- 1Department of Pharmacy, Shenyang Pharmaceutical University, Shenyang, China
- 2Department of Veterinary Medicine, Northeast Agricultural University, Harbin, China
- 3School of Pharmacy, Nankai University, Tianjin, China
Streptococcus suis is one of the most important swine pathogens, which can cause persistent infection by forming biofilms. In this study, sub-minimum inhibitory concentration (sub-MIC) of rhubarb water extracts were found to inhibit biofilm formation. Two-component signal transduction systems (TCSs), transcriptional regulators, and DNA binding proteins were compared under two conditions: (1) cells treated with sub-MIC rhubarb water extracts and (2) untreated cells. Using an isobaric tags for relative and absolute quantitation (iTRAQ) strategy, we found that TCSs constituent proteins of histidine kinase and response regulator were significantly down-regulated. This down-regulation can affect the transfer of information during biofilm formation. The transcriptional regulators and DNA binding proteins that can interact with TCSs and interrupt gene transcription were also significantly altered. For these reasons, the levels of protein expressions varied in different parts of the treated vs. untreated cells. In summary, rhubarb water extracts might serve as potential inhibitor for the control of S. suis biofilm formation. The change in TCSs, transcriptional regulators, and DNA binding proteins may be important factors in S. suis biofilm inhibition.
Introduction
Streptococcus suis (S. suis) is a Gram-positive bacterium and it is considered to be one of the most important swine pathogens worldwide. It can cause meningitis, arthritis, septicemia, bronchopneumonia, and other pathological conditions (Zhao et al., 2015). At present, at least 29 S. suis serotypes have been identified. S. suis serotype 2 (SS2) is considered to be the most virulent for both humans and swine. In China, up to 70% of S. suis isolates accounting for systemic diseases in piglets are serotype 2. The diseases caused by S. suis 2 are difficult to cure, as S. suis serotype 2 can induce persistent in vivo infections as a result of biofilm formation (Zhao et al., 2015).
Biofilm is a community of microorganisms adhering to each other on biotic or abiotic surfaces, and the bacteria in biofilm are surrounded by self-produced matrix of extracellular polymers that can offer advantages for the microorganisms (Khoramian et al., 2015). Biofilm can reduce the penetration of antimicrobial agents and increase protection against the host immune system. Compared with free-floating bacteria of the same species, biofilm can induce as much as 1000-fold resistance to detergents, antiseptics, and antibiotics (Rasamiravaka et al., 2015). Bacterial biofilm plays an important role in persistent infections, which are rarely eradicated by antimicrobials (Costerton et al., 1999). Thus, inhibiting biofilm formation may be an important strategy for eradicating persistent bacterial infections.
Rhubarb is a drug described in the Chinese Pharmacopoeia, and it has important clinicial applications. The active ingredients of rhubarb are anthraquinones, which have been reported to inhibit the growth of viruses, bacteria, and biofilm. These mainly comprise emodin, rhein, chrysophanol, aloe-emodin, and physcion (Lin et al., 2008; Wierzchacz et al., 2009). Rhein can impair the pathogenicity of Porphyromonas gingivalis pathogenicity by intervening in the transcription of genes, these genes code important virulence factors which affect the bacterial proteolytic activity of the bacterium (Liao et al., 2013). Aloe-emodin can disrupt bacterial membranes by interposing between the two major phospholipids (phosphatidylethanolamine and phosphatidylglycerol) (Liao et al., 2013) that are present in bacterial membranes. It also shows antacid activity against S. mutans biofilm (Kim et al., 2011). Emodin can inhibit of Pseudomonas aeruginosa and Stenotrophomonas maltophilia biofilm formation (Alves et al., 2004). These anthraquinone monomers are the principal components of the rhubarb water extracts, and their effects on S. suis biofilm are currently unknown; therefore, in our study, we aimed to characterize the inhibitory effects of rhubarb water extracts on the formation of S. suis biofilm.
Two-component signal transduction systems (TCSs) are some of the most important cell-to-cell communication systems, and they play a key role in biofilm formation (Rasamiravaka et al., 2015). They consist mainly of a membrane-bound sensor histidine kinase (HK), which perceives a stimulus, and a cytoplasmic response regulator (RR), which mediates the response to the stimulus (Nobile et al., 2012). Biofilm is a multicellular aggregate of micro-organism, and its formation has been shown to be specifically involved with recognizing and responding to small self-generated secreted molecules (Rasamiravaka et al., 2015). The TCSs can recognize small molecules via HK and transmit information through the RR via conserved phosphorylation and dephosphorylation reactions (Jung et al., 2012; Parellada et al., 2013; Siddiqui et al., 2015). The final activated RR can interact with transcriptional regulators and DNA binding proteins, thereby interrupting gene transcription (Siddiqui et al., 2015; Zhao et al., 2015).
In our study, the inhibitory effects of the rhubarb water extracts on S. suis biofilm formation and the mechanisms responsible for this inhibition were investigated. To gain insight into these processes, tissue culture plate (TCP) and scanning electron microscopy (SEM) assessments were used to test the inhibitory effects of rhubarb water extracts on biofilm formation, and the iTRAQ technique was applied to measure variations in TCSs, transcriptional regulators, and DNA binding proteins between untreated and rhubarb water extracts-treated cells.
Materials and Methods
Bacterial Cultures and Biofilm Formation
Streptococcus suis strain ATCC 700794 was grown at 37°C with constant shaking in Todd–Hewitt broth (THB; Sigma–Aldrich, St. Louis, MO, United States) containing (w/v) 0.5% beef extract, 0.3% yeast extract, 2% peptone, 2% calf serum, 0.5% glucose, 0.04% Na2HPO4, 0.25% Na2CO3 and 0.2% NaCl. For biofilm cultures, S. suis was grown in THB medium (Oxoid, Wesel, Germany) supplemented with 1% fibrinogen in 200 mm polystyrene petri dishes at 37°C for 72 h. The supernatant was then removed and the petri dishes were rinsed twice with 50 mM Tris/HC1 (pH 7.5). Biofilm was scraped, and cells were sonicated for 5 min (Bransonic 220; Branson Consolidated Ultrasonic Pvt Ltd., Australia) followed by centrifugation at 4°C for 10 min at 12,000 × g. The supernatant was then discarded. Cell pellets were rinsed twice with 50 mM Tris–HC1 (pH 7.5) and collected by centrifugation (14,000 rpms). S. suis planktonic cells were grown at 37°C for 24 h. The cells were collected and washed as described above.
Rhubarb Water Extracts Preparation and Anthraquinones Analysis
Rhubarb (Tongren Tang Co., Ltd., Beijing, China) 200 g was smashed and soaked at room temperature in 2 L water for 12 h. The suspension was then boiled for 45 min and filtered through gauze. The previous procedures were then repeated but 1.5 L water was added, and the suspension was boiled for 30 min. The two filtrates were combined and concentrated to 100 mL, and then lyophilized.
The anthraquinones content of rhubarb water extracts was analyzed by high-performance liquid chromatography (HPLC) using an RP-C18 column. The mobile phase was methanol–0.1% phosphoric acid water (85:15, v/v) at a flow rate of 1.0 ml/min at room temperature (25°C). Twenty microliters of sample was injected and monitored at 254 nm after they were diluted by the mobile phase. All assays were performed in triplicate.
Effect of Rhubarb Water Extracts on Biofilm Formation Determined by the TCP Assay
The minimum inhibitory concentration (MIC) of rhubarb water extracts against S. suis was determined by the microtiter method as described in the Clinical and Laboratory Standards Institute guidelines, but with Mueller–Hinton broth was replaced by THB.
Streptococcus suis ATCC 700794 in mid-exponential growth phase was adjusted to an optical density of 0.2 at 600 nm (OD600). One-hundred microliters of culture and 100 μL of rhubarb water extracts solution were added to each well of 96-well microplate, and the final concentrations of rhubarb water extracts were 1/2, 1/4, 1/8, or 1/16 × MIC. The cells suspension without rhubarb water extracts was added for the negative controls. After incubation for 72 h without shaking, wells were rinsed with 50 mM phosphate-buffered saline (PBS; pH 7.2), fixed for 30 min with 200 μL of methanol, stained with 200 μL of 1% crystal violet (w/v) for 30 min, rinsed three times with PBS (pH 7.2), and dried for 2 h at 37°C. Finally, 200 μL of 33% acetic acid (v/v) was added, and the plate was shaken for 10 min. All wells were measured using a Tecan GENios Plus microplate reader (Tecan, Austria) at 595 nm (Zhao et al., 2015).
Sub-MIC rhubarb water extracts were added into cell suspension at 0, 24, or 48 h, and cell suspension without rhubarb water extracts served as negative controls. After incubation for 72 h without shaking, the wells were monitored, using a Tecan GENios Plus microplate reader (Tecan, Austria) at 600 nm. All assays were performed in triplicate.
Scanning Electron Microscopy (SEM)
The S. suis ATCC 700794 biofilm was tested by SEM as described by Zhao et al. (2015). Briefly, a mid-exponential S. suis growth culture was diluted to an optical density of 0.1 at 600 nm (OD600), and 1 mL cell suspension was added to 6-well microplate (Corning Co., Ltd., America) in which each well contained an 11 mm × 11 mm sterilized rough organic membrane (Mosutech Co., Ltd., Shanghai, China) on the bottom. The plates were incubated at 37°C for 72 h without shaking, the supernatant was removed, and the organic membranes were rinsed with sterile PBS. Both untreaded and 1/2 MIC rhubarb water extracts-treaded bifilm were fixed with 4% (w/v) glutaraldehyde for 6 h, then washed three times with 0.1 M PBS, and fixed to black transparency with 2% (w/v) osmium tetroxide. After dehydration and critical point drying, samples were sputtered gold-sputtered using an ion sputtering instrument (current 15 mA, 2 min) and examined by scanning electron microscopy (FEI Quanta, The Netherlands).
Determination of the Growth Inhibition Activity of Rhubarb Water Extracts
The growth of both untreated and S. suis treated with 1/2 MIC of rhubarb water extracts was monitored. The samples were incubated at 37°C for 48 h, and OD600 measurements were taken at 4, 6, 8, 10, 12, 24, and 48 h. All assays were performed in triplicate (Wisniewski et al., 2009).
Protein Digestion and iTRAQ Labeling
Protein digestion was carried out according to the filter-aided sample preparation (FASP) procedure. Briefly, 230 μg of protein was mixed with dithiothreitol and boiled for 5 min, and proteins were collected by ultrafiltration (Microcon units, 10 kD). The sample pellets were resuspended and incubated for 30 min in darkness with 100 μL of IAA (50 mM IAA in UA), samples were collected by centrifugation at 4°C for 15 min at 14,000 × g, and 5 μg of trypsin (Promega) in 40 μl of DS buffer was added overnight at 37°C. Finally, the resulting peptides were collected by filtration, and the UV light spectral density at 280 nm using an extinction coefficient of 1.1 of 0.1% solution was estimated.
For labeling, 30 μg of peptides from each sample were labeled with iTRAQ reagents that were dissolved in 70 μl of ethanol and labeled as (Sample1)-117 and (Sample2)-118. The labeled samples were combined and vacuum dried.
Strong Cation Exchange (SCX) Chromatography Separation of iTRAQ Reagent-Labeled Peptides
The labeled samples were separated by SCX chromatography with a PolySULFOETHYL column (4.6 mm × 100 mm, 5 μm, 200 Å, PolyLC Inc., Columbia, MD, United States). The column was equilibrated with buffer A (10 mM KH2PO4, 25% (v/v) acetonitrile, pH 2.7) for 25 min, and the samples were separated at a flow rate of 1 mL/min, using a step gradient of 0–10% (v/v) buffer B (500 mM KCl, 25% (v/v) acetonitrile, 10 mM KH2PO4, pH 2.7) for 2 min; 10–20% (v/v) buffer B for 25 min; 20–45% (v/v) buffer B for 5 min; 45–100% (v/v) buffer B for 5 min; and 100% (v/v) buffer B for 8 min. The elution was monitored at 214 nm, and the collected fractions were desalted on a C18 Cartridge (Sigma).
Liquid Chromatography (LC)–Electrospray Ionization (ESI) Tandem Mass Spectrometry (MS/MS) Analysis
Sample fractions were analyzed on a Q Exactive mass spectrometer (Thermo Finnigan) that was coupled to Easy nLC (Proxeon Biosystems, now Thermo Fisher Scientific). Ten microliters of each fractions were injected and loaded onto a C18-reversed phase column (100 mm × 75 μm, 3 μm) in buffer A (0.1% formic acid). The peptide mixture was separated at a flow rate of 250 nL/min with a linear gradient of buffer B (80% acetonitrile and 0.1% formic acid) controlled by IntelliFlow technology over 140 min. A full scan range from 300 to 1800 m/z was recorded in profile mode with a resolution of 70,000 at m/z for 120 min with resolution for higher-energy collisional dissociation (HCD) spectra set at 17,500 at m/z. Normalized collision energy was set to 30 eV and the underfill ratio was 0.1%. The instrument was run with the peptide recognition mode in the enabled position.
Data Analysis and Statistics
The acquired peak-lists of all MS/MS spectra were found using MASCOT engine (Matrix Science, London, United Kingdom; version 2.2) inset with Proteome Discoverer 1.4 (Thermo Electron, San Jose, CA, United States) against the Uniprot_streptococcus_suis.fasta (38369 sequences, downloaded on November 4th, 2013) and the decoy database. The following parameters were set: Missed cleavage = 2; Enzyme = Trypsin; Peptide mass tolerance = 20 ppm; MS/MS tolerance = 0.1 Da; Fixed modification: Carbamidomethyl (C); iTRAQ8plex(N-term); iTRAQ8plex(K); Variable modification: Oxidation(M); False discovery rate ≤ 0.01.
The values were calculated as the mean of individual experiments in triplicate and compared with those of the control groups. Differences between two mean values were calculated by Student’s t-test using SPSS 11.0 statistical software.
Results
Quantitative Analysis of the Anthraquinones in Rhubarb Extracts
The rhubarb water extracts were analyzed by HPLC, and the mean contents of aloe-emodin, rhein, emodin, chrysophanol and physcion were 0.96 ± 0.0011 mg/g, 2.63 ± 0.0033 mg/g, 1.08 ± 0.0058 mg/g, 2.29 ± 0.0137 mg/g, and 1.82 ± 0.0083 mg/g, respectively (Table 1).

TABLE 1. Contents of aloe-emodin, rhein, emodin, chrysophanol, and physcion in rhubarb water extracts.
Growth Inhibitory Activity and Biofilm Formation Inhibition Assay
First, the MIC of rhubarb water extracts against S. suis ATCC 700794 was determined as 1.56 mg.mL-1. Second, sub-inhibitory concentrations of rhubarb water extracts corresponding to 1/2, 1/4, 1/8, and 1/16 MIC (0.78, 0.39, 0.20, and 0.1 mg/ml of rhubarb water extracts, respectively) were assayed by TCP to assess the inhibitory effects of biofilm formation. The results demonstrated that the rhubarb water extracts could obviously reduce biofilm formation, especially 1/2 MIC (Figures 1, 2). Finally, 1/2 MIC of rhubarb water extracts were added to the wells at 0, 24, or 48 h, and then the wells were assayed after incubation for 72 h without shaking. The results showed that biofilm formation was inhibited in a time-dependent manner (Figure 3). There was almost no biofilm formation when rhubarb water extracts were added at 0 h, and the results from SEM can be seen in Figure 1.
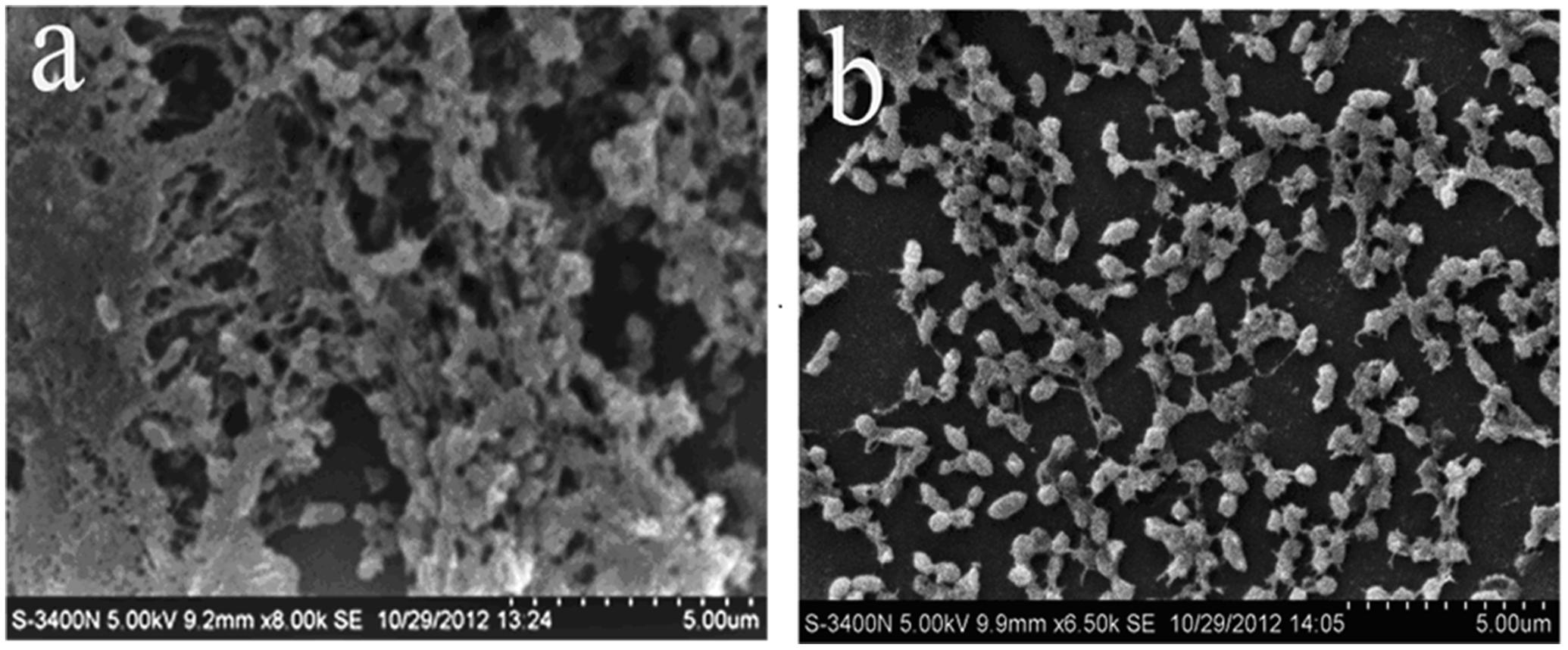
FIGURE 1. Scanning electron microscope observation. (a) Untreated Streptococcus suis ATCC700794. (b) S. suis ATCC700794 treated with 1/2 MIC rhubarb water extracts.
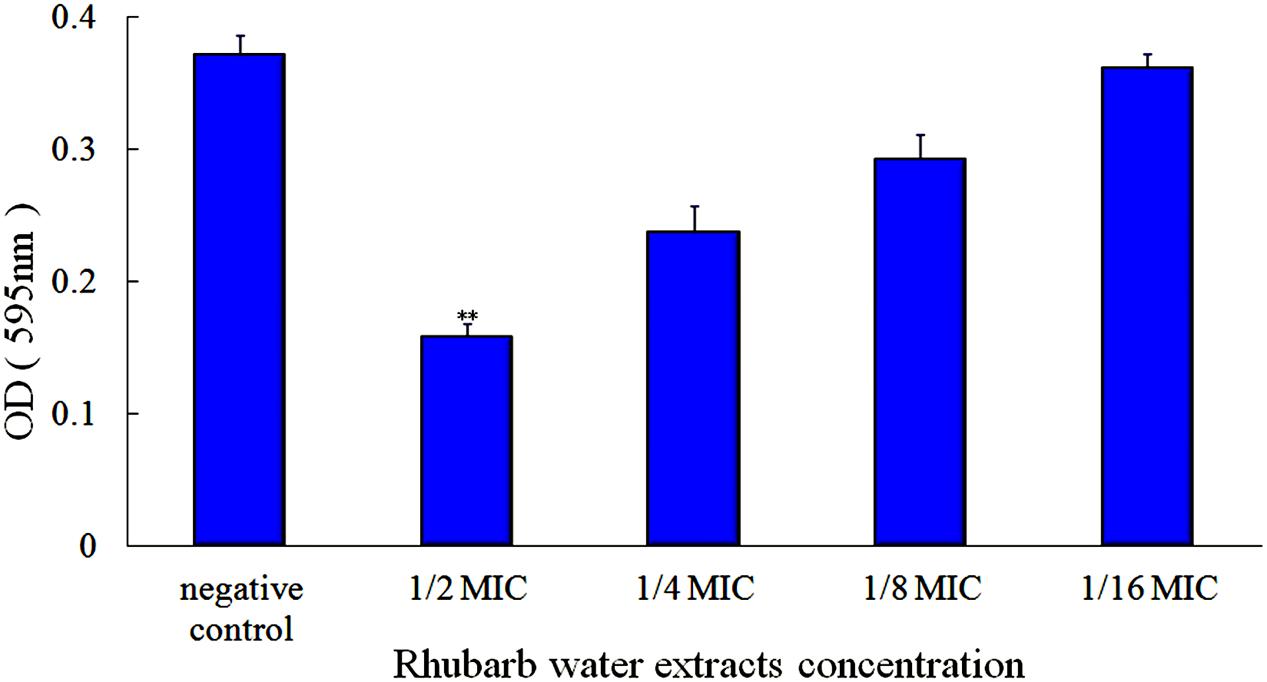
FIGURE 2. Effects of rhubarb water extracts on S. suis ATCC700794 biofilm formation at different concentrations. Data are expressed as means ± standard deviations (∗∗p < 0.05).
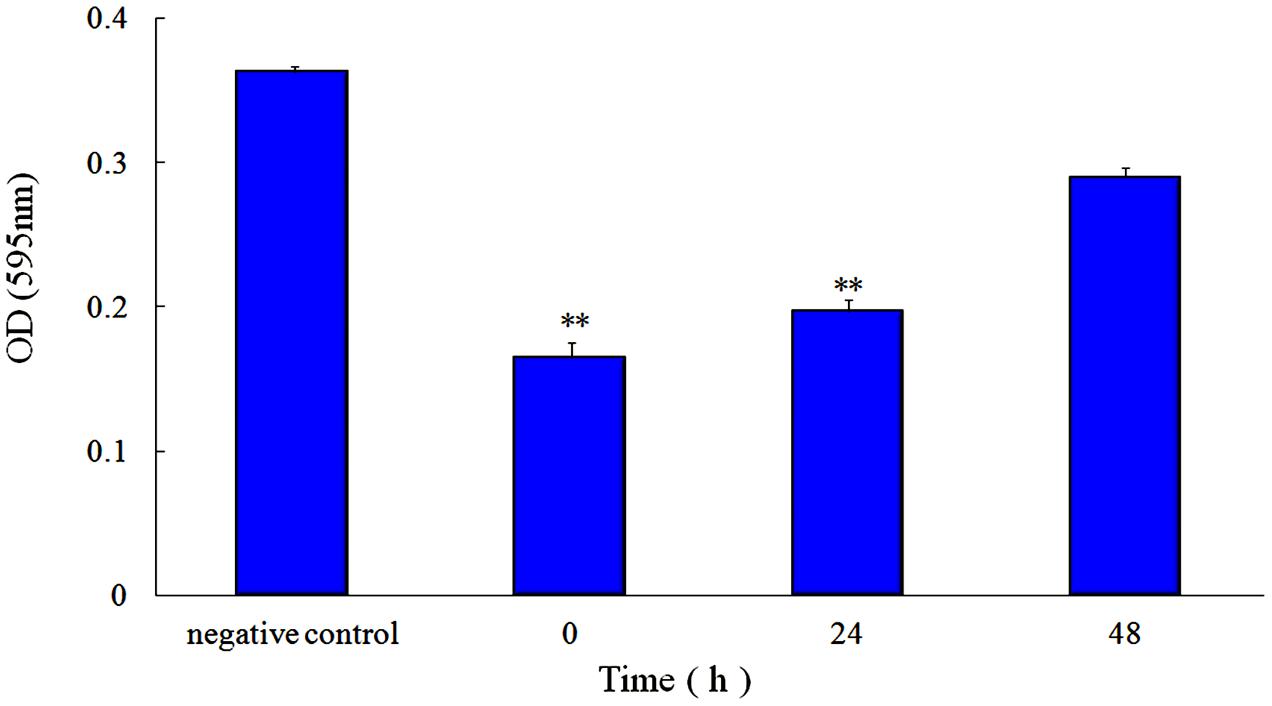
FIGURE 3. Effect of 1/2 MIC rhubarb water extracts on S. suis ATCC700794 biofilm formation. Data are expressed as means ± standard deviations (∗∗p < 0.05).
In addition, the extent of growth inhibition of S. suis by 1/2 MIC rhubarb extracts was determined. S. suis was incubated at 37°C for 48 h, and growth was then monitored at 4, 6, 8, 10, 12, 24, and 48 h by measuring OD600. The data indicated that the rhubarb water extracts had no obvious effects on the growth rate of S. suis (Figure 4).
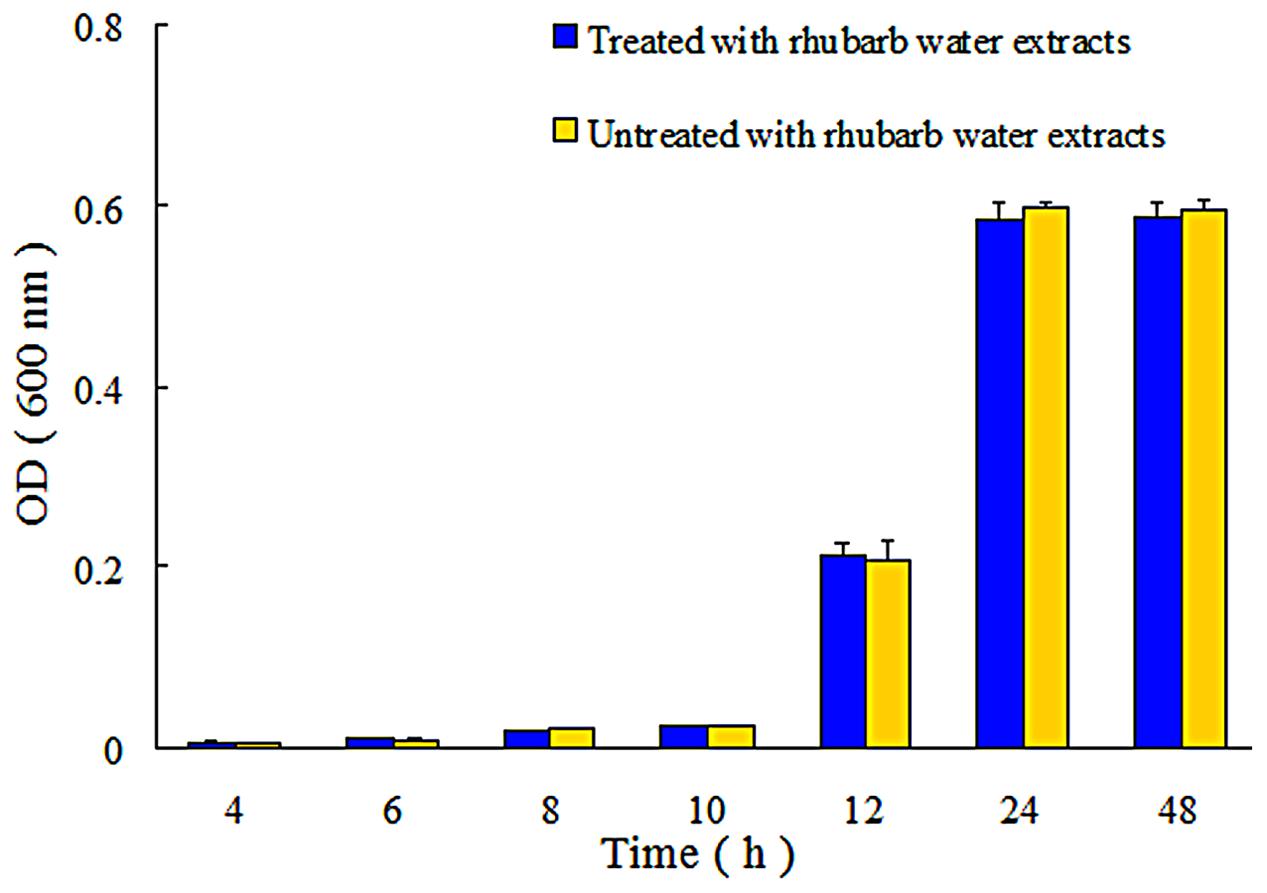
FIGURE 4. Growth of S. suis ATCC700794 in treated and untreated with 1/2 MIC rhubarb water extracts. Data are expressed as means ± standard deviations.
Determination of Protein Expression of TCSs, Transcriptional Regulators, and DNA Binding Proteins Using iTRAQ
The expression of TCSs, consisting of HKs and RRs, was measured using an iTRAQ technique (Wang et al., 2011). TCSs were extracted from S. suis that was either untreated or treated with 1/2 MIC rhubarb water extracts in vitro. HKs were down-regulated in the treated group, especially for G7SK03 and C6GVK0 (Table 2). Moreover, most of the RRs were also down-regulated, and B9WXL3 and A4W3Y3 showed a sharp decline (Table 3). DNA binding proteins and transcriptional regulators were tested using the iTRAQ technique. Most of the DNA binding proteins were decreased, especially G5KZN4 and G7S463, which sharply declined, in contrast, A4W104 and J7KEW5 showed a significant increase (Table 4). There were also some differences for the transcriptional regulators, G5L098 and B9WUV5 were obviously up-regulated, whereas J7KGR5 and G7S5V2 were significantly down-regulated (Table 5).

TABLE 2. Changes in histidine kinase proteins expression levels, which were treated with 1/2 MIC rhubarb water extracts.

TABLE 3. Changes in response regulator proteins expression levels, which were treated with 1/2 MIC rhubarb water extracts.
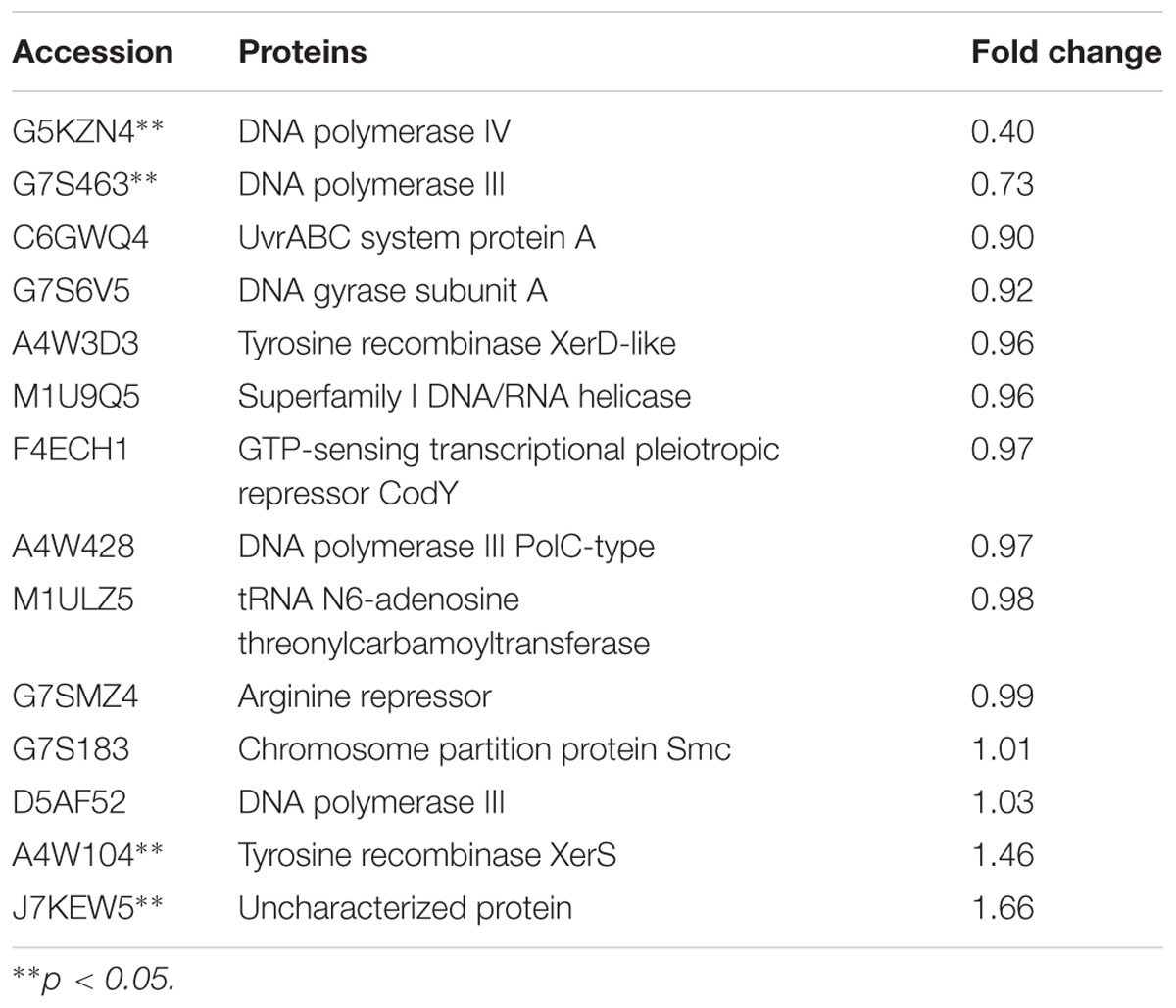
TABLE 4. Changes in DNA binding proteins expression levels, which were treated with 1/2 MIC rhubarb water extracts.
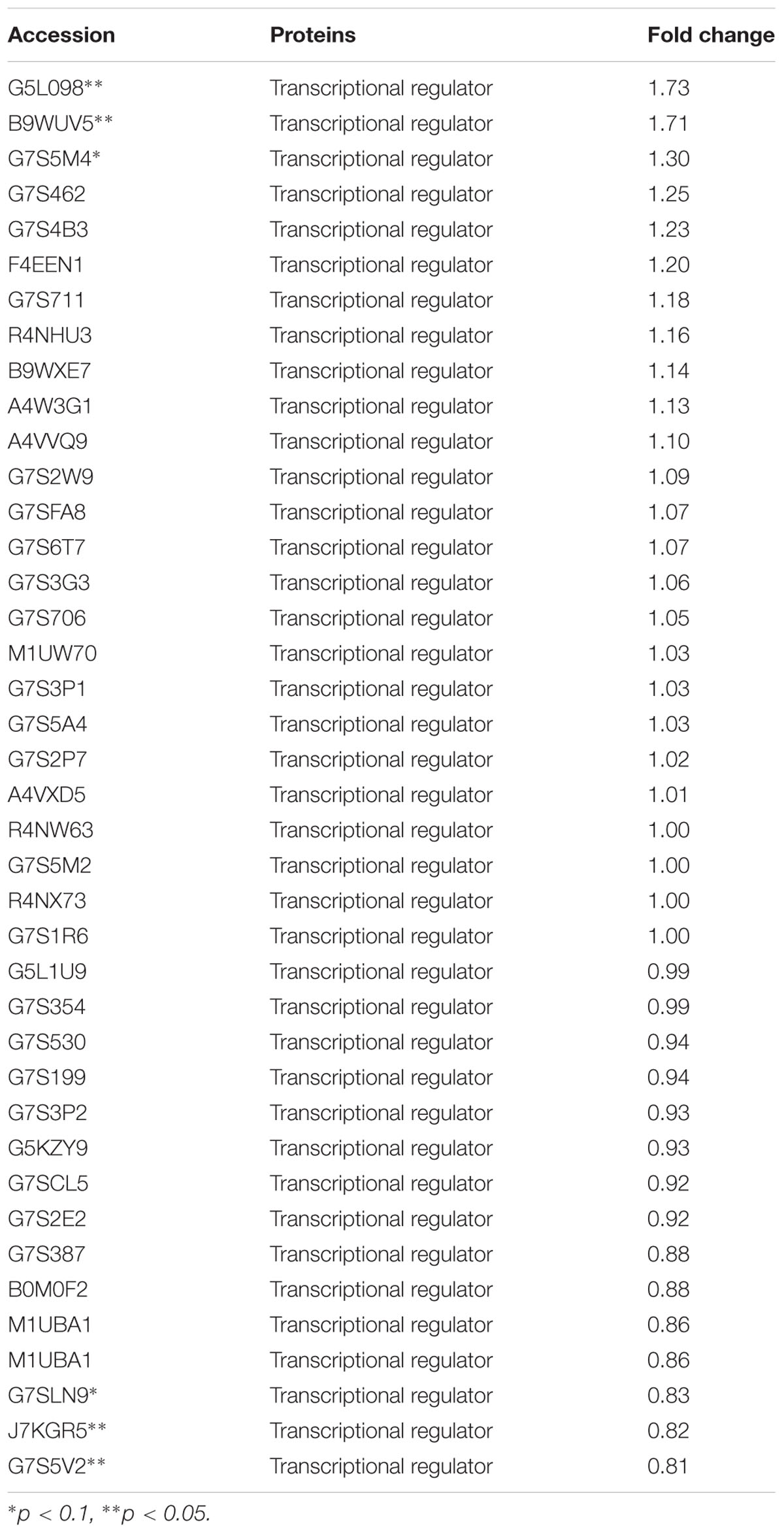
TABLE 5. Changes in transcriptional regulators expression levels, which were treated with 1/2 MIC rhubarb water extracts.
Determination of Changes in Protein Expression in Different Parts of the Cell
The proteins in different parts of the cell were also investigated in our study, and the results indicated that there were significant changes between rhubarb water extracts-treated and untreated cells. About 36 cellular membrane proteins were down-regulated, and 14 proteins were up-regulated, in treated cells as compared to untreated cells. In contrast, about 38 cytoplasmic proteins were up-regulated and 9 proteins were down-regulated (Figure 5).
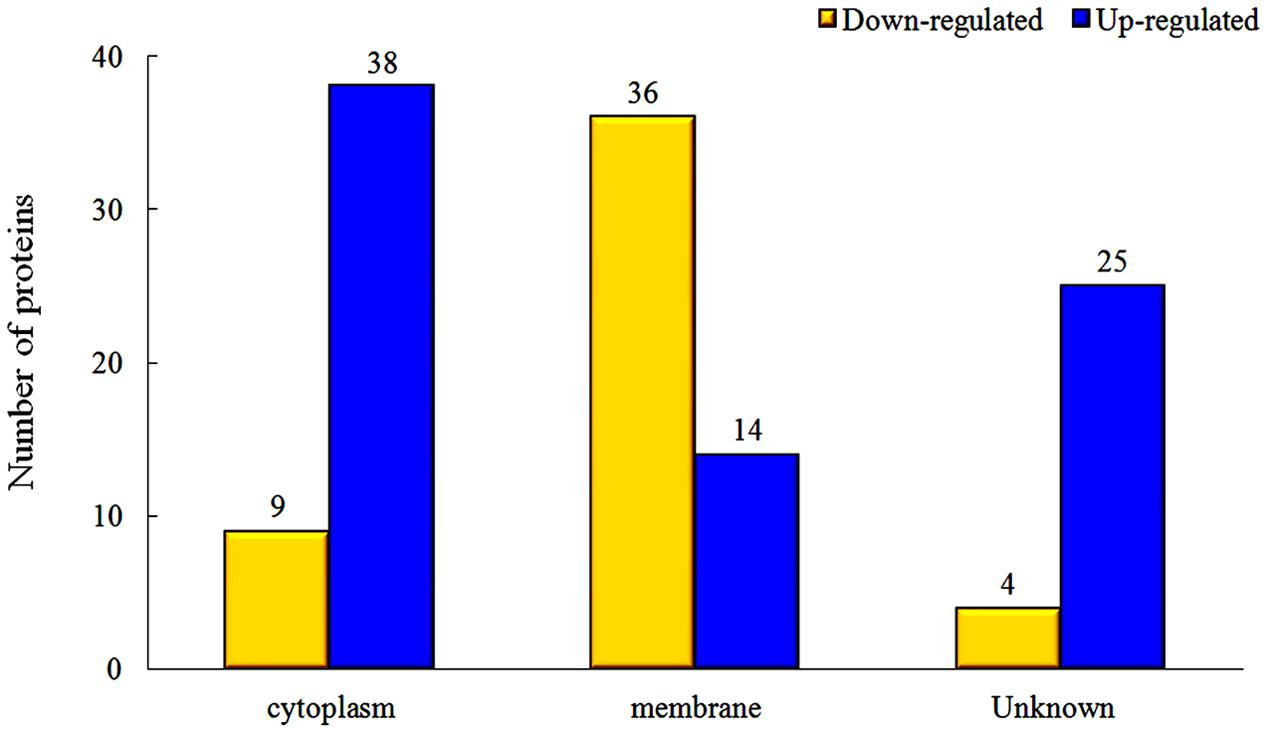
FIGURE 5. Changes of protein expression levels in different parts of cells, which were treated with 1/2 MIC rhubarb water extracts.
Discussion
Rhubarb is a common Chinese herbal medication, and its water extracts are often used in Chinese clinics. Anthraquinones are essential and effective constituent of rhubarb, they can inhibit the growth of viruses, bacteria, and biofilm (Lin et al., 2008; Wierzchacz et al., 2009), so the content of anthraquinones in the rhubarb water extracts were determined. Here, the effect of the rhubarb water extracts on S. suis ATCC 700794 biofilm formation inhibition was studied. Both TCP and SEM assays were carry out to test the effects of 1/2 MIC rhubarb extract on inhibition of S. suis biofilm formation, and the results demonstrated that the rhubarb water extracts could obviously interrupt S. suis biofilms formation (Figures 1–3); the data from Figures 2, 3 further indicated that the biofilm formation inhibition rate was dose- and time-dependent. These results were similar to the interruption of Pseudomonas aeruginosa biofilm formation by rhein, chrysophanol, and emodin, all of which have been reported to inhibit the bacterial quorum sensing (QS) system and reduce biofilm formation (Ding et al., 2011).
Biofilm formation can be affected by the number of bacteria, and reducing the bacterial number can thus inhibit biofilm formation. In our study, S. suis growth inhibition was tested in both untreated and 1/2 MIC rhubarb water extracts-treated cells, and the results indicated that the S. suis growth rate was not influenced by the rhubarb water extracts. It appears that inhibition of biofilm formation does not occur through the death of cells.
Transduction system play an important role in biofilm formation (Bhagwat et al., 2001; Parkins et al., 2001; Rasamiravaka et al., 2015; Kim et al., 2016). TCSs can affect autolytic activities, including the synthesis of Atl A, which involved in biofilm formation (Houston et al., 2011; Haghighat et al., 2017). Autolysin can affect bacteria adhesion, which is an essential process in biofilm formation (Heilmann et al., 2003; Biswas et al., 2006; Hussain et al., 2014). Thus the depletion of TCSs can leads to phenotypes directly related to a lack of autolysins, and impairment of both cell aggregation and biofilm formation (Dubrac et al., 2007). In addition, TCSs can receive stimulus signaling [such as autoinducers (AI)] and regulate the QS system, which plays a key role in pathogenic biofilm formation regulation (Rahman et al., 2015; Vasavi et al., 2017). The QS system is a widespread signaling system used by bacteria for cell-to-cell communication. It operates via production, release, and detection of small diffusible signaling molecules (AI), which can promote bacterial communication by interacting with TCSs (Bauer et al., 2015; Ma et al., 2017). Because of their importance in biofilm formation, TCSs have become attractive targets for the development of novel antibacterial, anti-biofilm drugs (Dan et al., 2014). In our study, TCSs were down-regulated expression (Tables 2, 3) when sub-MIC rhubarb water extracts were used to treat S. suis, and biofilm formation was significantly inhibited (Figure 2), this phenomenon agrees with the result from other literature.
The interaction of TCSs with signaling molecules (AI) can regulate transcriptional levels (Mascher et al., 2006; Wu et al., 2014). These functions are controlled by three phosphotransfer reactions: (1) when the signaling molecule is binding or reacting, HK will be autophosphorylated; (2) the actual transfer of the phosphorus group to the receiver domain of the RR (via the activity of the RR); and (3) dephosphorylation of RR, which returns the system back to the initial state (Figure 6) (Nobile et al., 2012; Connan and Popoff, 2015). In our study, TCSs were measured by the iTRAQ technique and the levels of most of the HKs and RRs were reduced by treatment with the rhubarb water extracts (Tables 2, 3). Compared with the control group, the interaction between TCSs and signaling molecules (AI) could be altered, and message transmission would subsequently be affected. Proteins expression may be different in the rhubarb water extracts-treated group (Figure 6). This is consistent with our experimental results as seen in Figure 5. There were about 126 altered proteins, 49 protein expression levels were decreased, and 77 protein expression levels were increased (Supplementary Table 1).
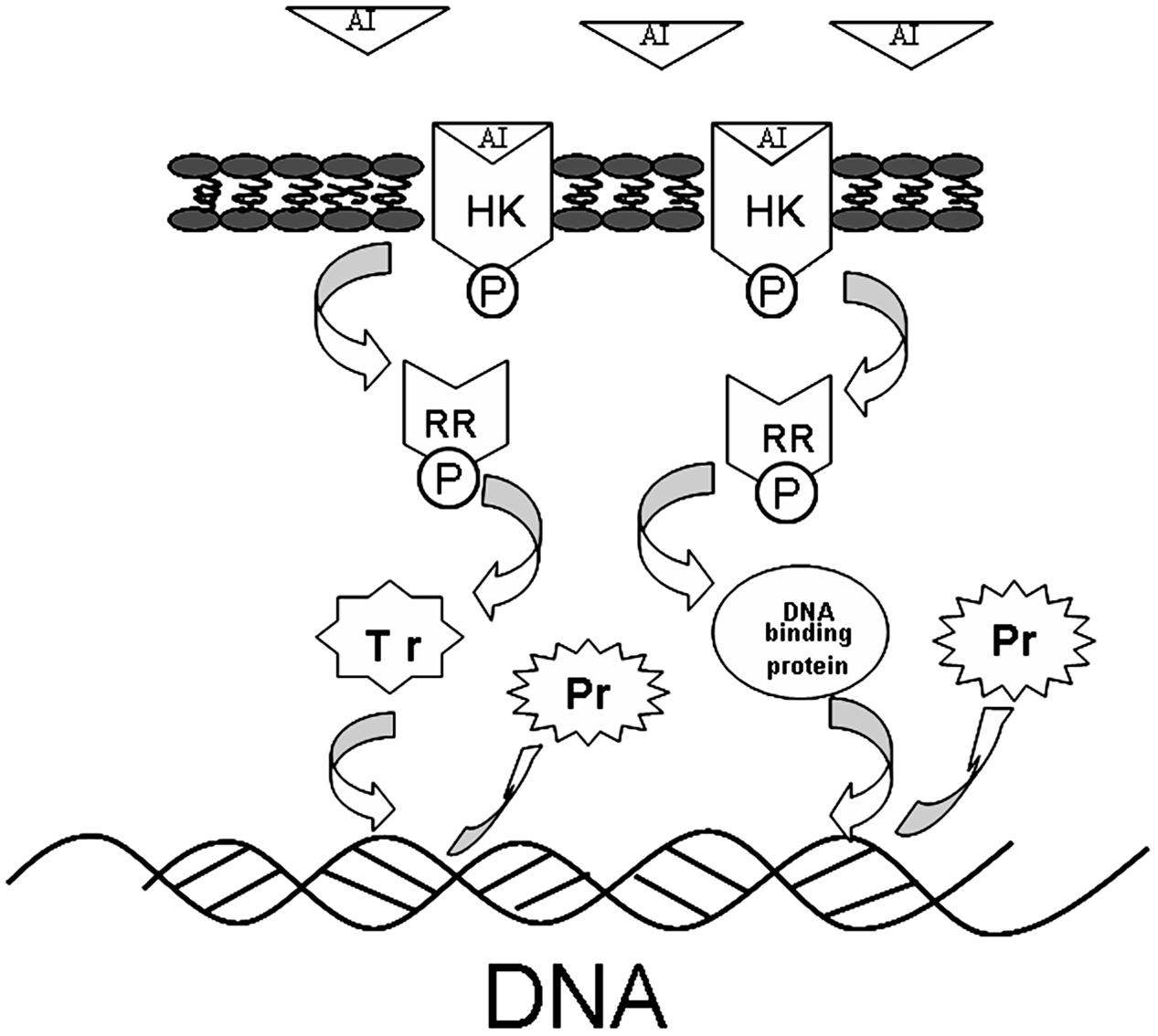
FIGURE 6. The protein expression influence of TCSs. When the AI is binding or reacting, histidine kinase (HK) will be autophosphorylated, the phosphorus will be transferred to receiver domain of the response regulator (RR), and RR will be dephosphorylated, after interacting with DNA binding proteins or transcriptional regulators. The system will return to its initial state after producing the appropriate cellular responses, which lead to differential protein expression.
Given that TCSs can interact with transcriptional regulators and DNA binding proteins to interrupt transcription (Figure 6), in our study, transcriptional regulators and DNA binding proteins were measured using iTRAQ technique. The data can be seen in Tables 4, 5, which indicated that both classes of proteins could be affected by the rhubarb extracts. Transcriptional regulators can define a group of coordinated target genes that act together to carry out specific cellular functions (Whiteson et al., 2007; Romero-Rodriguez et al., 2015). They not only regulate primary metabolic processes such as maintenance of fatty acids, amino acid catabolism, and regulation of carbon catabolism, but also affect secondary metabolism (Whiteson et al., 2007). Thus, if transcriptional regulators are perturbed, S. suis biofilm formation may be interrupted.
Expression levels of DNA binding proteins were affected in the group treated with the rhubarb water extracts (Table 4), especially DNA polymerases III, DNA polymerases IV and tyrosine recombinase XerS. Tyrosine recombinase XerS is a site-specific tyrosine recombinase, which acts by catalyzing the cutting and rejoining of recombining DNA molecules. It is essential for conversion of dimers of bacterial chromosomes into monomers and permitting their segregation at cell division (Ma et al., 2007; Poulter and Butler, 2015). Thus, up-regulating tyrosine recombinase XerS may affect cell division. DNA polymerase III performs coordinated high-fidelity synthesis of leading and lagging strands at the replication fork in a highly progressive manner. It is often induced in response to DNA damage. It plays an important role in DNA damage tolerance by performing translesion DNA synthesis (TLS) (Friedberg, 2005; Nohmi, 2006; Sidorenko et al., 2011). DNA polymerase III is exceptionally fast and accurate, but it is inefficient at synthesizing DNA on damaged templates. When DNA is significantly damaged, the replication fork will collapse and synthesis is difficult to carry out via DNA polymerase III (Robinson et al., 2015). This situation can be addressed by an SOS response, and DNA polymerase IV, as a part of the SOS regulon, can be induced to respond to DNA damage. DNA polymerase IV can lead accurate or error-prone DNA synthesis after replacing DNA polymerase III at the replication fork (Robinson et al., 2015). DNA polymerase IV belongs to Y-family polymerases that are known for their specialized ability to accommodate and bypass lesion-containing DNA (Scotland et al., 2015). They have lower DNA fidelity, are potentially mutagenic, and have a close relationship with bacterial resistance (Jacob and Eckert, 2007). In our study, DNA polymerases III and IV were distinctly down-regulated by the rhubarb water extracts, thus we can speculate that gene repair and bacterial resistance can be affected by the rhubarb extracts, and these results can be further studied in the future.
This work extends knowledge of the pharmacological effects of rhubarb on inhibition of S. suis biofilm formation. The inhibitory mechanism was investigated using the iTRAQ technique. The experimental data demonstrated that the S. suis biofilm formation could be obviously inhibited by the rhubarb water extracts. Anthraquinones, which are important active ingredient of rhubarb, were measured in the rhubarb water extracts studied here. The reason for the inhibition of biofilm formation was studied in our experiments. Firstly, this inhibition was not achieved by cell killing, which was proven by the experiments on growth inhibition. Secondly, the reduction of biofilm formation may be caused by TCS down-regulation because TCS can affect autolytic activities and QS, which play key role in biofilm formation (Parkins et al., 2001; Rasamiravaka et al., 2015). Thirdly, levels of DNA binding proteins and transcriptional regulators were affected by rhubarb water extracts in the cell. Finally, variations in protein expression levels were measured in different parts of the cell, and about 126 proteins were shown to be affected.
Author Contributions
Y-HL and ZH guided the experiment. Y-LZ, Y-BY, and QH Streptococcus suis experiment. XL, X-YC, HL, and X-YA edited the article. W-YD performed the experiments and wrote the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We would like to thank Shanghai Applied Protein Technology Co. Ltd. for the help with iTRAQ.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphar.2017.00425/full#supplementary-material
References
Alves, D. S., Perez-Fons, L., Estepa, A., and Micol, V. (2004). Membrane-related effects underlying the biological activity of the anthraquinones emodin and barbaloin. Biochem. Pharmacol. 68, 549–561. doi: 10.1016/j.bcp.2004.04.012
Bauer, W. J., Luthra, A., Zhu, G., Radolf, J. D., Malkowski, M. G., and Caimano, M. J. (2015). Structural characterization and modeling of the Borrelia burgdorferi hybrid histidine kinase Hk1 periplasmic sensor: A system for sensing small molecules associated with tick feeding. J. Struct. Biol. 192, 48. doi: 10.1016/j.jsb.2015.08.013
Bhagwat, S. P., Nary, J., and Burne, R. A. (2001). Effects of mutating putative two-component systems on biofilm formation by Streptococcus mutans UA159. FEMS Microbiol. Lett. 205, 225–230.
Biswas, R., Voggu, L., Simon, U. K., Hentschel, P., Thumm, G., and Götz, F. (2006). Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. 259, 260–268.
Connan, C., and Popoff, M. R. (2015). Two-component systems and toxinogenesis regulation in Clostridium botulinum. Res. Microbiol. 166, 332–343. doi: 10.1016/j.resmic.2014.12.012
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318.
Dan, Z., Chen, C., Liu, H., Zheng, L., Yao, T., Di, Q., et al. (2014). Biological evaluation of halogenated thiazolo[3,2-a]pyrimidin-3-one carboxylic acid derivatives targeting the YycG histidine kinase. Eur. J. Med. Chem. 87, 500–507. doi: 10.1016/j.ejmech.2014.09.096
Ding, X., Yin, B., Qian, L., Zeng, Z., Yang, Z., Li, H., et al. (2011). Screening for novel quorum-sensing inhibitors to interfere with the formation of Pseudomonas aeruginosa biofilm. J. Med. Microbiol. 60, 1827–1834. doi: 10.1099/jmm.0.024166-0
Dubrac, S., Boneca, I. G., Poupel, O., and Msadek, T. (2007). New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 189, 8257–8269.
Friedberg, E. C. (2005). Suffering in silence: the tolerance of DNA damage. Nat. Rev. Mol. Cell Biol. 6, 943–953. doi: 10.1038/rnm1781
Haghighat, S., Siadat, S. D., Sorkhabadi, S. M. R., Sepahi, A. A., and Mahdavi, M. (2017). Cloning, expression and purification of autolysin from methicillin-resistant Staphylococcus aureus: potency and challenge study in Balb/c mice. Mol. Immunol. 82, 10–18. doi: 10.1016/j.molimm.2016.12.013
Heilmann, C., Hussain, M., Peters, G., and Götz, F. (2003). Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24, 1013–1024.
Houston, P., Rowe, S. E., Pozzi, C., Waters, E. M., and O’Gara, J. P. (2011). Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect. Immun. 79, 1153. doi: 10.1128/IAI.00364-10
Hussain, M., Steinbacher, T., Peters, G., Heilmann, C., and Becker, K. (2014). The adhesive properties of the Staphylococcus lugdunensis multifunctional autolysin AtlL and its role in biofilm formation and internalization. Int. J. Med. Microbiol. 305, 129–139. doi: 10.1016/j.ijmm.2014.11.010
Jacob, K. D., and Eckert, K. A. (2007). Escherichia coli DNA polymerase IV contributes to spontaneous mutagenesis at coding sequences but not microsatellite alleles. Mutat. Res. 619, 93–103. doi: 10.1016/j.mrfmmm.2007.02.007
Jung, K., Fried, L., Behr, S., and Heermann, R. (2012). Histidine kinases and response regulators in networks. Curr. Opin. Microbiol. 15, 118–124. doi: 10.1016/j.mib.2011.11.009
Khoramian, B., Jabalameli, F., Niasari-Naslaji, A., Taherikalani, M., and Emaneini, M. (2015). Comparison of virulence factors and biofilm formation among Staphylococcus aureus strains isolated from human and bovine infections. Microb. Pathog. 88, 73–77. doi: 10.1016/j.micpath.2015.08.007
Kim, J. E., Kim, H. J., Pandit, S., Chang, K. W., and Jeon, J. G. (2011). Inhibitory effect of a bioactivity-guided fraction from Rheum undulatum on the acid production of Streptococcus mutans biofilms at sub-MIC levels. Fitoterapia 82, 352–356. doi: 10.1016/j.fitote.2010.11.005
Kim, T., Choi, J., Lee, S., Yeo, K. J., Cheong, H. K., and Kim, K. K. (2016). Structural studies on the extracellular domain of sensor histidine kinase YycG from Staphylococcus aureus and its functional implications. J. Mol. Biol. 428, 3074–3089. doi: 10.1016/j.jmb.2016.06.019
Liao, J., Zhao, L., Yoshioka, M., Hinode, D., and Grenier, D. (2013). Effects of Japanese traditional herbal medicines (Kampo) on growth and virulence properties of Porphyromonas gingivalis and viability of oral epithelial cells. Pharm. Biol. 51, 1538–1544. doi: 10.3109/13880209.2013.801995
Lin, C. W., Wu, C. F., Hsiao, N. W., Chang, C. Y., Li, S. W., Wan, L., et al. (2008). Aloe-emodin is an interferon-inducing agent with antiviral activity against Japanese encephalitis virus and enterovirus 71. Int. J. Antimicrob. Agents 32, 355–359. doi: 10.1016/j.ijantimicag.2008.04.018
Ma, C. H., Kwiatek, A., Bolusani, S., Voziyanov, Y., and Jayaram, M. (2007). Unveiling hidden catalytic contributions of the conserved His/Trp-III in tyrosine recombinases: assembly of a novel active site in Flp recombinase harboring alanine at this position. J. Mol. Biol. 368, 183–196. doi: 10.1016/j.jmb.2007.02.022
Ma, R., Qiu, S., Jiang, Q., Sun, H., Xue, T., Cai, G., et al. (2017). AI-2 quorum sensing negatively regulates rbf expression and biofilm formation in Staphylococcus aureus. Int. J. Med. Microbiol. 307, 257–267. doi: 10.1016/j.ijmm.2017.03.003
Mascher, T., Helmann, J. D., and Unden, G. (2006). Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70, 910–938.
Nobile, C. J., Fox, E. P., Nett, J. E., Sorrells, T. R., Mitrovich, Q. M., Hernday, A. D., et al. (2012). A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148, 126–138. doi: 10.1016/j.cell.2011.10.048
Nohmi, T. (2006). Environmental stress and lesion-bypass DNA polymerases. Annu. Rev. Microbiol. 60, 231.
Parellada, E. A., Ferrero, M., Cartagena, E., Bardon, A., and Neske, A. (2013). Laherradurin, a natural stressor, stimulates QS mechanism involved in biofilm formation of a PAHs degrading bacterium. Int. Biodeterior. Biodegradation 85, 78–84. doi: 10.1016/j.ibiod.2013.06.009
Parkins, M. D., Ceri, H., and Storey, D. G. (2001). Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 40, 1215–1226.
Poulter, R. T., and Butler, M. I. (2015). Tyrosine Recombinase Retrotransposons and Transposons. Microbiol. Spectr. 3, MDNA3–0036–2014. doi: 10.1128/microbiolspec.MDNA3-0036-2014
Rahman, M. R., Lou, Z., Yu, F., Wang, P., and Wang, H. (2015). Anti-quorum sensing and anti-biofilm activity of Amomum tsao-ko (Amomum tsao-ko Crevost et Lemarie) on foodborne pathogens. Saudi J. Biol. Sci. 24, 324. doi: 10.1016/j.sjbs.2015.09.034
Rasamiravaka, T., Labtani, Q., Duez, P., and El, J. M. (2015). The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed. Res. Int. 2015:759348. doi: 10.1155/2015/759348
Robinson, A., Mcdonald, J. P., Caldas, V. E. A., Patel, M., Wood, E. A., Punter, C. M., et al. (2015). Regulation of mutagenic DNA polymerase V activation in space and time. PLoS Genet. 11:e1005482. doi: 10.1371/journal.pgen.1005482
Romero-Rodriguez, A., Robledo-Casados, I., and Sanchez, S. (2015). An overview on transcriptional regulators in Streptomyces. Biochimi. Biophys. Acta 1849, 1017–1039. doi: 10.1016/j.bbagrm.2015.06.007
Scotland, M. K., Heltzel, J. M. H., Kath, J. E., Choi, J. S., Berdis, A. J., Loparo, J. J., et al. (2015). A genetic selection for dinB Mutants reveals an interaction between DNA polymerase IV and the replicative polymerase that is required for translesion synthesis. PLoS Genet. 11:e1005507. doi: 10.1371/journal.pgen.1005507
Siddiqui, M. F., Rzechowicz, M., Harvey, W., Zularisam, A. W., and Anthony, G. F. (2015). Quorum sensing based membrane biofouling control for water treatment: a review. J. Water Process Eng. 7, 112–122.
Sidorenko, J., Jatsenko, T., Saumaa, S., Teras, R., Tark-Dame, M., Horak, R., et al. (2011). Involvement of specialized DNA polymerases Pol II, Pol IV and DnaE2 in DNA replication in the absence of Pol I in Pseudomonas putida. Mutat. Res. 714, 63–77. doi: 10.1016/j.mrfmmm.2011.06.013
Vasavi, H. S., Sudeep, H. V., Lingaraju, H. B., and Prasad, K. S. (2017). Bioavailability-enhanced ResveramaxTM modulates quorum sensing and inhibits biofilm formation in Pseudomonas aeruginosa PAO1. Microb. Pathog. 104, 64–71. doi: 10.1016/j.micpath.2017.01.015
Wang, Y., Zhang, W., Wu, Z., Zhu, X., and Lu, C. (2011). Functional analysis of luxS in Streptococcus suis reveals a key role in biofilm formation and virulence. Vet. Microbiol. 152, 151–160. doi: 10.1016/j.vetmic.2011.04.029
Whiteson, K. L., Chen, Y., Chopra, N., Raymond, A. C., and Rice, P. A. (2007). Identification of a potential general acid/base in the reversible phosphoryl transfer reactions catalyzed by tyrosine recombinases: Flp H305. Chem. Biol. 14, 121–129. doi: 10.1016/j.chembiol.2007.01.011
Wierzchacz, C., Su, E. J., and Gebhardt, R. (2009). Differential inhibition of matrix metalloproteinases-2, -9, and -13 activities by selected anthraquinones. Planta Med. 75, 327–329. doi: 10.1055/s-0028-1112205
Wisniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. doi: 10.1038/nmeth.1322
Wu, Y., Liu, J., Jiang, J., Hu, J., Xu, T., Wang, J., et al. (2014). Role of the two-component regulatory system arlRS in ica operon and aap positive but non-biofilm-forming Staphylococcus epidermidis isolates from hospitalized patients. Microb. Pathog. 76, 89. doi: 10.1016/j.micpath.2014.09.013
Keywords: Streptococcus suis, biofilm, rhubarb water extracts, two-component signal transduction system (TCSs), transcriptional regulator, DNA binding protein
Citation: Ding W-Y, Li Y-H, Lian H, Ai X-Y, Zhao Y-L, Yang Y-B, Han Q, Liu X, Chen X-Y and He Z (2017) Sub-Minimum Inhibitory Concentrations of Rhubarb Water Extracts Inhibit Streptococcus suis Biofilm Formation. Front. Pharmacol. 8:425. doi: 10.3389/fphar.2017.00425
Received: 02 December 2016; Accepted: 14 June 2017;
Published: 07 July 2017.
Edited by:
Aiping Lu, Hong Kong Baptist University, Hong KongReviewed by:
Rene Cardenas, National Autonomous University of Mexico, MexicoRoopali Mittal, University of Oklahoma Health Sciences Center, United States
Copyright © 2017 Ding, Li, Lian, Ai, Zhao, Yang, Han, Liu, Chen and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhonggui He, ZXpob25nZ3VpQHZpcC4xNjMuY29t; aGV6aGd1aV9zdHVkZW50QGFsaXl1bi5jb20= Yan-Hua Li, bGl5YW5odWExOTcwQDE2My5jb20= Wen-Ya Ding, ZGluZ3dlbnlhMzA2QDE2My5jb20=
 Wen-Ya Ding
Wen-Ya Ding Yan-Hua Li2*
Yan-Hua Li2* Yu-Lin Zhao
Yu-Lin Zhao Yan-Bei Yang
Yan-Bei Yang