- 1Department of Thoracic Surgery, Huazhong University of Science and Technology Tongji Medical College Union Hospital, Wuhan, Hubei, China
- 2School of Computer Science and Engineering, Sun Yat-Sen University, Guangzhou, Guangdong, China
Lung adenocarcinoma (LUAD) is one of the main causes of cancer-related mortality worldwide. Pathological risk factors such as spreading through air spaces, high-risk pathological subtypes, occult lymph nodes, and visceral pleural invasion have significant impact on patient prognosis. In recent years, there has been significant progress in the application of artificial intelligence (AI) technology, e.g., deep learning (DL), in medical image analysis and pathological diagnosis of lung cancer, offering novel approaches for predicting the aforementioned pathological risk factors. This article reviews recent advancements in AI-based analysis and prediction of pathological risk factors in lung adenocarcinoma, with a focus on the applications and limitations of DL models, focusing on studies aimed at improving diagnostic accuracy and efficiency for specific high-risk pathological subtypes. Finally, we summarize current challenges and future directions, emphasizing the need to expand dataset diversity and scale, improve model interpretability, and enhance the clinical applicability of AI models. This article aims to provide a reference for future research on the analysis and prediction of pathological risk factors of LUAD and to promote the development and application of AI, especially DL, in this field.
1 Introduction
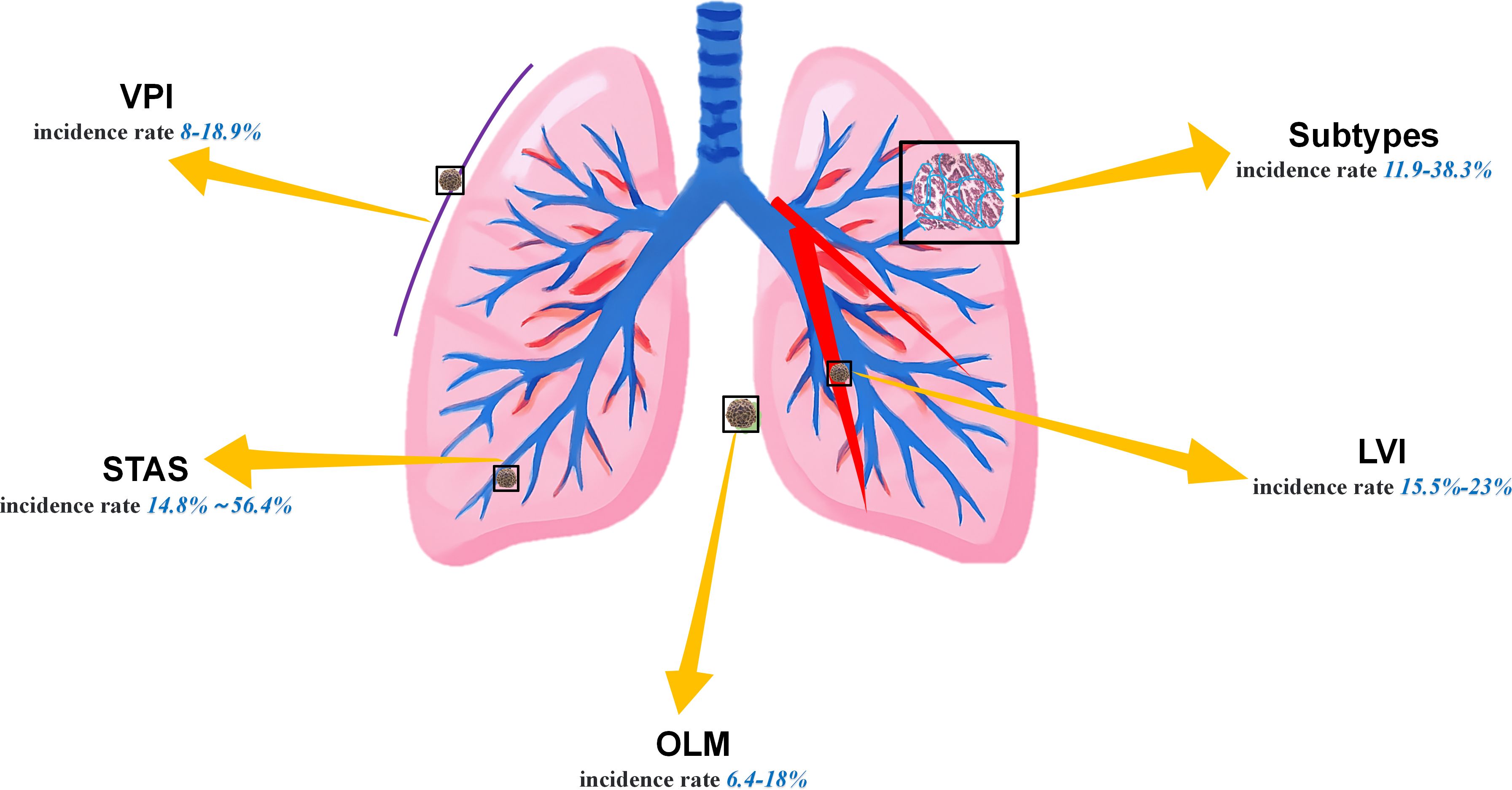
Lung cancer remains the leading cause of cancer-related mortality worldwide. Lung adenocarcinoma (LUAD), accounting for over 50% of newly diagnosed lung cancer cases (1, 2), is particularly prevalent. Although surgical resection, as the standard treatment, has significantly improved the prognosis of patients with early-stage LUAD, specific pathological risk factors still contribute to recurrence in certain patients. The currently widely recognized LUAD pathological risk factors primarily include spreading through air spaces (STAS), visceral pleural invasion (VPI), lymphovascular invasion (LVI), occult lymph node metastasis (OLM), and presence of micropapillary and solid subtypes (Figure 1) (3, 4). Owing to the presence of these high-risk factors, some subtypes of adenocarcinoma exhibit greater aggressiveness. Although the specific mechanisms remain unclear, the factors may be associated with higher tumor mutation burdens or the presence of microscopic tumor residues (5, 6). Simultaneously, the disruption of intercellular adhesion complexes in the lungs exacerbates the invasive spread of tumors (7). More critically, STAS and subtypes such as micropapillary or solid subtypes often coexist. Patients with a larger number of concomitant high-risk factors are more prone to recurrence and exhibit poorer long-term survival rates (4, 8–10). For such patients, lobectomy with radical lymph node dissection is recommended over segmentectomy (11). Unfortunately, it is not possible to diagnose these factors by visually examining computed tomography (CT) images (12), and invasive preoperative procedures such as bronchoscopy or percutaneous transthoracic needle biopsy (PTNB) may cause harm to patients and offer low sensitivity. Therefore, how to effectively enhance the accuracy of screening for LUAD risk factors through noninvasive methods before surgery, reduce the rate of missed diagnoses, and alleviate the workload of clinicians have become an urgent issue requiring resolution nowadays.
Owing to recent advancements in computer technology and statistics, intelligent clinical diagnosis and treatment models for lung cancer, based on tumor imaging, have begun to emerge (13–17). Machine learning (ML) is a primary form of artificial intelligence (AI) technology. Instead of explicit programming, computing systems learn from small- and medium-sized datasets to make predictions or decisions; examples include linear regression and decision trees (18). Deep learning (DL) is a specialized subset of ML, which offers enhanced data volume and model complexity, sacrificing interpretability to achieve extraordinary predictive capabilities from massive datasets (19), e.g., convolutional neural networks (CNNs) (19, 20) and visual transformer (ViTs) (21, 22). The CNNs automatically learn complex features from images through multiple convolutions and pooling operations, thereby enhancing the accuracy and efficiency of lung cancer subtype analysis. Specifically, ResNet is a great landmark architecture of CNN that introduced residual blocks to overcome degradation issues, enabling the training of extremely deep networks (Figure 2) (23). Furthermore, ViTs have emerged as another powerful deep learning model for leveraging self-attention mechanisms to capture long-range dependencies and global contextual information within images. The current assumption is that the primary objective of research is to compare the diagnostic capabilities of AI with those of human experts. However, it is evident that classical ML and DL, as complementary AI paradigms, have already demonstrated superior performance and immense potential in terms of speed, reproducibility, and accuracy, surpassing human standards. Given the rapid changes and advancements in this field, this review attempts to summarize the status and trends in the application of AI, especially DL, in the diagnosis and prediction of LUAD risk factors and to discuss current challenges and future perspectives.
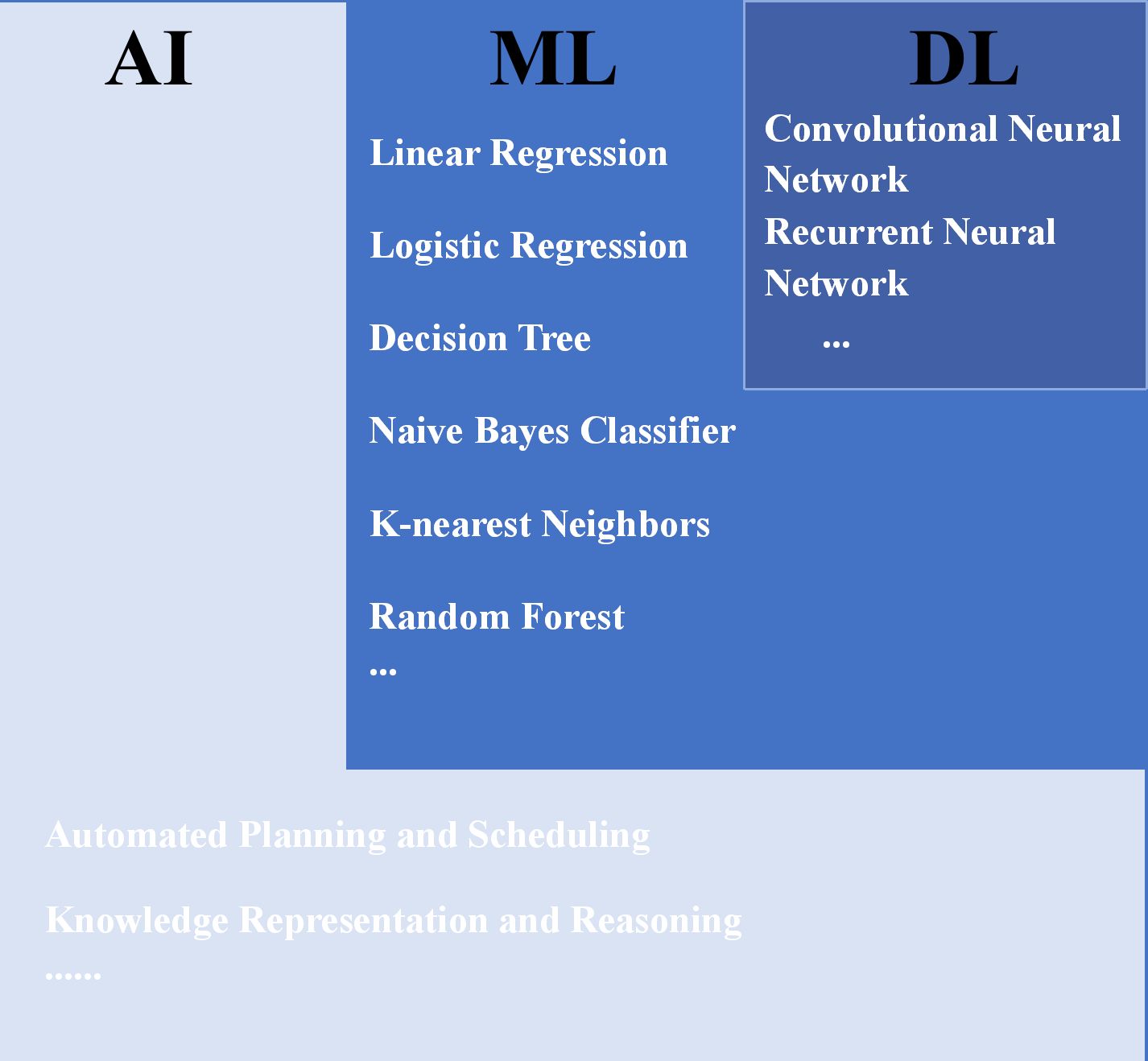
Figure 2. Relationship between AI, ML, DL, and some of their algorithms (14, 24). AI, artificial intelligence; ML, machine learning; DL, deep learning.
2 Assessment of high-risk factors in preoperative diagnosis
The abovementioned risk factors can only be diagnosed by experienced pathologists based on post-operative slides. However, at this stage, surgical treatment has already concluded, and the surgical approach cannot be altered. Patients with high-risk factors can only receive drug-assisted therapy post-operatively. Therefore, our primary concern is how to enable surgeons to achieve the goals of preoperative visualization of lesions, precise intraoperative resection, and minimized postoperative complications through imaging examinations. Preoperative CT or PET/CT images represent a widely available, easily accessible, and non-invasive source of data. ML is an initial attempt in this area, and DL has become ever more popular in the recent few years (15, 20). Figure 3 illustrates the fundamental workflow and key components for lung cancer diagnosis of clinician (A), radiomics (B), and deep learning (C).
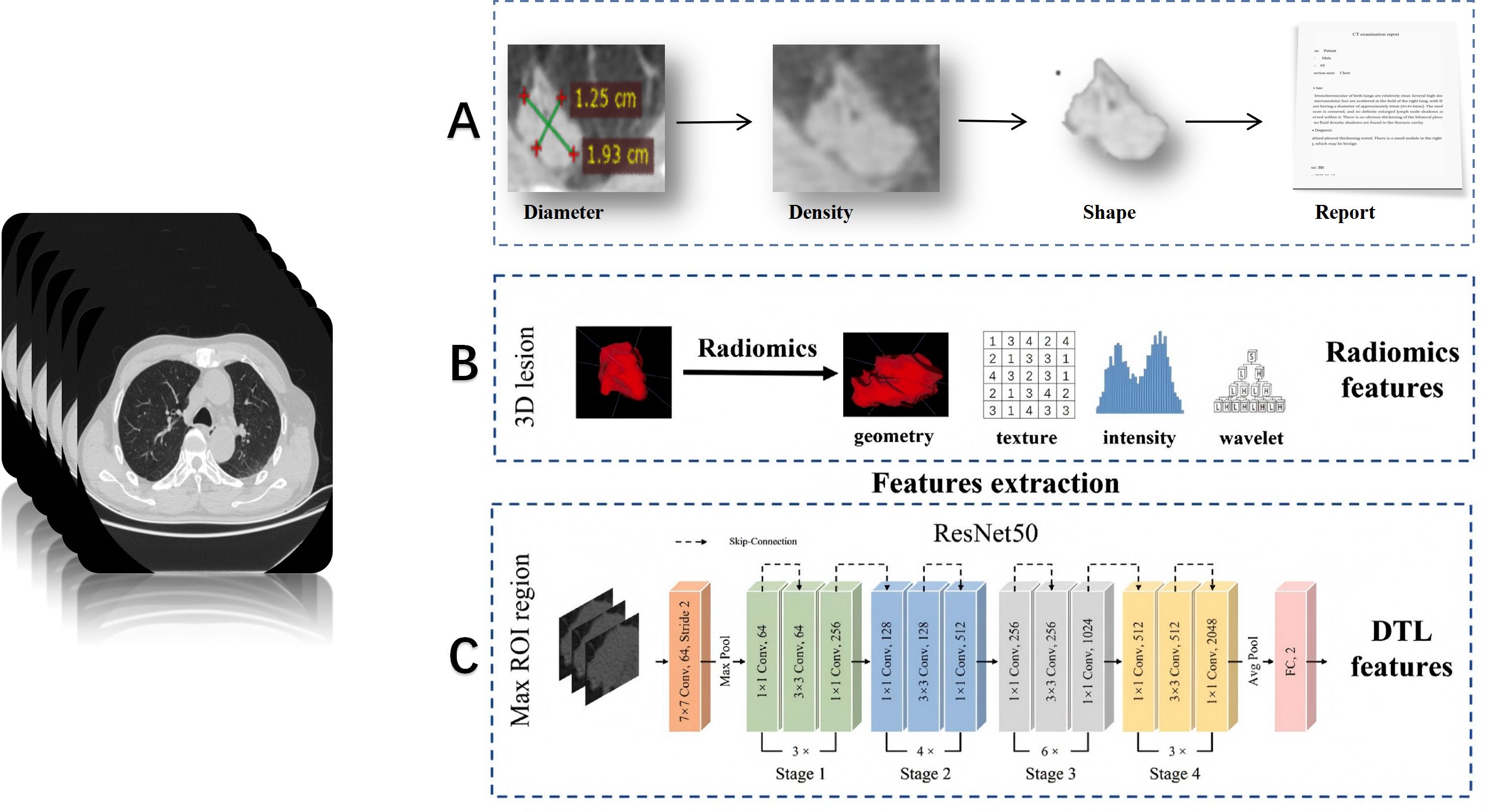
Figure 3. Typical process and differences between clinician (A), radiomics (B), and deep learning (C) for lung cancer diagnostic applications (25, 26).
2.1 Spreading through air spaces
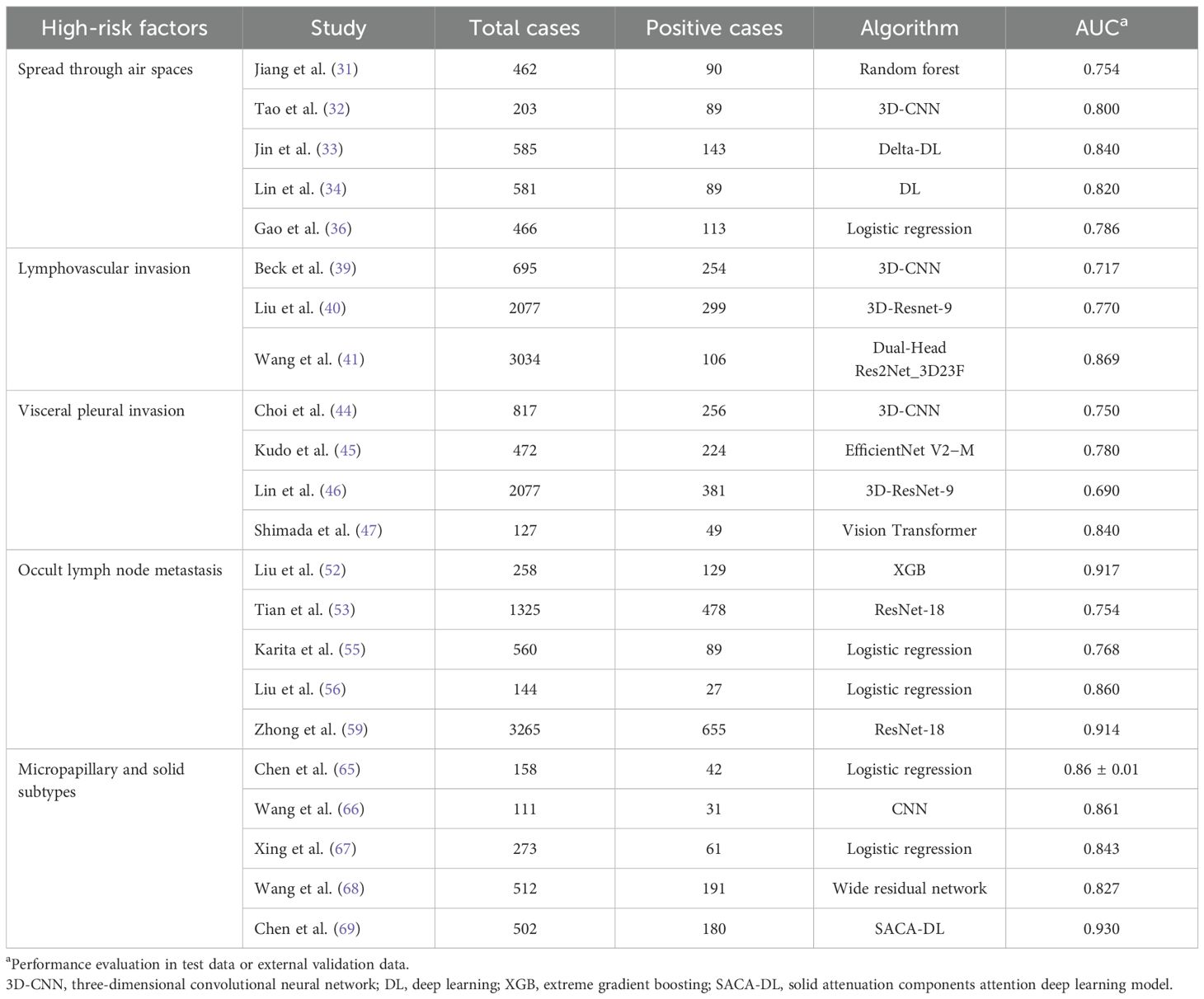
STAS is defined as the presence of tumor cells in the air space of the lungs beyond the margins of the existing tumor. It consists of three main forms, namely (1): bronchus filled by microstructures without a central fibrovascular core (2), solid nests in which the air space is filled by solid components of the tumor (3), and bronchus filled by multiple discrete and discrete continuous single tumor cell filling (27). STAS is always indicative of worse prognosis for patients with LUAD (11, 28). Given the significant implications of STAS for lung cancer progression, it has been proposed as a reference for TNM staging (29). Studies said that leveraging AI by interpreting the clinical significance of STAS can enhance diagnostic accuracy or enable risk stratification for STAS-positive patients. This approach will improve clinical workflows and assist physicians in developing appropriate treatment plans (28, 30). Jiang et al. (31) conducted the first study predicting STAS in LUAD using a ML model based on CT imaging. Their results indicated that quantitative parameters (e.g., tumor size, solid composition, and homogeneity) could serve as predictive biomarkers. However, the study’s limited STAS-positive cohort (19.5%) suggests potential data imbalance issues that may compromise the accuracy of the results. Owing to its stronger feature extraction ability and generalization ability, three-dimensional CNN (3D-CNN) offers superior performance to traditional imaging histological models and computer vision models in predicting STAS of non-small cell lung cancer (NSCLC) (32). Features extracted by CNNs from aligned images can be combined with delta-imaging genomics to generate a double delta model (33). Thus, the importance of stereoscopic features and their corresponding delta features in model construction should be confirmed, while extracting a large number of fine features is precisely where DL excels (34). Regarding data sources, unlike the traditional thin-slice CT, PET/CT can simultaneously reflect the morphological and metabolic characteristics of the lesion, thus offering more abundant information (35). Furthermore, maximum standard uptake value (SUVmax) exhibits a higher predictive effect than the maximum tumor diameter in the STAS (36, 37). Therefore, exploration based on DL technology and 18F-FDG PET/CT for predicting the existence of STAS is expected (28). Retrospective analyses predicting STAS using AI have become increasingly common, with multiple features such as high-density images confirmed to show a significant association with STAS; yet, how to predict STAS in the patient population with non-solid nodules remains under-explored. We call for more prospective studies on STAS, ideally encompassing a broader cohort of patients with lung cancer.
2.2 Lymphovascular invasion
LVI is characterized by the invasion of tumor emboli into the peritumoral lymphatic and/or vascular systems. Consequently, even with complete resection of the primary lesion, the presence of lung cancer cells may still promote recurrence or metastasis via peripheral lymphatics and arteries/veins (8). This explains why LVI is associated with recurrence-free survival (RFS) and overall survival (OS) in NSCLC (8, 38). In 2021, Kyongmin et al. (39) developed and validated a deep cubical nodule transfer learning algorithm (DeepCUBIT) that utilizes transfer learning and 3D-CNN to predict LVI in CT images. In the external validation cohort, DeepCUBIT significantly outperformed single support vector machine (SVM) models that rely solely on tumor size or C/T ratio, demonstrating the informational value of peritumoral regions in CT images. Specifically, in some nodules with C/T ratios below 1.0, the main feature for the prediction comes from the tumor margins—the interface between the tumor and adjacent lung parenchyma. Therefore, we suggest that future studies should consider reasonable expansion in lung nodule image segmentation to incorporate more information. Furthermore, integrating clinical and radiological data can also enhance the accuracy of LVI prediction (40). Compared with simple models, two-dimensional and three-dimensional CT imaging features and clinical radiological data can more accurately reflect the true presence of lung cancer. Of course, the integration of expanded multi-center datasets is equally essential to optimize the model’s learning capabilities (41).
Moreover, the analysis of LVI prediction relevance and interpretability is crucial, as a model that merely reports LVI positivity, without disclosing the reason for the result, cannot deliver satisfactory clinical performance. This necessitates further validation to confirm the efficacy of customized deep network features in capturing imaging phenotypes beyond known histological characteristics (39, 42).
2.3 Visceral pleural invasion
VPI can reduce the 5-year survival rate of patients in stage IA by 16% (8, 10). Therefore, VPI not only reflects the severity of tumor invasiveness but is also regarded as an independent prognostic indicator (10). Regrettably, when thoracic surgeons combined clinical information, preoperative CT scans, and intraoperative pleural biopsies, the diagnostic accuracy rate of VPI was found to be no more than 60% (43). In contrast, DL can automatically learn useful feature representations from lung nodes of raw data, significantly improving the efficiency and accuracy of diagnosis (44, 45). Choi et al. developed a VGG-16 model that achieved an area under the curve (AUC) of 0.81, although its accuracy was constrained by the small sample size (n = 212) (44). In addition to the issue of sample size, the heterogeneity of diagnosis also affects the stability of the model (46). However, other researchers remain optimistic about the future of this field—for instance, one study found that images based on high-resolution computed tomography (HRCT) could effectively predict VPI (45). Another study developed a 3D-ResNet DL model and found that pleural traction angle >30° and disappearance of subpleural fat space were important predictive indicators. The AUC of this model in the training, internal validation, and external validation queues was around 0.70 (46). Tsuchiya’s team used thoracoscopic images and AI to identify the presence of VPI in clinical stage I LUAD and found that the AI model based on ViTs achieved a sensitivity of 0.85 in real-time VPI recognition (47). This study therefore suggests another approach to acquiring the ability to intraoperatively diagnose high-risk factors from images taken by intraoperative thoracoscopy as well, rather than relying solely on preoperative CT. The VPI prediction model holds great promise and significant clinical value, as relying solely on the distance between nodules and the pleura in CT images to diagnose VPI carries risks; in other words, the presence of pleural–tumor contact does not always indicate VPI. Conversely, tumors can invade the pleura through retraction even without direct contact (48). Therefore, a stronger collaboration between radiologists, surgeons, and pathologists is recommended to foster multidisciplinary consensus and address challenges in this field.
2.4 Occult lymph node metastasis
OLM refers to the situation where preoperative CT is not showing lymph node enlargement, but postoperative pathological results do confirm the existence of tumor metastasis (49). However, the fact that radiologists may not detect lymph node abnormalities based on the images does not necessarily indicate that no abnormal manifestations exist on the images—in many cases, these abnormalities are too small to be observed by eye. Therefore, at present, experienced radiologists spend a considerable amount of time carefully examining tumor morphology and lymph node manifestations before diagnosing few OLM cases (50). In addition, by integrating parameters such as carcinoma embryonic antigen (CEA) (>5 ng/mL) and the proportion of nodular solid components (consolidation-to-tumor ratio, CTR ≥0.7), 83% of N1 metastatic cases can be identified (51). AI models based on architectures such as XGB have demonstrated excellent performance in predicting OLM in patients with clinical stage IA LUAD (52–54). The pulmonary nodule volume-doubling time (VDT) of patients with LUAD is shorter when solid/microform components are dominant, which leads to a higher metastasis rate; therefore, VDT or mass doubling time (MDT) may be effective predictors of OLM among NSCLC (55, 56). In other words, when establishing a DL model, considering CT follow-up results for more than 1 year is conducive to a better model (55).
PET/CT has high sensitivity in detecting hilar and mediastinal lymph nodes. Cytokeratin fragment (CYFRA) 21-1 (>2.46) and liver SUVmax (L-SUR) (>2.68) have been proven to be independent risk factors for OLM (57). Furthermore, the use of PET/CT data to build AI models is feasible in predicting OLM and offers high sensitivity and specificity in prospective tests (58). PET/CT-DL models have achieved excellent performance, respectively, in the prediction of latent N1 and N2. High-risk patients identified by this model can benefit from lymph node biopsy, lobectomy, and adjuvant therapy (59). It is important to note that although the images are necessary for analysis, not all PET/CT devices have the ability to output such high-quality images. Furthermore, PET/CT is not commonly used for the diagnosis of ground-glass lung nodules ≤2 cm (35, 60) and is often not the preferred test in developing countries, such as China, owing to its high cost. This may reduce the clinical applicability of such models in some institutions. Because of the low incidence of occult lymph nodes (approximately 5%–15%) (6), the imbalance in samples caused by a low number of positive cases limits AI’s learning capacity. Furthermore, while the study targets metastatic lymph nodes, preoperative CT scans struggle to accurately diagnose and delineate them. Consequently, recent AI predictions still focus on pulmonary nodules as regions of interest—a contradiction that appears difficult to resolve. Additionally, an increasing number of studies are shifting their focus to N1 and N2 metastases rather than simple binary classification problems, which better align with actual clinical needs regarding lung cancer diagnosis and treatment.
2.5 High-grade pathological subtype (micropapillary or solid pattern)
During the development of LUAD, various growth patterns (including squamous, acinar, micropapillary, and solid types) occur. Generally, various pathological subtypes are calculated in increments of 5%, and the proportion of the main subtypes determines the dominance of the pathological types (3). We focused on the micropapillary subtypes and solid types in LUAD, which are closely related to DFS and OS after surgery (61) and accompanied by a higher postoperative recurrence rate (9, 62). More researchers are exploring the application of DL techniques in predicting LUAD pathotypes due to DL detection of details that cannot be observed by the human eye (63). Dong et al. developed two deep learning models based on LeNet and DenseNet, discovering that deep learning techniques can not only be applied to the classification of invasive LUAD but also used to predict micropapillary patterns—marking the first attempt at utilizing deep learning technology in this field (64). Chang et al. used an advanced LUAD subtype detection algorithm to analyze near-pure lung adenocarcinoma imaging with histological features and plaque images and obtained high sensitivity and moderate specificity (65). As intratumoral heterogeneity is manifested in the pathological components and tumor imaging features, a single modality may not be able to accurately identify the solid components within the tumor (66–68). Thus, the establishment of a comprehensive model combining DL-based scoring and clinical imaging features to distinguish LUAD with micropapillary and solid-type structure enhanced the classification performance (68). Chen et al. developed a DL model based on the data of 502 patients with pathologically diagnosed high-grade adenocarcinoma within 4 years. They applied a solid-attenuation-component-like subregion masks (tumor area ≥-190 HU) to guide the DL model in analyzing high-grade subtypes. This is another promising preoperative prediction method for high-grade adenocarcinoma subtypes (69). Similar to the assessment of other high-risk factors, 18F-FDG PET/CT data are increasingly being applied to DL because of the high metabolic rate of tumor cells and have demonstrated decent predictive accuracy for microscopic or solid components within LUAD (70, 71). A new study demonstrated the combined DL of texture features for ultra-short echo time MRI (UTE-MRI) to avoid radiation exposure while maintaining comparable diagnostic performance with CT for subtypes such as micropapillary (AUC = 0.82 vs. 0.79) (72). Despite the small sample size of the study (74 lesions), this implies the feasibility of the UTE-MRI DL model with the significant advantage of UTE-MRI in avoiding radiation hazards, especially for patients requiring multiple follow-up visits over time and for children with radiation sensitivity. Furthermore, various imaging examinations and large models that integrate imaging and genomics are providing new perspectives and tools for personalized treatment and surgical decisions. Both clinical and quantitative radiomics features, along with DL-based scoring models, can decode LUAD phenotypes. However, a notable issue arises: LUAD with multiple subtypes may exhibit mixed subtype characteristics rather than distinct information from the three subtypes, potentially limiting the discriminative capability of AI processing. Most studies rely on quantitative analysis of specific subtypes using thresholds such as 5%, 10%, or 20%. Future DL models and related fusion models require a more detailed stratified analysis beyond qualitative predictions (Table1).
3 Evaluation of high-risk factors combining pathological tissue and CT
Multimodal fusion has become an inevitable direction for AI-assisted lung cancer diagnosis. Pathological images serve as another critical information source alongside radiological imaging—for example, the ResNet model trained on whole-slide images demonstrates recognition capabilities comparable to those of senior pathologists in identifying pathological subtypes of LUAD (73). In recent years, the integration of CT and pathological information is more commonly used to predict patient prognosis. However, Wu and colleagues constructed a predictive model using preoperative CT images and intraoperative frozen section results to assess the extent of lung cancer invasion (74). This represents a novel clinical issue: although diagnosis based on intraoperative frozen section offers high specificity to identify high-risk factors in LUAD, its sensitivity remains limited (75). DL models analyze feature maps and perform logical reasoning, exhibiting decision-making processes consistent with the diagnostic reasoning of pathologists. They even demonstrate superior capabilities in identifying minute lesions (76). This demonstrates that AI models can now automatically detect and quantitatively analyze STAS, acinar, micropapillary, papillary, solid subtypes, and normal tissue in final histopathological sections, achieving satisfactory results (77). Thus, leveraging AI to further enhance the diagnostic accuracy of frozen section analysis has a solid practical foundation. Going a step further, the question of whether integrating preoperative CT imaging with preoperative needle biopsy specimens, bronchoscopic biopsy specimens, or intraoperative frozen section results to construct a more comprehensive and integrated intelligent model could achieve more precise predictions warrants in-depth exploration.
4 The future of prediction of high-risk factors for LUAD based on DL
DL offers advantages such as high efficiency, accuracy, and automation, making its potential self-evident. However, several key challenges remain, including technical issues such as sparse datasets and model generalization. In medical applications, AI models must also demonstrate high interpretability to gain the trust and acceptance of clinicians. Therefore, we believe that future research directions may include the following:
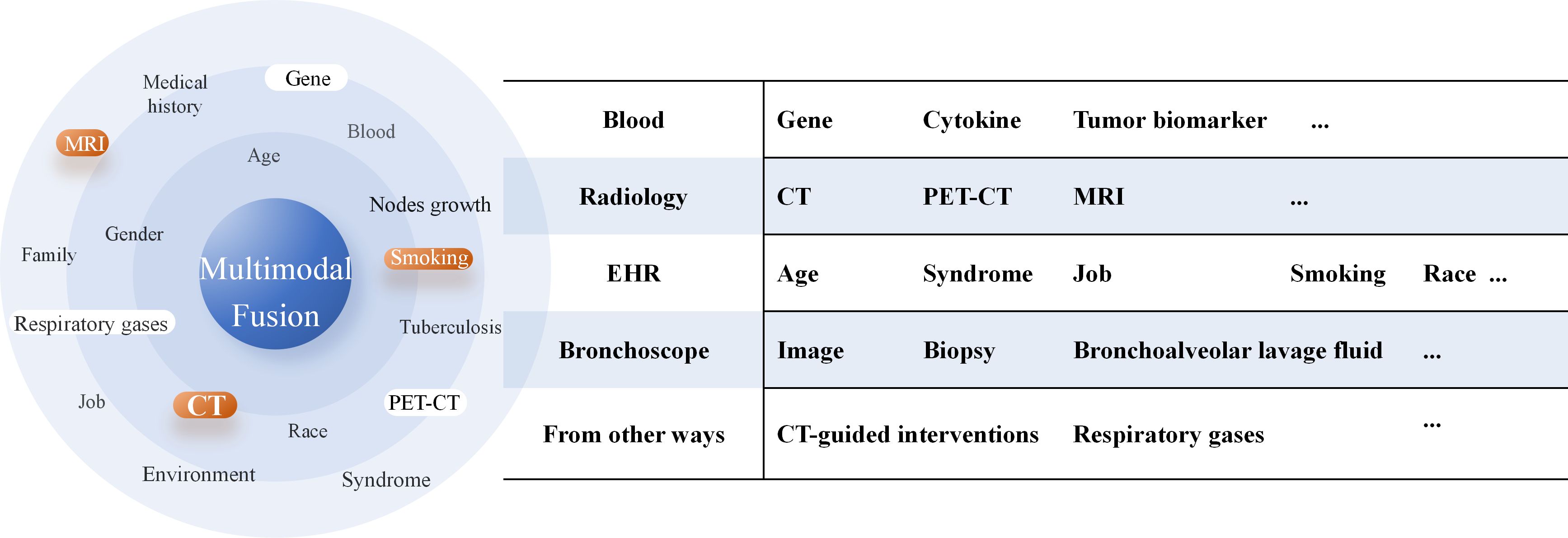
1. In predicting high-risk factors for LUAD, the core value of multimodal fusion models lies in their ability to integrate heterogeneous medical data, break down information barriers, and ultimately enable more precise and personalized prognostic assessments. By integrating imaging data such as CT, PET/CT, and pathological images from the puncture surgery or bronchoscopy and electronic health records (EHRs)—including the age, symptoms, and smoking history (Figure 4)— multimodal models can construct multidimensional disease views, providing a more comprehensive reflection of lung cancer progression stages and individual patient condition. The adoption of large language models further facilitates faster and more standardized generation of EHRs in clinical practice, enhancing the input quality for multimodal systems (78), and significant progress has been made in validating the feasibility and potential of multimodal strategies—for instance, She et al. (59) developed a multimodal DL model based on PET/CT that demonstrated a significantly higher accuracy in diagnosing OLM relative to single-modality DL models, clinical models, and physicians. Chen et al. (79) established a multiomics model (clinic-RadmC) integrating clinical and radiomic data with circulating cell-free DNA fragmentomic features in 5-methylcytosine (5mC)-enriched regions to predict the malignancy risk of indeterminate pulmonary nodules. Moreover, a combined DL model incorporating clinical, imaging, and cell-free DNA methylation biomarkers has been shown to accurately classify pulmonary nodules, potentially reducing approximately 80% of unnecessary surgeries and delayed treatments (80).However, the construction and application of multimodal models still face several critical challenges. First, significant differences in format, dimensionality, and semantic levels exist between different modal data, making effective modal alignment and deep fusion a persistent technical hurdle. Second, the inherent “black-box” nature of DL models makes it difficult for clinicians to understand which features—such as image regions or gene mutations—underlie the model’s high-risk assessments.
2. In the field of medical imaging, some phenomena, such as occult lymph nodes whose incidence rate is approximately 10%, are not common. Moreover, the manual annotation of CT and pathological images is costly and time-consuming, and the limited availability of annotated datasets makes small-sample learning a key challenge. To address this issue, researchers have explored various strategies. Data augmentation techniques, including geometric transformations, intensity perturbations, and advanced methods such as MixUp and CutMix, enrich the diversity of training and reduce overfitting (81, 82). Few-shot and meta-learning paradigms offer another promising direction, wherein models are trained in a batch manner to learn transferable representations, enabling rapid adaptation to new cohorts or rare subtypes with only a few examples (83, 84). Knowledge distillation from large teacher models to lightweight student models has also been used to mitigate overfitting and improve sample efficiency (85). Overall, these methods aim to overcome the inherent limitations of small-sample medical datasets and support more robust and generalizable DL models.
Furthermore, the latest advancements in large-scale pre-trained models show great promise. Models such as CLIP, SAM, and DINOv2, which were initially developed for natural images, have demonstrated significant potential in the field of medical imaging, particularly in few-shot adaptation, interactive annotation, and cross-modal integration (24, 86, 87). These developments collectively suggest that customizing model designs for the characteristics of medical data and transferring from foundational models will also play a crucial role in overcoming the inherent limitations of medical datasets.
3. The application of AI in clinical practice relies heavily on evidence-based medical support obtained through rigorous clinical trials to validate its efficacy, safety, and reliability. Most previously mentioned studies are predominantly retrospective analyses, although some high-quality studies have included prospective validation cohorts. Most published studies have focused on the East Asian populations (such as patients from China, Japan, and South Korea), thereby presenting geographical and demographic limitations. To address the issue of generalization of the capabilities of AI models across diverse populations, regulatory bodies such as the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) are actively promoting multicenter trials and external validation mechanisms to assess their actual efficacy in heterogeneous populations (88, 89). However, it is encouraging that the clinical research findings on AI published to date have been almost entirely positive. A systematic review of 39 randomized controlled trials (RCTs) conducted as of July 2021 revealed that 77% (30/39) of AI interventions demonstrated superior efficacy relative to standard clinical care and 70% (21/30) showed clinically meaningful improvements in outcomes, with radiology studies constituting the majority of these findings (90). Other prospective studies have also demonstrated that CT-based AI tools can enhance the sensitivity of lung nodule diagnosis (90–92). However, no further studies with higher levels of evidence have been published on the prediction of high-risk factors for LUAD. Despite its promise, AI still faces multiple challenges in clinical applications, including data privacy, system security, and interoperability. Future research should include multicenter, large-scale, rigorously designed clinical trials to fully validate the effectiveness of AI—particularly DL models—in real-world clinical settings. Simultaneously, collaboration between clinicians, AI companies, and regulatory bodies must be enhanced to jointly address complex ethical and legal issues, thereby promoting the robust and responsible integration of AI within the healthcare sector.
5 Conclusions
AI-based research on the prediction of high-risk factors for LUAD shows great potential in medical image analysis and pathological diagnosis. DL models significantly improve the accuracy and efficiency of early prediction and diagnosis of lung cancer by automatically identifying and classifying high-risk factors in CT images. However, current techniques still face challenges such as insufficient datasets, high model complexity, poor interpretability, and data privacy. Future research should focus on translating cutting-edge technologies into clinical practice through standardized data collection, lightweight model architectures, and rigorous evidence-based validation. With the introduction of new architectures such as transformers, AI is expected to become an important method to support decision-making in the diagnosis of high-risk factors and treatment of LUAD.
Author contributions
YH: Writing – review & editing, Writing – original draft, Validation, Investigation. BZ: Writing – original draft, Writing – review & editing. RY: Writing – original draft, Writing – review & editing. CZ: Writing – original draft. ZG: Writing – original draft. PM: Writing – review & editing. KL: Funding acquisition, Project administration, Conceptualization, Supervision, Writing – review & editing. YL: Writing – review & editing, Funding acquisition, Conceptualization, Supervision, Project administration.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. Funded by National Key Research and Development Program (No.2023YFC2508603), National Science Foundation for Young Scientists of China (No.62406122) and Xiong’an New Area Science and Technology Innovation Special Project (No.2023XAGG0071).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. Generative AI and AI-assisted technologies can only be used during the polish process to enhance the readability and language of the manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AI, artificial intelligence; AUC, area under the curve; CNN, convolutional neural networks; CT, computed tomography; DL, deep learning; EMA, European Medicines Agency; FDA, Food and Drug Administration; HRCT, high-resolution computed tomography; LUAD, lung adenocarcinoma; MDT, mass doubling time; ML, machine learning; OLM, occult lymph node metastasis; OS, overall survival; PTNB, percutaneous transthoracic needle biopsy; RCT, randomized controlled trials; RFS, recurrence-free survival; STAS, spreading through air spaces; SVM, support vector machine; VDT, volume-doubling time; VPI, visceral pleural invasion.
References
1. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2024) 74:229–63. doi: 10.3322/caac.21834
2. Zhang Y, Vaccarella S, Morgan E, Li M, Etxeberria J, Chokunonga E, et al. Global variations in lung cancer incidence by histological subtype in 2020: a population-based study. Lancet Oncol. (2023) 24:1206–18. doi: 10.1016/S1470-2045(23)00444-8
3. Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, et al. International association for the study of lung cancer american thoracic society european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. (2011) 6:244–85. doi: 10.1097/JTO.0b013e318206a221
4. Bains S, Eguchi T, Warth A, Yeh YC, Nitadori JI, Woo KM, et al. Procedure-specific risk prediction for recurrence in patients undergoing lobectomy or sublobar resection for small (</=2 cm) lung adenocarcinoma: an international cohort analysis. J Thorac Oncol. (2019) 14:72–86. doi: 10.1016/j.jtho.2018.09.008
5. Caso R, Sanchez-Vega F, Tan KS, Mastrogiacomo B, Zhou J, Jones GD, et al. The underlying tumor genomics of predominant histologic subtypes in lung adenocarcinoma. J Thorac Oncol. (2020) 15:1844–56. doi: 10.1016/j.jtho.2020.08.005
6. Beyaz F, Verhoeven RLJ, Schuurbiers OCJ, Verhagen A, and van der Heijden E. Occult lymph node metastases in clinical N0/N1 NSCLC; A single center in-depth analysis. Lung Cancer. (2020) 150:186–94. doi: 10.1016/j.lungcan.2020.10.022
7. Miyoshi T, Shirakusa T, Ishikawa Y, Iwasaki A, Shiraishi T, Makimoto Y, et al. Possible mechanism of metastasis in lung adenocarcinomas with a micropapillary pattern. Pathol Int. (2005) 55:419–24. doi: 10.1111/j.1440-1827.2005.01847.x
8. Wang S, Zhang B, Qian J, Qiao R, Xu J, Zhang L, et al. Proposal on incorporating lymphovascular invasion as a T-descriptor for stage I lung cancer. Lung Cancer. (2018) 125:245–52. doi: 10.1016/j.lungcan.2018.09.024
9. Takeno N, Tarumi S, Abe M, Suzuki Y, Kinoshita I, and Kato T. Lung adenocarcinoma with micropapillary and solid patterns: Recurrence rate and trends. Thorac Cancer. (2023) 14:2987–92. doi: 10.1111/1759-7714.15087
10. Shimizu K, Yoshida J, Nagai K, Nishimura M, Ishii G, Morishita Y, et al. Visceral pleural invasion is an invasive and aggressive indicator of non-small cell lung cancer. J Thorac Cardiovasc Surg. (2005) 130:160–5. doi: 10.1016/j.jtcvs.2004.11.021
11. Eguchi T, Kameda K, Lu S, Bott MJ, Tan KS, Montecalvo J, et al. Lobectomy Is Associated with Better Outcomes than Sublobar Resection in Spread through Air Spaces (STAS)-Positive T1 Lung Adenocarcinoma: A Propensity Score-Matched Analysis. J Thorac Oncol. (2019) 14:87–98. doi: 10.1016/j.jtho.2018.09.005
12. Kukhareva PV, Li H, Caverly TJ, Fagerlin A, Del Fiol G, Hess R, et al. Lung cancer screening before and after a multifaceted electronic health record intervention: A nonrandomized controlled trial. JAMA Netw Open. (2024) 7:e2415383. doi: 10.1001/jamanetworkopen.2024.15383
13. Cheng DO, Khaw CR, McCabe J, Pennycuick A, Nair A, Moore DA, et al. Predicting histopathological features of aggressiveness in lung cancer using CT radiomics: a systematic review. Clin Radiol. (2024) 79:681–9. doi: 10.1016/j.crad.2024.04.022
14. Huang S, Yang J, Shen N, Xu Q, and Zhao Q. Artificial intelligence in lung cancer diagnosis and prognosis: Current application and future perspective. Semin Cancer Biol. (2023) 89:30–7. doi: 10.1016/j.semcancer.2023.01.006
15. de Margerie-Mellon C and Chassagnon G. Artificial intelligence: A critical review of applications for lung nodule and lung cancer. Diagn Interv Imaging. (2023) 104:11–7. doi: 10.1016/j.diii.2022.11.007
16. Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. (2014) 5:4006. doi: 10.1038/ncomms5006
17. Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. (2017) 14:749–62. doi: 10.1038/nrclinonc.2017.141
18. Li J, Li Z, Wei L, and Zhang X. Machine learning in lung cancer radiomics. Mach Intell Res. (2023) 20:753–82. doi: 10.1007/s11633-022-1364-x
19. Thanoon MA, Zulkifley MA, Mohd Zainuri MAA, and Abdani SR. A Review of deep learning techniques for lung cancer screening and diagnosis based on CT Images. Diagnostics (Basel). (2023) 13:2617. doi: 10.3390/diagnostics13162617
20. Yamashita R, Nishio M, Do RKG, and Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. (2018) 9:611–29. doi: 10.1007/s13244-018-0639-9
21. Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, et al. An image is worth 16×16 words: transformers for image recognition at scale. Proceedings of the International Conference on Learning Representations (ICLR). (2021). doi: 10.48550/arXiv.2010.11929
22. Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, et al.Swin transformer: hierarchical vision transformer using shifted windows. Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV). (2021):10012–10022. doi: 10.1109/ICCV48922.2021.00986
23. He K, Zhang X, Ren S, and Sun J. (2016). Deep residual learning for image recognition, in: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–8. doi: 10.1109/CVPR.2016.90
24. Fang M, Wang Z, Pan S, Feng X, Zhao Y, Hou D, et al. Large models in medical imaging: Advances and prospects. Chin Med J (Engl). (2025) 138:1647–64. doi: 10.1097/CM9.0000000000003699
25. Ye G, Wu G, Li K, Zhang C, Zhuang Y, Liu H, et al. Development and validation of a deep learning radiomics model to predict high-risk pathologic pulmonary nodules using preoperative computed tomography. Acad Radiol. (2024) 31:1686–97. doi: 10.1016/j.acra.2023.08.040
26. Ye G, Wu G, Li Y, Zhang C, Qin L, Wu J, et al. Advancing presurgical non-invasive spread through air spaces prediction in clinical stage IA lung adenocarcinoma using artificial intelligence and CT signatures. Front Surg. (2024) 11:1511024. doi: 10.3389/fsurg.2024.1511024
27. Kadota K, Nitadori JI, Sima CS, Ujiie H, Rizk NP, Jones DR, et al. Tumor Spread through Air Spaces is an Important Pattern of Invasion and Impacts the Frequency and Location of Recurrences after Limited Resection for Small Stage I Lung Adenocarcinomas. J Thorac Oncol. (2015) 10:806–14. doi: 10.1097/JTO.0000000000000486
28. Wang Y, Lyu D, Fan L, and Liu S. Advances in the prediction of spread through air spaces with imaging in lung cancer: a narrative review. Transl Cancer Res. (2023) 12:624–30. doi: 10.21037/tcr-22-2593
29. Travis WD, Eisele M, Nishimura KK, Aly RG, Bertoglio P, Chou TY, et al. The international association for the study of lung cancer (IASLC) staging project for lung cancer: recommendation to introduce spread through air spaces as a histologic descriptor in the ninth edition of the TNM classification of lung cancer. Analysis of 4061 Pathologic Stage I NSCLC. J Thorac Oncol. (2024) 19:1028–51. doi: 10.1016/j.jtho.2024.03.015
30. Feng Y, Ding H, Huang X, Zhang Y, Lu M, Zhang T, et al. Deep learning-based detection and semi-quantitative model for spread through air spaces (STAS) in lung adenocarcinoma. NPJ Precis Oncol. (2024) 8:173. doi: 10.1038/s41698-024-00664-0
31. Jiang C, Luo Y, Yuan J, You S, Chen Z, Wu M, et al. CT-based radiomics and machine learning to predict spread through air space in lung adenocarcinoma. Eur Radiol. (2020) 30:4050–7. doi: 10.1007/s00330-020-06694-z
32. Tao J, Liang C, Yin K, Fang J, Chen B, Wang Z, et al. 3D convolutional neural network model from contrast-enhanced CT to predict spread through air spaces in non-small cell lung cancer. Diagn Interv Imaging. (2022) 103:535–44. doi: 10.1016/j.diii.2022.06.002
33. Jin W, Shen L, Tian Y, Zhu H, Zou N, Zhang M, et al. Improving the prediction of Spreading Through Air Spaces (STAS) in primary lung cancer with a dynamic dual-delta hybrid machine learning model: a multicenter cohort study. Biomark Res. (2023) 11:102. doi: 10.1186/s40364-023-00539-9
34. Lin MW, Chen LW, Yang SM, Hsieh MS, Ou DX, Lee YH, et al. CT-based deep-learning model for spread-through-air-spaces prediction in ground glass-predominant lung adenocarcinoma. Ann Surg Oncol. (2024) 31:1536–45. doi: 10.1245/s10434-023-14565-2
35. Greenspan BS. Role of PET/CT for precision medicine in lung cancer: perspective of the Society of Nuclear Medicine and Molecular Imaging. Transl Lung Cancer Res. (2017) 6:617–20. doi: 10.21037/tlcr.2017.09.01
36. Gao Z, An P, Li R, Wu F, Sun Y, Wu J, et al. Development and validation of a clinic-radiological model to predict tumor spread through air spaces in stage I lung adenocarcinoma. Cancer Imaging. (2024) 24:25. doi: 10.1186/s40644-024-00668-w
37. Li H, Li L, Liu Y, Deng Y, Zhu Y, Huang L, et al. Predictive value of CT and (18)F-FDG PET/CT features on spread through air space in lung adenocarcinoma. BMC Cancer. (2024) 24:434. doi: 10.1186/s12885-024-12220-x
38. Mollberg NM, Bennette C, Howell E, Backhus L, Devine B, and Ferguson MK. Lymphovascular invasion as a prognostic indicator in stage I non-small cell lung cancer: a systematic review and meta-analysis. Ann Thorac Surg. (2014) 97:965–71. doi: 10.1016/j.athoracsur.2013.11.002
39. Beck KS, Gil B, Na SJ, Hong JH, Chun SH, An HJ, et al. DeepCUBIT: predicting lymphovascular invasion or pathological lymph node involvement of clinical T1 stage non-small cell lung cancer on chest CT scan using deep cubical nodule transfer learning algorithm. Front Oncol. (2021) 11:661244. doi: 10.3389/fonc.2021.661244
40. Liu K, Lin X, Chen X, Chen B, Li S, Li K, et al. Development and validation of a deep learning signature for predicting lymphovascular invasion and survival outcomes in clinical stage IA lung adenocarcinoma: A multicenter retrospective cohort study. Transl Oncol. (2024) 42:101894. doi: 10.1016/j.tranon.2024.101894
41. Wang J, Yang Y, Xie Z, Mao G, Gao C, Niu Z, et al. Predicting lymphovascular invasion in non-small cell lung cancer using deep convolutional neural networks on preoperative chest CT. Acad Radiol. (2024) 31:5237–47. doi: 10.1016/j.acra.2024.05.010
42. Zhang X, Zhang G, Qiu X, Yin J, Tan W, Yin X, et al. Exploring non-invasive precision treatment in non-small cell lung cancer patients through deep learning radiomics across imaging features and molecular phenotypes. biomark Res. (2024) 12:12. doi: 10.1186/s40364-024-00561-5
43. Takizawa H, Kondo K, Kawakita N, Tsuboi M, Toba H, Kajiura K, et al. Autofluorescence for the diagnosis of visceral pleural invasion in non-small-cell lung cancer. Eur J Cardiothorac Surg. (2018) 53:987–92. doi: 10.1093/ejcts/ezx419
44. Choi H, Kim H, Hong W, Park J, Hwang EJ, Park CM, et al. Prediction of visceral pleural invasion in lung cancer on CT: deep learning model achieves a radiologist-level performance with adaptive sensitivity and specificity to clinical needs. Eur Radiol. (2021) 31:2866–76. doi: 10.1007/s00330-020-07431-2
45. Kudo Y, Saito A, Horiuchi T, Murakami K, Kobayashi M, Matsubayashi J, et al. Preoperative evaluation of visceral pleural invasion in peripheral lung cancer utilizing deep learning technology. Surg Today. (2025) 55:18–28. doi: 10.1007/s00595-024-02869-z
46. Lin X, Liu K, Li K, Chen X, Chen B, Li S, et al. A CT-based deep learning model: visceral pleural invasion and survival prediction in clinical stage IA lung adenocarcinoma. iScience. (2024) 27:108712. doi: 10.1016/j.isci.2023.108712
47. Shimada Y, Ojima T, Takaoka Y, Sugano A, Someya Y, Hirabayashi K, et al. Prediction of visceral pleural invasion of clinical stage I lung adenocarcinoma using thoracoscopic images and deep learning. Surg Today. (2024) 54:540–50. doi: 10.1007/s00595-023-02756-z
48. Lim WH, Lee KH, Lee JH, Park H, Nam JG, Hwang EJ, et al. Diagnostic performance and prognostic value of CT-defined visceral pleural invasion in early-stage lung adenocarcinomas. Eur Radiol. (2024) 34:1934–45. doi: 10.1007/s00330-023-10204-2
49. Yoon DW, Kang D, Jeon YJ, Lee J, Shin S, Cho JH, et al. Computed tomography characteristics of cN0 primary non-small cell lung cancer predict occult lymph node metastasis. Eur Radiol. (2024) 34:7817–28. doi: 10.1007/s00330-024-10835-z
50. Guglielmo P, Marturano F, Bettinelli A, Sepulcri M, Pasello G, Gregianin M, et al. Additional value of PET and CT image-based features in the detection of occult lymph node metastases in lung cancer: A systematic review of the literature. Diagnostics (Basel). (2023) 13:2153. doi: 10.3390/diagnostics13132153
51. Ai J GH, Shi G, Lan Y, Hu S, Wang Z, Liu L, et al. A clinical nomogram for predicting occult lymph node metastasis in patients with non-small-cell lung cancer ≤2 cm. Interdiscip Cardiovasc Thorac Surg. (2024) 39:ivae098. doi: 10.1093/icvts/ivae098
52. Liu MW, Zhang X, Wang YM, Jiang X, Jiang JM, Li M, et al. A comparison of machine learning methods for radiomics modeling in prediction of occult lymph node metastasis in clinical stage IA lung adenocarcinoma patients. J Thorac Dis. (2024) 16:1765–76. doi: 10.21037/jtd-23-1578
53. Tian W, Yan Q, Huang X, Feng R, Shan F, Geng D, et al. Predicting occult lymph node metastasis in solid-predominantly invasive lung adenocarcinoma across multiple centers using radiomics-deep learning fusion model. Cancer Imaging. (2024) 24:8. doi: 10.1186/s40644-024-00654-2
54. Ye G, Zhang C, Zhuang Y, Liu H, Song E, Li K, et al. An advanced nomogram model using deep learning radiomics and clinical data for predicting occult lymph node metastasis in lung adenocarcinoma. Transl Oncol. (2024) 44:101922. doi: 10.1016/j.tranon.2024.101922
55. Karita R, Suzuki H, Onozato Y, Kaiho T, Inage T, Ito T, et al. A simple nomogram for predicting occult lymph node metastasis of non-small cell lung cancer from preoperative computed tomography findings, including the volume-doubling time. Surg Today. (2024) 54:31–40. doi: 10.1007/s00595-023-02695-9
56. Liu M, Mu J, Song F, Liu X, Jing W, and Lv F. Growth characteristics of early-stage (IA) lung adenocarcinoma and its value in predicting lymph node metastasis. Cancer Imaging. (2023) 23:115. doi: 10.1186/s40644-023-00631-1
57. Shi YM, Niu R, Shao XL, Zhang FF, Shao XN, Wang JF, et al. Tumor-to-liver standard uptake ratio using fluorine-18 fluorodeoxyglucose positron emission tomography computed tomography effectively predict occult lymph node metastasis of non-small cell lung cancer patients. Nucl Med Commun. (2020) 41:459–68. doi: 10.1097/MNM.0000000000001173
58. Ouyang ML, Zheng RX, Wang YR, Zuo ZY, Gu LD, Tian YQ, et al. Deep learning analysis using (18)F-FDG PET/CT to predict occult lymph node metastasis in patients with clinical N0 lung adenocarcinoma. Front Oncol. (2022) 12:915871. doi: 10.3389/fonc.2022.915871
59. Zhong Y, Cai C, Chen T, Gui H, Deng J, Yang M, et al. PET/CT based cross-modal deep learning signature to predict occult nodal metastasis in lung cancer. Nat Commun. (2023) 14:7513. doi: 10.1038/s41467-023-42811-4
60. MacMahon H, Naidich DP, Goo JM, Lee KS, Leung ANC, Mayo JR, et al. Guidelines for management of incidental pulmonary nodules detected on CT images: from the fleischner society 2017. Radiology. (2017) 284:228–43. doi: 10.1148/radiol.2017161659
61. Cha MJ, Lee HY, Lee KS, Jeong JY, Han J, Shim YM, et al. Micropapillary and solid subtypes of invasive lung adenocarcinoma: clinical predictors of histopathology and outcome. J Thorac Cardiovasc Surg. (2014) 147:921–928.e922. doi: 10.1016/j.jtcvs.2013.09.045
62. Wang W, Hu Z, Zhao J, Huang Y, Rao S, Yang J, et al. Both the presence of a micropapillary component and the micropapillary predominant subtype predict poor prognosis after lung adenocarcinoma resection: a meta-analysis. J Cardiothorac Surg. (2020) 15:154. doi: 10.1186/s13019-020-01199-8
63. Choi Y, Aum J, Lee SH, Kim HK, Kim J, Shin S, et al. Deep learning analysis of CT images reveals high-grade pathological features to predict survival in lung adenocarcinoma. Cancers (Basel). (2021) 13:4077. doi: 10.3390/cancers13164077
64. Ding H, Xia W, Zhang L, Mao Q, Cao B, Zhao Y, et al. CT-based deep learning model for invasiveness classification and micropapillary pattern prediction within lung adenocarcinoma. Front Oncol. (2020) 10:1186. doi: 10.3389/fonc.2020.01186
65. Chen LW, Yang SM, Wang HJ, Chen YC, Lin MW, Hsieh MS, et al. Prediction of micropapillary and solid pattern in lung adenocarcinoma using radiomic values extracted from near-pure histopathological subtypes. Eur Radiol. (2021) 31:5127–38. doi: 10.1007/s00330-020-07570-6
66. Wang X, Zhang L, Yang X, Tang L, Zhao J, Chen G, et al. Deep learning combined with radiomics may optimize the prediction in differentiating high-grade lung adenocarcinomas in ground glass opacity lesions on CT scans. Eur J Radiol. (2020) 129:109150. doi: 10.1016/j.ejrad.2020.109150
67. Xing X, Li L, Sun M, Zhu X, and Feng Y. A combination of radiomic features, clinic characteristics, and serum tumor biomarkers to predict the possibility of the micropapillary/solid component of lung adenocarcinoma. Ther Adv Respir Dis. (2024) 18:17534666241249168. doi: 10.1177/17534666241249168
68. Wang F, Wang CL, Yi YQ, Zhang T, Zhong Y, Zhu JJ, et al. Comparison and fusion prediction model for lung adenocarcinoma with micropapillary and solid pattern using clinicoradiographic, radiomics and deep learning features. Sci Rep. (2023) 13:9302. doi: 10.1038/s41598-023-36409-5
69. Chen LW, Yang SM, Chuang CC, Wang HJ, Chen YC, Lin MW, et al. Solid attenuation components attention deep learning model to predict micropapillary and solid patterns in lung adenocarcinomas on computed tomography. Ann Surg Oncol. (2022) 29:7473–82. doi: 10.1245/s10434-022-12055-5
70. Choi W, Liu CJ, Alam SR, Oh JH, Vaghjiani R, Humm J, et al. Preoperative (18)F-FDG PET/CT and CT radiomics for identifying aggressive histopathological subtypes in early stage lung adenocarcinoma. Comput Struct Biotechnol J. (2023) 21:5601–8. doi: 10.1016/j.csbj.2023.11.008
71. Zhou L, Sun J, Long H, Zhou W, Xia R, Luo Y, et al. Imaging phenotyping using (18)F-FDG PET/CT radiomics to predict micropapillary and solid pattern in lung adenocarcinoma. Insights Imaging. (2024) 15:5. doi: 10.1186/s13244-023-01573-9
72. Lee S, Lee CY, Kim NY, Suh YJ, Lee HJ, Yong HS, et al. Feasibility of UTE-MRI-based radiomics model for prediction of histopathologic subtype of lung adenocarcinoma: in comparison with CT-based radiomics model. Eur Radiol. (2024) 34:3422–30. doi: 10.1007/s00330-023-10302-1
73. Ding H, Feng Y, Huang X, Xu J, Zhang T, Liang Y, et al. Deep learning-based classification and spatial prognosis risk score on whole-slide images of lung adenocarcinoma. Histopathology. (2023) 83:211–28. doi: 10.1111/his.14918
74. Wu G, Woodruff HC, Sanduleanu S, Refaee T, Jochems A, Leijenaar R, et al. Preoperative CT-based radiomics combined with intraoperative frozen section is predictive of invasive adenocarcinoma in pulmonary nodules: a multicenter study. Eur Radiol. (2020) 30:2680–91. doi: 10.1007/s00330-019-06597-8
75. Cao H, Zheng Q, Deng C, Fu Z, Shen X, Jin Y, et al. Prediction of spread through air spaces (STAS) by intraoperative frozen section for patients with cT1N0M0 invasive lung adenocarcinoma: A multi-center observational study (ECTOP-1016). Ann Surg. (2024) 281:187–192. doi: 10.1097/SLA.0000000000006525
76. Zhao Y, He S, Zhao D, Ju M, Zhen C, Dong Y, et al. Deep learning-based diagnosis of histopathological patterns for invasive non-mucinous lung adenocarcinoma using semantic segmentation. BMJ Open. (2023) 13:e069181. doi: 10.1136/bmjopen-2022-069181
77. Qiao B, Jumai K, Ainiwaer J, Niyaz M, Zhang Y, Ma Y, et al. A novel transfer-learning based physician-level general and subtype classifier for non-small cell lung cancer. Heliyon. (2022) 8:e11981. doi: 10.1016/j.heliyon.2022.e11981
78. Ding H, Xia W, Zhou Y, Wei L, Feng Y, Wang Z, et al. Evaluation and practical application of prompt-driven ChatGPTs for EMR generation. NPJ Digit Med. (2025) 8:77. doi: 10.1038/s41746-025-01472-x
79. Zhao M, Xue G, He B, Deng J, Wang T, Zhong Y, et al. Integrated multiomics signatures to optimize the accurate diagnosis of lung cancer. Nat Commun. (2025) 16:84. doi: 10.1038/s41467-024-55594-z
80. He J, Wang B, Tao J, Liu Q, Peng M, Xiong S, et al. Accurate classification of pulmonary nodules by a combined model of clinical, imaging, and cell-free DNA methylation biomarkers: a model development and external validation study. Lancet Digit Health. (2023) 5:e647–56. doi: 10.1016/S2589-7500(23)00125-5
81. Zhang H, Cisse M, Dauphin YN, and Lopez-Paz DMixup: beyond empirical risk minimization.Proceedings of the International Conference on Learning Representations (ICLR). (2018). doi: 10.48550/arXiv.1710.09412
82. Yun S, Han D, Chun S, Oh SJ, Yoo Y, and Choe J. (2019). CutMix: regularization strategy to train strong classifiers with localizable features, in: 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Korea (South). pp. 6022–31.
83. Ravi Sachin and Larochelle H. Optimization as a model for few-shot learning.Proceedings of the International Conference on Learning Representations (ICLR) (2017). Available online at: https://openreview.net/forum?id=rJY0-Kcll.
84. Pachetti E and Colantonio S. A systematic review of few-shot learning in medical imaging. Artif Intell Med. (2024) 156:102949. doi: 10.1016/j.artmed.2024.102949
85. Meng H, Lin Z, Yang F, Xu Y, and Cui L. (2021). Knowledge distillation in medical data mining: A survey, in: 5th International Conference on Crowd Science and Engineering, New York, USA. pp. 175–82. doi: 10.1145/3503181.3503211
86. Oquab M, Darcet T, Moutakanni T, Maneau J, Ramé M, Henaff M, et al.(2018). DINOv2: Learning robust visual features without supervision.Proceedings of the International Conference on Learning Representations (ICLR), doi: 10.48550/arXiv.2304.07193
87. Zhao Z, Liu Y, Wu H, Wang M, Li Y, Wang S, et al. CLIP in medical imaging: A survey. Med Image Anal. (2025) 102:103551. doi: 10.1016/j.media.2025.103551
88. Aravazhi PS, Gunasekaran P, Benjamin NZY, Thai A, Chandrasekar KK, Kolanu ND, et al. The integration of artificial intelligence into clinical medicine: Trends, challenges, and future directions. Dis Mon. (2025) 71:101882. doi: 10.1016/j.disamonth.2025.101882
89. He J, Baxter SL, Xu J, Xu J, Zhou X, and Zhang K. The practical implementation of artificial intelligence technologies in medicine. Nat Med. (2019) 25:30–6. doi: 10.1038/s41591-018-0307-0
90. Lam TYT, Cheung MFK, Munro YL, Lim KM, Shung D, and Sung JJY. Randomized controlled trials of artificial intelligence in clinical practice: systematic review. J Med Internet Res. (2022) 24:e37188. doi: 10.2196/37188
91. Lee HW, Jin KN, Oh S, Kang SY, Lee SM, Jeong IB, et al. Artificial intelligence solution for chest radiographs in respiratory outpatient clinics: multicenter prospective randomized clinical trial. Ann Am Thorac Soc. (2023) 20:660–7. doi: 10.1513/AnnalsATS.202206-481OC
Keywords: artificial intelligence, pathology, deep learning, high risk factors, lung adenocarcinoma
Citation: Huang Y, Zhao B, Yan R, Zhang C, Geng Z, Mei P, Li K and Liao Y (2025) AI-based prediction of pathological risk factors in lung adenocarcinoma from CT imaging: bridging innovation and clinical practice. Front. Oncol. 15:1687360. doi: 10.3389/fonc.2025.1687360
Received: 17 August 2025; Accepted: 17 October 2025;
Published: 13 November 2025.
Edited by:
Xiaonan Cui, Tianjin Medical University Cancer Institute and Hospital, ChinaReviewed by:
Hanlin Ding, Jiangsu Cancer Hospital, ChinaCopyright © 2025 Huang, Zhao, Yan, Zhang, Geng, Mei, Li and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kuo Li, bGt0ajEyNkAxMjYuY29t; Yongde Liao, MjAxOXhoMDI0OUBodXN0LmVkdS5jbg==
†These authors have contributed equally to this work
 Yu Huang1†
Yu Huang1† Zuhan Geng
Zuhan Geng Yongde Liao
Yongde Liao