- 1Nursing Department, The First Affiliated Hospital of Chongqing Medical University, Chongqing, China
- 2College of Stomatology, Chongqing Medical University, Chongqing, China
Biliary tract cancers (BTCs), a group of rare aggressive malignancies, posed significant clinical challenges due to late diagnosis and limited therapies. While gut microbiota had been extensively studied in gastrointestinal cancers, the role of oral microbiota—a primary microbial reservoir entering the digestive system—remained poorly understood. Emerging evidence indicated that oral bacteria might affect biliary carcinogenesis through direct colonization, immune modulation, and metabolic interactions via the oral-gut-liver axis. This narrative review analyzed current research connecting oral microbial imbalance with BTCs. It explored how bacterial translocation, inflammatory metabolites, and immune alterations could promote cancer development. Established BTC risk factors—including gallstones, primary sclerosing cholangitis, cirrhosis, and H. pylori infection—were evaluated for their associations with oral microbiota changes. Epidemiological studies revealed that periodontal disease and poor oral hygiene elevated BTC risk. Sequencing analyses identified oral-origin bacteria (Prevotella, Fusobacterium, Streptococcus) in bile and tumor tissues, suggesting microbial migration through swallowing or bloodstream. Mechanistic investigations showed microbial components (e.g., lipopolysaccharides, membrane vesicles) activated inflammatory pathways (TLR4/NF-κB, STAT3) and modified immune checkpoints, while metabolites potentially altered biliary cell metabolism. Different studies have found variable changes in oral microbiota in the presence of BTCs, thus a novel “biphasic dysbiosis” hypothesis was proposed to explain differing oral microbial diversity patterns across BTC subtypes. Despite progress, critical knowledge gaps persisted regarding causality, spatial microbial variations, and functional impacts of metabolites in BTCs. Future research was recommended to employ multi-omics approaches, single-cell analysis, and AI tools to enhance early detection and prevention strategies.
1 Introduction
The human microbiota encompassed diverse communities of microorganisms that exist in association with human hosts, comprising microbes from various domains of life (1). These microbial populations, spanning bacteria, fungi, viruses, and archaea, play a dual role in host biology. On one hand, it played essential roles in maintaining host homeostasis through immune modulation, nutrient metabolism, and epithelial barrier integrity. On the other, dysbiosis—an imbalance in microbial composition—has been implicated in oncogenesis via chronic inflammation, metabolic reprogramming, and immune evasion (2). Microbiota composition is highly site-specific, varying considerably across different anatomical locations such as the skin, oral cavity, gut, and biliary tract. Each niche harbors a unique microbial ecosystem influenced by age, environment, diet, genetics, and disease states. The interplay between microbiota and host immunity represents a critical determinant of health status, with alterations in local microbial compositions being extensively associated with various pathological conditions (3). Of these, the oral cavity is of particular interest as it constitutes the second largest microbial habitat in the body and serves as the initial gateway to the gastrointestinal system. The oral microbiota comprises more than 700 bacterial species, alongside fungi and viruses, and its balance can be disrupted by factors such as tobacco use, alcohol consumption, poor dental hygiene, periodontitis, and prolonged antibiotic exposure (4).
Biliary tract cancers (BTCs) represented a heterogeneous group of malignant neoplasms originating from the epithelial cells of the biliary system, including intrahepatic cholangiocarcinoma (iCCA), perihilar cholangiocarcinoma (pCCA), distal cholangiocarcinoma (dCCA) and gallbladder cancer. Despite their relatively low overall incidence in the general population, BTCs demonstrate an increasing global trend, with notably higher prevalence in Asian regions (5). These malignancies presented significant clinical challenges due to their asymptomatic early stages, frequent late-stage diagnosis post-metastasis, high chemotherapy resistance, and limited targeted therapeutic options. As a result, their five-year survival rate remains below 30%, underscoring the urgent need for improved prevention and early diagnostic strategies (6, 7).
The tumor microenvironment constitutes a complex ecosystem comprising both host and microbial cells associated with neoplastic tissue, in which resident microbiota actively participated in modulating cancer progression. Emerging evidence illustrated how microbiota shaped the tumor microenvironment through direct contact with host tissues or via secreted metabolites (8, 9). For example, butyrate, a short-chain fatty acid produced by bacterial fermentation of dietary fiber, demonstrated anti-tumor properties. It promoted cancer cell apoptosis and inhibited tumor growth. These effects were mediated through multiple mechanisms, including histone deacetylase (HDAC)—the enzymes that control gene expression through chromatin remodeling—inhibition, G protein-coupled receptor activation, and cellular metabolism regulation (10).
While most microbiome-oncology studies have centered on the gut, growing evidence suggested that oral microbes might also influence systemic disease processes. With evidence has established that various pathophysiological factors can induce oral microbial dysbiosis, which include tobacco use, alcohol consumption, prolonged antibiotic administration, dental caries, and periodontitis. Such disruptions to the oral microbiome have significant implications for oral health (11). As the initial segment of the digestive tract, oral microbiota can translocate to downstream digestive organs via swallowing (the oral-gut axis), entering systemic circulation through inflamed or damaged oral tissues (the oral-blood axis) or other ways (12, 13). These translocated microbes and their metabolites could affect distant organs, including the liver and biliary system, by disrupting local microbial communities, triggering inflammatory responses, or altering metabolic pathways. Recent investigations have increasingly demonstrated both direct and indirect associations between oral microbiota (particularly Porphyromonas gingivalis and Fusobacterium nucleatum) and various digestive system disorders, including colorectal cancer, inflammatory bowel disease, and chronic liver diseases (14–17).
While significant advances have been made in understanding the relationship between microbiota and digestive system malignancies, the potential connection between oral microbiota and BTCs-a significant component of digestive system cancers-remained incompletely elucidated. Understanding this connection could open new avenues for early diagnosis and intervention, particularly given the oral cavity’s accessibility for non-invasive sampling.
This narrative review aimed to comprehensively examine the mechanistic and clinical associations between the oral microbiota and BTCs. We focused on the translocation pathways, metabolic and immunological interactions, and the impact of oral microbial dysbiosis on BTC risk and progression. We further evaluated the relationship between established BTC risk factors—such as cholelithiasis, primary sclerosing cholangitis (PSC), liver cirrhosis, and Helicobacter pylori infection—and oral microbial alterations. Our review synthesized findings from observational studies, microbiome sequencing analyses, and mechanistic investigations. The study was conducted with comprehensive searches performed in PubMed and Web of Science databases from 2000 to April 17, 2025. The search terms included: (“oral microbiota” OR “oral microbiome” OR “oral flora” OR “oral bacteria”) AND (“biliary tract cancer” OR “biliary tract neoplasm” OR “cholangiocarcinoma” OR “bile duct cancer” OR “gallbladder cancer”). Additional searches were conducted using terms related to established risk factors: (“oral microbiota” OR “oral microbiome” OR “oral flora” OR “oral bacteria”) AND (“cholelithiasis” OR “primary sclerosing cholangitis” OR “liver cirrhosis” OR “Helicobacter pylori”).
By consolidating and critically interpreting the current evidence, this review aims to clarify the biological relevance of the oral microbiota in BTC development and highlight future research directions. In doing so, it seeks to make this emerging field more accessible to interdisciplinary audiences spanning oncology, microbiology, hepatology, and oral medicine.
2 Normal oral microbiota
The oral cavity represents a complex microbial ecosystem, serving as the second largest microbial reservoir in the human body. Advanced DNA analysis of the oral microbiome has revealed the presence of over 700 distinct bacterial species. Oral microbial genera and species vary between individuals due to environmental factors. However, the predominant bacterial phyla remain relatively consistent in healthy individuals. This conservation of main microbiota composition in health states provided valuable insights into understanding the relationship between oral and systemic health (11, 18).
The microbiota comprises various microorganisms, including bacteria, fungi, viruses, archaea, and prokaryotes, but current research predominantly focused on bacterial communities. Contemporary technological advances, including 16S rRNA high-throughput sequencing, metagenomics, single-cell genomics, and integrated multi-omics analysis, enabled comprehensive and precise characterization of oral microbiota structure and function. Current evidence established the major bacterial phyla in the oral cavity, in order of prevalence: Firmicutes (including Streptococcus, Gemella, Eubacterium, Selenomonas, Veillonella), Actinobacteria (including Actinomyces, Atopobium, Rothia), Proteobacteria (including Neisseria, Eikenella, Campylobacter), Bacteroidetes (including Porphyromonas, Prevotella, Capnocytophaga), Fusobacteria (including Fusobacterium and Leptotrichia), TM7, Spirochaetes, OD2, and Synergistetes. The predominant genera include Streptococcus, Haemophilus, Neisseria, Prevotella, Veillonella, and Rothia (19, 20).
3 Oral microbiota and BTCs
The oral cavity, serving as the initial segment of the digestive tract, functioned as an endogenous reservoir for gut microbiota. Oral microorganisms demonstrated multiple pathways of translocation to the digestive tract through direct colonization, immune mediator regulation, and metabolite diffusion. Through the oral-gut axis, oral microbiota directly migrated and colonized the intestinal mucosa via swallowing and blood circulation (21). Upon entering the intestinal tract, oral microbiota interact with resident gut microbes and alter the intestinal microbial composition and immune microenvironment through mechanisms such as competition, symbiosis, and nutrient sharing (21, 22). Once established in the gut, oral microbiota can modulate the enterohepatic circulation of bile acids, influencing their conversion into secondary bile acids by the intestinal microbiota. These secondary bile acids affect liver metabolism and immune responses, leading to altered bile acid signaling through FXR and TGR5, which may contribute to chronic inflammation and immune dysregulation, potentially promoting the development of BTCs (23). Moreover, gut microbiota and their metabolites, including short-chain fatty acids, secondary bile acids, and lipopolysaccharides (LPS), exert significant effects on the hepatobiliary system through gut-liver axis signaling mechanism (24). (See Figure 1 for schematic representation of these pathways).
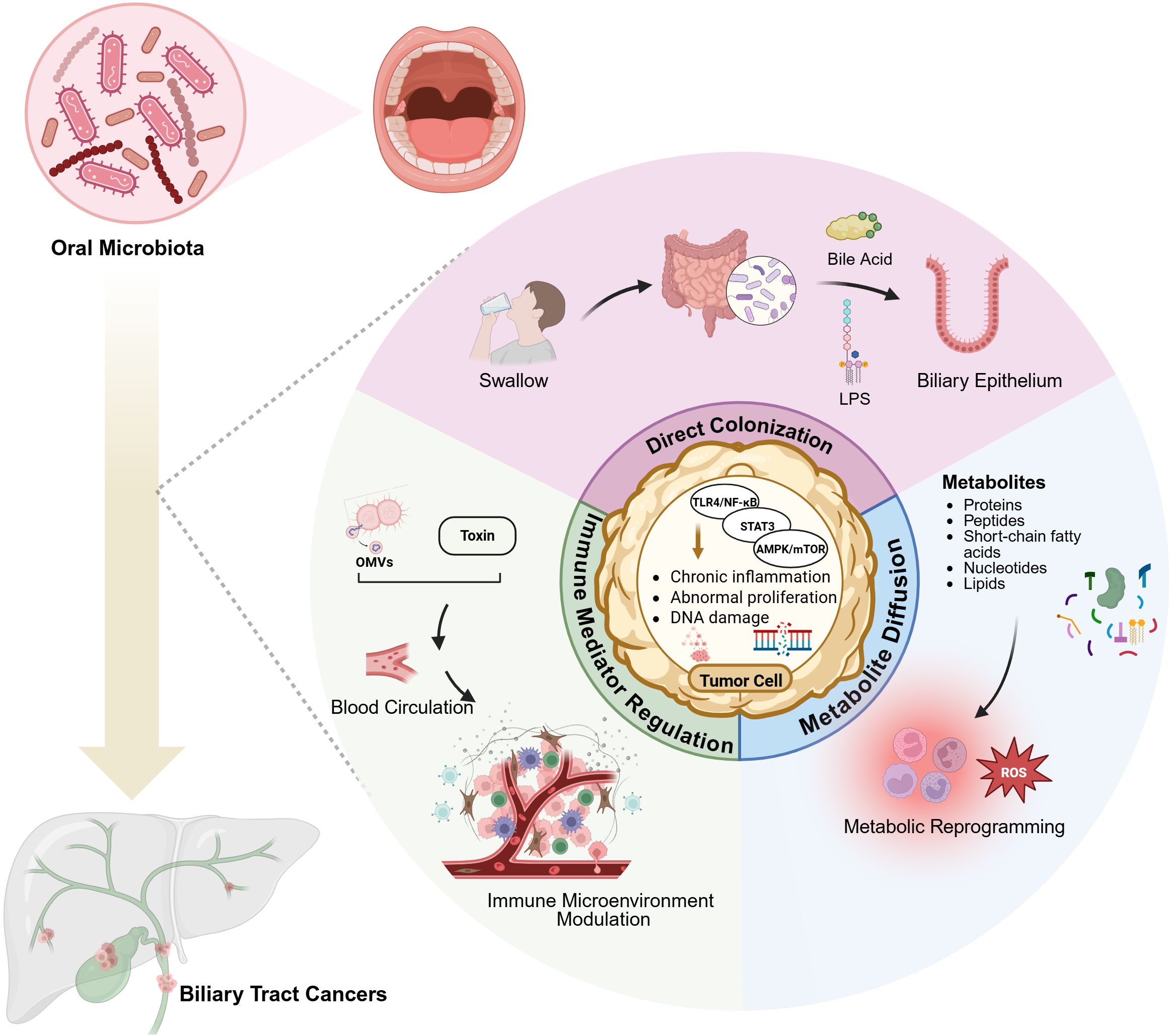
Figure 1. Possible oral microbiota-mediated pathways in biliary tract cancer development. Through the oral-gut axis, oral microorganisms directly colonize intestinal mucosa via swallowing. Their metabolites, including secondary bile acids and LPS, influence the hepatobiliary system through the gut-liver axis. Microbial metabolites interact with host cell receptors to modulate local inflammatory responses and cellular metabolism through metabolic reprogramming. Bacterial genera such as Streptococcus and Prevotella survive in the bloodstream, where their secreted toxins and bacterial outer membrane vesicles (OMVs) modulate host immune responses and reshape the cancer immune microenvironment. Created in https://BioRender.com.
Oral microbiota exerted indirect influence on digestive organs through the production of diverse metabolites (e.g., proteins, peptides, short-chain fatty acids, nucleotides, lipids). Through interactions with host cell receptors, these metabolites can modulate local inflammatory responses, metabolic functions, and even carcinogenic processes. For instance, butyrate produced by Prevotella was believed to regulate intestinal immune barriers, while LPS produced by Veillonella could exacerbate chronic inflammatory conditions in the biliary tract (13).
Poor periodontal health is associated with alterations in oral microbial diversity (25). And in a prospective study of 65,869 women, a history of periodontal disease was significantly associated with an elevated risk of BTCs (hazard ratio, 1.73; 95% confidence interval, 1.01-2.95) (26). Additionally, a United Kingdom-based cohort study involving 286 BTCs cases revealed that self-reported poor oral health correlated with increased risk of biliary system malignancy in unadjusted analyses (1.32; 95% CI, 0.95-1.80) (27). These findings suggested a potential relevance between oral microbiota, as key determinants of oral health, and increased BTCs risk.
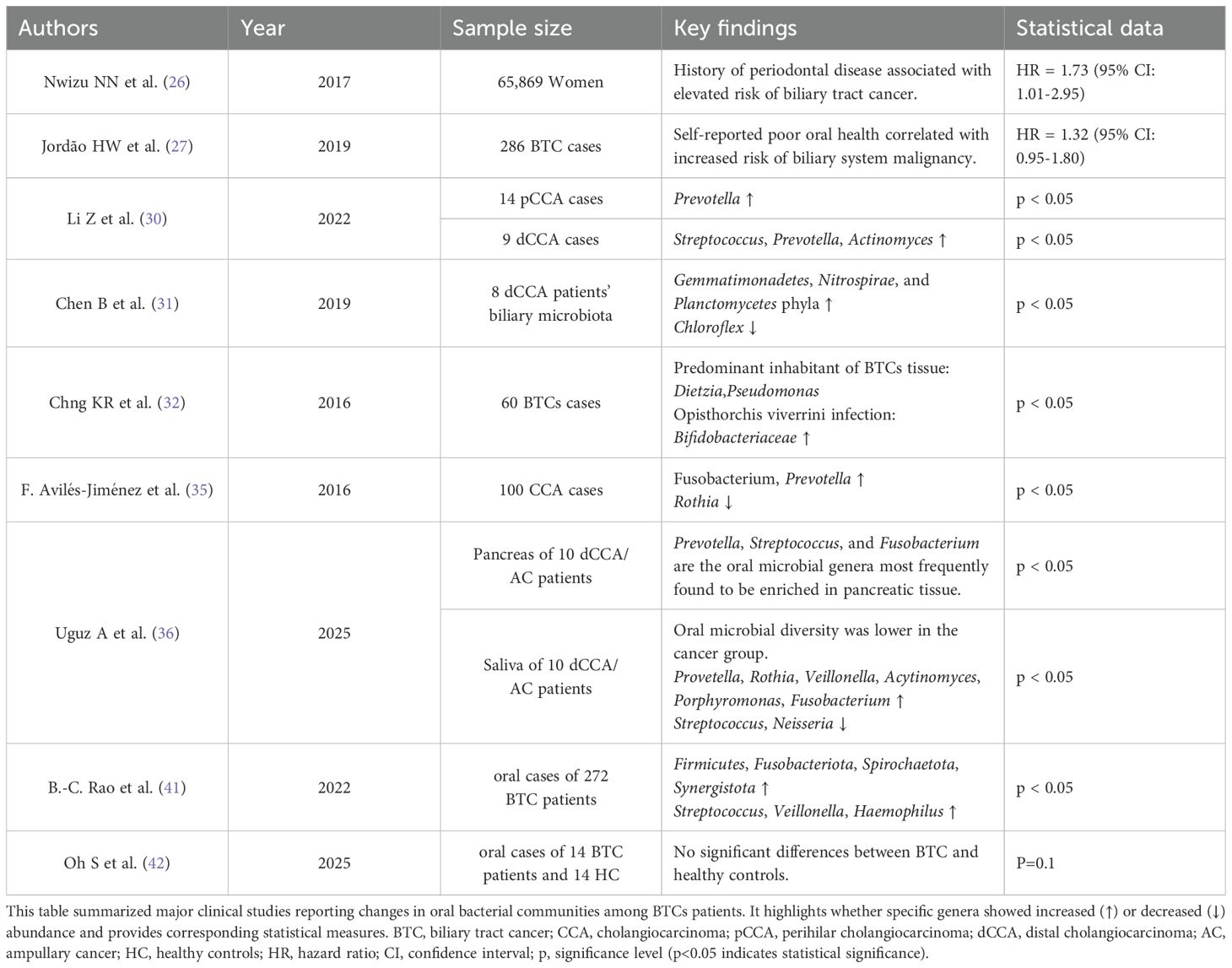
Bile, a light yellow fluid synthesized by hepatocytes, comprises bile acids (BA), cholesterol, phospholipids, and proteins, and undergoes transport from the liver and gallbladder to the intestine via bile ducts (28). The biliary microbiota predominantly consists of Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria phyla, with additional phyla including Verrucomicrobia, Chlamydiae, Acidobacteria, Planctomycetes, Cyanobacteria, Spirochaetes, and Fusobacteria present in lower proportions (0.05-0.5%) (29). In a study of 14 patients with pCCA, increased abundance of oral-associated Prevotella genera was observed. Analysis of 9 patients with dCCA revealed elevated levels of Streptococcus, Prevotella, and Actinomyces genera (30). Through 16S rRNA sequencing analysis of biliary microbiota from 8 dCCA patients, Chen et al. demonstrated that while overall phylum-level composition remained relatively stable, significant increases occurred in Gemmatimonadetes, Nitrospirae, and Planctomycetes phyla abundance, accompanied by notable reduction in Chloroflexi (31). In another 16S rRNA sequencing analysis involving 60 BTCs patients, Dietzia and Pseudomonas genera were identified as the predominant inhabitants of the biliary tract tumor tissue. Additionally, enrichment of Bifidobacteriaceae was observed in cases with Opisthorchis viverrini infection (32). Notably, Dietzia genera primarily constitute skin and oral microbiota components, while Pseudomonas and Bifidobacteriaceae, as opportunistic pathogens, may colonize the oral cavity through environmental exposure before subsequent biliary tract translocation (33, 34). In a multicenter case-control study encompassing 100 BTCs patients, Avilés-Jiménez et al. documented significant increases in Fusobacterium and Prevotella genera abundance, accompanied by decreased levels of Rothia, typically considered a normal oral microbiota constituent (35).
Uguz A et al. examined pancreatic samples from 10 patients with dCCA or ampullary cancer (AC). The microbiota was dominated by the phyla Firmicutes, Actinobacteria, Proteobacteria, Bacteroidetes, and Acidobacteria. Firmicutes were the most abundant, while Bacteroidetes and Acidobacteria were significantly enriched. At the genus level, microbial profiles varied markedly between individuals. Sequence alignments of saliva and pancreatic samples identified Prevotella, Streptococcus, and Fusobacterium as the oral genera most frequently enriched in pancreatic tissue (36). These findings further suggest the potential impact of oral microbiota on peritumoral BTC tissues.
Based on the gut-liver axis theory, intestinal microbiota and their metabolites can directly affect the hepatobiliary system through the portal venous circulation, participating in the development and progression of various diseases (37). In BTCs patients, increased abundances of Veillonella, Parabacteroides, and Enterobacter genera were observed, along with elevated levels of Firmicutes and Actinobacteria phyla, revealing the correlation between dysbiosis and carcinogenesis. Further studies demonstrated that the intestinal microbiota can drive BTCs progression through key regulatory points, including energy metabolism reprogramming and cell proliferation control via the AMPK and mTOR signaling pathways (38).
The mechanisms of oral bacterial metabolites in BTCs development require further investigation. For instance, P. gingivalis-derived LPS activates the TLR4/NF-κB pathway in microglial cells, triggering inflammatory cascades (39). And sustained inflammatory microenvironment promotes aberrant cell proliferation and carcinogenesis through multiple signaling pathways, including NF-κB and STAT3 (40). While it was relatively clear that the aforementioned oral microbiota metabolites might have influenced the development and progression of BTCs by modulating inflammatory pathways and metabolic reprogramming, their temporal distribution, precise concentrations, and specific targets within the biliary microenvironment had not been adequately clarified (41). With the advancement of relevant technologies, future studies urgently need to employ techniques such as isotope labeling-based metabolic analysis and targeted metabolomics analysis in both vivo and vitro experiments. These approaches are expected to clarify the local dynamic changes and specific mechanisms of these metabolites, thereby defining their precise roles in the pathogenesis of BTCs.
During BTCs development, changes occur not only in the biliary and gastrointestinal microbiota but also in the oral microbiome composition. A study of gallbladder cancer patients (n=272) demonstrated that these patients exhibited significantly higher α-diversity and abundance of rare species in their oral microbiota compared to healthy controls. They also successfully developed a high-reliability predictive model for BTCs’ probability using a marker set comprising three genera—Actinomyces, Alloprevotella, and Lautropia. This model revealed the feasibility of utilizing changes in the oral microbiota as an auxiliary diagnostic indicator for BTCs (42). However, another study of 10 patients with dCCA or ampullary cancer found that oral microbial diversity was lower in the cancer group than in healthy controls (36). Peculiarly, Oh S. et al. compared the oral microbiota of 14 BTC patients with 14 healthy controls and found no significant difference (p = 0.1) (43). Although variations in sample size, sequencing depth, and control of confounding factors may have caused these differences, Oh S. et al. observed that cancers at different anatomical sites produced distinct shifts in the oral microbiota. This finding demonstrated that the anatomical site determines the pattern of microbial change (43). It is acknowledged that iCCA is propelled by chronic inflammation and is characterized by a highly immunosuppressive tumor microenvironment, which includes elevated PD-L1 expression and MDSC accumulation (44). In this context, it is hypothesized that these mechanisms could weaken immune surveillance and can contribute to systemic immune dysregulation, potentially affecting sites such as the oral mucosa. Weakended immune surveillance allows rare oral bacteria taxa to flourish, increasing overall oral microbiota diversity. In contrast, dCCA is usually accompanied by bile duct obstruction and repeated biliary inflammation. These conditions often require early and frequent antimicrobial treatment, which suppresses some species and lets a few dominant pathogens take over, thereby reducing diversity (45, 46). In addition, factors such as malnutrition, oral dryness, reduced food intake, and preoperative antibiotic use in some advanced dCCA patients lowered oral microbial diversity. These findings also underscore the shortage of studies examining heterogeneity driven by anatomical subtypes. Future work should clarify the underlying pathways and involve large-scale, multicenter cohorts. (See Table 1 for summary of clinical evidence regarding oral microbiota changes in BTC patients).
4 The oral microbiota and risk factors for BTCs
BTCs encompasses two main subtypes: bile duct cancer and gallbladder cancer. Established risk factors for bile duct cancer included primary sclerosing cholangitis (PSC), Caroli’s disease, intrahepatic bile duct stones, and liver fluke infection. Primary risk factors for gallbladder cancer included cholelithiasis and primary sclerosing cholangitis, among others (5). (See Figure 2 for microbial alterations associated with established BTC risk factors).
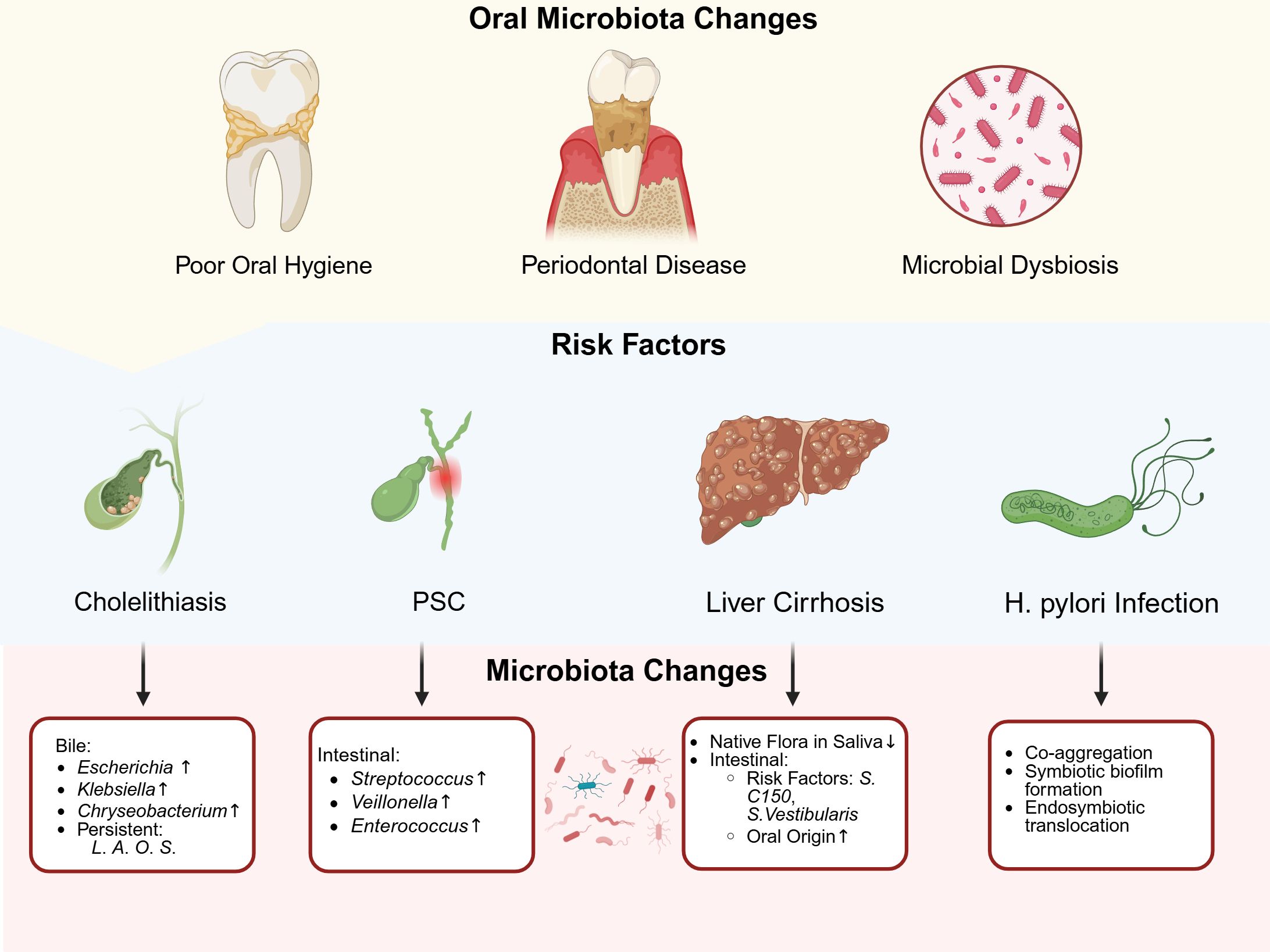
Figure 2. The association between oral microbiota and risk factors in biliary tract cancer. Four established risk factors-gallstones, PSC, liver cirrhosis and Helicobacter pylori infection-were each linked to distinct shifts in oral microbial communities. Gallstones and poor oral health correlate with higher levels of oral bacteria such as Escherichia, Klebsiella and Chryseobacterium in biliary samples, while there was a persistence of Lachnoanerobaculum (L), Atopobium (A), Oribacterium (O) and Stomatobaculum (S). PSC patients exhibit increased Streptococcus salivarius, Veillonella parvula and Enterococcus in intestinal microbiota. Cirrhosis−related gum disease showed loss of native flora in saliva (e.g., Streptococcus) alongside a rise of risk factors (e.g., S. C150, S. Vestibularis) and oral oringin microbiota in the gut. H. pylori infection reshaped oral communities by promoting co-aggregation and the formation of biofilms by species such as Gemella and Holdemanella, as well as endosymbiotic translocation. These microbial changes may contribute to cancer development through direct colonization, immune modulation and chronic inflammation. Created in https://BioRender.com.
4.1 Cholelithiasis
Cholelithiasis can occur at any location within the biliary system, encompassing the gallbladder and both intrahepatic and extrahepatic bile ducts (47). Current evidence demonstrated a positive correlation between cholelithiasis and increased risk of biliary tract and gallbladder carcinoma, with notably 70-90% of gallbladder cancer patients presenting with antecedent gallstone disease (48).
Multiple investigations have elucidated the role of microorganisms in cholelithiasis pathogenesis (49). As previously established, oral microbiota maintained intimate associations with oral hygiene status. A population-based survey in the United States demonstrated a significant positive correlation between poor oral hygiene and ultrasonographically confirmed cholelithiasis, further supporting the connection between oral microbiota and gallstone disease (50). Using whole-metagenome shotgun (WMS) and 16S sequencing, Shen et al. identified 25 oral/respiratory tract-derived microorganisms among 54 microbial species in bile samples from patients with common bile duct stones. Notably, these oral-derived bacterial species exceeded the quantity of intestinal-derived species. Furthermore, among 13 newly discovered species in bile, 8 belonged to oral microbial groups, suggesting a substantial association between oral microbiota and cholelithiasis (51). Additional studies using 16S sequencing revealed elevated levels of Escherichia and Klebsiella in bile samples from cholelithiasis patients, with Chryseobacterium also being detected in the bile (52). Moreover, oral microbiota-associated genera including Lachnoanerobaculum, Atopobium, Oribacterium, and Stomatobaculum demonstrated persistent presence in the bile of cholelithiasis patients, with their abundance increasing during disease onset and progression (53).
4.2 Primary sclerosing cholangitis
PSC represents a chronic inflammatory liver condition characterized by progressive scarring of bile ducts and intrahepatic biliary system, potentially predisposing to BTCs development (54).
Multiple studies have confirmed increased abundances of Streptococcus, Veillonella, and Enterococcus genera in the intestinal microbiota of PSC patients (55–57). Analysis of saliva and fecal samples from PSC patients revealed significant enrichment of eight bacterial species, including Streptococcus salivarius, Veillonella parvula, Actinomyces, and Bifidobacterium, suggesting potential oral microbiota translocation and colonization as crucial factors in PSC pathogenesis (57).
4.3 Liver cirrhosis
Evidence and mechanisms supporting liver cirrhosis as a premalignant condition for BTCs have been established (58).
As previously noted, periodontal status is closely associated with the oral microbiome (25). A direct causal relationship existed between liver cirrhosis and periodontitis (59). The study involving 164 non-alcoholic fatty liver disease patients demonstrated that those with P. gingivalis infection in saliva and deeper periodontal pockets exhibited higher liver stiffness values compared to patients without periodontal disease. Patients with increased liver stiffness showed elevated serum antibody titers against P. gingivalis strains FDC381 and SU63. Logistic regression analysis confirmed the correlation between periodontal disease and liver stiffness (60). The subgingival microbiota in cirrhosis-associated periodontitis consisted of a distinct bacterial community that was typically unrelated to conventional periodontitis, likely resulting from dysbiosis due to compromised immune function (61). The oral cavity represented a significant source of inflammation in liver cirrhosis. Periodontal treatment can improve endotoxemia, salivary inflammation, and systemic inflammation in cirrhotic patients, while also regulating dysbiosis in salivary and fecal microbial communities (62).
Regarding the characteristic changes in salivary microbiota of patients with liver cirrhosis, there is a decreased relative abundance of indigenous bacteria (such as Streptococcus), while potentially pathogenic families (Enterobacteriaceae and Enterococcaceae) show significantly increased relative abundance. These findings suggested that oral dysbiosis could be associated with the progression of liver disease (63). In oropharyngeal swabs from cirrhotic patients with pneumonia, compared to those without pneumonia, there were increased populations of Bacteroides, Neisseria, and Actinomyces, while the Streptococcus population was decreased (64).
Due to the persistent presence and high abundance of lantibiotics genes in the gut microbiome of cirrhotic patients, oral Streptococcus C150 and Streptococcus vestibularis, which encode lantibiotics before their translocation to the intestine, are potential risk factors for liver cirrhosis (65).
In a quantitative metagenomic analysis of gut microbiome from 98 cirrhotic patients and 83 healthy controls, there were 66 clusters representing homologous bacterial species that differed between the cirrhotic and healthy groups. Among the 28 bacterial species enriched in cirrhotic patients, the majority originated from the oral cavity (66). Japanese researchers have demonstrated that patients with liver fibrosis show increased relative abundance of Clostridium strains and decreased relative abundance of Faecalibacterium in their gut microbiota (67).
4.4 Helicobacter pylori
Specific H. pylori strains clearly associate with increased BTCs risk (35, 68).
The oral microbiota, being upstream of the stomach, serves as a primary source of gastric microbiota. Research showed that H. pylori infection was not only associated with the degree of coexistence between oral and gastric mucosal microbiota, but H. pylori infection itself can also influence the composition of oral bacterial communities through altering local pH or competitive colonization (69, 70). The oral microbiota influenced the transmission and colonization of H. pylori through co-aggregation, symbiotic biofilm formation, and endosymbiotic translocation colonization (71).
Interestingly, recent research has shown that abnormal abundances of specific oral bacteria—such as Gemella and Holdemanella—significantly affect the risk of gastric cancer (43). These species were not only closely linked to gastric cancer and may, through their complex interactions with H.pylori, alter the bacterium’s growth and ability to cause disease. Consequently, this could influence the risk of BTCs. Such dysbiosis may impair both oral and gastric mucosal barriers. It can also modulate H. pylori’s pathogenicity through bacterial coaggregation, co−formation of biofilms, and internal translocation. These processes could contribute to BTCs initiation and progression. Future research should clarify how oral microbiota and H. pylori interact and determine how these microbial interactions influence BTC risk and disease course.
5 Conclusions and future directions
Accumulating evidence indicates that the oral microbiota can influence biliary tract carcinogenesis via three principal mechanisms: direct bacterial colonization of biliary tissues, modulation of the local immune microenvironment, and microbial metabolic interactions (21–24). Opportunistic oral bacteria such as Porphyromonas gingivalis are capable of ectopically colonizing the bile ducts and triggering chronic biliary inflammation (e.g., through TLR4/NF-κB activation), which in turn promotes abnormal epithelial proliferation and DNA damage. Additionally, bacterial products like OMVs can dampen anti-tumor immune responses for instance, by upregulating the PD-1/PD-L1 immune checkpoint, and can act in concert with host pro-inflammatory cytokines to create a tumor-promoting immunosuppressive niche Under gut-liver axis regulation, Veillonella-derived LPS and secondary bile acids cooperatively drived biliary cell metabolic reprogramming through oxidative stress and FXR receptor signaling. These mechanistic links are supported by clinical observations: epidemiological studies have correlated poor oral health (e.g., periodontitis) with elevated risk of BTC, and oral bacterial taxa such as Fusobacterium and Prevotella have been detected within bile and tumor tissue of BTCs patients.
Contemporary research substantiates a significant correlation between oral microbiota dysbiosis and BTCs risk, with periodontal disease history and poor oral health linked to 73% (HR=1.73) and 32% (HR=1.32) increased risk, respectively (26, 27). Molecular biological evidence has revealed enrichment of oral-origin bacteria genera in bile and biliary tissues of BTCs patients, including Fusobacterium, Prevotella, and Streptococcus (30, 35). Opportunistic pathogens such as Deinococcus and Pseudomonas may contribute to carcinogenesis through oral-biliary ectopic colonization. Further mechanistic studies indicate that oral bacterial metabolites (such as P. gingivalis LPS) can induce chronic inflammation by activating the TLR4/NF-κB pathway, working synergistically with pathways like STAT3 to drive abnormal cell proliferation (39, 41). According to gut-liver axis theory, BTCs patients show intestinal dysbiosis. This condition, marked by increased Veillonella and Enterobacteriaceae, may promote tumors through AMPK/mTOR-mediated metabolic reprogramming (38). Given the significantly increased oral microbial α-diversity and abnormally elevated abundance of oral dominant bacteria (Firmicutes, Streptococcus) in gallbladder cancer patients, dynamic changes in oral microbiota showed promise as biomarkers for BTCs development.
Overall, we proposed a novel mechanistic hypothesis in section 3 to explain the variations in oral microbiota diversity observed across different types of BTCs. Dysbiosis of the oral microbiota associated with biliary cancer may have shown a “biphasic pattern”. Specifically, this biphasic pattern refers to the observation that, although both iCCA and dCCA are subtypes of BTCs, oral microbial diversity is higher in iCCA patients than in healthy controls, whereas it is lower in dCCA patients (36, 42). In some cases (e.g., iCCA), weakened immune surveillance and prolonged low-grade inflammation supported the coexistence and proliferation of multiple opportunistic pathogens in the oral cavity, leading to increased microbial diversity. This enriched microbial community continuously released bioactive molecules, such as LPS and metabolic byproducts, which affected liver immune responses. These changes contributed to the development of a chronic inflammatory microenvironment that promoted malignant transformation of biliary epithelial cells (44). In contrast, in other cases (e.g., dCCA), repeated acute inflammation and medical interventions led to colonization by only a few dominant pathogenic species, resulting in reduced diversity (45, 46). Though these dominant microbes had stronger pro-inflammatory and carcinogenic potential. Both two types of dysbiosis eventually contributed to cholangiocarcinogenesis through both direct and indirect pathways mentioned above.
Despite significant progress, current researches exhibits several limitations. The predominance of cross-sectional study designs precluded establishment of clear temporal relationships between oral dysbiosis and BTCs development. Limited sample sizes in some studies (n<50) constrain the identification of specific bacterial signatures (30, 36, 43). Analysis of specific metabolic regulatory networks remains incomplete, particularly regarding the quantification of dynamic concentrations and targets of oral bacterial metabolites in the biliary microenvironment (41). Additionally, spatial heterogeneity of microbiota across gallbladder anatomical subsites requires further investigation.
Future research should validate causality through multicenter longitudinal cohort studies and humanized mouse models. These studies could lead to novel therapeutic approaches targeting metabolic pathways and microbiota. One potential strategy is to leverage machine learning on microbiota data to enable early cancer prediction and diagnosis. Large-scale analyses of oral and gut microbiota from gastrointestinal cancer patients have already enabled machine learning models to distinguish cancer cases with high accuracy (AUC>0.8) (72, 73). These findings encourage the development of AI-driven oral microbiome diagnostics for GI and hepatobiliary cancers as a complement to or even replacement for more invasive screening methods. And multi-omics AI models are being explored: an integrative graph convolutional network that combined microbiome features with exposome data achieved about 90% accuracy (AUROC ~0.89) in detecting pancreatic cancer (74). Larger, long−term studies and validations across diverse populations are needed to bring these AI−powered microbiome models into routine cancer screening. Saccharomyces cerevisiae has been validated as a novel microbial platform for delivering agents to gastrointestinal tumors (75). Future work could engineer strains to express BTCs-specific antigens. This strategy may overcome the poor mucosal penetration and high systemic toxicity of conventional therapies. Additionally, integrating single-cell and spatial omics with metabolic tracing and intelligent delivery systems could accelerate the discovery of tumor–microbe interaction mechanisms. It could also enable microbiome-based early screening and targeted interventions, offering new translational avenues for BTCs that have poor prognosis and lack reliable biomarkers.
Author contributions
YZ: Writing – original draft, Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Software, Visualization. SZ: Writing – review & editing, Conceptualization, Data curation, Formal Analysis, Funding acquisition, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by Research and Innovation Projects in Nursing Science at the First Affiliated Hospital of Chongqing Medical University in 2024 (Project number: HLYB2024-05).
Acknowledgments
We acknowledge all the authors whose publications are referred in our article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
BTCs, Biliary tract cancers; iCCA, Intrahepatic cholangiocarcinoma; dCCA, Distal cholangiocarcinoma; pCCA, Perihilar cholangiocarcinoma; LPS, Lipopolysaccharide; HDAC, Histone deacetylase; BA, Bile acids; PSC, Primary sclerosing cholangitis; O. viverrini, Opisthorchis viverrini; H. pylori, Helicobacter pylori; WMS, Whole-metagenome shotgun; HR, Hazard ratio; OMVs, Outer membrane vesicles; CI, Confidence interval; L., Lachnoanerobaculum; A., Atopobium; O., Oribacterium; S., Stomatobaculum.
References
1. Berg G, Rybakova D, Fischer D, Cernava T, Vergès M-CC, Charles T, et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome. (2020) 8:103. doi: 10.1186/s40168-020-00875-0
2. Rajagopala SV, Vashee S, Oldfield LM, Suzuki Y, Venter JC, Telenti A, et al. The Human Microbiome and Cancer. Cancer Prev Res. (2017) 10:226–34. doi: 10.1158/1940-6207.CAPR-16-0249
3. Cho I and Blaser MJ. The Human Microbiome: at the interface of health and disease. Nat Rev Genet. (2012) 13:260–70. doi: 10.1038/nrg3182
4. Caselli E, Fabbri C, D’Accolti M, Soffritti I, Bassi C, Mazzacane S, et al. Defining the oral microbiome by whole-genome sequencing and resistome analysis: the complexity of the healthy picture. BMC Microbiol. (2020) 20:120. doi: 10.1186/s12866-020-01801-y
5. Valle JW, Kelley RK, Nervi B, Oh D-Y, and Zhu AX. Biliary tract cancer. Lancet. (2021) 397:428–44. doi: 10.1016/S0140-6736(21)00153-7
6. Shroff RT, Kennedy EB, Bachini M, Bekaii-Saab T, Crane C, Edeline J, et al. Adjuvant Therapy for Resected Biliary Tract Cancer: ASCO Clinical Practice Guideline. JCO. (2019) 37:1015–27. doi: 10.1200/JCO.18.02178
7. Blechacz B, Komuta M, Roskams T, and Gores GJ. Clinical diagnosis and staging of cholangiocarcinoma. Nat Rev Gastroenterol Hepatol. (2011) 8:512–22. doi: 10.1038/nrgastro.2011.131
8. Swartz MA, Iida N, Roberts EW, Sangaletti S, Wong MH, Yull FE, et al. Tumor microenvironment complexity: emerging roles in cancer therapy. Cancer Res. (2012) 72:2473–80. doi: 10.1158/0008-5472.CAN-12-0122
9. Louis P, Hold GL, and Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. (2014) 12:661–72. doi: 10.1038/nrmicro3344
10. Maiuolo J, Bulotta RM, Ruga S, Nucera S, Macrì R, Scarano F, et al. The Postbiotic Properties of Butyrate in the Modulation of the Gut Microbiota: The Potential of Its Combination with Polyphenols and Dietary Fibers. Int J Mol Sci. (2024) 25:6971. doi: 10.3390/ijms25136971
11. Denefil O, Chorniy S, Boitsaniuk S, Chornij N, Levkiv M, Patskan L, et al. Comparative analysis of dysbiotic changes in the oral cavity of patients with periodontal diseases and systemic pathologies. Explor Med. (2024) 5:574–83. doi: 10.37349/emed.2024.00241
12. Kuraji R, Sekino S, Kapila Y, and Numabe Y. Periodontal disease–related nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: An emerging concept of oral-liver axis. Periodontol. (2021) 2000:87. doi: 10.1111/prd.12387
13. Freire M, Nelson KE, and Edlund A. The Oral Host-Microbial Interactome: An Ecological Chronometer of Health? Trends Microbiol. (2021) 29:551–61. doi: 10.1016/j.tim.2020.11.004
14. Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. (2016) 65:1973–80. doi: 10.1136/gutjnl-2015-310101
15. Wang A, Ding Y, and Cao H. DOP026 Porphyromonas gingivalis secreted htpG disrupts TLR4/PAPSS2-mediated mucin sulfation and aggravates ulcerative colitis. J Crohn’s Colitis. (2025) 19:i136. doi: 10.1093/ecco-jcc/jjae190.0065
16. Díaz-Basabe A, Lattanzi G, Perillo F, Amoroso C, Baeri A, Farini A, et al. Porphyromonas gingivalis fuels colorectal cancer through CHI3L1-mediated iNKT cell-driven immune evasion. Gut Microbes. (2024) 16:2388801. doi: 10.1080/19490976.2024.2388801
17. Yao C, Lu L, Lan D, Zhu X, Li X, Gao Y, et al. Porphyromonas gingivalis as a promotor in the development of the alcoholic liver disease via ferroptosis. Microbes Infect. (2024) 26:105250. doi: 10.1016/j.micinf.2023.105250
18. Eren AM, Borisy GG, Huse SM, and Mark Welch JL. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci. (2014) 111:E2875–84. doi: 10.1073/pnas.1409644111
19. Aas JA, Paster BJ, Stokes LN, Olsen I, and Dewhirst FE. Defining the Normal Bacterial Flora of the Oral Cavity. J Clin Microbiol. (2005) 43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005
20. Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of ten healthy individuals. ISME J. (2010) 4:962–74. doi: 10.1038/ismej.2010.30
21. Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science. (2017) 358:359–65. doi: 10.1126/science.aan4526
22. Ohtsu A, Takeuchi Y, Katagiri S, Suda W, Maekawa S, Shiba T, et al. Influence of Porphyromonas gingivalis in gut microbiota of streptozotocin-induced diabetic mice. Oral Dis. (2019) 25:868–80. doi: 10.1111/odi.13044
23. Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. (2018) 15:397–411. doi: 10.1038/s41575-018-0011-z
24. Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. (2018) 360:eaan5931. doi: 10.1126/science.aan5931
25. Yu G, Dye BA, Gail MH, Shi J, Klepac-Ceraj V, Paster BJ, et al. The association between the upper digestive tract microbiota by HOMIM and oral health in a population-based study in Linxian, China. BMC Public Health. (2014) 14:1110. doi: 10.1186/1471-2458-14-1110
26. Nwizu NN, Marshall JR, Moysich K, Genco RJ, Hovey KM, Mai X, et al. Periodontal Disease and Incident Cancer Risk among Postmenopausal Women: Results from the Women’s Health Initiative Observational Cohort. Cancer Epidemiol Biomarkers Prev. (2017) 26:1255–65. doi: 10.1158/1055-9965.EPI-17-0212
27. Jordão HW, McKenna G, McMenamin ÚC, Kunzmann AT, Murray LJ, and Coleman HG. The association between self-reported poor oral health and gastrointestinal cancer risk in the UK Biobank: A large prospective cohort study. U Eur Gastroenterol J. (2019) 7:1241–9. doi: 10.1177/20506406198580433
28. Boyer JL and Soroka CJ. Bile formation and secretion: An update. J Hepatol. (2021) 75:190–201. doi: 10.1016/j.jhep.2021.02.011
29. Molinero N, Ruiz L, Milani C, Gutiérrez-Díaz I, Sánchez B, Mangifesta M, et al. The human gallbladder microbiome is related to the physiological state and the biliary metabolic profile. Microbiome. (2019) 7:100. doi: 10.1186/s40168-019-0712-8
30. Li Z, Chu J, Su F, Ding X, Zhang Y, Dou L, et al. Characteristics of bile microbiota in cholelithiasis, perihilar cholangiocarcinoma, distal cholangiocarcinoma, and pancreatic cancer. Am J Transl Res. (2022) 14:2962–71.
31. Chen B, Fu SW, Lu L, and Zhao H. A Preliminary Study of Biliary Microbiota in Patients with Bile Duct Stones or Distal Cholangiocarcinoma. BioMed Res Int. (2019) 2019:1092563. doi: 10.1155/2019/1092563
32. Chng KR, Chan SH, Ng AHQ, Li C, Jusakul A, Bertrand D, et al. Tissue Microbiome Profiling Identifies an Enrichment of Specific Enteric Bacteria in Opisthorchis viverrini Associated Cholangiocarcinoma. EBioMedicine. (2016) 8:195–202. doi: 10.1016/j.ebiom.2016.04.034
33. Leão I, de Carvalho TB, Henriques V, Ferreira C, Sampaio-Maia B, and Manaia CM. Pseudomonadota in the oral cavity: a glimpse into the environment-human nexus. Appl Microbiol Biotechnol. (2023) 107:517–34. doi: 10.1007/s00253-022-12333-y
34. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu W-H, et al. The human oral microbiome. J Bacteriol. (2010) 192:5002–17. doi: 10.1128/JB.00542-10
35. Avilés-Jiménez F, Guitron A, Segura-López F, Méndez-Tenorio A, Iwai S, Hernández-Guerrero A, et al. Microbiota studies in the bile duct strongly suggest a role for Helicobacter pylori in extrahepatic cholangiocarcinoma. Clin Microbiol Infect. (2016) 22:178.e11–178.e22. doi: 10.1016/j.cmi.2015.10.008
36. Uguz A, Muftuoglu C, Mert U, Gumus T, Ece D, Asadi M, et al. Unveiling Microbiota Profiles in Saliva and Pancreatic Tissues of Patients with Pancreatic Cancer. Microorganisms. (2025) 13:119. doi: 10.3390/microorganisms13010119
37. Mehal WZ. The gut-liver axis: A busy two-way street. Hepatology. (2012) 55:1647. doi: 10.1002/hep.25704
38. Chen Z, Shi W, Chen K, Lu C, Li X, and Li Q. Elucidating the causal association between gut microbiota and intrahepatic cholangiocarcinoma through Mendelian randomization analysis. Front Microbiol. (2023) 14:1288525. doi: 10.3389/fmicb.2023.1288525
39. Qiu C, Yuan Z, He Z, Chen H, Liao Y, Li S, et al. Lipopolysaccharide Preparation Derived From Porphyromonas gingivalis Induces a Weaker Immuno-Inflammatory Response in BV-2 Microglial Cells Than Escherichia coli by Differentially Activating TLR2/4-Mediated NF-κB/STAT3 Signaling Pathways. Front Cell Infect Microbiol. (2021) 11:606986. doi: 10.3389/fcimb.2021.606986
40. Zhou Y, Xia L, Liu Q, Wang H, Lin J, Oyang L, et al. Induction of Pro-Inflammatory Response via Activated Macrophage-Mediated NF-κB and STAT3 Pathways in Gastric Cancer Cells. Cell Physiol Biochem. (2018) 47:1399–410. doi: 10.1159/000490829
41. Zhu C, Wang Y, Zhu R, Wang S, Xue J, Zhang D, et al. Gut microbiota and metabolites signatures of clinical response in anti-PD-1/PD-L1 based immunotherapy of biliary tract cancer. Biomark Res. (2024) 12:56. doi: 10.1186/s40364-024-00607-8
42. Rao B-C, Zhang G-Z, Zou Y-W, Ren T, Ren H-Y, Liu C, et al. Alterations in the human oral microbiome in cholangiocarcinoma. Military Med Res. (2022) 9:62. doi: 10.1186/s40779-022-00423-x
43. Oh S, Kim J, Shin CM, Lee H-J, Lee HS, and Park KU. Metagenomic characterization of oral microbiome signatures to predict upper gastrointestinal and pancreaticobiliary cancers: a case-control study. J Transl Med. (2025) 23:20. doi: 10.1186/s12967-024-05989-9
44. Donato F, Gelatti U, Tagger A, Favret M, Ribero ML, Callea F, et al. Intrahepatic cholangiocarcinoma and hepatitis C and B virus infection, alcohol intake, and hepatolithiasis: a case–control study in Italy. Cancer Causes Control. (2001) 12:959–64. doi: 10.1023/A:1013747228572
45. Tsen A, Barbara M, and Rosenkranz L. Dilemma of elevated CA 19–9 in biliary pathology. Pancreatology. (2018) 18:862–7. doi: 10.1016/j.pan.2018.09.004
46. Cai L, Zhu H, Mou Q, Wong PY, Lan L, Ng CWK, et al. Integrative analysis reveals associations between oral microbiota dysbiosis and host genetic and epigenetic aberrations in oral cavity squamous cell carcinoma. NPJ Biofilms Microb. (2024) 10:1–16. doi: 10.1038/s41522-024-00511-x
47. Shaffer EA. Epidemiology and risk factors for gallstone disease: Has the paradigm changed in the 21st century? Curr Gastroenterol Rep. (2005) 7:132–40. doi: 10.1007/s11894-005-0051-8
48. Massarweh NN and El-Serag HB. Epidemiology of Hepatocellular Carcinoma and Intrahepatic Cholangiocarcinoma. Cancer Control. (2017) 24:1073274817729245. doi: 10.1177/1073274817729245
49. Stewart L, Grifiss JM, Jarvis GA, and Way LW. Biliary bacterial factors determine the path of gallstone formation. Am J Surg. (2006) 192:598–603. doi: 10.1016/j.amjsurg.2006.08.001
50. Bhandari S, Reddy M, and Shahzad G. Association between oral hygiene and ultrasound-confirmed gallstone disease in US population. Eur J Gastroenterol Hepatol. (2017) 29:861. doi: 10.1097/MEG.0000000000000885
51. Shen H, Ye F, Xie L, Yang J, Li Z, Xu P, et al. Metagenomic sequencing of bile from gallstone patients to identify different microbial community patterns and novel biliary bacteria. Sci Rep. (2015) 5:17450. doi: 10.1038/srep17450
52. Ye F, Shen H, Li Z, Meng F, Li L, Yang J, et al. Influence of the Biliary System on Biliary Bacteria Revealed by Bacterial Communities of the Human Biliary and Upper Digestive Tracts. PLoS One. (2016) 11:e0150519. doi: 10.1371/journal.pone.0150519
53. Cai X, Peng Y, Gong Y, Huang X, Liu L, Chen Y, et al. Variations of bile bacterial community alongside gallstone disease progression and key taxa involved in poor outcomes after endoscopic surgery. Eur J Med Res. (2023) 28:313. doi: 10.1186/s40001-023-01308-y
54. Hov JR and Karlsen TH. The Microbiome in Primary Sclerosing Cholangitis: Current Evidence and Potential Concepts. Semin Liver Dis. (2017) 37:314–31. doi: 10.1055/s-0037-1608801
55. Rühlemann M, Liwinski T, Heinsen F-A, Bang C, Zenouzi R, Kummen M, et al. Consistent alterations in faecal microbiomes of patients with primary sclerosing cholangitis independent of associated colitis. Aliment Pharmacol Ther. (2019) 50:580–9. doi: 10.1111/apt.15375
56. Bajer L, Kverka M, Kostovcik M, Macinga P, Dvorak J, Stehlikova Z, et al. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J Gastroenterol. (2017) 23:4548–58. doi: 10.3748/wjg.v23.i25.4548
57. Lapidot Y, Amir A, Ben-Simon S, Veitsman E, Cohen-Ezra O, Davidov Y, et al. Alterations of the salivary and fecal microbiome in patients with primary sclerosing cholangitis. Hepatol Int. (2021) 15:191–201. doi: 10.1007/s12072-020-10089-z
58. Desjonqueres E, Campani C, Marra F, Zucman-Rossi J, and Nault J-C. Preneoplastic lesions in the liver: Molecular insights and relevance for clinical practice. Liver Int. (2022) 42:492–506. doi: 10.1111/liv.15152
59. Qiao F, Li X, Liu Y, Zhang S, Liu D, and Li C. Periodontitis and NAFLD-related diseases: A bidirectional two-sample Mendelian randomization study. Oral Dis. (2024) 30:3452–61. doi: 10.1111/odi.14785
60. Sato S, Kamata Y, Kessoku T, Shimizu T, Kobayashi T, Kurihashi T, et al. A cross-sectional study assessing the relationship between non-alcoholic fatty liver disease and periodontal disease. Sci Rep. (2022) 12:13621. doi: 10.1038/s41598-022-17917-2
61. Jensen A, Ladegaard Grønkjær L, Holmstrup P, Vilstrup H, and Kilian M. Unique subgingival microbiota associated with periodontitis in cirrhosis patients. Sci Rep. (2018) 8:10718. doi: 10.1038/s41598-018-28905-w
62. Bajaj JS, Matin P, White MB, Fagan A, Deeb JG, Acharya C, et al. Periodontal therapy favorably modulates the oral-gut-hepatic axis in cirrhosis. Am J Physiol Gastrointest Liver Physiol. (2018) 315:G824–37. doi: 10.1152/ajpgi.00230.2018
63. Bajaj JS, Betrapally NS, Hylemon PB, Heuman DM, Daita K, White MB, et al. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology. (2015) 62:1260. doi: 10.1002/hep.27819
64. Lu H, Qian G, Ren Z, Zhang C, Zhang H, Xu W, et al. Alterations of Bacteroides sp., Neisseria sp., Actinomyces sp., and Streptococcus sp. populations in the oropharyngeal microbiome are associated with liver cirrhosis and pneumonia. BMC Infect Dis. (2015) 15:239. doi: 10.1186/s12879-015-0977-x
65. Jia B, Kim KH, Ruan W, Kim HM, and Jeon CO. Lantibiotic-encoding Streptococcus in the human microbiome are underlying risk factors for liver diseases. J Infect. (2022) 84:e70–2. doi: 10.1016/j.jinf.2022.02.020
66. Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. (2014) 513:59–64. doi: 10.1038/nature13568
67. Sato S, Iino C, Chinda D, Sasada T, Tateda T, Kaizuka M, et al. Effect of Liver Fibrosis on Oral and Gut Microbiota in the Japanese General Population Determined by Evaluating the FibroScan-Aspartate Aminotransferase Score. Int J Mol Sci. (2023) 24:13470. doi: 10.3390/ijms241713470
68. Zhou D, Wang J, Weng M, Zhang Y, Wang X, Gong W, et al. Infections of Helicobacter spp. in the biliary system are associated with biliary tract cancer: a meta-analysis. Eur J Gastroenterol Hepatol. (2013) 25:447. doi: 10.1097/MEG.0b013e32835c0362
69. Wu Z-F, Zou K, Xiang C-J, Jin Z-J, Ding H-H, Xu S, et al. Helicobacter pylori infection is associated with the co-occurrence of bacteria in the oral cavity and the gastric mucosa. Helicobacter. (2021) 26:e12786. doi: 10.1111/hel.12786
70. Schulz C, Schütte K, Koch N, Vilchez-Vargas R, Wos-Oxley ML, Oxley APA, et al. The active bacterial assemblages of the upper GI tract in individuals with and without Helicobacter infection. Gut. (2018) 67:216–25. doi: 10.1136/gutjnl-2016-312904
71. Chen X, Wang N, Wang J, Liao B, Cheng L, and Ren B. The interactions between oral-gut axis microbiota and Helicobacter pylori. Front Cell Infect Microbiol. (2022) 12:914418. doi: 10.3389/fcimb.2022.914418
72. Yang J, He Q, Lu F, Chen K, Ni Z, Wang H, et al. A distinct microbiota signature precedes the clinical diagnosis of hepatocellular carcinoma. Gut Microbes. (2023) 15:2201159. doi: 10.1080/19490976.2023.2201159
73. Ma Y, Jiang Z, Wang Y, Pan L, Liu K, Xia R, et al. Tongue coating microbiota-based machine learning for diagnosing digestive system tumours. J Oral Microbiol. (2025) 17:2487645. doi: 10.1080/20002297.2025.2487645
74. Zhang Y, Zhang H, Liu B, and Ning K. Highly accurate diagnosis of pancreatic cancer by integrative modeling using gut microbiome and exposome data. iScience. (2024) 27:109294. doi: 10.1016/j.isci.2024.109294
Keywords: oral microbiota, oncogenic bacteria, biliary tract cancer, bacterial translocation, mechanisms
Citation: Zhang Y and Zhang S (2025) Oral microbiota and biliary tract cancers: unveiling hidden mechanistic links. Front. Oncol. 15:1585923. doi: 10.3389/fonc.2025.1585923
Received: 01 March 2025; Accepted: 26 May 2025;
Published: 11 June 2025.
Edited by:
Soumyadev Sarkar, Arizona State University, United StatesReviewed by:
Shintaro Nakajima, California Institute of Technology, United StatesOriana Lo Re, Mediterranean Institute for Transplantation and Highly Specialized Therapies (ISMETT), Italy
Zhengqi Li, China-Japan Friendship Hospital, China
Copyright © 2025 Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shu Zhang, c3Vlc3VlMDgyNUAxNjMuY29t
 Yuhan Zhang
Yuhan Zhang Shu Zhang1*
Shu Zhang1*