- 1Affiliated Cancer Hospital & Institute of Guangzhou Medical University, Guangzhou Municipal and Guangdong Provincial Key Laboratory of Protein Modification and Degradation, School of Basic Medical Science, Guangzhou Medical University, Guangzhou, China
- 2Department of Chemistry, Brown University, Providence, RI, United States
- 3Department of Chemistry, Washington State University, Pullman, WA, United States
Metabolic reprogramming is a cancer hallmark. Although the reprogramming of central carbon has been well documented, the role of sulfur metabolism has been largely overlooked. Additionally, the effects of sulfur are sometimes contradictory in tumorigenesis. In this study, we aimed to investigate the gene expression profile in hepatocellular carcinoma (HCC) and the effects of reactive sulfur species (RSS) on HCC tumor cells. Furthermore, the cell imaging technology was applied to discover some potential anti-cancer compounds. Gene Set Enrichment Analysis (GSEA) of Gene Expression Omnibus (GEO) dataset (GSE102083) revealed that sulfur amino acid-related metabolism and vitamin B6 binding activity in HCC tissues were downregulated. Calculation of the interaction network identified nine hub genes, among which eight were validated by differential expression and survival analysis in the TCGA_LIHC cohort, and two (CSE and CBS) had the highest enrichment degree. The metabolomics analysis suggested that the hub genes were associated with RSS metabolism including H2S, H2S2, cystine, cysteine, homocysteine, cystathionine, and methionine. The cell viability assay demonstrated that H2S2 had significant anti-cancer effects in HCC SNU398 tumor cells. The cell imaging assay showed that treatment with H2S2 remarkably increased intracellular sulfane sulfur content. On this basis, the anti-cancer activity of some other sulfane sulfur compounds, such as DATS and DADS, was further verified. Lastly, according to the fact that HCC tumor cells preferentially take in cystine due to high expression of SLC7A11 (a cystine/glutamate transporter), persulfided cysteine precursor (PSCP) was tested for its sulfane sulfur release capability and found to selectively inhibit HCC tumor cell viability. Collectively, this study uncovered sulfur metabolism in HCC was reprogrammed, and provided a potential therapeutic strategy for HCC by donating sulfane sulfur.
Introduction
In liver cancer, hepatocellular carcinoma (HCC) is the most common primary malignancy, which has been the leading cause of cancer-related death globally. Surgical resection was considered to be the standard and first option treatment of HCC, because of its resistance to radiotherapy or chemotherapy (Lohitesh et al., 2018). Unfortunately, many HCC patients may not have the chance of surgical resection because the diagnosis usually occurs at the late stage. Therefore, revealing the altered cell biological behavior with the gene expression profile at the early stage will contribute to the discovery of potential targets that are of therapeutic significance.
It has been known for decades that plants carrying organosulfur-containing compounds, like garlic, have anti-cancer effects (Weisberger and Pensky, 1957). Studies supported that many of garlic’s effects are attributed to the generation of sulfane sulfur, a reactive sulfur species (RSS) (Iciek et al., 2001; Iciek et al., 2012). For human beings, persulfide (RSSH), polysulfide (RSSnR), and thiosulfate (RSSO3) are major forms of sulfane sulfur in the body (Yuan et al., 2017; Akaike et al., 2017; Alvarez et al., 2017; Yang et al., 2019a; Liu H. et al., 2019). Additionally, some endogenous RSS, like cystine, cysteine, homocysteine, methionine, glutathione, and hydrogen sulfide (H2S), are not sulfane sulfur per se, but can be metabolized to produce sulfane sulfur (Jackson et al., 2012). However, the effects of these RSS on cancer therapy including HCC are contradictory, as both anti-cancer (Iciek et al., 2001; Iciek et al., 2012) and pro-cancer (Pan et al., 2014; Zhen et al., 2018; Yang et al., 2019b) activities have been reported, which hinders the sulfur-based HCC therapy.
Metabolic reprogramming is an important cancer hallmark that has been studied over a century but is still of research significance (DeBerardinis et al., 2008; Faubert et al., 2020; Zhou et al., 2020). In tumor cells, a variety of metabolic processes, such as aerobic glycolysis, glutamine catabolism, macromolecular synthesis, and redox homeostasis, are distinctively different from that in normal cells. This metabolic flexibility is believed to fuel tumor fast proliferation (Kim and DeBerardinis, 2019). However, the metabolism of RSS in HCC tissues has often been overlooked. Additionally, it is unclear whether the different therapeutic outcomes of sulfur compounds are associated with the reprogramed RSS metabolism. A recent study showed that the diet restriction of methionine (a sulfur amino acid) was able to inhibit tumor growth in colorectal cancer (Gao et al., 2019). On the other hand, the elevated level of homocysteine (Wu and Wu, 2002), or decreased level of vitamin B6 (Galluzzi et al., 2013), could promote tumorigenesis. As known, vitamin B6 (pyridoxal 5’-phosphate) is a cofactor of cystathionine-β-synthase (CBS) or cystathionine-γ-lyase (CSE/CTH) in the generation of RSS including H2S and H2S2 (Renga, 2011; Gregory et al., 2016). These studies inspired us to explore the overall difference between HCC and normal liver tissues in the RSS metabolism, by which some useful therapeutic strategies may be uncovered.
Transcriptomics analysis of high throughput RNA sequencing is a highly efficient means to discover the potential causes of unique HCC phenotype. Gene Set Enrichment Analysis (GSEA) is one of the most popular computational tools for transcriptomics analysis. Therefore, in this work, GSEA was used to demonstrate whether the sulfur metabolism was reprogrammed in HCC tissues. Then, through the bioinformatics analysis, the involved hub genes were revealed, their significance was verified, and the interacted RSS were discovered. With the cell imaging and viability assay, some RSS that have anti-HCC activity were suggested. Lastly, based on the enhanced cystine transport capacity in HCC tumor cells, persulfided cysteine precursor (PSCP) and a series of esterase-triggered sulfane sulfur donors were synthesized and their anti-HCC effects were evaluated.
Materials and Methods
Materials
Sodium sulfide (Na2S), sodium persulfide (Na2S2), sodium trisulfide (Na2S3), and Cell Counting Kit (CCK)-8 were purchased from Dojindo Lab (Kyushu, Japan). N-acetyl cysteine (NAC), cystine, cystathionine, and GSSG were bought from Thermo Fisher Scientific Co. (Pittsburgh, PA, USA). Gemcell™ fetal bovine serum (FBS) was supplied by Gemini Company (Woodland, CA, USA). SSP4, PSCP, and the controllable sulfane sulfur donors were synthesized following the literature procedure and their characterization matches reported data (Rietman et al., 1994; Chen et al., 2013; Artaud and Galardon, 2014).
Cell Culture and Viability Assay
HCC (SNU387 and SNU398) tumor cells, lung adenocarcinoma A549 cells, prostate cancer PC3 cells, cervical cancer Hela cells, breast MDA-MB-231 cells, raw-blue macrophages, LO2 liver cells, and H9c2 cardiomyocytes were obtained from ATCC. The cells were maintained in DMEM-F12 medium supplemented with 10% FBS at 37°C under an atmosphere of 5% CO2 and 95% air. They were passaged and harvested with 0.25% trypsin.
Cell viability was measured with the CCK-8 kit. The cells were plated in 96-well plates at a density of 7,000 cells/well. When grown to approximately 60~70% confluence, they were treated correspondingly for 48 h. Then, the plates were washed and 100 μl of CCK-8 solution diluted with FBS-free medium was added. The cells were incubated for a further 2 h at 37°C. The absorbance was measured at 450 nm with a microplate reader (Molecular Devices, USA).
Data Collection
The microarray data of four normal mouse liver tissues and four HCC tumor tissues were downloaded from Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=%20GSE102081) (Chiyonobu et al., 2018). For the Cancer Genome Atlas (TCGA) data, the analysis of differential expression was downloaded from GEPIA2 (http://gepia2.cancer-pku.cn/#analysis), and the survival data were downloaded from the Human Protein Atlas (https://www.proteinatlas.org/).
Gene Set Enrichment Analysis
The expression dataset from GSE102081 was converted as follows: According to the probes of Agilent Sureprint G3ge8x60k in the first column, a column of gene symbol (mgi_symbol) was added. The subsequent columns were named C1, C2, C3, and C4 for the normal liver tissues, while H1, H2, H3, and H4 for the HCC tumor tissues. The data file was imported into Gene Set Enrichment Analysis (GSEA) 4.0.3 software for further calculation and illustration according to the protocol (Subramanian et al., 2005).
Prediction of Hub Genes
The interaction of the leading-edge genes identified by GSEA was subjected to STRING analysis (https://string-db.org/) by constructing a protein-protein interaction (PPI) network. The disconnected nodes were not involved in this network. The data were downloaded and imported to Cytoscape 3.7 software for further calculation, visual analysis, and hub gene prediction.
Observation of Intracellular Sulfane Sulfur Levels
HCC SNU398 tumor cells were inoculated in 24-well plates and grown to 60∼70% confluence. After treatment with the indicated RSS for 1 h, intracellular sulfane sulfur was measured with SSP4 fluorescent probe (Chen et al., 2013; Bibli et al., 2018). Briefly, the treated cells were incubated with 10 μM SSP4 in the presence of 20 μM cetyl trimethyl ammonium bromide (CTAB) in 1% FBS medium for 20 min in the dark. The cell imaging was carried out after a slight wash with PBS buffer. The intracellular fluorescence signal was visualized under the Nikon E600 Fluorescence microscope (Walpole, MA, US).
Chemical Synthesis and Characterization
PSCP was synthesized following reported procedure (Rietman et al., 1994). NMR matches the reported data. 1H NMR (400 MHz, Methanol-d4) 4.24 (m, 1H), 3.93 (s, 3H), 3.44–3.47 (dd, 2H), 3.21–3.27 (m, 2H).
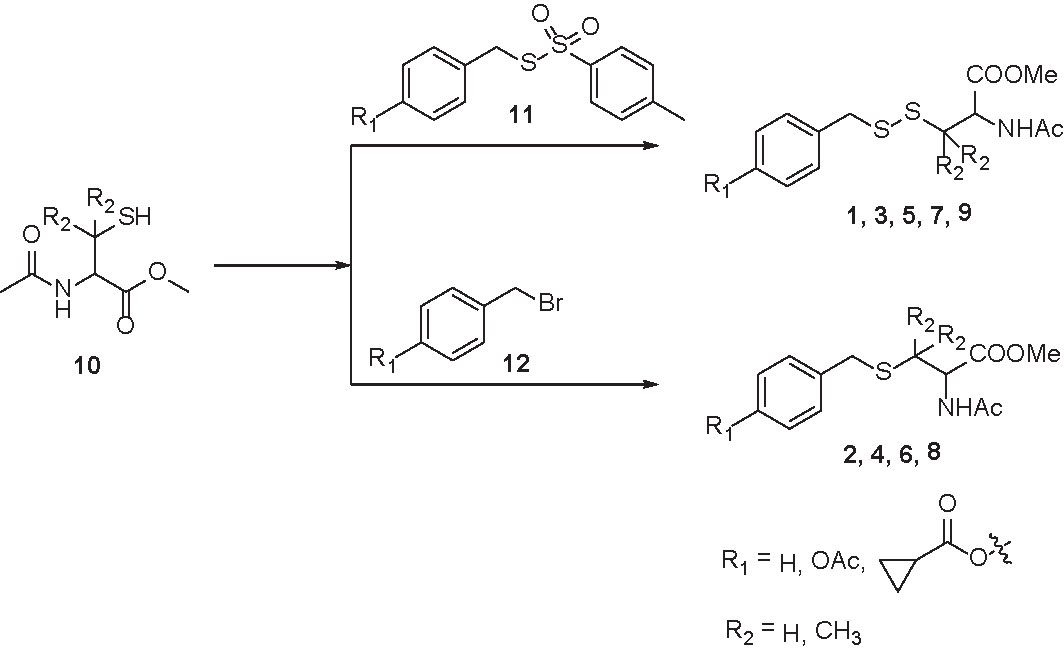
Compounds 1, 3, 5, 7, and 9 were synthesized by treating protected cysteine/penicillamine (10) with the corresponding tosylated para-substituted benzyl mercaptans (11). Briefly, to a solution of 10 (2 mM) in dichloromethane (10 ml) was added 11 (1.8 mM) and triethylamine (4 mM). Reaction was stirred under room temperature overnight. The mixture was then purified by column chromatography (Scheme 1).
Compounds 2, 4, 6, and 8 were synthesized by (10) with the corresponding para-substituted benzyl bromides (12). Briefly, under argon, to a solution of 12 (1.9 mM) and triethylamine (5 mM) in dichloromethane (5 ml) was added a solution of 10 (2 mM) in dichloromethane dropwise. Reaction was stirred under argon overnight. The mixture was then purified by column chromatography (Scheme 1).
Measurement of Sulfane Sulfur Content
SSP4 and CTAB were dissolved in DMSO and ethanol respectively to make 1 mM and 5 mM stock solutions. To initiate the experiment, 2 mM PSCP was prepared with PBS buffer (pH 7.4, 50 mM, 10 ml). At 5 min, 10 min, 20 min, 1 h, 1.5 h, 2 h, 4 h, 6 h, and 12 h, 80 µl of the above solution was aliquoted into a testing solution containing CTAB stock solution (80 µl), SSP4 stock solution (40 µl) and PBS buffer (pH 7.4, 50 mM, 3,800 µl), making the concentration of PSCP, SSP4, and CTAB 40, 10, and 100 µM, respectively. The resulting solution was incubated under room temperature for 10 min and fluorescence was analyzed by a fluorometer (Cary Eclipse, Agilent, USA).
Statistical Analysis
The experiment data are presented as mean ± standard deviation (SD). Significance between groups was evaluated by one-way analysis of variance (ANOVA) followed by Student-Newman-Keuls Test using GraphPad Prism 8 software (SanDiego, US). A probability <0.05 was considered statistically significant.
Result
Sulfur Metabolism Is Reprogrammed in HCC Tissues
To dissect the difference of sulfur metabolism between normal liver and HCC tumor tissues, the gene expression profile was screened using the GSEA tool. As presented in Figure 1, compared with the normal liver tissues, in HCC tumor tissues 12 out of 16 genes were negatively expressed in sulfur amino acid metabolism GO geneset (Figures 1A, D), 17 out of 32 genes were negatively expressed in cysteine and methionine metabolism KEGG geneset (Figures 1B, E), and 26 out of 48 genes were negatively expressed in vitamin B6 binding activity GO geneset (Figures 1C, F). The result indicates that sulfur-related metabolism is distinctively disturbed in the HCC phenotype.
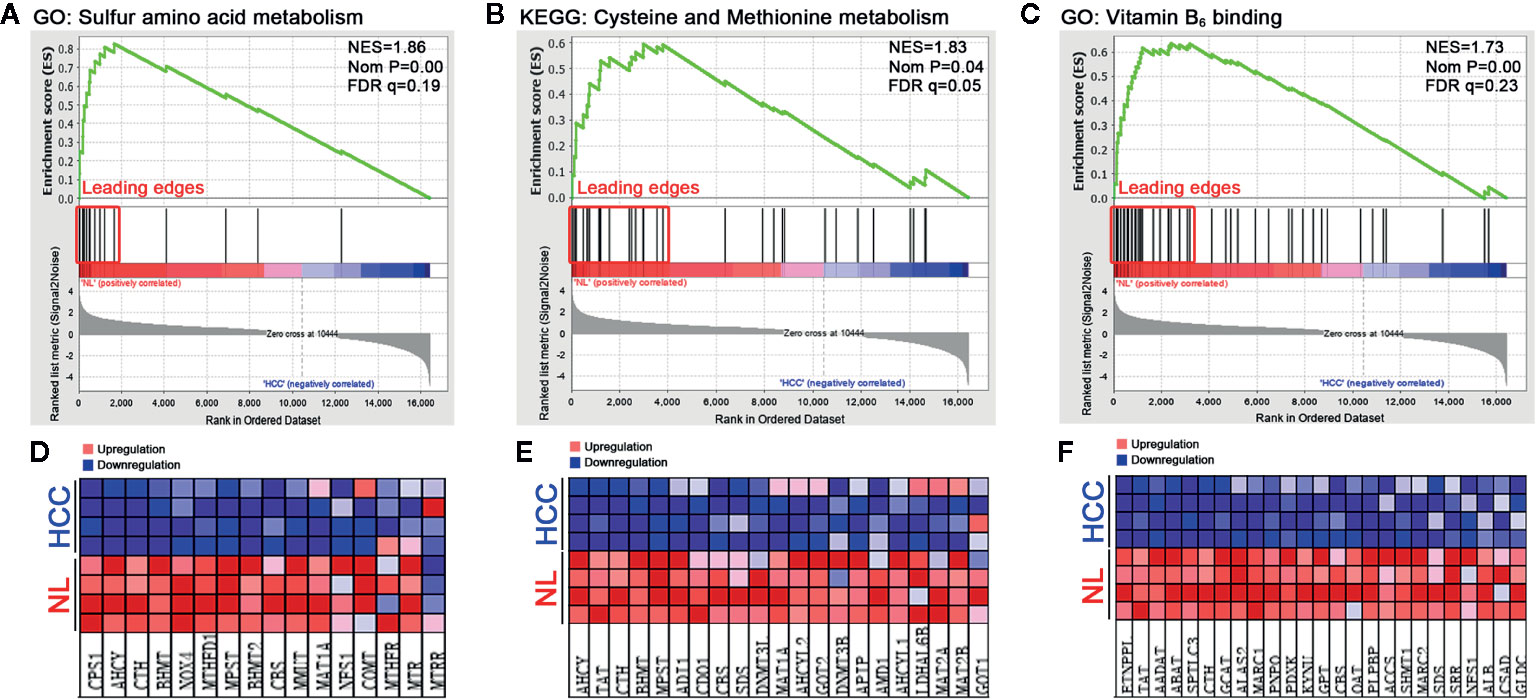
Figure 1 Functional enrichment analysis of genesets in HCC tumor tissues. A microarray dataset (GSE102081) of HCC tumor tissues and normal liver (NL) tissues was analyzed using the GSEA tool. (A–C) The enrichment indexes of three genesets are shown in (A) (sulfur amino acid metabolism), (B) (cysteine and methionine metabolism), and (C) (vitamin B6 binding activity), respectively. The genesets were considered to be significantly enriched at NES (normalized enrichment score) >1, Nom P (nominal p-value) <0.05, and FDR q-value <0.25. (D–F) Heatmaps of the enriched genes in (A–C) were created to display their relative expression.
Prediction of Hub Genes in Leading-Edge Sets
To visualize the inter-relationships between these enriched genesets, a chord diagram was drawn. As shown in Figure 2A, 10 mutual genes were found, i.e. AHCY, BHMT, GOT2, MAT1A, MPST, NFS1, SDS, TAT, CBS, and CTH, among which CTH and CBS were shared by three genesets. Additionally, after the genes were sorted by expression level, which was displayed as log2 (Fold of Change), CTH was found to be dramatically downregulated.
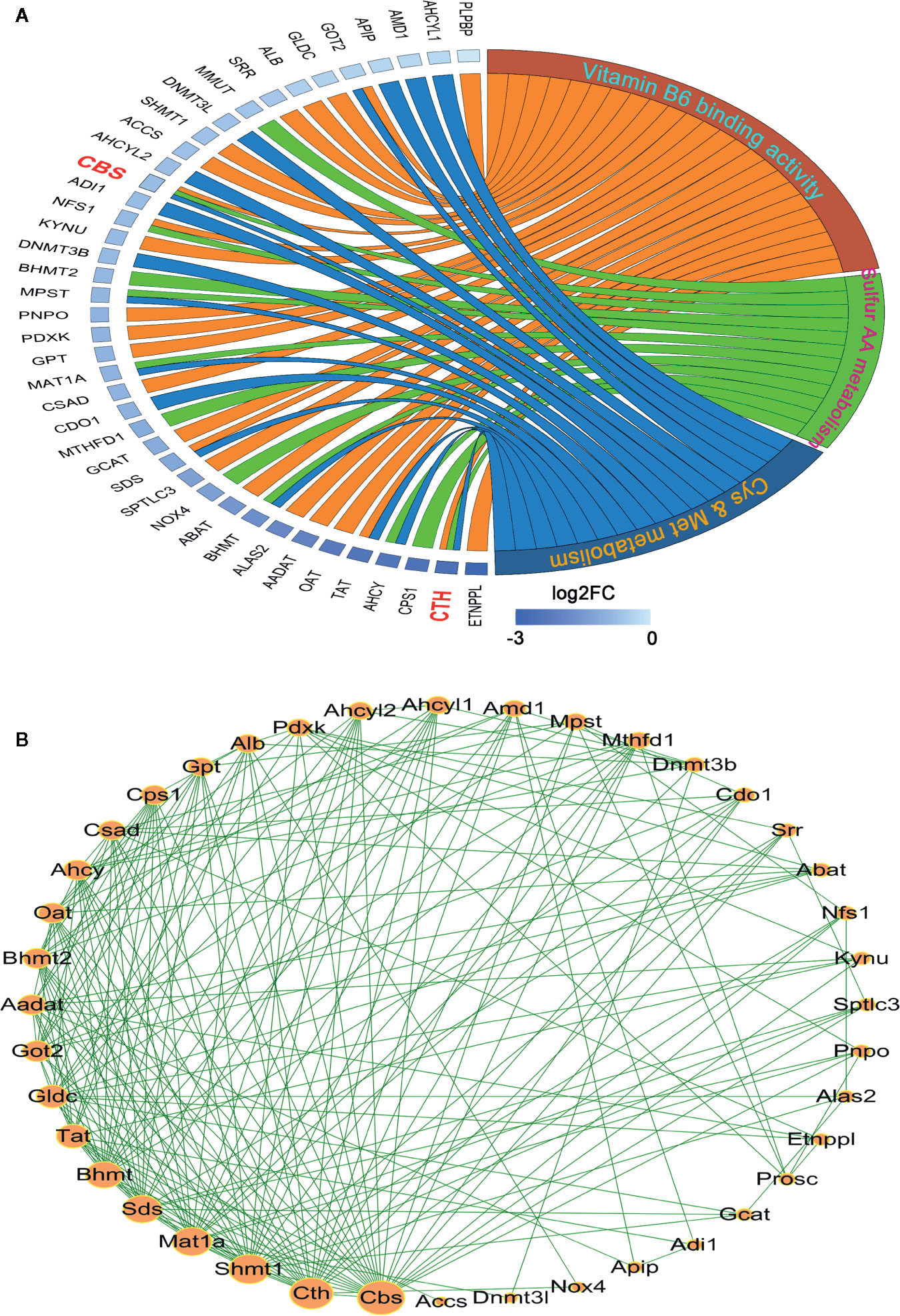
Figure 2 Analysis of the interaction among the leading-edge genes. (A) The ownership and relative expression of the leading-edge genes from three genesets (sulfur amino acid metabolism GO set, cysteine and methionine metabolism KEGG set, and vitamin B6 binding activity GO set) were presented through a chord diagram, which was drawn using the GOPlot R package. (B) The interaction of the genes was analyzed with the STRING online tool. The PPI network was constructed and the interaction degrees were calculated with Cytoscape 3.7 software.
In order to reveal the interaction of leading-edge genes, 12 enriched genes in sulfur amino acid metabolism GO set, 17 enriched genes in cysteine and methionine metabolism KEGG set, and 26 enriched genes in vitamin B6 binding activity GO geneset were subjected to the PPI analysis. As shown in Figure 2B, nine genes, i.e. CBS, CTH, SHMT1, MAT1A, SDS, BHMT, TAT, GLDC, and GOT2 had significantly high interaction degrees (>15), thereby being considered as hub genes responsible for the unique HCC phenotype.
Expression and Significance of Hub Genes in HCC Patients
To verify whether the hub genes were downregulated in HCC tumor tissues, the TCGA cohort (368 tumor samples vs. 50 normal liver samples) and GTEx dataset (110 normal liver samples) were applied for differential expression analysis. As shown in Figures 3A–H, all the hub genes except GOT2 (Figure 3I) were significantly downregulated in HCC tumor tissues compared with normal tissues (P < 0.001). Additionally, importance of the hub genes was evaluated by observing their influence on HCC patient survival in the TCGA cohort. As shown in Figures 3J–R, the downregulation of CBS, CTH, SHMT1, MAT1A, SDS, BHMT, TAT, and GOT2 could significantly reduce the survival rate, instead of GLDC (Figure 3Q). Therefore, GLDC was not involved in the following investigation.
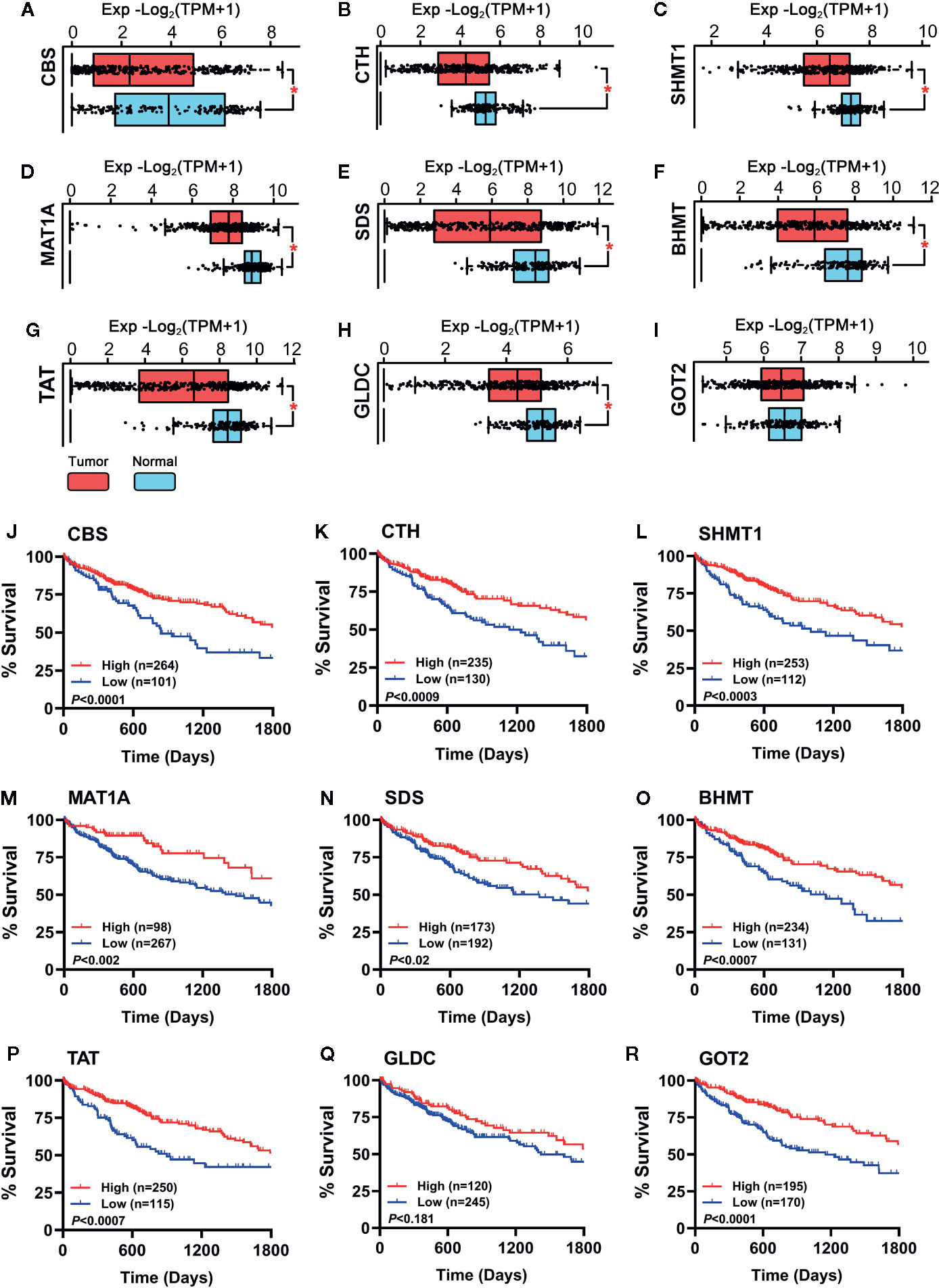
Figure 3 (A–I) Expression of the hub genes between 160 normal liver tissues (50 samples of TCGA_LIHC cohort and 110 samples in GTEx dataset) and 368 HCC tumor tissues of TCGA_LIHC cohort were observed. Data are shown as median ± quartile, *P < 0.001 vs. Normal liver tissues. (J–R) Analysis of the survival rate of HCC patients in TCGA_LIHC database between high expression (red lines) and low expression (blue lines) of the hub genes. The number of patients in either group and the Log-rank test’s P are shown in each panel.
Identification of RSS That Interact With Hub Genes
To find out which RSS were potentially affected by the aforementioned hub genes, the metabolomics was investigated with the Metscape 3.0 tool, which is used to show the interaction of genes and compounds. As presented in Figure 4, the metabolism of methionine, cystine, cysteine, homocysteine, cystathionine, H2S, and H2S2 was found to be closely associated with the above hub genes. Therefore, the imbalance of these compounds is probably responsible for the development of HCC, and their application may affect the growth of HCC tumor cells.
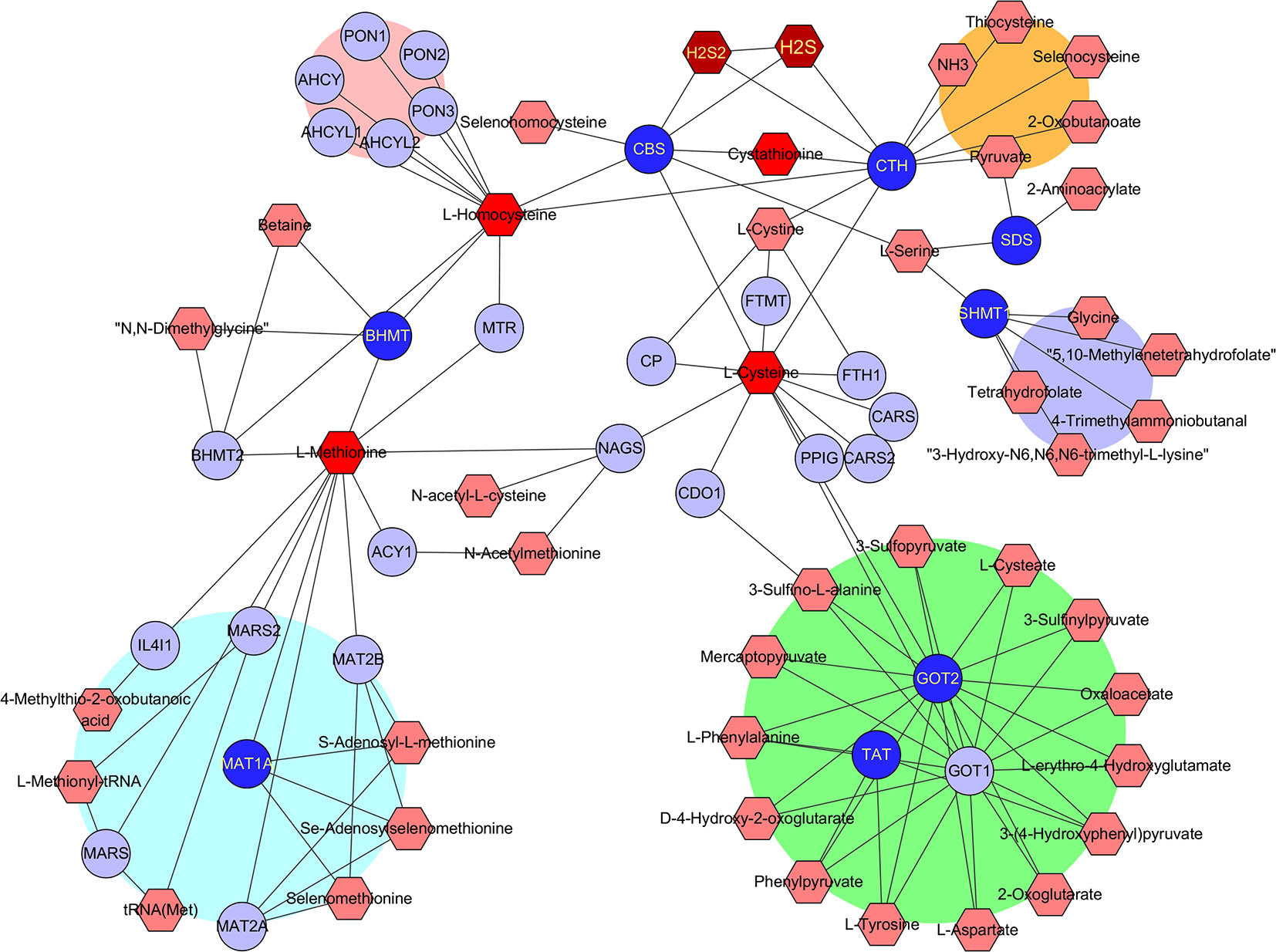
Figure 4 Identification of RSS affected by the hub genes. The hub genes and RSS compounds were imported into Metscape 3.0 software. The construction of network type was set as compound-gene, and the network was created. Hub genes (Dark blue ○), Related genes (Light blue ○), Interacted RSS compounds (Dark red hexagon).
Effects of Identified RSS on Viability in HCC SNU398 Tumor Cells
With the identified RSS compounds that can interact with the hub genes, we investigated their influence on the viability in HCC SNU398 tumor cells. This cell line has relatively low CTH expression among 25 HCC tumor cell lines (Figure 5A), which is consistent with the discovery of reprogrammed sulfur metabolism in HCC tissues. Therefore, SNU398 tumor cells were applied in the following experiments. As shown in Figure 5, comparing with H2S (in the form of Na2S) (Figure 5B), treatment of the cells with H2S2 (in the form of Na2S2) (Figure 5C) or H2S3 (in the form of Na2S3) (Figure 5D) for 48 h remarkably decreased SNU398 cell viability, and the effect of H2S3 was much stronger. On the contrary, treatment with 200 μM H2S increased cell viability (Figure 5B). Additionally, RSS, such as sulfur-containing amino acids (NAC, homocysteine, and cystine), as well as derived peptide (cystathionine and GSSG), did not show inhibitory effects on the cell viability (Figures 5E–I).
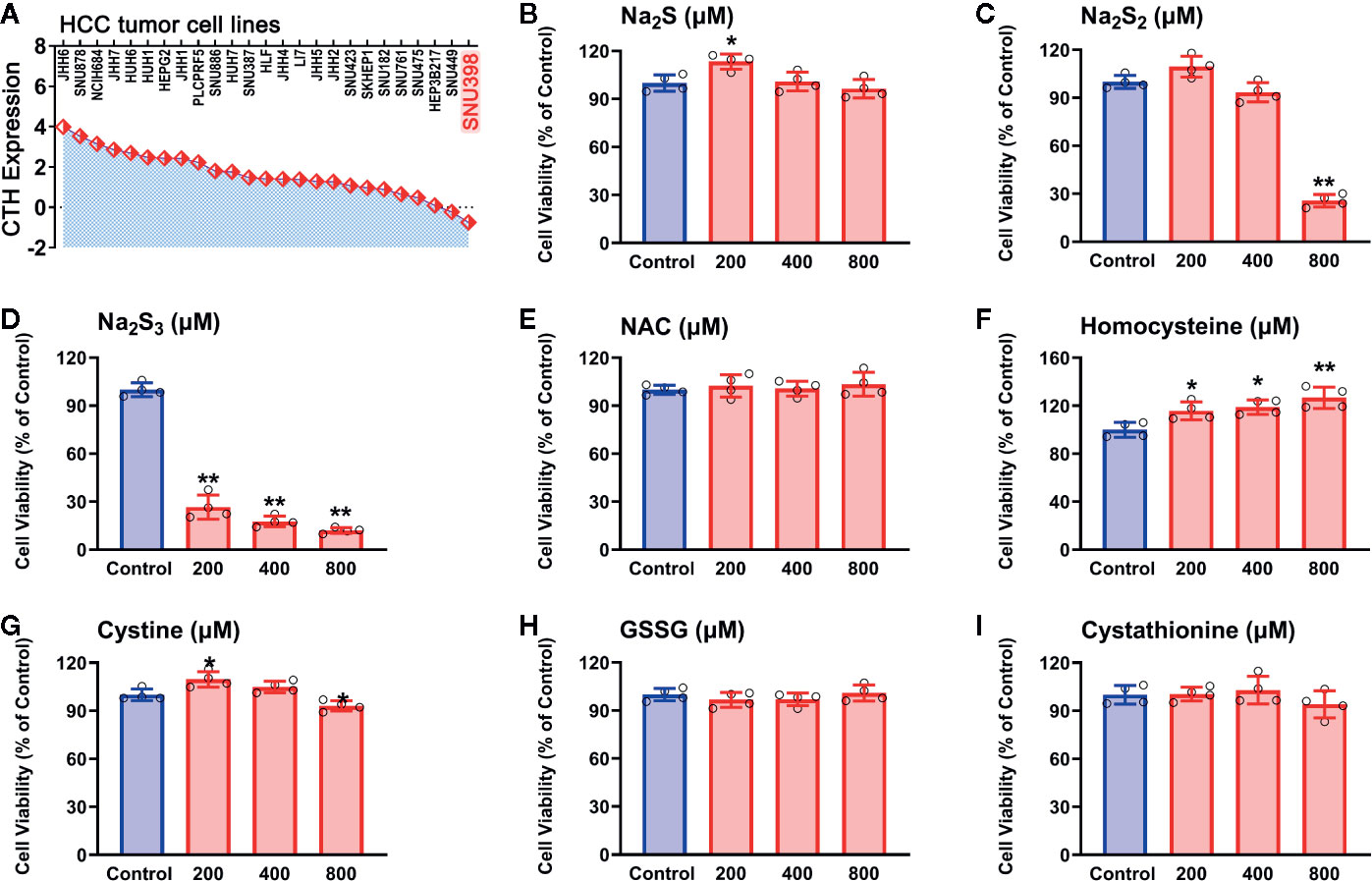
Figure 5 Effects of RSS on viability in HCC SNU398 tumor cells. (A) Expression of CTH in various common HCC liver cancer cell lines. (B–I) After treatment with increasing concentrations of Na2S (B), Na2S2 (C), Na2S3 (D), NAC (E), Homocysteine (F), Cystine (G), GSSG (H), or Cystathionine (I) for 48 h, the cell viability was tested with CCK-8 assay. Data are expressed as mean ± SD of four independent experiments. *P < 0.05, **P < 0.01 vs. Control group.
To examine whether the effects of H2S2 and H2S3 were associated with reactive sulfane sulfur, the cell imaging technology was applied to observe intracellular sulfane sulfur content. As shown in Figure 6, the treatment with H2S2 or H2S3 could clearly enhance the intracellular sulfane sulfur content, while other RSS compounds did not show significant effects (Figure 6). Moreover, scavenging sulfane sulfur with excess GSH remarkably attenuated Na2S3-induced anti-HCC effects (Supplementary Figure 1). The data indicate that among RSS, sulfane sulfur may be important for HCC therapy.
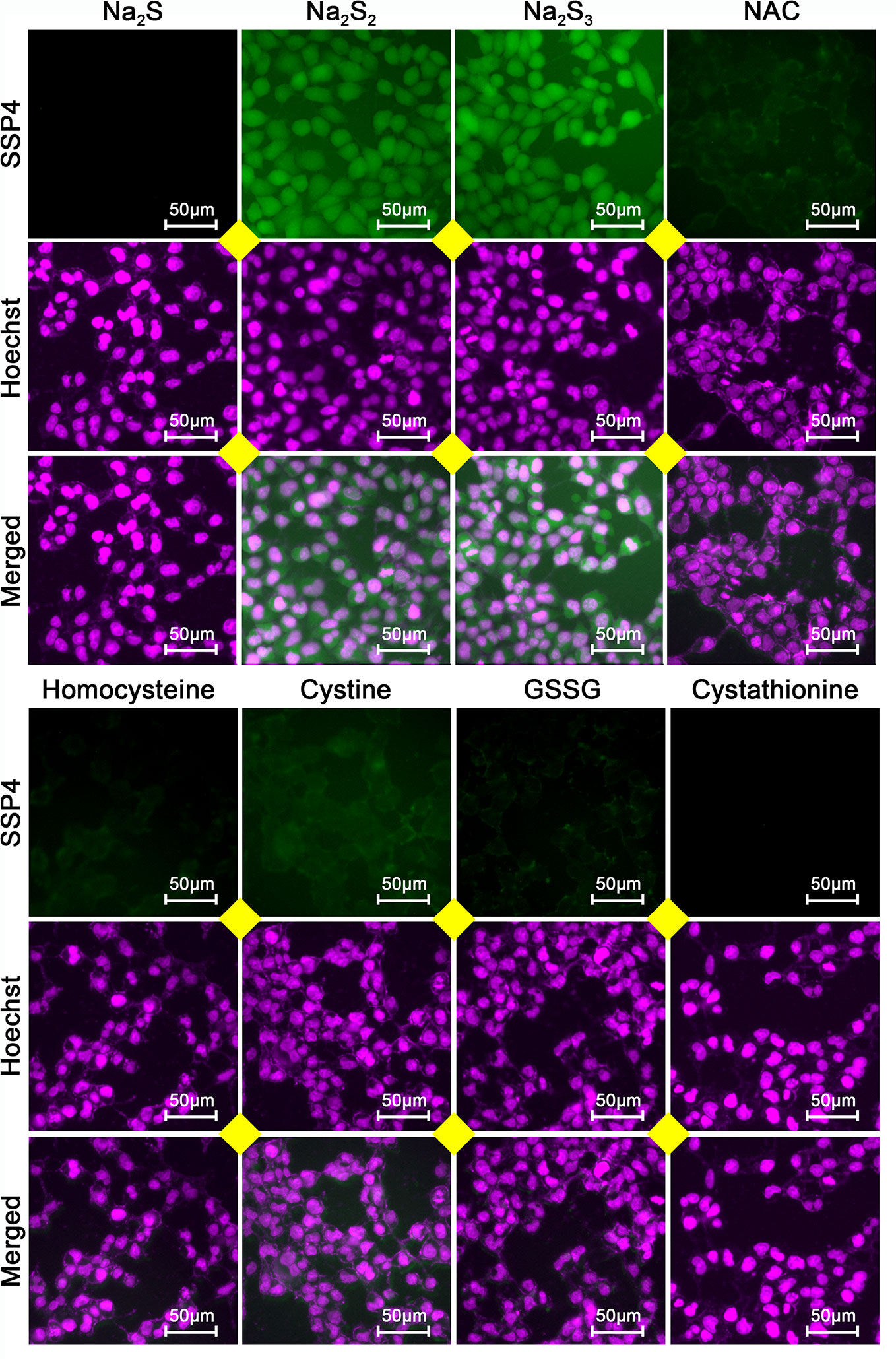
Figure 6 Effects of RSS on intracellular sulfane sulfur content in HCC SNU398 cells. After treatment with 400 μM of Na2S, Na2S2, Na2S3, NAC, Homocysteine, Cystine, GSSG, or Cystathionine for 1 h, the intercellular sulfane sulfur was observed with fluorescent probe SSP4 staining followed by photofluorography, and the cell nuclei were labeled through Hoechst 33324 staining.
Sulfane Sulfur Mediates RSS-Induced Anti-HCC Effects in SNU398 Tumor Cells
Since H2S2 and H2S3 could reduce SNU398 cell viability and increase intracellular sulfane sulfur content, and scavenging sulfane sulfur could attenuate H2S3-induced anti-HCC effects, we further validated this finding by sulfane sulfur-containing compounds. As shown in Figure 7A, exposure of SNU398 cells to diallyl trisulfide (DATS) significantly reduced cell viability. Such inhibitory effects were also observed in dimethyl trisulfide (DMTS), although only under high concentrations (Figure 7B). Additionally, the treatment with DATS or DMTS could increase intracellular sulfane sulfur contents (Figure 7C). The result supports the significance of sulfane sulfur in HCC therapy.
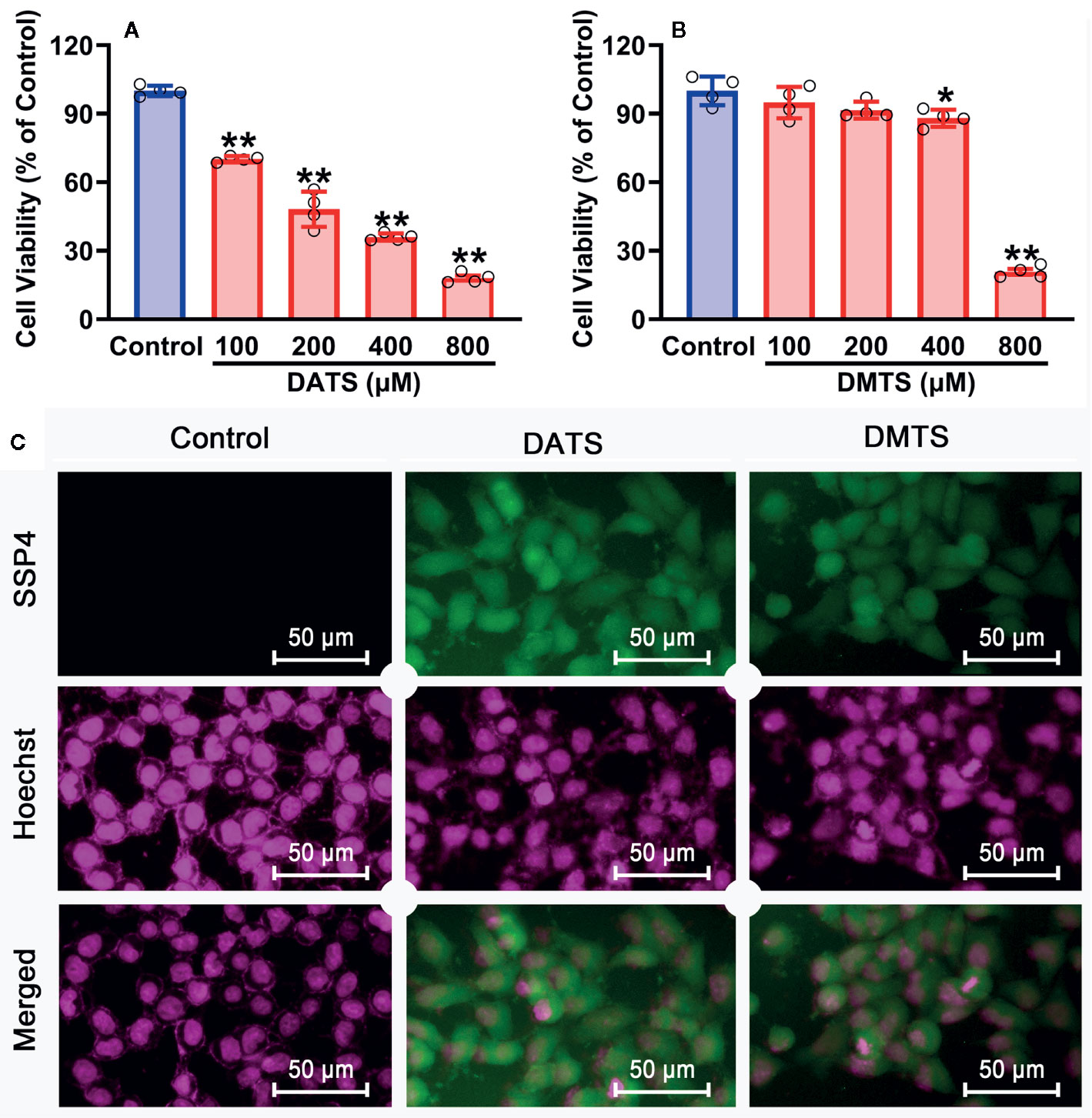
Figure 7 Anti-cancer effects of sulfane sulfurs in HCC SNU398 cells. (A, B) The cells were treated with increasing concentrations of DATS or DMTS for 48 h. The cell viability was measured with CCK-8 assay. Data are expressed as mean ± SD of four independent experiments. *P < 0.05, **P < 0.01 vs. Control group. (C) After the treatment with 400 μM DATS or DMTS, the intercellular content of sulfane sulfur was observed with SSP4 staining followed by photofluorography, and the cell nuclei were labeled using Hoechst 33324.
Design, Synthesis, and Evaluation of Controllable Sulfane Sulfur Donors
Although Na2S2 and Na2S3 could inhibit the viability of SNU398 cells by producing sulfane sulfur, their usage probably has some problems. First, they are difficult to handle due to fast decomposition. Additionally, they may contain impurities like Na2S, further complicating the situation. Especially, Na2S2 and Na2S3 have limited trans-membrane ability. In the TCGA dataset, SLC7A11 was found to be a pivotal risk for the poor survival of HCC patients (Figure 8A). This gene is highly expressed in HCC tumor tissues, as well as some tumor cell strains including SNU398 and SNU387 liver cancer cells (Figure 8B). Since SLC7A11 protein is responsible for the transport of cystine or cysteine, we envisioned sulfane sulfur donors with a structure similar to cystine/cysteine might have better efficiency for HCC’s therapy. Additionally, they would be more stable and flexible due to the absence of free persulfide (R-SSH). As such, we prepared and tested S-(methoxycarbonylsulfenyl) cysteine hydrochloride, a persulfided cysteine precursor (PSCP). Under physiological pH, PSCP can undergo an intramolecular acyl transfer reaction to release cysteine persulfide (Cys-SSH), which eventually degrades to form polysulfides (Figure 8C), similar to a known design (Artaud and Galardon, 2014). Additionally, mass spectroscopy identified cysteine di-, tri-, and tetrasulfide derivatives in the solution (Supplementary Figure 2). These compounds are not only evidence of the persulfide formation, but also are sulfane sulfur themselves for tri- and tetrasulfide. Both persulfides and polysulfides belong to sulfane sulfur, so PSCP can be considered as the precursor of cysteine-derived sulfane sulfur. As expected, under physiological pH, sulfane sulfur formed from PSCP (2 mM) reached the maximum concentration in 120 min. Significant amounts of sulfane sulfur were still detectable after 720 min (Figure 8D). The cell imaging assay showed that treatment with PSCP was able to increase intracellular sulfane sulfur contents in SNU 398 cells (Figure 8E).
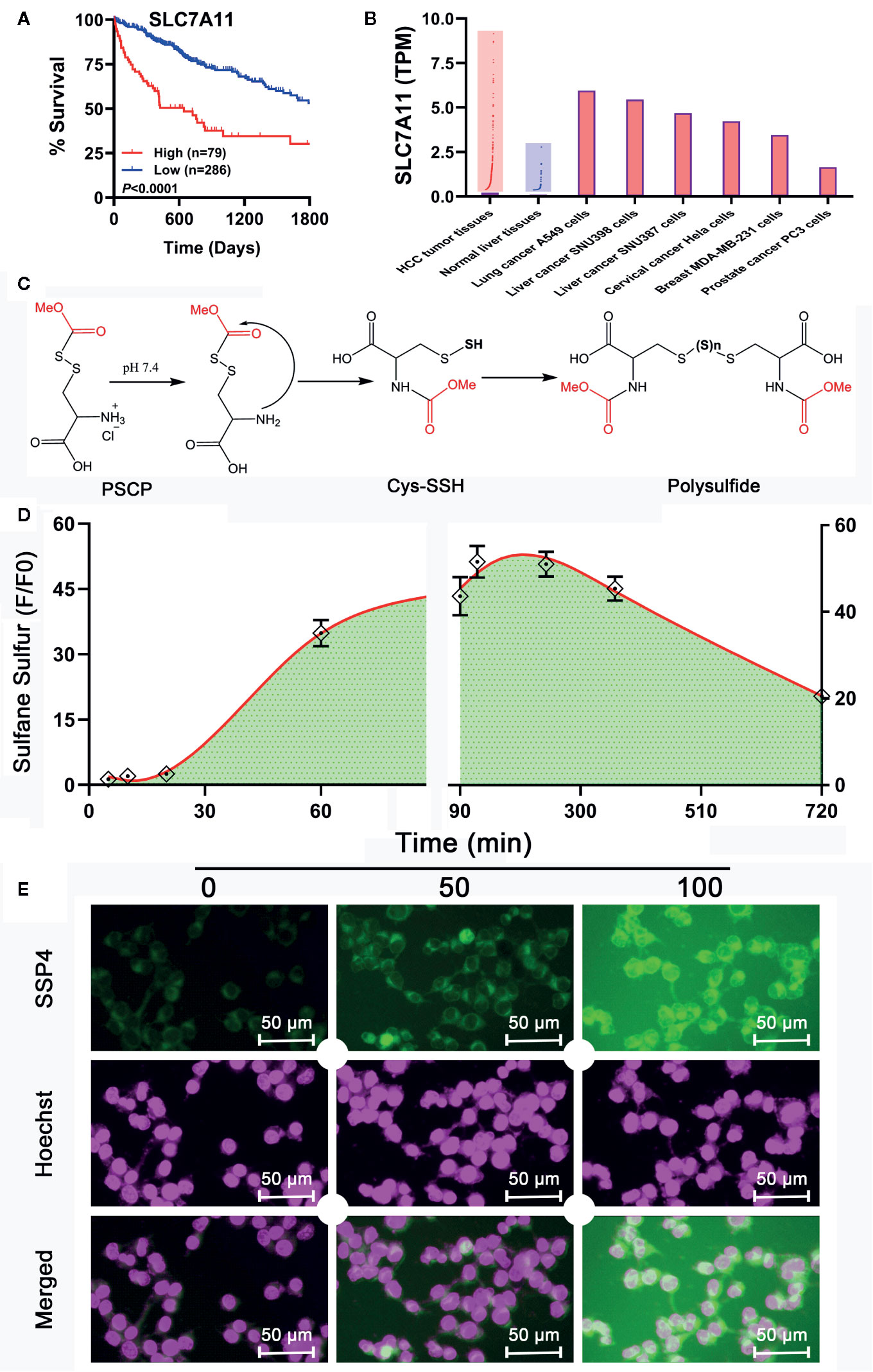
Figure 8 Development of a sulfane sulfur donor-specific to tumor tissues. (A) With TCGA cohort, effects of SLC7A11 on HCC patient survival between high (red line) and low (blue line) expression groups were examined using Log-rank test. (B) Analysis of TCGA cohort and Cancer Cell Line Encyclopedia for SLC7A11 expression in HCC patient tissues and tumor cell lines, respectively. (C) Mechanism underlying sulfane sulfur generation from PSCP. (D) The generated sulfane sulfur in PBS buffer was measured with a spectrophotometer. (E) After treatment of SNU398 cells with 200 μM PSCP for different times, intracellular sulfane sulfur was observed with SSP4 staining followed by photofluorography, and the cell nuclei were labeled using Hoechst 33324 staining.
However, as found in Figures 8C, D and Supplementary Figure 1, the generation of sulfane sulfur from PSCP was spontaneous and the effect of sulfane sulfur was dramatically attenuated by GSH, which may impede its future application. Therefore, we have attempted to develop donors triggered by physiological stimuli, like enzymes. As such, four esterase-triggered donors were designed and synthesized (1, 3, 5, and 7), which consisted of a cleavable ester trigger, a self-immolative linker, and a sulfane sulfur-releasing moiety. These donors can theoretically release protected cysteine or penicillamine persulfide, a form of sulfane sulfur, upon reaction with intracellular esterase. Meanwhile, four control compounds (2, 4, 6, and 8), which should only release protected cysteine/penicillamine, were also prepared. The characterizations of Compound 1-8 can be found in Supplementary Figure 3. Additionally, we have made a compound, which has no ester trigger and is therefore not able to release persulfide (9). As shown in Figure 9, treatment with these esterase-triggered donors (1, 3, 5, and 7) significantly inhibited SNU398 cell viability (Figures 9A, C, E, G), while no obvious inhibitory effects in the control compounds (2, 4, 6, and 8) was found (Figures 9B, D, F, H). Although these results are promising, compound 9, which theoretically cannot generate sulfane sulfur, also reduced cell viability. Additionally, we were not able to detect sulfane sulfur generation from these donors using SSP4. The data suggest that inhibitory effects of (1, 3, 5, and 7) may not come from sulfane sulfur. Although it is still not clear how these compounds achieved inhibitor effects, the present design of esterase-triggered sulfane sulfur donor should be improved.
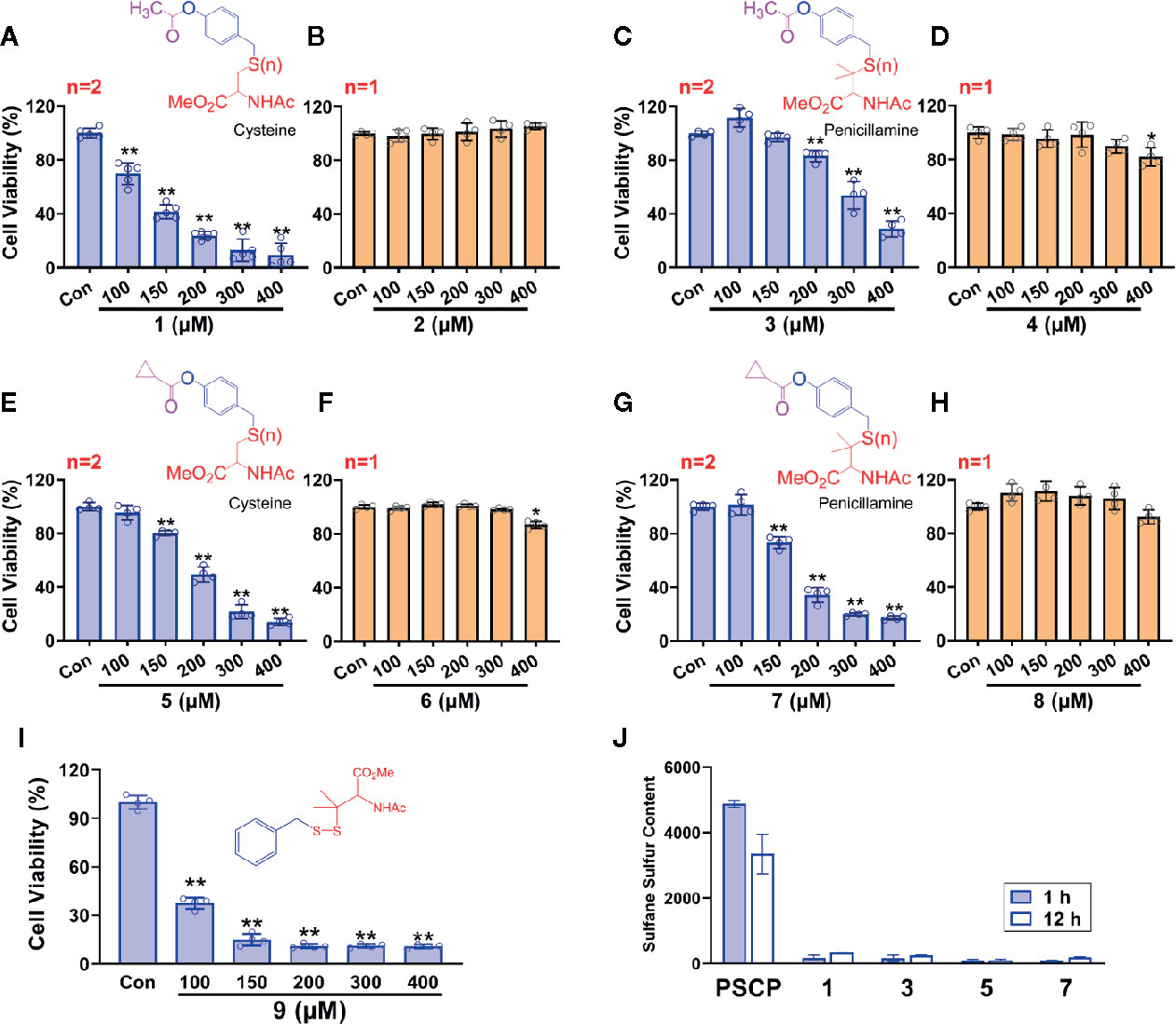
Figure 9 Controllable sulfane sulfur compounds-mediated inhibition of cell viability. HCC SNU398 cells were treated with esterase-triggered sulfane sulfur donor 1 (A), 3 (C), 5 (E), or 7 (G), their control compounds 2 (B), 4 (D), 6 (F), or 8 (H), as well as 9 (I), which does not generate sulfane sulfur. After 48 h, the cell viability was tested with CCK-8 assay. (J) Sulfane sulfur generated from the 1, 3, 5, and 7 was measured at 1 and 12 h with a fluorescence probe SSP4. Data are expressed as mean ± SD of four independent experiments. *P < 0.05, **P < 0.01 vs. Control group.
Evaluation of PSCP’s Anti-Cancer Effects in Various Cells
As shown in Figure 10, in HCC SNU398 cells (Figure 10A) and another liver cancer cell line SNU387 (Figure 10B), PSCP could exert obvious anti-cancer effects. However, in other tumor cell strains, including lung cancer A549 cells (Figure 10C), prostate cancer PC3 cells (Figure 10D), cervical cancer Hela cells (Figure 10E), and breast MDA-MB-231 cells (Figure 10F), remarkable anti-cancer effects were not observed. Notably, in normal cells, such as Raw-blue macrophages, H9c2 cardiomyocytes, as well as normal liver cells, PSCP did not show severe toxicity. The result suggests that sulfane sulfur (PSCP) is able to inhibit tumor cell viability in a liver cancer-specific manner.
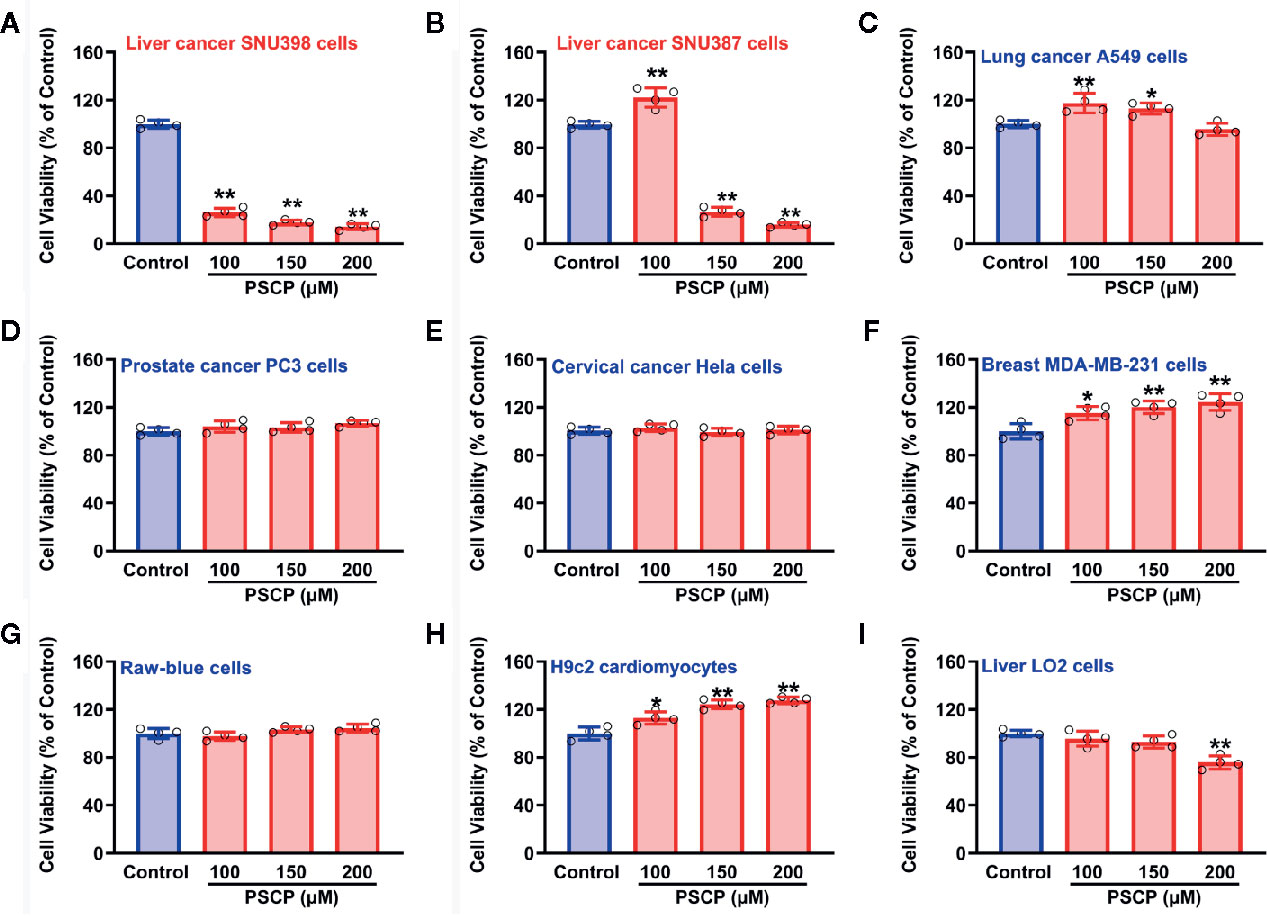
Figure 10 Effects of PSCP on viability in various cell strains. (A) Liver cancer SNU398 cells, (B) Liver cancer SNU387 cells, (C) Lung cancer A549 cells, (D) Prostate cancer PC3 cells, (E) Cervical cancer Hela cells, (F) Breast MDA-MB-231 cells, (G) Raw-blue macrophages, (H) H9c2 cardiomyocytes, and (I) Liver LO2 cells. Different types of cell strains were treated with increasing concentrations of PSCP for 48 h. The cell viability was tested with CCK-8 assay. Data are expressed as mean ± SD of four independent experiments. *P < 0.05, **P < 0.01 vs. Control group.
Discussion
Metabolic reprogramming is a distinctive hallmark of cancer. The reprogrammed glucose metabolism in tumorigenesis has been studied for many years (Kato et al., 2018; Lin et al., 2019). Additionally, the metabolism of amino acids, such as cysteine, homocysteine, glutamine, and methionine, are attracting increased attention in recent years (Gao et al., 2019; Lieu et al., 2020). Through metabolic reprogramming, tumor cells can generate large amounts of acids, reduced compounds, as well as one-carbon units, to support their rapid growth (Pavlova and Thompson, 2016; Sun et al., 2020).
Sulfur metabolism in HCC tumor tissues is still a novel field of research. In this work, we observed the sulfur metabolism was reprogrammed in HCC through the bioinformatic GSEA screening. The scanning revealed that the gene sets of sulfur amino acid metabolism, cysteine and methionine metabolism, and vitamin B6 binding activity were remarkably impaired in HCC tissues. The further PPI analysis suggested that CBS, CTH, SHMT1, MAT1A, SDS, BHMT, TAT, and GOT2 were significant hub genes. Importantly, the TCGA analysis validated their significance in HCC patients. Through these data, we believe that the genes related to sulfur metabolism should be dysregulated in HCC tissues, which may be responsible for the low survival of patients. Recently, some studies demonstrated that high levels of methionine and homocysteine were able to promote tumorigenesis (Sun et al., 2002; Gao et al., 2019), while the methionine-restricted diet arrested tumor growth and increased chemotherapy sensitivity (Hoffman, 2019). Moreover, a previous study showed that the content of vitamin B6 pyridoxal phosphate and the activity of pyridoxine kinase in Morris hepatomas were lower than those in normal liver tissues (Meisler et al., 1982). The antioxidant NAC, a cysteine equivalent, was reported to accelerate lung cancer progression (Sayin et al., 2014). These studies support that the increased methionine, homocysteine, and cysteine, as well as the decreased vitamin B6 binding activity, are probably important causes for HCC tumor growth, which is consistent with the present analyses. For this reason, metabonomic analysis can bring to light the potential interaction between RSS and the hub genes in HCC tissues. Meanwhile, the findings provide a reasonable explanation that through sulfur metabolic reprogramming, HCC tumor cells can raise the content of methionine, homocysteine, or cysteine to fuel their fast proliferation.
Reprogrammed sulfur metabolism is likely to make HCC tumor cells adapt to a new sulfur environment. Since the metabolism of RSS was impaired, we scanned the effects of RSS on HCC tumor cell viability. It was found that Na2S2 and Na2S3 remarkably inhibited cell viability, while the other RSS compounds did not exert such effects. It is worth noting that Na2S2 and Na2S3 are sulfane sulfur donors (Jiang et al., 2019). Also, DATS and DMTS belong to sulfane sulfurs, which could also attenuate HCC tumor cell viability in the present study, consistent with the previous reports (Weisberger and Pensky, 1957; Liu X. et al., 2019). Some researchers considered that anti-cancer effects from garlic-derived compounds were attributed to H2S generation. However, in this study, we did not observe that H2S had anti-cancer effects at the same treatment profile. In recent years, the studies of H2S on HCC tumorigenesis are conflicting, i.e. anti-cancer vs. pro-cancer (Wu et al., 2017; Yang et al., 2019b; Wu et al., 2019). Those studies on H2S’s anti-HCC activity, we think, are probably due to the generation of sulfane sulfur from Na2S/NaHS.
To support the above hypothesis, the cell imaging assay was performed and the results demonstrated that the treatment with Na2S2 or Na2S3 was able to enhance the intracellular sulfane sulfur levels, so did DATS and DMTS. Importantly, their treatment all showed anti-HCC effects. Moreover, scavenging of sulfane sulfur with excess GSH significantly attenuated sulfane sulfur-induced anti-HCC effects. These findings support the notion that the application of sulfane sulfurs may be an effective therapeutic strategy, especially for the HCC tumor cells with reprogrammed sulfur metabolism.
Notably, Na2S2 and Na2S3 are unstable chemicals, while commercial DATS is highly volatile, which will hinder their applications. Through the TCGA cohort analysis, it was shown that SLC7A11 (xCT) is upregulated in HCC tissues, indicating an increased cystine transportability. We then synthesized PSCP and found it was able to produce cystine sulfane sulfurs including per-, tri-, and tetrasulfide. The continuous monitoring indicated that the content of sulfane sulfur could keep stable for at least ten hours in aqueous solution. Additional cell experiments showed that the treatment with PSCP did not only enhance intracellular sulfane sulfur, but also inhibited cell viability. Furthermore, we tested the effects of PSCP in other cancer cells like lung cancer A549 cells, prostate cancer PC3 cells, cervical cancer Hela cells, and breast MDA-MB-231 cells, and did not find remarkable inhibitory activity. We surmise that the sulfur metabolism may not be reprogrammed, although some of these cells have high cystine transportability, for example the increased SLC7A11 expression in A549 cells. Interestingly, in H9c2 cardiomyocytes and normal liver LO2 cells, the cytotoxicity was not very high, indicating its safety for the future in vivo application. Lastly, because of the spontaneous release of sulfane sulfur from PSCP and the influence of GSH, we synthesized esterase-triggered sulfane sulfur donors. Although the donors may produce sulfane sulfur theoretically and showed promising inhibition on cell viability, we were not able to observe the generation of sulfane sulfur from them. Therefore, the present design of enzyme-triggered sulfane sulfur donors should be improved in the future.
In conclusion, we have discovered that sulfur-related metabolism in HCC is reprogrammed. The RSS screening indicates that HCC tumor cells are sensitive to sulfane sulfur. The selective inhibition of HCC cell viability from PSCP, a sulfane sulfur donor, further validates this finding. This study provides a basic evidence for the treatment of HCC that has sulfur metabolic reprogramming.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
CY, SX, and XMZ designed the experiments and wrote the manuscript. MC, XN, YW, and HZ performed all the experiments and statistical analyses. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Natural Science Foundation of Guangdong Province (2017A030313892) and Guangzhou Key Medical Discipline Construction Project.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Kaoru Mogushi at Juntendo University for the mRNA expression microarray.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2020.571143/full#supplementary-material
References
Akaike, T., Ida, T., Wei, F. Y., Nishida, M., Kumagai, Y., Alam, M. M., et al. (2017). Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat. Commun. 8, 1177. doi: 10.1038/s41467-017-01311-y
Alvarez, L., Bianco, C. L., Toscano, J. P., Lin, J., Akaike, T., Fukuto, J. M. (2017). Chemical biology of hydropersulfides and related species: Possible roles in cellular protection and redox signaling. Antioxid. Redox Signal. 27, 622–633. doi: 10.1089/ars.2017.7081
Artaud, I., Galardon, E. (2014). A persulfide analogue of the nitrosothiol SNAP: formation, characterization and reactivity. Chembiochem 15, 2361–2364. doi: 10.1002/cbic.201402312
Bibli, S. I., Luck, B., Zukunft, S., Wittig, J., Chen, W., Xian, M., et al. (2018). A selective and sensitive method for quantification of endogenous polysulfide production in biological samples. Redox Biol. 18, 295–304. doi: 10.1016/j.redox.2018.07.016
Chen, W., Liu, C., Peng, B., Zhao, Y., Pacheco, A., Xian, M. (2013). New fluorescent probes for sulfane sulfurs and the application in bioimaging. Chem. Sci. 4, 2892–2896. doi: 10.1039/C3SC50754H
Chiyonobu, N., Shimada, S., Akiyama, Y., Mogushi, K., Itoh, M., Akahoshi, K., et al. (2018). Fatty acid binding protein 4 (FABP4) overexpression in intratumoral hepatic stellate cells within hepatocellular carcinoma with metabolic risk factors. Am. J. Pathol. 188, 1213–1224. doi: 10.1016/j.ajpath.2018.01.012
DeBerardinis, R. J., Lum, J. J., Hatzivassiliou, G., Thompson, C. B. (2008). The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7, 11–20. doi: 10.1016/j.cmet.2007.10.002
Faubert, B., Solmonson, A., DeBerardinis, R. J. (2020). Metabolic reprogramming and cancer progression. Science 368, eaaw5473. doi: 10.1126/science.aaw5473
Galluzzi, L., Vacchelli, E., Michels, J., Garcia, P., Kepp, O., Senovilla, L., et al. (2013). Effects of vitamin B6 metabolism on oncogenesis, tumor progression and therapeutic responses. Oncogene 32, 4995–5004. doi: 10.1038/onc.2012.623
Gao, X., Sanderson, S. M., Dai, Z., Reid, M. A., Cooper, D. E., Lu, M., et al. (2019). Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 572, 397–401. doi: 10.1038/s41586-019-1437-3
Gregory, J. F., DeRatt, B. N., Rios-Avila, L., Ralat, M., Stacpoole, P. W. (2016). Vitamin B6 nutritional status and cellular availability of pyridoxal 5’-phosphate govern the function of the transsulfuration pathway’s canonical reactions and hydrogen sulfide production via side reactions. Biochimie 126, 21–26. doi: 10.1016/j.biochi.2015.12.020
Hoffman, R. M. (2019). Clinical studies of methionine-restricted diets for cancer patients. Methods Mol. Biol. 1866, 95–105. doi: 10.1007/978-1-4939-8796-2_9
Iciek, M. B., Rokita, H. B., Wlodek, L. B. (2001). Effects of diallyl disulfide and other donors of sulfane sulfur on the proliferation of human hepatoma cell line (HepG2). Neoplasma 48, 307–312.
Iciek, M., Kwiecien, I., Chwatko, G., Sokolowska-Jezewicz, M., Kowalczyk-Pachel, D., Rokita, H. (2012). The effects of garlic-derived sulfur compounds on cell proliferation, caspase 3 activity, thiol levels and anaerobic sulfur metabolism in human hepatoblastoma HepG2 cells. Cell Biochem. Funct. 30, 198–204. doi: 10.1002/cbf.1835
Jackson, M. R., Melideo, S. L., Jorns, M. S. (2012). Human sulfide: quinone oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite. Biochemistry 51, 6804–6815. doi: 10.1021/bi300778t
Jiang, G., Li, M., Wen, Y., Zeng, W., Zhao, Q., Chen, C., et al. (2019). Visualization of sulfane sulfur in plants with a near-infrared fluorescent probe. ACS Sens. 4, 434–440. doi: 10.1021/acssensors.8b01423
Kato, Y., Maeda, T., Suzuki, A., Baba, Y. (2018). Cancer metabolism: New insights into classic characteristics. Jpn. Dent. Sci. Rev. 54, 8–21. doi: 10.1016/j.jdsr.2017.08.003
Kim, J., DeBerardinis, R. J. (2019). Mechanisms and implications of metabolic heterogeneity in cancer. Cell Metab. 30, 434–446. doi: 10.1016/j.cmet.2019.08.013
Lieu, E. L., Nguyen, T., Rhyne, S., Kim, J. (2020). Amino acids in cancer. Exp. Mol. Med. 52, 15–30. doi: 10.1038/s12276-020-0375-3
Lin, J., Xia, L., Liang, J., Han, Y., Wang, H., Oyang, L., et al. (2019). The roles of glucose metabolic reprogramming in chemo- and radio-resistance. J. Exp. Clin. Cancer Res. 38, 218. doi: 10.1186/s13046-019-1214-z
Liu, H., Radford, M. N., Yang, C. T., Chen, W., Xian, M. (2019). Inorganic hydrogen polysulfides: chemistry, chemical biology and detection. Br. J. Pharmacol. 176, 616–627. doi: 10.1111/bph.14330
Liu, X., Baecker, A., Wu, M., Zhou, J. Y., Yang, J., Han, R. Q., et al. (2019). Raw garlic consumption and risk of liver cancer: A population-based case-control study in Eastern China. Nutrients 11, 2038. doi: 10.3390/nu11092038
Lohitesh, K., Chowdhury, R., Mukherjee, S. (2018). Resistance a major hindrance to chemotherapy in hepatocellular carcinoma: an insight. Cancer Cell Int. 18, 44. doi: 10.1186/s12935-018-0538-7
Meisler, N. T., Nutter, L. M., Thanassi, J. W. (1982). Vitamin B6 metabolism in liver and liver-derived tumors. Cancer Res. 42, 3538–3543.
Pan, Y., Ye, S., Yuan, D., Zhang, J., Bai, Y., Shao, C. (2014). Hydrogen sulfide (H2S)/cystathionine gamma-lyase (CSE) pathway contributes to the proliferation of hepatoma cells. Mutat. Res. 763–764, 10–18. doi: 10.1016/j.mrfmmm.2014.03.002
Pavlova, N. N., Thompson, C. B. (2016). The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47. doi: 10.1016/j.cmet.2015.12.006
Renga, B. (2011). Hydrogen sulfide generation in mammals: the molecular biology of cystathionine-beta- synthase (CBS) and cystathionine-gamma-lyase (CSE). Inflamm. Allergy Drug Targets. 10, 85–91. doi: 10.2174/187152811794776286
Rietman, B. H., Peters, R. F. R., Tesser, G. I. (1994). A facile method for the preparation of S-(Alkylsulfenyl)cysteines. Synthetic Commun. 24, 1323–1332. doi: 10.1080/00397919408011734
Sayin, V. I., Ibrahim, M. X., Larsson, E., Nilsson, J. A., Lindahl, P., Bergo, M. O. (2014). Antioxidants accelerate lung cancer progression in mice. Sci. Transl. Med. 6, 221ra15. doi: 10.1126/scitranslmed.3007653
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550. doi: 10.1073/pnas.0506580102
Sun, C. F., Haven, T. R., Wu, T. L., Tsao, K. C., Wu, J. T. (2002). Serum total homocysteine increases with the rapid proliferation rate of tumor cells and decline upon cell death: a potential new tumor marker. Clin. Chim. Acta 321, 55–62. doi: 10.1016/s0009-8981(02)00092-x
Sun, H., Zhou, Y., Skaro, M. F., Wu, Y., Qu, Z., Mao, F., et al. (2020). Metabolic reprogramming in cancer is induced to increase proton production. Cancer Res. 80, 1143–1155. doi: 10.1158/0008-5472.CAN-19-3392
Weisberger, A. S., Pensky, J. (1957). Tumor-inhibiting effects derived from an active principle of garlic (Allium sativum). Science 126, 1112–1114. doi: 10.1126/science.126.3283.1112-a
Wu, L. L., Wu, J. T. (2002). Hyperhomocysteinemia is a risk factor for cancer and a new potential tumor marker. Clin. Chim. Acta 322, 21–28. doi: 10.1016/s0009-8981(02)00174-2
Wu, D., Li, M., Tian, W., Wang, S., Cui, L., Li, H., et al. (2017). Hydrogen sulfide acts as a double-edged sword in human hepatocellular carcinoma cells through EGFR/ERK/MMP-2 and PTEN/Akt signaling pathways. Sci. Rep. 7, 5134. doi: 10.1038/s41598-017-05457-z
Wu, D. D., Wang, D. Y., Li, H. M., Guo, J. C., Duan, S. F., Ji, X. Y. (2019). Hydrogen sulfide as a novel regulatory factor in liver health and disease. Oxid. Med. Cell Longev. 2019, 3831713. doi: 10.1155/2019/3831713
Yang, C. T., Devarie-Baez, N. O., Hamsath, A., Fu, X. D., Xian, M. (2019a). S-Persulfidation: chemistry, chemical biology, and significance in health and disease. Antioxid. Redox Signal. doi: 10.1089/ars.2019.7889
Yang, C. T., Wang, Y., Marutani, E., Ida, T., Ni, X., Xu, S., et al. (2019b). Data-driven identification of hydrogen sulfide scavengers. Angew. Chem. Int. Ed. Engl. 58, 10898–10902. doi: 10.1002/anie.201905580
Yuan, S., Shen, X., Kevil, C. G. (2017). Beyond a gasotransmitter: Hydrogen sulfide and polysulfide in cardiovascular health and immune response. Antioxid. Redox Signal. 27, 634–653. doi: 10.1089/ars.2017.7096
Zhen, Y., Wu, Q., Ding, Y., Zhang, W., Zhai, Y., Lin, X., et al. (2018). Exogenous hydrogen sulfide promotes hepatocellular carcinoma cell growth by activating the STAT3-COX-2 signaling pathway. Oncol. Lett. 15, 6562–6570. doi: 10.3892/ol.2018.8154
Keywords: bioinformatics, hepatocellular carcinoma, metabolic reprogramming, reactive sulfur species, sulfane sulfur
Citation: Zhang X, Chen M, Ni X, Wang Y, Zheng X, Zhang H, Xu S and Yang CT (2020) Metabolic Reprogramming of Sulfur in Hepatocellular Carcinoma and Sulfane Sulfur-Triggered Anti-Cancer Strategy. Front. Pharmacol. 11:571143. doi: 10.3389/fphar.2020.571143
Received: 10 June 2020; Accepted: 01 September 2020;
Published: 25 September 2020.
Edited by:
Qi Zeng, Xidian University, ChinaReviewed by:
Hui Xie, Xidian University, ChinaWen Zhou, Guangzhou University of Chinese Medicine, China
Copyright © 2020 Zhang, Chen, Ni, Wang, Zheng, Zhang, Xu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chun-tao Yang, Y3lhbmdAZ3pobXUuZWR1LmNu, orcid.org/0000-0002-5640-0930; Shi Xu, c2hpX3h1QGJyb3duLmVkdQ==, orcid.org/0000-0002-4325-6503
 Ximing Zhang1
Ximing Zhang1 Xue Zheng
Xue Zheng Shi Xu
Shi Xu Chun-tao Yang
Chun-tao Yang