- 1Section of Genomics and Transcriptomics, Fondazione Pisana per la Scienza, San Giuliano Terme, Pisa, Italy
- 2Department of Clinical and Experimental Medicine, University of Pisa, Pisa, Italy
- 3Department of Biology, University of Pisa, Pisa, Italy
- 4Department of Neurosurgery, Azienda Ospedaliera Universitaria Pisana, Pisa, Italy
- 5Neurosurgical Department of Spedali Riuniti di Livorno, Livorno, Italy
- 6Department of Pathology, New York University (NYU) Langone Medical Center, New York City, NY, United States
- 7Department of Translational Research and New Technologies in Medicine and Surgery, University of Pisa, Pisa, Italy
- 8Anatomical Pathology Department, Azienda Ospedaliera Toscana Nord-ovest, Livorno, Italy
- 9National Enterprise for nanoScience and nanoTechnology (NEST), Scuola Normale Superiore and Istituto Nanoscienze-CNR, Pisa, Italy
- 10Section of Nanomedicine, Fondazione Pisana per la Scienza, San Giuliano Terme, Pisa, Italy
- 11Department of Oncology, University College London Hospitals, London, United Kingdom
- 12Department of Radiation Oncology, Azienda Ospedaliera Universitaria Pisana, University of Pisa, Pisa, Italy
- 13Department of Oncology, University of Oxford, Oxford, United Kingdom
- 14Section of Bioinformatics, Fondazione Pisana per la Scienza, San Giuliano Terme, Pisa, Italy
A Corrigendum on
Metabolic-imaging of human glioblastoma live tumors: a new precision-medicine approach to predict tumor treatment response early
by Morelli M, Lessi F, Barachini S, Liotti R, Montemurro N, Perrini P, Santonocito OS, Gambacciani C, Snuderl M, Pieri F, Aquila F, Farnesi A, Naccarato AG, Viacava P, Cardarelli F, Ferri G, Mulholland P, Ottaviani D, Paiar F, Liberti G, Pasqualetti F, Menicagli M, Aretini P, Signore G, Franceschi S and Mazzanti CM (2022). Front. Oncol. 12:969812. doi: 10.3389/fonc.2022.969812
In the published article, there was an error in Figure 1 as published. The two figures above and below in Figures 1K-L, related to SOX2 staining, are part of a series of photographs aimed at demonstrating that GB explants—small tissue fragments approximately 300 µm in size—retain the original cytoarchitecture of the tissue from which they are derived (surgery tissue). The upper figure represents an image of the surgery tissue, while the lower figure shows the corresponding small explant (GB-EXP) derived from it. The SOX2 staining was specifically performed to add further evidence of the preserved representation of tumor components within the GB explant, a conclusion also supported by other images in the series (GFAP; CD105,CD33).
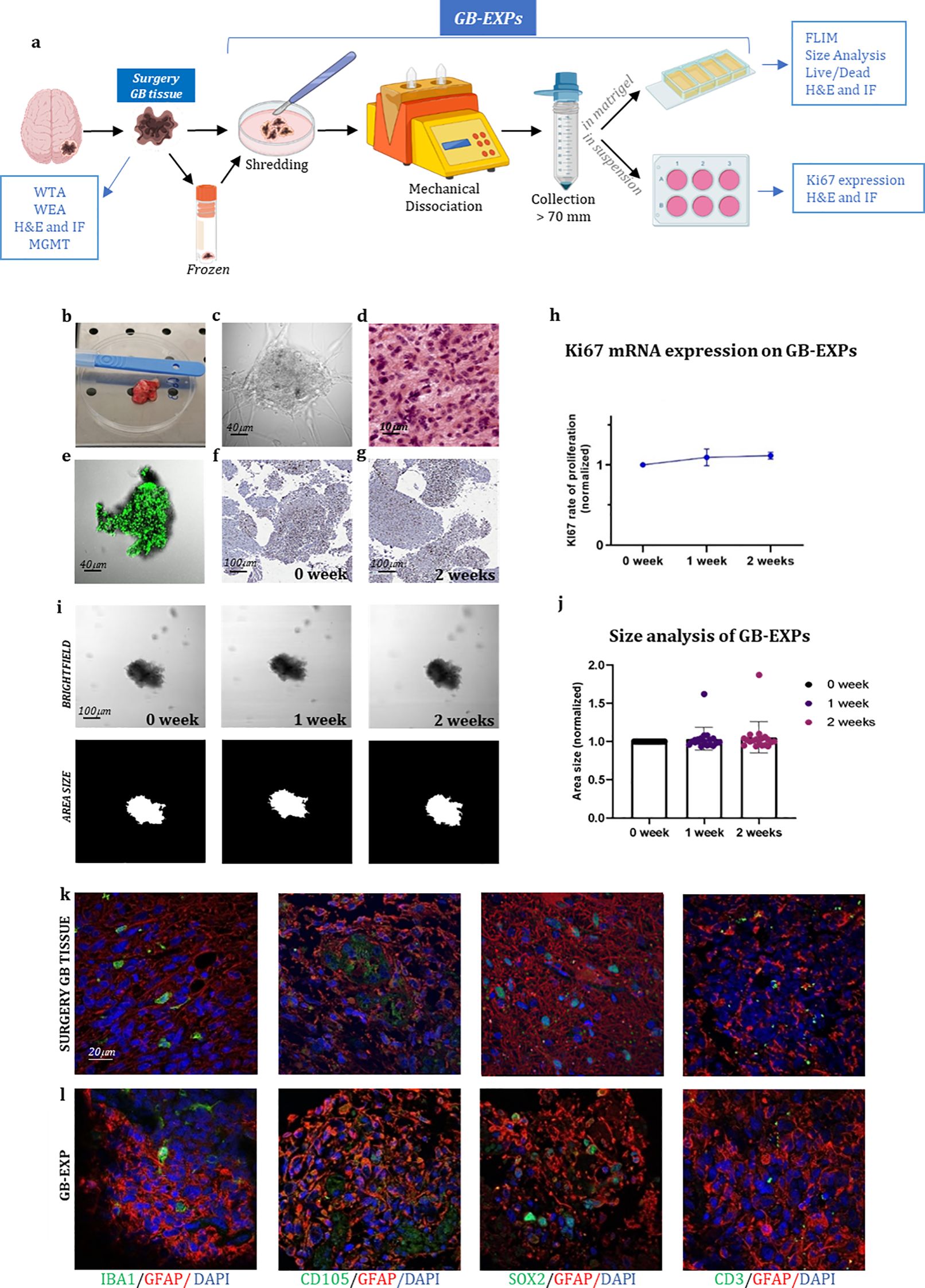
Figure 1. GB-EXPs cultures (A) Experimental design. (B) A surgery tumor sample. (C) GB-EXP embedded in matrigel. (D) H&E staining of a surgery GB tissue. (E) Live/dead staining of a GB-EXP at 2 weeks after culturing. (F, G) Ki67 immunostaining of GB-EXPs in suspension at 0 week and 2 weeks. (H) Ki67 mRNA expression analysis of 13 GB case-derived GB-EXPs at 0 and 2 weeks. Graph represents mean ± s.d. of triplicated measures. (I) Representative brightfield and area size images of a GB-EXP in matrigel at 0 week, 1 week and 2 weeks. (J) Size analysis 20 GB case-derived GB-EXPs at 0, 1 and 2 weeks. (K, L) Immunofluorescence assays of surgery GB tissue (K) and GB-EXPs (L). GFAP (astrocytes), IBA1 (microglia), SOX2 (stem cells), CD105 (endothelial cells) and CD3 (lymphocytes). WTA, Whole Transcriptome Analysis; WEA, Whole Exome Analysis; FC, Flow Cytometry; H&E, Hematoxylin, and Eosin; MGMT, MGMT promoter methylation analysis.
We acknowledge an error of duplication in the figures related to SOX2, as the upper image is, in fact, a photo of the same explant but with a slightly shifted area. We deeply regret this misplacement and are providing the correct image of the tumor stained with SOX2.The corrected Figure 1 and its legend appear below.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: glioblastoma, metabolic imaging, drug response assay, predictive model, FLIM (fluorescence lifetime imaging microscopy)
Citation: Morelli M, Lessi F, Barachini S, Liotti R, Montemurro N, Perrini P, Santonocito OS, Gambacciani C, Snuderl M, Pieri F, Aquila F, Farnesi A, Giuseppe Naccarato A, Viacava P, Cardarelli F, Ferri G, Mulholland P, Ottaviani D, Paiar F, Liberti G, Pasqualetti F, Menicagli M, Aretini P, Signore G, Franceschi S and Mazzanti CM (2025) Corrigendum: Metabolic-imaging of human glioblastoma live tumors: a new precision-medicine approach to predict tumor treatment response early. Front. Oncol. 15:1588290. doi: 10.3389/fonc.2025.1588290
Received: 05 March 2025; Accepted: 04 April 2025;
Published: 24 April 2025.
Edited and Reviewed by:
Ubaldo Emilio Martinez-Outschoorn, Thomas Jefferson University, United StatesCopyright © 2025 Morelli, Lessi, Barachini, Liotti, Montemurro, Perrini, Santonocito, Gambacciani, Snuderl, Pieri, Aquila, Farnesi, Giuseppe Naccarato, Viacava, Cardarelli, Ferri, Mulholland, Ottaviani, Paiar, Liberti, Pasqualetti, Menicagli, Aretini, Signore, Franceschi and Mazzanti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chiara Maria Mazzanti, Yy5tYXp6YW50aUBmcHNjaWVuY2UuaXQ=; Mariangela Morelli, bS5tb3JlbGxpQGZwc2NpZW5jZS5pdA==
 Mariangela Morelli
Mariangela Morelli Francesca Lessi
Francesca Lessi Serena Barachini
Serena Barachini Romano Liotti
Romano Liotti Nicola Montemurro
Nicola Montemurro Paolo Perrini
Paolo Perrini Orazio Santo Santonocito
Orazio Santo Santonocito Carlo Gambacciani
Carlo Gambacciani Matija Snuderl6
Matija Snuderl6 Francesco Cardarelli
Francesco Cardarelli Gianmarco Ferri
Gianmarco Ferri Fabiola Paiar
Fabiola Paiar Francesco Pasqualetti
Francesco Pasqualetti Paolo Aretini
Paolo Aretini Giovanni Signore
Giovanni Signore Sara Franceschi
Sara Franceschi Chiara Maria Mazzanti
Chiara Maria Mazzanti