- 1Colorectal Surgery of Huzhou Central Hospital, Affiliated Central Hospital of Huzhou University, Huzhou, China
- 2Central Laboratory of Huzhou Central Hospital, Affiliated Central Hospital of Huzhou University, Huzhou, China
- 3Pathologic Department of Huzhou Central Hospital, Affiliated Central Hospital of Huzhou University, Huzhou, China
- 4General Surgery of Huzhou Central Hospital, Affiliated Central Hospital of Huzhou University, Huzhou, China
Aim: To explore the effects of two combined methods—P53 expression and preoperative serum carcinoembryonic antigen (S-CEA) detection—on the prognosis of colorectal cancer (CRC).
Methods: Two classified combinations of tissue P53 and S-CEA were utilized: Combined P53 groups (normal P53 and S-CEA, or one or both elevated) and Recombined groups (P53 normal & S-CEA normal, P53 normal & S-CEA high, P53 high & S-CEA normal, P53 high & S-CEA high). Clinicopathologic features were analyzed by P53, S-CEA, Combined P53, and Recombined P53. Correlations between them were examined. Overall survival (OS) and disease-free survival (DFS) were evaluated using the Kaplan-Meier method and Log-Rank test. Univariate and multivariate analyses were performed for Combined P53 and Recombined P53 to determine independent factors. Three-year, two-year, and one-year OS and DFS were further analyzed using multimeROC. SPSS 27 and R 4.4.1 were used for analysis.
Results: TNM stage, CA199, differentiation, tumor maximum size, and minimum size showed significant differences between the single P53 and S-CEA groups (all P < 0.05). TNM stage, CA199, and chemotherapy differed in both Combined P53 and Recombined P53 groups (all P < 0.05). Significant correlations were found between P53, S-CEA, Combined P53, and Recombined P53 (all P < 0.001). No significant differences in OS and DFS were observed with P53 and Combined P53 (all P > 0.05), but differences were noted with S-CEA and Recombined P53 (all P < 0.05). Univariate and multivariate analyses identified laparoscopy, chemotherapy, differentiation, TNM stage, and Recombined P53 as independent factors for OS and DFS, while P53, S-CEA, and Combined P53 were not. Further multimeROC analysis showed that 3-year OS had better sensitivity and specificity (Area Under Curve [AUC] = 0.54), and 1-year DFS was better (AUC = 0.59).
Conclusions: Recombined P53 classification was more effective than traditional Combined P53 classification for assessing CRC prognosis and was an independent factor. Additionally, the 3-year OS and 1-year DFS analysis demonstrated higher sensitivity and specificity with Recombined P53.
1 Introduction
Colorectal carcinoma (CRC) is the fourth leading cause of cancer-related death, accounting for 600,000 deaths annually (1). Clinical factors such as tumor stage, tumor necrosis, vascular invasion, differentiation, Ki67, serum CEA, and inflammation have been reported to influence the prognosis of CRC patients (2–5). Although significant progress has been made in understanding CRC pathogenesis and clinical treatment, tumor resection remains the preferred option, with a 5-year survival rate of less than 65% (6). The p53 gene is a critical tumor suppressor activated by DNA damage, oxidative stress, and oncogene activation to produce p53 protein, which induces DNA repair, apoptosis, and regulates cell cycle checkpoints. TP53 mutations result in the loss of tumor suppressor function, enhancing tumor invasiveness and metastasis, leading to reduced survival rates (7–10). Patients with metastatic right-sided CRC (RCC) tend to have poorer survival compared to those with left-sided CRC (LCC), particularly among those with non-gain-of-function (non-GOF) mutp53. Conversely, gain-of-function (GOF) mutp53 is associated with worse survival only in patients with LCC (11). However, a recent study found no correlation between the TP53 Arg72Pro polymorphism and CRC risk, with no significant differences in genotype and allele frequencies across sex, age, histological grade, tumor stage, smoking status, or alcohol consumption (12). Basic studies have shown that p53 suppresses tumor progression through the p53 gene and signaling pathway (13, 14). Clinical studies on p53 expression in CRC are limited and often controversial, with available evidence failing to support p53 as a prognostic marker in metastatic CRC. Prospective studies with larger sample sizes and standardized methodologies are needed to explore the prognostic role of p53 in metastatic CRC patients (15–17). Consequently, combining p53 with other tumor markers in CRC, such as S-CEA, Ki67, MLH1, p16INK4a, and Kras, has been suggested (18–22). However, the outcomes remain controversial. Huang et al. found that patients with high preoperative serum CEA levels and P53 gene mutations had poor prognoses (23). Due to CRC’s highly heterogeneous nature, a single tumor marker is unlikely to serve as a standalone diagnostic tool due to its insufficient sensitivity and/or specificity. A combined approach using multiple tumor markers for CRC diagnosis holds potential as an effective strategy. The use of serum protein biomarkers may lead to the development of inexpensive, noninvasive tests for CRC detection, and combining tumor markers could improve screening effectiveness. If serum p53 antibody levels remain elevated long after resection, the case warrants intensive follow-up (24, 25). Given the importance of joint detection of P53 in CRC patients and the ongoing controversy surrounding related research, this study aims to explore the prognostic roles of CRC through two combined methods—tissue p53 and S-CEA.
2 Materials and methods
2.1 Patients
Data were collected from 750 CRC surgery patients at our hospital between January 2017 and December 2019. A total of 265 cases were excluded due to missing clinical, pathological, or follow-up data, endoscopic resection (EMR), or death from non-tumor-related causes. Ultimately, 485 patients were included in this study. The inclusion criteria were as follows: patients diagnosed with CRC through colonoscopy, computed tomography (CT), and pathological tests, either in our hospital or elsewhere; no preoperative adjuvant treatment; surgery performed in our department; routine preoperative S-CEA detection; lymph node dissection with ≥12 lymph nodes detected (although a small number of samples with 8–11 lymph nodes were included); CRC-related death as the termination event; and postoperative routine immunohistochemical (IHC) analysis and pathological examination for P53, with postoperative chemotherapy determined according to AJCC-8 guidelines (2). The exclusion criteria included serious diseases of the heart, brain, liver, or lungs that contraindicated surgery, non-CRC tumors leading to patient death, and missing follow-up or clinicopathological data. Data bias was minimized as much as possible through these inclusion and exclusion criteria.
2.2 Follow up
Patients were followed up every 3 months during the first year after primary CRC surgery, then every 6 months during the second year, and annually for the remaining 3 years, for a total of 5 years. All follow-up data were obtained from our records, either by phone or through the inpatient electronic medical record system (Haitai Software Version 3.0, Nanjing). Survival time was calculated from the date of primary surgery to the date of death or the end of the follow-up period, which lasted at least 5 years. If survival exceeded 60 months, it was capped at 60 months. Death due to the primary tumor or tumor-related disease was considered a positive event, while other causes were treated as censoring events. Thus, OS and DFS were analyzed.
2.3 Detection of S-CEA
Venous blood was drawn from each patient before surgery, and a fully automated chemiluminescence microsphere immunoassay device (ARCHITECT) was used to detect S-CEA. A reference range of 0–5 ng/ml was considered normal, while levels >5 ng/ml were classified as high.
2.4 Detection of tumor P53 by immunohistochemistry
Slicing - dewaxing to water - antigen repair (citric acid thermal repair) -4% hydrogen peroxide - addition of 1 antibody (P53) - incubation at 37°C for 30 minutes - universal type 2 antibody - incubation at 37°C for 30 minutes - DAB staining for 5 minutes - dehydration and sealing. Kits were purposed from Fuzhou Maixin Biotechnology Development Co., Ltd (Kits Number: MAB0674). The criterion for p53 positive staining results is the presence of yellow or brown or tan particles in the nucleus. No positive cells or positive cells <5% are negative, positive cells 5%– 25% are weakly positive (+), 25% ~75% is positive (++), >75% is strong positive (+++). Patients with p53 staining greater than 75% were classified as the p53 high-expression group, while the remaining patients were classified as the normal group (6). Figure 1A shows normal p53 expression (left ×200, right ×400), and Figure 1B shows high p53 expression (left ×200, right ×400).
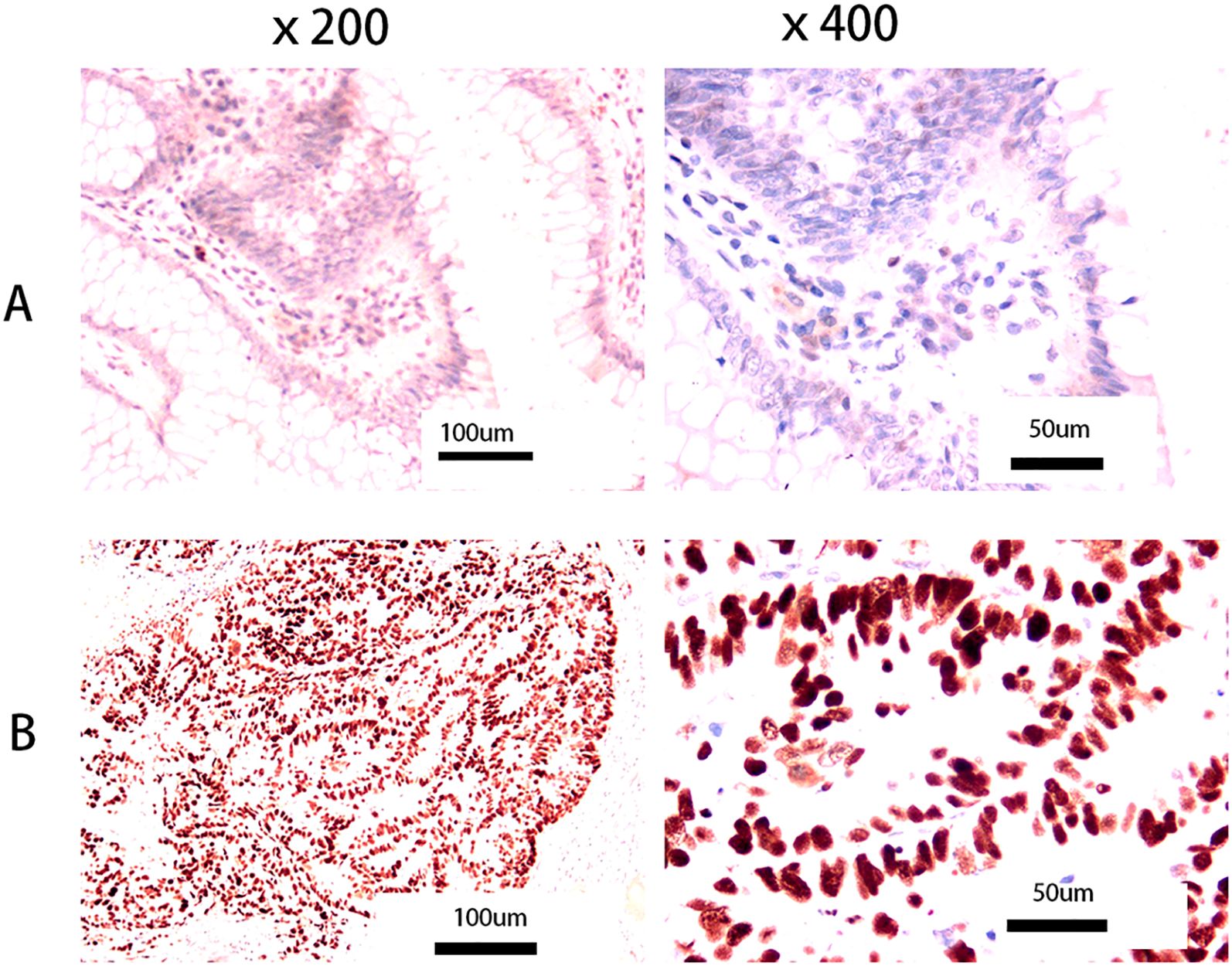
Figure 1. Pictures of p53 tumor expression. (A) Normal p53 expression (left: × 200, right: ×400); (B) High p53 expression (left: ×200, right: ×400). No positive cells or positive cells <5% are negative, positive cells 5%– 25% are weakly positive (+), 25% ~75% is positive (++), >75% is strong positive (+++). We refer to patients with p53 staining greater than 75% as the p53 high- expression group, and the rest as the normal group (6).
2.5 Two methods of combined P53 and S-CEA Classification (Combined P53 and Recombined P53)
Based on previous literature (26), when both p53 and S-CEA are normal, the classification is considered normal; otherwise, it is considered high, resulting in two groups: Combined P53: normal and high. Another method, as defined in two previously published studies (27, 28) (including our own), is the Recombined P53 method, which divides patients into four groups: p53 normal & S-CEA normal, p53 normal & S-CEA high, p53 high & S-CEA normal, and p53 high & S-CEA high.
2.6 Receiver operating characteristic curve analysis
OS and DFS were analyzed using ROC curves for Recombined P53. ROC curves were carried out for 1, 2, and 3 years of OS and DFS, and the sensitivity and specificity for these time periods were analyzed based on the AUC values.
2.7 Statistical analysis
All clinicopathological features were analyzed using SPSS 27. ANOVA and crosstab methods were employed to analyze continuous and categorical variables, respectively. Means and standard deviations were calculated. Comparisons of clinicopathological features between p53, S-CEA, combined p53, and recombined p53 were performed using the Tukey and χ² tests. Kaplan–Meier and log-rank tests were used for survival analysis between groups. Cox regression analysis was performed for univariate and multivariate analyses. MultiTimeRoc and five-year OS and DFS survival curves with numbers at risk were generated using R software (version 4.4.1) with the “ggplot2,” “survival,” “survminer,” and “timeROC” packages.
3 Results
3.1 Clinicopathological features in p53, S-CEA, combined p53, recombined p53
A total of 485 cases were included, with a mean age of 65.13 years and a standard deviation (SD) of 10.84 (ranging from 25 to 90). Significant differences in gender, TNM stage, Carbohydrate Antigen 199 (CA199), differentiation, and both minimum and maximum tumor size were observed in the single p53 groups (all P<0.05). However, no significant differences were found for age, laparoscopy, tumor location, duration (days), lymph node harvest, costs, preoperative C-reactive protein (CRP), preoperative albumin, postoperative chemotherapy, or complications (all P>0.05) in the single p53 groups. Significant differences in TNM stage, CA199, postoperative chemotherapy, differentiation, and both minimum and maximum tumor size were observed in the single S-CEA groups (all P<0.05), while other variables showed no significant differences (all P>0.05). Significant differences in TNM stage, CA199, and postoperative chemotherapy were found in the combined p53 groups (all P<0.05), while other variables showed no significant differences (all P>0.05). In the recombined p53 groups, significant differences were noted for tumor location, TNM stage, CA199, postoperative chemotherapy, and both minimum and maximum tumor size (all P<0.05), while other variables showed no significant differences (all P>0.05). The detailed values and comparisons are shown in Table 1.
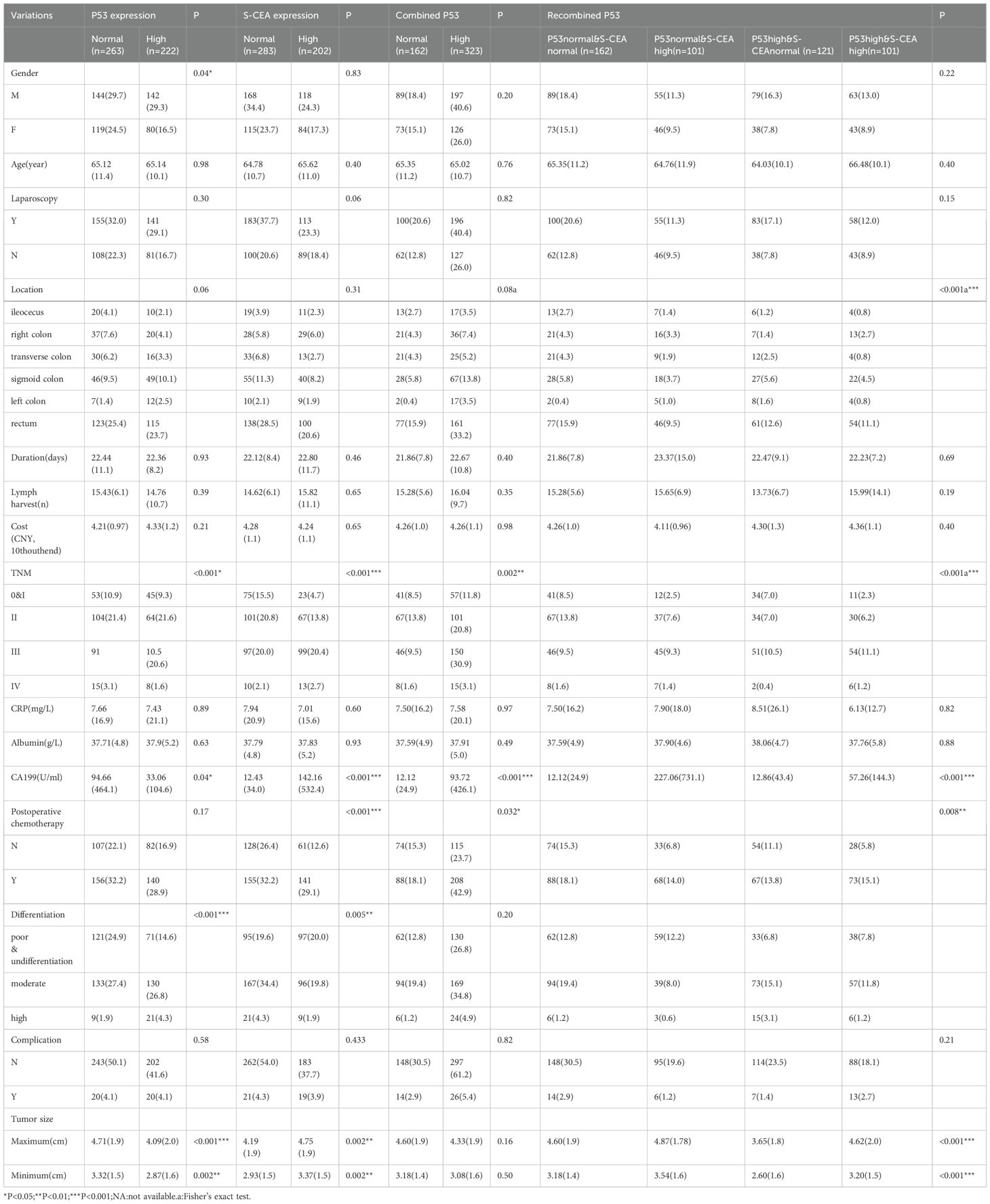
Table 1. Clinicopathologic features compared by P53, S-CEA, combined P53, recombined P53(N,%,Mean,SD).
3.2 Comparisons of 5 year overall survival rate, disease-free survival rate by p53,S-CEA,combined p53, recombined p53 groups
There were no significant differences in OS and DFS rates between the single p53 groups (P=0.65 and P=0.90, respectively, Figure 2A). Significant differences in OS and DFS rates were observed in the single S-CEA groups (both P<0.001, Figure 2B). No significant differences in OS and DFS rates were found in the combined p53 groups (P=0.16 and P=0.087, respectively, Figure 2C). However, significant differences in OS and DFS rates were observed in the recombined p53 groups (P=0.0073 and P=0.0022, Figure 2D).
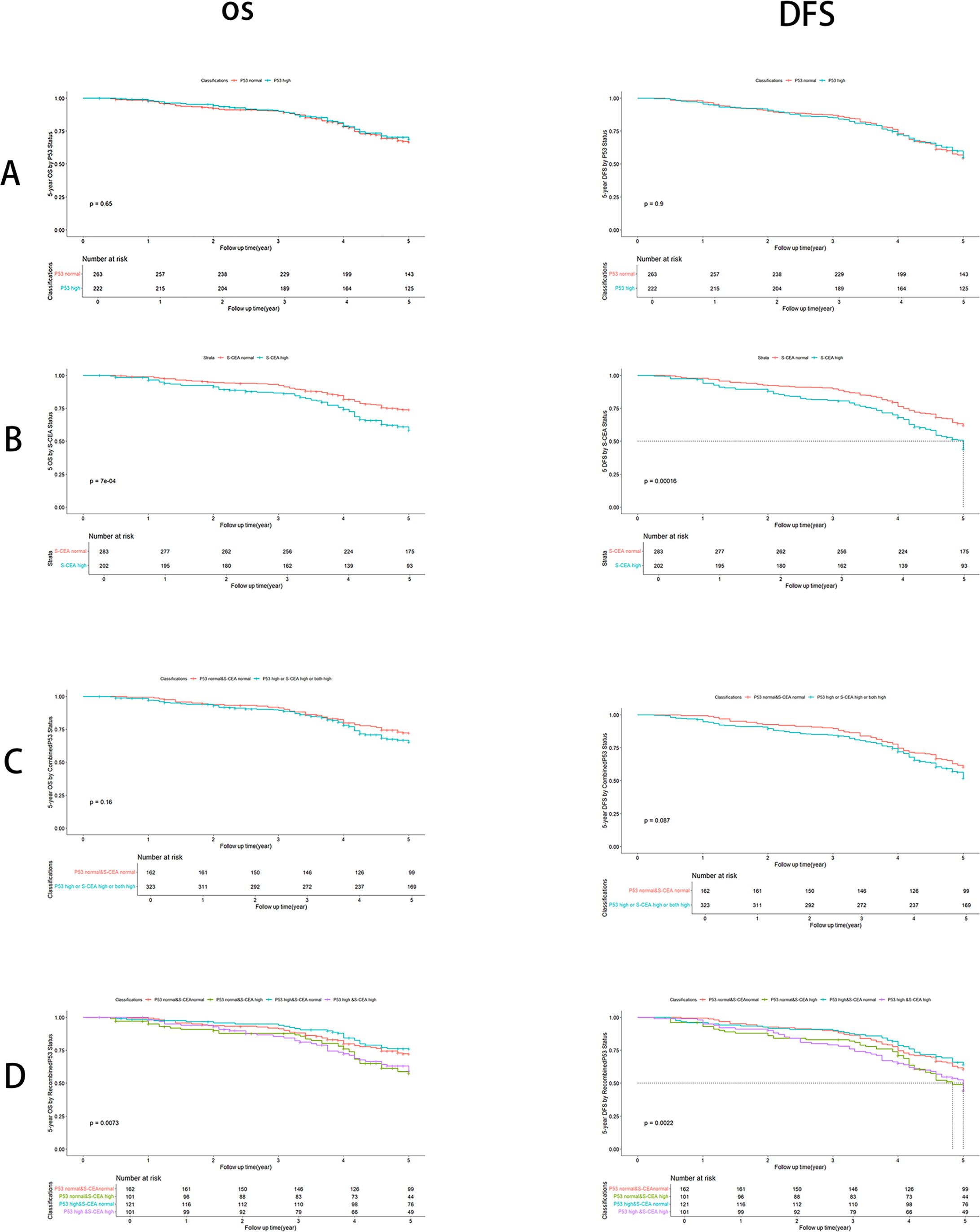
Figure 2. Overall survival (OS) and Disease free survival (DFS) analysis with number at risk by Kaplan-Meier and Log-rank test using R 4.4.1. (A) By p53 groups, there are no significant differences about OS (P=0.65) and DFS (P=0.9); (B) By S-CEA groups, there are significant differences about OS (P<0.001) and DFS (P<0.001); (C) By combined p53 groups, there are no significant differences about OS (P=0.16) and DFS (P=0.087); (D) By recombined p53, there are significant differences about OS (P=0.0073) and DFS (P=0.0022).
3.3 Correlations between P53, S-CEA, combined P53 and recombined P53
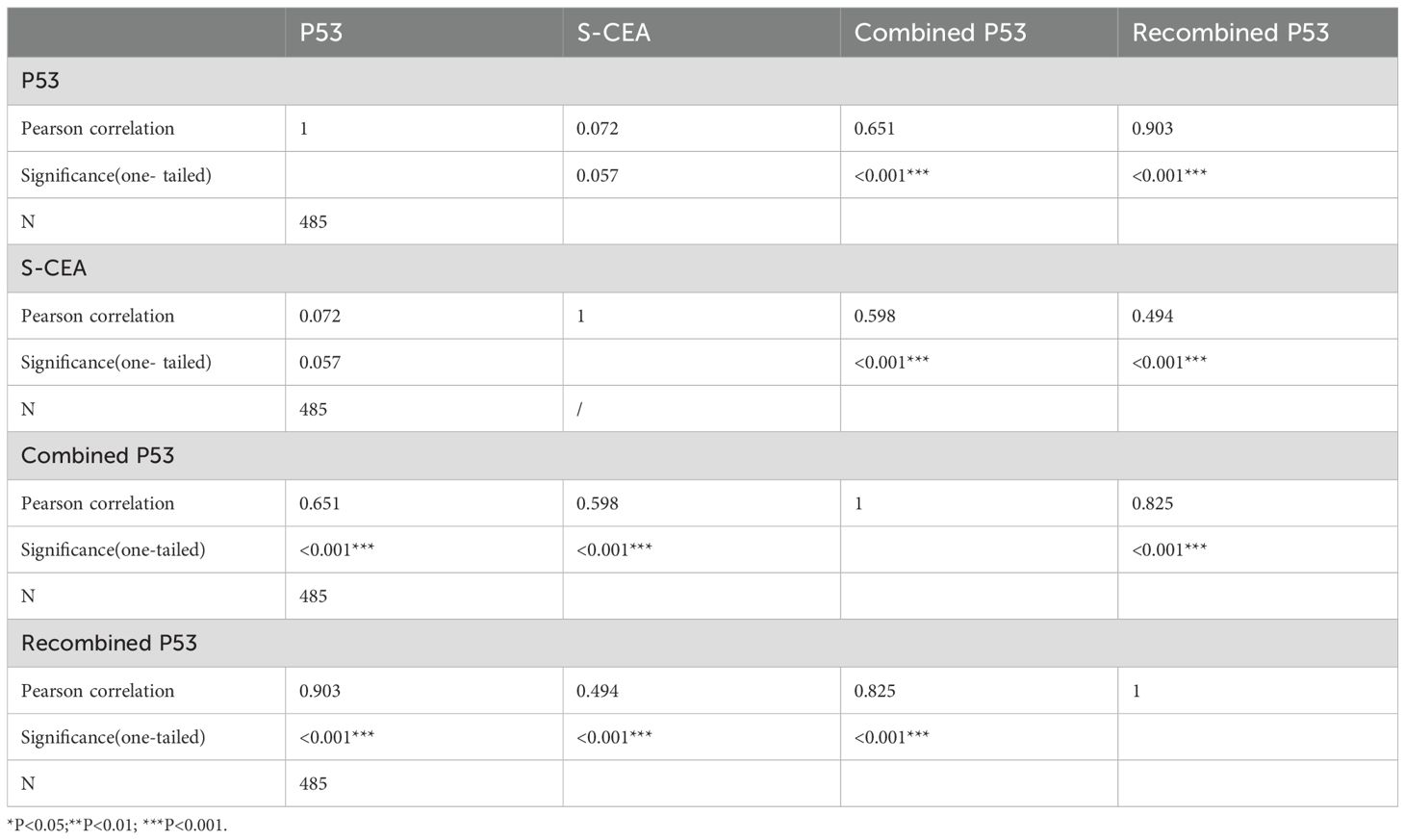
The Pearson correlation between p53 and S-CEA was 0.072 (P=0.057, one-tailed), with no significant difference. The Pearson correlations between p53 and combined p53, as well as p53 and recombined p53, were 0.651 and 0.903, respectively, both showing significant differences (both P<0.001, one-tailed). The Pearson correlations between S-CEA and combined p53, as well as S-CEA and recombined p53, were 0.598 and 0.494, respectively, both significant (P<0.001, one-tailed). The Pearson correlation between combined p53 and recombined p53 was 0.825 (P<0.001, one-tailed). The details are shown in Table 2.
3.4 Univariate and multivariate analysis of overall survival and disease free survival
The categorical variables were analyzed by univariate and multivariate analyses for OS and DFS using Cox regression. Variables that showed significant differences in univariate analysis were further analyzed by multivariate analysis. The method used was the input approach, with the first factor as the reference. For OS, significant differences were found in location, laparoscopy, postoperative chemotherapy, differentiation, TNM stage, S-CEA, and recombined p53 (all P<0.05). However, no significant differences were observed for gender, p53, or combined p53 in the univariate analysis. In the multivariate analysis, only laparoscopy, postoperative chemotherapy, differentiation, TNM stage, and recombined p53 showed significant differences, indicating that they are independent factors for OS. For DFS, significant differences were found in laparoscopy, postoperative chemotherapy, differentiation, TNM stage, S-CEA, and recombined p53 in the univariate analysis. In the multivariate analysis, only laparoscopy, postoperative chemotherapy, differentiation, TNM stage, and recombined p53 remained significant, indicating that they are independent prognostic factors for CRC. Based on these analyses, recombined p53 emerged as an independent factor for CRC prognosis in both OS and DFS, while single p53, S-CEA, and combined p53 were not. Table 3 shows the details.
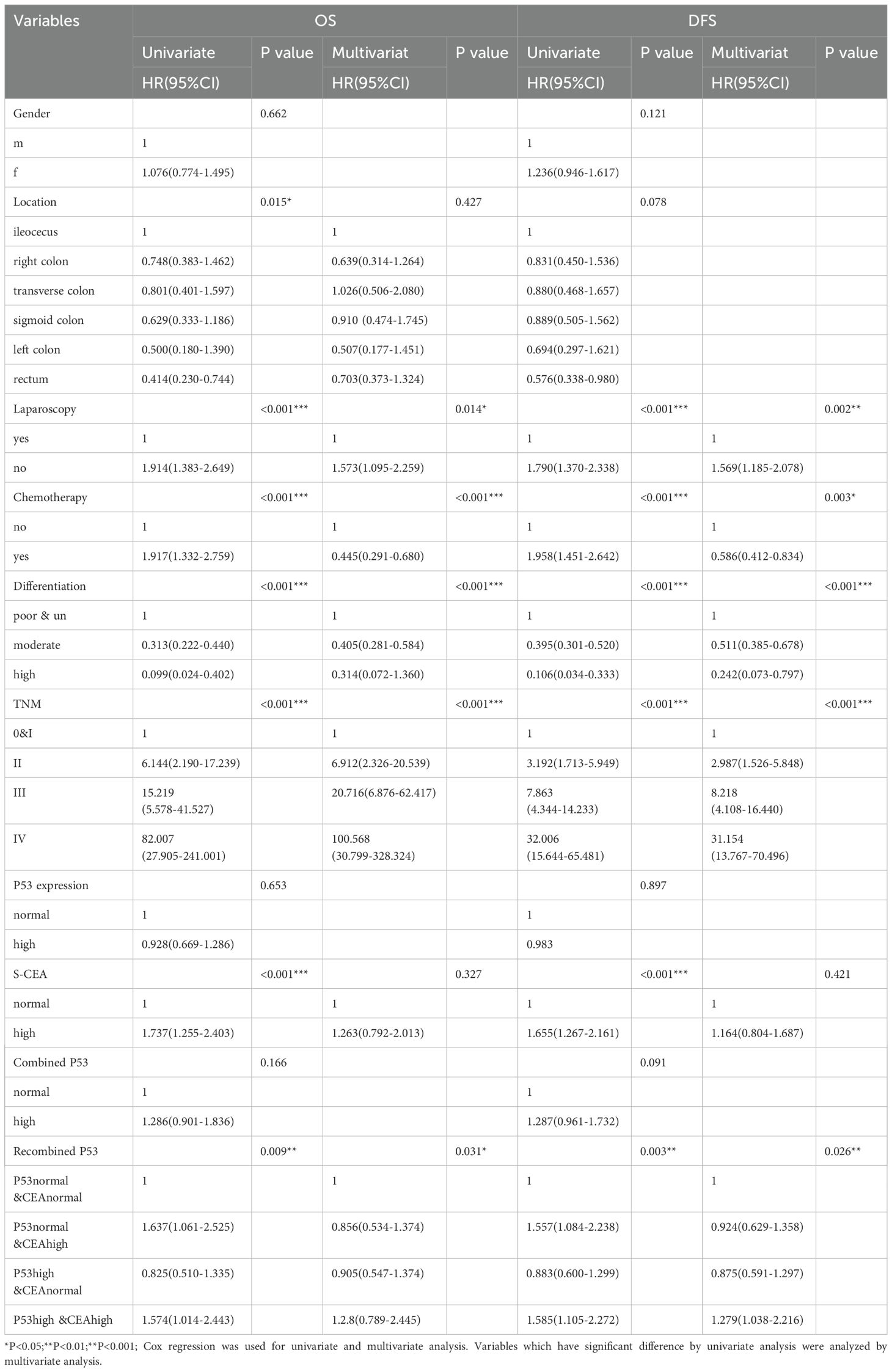
Table 3. Univariate and Multivariate analysis of overall survival (OS)and disease free survival(DFS).
3.5 1,2,3 years of receiver operating characteristic curve analysis by recombined p53
After identifying recombined p53 as an independent prognostic factor, we performed ROC analysis for OS and DFS at 1, 2, and 3 years. The results showed that recombined p53 demonstrated superiority in predicting 3-year OS (AUC=0.54, Figure 3A) and 1-year DFS (AUC=0.59, Figure 3B). We know that the closer the AUC value is to 1, the higher the sensitivity and specificity (29, 30).
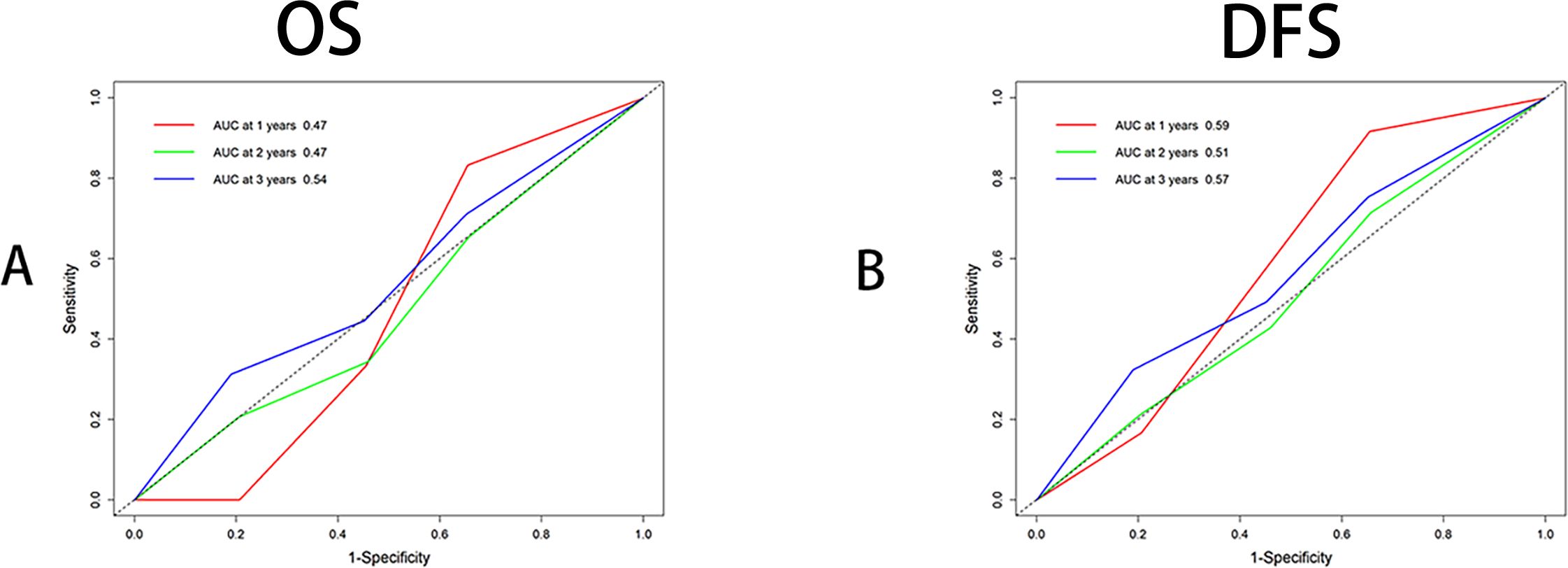
Figure 3. Receiver operating characteristic curve analysis (ROC) by multitimeROC using R4.4.1. (A) ROC analysis using OS (Binary variable), Area Under Curve (AUC) were 0.47,0.47 and 0.54 respectively for 1,2,3 years indicating 3 years has superior. (B) ROC analysis using DFS (Binary variable), Area Under Curve (AUC) were 0.59, 0.57 and 0.51 respectively for 1,2,3 years indicating 1 year has superior.
4 Discussion
As we know, p53 is a tumor suppressor gene for CRC and other malignant tumors. Its mutation and deficiency often promote tumor invasion, progression, and metastasis (31–36). Many genes, such as ubiquitin-specific protease 36 (USP36) and carnitine palmitoyltransferase-2 (CPT2), affect the progression of CRC through the p53 signaling pathway (37–43). However, clinical studies on p53 have shown varying prognostic efficacy for CRC (31, 44). This study demonstrated that single p53 detection has no effect on the prognosis of CRC. Therefore, combining p53 with other genes has been a hot topic recently, such as combining p53 with mitochondrial translation elongation factor Tu (TUFM) and metastasis suppressor 23-H1 (Nm23-H1) (20, 25, 45). Preoperative and postoperative S-CEA detection are commonly used for CRC screening and prognostic recurrence assessment as reference indicators, but many CRC patients do not show elevated preoperative S-CEA levels (46–48). This study found that the preoperative S-CEA elevation rate is 41.6% (202/485), indicating its limited sensitivity. While this study showed that single S-CEA has a prognostic role in CRC, it is not an independent factor for OS and DFS, which is consistent with some published literature (49–52). Due to the controversy surrounding the limitations and prognostic impact of detecting tumor p53 and preoperative S-CEA separately for CRC, we aimed to explore the effect of combined detection on CRC prognosis using two different combinations from previous studies (26, 28, 46, 53, 54). Despite the findings regarding p53 vs S-CEA, significant pairwise associations were found among p53, S-CEA, combined p53, and recombined p53. This study showed that gender, TNM stage, Carbohydrate Antigen 199 (CA199), differentiation, and tumor size (both minimum and maximum) had significant differences in the single p53 group. TNM, CA199, postoperative chemotherapy, differentiation, and tumor size (both minimum and maximum) had significant differences in the single S-CEA group. In the combined p53 group, TNM, CA199, and postoperative chemotherapy showed significant differences, while in the recombined p53 group, tumor location, TNM, CA199, postoperative chemotherapy, and tumor size (both minimum and maximum) showed significant differences. Kaplan-Meier and Log-rank tests showed that S-CEA and recombined p53 had significant differences for OS and DFS of CRC, whereas single p53 and combined p53 showed limited effectiveness for prognostic values, which is consistent with some published literature (15, 31). However, some studies indicated that loss of p53 expression predicted a worse prognosis in CRC (55–57). Through Cox regression analysis, this study found that p53, S-CEA, and combined p53 were not independent factors for OS and DFS of CRC, while recombined p53 was, emphasizing the importance of recombined p53 for CRC prognosis. These findings underscore the limitations of single p53, S-CEA detection, and the combined p53 method, aligning with results from previous literature (11, 15, 17, 58). After identifying recombined p53 as an independent prognostic factor, we performed ROC analysis for OS and DFS at 1, 2, and 3 years. The results showed that recombined p53 demonstrated superiority in predicting 3-year OS.Du to no longer follow-up data than 5 years, we can’t analyze 1,3,5 years ROC about OS and DFS which we may mention in the limitations meanwhile the AUC value in this article is around 0.5, which may lead to insufficient persuasiveness in the AUC analysis section while one similar study showed higher AUC value (59). TNM staging remains a key factor in the prognosis of CRC, and this study further evaluated the TNM staging in AJCC-8. We did not overlook the role of TNM staging. On the contrary, this study found significant differences in TNM among CEA, P53, Combined P53, and Recombined P53 (Tables 1, 3).
Due to the fact that only recombined p53 is an independent factor for OS and DFS in CRC, we performed 1-, 2-, and 3-year time ROC analysis. The results showed that recombined p53 demonstrated superior performance at 3 years for OS and at 1 year for DFS.
5 Limitations
This study has several limitations. These include the absence of genetic analysis, the reliance on outdated data, and the fact that it is a single-center, retrospective study. It is also important to note that Stage IV refers only to patients with resectable Stage IV colorectal carcinoma, excluding all Stage IV patients, which limits the generalizability of the findings. Additionally, the study is based on data from a single medical center, which could limit the applicability of the findings to a broader population. According to statistical principles, the follow-up period is 5 years instead of 10 years, so we can only perform ROC analysis on OS and DFS for 1, 2, and 3 years. If the follow-up period is 10 years, we can perform ROC analysis on OS and DFS for 1, 3, and 5 years. This is a limitation of our study. Genetic typing or molecular analysis of p53 is more precise than IHC staining. This study is a retrospective study in which the data came from pathological department using ICH method to type the analysis of p53 that is more general and more affordable. The AUC value in this article is around 0.5, which may lead to insufficient persuasiveness in the AUC analysis section. However, the data collected in this study shows these results. These may be another limitations in this paper.
6 Conclusions
Recombined p53 classification proved to be superior to traditional combined p53 classification for assessing the prognosis of CRC and was found to be an independent factor. Furthermore, the 3-year OS and 1-year DFS analysis demonstrated greater sensitivity and specificity for recombined p53.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The current study was carried out according to the ethical guidelines of the 2013 Declaration of Helsinki and was approved by the ethics committee of Huzhou Central Hospital (No.202503001). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
GT: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. YW: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing. HQ: Conceptualization, Methodology, Supervision, Writing – review & editing. ZT: Conceptualization, Methodology, Supervision, Writing – review & editing. YS: Conceptualization, Methodology, Supervision, Writing – review & editing. HL: Conceptualization, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors thank Professor Zhaozheng Zheng and Yan Chen (Colorectal department of Huzhou Central Hospital, China) for providing valuable clinic data on our manuscript and the authors also thank MedSci (Shanghai) for editing our paper.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Luo Z-W, Zhu M-G, Zhang Z-Q, Ye F-J, Huang W-H, and Luo X-Z. Increased expression of Ki-67 is a poor prognostic marker for colorectal cancer patients: a meta analysis. BMC Cancer. (2019) 19:123–3.
2. Tong G-J, Zhang G-Y, Liu J, Zheng Z-Z, Chen Y, Niu P-P, et al. Comparison of the eighth version of the American Joint Committee on Cancer manual to the seventh version for colorectal cancer: A retrospective review of our data. World J Clin Oncol. (2018) 9:148–61.
3. Tong G, Zhang G, Liu J, Zheng Z, Chen Y, Niu P, et al. Cutoff of 25% for Ki67 expression is a good classification tool for prognosis in colorectal cancer in the AJCC−8 stratification. Oncol Rep. (2020).
4. Tong G, Xu W, Zhang G, Liu J, Zheng Z, Chen Y, et al. The role of tissue and serum carcinoembryonic antigen in stages I to III of colorectal cancer-A retrospective cohort study. Cancer Med. (2018) 7:5327–38.
5. Stidham RW and Higgins PDR. Colorectal cancer in inflammatory bowel disease. Clinics Colon Rectal Surg. (2018) 31:168–78. doi: 10.1055/s-0037-1602237
6. Wang L, Lin S, Yang C, Cai S, and Li W. Effect of KRAS mutations and p53 expression on the postoperative prognosis of patients with colorectal cancer. Mol Genet Genom Med. (2022) 10:e1905. doi: 10.1002/mgg3.1905
7. Zhang C, Liu J, Xu D, Zhang T, Hu W, and Feng Z. Gain-of-function mutant p53 in cancer progression and therapy. J Mol Cell Biol. (2020) 12:674–87. doi: 10.1093/jmcb/mjaa040
8. Roszkowska KA, Gizinski S, Sady M, Gajewski Z, and Olszewski MB. Gain-of-function mutations in p53 in cancer invasiveness and metastasis. Int J Mol Sci. (2020) 21. doi: 10.3390/ijms21041334
9. Schulz-Heddergott R, Stark N, Edmunds SJ, Li J, Conradi LC, Bohnenberger H, et al. Therapeutic ablation of gain-of-function mutant p53 in colorectal cancer inhibits stat3-mediated tumor growth and invasion. Cancer Cell. (2018) 34:298–314.e297. doi: 10.1016/j.ccell.2018.07.004
10. Giacomelli AO, Yang X, Lintner RE, McFarland JM, Duby M, Kim J, et al. Mutational processes shape the landscape of TP53 mutations in human cancer. Nat Genet. (2018) 50:1381–7. doi: 10.1038/s41588-018-0204-y
11. Pan M, Jiang C, Tse P, Achacoso N, Alexeeff S, Solorzano AV, et al. TP53 gain-of-function and non-gain-of-function mutations are differentially associated with sidedness-dependent prognosis in metastatic colorectal cancer. J Clin Oncol: Off J Am Soc Clin Oncol. (2022) 40:171–9. doi: 10.1200/JCO.21.02014
12. Yagublu V, Bayramov B, Yuce M, and Aslanov H. TP53 codon 72 polymorphism and the risk of colorectal cancer in an Azerbaijani population. Turkish J Gastroenterol: Off J Turkish Soc Gastroenterol. (2022) 33:485–90.
13. Zhang W, Sun R, Zhang Y, Hu R, Li Q, Wu W, et al. Cabazitaxel suppresses colorectal cancer cell growth via enhancing the p53 antitumor pathway. FEBS Open Bio. (2021) 11:3032–50. doi: 10.1002/2211-5463.13290
14. Rong Z, Luo Z, Fu Z, Zhang P, Li T, Zhang J, et al. The novel circSLC6A6/miR-1265/C2CD4A axis promotes colorectal cancer growth by suppressing p53 signaling pathway. J Exp Clin Cancer Res: CR. (2021) 40:324. doi: 10.1186/s13046-021-02126-y
15. Ottaiano A, Santorsola M, Capuozzo M, Perri F, Circelli L, Cascella M, et al. The prognostic role of p53 mutations in metastatic colorectal cancer: A systematic review and meta-analysis. Crit Rev Oncol/Hematol. (2023) 186:104018. doi: 10.1016/j.critrevonc.2023.104018
16. Zheng H, Liu H, Li H, Dou W, Wang J, Zhang J, et al. Characterization of stem cell landscape and identification of stemness-relevant prognostic gene signature to aid immunotherapy in colorectal cancer. Stem Cell Res Ther. (2022) 13:244. doi: 10.1186/s13287-022-02913-0
17. Kim KM, Ahn AR, Park HS, Jang KY, Moon WS, Kang MJ, et al. Clinical significance of p53 protein expression and TP53 variation status in colorectal cancer. BMC Cancer. (2022) 22:940. doi: 10.1186/s12885-022-10039-y
18. Ko HJ, Chiou SJ, Tsai CY, Loh JK, Lin XY, Tran TH, et al. BMX, a specific HDAC8 inhibitor, with TMZ for advanced CRC therapy: a novel synergic effect to elicit p53-, β-catenin- and MGMT-dependent apoptotic cell death. Cell Commun Signal. (2022) 20:200. doi: 10.1186/s12964-022-01007-x
19. Wang L, Liu Z, Fisher KW, Ren F, Lv J, Davidson DD, et al. Prognostic value of programmed death ligand 1, p53, and Ki-67 in patients with advanced-stage colorectal cancer. Hum Pathol. (2018) 71:20–9. doi: 10.1016/j.humpath.2017.07.014
20. Wu Y, Li Y, Zhao X, Dong D, Tang C, Li E, et al. Combined detection of the expression of Nm23-H1 and p53 is correlated with survival rates of patients with stage II and III colorectal cancer. Oncol Lett. (2017) 13:129–36. doi: 10.3892/ol.2016.5425
21. Perraud A, Akil H, Nouaille M, Petit D, Labrousse F, Jauberteau MO, et al. Expression of p53 and DR5 in normal and Malignant tissues of colorectal cancer: correlation with advanced stages. Oncol Rep. (2011) 26:1091–7. doi: 10.3892/or.2011.1404
22. Miladi-Abdennadher I, Abdelmaksoud-Damak R, Ayadi L, Khabir A, Amouri A, Frikha F, et al. Expression of p16INK4a, alone or combined with p53, is predictive of better prognosis in colorectal adenocarcinoma in Tunisian patients. Appl Immunohistochem Mol Morphol: AIMM. (2011) 19:562–8. doi: 10.1097/PAI.0b013e3182143380
23. Huang Liang WH, Yu B, and Zheng M. The prognostic significance of P53gene mutation and preoperative serum levels of tumor markers in colorectal carcinoma. Chin J Gastrointestinal Surg. (1999) 2:171–3.
24. Kobayashi S, Hiwasa T, Kitamura K, Kano M, Hoshino T, Hirano S, et al. Combinational antibody detection approach increases the clinical validity of colorectal cancer screening. J Clin Lab Anal. (2023) 37:e24978. doi: 10.1002/jcla.24978
25. Kawahara D, Fujita F, Hayashi H, Matsuyama T, and Eguchi S. The Significance of Combined Measurement of p53 Antibody and other Tumor Markers for Colorectal Cancer after Curative Resection. Hepato-gastroenterology. (2015) 62:624–8.
26. Wang J, Wang X, Yu F, Chen J, Zhao S, Zhang D, et al. Combined detection of preoperative serum CEA, CA19–9 and CA242 improve prognostic prediction of surgically treated colorectal cancer patients. Int J Clin Exp Pathol. (2015) 8:14853–63.
27. Li Z, Zhu H, Pang X, Mao Y, Yi X, Li C, et al. Preoperative serum CA19–9 should be routinely measured in the colorectal patients with preoperative normal serum CEA: a multicenter retrospective cohort study. BMC Cancer. (2022) 22:962. doi: 10.1186/s12885-022-10051-2
28. Tong G, Li H, Shen Y, Tan Z, and Qian H. The combined evaluation of preoperative serum CEA and postoperative tissue CEA as a prognostic factor in stages 0–IV colorectal cancer: a retrospective cohort study. Front Med. (2025) 11. doi: 10.3389/fmed.2024.1447041
29. Coker OO, Liu C, Wu WKK, Wong SH, Jia W, Sung JJY, et al. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome. (2022) 10:35. doi: 10.1186/s40168-021-01208-5
30. Chen F, Dai X, Zhou CC, Li KX, Zhang YJ, Lou XY, et al. Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut. (2022) 71:1315–25. doi: 10.1136/gutjnl-2020-323476
31. Yuan C, Huang J, Wang Y, and Xiao H. Exploration and validation of the Ki67, Her-2, and mutant P53 protein-based risk model, nomogram and lymph node metastasis model for predicting colorectal cancer progression and prognosis. Front Oncol. (2023) 13:1236441. doi: 10.3389/fonc.2023.1236441
32. Manzoor S, Muhammad JS, Maghazachi AA, and Hamid Q. Autophagy: A versatile player in the progression of colorectal cancer and drug resistance. Front Oncol. (2022) 12:924290. doi: 10.3389/fonc.2022.924290
33. Neufert C, Heichler C, Brabletz T, Scheibe K, Boonsanay V, Greten FR, et al. Inducible mouse models of colon cancer for the analysis of sporadic and inflammation-driven tumor progression and lymph node metastasis. Nat Protoc. (2021) 16:61–85. doi: 10.1038/s41596-020-00412-1
34. Liebl MC and Hofmann TG. The role of p53 signaling in colorectal cancer. Cancers (Basel). (2021) 13. doi: 10.3390/cancers13092125
35. Malki A, ElRuz RA, Gupta I, Allouch A, Vranic S, and Al Moustafa AE. Molecular mechanisms of colon cancer progression and metastasis: recent insights and advancements. Int J Mol Sci. (2020) 22. doi: 10.3390/ijms22010130
36. Liu D, Ertay A, Hill C, Zhou Y, Li J, Zou Y, et al. ASPP1 deficiency promotes epithelial-mesenchymal transition, invasion and metastasis in colorectal cancer. Cell Death Dis. (2020) 11:224. doi: 10.1038/s41419-020-2415-2
37. Lin X, Zhao X, Chen Q, Wang X, Wu Y, and Zhao H. Quercetin ameliorates ferroptosis of rat cardiomyocytes via activation of the SIRT1/p53/SLC7A11 signaling pathway to alleviate sepsis−induced cardiomyopathy. Int J Mol Med. (2023) 52. doi: 10.3892/ijmm.2023.5319
38. Lin H, Ren TH, Tong YT, Wu GF, Zhang T, Chen TX, et al. p53 regulates primordial follicle activation through the mTOR signaling pathway. Sheng Li Xue Bao: Acta Physiol Sinica. (2023) 75:339–50.
39. Kennedy MC and Lowe SW. Mutant p53: it’s not all one and the same. Cell Death Differ. (2022) 29:983–7. doi: 10.1038/s41418-022-00989-y
40. Rahman N, Khan H, Zia A, Khan A, Fakhri S, Aschner M, et al. Bcl-2 Modulation in p53 Signaling Pathway by Flavonoids: A Potential Strategy towards the Treatment of Cancer. Int J Mol Sci. (2021) 22. doi: 10.3390/ijms222111315
41. Huang J. Current developments of targeting the p53 signaling pathway for cancer treatment. Pharmacol Ther. (2021) 220:107720. doi: 10.1016/j.pharmthera.2020.107720
42. Xu H, Wang T, Nie H, Sun Q, Jin C, Yang S, et al. USP36 promotes colorectal cancer progression through inhibition of p53 signaling pathway via stabilizing RBM28. Oncogene. (2024) 43:3442–55. doi: 10.1038/s41388-024-03178-y
43. Liu F, Li X, Yan H, Wu J, Yang Y, He J, et al. Downregulation of CPT2 promotes proliferation and inhibits apoptosis through p53 pathway in colorectal cancer. Cell Signalling. (2022) 92:110267. doi: 10.1016/j.cellsig.2022.110267
44. Niloofa R, De Zoysa MI, and Seneviratne LS. Autoantibodies in the diagnosis, prognosis, and prediction of colorectal cancer. J Cancer Res Ther. (2021) 17:819–33. doi: 10.4103/jcrt.JCRT_64_19
45. Xi HQ, Zhang KC, Li JY, Cui JX, Zhao P, and Chen L. Expression and clinicopathologic significance of TUFM and p53 for the normal-adenoma-carcinoma sequence in colorectal epithelia. World J Surg Oncol. (2017) 15:90. doi: 10.1186/s12957-017-1111-x
46. Hong J, Chen X, Chen L, Wang Y, Huang B, and Fang H. Clinical value of combined detection of serum sTim-3 and CEA or CA19–9 for postoperative recurrence of colorectal cancer diagnosis. Cancer Manag Res. (2023) 15:563–72. doi: 10.2147/CMAR.S407930
47. Li C, Zhang D, Pang X, Pu H, Lei M, Fan B, et al. Trajectories of perioperative serum tumor markers and colorectal cancer outcomes: A retrospective, multicenter longitudinal cohort study. EBioMedicine. (2021) 74:103706. doi: 10.1016/j.ebiom.2021.103706
48. Fenqi D, Yupeng L, Qiuju Z, Chao Y, Wenjie S, Tianyi X, et al. Early postoperative serum carcinoembryonic antigen is a stronger independent prognostic factor for stage II colorectal cancer patients than T4 stage and preoperative CEA. Front Oncol. (2021) 11:758509. doi: 10.3389/fonc.2021.758509
49. Sun Q and Long L. Diagnostic performances of methylated septin9 gene, CEA, CA19–9 and platelet-to-lymphocyte ratio in colorectal cancer. BMC Cancer. (2024) 24:906. doi: 10.1186/s12885-024-12670-3
50. Su YT, Chen JW, Chang SC, Jiang JK, and Huang SC. The clinical experience of the prognosis in opposite CEA and image change after therapy in stage IV colorectal cancer. Sci Rep. (2022) 12:20075. doi: 10.1038/s41598-022-24187-5
51. Rao H, Wu H, Huang Q, Yu Z, and Zhong Z. Clinical value of serum CEA, CA24–2 and CA19–9 in patients with colorectal cancer. Clin Lab. (2021) 67. doi: 10.7754/Clin.Lab.2020.200828
52. Campos-da-Paz M, Dórea JG, Galdino AS, and Lacava ZGM. de fatima menezes almeida santos M: carcinoembryonic antigen (CEA) and hepatic metastasis in colorectal cancer: update on biomarker for clinical and biotechnological approaches. Recent Patents Biotechnol. (2018) 12:269–79. doi: 10.2174/1872208312666180731104244
53. Chen S, Zhang J, Qian C, Qi X, Mao Y, and Lu T. Prognostic value of combined LMR and CEA dynamic monitoring in postoperative colorectal cancer patients. J Inflammation Res. (2023) 16:4229–50. doi: 10.2147/JIR.S422500
54. Lakemeyer L, Sander S, Wittau M, Henne-Bruns D, Kornmann M, and Lemke J. Diagnostic and prognostic value of CEA and CA19–9 in colorectal cancer. Dis (Basel Switzerland). (2021) 9. doi: 10.3390/diseases9010021
55. Nagao K, Koshino A, Sugimura-Nagata A, Nagano A, Komura M, Ueki A, et al. The complete loss of p53 expression uniquely predicts worse prognosis in colorectal cancer. Int J Mol Sci. (2022) 23. doi: 10.3390/ijms23063252
56. Jamai D, Kallel I, Mekrezi S, Walha M, Gharsallah M, Khabir A, et al. Prognostic value of combining E-cadherin, p53, Bcl-2 and Bcl-xL expression and survival in Tunisian colorectal adenocarcinoma patients. Cell Mol Biol (Noisy-le-Grand France). (2022) 68:93–107. doi: 10.14715/cmb/2022.68.4.12
57. Tomicic MT, Dawood M, and Efferth T. Epigenetic alterations upstream and downstream of p53 signaling in colorectal carcinoma. Cancers (Basel). (2021) 13. doi: 10.3390/cancers13164072
58. Ottaiano A, Santorsola M, Circelli L, Perri F, Cascella M, Sabbatino F, et al. Hypertension, type 2 diabetes, obesity, and p53 mutations negatively correlate with metastatic colorectal cancer patients’ survival. Front Med. (2023) 10:1091634. doi: 10.3389/fmed.2023.1091634
Keywords: p53, CEA, CRC, OS, DFS, prognosis, ROC
Citation: Tong G, Wang Y, Qian H, Tan Z, Shen Y and Li H (2025) The effects of two combined methods of P53 expression and preoperative serum CEA detection on the prognosis of colorectal cancer. Front. Oncol. 15:1590836. doi: 10.3389/fonc.2025.1590836
Received: 10 March 2025; Accepted: 07 July 2025;
Published: 21 July 2025.
Edited by:
Yu-Chan Chang, National Yang Ming Chiao Tung University, TaiwanReviewed by:
Jianlong Wang, Second Hospital of Hebei Medical University, ChinaArjun Katailiha, University of Texas MD Anderson Cancer Center, United States
Copyright © 2025 Tong, Wang, Qian, Tan, Shen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanyan Wang, Y2VsbDAyMjhAMTYzLmNvbQ==
 Guojun Tong
Guojun Tong Yanyan Wang3*
Yanyan Wang3*