- 1Department of Restorative Dentistry, Faculty of Dentistry, Universiti Malaya, Kuala Lumpur, Malaysia
- 2Department of Oral Biology, Faculty of Dentistry, University of Jenderal Achmad Yani, Cimahi, Indonesia
- 3Faculty of Dentistry, Kimyo International University in Tashkent, Tashkent, Uzbekistan
- 4Department of Oral & Craniofacial Sciences, Faculty of Dentistry, Universiti Malaya, Kuala Lumpur, Malaysia
- 5Department of Microbiology, School of Life Sciences and Technology, Institut Teknologi Bandung, Bandung, Indonesia
- 6Department of Clinical Sciences, College of Dentistry, Ajman University, Ajman, United Arab Emirates
- 7Centre of Medical and Bio-allied Health Sciences Research, Ajman University, Ajman, United Arab Emirates
Introduction: Streptococcus mutans is central to plaque-induced oral diseases due to its robust biofilm-forming ability. Understanding the genetic and regulatory basis of this process is critical for developing targeted anti-virulence strategies that preserve the balance of the oral microbiome. This systematic review aims to gather and evaluate existing evidence on the virulence genes associated with Streptococcus mutans biofilm formation.
Methods: A comprehensive search of PubMed, Scopus, and Web of Science was conducted in accordance with PRISMA guidelines. Studies investigating the genetic and regulatory mechanisms of biofilm formation, as well as the effects of experimental treatments, were included, and the risk of bias was assessed using the QUIN tool.
Results: Key virulence genes were identified, including glucosyltransferases (gtfB, gtfC, gtfD), glucan-binding proteins (gbpB, gbpC), and two-component systems (vicRK, liaSR). These genes contribute to adhesion, extracellular polysaccharide synthesis, and environmental adaptation, processes critical for biofilm development. Various anti-virulence strategies, such as quorum sensing inhibitors and gene-targeted compounds, show promise in controlling biofilm formation without compromising bacterial viability, thereby preserving the homeostasis of the normal oral flora, which is essential for maintaining overall oral health.
Conclusion: While key virulence genes have been well characterized, further research is needed to clarify how their regulation is influenced by environmental conditions. Insights from this review may support the development of novel therapeutic approaches that reduce Streptococcus mutans pathogenicity while maintaining oral microbial balance.
Systematic Review Registration: https://www.crd.york.ac.uk/PROSPERO/view/CRD42024577977, PROSPERO CRD42024577977.
1 Introduction
Dental caries is recognized as one of the most widespread global oral health issues, affecting an estimated 2 billion people according to the World Health Organization (WHO) (1). Oral health disparities remain significant, particularly in low- and middle-income countries (2). Streptococcus mutans is among the pathogens of concern for WHO due to its increasing multidrug resistance (MDR) and its key role in oral infections (3). Addressing the virulence mechanisms of S. mutans, particularly its biofilm formation capabilities, is thus critical not only for individual patient health but also for public health initiatives aimed at improving global oral health outcomes (4). Targeting biofilm formation through advanced strategies, including the integration of Artificial Intelligence driven screening for biofilm inhibitors, fluoride-based interventions, or bioactive dental materials, represents promising avenues for future preventive strategies in dental caries management at the global scale (5–8).
Dental plaque is a biofilm that forms on the tooth surface through interactions between oral bacteria, their metabolic byproducts, saliva and diet (9, 10). The structural organization of a biofilm provides several advantages to bacteria such as protection from antimicrobial agents and the host immune system, enhanced co-aggregation, and specific interaction preferences. These protective mechanisms make the biofilm a challenging target for therapeutic interventions (11, 12).
Biofilm formation generally progresses through four distinct stages: 1. the adhesion of bacteria to a surface, 2. the development of microcolonies, 3. the maturation of the biofilm structure, and 4. detachment or dispersal, which facilitates bacterial colonization of new environments (13). The initial adhesion of bacterial cells is a critical stage in biofilm formation. After adhesion, bacteria may follow one of two pathways influenced by environmental conditions: they may remain attached and progress to biofilm development, or they may revert to a planktonic state (11). Biofilms are highly dynamic ecosystems, with cells continuously detaching from the main structure either actively or passively. These dispersed cells can colonize new surfaces and form fresh bacterial colonies. Bacteria within the biofilm, known as sessile bacteria, typically exist in a stationary or dormant growth phase and exhibit phenotypic traits distinct from those of planktonic bacteria (13, 14). Bacteria within biofilms exhibit exceptional resistance to environmental stresses, particularly antibiotics. This resistance makes biofilms a significant public health concern, as they are responsible for 60%–80% of human microbial infections (12, 14).
Streptococcus mutans plays a central role in dental plaque formation and is closely associated with the development of oral diseases such as dental (15). The plaque-forming and cariogenic potential of S. mutans is widely recognized to stem from three key attributes: 1. its ability to synthesize large amounts of extracellular glucan polymers from sucrose, which facilitate permanent colonization of hard surfaces and the formation of the extracellular polymeric matrix in situ, 2. its capacity to transport and metabolize a broad range of carbohydrates into organic acids (acidogenicity), and 3. its ability to survive under environmental stress, particularly in low pH conditions (aciduricity) (15, 16). Key contributors to its biofilm-forming capability include glucosyltransferases (gtfB, gtfC, gtfD), which synthesize extracellular glucans that promote adhesion and structural integrity of the plaque biofilm, as well as regulatory systems like vicRK, which influence biofilm maturation and stress response.
Recent studies have explored the potential of anti-virulence agents, such as shikimic acid and betulin, to downregulate key virulence genes involved in biofilm formation without affecting bacterial viability (17, 18). These findings underscore the need for a comprehensive understanding of the specific virulence genes involved in biofilm formation and their regulatory mechanisms in S. mutans biofilm formation. This systematic review aims to gather and evaluate current evidence on the virulence genes associated with S. mutans biofilm formation. By examining their regulatory mechanisms and functional roles, this review seeks to provide insight into potential therapeutic targets that could reduce biofilm-associated pathogenicity while maintaining the ecological balance of the oral microbiome.
2 Methods
2.1 Research strategy
This systematic review was registered in PROSPERO with the registration number CRD42024577977 and was carried out following the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA). Article selection and data extraction were performed by two independent reviewers (D.K.F and N.T) comprehensively, using three electronic databases: Scopus, Web of Science, and PubMed MEDLINE. Free-text and MeSH terms were applied to the search, using Boolean operators (OR, AND) to optimize term combinations, as outlined in Table 1. Manual searches were additionally performed to ensure comprehensive coverage of relevant literature. All reviewers independently screened the titles and abstracts of the search results, with any discrepancies discussed and resolved collaboratively.
2.2 Eligibility criteria
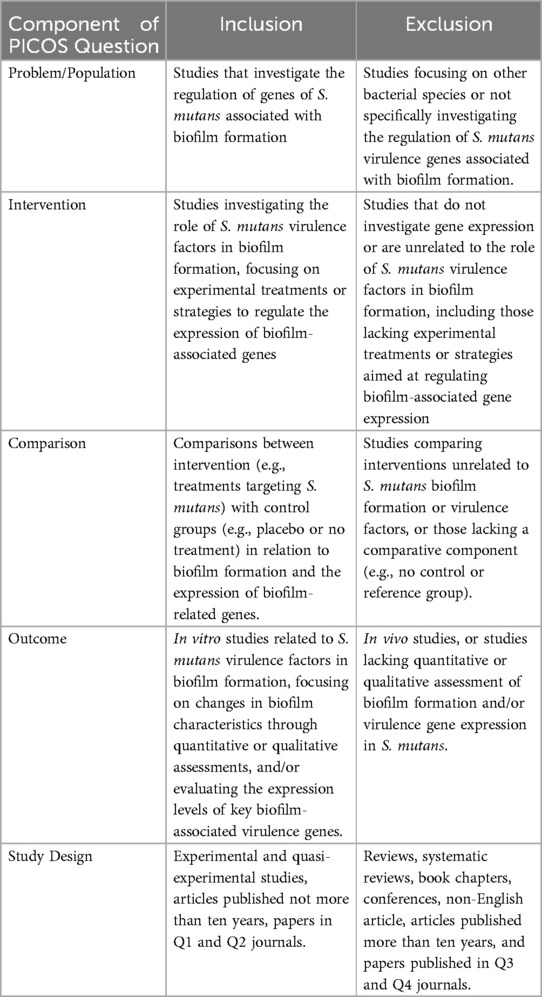
The eligibility criteria for each study type were defined using the PICOS framework, which considers Population/Problem, Intervention, Comparison, Outcome, and Study design. This structured approach, outlined in Table 2, was designed to ensure the inclusion of studies that are both reliable and relevant to the objectives of the review.
2.3 Data extraction
The primary and secondary reviewers reached a consensus to extract the necessary data from reputable scientific databases, such as Scopus, Web of Science, and PubMed MEDLINE. Before data extraction, the keywords for the search were clarified and approved by the third and fourth authors. Reviewer 1 collects the data in.csv format and imports it into an Excel file to create a table. The table includes seven columns: authors, title, publication year, source title, abstract, link (or DOI), and comments. In parallel, Reviewer 2 independently performs a similar task, adhering to the same inclusion/exclusion criteria. This process ensures the accurate selection of papers and minimizes the risk of errors.
2.4 Risk of bias assessment
Two reviewers (D.K.F and N.T) used the Quality Assessment Tool for In vitro Studies (QUIN tool) to assess the risk of bias in the selected studies. The QUIN tool provides a standardized approach to evaluating the risk of bias in in vitro studies included in systematic reviews and meta-analyses. It has been tested for content validity and includes 12 criteria. Each criterion is given a score: 2 points for adequately specified, 1 point for inadequately specified, 0 points for not specified, and N/A (not applicable) for criteria excluded from the calculation. The scores for all 12 criteria are then added up to give a total score for the study. Based on this total score, studies are categorized into three risk levels: high (<50%), medium (50%–70%), or low (>70%) risk. The categorization is determined by the formula: Final score = (Total score × 100)/(2 × number of applicable criteria) (19).
3 Results
3.1 Search result
The study selection process adhered to a predefined framework of inclusion and exclusion criteria based on the PICOS scheme, targeting research on virulence genes involved in biofilm formation by Streptococcus mutans. An initial search identified 272 studies through keyword screening from three databases: Scopus (n = 106), Web of Science (n = 68), Pubmed/Medline (n = 91) and seven articles were selected from hand searching. Following a detailed evaluation, 11 in vitro studies met the eligibility criteria and published in Q1 and Q2 indexed journals within the last decade (2014–2024). The selection process for this review is presented in Figure 1 (20).
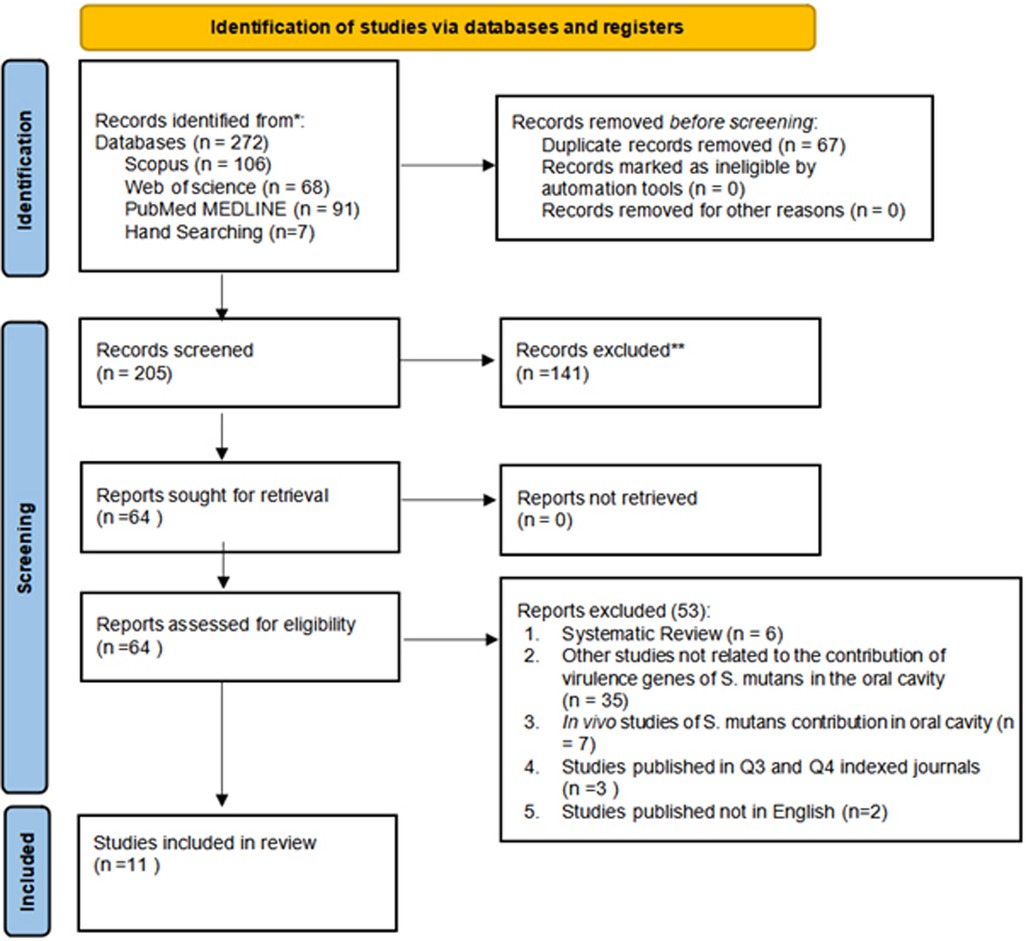
Figure 1. PRISMA 2020 flow diagram illustrating the study selection process. Adapted from Page MJ, Moher D, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. PRISMA 2020 Explanation and elaboration: updated guidance and exemplars for reporting systematic reviews (20). Licensed under CC BY 4.0.
3.2 Risk of bias and quality assessment
The risk of bias (RoB) assessment was independently conducted by two reviewers (D.K.F and N.T) using the QUIN tool for in vitro studies (19). Six studies were classified as having low risk of bias (21–26), while the remaining four studies were classified as having medium risk of bias as presented in Figure 2 (17, 18, 27, 28). As a result, all studies included in this review surpassed 50% of the evaluated criteria. However, the assessment using the QUIN tool highlighted a consistent limitation across all selected studies: none provided details regarding the method used to calculate the sample size or operator details. Figure 2 illustrates the results of the methodological evaluation of in vitro assays according to the Quality Assessment Tool for In vitro Studies (QUIN).
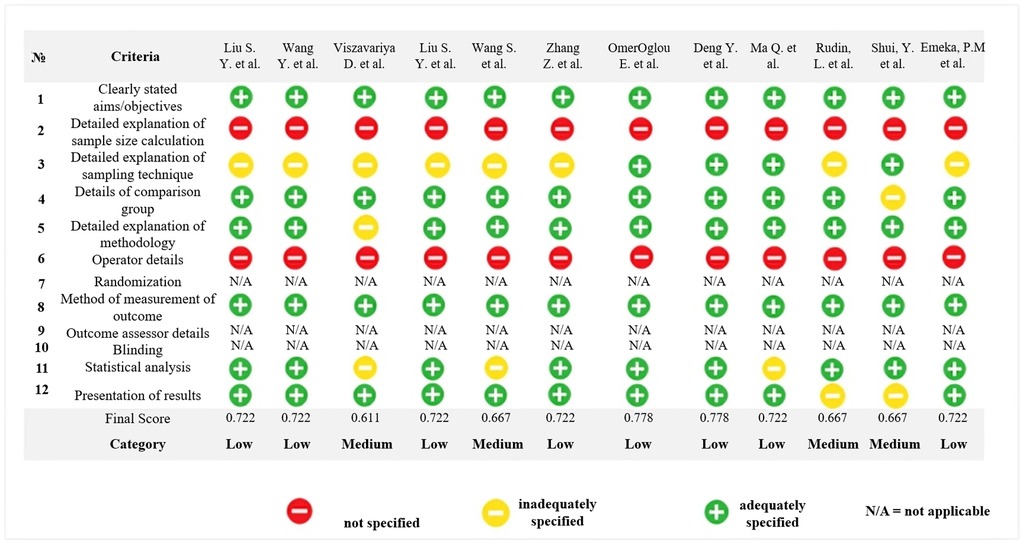
Figure 2. Methodological evaluation of in vitro assays according to the quality assessment tool for in vitro studies (QUIN).
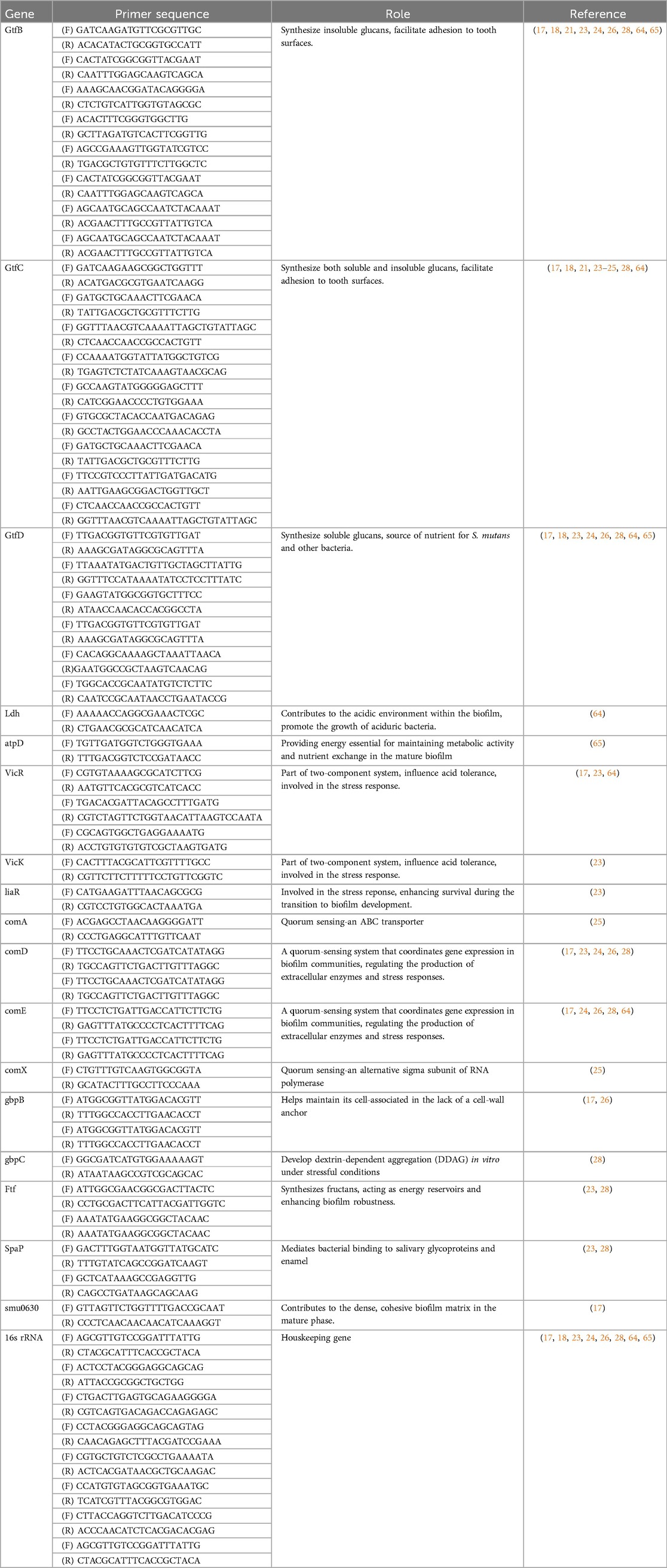
All eleven studies examined the gene expression of S. mutans related to biofilm formation, quorum sensing, adherence, EPS regulation, and virulence using RT-qPCR method with various tested substances. Four of these studies used broth supplemented with different sucrose concentrations in the biofilm formation process (17, 18, 21, 28). Most of the studies reported downregulation of the targeted genes when treated with the respective test materials compared to the untreated group as the negative control. The virulence genes, experimental methods, and key findings from the included studies on Streptococcus mutans biofilm formation are summarized in Table 3. Table 4 presents the corresponding primer sequences for the identified genes.
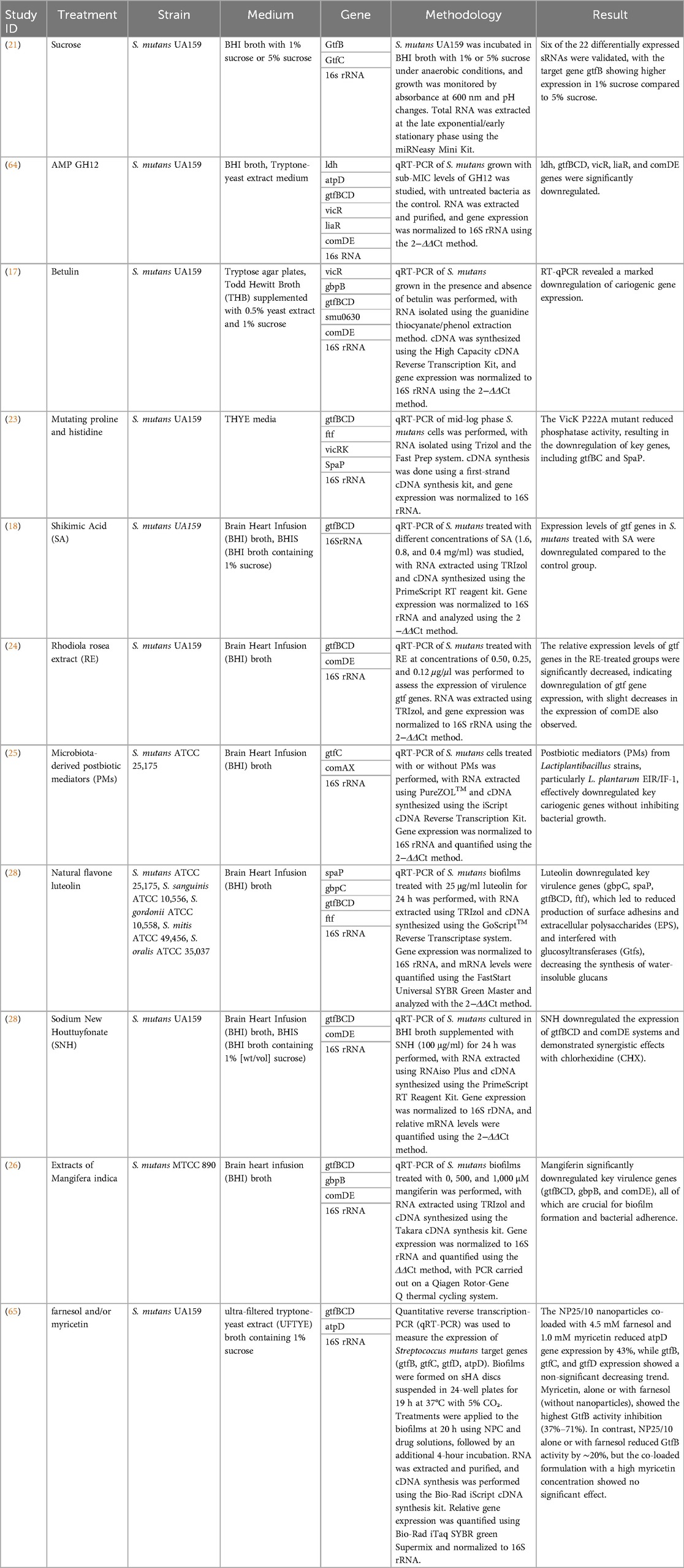
Table 3. Overview of included studies on virulence genes associated with Streptococcus mutans biofilm formation.
4 Discussion
4.1 Biofilm formation
The oral cavity is a dynamic environment constantly exposed to external factors such as foods and drinks, which relies on its normal oral microbiota to maintain microbial homeostasis and overall oral health. Disruption of this balance can lead to dysbiosis of the oral microbiota and promote the formation of biofilm on dental enamel. This biofilm, known as dental plaque, provides an environment conducive to the proliferation of acidogenic and aciduric bacteria. Their metabolic interaction with fermentable carbohydrates leads to the progressive demineralization of calcified dental tissues, marking the onset and progression of dental caries, one of the most prevalent oral diseases worldwide (9, 10). Among the cariogenic bacteria, S. mutans is recognized as the most significant contributor to dental caries. A key virulence factor of S. mutans is its ability to form biofilms on tooth surfaces through the production of glucosyltransferases (GTFs) and fructosyltransferases (Ftf), which are essential for sucrose-dependent adhesion. The key GTF enzymes, encoded by gtfB, gtfC, and gtfD, synthesize extracellular glucans from fermentable carbohydrates. These glucans create specific binding sites that facilitate bacterial colonization and contribute to the formation and stability of cariogenic biofilms. These processes underscore the critical role of S. mutans in the development of dental caries (11, 12).
The adherence of S. mutans to the tooth surface involves both sucrose-dependent and sucrose-independent mechanisms. In sucrose-dependent adhesion, glucosyltransferases (Gtfs), particularly gtfB and gtfC, synthesize extracellular glucans from sucrose, forming a sticky matrix that anchors bacteria to the enamel and to each other (13, 14). These glucans also serve as a scaffold for other microbes and enhance interbacterial cohesion, enabling the maturation of a structurally stable biofilm. Moreover, these enzymes contribute to the formation of water-insoluble glucans that provide mechanical strength and acid retention capability within the plaque. The Gtfs are regulated by environmental conditions such as carbohydrate availability, pH, and stress signals. In contrast, sucrose-independent adhesion involves specific bacterial surface proteins like SpaP (also known as P1 or antigen I/II) that bind to salivary agglutinin glycoproteins (SAGs) coating the tooth surface. This interaction facilitates initial colonization even in the absence of dietary sucrose and primes the biofilm site for later glucan-mediated accumulation. Additionally, this process is aided by lipoteichoic acids (LTAs) and other surface adhesins that stabilize weak initial bonds, allowing S. mutans to persist during fluctuating nutrient conditions. Together, these mechanisms allow S. mutans to establish residence in the oral cavity under both sucrose-rich and sucrose-limited conditions, providing a dual advantage in biofilm initiation and progression (15). Sucrose-independent mechanisms support early-stage biofilm formation by enabling initial bacterial attachment via SpaP-mediated interaction with salivary glycoproteins. This establishes a foundational layer for subsequent polysaccharide-mediated aggregation in the presence of sucrose, facilitating a more robust biofilm structure (29, 30). The research by Fujiwara et al. (1996) reported that deletion of gtfB and gtfC led to a significant reduction in biofilm formation, characterized by minimal bacterial accumulation and limited polysaccharide synthesis in vitro (16). This finding underscores the essential role of these genes in synthesizing water-insoluble extracellular polysaccharides (EPS), which contribute to the structural integrity of cariogenic biofilms. Supporting this, inhibition of gtfB and gtfC by ActG, a protein acetyltransferase (KAT) in Streptococcus mutans, also resulted in markedly reduced EPS production and biofilm formation. Notably, ActG inhibition did not affect planktonic growth in either the mid-logarithmic or stationary phases, aligning with earlier findings that GTF deletion impairs biofilm development without compromising planktonic viability (16–18). Collectively, these observations suggest that targeting GTF activity is a promising strategy to inhibit biofilm development without adversely affecting the overall growth of S. mutans in its planktonic state (31).
Another critical component in S. mutans sucrose-dependent biofilm formation is the group of glucan-binding proteins (Gbps), play a key role in mediating the interaction between S. mutans and glucans. Gbps facilitate the binding of S. mutans to glucans synthesized in situ, thereby complementing the role of cell-associated glucosyltransferase (GTF) enzymes. This cooperative interaction between GTFs and non-GTF Gbps is essential for efficient adherence, colonization, and stabilization of the biofilm matrix, underscoring their importance in sucrose-dependent biofilm development (19, 20).
Mattos-Graner et al. (2001) demonstrated that the depletion of gbpB significantly disrupted the early stages of sucrose-dependent biofilm formation, impairing processes such as cell division and other physiological mechanisms critical for the transition from planktonic growth to biofilm establishment (21). These findings underscore the pivotal role of Gbps, particularly gbpB, in the initial organization and structural development of S. mutans biofilms (21, 22). Moreover, the gbpC gene encodes a surface-associated protein that mediates dextran-induced aggregation. The absence of gbpC led to reduced biofilm biomass and bacterial aggregation, indicating its role as a key receptor for glucans. This highlights the critical contribution of gbpC in maintaining the structural integrity and aggregation of S. mutans within the biofilm matrix, further reinforcing the essential roles of Gbps in biofilm formation (23, 24).
VicK is a histidine protein kinase in S. mutans that plays a pivotal role in biofilm formation by sensing and transmitting chemical signals to downstream regulatory proteins, such as VicR and CovR (25). These regulatory proteins influence the transcription of biofilm-associated genes, including gtfB, gtfC, ftf, and gbpB. Deng et al. (2021) reported that deletion of the vicK gene leads to a significant reduction in biofilm formation and exopolysaccharide (EPS) production, with EPS primarily accumulating around the bacterial cells (26). Additionally, the molecular weight and monosaccharide composition of EPS were markedly altered. In vivo, the resulting biofilms were sparse and associated with reduced dental caries severity. Most EPS synthesis-related genes showed downregulated expression following vicK deletion, with the exception of gtfB. These findings highlight the crucial role of the vicK gene in promoting biofilm development and caries progression, potentially through its regulatory effects on EPS metabolism, including synthesis and structural modification (27, 28).
S. mutans has evolved multiple regulatory systems to adapt to environmental stress, among which. the LiaSR two-component signal transduction system plays a pivotal role, particularly in biofilm formation and stress adaptation. The liaR gene products are essential for cellular response to stressors, including those that damage the cell envelope (32). This system controls related pathways that enable S. mutans to tolerate acidic environments, resist antibiotics, and withstand exposure to detergents. Within the oral environment, LiaSR contributes to the growth and survival of the bacterium, providing robust defenses against acid-induced stress, antimicrobial agents, and structural damage (33, 34). Additionally, the phosphotransfer from LiaS to LiaR is crucial for the activation of the vicRKX genes. Studies have shown that the VicRKX two-component signal transduction system (TCSTS) influences the ComCDE system and the expression of genes such as gtfB, gtfC, and gtfD. comD encodes a histidine kinase receptor that responds to competence-stimulating peptide (CSP), while comE encodes an intracellular response regulator that mediates the expression of related genes (32). Upon activation, ComE promotes the synthesis of mutacins IV and V and enhances genetic competence, thereby contributing to the pathogenicity and biofilm formation of S. mutans (35). Suppression of these regulatory systems compromises S. mutans' ability to respond to environmental stress, resulting in decreased cell persistence and disrupted biofilm development. Ultimately, this impairs the bacterium's capacity to maintain its virulence and long-term survival in the oral cavity (34). Figure 3 depicts the biofilm formation cycle, transitioning from reversible to irreversible phases. In the reversible stages, bacteria first adhere to the acquired pellicle, followed by attachment and early aggregation. The irreversible phases involve stable coaggregation, where the extracellular polymeric matrix forms, leading to biofilm maturation. The cycle concludes with bacterial dispersal, enabling colonization of new surfaces. This figure highlights the key stages critical to biofilm stability and pathogenicity. The LiaSR system is enhanced by upstream activation of the VicRKX two-component system. Environmental stimuli sensed by VicK initiate phosphorylation cascades that upregulate liaSR expression, promoting resistance to envelope stress and supporting robust biofilm architecture (36–38).
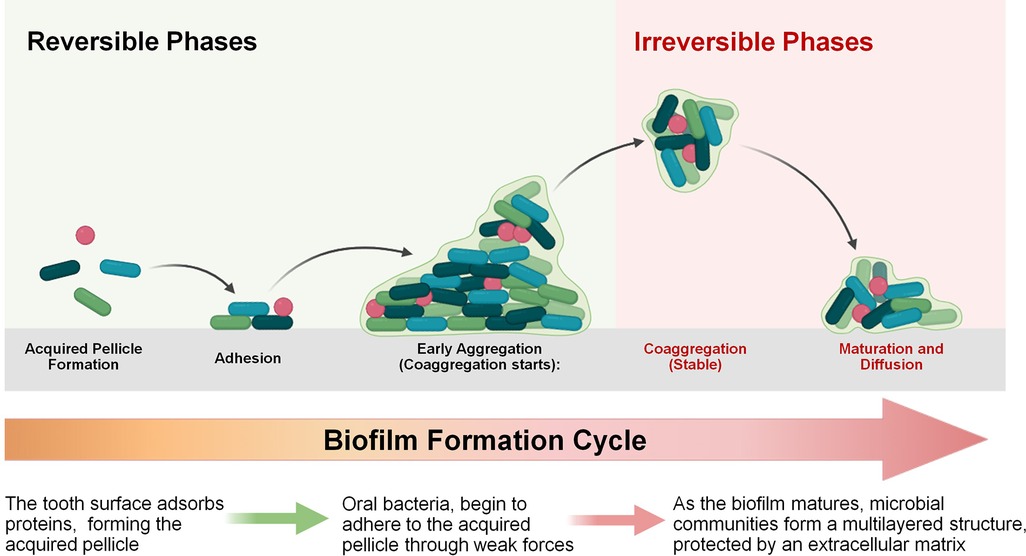
Figure 3. Biofilm formation cycle. The process begins with the formation of the acquired pellicle and initial bacterial adhesion, which are reversible. As the biofilm matures, bacteria coaggregate and develop a stable, multilayered structure protected by an extracellular matrix, marking the irreversible phase.
In addition, the increasing prevalence of multidrug-resistant (MDR) strains of S. mutans aligns with global concerns raised by WHO regarding antimicrobial resistance (39, 40). The WHO's Global Antimicrobial Resistance Surveillance System (GLASS) emphasizes the urgency of addressing antimicrobial resistance across pathogens, including those implicated in oral infections such as dental caries (41). Biofilm-mediated resistance mechanisms in S. mutans further complicate treatment approaches, underscoring the importance of alternative therapeutic strategies like smart materials, and advanced computational approaches including AI-driven identification of anti-biofilm compounds (5–8, 42, 43). Such measures are important to mitigating the rise of MDR strains and enhancing oral health care globally.
4.2 Quorum sensing
Quorum sensing is a fundamental communication mechanism that enable bacteria to adapt to their environment by facilitating communication within a population. This process allows bacterial cells to synchronize gene expression in a cell density-dependent manner, enhancing their survival and adaptability. It operates by producing, releasing, detecting, and responding to signaling molecules known as autoinducers or self-inducers, which act similarly to hormones (44). In S. mutans, the quorum-sensing Com system plays a critical role in regulating biofilm formation and structural organization. This system functions most efficiently during active cell growth within the biofilm and involves the synthesis of competence-stimulating peptides (CSP), which are detected via the two-component signaling system ComDE. This intercellular signaling highlights the essential role of quorum sensing in the development, stability, and maintenance of S. mutans biofilms (45).
The ABC transporter complex, particularly ComAB, is responsible for processing the comC-encoded propeptide into a 21-amino acid competence-stimulating peptide (21-CSP), which is then cleaved by SepM into its active 18-amino acid form. This active CSP binds to the membrane-bound histidine kinase receptor ComD, initiating a phosphorylation cascade through the response regulator ComE, which in turn activates the transcription of virulence and competence genes involved in biofilm development (46). The activity of this ABC transporter system is favored during exponential bacterial growth, under nutrient-rich conditions with sufficient sucrose availability and high cell density. These environmental factors stimulate quorum sensing by enhancing CSP production and release. Additionally, acidic stress and oxidative stress conditions in mature plaque biofilms can further enhance CSP-mediated signaling, allowing S. mutans to coordinate biofilm-specific gene expression and genetic competence (47–50). Pourhajibagher et al. (2022) demonstrated that quorum quenching using blue laser and N-QCT suppressed the expression of QS-related genes (comA, comB, comDE), alongside the virulence gene gtfB, confirming the role of this signaling system in biofilm-associated virulence regulation (51).
4.3 Acidogenicity and acid tolerance
S. mutans is a highly acidogenic bacterium, meaning it can rapidly ferment dietary carbohydrates, particularly sucrose, glucose, and fructose into organic acids such as lactic acid (15). This acid production leads to a significant reduction in local pH, contributing to the demineralization of enamel and the initiation of dental caries. Key enzymes involved in acidogenesis include lactate dehydrogenase (ldh) and pyruvate formate lyase, which enable the bacterium to metabolize sugars under both aerobic and anaerobic conditions. Acidogenic activity is further enhanced by the presence of extracellular glucans that retain fermentable substrates within the biofilm matrix, creating a localized reservoir for sustained acid production. S. mutans is also aciduric, meaning it can tolerate and thrive in low-pH environments where many other oral commensals fail to survive (52). This aciduric property is critical for its dominance in cariogenic biofilms (53). The ability to maintain intracellular pH homeostasis under acidic conditions is regulated by several stress response systems, including F-ATPase proton pumps, two-component systems such as liaSR and vicRK, and membrane-associated proteins that reinforce cell envelope integrity. Additional protective systems, including the arginine deiminase pathway and malolactic fermentation, generate alkaline by-products that counteract cytoplasmic acidification. These mechanisms allow S. mutans to sustain intracellular pH stability, metabolic activity, and genetic competence within the acidic microenvironment of the dental plaque (54). Upregulation of acid tolerance genes is often observed under low pH, confirming their role in long-term survival. Moreover, environmental stress conditions such as acidity and nutrient limitation can further stimulate the expression of virulence genes, enhancing the pathogen's persistence and contributing to its role in dental caries development (55). Understanding these acidogenic and aciduric mechanisms is crucial for designing effective anti-caries strategies that disrupt the ecological advantages of S. mutans without affecting the balance of the oral microbiota (56, 57). Emerging technologies, including AI-based drug discovery, fluoride-releasing smart materials, silver diamine fluoride, and bioactive restorative agents, are being investigated to neutralize acidic microenvironments and impair biofilm persistence in caries-prone sites (5–8, 58–62).
5 Conclusion
This systematic review underscores the critical role of specific virulence genes in biofilm formation by S. mutans, a key contributor to dental caries. Genes involved in glucan synthesis (gtfB, gtfC, gtfD), glucan-binding (gbpB, gbpC), and regulatory systems like vicRK and liaSR are pivotal in adhesion, extracellular polysaccharide production, and environmental stress adaptation. Targeting these genes through emerging strategies, such as quorum-sensing inhibitors and anti-virulence agents, offers promising potential to reduce biofilm formation and pathogenicity. Importantly, these approaches can disrupt pathogenicity without compromising bacterial viability, thereby preserving the oral microbiome's ecological balance and providing novel pathways for therapeutic intervention in dental caries management.
Future research should focus on validating the in vitro findings through in vivo studies and clinical trials to assess the real-world effectiveness of targeting S. mutans virulence genes. Exploring innovative anti-virulence strategies, such as gene-editing technologies and quorum-sensing inhibitors, is essential to evaluate their safety and long-term impact on biofilm formation. Additionally, integrating omics approaches, including transcriptomics and proteomics, will provide a deeper understanding of the mechanisms driving S. mutans pathogenicity and biofilm formation. Collaborative studies combining these strategies with conventional treatments could improve therapeutic outcomes while preserving the oral microbiome's ecological balance. Furthermore, addressing potential resistance to these therapies will be key to ensuring their future success in preventing dental caries.
5.1 Limitations of the study
Most included studies focused narrowly on specific virulence genes or isolated pathways, limiting comprehensive insights into broader gene networks involved in S. mutans biofilm formation (19). Additionally, variability in experimental conditions (such as culture media, sucrose concentration, methods for gene expression analysis, and duration of biofilm maturation) introduced heterogeneity, potentially affecting the consistency and applicability of results. Another limitation highlighted by QUIN assessment was the absence of standardized sample size calculations and operator information, potentially affecting reproducibility and generalizability of findings (63). Furthermore, all studies included were in vitro, which restricts direct applicability to clinical practice. Hence, validation through clinical trials and in vivo studies is necessary for more robust and applicable conclusions.
Data availability statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
Author contributions
DF: Writing – original draft, Data curation, Methodology, Investigation. NT: Conceptualization, Methodology, Writing – original draft. WW: Writing – review & editing, Methodology, Conceptualization. IP: Writing – review & editing, Methodology, Conceptualization. AC: Methodology, Conceptualization, Writing – review & editing. MZ: Conceptualization, Writing – review & editing, Methodology.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors would like to thank the Universiti Malaya for supporting this study and providing the data via the Central Library of the Universiti Malaya.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Jain N, Dutt U, Radenkov I, Jain S. WHO’s global oral health status report 2022: actions, discussion and implementation. Oral Dis. (2024) 30:73–9. doi: 10.1111/odi.14516
2. Bhandari B, Newton JT, Bernabé E. Social inequalities in adult oral health in 40 low- and middle-income countries. Int Dent J. (2016) 66:295–303. doi: 10.1111/idj.12243
3. Li X, Wang Y, Jiang X, Zeng Y, Zhao X, Washio J, et al. Investigation of drug resistance of caries-related streptococci to antimicrobial peptide GH12. Front Cell Infect Microbiol. (2022) 12:991938. doi: 10.3389/fcimb.2022.991938
4. Krzyściak W, Jurczak A, Kościelniak D, Bystrowska B, Skalniak A. The virulence of Streptococcus mutans and the ability to form biofilms. Eur J Clin Microbiol Infect Dis. (2014) 33:499–515. doi: 10.1007/s10096-013-1993-7
5. Samaranayake L, Tuygunov N, Schwendicke F, Osathanon T, Khurshid Z, Boymuradov SA, et al. The transformative role of artificial intelligence in dentistry: a comprehensive overview. Part 1: fundamentals of AI, and its contemporary applications in dentistry. Int Dent J. (2025) 75:383–96. doi: 10.1016/j.identj.2025.02.005
6. Tuygunov N, Samaranayake L, Khurshid Z, Rewthamrongsris P, Schwendicke F, Osathanon T, et al. The transformative role of artificial intelligence in dentistry: a comprehensive overview part 2: the promise and perils, and the international dental federation communique. Int Dent J. (2025) 75:397–404. doi: 10.1016/j.identj.2025.02.006
7. Samaranayake L, Porntaveetus T, Tsoi J, Tuygunov N. Facts and fallacies of the fluoride controversy: a contemporary perspective. Int Dent J. (2025) 75:100833. doi: 10.1016/j.identj.2025.04.013
8. Al Zangana T, Tuygunov N, Yahya NA, Abdul Aziz A. The impact of resin coatings on the properties and performance of glass ionomer cements: a systematic review. J Mech Behav Biomed Mater. (2025) 169:107044. doi: 10.1016/j.jmbbm.2025.107044
9. Hajishengallis E, Parsaei Y, Klein MI, Koo H. Advances in the microbial etiology and pathogenesis of early childhood caries. Mol Oral Microbiol. (2017) 32:24–34. doi: 10.1111/omi.12152
10. Bowen WH, Burne RA, Wu H, Koo H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. (2018) 26:229–42. doi: 10.1016/j.tim.2017.09.008
11. Berger D, Rakhamimova A, Pollack A, Loewy Z. Oral biofilms: development, control, and analysis. High Throughput. (2018) 7:24. doi: 10.3390/ht7030024
12. Bertolini M, Costa RC, Barão VAR, Cunha Villar C, Retamal-Valdes B, Feres M, et al. Oral microorganisms and biofilms: new insights to defeat the main etiologic factor of oral diseases. Microorganisms. (2022) 10:2413. doi: 10.3390/microorganisms10122413
13. Crouzet M, Le Senechal C, Brözel VS, Costaglioli P, Barthe C, Bonneu M, et al. Exploring early steps in biofilm formation: set-up of an experimental system for molecular studies. BMC Microbiol. (2014) 14:253. doi: 10.1186/s12866-014-0253-z
14. Kumar L, Bisen M, Harjai K, Chhibber S, Azizov S, Lalhlenmawia H, et al. Advances in nanotechnology for biofilm inhibition. ACS Omega. (2023) 8:21391–409. doi: 10.1021/acsomega.3c02239
15. Lemos JA, Palmer SR, Zeng L, Wen ZT, Kajfasz JK, Freires IA, et al. The biology of Streptococcus mutans. Microbiol Spectr. (2019) 7:10–128. doi: 10.1128/microbiolspec.GPP3-0051-2018
16. Zheng T, Jing M, Gong T, Yan J, Wang X, Xu M, et al. Regulatory mechanisms of exopolysaccharide synthesis and biofilm formation in Streptococcus mutans. J Oral Microbiol. (2023) 15:2225257. doi: 10.1080/20002297.2023.2225257
17. Viszwapriya D, Subramenium GA, Radhika S, Pandian SK. Betulin inhibits cariogenic properties of Streptococcus mutans by targeting vicRK and gtf genes. Antonie Van Leeuwenhoek. (2017) 110:153–65. doi: 10.1007/s10482-016-0785-3
18. Zhang Z, Yang Y, Sun Q, Zeng W, Li Y. Inhibition of biofilm formation and virulence factors of cariogenic oral pathogen Streptococcus mutans by shikimic acid. Microbiol Spectr. (2022) 10:e0119922. doi: 10.1128/spectrum.01199-22
19. Sheth VH, Shah NP, Jain R, Bhanushali N, Bhatnagar V. Development and validation of a risk-of-bias tool for assessing in vitro studies conducted in dentistry: the QUIN. J Prosthet Dent. (2024) 131:1038–42. doi: 10.1016/j.prosdent.2022.05.019
20. Page MJ, Moher D, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. PRISMA 2020 Explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. Br Med J. (2021) 372:n160. doi: 10.1136/bmj.n160
21. Liu SS, Zhu WH, Zhi QH, Liu J, Wang Y, Lin HC. Analysis of sucrose-induced small RNAs in Streptococcus mutans in the presence of different sucrose concentrations. Appl Microbiol Biotechnol. (2017) 101:5739–48. doi: 10.1007/s00253-017-8346-x
22. Wang Y, Wang X, Jiang W, Wang K, Luo J, Li W, et al. Antimicrobial peptide GH12 suppresses cariogenic virulence factors of Streptococcus mutans. J Oral Microbiol. (2018) 10:1442089. doi: 10.1080/20002297.2018.1442089
23. Wang S, Long L, Yang X, Qiu Y, Tao T, Peng X, et al. Dissecting the role of VicK phosphatase in aggregation and biofilm formation of Streptococcus mutans. J Dent Res. (2021) 100:631–38. doi: 10.1177/0022034520979798
24. Zhang Z, Liu Y, Lu M, Lyu X, Gong T, Tang B, et al. Rhodiola rosea extract inhibits the biofilm formation and the expression of virulence genes of cariogenic oral pathogen Streptococcus mutans. Arch Oral Biol. (2020) 116:104762. doi: 10.1016/j.archoralbio.2020.104762
25. OmerOglou E, Karaca B, Kibar H, Haliscelik O, Kiran F. The role of microbiota-derived postbiotic mediators on biofilm formation and quorum sensing-mediated virulence of Streptococcus mutans: a perspective on preventing dental caries. Microb Pathog. (2022) 164:105390. doi: 10.1016/j.micpath.2022.105390
26. Emeka PM, Badger-Emeka LI, Ibrahim H-IM, Thirugnanasambantham K, Hussen J. Inhibitory potential of mangiferin on glucansucrase producing Streptococcus mutans biofilm in dental plaque. Appl Sci. (2020) 10(22):8297. doi: 10.3390/app10228297
27. Rudin L, Roth N, Kneubühler J, Dubey BN, Bornstein MM, Shyp V. Inhibitory effect of natural flavone luteolin on Streptococcus mutans biofilm formation. Microbiol Spectr. (2023) 11:e0522322. doi: 10.1128/spectrum.05223-22
28. Shui Y, Jiang Q, Lyu X, Wang L, Lin Y, Ma Q, et al. Inhibitory effects of sodium new houttuyfonate on growth and biofilm formation of Streptococcus mutans. Microb Pathog. (2021) 157:104957. doi: 10.1016/j.micpath.2021.104957
29. Ahn SJ, Ahn SJ, Wen ZT, Brady LJ, Burne RA. Characteristics of biofilm formation by Streptococcus mutans in the presence of saliva. Infect Immun. (2008) 76:4259–68. doi: 10.1128/IAI.00422-08
30. Lin Y, Chen J, Zhou X, Li Y. Inhibition of Streptococcus mutans biofilm formation by strategies targeting the metabolism of exopolysaccharides. Crit Rev Microbiol. (2021) 47:1–11. doi: 10.1080/1040841X.2021.1915959
31. Kashi M, Varseh M, Hariri Y, Chegini Z, Shariati A. Natural compounds: new therapeutic approach for inhibition of Streptococcus mutans and dental caries. Front Pharmacol. (2025) 16:1548117. doi: 10.3389/fphar.2025.1548117
32. Ray R. Dental biofilm: risks, diagnostics and management. Biocatal Agric Biotechnol. (2022) 43:102381. doi: 10.1016/j.bcab.2022.102381
33. Aytac Bal F, Ozkocak I, Cadirci BH, Sirin Karaarslan E, Cakdinleyen M, Agaccioglu M. Effects of photodynamic therapy with indocyanine green on Streptococcus mutans biofilm. Photodyn Photodyn Ther. (2019) 26:229–34. doi: 10.1016/j.pdpdt.2019.04.005
34. Yue J, Yang H, Liu S, Song F, Guo J, Huang C. Influence of naringenin on the biofilm formation of Streptococcus mutans. J Dent. (2018) 76:24–31. doi: 10.1016/j.jdent.2018.04.013
35. Chen L, Ren Z, Zhou X, Zeng J, Zou J, Li Y. Inhibition of Streptococcus mutans biofilm formation, extracellular polysaccharide production, and virulence by an oxazole derivative. Appl Microbiol Biotechnol. (2016) 100:857–67. doi: 10.1007/s00253-015-7092-1
36. Huang S, Du J, Li Y, Wu M, Chen S, Jiang S, et al. LiaSR two-component system modulates the oxidative stress response in Streptococcus mutans. Microb Pathog. (2023) 185:106404. doi: 10.1016/j.micpath.2023.106404
37. Cho THS, Pick K, Raivio TL. Bacterial envelope stress responses: essential adaptors and attractive targets. Biochimica et Biophysica Acta (BBA). (2023) 1870:119387. doi: 10.1016/j.bbamcr.2022.119387
38. Suntharalingam P, Senadheera MD, Mair RW, Lévesque CM, Cvitkovitch DG. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J Bacteriol. (2009) 191:2973–84. doi: 10.1128/JB.01563-08
39. Salam MA, Al-Amin MY, Salam MT, Pawar JS, Akhter N, Rabaan AA, et al. Antimicrobial resistance: a growing serious threat for global public health. Healthcare (Basel). (2023) 11(13):1946. doi: 10.3390/healthcare11131946
40. Ali M, Garg A, Srivastava A, Arora PK. The role of antimicrobial peptides in overcoming antibiotic resistance. The Microbe. (2025) 7:100337. doi: 10.1016/j.microb.2025.100337
41. Alam M, Saleem Z, Haseeb A, Qamar MU, Sheikh A, Almarzoky Abuhussain SS, et al. Tackling antimicrobial resistance in primary care facilities across Pakistan: current challenges and implications for the future. J Infect Public Health. (2023) 16:97–110. doi: 10.1016/j.jiph.2023.10.046
42. Liu HY, Prentice EL, Webber MA. Mechanisms of antimicrobial resistance in biofilms. npj Antimicrob Resist. (2024) 2:27. doi: 10.1038/s44259-024-00046-3
43. Gao Z, Chen X, Wang C, Song J, Xu J, Liu X, et al. New strategies and mechanisms for targeting Streptococcus mutans biofilm formation to prevent dental caries: a review. Microbiol Res. (2024) 278:127526. doi: 10.1016/j.micres.2023.127526
44. Shanker E, Federle MJ. Quorum sensing regulation of competence and bacteriocins in Streptococcus pneumoniae and mutans. Genes (Basel). (2017) 8:15. doi: 10.3390/genes8010015
45. Suzuki Y, Nagasawa R, Senpuku H. Inhibiting effects of fructanase on competence-stimulating peptide-dependent quorum sensing system in Streptococcus mutans. J Infect Chemother. (2017) 23:634–41. doi: 10.1016/j.jiac.2017.06.006
46. Bikash CR, Hamry SR, Tal-Gan Y. Structure-activity relationships of the competence stimulating peptide in Streptococcus mutans reveal motifs critical for membrane protease SepM recognition and ComD receptor activation. ACS Infect Dis. (2018) 4:1385–94. doi: 10.1021/acsinfecdis.8b00115
47. Oda M, Kurosawa M, Yamamoto H, Domon H, Takenaka S, Ohsumi T, et al. Sulfated vizantin inhibits biofilm maturation by Streptococcus mutans. Microbiol Immunol. (2020) 64:493–501. doi: 10.1111/1348-0421.12797
48. Ma Q, Pan Y, Chen Y, Yu S, Huang J, Liu Y, et al. Acetylation of glucosyltransferases regulates Streptococcus mutans biofilm formation and virulence. PLoS Pathog. (2021) 17:e1010134. doi: 10.1371/journal.ppat.1010134
49. Zhang Q, Nijampatnam B, Hua Z, Nguyen T, Zou J, Cai X, et al. Structure-based discovery of small molecule inhibitors of cariogenic virulence. Sci Rep. (2017) 7:5974. doi: 10.1038/s41598-017-06168-1
50. Matsumoto-Nakano M. Role of Streptococcus mutans surface proteins for biofilm formation. Jpn Dent Sci Rev. (2018) 54:22–9. doi: 10.1016/j.jdsr.2017.08.002
51. Pourhajibagher M, Alaeddini M, Etemad-Moghadam S, Rahimi Esboei B, Bahrami R, Miri Mousavi RS, et al. Quorum quenching of Streptococcus mutans via the nano-quercetin-based antimicrobial photodynamic therapy as a potential target for cariogenic biofilm. BMC Microbiol. (2022) 22:125. doi: 10.1186/s12866-022-02544-8
52. Baker JL, Faustoferri RC, Quivey RG Jr. Acid-adaptive mechanisms of Streptococcus mutans-the more we know, the more we don't. Mol Oral Microbiol. (2017) 32:107–17. doi: 10.1111/omi.12162
53. Chen X, Daliri EB, Tyagi A, Oh DH. Cariogenic biofilm: pathology-related phenotypes and targeted therapy. Microorganisms. (2021) 9:1311. doi: 10.3390/microorganisms9061311
54. Jurakova V, Farková V, Kucera J, Dadakova K, Zapletalova M, Paskova K, et al. Gene expression and metabolic activity of Streptococcus mutans during exposure to dietary carbohydrates glucose, sucrose, lactose, and xylitol. Mol Oral Microbiol. (2023) 38:424–41. doi: 10.1111/omi.1242837440366
55. Motsay M, Saputo S. Acid adaptation alters Streptococcus mutans drug susceptibility profile. The Microbe. (2024) 2:100028. doi: 10.1016/j.microb.2023.100028
56. Jin P, Wang L, Chen D, Chen Y. Unveiling the complexity of early childhood caries: candida albicans and Streptococcus mutans cooperative strategies in carbohydrate metabolism and virulence. J Oral Microbiol. (2024) 16:2339161. doi: 10.1080/20002297.2024.233916138606339
57. Spatafora G, Li Y, He X, Cowan A, Tanner ACR. The evolving microbiome of dental caries. Microorganisms. (2024) 12:121. doi: 10.3390/microorganisms12010121
58. Christie B, Musri N, Djustiana N, Takarini V, Tuygunov N, Zakaria MN, et al. Advances and challenges in regenerative dentistry: a systematic review of calcium phosphate and silicate-based materials on human dental pulp stem cells. Materials Today Bio. (2023) 23:100815. doi: 10.1016/j.mtbio.2023.100815
59. Tuygunov N, Khairunnisa Z, Yahya NA, Aziz AA, Zakaria MN, Israilova NA, et al. Bioactivity and remineralization potential of modified glass ionomer cement: a systematic review of the impact of calcium and phosphate ion release. Dent Mater J. (2024) 43:1–10. doi: 10.4012/dmj.2023-132
60. Tuygunov N, Zakaria MN, Yahya NA, Abdul Aziz A, Cahyanto A. Efficacy and bone-contact biocompatibility of glass ionomer cement as a biomaterial for bone regeneration: a systematic review. J Mech Behav Biomed Mater. (2023) 146:106099. doi: 10.1016/j.jmbbm.2023.106099
61. Kaewkamchai S, Thanyasrisung P, Sukarawan W, Samaranayake L, Tuygunov N, Songsiripradubboon S. Efficacy of silver diamine fluoride (SDF) in arresting dentin caries against inter-kingdom biofilms of Streptococcus mutans and Candida albicans. PLoS One. (2024) 19:e0308656. doi: 10.1371/journal.pone.0308656
62. Khairunnisa Z, Tuygunov N, Cahyanto A, Aznita WH, Purwasena IA, Noor NSM, et al. Potential of microbial-derived biosurfactants for oral applications–a systematic review. BMC Oral Health. (2024) 24:707. doi: 10.1186/s12903-024-04479-038898470
63. Magana M, Sereti C, Ioannidis A, Mitchell CA, Ball AR, Magiorkinis E, et al. Options and limitations in clinical investigation of bacterial biofilms. Clin Microbiol Rev. (2018) 31:10–1128. doi: 10.1128/CMR.00084-16
64. Sims KR Jr., Maceren JP, Liu Y, Rocha GR, Koo H, Benoit DSW. Dual antibacterial drug-loaded nanoparticles synergistically improve treatment of Streptococcus mutans biofilms. Acta Biomater. (2020) 115:418–31. doi: 10.1016/j.actbio.2020.08.032
65. Smith EG, Spatafora GA. Gene regulation in S. mutans: complex control in a complex environment. J Dent Res. (2012) 91:133–41. doi: 10.1177/002203451141541521743034
Keywords: Streptococcus mutans, virulence genes, biofilm formation, dental caries, dental plaque, glucosyltransferase
Citation: Fitri DK, Tuygunov N, Wan Harun WHA, Purwasena IA, Cahyanto A and Zakaria MN (2025) Key virulence genes associated with Streptococcus mutans biofilm formation: a systematic review. Front. Oral Health 6:1654428. doi: 10.3389/froh.2025.1654428
Received: 26 June 2025; Accepted: 12 August 2025;
Published: 26 August 2025.
Edited by:
Katarzyna Garbacz, Medical University of Gdansk, PolandReviewed by:
Oleksandr Nazarchuk, National Pirogov Memorial Medical University, UkraineThayumanavan Thangavelu, Kalaignarkarunanidhi Institute of Technology (KIT), India
Copyright: © 2025 Fitri, Tuygunov, Wan Harun, Purwasena, Cahyanto and Zakaria. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Myrna Nurlatifah Zakaria, bXlybmEubi56YWthcmlhQHVtLmVkdS5teQ==
 Dinda Kurnia Fitri
Dinda Kurnia Fitri Nozimjon Tuygunov
Nozimjon Tuygunov Wan Himratul Aznita Wan Harun4
Wan Himratul Aznita Wan Harun4 Arief Cahyanto
Arief Cahyanto