- 1College of Pharmacy, Inner Mongolia Medical University, Hohhot, Inner Mongolia Autonomous Region, China
- 2School of Basic Medicine, Qingdao University, Qingdao, Shandong, China
- 3School of Stomatology, Zhangzhou Health Vocational College, Zhangzhou, Fujian, China
- 4College of Life Sciences, Inner Mongolia Agricultural University, Hohhot, Inner Mongolia Autonomous Region, China
Circular RNAs (circRNAs) are a new category of non-coding RNAs implicated in the molecular pathology of cancer, such as oral squamous cell carcinoma (OSCC). Circular intronic RNAs (ciRNAs), exonic circRNAs (ecircRNAs), and exon-intron circRNAs (EIciRNAs) are three primary types of circRNAs resulted from the circularization of extron and intron which give rise to the distinct biology of circRNAs. Due to their unique structure and biogenesis, circRNAs exhibit tissue- and cell-specific expression profiles. Recent studies have highlighted that in OSCC, some circRNAs are differentially expressed compared with adjacent normal tissues, with these variables potentially influencing OSCC initiation and progression through diverse mechanisms. Furthermore, earlier clinical trials have indicated that circRNAs could be considered as potential therapeutic targets and biomarkers for OSCC. It should be noted that many circRNAs modulate tumor cells proliferation/apoptosis and metastasis via regulating gene transcription and post transcriptional expressions. Furthermore, certain circRNAs function as effective microRNA sponges, thereby inhibiting oncogenic pathways in OSCC. In summary, the discovery of circRNAs has unveiled new avenues for cancer research, particularly in OSCC. This review provides an overview of circRNA biogenesis, their biological functions, and their roles as differentially expressed molecules in OSCC, emphasizing their potential for clinical application and warranting further investigation into their functional and therapeutic relevance.
1 Introduction
Oral squamous cell carcinoma (OSCC) is a malignant tumor that is prevalent in the head and neck region, ranking eleventh among the most common cancers worldwide (1). OSCC arises from various risk factors, including smoking, chronic betel nut chewing, human papillomavirus (HPV) infection, and poor oral hygiene, typically affecting areas such as the tongue, gums, oropharynx, and buccal mucosa (2, 3). Due to its asymptomatic and non-specific early-stage presentation, OSCC is often diagnosed at advanced stages, frequently with lymph node metastasis (LNM) or distant metastasis (DM) (4). Conventional surgical approaches often fail to achieve complete remission, necessitating adjunctive therapies like radiotherapy or chemotherapy to improve survival and prognosis (5). However, the inherent tumor heterogeneity allows cancer cells to frequently develop resistance to these treatments, with approximately 60% of patients with OSCC showing resistance to radiation or chemotherapy in clinical settings (6). Given these complexities, the current clinical strategies for OSCC remain suboptimal, with a five-year survival rate below 60% (7). This underscores the urgent need to elucidate the molecular mechanisms driving OSCC in order to identify effective biomarkers and novel therapeutic targets for clinical application.
The discovery of new disease targets and diagnostic technologies has brought the prospect of tumor eradication closer to reality due to recent advances in biology (8). For instance, identify the non-coding RNAs (ncRNAs), verify their specific biological functions has challenged the previous notion of these RNAs as mere ‘genetic byproducts’ (9). Circular RNAs (circRNAs) are endogenous non-coding RNAs that are covalently closed loop structures and are expressed everywhere and specifically in eukaryotic cells. Initially considered byproducts of aberrant splicing (10), circRNAs are now recognized for their stability, as their unique closed-loop structure protects them from degradation by nucleases (11). Furthermore, the circRNAs that are expressed differently in different diseases and tumor tissues indicate their tissue- and time-specific functions (10, 12). In addition, exosomes secreted by oral cancer stem cells also contain circRNAs, which may similarly influence OSCC progression through intercellular communication (13). Although many aspects of circRNA biology remain unclear, accumulating evidence suggests that certain circRNAs may serve as promising therapeutic targets and diagnostic/prognostic biomarkers in cancers. This review provides an overview of the biological activities and mechanisms of circRNAs, highlighting their vital roles in the tumorigenesis of OSCC, examining their regulatory mechanisms, and exploring their potential clinical applications.
2 CircRNAs synthesis (biogenesis)
In eukaryotes, newly synthesized precursor mRNAs consist of introns and exons that undergo splicing to produce various RNA forms (14). In classical RNA splicing, the upstream 5’-donor site is linked to the downstream 3’-acceptor site, where introns are removed and exons are ligated to form mature linear mRNAs (15). In contrast, circRNA formation involves reverse splicing, where the 3’ end of an exon joins the 5’ end of an intron, creating a covalently closed circular structure composed of one or more exons, devoid of a 5’-cap or 3’-poly(A) tail (16, 17). After splicing, circRNAs can be formed three main types based on their sequence features: exon-derived circRNAs (ecRNAs), intronic circRNAs (ciRNAs), and exon-intron complex circRNAs (EIciRNAs). Most circRNAs originate from exons and are principally distributed in the cytoplasm, while circRNAs containing intronic sequences are more abundant in the nucleus (18, 19). Their formation mechanisms can be categorized as follows (Figure 1).
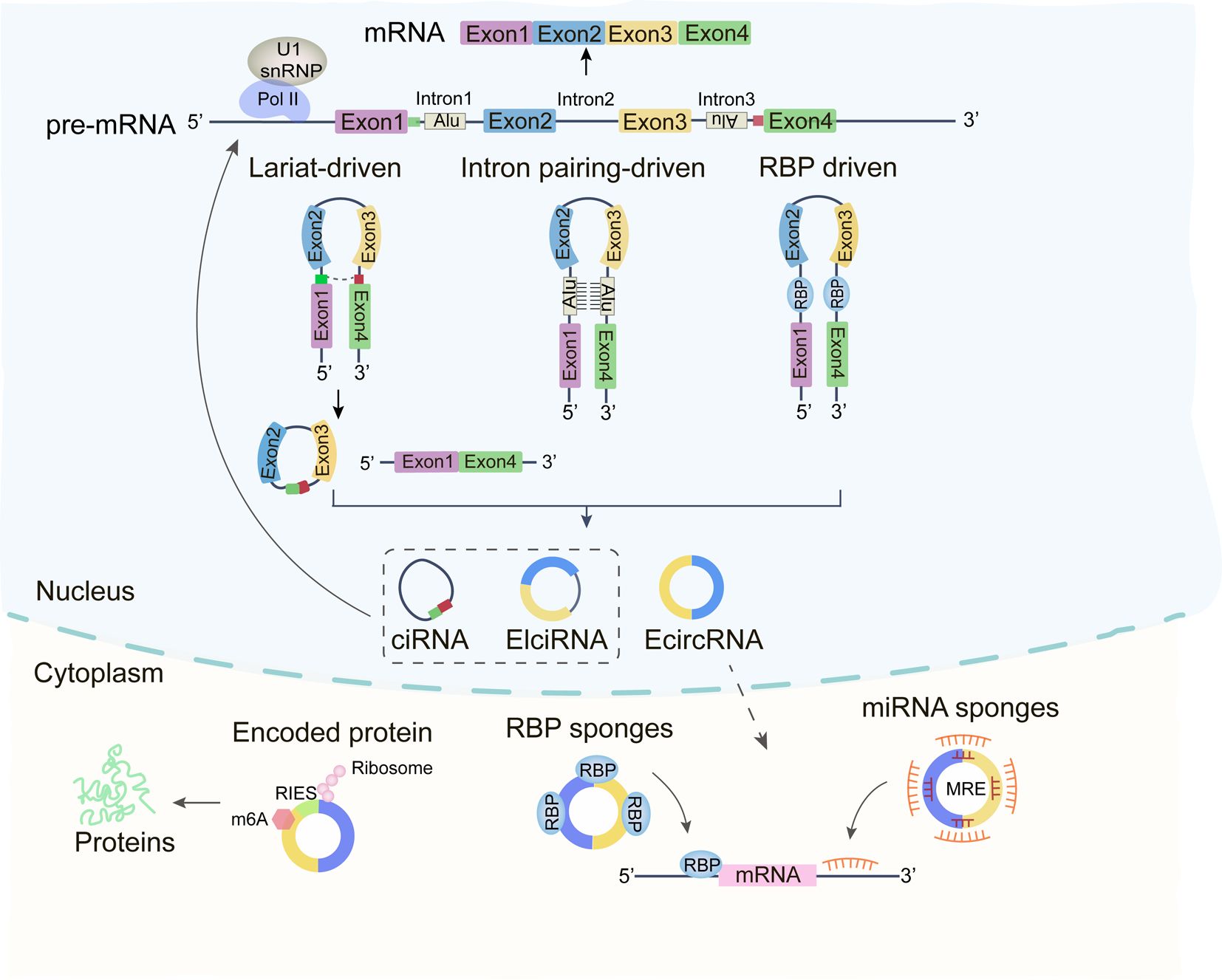
Figure 1. Depiction of the biogenesis and functions of circRNAs. Circular RNAs (circRNA) are synthesized via back-splicing, culminating in a covalently bonded, closed cyclic nucleotide structure. These circRNAs play a pivotal role in interplaying with RNA Binding Proteins (RBP), acting as sponges for miRNAs, and vying with mRNAs in the modulation of gene expression and protein translation.
2.1 Circularization driven by exon skipping
Exon skipping leads to the generation of both linear mRNA and a lasso-like structure comprising skipped exons and introns (17). Reverse splicing of this lasso can produce three types of circRNAs: EIciRNA, ecRNA, and ciRNA (20).
2.2 Intron base pairing
Some circRNAs arise from matching complementary sequences by base-pairing in the flanking intronic regions of exons, followed by alternative splicing to form EIciRNAs or ecRNAs (21).
2.3 RBP-mediated circularization
RNA-binding proteins (RBPs) binding to sites within the flanking intronic regions of exons can bring the splice donor and acceptor sites into proximity, facilitating circRNA circularization (22).
3 Functions of circRNAs
The subcellular localization of circRNAs determines their functional diversity (23). Most circRNAs reside in the cytoplasm, where they function as miRNA sponges, regulating target mRNA expression through competitive binding to miRNAs at specific 3’UTR regions (24). Some cytoplasmic circRNAs also recruit ribosomes for translation into proteins or peptides (16). In contrast, nuclear circRNAs are involved in regulating gene transcription (18).
3.1 miRNA sponges
Cytoplasmic circRNAs, which contain multiple miRNA response elements (MREs), competitively sequester miRNAs, preventing them from binding to mRNA 3’UTRs and thus modulating mRNA stability (25). For instance, CircHIPK3, widely expressed in human cells, sponges multiple miRNAs (e.g., miR-30a-3p, miR-7, and miR-124-3p), influencing processes such as tumor growth, metastasis, and angiogenesis (26–28).
3.2 Interaction with RBPs
RBPs are integral to the regulation of gene expression. Certain circRNAs, characterized by RBP-binding motifs, can form stable loops through complementary sequences, interacting with proteins to exert their functions (29, 30). For example, the RBP human antigen R (HuR), a key player in colorectal cancer, associates with circRHOBTB3, promoting its ubiquitination and degradation, which in turn reduces PTBP1 mRNA levels and suppresses tumor metastasis (31). In LCC and LLN cells, circMTCL1 interacts with the RBP protein C1Q binding protein (C1QBP), inhibiting its ubiquitination and degradation, therefore regulating the tumor progression through Wnt/β-catenin signaling pathway (32).
3.3 Translation (coding)
The cap-dependent ribosome scanning mechanism is the primary mode of translation in eukaryotes (33). While circRNAs were once thought to be non-translatable but recent findings imply that some circRNAs possess internal ribosome entry sites (IRES) or m6A modifications in the 5’-UTR, enabling translation (34). In breast cancer, circSEMA4B both encodes the protein SEMA4B-211aa and sponge miR-330-3p, inhibiting the phosphorylation of the PI3K/AKT pathway in tumors (35). In stomach cancer, circMAPK1 encodes a peptide of 109 amino acids, which binds to and inhibits MEK1 function (36).
3.4 Regulation of parental gene expressions
Nuclear-localized EIciRNAs and ciRNAs interact with Pol II (RNA polymerase II) or U1 snRNP (small nuclear proteins) and regulating parental gene expression (22). For instance, circSMARCA5 is downregulated in breast cancer while its parental gene SMARCA is upregulated. circSMARCA5 binds to its parental gene to form an R-loop, inhibiting transcription and regulating DNA damage repair and cisplatin resistance (37). Similarly, CircME1 binds to U1 snRNP at the promoter of its parental gene ME1, positively regulating its expression and promoting renal cell carcinoma progression (38).
4 CircRNAs in OSCC
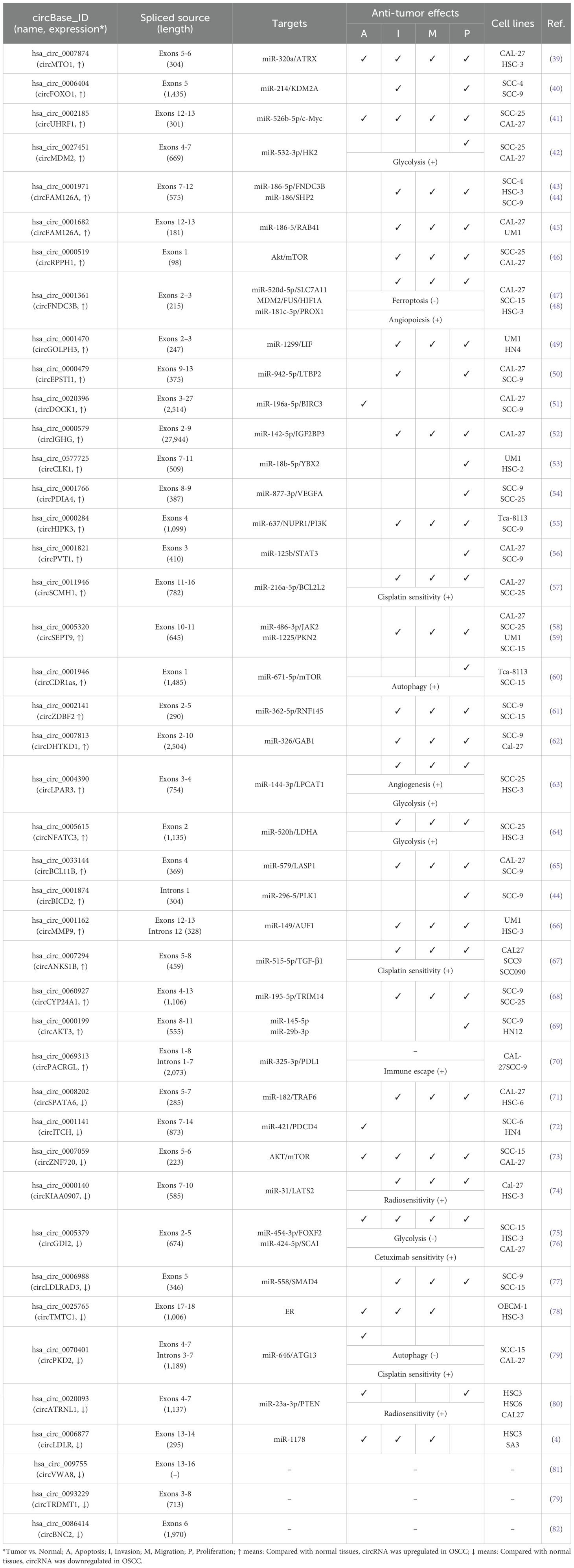
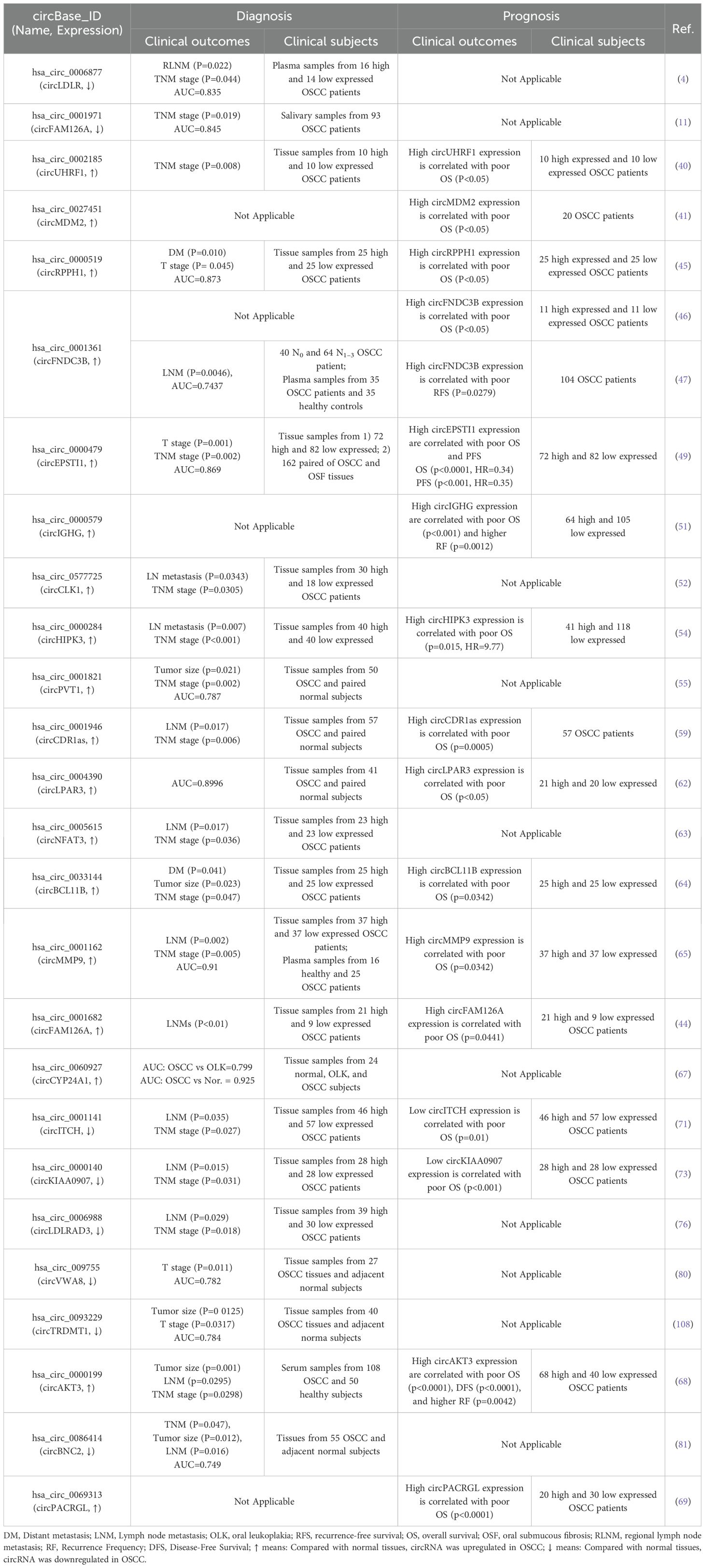
Commonly used methods for detecting circRNAs in tissues include microarray analysis and RNA sequencing. In addition, single-cell sequencing technology enables the detection of circRNAs at the single-cell level, making it particularly suitable for uncovering cellular heterogeneity. Studies on OSCC have identified 43 differentially expressed circRNAs (30 upregulated and 13 downregulated), elucidating their roles in the tumorigenesis and progression of OSCC (Table 1). Mechanistically, many circRNAs act as miRNA sponges, modulating gene expression across various stages of OSCC development, including cell proliferation, apoptosis, cycle arrest, invasion, metastasis, angiogenesis, immune response, and drug resistance (Figure 2).
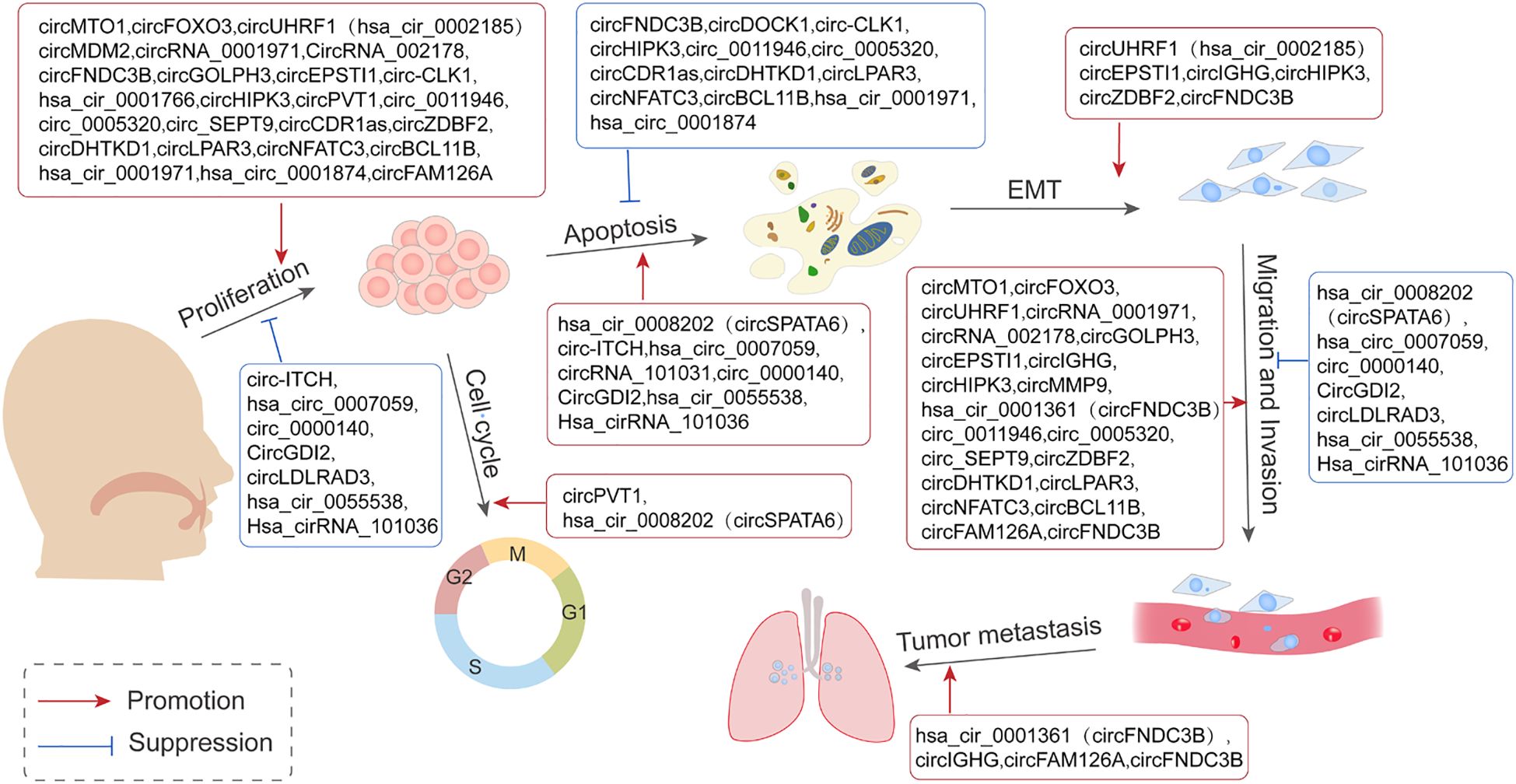
Figure 2. Representation of the contribution of circRNAs in the mediation of various stages of Oral Squamous Cell Carcinoma (OSCC) tumorigenesis. This is achieved primarily through the mechanism of acting as sponges for miRNAs and in turn regulating the expression of pertinent genes involved in cell proliferation, apoptosis, cycle arrest, invasion, and metastasis.
5 CircRNAs promote the proliferation, invasion, and migration in OSCC
Metastasis represents the final stage of cancer progression, including in OSCC (5). Factors such as the tumor microenvironment, genetic predispositions, and mutations contribute to the transformation of normal epithelial cells into tumor cells with excessive proliferative capacities. These transformed cells form clonal subtypes and acquire the ability to invade, spread, and colonize distant organs (83). Excessive proliferation thus serves as a critical driver of tumor development. Lymphatic and distal metastases resulting from unchecked proliferation are major contributors to the high recurrence and mortality rates associated with OSCC (84). Previous studies have pointed out the anti-tumorigenesis effects of circRNA-miRNA axis in OSCC through regulating the downstream target gene expressions (85). Key signaling pathways influenced by this axis are as follows.
5.1 Transforming growth factor beta-SMADs signal pathway
The TGFβ signaling pathway, which includes extracellular cytokines, cytomembrane receptors, and intracellular signal messengers, is essential for regulating cell behavior. TGFβ receptors (TGFβ R) recruit and activate downstream transcription factors, SMAD family member 2/3 (SMAD2/3), which, in conjunction with SMAD4, translocate to the nucleus. There, they regulate gene transcription, influencing the cell cycle distributions, cell proliferation/apoptosis, adhesion, and metastasis. In OSCC, overexpressed circRNAs, such as circEPSTI1 (50) and circCYPANSK1B (67), promote LTBP2 (Latent Transforming Growth Factor Beta Binding Protein 2) and TGFβ1 expressions by sponging miR-942-5p and miR-515-5p, respectively. This activation of the TGFβ pathway facilitates OSCC cell proliferation, metastasis, and invasion. Conversely, circKIAA0907 (74) and circLDLRAD3 (77) act as potential suppressors of OSCC. Their low expression in OSCC tissues leads to a failure to sponge miR-31 and miR-558, resulting in the indirect inhibition of LATS2 (Large Tumor Suppressor Kinase 2) and SMAD4. TAK1 (TGF-β activated kinase 1), a pivotal regulatory factor in cell death by activating downstream effectors, including NF-κB (Nuclear Factor Kappa B) and MAPKs (mitogen-activated protein kinases). Over-expressed circSPATA6/miR-182 axis has been shown to activate TAK1/NF-κB pathway by upregulating TRAF6 (TNF Receptor Associated Factor 6) expression, thereby promoting the tumorigenesis in OSCC cells (71). As a key transcription factor, NF-κB expression and/or activity is also indirectly regulated by circRNAs in OSCC, including through the circDOCK1/miR-196a-5p/BIRC3 (51), circZDBF2/miR-362-5p/RNF145 (61), and circITCH/miR-421/PDCD4 (72) axes, all of which contribute to OSCC tumorigenesis (Figure 3).
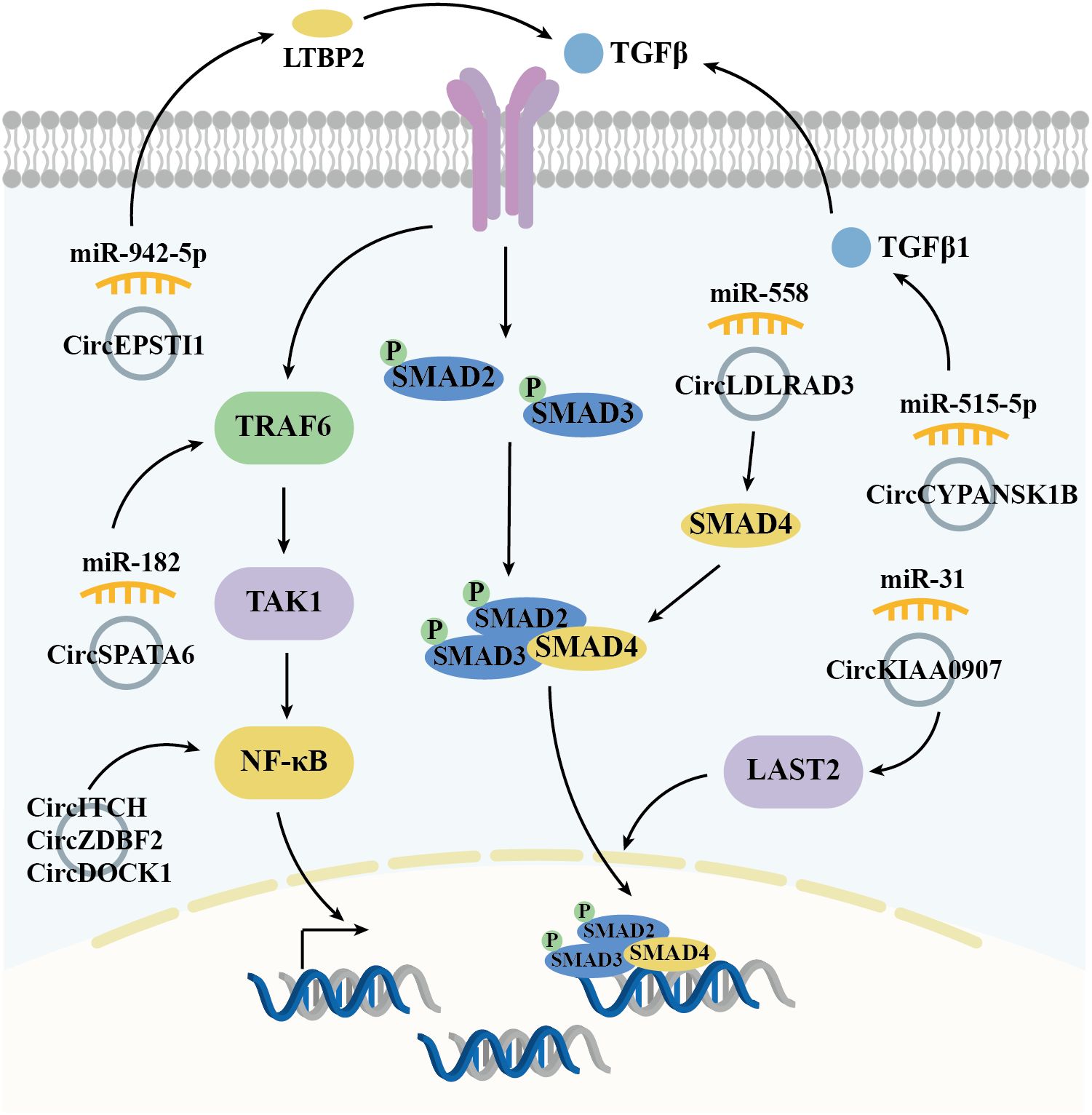
Figure 3. Diagrammatic illustration of how circRNAs regulate the TGFβ-SMADs signaling pathway in the mediation of OSCC.
5.2 Phosphatidylinositol-3-kinase-AKT signal pathway
The PI3K/Akt signaling pathway, activated by RTKs (receptor tyrosine kinases), is essential in controlling the onset and progression of numerous cancers, including OSCC, by altering metabolism, cell proliferation, survival, and angiogenesis (Figure 4). Studies have demonstrated that the circPDIA4/miR-877-3p axis, which is overexpressed in OSCC, promotes VEGFA (vascular endothelial growth factor A) expression. This, in turn, activates PI3K through its receptor, driving the proliferation of OSCC cell lines SCC9 and SCC25 (54). Similarly, circHIPK3, which is highly expressed in OSCC, sponges miR-637 to induce the expression of NUPR1 (nuclear protein 1), thereby activating PI3K and promoting OSCC cell proliferation, metastasis, and invasion (55). PTEN (phosphatase and tensin homolog) is a PI3K pathway negative regulatory factor by inhibiting PI3K activity. In OSCC tissues, the expression of circATRNL1 is significantly lower compared to adjacent tissues, which impairs its ability to sponge miR-23a-3p. This results in reduced PTEN expression, leading to increased PI3K activation and enhanced tumor progression (80). In OSCC, PI3K activation leads to phosphorylation of AKT (protein kinase B, PKB), facilitating downstream signaling. CircRPPH1 (46) and circZNF720 (73) directly upregulate AKT expression, promoting cell proliferation and metastasis in OSCC tissues and cell lines. LASP1 (LIM and SH3 protein 1), a gene associated with lymph node metastasis and poor clinical prognosis, is upregulated in several malignant tumors, underscoring its oncological significance. In OSCC, circBCL11B/miR-579 regulates LASP1 expression, which activates AKT, thereby influencing OSCC tumorigenesis and progression (65). c-MYC, a PI3K/AKT pathway downstream transcription factor, is indirectly upregulated by the circUHRF1/miR-526b-5p axis, contributing to cell cycle distribution, proliferation, apoptosis, and cell differentiation (41). In addition to c-MYC, mTOR (mammalian target of rapamycin) is another critical kinase, is activated by the PI3K-AKT pathway. mTOR regulates metabolism, immune response, autophagy, and cell survival. In OSCC, circRPPH1 and circCDR1as, both highly expressed in tumor tissues, modulate mTOR activity by activating AKT and sponging miR-671-5p, respectively, thereby promoting OSCC cell proliferation, metastasis, and invasion (46, 60).
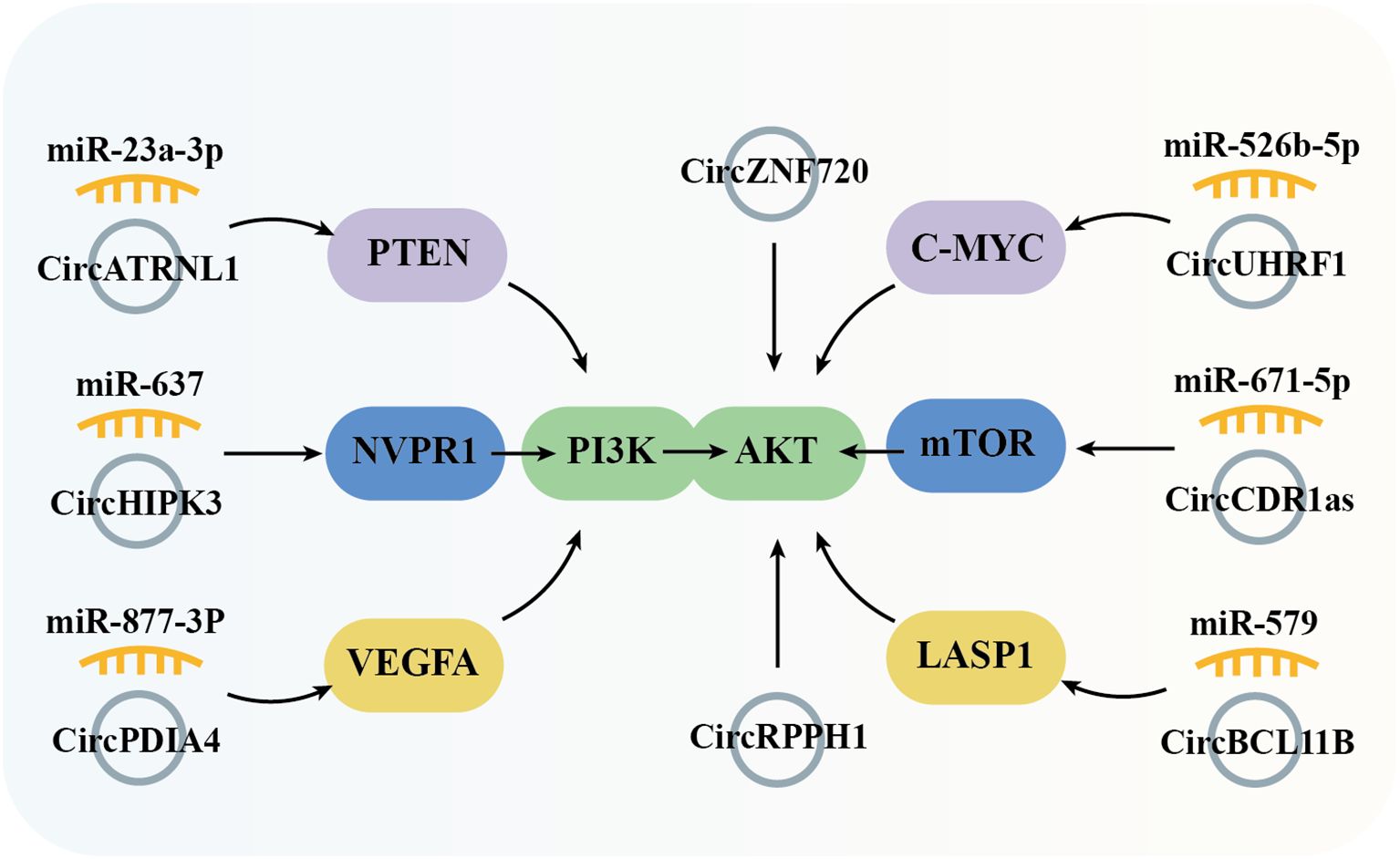
Figure 4. Graphical representation of circRNA-mediated OSCC through the regulation of the PI3K/Akt signaling pathway.
5.3 Janus kinase-signal transducer and activator of transcription signal pathway
The JAK/STAT signaling pathway plays an important role in hematopoiesis, immunity, tissue repair, inflammation, apoptosis, and adipogenesis. This signaling pathway encompasses over 50 members, including IFNs (Interferons), ILs (Interleukins), CSFs (Colony stimulating factors), and hormones. Dysregulation or mutations in JAK/STAT components are implicated in numerous human diseases, including OSCC. Studies have shown that circSEPT9 (56) and circPVT1 (58) are highly expressed in OSCC tissues and regulate the expression of JAK2 and STAT3 through miR-486-3p and miR-125b, respectively, promoting cell proliferation and metastasis. Additionally, the circGOLPH3/miR-1299 axis induces the expression of Leukemia inhibitory factor (LIF), which activates JAK signaling and contributes to OSCC progression (49) (Figure 5).
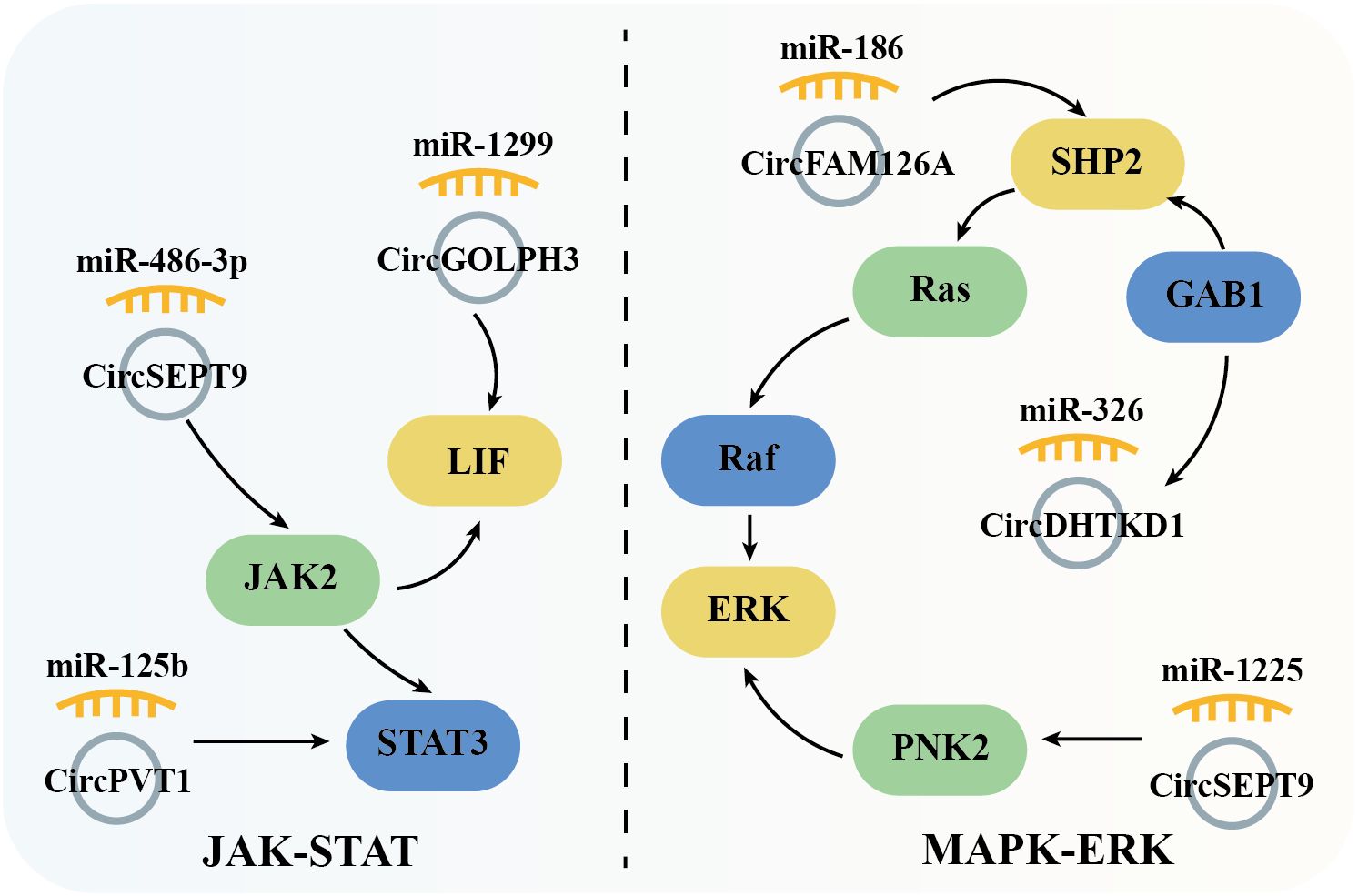
Figure 5. Schematic elucidation of circRNAs’ regulation of the JAK-STAT and MAPK-ERK signaling pathway and their consequential mediation of OSCC.
5.4 Mitogen-activated protein kinase-ERK signal pathway
The MAPK cascade is critical for regulating normal cell proliferation, survival, and differentiation, with dysregulation often leading to cancer and other diseases. Specifically, as a crucial member of MAPK pathway, ERK (extracellular signal-regulated kinase) is activated by Raf serine/threonine kinases, which serve as downstream effectors of the commonly mutated oncogene Ras small GTPase. This forms the Ras/Raf/ERK signaling network, which is further modulated by the tyrosine phosphatase SHP2 to regulate cell proliferation, survival, and differentiation. In OSCC, overexpressed circFAM126A promotes SHP2 expression by sponging miR-186 [54]. Similarly, circDHTKD1 and circSEPT9, which are also highly expressed in OSCC, upregulate GAB1 (GRB2-associated binding protein 1) and PKN2 (protein kinase N2) through miR-326 and miR-1225, respectively. GAB1 directly activates SHP2, while PKN2 activates ERK, thus driving OSCC cell proliferation, metastasis, and invasion (59, 62) (Figure 5).
5.5 Glycolysis
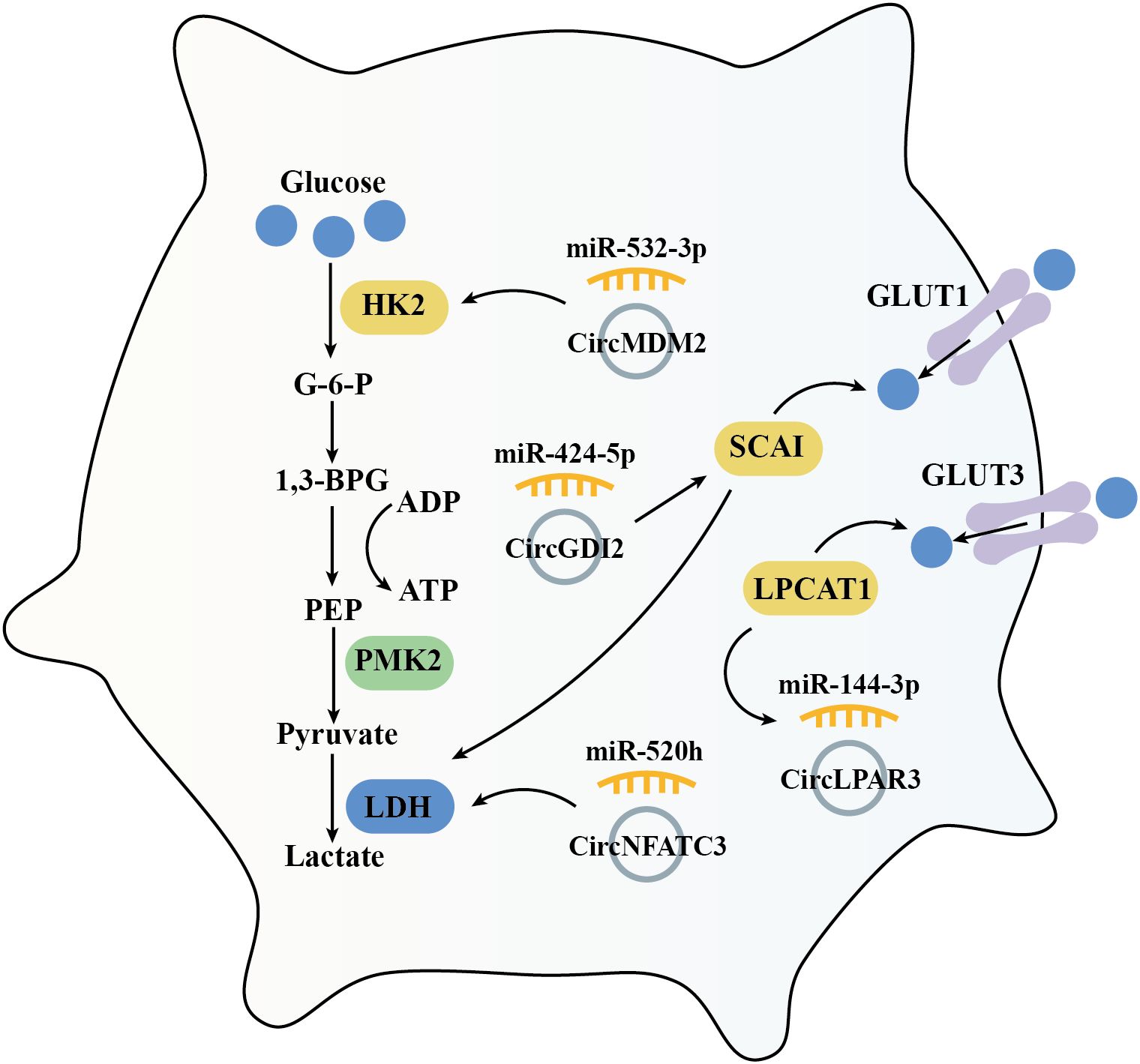
Cancer cells reprogram their metabolism to support cell growth, metastasis, and survival. Increased glucose uptake and reliance on glycolysis are essential for meeting the synthetic metabolic demands of these cells (86). This tumor cell specific metabolic characteristics is defined as ‘Warburg effect,’ persists even in the presence of fully functional mitochondria (87). Hexokinase 2 (HK2), a rate-limiting enzyme in glycolysis, plays a key role in tumorigenesis. HK2 is high expressed in OSCC compared with adjacent normal tissues, and has been shown to promote OSCC cell growth both in vitro and in vivo (88). Previous investigation suggested that circMDM2 is significantly upregulated in OSCC, promote OSCC proliferation and glycolysis through regulating the miR-532-3p/HK2 axis (42). In addition, knockout of LDHA (lactate dehydrogenase A) inhibits cell proliferation and EMT process in OSCC cells (89). Similarly, circNFATC3 is over-expressed in OSCC which acts as a miR-520h sponge. This interaction induces LDHA expression and promotes glycolysis, proliferation, and invasion in OSCC cells (64). Conversely, circGDI2 is low expressed in OSCC. When overexpressed, circGDI2 targets miR-424-5p/SCAI axis, regulates glycolytic proteins like GLUT1 and LDHA, and therefore inhibiting OSCC cell reproduction and metastasis (76).
LPCAT1 (Lysophosphatidylcholine acyltransferase 1) is an enzyme involved in phospholipid biosynthesis and remodeling, playing a pivotal role in the lipid remodeling and various cancers, including OSCC (90, 91). In OSCC, LPCAT1 promotes tumorigenesis via regulating PAF (platelet-activating factor) and its receptor, PAFR (92). Furthermore, LPCAT1 has been shown to activate the NF-κB/STAT3 signal pathway and then increased GLUT3 expression, enhanced glycolysis, and increased proliferation in keratinocytes (93). In OSCC tissues and cell lines, overexpressed circLPAR3 via miR-144-3p promotes LPCAT1 which enhances OSCC cell proliferation, migration, invasion, angiogenesis, and glycolysis (63) (Figure 6).
5.6 Autophagy and Ferroptosis
Autophagy is a highly conserved cellular process essential for maintaining homeostasis through self-digestion and catabolism (94). In tumor cells, autophagy involved in regulating both cell survival and apoptosis signaling (95). ATG13 (autophagy-related protein 13) is a key component of the ULK1 complex, whom could activate ULK1 kinase activity (96). In tumor cells, ATG13 knockout impedes cell cycle progression and inhibits proliferation in both in vitro and in vivo models (97). Furthermore, downregulation of circPKD2 fails to sponge miR-646, leading to suppressed ATG13 expression and reduced tumorigenesis in OSCC tissues and cell lines (79). In addition to ATG13, TRIM14 (tripartite motif 14) has been implicated in mediating HCC cells proliferation, autophagy, and metastasis, with TRIM14 knockdown resulting in the opposite effects (98). In OSCC, the circCYP24A1/miR-195-5p axis upregulates TRIM14 expression, driving carcinogenesis (68) (Figure 7).
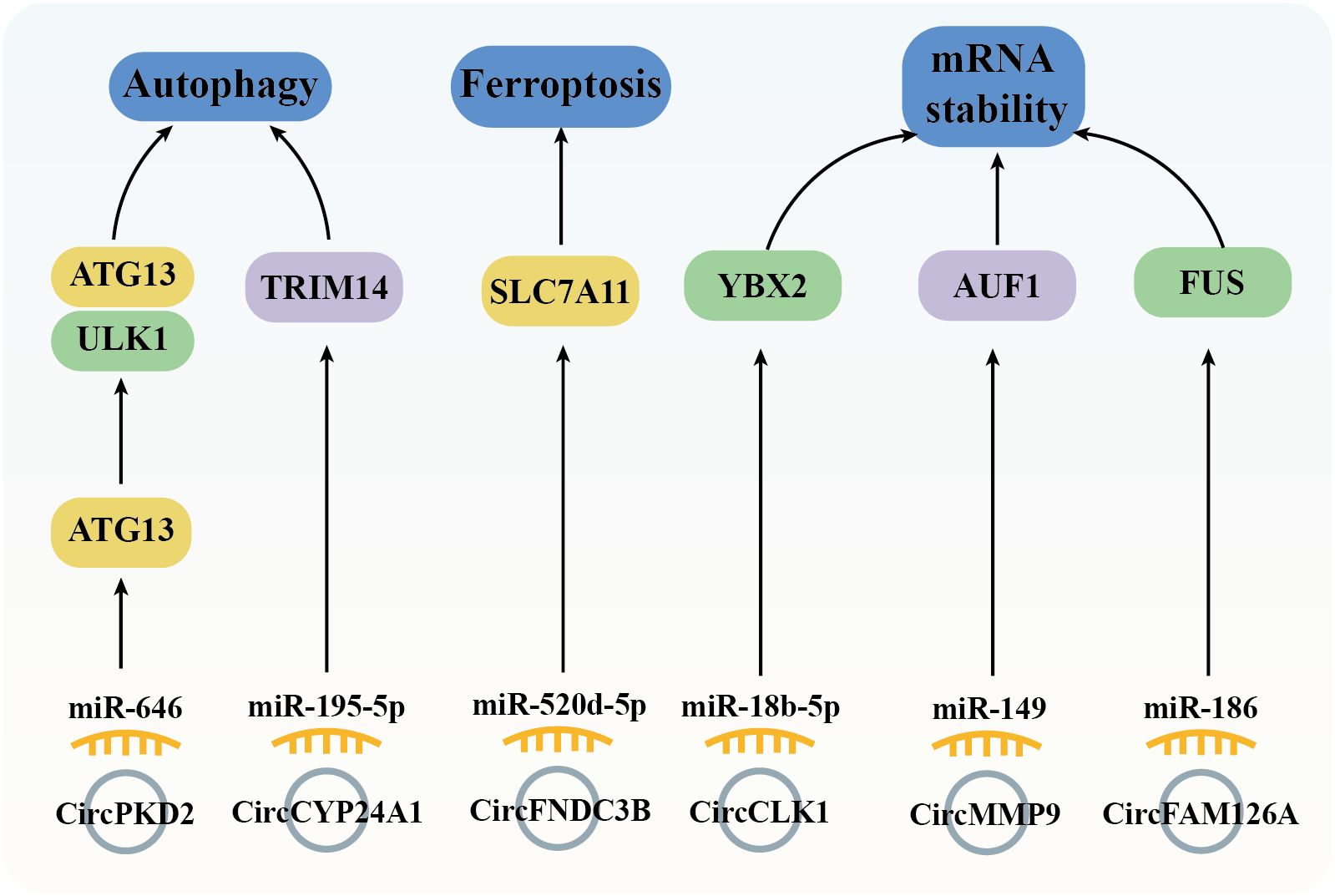
Figure 7. Diagram depicting the regulation of autophagy, ferroptosis, and mRNA stability by circRNAs in the mediation of OSCC.
Ferroptosis, a regulated cell death (RCD) process, is triggered by the toxic lipid peroxidation products (99). The evasion of ferroptosis through oncogenes and oncogenic signals promotes tumor initiation, progression, metastasis, and drug resistance (100). In cancer cells, dysregulated expression of the cysteine transporter SLC7A11 (Solute Carrier Family 7 Member 11) imports cystine, biosynthesis GSH, thereby inhibiting ferroptosis (101). In OSCC tissues, high expression of circFNDC3B modulates the miR-520d-5p/SLC7A11 axis to prevent ferroptosis and promote tumor survival (48) (Figure 7).
5.7 mRNA stability
Genetic variations are widely recognized as drivers of cancer; however, post-transcriptional events also significantly influence cancer development by regulating mRNA cycling and translation, including mRNA stability, which is controlled by RBPs (102). Notable RBPs such as FUS (Fused in Sarcoma), YBX2 (Y-box Binding Protein 2), and AUF1 (AU-rich Element RNA-binding Protein 1) could regulate proliferation, migration, and invasion across various tumor cell types (102–104). Previous studies have demonstrated that overexpression of circCLK1 (53), circMMP9 (66), and circFAM126A (45), through sponging specific miRNAs (miR-18b-5p, miR-149, and miR-186-5p, respectively), enhances the expression of YBX2, AUF1, and FUS, thereby promoting OSCC tumorigenesis (Figure 7).
5.8 Other factors
In addition to regulating the tumorigenesis related signal pathways, dysregulated circRNAs in OSCC tissues and/or cell lines have also been implicated could modulate transcription factor expressions. For example, circIGHG is overexpressed in OSCC tissues and is associated with poor prognosis in OSCC patients. In terms of mechanism, circIGHG induces IGF2BP3 expression via binding to miR-142-5p, thereby promoting OSCC cell invasiveness (52). Furthermore, the oncogenic circFNDC3B serves multiple roles in OSCC. In the nucleus, circFNDC3B co-localizes with FUS, facilitating FUS ubiquitination and degradation. Through regulation of MDM2 and HIF1A expression, it positively influences VEGF expression and angiogenesis. In the cytoplasm, circFNDC3B regulates the miR-181c-5p/PROX1 axis to promote lymphangiogenesis and tumor metastasis (47).
6 CircRNAs and radiotherapy/chemotherapy resistance in OSCC cells
Radiation therapy and chemotherapy are key treatment strategies for OSCC; however, the development of drug resistance significantly worsens patient prognosis. Notably, compared to normal tissues, dysregulated circRNAs in OSCC tissues may contribute to the regulation of tumor sensitivity to chemoradiotherapy (Table 2, Figure 8).
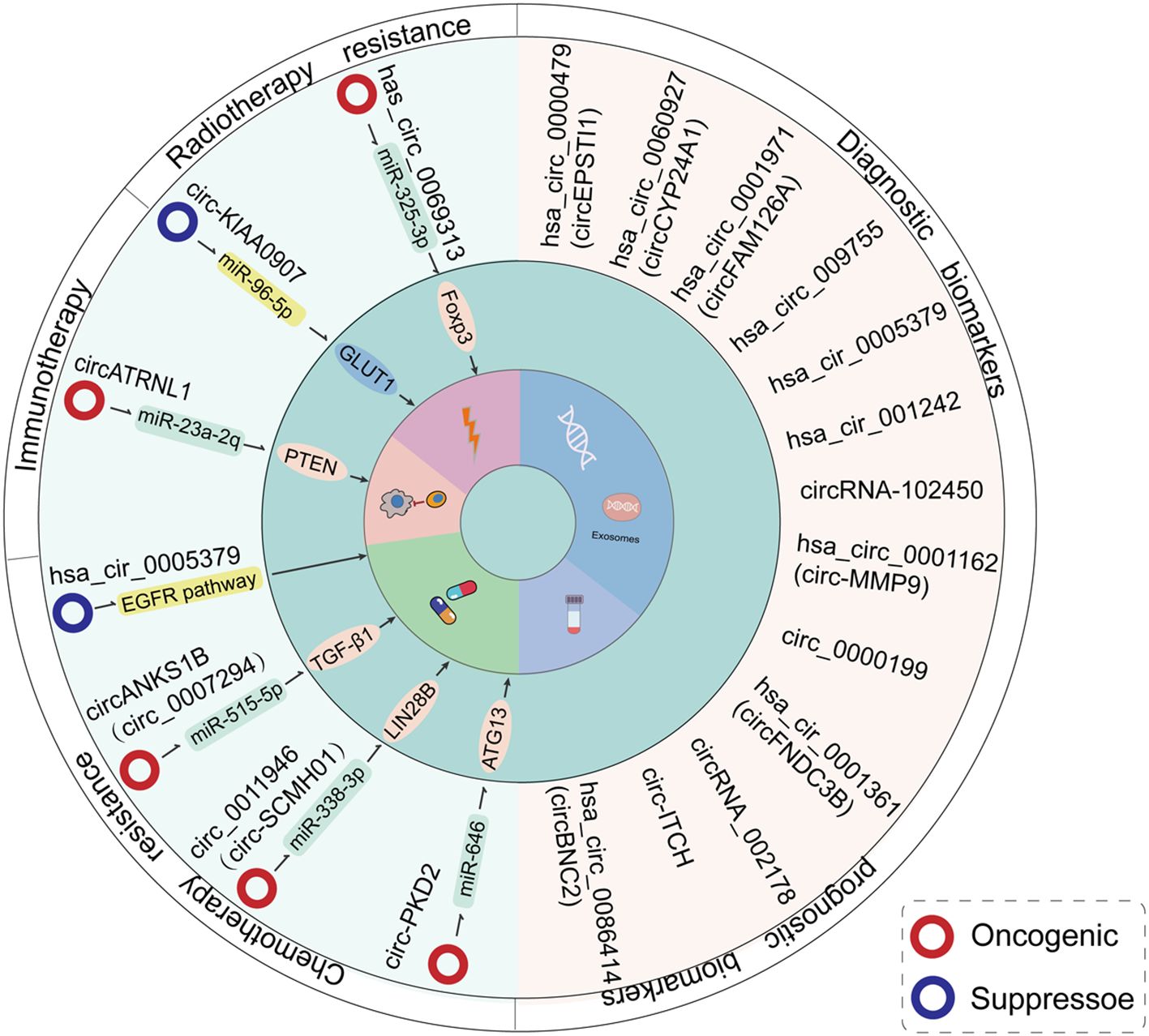
Figure 8. An overview of the clinical functions of circRNAs in the context of OSCC treatment. The expression levels of circRNAs are associated with resistance to radiotherapy/chemotherapy in OSCC patients. Additionally, certain circRNAs may constitute effective diagnostic and prognostic markers for the disease.
Cisplatin (CDDP), a first-generation platinum-based anticancer drug approved by the US FDA in 1978, remains a cornerstone of clinical treatment (108). Despite its broad anticancer efficacy, drug resistance driven by tumor heterogeneity significantly limits its clinical effectiveness (67). Recent studies have highlighted the role of circRNAs, acting as sponges for microRNAs, in mediating CDDP sensitivity in OSCC by regulating various target genes or biological processes. For instance, the oncogenic circANKS1B promotes OSCC metastasis and CDDP resistance by upregulating TGFβ1 via sponging miR-515-5p. Knockdown of circANKS1B using siRNA increases OSCC cell sensitivity to CDDP (67). Additionally, circSCMH1 is high expressed in CDDP-resistant OSCC tissues and cell lines. CircSCMH1 can be transferred to surrounding tumor cells via exosomes, where it regulates the miR-338-3p/LIN28B axis, driving malignant progression and CDDP resistance in OSCC (105). Another study found that CDDP treatment significantly increases circ-PKD2 expression in OSCC cells. Mechanistically, after overexpression, circPKD2 sponge miR-646 and thus promoting ATG13 expression, triggering autophagy, inducing tumor cell apoptosis, and enhancing chemotherapy sensitivity (79).
Cetuximab is an effective therapy for advanced OSCC. Su et al. demonstrated that circGD12 is involved in Cetuximab resistance, and overexpression of circGD12 enhances cell sensitivity to Cetuximab and induces tumor cell apoptosis through regulation of the EGFR pathway (106).
Furthermore, circRNAs have emerged as potential biological targets to enhance the clinical efficacy of radiation therapy. High-throughput sequencing identified circATRNL1, derived from ATRNL1, as downregulated in OSCC cells after 4Gy ionizing radiation treatment. Upregulation of circATRNL1 induces PTEN expression via miR-23a-2p, promoting cell apoptosis, cell cycle arrest, and enhancing OSCC cell sensitivity to radiotherapy (80). Additionally, Dong et al. found that circKIAA0907 may serve as a potential target to improve OSCC radiation resistance. By targeting the circKIAA0907/miR-96-5p/GLUT1 axis, circKIAA0907 enhances OSCC radiation resistance and promotes tumor cell apoptosis (107).
7 CircRNAs can be used as a diagnostic marker for OSCC
The highly stable, covalently closed structure, along with the tissue-specific and disease-specific expression patterns of circRNAs, positions them as promising diagnostic biomarkers (15, 109). Several studies have confirmed that aberrantly expressed circRNAs in OSCC may serve as biomarkers for both diagnosis and prognosis (Table 3, Figure 8).
7.1 Screening and diagnosis
Screening and early diagnosis are essential for improving the survival rates and reducing the mortality of patients with OSCC. Oral leukoplakia (OLK) and submucosal fibrosis (OSF) are common precancerous lesions in the oral cavity. Table 3 summarizes circRNAs that may serve as potential markers for OSCC (Figure 8). In general, 18 circRNAs were considered as OSCC candidate biomarkers for clinical screening and diagnosis, including 12 overexpressed and 6 downregulated circRNAs in OSCC tissues compared to adjacent normal or healthy control samples. For instance, high expression of circUHRF1 [40] and circFAM126A (45) in OSCC tissues correlates with the TNM stage (Tumor-Node-Metastasis) (p = 0.008) and LNM (lymph node metastasis, p < 0.01), respectively. The dysregulated expression of these circRNAs in OSCC tissues is associated with TNM stage [circUHRF1 (41)], LNM [circFAM126A (45)], or both [circCLK1 (53), circHIPK3 (55), circCDR1as (60), circNFAT3 (64), circITCH (72), circKIAA0907 (74), and circLDLRAD3 (77)]. In addition to their correlation with tumor stage and metastasis, the tumor expression levels of circPVT1 (56) and circTRDMT1 (80) are associated with tumor size. A study examining the correlation between circRNA expression and early OSCC lesions (including normal buccal mucosa, OSF, and OSCC tissues) found that high expression of circEPSTI1 significantly correlates with T stage (p = 0.001) and advanced TNM stage (p = 0.002). ROC curve analysis (AUC) demonstrated that circEPSTI1 could sensitively differentiate OSCC from OSF (AUC = 0.869) (39). Moreover, circCYP24A1 expression is increased in OSCC, with ROC analysis showing an AUC of 0.799 (95% CI = 0.633-0.916) for OSCC versus OLK, and an AUC of 0.925 (95% CI = 0.846-1.0) for OSCC versus normal tissue, suggesting its potential as an early diagnostic biomarker for OSCC (78). Additionally, circBNC2 expression is negatively associated with TNM stage, LNM, and tumor size (all of the p-values were below 0.05), with a potential diagnostic value shown by an ROC AUC of 0.749 (p < 0.0001) (83).
7.2 Liquid biopsy
Compared to surgical biopsy, liquid biopsy has gained significant attention in cancer diagnosis and prognosis research due to its real-time, rapid, and minimally invasive nature. Recent studies have explored the correlation between clinical characteristics of OSCC and circRNA expression in body fluids, including saliva, plasma, and serum. CircRNAs in these fluids demonstrate higher sensitivity and specificity compared to tissue samples (110). A circRNA microarray combined with droplet digital PCR (RT ddPCR) identified that circLDLR was a tumor suppressor whom simultaneously related to TNM stage (p = 0.044) and LNM (p = 0.022) in OSCC (4). Similarly, circMMP9 levels were significantly elevated in OSCC plasma samples (the AUC value was 0.91, 95% CI: 0.8216–0.9984) and its expression was associated with TNM stage (p = 0.005) and LNM (p = 0.002) (66). Additionally, circFNDC3B expression was markedly higher in OSCC patients’ serum, with an AUC of 0.7437. High circFNDC3B expression correlated with LNM in 104 patients with OSCC (p = 0.0046) (48). After analyzed the circRNA expressions in serum exosomes from 108 OSCC patients, circAKT3 was screened out due to high expressed circAKT3 in serum was associated with tumor size, TNM stage, and LNM (all of the p-values were below 0.05) (69). Moreover, in clinical salivary samples from 93 patients with OSCC, the levels of circFAM126A were closely related to TNM stage (p = 0.019), and the AUC value was 0.845 (95% CI: 0.784–0.905, p < 0.001) (11).
8 CircRNA can be used to analyze the prognosis of OSCC patients
In addition to their diagnostic potential, aberrant circRNA expression in OSCC also serves as a prognostic marker (Table 3, Figure 8). Previous studies have identified 11 overexpressed and 2 underexpressed circRNAs in OSCC, which correlate with poor overall survival (OS). These include overexpressed circUHRF1 (33), circMDM2 (34), circRPPH1 (36), circFNDC3B (37), circHIPK3 (44), circCDR1as (49), circLPAR3 (52), circBCL11B (54), circMMP9 (56), circFAM126A (57), and circPACRGL (77); and downregulated circITCH (60) and circKIAA0907 (62). In addition to OS, Kaplan-Meier analysis revealed that high expression of circIGHG and circFNDC3B was associated with higher recurrence frequency (RF, p = 0.0012) (41) and poor recurrence-free survival (RFS, p = 0.0279) (58), respectively. Furthermore, compared to low-circAKT3-expressed OSCC patients, high circAKT3 expression in serum exosomes was linked to lower OS and DFS (p-values were below 0.0001) (82).
9 Conclusion
The difficulty in early diagnosis, coupled with a high incidence of metastasis and recurrence, results in poor clinical outcomes for patients with OSCC. CircRNAs were initially considered as mere “by-products” of gene transcription. However, accumulating evidence implies that circRNAs act as a crucial factor in regulating the pathological and physiological processes of various diseases, prompting a re-evaluation of these previously overlooked “wastes” (50, 52, 111).
As research on circRNAs in OSCC deepens, multiple studies have highlighted that differentially expressed circRNAs function as either inhibitors or promoters involved in various stages of OSCC tumorigenesis. These processes include sustained cell proliferation, apoptosis resistance, angiogenesis, invasion, metastasis, metabolic reprogramming, immune escape, and acquisition of drug resistance. These findings suggest that circRNAs hold potential as therapeutic targets. Indeed, previous studies have demonstrated that overexpression or knockdown of oncogenic circRNAs has significant anti-tumorigenic effects in OSCC by modulating the expression of target genes (53, 57, 61). Moreover, the stable expression and potential to encode proteins or peptides make circRNAs promising for therapeutic applications. According to recent research, RNA-targeting type VI CRISPR systems (Cas13a, Cas13b, and Cas13d) are capable of directly cleaving single-stranded RNA in mammalian cells. Among them, RfxCas13d has been identified as the most effective system for circRNA interference (112). Artificial circRNAs expressing relevant antigens have shown therapeutic and prophylactic effects in several malignancies (113).
Screening and early diagnosis are critical for improving the survival rate of patients with OSCC (4, 106). However, current diagnostic methods, such as surgical biopsy, are often used to confirm the presence of pathological changes but are unsuitable for early screening. Additionally, established tumor markers such as CEA (carcinoembryonic antigen), ESM-1 (endothelial cell-specific molecule-1), and SCC (squamous cell carcinoma antigen) are not fully applicable for clinical diagnosis of OSCC due to their limited sensitivity and specificity (114, 115). Several studies have shown that dysregulated circRNAs in tumors and body fluids exhibit stage-specific expression in OSCC, closely correlating with clinical features such as tumor size, distant metastasis, and TNM stage. Notably, circRNAs in body fluids, due to their ease of collection, rapid analysis, and minimally invasive nature, are poised to become a primary focus for clinical translation in the future (116). While current circRNA validation remains limited to cellular and animal models due to technological constraints and high costs, continued advancements in molecular biology technology hold promise for substantial progress in the clinical application of circRNAs. Although current RNA sequencing technologies still face challenges in detecting circRNAs in large-scale samples—particularly those with low abundance—the advancements in third-generation sequencing technologies hold promise in overcoming these limitations. These improvements may accelerate the clinical application of circRNAs as reliable biomarkers.
Author contributions
J-LL: Data curation, Writing – original draft. RM: Data curation, Writing – original draft. Y-SR: Data curation, Writing – original draft. Q-YL: Data curation, Writing – original draft. H-MZ: Data curation, Writing – original draft. G-CD: Conceptualization, Writing – review & editing. JL: Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the earmarked fund for Inner Mongolia Agriculture Research System (IMARS-11), The 12th batch of “grassland talents” innovation and entrepreneurship talent team in Inner Mongolia (No. 1 of Nei Ren Cai Fa [2023]) No.283, The rolling support talent project of “grassland talents” in 2022 (No. 15 of Nei Zu Tong Zi [2022]) No. 139, Talent Development Fund of Inner Mongolia 2022 (the 22nd batch in total) High level Talent Personal Project (No. 110 of Nei Ren She Ban Fa [2022]) No. 66, The basic research funds project for directly affiliated universities in Inner Mongolia Autonomous Region—special project for building research innovation teams (BR251302).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Chi AC, Day TA, and Neville BW. Oral cavity and oropharyngeal squamous cell carcinoma–an update. CA Cancer J Clin. (2015) 65:401–21. doi: 10.3322/caac.21293
2. Ford PJ and Rich AM. Tobacco use and oral health. Addiction. (2021) 116:3531–40. doi: 10.1111/add.15513
3. Kolegova ES, Patysheva MR, Larionova IV, Fedorova IK, Kulbakin DE, Choinzonov EL, et al. Early-onset oral cancer as a clinical entity: aetiology and pathogenesis. Int J Maxillofac Surg. (2022) 51:1497–509. doi: 10.1016/j.ijom.2022.04.005
4. Ando T, Kasamatsu A, Kawasaki K, Hiroshima K, Fukushima R, Iyoda M, et al. Tumor suppressive circular rna-102450: development of a novel diagnostic procedure for lymph node metastasis from oral cancer. Cancers (Basel). (2021) 13:5708. doi: 10.3390/cancers13225708
5. Tan Y, Wang Z, Xu M, Li B, Huang Z, Qin S, et al. Oral squamous cell carcinomas: state of the field and emerging directions. Int J Sci. (2023) 15:44. doi: 10.1038/s41368-023-00249-w
6. Cheng Y, Li S, Gao L, Zhi K, and Ren W. The molecular basis and therapeutic aspects of cisplatin resistance in oral squamous cell carcinoma. Front Oncol. (2021) 11:761379. doi: 10.3389/fonc.2021.761379
7. Ling Z, Cheng B, and Tao X. Epithelial-to-mesenchymal transition in oral squamous cell carcinoma: challenges and opportunities. Int J Cancer. (2021) 148:1548–61. doi: 10.1002/ijc.33352
8. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, and Kjems J. The biogenesis, biology and characterization of circular rnas. Nat Rev Genet. (2019) 20:675–91. doi: 10.1038/s41576-019-0158-7
9. Zhang F, Jiang J, Qian H, Yan Y, and Xu W. Exosomal circrna: emerging insights into cancer progression and clinical application potential. J Hematol Oncol. (2023) 16:67. doi: 10.1186/s13045-023-01452-2
10. Liu CX and Chen LL. Circular rnas: characterization, cellular roles, and applications. Cell. (2022) 185:2016–34. doi: 10.1016/j.cell.2022.04.021
11. Zhao SY, Wang J, Ouyang SB, Huang ZK, and Liao L. Salivary circular rnas hsa_Circ_0001874 and hsa_Circ_0001971 as novel biomarkers for the diagnosis of oral squamous cell carcinoma. Cell Physiol Biochem. (2018) 47:2511–21. doi: 10.1159/000491624
12. Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. Circrna: functions and properties of a novel potential biomarker for cancer. Mol Cancer. (2017) 16:94. doi: 10.1186/s12943-017-0663-2
13. Wu L, Ye S, Yao Y, Zhang C, and Liu W. Oral cancer stem cell-derived small extracellular vesicles promote M2 macrophage polarization and suppress cd4+ T-cell activity by transferring uca1 and targeting lamc2. Stem Cells Int. (2022) 2022:5817684. doi: 10.1155/2022/5817684
14. Darnell JE. Implications of Rna·Rna Splicing in Evolution of Eukaryotic Cells. Science. (1978) 202(4374):1257-60. doi: 10.1126/science.364651
15. Chen LL. The expanding regulatory mechanisms and cellular functions of circular rnas. Nat Rev Mol Cell Biol. (2020) 21:475–90. doi: 10.1038/s41580-020-0243-y
16. Ma S, Kong S, Wang F, and Ju S. Circrnas: biogenesis, functions, and role in drug-resistant tumours. Mol Cancer. (2020) 19:119. doi: 10.1186/s12943-020-01231-4
17. Xiao MS, Ai Y, and Wilusz JE. Biogenesis and functions of circular rnas come into focus. Trends Cell Biol. (2020) 30:226–40. doi: 10.1016/j.tcb.2019.12.004
18. Yang X, Ye T, Liu H, Lv P, Duan C, Wu X, et al. Expression profiles, biological functions and clinical significance of circrnas in bladder cancer. Mol Cancer. (2021) 20:4. doi: 10.1186/s12943-020-01300-8
19. Obi P and Chen YG. The design and synthesis of circular rnas. Methods. (2021) 196:85–103. doi: 10.1016/j.ymeth.2021.02.020
20. Yuan Y, Zhang X, Fan X, Peng Y, and Jin Z. The emerging roles of circular rna-mediated autophagy in tumorigenesis and cancer progression. Cell Death Discov. (2022) 8:385. doi: 10.1038/s41420-022-01172-5
21. Pisignano G, Michael DC, Visal TH, Pirlog R, Ladomery M, and Calin GA. Going circular: history, present, and future of circrnas in cancer. Oncogene. (2023) 42:2783–800. doi: 10.1038/s41388-023-02780-w
22. Du WW, Zhang C, Yang W, Yong T, Awan FM, and Yang BB. Identifying and characterizing circrna-protein interaction. Theranostics. (2017) 7:4183–91. doi: 10.7150/thno.21299
23. Hwang HJ and Kim YK. Molecular mechanisms of circular rna translation. Exp Mol Med. (2024) 56:1272–80. doi: 10.1038/s12276-024-01220-3
24. Kristensen LS, Jakobsen T, Hager H, and Kjems J. The emerging roles of circrnas in cancer and oncology. Nat Rev Clin Oncol. (2022) 19:188–206. doi: 10.1038/s41571-021-00585-y
25. Zhang Y, Luo J, Yang W, and Ye WC. Circrnas in colorectal cancer: potential biomarkers and therapeutic targets. Cell Death Dis. (2023) 14:353. doi: 10.1038/s41419-023-05881-2
26. Xu Q, Cheng D, Li G, Liu Y, Li P, Sun W, et al. Circhipk3 regulates pulmonary fibrosis by facilitating glycolysis in mir-30a-3p/foxk2-dependent manner. Int J Biol Sci. (2021) 17:2294–307. doi: 10.7150/ijbs.57915
27. Li X, Yang Y, Lai Z, Lin X, Fu X, and He X. Circhipk3 Targets Drp1 to Mediate Hydrogen Peroxide-Induced Necroptosis of Vascular Smooth Muscle Cells and Atherosclerotic Vulnerable Plaque Formation. J Adv Res. (2025) 69:329-41. doi: 10.1016/j.jare.2024.04.011
28. Shi P, Liu Y, Yang H, and Hu B. Breast cancer derived exosomes promoted angiogenesis of endothelial cells in microenvironment via circhipk3/mir-124-3p/mtdh axis. Cell Signal. (2022) 95:110338. doi: 10.1016/j.cellsig.2022.110338
29. Lu Y, Yan Y, Li B, Liu M, Liang Y, Ye Y, et al. A novel prognostic model for oral squamous cell carcinoma: the functions and prognostic values of rna-binding proteins. Front Oncol. (2021) 11:592614. doi: 10.3389/fonc.2021.592614
30. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The rna binding protein quaking regulates formation of circrnas. Cell. (2015) 160:1125–34. doi: 10.1016/j.cell.2015.02.014
31. Chen J, Wu Y, Luo X, Jin D, Zhou W, Ju Z, et al. Circular rna circrhobtb3 represses metastasis by regulating the hur-mediated mrna stability of ptbp1 in colorectal cancer. Theranostics. (2021) 11:7507–26. doi: 10.7150/thno.59546
32. Wang Z, Sun A, Yan A, Yao J, Huang H, Gao Z, et al. Circular rna mtcl1 promotes advanced laryngeal squamous cell carcinoma progression by inhibiting C1qbp ubiquitin degradation and mediating beta-catenin activation. Mol Cancer. (2022) 21:92. doi: 10.1186/s12943-022-01570-4
33. Diallo LH, Tatin F, David F, Godet AC, Zamora A, Prats AC, et al. How are circrnas translated by non-canonical initiation mechanisms? Biochimie. (2019) 164:45–52. doi: 10.1016/j.biochi.2019.06.015
34. Kong S, Tao M, Shen X, and Ju S. Translatable circrnas and lncrnas: driving mechanisms and functions of their translation products. Cancer Lett. (2020) 483:59–65. doi: 10.1016/j.canlet.2020.04.006
35. Wang X, Jian W, Luo Q, and Fang L. Circsema4b inhibits the progression of breast cancer by encoding a novel protein sema4b-211aa and regulating akt phosphorylation. Cell Death Dis. (2022) 13:794. doi: 10.1038/s41419-022-05246-1
36. Jiang T, Xia Y, Lv J, Li B, Li Y, Wang S, et al. A novel protein encoded by circmapk1 inhibits progression of gastric cancer by suppressing activation of mapk signaling. Mol Cancer. (2021) 20:66. doi: 10.1186/s12943-021-01358-y
37. Xu X, Zhang J, Tian Y, Gao Y, Dong X, Chen W, et al. Circrna inhibits DNA damage repair by interacting with host gene. Mol Cancer. (2020) 19:128. doi: 10.1186/s12943-020-01246-x
38. Zhang MX, Wang JL, Mo CQ, Mao XP, Feng ZH, Li JY, et al. Circme1 promotes aerobic glycolysis and sunitinib resistance of clear cell renal cell carcinoma through cis-regulation of me1. Oncogene. (2022) 41:3979–90. doi: 10.1038/s41388-022-02386-8
39. Zou C, Li X, Lv X, Wu S, Song J, Tang Z, et al. Circular rna mitochondrial translation optimization 1 homologue (Circmto1) induced by zinc finger protein 460 (Znf460) promotes oral squamous cell carcinoma progression through the microrna mir-320a/alpha thalassemia/mental retardation, X-linked (Atrx) axis. Bioengineered. (2021) 12:9585–97. doi: 10.1080/21655979.2021.1997699
40. Ai Y, Wu S, Zou C, and Wei H. Circular rna circfoxo3 regulates kdm2a by targeting mir-214 to promote tumor growth and metastasis in oral squamous cell carcinoma. J Cell Mol Med. (2022) 26:1842–52. doi: 10.1111/jcmm.16533
41. Zhao W, Cui Y, Liu L, Qi X, Liu J, Ma S, et al. Splicing factor derived circular rna circuhrf1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ. (2020) 27:919–33. doi: 10.1038/s41418-019-0423-5
42. Zheng Z, Ma X, and Li H. Circular rna circmdm2 accelerates the glycolysis of oral squamous cell carcinoma by targeting mir-532-3p/hk2. J Cell Mol Med. (2020) 24:7531–7. doi: 10.1111/jcmm.15380
43. Zhang J, Peng Y, Jiang S, and Li J. Hsa_Circrna_0001971 contributes to oral squamous cell carcinoma progression via mir-186-5p/fibronectin type iii domain containing 3b axis. J Clin Lab Anal. (2022) 36:e24245. doi: 10.1002/jcla.24245
44. Jun W, Shaobo O, Xianhua Z, Siyu Z, Mingyang C, Xin F, et al. Deregulation of hsa_Circ_0001971/mir-186 and hsa_Circ_0001874/mir-296 signaling pathways promotes the proliferation of oral squamous carcinoma cells by synergistically activating shp2/plk1 signals. Sci Rep. (2021) 11:20561. doi: 10.1038/s41598-021-99488-2
45. Wang J, Ouyang S, Zhao S, Zhang X, Cheng M, Fan X, et al. Sp1-mediated upregulation of circfam126a promotes proliferation and epithelial-mesenchymal transition of oral squamous cell carcinoma via regulation of rab41. Front Oncol. (2022) 12:715534. doi: 10.3389/fonc.2022.715534
46. Yang Y, Ci HS, Mao YL, Li JW, and Zuo JH. Circrna_002178 promotes the proliferation and migration of oral squamous cell carcinoma cells by activating the akt/mtor pathway. Eur Rev Med Pharmacol Sci. (2020) 24:6122–30. doi: 10.26355/eurrev_202006_21507
47. Yang J, Cao XH, Luan KF, and Huang YD. Circular rna fndc3b protects oral squamous cell carcinoma cells from ferroptosis and contributes to the Malignant progression by regulating mir-520d-5p/slc7a11 axis. Front Oncol. (2021) 11:672724. doi: 10.3389/fonc.2021.672724
48. Li X, Wang C, Zhang H, Li Y, Hou D, Liu D, et al. Circfndc3b accelerates vasculature formation and metastasis in oral squamous cell carcinoma. Cancer Res. (2023) 83:1459–75. doi: 10.1158/0008-5472.Can-22-2585
49. Zou C, Li X, Wei H, Wu S, Song J, Tang Z, et al. Circular golph3 rna exerts oncogenic effects in vitro by regulating the mirna-1299/lif axis in oral squamous cell carcinoma. Bioengineered. (2022) 13:11012–25. doi: 10.1080/21655979.2022.2067288
50. Wang J, Jiang C, Li N, Wang F, Xu Y, Shen Z, et al. The circepsti1/mir-942-5p/ltbp2 axis regulates the progression of oscc in the background of osf via emt and the pi3k/akt/mtor pathway. Cell Death Dis. (2020) 11:682. doi: 10.1038/s41419-020-02851-w
51. Wang L, Wei Y, Yan Y, Wang H, Yang J, Zheng Z, et al. Circdock1 suppresses cell apoptosis via inhibition of mir−196a−5p by targeting birc3 in oscc. Oncol Rep. (2018) 39:951–66. doi: 10.3892/or.2017.6174
52. Liu J, Jiang X, Zou A, Mai Z, Huang Z, Sun L, et al. Circighg-induced epithelial-to-mesenchymal transition promotes oral squamous cell carcinoma progression via mir-142-5p/igf2bp3 signaling. Cancer Res. (2021) 81:344–55. doi: 10.1158/0008-5472.Can-20-0554
53. Guo L, Lin Q, Zhao X, and Xu J. Circular cdc like kinase 1 suppresses cell apoptosis through mir-18b-5p/Y-box protein 2 axis in oral squamous cell carcinoma. Bioengineered. (2022) 13:4226–34. doi: 10.1080/21655979.2022.2027174
54. Shao Y, Song Y, Xu S, Li S, and Zhou H. Expression profile of circular rnas in oral squamous cell carcinoma. Front Oncol. (2020) 10:533616. doi: 10.3389/fonc.2020.533616
55. Jiang W, Zhang C, Zhang X, Sun L, Li J, and Zuo J. Circrna hipk3 promotes the progression of oral squamous cell carcinoma through upregulation of the nupr1/pi3k/akt pathway by sponging mir-637. Ann Transl Med. (2021) 9:860. doi: 10.21037/atm-21-1908
56. He T, Li X, Xie D, and Tian L. Overexpressed circpvt1 in oral squamous cell carcinoma promotes proliferation by serving as a mirna sponge. Mol Med Rep. (2019) 20:3509–18. doi: 10.3892/mmr.2019.10615
57. Zhou Y, Zhang S, Min Z, Yu Z, Zhang H, and Jiao J. Knockdown of circ_0011946 targets mir-216a-5p/bcl2l2 axis to regulate proliferation, migration, invasion and apoptosis of oral squamous cell carcinoma cells. BMC Cancer. (2021) 21:1085. doi: 10.1186/s12885-021-08779-4
58. Zheng X, Du F, Gong X, and Xu P. Circ_0005320 promotes oral squamous cell carcinoma tumorigenesis by sponging microrna-486-3p and microrna-637. Bioengineered. (2022) 13:440–54. doi: 10.1080/21655979.2021.2009317
59. Ai Y, Tang Z, Zou C, Wei H, Wu S, and Huang D. Circ_Sept9, a newly identified circular rna, promotes oral squamous cell carcinoma progression through mir-1225/pkn2 axis. J Cell Mol Med. (2020) 24:13266–77. doi: 10.1111/jcmm.15943
60. Gao L, Dou ZC, Ren WH, Li SM, Liang X, and Zhi KQ. Circcdr1as upregulates autophagy under hypoxia to promote tumor cell survival via akt/erk(½)/mtor signaling pathways in oral squamous cell carcinomas. Cell Death Dis. (2019) 10:745. doi: 10.1038/s41419-019-1971-9
61. Rong L, Chen B, Liu K, Liu B, He X, Liu J, et al. Circzdbf2 up-regulates rnf145 by cerna model and recruits cebpb to accelerate oral squamous cell carcinoma progression via nfκb signaling pathway. J Transl Med. (2022) 20:148. doi: 10.1186/s12967-022-03347-1
62. Wu Z, He X, and Chen S. Oncogenic circdhtkd1 promotes tumor growth and metastasis of oral squamous cell carcinoma in vitro and in vivo via upregulating mir-326-mediated gab1. Braz J Med Biol Res. (2021) 54:e10837. doi: 10.1590/1414-431X2020e10837
63. Su Z, Pan C, Xie H, Ning Y, Li S, and Xiao H. Downregulation of circlpar3 inhibits tumor progression and glycolysis by liberating mir-144-3p and upregulating lpcat1 in oral squamous cell carcinoma. Laryngoscope Invest Otolaryngol. (2022) 7:425–36. doi: 10.1002/lio2.771
64. Xie H and Lu X. Circnfatc3 facilitated the progression of oral squamous cell carcinoma via the mir-520h/ldha axis. Open Med (Wars). (2023) 18:20230630. doi: 10.1515/med-2023-0630
65. Zeng W, Guo M, Yao L, and Deng Z. Circular rna hsa_Circ_0033144 (Circbcl11b) regulates oral squamous cell carcinoma progression via the mir-579/lasp1 axis. Bioengineered. (2021) 12:4111–22. doi: 10.1080/21655979.2021.1953214
66. Xia B, Hong T, He X, Hu X, and Gao Y. A circular rna derived from mmp9 facilitates oral squamous cell carcinoma metastasis through regulation of mmp9 mrna stability. Cell Transplant. (2019) 28:1614–23. doi: 10.1177/0963689719875409
67. Yan J and Xu H. Regulation of transforming growth factor-beta1 by circanks1b/mir-515-5p affects the metastatic potential and cisplatin resistance in oral squamous cell carcinoma. Bioengineered. (2021) 12:12420–30. doi: 10.1080/21655979.2021.2005221
68. Xu S, Song Y, Shao Y, and Zhou H. Hsa_Circ_0060927 is a novel tumor biomarker by sponging mir-195-5p in the Malignant transformation of olk to oscc. Front Oncol. (2021) 11:747086. doi: 10.3389/fonc.2021.747086
69. Luo Y, Liu F, Guo J, and Gui R. Upregulation of circ_0000199 in circulating exosomes is associated with survival outcome in oscc. Sci Rep. (2020) 10:13739. doi: 10.1038/s41598-020-70747-y
70. Chen Y, Li Z, Liang J, Liu J, Hao J, Wan Q, et al. Circrna has_Circ_0069313 induced oscc immunity escape by mir-325-3p-foxp3 axes in both oscc cells and treg cells. Aging (Albany NY). (2022) 14:4376–89. doi: 10.18632/aging.204068
71. Fan X and Wang Y. Circular rna circspata6 inhibits the progression of oral squamous cell carcinoma cells by regulating traf6 via mir-182. Cancer Manag Res. (2021) 13:1817–29. doi: 10.2147/cmar.S292074
72. Hao C, Wangzhou K, Liang Z, Liu C, Wang L, Gong L, et al. Circular rna itch suppresses cell proliferation but induces apoptosis in oral squamous cell carcinoma by regulating mir-421/pdcd4 axis. Cancer Manag Res. (2020) 12:5651–8. doi: 10.2147/cmar.S258887
73. Su W, Wang Y, Wang F, Zhang B, Zhang H, Shen Y, et al. Circular rna hsa_Circ_0007059 indicates prognosis and influences Malignant behavior via akt/mtor in oral squamous cell carcinoma. J Cell Physiol. (2019) 234:15156–66. doi: 10.1002/jcp.28156
74. Peng QS, Cheng YN, Zhang WB, Fan H, Mao QH, and Xu P. Circrna_0000140 suppresses oral squamous cell carcinoma growth and metastasis by targeting mir-31 to inhibit hippo signaling pathway. Cell Death Dis. (2020) 11:112. doi: 10.1038/s41419-020-2273-y
75. Shi D, Li H, Zhang J, and Li Y. Circgdi2 regulates the proliferation, migration, invasion and apoptosis of oscc via mir-454-3p/foxf2 axis. Cancer Manag Res. (2021) 13:1371–82. doi: 10.2147/CMAR.S277096
76. Zhang Y, Tang K, Chen L, Du M, and Qu Z. Exosomal circgdi2 suppresses oral squamous cell carcinoma progression through the regulation of mir-424-5p/scai axis. Cancer Manag Res. (2020) 12:7501–14. doi: 10.2147/cmar.S255687
77. Zhang X, Guo GY, Liu RY, Wu T, Wang ZH, and Zhang ZT. Circldlrad3 inhibits oral squamous cell carcinoma progression by regulating mir-558/smad4/tgf-B. J Cell Mol Med. (2023) 27:3271–85. doi: 10.1111/jcmm.17898
78. Deng W, Fu J, Wang T, Chen JX, Fu LB, and Peng W. Hsa_Circrna_101036 acts as tumor-suppressor in oral squamous cell carcinoma cells via inducing endoplasmic reticulum stress. Eur Rev Med Pharmacol Sci. (2020) 24:6111–21. doi: 10.26355/eurrev_202006_21506
79. Gao L, Zhang Q, Li S, Zheng J, Ren W, and Zhi K. Circ-pkd2 promotes atg13-mediated autophagy by inhibiting mir-646 to increase the sensitivity of cisplatin in oral squamous cell carcinomas. Cell Death Dis. (2022) 13:192. doi: 10.1038/s41419-021-04497-8
80. Chen G, Li Y, He Y, Zeng B, Yi C, Wang C, et al. Upregulation of circular rna circatrnl1 to sensitize oral squamous cell carcinoma to irradiation. Mol Ther Nucleic Acids. (2020) 19:961–73. doi: 10.1016/j.omtn.2019.12.031
81. Wang Z, Tang J, Wang Y, Sun S, Chen Y, Shen Y, et al. Circular rna hsa_Circ_009755 downregulation correlates with clinicopathology in oral squamous cell carcinoma. Onco Targets Ther. (2019) 12:4025–31. doi: 10.2147/OTT.S196618
82. Li L and Zhang ZT. Hsa_Circ_0086414 might be a diagnostic biomarker of oral squamous cell carcinoma. Med Sci Monit. (2020) 26:e919383. doi: 10.12659/MSM.919383
83. Chin L and Gray JW. Translating Insights from the Cancer Genome into Clinical Practice. Nature. (2008) 452(7187):553-63. doi: 10.1038/nature06914
84. Gruner BM and Fendt SM. Cancer cells stock up in lymph vessels to survive. Nature. (2020) 585:36–7. doi: 10.1038/d41586-020-02383-5
85. Saikishore R, Velmurugan P, Ranjithkumar D, Latha R, Sathiamoorthi T, Arun A, et al. The circular rna-mirna axis: A special rna signature regulatory transcriptome as a potential biomarker for oscc. Mol Ther - Nucleic Acids. (2020) 22:352–61. doi: 10.1016/j.omtn.2020.09.001
86. Paul S, Ghosh S, and Kumar S. Tumor glycolysis, an essential sweet tooth of tumor cells. Semin Cancer Biol. (2022) 86:1216–30. doi: 10.1016/j.semcancer.2022.09.007
87. Ganapathy-Kanniappan S and Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer. (2013) 12:152. doi: 10.1186/1476-4598-12-152
88. Li M, Gao F, Zhao Q, Zuo H, Liu W, and Li W. Tanshinone iia inhibits oral squamous cell carcinoma via reducing akt-C-myc signaling-mediated aerobic glycolysis. Cell Death Dis. (2020) 11:381. doi: 10.1038/s41419-020-2579-9
89. Cai H, Li J, Zhang Y, Liao Y, Zhu Y, Wang C, et al. Ldha promotes oral squamous cell carcinoma progression through facilitating glycolysis and epithelial-mesenchymal transition. Front Oncol. (2019) 9:1446. doi: 10.3389/fonc.2019.01446
90. Wang B and Tontonoz P. Phospholipid remodeling in physiology and disease. Annu Rev Physiol. (2019) 81:165–88. doi: 10.1146/annurev-physiol-020518-114444
91. Du Y, Wang Q, Zhang X, Wang X, Qin C, Sheng Z, et al. Lysophosphatidylcholine acyltransferase 1 upregulation and concomitant phospholipid alterations in clear cell renal cell carcinoma. J Exp Clin Cancer Res. (2017) 36:66. doi: 10.1186/s13046-017-0525-1
92. Shida-Sakazume T, Endo-Sakamoto Y, Unozawa M, Fukumoto C, Shimada K, Kasamatsu A, et al. Lysophosphatidylcholine acyltransferase1 overexpression promotes oral squamous cell carcinoma progression via enhanced biosynthesis of platelet-activating factor. PloS One. (2015) 10:e0120143. doi: 10.1371/journal.pone.0120143
93. Huang Y, Wang Y, Zhen Y, Liu W, Wang Y, Wang R, et al. Lpcat1 facilitates keratinocyte hyperproliferation and skin inflammation in psoriasis by regulating glut3. J Invest Dermatol. (2024) 144:1479–90 e14. doi: 10.1016/j.jid.2024.01.004
94. Gao W, Wang X, Zhou Y, Wang X, and Yu Y. Autophagy, ferroptosis, pyroptosis, and necroptosis in tumor immunotherapy. Signal Transduct Target Ther. (2022) 7:196. doi: 10.1038/s41392-022-01046-3
95. Assi M and Kimmelman AC. Impact of context-dependent autophagy states on tumor progression. Nat Cancer. (2023) 4:596–607. doi: 10.1038/s43018-023-00546-7
96. Li Z and Zhang X. Phospho-regulation and function of ulk1-atg13 during the cell cycle. Autophagy. (2021) 17:1054–6. doi: 10.1080/15548627.2021.1898750
97. li Z, Tian X, Ji X, Wang D, and Zhang X. Cdk1 phosphorylates ulk1-atg13 complex to regulate mitotic autophagy and taxol chemosensitivity. PLOS Biology (2019) 18:. doi: 10.1101/634733
98. Xu W, Zhuang L, Zhu H, Mao A, Zhou J, and Wang L. Trim14 overexpression induces chemoresistance and Malignant behaviors of hepatocellular carcinoma cells by activating the stat3/hif-1alpha pathway. Int J Mol Sci. (2023) 24:12589. doi: 10.3390/ijms241612589
99. Tang D, Chen X, Kang R, and Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. (2021) 31:107–25. doi: 10.1038/s41422-020-00441-1
100. Lei G, Zhuang L, and Gan B. Targeting ferroptosis as a vulnerability in cancer. Nat Rev Cancer. (2022) 22:381–96. doi: 10.1038/s41568-022-00459-0
101. Liu X, Nie L, Zhang Y, Yan Y, Wang C, Colic M, et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat Cell Biol. (2023) 25:404–14. doi: 10.1038/s41556-023-01091-2
102. Li W, Deng X, and Chen J. Rna-binding proteins in regulating mrna stability and translation: roles and mechanisms in cancer. Semin Cancer Biol. (2022) 86:664–77. doi: 10.1016/j.semcancer.2022.03.025
103. Ward CL, Boggio KJ, Johnson BN, Boyd JB, Douthwright S, Shaffer SA, et al. A loss of fus/tls function leads to impaired cellular proliferation. Cell Death Dis. (2014) 5:e1572. doi: 10.1038/cddis.2014.508
104. Cai Y, Li N, and Li H. Ybx2 modulates mrna stability via interaction with ythdf2 in endometrial cancer cells. Exp Cell Res. (2023) 427:113586. doi: 10.1016/j.yexcr.2023.113586
105. Qiu F, Qiao B, Zhang N, Fang Z, Feng L, Zhang S, et al. Blocking circ-scmh1 (Hsa_Circ_0011946) suppresses acquired ddp resistance of oral squamous cell carcinoma (Oscc) cells both in vitro and in vivo by sponging mir-338-3p and regulating lin28b. Cancer Cell Int. (2021) 21:412. doi: 10.1186/s12935-021-02110-8
106. Su W, Wang Y, Wang F, Sun S, Li M, Shen Y, et al. Hsa_Circ_0005379 regulates Malignant behavior of oral squamous cell carcinoma through the egfr pathway. BMC Cancer. (2019) 19:400. doi: 10.1186/s12885-019-5593-5
107. Dong W, Zhao L, Zhang S, Zhang S, and Si H. Circ-kiaa0907 inhibits the progression of oral squamous cell carcinoma by regulating the mir-96-5p/unc13c axis. World J Surg Oncol. (2021) 19:75. doi: 10.1186/s12957-021-02184-8
108. Dai F, Dai L, Zheng X, Guo Y, Zhang Y, Niu M, et al. Non-coding rnas in drug resistance of head and neck cancers: A review. BioMed Pharmacother. (2020) 127:110231. doi: 10.1016/j.biopha.2020.110231
109. Sun S, Li B, Wang Y, Li X, Wang P, Wang F, et al. Clinical significance of the decreased expression of hsa_Circ_001242 in oral squamous cell carcinoma. Dis Markers. (2018) 2018:6514795. doi: 10.1155/2018/6514795
110. Wang M, Zhang L, Ren W, Li S, Zhi K, Zheng J, et al. Diagnostic value of circrnas as potential biomarkers in oral squamous cell carcinoma: A meta-analysis. Front Oncol. (2021) 11:693284. doi: 10.3389/fonc.2021.693284
111. Zhang Z, Yang T, and Xiao J. Circular rnas: promising biomarkers for human diseases. EBioMedicine. (2018) 34:267–74. doi: 10.1016/j.ebiom.2018.07.036
112. Li S, Wu H, and Chen L-L. Screening circular rnas with functional potential using the rfxcas13d/bsj-grna system. Nat Protoc. (2022) 17:2085–107. doi: 10.1038/s41596-022-00715-5
113. Niu D, Wu Y, and Lian J. Circular rna vaccine in disease prevention and treatment. Signal Transduct Target Ther. (2023) 8:341. doi: 10.1038/s41392-023-01561-x
114. Holthoff JH, Chandrashekar K, and Juncos LA. The role of esm-1 in diabetic kidney disease: more than just a biomarker. Kidney360. (2022) 3:1998–2000. doi: 10.34067/KID.0004952022
115. Yu SS and Cirillo N. The molecular markers of cancer stem cells in head and neck tumors. J Cell Physiol. (2020) 235:65–73. doi: 10.1002/jcp.28963
Keywords: circular RNA, oral squamous cell carcinoma, tumorigenesis, mechanism, biomarkers
Citation: Lv J-L, Ma R, Ren Y-S, Liang Q-Y, Zhang H-M, Dong G-C and Li J (2025) CircRNA: the potential biomarkers and therapeutic targets in oral squamous cell carcinoma (OSCC). Front. Oncol. 15:1555002. doi: 10.3389/fonc.2025.1555002
Received: 03 January 2025; Accepted: 19 May 2025;
Published: 05 June 2025.
Edited by:
Sharon R. Pine, University of Colorado Anschutz Medical Campus, United StatesReviewed by:
Shanchun Guo, Xavier University of Louisiana, United StatesRuma Dey Ghosh, Tata Translational Cancer Research Center (TTCRC), India
Copyright © 2025 Lv, Ma, Ren, Liang, Zhang, Dong and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gui-Cheng Dong, Z2NkaG9oaG90QHNvaHUuY29t; Jie Li, bGlqaWU1NzY3QHNvaHUuY29t
†These authors have contributed equally to this work
 Jun-Lin Lv
Jun-Lin Lv Ru Ma1,3†
Ru Ma1,3† Yu-Shan Ren
Yu-Shan Ren Jie Li
Jie Li