- 1College of Horticulture, Hunan Agricultural University, Changsha, China
- 2Changsha Central Hospital, Changsha, China
Introduction: Pancreatic cancer (PC) is a particularly aggressive malignancy with limited therapeutic options. The search for innovative treatments has focused on traditional Chinese medicine, specifically epimedium. This research investigates epimedium’s active ingredients, potential targets, and underlying mechanisms in treating PC.
Methods: High-performance liquid chromatography (HPLC) was used to quantify the active components of epimedium and HPLC-Q-TOF-MS was employed for qualitative identification. Potential targets of epimedium’s active ingredients were identified using the TCMSP, ETCM, CTD, and Swiss Target Prediction databases. Potential PC-related targets were sourced from DisGeNET, GeneCards, and OMIM databases. A Venn diagram was utilized to identify overlapping PC-related and epimedium targets. Core targets and pathways were elucidated through protein-protein interaction (PPI) network analysis, Gene Ontology (GO) assessments, and Reactome pathway enrichment analyses. Molecular docking techniques investigated interactions between active compounds and these targets. The expression and prognostic implications of target genes were evaluated using GEPIA2 and the Human Protein Atlas (HPA) databases. In vitro studies assessed the impact of epimedium extract (EPE) on Panc-1 cell viability, and Western blot analysis examined the expression levels of key targets.
Results: Network pharmacological indicate that epimedium econtains active components such as baohuoside I, icariin, hyperoside, and epimedin B, which have potential therapeutic effects against PC. In vitro assays confirmed that EPE significantly reduced the viability of Panc-1 cells. Western blot analysis revealed a considerable decrease in the expression of key targets in EPE-treated cells, including AKT1, EGFR, p-EGFR, JUN, BCL2, IL6, and SRC. The R-HSA-1280215: Interleukin-4 and Interleukin-13 signaling pathways involving these genes were identified as potential therapeutic targets.
Discussion: Epimedium holds promise as a candidate for treating PC. The modulation of interleukin-4 and interleukin-13 signaling pathways could be a pivotal mechanism by which epimedium impedes tumor development. Further research is warranted to validate these findings and explore the clinical applicability of epimedium in PC treatment.
Introduction
Pancreatic cancer (PC) is a highly aggressive malignancy that has been on the rise over the past decade, with its incidence increasing from 43,140 to 60,430 cases (Jemal et al., 2010; Siegel et al., 2021). As a result, it is now the third leading cause of cancer-related deaths in the United States, after lung and breast cancers (Grossberg et al., 2020). While established risk factors for PC include age, gender, race/ethnicity, family history of the disease, smoking habits, diabetes, and obesity (Yamaguchi et al., 2017), its prevalence is still increasing due to the aging global population (Goral, 2015; Jia et al., 2018; Klein, 2021). Despite this alarming trend, there is still hope for the development of new treatments and therapies to combat this deadly disease. PC is typically asymptomatic at onset and progresses rapidly with early metastasis, resulting in many patients being diagnosed too late for surgery and a high mortality rate (Song et al., 2020; Ye et al., 2021).
Surgery with negative margin resection under a microscope is challenging and only benefits a few patients (Kamisawa et al., 2016). Therefore, radiotherapy or chemotherapy is necessary for most patients. In recent years, targeted therapies and immunotherapies have emerged as novel treatments, offering more options for those with advanced disease. However, they can be associated with adverse effects such as rash, liver dysfunction, or diarrhea (Rudin et al., 2008; Suzumura et al., 2012), which can affect the patient’s quality of life or lead to discontinuation of therapy. Furthermore, these costly drugs are effective only for certain patients, highlighting the need to find more effective yet less toxic new drugs.
Chinese herbal medicine has a long history of use in the treatment of critically ill patients. Traditional Chinese herbs have been used for centuries to treat various ailments and diseases, including cancer (Zhang et al., 2014; Zhang et al., 2016). However, it is essential to note that some whole herbs may also contain toxins and non-medicinal chemicals. Further research is needed to understand the mechanisms and pathways involved in cancer treatment. Investigating active substances with precise chemical structures separated from herbs may be beneficial. Many natural compounds, such as paclitaxel and vinblastine, have been found to possess anticancer activity, either directly or indirectly. Even at high concentrations, multiple natural chemicals can be effectively tolerated by patients (Jiang et al., 2016; Rejhová et al., 2018). Thus, novel natural substances offer great promise for developing effective yet less toxic anticancer drugs.
Epimedium, guided by Traditional Chinese Medicine (TCM) principles of reinforcing positive and eliminating negative energy, employed in cancer treatment for an extensive period (Liu et al., 2021; Zhao et al., 2022). The polysaccharide (EPS) derived from the epimedium plant has been demonstrated to enhance the secretion of immunomodulatory cytokines by peritoneal macrophages and T cells, thereby reducing tumor burden. Moreover, icariin, a derivative of epimedium known as a prenylflavonoid, has demonstrated antitumor effects on diverse human cell lines, including breast, prostate, endometrial cancer, renal cell carcinoma, and leukemia (Guo et al., 2011; Tong et al., 2011). Furthermore, Wang et al. (2019) discovered its capability to inhibit tumor cell growth by generating reactive oxygen species and inducing DNA damage.
In 2007, Hopkins and colleagues introduced the concept of “network pharmacology,” which involves investigating the effects of drugs on disease networks. This approach established a “drug-target-disease” network, offering an innovative approach to drug development. This approach enabled researchers to understand better the complex interactions among drugs, targets, and diseases while identifying new drug targets and potential drug combinations. Through the aid of network pharmacology, researchers can now enhance their ability to predict the efficacy and safety of drugs and develop new drugs that are more effective and possess fewer side effects (Liu and Du, 2009). In recent times, network pharmacology has been successfully employed in numerous studies to investigate the mechanisms of TCM (Wu et al., 2020). To advance our understanding of the mechanisms and pathways involved in the therapeutic effects of epimedium on PC, we applied this approach to examine its impact on PC and confirm its regulatory associations with key signaling pathways.
Materials and methods
Cells and reagents
Epimedium was obtained from Hunan Zirantang Traditional Chinese Medicine Pieces Co., Ltd. (production batch number: 230401). Hyperoside, Epimedin A, epimedin B, epimedin C, icariin, and baohuoside I standards (Purity ≥98%) were purchased from Shanghai Yuanye Co., Ltd. (Shanghai, China). The human pancreatic cancer cell line PANC-1 was acquired from Procell Co., Ltd. (Wuhan, China). DEME medium (#C11995500CP), 10% fetal bovine serum (#FSP500), and penicillin-streptomycin were obtained from Gibco. The ELISA kit was purchased from BOSTER Technology Co., Ltd. (Wuhan, China). DMSO, CCK-8 kit, RIPA lysis buffer (#PC0020), and BCA protein assay kit (#PC0020) were purchased from Solarbio Technology Co., Ltd. (Beijing, China). The antibodies used are as follows: EGFR (#ab52894), p-EGFR (#ab40815), AKT1 (#ab81283), JUN (#ab32385), IL6 (#ab214429), SRC (#ab109381), BCL2 (#ab182858) and β-actin (#ab8227) were purchased from Abcam (Cambridge, United Kingdom) and Cell Signaling Technology (Boston, MA, United States).
Preparation of epimedium extract
Epimedium sagittatum (Sieb.etZucc.) Maxim., a Berberidaceae plant, was purchased from Hunan Zirantang Traditional Chinese Medicine Pieces Co., Ltd. (production batch number: 230401) and identified by Professor Xiaohong Zhong. Epimedium extract (EPE) preparation method involved taking 100 g of epimedium, adding 8 times 70% ethanol, soaking for 30 min, boiling for 1.5 h, centrifuging at 10,000 RPM for 30 min, and collecting the supernatant. This extraction process was repeated twice, and the supernatants were mixed and evaporated to obtain a powder and stored at −20°C for later use.
Chemical determination of extract contents
Flavonoids are regarded as the main active ingredient in epimedium (Yang et al., 2020; Zhao et al., 2018). Flavonoids were detected and identified, and the contents were determined by liquid chromatography-ultraviolet/quadrupole-time-of-flight mass spectrometry (LC-Q-TOF-MS) (Wang et al., 2010; Zhao et al., 2018).
The preparation of the reference solution was carried out as follows: Accurately weigh an appropriate amount of each reference substance (hyperoside, epimedin A, epimedin B, epimedin C, icariin, baohuoside I) (5–10 mg) into a 50 mL volumetric flask, add 50% methanol to dissolve and dilute to the mark, filtered through a 0.45 μm microporous membrane. Preparation of test solution: epimedium extract was accurately weighed and placed in a conical flask with a stopper. 70% ethanol (20 mL) was added, the weight was accurately measured, and the sample was sonicated for 10 min. The samples were then removed and left to cool down to room temperature, and the loss in weight was made up with 70% ethanol. After filtering through a 0.45 μm microporous membrane, the filtrate was used as the test solution.
In brief, an ultra-performance liquid chromatograph, Agilent 1290, coupled with an Agilent 6495 tandem mass spectrometer (Agilent Technologies, Wilmington, DE, United States) was used. The chromatographic conditions were as follows: The chromatographic column was Hypersil BDS C18 (250 mm × 4.6 mm, 5 μm), column temperature was 25°C. The mobile phases of HPLC were composed of acetonitrile (A) and water (B). The linear gradient was adopted as follows: 24%–26% A at 0–20 min, 26%–30% A at 20–30 min, 30%–45% A at 30–45 min, 45%–60% A at 45–50 min, 60% to 24%A at 50–60 min. The flow rate was 1.0 mL/min with detection at 270 nm. The injection volume was 10.0 μL. MS conditions: electrospray ion source (ESI), positive and negative ion modes, and scanning range of m/z 100–2,000; dry gas temperature 350°C, dry gas flow rate 10 L·min−1, spray voltage 30 psi, fragmentor voltage 135 V, cone voltage 65 V, ion spray voltage (IS) 3500 V (positive ion mode)/-4000 V (negative ion mode). For MS/MS analyses, auto-MS/MS was performed with a collision energy between 10 and 55 eV.
Screening of related targets
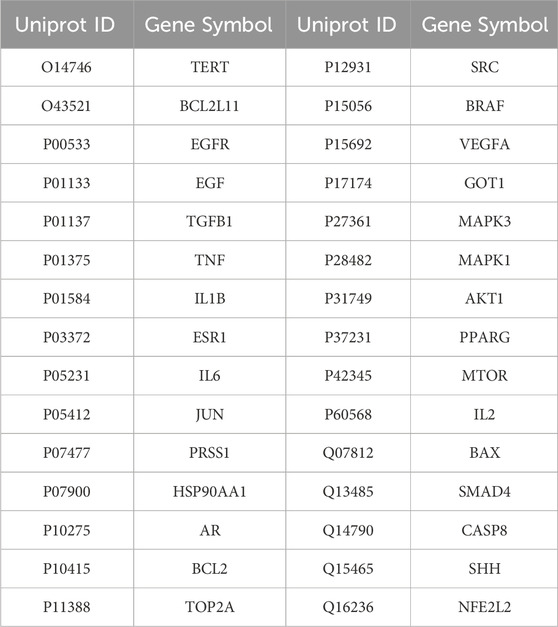
The potential targets of active ingredients of epimedium were screened from the following databases: Traditional Chinese Medicine Systems Pharmacology (TCMSP) database (https://www.tcmsp-e.com/) (Ye et al., 2022) Encyclopedia of Traditional Chinese Medicine (ETCM, http://www.tcmip.cn/ETCM2/front/#/) (Xu et al., 2019), Swiss Target Prediction (http://www.swisstargetprediction.ch/), Comparative Toxicogenomics Database (CTD) (https://ctdbase.org/). Ultimately, the potential targets of epimedium were identified by standardizing and removing duplicates using the Uniprot database (https://www.uniprot.org/).
Acquisition of potential anti-pancreatic cancer targets and their shared targets with epimedium
To identify genes associated with pancreatic cancer, we conducted a thorough search of the GeneCards, GeneCards, and OMIM databases, focusing on the “Homo sapiens” species and using the keywords “pancreatic cancer.” As a result, we were able to pinpoint genes that have been previously reported to be linked with pancreatic cancer. Subsequently, we carefully eliminated any duplicate entries and false positives from the gene list, cross-referencing it with previously identified potential targets of epimedium. Consequently, we successfully identified potential targets for combating pancreatic cancer using epimedium.
Protein-protein interaction construction
By inputting the potential anti-pancreatic cancer targets of epimedium into the STRING database (https://cn.string-db.org/), restricted to the “H. sapiens” species and filtered with a minimum interactome requirement score of >0.7 (Von Mering et al., 2005), we acquired protein-protein interaction relationships. Subsequently, we employed Cytoscape (version 3.7.1) to visually represent the PPI network. Moreover, we installed the CytoNCA package to conduct network topology analysis, aiming to identify crucial target genes associated with the mechanism of icariin. In order to identify highly interconnected subnetworks within the PPI network, we employed Cytoscape’s Molecular Complex Detection technique (MCODE) (Bader and Hogue, 2003).
Protein functional enrichment analysis
To gain a deeper understanding of the potential biological functions of epimedium in the treatment of pancreatic cancer, we carried out gene ontology (GO), biological processes (BPs), and Reactome pathway enrichment analyses using the DAVID Database (Sherman et al., 2022) and Reactome Database (Wright et al., 2022). The resulting data was organized according to their adjusted p values.
Ingredient-target-pathway network construction
An Ingredient-Target-Pathway (I-T-P) network was established in Cytoscape (version 3.7.1), encompassing overlapping genes, pathways, and selected active components. Nodes of diverse colors represented various clusters, while edges delineated node relationships.
Molecular docking verification of core active ingredients and core targets
Molecular docking was performed to validate the interaction between the core active ingredient and the core target identified in the PPI network. The three-dimensional structures of the core targets were obtained from the Protein Data Bank (PDB, www.rcsb.org), and the two-dimensional structures of the top five core compounds were acquired from PubChem in SDF format. PyMOL v2.4, an open-source molecular graphics tool, was used to remove solvent and organic components to prepare for molecular docking. Subsequently, AutoDock Tools (version 1.5.6) were employed for molecular docking, and the resulting data was analyzed and interpreted using PyMOL v2.4.
Validation of core targets in GEPIA2 and HPA database
The Gene Expression Profiling Interactive Analysis (GEPIA 2; http://gepia2.cancer-pku.cn/) web server is a publicly available database that analyzes gene expression in tumor and normal samples from TCGA and GTEx. We used GEPIA two to analyze the expression levels of core targets relevant to our research question in normal pancreatic and pancreatic adenocarcinoma (PAAD) tissues. Additionally, we investigated the overall survival data of these core targets using the Human Protein Atlas (HPA; https://www.proteinatlas.org/), an open-access database that employs various omics technologies. HPA provides comprehensive mappings of human protein expression patterns in cells, tissues, and organs through antibody-based imaging, mass spectrometry-based proteomics, and transcriptomics.
Experimental validation
Cell culture and viability assays
Cell culture and viability assays were performed as described previously (Shen et al., 2020). Briefly, Panc-1 cells were cultured in 10% fetal calf serum and 100 U/mol penicillin-streptomycin solution at 37°C, 5% CO2, and 95% relative humidity. After the intervention, each cell was in the logarithmic growth phase. EPE was dissolved in DMSO at a concentration of 200 mg/mL and then diluted to different concentrations of EPE (0, 100, 200, 300, 400, 500, and 600 μg/mL) for pretreatment. The pretreatment time was 48 h. Cell viability was assessed using a Thermo Fisher microplate spectrophotometer with the CCK-8 assay, following the manufacturer’s instructions. The optical density (OD) of the solution at 450 nm in each well was measured. All data were normalized to control wells without cells and expressed as mean ± standard deviation (SD).
Western blot
Panc-1 cells were seeded in 6-well plates at a density of 3 × 105 cells per well. After incubating for 48 h, EPE (200 μg/mL or 400 μg/mL) was added and treated for 48 h. The cells were then collected, and total protein was extracted using radioimmunoprecipitation assay (RIPA) buffer, which contained phenylmethylsulfonyl fluoride (PMSF), aprotinin, and phosphatase inhibitors. The supernatant was collected after centrifugation (14,000 × g, 4°C for 15 min), and the protein concentration was determined using a BCA protein assay kit (Genview, United States). 25 μg of each protein sample was separated by SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA, United States). The membranes were then blocked with 5% bovine serum albumin (BSA) for 2 h at room temperature. Primary antibodies for AKT1, EGFR, p-EGFR, JUN, BCL2, IL6, and SRC were added and incubated overnight at 4°C. After three washes, an enzyme-labeled secondary antibody IgG (1:2000) was used to incubate for 2 h at room temperature under dark conditions. Finally, a secondary antibody (1:5000, ab clone) was added and incubated at room temperature for 2 h. Finally, membranes were washed and developed by adding enhanced chemiluminescence (ECL) substrate (Thermo Fisher Scientific, Rockford, United States). Proteins were visualized using the intelligent gel imaging system iBright FL1000 (Thermofisher, Rockford, United States).
Statistical analyses
Statistical analysis was conducted on SPSS 22.0 (SPSS, Chicago, IL, United States), with a data-processing method of independent-sample t-test. Comparisons between groups were statistically analyzed, and *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 was considered to be statistically significant.
Results
Identification and content of active ingredients in epimedium
All the calibration curves showed a good linear regression relationship in the test range (r > 0.999). RSDs of precision, stability, and repeatability were all less than 3.0%. Therefore, the HPLC method was precise, accurate, and sensitive enough for simultaneous quantitative analysis of the six flavonoids in epimedium extract. The HPLC analysis identified hyperoside, epimedin A, epimedin B, epimedin C, icariin, and baohuoside I as the primary compounds, with EPE at 200 μg/mL containing 1.6 μg/mL of hyperoside, 5.8 μg/mL of icariin, 5.9 μg/mL of epimedin A, 20.3 μg/mL of epimedin B, and 3.4 μg/mL of epimedin C, 4.8 μg/mL of baohuoside I.
Based on the optimization of the elution program, we used high-performance HPLC-Q-TOF-MS/MS to perform a qualitative analysis of compounds in epimedium extract. By comparing the retention time (tR) and characteristic fragment ions with reference compounds and literature data, a total of six compounds were identified. See Figure 1 and Table 1 for details.
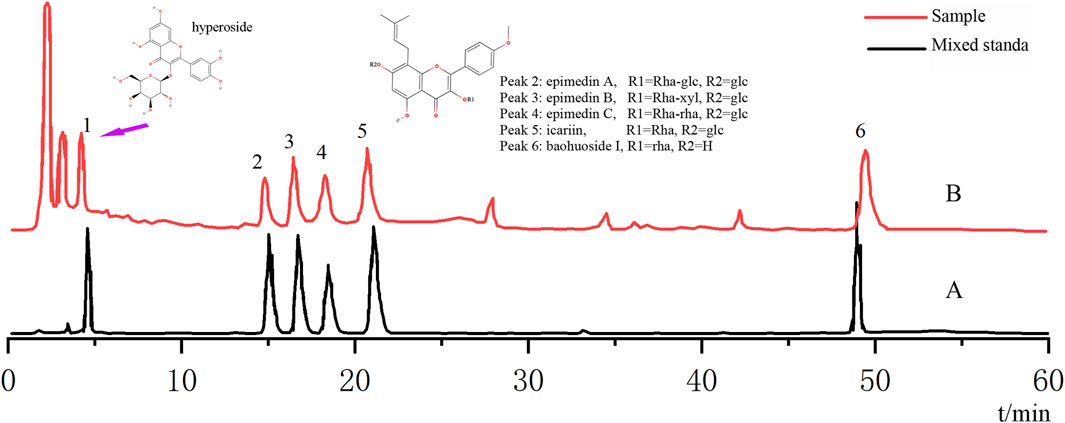
Figure 1. Analysis of the chemical composition of epimedium extract by HPLC. (A) HPLC chromatogram of mixed standard; (B) HPLC chromatogram of the sample.
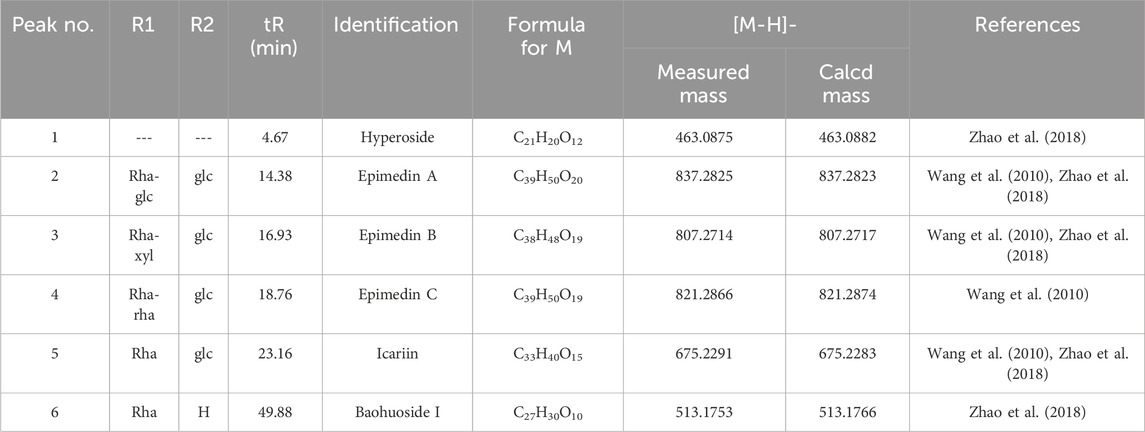
Table 1. Retention time (tR), MS data, and UV spectra for identifying six compounds in epimedium extract by HPLC-Q-TOF-MS/MS.
Acquisition of potential targets of epimedium against PC
Based on TCMSP, ETCM, CTD database, and Swiss Target Prediction, a total of 174 potential targets were identified for the six active ingredients from epimedium, including hyperoside, epimedin A, epimedin B, and epimedin C, icariin, and baohuoside I (Supplementary Table S1).
The GeneCards, GeneCards, and OMIM databases revealed 316 targets associated with PC. Subsequently, a Venn diagram was utilized to identify 30 overlapping targets between the PC-related targets and the potential targets of epimedium (Figure 2; Table 2).
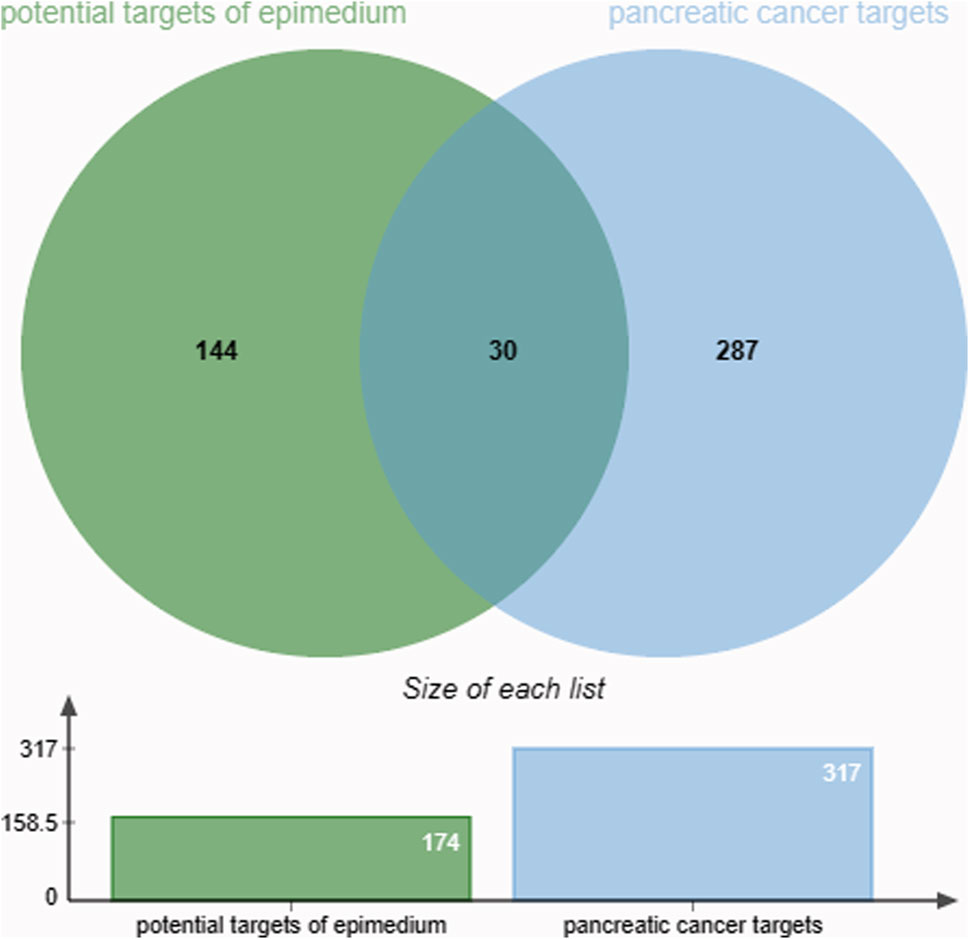
Figure 2. The intersection of the active ingredient and pancreatic cancer targets is represented in 30 overlapping targets.
Construction of a protein-protein network
The genes related to the 30 overlapping targets were used to establish protein-protein interaction (PPI) relationships through the String database. The resultant PPI network was visualized using Cytoscape 3.7.1, revealing 26 protein nodes and 168 edges representing their interactions. Nodes with a deeper shade of red indicated higher degree values, signifying a strong association between these targets and PC (Figure 3A).
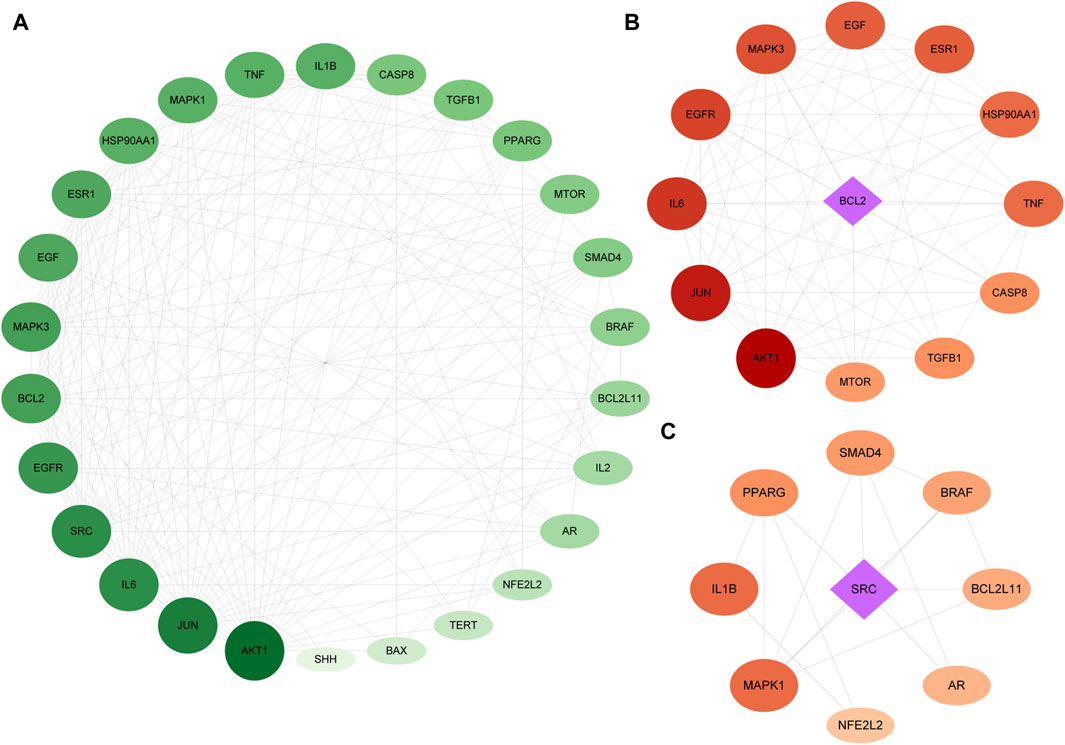
Figure 3. PPI of the intersection targets. The node’s size indicates the degree of the target protein in the network. Panel (A) is a PPI network diagram arranged according to a degree, with two clusters detected in the epimedium-anti-PC PPI network. Panels (B,C) show clusters 1 and 2, respectively, with diamonds representing the seed nodes of each cluster.
We utilized MCODE analysis to identify three functionally related protein clusters based on topology within the PPI network. The cluster attributes are presented in Figures 3B, C. In PPI networks, clusters formed by interlinked nodes often represent protein complexes or pathways. Similarly, clusters in protein similarity networks typically indicate protein families, where the proteins within a cluster exhibit similar structures and functions.
GO enrichment and KEGG enrichment
Upon uploading the 30 overlapped targets to the DAVID database for GO enrichment analysis, we identified 129 biological processes (BP). The top five processes included positive regulation of transcription from RNA polymerase II promoter, positive regulation of pri-miRNA transcription from RNA polymerase II promoter, positive regulation of apoptotic process, positive regulation of transcription, DNA-templated, positive regulation of peptidyl-serine phosphorylation. Additionally, 32 cellular components (CC) were identified, with the top five being cytoplasm, macromolecular complex, nucleus, Bcl-2 family protein complex, and mitochondrial outer membrane. Moreover, 32 molecular functions (MF) were identified, with the top five being identical protein binding, nitric-oxide synthase regulator activity, nitric-oxide synthase regulator activity, transcription coactivator binding, and enzyme binding (Figure 4).
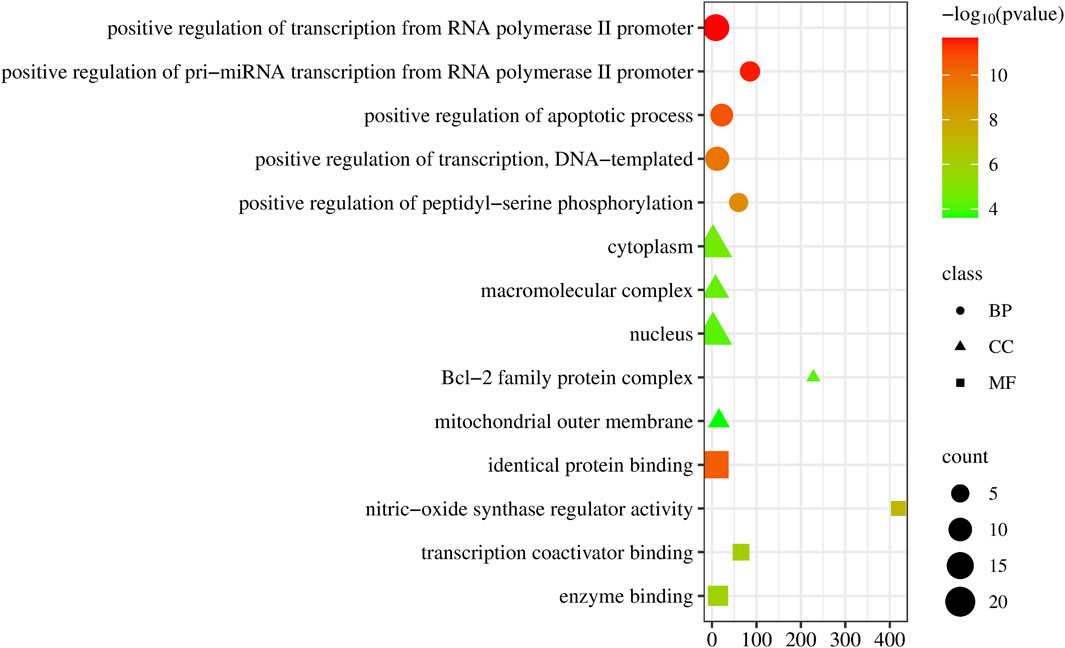
Figure 4. Dot plot of the GO enrichment of epimedium against pancreatic cancer. The horizontal axis represents the enrichment rate of the input genes in the GO Term, while the vertical axis represents the GO term name. The color scale indicates different p-value thresholds, and the dot size indicates the number of genes corresponding to each term.
The Reactome pathway enrichment analysis identified 657 pathways, with the top 10 pathways visually represented in Figure 5. These pathways exhibit notable enrichment in processes associated with anti-PC effects, encompassing Interleukin-4 and Interleukin-13 signaling, Signaling by Interleukins, Diseases of signal transduction by growth factor receptors and second messengers, Cytokine Signaling in Immune system, and Extra-nuclear estrogen signaling.
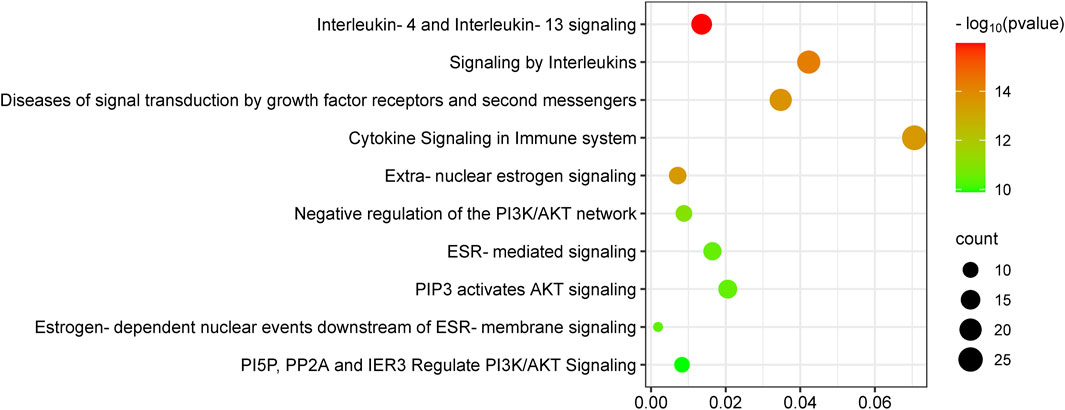
Figure 5. KEGG enrichment analysis of target genes of epimedium against pancreatic cancer. The number of genes enriched in each KEGG pathway term is shown in different colors, showing the circle size and the p-value.
Ingredient-target-pathway network construction
The analysis of the Ingredients-Targets-Pathways network revealed 44 nodes and 152 edges, consisting of four active constituents of epimedium (baohuoside I, icariin, hyperoside, epimedin B), 30 potential anti-PC targets, and 10 pathways (Figure 6). Our findings illustrated that baohuoside I had the highest level of connectivity with 18 targets, followed by icariin (15), hyperoside (13), and epimedin B (5), indicating their potential involvement in regulating pathways associated with PC.
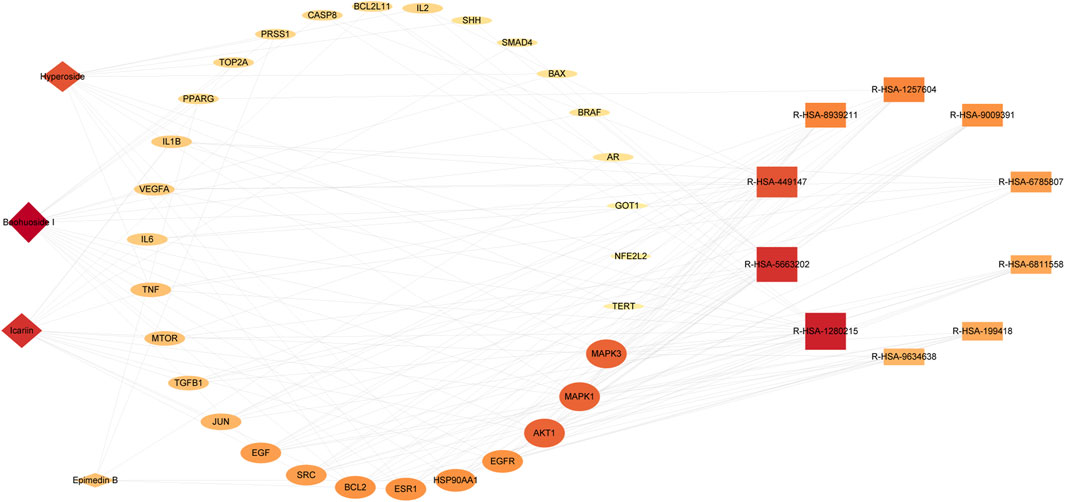
Figure 6. Ingredient-target-pathway network diagram. The active ingredient is displayed as a diamond shape, the targets as an ellipse, and the pathway as a rectangle.
Molecular docking verification of core active ingredients and core protein targets
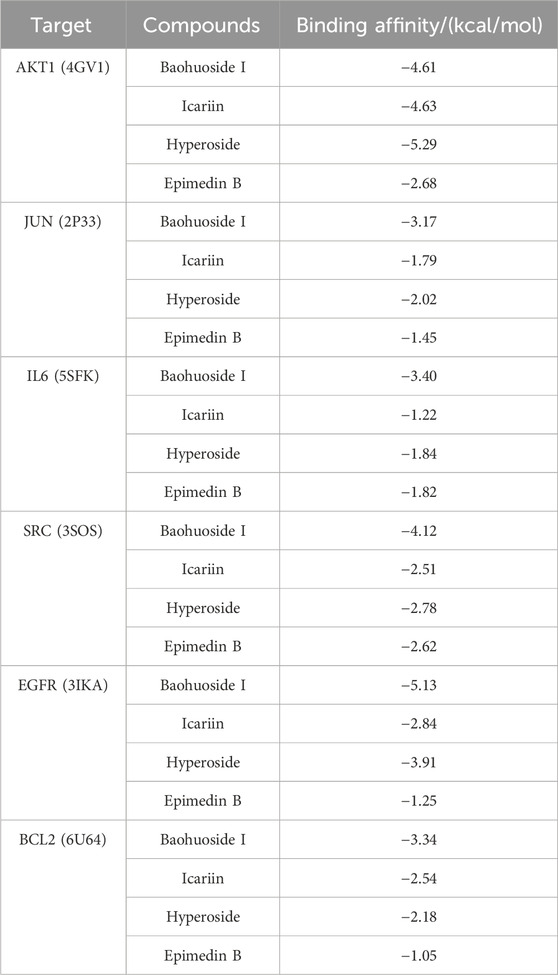
The results of molecular docking studies have revealed the binding strength of the essential active constituents present in epimedium with their specific protein targets. After conducting I-T-P network screening, we identified baohuoside I, icariin, hyperoside, and epimedin B as the core compounds. The hub protein targets comprise the top five node degree proteins (AKT1, JUN, IL6, SRC, and EGFR) and two seed nodes in clusters (SRC and BCL2).
The binding affinities of the six core active ingredients to their respective target proteins were investigated. Baohuoside I displayed solid binding affinities, ranging from −3.17 kcal/mol to −5.13 kcal/mol, with JUN, IL6, SRC, EGFR, and BCL2, respectively, while icariin and hyperoside exhibited a high affinity of −4.63 and −5.29 kcal/mol with AKT1. The strong binding of the small-molecule active ingredients was attributed to various interactions (as shown in Figure 7; Table 3).
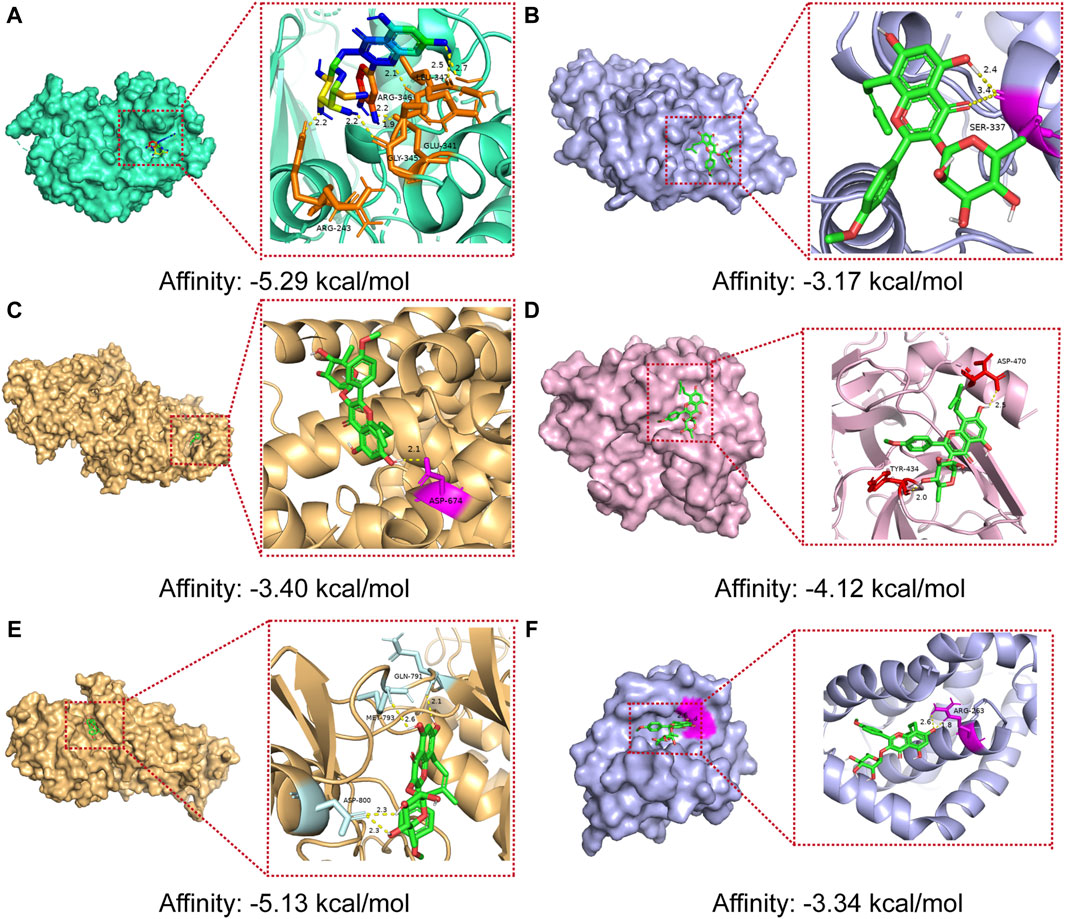
Figure 7. Molecular docking models of core compounds and core protein targets (A) AKT1 - Hyperoside, (B) JUN - Baohuoside I, (C) IL6 - Baohuoside_I, (D) SRC - Baohuoside _I, (E) EGFR - Baohuoside _I, and (F) BCL2 - Baohuoside I.
Validation of key targets in GEPIA2 and HPA database
The analysis using the GEPIA two database revealed that the expression of core targets (AKT1, IL6, and SRC) was higher in PAAD samples compared to normal pancreas tissues (p < 0.05; Figure 8). Moreover, the examination of overall survival data from the HPA database indicated that high expression of EGFR and IL6 was correlated with a significantly poorer prognosis (p < 0.05; Figure 9).
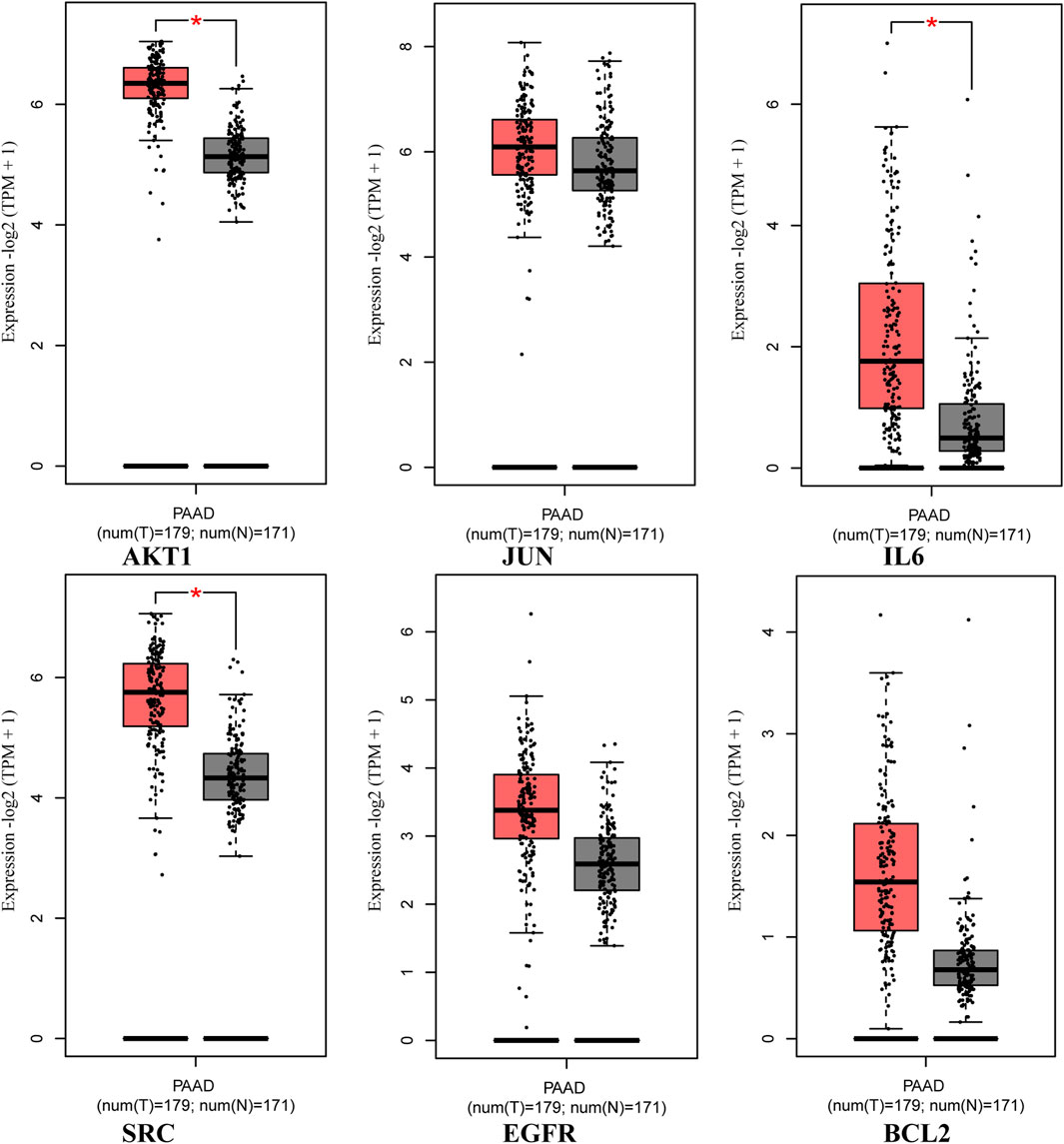
Figure 8. Expression of core targets in the GEPIA two database. The red box plots represent the tumor, and the gray box plots represent normal. Match TCGA normal and GTEx data.
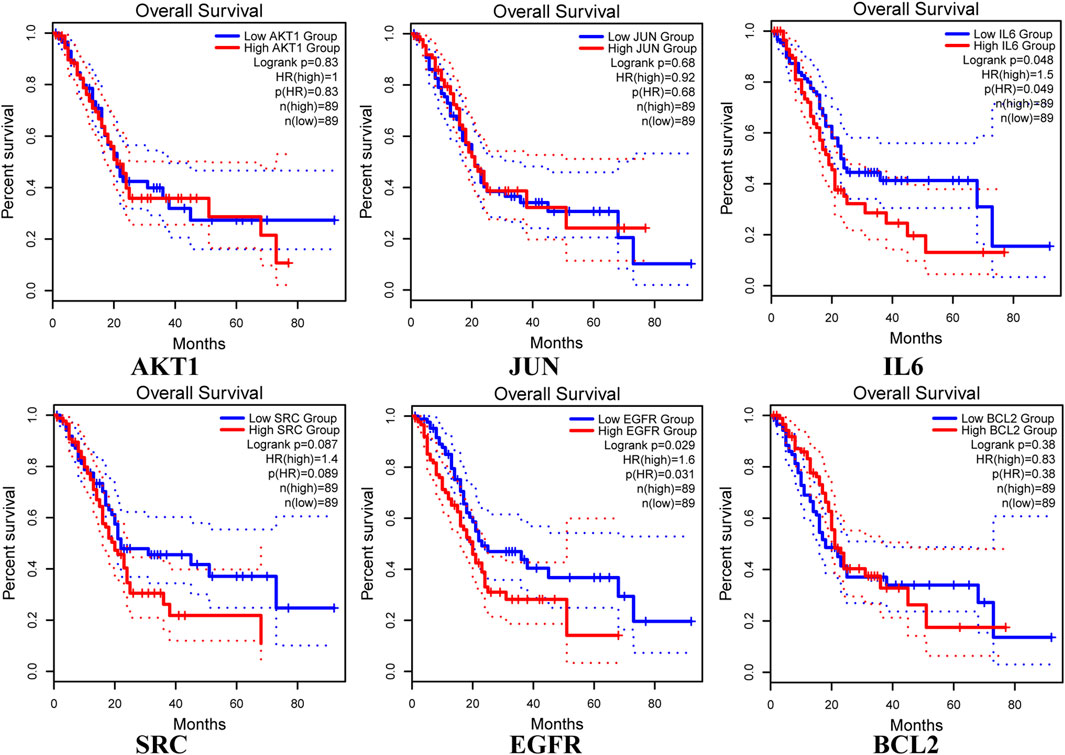
Figure 9. Overall survival of hub genes in the HPA database, with blue representing low expressions and red representing high expressions. Kaplan-Meier survival curves were independently plotted for each gene.
In vitro experimental validation
To investigate the impact of EPE on pancreatic cancer cells, we employed Cell Counting Kit-8 (CCK-8) to assess the cell viability of Panc-1 cells following treatment with varying concentrations of EPE (Figure 10). Our findings revealed a noteworthy effect of EPE on the viability of Panc-1 cells in a dose-dependent manner with a half-maximal inhibitory concentration (IC50) of 207.0 μg/mL.
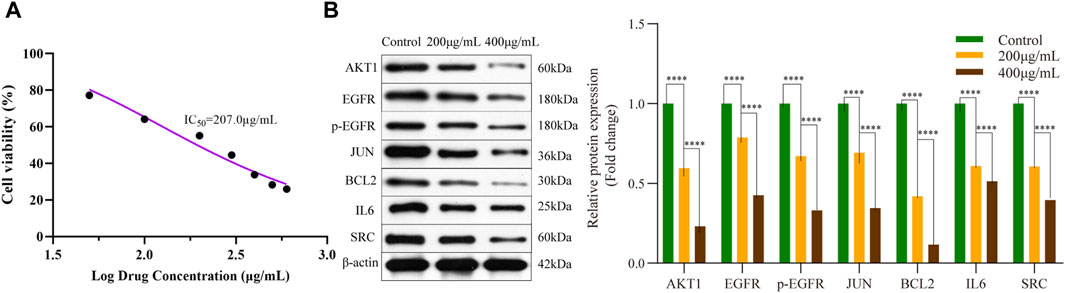
Figure 10. Epimedium extract suppressed R-HSA-1280215: Interleu in-4 and Interleukin-13 signaling pathway in PANC-1. (A) Cell viability of PANC-1 cells treated with different concentrations of epimedium extract; (B) Effect of AKT1, EGFR, p-EGFR, JUN, BCL2, IL6 and SRC expression in PANC-1 cells analyzed by Western blot (****p < 0.0001); IC50, half maximal inhibitory concentration.
The R-HSA-1280215: Interleu in-4 and Interleukin-13 signaling was the first pathway obtained in the KEGG pathway enrichment analysis, involving downstream proteins. To verify the effect of EPE on key targets, we utilized the Western blot method to measure the expression of AKT1, EGFR, p-EGFR, JUN, BCL2, IL6 and SRC proteins in Panc-1 cells. The results, shown in Figure 10B, indicate a significant decrease in the expression of AKT1, EGFR, p-EGFR, JUN, BCL2, IL6, and SRC in EPE-treated Panc-1 cells.
Discussion
PC exhibits a high mortality rate and is frequently diagnosed at advanced stages, posing significant challenges for effective treatment. Therefore, it is essential to develop novel therapeutics and drugs to improve the quality of life and extend the lifespan of affected individuals. The progression of PC involves intricate biological processes, such as metabolic disorders, local inflammation, and aberrant molecular pathways (Motoo et al., 2011; Ansari et al., 2019). As a result, single-drug therapies targeting a single pathway may not be sufficient to achieve optimal therapeutic efficacy (Li et al., 2019).
Studies have indicated that various components of TCM, including the Qingyi Huaji formula (Hua et al., 2014; Zhang et al., 2013) and Huang-Qin-Tang (Saif et al., 2014), can effectively boost the immune system and suppress the proliferation of PC cells. These components play a beneficial role in enhancing immunity against infectious diseases and mitigating factors that disrupt normal physiological functions (Li et al., 2015). Moreover, single-herb TCM or herbal formulas have demonstrated the ability to engage multiple targets and pathways of action.
In this study, network pharmacology was employed to investigate the mechanism of epimedium’s action against PC. By analyzing the PPI network, we identified six potential therapeutic core targets (AKT1, EGFR, JUN, BCL2, IL6, and SRC) that exhibited significant correlations with PC. Subsequently, we performed molecular docking calculations to evaluate the binding affinities of epimedium’s active ingredients to these proteins. The results revealed that baohuoside I, icariin, hyperoside, and epimedin B displayed high binding affinities to AKT1, EGFR, JUN, BCL2, IL6 and SRC, indicating their potential relevance in PC treatment.
The validation of key targets using the GEPIA2 and HPA databases revealed significantly higher expressions of AKT1, IL6, and SRC in PAAD tissues compared to normal pancreatic tissues. Furthermore, high expressions of EGFR and IL6 were associated with significantly poorer survival rates.
In the development and progression of cancer, AKT1, JUN, IL6, SRC, EGFR and BCL2 plays a critical role. Akt1 (serine-threonine protein kinase) is a part of the PI3K-dependent signaling pathway and is a key cell survival protein functionally involved in antiapoptosis in various cancers (Goswami et al., 2006). Epidermal growth factor receptor (EGFR) is a trans-membrane receptor tyrosine kinase. It plays a critical oncogenic role in cancer growth, survival, migration, and invasion (Jiang et al., 2017). The study demonstrated that epimedium extract inhibits the expression and phosphorylation of AKT1 and EGFR in the pancreatic cancer cell line PANC-1. Similar findings have been previously documented. For instance, Geng et al. (2014) reported that icariside II inhibits the proliferation of transplantable tumors and the EGFR/mTOR signaling pathway, leading to reduced phosphorylation of EGFR and AKT1 proteins. In addition, Ding et al. (2018) indicated that Icariin enhances the sexual function of male mice by activating the PI3K/Akt/eNos/No signaling pathway. Furthermore, Li et al. (2020) demonstrated that icariin, via the EGFR/PI3K/mTOR signaling pathways, decreases the expression and phosphorylation of EGFR and Akt proteins. Moreover, Sun et al. (2022) found that icariin interacts with the estrogen receptor on the cell membrane, resulting in elevated phosphorylation levels of Akt and Creb proteins and increased transcription of genes encoding steroidogenic enzymes and testosterone synthesis.
EGFRY1068 phosphorylation can be characterized by its association with the recruitment of specific signaling proteins and its potential role as a predictive biomarker for treatment response in certain cancer patients (Sette et al., 2015; Wirth et al., 2023). EGFRY1068 phosphorylation induces the recruitment of Grb2 and Gab1, leading to the activation of the AKT and STAT3/5 signaling pathways (Wirth et al., 2023). Prior research indicates that Icariin suppresses the epithelial-mesenchymal transition process induced by epidermal growth factor by downregulating the PI3K/Akt signaling pathway (Wang et al., 2021). Icariin can diminish the expression of ERα36 and hinder E2β-triggered Akt phosphorylation, showing heightened efficacy in suppressing TNBC cell proliferation and facilitating apoptosis when combined with the EGFR inhibitor Cetuximab (Yin et al., 2020). Additionally, it inhibits the activation of ERK1/2 and Akt induced by ionizing radiation (Hong et al., 2013), diminishes STAT3, and Akt in MDSCs, leading to the differentiation of MDSCs into dendritic cells and macrophages (Zhou et al., 2011). In our study, epimedium extract was found to inhibit EGFRY1068 phosphorylation in the pancreatic cancer cell line, PANC-1. However, it should be noted that the epimedium extract used in this study consisted of various components, including hyperoside, epimedin A, epimedin B, epimedin C, icariin, and baohuoside I. Further investigation is required to discern the specific active ingredients and elucidate the underlying mechanisms responsible for the inhibitory effects of epimedium extract on EGFRY1068 phosphorylation.
BCL2 is widely recognized as a multifunctional anti-apoptotic protein in various types of cancer. The overexpression of the BCL2 is linked to tumor initiation, progression, and resistance to current anticancer therapies (Das et al., 2020). Icariin has been shown to enhance its antioxidant and anti-apoptotic functions by inhibiting the phosphorylation of BCL2 (Zhang et al., 2016). Additionally, hyperoside relieved H2O2-induced oxidative stress and attenuated H2O2-induced apoptosis of granulosa cells, resulting in decreased expression of the BCL2 protein (Wang et al., 2019). Icariin and hyperoside are critical active components of epimedium extract in our study.
Conclusion
This study employed bioinformatics, network pharmacology, database verification, and in vitro validation to identify potential active compounds and pathways of epimedium in the treatment of PC. Through the regulation of genes such as AKT1, EGFR, JUN, BCL2, IL6 and SRC, epimedium was found to modulate the R-HSA-1280215: Interleu in-4 and Interleukin-13 signaling pathway. The findings suggest that further investigation of epimedium as a subsequent treatment for PC is warranted.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
YC: Conceptualization, Formal Analysis, Investigation, Methodology, Validation, Writing–original draft, Writing–review and editing. HX: Investigation, Methodology, Visualization, Writing–review and editing. XZ: Conceptualization, Supervision, Validation, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Science and Technology Innovation Program of Hunan Province (Grant no. 142203900999).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1389221/full#supplementary-material
References
Ansari, D., Ohlsson, H., Althini, C., Bauden, M., Zhou, Q., Hu, D., et al. (2019). The hippo signaling pathway in pancreatic cancer. Anticancer Res. 39 (7), 3317–3321. doi:10.21873/anticanres.13474
Bader, G. D., and Hogue, C. W. V. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinforma. 4 (1), 2. doi:10.1186/1471-2105-4-2
Das, J., Chakraborty, S., and Maiti, T. K. (2020). Mechanical stress-induced autophagic response: a cancer-enabling characteristic? Semin. Cancer Biol. 66, 101–109. doi:10.1016/j.semcancer.2019.05.017
Ding, J., Tang, Y., Tang, Z., Zu, X., Qi, L., Zhang, X., et al. (2018). Icariin improves the sexual function of male mice through the PI3K/AKT/eNOS/NO signalling pathway. Andrologia 50 (1), e12802. doi:10.1111/and.12802
Geng, Y.-D., Yang, L., Zhang, C., and Kong, L.-Y. (2014). Blockade of epidermal growth factor receptor/mammalian target of rapamycin pathway by Icariside II results in reduced cell proliferation of osteosarcoma cells. Food Chem. Toxicol. 73, 7–16. doi:10.1016/j.fct.2014.08.002
Goral, V. (2015). Pancreatic cancer: pathogenesis and diagnosis. Asian Pac. J. Cancer Prev. 16 (14), 5619–5624. doi:10.7314/apjcp.2015.16.14.5619
Goswami, A., Ranganathan, P., and Rangnekar, V. M. (2006). The phosphoinositide 3-kinase/Akt1/Par-4 axis: a cancer-selective therapeutic target. Cancer Res. 66 (6), 2889–2892. doi:10.1158/0008-5472.Can-05-4458
Grossberg, A. J., Chu, L. C., Deig, C. R., Fishman, E. K., Hwang, W. L., Maitra, A., et al. (2020). Multidisciplinary standards of care and recent progress in pancreatic ductal adenocarcinoma. CA Cancer J. Clin. 70 (5), 375–403. doi:10.3322/caac.21626
Guo, Y., Zhang, X., Meng, J., and Wang, Z. Y. (2011). An anticancer agent icaritin induces sustained activation of the extracellular signal-regulated kinase (ERK) pathway and inhibits growth of breast cancer cells. Eur. J. Pharmacol. 658 (2-3), 114–122. doi:10.1016/j.ejphar.2011.02.005
Hong, J., Zhang, Z., Lv, W., Zhang, M., Chen, C., Yang, S., et al. (2013). Icaritin synergistically enhances the radiosensitivity of 4T1 breast cancer cells. PLoS One 8 (8), e71347. doi:10.1371/journal.pone.0071347
Hua, Y. Q., Chen, Z., Meng, Z. Q., Chen, H., Shen, J. G., Wang, K., et al. (2014). High expression of erythropoietin-producing hepatoma cell line-B2 (EphB2) predicts the efficiency of the Qingyihuaji formula treatment in pancreatic cancer CFPAC-1 cells through the EphrinB1-EphB2 pathway. Oncol. Lett. 8 (1), 17–24. doi:10.3892/ol.2014.2134
Jemal, A., Siegel, R., Xu, J., and Ward, E. (2010). Cancer statistics, 2010. CA Cancer J. Clin. 60 (5), 277–300. doi:10.3322/caac.20073
Jia, X., Du, P., Wu, K., Xu, Z., Fang, J., Xu, X., et al. (2018). Pancreatic cancer mortality in China: characteristics and prediction. Pancreas 47 (2), 233–237. doi:10.1097/mpa.0000000000000976
Jiang, R., Tang, J., Chen, Y., Deng, L., Ji, J., Xie, Y., et al. (2017). The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat. Commun. 8, 15129. doi:10.1038/ncomms15129
Jiang, Y., Liu, L. S., Shen, L. P., Han, Z. F., Jian, H., Liu, J. X., et al. (2016). Traditional Chinese Medicine treatment as maintenance therapy in advanced non-small-cell lung cancer: a randomized controlled trial. Complement. Ther. Med. 24, 55–62. doi:10.1016/j.ctim.2015.12.006
Kamisawa, T., Wood, L. D., Itoi, T., and Takaori, K. (2016). Pancreatic cancer. Lancet 388 (10039), 73–85. doi:10.1016/s0140-6736(16)00141-0
Klein, A. P. (2021). Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat. Rev. Gastroenterol. Hepatol. 18 (7), 493–502. doi:10.1038/s41575-021-00457-x
Li, B., Gan, R., Yang, Q., Huang, J., Chen, P., Wan, L., et al. (2015). Chinese herbal medicines as an adjunctive therapy for unresectable pancreatic cancer: a systematic review and meta-analysis. Evid. Based Complement. Altern. Med. 2015, 350730. doi:10.1155/2015/350730
Li, J., Ma, J., Tian, Y., Zhao, P., Liu, X., Dong, H., et al. (2020). Effective-component compatibility of Bufei Yishen formula II inhibits mucus hypersecretion of chronic obstructive pulmonary disease rats by regulating EGFR/PI3K/mTOR signaling. J. Ethnopharmacol. 257, 112796. doi:10.1016/j.jep.2020.112796
Li, S., Xu, H. X., Wu, C. T., Wang, W. Q., Jin, W., Gao, H. L., et al. (2019). Angiogenesis in pancreatic cancer: current research status and clinical implications. Angiogenesis 22 (1), 15–36. doi:10.1007/s10456-018-9645-2
Liu, A. L., and Du, G. H. (2009). Research progress of virtual screening aided drug discovery. Acta Pharm. Sin. 44 (6), 566–570. doi:10.3321/j.issn:0513-4870.2009.06.002
Liu, X., Wang, S., Zheng, H., Liu, Q., Shen, T., Wang, X., et al. (2021). Epimedokoreanin C, a prenylated flavonoid isolated from Epimedium koreanum, induces non-apoptotic cell death with the characteristics of methuosis in lung cancer cells. Am. J. Cancer Res. 11 (7), 3496–3514.
Motoo, Y., Shimasaki, T., Ishigaki, Y., Nakajima, H., Kawakami, K., and Minamoto, T. (2011). Metabolic disorder, inflammation, and deregulated molecular pathways converging in pancreatic cancer development: implications for new therapeutic strategies. Cancers (Basel) 3 (1), 446–460. doi:10.3390/cancers3010446
Rejhová, A., Opattová, A., Čumová, A., Slíva, D., and Vodička, P. (2018). Natural compounds and combination therapy in colorectal cancer treatment. Eur. J. Med. Chem. 144, 582–594. doi:10.1016/j.ejmech.2017.12.039
Rudin, C. M., Liu, W., Desai, A., Karrison, T., Jiang, X., Janisch, L., et al. (2008). Pharmacogenomic and pharmacokinetic determinants of erlotinib toxicity. J. Clin. Oncol. 26 (7), 1119–1127. doi:10.1200/jco.2007.13.1128
Saif, M. W., Li, J., Lamb, L., Kaley, K., Elligers, K., Jiang, Z., et al. (2014). First-in-human phase II trial of the botanical formulation PHY906 with capecitabine as second-line therapy in patients with advanced pancreatic cancer. Cancer Chemother. Pharmacol. 73 (2), 373–380. doi:10.1007/s00280-013-2359-7
Sette, G., Salvati, V., Mottolese, M., Visca, P., Gallo, E., Fecchi, K., et al. (2015). Tyr1068-phosphorylated epidermal growth factor receptor (EGFR) predicts cancer stem cell targeting by erlotinib in preclinical models of wild-type EGFR lung cancer. Cell Death Dis. 6 (8), e1850. doi:10.1038/cddis.2015.217
Shen, J., Li, L., Yang, T., Cohen, P. S., and Sun, G. (2020). Biphasic mathematical model of cell-drug interaction that separates target-specific and off-target inhibition and suggests potent targeted drug combinations for multi-driver colorectal cancer cells. Cancers (Basel) 12 (2), 436. doi:10.3390/cancers12020436
Sherman, B. T., Hao, M., Qiu, J., Jiao, X., Baseler, M. W., Lane, H. C., et al. (2022). DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50 (W1), W216–W221. doi:10.1093/nar/gkac194
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2021). Cancer statistics, 2021. CA Cancer J. Clin. 71 (1), 7–33. doi:10.3322/caac.21654
Song, Z., Xiang, X., Li, J., Deng, J., Fang, Z., Zhang, L., et al. (2020). Ruscogenin induces ferroptosis in pancreatic cancer cells. Oncol. Rep. 43 (2), 516–524. doi:10.3892/or.2019.7425
Sun, J., Xu, W., Zheng, S., Lv, C., Lin, J., Chen, S., et al. (2022). Icariin promotes mouse Leydig cell testosterone synthesis via the Esr1/Src/Akt/Creb/Sf-1 pathway. Toxicol. Appl. Pharmacol. 441, 115969. doi:10.1016/j.taap.2022.115969
Suzumura, T., Kimura, T., Kudoh, S., Umekawa, K., Nagata, M., Matsuura, K., et al. (2012). Reduced CYP2D6 function is associated with gefitinib-induced rash in patients with non-small cell lung cancer. BMC Cancer 12, 568. doi:10.1186/1471-2407-12-568
Tong, J. S., Zhang, Q. H., Huang, X., Fu, X. Q., Qi, S. T., Wang, Y. P., et al. (2011). Icaritin causes sustained ERK1/2 activation and induces apoptosis in human endometrial cancer cells. PLoS One 6 (3), e16781. doi:10.1371/journal.pone.0016781
Von Mering, C., Jensen, L. J., Snel, B., Hooper, S. D., Krupp, M., Foglierini, M., et al. (2005). STRING: known and predicted protein–protein associations, integrated and transferred across organisms. Nucleic Acids Res. 33 (Suppl. l_1), D433–D437. doi:10.1093/nar/gki005
Wang, M., Wan, Y., Liang, Z., Zhu, D., Huang, Y., Chen, M., et al. (2021). Effects of icariin on EGF-induced breast cancer cell MCF-7 epithelial mesenchymal regulation process and its mechanism. China J. Tradit. Chin. Med. Pharm. 36 (5), 2545–2549.
Wang, S., Wang, Q., Wang, H., Qin, C., Cui, X., Li, L., et al. (2019). Induction of ROS and DNA damage-dependent senescence by icaritin contributes to its antitumor activity in hepatocellular carcinoma cells. Pharm. Biol. 57 (1), 424–431. doi:10.1080/13880209.2019.1628073
Wang, Y., Guo, Z., Jin, Y., Zhang, X., Wang, L., Xue, X., et al. (2010). Identification of prenyl flavonoid glycosides and phenolic acids in Epimedium koreanum Nakai by Q-TOF-MS combined with selective enrichment on “click oligo (ethylene glycol)” column. J. Pharm. Biomed. Anal. 51 (3), 606–616. doi:10.1016/j.jpba.2009.09.033
Wirth, D., Özdemir, E., and Hristova, K. (2023). Quantification of ligand and mutation-induced bias in EGFR phosphorylation in direct response to ligand binding. Nat. Commun. 14 (1), 7579. doi:10.1038/s41467-023-42926-8
Wright, A. J., Orlic-Milacic, M., Rothfels, K., Weiser, J., Trinh, Q. M., Jassal, B., et al. (2022). Evaluating the predictive accuracy of curated biological pathways in a public knowledgebase. Database 2022, baac009. doi:10.1093/database/baac009
Wu, W., Zhang, Z., Li, F., Deng, Y., Lei, M., Long, H., et al. (2020). A network-based approach to explore the mechanisms of uncaria alkaloids in treating hypertension and alleviating alzheimer’s disease. Int. J. Mol. Sci. 21 (5), 1766. doi:10.3390/ijms21051766
Xu, H. Y., Zhang, Y. Q., Liu, Z. M., Chen, T., Lv, C. Y., Tang, S. H., et al. (2019). ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 47 (D1), D976–D982. doi:10.1093/nar/gky987
Yamaguchi, K., Okusaka, T., Shimizu, K., Furuse, J., Ito, Y., Hanada, K., et al. (2017). Clinical practice guidelines for pancreatic cancer 2016 from the Japan pancreas society: a synopsis. Pancreas 46 (5), 595–604. doi:10.1097/mpa.0000000000000816
Yang, X.-H., Li, L., Xue, Y.-B., Zhou, X.-X., and Tang, J.-H. (2020). Flavonoids from Epimedium pubescens: extraction and mechanism, antioxidant capacity and effects on CAT and GSH-Px of Drosophila melanogaster. PeerJ 8, e8361. doi:10.7717/peerj.8361
Ye, X. W., Wang, H. L., Cheng, S. Q., Xia, L. J., Xu, X. F., and Li, X. R. (2022). Network pharmacology-based strategy to investigate the pharmacologic mechanisms of coptidis rhizoma for the treatment of alzheimer's disease. Front. Aging Neurosci. 14, 890046. doi:10.3389/fnagi.2022.890046
Ye, Z., Zhuo, Q., Hu, Q., Xu, X., Mengqi, L., Zhang, Z., et al. (2021). FBW7-NRA41-SCD1 axis synchronously regulates apoptosis and ferroptosis in pancreatic cancer cells. Redox Biol. 38, 101807. doi:10.1016/j.redox.2020.101807
Yin, L., Qi, X. W., Liu, X. Z., Yang, Z. Y., Cai, R. L., Cui, H. J., et al. (2020). Icaritin enhances the efficacy of cetuximab against triple-negative breast cancer cells. Oncol. Lett. 19 (6), 3950–3958. doi:10.3892/ol.2020.11496
Zhang, J., Wang, P., Ouyang, H., Yin, J., Liu, A., Ma, C., et al. (2013). Targeting cancer-related inflammation: Chinese herbal medicine inhibits epithelial-to-mesenchymal transition in pancreatic cancer. PLoS One 8 (7), e70334. doi:10.1371/journal.pone.0070334
Zhang, L., Wu, C., Zhang, Y., Liu, F., Wang, X., Zhao, M., et al. (2014). Comparison of efficacy and toxicity of traditional Chinese medicine (TCM) herbal mixture LQ and conventional chemotherapy on lung cancer metastasis and survival in mouse models. PLoS One 9 (10), e109814. doi:10.1371/journal.pone.0109814
Zhang, Q. B., Meng, X. T., Jia, Q. A., Bu, Y., Ren, Z. G., Zhang, B. H., et al. (2016). Herbal compound songyou yin and moderate swimming suppress growth and metastasis of liver cancer by enhancing immune function. Integr. Cancer Ther. 15 (3), 368–375. doi:10.1177/1534735415622011
Zhao, Y., Chen, S., Wang, Y., Lv, C., Wang, J., and Lu, J. (2018). Effect of drying processes on prenylflavonoid content and antioxidant activity of Epimedium koreanum Nakai. J. Food Drug Anal. 26 (2), 796–806. doi:10.1016/j.jfda.2017.05.011
Zhao, Y.-D., Zhang, X., Yang, W.-Y., Zhang, R.-Q., Mu, L.-T., Han, L., et al. (2022). New anti-pulmonary fibrosis prenylflavonoid glycosides from Epimedium koreanum. Chin. J. Nat. Med. 20 (3), 221–228. doi:10.1016/S1875-5364(21)60116-4
Zhou, J., Wu, J., Chen, X., Fortenbery, N., Eksioglu, E., Kodumudi, K. N., et al. (2011). Icariin and its derivative, ICT, exert anti-inflammatory, anti-tumor effects, and modulate myeloid derived suppressive cells (MDSCs) functions. Int. Immunopharmacol. 11 (7), 890–898. doi:10.1016/j.intimp.2011.01.007
Keywords: pancreatic cancer, epimedium, hyperoside, icariin, baohuoside I, interleukin-4, interleukin-13
Citation: Chen Y, Xia H and Zhong X (2024) In Vitro evaluation of the anti-pancreatic cancer activity of epimedium herb. Front. Pharmacol. 15:1389221. doi: 10.3389/fphar.2024.1389221
Received: 21 February 2024; Accepted: 13 June 2024;
Published: 01 July 2024.
Edited by:
Raghuveera Kumar Goel, Boston University, United StatesReviewed by:
Xiangxuan Zhao, China Medical University, ChinaAmit Kumar Tripathi, University of North Texas Health Science Center, United States
Copyright © 2024 Chen, Xia and Zhong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yangfeng Chen, Y2hlbnlmQGh1bmF1LmVkdS5jbg==; Xiaohong Zhong, eGgtemhvbmdAMTYzLmNvbQ==
 Yangfeng Chen
Yangfeng Chen Han Xia
Han Xia Xiaohong Zhong
Xiaohong Zhong