of thyroid cancer, thereby paving the way for the optimization and improvement of clinical diagnosis and treatment of thyroid cancer.
Author contributions
YH: Data curation, Investigation, Methodology, Resources, Writing – original draft. BW: Supervision, Writing – review & editing. JT: Supervision, Writing – review & editing. RW: Writing – review & editing. JW: Resources, Writing – review & editing. XR: Data curation, Writing – review & editing. YZ: Funding acquisition, Supervision, Writing – review & editing. ZG: Funding acquisition, Investigation, Supervision, Writing – review & editing. XX: Funding acquisition, Investigation, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (82102102 and 81771866) and Hubei Provincial Natural Science Foundation Project (2024AFB633).
Acknowledgments
The authors acknowledge the use of Medpeer and Figdraw for creating the schematic figures; Figure 1 was created with Medpeer.cn; Figure 2 was created with Figdraw.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, et al. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal transduction targeted Ther. (2019) 4:62. doi: 10.1038/s41392-019-0095-0
2. Morgan TH. SEX LIMITED INHERITANCE IN DROSOPHILA. Sci (New York NY). (1910) 32:120–2. doi: 10.1126/science.32.812.120
4. Hemberger M, Dean W, and Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat Rev Mol Cell Biol. (2009) 10:526–37. doi: 10.1038/nrm2727
5. Crews FT, Coleman LG Jr., Macht VA, and Vetreno RP. Targeting persistent changes in neuroimmune and epigenetic signaling in adolescent drinking to treat alcohol use disorder in adulthood. Pharmacol Rev. (2023) 75:380–96. doi: 10.1124/pharmrev.122.000710
6. Cheng H, Shang D, and Zhou R. Germline stem cells in human. Signal transduction targeted Ther. (2022) 7:345. doi: 10.1038/s41392-022-01197-3
7. Sharples AP, Stewart CE, and Seaborne RA. Does skeletal muscle have an ‘epi’-memory? The role of epigenetics in nutritional programming, metabolic disease, aging and exercise. Aging Cell. (2016) 15:603–16. doi: 10.1111/acel.2016.15.issue-4
8. Shen H and Laird PW. Interplay between the cancer genome and epigenome. Cell. (2013) 153:38–55. doi: 10.1016/j.cell.2013.03.008
9. Cui W, Huang Z, Jin SG, Johnson J, Lau KH, Hostetter G, et al. Deficiency of the polycomb protein RYBP and TET methylcytosine oxidases promotes extensive cpG island hypermethylation and Malignant transformation. Cancer Res. (2023) 83:2480–95. doi: 10.1158/0008-5472.CAN-23-0269
10. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2024) 74:229–63. doi: 10.3322/caac.21834
11. Pizzato M, Li M, Vignat J, Laversanne M, Singh D, La Vecchia C, et al. The epidemiological landscape of thyroid cancer worldwide: GLOBOCAN estimates for incidence and mortality rates in 2020. Lancet Diabetes endocrinology. (2022) 10:264–72. doi: 10.1016/S2213-8587(22)00035-3
12. Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, and Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. (2014) 74:2913–21. doi: 10.1158/0008-5472.CAN-14-0155
13. Chen DW, Lang BHH, McLeod DSA, Newbold K, and Haymart MR. Thyroid cancer. Lancet (London England). (2023) 401:1531–44. doi: 10.1016/S0140-6736(23)00020-X
14. Miller KD, Nogueira L, Devasia T, Mariotto AB, Yabroff KR, Jemal A, et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin. (2022) 72:409–36. doi: 10.3322/caac.21731
15. Tesselaar MH, Smit JW, Nagarajah J, Netea-Maier RT, and Plantinga TS. Pathological processes and therapeutic advances in radioiodide refractory thyroid cancer. J Mol Endocrinol. (2017) 59:R141–r54. doi: 10.1530/JME-17-0134
16. Nagaiah G, Hossain A, Mooney CJ, Parmentier J, and Remick SC. Anaplastic thyroid cancer: a review of epidemiology, pathogenesis, and treatment. J Oncol. (2011) 2011:542358. doi: 10.1155/2011/542358
17. Jannin A, Escande A, Al Ghuzlan A, Blanchard P, Hartl D, Chevalier B, et al. Anaplastic thyroid carcinoma: an update. Cancers. (2022) 14:1–25. doi: 10.3390/cancers14041061
18. Tamhane S and Gharib H. Thyroid nodule update on diagnosis and management. Clin Diabetes endocrinology. (2016) 2:17. doi: 10.1186/s40842-016-0035-7
19. Durante C, Grani G, Lamartina L, Filetti S, Mandel SJ, and Cooper DS. The diagnosis and management of thyroid nodules: A review. Jama. (2018) 319:914–24. doi: 10.1001/jama.2018.0898
20. Gharib H, Goellner JR, and Johnson DA. Fine-needle aspiration cytology of the thyroid. A 12-year experience with 11,000 biopsies. Clinics Lab Med. (1993) 13:699–709. doi: 10.1016/S0272-2712(18)30434-7
21. Baloch ZW, LiVolsi VA, Asa SL, Rosai J, Merino MJ, Randolph G, et al. Diagnostic terminology and morphologic criteria for cytologic diagnosis of thyroid lesions: a synopsis of the National Cancer Institute Thyroid Fine-Needle Aspiration State of the Science Conference. Diagn cytopathology. (2008) 36:425–37. doi: 10.1002/dc.20830
22. Fisher SB and Perrier ND. The incidental thyroid nodule. CA Cancer J Clin. (2018) 68:97–105. doi: 10.3322/caac.21447
23. Filetti S, Durante C, Hartl D, Leboulleux S, Locati LD, Newbold K, et al. Thyroid cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann oncology: Off J Eur Soc Med Oncol. (2019) 30:1856–83. doi: 10.1093/annonc/mdz400
24. Fugazzola L, Elisei R, Fuhrer D, Jarzab B, Leboulleux S, Newbold K, et al. European thyroid association guidelines for the treatment and follow-up of advanced radioiodine-refractory thyroid cancer. Eur Thyroid J. (2019) 8:227–45. doi: 10.1159/000502229
25. Smallridge RC and Copland JA. Anaplastic thyroid carcinoma: pathogenesis and emerging therapies. Clin Oncol (Royal Coll Radiologists (Great Britain)). (2010) 22:486–97. doi: 10.1016/j.clon.2010.03.013
26. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Integrative analysis of 111 reference human epigenomes. Nature. (2015) 518:317–30. doi: 10.1038/nature14248
27. Zhao J, Tian Y, Jia Z, Yao J, Liao L, and Dong J. Abnormal glucose metabolism parameters and the aggressiveness of differentiated thyroid carcinoma: A hospital-based cross-section study in China. Front endocrinology. (2022) 13:806349. doi: 10.3389/fendo.2022.806349
28. Dong WW, Zhang DL, Wang ZH, Lv CZ, Zhang P, and Zhang H. Different types of diabetes mellitus and risk of thyroid cancer: A meta-analysis of cohort studies. Front endocrinology. (2022) 13:971213. doi: 10.3389/fendo.2022.971213
29. Cho BY, Choi HS, Park YJ, Lim JA, Ahn HY, Lee EK, et al. Changes in the clinicopathological characteristics and outcomes of thyroid cancer in Korea over the past four decades. Thyroid: Off J Am Thyroid Assoc. (2013) 23:797–804. doi: 10.1089/thy.2012.0329
30. Zhang W, Ruan X, Huang Y, Zhang W, Xu G, Zhao J, et al. SETMAR facilitates the differentiation of thyroid cancer by regulating SMARCA2-mediated chromatin remodeling. Advanced Sci (Weinheim Baden-Wurttemberg Germany). (2024) 11:e2401712. doi: 10.1002/advs.202401712
31. Tsai HC and Baylin SB. Cancer epigenetics: linking basic biology to clinical medicine. Cell Res. (2011) 21:502–17. doi: 10.1038/cr.2011.24
32. Zafon C, Gil J, Pérez-González B, and Jordà M. DNA methylation in thyroid cancer. Endocrine-related Cancer. (2019) 26:R415–r39. doi: 10.1530/ERC-19-0093
33. Zhou S, Liu J, Wan A, Zhang Y, and Qi X. Epigenetic regulation of diverse cell death modalities in cancer: a focus on pyroptosis, ferroptosis, cuproptosis, and disulfidptosis. J Hematol Oncol. (2024) 17:22. doi: 10.1186/s13045-024-01545-6
34. Jones PA and Baylin SB. The epigenomics of cancer. Cell. (2007) 128:683–92. doi: 10.1016/j.cell.2007.01.029
35. Gao X, Reid MA, Kong M, and Locasale JW. Metabolic interactions with cancer epigenetics. Mol aspects Med. (2017) 54:50–7. doi: 10.1016/j.mam.2016.09.001
36. Ferry L, Fournier A, Tsusaka T, Adelmant G, Shimazu T, Matano S, et al. Methylation of DNA ligase 1 by G9a/GLP recruits UHRF1 to replicating DNA and regulates DNA methylation. Mol Cell. (2017) 67:550–65.e5. doi: 10.1016/j.molcel.2017.07.012
37. Esteller M. Epigenetics in cancer. New Engl J Med. (2008) 358:1148–59. doi: 10.1056/NEJMra072067
38. Bird AP. CpG-rich islands and the function of DNA methylation. Nature. (1986) 321:209–13. doi: 10.1038/321209a0
39. Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. (2009) 1:239–59. doi: 10.2217/epi.09.33
40. Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. (2002) 21:5427–40. doi: 10.1038/sj.onc.1205600
41. Hu S, Ewertz M, Tufano RP, Brait M, Carvalho AL, Liu D, et al. Detection of serum deoxyribonucleic acid methylation markers: a novel diagnostic tool for thyroid cancer. J Clin Endocrinol Metab. (2006) 91:98–104. doi: 10.1210/jc.2005-1810
42. Smith JA, Fan CY, Zou C, Bodenner D, and Kokoska MS. Methylation status of genes in papillary thyroid carcinoma. Arch otolaryngology–head Neck surgery. (2007) 133:1006–11. doi: 10.1001/archotol.133.10.1006
43. Hoque MO, Rosenbaum E, Westra WH, Xing M, Ladenson P, Zeiger MA, et al. Quantitative assessment of promoter methylation profiles in thyroid neoplasms. J Clin Endocrinol Metab. (2005) 90:4011–8. doi: 10.1210/jc.2005-0313
44. Hu S, Liu D, Tufano RP, Carson KA, Rosenbaum E, Cohen Y, et al. Association of aberrant methylation of tumor suppressor genes with tumor aggressiveness and BRAF mutation in papillary thyroid cancer. Int J cancer. (2006) 119:2322–9. doi: 10.1002/ijc.22110
45. Venkataraman GM, Yatin M, Marcinek R, and Ain KB. Restoration of iodide uptake in dedifferentiated thyroid carcinoma: relationship to human Na+/I-symporter gene methylation status. J Clin Endocrinol Metab. (1999) 84:2449–57. doi: 10.1210/jc.84.7.2449
46. Xing M, Usadel H, Cohen Y, Tokumaru Y, Guo Z, Westra WB, et al. Methylation of the thyroid-stimulating hormone receptor gene in epithelial thyroid tumors: a marker of Malignancy and a cause of gene silencing. Cancer Res. (2003) 63:2316–21.
47. Toraño EG, Petrus S, Fernandez AF, and Fraga MF. Global DNA hypomethylation in cancer: review of validated methods and clinical significance. Clin Chem Lab Med. (2012) 50:1733–42. doi: 10.1515/cclm-2011-0902
48. Klein Hesselink EN, Zafon C, Villalmanzo N, Iglesias C, van Hemel BM, Klein Hesselink MS, et al. Increased global DNA hypomethylation in distant metastatic and dedifferentiated thyroid cancer. J Clin Endocrinol Metab. (2018) 103:397–406. doi: 10.1210/jc.2017-01613
49. Esteller M and Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J pathology. (2002) 196:1–7. doi: 10.1002/path.v196:1
50. Dillinger T, Sheibani-Tezerji R, Pulverer W, Stelzer I, Hassler MR, Scheibelreiter J, et al. Identification of tumor tissue-derived DNA methylation biomarkers for the detection and therapy response evaluation of metastatic castration resistant prostate cancer in liquid biopsies. Mol cancer. (2022) 21:7. doi: 10.1186/s12943-021-01445-0
51. Hong S, Lin B, Xu M, Zhang Q, Huo Z, Su M, et al. Cell-free DNA methylation biomarker for the diagnosis of papillary thyroid carcinoma. EBioMedicine. (2023) 90:104497. doi: 10.1016/j.ebiom.2023.104497
52. Ponnaluri VK, Ehrlich KC, Zhang G, Lacey M, Johnston D, Pradhan S, et al. Association of 5-hydroxymethylation and 5-methylation of DNA cytosine with tissue-specific gene expression. Epigenetics. (2017) 12:123–38. doi: 10.1080/15592294.2016.1265713
53. Oishi N, Vuong HG, Mochizuki K, and Kondo T. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of thyroid carcinomas with TERT promoter mutations. Endocrine pathology. (2020) 31:359–66. doi: 10.1007/s12022-020-09652-z
54. Tong M, Gao S, Qi W, Shi C, Qiu M, Yang F, et al. 5-Hydroxymethylcytosine as a potential epigenetic biomarker in papillary thyroid carcinoma. Oncol letters. (2019) 18:2304–9. doi: 10.3892/ol.2019.10531
55. Emran AA, Marzese DM, Menon DR, Hammerlindl H, Ahmed F, Richtig E, et al. Commonly integrated epigenetic modifications of differentially expressed genes lead to adaptive resistance in cancer. Epigenomics. (2019) 11:732–7. doi: 10.2217/epi-2018-0173
56. Teodoridis JM, Strathdee G, and Brown R. Epigenetic silencing mediated by CpG island methylation: potential as a therapeutic target and as a biomarker. Drug resistance updates: Rev commentaries antimicrobial Anticancer chemotherapy. (2004) 7:267–78. doi: 10.1016/j.drup.2004.06.005
57. Phi LTH, Sari IN, Yang YG, Lee SH, Jun N, Kim KS, et al. Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. (2018) 2018:5416923. doi: 10.1155/2018/5416923
58. Miasaki FY, Vivaldi A, Ciampi R, Agate L, Collecchi P, Capodanno A, et al. Retinoic acid receptor beta2 re-expression and growth inhibition in thyroid carcinoma cell lines after 5-aza-2’-deoxycytidine treatment. J endocrinological Invest. (2008) 31:724–30. doi: 10.1007/BF03346422
59. Vivaldi A, Miasaki FY, Ciampi R, Agate L, Collecchi P, Capodanno A, et al. Re-differentiation of thyroid carcinoma cell lines treated with 5-Aza-2’-deoxycytidine and retinoic acid. Mol Cell Endocrinol. (2009) 307:142–8. doi: 10.1016/j.mce.2009.03.020
60. Hingorani M, Spitzweg C, Vassaux G, Newbold K, Melcher A, Pandha H, et al. The biology of the sodium iodide symporter and its potential for targeted gene delivery. Curr Cancer Drug targets. (2010) 10:242–67. doi: 10.2174/156800910791054194
61. Gulec SA, Benites C, and Cabanillas ME. Molecular perspectives in radioactive iodine theranostics: current redifferentiation protocols for mis-differentiated thyroid cancer. J Clin Med. (2024) 13:1–17. doi: 10.3390/jcm13133645
62. Allfrey VG, Faulkner R, and Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci United States America. (1964) 51:786–94. doi: 10.1073/pnas.51.5.786
63. Kouzarides T. Chromatin modifications and their function. Cell. (2007) 128:693–705. doi: 10.1016/j.cell.2007.02.005
64. Chi P, Allis CD, and Wang GG. Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. (2010) 10:457–69. doi: 10.1038/nrc2876
65. Berger SL. The complex language of chromatin regulation during transcription. Nature. (2007) 447:407–12. doi: 10.1038/nature05915
66. Barnes CE, English DM, and Cowley SM. Acetylation & Co: an expanding repertoire of histone acylations regulates chromatin and transcription. Essays Biochem. (2019) 63:97–107. doi: 10.1042/EBC20180061
67. Millán-Zambrano G, Burton A, Bannister AJ, and Schneider R. Histone post-translational modifications - cause and consequence of genome function. Nat Rev Genet. (2022) 23:563–80. doi: 10.1038/s41576-022-00468-7
68. Puppin C, Passon N, Lavarone E, Di Loreto C, Frasca F, Vella V, et al. Levels of histone acetylation in thyroid tumors. Biochem Biophys Res Commun. (2011) 411:679–83. doi: 10.1016/j.bbrc.2011.06.182
69. Hou P, Bojdani E, and Xing M. Induction of thyroid gene expression and radioiodine uptake in thyroid cancer cells by targeting major signaling pathways. J Clin Endocrinol Metab. (2010) 95:820–8. doi: 10.1210/jc.2009-1888
70. Furuya F, Shimura H, Suzuki H, Taki K, Ohta K, Haraguchi K, et al. Histone deacetylase inhibitors restore radioiodide uptake and retention in poorly differentiated and anaplastic thyroid cancer cells by expression of the sodium/iodide symporter thyroperoxidase and thyroglobulin. Endocrinology. (2004) 145:2865–75. doi: 10.1210/en.2003-1258
71. Pugliese M, Fortunati N, Germano A, Asioli S, Marano F, Palestini N, et al. Histone deacetylase inhibition affects sodium iodide symporter expression and induces 131I cytotoxicity in anaplastic thyroid cancer cells. Thyroid: Off J Am Thyroid Assoc. (2013) 23:838–46. doi: 10.1089/thy.2012.0359
72. Fu H, Cheng L, Jin Y, Cheng L, Liu M, and Chen L. MAPK inhibitors enhance HDAC inhibitor-induced redifferentiation in papillary thyroid cancer cells harboring BRAF (V600E): an in vitro study. Mol Ther oncolytics. (2019) 12:235–45. doi: 10.1016/j.omto.2019.01.007
73. Perona M, Ibañez IL, Thomasz L, Villaverde MS, Oglio R, Rosemblit C, et al. Valproic acid radiosensitizes anaplastic thyroid cells through a decrease of the DNA damage repair capacity. J endocrinological Invest. (2023) 46:2353–65. doi: 10.1007/s40618-023-02092-6
74. Wächter S, Damanakis AI, Elxnat M, Roth S, Wunderlich A, Verburg FA, et al. Epigenetic modifications in thyroid cancer cells restore NIS and radio-iodine uptake and promote cell death. J Clin Med. (2018) 7:1–15. doi: 10.3390/jcm7040061
75. Nilubol N, Merkel R, Yang L, Patel D, Reynolds JC, Sadowski SM, et al. A phase II trial of valproic acid in patients with advanced, radioiodine-resistant thyroid cancers of follicular cell origin. Clin endocrinology. (2017) 86:128–33. doi: 10.1111/cen.13154
76. Luong QT, O’Kelly J, Braunstein GD, Hershman JM, and Koeffler HP. Antitumor activity of suberoylanilide hydroxamic acid against thyroid cancer cell lines in vitro and in vivo. Clin Cancer research: an Off J Am Assoc Cancer Res. (2006) 12:5570–7. doi: 10.1158/1078-0432.CCR-06-0367
77. Ravasio R, Ceccacci E, Nicosia L, Hosseini A, Rossi PL, Barozzi I, et al. Targeting the scaffolding role of LSD1 (KDM1A) poises acute myeloid leukemia cells for retinoic acid-induced differentiation. Sci Adv. (2020) 6:eaax2746. doi: 10.1126/sciadv.aax2746
78. Jambhekar A, Dhall A, and Shi Y. Roles and regulation of histone methylation in animal development. Nat Rev Mol Cell Biol. (2019) 20:625–41. doi: 10.1038/s41580-019-0151-1
79. Zhang W, Ruan X, Li Y, Zhi J, Hu L, Hou X, et al. KDM1A promotes thyroid cancer progression and maintains stemness through the Wnt/β-catenin signaling pathway. Theranostics. (2022) 12:1500–17. doi: 10.7150/thno.66142
80. Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. (2005) 437:436–9. doi: 10.1038/nature04020
81. Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. (2004) 119:941–53. doi: 10.1016/j.cell.2004.12.012
82. Borbone E, Troncone G, Ferraro A, Jasencakova Z, Stojic L, Esposito F, et al. Enhancer of zeste homolog 2 overexpression has a role in the development of anaplastic thyroid carcinomas. J Clin Endocrinol Metab. (2011) 96:1029–38. doi: 10.1210/jc.2010-1784
83. Fu H, Cheng L, Sa R, Jin Y, and Chen L. Combined tazemetostat and MAPKi enhances differentiation of papillary thyroid cancer cells harbouring BRAF(V600E) by synergistically decreasing global trimethylation of H3K27. J Cell Mol Med. (2020) 24:3336–45. doi: 10.1111/jcmm.15007
84. Ptak C and Wozniak RW. Nucleoporins and chromatin metabolism. Curr Opin Cell Biol. (2016) 40:153–60. doi: 10.1016/j.ceb.2016.03.024
85. Clapier CR and Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. (2009) 78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223
86. Wilson BG and Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. (2011) 11:481–92. doi: 10.1038/nrc3068
87. Ho L and Crabtree GR. Chromatin remodelling during development. Nature. (2010) 463:474–84. doi: 10.1038/nature08911
88. Venkatesh S and Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. (2015) 16:178–89. doi: 10.1038/nrm3941
89. Kumar R, Li DQ, Müller S, and Knapp S. Epigenomic regulation of oncogenesis by chromatin remodeling. Oncogene. (2016) 35:4423–36. doi: 10.1038/onc.2015.513
90. Clapier CR, Iwasa J, Cairns BR, and Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. (2017) 18:407–22. doi: 10.1038/nrm.2017.26
91. Mittal P and Roberts CWM. The SWI/SNF complex in cancer - biology, biomarkers and therapy. Nat Rev Clin Oncol. (2020) 17:435–48. doi: 10.1038/s41571-020-0357-3
92. Saqcena M, Leandro-Garcia LJ, Maag JLV, Tchekmedyian V, Krishnamoorthy GP, Tamarapu PP, et al. SWI/SNF complex mutations promote thyroid tumor progression and insensitivity to redifferentiation therapies. Cancer discovery. (2021) 11:1158–75. doi: 10.1158/2159-8290.CD-20-0735
93. Frye M, Harada BT, Behm M, and He C. RNA modifications modulate gene expression during development. Sci (New York NY). (2018) 361:1346–9. doi: 10.1126/science.aau1646
94. Wiener D and Schwartz S. The epitranscriptome beyond m(6)A. Nat Rev Genet. (2021) 22:119–31. doi: 10.1038/s41576-020-00295-8
95. Wei K, Gao Y, Wang B, and Qu YX. Methylation recognition protein YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) regulates the proliferation, migration and invasion of osteosarcoma by regulating m6A level of CCR4-NOT transcription complex subunit 7 (CNOT7). Bioengineered. (2022) 13:5236–50. doi: 10.1080/21655979.2022.2037381
96. An Y and Duan H. The role of m6A RNA methylation in cancer metabolism. Mol cancer. (2022) 21:14. doi: 10.1186/s12943-022-01500-4
97. Zhu Y, Peng X, Zhou Q, Tan L, Zhang C, Lin S, et al. METTL3-mediated m6A modification of STEAP2 mRNA inhibits papillary thyroid cancer progress by blocking the Hedgehog signaling pathway and epithelial-to-mesenchymal transition. Cell Death disease. (2022) 13:358. doi: 10.1038/s41419-022-04817-6
98. Allegri L, Baldan F, Molteni E, Mio C, and Damante G. Role of m6A RNA methylation in thyroid cancer cell lines. Int J Mol Sci. (2022) 23:1–17. doi: 10.3390/ijms231911516
99. Zhou X, Chang L, Liang Q, Zhao R, Xiao Y, Xu Z, et al. The m6A methyltransferase METTL3 drives thyroid cancer progression and lymph node metastasis by targeting LINC00894. Cancer Cell Int. (2024) 24:47. doi: 10.1186/s12935-024-03240-5
100. Xue C, Chu Q, Zheng Q, Jiang S, Bao Z, Su Y, et al. Role of main RNA modifications in cancer: N(6)-methyladenosine, 5-methylcytosine, and pseudouridine. Signal transduction targeted Ther. (2022) 7:142. doi: 10.1038/s41392-022-01003-0
101. Li P, Wang W, Zhou R, Ding Y, and Li X. The m(5) C methyltransferase NSUN2 promotes codon-dependent oncogenic translation by stabilising tRNA in anaplastic thyroid cancer. Clin Trans Med. (2023) 13:e1466. doi: 10.1002/ctm2.v13.11
102. Ning J, Hou X, Hao J, Zhang W, Shi Y, Huang Y, et al. METTL3 inhibition induced by M2 macrophage-derived extracellular vesicles drives anti-PD-1 therapy resistance via M6A-CD70-mediated immune suppression in thyroid cancer. Cell Death differentiation. (2023) 30:2265–79. doi: 10.1038/s41418-023-01217-x
103. Hou X, Dong Q, Hao J, Liu M, Ning J, Tao M, et al. NSUN2-mediated m(5)C modification drives alternative splicing reprogramming and promotes multidrug resistance in anaplastic thyroid cancer through the NSUN2/SRSF6/UAP1 signaling axis. Theranostics. (2025) 15:2757–77. doi: 10.7150/thno.104713
104. Goodall GJ and Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. (2021) 21:22–36. doi: 10.1038/s41568-020-00306-0
105. Schmitt AM and Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. (2016) 29:452–63. doi: 10.1016/j.ccell.2016.03.010
106. Sahu A, Singhal U, and Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends cancer. (2015) 1:93–109. doi: 10.1016/j.trecan.2015.08.010
107. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. (2011) 25:1915–27. doi: 10.1101/gad.17446611
108. Ntziachristos P, Abdel-Wahab O, and Aifantis I. Emerging concepts of epigenetic dysregulation in hematological Malignancies. Nat Immunol. (2016) 17:1016–24. doi: 10.1038/ni.3517
109. Yu Y, Yarrington RM, Chuong EB, Elde NC, and Stillman DJ. Disruption of promoter memory by synthesis of a long noncoding RNA. Proc Natl Acad Sci United States America. (2016) 113:9575–80. doi: 10.1073/pnas.1601793113
110. Yoon JH, Abdelmohsen K, and Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. (2013) 425:3723–30. doi: 10.1016/j.jmb.2012.11.024
111. Samimi H, Sajjadi-Jazi SM, Seifirad S, Atlasi R, Mahmoodzadeh H, Faghihi MA, et al. Molecular mechanisms of long non-coding RNAs in anaplastic thyroid cancer: a systematic review. Cancer Cell Int. (2020) 20:352. doi: 10.1186/s12935-020-01439-w
112. Li L, Lin X, Xu P, Jiao Y, and Fu P. LncRNA GAS5 sponges miR-362-5p to promote sensitivity of thyroid cancer cells to (131) I by upregulating SMG1. IUBMB Life. (2020) 72:2420–31. doi: 10.1002/iub.v72.11
113. Lin QY, Qi QL, Hou S, Chen Z, Zhang L, Zhao HG, et al. LncRNA PVT1 acts as a tumor promoter in thyroid cancer and promotes tumor progression by mediating miR-423-5p-PAK3. Cancer Manage Res. (2020) 12:13403–13. doi: 10.2147/CMAR.S283443
114. Zhang Y, Yu S, Jiang L, Wang X, and Song X. HOTAIR is a promising novel biomarker in patients with thyroid cancer. Exp Ther Med. (2017) 13:2274–8. doi: 10.3892/etm.2017.4231
115. Batista PJ and Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. (2013) 152:1298–307. doi: 10.1016/j.cell.2013.02.012
116. Oki Y, Kantarjian HM, Gharibyan V, Jones D, O’Brien S, Verstovsek S, et al. Phase II study of low-dose decitabine in combination with imatinib mesylate in patients with accelerated or myeloid blastic phase of chronic myelogenous leukemia. Cancer. (2007) 109:899–906. doi: 10.1002/cncr.v109:5
117. Dobosy JR and Selker EU. Emerging connections between DNA methylation and histone acetylation. Cell Mol Life sciences: CMLS. (2001) 58:721–7. doi: 10.1007/PL00000895
118. Dawson MA and Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. (2012) 150:12–27. doi: 10.1016/j.cell.2012.06.013
119. Saini S, Tulla K, Maker AV, Burman KD, and Prabhakar BS. Therapeutic advances in anaplastic thyroid cancer: a current perspective. Mol cancer. (2018) 17:154. doi: 10.1186/s12943-018-0903-0
120. Alzahrani AS. Clinical use of molecular data in thyroid nodules and cancer. J Clin Endocrinol Metab. (2023) 108:2759–71. doi: 10.1210/clinem/dgad282
121. Rodríguez-Rodero S, Delgado-Álvarez E, Díaz-Naya L, Martín Nieto A, and Menéndez Torre E. Epigenetic modulators of thyroid cancer. Endocrinologia Diabetes y nutricion. (2017) 64:44–56. doi: 10.1016/j.endinu.2016.09.006
122. Averbeck D and Rodriguez-Lafrasse C. Role of mitochondria in radiation responses: epigenetic, metabolic, and signaling impacts. Int J Mol Sci. (2021) 22:1–59. doi: 10.3390/ijms222011047
123. Mitmaker EJ, Griff NJ, Grogan RH, Sarkar R, Kebebew E, Duh QY, et al. Modulation of matrix metalloproteinase activity in human thyroid cancer cell lines using demethylating agents and histone deacetylase inhibitors. Surgery. (2011) 149:504–11. doi: 10.1016/j.surg.2010.10.007
124. Simon D, Koehrle J, Reiners C, Boerner AR, Schmutzler C, Mainz K, et al. Redifferentiation therapy with retinoids: therapeutic option for advanced follicular and papillary thyroid carcinoma. World J surgery. (1998) 22:569–74. doi: 10.1007/s002689900436
125. Kebebew E, Greenspan FS, Clark OH, Woeber KA, and McMillan A. Anaplastic thyroid carcinoma. Treat outcome prognostic factors. Cancer. (2005) 103:1330–5. doi: 10.1002/cncr.20936
126. Woyach JA, Kloos RT, Ringel MD, Arbogast D, Collamore M, Zwiebel JA, et al. Lack of therapeutic effect of the histone deacetylase inhibitor vorinostat in patients with metastatic radioiodine-refractory thyroid carcinoma. J Clin Endocrinol Metab. (2009) 94:164–70. doi: 10.1210/jc.2008-1631
127. Ueda H, Manda T, Matsumoto S, Mukumoto S, Nishigaki F, Kawamura I, et al. FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum No. 968. III. Antitumor activities on experimental tumors in mice. J antibiotics. (1994) 47:315–23. doi: 10.7164/antibiotics.47.315
128. Ueda H, Nakajima H, Hori Y, Fujita T, Nishimura M, Goto T, et al. FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum No. 968. I. Taxonomy, fermentation, isolation, physico-chemical and biological properties, and antitumor activity. J antibiotics. (1994) 47:301–10. doi: 10.7164/antibiotics.47.301
129. Ueda H, Nakajima H, Hori Y, Goto T, and Okuhara M. Action of FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum no. 968, on Ha-ras transformed NIH3T3 cells. Bioscience biotechnology Biochem. (1994) 58:1579–83. doi: 10.1271/bbb.58.1579
130. Leboulleux S, Bastholt L, Krause T, de la Fouchardiere C, Tennvall J, Awada A, et al. Vandetanib in locally advanced or metastatic differentiated thyroid cancer: a randomised, double-blind, phase 2 trial. Lancet Oncol. (2012) 13:897–905. doi: 10.1016/S1470-2045(12)70335-2
131. Britten CD, Kabbinavar F, Hecht JR, Bello CL, Li J, Baum C, et al. A phase I and pharmacokinetic study of sunitinib administered daily for 2 weeks, followed by a 1-week off period. Cancer chemotherapy Pharmacol. (2008) 61:515–24. doi: 10.1007/s00280-007-0498-4
132. Bikas A, Kundra P, Desale S, Mete M, O’Keefe K, Clark BG, et al. Phase 2 clinical trial of sunitinib as adjunctive treatment in patients with advanced differentiated thyroid cancer. Eur J endocrinology. (2016) 174:373–80. doi: 10.1530/EJE-15-0930
133. Worden F, Fassnacht M, Shi Y, Hadjieva T, Bonichon F, Gao M, et al. Safety and tolerability of sorafenib in patients with radioiodine-refractory thyroid cancer. Endocrine-related cancer. (2015) 22:877–87. doi: 10.1530/ERC-15-0252
134. Brose MS, Nutting CM, Jarzab B, Elisei R, Siena S, Bastholt L, et al. Sorafenib in radioactive iodine-refractory, locally advanced or metastatic differentiated thyroid cancer: a randomised, double-blind, phase 3 trial. Lancet (London England). (2014) 384:319–28. doi: 10.1016/S0140-6736(14)60421-9
135. Brose MS, Nutting CM, Sherman SI, Shong YK, Smit JW, Reike G, et al. Rationale and design of decision: a double-blind, randomized, placebo-controlled phase III trial evaluating the efficacy and safety of sorafenib in patients with locally advanced or metastatic radioactive iodine (RAI)-refractory, differentiated thyroid cancer. BMC cancer. (2011) 11:349. doi: 10.1186/1471-2407-11-349
136. Capdevila J, Matos I, Mancuso FM, Iglesias C, Nuciforo P, Zafon C, et al. Identification of expression profiles defining distinct prognostic subsets of radioactive-iodine refractory differentiated thyroid cancer from the DECISION trial. Mol Cancer Ther. (2020) 19:312–7. doi: 10.1158/1535-7163.MCT-19-0211
137. Busaidy NL, Konda B, Wei L, Wirth LJ, Devine C, Daniels GA, et al. Dabrafenib versus dabrafenib + Trametinib in BRAF-mutated radioactive iodine refractory differentiated thyroid cancer: results of a randomized, phase 2, open-label multicenter trial. Thyroid: Off J Am Thyroid Assoc. (2022) 32:1184–92. doi: 10.1089/thy.2022.0115
138. Owen DH, Konda B, Sipos J, Liu T, Webb A, Ringel MD, et al. KRAS G12V mutation in acquired resistance to combined BRAF and MEK inhibition in papillary thyroid ancer. J Natl Compr Cancer Network: JNCCN. (2019) 17:409–13. doi: 10.6004/jnccn.2019.7292
139. Basté N, Mora M, and Grau JJ. Emerging systemic antitarget treatment for differentiated thyroid carcinoma. Curr Opin Oncol. (2021) 33:184–95. doi: 10.1097/CCO.0000000000000727
140. Choueiri TK, Pal SK, McDermott DF, Morrissey S, Ferguson KC, Holland J, et al. A phase I study of cabozantinib (XL184) in patients with renal cell cancer. Ann oncology: Off J Eur Soc Med Oncol. (2014) 25:1603–8. doi: 10.1093/annonc/mdu184
141. Brose MS, Robinson B, Sherman SI, Krajewska J, Lin CC, Vaisman F, et al. Cabozantinib for radioiodine-refractory differentiated thyroid cancer (COSMIC-311): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. (2021) 22:1126–38. doi: 10.1016/S1470-2045(21)00332-6
142. Wittig A, Sheu-Grabellus SY, Collette L, Moss R, Brualla L, and Sauerwein W. BPA uptake does not correlate with LAT1 and Ki67 expressions in tumor samples (results of EORTC trial 11001). Appl Radiat isotopes: including data instrumentation Methods Use agriculture industry Med. (2011) 69:1807–12. doi: 10.1016/j.apradiso.2011.02.018
143. Bendel P, Wittig A, Basilico F, Mauri PL, and Sauerwein W. Metabolism of borono-phenylalanine-fructose complex (BPA-fr) and borocaptate sodium (BSH) in cancer patients–results from EORTC trial 11001. J Pharm Biomed analysis. (2010) 51:284–7. doi: 10.1016/j.jpba.2009.08.018
144. Wittig A, Collette L, Appelman K, Bührmann S, Jäckel MC, Jöckel KH, et al. EORTC trial 11001: distribution of two 10B-compounds in patients with squamous cell carcinoma of head and neck, a translational research/phase 1 trial. J Cell Mol Med. (2009) 13:1653–65. doi: 10.1111/j.1582-4934.2009.00856.x
145. Wittig A, Collette L, Moss R, and Sauerwein WA. Early clinical trial concept for boron neutron capture therapy: a critical assessment of the EORTC trial 11001. Appl Radiat isotopes: including data instrumentation Methods Use agriculture industry Med. (2009) 67:S59–62. doi: 10.1016/j.apradiso.2009.03.012
146. Wittig A, Malago M, Collette L, Huiskamp R, Bührmann S, Nievaart V, et al. Uptake of two 10B-compounds in liver metastases of colorectal adenocarcinoma for extracorporeal irradiation with boron neutron capture therapy (EORTC Trial 11001). Int J cancer. (2008) 122:1164–71. doi: 10.1002/ijc.v122:5
Keywords: epigenetics, follicular cell-derived thyroid cancer, biomarkers, inhibitors, therapy
Citation: Han Y, Wu B, Tan J, Wu R, Wang J, Ren X, Zhang Y, Gao Z and Xia X (2025) Epigenetic modifications in follicular cell-derived thyroid cancer: new dimensions in pathogenesis and treatment. Front. Oncol. 15:1549477. doi: 10.3389/fonc.2025.1549477
Received: 21 December 2024; Accepted: 25 April 2025;
Published: 20 May 2025.
Edited by:
Priya Mondal, Central Food Technological Research Institute (CSIR), IndiaReviewed by:
Angelika Buczyńska, Medical University of Bialystok, PolandPraveen Koganti, Sanford Burnham Prebys Medical Discovery Institute, United States
Copyright © 2025 Han, Wu, Tan, Wu, Wang, Ren, Zhang, Gao and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yajing Zhang, emhhbmd5anl5eTE5ODZAMTYzLmNvbQ==; Zairong Gao, Z2FvYm9ubkBodXN0LmVkdS5jbg==; Xiaotian Xia, eGlhb3RpYW5feGlhQGh1c3QuZWR1LmNu
 Yanmei Han
Yanmei Han Bian Wu
Bian Wu Jie Tan
Jie Tan Ruolin Wu
Ruolin Wu Jingjing Wang
Jingjing Wang Xiaojing Ren
Xiaojing Ren Yajing Zhang
Yajing Zhang Zairong Gao
Zairong Gao Xiaotian Xia
Xiaotian Xia