- 1Respiratory and Critical Care Medicine Center, Weifang People’s Hospital, Weifang, China
- 2The First Affiliated Hospital, Shandong Second Medical University, Weifang, China
- 3Precision Pathology Diagnosis Center, Weifang People’s Hospital, Weifang, China
- 4Critical care medicine, Weifang People’s Hospital, Weifang, China
- 5College of Mechanical Engineering and Automation, Weifang University, Weifang, China
Background: With the rapid advances in artificial intelligence—particularly convolutional neural networks—researchers now exploit CT, PET/CT and other imaging modalities to predict epidermal growth factor receptor (EGFR) mutation status in non-small-cell lung cancer (NSCLC) non-invasively, rapidly and repeatably. End-to-end deep-learning models simultaneously perform feature extraction and classification, capturing not only traditional radiomic signatures such as tumour density and texture but also peri-tumoural micro-environmental cues, thereby offering a higher theoretical performance ceiling than hand-crafted radiomics coupled with classical machine learning. Nevertheless, the need for large, well-annotated datasets, the domain shifts introduced by heterogeneous scanning protocols and preprocessing pipelines, and the “black-box” nature of neural networks all hinder clinical adoption. To address fragmented evidence and scarce external validation, we conducted a systematic review to appraise the true performance of deep-learning and radiomics models for EGFR prediction and to identify barriers to clinical translation, thereby establishing a baseline for forthcoming multicentre prospective studies.
Methods: Following PRISMA 2020, we searched PubMed, Web of Science and IEEE Xplore for studies published between 2018 and 2024. Fifty-nine original articles met the inclusion criteria. QUADAS-2 was applied to the eight studies that developed models using real-world clinical data, and details of external validation strategies and performance metrics were extracted systematically.
Results: The pooled internal area under the curve (AUC) was 0.78 for radiomics–machine-learning models and 0.84 for deep-learning models. Only 17 studies (29%) reported independent external validation, where the mean AUC fell to 0.77, indicating a marked domain-shift effect. QUADAS-2 showed that 31% of studies had high risk of bias in at least one domain, most frequently in Index Test and Patient Selection.
Conclusion: Although deep-learning models achieved the best internal performance, their reliance on single-centre data, the paucity of external validation and limited code availability preclude their use as stand-alone clinical decision tools. Future work should involve multicentre prospective designs, federated learning, decision-curve analysis and open sharing of models and data to verify generalisability and facilitate clinical integration.
1 Introduction
Non-small cell lung cancer (NSCLC), the most prevalent subtype of lung cancer, accounts for approximately 85% of all lung cancer cases (1). Within NSCLC, adenocarcinoma and squamous cell carcinoma represent the two most common histopathological subtypes. With the evolution of personalized and precision medicine, the detection of specific genetic mutations in NSCLC has become pivotal for stratifying patients based on therapeutic responsiveness. Among these, epidermal growth factor receptor (EGFR) mutation profiling is particularly critical, as EGFR a cell-surface receptor driving cellular proliferation and survival exhibits mutations that enhance sensitivity to tyrosine kinase inhibitors (TKIs). Clinically, EGFR-mutant patients are frequently characterized by non-smoking status, adenocarcinoma histology, female sex, and East Asian ethnicity (2–4).
In clinical practice, histopathological biopsy remains the gold standard for procuring tissue specimens and conducting mutational analysis to guide treatment planning. However, obtaining sufficient biopsy material for molecular profiling is not always feasible, particularly in high-risk patients with coagulopathies or comorbidities contraindicating invasive procedures. Furthermore, biopsy-derived tumor cells may inadequately capture intratumoral heterogeneity, reflecting only a limited spatial sampling of the tumor’s genomic landscape (5, 6). For instance, Taniguchi et al. (7) demonstrated that among 50–60 tumor regions analyzed in 21 EGFR-mutant patients, 28.6% exhibited intratumoral heterogeneity harboring both EGFR-mutated and wild-type subclones. Given that all patients with pulmonary masses undergo pre-treatment computed tomography (CT), these images serve as a rich data source for supplementary genomic interrogation, potentially identifying EGFR mutation carriers. Discordant findings between biopsy and CT-based analyses may warrant tumor re-sampling, thereby reducing the likelihood of missing actionable EGFR mutations. Consequently, quantitative characterization of CT-derived features has emerged as a critical adjunct for refining EGFR mutation status assessment.
Medical imaging has emerged as a pivotal platform for discovering and applying biomarkers in lung cancer. Recent investigations have employed artificial‐intelligence algorithms to quantify the biological, phenotypic and functional information embedded in imaging data, with radiomics and deep-learning approaches representing the two most prominent paradigms (8, 9). Radiomics relies on manual lesion delineation followed by extraction of high-dimensional texture and morphological features, enabling rapid screening and preliminary subtyping, yet it remains constrained in characterising tumour margins and the peritumoral micro-environment. By contrast, deep learning employs end-to-end networks that automatically learn image features without precise segmentation, capturing latent cues strongly associated with key outcomes—such as EGFR mutation status—while reducing labour costs. Moreover, deep networks can identify intratumoural subregions linked to genetic heterogeneity, thereby providing targets for image-guided biopsy. Each method offers distinct advantages, and both have demonstrated clinical potential in supporting diagnosis, response assessment and therapeutic decision-making, thus furnishing new imaging-based pillars for precision oncology in lung cancer (10–13).
This review seeks to advance understanding of AI applications in oncology by categorising current algorithmic strategies and summarising recent advances in predicting EGFR mutation status and subtypes in lung cancer.
2 Deep learning-based artificial intelligence technologies
2.1 Applications of deep learning in lung cancer diagnosis
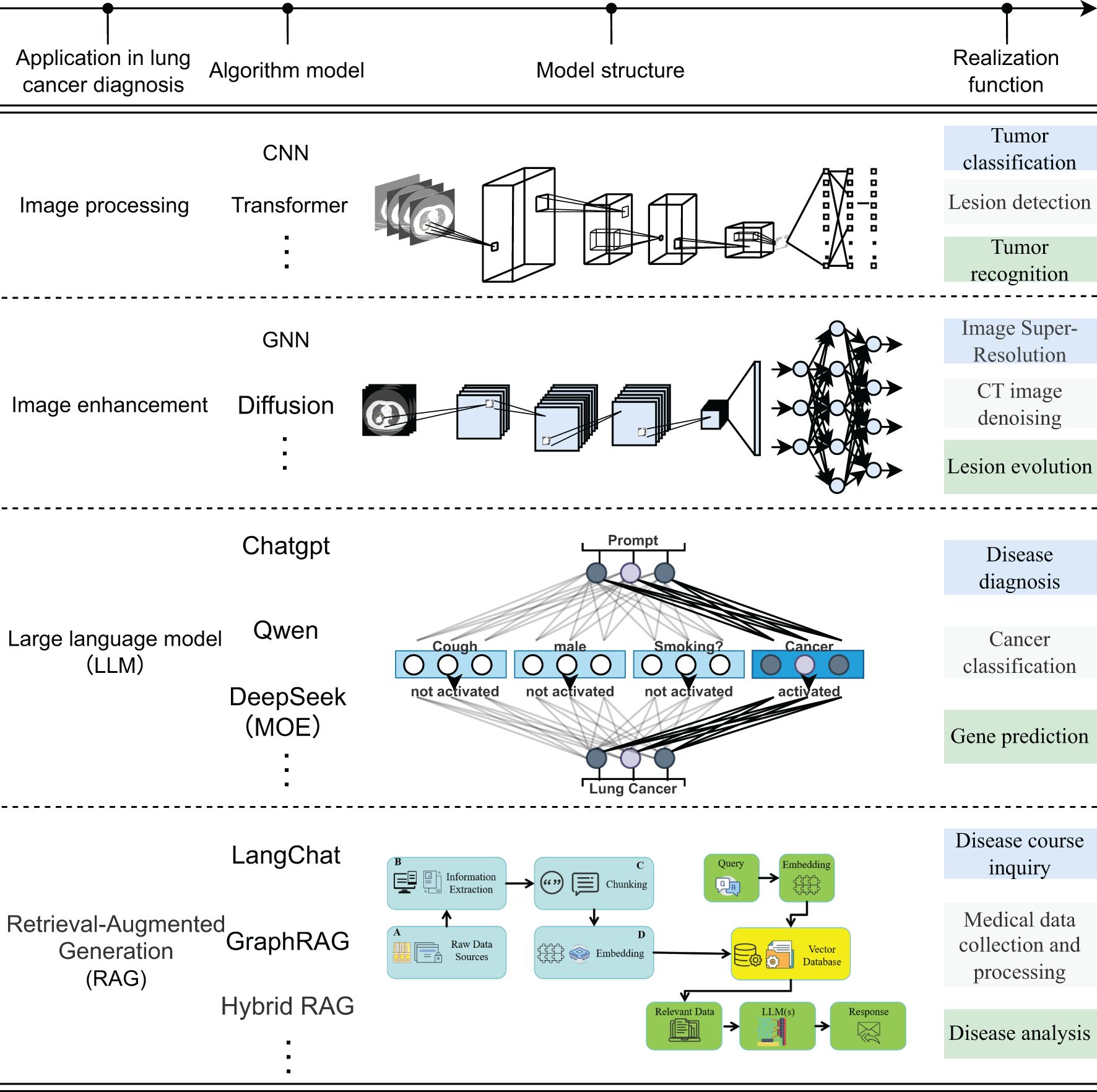
The current application of deep learning technology in the field of artificial intelligence for lung cancer images can be summarized into four aspects: ① Image processing: Convolutional neural networks focus on capturing local texture and morphology, while the Transformer relies on the self-attention mechanism to achieve cross-scale global feature integration; ② Image enhancement: Graph neural networks improve the contrast and consistency of CT/MR Images by modeling the topological relationship between pixels, and diffusion models significantly improve the resolution in low-dose or low-signal-to-noise scenes through iterative denoising. ③ Large language model: With the ability of deep semantic reasoning and context modeling, it integrates radiological images, pathological images and clinical texts to generate interpretable comprehensive diagnosis and genetic variation prediction reports; ④ Retrieval enhanced generation model: By integrating information retrieval and generation mechanisms, it enables the rapid aggregation of multimodal case knowledge and longitudinal disease course analysis, as shown in Figure 1. The four types of methods jointly demonstrate the potential of deep learning in automatic feature learning, multimodal data fusion, and intelligent decision support, and also highlight the ongoing challenges in terms of computing resources, data quality, and model interpretability (14–28).
2.2 Generalized framework for AI-driven prediction of genetic mutations in non-small cell lung cancer
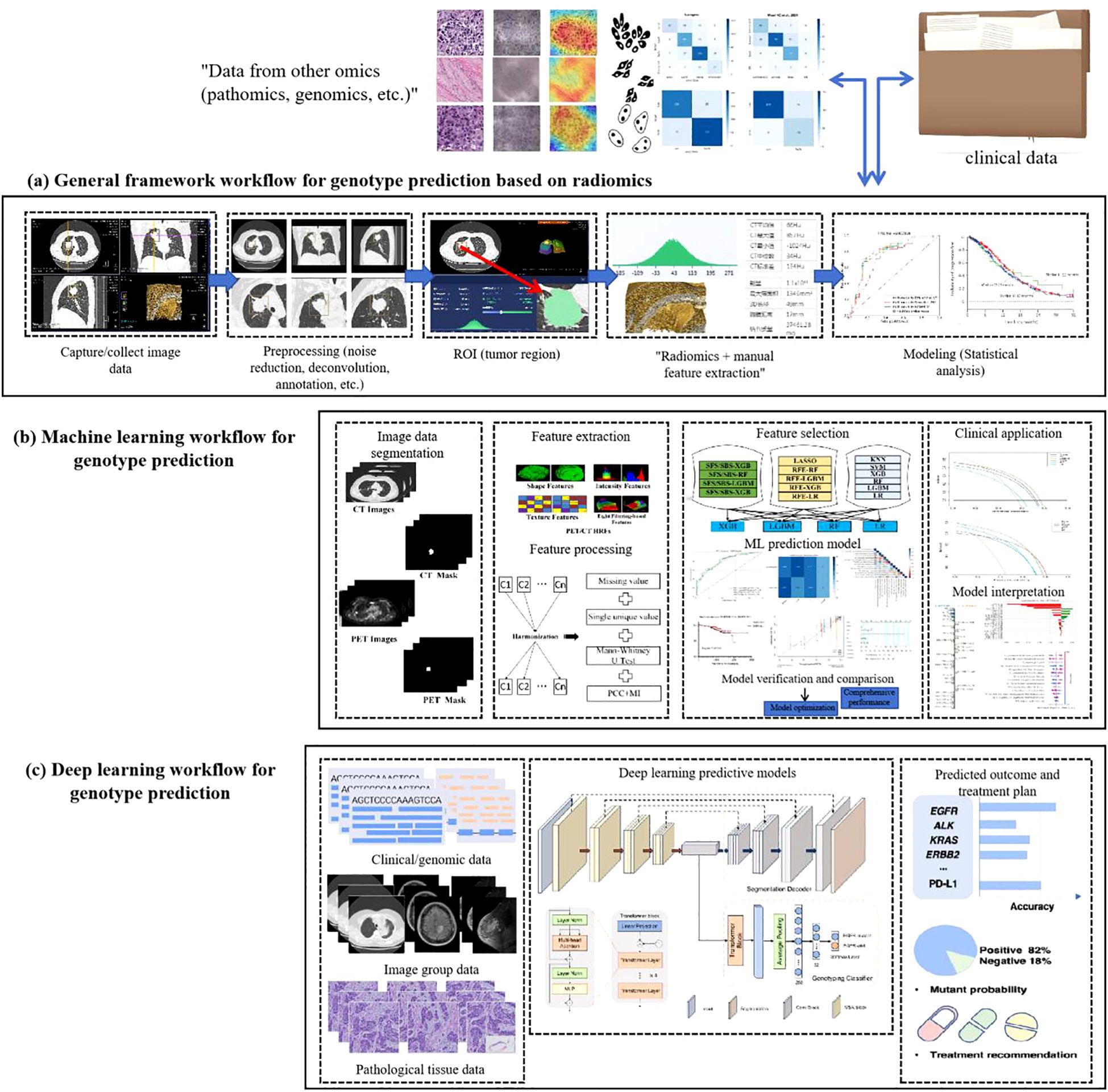
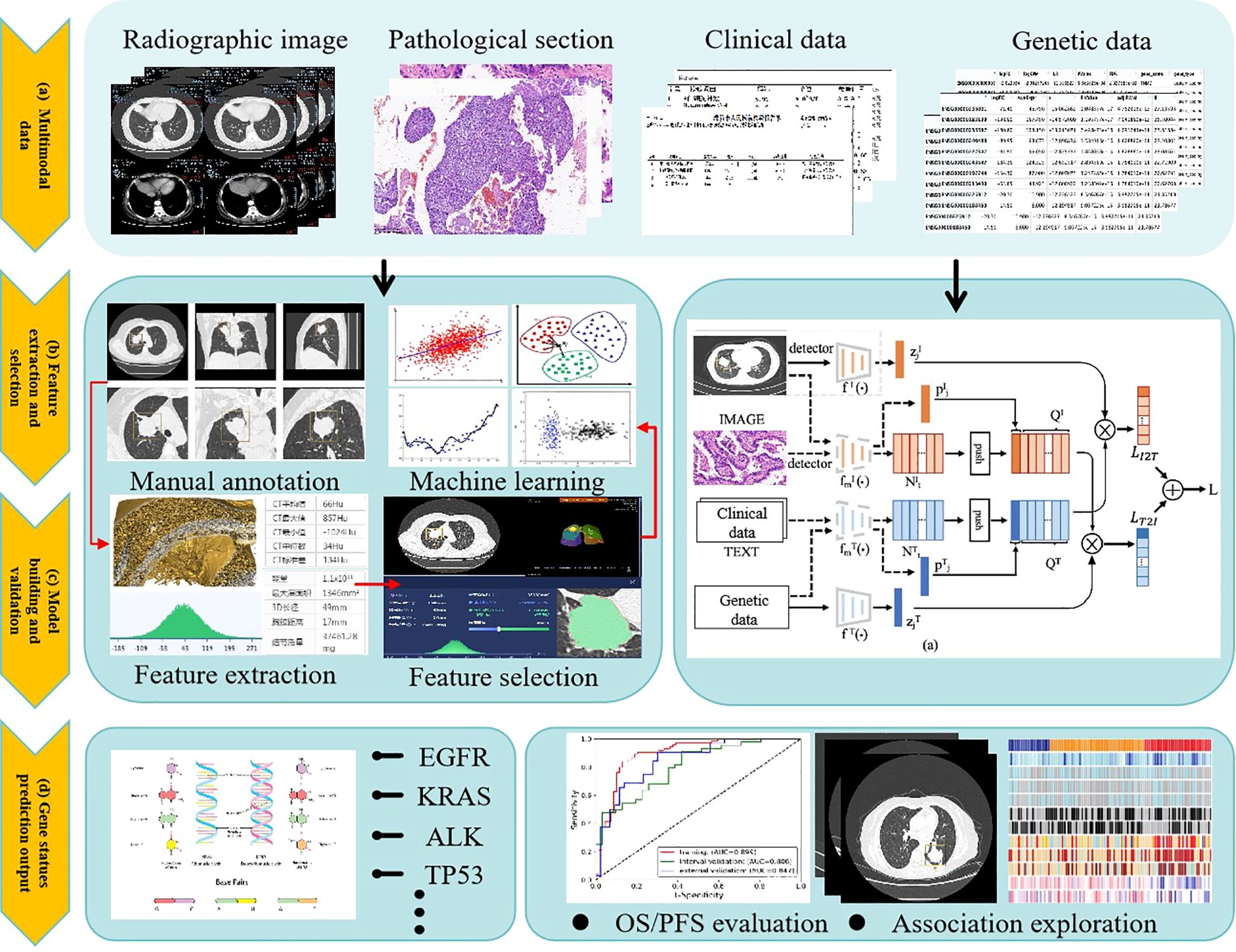
The evolution of artificial intelligence (AI) has necessitated the integration of multimodal data to improve the accuracy and reliability of predicting genetic mutations in non-small cell lung cancer (NSCLC). A standardized analytical workflow typically comprises four critical phases: (a) data acquisition and preprocessing, (b) lesion segmentation and radiomic feature extraction, (c) predictive model development and training optimization, and (d) prediction validation and clinical interpretation (29, 30), as illustrated in Figure 2.
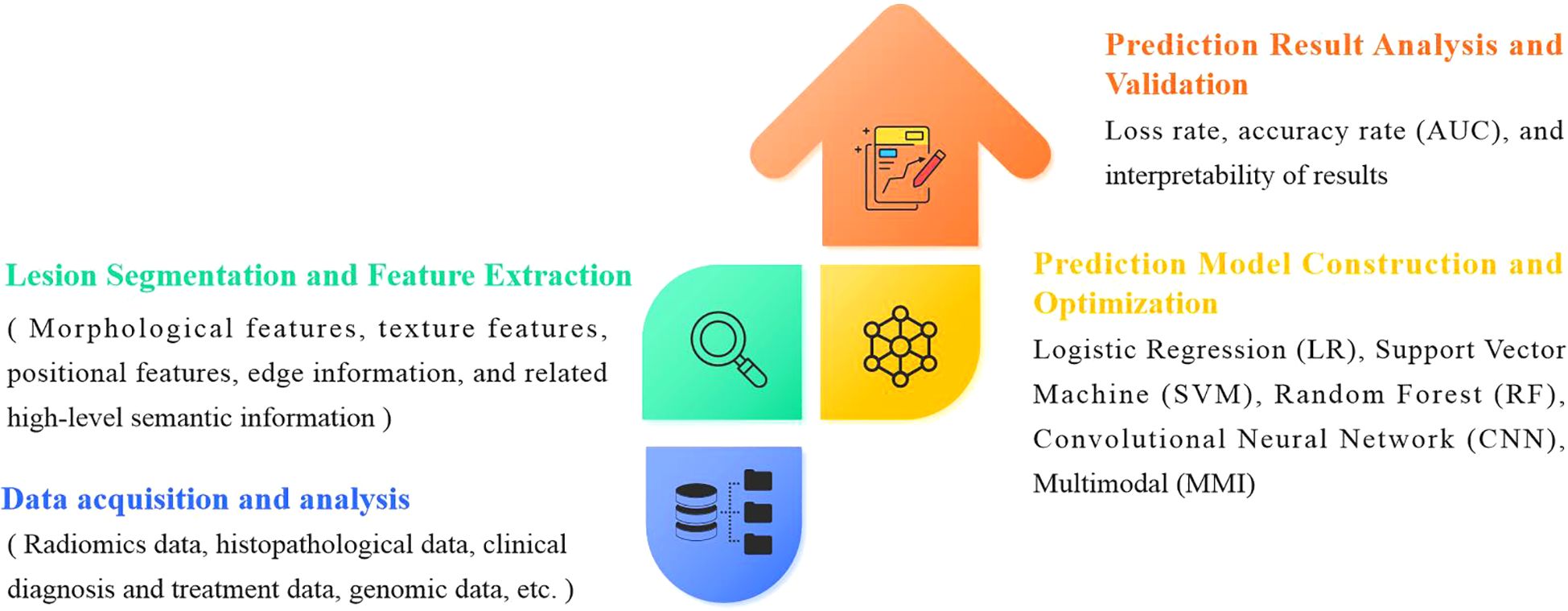
Figure 2. General framework for AI techniques to predict gene mutations in non-small cell lung cancer.
During the data collection phase, case selection is restricted to pathologically confirmed lung adenocarcinoma patients with EGFR mutations (exon 19 deletions or L858R substitutions) verified by molecular testing. Corresponding radiological (CT) and histopathological imaging data are acquired concurrently. Exclusion criteria include: (1) prior antitumor therapy before CT imaging or EGFR genotyping, and (2) temporal misalignment between imaging, biopsy, and molecular testing procedures.
Data preprocessing forms the foundation for ensuring analytical validity in predictive modeling. In the early 2000s, AI applications in medical imaging predominantly employed supervised learning models relying on manually engineered features. These required domain experts to annotate imaging characteristics (e.g., tumor texture, margins) and construct fully labeled datasets for model training. Circa 2012, deep learning paradigms particularly convolutional neural networks (CNNs) revolutionized the field by autonomously learning hierarchical features directly from raw imaging data through end-to-end training on large annotated datasets, significantly enhancing model performance and generalizability. Post-2020, self-supervised learning emerged as a transformative approach, enabling feature extraction from unlabeled data without external annotations (31), as shown in Figure 3.
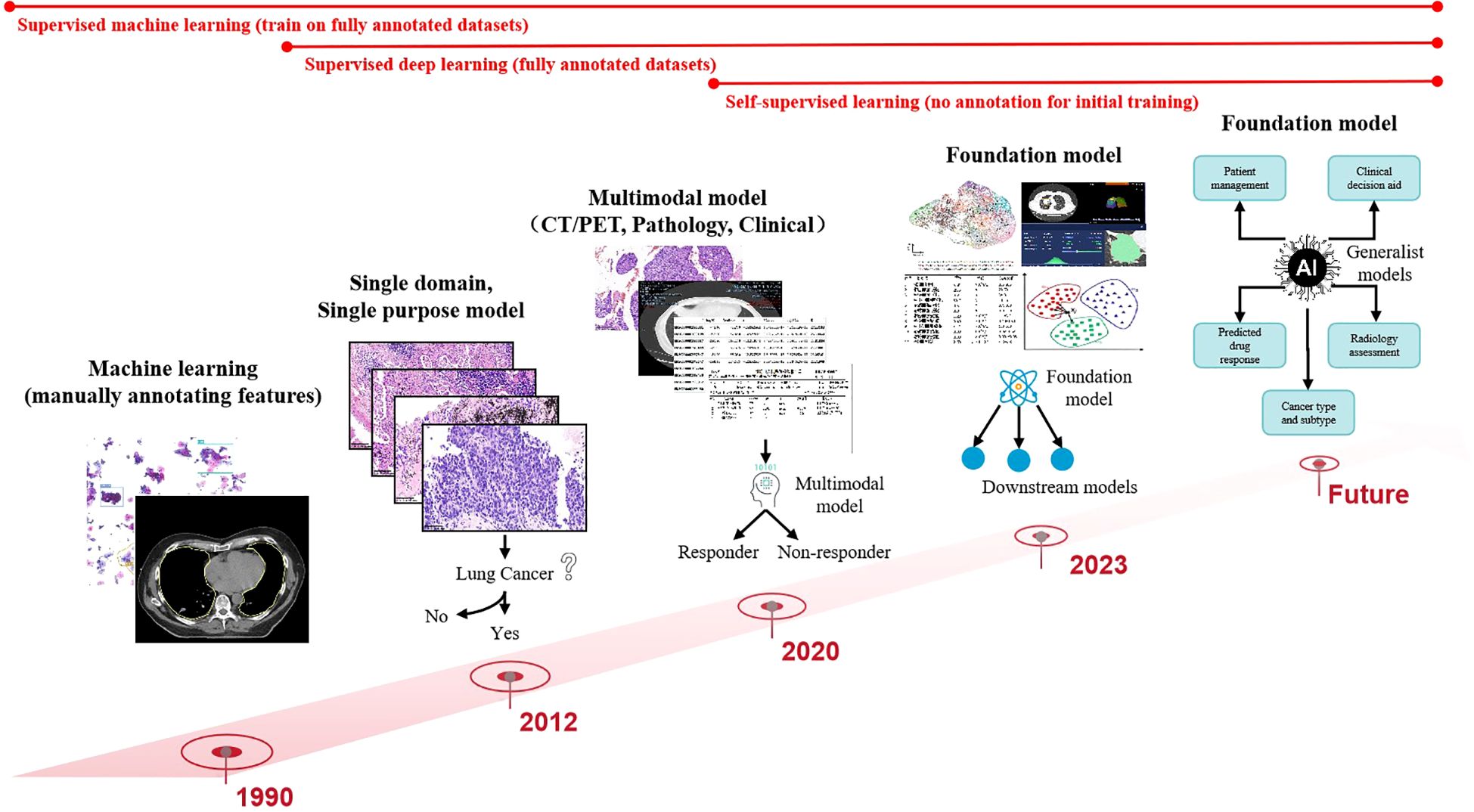
Figure 3. Computer vision has evolved from simple, specialized, shallow models to deep, multimodal, general-purpose models.
While manual annotation ensures high interpretability and accuracy, its labor-intensive nature limits scalability, making it suitable only for small, homogeneous datasets. In contrast, contemporary multimodal predictive models integrate radiomics (CT/PET), histopathology (whole-slide imaging), clinical metadata (e.g., ECOG status), and genomic profiles (e.g., EGFR variant allele frequency), achieving superior predictive accuracy for NSCLC mutations. Future advancements aim to develop large-scale, self-supervised foundation models pretrained on cross-modal unlabeled datasets. These models could be efficiently fine-tuned for diverse downstream tasks (e.g., mutation prediction, treatment response) with minimal task-specific training data. The ultimate objective is to create multifunctional AI systems capable of analytical reasoning, clinical interpretation, predictive modeling, and interactive decision support for patients and clinicians (32).
(1) Manual Tumor Feature Extraction
In the 1990s, early applications of machine learning for pulmonary tumor characterization predominantly relied on manual annotation by experienced radiologists or pathologists to delineate tumor regions. Radiomics-driven approaches enabled precise extraction of diverse tumor features, including morphological characteristics (e.g., size, shape, margin spiculation), textural patterns (e.g., gray-level distribution, heterogeneity), and locational attributes (e.g., lobar positioning). Post-extraction, feature selection techniques such as correlation coefficient analysis, least absolute shrinkage and selection operator (LASSO) regression, and principal component analysis (PCA) were applied to identify features most predictive of clinical endpoints. Selected feature sets were then input into machine learning models including logistic regression (LR), support vector machines (SVMs), and random forests (RFs) for malignancy grading, therapeutic response prediction, or prognostic stratification. Manual annotation leverages clinical expertise to capture nuanced visual patterns, particularly in cases with ill-defined tumor boundaries or irregular morphology, often outperforming automated methods in accuracy. Furthermore, manually derived features offer high interpretability for clinical decision-making. However, this approach suffers from inherent limitations: inter-observer variability, time-intensive workflows, and poor scalability to large datasets, rendering it impractical for modern precision medicine demands (33–35).
(2) Deep Learning-Based Feature Extraction
Deep learning automates tumor feature extraction through artificial neural networks (e.g., convolutional neural networks, CNNs) trained to hierarchically learn imaging signatures from radiomic data. These models capture both low-level textural/edge features and high-level semantic patterns (e.g., tumor shape, intralesional heterogeneity, peritumoral tissue interactions). Extracted features, represented as high-dimensional vectors, facilitate tasks such as tumor classification (benign vs. malignant), staging, treatment efficacy evaluation, and survival analysis. Dimensionality reduction techniques (e.g., PCA, t-distributed stochastic neighbor embedding [t-SNE]) may visualize these features before downstream model integration (36).
Deep learning offers advantages in processing large-scale data with minimal human intervention, ensuring feature consistency and efficiency. It excels at identifying subvisual patterns imperceptible to human observers, enhancing predictive performance. Nevertheless, critical challenges persist: the “black-box” nature of deep neural networks limits feature interpretability and clinical translatability; high-quality annotated datasets costly to acquire in medical domains are required for robust training; and overfitting risks escalate with limited data. Thus, while deep learning demonstrates transformative potential, its clinical adoption necessitates rigorous validation against domain knowledge and multimodal integration (37, 38).
3 Data sources and literature search strategy
This review was prepared following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA 2020) guidelines and includes a systematic literature search with quality assessment, as well as an integrated evaluation of study quality and risk of bias. We provide a comprehensive analysis of recent artificial intelligence–based approaches for predicting epidermal growth factor receptor (EGFR) mutation status in non-small cell lung cancer (NSCLC), detailing the research methods, data modalities, and model performance (39, 40).
3.1 Search strategy
A systematic literature search was conducted up to December 2024 in PubMed (Medline), Web of Science, and IEEE Xplore. The search strategy combined controlled vocabulary (e.g., MeSH) and free‐text terms, including “EGFR,” “epidermal growth factor receptor,” “mutation,” “deep learning,” “artificial intelligence,” “radiomics,” “machine learning,” and “non‐small cell lung cancer,” with Boolean operators. To ensure comprehensiveness, we also performed supplementary screening by examining the reference lists of all relevant articles.
3.2 Inclusion and exclusion criteria
Study selection was performed according to the following inclusion and exclusion criteria: as shown in Table 1.
3.3 Study selection process
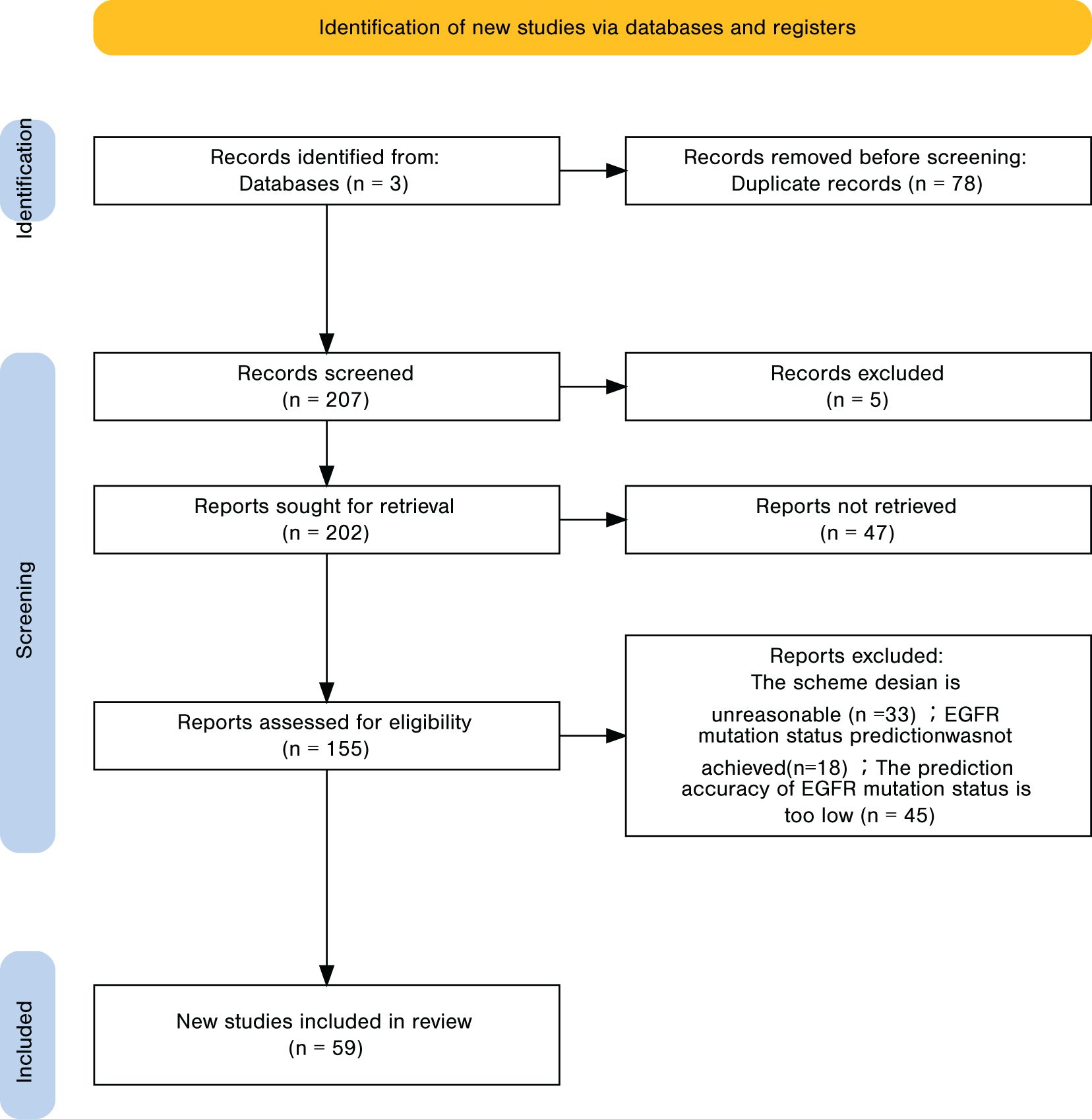
A total of 285 records were identified, of which 78 duplicates were removed, leaving 207 unique records. Title and abstract screening excluded 55 irrelevant studies, resulting in 155 articles for full-text review. Applying the predefined inclusion and exclusion criteria led to the exclusion of 96 articles, and 59 studies were ultimately included in the analysis. The detailed screening workflow is presented in Figure 4. For every included study, we recorded the point estimate of the AUC; when a 95% confidence interval (CI) was provided, it was extracted alongside the point estimate. If the CI was absent, the entry was labelled “NR (Not Reported)”, as noted in the footnote of the results table.
3.4 Risk of bias and applicability assessment
To systematically evaluate the methodological quality, risk of bias, and clinical applicability of the included studies, we applied the QUADAS-2 tool (Quality Assessment of Diagnostic Accuracy Studies 2). Eight original studies that developed AI models for EGFR mutation prediction using real-world clinical data were selected for this analysis.
QUADAS-2 evaluates:
1. Risk of Bias, across four domains:
● Patient Selection
● Index Test (i.e., the AI model)
● Reference Standard (EGFR mutation detection method)
● Flow and Timing (appropriateness of the sequence and interval between data collection and model evaluation)
2. Applicability Concerns, addressing the first three domains (Patient Selection, Index Test, Reference Standard) in terms of clinical relevance and generalizability.
Two reviewers performed all assessments independently, and any discrepancies were resolved through discussion. The summary of these evaluations is presented in Figure 5.
4 Radiomics-based prediction of genetic mutation status in non-small cell lung cancer
Radiomics is a systematic methodology encompassing the entire workflow from image acquisition to predictive performance evaluation. This approach involves critical steps: (1) image acquisition and reconstruction, (2) tumor segmentation, (3) feature extraction and filtering, (4) predictive model development, and (5) validation and performance assessment. During model construction, researchers commonly employ diverse classifiers for data analysis. Radiomics has evolved through three distinct phases based on classifier technologies: Traditional Statistical Radiomics (TSR), Machine Learning-based Radiomics (MLR) (particularly shallow learning algorithms), and Deep Learning (DL). These subcategories reflect iterative advancements in radiomics, each with unique characteristics and applicable clinical scenarios (Figure 6). TSR relies on statistical hypothesis testing (e.g., t-tests, ANOVA) to identify imaging biomarkers, while MLR leverages classical algorithms (e.g., SVM, RF) to map radiomic features to mutation probabilities. DL further automates feature engineering through hierarchical representation learning. Methodological divergences in data processing and feature analysis underscore both technological progress and context-specific problem-solving strategies (41, 42).
Traditional Statistical Radiomics (TSR) (43, 44) employs radiomic feature extraction (e.g., shape-, histogram-, and texture-based features) combined with statistical methodologies such as the least absolute shrinkage and selection operator (LASSO) to identify key features with non-zero coefficients. These features are then weighted to compute a radiomics score (Rad-score) for each lesion, representing a linear combination of selected features. In TSR, classical logistic regression (LR) serves as the primary classifier for model construction. Renowned for its simplicity and interpretability, TSR remains a foundational and clinically transparent approach in radiomics, enabling quantitative lesion characterization to inform diagnostic and therapeutic decision-making.
Machine Learning-based Radiomics (MLR) (41, 45) represents a mature, mainstream methodology for building classification/prediction models following feature extraction and optimization. Commonly used classifiers include random forests (RF), support vector machines (SVM), decision trees (DT), Bayesian networks (BN), and k-nearest neighbors (KNN). Subsequent to MLR, deep learning radiomics (DLR) (42) has emerged, leveraging artificial neural networks (ANNs) such as convolutional neural networks (CNNs) to extract deep features and construct predictive models. Unlike TSR and MLR which rely on manual feature engineering DL-based approaches utilize multi-layered nonlinear neural networks to automate feature learning directly from images via end-to-end workflows, eliminating human intervention.
4.1 Statistical prediction methods
Prior to the advent of radiomics, the prediction of EGFR mutation status in lung adenocarcinoma predominantly relied on clinical characteristics. Multiple studies have demonstrated significant associations between EGFR mutations and female sex, non-smoking status, and specific adenocarcinoma histologic subtypes. Additionally, CT imaging features including tumor maximal diameter, location, density, ground-glass opacity, pleural retraction, and air bronchogram have been validated as predictive biomarkers for EGFR mutations. Recent investigations further suggest that reduced tumor long-axis diameter correlates with increased EGFR mutation risk, while ground-glass opacity patterns are strongly linked to EGFR-mutant tumors. Yip et al. (43) highlighted the potential of radiomic features to quantify metabolic phenotypes for EGFR mutation prediction.
However, radiomic features primarily reflect imaging data and may insufficiently capture comprehensive disease profiles. To address this, researchers increasingly integrate clinical variables (including CT features) into radiomic models to enhance accuracy in identifying EGFR mutation status and subtypes. Recent studies indicate that models combining clinical and PET-derived features exhibit superior diagnostic performance and goodness-of-fit. For example, Li et al. (46) extracted 2,632 radiomic features from PET/CT images of 179 lung adenocarcinomas, randomly splitting the cohort into training (n=125) and testing (n=54) sets. Their models achieved AUCs of 0.708 and 0.652 for predicting exon 19 deletions and L858R mutations, respectively. Zhang et al. (45) retrospectively analyzed 18F-FDG PET/CT data from 173 NSCLC patients (71 EGFR+, 102 EGFR−), with 39% (68/173) at stages I/II and 61% (105/173) at stages III/IV. A combined PET/CT radiomics-clinical model achieved modest predictive performance (AUC=0.661). Nair et al. (47) demonstrated that PET/CT features outperformed CT alone in discriminating exon 19 and 21 mutations (AUC=0.86) using 326 features from 50 NSCLC patients, though limited sample size and absence of independent validation constrained generalizability.
Despite the rapid evolution of radiomics, traditional statistical regression (TSR) remains widely utilized due to its interpretability and clinical accessibility. TSR effectively transforms complex data into interpretable scores, often visualized via nomograms. However, TSR is limited by lower predictive efficiency compared to machine learning classifiers, driving the adoption of multivariate logistic regression (MLR) approaches.
4.2 Machine learning-based prediction methods
Machine learning (ML), a pivotal artificial intelligence (AI) methodology, involves constructing probabilistic statistical models from data to enable predictive analytics (48). Widely applied in medical imaging (MRI, CT, ultrasound), ML classifier selection requires careful consideration of data characteristics, model performance, computational resources, and clinical utility, with optimal algorithms typically identified through empirical validation.
Duan Yanan et al. (49) evaluated CT radiomics-driven ML models for EGFR mutation prediction in NSCLC. Their study enrolled 198 patients, extracting 1,050 radiomic features per case, ultimately selecting 16 features through dimensionality reduction. Seven classifiers logistic regression (LR), decision tree (DT), random forest (RF), neural network (NN), support vector machine (SVM), naïve Bayes (NB), and k-nearest neighbors (KNN) were tested. RF demonstrated superior performance, achieving AUC and F1 scores of 0.988/0.983 in the training cohort and 0.793/0.653 in the validation cohort. Delzell et al. (50) analyzed 416 quantitative imaging biomarkers from 200 lung nodule CT scans, employing three feature selection methods (linear combination filter, pairwise correlation filter, PCA) and three classifiers (linear, nonlinear, ensemble). Elastic net and SVM with linear/correlation-based feature selection yielded optimal tumor classification accuracy, while RF and bagged trees underperformed. Their findings underscore the efficacy of radiomic-ML integration in reducing false-positive rates. Naïve et al. (51) utilized gene expression (GDS3257) and DNA methylation β-values from The Cancer Genome Atlas (TCGA) to classify LUAD and LUSC subtypes. Bayesian and ReliefF/Limma feature selection identified 19 predictive genes, achieving AUC=0.89 across datasets. However, the absence of prospective validation limited clinical applicability. Key studies leveraging ML classifiers for NSCLC EGFR mutation prediction are summarized in Table 2, providing a comparative overview of recent advancements.
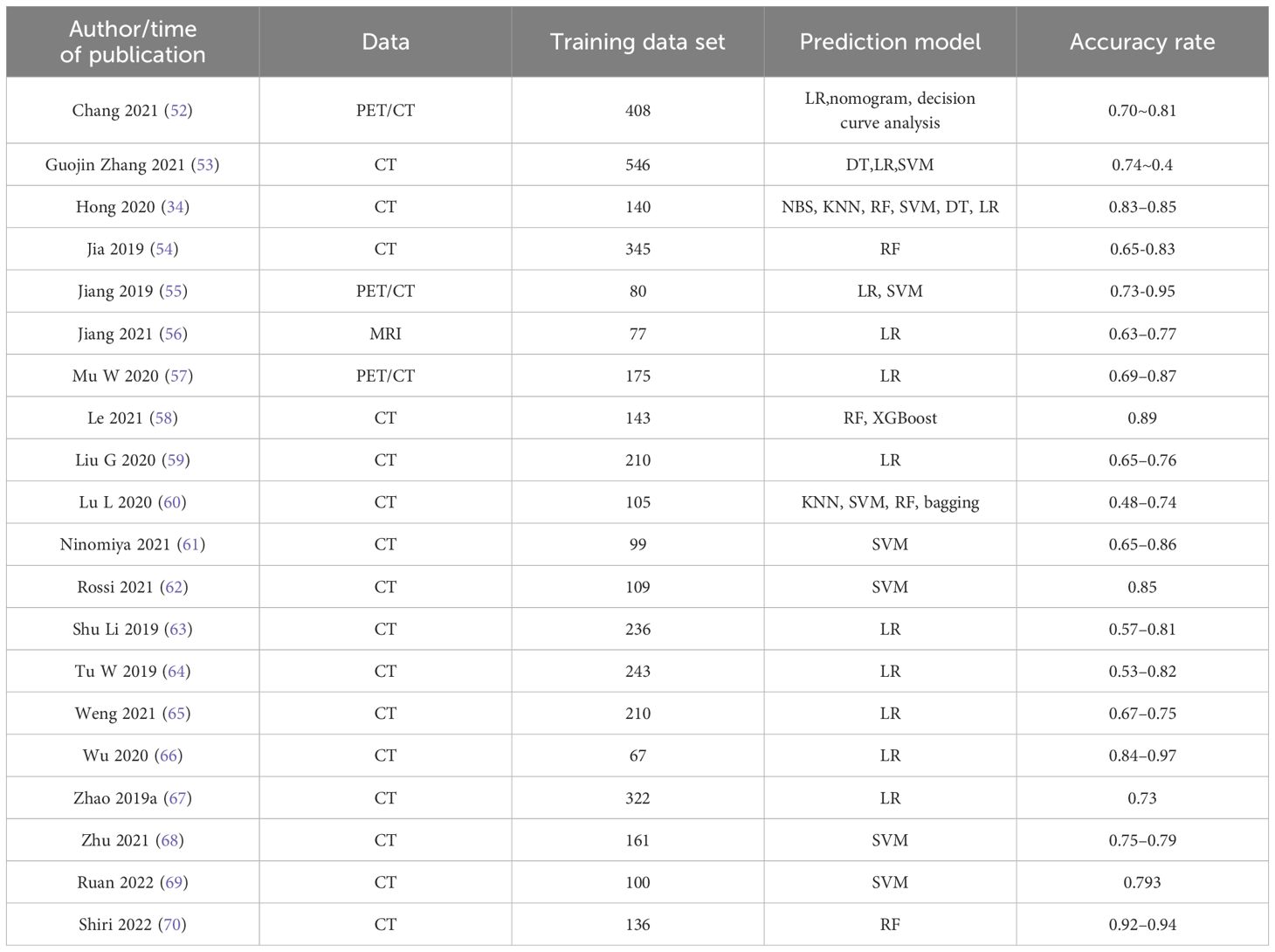
Table 2. Research on EGFR mutation prediction of lung cancer based on different machine learning classifiers.
Based on the summary analysis of Table 2, traditional machine learning (ML) demonstrates clear advantages, primarily due to the relative simplicity of the models, ease of implementation, and strong clinical interpretability. Commonly used algorithms, such as logistic regression (LR) and decision trees (DT), can explicitly highlight feature importance and decision pathways, making them particularly understandable and trustworthy for clinicians; consequently, they have been widely adopted in studies like those by Chang (2021), Wu (2020), Tu (2019), and Weng (2021). Furthermore, certain studies, including that by Duan et al., utilized random forest (RF) algorithms, achieving notably high training performance (AUC: 0.988, accuracy: 0.983) by effective feature selection (reducing 1050 features down to 16), indicating that traditional ML can provide excellent internal predictive performance under suitable data scales and optimized feature engineering.
However, significant limitations are also evident in traditional ML-based EGFR mutation prediction studies. First, the predictive performance of these models varies substantially among different studies, even when applying the same algorithm. For instance, the accuracy of LR reached as high as 0.97 in Wu (2020), whereas Lu (2020) reported accuracies ranging from only 0.48 to 0.74 using algorithms such as KNN, SVM, and RF. This highlights significant instability, potentially attributable to differences in imaging protocols, feature selection methodologies, and inherent dataset heterogeneity across studies. Second, most of these studies included relatively small sample sizes (generally between 100–300 cases), predominantly from single-center retrospective cohorts, which may not adequately represent broader population characteristics or variations in imaging protocols, thereby limiting generalizability. Additionally, very few studies (e.g., Duan et al.) have reported independent external validation results, where external validation performance (AUC: 0.793) was substantially lower compared to internal training results (AUC: 0.988), reflecting significant overfitting risks and limited cross-institution applicability.
Overall, although traditional ML models hold advantages in terms of interpretability and implementation costs, the insufficient generalizability, sensitivity to data quality and feature selection, and instability across diverse clinical settings remain critical issues requiring further attention. Thus, future research should prioritize increasing sample sizes and data diversity, optimizing feature selection strategies, enhancing model generalizability, and emphasizing multicenter and independent external validations to facilitate the broader clinical implementation and application of traditional ML-based models.
4.3 Deep learning-based prediction of EGFR Gene mutations
Deep learning-based approaches for analyzing lung tumors and predicting genetic mutations hold broad prospects and significant clinical value, offering critical implications for improving therapeutic outcomes and patient survival rates. Research on constructing classification and predictive models for lung cancer using deep learning techniques has emerged as a global research focus. Deep learning demonstrates substantial potential for automating feature extraction processes from medical images, thereby streamlining workflows and enhancing predictive analyses (71). By autonomously learning abstract, high-level features from datasets and continuously improving model performance through iterative training, deep learning has enabled researchers to develop models for predicting EGFR mutation status with promising results. These advancements bridge fundamental, translational, and clinical research in non-small cell lung cancer (NSCLC).
Xiao et al. (72) collected PET/CT imaging data from 150 EGFR-mutant patients between 2016 and 2019, generating 3,794 PET/CT fusion datasets after 2D slicing (1,913 wild-type and 1,881 EGFR-mutant samples). Their study proposed a deep learning framework based on the EfficientNet-V2 model. First, 32 two-dimensional views were extracted from 3D cubic volumes of each pulmonary nodule. Deep features from these views were then utilized to predict EGFR mutation status. The deep learning model achieved AUCs of 83.64% and 82.41%, respectively, demonstrating promising efficacy in EGFR mutation prediction.
Seonhwa Kim et al. (73) retrospectively analyzed CT scans and clinical data from 1,280 NSCLC patients tested for EGFR mutations (454 mutant-type and 826 wild-type). The team developed a novel hybrid method integrating deep learning and radiomics to predict EGFR mutations. Radiomic features were extracted from preprocessed CT images of NSCLC tumors and combined with tumor images and clinical data as input for the predictive model. This approach achieved AUCs of approximately 0.81 and 0.78 in the initial cohort and external validation, respectively, highlighting the feasibility of combining radiomic analysis with deep learning for EGFR mutation prediction.
Chengdi Wang et al. (74) collected clinical information, histopathology reports, CT imaging data, and genetic testing results from 1,262 patients. The dataset was partitioned into training (N=882), validation (N=125), and test (N=255) sets at a 7:1:2 ratio. They proposed a novel deep learning method to predict EGFR mutation and PD-L1 expression status in NSCLC patients, integrating selected features to construct a prognostic model. A 3D convolutional neural network (CNN) was employed, achieving AUCs of 0.96 (95% CI: 0.94–0.98), 0.80 (95% CI: 0.72–0.88), and 0.73 (95% CI: 0.63–0.83) in the training, validation, and test cohorts, respectively.
Abhishek Mahajan et al. (48) analyzed CT imaging data from 990 patients with primary lung adenocarcinoma confirmed by genetic testing. The team developed and validated a deep learning-based radiogenomics (DLR) model combined with radiomic features to predict EGFR mutations in NSCLC, while evaluating semantic and clinical features associated with mutation detection. An end-to-end pipeline was applied to CT images from two NSCLC trials without precise segmentation. Two 3D CNNs were used to segment lung masses and nodules. The combined radiomic-DLR model achieved an AUC of 0.88 ± 0.03 for EGFR mutation prediction, outperforming individual models. Integration of semantic features further improved accuracy, yielding an AUC of 0.88 ± 0.05.
Shuo Wang et al. (75) compiled CT imaging and EGFR sequencing data from 18,232 lung cancer patients across nine cohorts in China and the U.S., including a prospective Asian cohort (n=891) and The Cancer Imaging Archive (TCIA) cohorts of White populations stratified into thick-slice and thin-slice CT groups. The authors proposed a fully automated artificial intelligence system (FAIS) to extract whole-lung information from CT scans for predicting EGFR genotype and prognosis of EGFR-TKI therapy. FAIS was evaluated using AUC for EGFR genotype prediction and Kaplan-Meier analysis for progression-free survival (PFS) in EGFR-TKI-treated patients.
Wentao Zhu et al. (76) analyzed CT scans from 191 patients with biopsy-confirmed lung adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC). They introduced a self-generating hybrid feature network (SGHF-Net) to classify lung cancer subtypes on CT images. A pathological feature synthesis module (PFSM) was innovatively designed to quantify cross-modal correlations via deep neural networks, deriving “gold-standard” pathological information from CT images. Simultaneously, a radiomic feature extraction module (RFEM) fused CT-derived features with pathological priors under an optimized framework, enhancing the model’s ability to generate discriminative and subtype-specific features for accurate prediction.
Le NQK et al. (77–79) compiled CT imaging and bed data from 576 patients diagnosed with non-small cell lung cancer (NSCLC). The dataset was partitioned into a training set (N = 420) and a test set (N = 156). A multimodal deep learning framework was subsequently developed, integrating 3D CNN survival analysis and DeepSurv methodologies. By combining deep radiomics, traditional radiomics, and clinical parameters, the model predicted the survival status of NSCLC patients. The findings indicate that the DeepSurv CT deep radiomics model outperforms the conventional Cox-PH model, and the integration of multiple parameters enhances prediction accuracy.
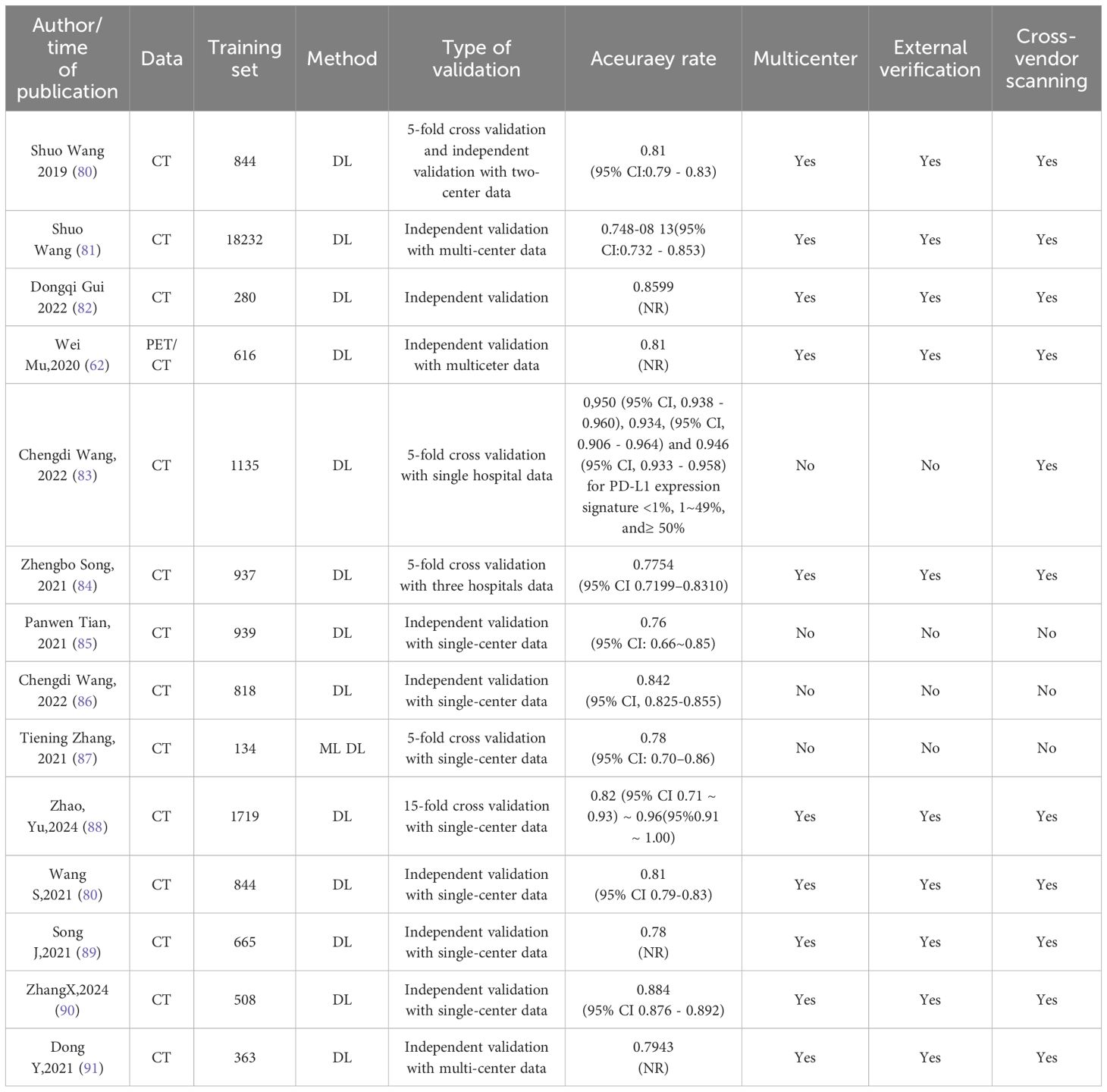
Other research findings regarding the prediction of EGFR mutations in lung cancer using deep learning methods, along with detailed information on data sources, scanner heterogeneity, and validation designs, are presented in Table 3.
In medical imaging, acquiring large-scale training datasets remains challenging. Transfer learning (TL) strategies theoretically address this limitation by leveraging visual feature similarities across domains (92). However, the scarcity of pre-trained models in medical imaging restricts TL applications. Recent advances propose semi-supervised learning techniques, such as pseudo-labeling and generative adversarial networks (GANs), to utilize unlabeled data and mitigate sample size constraints (93). These innovations hold significant potential for EGFR mutation prediction and warrant further exploration (94). Additionally, training deep learning models demands substantial computational resources (e.g., CPUs, GPUs, and memory). In resource-limited settings, traditional regression (TR) may offer a pragmatic alternative. The “black box” nature of deep neural networks also poses interpretability challenges, as their high complexity and multi-parametric architecture obscure internal decision-making processes (95).
Through comprehensive analysis, deep learning (DL) models exhibit remarkable advantages in the automatic extraction of medical imaging features and classification accuracy, with most studies reporting predictive accuracies (AUC values) ranging between 0.76 and 0.96. For instance, the DL-based approach proposed by Wang et al. demonstrated high stability across a multicenter independent validation dataset, achieving an AUC ranging from 0.748 to 0.813, indicating potential cross-institutional generalizability. Additionally, several studies (e.g., Gui, 2022; Zhang, 2024) highlight that deep learning techniques, particularly through advanced feature extraction and multimodal data integration (e.g., PET/CT combined with clinical information), significantly enhance prediction performance, reaching AUC values as high as 0.86 to 0.88.
However, current DL models for EGFR mutation prediction still encounter notable limitations and challenges. Firstly, most studies predominantly rely on single-center datasets (more than half conducted validations solely within single-center cohorts), with limited multicenter and cross-device validation, potentially restricting the model’s true generalizability. Specifically, among the published studies, only 17 out of 59 (29%) conducted external validation using independent datasets, and merely 10 out of 59 (17%) involved multicenter validations using scanners from different manufacturers. Notably, the pooled AUC was 0.86 ± 0.07 on internal test sets, yet this performance decreased to 0.77 ± 0.06 during external validations. Secondly, substantial variations in sample size exist among studies, ranging from several hundred to tens of thousands of cases (e.g., Wang et al. included a sample size of 18,232), highlighting the lack of consensus on data diversity and standardization across research efforts. Additionally, the inherent “black box” nature of DL models significantly limits their clinical interpretability.
In conclusion, future research should prioritize expanding multicenter validation datasets, standardizing imaging protocols and data quality control, and enhancing model interpretability studies, ultimately promoting the practical application and widespread adoption of deep learning models in clinical practice.
5 Novel artificial intelligence approaches for predicting EGFR mutations in non-small cell lung cancer
Although radiomics-based machine learning (ML) and deep learning (DL) models have demonstrated potential in EGFR mutation detection (e.g., CT radiomics models achieving AUCs of 0.82–0.88) (96, 97), current studies remain constrained by several limitations. First, single-modality imaging data primarily reflect tumor morphology or functional characteristics, failing to capture molecular dynamics or microenvironmental heterogeneity linked to EGFR mutations. Second, traditional models rely on manual feature engineering or isolated DL architectures, limiting their capacity to model cross-scale biological correlations and resulting in poor interpretability (only ~35% of key feature contributions are explainable). Third, most studies utilize static imaging data, lacking dynamic tracking of EGFR mutation evolution during treatment. To address these challenges, novel artificial intelligence (AI)-driven multimodal data fusion strategies are emerging as pivotal solutions. By integrating radiomics, liquid biopsy, pathomics, and dynamic clinical data, next-generation models can construct cross-dimensional feature association networks (e.g., spatiotemporal coupling of imaging texture features and ctDNA methylation profiles). Leveraging graph neural networks (GNNs) and federated learning, these models enable collaborative optimization of multicenter heterogeneous data, enhancing predictive performance (AUC projected to exceed 0.95) while elucidating EGFR mutation-driven mechanisms and drug resistance evolution, thereby supporting precision therapeutic decision-making across the treatment continuum (15, 98).
5.1 Multimodal data fusion for predicting lung cancer gene mutations
Recent advances in multimodal data fusion have significantly improved EGFR mutation prediction in non-small cell lung cancer (NSCLC). Researchers worldwide have developed innovative predictive models by integrating radiomics, genomics, pathology, and clinical data. The core advantage of multimodal models lies in their ability to fuse heterogeneous, multidimensional data for comprehensive and precise EGFR mutation prediction (99–102). The analytical workflow typically involves four key stages:
1. Data Preprocessing: Standardization of heterogeneous data sources, including CT image resampling and normalization, genomic variant annotation and feature selection, and clinical data imputation and encoding.
2. Feature Extraction: High-dimensional feature extraction from multimodal data using DL or traditional ML methods. Examples include texture feature extraction from CT images via convolutional neural networks (CNNs) and semantic feature extraction from pathology reports using natural language processing (NLP).
3. Feature Fusion: Alignment and integration of multimodal features through early fusion, late fusion, or intermediate fusion strategies. Common techniques include graph neural networks (GNNs), attention mechanisms, and multimodal Transformer architectures.
4. Model Training and Validation: Performance evaluation via cross-validation or independent test sets, coupled with interpretability tools (e.g., SHAP values or Gradient-weighted Class Activation Mapping [Grad-CAM]) to quantify key feature contributions.
This approach not only enhances EGFR mutation prediction accuracy (AUC improvements of 10%–15%) but also provides novel insights into the interplay between imaging features and molecular mechanisms.
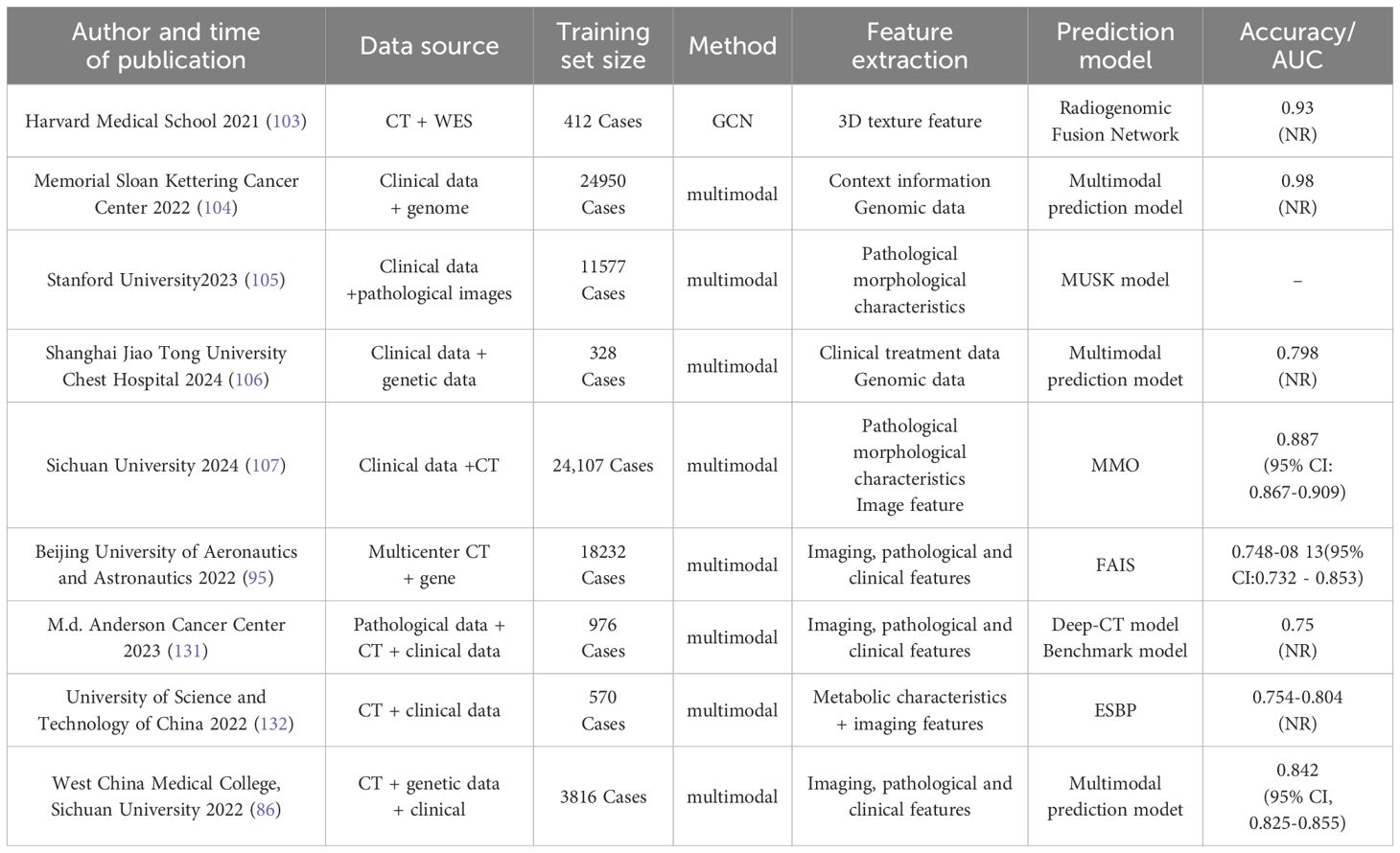
Internationally, research consortia have demonstrated leading expertise in imaging-genomics joint modeling, multi-omics integration, and dynamic predictive system development. The Harvard Medical School team (2021) proposed the “Radiogenomic Fusion Network” (103), which employs graph convolutional networks (GCNs) to achieve deep integration of high-resolution CT imaging features with whole-exome sequencing (WES) data. Validated in a cohort of 412 NSCLC patients, the model achieved an AUC of 0.93 for EGFR mutation prediction and first identified a specific association between “ground-glass opacity” on CT imaging and the EGFR L858R mutation. The Memorial Sloan Kettering Cancer Center (MSK) Cancer Data Science Initiative Group (2024) (104) developed a framework integrating patient-reported clinical genomic data with natural language processing (NLP) techniques and multimodal biomarkers to improve the accuracy of overall survival (OS) prediction in cancer patients. The research team constructed a comprehensive dataset encompassing patients with non-small cell lung cancer (NSCLC), breast cancer, colorectal cancer, and other malignancies. NLP methodologies were employed to extract critical information from unstructured textual data, including clinical notes and diagnostic reports, while structured data—such as treatment histories, survival outcomes, and tumor characteristics—were systematically integrated to build a multimodal predictive model. Through rigorous cross-validation and external validation, the NLP component demonstrated stable performance across cancer subtypes. Notably, in NSCLC cohorts, the model achieved a precision of 0.78, an AUC of 0.98, and a recall of 0.92 for identifying prior treatment histories. The multimodal fusion approach effectively processed heterogeneous data types (unstructured text and structured clinical parameters), enabling robust survival prediction. This methodology provides clinicians with enhanced capabilities for personalized treatment planning, potentially improving therapeutic outcomes and patient survival rates.The research team led by Rui-Jiang Li and Sen Yang at Stanford University developed MUSK(2025) (105), a pre-trained foundational model based on the BeiT3 architecture. MUSK effectively leverages unlabeled and unpaired image-text data through a unified masked modeling approach. The model was trained on an extensive dataset comprising 50 million pathological image patches and 1 billion text tokens. To address the distinct visual characteristics and data distribution differences between pathological images and natural images, the team implemented several tailored optimizations: a multi-scale training strategy, pathological staining data augmentation, noisy data bootstrapping enhancement, and fine-grained multimodal alignment techniques. These methodological adaptations significantly improved the model’s learning capability for pathological data, resulting in enhanced clinical prediction accuracy.
Domestically, multimodal modeling research has prioritized clinical translation and optimization for region-specific healthcare contexts. The team led by Professor Lu Shun at Shanghai Chest Hospital, affiliated with Shanghai Jiao Tong University (2024) (106), innovatively integrated multiple non-invasive biomarkers to explore the potential for early identification of non-small cell lung cancer (NSCLC) patients who may derive durable clinical benefits from immune checkpoint inhibitor (ICI) therapy. The team developed a multiparameter predictive model incorporating standardized bTMB, dynamic changes in ctDNA during early treatment, and RECIST response to predict durable clinical benefit (DCB) from ICI therapy. This model demonstrated robust predictive performance in both the training and validation cohorts, with AUC values of 0.854 and 0.798, and accuracy rates of 79.5% and 74.7%, respectively. The inclusion of RECIST response further enhanced the model’s predictive capability, particularly in the validation cohort, where both sensitivity and specificity showed improvement.The research team led by Professor Li Weimin at Sichuan University (2024) (107) developed a multimodal artificial intelligence (MMI) system that integrates multidimensional clinical data—including clinical texts, imaging data, and laboratory indicators—to achieve accurate prediction of pulmonary infectious diseases, pathogen types, and timely identification of critical illness, thereby providing robust support for clinical decision-making. The MMI model was trained on 24,107 patient records comprising clinical texts and CT images to distinguish bacterial, fungal, viral pneumonia, and tuberculosis. The system demonstrated exceptional performance in both internal and external validation datasets, achieving AUC values of 0.910 (95% CI: 0.904–0.916) and 0.887 (95% CI: 0.867–0.909), respectively, comparable to the diagnostic accuracy of experienced clinicians. Furthermore, the MMI system rapidly differentiated viral and bacterial subtypes, with mean AUCs of 0.822 (95% CI: 0.805–0.837) for viral subtypes and 0.803 (95% CI: 0.775–0.830) for bacterial subtypes. Notably, the system also facilitated personalized medication recommendations to mitigate antibiotic misuse and exhibited significant advantages in predicting critical illness risks, offering a promising tool to optimize clinical workflows.A research team led by Beihang University (2022) (95) developed a fully automated artificial intelligence system (FAIS) that leverages whole-lung CT imaging information to predict EGFR genotype and evaluate prognosis in patients receiving EGFR-TKIs therapy. This multicenter study encompassed 18,232 lung cancer patients across nine Chinese and American cohorts, incorporating both prospective and retrospective data from Asian and Caucasian populations. Participants were stratified into thick- and thin-section CT groups. FAIS demonstrated capability in predicting EGFR mutation status and progression-free survival for EGFR-TKIs treated patients, with performance validated through AUC metrics and Kaplan-Meier analysis. Compared with two tumor region-based deep learning models, FAIS showed superior performance across multiple test cohorts, achieving AUC values ranging from 0.748 to 0.813. The FAIS-C model integrating clinical factors exhibited significant correlation with EGFR-TKIs therapeutic outcomes (log-rank p < 0.05), suggesting its potential as an effective complement to genetic sequencing methods.
5.2 Applications of large language models
Large Language Models (LLMs) are deep learning-based natural language processing models trained on massive textual datasets, enabling comprehension and generation of human language. Their core architecture typically relies on the Transformer framework, which utilizes self-attention mechanisms to capture long-range dependencies in text, facilitating deep contextual semantic understanding. Recent advances in computational power and data scalability have demonstrated the robust capabilities of LLMs across diverse domains, exemplified by models such as the GPT (Generative Pre-trained Transformer) series, DeepSeek, Qwen series, BERT (Bidirectional Encoder Representations from Transformers), and ChatGLM. These models excel not only in general natural language tasks (e.g., text generation, translation, and question answering) but also show significant potential in medical applications (108, 109).
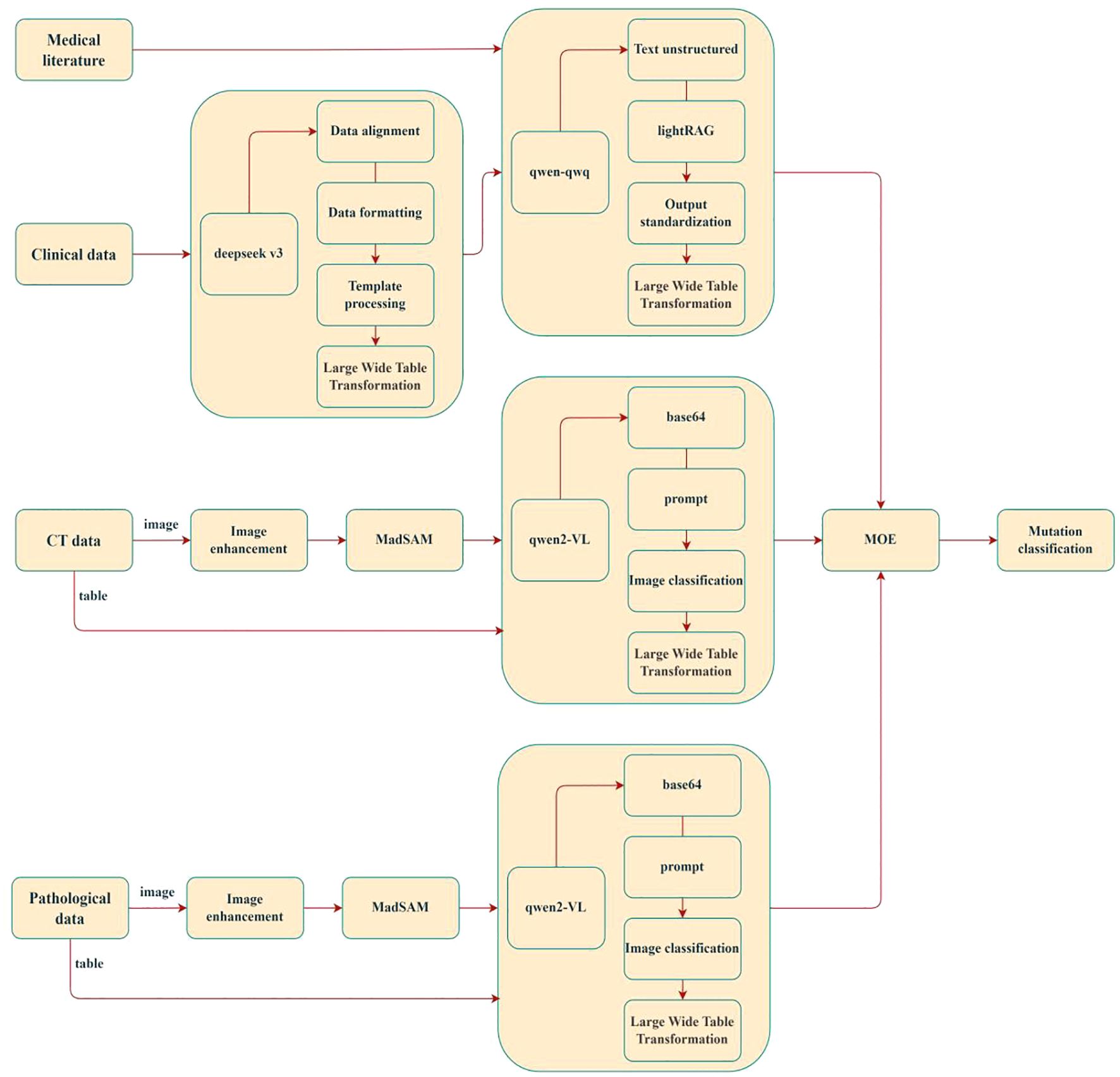
In recent years, the utility of LLMs in predicting epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer (NSCLC) has garnered substantial interest. Leveraging their advanced natural language processing and contextual reasoning capabilities, LLMs can effectively integrate textual information from multimodal data (e.g., pathology reports, clinical notes, and genomic annotations) to enhance predictive performance. Researchers have proposed novel technical approaches to optimize LLMs for lung cancer mutation prediction, achieving promising results (27, 110, 111). As illustrated in Figure 7, the LMOE (Large Mixture of Experts) framework exemplifies such innovation. This model integrates multimodal inputs—including medical imaging (CT scans, histopathology images) and clinical data—through image enhancement, data alignment, and structured processing to construct a unified high-dimensional feature space (wide-table transformation). It employs a lightRAG module for cross-modal semantic association and utilizes Qwen-series algorithms for final mutation classification. The LMOE framework directly addresses critical challenges in integrating multi-source heterogeneous clinical data, offering the following advantages:
1. Depth of Data Integration: Moving beyond traditional single-modality analyses, the model fuses histopathological texture features (via the MadSAM module), 3D spatial information from CT imaging, and temporal clinical parameters to better reflect the biological heterogeneity of lung cancer. For example, dynamic correlations between locally enhanced histopathological features (e.g., H&E-stained regions) and volumetric changes in CT-detected pulmonary nodules may reveal radiomic biomarkers specific to mutations such as EGFR or ALK.
2. Clinical Interpretability: A standardized output module maps predictions to mutation classification systems in clinical guidelines (e.g., NCCN criteria), ensuring model outputs directly inform targeted therapy decisions. This end-to-end clinical alignment significantly outperforms traditional “black-box” models.
3. Technical Extensibility: The Mixture of Experts (MOE) architecture supports phased validation, where clinical data serve as prior knowledge to constrain model training. Additionally, the Qwen2-VL module’s vision-language alignment capability provides scalable interfaces for integrating pathology report text with image features, as shown in Figure 8.
Multiple international and domestic research teams have conducted in-depth investigations into the application of large language models (LLMs) for predicting lung cancer gene mutations, achieving notable progress. For instance, the Google Health team (2022) (112) proposed the “Med-PaLM” model, which fine-tunes LLMs (e.g., GPT-3) to integrate pathology reports with radiomics data, achieving an AUC of 0.89(NR) in NSCLC EGFR mutation prediction, significantly outperforming conventional methods. The “BB-TEN” model, developed by a Columbia University research team (2025) (113), enables automated TNM (tumor size, regional lymph node involvement, and distant metastasis) classification from pathology report text. This framework employs a BERT architecture for semantic analysis of pathology reports combined with CT imaging features. Evaluated on nearly 8,000 pathology reports from Columbia University Medical Center, the model demonstrated robust performance with AUC values ranging from 0.815 to 0.942(NR). Harvard Medical School(2024) has developed PathChat (114), a vision-language general-purpose AI assistant for human pathology slide analysis. The system was pre-trained via self-supervised learning on image patches derived from over 1 million histopathological slides, enabling accurate disease identification from biopsy specimens with an accuracy rate approaching 90%. This performance surpasses that of GPT-4V, demonstrating its advanced diagnostic capabilities in computational pathology. In the same year, Professor Kunxing Yu at Harvard University developed the Clinical Histopathology Imaging Evaluation Foundation (CHIEF) model (115), a foundational framework for histopathological image analysis. The CHIEF model demonstrates diagnostic capabilities for 19 cancer types originating from pulmonary, breast, prostate, colorectal, gastric, esophageal, renal, cerebral, hepatic, thyroid, pancreatic, cervical, uterine, ovarian, testicular, cutaneous, soft tissue, adrenal, and bladder tissues, achieving a diagnostic accuracy approaching 94%. The research team led by Guangyu Wang at Beijing University of Posts and Telecommunications has developed “MedFound,” (116) a 176-billion-parameter medical large language model (LLM). This general-purpose model was pre-trained on a large-scale corpus comprising diverse medical texts and real-world clinical records. Through fine-tuning, MedFound employs a self-bootstrapping chain-of-thought approach to emulate physicians’ diagnostic reasoning processes, while incorporating a unified preference alignment framework to ensure consistency with standardized clinical practices. A study conducted by the First Affiliated Hospital of Sun Yat-sen University on the imaging and pathological evaluation of thyroid nodules demonstrated that in a comparison of 1,161 thyroid nodule imaging diagnoses from 725 patients, ChatGPT 4.0 and Bard exhibited significant to nearly perfect internal consistency. This performance was comparable to the human-machine interaction strategies employed by two senior radiologists and one junior radiologist, and surpassed the strategy involving only a single junior radiologist (117). Additionally, a large diagnostic model for pneumoconiosis, named “PneumoLLM” (118), developed by Chinese researchers, has established a novel paradigm for applying Large Language Models (LLMs) to data-scarce occupational diseases, as shown in Table 4. Extensive experiments have demonstrated the superior diagnostic capabilities of this large model in identifying pneumoconiosis.
Analysis of these studies reveals that LLMs excel in NSCLC EGFR mutation prediction by efficiently utilizing unstructured textual data (e.g., pathology reports, clinical notes), addressing the underutilization of such data in traditional approaches. Furthermore, LLMs demonstrate superior contextual understanding, enabling robust cross-modal feature alignment and fusion across imaging, text, and genomic data. Their capacity for temporal data modeling allows tracking of dynamic EGFR mutation changes during treatment, while attention mechanisms and feature contribution analyses provide biologically interpretable insights. However, despite their promise, challenges persist in medical applications of LLMs. Key limitations include the reliance on massive annotated datasets, which are scarce in medicine; high computational demands for training and inference, hindering deployment in resource-limited settings; suboptimal domain-specific terminology comprehension by general-purpose LLMs; and privacy risks during multi-center data sharing and model training.
As artificial intelligence advances in NSCLC mutation prediction, LLMs are driving dual evolutionary pathways: technological paradigm restructuring and clinical value enhancement. Current trends indicate future LLM development will focus on synergistic optimization of efficiency, dynamism, security, and interpretability, bridging the gap from algorithmic validation to clinical implementation. On one front, lightweight architectures (e.g., Med-GPT) leveraging domain-specific knowledge distillation—via parameter pruning and attention mechanism optimization—are reducing computational costs while enhancing analysis of radiomic features (e.g., CT texture heterogeneity) and temporal clinical data (e.g., treatment response dynamics), offering accessible molecular subtyping tools for primary care settings. Concurrently, multimodal dynamic modeling, integrating time-series Transformer architectures with liquid biopsy data (e.g., ctDNA mutation burden), enables construction of spatial-temporal models of EGFR mutation evolution. These models capture dynamic correlations between tumor heterogeneity (e.g., lung nodule volume growth rates) and molecular biomarkers, predicting tyrosine kinase inhibitor (TKI) resistance trajectories. Privacy-preserving technologies, such as federated learning frameworks (FedLLM) with homomorphic encryption and differential privacy, are overcoming multi-center data silos, enabling collaborative training on cross-institutional pathology and genomic datasets. Notably, enhanced interpretability methods (e.g., medical-specific SHAP analysis) are quantifying associations between radiomic features (e.g., ground-glass nodule CT value distributions) and EGFR mutation subtypes (e.g., L858R or exon 19 deletions), building trust between AI predictions and clinical decision-making. This technological evolution not only addresses disparities in genetic testing resource allocation but also promises seamless integration with PACS systems, bridging radiological diagnosis to molecular pathology inference and advancing precision medicine toward preemptive intervention and dynamic monitoring.
6 Future prospects and challenges
6.1 Data sources and standardization
The current performance bottlenecks of artificial intelligence (AI) models in predicting EGFR mutations in non-small cell lung cancer (NSCLC) primarily stem from fragmented data ecosystems and a lack of standardization. Although multimodal data integration—including CT imaging, histopathology slides, liquid biopsies, and clinical narratives—has emerged as the mainstream paradigm, significant heterogeneity persists in data acquisition protocols. For instance, in medical imaging, variations in CT scanner parameters (e.g., kVp, slice thickness, and reconstruction algorithms) across manufacturers (e.g., Siemens, GE Healthcare, Philips) induce texture feature distribution shifts (domain shift). Studies demonstrate that increasing slice thickness from 1 mm to 5 mm elevates radiomic feature variability by 27%. In genomic profiling, differences in sensitivity between next-generation sequencing (NGS, 0.1%) and digital PCR (1%) introduce substantial labeling noise for low-abundance mutations (e.g., EGFR T790M) (119, 120).
Future efforts must prioritize the development of cross-modal data standardization frameworks. For example, domain adaptation techniques using generative adversarial networks (CycleGAN) could harmonize CT images across scanner vendors, while ISO/IEC 20547-compliant biomedical data lake architectures may enable dynamic alignment and version control of multicenter data.
In data processing, conventional manual annotations (e.g., tumor ROI delineation) suffer from subjectivity and poor reproducibility. Recent advancements propose “hybrid annotation” strategies that integrate expert annotations (following RECIST 1.1 criteria) with weakly supervised learning (e.g., text-image alignment using pathology reports) to extract cross-modality consistent features via contrastive learning. For small-sample mutation subtypes (e.g., EGFR exon 20 insertion mutations, <10% of EGFR mutations), diffusion models (DMs) show promise in synthetic data generation. Experiments reveal that synthetic CT images generated via Stable Diffusion architectures improve model AUC for rare mutations by 12% (121–123).
Furthermore, breakthroughs in federated learning (FL), such as the FedMA algorithm (an enhanced variant of FedAvg), will facilitate cross-institutional collaboration. These frameworks enable distributed model training on global multicenter datasets (e.g., the NSCLC-Radiomics-Genomics Consortium) while ensuring compliance with patient privacy regulations (GDPR/HIPAA).
Cross-population generalisability remains the principal bottleneck. More than 60% of published cohorts originate from single-centre East-Asian datasets, while African-American and Hispanic patients are virtually absent. Scanner-parameter variability is seldom quantified; for example, increasing slice thickness from 1 mm to 5 mm can alter radiomic features by up to 27%. Although the multinational, nine-centre FAIS dataset demonstrates that broader sampling is feasible, such resources remain exceptional. Future work should employ prospective PROBE designs that cover multiple ethnic groups and acquisition protocols, and integrate domain-adaptation or federated-learning strategies to improve out-of-domain performance.
6.2 Model interpretability and repeatability
The limited interpretability of artificial intelligence (AI) models has become a critical barrier to their clinical adoption, despite their expanding applications in medicine. While existing methods (e.g., Grad-CAM, LIME) can visualize critical imaging regions (e.g., ground-glass opacity contributing to EGFR mutation predictions), they fail to provide biologically meaningful explanations of molecular mechanisms. Recent research trends focus on cross-modal causal inference to bridge this gap (124). For example, integrating radiomic features with spatial transcriptomic atlases of the tumor microenvironment (TME): spatial transcriptomic sequencing (10x Visium) generates gene expression matrices of tumor regions, and graph attention networks (GATs) establish quantitative models linking CT imaging texture features (e.g., gray-level co-occurrence matrix entropy) to EGFR signaling pathway activation (e.g., PI3K-Akt-mTOR). Such studies have demonstrated that peritumoral vascular tortuosity on CT scans (quantified by fractal dimension) strongly correlates with VEGF overexpression (r=0.73, p<0.001), offering mechanistic insights into imaging-based predictions of EGFR-TKI resistance (125).
For dynamic interpretability, Transformer-based time-series models (e.g., TimeSformer) enable tracking of EGFR mutation evolution during treatment. By integrating serial CT imaging (3-month follow-ups) with ctDNA monitoring data, these models can detect imaging phenotype shifts in EGFR L858R mutations (e.g., reduced ground-glass opacity with increased solid components) and predict the risk of T790M resistance mutations (HR=2.34, 95% CI 1.87–2.93).
Additionally, knowledge graphs enhance model transparency by embedding clinical guidelines (e.g., NCCN), databases (e.g., OncoKB), and imaging features into unified graph structures. This approach renders model decision pathways traceable to evidence-based clinical linkages, such as associations between specific imaging patterns and EGFR-TKI response rates (126, 127).
Our pooled analysis shows that, among the 59 eligible studies, only 11 prediction models (18.6%) have made their source code or executable software publicly available, and none of these releases include the corresponding trained weights or underlying datasets. Furthermore, just one study has employed a prospective, PROBE-style multicentre design and is prospectively registered on ClinicalTrials.gov. The scarcity of shared code and weights makes it impossible to quantify the net clinical benefit, potential risks, and reproducibility of the vast majority of proposed models.
6.3 Clinical translation trends
The clinical translation of AI technologies continues to face critical challenges (128), including:
1. Technical Validation Frameworks: Most current studies rely on retrospective single-center data. Future efforts must prioritize prospective multicenter validation platforms (e.g., NCT04253679 trial) employing a Prospective Randomized Open Blinded Endpoint (PROBE) design to evaluate AI models’ impact on clinical endpoints. For example, comparing the detection rate differences (targeting ≥15% improvement) between AI-guided biopsy localization (targeting regions with high predicted mutation probabilities) and conventional random biopsy.
2. Workflow Integration: Developing embedded AI diagnostic systems that seamlessly interface with hospital Picture Archiving and Communication Systems (PACS). The Philips IntelliSpace AI platform has demonstrated real-time CT image analysis capabilities (latency <3 seconds), but its EGFR mutation prediction module still requires FDA’s Center for Devices and Radiological Health (CDRH) certification. A key breakthrough lies in constructing lightweight models (e.g., MobileNetV3-enhanced architectures) that maintain AUC >0.85 while compressing parameter counts to <5M, enabling deployment on edge computing devices (e.g., CT scanner-integrated GPUs).
Future clinical applications of AI will likely follow two trajectories (126, 129, 130):
▪ Dynamic Precision Monitoring: AI-driven “digital twin” models simulating tumor evolution. For instance, generating virtual clones based on baseline CT imaging and genomic data to predict spatiotemporal changes in EGFR mutation abundance under different therapeutic strategies (e.g., osimertinib vs. chemotherapy), thereby guiding personalized treatment planning.
▪ Health Economics Optimization: Markov decision models quantifying AI’s cost-effectiveness ratios. Preliminary studies indicate that AI-guided EGFR mutation screening reduces genomic testing costs by 38% (via minimizing unnecessary NGS assays) while improving targetable population identification rates by 22%.
Ethical and regulatory challenges remain critical. Robust accountability mechanisms must be established to address diagnostic errors (e.g., false negatives causing treatment delays), clarifying legal liabilities. Furthermore, mitigating model bias is paramount—transfer learning strategies tailored to ethnic groups (Asian vs. Caucasian populations) can reduce AUC disparities in EGFR mutation prediction from 0.12 to 0.04.
7 Conclusion
Analysis indicates that radiomics-based machine-learning models, which rely on handcrafted feature engineering, require smaller sample sizes and offer high interpretability—advantages in data-limited settings or when traceable decision paths are needed. However, handcrafted features cannot fully characterise the tumour micro-environment, thus capping predictive performance. Deep-learning models, by contrast, learn end-to-end representations that automatically capture high-dimensional textures and contextual cues, uncovering richer EGFR-associated patterns in heterogeneous images and therefore hold a higher theoretical ceiling. Yet, despite encouraging internal results (pooled AUC ≈ 0.84), the absence of external validation, heightened risk of bias, and variability in scanning protocols and patient demographics continue to impede clinical translation.
To advance these models toward bedside application, we advocate: (i) multicentre, prospective PROBE-style studies supplemented by federated learning to broaden data coverage; (ii) domain-adaptation strategies to mitigate scanner and protocol discrepancies; (iii) routine release of model code and weights, decision-curve and cost–benefit analyses; and (iv) enhanced explainability and seamless integration into clinical workflows. Only through sustained verification across diverse populations and devices—and adherence to STARD-AI and CLAIM reporting standards—can AI-based predictors evolve from research prototypes into reliable tools for precision management of lung cancer.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. further inquiries can be directed to the corresponding authors.
Author contributions
LH: Data curation, Funding acquisition, Writing – original draft, Writing – review & editing. PS: Data curation, Formal Analysis, Writing – review & editing. LZ: Data curation, Resources, Writing – original draft. LC: Data curation, Methodology, Software, Writing – review & editing. LL: Funding acquisition, Methodology, Project administration, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported in part by the Shandong Province medical health science and technology project (202403020298) and Natural Science Foundation of Shandong Province under Grant ZR2024MA055 and ZR2023MF047.
Acknowledgments
We would like to express our gratitude to the radiologists and laboratory physicians at Weifang People’s Hospital for their invaluable assistance in data collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Siegel RL, Miller KD, Wagle NS, and Jemal A. Cancer statistics, 2023. CA A Cancer J Clin. (2023) 73:17–48. doi: 10.3322/caac.21763
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
3. Majeed U, Manochakian R, Zhao Y, and Lou Y. Targeted therapy in advanced non-small cell lung cancer: current advances and future trends. J Hematol Oncol. (2021) 14:1–20. doi: 10.1186/s13045-021-01121-2
4. Reck M, Remon J, and Hellmann MD. First-line immunotherapy for non–small-cell lung cancer. J Clin Oncol. (2022) 40:586–97. doi: 10.1200/JCO.21.01497
5. Zhang X, Zhang Y, Zhang G, Qiu X, Tan W, Yin X, et al. Deep learning with radiomics for disease diagnosis and treatment: challenges and potential. Front Oncol. (2022) 12:773840. doi: 10.3389/fonc.2022.773840
6. Tomaszewski MR and Gillies RJ. The biological meaning of radiomic features. Radiology. (2021) 298:505–16. doi: 10.1148/radiol.2021202553
7. Taniguchi K, Okami J, Kodama K, Higashiyama M, and Kato K. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci. (2008) 99:929–35. doi: 10.1111/j.1349-7006.2008.00782.x
8. Chakrabarty N and Mahajan A. Imaging analytics using artificial intelligence in oncology: A comprehensive review. Clin Oncol R Coll Radiol. (2023) 36:498–513. doi: 10.1016/j.clon.2023.09.013
9. Mahajan A, Gurukrishna B, Wadhwa S, Agarwal U, Baid U, Talbar S, et al. Deep learning based automated epidermal growth factor receptor and anaplastic lymphoma kinase status prediction of brain metastasis in non-small cell lung cancer. Explor Targeting Anti-Tumor Ther. (2023) 4:657–68. doi: 10.37349/etat
10. Wang K, Lu X, Zhou H, Gao Y, Zheng J, Tong M, et al. Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: A prospective multicentre study. Gut. (2019) 68:729–41. doi: 10.1136/gutjnl-2018-316204
11. Yoon HJ, Choi J, Kim E, Um S-W, Kang N, Kim W, et al. Deep learning analysis to predict EGFR mutation status in lung adenocarcinoma manifesting as pure ground-glass opacity nodules on CT. Front Oncol. (2022) 12:951575. doi: 10.3389/fonc.2022.951575
12. Ting DSW, Cheung CY-L, Lim G, Tan GSW, Quang ND, Gan A, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinalImages from multiethnic populations with diabetes. JAMA. (2017) 318:2211–23. doi: 10.1001/jama.2017.18152
13. Lakhani P and Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. (2017) 284:574–82. doi: 10.1148/radiol.2017162326
14. Atmakuru A, Chakraborty S, Faust O, Salvi M, Datta Barua P, Molinari F, et al. Deep learning in radiology for lung cancer diagnostics: A systematic review of classification, segmentation, and predictive modeling techniques. Expert Syst With Appl. (2024) 255. doi: 10.1016/j.eswa.2024.124665
15. Yang D, Miao Y, Liu C, Zhang N, Zhang D, Guo Q, et al. Advances in artificial intelligence applications in the field of lung cancer. Front Oncol. (2024) 14. doi: 10.3389/fonc.2024.1449068
16. Javed R, Abbas T, Khan AH, Daud A, Bukhari A, and Alharbey R. Deep learning for lungs cancer detection: a review. Artif Intell Rev. (2024) 57. doi: 10.1007/s10462-024-10807-1
17. Ferro A, Bottosso M, Dieci MV, Scagliori E, Miglietta F, Aldegheri V, et al. Clinical applications of radiomics and deep learning in breast and lung cancer: A narrative literature review on current evidence and future perspectives. Crit Rev Oncol Hematol. (2024) 203:104479. doi: 10.1016/j.critrevonc.2024.104479
18. Quanyang W, Yao H, Sicong W, Linlin Q, Zewei Z, Donghui H, et al. Artificial intelligence in lung cancer screening: Detection, classification, prediction, and prognosis. Cancer Med. (2024) 13:e7140. doi: 10.1002/cam4.7140
19. Hephzibah R, Anandharaj HC, Kowsalya G, Jayanthi R, and Chandy DA. Review on deep learning methodologies in medical image restoration and segmentation. Curr Med Imaging. (2023) 19:844–54. doi: 10.2174/1573405618666220407112825
20. Yao W, Bai J, Liao W, Chen Y, Liu M, and Xie Y. From CNN to transformer: A review of medical image segmentation models. J Imaging Inform Med. (2024) 37:1529–47. doi: 10.1007/s10278-024-00981-7
21. Xu J, Bian Q, Li X, Zhang A, Ke Y, Qiao M, et al. Contrastive graph pooling for explainable classification of brain networks. IEEE Trans Med Imaging. (2024) 43:3292–305. doi: 10.1109/TMI.2024.3392988
22. Ahmed S, Jinchao F, Ferzund J, Ali MU, Yaqub M, Manan MA, et al. GraFMRI: A graph-based fusion framework for robust multi-modal MRI reconstruction. Magn Reson Imaging. (2025) 116:110279. doi: 10.1016/j.mri.2024.110279
23. Lim ZW, Pushpanathan K, Yew SME, Lai Y, Sun CH, Lam JSH, et al. Benchmarking large language models’ performances for myopia care: a comparative analysis of ChatGPT-3.5, ChatGPT-4.0, and Google Bard. EBioMedicine. (2023) 95:104770. doi: 10.1016/j.ebiom.2023.104770
24. Shi R, Liu S, Xu X, Ye Z, Yang J, Le Q, et al. Benchmarking four large language models’ performance of addressing Chinese patients’ inquiries about dry eye disease: A two-phase study. Heliyon. (2024) 10:e34391. doi: 10.1016/j.heliyon.2024.e34391
25. Ong JCL, Seng BJJ, Law JZF, Low LL, Kwa ALH, Giacomini KM, et al. Artificial intelligence, ChatGPT, and other large language models for social determinants of health: Current state and future directions. Cell Rep Med. (2024) 5:101356. doi: 10.1016/j.xcrm.2023.101356
26. Matsuo H, Nishio M, Matsunaga T, Fujimoto K, and Murakami T. Exploring multilingual large language models for enhanced TNM classification of radiology report in lung cancer staging. Cancers (Basel). (2024) 16:3621. doi: 10.3390/cancers16213621
27. Tozuka R, Johno H, Amakawa A, Sato J, Muto M, Seki S, et al. Application of NotebookLM, a large language model with retrieval-augmented generation, for lung cancer staging. Jpn J Radiol. (2024) 43:706–712. doi: 10.1007/s11604-024-01705-1
28. Wang CK, Ke CR, Huang MS, Chong IW, Yang YH, Tseng VS, et al. Using large language models for efficient cancer registry coding in the real hospital setting: A feasibility study. Pac Symp Biocomput. (2025) 30:121–37. doi: 10.1142/9789819807024_0010
29. Megyesfalvi Z, Gay CM, Popper H, Pirker R, Ostoros G, Heeke S, et al. Clinical insights into small cell lung cancer: Tumor heterogeneity, diagnosis, therapy, and future directions. CA Cancer J Clin. (2023) 73:620–52. doi: 10.3322/caac.21785
30. Ferber D, Wölflein G, Wiest IC, Ligero M, Sainath S, Ghaffari Laleh N, et al. In-context learning enables multimodal large language models to classify cancer pathology images. Nat Commun. (2024) 15:10104. doi: 10.1038/s41467-024-51465-9
31. Xiao H, Zhou F, Liu X, Liu T, Li Z, Liu X, et al. A comprehensive survey of large language models and multimodal large language models in medicine. Inf Fusion. (2024). doi: 10.2139/ssrn.5031720
32. Lei L, Xiaoyan Y, Junchi L, Shen Y, Wang J, Wei P, et al. A survey on medical large language models: technology, application, trustworthiness, and future directions. arXiv preprint arXiv:2406.03712. (2024) 14. doi: 10.48550/arXiv.2406.03712
33. Shmatko A, Ghafari Laleh N, Gerstung M, and Kather JN. Artificial intelligence in histopathology: enhancing cancer research and clinical oncology. Nat Cancer. (2022) 3:1026–38. doi: 10.1038/s43018-022-00436-4
34. Unger M and Kather JN. A systematic analysis of deep learning in genomics and histopathology for precision oncology. BMC Med Genomics. (2024) 17:48. doi: 10.1186/s12920-024-01796-9
35. Echle A, Rindtorff NT, Brinker TJ, Luedde T, Pearson AT, and Kather JN. Deep learning in cancer pathology: a new generation of clinical biomarkers. Br J Cancer. (2021) 124:686–96. doi: 10.1038/s41416-020-01122-x
36. Jiang X, Hoffmeister M, Brenner H, Muti HS, Yuan T, Foersch S, et al. End-to-end prognostication in colorectal cancer by deep learning: a retrospective, multicentre study. Lancet Digit Health. (2024) 6:e33–e43. doi: 10.1016/S2589-7500(23)00208-X
37. Khader F, Müller-Franzes G, Wang T, Han T, Tayebi Arasteh S, Haarburger C, et al. Multimodal deep learning for integrating chest radiographs and clinical parameters: a case for transformers. Radiology. (2023) 309:e230806. doi: 10.1148/radiol.230806
38. Yu AC, Mohajer B, and Eng J. External validation of deep learning algorithms for radiologic diagnosis: a systematic review. Radiol Artif Intell. (2022) 4:e210064. doi: 10.1148/ryai.210064
39. Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ. (2021) 372:n71. doi: 10.1016/j.ijsu.2021.105906
40. Page MJ, Moher D, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. PRISMA 2020 explanation and elaboration: Updated guidance and exemplars for reporting systematic reviews. BMJ. (2021) 372:n160. doi: 10.1136/bmj.n160
41. Mayerhoefer ME, Materka A, Langs G, Haggstrom I, Szczypinski P, Gibbs P, et al. Introduction to radiomics. JNuclMed. (2020) 61:488–95. doi: 10.2967/jnumed.118.222893
43. Zhou JY, Zheng J, Yu ZF, Xiao WB, Zhao J, Sun K, et al. Comparative analysis of clinicoradiologic characteristics of lung adenocarcinomas with alk rearrangements or egfr mutations. Eur Radiol. (2015) 25:1257–66. doi: 10.1007/s00330-014-3516-z
44. Rizzo S, Petrella F, Buscarino V, De Maria F, Raimondi S, Barberis M, et al. Ct radiogenomic characterization of egfr, K-ras, and alk mutations in non-small cell lung cancer. Eur Radiol. (2016) 26:32–42. doi: 10.1007/s00330-015-3814-0
45. Yin X, Liao H, Yun H, Lin N, Li S, Xiang Y, et al. Artificial intelligence-based prediction of clinical outcome in immunotherapy and targeted therapy of lung cancer. Semin Cancer Biol. (2022) 86:146–59. doi: 10.1016/j.semcancer.2022.08.002
46. Li S, Li Y, Zhao M, Wang P, and Xin J. Combination of (18)F-fluorodeoxyglucose pet/ct radiomics and clinical features for predicting epidermal growth factor receptor mutations in lung adenocarcinoma. Korean J Radiol. (2022) 23:921–30. doi: 10.3348/kjr.2022.0295
47. Zhao W, Yang J, Ni B, Bi D, Sun Y, Xu M, et al. Toward automatic prediction of egfr mutation status in pulmonary adenocarcinoma with 3d deep learning. Cancer Med. (2019) 8:3532–43. doi: 10.1002/cam4.2019.8.issue-7
48. Huang W, Wang J, Wang H, Zhang Y, Zhao F, Li K, et al. Pet/ct based egfr mutation status classification of nsclc using deep learning features and radiomics features. Front Pharmacol. (2022) 13:898529. doi: 10.3389/fphar.2022.898529
49. Zhang M, Bao Y, Rui W, Shangguan C, Liu J, Xu J, et al. Performance of (18)F-fdg pet/ct radiomics for predicting egfr mutation status in patients with non-small cell lung cancer. Front Oncol. (2020) 10:568857. doi: 10.3389/fonc.2020.568857
50. Nair JKR, Saeed UA, McDougall CC, Sabri A, Kovacina B, Raidu BVS, et al. Radiogenomic models using machine learning techniques to predict egfr mutations in non-small cell lung cancer. Can Assoc Radiol J. (2021) 72:109–19. doi: 10.1177/0846537119899526
51. Touvron H, Lavril Thibaut, Izacard Gautier, Martinet Xavier, Lachaux Marie-Anne, Lacroix Timothee, et al. LLaMA: open and efficient foundation language models. arXiv. (2023). doi: 10.48550/arXiv.2302.13971
52. Foersch S, Glasner C, Woerl AC, Eckstein M, Wagner DC, Schulz S, et al. Multistain deep learning for prediction of prognosis and therapy response in colorectal cancer. Nat Med. (2023) 29:430–9. doi: 10.1038/s41591-022-02134-1
53. Boehm KM, Khosravi P, Vanguri R, Gao J, and Shah SP. Harnessing multimodal data integration to advance precision oncology. Nat Rev Cancer. (2022) 22:114–26. doi: 10.1038/s41568-021-00408-3
54. Delzell D, Magnuson S, Peter T, Smith M, and Smith BJ. Machine learning and feature selection methods for disease classification with application to lung cancer screening image data. Front Oncol. (2019) 9:1393. doi: 10.3389/fonc.2019.01393
55. Pineda AL, Ogoe HA, Balasubramanian JB, Escareño CR, Visweswaran S, Herman JG, et al. ‘‘On predict-ing lung cancer subtypes using ‘omic’ data from tumor and tumor-adjacent histologically-normal tissue,’’. BMCCancer. (2016) 16:184. doi: 10.1186/s12885-016-2223-3
56. Chang C, Zhou S, Yu H, Zhao W, Ge Y, Duan S, et al. A clinically practical radiomics-clinical combined model based on PET/CT data and nomogram predicts EGFR mutation in lung adenocarcinoma. Eur Radiol. (2021) 31:6259–68. doi: 10.1007/s00330-020-07676-x
57. Huang L, Cao Y, Zhou F, Li J, Ren J, Zhang G, et al. Lung adenocarcinoma: development of nomograms based on PET/CT images for prediction of epidermal growth factor receptor mutation status and subtypes. Nucl Med Commun. (2022) 43:310–22. doi: 10.1097/MNM.0000000000001519
58. Hong D, Xu K, Zhang L, Wan X, and Guo Y. Radiomics signature as a predictive factor for EGFR mutations in advanced lung adenocarcinoma. Front Oncol. (2020) 10:28. doi: 10.3389/fonc.2020.00028
59. Jia TY, Xiong JF, Li XY, Yu W, Xu ZY, Cai XW, et al. Identifying EGFR mutations in lung adenocarcinoma by noninvasive imaging using radiomics features and random forest modeling. Eur Radiol. (2019) 29:4742–50. doi: 10.1007/s00330-019-06024-y
60. Jiang M, Zhang Y, Xu J, et al. Assessing EGFR gene mutation status in non-small cell lung cancer with imaging features from PET/CT. Nucl Med Commun. (2019) 40:842–9. doi: 10.1097/MNM.0000000000001043
61. Jiang X, Ren M, Shuang X, Yang H, Shi D, Lai Q, et al. Multiparametric MRI-based radiomics approaches for preoperative prediction of EGFR mutation status in spinal bone metastases in patients with lung adenocarcinoma. J Magn Reson Imaging. (2021) 54:497–507. doi: 10.1002/jmri.27579
62. Mu W, Jiang L, Zhang J, Shi Y, Gray JE, Tunali I, et al. Non-invasive decision support for NSCLC treatment using PET/CT radiomics. Nat Commun. (2020) 11:5228. doi: 10.1038/s41467-020-19116-x
63. Le NQK, Kha QH, Nguyen VH, Chen YC, Cheng SJ, and Chen CY. Machine learning-based radiomics signatures for EGFR and KRAS mutations prediction in non-small-cell lung cancer. Int J Mol Sci. (2021) 22:17. doi: 10.3390/ijms22179254
64. Liu G, Xu Z, Ge Y, Jiang B, Groen H, Vliegenthart R, et al. 3D radiomics predicts EGFR mutation, exon-19 deletion and exon-21 L858R mutation in lung adenocarcinoma. Transl Lung Cancer Res. (2020) 9:1212–24. doi: 10.21037/tlcr-20-122
65. Lu L, Sun SH, Yang H, E L, Guo P, Schwartz LH, et al. Radiomics prediction of EGFR status in lung cancer – our experience in using multiple feature extractors and the cancer imaging archive data. Tomography. (2020) 6:223–30. doi: 10.18383/j.tom.2020.00017
66. Ninomiya K, Arimura H, Chan WY, Tanaka K, Mizuno S, Muhammad Gowdh NF, et al. Robust radiogenomics approach to the identification of EGFR mutations among patients with NSCLC from three different countries using topologically invariant Betti numbers. PloS One. (2021) 16:e0244354. doi: 10.1371/journal.pone.0244354
67. Rossi G, Barabino E, Fedeli A, Ficarra G, Coco S, Russo A, et al. Radiomic detection of EGFR mutations in NSCLC. Cancer Res. (2021) 81:724–31. doi: 10.1158/0008-5472.CAN-20-0999
68. Li S, Ding C, Zhang H, Song J, and Wu L. Radiomics for the prediction of EGFR mutation subtypes in non-small cell lung cancer. Med Phys. (2019) 46:4545–52. doi: 10.1002/mp.v46.10
69. Tu W, Sun G, Fan L, Wang Y, Xia Y, Guan Y, et al. Radiomics signature: a potential and incremental predictor for EGFR mutation status in NSCLC patients, comparison with CT morphology. Lung Cancer. (2019) 132:28–35. doi: 10.1016/j.lungcan.2019.03.025
70. Weng Q, Hui J, Wang H, Lan C, Huang J, Zhao C, et al. Radiomic feature-based nomogram: a novel technique to predict EGFR-activating mutations for EGFR tyrosin kinase inhibitor therapy. Front Oncol. (2021) 11:590937. doi: 10.3389/fonc.2021.590937
71. Ruan D, Fang J, and Teng X. Efficient 18f-fluorodeoxyglucose positron emission tomography/computed tomography-based machine learning model for predicting epidermal growth factor receptor mutations in non-small cell lung cancer. Q J Nucl Med Mol Imaging. (2022) 68:70–83. doi: 10.23736/S1824-4785.22.03441-0
72. Shiri I, Amini M, Nazari M, Hajianfar G, Haddadi Avval A, Abdollahi H, et al. Impact offeature harmonization on radiogenomics analysis: prediction of egfr and kras mutations from non-small cell lung cancer pet/ct images. Comput Biol Med. (2022) 142:105230. doi: 10.1016/j.compbiomed.2022.105230
73. Naimi AI and Balzer LB. Stacked generalization: an introduction to super learning. Eur J Epidemiol. (2018) 33:459–64. doi: 10.1007/s10654-018-0390-z
74. Papadimitroulas P, Brocki L, Christopher Chung N, MarChadour W, Vermet F, Gaubert L, et al. Artificial intelligence: deep learning in oncological radiomics and challenges of interpretability and data harmonization. Physica Med PM. (2021) 83:108–21. doi: 10.1016/j.ejmp.2021.03.009
75. Mahajan A, Chakrabarty N, Majithia J, Ahuja A, Agarwal U, Suryavanshi S, et al. Multisystem imaging recommendations/guidelines: in the pursuit of precision oncology. Indian J Med Paediatr Oncol. (2023) 44:2–25. doi: 10.1055/s-0043-1761266
76. Xiao Z, Cai H, Wang Y, Cui R, Huo L, Lee EY, et al. Deep learning for predicting epidermal growth factor receptor mutations of non-small cell lung cancer on pet/ct images. Quant Imaging Med Surg. (2023) 13:1286–99. doi: 10.21037/qims-22-760
77. Le VH, Minh TNT, Kha QH, and Le NQK. Deep learning radiomics for survival prediction in non-small-cell lung cancer patients from CT images. J Med Syst. (2025)49:22. doi: 10.1007/s10916-025-02156-5
78. Le NQK. Hematoma expansion prediction: still navigating the intersection of deep learning and radiomics. Eur Radiol. (2024) 34:2905–7. doi: 10.1007/s00330-024-10586-x
79. Nguyen HS, Ho DKN, Nguyen NN, Tran HM, Tam KW, and Le NQK. Predicting EGFR mutation status in non-small cell lung cancer using artificial intelligence: A systematic review and meta-analysis. Acad Radiol. (2024) 31:660–83. doi: 10.1016/j.acra.2023.03.040
80. Wang S, Shi J, Ye Z, Dong D, Yu D, Zhou M, et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J. (2019) 53. doi: 10.1183/13993003.00986-2018
81. Wang S, Yu H, Gan Y, Wu Z, Li E, Li X, et al. Mining whole-lung information by artificial intelligence for predicting EGFR genotype and targeted therapy response in lung cancer: a multicohort study. Lancet Digit Health. (2022) 4:e309–e319. doi: 10.1016/S2589-7500(22)00024-3
82. Gui D, Song Q, Song B, Li H, Wang M, Min X, et al. AIR-Net: A novel multi-task learning method with auxiliary image reconstruction for predicting EGFR mutation status on CT images of NSCLC patients. Comput Biol Med. (2022) 141:105157. doi: 10.1016/j.compbiomed.2021.105157
83. Wang C, Ma J, Shao J, Zhang S, Li J, Yan J, et al. Non-invasive measurement using deep learning algorithm based on multi-source features fusion to predict PD-L1 expression and survival in NSCLC. Front Immunol. (2022) 13:828560. doi: 10.3389/fimmu.2022.828560
84. Song Z, Liu T, Shi L, Yu Z, Shen Q, Xu M, et al. The deep learning model combining CT image and clinicopathological information for predicting ALK fusion status and response to ALK-TKI therapy in non-small cell lung cancer patients. Eur J Nucl Med Mol Imaging. (2021) 48:361–71. doi: 10.1007/s00259-020-04986-6
85. Tian P, He B, Mu W, Liu K, Liu L, Zeng H, et al. Assessing PD-L1 expression in non-small cell lung cancer and predicting responses to immune checkpoint inhibitors using deep learning on computed tomography images. Theranostics. (2021) 11:2098–107. doi: 10.7150/thno.48027
86. Wang C, Ma J, Shao J, Zhang S, Liu Z, Yu Y, et al. Predicting EGFR and PD-L1 status in NSCLC patients using multitask AI system based on CT images. Front Immunol. (2022) 13:813072. doi: 10.3389/fimmu.2022.813072
87. Zhang T, Xu Z, Liu G, Jiang B, de Bock GH, Groen HJM, et al. Simultaneous identification of EGFR,KRAS,ERBB2, and TP53 mutations in patients with non-small cell lung cancer by machine learning-derived three-dimensional radiomics. Cancers (Basel). (2021) 13. doi: 10.3390/cancers13081814
88. Zhao Y, Xiong S, Ren Q, Wang J, Li M, Yang L, et al. Deep learning using histological images for gene mutation prediction in lung cancer: a multicentre retrospective study. Lancet Oncol. (2025) 26:136–46. doi: 10.1016/S1470-2045(24)00599-0
89. Song J, Ding C, Huang Q, Luo T, Xu X, Chen Z, et al. Deep learning predicts epidermal growth factor receptor mutation subtypes in lung adenocarcinoma. Med Phys. (2021) 48:7891–9. doi: 10.1002/mp.15307
90. Zhang X, Zhang G, Qiu X, Yin J, Tan W, Yin X, et al. Exploring non-invasive precision treatment in non-small cell lung cancer patients through deep learning radiomics across imaging features and molecular phenotypes. biomark Res. (2024)12:12. doi: 10.1186/s40364-024-00561-5
91. Dong Y, Hou L, Yang W, Han J, Wang J, Qiang Y, et al. Multi-channel multi-task deep learning for predicting EGFR and KRAS mutations of non-small cell lung cancer on CT images. Quant Imaging Med Surg. (2021) 11:2354–75. doi: 10.21037/qims-20-600
92. Kim S, Lim JH, Kim CH, Roh J, You S, Choi JS, et al. Deep learning-radiomics integrated noninvasive detection of epidermal growth factor receptor mutations in non-small cell lung cancer patients. Sci Rep. (2024) 14:922. doi: 10.1038/s41598-024-51630-6
93. Wang C, Xu X, Shao J, Zhou K, Zhao K, He Y, et al. Deep learning to predict EGFR mutation and PD-L1 expression status in non-small-cell lung cancer on computed tomography images. J Oncol. (2021) 2021:5499385. doi: 10.1155/2021/5499385
94. Mahajan A, Kania V, Agarwal U, Ashtekar R, Shukla S, Patil VM, et al. Deep-learning-based predictive imaging biomarker model for EGFR mutation status in non-small cell lung cancer from CT imaging. Cancers (Basel). (2024) 16:1130. doi: 10.3390/cancers16061130
95. Wang S, Yu H, Gan Y, Wu Z, Li E, Li X, et al. Mining whole-lung information by artificial intelligence for predicting EGFR genotype and targeted therapy response in lung cancer: a multicohort study. Lancet Digit Health. (2022) 4:e309-e319. doi: 10.1016/S2589-7500(22)00024-3
96. Wu J, Meng H, Zhou L, Wang M, Jin S, Ji H, et al. Habitat radiomics and deep learning fusion nomogram to predict EGFR mutation status in stage I non-small cell lung cancer: a multicenter study. Sci Rep. (2024) 14:15877. doi: 10.1038/s41598-024-66751-1
97. Ashayeri H, Sobhi N, Pławiak P, Pedrammehr S, Alizadehsani R, Jafarizadeh A, et al. Transfer learning in cancer genetics, mutation detection, gene expression analysis, and syndrome recognition. Cancers (Basel). (2024) 16:2138. doi: 10.3390/cancers16112138
98. Lococo F, Ghaly G, Flamini S, Campanella A, Chiappetta M, Bria E, et al. Artificial intelligence applications in personalizing lung cancer management: state of the art and future perspectives. J Thorac Dis. (2024) 16:7096–110. doi: 10.21037/jtd-24-244
99. Wu G, Jochems A, Refaee T, Ibrahim A, Yan C, Sanduleanu S, et al. Structural and functional radiomics for lung cancer. Eur J Nucl Med Mol Imaging. (2021) 48:3961–74. doi: 10.1007/s00259-021-05242-1
100. Zhang J, Yao H, Lai C, Sun X, Yang X, Li S, et al. A novel multimodal prediction model based on DNA methylation biomarkers and low-dose computed tomography images for identifying early-stage lung cancer. Chin J Cancer Res. (2023) 35:511–25. doi: 10.21147/j.issn.1000-9604.2023.05.08
101. Tong S, Huang K, Xing W, Chu Y, Nie C, Ji L, et al. Unveiling the distinctive variations in multi-omics triggered by TP53 mutation in lung cancer subtypes: An insight from interaction among intratumoral microbiota, tumor microenvironment, and pathology. Comput Biol Chem. (2024) 113:108274. doi: 10.1016/j.compbiolchem.2024.108274
102. Wu S, Shen G, Mao J, and Gao B. CT radiomics in predicting EGFR mutation in non-small cell lung cancer: a single institutional study. Front Oncol. (2020) 10:542957. doi: 10.3389/fonc.2020.542957
103. Lu MY, Chen TY, Williamson DFK, Zhao M, Shady M, Lipkova J, et al. AI-based pathology predicts origins for cancers of unknown primary. Nature. (2021) 594:106–10. doi: 10.1038/s41586-021-03512-4
104. Jee J, Fong C, Pichotta K, Tran TN, Luthra A, Waters M, et al. Automated real-world data integration improves cancer outcome prediction. Nature. (2024) 636:728–36. doi: 10.1038/s41586-024-08167-5
105. Xiang J, Wang X, Zhang X, Xi Y, Eweje F, Chen Y, et al. A vision-language foundation model for precision oncology. Nature. (2025) 638(8051):769–78. doi: 10.1038/s41586-024-08378-w
106. Ai X, Jia B, He Z, Zhang J, Zhuo M, Zhao J, et al. Noninvasive early identification of durable clinical benefit from immune checkpoint inhibition: a prospective multicenter study (NCT04566432). Signal Transduct Target Ther. (2024) 9:350. doi: 10.1038/s41392-024-02060-3
107. Shao J, Ma J, Yu Y, Zhang S, Wang W, Li W, et al. A multimodal integration pipeline for accurate diagnosis, pathogen identification, and prognosis prediction of pulmonary infections. Innovation (Camb). (2024) 5:100648. doi: 10.1016/j.xinn.2024.100648
108. Lee JE, Park KS, Kim YH, Song HC, Park B, and Jeong YJ. Lung cancer staging using chest CT and FDG PET/CT free-text reports: comparison among three chatGPT large language models and six human readers of varying experience. AJR Am J Roentgenol. (2024) 223:e2431696. doi: 10.2214/AJR.24.31696
109. Kim S, Jang S, Kim B, Sunwoo L, Kim S, Chung JH, et al. Automated pathologic TN classification prediction and rationale generation from lung cancer surgical pathology reports using a large language model fine-tuned with chain-of-thought: algorithm development and validation study. JMIR Med Inform. (2024) 12:e67056. doi: 10.2196/67056
110. Park HJ, Huh JY, Chae G, and Choi MG. Extraction of clinical data on major pulmonary diseases from unstructured radiologic reports using a large language model. PloS One. (2024) 19:e0314136. doi: 10.1371/journal.pone.0314136
111. Yamagishi Y, Nakamura Y, Hanaoka S, and Abe O. Large language model approach for zero-shot information extraction and clustering of Japanese radiology reports: algorithm development and validation. JMIR Cancer. (2025) 11:e57275. doi: 10.2196/57275
112. Singhal K, Azizi S, Tu T, Mahdavi SS, Wei J, Chung HW, et al. Large language models encode clinical knowledge. Nature. (2023) 620:172–80. doi: 10.1038/s41586-023-06291-2
113. Kefeli J and Tatonetti N. Generalizable and automated classification of TNM stage from pathology reports with external validation. medRxiv. (2023) 15:8916. doi: 10.1101/2023.06.26.23291912
114. Lu MY, Chen B, Williamson DFK, Chen RJ, Zhao M, Chow AK, et al. A multimodal generative AI copilot for human pathology. Nature. (2024) 634:466–73. doi: 10.1038/s41586-024-07618-3
115. Wang X, Zhao J, Marostica E, Yuan W, Jin J, Zhang J, et al. A pathology foundation model for cancer diagnosis and prognosis prediction. Nature. (2024) 634:970–8. doi: 10.1038/s41586-024-07894-z
116. Liu X, Liu H, Yang G, Jiang Z, Cui S, Zhang Z, et al. A generalist medical language model for disease diagnosis assistance. Nat Med. (2025) 31:932–42. doi: 10.1038/s41591-024-03416-6.
117. Wu SH, Tong WJ, Li MD, Hu HT, Lu XZ, Huang ZR, et al. Collaborative enhancement of consistency and accuracy in US diagnosis of thyroid nodules using large language models. Radiology. (2024) 310:e232255. doi: 10.1148/radiol.232255
118. Song M, Wang J, Yu Z, Wang J, Yang L, Lu Y, et al. PneumoLLM: Harnessing the power of large language model for pneumoconiosis diagnosis. Med image Anal. (2024) 97:103248. doi: 10.1016/j.media.2024.103248
119. Zhao W, Wu Y, Xu Y, Sun Y, Gao P, Tan M, et al. The potential of radiomics nomogram in non- invasively prediction of epidermal growth factor receptor mutation status and subtypes in lung adenocarcinoma. Front Oncol. (2019) 9:1485. doi: 10.3389/fonc.2019.01485
120. Zhu Y, Guo YB, Xu D, Zhang J, Liu ZG, Wu X, et al. A computed tomography (CT)-derived radiomics approach for predicting primary co-mutations involving TP53 and epidermal growth factor receptor (EGFR) in patients with advanced lung adenocarcinomas (LUAD). Ann Transl Med. (2021) 9:545. doi: 10.21037/atm-20-6473
121. Acosta JN, Falcone GJ, Rajpurkar P, and Topol EJ. Multimodal biomedical AI. Nat Med. (2022) 28:1773–84. doi: 10.1038/s41591-022-01981-2
122. Warnat-Herresthal S, Schultze H, Shastry KL, Manamohan S, Mukherjee S, Garg V, et al. Swarm Learning for decentralized and confidential clinical machine learning. Nature. (2021) 594:265–70. doi: 10.1038/s41586-021-03583-3
123. Thomasian NM, Kamel IR, and Bai HX. Machine intelligence in non-invasive endocrine cancer diagnostics. Nat Rev Endocrinol. (2022) 18:81–95. doi: 10.1038/s41574-021-00543-9
124. London AJ. Artificial intelligence and black-box medical decisions: accuracy versus explainability. Hastings Cent Rep. (2019) 49:15–21. doi: 10.1002/hast.2019.49.issue-1
125. Zhao T, Chiang ZD, Morriss JW, LaFave LM, Murray EM, Del Priore I, et al. Spatial genomics enables multi-modal study of clonal heterogeneity in tissues. Nature. (2022) 601:85–91. doi: 10.1038/s41586-021-04217-4
126. Pich O, Bailey C, Watkins TBK, Zaccaria S, Jamal-Hanjani M, and Swanton C. The translational challenges of precision oncology. Cancer Cell. (2022) 40:458–78. doi: 10.1016/j.ccell.2022.04.002
127. Ricci Lara MA, Echeveste R, and Ferrante E. Addressing fairness in artificial intelligence for medical imaging. Nat Commun. (2022) 13:4581. doi: 10.1038/s41467-022-32186-3
128. Perez-Lopez R, Ghaffari Laleh N, Mahmood F, and Kather JN. A guide to artificial intelligence for cancer researchers. Nat Rev Cancer. (2024) 24:427–41. doi: 10.1038/s41568-024-00694-7
129. Castelo-Branco L, Pellat A, Martins-Branco D, Valachis A, Derksen JWG, Suijkerbuijk KPM, et al. ESMO guidance for reporting oncology real-world evidence (GROW). Ann Oncol. (2023) 34:1097–112. doi: 10.1016/j.annonc.2023.10.001
130. Huang X, Rymbekova A, Dolgova O, Lao O, and Kuhlwilm M. Harnessing deep learning for population genetic inference. Nat Rev Genet. (2024) 25:61–78. doi: 10.1038/s41576-023-00636-3
131. Saad MB, Hong L, Aminu M, Vokes NI, Chen P, Salehjahromi M, et al. Predicting benefit from immune checkpoint inhibitors in patients with non-small-cell lung cancer by CT-based ensemble deep learning: a retrospective study. Lancet Digit Health. (2023) 5:e404-e420. doi: 10.1016/S2589-7500(23)00082-1
132. Deng K, Wang L, Liu Y, Li X, Hou Q, Cao M, et al. A deep learning-based system for survival benefit prediction of tyrosine kinase inhibitors and immune checkpoint inhibitors in stage IV non-small cell lung cancer patients: A multicenter, prognostic study. EClinicalMedicine. (2022) 51:101541. doi: 10.1016/j.eclinm.2022.101541
Keywords: artificial intelligence, Non-small cell lung cancer (NSCLC), EGFR mutation, deep learning, medical imaging
Citation: Haixian L, Shu P, Zhao L, Chunfeng L and Lun L (2025) Machine learning approaches for EGFR mutation status prediction in NSCLC: an updated systematic review. Front. Oncol. 15:1576461. doi: 10.3389/fonc.2025.1576461
Received: 17 February 2025; Accepted: 19 June 2025;
Published: 10 July 2025.
Edited by:
Jinshuai Song, School of Chemistry, Zhengzhou University, ChinaReviewed by:
Nguyen Quoc Khanh Le, Taipei Medical University, TaiwanYuan Zhao, Henan University, China
Copyright © 2025 Haixian, Shu, Zhao, Chunfeng and Lun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liu Haixian, MTg3NjU2OTIzNzVAMTYzLmNvbQ==; Li Lun, bGw0MDg5MDc2NTJAMTYzLmNvbQ==
 Liu Haixian
Liu Haixian Pang Shu2,3
Pang Shu2,3 Li Lun
Li Lun