- 1Guangdong Provincial Key Laboratory of Fermentation and Enzyme Engineering, Guangdong Provincial Engineering and Technology Research Center of Biopharmaceuticals, School of Biology and Biological Engineering, South China University of Technology, Guangzhou, China
- 2Joint International Research Laboratory of Synthetic Biology and Medicine, Guangdong Provincial Engineering and Technology Research Center of Biopharmaceuticals, School of Biology and Biological Engineering, South China University of Technology, Guangzhou, China
- 3Center for Certification and Evaluation, Guangdong Drug Administration, Guangzhou, China
- 4State Key Laboratory of Functions and Applications of Medicinal Plants, College of Pharmacy, Guizhou Provincial Engineering Technology Research Center for Chemical Drug R&D, Guizhou Medical University, Guiyang, China
by He, S., Zhao, D., Ling, Y., Cai, H., Cai, Y., Zhang, J., and Wang, L. (2021). Front. Pharmacol. 12:796534. doi:10.3389/fphar.2021.796534
In the original article, there was a mistake in Figures 4 and 5 as published. There are some errors in the figure insertion, Figure 4 is repeated with Figure 3, and Figure 5 is the result of Figure 4. The corrected Figures 4 and 5 appear below.

FIGURE 4. Performance of fingerprint-based BC prediction models. (A) AUC results of the AtomPairs-based models. (B) AUC results of the MACCS-based models. (C) AUC results of the Morgan-based models. (D) AUC results of the PharmacoPFP-based models.
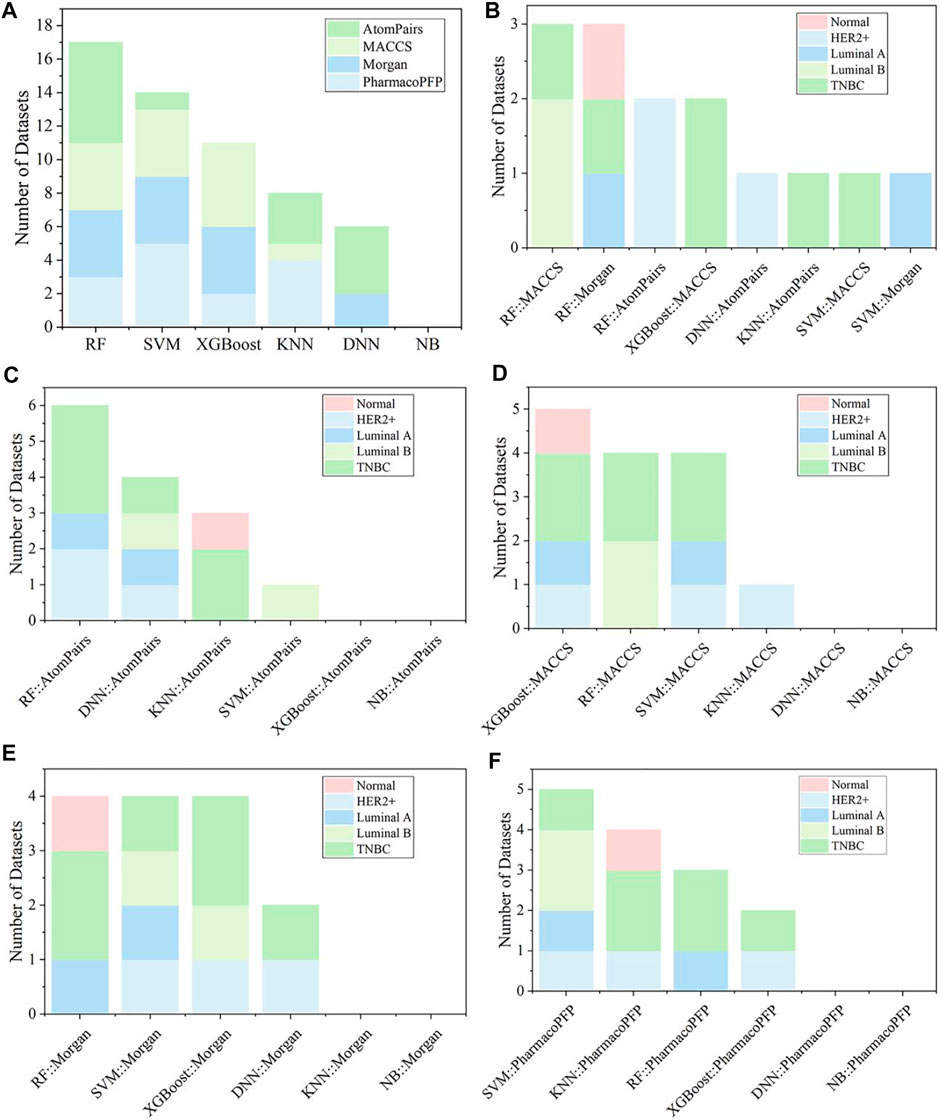
FIGURE 5. (A) Summary of the optimal models for each fingerprint-based feature. (B) The best models among various fingerprint-based models for different kinds of breast cell lines. The optimal models based on (C) AtomPairs, (D) MACCS, (E) Morgan, and (F) PharmacoPFP for different subtypes of breast cell lines.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: breast cancer, machine learning, graph neural networks, molecular fingerprints, structural fragments
Citation: He S, Zhao D, Ling Y, Cai H, Cai Y, Zhang J and Wang L (2022) Corrigendum: Machine Learning Enables Accurate and Rapid Prediction of Active Molecules Against Breast Cancer Cells. Front. Pharmacol. 13:901513. doi: 10.3389/fphar.2022.901513
Received: 22 March 2022; Accepted: 12 May 2022;
Published: 30 May 2022.
Edited and reviewed by:
Adriano D. Andricopulo, University of Sao Paulo, BrazilCopyright © 2022 He, Zhao, Ling, Cai, Cai, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiquan Zhang, empxZ21jQDE2My5jb20=; Ling Wang, bGluZ3dhbmdAc2N1dC5lZHUuY24=
†These authors have contributed equally to this work
 Shuyun He
Shuyun He Duancheng Zhao
Duancheng Zhao Yanle Ling1,2
Yanle Ling1,2 Hanxuan Cai
Hanxuan Cai Jiquan Zhang
Jiquan Zhang Ling Wang
Ling Wang