- 1Department of Hepatobiliary and Pancreatic Surgery, The Third Affiliated Hospital of Chongqing Medical University, Chongqing, China
- 2Department of Pharmacy, The Third Affiliated Hospital of Chongqing Medical University, Chongqing, China
- 3Department of Pharmacology, College of Pharmacy, Chongqing Medical University, Chongqing, China
- 4Department of Nephrology, The Third Affiliated Hospital of Chongqing Medical University, Chongqing, China
- 5Department of Medicine Huddinge, Karolinska Institute, Stockholm, Sweden
Background: Pancreatic cancer (PC) is a highly malignant type of cancer characterized by poor prognosis and high mortality rate. The role of gap junction protein beta 4 (GJB4) in PC has been scarcely reported. Therefore, the aim of this study was to investigate the function of GJB4 in PC cells and its potential as a therapeutic target.
Methods: The expression of GJB4 in patients with PC was examined using data from The Cancer Genome Atlas (TCGA), The Human Protein Atlas, Gene Expression Omnibus (GEO), Tumor Immune Estimation Resource (TIMER), microRNA Target Prediction Database (miRDB), and Encyclopedia of RNA Interactomes (ENCORI). A short hairpin-RNA lentiviral vector was employed to downregulate GJB4 in PC cells. The effects on tumor cell proliferation, invasion, and metastasis were assessed using cell proliferation assays, Transwell migration assays, wound healing assays, and subcutaneous xenograft models. Bioinformatics analysis was conducted to explore the function of GJB4 in PC cells. Changes in ferroptosis in PC cells following GJB4 downregulation were detected by immunofluorescence staining, transmission electron microscopy, and western blotting.
Results: GJB4 expression was found to be upregulated in PC tissues and positively correlated with patient survival. The expression of GJB4 was related to immune cell infiltration, tumor mutational burden expression and miRNAs. Downregulation of GJB4 inhibited the proliferation, metastasis, and invasion of PC cells, as well as tumor growth in nude mouse subcutaneous xenografts. Knockdown of GJB4 in PC cells disrupted oxidative and iron ion balance, and promoted ferroptosis, thereby inhibiting tumorigenic properties.
Conclusion: The findings of this study indicate that downregulation of GJB4 may suppress tumor characteristics by promoting ferroptosis in PC cells. Therefore, GJB4 may be a promising therapeutic target for the treatment of PC.
1 Introduction
Pancreatic cancer (PC), a highly malignant type of cancer, is characterized by poor prognosis and high mortality rates (1). Despite early detection, prompt diagnosis, and standardized treatment, the probability of survival for patients with PC remains dismally low (2). This underscores the critical need for thorough research into the mechanisms underlying the development and progression of the disease to discover potential therapeutic targets.
Ferroptosis is a unique form of cell death, different from apoptosis, and is marked by oxidative damage to cell membranes (3). This process is initiated by perturbations in iron homeostasis, leading to the accumulation of ferrous ions (Fe2+) with potent oxidative capabilities (4). These ions trigger iron-dependent Fenton reactions, producing high levels of reactive oxygen species (ROS) inside cells (2). Subsequently, ROS cause oxidative damage to cell membranes, significantly increasing phospholipid hydroperoxides, particularly lipid hydroperoxides (5–7). The accumulation of lipid hydroperoxides within the cell ultimately leads to ferroptosis. Recent research indicates that inducing ferroptosis in tumor cells may be a promising strategy for improving the effectiveness of cancer drugs (8, 9). However, the use of ferroptosis in treating PC is still not widely established.
Gap junction proteins (connexins), encoded by the GJB gene family, comprise 21 homologs (GJB1-21). These are highly sensitive to microenvironmental alterations, which can significantly modulate their expression patterns (10–12). Given the well-established influence of the tumor microenvironment on cancer progression (13) and the critical role of gap junctions in facilitating intercellular communication – both among tumor cells and between tumor and immune cells (14) – connexins have emerged as important regulators in oncology.
The purpose of this study was to further elucidate the role of GJB4 in PC and to investigate the functional role of GJB4 and its underlying molecular mechanisms in ferroptosis. Our findings contribute to the understanding of the mechanisms underlying the potential of GJB4 as a therapeutic target for PC.
2 Materials and methods
2.1 Cell culture
The human PC cell lines PANC-1 (Catalog number: CL-0184, hereinafter referred to as PANC) and HPAC (Catalog number: CL-0361) were purchased from Wuhan Procell Life Science and Technology Co., Ltd. PANC cells were cultured in Dulbecco’s Modified Eagle Medium (Procell PM150210) containing 10% calf serum. HPAC was cultured in medium (Procell CM-0361) for HPAC cells.
2.2 Analysis of GJB4 expression in single-cell sequencing
The data were downloaded from the Gene Expression Omnibus (GEO) dataset (GSE154778) (15). Quality control and reduction of dimensionality were performed with R package Seurat (16). SingleR was used to annotate cell groups (17).
2.3 Immune cell infiltration investigation
Tumor Immune Estimation Resource (TIMER) (http://timer.cistrome.org/) was used to analyze the relationship between immune cells and GJB4 expression (18).
2.4 Tumor mutational burden expression with GJB4
R package maftools was loaded to map the waterfall plots of gene mutations in the high and low GJB4 expression groups, respectively (19). The data were downloaded from The Cancer Genome Atlas (TCGA) dataset by R package TCGAbiolinks (20).
2.5 Competing endogenous RNA network of GJB4
The relevant miRNAs were predicted by miRDB (https://mirdb.org/). The ENCORI database (https://rnasysu.com/encori/index.php) was used to analyze the correlation between miRNAs and long noncoding RNAs with GJB4 in pancreatic adenocarcinoma (PAAD) (21).
2.6 Steady knockdown of GJB4
A lentiviral system was used to knock down GJB4. The GJB4 knockdown sequence, shGJB4: 5’-GGGATTATGACATGCCCCGCG-3’, and negative control sequence, NC: 5’-GACTCGGCGTGCGCTGCATAA-3’, were inserted into the backbone plasmid pLVX-shRNA1 separately. The 293T cells were co-transfected with the backbone plasmid, pMD2.G, and psPAX2. Cell supernatants were collected and used to transfect PANC or HPAC cells. After transfection, puromycin was used to screen stable cell lines.
2.7 Propidium iodide assay
Cells were seeded into 24-well plates at a density of approximately 1×104 cells per well. The next day, the medium was aspirated. Cells were washed once in 1× phosphate buffered saline (PBS) and incubated in 500 nM PI dye (Beyotime; ST511) at 37°C for 20 min. After removing the PI dye, the cells were washed thrice in PBS, fixed using 5% paraformaldehyde, and stained with 4’,6-diamidino-2-phenylindole (DAPI). Subsequently, images were captured by a fluorescence microscope (Nikon, Japan).
2.8 Calcein-acetoxymethyl assay
Calcein-AM solutions (Beyotime; C2013S) were incubated at 37°C for 30 min in the dark according to the instructions provided by the manufacturer. Images were captured by fluorescence microscopy (Nikon, Japan).
2.9 ROS assay
Cells were seeded into 24-well plates at a density of approximately 1×104 cells per well. The next day, the medium was removed. The cells were washed once in 1×PBS, incubated in dihydroethidium (DHE) or dichloro-dihydro-fluorescein-diacetate (DCFH-DA) (10 μM) at 37°C for 20 min, and washed thrice in 1×PBS. Images were captured by fluorescence microscopy (Nikon, Japan).
2.10 Iron assay
The iron concentration in the cells was assessed using the Iron Content Determination Kit (Sigma–Aldrich LLC, Shanghai, China; MAK025) according to the instructions provided by the manufacturer. Cells (2×106) were rapidly homogenized in iron test buffer and centrifuged to remove insoluble material. Samples (30 μL) were added to a 96-well plate and incubated with 5 μL of iron assay buffer (ferrous iron) or iron reducing agent (total iron) at 25°C to determine ferrous or total iron content, respectively. After 30 min, iron probe (100 μL) was added to each well, and the plate was incubated at 25°C for 60 min. Finally, the absorbance was determined with a microplate reader (BioTek, USA) at 593 nm.
2.11 Malondialdehyde assay
MDA content in cells was detected using the Lipid Peroxide MDA Assay Kit (Beyotime; S0131S). The test was performed according to the instructions provided by the manufacturer, and the absorbance was measured at 532 nm with a microplate reader.
2.12 Western blotting analysis
Cells were lysed using RIPA (Beyotime; P0013B) to obtain total cell protein. The protein concentration was determined by the BCA Protein Concentration Assay Kit (Beyotime; P0010). After mixing with 5× SDS-PAGE Sample Loading Buffer, samples were denatured at 100°C for 10 min. Next, the proteins were separated by electrophoresis in sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride (Millipore, Billerica, MA, USA) membranes. After blocking with 5% skim milk for 2 h at room temperature, the samples were incubated overnight at 4°C with the following primary antibodies purchased from Abmart: GJB4 (PS07587) and Proteintech: ferritin heavy chain (FTH; 11682-1-AP), transferrin receptor (TfRC; 65236-1-Ig), ferroportin (Fpn; 26601-1-AP), Cystine/glutamate transporter (xCT; 26864-1-AP), glutathione peroxidase 4 (GPX4; 30388-1-AP), ferroptosis suppressor protein 1 (FSP1; 20886-1-AP). Membranes were washed thrice with 1×tris-buffered saline with Tween 20 and incubated with the secondary antibody at room temperature for 2 h. Photographs were captured using the Odyssey® CLx Imaging System (LI-COR Biosciences, USA) or the Tanon 5200 luminescence imaging system.
2.13 Transmission electron microscopy imaging
Knockdown and control PANC cells were seeded in 6-cm dishes and treated with RSL3 (0.1 μM) for 24 h. Cells were fixed with 2.5% glutaraldehyde and collected. Transmission electron microscopy imaging was performed at Lilai Biomedicine Experiment Center (Chengdu, China).
2.14 Mouse xenograft model
All animal procedures performed in this study were approved by the Animal Welfare Committee of Chongqing Medical University (Chongqing, China). To establish a tumor xenograft model, 5×106 PANC cells were injected subcutaneously into the left hind leg. One week after injection, the body weight of the mice and tumor size were measured every 3 days. After 3 weeks, nude mice were executed by carbon dioxide asphyxiation, tumors were removed, and tumors were measured and weighed. Tumor tissues were embedded in paraffin and sectioned. Hematoxylin and eosin (H&E) (Hematoxylin and Eosin Staining Kit; Beyotime; C0105S) and Masson staining (Servicebio; G1006-20ML) were conducted based on the instructions provided by the manufacturer.
2.15 Statistical analysis
Statistical analyses were performed by R, version 4.4.1 and GraphPad 9.0.0. The differential expression between the two groups was analyzed by Student’s t-test, Wilcoxon signed-rank test, and Fisher’s exact test. The threshold of significant difference was set at P-values <0.05.
3 Results
3.1 High expression of GJB4 is associated with poor prognosis in patients with PC
TCGA database was used to investigate the expression of GJB4 in PC tissues. In PC, the expression of GJB4 in cancerous tissues and adjacent tissues was significantly elevated (Figure 1A). Survival analysis showed that the survival curve of patients with high expression of GJB4 was lower than that of patients with low expression (Figure 1B). This finding suggests that patients with high expression of GJB4 had a shorter survival time. We investigated the expression of GJB4 in a total of 24 tumors included in TCGA database. GJB4 expression markedly varied in different tissues; for example, low expression was detected in normal tissues, such as liver hepatocellular carcinoma, thyroid carcinoma, and kidney renal papillary cell carcinoma, while high expression was observed in PAAD compared with other tumors (Figures 1C, D). These results suggest that the use of GJB4 as a therapeutic target for PC is associated with limited side effects on the liver and thyroid gland, as well as high tissue specificity. Subsequently, the expression of GJB4 in PC tumors was investigated in The Human Protein Atlas (https://www.proteinatlas.org/) database (Figure 1E). The data showed that GJB4 was highly expressed in PC and mainly expressed in PC tumor glandular tissues. These results suggest that GJB4 is associated with pancreatic tumor tissue glands. The differential expression of GJB4 in PC is obvious compared with other tumors. This result suggests that GJB4 has research value in PC.
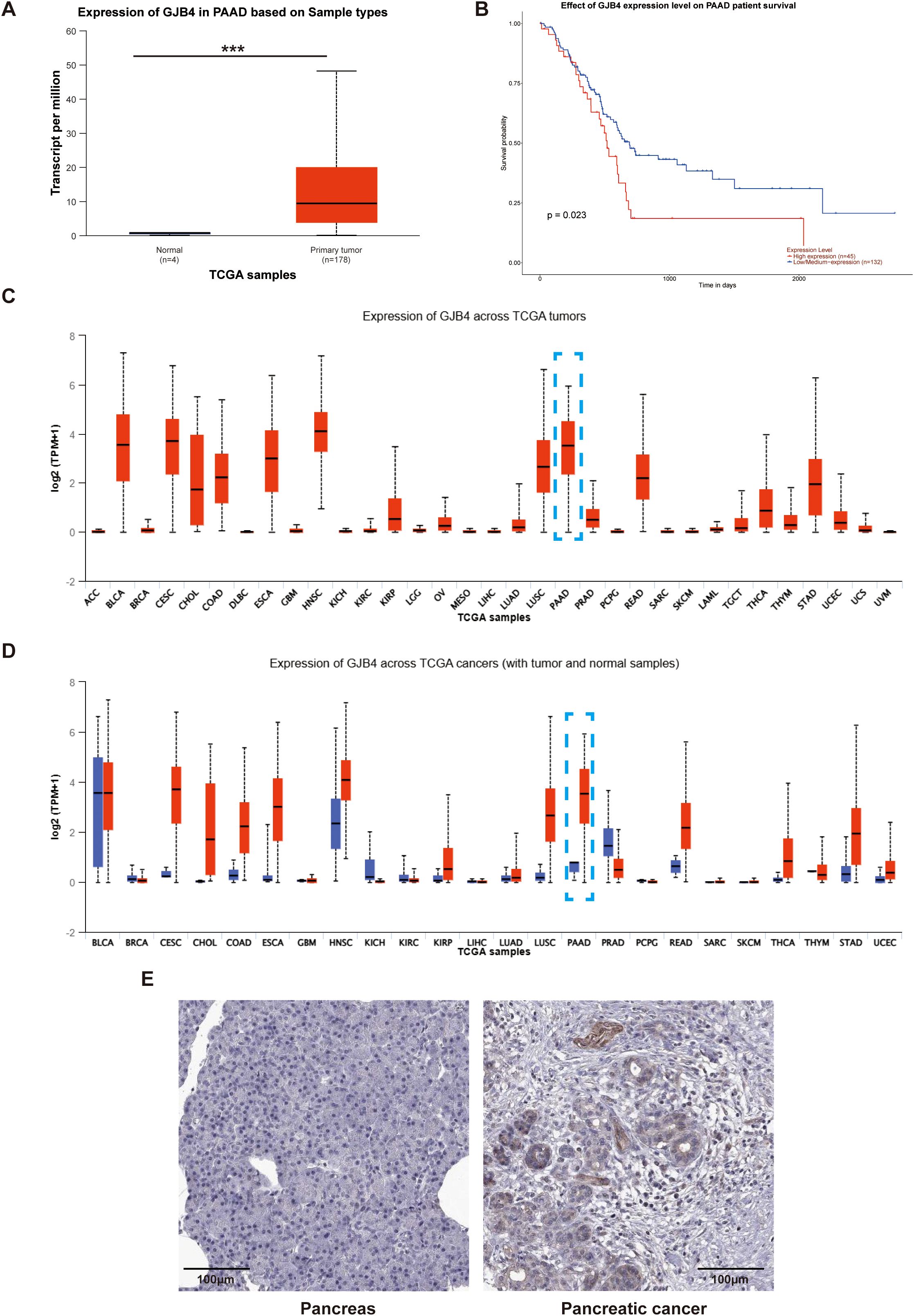
Figure 1. High expression of GJB4 is associated with poor prognosis in patients with PC. (A) GJB4 expression in PAAD in TCGA database. (B) Survival curves of patients grouped according to the expression of GJB4. (C) Expression of GJB4 in different tumors in TCGA database (blue box indicates PAAD). (D) Expression of GJB4 versus normal tissues in different tumors in TCGA database. (E) Expression of GJB4 in pancreatic cancer tumor tissue and normal pancreatic tissue in The Human Protein Atlas database. ***P < 0.001. GJB4, gap junction protein beta 4; TCGA, The Cancer Genome Atlas.
3.2 Analysis of GJB4 expression across various cell types
Single-cell sequencing data from PC cells (GSE154778) were downloaded, and SingleR was utilized for cellular type annotation. Following the processing methodology provided by the dataset developers (15), a heatmap was generated depicting the top 10 expressed genes across all cell types (Figure 2A). In accordance with the manuscript accompanying the dataset, epithelial cells were annotated as tumor cells (Figure 2B). Compared with all other cell types, GJB4 expression was observed exclusively in tumor cells (Figure 2C).
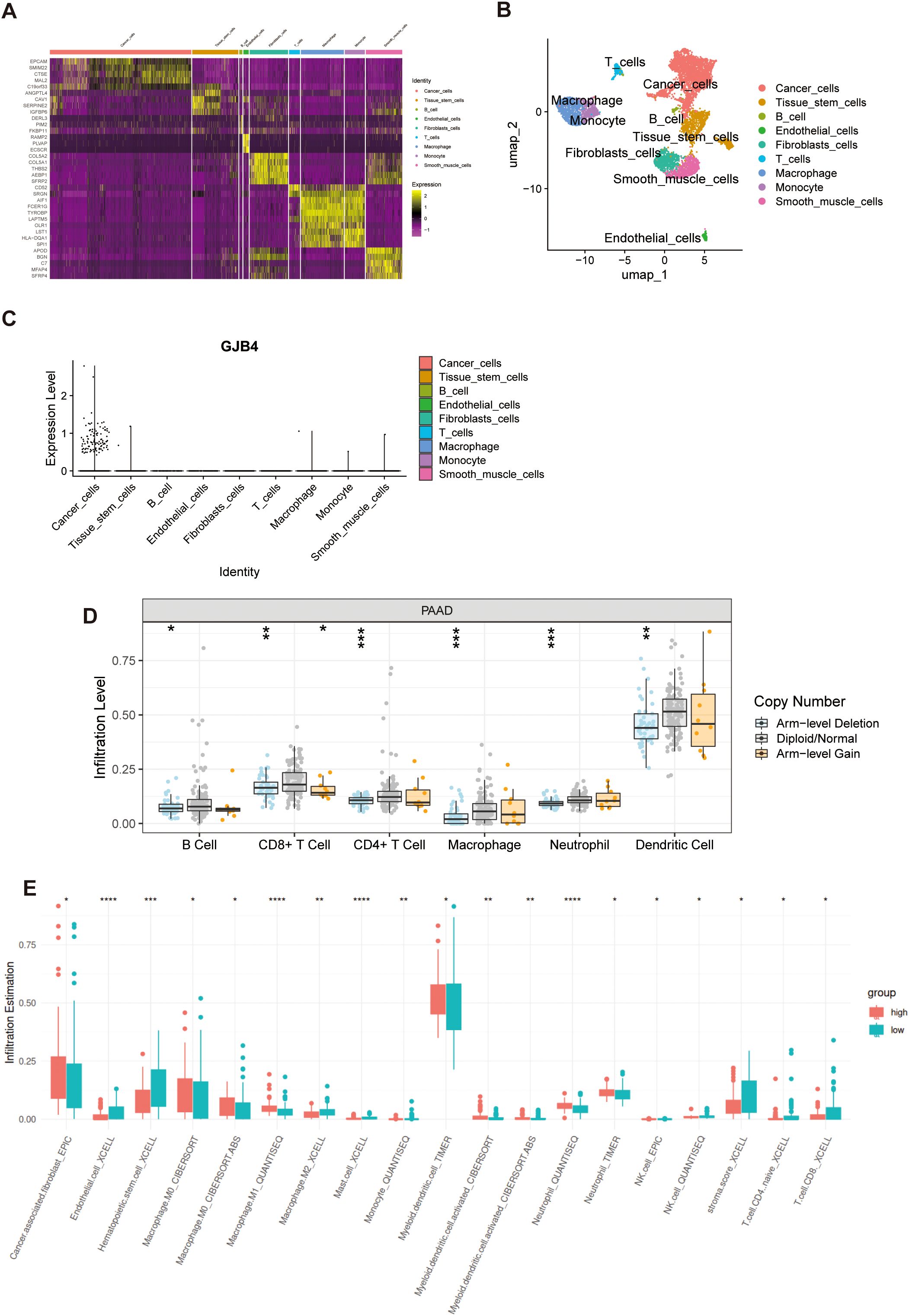
Figure 2. Single-cell and immune cell infiltration investigation of GJB4. (A) Heat map of the expression of marker genes. (B) UMAP map with cell annotations. (C) GJB4 expression in multiple cells. (D) The immune cell infiltration levels with different GJB4 CNV in PAAD. (E) Immune cell infiltration in the high and low GJB4 groups was analyzed using various computational methods. *P < 0.05, **P < 0.01, and ***P < 0.001. CNV, copy number variation; GJB4, gap junction protein beta 4; PAAD, pancreatic adenocarcinoma; GJB4, gap junction protein beta 4; TCGA, The Cancer Genome Atlas.
3.3 Immune cell infiltration based on the differential expression of GJB4
Samples in the arm-level deletion of GJB4 group showed decreased infiltration of immune cells, including B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells (Figure 2D). Samples with high expression of GJB4 showed lower immune infiltration in seven immune cell groups (endothelial, hematopoietic stem cells, M2 macrophages, mast cells, monocytes, CD4 T cells, and CD8 T cells) and higher immune infiltration in six immune groups (cancer-associated fibroblasts, M0 macrophages, M1 macrophages, myeloid dendritic cells, activated myeloid dendritic cells, and neutrophils) (Figure 2E). In addition, the sample with high GJB4 expression had lower stromal scores (Figure 2E).
3.4 Differences in TMB between high and low GJB4 expression groups
In patients with high expression of GJB4, 94.05% (79 of 84) exhibited tumor genetic mutations (Figure 3A). In contrast, among patients with low expression of GJB4, 65% (52 of 80) had genetic mutations (Figure 3B). Patients with high expression of GJB4 exhibited a higher TMB than those with low expression (Figure 3C). Specifically, within the high GJB4 expression group, 77% and 74% of patients had Ki-ras2 Kirsten rat sarcoma viral oncogene homolog gene and tumor protein p53 gene mutations, respectively. In the low GJB4 expression group, these values were 42% and 41%, respectively (Figure 3D). These differences were statistically significant (Figure 3E).
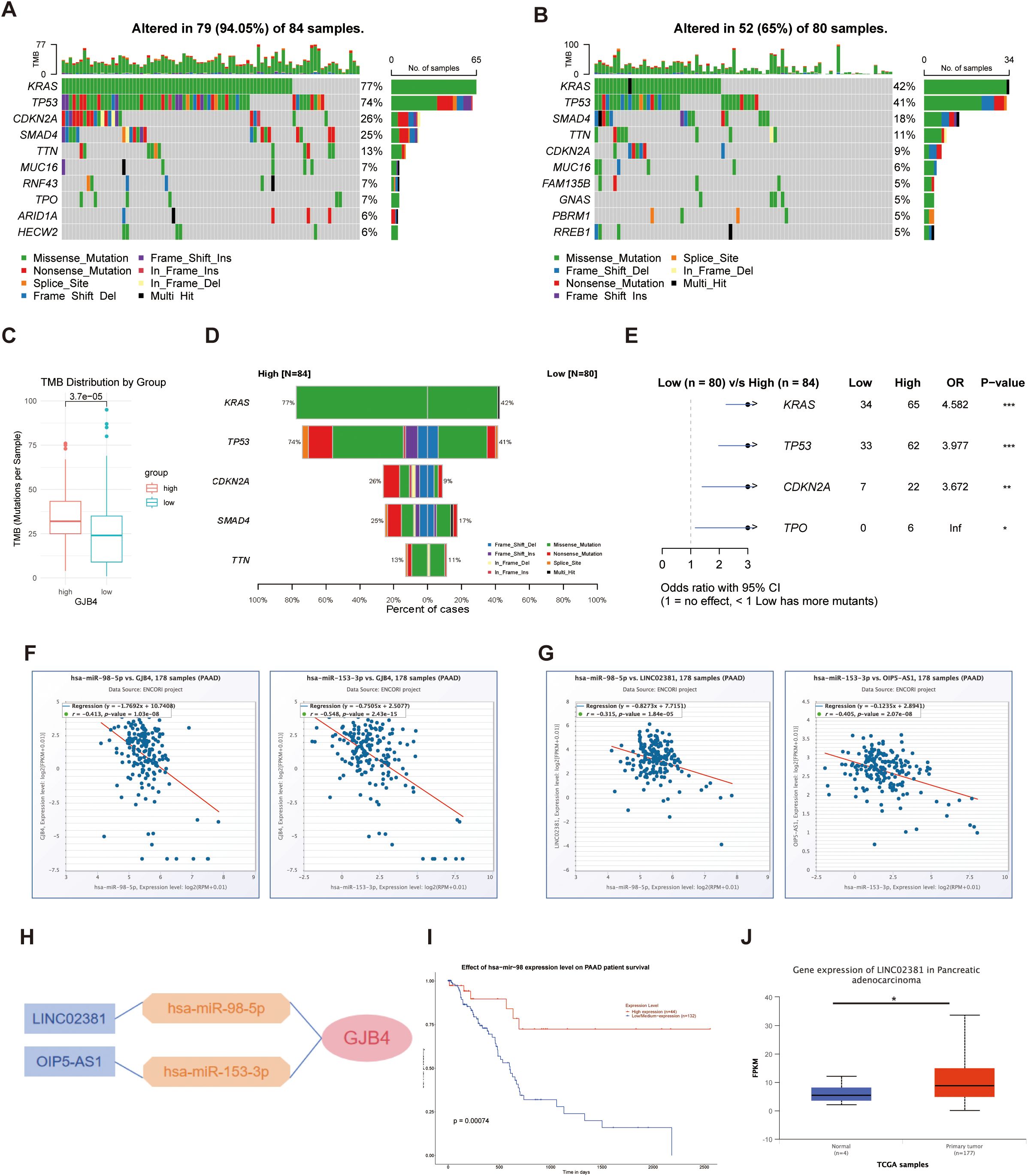
Figure 3. Investigating TMB and ceRNAs for GJB4. (A, B) Waterfall plots of genetic mutations in samples with high (A) and low (B) GJB4 expression. (C) Total TMB between the high and low GJB4 expression groups. Wilcoxon signed-rank test. (D) Comparisons between the high and low GJB4 expression groups. (E) Statistical results. Fisher’s exact test. (F) Correlation analysis between GJB4 and miRNA, has-miR-98-5p, and has-miR-153-3p. (G) Correlation analysis between has-miR-98-5p and has-miR-153-3p with lncRNA LINC02381 and OIP5-AS1, respectively. (H) The ceRNA regulation network. (I) Survival curve among patients with higher or lower has-miR-98 expression. (J) Different expression in patients with PAAD. *P < 0.05, **P < 0.01, and ***P < 0.001. GJB4, gap junction protein beta 4; TMB, tumor mutational burden, ceRNA, competing endogenous RNA; LINC02381, long intergenic non-protein coding RNA 2381; lncRNA, long noncoding RNA; OIP5-AS1, Opa interacting protein 5-antisense RNA 1; PAAD, pancreatic adenocarcinoma.
3.5 Investigating ceRNAs for GJB4
The predicted miRNAs has-miR-98-5p and has-miR-153-3p were negatively correlated with GJB4 (Figure 3F). The predicted long noncoding RNAs long intergenic non-protein coding RNA 2381 (LINC02381) and Opa interacting protein 5-antisense RNA 1 (OIP5-AS1) were negatively correlated with the expression levels of has-miR-98-5p and has-miR-153-3p, respectively (Figure 3G). The regulation network was predicted according to the co-expression relationships (Figure 3H). Among them, patients with higher has-miR-98 expression showed a better survival curve than other patients (Figure 3I), and LINC02381 was overexpressed in the PAAD group (Figure 3J).
3.6 Knockdown of GJB4 reduced the tumor phenotype of PC cells
To investigate the function of GJB4 in PC, the lentiviral systems carrying shGJB4 and NC were introduced into PANC and HPAC cell lines, and the protein expression in the shGJB4 group was detected by WB. The results confirmed the knockdown of GJB4 protein in the cells (Figures 4A, B).

Figure 4. Knockdown of GJB4 reduced the tumor phenotype of PC cells. (A) WB detection of GJB4 expression in shGJB4-transfected PC cells. (B) WB grayscale chart. (C) Wound healing assay to detect the effect of shGJB4 on the migratory ability of PC cell lines. (D) Statistical chart of the results of the rate of migration. (E) CCK-8 assay to detect the effect of shGJB4 on PC cell proliferation. (F) Transwell assay to detect changes in the migratory ability of shGJB4-transfected cells. (G) Statistics of the Transwell experiment. *P < 0.05, **P < 0.01, and ***P < 0.001. CCK-8, Cell Counting Kit-8; GJB4, gap junction protein beta 4; WB, western blotting.
Subsequently, the effect of shGJB4 on the tumor phenotype was further examined. The changes in cell viability were continuously detected for four days with Cell Counting Kit-8 assay (Figure 4E), and the cell proliferation rate was observed. The results showed that the proliferation rate of the GJB4 knockdown group was slower than that of the control group, and the difference was statistically significant from the second day of observation.
The scratch assay was used to detect the effect of shGJB4 on the migratory ability of PC cells. At 24 h after scratching, PC cells in the shGJB4 group exhibited less healing than those in the control group (Figures 4C, D). The results suggested that the migratory ability of PC cells was reduced after transfection with shGJB4 and knockdown of GJB4. The Transwell experiment also revealed that the cells transfected with shGJB4 had reduced invasiveness compared with control group cells (Figures 4F, G). These data indicate that through enhance the proliferation and migration of PC cells, GJB4 can promote pancreatic tumor characteristics.
3.7 Knockdown of GJB4 inhibits the tumor phenotype of PC in vivo
A mouse subcutaneous tumorigenesis model was used to evaluate the role of GJB4 in vivo (Figures 5A, B). The results showed that the tumor volume produced was significantly reduced in the shGJB4 group compared with the control group without statistically significant difference in body weight between the two groups (Figure 5C). Moreover, the volume and weight of shGJB4 tumors removed from the mice were reduced compared with those removed from the control group (Figure 5D). In addition, H&E staining revealed that the widely distributed gaps in the control group were reduced in the shGJB4 group (Figure 5E). To further distinguish whether these gaps are present in tumor cells or connective tissue, we performed Masson staining of tumor tissue (Figure 5F). The results showed that the cavity originally surrounded by tumor cells was significantly reduced in the shGJB4 group compared with the control group. This observation suggested that the original tumor structure of PC was destroyed after knockdown of GJB4, indicating the involvement of GJB4 in the structural formation of PC. These findings imply that knockdown of GJB4 reduces the tumor phenotype of PC in vivo.
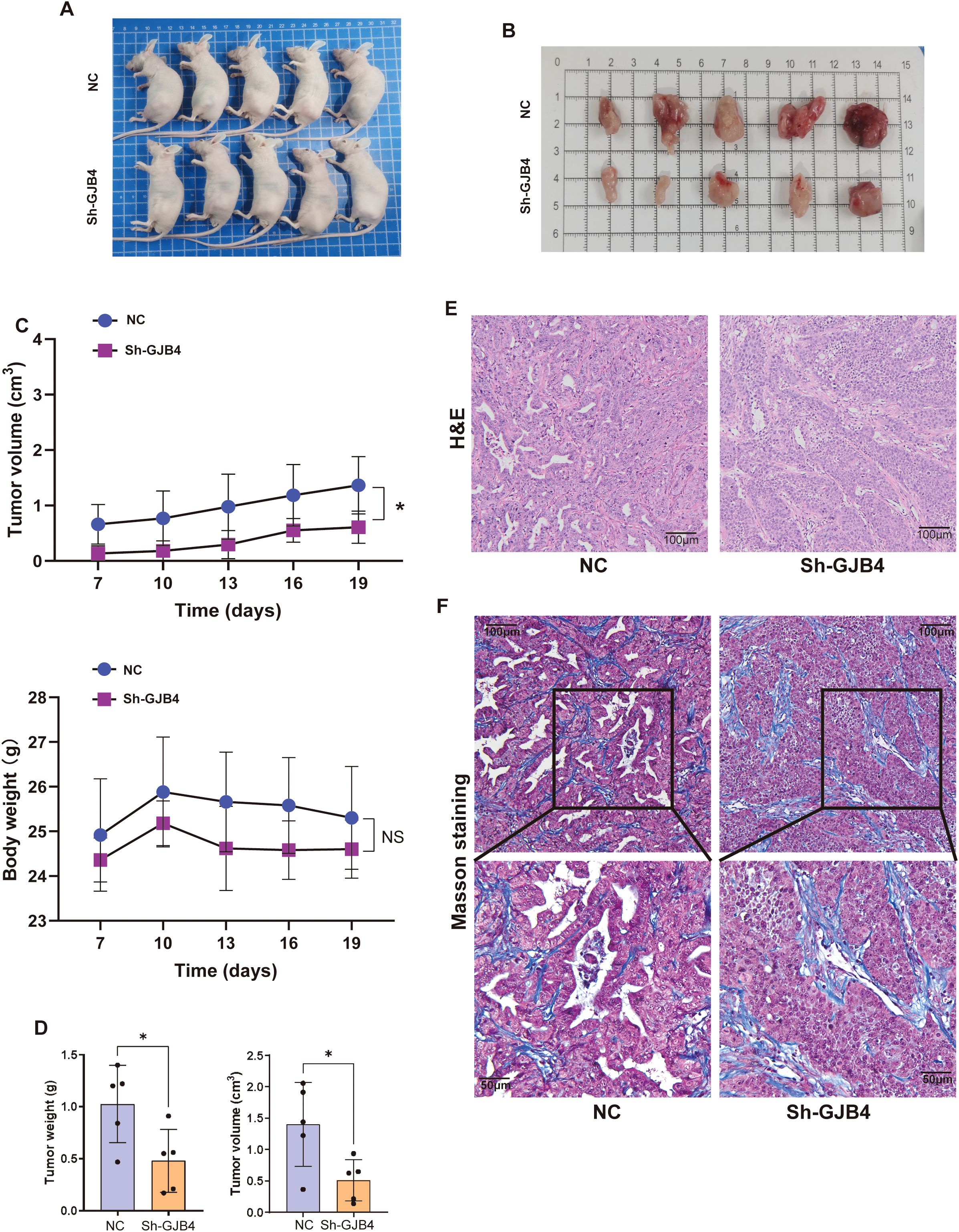
Figure 5. Knockdown of GJB4 inhibited the tumor phenotype of PC in vivo. (A) Cells transfected with shGJB4 or control vector were subcutaneously inoculated into the left flanks of nude mice. (B) Mice were sacrificed, and tumors were harvested. (C) The weight and volumes of harvested tumors were calculated. (D) The growth of tumors and body weight were measured once every 3 days. (E) H&E staining of harvested tumors. (F) Masson staining to distinguish tumor tissue from connective tissue. *P < 0.05. GJB4, gap junction protein beta 4; H&E, hematoxylin and eosin.
3.8 Bioinformatics analysis suggested that GJB4 is associated with iron metabolism in PC
To further investigate the mechanism through which GJB4 enhances tumor characteristics, we extracted the sequencing data of PAAD PC from TCGA database, divided the data into high and low expression groups based on the median expression of GJB4, and analyzed the differential gene expression between the two groups (Figure 6A). Differentially expressed genes with fold-change ≥1 and P-values <0.05 were selected for enrichment analysis (Figure 6B). In the analysis of cellular component, biological process, and molecular function (Figure 6C), we found that differentially expressed genes were enriched for multiple functions related to ion metabolism and transport, such as ion channel complex in cellular component and ion channel. Further analysis was conducted on genes extracted from UALCAN (22) that are associated with GJB4 expression. The study focused on genes with a Pearson correlation coefficient absolute value >0.3, which led to the identification of ferroptosis-related genes (as cataloged in FerrDb) (Figure 6D). We hypothesized that GJB4 may be associated with iron transport metabolism in PC cells. Recently, ferroptosis in PC has been identified as a key mechanism influencing the tumor characteristics of PC cells. Therefore, we suggest that GJB4 affects the phenotype of PC cells by regulating iron metabolism.
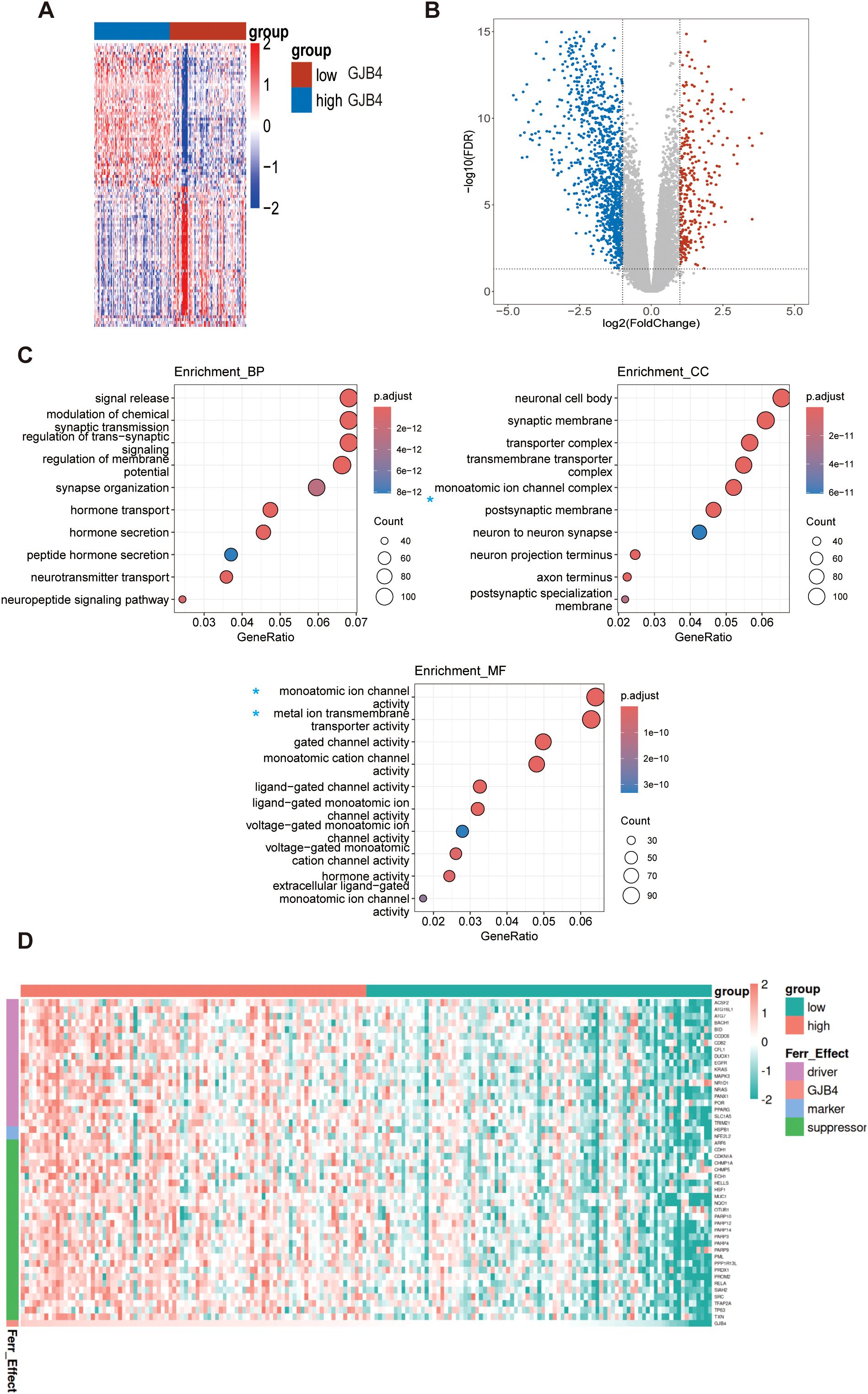
Figure 6. Bioinformatics analysis suggested that GJB4 is associated with iron metabolism in PC. (A, B) Differentially expressed gene heat maps and volcano maps of the high and low expression groups. (C) Enrichment analysis revealed that the differentially expressed genes were enriched in relation to the relevant iron metabolic pathway (blue asterisk). (D) Correlated gene heat map. A subset of genes, extracted from UALCAN and characterized by a Pearson correlation coefficient absolute value >0.3, corresponds to the ferroptosis genes recorded in FerrDb. The expression levels of these genes were utilized to construct the heat map. Driver, ferroptosis driver genes; Ferr_effect, ferroptosis effects; GJB4, gap junction protein beta 4; Marker, ferroptosis marker genes; Suppressor, ferroptosis suppressor genes.
3.9 Knockdown of GJB4 promoted cell death and disrupted the balance of ROS and metal ions in PC cells
To verify that GJB4 affects tumor cells by regulating ferroptosis in PC cells, we first examined the effect of GJB4 on PC cell death. PI staining assay was used to detect the effect of shGJB4 on cell death (Figure 7A). The results showed that the shGJB4 group had stronger fluorescence staining than the negative control group, indicating the presence of a greater number of dead cells. These results indicated that GJB4 may be involved in the regulation of cell death. Next, the concentration of endogenous ROS was detected using two immunoprobes, DHE and DCFH-DA (Figure 7B). The fluorescence intensity of DHE and DCFH-DA was higher in the shGJB group compared with the negative control group, indicating an increase in intracellular ROS concentration after knockdown of GJB4. These results suggest that GJB4 also stabilizes the role of intracellular ROS. Furthermore, the intracellular MDA content was also increased after knockdown of GJB4 (Figure 7C). Finally, the Calcein-AM was used to detect the concentration of intracellular metal ions (Figure 7D). The results showed that the fluorescence intensity of the shGJB4 group decreased, suggesting that the intracellular metal ions increased after knocking down GJB4. These results suggest that PC cell death, intracellular ROS, and metal ions increase after GJB4 knockdown, further denoting that GJB4 may regulate the tumor phenotypes of PC cells through ferroptosis.
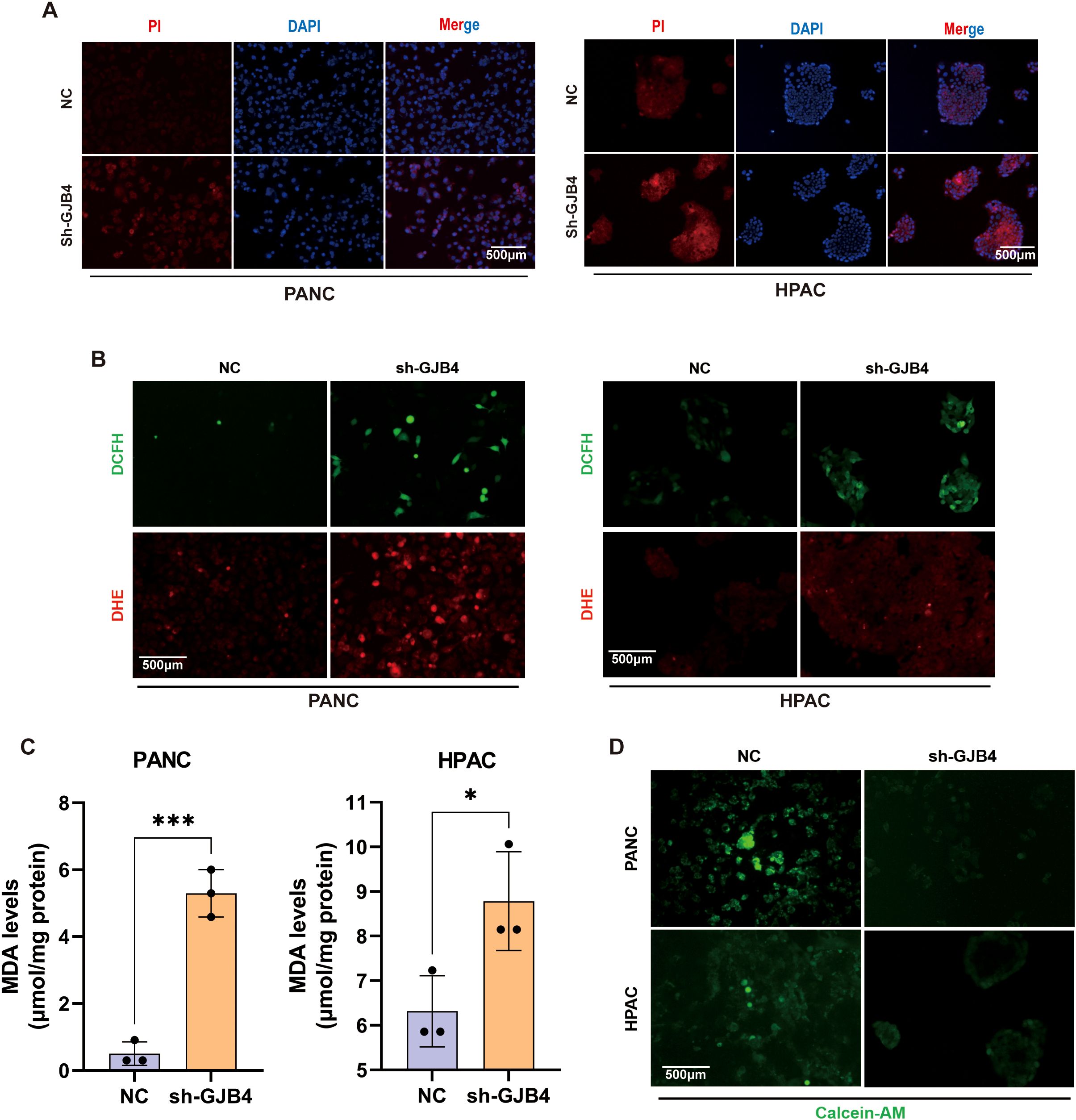
Figure 7. Knockdown of GJB4 promoted cell death and disrupted the balance of ROS and metal ions in PC cells. (A) Fluorescence intensity of PI staining was enhanced after knockdown of GJB4. (B) DCFH and DHE staining showed an increase in intracellular ROS after knockdown of GJB4. (C) MDA detection. (D) Calcein-AM staining. Knockdown of GJB4 and weakening of intracellular staining suggest that reducing the expression of GJB4 can disrupt the intracellular metal ion balance. *P < 0.05 and ***P < 0.001. Calcein-AM, Calcein-acetoxymethyl; DCFH, dichloro-dihydro-fluorescein-diacetate; DHE, dihydroethidium; GJB4, gap junction protein beta 4; MDA, malondialdehyde; PI, propidium iodide; ROS, reactive oxygen species.
3.10 Knockdown of GJB4 promotes ferroptosis in PC cells
To further validate the effect of GJB4 on ferroptosis in PC cells, intracellular iron concentrations, including total iron and ferrous ion concentrations, were measured (Figures 8A, B). The results showed that both the total ferritin and ferrous ion concentrations increased significantly after knockdown of GJB4, suggesting that GJB4 could regulate the transport of iron ions in cells.
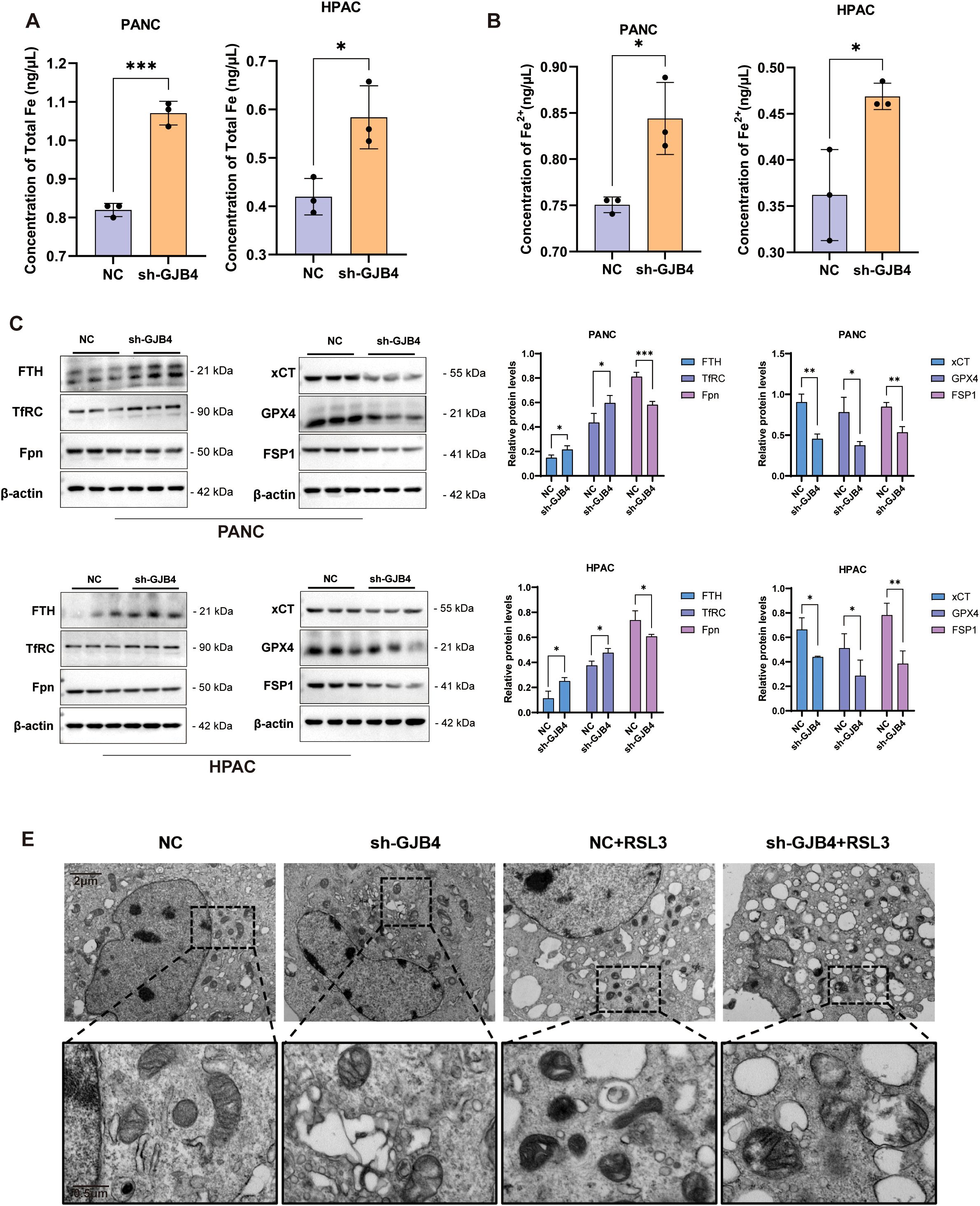
Figure 8. Knockdown of GJB4 promoted ferroptosis in PC cells. (A, B) Intracellular total iron content (A) and ferrous ion content (B) increased after knockdown of GJB4. (C) WB to detect ferritin changes. (D) Protein grayscale statistics. (E) Transmission electron microscopy to observe mitochondria within cells. *P < 0.05, **P < 0.01, and ***P < 0.001. GJB4, gap junction protein beta 4; WB, western blotting.
Subsequently, WB was used to detect the expression of iron transport-related proteins (Figures 8C, D). In the shGJB4 group, the expression of the protein FTH (which stores iron function) and the transferrin TfRC (which transfers iron from outside the cell) increased and decreased the expression of Fpn protein (which transfers iron ions out of the cell), indicating that the iron homeostasis of the cells was disrupted. WB detection of the core anti-ferroptosis protein of ferroptosis showed that the expression of xCT, GPX4, and FSP1 was reduced in the shGJB4 group, suggesting that intracellular ferroptosis increased after knockdown of GJB4.
Following the occurrence of ferroptosis, the mitochondrial volume of the cell decreases, the mitochondrial ridge decreases, the outer membrane is broken or thickened, the mitochondrial density increases, etc. (23, 24). The mitochondrial changes in PC cells PANC were detected by transmission electron microscopy, and the control cells were treated with the ferroptosis inducer RSL3 as a positive reference (Figure 8E). The results showed that the mitochondrial volume of the cells was reduced and the membrane was thickened by treatment with RSL3. In the shGJB4 group, the mitochondrial ridge became disordered; after adding RSL3, the mitochondrial was destroyed, suggesting that shGJB4 enhanced the effect of RSL3 on ferroptosis.
In summary, GJB4 knockdown can promote ferroptosis in PC cells. GJB4 maintains the tumor properties of PC cells by inhibiting ferroptosis.
4 Discussion
In this study, we found that high expression of GJB4 is associated with a poor prognosis in patients with PC. Knocking down GJB4 in PC cells inhibited the tumor phenotype of these cells. Bioinformatics analysis and subsequent experiments revealed that GJB4 was associated with iron metabolism in PC cells. Furthermore, the knockdown of GJB4 resulted in an increase in ROS and an enhanced susceptibility of PC cells to ferroptosis.
Members of the GJB family have important functions in a variety of tumors. GJB1 and GJB2 are highly expressed in glioblastoma and PC, which are closely related to chemotherapy resistance (25, 26). High expression of GJB1 and GJB2 was also associated with lymph node metastasis and prognosis in carcinoma of the pancreas (27). GJB2 has been shown to drive hepatocellular carcinoma progression by fostering an immunosuppressive tumor microenvironment (28). GJB1 enhanced the metastasis and invasion of lung cancer cells (29). GJB4, which has close relationship with tumors, increased the proliferation and metastasis of gastric cancer cells through Wnt/catenin beta 1 (Wnt/CTNNB1) (30). Moreover, it has been used as a tumor marker for lung cancer, because it promotes metastasis and chemotherapy resistance (31). In bladder cancer, GJB4 (target protein of miR-492) promoted the proliferation and metastasis of bladder cancer cells (32). In pancreatic tissue, GJB4 can increase insulin secretion (33). Consistent with our results, a recent study on GJB4 in PC also found a negative relationship with poor outcome of patients (26). Finally, our study suggests that GJB4 inhibited ferroptosis in PC cells, thereby preserving tumor properties. The present evidence enriches the understanding of the role of GJB family proteins in tumor diseases.
The treatment of tumors through the ferroptosis mechanism is increasingly being confirmed by research. For example, promoting ferroptosis in cells prevented the development of resistance to dabrafenib and trametinib during treatment, thereby enhancing the efficacy of melanoma therapy (23, 34). During immunotherapy, activated CD8+ T cells increased lipid peroxidation in tumor cells, which in turn promoted ferroptosis and enhanced the effectiveness of immunotherapy (35, 36). This approach targeted the tumor more effectively, and overcame the challenges associated with drug resistance and immune evasion (37). 2-imino-6-methoxy-2H-chromene-3-carbothioamide (IMCA) inhibited PC by inducing ferroptosis, indicating that suppression of ferroptosis pathways may be a major reason for the development of drug resistance in PC cells (38). Therefore, our findings on the molecular mechanisms of inhibition of ferroptosis in PC may provide a promising target for the treatment of PC.
GJB4 may cause ferroptosis by perturbing the tumor immune environment. In human tissue specimens, we discovered that the expression of GJB4 correlated with the location of PC glands. According to this result, the expression of GJB4 may be associated with the glandular structure. In the animal subcutaneous tumorigenesis experiment, we found that the tumor original cavity surrounded by tumor cells decreased in the GJB4 downregulation group, implying that GJB4 may be related to the tumor structure of PC. PC tumor structure is closely related to the tumor microenvironment (39). The conclusion that abnormalities in the cancer microenvironment cause ferroptosis in cancers (35, 40) hints that GJB4 caused ferroptosis in PC cells by disrupting the balance of the immune environment. In the future, we plan to investigate whether GJB4 changes tumor immunity by changing the tumor structure and altering the tumor environment.
5 Conclusions
GJB4 may be a therapeutic target for PC by regulating the molecular mechanisms in ferroptosis.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. The animal study was approved by Institutional Animal Care and Use of Chongqing medical university. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
XZ: Data curation, Formal Analysis, Funding acquisition, Software, Writing – original draft. WL: Data curation, Methodology, Writing – original draft. XL: Data curation, Writing – original draft. QY: Data curation, Writing – original draft. LZ: Resources, Writing – original draft. RF: Writing – review & editing. PB: Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the Third Affiliated Hospital of Chongqing Medical University Project (KY23042), the Chongqing Natural Science Foundation Postdoctoral Science Foundation Project (cstc2021jcyj-bshX0110), and the Chongqing Natural Science Foundation (CSTB2023NSCQ-MSX0320).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. (2016) 66:115–32. doi: 10.3322/caac.21338
2. Liu K, Huang L, Qi S, Liu S, Xie W, Du L, et al. Ferroptosis: the entanglement between traditional drugs and nanodrugs in tumor therapy. Adv Healthc Mater. (2023) 12:e2203085. doi: 10.1002/adhm.202203085
3. Chen X, Kang R, Kroemer G, and Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. (2021) 18:280–96. doi: 10.1038/s41571-020-00462-0
4. Wang D, Tang L, Zhang Y, Ge G, Jiang X, Mo Y, et al. Regulatory pathways and drugs associated with ferroptosis in tumors. Cell Death Dis. (2022) 13:544. doi: 10.1038/s41419-022-04927-1
5. Saraev DD and Pratt DA. Reactions of lipid hydroperoxides and how they may contribute to ferroptosis sensitivity. Curr Opin Chem Biol. (2024) 81:102478. doi: 10.1016/j.cbpa.2024.102478
6. Su F, Descher H, Bui-Hoang M, et al. Iron(III)-salophene catalyzes redox cycles that induce phospholipid peroxidation and deplete cancer cells of ferroptosis-protecting cofactors. Redox Biol. (2024) 75:103257. doi: 10.1016/j.redox.2024.103257
7. Wang Z, Ren X, Li Y, Qiu L, Wang D, Liu A, et al. Reactive oxygen species amplifier for apoptosis-ferroptosis mediated high-efficiency radiosensitization of tumors. ACS Nano. (2024) 18:10288–301. doi: 10.1021/acsnano.4c01625
8. Wu Y, Zhang S, Gong X, Tam S, Xiao D, Liu S, et al. The epigenetic regulators and metabolic changes in ferroptosis-associated cancer progression. Mol Cancer. (2020) 19:39. doi: 10.1186/s12943-020-01157-x
9. Zhang C, Liu X, Jin S, Chen Y, and Guo R. Ferroptosis in cancer therapy: a novel approach to reversing drug resistance. Mol Cancer. (2022) 21:47. doi: 10.1186/s12943-022-01530-y
10. Lampe PD and Laird DW. Recent advances in connexin gap junction biology. Fac Rev. (2022) 11:14. doi: 10.12703/r/11-14
11. Spray DC. Gap junction proteins: where they live and how they die. Circ Res. (1998) 83:679–81. doi: 10.1161/01.RES.83.6.679
12. Losa D, Chanson M, and Crespin S. Connexins as therapeutic targets in lung disease. Expert Opin Ther Targets. (2011) 15:989–1002. doi: 10.1517/14728222.2011.584875
13. Yuan S, Almagro J, and Fuchs E. Beyond genetics: driving cancer with the tumour microenvironment behind the wheel. Nat Rev Cancer. (2024) 24:274–86. doi: 10.1038/s41568-023-00660-9
14. Kutova OM, Pospelov AD, and Balalaeva IV. The multifaceted role of connexins in tumor microenvironment initiation and maintenance. Biol (Basel). (2023) 12:204. doi: 10.3390/biology12020204
15. Lin W, Noel P, Borazanci EH, Lee J, Amini A, Han IW, et al. Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med. (2020) 12:80. doi: 10.1186/s13073-020-00776-9
16. Hao Y, Stuart T, Kowalski MH, Choudhary S, Hoffman P, Hartman A, et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat Biotechnol. (2024) 42:293–304. doi: 10.1038/s41587-023-01767-y
17. Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. (2019) 20:163–72. doi: 10.1038/s41590-018-0276-y
18. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. (2020) 48:W509–14. doi: 10.1093/nar/gkaa407
19. Mayakonda A, Lin DC, Assenov Y, Plass C, and Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. (2018) 28:1747–56. doi: 10.1101/gr.239244.118
20. Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. (2016) 44:e71. doi: 10.1093/nar/gkv1507
21. Tambuzzi S, Gentile G, Bilardo G, Boracchi M, Bailo P, Casalino T, et al. Technical note: A comparison between rehydrating solutions in the pretreatment of mummified and corified skin for forensic microscopic examination. Int J Legal Med. (2022) 136:997–1007. doi: 10.1007/s00414-022-02833-x
22. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. (2017) 19:649–58. doi: 10.1016/j.neo.2017.05.002
23. Jiang X, Stockwell BR, and Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. (2021) 22:266–82. doi: 10.1038/s41580-020-00324-8
24. Tang D, Chen X, Kang R, and Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. (2021) 31:107–25. doi: 10.1038/s41422-020-00441-1
25. Jia Y, Guo B, Zhang W, Wang F, Zhang Y, Zhang Q, et al. Pan-cancer analysis of the prognostic and immunological role of GJB2: a potential target for survival and immunotherapy. Front Oncol. (2023) 13:1110207. doi: 10.3389/fonc.2023.1110207
26. Muramatsu J, Arihara Y, Yoshida M, Kubo T, Nakamura H, Ishikawa K, et al. Gap junction beta-4 accelerates cell cycle progression and metastasis through MET-AKT activation in pancreatic cancer. Cancer Sci. (2024) 115:1564–75. doi: 10.1111/cas.16101
27. Kyo N, Yamamoto H, Takeda Y, Ezumi K, Ngan CY, Terayama M, et al. Overexpression of connexin 26 in carcinoma of the pancreas. Oncol Rep. (2008) 19:627–31. doi: 10.3892/or.19.3.627
28. Liu H, Li X, Zhang C, Hao X, Cao Y, Wang Y, et al. GJB2 promotes HCC progression by activating glycolysis through cytoplasmic translocation and generating a suppressive tumor microenvironment based on single cell RNA sequencing. Adv Sci (Weinh). (2024) 11:e2402115. doi: 10.1002/advs.202402115
29. Elzarrad MK, Haroon A, Willecke K, Dobrowolski R, Gillespie MN, and Al-Mehdi AB. Connexin-43 upregulation in micrometastases and tumor vasculature and its role in tumor cell attachment to pulmonary endothelium. BMC Med. (2008) 6:20. doi: 10.1186/1741-7015-6-20
30. de Oliveira R, Christofolini DM, Criado PR, Lacaz Martins E, Kelsell D, and Machado Filho . Clinical variability of the GJB4:c.35G > A gene variant: a study of a large Brazilian erythrokeratodermia pedigree. Int J Dermatol. (2020) 59:722–5. doi: 10.1111/ijd.14894
31. Lin YP, Wu JI, Tseng CW, Chen HJ, and Wang LH. Gjb4 serves as a novel biomarker for lung cancer and promotes metastasis and chemoresistance via Src activation. Oncogene. (2019) 38:822–37. doi: 10.1038/s41388-018-0471-1
32. Wang K, Lü H, Qu H, Xie Q, Sun T, Gan O, et al. miR-492 promotes cancer progression by targeting GJB4 and is a novel biomarker for bladder cancer. Onco Targets Ther. (2019) 12:11453–64. doi: 10.2147/OTT.S223448
33. Gässler A, Quiclet C, Kluth O, Gottmann P, Schwerbel K, Helms A, et al. Overexpression of Gjb4 impairs cell proliferation and insulin secretion in primary islet cells. Mol Metab. (2020) 41:101042. doi: 10.1016/j.molmet.2020.101042
34. Nyakas M, Fleten KG, Haugen MH, Engedal N, Sveen C, Farstad IN, et al. AXL inhibition improves BRAF-targeted treatment in melanoma. Sci Rep. (2022) 12:5076. doi: 10.1038/s41598-022-09078-z
35. Hou CY, Lv P, Yuan HF, Zhao LN, Wang YF, Zhang HH, et al. Bevacizumab induces ferroptosis and enhances CD8(+) T cell immune activity in liver cancer via modulating HAT1 and increasing IL-9. Acta Pharmacol Sin. (2024) 45:1951–63. doi: 10.1038/s41401-024-01299-4
36. Lin Z, Zou S, and Wen K. The crosstalk of CD8+ T cells and ferroptosis in cancer. Front Immunol. (2023) 14:1255443. doi: 10.3389/fimmu.2023.1255443
37. Hangauer MJ, Viswanathan VS, Ryan MJ, Bole D, Eaton JK, Matov A, et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. (2017) 551:247–50. doi: 10.1038/nature24297
38. Zhang L, Liu W, Liu F, Wang Q, Song M, Yu Q, et al. IMCA induces ferroptosis mediated by SLC7A11 through the AMPK/mTOR pathway in colorectal cancer. Oxid Med Cell Longev. (2020) 2020:1675613. doi: 10.1155/2020/6901472
39. Yang M, Li J, Gu P, and Fan X. The application of nanoparticles in cancer immunotherapy: Targeting tumor microenvironment. Bioact Mater. (2021) 6:1973–87. doi: 10.1016/j.bioactmat.2020.12.010
Keywords: pancreatic cancer, GJB4, reactive oxygen species, ferroptosis, bioinformactics
Citation: Zheng X, Li W, Li X, Yao Q, Zheng L, Fan R and Bie P (2025) Targeting GJB4 to inhibit tumor growth and induce ferroptosis in pancreatic cancer. Front. Oncol. 15:1585236. doi: 10.3389/fonc.2025.1585236
Received: 28 February 2025; Accepted: 27 May 2025;
Published: 12 June 2025.
Edited by:
Hongbing Zhang, Tianjin Medical University General Hospital, ChinaReviewed by:
Xiaochun Yang, Feinstein Institute for Medical Research, United StatesJyoti Bala Kaushal, University of Nebraska Medical Center, United States
Copyright © 2025 Zheng, Li, Li, Yao, Zheng, Fan and Bie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Bie, NjUxNzY2QGhvc3BpdGFsLmNxbXUuZWR1LmNu
 Xiangru Zheng
Xiangru Zheng Wenjun Li
Wenjun Li Xiaoli Li3
Xiaoli Li3 Rongrong Fan
Rongrong Fan Ping Bie
Ping Bie