- 1Department of Bio-Resources, School of Biological Sciences, University of Kashmir, Srinagar, Jammu and Kashmir, India
- 2Department of Life Science, National Institute of Technology, Rourkela, Odisha, India
- 3Department of Family and Community Medicine, Faculty of Medicine, Al Baha University, Al Bahah, Saudi Arabia
- 4Department of Biology, College of Science, Hafr Al Batin University of Hafr Al-Batin, Hafar Al Batin, Saudi Arabia
- 5Regional Research Institute of Unani Medicine (RRIUM), University of Kashmir, Srinagar, Jammu and Kashmir, India
- 6Department of Clinical Biochemistry, School of Biological Sciences, University of Kashmir, Srinagar, Jammu and Kashmir, India
Delphinium roylei Munz is an indigenous medicinal plant to India where its activity against cancer has not been previously investigated, and its specific interactions of bioactive compounds with vulnerable breast cancer drug targets remain largely unknown. Therefore, in the current study, we aimed to evaluate the anti-breast cancer activity of different extracts of D. roylei against breast cancer and deciphering the molecular mechanism by Network Pharmacology combined with Molecular Docking and in vitro verification. The experimental plant was extracted with various organic solvents according to their polarity index. Phytocompounds were identified by High resolution-liquid chromatography-mass spectrometry (HR-LC/MS) technique, and SwissADME programme evaluated their physicochemical properties. Next, target(s) associated with the obtained bioactives or breast cancer-related targets were retrieved by public databases, and the Venn diagram selected the overlapping targets. The networks between overlapping targets and bioactive were visualized, constructed, and analyzed by STRING programme and Cytoscape software. Finally, we implemented a molecular docking test (MDT) using AutoDock Vina to explore key target(s) and compound(s). HR-LC/MS detected hundreds of phytocompounds, and few were accepted by Lipinski’s rules after virtual screening and therefore classified as drug-like compounds (DLCs). A total of 464 potential target genes were attained for the nine quantitative phytocompounds and using Gene Cards, OMIM and DisGeNET platforms, 12063 disease targets linked to breast cancer were retrieved. With Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway enrichment, a total of 20 signalling pathways were manifested, and a hub signalling pathway (PI3K-Akt signalling pathway), a key target (Akt1), and a key compound (8-Hydroxycoumarin) were selected among the 20 signalling pathways via molecular docking studies. The molecular docking investigation revealed that among the nine phytoconstituents, 8-hydroxycoumarin showed the best binding energy (−9.2 kcal/mol) with the Akt1 breast cancer target. 8-hydroxycoumarin followed all the ADME property prediction using SwissADME, and 100 nanoseconds (ns) MD simulations of 8-hydroxycoumarin complexes with Akt1 were found to be stable. Furthermore, D. roylei extracts also showed significant antioxidant and anticancer activity through in vitro studies. Our findings indicated for the first time that D. roylei extracts could be used in the treatment of BC.
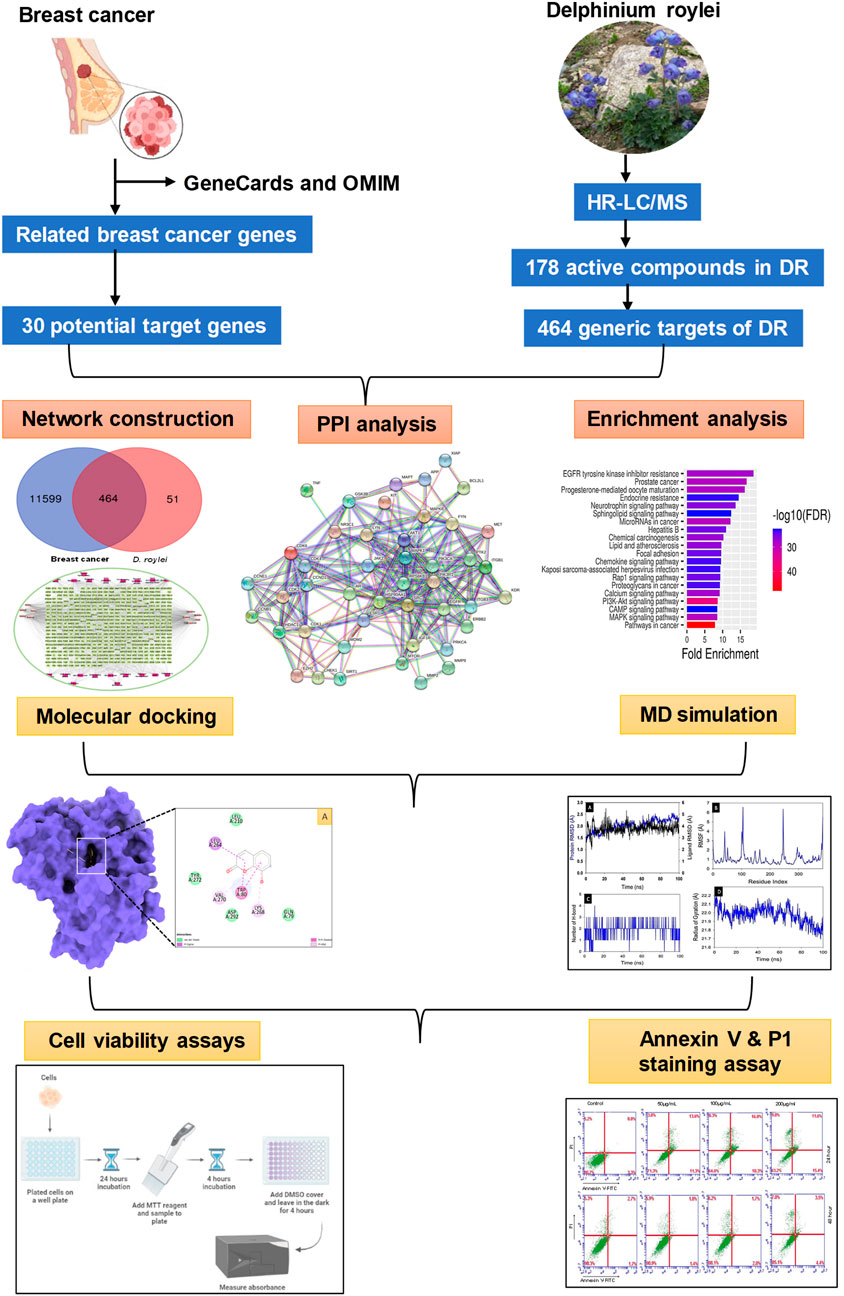
GRAPHICAL ABSTRACT | The graphical abstract illustrates a multidisciplinary approach to uncover the therapeutic promise of Delphinium roylei Munz, a medicinal plant, against breast carcinoma.
Introduction
Breast cancer (BC) is the most commonly diagnosed cancer worldwide, and its burden has been rising over the past decades (Pistelli et al., 2021; Pospelova et al., 2022). Having replaced lung cancer as the most commonly diagnosed cancer globally, breast cancer today accounts for 1 in 8 cancer diagnoses and a total of 2.3 million new cases in both sexes combined (Sung et al., 2021). Representing a quarter of all cancer cases in females, it was by far the most commonly diagnosed cancer in women in 2020, and its burden has been growing in many parts of the world, particularly in transitioning countries (Heer et al., 2020). An estimated 685,000 women died from breast cancer in 2020, corresponding to 16% or 1 in every 6 cancer deaths in women (Deo et al., 2022). The discovery of new cancer treatments is still regarded as an active research area despite the great advance in the area of chemotherapy. Medicinal plants are regarded as a renewable source of bioactive compounds that can be exploited in the treatment of various ailments including cancer (Tariq et al., 2021; Mir et al., 2022a; Mir et al., 2022b). Phytochemicals play an important role in the initiation, development, and advancement of carcinogenesis, as well as in suppressing or reversing the early stages of cancer or the invading potential of premalignant cells and also regulate cell proliferation and apoptosis signalling pathways in transformed cells (George et al., 2021; Bhat et al., 2022a; Bhat et al., 2022b).
Delphinium roylei Munz. is an important medicinal herb of the Delphinium genus. The ethnomedicinal uses of this plant include treating the liver and persistent lower back discomfort (Sharma and Singh, 1989; Kumar and Hamal, 2011). Diterpenoid alkaloid and flavanols, which constitute most of the compounds isolated from Delphinium plants, have been tested for various biological activities, such as effects on cholinesterase inhibition, antimicrobial, antineoplastic, insecticidal, anti-inflammatory, and anticancer activities (Yin et al., 2021). Delphinidin is a type of anthocyanin isolated from genus Delphinium, which has anticancer, anti-inflammatory, and anti-angiogenic properties. Recent in-vitro studies showed that delphinidin can inhibit the invasion of HER-2-positive MDA-MB-453 breast cancer cell line, with low cytotoxicity on normal breast cells (Wu et al., 2021) and ovarian cancer cells (Lim et al., 2017) and can also induce autophagy in breast cancer cells (Chen et al., 2018).
Due to the complex nature of plant extracts and their chemical constituents, it is difficult to understand the molecular mechanism by which they act on certain molecular targets due to the synergistic effects of their chemical constituents and the fact that they could interact with many targets simultaneously. In recent years, network pharmacological analysis has been effectively applied for prediction of the protein targets and the related disease pathways of plant active constituents.
Network pharmacology, based on system biology, includes network database retrieval, virtual computing, and high throughput omics data analysis. This approach breaks the ancient limitation of one drug–one biological target research and is applied mainly to explaining effective mechanisms, active ingredient screening, and pathogenesis research (Yu et al., 2021).
The versatile approach of molecular docking, which is based on the theory of ligand-receptor interactions, is widely used in drug discovery to understand how compounds bind with their molecular targets (Tiwari and Singh, 2022; Saifi et al., 2023).
Here, in the present study, we carried out a HR/LC-MS analysis to identify the phytoconstituents present in the D. roylei and screened to find drug-likeness compounds by ADMET analysis. Secondly, we used network pharmacology to predict the potential effective components, corresponding target genes, and pathways of phytocompounds of D. roylei against breast cancer. Lastly, we explore the molecular mechanism of the most potent bioactive constituent of D. roylei and a hub therapeutic target to alleviate the breast cancer based on molecular docking, molecular dynamic (MD) simulation and in vitro experimental analysis.
Materials and methods
Collection of plant material
The root parts of the Delphinium roylei plant were taken from high-altitude areas of Kashmir Himalaya. The collected samples were identified and confirmed by Akhtar Hussain Malik, Taxonomist, Department of Botany, the University of Kashmir, with voucher specimen No.2954-(KASH).
Extraction
Various solvents such as methanol, ethanol, ethyl acetate, and petroleum ether were selected as extraction solvents according to their polarity index to obtain plant extract using the Soxhlet equipment technique. About 200 g of the D. roylei plant material was powdered using a mechanical grinder after being cleaned with deionized water, dried in the shade for 15 days, pulverized with a mechanical grinder, and placed in an airtight container. The extracts were concentrated using a rotating vacuum evaporator after being filtered using Whatman no. 1 filter paper. They were then kept at 4°C for further use.
High resolution-liquid chromatography-mass spectrometry (HR/LC-MS)
The HR/LC-MS analysis of the ethanolic extract was carried out by a UHPLC-PDA-Detector Mass Spectrophotometer (HR/LC-MS 1290 Infinity UHPLC System), Agilent Technologies®, Santa Clara, CA, USA (Noumi et al., 2020). It consisted of a HiP sampler, binary gradient solvent pump, column compartment, and Quadrupole Time of Flight Mass Spectrometer (MS Q-TOF) with a dual Agilent Jet Stream Electrospray (AJS ES) ion source. A total of 1% formic acid was used as a solvent in deionized water (solvent A) and acetonitrile (solvent B). The flow rate of 0.350 mL/min was used, while MS detection was performed in MS Q-TOF. Compounds were identified via their mass spectra and their unique mass fragmentation patterns. Compound Discoverer 2.1, ChemSpider, and PubChem were the main tools for identifying the phytochemical constituents.
Network pharmacology-based analysis
Eight 8) compounds were identified according to UPLC-MS/MS analysis and were subjected to network pharmacology-based analysis. The identification of the target genes linked to the selected constituents was performed using the database STITCH DB (http://stitch.embl.de/,ver.5.0) and the obtained results were utilized for construction of compound-target (C-T) network using Cytoscape 3.5.1. Cytoscape combined score of interactions was adopted for judging the importance of nodes in each given network. Information about functional annotation and the pathways that were highly associated with the target proteins were retrieved from DAVID ver. 6.8 (Database for Annotation, Visualization and Integrated Discovery) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Relevant pathways with p-values <0.05 were selected. Target-pathway and constituent-Pathway networks were constructed to visualize the interactions between compounds, targets and cancer-related pathways.
In-silico drug-likeness and toxicity predictions
In silico drug-likeness and toxicity of top hit compounds in the database based on were carried out using the SwissADME web browser (http://www.swissdme.com) (Daina et al., 2017). Drug-likeness and toxicity filtering was based on Lipinski’s rule of five (Lipinski, 2004). For example, constituents with predicted oral bioavailability (OB) ≥30 were considered active. Constituents that satisfied less than three criteria were considered inactive.
Molecular docking
The ligand molecule (8-hydroxycoumarin) are retrieved in the form of 3D Standard Data Format from the PubChem database (3D SDF) (Bolton et al., 2011). The ligand was then converted from 3D SDF to Protein Data Bank (PDB) format using Avogadro software. The crystal structures of breast cancer target proteins (Akt1, SRC, EGFR, IL6, HSP90AA and ESR1) were retrieved from the Protein Data Bank. Biovia Discovery studio was used to eliminate undesirable bindings, ligand molecules, water molecules, and other contaminants from the macromolecule. After that polar hydrogens were added to the protein throughout the preparation process for improved interactions, followed by Kollman charges and other modifications. Molecular docking studies were performed between the target proteins of breast cancer and compounds of the selected plant using AutoDock version 4.2.1 (Trott and Olson, 2010). All other parameters were left at their default values. the grid box was generated centered at X = 14.09, Y = 15.47, Z = 15.48 with dimensions X:3.53, Y:0.58, Z: 12.41. The Lamarckian Genetic Algorithm (LGA) was used for docking studies on the protein and ligand complexes (Fuhrmann et al., 2010). The RMSD clustering maps were developed after the docking process was complete by re-clustering with the clustering tolerances of 1, 0.5, and 0.25 to identify the best cluster with the most populations and lowest energy score.
Molecular dynamics (MD) simulation study
MD simulations were performed using the Desmond 2020.1 from Schrodinger, LLC (Chow et al., 2008; Release, 2017) on the dock complex of Akt1 and 8-hydroxycoumarin ligand. This system used the OPLS4. force field (Mazurek et al., 2021) and an explicit solvent model containing TIP3P water molecules in a period boundary salvation box of 10 Å × 10 Å x 10 Å dimensions. The system was initially equilibrated to retrain over the protein-ligand complexes using an NVT ensemble for 10 ns. Following the preceding phase, an NPT ensemble was used to carry out the brief run of minimization and equilibration for 12 ns. The NPT ensemble was assembled using the Nose–Hoover chain coupling method and operated at 27°C for 1.0 ps under a pressure of 1 bar for the duration of the investigation. The time step was 2 fs. The Martyna–Tuckerman–Klein barostat method with a 02 ps relaxation time was adopted for pressure regulation. The radius for coulomb interactions was set at 9 nm, and long-range electrostatic interactions were calculated using Ewald’s particle mesh approach. With each trajectory, the bonded forces were estimated using the RESPA integrator for the time step of 2 fs. Root mean square fluctuation (RMSF), root mean square deviation (RMSD), solvent accessible surface area (SAS Area), and radius of gyration (Rg) were estimated to monitor the stability of molecular docking simulations (Rapaport and Rapaport, 2004).
In vitro antioxidant activity
1, 1-diphenyl-2-picrylhydrazyl radical scavenging activity
The capacity of the D. roylei extracts to scavenge the DPPH radicals were assessed by using the Gyamfi et al. method with slight modification (Obi et al., 2022). 0.5 mL of a test extract aliquot at various concentrations of 20–160 μg/mL in methanol was dissolved with 0.5 mL of a 100 mM DPPH solution. The resulting absorbance was measured at 517 nm after a 30-min incubation period in complete darkness and at room temperature. The following formula was used to compute the percentage inhibition:
Hydroxyl radical scavenging activity
Hydroxyl radical scavenging activity was determined by Elizabeth and Rao with a bit of modification (Nwakaego et al., 2019). The assay measures the 2-deoxyribose breakdown product by condensing it with Thiobarbituric acid. Hydroxyl radicals are produced by the Fenton reaction, which includes a ferric ions-EDTA-ascorbic acid-H2O2system. The reaction mixture contains these above components and different plant extract concentrations (10–80 μg/mL). 0.5 mL of the reaction mixture was dissolved in 1 mL of 2.8 percent of TCA after 1-h incubation at 37°C, then 1 mL of 1 percent aqueous TBA was poured, and the mixture was then incubated for 15 min at 90°C to develop the color. The absorbance was calculated at 532 nm. Butylated hydroxytoluene (BHT) was used as standard.
Reducing power
The assay was carried out using Oyaizu’s method (Zhang et al., 2021). This method estimated the reduction of Fe3+-Fe2+ by measuring the absorbance of Pearl’s Prussian blue complex. This procedure relies on the stoichiometric reduction of (Fe3+) ferricyanide relative to antioxidants. Various concentrations of the plant extracts (1–200 μg/mL) were added to 2.5 mL of 1% potassium ferricyanide [K3Fe (CN)6] and 2.5 mL of 0.2 M phosphate buffer with a pH of 6.6. Then 2.5 mL of 10% trichloroacetic acid was added to the mixture after 20 min of incubation at 50°C, and the mixture was then centrifuged at 3,000 rpm for 10 min. The upper layer (2.5 mL) was dissolved in 2.5 mL of distilled water and 0.5 mL of 0.1% FeCl3, and the absorbance was determined at 700 nm. Rutin was taken as a positive antioxidant, and the reducing power was calculated according to the absorbance values.
Superoxide radical scavenging activity
This assay was based on the extract’s capacity to reduce formazan production by scavenging the radicals produced by the riboflavin-NBT system (Jaganathan et al., 2018). The reaction mixture consists of 20 μg riboflavin, 50 mM phosphate buffer with a pH of 7.6, 0.1 mg/3mL NBT, and 12 mM EDTA. The reaction was initiated by illuminating the above reaction mixture with various concentrations of plant extracts (10–80 μg/mL) for 90 s. Then the absorbance was estimated at 590 nm. BHT was used standard antioxidant. The %age of scavenging of superoxide anion s was calculated using the equation.
Where AS is the absorbance of the test sample, and AC is the absorbance of the control used.
In vitro anticancer activity
Cell culture
MCF-7, MDA-MB-231, and MDA-MB-468 cell lines were purchased from the National Centre for Cell Science (NCCS) in Pune. Prof. Annapoorni Rangarajan, IISC, Bangalore, kindly supplied the 4T1 cell line. Cells were grown in high glucose media DMEM using 10% FBS and 1% penicillin/streptomycin. The cells were cultured in a CO2 incubator (5%) at 37°C.
Cytotoxicity assay
The MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was utilized to determine the cell cytotoxicity (Rezadoost et al., 2019). Breast cancer cells were seeded in 96 well plates with the cell number of 3 × 103 cells in each well and allowed to adhere overnight. To prepare the stock solution, From the stock solution 10 mg/mL, various concentrations (12.5–400 μg/mL) of various extracts of Delphinium roylei were obtained in fresh media. Then the breast cancer cells (MCF-7, MDA-MB-231, MDA-MB-468 and 4T1) were treated with extracts of Delphinium roylei for 72 h and the plate system was placed into the incubator. MTT solution was applied to every well after incubation. The plate was kept in the incubator at 37° C for 4 h under dim lighting. The supernatant was removed after 4 h, and 100 μL of DMSO was added to the purple formazan to dilute it. The ELISA plate reader was used to read the plate at 595 nm. The percentage of cell cytotoxicity was determined using an optical density.
Annexin V/PI apoptosis detection
We used a BD Biosciences Annexin V apoptosis detection kit to examine the mechanism behind the anticancer effect of D. roylei ethyl acetate extract. MDA-MB-231 was treated with acetate extract for 24 and 48 h. As instructed by the manufacturer, adherent and free-floating cells were all collected and stained with the fluorescent dyes FITC-Annexin V and PI. Flow cytometry was performed at the Department of Biotechnology, National Institute of Technology, Rourkela, Odisha, India, on a BD Accuri™ C6 flow cytometer (Mehraj et al., 2022)
Results
Quantification of chemical components of HR/LC-MS
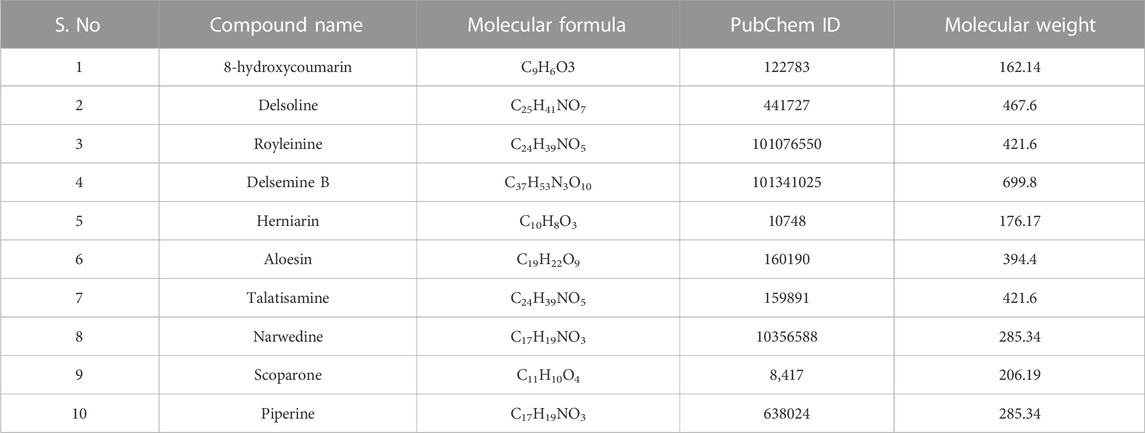
In our previous investigation, 168 phytocompounds were tentatively identified using the chromatography-mass spectrometry (LC/MS) approach in both negative and positive ionization modes (Mir et al., 2022b). A few more compounds were identified by performing the advanced technique like HR-LC/MS) and chromatograms in TOF MS ES + shown in (Figures 1A–D). Figure S1 demonstrates the total ion chromatogram (TIC) of the studied plant’s methanolic, ethanolic, ethyl acetate, and petroleum ether extracts. The examples of a few phytoconstituents identified through HR-LC/MS are 8-hydroxycoumarin, Delsoline, Royleinine, Delsemine B, Herniarin, Aloesin, Talatisamine, Narwedine, Scoparone, and Piperine in D. roylei extracts by contrasting their output mass data and retention time relatives to external standards with a reference database and already published data using different software’s. Some identified phytocompounds from chromatograms that possess important pharmacological properties are listed in Table 1. These nine compounds were subjected to network pharmacology, in silico docking, and MD simulations analysis. These phytocompounds were identified as chemical markers and listed as potential candidates for further network pharmacology analysis.
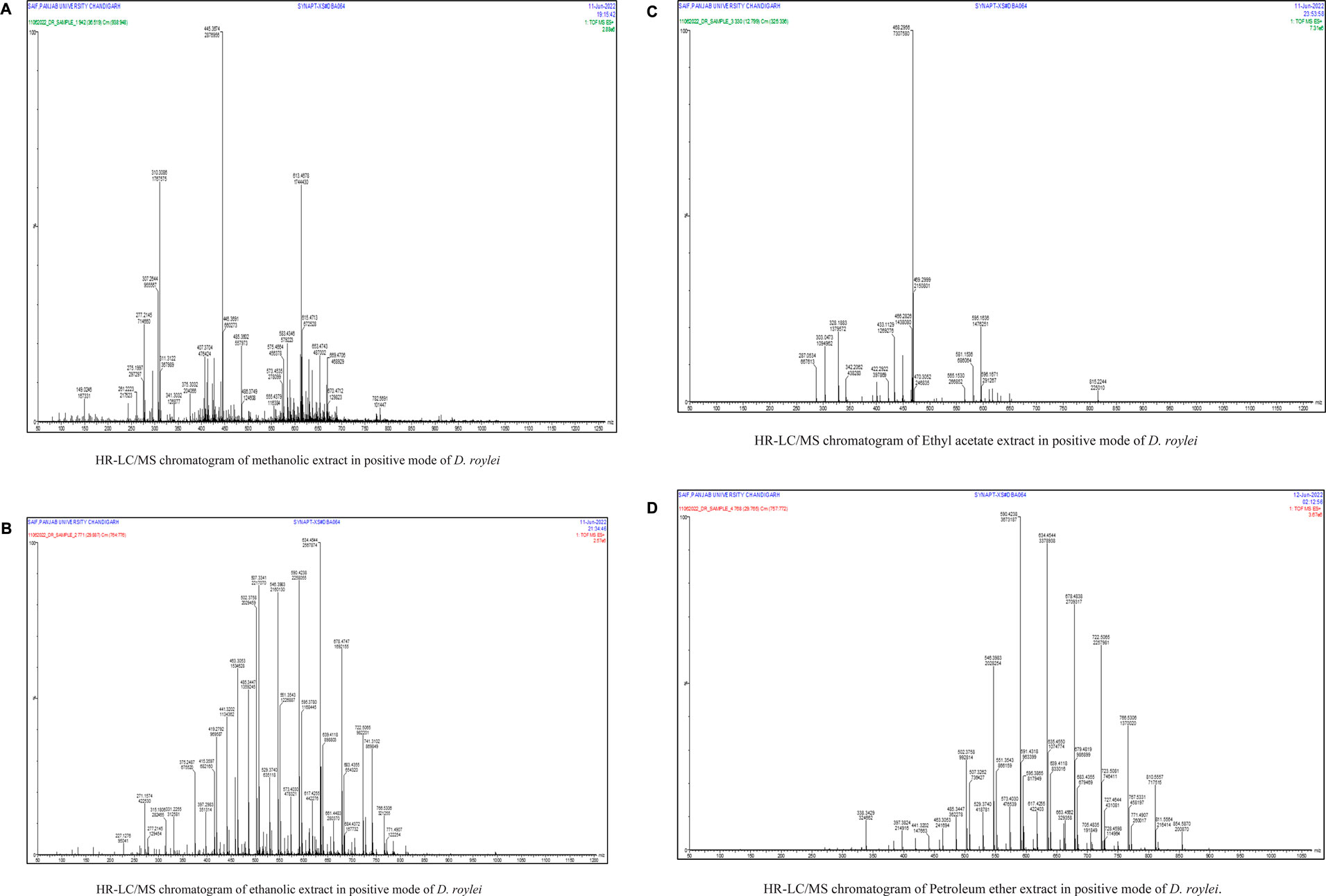
FIGURE 1. (Continued). (A). HR-LC/MS chromatogram of methanolic extract in positive mode of D. roylei (B) HR-LC/MS chromatogram of ethanolic extract in positive mode of D. roylei (C) HR-LC/MS chromatogram of Ethyl acetate extract in positive mode of D. roylei (D) HR-LC/MS chromatogram of Petroleum ether extract in positive mode of D. roylei.
Network pharmacology
Target gene screening and interaction network construction
Total of 514 potential target genes were obtained for the 10 quantitative components of HR/LC-MS (shown in Figure 2). Meanwhile, 12063 disease target genes associated with breast cancer were retrieved using GeneCards, OMIM and DisGeNET platforms. 464 shared common target genes were identified between the quantitative components of HR/LC-MS and breast cancer. All 10 components in D. roylei, namely, 8-hydroxycoumarin, Delsoline, Royleinine, Delsemine B, Herniarin, Aloesin, Talatisamine, Narwedine, Scoparone, Piperine in D. roylei were targeted for further analysis. The common target genes PPI diagram indicated that there were 464 nodes and 6,035 edges in PPI (Figure 3A). The frequency of occurrence of the top 30 common target genes was shown in Figure 3B. Akt1, SRC, EGFR, IL6, HSP90AA, and other target genes exhibited a high frequency of protein interaction, which may be the node protein of the whole network. The results showed that the selected components of D. roylei had a high binding activity with these target proteins and could be used as the potential target genes of HR/LC-MS for treating breast cancer.
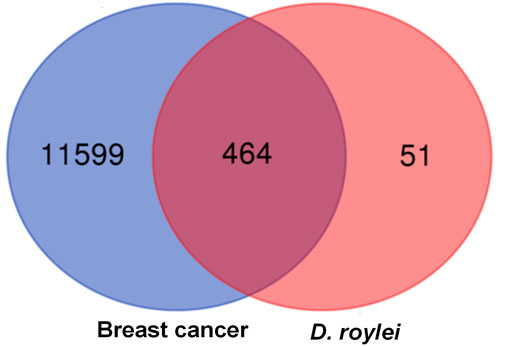
FIGURE 2. Venn diagram represents common target genes for compound prescription and disease. The size denotes the number of the target genes, the blue circle symbolizes the target genes of breast cancer, the red circle represents the target genes of 10 quantitative components in HR/LC-MS, and the coincident part depicts the common target genes.
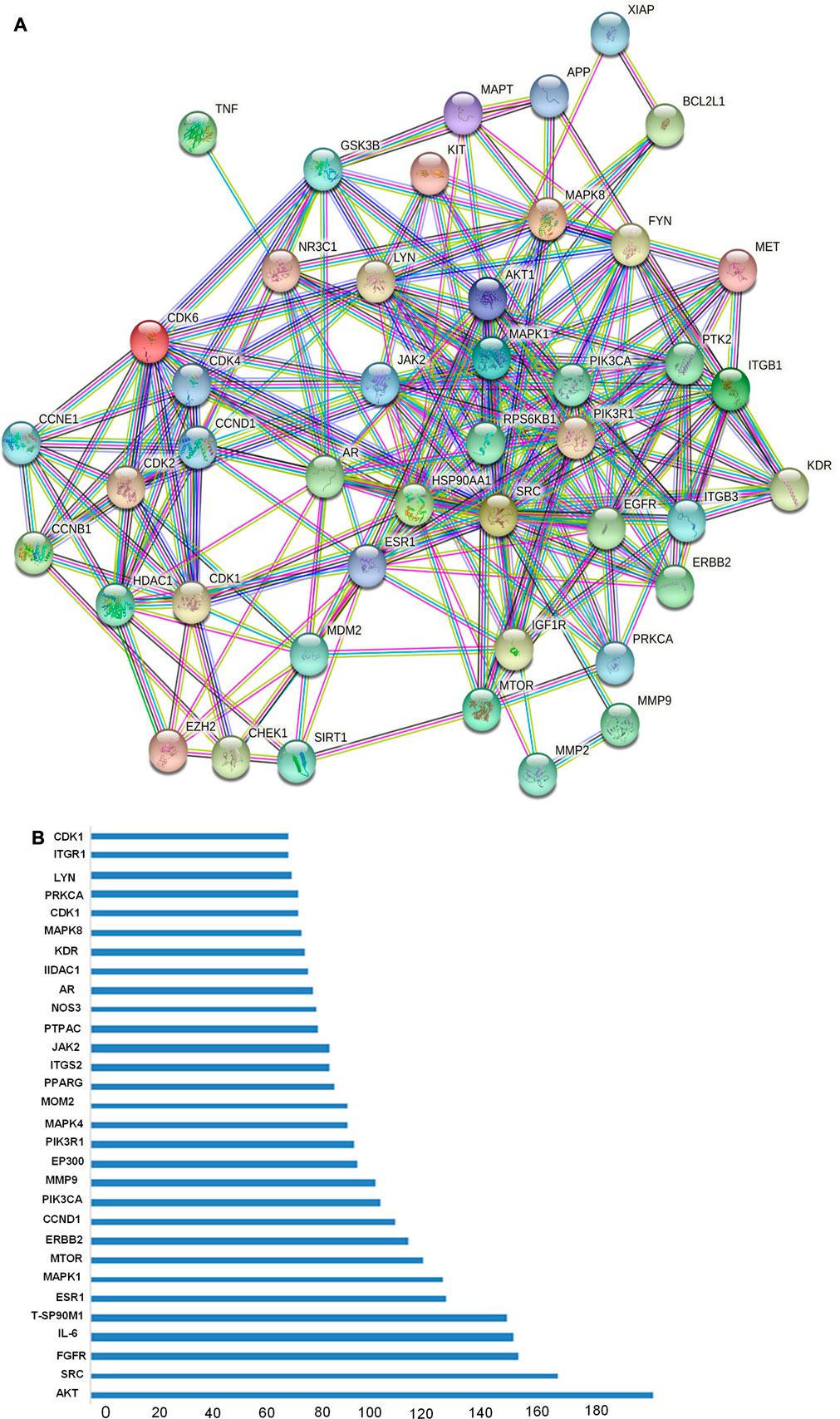
FIGURE 3. (Continued). The common target genes network interaction results (A) PPI network of the common target genes. The nodes represent target genes; the stuffing of the nodes represents the 3D structure of target genes; the edges represent target genes-target genes associations; the colors of the edges represent different interactions; cyan and purple represent known interactions; green, red, and blue-purple represent predicted interactions; chartreuse, black, and light blue represent others.
Screening of key pathways of HR-LC/MS for treating breast cancer
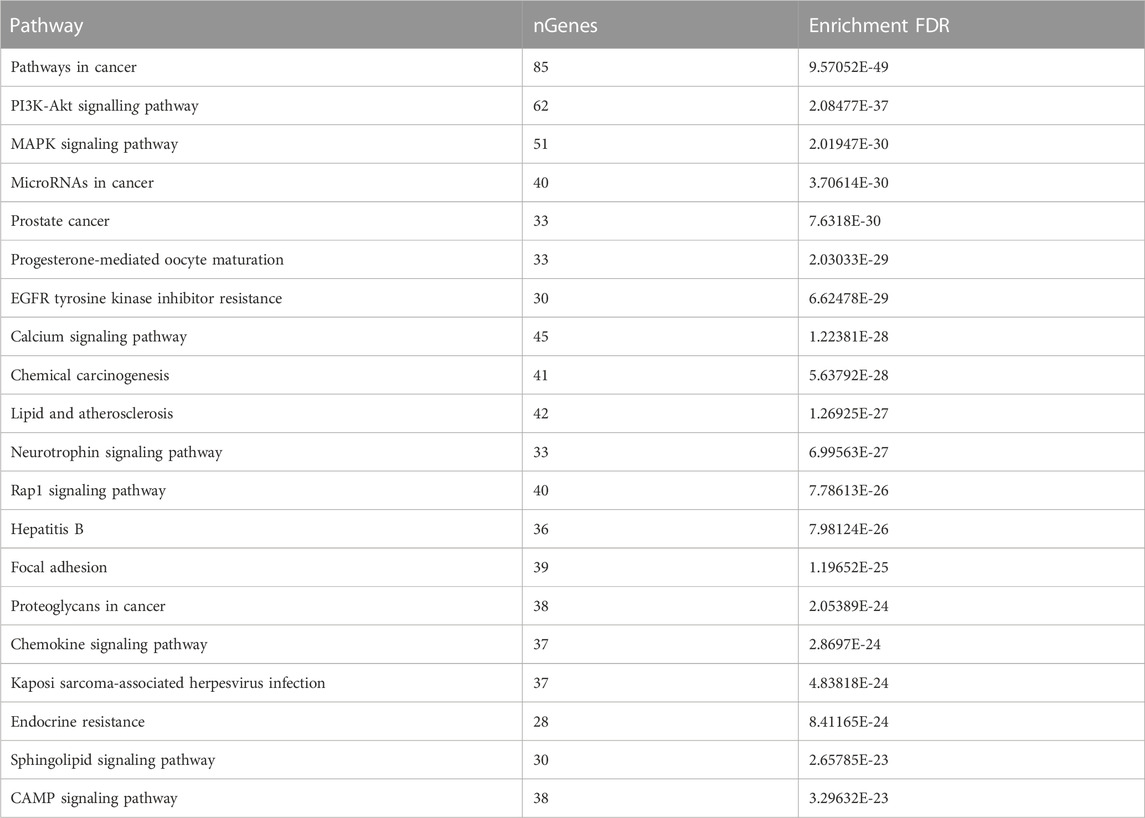
GO analysis of the common target genes showed that the biological process was mainly involved in Protein autophosphorylation, peptidyl-tyrosine modification, and pos. neg. of phosphorylation (Figure 4) and also given as S-2 to S-4. The molecular functions non-membrane spanning protein tyrosine kinase activity and protein serine/threonine/tyrosine kinase activity were leading. The cyclin-dependent protein kinase holoenzyme complex and protein-kinase complex were observed in the cellular component. KEGG pathway enrichment analysis of the aforementioned common target genes is shown in Figure 5 and also given as S-5. After the exclusion of broad pathways, the top 20 signalling pathways are listed in Table 2. This suggested that the effect of HR-LC/MS for treating breast cancer may act on multiple pathways, as well as complex interactions among these pathways.
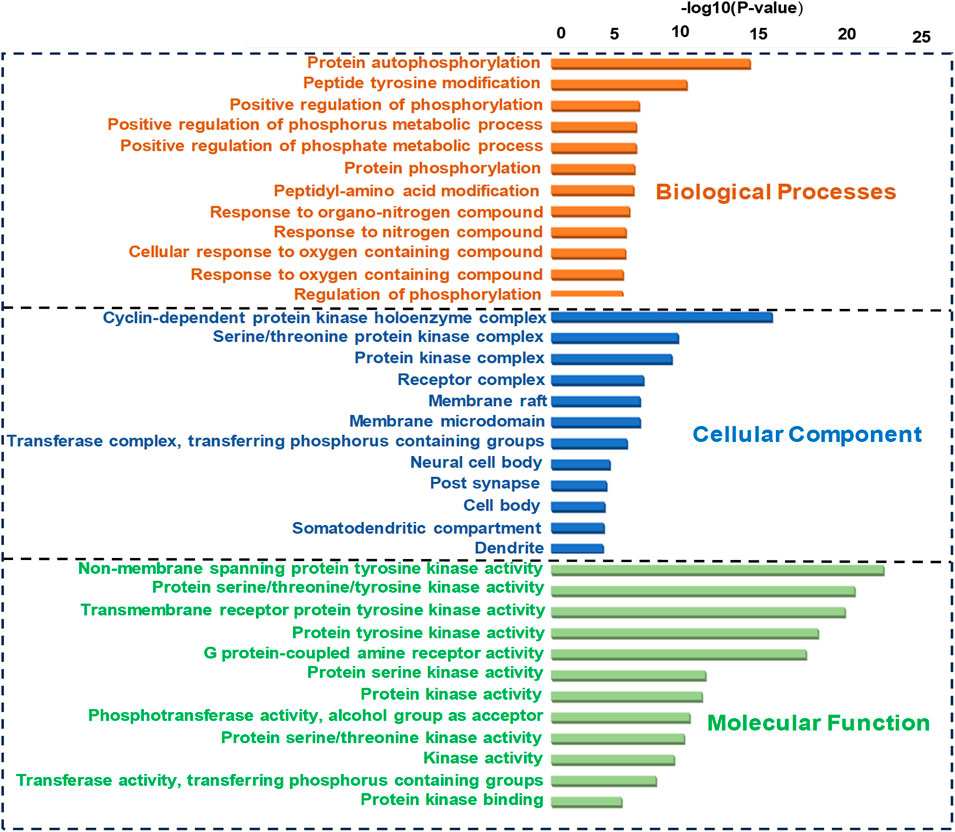
FIGURE 4. GO (Biological process, molecular function, and cellular component) analysis (top 20). The node length represents the number of target genes enriched, and the node color from blue to red represents the p-value from large to small.
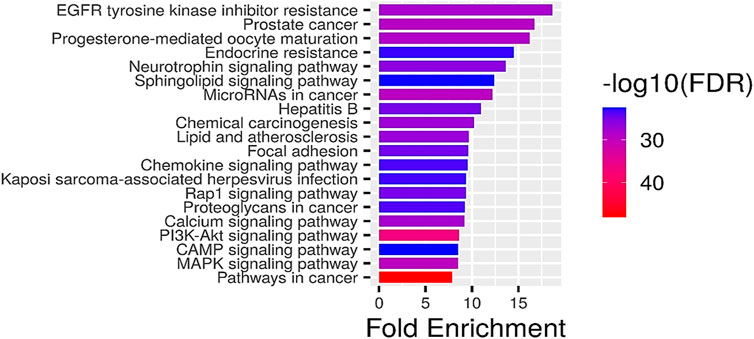
FIGURE 5. KEGG pathway enrichment analysis (top 20). The node size represents the number of target genes enriched, and the node color from blue to red represents the p-value from large to small.
Compound prescription-active component-disease target gene-pathway interaction network
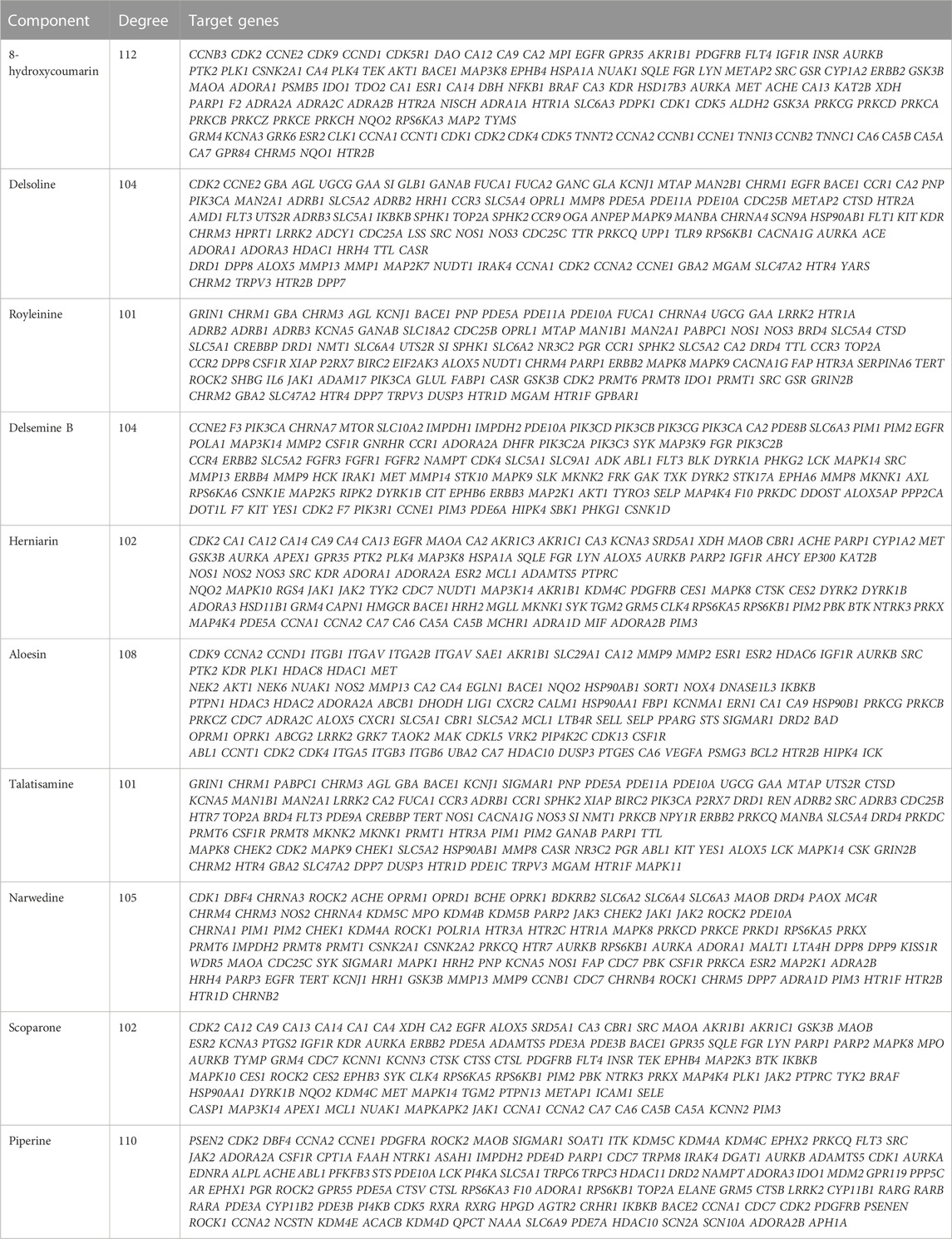
Compound prescription-active component-disease-target gene-pathway interaction network finding is shown in Figure 6. The network contained 474 nodes (464 target genes, 10 active components. Besides, the interaction network results of 10 active compounds are shown in Table 3. The degree of 8-hydroxycoumarin, Delsoline, Royleinine, Delsemine B, Herniarin, Aloesin, Talatisamine, Narwedine, Scoparone, and Piperine were 112, 104, 101, 104, 102, 108, 101, 105, 102 and 110 respectively. The results above show that quality markers in LC-MS may act on the whole biological network system rather than on a single target gene.

FIGURE 6. Compound prescription-active component-disease-target gene. The node colors represent different groups: purple represents the components, and blue represents target genes. Component node size represents the degree; low to high degree represents the node size from small to large.
Molecular docking studies
All the binding energy scores are calculated from the best cluster (95%) that falls within the lowest RMSD 0.25 Å. With the lowest binding energy (ΔG–9.2 kcal/mol) and inhibitory concentration, Ki (1.14 µM), 8-hydroxycoumarin showed a considerable binding affinity for the Akt1 (Figure 7A). During the interaction of the ligand 8-hydroxycoumarin, Trp80 is involved in pi-pi stacking, and Lys268 and Val270 residues are involved in pi-alkyl interaction at the binding cavity of the protein. Besides hydrogen bonding, Leu264 is involved in pi-sigma interaction, and the rest residues are van der Waal’s interactions by amino acid residues formed weak non-bonded interaction with the ligand (Figure 7A and right panel). Molecular docking studies were performed to verify the affinity of target protein(s) and bioactive phytocompounds. The 3D interactions of 8-Hydroxycoumarin with various target proteins; SRC, EGFR, IL-6, Hsp90aa1 and ESR-1 and 2D structure of 8-Hydroxycoumarin interacted with respective amino acids respectively are shown in Figures 7B–F. Table 4 depicts the docking and scores of the D. roylei compounds; 8-hydroxycoumarin, Delsoline, Royleinine, Delsemine B, Herniarin, Aloesin, Talatisamine, Narwedine, Scoparone, and Piperine against the active sites of the identified protein targets, Akt1, SRC, EGFR, IL-6, Hsp90aa1and ESR-1 performed using the AutoDock Vina software.
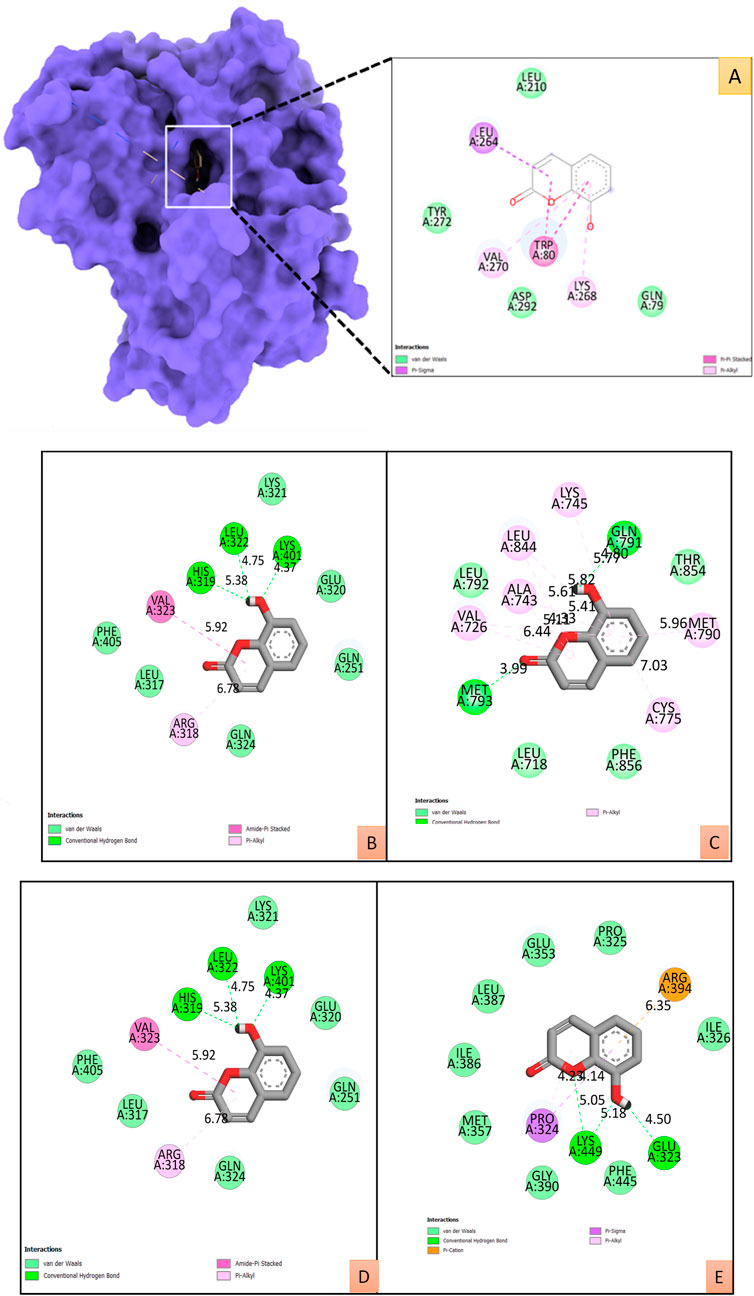
FIGURE 7. (A). The surface view of the best pose of the Akt1 (PDB ID:4EKL) +8-Hydroxycoumarin complex displays the surface view on the left panel and the 2D interaction profile of the ligand with binding cavity residues. B-E: 2D and 3D interactions of 8-Hydroxycoumarin with various target proteins (B). SRC (PDB ID:2H8H), (C). EGFR (PDB ID: 6DUK), (D). IL-6 (PDB ID:1P9M), (E). ESR-1 (PDB ID: 1GWQ) with respective amino acids respectively.
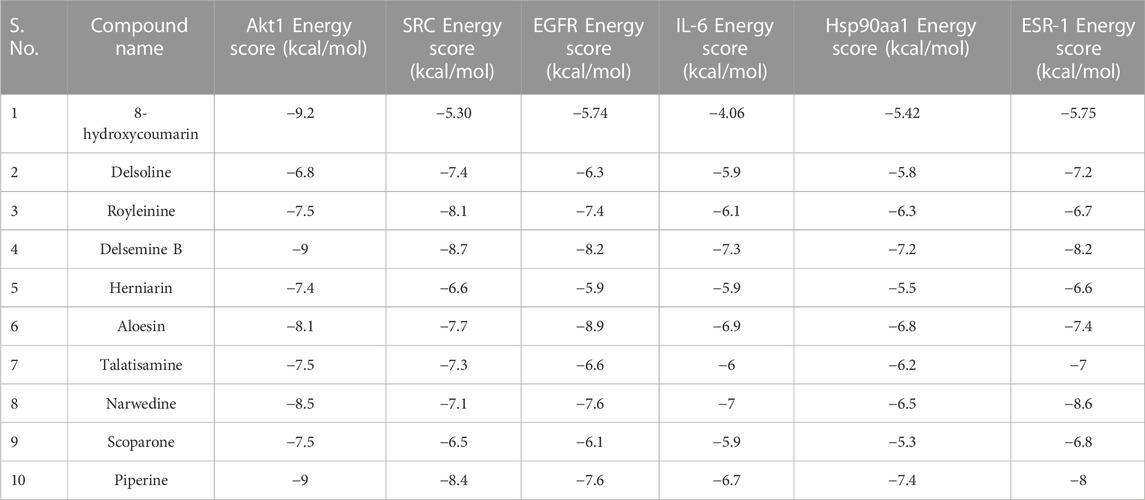
TABLE 4. Binding energy obtained from the docking calculations of bioactive phytoconstituents with target proteins.
Drug-likeness prediction of D. roylei phytoconstituents
The compounds retrieved from PubChem were assessed for Lipinski’s Rule of 5, with drug-likeliness properties. Furthermore, ADMET evaluation was applied and was selected for molecular docking to determine the binding affinity with protein at the active site. Almost all the nine phytoconstituents accept Lipinski’s rule with few limitations, as shown in Table 5.
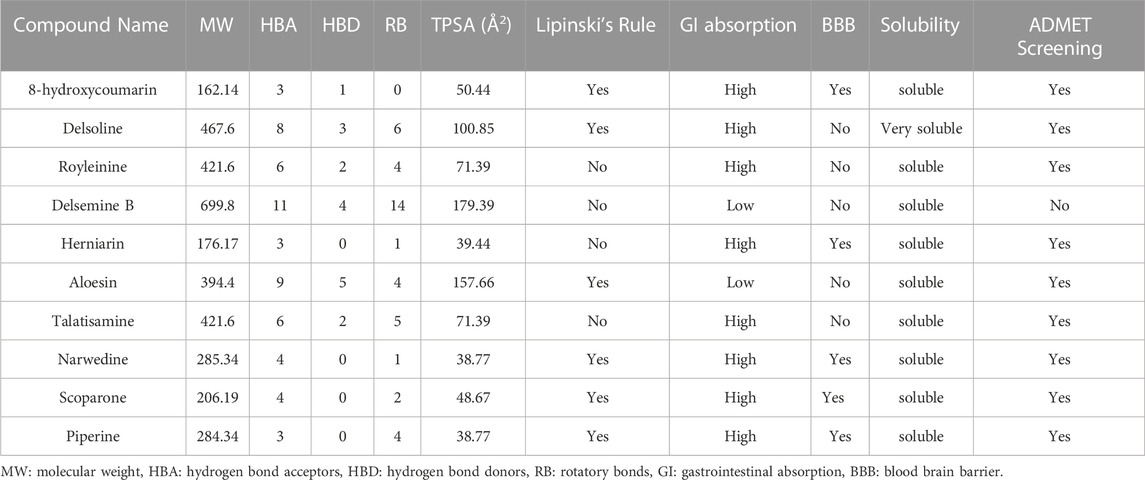
TABLE 5. Drug-likeness prediction of D. roylei phytoconstituents by ADMET evaluation using SwissADME Software.
Molecular dynamic (MD) simulation study
Studies using molecular dynamics and simulation (MD) were conducted to determine the convergence and stability of the Akt1+8-Hydroxycoumarin complex. Comparing the root mean square deviation (RMSD) values, the simulation of 100 ns showed stable conformation. The RMSD of the Cα-backbone of Akt1 bound to 8-Hydroxycoumarin showed a deviation of 2.1 Å (Figure 8A), while the ligand showed an RMSD deviation of 3.8.0 Å. Stable RMSD plots indicate good convergence and stable conformations throughout the simulation. As a result, it may be inferred that 8-Hydroxycoumarin bound with Akt1 is quite stable in complex due to the ligand’s increased affinity. The plot for root mean square fluctuations (RMSF) indicates the residual fluctuations due to conformational variations in different secondary structures. Here RMSF plot displayed fluctuating residues while high fluctuations were observed among 50–60, 110–130, and 240–260 residual positions (Figure 8B). The highest fluctuating peaks comprised of 3.8 Å to 6.7–6.8 Å, might be due to higher ordered flexibility conforming into loops (Figure 2B). Therefore, the protein Akt1 has significant flexibility to conform to specific secondary structures to accommodate the ligand. The radius of gyration (Rg) quantifies how compact a protein is with a ligand molecule (Miu et al., 2008). In this investigation, the Akt1 Cα -backbone linked to 8-hydroxycourmarin demonstrated a stable radius of gyration (Rg) values between 22.1 and 21.8 Å, hence there are no sudden changes in radius of gyration as shown in (Figure 8D). This steady values of Rg suggest that despite the structural changes caused by the compounds, the protein remains folded. Significant stable gyration (Rg) suggests that the protein is oriented in a highly compact manner when it is attached to a ligand. The quantity of hydrogen bonds between the ligand and protein indicates the complex’s interaction’s complex stability and depth. During the 100 ns of the simulation, there were significant numbers of hydrogen bonds between Akt1 and 8-hydroxycourmarin (Figure 8C). Between Akt1 and 8-hydroxycourmarin-ligand, the average number of hydrogen bonds is two. The overall analysis of Rg indicates that the binding of the various ligands causes the corresponding proteins to become less flexible and more compact.
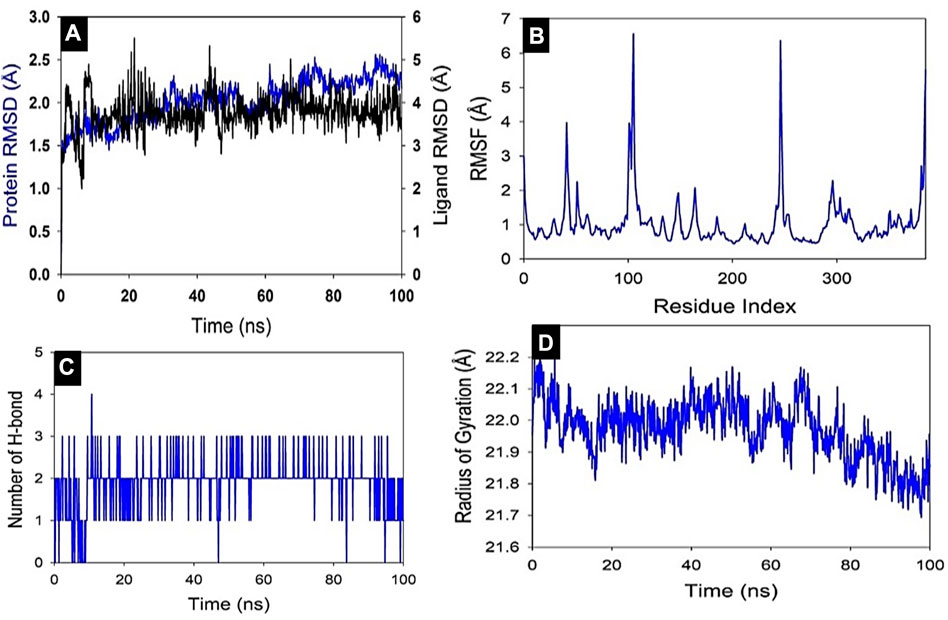
FIGURE 8. MD simulation analysis of 100 ns trajectories of (A) Cα backbone of Akt1 bound to 8-Hydroxycoumarin ligand, (B) RMSF of Cα backbone of Akt1 bound to 8-Hydroxycoumarin-ligand (C) Radius of gyration (Rg) of Cα backbone of Akt1 bound to 8-Hydroxycoumarin (D) Formation of hydrogen bonds in Akt1 bound to 8-Hydroxycoumarin.
Molecular mechanics generalized born surface area (MM-GBSA) calculations
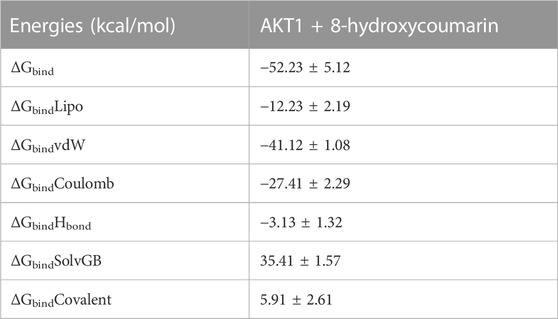
Utilizing the MD simulation trajectory, the binding free energy and other contributing energy in the form of MM-GBSA is determined for Akt1 bound to 8-Hydroxycoumarin complexes. The results (Table 6) suggested that the maximum contribution to ΔGbind in the stability of the simulated complexes was due to ΔGbindCoulomb, ΔGbindvdW, ΔGbindHbond, and ΔGbindLipo, while ΔGbindCovalent and ΔGbindSolvGB contributed to the instability of the corresponding complex. Akt1 bound to 8-Hydroxycoumarin complex has significantly higher binding free energies dGbind = −52.23 ± 5.12 kcal/mol (Table 6). These results supported the potential of AKT1 bound to 8-Hydroxycoumarin having a high affinity of binding to the protein, efficiency in binding to the selected protein, and the ability to form stable protein-ligand complexes (Balasubramaniam et al., 2021).
In vitro antioxidant activity
DPPH assay
All the extracts showed different levels of DPPH radical scavenging activity over the range of 20–160 μg/mL concentration, as shown in Figure 9A. The methanolic extract exhibited the strongest DPPH radical scavenging activity compared to other extracts. The extract’s radical scavenging activity was effective in the order Methanolic > ethanol > Ethyl acetate > Petroleum ether. A maximum of 82.46% ± 0.2% radical scavenging potential was observed at 200 μg/mL of methanolic extract used, whereas for ascorbic acid, the scavenging activity was 89.57% ± 0.25%. A minimum of 66.35% ± 0.32% scavenging potential was observed at 200 μg/mL of petroleum ether extract. Standards and all the extracts showed a dose-dependent inhibition of the DPPH radicals.
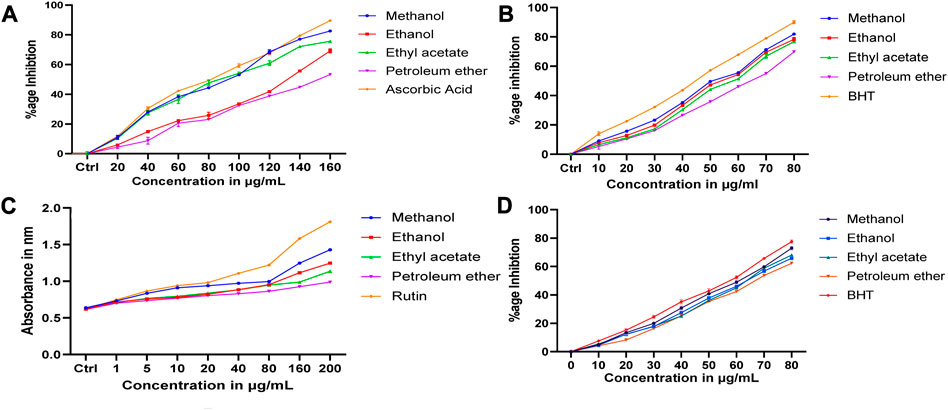
FIGURE 9. Antioxidant results of D. roylei (A). DPPH radical scavenging assay (B). Hydroxyl radical scavenging assay (C). Reducing power assay, and (D). Superoxide radical scavenging assay.
Hydroxyl radical scavenging assay
In this assay, results showed that the methanolic extract of D. roylei had the highest potential to scavenge hydroxyl radicals than the ethanolic extracts, ethyl acetate, and petroleum ether, as shown in Figure 9B. At a concentration of 80 μg/mL, the methanolic, ethyl acetate, ethanolic, and petroleum ether extract showed the maximum scavenging effect of 81.92% ± 0.49%, 78.72% ± 0.8%, 73% ± 0.7% and 69.88% ± 0.55% inhibition on hydroxyl radicals. Butylated hydroxytoluene (BHT) taken as a control had shown a more scavenging effect (90.07% ± 1%) than plant extracts.
Reducing power assay
As illustrated in Figure 9C, Fe3+ was transformed to Fe2+ in the presence of D. roylei extract and the reference compound Rutin to measure the reductive capability. The reducing power increased with an increase in the concentration of plant extracts. The methanolic extract of D. roylei showed significant reducing power when compared with standard Rutin. The reducing power demonstrated by the methanolic extract of the plant was 1.429 ± 0.005 at the concentration of 200 μg/ml as compared to 1.811 ± 0.0035 shown by standard Rutin at the same concentration. Ethanolic, ethyl acetate, and petroleum ether extracts showed less reducing power (1.246 ± 0.0025, 1.136 ± 003, and 0.987 ± 0.005, respectively) compared to methanolic extracts at 200 μg/mL concentration.
Superoxide radical scavenging (SARS) assay
All the test extracts exhibited effective O2−* scavenging activity in a concentration-dependent manner (10–80 μg/mL), as shown in Figure 9D. The highest activity (Scavenging effect of 72.91 ± 0.76 was shown by methanolic extracts of D. roylei at a concentration of 80 μg/mL, followed by ethyl acetate, ethanolic, and petroleum ether extracts with a scavenging effect of 68.125% ± 0.45%, 65.84% ± 0.41% and 62.27% ± 0.5% respectively, which is least as compared to methanolic extract and standard Rutin. Standard Rutin showed the highest scavenging potential of 77.46% ± 0.7% showed the highest potential at 80 μg/mL concentration as compared to all four extracts of the plant.
In vitro anticancer activity
MTT results
Figures 10A,B shows the cell viability (%) of various breast cancer cell lines; MDA-MB-231, MCF-7, MDA-MB-468, and 4T1 when treated with different concentrations of methanolic, ethanolic, ethyl acetate, and petroleum ether extracts of D. roylei and doxorubicin drug. The IC50 values of four different extracts and positive control against various breast cancer cell lines are shown in Table 7. The ethyl acetate extract of the plant showed a maximum cytotoxic effect on MDA-MB-231 with an IC50 value of 116.7 μg/mL, followed by methanolic, petroleum ether, and ethanolic extract with IC50 values of 274.1, 317.1, and 549.3 μg/mL respectively. The Methanolic extract of the plant showed maximum reduction in the growth of MD-MB-468 breast cancer cell line with the lowest IC50 value of 166.3 μg/mL, followed by ethyl acetate, ethanolic, and petroleum ether extract with IC50 values of 296.1, 335.5 and 415.5 μg/mL respectively. The ethyl acetate extract showed maximum anti-proliferative potential against the 4T1 cell line with an IC50 value of 157.9 μg/mL. The ethanolic, petroleum ether and methanolic extracts have shown less anti-proliferative potential against 4T1with IC50 values of 420.7, 598.9, and 689.4 μg/mL, respectively. The ethyl acetate extract is highly specific to MCF-7 cell lines with an IC50 value of 125.5 μg/mL, followed by methanolic, ethanolic, and petroleum ether extracts with IC50 values of 213, 299.3, and 498.6 μg/mL, respectively. The IC50 values of standard doxorubicin are shown in Table 4. Hence ethyl acetate extract of D. roylei showed the highest anticancer activity against the MDA-MB-231, MCF-7 and 4T1 cell lines compared to the other three extracts.
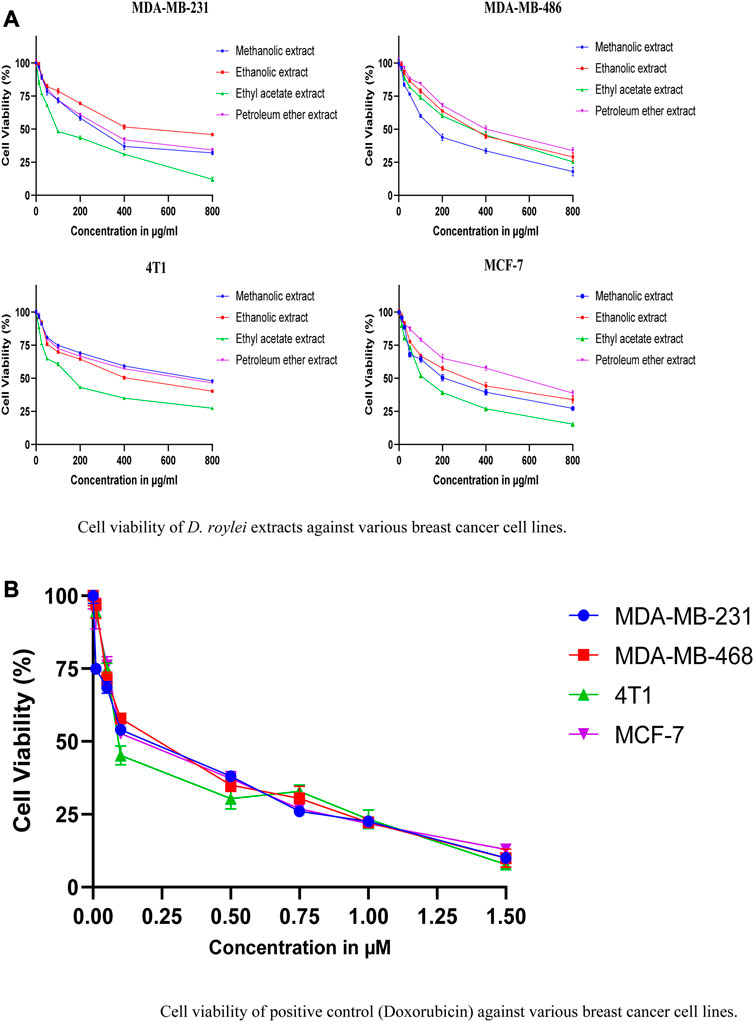
FIGURE 10. (A). Cell viability of D. roylei extracts against various breast cancer cell lines. (B) Cell viability of positive control (Doxorubicin) against various breast cancer cell lines.
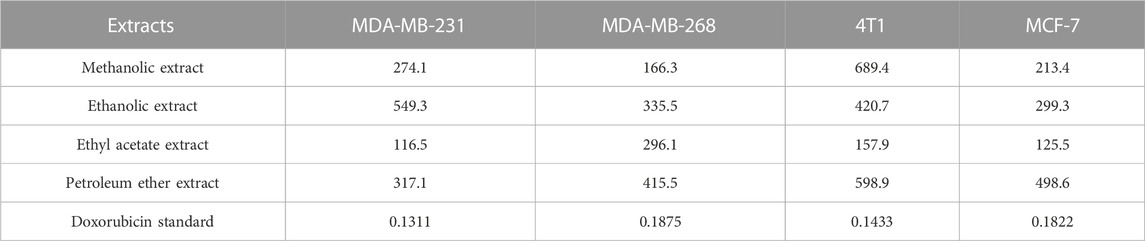
TABLE 7. IC50 values in μg/mL of various extracts of D. roylei against different breast cancer cell lines.
Furthermore, we utilized Annexin V and PI staining to assess the apoptosis induction potential of ethyl acetate extract of D. roylei. MDA-MB-231 cells were treated for 24 and 48 h, followed by staining with Annexin V and PI. Flow cytometry analysis revealed that plant extract induction tumour cell death via induction of apoptosis and apoptosis enhanced significantly upon higher concentration are shown in Figure 11.
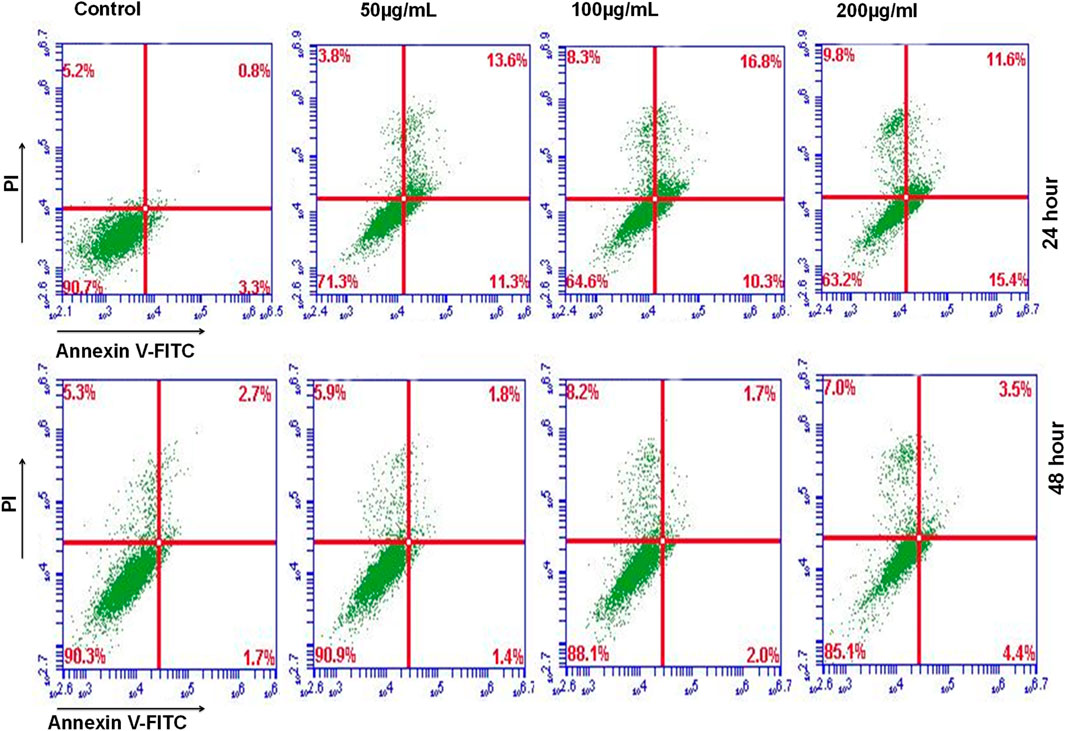
FIGURE 11. Annexin V & P1 staining showed increased apoptotic cells in plates treated with the ethyl acetate extract of D. roylei after 24 h or 48 h periods.
Discussion
In the current study, many secondary metabolites were identified using HR-LC/MS of D. roylei, out of which few phytocompounds were selected to propose a possible mechanism against breast cancer treatment using network pharmacology, molecular docking, molecular dynamic simulation and in vitro studies. The network pharmacology analysis suggested that the therapeutic efficacy of the D. roylei phytoconstituents against breast cancer was mainly associated with 20 signalling pathways, 30 potential target genes, and 10 bioactives. Through the network pharmacology, we identified the most significant protein (Akt1) associated with the occurrence and development of cancer and a bioactive 8-hydroxycoumarin (8-hydroxychromen-2-one) from the D. roylei. We identified a hub signaling pathway (PI3K-Akt signaling pathway, indicating the lowest rich factor among 20 signaling pathways. Akt1 kinase is a protein made according to instructions from the Akt1 gene (Lv et al., 2020). This protein is present in many different cell types throughout the body and plays a crucial role in numerous signaling pathways. For instance, Akt1 kinase is vital in controlling cell survival, apoptosis, proliferation, and differentiation (Jafari et al., 2019). Recent research has demonstrated that the PI3K/Akt signalling pathways, which play a role in the above-mentioned processes, are frequently disrupted in various human malignancies (Xu et al., 2019). This pathway is crucial for tumor growth and potential responsiveness to cancer treatments.
Many new targeted agents have been created, especially to target P13K/Akt-related targets. Therefore, having a better understanding of the PI3K/Akt signaling pathway may help to enhance the oncologist’s accuracy of prediction as to response to treatment.
Based on the degree value of compounds in the network, we obtained the 8-hydroxycoumarin compound as the most active ingredient of D. roylei. Previous studies have revealed that 7-and 4-hydroxycoumarin and its derivatives have numerous therapeutic benefits (Gaber et al., 2021). These are employed as drug intermediates and as antitumor drugs, anti-inflammatory, anti-HIV, antimicrobial, anti-coagulant, antioxidant, and anti-viral agents (Gaber et al., 2021). However, the degree of other compounds was also high, indicating that quality markers may affect the entire biological network system instead of only one target gene.
Furthermore, the KEGG pathway enrichment analysis of 30 targets suggested that 20 top signaling pathways were involved in breast cancer occurrence and development. This indicated that the effect of phytocompounds acts on multiple pathways for treating breast cancer and complex interactions among these pathways. Based on the frequency of each gene in the compound-gene network, Akt1 showed the highest frequency of protein interaction, followed by SRC, MAPK3, EGFR, IL-6, HSP90AA1, ESR-1, and other target genes.
Furthermore, an in silico docking analysis of the nine most prevalent compounds was carried out against the 30 potential targets using AutoDock version 4.2.6 software (Morris et al., 2008). All tested compounds showed promising results against Akt1 protein based on docking scores. Among the all, 8-hydroxycoumarin bioactive showed the highest energy score with Akt1. 8-hydroxycoumarin fits comfortably into the binding sites on Akt1 protein and interacts favourably with critical amino acid residues (Figure 7). During the interaction of the ligand 8-hydroxycoumarin, Trp80 is involved in pi-pi stacking, and Lys268 and Val270 residues are involved in pi-alkyl interaction at the binding cavity of the protein. Besides hydrogen bonding, Leu264 is involved in pi-sigma interaction, and the rest residues are van der Waal’s interactions by amino acid residues formed weak non-bonded interaction with the ligand (Figure 7A and right panel). All the binding energy scores are determined from the best cluster (95 percent) that falls within the lowest RMSD 0.25Å. Therefore, it can be inferred from the molecular docking studies that 8-hydroxycoumarin has a high affinity for the protein Akt1.
Also, the stability of the representative Akt1 and 8-hydroxycoumarin complex was further explored using molecular dynamics simulations. The RMSD plots show that the MD results showed stable patterns throughout the entire simulation run (Figures 8A,B). The RMSF graphs show that Akt1 has high flexibility to accommodate the ligand at the binding pocket. The Rg plots demonstrate that the protein stayed compact throughout the simulation. According to the average results observed, the protein backbone was compact. To understand how the residues behaved throughout the simulation run, the fluctuations of the residues were analyzed. After ligand binding, the target’s surface area that was accessible to solvent decreased.
Additionally, our research aimed to find the antioxidant and anticancer potential of D. roylei active extracts by in-vitro approach. In this investigation, DPPH radical scavenging, Hydroxyl scavenging effect, reducing power, and superoxide radical anion scavenging have shown the antioxidant potential of D. roylei extracts and were observed to be significant when compared to positive controls such as Ascorbic acid, BHT, Rutin, and BHT respectively. These observations are in accordance with the previous studies on the antioxidant potential of Delphinium malabaricum extracts that the DPPH radical scavenging assay has investigated and the Ferric reducing antioxidant power (FRAP) assay (Lotfaliani et al., 2021)
Further, the petroleum ether, ethyl acetate, methanol, and ethanolic extracts were subjected to cytotoxicity assay against various breast cancer cell lines. The results shown in Figure 10 demonstrated that ethyl acetate extract of D. roylei showed the highest anticancer activity against the 4T1, MCF-7, MD-MB-468, and MDA-MB-231 breast cell lines as compared to the other three extracts. As a result, the phytoconstituents found in plant extracts might have a greater propensity to suppress the proliferation of cancer cells. D. roylei extracts had relatively lower IC50 values against breast cancer cell lines, which may be due to phytocomponents with stronger binding affinities or altering proteins or pathways implicated in tumor development. It can be observed that the ethyl acetate extract had the highest content of the compounds verifying the above-given conclusions about these compounds being the key constituents responsible for the cytotoxic activity of the studied plant. Previous studies by (Yin et al., 2020) revealed that Siwanine E, Uraphine, Delpheline, Delcorinine, Nordhagenine A, Delbrunine, and Delbrunine from D. honanense and D. chrysotrichum exhibited anticancer potential against MCF-7 and cells with IC50 values of 9.62–35.32 μM. Flow cytometry analysis revealed that plant extract induction tumor cell death via induction of apoptosis and apoptosis enhanced significantly upon higher concentration are shown in Figure 11.
Conclusion
In the present study, we explored the potential mechanisms of phytocompounds present in D. roylei in suppressing breast cancer by network pharmacology-based analysis in combination with chemical profiling, molecular docking, MD simulation, and in vitro studies. HR-LC/MS identified some important phytoconstituents followed by network pharmacology analysis which revealed that 8-hydroxycoumarin, Delsoline, Royleinine, Delsemine B, Herniarin, Aloesin, Talatisamine, Narwedine, Scoparone, and Piperine were the main constituents related to breast cancer targets while Akt1, SRC, EGFR, IL-6, Hsp-90AA1, and ESR-1 were the main breast cancer-related molecular targets. 20 cancer-related pathways were identified where neuroactive ligand-receptor interaction was the most enriched with the highest number of observed genes and lowest false discovery rate, followed by non-small-cell lung cancer. Molecular docking studies showed that 8-hydroxycoumarin possessed the highest binding energies towards all the target proteins, followed by other compounds against studied targets. Furthermore, in vitro studies showed that ethyl acetate extract possess the highest anticancer activity and methanolic extract showed significant antioxidant activity compared to other extracts in the studied plant. The study provided a comprehensive understanding of the suggested mechanism of action of Delphinium roylei that may have potential use in breast cancer treatment.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
MM conceived and designed the study; WM and BB performed the study and wrote the manuscript; AK, RD, MA, AA, MD, and SG analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Jammu Kashmir Science Technology & Innovation Council Department of Science and Technology JKST&IC India with grant No. JKST&IC/SRE/885-87 to MM.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AKT1; RAC-alpha serine/threonine-protein kinase, HR-LC/MS; High Resolution Liquid Chromatography Mass Spectrometry, STRING; Search Tool for the Retrieval of Interacting Genes/Proteins, CDKs; Cyclin Dependent Kinases, RCSB; Research Collaboratory for Structural Bioinformatics, PDB; Protein Data Bank, LGA; Lamarckian Genetic Algorithm, MD; Molecular Dynamic Simulation, RMSD; Root Mean Square Deviation, RMSF; Root Mean Square Fluctuation, OPLS; Optimized Potentials for Liquid Simulations, RG; Radius of Gyration, SASA; Solvent Accessible Surface Area, DPPH; 2,2-diphenyl-1-picrylhydrazyl, MTT; (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide), GO; Gene ontology, KEGG; (Kyoto Encyclopaedia of Genes and Genomes).
References
Balasubramaniam, M., Lakkaniga, N. R., Dera, A. A., Fayi, M. A., Abohashrh, M., Ahmad, I., et al. (2021). FCX-146, a potent allosteric inhibitor of Akt kinase in cancer cells: lead optimization of the second-generation arylidene indanone scaffold. Biotechnol. Appl. Biochem. 68, 82–91. doi:10.1002/bab.1896
Bhat, B. A., Almilaibary, A., Mir, R. A., Aljarallah, B. M., Mir, W. R., Ahmad, F., et al. (2022a). Natural therapeutics in aid of treating alzheimer’s disease: a green gateway toward ending quest for treating neurological disorders. Front. Neurosci. 16, 884345. doi:10.3389/fnins.2022.884345
Bhat, B. A., Mir, W. R., Sheikh, B. A., Alkanani, M., and Mir, M. A. (2022b). Metabolite fingerprinting of phytoconstituents from Fritillaria cirrhosa D. Don and molecular docking analysis of bioactive peonidin with microbial drug target proteins. Sci. Rep. 12, 7296. doi:10.1038/s41598-022-10796-7
Bolton, E. E., Chen, J., Kim, S., Han, L., He, S., Shi, W., et al. (2011). PubChem3D: a new resource for scientists. J. cheminformatics 3, 32–15. doi:10.1186/1758-2946-3-32
Chen, J., Zhu, Y., Zhang, W., Peng, X., Zhou, J., Li, F., et al. (2018). Delphinidin induced protective autophagy via mTOR pathway suppression and AMPK pathway activation in HER-2 positive breast cancer cells. BMC cancer 18, 342–413. doi:10.1186/s12885-018-4231-y
Chow, E., Rendleman, C. A., Bowers, K. J., Dror, R. O., Hughes, D. H., Gullingsrud, J., et al. (2008). Desmond performance on a cluster of multicore processors. Report DESRES/TR--2008-01. New York: DE Shaw Research Technical.
Daina, A., Michielin, O., and Zoete, V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 42717–42813. doi:10.1038/srep42717
Deo, S. V. S., Sharma, J., and Kumar, S. (2022). GLOBOCAN 2020 report on global cancer burden: challenges and opportunities for surgical oncologists. Ann. Surg. Oncol. 29, 6497–6500. doi:10.1245/s10434-022-12151-6
Fuhrmann, J., Rurainski, A., Lenhof, H. P., and Neumann, D. (2010). A new Lamarckian genetic algorithm for flexible ligand-receptor docking. J. Comput. Chem. 31, 1911–1918. doi:10.1002/jcc.21478
Gaber, A., Alsanie, W. F., Alhomrani, M., Alamri, A. S., El-Deen, I. M., and Refat, M. S. (2021). Synthesis and characterization of some new coumarin derivatives as probable breast anticancer MCF-7 drugs. Crystals 11, 565. doi:10.3390/cryst11050565
George, B. P., Chandran, R., and Abrahamse, H. (2021). Role of phytochemicals in cancer chemoprevention: insights. Antioxidants 10, 1455. doi:10.3390/antiox10091455
Heer, E., Harper, A., Escandor, N., Sung, H., McCormack, V., and Fidler-Benaoudia, M. M. (2020). Global burden and trends in premenopausal and postmenopausal breast cancer: a population-based study. Lancet Glob. Health 8, e1027–e1037. doi:10.1016/S2214-109X(20)30215-1
Jafari, M., Ghadami, E., Dadkhah, T., and Akhavan-Niaki, H. (2019). PI3k/AKT signaling pathway: erythropoiesis and beyond. J. Cell. physiology 234, 2373–2385. doi:10.1002/jcp.27262
Jaganathan, P., Rajan, M., Sathyanarayanan, S., Murugaiyan, I., Jeyalakshmi, G. S. S., and Thangaraj, P. (2018). 17 antioxidant potential. Medicinal plants: promising future for health and new drugs, 317.
Kumar, S., and Hamal, I. A. (2011). Herbal remedies used against arthritis in Kishtwar high altitude National Park. India: NISCAIR-CSIR.
Lim, W-C., Kim, H., Kim, Y-J., Park, S-H., Song, J-H., Lee, K. H., et al. (2017). Delphinidin inhibits BDNF-induced migration and invasion in SKOV3 ovarian cancer cells. Bioorg. Med. Chem. Lett. 27, 5337–5343. doi:10.1016/j.bmcl.2017.09.024
Lipinski, C. A. (2004). Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov. today Technol. 1, 337–341. doi:10.1016/j.ddtec.2004.11.007
Lotfaliani, M., Ayatollahi, S. A., Kobarfard, F., Pour, P. M., and Mohammadi Pour, P. (2021). Chemistry, biological activities and toxic effects of alkaloidal constituents of genus Delphinium-A mini review. J. Herbmed Pharmacol. 10, 486–499. doi:10.34172/jhp.2021.56
Lv, P., Gao, P., Tian, G., Yang, Y., Mo, F., Wang, Z-H., et al. (2020). Osteocyte-derived exosomes induced by mechanical strain promote human periodontal ligament stem cell proliferation and osteogenic differentiation via the miR-181b-5p/PTEN/AKT signaling pathway. Stem Cell Res. Ther. 11, 295–315. doi:10.1186/s13287-020-01815-3
Mazurek, A. H., Szeleszczuk, Ł., and Gubica, T. (2021). Application of molecular dynamics simulations in the analysis of cyclodextrin complexes. Int. J. Mol. Sci. 22, 9422. doi:10.3390/ijms22179422
Mehraj, U., Mir, I. A., ul Hassan, M., Mir, N. A., and Mir, M. A. (2022). Adapalene synergistically with doxorubicin promotes apoptosis of TNBC Cells by hyperactivation of the ERK1/2 pathway through ROS induction. Preprint.
Mir, W. R., Bhat, B. A., Almilaibary, A., Asdaq, S. M. B., and Mir, M. A. (2022a). Evaluation of the in vitro antimicrobial activities of Delphinium roylei: an insight from molecular docking and MD-simulation studies. Med. Chem. 18, 1109–1121. doi:10.2174/1573406418666220429093956
Mir, W. R., Bhat, B. A., Rather, M. A., Muzamil, S., Almilaibary, A., Alkhanani, M., et al. (2022b). Molecular docking analysis and evaluation of the antimicrobial properties of the constituents of Geranium wallichianum D. Don ex Sweet from Kashmir Himalaya. Sci. Rep. 12, 12547. doi:10.1038/s41598-022-16102-9
Miu, L., Bogatyreva, N. S., and Galzitskaia, O. V. (2008). Radius of gyration is indicator of compactness of protein structure. Mol. Biol. 42, 701–706.
Morris, G. M., Huey, R., and Olson, A. J. (2008). Using autodock for ligand-receptor docking. Curr. Protoc. Bioinforma. 24, Unit 8.14–14. doi:10.1002/0471250953.bi0814s24
Noumi, E., Snoussi, M., Anouar, E. H., Alreshidi, M., Veettil, V. N., Elkahoui, S., et al. (2020). HR-LCMS-based metabolite profiling, antioxidant, and anticancer properties of teucrium polium L. Methanolic extract: computational and in vitro study. Antioxidants 9, 1089. doi:10.3390/antiox9111089
Nwakaego, N. L., Chibuike, O. K., Chukwugekwu, E. M., Marylyn, A. C., Ngozi, E. I., and Chukwunonye, E. R. (2019). In vitro antioxidant and free radical scavenging potential of methanolic extracts of Uvaria chamae leaves and roots. Int. J. Pharm. Pharm. Sci. 11, 67–71. doi:10.22159/ijpps.2019v11i1.29330
Obi, K. N., Onyeike, E. N., and Anacletus, F. C. (2022). Studies on in vitro Radical Scavenging Potentials of Methanol Leaf Extract of Ficus sur and its Fractions. Free Radicals Antioxidants 12, 15–21. doi:10.5530/fra.2022.1.3
Pistelli, M., Natalucci, V., Scortichini, L., Agostinelli, V., Lenci, E., Crocetti, S., et al. (2021). The impact of lifestyle interventions in high-risk early breast cancer patients: a modeling approach from a single institution experience. Cancers 13, 5539. doi:10.3390/cancers13215539
Pospelova, M., Krasnikova, V., Fionik, O., Alekseeva, T., Samochernykh, K., Ivanova, N., et al. (2022). Adhesion molecules ICAM-1 and PECAM-1 as potential biomarkers of central nervous system damage in women breast cancer survivors. Pathophysiology 29, 52–65. doi:10.3390/pathophysiology29010006
Rapaport, D. C., and Rapaport, D. C. R. (2004). The art of molecular dynamics simulation. Cambridge: Cambridge University Press.
Release, S. (2017). 3: desmond molecular dynamics system. New York, NY: DE Shaw research, Maestro-Desmond Interoperability Tools, Schrödinger.
Rezadoost, M. H., Kumleh, H. H., and Ghasempour, A. (2019). Cytotoxicity and apoptosis induction in breast cancer, skin cancer and glioblastoma cells by plant extracts. Mol. Biol. Rep. 46, 5131–5142. doi:10.1007/s11033-019-04970-w
Saifi, I., Bhat, B. A., Hamdani, S. S., Bhat, U. Y., Lobato-Tapia, C. A., Mir, M. A., et al. (2023). Artificial intelligence and cheminformatics tools: a contribution to the drug development and chemical science. J. Biomol. Struct. Dyn. 2023, 1–19. doi:10.1080/07391102.2023.2234039
Sharma, P. K., and Singh, V. (1989). Ethnobotanical studies in northwest and Trans-Himalaya. V. Ethno-veterinary medicinal plants used in Jammu and Kashmir, India. J. Ethnopharmacol. 27, 63–70. doi:10.1016/0378-8741(89)90078-0
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: gLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA a cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Tariq, L., Bhat, B. A., Hamdani, S. S., and Mir, R. A. (2021). Phytochemistry, pharmacology and toxicity of medicinal plants. Med. Aromatic Plants Healthc. Industrial Appl. 2021, 217–240.
Tiwari, A., and Singh, S. (2022). “Computational approaches in drug designing,” in Bioinformatics (Amsterdam, Netherlands: Elsevier), 207–217.
Trott, O., and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. doi:10.1002/jcc.21334
Wu, A., Zhu, Y., Han, B., Peng, J., Deng, X., Chen, W., et al. (2021). Delphinidin induces cell cycle arrest and apoptosis in HER-2 positive breast cancer cell lines by regulating the NF-κB and MAPK signaling pathways. Oncol. Lett. 22, 832–911. doi:10.3892/ol.2021.13093
Xu, X., Yu, Y., Zong, K., Lv, P., and Gu, Y. (2019). Up-regulation of IGF2BP2 by multiple mechanisms in pancreatic cancer promotes cancer proliferation by activating the PI3K/Akt signaling pathway. J. Exp. Clin. Cancer Res. 38, 497–514. doi:10.1186/s13046-019-1470-y
Yin, T., Cai, L., and Ding, Z. (2020). An overview of the chemical constituents from the genus Delphinium reported in the last four decades. RSC Adv. 10, 13669–13686. doi:10.1039/d0ra00813c
Yin, T., Zhang, H., Zhang, W., and Jiang, Z. (2021). Chemistry and biological activities of hetisine-type diterpenoid alkaloids. RSC Adv. 11, 36023–36033. doi:10.1039/d1ra07173d
Yu, S., Gao, W., Zeng, P., Chen, C., Zhang, Z., Liu, Z., et al. (2021). Exploring the effect of Gupi Xiaoji Prescription on hepatitis B virus-related liver cancer through network pharmacology and in vitro experiments. Biomed. Pharmacother. 139, 111612. doi:10.1016/j.biopha.2021.111612
Keywords: Delphinium roylei, breast cancer, HR/LC-MS, 8-hydroxycoumarin, anticancer activity, network pharmacology, molecular docking and MD simulations
Citation: Mir WR, Bhat BA, Kumar A, Dhiman R, Alkhanani M, Almilaibary A, Dar MY, Ganie SA and Mir MA (2023) Network pharmacology combined with molecular docking and in vitro verification reveals the therapeutic potential of Delphinium roylei munz constituents on breast carcinoma. Front. Pharmacol. 14:1135898. doi: 10.3389/fphar.2023.1135898
Received: 02 January 2023; Accepted: 09 August 2023;
Published: 01 September 2023.
Edited by:
Olivier Feron, Université catholique de Louvain, BelgiumReviewed by:
Udhaya Kumar. S, Baylor College of Medicine, United StatesYoung Hoon Son, Emory University, United States
Hassan Bardania, Yasuj University of Medical Sciences, Iran
Copyright © 2023 Mir, Bhat, Kumar, Dhiman, Alkhanani, Almilaibary, Dar, Ganie and Mir. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manzoor Ahmad Mir, ZHJtYW56b29yQGthc2htaXJ1bml2ZXJzaXR5LmFjLmlu
†These authors have contributed equally to this work
 Wajahat Rashid Mir1†
Wajahat Rashid Mir1† Rohan Dhiman
Rohan Dhiman Mustfa Alkhanani
Mustfa Alkhanani Showkat Ahmad Ganie
Showkat Ahmad Ganie Manzoor Ahmad Mir
Manzoor Ahmad Mir