- 1Department of Periodontology, School of Dental Medicine, University of Bern, Bern, Switzerland
- 2Perio Trap Pharmaceutical GmbH, Halle/Saale, Germany
- 3Department of Molecular Drug Design and Target Validation, Fraunhofer Institute for Cell Therapy and Immunology, Halle (Saale), Germany
- 4Department of Microbiology, Faculty of Biochemistry Biophysics and Biotechnology, Jagiellonian University, Kraków, Poland
- 5Department of Oral Immunology and Infectious Diseases, School of Dentistry, University of Louisville, Louisville, KY, United States
Introduction: Modifying bacterial virulence could be an interesting alternative to antibiotics. The study aimed to examine the effects of an inhibitor targeting bacterial glutaminyl cyclase [which is selectively present in Porphyromonas gingivalis (Pg), Tannerella forsythia (Tf), and Prevotella intermedia (Pi)] on various multispecies biofilms.
Methods: Two multi-species biofilms—one containing four species (including Tf) and another with 12 species (including Tf, Pg, and Pi)—were cultured in the presence of 31.25–500 µM of a [4,5-c]pyridine-based inhibitor. After 24 h, bacterial counts, biofilm biomass, metabolic activity, and, when Pg was included, Arg-gingipain activity were measured. Additionally, the biofilms were exposed to monocytic cells; here, the release of interleukin (IL)-1β and IL-10 was analyzed. The data were analyzed using a one-way analysis of variance (ANOVA) with a post-hoc comparison performed using the Bonferroni correction.
Results and Discussion: In all biofilms, total bacterial counts and those of Pg and Tf remained unaffected by the inhibitor. In the 12-species biofilm, both biomass and total metabolic activity decreased at high inhibitor concentrations (500 µM to 75.2 ± 6.5% and 87.2 ± 5.8%, respectively; each p < 0.001). The arginine-specific amidolytic activities of Rgp declined dose-dependently, down to 60.4 ± 10.2% (p < 0.001) at 500 µM of the inhibitor. Consequently, Pg colonies lost pigmentation as inhibitor concentrations increased. The inhibitor also reduced IL-1β release from monocytic cells stimulated by the 12-species biofilm. The studied [4,5-c]pyridine-based inhibitor is able to modify virulence of a multispecies biofilm. It might have the potential to be a promising approach in periodontal prevention and therapy.
1 Introduction
Periodontitis is a chronic disease that affects the tissues surrounding the teeth and can lead to tooth loss if left untreated. It is one of the most prevalent diseases worldwide; a recent systematic review estimated a prevalence of 61.6% among adult dentate individuals (1). The disease is triggered by a dysbiotic microbiota, which causes an increase in subtypes of epithelial cells and fibroblasts with an inflammatory signature, a dysregulated neutrophil response, an increase in pro-inflammatory (M1) macrophages, and lymphocytes with innate immune properties. All these factors together activate NF-κB signaling, which promotes bone resorption and suppresses the expression of bone matrix proteins (48). The transformation of the eubiotic microbiota into a dysbiotic state associated with periodontal disease has been linked to Porphyromonas gingivalis. This keystone pathogen can impair the host's protective response while promoting inflammation (2). Major virulence factors of P. gingivalis include its two arginine-specific (RgpA, RgpB) and one lysine-specific (Kgp) cysteine proteases, also known as gingipains (3). Other bacterial species, such as Tannerella forsythia and Treponema denticola, are designated as inflammatory pathobionts, as their virulence genes are upregulated only when tissue destruction has already occurred (2). The subgingival microbiota consists of hundreds of species. Among those enriched in periodontitis compared to periodontal health are, in addition to the previously mentioned three species, Prevotella intermedia, various Fusobacterium nucleatum subspecies, Filifactor alocis, Fretibacterium species, and Saccharibacteria TM7 (4).
According to the European Federation of Periodontology (EFP) S3 level clinical practice guideline (5), the treatment of periodontal disease involves several steps. The first step focuses on controlling supragingival biofilm, gingival inflammation, and risk factors. The second step includes instrumentation, with or without adjuncts. Patients who do not respond adequately receive additional treatment, such as periodontal surgery, in the third step. Finally, all patients should participate in a supportive care program at intervals of 3–12 months. As before, controlling supragingival biofilm, gingival inflammation, and risk factors remain essential parts of this stage of therapy (5).
As the dysbiotic biofilm plays a crucial role in disease development, the use of antiseptics and/or antibiotics is discussed in periodontal treatment. According to the guidelines mentioned earlier, the adjunctive use of antiseptics (chlorhexidine) may be considered in the second step of therapy (5). Chlorhexidine is a bisbiguanide and is widely used in dental practice (6). It acts quickly to kill many bacteria (7). Specifically, when included in a mouthwash, it effectively prevents new biofilm formation (8). However, adverse effects like tooth discoloration, taste changes, irritation of the oral mucosa or tongue are commonly reported (8, 9).
Using systemic antibiotics as an adjunct improves clinical outcomes (10). However, routine use is not advised; it should only be considered for severe periodontitis in young adults (5). A recent analysis supports this, showing that a combination of amoxicillin and metronidazole effectively halts disease progression, especially in patients with generalized periodontitis at stage III/grade C (11). Antibiotics can cause side effects like gastrointestinal issues, metallic taste, and nausea (10). Additionally, the rise of antimicrobial resistance has become a growing global concern, with nearly 5 million deaths estimated in 2019 linked to infections caused by antibiotic-resistant bacteria (12). One of the key factors contributing to this increase is the widespread use of antibiotics (13).
This suggests that antibiotic use should be limited and alternative options explored. Altering bacterial virulence factors, such as gingipains, presents an interesting approach (14). One potential target could be glutaminyl cyclase (QC) in P. gingivalis. This enzyme is a mammalian-like type II enzyme that catalyzes the cyclization of glutamine (Q) to pyroglutamate (15, 16). Most proteins secreted by P. gingivalis via the type 9 secretion system (T9SS), including gingipains, and those exported into the periplasm are transported through the inner membrane via the Sec pathway, where the signal peptide is cleaved by a signal peptidase (17). Downstream of this cleavage site, the proteins often carry a Q, which seems to be cyclized by QC attached to the inner membrane on the periplasmic side (15). Besides P. gingivalis, a QC is also found in T. forsythia and P. intermedia (15, 16). The first tested QC inhibitor was able to suppress growth in these three species but did not impact other oral bacteria (16). Further development led to the discovery of a new class of P. gingivalis QC inhibitors based on a tetrahydroimidazo[4,5-c]pyridine scaffold (18). Selected compounds could inhibit the QC of P. gingivalis and P. intermedia at concentrations around 0.1 µM, and those of T. forsythia at 0.2 µM (18).
The purpose of the experiments outlined below was to determine the effects of a newly developed [4,5-c]pyridine-based QC inhibitor (S-0636) on two different multispecies biofilms. It was shown that the inhibitor does not affect viability of P. gingivalis, that it inhibits hemagglutination and invasion of P. gingivalis into keratinocytes and that it is potentially safe to host cells (Taudte et al. manuscript sumitted (19)). The key questions were: a) Does the inhibitor impact the biofilm overall structure, including biofilm biomass covering bacteria and matrix, total bacterial counts, and metabolic activity? b) Does it influence specific bacterial populations (such as commensals and pathobionts like T. forsythia, P. gingivalis, P. intermedia)? c) Does it affect bacterial virulence, specifically the Arg-gingipain activity of P. gingivalis? d) What concentration of the inhibitor disrupts the biofilm? e) Are there differences between the different types of biofilms (4-species without P. gingivalis, and 12-species)? and f) Does pretreating the biofilm with the inhibitor modulate the immune response?
2 Materials and methods
2.1 Inhibitors
From a panel of different compounds, a [4,5-c]pyridine-based inhibitor was selected: S-0636. Its synthesis is described elsewhere (19). It was dissolved in phosphate buffered saline (PBS, Gibco™ PBS pH 7.4,Thermofisher Scientific, Waltham, MA, USA). The inhibitor was used in at final concentrations of 500 µM, 250 µM, 125 µM, 62.5 µM, 31.25 µM in the assays. PBS served as a negative control.
2.2 Microorganisms in defined biofilm
Two different biofilms were used. The four-species biofilm included Streptococcus gordonii ATCC 10558, Actinomyces naeslundii ATCC 12104, Fusobacterium nucleatum ATCC 25586, and Tannerella forsythia ATCC 43037. For the 12-species biofilm, the following species were additionally included: Porphyromonas gingivalis ATCC 33277, Prevotella intermedia ATCC 25611, Campylobacter rectus ATCC 33238, Capnocytophaga gingivalis ATCC 33624, Eikenella corrodens ATCC 23834, Filifactor alocis ATCC 33238, Parvimonas micra ATCC 33270, and Treponema denticola ATCC 35405.
The strains were passaged on tryptic-soy agar plates (Oxoid, Basingstoke, GB) with 5% sheep blood (and with 10 mg/L N-acetylic muramic acid for T. forsythia). T. denticola ATCC 35405 was maintained in modified mycoplasma broth (BD, Franklin Lake, NJ) enriched with 1 g/mL cysteine, and 5 μg/mL cocarboxylase. All the strains were cultured at 37 °C, streptococci, and A. naeslundii ATCC 12104 in 10% of CO2, the other strains under anaerobic conditions.
2.3 Biofilm formation
The biofilm formation followed our recently published protocols (20, 21). Ninety-six-well plates were coated with 10 µL/well of a protein solution containing 0.67% mucin (Merck KGaA, Darmstadt, Germany) and 1.5% (w/v) bovine serum albumin (SERVA Electrophoresis GmbH, Heidelberg, Germany) for 15 min.
Twenty-four hours cultures of bacterial strains were scraped from agar-plates suspended in 0.9% w/v NaCl solution based on OD600nm = 1 (about 1.2 × 109 bacteria/mL). Then, the bacterial mixture was prepared using 1 part S. gordonii, 2 parts A. naeslundii and 4 parts of the other strains except T. forsythia, P. gingivalis and T. denticola (in case of the 4-species biofilm 1 part S. gordonii, 2 parts A. naeslundii, each 4 parts F. nucleatum, T. forsythia). This mixture was added in a ratio 1: 9 to the culture medium [Wilkins-Chalgren broth (Oxoid, Basingstoke, GB)] with 10 mg/L β-NAD + 10 mg/L N-acetylic muramic acid. From this suspension, 200 µL was pipetted per well and the plate was incubated for 6 h under anaerobic conditions. Afterward, 50 µL of a suspension (prepared as before for the other strains) containing T. forsythia (P. gingivalis and T. denticola according to the experiments) along with the inhibitor at the respective concentration or a control, was added (final volume 250 µL). Following an additional 18 h of incubation (total 24 h) in anaerobic conditions, the samples were analyzed.
2.4 Analysis of biofilms
The supernatant of the biofilm was removed and the biofilms were briefly and carefully washed once. Then, 250 µL of 0.9% w/v NaCl was added. The biofilms were scraped from the surface and thoroughly mixed. From that suspension, 25 µL were used to determine colony forming unit counts (CFU), 25 µL for measuring metabolic activity, 25 µL for biofilm biomass determination, 25 µL for qPCR, and 100 µL for quantifying the arginine-specific amidolylic activity (BApNA).
A 10-fold dilution series was performed on the aliquot designated for CFU determination. Each 25 µL sample was plated on tryptic-soy-agar plates (Oxoid) supplemented with 5% sheep blood. One series was incubated under anerobic conditions (representing total CFU counts), while the other was incubated with 10% CO2 to count of streptococci and actinomyces (as commensals)). Measurement of metabolic activity and of biofilm omass followed recently published methods (22–24). For measuring metabolic activity, 200 mg/L resazurine (Merck KGaA) was mixed with nutrient media at a ratio 1:10, each 100 µL per well was added to the biofilm suspension in a 96-well-plate. After 1-hour incubation at 37 °C, the plate was read at 570 nm against 600 nm. For quantification the biofilm biomass, biofilm aliquots were pipetted into a new 96-well-plate and fixed on the surface at 60 °C for 60 min. Next, 50 µL of a 0.06% (w/v) crystal violet solution was added per well. After washing, 50 µL of 30% acetic acid were added, and after 10 min the plate was read at 600 nm.
The counts of T. forsythia, P. gingivalis and P. intermedia were determined by using qPCR (25). Additionally, in a 12-species biofilm (including P. gingivalis), the arginine-specific amidolytic activity was measured. The respective suspension was centrifuged for 10 min with 5,000 g at 20 °C. To the sediment, 200 µL NaCl was added, the mixture was exposed to ultrasonication to breakdown the cell wall of bacteria, ensuring that cell-wall activity was also included the measurements. The amidolytic activity was monitored at 405 nm for 1 h at 37 °C after adding 0.5 mM N-a-benzoyl-DLarginine-p-nitroanilide (BApNA; Merck KgaA) in 1.0 mL of 0.2 M Tris–HCl, 0.1 M NaCl, 5 mM CaCl2, 10 mM cysteine, pH 7.6.
Additionally, in selected samples (12-species biofilm, 500 µM of inhibitor and control), the expression of gingipains was quantified and the location of P. gingivalis and T. forsythia was visualized. To quantify gingipain expression, RNA was extracted, cDNA was synthesized, and qPCR was performed as described previously (26).
For visualization by FISH technology, biofilms were cultured as before but on glass slides in 24-well plates. Samples have been were fixed in 4% paraformaldehyde/PBS for 1 h. After washing in PBS and preincubating in hybridization buffer (20% formamide, 0.9 M NaCl, 20 mM Tris-HCl, 0.01% SDS) at 46 °C for 15 min, the probes for P. gingivalis [Pg-Cy3 (27)], T. forsythia (Tf-Atto-488, sequence (27) and all bacteria [EUB 338-Atto-425, sequence (28)] in concentrations of 1 µM, 1.5 µM and 0.5 µM were added to the hybridization buffer and incubated for 3 h at 46 °C. After several washing steps, samples were embedded and visualized using a Zeiss LSM 710 confocal microscope (Carl Zeiss). Additional visualization was conducted with Imarisviewer software (Bitplane, IMARIS 10.0.0).
2.5 Interaction of monocytic cells with biofilm
The biofilms were formed as described above for 24 h.
MONO-MAC-6-cells (DSMZ no. ACC 124), a monocytic cell line of human origin, were cultured with RPMI 1640 medium (Invitrogen; ThermoFisher Scientific, Waltham, MA, USA) supplemented with 10% fetale bovine serum (FBS; Invitrogen). Before use in the experiments, MONO-MAC-6 cells were washed once and adjusted to 105 cells/mL in RPMI 1640 medium with 0.5% v/v FBS and the inhibitor at the respective concentration.
The supernatants of the biofilms were carefully removed and replaced with 200 µL/well of MONO-MAC-6 cell suspension, with and without inhibitor. There is always also a cell control w/o biofilm. After 4 h of incubation, the media were taken out, centrifuged at 250 g at 20 °C for 10 min. The supernatants were stored at −80 °C until analysis. Commercially ELISA kits (R&D Systems, Minnesota, MN, USA) were used to measure the levels of released interleukin IL-1β and IL-10 according to the manufacturer's instructions. The detection limit was 1 pg/mL for both cytokines.
An influence on cell vitality by the experimental settings was screened using the MTT assay (29).
2.6 Statistical analysis
In all biofilm experiments, each of eight independent biological samples (obtained in two series) was included in the statistical analysis. Bacterial counts were recorded as log10. In presentations, the arbitrary units of activity and the mass data of the biofilm were related to the means of the untreated controls. The data were compared using a one-way analysis of variance (ANOVA) with a post-hoc comparison using Bonferroni correction. The statistical analysis was performed with SPSS 29.0 (IBM, Armonk, NY, USA).
3 Results
Of the post-hoc analyses, only statistically significant differences vs. control are presented.
3.1 Four-species biofilm
The total CFU counts of the 4-species biofilm without the addition of an inhibitor were 7.42 ± 0.10 log10, and the counts of T. forsythia were 6.65 ± 0.15 log10. There was no statistically significant difference in bacterial counts, biofilm activity, or biofilm biomass when any concentration of the inhibitor was added (Figure 1).
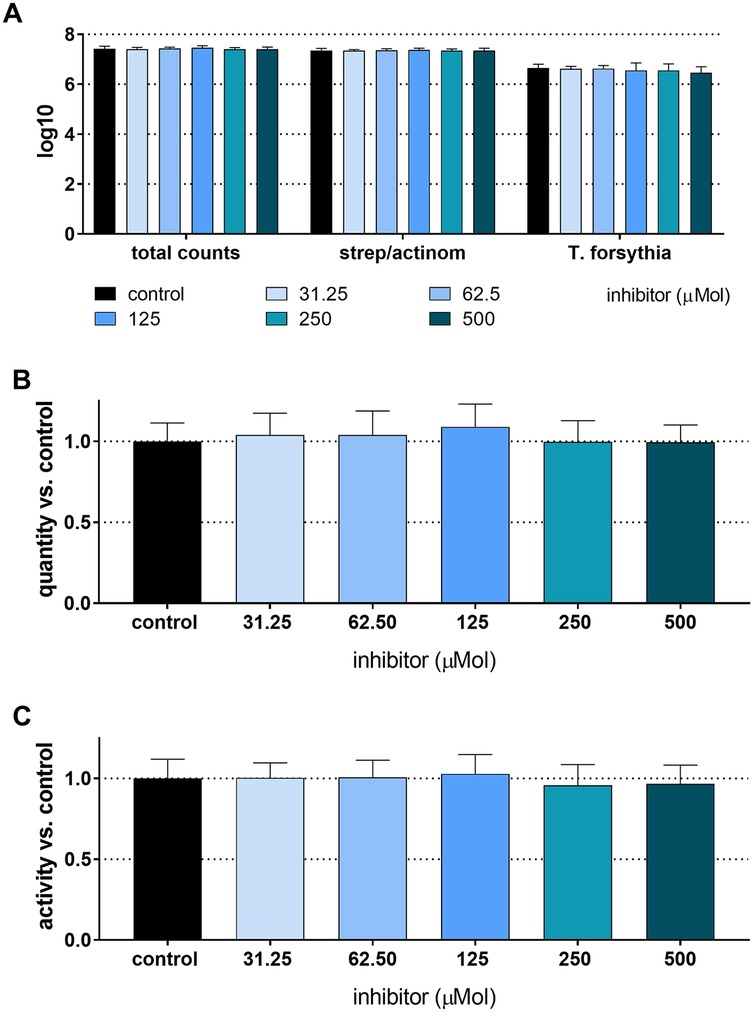
Figure 1. Total counts and counts of streptococci/actinomyces and Tannerella forsythia (A), biomass (B), and metabolic activity (C) of 24 h 4-species biofilms cultured with and without different concentrations of S-0636.
3.2 Twelve-species biofilm
The total CFU counts in the 12-species biofilm without added inhibitor were 7.75 ± 0.05 log10. T. forsythia counts were 6.05 ± 0.12 log10, P. gingivalis counts were 6.37 ± 0.23 log10, and P. intermedia counts were 6.55 ± 0.09 log10. No bacterial counts showed a statistically significant difference, except for P. intermedia, which decreased in a concentration-dependent manner. The lowest counts occured at 500 µM of the inhibitor (5.84 ± 0.11 log10). The biofilm biomass was statistically significantly reduced, when treated with 250 µM (82.7 ± 7%, p < 0.001) and 500 µM of the inhibitor (75.2 ± 6.5%, p < 0.001). Additionally, the total biofilm metabolic activity decreased statistically significantly at inhibitor concentrations of 125 µM and higher (125 µM to 93.3 ± 3.7%, p = 0.033; 250 µM to 90.3 ± 4.8%, p < 0.001; 500 µM to 87.2 ± 5.8%, p < 0.001). The arginine-specific amidolytic activities of the biofilm decreased in a concentration-dependent manner with the inhibitor, and the results were statistically significant compared to control at 62.5 µM (85.2 ± 7.0%, p = 0.009), 125 µM (77.1 ± 5.9%, p < 0.001), 250 µM (70.7 ± 6.2%, p < 0.001) and 500 µM (60.4 ± 10.2%, p < 0.001) (Figure 2).
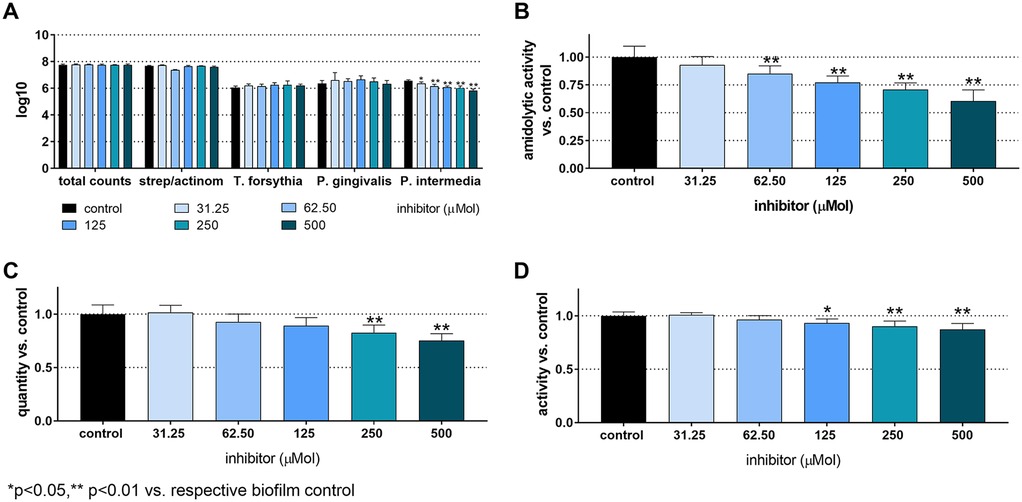
Figure 2. Total counts and counts of streptococci/actinomyces, Tannerella forsythia, Porphyromonas gingivalis, and Prevotella intermedia (A), arginine-specific amidolytic activity (B), biomass (C), and metabolic activity (D) of 24 h 12-species biofilms cultured without and with different concentrations of S-0636. *p < 0.05, **p < 0.01 vs. control.
It is of interest to note that P. gingivalis colonies lost pigmentation after high concentrations of the inhibitors (Figure 3).
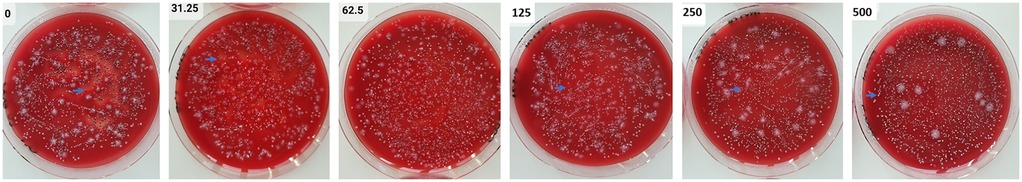
Figure 3. Agar plates with samples from 24 h 12-species biofilms were tested both without and with varying concentrations of S-0636. After performing a dilution series, aliquots of the biofilms were spread on agar plates and incubated anaerobically for 8 days. P. gingivalis (→) forms black colonies without the inhibitor; with increasing concentrations of the inhibitor, its colonies gradually lose their pigmentation.
3.3 mRNA expression of gingipains and FISH staining of biofilms
The analysis of mRNA expression of the gingipains after culturing the biofilm with 500 µM of the inhibitor resulted in nearly no difference for kgp (0.84 ± 0.17-fold) and a slight increase for rgpA (1.81 ± 0.47-fold) and rgpB (3.54 ± 0.17-fold) comparted to the untreated biofilm.
Confocal laser scanning microscopy (CLSM) photographs were taken from biofilms stained with three different FISH-probes. The images confirm the greater thickness of the control biofilms compared to the biofilm exposed to 500 µM of the [4,5-c]pyridine-based inhibitor (Figure 4).
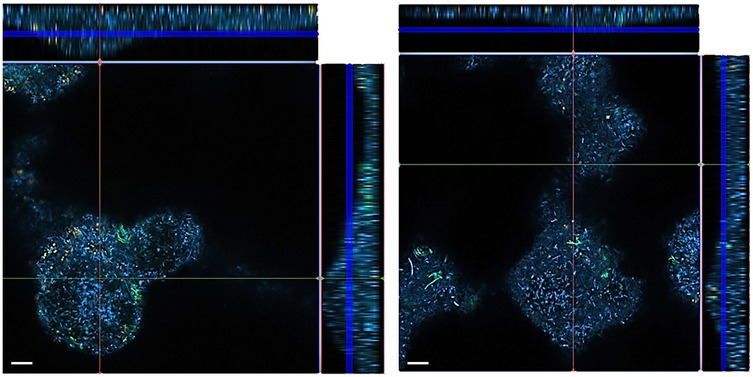
Figure 4. FISH images of 12-species biofilms cultured without (left) and with 500 µM of S-0636 (right). P. gingivalis appears yellow-orange, T. forsythia green, and all other bacteria are blue.
The photographs taken from different layers of the biofilm suggest that P. gingivalis is primarily located at the bottom, T. forsythia is found more in the middle layers of the biofilm, and it appears that T. forsythia forms clusters within the biofilms. Clear differences between the control biofilm and those cultured with 500 µM of the inhibitor are not visible (Figure 5).
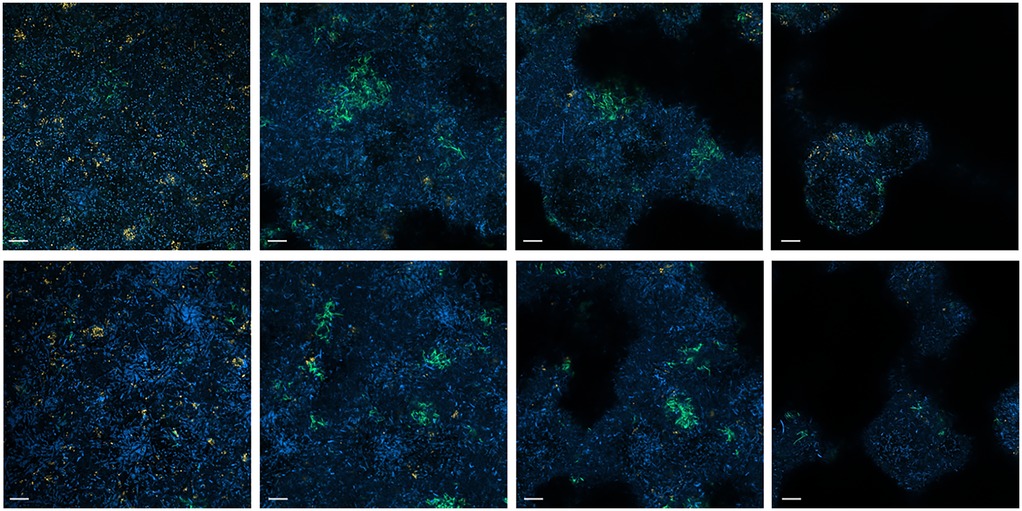
Figure 5. FISH images (630-fold magnification) of 12-species biofilms cultured without (upper row) and with 500 µM of S-0636 (lower row). Different layers of the biofilm from bottom to top (left to right) are visualized. P. gingivalis appears yellow-orange, T. forsythia green, and all other bacteria are blue. Bar 10 µm.
3.4 Release of IL-1β and IL-10
Without bacterial stimulus, the MONO-MAC-6 cells did not release detectable IL-1β, but they did produce moderate amounts of IL-10. With the influence of biofilms, IL-1β levels increased and were highest in the case of the 12-species biofilm without inhibitor. The [4,5-c]pyridine-based inhibitor reduced the IL-1β release stimulated by the 12-species biofilm. In case of the 4-species biofilm, the IL-1β levels in the presence of the compound did not change statistically significantly compared to the 4-species biofilm without inhibitor (Figure 6A). Exposure to biofilms (both 4- and 12-species) decreased IL-10 levels. The inhibitor had no additional effect (Figure 6B). The MTT-test confirmed that S-0636 did not cause any cytotoxic reaction in the MONO-MAC-6 cells.
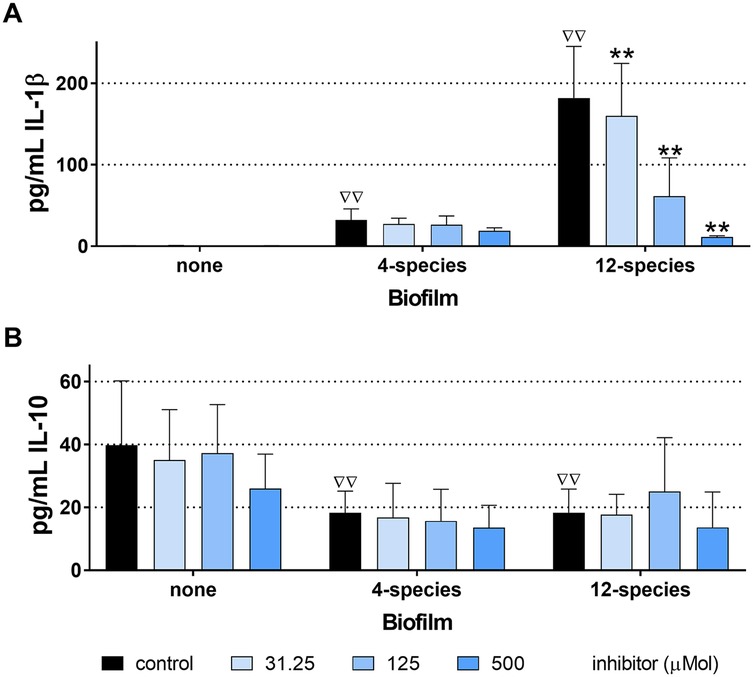
Figure 6. Levels of interleukin (IL)-1β (A) and IL-10 (B) released from MONO-MAC-6 cells after 4 h exposure to biofilms cultured without and with various concentrations of S-0636, ΔΔp < 0.01 compared to no biofilm (comparison only made for controls).**p < 0.01 vs. respective biofilm control.
4 Discussion
The experiments focused on the effect of a [4,5-c]pyridine-based inhibitor on two defined biofilms. Two biofilms were included, a 4-species biofilm containing bacteria with a QC only T. forsythia, and a 12-species biofilm that also included P. gingivalis and P. intermedia.
One of the questions to be answered was whether there was a difference between the two biofilms. The 4-species biofilm was not affected by the inhibitor. Statistically significant differences were found only when using the 12-species biofilm. It might be linked to a stronger inhibition (lower Ki-value) of P. gingivalis QC compared to T. forsythia QC (30).
Other questions were whether the inhibitor affects the biofilm as a whole and whether it alters the counts of specific bacteria. The total bacterial counts of the biofilm did not change. The counts of individual species, including P. gingivalis, T. forsythia, and the commensals (streptococci/actinomyces) remained stable. This is supported by the FISH images which clearly showed no differences in the counts of P. gingivalis and T. forsythia.
Only P. intermedia showed a concentration-dependent decrease in the number of cells in the biofilm treated with the inhibitor. A direct bactericidal activity of the inhibitor via QC can likely be ruled out, as the inhibitor's potency was higher compared to P. gingivalis QC (30). A possible reason for the reduction of P. intermedia numbers in the biofilm may be a decrease in the coaggregation ability between P. gingivalis and P. intermedia. An important coaggregation factor of P. gingivalis with P. intermedia is a protein from the outer membrane vesicles encoded by gingipain genes (31), whose secretion can be hindered.
The lower metabolic activity and biofilm biomass could indicate that impaired synergy in the biofilm formation. Since total bacterial counts were unaffected, the reduced biofilm biomass suggests an effect on the biofilm's matrix. Synergistic effects among periodontal bacteria have been described for P. gingivalis. Diffusible signaling molecules from P. gingivalis can interfere with F. nucleatum's metabolism and promote increased polysaccharide synthesis (32). A strong synergy in biofilm formation was observed for P. gingivalis and T. denticola, with gingipains playing a key role (33). RgpA appears to facilitate the growth of other oral species within a community, including P. gingivalis, P. micra, F. nucleatum and Streptococcus constellatus (34). Considering the specificity of the inhibitor, molecules being transported by the T9SS may also contribute to the formation of the biofilm matrix. According to functional categories, a high percentage of proteins involved in T9SS, outer membrane functions, and peptidoglycan biosynthesis carry an N-terminal Q (35). Pyroglutaminylation of the proteins by QC appears to stabilize them and protects them from degradation (35). It can be assumed that fewer proteins contributing to matrix formation are available; however, knowledge about proteins produced by P. gingivalis, T. forsythia and P. intermedia that contribute to biofilm matrix synthesis is remains limited.
The Arg-gingipain specific amidolytic activity, which correlates with P. gingivalis virulence, was clearly reduced by the QC inhibitor in a concentration-dependent manner. Our data suggest limited synthesis or processing of the gingipains. However, limited synthesis can be ruled out at least at the mRNA level. Synthesized prepro-gingipains undergo extensive post-translational proteolytic processing, similar to many other virulence factors, as they are first transported by the Sec system to periplasm and then through the T9SS (36). Blocking specific components of T9SS caused a loss of cell-associated gingipain activity (Kgp or total) and colony pigmentation (37, 38). In our study, we also observed a loss of colony pigmentation in P. gingivalis was observed. It has been shown that glutamine cyclization to pyro-glutamate plays a role in cell-associated activity of RgpA (15). The underlying mechanisms might be explained by the fact that proteins with N-terminal Q involved in T9SS (35) are either not functional or only partially functional when a QC inhibitor is present.
Furthermore, we evaluated the potential interaction between the QC inhibitor's effect on biofilm and the host response. For this, the pre-cultivated biofilm, with and without the inhibitor, was exposed to monocytic cells for 4 h, after which the release of interleukin (IL)-1β, a pro-inflammatory cytokine and IL-10, an anti-inflammatory cytokine was measured. In periodontitis, macrophages can be polarized into M1 or M2 types. M1-macrophages, which promote inflammation are characterized by high levels of pro-inflammatory cytokines, such as IL-1. In contrast, M2-macrophages, which facilitate anti-inflammatory functions, are marked by high levels of IL-10 (39). The initiation and progression of periodontal disease are driven by a dysregulated inflammatory response to a dysbiotic biofilm (40). This is supported by the in vitro-result showing high levels of IL-1β and sustained levels of IL-10 after exposure of the 12-species biofilm to monocytic cells. IL-1β is known to limit bacterial dissemination but mainly mediates periodontal inflammation, activation of osteoclasts and tissue destruction (41). A high level of IL-1β in saliva is an indicator for an occurring inflammation in periodontal tissue (42). Clinically, high IL-1β-level decrease after periodontal therapy, coinciding with improvements in periodontal clinical parameters (43). In the present study, IL-1β levels also decreased following application of the inhibitor.
Results in the 12-species biofilm were clearly dependent on the concentration of the inhibitor used. Since the compound will be incorporated into an oral health-care product for local application, it should be available at a sufficiently high concentration. It should be noted that in this in vitro study, the concentration of the inhibitor remained stable within the experimental setup. In vivo, depending on the application site, the flow of saliva or gingival crevicular fluid must be considered. The mean unstimulated salivary flow rate was measured at 287 µL/min (44), while stimulated salivary flow can reach up to 640 µL/min (45). Gingival crevicular fluid flow was approximately 0.5 µL/min (46). To address the issue of immediate dilution of the compound, it may be necessary to incorporate it into a slow-release device that can maintain the active concentration over an extended period (47).
The advantage of the study was using a clearly defined study design that allowed comparison of the two biofilms and different inhibitor concentrations. The studied variables enabled conclusions about the anti-biofilm, anti-virulence, and immunomodulatory properties of the inhibitor. However, the models only reflected part of the situation in the oral cavity; the biofilm consisted of up to 12 species, the interaction with host cells included only a monocytic cell line, and salivary or gingival crevicular fluid flow was not considered. These could be limitations of the study, highlighting the need for further in vitro research with more complex models. Finally, the efficacy of the inhibitor must be demonstrated through randomized clinical trials.
Overall, the compound might have potential for inclusion in oral health care products. It may alter the virulence of a dysbiotic biofilm, leading to a balanced pro- and anti-inflammatory responses. Since the inhibitor clearly acts in a concentration-dependent manner, it should be included at a sufficiently high concentration in an oral health care product.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
SE: Conceptualization, Data curation, Methodology, Supervision, Writing – original draft, Writing – review & editing. NT: Conceptualization, Writing – original draft, Writing – review & editing. DR: Conceptualization, Writing – review & editing. AM: Formal analysis, Investigation, Writing – review & editing. AS: Resources, Writing – review & editing. JP: Conceptualization, Writing – review & editing. MB: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The study was funded by the Federal Ministry of Education and Research, Germany; Grant No: 16LW0362K.
Acknowledgement
We are grateful to Xilei Zhu for help in taking the CLSM photographs. We dedicate this work to Professor Hans-Ulrich Demuth, one of the initiators of this project, whose visionary ideas and scientific insights laid the foundation for this research. His pioneering contributions continue to inspire and help us to guide our work in this field.
Conflict of interest
MB is shareholder of PerioTrap Pharmaceutical GmbH. SE and JP are member of the Scientific Advisory board of PerioTrap Pharmaceutical GmbH.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Trindade D, Carvalho R, Machado V, Chambrone L, Mendes JJ, Botelho J. Prevalence of periodontitis in dentate people between 2011 and 2020: a systematic review and meta-analysis of epidemiological studies. J Clin Periodontol. (2023) 50(5):604–26. doi: 10.1111/jcpe.13769
2. Hajishengallis G, Lamont RJ. Polymicrobial communities in periodontal disease: their quasi-organismal nature and dialogue with the host. Periodontol 2000. (2021) 86(1):210–30. doi: 10.1111/prd.12371
3. Hocevar K, Potempa J, Turk B. Host cell-surface proteins as substrates of gingipains, the main proteases of Porphyromonas gingivalis. Biol Chem. (2018) 399(12):1353–61. doi: 10.1515/hsz-2018-0215
4. Abusleme L, Hoare A, Hong BY, Diaz PI. Microbial signatures of health, gingivitis, and periodontitis. Periodontol 2000. (2021) 86(1):57–78. doi: 10.1111/prd.12362
5. Sanz M, Herrera D, Kebschull M, Chapple I, Jepsen S, Beglundh T, et al. Treatment of stage I-III periodontitis-the EFP S3 level clinical practice guideline. J Clin Periodontol. (2020) 47(Suppl 22):4–60. doi: 10.1111/jcpe.13290
6. Thangavelu A, Kaspar SS, Kathirvelu RP, Srinivasan B, Srinivasan S, Sundram R. Chlorhexidine: an elixir for periodontics. J Pharm Bioallied Sci. (2020) 12(Suppl 1):S57–s59. doi: 10.4103/jpbs.JPBS_162_20
7. Eick S, Radakovic S, Pfister W, Nietzsche S, Sculean A. Efficacy of taurolidine against periodontopathic species–an in vitro study. Clin Oral Investig. (2012) 16(3):735–44. doi: 10.1007/s00784-011-0567-2
8. Supranoto SC, Slot DE, Addy M, Van der Weijden GA. The effect of chlorhexidine dentifrice or gel versus chlorhexidine mouthwash on plaque, gingivitis, bleeding and tooth discoloration: a systematic review. Int J Dent Hyg. (2015) 13(2):83–92. doi: 10.1111/idh.12078
9. James P, Worthington HV, Parnell C, Harding M, Lamont T, Cheung A, et al. Chlorhexidine mouthrinse as an adjunctive treatment for gingival health. Cochrane Database Syst Rev. (2017) 3(3):Cd008676. doi: 10.1002/14651858.CD008676.pub2
10. Teughels W, Feres M, Oud V, Martín C, Matesanz P, Herrera D. Adjunctive effect of systemic antimicrobials in periodontitis therapy: a systematic review and meta-analysis. J Clin Periodontol. (2020) 47(Suppl 22):257–81. doi: 10.1111/jcpe.13264
11. Eickholz P, Koch R, Göde M, Nickles K, Kocher T, Lorenz K, et al. Clinical benefits of systemic amoxicillin/metronidazole may depend on periodontitis stage and grade: an exploratory sub-analysis of the ABPARO trial. J Clin Periodontol. (2023) 50(9):1239–52. doi: 10.1111/jcpe.13838
12. Collaborators AR. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. (2022) 399(10325):629–55. doi: 10.1016/s0140-6736(21)02724-0
13. Chatterjee A, Modarai M, Naylor NR, Boyd SE, Atun R, Barlow J, et al. Quantifying drivers of antibiotic resistance in humans: a systematic review. Lancet Infect Dis. (2018) 18(12):e368–78. doi: 10.1016/s1473-3099(18)30296-2
14. Pedrosa ME, Martin V, Fernandes MH, Gomes PS. Gingipain inhibitors as an innovative therapy for periodontal and associated-systemic diseases: a systematic review. Clin Oral Investig. (2025) 29(9):418. doi: 10.1007/s00784-025-06472-5
15. Bochtler M, Mizgalska D, Veillard F, Nowak ML, Houston J, Veith P, et al. The bacteroidetes Q-rule: pyroglutamate in signal peptidase I substrates. Front Microbiol. (2018) 9:230. doi: 10.3389/fmicb.2018.00230
16. Taudte N, Linnert M, Rahfeld JU, Piechotta A, Ramsbeck D, Buchholz M, et al. Mammalian-like type II glutaminyl cyclases in Porphyromonas gingivalis and other oral pathogenic bacteria as targets for treatment of periodontitis. J Biol Chem. (2021) 296:100263. doi: 10.1016/j.jbc.2021.100263
17. Lasica AM, Ksiazek M, Madej M, Potempa J. The type IX secretion system (T9SS): highlights and recent insights into its structure and function. Front Cell Infect Microbiol. (2017) 7:215. doi: 10.3389/fcimb.2017.00215
18. Ramsbeck D, Taudte N, Janckel N, Strich S, Rahfeld JU, Buchholz M. Tetrahydroimidazo[4,5-c]pyridine-based inhibitors of Porphyromonas gingivalis glutaminyl cyclase. Pharmaceuticals (Basel). (2021) 14(12):1206. doi: 10.3390/ph14121206
19. Taudte N, Liebe L, Jänckel N, Ramsbeck D, Schilling S, Potempa J, et al. (2025). Inhibition of PgQC by a novel small molecule diminishes pathogenic activity of Porphyromonas gingivalis Frontiers in Microbiology, submitted.
20. Zollinger L, Schnyder S, Nietzsche S, Sculean A, Eick S. In vitro activity of taurolidine on single species and a multispecies population associated with periodontitis. Anaerobe. (2015) 32:18–23. doi: 10.1016/j.anaerobe.2014.11.008
21. Zhu X, Sculean A, Eick S. In vitro effects of different hyaluronic acids on periodontal biofilm-immune cell interaction. Front Cell Infect Microbiol. (2024) 14:1414861. doi: 10.3389/fcimb.2024.1414861
22. Pettit RK, Weber CA, Kean MJ, Hoffmann H, Pettit GR, Tan R, et al. Microplate alamar blue assay for Staphylococcus epidermidis biofilm susceptibility testing. Antimicrob Agents Chemother. (2005) 49(7):2612–7. doi: 10.1128/AAC.49.7.2612-2617.2005
23. Kwasny SM, Opperman TJ. Static biofilm cultures of gram-positive pathogens grown in a microtiter format used for anti-biofilm drug discovery. Curr Protoc Pharmacol. (2010) 50(1):13A-8. doi: 10.1002/0471141755.ph13a08s50. Chapter 13, Unit 13A 18.
24. Pirracchio L, Joos A, Luder N, Sculean A, Eick S. Activity of taurolidine gels on ex vivo periodontal biofilm. Clin Oral Investig. (2018) 22(5):2031–7. doi: 10.1007/s00784-017-2297-6
25. Jentsch HFR, Heusinger T, Weickert A, Eick S. Professional tooth cleaning prior to non-surgical periodontal therapy: a randomized clinical trial. J Periodontol. (2020) 91(2):174–82. doi: 10.1002/JPER.19-0023
26. Eick S, Gadzo N, Tacchi M, Sculean A, Potempa J, Stavropoulos A. Gingipains impair attachment of epithelial cell to dental titanium abutment surfaces. J Biomed Mater Res B Appl Biomater. (2019) 107(8):2549–56. doi: 10.1002/jbm.b.34345
27. Sunde PT, Olsen I, Göbel UB, Theegarten D, Winter S, Debelian GJ, et al. Fluorescence in situ hybridization (FISH) for direct visualization of bacteria in periapical lesions of asymptomatic root-filled teeth. Microbiology (Reading). (2003) 149(Pt 5):1095–102. doi: 10.1099/mic.0.26077-0
28. Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. (1990) 172(2):762–70. doi: 10.1128/jb.172.2.762-770.1990
29. Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. (1983) 65(1–2):55–63. doi: 10.1016/0022-1759(83)90303-4
30. Jäger C, Liebe L, Ramsbeck D, Linnert M, Geissler S, Piechotto A, et al. Novel inhibitors of bacterial glutaminyl cyclases for use in the treatment of periodontal and related diseases. International Patent Application WIPO WO20197162458 A1 (2019).
31. Kamaguchi A, Ohyama T, Sakai E, Nakamura R, Watanabe T, Baba H, et al. Adhesins encoded by the gingipain genes of Porphyromonas gingivalis are responsible for co-aggregation with Prevotella intermedia. Microbiology (Reading). (2003) 149(Pt 5):1257–64. doi: 10.1099/mic.0.25997-0
32. Yamaguchi-Kuroda Y, Kikuchi Y, Kokubu E, Ishihara K. Porphyromonas gingivalis diffusible signaling molecules enhance Fusobacterium nucleatum biofilm formation via gene expression modulation. J Oral Microbiol. (2023) 15(1):2165001. doi: 10.1080/20002297.2023.2165001
33. Zhu Y, Dashper SG, Chen YY, Crawford S, Slakeski N, Reynolds EC. Porphyromonas gingivalis and Treponema denticola synergistic polymicrobial biofilm development. PLoS One. (2013) 8(8):e71727. doi: 10.1371/journal.pone.0071727
34. Davies JR, Kad T, Neilands J, Kinnby B, Prgomet Z, Bengtsson T, et al. Polymicrobial synergy stimulates Porphyromonas gingivalis survival and gingipain expression in a multi-species subgingival community. BMC Oral Health. (2021) 21(1):639. doi: 10.1186/s12903-021-01971-9
35. Szczesniak K, Veillard F, Scavenius C, Chudzik K, Ferenc K, Bochtler M, et al. The Bacteroidetes Q-rule and glutaminyl cyclase activity increase the stability of extracytoplasmic proteins. mBio. (2023) 14(5):e0098023. doi: 10.1128/mbio.00980-23
36. Veith PD, Glew MD, Gorasia DG, Cascales E, Reynolds EC. The type IX secretion system and its role in bacterial function and pathogenesis. J Dent Res. (2022) 101(4):374–83. doi: 10.1177/00220345211051599
37. Heath JE, Seers CA, Veith PD, Butler CA, Nor Muhammad NA, Chen YY, et al. PG1058 Is a novel multidomain protein component of the bacterial type IX secretion system. PLoS One. (2016) 11(10):e0164313. doi: 10.1371/journal.pone.0164313
38. Naito M, Tominaga T, Shoji M, Nakayama K. PGN_0297 is an essential component of the type IX secretion system (T9SS) in Porphyromonas gingivalis: tn-seq analysis for exhaustive identification of T9SS-related genes. Microbiol Immunol. (2019) 63(1):11–20. doi: 10.1111/1348-0421.12665
39. Sun X, Gao J, Meng X, Lu X, Zhang L, Chen R. Polarized macrophages in periodontitis: characteristics, function, and molecular signaling. Front Immunol. (2021) 12:763334. doi: 10.3389/fimmu.2021.763334
40. Nguyen T, Sedghi L, Ganther S, Malone E, Kamarajan P, Kapila YL. Host-microbe interactions: profiles in the transcriptome, the proteome, and the metabolome. Periodontol 2000. (2020) 82(1):115–28. doi: 10.1111/prd.12316
41. Neurath N, Kesting M. Cytokines in gingivitis and periodontitis: from pathogenesis to therapeutic targets. Front Immunol. (2024) 15:1435054. doi: 10.3389/fimmu.2024.1435054
42. Abdullameer MA, Abdulkareem AA. Salivary interleukin-1β as a biomarker to differentiate between periodontal health, gingivitis, and periodontitis. Minerva Dent Oral Sci. (2023) 72(5):221–9. doi: 10.23736/s2724-6329.23.04778-2
43. Nanakaly HT, Nouri Ahmed S, Warya Azeez H. Effect of periodontal therapy on serum and salivary interleukin-1 beta (IL-1β) and malondialdehyde levels in chronic periodontitis. Cell Mol Biol (Noisy-le-grand). (2024) 70(10):167–73. doi: 10.14715/cmb/2024.70.10.22
44. Gill SK, Price M, Costa RJ. Measurement of saliva flow rate in healthy young humans: influence of collection time and mouthrinse water temperature. Eur J Oral Sci. (2016) 124(5):447–53. doi: 10.1111/eos.12294
45. Ligtenberg AJM, Meuffels M, Veerman ECI. Effects of environmental temperature on saliva flow rate and secretion of protein, amylase and mucin 5B. Arch Oral Biol. (2020) 109:104593. doi: 10.1016/j.archoralbio.2019.104593
46. Lima SNL, Ribeiro IS, Grisotto MA, Fernandes ES, Hass V, de Jesus Tavarez RR, et al. Evaluation of several clinical parameters after bleaching with hydrogen peroxide at different concentrations: a randomized clinical trial. J Dent. (2018) 68:91–7. doi: 10.1016/j.jdent.2017.11.008
47. Schmid JL, Kirchberg M, Sarembe S, Kiesow A, Sculean A, Mäder K, et al. In vitro evaluation of antimicrobial activity of minocycline formulations for topical application in periodontal therapy. Pharmaceutics. (2020) 12(4):352. doi: 10.3390/pharmaceutics12040352
Keywords: modifying bacterial virulence, Porphyromonas gingivalis, alternatives antibiotics, biofilm modulation, periodontitis
Citation: Eick S, Taudte N, Ramsbeck D, Magdoń A, Sculean A, Potempa J and Buchholz M (2025) Effect of a bacterial glutaminyl cyclase inhibitor on multi-species-biofilms. Front. Oral Health 6:1716625. doi: 10.3389/froh.2025.1716625
Received: 30 September 2025; Revised: 3 November 2025;
Accepted: 6 November 2025;
Published: 1 December 2025.
Edited by:
George Grant, University of Aberdeen (retired)/now independent researcher, Aberdeen, United KingdomReviewed by:
Zaleha Shafiei, Universiti Kebangsaan Malaysia, MalaysiaRui Li, China Medical University, China
Copyright: © 2025 Eick, Taudte, Ramsbeck, Magdoń, Sculean, Potempa and Buchholz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sigrun Eick, c2lncnVuLmVpY2tAdW5pYmUuY2g=
†Present Address: Daniel Ramsbeck, Department of Applied Biosciences and Process Engineering, Anhalt University of Applied Sciences, Köthen (Anhalt), Germany
 Sigrun Eick
Sigrun Eick Nadine Taudte2
Nadine Taudte2 Daniel Ramsbeck
Daniel Ramsbeck Jan Potempa
Jan Potempa Mirko Buchholz
Mirko Buchholz