- 1Department of Thoracic Surgery, Fujian Institute of Thoracic and Cardiac Surgery, Fujian Medical University Union Hospital, Fuzhou, China
- 2Medical Department, Nanjing Geneseeq Technology Inc., Nanjing, China
- 3Department of Pathology, Fujian Medical University Union Hospital, Fuzhou, China
Background: Lung adenocarcinoma with a micropapillary component (LMPC) is an aggressive histologic subtype of lung cancer characterized by unique pathological features and poor prognosis. While previous studies have identified driver mutations in LMPC, its comprehensive molecular profile and prognosis-related biomarkers in the Chinese population remain poorly understood.
Methods: We conducted a retrospective study of 54 stage I-III LMPC patients who underwent complete resection. Tumor samples from these patients were analyzed using broad-panel next-generation sequencing of 425 cancer-related genes. We explored the associations among clinicopathologic factors, genomic characteristics, and post-operative recurrence risk.
Results: Compared to a reference cohort of 113 LADC patients, LMPC exhibited a distinct genetic profile, with a greater diversity of targetable mutations, an increased number of oncogenic pathway alterations (NPA), and more oncogenic pathway-related alterations. The mutation frequencies of ERBB4 (11.1% vs. 1.8%, P=0.015), BRAF (9.3% vs. 1.8%, P=0.037), PIK3CA (14.8% vs. 4.4%, P=0.029), RPTOR (P=0.033), and NOTCH2 (P=0.033) were significantly higher in LMPC. Additionally, LMPC patients had significantly more alterations in three oncogenic pathways (PI3K, Wnt, and TGF-β) and a significantly increased NPA (P<0.001). In stage II-III LMPC patients, SMARCA4 mutations (13.9 months vs. not reached (NR), P=0.013) and alterations in the SWI/SNF (16.3 months vs. NR, P=0.003) and Nrf2 (17.0 months vs. NR, P=0.046) pathways were significantly associated with higher postoperative recurrence risk. Furthermore, tumor mutation burden (TMB) was significantly correlated with postoperative disease-free survival (DFS), with patients having low TMB showing prolonged median DFS compared to those with high TMB (NR vs. 16.8 months, P=0.021).
Conclusion: Our study elucidates the unique genetic landscape of Chinese resectable LMPC patients and highlights high TMB and mutations in SMARCA4, SWI/SNF, and Nrf2 pathways as potential prognostic indicators in stage II-III disease. As these factors were not confirmed in multivariate models, they should be validated in larger, multi-center cohorts to guide future risk stratification and treatment decisions.
Introduction
Lung cancer remains the leading cause of cancer-related mortality worldwide, with non-small-cell lung cancer (NSCLC) accounting for over 80% of all cases (1, 2). Lung adenocarcinoma (LADC) is the most prevalent NSCLC subtypes (2). The 2011 classification system proposed by the International Association for the Study of Lung Cancer (IASLC), the American Thoracic Society (ATS), and the European Respiratory Society (ERS) categorizes LADC into several histologic patterns: lepidic, acinar, papillary, solid, and micropapillary (3, 4). More recently, deep learning-based approaches have demonstrated high accuracy in classifying these LADC subtypes using whole-slide images or computed tomography (CT) images (5–7). Notably, the presence of a micropapillary component (MPC) in LADC is linked to a poor prognosis. Even a minor proportion of MPC in adenocarcinoma significantly increases the risk of disease recurrence (8–12).
Several studies have identified oncogenic mutations in LADC with a micropapillary component (LMPC), revealing a diverse range of driver mutations. For instance, a meta-analysis by Pyo and Kim reported estimated mutation rates in LMPC: 62.0% for EGFR, 11.8% for KRAS, and 10.2% for ALK (13). Another study involving 121 LMPC patients found EGFR mutations to be the most prevalent (76.9%), followed by ALK translocations (3.3%), KRAS mutations (2.5%), HER2 mutations (1.7%), and RET fusions (1.7%) (14). Additionally, Ou et al. conducted both RNA-sequencing and whole-exome sequencing in 101 LMPC patients and found MACF1, PCLO, ADGRV1, and Fanconi Anemia pathway mutations as negative indicators for recurrence-free survival (15). Despite these insights, the molecular mechanisms underlying the poor prognosis of LMPC compared to other LADC subtypes remain poorly understood, particularly in Chinese populations. Moreover, the association between specific molecular alterations and clinical outcomes has not been comprehensively investigated. Although surgical resection remains the standard treatment for early-stage LMPC, approximately 30-50% of patients experience disease recurrence within five years postoperatively (16). Thus, identifying molecular biomarkers to stratify postoperative recurrence risk in patients diagnosed with LMPC is clinically significant, as it may inform adjuvant treatment decisions and follow-up strategies beyond conventional histopathologic staging.
In this study, we aimed to investigate the molecular characteristics and potential prognostic biomarkers in Chinese patients with resectable LMPC. We conducted a comprehensive retrospective analysis of the clinicopathological features and oncogenic mutation profiles of 54 Chinese LMPC patients who underwent surgical resection, with tumors containing at least 5% MPC. For comparison, we used an external cohort of 113 Chinese LADC patients from a previous study (17). Our research focused on comparing key genetic features and aberrant signaling pathways between LADC patients with and without MPC, and analyzing the prognostic implications of these molecular features in stage II-III LMPC patients.
Materials and methods
Patients and samples
We retrospectively reviewed all patients with LADC who underwent radical surgical resection at the Department of Thoracic Surgery, Fujian Medical University Union Hospital between February 2017 and December 2019. Cases with an MPC of at least 5% were initially identified through pathology reports and reviewed independently by two experienced pathologists to confirm eligibility. From this initial pool, 54 patients met all criteria for final inclusion (Supplementary Figure S1). Inclusion criteria were as follows: 1) Histologically confirmed LADC with ≥5% MPC. 2) Negative surgical margins (R0 resection). 3) Pathological stage I-III based on the 8th edition of the TNM staging system (18). 4) Surgical procedure was lobectomy or sublobectomy with systematic mediastinal lymph node dissection. 5) No significant cardiopulmonary comorbidities or major postoperative complications. Exclusion criteria included: 1) Death from other primary malignancies or non-cancer causes. Incomplete clinicopathological or follow-up data. 2) Prior neoadjuvant therapy. 3) NGS failure or inadequate tumor tissue for sequencing. This study received approval from the Ethics Committees of Fujian Medical University Union Hospital (No. 2019JYKY017). All patients provided written informed consent, and authors had access to information that could identify individual participants during or after data collection. Tumor tissue samples from these 54 patients were collected and prepared as formalin-fixed paraffin-embedded (FFPE) specimens. These FFPE slides were reviewed by two certified pathologists to classify the histologic subtypes of LADC according to the IASLC/ATS/ERS multidisciplinary classification system. LMPC was defined as LADC with at least 5% MPC. Based on a previous study (17), LMPC cases were further divided into MPC-low (5%≤MPC<20%) and MPC-high (MPC≥20%) groups according to the proportion of MPC content.
All patients underwent R0 resection, defined as complete tumor resection with negative margins. Surgical procedures included either lobectomy or sublobectomy, both performed with systematic mediastinal lymph node dissection. Intraoperative frozen section analysis was routinely performed on bronchial margins to confirm tumor-free status. For sublobectomy cases, an adequate surgical margin was ensured, defined as either a margin >2 cm or greater than the diameter of the tumor nodule. Systematic mediastinal lymph node dissection was defined as the complete removal of lymph nodes from at least three mediastinal stations. Specifically, for right-sided tumors, nodal stations 2R, 4R, 7, and 8 were routinely dissected, while for left-sided tumors, stations 5, 6, 7, and 8 were removed. All FFPE samples were sent to Nanjing Geneseeq Technology Inc. (Nanjing, China) for targeted next-generation sequencing (NGS) of 425 cancer-relevant genes.
Follow-up assessments were conducted every three months during the first two years after surgery, every six months in the third and fourth years, and annually thereafter. Postoperative outcomes were documented during follow-up visits and supplemented through telephone interviews. Regular evaluations included physical examinations and chest computed tomography (CT), while additional imaging modalities, such as positron emission tomography-computed tomography (PET-CT), ultrasound, endoscopy, magnetic resonance imaging (MRI), or whole-body bone scans, were utilized as clinically indicated. The follow-up period concluded at the end of June 2021, with a median follow-up duration of 30.6 months (range: 6.6-54.6 months) across all patients. Disease-free survival was defined as the interval from the date of surgery to the first occurrence of disease recurrence, cancer-related death, or the last follow-up. Local recurrence was defined as tumor recurrence within the ipsilateral chest cavity, including the surgical margin of the lung or bronchus, hilar or mediastinal lymph nodes, and malignant pleural effusion.
DNA extraction and library construction
Genomic DNA was extracted from FFPE sections using the QIAamp DNA FFPE Tissue kit (Qiagen). For normal control, genomic DNA was extracted from white blood cells (WBCs) using the DNeasy Blood & Tissue Kit (Qiagen) to remove germline variations. DNA concentration was measured with a Qubit 3.0 fluorometer using the dsDNA HS Assay Kit (Life Technologies), and quality assessment was conducted using a Nanodrop 2000 spectrophotometer (Thermo Fisher).
Approximately 0.5-1 μg of fragmented genomic DNA from each sample was used for library preparation. Library preparation and targeted capture enrichment were performed as previously described with some modifications (19). A customized xGen lockdown probe panel encompassing 425 predefined cancer-related genes was employed for selective enrichment. The captured libraries were then amplified, purified, and quantified.
Sequencing and bioinformatics analysis
Target-enriched libraries were sequenced on the HiSeq4000 platform (Illumina) using 2×150 bp paired-end reads. Sequencing data were demultiplexed using bcl2fastq (v2.19) and processed with Trimmomatic to remove low-quality bases (quality<15) or N bases. The cleaned data were then aligned to the hg19 reference human genome using the Burrows-Wheeler Aligner (bwa-mem). Further processing was conducted with the Picard suite (available at: https://broadinstitute.github.io/picard/) and the Genome Analysis Toolkit (GATK).
Single-nucleotide polymorphisms (SNPs) and insertions/deletions (indels) were called using VarScan2 and the HaplotypeCaller/UnifiedGenotyper in GATK. The mutant allele frequency (MAF) cutoff for single-nucleotide variants (SNVs) and indels was set at 1%. Common variants were filtered out using dbSNP and the 1000 Genomes Project. Germline mutations were excluded by comparing the results to the patient’s WBC controls.
Gene fusions were identified using FACTERA and copy number variations (CNVs) were analyzed with ADTEx. The log2 ratio cutoff for copy number gain was defined as 2.0 for tissue samples, while a log2 ratio cutoff of 0.6 was used for copy number loss. Allele-specific CNVs were analyzed by FACETS with a 0.2 drift cutoff for unstable joint segments. Tumor mutational burden (TMB) was defined as the number of nonsynonymous mutations per sample, as previously described (20). The number of oncogenic pathway alterations (NPA) was quantified as the cumulative count of somatically altered pathways per patient, encompassing ten canonical pathways profiled by The Cancer Genome Atlas (TCGA) (21) and the SWI/SNF chromatin remodeling complex pathway.
Statistical analysis
Quantitative data are presented as the median (range) or as the number of patients (percentage). Comparisons of proportions between groups were conducted using Fisher’s exact test. Survival analysis was performed using Kaplan-Meier curves, with the P value determined by the log-rank test. Hazard ratio (HR) were calculated using the Cox proportional hazards model. Both univariate and multivariate analyses were conducted to investigate the association between various variables and disease-free survival (DFS). A two-sided P value of less than 0.05 was considered statistically significant for all tests unless otherwise specified. All analyses were conducted using R version 3.6.0.
Results
Patient characteristics
The clinicopathological features of the 54 Chinese patients with resectable LMPC are summarized in Table 1. Paired treatment-naive tumor tissue and corresponding WBCs control samples were available for each patient. The cohort consisted of 20 females (37.0%) and 34 males (63.0%), with a median age of 60 years. The study included an equal number of stage I and stage II-III patients, and 25 patients (46.3%) had a history of smoking. Based on the percentage of MPC in the tumor, 29 patients (53.7%) were classified as MPC-low (5%≤MPC<20%) and 25 patients (46.3%) as MPC-high (MPC≥20%). Lymph node metastases were observed in 18 patients (33.3%) while lymphovascular invasion and pleural invasion were detected in 6 patients (11.1%) and 8 patients (14.8%) respectively. Following surgical resection, 24 patients (44.4%) received adjuvant chemotherapy and/or adjuvant EGFR or ALK tyrosine kinase inhibitors (TKIs) treatment. Additionally, approximately 26% (7/27) of stage II-III patients received adjuvant EGFR or ALK TKIs treatment, while none of the patients in our cohort received immunotherapy. No statistically significant differences in clinicopathological features were observed between the 54 LMPC patients and the 113 reference LADC patients (Table 1).
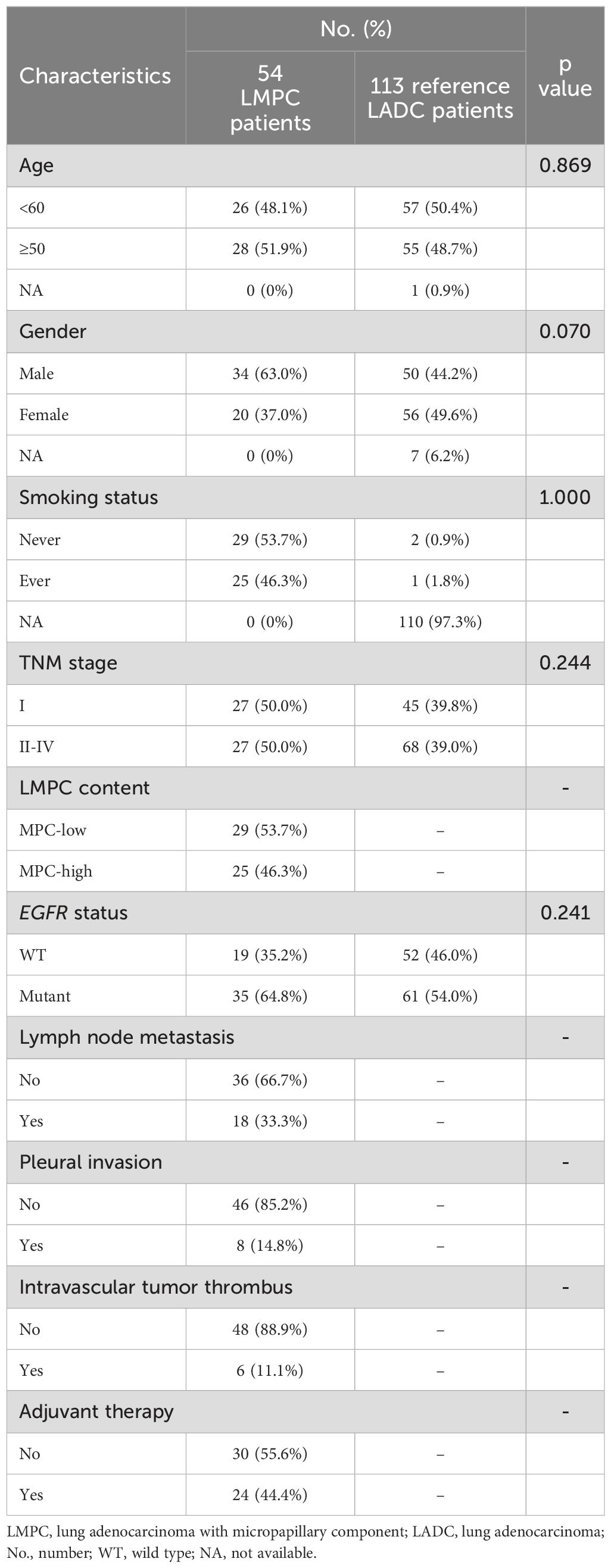
Table 1. Clinicopathologic characteristics of LMPC patients enrolled in this study compared to the reference cohort of 113 LADC patients.
Driver gene characteristics of LMPC
As shown in Figure 1, the most commonly mutated genes in LMPC were EGFR (75.9%), TP53 (57.4%), KRAS (14.8%), ALK (14.8%), and PIK3CA (14.8%). Driver mutations, including EGFR, ALK, KRAS, ERBB2, BRAF, and HRAS, were detected in over 90% of patients (Figure 1, Supplementary Figure S2). ALK fusions were detected in 4 patients (7.4%), and KRAS mutations were present in 8 patients (14.8%), with 3 cases involving the KRAS p.G12C mutation (Figure 1, Supplementary Figure S2). RTK/RAS was the most frequently mutated signaling pathway, primarily due to the high frequency of EGFR mutations. Specifically, 64.8% of the patients had EGFR driver mutations, with an additional 11.1% harboring other EGFR mutations. Other RTK/RAS-related gene mutations were also prevalent, including ALK (14.8%), KRAS (14.8%), BRAF (9.3%), and ERBB2 (5.6%) (Figure 1).
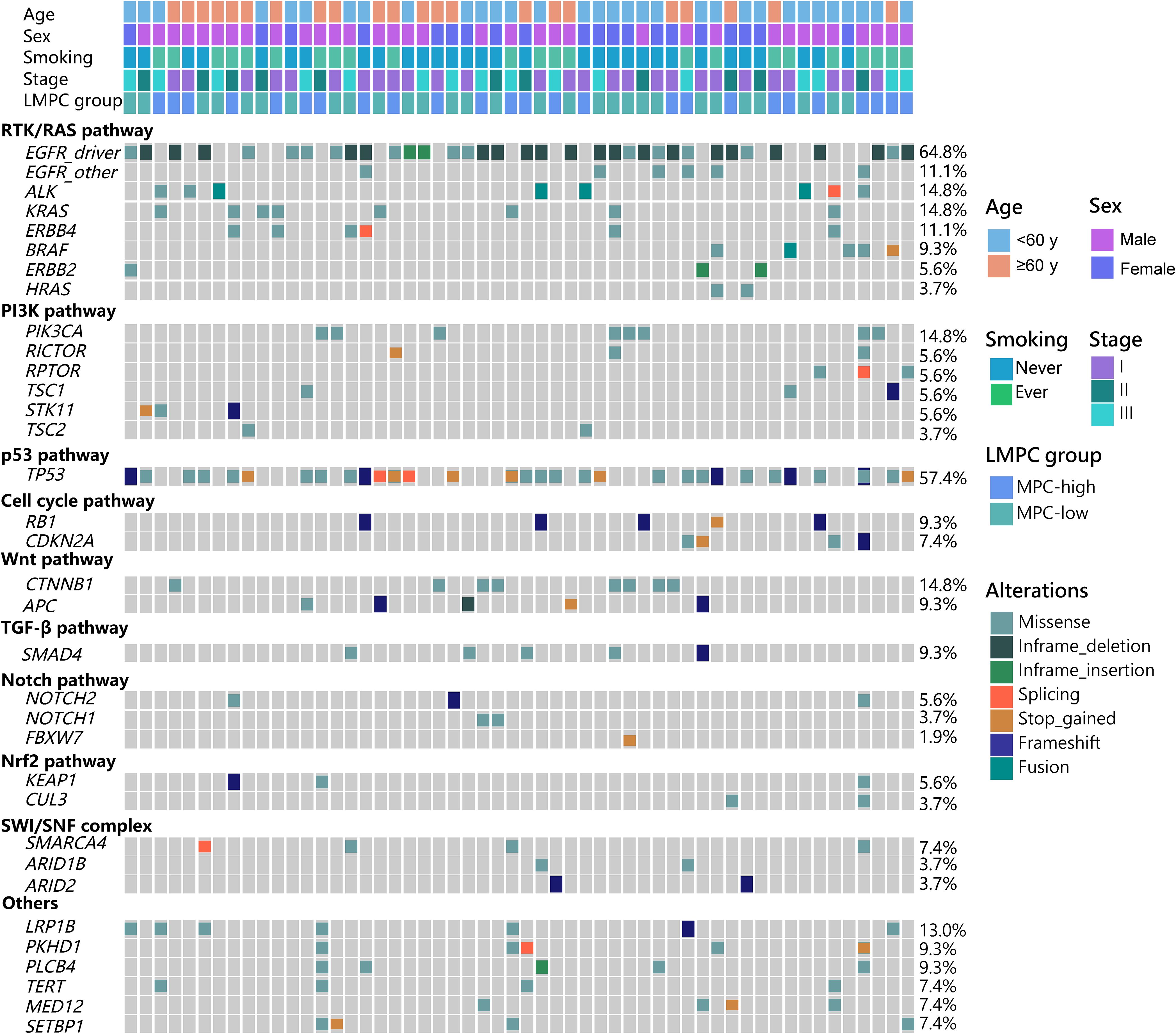
Figure 1. The genetic landscape of the 54 stage I-III LMPC patients enrolled in this study. The demographic and clinical features of the patients were listed at the top of the graph while various oncogenic pathway-associated mutations were listed below. LMPC, lung adenocarcinoma with micropapillary component; MPC, micropapillary component.
Genetic alterations in the PI3K pathway were commonly detected, with mutations in PIK3CA (14.8%), RICTOR (5.6%), RPTOR (5.6%), STK11 (5.6%), TSC1 (5.6%), and TSC2 (3.7%). In the Notch pathway, mutation frequencies were 1.9% for FBXW7, 3.7% for NOTCH1, and 5.6% for NOTCH2. For the Nrf2 pathway, KEAP1 and CUL3 had mutation rates of 5.6% and 3.7%, respectively. Alterations were also observed in the SWI/SNF pathway, with mutations in SMARCA4 (7.4%), ARID1B (3.7%), and ARID2 (3.7%) (Figure 1). Overall, we observed a relatively high frequency of oncogenic driver mutations and aberrant signaling pathways in Chinese resectable LMPC patients, highlighting the complexity and heterogeneity of this disease.
Comparison of mutation profiles between LMPC and LADC patients
Given that LMPC and the reference cohort of 113 LADC patients exhibited distinct clinical outcomes, we hypothesized that underlying molecular and genetic differences might exist between these two patient populations. Therefore, we compared the mutational characteristics of our LMPC patients with those of the 113 Chinese LADC patients described in recent literature (17). The mutation frequencies of EGFR, ALK, ERBB2, KRAS, and HRAS were similar between the LMPC and LADC groups. However, mutations in ERBB4 (11.1% vs. 1.8%, P=0.015) and BRAF (9.3% vs. 1.8%, P=0.037) were significantly more enriched in the LMPC group (Figure 2A).
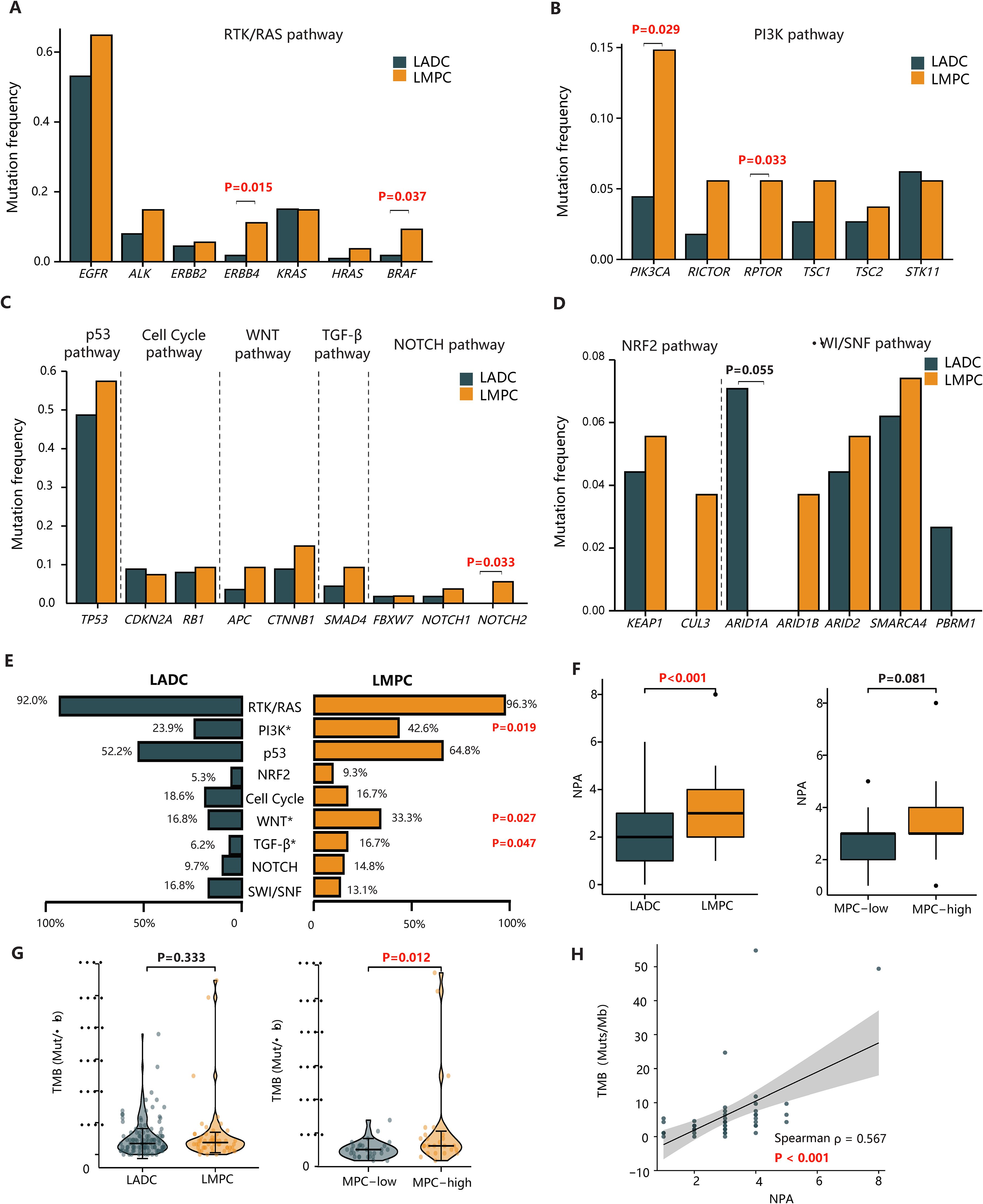
Figure 2. The comparison of genetic features between our LMPC patients and the reference cohort. (A–D) The bar plots of the mutation frequency of significantly altered genes between LMPC and LADC groups in the RTK/RAS, PI3K, p53, Cell cycle, Wnt, TGF-β, Notch, Nrf2, and SWI/SNF pathways. (E) The comparison of the frequency of aberrant oncogenic pathways between LMPC and LADC groups. (F) The comparison of NPA in tumors categorized based on the presence of MPC (LMPC vs. LADC) or the MPC content (MPC-low vs. MPC-high). (G) The violin plot of the differential TMB levels in tumors categorized based on the presence of MPC (LMPC vs. LADC) or the MPC content (MPC-low vs. MPC-high). (H) The scatterplot of TMB versus NPA in the LMPC group. The Spearman’s correlation coefficient(ρ) and the associated P value were labeled on the figure. LADC, lung adenocarcinoma; LMPC, lung adenocarcinoma with micropapillary component; MPC, micropapillary component; TMB, tumor mutation burden; NPA, the number of oncogenic pathway alterations.
Beyond the RTK/RAS pathway, mutations in the PI3K pathway (PIK3CA: 14.8% vs. 4.4%, P=0.029; RPTOR: 5.6% vs. 0%, P=0.033) and the Notch pathway (NOTCH2: 5.6% vs. 0%, P=0.033) were more frequently observed in LMPC patients than in LADC patients (Figures 2B, C). In contrast, the mutation frequency of ARID1A (7.1% vs. 0%, P=0.055) in the SWI/SNF pathway tended to be higher in LADC than in LMPC, although this result was not statistically significant (Figure 2D).
We further compared differences in oncogenic pathways between the LADC and LMPC groups, analyzing nine classical oncogenic pathways altered in at least five LMPC patients (Figure 2E). Consistent with the gene-level mutational analysis, LMPC patients harbored more genetic alterations in several oncogenic pathways, including PI3K (42.6% vs. 23.9%, P=0.019), Wnt (33.3% vs. 16.8%, P=0.027), and TGF-β (16.7% vs. 6.2%, P=0.047). Additionally, the NPA in the LMPC group was significantly higher than in the LADC group (P<0.001) (Figure 2F). Similarly, the NPA in the MPC-high group tended to be higher than in the MPC-low group (P=0.081) (Figure 2F).
We then compared the TMB levels between LADC and LMPC patients and found no significant difference between the two groups (3.44 vs. 3.76, P=0.333) (Figure 2G). However, a further comparison of TMB levels between MPC-low and MPC-high patients revealed that the TMB levels were significantly higher in the MPC-high group (3.22 vs. 4.30, P=0.012) (Figure 2G). Moreover, there was a positive correlation between TMB and NPA in LMPC patients (Spearman’s correlation coefficient, r=0.567, P<0.001) (Figure 2H). Overall, LMPC patients, especially those with high MPC, tended to carry more oncogenic alterations, signaling pathway aberrations, and higher mutational burdens.
The association between clinicopathological characteristics and prognosis in LMPC patients
As resectable LMPC patients often experience poor prognosis, we explored the association between post-surgical DFS and various clinical characteristics. Univariate Cox regression analysis indicated that clinicopathological factors such as age, sex, smoking history, MPC content, pleural invasion, lymphovascular invasion, and adjuvant therapy did not significantly affect patients’ prognosis. However, tumor stage and lymph node metastasis status were significantly correlated with postoperative DFS in LMPC patients (Figure 3A).
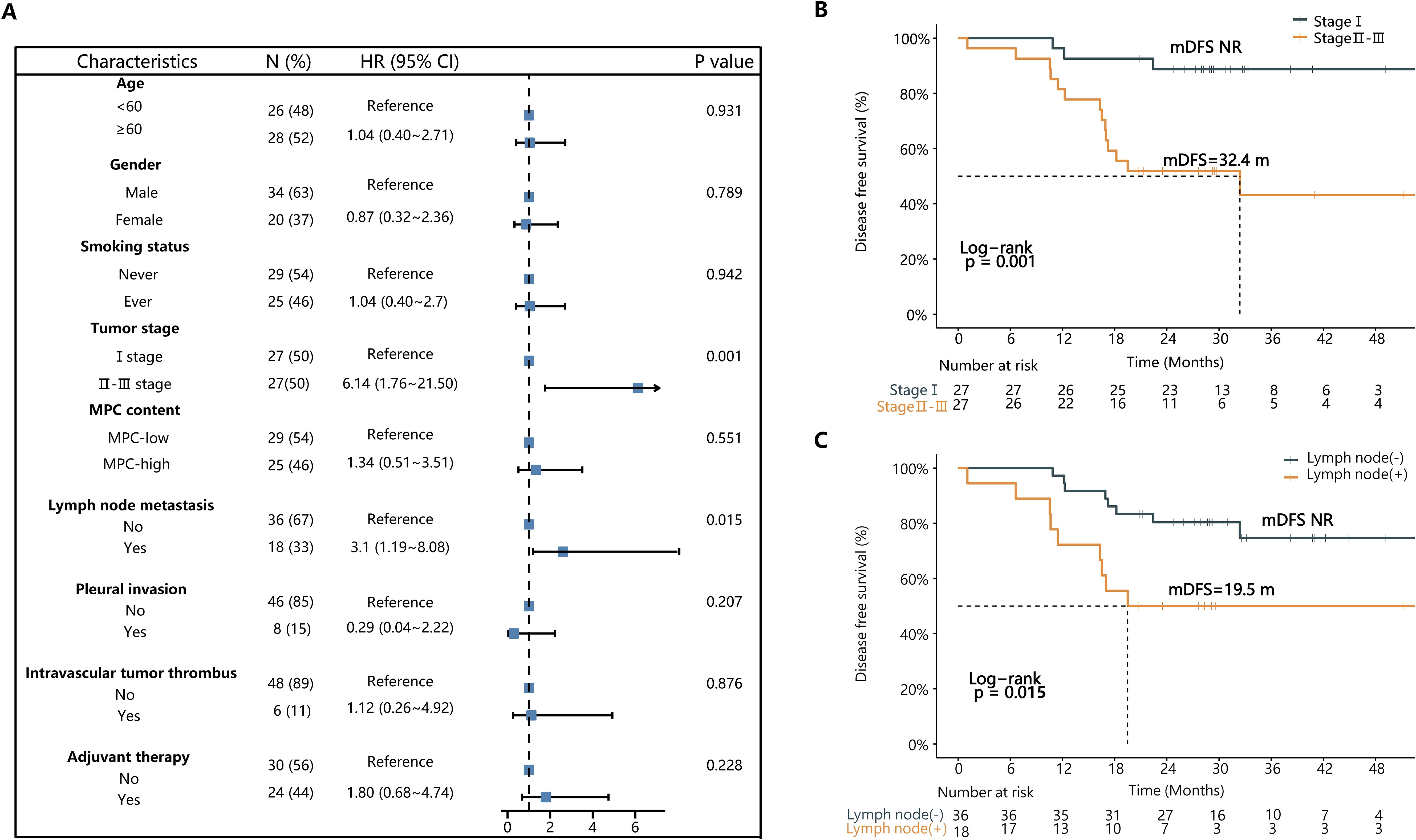
Figure 3. The univariate Cox regression analysis of clinicopathologic characteristics associated with DFS in the 54 patients. (A) The forest plot presents hazard ratios (HRs) of various clinicopathological characteristics associated with DFS. (B) The Kaplan-Meier curve of DFS in stage I vs stage II-III patients. (C) The Kaplan-Meier curve of DFS in patients with or without lymph node metastasis. MPC, micropapillary component; mDFS, median disease-free survival; NR, not reached.
Specifically, stage I patients had significantly better postoperative median DFS (mDFS) compared to stage II and III patients (Not reached (NR) vs. NR vs. 18.2 months, P=0.004, HR = 6.14) (Figure 3B; Supplementary Figure S3A). There was no significant difference in mDFS between stage II and stage III patients (NR vs. 18.2 months, P=0.520) (Supplementary Figure S3A). In univariate Cox regression analysis for stage II-III LMPC patients, none of the tested clinicopathological factors had a significant effect on mDFS (Supplementary Table S1).
Among the 54 LMPC cases, patients without lymph node metastasis demonstrated significantly better DFS compared to those with lymph node metastasis (NR vs. 19.5 months, P=0.015, HR=3.10) (Figure 3C). Furthermore, no significant difference in DFS was observed between MPC-high and MPC-low patients (P=0.550), indicating that micropapillary content alone may not independently predict prognosis in this cohort (Supplementary Figures S3B, C).
Correlation analysis of gene mutations, signaling pathways, TMB and recurrence in surgically resected stage II-III LMPC patients
Considering that all the investigated clinicopathological factors failed to stratify DFS in stage II-III LMPC patients (Supplementary Table S1), and recognizing the clinical importance of estimating prognosis in patients with more advanced diseases, we decided to explore the prognostic effects of tumor mutations and aberrant signaling pathways in these patients. Genes or pathways that were altered in ≥3 patients were included in the analysis, and their correlations with mDFS were examined.
Among the 27 stage II-III LMPC patients, SMARCA4 mutations were significantly associated with poorer mDFS (13.9 months vs. NR, P=0.013) (Figure 4A; Supplementary Table S2). Additionally, patients harboring KEAP1 mutations tended to have poorer mDFS, although this difference was not statistically significant (18.2 months vs. NR, P=0.159) (Supplementary Table S2).
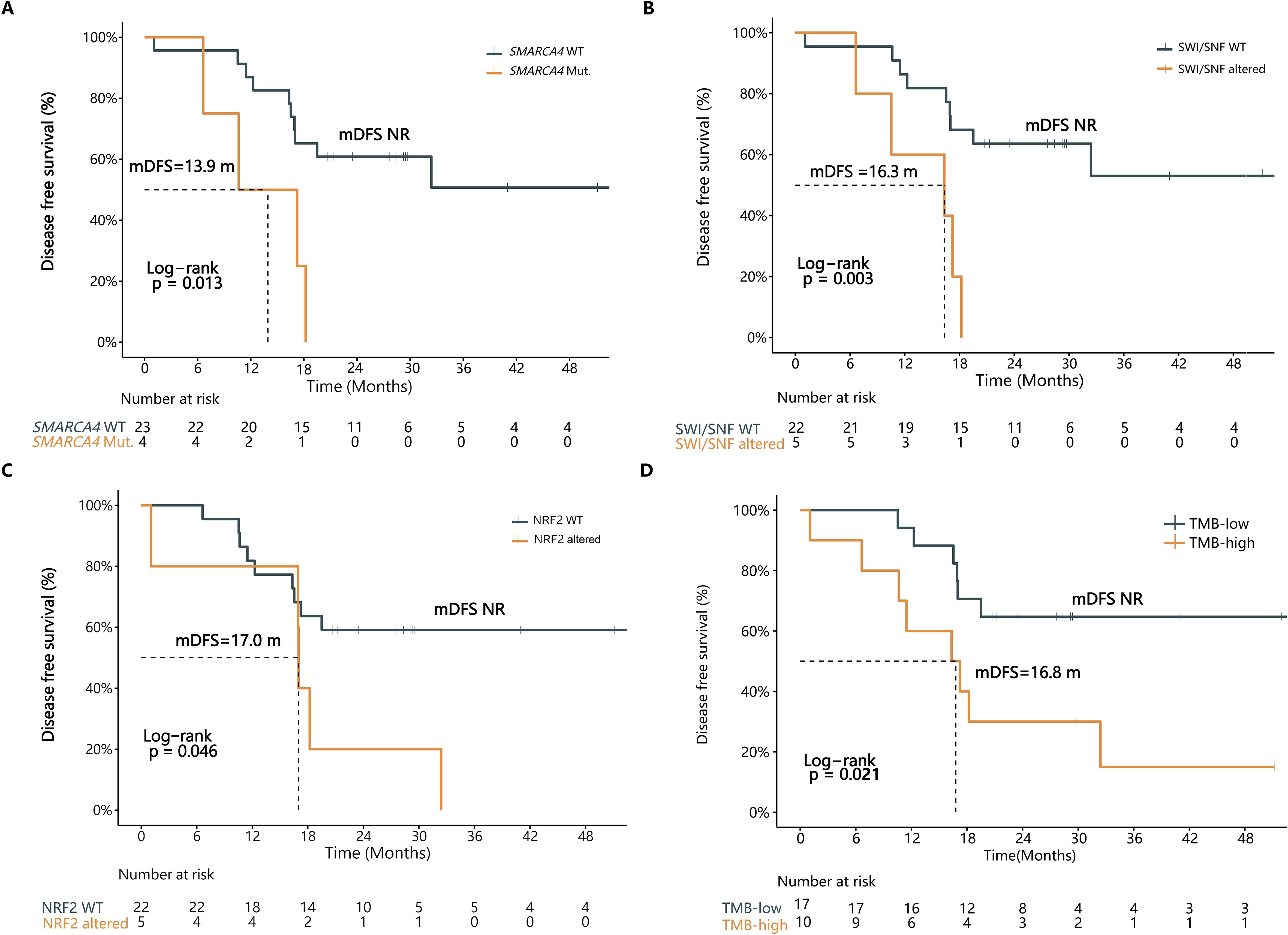
Figure 4. Multiple genetic features were associated with DFS in stage II-III LMPC patients. The Kaplan-Meier curve of DFS in stage II-III LMPC patients in strata of (A) SMARCA4 mutation, (B) SWI/SNF pathway alteration, (C) Nrf2 pathway alteration, or (D) TMB status. WT, wild type; Mut., mutation; mDFS, median disease-free survival; NR, not reached; TMB, tumor mutation burden.
Patients with aberrations in the SWI/SNF pathway exhibited significantly shorter mDFS (16.3 months vs. NR, P=0.003) (Figure 4B; Supplementary Table S2). Similarly, patients with Nrf2 pathway-related mutations had significantly reduced mDFS (17.0 months vs. NR, P=0.046) (Figure 4C; Supplementary Table S2).
Next, we analyzed the correlation between TMB and DFS in the 27 stage II-III LMPC patients. The findings suggest that mutations in specific genes and pathways could serve as prognostic indicators in more advanced LMPC cases, providing valuable insights for clinical management and therapeutic strategies.
In this study, the highest tertile of TMB was defined as TMB-high, reflecting a TMB≥6.44 mutations per megabase (mut/Mb) in the 27 stage II-III LMPC patients. The intermediate and low tertiles were defined as TMB-low. Patients with TMB-low experienced a significant DFS benefit compared to those with TMB-high (Not reached (NR) vs. 16.8 months, P=0.021, HR=3.26) (Figure 4D; Supplementary Table S2).
Therefore, several genetic features, including specific gene mutations, pathway aberrations, and TMB, can potentially serve as postoperative prognostic biomarkers in stage II-III LMPC patients. These findings highlight the importance of molecular profiling in predicting clinical outcomes and guiding treatment decisions for patients with advanced LMPC.
Multivariate cox regression analysis of DFS and molecular characteristics in stage II-III LMPC patients
We conducted a multivariate Cox regression analysis using the molecular features that were significantly associated with DFS in stage II-III LMPC patients. Surprisingly, none of the selected molecular features were statistically significant in the multivariate analysis, although the insignificance of the SWI/SNF pathway (P=0.147) might be attributed to the limited sample size (Supplementary Table S3). This implies that the SWI/SNF pathway, the Nrf2 pathway, and TMB are not independent prognostic factors in stage II-III LMPC, suggesting possible correlations among these features.
To test this hypothesis, we performed correlation analyses between various molecular features. The mutation status of multiple genes, including TP53, LRP1B, SMARCA4, KEAP1, and PKHD1, was significantly associated with increased TMB (2.7 vs. 4.3, P=0.023; 3.2 vs. 8.6, P=0.019; 3.2 vs. 16.6, P=0.016; 3.2 vs. 49.4, P<0.001; 3.2 vs. 37.1, P=0.011) (Supplementary Figures S4A, B). Additionally, patients with altered p53, Nrf2, SWI/SNF, and TGF-β pathways showed a significant or close-to-significant association with higher TMB (Supplementary Figures S4C, D). Overall, the alterations in the SWI/SNF and Nrf2 pathways were correlated with increased TMB, which may explain the insignificant results in the multivariate analysis. These findings indicate that while individual molecular features may not independently predict prognosis, their interactions and combined effects on TMB could play a crucial role in the clinical outcomes of stage II-III LMPC patients.
Lastly, we conducted a preliminary prognostic modeling analysis using the 27 stage II-III LMPC patients in our cohort as a training set. We applied the least absolute shrinkage and selection operator (LASSO) regression for feature selection from a panel of over ten clinical and molecular features, including TMB, pathway alterations, and NPA classification. This analysis identified the Nrf2 and SWI/SNF pathways as the most predictive combination of features for DFS (Figure 5A). Subsequently, we constructed a nomogram based on a Cox proportional hazards model integrating these two molecular features. The performance of the model was evaluated using the concordance index (C-index), which was 0.792 (95% CI: 0.676-0.908, P = 7.73e-07) (Figure 5B), indicating good discriminative ability within this cohort.
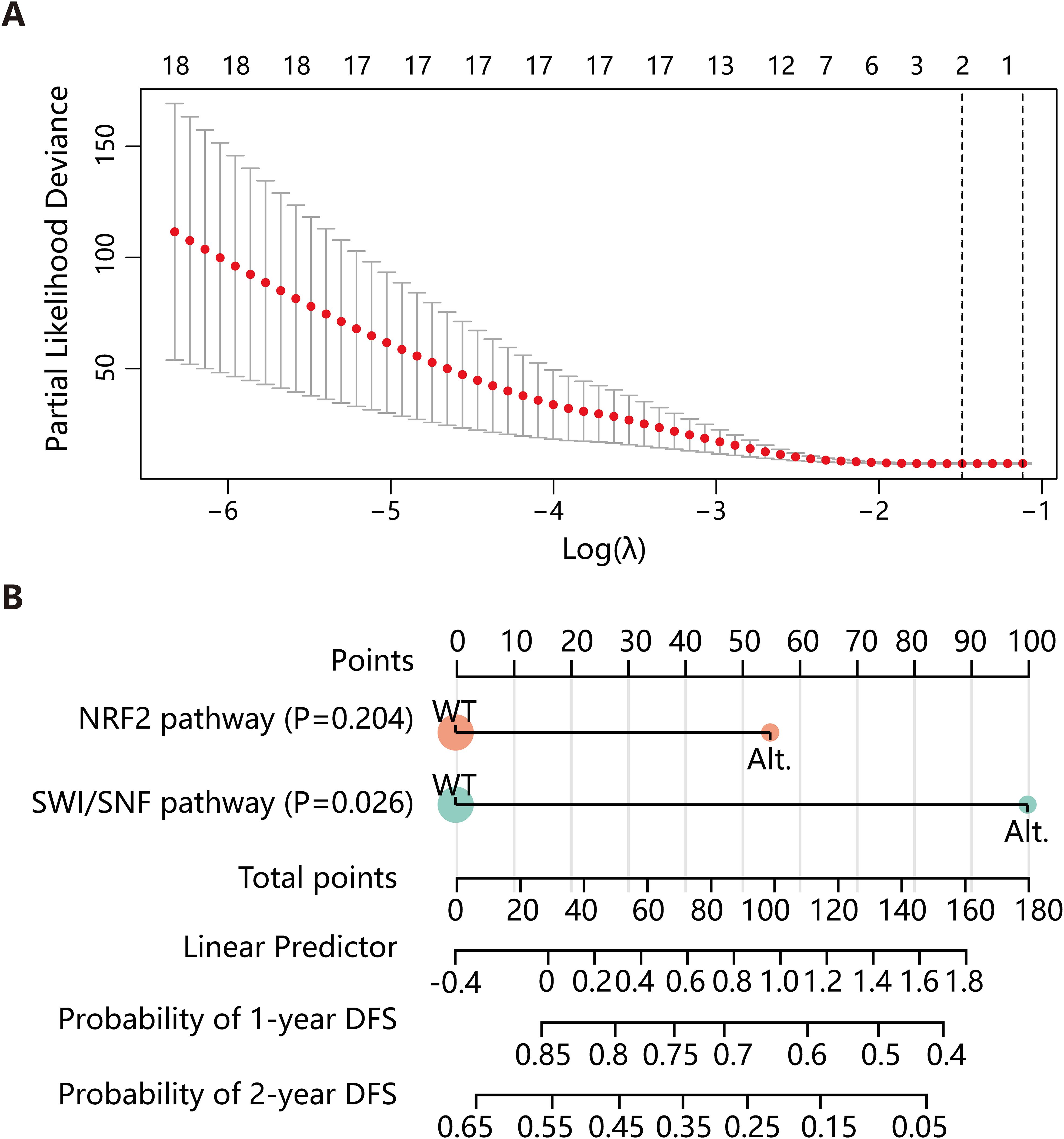
Figure 5. Construction of the DFS predictive model. (A) LASSO regression was employed to select variables for constructing the DFS prediction model. (B) A nomogram was developed based on a Cox proportional hazards model, incorporating the Nrf2 and SWI/SNF pathways. DFS, disease-free survival; LASSO, least absolute shrinkage and selection operator; Alt., alteration; WT, wild type.
Discussion
LMPC, a histologic subtype of LADC, is generally associated with poor prognosis. Despite its clinical significance, the molecular characteristics and associated prognosis of LMPC have been under-investigated, particularly in the Chinese population. To address this gap, we conducted a systematic analysis to characterize the molecular and clinical features of 54 Chinese resectable LMPC patients. Our findings revealed that LMPC tumors possess a unique genetic profile, featuring more diverse targetable mutations, an increased NPA, and more extensive oncogenic pathway alterations compared to LADC tumors. Specifically, the mutational frequencies of ERBB4, BRAF, PIK3CA, RPTOR, and NOTCH2 were significantly higher in LMPC than in LADC. LMPC patients were also more likely to harbor genetic alterations in multiple oncogenic pathways, including PI3K, Wnt, and TGF-β. Among stage II-III LMPC patients, molecular features such as SMARCA4 mutations, SWI/SNF pathway alterations, Nrf2 pathway alterations, and TMB were significantly associated with postoperative recurrence risk.
We discovered that driver gene mutations, including EGFR, KRAS, ALK, ERBB2, and BRAF, occurred in 91% of LMPC tumors. Previous studies on Asian LMPC patients have shown that mutations in EGFR, ALK, and KRAS occur in approximately 65-75%, 4-7%, and 3-6% of cases, respectively (13, 14, 22). These rates are higher than those observed in other LADC subtypes, which is generally consistent with our results but with some discrepancies. In this study, all exon regions of the driver genes in the predefined gene panel (425 cancer-related genes) were sequenced using NGS technology. Compared to previous RT-PCR hotspot sequencing technologies that focused on a few driver mutations, NGS provides a comprehensive characterization of common and rare driver gene mutations in LMPC. For example, five patients with CHMP3-ALK fusion, SND1-BRAF fusion, and multiple driver gene mutations were identified in our LMPC cohort.
To better understand the differences in signaling pathways between LADC patients with and without an MPC, we conducted a similar analysis. The LMPC group exhibited a higher NPA than the reference Chinese LADC patients, with three oncogenic pathways (PI3K, Wnt, and TGF-β) frequently altered in LMPC. Caso et al., who performed NGS analysis on 604 LADC patients, reported that NPA and TMB were associated with increasing subtype invasiveness and significantly higher frequencies of the micropapillary or solid subtypes (23). In our study, the mutation frequencies of altered oncogenic pathways in LMPC patients were significantly higher than those in LADC patients, and a similar trend was observed when comparing MPC-high with MPC-low tumors. Although there was no significant difference in TMB between the LMPC and LADC groups, the MPC-high group had significantly higher TMB levels than the MPC-low group (P=0.012), consistent with previous reports showing that TMB tended to increase with elevated MPC percentage (17).
In our study, SMARCA4 mutations and alterations in the SWI/SNF signaling pathway were significantly associated with poor outcomes in stage II-III LMPC patients. SMARCA4 mutations, reported as the most frequent mutations in the SWI/SNF complex, are associated with poor prognosis in lung cancer, although SMARCA4-mutated lung cancer may be more sensitive to immunotherapy (24). Two other studies showed that SMARCA4 mutations were related to significantly shorter overall survival and that the presence of SMARCA4 mutations might lead to poorer immunotherapy outcomes in NSCLC patients with KRAS co-mutation (25, 26). Another SWI/SNF complex gene, ARID1A, has been reported to contribute to better immunotherapy outcomes (25, 27). A similar trend was observed in our LMPC cohort, although the result was not statistically significant (P=0.055). Therefore, SMARCA4 mutation could potentially serve as a prognostic and/or predictive biomarker in NSCLC. Given that immunotherapy efficacy may not be ideal in LMPC patients with SMARCA4 and KRAS co-mutations, it is worth exploring the clinical utility of testing SMARCA4, KRAS, and ARID1A mutations in LMPC patients to predict their eligibility for immunotherapy.
We found that genetic alterations in KEAP1, a key component of the Nrf2 pathway, were correlated with poor prognosis in stage II-III LMPC patients. Mutations in the Nrf2 pathway, an important regulator of redox balance and cell homeostasis, are common in NSCLC and are associated with increased tumor growth and aggressiveness (28). Recent studies suggest that KEAP1/NRF2 alterations in NSCLC serve as biomarkers of poor prognosis and contribute to resistance to various cancer treatments, such as chemotherapy, radiotherapy, immunotherapy, and TKI therapy (29). Results from two multicenter randomized clinical trials showed that advanced NSCLC patients with KEAP1/NFE2L2 mutations had worse clinical outcomes than wild-type patients when treated with immunotherapy and chemotherapy (30). Therefore, the poor prognosis of LMPC patients might be at least partially due to the relatively high frequency of Nrf2 pathway-related aberrations, warranting further research into the efficacy of anti-NRF2 drugs in LMPC.
TMB may also help predict the postoperative prognosis of early-stage NSCLC patients, although its predictive value remains controversial. Previous studies have shown that high TMB is a biomarker of good prognosis in resectable early-stage LADC and NSCLC (31, 32), with patients in the TMB-high group having better DFS and overall survival. However, several studies hold the opposite opinion. A study on Chinese LADC patients reported that high TMB was associated with shorter DFS, and high TMB was more likely to occur in older patients with a smoking history (33). Another study involving 90 patients with early-stage lung cancer reported that high TMB was a poor prognostic factor (34). Besides, higher TMB status conferred a worse implication on OS among patients with non-squamous NSCLC who received platinum-based adjuvant chemotherapy (35). In our study, low TMB was significantly associated with improved DFS in postoperative stage II-III LMPC patients, and there was a high correlation between NPA and TMB. This finding is consistent with previous observations that TMB and NPA are primarily enriched in histologic subtypes of lung cancer with poor prognosis (23). Specifically, we found high TMB correlated significantly with mutations in TP53, KEAP1, and SMARCA4, which have been shown to promote aggressive tumor behavior, and immunosuppressive tumor microenvironments (36–39). These factors may drive early recurrence post-surgery, leading to the observed worse DFS in TMB-high patients in our cohort.
Notably, all genetic profiling in this study was performed on resected tumor tissue. Thus, surgical management was independent of mutation status. However, as liquid biopsy becomes increasingly accurate and available, integrating preoperative genomic data into surgical and perioperative planning may represent a valuable future direction for precision care in early-stage lung cancer.
There were some limitations to this study. Firstly, because the incidence rate of the LMPC subtype is only about 5%, the number of LMPC patients included in this study was relatively small. The small sample size may limit the statistical analyses, impairing the power to detect statistically significant differences in the mutation profiles between the MPC-low and MPC-high groups. Specifically, we acknowledge that the relatively small size of our LMPC cohort may limit the statistical power of multivariate analyses. The loss of statistical significance for SWI/SNF pathway alterations may reflect this limitation rather than a lack of true biological relevance. Further research using larger patient cohorts is needed to fully characterize the LMPC genetic profile. Secondly, as most LADCs are composed of a mixture of multiple histologic subtypes, microdissection of the micropapillary components was not carried out in this study, and the molecular characteristics of the analyzed genes may be affected by the presence of other histologic subtypes. Thirdly, another limitation of our study is that survival data were not available for the reference cohort of 113 LADC patients, which prevented a direct comparison of disease-free survival (DFS) between LMPC and LADC. Future studies integrating molecular and survival data across histologic subtypes will be necessary to fully understand how LMPC differs from conventional lung adenocarcinoma in both biology and clinical behavior. Fourthly, as with many genomic analyses involving multiple comparisons, there is a risk of false-positive (Type I) findings due to the absence of formal adjustment for multiple testing. While we limited our analysis to genes/pathways altered in at least 3 patients, we acknowledge that some associations, particularly those with borderline significance, may not withstand correction for false discovery rate. These findings should therefore be considered exploratory and warrant validation in larger, prospectively designed cohorts. Lastly, all patients included in this study were of East Asian (Chinese) ancestry. Given known ethnic differences in mutational patterns and lung cancer biology, caution should be exercised when generalizing these results to Western or other non-East Asian populations. Future validation in multi-ethnic, geographically diverse cohorts is warranted.
In summary, our study systematically delineated the genetic profile of LMPC and characterized multiple molecular features that differentiate LMPC from other LADC subtypes. Our findings suggest that alterations in SMARCA4, KEAP1, and components of the SWI/SNF and Nrf2 pathways may be associated with prognosis in stage II-III LMPC. However, these associations were not statistically significant in multivariate analyses and should therefore be validated in larger, multi-center studies. Our study provides a more comprehensive understanding of the molecular characteristics and underlying mechanisms of the poor prognosis of LMPC, aiding in prognostic estimation and treatment decisions in resectable LMPC.
Data availability statement
The sequencing data presented in the study are deposited in the Genome Sequence Archive for Human (GSA-Human) repository (https://ngdc.cncb.ac.cn/gsa-human/), with accession number HRA011575 (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA011575).
Ethics statement
The studies involving humans were approved by the Ethics Committees of Fujian Medical University Union Hospital (No.2019JYKY017). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
JZ: Data curation, Formal Analysis, Investigation, Resources, Writing – review & editing. XS: Data curation, Project administration, Resources, Writing – review & editing. JH: Data curation, Formal Analysis, Writing – original draft. WL: Data curation, Resources, Writing – review & editing. YZ: Formal Analysis, Writing – review & editing. JY: Writing – review & editing. YX: Formal Analysis, Investigation, Resources, Writing – review & editing. LW: Formal Analysis, Investigation, Resources, Writing – review & editing. YL: Formal Analysis, Investigation, Resources, Writing – review & editing. ZZ: Conceptualization, Funding acquisition, Investigation, Project administration, Supervision, Writing – review & editing. JL: Conceptualization, Funding acquisition, Investigation, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
We would like to thank the patients and family members who gave their consent on presenting the data in this study.
Conflict of interest
Authors YZ, JY, and YX are employees of the company Nanjing Geneseeq Technology Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1574817/full#supplementary-material
References
1. Thai AA, Solomon BJ, Sequist LV, Gainor JF, and Heist RS. Lung cancer. Lancet. (2021) 398:535–54. doi: 10.1016/S0140-6736(21)00312-3
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
3. Russell PA, Barnett SA, Walkiewicz M, Wainer Z, Conron M, Wright GM, et al. Correlation of mutation status and survival with predominant histologic subtype according to the new IASLC/ATS/ERS lung adenocarcinoma classification in stage III (N2) patients. J Thorac Oncol. (2013) 8:461–8. doi: 10.1097/JTO.0b013e3182828fb8
4. Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger K, Yatabe Y, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society: international multidisciplinary classification of lung adenocarcinoma: executive summary. Proc Am Thorac Soc. (2011) 8:381–5. doi: 10.1513/pats.201107-042ST
5. Ding H, Feng Y, Huang X, Xu J, Zhang T, Liang Y, et al. Deep learning-based classification and spatial prognosis risk score on whole-slide images of lung adenocarcinoma. Histopathology. (2023) 83:211–28. doi: 10.1111/his.14918
6. Xing X, Li L, Sun M, Yang J, Zhu X, Peng F, et al. Deep-learning-based 3D super-resolution CT radiomics model: Predict the possibility of the micropapillary/solid component of lung adenocarcinoma. Heliyon. (2024) 10:e34163. doi: 10.1016/j.heliyon.2024.e34163
7. Li Y, Zhao J, Li R, Yao X, Dong X, Zhao Y, et al. Predicting prognosis in patients with stage IA lung adenocarcinoma with a micropapillary component using a nomogram based on computed tomography radiomics and clinicopathologic factors: a retrospective analysis. Transl Lung Cancer Res. (2024) 13:2585–602. doi: 10.21037/tlcr-24-544
8. Motono N, Mizoguchi T, Ishikawa M, Iwai S, Iijima Y, and Uramoto H. Predictive value of recurrence of solid and micropapillary subtypes in lung adenocarcinoma. Oncology. (2024) 102:366–73. doi: 10.1159/000530528
9. Wang W, Hu Z, Zhao J, Huang Y, Rao S, Yang J, et al. Both the presence of a micropapillary component and the micropapillary predominant subtype predict poor prognosis after lung adenocarcinoma resection: a meta-analysis. J Cardiothorac Surg. (2020) 15:154. doi: 10.1186/s13019-020-01199-8
10. Watanabe K, Sakamaki K, Ito H, Yokose T, Yamada K, Nakayama H, et al. Impact of the micropapillary component on the timing of recurrence in patients with resected lung adenocarcinoma. Eur J Cardiothorac Surg. (2020) 58:1010–8. doi: 10.1093/ejcts/ezaa138
11. Zhao Y, Wang R, Shen X, Pan Y, Cheng C, Li Y, et al. Minor components of micropapillary and solid subtypes in lung adenocarcinoma are predictors of lymph node metastasis and poor prognosis. Ann Surg Oncol. (2016) 23:2099–105. doi: 10.1245/s10434-015-5043-9
12. Yanagawa N, Shiono S, Abiko M, Katahira M, Osakabe M, and Ogata SY. The clinical impact of solid and micropapillary patterns in resected lung adenocarcinoma. J Thorac Oncol. (2016) 11:1976–83. doi: 10.1016/j.jtho.2016.06.014
13. Pyo JS and Kim JH. Clinicopathological significance of micropapillary pattern in lung adenocarcinoma. Pathol Oncol Res. (2018) 24:547–55. doi: 10.1007/s12253-017-0274-7
14. Zhang Y, Wang R, Cai D, Li Y, Pan Y, Hu H, et al. A comprehensive investigation of molecular features and prognosis of lung adenocarcinoma with micropapillary component. J Thorac Oncol. (2014) 9:1772–8. doi: 10.1097/JTO.0000000000000341
15. Qu Y, Feng X, Chen H, Tan F, Shao A, Pang J, et al. Multi-omics analyses reveal distinct molecular characteristics and transformation mechanisms of stage I-III micropapillary lung adenocarcinoma. J Pathol. (2025) 266:204–16. doi: 10.1002/path.6416
16. Ma M, She Y, Ren Y, Dai C, Zhang L, Xie H, et al. Micropapillary or solid pattern predicts recurrence free survival benefit from adjuvant chemotherapy in patients with stage IB lung adenocarcinoma. J Thorac Dis. (2018) 10:5384–93. doi: 10.21037/jtd.2018.08.64
17. Zhang S, Xu Y, Zhao P, Bao H, Wang X, Liu R, et al. Integrated analysis of genomic and immunological features in lung adenocarcinoma with micropapillary component. Front Oncol. (2021) 11:652193. doi: 10.3389/fonc.2021.652193
18. Chansky K, Detterbeck FC, Nicholson AG, Rusch VW, Vallières E, Groome P, et al. The IASLC lung cancer staging project: external validation of the revision of the TNM stage groupings in the eighth edition of the TNM classification of lung cancer. J Thorac Oncol. (2017) 12:1109–21. doi: 10.1016/j.jtho.2017.04.011
19. Tong L, Ding N, Tong X, Li J, Zhang Y, Wang X, et al. Tumor-derived DNA from pleural effusion supernatant as a promising alternative to tumor tissue in genomic profiling of advanced lung cancer. Theranostics. (2019) 9:5532–41. doi: 10.7150/thno.34070
20. Fang W, Ma Y, Yin JC, Hong S, Zhou H, Wang A, et al. Comprehensive genomic profiling identifies novel genetic predictors of response to anti-PD-(L)1 therapies in non-small cell lung cancer. Clin Cancer Res. (2019) 25:5015–26. doi: 10.1158/1078-0432.CCR-19-0585
21. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. (2018) 173:321–37.e310. doi: 10.1016/j.cell.2018.03.035
22. Zhang J, Sun J, Zhang Z, Wang A, Liang X, Lu J, et al. Driver mutation profiles and clinicopathological correlation in pulmonary adenocarcinoma with a micropapillary component. Hum Pathol. (2019) 85:242–50. doi: 10.1016/j.humpath.2018.11.008
23. Caso R, Sanchez-Vega F, Tan KS, Mastrogiacomo B, Zhou J, Jones GD, et al. The underlying tumor genomics of predominant histologic subtypes in lung adenocarcinoma. J Thorac Oncol. (2020) 15:1844–56. doi: 10.1016/j.jtho.2020.08.005
24. Schoenfeld AJ, Bandlamudi C, Lavery JA, Montecalvo J, Namakydoust A, Rizvi H, et al. The genomic landscape of SMARCA4 alterations and associations with outcomes in patients with lung cancer. Clin Cancer Res. (2020) 26:5701–8. doi: 10.1158/1078-0432.CCR-20-1825
25. Alessi JV, Ricciuti B, Spurr LF, Gupta H, Li YY, Glass C, et al. SMARCA4 and other SWItch/sucrose nonFermentable family genomic alterations in NSCLC: clinicopathologic characteristics and outcomes to immune checkpoint inhibition. J Thorac Oncol. (2021) 16:1176–87. doi: 10.1016/j.jtho.2021.03.024
26. Liu L, Ahmed T, Petty WJ, Grant S, Ruiz J, Lycan TW, et al. SMARCA4 mutations in KRAS-mutant lung adenocarcinoma: a multi-cohort analysis. Mol Oncol. (2021) 15:462–72. doi: 10.1002/1878-0261.12831
27. Zhu G, Shi R, Li Y, Zhang Z, Xu S, Chen C, et al. ARID1A, ARID1B, and ARID2 mutations serve as potential biomarkers for immune checkpoint blockade in patients with non-small cell lung cancer. Front Immunol. (2021) 12:670040. doi: 10.3389/fimmu.2021.670040
28. Hellyer JA, Padda SK, Diehn M, and Wakelee HA. Clinical implications of KEAP1-NFE2L2 mutations in NSCLC. J Thorac Oncol. (2021) 16:395–403. doi: 10.1016/j.jtho.2020.11.015
29. Dempke WCM and Reck M. KEAP1/NRF2 (NFE2L2) mutations in NSCLC - Fuel for a superresistant phenotype? Lung Cancer. (2021) 159:10–7. doi: 10.1016/j.lungcan.2021.07.006
30. Zhu H, Xie D, Yu Y, Yao L, Xu B, Huang L, et al. KEAP1/NFE2L2 mutations of liquid biopsy as prognostic biomarkers in patients with advanced non-small cell lung cancer: results from two multicenter, randomized clinical trials. Front Oncol. (2021) 11:659200. doi: 10.3389/fonc.2021.659200
31. Talvitie EM, Vilhonen H, Kurki S, Karlsson A, Orte K, Almangush A, et al. High tumor mutation burden predicts favorable outcome among patients with aggressive histological subtypes of lung adenocarcinoma: A population-based single-institution study. Neoplasia. (2020) 22:333–42. doi: 10.1016/j.neo.2020.05.004
32. Devarakonda S, Rotolo F, Tsao MS, Lanc I, Brambilla E, Masood A, et al. Tumor mutation burden as a biomarker in resected non-small-cell lung cancer. J Clin Oncol. (2018) 36:2995–3006. doi: 10.1200/JCO.2018.78.1963
33. Wang C, Liang H, Lin C, Li F, Xie G, Qiao S, et al. Molecular subtyping and prognostic assessment based on tumor mutation burden in patients with lung adenocarcinomas. Int J Mol Sci. (2019) 20:4251. doi: 10.3390/ijms20174251
34. Owada-Ozaki Y, Muto S, Takagi H, Inoue T, Watanabe Y, Fukuhara M, et al. Prognostic impact of tumor mutation burden in patients with completely resected non-small cell lung cancer: brief report. J Thorac Oncol. (2018) 13:1217–21. doi: 10.1016/j.jtho.2018.04.003
35. Shen WX, Li GH, Li YJ, Zhang PF, Yu JX, Shang D, et al. Prognostic significance of tumor mutation burden among patients with non-small cell lung cancer who received platinum-based adjuvant chemotherapy: an exploratory study. J Cancer Prev. (2023) 28:175–84. doi: 10.15430/JCP.2023.28.4.175
36. Barrera-Rodríguez R. Importance of the Keap1-Nrf2 pathway in NSCLC: Is it a possible biomarker? BioMed Rep. (2018) 9:375–82. doi: 10.3892/br.2018.1143
37. Mogi A and Kuwano H. TP53 mutations in nonsmall cell lung cancer. J BioMed Biotechnol. (2011) 2011:583929. doi: 10.1155/2011/583929
38. Manolakos P, Boccuto L, and Ivankovic DS. A critical review of the impact of SMARCA4 mutations on survival outcomes in non-small cell lung cancer. J Pers Med. (2024) 14:684. doi: 10.3390/jpm14070684
Keywords: lung adenocarcinoma, micropapillary component, disease-free survival, genomic features, prognostic biomarkers
Citation: Zhu J, Sun X, Huang J, Lin W, Zhan Y, Yan J, Xu Y, Wu L, Lian Y, Zhang Z and Lin J (2025) Genomic profiles and prognostic biomarkers of resectable lung adenocarcinoma with a micropapillary component. Front. Oncol. 15:1574817. doi: 10.3389/fonc.2025.1574817
Received: 28 February 2025; Accepted: 08 May 2025;
Published: 29 May 2025.
Edited by:
Yiming Meng, China Medical University, ChinaReviewed by:
Yu Peng, Zhejiang University, ChinaHanlin Ding, Jiangsu Cancer Hospital, China
Devanish Kamtam, Stanford University, United States
Zenghua Deng, Capital Medical University, China
Copyright © 2025 Zhu, Sun, Huang, Lin, Zhan, Yan, Xu, Wu, Lian, Zhang and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiangbo Lin, WmhhbmcxNTk4MDExNzk4NUAxNjMuY29t; Zhenyang Zhang, enp5Y3VycnlAc2luYS5jb20=
†These authors have contributed equally to this work and share first authorship
 Jiafu Zhu1†
Jiafu Zhu1† Xinhai Sun
Xinhai Sun Jiangshan Huang
Jiangshan Huang Junrong Yan
Junrong Yan Yang Xu
Yang Xu Jiangbo Lin
Jiangbo Lin