- 1Department of Pathology, The Second Xiangya Hospital, Central South University, Changsha, China
- 2Hunan Clinical Medical Research Center for Cancer Pathogenic Genes Testing and Diagnosis, Changsha, China
- 3Geneplus-Beijing, Beijing, China
ELOC(TCEB1)-mutant renal cell carcinoma [ELOC(TCEB1)-RCC] is a newly recognized type of RCC characterized by clear cell morphology and ELOC(TCEB1) gene mutation. We analyzed one case with a point mutation in TCEB1 c.218T>A (p.V73E), which is a novel mutation site and has not been reported in ELOC(TCEB1)-RCC. The case involved a male individual of age 48, whose computed tomography scan of the abdomen indicated the presence of a solid nodule located in the kidney. The tumor cells showed expression of PAX8, CA9, AMACR/P504S, Vimentin, CK7, CD10, FH, INI1(SMARCB1) and ELOC(TCEB1), and ELOC was mainly located in the nucleus. CD117, TFE3, HMB45, and SDHB were not express, and the expression rate of Ki67 was <5%. The novel variant in ELOC(TCEB1) gene was identified by the next-generation sequencing (NGS) test, subsequently also confirmed by Sanger sequencing. The ELOC(TCEB1) gene mutation testing is helpful for the diagnosis of this type of RCC. The case further expands our knowledge of the spectrum of TCEB1 gene mutation in ELOC(TCEB1)-RCC and enhances the optimization of clinical decision-making.
1 Introduction
The molecular features that define clear cell renal cell carcinoma (ccRCC) initiation and progression are being increasingly defined (1). The molecular features that arise from these defects enable categorization of ccRCC into clinically and therapeutically relevant subtypes. The main change in the WHO 2022 classification is the introduction of a new category of molecularly-defined RCC, which includes TFE3-rearranged RCC, TFEB-rearranged RCC, and TFEB-amplified RCC, FH-deficient RCC, SDH-deficient RCC, ALK-rearranged RCC, ELOC(TCEB1)-mutated RCC, INI1(SMARCB1)-deficient RCC (2). The transcription elongation factor B (TCEB1) gene, encoding the protein elongin C (ELOC), contributes to the Von Hippel-Lindau (VHL) complex to ubiquitinate hypoxia-inducible factor (HIF). Integrated sequencing analysis identified a group of tumors among RCCs characterized by hotspot mutations in TCEB1 gene S23L, Y79C/S/F/N, I95N or A100P, A106D (3–5). ELOC(TCEB1)-mutated RCC [ELOC(TCEB1)-RCC] is a newly recognized type of RCC characterized by clear cell morphology and TCEB1 gene hotspot mutations, which has been classified as RCC with leiomyomatous stroma (RCCLMS) (1, 6). RCCLMS is a novel subtype of RCC with unique morphologic, immunohistochemical, and molecular characteristics that is distinct from ccRCC and clear cell-papillary RCC. RCCLMS harbors recurrent mutations of TSC1/TSC2, MTOR, and/or ELOC(TCEB1) genes, consistent with hyperactive MTOR complex; while ccRCC demonstrates primary alterations in VHL gene (7). With the increasing application of next generation sequencing (NGS) and other molecular biological detection technologies, it has been possible to determine the oncogenic activation alterations of many solid tumors (8). ELOC(TCEB1)-RCC is a distinct entity with recurrent hotspot mutations, specific copy number alterations, pathway activation and characteristic morphologic features (2). Here, we find a rare case of ELOC(TCEB1)-RCC, harboring a novel mutation site in TCEB1 gene c.218T>A (p.V73E), which can broaden our understanding of RCC genotype and maybe provide treatment options.
2 Case presentation
2.1 Clinical data
A 48-year-old man presented to the urology department with an incidentally discovered renal lesion on screening CT scan (Figure 1A), showing rounded nodular low-density shadows in renal parenchyma with a clear boundary, and uneven enhancement was observed on the enhanced CT scan. The TNM stage was observed to be stage I. The patient underwent radical nephrectomy and were followed up for 14 months without any other treatment after surgery. The case has survived till now and showed no evidence of recurrence or metastasis.
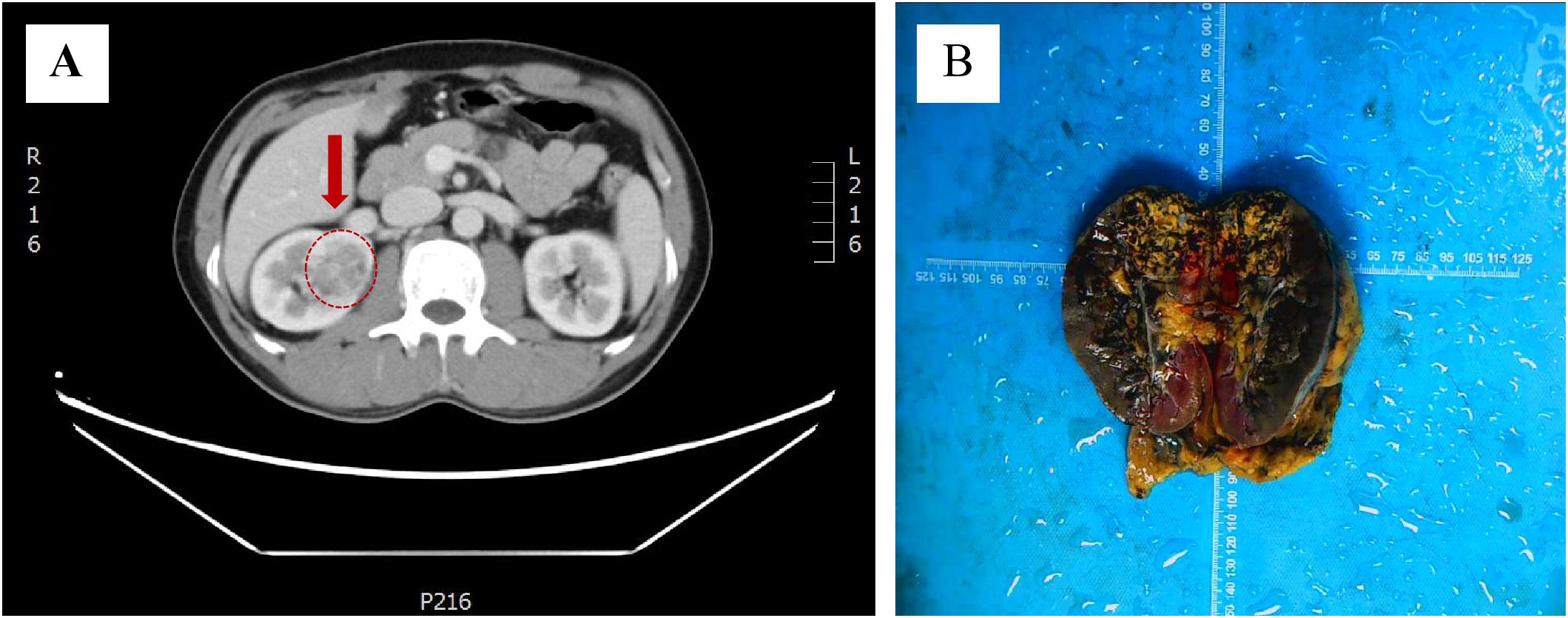
Figure 1. Discovery of ELOC(TCEB1)-RCC. (A) CT scans showed rounded low-density shadows in renal parenchyma. (B) Gross observation of ELOC(TCEB1)-RCC.
2.2 Pathological examination
The tumor was located in the renal parenchyma, approximately 3.5 cm in diameter, nodular, gray-yellow or gray-brown, solid in texture and with clear boundaries (Figure 1B). All hematoxylin and eosin (H&E) slides from the case of ELOC(TCEB1)-RCC from Department of Pathology, The Second Xiangya Hospital, were reviewed by two experienced genitourinary pathologists. The tumor of the patient was nodular (Figure 2A); a thick fibrous pseudocapsule rich in smooth muscle was visible (Figure 2B). The tumor cells were mainly arranged in dense medium-sized acini (Figure 2C) or short papillae (Figure 2D). The tumor cells had clear boundaries, transparent cytoplasm, irregular or short fusiform nuclei, dense chromatin, unclear nucleoli, slight atypia, and WHO/ISUP nuclear grade 1 (Figure 2E). In the tumor of this patient, multifocal lymphocyte aggregation (Figure 2F, G) was observed, accompanied by hemorrhage and hemosiderin deposition (Figure 2H).
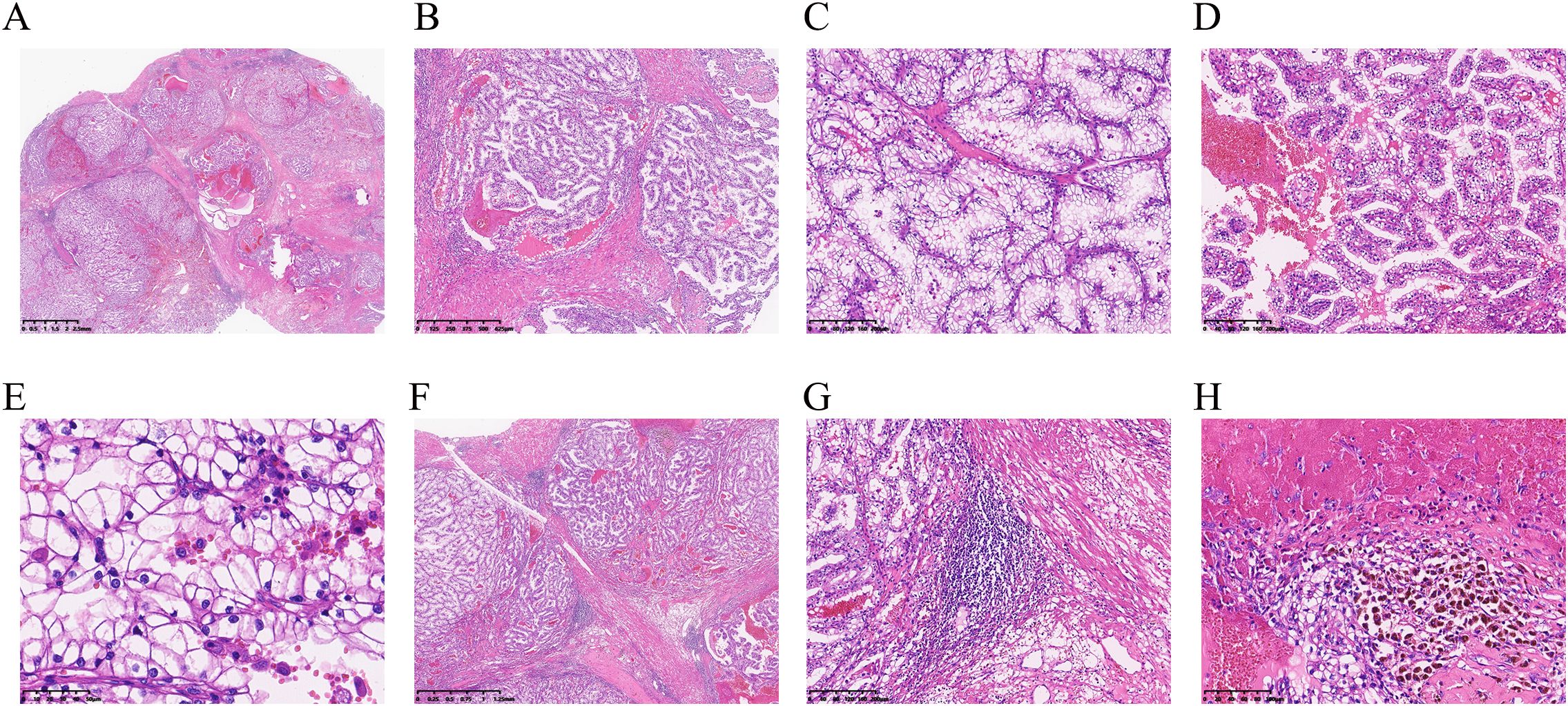
Figure 2. Microscopy of ELOC(TCEB1)-RCC. (A) Under low-power magnification, the tumor is separated into multinodular masses by thick fibromuscular stroma (×6.6). (B) A thick fibrous pseudocapsule rich in smooth muscle can be seen within the tumor (×40). (C) Tumor cells show dense arrangement in medium-sized acinar patterns (×100). (D) The tumor cells were arranged in short papillary shapes (×100). (E) WHO/ISUP nuclear grading for the tumor was grade 1 in the patient, and nuclei were densely stained and irregular in shape (×400). (F, G) In this case, focal lymphocyte aggregation (F, ×20) and lymphoid follicle formation (G, ×100) were observed. (H) Intratumoral hemorrhage and hemosiderin deposition are observed (×200).
2.3 Immunohistochemical staining
The immunohistochemical stains for carbonic anhydrase-IX (CA9), Paired box 8 (PAX8), Cytokeratin 7 (CK7), common acute lymphocytic leukemia antigen/CD10 (CALLA/CD10) and α-methylacyl CoA racemase/P504S (AMACR/P504S) were performed. The choice of immunohistochemical stains was based on the utility of these markers among certain renal cell carcinomas that may be confused with these TCEB1-mutated tumors because of some morphologic overlaps. The immunohistochemical results showed that the tumor was positive for PAX8, CA9 (diffuse box-like positivity), CK7, CD10, P504S, and vimentin. Among them, tumor cells showed moderate positivity for CK7 (Figure 3A, B); CA9 showed diffuse strong positive membrane staining, completely outlining the cell membrane (Figure 3C, D); ELOC was moderate positive, mainly localized in the nucleus, with varying degrees in staining intensity and range (Figure 3E, F). Additionally, in this case, the tumor cells showed Ki67<5%, and did not express CD117, TFE3, HMB45, or SDHB, while expressing FH and INI1(SMARCB1).
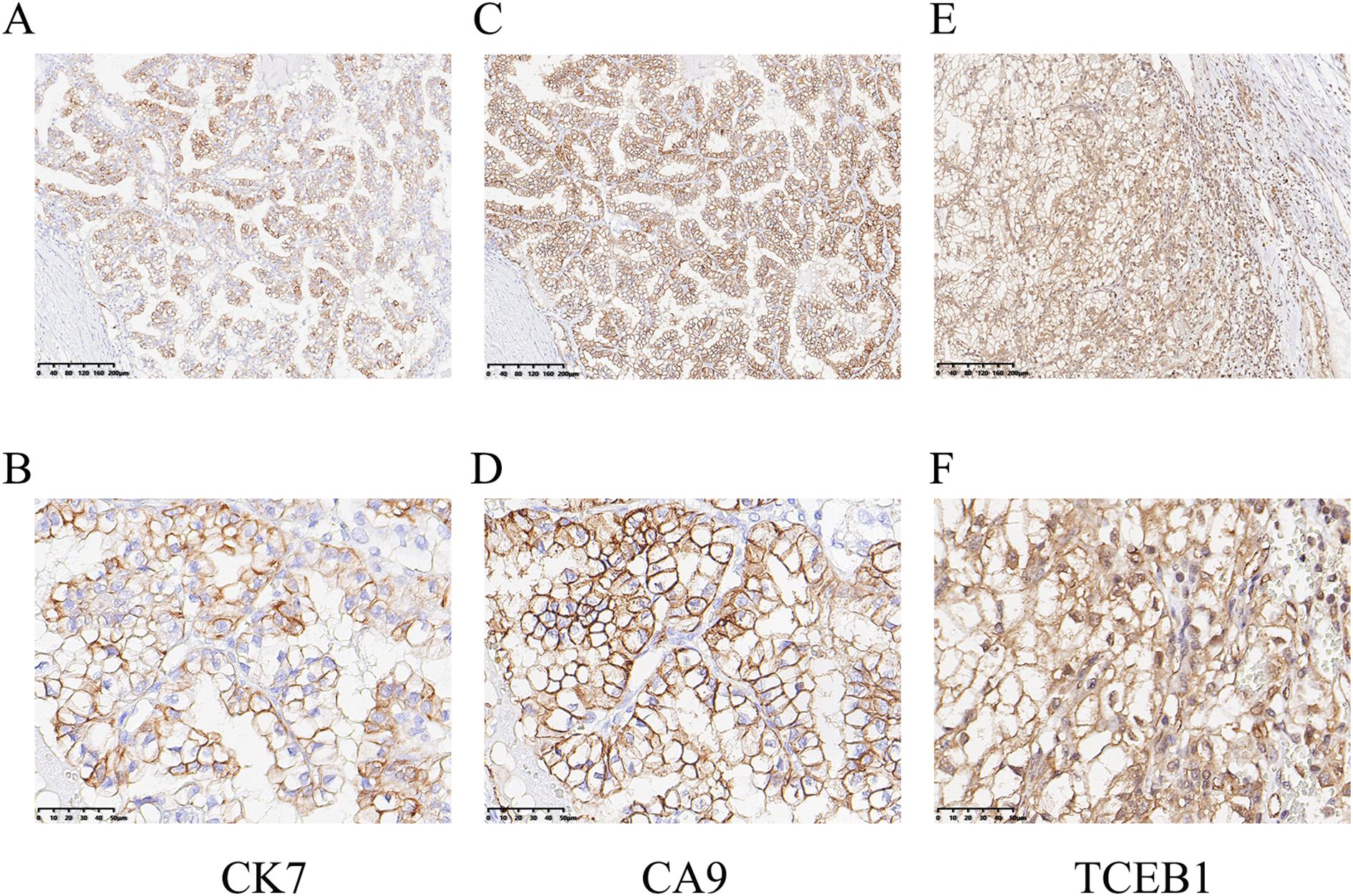
Figure 3. Immunohistochemical staining of ELOC(TCEB1)-RCC. (A, B) The tumor cells were positive for CK7 (A, ×100; B, ×400). (C, D) Diffuse membrane strong positive for CA9 (C, ×100; D, ×400). (E, F) ELOC (TCEB1) was mainly located in the nucleus of tumor cells (E, ×100; F, ×400).
2.4 TCEB1 gene mutation detection
The tumor tissues from lung puncture were made into FFPE samples and used for making pathological sections. Genomic DNA was extracted from FFPE tumor samples using the QIAamp DNA FFPE tissue kit (Qiagen, Hilden, Germany). Mutational hot spots were analyzed using targeted deep sequencing with a capture-based NGS panel obtained from GenePlus Technology Co., Ltd. (Beijing, China). The NGS panel included an assay targeting 1021 genes (EGFR/KRAS/ALK etc.) known to be involved in solid tumors. The 1021 panel covers full exonic coverage for VHL, BAP1, PBRM1, SETD2, FLCN, MET, PTEN, TP53, FH, SDH, ALK, TSC1, TSC2, and MTOR genes; for TFE3 gene, it covers partial exons and partial introns; while TFEB gene is not covered. Therefore, there may be some limitations in the detection of rearrangement variants of TFE3 and TFEB. DNA sequencing was then performed on the GenePlus Seq-2000 system. The assay can identify various types of genomic alterations, including single base substitutions, insertions/deletions of different lengths, copy number variations, gene fusions, and rearrangements. A point mutation variant in TCEB1 gene NM_005648.3: c.218T>A (p.Val73Glu) was identified, resulting in a change in the codon 73 from Valine to Glutamic acid (Figure 4A). The p.Val73Glu of TCEB1 gene is a novel mutation site, which has not been reported. In this case, the copy number alterations and loss of heterozygosity of chromosome 8 were not observed.
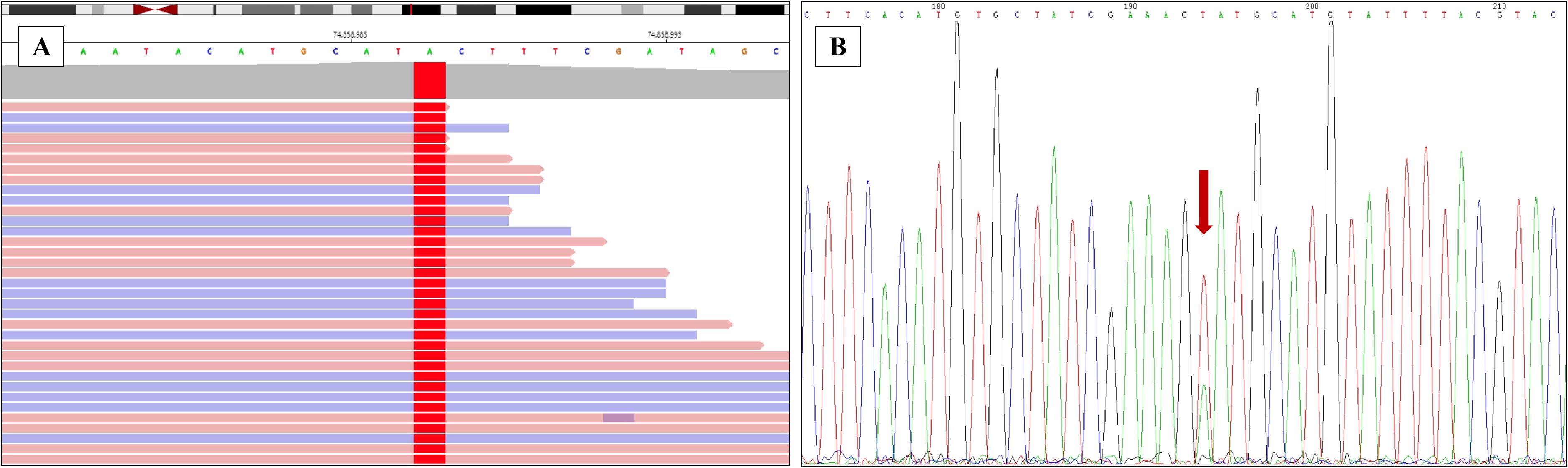
Figure 4. Validating the novel mutation of ELOC(TCEB1) gene. (A) The point mutation of ELOC(TCEB1) gene in the 73th amino acid was identified by the next-generation sequencing (NGS). (B) The point mutation of TCEB1 gene in the 73th amino acid was verified by Sanger sequencing.
This novel mutation site was also identified with Sanger Sequencing. TCEB1 gene was amplified using polymerase chain reaction (PCR). The Primer 5 software was used to design the primers, and the primer sequences are presented below: upstream primer: 5′-TGGATTGCCACCCTAATGAC-3′, downstream primer: 5′-GAATTCAGGAATCTCGGTGGAG-3′. The following PCR reaction conditions were used: pre-denaturation for 1 min at 98°C; (i) denaturation at 98°C for 10 s; (ii) annealing at 55°C for 5 s; (iii) extension at 72°C for 15 s; (iv) repeat (i)–(iii) for 35 times and (v) incubate at 72°C for 10 min. PCR products were identified using agarose gel electrophoresis and then purified. The purified amplification products were sent to Sheng Gong bioengineering (Shanghai, China) co., LTD. After purification, the 3730XL (ABI, Singapore) sequencer was used for sequencing (Figure 4B).
3 Discussion
ELOC(TCEB1)-RCC was first reported in 2015, with more than 90% of patients being male (9). Most cases exhibit low-grade malignancy, but 10% of cases show metastasis. This case involves a 48-year-old male, with the tumor located in the right kidney and no metastasis observed (Figure 1). ELOC(TCEB1)-RCC has a broad morphological spectrum, mainly characterized by fibrous smooth muscle tissue separating the tumor into nodular structures (10). The tumor cells have clear cytoplasm and fibrous/fibromuscular stroma (FMS) (or leiomyoma-like stroma), making the tumor appear nodular under low-power microscopy. The architecture is diverse, including solid, acinar, and nested patterns, occasionally with cystic and tubulopapillary structures (11). Importantly, this tumor shows positive CK7 expression with significant variation—expression results differ in some tumors, with as low as 10% to 15% of tumor cells stained, ranging from patchy to diffuse. CA9 (typically with complete membranous staining) and CD10 are consistently immunoreactive. The positivity of CK7 and CA9 is typical and is required for the diagnosis of RCC with FMS (10). These morphological and immunological features are essential criteria for the diagnosis of ELOC(TCEB1)-RCC. The case is also characterized by fibromuscular tissue dividing the tumor into nodular shapes. Morphologically, it is characterized by branched acinar or tubular structures, with focal scattered short papillary structures (Figure 2). Immunologically, tumor cells are positive for CA9 and CK7, and ELOC shows nuclear positivity (Figure 3). Additionally, in this case, tumor cells don’t express CD117, TFE3, HMB45, or SDHB, but express FH and INI1(SMARCB1). Thus, we can exclude other molecularly-defined RCC categories, including TFE3-rearranged RCC, FH-deficient RCC, SDH-deficient RCC, and SMARCB1-deficient RCC.
ELOC(TCEB1)-RCC is a subtype of RCC first recognized by the World Health Organization (WHO) in 2022, molecularly characterized by the presence of TCEB1 gene mutations and the absence of VHL gene mutations (12). Most patients with ELOC(TCEB1)-RCC have a good prognosis, while nuclear pleomorphism and multifocal necrosis may indicate adverse biological behavior (9). The researchers proposed for the first time the significance of immunohistochemical nuclear positive for ELOC and Sanger sequencing in the diagnosis of ELOC(TCEB1)-RCC (3). TCEB1-mutated tumors also did not possess any additional recurrent copy number events such as 5q amplifications or 14q or 9p losses which are common in ccRCC, papillary renal cell carcinoma (PRCC), and collecting duct carcinoma (CDC) (3, 13). 5q amplification is one of the most common copy number variations in ccRCC, present in approximately 65%~70% of patients with ccRCC (13). 14q, harboring HIF-1α gene, loss is present in ccRCC (14), and type 1 PRCC with a relatively low incidence (<10%) (15). CDKN2A/2B (9p) deletions are present in ccRCC, type 2 PRCC, and CDC (16). ELOC(TCEB1)-RCCs are mostly low grade, lack the common chromosomal alterations or gene mutations seen in RCC, including PBRM1, SETD2, BAP1, TSC1, TSC2, or mTOR (11, 17). The TCEB1 hotspot mutations (Y79C/S/F/N, E92K, A100P, A106D, and C112Vfs∗3) were all located within or close to the VHL-binding domains in ELOC protein (9, 18). In this case, a novel mutation site (p.V73E) in TCEB1 gene was identified in the tumor by NGS and Sanger Sequencing (figure 4). Researchers have reported the crystal structure of VHL bound to a Cul2 N-terminal domain and ELOC protein (residues 17-112), which is the minimal domain required for VHL binding (18). The mutation site of TCEB1 gene V73E is exactly located within the VHL-binding domain. We believe that the novel variant of TCEB1 c.218T>A (p.V73E) is a driving mutation and harbors oncogenic potential, which is one of the potential pathogeneses in ELOC(TCEB1)-RCC. However, it remains to be further validated.
ELOC(TCEB1)-RCC exhibits an indolent biological behavior with limited metastatic potential. If no clinical aggressiveness, surgical resection is usually curative. For tumors in stages T1-T2, adjuvant therapy is not required after surgery, while adjuvant treatment regimens for advanced-stage tumors are the same as those for non-ccRCC (12). Currently, no record of the novel mutation site c.218T>A (p.V73E) in TCEB1 gene has been found in public population databases (e.g., 1000 Genomes, GO ESP, Gnomad) or in COSMIC, a database of human cancer driver genes. This mutation scores 0.9987 in AlphaMissense and is predicted to be a Strong Pathogenic mutation. The codon altered by this mutation site is located in the VHL-binding domain of ELOC, which belongs to a relatively conserved region of the protein. The functional prediction software SIFT indicates that this mutation is deleterious. Overall, the case was diagnosed as ELOC(TCEB1)-RCC through morphological features, immunophenotypic characteristics, and molecular pathological analyses. The patient has been free of recurrence or metastasis for more than a dozen months after undergoing radical nephrectomy.
In conclusion, we report a very rare case of ELOC(TCEB1)-RCC in an adult with a novel variation in TCEB1: c.218T>A (p.V73E). The presence of FMS, and immunohistochemical positive for PAX8, CK7, CA9, CD10, and AMACR/P504S, and positive ELOC nuclear staining may be an important indication for the diagnosis of this disease. The detection of TCEB1 gene mutation is helpful for the diagnosis of ELOC(TCEB1)-RCC through Sanger sequencing or next generation sequencing.
Data availability statement
Datasets are available on request. The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by the Ethics Committee of the Second Xiangya Hospital of Central South University. Written informed consent was obtained from the patient for the publication of any potentially identifiable images or data included in this article. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
HC: Data curation, Funding acquisition, Writing – original draft. NL: Data curation, Validation, Writing – review & editing. KZ: Data curation, Validation, Writing – review & editing. PZ: Supervision, Validation, Writing – review & editing. XL: Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundations of China (81900070 for LXH, and 82200086 for CHP). The funders have roles in the design and conduct of the study, the analysis and interpretation of the data, and preparation of the manuscript.
Conflict of interest
Author KZ was employed by Geneplus-Beijing.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Jonasch E, Walker CL, and Rathmell WK. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat Rev Nephrol. (2021) 17:245–61. doi: 10.1038/s41581-020-00359-2
2. Alaghehbandan R, Siadat F, and Trpkov K. What’s new in the who 2022 classification of kidney tumours? Pathologica. (2022) 115:8–22. doi: 10.32074/1591-951x-818
3. Hakimi AA, Tickoo SK, Jacobsen A, Sarungbam J, Sfakianos JP, Sato Y, et al. Tceb1-mutated renal cell carcinoma: A distinct genomic and morphological subtype. Mod Pathol. (2015) 28:845–53. doi: 10.1038/modpathol.2015.6
4. DiNatale RG, Gorelick AN, Makarov V, Blum KA, Silagy AW, Freeman B, et al. Putative drivers of aggressiveness in tceb1-mutant renal cell carcinoma: an emerging entity with variable clinical course. Eur Urol Focus. (2021) 7:381–9. doi: 10.1016/j.euf.2019.11.013
5. Parilla M, Alikhan M, Al-Kawaaz M, Patil S, Kadri S, Ritterhouse LL, et al. Genetic underpinnings of renal cell carcinoma with leiomyomatous stroma. Am J Surg Pathol. (2019) 43:1135–44. doi: 10.1097/pas.0000000000001255
6. Andreou A, Yngvadottir B, Bassaganyas L, Clark G, Martin E, Whitworth J, et al. (Eloc/tceb1)-associated von hippel-lindau disease. Hum Mol Genet. (2022) 31:2728–37. doi: 10.1093/hmg/ddac066
7. Shah RB, Stohr BA, Tu ZJ, Gao Y, Przybycin CG, Nguyen J, et al. Renal cell carcinoma with leiomyomatous stroma” Harbor somatic mutations of tsc1, tsc2, mtor, and/or eloc (Tceb1): clinicopathologic and molecular characterization of 18 sporadic tumors supports a distinct entity. Am J Surg Pathol. (2020) 44:571–81. doi: 10.1097/pas.0000000000001422
8. Li NM, Jiang SH, Zhou P, and Li XH. Case report: an ntrk1 fusion-positive embryonal rhabdomyosarcoma: clinical presentations, pathological characteristics and genotypic analyses. Front Oncol. (2023) 13:1178945. doi: 10.3389/fonc.2023.1178945
9. Wang JJ, Huang RR, Cone BD, Kang SL, Setoodeh R, Sisk AE, et al. Eloc-mutated renal cell carcinoma is a rare indolent tumor with distinctive genomic characteristics. Mod Pathol. (2025) 38:100777. doi: 10.1016/j.modpat.2025.100777
10. Shah RB. Renal cell carcinoma with fibromyomatous stroma-the whole story. Adv Anat Pathol. (2022) 29:168–77. doi: 10.1097/pap.0000000000000337
11. Batavia AA, Rutishauser D, Sobottka B, Schraml P, Beerenwinkel N, and Moch H. Biallelic eloc-inactivated renal cell carcinoma: molecular features supporting classification as a distinct entity. Mod Pathol. (2023) 36:100194. doi: 10.1016/j.modpat.2023.100194
12. Rizzo M, Caliò A, Brunelli M, Pezzicoli G, Ganini C, Martignoni G, et al. Clinico-pathological implications of the 2022 who renal cell carcinoma classification. Cancer Treat Rev. (2023) 116:102558. doi: 10.1016/j.ctrv.2023.102558
13. Diez-Calzadilla NA, Noguera Salvá R, Soriano Sarrió P, and Martínez-Jabaloyas JM. Genetic profile and immunohistochemical study of clear cell renal carcinoma: pathological-anatomical correlation and prognosis. Cancer Treat Res Commun. (2021) 27:100374. doi: 10.1016/j.ctarc.2021.100374
14. Moore LE, Jaeger E, Nickerson ML, Brennan P, De Vries S, Roy R, et al. Genomic copy number alterations in clear cell renal carcinoma: associations with case characteristics and mechanisms of vhl gene inactivation. Oncogenesis. (2012) 1:e14. doi: 10.1038/oncsis.2012.14
15. Sanders ME, Mick R, Tomaszewski JE, and Barr FG. Unique patterns of allelic imbalance distinguish type 1 from type 2 sporadic papillary renal cell carcinoma. Am J Pathol. (2002) 161:997–1005. doi: 10.1016/s0002-9440(10)64260-5
16. Trpkov K, Hes O, Williamson SR, Adeniran AJ, Agaimy A, Alaghehbandan R, et al. New developments in existing who entities and evolving molecular concepts: the genitourinary pathology society (Gups) update on renal neoplasia. Mod Pathol. (2021) 34:1392–424. doi: 10.1038/s41379-021-00779-w
17. Martignoni G, Brunelli M, Segala D, Gobbo S, Borze I, Atanesyan L, et al. Renal cell carcinoma with smooth muscle stroma lacks chromosome 3p and vhl alterations. Mod Pathol. (2014) 27:765–74. doi: 10.1038/modpathol.2013.180
Keywords: renal cell carcinoma, ELOC(TCEB1) gene, novel variant, case report, next generation sequencing (NGS)
Citation: Cheng H-P, Li N-M, Zeng K-X, Zhou P and Li X-H (2025) A case report: identifying a novel variant in ELOC(TCEB1)-mutant renal cell carcinoma. Front. Oncol. 15:1661834. doi: 10.3389/fonc.2025.1661834
Received: 08 July 2025; Accepted: 27 October 2025;
Published: 11 November 2025.
Edited by:
Mottaran Angelo, University of Bologna, ItalyReviewed by:
Cherry Bansal, Tantia University, IndiaMelissa Tjota, University of Chicago, United States
Copyright © 2025 Cheng, Li, Zeng, Zhou and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Hong Li, bGkxOTg5QGNzdS5lZHUuY24=
†ORCID: Hai-Peng Cheng, orcid.org/0000-0001-8422-6223
Na-Mei Li, orcid.org/0000-0002-0137-4739
Ke-Xin Zeng, orcid.org/0009-0000-5603-1189
Peng Zhou, orcid.org/0000-0001-8328-6320
Xiao-Hong Li, orcid.org/0000-0002-6953-986X
 Hai-Peng Cheng
Hai-Peng Cheng Na-Mei Li
Na-Mei Li Ke-Xin Zeng3†
Ke-Xin Zeng3† Peng Zhou
Peng Zhou Xiao-Hong Li
Xiao-Hong Li