- 1Department of Gynecology and Obstetrics, First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, Shaanxi, China
- 2School of Medicine, Northwest University, Xi’an, Shaanxi, China
Ovarian cancer (OC) is a highly lethal gynecologic malignancy because of the absence of specific early symptoms and reliable biomarkers, most OC patients are often diagnosed at advanced stages, resulting in poor prognosis. Traditional tissue biopsy and serological biomarkers like CA125 have limited clinical application. Therefore, there is an urgent demand for effective diagnostic and screening tools in clinical practice. Liquid biopsy is a non-invasive method for early cancer detection by analyzing tumor-associated components shed into different body fluids, for example, circulating tumor DNA (ctDNA), circulating tumor cells (CTCs), cell-free RNA, proteins, and metabolites. Increasing evidence has demonstrated that liquid biopsy is promising for detecting cancer at an early stage. In this review, we outline the results for the utility of each liquid biopsy fluid, including serum/plasma, urine, cervical/vaginal sample, uterine lavage, and summarize the advantages and current constraints associated with their application in clinical settings. Future directions and challenges are also highlighted, along with areas where more research is warranted.
1 Introduction
Global Cancer Statistics reported that ovarian cancer (OC) was the 8th most frequently occurring cancer and the leading cause of cancer-related death in 2022, approximately 324,398 new ovarian cancer cases and 206,839 deaths occurred (1). The prognosis for ovarian cancer is poor because most OC (58%) are diagnosed at an advanced stage (III or IV;) (2).
OC is an extremely heterogeneous cancer. Epithelial ovarian cancer (EOC) is the most predominate pathological type, accounting for 90% of cases, which are classified into high-grade serous (up to 75%), low-grade serous (<5%), endometrioid (~10%), clear cell (~6%), mucinous (<5%) (3). A binary classification system divides invasive cancer into two types. Type I tumors are low-grade, some (endometrioid, mucinous, and clear cell types) harbor mutations in BRAF, KRAS, and PTEN with microsatellite instability, which are more indolent, less invasive. These tumors can be diagnosed at earlier stages of the disease (stage I-II). In contrast, Type II tumors included high-grade serous ovarian cancer (HGSOC), carcinosarcoma, and undifferentiated carcinomas, which are aggressive, highly genetic instability, and are mostly diagnosed at advanced stages (stage III-IV;). They are associated with high TP53 mutations, somatic and germline BRCA1/2 mutations, and other homozygous recombination genes mutations (4, 5).
Ovarian cancer has a high mortality rate primarily due to its asymptomatic nature during early stages. Most patients are diagnosed at an advanced stage when the tumor has already spread extensively. Thus, the survival rate of ovarian cancer is highly correlated with the stage at primary diagnosis. According to studies, the 5-year survival rate for early-stage disease is 92%, whereas for late-stage disease it is only 29% (6). The absence of effective screening methods and reliable biomarkers hampers early detection. Cancer antigen 125 (CA125) is considered the most useful diagnostic serum biomarker for ovarian cancer and is often used in combination with transvaginal ultrasound (TVUS) as a screening tool for detecting the disease. Currently, general population screening for ovarian cancer is not recommended because the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS), the largest multicenter, randomized, controlled ovarian cancer screening trial to date, did not have a mortality benefit (2). However, this trial provides the first evidence that screening can detect high-grade serous tubo-ovarian cancer earlier than no screening and improve short-term treatment outcomes. The potential survival benefit was small, most likely attributed to modest gains in early detection and treatment improvements (7). This indicates that novel techniques that can detect more women with high-grade serous cancers earlier, combined with treatment improvements and a better understanding of tumor biology, may achieve a mortality benefit.
Histopathologic examination is the gold standard for OC diagnosis. However, the tumor size is usually small in early-stage patients, making puncture difficult. Additionally, transabdominal tissue biopsy is highly invasive and may lead to intra-abdominal dissemination of tumor cells. Liquid biopsy has developed rapidly over the past decades. It involves using certain biological fluids as samples to analyze and identify tumor-specific components through various omics-based detection methods. The most important advantages of liquid biopsy over traditional tissue biopsy are less invasive and can be repeated during follow-up, providing a systematic and complete response to the disease by obtaining consecutive samples for dynamic monitoring. This review systematically evaluates the evolution of biomarkers for early OC diagnosis based on different sample types used in liquid biopsy, provides a comprehensive comparison of their respective advantages and limitations across multiple dimensions, and offers theoretical foundations for optimizing early OC screening and detection strategies.
2 Liquid biopsy in the early detection of ovarian cancer
Liquid biopsy is gaining attention as a less invasive and more efficient alternative to traditional tissue biopsy for cancer monitoring due to its real-time results and shorter reporting time, which helps in cancer diagnosis, prognosis, monitoring disease progression, selecting appropriate treatment, and identifying drug resistance (4, 8).
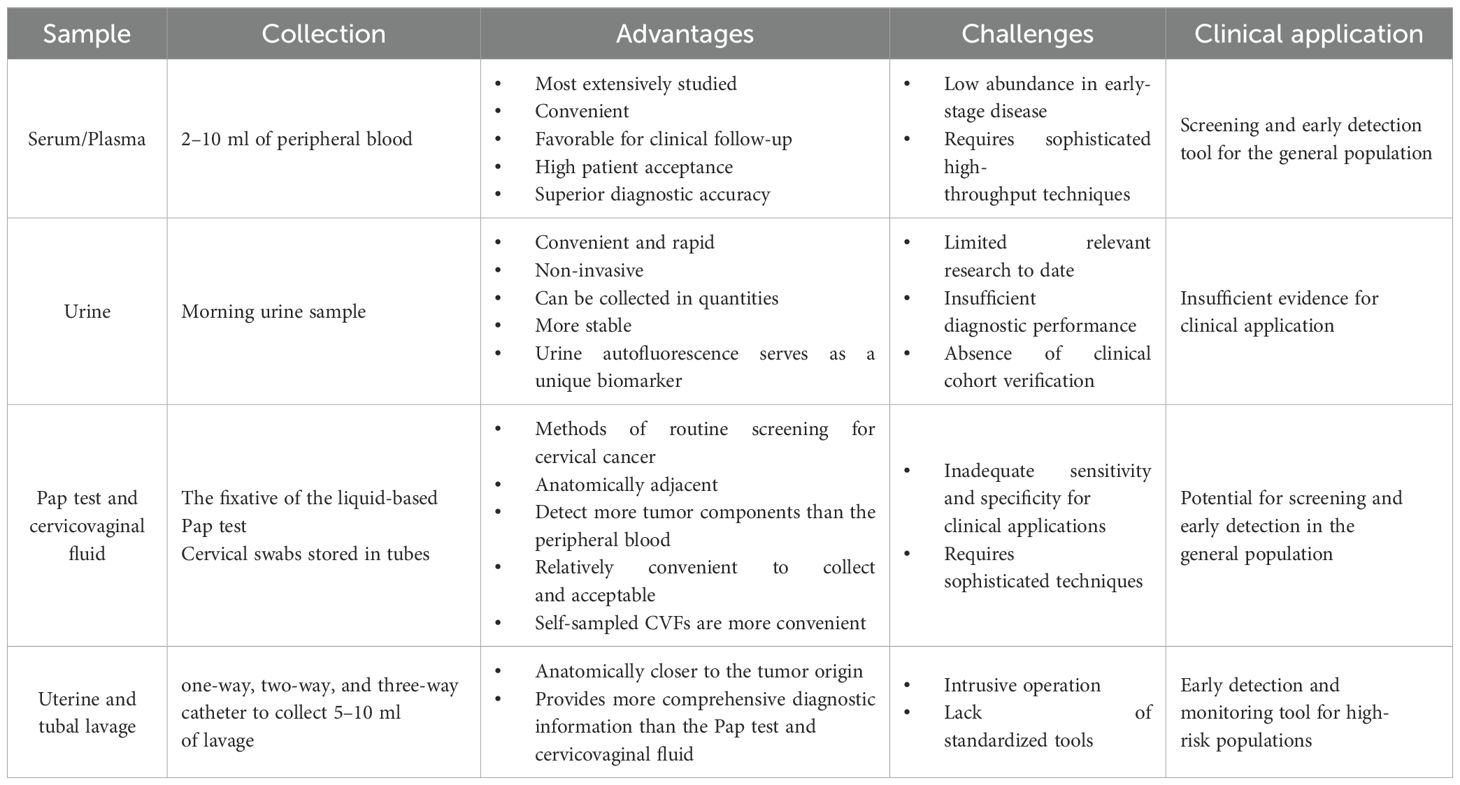
Various types of samples used for liquid biopsies are related to early diagnosis of OC, such as conventional serum and plasma, urine, Pap smears, cervicovaginal mucus, and uterine lavage fluid. Each of the different liquid biopsy specimens has its characteristics. Figure 1 provides a comprehensive list of samples utilized in the current research on ovarian cancer diagnostics, highlighting their various advantages and disadvantages. In these different body fluid samples, researchers have identified many tumor-associated components using multiple omics technologies, that is, genomics, transcriptomics, proteomics, and metabolomics. These tumor-associated constituents can be used as biomarkers for the early detection of OC, including circulating tumor DNA (ctDNA), circulating tumor cells (CTCs), cell-free RNA (cfRNA), tumor-specific proteins, and metabolites. Recently, several novel components have been identified for early diagnosis through liquid biopsy, such as tumor-educated platelets (TEPs) and nano-biosensor-detected immune signals from tumor-associated neutrophils (TANs) (9). Starting from different fluid samples, we describe the collection methods of these samples and summarize the research progress of different kinds of biomarkers in different fluid samples. Then, we compare the diagnostic performance of different biomarkers from the perspective of detection technology and sample source. The aim is to identify the most appropriate method to be used for the early management of ovarian cancer.
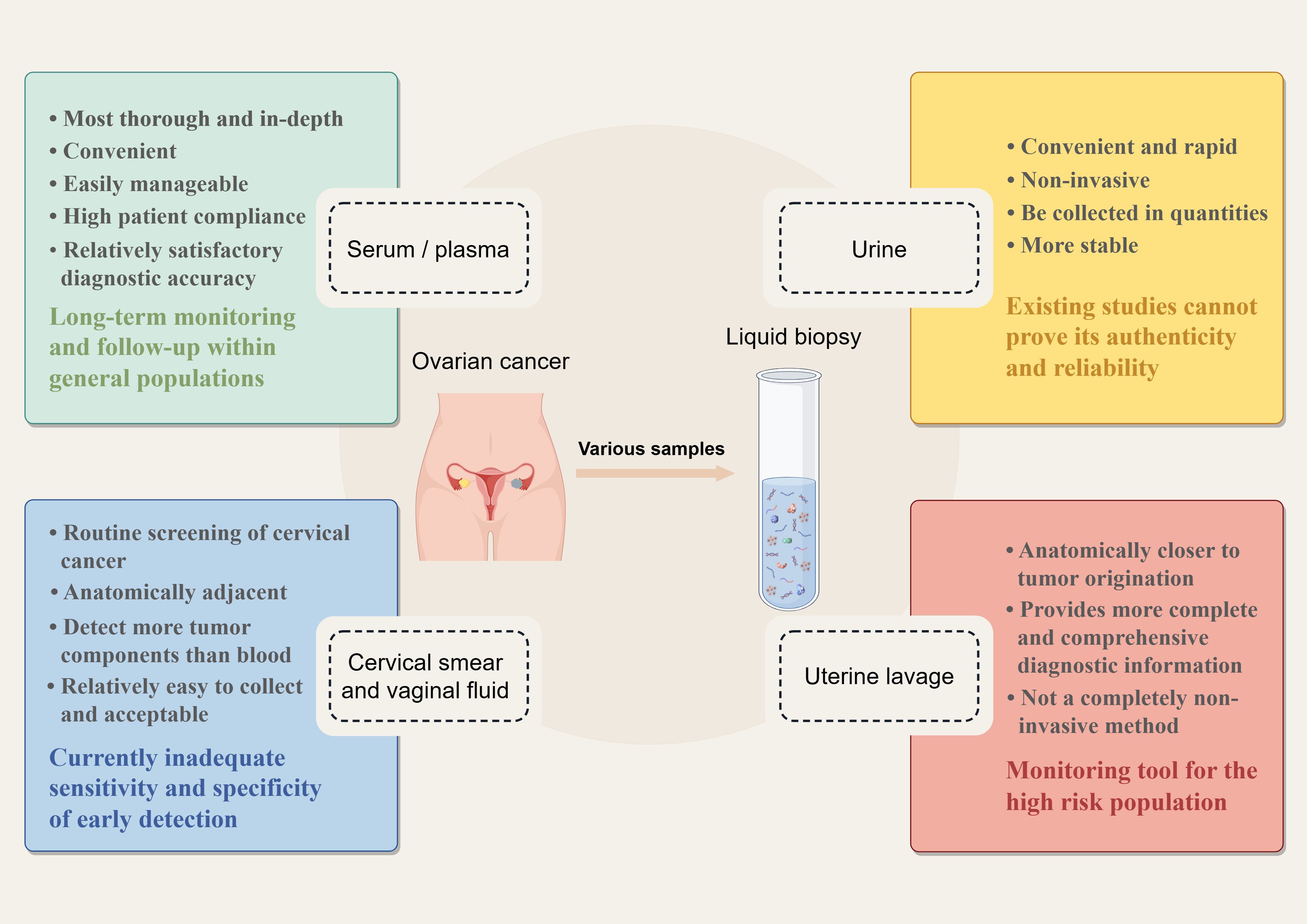
Figure 1. Liquid biopsy samples in the early detection of ovarian cancer (By Figdraw). The most important merits of liquid biopsy over traditional tissue biopsy are that it is minimally invasive and can be repeated several times during follow-up. For ovarian cancer, many kinds of samples have been used to detect ovarian cancer early, including serum/ plasma, urine, cervical smears, cervicovaginal mucus, and novel uterine lavage fluid. Each sample has its unique characteristics.
2.1 Conventional serum/plasma
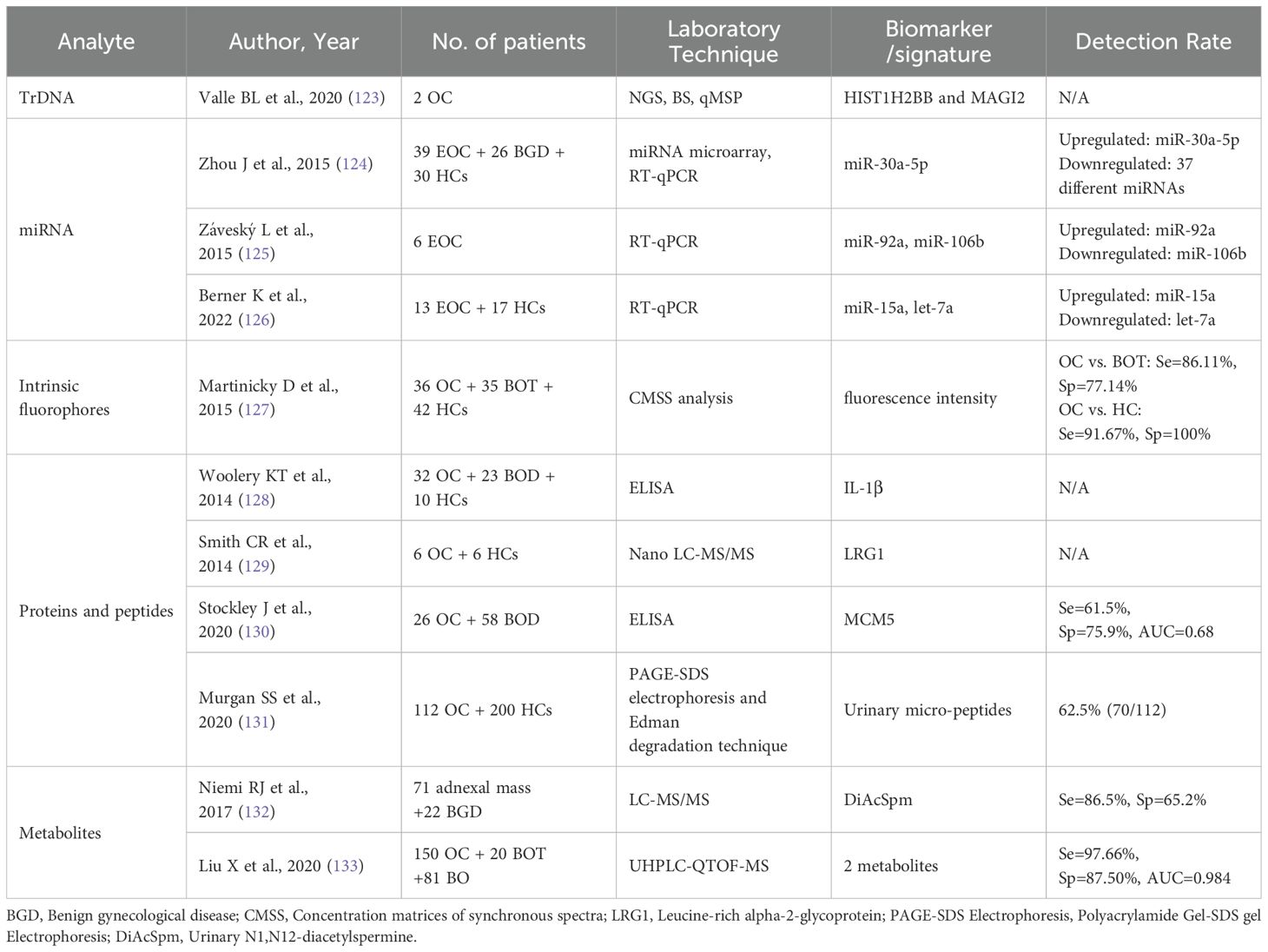
Blood samples have been widely and intensively studied as a conventional source of liquid biopsies over the past decades. With its convenience and accessibility, it has become more acceptable to patients. Approximately 2–10 ml of peripheral blood is collected from cancer patients, after which the plasma or serum is isolated for further study. With advances in molecular biology testing, technological innovations have emerged in genomic, transcriptomic, proteomic, and metabolomic assays, leading to the discovery of numerous novel biomarkers. Table 1 presents the diverse biomarkers and their diagnostic efficacy in various studies using serum/plasma-based liquid biopsies.
2.1.1 Cell-free DNA/circulating tumor DNA
Cell-free DNA (cfDNA) is released into the bloodstream mainly through apoptosis or necrosis. Elevated levels of cfDNA are observed in OC patients compared to healthy individuals, making it a valuable tool for early cancer diagnosis and screening (33, 80). Circulating tumor DNA (ctDNA), a subset of cfDNA, carries genetic and epigenetic information specific to the tumor, providing a "real-time" snapshot of the disease burden (81). With the advancement of sequencing technologies, researchers have shifted their focus toward identifying tumor-associated genetic mutations (29, 30, 34, 37), analyzing their methylation status, and performing DNA fragment mapping analysis (24) as well as CNV-based genomic instability screening (12, 13, 28, 31).
The detection rate of TP53 mutant-ctDNA in HGSOC patients is relatively high, ranging from 75% to 100% (82). Previous studies of TP53 mutations in EOC patients have demonstrated high sensitivity (>75%) and specificity (>80%) for recognizing ctDNA mutations (30, 34, 37). However, a majority of research on tumor-associated mutations involves the prior identification of tumor-specific mutations through tumor or formalin-fixed paraffin-embedded (FFPE) tissue, followed by the development of assays for these specific mutations. This approach limits such studies to the theoretical stage, as tumor tissue-specific mutations cannot be determined before diagnosis. For this reason, Phallen et al. developed targeted error correction sequencing (TEC-Seq) to allow ultra-sensitive direct assessment of serial changes in cfDNA by massively parallel sequencing, without the need for prior knowledge of genetic alterations in the tumor. This platform increases the sensitivity to 97.4% and the specificity to 100% (29).
Increased methylation of promoter regions is thought to be an early epigenetic event during tumorigenesis that can transform the expression of tumor suppressor genes. Methylation of cytosine occurs at relatively stable modified cytosine-phosphate-guanine (CpG)-rich regions (CpG islands) of DNA (83, 84). Several studies found that DNA methylation can be a promising biomarker for OC diagnostic, therapeutic, and prognostic (85). A comparative systematic review of 29 articles identified RASSF1A, BRCA1 (23, 35), and OPCML as common gene-specific methylation biomarkers, with OPCML showing the best diagnostic performance and an optimal sensitivity of 97.8% (86). While methylation panels consisting of 2 or more genes, the combination of different regions and CpGs had better diagnostic performance, with sensitivity ranging from 84.2% to 94.7%, and specificity ranging from 86.7% to 100% (15, 16, 19, 32, 36, 86, 87). Among these studies, Zhang et al. identified seven candidate genes using multiplex methylation-specific PCR (MSP), with 85.3% sensitivity and 90.5% specificity for stage I EOC (36). OvaPrint™ is a cfDNA methylation-based liquid biopsy platform to discriminate benign pelvic masses from HGSOC preoperatively. By leveraging machine learning to analyze sequencing data, researchers constructed this classifier (OvaPrint™), which achieves a positive predictive value of 95% (14). As for the combination of AI with several different CpG markers, statistical and bioinformatics approaches yielded high diagnostic accuracy with an AUC close to or equal to 1.0 (17). Li et al. employed transformer-based AI technology to learn genome-wide methylation patterns among different CpG sites from 110,000 cancer samples. These features were then applied to analyze large-scale cfDNA methylation markers in 754 EOC patients (including 205 early-stage EOC patients) and 1,118 healthy female controls. Using a pretrained AI transformer system named MethylBERT, they developed an EOC diagnostic model, which achieved a sensitivity of 80% and a specificity of 95% in early-stage EOC detection (10). Unlike most previous studies that relied on conventional modeling strategies, where genetic or epigenetic differences in cfDNA between cancer patients and healthy controls were directly analyzed to build diagnostic models, this study employed a pretrained AI methylation transformer system. Traditional methods like LASSO regression were constrained by the events-per-variable (EPV) rule, limiting the number of input markers that could be incorporated. In contrast, this pretrained AI transformer system is unrestricted by input feature, making it an ideal choice for constructing cfDNA-based diagnostic models. Sequencing data combined with artificial intelligence algorithm analysis could be a big step towards the early detection of ovarian cancer.
Recent advancements in detection technology have produced highly sensitive methods such as digital PCR (dPCR) and droplet digital PCR (ddPCR). Next-generation sequencing (NGS) allows for the detection of multiple genomic regions in a single assay, facilitating DNA mutation profiling and tumor mutation load assessment. Notably, whole genome sequencing (WGS) has also significantly improved the diagnosis of copy number variations (CNV). HGSOC is characterized by high chromosomal instability. Using CNV profiles from cfDNA as biomarkers could enhance the detection of malignancy in patients with adnexal masses (28). The DrCf10CNV predictive model was formulated using a combination of the CNV panel and machine learning algorithms (12). It has a sensitivity of 89% for distinguishing between malignant and non-malignant ovarian tumors. Another integrated scoring system, termed the OC score, was developed to classify OC patients from healthy controls based on four genomic features: CNV, 5'-end motifs, fragmentation profiles, and nucleosome footprinting (NF). The system has a high precision (AUC 97.7%; sensitivity 94.7%; specificity 98.0%) as a new comprehensive diagnostic method and a satisfactory sensitivity (85.7%) for early-stage OC (13). Compared with single-dimensional methylation sequencing technology, this study used multi-dimensional variation indicators to make the OC score system with stable performance, wider coverage, and greater overall accuracy.
Despite advancements in detection technology, the biological characteristics of ctDNA hinder its ability to detect small tumors or pre-cancerous lesions. Firstly, the limited detection of low-frequency ctDNA alleles may be attributed to the fact that ctDNA is diluted in higher concentrations of non-tumor cfDNA and contaminated with high molecular weight DNA (88). Additionally, it is possible that cancers may not shed enough ctDNA to detect early-stage or micrometastatic disease due to low disease burden. It is important to note that false-positive ctDNA mutations may be caused by non-cancerous mutations. Therefore, it is necessary to have superior sensitivity and precision in detecting ctDNA in early-stage disease. Excitingly, Thusgaard CF et al. recently reported for the first time a highly sensitive and transparent tumor-informed ctDNA single nucleotide variant detection method (89). This approach combines various allelic and read depth filters with different cut-offs, introduces an additional panel of normals to eliminate background noise, and utilizes a new filtering method to enhance the detection of ctDNA signals in plasma. These advancements significantly enhance the reliability of ctDNA-based approaches for the early diagnosis of OC.
2.1.2 Circulating tumor cells
Circulating tumor cells (CTCs) are thought to be detached from the primary tumor site, undergo the process of epithelial-mesenchymal transition (EMT), pass through the bloodstream, and colonize distant sites, leading to regional or distant metastasis (90). The molecular characterization and analysis of CTCs in different solid tumors exhibit variations (91). Most studies regarding CTCs in OC have focused on more advanced staging and have mainly investigated the relationship with prognosis, with fewer studies related to early diagnosis. For stage IA-IB patients, the positive rate of CTCs was 93%, which was significantly higher than the positive rate of CA125 in the same patients. However, the number of CTCs found in stage I was significantly lower than those in stage III and IV (40). Similarly, CTCs were not only found at a higher rate in advanced stages compared to early stages but also revealed a higher CTC count, 41.3 CTCs/ml versus 6.0 CTCs/ml (92).
Due to variations in CTC isolation methods and detection protocols across studies, including differences in sampling timepoints and cohort sizes, the reported CTC positivity rates in OC patients varied widely, ranging from 14% to 100% (90). Immunoaffinity-based biotechnology is the most common method for CTC enrichment. The CellSearch® system, approved by the FDA in 2004, utilizes EpCAM-targeted immunomagnetic beads to isolate CTCs from peripheral blood samples. However, the CellSearch® system demonstrates relatively low overall detection rates in ovarian cancer patients, ranging between 14.4% to 26% (93). The biggest limitation of this detection method is that EpCAM is a marker of epithelial cells and not a tumor-specific marker. Subsequently, a series of novel CTC detection technologies have emerged, including: invasive CTC subset marker detection techniques (42), high-throughput microfluidic systems integrating both physical and biological characteristics (41, 94), fluorescent-magnetic nanoparticles modified with folic acid and antifouling hydrogel (95), and flexible graphene-based biosensor for sensitive and rapid detection (96). These novel methods have significantly improved the detection rate of CTCs to over 70%. The Cell Adhesion Matrix (CAM)-based platform for detecting invasive CTCs (iCTCs) achieved a positive predictive value (PPV) of 77.8% in identifying early-stage epithelial ovarian cancer (EOC) patients (42). An optimized CTCs detection model incorporating three markers (EpCAM, MUC1, and WT1) achieved 79.4% sensitivity and 92.2% specificity (38).
Despite promising results from some studies, these findings are not yet recommended for clinical application. Some of the aforementioned novel technologies remain confined to theoretical exploration or have only been validated in small patient cohorts, limiting their generalizability to broader populations. Further clinical validation remains imperative. Research on early diagnosis is still in the initial phase, and further work is necessary for rapid, simple, and standardized assays, as well as studies targeting different subgroups with heterogeneity of CTCs. Further assessment is pivotal to determine the diagnostic performance of CTCs or specific subgroups.
2.1.3 Cell-free mRNA and EVs miRNA
Rapid tumor progression leads to increased gene transcription and the release of cell-free RNAs (cfRNAs), including mRNAs and miRNAs, into circulation. MiRNAs are stable in body fluids, and their altered expression is closely associated with tumor progression, invasion, metastasis, and drug resistance (97). It was reported that miRNA expression was dysregulated in the blood of ovarian cancer patients (46, 98–101).
Cf-miRNAs are highly stable in body fluids. Early studies used qRT-PCR and miRNA microarrays for expression analysis to detect aberrantly expressed miRNAs, with AUC ranging from 0.784-0.843 (46, 47). After genome-wide analysis, more tumor-associated cf-miRNAs were identified, and detection performance could be greatly improved by constructing a miRNA panel. The three miRNA panels can achieve 90.5% sensitivity and 100% specificity for their overall diagnostic potential in early serum samples (44). The other 7 cf-miRNAs panel and the 14 miRNAs neural network model were able to achieve the AUC of 0.87 and 0.93, respectively (43, 45, 102).
Research has predominantly focused on secreted miRNAs within extracellular vesicles (EVs), primarily produced by cells, notably cancer cells. EVs, categorized as exosomes, microvesicles, and apoptotic vesicles (103), facilitate intercellular communication and influence cancer development, progression, and metastasis by transporting bioactive components like nucleic acids, proteins, metabolites, and lipids. These substances are found in circulating blood and various biofluids, and have emerged as promising non-invasive biomarkers (104). EVs carry abundant miRNAs, which are well-protected from RNase degradation. Consequently, EV-miRNAs, such as the miR-200 family (47, 57), miR-21, miR-100 (56), miR-99a-5p (54), and miR-1290 (55), are more frequently studied as potential biomarkers compared to non-exosome circulating miRNAs, for early detection of OC (105). EVs' miRNAs can facilitate paracrine and endocrine communication between different tissues, regulating gene expression and remotely modulating cellular functions (106). The sEVmiR-EOC score constructed from seven EVs miRNAs had superior diagnostic performance, especially in distinguishing patients with stage I EOC from benign ovarian tumors, with an AUC of 0.903 (specificity, 100%; sensitivity, 80%) (49). The OCEM signature, composed of eight EV mRNAs, achieved an AUC of 0.976 (51). Candidate exosomal miRNAs as biomarkers were mostly selected based on miRNA profiles (53, 56, 57) of tumors or exosomes secreted by ovarian cancer cell lines (54, 55), without a large-scale screening of patient plasma exosomal miRNAs. Therefore, the expression of miRNAs in tumor tissues might be inconsistent with the expression of circulating exosomal miRNAs. For example, miR-145 has been reported to be significantly down-regulated in EOC tissues, especially in HGSOC (107, 108). However, it was significantly up-regulated in serum exosomes from EOC patients and showed promising accuracy for EOC detection (sensitivity of 91.7%, specificity of 75%, AUC of 0.910) (53). The researchers employed a “banishing theory” to state that because miR-145 overexpression in ovarian cancer cells inhibits cancer progression, it is released from the cancer cells as exosomes. This implies that there may be undiscovered selection and sorting mechanisms that control the preferential encapsulation of specific miRNAs into exosomes before releasing them into the tumor microenvironment for intercellular communication (105).
For both cf-miRNA and EVs miRNA, the diagnostic accuracy of single miRNA is limited, but combining these miRNAs with traditional serological markers can increase the sensitivity of the assay (43). Combining exosomal miR-205 with CA125 and HE4 raised the AUC to 0.951, with sensitivity and specificity of 100% and 86.1%, respectively (50). Additionally, pairing CA125 with miR-99a-5p resulted in an AUC of 0.91 for differentiating early EOC from healthy controls (54). By combining next-generation sequencing of serum miRNAs with machine learning techniques, neural network analysis has the ability to identify more stage I/II ovarian cancers with a significantly lower false-positive rate, as well as identifying borderline tumors much better than CA125 (45). EVs' miRNA/cf-miRNA profiles may be influenced by a variety of factors. These comprise individual genetic variation, specimen source, various pre-analytical factors (including the degree of hemolysis), miRNA isolation methods, different assay platforms (e.g., RT-qPCR or NGS), different qPCR techniques (e.g., Taqman and LNA assays), and selection of standard reference genes. Similar to ctDNA, exosomes pose similar hurdles. Tumor-derived vesicles often account for less than 2% of circulating exosomes and undergo rapid clearance, necessitating high-throughput, high-sensitivity analytical methods for accurate detection. To successfully apply EVs or cf-miRNAs as biomarkers for the early diagnosis of OC in the clinical setting, these factors need to be carefully considered and standardized.
2.1.4 Tumor-educated platelets
Platelets display reactive responses during tumor progression and treatment. Tumor cells can directly or indirectly alter platelet RNA and protein content, resulting in the transfer of tumor-associated biomolecules to platelets. These tumor-educated platelets (TEPs) can promote cancer cell survival and metastasis and are considered potential diagnostic tools for cancer (109).
TEPs may also undergo queue-specific splicing events in response to signals released by cancer cells and the tumor microenvironment. The combination of specific splice events in response to external signals and the ability of platelets to directly uptake circulating mRNA could provide TEPs with a highly dynamic mRNA library, with potential applicability to cancer diagnosis (109). RNA sequencing of TEPs has become the latest component of liquid biopsy for cancer detection. Through mRNA sequencing of TEPs from 283 platelet samples, Best et al. achieved 96% accuracy in distinguishing 228 patients with localized or metastatic tumors from 55 healthy controls, along with 71% accuracy in identifying primary tumor locations (110). Although this study included samples from six different types of cancer, the results proved remarkably robust and suggest potential applications for similar technology in ovarian cancer patients. Subsequently, Piek et al. demonstrated the advantages of TEPs in both diagnosing and differentiating early-stage ovarian cancer from benign tumors, achieving an accuracy of 80% (111). Researchers developed an enhanced bioinformatics approach using deep learning, termed imPlatelet. The imPlatelet classifier converts platelet RNA sequencing data into images, where each pixel corresponds to the expression level of a certain gene. This method achieved 95% accuracy in distinguishing non-cancer patients from those with ovarian cancer (78). In addition to mRNA, proteins in platelets can also serve as potential biomarkers. An extensive proteomic approach identified a 9-protein panel in TEPs, yielding an AUC of 0.831 for early OC diagnosis (79).
While the aforementioned studies were conducted in limited patient cohorts, an intercontinental, hospital-based, diagnostic study enrolled 761 treatment-naïve inpatients with histologically confirmed adnexal masses and 167 healthy controls from nine medical centers (77). The TEPOC classifier, comprising 102 platelet RNAs, demonstrated robust diagnostic performance across diverse populations and OC subtypes, achieving an AUC of 0.858 for early-stage OC detection. This study confirms the potential of OC early detection by platelet RNA. In the future, TEPs analysis with complementary ctDNA/CTC analysis and platelet quantification may become a blood-based cancer diagnostic method.
2.1.5 Protein
Proteins are integral to numerous biological processes, and their post-translational modifications (PTMs), such as phosphorylation, acetylation, and glycosylation, play critical roles in cancer development and progression (112). The tumor biomarker CA125, which is routinely used in clinical practice, is a highly glycosylated mucin. Although it is the most sensitive and accurate serum biomarker in practical implementation, CA125 is still insufficient for early detection of ovarian cancer. Moreover, some benign diseases can also cause elevated CA125, such as endometriosis and pelvic inflammatory diseases. Many new biomarkers have been identified in the ongoing progress of MS-based proteomics, facilitating the development of OC multivariate index assays such as OVA1, Risk of Ovarian Malignancy Algorithm (ROMA), and Overa (113–116). These tests have greatly improved the sensitivity of OC diagnosis compared to single CA125, but have also reduced specificity to some extent. Proteomic analysis of exosomes isolated from the peripheral blood of patients with early-stage EOC and non-cancer controls identified eight proteins that could serve as potential ovarian cancer markers (63). The study initially identified upregulation of 35 proteins in both serum exosomes and tumor tissues from OC patients. Among these 35 proteins, eight of these proteins were confirmed in both exosome databases and other studies. However, validation in clinical cohorts is missing. Jo et al. developed a high-throughput EV screening platform (SAViA) to construct an EVHGSOC score model containing five proteins (EpCAM, CD24, VCAN, HE4, and TNC). This model demonstrated a sensitivity of 89%, a specificity of 93%, and an AUC of 0.95. The panel was able to differentiate early-stage HGSOC from the advanced-stage and non-cancer groups with a specificity of 91% and a sensitivity of 76% (62). The Proximity Extension Assay (PEA) Explore is an ultra-sensitive proteomics technology capable of characterizing much more of the plasma proteome with very little input material. PEA technology greatly improves the detection rate by integrating the high specificity of antibody immunoassay with the high sensitivity and throughput of genomics. Combining PEA analysis and machine learning identified multi-protein panels with AUCs exceeding 0.96 (60, 61). Most proteins in these models were associated with OC. All the protein panels mentioned above include WFDC2 (HE4), which is a clinically used biomarker for ovarian cancer.
Despite methodological differences, Jo et al. established murine fallopian tube (mFT) cells with oncogenic mutations and performed proteomic analysis on mFT-derived EVs (62). In contrast, Gyllensten et al. employed PEA Explore to compare plasma proteins between benign and malignant tumor patients (60, 61). All three studies used high-throughput analytical methods and obtained robust results, suggesting that novel plasma-based biomarker candidates for ovarian cancer screening can be discovered by harnessing the power of high-precision proteomics. The integration of high-throughput sample preparation technologies and automated systems, advanced MS-based glycoproteomic research methods, AI-driven data analysis techniques, and the establishment of more comprehensive and complete databases can accelerate the discovery and application of protein-based OC biomarkers (117).
2.1.6 Metabolites
Metabolites constitute the endpoints of many biofunctional molecular processes, and disturbances at the level of metabolism in the blood and/or other body fluids have long been recognized as promising indicators of cancer. Metabolic profiles have been proposed as molecular phenotypes of biological systems, reflecting pooled information encoded at the genomic level as well as responses at the transcriptomic and proteomic levels (65). Exploring the metabolic profile of OC can both assist in early diagnosis and investigate the underlying biological mechanisms of ovarian cancer (68, 118). Abnormal lipid metabolism (70, 71), fatty acid β-oxidation (67, 72), and amino acid catabolism (72, 118) are among the metabolic pathways associated with ovarian cancer progression. Metabolomics studies of ovarian cancer have mainly used nuclear magnetic resonance (NMR) (76, 119) and mass spectrometry (MS)-based methods (67, 68, 73–75). Garcia et al. applied NMR spectroscopy-based metabolomics to discriminate ovarian cancer patients at early stages from healthy controls, with an AUC of 0.949 (76). Chen et al. discovered 27-nor-5β-cholestane-3,7,12,24,25 pentol glucuronide (CPG) as a complementary diagnostic marker to CA125 with an AUC of 0.747 (75). Zhang et al. also used UPLC/Q-TOF/MS for the test, and the six metabolites model could reach an AUC of 0.86 (73), and the other eight metabolites model had an AUC of 0.941 (74). Ke et al. conducted a large-scale metabolic study of 448 plasma samples with a UPLC/MS platform. The study results identified metabolic profiles and potential biomarkers, distinguishing between EOC or early-stage EOC and benign ovarian tumors (BOT), with AUC values of 0.9100 and 0.8385, respectively (72). Combining metabolomics profiling with machine learning enables more accurate analysis of large datasets, facilitating the understanding of disease-specific variants and further biomarker discovery. A linear support vector machine (SVM) model consisting of 16 diagnostic metabolites was able to identify early OC in a patient cohort with 100% accuracy (71). Using recursive feature elimination (RFE) coupled with repeated cross-validation (CV) based on UPLC-MS metabolomics, the developed model was able to distinguish OC cases from controls with 93% accuracy. Importantly, the overall predictive accuracy of the consensus classifier was better for early-stage patients compared to advanced-stage patients (65).
2.1.7 Advantages and challenges
Plasma and serum, the traditional liquid biopsy samples, have the most abundant relevant studies, demonstrating relatively satisfactory diagnostic performance. A series of sophisticated high-throughput techniques has also been prioritized in plasma/serum, enabling further enhancement of the precision of the assay. To sum up, serum/plasma testing is a convenient and readily acceptable method for the dynamic monitoring of a patient's disease progression.
However, tumor-associated components are shed from the primary tumor site, enter the bloodstream, and circulate systemically. During this process, their detection rate may be affected by the significant reduction of their number, together with the presence of non-tumor-associated components in the blood. Therefore, high-performing single biomarker and/or biomarker panels in blood samples currently rely on advanced and high-throughput technologies, most of which are expensive and do not meet the requirements of healthy economics. There is still a need for further research into potential biomarkers which would be practical and could be used for census purposes. Ahn et al. proposed that combining plasma proteomics and metabolomics can identify emerging features that are difficult to detect using a single omics approach (120). This indicates that in the future, the combination of multi-omics methods and machine learning will better facilitate the early detection of OC.
2.2 Urine
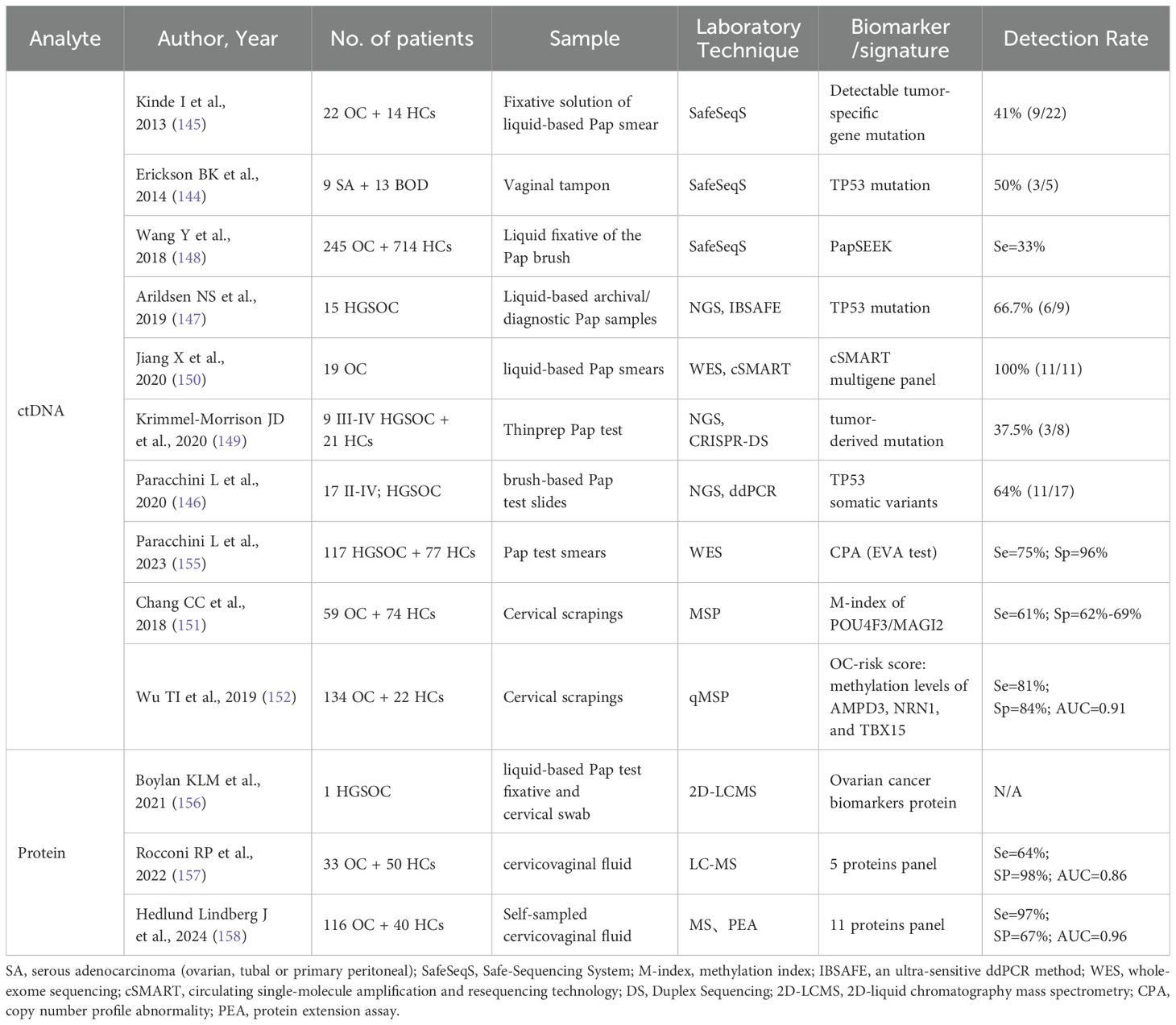
Urine can be used as a convenient, rapid, and completely non-invasive method of liquid biopsy for nearly all patients. Since it is a product of normal metabolism and secretion, it can be collected in large quantities. Urine has been shown to contain a variety of proteins/peptides, with approximately 70% of urinary protein derived from the kidney and 30% from plasma (121), making it a potential biomarker. Compared to blood, urine is more stable, and protein-related degradation pathways are completed by the time of excretion. In contrast, many protein hydrolysis degradation products are produced by the activation of proteases in blood samples (especially the coagulation cascade), which may interfere with the results (122). Table 2 presents studies related to the early detection of ovarian cancer with urine samples, and we can observe that most analyses focused on proteomics and metabolomics. With regard to genomics, there are fewer reports about urine. Only one team found that transrenal DNA (TrDNA) was more indicative of DNA methylation status than serum/plasma. For the methylation of HIST1H2BB and MAGI2, the correlation between tumor DNA and TrDNA methylation measurements was stronger (123).
Urinary miRNAs are usually isolated via total RNA from extracellular vesicles and cellular components, and then small RNA molecules (<200 nt) are amplified for qRT-PCR analysis (134). The current study only identified up- or down-regulated miRNAs in the urine of OC patients and did not conduct further trials on whether these miRNAs could detect OC earlier. Zhou et al. provided the first evidence of elevated miR-30a-5p in the urine of OC patients and confirmed that the up-regulation of miR-30a-5p may be closely related to the early stage and lymphatic metastasis (124). Záveský et al. analyzed and compared preoperative and postoperative specimens from the same EOC patients, finding that urinary miR-92a was upregulated while miR-106b was downregulated (125). However, subsequent studies by this team showed that miRNAs from extracellular urine sources did not show significant differences compared to tissue and ascites samples (135). The lower levels of detectable miRNAs in urine compared to blood may account for this. It is postulated that most circulating miRNAs are reabsorbed by the kidneys through an as yet unknown mechanism and destroyed by high levels of RNase in the urinary tract (134).
Since the first analysis of serum peptidomics in OC (136), several studies have found that low molecular weight proteins and peptides in different body fluids can provide specific diagnostic information for different types of cancer (137, 138). The first study of OC-associated urinary peptidomes was performed by Smith et al. They identified several endogenous urinary peptides that could serve as potential biomarkers, the most promising of which was leucine-rich alpha-2-glycoprotein (LRG1) (129). A prospective, longitudinal, case-control study assayed and sequenced urinary micropeptides from OC patients and their age-matched volunteers, revealing that urinary catalase (CAT), alpha-1 acid glycoprotein (AAG), and peroxidase-2 (Prx-2) could serve as biomarkers for early detection of ovarian cancer and response to treatment (131). Studies have also found protein expression in the urine of OC patients that correlates with cancer cell proliferation and invasion. For instance, IL-1β and MCM5 have been identified (128, 130). In vitro trials have also shown differences in urinary protein expression between OC tumorigenic and non-tumorigenic rats, which have been hypothesized to be related to ovarian cancer metastasis (139). But the studies above either lacked evaluation of diagnostic tests or had insufficient diagnostic performance.
Slupsky et al. demonstrated for the first time that changes in metabolite concentrations displayed by urinary metabolic profiling may be associated with ovarian cancer specificity (140). Metabolite polyamines are present in elevated levels during the process of active cell proliferation, such as in cancer patients. Urinary N1, N12-diacetylspermine (DiAcSpm) serves as a significant diagnostic and prognostic marker in various types of cancer. A proof-of-concept study utilizing LC-MS/MS revealed elevated levels of urinary DiAcSpm in patients with malignant ovarian tumors, including those with low malignant potential and early-stage disease. DiAcSpm had a better sensitivity (86.5%) but lower specificity (65.2%) (132). One metabolomics study utilized non-targeted techniques to detect metabolite profiles in urine, and developed a model of 2 urinary putative metabolites for ovarian cancer diagnosis by the support vector machine algorithm. The AUC of the model was 0.984, with a sensitivity of 97.66% and a specificity of 87.50% (133).
Urine samples have a special feature of containing several intrinsic fluorophores that can be detected through fluorescence analysis, which is rapid, safe, and highly sensitive. Therefore, the analysis and surveillance of autofluorescence in urine present a new opportunity for ovarian cancer screening methods. Blue-fluorescing pteridines dominate in the excitation-emission matrices of cancer urine samples (141). The study found that using Concentration Matrices of Synchronous Spectra (CMSS) resulted in high sensitivity (91.67%) and specificity (100%) in differentiating between patients with ovarian malignancy and healthy women (127). There are fewer studies related to cancer with urine fluorescence. Urine autofluorescence at 295 nm was found to be significantly higher than that of healthy controls in the urine of patients with malignant melanoma at each clinical stage (142). A combination of urine fluorescence spectroscopy with machine learning algorithms has shown promising capabilities in screening for endometrial cancer (143). OC is a near-urinary tumor, along with endometrial cancer, and perhaps urine autofluorescence could also be used as a tool for OC screening.
Current studies on relevant urinary biomarkers for the detection of OC are mostly limited to a theoretical level. The authenticity and reliability of these potential biomarkers have not been evaluated. Available clinical findings are also insufficient to effectively detect early OC, though they may have greater diagnostic power when combined with other non-urinary biomarkers and imaging tests. Thus, further validation is required for urine as a liquid biopsy sample to detect OC.
2.3 Pap test and cervicovaginal fluid
Since the advent of the Papanicolaou (Pap) test as a routine screening tool for cervical cancer, the mortality of cervical cancer has dramatically decreased. The ovaries, fallopian tubes, uterus, and vagina are anatomically adjacent, it has been proven that tumor DNA could be detected in the vaginal tract of OC patients (144). In recent years, researchers have continued to evaluate the potential role of the Pap test in the early detection of OC. Because the cells in such samples are shed directly from the primary tumor, they harbor tumor DNA in both greater quantities and higher concentrations than the DNA in circulating in the bloodstream.
In 2013, Kinde et al. first discovered ovarian cancer mutated DNA molecules in Pap smear specimens (145). This indicated that ovarian cancer cells and cell debris were present in the cervix and could be detected by molecular genetic techniques, providing early evidence of the viability of detecting gynecologic cancers by intracervical DNA detection. This was the beginning of a chapter in the study of early diagnosis of OC using Pap test. The most commonly mutated gene in epithelial ovarian cancers was TP53 (145). TP53 has been identified as the most common gene associated with OC in purified DNA from Pap test samples. These samples were collected approximately 2.5–7 years before OC diagnosis in pre-symptomatic women (146, 147). Despite the low DNA abundance in these samples, ultrasensitive ddPCR was capable of identifying tumorigenic TP53 mutations. The mutation detection rate among tumor-associated genes, such as TP53, in Pap smear specimens from both OC patients and those with presymptomatic OC was inadequate to fulfill the screening standards. The sensitivity of detecting ovarian cancer by tumor-specific mutations in Pap test sample ranged from 33% to 75% (145–149). Only Jiang et al. showed that somatic mutations carrying tumor genomic information were tested in all Pap smears (150). The reason for this phenomenon may be related to small patient cohorts, most of which had only approximately 20 OC patients. A large cohort study with 245 OC patients and 714 healthy controls used PapSEEK. This test incorporates assays for mutations in 18 genes as well as an aneuploidy assay, has shown a sensitivity of 33%, including 34% of patients with early-stage disease (148). Deep sequencing of the TP53 gene using Duplex Sequencing (DS) on all 30 Pap test specimens only detected tumor-derived TP53 mutations in 38% of OC cancer patients (149). Combining plasma circulating tumor DNA with a Pap test has been shown to increase sensitivity (148), but it is insufficient for clinical application. Despite the use of more sophisticated techniques, higher sensitivity assays, and larger cohorts, the mutation detection rate has not substantially improved. This may be due to the anatomical distance between the collection site (cervical canal) and the fallopian tube, which is thought to be the origin of serous ovarian cancer, rather than technical limitations. The improved detection sensitivity achieved with Tao brushes further supports this hypothesis (148).
Besides detecting DNA mutations, Chang et al. demonstrated the feasibility through DNA methylation analysis of cervical scrapings. The hypermethylation of POU4F3/MAGI2 was observed in both OC tissue and cervical scrapings, with a sensitivity and specificity of 61% and 62%-69%, respectively (151). Then, the same team thoroughly investigated OC-specific DNA methylation biomarkers in conventional Pap tests, analyzed the methylomes of tissues and cervical scrapings, and integrated public methylomics datasets to depict methylomics profiles. An OC risk prediction model comprising AMPD3, NRN1, and TBX15, achieved a sensitivity of 81%, a specificity of 84%, and OC detection accuracy of 91% (152).
In the current study, the investigators chose to focus on genomic instability instead of somatic gene mutations. A pattern of genomic instability has been demonstrated in the early stages of HGSOC progression (153). Copy number aberrations (CNA), a form of chromosomal instability, are the most prevalent structural variation in the genome. Unlike single nucleotide variants, somatic copy number alterations (SCNAs) are rarely found in normal tissues, though they are common in cancer (especially in HGSOC) (154). A recent retrospective and multicenter cohort study analyzed 250 archived Pap test DNA collected before diagnosis. Researchers derived the copy number profile abnormality (CPA) from Pap test samples using low-pass whole-genome sequencing. They integrated the CPA score into the EVA (early ovarian cancer) test, achieving sensitivity of 75%, specificity of 96%, and accuracy of 81% (155). The detection of characteristic DNA methylation and genomic instability in Pap test specimens has greatly improved the sensitivity and specificity of ovarian cancer screening. The number of such studies is limited; larger and numerous studies are necessary to further confirm the diagnostic capability.
Through fallopian tube peristalsis, protein-rich fluid from the endometrial cavity is transported into the vagina via the cervix. Protein analysis of both Pap test fluid and cervical swabs revealed similarities between these samples and tumor extracts. This establishes the first step in the feasibility of detecting ovarian cancer protein biomarkers in Pap test fluid or cervical swabs (156). Cervicovaginal fluid collected by inserting a cytobrush, similar to a Pap test into the cervix, is a useful method of detecting early changes in the fallopian tubes and their microenvironment. Rocconi et al. evaluated cervicovaginal fluid specimens by LC-MS and constructed a protein panel containing five proteins: serine proteinase inhibitor A1; periplakin; profilin1; apolipoprotein A1; and thymosin beta4-like protein. This panel was used as a biomarker for ovarian cancer screening to distinguish ovarian cancer patients from control groups, with an AUC of 0.86 (157).
Samples from the cervix and vagina are relatively easy to collect and acceptable to patients. Similar to cervical cancer screening, it can be integrated into routine gynecological screening. Table 3 lists all studies that used cervical and vaginal samples for analysis. There are two main kinds of cervical cytology samples used in early detection of OC. The first is the fixative of liquid-based Pap test (145, 147, 148, 150), such as the Thinprep Cytologic Test (TCT) applied for routine screening of cervical cancer (149). The other is the brush-based Pap test (146). The obtained cervical swabs were stored in conical tubes and soaked in phosphate-buffered saline (PBS) to elute their supra-components when needed (152, 156). The prevalence of liquid-based TCT let us to hypothesize that samples extracted from the liquid-based vial are suitable for ancillary testing, ensuring both accuracy and convenience. Pap tests and cervicovaginal fluid are theoretically feasible, but their sensitivity and specificity have yet to reach expected clinical standards. In a recent study, MS and protein extension assay (PEA) were used in combination with artificial intelligence to create an 11-protein panel. This panel had a sensitivity of 97%, a specificity of 67%, and an AUC of 0.96 (158). Although this outcome did not align with the optimal differentiation achieved by the multivariate model in plasma, the researchers utilized a sample of dried, self-sampled cervicovaginal fluid (CVF) deposited on elute filter paper cards, which has been shown to provide accurate and cost-efficient screening of cervical cancer (159, 160). While the specificity of CVF compared to plasma still needs to be increased, the results offer the possibility of an ovarian cancer screening program based on self-collected CVF samples. In addition, this study also included specimens collected prior to the diagnosis of OC (before the onset of symptoms), in which the signal could also be tested, indicating that this model could provide information about future disease risk. In summary, utilizing Pap tests and cervico-vaginal fluid as liquid biopsy samples for early ovarian cancer detection offers distinct operational advantages. The diagnostic accuracy has significantly improved with advances in multi-omics technologies, demonstrating considerable clinical potential.
2.4 Uterine and tubal lavage
Progressing in parallel with studies using Pap tests for early detection of OC is uterine cervical cells. A study detected malignant cells shed from ovarian, fallopian tubes, and peritoneal cancers by examining endometrial cytologic samples from the endometrial cavity (endometrial sampler). It is a concept similar to the detection of cervical cancer by Pap smear, but this research had a low positive detection rate of only 12% (161). Then, investigators found that using Tao brush for intrauterine sampling increased the detection rate of the diagnostic multiplex PCR-based test named PapSEEK, 45% of 51 ovarian cancer patients tested positive (148). Tumor cells shed from ovarian or endometrial cancers are carried into the uterine cavity, where the Tao brush can collect them. Consequently, the detection of exfoliated cells from high-grade-serous ovarian cancer, or precursor lesions, is a promising concept for earlier diagnosis. Table 4 shows the studies currently used to diagnose OC by uterine lavage.
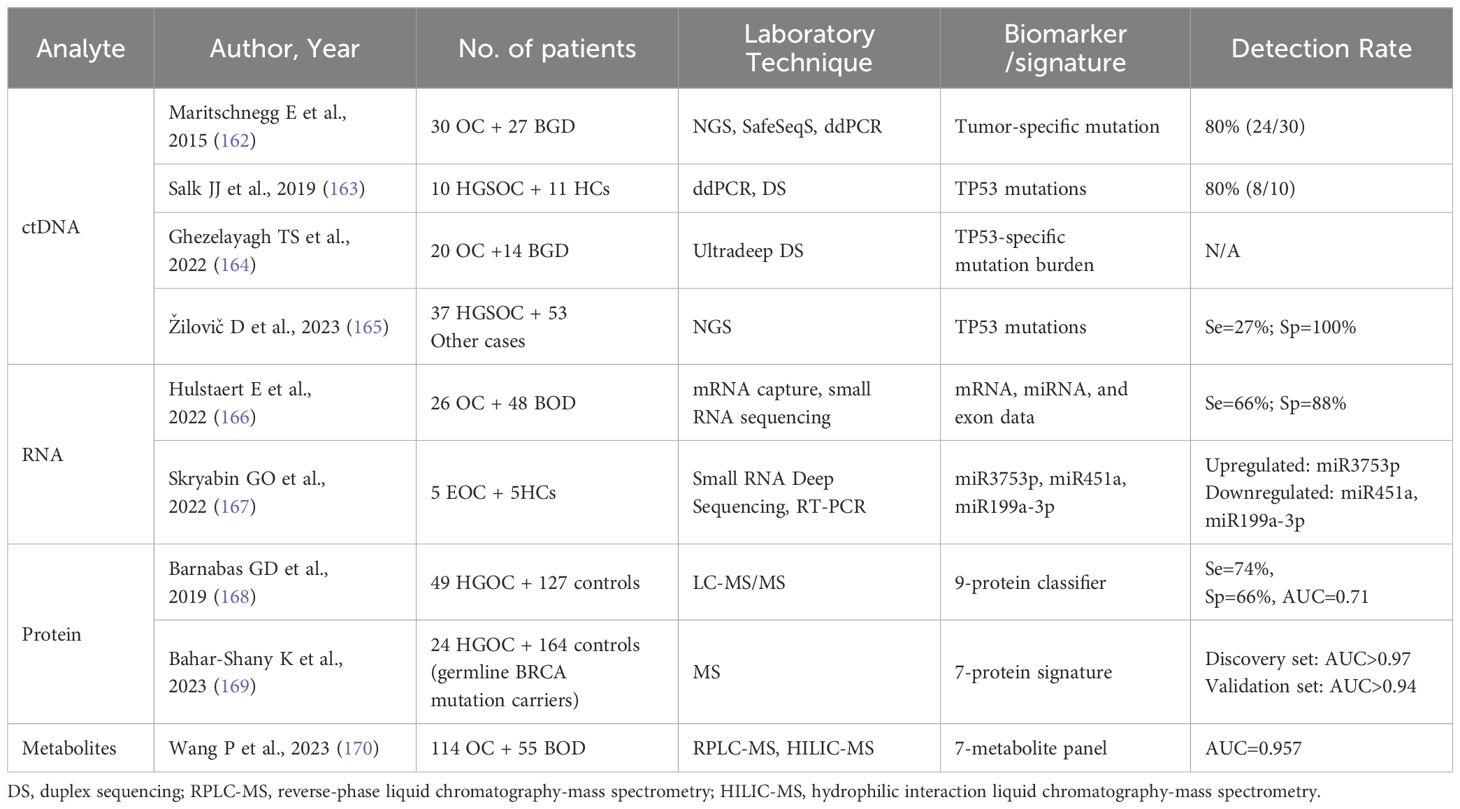
In 2015, Maritschnegg et al. firstly demonstrated that cancerous cells originating from ovarian tumors could be shed and collected through uterine lavage. Using NGS and singleplex analysis, researchers detected mutations that were consistent with those found in primary tumor tissue in 80% of OC uterine lavage samples (162). This proof-of-concept study has shown the potentially diagnostic power of the uterine lavage method for OC detection, especially for early detection in high-risk populations. Analysis of uterine lavage samples using NGS reveals TP53 mutations in about 60% of OC patients (162, 165). Since previous studies have been performed based on known mutated genes in the primary tumor lesion, Salk et al. combined Uterine and tubal lavage (Utl) with Duplex Sequencing (DS), which has a higher sensitivity (80%) for detecting true-positive cancer-derived TP53 mutations in HGSOC without prior knowledge of the tumor mutation (163).
Current transcriptomic studies utilizing anatomically proximate fluids remain theoretical. A proof-of-principle extracellular transcriptomic analysis utilizing messenger RNA capture and small RNA sequencing revealed that the lavage fluid had ovarian and fallopian tube-specific mRNA enrichment (166). This study first applied RNA-seq to utero-tubal lavage samples, yielding a multi-omics classifier based on combined mRNA, miRNA and exon data. With 66% sensitivity and 88% specificity, the model demonstrates technical feasibility for RNA isolation and sequencing from utero-tubal lavage fluid, revealing ovarian and fallopian tube-specific mRNA signatures useful for early diagnosis. Skryabin et al. also confirmed the differences in the expression levels of miRNAs between healthy individuals and EOC patients are particularly associated with cancer, such as miR-200 family members (167). Despite the lack of reliable cohort study data demonstrating the value of its application, RNA in utero-tubal lavage fluid is 8 times higher than in platelet-free plasma (166) and remains a potential biomarker that can be used for the early detection.
Also, the protein and metabolites in the uterine fluid have the potential to provide a broader range of biomarkers for early detection. Combining deep microvesicle proteomics with gynecologic intracanal fluid biopsy, support vector machine algorithms were applied to generate a 9-protein classifier with 70% sensitivity and 76% specificity. The signature correctly identified all Stage I lesions (168). As the expression profile of BRCA-mutated Müllerian epithelium is significantly different from the WT pattern, the team further characterized the proteomic signatures to identify HGSOC in BRCA carriers. The 7-protein panel discriminated between patients with high-risk germline BRCA mutations and controls with an AUC >0.97 and a negative predictive value of 100%. In addition, the questionnaire results reported that this sampling method is clinically acceptable with favorable pain scores and safety (169). As for metabolomics, Wang et al. revealed the metabolomic profile of uterine fluid and developed a panel of 7 metabolites that can discriminate women with benign gynecological diseases from those with early-stage OC with obscure symptoms. The AUC of this panel was 0.957, significantly higher than 0.817 for CA125 and 0.841 for ROMA, providing an accurate and sensitive strategy for the early diagnosis of OC (170).
Depending on the different types of catheters, current UtL collection approaches can be categorized into three types: one-way, two-way, and three-way catheters. One-way catheter, such as intrauterine insemination catheter and rigid pipelle uterine samplers (166, 168, 169), was inserted transcervically into the uterine cavity. The saline was flushed directly and retrieved immediately. The two-way catheter is primarily composed of existing catheters, such as dual-channel catheters (167), two-way hysterosalpingography catheter (165), and size ten (10 F) Foley catheter (170), that are primarily used for other purposes. The balloon was inflated with saline to seal the cervical canal and prevent retrograde leakage of saline. The normal saline was slowly infused into the uterine cavity through the catheter tube, and left for a while before gently suctioning to collect the liquid. The uterine cavity is very small and the anterior and posterior walls lie on top of each other. When using a single-channel catheter, it was removed immediately after saline injection, the saline could not adequately and completely irrigate the uterine cavity. Besides, the injected saline tended to flow back into the vagina. The use of a two-channel catheter with a balloon solved the problem of saline reflux, but did not ensure that the sample collected could be fully representative of the internal environment in the uterine cavity. In order to solve the above problems, the novel three-way catheter was designed and developed (171). There are two lavage channels, each with two openings, one on the tip of the catheter facing forward and one at the side. The third tube is the balloon channel carrying a valve. Two syringes, one of them containing saline, are connected to the two lavage tubes. By pushing on the plunger of the syringe containing saline, the uterine cavity and fallopian tubes were slowly perfused. Simultaneously, the plunger of the empty syringe was gently pulled out. The clinical utility of the three-way catheter was evaluated on whether lavage solution could be successfully obtained, how easy it was to insert, whether cervical dilation was required, the volume of lavage fluid collected, and the amount of DNA extracted. Moreover, the study assessed the pain level, time required for placement, and other complications in patients compared to intrauterine device (IUD) placement. It has been proven that this three-way catheter caused minimal pain, both in terms of intensity and duration, making it a practical and safe option.
Currently, using uterine lavage fluid as a liquid biopsy sample, the sensitivity of OC early detection ranges from 70% to 80%. The detection accuracy for tumor-associated TP53 mutations in uterine lavage reached approximately 80%, higher than the Pap test. These findings support the clinical value of proximal liquid biopsy for improving detection rates. Recent research has established that most HGSOCs originate from epithelial precursor lesions on the fallopian tubes rather than ovarian tissue (172). Serous tubal intraepithelial carcinoma (STIC) is now recognized as the direct precancerous lesion preceding HGSOC development. Mutational evolutionary analyses identify a 6-year interval between the TP53-mutated precursor emergence and the initiation of HGSOC (172). Tumor cells and related substances from precancerous or early OC lesions can transit through the tubal ducts into the uterine cavity. These can be detected directly in utero-tubal lavage fluid, where they are more abundant than circulating tumor cells in the blood, facilitating early OC detection and diagnosis. In summary, there are limited studies on the detection of ovarian cancer using uterine lavage, which predominantly employ sequencing to analyze tumor-associated genes, such as TP53, yet the detection accuracy remains unsatisfactory. The application of multiple MS-based assays of proteins or metabolites in uterine lavage fluid, in conjunction with machine learning algorithms, has markedly enhanced the diagnostic efficacy. This may prove to be a fruitful avenue for future research and development.
3 Discussion
With the accelerating development of multi-omics technologies, especially in combination with the application of machine learning algorithms, early detection of cancer has evolved from a single level of tumor-associated gene mutations to an integrated multi-omics analysis. More and more studies have supported the potential of liquid biopsy in the discovery of candidate biomarkers. This has facilitated the progression of the early detection of OC. There are commercially available liquid biopsy-based platforms for early detection of ovarian cancer, such as CancerSEEK, PapSEEK, OvaPrintTM, and MCED. CancerSEEK is a liquid biopsy platform designed for early detection of eight cancer types. It demonstrates 98% sensitivity for ovarian cancer (25). PapSEEK is a diagnostic multiplex PCR-based test. It utilizes Pap brush and Tao brush samples to detect 18 mutations highly associated with endometrial and ovarian cancers for early diagnosis (148). OvaPrintTM is a cfDNA mehylation liquid biopsy platform to discriminate benign pelvic masses from HGSOC preoperatively (14). Multicancer early detection (MCED) blood tests can detect cancer signals from cfDNA through detection of cancer-specific DNA methylation. PATHFINDER, a prospective cohort study, enrolled 6,662 participants aged 50 years or older without signs or symptoms of cancer in MCED testing (173). In cases with positive MCED results and confirmed cancer diagnoses, the testing accurately predicted tumor origin and significantly reduced time to diagnostic confirmation, demonstrating the clinical feasibility of this approach. Among the 35 true-positive participants, there was only one case of ovarian cancer, which was stage III. CancerSEEK and MCED are platforms for pan-cancer early detection. The updated results from CancerSEEK are still pending. Researchers are currently conducting larger-scale trials based on the PATHFINDER study to evaluate MCED testing. Regarding PapSEEK and OvaPrint™, validation studies for ovarian cancer early detection in wider populations are still lacking. Thus, whether these platforms are applicable for early detection of OC remains unknown.
It is not easy to make side-by-side comparisons between samples, as differences in the baseline of the different samples, the use of the detection assays, and the discovery of different potential markers will inevitably lead to differences in the accuracy of the assays. Table 5 summarizes the collection methods, respective advantages, current challenges, and target populations of different liquid biopsy samples for early detection of ovarian cancer. The use of plasma/serum for early detection of ovarian cancer leads to relatively satisfactory performance. Peripheral blood represents a convenient and easily manageable biospecimen with high patient compliance, making it particularly suitable for long-term monitoring in general populations. However, it remains debatable whether blood-based assays can truly detect OC at an early stage and reduce mortality. Technical challenges in analyzing low-concentration circulating tumor components drive the need for sophisticated detection platforms, with consequent economic implications for diagnostic implementation. As for urine, the clinical manifestations of relevant potential biomarkers are not sufficient for the effective detection of early OC, and existing studies cannot prove their authenticity and reliability for OC early screening. Interestingly, the fluorescent sites in urine are unique, and monitoring urine autofluorescence may offer new opportunities for the development of ovarian cancer screening methods (127).
Proximal liquid biopsy is an assay that takes the sample directly from the body cavity where the tumor is located, increasing the likelihood of detecting early or even precancerous lesions. The entire female genital tract is a connected lumen. Collection of biospecimens from cervicovaginal fluid and uterine lavage is clinically termed proximal fluid biopsy. Using cervical smears to diagnose OC is a relatively new approach that has emerged in the last decade, similar to using cervicovaginal fluid. Figure 2 illustrates a chronological timeline from 2013 to the present, demonstrating landmark studies of proximal liquid biopsy in ovarian cancer detection. To meet the high sensitivity and specificity required for early diagnosis of OC, the performance of such specimens is currently inadequate. It is encouraging to note that the latest studies use more accurate high-throughput technology combined with artificial intelligence to develop tests that could significantly improve the diagnostic performance of cervical cells and cervicovaginal fluid. For example, Hedlund Lindberg J et al. achieved a sensitivity of 97% using self-collected cervicovaginal fluid on paper cards as the sample. Although the specificity is not the most satisfactory, it offers the possibility of using self-collected samples for ovarian cancer screening programs (158). With further technological advancements, we believe these specimens have promising potential to be used for OC early diagnosis in the future, similar to cervical cancer screening. The uterine cavity is anatomically closer to where the tumor originates, and components obtained from it could theoretically provide more complete and abundant diagnostic information, further increasing the likelihood of detecting early-stage or even precancerous lesions. Practically, uterine lavage has also demonstrated superiority and accuracy over cervical/vaginal cytology. This assay has a high diagnostic potential, but is not a completely non-invasive sampling method. It is uncomfortable for patients and carries a theoretical risk of infection to some extent, making it unsuitable for universal screening of healthy populations. For carriers of germline BRCA mutations, uterine lavage could be a viable monitoring option. The general population of women should be divided into the population with high risk and the population with an average risk of ovarian cancer. Because of the absence of screening tools with excellent sensitivity and specificity at present, bilateral salpingo-oophorectomy remains the standard risk-reduction strategy for high-risk women, particularly BRCA1/2 mutation carriers, typically recommended between the ages of 35–45 after completing their reproductive plans. Although risk-reduction bilateral salpingo-oophorectomy (RRSO) has been confirmed to significantly decrease the risk of BRCA1/2-associated ovarian or fallopian tube cancer, and consequently reduce mortality, many high-risk individuals have declined or postponed the procedure due to menopause and the subsequent health consequences of early surgery. For this population, uterine lavage, combined with a superior diagnostic performance biomarker panel, can be administered semiannually as a tool for long-term surveillance when a deferred RRSO is requested or required (169).
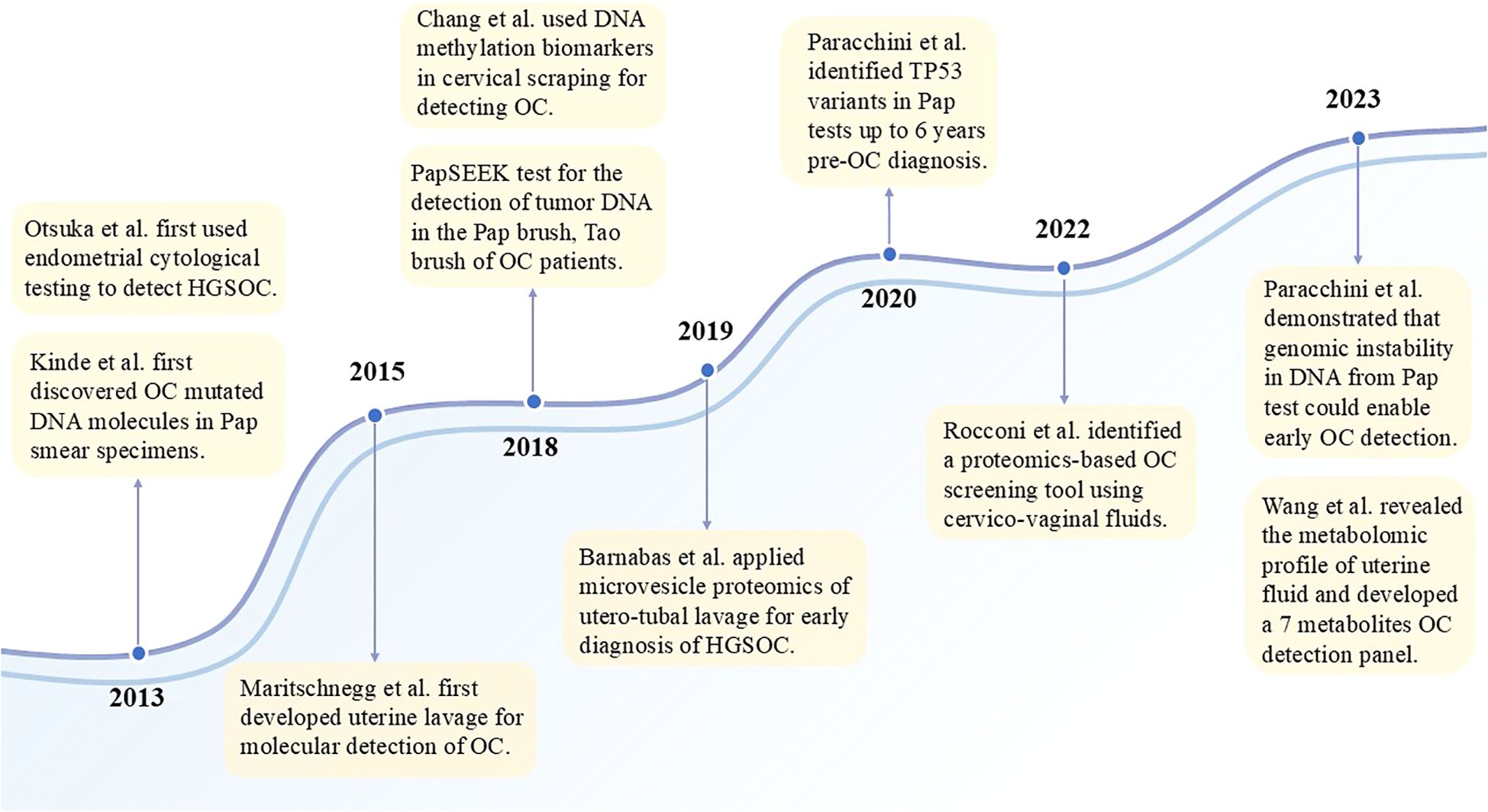
Figure 2. Chronological development of proximal liquid biopsy in ovarian cancer research. Since 2013, when Pap tests and uterine lavage were first shown to detect ovarian cancer, multi-omics studies of proximal liquid biopsies have aimed to develop practical early detection tools with high sensitivity.
However, a gap persists between discovery research and practical clinical application, with several challenges need to be addressed. First, ovarian cancer is highly heterogeneous. Because of the predominance of EOC, current studies either focus only on EOC, even HGSOC, or the study cohorts have a small percentage of other OC subtypes, which are not well represented. OC is typically asymptomatic in its early stages, with most patients presenting at advanced stages when they come for the initial consultation. Consequently, advanced-stage OC accounted for a large proportion of the cohorts, and the majority of early-stage OC are type I or borderline tumors. Importantly, the paucity of stage I and STIC lesions hinders the evaluation of liquid biopsy techniques and classifiers for genuinely detecting clinically latent OC. Also, there is a lack of multicenter prospective studies in large, multi-ethnic populations. Most of the research has only evaluated the diagnostic performance of candidate biomarkers/biomarker panels, and a small proportion of which has examined the relationship between candidate panels and age. Further work is needed to explore the correlations and interactions of candidate biomarkers with other ovarian cancer risk factors, such as prior chemotherapy, endometriosis, or germline BRCA mutations. Lastly, both the collection of uterine lavage fluid and the extraction of tumor-associated fractions in the laboratory do not have standardized workflows, which severely limits the reproducibility of assays. Future efforts should develop optimal standardized procedures and analysis platforms to validate new technologies and prospective biomarkers in robustness and reproducibility.
In conclusion, liquid biopsy has emerged as a promising option for screening and early detection of OC. The minimally invasive and rapid nature meets the requirements for screening in healthy populations. Various body fluid specimens have their strengths and weaknesses; blood, cervical cytology, cervicovaginal fluid, and uterine lavage could be potential specimen sources for screening and early diagnosis of OC in different risk groups. Hopefully, the selection of appropriate liquid biopsy samples, the application of multi-omics technology for analysis, and the combination of artificial intelligence and machine learning algorithms will improve OC early detection and contribute to management.
Author contributions
YF: Writing – original draft, Conceptualization, Writing – review & editing. WY: Writing – original draft. JZ: Writing – review & editing, Data curation. SW: Writing – review & editing, Data curation. NW: Writing – review & editing. HZ: Writing – review & editing. XY: Validation, Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Funded by Clinical Research Center, First Affiliated Hospital of Xi'an Jiaotong University (grants XJTU1AF2021CRF-025).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2024) 74:229–63. doi: 10.3322/caac.21834
2. Menon U, Gentry-Maharaj A, Burnell M, Singh N, Ryan A, Karpinskyj C, et al. Ovarian cancer population screening and mortality after long-term follow-up in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet. (2021) 397:2182–93. doi: 10.1016/S0140-6736(21)00731-5
3. Webb PM and Jordan SJ. Global epidemiology of epithelial ovarian cancer. Nat Rev Clin Oncol. (2024) 21:389–400. doi: 10.1038/s41571-024-00881-3
4. Zhu JW, Charkhchi P, and Akbari MR. Potential clinical utility of liquid biopsies in ovarian cancer. Mol Cancer. (2022) 21:114. doi: 10.1186/s12943-022-01588-8
5. Jayson GC, Kohn EC, Kitchener HC, and Ledermann JA. Ovarian cancer. Lancet. (2014) 384:1376–88. doi: 10.1016/S0140-6736(13)62146-7
6. Zapardiel I, Diestro MD, and Aletti G. Conservative treatment of early stage ovarian cancer: Oncological and fertility outcomes. Eur J Surg Oncol (EJSO). (2014) 40:387–93. doi: 10.1016/j.ejso.2013.11.028
7. Menon U, Gentry-Maharaj A, Burnell M, Ryan A, Singh N, Manchanda R, et al. Tumour stage, treatment, and survival of women with high-grade serous tubo-ovarian cancer in UKCTOCS: an exploratory analysis of a randomised controlled trial. Lancet Oncol. (2023) 24:1018–28. doi: 10.1016/S1470-2045(23)00335-2
8. Della Corte L, Russo G, Pepe F, Pisapia P, Dell’Aquila M, Malapelle U, et al. The role of liquid biopsy in epithelial ovarian cancer: state of the art. Crit Rev Oncol Hematol. (2024) 194:104263. doi: 10.1016/j.critrevonc.2024.104263
9. Sekar Y, Ishwar D, Tan B, and Venkatakrishnan K. Nano biosensor unlocks tumor derived immune signals for the early detection of ovarian cancer. Biosens Bioelectron. (2025) 278:117368. doi: 10.1016/j.bios.2025.117368
10. Li G, Zhang Y, Li K, Liu X, Lu Y, Zhang Z, et al. Transformer-based AI technology improves early ovarian cancer diagnosis using cfDNA methylation markers. Cell Rep Med. (2024) 5:101666. doi: 10.1016/j.xcrm.2024.101666
11. Gaillard DHK, Lof P, Sistermans EA, Mokveld T, Horlings HM, Mom CH, et al. Evaluating the effectiveness of pre-operative diagnosis of ovarian cancer using minimally invasive liquid biopsies by combining serum human epididymis protein 4 and cell-free DNA in patients with an ovarian mass. Int J Gynecol Cancer. (2024) 34(5):713–21. doi: 10.1136/ijgc-2023-005073
12. Chen L, Ma R, Luo C, Xie Q, Ning X, Sun K, et al. Noninvasive early differential diagnosis and progression monitoring of ovarian cancer using the copy number alterations of plasma cell-free DNA. Transl Res. (2023) 262:12–24. doi: 10.1016/j.trsl.2023.07.005
13. Zhou H, Zhang X, Liu Q, Yang J, Bai J, Yin M, et al. Can circulating cell free DNA be a promising marker in ovarian cancer? - a genome-scale profiling study in a single institution. J Ovarian Res. (2023) 16:11. doi: 10.1186/s13048-022-01068-z
14. Buckley DN, Lewinger JP, Gooden G, Spillman M, Neuman M, Guo XM, et al. OvaPrint-A cell-free DNA methylation liquid biopsy for the risk assessment of high-grade serous ovarian cancer. Clin Cancer Res. (2023) 29:5196–206. doi: 10.1158/1078-0432.CCR-23-1197
15. Marinelli LM, Kisiel JB, Slettedahl SW, Mahoney DW, Lemens MA, Shridhar V, et al. Methylated DNA markers for plasma detection of ovarian cancer: Discovery, validation, and clinical feasibility. Gynecol Oncol. (2022) 165:568–76. doi: 10.1016/j.ygyno.2022.03.018
16. Liang L, Zhang Y, Li C, Liao Y, Wang G, Xu J, et al. Plasma cfDNA methylation markers for the detection and prognosis of ovarian cancer. EBioMedicine. (2022) 83:104222. doi: 10.1016/j.ebiom.2022.104222
17. Bahado-Singh RO, Ibrahim A, Al-Wahab Z, Aydas B, Radhakrishna U, Yilmaz A, et al. Precision gynecologic oncology: circulating cell free DNA epigenomic analysis, artificial intelligence and the accurate detection of ovarian cancer. Sci Rep. (2022) 12:18625. doi: 10.1038/s41598-022-23149-1
18. Faaborg L, Fredslund Andersen R, Waldstrøm M, Høgdall E, Høgdall C, Adimi P, et al. Analysis of HOXA9 methylated ctDNA in ovarian cancer using sense-antisense measurement. Clin Chim Acta. (2021) 522:152–7. doi: 10.1016/j.cca.2021.08.020
19. Singh A, Gupta S, Badarukhiya JA, and Sachan M. Detection of aberrant methylation of HOXA9 and HIC1 through multiplex MethyLight assay in serum DNA for the early detection of epithelial ovarian cancer. Int J Cancer. (2020) 147:1740–52. doi: 10.1002/ijc.32984
20. Ogasawara A, Hihara T, Shintani D, Yabuno A, Ikeda Y, Tai K, et al. Evaluation of circulating tumor DNA in patients with ovarian cancer harboring somatic PIK3CA or KRAS mutations. Cancer Res Treat. (2020) 52:1219–28. doi: 10.4143/crt.2019.688
21. Miller BF, Pisanic Ii TR, Margolin G, Petrykowska HM, Athamanolap P, Goncearenco A, et al. Leveraging locus-specific epigenetic heterogeneity to improve the performance of blood-based DNA methylation biomarkers. Clin Epigenet. (2020) 12:154. doi: 10.1186/s13148-020-00939-w
22. Li S, Huang W, Li Y, Chen B, and Li D. A study of hTERT promoter methylation in circulating tumour DNAs of patients with ovarian magnificent tumour. Onco Targets Ther. (2020) 13:12317–23. doi: 10.2147/OTT.S274743
23. SK S, Swamy SN, Premalatha CS, Pallavi VR, and Gawari R. Aberrant promoter hypermethylation of RASSF1a and BRCA1 in circulating cell-free tumor DNA serves as a biomarker of ovarian carcinoma. Asian Pac J Cancer Prev. (2019) 20:3001–5. doi: 10.31557/APJCP.2019.20.10.3001
24. Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. (2019) 570:385–9. doi: 10.1038/s41586-019-1272-6
25. Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. (2018) 359:926–30. doi: 10.1126/science.aar3247
26. Widschwendter M, Zikan M, Wahl B, Lempiäinen H, Paprotka T, Evans I, et al. The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer. Genome Med. (2017) 9:116. doi: 10.1186/s13073-017-0500-7
27. Wang B, Yu L, Luo X, Huang L, Li Q-S, Shao X-S, et al. Detection of OPCML methylation, a possible epigenetic marker, from free serum circulating DNA to improve the diagnosis of early-stage ovarian epithelial cancer. Oncol Lett. (2017) 14:217–23. doi: 10.3892/ol.2017.6111
28. Vanderstichele A, Busschaert P, Smeets D, Landolfo C, Van Nieuwenhuysen E, Leunen K, et al. Chromosomal instability in cell-free DNA as a highly specific biomarker for detection of ovarian cancer in women with adnexal masses. Clin Cancer Res. (2017) 23:2223–31. doi: 10.1158/1078-0432.CCR-16-1078
29. Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. (2017) 9:eaan2415. doi: 10.1126/scitranslmed.aan2415
30. Parkinson CA, Gale D, Piskorz AM, Biggs H, Hodgkin C, Addley H, et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: A retrospective study. PLoS Med. (2016) 13:e1002198. doi: 10.1371/journal.pmed.1002198
31. Cohen PA, Flowers N, Tong S, Hannan N, Pertile MD, and Hui L. Abnormal plasma DNA profiles in early ovarian cancer using a non-invasive prenatal testing platform: implications for cancer screening. BMC Med. (2016) 14:126. doi: 10.1186/s12916-016-0667-6
32. Wang B, Yu L, Yang G-Z, Luo X, and Huang L. Application of multiplex nested methylated specific PCR in early diagnosis of epithelial ovarian cancer. Asian Pac J Cancer Prev. (2015) 16:3003–7. doi: 10.7314/apjcp.2015.16.7.3003
33. Shao X, He Y, Ji M, Chen X, Qi J, Shi W, et al. Quantitative analysis of cell-free DNA in ovarian cancer. Oncol Lett. (2015) 10:3478–82. doi: 10.3892/ol.2015.3771
34. Pereira E, Camacho-Vanegas O, Anand S, Sebra R, Catalina Camacho S, Garnar-Wortzel L, et al. Personalized circulating tumor DNA biomarkers dynamically predict treatment response and survival in gynecologic cancers. PLoS One. (2015) 10:e0145754. doi: 10.1371/journal.pone.0145754
35. Wu Y, Zhang X, Lin L, Ma X-P, Ma Y-C, and Liu P-S. Aberrant methylation of RASSF2A in tumors and plasma of patients with epithelial ovarian cancer. Asian Pac J Cancer Prev. (2014) 15:1171–6. doi: 10.7314/apjcp.2014.15.3.1171
36. Zhang Q, Hu G, Yang Q, Dong R, Xie X, Ma D, et al. A multiplex methylation-specific PCR assay for the detection of early-stage ovarian cancer using cell-free serum DNA. Gynecol Oncol. (2013) 130:132–9. doi: 10.1016/j.ygyno.2013.04.048
37. Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DWY, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. (2012) 4:136ra68. doi: 10.1126/scitranslmed.3003726
38. Wang T, Gao Y, Wang X, Tian J, Li Y, Yu B, et al. Establishment of an optimized CTC detection model consisting of EpCAM, MUC1 and WT1 in epithelial ovarian cancer and its correlation with clinical characteristics. Chin J Cancer Res. (2022) 34:95–108. doi: 10.21147/j.issn.1000-9604.2022.02.04
39. Ma J, Yang J, Jin Y, Cheng S, Huang S, Zhang N, et al. Artificial intelligence based on blood biomarkers including CTCs predicts outcomes in epithelial ovarian cancer: A prospective study. Onco Targets Ther. (2021) 14:3267–80. doi: 10.2147/OTT.S307546
40. Zhang X, Li H, Yu X, Li S, Lei Z, Li C, et al. Analysis of circulating tumor cells in ovarian cancer and their clinical value as a biomarker. Cell Physiol Biochem. (2018) 48:1983–94. doi: 10.1159/000492521
41. Rao Q, Zhang Q, Zheng C, Dai W, Zhang B, Ionescu-Zanetti C, et al. Detection of circulating tumour cells in patients with epithelial ovarian cancer by a microfluidic system. Int J Clin Exp Pathol. (2017) 10:9599–606.
42. Pearl ML, Zhao Q, Yang J, Dong H, Tulley S, Zhang Q, et al. Prognostic analysis of invasive circulating tumor cells (iCTCs) in epithelial ovarian cancer. Gynecol Oncol. (2014) 134:581–90. doi: 10.1016/j.ygyno.2014.06.013
43. Gahlawat AW, Witte T, Haarhuis L, and Schott S. A novel circulating miRNA panel for non-invasive ovarian cancer diagnosis and prognosis. Br J Cancer. (2022) 127:1550–6. doi: 10.1038/s41416-022-01925-0
44. Kumar V, Gupta S, Chaurasia A, and Sachan M. Evaluation of diagnostic potential of epigenetically deregulated miRNAs in epithelial ovarian cancer. Front Oncol. (2021) 11:681872. doi: 10.3389/fonc.2021.681872
45. Elias KM, Fendler W, Stawiski K, Fiascone SJ, Vitonis AF, Berkowitz RS, et al. Diagnostic potential for a serum miRNA neural network for detection of ovarian cancer. eLife. (2017) 6:e28932. doi: 10.7554/eLife.28932
46. Zheng H, Zhang L, Zhao Y, Yang D, Song F, Wen Y, et al. Plasma miRNAs as diagnostic and prognostic biomarkers for ovarian cancer. PLoS One. (2013) 8:e77853. doi: 10.1371/journal.pone.0077853
47. Kan CWS, Hahn MA, Gard GB, Maidens J, Huh JY, Marsh DJ, et al. Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer. BMC Cancer. (2012) 12:627. doi: 10.1186/1471-2407-12-627
48. Liu C-N and Zhang H-Y. Serum lncRNA LOXL1-AS1 is a diagnostic and prognostic marker for epithelial ovarian cancer. J Gene Med. (2020) 22:e3233. doi: 10.1002/jgm.3233
49. Li L, Zhang F, Zhang J, Shi X, Wu H, Chao X, et al. Identifying serum small extracellular vesicle microRNA as a noninvasive diagnostic and prognostic biomarker for ovarian cancer. ACS Nano. (2023) 17:19197–210. doi: 10.1021/acsnano.3c05694
50. Zhu Z, Chen Z, Wang M, Zhang M, Chen Y, Yang X, et al. Detection of plasma exosomal miRNA-205 as a biomarker for early diagnosis and an adjuvant indicator of ovarian cancer staging. J Ovarian Res. (2022) 15(1):27. doi: 10.1186/s13048-022-00961-x
51. Wang W, Jo H, Park S, Kim H, Kim SI, Han Y, et al. Integrated analysis of ascites and plasma extracellular vesicles identifies a miRNA-based diagnostic signature in ovarian cancer. Cancer Lett. (2022) 542:215735. doi: 10.1016/j.canlet.2022.215735
52. Su YY, Sun L, Guo ZR, Li JC, Bai TT, Cai XX, et al. Upregulated expression of serum exosomal miR-375 and miR-1307 enhance the diagnostic power of CA125 for ovarian cancer. J Ovarian Res. (2019) 12:6. doi: 10.1186/s13048-018-0477-x
53. Kim S, Choi MC, Jeong J-Y, Hwang S, Jung SG, Joo WD, et al. Serum exosomal miRNA-145 and miRNA-200c as promising biomarkers for preoperative diagnosis of ovarian carcinomas. J Cancer. (2019) 10:1958–67. doi: 10.7150/jca.30231
54. Yoshimura A, Sawada K, Nakamura K, Kinose Y, Nakatsuka E, Kobayashi M, et al. Exosomal miR-99a-5p is elevated in sera of ovarian cancer patients and promotes cancer cell invasion by increasing fibronectin and vitronectin expression in neighboring peritoneal mesothelial cells. BMC Cancer. (2018) 18:1065. doi: 10.1186/s12885-018-4974-5
55. Kobayashi M, Sawada K, Nakamura K, Yoshimura A, Miyamoto M, Shimizu A, et al. Exosomal miR-1290 is a potential biomarker of high-grade serous ovarian carcinoma and can discriminate patients from those with Malignancies of other histological types. J Ovarian Res. (2018) 11:81. doi: 10.1186/s13048-018-0458-0
56. Pan C, Stevic I, Müller V, Ni Q, Oliveira-Ferrer L, Pantel K, et al. Exosomal microRNAs as tumor markers in epithelial ovarian cancer. Mol Oncol. (2018) 12:1935–48. doi: 10.1002/1878-0261.12371
57. Meng X, Müller V, Milde-Langosch K, Trillsch F, Pantel K, and Schwarzenbach H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget. (2016) 7:16923–35. doi: 10.18632/oncotarget.7850
58. Lyu W, Cheng X, Yu Z, Dong R, Sheng Z, Zhang T, et al. Early-stage diagnosis of ovarian cancer via digital immunoassay on a SlipChip. Talanta. (2024) 280:126782. doi: 10.1016/j.talanta.2024.126782
59. Galan A, Papaluca A, Nejatie A, Matanes E, Brahimi F, Tong W, et al. GD2 and GD3 gangliosides as diagnostic biomarkers for all stages and subtypes of epithelial ovarian cancer. Front Oncol. (2023) 13:1134763. doi: 10.3389/fonc.2023.1134763
60. Gyllensten U, Hedlund-Lindberg J, Svensson J, Manninen J, Öst T, Ramsell J, et al. Next generation plasma proteomics identifies high-precision biomarker candidates for ovarian cancer. Cancers (Basel). (2022) 14:1757. doi: 10.3390/cancers14071757
61. Enroth S, Berggrund M, Lycke M, Broberg J, Lundberg M, Assarsson E, et al. High throughput proteomics identifies a high-accuracy 11 plasma protein biomarker signature for ovarian cancer. Commun Biol. (2019) 2:221. doi: 10.1038/s42003-019-0464-9
62. Jo A, Green A, Medina JE, Iyer S, Ohman AW, McCarthy ET, et al. Inaugurating high-throughput profiling of extracellular vesicles for earlier ovarian cancer detection. Adv Sci (Weinh). (2023) 10(27):e2301930. doi: 10.1002/advs.202301930
63. Peng P, Zhang W, Cao D, Yang J, and Shen K. The proteomic comparison of peripheral circulation-derived exosomes from the epithelial ovarian carcinoma (EOC) patients and non-EOC subjects. Trans Cancer Res. (2019) 8(2):452–65. doi: 10.21037/tcr.2019.03.06
64. Fahrmann JF, Ghasemi SM, Han CY, Wu R, Dennison JB, Vykoukal J, et al. A metabolite-based liquid biopsy for detection of ovarian cancer. Biomark Res. (2024) 12:91. doi: 10.1186/s40364-024-00629-2
65. Ban D, Housley SN, Matyunina LV, McDonald LD, Bae-Jump VL, Benigno BB, et al. A personalized probabilistic approach to ovarian cancer diagnostics. Gynecol Oncol. (2024) 182:168–75. doi: 10.1016/j.ygyno.2023.12.030
66. Irajizad E, Han CY, Celestino J, Wu R, Murage E, Spencer R, et al. A blood-based metabolite panel for distinguishing ovarian cancer from benign pelvic masses. Clin Cancer Res. (2022) 28:4669–76. doi: 10.1158/1078-0432.CCR-22-1113
67. Yang W, Mu T, Jiang J, Sun Q, Hou X, Sun Y, et al. Identification of potential biomarkers and metabolic profiling of serum in ovarian cancer patients using UPLC/Q-TOF MS. Cell Physiol Biochem. (2018) 51:1134–48. doi: 10.1159/000495492
68. Fan L, Yin M, Ke C, Ge T, Zhang G, Zhang W, et al. Use of plasma metabolomics to identify diagnostic biomarkers for early stage epithelial ovarian cancer. J Cancer. (2016) 7:1265–72. doi: 10.7150/jca.15074
69. Cheng Y, Li L, Zhu B, Liu F, Wang Y, Gu X, et al. Expanded metabolomics approach to profiling endogenous carbohydrates in the serum of ovarian cancer patients. J Sep Sci. (2016) 39:316–23. doi: 10.1002/jssc.201500964
70. Buas MF, Gu H, Djukovic D, Zhu J, Drescher CW, Urban N, et al. Identification of novel candidate plasma metabolite biomarkers for distinguishing serous ovarian carcinoma and benign serous ovarian tumors. Gynecol Oncol. (2016) 140:138–44. doi: 10.1016/j.ygyno.2015.10.021
71. Gaul DA, Mezencev R, Long TQ, Jones CM, Benigno BB, Gray A, et al. Highly-accurate metabolomic detection of early-stage ovarian cancer. Sci Rep. (2015) 5:16351. doi: 10.1038/srep16351
72. Ke C, Hou Y, Zhang H, Fan L, Ge T, Guo B, et al. Large-scale profiling of metabolic dysregulation in ovarian cancer. Int J Cancer. (2015) 136:516–26. doi: 10.1002/ijc.29010
73. Zhang T, Wu X, Yin M, Fan L, Zhang H, Zhao F, et al. Discrimination between Malignant and benign ovarian tumors by plasma metabolomic profiling using ultra performance liquid chromatography/mass spectrometry. Clin Chim Acta. (2012) 413:861–8. doi: 10.1016/j.cca.2012.01.026
74. Fan L, Zhang W, Yin M, Zhang T, Wu X, Zhang H, et al. Identification of metabolic biomarkers to diagnose epithelial ovarian cancer using a UPLC/QTOF/MS platform. Acta Oncol. (2012) 51:473–9. doi: 10.3109/0284186X.2011.648338
75. Chen J, Zhang X, Cao R, Lu X, Zhao S, Fekete A, et al. Serum 27-nor-5β-cholestane-3,7,12,24,25 pentol glucuronide discovered by metabolomics as potential diagnostic biomarker for epithelium ovarian cancer. J Proteome Res. (2011) 10:2625–32. doi: 10.1021/pr200173q
76. Garcia E, Andrews C, Hua J, Kim HL, Sukumaran DK, Szyperski T, et al. Diagnosis of early stage ovarian cancer by 1H NMR metabonomics of serum explored by use of a microflow NMR probe. J Proteome Res. (2011) 10:1765–71. doi: 10.1021/pr101050d
77. Gao Y, Liu C-J, Li H-Y, Xiong X-M, Li G-L, In ’t Veld SGJG, et al. Platelet RNA enables accurate detection of ovarian cancer: an intercontinental, biomarker identification study. Protein Cell. (2023) 14:579–90. doi: 10.1093/procel/pwac056
78. Pastuszak K, Supernat A, Best MG, In ’t Veld SGJG, Łapińska-Szumczyk S, Łojkowska A, et al. ImPlatelet classifier: image-converted RNA biomarker profiles enable blood-based cancer diagnostics. Mol Oncol. (2021) 15:2688–701. doi: 10.1002/1878-0261.13014
79. Lomnytska M, Pinto R, Becker S, Engström U, Gustafsson S, Björklund C, et al. Platelet protein biomarker panel for ovarian cancer diagnosis. Biomark Res. (2018) 6:2. doi: 10.1186/s40364-018-0118-y
80. Zachariah RR, Schmid S, Buerki N, Radpour R, Holzgreve W, and Zhong X. Levels of circulating cell-free nuclear and mitochondrial DNA in benign and Malignant ovarian tumors. Obstet Gynecol. (2008) 112:843–50. doi: 10.1097/AOG.0b013e3181867bc0
81. Alix-Panabières C and Pantel K. Liquid biopsy: from discovery to clinical application. Cancer Discov. (2021) 11:858–73. doi: 10.1158/2159-8290.CD-20-1311
82. Asante D-B, Calapre L, Ziman M, Meniawy TM, and Gray ES. Liquid biopsy in ovarian cancer using circulating tumor DNA and cells: Ready for prime time? Cancer Lett. (2020) 468:59–71. doi: 10.1016/j.canlet.2019.10.014
83. Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV, and CCGA Consortium. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. (2020) 31:745–59. doi: 10.1016/j.annonc.2020.02.011
84. Singh A, Gupta S, and Sachan M. Epigenetic biomarkers in the management of ovarian cancer: current prospectives. Front Cell Dev Biol. (2019) 7:182. doi: 10.3389/fcell.2019.00182
85. Barton CA, Hacker NF, Clark SJ, and O’Brien PM. DNA methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecol Oncol. (2008) 109:129–39. doi: 10.1016/j.ygyno.2007.12.017
86. Terp SK, Stoico MP, Dybkær K, and Pedersen IS. Early diagnosis of ovarian cancer based on methylation profiles in peripheral blood cell-free DNA: a systematic review. Clin Epigenet. (2023) 15:24. doi: 10.1186/s13148-023-01440-w
87. Liggett TE, Melnikov A, Yi Q, Replogle C, Hu W, Rotmensch J, et al. Distinctive DNA methylation patterns of cell-free plasma DNA in women with Malignant ovarian tumors. Gynecol Oncol. (2011) 120:113–20. doi: 10.1016/j.ygyno.2010.09.019
88. Lee J-S, Kim M, Seong M-W, Kim H-S, Lee YK, Kang HJ, et al. Serum in circulating tumor DNA measurement: characterization by DNA fragment sizing and digital droplet polymerase chain reaction. Clin Chem Lab Med. (2020) 58:527–32. doi: 10.1515/cclm-2019-0896
89. Thusgaard CF, Sadegh S, Jochumsen KM, Kruse TA, and Thomassen M. A sensitive and transparent method for tumor-informed detection of circulating tumor DNA in ovarian cancer using whole-genome sequencing. Int J Mol Sci. (2024) 25:13349. doi: 10.3390/ijms252413349
90. Yang J, Cheng S, Zhang N, Jin Y, and Wang Y. Liquid biopsy for ovarian cancer using circulating tumor cells: Recent advances on the path to precision medicine. Biochim Biophys Acta Rev Cancer. (2022) 1877:188660. doi: 10.1016/j.bbcan.2021.188660
91. Lianidou ES, Strati A, and Markou A. Circulating tumor cells as promising novel biomarkers in solid cancers. Crit Rev Clin Lab Sci. (2014) 51:160–71. doi: 10.3109/10408363.2014.896316
92. Fan T, Zhao Q, Chen JJ, Chen W-T, and Pearl ML. Clinical significance of circulating tumor cells detected by an invasion assay in peripheral blood of patients with ovarian cancer. Gynecol Oncol. (2009) 112:185–91. doi: 10.1016/j.ygyno.2008.09.021
93. Jou H-J, Ling P-Y, and Hsu H-T. Circulating tumor cells as a “real-time liquid biopsy”: Recent advances and the application in ovarian cancer. Taiwan J Obstet Gynecol. (2022) 61:34–9. doi: 10.1016/j.tjog.2021.11.008
94. Obermayr E, Maritschnegg E, Agreiter C, Pecha N, Speiser P, Helmy-Bader S, et al. Efficient leukocyte depletion by a novel microfluidic platform enables the molecular detection and characterization of circulating tumor cells. Oncotarget. (2018) 9:812–23. doi: 10.18632/oncotarget.22549
95. Pan Y, Wang Z, Ma J, Zhou T, Wu Z, Ding P, et al. Folic acid-modified fluorescent-magnetic nanoparticles for efficient isolation and identification of circulating tumor cells in ovarian cancer. Biosens (Basel). (2022) 12:184. doi: 10.3390/bios12030184
96. Han L, Wan Q, Zheng A, Guo Y, and Chen Y. Demonstration of a flexible graphene-based biosensor for sensitive and rapid detection of ovarian cancer cells. Nanoscale Res Lett. (2021) 16:181. doi: 10.1186/s11671-021-03633-9
97. Dwivedi SKD, Rao G, Dey A, Mukherjee P, Wren JD, and Bhattacharya R. Small non-coding-RNA in gynecological Malignancies. Cancers (Basel). (2021) 13:1085. doi: 10.3390/cancers13051085
98. Zhang H, Xu S, and Liu X. MicroRNA profiling of plasma exosomes from patients with ovarian cancer using high-throughput sequencing. Oncol Lett. (2019) 17:5601–7. doi: 10.3892/ol.2019.10220
99. Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, and Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. (2009) 112:55–9. doi: 10.1016/j.ygyno.2008.08.036
100. Langhe R, Norris L, Saadeh FA, Blackshields G, Varley R, Harrison A, et al. A novel serum microRNA panel to discriminate benign from Malignant ovarian disease. Cancer Lett. (2015) 356:628–36. doi: 10.1016/j.canlet.2014.10.010
101. Hulstaert E, Morlion A, Levanon K, Vandesompele J, and Mestdagh P. Candidate RNA biomarkers in biofluids for early diagnosis of ovarian cancer: A systematic review. Gynecol Oncol. (2021) 160:633–42. doi: 10.1016/j.ygyno.2020.11.018
102. Gahlawat AW, Witte T, Sinn P, and Schott S. Circulating cf-miRNA as a more appropriate surrogate liquid biopsy marker than cfDNA for ovarian cancer. Sci Rep. (2023) 13:5503. doi: 10.1038/s41598-023-32243-x
103. Liu Y-J and Wang C. A review of the regulatory mechanisms of extracellular vesicles-mediated intercellular communication. Cell Commun Signal. (2023) 21:1–12. doi: 10.1186/s12964-023-01103-6
104. Kalluri R and McAndrews KM. The role of extracellular vesicles in cancer. Cell. (2023) 186:1610–26. doi: 10.1016/j.cell.2023.03.010
105. Shiao M-S, Chang J-M, Lertkhachonsuk A-A, Rermluk N, and Jinawath N. Circulating exosomal miRNAs as biomarkers in epithelial ovarian cancer. Biomedicines. (2021) 9:1433. doi: 10.3390/biomedicines9101433
106. Mori MA, Ludwig RG, Garcia-Martin R, Brandão BB, and Kahn CR. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. (2019) 30:656–73. doi: 10.1016/j.cmet.2019.07.011
107. Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. (2008) 14:2690–5. doi: 10.1158/1078-0432.CCR-07-1731
108. Li Y, Yao L, Liu F, Hong J, Chen L, Zhang B, et al. Characterization of microRNA expression in serous ovarian carcinoma. Int J Mol Med. (2014) 34:491–8. doi: 10.3892/ijmm.2014.1813
109. Sol N and Wurdinger T. Platelet RNA signatures for the detection of cancer. Cancer Metastasis Rev. (2017) 36:263–72. doi: 10.1007/s10555-017-9674-0
110. Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, et al. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. (2015) 28:666–76. doi: 10.1016/j.ccell.2015.09.018
111. Piek JMJ, In't Veld SGJG, Best MG, Tannous B, Supernat A, and Lok CAR. EP457 Assessment of ovarian tumors with tumor educated platelets (TEPs) | Request PDF. ResearchGate. (2019) 29:A291–2. doi: 10.1136/ijgc-2019-ESGO.516
112. Pan S and Chen R. Pathological implication of protein post-translational modifications in cancer. Mol Aspects Med. (2022) 86:101097. doi: 10.1016/j.mam.2022.101097
113. Coleman RL, Herzog TJ, Chan DW, Munroe DG, Pappas TC, Smith A, et al. Validation of a second-generation multivariate index assay for Malignancy risk of adnexal masses. Am J Obstet Gynecol. (2016) 215:82.e1–82.e11. doi: 10.1016/j.ajog.2016.03.003
114. Ueland FR, Desimone CP, Seamon LG, Miller RA, Goodrich S, Podzielinski I, et al. Effectiveness of a multivariate index assay in the preoperative assessment of ovarian tumors. Obstetr Gynecol. (2011) 117(6):1289–97. doi: 10.1097/AOG.0b013e31821b5118
115. Zhang Z and Chan DW. The road from discovery to clinical diagnostics: lessons learned from the first FDA-cleared in vitro diagnostic multivariate index assay of proteomic biomarkers. Cancer Epidemiol Biomarkers Prev. (2010) 19:2995–9. doi: 10.1158/1055-9965.EPI-10-0580
116. Moore RG, Jabre-Raughley M, Brown AK, Robison KM, Miller MC, Allard WJ, et al. Comparison of a novel multiple marker assay vs the Risk of Malignancy Index for the prediction of epithelial ovarian cancer in patients with a pelvic mass. Am J Obstet Gynecol. (2010) 203:228.e1–6. doi: 10.1016/j.ajog.2010.03.043
117. Lee J, Park JE, Lee D, Seo N, and An HJ. Advancements in protein glycosylation biomarkers for ovarian cancer through mass spectrometry-based approaches. Expert Rev Mol Diagn. (2023) 24(4):249–58. doi: 10.1080/14737159.2023.2297933
118. Li J, Wang Z, Liu W, Tan L, Yu Y, Liu D, et al. Identification of metabolic biomarkers for diagnosis of epithelial ovarian cancer using internal extraction electrospray ionization mass spectrometry (iEESI-MS). Cancer Biomark. (2023) 37:67–84. doi: 10.3233/CBM-220250
119. Nunes SC, Sousa J, Silva F, Silveira M, Guimarães A, Serpa J, et al. Peripheral blood serum NMR metabolomics is a powerful tool to discriminate benign and Malignant ovarian tumors. . Metabol. (2023) 13(9):989. doi: 10.3390/metabo13090989
120. Ahn H-S, Yeom J, Yu J, Kwon Y-I, Kim J-H, and Kim K. Convergence of plasma metabolomics and proteomics analysis to discover signatures of high-grade serous ovarian cancer. Cancers (Basel). (2020) 12:3447. doi: 10.3390/cancers12113447
121. Good DM, Thongboonkerd V, Novak J, Bascands J-L, Schanstra JP, Coon JJ, et al. Body fluid proteomics for biomarker discovery: lessons from the past hold the key to success in the future. J Proteome Res. (2007) 6:4549–55. doi: 10.1021/pr070529w
122. Davis MT, Auger PL, and Patterson SD. Cancer biomarker discovery via low molecular weight serum profiling–are we following circular paths? Clin Chem. (2010) 56:244–7. doi: 10.1373/clinchem.2009.127951
123. Valle BL, Rodriguez-Torres S, Kuhn E, Díaz-Montes T, Parrilla-Castellar E, Lawson FP, et al. HIST1H2BB and MAGI2 methylation and somatic mutations as precision medicine biomarkers for diagnosis and prognosis of high-grade serous ovarian cancer. Cancer Prev Res (Phila). (2020) 13:783–94. doi: 10.1158/1940-6207.CAPR-19-0412
124. Zhou J, Gong G, Tan H, Dai F, Zhu X, Chen Y, et al. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol Rep. (2015) 33:2915–23. doi: 10.3892/or.2015.3937
125. Záveský L, Jandáková E, Turyna R, Langmeierová L, Weinberger V, Záveská Drábková L, et al. Evaluation of cell-free urine microRNAs expression for the use in diagnosis of ovarian and endometrial cancers. A pilot study. Pathol Oncol Res. (2015) 21:1027–35. doi: 10.1007/s12253-015-9914-y
126. Berner K, Hirschfeld M, Weiß D, Rücker G, Asberger J, Ritter A, et al. Evaluation of circulating microRNAs as non-invasive biomarkers in the diagnosis of ovarian cancer: a case-control study. Arch Gynecol Obstet. (2022) 306:151–63. doi: 10.1007/s00404-021-06287-1
127. Martinicky D, Zvarik M, Sikurova L, Lajdova I, and Hunakova L. Fluorescence analysis of urine and its potential for ovarian cancer screening. Neoplasma. (2015) 62:500–6. doi: 10.4149/neo_2015_060
128. Woolery KT, Hoffman MS, Kraft J, Nicosia SV, Kumar A, and Kruk PA. Urinary interleukin-1β levels among gynecological patients. J Ovarian Res. (2014) 7:104. doi: 10.1186/s13048-014-0104-4
129. Smith CR, Batruch I, Bauça JM, Kosanam H, Ridley J, Bernardini MQ, et al. Deciphering the peptidome of urine from ovarian cancer patients and healthy controls. Clin Proteomics. (2014) 11:23. doi: 10.1186/1559-0275-11-23
130. Stockley J, Akhand R, Kennedy A, Nyberg C, Crosbie EJ, and Edmondson RJ. Detection of MCM5 as a novel non-invasive aid for the diagnosis of endometrial and ovarian tumours. BMC Cancer. (2020) 20:1000. doi: 10.1186/s12885-020-07468-y
131. Murgan SS, Abd Elaziz FJ, Nasr AMA, Elfaki MEE, and Khalil EAG. Ovarian cancer: tumor-specific urinary micro-peptides profiling as potential biomarkers for early diagnosis. Proteomes. (2020) 8:32. doi: 10.3390/proteomes8040032
132. Niemi RJ, Roine AN, Häkkinen MR, Kumpulainen PS, Keinänen TA, Vepsäläinen JJ, et al. Urinary polyamines as biomarkers for ovarian cancer. Int J Gynecol Cancer. (2017) 27:1360–6. doi: 10.1097/IGC.0000000000001031
133. Liu X, Liu G, Chen L, Liu F, Zhang X, Liu D, et al. Untargeted metabolomic characterization of ovarian tumors. Cancers (Basel). (2020) 12:3642. doi: 10.3390/cancers12123642
134. Gasparri ML, Casorelli A, Bardhi E, Besharat AR, Savone D, Ruscito I, et al. Beyond circulating microRNA biomarkers: Urinary microRNAs in ovarian and breast cancer. Tumour Biol. (2017) 39:1010428317695525. doi: 10.1177/1010428317695525
135. Záveský L, Jandáková E, Weinberger V, Minář L, Hanzíková V, Dušková D, et al. Ovarian cancer: differentially expressed microRNAs in tumor tissue and cell-free ascitic fluid as potential novel biomarkers. Cancer Invest. (2019) 37:440–52. doi: 10.1080/07357907.2019.1663208
136. Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. (2002) 359:572–7. doi: 10.1016/S0140-6736(02)07746-2
138. Li J, Zhang Z, Rosenzweig J, Wang YY, and Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. (2002) 48:1296–304. doi: 10.1093/clinchem/48.8.1296
139. Wei J, Ni N, Meng W, Huan Y, and Gao Y. Early urinary protein changes during tumor formation in a NuTu-19 tail vein injection rat model. Sci Rep. (2020) 10:11709. doi: 10.1038/s41598-020-68674-z
140. Slupsky CM, Steed H, Wells TH, Dabbs K, Schepansky A, Capstick V, et al. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin Cancer Res. (2010) 16:5835–41. doi: 10.1158/1078-0432.CCR-10-1434
141. Zvarik M, Martinicky D, Hunakova L, Lajdova I, and Sikurova L. Fluorescence characteristics of human urine from normal individuals and ovarian cancer patients. Neoplasma. (2013) 60:533–7. doi: 10.4149/neo_2013_069
142. Birková A, Valko-Rokytovská M, Hubková B, Zábavníková M, and Mareková M. Strong dependence between tryptophan-related fluorescence of urine and Malignant melanoma. Int J Mol Sci. (2021) 22:1884. doi: 10.3390/ijms22041884
143. Švecová M, Dubayová K, Birková A, Urdzík P, and Mareková M. Non-invasive endometrial cancer screening through urinary fluorescent metabolome profile monitoring and machine learning algorithms. Cancers (Basel). (2024) 16:3155. doi: 10.3390/cancers16183155
144. Erickson BK, Kinde I, Dobbin ZC, Wang Y, Martin JY, Alvarez RD, et al. Detection of somatic TP53 mutations in tampons of patients with high-grade serous ovarian cancer. Obstetr Gynecol. (2014) 124:881–5. doi: 10.1097/AOG.0000000000000484
145. Kinde I, Bettegowda C, Wang Y, Wu J, Agrawal N, Shih I-M, et al. Evaluation of DNA from the papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med. (2013) 5(167):167ra4. doi: 10.1126/scitranslmed.3004952
146. Paracchini L, Pesenti C, Delle Marchette M, Beltrame L, Bianchi T, Grassi T, et al. Detection of TP53 clonal variants in papanicolaou test samples collected up to 6 years prior to high-grade serous epithelial ovarian cancer diagnosis. JAMA Netw Open. (2020) 3:e207566. doi: 10.1001/jamanetworkopen.2020.7566
147. Arildsen NS, Martin de la Fuente L, Måsbäck A, Malander S, Forslund O, Kannisto P, et al. Detecting TP53 mutations in diagnostic and archival liquid-based Pap samples from ovarian cancer patients using an ultra-sensitive ddPCR method. Sci Rep. (2019) 9:15506. doi: 10.1038/s41598-019-51697-6
148. Wang Y, Li L, Douville C, Cohen JD, Yen T-T, Kinde I, et al. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci Transl Med. (2018) 10:eaap8793. doi: 10.1126/scitranslmed.aap8793
149. Krimmel-Morrison JD, Ghezelayagh TS, Lian S, Zhang Y, Fredrickson J, Nachmanson D, et al. Characterization of TP53 mutations in Pap test DNA of women with and without serous ovarian carcinoma. Gynecol Oncol. (2020) 156:407–14. doi: 10.1016/j.ygyno.2019.11.124
150. Jiang X, Li W, Yang J, Wang S, Cao D, Yu M, et al. Identification of somatic mutations in papanicolaou smear DNA and plasma circulating cell-free DNA for detection of endometrial and epithelial ovarian cancers: A pilot study. Front Oncol. (2020) 10:582546. doi: 10.3389/fonc.2020.582546
151. Chang CC, Wang HC, Liao YP, Chen YC, Weng YC, Yu MH, et al. The feasibility of detecting endometrial and ovarian cancer using DNA methylation biomarkers in cervical scrapings. J Gynecol Oncol. (2018) 29:e17. doi: 10.3802/jgo.2018.29.e17
152. Wu T-I, Huang R-L, Su P-H, Mao S-P, Wu C-H, and Lai H-C. Ovarian cancer detection by DNA methylation in cervical scrapings. Clin Epigenet. (2019) 11:166. doi: 10.1186/s13148-019-0773-3
153. Pesenti C, Beltrame L, Velle A, Fruscio R, Jaconi M, Borella F, et al. Copy number alterations in stage I epithelial ovarian cancer highlight three genomic patterns associated with prognosis. Eur J Cancer. (2022) 171:85–95. doi: 10.1016/j.ejca.2022.05.005
154. Killcoyne S, Yusuf A, and Fitzgerald RC. Genomic instability signals offer diagnostic possibility in early cancer detection. Trends Genet. (2021) 37:966–72. doi: 10.1016/j.tig.2021.06.009
155. Paracchini L, Mannarino L, Romualdi C, Zadro R, Beltrame L, Nerini IF, et al. Genomic instability analysis in DNA from Papanicolaou test provides proof-of-principle early diagnosis of high-grade serous ovarian cancer. Sci Trans Med. (2023) 15(725):eadi2556. doi: 10.1126/scitranslmed.adi2556
156. Boylan KLM, Afiuni-Zadeh S, Geller MA, Argenta PA, Griffin TJ, and Skubitz APN. Evaluation of the potential of Pap test fluid and cervical swabs to serve as clinical diagnostic biospecimens for the detection of ovarian cancer by mass spectrometry-based proteomics. Clin Proteomics. (2021) 18:4. doi: 10.1186/s12014-020-09309-3
157. Rocconi RP, Wilhite AM, Schambeau L, Scalici J, Pannell L, and Finan MA. A novel proteomic-based screening method for ovarian cancer using cervicovaginal fluids: A window into the abdomen. Gynecol Oncol. (2022) 164:181–6. doi: 10.1016/j.ygyno.2021.10.083
158. Hedlund Lindberg J, Widgren A, Ivansson E, Gustavsson I, Stålberg K, Gyllensten U, et al. Toward ovarian cancer screening with protein biomarkers using dried, self-sampled cervico-vaginal fluid. iScience. (2024) 27:109001. doi: 10.1016/j.isci.2024.109001
159. Aarnio R, Östensson E, Olovsson M, Gustavsson I, and Gyllensten U. Cost-effectiveness analysis of repeated self-sampling for HPV testing in primary cervical screening: a randomized study. BMC Cancer. (2020) 20:645. doi: 10.1186/s12885-020-07085-9
160. Gustavsson I, Aarnio R, Berggrund M, Hedlund-Lindberg J, Strand A-S, Sanner K, et al. Randomised study shows that repeated self-sampling and HPV test has more than two-fold higher detection rate of women with CIN2+ histology than Pap smear cytology. Br J Cancer. (2018) 118:896–904. doi: 10.1038/bjc.2017.485
161. Otsuka I, Kameda S, and Hoshi K. Early detection of ovarian and fallopian tube cancer by examination of cytological samples from the endometrial cavity. Br J Cancer. (2013) 109:603–9. doi: 10.1038/bjc.2013.402
162. Maritschnegg E, Wang Y, Pecha N, Horvat R, Van Nieuwenhuysen E, Vergote I, et al. Lavage of the uterine cavity for molecular detection of müllerian duct carcinomas: A proof-of-concept study. J Clin Oncol. (2015) 33:4293–300. doi: 10.1200/JCO.2015.61.3083
163. Salk JJ, Loubet-Senear K, Maritschnegg E, Valentine CC, Williams LN, Higgins JE, et al. Ultra-sensitive TP53 sequencing for cancer detection reveals progressive clonal selection in normal tissue over a century of human lifespan. Cell Rep. (2019) 28:132–144.e3. doi: 10.1016/j.celrep.2019.05.109
164. Ghezelayagh TS, Kohrn BF, Fredrickson J, Manhardt E, Radke MR, Katz R, et al. Uterine lavage identifies cancer mutations and increased TP53 somatic mutation burden in individuals with ovarian cancer. Cancer Res Commun. (2022) 2:1282–92. doi: 10.1158/2767-9764.crc-22-0314
165. Žilovič D, Vaicekauskaitė I, Čiurlienė R, Sabaliauskaitė R, and Jarmalaitė S. Uterine cavity lavage mutation analysis in Lithuanian ovarian cancer patients. Cancers (Basel). (2023) 15:868. doi: 10.3390/cancers15030868
166. Hulstaert E, Levanon K, Morlion A, Van Aelst S, Christidis A-A, Zamar R, et al. RNA biomarkers from proximal liquid biopsy for diagnosis of ovarian cancer. Neoplasia. (2022) 24:155–64. doi: 10.1016/j.neo.2021.12.008
167. Skryabin GO, Komelkov AV, Zhordania KI, Bagrov DV, Vinokurova SV, Galetsky SA, et al. Extracellular vesicles from uterine aspirates represent a promising source for screening markers of gynecologic cancers. Cells. (2022) 11:1064. doi: 10.3390/cells11071064
168. Barnabas GD, Bahar-Shany K, Sapoznik S, Helpman L, Kadan Y, Beiner M, et al. Microvesicle proteomic profiling of uterine liquid biopsy for ovarian cancer early detection. Mol Cell Proteomics. (2019) 18:865–75. doi: 10.1074/mcp.RA119.001362
169. Bahar-Shany K, Barnabas GD, Deutsch L, Deutsch N, Glick-Saar E, Dominissini D, et al. Proteomic signature for detection of high-grade ovarian cancer in germline BRCA mutation carriers. Int J Cancer. (2023) 152:781–93. doi: 10.1002/ijc.34318
170. Wang P, Ma J, Li W, Wang Q, Xiao Y, Jiang Y, et al. Profiling the metabolome of uterine fluid for early detection of ovarian cancer. Cell Rep Med. (2023) 4:101061. doi: 10.1016/j.xcrm.2023.101061
171. Maritschnegg E, Heitz F, Pecha N, Bouda J, Trillsch F, Grimm C, et al. Uterine and tubal lavage for earlier cancer detection using an innovative catheter: A feasibility and safety study. Int J Gynecol Cancer. (2018) 28:1692–8. doi: 10.1097/IGC.0000000000001361
172. Labidi-Galy SI, Papp E, Hallberg D, Niknafs N, Adleff V, Noe M, et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat Commun. (2017) 8:1093. doi: 10.1038/s41467-017-00962-1
Keywords: ovarian cancer, early detection, liquid biopsy, biomarkers, multi-omics
Citation: Feng Y, Yang W, Zhu J, Wang S, Wu N, Zhao H and Yang X (2025) Clinical utility of various liquid biopsy samples for the early detection of ovarian cancer: a comprehensive review. Front. Oncol. 15:1594100. doi: 10.3389/fonc.2025.1594100
Received: 15 March 2025; Accepted: 10 June 2025;
Published: 01 July 2025.
Edited by:
Carolina Reduzzi, NewYork-Presbyterian, United StatesReviewed by:
Vasudha Mishra, The University of Chicago, United StatesBarbara Frossi, University of Udine, Italy
Tasaduq H. Wani, University of Oxford, United Kingdom
Copyright © 2025 Feng, Yang, Zhu, Wang, Wu, Zhao and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofeng Yang, eXhmNzNAMTYzLmNvbQ==
 Yueqi Feng
Yueqi Feng Wanjun Yang
Wanjun Yang Jingyu Zhu
Jingyu Zhu Shirui Wang
Shirui Wang Ningjuan Wu
Ningjuan Wu Hanzhi Zhao
Hanzhi Zhao Xiaofeng Yang
Xiaofeng Yang