- 1Frontiers Science Center for Disease-related Molecular Network, Department of Orthopedic Surgery and Orthopedic Research Institute, West China Hospital, Sichuan University, Chengdu, Sichuan, China
- 2Department of Orthopedic Surgery and Orthopedic Research Institute, West China Hospital, Sichuan University, Chengdu, Sichuan, China
- 3Department of Urology, Chongqing University Fuling Hospital, Chongqing, China
- 4Department of Urology, The First Affiliated Hospital of Chongqing Medical University, Chongqing, China
- 5The First Affiliated Hospital of Chongqing Medical University, Chongqing, China
Introduction: Bladder cancer (BCa) is one of the most prevalent genitourinary malignancies with high recurrence worldwide. A lack of reliable prognostic biomarkers and effective therapeutic targets hinders its treatment. Emerging evidence indicates that long noncoding RNAs (lncRNAs) are involved in human cancers, including BCa. While lncRNAs hold enormous promise, their specific roles and mechanisms in BCa remain largely unexplored. Here, we identify the lncRNA ELDR as a pivotal oncogenic driver in BCa.
Methods: RT-qPCR was used to analyze the expression patterns of ELDR, miR-1343-3p and TRIM44. CCK-8, colony formation, EdU, and Transwell assays were used to detect the effect of ELDR on cell proliferation, migration, and invasion. The association between ELDR, miR-1343-3p and TRIM44 was analyzed by bioinformatics analysis and dual-luciferase reporter assay. Finally, the role of the ELDR-miR-1343-3p-TRIM44 axis in bladder cancer cell behavior was demonstrated.
Results and Discussion: ELDR is significantly upregulated in BCa tissues, and its high expression correlates with aggressive clinicopathological features and predicts poor prognosis in BCa patients. Functional experiments demonstrate that ELDR enhances BCa cell proliferation, colony formation, migration, and invasion in vitro and accelerates tumor growth in vivo. Mechanistically, ELDR functions as a competitive endogenous RNA (ceRNA) by sequestering tumor-suppressive miR-1343-3p in the cytoplasm, which consequently leads to the upregulation of the oncogene TRIM44. Our findings unveil the ELDR/miR-1343-3p/TRIM44 axis as a crucial pathway in BCa progression, establishing ELDR as a promising prognostic biomarker and an attractive candidate for the development of targeted therapies.
1 Introduction
Bladder cancer, ranking as the second most prevalent malignancy of the genitourinary system, accounts for over 573,000 new cases globally each year (1). The characteristics of frequent recurrence and metastasis are the main reasons for its poor prognosis (2). The past decade has been important for strides in BCa detection and management. However, the prognosis for most patients remains poor. High recurrence rates and the aggressive course of disease mean an undesirable 5-year survival rate of BCa (3). Therefore, identifying novel genes and pathways involved in BCa is critical for developing new diagnostic and therapeutic approaches.
Long non-coding RNAs (lncRNAs) are defined as transcripts longer than 200 nucleotides with limited protein-coding potential. They have emerged as critical regulators of gene expression and cellular homeostasis (4). Although initially considered “transcriptional noise”, recent studies reveal that lncRNAs participate in diverse biological processes, including chromatin remodeling, RNA stability modulation, and signal transduction cascades. Interactions with proteins, DNA, or other RNAs enable these functions (5, 6). The roles of lncRNAs in disease pathogenesis—particularly cancer—are increasingly recognized. Numerous dysregulated lncRNAs operate as competitive endogenous RNAs (ceRNAs). They contribute to tumor initiation, metastasis, and therapy resistance by sponging tumor-suppressive miRNAs and derepressing oncogenic target genes (7, 8). For example, Xue et al. revealed that lncRNA LUESCC is highly expressed in ESCC and acts as a ceRNA to promote tumor proliferation, invasion, and migration by targeting the miR-6785-5p/NRSN2 axis (9). Furthermore, Sheng et al. observed a clear upregulation of LINC01980 in HCC, which they correlated with a poor prognosis (10). Despite its association with various cancers, the role of lncRNA ELDR in BCa remains unclear. A previous study on oral cancer demonstrated that ELDR enhances tumor growth by promoting ILF3-cyclin E1 signaling (11). Additionally, ELDR is highly expressed and has potential as a biomarker of poor prognosis in the serum extracellular vesicles of breast cancer patients (12). These findings support the role of ELDR as an oncogenic molecule, suggesting the need for further studies on its mechanistic role in bladder carcinogenesis progression.
MicroRNAs (miRNAs) are small non-coding RNAs of 21–25 nucleotides. They are master regulators of carcinogenesis, modulating oncogenes and tumor suppressors at the post-transcriptional level (13). In BCa, dysregulated miRNAs drive malignant phenotypes—including proliferation, migration, invasion, and therapy resistance—through interactions with key genes (14, 15). For instance, Zhang et al. displayed that miR-15b-3p-mediated inhibition of ferroptosis could weaken bicalutamide sensitivity in prostate cancer (16). As a tumor suppressor gene, miR-1343-3p has been reported to be downregulated in expression in a variety of malignancies (17–19). Notably, Lai et al. revealed that miR-1343-3p can serve as an early screening marker for BCa (20). However, the potential mechanism of miR-1343-3p in BCa needs to be further explored.
Tripartite motif-containing 44 (TRIM44), a cytoplasmic and nuclear regulatory protein (21), is dysregulated in multiple human malignancies. Previous research indicated elevated TRIM44 levels in ovarian cancer drive tumor progression via activating the NF-kB pathway (22). It also correlates with aggressive clinical behavior in prostate cancer (23). A previous study demonstrated that TRIM44 was a risk factor affecting BCa (24); however, the underlying mechanism by which TRIM44 regulated BCa development remained unclear.
In this investigation, we demonstrate that highly expressed ELDR promotes malignant progression of BCa by targeting the miR-1343-3p/TRIM44 axis, which is correlated with the poor prognosis. Therefore, ELDR may serve as a diagnostic biomarker and therapeutic target for BCa patients.
2 Materials and methods
2.1 Clinical tissue specimens collection
Primary BCa tumor tissues and paired adjacent normal tissues (at least 3 cm from the edge of cancer tissues) were acquired from 58 treatment-naïve patients undergoing curative resection at the First Affiliated Hospital of Chongqing Medical University (2019-2021). All cases received histopathological confirmation by two independent pathologists, with exclusion criteria encompassing any preoperative anticancer therapy. Immediately following surgical excision, tissues were snap-frozen in liquid nitrogen and cryopreserved at -80°C for molecular analyses. The study protocol obtained formal approval from the Medical Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (2021-199), with written informed consent procured from all participants.
2.2 Cell culture and treatment
The human urothelial cell lines (SV-HUC-1, J82, T24, UM-UC-3, 5637, and RT4) were obtained from Wuhan Procell Biotechnology (Wuhan, China). Cells were maintained at 37°C in a humidified 5% CO2 incubator using the following media: SV-HUC-1 in F12K basal medium (Gibco, USA), J82 and UM-UC-3 in Dulbecco’s modified eagle media (DMEM, Gibco, USA), and T24/5637/RT4 in RPMI-1640 (Gibco, USA). All media contained 10% bovine serum (FBS; Procell, Wuhan, China) and 1% penicillin/streptomycin (Sangon, Shanghai, China).
2.3 Cell transfection, plasmids and oligonucleotides
Overexpression constructs for ELDR (pcDNA3.1-ELDR) and TRIM44 (pcDNA3.1-TRIM44), along with ELDR-targeting shRNA (sh-ELDR) and miR-1343-3p mimics/inhibitors, were procured from Tsingke Biotechnology (Beijing, China). Corresponding negative control vectors and oligonucleotides were included. For gene modulation, cells were transfected using Lipofectamine™ 3000 (Invitrogen, USA) per the manufacturer’s protocol, with plasmid validation via Sanger sequencing.
2.4 RNA isolation, reverse transcription, and RT-qPCR
Total RNA isolation from cells and clinical specimens was performed with TRIzol reagent (Abclonal, China). The purified RNA underwent reverse transcription using the PrimeScript qRT-PCR kit (Abclonal, China). Subsequently, qRT-PCR analysis was conducted using the SYBR(R) Prime-Script RT-PCR kit (Abclonal, China) on an ABI 7500 Real-Time PCR Platform (Applied Biosystems, USA). Actin and U6 were used as internal controls, and gene expression levels were normalized to internal controls and quantified via the 2-ΔCt method. All samples were run in triplicate.
2.5 Cell counting kit-8 and colony formation assay
Cells were plated in 96-well plates at a density of 3 × 10³ cells per well. After incubation for 6 h, 24 h, 48 h, 72 h, and 96 h, the CCK-8 reagent (10 μL, Sangon) was incubated with each well for 1 h at 37°C, 5% CO2. Absorbance was measured at 450 nm using a microplate reader (Thermo Fisher, USA). About the colony formation assay, cells (1.5 × 10³/well) were seeded in 6-well plates and incubated at 37°C, 5% CO2 until colonies were visible. The cells were then fixed with 4% paraformaldehyde and stained with 0.1% crystal violet, and the colonies were counted.
2.6 Transwell migrating and invasion assay
For migration assays, cells were seeded in RPMI-1640 (500 μL) at 1×104density in the upper chamber of Transwell inserts, with DMEM containing 10% FBS (1000 μL) in the lower compartment. Following 12 h incubation, migrated cells were fixed and stained with 0.1% crystal violet (30 min), then imaged using a microscope (Thermo Fisher, USA). Invasion assays followed identical procedures except for Matrigel coating (Corning; 1:8 dilution in serum-free medium) applied to the upper membrane prior to cell seeding.
2.7 Proliferation assay
Cell proliferation was assessed using an EdU Cell Proliferation Kit (C0071S, Beyotime), following the manufacturer’s protocol. Briefly, cells 4y of 4×104 cells per well and pretreated for 24 hours. Subsequently, half of the medium was replaced with EdU-containing buffer (20 μM) for a 3-hour incubation period. Following fixation and permeabilization, the cells were incubated with click reaction solution and DAPI (1:1000 dilution). Three randomly selected fields per well were imaged, and the results were averaged for statistical analysis.
2.8 Western blot assay
Total protein was isolated from cells and tissues using RIPA lysis buffer (Beyotime) supplemented with phenylmethanesulfonyl fluoride (PMSF) at a 1:100 ratio. Following separation by SDS-PAGE, proteins were transferred onto PVDF membranes (EMD Millipore). The membranes were blocked for 1 hour in Tris-buffered saline (TBS) containing 5% skim milk and then incubated overnight at 4 °C with the following primary antibodies: PCNA (Proteintech, 10205-2-AP), GAPDH (Proteintech, 60004-1-Ig), TRIM44 (Abcam, ab236422), and β-actin (Proteintech, 66009-1-Ig). After washing, membranes were exposed to a species-matched secondary antibody for 1 hour at room temperature. Protein signals were finally detected using enhanced chemiluminescence (Cell Signaling Technology, USA).
2.9 Subcellular fractionation
Nuclear and cytoplasmic fractions were prepared from BCa cells cultured on 15 cm plates. Following two washes with ice-cold PBS, cells were gently scraped into a 15 mL Falcon tube. The resulting cell pellet was resuspended in 1 mL of hypotonic buffer (10 mM HEPES pH 8.0, 1.5 μM MgCl2, 10 mM KCl, 1 μM DTT) and incubated on ice for 15 minutes to induce cell swelling. After that, NP-40 was then added to the suspension to a final concentration of 1%, followed by brief vortexing (10 seconds) and centrifugation at 12,000 rpm for 2–3 minutes; the supernatant constituted the cytoplasmic fraction, while the pellet represented the nuclear fraction. Total RNA or protein from each compartment was subsequently extracted using Trizol (Abclonal, China) or RIPA lysis buffer (Beyotime), respectively, according to the manufacturers’ instructions. ELDR expression patterns across cellular fractions were assessed by RT-qPCR, using actin and U6 as internal controls for cytoplasmic and nuclear RNA, respectively. Finally, fraction purity was validated by immunoblotting with anti-GAPDH and anti-PCNA antibodies serving as cytoplasmic and nuclear markers, respectively.
2.10 Tumor xenograft in vivo
Six-week-old male BALB/c nude mice were randomly divided into three groups (n=5) and housed under SPF conditions. For xenograft experiments, T24 cells stably transfected with lentiviral vectors for shRNA against ELDR (1 × 10^6/100 μL) were injected subcutaneously into the back of nude mice. Tumor. Tumor size was assessed for three consecutive weeks, and after 21 days the mice received intraperitoneal pentobarbital (150 mg/kg). Death was confirmed by cervical dislocation, and the tumor tissue was weighed. The animal experiments were approved by the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University.
2.11 Immunohistochemistry
Tumor specimens were fixed in 10% paraformaldehyde, decalcified with formic acid, and paraffin-embedded. Consecutive 4-μm sections were cut and subjected to deparaffinization and antigen retrieval (Dako, CA, USA). After blocking with goat serum, avidin, and biotin solutions, sections were incubated overnight with anti-Ki67 primary antibody (Abcam, 1:200, ab15580), followed by 1-hour incubation with secondary antibody (Abcam, 1:500, ab150077). Stained sections were finally examined under an inverted microscope (Nikon, Tokyo, Japan).
2.12 Dual luciferase reporter assay
The potential binding site between ELDR and miR-1343-3p was predicted using the starBase online database. A fragment of the human ELDR gene containing this site was amplified by PCR. Site-directed mutagenesis was performed on the predicted miR-1343-3p binding site within this fragment using a dedicated kit (Stratagene, USA). The resulting wild-type (wt) and mutant (mut) ELDR sequences were cloned into the PGL3 vector (GenePharma) to generate ELDR-wt-luc and ELDR-mut-luc reporter plasmids. Cells were then co-transfected with either ELDR-wt-luc or ELDR-mut-luc plasmids together with miR-1343-3p mimics or negative control mimics (NC), using Lipofectamine™ 3000 (Invitrogen). Luciferase activity was measured 48 hours post-transfection using the Dual-Luciferase® Reporter Assay System (Promega, WI, USA), following the manufacturer’s instructions. This identical experimental approach was applied to confirm the interaction between miR-1343-3p and TRIM44.
2.13 AGO2-RIP and MS2-RIP
The Magna RIP Kit (Millipore, USA) was used for RNA immunoprecipitation (RIP) experiments in accordance with product guidelines. 5 µg of control IgG or anti-Argonaute 2 (AGO2) antibody (Abcam, USA) was added to pre-cleared BCa cell lysates in RIP lysis solution, which were then rotated and incubated for an entire night at 4 °C. After adding protein A/G magnetic beads to separate RNA-protein complexes, bound RNAs were released by proteinase K digestion. Using quantitative real-time PCR, the quantities of precipitated target RNA were measured.
For MS2-RIP validation of endogenous lncRNA-ELDR/miR-1343-3p binding, wild-type lncRNA-ELDR and its miR-1343-3p binding-site mutant were cloned into pcDNA3.1-MS2 (12×) to create pcDNA3.1-MS2-ELDR-WT and pcDNA3.1-MS2-ELDR-MUT constructs for MS2-RIP validation of endogenous lncRNA-ELDR/miR-1343-3p binding. In addition to pcDNA3.1-MS2/GFP (expressing MS2-GFP fusion protein), BCa cells were co-transfected with pcDNA3.1-MS2 (vector control), pcDNA3.1-MS2-ELDR-WT, or pcDNA3.1-MS2-ELDR-MUT. Using the previously indicated RIP technique, RNA complexes were immunoprecipitated with anti-GFP antibody (Abcam, USA) during a 48-hour incubation period.
2.14 Statistical analysis
Data from at least three independent experiments are expressed as mean ± SD and analyzed using GraphPad Prism 9.5.1. Group comparisons employed Student’s t-test (two groups) or one-way ANOVA (multiple groups). Associations between ELDR expression and clinicopathological features were assessed by the chi-square test. Survival distributions were analyzed with Kaplan-Meier curves and log-rank testing. Correlation analyses used Pearson’s coefficients. All tests were two-tailed, with *p* < 0.05 considered statistically significant.
3 Results
3.1 ELDR is significantly upregulated in BCa tissues and cell lines, correlating with poor prognosis
To explore the correlation of ELDR with BCa development, we analyzed its expression in TCGA-BCa data via the UALCAN web portal. As shown in Figure 1A, ELDR expression was substantially upregulated in BCa tissues compared to normal tissues. Subsequently, using the starBase database, we found that high ELDR levels were markedly associated with poor prognosis in BCa patients (Figure 1B). To validate these findings and further strengthen the clinical significance of ELDR, we obtained tumor tissues and paired adjacent normal tissues from 58 bladder cancer patients. The result of the qRT-PCR assay suggested that the ELDR expression was obviously elevated in BCa tissues (Figure 1C, Supplementary Table S1). Moreover, we found that higher ELDR levels predicted a worse prognosis and enhanced malignant progression (Figures 1D, E, Supplementary Table S1). Multivariate analysis of clinicopathological characteristics identified that high levels of ELDR were independently associated with tumor size, tumor invasion depth, and TNM stage within our cohort (Supplementary Table S2). Consistently, qRT-PCR analysis confirmed that ELDR levels were markedly higher in BCa cell lines (J82, T24, UM-UC-3, 5637, and RT4) than in the normal uroepithelial sv-HUC-1 line (Figure 1F). In summary, these results suggested that ELDR was pronouncedly elevated in BCa tissues and cell lines, correlating with clinicopathological features.
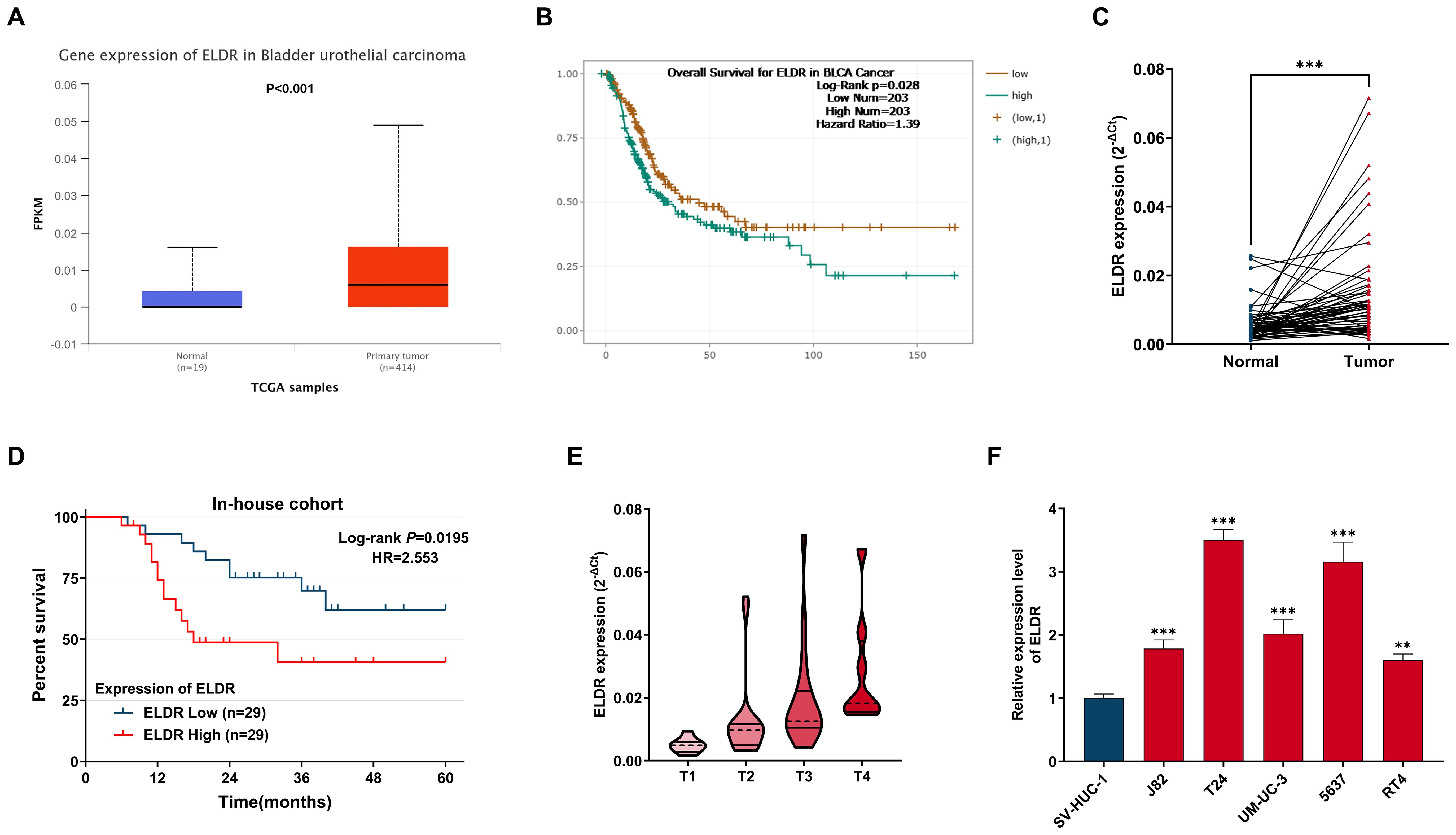
Figure 1. LncRNA ELDR expression is over-presented in BCa tumors tissues and cells, and predicts poor prognosis in BCa patients. (A) ELDR expression in tumor and normal tissues in UALCAN database. (B) The overall Survival for ELDR in BCa from starBase database. (C) The expression of ELDR in an in-house cohort of 58 paired BCa tumor and adjacent normal tissues is shown. (D) The correlation between the expression of ELDR and the prognosis of patients for the BCa in in-house cohort. (E) ELDR expression in BCa tissue of TNM stage 1-2 (n=33) and BCa tissues of TNM stage 3-4 (n=25) were determined using qRT-PCR assay. (F) ELDR expression in SV-HUC-1, J82, T24, UM-UC-3, 5637, RT4 cells were detected by qRT-PCR assay. Data were shown as mean ± SD of three. **P <0.01, ***P < 0.001.
3.2 ELDR promotes BCa cells malignant behaviors in vitro
To investigate the potential biological functions of ELDR in BCa progression, we performed gain- and loss-of-function experiments. Transfection with pcDNA3.1-ELDR significantly increased ELDR levels in both T24 and 5637 cells, while sh-ELDR transfection effectively reduced its expression (Figure 2A). Subsequently, we performed CCK-8, colony formation, transwells and EdU assays in vitro. As shown in Figures 2B-E, ELDR knockdown substantially inhibited proliferation, colony formation, migration, and invasion in both cell lines, whereas its overexpression potentiated these malignant phenotypes. Taken together, ELDR promotes the malignant behaviors of BCa cells.
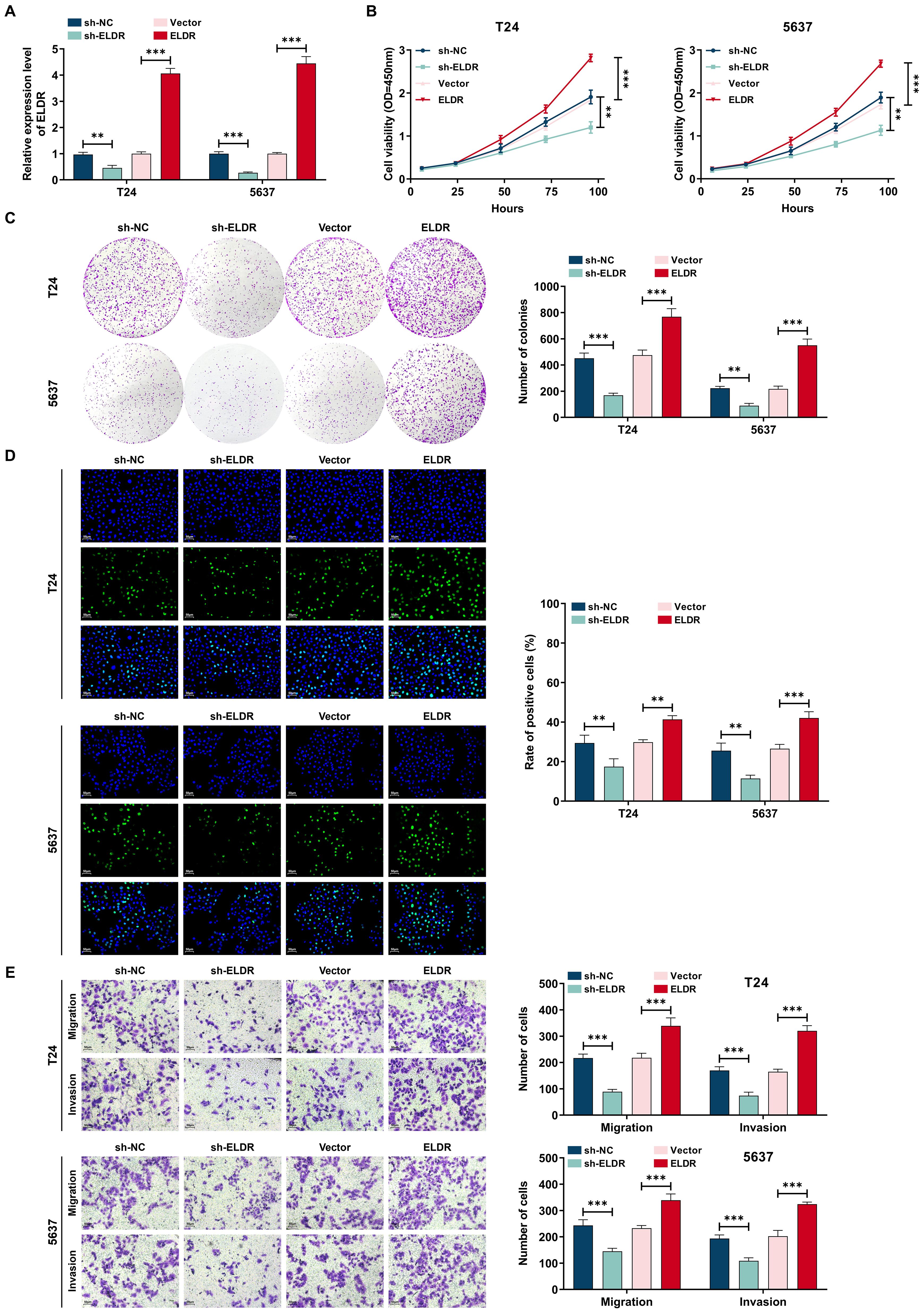
Figure 2. ELDR promotes cell proliferation, colony formation, migration, and invasion in BCa cells. T24 and 5637 were transfected with sh-NC, sh-ELDR, pcDNA3.1 or pc-ELDR. (A) ELDR expression was assessed using qRT-PCR assay. (B-D) CCK-8, colony formation, and EDU assays were employed to determine cell proliferation. (E) Cell migration and invasion were detected using transwell assay. Data were shown as mean ± SD. **P <0.01, ***P < 0.001.
3.3 ELDR knockdown suppresses tumor growth in vivo
Next, to further assess the effects of ELDR on BCa tumorigenesis in vivo, we established subcutaneous xenograft models by injecting nude mice with T24 cells stably expressing control shRNA or two independent shRNAs targeting ELDR (FigureS 3A, B). Tumors derived from sh-ELDR-infected T24 cells exhibited significantly reduced growth rates and smaller volumes compared to those from control shRNA-infected cells (Figures 3C-E). Additionally, immunohistochemical (IHC) analysis revealed a striking reduction in the expression of the proliferation marker Ki67 in tumors from the sh-ELDR group compared to the control group (Figure 3F). Collectively, these results demonstrate that ELDR inhibition could suppress BCa tumor growth in vivo.
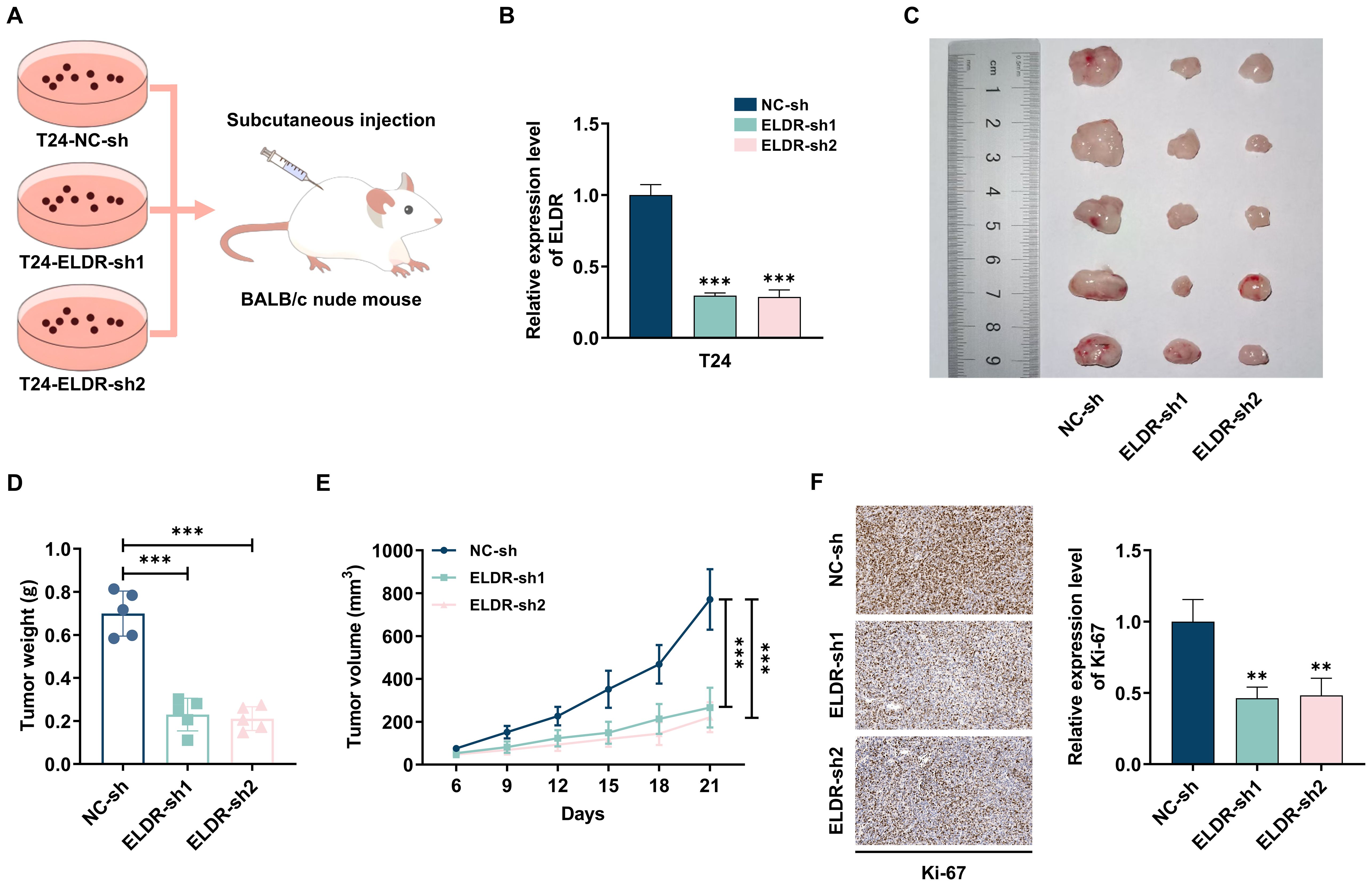
Figure 3. ELDR knockdown inhibited tumor formation in vivo. (A, B) T24 cells transfected with sh-ELDR and sh-NC were injected subcutaneously into the back of nude mice. (C-E) The tumors were collected, and the size and weight of tumors were measured. (F) The level of Ki67 in tumor tissues was evaluated using IHC. Data were shown as mean ± SD. **P <0.01, ***P < 0.001.
3.4 ELDR localizes to the cytoplasm and functions as a miR-1343-3p sponge
We explored the molecular mechanisms of ELDR-mediated BCa tumorigenesis. Considering that the mechanisms and functions are dictated by subcellular localization, we first defined the compartmental distribution of ELDR (25). Bioinformatics prediction using the iLoc-lncRNA database indicated its predominant cytoplasmic accumulation (Figure 4A). This distribution was confirmed experimentally through subcellular fractionation and qRT-PCR in both T24 and 5637 cells, with fraction purity verified by immunoblotting for GAPDH (cytoplasmic marker) and PCNA (nuclear marker) (Figures 4B, C). Based on the established role of cytoplasmic noncoding RNAs in miRNA sequestration and target derepression, we hypothesized that ELDR acts as a molecular sponge for specific miRNAs to regulate downstream oncogenic genes (26).
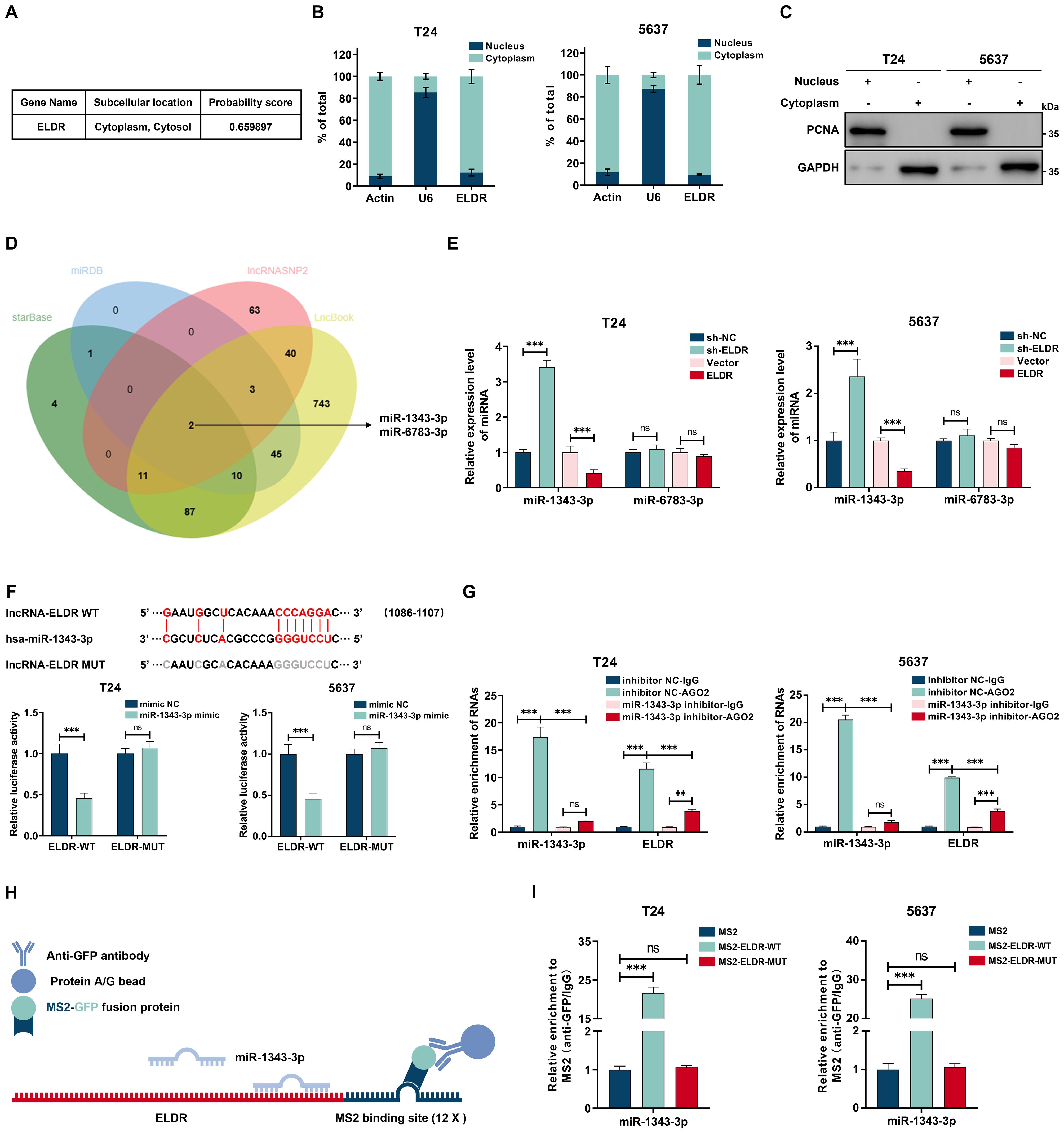
Figure 4. Cytoplasmic ELDR acted as a sponge of miR-1343-3p. (A) Subcellular localization of ELDR in cells predicted by iLoc-lncRNA databese. (B) The subcellular localization of ELDR in T24 (left panel) and 5637 (right panel) cells was determined by nuclear and cytoplasmic fractionation experiment followed by RT-qPCR analysis. (C) The nuclear and cytoplasmic fractionations as described in B were subjected to immunoblotting analysis. (D) Online predicting the ELDR target miRNAs. Venn gram showed the intersection miRNAs. (E) qRT-PCR assays were performed to screen target miRNAs of ELDR by silencing or overexpressing ELDR in BCa cell lines. (F) Online predicting of the interaction sites between ELDR and miR-1343-3p, T24 and 5637 cells were transfected with reporters containing wild-type or mutated ELDR in the presence or absence of negative control miRNA mimic or miR-1343-3p mimic followed by luciferase activity measurement. (G) RIP assays were used to pulldown the endogenous RNA associated with AGO2 in T24 and 5637 cells transfected with miR-1343-3p inhibitor or inhibitor NC, and the relative levels of ELDR and miR-1343-3p were measured and normalized according to the result in IgG group. (H) Schematic delineating the strategy of MS2-RIP. (I) MS2-RIP assays in T24 and 5637 cells followed by qRT-PCR examining the endogenous binding between ELDR and miR-1343-3p. Data were shown as mean ± SD, **P <0.01, ***P < 0.001.
To identify the underlying mechanism, potential miRNAs binding to ELDR were predicted using the starBase, miRDB, lncRNASNP2, and LncBook databases. By intersecting the four databases, we obtained two predicted miRNAs, miR-1343-3p and miR-6783-3p (Figure 4D). We performed qRT-PCR assays to confirm that miR-1343-3p is the direct downstream target of ELDR, as ELDR overexpression decreased miR-1343-3p levels while its knockdown upregulated them in both cell lines (Figure 4E). To ascertain the interaction between ELDR and miR-1343-3p, we predicted their binding sites using starBase and constructed the wild-type (WT) and mutant (MUT) ELDR luciferase reporter genes respectively. We performed the dual-luciferase assays, and the results of dual-luciferase indicated that the luciferase activity of ELDR-WT, but not MUT, was markedly attenuated by miR-1343-3p (Figure 4F, Supplementary Figure S1A). Furthermore, ELDR expression levels were significantly higher than those of miR-1343-3p in both cell lines, supporting its efficacy as a competing endogenous RNA in binding miR-1343-3p (Supplementary Figure S1B). It is well established that lncRNA-miRNA interactions depend on the AGO2 complex; therefore, we performed AGO2-RIP assays in both cell lines, which revealed that ELDR directly bound to the AGO2-containing miR-1343-3p ribonucleoprotein complex, but this interaction was obviously reduced upon inhibition of miR-1343-3p (Figure 4G, Supplementary Figure S1C). Subsequently, MS2-RIP assays demonstrated that endogenous miR-1343-3p was enriched by the MS2-ELDR-WT complex, further validating endogenous binding between ELDR and miR-1343-3p (Figures 4H, I). Besides, in BCa tissues from our in-house cohort, the miR-1343-3p expression level was significantly downregulated compared to paired adjacent normal tissues, and correlation analyses revealed that the ELDR expression level was negatively correlated with miR-1343-3p (Supplementary Figures S1D, E, Supplementary Table S1). Altogether, ELDR localized in the cytoplasm sponges miR-1343-3p to suppress its expression in BCa.
3.5 MiR-1343-3p inhibitor rescued malignant phenotypes suppressed by ELDR knockdown in BCa cells
To explore the functional significance of the ELDR/miR-1343-3p axis in BCa progression, we allocated T24 and 5637 cells into three experimental groups: shRNA negative control (sh-NC), ELDR-knockdown (sh-ELDR), and combined ELDR-knockdown with miR-1343-3p inhibitor (sh-ELDR + miR-1343-3p inhibitor), followed by CCK-8, colony formation, EdU and transwell assays (Figures 5A). As displayed in Figures 5B-E, ELDR knockdown markedly suppressed proliferation, colony formation, migration, and invasion capacities in both cell lines. Conversely, the miR-1343-3p inhibitor attenuated these suppressive effects, rescuing malignant phenotypes. Collectively, ELDR functions as a driver in BCa progression by targeting miR-1343-3p.
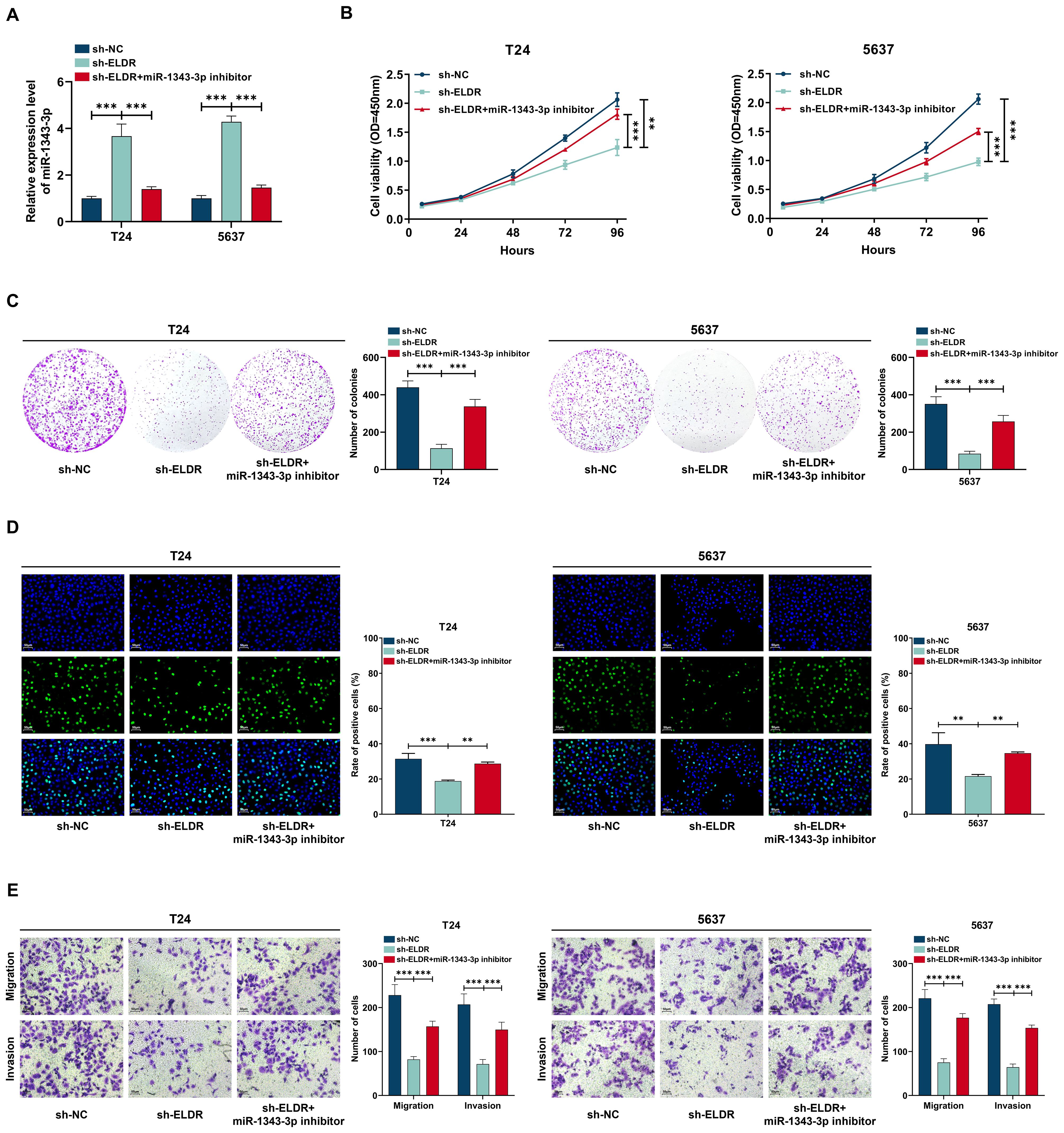
Figure 5. MiR-1343-3p inhibitor reversed the inhibitory effect of ELDR knockdown on the malignant phenotypes of BCa cells. T24 and 5637 cells were classified into: sh-NC group, sh-ELDR group, sh-ELDR+miR-1343-3p inhibitor group. (A) MiR-1343-3p was determined using qRT-PCR assay. (B-D) Cell proliferation was determined using CCK-8 assay, colony formation, and EDU assay. (E) Cell migration and invasion were determined using transwell assay. Data were expressed as mean ± SD, **P <0.01, ***P < 0.001.
3.6 ELDR functions as a ceRNA for miR-1343-3p to upregulate TRIM44 expression
Given that miRNAs exert their function via regulating the expression of target genes, we employed four bioinformatic databases—starBase, miRDB, miRTarBase, and TargetScan—to predict candidate targets of miR-1343-3p. As shown in Figure 6A, sixteen targets overlapped in the prediction results of these four databases. We performed qRT-PCR assays, and the results of showed that only the expression level of TRIM44 mRNA was sharply decreased after treatment with miR-1343-3p mimic in T24 and 5637 cells (Supplementary Figure S1F). Conversely, the miR-1343-3p inhibitor promoted the TRIM44 mRNA expression level in both cell lines (Figure 6B). A similar pattern of expression was observed for the TRIM44 protein by western blot assay (Figure 6C). Subsequently, we confirmed that miR-1343-3p directly bound to the 3’-UTR of TRIM44 via a dual-luciferase reporter assay. Transfection with the miR-1343-3p mimic significantly suppressed luciferase activity driven by the wild-type (WT) TRIM44 3’-UTR reporters relative to the miR-NC control in both cell lines, whereas activity of the mutant (MUT) reporter was unaffected (Figure 6D). We also detected TRIM44 expression level in our in-house cohort using western blot and qRT-PCR assays. The result showed that TRIM44 was markedly upregulated in the tumor tissues compared with paired adjacent normal tissues (Supplementary Figures S1G, SH). Moreover, TRIM44 mRNA expression level showed a positive relationship with ELDR and an inverse association with miR-1343-3p (Supplementary Figures SI, SJ, Supplementary Table S1). Finally, silencing ELDR expression substantially diminished TRIM44 protein levels in both cell lines, whereas this reduction was reversed by the miR-1343-3p inhibitor (Figure 6E). In summary, ELDR promotes TRIM44 expression via sponging miR-1343-3p.
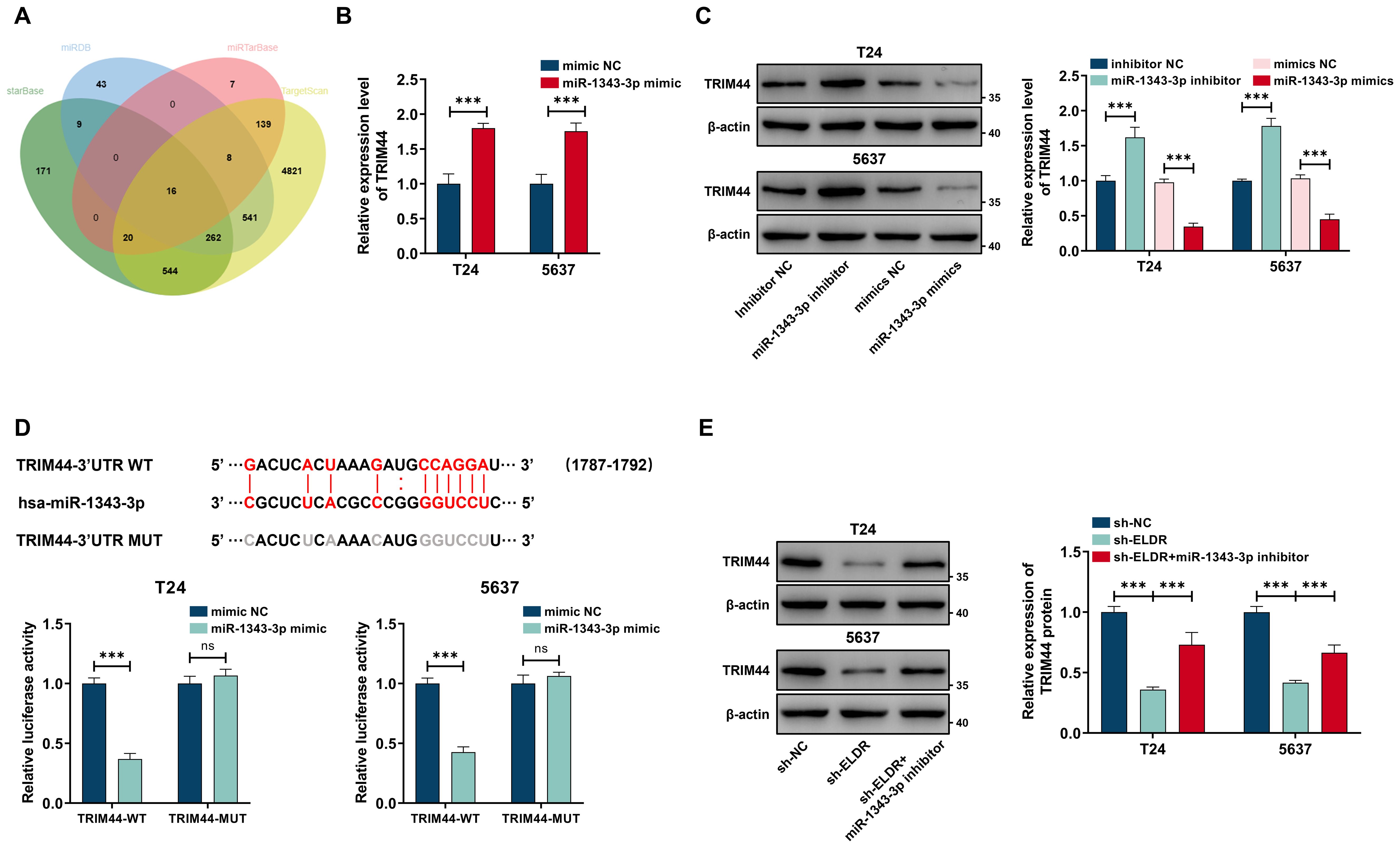
Figure 6. ELDR positively regulated TRIM44 expression through sponging miR-1343-3p. (A) Online predicting the miR-1343-3p target genes. Venn gram showed the intersection genes. (B) TRIM44 mRNA expression in T24 and 5637 cells after miR-1343-3p overexpression was determined using qRT-PCR. (C) TRIM44 proteins levels after transfection of miR-1343-3p mimic and inhibitor in the above two cell lines. (D) Online predicting of the interaction sites between miR-1343-3p and TRIM44, and luciferase reporter assays demonstrated TRIM44 was a direct target of miR-1343-3p. (E) TRIM44 expression in 5637 and T24 cells after sh-ELDR transfection or sh-ELDR and miR-1343-3p inhibitor co-transfection was detected by western blot. Data were expressed as mean ± SD, **P <0.01, ***P < 0.001.
3.7 TRIM44 overexpression rescued the inhibition of ELDR knockdown on the malignant phenotypes of BCa cells
To determine whether TRIM44 mediates ELDR oncogenic function in BCa, we performed rescue experiments via restoring TRIM44 expression. ELDR knockdown significantly reduced TRIM44 protein levels in T24 and 5637 cells, an effect rescued by pcDNA3.1-TRIM44 transfection (Figure 7A). Subsequent functional assays revealed that silencing ELDR inhibited BCa cell proliferation, colony formation, migration, and invasion in both cell lines (Figures 7B-E). Notably, TRIM44 restoration abolished ELDR-silencing-induced suppression of these malignant phenotypes (Figures 7B-E). Collectively, TRIM44 overexpression rescues the inhibitory effects of ELDR depletion on BCa oncogenicity.
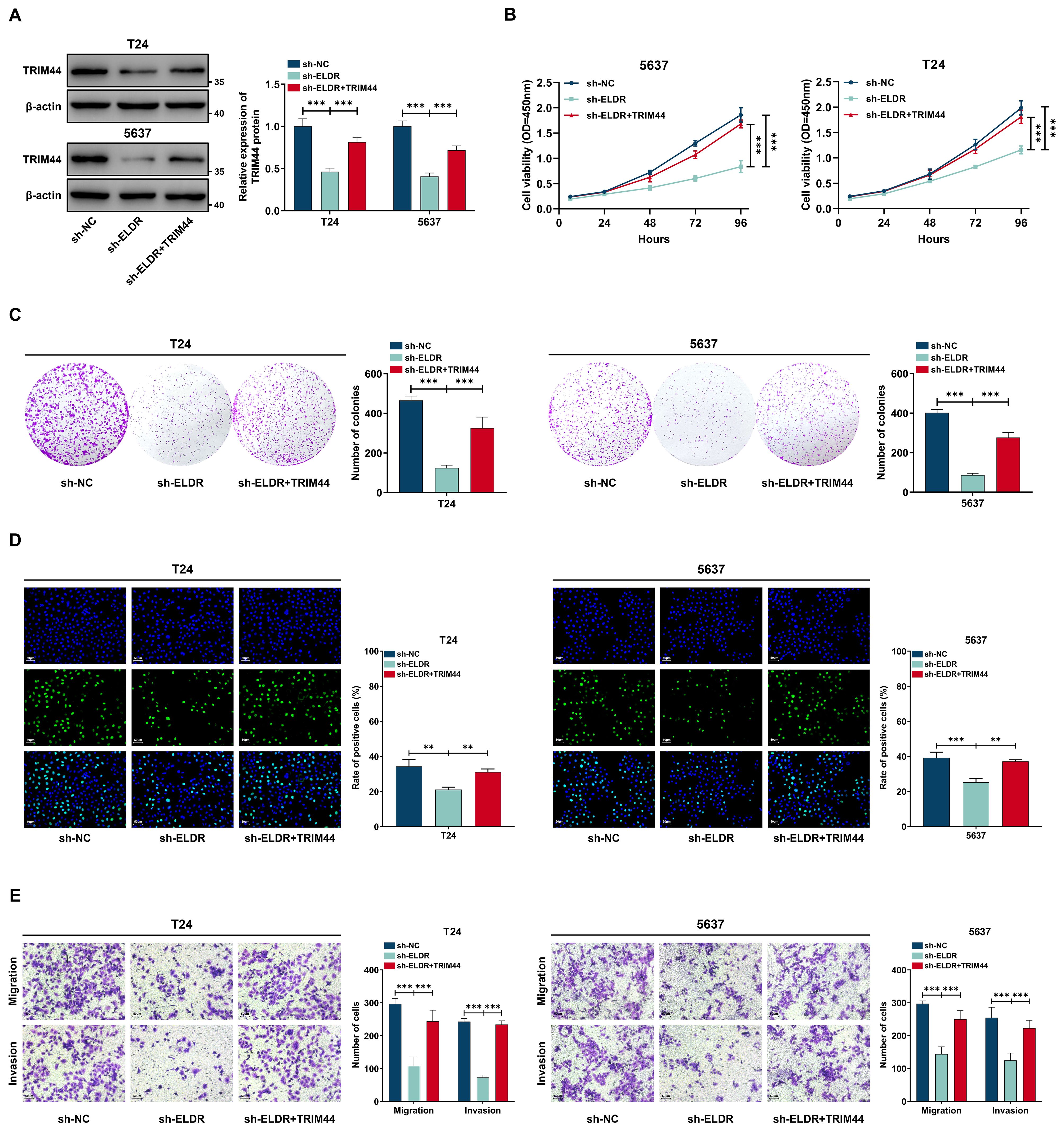
Figure 7. TRIM44 overexpression reversed the inhibitory effect of ELDR knockdown on the malignant phenotypes of BCa cells. T24 and 5637 cells were classified into: sh-NC group, sh-ELDR group, and sh-ELDR+TRIM44 group. (A) Western blot was employed to asses TRIM44 level in T24 and 5637 cells. (B-D) CCK-8 assay, colony information assay, and EDU assay were employed to determine cell proliferation. (E) Cell migration and invasion were determined using transwell assay. Data were expressed as mean ± SD, **P <0.01, ***P < 0.001.
4 Discussion
As the primary genitourinary malignancy, the prognosis of BCa remains poor due to a lack of effective diagnostic biomarkers and therapeutic drugs (27). It has been reported that abnormal proliferation is the main reason for the occurrence and malignant progression of BCa (28). Therefore, there is an urgent need to better understand the underlying molecular mechanisms of BCa progression and improve the survival of BCa patients. LncRNAs have received increasing attention due to their important roles in the development and progression of human cancers, including BCa (29–32). For example, TUG1 (33) and SNHG16 (34) showed overexpression in BCa tumor tissues and served as bad predictors of overall survival. However, the exact role of lncRNAs in BCa remains poorly understood. A comprehensive understanding of the mechanisms of lncRNAs will help reveal promising biomarkers and therapeutic targets for BCa patients. While the ceRNA paradigm and the oncogenic role of TRIM44 have been documented in various cancer types, the upstream regulators that dictate TRIM44 expression in BCa remain poorly characterized.
In this study, we focused our attention on a lncRNA ELDR that has not been reported in BCa and whose function is still unclear. Our work provides the first evidence that ELDR functions as a key oncogenic lncRNA in BCa by operating as a competitive endogenous RNA (ceRNA). We found that the expression level of ELDR was obviously upregulated in BCa tissues compared to the paired adjacent normal tissues, correlating with tumor size, invasion depth, TNM stage and poor prognosis in BCa patients. Functional experiments displayed that silencing ELDR inhibited the BCa cell proliferation, colony formation, migration, and invasion, whereas overexpression of ELDR showed the opposite effect. The tumor-promoting effect of ELDR in vivo was further validated. While other well-characterized oncogenic lncRNAs in BCa, such as UCA1 and MALAT1, have been associated with specific processes like chemoresistance and metastasis, respectively, our date defines a distinct role for ELDR. We demonstrate that ELDR directly sequesters miR-1343-3p, which in turn leads to the derepression of its target oncogene, TRIM44. These results motivate us to further explore the biological mechanism of ELDR in BCa.
TRIM44, a member of the tripartite motif (TRIM) family E3 ubiquitin ligases, is recognized for its involvement in protein stability regulation and signal transduction (35, 36). Accumulating evidence indicates that the crucial roles of TRIM44 in the development of various types of malignant tumors. TRIM44 overexpression was confirmed to be associated with the malignant phenotype in gastric cancer (37), lung adenocarcinoma (38), and ovarian cancer (22). The precise molecular effects triggered by the upregulation of TRIM44 in bladder cancer require further investigation. our study provides the first evidence of TRIM44 dysregulation in BCa. Moreover, TRIM44 expression was positively correlated with that of ELDR. Importantly, the inhibitory effect of ELDR silencing on the BCa malignant phenotype were significantly attenuated in the presence of TRIM44 overexpression. Based on these results, we concluded that ELDR promotes the progression of BCa, at least to some extent, by stimulating the expression of TRIM44. There is already a lot of proof that TRIM44 makes the PI3K protein more stable and speeds up the PI3K/AKT signaling pathways in different malignancies. This suggests that TRIM44 overexpression caused by ELDR probably acts in bladder cancer by making this essential cancer-causing pathway work too hard. This potential association provides a plausible explanation for the enhanced growth capacity observed in ELDR-overexpressing cells. Concurrently, the potent pro-migration and invasion phenotypes induced by ELDR strongly suggest its involvement in regulating epithelial-mesenchymal transition (EMT)—a core mechanism of cancer metastasis. Future research focusing on this pathway’s effects on key EMT transcription factors and biomarkers (such as E-cadherin, N-cadherin, and vimentin) will be crucial for comprehensively elucidating its role in bladder cancer dissemination.
Our findings indicate promising potential for translational applications. From a diagnostic perspective, the substantial increase of ELDR in BCa tissues and its association with aggressive disease highlight its potential as a predictive biomarker. Identifying ELDR levels in liquid biopsies, such as serum or urine extracellular vesicles, may aid in the creation of non-invasive diagnostics for early detection and risk assessment. From a therapeutic standpoint, the unique and heightened expression of ELDR in malignancies renders it a compelling target for targeted therapies. Targeting lncRNAs poses technical challenges; however, innovative methodologies, including the application of antisense oligonucleotides (ASOs) or small interfering RNAs (siRNAs) specifically engineered to inhibit ELDR, may provide a novel and precise therapeutic approach to disrupt this carcinogenic pathway.
It must be acknowledged that this study has limitations. First, our clinical relevance analysis is based solely on a single-center cohort with limited sample size. To establish the post-rain value of ELDR, validation through independent, multicenter, large-scale prospective cohorts is essential. Second, although in vitro and in vivo data strongly support the functional role of the ELDR/miR-1343-3p/TRIM44 axis, the direct clinical utility of ELDR and its feasibility for patient treatment remain to be verified. Finally, confirmatory validation of the proposed downstream pathways—particularly the direct activation mechanism of the PI3K/AKT pathway and the role of TRIM44 in inducing epithelial-mesenchymal transition (EMT) in bladder cancer—has become the core focus of our current research.
Collectively, these findings delineate the ELDR/miR-1343-3p/TRIM44 axis as a functionally independent pathway that expands the known regulatory network of lncRNAs in bladder cancer pathogenesis (Figure 8).
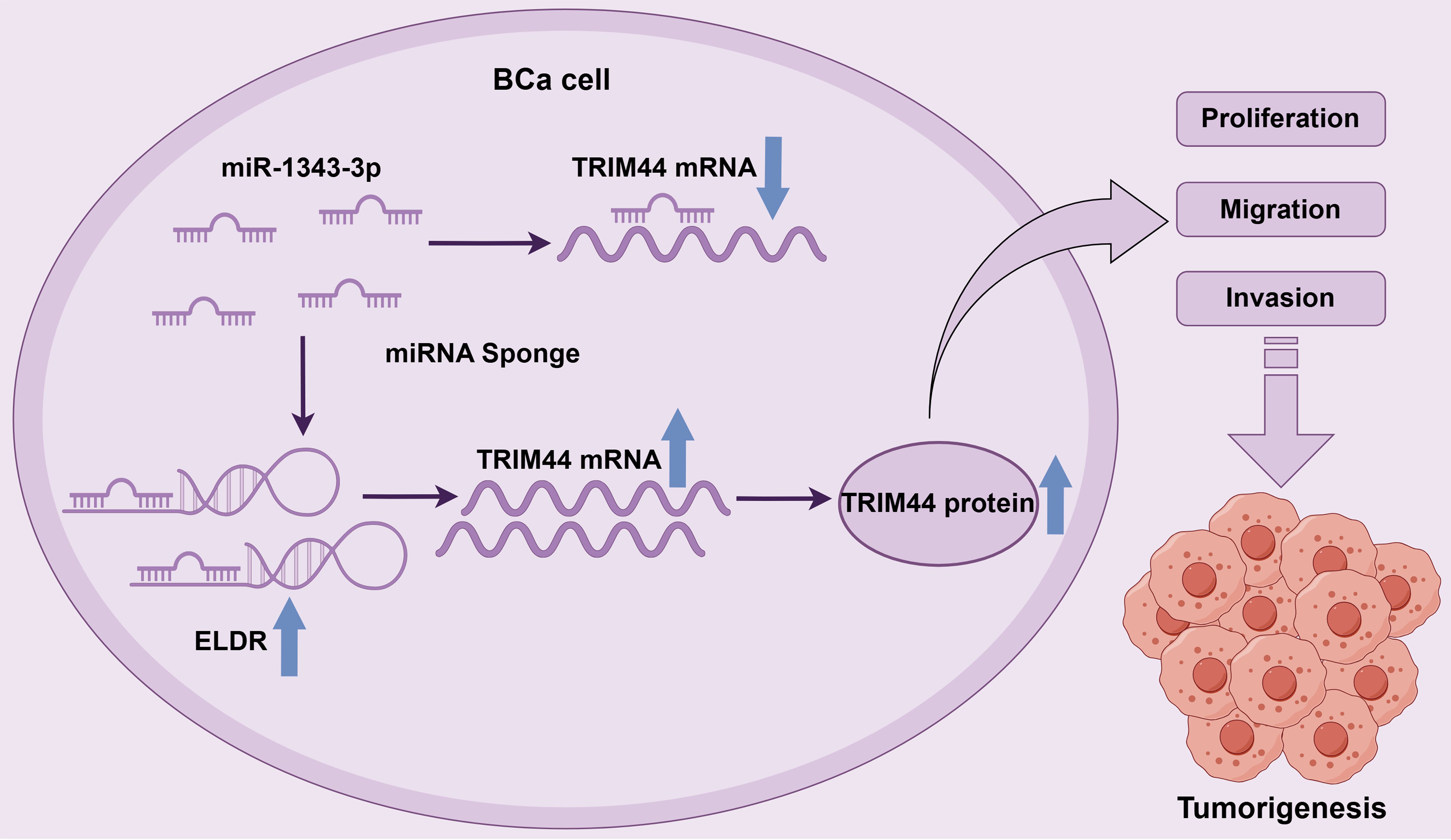
Figure 8. The schematic diagram of LncRNA ELDR promotes bladder cancer malignant progression by regulating the miR-1343-3p/TRIM44 axis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by the Medical Ethics Committee of the First Affiliated Hospital of Chongqing Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal study was approved by Chongqing Medical University Experimental Animal Center. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
XH: Data curation, Formal Analysis, Methodology, Investigation, Writing – review & editing. TX: Writing – review & editing, Formal Analysis, Data curation, Validation, Funding acquisition. XX: Methodology, Writing – review & editing, Funding acquisition, Supervision, Project administration. YM: Methodology, Visualization, Data curation, Formal Analysis, Investigation, Conceptualization, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Fuling District Science and Technology Research Program (FLKJ.2024AAN3066), National Natural Science Foundation of China (82372439); National Key Research and Development Program of China (2023YFC3604404, 2023YFC3604405).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1685792/full#supplementary-material
References
1. Leeming RC, Karagas MR, Zens MS, Schned AR, Seigne JD, and Passarelli MN. Bladder cancer risk variants and overall and disease-specific survival in two independent cohorts. BJU Int. (2022) 129:309–11. doi: 10.1111/bju.15640
2. Prout GR Jr., Wesley MN, Greenberg RS, Chen VW, Brown CC, Miller AW, et al. Bladder cancer: race differences in extent of disease at diagnosis. Cancer. (2000) 89:1349–58. doi: 10.1002/1097-0142(20000915)89:6<1349::AID-CNCR20>3.0.CO;2-D
3. Ploeg M, Aben KK, and Kiemeney LA. The present and future burden of urinary bladder cancer in the world. World J Urol. (2009) 27:289–93. doi: 10.1007/s00345-009-0383-3
4. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. (2015) 47:199–208. doi: 10.1038/ng.3192
5. Schloßhauer JL, Shamloo S, Schamrin K, and Imig J. Dual CRISPR-interference strategy for targeting synthetic lethal interactions between non-coding RNAs in cancer cells. J Vis Exp. (2025). doi: 10.3791/67752
6. Shi M, Zhang R, Lyu H, Xiao S, Guo D, Zhang Q, et al. Long non-coding RNAs: Emerging regulators of invasion and metastasis in pancreatic cancer. J Adv Res. (2025). doi: 10.1016/j.jare.2025.02.001
7. Liu P, Fan B, Othmane B, Hu J, Li H, Cui Y, et al. m(6)A-induced lncDBET promotes the Malignant progression of bladder cancer through FABP5-mediated lipid metabolism. Theranostics. (2022) 12:6291–307. doi: 10.7150/thno.71456
8. Dai C, Li Q, Wang L, Zhang J, Yang S, and Zhang X. LncRNA TRPM2-AS promotes cell proliferation, migration, and invasion by regulating the miR-195-5p/COP1 axis in bladder cancer. Naunyn Schmiedebergs Arch Pharmacol. (2025). doi: 10.1007/s00210-025-04377-4
9. Xue ST, Cao SQ, Ding JC, Li WJ, Hu GS, Zheng JC, et al. LncRNA LUESCC promotes esophageal squamous cell carcinoma by targeting the miR-6785-5p/NRSN2 axis. Cell Mol Life Sci. (2024) 81:121. doi: 10.1007/s00018-024-05172-9
10. Sheng J, Luo Y, Lv E, Liang H, Tao H, Yu C, et al. LINC01980 induced by TGF-beta promotes hepatocellular carcinoma metastasis via miR-376b-5p/E2F5 axis. Cell Signal. (2023) 112:110923. doi: 10.1016/j.cellsig.2023.110923
11. Sur S, Nakanishi H, Steele R, Zhang D, Varvares MA, and Ray RB. Long non-coding RNA ELDR enhances oral cancer growth by promoting ILF3-cyclin E1 signaling. EMBO Rep. (2020) 21:e51042. doi: 10.15252/embr.202051042
12. Zhao X, Guo X, Jiao D, Zhu J, Xiao H, Yang Y, et al. Analysis of the expression profile of serum exosomal lncRNA in breast cancer patients. Ann Transl Med. (2021) 9:1382. doi: 10.21037/atm-21-3483
13. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
14. Nikkhah O, Einollahi B, Asadi M, Heiat M, and Hushmandi K. Gene network changes after exposure to nanoliposomes containing antisense miR-21 and miR-373 in bladder cancer Cells: An in vitro model study. Biochem Biophys Rep. (2025) 42:102041. doi: 10.1016/j.bbrep.2025.102041
15. Yin C, Liufu C, Ye S, Zhu T, Jiang J, Wang M, et al. Tumor-derived exosomal KPNA2 activates fibroblasts and interacts with KIFC1 to promote bladder cancer progression, a process inhibited by miR-26b-5p. Cell Mol Biol Lett. (2025) 30:20. doi: 10.1186/s11658-025-00687-w
16. Zhang C, Yu H, Bai X, Zhou X, Feng Z, Li Y, et al. MiR-15b-3p weakens bicalutamide sensitivity in prostate cancer via targeting KLF2 to suppress ferroptosis. J Cancer. (2024) 15:2306–17. doi: 10.7150/jca.92379
17. Xu L, Yu S, and Li D. miR-1343-3p, mediated by LncRNA ASMTL-AS1, induces ferroptosis in osteosarcoma progression by targeting TYRO3. Gene. (2025) 963:149547. doi: 10.1016/j.gene.2025.149547
18. Zhang Y, Wang X, Liu W, Lei T, Qiao T, Feng W, et al. CircGLIS3 promotes gastric cancer progression by regulating the miR-1343-3p/PGK1 pathway and inhibiting vimentin phosphorylation. J Transl Med. (2024) 22:251. doi: 10.1186/s12967-023-04625-2
19. Wang X, Zhang Z, and Cao X. Salidroside inhibited the proliferation of gastric cancer cells through up-regulating tumor suppressor miR-1343-3p and down-regulating MAP3K6/MMP24 signal molecules. Cancer Biol Ther. (2024) 25:2322206. doi: 10.1080/15384047.2024.2322206
20. Lai C, Hu Z, Hu J, Li Z, Li L, Liu M, et al. Screening model for bladder cancer early detection with serum miRNAs based on machine learning: A mixed-cohort study based on 16,189 participants. Cancer Med. (2024) 13:e70338. doi: 10.1002/cam4.70338
21. Yu XZ, Yuan JL, Ye H, Yi K, Qie MR, and Hou MM. TRIM44 facilitates ovarian cancer proliferation, migration, and invasion by inhibiting FRK. Neoplasma. (2021) 68:751–9. doi: 10.4149/neo_2021_201128N1285
22. Yu Y, Li S, Sun J, Wang Y, Xie L, Guo Y, et al. Overexpression of TRIM44 mediates the NF-κB pathway to promote the progression of ovarian cancer. Genes Genomics. (2024) 46:689–99. doi: 10.1007/s13258-024-01517-7
23. Zhou S, Wu C, Tang Q, Zhou X, He B, Chang P, et al. Silencing LINC00491 inhibits prostate cancer development through the miR-384/TRIM44 axis. J Biochem Mol Toxicol. (2023) 37:e23370. doi: 10.1002/jbt.23370
24. Yamada Y, Kimura N, Maki K, Hakozaki Y, Urabe F, Kimura S, et al. The roles of tripartite motif proteins in urological cancers: A systematic review. Cancers (Basel). (2025) 17:2367. doi: 10.3390/cancers17142367
25. Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. (2016) 41:761–72. doi: 10.1016/j.tibs.2016.07.003
26. Svoronos AA, Engelman DM, and Slack FJ. OncomiR or tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res. (2016) 76:3666–70. doi: 10.1158/0008-5472.CAN-16-0359
27. Kurtova AV, Xiao J, Mo Q, Pazhanisamy S, Krasnow R, Lerner SP, et al. Blocking PGE2-induced tumour repopulation abrogates bladder cancer chemoresistance. Nature. (2015) 517:209–13. doi: 10.1038/nature14034
28. Olsson AY, Feber A, Edwards S, Te Poele R, Giddings I, Merson S, et al. Role of E2F3 expression in modulating cellular proliferation rate in human bladder and prostate cancer cells. Oncogene. (2007) 26:1028–37. doi: 10.1038/sj.onc.1209854
29. Ulitsky I and Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. (2013) 154:26–46. doi: 10.1016/j.cell.2013.06.020
30. Ma CC, Xiong Z, Zhu GN, Wang C, Zong G, Wang HL, et al. Long non-coding RNA ATB promotes glioma Malignancy by negatively regulating miR-200a. J Exp Clin Cancer Res. (2016) 35:90. doi: 10.1186/s13046-016-0367-2
31. Xiong J, Liu Y, Jiang L, Zeng Y, and Tang W. High expression of long non-coding RNA lncRNA-ATB is correlated with metastases and promotes cell migration and invasion in renal cell carcinoma. Jpn J Clin Oncol. (2016) 46:378–84. doi: 10.1093/jjco/hyv214
32. Ding J, Lu B, Wang J, Wang J, Shi Y, Lian Y, et al. Long non-coding RNA Loc554202 induces apoptosis in colorectal cancer cells via the caspase cleavage cascades. J Exp Clin Cancer Res. (2015) 34:100. doi: 10.1186/s13046-015-0217-7
33. Yu G, Zhou H, Yao W, Meng L, and Lang B. lncRNA TUG1 Promotes Cisplatin Resistance by Regulating CCND2 via Epigenetically Silencing miR-194-5p in Bladder Cancer. Mol Ther Nucleic Acids. (2019) 16:257–71. doi: 10.1016/j.omtn.2019.02.017
34. Zhou X, Zhang C, Yu H, Feng Z, Bai X, Mei Y, et al. The MEF2A/SNHG16/miR-425-5p/NOTCH2 axis induces gemcitabine resistance by inhibiting ferroptosis in the starving bladder tumor microenvironment. Cell Signal. (2024) 122:111337. doi: 10.1016/j.cellsig.2024.111337
35. Watanabe M and Hatakeyama S. TRIM proteins and diseases. J Biochem. (2017) 161:135–44. doi: 10.1093/jb/mvw087
36. Allen MD and Bycroft M. The solution structure of the ZnF UBP domain of USP33/VDU1. Protein Sci. (2007) 16:2072–5. doi: 10.1110/ps.072967807
37. Zhang X, Wu X, Sun Y, Chu Y, Liu F, and Chen C. TRIM44 regulates tumor immunity in gastric cancer through LOXL2-dependent extracellular matrix remodeling. Cell Oncol (Dordr). (2023) 46:423–35. doi: 10.1007/s13402-022-00759-5
Keywords: bladder cancer, ELDR, ceRNA, miR-1343-3p, TRIM44
Citation: Hu X, Xie T, Xiao X and Mei Y (2025) LncRNA ELDR promotes bladder cancer malignant progression by regulating the miR-1343-3p/TRIM44 axis. Front. Oncol. 15:1685792. doi: 10.3389/fonc.2025.1685792
Received: 14 August 2025; Accepted: 07 November 2025; Revised: 31 October 2025;
Published: 21 November 2025.
Edited by:
Steven Kregel, Loyola University Chicago, United StatesReviewed by:
Ritika Tiwari, University of Miami, United StatesPaula Dobosz, Poznan University of Medical Sciences, Poland
Copyright © 2025 Hu, Xie, Xiao and Mei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao Xiao, ZHIueGlhb3hpYW9Ac3R1LmNxbXUuZWR1LmNu; Yuhua Mei, bWVpeXVodWFAc3R1LmNxbXUuZWR1LmNu
†These authors have contributed equally to this work
 Xiao Hu
Xiao Hu Tianhang Xie2†
Tianhang Xie2† Xiao Xiao
Xiao Xiao Yuhua Mei
Yuhua Mei