- 1School of Laboratory Medicine and Biotechnology, Southern Medical University, Guangzhou, Guangdong, China
- 2Clinical Laboratory, Second Hospital of Tianjin Medical University, Tianjin, China
Breast cancer is the most common malignant tumor and the leading cause of mortality among women worldwide. Triple-negative breast cancer (TNBC) is recognized as t the most aggressive form of breast cancer, with a poor prognosis and a high mortality rate within two years. The role of tumor markers in facilitating the early diagnosis, treatment, and monitoring of therapeutic efficacy and prognosis in TNBC is well-established. Currently, tissue biopsy remains the standard clinical method for determining tumor histology and staging. However, the invasive nature of tissue biopsy often leads to poor patient compliance, especially when repeated biopsies are required. In contrast, liquid biopsy offers several advantages: it is non-invasive, sample collection is straightforward, it can reflect the overall tumor burden and heterogeneity, and it allows for real-time monitoring. The markers primarily encompass circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), microRNAs (miRNAs), long ncRNA (lncRNAs), exosome, and so forth. The present review aims to provide a comprehensive overview of the recent advancements and potential clinical applications of liquid biopsy technology in the context of TNBC.
1 Introduction
Breast cancer is one of the most common malignancies and the leading cause of death among women around the world (1). Triple-negative breast cancer(TNBC) is a tumor type characterized by the absence of estrogen receptor (ER), progesterone receptors (PR) and HER2 expression. They account for approximately 12%–17% of patients diagnosed with breast cancer (2, 3). A significant body of research has indicated patients with TNBC are notably younger than those diagnosed with other breast cancer subtypes (4). TNBC is the most aggressive subtype, with a poor prognosis and high mortality rate within two years. Moreover, TNBC is susceptible to central nervous system metastases during the initial and subsequent recurrences. This markedly poor prognosis serves to further reduce the survival rate (5). Neoadjuvant chemotherapy has been shown to increase the pathological complete response (pCR) rate in patients diagnosed with TNBC (6). However, this treatment approach has also been linked to a reduction in both three-year progression-free survival (PFS) and three-year survival rates in individuals with this specific form of breast cancer (7). In patients with advanced TNBC, the immunotherapy PD-1 inhibitor pembrolizumab has no significant therapeutic effect (8). Similarly, the use of carboplatin and docetaxel did not improve efficacy or overall survival in patients with metastatic or recurrent locally advanced TNBC (9).It can be reasonably deduced that the implementation of early diagnosis and treatment protocols represents an efficacious strategy for the reduction of mortality rates among patients diagnosed with primary breast cancer.
A clinical diagnosis requires tissue biopsy to ascertain the tumor’s histology and stage. Nevertheless, there are certain constraints and potential complications associated with the surgical procedure of collecting tissue biopsies. Additionally, this technique is limited by the spatial and temporal heterogeneity of tumors. For patients with solid tumors such as TNBC, the presence of microscopic metastatic lesions that cannot be detected by current imaging modalities, such as computed tomography (CT) or positron emission tomography (PET) significantly complicates the prevention and treatment of fully metastatic disease (10). Liquid biopsy, defined as a noninvasive method that detects cancer-related biomarkers in blood or other body fluids, represents a promising approach for the early detection of cancer in asymptomatic individuals and those at high risk, with the potential to be integrated into screening programs utilizing non-invasive methods based on the analysis of blood or body secretions (11). This may result in the earlier detection of cancer, which could in turn lead to improved cure and survival rates. For patients with confirmed tumors, liquid biopsy offers a straightforward method of obtaining blood samples and monitoring tumors in real time (12).
The field of liquid biopsy has witnessed significant advancements, evolving from the initial detection of circulating tumor cells (CTCs) (13) and circulating tumor DNA (ctDNA) (14) to encompass the detection of exosomes (15), microRNAs (miRNAs), peripheral blood circulating RNA, tumor-educated platelets (TEPs) (16) and circulating tumor vascular endothelial cells (CTECs) (17). The heterogeneity of TNBC patients places high demands on detection technology. Currently, TNBC markers can be captured and detected more sensitively using custom personalized ctDNA microarrays (18), liquid chromatography-tandem mass spectrometry (LC-MS/MS) lipid profiling (19), the c-TRAK TN trial (20), miRNA single-molecule sensors (21), near-infrared (NIR) probes (22) and other technologies. In light of the rapid advancements being made in the field of liquid biopsy at present, this review summarizes of the research progresses and clinical applications of liquid biopsy technology in the context of TNBC, including early diagnosis, personalized treatment, prognosis prediction and the advancement of related technologies.
2 CTCs
2.1 Biological significance of CTCs
The concept of CTCs was first described by an Australian doctor in 1869 during an autopsy on a patient who had died of advanced metastatic cancer (23). CTCs are cancer cells originating from solid tumor which are found circulating in the peripheral blood. It is generally accepted that these cells are shed into the bloodstream from the primary or secondary tumor of patients with advanced cancer, and studies have shown that the morphology of CTCs varies depending on the stage and/or type of tumor (24). Compared to traditional invasive tissue biopsies, CTCs are not only easy to sampled, but also offer real-time insights into the dynamic changes in CTC levels (25). As CTCs can originate from both primary and (micro)metastatic sites, they can better represent the entire heterogeneous tumor cell population (26).
2.2 Evolution of CTC detection technologies
In general, CTC detection involves two steps: enrichment or separation—using pathological and immunological techniques—and detection or identification—using cytometry and nucleic acid (27). Early CTC detection techniques based on nucleic acid detection of CTCs included purification of nucleated cells, followed by lysis and nucleic acid extraction (28), and sequencing of individual tumor-specific translocation breakpoints (29). However, this method of nucleic acid analysis from free plasma or unpurified blood cell components has relatively low sensitivity. Moreover, it is difficult to distinguish whether the absence of a clear molecular abnormality in the blood sample is due to the lack of such abnormalities or to an insufficient amount of tumor-derived DNA circulating in the bloodstream of an individual patient (30).
Currently, enrichment methods for CTC detection fall into two broad categories: label-free and affinity-based. Label-free methods for CTC enrichment are achieved by exploiting biophysical properties, as these methods are independent of biomarkers and can capture cells regardless of CTC expression profiles (31). These methods enable the isolation and enrichment of epithelial-mesenchymal transition (EMT) cells, cells with stem-like cancer properties (32), and fibroblasts that may be associated with cancer (33). Immunomagnetic CTC enrichment is the most widely used strategy, which relies on the use of cell surface-expressed biomarkers. Tumor cells are positively selected by anti-epithelial cell adhesion molecule (EpCAM). Alternatively, the leukocyte background is removed by targeting the CD45 antigen expressed on leukocytes, thereby achieving negative selection (31). As the technology evolves, a variety of techniques are available to selectively detect viable CTCs, such as the Epithelial ImmunoSPOT (EPISPOT) assay, which can detect single cells of CTCs and has been successfully used in a large cohorts of breast cancer patients (34). With the development of technology, the technique of isolating CTCs from patients has gradually matured and can provide more accurate information in real-time for early detection, treatment and prognosis of tumors.
2.3 Clinical utility of CTCs in TNBC
In patients with aggressive cancers, metastasis can occur at an early stage of tumor development and is the leading cause of cancer-related deaths (35). The invasion of cancer cells into the bloodstream and their spread to other parts of the body via the bloodstream is the first step in metastasis. Studies of disseminated tumor cells (DTCs) in the bone marrow of breast cancer patients have shown that even patients with small early tumors can have early spread to distant sites (36). Therefore, the primary indicator of the early-stage of cancer metastasis may be CTCs in the blood. In early-stage TNBC patients, CTCs were detected in tumors of histological grades 1–4 tumors, with a detection rate of 17.8% for grade 2 tumors and 60% for grade 3 tumors, indicating the high sensitivity of CTC testing in identifying TNBC patients (37). Therefore, CTCs have important value and research prospects for early detection of TNBC.
CTC testing can detect molecular disease recurrence prior to radiographic or clinical manifestations and predict treatment response prior to surgery. This ‘lead time’, allows clinicians to take pre-emptive action earlier in patients who are not responding well to traditional chemotherapy regimens (38). A study of 56 metastatic breast cancer patients (10 of whom had the TNBC subtype) found that the prognosis for PFS was worse in patients with positive CTCs at baseline. Moreover, a favorable prognosis was associated with a ≥70% reduction in CTC count from baseline or fewer than 5 CTCs in 7.5 mL of blood before the second cycle of third-line chemotherapy. These results suggest that early changes in CTCs during systemic therapy may have predictive value for treatment outcome (39). Similarly, Paoletti C. et al. analyzed the number of CTCs, the presence of CTC clusters and the dynamics of apoptotic CTCs in 64 mTNBC patients prior to chemotherapy with nanoparticle albumin-bound paclitaxel or chemotherapy without the addition of trastuzumab. This study showed that a reduction in the number of CTCs was associated with improved PFS in patients and indicated a response to treatment (40). Furthermore, longitudinal CTC assessment by Magbanua et al. revealed that post-treatment CTC counts (days 7-14) better predicted progression risk than baseline values in TNBC patients (41). Consequently, CTC represents a promising tumor marker that can be readily obtained from peripheral blood. It has the potential to become a valuable tool in the future, enabling the refinement of treatment plans for tumors.
Besides its established role in predicting treatment response, accumulating evidence confirms CTCs’ prognostic value in TNBC. Peeters D.J.E. et al. demonstrated no significant difference in CTC counts across breast cancer subtypes, The survival rate and prognosis of breast cancer patients with a CTC count of 5 or more in a 7.5 ml blood sample is significantly worse than that of patients with a CTC count of less than 5 (42). Tsai et al. corroborated this threshold, finding <5 CTCs conferred only 12.5% distant metastasis risk versus ≥5 CTCs (43). Importantly, Jansson et al. reported TNBC patients with CTC clusters within 3 months post-diagnosis exhibited significantly shorter PFS/OS versus cluster-negative cases, though this difference attenuated by 6 months (44). The ability of CTCs to predict the prognosis of TNBC has shown good results, allowing physicians to adjust treatment plans for TNBC patients to prolong their survival. The use of CTCs as a tumor marker, which can be readily obtained from peripheral blood, may become more prevalent in the future, particularly in guiding the diagnosis and treatment of tumors.
2.4 Integrated single-cell omics for CTC characterization
Single-cell technologies provide unprecedented resolution for profiling circulating tumor cells (CTCs) in breast cancer, enabling deep insights into tumor heterogeneity and metastasis mechanisms. Single-cell RNA sequencing (scRNA-seq) deciphers gene expression at cellular resolution, distinguishing breast cancer subtypes and identifying subpopulations linked to poor prognosis and drug resistance (45). Integrated genomic/epigenomic analyses track clonal evolution and genetic instability driving metastatic spread (46). Additionally, single-cell proteomics maps protein networks underlying functional heterogeneity and therapy resistance. These approaches synergistically enhance CTC detection sensitivity. CTC quantities and molecular features (e.g., EMT phenotypes) correlate with early metastasis risk and survival outcomes (47). Clinically, scRNA-seq of CTCs identifies resistant subpopulations, enabling real-time therapy adjustments, while CTC genomic profiles guide personalized regimens. Moving forward, enhancing detection specificity and advancing multi-omics integration will maximize clinical translation.
3 ctDNA
3.1 Biological origins of ctDNA
The existence of tumor-derived cell-free DNA (cfDNA) in the blood was proposed by Stroun et al. in 1989 (47). ctDNA is a type of cfDNA shed by apoptotic or necrotic tumor cells and is derived from the tumor cells themselves (48). Tumor cells consist of malignant cells and cells of the tumor microenvironment, such as stromal cells, endothelial cells, lymphocytes and other immune cells. All of these cells can be a potential source of ctDNA (49). ctDNA can even originate from CTCs in the blood (50). ctDNA fragments are predominantly double-stranded with a length of approximately 132–145 base pairs (51). They have a short half-life of approximately 2 hours. In addition, the tumor–molecular complex binding or the type and stage of the tumor, treatment and other factors can affect the amount and nature of ctDNA (52). ctDNA can collect tumor fragments from multiple lesions in the body, with less spatial variation than needle biopsy, and can better capture heterogeneity (53). Due to these characteristics, ctDNA has the potential to reflect the occurrence and evolution of tumors in the body in real time. The real-time detection of ctDNA has been the subject of considerable interest and has established ctDNA as a promising tumor marker.
3.2 Evolution of ctDNA detection technologies
ctDNA fragments are predominantly double-stranded fragments of approximately 132–145 base pairs with a short half-life of approximately 2 hours (54). In cancer patients, 1 mL of plasma contains approximately 1,500 diploid genome equivalents (GE), equivalent to ~10 ng of DNA. From 10 mL of blood, roughly 4 mL of plasma can be extracted, yielding about 6,000 GE. This sets a theoretical sensitivity limit of 0.1% for mutation detection (54), posing significant challenges for developing reliable assays. Digital PCR (dPCR) addresses this limitation. Pioneered by Vogelstein & Kinzler (55), it transforms exponential PCR amplification into linear quantification. The method partitions DNA into microdroplets for endpoint amplification, with target concentration calculated from ratios of fluorescence-positive droplets. Next-generation sequencing (NGS) offers distinct advantages as a non-targeted approach, detecting emerging alterations—including copy number variants, structural rearrangements, and single nucleotide substitutions. However, NGS faces sensitivity constraints: its typical 30-100x coverage in whole exome/genome sequencing struggles to identify mutations below 5% allele frequency (56). Although the non-targeted approach is less sensitive than the targeted approach, one of its potential advantages is that it can detect subclonal mutations that are less common in the primary tumor but selected during adjuvant therapy or the natural disease course after surgery (57).
3.3 Clinical utility of ctDNA in TNBC management
Currently, neoadjuvant chemotherapy remains the primary systemic pharmacological intervention for patients with TNBC. Given the favorable treatment response observed in patients with early-stage TNBC, it is of paramount importance to implement effective strategies for the early detection of this disease (58). A comparable study conducted by Olsson and colleagues demonstrated that serial ctDNA monitoring in patients with primary breast cancer can yield an average lead time of up to 11 months before the onset of metastatic disease (59). Garcia-Murillas et al. conducted a prospective study in 55 high-risk early-stage breast cancer patients treated with neoadjuvant chemotherapy and surgery. By designing patient-specific ddPCR assays for tumor mutations, they achieved an 80% sensitivity for relapse prediction on (60). In a larger multicenter cohort of 101 breast cancer patients (including TNBC), periodic ddPCR monitoring revealed that ctDNA detection during follow-up was strongly associated with relapse and preceded clinical relapse by a median of 10.7 months. Together, these studies underscore ctDNA’s ability to reveal minimal residual disease (MRD) and pre-empt clinical recurrence by many months, creating a potential window for therapeutic intervention (61).
Additionally, ctDNA represents a valuable tool for predicting the treatment effect, recurrence rate, and survival rate of TNBC patients. In metastatic TNBC, ctDNA demonstrates 81% sensitivity and 97% concordance with tissue biopsy for mutation profiling (62). Consequently, ctDNA testing can be employed as a non-invasive and real-time method for the detection of metastatic TNBC progression, thereby facilitating the timely adjustment of treatment plans. Furthermore, a study conducted by Francesca Riva and colleagues on 46 patients with non-metastatic TNBC demonstrated a significant correlation between ctDNA levels and several clinical parameters, including continuous variation, mitotic index, tumor grade and tumor stage. It is thus feasible to more accurately monitor the patient’s treatment response in real time during neoadjuvant chemotherapy (NCT). Furthermore, this method is also applicable to postoperative treatment (63). In a clinical study of 196 early TNBC patients, Milan Radovich and colleagues observed a significant decline in distant disease-free survival (DDFS) among patients with ctDNA detected in the blood following neoadjuvant chemotherapy. The 24-month DDFS rate for patients with ctDNA positivity was 56%, while that for patients with ctDNA negativity was as high as 81%. A similar trend was observed with respect to disease-free survival (DFS) and overall survival (OS) (64). Residual cancer burden (RCB) stratification further reveals escalating ctDNA positivity rates (RCB-I:14%, RCB-II:31%, RCB-III:57%) coupled with declining 3-year event-free survival (EFS: 100%/79%/23%) (65). Furthermore, digital pCR mutation tracking demonstrated that the median recurrence time for TNBC is 10.6 months, indicating a high propensity for recurrence in the short term (66). Consequently, the real-time detection of ctDNA enables the implementation of timely interventions to enhance the prognosis and prolong the survival period in instances where ctDNA is identified, showing the potential to link disease progression to one quantitative and non-invasive method in TNBC.
However, clonal hematopoiesis of indeterminate potential (CHIP) confounds cfDNA-based liquid biopsy analysis, as somatic mutations in genes including DNMT3A, TET2, ASXL1, PPM1D, and TP53 from age-associated hematopoietic clones can mimic tumor-derived variants (67). In breast cancer cohorts, ≈15% of patients harbor CHIP mutations at diagnosis, with chemotherapy promoting expansion of TP53-mutant clones (variant allele frequency ≥0.5%). Metastatic triple-negative breast cancer (TNBC) exhibits comparable CHIP prevalence without demonstrable survival impact, necessitating rigorous discrimination between leukocyte-derived DNA and true tumor ctDNA (68). Effective mitigation combines three core strategies: concurrent sequencing of matched peripheral blood leukocytes, application of <5% variant allele frequency (VAF) thresholds for CHIP exclusion, and bioinformatic classifiers trained on mutational signatures, collectively enhancing ctDNA analysis specificity for clinical deployment.
4 miRNAs
4.1 Biogenesis and functions of miRNAs
It is estimated that approximately 75% of the human genes are transcribed into RNA, of which only about 3% are subsequently translated into proteins (69). Non-coding RNAs (ncRNAs) are classified by their length, structure, and subcellular localization. The most notable types of ncRNA include miRNA, long ncRNA (lncRNA), circular RNA (circRNA) and PIWI-interacting RNA (piRNA); each has distinct functions in cancer (70). miRNA is a type of small non-coding RNA (sncRNA) that is approximately 22 nucleotides in length. It has the capacity to influence the progression of tumors by post-transcriptionally targeting oncogenes or tumor suppressors (71). It has been demonstrated that the upregulation of miRNA has a role in the promotion of carcinogenesis, while the downregulation of certain miRNA has been identified as a potential antitumor agent. Aberrant expression of these molecules has been linked to the regulation of critical biological processes, including the ABC transporter, cell death, cell cycle, and signaling pathways, which are associated with tumor development, including the progression, metastasis, and chemoresistance of TNBC (72).
4.2 Technical challenges in miRNA detection
miRNA is characterized by their short length and natural occurrence at low abundance in biological samples, which necessitates the employment of highly sensitive detection methods. Reverse transcription-quantitative PCR (RT-qPCR) is the gold standard for miRNA quantification (73). RT-qPCR is characterized by its high sensitivity and specificity, and is widely regarded as the gold standard for the detection of miRNAs (74). However, RT-qPCR analysis of miRNAs necessitates a multitude of sample preparation steps and the utilization of various enzymes, thereby augmenting the cost and complexity of the analysis (75). Currently, next-generation sequencing (NGS) and DNA microarrays are also available for the detection of miRNAs. NGS has the capacity to detect multiple targets, a property that is valuable for the discovery of novel miRNAs and comprehensive analysis. Nevertheless, due to the time-consuming and costly nature of next-generation sequencing technology, its use in rapid clinical testing and large-scale applications in early diagnosis is not feasible (76). DNA microarrays are extensively utilized for the detection and diagnosis of miRNAs, as well as target selection. The advantages of DNA microarrays include their relatively low cost, high throughput, and the capacity for multiplex detection. Nevertheless, traditional DNA microarrays are constrained by protracted detection times and intricate operating procedures, encompassing PCR amplification, target labelling and washing steps. This is particularly challenging for miRNAs, a type of short-chain RNA, due to the complex and time-consuming nature of amplification and fluorescent labelling (77). Chen et al. established a fluorescent detection method based on SCas12a RNP, which has been demonstrated to be capable of detecting miRNAs with high sensitivity, high selectivity and multiplexing, without reverse transcription or preamplification (78). Haruka et al. established a signal probe-based hybridization system for simple PCR, target and cleaning-free detection of miRNAs. The signal probe has been demonstrated to be capable of detecting a variety of different miRNAs, encompassing those of varying lengths and GC contents, including those with high GC content. The detection method has the potential for use in liquid biopsy and is expected to achieve economical, high-throughput and multiplex detection of miRNAs (79).
4.3 miRNA signatures in TNBC diagnosis
As a result of the application of high-throughput sequencing technology, the number of mature miRNAs discovered has now exceeded 28,000. This represents between 1% and 5% of the human genome. Kumar et al. conducted a study in which the expression of microRNAs was analyzed based on qRT-PCR. The results of this study demonstrated that the expression levels of miR-155 and miR-21 were significantly elevated in patients diagnosed with TNBC, while the expression levels of miRNA-205 were markedly reduced. The disruption of these miRNA expressions can thus be used as an independent indicator to distinguish TNBC from healthy patients (80). Kalani et al. developed a HJ and FRET-based single-molecule sensor capable of detecting TNBC-related miRNAs (miR-342-3p) with high sensitivity and specificity. It is anticipated that this sensor will serve as a novel approach for the early diagnosis of TNBC through the analysis of body fluids (21).
In comparison with other tissues, the high expression of miRNA in TNBC tissue can be utilized as a reference for the diagnosis of TNBC. Hou et al. found that the expression of miRNA-1207-5p was significantly increased in tissue samples of TNBC compared to adjacent normal tissue. Furthermore, they found that the expression of miR-1207-5p was significantly higher in TNBC tissue non-responsive to Taxol compared to Taxol-responsive (81). It is hypothesized that miR-1207-5p has the potential to serve as a novel biomarker for the diagnosis of TNBC.
4.4 Prognostic and therapeutic potential
miRNA has been demonstrated to be associated with the malignant characteristics of TNBC, including proliferation, migration, metastasis and drug resistance. Telomeric repeat-binding factor 2 (TRF2) has been observed to be overexpressed in a variety of human cancers, including breast cancer, and has been demonstrated to promote cancer cell escape (82). Dinami et al. found that the ectopic expression of miR-182-3p significantly reduced levels of the TRF2 protein, thereby activating DNA damage. Therefore, measuring miR-182-3p levels can serve as an indicator to assess apoptosis in TNBC cells. Furthermore, lipid nanoparticles (LNPs) containing miR-182-3p (LNPs-miR-182-3p) has been demonstrated to facilitate the crossing of the blood-brain barrier, resulting in the reduction of intracranial tumors. These finding offers novel therapeutic prospects for the management of metastatic brain disease (83).
Differential miRNA expression patterns stratify TNBC progression and treatment response. Berber et al. identified suppressed miR-200c and miR-205 as predictors of lymph node metastasis (84), while Song et al. associated miR-1-3p and miR-133a-3p with overall prognosis (85). For chemotherapy response, Li et al. identified miR-770 overexpression as a key sensitivity indicator through transcriptome analysis—a role further validated in TCGA datasets for prognostic stratification (86). Survival-linked miRNAs further refine risk assessment: Deng et al. reported extended median survival (77 vs. 70 months) with high miR-221-3p expression, independent of clinicopathological variables (87), and Svoboda et al. demonstrated negative correlations between miR-34b levels and disease-free/overall survival (88). It is evident from the aforementioned markers that miRNA has the potential to serve as valuable prognostic markers for TNBC. It has been established that a significant proportion of miRNAs are expressed at elevated levels in TNBC, with these molecules demonstrating a strong correlation with the malignant characteristics of tumors. Consequently, these miRNAs have the potential to serve as biomarkers for liquid biopsy TNBC.
5 lncRNAs
5.1 Biological roles of lncRNAs in TNBC
lncRNA constitute a subclass of non-coding ribonucleic acid (ncRNA) characterized by a sequence length of more than 200 nucleotides and little or no protein-coding potential (89). LncRNA has been found to be present in the cell membrane or nuclear region, interspersed and overlapping regions of coding and non-coding transcripts. These molecules possess a variety of molecular functions and roles, including acting as scaffolds, decoys, or guides; modulating signaling pathways; organizing chromatin architecture; and responding to developmental or environmental stimuli (90). Aberrant expression levels of lncRNA expression is associated with a multitude of malignant biological processes, including genes, proliferation, angiogenesis, EMT and distant metastasis (91). A plethora of studies have demonstrated that aberrant expression of lncRNA plays a pivotal role in the pathogenesis, progression and metastasis of TNBC. Thus, lncRNAs hold significant promise for TNBC liquid-biopsy applications.
5.2 lncRNA in TNBC diagnosis
LncRNAs demonstrate functional correlations with diverse regulatory mechanisms—including transcription factor modulation, epigenetic alterations, and post-translational modifications—alongside interactions with small peptides, collectively underpinning their clinical relevance for early TNBC diagnosis and therapeutic intervention (92). Detection of lncRNAs generally parallels miRNA workflows. Swellam et al. utilized qRT-PCR to detect the expression of X inactive specific transcript (XIST) and nu-clear paraspeckle assembly transcript 1 (NEAT1) in serum samples from breast cancer patients, patients with benign breast lesions and healthy volunteers, respectively. Their findings revealed significantly elevated levels in breast cancer patients versus benign/healthy controls, with further increases in TNBC subgroups, establishing these lncRNAs as diagnostic biomarkers (93). Furthermore, Lv et al. utilized microarray detection to identify 880 upregulated and 784 downregulated lncRNAs in TNBC versus non-TNBC with ROC validation confirming RP11-434D9.1, LINC00052, BC016831, and IGKV as diagnostic indicators (94).
5.3 lncRNA in the treatment and prognosis of TNBC
LncRNAs play pivotal roles in regulating TNBC proliferation, apoptosis, and drug resistance (95). Zheng et al. demonstrated that lncRNA MILIP is highly expressed in p53-mutant TNBC cells, where silencing suppresses cell viability and xenograft growth. Mechanistically, MILIP interacts with eukaryotic elongation factor 1 alpha 1 (eEF1a1) to form RNA-RNA duplex with the type II tRNAs tRNALeu and tRNASer through their variable loops. This interaction promotes tRNA binding and protein synthesis. Disruption of this interaction reduces cellular viability (96). Parallel studies revealed MALAT1 upregulation in TNBC tissues (43 paired samples) enhances proliferation, invasion and cell cycle progression via the MALAT1/miR-129-5p axis (97). Wang YF et al. found that HIF1A-AS2 expression was significantly higher in TNBC cell lines than in normal breast epithelial cell lines. The findings indicated that elevated HIF1A-AS2 expression was associated with lymph node metastasis, distant metastasis and poor histological grading in patients with TNBC. Furthermore, silencing HIF1A-AS2 expression led to a significant inhibition of the migration and invasion of TNBC cells (98). Emerging evidence reveals that LINK-A mediates drug resistance through PKA-TRIM71 axis activation, compromising immune checkpoint inhibitor efficacy (99). Complementary research shows dual silencing of HIF1A-AS2 and AK124454 in TNBC cells elevates paclitaxel half-maximal inhibitory concentration (IC50) >2-fold. This IC50 increase directly indicates reduced paclitaxel sensitivity, confirming taxane resistance association. Furthermore, the establishment of a drug resistance prediction model based on these two genes has enabled the development of personalized treatment regimens for patients with TNBC (100). The aforementioned markers facilitate the real-time monitoring of TNBC development and metastasis, with significant implications for the monitoring and adjustment of treatment efficacy.
He et al. found that OSTN-AS1 serves as an immune-related biomarker, while T376626 levels correlate with advanced stages, aggressive molecular subtypes, and poor survival (101). Subsequent analysis confirmed elevated LINK-A predicts reduced relapse-free survival (102). In summary, the detection of lncRNAs in blood samples has become a promising method for monitoring the progression of TNBC and for prognostic prediction, which can be incorporated into the clinical management of patients.
6 Other biomarkers
6.1 Exosomes
Exosomes are 40–160 nm in diameter and consist of a lipid bilayer. These vesicles are released by most cells and circulate stably in body fluids (103). Exosomes are rich in a variety of biologically active molecules, including nucleic acids, proteins and lipids, which can be transferred from donor to recipient cells, facilitating intracellular information transmission (104). Hoshino et al. demonstrated plasma-derived exosome analysis enables cancer-type identification and tumor-origin determination via proteomics (105). Yu et al. found that acetylated LAP-TGF-β1 enters cells via exosomes, accelerating lung metastasis and spread, thereby establishing exosomal TGF-β1 as a promising therapeutic target for metastasis (106). In the study by Zhao et al., it was established that cationic bovine serum albumin (CBSA) conjugated with siS100A4 and exosome membrane-coated biomimetic nanoparticles (CBSA/siS100A4@Exosome) can significantly inhibit the postoperative metastasis of triple-negative breast cancer (107). Consequently, the delivery of siS100A4 by exosome membrane-coated core-shell nanoparticles is anticipated to emerge as a promising clinical strategy for cancer prevention and treatment.
6.2 Circular RNA
Circular RNA (circRNA) is a class of endogenous RNA transcripts that feature covalently closed loop structures. In comparison with linear RNA, circRNA is devoid of 5’ caps or 3’ tails, and is distinguished by its enhanced half-life, elevated evolutionary conservation, and augmented resistance to digestion by RNase R (108). In the study by Wang et al., it was found that the expression of circRNA-CREIT (hsa_circ_0001798) was significantly reduced in breast cancer, particularly in chemo resistant breast cancer cells and the TNBC subtype. Additionally, low expression of circRNA-CREIT was found to be associated with higher grade, increased lymph node metastasis, larger tumor volume, and poorer prognosis in breast cancer patients. Furthermore, circRNA-CREIT has been demonstrated to possess the capability to impede the proliferation and migration of TNBC cells, whilst concomitantly inducing apoptosis (109). It was previously reported that circHIF1A promotes the proliferation and metastasis of TNBC by upregulating NFIB (110). The aforementioned markers indicate a strong association between circRNA and the metastasis and prognosis of TNBC, suggesting its potential as a novel and valuable marker.
6.3 Tumor-associated neutrophils
Neutrophils, also referred to as polymorphonuclear cells, are a type of granulocyte belonging to the myeloid lineage. These cells represent the most abundant white blood cell type, and they are the most significant immune cells, comprising 50–70% of white blood cells in adults (111). Neutrophils have been shown to play a role in bacterial infections and are also actively involved in various aspects of breast cancer development, including growth, migration/invasion, angiogenesis and metastasis (112). Tumor-associated neutrophils (TANs) are gaining recognition as a significant functional attribute that may influence the prognosis of breast cancer, particularly in the context of the TNBC subtype (113). Breast cancer patients, including TNBC patients, with CTC-neutrophil clusters have worse progression-free survival than those without clustered CTC. Circulating neutrophils confer higher malignant potential to CTCs (114). Furthermore, Wang et al. discovered that elevated levels of tumor-infiltrating neutrophils (TINs) were associated with advanced histological grade, tumor stage, and the TNBC subtype. Mechanistically, parenchymal TINs promote breast cancer cell migration, invasion, and EMT through TIMP-1 secretion in a CD90-mediated cell-contact dependent manner (115). Consequently, CTC-neutrophil clusters show diagnostic potential, while parenchymal TIN density serves as a validated prognostic indicator for TNBC.
7 Conclusion and future perspectives
Liquid biopsy represents a paradigm-shifting approach in triple-negative breast cancer (TNBC) management, offering non-invasive access to molecular information through blood/urine biomarkers including CTCs, ctDNA, miRNAs, and lncRNAs (Figure 1). This technology enables dynamic monitoring of tumor evolution, facilitating early diagnosis, treatment response assessment, recurrence surveillance, and personalized therapy selection—addressing TNBC’s critical unmet need for targeted therapeutic strategies.
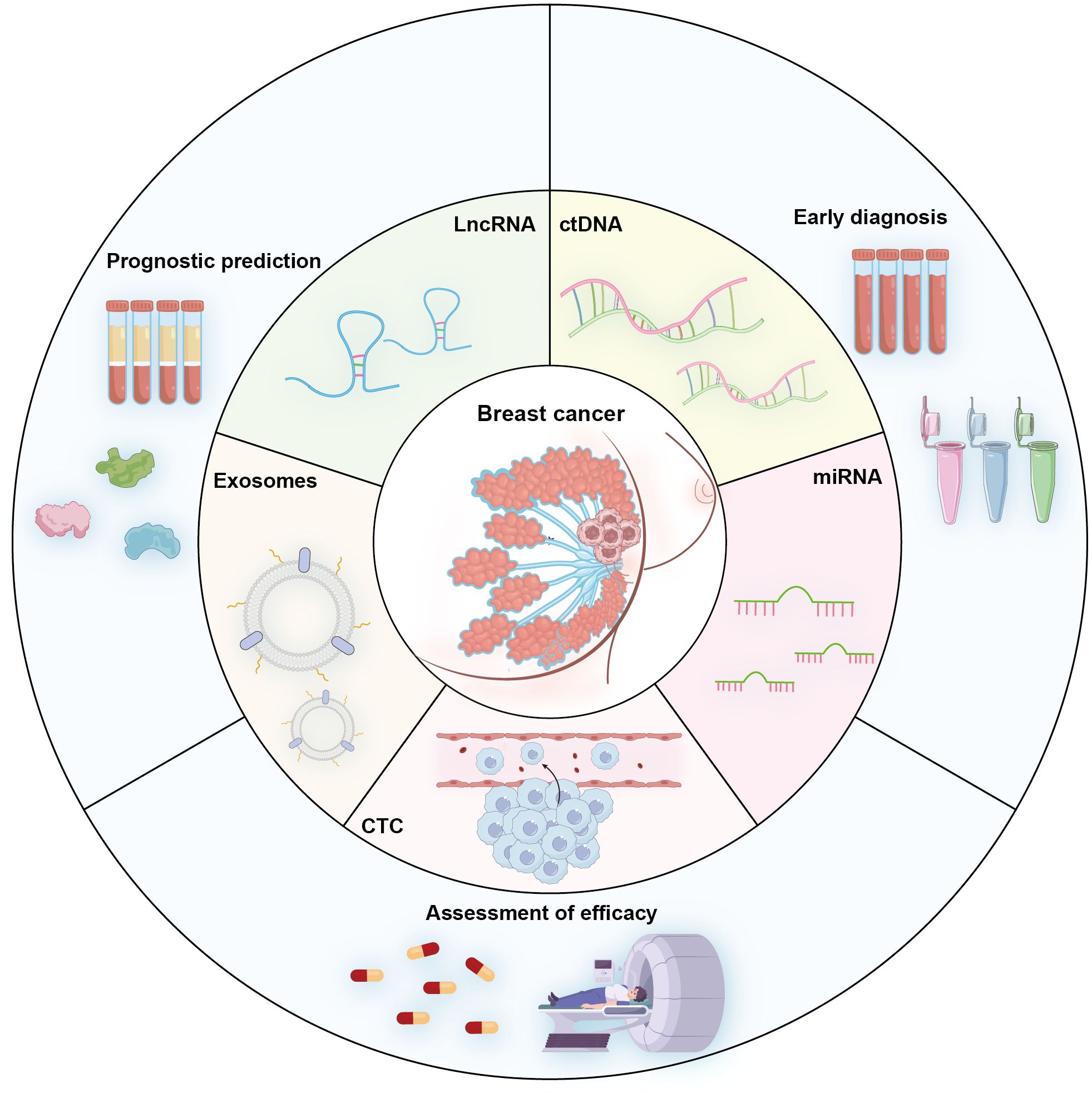
Figure 1. Schematic diagram illustrating key circulating analytes used in TNBC liquid biopsy and their primary clinical utilities including Early diagnosis (screening and detection of initial disease), Prognostic prediction (risk stratification and outcome forecasting), and Evaluation of Treatment Efficacy (monitoring response to therapy and minimal residual disease).
The future development of TNBC liquid biopsy centers on four interconnected priorities. First, integrating multi-analyte signatures through multiplexed detection platforms will enhance diagnostic precision by combining complementary biomarkers into comprehensive panels (Table 1). Second, technological breakthroughs in ultrasensitive detection methodologies—such as advanced digital PCR systems, nanopore sequencing, and CRISPR-based diagnostics—promise to overcome current sensitivity limitations by identifying rare variants below 0.1% frequency thresholds, enabling earlier interception of minimal residual disease and pre-symptomatic recurrence.
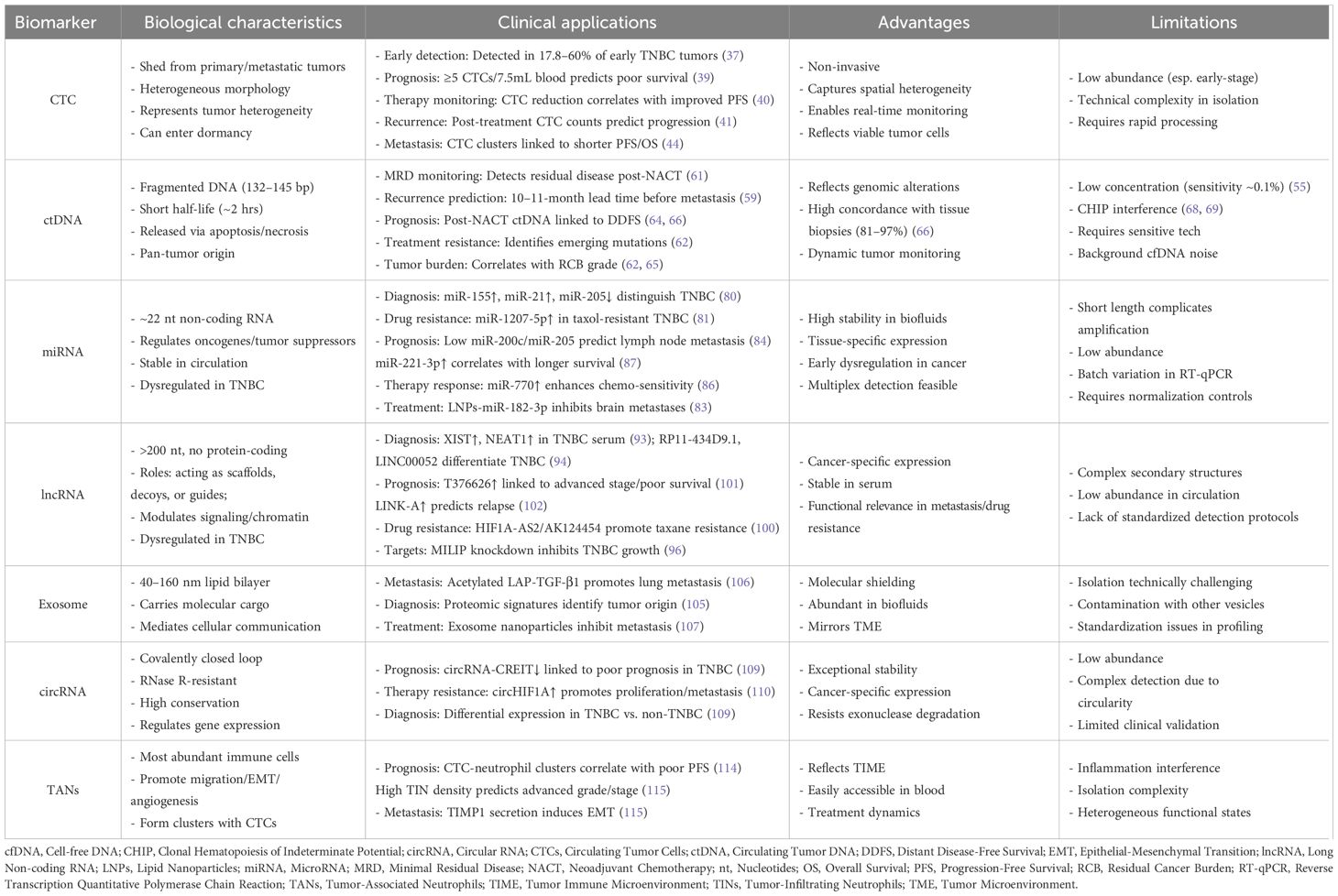
Table 1. Liquid biopsy biomarkers in TNBC: biological characteristics, clinical applications, and technical considerations.
Third, expanding the precision medicine framework requires exploring population-specific biomarker signatures across diverse ethnicities, geographic regions, and age cohorts to address TNBC’s intrinsic molecular heterogeneity. This necessitates merging artificial intelligence with liquid biopsy data to decode complex biomarker patterns while integrating radiomic features from MRI, CT, and PET imaging to quantify tumor burden and spatial heterogeneity.
Finally, accelerating clinical translation demands validating liquid biopsy in prospective trials comparing biopsy-guided interventions versus standard care protocols, developing point-of-care platforms for global accessibility, and establishing international standardization for analytical validation. These foundations will enable liquid biopsy to evolve as an essential companion diagnostic, particularly as novel TNBC therapeutics including immune checkpoint inhibitors, PARP inhibitors, and antibody-drug conjugates enter clinical practice.
Ultimately, the convergence of these advances will transform TNBC management from episodic tissue sampling to continuous molecular monitoring, ushering in an era where real-time liquid biopsy data dynamically navigates precision treatment decisions throughout the patient journey.
Author contributions
JS: Writing – original draft. XZ: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Siegel RL, Miller KD, Fuchs HE, and Jemal A. Cancer statistics, 2022. CA Cancer J Clin. (2022) 72:7–33. doi: 10.3322/caac.21708
2. Foulkes WD, Smith IE, and Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. (2010) 363:1938–48. doi: 10.1056/NEJMra1001389
3. Derakhshan F and Reis-Filho JS. Pathogenesis of triple-negative breast cancer. Annu Rev Pathol. (2022) 17:181–204. doi: 10.1146/annurev-pathol-042420-093238
4. Agarwal G, Nanda G, Lal P, Mishra A, Agarwal A, Agrawal V, et al. Outcomes of triple-negative breast cancers (Tnbc) compared with non-tnbc: does the survival vary for all stages? World J Surg. (2016) 40:1362–72. doi: 10.1007/s00268-016-3422-4
5. Lin NU, Vanderplas A, Hughes ME, Theriault RL, Edge SB, Wong YN, et al. Clinicopathologic features, patterns of recurrence, and survival among women with triple-negative breast cancer in the national comprehensive cancer network. Cancer. (2012) 118:5463–72. doi: 10.1002/cncr.27581
6. von Minckwitz G, Untch M, Blohmer JU, Costa SD, Eidtmann H, Fasching PA, et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol. (2012) 30:1796–804. doi: 10.1200/JCO.2011.38.8595
7. Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. (2008) 26:1275–81. doi: 10.1200/JCO.2007.14.4147
8. Nanda R, Chow LQ, Dees EC, Berger R, Gupta S, Geva R, et al. Pembrolizumab in patients with advanced triple-negative breast cancer: phase ib keynote-012 study. J Clin Oncol. (2016) 34:2460–7. doi: 10.1200/JCO.2015.64.8931
9. Leon-Ferre RA and Goetz MP. Advances in systemic therapies for triple negative breast cancer. BMJ. (2023) 381:e071674. doi: 10.1136/bmj-2022-071674
10. Bardelli A and Pantel K. Liquid biopsies, what we do not know (yet). Cancer Cell. (2017) 31:172–9. doi: 10.1016/j.ccell.2017.01.002
11. Riethdorf S, O’Flaherty L, Hille C, and Pantel K. Clinical applications of the cellsearch platform in cancer patients. Adv Drug Delivery Rev. (2018) 125:102–21. doi: 10.1016/j.addr.2018.01.011
12. Salati M, Orsi G, Smyth E, Aprile G, Beretta G, De Vita F, et al. Gastric cancer: translating novels concepts into clinical practice. Cancer Treat Rev. (2019) 79:101889. doi: 10.1016/j.ctrv.2019.101889
13. Kilgour E, Rothwell DG, Brady G, and Dive C. Liquid biopsy-based biomarkers of treatment response and resistance. Cancer Cell. (2020) 37:485–95. doi: 10.1016/j.ccell.2020.03.012
14. Luo H, Wei W, Ye Z, Zheng J, and Xu RH. Liquid biopsy of methylation biomarkers in cell-free DNA. Trends Mol Med. (2021) 27:482–500. doi: 10.1016/j.molmed.2020.12.011
15. Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. Microrna mir-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. (2008) 14:2348–60. doi: 10.1261/rna.1034808
16. Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, et al. Rna-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. (2015) 28:666–76. doi: 10.1016/j.ccell.2015.09.018
17. Lin PP. Aneuploid circulating tumor-derived endothelial cell (Ctec): A novel versatile player in tumor neovascularization and cancer metastasis. Cells. (2020) 9. doi: 10.3390/cells9061539
18. Barnell EK, Fisk B, Skidmore ZL, Cotto KC, Basu A, Anand A, et al. Personalized ctdna micro-panels can monitor and predict clinical outcomes for patients with triple-negative breast cancer. Sci Rep. (2022) 12:17732. doi: 10.1038/s41598-022-20928-8
19. Eghlimi R, Shi X, Hrovat J, Xi B, and Gu H. Triple negative breast cancer detection using lc-ms/ms lipidomic profiling. J Proteome Res. (2020) 19:2367–78. doi: 10.1021/acs.jproteome.0c00038
20. Turner NC, Swift C, Jenkins B, Kilburn L, Coakley M, Beaney M, et al. Results of the C-trak tn trial: A clinical trial utilising ctdna mutation tracking to detect molecular residual disease and trigger intervention in patients with moderate- and high-risk early-stage triple-negative breast cancer. Ann Oncol. (2023) 34:200–11. doi: 10.1016/j.annonc.2022.11.005
21. Wijesinghe KM, Kanak MA, Harrell JC, and Dhakal S. Single-molecule sensor for high-confidence detection of mirna. ACS Sens. (2022) 7:1086–94. doi: 10.1021/acssensors.1c02748
22. Zhang K, Liu Z, Yao Y, Qiu Y, Li F, Chen D, et al. Structure-based design of a selective class I histone deacetylase (Hdac) near-infrared (Nir) probe for epigenetic regulation detection in triple-negative breast cancer (Tnbc). J Med Chem. (2021) 64:4020–33. doi: 10.1021/acs.jmedchem.0c02161
23. Oliveira MM and Klann E. A deep dive into local mrna translation in neurons. Proc Natl Acad Sci U S A. (2021) 118. doi: 10.1073/pnas.2117116118
24. Parkinson DR, Dracopoli N, Petty BG, Compton C, Cristofanilli M, Deisseroth A, et al. Considerations in the development of circulating tumor cell technology for clinical use. J Transl Med. (2012) 10:138. doi: 10.1186/1479-5876-10-138
25. de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. (2008) 14:6302–9. doi: 10.1158/1078-0432.CCR-08-0872
26. Lawrence R, Watters M, Davies CR, Pantel K, and Lu YJ. Circulating tumour cells for early detection of clinically relevant cancer. Nat Rev Clin Oncol. (2023) 20:487–500. doi: 10.1038/s41571-023-00781-y
27. Sun YF, Yang XR, Zhou J, Qiu SJ, Fan J, and Xu Y. Circulating tumor cells: advances in detection methods, biological issues, and clinical relevance. J Cancer Res Clin Oncol. (2011) 137:1151–73. doi: 10.1007/s00432-011-0988-y
28. Igetei R, Otegbayo JA, Ndububa DA, Lesi OA, Anumudu CI, Hainaut P, et al. Detection of P53 codon 249 mutation in Nigerian patients with hepatocellular carcinoma using a novel evaluation of cell-free DNA. Ann Hepatol. (2008) 7:339–44. doi: 10.1016/s1665-2681(19)31834-4
29. Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. (2010) 2:20ra14. doi: 10.1126/scitranslmed.3000702
30. Yu M, Stott S, Toner M, Maheswaran S, and Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. (2011) 192:373–82. doi: 10.1083/jcb.201010021
31. Rawal S, Yang YP, Cote R, and Agarwal A. Identification and quantitation of circulating tumor cells. Annu Rev Anal Chem (Palo Alto Calif). (2017) 10:321–43. doi: 10.1146/annurev-anchem-061516-045405
32. Gkountela S and Aceto N. Stem-like features of cancer cells on their way to metastasis. Biol Direct. (2016) 11:33. doi: 10.1186/s13062-016-0135-4
33. Ao Z, Shah SH, Machlin LM, Parajuli R, Miller PC, Rawal S, et al. Identification of cancer-associated fibroblasts in circulating blood from patients with metastatic breast cancer. Cancer Res. (2015) 75:4681–7. doi: 10.1158/0008-5472.CAN-15-1633
34. Alix-Panabieres C and Pantel K. Liquid biopsy in cancer patients: advances in capturing viable ctcs for functional studies using the epispot assay. Expert Rev Mol Diagn. (2015) 15:1411–7. doi: 10.1586/14737159.2015.1091729
35. Suhail Y, Cain MP, Vanaja K, Kurywchak PA, Levchenko A, Kalluri R, et al. Systems biology of cancer metastasis. Cell Syst. (2019) 9:109–27. doi: 10.1016/j.cels.2019.07.003
36. Braun S, Vogl FD, Naume B, Janni W, Osborne MP, Coombes RC, et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. (2005) 353:793–802. doi: 10.1056/NEJMoa050434
37. Agelaki S, Dragolia M, Markonanolaki H, Alkahtani S, Stournaras C, Georgoulias V, et al. Phenotypic characterization of circulating tumor cells in triple negative breast cancer patients. Oncotarget. (2017) 8:5309–22. doi: 10.18632/oncotarget.14144
38. Chen JH, Addanki S, Roy D, Bassett R, Kalashnikova E, Spickard E, et al. Monitoring response to neoadjuvant chemotherapy in triple negative breast cancer using circulating tumor DNA. BMC Cancer. (2024) 24:1016. doi: 10.1186/s12885-024-12689-6
39. Helissey C, Berger F, Cottu P, Dieras V, Mignot L, Servois V, et al. Circulating tumor cell thresholds and survival scores in advanced metastatic breast cancer: the observational step of the circe01 phase iii trial. Cancer Lett. (2015) 360:213–8. doi: 10.1016/j.canlet.2015.02.010
40. Paoletti C, Li Y, Muniz MC, Kidwell KM, Aung K, Thomas DG, et al. Significance of circulating tumor cells in metastatic triple-negative breast cancer patients within a randomized, phase ii trial: tbcrc 019. Clin Cancer Res. (2015) 21:2771–9. doi: 10.1158/1078-0432.CCR-14-2781
41. Magbanua MJ, Carey LA, DeLuca A, Hwang J, Scott JH, Rimawi MF, et al. Circulating tumor cell analysis in metastatic triple-negative breast cancers. Clin Cancer Res. (2015) 21:1098–105. doi: 10.1158/1078-0432.CCR-14-1948
42. Peeters DJ, van Dam PJ, Van den Eynden GG, Rutten A, Wuyts H, Pouillon L, et al. Detection and prognostic significance of circulating tumour cells in patients with metastatic breast cancer according to immunohistochemical subtypes. Br J Cancer. (2014) 110:375–83. doi: 10.1038/bjc.2013.743
43. Tsai WS, Chen JS, Shao HJ, Wu JC, Lai JM, Lu SH, et al. Circulating tumor cell count correlates with colorectal neoplasm progression and is a prognostic marker for distant metastasis in non-metastatic patients. Sci Rep. (2016) 6:24517. doi: 10.1038/srep24517
44. Jansson S, Bendahl PO, Larsson AM, Aaltonen KE, and Ryden L. Prognostic impact of circulating tumor cell apoptosis and clusters in serial blood samples from patients with metastatic breast cancer in a prospective observational cohort. BMC Cancer. (2016) 16:433. doi: 10.1186/s12885-016-2406-y
45. Yu T, Wang C, Xie M, Zhu C, Shu Y, Tang J, et al. Heterogeneity of ctc contributes to the organotropism of breast cancer. BioMed Pharmacother. (2021) 137:111314. doi: 10.1016/j.biopha.2021.111314
46. Guan X, Ma F, Li C, Wu S, Hu S, Huang J, et al. The prognostic and therapeutic implications of circulating tumor cell phenotype detection based on epithelial-mesenchymal transition markers in the first-line chemotherapy of her2-negative metastatic breast cancer. Cancer Commun (Lond). (2019) 39:1. doi: 10.1186/s40880-018-0346-4
47. Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, and Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. (1989) 46:318–22. doi: 10.1159/000226740
48. Kumaki Y, Olsen S, Suenaga M, Nakagawa T, Uetake H, and Ikeda S. Comprehensive genomic profiling of circulating cell-free DNA distinguishes focal met amplification from aneuploidy in diverse advanced cancers. Curr Oncol. (2021) 28:3717–28. doi: 10.3390/curroncol28050317
49. Thierry AR, El Messaoudi S, Gahan PB, Anker P, and Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. (2016) 35:347–76. doi: 10.1007/s10555-016-9629-x
50. Chen W, Yan H, Li X, Ge K, and Wu J. Circulating tumor DNA detection and its application status in gastric cancer: A narrative review. Transl Cancer Res. (2021) 10:529–36. doi: 10.21037/tcr-20-2856
51. Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, et al. Fragment length of circulating tumor DNA. PloS Genet. (2016) 12:e1006162. doi: 10.1371/journal.pgen.1006162
52. Yao W, Mei C, Nan X, and Hui L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: A qualitative study. Gene. (2016) 590:142–8. doi: 10.1016/j.gene.2016.06.033
53. Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. (2012) 4:136ra68. doi: 10.1126/scitranslmed.3003726
54. Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. (2016) 34:547–55. doi: 10.1038/nbt.3520
55. Holm M, Andersson E, Osterlund E, Ovissi A, Soveri LM, Anttonen AK, et al. Detection of kras mutations in liquid biopsies from metastatic colorectal cancer patients using droplet digital pcr, idylla, and next generation sequencing. PloS One. (2020) 15:e0239819. doi: 10.1371/journal.pone.0239819
56. Pessoa LS, Heringer M, and Ferrer VP. Ctdna as a cancer biomarker: A broad overview. Crit Rev Oncol Hematol. (2020) 155:103109. doi: 10.1016/j.critrevonc.2020.103109
57. Pantel K and Alix-Panabieres C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol. (2019) 16:409–24. doi: 10.1038/s41571-019-0187-3
58. La Belle A, Khatib J, Schiemann WP, and Vinayak S. Role of platinum in early-stage triple-negative breast cancer. Curr Treat Options Oncol. (2017) 18:68. doi: 10.1007/s11864-017-0506-9
59. Olsson E, Winter C, George A, Chen Y, Howlin J, Tang MH, et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med. (2015) 7:1034–47. doi: 10.15252/emmm.201404913
60. Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. (2015) 7:302ra133. doi: 10.1126/scitranslmed.aab0021
61. Cailleux F, Agostinetto E, Lambertini M, Rothé F, Wu H-T, Balcioglu M, et al. Circulating tumor DNA after neoadjuvant chemotherapy in breast cancer is associated with disease relapse. JCO Precis Oncol. (2022) 6):e2200148. doi: 10.1200/po.22.00148
62. Madic J, Kiialainen A, Bidard FC, Birzele F, Ramey G, Leroy Q, et al. Circulating tumor DNA and circulating tumor cells in metastatic triple negative breast cancer patients. Int J Cancer. (2015) 136:2158–65. doi: 10.1002/ijc.29265
63. Riva F, Bidard FC, Houy A, Saliou A, Madic J, Rampanou A, et al. Patient-specific circulating tumor DNA detection during neoadjuvant chemotherapy in triple-negative breast cancer. Clin Chem. (2017) 63:691–9. doi: 10.1373/clinchem.2016.262337
64. Radovich M, Jiang G, Hancock BA, Chitambar C, Nanda R, Falkson C, et al. Association of circulating tumor DNA and circulating tumor cells after neoadjuvant chemotherapy with disease recurrence in patients with triple-negative breast cancer: preplanned secondary analysis of the bre12–158 randomized clinical trial. JAMA Oncol. (2020) 6:1410–5. doi: 10.1001/jamaoncol.2020.2295
65. Stecklein SR, Kimler BF, Yoder R, Schwensen K, Staley JM, Khan QJ, et al. Ctdna and residual cancer burden are prognostic in triple-negative breast cancer patients with residual disease. NPJ Breast Cancer. (2023) 9:10. doi: 10.1038/s41523-023-00512-7
66. Garcia-Murillas I, Chopra N, Comino-Mendez I, Beaney M, Tovey H, Cutts RJ, et al. Assessment of molecular relapse detection in early-stage breast cancer. JAMA Oncol. (2019) 5:1473–8. doi: 10.1001/jamaoncol.2019.1838
67. Marshall CH, Gondek LP, Luo J, and Antonarakis ES. Clonal hematopoiesis of indeterminate potential in patients with solid tumor Malignancies. Cancer Res. (2022) 82:4107–13. doi: 10.1158/0008-5472.Can-22-0985
68. Morganti S, Gibson CJ, Jin Q, Santos K, Patel A, Wilson A, et al. Prevalence, dynamics, and prognostic role of clonal hematopoiesis of indeterminate potential in patients with breast cancer. J Clin Oncol. (2024) 42:3666–79. doi: 10.1200/JCO.23.01071
69. Kimura T. Non-coding natural antisense rna: mechanisms of action in the regulation of target gene expression and its clinical implications. Yakugaku Zasshi. (2020) 140:687–700. doi: 10.1248/yakushi.20-00002
70. Ghildiyal M and Zamore PD. Small silencing rnas: an expanding universe. Nat Rev Genet. (2009) 10:94–108. doi: 10.1038/nrg2504
71. Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, et al. E2f1-regulated micrornas impair tgfβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. (2008) 13:272–86. doi: 10.1016/j.ccr.2008.02.013
72. Iorio MV and Croce CM. Microrna dysregulation in cancer: diagnostics, monitoring and therapeutics. A Compr Rev EMBO Mol Med. (2012) 4:143–59. doi: 10.1002/emmm.201100209
73. Han JG, Jiang YD, Zhang CH, Yang YM, Pang D, Song YN, et al. A novel panel of serum mir-21/mir-155/mir-365 as a potential diagnostic biomarker for breast cancer. Ann Surg Treat Res. (2017) 92:55–66. doi: 10.4174/astr.2017.92.2.55
74. Jinek M and Doudna JA. A three-dimensional view of the molecular machinery of rna interference. Nature. (2009) 457:405–12. doi: 10.1038/nature07755
75. Jayaprakash AD, Jabado O, Brown BD, and Sachidanandam R. Identification and remediation of biases in the activity of rna ligases in small-rna deep sequencing. Nucleic Acids Res. (2011) 39:e141. doi: 10.1093/nar/gkr693
76. Ouyang T, Liu Z, Han Z, and Ge Q. Microrna detection specificity: recent advances and future perspective. Anal Chem. (2019) 91:3179–86. doi: 10.1021/acs.analchem.8b05909
77. Calvo-Lozano O, Garcia-Aparicio P, Raduly LZ, Estevez MC, Berindan-Neagoe I, Ferracin M, et al. One-step and real-time detection of microrna-21 in human samples for lung cancer biosensing diagnosis. Anal Chem. (2022) 94:14659–65. doi: 10.1021/acs.analchem.2c02895
78. Chen Y, Wang X, Zhang J, Jiang Q, Qiao B, He B, et al. Split crrna with crispr-cas12a enabling highly sensitive and multiplexed detection of rna and DNA. Nat Commun. (2024) 15:8342. doi: 10.1038/s41467-024-52691-x
79. Uno H, Takeuchi H, Abe I, Yoshino T, Taguchi T, Hirakawa Y, et al. Pcr- and wash-free detection of serum mirna via signaling probe hybridization. Biotechnol Bioeng. (2024) 122:159–66. doi: 10.1002/bit.28859
80. Kumar V, Gautam M, Chaudhary A, and Chaurasia B. Impact of three mirna signature as potential diagnostic marker for triple negative breast cancer patients. Sci Rep. (2023) 13:21643. doi: 10.1038/s41598-023-48896-7
81. Hou X, Niu Z, Liu L, Guo Q, Li H, Yang X, et al. Mir-1207-5p regulates the sensitivity of triple-negative breast cancer cells to taxol treatment via the suppression of lzts1 expression. Oncol Lett. (2019) 17:990–8. doi: 10.3892/ol.2018.9687
82. El Mai M, Janho Dit Hreich S, Gaggioli C, Roisin A, Wagner N, Ye J, et al. A novel screen for expression regulators of the telomeric protein trf2 identified small molecules that impair trf2 dependent immunosuppression and tumor growth. Cancers (Basel). (2021) 13. doi: 10.3390/cancers13122998
83. Dinami R, Pompili L, Petti E, Porru M, D’Angelo C, Di Vito S, et al. Mir-182-3p targets trf2 and impairs tumor growth of triple-negative breast cancer. EMBO Mol Med. (2023) 15:e16033. doi: 10.15252/emmm.202216033
84. Berber U, Yilmaz I, Narli G, Haholu A, Kucukodaci Z, and Demirel D. Mir-205 and mir-200c: predictive micro rnas for lymph node metastasis in triple negative breast cancer. J Breast Cancer. (2014) 17:143–8. doi: 10.4048/jbc.2014.17.2.143
85. Song X, Zhang C, Liu Z, Liu Q, He K, and Yu Z. Characterization of cerna network to reveal potential prognostic biomarkers in triple-negative breast cancer. PeerJ. (2019) 7:e7522. doi: 10.7717/peerj.7522
86. Li Y, Liang Y, Sang Y, Song X, Zhang H, Liu Y, et al. Mir-770 suppresses the chemo-resistance and metastasis of triple negative breast cancer via direct targeting of stmn1. Cell Death Dis. (2018) 9:14. doi: 10.1038/s41419-017-0030-7
87. Deng L, Lei Q, Wang Y, Wang Z, Xie G, Zhong X, et al. Downregulation of mir-221-3p and upregulation of its target gene parp1 are prognostic biomarkers for triple negative breast cancer patients and associated with poor prognosis. Oncotarget. (2017) 8:108712–25. doi: 10.18632/oncotarget.21561
88. Svoboda M, Sana J, Redova M, Navratil J, Palacova M, Fabian P, et al. Mir-34b is associated with clinical outcome in triple-negative breast cancer patients. Diagn Pathol. (2012) 7:31. doi: 10.1186/1746-1596-7-31
89. Mercer TR, Dinger ME, and Mattick JS. Long non-coding rnas: insights into functions. Nat Rev Genet. (2009) 10:155–9. doi: 10.1038/nrg2521
90. Volovat SR, Volovat C, Hordila I, Hordila DA, Mirestean CC, Miron OT, et al. Mirna and lncrna as potential biomarkers in triple-negative breast cancer: A review. Front Oncol. (2020) 10:526850. doi: 10.3389/fonc.2020.526850
91. Fu J, Dong G, Shi H, Zhang J, Ning Z, Bao X, et al. Lncrna mir503hg inhibits cell migration and invasion via mir-103/olfm4 axis in triple negative breast cancer. J Cell Mol Med. (2019) 23:4738–45. doi: 10.1111/jcmm.14344
92. Mofed D, Omran JI, Sabet S, Baiomy AA, Emara M, and Salem TZ. The regulatory role of long non- coding rnas as a novel controller of immune response against cancer cells. Mol Biol Rep. (2022) 49:11775–93. doi: 10.1007/s11033-022-07947-4
93. Swellam M, El Magdoub HM, Shawki MA, Adel M, Hefny MM, and El-Shazly SS. Clinical impact of lncrna xist and lncrna neat1 for diagnosis of high-risk group breast cancer patients. Curr Probl Cancer. (2021) 45:100709. doi: 10.1016/j.currproblcancer.2021.100709
94. Lv M, Xu P, Wu Y, Huang L, Li W, Lv S, et al. Lncrnas as new biomarkers to differentiate triple negative breast cancer from non-triple negative breast cancer. Oncotarget. (2016) 7:13047–59. doi: 10.18632/oncotarget.7509
95. Ma J, Zhao W, Zhang H, Chu Z, Liu H, Fang X, et al. Long non-coding rna anril promotes chemoresistance in triple-negative breast cancer via enhancing aerobic glycolysis. Life Sci. (2022) 306:120810. doi: 10.1016/j.lfs.2022.120810
96. Zheng SM, Feng YC, Zhu Q, Li RQ, Yan QQ, Teng L, et al. Milip binding to trnas promotes protein synthesis to drive triple-negative breast cancer. Cancer Res. (2024) 84:1460–74. doi: 10.1158/0008-5472.CAN-23-3046
97. Zuo Y, Li Y, Zhou Z, Ma M, and Fu K. Long non-coding rna malat1 promotes proliferation and invasion via targeting mir-129-5p in triple-negative breast cancer. BioMed Pharmacother. (2017) 95:922–8. doi: 10.1016/j.biopha.2017.09.005
98. Wang Y, Zhang G, and Han J. Hif1a-as2 predicts poor prognosis and regulates cell migration and invasion in triple-negative breast cancer. J Cell Biochem. (2019) 120:10513–8. doi: 10.1002/jcb.28337
99. Hu Q, Ye Y, Chan LC, Li Y, Liang K, Lin A, et al. Oncogenic lncrna downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol. (2019) 20:835–51. doi: 10.1038/s41590-019-0400-7
100. Jiang YZ, Liu YR, Xu XE, Jin X, Hu X, Yu KD, et al. Transcriptome analysis of triple-negative breast cancer reveals an integrated mrna-lncrna signature with predictive and prognostic value. Cancer Res. (2016) 76:2105–14. doi: 10.1158/0008-5472.CAN-15-3284
101. He Y, Xiao B, Lei T, Xuan J, Zhu Y, Kuang Z, et al. Lncrna T376626 is a promising serum biomarker and promotes proliferation, migration, and invasion via binding to lamc2 in triple-negative breast cancer. Gene. (2023) 860:147227. doi: 10.1016/j.gene.2023.147227
102. Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, et al. The link-a lncrna activates normoxic hif1alpha signalling in triple-negative breast cancer. Nat Cell Biol. (2016) 18:213–24. doi: 10.1038/ncb3295
103. Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, and Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. (2005) 17:879–87. doi: 10.1093/intimm/dxh267
104. Shao H, Im H, Castro CM, Breakefield X, Weissleder R, and Lee H. New technologies for analysis of extracellular vesicles. Chem Rev. (2018) 118:1917–50. doi: 10.1021/acs.chemrev.7b00534
105. Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. (2020) 182:1044–61 e18. doi: 10.1016/j.cell.2020.07.009
106. Yu P, Han Y, Meng L, Tang Z, Jin Z, Zhang Z, et al. The incorporation of acetylated lap-tgf-β1 proteins into exosomes promotes tnbc cell dissemination in lung micro-metastasis. Mol Cancer. (2024) 23:82. doi: 10.1186/s12943-024-01995-z
107. Zhao L, Gu C, Gan Y, Shao L, Chen H, and Zhu H. Exosome-mediated sirna delivery to suppress postoperative breast cancer metastasis. J Control Release. (2020) 318:1–15. doi: 10.1016/j.jconrel.2019.12.005
108. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular rnas are a large class of animal rnas with regulatory potency. Nature. (2013) 495:333–8. doi: 10.1038/nature11928
109. Wang X, Chen T, Li C, Li W, Zhou X, Li Y, et al. Circrna-creit inhibits stress granule assembly and overcomes doxorubicin resistance in tnbc by destabilizing pkr. J Hematol Oncol. (2022) 15:122. doi: 10.1186/s13045-022-01345-w
110. Chen T, Wang X, Li C, Zhang H, Liu Y, Han D, et al. Circhif1a regulated by fus accelerates triple-negative breast cancer progression by modulating nfib expression and translocation. Oncogene. (2021) 40:2756–71. doi: 10.1038/s41388-021-01739-z
111. Lecot P, Sarabi M, Pereira Abrantes M, Mussard J, Koenderman L, Caux C, et al. Neutrophil heterogeneity in cancer: from biology to therapies. Front Immunol. (2019) 10:2155. doi: 10.3389/fimmu.2019.02155
112. Hajizadeh F, Aghebati Maleki L, Alexander M, Mikhailova MV, Masjedi A, Ahmadpour M, et al. Tumor-associated neutrophils as new players in immunosuppressive process of the tumor microenvironment in breast cancer. Life Sci. (2021) 264:118699. doi: 10.1016/j.lfs.2020.118699
113. Soto-Perez-de-Celis E, Chavarri-Guerra Y, Leon-Rodriguez E, and Gamboa-Dominguez A. Tumor-associated neutrophils in breast cancer subtypes. Asian Pac J Cancer Prev. (2017) 18:2689–93. doi: 10.22034/APJCP.2017.18.10.2689
114. Szczerba BM, Castro-Giner F, Vetter M, Krol I, Gkountela S, Landin J, et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature. (2019) 566:553–7. doi: 10.1038/s41586-019-0915-y
Keywords: TNBC, diagnostics, liquid biopsy, CTCs, CtDNA, miRNAs, lncRNAs
Citation: Sheng J and Zong X (2025) Liquid biopsy in TNBC: significance in diagnostics, prediction, and treatment monitoring. Front. Oncol. 15:1607960. doi: 10.3389/fonc.2025.1607960
Received: 08 April 2025; Accepted: 17 July 2025;
Published: 04 August 2025.
Edited by:
Sharon R. Pine, University of Colorado Anschutz Medical Campus, United StatesReviewed by:
Elena Lastraioli, University of Florence, ItalyViola Bianca Serio, University of Siena, Italy
Vasyl Chekhun, National Academy of Sciences of Ukraine (NAN Ukraine), Ukraine
Copyright © 2025 Sheng and Zong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolong Zong, ZnJlZWRvbVpYTEAxNjMuY29t
 Jiayi Sheng
Jiayi Sheng Xiaolong Zong
Xiaolong Zong