- 1Department of Clinical Genetics and Genomics, Karolinska University Hospital, Solna, Sweden
- 2Department of Clinical Pathology and Cancer Diagnostics, Karolinska University Hospital and Institute, Stockholm, Sweden
- 3Center for Hematology and Regenerative Medicine, Department of Medicine Huddinge, Karolinska Institute, Karolinska University Hospital, Stockholm, Sweden
Platelet-derived growth factor receptor beta (PDGFRB)-rearranged myeloid/lymphoid neoplasms (MLNs) are rare hematologic malignancies typically responsive to tyrosine kinase inhibitors (TKIs) such as imatinib. However, resistance—particularly in the context of co-occurring high-risk mutations—is uncommon and poorly characterized. We report a case of a 65-year-old man diagnosed with a ETV6::PDGFRB-translocated MLN, presenting as atypical chronic myeloid leukemia (aCML), who exhibited a brief response with development of resistance to imatinib. Although the patient initially achieved hematologic and partial cytogenetic remission, residual fibrosis and cytogenetic abnormalities persisted despite dose escalation. Molecular profiling revealed high-risk mutations in ASXL1, KRAS, NRAS, SETBP1, and SRSF2, along with a variant of uncertain significance (VUS) in IDH2. The patient progressed to acute myeloid leukemia (AML) within 11 months despite sequential therapies including dasatinib and azacitidine-venetoclax, ultimately succumbing to sepsis. This case highlights the limitations of TKI monotherapy in MLNs with PDGFRB rearrangements and co-existing high-risk mutations, underscoring the importance of early molecular profiling and consideration of allogeneic hematopoietic stem cell transplantation in cases with poor risk features.
Introduction
Platelet-derived growth factor receptors alpha (PDGFRα) and beta (PDGFRß) are members of the class III receptor tyrosine kinase family, playing central roles in cellular growth, differentiation, and proliferation (1). Myeloid/lymphoid neoplasms with eosinophilia and PDGFRB rearrangements typically present as chronic myeloid neoplasms and are classified as a distinct entity in the World Health Organization (WHO) classification. The ETV6 gene is the most common fusion partner (2).
These rearrangements result in constitutive tyrosine kinase activation, rendering most cases highly sensitive to imatinib (3, 4). Over 40 different fusion partners have been reported, with ETV6 being the most frequent (5).
Patients with PDGFRB translocations may lack eosinophilia, only low grade in this case, and rarely present with additional cytogenetic or molecular abnormalities (4, 6–8).
Although most cases respond well to TKIs, resistance—either primary or acquired—remains poorly understood. Proposed mechanisms include secondary kinase domain mutations or disease progression to AML (9). TKI resistance has also been described in lymphoid malignancies with PDGFRB rearrangements (10, 11), though B-cell acute lymphatic leukemia with such fusions appears highly TKI-sensitive (12).
Here we describe a patient with a ETV6::PDGFRB translocation presenting as aCML, developing resistance to imatinib. The presence of high-risk mutations likely contributed to the resistance and rapid progression to AML. This case emphasizes the need for comprehensive molecular profiling, clonal monitoring, and early consideration of allogeneic stem cell transplantation in MLN cases with PDGFRB rearrangements that do not respond as expected to treatment.
Case description
A 65-year-old man with a history of prostate cancer treated by radical prostatectomy 12 years prior, without adjuvant therapy, presented with splenomegaly (12 cm below the left costal margin), 8-kg weight loss over 2 months, and gout. Laboratory studies revealed leukocytosis (69 × 109/L), mild peripheral eosinophilia (2%), and mild anemia. Bone marrow biopsy showed hypercellularity with myeloid proliferation, dysgranulopoiesis, and grade 2 fibrosis, without increased blasts or significant eosinophilia (Figure 1). Cytogenetics revealed t(5;12)(q33;p13) as the sole abnormality. Fluorescence in situ hybridization (FISH) examination confirmed PDGFRB rearrangement. RNA sequencing confirmed the presence of an ETV6::PDGFRB fusion transcript involving exon 4 of ETV6 (NM_001987.5) and exon 9 of PDGFRB (NM_002609.4). Accordingly, whole genome sequencing identified the translocation breakpoint within intron 4 of ETV6 (NM_001987.5) and intron 8 of PDGFRB (NM_001355017.2). BCR::ABL1 was negative. Targeted sequencing revealed pathogenic mutations in ASXL1, KRAS, NRAS, SETBP1, SRSF2, and a VUS in IDH2 (13).
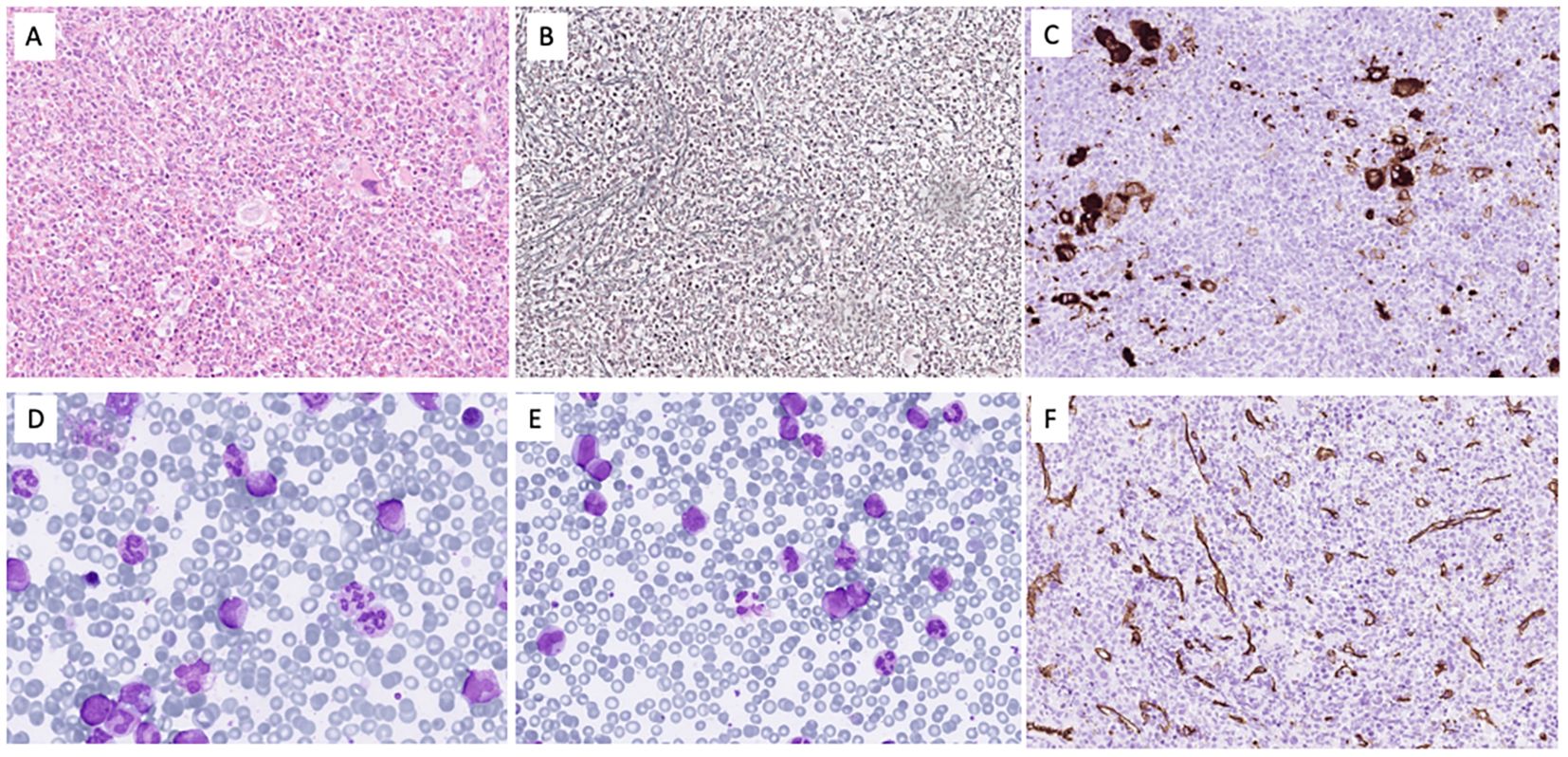
Figure 1. The bone marrow was hypercellular for age due to expansion of the granulocytopoiesis (A) with fibrosis grade II (B) and clusters of small megakaryocytes with monolobated nuclei (C). In the bone marrow smears (D), the M/E ratio was 8.3 with 1.8% blasts, 3% promyelocytes, and 22.6% myelocytes/metamyelocytes. The eosinophils were 1.6% and the monocytes 4%. In blood (E), blasts constituted 1.5%, promyelocytes 3%, and myelocytes/metamyelocytes 14%. CD34 staining (F) showed no increase in blasts.
Imatinib at 100 mg/day was initiated. Hematologic remission and spleen size normalization occurred within 2 months. However, at 10 months, a follow-up marrow showed only partial response, persistent patchy fibrosis, and 17% FISH positivity for the PDGFRB rearrangement. Imatinib was escalated to 400 mg/day, and transplantation workup was initiated.
Repeat marrow and FISH at 3 months later showed no further cytogenetic improvement and no blast increase. PDGFRB kinase domain sequencing showed no secondary mutations. Dasatinib at 100 mg/day was briefly trialed, but increasing leukocytosis necessitated azacitidine plus venetoclax. Despite therapy, the patient progressed to AML 15 months after diagnosis and died of sepsis with multiorgan failure during a neutropenic episode.
Serial sampling using the same targeted sequencing panel (13) in parallel with FISH for PDGFRB-rearrangement showed a decrease in PDGFRB-rearranged clone burden and parallel decreases in KRAS/NRAS variant allele frequency (VAF) (see Figure 2). In contrast, ASXL1, SETBP1, and SRSF2 VAFs remained stable. Interestingly, the IDH2 VUS VAF declined with the PDGFRB clone, suggesting that the variant was part of that clone. The two NRAS variants were consistently detected at low VAFs. Cytogenetics at AML transformation revealed monosomy 7 in 19/21 metaphases and a 17p11.2 locus loss in seven metaphases, alongside the persistent t(5;12), indicating clonal evolution. No TP53 mutations or new molecular abnormalities were found (Figure 2).
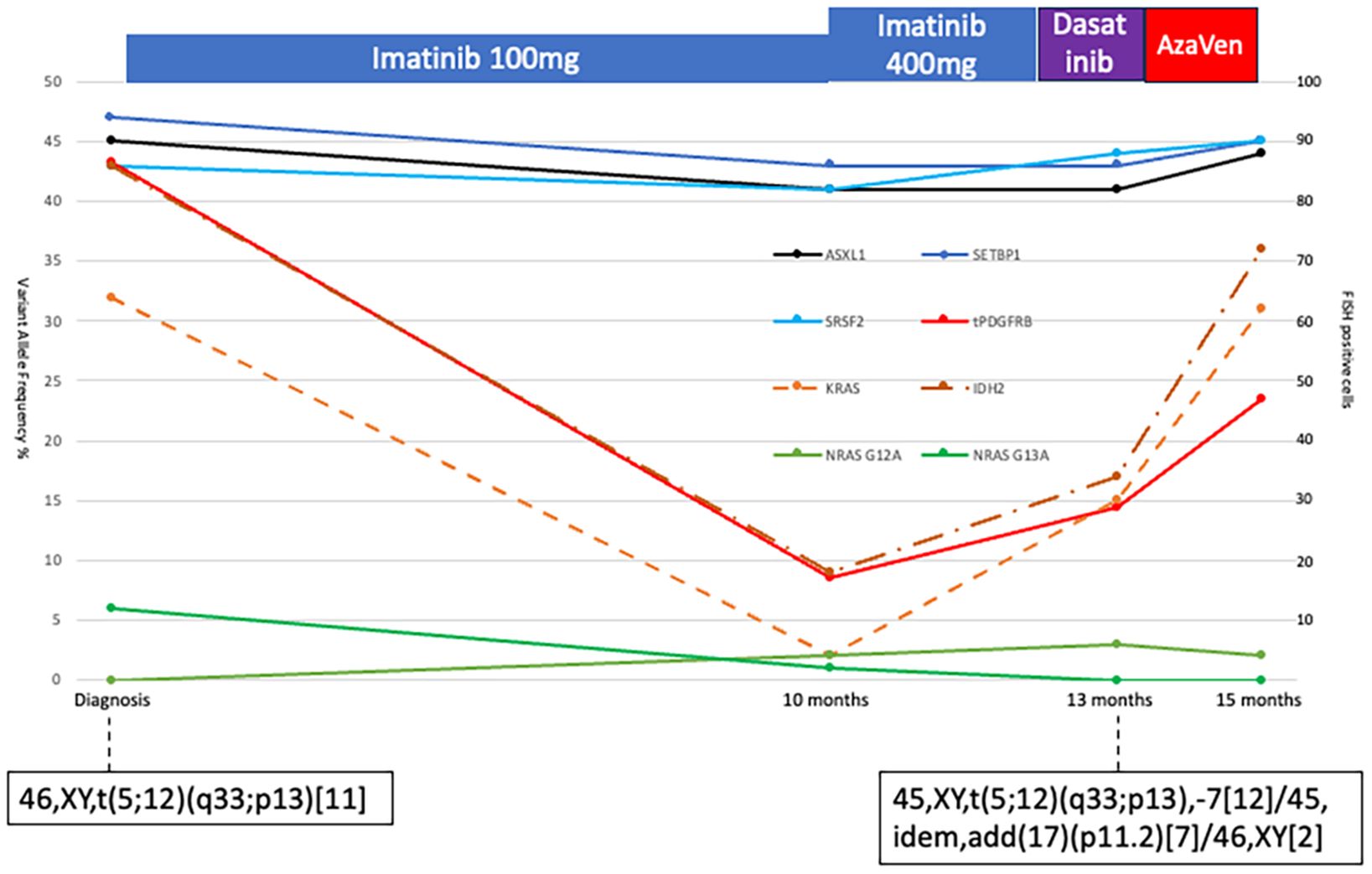
Figure 2. Diagram of bone marrow allele frequencies of ASXL1, SETBP1, SRSF2, KRAS, two NRAS, and IDH2 variants at different time points (scale on the left vertical). The PDGFB-translocation was measured with FISH at the same timepoints. A total of 200 cells were assessed, and the number of PDGFB-translocation-positive cells is plotted in the figure (red solid line, scale on the right vertical). Treatment is indicated above the diagram. Cytogenetic analyses were performed twice, and the results are indicated in the figure.
Discussion
Chronic-phase PDGFRB-rearranged neoplasms are typically sensitive to TKIs such as imatinib. Resistance is rare. Byrgazov et al. reported one imatinib-resistant NDEL1::PDGFRB case with a D850E mutation in the activation loop—a mechanism excluded in our case (9). Other reports describe TKI resistance predominantly in advanced-stage AML or lymphoid malignancies (7, 10, 11).
PDGFRB translocation alone appears to be a leukemogenic driver (14), explaining the usual absence of additional mutations or cytogenetic anomalies—similar to BCR::ABL1 in CML. Among the rare cases of TKI failure, few underwent molecular profiling (15, 16), and no previously identified mutations have been clearly linked to resistance (7, 17).
An exception is a case by Gou et al., who described an ETV6::PDGFRB-positive patient with concurrent NPM1, TET2, and NOTCH3 mutations and 46,XY,del(12)(p13p11.2), who developed a PDGFR-translocation positive T-lymphoblastic lymphoma while on imatinib (10).
In our case, ASXL1, SETBP1, and SRSF2 VAFs remained unchanged during treatment, indicating their independence from the PDGFRB-positive clone. While ASXL1 and SRSF2 mutations may reflect age-related clonal hematopoiesis, SETBP1 is more strongly associated with secondary AML, CMML, and aCML—often co-occurring with ASXL1 and SRSF2 in those neoplasms (18, 19). Given the unusual clinical course with an initial partial response to TKI followed by TKI-insensitive relapse and progression to AML, it is difficult not to think that the co-occurring mutations, in some way, contributed to the dismal outcome, even if it is impossible to conclude this in a single observational study such as this.
This case report has certain limitations, particularly in terms of generalizability—for example, the possibility of a pre-existing myeloproliferative neoplasm (MPN) with adverse mutations preceding the acquisition of the PDGFRB rearrangement cannot be excluded. Therefore, these findings should be interpreted with caution and may serve as a basis for further investigation.
The key message is to consider broad mutational profiling—even in the presence of targetable lesions like PDGFRB rearrangements and especially if signs of poor treatment response occur. In cases with poor molecular risk, rapid TKI response should not be assumed. While similar concerns have been raised in CML (20), this is, to our knowledge, the first report of primary TKI resistance in chronic-phase PDGFRB-rearranged MLN associated with high-risk mutations and adverse outcome.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The requirement of ethical approval was waived by Etikprövningsmyndigheten for the studies involving humans because Case reports are waived from ethical approval in Sweden. The patients closest relative (only daughter) approved the case study in writing and the patient approved it orally previous to his demise. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin because Please see the previous answer (8). Written informed consent was not obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article because The patient provided oral consent to publish his case as a case report prior to his demise. His closest relative (the only daughter) was present at the time and has provided written informed consent in the patients absence. Written informed consent was obtained from the participant/patient(s) for the publication of this case report.
Author contributions
LC: Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. LS: Formal Analysis, Investigation, Supervision, Validation, Writing – original draft, Writing – review & editing. BS: Data curation, Visualization, Writing – original draft, Writing – review & editing. SD: Conceptualization, Data curation, Funding acquisition, Investigation, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Guérit E, Arts F, Dachy G, Boulouadnine B, and Demoulin JB. PDGF receptor mutations in human diseases. Cell Mol Life Sci. (2021) 78:3867–81. doi: 10.1007/s00018-020-03753-y
2. Shomali W and Gotlib J. World Health Organization and International Consensus Classification of eosinophilic disorders: 2024 update on diagnosis, risk stratification, and management. Am J Hematol. (2024) 99:946–68. doi: 10.1002/ajh.27287
3. Apperley JF, Gardembas M, Melo JV, Russell-Jones R, Bain BJ, Baxter EJ, et al. Response to imatinib mesylate in patients with chronic myeloproliferative diseases with rearrangements of the platelet-derived growth factor receptor beta. N Engl J Med. (2002) 347:481–7. doi: 10.1056/NEJMoa020150
4. David M, Cross NC, Burgstaller S, Chase A, Curtis C, Dang R, et al. Durable responses to imatinib in patients with PDGFRB fusion gene-positive and BCR-ABL-negative chronic myeloproliferative disorders. Blood. (2007) 109:61–4. doi: 10.1182/blood-2006-05-024828
5. Saft L, Kvasnicka HM, Boudova L, Gianelli U, Lazzi S, and Rozman M. Myeloid/lymphoid neoplasms with eosinophilia and tyrosine kinase fusion genes: A workshop report with focus on novel entities and a literature review including paediatric cases. Histopathology. (2023) 83:829–49. doi: 10.1111/his.15021
6. Patnaik MM, Lasho TL, Finke CM, Pardanani A, and Tefferi A. Targeted next generation sequencing of PDGFRB rearranged myeloid neoplasms with monocytosis. Am J Hematol. (2016) 91:E12–4. doi: 10.1002/ajh.24267
7. Baer C, Muehlbacher V, Kern W, Haferlach C, and Haferlach T. Molecular genetic characterization of myeloid/lymphoid neoplasms associated with eosinophilia and rearrangement of PDGFRA, PDGFRB, FGFR1 or PCM1-JAK2. Haematologica. (2018) 103:e348–e50. doi: 10.3324/haematol.2017.187302
8. Metzgeroth G, Steiner L, Naumann N, Lübke J, Kreil S, Fabarius A, et al. Myeloid/lymphoid neoplasms with eosinophilia and tyrosine kinase gene fusions: reevaluation of the defining characteristics in a registry-based cohort. Leukemia. (2023) 37:1860–7. doi: 10.1038/s41375-023-01958-1
9. Byrgazov K, Kastner R, Gorna M, Hoermann G, Koenig M, Lucini CB, et al. NDEL1-PDGFRB fusion gene in a myeloid Malignancy with eosinophilia associated with resistance to tyrosine kinase inhibitors. Leukemia. (2017) 31:237–40. doi: 10.1038/leu.2016.250
10. Gou Y, Tang Y, Liu S, Cheng S, Deng X, Wen Q, et al. Myeloid/lymphoid neoplasms with ETV6::PDGFRB fusion gene: A rare case of poor response to imatinib and possible transformation mechanisms from myeloid neoplasms of bone marrow to T-cell lymphoblastic lymphoma invasion in lymph nodes. J Inflammation Res. (2023) 16:5163–70. doi: 10.2147/JIR.S427995
11. Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. (2014) 371:1005–15. doi: 10.1056/NEJMoa1403088
12. Roberts KG. Why and how to treat Ph-like ALL? Best Pract Res Clin Haematol. (2018) 31:351–6. doi: 10.1016/j.beha.2018.09.003
13. Orsmark-Pietras C, Lyander A, Ladenvall C, Hallström B, Staffas A, Awier H, et al. Precision diagnostics in myeloid Malignancies: development and validation of a national capture-based gene panel. Genes Chromosomes Cancer. (2024) 63:e23257. doi: 10.1002/gcc.23257
14. Dobbin E, Graham C, Corrigan PM, Thomas KG, Freeburn RW, and Wheadon H. Tel/PDGFRbeta induces stem cell differentiation via the Ras/ERK and STAT5 signaling pathways. Exp Hematol. (2009) 37:111–21. doi: 10.1016/j.exphem.2008.09.012
15. Cheah CY, Burbury K, Apperley JF, Huguet F, Pitini V, Gardembas M, et al. Patients with myeloid Malignancies bearing PDGFRB fusion genes achieve durable long-term remissions with imatinib. Blood. (2014) 123:3574–7. doi: 10.1182/blood-2014-02-555607
16. Grand FH, Burgstaller S, Kühr T, Baxter EJ, Webersinke G, Thaler J, et al. p53-Binding protein 1 is fused to the platelet-derived growth factor receptor beta in a patient with a t(5;15)(q33;q22) and an imatinib-responsive eosinophilic myeloproliferative disorder. Cancer Res. (2004) 64:7216–9. doi: 10.1158/0008-5472.CAN-04-2005
17. Ross DM, Altamura HK, Hahn CN, Nicola M, Yeoman AL, Holloway MR, et al. Delayed diagnosis leading to accelerated-phase chronic eosinophilic leukemia due to a cytogenetically cryptic, imatinib-responsive TNIP1-PDFGRB fusion gene. Leukemia. (2016) 30:1402–5. doi: 10.1038/leu.2015.301
18. Meggendorfer M, Bacher U, Alpermann T, Haferlach C, Kern W, Gambacorti-Passerini C, et al. SETBP1 mutations occur in 9% of MDS/MPN and in 4% of MPN cases and are strongly associated with atypical CML, monosomy 7, isochromosome i(17)(q10), ASXL1 and CBL mutations. Leukemia. (2013) 27:1852–60. doi: 10.1038/leu.2013.23
19. Makishima H. Somatic SETBP1 mutations in myeloid neoplasms. Int J Hematol. (2017) 105:732–42. doi: 10.1007/s12185-017-2241-1
Keywords: platelet-derived growth factor receptor beta-rearranged myeloid/lymphoid neoplasms, treatment resistance, imatinib, case report, acute myeloid leukemia
Citation: Cavelier L, Saft L, Sander B and Deneberg S (2025) Resistance to imatinib in a ETV6::PDGFRB rearranged myeloid/lymphoid neoplasm with high-risk mutations: a case report. Front. Oncol. 15:1611747. doi: 10.3389/fonc.2025.1611747
Received: 14 April 2025; Accepted: 21 October 2025;
Published: 07 November 2025.
Edited by:
Anjali Mishra, Sidney Kimmel Cancer Center, United StatesReviewed by:
Nicholas Cross, University of Southampton, United KingdomLiqiong Liu, Affiliated Nanshan Hospital of Shenzhen University, China
Copyright © 2025 Cavelier, Saft, Sander and Deneberg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefan Deneberg, c3RlZmFuLmRlbmViZXJnQHJlZ2lvbnN0b2NraG9sbS5zZQ==
 Lucia Cavelier
Lucia Cavelier Leonie Saft2
Leonie Saft2 Stefan Deneberg
Stefan Deneberg