- 1Department of Obstetrics and Gynecology, Faculty of Medicine, Universiti Kebangsaan Malaysia, Kuala Lumpur, Malaysia
- 2Faculty of Medicine, Universiti Kebangsaan Malaysia, Kuala Lumpur, Malaysia
- 3Department of Obstetrics and Gynecology, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
Background: Increasing evidence shows the that gut microbiome (GM) plays a crucial role in ovarian cancer (OC) progression, offering potential avenues for microbiome-based intervention strategies. However, research in this area remains limited. This systematic review aimed to synthesize current evidence on microbiome composition and diversity in OC, focusing on its association with disease diagnosis, postoperative changes, and responses to chemotherapy or PARP inhibitor (PARPi) therapy.
Methods: A literature search was performed in PubMed and Web of Science up to October 2025 using keywords: (gut microb* OR gut bacteri* OR intestinal microb* OR intestinal bacteri*) AND (ovarian cancer OR ovarian carcinoma OR carcinoma of ovary). Only original research articles involving human subjects were included. Data on GM alterations in OC patients, postoperative changes, and responses to chemotherapy or PARP inhibitor (PARPi) therapy were extracted and analysed.
Results: Nine eligible studies, comprising longitudinal and case-control studies were reviewed. At diagnosis, OC patients displayed gut dysbiosis characterised by an increase in Proteobacteria and a decrease in Firmicutes. Genus-level analysis revealed lower levels of Akkermansia and elevated levels of Bacteroides and Prevotella, suggesting disrupted microbial homeostasis. Following surgery, both Firmicutes and Proteobacteria declined, indicating significant microbiome shifts. During chemotherapy, especially neoadjuvant treatment, Firmicutes re-emerged as the dominant phylum. Family-level analyses identified increased Coriobacteriaceae and decreased Ruminococcaceae. Platinum-sensitive patients demonstrated more stable GM profiles than those with platinum resistance Genera such as Angelakisella, Arenimonas, and Roseburia emerged as potential candidates for diagnostic or prognostic markers of chemotherapy resistance. Meanwhile, Phascolarbacterium is identified as a PARPi response in BRCA1/2-negative OC, with higher levels linked to longer progression-free survival.
Conclusion: This review highlights a dynamic GM composition in OC across disease stages and treatments, underscoring the need for further research on microbiome-targeted therapeutic strategies.
1 Introduction
Ovarian cancer (OC) is among the most common malignancies worldwide. Although it ranks third in incidence among gynecological cancers, following cervix and uterine cancers, it is the second leading cause of cancer-related mortality in women. According to cancer statistics published in 2022, OC accounts for 324,402 new cases and 206,839 deaths annually (1). Meanwhile, the number of women diagnosed with ovarian cancer is projected to exceed 503,448 by 2050 (1).
OC comprises several subtypes, with epithelial ovarian cancer (EOC) being the most prevalent, accounting for over 90% of cases, followed by germ cell tumors and sex cord-stromal tumors (2). EOC is widely considered a heterogeneous disease. Therefore, it has been classified into five main subtypes based on their clinicopathological features, which are high-grade serous ovarian cancer (HGSOC), low-grade serous carcinoma (LGSC), endometrioid ovarian carcinoma (ENOC), mucinous carcinoma (MC), and clear cell ovarian carcinoma (CCOC). Currently, primary screening methods for OC include histopathological examination, transvaginal ultrasound, and detection of tumor markers such as cancer antigen 125 (CA125) and human epididymis protein 4 (HE4). Survival rates in OC are strongly influenced by the stage at diagnosis, with a 5-year survival rate of approximately 20% in stage IV, 40% in stage III, 70% in stage II, and 90% in stage I (3). However, statistics show that less than 30% of patients are diagnosed at an early stage of OC, largely due to the limitations of current screening methods, which contribute to late diagnosis and poorer outcomes.
In recent years, the role of gut microbiome (GM) in cancer has received significant attention. The GM consists of bacteria, viruses, fungi, archaea, and protozoa in the digestive tract. It has been proven that GM plays a crucial role in maintaining host homeostasis and modulating disease development. The alteration in GM composition, known as dysbiosis, contributes to cancer pathogenesis through interrelated mechanisms involving chronic inflammation, immune modulation, altered estrogen metabolism and microbial metabolites production (4). Dysbiosis can compromise intestinal barrier integrity, allowing bacterial components such as lipopolysaccharides to enter systemic circulation and activate pattern recognition receptors, including Toll-like receptors, thereby triggering persistent inflammation that promotes cellular proliferation, angiogenesis and tumor progression (5, 6). Moreover, GM influences estrogen metabolism by increasing β-glucuronidase-producing bacteria, which enhance estrogen deconjugation and reabsorption, leading to elevated circulating estrogen levels that stimulate hormone-dependent tumor growth (7). Microbial metabolites also play dual roles in tumorigenesis, where secondary bile acids and lactate can promote DNA damage, pro-inflammatory signaling and metastasis, while short-chain fatty acids such as butyrate exhibit anti-inflammatory and tumor-suppressive effects (8, 9). Specific bacterial taxa, including Fusobacterium nucleatum and Escherichia coli have been implicated in promoting DNA damage, immune evasion and inflammatory responses (10, 11), whereas Coriobacteriaceae and Bifidobacterium have been associated with enhanced lactate metabolism, particularly in chemotherapy-resistant malignancies (12). In OC, elevated estrogen levels may bind to estrogen receptors on ovarian epithelial cells, promoting DNA synthesis, cell proliferation, and resistance to apoptosis. Oxidative estrogen metabolites can induce mutagenic DNA damage that facilitates malignant transformation (13).
Cancer patients often exhibit reduced gut bacterial diversity and abundance compared to healthy individuals, suggesting a link between gut dysbiosis and tumorigenesis. Specific bacterial taxa have been associated with the initiation and progression of various malignancies. For example, decreased levels of Ruminococcus 2 in the GM have been correlated with cervical cancer (14). Meanwhile, excessive aggregation of Clostridium nucleatum has been associated with poor prognosis in metastatic colorectal cancer (CRC) (15). In endometrial cancer, Ruminococcus sp. N15.MGS-57 and C16:1 have been identified as potential biomarkers associated with distinct clinical features and outcomes (16). GM imbalance may promote tumorigenesis by altering metabolic functions and estrogen levels, thus influencing the onset and progression of endometrial cancer (17). Beyond carcinogenesis, the GM also modulates host responses to cancer therapy. In patients with metastatic melanoma treated with ipilimumab, enrichment of Faecalibacterium and other Firmicutes was associated with longer progression-free and overall survival, while members of the Bacteroidetes phylum were enriched in patients with colitis resistance (18).
Overall, these findings suggest that the GM may contribute to the development and progression of OC. However, the specific compositions and influence of GM in OC remain poorly understood, particularly regarding disease progression and treatment outcomes. Therefore, this systematic review aims to comprehensively integrate current evidence on alterations in the human microbiome associated with OC or EOC and its treatment outcomes, focusing on microbial composition and diversity. The review includes studies investigating GM alterations in OC broadly, as well as those focusing specifically on EOC, to provide a comprehensive understanding of microbial diversity and its potential associations with disease development and outcomes.
2 Methods
2.1 Search strategy
This systematic literature review complied with guidelines outlined by the Preferred Reported Items for Systematic Reviews and Meta-Analysis (PRISMA) (19). A comprehensive literature search was systematically carried out using the two electronic databases, PubMed and Web of Science, to identify all relevant articles related to GM and OC. Z.B., C.K.T., D.W., and M.N.S. discussed these search strategies for different databases. A systematic literature search was performed until 31st January 2024 and all data were pooled and kept in EndNote software (Version 20) software (Clarivate, UK). Detailed search string for each database, including Boolean operators, MeSH terms, and an applied filter, is presented in Supplementary Table 1. An additional search was conducted on 12 October 2025, using the same search strategy to include any recent articles. The search was limited to English-language articles involving human participants published up to October 2025.
2.2 Inclusion and exclusion criteria
Studies were included based on the following criteria: (i) original research article, (ii) studies on GM and OC, and (iii) human studies. However, studies were excluded if they met any of the following conditions: (i) gray literature included meeting abstracts, editorials, case reports, and review articles, (ii) the study did not involve the GM and OC, (iii) a lack of baseline characteristics data, and (iv) non-English articles or not available. This systematic review included studies focusing on either ovarian cancer in general or specifically on EOC, which is the most common histological subtype.
2.3 Screening of articles for eligibility
Z.B., C.K.T., and D.W. were involved in the data extraction process, with disagreements resolved by a fourth reviewer (M.N.S.). The manual screening of the reference lists of all included articles was conducted to enhance the robustness of the search strategy. After removing all duplicates, these articles’ titles and abstracts were reviewed based on inclusion and exclusion criteria. A data collection form was used to standardize the data collection, and each researcher performed all data extraction independently. Any disagreements were resolved through discussion, with final decisions based on majority consensus. Additionally, records of the reasons for excluding articles were documented for reference.
Our database search identified a total of 439 articles. After 112 duplicates had been removed, 327 articles were left for further screening. A total of 133 articles were chosen for a full-text screening. Following a full-text review, 124 articles were excluded for the following reasons: meeting abstracts and editorials (n = 9), review papers (n = 63), case reports (n = 5), unrelated studies (n = 46), and studies with fewer than 10 cases (n =1). Only nine studies (12, 20–27) met the inclusion criteria and were included in this systematic study. The study flowchart is illustrated in Figure 1.
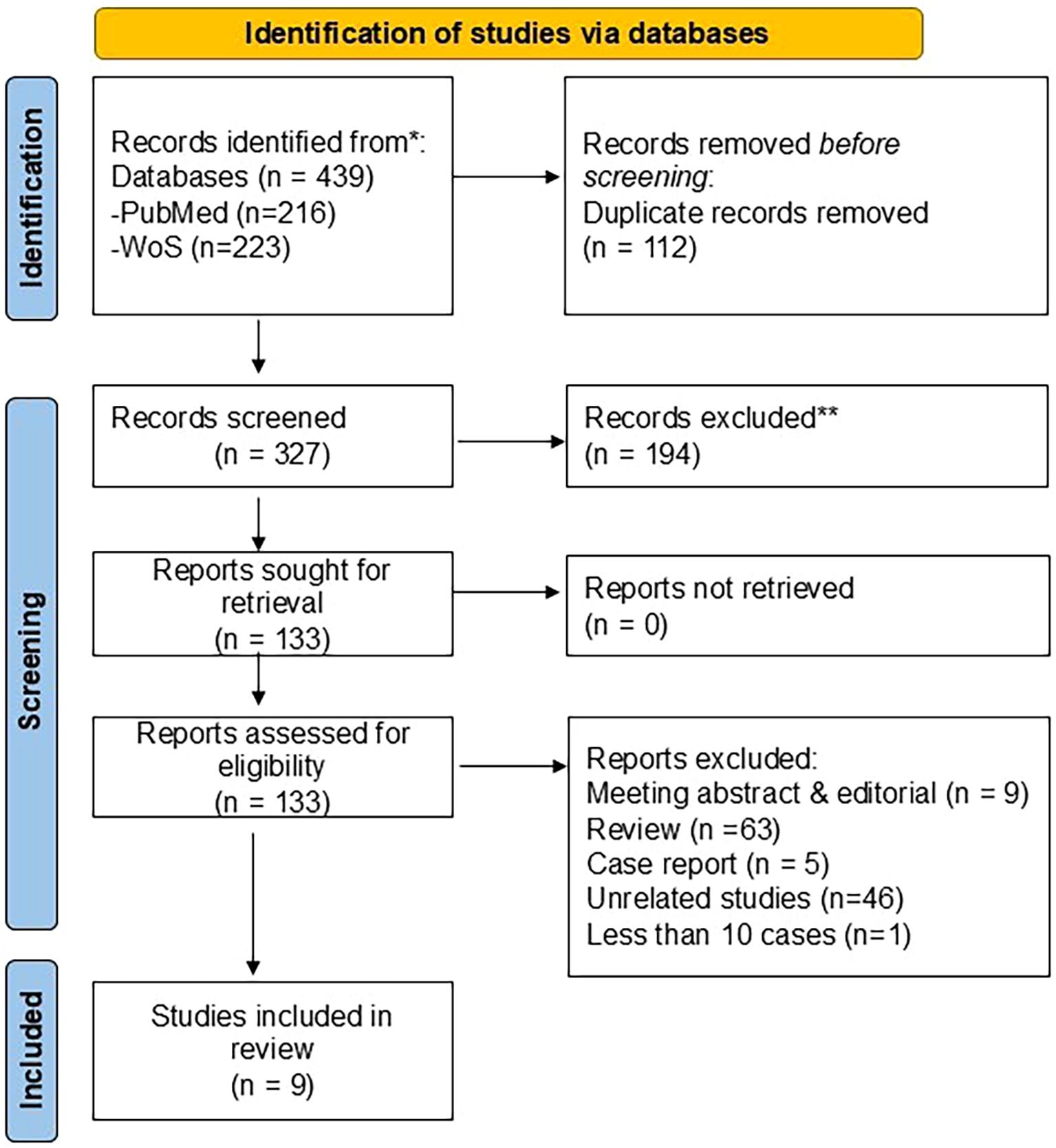
Figure 1. Flowchart of the process of literature search and extraction of studies meeting the inclusion criteria.
2.4 Risk of bias assessment
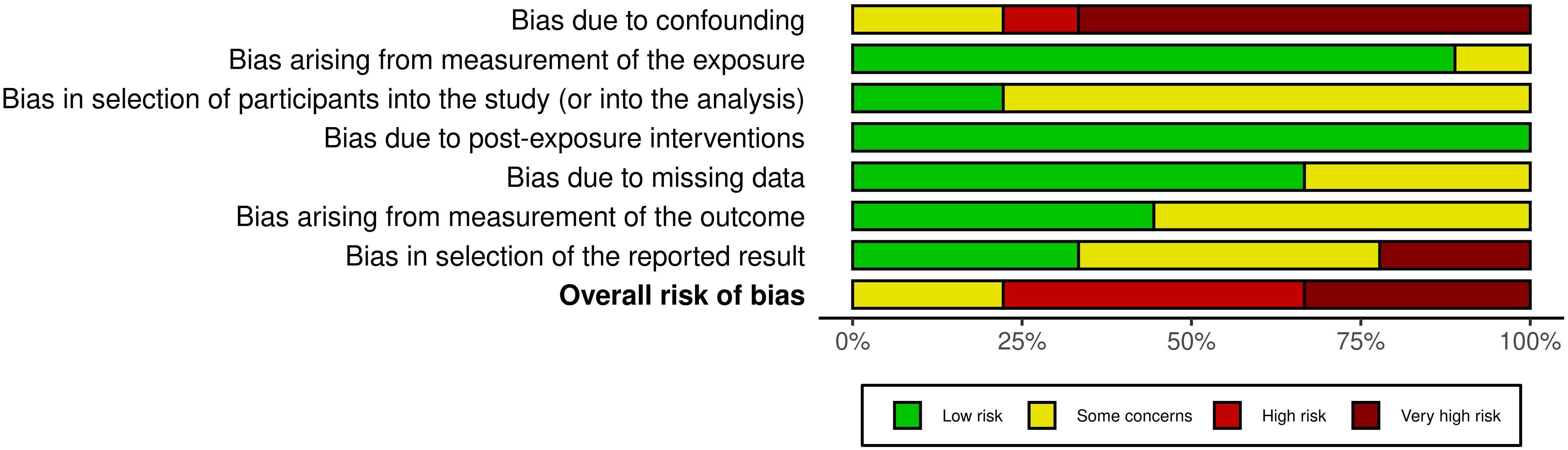
The risk of bias was independently assessed by two authors (Z.B. and N.F.N.M.S.), with any disagreements consulted and resolved by a third reviewer (M.N.S). The Risk of Bias in Non-Randomized Studies of Exposure (ROBINS-E) tool was used to evaluate the included studies (28). This tool assessed bias across seven domains: (1) confounding, (2) selection of participants, (3) measurement of exposure, (4) post-exposure interventions, (5) missing data, (6) measurement of outcomes and (7) selection of the reported result. Each study was rated as “Low”, “Some concerns”, “High” or “Very high” risk of bias. The findings were visualized in a summary plot using the Risk-of-bias VISualization (robvis) (29) to provide a clear overview of the risk of bias in the included studies.
3 Results
3.1 Characteristics of reviewed studies
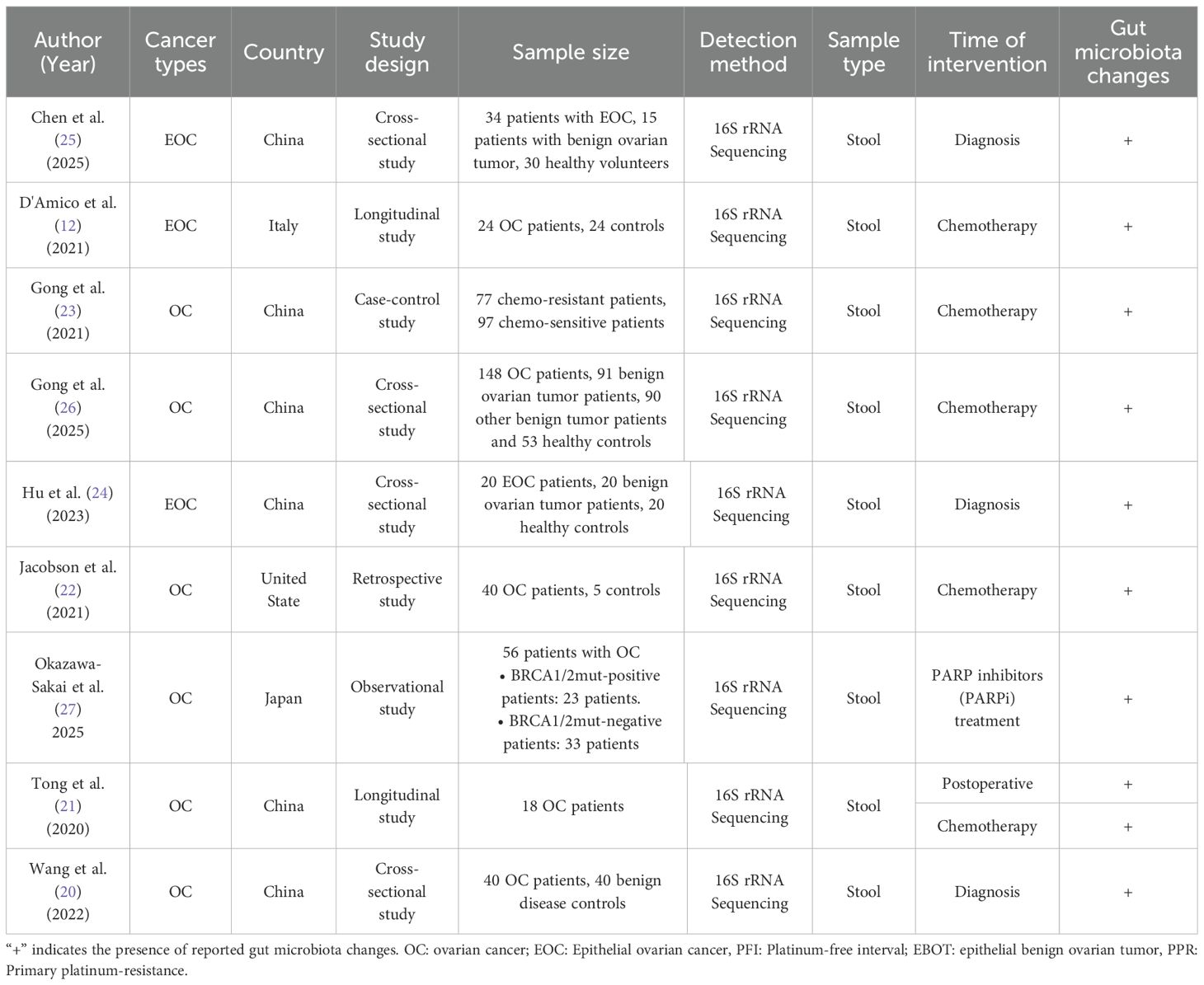
The extracted data from these studies include (1) author and publication year, (2) country of origin, (3) study design, (4) number of sample size, (5) detection method, (6) sample type, (7) time of intervention, and (8) reported GM alteration in OC patients, as summarized in Table 1. All nine studies were published between 2020 and 2025 and were conducted across multiple countries. This review includes cross-sectional (24–26), longitudinal (12), observational (27), retrospective (22) and case-control designs (23). All of these studies used 16S rRNA gene sequencing to profile microbiota in stool samples. Most of the articles included in this review investigated the association between the GM and OC, while only three studies specifically focused on EOC (12, 24, 25).
Across all studies, notable GM alterations were reported. These changes were observed at different clinical time points, including at diagnosis, during the postoperative period, throughout or after chemotherapy and during PARP inhibitor therapy. Three studies specifically compared chemotherapy-sensitive and chemotherapy-resistant patients (12, 22, 23), while one study evaluated the association between GM composition and the efficacy of PARP inhibitors in patients with OC (27). The remaining studies investigated differences in GM composition between OC or EOC, benign tumors, and healthy controls (20, 21, 24–26).
3.2 Risk of bias studies
The risk of bias for the included studies was assessed using the ROBINS-E tool, as shown in Figure 2. Most studies demonstrated a low to some concern risk of bias in the domains related to selection of participants, exposure classification, and missing data (Supplementary Table 2). However, high to very high risk of bias was observed in the domains of confounding and selection of reported results, particularly due to limited adjustment for key covariates, such as diet, antibiotic use or comorbidities. The measurement of the outcomes domain generally showed some concerns because blinding of outcome assessors was seldom reported. Overall, the included studies were rated as some concern to high risk, indicating that the results should be interpreted with caution.
3.3 GM compositions of OC at diagnosis
There was no significant difference in α-diversity between OC or EOC patients and healthy women before surgery, with both groups predominantly harboring Firmicutes, Bacteroidetes, Actinobacteria and, Proteobacteria (12, 20, 21, 24). However, β-diversity showed a significant difference between the two groups.
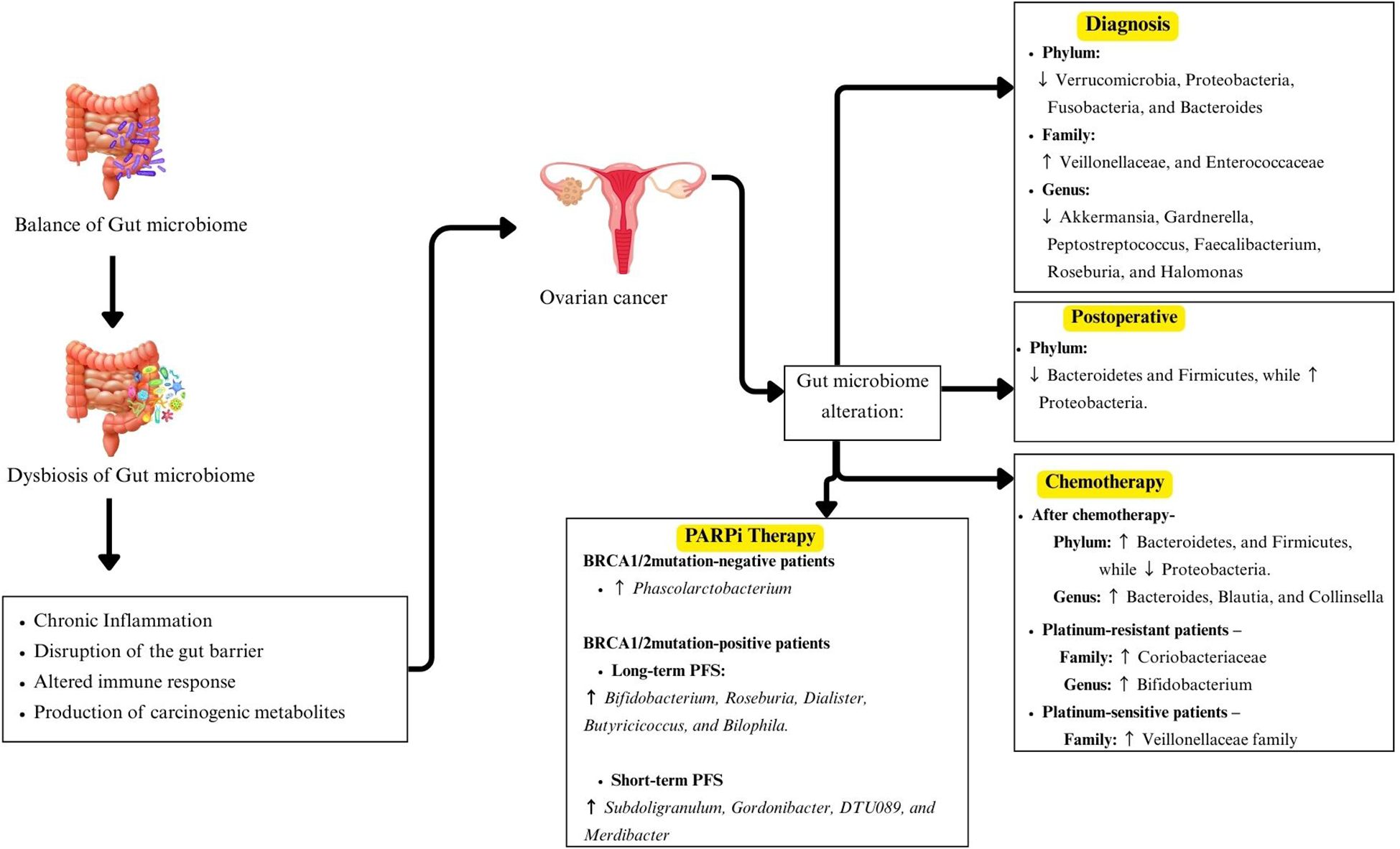
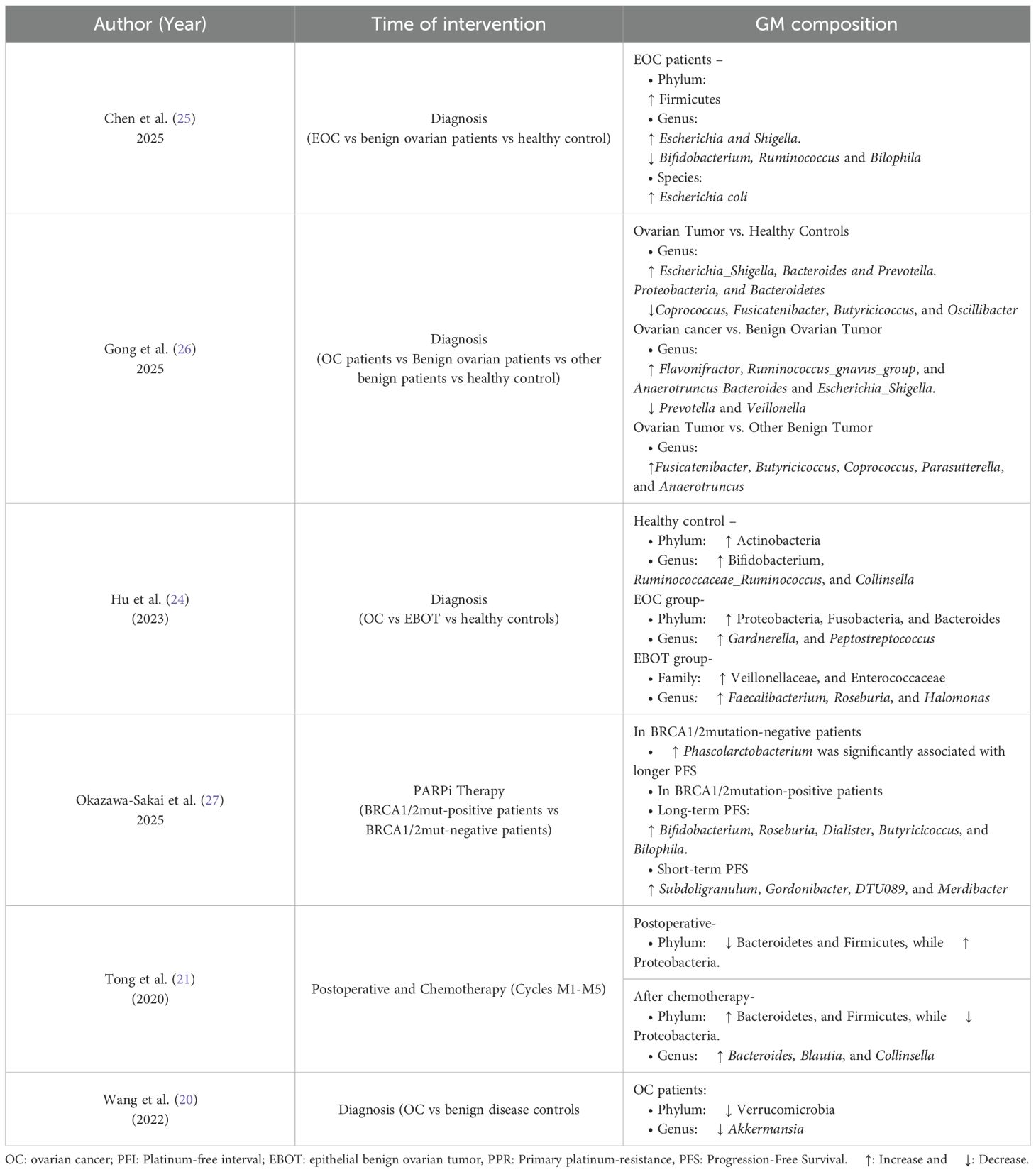
At the phylum level, EOC patients exhibited a significant increase in Proteobacteria, accompanied by a decrease in Firmicutes, Actinobacteria and Verrucomicrobia (20, 21, 24). In advanced EOC cases, the GM profile deviated further from healthy controls, with a more pronounced increase in Proteobacteria and a further decline in Actinobacteria (24). Conversely, Chen et al. (25) reported enrichment of Firmicutes, indicating possible compositional variation across cohorts. At the family level, EOC patients showed reduced abundances of Lachnospiraceae, Bifidobacteriaceae, Clostridiaceae, Rikenellaceae, and Porphyromonadaceae, while Coriobacteriaceae was significantly increased (12).
At the genus level, Akkermansia, the only genus within Verrucomicrobia, significantly declined in OC patients. Linear discriminant analysis (LDA) supported this finding, suggesting that the absence of Akkermansia may be a key microbial signature of OC (20). Additionally, EOC patients had increased levels of Prevotella, Bacteroides, Adlercreutzia, Collinsella, Lactococcus, Lachnobacterium, Escherichia-Shigella and tumor-associated genera such as Flavonifractor, Ruminococcus-gnavus-group and Anaerotruncus, whereas beneficial or probiotic genera including Bifidobacterium, Ruminococcaceae_Ruminococcus, Coprococcus, Blautia, Dorea, Lachnospira, Roseburia, and Bifidobacterium were either reduced or absent in EOC patients (22, 25, 26). At the species level, Escherichia coli was increased in EOC patients with the enrichment of opportunistic genera such as Escherichia and Shigella (25).
3.4 Perioperative changes in GM composition
Only one study has examined GM differences before and after radical surgery in OC patients (21). The study found significant differences in the intestinal microbiota at all taxonomic levels. At the phylum level, Firmicutes was the dominant phylum in preoperative fecal samples (Group B). However, in postoperative samples (Group M0), the dominant phylum shifted to Proteobacteria. Despite this shift, both Firmicutes and Proteobacteria showed a significant overall decrease post-surgery. Additionally, Actinobacteria exhibited a decrease after surgery, although the change was not statistically significant.
The relative abundance of several genera was markedly reduced in Group M0 compared to Group B, including Bacteroides, Bilophila, Faecalibacterium, Collinsella, and Coprococcus. Conversely, Klebsiella, Enterobacter, and Enterococcus were significantly higher in post-surgery. Other genera such as Lachnospiraceae_Ruminococcus, Blautia, Roseburia, Prevotella, and Collinsella also showed decreased trends postoperatively, though these changes were not statistically significant.
To further explore these microbial shifts, Linear Discriminant Analysis Effect Size (LEfSe) analysis was employed. The analysis revealed that Firmicutes and Proteobacteria were dominant phyla in Group B and M0, respectively. At the genus level, Bilophila and Faecalibacterium were key genera of Group B, while Klebsiella and Enterococcus were predominant in Group M0.
3.5 GM compositions associated with targeted therapy (PARP inhibitor)
The composition of the GM may influence the efficacy of PARP inhibitor (PARPi) therapy in EOC patients, potentially affecting progression-free survival (PFS) in a BRCA mutation-dependent manner (27). A higher abundance of Phascolartobacterium was significantly associated with longer PFS in patients without BRCA1/2 mutation. In BRCA1/2 mutation-positive patients, long-term PFS correlated with increased levels of Bifidobacterium, Roseburia, Dialister, Butyricoccus, and Bilophila. Conversely, short-term PFS in this group was associated with higher abundance of Subdoligranulum, Gordonibacter, DTU089 and Merdibacter. Table 2 displayed a summary of reported gut microbiota alterations observed in ovarian cancer patients across all included studies.
3.6 GM compositions associated with chemotherapy
In OC or EOC patients undergoing chemotherapy, the dominant bacterial phyla identified as Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, and Verrucomicrobia (12, 21–23). Among these, Firmicutes consistently remained the most prevalent across different treatment stages (12, 21, 22). Chemotherapy impacted the relative abundance of several phyla, with some fluctuations observed across cycles. For example, in platinum-sensitive (PS) patients receiving neoadjuvant chemotherapy, members of Bacteroidetes, such as Bacteroides, Prevotella, and Parabacteroides, were notably enriched (12).
At the family level, significant changes were observed following chemotherapy. Coriobacteriaceae increased progressively during treatment, particularly in platinum-resistant (PR) patients, and was associated with lower survival probability. This family includes genera such as Eggerthella and Collinsella. In contrast, Veillonellaceae was more abundant in PS patients and linked to improved survival outcomes. Other key families affected by chemotherapy included Lachnospiraceae, Enterobacteriaceae, Bifidobacteriaceae, Lactobacillaceae, Erysipelotrichaceae, Ruminococcaceae, and Mogibacteriaceae, all of which showed treatment-related changes in relative abundance.
At the genus level, the dominant or frequently observed genera during chemotherapy included Lachnospiraceae_unclassified, Blautia, Ruminococcus, Clostridiaceae_Unclassified, Coriobacteriaceae, Enterobacteriaceae_unclassified, Bifidobacterium, Bacteroides, Faecalibacterium, Collinsella, Bilophila, Coprococcus, Klebsiella, Enterobacter, Enterococcus, Veillonella, Akkermansia, Prevotella, and Lactobacillaceae_unclassified (12, 21, 23). Chemotherapy led to a significant reduction in the relative abundance of certain genera, such as Enterobacteriaceae_unclassified, Klebsiella, and Enterobacter (21). Conversely, other genera, including Bacteroides, Blautia, Collinsella, Bilophila, Faecalibacterium, and Coprococcus showed increased abundance during treatment (12, 21, 23). Interestingly, some genera (Enterobacteriaceae_unclassified, Klebsiella, Enterobacter, Bilophila, Enterococcus, Coprococcus, Veillonella, Bifidobacterium, Akkermansia and Lactobacillaceae_unclassified) returned to near pre-chemotherapy levels over multiple treatment cycles (21).
Differences in GM composition were also observed between PR and PS patients in EOC (12). PR patients had significantly higher in Coriobacteriaceae, especially Eggerthella and Bifidobacterium, while PS patients exhibited increased levels of Veillonellaceae, Catenibacterium, and Anaerotruncus (12). These compositional shifts were associated with clinical outcomes, with higher Coriobacteriaceae levels correlating with reduced survival and higher Veillonellaceae levels with improved survival (12). The type of chemotherapy further influenced microbial profiles. In PS patients receiving adjuvant chemotherapy, enriched taxa included Bacteroides, Prevotella, and Parabacteroides, Faecalibacterium, and Acidaminococcus, while PS patients on neoadjuvant chemotherapy had higher levels of Desulfovibrio, Paraprevotella, Anaerostipes, Sutterella, and Pseudoramibacter_Eubacterium. Notably, in neoadjuvant-treated patients, Coriobacteriaceae abundance increased over time, while Ruminococcaceae, especially Faecalibacterium and Ruminococcus, decreased (12).
Longitudinal profiling using Spearman Correlation Analysis across five chemotherapy cycles (M1-5) revealed stage-specific dominant genera. Before chemotherapy, Corynebacterium and Klebsiella were dominant. During the cycles, Erysipelotrichaceae_Unclassified and Ruminococcus were most relevant in M1; Clostridiaceae_Unclassified and SMB53 in M2; Lactococcus and Prevotella in M3, Prevotella and Rothia in M4; and Gemella and Mogibacteriaceae_Unclassified in M5 (21). The correlation coefficients for these associations were higher (R values = 0.89 to 0.99), indicating strong temporal relationships. According to the random forest model identified Angelakisella, Arenimonas, and Roseburia were identified as the top three most important predictors associated with OC resistance (23). Chemotherapy-related alterations in GM composition and their clinical associations are summarized in Table 3.
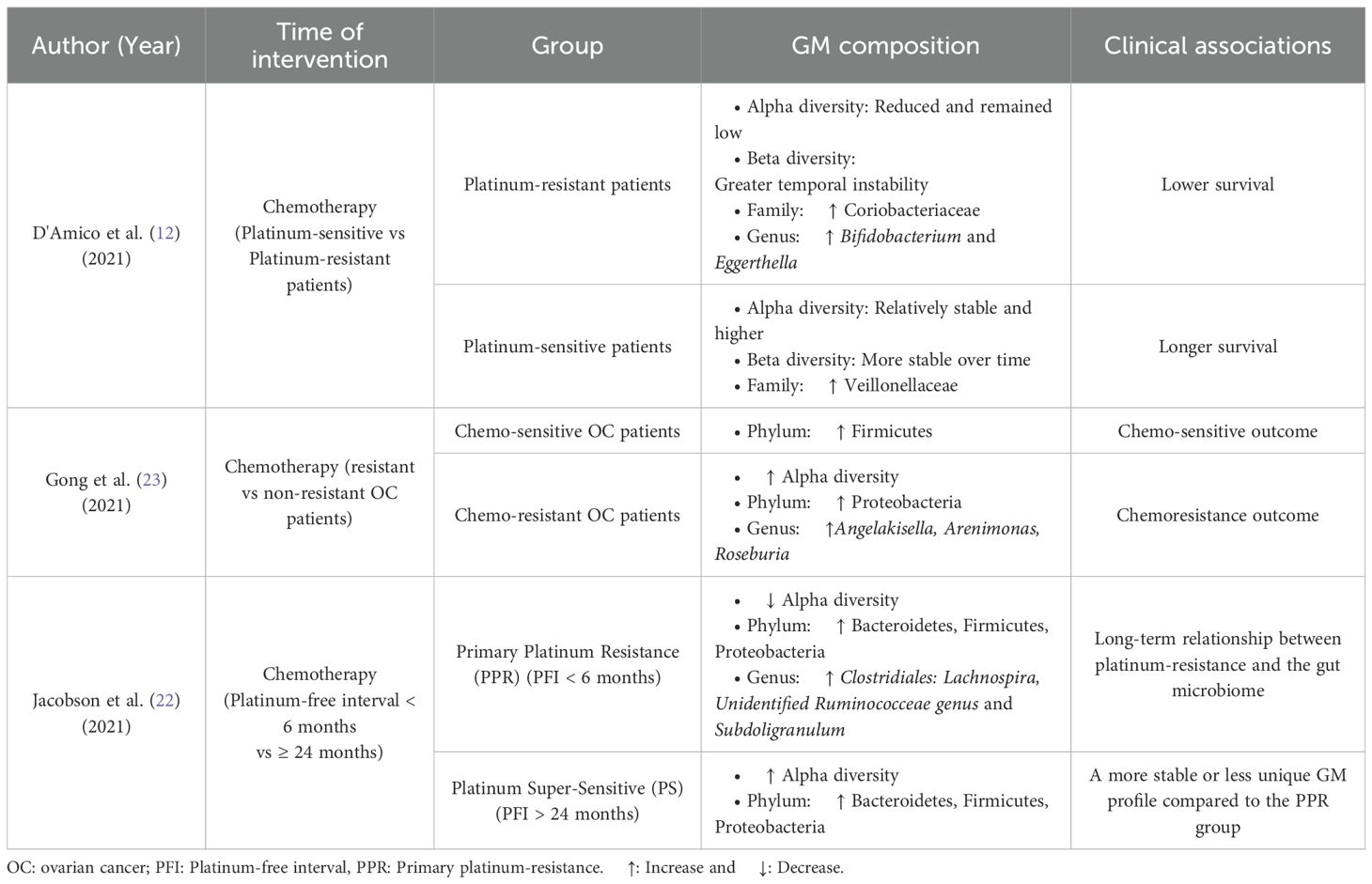
Table 3. Summary of gut microbiota (GM) compositional changes and clinical associations in ovarian cancer patients following chemotherapy.
4 Discussion
4.1 GM compositions of OC at diagnosis
This systematic review identified nine original research articles that examined GM compositions in stool samples from OC or EOC patients. Despite a broadly similar composition of dominant microbial phyla between OC patients and healthy individuals, several key changes in features were consistently observed. Notably, a significant increase in the relative abundance of Proteobacteria and a marked decrease in Firmicutes were observed in OC patients. This indicates a clear shift in microbial equilibrium. Similar results are consistent with a meta-analysis by Pammi et al. (30), which analyzed 14 studies. They concluded that GM dysbiosis preceding necrotizing enterocolitis in preterm infants was characterized by increased relative abundances of Proteobacteria and a decreased relative abundance of Firmicutes and Bacteroidetes. Similarly, the stage and subtype of EOC appear to exhibit a significant impact on GM composition. This includes reduced microbial diversity, as indicated by lower alpha diversity indices, and compositional shifts such as an increased abundance of Proteobacteria and decreased levels of Actinobacteria, Bifidobacterium, and Ruminococcaceae_Ruminococcus.
Mechanistically, the overrepresentation of Proteobacteria, a phylum of Gram-negative bacterium that includes pathogenic genera like Salmonella, helicobacter, Enterobacter and Escherichia (31), has been strongly linked to immune dysregulation, and carcinogenesis (32, 33). Lipopolysaccharide (LPS) derived from Proteobacteria activates Toll-like receptor 4 (TLR4), triggering the TLR4/MyD88/NF-Кβ signaling cascade (34). These conditions can promote pro-inflammatory cytokine release (IL-1β, IL-6 and TNF-α), angiogenesis and tumor proliferation (35). Persistent TLR4 activation enhances immune evasion and epithelial-mesenchymal transition (EMT), processes associated with OC progression and metastasis (36). Increased Proteobacteria has also been linked to oxidative stress and production of reactive oxygen species (ROS), which can induce DNA damage and activate oncogenic pathways such as PI3K/Akt and STAT3, further emphasizing tumor-promoting conditions (37, 38). Elevated Proteobacteria levels have also been significantly associated with other malignancies, including thyroid cancer, as demonstrated in both a systematic review and a Mendelian randomization analysis (39). Overall, an elevated abundance of Proteobacteria may reflect GM imbalance and may be associated with disease development and progression.
In contrast, the decline of beneficial taxa such as Akkermansia muciniphila, a mucin-degrading bacterium from the phylum Verrucomicrobiota, may compromise intestinal integrity and immunological balance (40). A. muciniphila maintains mucosal barrier function and produces beneficial short-chain fatty acids (SCFAs), including acetate and propionate, which exhibit anti-inflammatory effects via G-protein-coupled receptor (GPR43/109A) activation and suppression in NF-Кβ signaling (41). Besides, A. muciniphila has been shown to exhibit anti-tumor effects by activating and interacting with dendritic cells, which trigger the production of interleukin-12 (IL-12) (42). This process promotes the activation of cytotoxic T lymphocytes (CD8+), thereby enhancing the body’s immune response against tumors. Clinical studies have further associated its increased abundance with improved metabolic health, reduced atherosclerotic risk (43), enhanced GM diversity during weight loss (44), and protection against liver injury (45). Hence, reduced A. muciniphila levels have been associated with decreased mucus layer thickness, increased intestinal permeability and endotoxin leakage (40). The endotoxin leakage is a conducive condition to systemic inflammation and a tumor-promoting microenvironment.
OC patients also showed lower levels of certain bacterial families, such as Lachnospiraceae and Clostridiaceae (from the Firmicutes phylum), and Rikenellaceae and Porphyromonadaceae (from the Bacteroidetes phylum). This differs from the usual pattern observed in healthy individuals, where Firmicutes and Bacteroidetes are the most dominant and balanced groups in the GM (46). The depletion of these taxa may reflect impaired SCFA biosynthesis particularly of butyrate. Butyrate acts as a histone deacetylase (HDAC) inhibitor, promoting tumor suppressor gene expression, cell cycle arrest and apoptosis in malignant cells. Therefore, the loss of butyrate-producing bacteria such as Lachnospiraceae and Clostridiaceae removes an essential anti-tumor regulatory mechanism (47). This disruption promotes a pro-oncogenic niche associated with low-grade inflammation and immune dysregulation.
In addition, Bifidobacteriaceae, a well-known probiotic family, also exhibited variation, while a surprising increase in Bacteroides was observed. In another study, Bacteroides thetaiotaomicron is one of the species of Bacteroides is recognized for its role in carbohydrate fermentation and mucin degradation (48). It can disrupt mucosal integrity when overgrown, particularly under antibiotic pressure, leading to bacterial translocation, and increasing inflammation as seen in -versus-host disease (49). Other species of the Bacteroides have been implicated in systemic inflammatory disease, including malaria (50) and polycystic ovary syndrome (Bacteroides vulgatus) (51). Furthermore, alterations in Bacteroides abundance may influence cytokine signaling, as seen in colorectal cancer, where an inverse correlation was noted between Bacteroides spp. and interleukin 9 (IL-9) levels (52). Hence, these findings indicate that the depletion of SCFA-producing taxa together with the enrichment of Bacteroides disrupts intestinal immune homeostasis and may contribute to the pro-inflammatory microenvironment that facilitates ovarian tumorigenesis.
An increased abundance of Prevotella was another notable finding in OC patients. This genus has been previously associated with non-small cell lung cancer (53) as well as pre-hypertension and hypertension (54). Although Prevotella is a commensal organism, it has pathogenic potential, especially in the context of female genital tract infections, including endometritis, bacterial vaginosis, and chorioamnionitis (55). Mechanistically, Prevotella overgrowth activates the TLR2/TLR4 signaling pathway, leading to the upregulation of pro-inflammatory cytokines such as interleukin-1β (IL-1β), interleukin-6 (IL-6), and interleukin-23 (IL-23) (56). Furthermore, the secretion of virulence factors including ammonia, hydrolases, and sialidase, enhances mucin degradation, bacterial adherence, and impairs host immune defenses (55). These immunopathogenic mechanisms suggest a potential role for Prevotella in promoting epithelial invasion, immune evasion and oncogenic inflammation within the female reproductive tract, thereby contributing to the pathophysiology of OC.
Collinsella, another genus elevated in OC patients, has shown similar patterns of dysbiosis in conditions such as autism spectrum disorders (57) and childhood-onset asthma (17). Collinsella has been associated with increased intestinal permeability and enhanced disease severity, as demonstrated in arthritis models (58). However, some evidence suggests a potentially beneficial role for Collinsella aerofaciens, which was found in higher abundance among responders to anti-PD 1-based immunotherapy in metastatic melanoma (59). Interestingly, Collinsella and other genera such as Roseburia, Blautia, and Lachnospiraceae-unclassified were found to be depleted in adolescent depression but restored following sertraline treatment, with Roseburia in particular exhibiting high predictive potential for therapeutic response (60). While Roseburia is decreased in OC, it was found to be enriched in NSCLC, further illustrating the disease-specific context of GM shifts (53).
Besides, the genus Ruminococcus was significantly reduced in OC patients, a trend similarly observed in individuals with major depressive disorder, infantile cholestasis and cardiovascular diseases (61). Ruminococcus-dominated enterotypes are enriched in Ruminococcus and Akkermansia, play an important role in breaking down complex carbohydrates and supporting the gut’s protective barrier (62). Mechanistically, Ruminococcus species contribute to gut homeostasis through the fermentation of dietary polysaccharides and resistant starch into SCFAs, particularly butyrate (63). Butyrate serves as a primary source for colonocytes and enhances mucosal integrity. Moreover, Ruminococcus may facilitate mucin degradation and cross-feeding interactions with other commensals, thereby maintaining microbial diversity and epithelial health (64). Its depletion may be a signal of a loss of metabolic and structural elasticity within the gut ecosystem.
4.2 GM compositions of OC after surgery
There is growing evidence that surgical interventions have a significant impact on GM. In line with observations in OC, a study by Yu et al. (65) found that Klebsiella levels increased post-bariatric surgery, whereas beneficial genera such as Bacteroides, Coprococcus, and Faecalibacterium significantly declined. Moreover, they reported a notable rise in alpha diversity, particularly at the 3 months after surgery, accompanied by proliferation of Streptococcus, Akkermansia, and Prevotella. These genera are often associated with mucosal health and metabolic regulation.
In line with these findings, Özdemir et al. (66) observed a significant enhancement in GM alpha diversity in individuals with morbid obesity following bariatric surgery, eventually reaching levels similar to non-obese controls by 6 months post-operation. At the phylum level, there was a marked increase in Bacteroidetes and a concurrent reduction in Firmicutes, resulting in a significant reduction in the Firmicutes/Bacteroidetes ratio. This ratio is often associated with improved metabolic outcomes. Notably, genera such as Lactobacillus and Bifidobacterium decreased, while Akkermansia, a mucin-degrading bacterium linked to gut barrier integrity, significantly increased.
Ben Izhak et al. (67) also showed an increase in the phyla Proteobacteria and Fusobacteria abundance, and a decrease in Firmicutes after bariatric surgery. At the class level, Beta- and Gamma- proteobacteria were dramatically elevated, underscoring a shift toward potentially pro-inflammatory microbial populations.
Beyond metabolic surgeries, other surgical procedures such as appendectomy have also been impacted in long-term GM alterations and disease risk. A 20-year longitudinal study by Shi et al. (68) revealed that individuals who underwent appendectomy had a 73% increased risk of developing colorectal cancer (CRC). This was accompanied by an enrichment of seven CRC-promoting bacteria, including Bacteroides fragilis and a reduction in five commensal species. Interestingly, Fusobacterium nucleatum, a well-known CRC-associated species, was decreased after radical surgery in CRC patients. However, Clostridium scindens, a bile acid-transforming species linked to carcinogenesis via production of deoxycholate (DCA), along with upregulation of its associated bile acid-inducing genes (bai operon) (69). These findings highlight the substantial and surgery-specific reshaping of the GM. Such shifts not only reflect physiological adaptation but may also influence postoperative outcomes, including immune response modulation, metabolic improvement, and even oncogenesis.
4.3 GM compositions associated with targeted therapy
The study identified a specific GM, Phascolarctobacterium, whose high baseline abundance was significantly associated with longer PFS in OC patients receiving PARPi maintenance therapy, especially among those who were BRCA1/2 mutation-negative (27). Phascolarbacterium is a commensal bacterium known to produce SCFAs, particularly propionate and acetate (70). These SCFAs can enhance CD8+ T cells (71), which aligns mechanistically with PARPi-induced activation of the DNA-sensing type 1 interferon pathway (71, 72). The presence of Phascolarbacterium may synergistically enhance immunostimulatory effects of PARPi therapy, improving tumor control and delaying progression. These findings highlight the potential utility of GM profiling as a predictive biomarker for PARPi response, particularly in BRCA1/2 mutation-negative patients. The consistent association of Phascolarbacterium abundance with favorable outcomes suggests that microbiome modulation through dietary strategies, probiotic supplementation or FMT may enhance therapeutic efficacy.
In contrast, among BRCA1/2 mutation-positive patients, Linear Discriminant Analysis Effect Size (LEfSe) identified several SCFA-producing genera, including Bifidobacterium, Roseburia, Dialister, Butyricicoccus, and Bilophila, that were enriched in long-term responders. These bacteria are known for their immunomodulatory and anti-inflammatory functions, which may support a favorable response to therapy (73, 74). Meanwhile, higher abundances of Subdoligranulum, Gordonibacter, DTU089, and Merdibacter were associated with shorter PFS, suggesting potential roles as resistance-associated taxa, although this requires further validation.
4.4 GM composition associated with chemotherapy
Although research on the impact of chemotherapy on the GM is limited, available evidence highlights its significant impact on GM composition and diversity. Post-chemotherapy changes often mirror microbial imbalances observed at diagnosis. For instance, elevated levels of Bacteroides and Collinsella were reported after chemotherapy, reflecting their resilience or proliferation in response to treatment-induced perturbations. Similarly, a study in CRC patients demonstrated an increase in Bacteroides plebeius following chemotherapy, while another study reported minimal changes in Bacteroides and bifidobacteria (75). In contrast, Stringer et al. (76) observed that chemotherapy-induced diarrhea in cancer patients was associated with reduced levels of Bacteroides, Bifidobacterium, Lactobacillus, and Enterococcus, alongside increased abundance of opportunistic pathogens such as Escherichia coli and Staphylococcus spp. reflecting a shift towards a dysbiosis state.
In OC, PS patients displayed reduced GM diversity, with a notable increase in Coriobacteriaceae, Bifidobacterium, and Eggerthella (12). Coriobacteriaceae and Bifidobacterium are known as lactate-producing bacteria (77). Elevated lactate levels contribute to tumor progression via the Warburg effect. This effect is a metabolic reprogramming in cancer cells characterized by aerobic glycolysis and lactate overproduction. This metabolic shift triggers tumor growth, immune evasion, angiogenesis, and metastasis and reduced chemotherapeutic efficacy (77). Furthermore, Coriobacteriaceae has been affected in the promotion of colorectal tumorigenesis in high-fat diet-fed mice and has shown enrichment in acute myeloid leukemia, tuberculosis, and carcinoid syndrome patients (78–80). Coriobacteriaceae correlates with elevated of lipid-associated markers such as hydroxypropyl-hydroxyproline, prolyltyrosine, tyrosyl-proline, total cholesterol, and low-density lipoprotein-1 (LDL-1), and low-density lipoprotein-2 (LDL-2) (79). Interestingly, a contradictory role was reported in allergic rhinitis, where Coriobacteriaceae showed a potential protective effect (81).
Eggerthella, another genus overrepresented in PS OC patients, has been consistently linked with various conditions, including psychiatric disorders (major depressive disorder, bipolar disorder, psychosis and schizophrenia), autoimmune disease (Crohn’s disease, ulcerative colitis and rheumatoid arthritis), and multiple sclerosis, reflecting its potential role in systemic inflammation and host-microbiome interactions (82).
In contrast, Veillonellaceae was found to be enriched in PS OC patients, suggesting a potentially beneficial role. This family comprises lactate-utilizing bacteria, which may reduce excess lactate accumulation in the tumor microenvironment. Veillonella atypica, for example, metabolizes lactate to propionate. Scheiman et al. (83) demonstrated that V. atypica supplementation improved athletic performance in marathon runners, and similar mechanisms may support host resilience during chemotherapy. However, increased levels of Veillonellaceae have also been associated with a higher intrahepatic cholangiocarcinoma.
Notably, both Faecalibacterium and Ruminococcus were key genera associated with gut barrier integrity and anti-inflammatory function and found to decrease in OC patients undergoing neoadjuvant chemotherapy (12). Faecalibacterium prausnitzii, in particular, is considered a hallmark of gut health, and its reduction has been linked to inflammatory diseases and postoperative Crohn’s disease recurrence. In vitro studies show that Faecalibacterium prausnitzii can suppress the production of proinflammatory cytokines such as interleukin-8 (IL-8) and inhibit the nuclear factor kappa beta (NF-Кβ) signaling pathway, thereby exerting immunomodulatory effects (84). However, a different species, Fusobacterium nucleatum, was found to be enriched in relapsed CRC patients post-chemotherapy. Fusobacterium nucleatum has been affected in chemoresistance by activating the autophagy pathway and modulating host responses through the TLR4-MYD88 axis and specific microRNA signaling, thus impacting therapeutic efficacy (85).
These microbial changes are closely linked with clinical parameters. Shorter survival correlates with higher Bifidobacterium, Megamonas, and Pseudomonas, whereas longer survival associates with lower Klebsiella and Fusobacterium abundance (21). Random forest modeling demonstrates that GM profiles can predict chemotherapy response with an AUC of 0.909, underscoring the prognostic potential of GM signatures (23). Furthermore, taxa such as Ruminococcus, Desulfovibrio, and Lactobacillus are associated with gastrointestinal adverse effects, highlighting the dual role of GM in therapeutic outcomes and patient quality of life (21).
The higher abundance of Roseburia from the Firmicutes phylum has emerged as a notable microbial signature in chemotherapy-resistant OC patients, even though Roseburia is typically regarded as a beneficial commensal that supports gut health. Mechanistically, Roseburia produces SCFAs, particularly butyrate, which modulate host immune responses by promoting regulatory T cell (Treg) expansion and exhibiting anti-inflammatory effects (73, 86). While these actions are generally protective, during cancer treatment, they might unintentionally weaken the body’s anti-tumor response. This can allow cancer cells to survive and make chemotherapy less effective. Metabolites produced by Roseburia can also affect cellular metabolism and oxidative stress, potentially reducing the levels of ROS that are crucial for chemotherapy-induced cancer cell death (87). Therefore, higher Roseburia levels in platinum-resistant patients may indicate a gut-driven immune and metabolic change that promotes resistance, highlighting its potential as a biomarker for treatment response and a target for microbiome-based therapy.
Overall, chemotherapy-induced shifts in GM are multifaceted and potentially bidirectional, either attenuating and exacerbating treatment outcomes depending on the microbial signatures involved.
4.5 Clinical significance between OC and GM
The observed changes in GM composition across disease stages and treatment phases highlight the potential clinical significance of the microbiome in OC. Specific taxa such as Proteobacteria, Prevotella, and Collinsella, which are enriched in OC, may serve as microbial signatures indicative of dysbiosis, systemic inflammation, and tumor-promoting conditions. In contrast, the reduction of beneficial taxa like Akkermansia muciniphila, Faecalibacterium Prausnitzii, and Ruminococcus underscores a loss of gut barrier integrity and anti-inflammatory capacity, which may influence disease progression and treatment outcomes.
These microbial shifts may have diagnostic and prognostic implications, as they could be developed into non-invasive biomarkers for early detection or treatment response. Besides, modulation of GM through probiotics, dietary intervention, or fecal microbiota transplantation (FMT) may present a promising therapeutic strategy to enhance chemotherapy efficacy, reduce toxicity and improve overall patient outcomes. Future longitudinal and interventional studies are warranted to validate these associations and to explore the translational potential of microbiome-targeted strategies in clinical oncology.
4.6 Limitations
This review has several limitations that should be considered. Firstly, individual differences in diet, genetic background and environmental exposure were not adequately addressed in the included studies. These factors are well-established modulators of microbial diversity and function and may have contributed to the heterogeneity of findings observed between studies. Their omission limits the ability to attribute microbiome alteration solely to OC or its treatment effects.
Secondly, this review included only nine studies, which is a relatively small number for a systematic review. Moreover, most studies had small sample sizes and were geographically concentrated, which may limit the generalizability of the findings. The small evidence base restricts the strength of conclusions that can be drawn and highlights the need for more well-designed, multi-center investigations in this field.
Thirdly, there was notable heterogeneity in sequencing platforms, bioinformatic pipelines, and analytical methods used across studies, which may have influenced the reported taxonomic and functional outcomes. Consequently, the certainty of evidence remains low, and current findings should be interpreted as hypothesis-generating rather than definitive.
Fourthly, the findings of this systematic review should be interpreted in light of the methodological limitations of the included studies. Several studies were rated as having some concerns to very high risks of bias, especially in participant selection, selection of reported result, confounding control, and outcome measurement. These biases may have affected the internal validity and overall strength of the synthesized evidence, potentially leading to over- or underestimation of associations between GM profiles and OC characteristics. Therefore, the conclusions should be interpreted with caution, and future research with more rigorous and standardized designs are needed to validate these findings.
Fifthly, these review protocols were not prospectively registered in a public database such as PROSPERO. Although every effort was made to conduct the review systematically and meticulously. We recognize this limitation and propose that future reviews register techniques in advance to improve transparency and minimize possible bias.
Finally, an important gap in the literature is the limited exploration of how GM composition may relate to disease prognosis, including its potential impacts on disease progression, treatment response, and long-term survival outcomes in OC patients. Given that specific microbiota profiles may influence tumor microenvironment modulation, immune response and systemic inflammation, this represents a promising avenue for future research aimed at identifying prognostic microbiome biomarkers and therapeutic targets.
5 Conclusion
In conclusion, this review highlights emerging evidence suggesting that alterations in GM composition may play a role in OC (Figure 3). Several studies report a relative increase in Proteobacteria and a reduction in Akkermansia, potentially indicating an association with disease risk and progression. However, these observations remain preliminary and inconclusive. Differences in GM profiles between OC patients and healthy individuals appear more distinct with advanced disease stages, and chemotherapy seems to induce notable shifts in microbial composition, with Firmicutes often becoming dominant. Some taxa, including Angelakisella, Arenimonas and Roseburia, have been proposed as potential biomarkers for future investigation. But their diagnostic or prognostic efficacy remains to be validated. Moreover, recent evidence indicates that a higher abundance of Phascolartobacterium is significantly associated with improved progression-free survival in BRCA1/2 mutation-negative patients undergoing PARPi therapy. This finding suggests that Phascolarctobacterium may serve as a promising predictive biomarker for treatment response, especially in guiding PARPi therapy.
These observations highlight the potential role of microbiome-targeted therapies, such as probiotics, prebiotics, or faucal microbiota transplantation, as complementary strategies in OC management. However, the evidence is currently limited by small sample sizes, methodological heterogeneity, and unaddressed confounding factors across studies. Further well-designed, longitudinal research is needed to clarify causal relationships, validate microbial biomarkers, and determine whether personalized microbiome-based intervention can improve treatment outcomes in OC patients.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
ZB: Conceptualization, Data curation, Formal analysis, Investigation, Writing – original draft. NFNMS: Formal analysis, Investigation, Writing – review & editing. CKT: Writing – review & editing. DW: Writing – review & editing. MNS: Conceptualization, Data curation, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, and/or publication of this article.
Acknowledgments
The authors would like to express their deepest appreciation to all the parties who contributed to the preparation of this systematic review paper.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. The authors used an AI-based language editing tool (ChatGPT, OpenAI, USA) to improve readability and grammar. All substantive revisions were made and verified by the authors.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1690541/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Höhn AK, Brambs CE, Hiller GGR, May D, Schmoeckel E, and Horn LC. 2020 WHO classification of female genital tumors. Geburtshilfe Frauenheilkd. (2021) 81:1145–53. doi: 10.1055/a-1545-4279
3. Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, et al. Ovarian cancer statistic. CA Cancer J Clin. (2018) 68:284–96. doi: 10.3322/caac.21456
4. Zhang B, Mohd Sahardi NFN, Di W, Long X, and Shafiee MN. The gut–endometrium axis: exploring the role of microbiome in the pathogenesis and treatment of endometrial cancer—A narrative review. Cancers. (2025) 17:1044. doi: 10.3390/cancers17061044
5. Wang R, Yang X, Liu J, Zhong F, Zhang C, Chen Y, et al. Gut microbiota regulates acute myeloid leukaemia via alteration of intestinal barrier function mediated by butyrate. Nat Commun. (2022) 13:2522. doi: 10.1038/s41467-022-30240-8
6. Candelli M, Franza L, Pignataro G, Ojetti V, Covino M, Piccioni A, et al. Interaction between lipopolysaccharide and gut microbiota in inflammatory bowel diseases. Int J Mol Sci. (2021) 22:6242. doi: 10.3390/ijms22126242
7. Hu S, Ding Q, Zhang W, Kang M, Ma J, and Zhao L. Gut microbial beta-glucuronidase: a vital regulator in female estrogen metabolism. Gut Microbes. (2023) 15:2236749. doi: 10.1080/19490976.2023.2236749
8. Zeng H, Umar S, Rust B, Lazarova D, and Bordonaro M. Secondary bile acids and short chain fatty acids in the colon: A focus on colonic microbiome, cell proliferation, inflammation, and cancer. Int J Mol Sci. (2019) 20:1214. doi: 10.3390/ijms20051214
9. Di Lorenzo A, Bolli E, Tarone L, Cavallo F, and Conti L. Toll-like receptor 2 at the crossroad between cancer cells, the immune system, and the microbiota. Int J Mol Sci. (2020) 21:9418. doi: 10.3390/ijms21249418
10. Wu J, Li Q, and Fu X. Fusobacterium nucleatum contributes to the carcinogenesis of colorectal cancer by inducing inflammation and suppressing host immunity. Transl Oncol. (2019) 12:846–51. doi: 10.1016/j.tranon.2019.03.003
11. Li S, Liu J, Zheng X, Ren L, Yang Y, Li W, et al. Tumorigenic bacteria in colorectal cancer: mechanisms and treatments. Cancer Biol Med. (2022) 19:147–62. doi: 10.20892/j.issn.2095-3941.2020.0651
12. D'Amico F, Perrone AM, Rampelli S, Coluccelli S, Barone M, Ravegnini G, et al. Gut microbiota dynamics during chemotherapy in epithelial ovarian cancer patients are related to therapeutic outcome. Cancers (Basel). (2021) 13:3999. doi: 10.3390/cancers13163999
13. Kozieł MJ and Piastowska-Ciesielska AW. Estrogens, estrogen receptors and tumor microenvironment in ovarian cancer. Int J Mol Sci. (2023) 24:14673. doi: 10.3390/ijms241914673
14. Chang L, Qiu L, Lei N, Zhou J, Guo R, Gao F, et al. Characterization of fecal microbiota in cervical cancer patients associated with tumor stage and prognosis. Front Cell Infect Microbiol. (2023) 13:1145950. doi: 10.3389/fcimb.2023.1145950
15. Liu K, Yang X, Zeng M, Yuan Y, Sun J, He P, et al. The Role of Fecal Fusobacterium nucleatum and pks(+) Escherichia coli as Early Diagnostic Markers of Colorectal Cancer. Dis Markers. (2021) 2021:1171239. doi: 10.1155/2021/1171239
16. Zhao SS, Chen L, Yang J, Wu ZH, Wang XY, Zhang Q, et al. Altered gut microbial profile accompanied by abnormal fatty acid Metabolism activity exacerbates endometrial cancer progression. Microbiol Spectr. (2022) 10:e0261222. doi: 10.1128/spectrum.02612-22
17. Li R, Guo Q, Zhao J, Kang W, Lu R, Long Z, et al. Assessing causal relationships between gut microbiota and asthma: evidence from two sample Mendelian randomization analysis. Front Immunol. (2023) 14:1148684. doi: 10.3389/fimmu.2023.1148684
18. Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. (2017) 28:1368–79. doi: 10.1093/annonc/mdx108
19. Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. (2021) 372:n71. doi: 10.1136/bmj.n71
20. Wang Z, Qin X, Hu D, Huang J, Guo E, Xiao R, et al. Akkermansia supplementation reverses the tumor-promoting effect of the fecal microbiota transplantation in ovarian cancer. Cell Rep. (2022) 41:111890. doi: 10.1016/j.celrep.2022.11189
21. Tong J, Zhang X, Fan Y, Chen L, Ma X, Yu H, et al. Changes of intestinal microbiota in ovarian cancer patients treated with surgery and chemotherapy. Cancer Manag Res. (2020) 12:8125–35. doi: 10.2147/CMAR.S265205
22. Jacobson D, Moore K, Gunderson C, Rowland M, Austin R, Honap TP, et al. Shifts in gut and vaginal microbiomes are associated with cancer recurrence time in women with ovarian cancer. PeerJ. (2021) 9:e11574. doi: 10.7717/peerj.11574
23. Gong TT, He XH, Gao S, and Wu QJ. Application of machine learning in prediction of Chemotherapy resistant of Ovarian Cancer based on Gut Microbiota. J Cancer. (2021) 12:2877–85. doi: 10.7150/jca.46621
24. Hu X, Xu X, Zeng X, Jin R, Wang S, Jiang H, et al. Gut microbiota dysbiosis promotes the development of epithelial ovarian cancer via regulating Hedgehog signaling pathway. Gut Microbes. (2023) 15:2221093. doi: 10.1080/19490976.2023.2221093
25. Chen C, Deng C, Li Y, He S, Liu Y, Pan S, et al. Machine learning-derived diagnostic model of epithelial ovarian cancer based on gut microbiome signatures. J Transl Med. (2025) 23:319. doi: 10.1186/s12967-025-06339-z
26. Gong W, Jin G, Bao Y, Liu Q, Ni M, Wang J, et al. Characteristics and potential diagnostic value of gut microbiota in ovarian tumor patients. Sci Rep. (2025) 15:16504. doi: 10.1038/s41598-025-99912-x
27. Okazawa-Sakai M, Sakai SA, Hyodo I, Horasawa S, Sawada K, Fujisawa T, et al. Gut microbiome associated with PARP inhibitor efficacy in patients with ovarian cancer. J Gynecol Oncol. (2025) 36:e38. doi: 10.3802/jgo.2025.36.e38
28. Higgins JPT, Morgan RL, Rooney AA, Taylor KW, Thayer KA, Silva RA, et al. A tool to assess risk of bias in non-randomized follow-up studies of exposure effects (ROBINS-E). Environ Int. (2024) 186:108602. doi: 10.1016/j.envint.2024.108602
29. McGuinness LA and Higgins JPT. Risk-of-bias VISualization (robvis): An R package and Shiny web app for visualizing risk-of-bias assessments. Res Synth Methods. (2021) 12:55–61. doi: 10.1002/jrsm.1411
30. Pammi M, Cope J, Tarr PI, Warner BB, Morrow AL, Mai V, et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: a systematic review and meta-analysis. Microbiome. (2017) 5:31. doi: 10.1186/s40168-017-0248-8
31. Kim H, Kim S, Shin SJ, Park T, Nam Y, Kim CW, et al. Gram-negative bacteria and their lipopolysaccharides in alzheimer’s disease: pathologic roles and therapeutic implications. Transl Neurodegener. (2021) 10:49. doi: 10.1186/s40035-021-00273-y
32. Rizzatti G, Lopetuso LR, Gibiino G, Binda C, and Gasbarrini A. Proteobacteria: A common factor in human diseases. BioMed Res Int. (2017) 2017:1–7. doi: 10.1155/2017/9351507
33. Liu F, Liu A, Lü X, Zhang Z, Xue Y, Xu J, et al. Dysbiosis signatures of the microbial profile in tissue from bladder cancer. Cancer Med. (2019) 8:6904–14. doi: 10.1002/cam4.2419
34. Marongiu L, Gornati L, Artuso I, Zanoni I, and Granucci F. Below the surface: The inner lives of TLR4 and TLR9. J Leukoc Biol. (2019) 106:147–60. doi: 10.1002/jlb.3mir1218-483rr
35. Ma H, Wang L, Lv W, and Lv Z. Effects of miR-7 on Hcy-induced rat cerebral arterial vascular smooth muscle cell proliferation, migration and inflammatory factor expression by targeting MMP-14 to regulate TLR4/NF-κB signaling pathway. Cell Mol Biol (Noisy-le-grand). (2020) 66:12–7. doi: 10.14715/cmb/2020.66.7.3
36. Alshahrani MY, Oghenemaro EF, Rizaev J, Kyada A, Roopashree R, Kumar S, et al. Exploring the modulation of TLR4 and its associated ncRNAs in cancer immunopathogenesis, with an emphasis on the therapeutic implications and mechanisms underlying drug resistance. Hum Immunol. (2025) 86:111188. doi: 10.1016/j.humimm.2024.111188
37. Brandl N, Seitz R, Sendtner N, Müller M, and Gülow K. Living on the edge: ROS homeostasis in cancer cells and its potential as a therapeutic target. Antioxidants. (2025) 14:1002. doi: 10.3390/antiox14081002
38. Seixas AF, Quendera AP, Sousa JP, Silva AFQ, Arraiano CM, and Andrade JM. Bacterial response to oxidative stress and RNA oxidation. Front Genet. (2022) 12:821535. doi: 10.3389/fgene.2021.821535
39. Hou T, Wang Q, Dai H, Hou Y, Zheng J, Wang T, et al. Interactive association between gut microbiota and thyroid cancer. Endocrinology. (2023) 165:bqad184. doi: 10.1210/endocr/bqad184
40. Shaheen N, Khursheed W, Gurung B, and Wang S. Akkermansia muciniphila: A key player in gut microbiota-based disease modulation. Microbiol Res. (2025) 301:128317. doi: 10.1016/j.micres.2025.128317
41. Gao F, Cheng C, Li R, Chen Z, Tang K, and Du G. The role of Akkermansia muciniphila in maintaining health: a bibliometric study. Front Med (Lausanne). (2025) 12:1484656. doi: 10.3389/fmed.2025.1484656
42. Smith PL, Piadel K, and Dalgleish AG. Directing T-cell immune responses for cancer vaccination and immunotherapy. Vaccines. (2021) 9:1392. doi: 10.3390/vaccines9121392
43. Guevara-Cruz M, Flores-López AG, Aguilar-López M, Sánchez-Tapia M, Medina-Vera I, Díaz D, et al. Improvement of lipoprotein profile and metabolic endotoxemia by a lifestyle intervention that modifies the gut microbiota in subjects with metabolic syndrome. J Am Heart Assoc. (2019) 8:e012401. doi: 10.1161/JAHA.119.012401
44. Koutoukidis DA, Jebb SA, Zimmerman M, Otunla A, Henry JA, Ferrey A, et al. The association of weight loss with changes in the gut microbiota diversity, composition, and intestinal permeability: a systematic review and meta-analysis. Gut Microbes. (2022) 14:2020068. doi: 10.1080/19490976.2021.2020068
45. Xia J, Lv L, Liu B, Wang S, Zhang S, Wu Z, et al. Akkermansia muciniphila ameliorates acetaminophen-induced liver injury by regulating gut microbial composition and metabolism. Microbiol Spectr. (2022) 10:e0159621. doi: 10.1128/spectrum.01596-21
46. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, and Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. (2012) 489:220–30. doi: 10.1038/nature11550
47. Recharla N, Geesala R, and Shi X-Z. Gut microbial metabolite butyrate and its therapeutic role in inflammatory bowel disease: A literature review. Nutrients. (2023) 15:2275. doi: 10.3390/nu15102275
48. Porter NT, Luis AS, and Martens EC. Bacteroides thetaiotaomicron. Trends Microbiol. (2018) 26:966–7. doi: 10.1016/j.tim.2018.08.005
49. Hayase E, Hayase T, Jamal MA, Miyama T, Chang CC, Ortega MR, et al. Mucus-degrading Bacteroides link carbapenems to aggravated graft-versus-host disease. Cell. (2022) 185:3705–3719.e14. doi: 10.1016/j.cell.2022.09.007
50. Mandal RK, Mandal A, Denny JE, Namazii R, John CC, and Schmidt NW. Gut Bacteroides act in a microbial consortium to cause susceptibility to severe malaria. Nat Commun. (2023) 14:6465. doi: 10.1038/s41467-023-42235-0
51. Qi X, Yun C, Sun L, Xia J, Wu Q, Wang Y, et al. Gut microbiota-bile acid-interleukin-22 axis orchestrates polycystic ovary syndrome. Nat Med. (2019) 25:1225–33. doi: 10.1038/s41591-019-0509-0
52. Niccolai E, Russo E, Baldi S, Ricci F, Nannini G, Pedone M, et al. Significant and conflicting correlation of IL-9 with prevotella and bacteroides in human colorectal cancer. Front Immunol. (2020) 11:573158. doi: 10.3389/fimmu.2020.573158
53. Qian X, Zhang HY, Li QL, Ma GJ, Chen Z, Ji XM, et al. Integrated microbiome, metabolome, and proteome analysis identifies a novel interplay among commensal bacteria, metabolites and candidate targets in non-small cell lung cancer. Clin Transl Med. (2022) 12:e947. doi: 10.1002/ctm2.947
54. Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. (2017) 5:14. doi: 10.1186/s40168-016-0222-x
55. George SD, Van Gerwen OT, Dong C, Sousa LGV, Cerca N, Elnaggar JH, et al. The role of prevotella species in female genital tract infections. Pathogens. (2024) 13:364. doi: 10.3390/pathogens13050364
56. Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. (2017) 151:363–74. doi: 10.1111/imm.12760
57. Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J, et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome. (2017) 5:24. doi: 10.1186/s40168-017-0242-1
58. Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. (2016) 8:43. doi: 10.1186/s13073-016-0299-7
59. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. (2018) 359:104–8. doi: 10.1126/science.aao3290
60. Zhou M, Fan Y, Xu L, Yu Z, Wang S, Xu H, et al. Microbiome and tryptophan metabolomics analysis in adolescent depression: roles of the gut microbiota in the regulation of tryptophan-derived neurotransmitters and behaviors in human and mice. Microbiome. (2023) 11:145. doi: 10.1186/s40168-023-01589-9
61. Maes M, Vasupanrajit A, Jirakran K, Klomkliew P, Chanchaem P, Tunvirachaisakul C, et al. Exploration of the gut microbiome in thai patients with major depressive disorder shows a specific bacterial profile with depletion of the ruminococcus genus as a putative biomarker. Cells. (2023) 12:1240. doi: 10.3390/cells12091240
62. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. (2011) 473:174–80. doi: 10.1038/nature09944
63. Kim YJ, Jung DH, and Park CS. Important roles of Ruminococcaceae in the human intestine for resistant starch utilization. Food Sci Biotechnol. (2024) 33:2009–19. doi: 10.1007/s10068-024-01621-0
64. Crost EH, Le Gall G, Laverde-Gomez JA, Mukhopadhya I, Flint HJ, and Juge N. Mechanistic Insights Into the Cross-Feeding of Ruminococcus gnavus and Ruminococcus bromii on Host and Dietary Carbohydrates. Front Microbiol. (2018) 9:2558. doi: 10.3389/fmicb.2018.02558
65. Yu D, Shu XO, Howard EF, Long J, English WJ, and Flynn CR. Fecal metagenomics and metabolomics reveal gut microbial changes after bariatric surgery. Surg Obes Relat Dis. (2020) 16:1772–82. doi: 10.1016/j.soard.2020.06.032
66. Özdemir A, Yozgat A, Işgın-Atıcı K, Avcı E, Yıldız BD, Gündoğdu A, et al. Potential associations between alterations in gut microbiome and obesity-related traits after the bariatric surgery. J Hum Nutr Diet. (2023) 36:981–96. doi: 10.1111/jhn.13087
67. Ben Izhak M, Eshel A, Cohen R, Madar-Shapiro L, Meiri H, Wachtel C, et al. Projection of gut microbiome pre- and post-bariatric surgery to predict surgery outcome. mSystems. (2021) 6:e0136720. doi: 10.1128/mSystems.01367-20
68. Shi F, Liu G, Lin Y, Guo CL, Han J, Chu ESH, et al. Altered gut microbiome composition by appendectomy contributes to colorectal cancer. Oncogene. (2023) 42:530–40. doi: 10.1038/s41388-022-02569-3
69. Shiroma H, Shiba S, Erawijantari PP, Takamaru H, Yamada M, Sakamoto T, et al. Surgical treatment for colorectal cancer partially restores gut microbiome and metabolome traits. mSystems. (2022) 7:e0001822. doi: 10.1128/msystems.00018-22
70. Wu F, Guo X, Zhang J, Zhang M, Ou Z, and Peng Y. Phascolarctobacterium faecium abundant colonization in human gastrointestinal tract. Exp Ther Med. (2017) 14:3122–6. doi: 10.3892/etm.2017.4878
71. Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. (2019) 565:600–5. doi: 10.1038/s41586-019-0878-z
72. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. (2018) 359:97–103. doi: 10.1126/science.aan4236
73. Singh V, Lee G, Son H, Koh H, Kim ES, Unno T, et al. Butyrate producers, "The Sentinel of Gut": Their intestinal significance with and beyond butyrate, and prospective use as microbial therapeutics. Front Microbiol. (2022) 13:1103836. doi: 10.3389/fmicb.2022.1103836
74. Fusco W, Lorenzo MB, Cintoni M, Porcari S, Rinninella E, Kaitsas F, et al. Short-chain fatty-acid-producing bacteria: key components of the human gut microbiota. Nutrients. (2023) 15:2211. doi: 10.3390/nu15092211
75. Deng X, Li Z, Li G, Li B, Jin X, and Lyu G. Comparison of microbiota in patients treated by surgery or chemotherapy by 16S rRNA sequencing reveals potential biomarkers for colorectal cancer therapy. Front Microbiol. (2018) 9:1607. doi: 10.3389/fmicb.2018.01607
76. Stringer AM, Al-Dasooqi N, Bowen JM, Tan TH, Radzuan M, Logan RM, et al. Biomarkers of chemotherapy-induced diarrhoea: a clinical study of intestinal microbiome alterations, inflammation and circulating matrix metalloproteinases. Support Care Cancer. (2013) 21:1843–52. doi: 10.1007/s00520-013-1741-7
77. Sharma D, Gajjar D, and Seshadri S. Understanding the role of gut microfloral bifidobacterium in cancer and its potential therapeutic applications. Microb Res Rep. (2024) 3:3. doi: 10.20517/mrr.2023.51
78. Tang Q, Huang H, Xu H, Xia H, Zhang C, Ye D, et al. Endogenous Coriobacteriaceae enriched by a high-fat diet promotes colorectal tumorigenesis through the CPT1A-ERK axis. NPJ Biofilms Microb. (2024) 10:5. doi: 10.1038/s41522-023-00472-7
79. Xu J, Kang Y, Zhong Y, Ye W, Sheng T, Wang Q, et al. Alteration of gut microbiome and correlated amino acid metabolism are associated with acute myelocytic leukemia carcinogenesis. Cancer Med. (2023) 12:16431–43. doi: 10.1002/cam4.6283
80. Zhang Z, Li D, Xie F, Muhetaer G, and Zhang H. The cause-and-effect relationship between gut microbiota abundance and carcinoid syndrome: a bidirectional Mendelian randomization study. Front Microbiol. (2023) 14:1291699. doi: 10.3389/fmicb.2023.1291699
81. Jin Q, Ren F, Dai D, Sun N, Qian Y, and Song P. The causality between intestinal flora and allergic diseases: Insights from a bi-directional two-sample Mendelian randomization analysis. Front Immunol. (2023) 14:1121273. doi: 10.3389/fimmu.2023.1121273
82. Nikolova VL, Smith MRB, Hall LJ, Cleare AJ, Stone JM, and Young AH. Perturbations in gut microbiota composition in psychiatric disorders: A review and meta-analysis. JAMA Psychiatry. (2021) 78:1343–54. doi: 10.1001/jamapsychiatry.2021.2573
83. Scheiman J, Luber JM, Chavkin TA, MacDonald T, Tung A, Pham LD, et al. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat Med. (2019) 25:1104–9. doi: 10.1038/s41591-019-0485-4
84. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U.S.A. (2008) 105:16731–6. doi: 10.1073/pnas.0804812105
85. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. (2017) 170:548–563.e16. doi: 10.1016/j.cell.2017.07.008
86. Shen Z, Zhu C, Quan Y, Yang J, Yuan W, Yang Z, et al. Insights into Roseburia intestinalis which alleviates experimental colitis pathology by inducing anti-inflammatory responses. J Gastroenterol Hepatol. (2018) 33:1751–60. doi: 10.1111/jgh.14144
Keywords: gut microbiome, ovarian cancer, cancer biomarkers, microbiome-based therapies, dysbiosis
Citation: Zhang B, Mohd Sahardi NFN, Chew KT, Di W and Shafiee MN (2025) Gut microbiome alterations and their clinical and biological implications in ovarian cancer: a systematic review. Front. Oncol. 15:1690541. doi: 10.3389/fonc.2025.1690541
Received: 22 August 2025; Accepted: 07 November 2025; Revised: 31 October 2025;
Published: 28 November 2025.
Edited by:
Diogo Alpuim Costa, Hospital de Cascais Dr. José de Almeida, PortugalReviewed by:
Kwanchayanawish Machana, Nakhonratchasima College, ThailandSong He, Fudan University, China
Diane Mahoney, University of Kansas Medical Center, United States
Copyright © 2025 Zhang, Mohd Sahardi, Chew, Di and Shafiee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohamad Nasir Shafiee, bmFzaXJzaGFmaWVlQHVrbS5lZHUubXk=
 Beibei Zhang
Beibei Zhang Nur Fatin Nabilah Mohd Sahardi
Nur Fatin Nabilah Mohd Sahardi Kah Teik Chew
Kah Teik Chew Wen Di
Wen Di Mohamad Nasir Shafiee
Mohamad Nasir Shafiee