- 1Division of Breast Surgery, Department of General Surgery, Shenzhen People’s Hospital (The Second Clinical Medical College, Jinan University; The First Affiliated Hospital, Southern University of Science and Technology), Shenzhen, Guangdong, China
- 2Key Laboratory of Biomedical Engineering of Guangdong Province, South China University of Technology, Guangzhou, China
- 3Shenzhen International Graduate School, Tsinghua University, Shenzhen, Guangdong, China
- 4Department of Pathology, The Second Affiliated Hospital of Jinan University, Shenzhen People’s Hospital, Shenzhen, China
- 5Department of Anatomical and Cellular Pathology, Prince of Wales Hospital, The Chinese University of Hong Kong, Hong Kong, Hong Kong SAR, China
- 6Department of Radiology, Shenzhen People’s Hospital, The Second Clinical Medical College of Jinan University, Shenzhen, China
Background: Current management of ductal carcinoma in situ lacks robust risk stratification tools, leading to universal surgical and radiotherapy interventions despite heterogeneous progression risks. Optimizing therapeutic balance remains a critical unmet clinical need.
Materials and methods: We retrospectively analyzed two patient cohorts. The first included 173 cases with BI-RADS category 3 or higher findings, used to compare the diagnostic accuracy of four abbreviated MRI protocols against the full diagnostic MRI. The second cohort involved 210 patients who had both mammography and abbreviated MRI. We developed two separate predictive models—one for pure ductal carcinoma in situ and another for invasive ductal carcinoma with associated ductal carcinoma in situ—by integrating clinical, imaging, and pathological features. Deep learning and natural language processing techniques were used to extract relevant features, and model performance was assessed using bootstrap validation.
Results: Abbreviated Magnetic Resonance Imaging protocols demonstrated similar diagnostic accuracy to the full protocol (P > 0.05), offering a faster yet effective imaging option. The pure group incorporated features like nuclear grade, calcification morphology, and lesion size, achieving an Area Under the Curve of 0.905, with 86.8% accuracy and an F1 score of 0.853. The model for invasive cases incorporated features Ki-67 status, lymph vascular invasion, and enhancement patterns, achieved an Area Under the Curve of 0.880, with 86.2% accuracy and an F1 score of 0.834. Both models showed good calibration and clinical utility, as confirmed by bootstrap resampling and decision curve analysis.
Conclusion: Deep Learning-driven multimodal models enable precise ductal carcinoma in situ risk stratification, addressing overtreatment challenges. abbreviated Magnetic Resonance Imaging achieves diagnostic parity with full diagnostic protocol, positioning Magnetic Resonance Imaging as a viable ductal carcinoma in situ screening modality.
1 Introduction
Ductal carcinoma in situ (DCIS) is an early non-invasive lesion of breast cancer, accounting for approximately 18.6% of all newly diagnosed breast cancers (1). DCIS is generally considered a direct precursor to invasive ductal carcinoma (IDC), with approximately 25%-60% of untreated DCIS cases progressing to IDC over a follow-up period of 9–24 years (2). Due to the difficulty in distinguishing high-risk from low-risk DCIS, nearly all DCIS patients undergo either mastectomy or breast-conserving surgery, followed by radiotherapy or endocrine treatment (3). However, treatment of DCIS has not significantly reduced the incidence of invasive breast cancer, suggesting that some DCIS cases may not progress to invasive cancer (4, 5). Therefore, balancing overtreatment and undertreatment of DCIS has become a key clinical concern (6). Mammography is the conventional method for diagnosing DCIS (7), but its diagnostic efficacy is significantly affected by glandular density and tissue overlap (8, 9). Therefore, MRI, which is not influenced by glandular density and has higher sensitivity, is gradually being used as an adjunctive imaging tool for DCIS (10–13).
Artificial intelligence (AI), particularly deep learning (DL) technology, has shown great potential in the detection, diagnosis, and risk assessment of DCIS (14–18). AI algorithms based on mammography have been used to automatically identify calcified lesions, increasing the detection rate of DCIS and reducing the workload of radiologists (16, 19, 20). Furthermore, combining magnetic resonance imaging (MRI) with AI analysis can effectively overcome the limitations of traditional imaging caused by breast density, improving the sensitivity and diagnostic consistency of DCIS (21). However, due to the high time cost of MRI, it is not suitable for routine screening. Therefore, abbreviated breast MRI (Ab-MRI), which significantly reduces examination time, has gradually gained clinical acceptance (22). Nevertheless, current research on Ab-MRI mainly focuses on high-risk breast cancer populations and breast cancer screening, with limited studies using AI to analyze mammography and Ab-MRI image features for DCIS.
This study will combine clinical information with mammography and Ab-MRI image features to develop a prognostic risk prediction model for breast DCIS based on deep learning and multimodal imaging. It will also explore the value of Ab-MRI in the diagnosis and risk stratification of breast DCIS.
2 Materials and methods
2.1 Data set
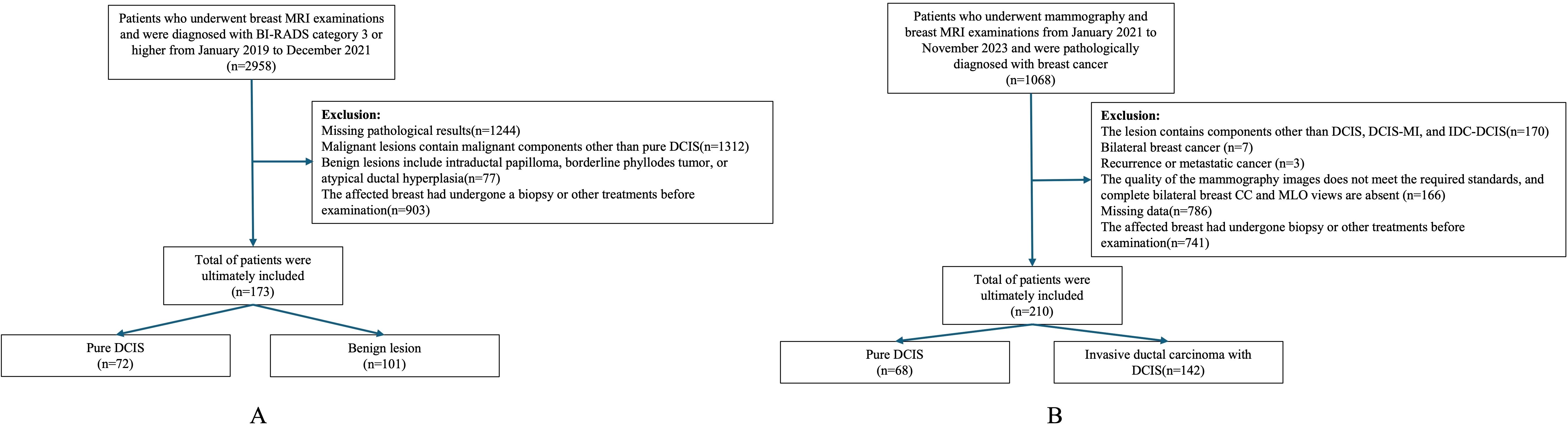
In Dataset One, We retrospectively collected data on 173 patients who underwent breast MRI examinations at our institution between January 2019 and December 2021 and were diagnosed as BI-RADS category 3 or higher. Among them, 72 cases were pure DCIS, and 101 cases were benign lesions. All patients were female, aged 23 to 77 years, with a median age of 46.4 ± 9.3 years. Inclusion criteria (1): Patients with pure DCIS who underwent breast-conserving surgery or total mastectomy after MRI examination (2). Patients with benign lesions who underwent at least core needle biopsy after MRI and had complete postoperative pathological data. Exclusion criteria (1): Malignant lesions containing components other than pure DCIS (2). Benign lesions associated with intraductal papilloma, borderline phyllodes tumor, or atypical ductal hyperplasia (3). Patients who underwent biopsy or other treatments on the affected breast before imaging examination (Figure 1A).
In Dataset Two, we collected data from 210 patients who underwent both mammography and breast MRI examinations at our institution between January 2021 and November 2023 and were pathologically confirmed to have breast cancer. Among them, 37 cases were pure DCIS, 31 cases were DCIS with microinvasion (DCIS-MI), and 142 cases were invasive ductal carcinoma with DCIS (IDC-DCIS). All patients were female, aged 29 to 71 years, with a median age of 49.0 ± 9.8 years. Given the similarity in clinical presentation and management strategies between pure DCIS and DCIS-MI, these two groups were combined for analysis. IDC-DCIS cases were characterized by predominant DCIS lesions with accompanying invasive components. Inclusion criteria (1): Pathologically confirmed DCIS-containing lesions (2); Underwent breast-conserving surgery or total mastectomy after mammography (3); Had complete clinical, imaging, pathological, and immunohistochemistry (IHC) data. Exclusion criteria (1): Lesions containing components other than DCIS, DCIS-MI, or IDC-DCIS (2); Bilateral breast cancer (3); Recurrent or metastatic breast cancer (4); Poor imaging quality or missing complete craniocaudal (CC) and mediolateral oblique (MLO) mammographic views for both breasts (5); Prior biopsy or other treatments on the affected breast before imaging examination (Figure 1B).
2.2 Examination methods
Breast mammography examinations were performed the following equipment: Siemens Mammomat Inspiration digital mammography system, GIOTTO IMAGE MD dual flat-panel digital mammography system, GE Senographe Pristina digital mammography system, and Hologic Selenia Dimensions mammography system. Standard CC and MLO views were acquired using a fully automatic compression system and automatic exposure control mode to optimize image quality (Figures 2A, B).
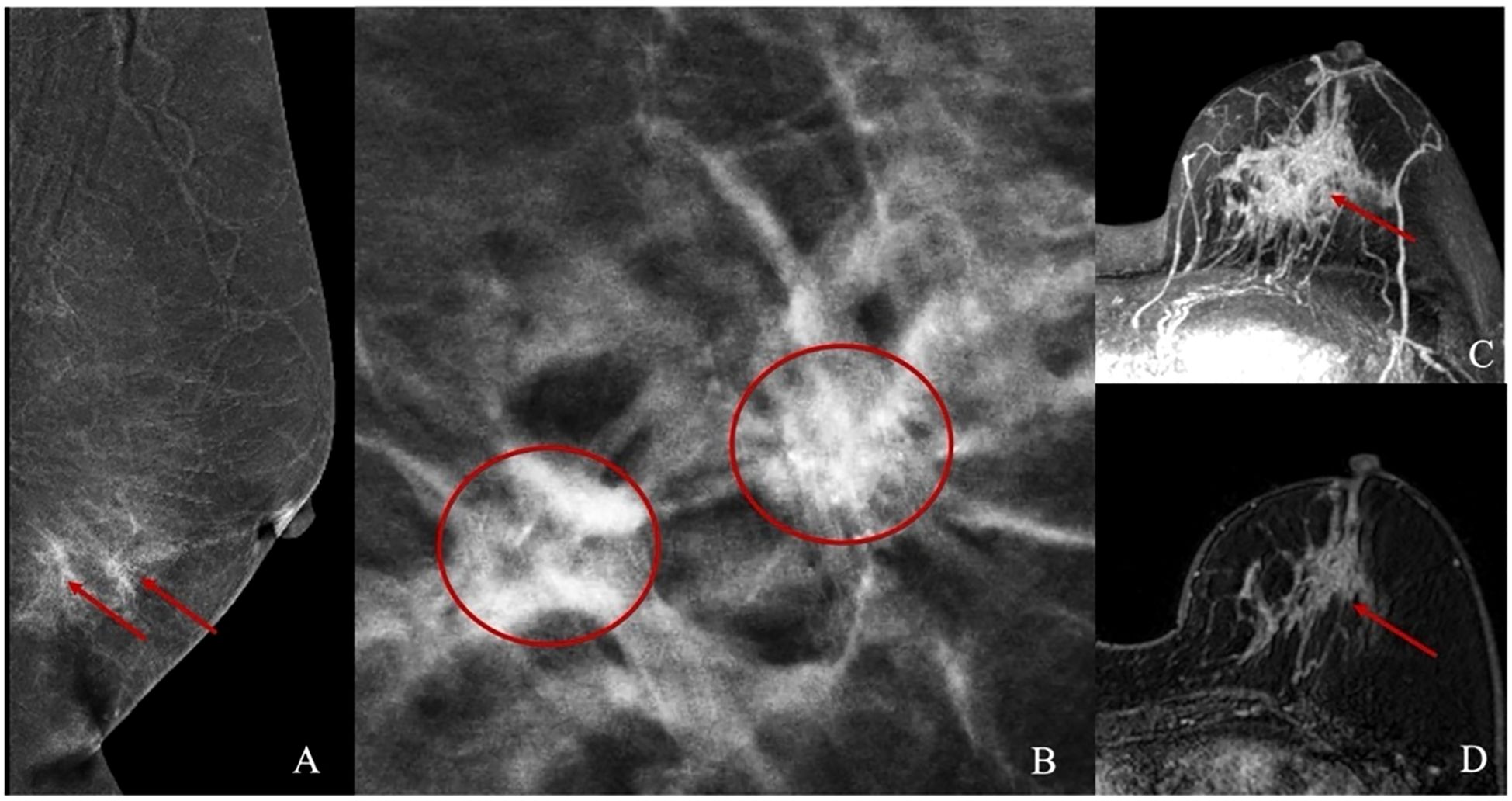
Figure 2. Illustrations of Mammography and Abbreviated Breast MRI. (A, B) present the mammographic images of a patient with non-mass enhancement and the corresponding calcification location. (C, D) depict the non-mass enhancement characteristics observed in the Abbreviated Breast MRI images.
MRI exams were conducted on Siemens Skyra 3.0T and Avanto 1.5T scanners with a dedicated breast coil, with patients positioned prone and both breasts naturally hanging. Unenhanced sequences included: T2-weighted imaging with fat suppression (T2WI-FS): Axial imaging with repetition time (TR) 3400/5000ms, echo time (TE) 54/58ms, slice thickness 4 mm, and slice gap 0.8/0.4 mm. Diffusion-weighted imaging (DWI): TR 5700/6400ms, TE 59/97 ms, slice thickness 4 mm, and slice gap 0.8/2 mm. Contrast-enhanced imaging was done 90 seconds after unenhanced imaging, with the first postcontrast phase using the following parameters: TR: 4.66/5.16ms. TE: 1.68/2.39ms. Slice thickness: 1.6/1.1 mm. Slice gap: 0.32/0.22 mm Contrast agent: Gadobenate dimeglumine, administered at 2.0 ml/s with a dose of 0.2 mmol/kg. Post-processing included maximum intensity projection (MIP) and first postcontrast subtracted (FAST) image reconstruction (Figures 2C, D).
2.3 Abbreviated breast MRI program screening
Given the unique biological characteristics of malignant tumors, this study incorporated DWI and early post-contrast images into the dynamic imaging protocol, with four imaging combinations designed as follows (1): T2WI-FS + DWI (2), T2WI-FS + MIP (3), DWI + MIP + FAST, and (4) T2WI-FS + DWI + MIP + FAST (12). A blinded reading approach was employed, where two radiologists with over five years of experience in breast MRI diagnosis independently evaluated cases from Dataset One using the four Ab-MRI protocols alongside the full diagnostic protocol (FDP). Each case was classified according to the BI-RADS system, categorizing them as BI-RADS 1, 2, 3 or BI-RADS 4, 5, without access to clinical data or other examination results. In cases of disagreement, a final classification was determined through consensus discussion. To minimize bias from repeated readings, a one-month interval was maintained between assessments for each protocol. Finally, the diagnostic performance of the four Ab-MRI protocols was compared with FDP, and the most effective Ab-MRI protocol was selected for further study.
2.4 Radiological feature extraction
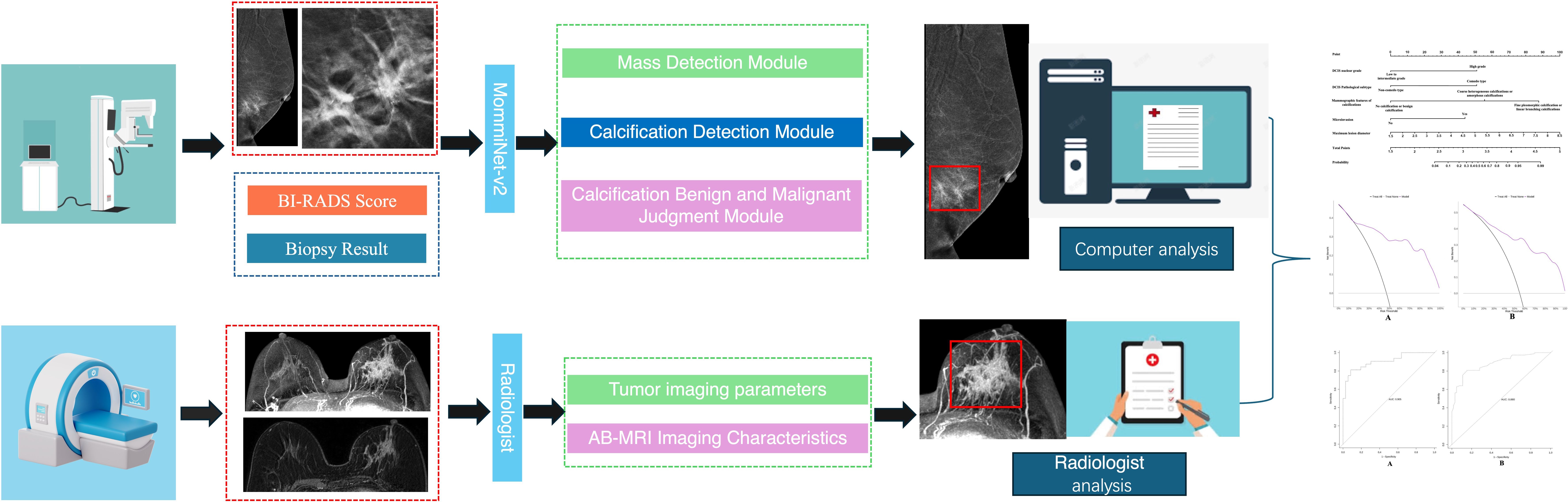
In the mammographic image feature analysis, 273 patients were randomly divided into a training set of 162 cases (simple group: 66 cases, invasive group: 96 cases) and a validation set (simple group: 44 cases, invasive group: 67 cases). This study adopted the AI model framework from the research team’s previous work (23) to analyze and extract both imaging and textual features, including breast tissue density, BI-RADS classification, and mammographic features. Building upon this, the MommiNet-v2 model was employed as a multi-view breast mass detection and classification system that simulates the radiologist’s clinical practice of comparing ipsilateral and contralateral views. It takes three-view mammograms along with clinical information (BI-RADS scores and biopsy results) as input. Structural and symmetry features are extracted through IpsiDualNet-v2 and BiDualNet-v2, then fused in an integrated network to generate final predictions. The training process incorporates nipple-based alignment, Focal Loss, and Distance-IoU Loss to enhance detection and classification accuracy. MommiNet-v2 has demonstrated strong performance on both public and real-world datasets, indicating its potential value for clinical implementation. According to the ACR BI-RADS guidelines (24), suspicious calcification shapes in mammography include amorphous, coarse heterogeneous, fine pleomorphic, and linear branching calcifications, while associated signs include skin retraction, nipple inversion, skin thickening, trabecular thickening, and axillary lymphadenopathy.
In the MRI image feature analysis, two radiologists with over five years of experience in breast MRI diagnosis evaluated Dataset Two using the selected optimal Ab-MRI protocol, with prior knowledge of patient clinical information, and recorded the degree of breast parenchymal enhancement, type of lesion enhancement, maximum lesion diameter, apparent diffusion coefficient (ADC) at high b-values, and the presence or absence of clustered ring enhancement within the lesion. If there was any discrepancy between the two radiologists, a final interpretation was reached through discussion. Figure 3 shows the flow chart of the study.
2.5 Pathological feature extraction
In the pathological analysis, the DCIS nuclear grade (low-intermediate/high) and pathological subtype (non-comedo/comedo) were recorded. Additionally, for the pure DCIS group, the presence or absence of microinvasion was noted, while for the invasive group, the proliferation index (Ki67), DCIS component proportion (using 25% as the threshold based on the concept of extensive intraductal component, EIC), and the presence or absence of neural/vascular invasion were also assessed. All specimens underwent IHC staining to evaluate the expression of Estrogen Receptor(ER), Progesterone Receptor(PR), Human Epidermal Growth Factor Receptor 2 (HER2), and Ki67. For patients in the invasive group, if discrepancies in IHC expression levels were observed between IDC and DCIS, the DCIS results were used as the reference. ER and PR positivity were defined as ≥1% nuclear staining. HER2 positivity was defined as 3+ (strong and complete membrane staining in >10% of tumor cells), while HER2 1+/0 was considered negative. For HER2 2+ cases, fluorescence in situ hybridization (FISH) was performed to confirm HER2 status, with HER2 gene amplification or FISH positivity indicating HER2 positivity. Ki67-positive cells ≤14% were classified as low proliferative activity, while >14% indicated high proliferative activity (25). All pathological results were independently reviewed by two attending pathologists with over 10 years of experience in breast pathology diagnosis. In cases of discrepancies, a panel of expert pathologists conducted a consensus review to determine the final interpretation.
2.6 Prediction model construction
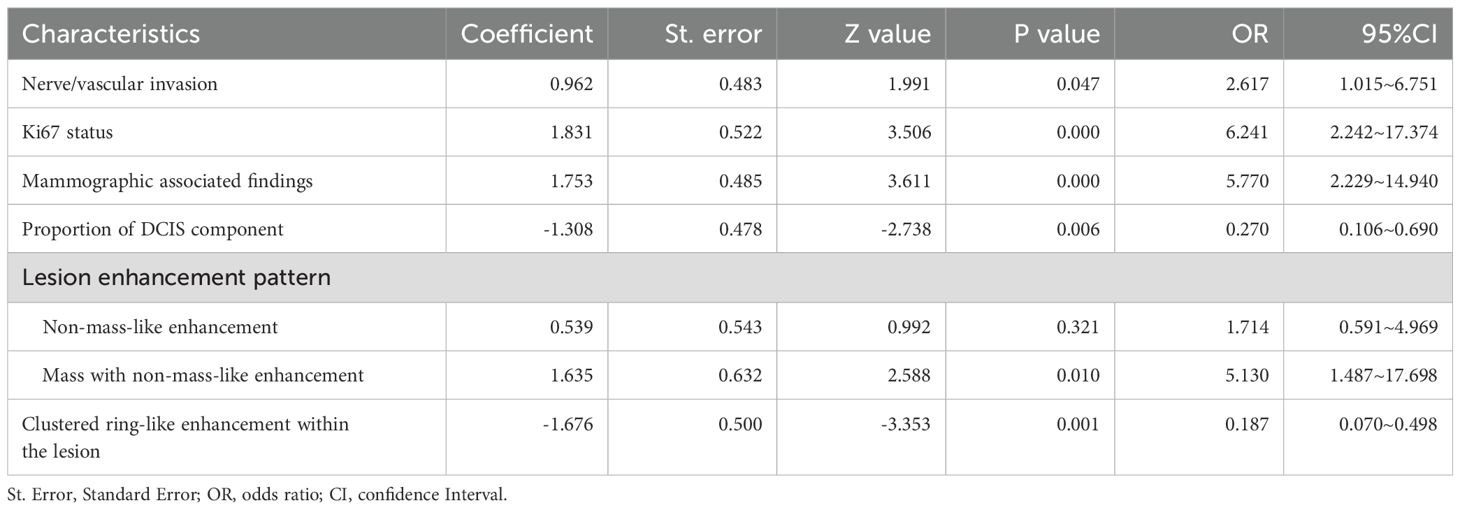
In the pure DCIS group prediction model, a total of eight features were extracted, including Ab-MRI image features, mammography image features, and non-imaging clinical features. These specific features were DCIS nuclear grade, DCIS pathological subtype, Mammographic features of calcifications, Presence of microinvasion, Degree of background parenchymal enhancement, Lesion enhancement pattern (derived from DCE-MRI), Maximum lesion diameter, Apparent diffusion coefficient value of the lesion. In the invasive group prediction model, nine features were extracted across the same three categories. These features included: Nerve/vascular invasion, Ki-67 proliferation index, Mammographic associated findings, Proportion of DCIS component, Degree of background parenchymal enhancement, Lesion enhancement pattern, Maximum lesion diameter, Apparent diffusion coefficient value, Clustered ring-like enhancement within the lesion.
All extracted features were initially subjected to univariate analysis, and those with statistical significance (P < 0.05) were subsequently included in multivariate logistic regression analysis. Features that remained significant in the multivariate analysis (P < 0.05) were identified as independent predictors of prognosis in both the pure DCIS and invasive cohorts.
To assess the performance of different model construction strategies, we used the Akaike Information Criterion (AIC) to evaluate model fit. The final models—the pure DCIS + DCIS-MI prediction model and the IDC-DCIS prediction model—were selected based on achieving the lowest AIC values, ensuring optimal balance between goodness-of-fit and model complexity.
2.7 Prognostic evaluation criteria
ER-negative or HER2-positive was selected as the reference standard for poor prognosis in the pure DCIS group (26–28). For the invasive group, the Nottingham Prognosis Index (NPI) (29) was used as the reference standard for poor prognosis, with NPI < 4.4 defined as good prognosis and NPI ≥ 4.4 as poor prognosis.
2.8 Statistical analysis
In the Ab-MRI protocol selection, SPSS 26.0 was used for statistical analysis to calculate diagnostic metrics for each MRI protocol, including positive predictive value (PPV), negative predictive value (NPV), sensitivity (SEN), specificity (SPE), and accuracy (ACC). The McNemar test was applied to compare the diagnostic performance of the Ab-MRI protocol with the full diagnostic protocol (FDP), with P < 0.05 indicating statistical significance. For predictive model establishment, R 4.1.2 was used for data organization and statistical analysis. Categorical data were presented as counts (percentages), and Pearson χ² test or Fisher’s exact test was used to compare differences between groups. For continuous data, if normally distributed, mean value was used, and independent t-test was applied for between-group comparisons; if not normally distributed, median value was used, and Mann-Whitney U test was employed for group comparisons. Univariate analysis was used to identify variables with P < 0.05, which were then included in multivariate logistic regression to establish the prediction model. The R “pROC” package was used to plot the ROC curve, and the “rms” package was used to create a nomogram for model visualization, along with calibration curves to assess model calibration. Additionally, the “dcurves” package was used to plot decision curves, and DCA was applied to assess the clinical net benefit of the model. To further investigate the stability of the model and ensure its fit, Bootstrap resampling (1000 iterations) was performed. P < 0.05 indicates statistical significance.
3 Results
3.1 Cohort characteristics and abbreviated breast MRI protocol
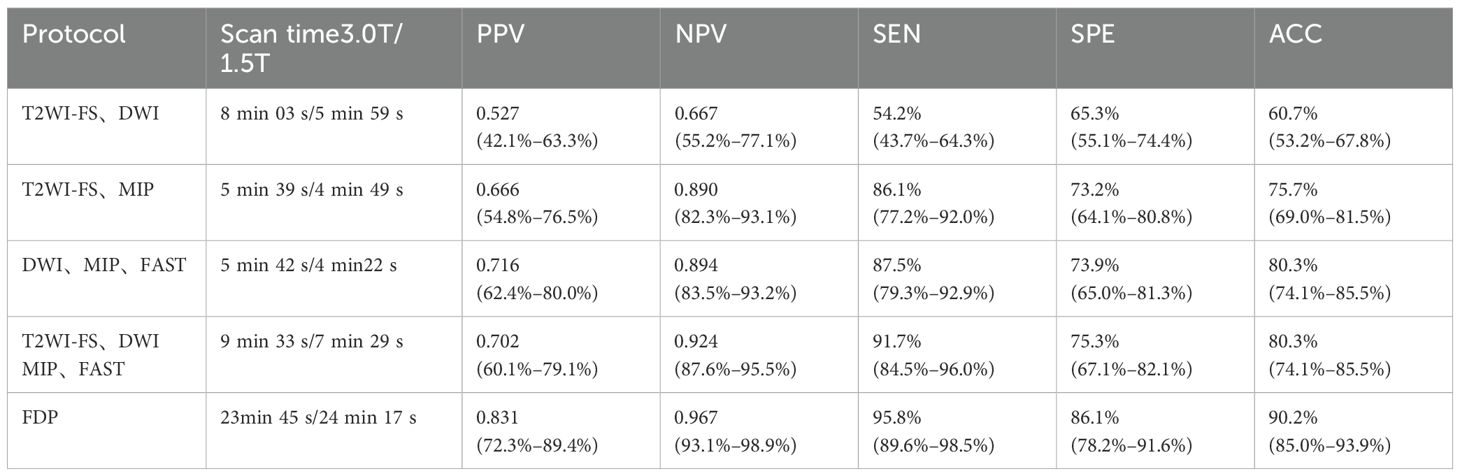
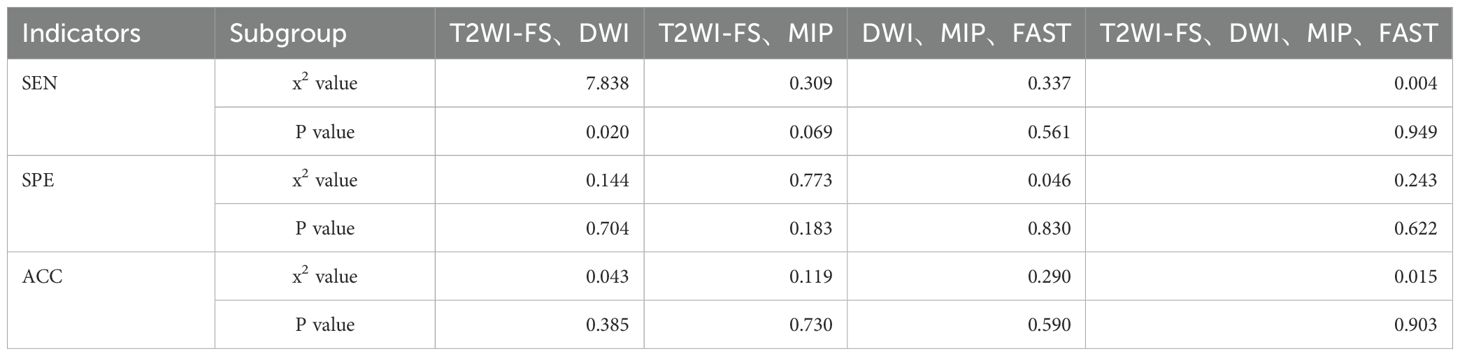
In Dataset One, this study selected 72 cases of pure DCIS and 101 cases of mixed benign lesions to evaluate the diagnostic performance of the Ab-MRI protocol. Among the benign lesions, there were 19 cases of fibroadenoma, 14 cases of fibrocystic breast disease, 3 cases of benign phyllodes tumor, 1 case of ductal adenoma, 1 case of hamartoma, and 1 case of breast tuberculosis, as well as 62 cases with no pathological result but no disease progression after 2 years of follow-up. The diagnostic performance and comparison of the four Ab-MRI protocols with FDP are shown in Tables 1, 2. In terms of scanning time, T2WI-FS was not affected by the device model, with scanning times exceeding 3 minutes; however, its diagnostic value was limited. In contrast, Protocol 3 (DWI, MIP, FAST) had a shorter scanning time and showed higher PPV, NPV, SEN and ACC. When comparing the four Ab-MRI protocols with the FDP, only Protocol 1 showed a significant difference in SEN compared to the FDP (P = 0.020), while no significant differences were found in sensitivity, specificity, or accuracy for the other protocols (P > 0.05). Therefore, the “DWI, MIP, FAST” protocol was selected for further study.
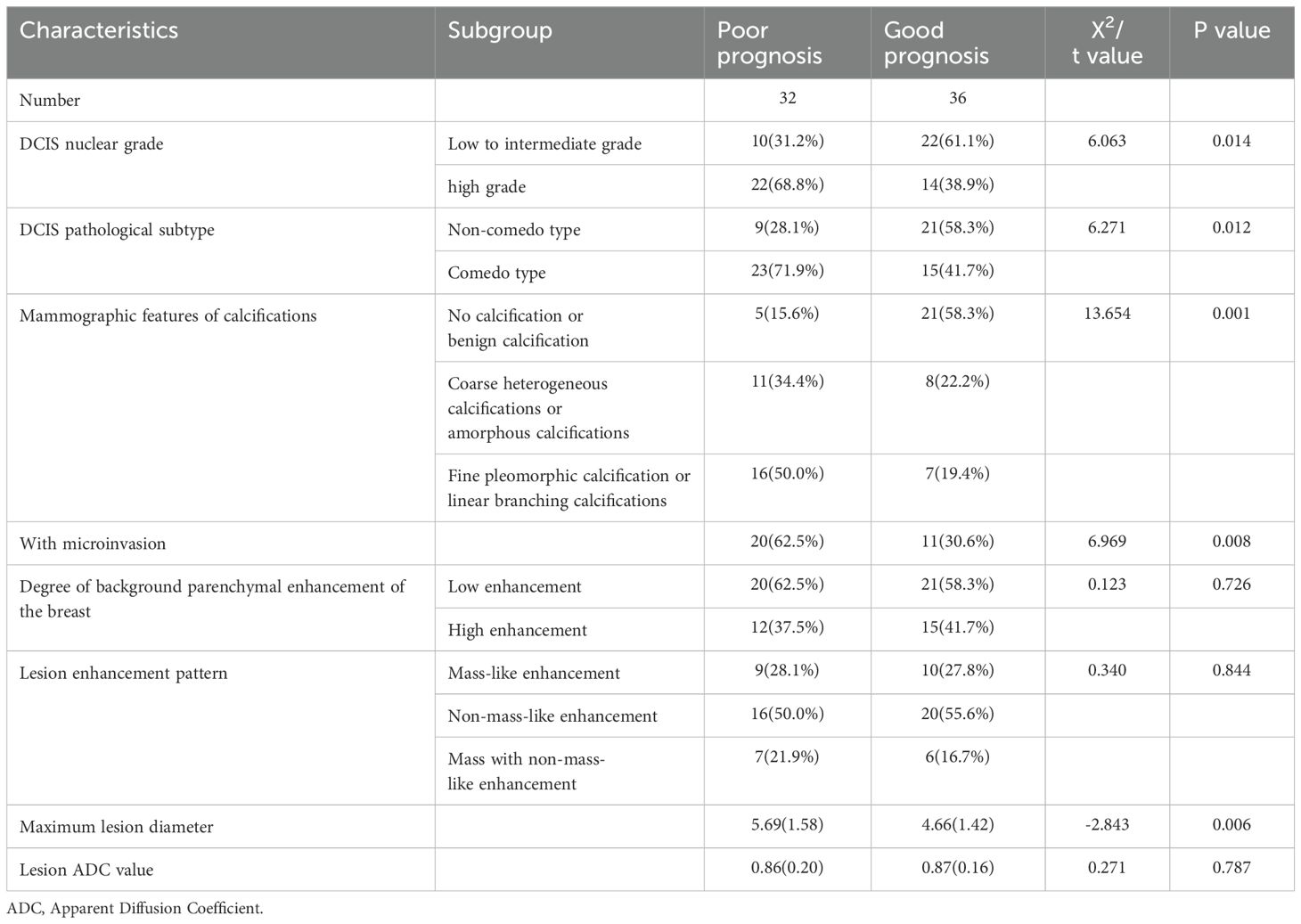
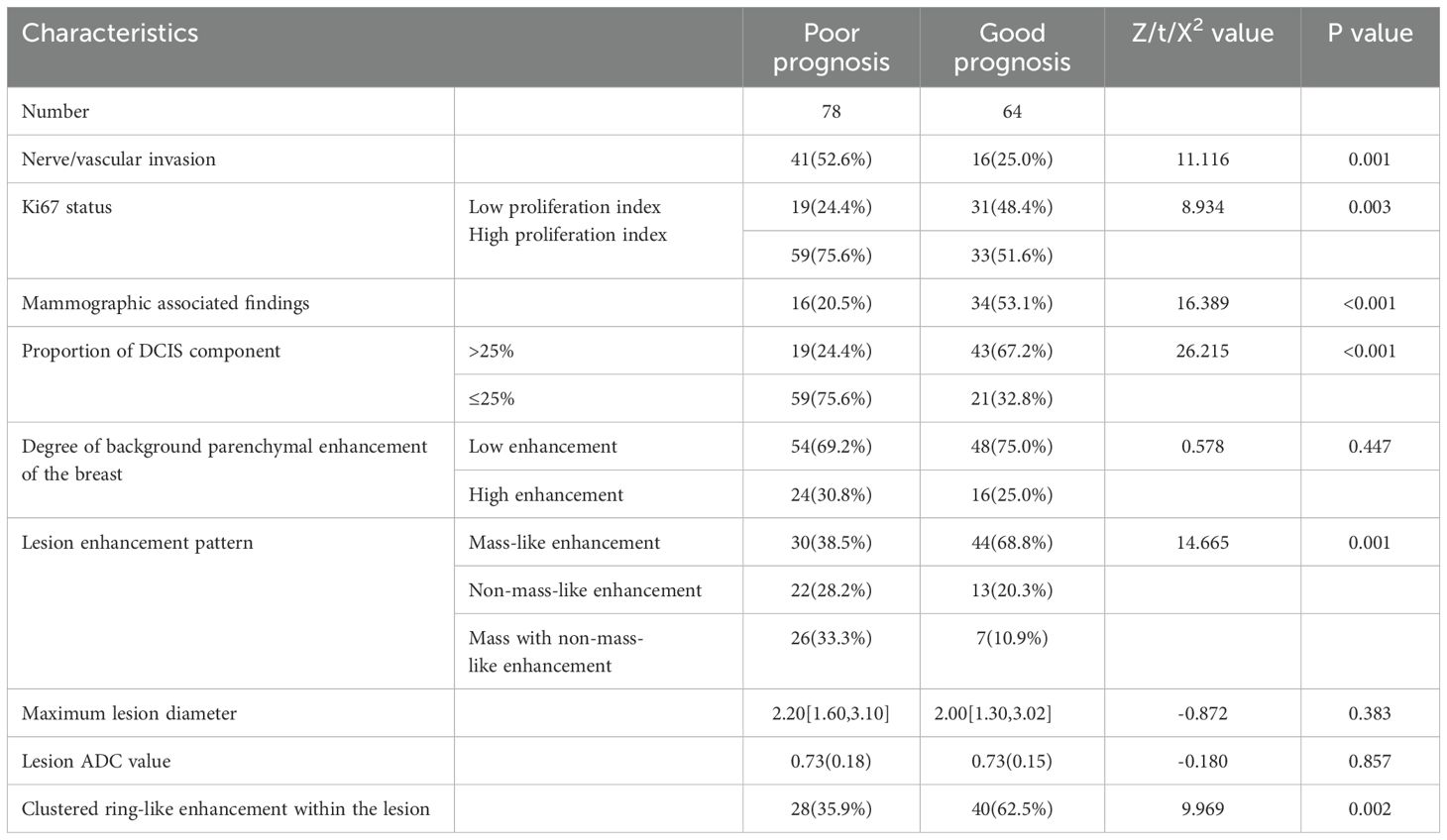
In Dataset Two, a total of 210 breast cancer patients were included in this study, with 68 patients in the pure DCIS group, of which 32 had poor prognosis and 36 had good prognosis. Among the 37 pure DCIS patients, 12 had poor prognosis and 25 had good prognosis; among the 31 DCIS-MI patients, 20 had poor prognosis and 11 had good prognosis. The invasive group included 142 patients, with 78 having poor prognosis and 64 having good prognosis. The baseline characteristics of patients in both the pure DCIS and invasive groups, as well as the results of univariate analysis, are presented in Tables 3, 4.
3.2 Development of predictive models
Through univariate comparisons between groups, factors with P < 0.05 were selected to construct the predictive models. In the pure DCIS group, 5 features, including DCIS nuclear grade, DCIS pathological subtype, mammographic calcification morphology, microinvasion, and maximum lesion diameter, were used to construct the predictive model, with an AIC value of 68. In the invasive group, 6 features, including neural/vascular invasion, Ki67 status, DCIS component proportion, mammographic associated findings, lesion enhancement type, and clustered ring enhancement within the lesion, were used to construct the predictive model, with an AIC value of 139.
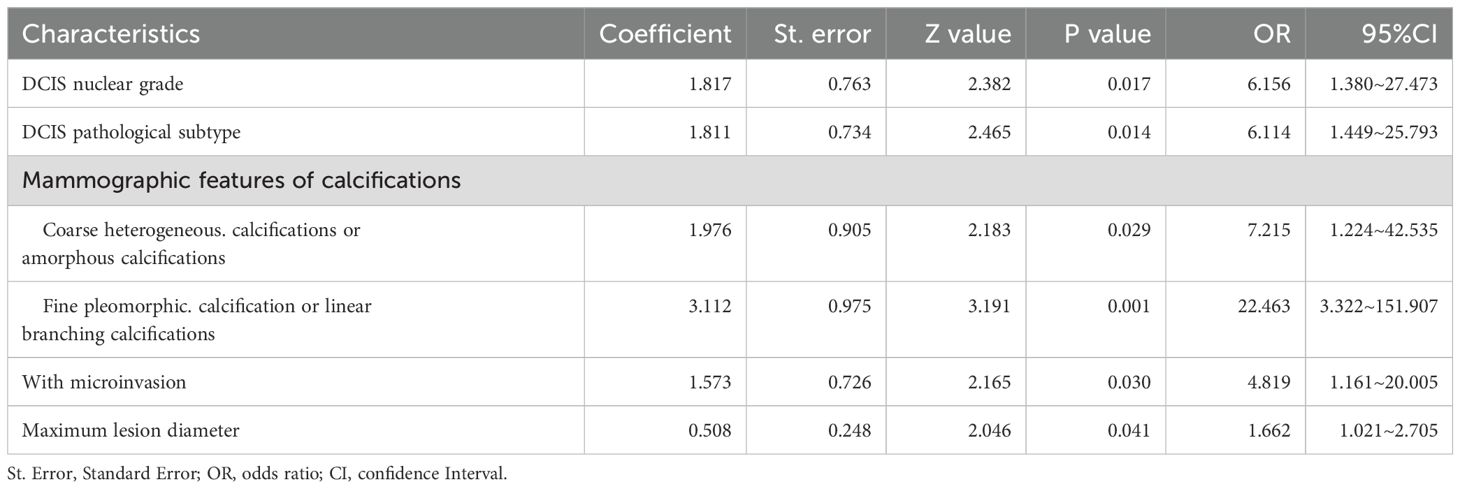
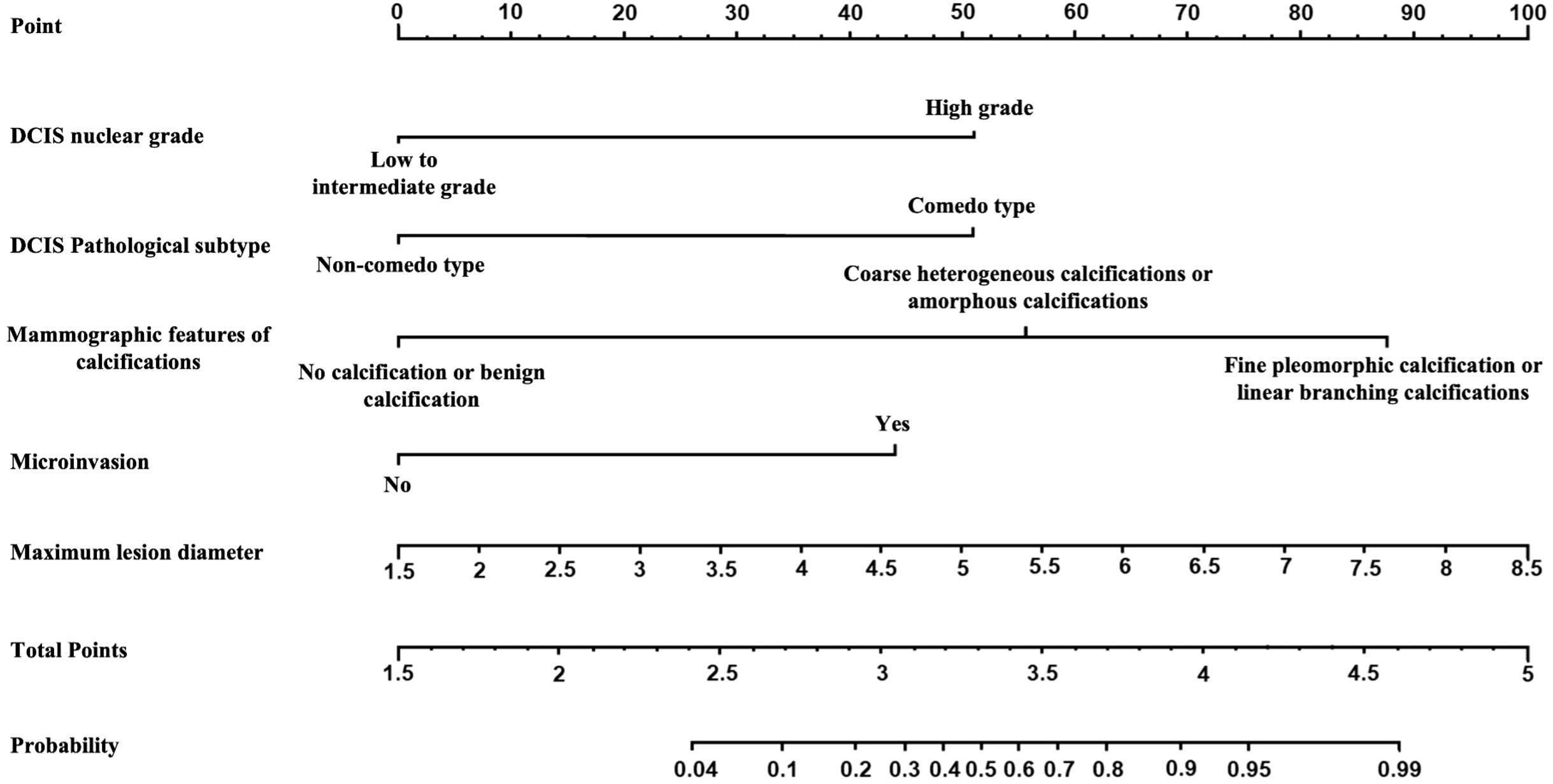
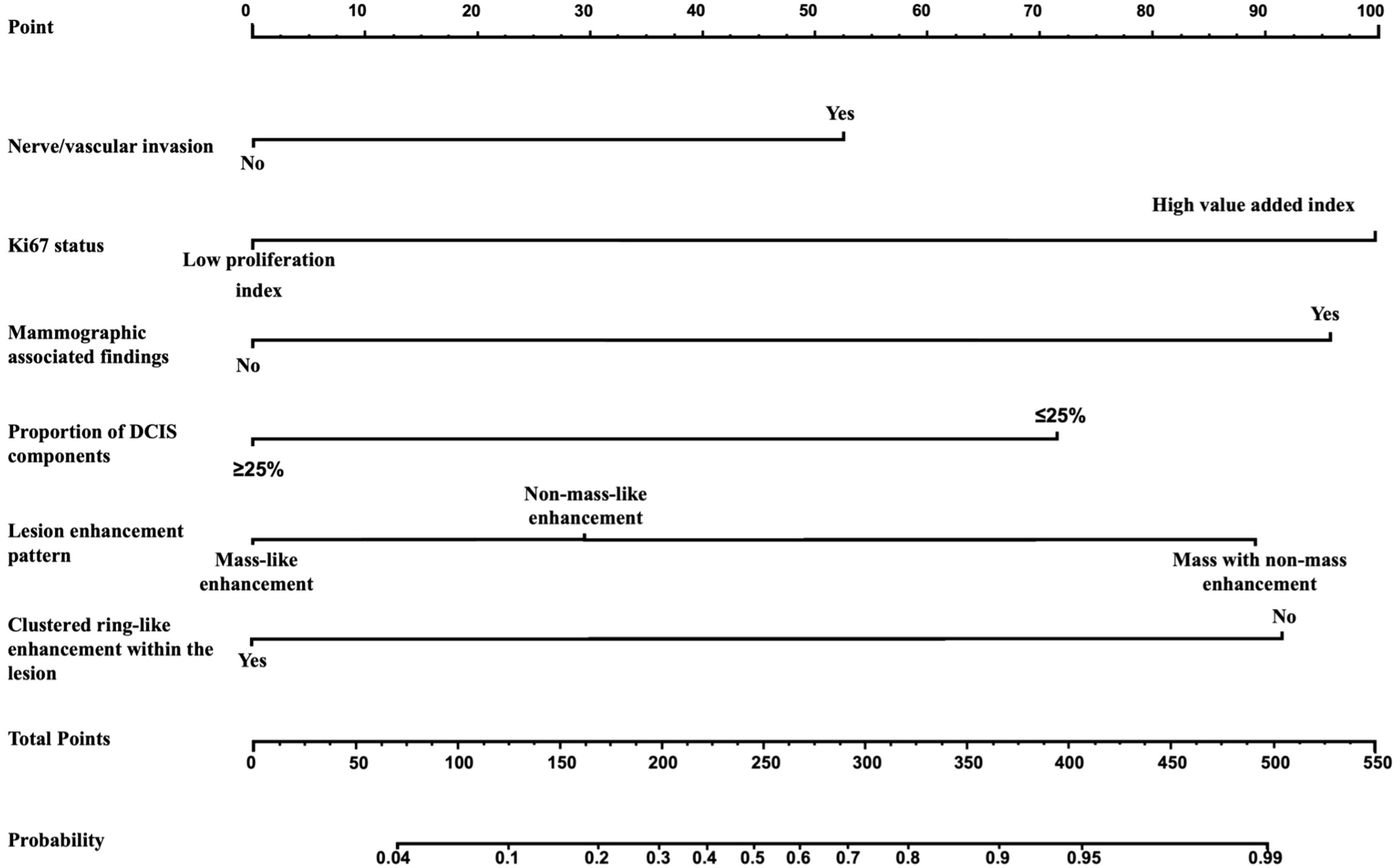
The multivariate analysis results for the pure DCIS group are shown in Table 5. DCIS nuclear grade, DCIS pathological subtype, mammographic calcification morphology, microinvasion, and maximum lesion diameter were identified as independent predictive factors for poor prognosis in the pure DCIS group. The nomogram for this predictive model is shown in Figure 4. The multivariate analysis results for the invasive group are shown in Table 6. Neural/vascular invasion, Ki67 status, mammographic associated findings, DCIS component proportion, lesion enhancement type, and clustered ring enhancement within the lesion were identified as independent predictive factors for poor prognosis in the invasive group. Among these, neural/vascular invasion, Ki67 status, mammographic associated findings, and lesion enhancement type had Odds Ratio(OR) values greater than 1, indicating these are risk factors, while DCIS component proportion and clustered ring enhancement had OR values less than 1, indicating these are protective factors. The nomogram for this predictive model is shown in Figure 5.
3.3 Evaluation of predictive model performance
ROC curve analysis (Figures 6A, B) showed that the pure DCIS group model for predicting the prognosis of pure DCIS + DCIS-MI had an AUC of 0.905 (95% CI 0.833–0.978). When the cutoff value was T = 0.642, the Youden index was maximized at 0.729, with a specificity of 91.7%(95%CI 82.4–97.5) and a sensitivity of 81.3%(95%CI 70.2–89.8). The corresponding prediction model achieved an accuracy of 86.76%(95%CI 82.1–90.8) and an F1 score of 0.853. The invasive group model for predicting the prognosis of IDC-DCIS had an AUC of 0.880 (95% CI 0.825–0.935). When the cutoff value was T = 0.573, the Youden index was maximized at 0.651, with a specificity of 84.4%(95%CI 75.9–91.2) and a sensitivity of 80.8%(95%CI 71.6–88.3). This model achieved an accuracy of 86.2%(95%CI 81.5–90.1) and an F1 score of 0.834.
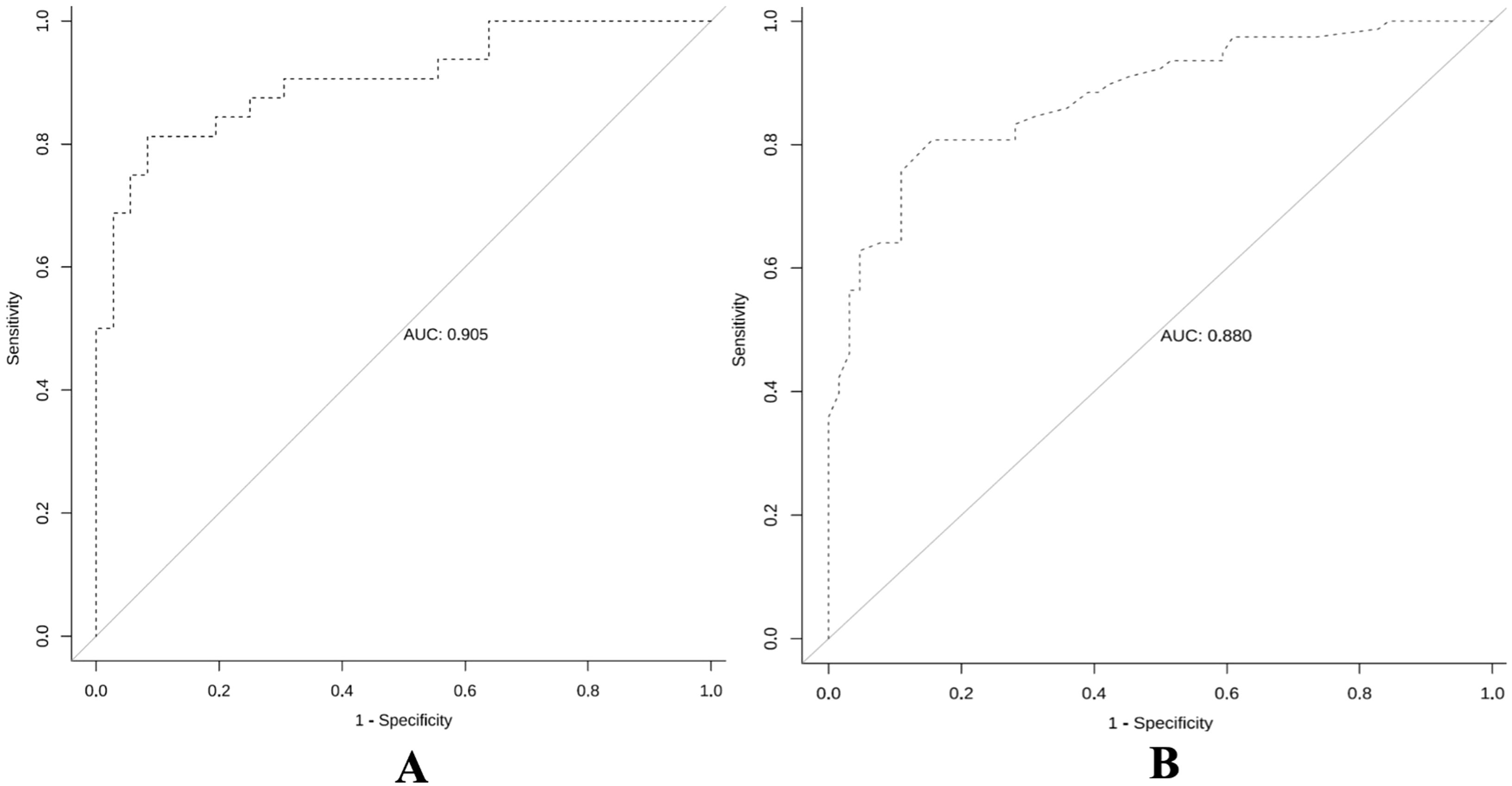
Figure 6. Receiver operating characteristic curves of the predictive models for the pure DCIS group (A) and the invasive group (B).
To further validate the stability of the model, an internal validation using the Bootstrap resampling method was performed with 1000 repetitions, and the mean AUC and 95% CI of the ROC curve were calculated. The results showed that the mean AUC for the pure DCIS group model was 0.906 (95% CI 0.801–0.957), and the mean AUC for the invasive group model was 0.879 (95% CI 0.817–0.927).
In the calibration curve analysis (Figures 7A, B), the X-axis represents the predicted risk probability, and the Y-axis represents the actual risk probability. The deviation correction curves of both prediction models fit well with the 45° ideal curve, indicating that the predicted malignant risk probability closely matches the actual malignant risk probability. According to the Hosmer and Lemeshow test, the P value for the pure DCIS model was 0.523, and the P value for the invasive group model was 0.127, both greater than 0.05, suggesting that the model fit is good. Decision curve analysis (Figures 8A, B) showed that the prediction model curves for both the pure DCIS group and the invasive group were significantly better than the two extreme lines. When the risk threshold was greater than 0.15 for the pure DCIS group and greater than 0.10 for the invasive group, net benefit was achieved, indicating that the model is beneficial for clinical decision-making.
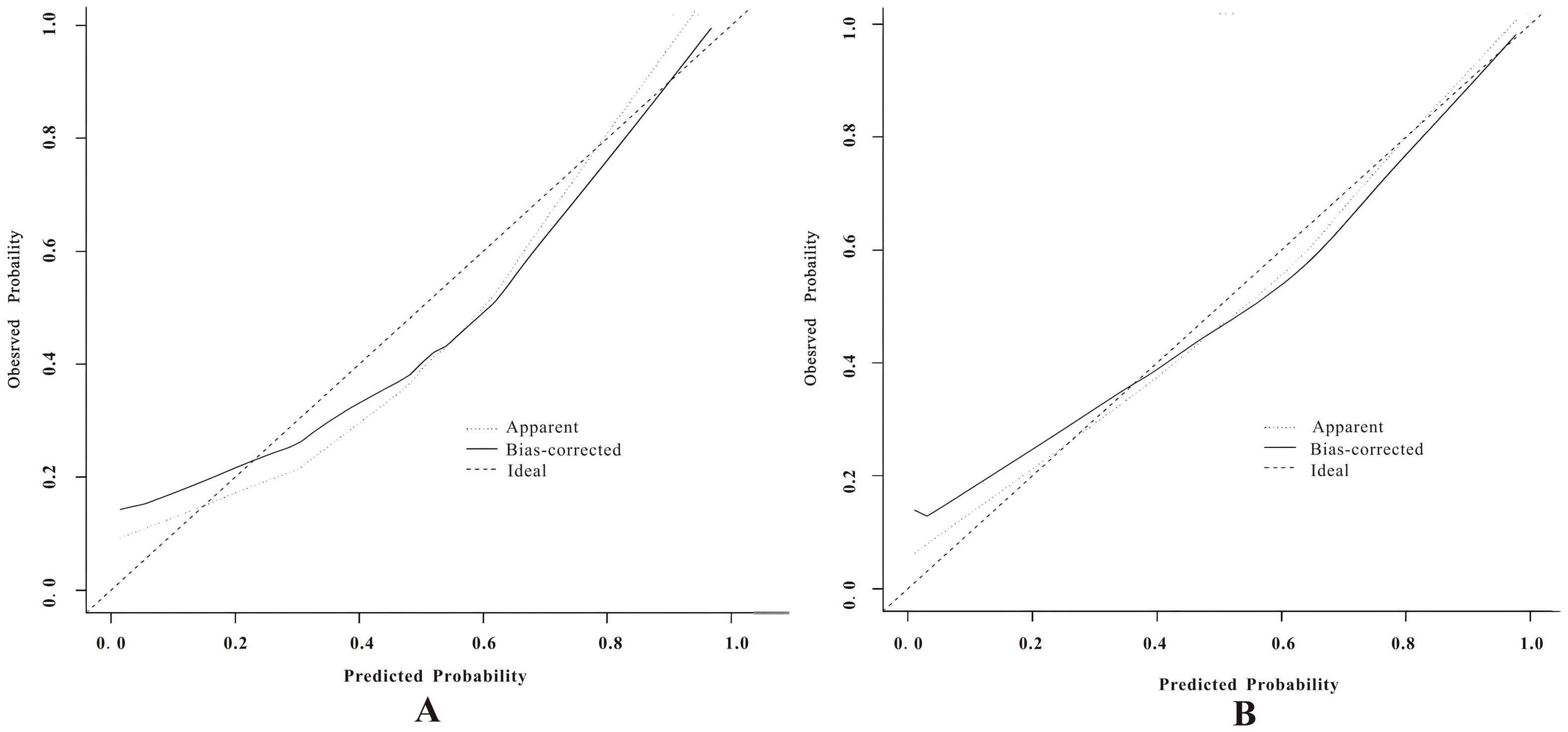
Figure 7. Calibration curves of the predictive models for the pure DCIS group (A) and the invasive group (B).
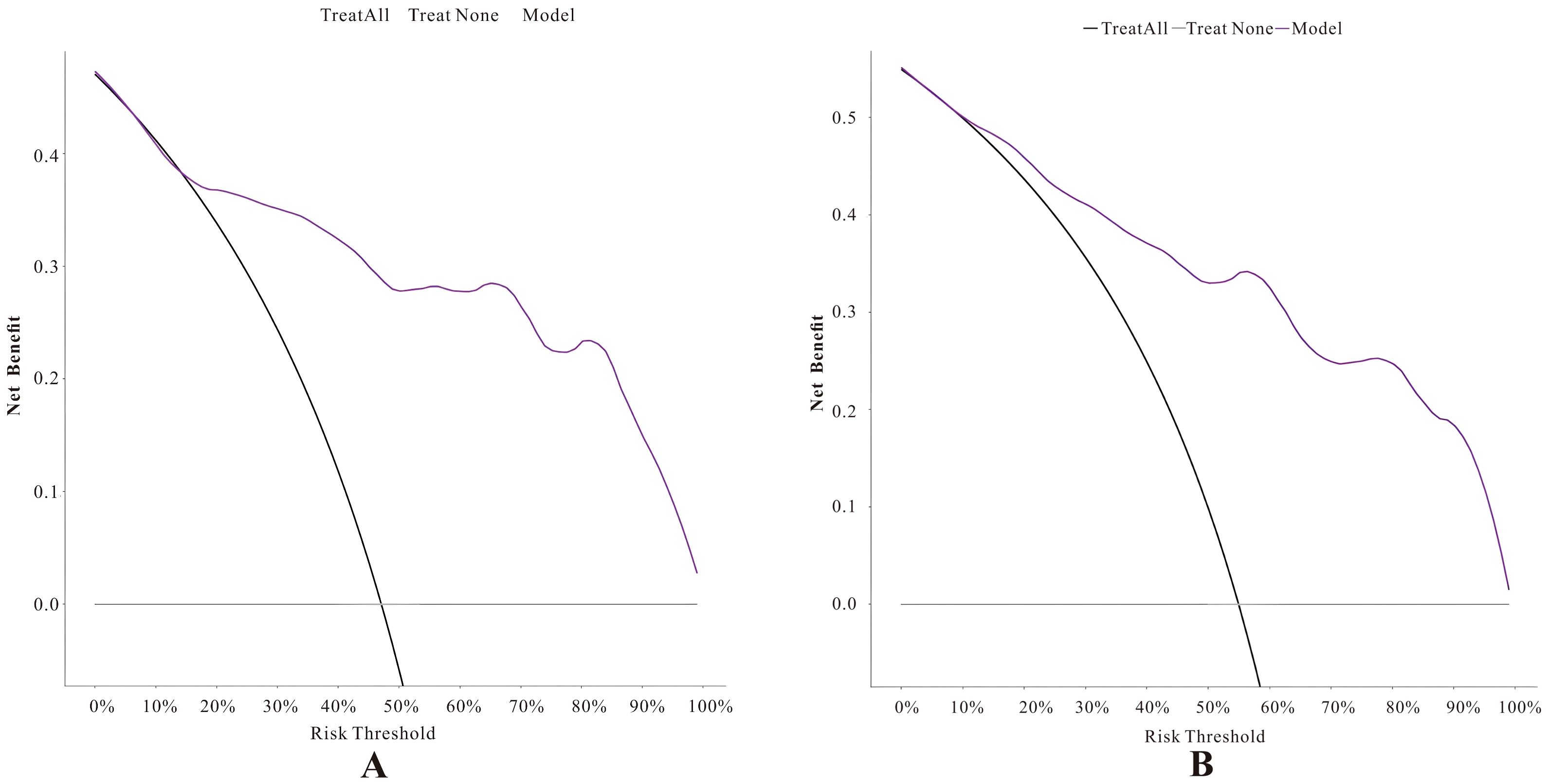
Figure 8. Decision curves of the predictive models for the pure DCIS group (A) and the invasive group (B).
This study compared the diagnostic performance of four Ab-MRI protocols with the full MRI protocol for DCIS. The DWI, MIP, and FAST protocols were selected for image feature extraction. In the pure DCIS group, the maximum lesion diameter was included, while in the invasive group, the lesion enhancement type and clustered ring enhancement within the lesion were incorporated. The first section’s predictive factors were combined to establish a multimodal prediction model. The results showed that the diagnostic performance of the Ab-MRI protocol for DCIS was not significantly different from the full MRI protocol, and the examination time was notably shorter. Additionally, the Ab-MRI protocol effectively performed risk stratification for DCIS at different pathological stages, with the established prediction model demonstrating good performance in ROC curve analysis, calibration curve analysis, and decision curve analysis.
4 Discussion
The extent and distribution of DCIS lesions are difficult to assess, posing a significant challenge for clinicians and patients in determining the appropriate surgical treatment. In the context of approximately 20% of DCIS cases developing invasive breast cancer in the same breast after treatment (30, 31). Therefore, accurate risk stratification can help guide the precise extent of surgery for patients who wish to preserve their breast (32, 33). We evaluated the diagnostic performance of different imaging features by two datasets, employing Ab-MRI protocols including DWI, MIP, and FAST. Our results demonstrate that the model effectively stratifies the risk of DCIS at different pathological stages and shows good performance in ROC, calibration, and decision curve analyses, providing accurate prognostic predictions for clinical use.
DCIS is a localized lesion, it exhibits considerable biological heterogeneity across patients, and some cases may progress to invasive breast cancer. Therefore, accurate diagnosis is essential for guiding appropriate treatment decisions. Mammography remains the primary screening tool for DCIS due to its ability to detect microcalcifications. However, its diagnostic performance is limited by breast density, which can lead to underdiagnosis or misclassification, particularly in non-calcified lesions (34, 35).This limitation may result in incomplete excision during breast-conserving surgery, thereby increasing the risk of recurrence and sometimes necessitating wider surgical resection. In contrast, MRI offers superior spatial resolution and contrast-enhanced imaging capabilities, enabling earlier and more accurate detection of DCIS and holding promise for improved long-term patient outcomes (36, 37). Furthermore, MRI has demonstrated higher sensitivity in detecting additional malignancies, with a detection rate of 87.9% compared to 63.6% for mammography, providing a more comprehensive assessment of disease burden (38).
However, despite its superior diagnostic capabilities in DCIS detection, MRI is not suitable for routine screening due to its high cost and lengthy examination time. As a potential alternative, Ab-MRI has emerged as a promising technique that significantly reduces scan time while maintaining diagnostic accuracy. Several studies comparing the diagnostic performance of Ab-MRI with that of standard MRI have demonstrated that, despite improving examination efficiency, Ab-MRI shows no significant differences in key diagnostic metrics, including recall rate, cancer detection rate, false-positive biopsy recommendation rate, PPV, SEN and SPE (13, 22). Our study further confirms that Ab-MRI provides comparable diagnostic performance to full-protocol MRI while significantly reducing scan time. However, compared with the reference FDP, the Ab−MRI acquisition protocol exhibits wider confidence intervals, likely due to three inherent limitations. First, the accelerated acquisition reduces temporal resolution, amplifying biological noise arising from intertumoral heterogeneity—especially in DCIS cases with variable necrosis patterns. Second, the lack of a compensation mechanism for dynamic contrast–enhanced phases prevent effective correction of motion artifacts, thereby increasing measurement uncertainty. Third, many low− and intermediate−grade DCIS lesions appear isointense on DWI sequences, which diminishes lesion–to–background contrast and further exacerbates data variability. Although FDP remains the diagnostic gold standard, in a screening context our Ab−MRI protocol—with a clinically acceptable specificity of 85%—demonstrates sufficient reliability and practical value.
By integrating mammographic imaging, MRI features, and pathological data, Ab-MRI allows for accurate prognostic risk assessment in DCIS patients. In addition to offering higher sensitivity and accuracy in imaging evaluation, Ab-MRI enhances diagnostic efficiency, making it a valuable tool for early breast cancer screening and diagnosis.
The novelty of this study lies in the first-ever integration of the Ab-MRI protocol with mammography to stratify the risk of DCIS at different pathological progression stages and explore the potential applications of this technology in breast precancerous lesions. By optimizing MRI sequences, Ab-MRI not only enhances diagnostic efficiency but also provides critical support for precise risk stratification and personalized treatment of DCIS. Despite its clinical value, this study has certain limitations. First, biopsy pathology results indicating intraductal papilloma, borderline phyllodes tumor, or atypical ductal hyperplasia often led to surgical excision in clinical practice. Since final surgical pathology may upgrade these lesions to DCIS or invasive carcinoma, we did not categorize them as benign lesions in our study, which may introduce selection bias. Second, as a single-center retrospective study with a relatively small sample size, the generalizability of our findings may be limited. Furthermore, considering the generally favorable prognosis and long survival of breast cancer patients, this study did not include disease-free survival or overall survival as prognostic endpoints. Lastly, as our study retrospectively analyzed imaging using the Ab-MRI protocol rather than simulating a real-world abbreviated MRI examination, potential bias in the results cannot be excluded.
In conclusion, this study explored the potential of Ab-MRI in the diagnosis of DCIS and utilized the Nottingham grading system for prognostic stratification. By integrating deep learning and natural language processing techniques, along with mammographic and Ab-MRI imaging features, we developed a DCIS prognostic risk prediction model to guide clinical decision-making and support precision medicine.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki. The study was approved by Shenzhen Peoples Hospital ethics board (LL-KY-2021624) and individual consent for this retrospective analysis was waived. The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’ legal guardians/next of kin because this research was retrospective analysis.
Author contributions
TZ: Data curation, Formal Analysis, Investigation, Methodology, Project administration, Writing – original draft, Writing – review & editing. TC: Resources, Validation, Writing – review & editing. ZC: Formal Analysis, Methodology, Software, Supervision, Validation, Visualization, Writing – review & editing. JH: Methodology, Supervision, Validation, Writing – review & editing. JM: Conceptualization, Data curation, Funding acquisition, Project administration, Resources, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Guangdong Provincial Natural Science Foundation (grant number 2025A1515012849).
Acknowledgments
The authors would like to thank all those involved in the study for dedicating their time and skills to the completion of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviation
DCIS, Ductal carcinoma in situ; IDC, invasive ductal carcinoma; DL, deep learning; AI, Artificial intelligence; MRI, magnetic resonance imaging; Ab-MRI, abbreviated breast MRI; DCIS-MI, Ductal carcinoma in situ with microinvasion; IDC-DCIS, invasive ductal carcinoma with Ductal carcinoma in situ; IHC, immunohistochemistry; CC, complete craniocaudal; MLO, mediolateral oblique; T2WI-FS, T2-weighted imaging with fat suppression; TR, repetition time; TE, echo time; DWI, Diffusion-weighted imaging; MIP, maximum intensity projection; FAST, first postcontrast subtracted; ADC, apparent diffusion coefficient; EIC, extensive intraductal component; FISH, fluorescence in situ hybridization; ER, Estrogen Receptor; PR, Progesterone Receptor; HER2, Human Epidermal Growth Factor Receptor 2; OR, Odds Ratio; AIC, Akaike Information Criterion; NPI, Nottingham Prognosis Index; PPV, positive predictive value; NPV, negative predictive value; SEN, sensitivity; SPE, specificity; ACC, accuracy; FDP, full diagnostic protocol.
References
1. Siegel RL, Kratzer TB, Giaquinto AN, Sung H, and Jemal A. Cancer statistics, 2025. CA Cancer J Clin. (2025) 75:10–45. doi: 10.3322/caac.21871
2. Wang J, Li B, Luo M, Huang J, Zhang K, Zheng S, et al. Progression from ductal carcinoma in situ to invasive breast cancer: molecular features and clinical significance. Signal Transduct Target Ther. (2024) 9:83. doi: 10.1038/s41392-024-01779-3
3. Schmitz R, van den Belt-Dusebout AW, Clements K, Ren Y, Cresta C, Timbres J, et al. Association of DCIS size and margin status with risk of developing breast cancer post-treatment: multinational, pooled cohort study. Bmj. (2023) 383:e076022. doi: 10.1136/bmj-2023-076022
4. Narod SA, Iqbal J, Giannakeas V, Sopik V, and Sun P. Breast cancer mortality after a diagnosis of ductal carcinoma in situ. JAMA Oncol. (2015) 1:888–96. doi: 10.1001/jamaoncol.2015.2510
5. Bleyer A and Welch HG. Effect of three decades of screening mammography on breast-cancer incidence. N Engl J Med. (2012) 367:1998–2005. doi: 10.1056/NEJMoa1206809
6. Delaloge S, Khan SA, Wesseling J, and Whelan T. Ductal carcinoma in situ of the breast: finding the balance between overtreatment and undertreatment. Lancet. (2024) 403:2734–46. doi: 10.1016/S0140-6736(24)00425-2
7. Rauch GM, Kuerer HM, Scoggins ME, Fox PS, Benveniste AP, Park YM, et al. Clinicopathologic, mammographic, and sonographic features in 1,187 patients with pure ductal carcinoma in situ of the breast by estrogen receptor status. Breast Cancer Res Treat. (2013) 139:639–47. doi: 10.1007/s10549-013-2598-7
8. Lee SH, Ryu HS, Jang MJ, Yi A, Ha SM, Kim SY, et al. Glandular tissue component and breast cancer risk in mammographically dense breasts at screening breast US. Radiology. (2021) 301:57–65. doi: 10.1148/radiol.2021210367
9. Ha SM, Jang MJ, Youn I, Yoen H, Ji H, Lee SH, et al. Screening outcomes of mammography with AI in dense breasts: A comparative study with supplemental screening US. Radiology. (2024) 312:e233391. doi: 10.1148/radiol.233391
10. Patel BK, Carnahan MB, Northfelt D, Anderson K, Mazza GL, Pizzitola VJ, et al. Prospective study of supplemental screening with contrast-enhanced mammography in women with elevated risk of breast cancer: results of the prevalence round. J Clin Oncol. (2024) 42:3826–36. doi: 10.1200/JCO.22.02819
11. Kuhl CK, Schrading S, Strobel K, Schild HH, Hilgers RD, and Bieling HB. Abbreviated breast magnetic resonance imaging (MRI): first postcontrast subtracted images and maximum-intensity projection-a novel approach to breast cancer screening with MRI. J Clin Oncol. (2014) 32:2304–10. doi: 10.1200/JCO.2013.52.5386
12. Comstock CE, Gatsonis C, Newstead GM, Snyder BS, Gareen IF, Bergin JT, et al. Comparison of abbreviated breast MRI vs digital breast tomosynthesis for breast cancer detection among women with dense breasts undergoing screening. Jama. (2020) 323:746–56. doi: 10.1001/jama.2020.0572
13. Jones LI, Marshall A, Elangovan P, Geach R, McKeown-Keegan S, Vinnicombe S, et al. Evaluating the effectiveness of abbreviated breast MRI (abMRI) interpretation training for mammogram readers: a multi-centre study assessing diagnostic performance, using an enriched dataset. Breast Cancer Res. (2022) 24:55. doi: 10.1186/s13058-022-01549-5
14. Zhang YD, Satapathy SC, Wu D, Guttery DS, Górriz JM, and Wang SH. Improving ductal carcinoma in situ classification by convolutional neural network with exponential linear unit and rank-based weighted pooling. Complex Intell Systems. (2021) 7:1295–310. doi: 10.1007/s40747-020-00218-4
15. Narayanan PL, Raza SEA, Hall AH, Marks JR, King L, West RB, et al. Unmasking the immune microecology of ductal carcinoma in situ with deep learning. NPJ Breast Cancer. (2021) 7:19. doi: 10.1038/s41523-020-00205-5
16. Liu C, Sun M, Arefan D, Zuley M, Sumkin J, and Wu S. Deep learning of mammogram images to reduce unnecessary breast biopsies: a preliminary study. Breast Cancer Res. (2024) 26:82. doi: 10.1186/s13058-024-01830-9
17. Fulawka L, Blaszczyk J, Tabakov M, and Halon A. Assessment of Ki-67 proliferation index with deep learning in DCIS (ductal carcinoma in situ). Sci Rep. (2022) 12:3166. doi: 10.1038/s41598-022-06555-3
18. Huang CY, Chang RF, Lin CY, Hsieh MS, Liao PC, Wang YJ, et al. Deep-learning model to improve histological grading and predict upstaging of atypical ductal hyperplasia/ductal carcinoma in situ on breast biopsy. Histopathology. (2024) 84:983–1002. doi: 10.1111/his.15144
19. Alaeikhanehshir S, Voets MM, van Duijnhoven FH, Lips EH, Groen EJ, van Oirsouw MCJ, et al. Application of deep learning on mammographies to discriminate between low and high-risk DCIS for patient participation in active surveillance trials. Cancer Imaging. (2024) 24:48. doi: 10.1186/s40644-024-00691-x
20. Wu H, Jiang Y, Tian H, Ye X, Cui C, Shi S, et al. Sonography-based multimodal information platform for identifying the surgical pathology of ductal carcinoma in situ. Comput Methods Prog BioMed. (2024) 245:108039. doi: 10.1016/j.cmpb.2024.108039
21. Zhu Z, Harowicz M, Zhang J, Saha A, Grimm LJ, Hwang ES, et al. Deep learning analysis of breast MRIs for prediction of occult invasive disease in ductal carcinoma in situ. Comput Biol Med. (2019) 115:103498. doi: 10.1016/j.compbiomed.2019.103498
22. Lawson MB, Partridge SC, Hippe DS, Rahbar H, Lam DL, Lee CI, et al. Comparative performance of contrast-enhanced mammography, abbreviated breast MRI, and standard breast MRI for breast cancer screening. Radiology. (2023) 308:e230576. doi: 10.1148/radiol.230576
23. Yang Z, Cao Z, Zhang Y, Tang Y, Lin X, Ouyang R, et al. MommiNet-v2: Mammographic multi-view mass identification networks. Med Image Anal. (2021) 73:102204. doi: 10.1016/j.media.2021.102204
24. Sickles EA. ACR BI-RADS® Atlas, Breast imaging reporting and data system. Am Coll Radiol. (2013) 39.
25. Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, and Senn HJ. Strategies for subtypes–dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. (2011) 22:1736–47. doi: 10.1093/annonc/mdr304
26. Thorat MA, Levey PM, Jones JL, Pinder SE, Bundred NJ, Fentiman IS, et al. Prognostic value of ER and pgR expression and the impact of multi-clonal expression for recurrence in ductal carcinoma in situ: results from the UK/ANZ DCIS trial. Clin Cancer Res. (2021) 27:2861–7. doi: 10.1158/1078-0432.CCR-20-4635
27. Kuerer HM, Albarracin CT, Yang WT, Cardiff RD, Brewster AM, Symmans WF, et al. Ductal carcinoma in situ: state of the science and roadmap to advance the field. J Clin Oncol. (2009) 27:279–88. doi: 10.1200/JCO.2008.18.3103
28. Holmes P, Lloyd J, Chervoneva I, Pequinot E, Cornfield DB, Schwartz GF, et al. Prognostic markers and long-term outcomes in ductal carcinoma in situ of the breast treated with excision alone. Cancer. (2011) 117:3650–7. doi: 10.1002/cncr.25942
29. Lee AH and Ellis IO. The Nottingham prognostic index for invasive carcinoma of the breast. Pathol Oncol Res. (2008) 14:113–5. doi: 10.1007/s12253-008-9067-3
30. Lips EH, Kumar T, Megalios A, Visser LL, Sheinman M, Fortunato A, et al. Genomic analysis defines clonal relationships of ductal carcinoma in situ and recurrent invasive breast cancer. Nat Genet. (2022) 54:850–60. doi: 10.1038/s41588-022-01082-3
31. Gorringe KL, Hunter SM, Pang JM, Opeskin K, Hill P, Rowley SM, et al. Copy number analysis of ductal carcinoma in situ with and without recurrence. Mod Pathol. (2015) 28:1174–84. doi: 10.1038/modpathol.2015.75
32. de Boniface J, Szulkin R, and Johansson ALV. Survival after breast conservation vs mastectomy adjusted for comorbidity and socioeconomic status: A swedish national 6-year follow-up of 48 986 women. JAMA Surg. (2021) 156:628–37. doi: 10.1001/jamasurg.2021.1438
33. Clarke M, Collins R, Darby S, Davies C, Elphinstone P, Evans V, et al. Effects of radiotherapy and of differences in the extent of surgery for early breast cancer on local recurrence and 15-year survival: an overview of the randomised trials. Lancet. (2005) 366:2087–106. doi: 10.1016/S0140-6736(05)67887-7
34. Monticciolo DL, Helvie MA, and Hendrick RE. Current issues in the overdiagnosis and overtreatment of breast cancer. AJR Am J Roentgenol. (2018) 210:285–91. doi: 10.2214/AJR.17.18629
35. Neal CH, Joe AI, Patterson SK, Pujara AC, and Helvie MA. Digital mammography has persistently increased high-grade and overall DCIS detection without altering upgrade rate. AJR Am J Roentgenol. (2021) 216:912–8. doi: 10.2214/AJR.20.23314
36. Chou SS, Romanoff J, Lehman CD, Khan SA, Carlos R, Badve SS, et al. Preoperative breast MRI for newly diagnosed ductal carcinoma in situ: imaging features and performance in a multicenter setting (ECOG-ACRIN E4112 trial). Radiology. (2021) 301:66–77. doi: 10.1148/radiol.2021219016
37. Grimm LJ, Rahbar H, Abdelmalak M, Hall AH, and Ryser MD. Ductal carcinoma in situ: state-of-the-art review. Radiology. (2022) 302:246–55. doi: 10.1148/radiol.211839
Keywords: abbreviated magnetic resonance imaging, deep learning, prognosis, ductal carcinoma in situ, mammography
Citation: Zhang T, Cui T, Cao Z, Hu J and Ma J (2025) Deep learning-based risk stratification of ductal carcinoma in situ using mammography and abbreviated breast magnetic resonance imaging. Front. Oncol. 15:1587882. doi: 10.3389/fonc.2025.1587882
Received: 05 March 2025; Accepted: 09 June 2025;
Published: 24 June 2025.
Edited by:
Izidor Mlakar, University of Maribor, SloveniaReviewed by:
Hongxiao Li, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaBabangida Lawal, Putra Malaysia University, Malaysia
Copyright © 2025 Zhang, Cui, Cao, Hu and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Ma, Y2pyLm1hamllQHZpcC4xNjMuY29t
 Tingfeng Zhang
Tingfeng Zhang Tingting Cui2
Tingting Cui2 Jie Ma
Jie Ma