- 1Department of Gastrointestinal Surgery, Beijing Tsinghua Changgung Hospital, School of Clinical Medicine, Tsinghua University, Beijing, China
- 2Department of Oncology Center, Peking University International Hospital, Beijing, China
- 3Geneplus-Beijing, Beijing, China
- 4Department of Oncology, Beijing Hospital, National Center of Gerontology, Beijing, China
Background: Heterogeneity of colorectal cancer (CRC) leads to significant differences in Overall Survival (OS). RNF43 is a new predictive marker for prognosis and anti-BRAF/EGFR combinatory therapies of CRC recently. However, few studies focused on the relationship between RNF43 and co-mutation characteristics and prognosis. This study aims to explore the different prognostic subtypes of RNF43-mutated CRC by analyzing the association of clinicopathological and genomic characteristics with survival outcomes.
Methods: The clinical characteristics, mutational characteristics, and survival data of CRC patients were obtained for RNF43-mutated analysis from cBioPortal. All mutation data were filtered by the 1021-panel (Geneplus-Beijing, China), and the processed data were used to analyze the predictive value of RNF43-mutated to OS and concomitant co-mutations. Cox regression analysis was selected to explore prognostic biomarkers, and finally, BRAF and MSI were selected for subgroup analysis. The independent validation cohort comprised 339 cases of stage IV CRC from Beijing Hospital.
Results: 11 datasets with 4028 patient data were screened for this study. The most common variant was frameshift, which occurred in codon 659-mutated of exon 9, including RNF43 p.G659Vfs*41 (N=116) and RNF43 p.G659Sfs*87 (N=2). RNF43 codon 659-mutated occurred frequently in right-sided CRC (59.32%, N=70, P<0.0001), and rarely in the left-sided (11.02%, N=13). The incidence of TMB-H in the RNF43 codon 659-mutated group was 93.22% (110/118), and MSI-H was 78.81% (93/118). Univariate Cox analysis and multivariate Cox analysis showed that MSI-H was the most significantly different biomarker for better prognosis (P=0.004, HR=3, CI 1.4-6.4), and Class 1 BRAF V600E was the most different biomarker for worse prognosis (P<0.001, HR=0.3, CI 0.21-0.42). RNF43 codon 659-mutated with non-class 1 BRAF-mutated or MSI-H suggests a better prognosis in CRC. We found that G1 (RNF43 codon 659-mutated, non-class 1 BRAF-mutated, and MSl-H) had a better PFS and OS. The mutation difference analysis showed that the core genes related to the cancer signaling pathway (PI3K-Akt signaling pathway, MicroRNAs pathway, DNA damage repair, and tumor suppressor genes) were highly frequent in G1. The analysis comparing the core gene mutation difference between RNF43-mutated and wild-type in the validation cohort yielded consistent conclusions.
Conclusions: In CRC, we found that the G1 cohort had the best prognosis, and patients with RNF43 Non-codon 659-mutated, BRAF V600E and MSS had the worst prognosis. This may provide clinical value for patients’ further accurate prognosis prediction, curative effect prediction, and follow-up management of patients.
1 Introduction
Colorectal cancer (CRC) is the third most common cancer in the world and the second leading cause of cancer-related death (1). Despite considerable advances in treatment strategies and survival, the prognosis for patients with colorectal cancer remains poor, with 5-year overall survival (OS) for metastatic colorectal cancer of about 14%. The 5-year survival rate for all colorectal cancer patients is about 65% (2). Currently, the Tumor-Node-Metastasis system (TNM) classification at diagnosis is a major determinant of survival, but CRC is a highly heterogeneous disease with different molecular characteristics, including genetic and epigenetic changes (3, 4). Even when shared with the same pathological type or disease stage, there are also significant differences in treatment efficacy and survival, as well as substantial differences in the response of patients with different molecular characteristics to the same treatment strategy, leading to imprecise prognostic predictions (5–7). Therefore, predicting the survival of CRC needs further exploration.
The Wnt/β-catenin signaling pathway is a traditional pathway initiated by changes in Wnt ligand-dependent genes (RNF43/ZNRF3/RSPO) or ligand-independent genes (APC) and plays a key role in the initiation, advancement, and metastasis of CRC (8). RNF43 (Ring finger protein 43) is an E3 ubiquitin-protein ligase that inhibits overactivation of the Wnt pathway, and RNF43 mutations lead to permanent activation of the Wnt pathway in cancer cells (9, 10). Previous studies have reported the clinical significance of RNF43 mutations in colorectal cancer. However, the effect of RNF43-mutated in colorectal cancer remains controversial. It has been suggested that RNF43-mutated can be used as a predictive biomarker of anti-BRAF/EGFR combination therapy response in microsatellite-stabilized (MSS) BRAF V600E metastatic colorectal cancer patients, and RNF43-mutated is a better predictive biomarker of progression-free survival (PFS) and OS in BRAF-mutated CRC patients (9, 11). Other studies have associated RNF43-mutated with poor prognosis and a higher recurrence rate (12, 13). Therefore, the prognostic value of RNF43-mutated remains to be determined.
BRAF is the core gene of the Mitogen-Activated Protein Kinase (MAPK) signaling pathway, which regulates cell proliferation and apoptosis (14). The incidence of BRAF-mutated CRC is about 10-20% (15, 16). Class 1 BRAF V600E-mutated is caused by c.1799T>A, suggesting the worst tumor biological behavior and poor prognosis, accounting for 90% of all BRAF-mutated in CRC according to a deeper classification system of BRAF-mutated derived from pre-clinical models functional studies (17, 18). BRAF V600 CRC has previously been extensively studied, and tumors with RNF43-mutated are associated with a high frequency of BRAF V600E-mutated, and these co-mutations are associated with poor survival (19, 20).
High microsatellite instability (MSI-H) of colon cancer can indicate better clinical outcomes of immune checkpoint inhibitors (ICIs) in the early-stage to the advanced population, and its predictive value in advanced CRC has been approved by the National Comprehensive Cancer Network (NCCN) clinical guidelines (21, 22). Previous studies have shown that RNF43-mutated is most associated with MSI-H, and it has been reported that RNF43 p. G659fs* is enriched in MSI-H cancer (23, 24). These results suggest that RNF43 is a predictive prognostic marker for colorectal cancer, and limited data are available to predict the significance of individual changes. However, a few studies on the relationship between RNF43 and co-mutation characteristics and prognosis, and the clinical significance of RNF43-mutated and other biomarkers such as BRAF and MSI status in colorectal cancer are still worth exploring.
In this study, we explored prognostic biomarkers with predictive value based on clinicopathological and molecular characteristics of colorectal cancer patients in the cBioPortal database. We analyzed the association of RNF43-mutated, co-occurring mutations, genomic characteristics (including MSI, TMB), and OS, and found that RNF43 codon 659-mutated has prognostic value and is a special subtype. We then determined the predictive prognostic value of three indicators based on RNF43, BRAF, and MSI status and performed differential mutation analysis and pathway enrichment analysis. The results reveal that the RNF43 subtype, combined with other molecular characteristics, can be used as biomarkers to predict the clinical outcome of CRC.
2 Materials and methods
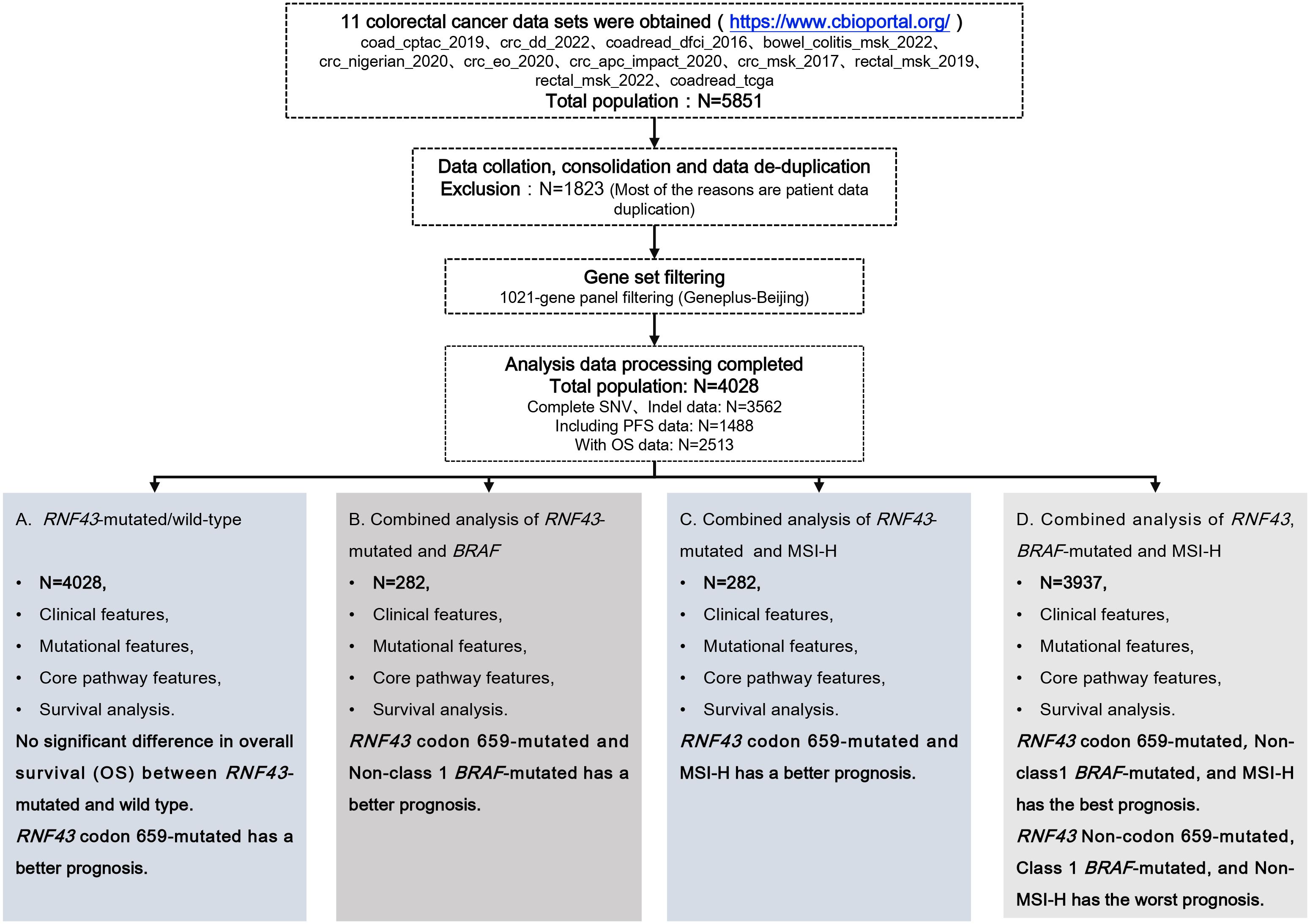
2.1 Data source and patient selection
The clinical characteristics, mutational characteristics, and survival data of CRC patients were recruited for RNF43-mutated analysis from the Cancer Genome Atlas (TCGA) database using the cBio Cancer Genomics Portal (cBioPortal), available at http://www.cbioportal.org (Supplementary Tables 1, 2) (25). We integrated all the data sets that have been published so far. We performed data consolidation and de-duplication, excluding a total of 1823 patient data, and finally obtained 4028 patients from 11 data sets (coad_cptac_2019,crc_dd_2022,coadread_dfci_2016,bowel_colitis_msk_2022,crc_nigerian_2020,crc_eo_2020,crc_apc_impact_2020,crc_msk_2017,rectal_msk_2019,rectal_msk_2022,coadread_tcga) for this study (Figure 1). We then filtered single-nucleotide variants (SNVs) through the 1021 panel (Geneplus-Beijing, China), a custom-designed biotinylated oligonucleotide probe (Roche NimbleGen, Madison, WI, USA) covering ~1.4 Mbp coding region of genomic sequence of 1,021 cancer-related genes to explore the relationship with tumor genomic characteristics and prognosis (Supplementary Table 3). The TMB was defined as the total number of mutations per megabase (1 Mb) of non-synonymous single-nucleotide variants (SNV), insertion/deletion (Indel), and splice ±2 (26). The upper quartile of tumor mutational burden (TMB) was deemed as high TMB (TMB-H), with a threshold of 8.87 mutations/Mb in this study (27, 28). MSI-H directly used the downloaded label with cBioPortal, and the total number of MSI-H and MSS patients was 296 and 2858, respectively. Before analyzing this study, we calculated the mutation frequencies in 11 cohorts to better understand the reproducibility and limitations of the research, as shown in Supplementary Table 4. We evaluated the overall research results based on the completeness of data for 100 genes, 200 genes, and 300 genes, with missing rates of 6.09%, 9.09%, and 14.91%, respectively. We believe that these data not only support the overall reliability of the data but also indicate that the data has certain stability and reproducibility. A total of 339 patients with advanced CRC who underwent 1021 panel NGS sequencing served as the validation cohort for this study (Supplementary Table 5) (29). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Beijing Hospital (2023BJYYEC-428-02).
2.2 Statistical analysis
All the data were analyzed using the R statistics package (R version 4.2.1, Austria) or GraphPad Prism version 8 (GraphPad Software, CA, USA). Differences between designed groups were analyzed based on the Fisher test or the t-test. Univariate Cox regression and Multivariate Cox regression analysis methods were used to analyze the correlation between mutation characteristics, genomic characteristics, and clinical outcomes. David 6.8 (https://david.ncifcrf.gov/) was used to carry out the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The log-rank test Kaplan-Meier (KM) survival curve was used to calculate prognostic differences between groups based on RNF43-mutated. Survival curves were calculated by the Kaplan-Meier method, and differences between groups based on RNF43-mutated status were tested by the log-rank test. P values < 0.05 were denoted as statistically significant.
3 Results
3.1 Patient characteristics
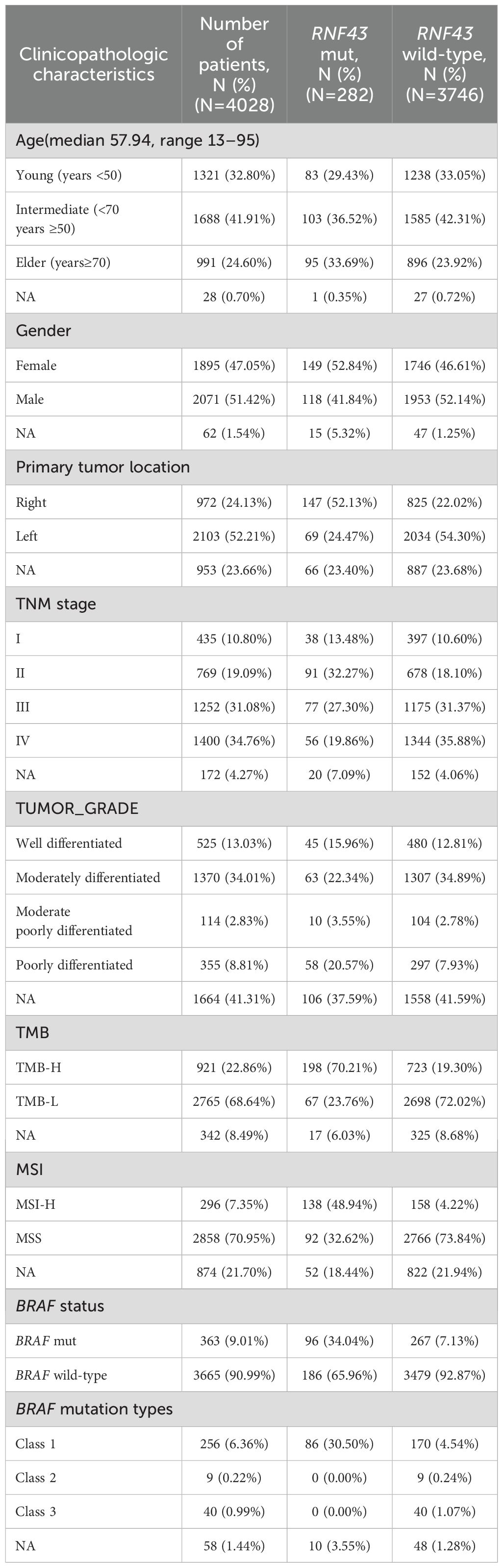
The clinicopathological and molecular characteristics of the enrolled patient population are shown in Table 1. The mean age of this cohort was 57.94 years, and most of the population (41.91%, N=1688) was between 50 and 70 years old. At the primary tumor site, the RNF43-mutated group had significantly more right-sided patients than left-sided patients (left-sided: 24.47%, N=69; right-sided: 52.13%, N=147), and the RNF43 wild-type cohort data were contrary (left-sided: 54.30%, N=2034; right-sided: 22.02%, N=825). A total of 65% of the patients in the whole cohort were stage III-IV patients, and the tumor grade was mainly moderately differentiated (34.01%, N=1370). The genomic markers TMB-H (198/282, 70.21%, P<0.001) and MSI-H (138/282, 48.94%, P<0.001) were significantly higher in the RNF43-mutated group compared to those in the RNF43 wild-type group. Class 1 BRAF-mutated and RNF43-mutated co-occurred frequently. In the RNF43-mutated group, the proportion of class 1 BRAF-mutated was 34.05%, and that in the RNF43 wild-type group was only 7.13%.
3.2 The Landscape of the RNF43-mutated CRC
In this study, 375 RNF43 variants were detected in 282 RNF43-mutated patients. The most common variant was frameshift, which occurred in codon 659-mutated of exon 9, including RNF43 p. G659Vfs*41 (N=116) and RNF43 p. G659Sfs*87 (N=2), as shown in Figure 2A, Supplementary Table 6. The distribution range of RNF43-mutated varies (1.89%-28.13%) in 11 cohorts. The frequency of RNF43-mutated in the vast majority of cohorts was between 7.34% and 17.27%. Additionally, RNF43 codon 659-mutated has a relatively high proportion in all queues, including coadread_dfci_2016, cro_eo_2020, rectal_msk_2022, and coad_cptac_2019 (Supplementary Figure 1). In RNF43-mutated cohorts, the most commonly mutated genes were ARIDIA (59%), CIC (45%), PIK3CA (43%), PTPRS (43%), APC (42%), FAT1 (40%), POLE (40%), NOTCH3 (39%), SPEN (39%), BRAF (38%) (Figure 2B). In contrast, the top10 mutated genes in RNF43 wild-type tumors were APC (74%), TP53 (71%), KRAS (41%), PIK3CA (17%), FBXW7 (14%), SMAD4 (13%), TCF7L2 (10%), SOX9 (10%), ARID1A (8%), BRAF (8%) (Supplementary Figure 2A). Survival analysis showed that RNF43-mutated had worse progression-free survival (PFS, P=0.0048) and overall survival (OS, P=0.18) (Supplementary Figures 2B, C). Considering the limitations of single-mutation data, we conducted a joint analysis using 106 mRNA data from the coad_cptac_2019 dataset. We found that the expression level of RNF43 in the RNF43-mutated was significantly lower than that in the RNF43 wild-type (P < 0.001, Supplementary Figure 2D). Meanwhile, we found that there were also cases of low RNF43 expression levels within the RNF43 wild-type. Given the relatively small size of the study cohort, we integrated two groups: the RNF43-mutated with expression values < 0, and the RNF43 wild-type with expression values > 0. We found that the RNF43-mutated/expression < 0 shared a similar mutation spectrum with the RNF43-mutated, and RNF43 wild-type/expression > 0 had a similar mutation spectrum to the RNF43 wild-type (Supplementary Figure 2E). This finding implies that DNA combined with RNA-based approaches for precise prognostic stratification represents a more optimal choice in the future.
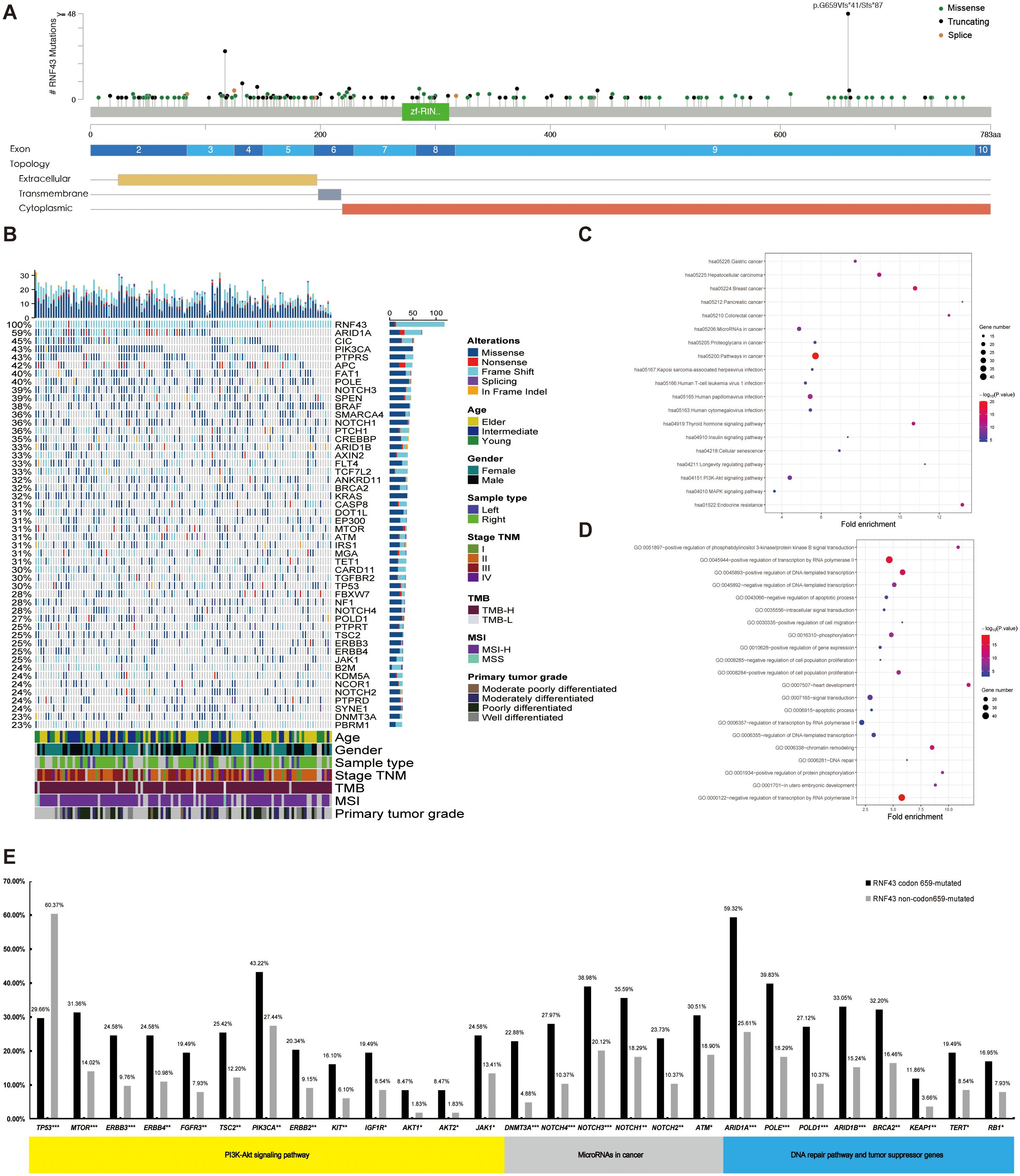
Figure 2. A panoramic analysis of the genomic and pathway characteristics of RNF43-mutated in CRC. (A) Lollipop plots (maps mutations on a linear protein and its domains) in this study. Truncating includes frameshift mutations and nonsense mutations. (B) Top 50 mutation spectrum in 283 RNF43-mutated patients. Each column represents a patient, and each row represents a gene. The table on the left represents the mutation rate of each gene. The top plot represents the overall number of mutations a patient carried. Different colors denote different types of mutations. KEGG (C) and GO (D) functional enrichment analyses of RNF43-mutated. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes. (E) The differences in core gene mutation of major signaling pathways (PI3K-Akt signaling pathway, MicroRNAs pathway, DNA damage repair, and tumor suppressor genes) between RNF43 codon 659-mutated and RNF43 Non-codon 659-mutated. CRC, Colorectal cancer; *p<0.05; **p<0.01; ***p<0.001.
In the validation cohort, we found consistent results in the RNF43-mutated: KRAS (32% vs 43%), APC (42% vs 35%), ARID1A (59% vs 35%), and NF1 (35% vs 28%). However, there were differences in TP53. The abundance in the validation cohort is as high as 70%. The top mutations of the RNF43 wild-type showed high consistency in both the analysis cohort and the validation cohort (Supplementary Figures 3A, B). Differential gene analysis showed that the RNF43-mutated group had significantly higher mutation frequency (Supplementary Table 7). The mutation differences between RNF43-mutated and RNF43 wild-type in validation cohort was also analyzed. We founf that NF1, ARID1A, BRAF, B2M, WRN were significantly enriched in RNF43-mutated group, while APC was significantly enriched in RNF43 wild-type, and the RNF43-mutated group had significantly higher mutation frequency (Supplementary Table 8, Supplementary Figure 3C). KEGG pathway enrichment analysis showed that hsa05206: MicroRNAs in cancer, and hsa04151: PI3K-Akt signaling pathway were significantly enriched in the RNF43-mutated group (Figure 2C). GO enrichment analysis showed that the RNF43-mutated group had higher enrichment of proliferative signaling pathway (GO: 0016310-Phosphorylation, GO: 0008284~positive regulation of cell population proliferation, GO: 0043066-negative regulation of apoptotic process, etc) (Figure 2D). The results of KEGG and GO enrichment analyses of the verification cohort were consistent (Supplementary Figures 3D, E).
3.3 RNF43 codon 659-mutated is a specific subtype of CRC
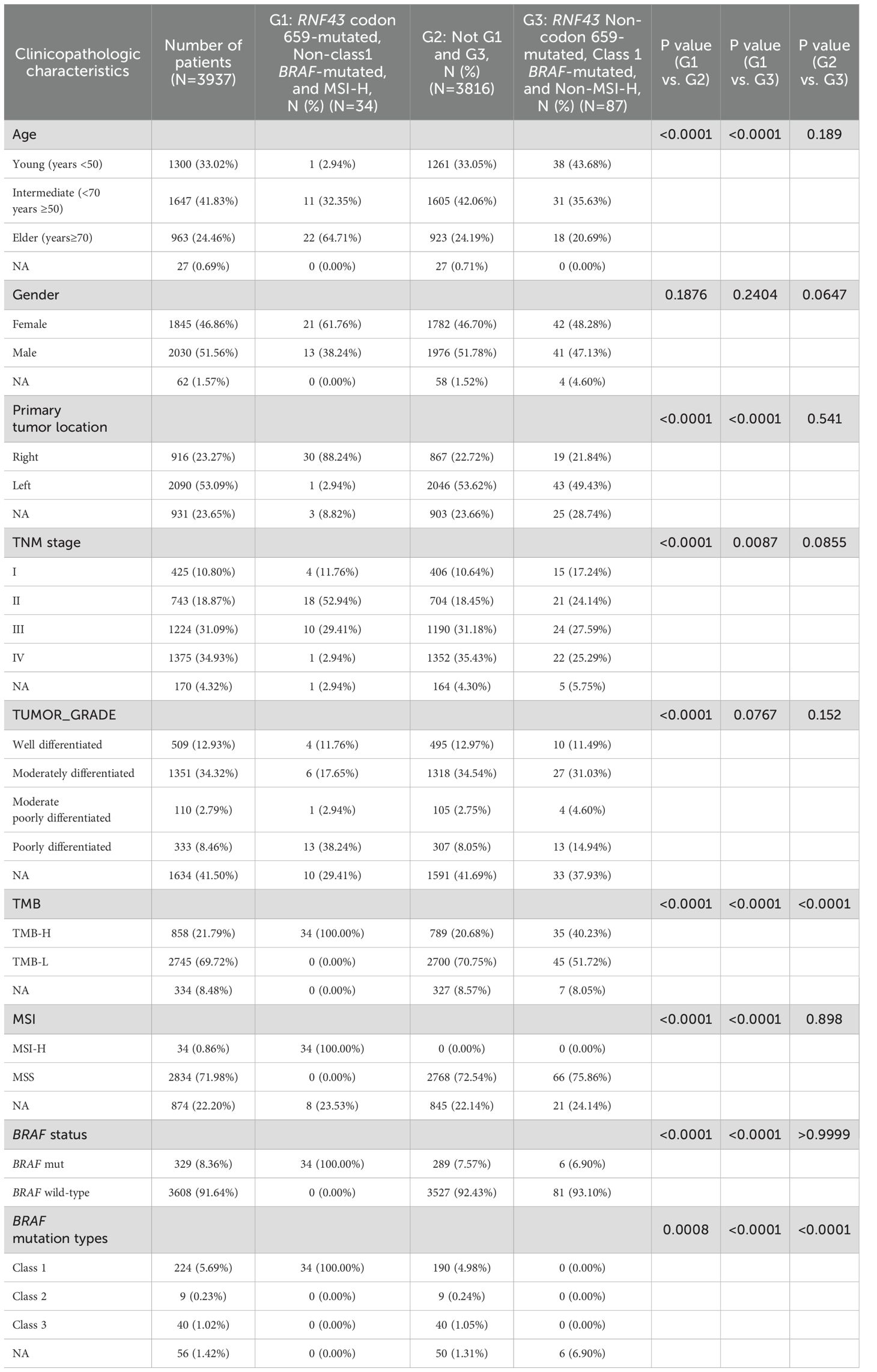
As the incidence of codon 659 mutation accounted for nearly half of the total RNF43-mutated and had unique clinical significance in predicting the efficacy of anti-BRAF/EGFR combinatory therapies (30), the p.G659Vfs*41 and p.G659Sfs*87 was defined as the RNF43 codon 659-mutated group and the other mutation types were defined as the RNF43 Non-codon 659-mutated group in this study. The clinicopathological and molecular characteristics of the two groups were different from the total population (Table 2). The proportion of RNF43 codon 659-mutated patients aged over 70 years was higher (38.98% vs. 29.88%, P=0.2249). The RNF43 Non-codon 659-mutated occurs most frequently in 50-70 years. There was no difference in gender between the two groups. RNF43 codon 659-mutated occurred frequently in right-sided CRC (59.32%, N=70, P<0.0001), and rarely in the left-sided (11.02%, N=13), while the left and right-sided were more balanced in the RNF43 Non-codon 659-mutated (34.15%, N=56; 46.95%, N=77). In terms of TNM stage, RNF43 codon 659-mutated mainly appeared in TNM II-III (66.1%, N=78), which was inconsistent with the total group staging concentrated in III-IV (65.85%, N=2652).
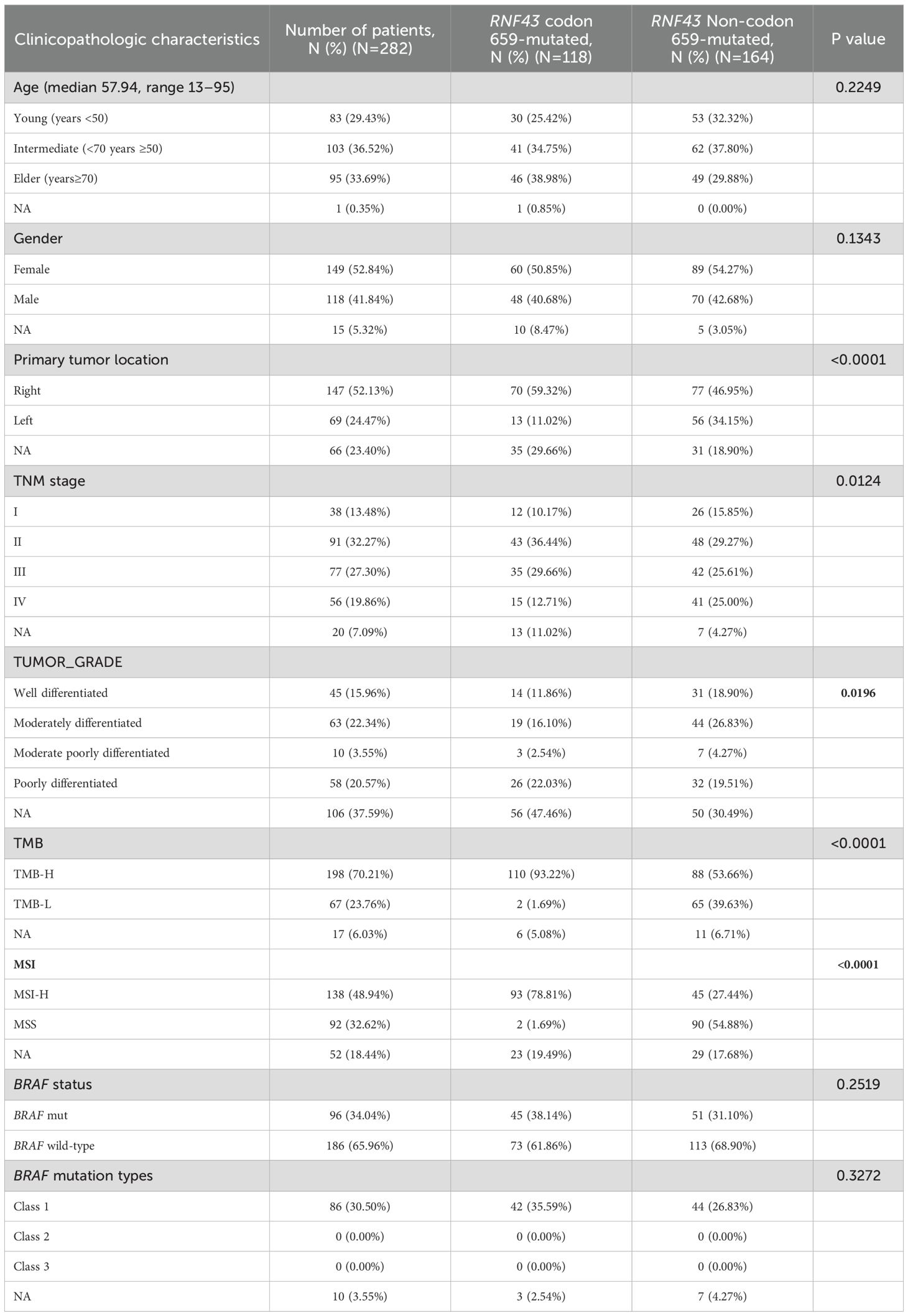
Table 2. Clinicopathological and molecular characteristics of RNF43 codon 659-mutated and RNF43 Non-codon 659-mutated.
Subsequently, we analyzed the mutation differences between RNF43 codon 659-mutated and RNF43 Non-codon 659-mutated and pathway enrichment results. CIC, ARID1A, PTCH1, SMARCA4, FLT4 were significantly enriched in RNF43 codon 659-mutated group, while TP53 was significantly enriched in RNF43 Non-codon 659-mutated (Supplementary Table 8). The differences in core gene mutation of major signaling pathways (PI3K-Akt signaling pathway, MicroRNAs pathway, DNA damage repair, and tumor suppressor genes) are shown in Figure 2E. Except for TP53 has the highest frequency in the RNF43 wild-type, other frequencies are significantly higher in RNF43-mutated. Considering that the population of RNF43 codon 659-mutated in the validation cohort is relatively small (only three cases), we are temporarily unable to carry out the validation work for this part.
3.4 Class 1 BRAF-mutated and MSI-H have strong prognostic value in CRC
Based on the differences in clinical features and mutational characteristics exhibited by RNF43 codon 659-mutated and RNF43 Non-codon 659-mutated. Our next step aims to screen biomarkers that predict prognosis. We conducted univariate Cox analysis and multivariate Cox analysis based on OS as clinical outcomes. Study results are shown in Figure 3A. We found age, sample_type (left-sided or right-sided), stage_TNM, MSI, KRAS, APC, and BRAF_V600E (Class1 BRAF-mutated) were biomarkers with significant prognostic differences. Subsequently, factors with P<0.05 were included in multivariate analysis, and it was found that MSI-H was the most significantly different biomarker for better prognosis (P=0.004, HR=3, CI 1.4-6.4), and Class 1 BRAF V600E was the most different biomarker for worse prognosis (P<0.001, HR=0.3, CI 0.21-0.42). We also found that KRAS-mutated was the second-highest predictor of poor prognosis (P<0.001, HR=0.68, CI 0.57-0.81).
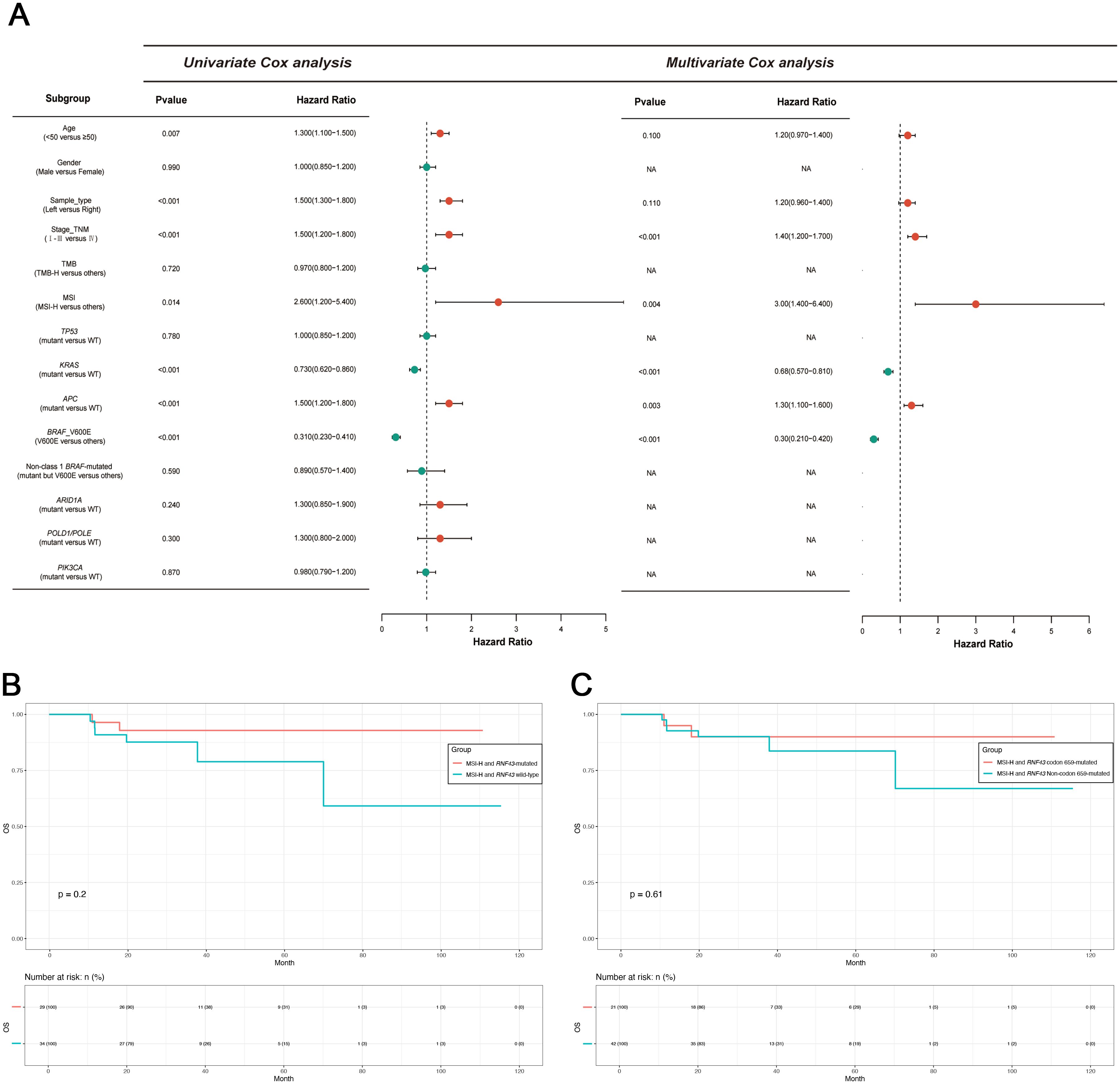
Figure 3. Univariate and multivariate Cox proportional hazards analysis of OS and survival analysis of the MSI-H subgroup in this study. (A) Univariate and multivariate Cox proportional hazards analysis of OS in this study. (B) Survival analysis of MSI-H with or without RNF43. (C) Survival analysis of MSI-H with or without RNF43 codon 659-mutated. OS, overall survival; TMB-H, patients with high TMB; MSI-H, patients with high MSI; WT, wild type.
MSI-H is a molecular marker that is included in the guidelines and serves as a biomarker indicating a favorable prognosis for CRC. Therefore, further exploration on the value of RNF43-mutated and RNF43 wild-type based on the MSI-H is necessary. Thus, we conducted two groups: MSI-H and RNF43-mutated (N=138), and MSI-H and RNF43 wild-type (N=158). We first conducted a statistical analysis of clinical information (Table 3). No significant differences in age, gender, stage, and tumor grade. However, the proportion of left-sided tumors in the MSI-H and RNF43-mutated group was significantly lower than that in the MSI-H and RNF43 wild-type group (15.22% vs. 28.48%, P=0.0235), and the TMB-H proportion in the MSI-H and RNF43-mutated group was higher (95.65% vs. 87.97%, P=0.0196). Next, we found the overall mutation frequency of MSI-H and RNF43-mutated was higher than that of MSI-H and RNF43 wild-type, with a difference in the distribution of mutations (Supplementary Table 9). In the MSI-H and RNF43-mutated group, the top 5 mutations were ARID1A, PTPRS, FAT1, PIK3CA, and SPEN (Supplementary Figure 4A). The top 5 mutations in the MSI-H and RNF43 wild-type group were APC, ARID1A, PIK3CA, PTPRS, and BRAF (Supplementary Figure 4B). Furthermore, we analyzed mutations specifically in the MSI-H and RNF43 codon 659-mutated group compared to the MSI-H and RNF43 Non-codon 659-mutated group. The results revealed that the high-frequency mutations in the MSI-H and RNF43 codon 659-mutated group included ARID1A, CIC, PTPRS, FAT1, and PIK3CA (Supplementary Figure 4C), with a higher mutation frequency than observed in the MSI-H and RNF43 Non-codon 659-mutated group (Supplementary Figure 4D). Finally, we conducted a prognostic analysis; the OS of the MSI-H and RNF43-mutated group was better than that of the MSI-H and RNF43 wild-type group, but there was no significant difference (P = 0.2, Figure 3B). The OS of the MSI-H and RNF43 codon 659-mutated group was better than that of the MSI-H and RNF43 Non-codon 659-mutated group, and there was also no significant difference (P = 0.61, Figure 3C). This lack of significant difference may be attributed to the high proportion of poorly differentiated individuals within the MSI-H and RNF43-mutated cohort.
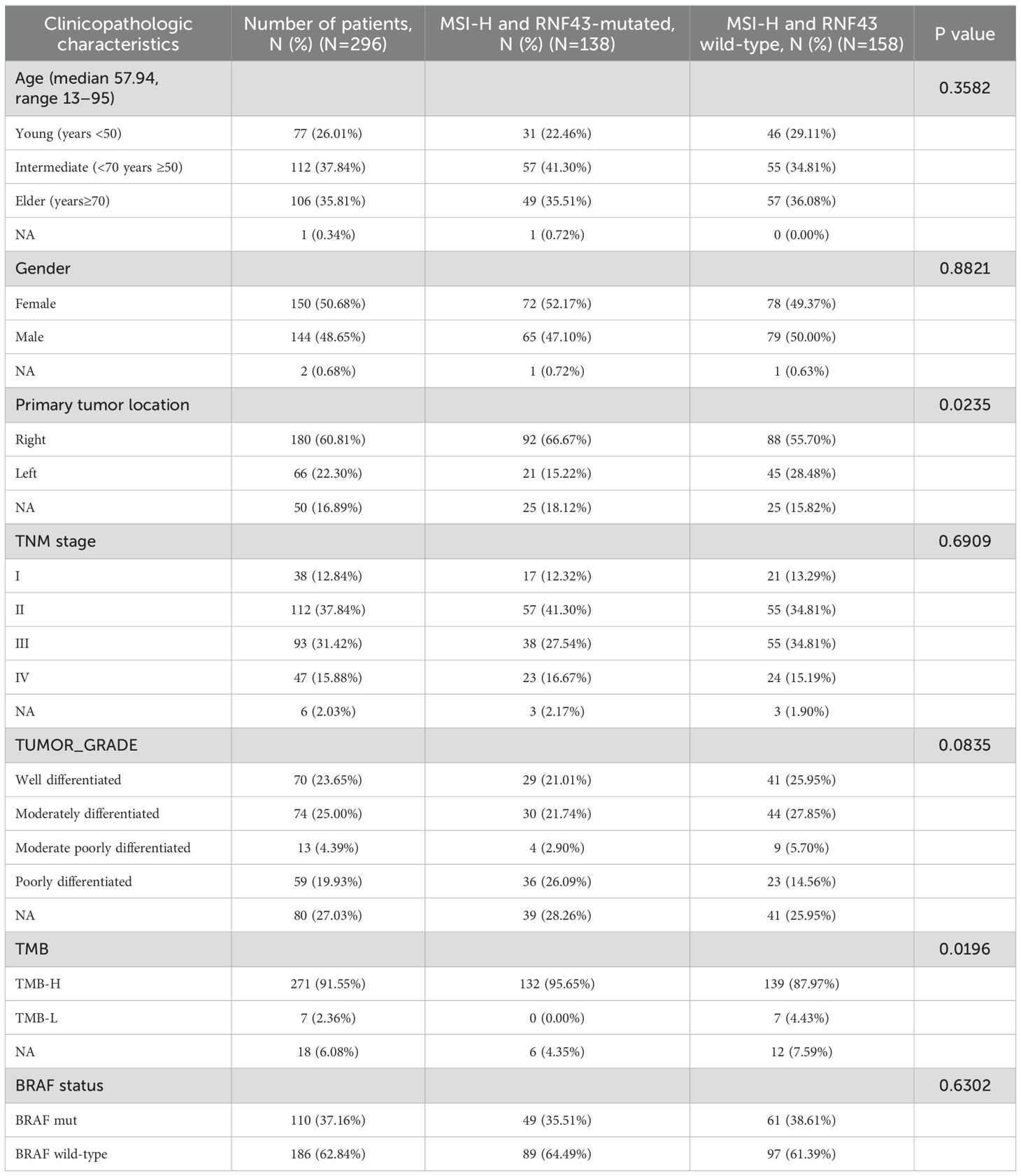
Table 3. Clinicopathological and molecular characteristics of MSI-H and RNF43-mutated or RNF43 wild-type.
3.5 RNF43 codon-659-mutated, class 1 BRAF-mutated, MSI-H has strong co-mutational characteristics
Further cluster analysis was performed for the prognostic markers BRAF and MSI, identified by Cox analysis before. We found high co-occurrence of the RNF43 mutation subtype and BRAF mutation, as well as strong associations with TMB and MSI. We conducted a multi-index UPSET correlation analysis. RNF43 codon 659-mutated was found to overlap with TMB-H, MSI-H, and BRAF V600E. The overlap degree of RNF43 Non-codon 659-mutated with TMB, MSI, and BRAF V600E is lower than that of RNF43 codon 659-mutated (Figure 4A). Class1 BRAF-mutated and RNF43 codon 659-mutated, RNF43 Non-codon 659-mutated, and RNF43 wild-type were 35.59%, 28.83% and 4.54%, respectively (P<0.0001). It is also worth noting that the incidence of TMB-H in the RNF43 codon 659-mutated group was 93.22% (110/118), and MSI-H was 78.81% (93/118). This suggests that the co-occurrence of RNF43 with TMB-H or MSI-H is mainly caused by RNF43 codon 659-mutated (Figure 4B, Table 2). Combined with the literature reporting that MSI-H is one of the factors with better prognosis in CRC (21, 22), we believe that multi-indicator association analysis may suggest a better prognosis CRC subgroup.
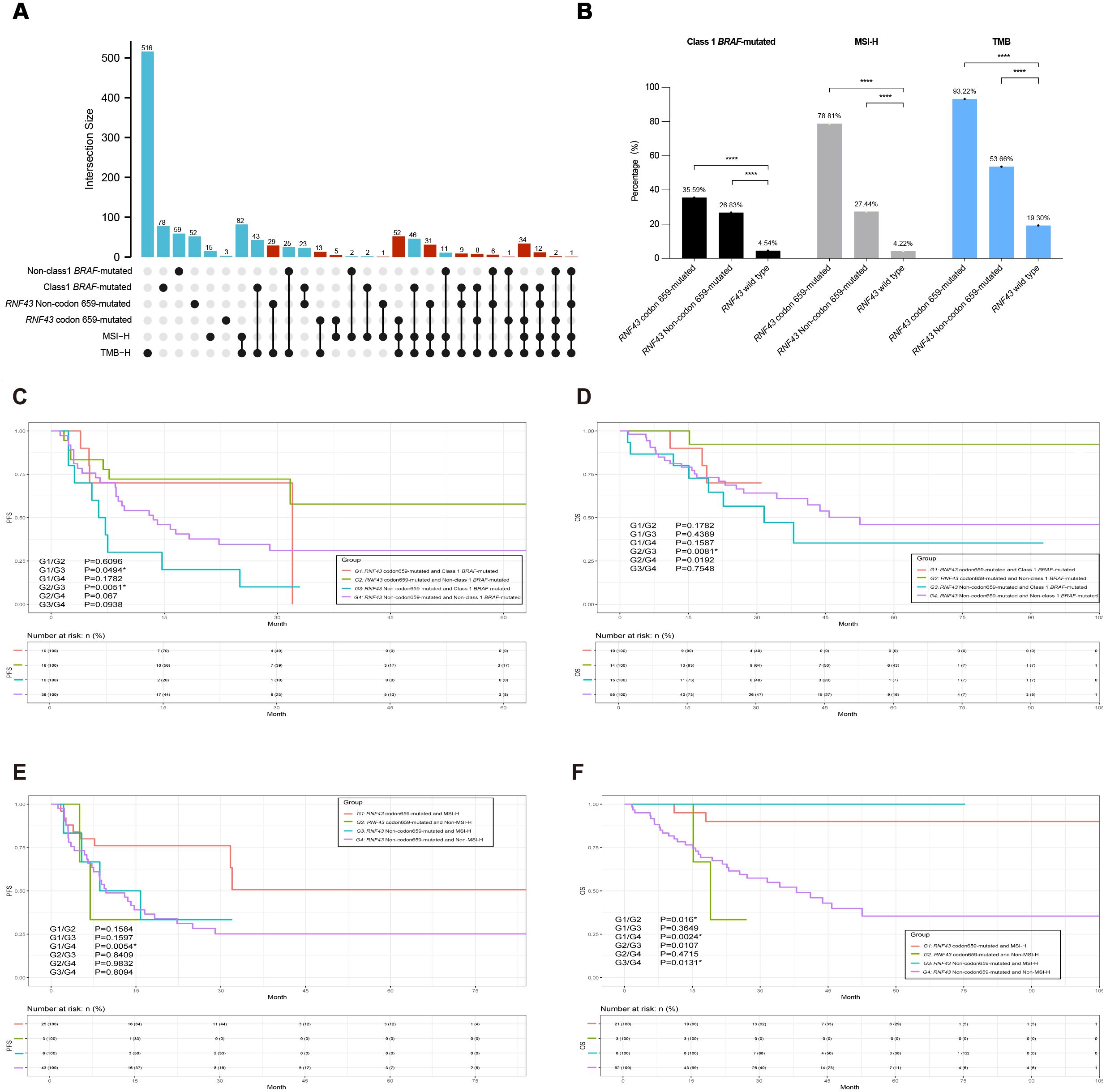
Figure 4. The integrated association’s analysis and survival analysis of RNF43 codon 659-mutated, MSI status, or BRAF-mutated in CRC. (A) UPSET plot showing the shared and unique marker numbers between RNF43, MSI, and BRAF in this study. (B) Correlation analysis bar chart of RNF43, MSI, and BRAF indicators. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. (C) KM analysis of PFS between RNF43-mutated and BRAF-mutated in this study. (D) KM analysis of OS between RNF43-mutated and BRAF-mutated in this study. (E) KM analysis of PFS between RNF43-mutated and MSI status in this study. (F) KM analysis of OS between RNF43-mutated and MSI status in this study. CRC, Colorectal cancer; PFS, progression-free survival; OS, overall survival; KM, Kaplan-Meier.
3.6 RNF43 codon 659-mutated with non-class 1 BRAF-mutated or MSI-H suggests a better prognosis in CRC
We then performed a joint analysis of the two indicators, starting with RNF43 combined with BRAF. A total of 282 patients with RNF43-mutated were enrolled and divided into four groups: G1 (N=42): RNF43 codon 659-mutated and Class 1 BRAF-mutated; G2 (N=76): RNF43 codon 659-mutated and Non-class 1 BRAF-mutated; G3 (N=44): RNF43 Non-codon 659-mutated and Class 1 BRAF-mutated; G4 (N=120): RNF43 Non-codon 659-mutated and Non-class 1 BRAF-mutated. The clinicopathological and molecular characteristics of G1-G4 groups are shown in Supplementary Table 10. Survival analysis of PFS results showed that the G1 group (P=0.0494) and G2 group (P=0.0051) had significantly better prognosis compared with G3 (Figure 4C), and OS analysis results showed that only the G2 group and G3 group had significant differences (P=0.0081, Figure 4D). Patients with RNF43 codon 659-mutated and Non-class 1 BRAF-mutated were found to have a better prognosis. Next, we analyzed the mutation difference between the G2 group and G3 group, and the TOP mutations of the two groups were shown in Supplementary Figure 5A (G2 group) and Supplementary Figure 5B (G3 group). The volcano map of mutation difference analysis showed that ARID1A, CIC, and other genes were significantly enriched in the G2 group (Supplementary Figure 5C), and the pathway enrichment results suggested that the microRNAs pathway, DNA damage repair, and tumor suppressive gene mutations were significantly enriched in G2 group: RNF43 codon 659-mutated and Non-class 1 BRAF-mutated. This is consistent with the results of the RNF43-mutated vs RNF43 wild-type analysis, suggesting that the combined detection of RNF43 and BRAF can help predict a better prognosis (Supplementary Figures 5D-F).
In order to match the clinical guidelines recommended, we only compared RNF43 combined with MSI and did not perform RNF43 combined with TMB. Therefore, four groups are assigned. G1 (N=93): RNF43 codon 659-mutated and MSI-H; G2 (N=25): RNF43 codon 659-mutated and Non-MSI-H; G3 (N=45): RNF43 Non-codon 659-mutated and MSI-H; G4 (N=119): RNF43 Non-codon 659-mutated and Non-MSI-H. This part also included 282 RNF43-mutated patients. The clinicopathological and molecular characteristics of the four groups are shown in Supplementary Table 11. As expected, G4 had worse PFS and OS (G1/G4-PFS: P=0.0054; G1/G4-OS: P=0.0024). The top mutations of group G1 and group G4 were shown in Supplementary Figure 6A (G1) and Supplementary Figure 6B (G4). Volcanic map analysis of mutation differences showed that ARID1A, CIC, PTPRS, PTCH1, and other genes were significantly enriched in group G1, and TP53 was significantly enriched in group G4 (Supplementary Figure 6C). This is consistent with the conclusion that CRC patients carrying TP53 mutations have a worse prognosis. The enrichment results were consistent with the results of RNF43-mutated vs RNF43 wild-type and RNF43 combined BRAF analysis (Supplementary Figures 6D–F).
3.7 RNF43 codon 659-mutated combined with non-class 1 BRAF-mutated and MSI-H has the best prognosis
Subsequently, we integrated the three indicators of RNF43, BRAF, and MSI found above for integrated analysis, to find the population with the best prognosis. Different from the previous analysis process, we also included RNF43 wild-type in this part, and a total of 3937 CRC patients with survival data were recorded and screened, which were divided into three groups: G1: RNF43 codon 659-mutated, Non-class 1 BRAF-mutated, and MSl-H, G3: RNF43 Non-codon 659-mutated (including RNF43 wild-type), Class 1 BRAF-mutated, and Non-MSl-H; G2: Non-G1 and Non-G3. The study found that the G1 had a better PFS and the G3 had a worse PFS (G1/G3: P=0.0005; G1/G2: P=0.3062; G2/G3: P<0.0001, Figure 5A). The results of the OS survival analysis were more significant: the OS of G1 was 100%. The G3 has the worst OS in this cohort (G1/G2: P=0.0155; G1/G3: 0.0022; G2/G3: 0.0267, Figure 5B). RNF43 codon 659-mutated, Non-class 1 BRAF-mutated, and MSl-H were found to have the best prognosis in the population.
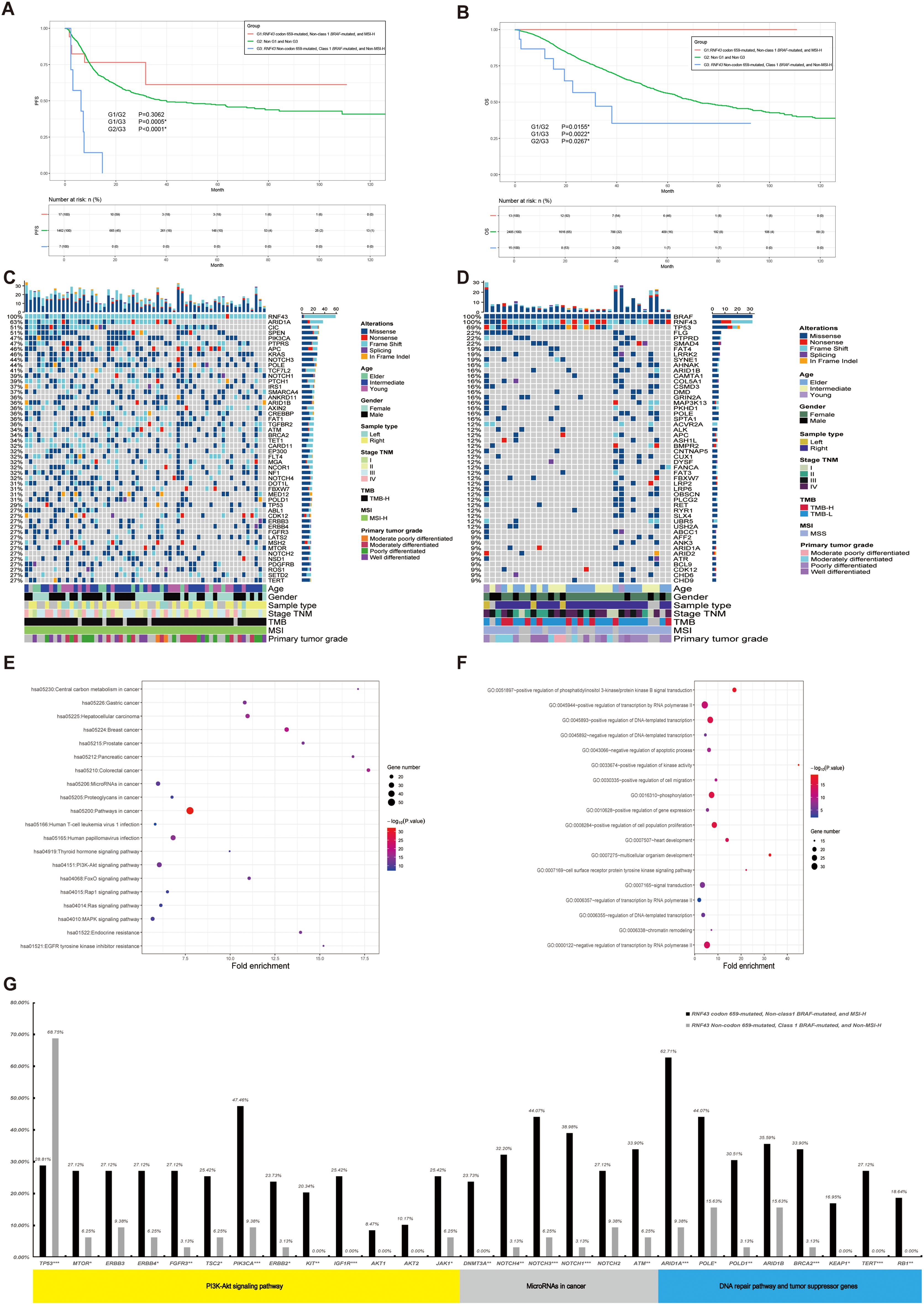
Figure 5. A panoramic analysis of the OS outcome, genomic and pathway characteristics of RNF43-mutated, MSI, and BRAF in CRC. (A) KM analysis of PFS between RNF43-mutated, MSI, and BRAF-mutated in this study. (B) KM analysis of OS between RNF43-mutated, MSI, and BRAF-mutated in this study. Top 50 mutation spectrum in G1 (C) and G3 (D) RNF43-mutated patients. Each column represents a patient, and each row represents a gene. The table on the left represents the mutation rate of each gene. The top plot represents the overall number of mutations a patient carried. Different colors denote different types of mutations. KEGG (E) and GO (F) functional enrichment analyses of G1 and G3. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes. G: The differences in core gene mutation of major signaling pathways (PI3K-Akt signaling pathway, MicroRNAs pathway, DNA damage repair, and tumor suppressor genes) between G1 and G3. G1: RNF43 codon 659-mutated, Non-class 1 BRAF-mutated, and MSI-H, G3: RNF43 Non-codon 659-mutated (including RNF43 wild-type), Class 1 BRAF-mutated, and Non-MSI-H; G2: Non-G1 and Non-G3. CRC, Colorectal cancer; *p<0.05; **p<0.01; ***p<0.001.
Meanwhile, we performed clinicopathological analysis and mutation characteristic analysis, as shown in Table 4. Compared with the G2 and G3 groups, the age of G1 was higher than that of the Elder group (64.71%; G1/G2: P<0.0001; G1/G3: P<0.0001). There was no significant difference in gender among the three groups. In terms of primary tumor location, contrary to previous conclusions, G1 was significantly enriched on the right side (88.24%; G1/G2: P<0.0001; G1/G3: P<0.0001). G1 was significantly enriched in stage II colorectal cancer (52.94%; G1/G2: P<0.0001; G1/G3: P = 0.0087). In terms of Tumor_Grade, the frequency of poorly differentiated tumors was higher in G1 (38.24%, N=13), but there was no statistical difference in G1/G3 group (P=0.0767). Finally, we show the mutation landscape of G1 (Figure 5C) and G3 (Figure 5D) and carry out mutation difference analysis (Supplementary Table 12) and pathway enrichment (Figures 5E–F). The enrichment results of major pathways were consistent with the results of RNF43-mutated/RNF43 wild-type and RNF43-mutated and BRAF/or MSI analysis (as shown in Figures 5E–F). The mutation frequency of core gene mutations of major signaling pathways (PI3K-Akt signaling pathway, MicroRNAs pathway, DNA damage repair, and tumor suppressor genes) in G1 was significantly higher than that in G3 (Figure 5G).
4 Discussion
Colorectal cancer (CRC) is highly heterogeneous and has significant prognostic differences (3, 4). Prognostic prediction based on molecular characteristics has been reported in some studies, but MSI-H is the only target that has been promoted to clinical treatment guidelines (21). In this study, we obtained data from 4,028 CRC patients for an in-depth analysis of RNF43 as a potential target. This analysis revealed significant differences in PFS and no significant differences in OS between patients with RNF43-mutated and RNF43 wild-type. DNA combined with RNA-based joint analysis in the coad_cptac_2019 cohort suggests that RNF43-mutated/expression < 0 shared a similar mutation spectrum with the RNF43-mutated, and RNF43 wild-type/expression > 0 had a similar mutation spectrum to the RNF43 wild-type, which represents a more optimal choice for precise prognostic stratification. RNF43 codon 659-mutated can be used as a prognostic indicator for CRC in this study, and RNF43 codon 659-mutated combined with Non-class1 BRAF-mutated and MSI-H has the best prognosis. RNF43 Non-codon 659-mutated combined with Class 1 BRAF-mutated and Non-MSI-H had the worst prognosis. We also found that RNF43 codon 659-mutated is highly correlated with MSI-H and TMB-H, which is consistent with previous studies (13), and indicates that RNF43 codon 659-mutated is a special subtype and may be an advantageous subtype for ICIs. This study integrates all published CRC cohorts with clinicopathological and mutational information in Cbioport. To our knowledge, this is the largest CRC research dataset to date.
RNF43 and BRAF are molecular events involved in the serrated tumor pathway during CRC development (31). Studies have reported that RNF43 (24), BRAF, and MSI status have clear clinical significance at present. RNF43-mutated patients are associated with improved survival in CRC patients receiving ICIs (30). RNF43-mutated often co-occur with BRAF V600E mutations. The combination of RNF43-mutated with BRAF V600E mutations was significantly associated with poorer survival (20, 30). However, the above study did not provide a more detailed analysis of RNF43-mutated types or characteristics. At present, only one study divided RNF43 into N-terminal and C-terminal based on codon 313 as a cutoff to demarcate the RING region, and found that RNF43 mutations in the N-terminal region showed a shorter overall survival (19). RNF43, a WNT signaling pathway negative regulator, can predict the response of BRAF V600E MSS metastatic colorectal cancer against BRAF/EGFR combination therapy, where MSI-H always carries RNF43 wildtype-like, encoding p.G659fs* and presents an intermediate response frequency (30). This suggests that the RNF43 codon 659-mutated is a special subtype that warrants further study. MSI-H/dMMR are identified as key biomarkers guiding treatment strategies and disease management in CRC, suggesting that mCRC patients benefit from immunotherapy. Furthermore, RNF43-mutated were frequent (12.9%) in precancerous lesions of ulcerative colitis (UC) patients and detectable in 24.4% of colitis-associated cancer patients. RNF43-mutated caused invasive CRC by aggravating and perpetuating inflammation due to impaired epithelial barrier integrity and pathogen control, and RNF43 inactivated mutation was even sufficient to cause spontaneous intestinal inflammation, resulting in subsequent invasive carcinoma development (32). Currently, there is no reference regarding the temporal sequence of RNF43 and MSI-H. In 2018, a study established a 20-gene panel that could distinguish CRC from adenomas (33). In 2024, a study compared the mutation characteristics of different precancerous lesions and stage I-IV CRC. However, the role of RNF43 in the process from precancerous lesions to the onset of CRC was not mentioned (34). Considering the high correlation between RNF43 and MSI-H found in this study, it is of great significance to conduct DNA and RNA multi-omics exploration through gastroscopy polyp screening and hereditary tumor screening to deeply analyze the role of RNF43 in the process from precancerous lesions to the onset of CRC. In this study, based on RNF43 codon 659-mutated combined with Non-class1 BRAF-mutated or MSI-H, CRC has a better prognosis. RNF43 codon 659-mutated combined with non-class 1 BRAF-mutated and MSI-H had the best prognosis, with OS reaching 100% in 13 patients, mainly stage III-IV CRC patients (11/13). It is also currently unreported that a combined biomarker can predict patient outcomes.
However, there are some limitations to this study. First of all, the data in this study came from a public database and only included SNV data, without collating CNV and SV data, which may provide obstacles for further findings, but the conclusions obtained in the current study will not be affected. Meanwhile, the completeness of the data we evaluated before the start of this study (Supplementary Table 4), the deletion rates further reinforces the reliability of the data and the conclusions drawn from it. Secondly, our study mainly provided cross-sectional data for the analysis of overall survival. Due to data limitations, we did not further analyze the subgroup of efficacy prediction based on the findings, which limited the innovation of this study. Subsequent studies should conduct efficacy prediction analysis in the cohort receiving targeted therapy and immunotherapy to verify the conclusions of this study and improve the depth of the overall study. Third, ethnic information was not available in this study, so the mutation heterogeneity among different ethnic groups was not deeply considered, which may limit the universality of the study’s conclusions. Fourth, the comparative analysis of RNF43 wild-type and RNF43-mutated on the basis of MSI-H showed that OS had a trend of prognostic prediction. However, no statistically significant difference was found. Therefore, we cannot conclusively determine that RNF43 is a key driver gene compared to MSI-H. Meanwhile, we only conducted independent cohort validations of the mutant and wild types of RNF43, and our findings were consistent with those from the analysis cohort. However, due to the limited number of validation codons, we have not yet performed cohort validations for the RNF43 codon 659-mutated, which may affect the credibility of our results. In the future, under the condition of sufficient sample size and relatively fewer confounding factors, the mutation difference and prognosis difference of different populations can be compared to make up for the shortcomings of this study. In conclusion, subsequent studies can set up independent subgroups based on the findings of this study to improve the statistical robustness of the conclusions.
5 Conclusions
In conclusion, our findings elucidated a good prognosis of RNF43 codon 659-mutated and concomitant Non-class 1 BRAF-mutated with MSI-H in CRC. Specifically, we found that RNF43 codon 659-mutated is a specific subtype that is more likely to benefit from ICIs in CRC, causing the incidence of TMB-H and MSI-H in the RNF43 codon 659-mutated group to be significantly higher. These results provided novel insights into the clinical applications based on mutation-based molecular typing that can help fine-screen populations with different prognoses and benefit from precision therapy.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by the institutional review boards of Beijing Hospital (2023BJYYEC-428-02). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
FW: Writing – original draft, Investigation. LL: Writing – review & editing, Investigation. ZL: Data curation, Writing – original draft, Writing – review & editing, Conceptualization. LQ: Writing – review & editing, Data curation. SZ: Writing – original draft. XH: Writing – original draft. YZ: Writing – review & editing, Funding acquisition. YH: Writing – review & editing, Conceptualization, Supervision.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
Authors ZL and LQ were employed by company Geneplus-Beijing.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1608664/full#supplementary-material
Supplementary Figure 1 | Lollipop plots (maps mutations on a linear protein and its domains) in each single cohort. Truncating includes frameshift mutations and nonsense mutations.
Supplementary Figure 2 | A panoramic analysis of the genomic characteristics of RNF43 wild-type in CRC. (A): Top 50 mutation spectrum in RNF43 wild-type patients. Each column represents a patient, and each row represents a gene. The table on the left represents the mutation rate of each gene. The top plot represents the overall number of mutations a patient carried. Different colors denote different types of mutations. (B): KM analysis of PFS between RNF43-mutated and RNF43 wild-type in this study. (C): KM analysis of OS between RNF43-mutated and RNF43 wild-type in this study. (D): RNF43 RNA expression levels in the RNF43-mutated and RNF43 wild-type. (E): The mutation landscape in the coad_cptac_2019 data set. CRC, Colorectal cancer; PFS: progression-free survival; OS, overall survival; KM, Kaplan-Meier.
Supplementary Figure 3 | A panoramic analysis of the genomic characteristics of RNF43 wild-type in validation cohort. A: Top 50 mutation spectrum in RNF43-mutated patients in validation cohort. B: Top 50 mutation spectrum in RNF43 wild-type patients in validation cohort. Each column represents a patient, and each row represents a gene. The table on the left represents the mutation rate of each gene. The top plot represents the overall number of mutations a patient carried. Different colors denote different types of mutations. C: The differences in core gene mutation of major signaling pathways (PI3K-Akt signaling pathway, MicroRNAs pathway, DNA damage repair, and tumor suppressor genes) between RNF43-mutated and RNF43 wild-type. D: KEGG functional enrichment analyses of RNF43-mutated and RNF43 wild-type. E: GO functional enrichment analyses of RNF43-mutated and RNF43 wild-type. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes. *, p<0.05; **, p<0.01; ***, p<0.001.
Supplementary Figure 4 | The mutation landscape analysis of the MSI-H subgroup with RNF43. A: The mutation landscape of the MSI-H and RNF43-mutated group. B: The mutation landscape of the MSI-H and RNF43 wild-type group. C: The mutation landscape of the MSI-H and RNF43 codon 659-mutated group. D: The mutation landscape of MSI-H and RNF43 Non-codon 659-mutated group.
Supplementary Figure 5 | A panoramic analysis of the survival outcome, genomic and pathway characteristics of RNF43-mutated, and BRAF in CRC. A: Top 50 mutation spectrum in RNF43 codon 659-mutated and Non-class 1 BRAF-mutated patients. B: Top 50 mutation spectrum in RNF43 Non-codon 659-mutated and Class 1 BRAF-mutated patients. Each column represents a patient, and each row represents a gene. The table on the left represents the mutation rate of each gene. The top plot represents the overall number of mutations a patient carried. Different colors denote different types of mutations. C: The volcanic maps for between RNF43 codon 659-mutated/Non-class 1 BRAF-mutated patients and RNF43 Non-codon 659-mutated/Class 1 BRAF-mutated. KEGG (D) and GO (E) functional enrichment analyses of RNF43 codon 659-mutated/Non-class 1 BRAF-mutated patients and RNF43 Non-codon 659-mutated/Class 1 BRAF-mutated patients. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes. F: The differences in core gene mutation of major signaling pathways (PI3K-Akt signaling pathway, MicroRNAs pathway, DNA damage repair, and tumor suppressor genes) between RNF43 codon 659-mutated/Non-class 1 BRAF-mutated patients and RNF43 Non-codon 659-mutated/Class 1 BRAF-mutated.
Supplementary Figure 6 | A panoramic analysis of the survival outcome, genomic and pathway characteristics of RNF43-mutated and BRAF-mutated in CRC. A: Top 50 mutation spectrum in RNF43 codon 659-mutated and MSI-H patients. B: Top 50 mutation spectrum in RNF43 Non-codon 659-mutated and Non-MSI-H patients. Each column represents a patient, and each row represents a gene. The table on the left represents the mutation rate of each gene. The top plot represents the overall number of mutations a patient carried. Different colors denote different types of mutations. MSI-H: patients with high MSI. C: The volcanic maps for between RNF43 codon 659-mutated/MSI-H patients and RNF43 Non-codon 659-mutated/Non-MSI-H patients. KEGG (D) and GO (E) functional enrichment analyses of RNF43 codon 659-mutated/MSI-H patients and RNF43 Non-codon 659-mutated/Non-MSI-H. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes. F: The differences in core gene mutation of major signaling pathways (PI3K-Akt signaling pathway, MicroRNAs pathway, DNA damage repair, and tumor suppressor genes) between RNF43 codon 659-mutated/MSI-H patients and RNF43 Non-codon 659-mutated/Non-MSI-H.
Glossary
CRC: Colorectal cancer
OS: Overall Survival
PFS: Progression-free survival
ICIs: Immune checkpoint inhibitors
NCCN: the National Comprehensive Cancer Network
RNF43: Ring finger protein 43
BRAF: B-Raf proto-oncogene, serine/threonine kinase
MAPK: the Mitogen-Activated Protein Kinase
MSI: Microsatellite instability
MSS: Microsatellite stable
TMB: Tumor mutational burden
TNM: The Tumor-Node-Metastasis system
HR: Hazard ratio
CI: Confidence interval
PI3K: Phosphoinositol-3 kinase
TCGA: The Cancer Genome Atlas
cBioPortal: cBio Cancer Genomics Portal
GO: Gene Ontology
KEGG: Kyoto Encyclopedia of Genes and Genomes
KM: Kaplan-Meier
ARID1A: AT-rich interaction domain 1A
CIC: Capicua transcriptional repressor
PIK3CA: Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
PTPRS: Protein tyrosine phosphatase receptor type S
APC: APC regulator of WNT signaling pathway
FAT1: FAT atypical cadherin 1
POLE: DNA polymerase epsilon, catalytic subunit
NOTCH3: Notch receptor 3
SPEN: Spen family transcriptional repressor
TP53: Tumor protein p53
KRAS: KRAS proto-oncogene, GTPase
FBXW7: F-box and WD repeat domain containing 7
SMAD4: SMAD family member 4
TCF7L2: Transcription factor 7 like 2
SOX9: SRY-box transcription factor 9
PTCH1: Patched 1
SMARCA4: SWI/SNF related BAF chromatin remodeling complex subunit ATPase 4
FLT4: Fms related receptor tyrosine kinase 4
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Morgan E, Arnold M, Gini A, Lorenzoni V, Cabasag CJ, Laversanne M, et al. Global burden of colorectal cancer in 2020 and 2040: incidence and mortality estimates from GLOBOCAN. Gut. (2023) 72:338–44. doi: 10.1136/gutjnl-2022-327736
3. Hjortborg M, Edin S, Böckelman C, Kaprio T, Li X, Gkekas I, et al. Systemic inflammatory response in colorectal cancer is associated with tumour mismatch repair and impaired survival. Sci Rep. (2024) 14:29738. doi: 10.1038/s41598-024-80803-6
4. Shi M, Yang Y, Huang N, Zeng D, Mo Z, Wang J, et al. Genetic and microenvironmental evolution of colorectal liver metastases under chemotherapy. Cell Rep Med. (2024) 5:101838. doi: 10.1016/j.xcrm.2024.101838
5. Guo H, Miao L, and Yu C. The efficacy of targeted therapy and/or immunotherapy with or without chemotherapy in patients with Colorectal Cancer: A Network Meta-Analysis. Eur J Pharmacol. (2024) 988:177219. doi: 10.1016/j.ejphar.2024.177219
6. Liang B, Tang M, Huang C, Yang Y, He Y, Liao S, et al. A systematic review and meta-analysis of the efficacy and safety of regorafenib in the treatment of metastatic colorectal cancer. J gastrointestinal Cancer. (2024) 56:36. doi: 10.1007/s12029-024-01158-9
7. Wang Z, Liu Y, Wang K, and Ma L. Efficacy and safety of PD-1 and PD-L1 inhibitors in advanced colorectal cancer: a meta-analysis of randomized controlled trials. BMC Gastroenterol. (2024) 24:461. doi: 10.1186/s12876-024-03554-8
8. Liu B, Liu R, Zhang X, Tian L, Li Z, and Yu J. Ubiquitin-conjugating enzyme E2T confers chemoresistance of colorectal cancer by enhancing the signal propagation of Wnt/β-catenin pathway in an ERK-dependent manner. Chemico-biological Interact. (2024) 406:111347. doi: 10.1016/j.cbi.2024.111347
9. Tsukiyama T, Fukui A, Terai S, Fujioka Y, Shinada K, Takahashi H, et al. Molecular role of RNF43 in canonical and noncanonical wnt signaling. Mol Cell Biol. (2015) 35:2007–23. doi: 10.1128/MCB.00159-15
11. Shang P, Lu J, Song F, Zhao Y, Hong W, He Y, et al. RNF43 is associated with genomic features and clinical outcome in BRAF mutant colorectal cancer. Front Oncol. (2023) 13:1119587. doi: 10.3389/fonc.2023.1119587
12. Eto T, Miyake K, Nosho K, Ohmuraya M, Imamura Y, Arima K, et al. Impact of loss-of-function mutations at the RNF43 locus on colorectal cancer development and progression. J Pathol. (2018) 245:445–55. doi: 10.1002/path.5098
13. Seeber A, Battaglin F, Zimmer K, Kocher F, Baca Y, Xiu J, et al. Comprehensive analysis of R-spondin fusions and RNF43 mutations implicate novel therapeutic options in colorectal cancer. Clin Cancer research: an Off J Am Assoc Cancer Res. (2022) 28:1863–70. doi: 10.1158/1078-0432.CCR-21-3018
14. Wu S, Deng Y, Sun H, Liu X, Zhou S, Zhao H, et al. BRAF inhibitors enhance erythropoiesis and treat anemia through paradoxical activation of MAPK signaling. Signal transduction targeted Ther. (2024) 9:338. doi: 10.1038/s41392-024-02033-6
15. Sanz-Garcia E, Argiles G, Elez E, and Tabernero J. BRAF mutant colorectal cancer: prognosis, treatment, and new perspectives. Ann oncology: Off J Eur Soc Med Oncol. (2017) 28:2648–57. doi: 10.1093/annonc/mdx401
16. Ros J, Matito J, Villacampa G, Comas R, Garcia A, Martini G, et al. Plasmatic BRAF-V600E allele fraction as a prognostic factor in metastatic colorectal cancer treated with BRAF combinatorial treatments. Ann oncology: Off J Eur Soc Med Oncol. (2023) 34:543–52. doi: 10.1016/j.annonc.2023.02.016
17. Yao Z, Yaeger R, Rodrik-Outmezguine VS, Tao A, Torres NM, Chang MT, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature. (2017) 548:234–8. doi: 10.1038/nature23291
18. Santarpia L, Lippman SM, and El-Naggar AK. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin Ther Targets. (2012) 16:103–19. doi: 10.1517/14728222.2011.645805
19. Huang ZY, Wen L, Ye LF, Lu YT, Pat Fong W, Zhang RJ, et al. Clinical and molecular characteristics of RNF43 mutations as promising prognostic biomarkers in colorectal cancer. Ther Adv Med Oncol. (2024) 16:17588359231220600. doi: 10.1177/17588359231220600
20. Matsumoto A, Shimada Y, Nakano M, Oyanagi H, Tajima Y, Nakano M, et al. RNF43 mutation is associated with aggressive tumor biology along with BRAF V600E mutation in right-sided colorectal cancer. Oncol Rep. (2020) 43:1853–62. doi: 10.3892/or.2020.7561
21. Benson AB, Venook AP, Adam M, Chang G, Chen YJ, Ciombor KK, et al. Colon cancer, version 3.2024, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Network: JNCCN. (2024) 22:e240029. doi: 10.6004/jnccn.2024.0029
22. Kong H, Yang Q, Wu C, Wu X, Yan X, Huang LB, et al. Spatial context of immune checkpoints as predictors of overall survival in patients with resectable colorectal cancer independent of standard tumor-node-metastasis stages. Cancer Res Commun. (2024) 4:3025–35. doi: 10.1158/2767-9764.CRC-24-0270
23. Bao X, Zhang H, Wu W, Cheng S, Dai X, Zhu X, et al. Analysis of the molecular nature associated with microsatellite status in colon cancer identifies clinical implications for immunotherapy. J Immunotherapy Cancer. (2020) 8:e001437. doi: 10.1136/jitc-2020-001437
24. Giannakis M, Hodis E, Jasmine Mu X, Yamauchi M, Rosenbluh J, Cibulskis K, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet. (2014) 46:1264–6. doi: 10.1038/ng.3127
25. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signaling. (2013) 6:pl1. doi: 10.1126/scisignal.2004088
26. Peng W, Li B, Li J, Chang L, Bai J, Yi Y, et al. Clinical and genomic features of Chinese lung cancer patients with germline mutations. Nat Commun. (2022) 13:1268. doi: 10.1038/s41467-022-28840-5
27. Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell. (2018) 33:125–136.e123. doi: 10.1016/j.ccell.2017.12.004
28. Mondaca S, Walch H, Nandakumar S, Chatila WK, Schultz N, and Yaeger R. Specific mutations in APC, but not alterations in DNA damage response, associate with outcomes of patients with metastatic colorectal cancer. Gastroenterology. (2020) 159:1975–1978.e1974. doi: 10.1053/j.gastro.2020.07.041
29. Huang Y, Jia W, Zhao G, Zhao Y, Zhang S, Li Z, et al. Clinical features and mutation analysis of class 1/2/3 BRAF mutation colorectal cancer. Chin Clin Oncol. (2024) 13:3. doi: 10.21037/cco-23-117
30. Elez E, Ros J, Fernández J, Villacampa G, Moreno-Cárdenas AB, Arenillas C, et al. RNF43 mutations predict response to anti-BRAF/EGFR combinatory therapies in BRAF(V600E) metastatic colorectal cancer. Nat Med. (2022) 28:2162–70. doi: 10.1038/s41591-022-01976-z
31. Mikaeel RR, Young JP, Li Y, Poplawski NK, Smith E, Horsnell M, et al. RNF43 pathogenic Germline variant in a family with colorectal cancer. Clin Genet. (2022) 101:122–6. doi: 10.1111/cge.14064
32. Dietl A, Ralser A, Taxauer K, Dregelies T, Sterlacci W, Stadler M, et al. RNF43 is a gatekeeper for colitis-associated cancer. bioRxiv. (2024) 130, 577936. doi: 10.1101/2024.01.30.577936
33. Lin S-H, Raju GS, Huff C, Ye Y, Gu J, Chen J-S, et al. The somatic mutation landscape of premalignant colorectal adenoma. Gut. (2018) 67:1299–305. doi: 10.1136/gutjnl-2016-313573
Keywords: colorectal cancer (CRC), cbioportal database, RNF43-mutated, mutation analysis, prognostic
Citation: Wang F, Lin L, Li Z, Qin L, Zhang S, Hu X, Zhao Y and Huang Y (2025) Define a good prognosis of RNF43 codon 659-mutated and concomitant genomic signatures in CRC: an analysis of the cBioPortal database. Front. Oncol. 15:1608664. doi: 10.3389/fonc.2025.1608664
Received: 09 April 2025; Accepted: 18 July 2025;
Published: 08 August 2025.
Edited by:
Tiziana Venesio, Institute for Cancer Research and Treatment (IRCC), ItalyCopyright © 2025 Wang, Lin, Li, Qin, Zhang, Hu, Zhao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingying Huang, eWluZ2hoQGhvdG1haWwuY29t
†These authors have contributed equally to this work
 Feng Wang
Feng Wang Li Lin2†
Li Lin2† Zhongkang Li
Zhongkang Li Shuai Zhang
Shuai Zhang Yingying Huang
Yingying Huang