- Department of Gynecological Oncology, Beijing Obstetrics and Gynecology Hospital, Capital Medical University, Beijing Maternal and Child Health Care Hospital, Beijing, China
Objective: To evaluate the mechanism of Kielin/chordin-like protein (KCP) in the resistance of cervical cancer cells to paclitaxel.
Method: A cervical squamous carcinoma cell line (SiHa) with KCP knockout was constructed and treated with paclitaxel. Key cell functions were assessed by colony formation assay, measurement of cell proliferation by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, and FACS-based detection of apoptosis. The downstream mechanism of KCP-mediated resistance to paclitaxel was then examined using human gene chip detection and IPA bioinformatics analysis, and qPCR analysis was used to validate its downstream genes.
Results: ①Functional studies of SiHa cells showed that KCP knockout (sgRNA) inhibited colony formation and proliferation of SiHa cells in the presence of paclitaxel (p<0.05). ②Using a whole human genome microarray, a total of 491 differentially expressed genes were identified in KCP knockout versus the NC SiHa cells. IPA-based bioinformatics analysis of upstream regulators showed that SPI1 was strongly activated and that SPI1 inhibited CCND1 and activated PML and CEBPA, which is consistent with results from gene chip analysis showing CCND1, PML, and CEBPA expression after KCP knockout. ③A total of 30 differentially expressed genes associated with tumor cell proliferation were identified by gene microarray and IPA analyses. The changes in the aforementioned genes after KCP knockout were verified by qPCR, and SERPINB3 and CEBPA expression were significantly lower and higher, respectively, compared to in the control group.
Conclusion: KCP increased resistance of cervical cancer to paclitaxel by enhancing cell proliferation and colony formation. We observed that KCP could act positively on the downstream gene SERPINB3 and negatively on the downstream gene CEBPA to affect the resistance of cervical carcinoma cells to paclitaxel.
Highlights
● KCP is associated with paclitaxel chemoresistance in cervical cancer.
● KCP can act as an upstream gene via SPI1 on downstream genes to participate in the regulatory network of replication of murine herpesvirus 4 and replication of vesicular stomatitis virus.
● KCP can also act positively on the downstream gene SERPINB3 and negatively on the downstream gene CEBPA to participate in the proliferative function of cervical squamous carcinoma cells and affect the resistance of cervical carcinoma to paclitaxel.
1 Introduction
Cervical cancer is the fourth most common malignancy affecting women worldwide. Approximately 90% of diagnosed cases are squamous cervical cancer (1). According to the data published by the International Agency for Research on Cancer (IARC) in 2020, approximately 604,000 new cervical cancer cases and 342,000 deaths were reported globally (2). China has the highest number of new cases and deaths (3), with 131,500 new cervical cancer cases and 53,000 deaths each year, accounting for 18.4% of female cancer-related mortality in China (4). In developing countries, the 5-year survival rate for patients with cervical cancer is < 50%. For patients diagnosed in stages III or IV, the 5-year survival rate is only 5% to 15%, thus posing a serious threat to the lives of women (2).
Paclitaxel, which is a first-line chemotherapy drug for cervical cancer, is widely used in patients with intermediate to advanced squamous cell cervical cancer and plays an important role in the chemotherapy of cervical squamous cell carcinoma. However, paclitaxel resistance is the main cause of treatment failure, and overcoming paclitaxel resistance has become the most important clinical problem for gynecological oncologists.
In 2002, Abreu’s research group (5) discovered that Kielin/chondrin like protein (KCP) can affect the progression of certain chronic and metabolic diseases by regulating extracellular signals. KCP is a secreted protein that is expressed at higher levels in embryonic germ and kidney tissues, and at lower levels in normal adult tissues. In the TCGA database, it was found that KCP is highly expressed in various tumor tissues, including bladder ureteral cancer, gallbladder cancer, colon cancer, head and neck tumors, esophageal cancer, and renal cancer (http://starbase.sysu.edu.cn/panGeneDiffExp.Php). KCP is also significantly highly expressed in cervical cancer, especially in young Asian squamous cell carcinoma patients, and affects prognosis. We previously successfully constructed a Kielin/chordin-like protein (KCP) knockout model of cervical squamous cell carcinoma cells in vitro using CRISPR/Cas9 gene editing technology, we designed a single lentiviral vector to deliver Cas9 sgRNA and a fluorescence selection marker to the target cells (6). SiHa is a cervical squamous cell carcinoma line containing human papillomavirus that is resistant to cisplatin. In this study, we preliminarily confirmed the regulatory function of KCP in mediating paclitaxel resistance of SiHa cells via cell function assays, whole human genome microarrays, and Information Process Analysis (IPA) bioinformatics analysis. This study is the first to explore the role of KCP in paclitaxel resistance in cervical cancer.
2 Materials and method
2.1 Materials
The human cervical squamous carcinoma cell line (SiHa) was purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). The following materials were used: DMEM medium (HyClone, 130701), fetal bovine serum (Hangzhou Sijiqing Company, VS500T), trypsin (GIBCO, 25200-072), paclitaxel (PTX) (PTX, Beijing Xiehe Pharmaceuticals, 141101), KCP-overexpressing lentivirus, KCP gene sgRNA lentivirus (Shanghai Genechem Co.,Ltd), MTT (Shanghai Dingguo Biotechnology Co., Ltd., DH343-2), apoptosis kit (eBioscience, 88-8007), human gene expression profiling chip GeneChip® PrimeView® human (Affymetrix, 902487), GeneChip® Hybridization Wash and Stain Kit (Shanghai GeneChem Co., Ltd), Agilent RNA 6000 Nano Kit (Agilent), TRIzol™ (Shanghai Pu Fei Company, 3101-100), M-mlV (Promega, M1705), dNTPs (Promega, U1240), RNase Inhibitor (Promega, N2115), oligo dT (Shanghai Biotech Company, B0205), Bulge-Loop™ miRNA qPCR Primer Set (Guangzhou Ruibo Company), KCP primary antibodies (Abcam, AB139956), β-actin (Abclonal, RP02968LQ), anti-rabbit secondary antibody (ZSGB-Bio, ZB2301) and SYBR® Green Master Mixture (TAKARA, DRR041B).
2.2 Equipment
The following instruments were used: fluorescence microscope (Olympus, IX71), CO2 incubator (Sanyo, MCO-15A), inverted microscope (Shanghai Cai Kang Optical Instruments Co., Ltd., XDS-100), centrifuge [Thermo Fisher Scientific (China) Co., Ltd., Fresco 21], biological safety cabinet (Shanghai Zhen Yang Chuang Air Purification Equipment Co., Bio-1200-II-A2), SDS-PAGE protein electrophoresis instrument (Shanghai Tianneng Company, VE-180), protein electrophoretic transfer instrument (Shanghai Tianneng Company, VE-186), high-speed refrigerated centrifuge (Thermo, Fresco 21), flow cytometer (Millipore, C6 PLUS), NanoDrop™ 2000 (Thermo), cellometer (Nexcelom Cellometer), 2100 Bioanalyzer (Agilent, Auto 1000), NanoDrop™ spectrophotometer (Thermo, 2000/2000C), GeneChip® Hybridization Oven 645 (Thermo, 00-0331), GeneChip® Fluidics Station 450 (Thermo, 00-0079), GeneChip® Scanner 3000 (Shanghai GeneChem Co., Ltd, Thermo 00-0362), stabilised flow electrophoresis instrument (Shanghai Tianneng Company, EPS-600), qPCR instrument (Roche, LightCycler® 480 II), and a microplate reader (Tecan infinite, M2009PR).
2.3 Methods
2.3.1 Cell culture
SiHa cells infected with KCP sgRNA lentivirus (sgRNA group; sgRNA sequence: GGACTCACCTTCCGCG- CCCG) and the control lentivirus (normal cells group, NC group; control lentivirussequence: CGCTTCCGCGGCCCGTTCAA) were incubated in DMEM medium containing 10% fetal bovine serum at 37°C in a 5% CO2 incubator. The medium was replaced every 2 to 3 day, and the cells in the logarithmic growth phase were extracted for the experiments. We applied Western Blot (WB) to verify whether KCP sgRNA was successfully transfected.
2.3.2 PTX concentrations for experiments
We determined the IC50 and concentration of PTX in earlier studies. PTX concentrations of 2 to 4 ng/ml inhibited cell growth by 23.34% to 74.12%, respectively (7), 2-4ng/ml can be chosen as the concentration for cell function experiments. After multiple repeated experimental evaluations, we finally chose 2ng/ml for colony formation assay experiments, 4ng/ml for FACS-based detection of apoptosis and MTT experiments.
2.3.3 Western blot
Approximately 1×106 siha cells were collected by centrifugation, and ripa lysate was added to lysis to release proteins. Protein concentration was determined using bicinchoninic acid (Pierce, Waltham, MA, USA). A protein sample of 3 ug/uL was prepared. Electrophoresis was carried out in 4% stacking gel at 90 V for 20 minutes and 8% separating gel at 120 V for 1 hour, then the proteins were transferred to a polyvinylidene fluoride membrane at 380 A for 90 minutes. The membrane was blocked in 5% non-fat milk for 1 hour at 25°C and incubated with primary antibodies (KCP antibody, Abcam, AB139956) at a ratio of 1:1,000 at 4°Covernight. Next, the membrane was incubated with secondary antibodies (anti-rabbit secondary antibody; Proteintech) at a ratio of 1:10,000 for 1 hour at 25°Cand protein bands were visualized using an Amersham Imager 600.
2.3.4 Colony formation assay
SiHa cells from the sgRNA and NC groups were harvested by trypsinization and counted, and the cell suspension was diluted before being inoculated in six-well plates at a density of 500 cells per well. The plates were then incubated in a cell incubator for 14 d until visible cell colonies appeared. On day 3, paclitaxel (2ng/ml) was added according to the experimental design, and no treatment was administered to the no-drug group. The medium was then discarded, and the cells were fixed in 4% paraformaldehyde for 15 min, followed by removal of the fixative. A total of 1 ml of crystal violet was added, and staining was performed for 10 min. The crystal violet was slowly washed away with running water. The colonies were counted directly with the naked eye or under a microscope for cell counts of more than 50, and the colony formation rate was calculated as follows: colony formation rate = (number of colonies/number of cells inoculated) × 100%.
2.3.5 Cell proliferation assay based on 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
SiHa cells derived from the sgRNA and NC groups were trypsinized and counted, and the cell density was adjusted to 2×103 cells/ml, followed by inoculation in 96-well plates at a volume of 100 ul/well. On day 2, paclitaxel (4ng/ml) was added according to the experimental design, and no treatment was administered to the no-drug group. The cells were continually incubated for a total of 5 days, after which a solution containing 20 ul of MTT (5mg/ml) was added to each well, followed by incubation for four hours. The culture medium was completely aspirated (care was taken not to aspirate the formazan granules at the bottom of the wells), and 100 ul of DMSO was added to each well to dissolve the formazan granules. After shaking with an oscillator for 3 to 5 min, the OD value was measured at 490 nm using a microplate reader. The relative viability of the cells was calculated from the OD values, and cell growth curves were generated. OD490/fold was the OD490 increase relative to day 1 for each experimental group from day 1 to day 6, representing the proliferation increase for each day. The inhibition rate was [1 - OD490/fold (sgKCP + paclitaxel)/OD490/fold(sgKCP)] × 100%.
2.3.6 FACS-based detection of apoptosis
Drug-induced apoptosis was assayed when the cells in six-well plates of each experimental group exhibited approximately 70% confluence (on day 7 after lentivirus infection). On day 4, paclitaxel (4ng/ml) was added according to the experimental design, and no treatment was administered to the no-drug group. The cells in the supernatant were collected and added to the adherent cell fraction. After trypsin digestion of the adherent cells, the complete medium was resuspended into a cell suspension and added to the supernatant cells in the same centrifuge tube (5ml), with three replicate wells per group (cell number≥5×105/process). They were then centrifuged at 1,300 rpm for 5 min, and the supernatant was discarded. The cell pellet was washed with pre-cooled D-Hanks buffer (pH=7.2~7.4) at 4°C. The cell pellet was washed once with 1× binding buffer, centrifuged at 1,300 rpm for 3 min, and then resuspended in a total of 200 ul of 1× binding buffer. A total of 10 ul of Annexin V-APC was added to the buffer for staining, and the solution was allowed to stain for 10–15 min at room temperature while protected from light. A total of 400 to 800 ul of 1×binding buffer was added depending on the number of cells, and the flow cytometry assay was performed with 1000 cells per run.
2.3.7 Gene chip detection and ingenuity pathway analysis bioinformatics analysis
Total RNA was extracted using the TRIzol™, and the samples were inspected using NanoDrop™ 2000 and Agilent Bioanalyzer 2100 instruments. Qualified samples were subjected to gene microarray testing according to the 3’ IVT PLUS kit instructions (Affymetrix item no. 902487). Based on the expression of differential genes detected by the gene microarray, we use the stats package in R language software (stats and graphics packages) to count the number of chip probe sets and plot them, IPA bioinformatics analysis was performed to predict the interaction relationship between the relevant detected genes and the target gene encoding KCP and to map the gene network (8, 9). Differentially expressed genes were defined as the FDR>1.3 or <-1.3, p-value<0.05.(URL:https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/).
2.3.8 q-PCR validation of downstream genes
Cells were collected (six-well plate, 80% cell density) and centrifuged at 2,000 rpm for 5 min. A total of 1 ml of TRIzol™ was added to the cell pellet and mixed thoroughly before allowing the solution to stand for 5 min at room temperature. The solution was then transferred to a new EP tube with a volume of 1.5 ml. A total of 200 ul of chloroform was added to each tube, which was turned upside down by hand before letting it stand for 8 min at room temperature. The solutions were then centrifuged at 12,800 rpm for 15 min at 4°C. The upper layer was transferred to a new EP tube with a volume of 1.5 ml. An equal volume of pre-cooled isopropanol was added, and the solution was mixed well before allowing it to stand for 10 min at 4°C. After centrifugation for 12 min at 12,800 rpm at 4°C, the supernatant was discarded, and the precipitate was washed with 1 ml of 75% ethanol. It was then centrifuged for 5 min at 11,800 rpm at 4°C, and most of the supernatant was discarded. The sample was centrifuged again for 5 min at 11,800 rpm at 4°C, and the supernatant was discarded, followed by drying at room temperature. When the RNA precipitate had turned clear, RNase-free water was added until it was completely dissolved, and the concentration and quality of the extracted RNA were determined with a NanoDrop™ 2000/2000C spectrophotometer. cDNA was obtained by reverse transcription, and real-time PCR was performed in two steps.
2.3.9 Statistical methods
Each experiment was repeated three times, and the average value was taken for analysis. SPSS 20.0 software was used for the statistical analysis and to generate plots. The measurement data are expressed as means ± the standard deviation (x ± s). The t-test was used for comparison of means between two groups. ANOVA was used for the comparison of means between multiple groups, and the chi-square test was used for comparison of rates between groups. Values with P < 0.05 were considered to indicate a statistically significant difference.
3 Results
3.1 WB were used to verify decreased expression after KCP knockout
We applied WB for validation of KCP-Cas9 knockout, using β-actin as an internal reference, in the SiHa control group (empty virus) vs SiHa-Cas9 KCP group, and it can be seen that KCP was successfully knocked out (Figures 1A, B) The experiment was repeated three times.
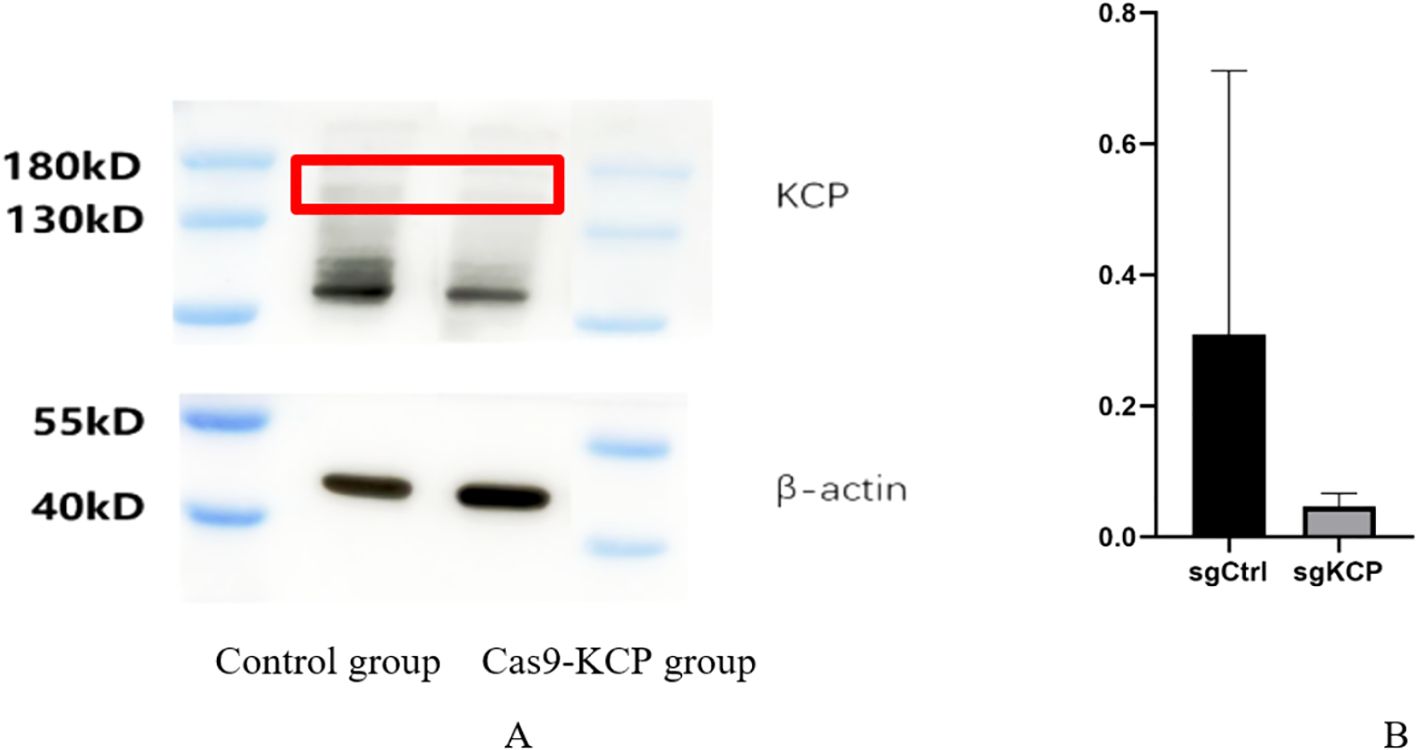
Figure 1. WB were used to verify expression after KCP knockout (A): Western blot demonstrating Cas9-KCP in SiHa cells. A lentivirus for the Cas9-KCP was constructed and co-cultured with SiHa cells. KCP expression was detected in the SiHa cells by western blotting. (B): KCP protein expression was signicantly lower in the Cas9-KCP group than in the control group.
3.2 Functional studies on the involvement of KCP in mediating paclitaxel resistance in cervical cancer cells
3.2.1 Use of the cervical squamous carcinoma cell line SiHa as an in vitro model of the influence of KCP on paclitaxel resistance
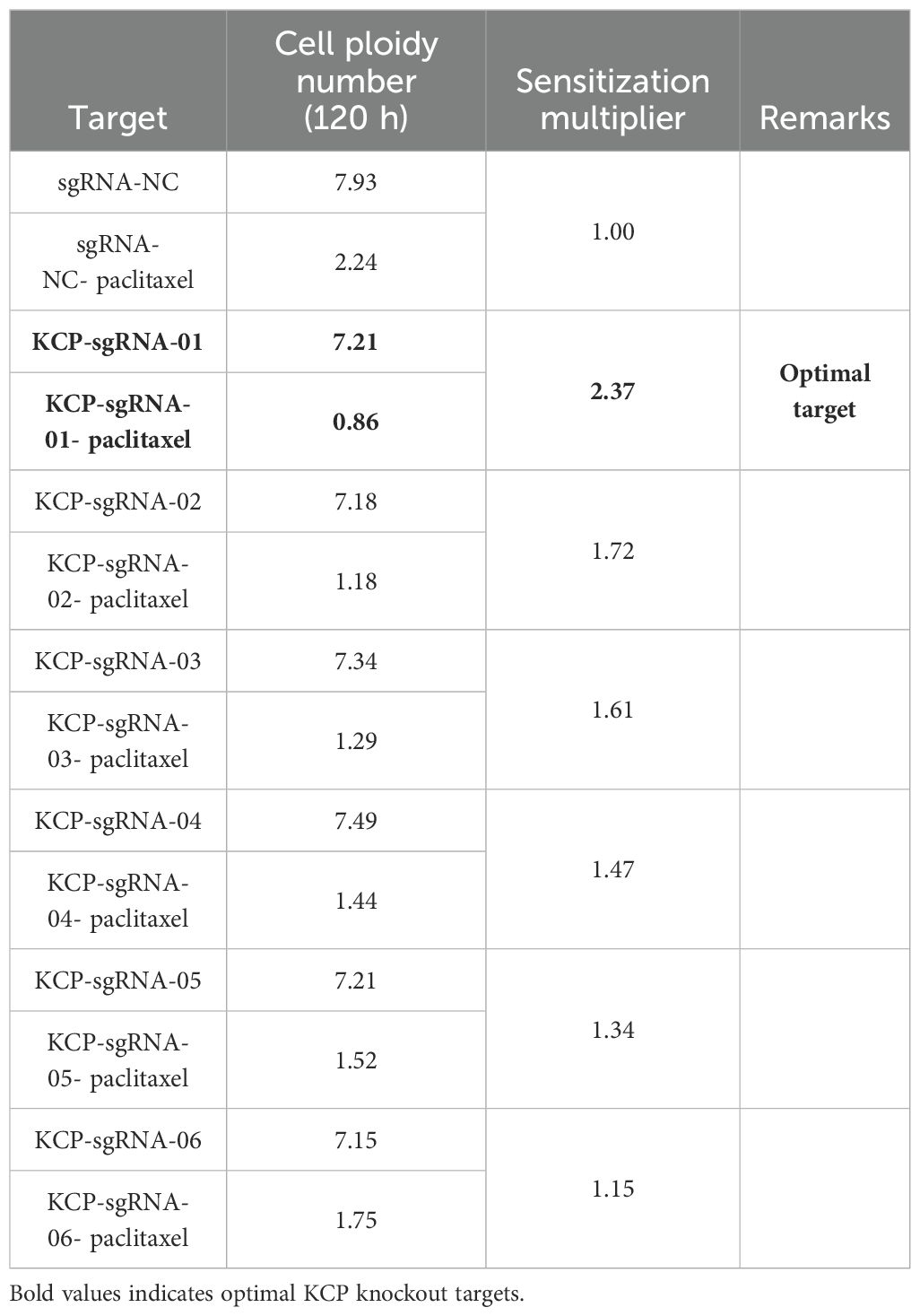
Using CRISPR/Cas9 gene editing, six sgRNAs were designed for the KCP gene. Plasmids were constructed, lentiviruses were packaged, and human cervical cancer cell line (SiHa) cells were infected to obtain the KCP knockout SiHa strain. We analyzed the ploidy data of each of the six SiHa strains after 120 hours of treatment with 4 ng/ml of paclitaxel or drug solvent (see Table 1). The optimal KCP knockout target was selected. KCP knockout significantly reduced the resistance of cervical squamous cell carcinoma SiHa cells to paclitaxel.
3.2.2 Effects on colony formation
The effect of KCP knockout on cervical squamous cell carcinoma SiHa colony formation was assayed. The cells were cultured for 9 days, and the number of cell colonies was then counted. The study group was incubated with or without paclitaxel (2ng/ml) on the fourth day. The results show that the number of SiHa cell colonies was as follows: 245 ± 11 in the sgCtrl group, 108 ± 8 in the sgCtrl+paclitaxel group, 233 ± 11 in the sgKCP group, and 56 ± 1 in the sgKCP + paclitaxel group. No statistically significant difference was observed between the sgCtrl and sgKCP groups (p=0.2755) and between the sgCtrl+paclitaxel and sgKCP+paclitaxel groups (p=0.0004). Compared to the sgCtrl+paclitaxel group, the sgKCP+paclitaxel group exhibited a significantly less potent SiHa cell colony formation ability, and the inhibition of SiHa cell colony formation after KCP knockdown increased the sensitivity to paclitaxel (Figure 2). The experiment was repeated three times.
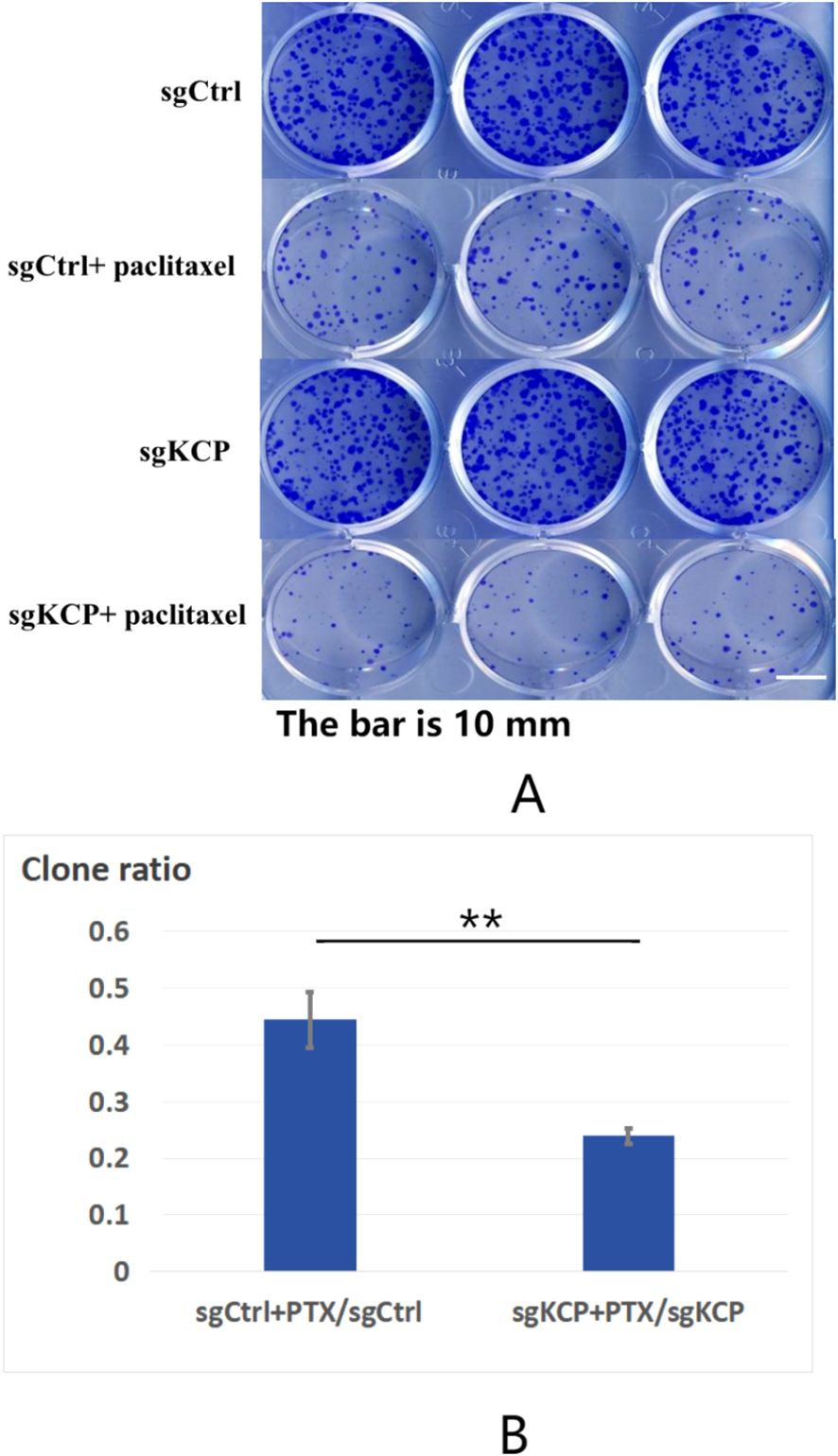
Figure 2. Effect of KCP knockout on SiHa cell colony formation, the experiment was repeated three times (A): The effect of KCP knockout on cervical squamous cell carcinoma SiHa colony formation. (B): The inhibition of SiHa cell colony formation after KCP knockdown increased the sensitivity to paclitaxel. sgCtrl: sgRNA-negative control virus-infected SiHa cells; sgCtrl+paclitaxel: paclitaxel-treated cells infected with sgRNA-negative control virus sgKCP: cells infected with sgRNA virus specific to the KCP gene; sgKCP+paclitaxel: paclitaxel-treated cells infected with the sgRNA virus specific to the KCP gene.
3.2.3 Effects on cell proliferation
To investigate the effect of KCP knockdown on cervical squamous carcinoma SiHa cell proliferation, we conducted experiments on SiHa cells infected with lentiviruses containing the target gene. After 72 hours of infection by the control or target lentivirus, observation of the cells by fluorescence microscopy showed that the infection efficiency was over 70% (Figure 3A). sgCtrl and sgKCP groups were incubated with or without paclitaxel (4ng/ml) for 6 days and treated with MTT for four hours. The changes in the light absorption at 490 nm were recorded separately for each group using a microplate reader. The OD490 reflects the number of viable cells. The results revealed a statistically significant difference between the sgCtrl and sgKCP groups (p=0.00219) and between the sgCtrl+paclitaxel and sgKCP+paclitaxel groups (p=0.000005). Compared to the sgCtrl+paclitaxel group, the cell proliferation in the sgKCP+paclitaxel group was significantly reduced, indicating that the cells in this group were more sensitive to paclitaxel. The experiment was repeated three times, the results are shown in Figure 3B.

Figure 3. MTT assay of SiHa cell proliferation after infection with sgKCP, the experiment was repeated three times (A): Comparison of changes in the light absorption measured at a wavelength of 490 nm using a microplate reader after treatment of sgRNA lentivirus-infected SiHa cells with MTT for four hours following incubation with paclitaxel. OD490/fold was the OD490 increase relative to day 1 for each experimental group from day 1 to day 6, representing the proliferation increase for each day. (B): Comparison of the inhibition of each cell group after the addition of paclitaxel, inhibition rate: [1-OD490/fold(sgKCP/sgKCP + paclitaxel)/OD490/fold(sgCtrl/sgCtrl + paclitaxel)] × 100. (C):The inhibition rate was significantly higher for the sgKCP group compared to that for the sgCtrl group after the addition of paclitaxel.
The inhibition rate of each cell group was compared after the addition of paclitaxel. The inhibition rate was significantly higher for the sgKCP group compared to that for the sgCtrl group, and the sensitivity to paclitaxel was increased after KCP knockout. The results are shown in Figure 3C.
3.2.4 Unaltered apoptosis rate of the cervical squamous cell carcinoma cell line SiHa after treatment with paclitaxel
The effect of KCP knockdown and control lentivirus on apoptosis in the cervical squamous carcinoma SiHa cell line was examined with and without paclitaxel (4ng/ml). No significant difference was observed between the apoptosis ratios of the groups. The results of the study revealed that the total apoptosis rate ratios were as follows: sgCtrl+paclitaxel/sgCtrl=3.6 and sgKCP+paclitaxel/sgKCP=3.57 (No significant, ns), the late apoptosis rate ratio of KCP knockout SiHa cells was not affected by paclitaxel treatment compared to the control group (ns), but the early apoptosis rate ratio of KCP knockout SiHa cells was lower compared to the control group (p<0.0001). The experiment was repeated three times, the results are shown in Figure 4.
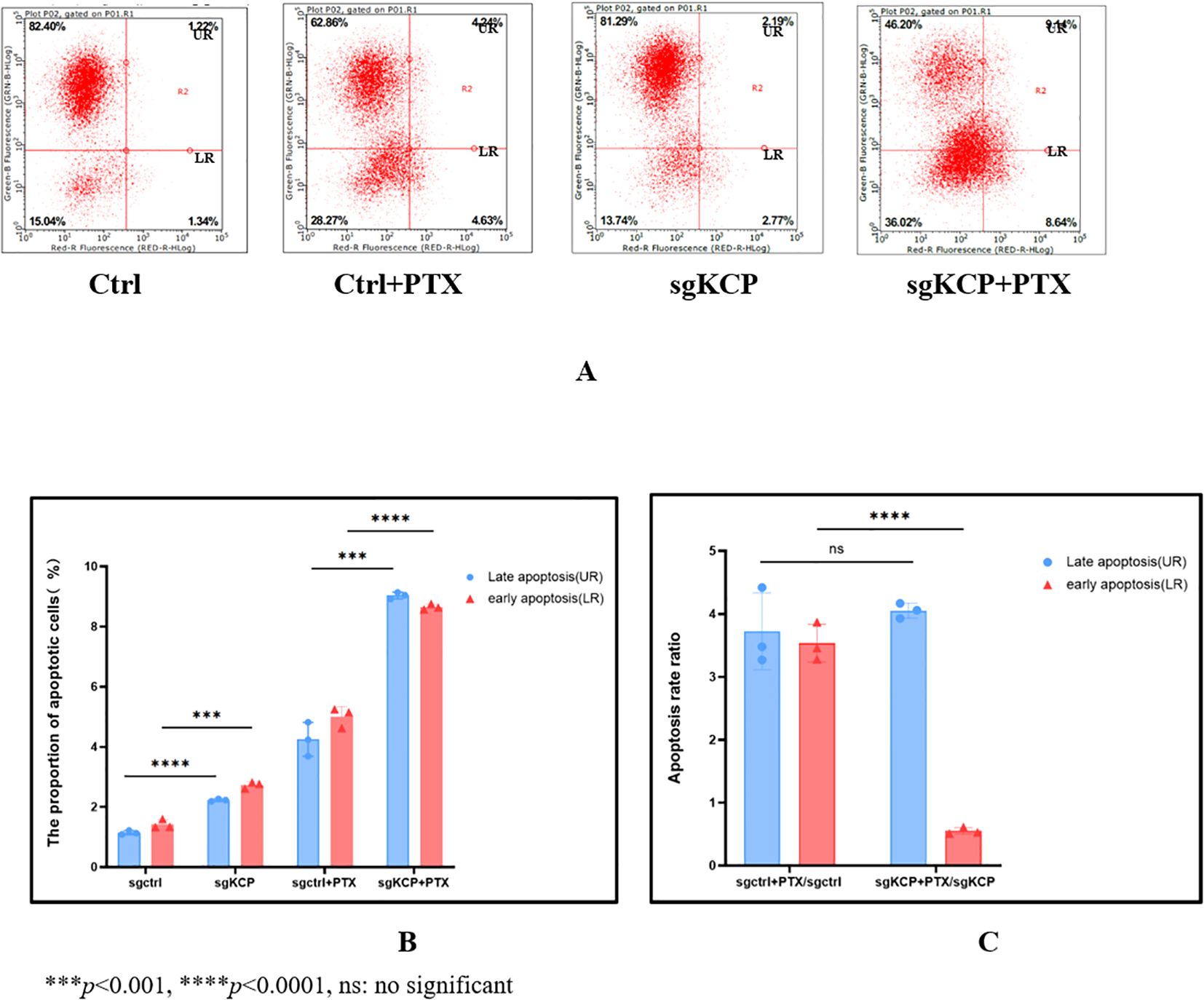
Figure 4. Effect of KCP knockout on SiHa cell apoptosis after treatment with paclitaxel, the experiment was repeated three times (A): Apoptosis and necrosis rates, UR: upper right (Late apoptosis), LR: lower right (Early apoptosis) (B): The proportion of early vs. late apoptosis in different group, the differences of early and late apoptosis between sgCtrl vs. sgKCP and sgCtrl+PTX vs. sgKCP+PTX were statistically significant. (C):The total apoptosis rate ratios were as follows: sgCtrl+paclitaxel/sgCtrl=3.6 and sgKCP+paclitaxel/sgKCP=3.57 (No significant, ns), the late apoptosis rate ratio of KCP knockout SiHa cells was not affected by paclitaxel treatment compared to the control group (ns), but the early apoptosis rate ratio of KCP knockout SiHa cells was lower compared to the control group (p<0.0001).
3.3 Human gene chip detection and IPA bioinformatics analysis
The total RNA of SiHa cells from the sgKCP and control lentivirus groups was extracted for gene chip assay. A total of 491 differentially expressed genes were screened by IPA bioinformatics analysis to examine if other factors may be related to the effects of KCP gene knockout. We also studied the mechanism of action underlying the effect of KCP on tumor cell proliferation. The IPA bioinformatics analysis included examination of regulatory effects, disease and function, classical pathways, upstream regulatory factors, and interaction networks. Ultimately, 30 possible KCP downstream genes were screened and validated using qPCR.
3.3.1 Classical pathways based on IPA bioinformatics analysis
Significant enrichment of differentially expressed genes in the classical pathways showed that 11 signaling pathways were significantly activated and three pathways were inhibited. The significantly activated signaling pathways included: the interferon (Z-score=2.0), role of pattern recognition receptors in recognition of bacteria and viruses (Z-score=2.0), corticotropin-releasing hormone g (Z-score=2.0), protein kinase A (Z-score=2.111), cardiac hypertrophy (Z-score=2.111), GPCR-mediated nutrient sensing in enteroendocrine cells (Z-score=2.236), P2Y purinergic receptor (Z-score=2.236), AMPK pathway (Z-score=2.236), dopamine-DARPP32 feedback in cAMP (Z-score=2.236), GNRH s (Z-score=2.236), and cardiac hypertrophy signaling pathways (Z-score=2.449). The significantly inhibited signaling pathways included the acute phase response (Z-score=-2.0), acute myeloid leukemia (Z-score=-2.0), and PI3K/AKT signaling pathways (Z-score=-2.0). Furthermore, the interferon signaling pathway was closely related to the tumor pathway (see Figure 5). The human gene microarray analysis showed that four differentially expressed genes, namely, IFIT1, OAS1, G1P2, and IFIT3, were important factors involved in mediating the interferon signaling pathway, suggesting that KCP may be involved in the interferon signaling pathway (Figure 6).
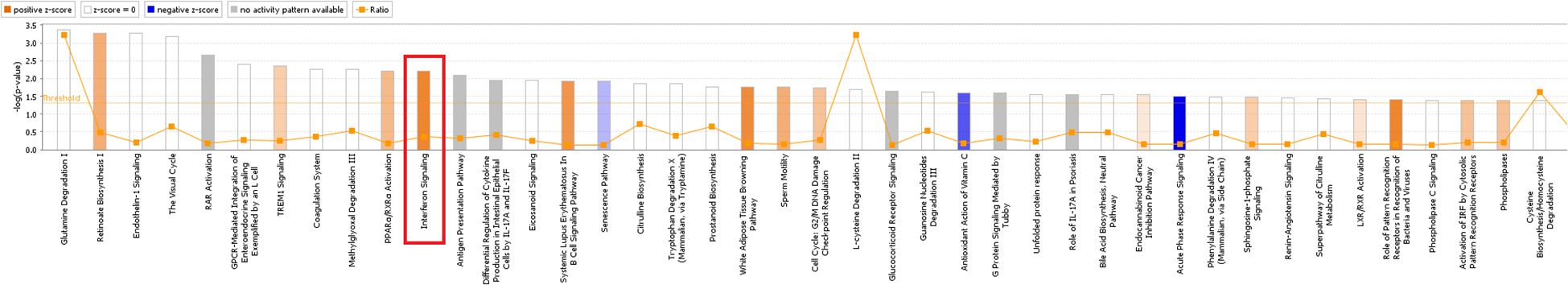
Figure 5. Classical pathway enrichment analysis. The abscissa indicates the name of the pathway, and the ordinate represents the enrichment significance level. The orange label indicates activation of the pathway. The blue label indicates inhibition of the pathway, and the depth of the orange and blue colors represents the degree of activation or inhibition. The ratio represents the number of differential genes in the signaling pathway to the number of all genes included in the pathway (The main genes are marked in red).
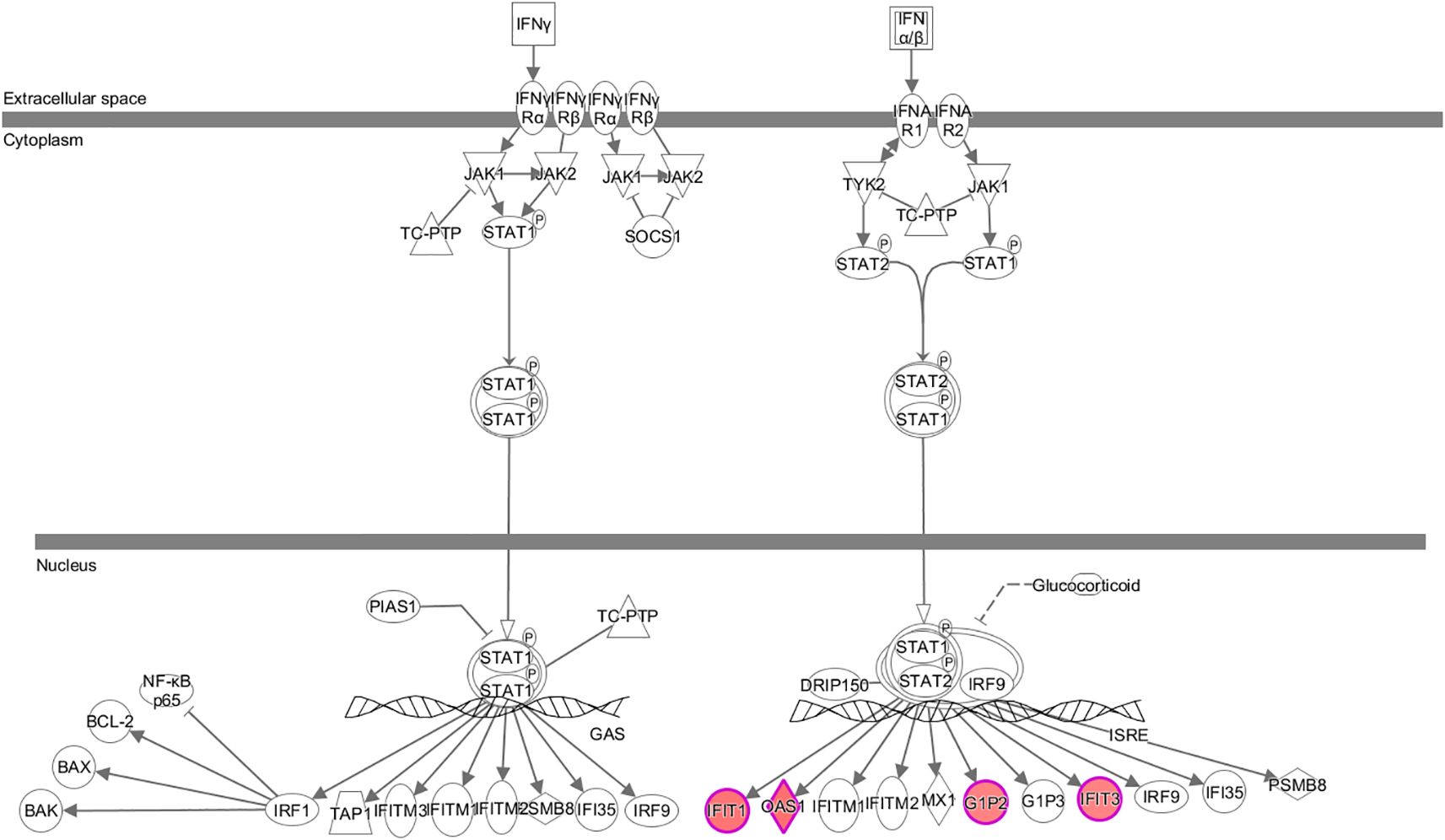
Figure 6. Expression trends of each molecule associated with the interferon signaling pathway based on gene chip analysis The interferon signaling pathway was significantly activated. Red color indicates that the gene was significantly upregulated, and green color indicates that the gene was significantly downregulated in the experimental results (The main genes are marked in red).
3.3.2 Upstream regulatory factors based on IPA bioinformatics analysis
Analysis of the upstream regulatory factors revealed that SPI1 was strongly activated, and 15 consistently activated genes were identified. SPI1-inhibited CCND1 activated both PML and CEBPA simultaneously, which is consistent with the observation of CCND1, PML, and CEBPA expression following KCP knockout (Figure 7).
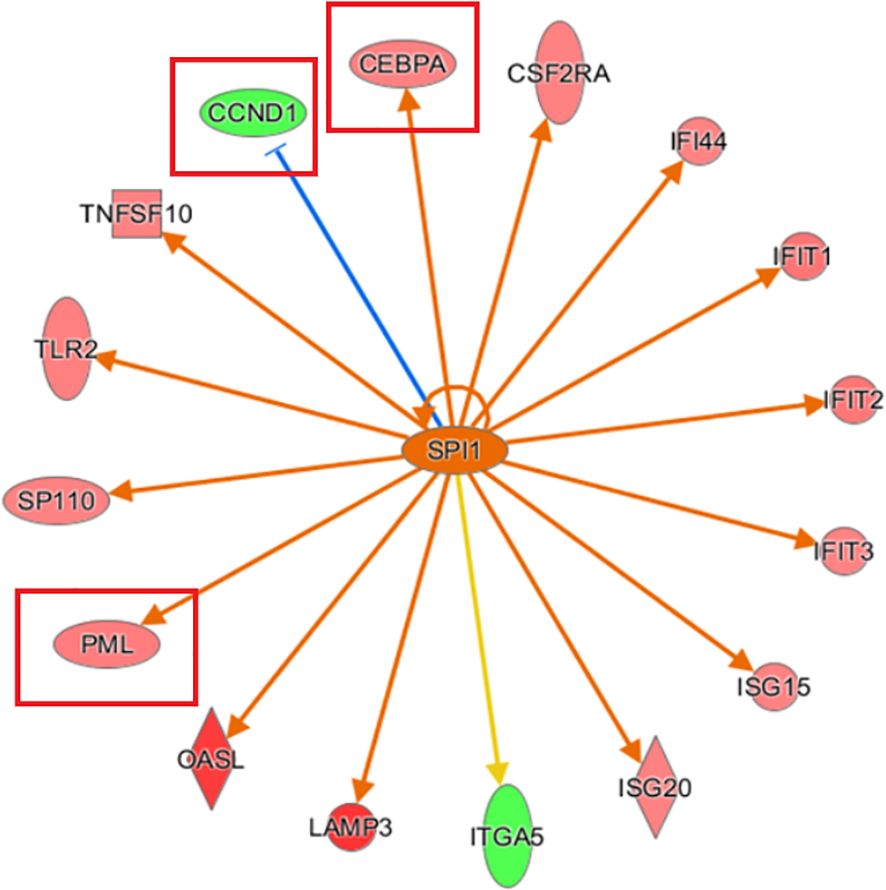
Figure 7. Network diagram of upstream regulatory factors based on IPA analysis Interaction relationships between the upstream regulatory factors based on IPA analysis, and directly related and co-existing downstream molecules in the dataset. The orange line indicates consistent activation of the upstream regulators and the gene. The blue line indicates consistent inhibition of the upstream regulators and the gene. The yellow line indicates inconsistent expression trends between the upstream regulators and the gene, and the grey line indicates the lack of predictive information related to the expression state in the dataset (The main genes are marked in red).
3.3.3 Diseases and functions examination by IPA bioinformatics analysis
Examination of diseases and functions based on IPA analysis showed significant activation of apoptosis of fibroblast cell lines (Z-score= 3.090) and cell death of connective tissue cells (Z-score= 2.998) and significant inhibition of replication of vesicular stomatitis virus (Z-score=-2.834) and abdominal carcinoma (Z-score=-2.337). PML and SERPINB3/4 (squamous cell carcinoma antigen 1, SCCA1) were key genes associated with the most significant activation of apoptosis of fibroblast cell lines (Figure 8).
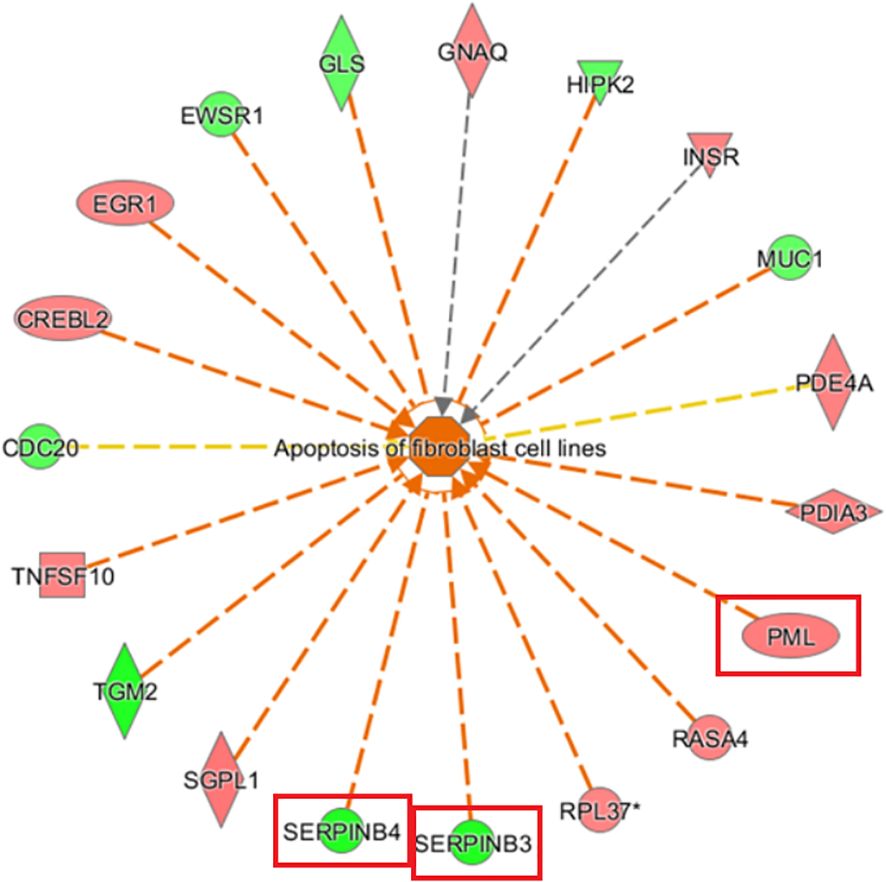
Figure 8. Gene disease or function network map based on IPA analysis The orange line indicates that changes in gene expression levels had an activating effect on this function. The blue line indicates that the changes in gene expression levels had an inhibiting effect on this function, and the yellow line indicates that changes in gene expression levels affected this gene function, which is inconsistent with existing literature reports. The grey line indicates that the regulatory relationship was unknown (The main genes are marked in red).
3.3.4 Analysis of regulatory effects by IPA bioinformatics
The top-ranked regulatory network in terms of regulatory effects according to the IPA-based analysis showed that it may involve the inhibitory effects of the differentially expressed genes ACKR2, Ifn, IFN Beta, IFNA2, Ifnar, IFNB1, IFNG, IFNl1, Il1RN, Interferon alpha, IRF1, IRF3, IRF5, IRF7, MAPK1 MAVS, PNPT1, SOCS3, SPI1, TRIM24 through DDX58, ISG20, MX2, OAS1, OASl, PlAAT4, PML, and SP110 on the regulators, replication of murine herpesvirus 4 and replication of vesicular stomatitis virus, thereby affecting the gene function of KCP (see Figure 9).
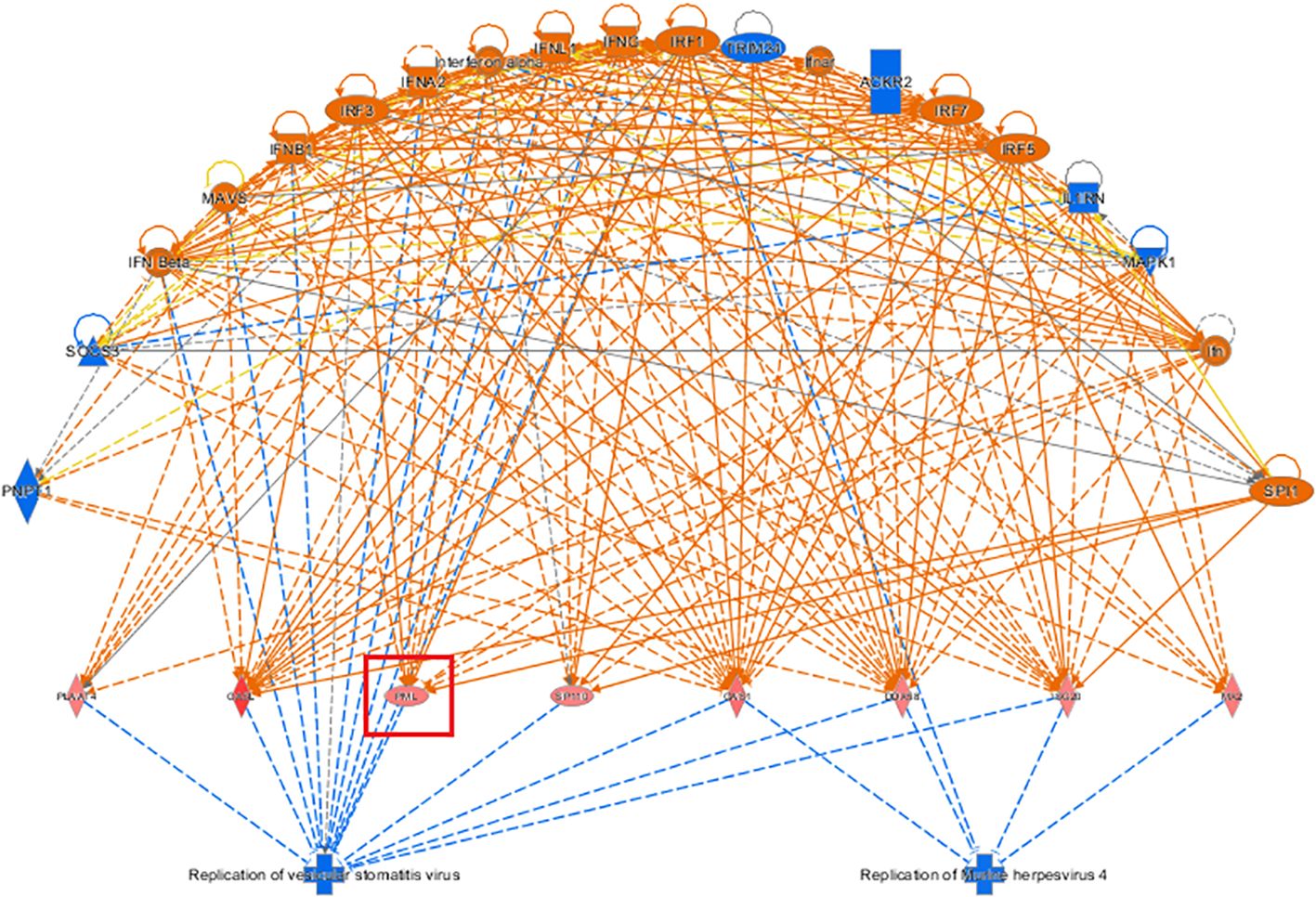
Figure 9. Network diagram of regulatory effects Showcased the interaction between genes, regulators, and functions in the dataset. The top-ranked regulatory network in terms of regulatory effects based on the IPA-based analysis showed that it may be due to the inhibitory effects of differentially expressed genes on the regulators, replication of murine herpesvirus 4 and replication of vesicular stomatitis virus, thereby affecting the gene function of KCP (The main genes are marked in red).
In the IPA analysis of differentially expressed genes in the KO vs. NC groups, genes related to tumor cell proliferation and apoptosis were identified in the disease function analysis results. Genes that inhibit tumor cell proliferation or that promote tumor cell apoptosis were selected, and KCP target genes were added and the gene relationship network diagram was drawn. The comprehensive results of the final IPA analysis indicate that KCP may be indirectly involved in the mechanism underlying paclitaxel resistance via the association of PML and CCND1 genes, thereby affecting the proliferation of SiHa cells (Figures 10).
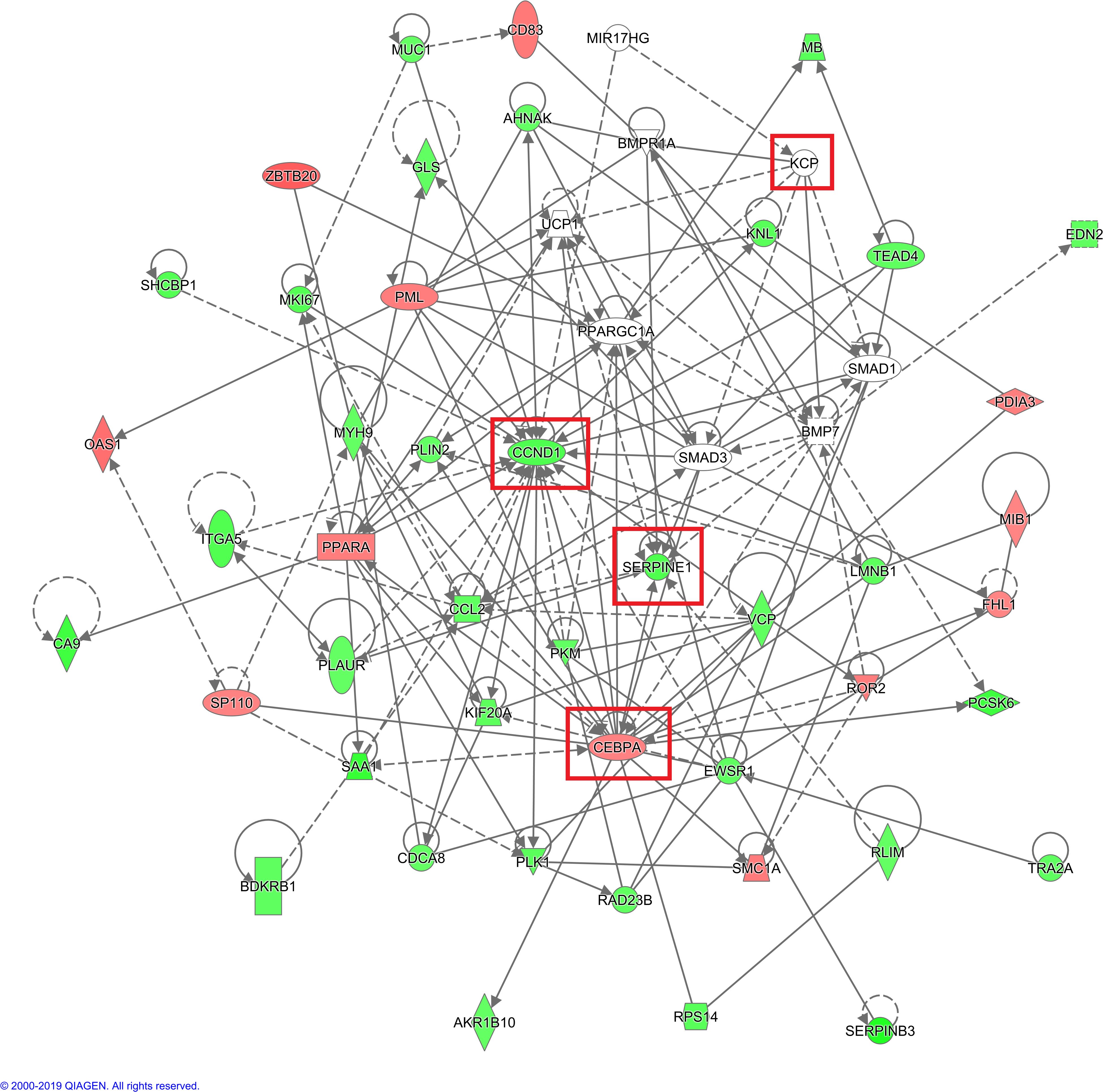
Figure 10. IPA analysis of gene interaction networks. Red color indicates that increased measurement, green color indicates that decreased measurement, darker color prompt more extreme increased measurement in dataset (The main genes are marked in red). Net work shapes: □ Cytokine, ◊ Enzyme, ▯ G-protein coupled receptor, ▽ Kinase, ▭ Ligand-dependent nuclear receptor,  microRNA,
microRNA,  other,
other,  Peptidase,
Peptidase,  Transcription regulator,
Transcription regulator,  Transmembrane receptor,
Transmembrane receptor,  Transporter.
Transporter.
3.4 Validation of the mechanism downstream of KCP knockout
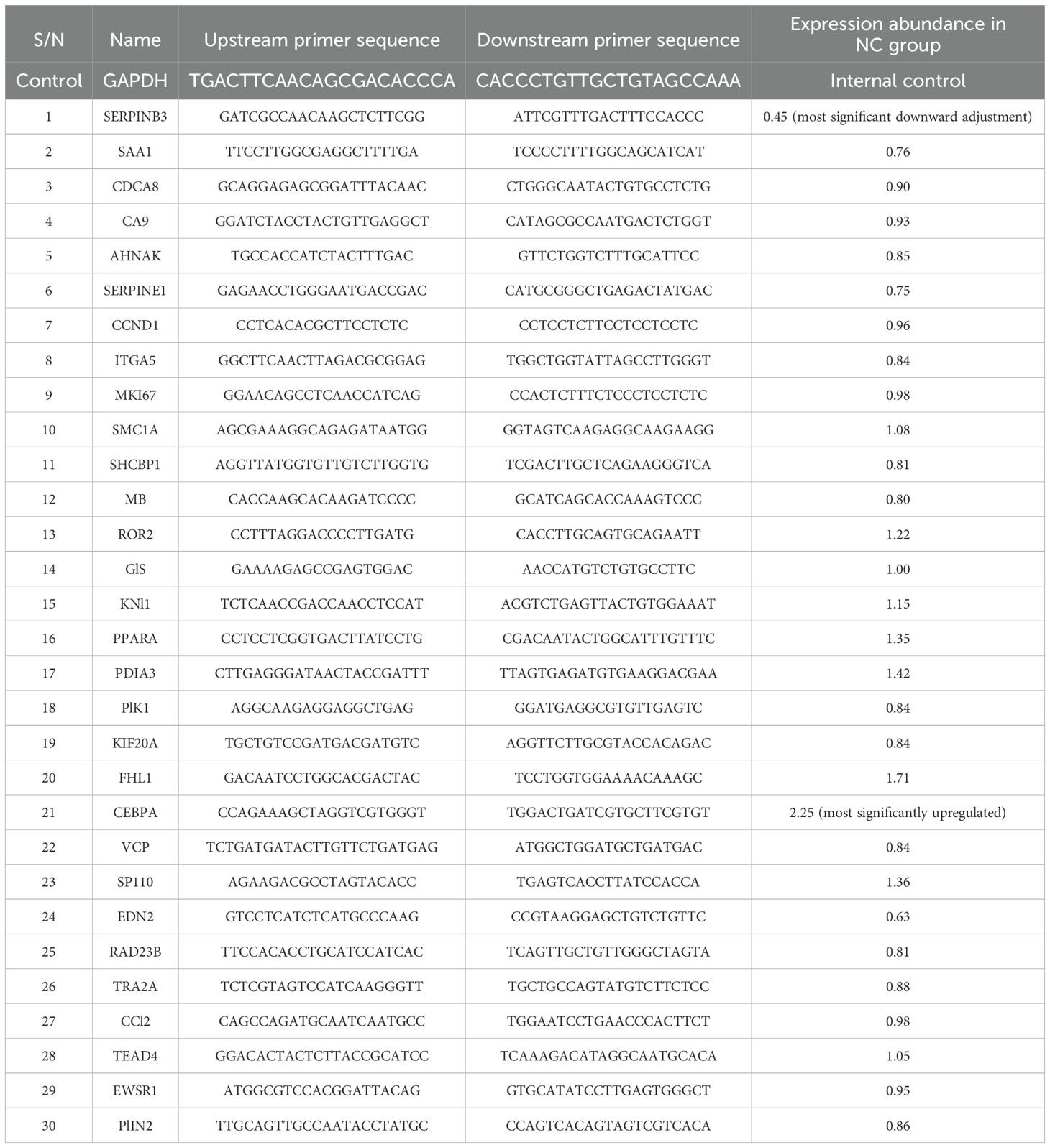
The differentially expressed genes screened based on the human gene chip were analyzed by IPA bioinformatics. A total of 52 genes associated with tumor cell proliferation were found by the disease function analysis. A total of 30 genes with the most significant differences were screened by qPCR for validation. Of these, SERPINB3 and CEBPA exhibited significantly lower and higher expression in KCP knockout group, respectively, compared to their expression in the control group. PML was not ranked among the top 30 genes due to its lesser difference, and hence, no follow-up validation was performed. The qPCR analysis verified that CCND1 had a slightly reduced expression. Therefore, we hypothesize that SERPINB3 and CEBPA may be forward and reverse downstream genes of KCP, respectively (see Table 2; Figure 11). CEBPA is a key factor of the acute myeloid leukemia signaling pathways, which may be significantly inhibited by KCP, as observed by the traditional IPA bioinformatics pathway analysis. SERPINB3 is the key gene that most significantly activates apoptosis of fibroblast cell lines, as shown by the IPA bioinformatics disease and functional analysis. As KCP is located in the extracellular matrix, further validation is required to elucidate its involvement in the transcriptional function of genes related to CCND1.
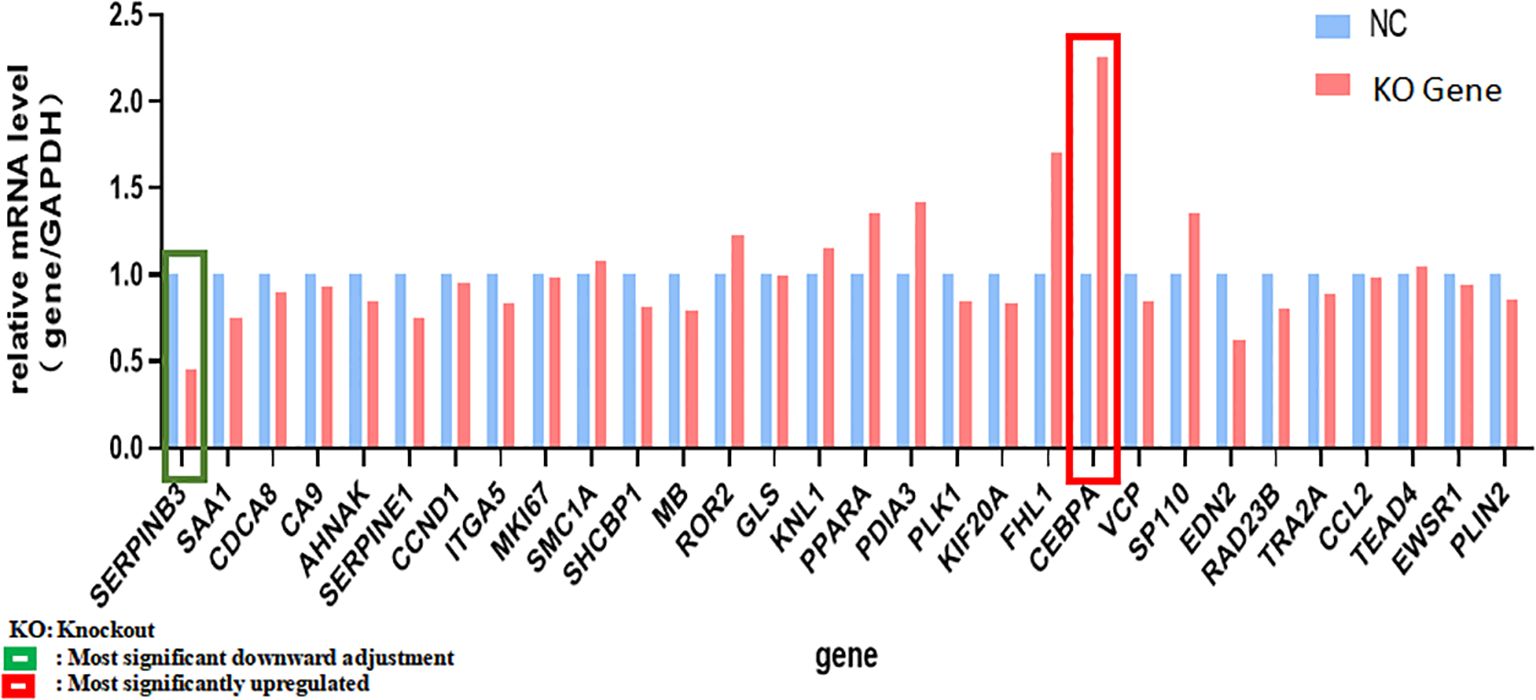
Figure 11. qPCR analysis verified genes in Table 2 qPCR was used to verified genes in Table 2. Green frame is the most significant downward adjustment, red frame is the most significantly upregulated (The main genes are marked in red).
4 Discussion
Cervical cancer is a serious health risk for women. Paclitaxel-based chemotherapy is pivotal in the treatment of advanced and recurrent cervical cancer as the primary treatment. Currently, paclitaxel-based chemotherapy regimens have been included in the guidelines of the National Comprehensive Cancer Network (NCCN) (10), the International Federation of Gynecology and Obstetrics and Gynecology (FIGO), and the European Society for Medical Oncology (ESMO) for the standard treatment of intermediate to advanced cervical cancer. However, in patients treated with paclitaxel as first-line chemotherapy, the drug resistance rates associated with intermediate to advanced squamous cervical cancers range from 20% to 44% (11). Paclitaxel resistance in patients with cervical squamous cell carcinoma leads to a progression of the disease, thus affecting the prognosis of patients and reducing the five-year survival rate by 20% to 30%. Studies in the USA and Japan have reported (12, 13) that 20% of patients with locally advanced squamous cell cervical cancer are resistant to paclitaxel-based neoadjuvant chemotherapy, resulting in treatment failure. Paclitaxel resistance is a major cause of treatment failure in patients with cervical squamous cell carcinoma. Currently, no specific markers or targets for drug resistance in cervical cancer are known.
In 2002, Abreu’s research group discovered that KCP could affect the progression of certain chronic and metabolic diseases via the regulation of extracellular signaling. KCP comprises a cysteine structural domain with a secretory function and is generally highly expressed in the germ and kidney structures during embryonic development, but there is little expression in normal adult tissues. Current research on KCP has focused on chronic liver and kidney fibrosis and metabolic diseases (14, 15), suggesting that it can affect cell proliferation, migration, and adhesion (11). Recent studies examining the pathogenic mechanisms underlying cervical cancer have focused on the Wnt, Notch, BMP, and Jak-Stat3 signaling pathways (16). The results show that the most common regulatory pathway involves binding of KCP to TGF-β receptors, thereby inhibiting their signaling and promoting receptor-ligand interactions in a paracrine manner. Furthermore, enhancement of the BMP signaling pathway has also been observed. The binding of KCP to BMP7 enhances the binding to type I receptors, which in turn increases Smad1-dependent transcription and phosphorylated Smad1 (P-Smad1) levels (17–19). TGF-β and BMP regulatory pathways play important roles in the proliferation and invasion of various malignant tumors.
In this study, we successfully generated a KCP knockout cervical squamous cell carcinoma in vitro model using CRISPR/Cas9 gene editing technology and preliminarily confirmed the regulatory function of KCP in SiHa cell line. Further testing confirmed the involvement of KCP in paclitaxel resistance of cervical cancer cells by examination of cell colony formation and proliferation. The study showed that KCP knockout significantly inhibited colony formation and proliferation of cervical cancer cells in the paclitaxel group, thereby increasing the sensitivity to paclitaxel. The differentially expressed genes after the KCP knockout were analyzed by human whole genome microarray and IPA bioinformatics analyses, and the downstream genes and mechanisms were explored.
The IPA bioinformatics analysis revealed that the most important classical pathway for KCP activation was the interferon signaling pathway, and the downstream factors included the JAK-STAT signaling pathway as well as BAX, BAK, BCl-2, and other possible key factors That is partially consistent with the recent report mentioned in the previous paragraph (20).The analysis of the IPA regulatory network showed that the aforementioned regulation may be associated with the regulatory network of the replication of the murine herpesvirus 4 and the replication of the vesicular stomatitis virus. This indicated that KCP may be indirectly involved in cell proliferation via PML and CCND1. further experiments are required to confirm the mechanism of CCND1 suppression and PML activation by observing the activation of SPI1 after KCP knockout. According to the IPA analysis of differentially expressed genes, qPCR was used to verify KCP could also act positively on the downstream gene of SERPINB3 and negatively on the downstream gene of CEBPA to affect the resistance of cervical carcinoma to paclitaxel. It has been reported that CEBPA is positively related to chemoresistance and that CEBPA overexpression can significantly enhance chemoresistance in oral squamous cell carcinoma (OSCC) (20, 21). The forward downstream gene, SERPINB3, as validated by qPCR, may be the key factor that significantly activates the disease and functions during the apoptosis of fibroblast cell lines. The endogenous lysosomal cysteine protease inhibitor SERPINB3 (SCCA1) is elevated in patients with cervical cancer and other malignancies. Cervical cancer cells lacking SERPINB3 are more sensitive to ionizing radiation (IR), suggesting that this protease inhibitor plays a role in the therapeutic response. t has been reported that SERPINB3 confers resistance to drug-induced apoptosis by inhibiting lysosomal cathepsin proteases in cancer cells (22). SERPINB3 enhances the survival ability of tumor cells by regulating cytokine activity and signaling pathways related to tumor development (23). Wang et al. demonstrated that SERPINB3-deficient cells had enhanced sensitivity to IR-induced cell death. Knockout of SERPINB3 sensitizes cells to a greater extent than cisplatin (24, 25). However, the reverse downstream gene, CEBPA, is a key factor involved in the significant inhibition of the disease and function-related signaling pathways in acute myeloid leukemia signaling. CEBPA participates in chemoresistance through METTL3/METTL14/BHLHB9 in vivo, which accelerated the tumor growth (26). Therefore, the involvement of KCP in related diseases and functions through the downstream factors SERPINB3 and CEBPA should also be studied in the future.
PTX resistance is the main cause of treatment failure in cervical cancer. Therefore, developing a strategy to overcome the PTX resistance has become the most critical clinical subject for gynecologic oncologists. Considering the close relationship between KCP and paclitaxel resistance, KCP can be used as a candidate gene for paclitaxel resistance markers and therapeutic targets in cervical cancer in the future.
5 Summary
The incidence and death rates of cervical cancer still rank it as the foremost malignant tumor of the female reproductive system in China. The process of onset and development is very complex, with involvement of multiple genes, molecular proteins, biological factors, and signaling pathways in its regulation. Chemotherapy resistance is a common cause of recurrence. In recent years, even though a degree of progress has been made in gene therapy and molecular targeted therapy for cervical cancer, it is still crucial to explore the molecular mechanism of chemotherapy resistance in cervical cancer in depth, to find effective therapeutic targets, to design new innovative drugs developed independently in China, to reduce the incidence of chemotherapy resistance, to prolong the OS and PFS of patients with tumor, and to improve their quality of life. This study suggests that KCP, which is an emerging protein, plays an important role in the proliferation of tumor cells, and its related mechanism has also been partially explored. We aim to explore more medical evidence, which is expected to provide a new direction to overcome the drug resistance of cervical cancer.
6 Limitations
This paper has some limitations:
1. Only one cell line of KCP knockout was established in this research. We discuss the paclitaxel resistance gene in cervical squamous cell carcinoma. SiHa is a human papilloma virus-related cervical squamous cell carcinoma line, resistant to cisplatin, we use this cell line to preliminarily explore the function of KCP in cervical squamous cell carcinoma. In future research, we will use various cell lines such as SiHa, Caski, HeLa, etc. to explore the mechanism of KCP’s involvement in paclitaxel resistance in cervical cancer.
2. A total of 30 genes with the most significant differences were screened for qPCR validation. However, it is not been confirmed that these differentially expressed genes are related to paclitaxel resistance.
3. The study of molecular mechanisms is mainly based on speculation of bioinformatics results and lacks cell and animal experimental confirmation.
7 Future perspective
1. We will conduct experiments on sensitive and resistant to paclitaxel squamous cervical cancer tumor tissues in the future to confirm the relationship of these genes with paclitaxel resistance.
2. The role of KCP in infiltration, invasion and metastasis in cervical cancer was not evaluated in the present study, we will conduct these experiments in the future.
3. We have preliminarily validated the differential genes that may be involved in paclitaxel resistance in cervical cancer screening, and our subsequent research will arrange experiments to clarify the downstream related genes of KCP.
4. We plan to perform animal experiments to supplement for the verification of KCP and related genes in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
YH: Writing – original draft, Writing – review & editing, Conceptualization, Supervision, Validation. J-QX: Investigation, Methodology, Writing – review & editing. J-JZ: Methodology, Resources, Writing – review & editing. Z-YL: Investigation, Methodology, Writing – review & editing. CJ: Resources, Software, Writing – review & editing. YL: Resources, Software, Writing – review & editing. Y-FW: Investigation, Writing – review & editing. MW: Data curation, Visualization, Writing – review & editing. Y-MW: Writing – review & editing, Supervision. YW: Writing – review & editing, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. 1.National Natural Science Foundation of China (Grant No. 82102711). 2. Beijing Municipal Science & Technology Commission (Grant No. Z221100007422087) 3. Talent Development Plan for High-level Public Health Technical Personnel Project(Subject Backbone-03-30) 4. Beijing Hospitals Authority Clinical Medicine Development of Special Funding Support (Grant No. ZLRK202529). 5. Laboratory for Clinical Medicine, Capital Medical University.
Acknowledgments
(1) We are grateful to all the reviewers and editors who provided comments that substantially improved the manuscript. (2) We are grateful to Shanghai GeneChem Co., Ltd for their support of the experimental procedures.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Podwika SE, Duska L. Top advances of the year: Cervical cancer. Cancer. (2023) 129:657–63. doi: 10.1002/cncr.v129.5
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
3. Ji L, Chen M, Yao L. Strategies to eliminate cervical cancer in China. Front Oncol. (2023) 13:1105468. doi: 10.3389/fonc.2023.1105468
4. Arbyn M, Weiderpass E, Bruni L, de Sanjosé S, Saraiya M, Ferlay J, et al. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob Health. (2020) 8:e191–203. doi: 10.1016/S2214-109X(19)30482-6
5. Abreu JG, Coffinier C, Larraín J, et al. Chordin-like CR domains and the regulation of evolutionarily conserved extracellular signaling systems. Gene. (2002) 287:39–47. doi: 10.1016/S0378-1119(01)00827-7
6. Wei W, He Y, Wu YM. Identification of genes associated with SiHa cell sensitivity to paclitaxel by CRISPR-Cas9 knockout screening. Int J Clin Exp Pathol. (2018) 11:1972–8.
7. He Y, Han SB, Geng YN, Yang SL, Wu YM. Quantitative analysis of proteins related to chemoresistance to paclitaxel and carboplatin in human SiHa cervical cancer cells via iTRAQ. J Gynecol Oncol. (2020) 31:e28. doi: 10.3802/jgo.2020.31.e28
8. Felciano RM, Bavari S, Richards DR, Billaud JN, Warren T, Panchal R, et al. Predictive systems biology approach to broad-spectrum, host-directed drug target discovery in infectious diseases. Pac Symp Biocomput. (2013) 2013:17–28. doi: 10.1038/s42003-021-02893-6
9. Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, et al. A network-based analysis of systemic inflammation in humans. Nature. (2005) 437:1032–7. doi: 10.1038/nature03985
10. Abu-Rustum NR, Yashar CM, Arend R, Barber E, Bradley K, Brooks R, et al. NCCN guidelines® Insights: Cervical Cancer, Version 1.2024. J Natl Compr Canc Netw. (2023) 21:1224–33. doi: 10.6004/jnccn.2023.0062
11. Kastritis E, Bamias A, Efstathiou E, Gika D, Bozas G, Zorzou P, et al. The outcome of advanced or recurrent non-squamous carcinoma of the uterine cervix after platinum-based combination chemotherapy. Gynecol Oncol. (2005) 99:376–82. doi: 10.1016/j.ygyno.2005.06.024
12. Mori T, Makino H, Okubo T, Fujiwara Y, Sawada M, Kuroboshi H, et al. Multi-institutional phase II study of neoadjuvant irinotecan and nedaplatin followed by radical hysterectomy and the adjuvant chemotherapy for locally advanced, bulky uterine cervical cancer: A Kansai Clinical Oncology Group study (KCOG-G1201). J Obstet Gynaecol Res. (2019) 45:671–8. doi: 10.1111/jog.2019.45.issue-3
13. de la Torre M. Neoadjuvant chemotherapy in woman with early or locally advanced cervical cancer. Rep Pract Oncol Radiother. (2018) 23:528–32. doi: 10.1016/j.rpor.2018.09.005
14. Garcia Abreu J, Coffinier C, Larraín J, Oelgeschläger M, De Robertis EM. Chordin-like CR domains and the regulation of evolutionarily conserved extracellular signaling systems. Gene. (2020) 287:39–47. doi: 10.1016/S0378-1119(01)00827-7
15. Bradford STJ, Ranghini EJ, Grimley E, Lee PH, Dressler GR. High-throughput screens for agonists of bone morphogenetic protein (BMP) signaling identify potent benzoxazole compounds. . J Biol Chem. (2019) 294:3125–36. doi: 10.1074/jbc.RA118.006817
16. Huang JM, Zhang GN, Shi Y, Zha X, Zhu Y, Wang MM, et al. Atractylenolide-I sensitizes human ovarian cancer cells to paclitaxel by blocking activation of TlR4/MyD88-dependent pathway. . Sci Rep. (2014) 4:3840. doi: 10.1038/srep03840
17. Federspiel JD, Tandon P, Wilczewski CM, Wasson L, Herring LE, Venkatesh SS, et al. Conservation and divergence of protein pathways in the vertebrate heart. PloS Biol. (2019) 17:e3000437. doi: 10.1371/journal.pbio.3000437
18. Soofi A, Wolf KI, Emont MP, Qi N, Martinez-Santibanez G, Grimley E, et al. The kielin/chordin-like protein (KCP) attenuates high-fat diet-induced obesity and metabolic syndrome in mice. J Biol Chemi. (2017) 292:9051–62. doi: 10.1074/jbc.M116.771428
19. Lin J, Patel SR, Cheng X, Cho EA, Levitan I, Ullenbruch M, et al. Kielin/chordin-like protein, a novel enhancer of BMP signaling, attenuates renal fibrotic disease. Nat Med. (2005) 11:387–93. doi: 10.1038/nm1217
20. Qiao X, Zhu L, Song R, Shang C, Guo Y. METTL3/14 and IL-17 signaling contribute to CEBPA-DT enhanced oral cancer cisplatin resistance. Oral Dis. (2023) 29:942–956. d2023. doi: 10.1111/odi.14083
21. Xu N, Chen WM, Li LD, Long LY, Wang X, Jiang Q, et al. High WT1 expression predicted induction chemotherapy failure in acute myeloid leukemia patients with non-favorable cytogenetic risk. Clin Exp Med. (2023) 23:2629–38. doi: 10.1007/s10238-023-00995-5
22. Suminami Y, Nagashima S, Vujanovic NL, Hirabayashi K, Kato H, Whiteside TL. Inhibition of apoptosis in human tumour cells by the tumour-associated serpin, SCC antigen-1. Br J Cancer. (2000) 82:981–9. doi: 10.1054/bjoc.1999.1028
23. Lim W, Kim HS, Jeong W, Ahn SE, Kim J, Kim YB, et al. SERPINB3 in the chicken model of ovarian cancer: a prognostic factor for platinum resistance and survival in patients with epithelial ovarian cancer. PloS One. (2012) 7:e49869. doi: 10.1371/journal.pone.0049869
24. Wang S, Luke CJ, Pak SC, Shi V, Chen L, Moore J, et al. SERPINB3 (SCCA1) inhibits cathepsin L and lysoptosis, protecting cervical cancer cells from chemoradiation. Commun Biol. (2022) 5(1):46. doi: 10.1038/s42003-021-02893-6
25. Markovina S, Wang S, Henke LE, Luke CJ, Pak SC, DeWees T, et al. Squamous cell carcinoma antigen mediates radiation resistance in cervical cancer. Br J Cancer. (2018) 118:72–8. doi: 10.1038/bjc.2017.390
Keywords: Kielin/chordin-like protein, paclitaxel resistance, cervical squamous cell carcinoma, mechanism, functions
Citation: He Y, Xu J-Q, Zhang J-J, Liu Z-Y, Ji C, Liu Y, Wang Y-F, Wang M, Wu Y-M and Wang Y (2025) Examination of the functions and mechanism of KCP in mediating paclitaxel resistance in cervical squamous carcinoma cells. Front. Oncol. 15:1550032. doi: 10.3389/fonc.2025.1550032
Received: 31 December 2024; Accepted: 26 March 2025;
Published: 14 April 2025.
Edited by:
Catalin Marian, Victor Babes University of Medicine and Pharmacy, RomaniaReviewed by:
Kulbhushan Thakur, University of Delhi, IndiaVarsha Karunakaran, University of Arkansas, United States
Copyright © 2025 He, Xu, Zhang, Liu, Ji, Liu, Wang, Wang, Wu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan-Wang, d3d5eTMxOTZAY2NtdS5lZHUuY24=; Yu-Mei Wu, d3ltNTk3MTE4QGNjbXUuZWR1LmNu
 Yue He
Yue He Ming Wang
Ming Wang Yu-Mei Wu
Yu-Mei Wu Yan Wang
Yan Wang