- 1Wuhan Kindstar Zhenyuan Medical Laboratory Co., Ltd., Wuhan, China
- 2Chest Hospital, Tianjin University, Tianjin, China
- 3Department of Thoracic Surgery, Tianjin Chest Hospital, Tianjin, China
- 4Department of Scientific Research, Kindstar Global Precision Medicine Institute, Wuhan, China
Lung cancer is the most prevalent and deadly malignant tumor in the world. Traditional treatment methods rely on histopathological analysis of cancer cells obtained through tissue biopsies, which carry risks due to their invasive nature. Thus, there is an urgent need to identify effective and non-invasive early screening methods for lung cancer. Exosomes, a crucial element of liquid biopsies, have emerged as a promising alternative due to their non-invasive collection, convenience and cost-effectiveness in diagnosing lung cancer. Research has underscored the role of exosomes in lung cancer invasion, metastasis, immune regulation, and the tumor microenvironment. Furthermore, the contents of exosomes, such as miRNAs, lncRNAs, circRNAs, and proteins, demonstrate considerable potential for the early diagnosis of lung cancer. This article provides a comprehensive review of the role and application of exosomes as liquid biopsy markers for early diagnosis of lung cancer, emphasizing their promise in improving patient outcomes through earlier detection and intervention.
1 Background
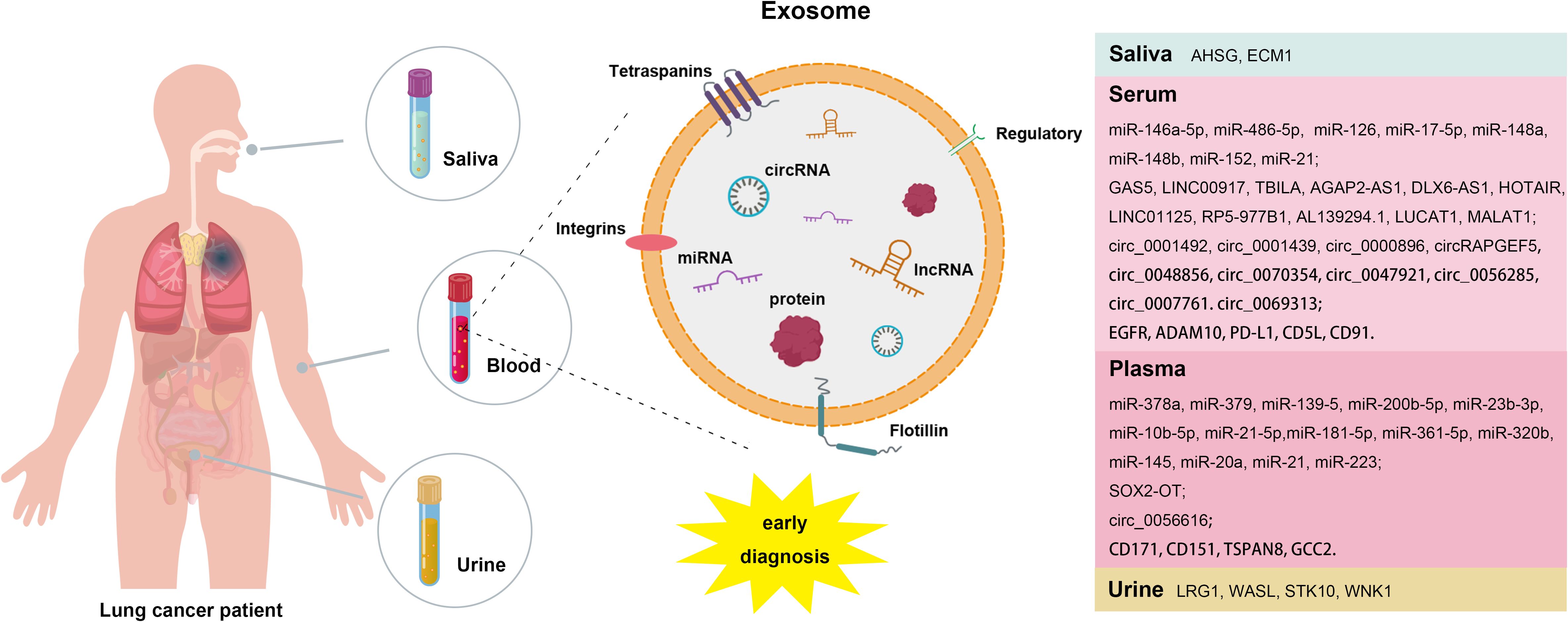
Lung cancer is a malignant tumor with a rapidly increasing incidence and mortality rare worldwide, representing a significant threat to human health. It encompasses various subtypes, including small cell lung cancer (SCLC), lung adenocarcinoma (LUAD), Lung squamous cell carcinoma (LUSC), and large-cell carcinoma (LCC) (1, 2). Currently, surgical resection remains the most effective treatment for early-stage lung cancer, however, the early diagnosis rate is only 15%, with fewer than 30% of patients meeting clear surgical criteria (3–6). Early screening is crucial for reducing mortality from non-small cell lung cancer (NSCLC) (7). Conventional clinical tests, such as magnetic resonance imaging (MRI) and computed tomography (CT), are often costly and can lead to false positives. While tumor markers can provide additional risk assessment for clinical decision making (8, 9), pathological biopsy, considered the gold standard for lung cancer diagnosis, is not suitable for early screening and follow-up due to its invasive nature. Liquid biopsy has emerged as a promising diagnostic technique characterized by its non-invasive approach, high specificity, sensitivity and early detection capabilities, with exosomes gaining attention as potential biomarkers for this method (10, 11). Exosomes are small lipid bilayer vesicles that contain a wealth of nucleic acids, proteins, and lipids, thereby facilitating liquid biopsy (12). Their stability is a key advantage, as the lipid bilayer structure protects their contents from degradation. Moreover, exosomes carry a complete set of genetic information from their parental cells, making them effective biomarkers that accurately reflect the physiological state of those cells. Notably, exosomes derived from tumor cells are enriched with tumor-specific DNA, RNA, and proteins (13–16). Recent studies have underscored the strong connection between exosomes and lung cancer, influencing various aspects of tumor progression, including proliferation, metastasis, drug resistance and angiogenesis. Consequently, exosomes are positioned as ideal biomarkers for the early diagnosis of lung cancer (17–21). This article provides an overview of research findings on exosomal miRNAs, lncRNAs, circRNAs and proteins as biomarkers, offering new insights for the early diagnosis and treatment of lung cancer (Figure 1).
2 Overview of exosomes
2.1 Biological characteristics of exosomes
The study of exosomes began in 1987, when Johnstone et al. (22) first observed that reticulocytes release vesicles into the extracellular space, coining the term “exosomes” based on their morphology and size. Exosomes are small vesicles with a diameter of 30-100nm and have a disc-like structure characterized by a bilayer lipid membrane (23). They are enriched proteins related to multivesicular body (MVB), including Flotillins, Annexins, GTPases, RABs, SNAREs, ALIX, TSG101, heat shock proteins (HSP70, HSP90) and transmembrane protein families such as CD9, CD63, CD81 and CD82 (23). These proteins play crucial roles in various biological processes of exosomes, including biogenesis, antigen presentation, membrane transport, and fusion (24, 25). In addition to proteins, exosomes contain a diverse array of nucleic acids, such as mRNAs, miRNAs, lncRNAs, circRNAs and snoRNAs (26–28). They also encompass various lipid components, including cholesterol (CHOL), sphingomyelin (SM), phosphatidylcholine (PC), phosphatidylserine (PS), phosphatidylethanolamine (PE), diacylglycerol (DAG), phosphatidic acid (PA), and phosphatidylinositol (PI) (29). These components contribute to the functional characteristics of exosomes, mirroring those of the parental cells and highlighting their potential as biomarkers (30–32).
2.2 Biogenesis of exosomes
Currently, the biogenesis of exosomes is primarily understood to occur through two pathways: the plasma membrane and endosome. The endosomal pathway, which has been extensively studies, involves the formation of early endosomes from the plasma membrane, the generation of MVBs, and their subsequent fusion with the plasma membrane to release exosomes (22). Additionally, exosomes can bud directly from the plasma membrane, a process referred to as the plasma membrane pathway. MVB formation involves two main mechanisms: ESCRT-dependent and ESCRT-independent mechanisms. The endosomal sorting complex required for transport (ESCRT) comprises four protein complexes, along with accessory proteins (ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III) (32). ESCRT plays a crucial role in MVB production by facilitating cargo aggregation, membrane invagination, and vesicle neck division (33). For instance, CD63 is directly involved in the ESCRT-independent sorting of premelanosome protein (PMEL) in human melanoma cells. In HEK293, CD82 and CD9 promote the excretion of exosomes enriched with β-catenin (34). Similarly, the tetraspanin Tspan8 is responsible for recruiting specific proteins and mRNAs to exosomes in pancreatic cancer (35). Notably, inhibiting these pathway dose not eliminate exosome production (36), suggesting that cells can utilize multiple pathways for MVB formation. Furthermore, the same MVB may arise from intracellular vesicles formed through different mechanisms.
The rate of exosome synthesis and secretion varies significantly among different cell types (37). For example, tumor cells exhibit increased exosome secretion during radiotherapy (38), leading to higher levels of exosomes in the blood of cancer patients compared to healthy individuals (39). Although the precise mechanisms governing exosome function are not fully understood, substantial research has explored their roles. Initially, exosomes were considered as cellular “garbage bag” responsible for exporting excess or non-functional components from cells (40). However, recent studies have revealed that exosomes originate from diverse sources and possess distinct biological functions. They play important roles in various physiological processes, including immune surveillance, neuroshaping, tissue repair, stem cell survival, and coagulation (41). Moreover, exosomes hold an irreplaceable role in pathological conditions, particularly in tumors development (30).
2.3 Extraction of exosomes
High purity of exosomes is essential for effective exosome research. Various extraction methods have been developed based on the characteristics of exosomes, such as size, density, and surface markers (42). Among these, ultracentrifugation is the most commonly used technique. This method first eliminates cell debris through low- and medium-speed centrifugation, followed by ultracentrifugation to separate and concentrate the exosomes. It is considered the gold standard due to its low cost, minimal risk of contamination, and high yield (43, 44). However, it requires a large sample volume and often results in lower purity of the isolated exosomes. Density gradient centrifugation is another method that utilizes biocompatible media, such as sucrose, to separate particles based on their different densities (45, 46). This approach can achieve higher purity of exosomes but is not suitable for small-volume samples. The immunocapture method (47) involves isolating and enriching exosomes by capturing their surface markers with specific antibodies. While this technique does not compromise the morphology of exosomes, it comes with high reagent costs and a reliance on the availability of specific antibodies.
In addition to these traditional methods, microfluidic technology has emerged as a promising approach for efficient exosome isolation from biological fluids. For instance, the automated centrifugal microfluidic disk system combined with functionalized membranes (Exo-CMDS) achieved a diagnostic accuracy of 91% for lung cancer detection in trace blood samples (15). Although microfluidic technology is user-friendly and offers a high capture rate, it typically requires integration with other instruments, and relying on a single separation method may impact both the purity and recovery of exosomes. Other extraction techniques, such as size exclusion chromatography (48), ultrafiltration (49), and polymer precipitation (50), also exist, each with its own advantages and disadvantages. No single method has yet proven capable of efficiently extracting exosomes from all types of samples. In summary, the extraction of exosomes continues to face significant challenges and requires further research and optimization to enhance its efficiency and purity.
3 Role of exosomes in lung cancer development
Immunosuppression: Tumor-derived exosomes actively interact with immune cells, transmitting inhibitory signals that result in a reduced number of antigen-presenting cells (APCs) and the suppression of T cell and natural killer (NK) cell activity. This interaction allows cancer cells to evade immune surveillance, induces immune tolerance, and further facilitates the growth, metastasis and invasion of tumor cells (51–57).
Proliferation: Platelet exosomes isolated from lung cancer patients have been observed to transfer CD41 to the surface of lung cancer cells. This transfer induces the expression of cyclin D2 in these cells, leading to the upregulation of MAPKp42/44 phosphorylation, which subsequently promotes lung cancer cell proliferation (58). Additionally, KLF9, a crucial factor for cell proliferation, differentiation, and tissue development, is targeted by plasma exosomal miR-660-5p, facilitating the progression of NSCLC (59). Furthermore, exosomal miR-96 has been shown to enhance the proliferation of LUAD H1299 cells by targeting LMO7 expression (60). Exosomal miR-29a and miR-21, derived from A549 cells, can bind to toll-like receptors (TLRs) on immune cells within the tumor microenvironment, thereby influencing lung cancer cell proliferation (61).
Metastasis: Metastasis is a complex process that involves the invasion, survival, attachment, and colonization of tumor cells in distant organs. Exosomes serve as key mediators of intercellular communication and play a crucial role in various stages of metastasis. Firstly, exosomes promote epithelial-mesenchymal transition (EMT), a phenomenon strongly linked to lung cancer metastasis. Specific exosomal miRNAs such as miR-193a-3p, miR-210-3p and miR-5100, active STAT3 signaling to induce EMT, thereby facilitating invasion and metastasis. Additionally, miR-23a regulates E-cadherin expression, maintaining EMT through the TGF-β pathway (62, 63). Secondly, exosomes stimulate angiogenesis by acting as messengers between tumor cells and vascular endothelial cells. For instance, overexpression of miR-210 in exosomes derived lung cancer patients activates the JAK2/STAT3 pathway, resulting in increased expression of pro-angiogenic factors such as MMP9, FGF2, and vascular endothelial growth factor (VEGF) (64). Abnormal expression of miRNA-497, miR-21 and miR-549a in exosomes from lung cancer cells has also been associated with angiogenesis (65–68). Lastly, exosomal RNA from mouse lung cancer cells has been shown to upregulate the expression of toll-like receptor 3 (TLR3) in type II alveolar epithelial cells through the NF-κB and MAPK pathways. This upregulation promotes the production of chemokines and neutrophil aggregation, ultimately contributing to the formation of a premetastatic niche (69).
4 Exosomes as biomarker for early diagnosis of lung cancer
4.1 Exosomal miRNAs
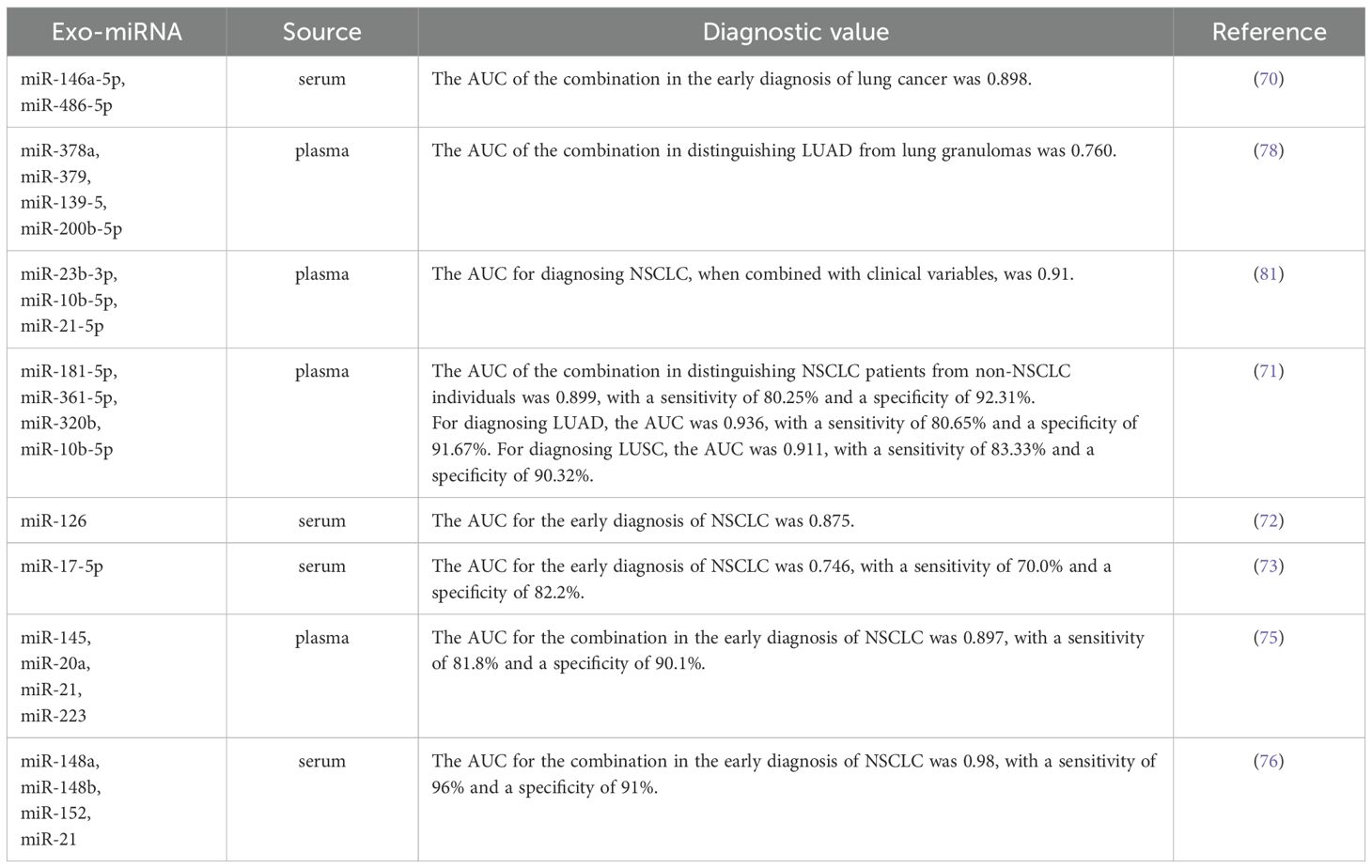
Due to their stability, accessibility and specificity, miRNAs encapsulated in exosomes are considered highly promising biomarkers for the early diagnosis of lung cancer. Numerous studies have identified specific miRNAs in exosomes that show diagnostic value for early-stage lung cancer. For instance, the study reported that the expression levels of four serum miRNAs (miR-21-5p, miR-1413p, miR-222-3p and miR-486-5p) and two serum exosomal miRNAs (miR-146a-5p and miR-486-5p) in patients with early-stage NSCLC were significant differences compared to those in patients with benign lung lesions and healthy individuals (70). The findings indicate that the area under the receiver operating characteristic curve (AUC) for these four serum miRNAs and the two serum exosomal miRNAs in early-stage NSCLC patients was ≥ 0.697, with the AUC for serum exosomal miRNAs exceeding that of serum miRNAs. This suggests that exosomal miRNAs hold considerable promise for application in the early diagnosis of lung cancer. Additionally, Jin et al. (71) conducted an analysis of plasma exosomal miRNAs from patients with early-stage NSCLC using miRNA sequencing and validated their results through qPCR. Their study identified two miRNAs (miR-181b-5p and miR-361-5p) specific to LUAD that were upregulated compared to healthy individuals and two that were downregulated (miR-30a-3p and miR-30e-3p). Furthermore, they also discovered three microRNAs specific to LUSC: miR-320b was found to be upregulated, while miR-10b-5p and miR-15b-5p were downregulated. The researchers also detected the expression levels of miR-378a, miR-379, miR-139-5p, miR-200b-5p, miR-19-3p, miR-21-5p, and miR-221-3p in the plasma exosomes, and miR-126, miR-17-5p, miR-21, miR-25, and miR-223 in serum exosomes of LUAD patients were significantly different from those in healthy individuals (72–79). Another study demonstrated that exosomes contribute to tumor growth and metastasis by delivering miR-1228-5p, positioning it as a potential biomarker for the diagnosis and prognosis of SCLC (80, 81). Therefore, exosomal miRNAs offer unique advantages and significant potential for the early diagnosis of lung cancer. More detailed information can be found in Table 1.
4.2 Exosomal lncRNAs
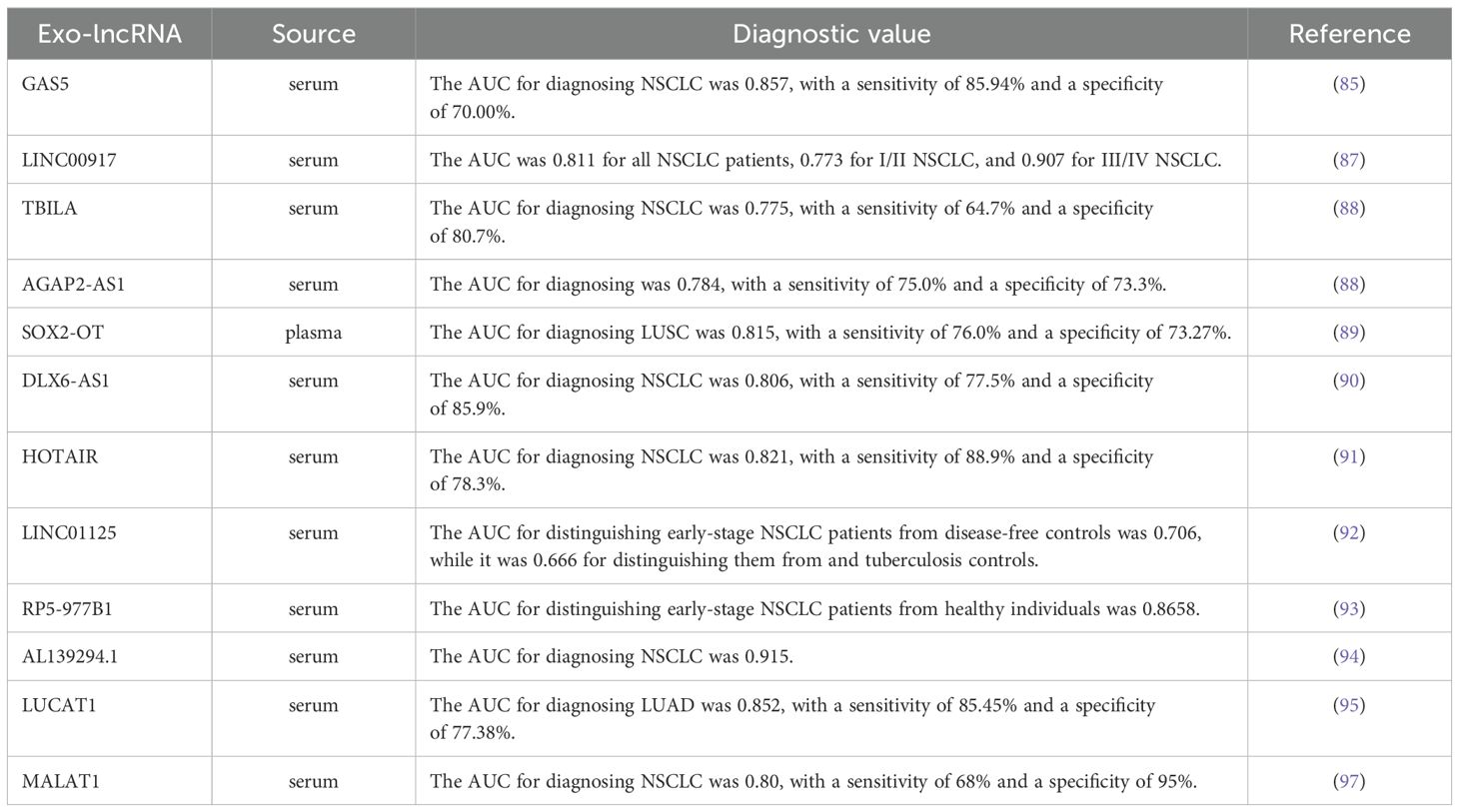
Long non-coding RNA (lncRNA), defined as RNA molecules longer than 200 nucleotides that do no encode proteins, play crucial roles in various important biological processes, including X chromosome silencing, chromatin modification, transcriptional activation and nuclear transport. They are closely associated with the occurrence, development and drug resistance of lung cancer (82–84). Recent studies have indicated that exosomal lncRNAs have promising potential as biomarkers for the early diagnosis of lung cancer. For example, the levels of serum exosomal lncRNA GAS5 in patients with early-stage NSCLC were found to be lower than those in healthy individuals, and with even lower levels observed in patients with advanced NSCLC. The AUC, sensitivity and specificity of exosomal lncRNA GAS5 for the diagnosis of NSCLC were 0.857, 85.94% and 70.00%, respectively. When combined with carcinoembryonic antigen (CEA), a commonly used nonspecific serum tumor biomarker for NSCLC, the AUC increased to 0.929 (85, 86). Additionally, another study revealed that serum exosome LINC00917 was expressed at higher levels in lung cancer patients compared to healthy controls, demonstrating significant diagnostic value for both early-stage and advanced lung cancer (87). Tao et al. (88) reported elevated expression of TGF-β induced lncRNA (TBILA) and AGAP2 antisense RNA 1 (AGAP2-AS1) in the serum exosomes of lung cancer patients when compared to healthy individuals. TBILA showed notable discriminatory capacity for all NSCLC patients, stage I NSCLC patients and adenocarcinoma (ADC) patients, while AGAP2-AS1 exhibited a higher AUC in distinguishing LUSC patients from healthy controls. Another study demonstrated significant up-regulation of exosomal lncRNA SOX2-OT in LUSC patients, with an AUC of 0.815, sensitivity of 76.00% and specificity of 73.17% for LUSC diagnosis (89). As research continues to advance, various exosomal lncRNAs, including growth-arrest specific protein 6 antisense RNA 1 (DLX6-AS1) (90), HOX transcript antisense RNA (HOTAIR) (91), LINC01125 (92), RP5-977B1 (93) (84), AL139294.1 (94), LUCAT1 (95, 96) and MALAT1 (97), have been identified as having clinical value in the diagnosis of lung cancer. More detailed information can be found in Table 2.
4.3 Exosomal circRNAs
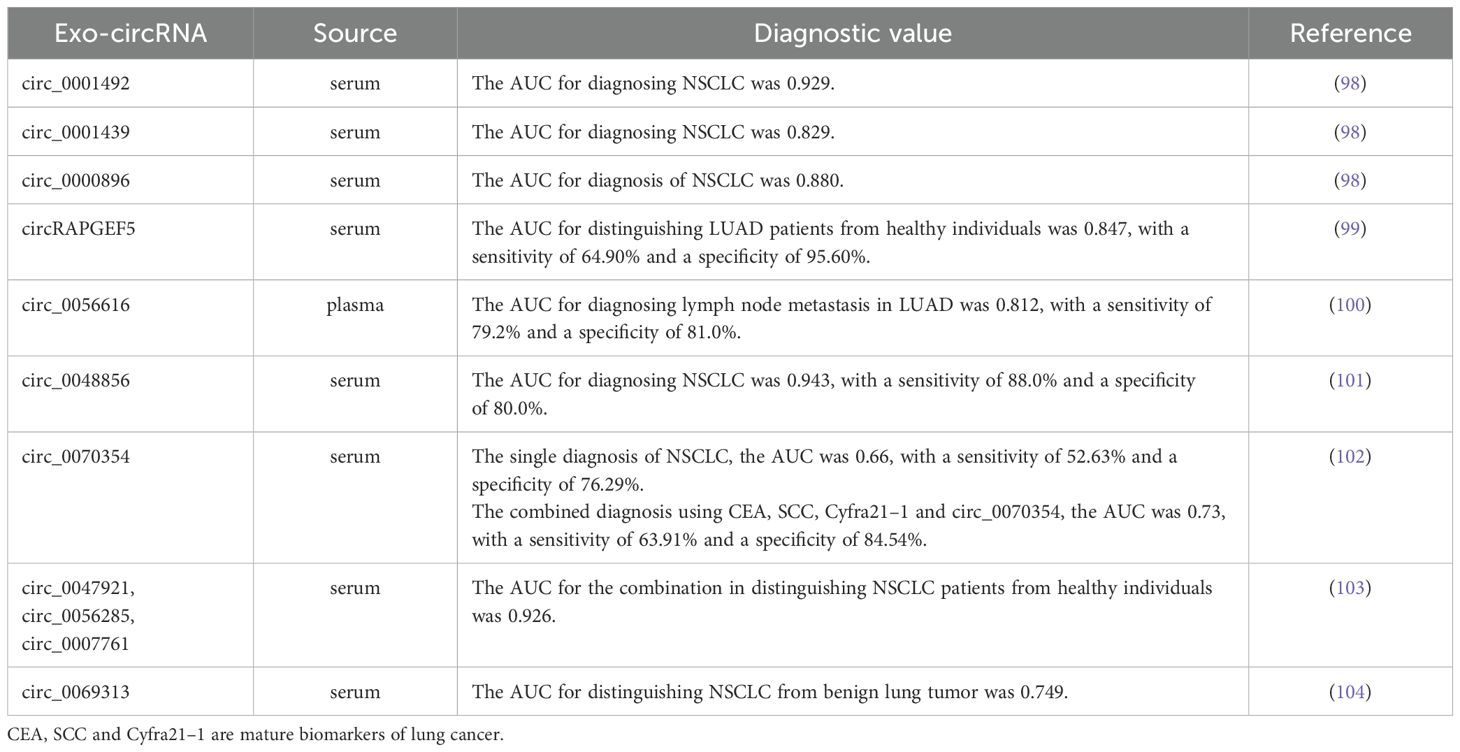
Exosomes-derived circular RNAs (circRNAs) have emerged as a new focus in non-coding RNA research, following the discoveries of miRNAs and lncRNAs. They hold significant promise as reliable biomarkers for the early diagnosis of lung cancer, potentially outperforming traditional tumor markers and serum circRNA. Analysis of serum exosomal circRNAs expression levels in LUAD patients before and after surgery revealed significant decrease in the expression of exosomal circ_0001492, circ_0001439 and circ_0000896 following the surgical procedure. ROC curve analysis demonstrated that the AUCs for these exosomal circRNAs were all greater than 0.75, and their combined diagnostic performance demonstrated higher sensitivity and specificity, with an AUC of 0.805 (98). Additionally, a study found that exosomal circRAPGEF5 exhibited strong diagnostic capability for LUAD, with an AUC, sensitivity and specificity of 0.847, 64.90% and 95.60%, respectively, and the combination of circRAPGEF5 with CEA further improved diagnostic efficacy (99). Furthermore, He et al. (100) identified exosomal circRNA_0056616 as being closely associated with lymph node and distant metastasis in LUAD, demonstrating high sensitivity and specificity for diagnosing lymph node metastasis, with an AUC of 0.812. Several other exosomal circRNAs have also demonstrated significant diagnostic value for NSCLC, including circ_0048856 (101), hsa_circ_0070354 (102), circ_0047921, circ_0056285, circ_0007761 (103), circ_0069313 (104) and circFARSA (105). More detailed information about these exosomal circRNAs can be found in Table 3. In summary, exosomal circRNAs has high specificity and sensitivity as biomarkers for the early diagnosis of lung cancer, potentially surpassing traditional diagnostic markers. The combined use of multiple exosomal circRNAs may provide greater specificity and sensitivity compared to individual exosomal circRNA.
4.4 Exosomal proteins
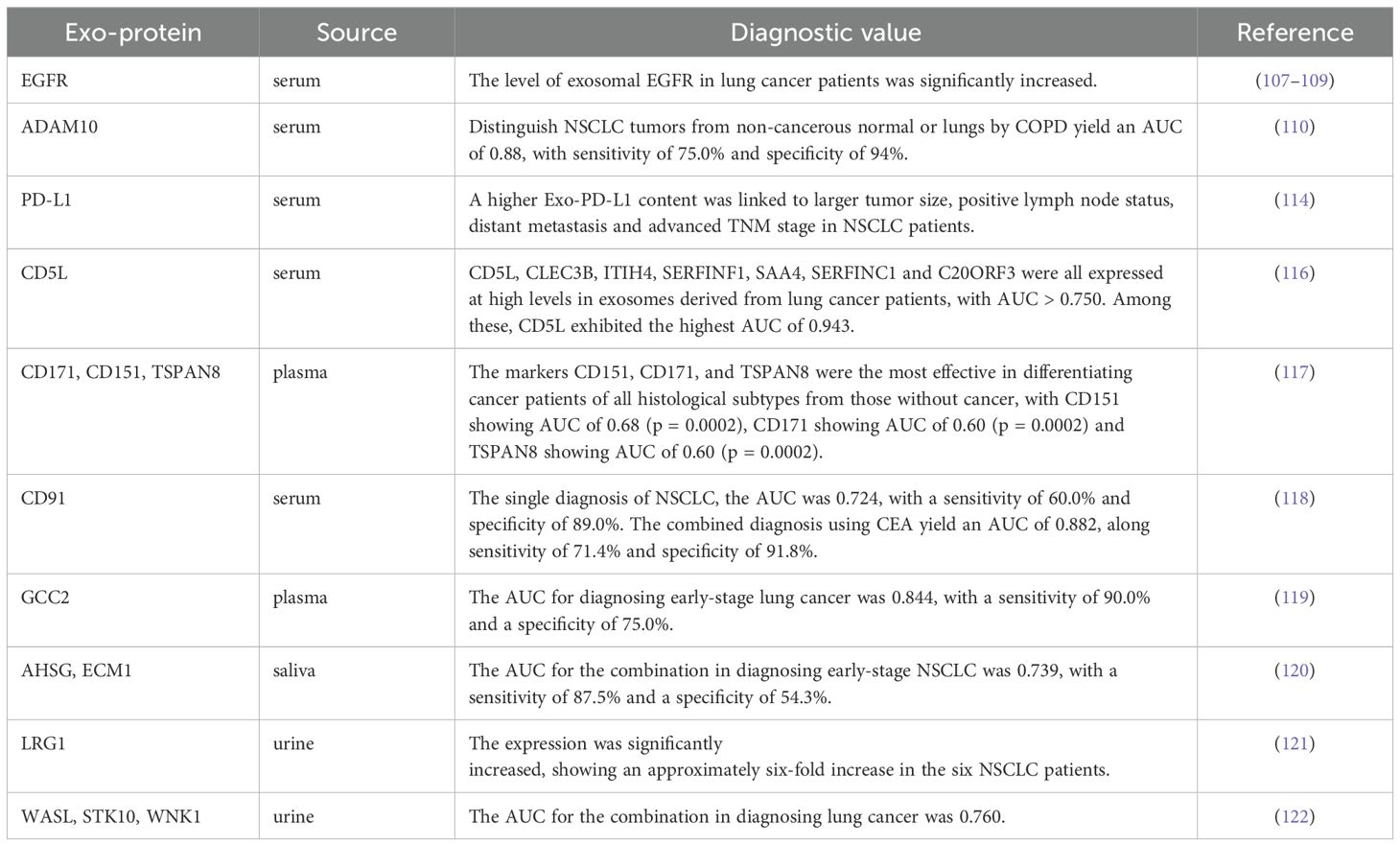
Exosomes contain a rich variety of proteins derived from their parental cells, making the detection of these exosomal proteins a valuable diagnostic approach for lung cancer (106). For example, epidermal growth factor receptor (EGFR), a crucial regulator of tumor growth, can be detected in the plasma of patients with early-stage lung cancer through exosomes (107). Approximately 80% of exosomes isolated from lung cancer samples contain EGFR, in stark contrast to only about 2% of exosomes from samples of chronic lung inflammation (108). While the levels of exosomal EGFR is significantly increased in lung cancer patients, the concentration of soluble EGFR in plasma does not show a notable difference (109). Moreover, Yoneyama et al. (110) found that active disintegrin and metalloproteinase domaincon-taining protein 10 (ADAM10) is significantly increased in the exosomes of NSCLC patients. This marker can effectively differentiate NSCLC patients from healthy individuals, establishing it as an important biomarker for NSCLC detection. Another important protein, programmed death-ligand 1 (PD-L1), has been identified in exosomes. PD-L1 binds to programmed death-1 (PD-1) on immune T cells and inhibit T-cell activation (111). The detection of PD-L1 in exosomes enables sampling from blood and other body fluids, providing an auxiliary diagnostic method for the early diagnosis of NSCLC (112). Studies have confirmed that exosomal PD-L1 levels in NSCLC patients are higher than those in normal controls, correlating with disease progression, clinicopathological characteristics, and TMN stage. Interestingly, no significant correlation has been found between PD-L1 levels in tumor tissue and the clinical characteristics of patients (113–115). Therefore, exosomal PD-L1 may offer greater clinical utility for diagnosing NSCLC compared to PD-L1 derived from tumor tissue. Choi et al. (116) demonstrated that the expression levels of seven proteins (CD5L, CLEC3B, ITIH4, SERFINF1, SAA4, SERFINC1 and C20-ORF3) were significantly increased in plasma exosomes of lung cancer patients. Further analysis revealed that only CD5L was significantly upregulated in cancer tissues, suggesting its potential as a biomarker for lung cancer diagnosis. Several studies have indicated that the expression levels of CD171, CD151, and TSPAN8 protein in plasma exosomes of lung cancer patients are significantly higher than healthy individuals, with CD151 and CD171 showing high expression in LUAD patients, while CD151 and TSPAN8 are elevated in LUSC patients (117). Ueda et al. (118) quantitatively identified CD91 as a LUAD-specific antigen on exosomes through a comprehensive analysis of 1369 exosome protein profiles, serving as a screening marker for lung cancer that is unaffected by gender or age. Compared to the CEA, exosomal CD91 exhibits improved detection sensitivity for early-stage lung cancer. High expression levels of GCC2 in the peripheral blood of patients demonstrate both specificity and sensitivity for early-stage lung cancer (119). Additionally, the proteins AHSG and ECM1 have been found to be significantly increased in the exosomes of NSCLC patients, suggesting their potential as diagnostic biomarkers for this condition (120).
In addition to exosomes derived from blood, several proteins have been reported to be significantly increased in the urine exosomes of NSCLC patients, including LRG1 (121), WASL, STK10 and WNK1 (122). One study identified twelve proteins, such as members of the annexin family (annexin A1, A2, A3, A5, A6, A11), along with PR-OM1, MUC1, BPIFA1, CRNN, MUC5B and IQGAP, which showed significant differences between lung cancer patients and healthy individuals. Furthermore, a comparative proteomics analysis revealed that MUC5B, IQGAP, ENO1 and SPARCL1 were identified in the salivary exosomes of lung cancer patients (123, 124). These exosomal proteins found in various body fluids show promise as potential biomarkers for lung cancer diagnosis. More detailed information can be found in Table 4. Given the diverse array of exosomal proteins, those sourced from different body fluids could serve as valuable biomarkers for lung cancer. It is also anticipated that combinations of various exosomal proteins may offer high clinical value as combined markers in the diagnosis and management of lung cancer.
5 Conclusion and prospects
Currently, the diagnostic technology for lung cancer has seen significant advancements compared to the past. However, most existing methods primarily focus on detecting advanced lung cancer, leaving early diagnosis as a considerable challenge. Current diagnostic techniques, such as endoscopic ultrasound-guided fine needle aspiration (EUS-FNA), MRI, and computed tomography (CT) (125), often involve invasive procedures that carry risks for patients. While histopathological examination is widely regarded as the gold standard, its invasive nature cannot be overlooked. Given that several years may elapse between the onset of lung cancer and the appearance of symptoms, liquid biopsy has emerged as a vital tool for early screening (126). This method provides a promising opportunity to improve treatment outcomes and enhance survival rates for patients. Exosomes derived from tumor cells, which encapsulate a wealth of biological information, have shown considerable potential as biomarkers for the early diagnosis of lung cancer (127). However, the standardization of exosome isolation and detection technologies is crucial for their effective integration into early diagnosis protocols. Additionally, conducting more extensive clinical studies to validate the efficacy of exosomes as biomarkers is a necessary step for advancing this field.
Authoritative studies have highlighted the potential of exosomal miRNAs as biomarkers due to their essential role in regulating lung cancer proliferation and invasion. Nonetheless, there remains a significant gap in the widespread clinical application of miRNAs for lung cancer diagnosis. While specific diagnostic models incorporating multiple biomarkers have shown promising predictive performance, their low repeatability and complex composition present major challenges for the practical use of miRNAs in early detection (128). Advancements in sequencing technology have facilitated the identification of various lncRNAs and circRNAs that exhibit differential expression in exosomes. Accumulating evidence suggests that the expression of specific lncRNAs and circRNAs may provide valuable insights into the clinical features of lung cancer (129). Currently, the analysis of lncRNAs and circRNAs largely relies on bioinformatics predictions, underscoring the need for further experimental and clinical studies to validate their diagnostic value (130, 131). Furthermore, while previous research has shown exosomal proteins can be utilized for lung cancer diagnosis, the upregulation of these proteins may be influenced by various factors, including smoking and inflammation. Thus, diagnosing tumor-associated proteins in exosomes must be grounded in precise isolation and identification methods. It is essential to explore optimal diagnostic indicators through multidisciplinary research that encompasses clinical medicine, immunology, and bioinformatics, providing new directions and insights for the realization of personalized lung cancer diagnosis.
In summary, while research on early diagnosis of lung cancer faces numerous challenges, exosomes present significant promise as potential biomarkers. Future studies should emphasize technical standardization and clinical validation to enhance the practical application of exosomes in the early detection of lung cancer.
Author contributions
TZ: Conceptualization, Writing – original draft, Methodology. HM: Methodology, Writing – original draft, Conceptualization. ZL: Methodology, Writing – original draft. YX: Writing – review & editing. LZ: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
TZ, ZL and LZ was employed by Wuhan Kindstar Zhenyuan Medical Laboratory Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Leiter A, Veluswamy RR, and Wisnivesky JP. The global burden of lung cancer: current status and future trends. Nat Rev Clin Oncol. (2023) 20:624–39. doi: 10.1038/s41571-023-00798-3
2. Feng R, Su Q, Huang X, Basnet T, Xu X, and Ye W. Cancer situation in China: what does the China cancer map indicate from the first national death survey to the latest cancer registration? Cancer Commun (Lond). (2023) 43:75–86. doi: 10.1002/cac2.12393
3. Galetta D. Advances in lung cancer therapy. Cancers (Basel). (2023) 15(10):2671. doi: 10.3390/cancers15102671
4. Sompallae RR, Dundar B, Guseva NV, Bossler AD, and Ma D. EGFR and ERBB2 exon 20 insertion/duplication in advanced non-small cell lung cancer: genomic profiling and clinicopathologic features. Front Oncol. (2023) 13:1163485. doi: 10.3389/fonc.2023.1163485
5. Jacobsen MM, Silverstein SC, Quinn M, Waterston LB, Thomas CA, Benneyan JC, et al. Timeliness of access to lung cancer diagnosis and treatment: A scoping literature review. Lung Cancer. (2017) 112:156–64. doi: 10.1016/j.lungcan.2017.08.011
6. Shimizu K, Ohtaki Y, Suzuki K, Date H, Yamashita M, Iizasa T, et al. Salvage surgery for non-small cell lung cancer after definitive radiotherapy. Ann Thorac Surg. (2021) 112:862–73. doi: 10.1016/j.athoracsur.2020.10.035
7. Wang Y, Li C, Wang Z, Wang Z, Wu R, Wu Y, et al. Comparison between immunotherapy efficacy in early non-small cell lung cancer and advanced non-small cell lung cancer: a systematic review. BMC Med. (2022) 20:426. doi: 10.1186/s12916-022-02580-1
8. Bustos García de Castro A, Ferreirós Domínguez J, Delgado Bolton R, Fernández Pérez C, Cabeza Martínez B, García García-Esquinas M, et al. PET-CT in presurgical lymph node staging in non-small cell lung cancer: the importance of false-negative and false-positive findings. Radiologia. (2017) 59:147–58. doi: 10.1016/j.rx.2016.12.001
9. Bodor JN, Boumber Y, and Borghaei H. Biomarkers for immune checkpoint inhibition in non-small cell lung cancer (NSCLC). Cancer. (2020) 126:260–70. doi: 10.1002/cncr.32468
10. Zhou B, Xu K, Zheng X, Chen T, Wang J, Song Y, et al. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct Target Ther. (2020) 5:144. doi: 10.1038/s41392-020-00258-9
11. Shen H, Jin Y, Zhao H, Wu M, Zhang K, Wei Z, et al. Potential clinical utility of liquid biopsy in early-stage non-small cell lung cancer. BMC Med. (2022) 20:480. doi: 10.1186/s12916-022-02681-x
12. Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, and Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. (2005) 17:879–87. doi: 10.1093/intimm/dxh267
13. Miyazaki K, Wada Y, Okuno K, Murano T, Morine Y, Ikemoto T, et al. An exosome-based liquid biopsy signature for pre-operative identification of lymph node metastasis in patients with pathological high-risk T1 colorectal cancer. Mol Cancer. (2023) 22:2. doi: 10.1186/s12943-022-01685-8
14. Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y, et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol. (2020) 13:156. doi: 10.1186/s13045-020-00991-2
15. Zhao L, Wang H, Fu J, Wu X, Liang XY, Liu XY, et al. Microfluidic-based exosome isolation and highly sensitive aptamer exosome membrane protein detection for lung cancer diagnosis. Biosens Bioelectron. (2022) 214:114487. doi: 10.1016/j.bios.2022.114487
16. He L, Zhu W, Chen Q, Yuan Y, Wang Y, Wang J, et al. Ovarian cancer cell-secreted exosomal miR-205 promotes metastasis by inducing angiogenesis. Theranostics. (2019) 9:8206–20. doi: 10.7150/thno.37455
17. Jiang Y, Wang K, Lu X, Wang Y, and Chen J. Cancer-associated fibroblasts-derived exosomes promote lung cancer progression by OIP5-AS1/miR-142-5p/PD-L1 axis. Mol Immunol. (2021) 140:47–58. doi: 10.1016/j.molimm.2021.10.002
18. Zhang X, Xu Y, Ma L, Yu K, Niu Y, Xu X, et al. Essential roles of exosome and circRNA_101093 on ferroptosis desensitization in lung adenocarcinoma. Cancer Commun (Lond). (2022) 42:287–313. doi: 10.1002/cac2.12275
19. Chen SW, Zhu SQ, Pei X, Qiu BQ, Xiong D, Long X, et al. Cancer cell-derived exosomal circUSP7 induces CD8(+) T cell dysfunction and anti-PD1 resistance by regulating the miR-934/SHP2 axis in NSCLC. Mol Cancer. (2021) 20:144. doi: 10.1186/s12943-021-01448-x
20. Song Z, Jia G, Ma P, and Cang S. Exosomal miR-4443 promotes cisplatin resistance in non-small cell lung carcinoma by regulating FSP1 m6A modification-mediated ferroptosis. Life Sci. (2021) 276:119399. doi: 10.1016/j.lfs.2021.119399
21. Wu S, Luo M, To KKW, Zhang J, Su C, Zhang H, et al. Intercellular transfer of exosomal wild type EGFR triggers osimertinib resistance in non-small cell lung cancer. Mol Cancer. (2021) 20:17. doi: 10.1186/s12943-021-01307-9
22. Johnstone RM, Adam M, Hammond JR, Orr L, and Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. (1987) 262:9412–20. doi: 10.1016/S0021-9258(18)48095-7
23. Raposo G and Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. (2013) 200:373–83. doi: 10.1083/jcb.201211138
24. Doyle LM and Wang MZ. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. (2019) 8(7):727. doi: 10.3390/cells8070727
25. Zhang W, He P, Wang S, Adili A, Chen Z, Zhang CY, et al. Characterization of protein profiling and mRNA expression of LLC exosomes. Protein J. (2019) 38:586–97. doi: 10.1007/s10930-019-09849-0
26. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, and Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. (2007) 9:654–9. doi: 10.1038/ncb1596
27. Gusachenko ON, Zenkova MA, and Vlassov VV. Nucleic acids in exosomes: disease markers and intercellular communication molecules. Biochem (Mosc). (2013) 78:1–7. doi: 10.1134/s000629791301001x
28. Sadik N, Cruz L, Gurtner A, Rodosthenous RS, Dusoswa SA, Ziegler O, et al. Extracellular RNAs: A new awareness of old perspectives. Methods Mol Biol. (2018) 1740:1–15. doi: 10.1007/978-1-4939-7652-2_1
29. Skotland T, Sandvig K, and Llorente A. Lipids in exosomes: Current knowledge and the way forward. Prog Lipid Res. (2017) 66:30–41. doi: 10.1016/j.plipres.2017.03.001
30. Zhang X, Yuan X, Shi H, Wu L, Qian H, and Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. (2015) 8:83. doi: 10.1186/s13045-015-0181-x
31. Yu W, Hurley J, Roberts D, Chakrabortty SK, Enderle D, Noerholm M, et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. (2021) 32:466–77. doi: 10.1016/j.annonc.2021.01.074
32. Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. (2013) 126:5553–65. doi: 10.1242/jcs.128868
33. Wollert T and Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. (2010) 464:864–9. doi: 10.1038/nature08849
34. Chairoungdua A, Smith DL, Pochard P, Hull M, and Caplan MJ. Exosome release of β-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol. (2010) 190:1079–91. doi: 10.1083/jcb.201002049
35. Nazarenko I, Rana S, Baumann A, McAlear J, Hellwig A, Trendelenburg M, et al. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. (2010) 70:1668–78. doi: 10.1158/0008-5472.Can-09-2470
36. Edgar JR, Eden ER, and Futter CE. Hrs- and CD63-dependent competing mechanisms make different sized endosomal intraluminal vesicles. Traffic. (2014) 15:197–211. doi: 10.1111/tra.12139
37. Kalluri. The biology R. and function of exosomes in cancer. J Clin Invest. (2016) 126:1208–15. doi: 10.1172/jci81135
38. Yang C and Robbins PD. The roles of tumor-derived exosomes in cancer pathogenesis. Clin Dev Immunol. (2011) 2011:842849. doi: 10.1155/2011/842849
39. Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. (2015) 523:177–82. doi: 10.1038/nature14581
40. Colombo M, Raposo G, and Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. (2014) 30:255–89. doi: 10.1146/annurev-cellbio-101512-122326
41. Lee Y, El Andaloussi S, and Wood MJ. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet. (2012) 21:R125–134. doi: 10.1093/hmg/dds317
42. Gandham S, Su X, Wood J, Nocera AL, Alli SC, Milane L, et al. Technologies and standardization in research on extracellular vesicles. Trends Biotechnol. (2020) 38:1066–98. doi: 10.1016/j.tibtech.2020.05.012
43. Gardiner C, Di Vizio D, Sahoo S, Théry C, Witwer KW, Wauben M, et al. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles. (2016) 5:32945. doi: 10.3402/jev.v5.32945
44. Li P, Kaslan M, Lee SH, Yao J, and Gao Z. Progress in exosome isolation techniques. Theranostics. (2017) 7:789–804. doi: 10.7150/thno.18133
45. Konoshenko MY, Lekchnov EA, Vlassov AV, and Laktionov PP. Isolation of extracellular vesicles: general methodologies and latest trends. BioMed Res Int. (2018) 2018:8545347. doi: 10.1155/2018/8545347
46. Greening DW, Xu R, Ji H, Tauro BJ, and Simpson RJ. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol Biol. (2015) 1295:179–209. doi: 10.1007/978-1-4939-2550-6_15
47. Willms E, Johansson HJ, Mäger I, Lee Y, Blomberg KE, Sadik M, et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep. (2016) 6:22519. doi: 10.1038/srep22519
48. Böing AN, van der Pol E, Grootemaat AE, Coumans FA, Sturk A, and Nieuwland R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles. (2014) 3(1):23430. doi: 10.3402/jev.v3.23430
49. Liu F, Vermesh O, Mani V, Ge TJ, Madsen SJ, Sabour A, et al. The exosome total isolation chip. ACS Nano. (2017) 11:10712–23. doi: 10.1021/acsnano.7b04878
50. Rider MA, Hurwitz SN, and Meckes DG Jr. ExtraPEG: A polyethylene glycol-based method for enrichment of extracellular vesicles. Sci Rep. (2016) 6:23978. doi: 10.1038/srep23978
51. Greening DW, Gopal SK, Xu R, Simpson RJ, and Chen W. Exosomes and their roles in immune regulation and cancer. Semin Cell Dev Biol. (2015) 40:72–81. doi: 10.1016/j.semcdb.2015.02.009
52. Whiteside. Exosomes TL. and tumor-mediated immune suppression. J Clin Invest. (2016) 126:1216–23. doi: 10.1172/jci81136
53. Gutiérrez-Vázquez C, Villarroya-Beltri C, Mittelbrunn M, and Sánchez-Madrid F. Transfer of extracellular vesicles during immune cell-cell interactions. Immunol Rev. (2013) 251:125–42. doi: 10.1111/imr.12013
54. Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. (2018) 560:382–6. doi: 10.1038/s41586-018-0392-8
55. Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, et al. Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell. (2019) 177:414–427.e413. doi: 10.1016/j.cell.2019.02.016
56. Kim JW, Wieckowski E, Taylor DD, Reichert TE, Watkins S, and Whiteside TL. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin Cancer Res. (2005) 11:1010–20. doi: 10.1158/1078-0432.1010.11.3
57. Berchem G, Noman MZ, Bosseler M, Paggetti J, Baconnais S, Le Cam E, et al. Hypoxic tumor-derived microvesicles negatively regulate NK cell function by a mechanism involving TGF-β and miR23a transfer. Oncoimmunology. (2016) 5:e1062968. doi: 10.1080/2162402x.2015.1062968
58. Janowska-Wieczorek A, Wysoczynski M, Kijowski J, Marquez-Curtis L, Machalinski B, Ratajczak J, et al. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int J Cancer. (2005) 113:752–60. doi: 10.1002/ijc.20657
59. Qi Y, Zha W, and Zhang W. Exosomal miR-660-5p promotes tumor growth and metastasis in non-small cell lung cancer. J buon. (2019) 24:599–607.
60. Wu H, Zhou J, Mei S, Wu D, Mu Z, Chen B, et al. Circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance by targeting LMO7. J Cell Mol Med. (2017) 21:1228–36. doi: 10.1111/jcmm.13056
61. Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. (2012) 109:E2110–2116. doi: 10.1073/pnas.1209414109
62. Cao M, Seike M, Soeno C, Mizutani H, Kitamura K, Minegishi Y, et al. MiR-23a regulates TGF-β-induced epithelial-mesenchymal transition by targeting E-cadherin in lung cancer cells. Int J Oncol. (2012) 41:869–75. doi: 10.3892/ijo.2012.1535
63. Zhang X, Sai B, Wang F, Wang L, Wang Y, Zheng L, et al. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol Cancer. (2019) 18:40. doi: 10.1186/s12943-019-0959-5
64. Fan J, Xu G, Chang Z, Zhu L, and Yao J. miR-210 transferred by lung cancer cell-derived exosomes may act as proangiogenic factor in cancer-associated fibroblasts by modulating JAK2/STAT3 pathway. Clin Sci (Lond). (2020) 134:807–25. doi: 10.1042/cs20200039
65. Jeong K, Yu YJ, You JY, Rhee WJ, and Kim JA. Exosome-mediated microRNA-497 delivery for anti-cancer therapy in a microfluidic 3D lung cancer model. Lab Chip. (2020) 20:548–57. doi: 10.1039/c9lc00958b
66. Ma Z, Wei K, Yang F, Guo Z, Pan C, He Y, et al. Tumor-derived exosomal miR-3157-3p promotes angiogenesis, vascular permeability and metastasis by targeting TIMP/KLF2 in non-small cell lung cancer. Cell Death Dis. (2021) 12:840. doi: 10.1038/s41419-021-04037-4
67. Liu Y, Luo F, Wang B, Li H, Xu Y, Liu X, et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. (2016) 370:125–35. doi: 10.1016/j.canlet.2015.10.011
68. Xuan Z, Chen C, Tang W, Ye S, Zheng J, Zhao Y, et al. TKI-resistant renal cancer secretes low-level exosomal miR-549a to induce vascular permeability and angiogenesis to promote tumor metastasis. Front Cell Dev Biol. (2021) 9:689947. doi: 10.3389/fcell.2021.689947
69. Liu Y, Gu Y, Han Y, Zhang Q, Jiang Z, Zhang X, et al. Tumor exosomal RNAs promote lung pre-metastatic niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell. (2016) 30:243–56. doi: 10.1016/j.ccell.2016.06.021
70. Wu Q, Yu L, Lin X, Zheng Q, Zhang S, Chen D, et al. Combination of Serum miRNAs with Serum Exosomal miRNAs in Early Diagnosis for Non-Small-Cell Lung Cancer. Cancer Manag Res. (2020) 12:485–95. doi: 10.2147/cmar.S232383
71. Jin X, Chen Y, Chen H, Fei S, Chen D, Cai X, et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res. (2017) 23:5311–9. doi: 10.1158/1078-0432.Ccr-17-0577
72. Grimolizzi F, Monaco F, Leoni F, Bracci M, Staffolani S, Bersaglieri C, et al. Exosomal miR-126 as a circulating biomarker in non-small-cell lung cancer regulating cancer progression. Sci Rep. (2017) 7:15277. doi: 10.1038/s41598-017-15475-6
73. Zhang Y, Zhang Y, Yin Y, and Li S. Detection of circulating exosomal miR-17-5p serves as a novel non-invasive diagnostic marker for non-small cell lung cancer patients. Pathol Res Pract. (2019) 215:152466. doi: 10.1016/j.prp.2019.152466
74. Rodríguez M, Silva J, López-Alfonso A, López-Muñiz MB, Peña C, Domínguez G, et al. Different exosome cargo from plasma/bronchoalveolar lavage in non-small-cell lung cancer. Genes Chromosomes Cancer. (2014) 53:713–24. doi: 10.1002/gcc.22181
75. Zhang H, Mao F, Shen T, Luo Q, Ding Z, Qian L, et al. Plasma miR-145, miR-20a, miR-21 and miR-223 as novel biomarkers for screening early-stage non-small cell lung cancer. Oncol Lett. (2017) 13:669–76. doi: 10.3892/ol.2016.5462
76. Yang JS, Li BJ, Lu HW, Chen Y, Lu C, Zhu RX, et al. Serum miR-152, miR-148a, miR-148b, and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumor Biol. (2015) 36:3035–42. doi: 10.1007/s13277-014-2938-1
77. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. (2008) 18:997–1006. doi: 10.1038/cr.2008.282
78. Cazzoli R, Buttitta F, Di Nicola M, Malatesta S, Marchetti A, Rom WN, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. (2013) 8:1156–62. doi: 10.1097/JTO.0b013e318299ac32
79. Zhou X, Wen W, Shan X, Zhu W, Xu J, Guo R, et al. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget. (2017) 8:6513–25. doi: 10.18632/oncotarget.14311
80. Mu X, Yu C, Zhao Y, Hu X, Wang H, He Y, et al. Exosomal miR-1228-5p down-regulates DUSP22 to promotes cell proliferation and migration in small cell lung cancer. Life Sci. (2024) 351:122787. doi: 10.1016/j.lfs.2024.122787
81. Liu Q, Yu Z, Yuan S, Xie W, Li C, Hu Z, et al. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget. (2017) 8:13048–58. doi: 10.18632/oncotarget.14369
82. Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. (2018) 33:706–720.e709. doi: 10.1016/j.ccell.2018.03.006
83. Hu WL, Jin L, Xu A, Wang YF, Thorne RF, Zhang XD, et al. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat Cell Biol. (2018) 20:492–502. doi: 10.1038/s41556-018-0066-7
84. Kopp F and Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. (2018) 172:393–407. doi: 10.1016/j.cell.2018.01.011
85. Li C, Lv Y, Shao C, Chen C, Zhang T, Wei Y, et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. (2019) 234:20721–7. doi: 10.1002/jcp.28678
86. Ma C, Shi X, Zhu Q, Li Q, Liu Y, Yao Y, et al. The growth arrest-specific transcript 5 (GAS5): a pivotal tumor suppressor long noncoding RNA in human cancers. Tumor Biol. (2016) 37:1437–44. doi: 10.1007/s13277-015-4521-9
87. Xiong D, Wang C, Yang Z, Han F, and Zhan H. Clinical significance of serum-derived exosomal LINC00917 in patients with non-small cell lung cancer. Front Genet. (2021) 12:728763. doi: 10.3389/fgene.2021.728763
88. Tao Y, Tang Y, Yang Z, Wu F, Wang L, Yang L, et al. Exploration of serum exosomal lncRNA TBILA and AGAP2-AS1 as promising biomarkers for diagnosis of non-small cell lung cancer. Int J Biol Sci. (2020) 16:471–82. doi: 10.7150/ijbs.39123
89. Teng Y, Kang H, and Chu Y. Identification of an exosomal long noncoding RNA SOX2-OT in plasma as a promising biomarker for lung squamous cell carcinoma. Genet Test Mol Biomarkers. (2019) 23:235–40. doi: 10.1089/gtmb.2018.0103
90. Zhang X, Guo H, Bao Y, Yu H, Xie D, and Wang X. Exosomal long non-coding RNA DLX6-AS1 as a potential diagnostic biomarker for non-small cell lung cancer. Oncol Lett. (2019) 18:5197–204. doi: 10.3892/ol.2019.10892
91. Chen L, Huang S, Huang J, Chen Q, and Zhuang Q. Role and mechanism of exosome-derived long noncoding RNA HOTAIR in lung cancer. ACS Omega. (2021) 6:17217–27. doi: 10.1021/acsomega.1c00905
92. Xian J, Zeng Y, Chen S, Lu L, Liu L, Chen J, et al. Discovery of a novel linc01125 isoform in serum exosomes as a promising biomarker for NSCLC diagnosis and survival assessment. Carcinogenesis. (2021) 42:831–41. doi: 10.1093/carcin/bgab034
93. Min L, Zhu T, Lv B, An T, Zhang Q, Shang Y, et al. Exosomal LncRNA RP5-977B1 as a novel minimally invasive biomarker for diagnosis and prognosis in non-small cell lung cancer. Int J Clin Oncol. (2022) 27:1013–24. doi: 10.1007/s10147-022-02129-5
94. Ma X, Chen Z, Chen W, Chen Z, Shang Y, Zhao Y, et al. LncRNA AL139294.1 can be transported by extracellular vesicles to promote the oncogenic behavior of recipient cells through activation of the Wnt and NF-κB2 pathways in non-small-cell lung cancer. J Exp Clin Cancer Res. (2024) 43:20. doi: 10.1186/s13046-023-02939-z
95. Wang L, Xie Y, Wang J, Zhang Y, Liu S, Zhan Y, et al. Characterization of a novel LUCAT1/miR-4316/VEGF-A axis in metastasis and glycolysis of lung adenocarcinoma. Front Cell Dev Biol. (2022) 10:833579. doi: 10.3389/fcell.2022.833579
96. Song X, Duan L, and Dong Y. Diagnostic accuracy of exosomal long noncoding RNAs in diagnosis of NSCLC: A meta-analysis. Mol Diagn Ther. (2024) 28:455–68. doi: 10.1007/s40291-024-00715-z
97. Wang MC, Gong GY, Wang CL, Ko HW, Weng RX, Chang PY, et al. Methods for collection of extracellular vesicles and their content RNA as liquid biopsy for lung cancer detection: application of differential centrifugation and annexin A5 coated beads. Curr Issues Mol Biol. (2022) 44:2374–86. doi: 10.3390/cimb44050162
98. Kang Y, You J, Gan Y, Chen Q, Huang C, Chen F, et al. Serum and Serum Exosomal CircRNAs hsa_circ_0001492, hsa_circ_0001439, and hsa_circ_0000896 as Diagnostic Biomarkers for Lung Adenocarcinoma. Front Oncol. (2022) 12:912246. doi: 10.3389/fonc.2022.912246
99. Zhou H, Huang X, Yang X, Jiang F, Shao F, Shi W, et al. CircRAPGEF5 Promotes the Proliferation and Metastasis of Lung Adenocarcinoma through the miR-1236-3p/ZEB1 Axis and Serves as a Potential Biomarker. Int J Biol Sci. (2022) 18:2116–31. doi: 10.7150/ijbs.66770
100. He F, Zhong X, Lin Z, Lin J, Qiu M, Li X, et al. Plasma exo-hsa_circRNA_0056616: A potential biomarker for lymph node metastasis in lung adenocarcinoma. J Cancer. (2020) 11:4037–46. doi: 10.7150/jca.30360
101. He Y, Liu Y, Cha N, Gao Y, Li F, Zhang M, et al. Exosomal circ_0048856 derived from non-small cell lung cancer contributes to aggressive cancer progression through downregulation of miR-1287-5p. Pathol Res Pract. (2022) 232:153659. doi: 10.1016/j.prp.2021.153659
102. Huang Y, Qin S, Gu X, Zheng M, Zhang Q, Liu Y, et al. Comprehensive assessment of serum hsa_circ_0070354 as a novel diagnostic and predictive biomarker in non-small cell lung cancer. Front Genet. (2021) 12:796776. doi: 10.3389/fgene.2021.796776
103. Xian J, Su W, Liu L, Rao B, Lin M, Feng Y, et al. Identification of three circular RNA cargoes in serum exosomes as diagnostic biomarkers of non-small-cell lung cancer in the Chinese population. J Mol Diag. (2020) 22:1096–108. doi: 10.1016/j.jmoldx.2020.05.011
104. Chen Y, Lou C, Ma X, Zhou C, Zhao X, Li N, et al. Serum exosomal hsa_circ_0069313 has a potential to diagnose more aggressive non-small cell lung cancer. Clin Biochem. (2022) 102:56–64. doi: 10.1016/j.clinbiochem.2022.01.005
105. Hang D, Zhou J, Qin N, Zhou W, Ma H, Jin G, et al. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. (2018) 7:2783–91. doi: 10.1002/cam4.1514
106. Li W, Li C, Zhou T, Liu X, Liu X, Li X, et al. Role of exosomal proteins in cancer diagnosis. Mol Cancer. (2017) 16:145. doi: 10.1186/s12943-017-0706-8
107. Mack PC, Miao J, Redman MW, Moon J, Goldberg SB, Herbst RS, et al. Circulating tumor DNA kinetics predict progression-free and overall survival in EGFR TKI-treated patients with EGFR-mutant NSCLC (SWOG S1403). Clin Cancer Res. (2022) 28:3752–60. doi: 10.1158/1078-0432.Ccr-22-0741
108. Huang SH, Li Y, Zhang J, Rong J, and Ye S. Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Invest. (2013) 31:330–5. doi: 10.3109/07357907.2013.789905
109. Xie X, Nie H, Zhou Y, Lian S, Mei H, Lu Y, et al. Eliminating blood oncogenic exosomes into the small intestine with aptamer-functionalized nanoparticles. Nat Commun. (2019) 10:5476. doi: 10.1038/s41467-019-13316-w
110. Yoneyama T, Gorry M, Sobo-Vujanovic A, Lin Y, Vujanovic L, Gaither-Davis A, et al. ADAM10 sheddase activity is a potential lung-cancer biomarker. J Cancer. (2018) 9:2559–70. doi: 10.7150/jca.24601
111. Ma Y, Marinkova R, Nenkov M, Jin L, Huber O, Sonnemann J, et al. Tumor-intrinsic PD-L1 exerts an oncogenic function through the activation of the wnt/β-catenin pathway in human non-small cell lung cancer. Int J Mol Sci. (2022) 23(19):11031. doi: 10.3390/ijms231911031
112. Kim DH, Kim H, Choi YJ, Kim SY, Lee JE, Sung KJ, et al. Exosomal PD-L1 promotes tumor growth through immune escape in non-small cell lung cancer. Exp Mol Med. (2019) 51:1–13. doi: 10.1038/s12276-019-0295-2
113. Wu Y, Zeng X, and Gan Q. A compact surface plasmon resonance biosensor for sensitive detection of exosomal proteins for cancer diagnosis. Methods Mol Biol. (2022) 2393:3–14. doi: 10.1007/978-1-0716-1803-5_1
114. Li C, Li C, Zhi C, Liang W, Wang X, Chen X, et al. Clinical significance of PD-L1 expression in serum-derived exosomes in NSCLC patients. J Transl Med. (2019) 17:355. doi: 10.1186/s12967-019-2101-2
115. Zhang Z, Jin W, Xu K, Zheng X, Zhou Y, Luo M, et al. Blood exosome PD-L1 is associated with PD-L1 expression measured by immunohistochemistry, and lymph node metastasis in lung cancer. Tissue Cell. (2022) 79:101941. doi: 10.1016/j.tice.2022.101941
116. Choi ES, Faruque HA, Kim JH, Kim KJ, Choi JE, Kim BA, et al. CD5L as an extracellular vesicle-derived biomarker for liquid biopsy of lung cancer. Diagnostics (Basel). (2021) 11(4):620. doi: 10.3390/diagnostics11040620
117. Sandfeld-Paulsen B, Jakobsen KR, Bæk R, Folkersen BH, Rasmussen TR, Meldgaard P, et al. Exosomal proteins as diagnostic biomarkers in lung cancer. J Thorac Oncol. (2016) 11:1701–10. doi: 10.1016/j.jtho.2016.05.034
118. Ueda K, Ishikawa N, Tatsuguchi A, Saichi N, Fujii R, and Nakagawa H. Antibody-coupled monolithic silica microtips for high throughput molecular profiling of circulating exosomes. Sci Rep. (2014) 4:6232. doi: 10.1038/srep06232
119. Jeong H, Choi BH, Park J, Jung JH, Shin H, Kang KW, et al. GCC2 as a new early diagnostic biomarker for non-small cell lung cancer. Cancers (Basel). (2021) 13(21):5482. doi: 10.3390/cancers13215482
120. Niu L, Song X, Wang N, Xue L, Song X, and Xie L. Tumor-derived exosomal proteins as diagnostic biomarkers in non-small cell lung cancer. Cancer Sci. (2019) 110:433–42. doi: 10.1111/cas.13862
121. Li Y, Zhang Y, Qiu F, and Qiu Z. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis. (2011) 32:1976–83. doi: 10.1002/elps.201000598
122. Jin S, Liu T, Wang W, Li T, Liu Z, and Zhang M. Lymphocyte migration regulation related proteins in urine exosomes may serve as a potential biomarker for lung cancer diagnosis. BMC Cancer. (2023) 23:1125. doi: 10.1186/s12885-023-11567-x
123. Sun Y, Xia Z, Shang Z, Sun K, Niu X, Qian L, et al. Facile preparation of salivary extracellular vesicles for cancer proteomics. Sci Rep. (2016) 6:24669. doi: 10.1038/srep24669
124. Sun Y, Huo C, Qiao Z, Shang Z, Uzzaman A, Liu S, et al. Comparative proteomic analysis of exosomes and microvesicles in human saliva for lung cancer. J Proteome Res. (2018) 17:1101–7. doi: 10.1021/acs.jproteome.7b00770
125. Wilson B, Becker A, Estes T, Keshavamurthy J, and Pucar D. Adrenal hemangioma definite diagnosis on CT, MRI, and FDG PET in a patient with primary lung cancer. Clin Nucl Med. (2018) 43:e192–4. doi: 10.1097/rlu.0000000000002069
126. Li P, Liu S, Du L, Mohseni G, Zhang Y, and Wang C. Liquid biopsies based on DNA methylation as biomarkers for the detection and prognosis of lung cancer. Clin Epigenetics. (2022) 14:118. doi: 10.1186/s13148-022-01337-0
127. Zhang W, Campbell DH, Walsh BJ, Packer NH, Liu D, and Wang Y. Cancer-derived small extracellular vesicles: emerging biomarkers and therapies for pancreatic ductal adenocarcinoma diagnosis/prognosis and treatment. J Nanobiotechnol. (2022) 20:446. doi: 10.1186/s12951-022-01641-0
128. Sun Y, Fang L, Yi Y, Feng A, Zhang K, and Xu JJ. Multistage nucleic acid amplification induced nano-aggregation for 3D hotspots-improved SERS detection of circulating miRNAs. J Nanobiotechnol. (2022) 20:285. doi: 10.1186/s12951-022-01500-y
129. Zuo YB, Zhang YF, Zhang R, Tian JW, Lv XB, Li R, et al. Ferroptosis in cancer progression: role of noncoding RNAs. Int J Biol Sci. (2022) 18:1829–43. doi: 10.7150/ijbs.66917
130. Liu Q and Li S. Exosomal circRNAs: Novel biomarkers and therapeutic targets for urinary tumors. Cancer Lett. (2024) 588:216759. doi: 10.1016/j.canlet.2024.216759
Keywords: lung cancer, exosomes, early diagnosis, biomarkers, liquid biopsy
Citation: Zhou T, Ma H, Li Z, Xu Y and Zhao L (2025) Exosomes in lung cancer: a role in early diagnosis. Front. Oncol. 15:1599608. doi: 10.3389/fonc.2025.1599608
Received: 25 March 2025; Accepted: 26 May 2025;
Published: 12 June 2025.
Edited by:
Lei Huang, University of Massachusetts Medical School, United StatesReviewed by:
Yangyang Zhu, Harvard Medical School, United StatesYue Liu, The University of Texas at Austin, United States
Copyright © 2025 Zhou, Ma, Li, Xu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingling Zhao, emhhb2xpbmdsaW5nQGtpbmRzdGFyLmNvbS5jbg==; Yijun Xu, dGpzeGt5eXh5akAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
 Tong Zhou1†
Tong Zhou1† Yijun Xu
Yijun Xu Lingling Zhao
Lingling Zhao