- 1School of Biomedical Engineering, Guangzhou Medical University, Guangzhou, China
- 2Department of Emergency, the Second Affiliated Hospital, Guangzhou Medical University, Guangzhou, China
Acute myeloid leukemia (AML) is a highly heterogeneous malignant hematological neoplasm. Although standard diagnostic procedures have been established, traditional methods still face limitations with regard to efficiency, accuracy, and standardization. In recent years, artificial intelligence (AI) has demonstrated notable advantages in medical image analysis, flow cytometry interpretation, and genetic data modeling, offering new approaches for adjunctive diagnosis of AML. This review systematically summarizes recent research advances in adjunctive diagnosis of AML, categorizing current AI-based approaches based on data modality into three groups: blood smear image analysis, flow cytometry data interpretation, and genetic data modeling. We focus on the application strategies, diagnostic performance, and limitations of these approaches. Studies have shown that AI not only enhances diagnostic efficiency and reduces subjective bias, but also holds promise in identifying novel biomarkers. Nevertheless, current models still suffer from limited generalizability and insufficient clinical interpretability. Future efforts should prioritize data standardization, improve model transparency, and facilitate the seamless integration of AI systems into clinical workflows to support precision diagnosis and treatment of AML.
1 Introduction
Acute myeloid leukemia (AML) is a hematopoietic malignancy characterized by marked molecular and clinical heterogeneity, accounting for approximately 80% of adult acute leukemia cases (1). According to data from the Global Burden of Disease Project, the global burden of AML has increased substantially between 1990 and 2021, with the annual incidence rising from 79,372 to 144,645 cases, and annual mortality increasing from 74,917 to 130,189 deaths (2). Pathologically, AML is driven by the accumulation of genetic alterations in myeloid progenitor cells, resulting in impaired differentiation and uncontrolled proliferation (see Figure 1), ultimately leading to hematopoietic failure (3). Clinically, AML often presents with nonspecific symptoms such as anemia, fever, and fatigue (4), yet progresses rapidly and is difficult to manage (5, 6). Notably, even after initial treatment, residual leukemic cells known as minimal residual disease (MRD) may persist, representing a key factor contributing to disease relapse (7). Overall, AML is associated with poor prognosis, with a 5-year survival rate of approximately 30%, and less than 10% in patients over the age of 65 (8). These challenges highlight the urgent need for more accurate diagnostic modalities, robust risk stratification frameworks, and individualized treatment strategies to improve clinical outcomes.
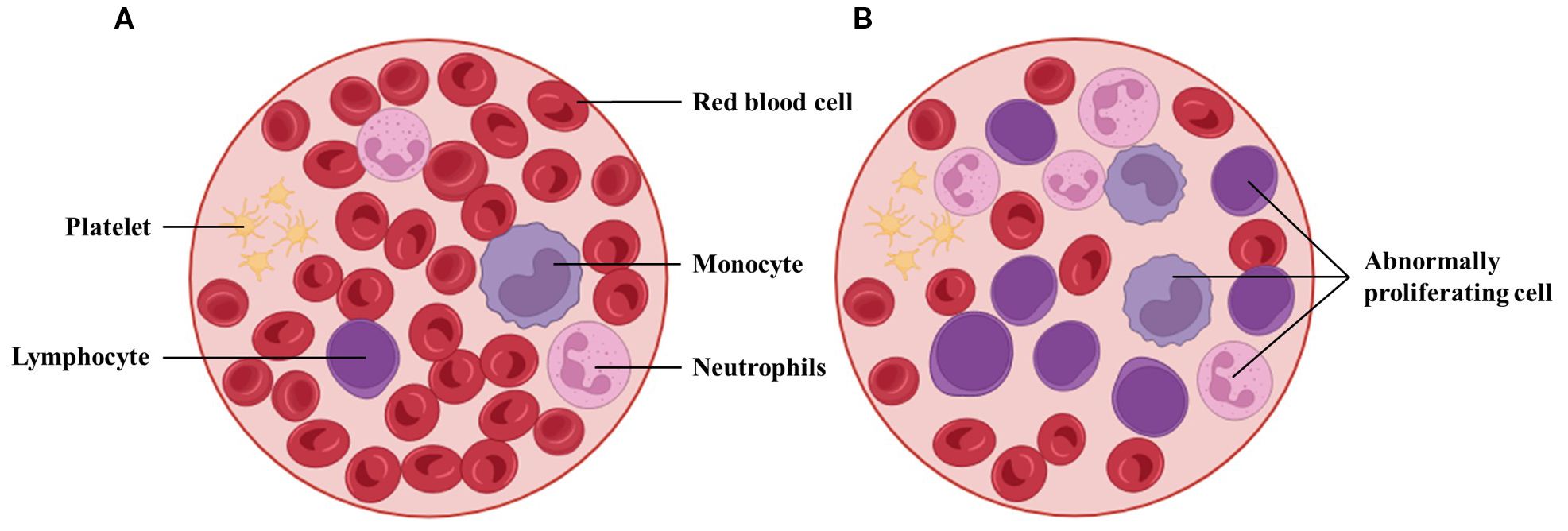
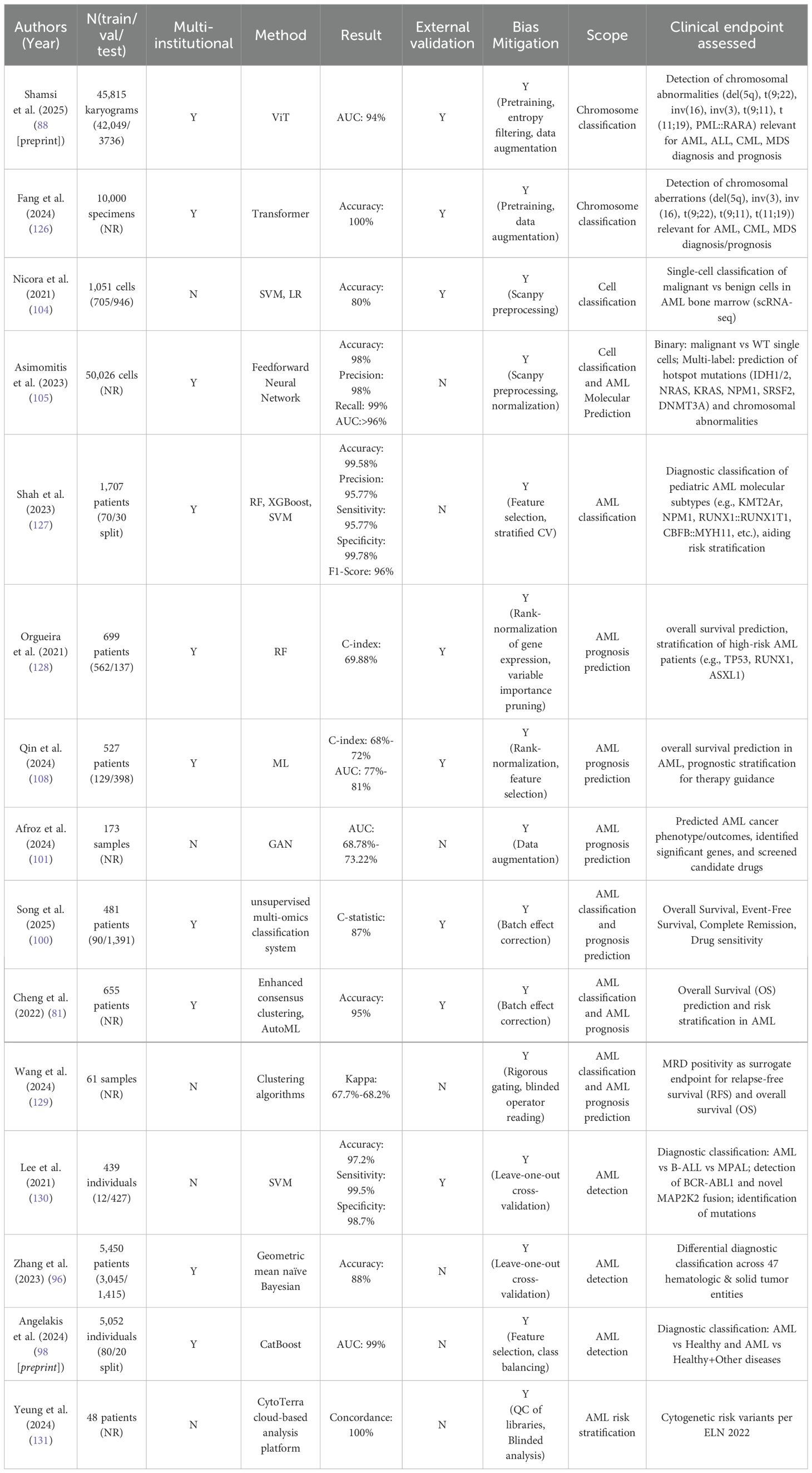
Figure 1. Morphological comparison between peripheral blood from a healthy individual and a patient with AML. (A) Normal smear with abundant mature erythrocytes and leukocyte subtypes. (B) AML smear with reduced mature cells and increased blasts. Such differences underpin AI-assisted blast detection and triage in hematology workflows, though performance depends on stain/scan standardization and external validation.
In recent years, artificial intelligence (AI) technologies, including machine learning and deep learning, have shown remarkable potential in recognizing complex patterns, analyzing high-dimensional data, and facilitating clinical decision-making (9, 10). Within the field of medical image analysis, AI algorithms have achieved significant success in tasks such as lesion detection, organ segmentation, and diagnostic assistance. In some scenarios, their performance has matched or even surpassed that of experienced physicians (11, 12). Beyond imaging, AI has also been extensively applied to the analysis of multidimensional datasets, such as flow cytometry and genomic profiles, to support disease prediction, classification, prognosis assessment, and evaluation of therapeutic responses (13, 14). The integration of AI into the automated analysis of images and flow cytometry data can greatly reduce diagnostic turnaround time, which is particularly critical for conditions requiring prompt intervention, such as acute promyelocytic leukemia (APL) (15), Additionally, AI systems help reduce subjectivity in morphological and flow cytometric interpretation, thereby improving reproducibility and standardization. More importantly, AI models are capable of identifying subtle features and potential novel biomarkers that may be imperceptible to human experts, contributing to a more profound understanding of disease pathogenesis.
In this review, “adjunctive” AI-assisted AML diagnosis refers to systems that do not render an autonomous final diagnosis. Instead, they function as decision-support tools that generate data-driven analyses—such as risk scores, classification recommendations, anomaly alerts, or triage prioritization—to enhance expert judgment, improve efficiency and consistency, and integrate with existing workflows. The final diagnostic decision remains with the treating physician.
Therefore, this review systematically summarizes recent advances in adjunctive diagnosis of AML, categorized by data modality into three major areas: blood smear image analysis, flow cytometry data interpretation, and genetic data modeling. For each modality, we examine the applied strategies, diagnostic performance, and inherent limitations. Finally, we discuss current challenges and outline future directions for the integration of AI-based adjunctive diagnostic techniques into routine clinical practice.
2 Review methods
This review was designed as a narrative survey of research on AI applied to adjunctive diagnosis of AML. Literature searches were conducted in PubMed and Web of Science, covering publications from January 2015 to March 2025, in order to capture both early applications and the most recent advances. The following keywords and their combinations were applied: “acute myeloid leukemia,” “artificial intelligence,” “machine learning,” “deep learning,” “blood smear,” “flow cytometry,” and “genomics.” Searches were limited to studies published in English.
Inclusion criteria were (1): original research applying AI or machine learning methods to AML diagnosis, subtype classification, MRD detection, or molecular feature prediction. (2) studies based on blood smear morphology, flow cytometry data, or genetic datasets. (3) reports providing quantitative outcomes such as accuracy, sensitivity, specificity, or AUC.
Exclusion criteria were: (1) studies focused solely on therapeutic prediction, drug screening, or treatment response without diagnostic relevance; and (2) narrative reviews, editorials, or conference abstracts lacking sufficient methodological detail.
Preprints (bioRxiv/medRxiv) were included when they presented AML-specific AI diagnostic research not yet available in peer-reviewed journals; these are clearly labeled as preprints in the References. Two authors independently screened titles/abstracts, reviewed full texts for eligibility, and extracted study design, data modality, sample size/splits, AI approach, and diagnostic performance metrics.
3 AML diagnosis: standards and clinical practice
3.1 Classification criteria
The clinical classification and diagnosis of AML are primarily based on three major systems. The first is the French-American-British (FAB) classification proposed in 1976 (16), which uses a threshold of >30% blast cells in the bone marrow for diagnosis. Based on cytomorphology and cytochemical staining, AML is subdivided into eight types (M0–M7).
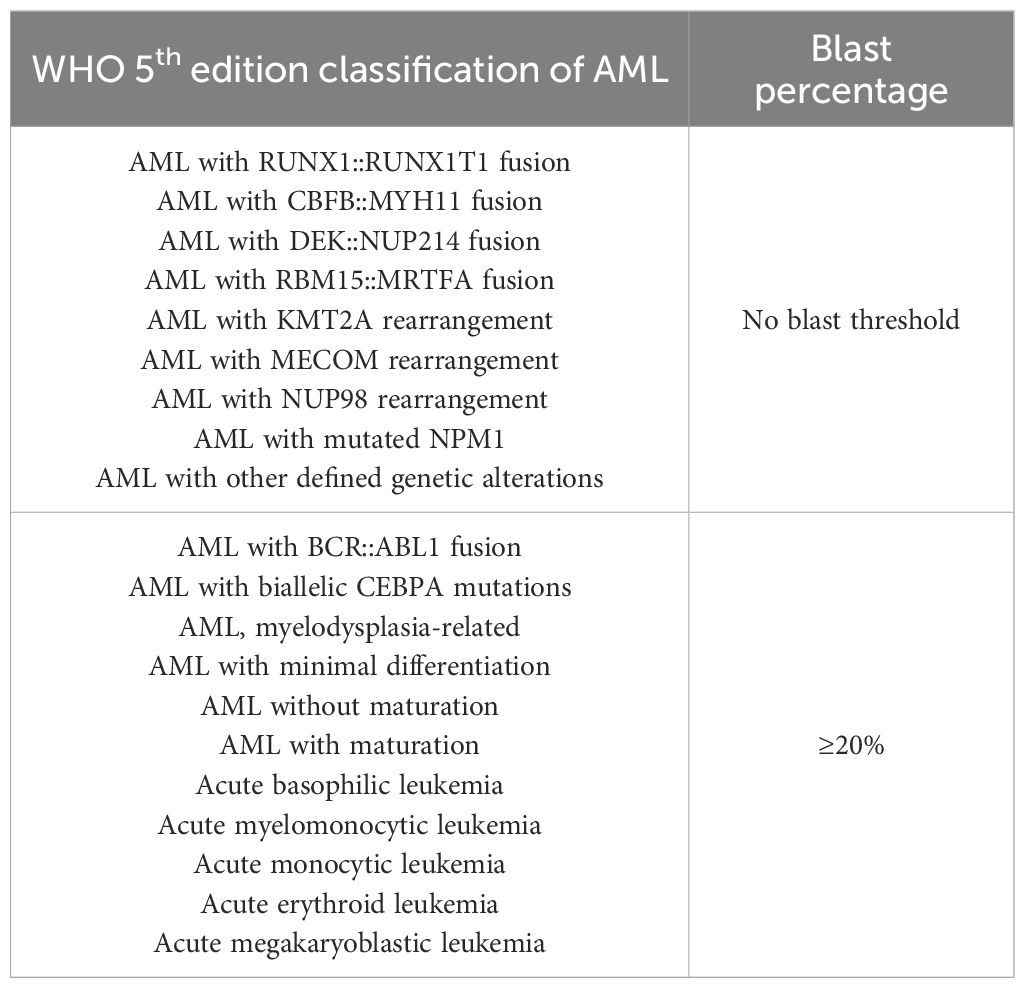
The second is the fifth edition of the World Health Organization (WHO) classification of hematologic malignancies (17), which lowers the diagnostic threshold to >20% blasts in the bone marrow or peripheral blood (see Table 1). It incorporates morphological, immunophenotypic, cytogenetic, and molecular genetic features into a comprehensive MICM (Morphology, Immunophenotype, Cytogenetics, Molecular abnormalities) framework. Notably, patients with specific genetic abnormalities such as PML::RARA or RUNX1::RUNX1T1 can be diagnosed with AML even if the blast percentage is below 20%.
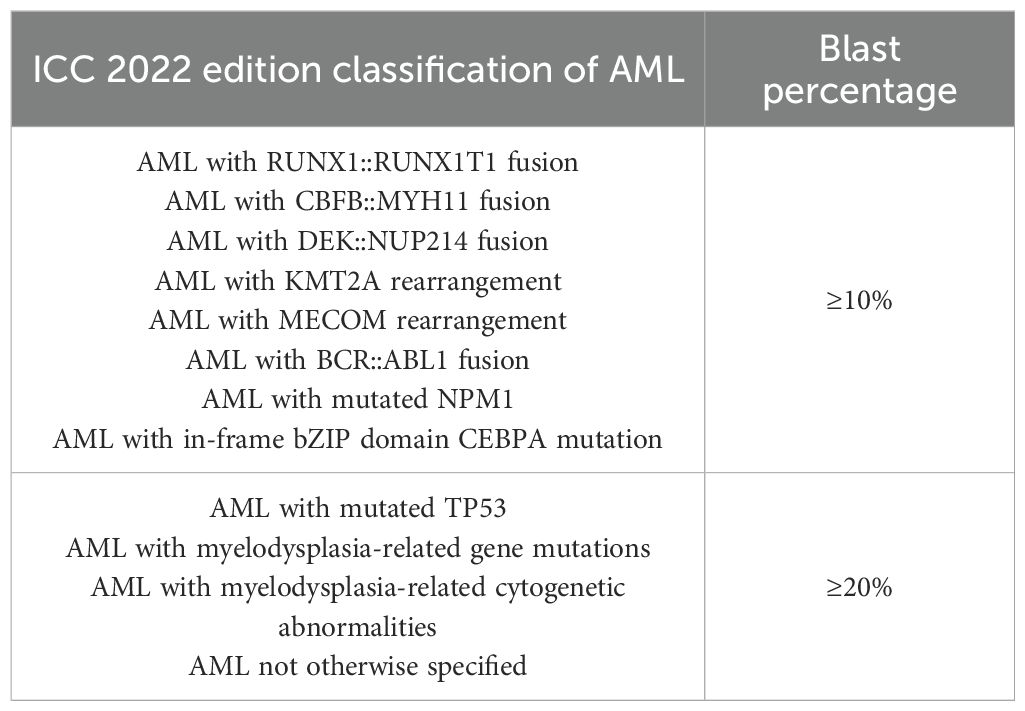
The third is the International Consensus Classification (ICC) released in 2022 (18), which largely aligns with the WHO system but introduces refinements (see Table 2). While retaining the 20% blast threshold as a general criterion, ICC allows for AML diagnosis at ≥10% blasts in certain clinical contexts, such as therapy-related or secondary AML, or in the presence of high-risk genetic mutations. Additionally, ICC delineates precursor states such as myelodysplasia-related AML, refines mutational criteria, and introduces several high-risk biomarkers to enhance diagnostic granularity.
In addition, the guidelines issued by the European LeukemiaNet (ELN) stratify patients into favorable, intermediate, and adverse risk groups (19). They also emphasize the importance of dynamically monitoring MRD using methods such as multiparameter flow cytometry (MFC) and quantitative PCR to support early prognostic evaluation and guide individualized treatment.
The FAB/WHO/ICC/ELN taxonomies define clinically accepted ground truth labels for supervised AI studies by specifying AML diagnostic categories and ELN risk strata. These frameworks also anchor clinically meaningful endpoints—such as overall/event-free survival, MRD status, and relapse—thereby aligning model outputs with prognostic relevance. Using these standardized labels and endpoints ensures cross-study comparability and enhances the translational validity of AI results.
3.2 Traditional diagnostic processes
Traditional diagnosis of AML typically involves the integrated application of multiple diagnostic modalities. Initial assessments include peripheral blood tests, such as complete blood count and morphological analysis of blood smears, to detect abnormalities in cell counts and morphology (20). This is followed by bone marrow aspiration and biopsy to evaluate blast cell percentage and cytomorphological features (21). Flow cytometry is then employed for immunophenotyping, enabling the detection of surface and cytoplasmic antigen expression patterns to assist in AML subtyping and MRD monitoring (22). For specific AML subtypes, additional cytogenetic and molecular genetic testing such as chromosomal aberrations and mutations in genes like FLT3 and NPM1 is often required for refined classification and risk stratification (23).
Despite the increased diagnostic accuracy achieved through multiple tests, several challenges remain in key steps. Morphological evaluation of peripheral blood smears (PBS) and bone marrow smears (BMS) depends heavily on experienced physicians for manual interpretation, which is labor-intensive, time-consuming, and prone to subjectivity (24). The diagnostic error rate in morphological assessments can be as high as 40% (25). Flow cytometry results may vary due to differences in detection protocols, antibody panel configurations, and analytical standards across laboratories, affecting reproducibility. Molecular testing, meanwhile, often requires expensive equipment and specialized reagents, with long turnaround times and high demands on data interpretation (26). Moreover, diagnostic workflows differ across clinical centers, and for AML patients, even a 24-hour delay in initiating treatment can significantly impact prognosis (27).
4 Adjunctive diagnostic of AML based on blood smear image data
4.1 Morphological analysis
Morphological examination of PBS and BMS is a fundamental and indispensable step in the diagnostic workflow of AML (28) (see Figure 2). Traditionally, this process relies on manual microscopic evaluation by hematologists, who assess various cellular features such as shape, size, color, and internal structures to determine the degree of differentiation, maturation status, and pathological abnormalities of blood cells (29). In AML, blood smears often reveal abnormal blast cells that are typically characterized by increased cell size, a high nucleocytoplasmic ratio, prominent nucleoli, reduced cytoplasmic volume, and abnormal granule distribution (30). These morphological abnormalities serve as critical indicators in the diagnosis of AML. Additionally, in certain subtypes such as APL, morphological cues may provide important subtype-specific diagnostic clues.
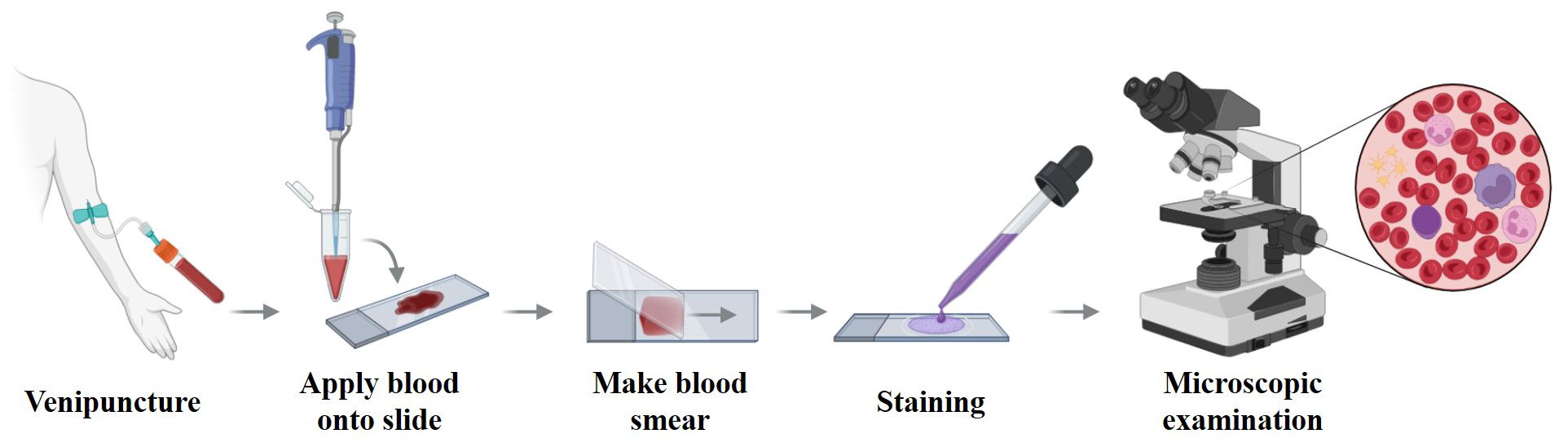
Figure 2. Workflow of peripheral blood smear preparation and microscopic examination. Venous blood is applied to a glass slide, spread to form a thin monolayer, air-dried, and stained (e.g., Wright–Giemsa) before microscopic review. This workflow yields the morphological cues used for AML screening and triage and provides the reference labels that many AI systems learn from; its standardization (smear quality, staining, scanning) is critical for model generalizability.
However, traditional morphological analysis is highly dependent on the observer’s expertise and is inherently subjective. The slide review process is labor-intensive and time-consuming (31), and even among experienced hematologists, inter-observer agreement on cell classification remains limited, with reported consistency rates of only around 60% (32).
With the advancement of automated hematological analysis, AI-driven models have been developed to automatically detect and classify leukemic cells, thereby assisting in the diagnosis of AML (33[preprint], 34). Recent studies have focused on the automated identification of leukemic cell morphology in PBS and BMS, subtype classification, and even the prediction of underlying genetic features.
4.2 Cell segmentation and feature extraction
Accurate segmentation of individual blood cells from complex smear backgrounds is a fundamental prerequisite for subsequent classification tasks. Traditional image processing methods have been widely employed for cell segmentation, including manual color thresholding (31), Otsu thresholding combined with morphological operations such as erosion and dilation for cytoplasm and nucleus segmentation (35), and K-means clustering for nucleus extraction (36). To address the challenge of overlapping cells, the watershed distance transform algorithm has proven effective for separating closely adherent leukocytes (37). In addition, more advanced techniques such as active contour models and fuzzy C-means clustering have been used to precisely delineate the boundaries of leukemic cells (38).
Compared with traditional image processing techniques, deep learning models are better equipped to handle complex backgrounds and cellular heterogeneity, thereby achieving superior performance in cell segmentation tasks. For example, Mask R-CNN has been widely applied for object detection and pixel-level segmentation of blood cells (39). introduced WBC-Net, a hybrid architecture that combines UNet++ and ResNet, significantly improving the precision of leukocyte boundary detection (40). Similarly, Roy et al. developed a semantic segmentation framework based on DeepLabv3+, which offers enhanced accuracy in delineating cell contours (41). In addition, some studies have proposed moment-based localization methods in the CMYK color space for extracting regions of interest, effectively balancing segmentation efficiency and accuracy (42).
Before inputting blood cell images into a classifier, it is necessary to extract features that can effectively distinguish between different cell types. Traditional approaches rely on manually engineered features, including geometric, color, and texture characteristics of the cells (43, 44). Geometric descriptors typically include parameters such as area, perimeter, nucleocytoplasmic ratio, and nuclear shape (45). Color features involve statistical measures such as the mean and variance of RGB or HSV color channels (46), while texture features describe the spatial distribution of structural patterns, commonly using gray level co-occurrence matrices and local binary patterns (LBP) (47).
In contrast, deep learning methods, particularly convolutional neural networks (CNN), can automatically learn hierarchical and task-specific representations directly from raw pixel data. For example, LeuFeatx, based on a fine-tuned VGG16 model, achieved a macro-average recall of 64.3% on an AML dataset, outperforming manual feature extraction methods (48). Wang et al. (49) utilized a ResNet model pretrained on ImageNet, which proved effective in extracting complex and robust features from medical images.
4.3 AML detection and subtype classification
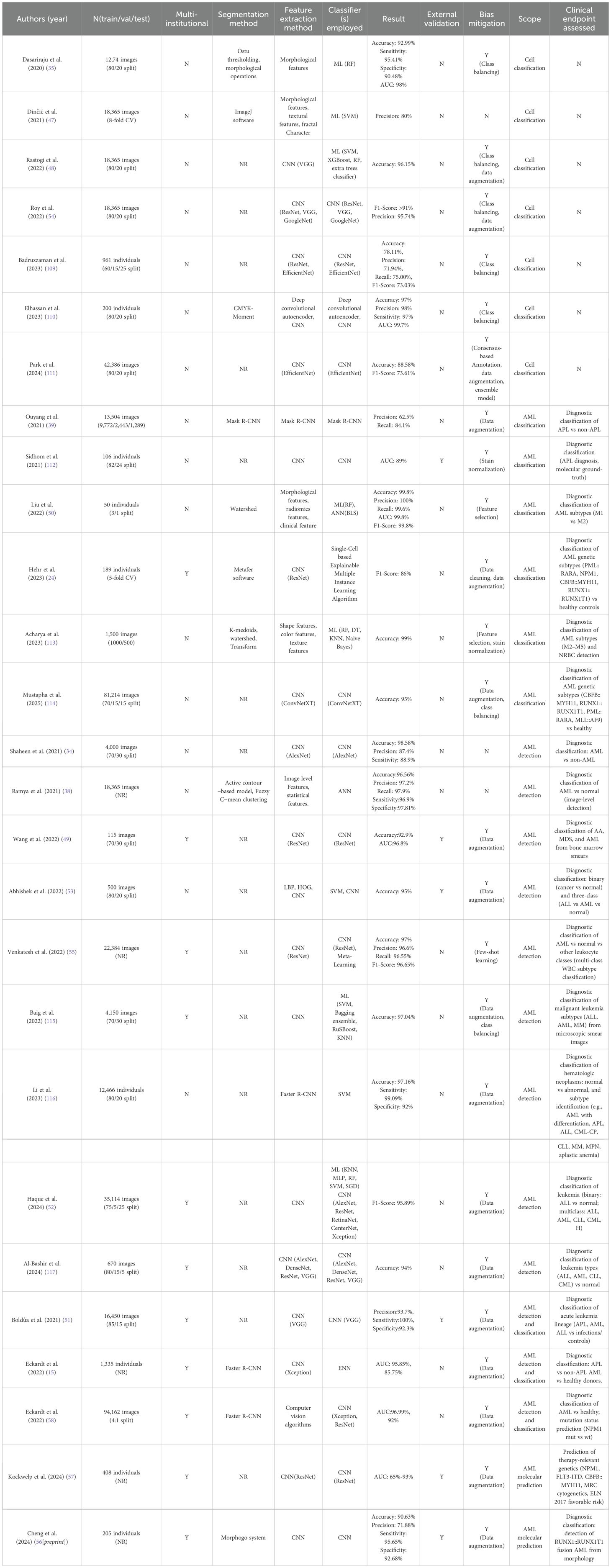
Increasing research attention has been directed toward developing automated models based on image data to distinguish leukocytes from AML patients and healthy individuals, and to further perform AML detection and subtype classification (see Table 3). For instance, Dinčić et al. (47) utilized support vector machines (SVMs) to classify mature and immature leukocytes using manually extracted morphological, fractal, and texture features, achieving an average classification accuracy of 80%. Liu et al. (50) analyzed bone marrow smear images obtained from the TCIA database and extracted two morphological features, six radiomic features, and one clinical feature. A random forest (RF) model was then used to classify AML subtypes.
In the realm of deep learning, CNN have demonstrated strong capabilities in automatically extracting high-dimensional discriminative features from peripheral blood or bone marrow smear images. These models have achieved sensitivity and specificity exceeding 90% in AML morphological recognition tasks (33[preprint]). For example, Shaheen et al. (34) used AlexNet to detect AML from bone marrow images with a classification accuracy of 98%.
However, training end-to-end deep learning models often requires large annotated datasets. To address this, several studies (51–54) have applied transfer learning, where CNNs are pretrained on large-scale general-purpose image datasets such as ImageNet and then fine-tuned for specific medical imaging tasks. In addition, Venkatesh et al. (55) proposed a few-shot learning approach by integrating a pretrained ResNet with meta-learning techniques, enabling accurate AML classification from limited samples.
Model interpretability is also a critical concern, particularly in clinical applications. Hehr et al. (24) introduced SCEMILA, an interpretable AI model for AML subtype classification from blood smears. The model’s highly attentive cells showed strong agreement with diagnostically relevant cells annotated by experts. Remarkably, SCEMILA could highlight subtype-specific cells and deconstruct blood smear composition without requiring single-cell annotations, offering a valuable example of explainable AI in hematologic diagnosis.
Furthermore, several studies have explored the use of image data to predict AML-related molecular alterations. Cheng et al. (56[preprint]) analyzed 60,000 bone marrow smear images from 205 AML patients and successfully predicted the presence of the RUNX1::RUNX1T1 fusion gene, achieving a sensitivity of 95% and specificity of 92% on the test set. Kockwelp et al. (57) trained a ResNet model using single-cell images derived from BMS to predict mutations such as CBFB::MYH11, NPM1, and FLT3-ITD, and employed sensitivity-based heatmaps for phenotype-genotype interpretability. Eckardt et al. (58) proposed a multi-step deep learning framework that performed cell segmentation, AML classification, and NPM1 mutation prediction, achieving an AUC of 0.92.
4.4 Limitations and future considerations
Despite encouraging results in automated cell segmentation, feature extraction, and classification, most blood smear studies remain limited by small, single-center datasets and substantial variability in staining protocols and imaging quality. Many models rely on retrospective data and lack external validation across institutions, raising concerns about generalizability (59). While some efforts, such as interpretable frameworks (e.g., SCEMILA), demonstrate potential to enhance transparency, the majority of CNN-based models still function as “black boxes.” (24). In addition, there is little evidence of integration into clinical workflows, where turnaround time, interpretability, and cross-platform robustness are essential. These limitations highlight the gap between promising algorithmic performance and actual clinical applicability in hematopathology.
To overcome these issues, future efforts should prioritize the development of large-scale, standardized, and cross-institutional image databases, along with the design of inherently interpretable network architectures to enhance both transparency and clinical utility in AML diagnosis.
5 Adjunctive diagnostic of AML based on flow cytometry
5.1 Flow cytometry
Flow cytometry is a critical technique for the diagnosis and monitoring of AML (see Figure 3). By detecting specific surface and intracellular antigen markers, it enables the identification of the lineage, differentiation stage, and aberrant immunophenotype of leukemic cells (60). MFC, which utilizes combinations of multiple antibodies, allows for the simultaneous analysis of antigen expression profiles in tens of thousands of cells. With single-cell resolution, MFC can detect rare abnormal cell populations in bone marrow, making it particularly valuable for MRD assessment (61).
Figure 3. Workflow of flow cytometry for detecting cell surface and intracellular antigens. Cells are first suspended in buffer and stained with fluorescently labeled antibodies. Surface antigens bind directly to the antibodies, while intracellular antigens require fixation and permeabilization to allow antibody access to the cytoplasm or nucleus. After staining, cells pass through the flow cytometer, where lasers excite the bound fluorochromes. Forward and side scatter, along with emitted fluorescence, are measured to analyze cellular characteristics. These readouts underpin AML diagnosis and MRD assessment and are targets for AI systems that automate gating and rare-population detection; panel standardization and external validation are critical for generalizability.
Moreover, flow cytometry also contributes to guiding targeted therapies. Antigens such as CD33 and CD123 serve not only as diagnostic markers for AML but also as therapeutic targets for antibody-based treatments (62). Compared to other techniques, flow cytometry offers rapid immunophenotyping within a few hours of sample processing and has the ability to distinguish viable cells from debris and dead cells (63).
However, the widespread clinical application of flow cytometry faces several challenges. Differences in antibody panels and data interpretation protocols across institutions hinder cross-center comparability and complicate standardization efforts. High-sensitivity detection depends on advanced cytometers and fluorescent-labeled antibodies, making individual assays relatively expensive. In addition, traditional manual gating used for data analysis is labor-intensive and subject to operator bias, especially when handling large volumes of multidimensional data (64).
5.2 Cell population analysis
A wide range of supervised and unsupervised machine learning algorithms have been applied to replace or assist the traditional manual gating process in flow cytometry, significantly improving the efficiency and accuracy of AML-related data analysis. Unsupervised learning techniques, in particular, have shown great value in dimensionality reduction and visualization. Nonlinear techniques such as t-SNE and UMAP project high-dimensional parameters into two-dimensional space, facilitating intuitive identification of cell subpopulations (65). Clustering algorithms like K-means and density-based methods (e.g., DBSCAN) have also been widely employed for cell classification tasks involving multiparametric data (66).
In recent years, self-organizing map (SOM) models have attracted growing attention due to their capabilities in visualization and adaptive pattern recognition. For example, one study combined SOM with XGBoost to construct a hybrid model for AML diagnosis, achieving 92.55% accuracy and 99.79% specificity on the validation dataset (67). Porwit et al. (66) further applied the FlowSOM algorithm to unsupervised clustering of erythroid precursor cells, successfully identifying 18 potentially abnormal subpopulations that provided new biological insights for diagnosis.
5.3 Automated diagnosis
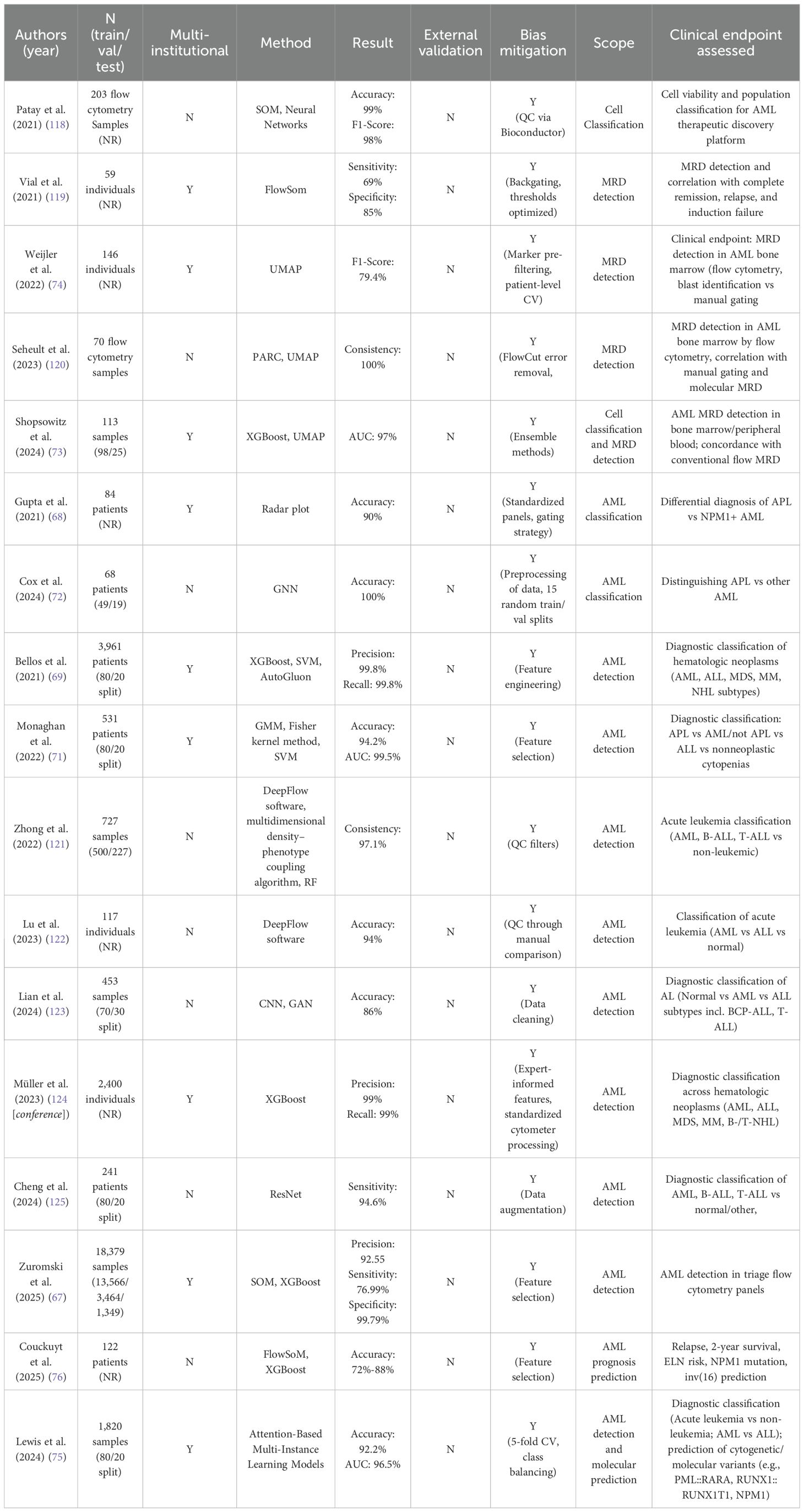
In the field of AML diagnosis and subtype classification, several studies have demonstrated high performance. Gupta et al. (68), using 10-color flow cytometry data, integrated key markers such as CD2, CD13, and CD64 into a radar plot to distinguish typical and variant APL, as well as NPM1-mutated AML, achieving 100% accuracy in identifying typical APL. Bellos et al. (69) conducted a large-scale study involving over 36,000 patients and built an AML diagnostic model combining XGBoost and SVM, attaining 99.9% accuracy in 82% of the cases, showcasing the potential of AI in large-cohort settings.
In addition to traditional shallow machine learning approaches, recent studies have increasingly explored multi-model fusion strategies and statistical modeling techniques to improve the extraction and classification of complex flow cytometry data. For instance, Ko et al. (70) combined SVM with Gaussian mixture models, achieving a classification accuracy of 92.4% in AML patient samples. Monaghan et al. (71) further introduced a Fisher kernel-based approach to extract multiparametric features, which were then classified using SVM to distinguish APL from non-APL cases. This method also identified key features associated with overall survival, offering new insights into prognostic modeling. Additionally, Cox et al. (72) structured cellular data into graph representations, demonstrating the potential of graph-based modeling techniques for detecting abnormal cell populations.
In terms of clinical translation, efforts have been made to integrate these AI-based models into routine workflows. Zuromski et al. (67) constructed a deployable AML diagnostic platform using flow cytometry data as input, enabling automatic report generation within hospital information systems. This work provides a valuable paradigm for the clinical implementation of AI-assisted diagnostic tools.
5.4 MRD detection and molecular feature prediction
MRD detection is widely recognized as a key metric for assessing treatment response and predicting relapse in AML. However, conventional flow-based MRD analysis requires high sensitivity and standardization, which are often difficult to maintain in routine practice. Recently, AI models have shown potential to complement or even replace manual assessment.
The MAGIC-DR framework (73), which integrates UMAP for dimensionality reduction and XGBoost for classification, achieved strong concordance with manual MRD assessments in 25 validation samples. Moreover, it identified immature monocytic populations that were often overlooked in manual analysis, thus enhancing overall detection sensitivity. Weijler et al. (74) proposed a semi-supervised strategy based on UMAP to separate abnormal populations in MRD samples, achieving an F1 score of 79.4%, suggesting its applicability in heterogeneous clinical data.
At a higher level of application, some studies have utilized flow cytometry data to predict molecular genetic features and patient prognosis. Lewis et al. (75) developed a multi-instance learning model with an attention mechanism using only flow cytometry data as input. The model achieved an AUC of 0.96 for diagnostic classification and was capable of predicting several WHO-defined genetic abnormalities in AML, such as t (8;21), t(15;17), and NPM1 mutations. Couckuyt et al. (76) further integrated flow cytometry data with machine learning algorithms to predict two-year survival, revealing significant associations between immune subtypes, genetic features, and patient outcomes.
5.5 Limitations and future considerations
AI-driven approaches to flow cytometry analysis have significantly reduced the reliance on manual gating and improved MRD detection. However, current studies are largely retrospective and often conducted on heterogeneous panels and protocols, reflecting the lack of international standardization. Only a few reports demonstrate prospective or real-world clinical validation, and cross-center reproducibility remains uncertain. Moreover, most machine learning models prioritize accuracy but do not adequately address class imbalance, operator bias, or rare subpopulation detection. While pilot platforms for automated reporting exist, their clinical readiness is still low-to-moderate, requiring regulatory approval, standardized antibody panels, and better interpretability tools before widespread adoption.
Overall, AI-assisted workflows have demonstrated strong consistency with expert assessments in various tasks, including cell population analysis, AML classification, and MRD detection (see Table 4). Future directions should focus on building standardized, multi-center flow cytometry data platforms to improve model generalizability. In addition, developing interpretable models and enhancing visualization capabilities will be critical for clinical integration, particularly in detecting rare subpopulations and tracking dynamic changes in MRD.
6 Adjunctive diagnostic of AML based on genetic data
6.1 Genetic analysis
Genetic analysis provides essential molecular insights that play a pivotal role in disease classification, risk stratification, and therapeutic decision-making in AML (77). Traditional approaches such as karyotyping and fluorescence in situ hybridization (FISH) have long been utilized in clinical diagnostics. In recent years, the emergence of advanced genomic technologies, including next-generation sequencing (NGS), single-cell sequencing, and transcriptome analysis, has continuously propelled the advancement of precision medicine in AML (78).
Karyotyping primarily uses G-banding to detect numerical and structural chromosomal abnormalities, such as trisomies, deletions, inversions, and translocations (79). FISH, on the other hand, utilizes fluorescent probes to identify specific gene rearrangements or chromosomal region abnormalities with higher resolution. It is commonly used to detect fusion genes such as PML::RARA and RUNX1::RUNX1T1 (26).
At the molecular level, NGS is a high-throughput sequencing technology (80) (see Figure 4) that enables comprehensive analysis of genomic or exonic mutations, insertions, and deletions, and has been widely applied in AML subtyping and prognostic evaluation. Single-cell sequencing allows the profiling of gene expression or genomic variations at the single-cell level, enabling the identification of leukemic subpopulations at different differentiation stages and offering insights into key subclones and relapse mechanisms (77). Transcriptome sequencing provides a global gene expression profile of AML patients, helping to identify expression signatures, dysregulated pathways, and prognostic biomarkers associated with AML development (81).
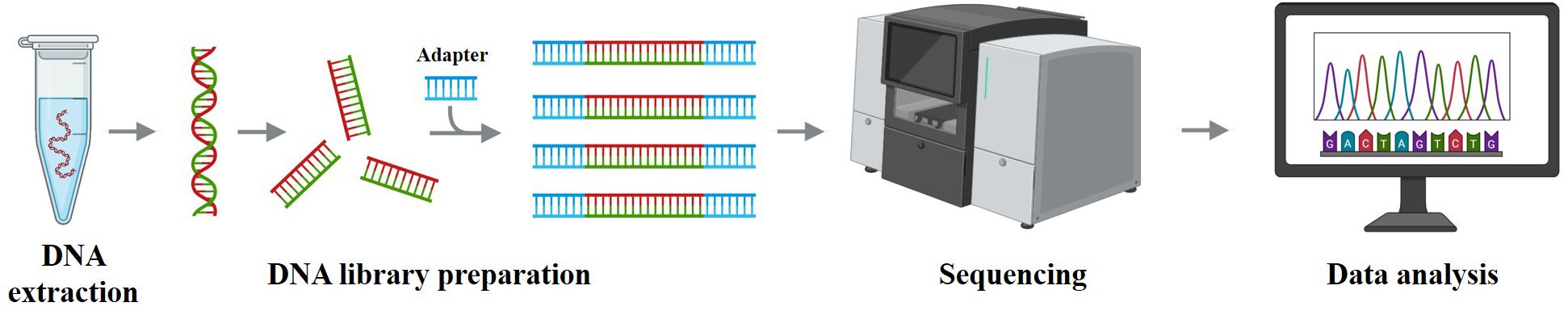
Figure 4. Schematic workflow of next-generation sequencing. Genomic DNA is extracted, fragmented, and ligated to adapters to build libraries, which are sequenced on a high-throughput platform; reads are then aligned and variants are called and annotated. These steps generate the molecular features used for AML diagnosis/risk stratification and for training AI models; pipeline harmonization (variant-calling/batch-effect control) and external multi-cohort validation are essential for generalizable results.
Despite advances in genetic testing methodologies, challenges remain. Traditional cytogenetic methods such as karyotyping and FISH are limited in throughput, sensitivity, and turnaround time (82), while modern techniques like NGS and single-cell sequencing are constrained by high costs, complex data interpretation, and a heavy reliance on bioinformatics expertise (26). With ongoing technological development, the integration of diverse genetic data combined with AI and machine learning holds promise for enhancing diagnostic accuracy and enabling more personalized treatment strategies for AML.
6.2 Karyotyping and FISH
Karyotyping is a standard technique for detecting chromosomal abnormalities. Traditionally, the interpretation of chromosome images requires highly experienced cytogeneticists. To enhance efficiency and reduce manual workload, recent studies have explored the use of AI to automate metaphase image recognition and chromosome classification (83), as well as to develop models for automated chromosome segmentation and pairing (84). For example, Hu et al. (85) proposed a multilayer CNN combined with a Softmax classifier, achieving 93.8% accuracy in pairing and identifying abnormal chromosomes. Similarly, Vajen et al. (86) developed a CNN-based tool that achieved 98.8% accuracy in chromosome classification and reduced manual analysis time by up to 42%.
Most current AI systems for karyotyping depend on large annotated datasets. To address this limitation, one study (87) proposed a machine learning strategy to simulate abnormal karyotype images from normal ones, combining this approach with a ResNet classifier that achieved over 95% accuracy. Furthermore, Shamsi et al. (88[preprint]) introduced the Vision Transformer (ViT) architecture into karyotype analysis for the first time, developing an end-to-end model that accurately identified clinically significant abnormalities such as t(9;22) from metaphase images. This approach significantly reduced the need for extensive labeled data by employing pre-training and fine-tuning strategies.
FISH is another widely used technique for detecting chromosomal number and structural abnormalities using DNA-targeted probes. However, traditional FISH analysis is labor-intensive and highly reliant on expert interpretation. To improve throughput and consistency, researchers have applied AI-based models to automate the FISH image analysis pipeline. Gudla et al. (89[conference]) developed a CNN-based system for automated detection of chromosomal abnormalities, achieving an accuracy rate exceeding 98%. Xue et al. (90) constructed an end-to-end detection model combining YOLOv3 with ResNet18 to assess gene amplification status, reaching 85% classification accuracy on the test set. Xu et al. (91) further proposed a multi-scale MobileNet-YOLOv4 framework for rapid detection of genetic abnormalities in circulating cells, achieving 93% accuracy and up to 500-fold improvement in detection speed. In addition, Bouilhol et al. (92) introduced DeepSpot, a deep learning tool designed to enhance the detection of fluorescent signals in single-molecule FISH images, attaining an accuracy of up to 97%.
6.3 Molecular analysis and prognostic prediction
Molecular genetics testing represents a core component of AML diagnostics, providing critical insights that inform classification, prognosis, and therapeutic decision-making. Mutations in genes such as NPM1 and FLT3-ITD, as well as fusion events like PML::RARA, are now incorporated into major clinical guidelines as essential molecular indicators for diagnosis and risk stratification (93).
With the rapid advancement of NGS, these techniques have increasingly been applied in clinical settings and have become indispensable tools for molecular subtyping of AML. NGS platforms can integrate whole-genome sequencing, exome sequencing, and RNA sequencing to simultaneously analyze hundreds of leukemia-related genes in a single assay, greatly improving detection efficiency and data richness (94). Wurm et al. (95) reported that the turnaround time for NGS-based analysis of AML samples decreased from 22 days in 2013 to just 10 days in 2023, reflecting substantial improvements in clinical feasibility. Zhang et al. (96) combined targeted RNA sequencing with a naïve Bayes classifier to perform differential diagnosis across 20 hematologic malignancies and 24 solid tumors, achieving an AUC of 88% for AML classification.
In large-scale applications, the integration of transcriptomic data with machine learning models has become an emerging trend in the adjunctive diagnosis of AML. Warnat-Herresthal et al. (97) integrated transcriptomic profiles from 12,029 samples across 105 studies and developed a machine learning model capable of distinguishing AML, MDS, and other myeloid neoplasms, achieving over 92% subtype classification accuracy across multiple datasets. Similarly, Angelakis et al. (98 [preprint]) used a CatBoost classifier on 12,708 transcriptomes from 5,052 individuals, reaching an AUC above 99% in distinguishing AML from healthy controls highlighting the synergy between big data and machine learning.
In the area of molecular subtyping and prognostic modeling, Awada et al. (99) applied Bayesian unsupervised learning to integrate mutation and immunophenotypic data, identifying four novel subtypes with distinct biological and prognostic characteristics. Song et al. (100) proposed an unsupervised multi-omics integration approach that stratified AML into three major subgroups using TCGA and clinical cohorts, showing strong generalizability. Afroz et al. (101) introduced the omicsGAN framework, which enhances predictive accuracy by synthesizing gene activity and DNA methylation profiles.
Given the high clonal heterogeneity of AML, single-cell RNA sequencing (scRNA-seq) has emerged as a powerful method to resolve cellular subpopulations and microenvironmental interactions (102). Galen et al. (103) combined scRNA-seq and genotyping data from 38,410 cells across 40 AML bone marrow samples and used machine learning to successfully classify distinct malignant subtypes and link them to specific genetic mutations. Nicora et al. (104) and Asimomitis et al. (105) applied supervised deep learning approaches to scRNA-seq data for cell state prediction and mutation status classification, respectively. Their models achieved classification AUCs of up to 98% and 84%, highlighting the significant potential of integrating single-cell omics with AI for clinically relevant analysis.
Recent studies (106) have further emphasized the role of transcriptomic changes in identifying novel therapeutic targets in AML, helping bridge the gap between genomic insights and clinical application. For instance, Gimeno et al. (107) employed a multidimensional module-optimized machine learning algorithm using RNA-seq data to predict gene mutations and drug response, providing valuable support for precision medicine. Qin et al. (108) integrated bulk RNA-seq, single-cell expression profiles, and matched clinicopathological data to construct a six-gene programmed cell death index capable of predicting chemotherapy resistance, drug sensitivity, and poor prognosis in AML patients.
With advances in high-throughput omics technologies and the integration of AI, molecular diagnostics for AML are evolving from single-marker identification toward a comprehensive, data-driven precision framework (see Table 5). NGS enables broad and efficient detection by combining genomic, exomic, and transcriptomic information. Transcriptomic data, when coupled with machine learning, have demonstrated exceptional performance in disease classification, subtype distinction, and prognostic assessment. Meanwhile, scRNA-seq offers unprecedented resolution of AML clonal heterogeneity and immune microenvironment features, enriching our understanding of disease mechanisms. More recently, the integration of omics data with drug response modeling has laid a foundation for individualized therapy and resistance prediction.
6.4 Limitations and future considerations
Genetic and transcriptomic studies represent one of the fastest-growing areas, especially with the advent of NGS and single-cell sequencing. Nonetheless, many published works face challenges of small sample size, high-dimensional data with risk of overfitting, and a heavy reliance on bioinformatics expertise. Several cited studies are still preprints, reflecting the novelty but also the limited peer-reviewed validation of these methods. External, multi-cohort validation is rare, and clinical integration of AI-based omics prediction frameworks is currently exploratory rather than routine. Furthermore, the complexity of multi-omics integration and the lack of interpretability hinder clinical decision-making.
Thus, omics-based AI models hold promise for risk stratification and prognostication. Looking ahead, intelligent clinical decision platforms that integrate omics, algorithmic inference, and clinical workflows are poised to facilitate the implementation of precision medicine in AML (78).
7 Conclusions and clinical integration
The pronounced heterogeneity of AML poses substantial challenges for both diagnosis and prognostic assessment. In recent years, AI has demonstrated remarkable potential to address these challenges by integrating flow cytometry data, medical images, and multi-omics information. Across diverse studies, AI models have achieved strong performance in key tasks such as cell-population identification, subtype classification, molecular-mutation prediction, and prognostic stratification. Research has also evolved from single-modal classification toward multimodal data fusion and molecular feature modeling, with increasing emphasis on interpretability and automated integration.
Despite these advances, AI still faces major barriers to clinical translation, including data heterogeneity, limited model generalizability, high annotation costs, and the scarcity of prospective validation. Future work should prioritize building standardized data platforms, enhancing model robustness and interpretability under real-world variability, and enabling seamless integration of AI systems into diagnostic and therapeutic processes. With continued innovation, AI is expected to become a core component of precision AML care, providing efficient and individualized decision support.
Clinical integration and workflow impact are essential for realizing this potential. In practice, deep learning models must be embedded into existing diagnostic and treatment pathways. During triage and diagnosis, they can rapidly screen peripheral blood smears and prioritize suspected AML cases, accelerating identification of high-risk patients. For urgent subtypes such as APL, models can trigger seconds-level alerts to facilitate timely intervention. Automated analysis substantially shortens morphological review time, a critical metric in emergency leukemia management. In measurable residual disease monitoring, sensitivity thresholds allow detection of very low abnormal-cell fractions, while cross-validation with flow cytometry or molecular assays reduces false positives and enhances longitudinal reliability. Model outputs—including calibrated confidence scores and visual heatmaps—should be standardized for interoperability with hospital information systems, supporting efficient hematopathologist review. When discrepancies arise between automated and manual interpretations, conflict-resolution strategies such as double-blind review, expert-panel adjudication, or weighted consensus voting ensure quality control. Within this closed-loop framework, AI can evolve into a “trustworthy, traceable, and controllable” collaborator that augments clinical expertise without replacing it.
Author contributions
WX: Investigation, Writing – original draft. XJ: Writing – review & editing, Investigation. JH: Writing – review & editing. MQ: Writing – review & editing. ZB: Supervision, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Siegel RL, Miller KD, Fuchs HE, and Jemal A. Cancer statistics, 2022. CA Cancer J Clin. (2022) 72:7–33. doi: 10.3322/caac.21708
2. Zhou Y, Huang G, Cai X, Liu Y, Qian B, and Li D. Global, regional, and national burden of acute myeloid leukemia, 1990–2021: a systematic analysis for the global burden of disease study 2021. biomark Res. (2024) 12:101. doi: 10.1186/s40364-024-00649-y
3. Long NA, Golla U, Sharma A, and Claxton DF. Acute myeloid leukemia stem cells: origin, characteristics, and clinical implications. Stem Cell Rev Rep. (2022) 18:1211–26. doi: 10.1007/s12015-021-10308-6
4. Wierzbowska A and Czemerska M. Clinical manifestation and diagnostic workup. In: Röllig C and Ossenkoppele GJ, editors. Acute myeloid leukemia. Springer International Publishing, Cham (2021). p. 119–26. doi: 10.1007/978-3-030-72676-8_6
5. Siegel RL, Miller KD, and Jemal A. Cancer statistics, 2017. CA Cancer J Clin. (2017) 67:7–30. doi: 10.3322/caac.21387
6. Shimony S, Stahl M, and Stone RM. Acute myeloid leukemia: 2023 update on diagnosis, risk-stratification, and management. Am J Hematol. (2023) 98:502–26. doi: 10.1002/ajh.26822
7. Kantarjian H, Kadia T, DiNardo C, Daver N, Borthakur G, Jabbour E, et al. Acute myeloid leukemia: current progress and future directions. Blood Cancer J. (2021) 11:41. doi: 10.1038/s41408-021-00425-3
8. DiNardo CD, Erba HP, Freeman SD, and Wei AH. Acute myeloid leukemia. Lancet. (2023) 401:2073–86. doi: 10.1016/S0140-6736(23)00108-3
9. Alzubaidi L, Zhang J, Humaidi AJ, Al-Dujaili A, Duan Y, Al-Shamma O, et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data. (2021) 8:53. doi: 10.1186/s40537-021-00444-8
10. Hunter B, Hindocha S, and Lee RW. The role of artificial intelligence in early cancer diagnosis. Cancers. (2022) 14:1524. doi: 10.3390/cancers14061524
11. Li M, Jiang Y, Zhang Y, and Zhu H. Medical image analysis using deep learning algorithms. Front Public Health. (2023) 11:1273253. doi: 10.3389/fpubh.2023.1273253
12. Chaurasia A, Namachivayam A, Koca-Ünsal RB, and Lee J-H. Deep-learning performance in identifying and classifying dental implant systems from dental imaging: a systematic review and meta-analysis. J Periodontal Implant Sci. (2024) 54:3. doi: 10.5051/jpis.2300160008
13. Yue S, Li S, Huang X, Liu J, Hou X, Zhao Y, et al. Machine learning for the prediction of acute kidney injury in patients with sepsis. J Transl Med. (2022) 20:215. doi: 10.1186/s12967-022-03364-0
14. Vadapalli S, Abdelhalim H, Zeeshan S, and Ahmed Z. Artificial intelligence and machine learning approaches using gene expression and variant data for personalized medicine. Brief Bioinform. (2022) 23:bbac191. doi: 10.1093/bib/bbac191
15. Eckardt J-N, Schmittmann T, Riechert S, Kramer M, Sulaiman AS, Sockel K, et al. Deep learning identifies Acute Promyelocytic Leukemia in bone marrow smears. BMC Cancer. (2022) 22:201. doi: 10.1186/s12885-022-09307-8
16. Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposals for the classification of the acute leukemias. French-American-British (FAB) co-operative group. Br J Haematol. (1976) 33:451–8. doi: 10.1111/j.1365-2141.1976.tb03563.x
17. Chandra DJ, Lachowiez CA, and Loghavi S. Practical considerations in clinical application of WHO 5th and ICC classification schemes for acute myeloid leukemia. Blood Rev. (2024) 64:101156. doi: 10.1016/j.blre.2023.101156
18. Nagler A, Labopin M, Versluis J, Sanz J, Gedde-Dahl T, Burns D, et al. Allogeneic stem cell transplantation for patients with acute myeloid leukemia with myelodysplasia-related features in first complete remission as per the international consensus classification (ICC) 2022: A study from the ALWP of the EBMT. Transplant Cell Ther Off Publ Am Soc Transplant Cell Ther. (2024) 30:S6–7. doi: 10.1016/j.jtct.2023.12.027
19. Bouligny IM, Maher KR, and Grant S. Secondary-type mutations in acute myeloid leukemia: updates from ELN 2022. Cancers. (2023) 15:3292. doi: 10.3390/cancers15133292
20. Lewis JE and Pozdnyakova O. Digital assessment of peripheral blood and bone marrow aspirate smears. Int J Lab Hematol. (2023) 45 Suppl 2:50–8. doi: 10.1111/ijlh.14082
21. Jamy O, Bourne G, Mudd TW, Thigpen H, and Bhatia R. Revisiting the role of day 14 bone marrow biopsy in acute myeloid leukemia. Cancers. (2025) 17:900. doi: 10.3390/cancers17050900
22. Rasheed HM, Donia HM, Nadwan EA, Mourad ZI, and Farahat N. Identifying leukemia-associated immunophenotypes in acute myeloid leukemia patients using multiparameter flow cytometry. Oman Med J. (2021) 36:e323. doi: 10.5001/omj.2021.108
23. Ali AM and Salih GF. Molecular and clinical significance of FLT3, NPM1, DNMT3A and TP53 mutations in acute myeloid leukemia patients. Mol Biol Rep. (2023) 50:8035–48. doi: 10.1007/s11033-023-08680-2
24. Hehr M, Sadafi A, Matek C, Lienemann P, Pohlkamp C, Haferlach T, et al. Explainable AI identifies diagnostic cells of genetic AML subtypes. PloS Digit Health. (2023) 2:e0000187. doi: 10.1371/journal.pdig.0000187
25. Amin MM, Kermani S, Talebi A, and Oghli MG. Recognition of acute lymphoblastic leukemia cells in microscopic images using k-means clustering and support vector machine classifier. J Med Signals Sens. (2015) 5:49–58. doi: 10.4103/2228-7477.150428
26. Jain H and Shetty D. Role of cytogenetics and fluorescence in situ hybridization in the laboratory workup of acute myeloid leukemias. Indian J Med Pediatr Oncol. (2023) 44:543–53. doi: 10.1055/s-0043-1768052
27. Franco S, Geng X, Korostyshevskiy V, Karp JE, and Lai C. Systematic review and meta-analysis: Prognostic impact of time from diagnosis to treatment in patients with acute myeloid leukemia. Cancer. (2023) 129:2975–85. doi: 10.1002/cncr.34894
28. Craig FE. The utility of peripheral blood smear review for identifying specimens for flow cytometric immunophenotyping. Int J Lab Hematol. (2017) 39:41–6. doi: 10.1111/ijlh.12651
29. Zini G. Hematological cytomorphology: Where we are. Int J Lab Hematol. (2024) 46:789–94. doi: 10.1111/ijlh.14330
30. Orazi A. Histopathology in the diagnosis and classification of acute myeloid leukemia, myelodysplastic syndromes, and myelodysplastic/myeloproliferative diseases. Pathobiology. (2007) 74:97–114. doi: 10.1159/000101709
31. Hegde RB, Prasad K, Hebbar H, and Singh BMK. Comparison of traditional image processing and deep learning approaches for classification of white blood cells in peripheral blood smear images. Biocybern BioMed Eng. (2019) 39:382–92. doi: 10.1016/j.bbe.2019.01.005
32. Zini G, Bain B, Bettelheim P, Cortez J, d’Onofrio G, Faber E, et al. A European consensus report on blood cell identification: terminology utilized and morphological diagnosis concordance among 28 experts from 17 countries within the European LeukemiaNet network WP10, on behalf of the ELN Morphology Faculty. Br J Haematol. (2010) 151:359–64. doi: 10.1111/j.1365-2141.2010.08366.x
33. Al-Obeidat F, Hafez W, Rashid A, Jallo MK, Gador M, Cherrez-Ojeda I, et al. Artificial intelligence for the detection of acute myeloid leukemia from microscopic blood images; a systematic review and meta-analysis. Front Big Data. (2025) 7:1402926. doi: 10.3389/fdata.2024.1402926
34. Shaheen M, Khan R, Biswal RR, Ullah M, Khan A, Uddin MI, et al. Acute myeloid leukemia (AML) detection using alexNet model. Complexity. (2021) 2021:6658192. doi: 10.1155/2021/6658192
35. Dasariraju S, Huo M, and McCalla S. Detection and classification of immature leukocytes for diagnosis of acute myeloid leukemia using random forest algorithm. Bioengineering. (2020) 7:120. doi: 10.3390/bioengineering7040120
36. Kazemi F, Najafabadi TA, and Araabi BN. Automatic recognition of acute myelogenous leukemia in blood microscopic images using K-means clustering and support vector machine. J Med Signals Sens. (2016) 6:183–93. doi: 10.4103/2228-7477.186885
37. Harjoko A, Ratnaningsih T, Suryani E, Palgunadi S, and Prakisya NPT. Classification of acute myeloid leukemia subtypes M1, M2 and M3 using active contour without edge segmentation and momentum backpropagation artificial neural network. MATEC Web Conf. (2018) 154:1041. doi: 10.1051/matecconf/201815401041
38. Ramya VJ and Lakshmi S. Acute myelogenous leukemia detection using optimal neural network based on fractional black-widow model. Signal Image Video Process. (2022) 16:229–38. doi: 10.1007/s11760-021-01976-5
39. Ouyang N, Wang W, Ma L, Wang Y, Chen Q, Yang S, et al. Diagnosing acute promyelocytic leukemia by using convolutional neural network. Clin Chim Acta Int J Clin Chem. (2021) 512:1–6. doi: 10.1016/j.cca.2020.10.039
40. Lu Y, Qin X, Fan H, Lai T, and Li Z. WBC-Net: A white blood cell segmentation network based on UNet++ and ResNet. Appl Soft Comput. (2021) 101:107006. doi: 10.1016/j.asoc.2020.107006
41. M.Roy R and P.m. A. Segmentation of leukocyte by semantic segmentation model: A deep learning approach. BioMed Signal Process Control. (2021) 65:102385. doi: 10.1016/j.bspc.2020.102385
42. Elhassan TAM, Rahim MSM, Swee TT, Hashim SZM, and Aljurf M. Feature extraction of white blood cells using CMYK-moment localization and deep learning in acute myeloid leukemia blood smear microscopic images. IEEE Access. (2022) 10:16577–91. doi: 10.1109/ACCESS.2022.3149637
43. Rodellar J, Alférez S, Acevedo A, Molina A, and Merino A. Image processing and machine learning in the morphological analysis of blood cells. Int J Lab Hematol. (2018) 40:46–53. doi: 10.1111/ijlh.12818
44. Puigví L, Merino A, Alférez S, Acevedo A, and Rodellar J. New quantitative features for the morphological differentiation of abnormal lymphoid cell images from peripheral blood. J Clin Pathol. (2017) 70:1038–48. doi: 10.1136/jclinpath-2017-204389
45. Merino A, Puigví L, Boldú L, Alférez S, and Rodellar J. Optimizing morphology through blood cell image analysis. Int J Lab Hematol. (2018) 40 Suppl 1:54–61. doi: 10.1111/ijlh.12832
46. Tavakoli S, Ghaffari A, Kouzehkanan ZM, and Hosseini R. New segmentation and feature extraction algorithm for classification of white blood cells in peripheral smear images. Sci Rep. (2021) 11:19428. doi: 10.1038/s41598-021-98599-0
47. Dinčić M, Popović TB, Kojadinović M, Trbovich AM, and Ilić AŽ. Morphological, fractal, and textural features for the blood cell classification: the case of acute myeloid leukemia. Eur Biophys J. (2021) 50:1111–27. doi: 10.1007/s00249-021-01574-w
48. Rastogi P, Khanna K, and Singh V. LeuFeatx: Deep learning–based feature extractor for the diagnosis of acute leukemia from microscopic images of peripheral blood smear. Comput Biol Med. (2022) 142:105236. doi: 10.1016/j.compbiomed.2022.105236
49. Wang M, Dong C, Gao Y, Li J, Han M, and Wang L. A deep learning model for the automatic recognition of aplastic anemia, myelodysplastic syndromes, and acute myeloid leukemia based on bone marrow smear. Front Oncol. (2022) 12:844978. doi: 10.3389/fonc.2022.844978
50. Liu K and Hu J. Classification of acute myeloid leukemia M1 and M2 subtypes using machine learning. Comput Biol Med. (2022) 147:105741. doi: 10.1016/j.compbiomed.2022.105741
51. Boldú L, Merino A, Acevedo A, Molina A, and Rodellar J. A deep learning model (ALNet) for the diagnosis of acute leukemia lineage using peripheral blood cell images. Comput Methods Programs BioMed. (2021) 202:105999. doi: 10.1016/j.cmpb.2021.105999
52. Haque R, Al Sakib A, Hossain MF, Islam F, Ibne Aziz F, Ahmed MR, et al. Advancing early leukemia diagnostics: A comprehensive study incorporating image processing and transfer learning. BioMedInformatics. (2024) 4:966–91. doi: 10.3390/biomedinformatics4020054
53. Abhishek A, Jha RK, Sinha R, and Jha K. Automated classification of acute leukemia on a heterogeneous dataset using machine learning and deep learning techniques. BioMed Signal Process Control. (2022) 72:103341. doi: 10.1016/j.bspc.2021.103341
54. Roy RM and Ameer PM. Identification of white blood cells for the diagnosis of acute myeloid leukemia. Intl J Imaging Sys Tech. (2022) 32(4):1307–17. doi: 10.1002/ima.22702
55. Venkatesh K, Pasupathy S, and Raja SP. Acute myeloid leukemia multi-classification using enhanced few-shot learning technique. Scalable Comput Pract Exp. (2022) 23:377–88. doi: 10.12694/scpe.v23i4.2048
56. Cheng H, Ding J, Wang J, Xiao Y, Jin X, Zhang Y, et al. Predicting RUNX1::RUNX1T1 genetic abnormalities in acute myeloid leukemia from bone marrow smears: Can artificial intelligence do better? iScience. (2025) 28(7):109388. doi: 10.21203/rs.3.rs-4019004/v1
57. Kockwelp J, Thiele S, Bartsch J, Haalck L, Gromoll J, Schlatt S, et al. Deep learning predicts therapy-relevant genetics in acute myeloid leukemia from Pappenheim-stained bone marrow smears. Blood Adv. (2024) 8:70–9. doi: 10.1182/bloodadvances.2023011076
58. Eckardt J-N, Middeke JM, Riechert S, Schmittmann T, Sulaiman AS, Kramer M, et al. Deep learning detects acute myeloid leukemia and predicts NPM1 mutation status from bone marrow smears. Leukemia. (2022) 36:111–8. doi: 10.1038/s41375-021-01408-w
59. Aby AE, Salaji S, Anilkumar KK, and Rajan T. A review on leukemia detection and classification using Artificial Intelligence-based techniques. Comput Electr Eng. (2024) 118:109446. doi: 10.1016/j.compeleceng.2024.109446
60. Robinson JP. Flow cytometry: past and future. BioTechniques. (2022) 72:159–69. doi: 10.2144/btn-2022-0005
61. Cai Q, Lan H, Yi D, Xian B, Zidan L, Li J, et al. Flow cytometry in acute myeloid leukemia and detection of minimal residual disease. Clin Chim Acta. (2025) 564:119945. doi: 10.1016/j.cca.2024.119945
62. Li W, Morgan R, Nieder R, Truong S, Habeebu SSM, and Ahmed AA. Normal or reactive minor cell populations in bone marrow and peripheral blood mimic minimal residual leukemia by flow cytometry. Cytometry B Clin Cytom. (2021) 100:590–601. doi: 10.1002/cyto.b.21968
63. Brooimans RA, van der Velden VHJ, Boeckx N, Slomp J, Preijers F, te Marvelde JG, et al. Immunophenotypic measurable residual disease (MRD) in acute myeloid leukemia: Is multicentric MRD assessment feasible? Leuk Res. (2019) 76:39–47. doi: 10.1016/j.leukres.2018.11.014
64. Manohar SM, Shah P, and Nair A. Flow cytometry: principles, applications and recent advances. Bioanalysis. (2021) 13:181–98. doi: 10.4155/bio-2020-0267
65. Ferrer-Font L, Mayer JU, Old S, Hermans IF, Irish J, and Price KM. High-dimensional data analysis algorithms yield comparable results for mass cytometry and spectral flow cytometry data. Cytometry A. (2020) 97:824–31. doi: 10.1002/cyto.a.24016
66. Porwit A, Violidaki D, Axler O, Lacombe F, Ehinger M, and Béné MC. Unsupervised cluster analysis and subset characterization of abnormal erythropoiesis using the bioinformatic Flow-Self Organizing Maps algorithm. Cytometry B Clin Cytom. (2022) 102:134–42. doi: 10.1002/cyto.b.22059
67. Zuromski LM, Durtschi J, Aziz A, Chumley J, Dewey M, English P, et al. Clinical validation of a real-time machine learning-based system for the detection of acute myeloid leukemia by flow cytometry. Cytometry B Clin Cytom. 27. doi: 10.1002/cyto.b.22229
68. Gupta M, Jafari K, Rajab A, Wei C, Mazur J, Tierens A, et al. Radar plots facilitate differential diagnosis of acute promyelocytic leukemia and NPM1+ acute myeloid leukemia by flow cytometry. Cytometry B Clin Cytom. (2021) 100:409–20. doi: 10.1002/cyto.b.21979
69. Bellos F, Pawelka K, Fortina E, Suriyalaksh M, Maschek S, Haferlach C, et al. Automated comprehensive diagnostics of hematologic neoplasms by artificial intelligence models using flow cytometric raw matrix data. Blood. (2021) 138:104. doi: 10.1182/blood-2021-150697
70. Ko B-S, Wang Y-F, Li J-L, Li C-C, Weng P-F, Hsu S-C, et al. Clinically validated machine learning algorithm for detecting residual diseases with multicolor flow cytometry analysis in acute myeloid leukemia and myelodysplastic syndrome. EBioMedicine. (2018) 37:91–100. doi: 10.1016/j.ebiom.2018.10.042
71. Monaghan SA, Li J-L, Liu Y-C, Ko M-Y, Boyiadzis M, Chang T-Y, et al. A machine learning approach to the classification of acute leukemias and distinction from nonneoplastic cytopenias using flow cytometry data. Am J Clin Pathol. (2022) 157:546–53. doi: 10.1093/ajcp/aqab148
72. Cox AM, Kim D, García R, Fuda FS, Weinberg OK, and Chen W. Automated prediction of acute promyelocytic leukemia from flow cytometry data using a graph neural network pipeline. Am J Clin Pathol. (2024) 161:264–72. doi: 10.1093/ajcp/aqad145
73. Shopsowitz K, Lofroth J, Chan G, Kim J, Rana M, Brinkman R, et al. MAGIC-DR: An interpretable machine-learning guided approach for acute myeloid leukemia measurable residual disease analysis. Cytometry B Clin Cytom. (2024) 106(4):239–51. doi: 10.1002/cyto.b.22168
74. Weijler L, Kowarsch F, Wödlinger M, Reiter M, Maurer-Granofszky M, Schumich A, et al. UMAP based anomaly detection for minimal residual disease quantification within acute myeloid leukemia. Cancers. (2022) 14:898. doi: 10.3390/cancers14040898
75. Lewis JE, Cooper LAD, Jaye DL, and Pozdnyakova O. Automated deep learning-based diagnosis and molecular characterization of acute myeloid leukemia using flow cytometry. Mod Pathol Off J U S Can Acad Pathol Inc. (2024) 37:100373. doi: 10.1016/j.modpat.2023.100373
76. Couckuyt A, Gassen SV, Emmaneel A, Janda V, Buysse M, Moors I, et al. Unraveling genotype–phenotype associations and predictive modeling of outcome in acute myeloid leukemia. Cytometry B Clin Cytom. (2025). doi: 10.1002/cyto.b.22230
77. Ediriwickrema A, Gentles AJ, and Majeti R. Single-cell genomics in AML: extending the frontiers of AML research. Blood. (2023) 141:345–55. doi: 10.1182/blood.2021014670
78. Teixeira A, Carreira L, Abalde-Cela S, Sampaio-Marques B, Areias AC, Ludovico P, et al. Current and emerging techniques for diagnosis and MRD detection in AML: A comprehensive narrative review. Cancers. (2023) 15:1362. doi: 10.3390/cancers15051362
79. Remani Sathyan R, Chandrasekhara Menon G, S H, Thampi R, and Duraisamy JH. Traditional and deep-based techniques for end-to-end automated karyotyping: A review. Expert Syst. (2022) 39:e12799. doi: 10.1111/exsy.12799
80. Qin D. Molecular testing for acute myeloid leukemia. Cancer Biol Med. (2022) 19:4–13. doi: 10.20892/j.issn.2095-3941.2020.0734
81. Cheng W-Y, Li J-F, Zhu Y-M, Lin X-J, Wen L-J, Zhang F, et al. Transcriptome-based molecular subtypes and differentiation hierarchies improve the classification framework of acute myeloid leukemia. Proc Natl Acad Sci. (2022) 119:e2211429119. doi: 10.1073/pnas.2211429119
82. Ikbal Atli E, Gurkan H, Onur Kirkizlar H, Atli E, Demir S, Yalcintepe S, et al. Pros and cons for fluorescent in situ hybridization, karyotyping and next generation sequencing for diagnosis and follow-up of multiple myeloma. Balk J Med Genet BJMG. (2021) 23:59–64. doi: 10.2478/bjmg-2020-0020
83. Moazzen Y, Çapar A, Albayrak A, Çalık N, and Töreyin BU. Metaphase finding with deep convolutional neural networks. BioMed Signal Process Control. (2019) 52:353–61. doi: 10.1016/j.bspc.2019.04.017
84. Saleh HM, Saad NH, and Isa NAM. Overlapping chromosome segmentation using U-net: convolutional networks with test time augmentation. Proc Comput Sci. (2019) 159:524–33. doi: 10.1016/j.procs.2019.09.207
85. Hu X, Yi W, Jiang L, Wu S, Zhang Y, Du J, et al. Classification of metaphase chromosomes using deep convolutional neural network. J Comput Biol J Comput Mol Cell Biol. (2019) 26:473–84. doi: 10.1089/cmb.2018.0212
86. Vajen B, Hänselmann S, Lutterloh F, Käfer S, Espenkötter J, Beening A, et al. Classification of fluorescent R-Band metaphase chromosomes using a convolutional neural network is precise and fast in generating karyograms of hematologic neoplastic cells. Cancer Genet. (2022) 260–261:23–9. doi: 10.1016/j.cancergen.2021.11.005
87. Deng J, Peng W, Lu Q, Wang Z, Fu Q, Zhou X, et al. Manually-established abnormal karyotype dataset based on normal chromosomes effectively train artificial intelligence model for better cytogenetic abnormalities prediction. Preprint (2023). doi: 10.21203/rs.3.rs-2913988/v1
88. Shamsi Z, Reid I, Bryant D, Wilson J, Qu X, Dubey A, et al. Automatic karyotyping: from metaphase image to diagnostic prediction. arXiv [q-bio.QM]. (2025) arXiv:2211.14312. doi: 10.48550/arXiv.2211.14312
89. Gudla PR, Nakayama K, Pegoraro G, and Misteli T. SpotLearn: convolutional neural network for detection of fluorescence in situ hybridization (FISH) signals in high-throughput imaging approaches. Cold Spring Harb Symp Quant Biol. (2017) 82:57–70. doi: 10.1101/sqb.2017.82.033761
90. Xue T, Chang H, Ren M, Wang H, Yang Y, Wang B, et al. Deep learning to automatically evaluate HER2 gene amplification status from fluorescence in situ hybridization images. Sci Rep. (2023) 13:9746. doi: 10.1038/s41598-023-36811-z
91. Xu C, Zhang Y, Fan X, Lan X, Ye X, and Wu T. An efficient fluorescence in situ hybridization (FISH)-based circulating genetically abnormal cells (CACs) identification method based on Multi-scale MobileNet-YOLO-V4. Quant Imaging Med Surg. (2022) 12:2961–76. doi: 10.21037/qims-21-909
92. Bouilhol E, Savulescu AF, Lefevre E, Dartigues B, Brackin R, and Nikolski M. DeepSpot: A deep neural network for RNA spot enhancement in single-molecule fluorescence in-situ hybridization microscopy images. Biol Imaging. (2022) 2:e4. doi: 10.1017/S2633903X22000034
93. Motyko E, Kirienko A, Kustova D, Gert T, Leppyanen I, Radjabova A, et al. Acute myeloid leukemia in the next-generation sequencing era: Real-world data from an Austrian tertiary cancer care center. Ann Hematol. (2024) 24:S311. doi: 10.1016/S2152-2650(24)01194-7
94. Guijarro F, Garrote M, Villamor N, Colomer D, Esteve J, and López-Guerra M. Novel tools for diagnosis and monitoring of AML. Curr Oncol. (2023) 30:5201–13. doi: 10.3390/curroncol30060395
95. Wurm S, Waltersdorfer M, Loindl S, Moritz JM, Herzog SA, Bachmaier G, et al. Acute myeloid leukemia in the next-generation sequencing era. Wien Klin Wochenschr. (2025) 137:504–16. doi: 10.1007/s00508-024-02463-w
96. Zhang H, Qureshi MA, Wahid M, Charifa A, Ehsan A, Ip A, et al. Differential diagnosis of hematologic and solid tumors using targeted transcriptome and artificial intelligence. Am J Pathol. (2023) 193:51–9. doi: 10.1016/j.ajpath.2022.09.006
97. Warnat-Herresthal S, Perrakis K, Taschler B, Becker M, Baßler K, Beyer M, et al. Scalable prediction of acute myeloid leukemia using high-dimensional machine learning and blood transcriptomics. iScience. (2020) 23:100780. doi: 10.1016/j.isci.2019.100780
98. Angelakis A, Nathoe R, and Filippakis M. Towards the gene profile of acute myeloid leukemia using machine learning and blood transcriptomics. Preprint. (2024). doi: 10.20944/preprints202402.0593.v1
99. Awada H, Durmaz A, Gurnari C, Kishtagari A, Meggendorfer M, Kerr CM, et al. Machine learning integrates genomic signatures for subclassification beyond primary and secondary acute myeloid leukemia. Blood. (2021) 138:1885–95. doi: 10.1182/blood.2020010603
100. Song Y, Wang Z, Zhang G, Hou J, Liu K, Wei S, et al. Classification of acute myeloid leukemia based on multi-omics and prognosis prediction value. Front Oncol. (2025) 19(6):1836–54. doi: 10.1002/1878-0261.70000
101. Afroz S. Multi-omics data integration and drug screening of AML cancer using Generative Adversarial Network. Methods. (2024) 226:138–50. doi: 10.1016/j.ymeth.2024.04.017
102. Khosroabadi Z, Azaryar S, Dianat-Moghadam H, Amoozgar Z, and Sharifi M. Single cell RNA sequencing improves the next generation of approaches to AML treatment: challenges and perspectives. Mol Med. (2025) 31:33. doi: 10.1186/s10020-025-01085-w
103. van Galen P, Hovestadt V, Wadsworth Ii MHW, Hughes TK, Griffin GK, Battaglia S, et al. Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell. (2019) 176:1265–1281.e24. doi: 10.1016/j.cell.2019.01.031
104. Nicora G and Bellazzi R. A reliable machine learning approach applied to single-cell classification in acute myeloid leukemia. AMIA Annu Symp Proc. (2021) 2020:925–32.
105. Asimomitis G, Sirenko M, Fotis C, Landau DA, Alexopoulos LG, and Papaemmanuil E. Predicting single cell genotypes from single cell expression profiles in AML using deep learning. In: Proceedings of the 2023 13th international conference on bioscience, biochemistry and bioinformatics. Association for Computing Machinery, New York, NY, USA (2023). p. 1–9. doi: 10.1145/3586139.3586140
106. Stetson LC, Balasubramanian D, Ribeiro SP, Stefan T, Gupta K, Xu X, et al. Single cell RNA sequencing of AML initiating cells reveals RNA-based evolution during disease progression. Leukemia. (2021) 35:2799–812. doi: 10.1038/s41375-021-01338-7
107. Gimeno M, San José-Enériz E, Villar S, Agirre X, Prosper F, Rubio A, et al. Explainable artificial intelligence for precision medicine in acute myeloid leukemia. Front Immunol. (2022) 13:977358. doi: 10.3389/fimmu.2022.977358
108. Qin Y, Pu X, Hu D, and Yang M. Machine learning-based biomarker screening for acute myeloid leukemia prognosis and therapy from diverse cell-death patterns. Sci Rep. (2024) 14:17874. doi: 10.1038/s41598-024-68755-3
109. Badruzzaman A and Arymurhty AM. A comparative study of convolutional neural network in detecting blast cells for diagnose acute myeloid leukemia. J Electron Electromed Eng Med Inform. (2023) 6:84–91. doi: 10.35882/jeeemi.v6i1.354
110. Elhassan TA, Mohd Rahim MS, Siti Zaiton MH, Swee TT, Alhaj TA, Ali A, et al. Classification of atypical white blood cells in acute myeloid leukemia using a two-stage hybrid model based on deep convolutional autoencoder and deep convolutional neural network. Diagnostics. (2023) 13:196. doi: 10.3390/diagnostics13020196
111. Park S, Park YH, Huh J, Baik SM, and Park DJ. Deep learning model for differentiating acute myeloid and lymphoblastic leukemia in peripheral blood cell images via myeloblast and lymphoblast classification. Digit Health. (2024) 10:1–11. doi: 10.1177/20552076241258079
112. Sidhom J-W, Siddarthan IJ, Lai B-S, Luo A, Hambley BC, Bynum J, et al. Deep learning for diagnosis of acute promyelocytic leukemia via recognition of genomically imprinted morphologic features. NPJ Precis Oncol. (2021) 5:1–8. doi: 10.1038/s41698-021-00179-y
113. Acharya V, Ravi V, Pham TD, and Chakraborty C. Peripheral blood smear analysis using automated computer-aided diagnosis system to identify acute myeloid leukemia. IEEE Trans Eng Manag. (2023) 70:2760–73. doi: 10.1109/TEM.2021.3103549
114. Mustapha MT and Ozsahin DU. Morphological analysis and subtype detection of acute myeloid leukemia in high-resolution blood smears using convNeXT. AI. (2025) 6:45. doi: 10.3390/ai6030045
115. Baig R, Rehman A, Almuhaimeed A, Alzahrani A, and Rauf HT. Detecting Malignant leukemia cells using microscopic blood smear images: A deep learning approach. Appl Sci. (2022) 12:6317. doi: 10.3390/app12136317
116. Li N, Fan L, Xu H, Zhang X, Bai Z, Li M, et al. An AI-aided diagnostic framework for hematologic neoplasms based on morphologic features and medical expertise. Lab Invest. (2023) 103:100055. doi: 10.1016/j.labinv.2022.100055
117. Al-Bashir AK, Khnouf RE, and Bany Issa LR. Leukemia classification using different CNN-based algorithms-comparative study. Neural Comput Appl. (2024) 36:9313–28. doi: 10.1007/s00521-024-09554-9
118. Patay BA, Zlotnicki AM, Shah S, Apilado R, and Andrews R. Machine learning algorithms accelerate throughput of a flow - sequencing cell based assay for an acute myeloid leukemia (AML) therapeutic discovery platform. Blood. (2021) 138:4930. doi: 10.1182/blood-2021-148224
119. Vial JP, Lechevalier N, Lacombe F, Dumas P-Y, Bidet A, Leguay T, et al. Unsupervised flow cytometry analysis allows for an accurate identification of minimal residual disease assessment in acute myeloid leukemia. Cancers. (2021) 13:629. doi: 10.3390/cancers13040629
120. Seheult JN, Shi M, Olteanu H, Otteson GE, Timm MM, Weybright MJ, et al. Machine learning enhancement of flow cytometry data accelerates the identification of minimal residual acute myeloid leukemia. Blood. (2023) 142:4339. doi: 10.1182/blood-2023-190275
121. Zhong P, Hong M, He H, Zhang J, Chen Y, Wang Z, et al. Diagnosis of acute leukemia by multiparameter flow cytometry with the assistance of artificial intelligence. Diagnostics. (2022) 12:827. doi: 10.3390/diagnostics12040827
122. Lu Z, Lyu YE, Gao A, Yeager TS, Morita M, Wang S, et al. B-142 development and clinical validation of artificial intelligence assisted flow cytometry diagnosis for acute leukemia. Clinical Chemistry. (2023) 69(Suppl_1):hvad097.475. doi: 10.1093/hvad097.4750i
123. Lian J-W, Wei C-H, Chen M-Y, and Lin C-C. Acute leukemia prediction and classification using convolutional neural network and generative adversarial network. Appl Soft Comput. (2024) 163:111819. doi: 10.1016/j.asoc.2024.111819
124. Müller M-L, Maschek S, Fortina E, Cunha M, Pagezy S, Koppelle A, et al. Conference abstract: Artificial intelligence to predict medical diagnosis from flow cytometric raw data in mature B-cell and T-cell neoplasms, AML, ALL, MDS and multiple myeloma. HemaSphere. (2023) 7:e350349d. doi: 10.1097/01.HS9.0000971748.35034.9d
125. Cheng F-M, Lo S-C, Lin C-C, Lo W-J, Chien S-Y, Sun T-H, et al. Deep learning assists in acute leukemia detection and cell classification via flow cytometry using the acute leukemia orientation tube. Sci Rep. (2024) 14:8350. doi: 10.1038/s41598-024-58580-z
126. Fang M, Shamsi Z, Bryant D, Wilson J, Qu X, Dubey A, et al. Karyotype AI for precision oncology. Blood. (2024) 144:1544. doi: 10.1182/blood-2024-211644
127. Shah K, Ma J, Djekidel M, Song G, Umeda M, Fan Y, et al. Gene expression machine learning models classify pediatric AML subtypes with high performance. Blood. (2023) 142:1570–0. doi: 10.1182/blood-2023-189450
128. Mosquera Orgueira A, Peleteiro Raíndo A, Cid López M, Díaz Arias JÁ, González Pérez MS, Antelo Rodríguez B, et al. Personalized survival prediction of patients with acute myeloblastic leukemia using gene expression profiling. Front Oncol. (2021) 11:657191. doi: 10.3389/fonc.2021.657191
129. Wang T. Molecular precision medicine: Multi-omics-based stratification model for acute myeloid leukemia. Heliyon. (2024) 10(17):e30432. doi: 10.1016/j.heliyon.2024.e36155
130. Lee J, Cho S, Hong S-E, Kang D, Choi H, Lee J-M, et al. Integrative analysis of gene expression data by RNA sequencing for differential diagnosis of acute leukemia: potential application of machine learning. Front Oncol. (2021) 11:717616. doi: 10.3389/fonc.2021.717616
Keywords: acute myeloid leukemia, blood smear image, flow cytometry, genetic analysis, artificial intelligence
Citation: Xie W, Jiang X, Huang J, Qin M and Bi Z (2025) Research advances in the adjunctive diagnosis of acute myeloid leukemia. Front. Oncol. 15:1634935. doi: 10.3389/fonc.2025.1634935
Received: 25 May 2025; Accepted: 22 September 2025;
Published: 07 October 2025.
Edited by:
Raffaele Palmieri, University of Rome Tor Vergata, ItalyReviewed by:
Michael Diamantidis, General Hospital of Larissa, GreeceNathalia Lopez Duarte, Rio de Janeiro Municipal Health Secretariat (SMS-RJ), Brazil
Copyright © 2025 Xie, Jiang, Huang, Qin and Bi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhisheng Bi, Yml2aWN0b3JAZ21haWwuY29t
†These authors have contributed equally to this work
 Wentao Xie
Wentao Xie Xinye Jiang1†
Xinye Jiang1† Jingying Huang
Jingying Huang Zhisheng Bi
Zhisheng Bi