- 1Department of Pharmacy, West China Hospital, Sichuan University, Chengdu, Sichuan, China
- 2West China School of Medicine, Sichuan University, Chengdu, Sichuan, China
- 3Key Laboratory of Drug-Targeting and Drug Delivery System of the Education Ministry, Sichuan Engineering Laboratory for Plant-Sourced Drug and Sichuan Research Center for Drug Precision Industrial Technology, West China School of Pharmacy, Sichuan University, Chengdu, Sichuan, China
Introduction: Adverse drug events (ADEs) pose a significant challenge in current clinical practice. Machine learning (ML) has been increasingly used to predict specific ADEs using electronic health record (EHR) data. This systematic review provides a comprehensive overview of the application of ML in predicting specific ADEs based on EHR data.
Methods: A systematic search of PubMed, Web of Science, Embase, and IEEE Xplore was conducted to identify relevant articles published from the inception to 20 May 2024. Studies that developed ML models for predicting specific ADEs or ADEs associated with particular drugs were included using EHR data.
Results: A total of 59 studies met the inclusion criteria, covering 15 drugs and 15 ADEs. In total, 38 machine learning algorithms were reported, with random forest (RF) being the most frequently used, followed by support vector machine (SVM), eXtreme gradient boosting (XGBoost), decision tree (DT), and light gradient boosting machine (LightGBM). The performance of the ML models was generally strong, with an average area under the curve (AUC) of 76.68% ± 10.73, accuracy of 76.00% ± 11.26, precision of 60.13% ± 24.81, sensitivity of 62.35% ± 20.19, specificity of 75.13% ± 16.60, and an F1 score of 52.60% ± 21.10. The combined sensitivity, specificity, diagnostic odds ratio (DOR), and AUC from the summary receiver operating characteristic (SROC) curve using a random effects model were 0.65 (95% CI: 0.65–0.66), 0.89 (95% CI: 0.89–0.90), 12.11 (95% CI: 8.17–17.95), and 0.8069, respectively. The risk factors associated with different drugs and ADEs varied.
Discussion: Future research should focus on improving standardization, conducting multicenter studies that incorporate diverse data types, and evaluating the impact of artificial intelligence predictive models in real-world clinical settings.
Systematic Review Registration: https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42024565842, identifier CRD42024565842.
Introduction
Medical treatment can pose potential risks associated with drug-induced impairments. Adverse drug events (ADEs) are defined as injuries resulting from medical interventions related to drugs and may manifest as clinical signs, symptoms, or laboratory abnormalities (Bates et al., 1995). ADEs include not only adverse drug reactions but also medication errors, therapeutic failures, adverse medication withdrawal events, and overdoses (Nebeker et al., 2004). ADEs have been recognized as a significant challenge in contemporary clinical practice. ADEs are estimated to contribute to up to 4.5 million ambulatory encounters, 1.3 million emergency department visits, 350,000 hospitalizations, and 106,000 deaths annually in the United States (Sarkar et al., 2011; Food and Drug Administration FDA, 2024). Although many of these events occur unintentionally, some are preventable (Hartigan-Go and Wong, 2000). Therefore, predicting ADEs has become an area of active research.
Various methods have been used to predict ADEs, such as drug-drug interactions (Dmitriev et al., 2019), the chemical structures of drugs (Mantripragada et al., 2021), spontaneous reporting systems (Bae et al., 2020), and health records (Lippenszky et al., 2024). The prediction of potential adverse events through drug-drug interactions and chemical structures is based on pharmacological mechanisms but does not account for individual patient physiological conditions or specific diseases. Although spontaneous reporting systems can predict ADEs using patient information, this information is often incomplete. Compared to these approaches, using health records for ADE prediction is more clinically practical as they encompass comprehensive data across a patient’s hospitalization period. Moreover, with the advent of electronic health record (EHR) systems, these data can be rapidly compiled and analyzed to achieve more accurate ADE prediction.
Specific ADEs are often associated with particular medications and risk factors, and under certain conditions, specific medications tend to induce distinct categories of ADEs. For instance, antihypertensive medications generally do not cause nausea and vomiting, while chemotherapy in cancer patients often results in these side effects (Adel, 2017). Additionally, opioid-induced injuries are more likely to occur in geriatric patients over the age of 60 (Sacerdote, 2008; Khanna et al., 2020; Horrigan et al., 2023). Consequently, multiple studies have focused on predicting specific ADEs and their associations with drugs. Traditional statistical methods have encountered limitations due to the large number of risk factors (Ji et al., 2018), leading to the introduction of novel statistical approaches.
Machine learning (ML) is an interdisciplinary field within the broader domain of artificial intelligence that integrates statistics and computer science. ML is adept at managing complex non-linear relationships between variables and outcomes, offering high generalization capabilities and precision (Deo, 2015). It has been widely used to predict specific ADEs based on EHR data. However, systematic evaluations of these applications remain limited. Therefore, this systematic review aims to provide a comprehensive overview of the application of ML in predicting specific ADEs using EHR data.
Method
This systematic review was conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Moher et al., 2009). The review protocol was registered with PROSPERO (CRD42024565842). As this study used publicly available data, ethical approval was not required.
Literature search and eligibility criteria
A systematic search was conducted across PubMed, Web of Science, Embase, and IEEE Xplore (containing a wealth of articles on biomedical engineering, medical devices, and health informatics) to identify relevant publications from their inception up to 20 May 2024. Search syntaxes were based on “machine learn” or “artificial intelligence” with “predict” and “adverse drug event or reaction”. Synonyms were determined based on MeSH terms and Emtree. The detailed study protocol on the search strategy and inclusion criteria were provided in the Supplementary Table S1.
Study selection
Studies were included if they met the following criteria: 1) predicting specific ADEs or ADEs related to specific drugs, 2) applying ML algorithms based on EHRs, 3) providing sufficient explanations for research findings, and 4) publishing in English. The exclusion criteria were as follows: 1) studies focused on medical safety events rather than ADEs, 2) studies aimed at identifying and warning ADEs rather than predicting them, 3) studies that predict all ADEs in specific populations without sufficient specificity, 4) studies lacked a full-text version, 5) studies published in languages other than English, 6) studies that employed conventional algorithms, and 7) studies not based on EHR data.
Screening process and data extraction
After removing duplicate studies, two independent reviewers (QZ Hu and YX Chen) evaluated the titles and abstracts to exclude studies that did not meet the eligibility criteria. The full texts of the potentially eligible studies were assessed. Any disagreements were resolved by consensus. The data extracted included the author, year, database, demographic characteristics, number of patients with ADEs, used ML algorithms, evaluation and validation performance metrics [e.g., accuracy, sensitivity, specificity, precision, F1 score, area under the curve of the receiver operating characteristic curve (AUC), and area under the precision-recall Curve (AUCPRC)], and risk factors before and after screening.
Quality evaluation
Two reviewers independently assessed the quality of the included studies. Given that these retrospective studies employed medical artificial intelligence, we utilized two assessment tools to thoroughly evaluate both the appropriateness of the retrospective study design and the rigor of the artificial intelligence methodologies applied. The Agency for Healthcare Research and Quality (AHRQ) (AHQR, 2024) tool was used to evaluate the methodological quality and risk of bias in primary cross-sectional studies, while the Checklist for the Assessment of Medical AI (ChAMAI) (Cabitza and Campagner, 2021) was applied to assess the quality of artificial intelligence studies in the medical field. AHRQ tool consists of 11 items, each rated as “yes” or “no”, with corresponding scores of 1 or 0, respectively (AHQR, 2024). The maximum AHRQ score is 11 points. ChAMAI includes 30 items in six dimensions: problem understanding, data understanding, data preparation, modeling, validation, and deployment (Cabitza and Campagner, 2021). Each question is classified as high-priority (scored 0, 1, or 2) or low-priority (scored 0, 0.5, or 1), with a maximum possible score of 50 points (Cabitza and Campagner, 2021).
Statistical analysis
The effects and 95% confidence intervals (CIs) were estimated using random effects models. Pooled sensitivity, specificity, diagnostic odds ratios (DORs), and their respective 95% CIs were calculated based on contingency tables. Overall performance was assessed using the summary receiver operating characteristic (SROC) curve and the area under the SROC curve (AUC). Publication bias was evaluated using the Deek funnel plot test. Pooled sensitivity, specificity, and DORs were calculated using Meta-DiSc 2.0, while publication bias was analyzed with Stata 16.0 software.
Results
Characteristics of the included studies
The database search yielded 7,480 relevant studies. After removing duplicates, the titles and abstracts were screened for relevance. Subsequently, the full texts of 80 studies were reviewed, with 59 studies (Liu et al., 2018; Dong et al., 2019; Imai et al., 2019; Imai et al., 2020; Lai et al., 2020; Sun et al., 2020; Yoo et al., 2020; Heilbroner et al., 2021; Herrin et al., 2021; Kim W. et al., 2021; Kim J. S. et al., 2021; Lewinson et al., 2021; Satheeshkumar et al., 2021; Simon et al., 2021; Zhou et al., 2021; 2023; Chang et al., 2022; Huang et al., 2022; Huang et al., 2023; Li et al., 2022; Mu et al., 2022; Okawa et al., 2022; On et al., 2022; Venäläinen et al., 2022; Wu et al., 2022; Xu et al., 2022; Asai et al., 2023; Chambers et al., 2023; Chen et al., 2023; Cheng et al., 2023; Gong et al., 2023; Goyal et al., 2023; Güven et al., 2023; Heo et al., 2023; Hu et al., 2023; Jeong et al., 2023; Jiang et al., 2023; Lee et al., 2023; Liao et al., 2023; Lu et al., 2023; Ma J. et al., 2023; Ma X. et al., 2023; Mao et al., 2023; Maray et al., 2023; Mora et al., 2023; Ruiz Sarrias et al., 2023; Yan et al., 2023; Zhang et al., 2023; Chiu et al., 2024; Choi et al., 2024; Lippenszky et al., 2024; Matsumoto et al., 2024; Nguyen et al., 2024; Noda et al., 2024; Patel et al., 2024; Surendran et al., 2024; Xiao et al., 2024; Yagi et al., 2024; Zhao et al., 2024) included in the qualitative synthesis. Among the 59 studies included, 33 studies that provided extractable contingency tables could been included in the quantitative synthesis, as illustrated in Figure 1.
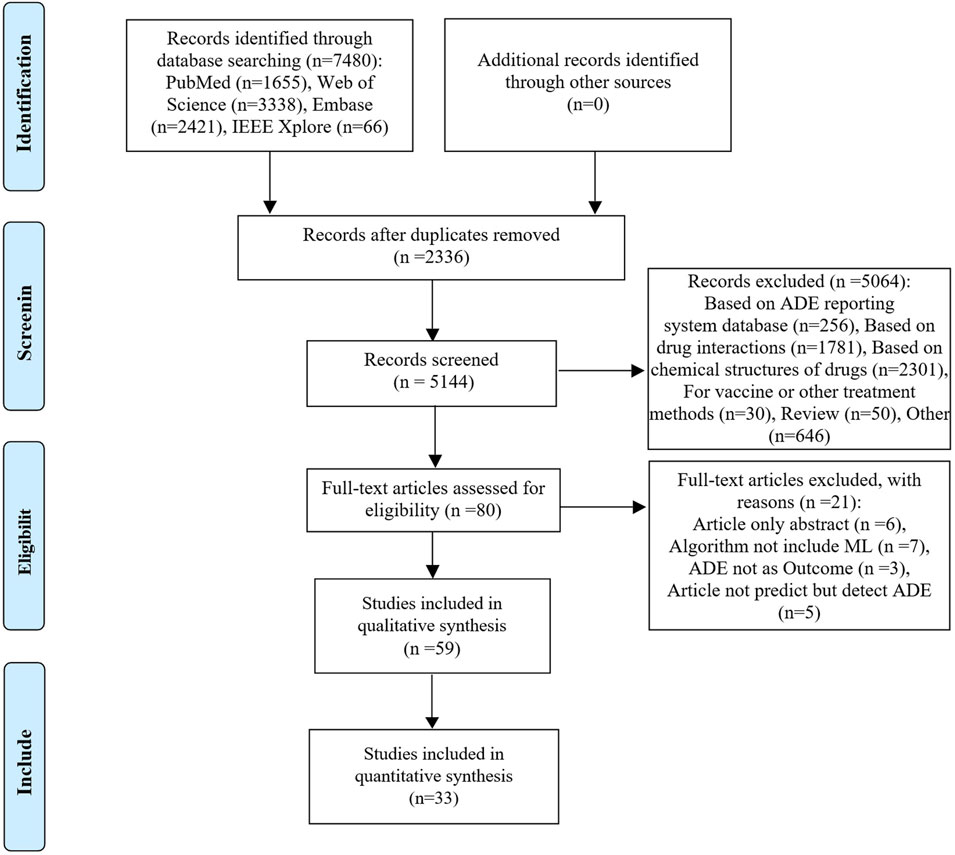
Figure 1. PRISMA flow diagram of the citation search and selection strategy. ADE: adverse drug event; PRISMA: Preferred Reporting Items for Systematic reviews and Meta-Analyses.
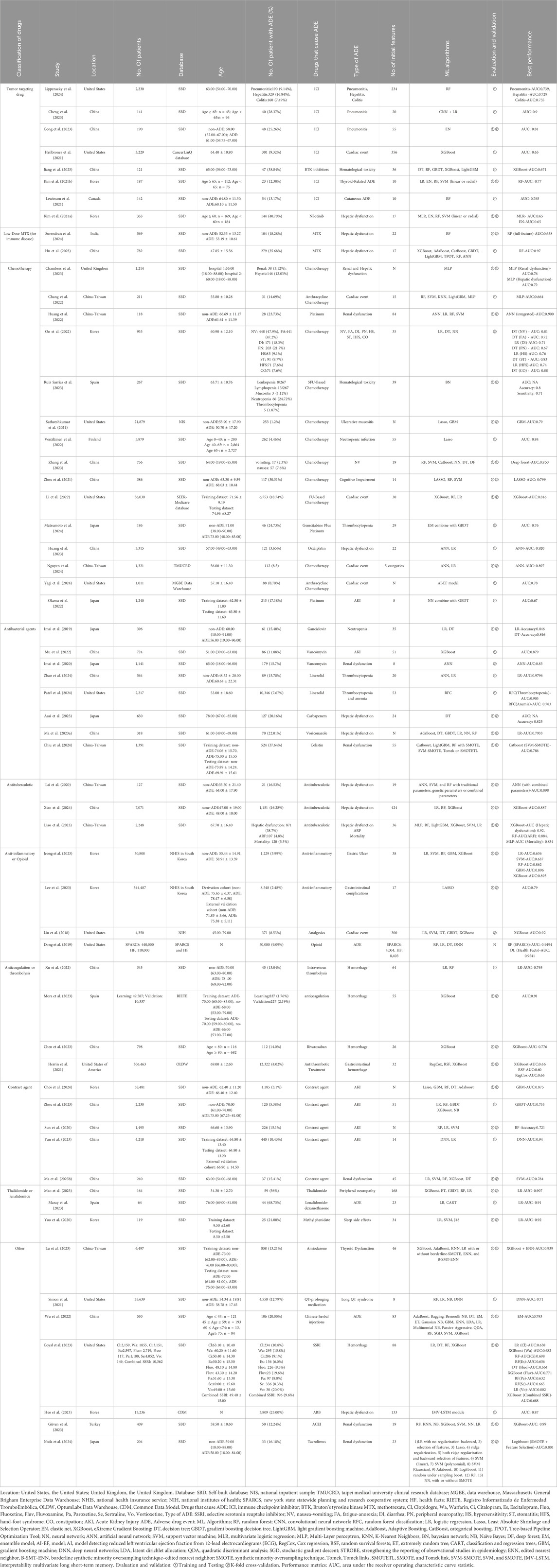
Most included studies were conducted in China (n = 19), followed by contributions from the United States, Korea, Taiwan, and Japan. These studies were published between 2018 and 2024, with most published in 2023. Ten studies were based on government or commercial databases from the United States, Korea, and Spain, while 49 used self-constructed databases based on hospital patient data. A total of 15 drug categories were identified, with most studies focusing on chemotherapy (n = 15), followed by tumor-targeting drugs, antibacterial agents, and contrast agents. ADEs in the included studies were classified into 15 categories, with the most predicted being renal dysfunction (n = 13), followed by hepatic dysfunction, gastrointestinal complications, cardiac events, and hematologic toxicity. 38 mL algorithms were reported, with random forest (RF) being the most frequently used, followed by support vector machine (SVM), eXtreme gradient boosting (XGBoost), decision tree (DT), and light gradient boosting machine (LightGBM). Additionally, logistic regression (LR), a traditional statistical method, was reported 37 times. Detailed information is provided in Table 1 and Figure 2.
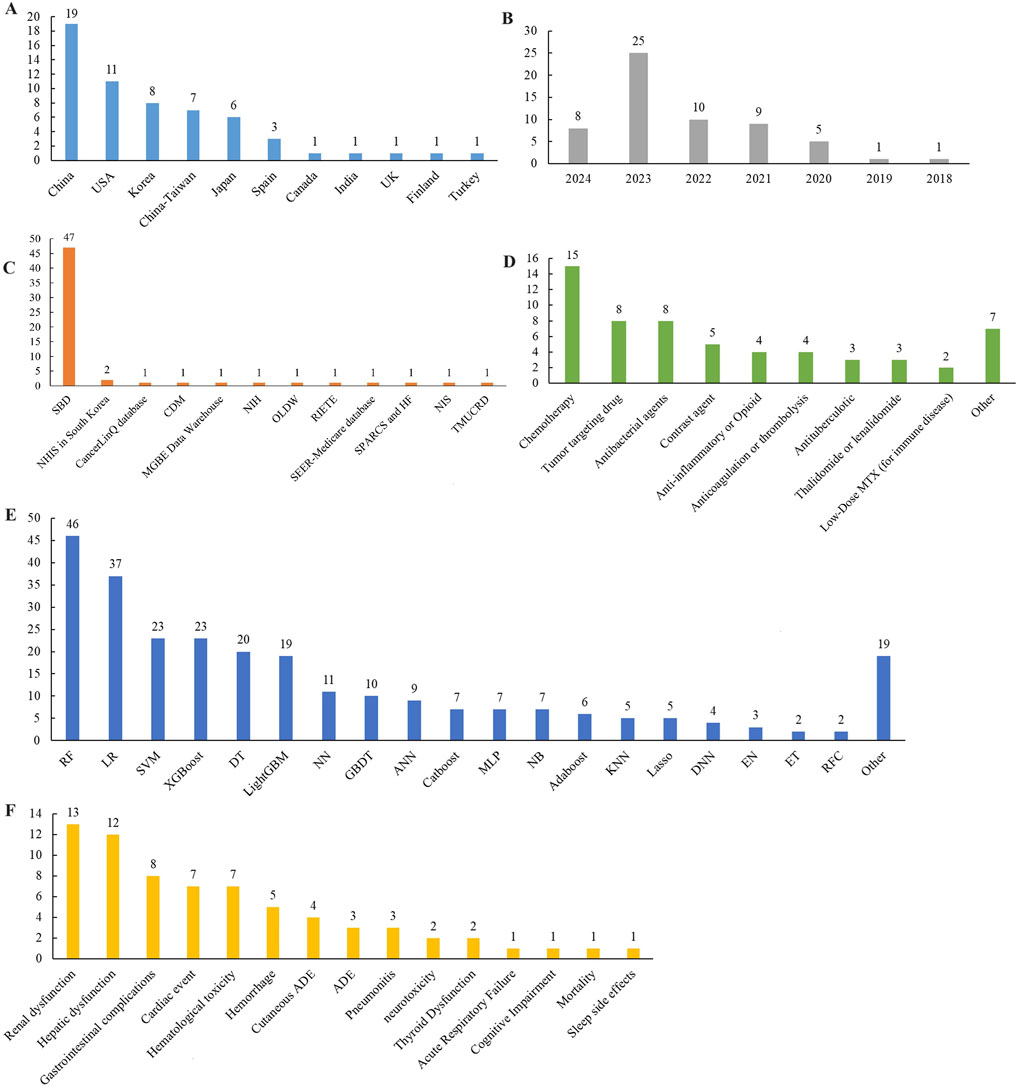
Figure 2. Distribution of included studies in (A) region, (B) publication year, (C) database, (D) medicine, (E) ML models, and (F) ADE. Location: United States, the United States, United Kingdom, the United Kingdom. Database: SBD, Self-built database; NIS, National Inpatient Sample; TMUCRD, Taipei Medical University Clinical Research Database; MGBE Data Warehouse, Massachusetts General Brigham Enterprise Data Warehouse; NHIS, National Health Insurance Service; NIH, National Institutes of Health; SPARCS, New York State Statewide Planning and Research Cooperative System; HF, Health Facts; RIETE, Registro Informatizado de Enfermedad TromboEmbólica; OLDW, OptumLabs Data Warehouse; CDM, Common Data Model. Drugs that cause ADE: MTX, methotrexate; ADE, Adverse drug event. ML Algorithms: RF, random forest; LR, Logistic regression; Lasso, Least Absolute Shrinkage and Selection Operator; EN, Elastic Net; XGBoost, eXtreme Gradient Boosting; DT, decision tree; GBDT, gradient boosting decision tree; LightGBM, light gradient boosting machine; AdaBoost, Adaptive Boosting, CatBoost, categorical boosting; NN, Neural network; ANN, Artificial Neural Network; SVM, Support vector machine; KNN, K-Nearest Neighbors; NB, Naïve Bayes; ET, extremely random tree; DNN, Deep neural networks; MLP, Multi-Layer perceptron; RFC, Random forest classification.
Among the 59 included studies, 6 (Sun et al., 2020; Satheeshkumar et al., 2021; Chambers et al., 2023; Ma J. et al., 2023; Choi et al., 2024; Yagi et al., 2024) did not provide any information on initial features, while 53 reported initial features associated with ADEs. Of these, 46 studies detailed the types and the number of initial features, while 7 studies (Dong et al., 2019; Heilbroner et al., 2021; Li et al., 2022; Güven et al., 2023; Zhou et al., 2023; Matsumoto et al., 2024; Xiao et al., 2024) reported only the number of initial features. The initial number of features used to build the models ranged from 8 to 8,403. The commonly identified features included demographic data, treatment information, laboratory results, and disease history. Additionally, genes, plasma concentration, and pharmacokinetics were considered risk factors in some studies (Lai et al., 2020; Huang et al., 2022; Ruiz Sarrias et al., 2023). Of the 59 included studies, 47 reported conducting feature selection to identify the most important predictors for their models, while 10 did not perform feature screening. Two studies (Chambers et al., 2023; Ma J. et al., 2023) did not provide details on initial or significant features. Further details are provided in Supplementary Table S2.
Evaluating the quality of studies
The risk of bias assessment using the AHRQ tool indicated that the overall quality of the studies was generally high, with an average score of 9.254 points. Among the included studies, 30 scored above 10 points, 28 scored between 7 and 9 points, and one study (Lewinson et al., 2021) scored 4 points. Items 8 and 11 had the lowest average scores, 0.424 and 0.322 points, respectively, while the average scores for the other items were considerably higher. Detailed information is provided in Supplementary Table S3.
According to the ChAMAI checklist, the overall average score of the included studies was 32.48, with scores ranging from 22.00 to 40.00 (Supplementary Table S4; Supplementary Figure S1). Of the included studies, 39 scored higher than 30 points, 19 scored between 25 and 29 points, and one study (Lewinson et al., 2021) scored below 25. Regarding specific categories, the highest scoring rates were observed in modeling and problem understanding, at 100% and 99.5%, respectively, while data preparation had the lowest scoring rate, at 37.25%.
Predictive performance for ML methods
AUC was a critical metric to assess model performance. Fifty-four studies reported AUC values, with an average AUC of 76.68% ± 10.73, indicating generally favorable performance in all models evaluated. Artificial neural networks (ANN), gradient boosting machines (GBM), CatBoost, and XGBoost demonstrated high AUC values, with average values exceeding 80%. The RF algorithm, though the most frequently reported, had a slightly lower average AUC of 77.00% ± 7.83. The impact of feature selection on model performance was examined in three studies (Sacerdote, 2008; Goyal et al., 2023; Noda et al., 2024), which showed that, in most cases, feature selection improved model performance.
In addition to AUC, model performance was evaluated using several other metrics: accuracy (76.00% ± 11.26), precision (60.13% ± 24.81), sensitivity (62.35% ± 20.19), specificity (75.13% ± 16.60), and F1 score (52.60% ± 21.10). Detailed metrics for the machine learning algorithms are presented in Figure 3 and Supplementary Table S5.
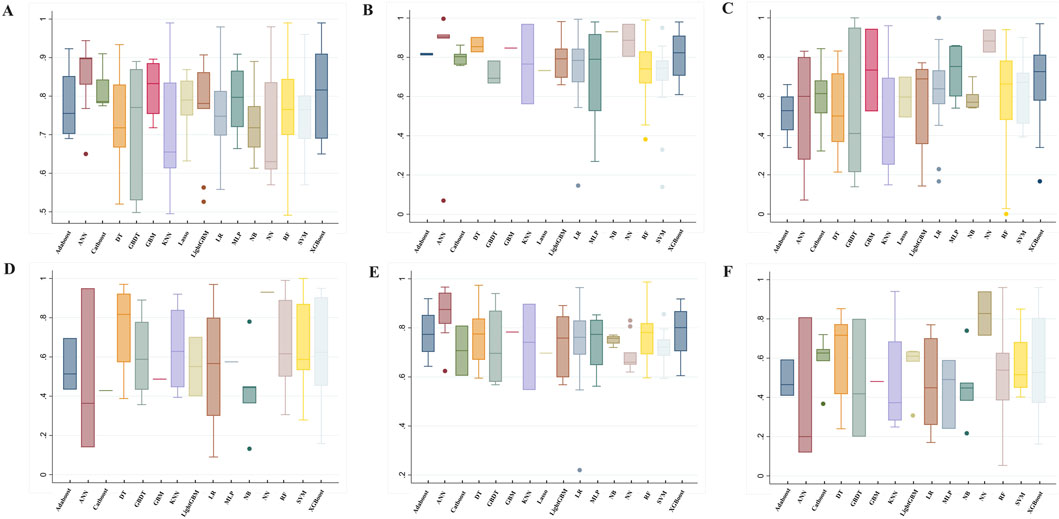
Figure 3. Summary of prediction model performance. (A) AUC, (B) Specificity, (C) Sensitivity (recall), (D) Precision, (E) Accuracy, (F) F1 ML Algorithms: RF, random forest; LR, Logistic regression; Lasso, Least Absolute Shrinkage and Selection Operator; XGBoost, eXtreme Gradient Boosting; DT, decision tree; GBDT, gradient boosting decision tree; LightGBM, light gradient boosting machine; AdaBoost, Adaptive Boosting; CatBoost, categorical boosting; NN, Neural network; ANN, Artificial Neural Network; SVM, Support vector machine; KNN, K-Nearest Neighbors; NB, Naïve Bayes; GBM, gradient boosting machine; MLP, Multi-Layer perceptron.
Meta-regression
Pooled analysis
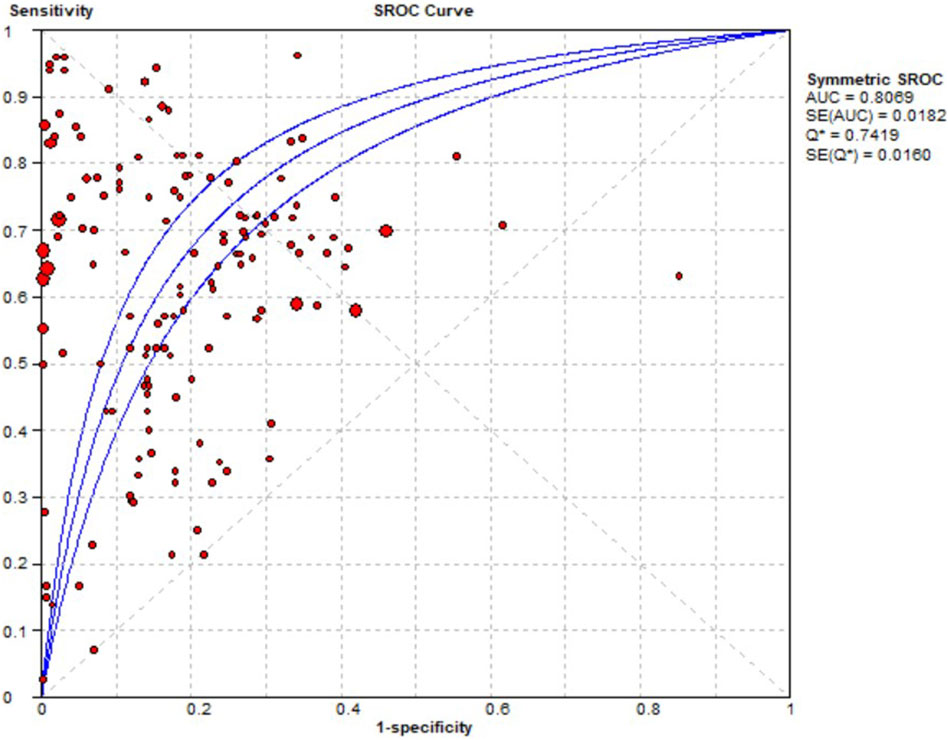
Contingency tables from 33 prediction studies (Liu et al., 2018; Dong et al., 2019; Sun et al., 2020; Yoo et al., 2020; Herrin et al., 2021; Li et al., 2022; Okawa et al., 2022; Wu et al., 2022; Chen et al., 2023; Cheng et al., 2023; Gong et al., 2023; Goyal et al., 2023; Güven et al., 2023; Heo et al., 2023; Huang et al., 2023; Hu et al., 2023; Jeong et al., 2023; Jiang et al., 2023; Lee et al., 2023; Liao et al., 2023; Lu et al., 2023; Ma X. et al., 2023; Mao et al., 2023; Mora et al., 2023; Ruiz Sarrias et al., 2023; Zhou et al., 2023; Chiu et al., 2024; Lippenszky et al., 2024; Matsumoto et al., 2024; Nguyen et al., 2024; Patel et al., 2024; Surendran et al., 2024; Xiao et al., 2024) were extracted, which included 13 ADEs, 33 ML models, and 15 drugs. Significant heterogeneity was detected in the combined results for sensitivity (I2 = 99.1%, p = 0.00), specificity (I2 = 100%, p = 0.00), and DOR (I2 = 99.8%, p = 0.00). Consequently, a random effects model was used to evaluate the prediction accuracy of ADEs. The combined sensitivity, specificity, DOR, and AUC were 0.65 (95% CI: 0.65–0.66), 0.89 (95% CI: 0.89–0.90), 12.11 (95% CI: 8.17–17.95), and 0.8069, respectively. These results indicate that the prediction accuracy for ADEs is relatively robust. The AUC of the SROC curve is shown in Figure 4.
Subgroup analysis and heterogeneity
A subgroup analysis was conducted to assess the predictive performance of ML across different ADEs. The results indicated that ML demonstrated superior performance in detecting peripheral neuropathy and acute respiratory failure compared to other ADEs. Specifically, for peripheral neuropathy, the sensitivity was 0.85 (95% CI: 0.83–0.93), specificity was 0.75 (95% CI: 0.65–0.84), the DOR was 18.13 (95% CI: 7.05–46.63), and the AUC was 0.8794. For acute respiratory failure, sensitivity was 0.78 (95% CI: 0.72–0.84), specificity was 0.80 (95% CI: 0.78–0.81), the DOR was 13.00 (95% CI: 9.23–18.31), and AUC was 0.8561.
A subgroup analysis comparing the performance of different models showed that the pooled sensitivity for LR was moderate at 0.59 (95% CI: 0.58–0.59). The performance of GBM and GBDT appeared strong, with pooled sensitivities of 0.92 (95% CI: 0.88–0.95) and 0.81 (95% CI: 0.77–0.84), respectively. Heterogeneity decreased somewhat when analyzing the same ADEs or ML models, as shown in Supplementary Figures S2–S5 and Supplementary Table S6.
Publication bias
The Deek funnel plot (Figure 5) was used to assess the presence of publication bias in the included studies. The results did not indicate significant publication bias in the studies included in this meta-analysis (p = 0.00).
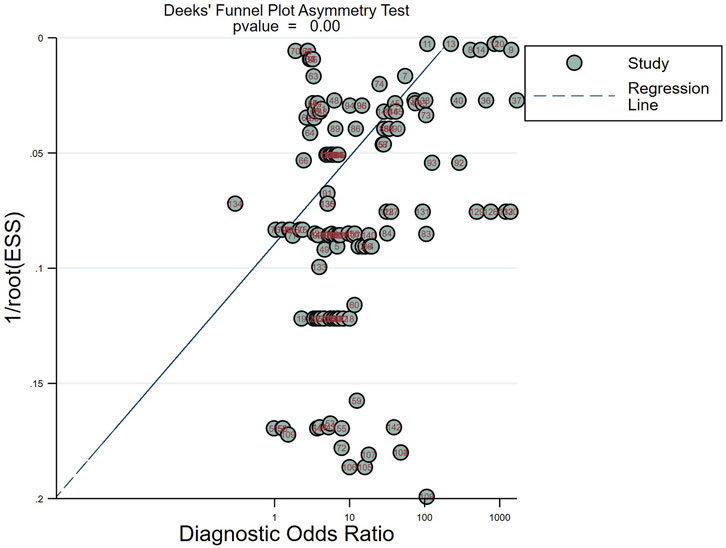
Figure 5. Deeks’ funnel plots for the assessment of potential bias in the meta-analysis for diagnosis.
Discussion
We present the first comprehensive systematic review and meta-analysis assessing the performance of ML models in predicting ADEs based on EHRs. This review demonstrates that ML algorithms can complement traditional clinical decision-making in ADE prediction. The included studies were of high quality, and the applications of ML were diverse, covering a wide range of ADEs.
ML has been applied to predict ADEs for various drugs, with antitumor drugs being the most frequently studied. A total of 23 studies (15 on chemotherapy and 8 on tumor-targeting drugs) reported predictive outcomes related to ADEs associated with antitumor medications. Cancer remains the leading cause of death worldwide, and the necessity for anti-cancer drugs contributes significantly to the financial burden on individuals and healthcare systems. Cytotoxic chemotherapy remains the gold standard and first-line treatment for many common cancers (Schirrmacher, 2019). Chemotherapy-related drug toxicity may also occur more frequently than with other drugs, further increasing the economic burden (Livshits et al., 2014). Our findings indicated that renal dysfunction, cardiac events, and gastrointestinal events were the most reported ADEs in predictive model studies, aligning with the common ADEs associated with chemotherapy drugs. The excretion of chemotherapeutic metabolites through the urinary system can lead to nephrotoxicity, which varies depending on the type of chemotherapy, the malignancy being treated, the patient’s age, and the underlying conditions (Sahni et al., 2009). Many chemotherapy agents, particularly platinum coordination complexes, are known to cause kidney damage (Madias and Harrington, 1978). And nephrotoxicity has emerged as a significant limiting factor in the therapeutic use of these compounds. Among the included studies, multi-layer perceptron (MLP), ANN, and GBDT were used to develop prediction models for chemotherapy-related renal dysfunction, demonstrating good performance with AUC values ranging from 0.67 to 0.90 (Huang et al., 2022; Okawa et al., 2022; Chambers et al., 2023). Factors such as age, sex, body surface area, serum creatinine levels, and genetic predispositions may influence the occurrence of chemotherapy-related renal dysfunction (Huang et al., 2022; Okawa et al., 2022).
The included studies also frequently reported cardiotoxicity associated with chemotherapy, a life-threatening side effect (Chang et al., 2022; Li et al., 2022; Nguyen et al., 2024; Yagi et al., 2024). Anthracyclines were identified as the main agents responsible for chemotherapy-induced cardiotoxicity (Chang et al., 2022; Yagi et al., 2024). These drugs have been widely used and continue to serve as the cornerstone of treatment for multiple solid tumors and hematologic malignancies (Henriksen, 2018). The predictive capacity of the ML models for chemotherapy-related cardiac events showed AUC values ranging from 0.66 to 0.90 (Chang et al., 2022; Li et al., 2022; Nguyen et al., 2024; Yagi et al., 2024). The results indicated that, in addition to aging and male sex, pre-existing heart disease was a significant risk factor. This included conditions such as hypertension, reduced left ventricular ejection fraction, ischemic heart disease, arrhythmia, and coronary artery disease (Chang et al., 2022; Li et al., 2022; Nguyen et al., 2024; Yagi et al., 2024). In addition to cardiotoxicity, gastrointestinal events were frequently reported as side effects of chemotherapy. These events, including nausea, vomiting, ulcerative mucositis, and constipation, can significantly impact patient quality of life, although they are generally not life-threatening. The AUC values for ML in predicting gastrointestinal events were promising, with nausea and vomiting ranging from 0.81 to 0.85, ulcerative mucositis from 0.79 to 0.83, and constipation at 0.88 (Satheeshkumar et al., 2021; On et al., 2022; Zhang et al., 2023). Factors contributing to nausea, vomiting, and constipation included the number of treatment cycles, line of treatment, reduction in chemotherapy doses, chemotherapy regimens, and creatinine clearance (Satheeshkumar et al., 2021; On et al., 2022; Zhang et al., 2023). For ulcerative mucositis, strategies to mitigate risk included addressing conditions such as pancytopenia, agranulocytosis, fluid and electrolyte imbalances, and chemotherapy-induced anemia (Satheeshkumar et al., 2021).
Recently, novel targeted cancer therapies have emerged, designed to block biological transduction pathways and/or target specific cancer proteins to induce cancer cell death through apoptosis, immune system stimulation, or precisely delivering chemotherapeutic agents to cancer cells (Pérez-Herrero and Fernández-Medarde, 2015). Although targeted therapies reduce specific undesirable side effects compared to traditional chemotherapy, they can still lead to specific ADEs. Among the studies reviewed, immune checkpoint inhibitors (ICIs) were the most reported, with immune-related adverse events (irAEs) being the main associated ADE. Previous research indicates that irAEs affect 20%–30% of patients undergoing ICI monotherapy and more than 50% of those receiving combination ICI therapies (Martins et al., 2019; Gumusay et al., 2022). Pneumonitis was commonly reported in included studies, with occurrence rates exceeding 25%. The AUC of the ML prediction models ranged from 0.74 to 0.90 (Cheng et al., 2023; Gong et al., 2023; Lippenszky et al., 2024). Other irAEs associated with ICIs included hepatitis, colitis, thyroid disorders, cutaneous reactions, and cardiac irAEs, with occurrence rates around 10% and AUC values for ML prediction models ranging from 0.65 to 0.77 (Heilbroner et al., 2021; Kim W. et al., 2021; Lewinson et al., 2021; Lippenszky et al., 2024). These irAEs were particularly prevalent among patients with pre-existing underlying conditions, and an increase in inflammatory cell counts (lymphocytes, neutrophils, or eosinophils) was often observed in these patients (Heilbroner et al., 2021; Kim W. et al., 2021; Lewinson et al., 2021; Gong et al., 2023; Lippenszky et al., 2024).
Antibacterial agents are commonly used in inpatient settings, and approximately 50% of hospitalized patients receive at least one antibiotic during their stay (Tamma et al., 2017). Common antimicrobials-associated ADEs include gastrointestinal, renal, hepatic, and hematologic abnormalities (Tamma et al., 2017). Among the included studies, ML models were developed to predict hepatic (Asai et al., 2023; Ma J. et al., 2023) and renal (Imai et al., 2020; Mu et al., 2022; Chiu et al., 2024) dysfunction, thrombocytopenia (Patel et al., 2024; Zhao et al., 2024), anemia (Patel et al., 2024), and neutropenia (Imai et al., 2019). Antibacterial-related liver and kidney dysfunction were more likely to occur in patients with abnormal blood concentrations of the drugs or those with pre-existing liver and kidney diseases (Imai et al., 2020; Mu et al., 2022; Asai et al., 2023; Ma J. et al., 2023; Chiu et al., 2024). Additionally, antibacterial-related thrombocytopenia and anemia were associated with baseline platelet counts, international normalized ratio, hemoglobin levels, and overall liver and kidney function (Patel et al., 2024; Zhao et al., 2024). Ganciclovir-related neutropenia was common among patients with impaired liver and kidney function, those who received prolonged or high doses of ganciclovir, and individuals with low body weight (Imai et al., 2019).
Antituberculosis drugs, a specific class of antibacterial agents, were associated with ADEs in three of the included studies (Lai et al., 2020; Liao et al., 2023; Xiao et al., 2024). Antituberculosis drug-induced hepatotoxicity is a serious ADE linked to first-line tuberculosis medications such as rifampin, isoniazid, and pyrazinamide (Tostmann et al., 2008). This condition often requires the temporary discontinuation of these drugs to prevent progression to fulminant liver disease or even death. In predictive modeling, the AUC values for ML models that assess the risk of antituberculosis drug-induced hepatotoxicity ranged from 0.89 to 0.90, indicating high accuracy in predicting which patients may be at risk (Lai et al., 2020; Liao et al., 2023; Xiao et al., 2024). Patients with a history of abnormal liver function, underlying liver disease, and elevated liver enzymes had a higher risk of developing liver damage induced by antituberculosis drugs (Lai et al., 2020; Liao et al., 2023; Xiao et al., 2024). Genetic factors also played a significant role, and polymorphisms such as NAT27, OATP1B11a/1a, OATP1B11a/15, and UGT1A127/*28 contributed to the risk (Lai et al., 2020). This information can help clinicians better manage treatment plans and monitor patients more effectively to mitigate the risk of liver damage.
Five studies reported contrast-induced nephropathy (CIN), a common complication arising from intravascular use of contrast media during arterial angiography, with an incidence rate ranging from 3% to 15% (Sun et al., 2020; Ma X. et al., 2023; Yan et al., 2023; Zhou et al., 2023; Choi et al., 2024). The incidence of CIN continues to increase, making it the third leading cause of hospital-acquired acute renal injury (Mehran et al., 2004). While typically transient, CIN significantly increases the need for dialysis among affected patients, with a poor prognosis that often leads to prolonged hospital stays and elevated mortality rates (Subramanian et al., 2007; Ma X. et al., 2023). Commonly reported ML models to predict CIN include RF, XGBoost, GBDT, and SVM, with AUC values ranging from 0.72 to 0.94 (Sun et al., 2020; Ma X. et al., 2023; Yan et al., 2023; Zhou et al., 2023; Choi et al., 2024). CIN was more likely to occur in patients with pre-existing kidney dysfunction, including chronic kidney disease, elevated serum creatinine levels, impaired creatinine clearance, and abnormal blood pressure (Sun et al., 2020; Ma X. et al., 2023; Yan et al., 2023; Zhou et al., 2023; Choi et al., 2024).
The application of ML algorithms in predicting ADEs induced by various medications, including anti-inflammatory or opioid drugs, anticoagulants or thrombolytics, antituberculosis agents, thalidomide or lenalidomide, and methotrexate, showed promising results. The AUC for these ML prediction models typically ranged from 0.75 to 0.95, indicating strong performance in ADE prediction. A total of 38 different ML algorithms were identified in the studies reviewed, with ensemble learning techniques being the most prevalent. Ensemble learning combines predictions from multiple weak learners to enhance predictive accuracy (Ahn et al., 2023). The primary ensemble learning methods include the bagging and boosting algorithms. RF, a classic bagging algorithm, was the most frequently reported in 30 studies. RF aggregates predictions from multiple decision trees, each built from a bootstrapped version of the training data set (Hu and Szymczak, 2023). Among the studies using RF, seven identified it as the optimal model, with an average AUC of 0.770 ± 0.104 and a sensitivity of 0.630 ± 0.215. Unlike bagging, where each weak learner operates in parallel, boosting algorithms train weak learners sequentially, with each iteration focusing on misclassified instances from the previous round (González-Recio et al., 2013). Boosting algorithms appeared in 29 studies, including models such as XGBoost, LightGBM, GBDT, AdaBoost, and CatBoost. XGBoost was frequently highlighted as the optimal model, achieving an average AUC of 0.810 ± 0.109 and a sensitivity of 0.680 ± 0.212. Other ML models, such as SVM, ANN, and K-nearest neighbors (KNN), were also reported; however, their performance did not surpass that of RF and boosting algorithms. These findings show the effectiveness of ML algorithms, particularly ensemble methods, in predicting ADEs associated with various medications.
Class-imbalanced data introduce bias, particularly pronounced in high-dimensional datasets (Blagus and Lusa, 2013). T This issue can be mitigated through resampling methods that generate class-balanced data. The resampling methods reported include oversampling techniques such as the Synthetic Minority Oversampling Technique (SMOTE), SVM-SMOTE, undersampling with Tomek links (Tomek), and a combination of oversampling and undersampling methods using SMOTE and Tomek links (SMOTETL) (Chiu et al., 2024). Imbalanced data analysis was discussed in 23 studies, three of which specifically compared the performance of models with and without resampling methods (Lu et al., 2023; Chiu et al., 2024; Noda et al., 2024). The results indicated that the boosting algorithms demonstrated strong performance when combined with the imbalance analysis. The AUC values improved significantly, increasing from 0.601–0.936 to 0.786–0.939 (Lu et al., 2023; Chiu et al., 2024; Noda et al., 2024). Additionally, whether imbalance analysis was applied was one of the evaluation criteria on the ChAMAI checklist. The findings also indicated that feature selection could improve the model performance. Therefore, we recommend the application of imbalance analysis and feature selection in developing ADE prediction models to reduce bias and improve efficiency.
The quality of the included studies was assessed as high based on the AHRQ tool and the ChAMAI. According to the AHRQ tool, all items were considered equally important (AHQR, 2024). The results showed that the included studies often did not score on the items related to the control of confounding factors and follow-up procedures. In the ChAMAI, the 30 items were categorized into 10 low-priority and 20 high-priority items (Cabitza and Campagner, 2021). As a tool for evaluating the rigor of artificial intelligence methodologies, this tool places greater emphasis on the quality of the data processing procedures and the model-building process. Key aspects of data processing include outlier detection, handling missing values, feature pre-processing, and addressing data imbalance. The model-building process involves reporting tasks, specifying outputs, detailing model architecture, data splitting, training, selection, calibration, and validation procedures. Sharing code and data is also a high-priority item, as it enhances transparency and adaptability. While studies with high scores had good data processing procedures and model building, there remains room for improvement in code and data sharing.
Limitations
This study has several limitations. First, although the overall quality of the included studies was high, there remains room for improvement. Specifically, items 8 and 11 of the AHRQ criteria received the lowest average scores, indicating the need for better control of confounding factors and better follow-up procedures. Additionally, the ChAMAI results suggested that improvements are needed in outlier detection and analysis and in code and data sharing. Furthermore, only two studies demonstrated the development and implementation of prediction systems in real-world applications (Jiang et al., 2023; Ruiz Sarrias et al., 2023). The limited practical application of these models may hinder both the studies’ quality and the models’ development. Second, significant heterogeneity was observed among the included studies. Although heterogeneity was somewhat reduced within the same ADE category, it was difficult to avoid completely. The variability was due to differences in the databases used, predictors, ML algorithms, hyperparameters, and populations studied, making it challenging to fully mitigate (Xie et al., 2022). Third, meta-regression was only conducted on the 33 studies that provided contingency tables, limiting our analysis. For the remaining studies, we only performed descriptive analyses of performance metrics. Fourth, we did not include studies that predict all ADEs in specific populations. We believe that the primary aim of these studies was to examine the epidemiology of ADEs within those populations, rather than to identify precise risk factors. Therefore, we recommend that these studies be comprehensively analyzed in a separate study. Finally, the included studies covered 15 different ADEs, 15 drug types, and 38 mL models, complicating the synthesis of results in these diverse studies.
Conclusion
This systematic review and meta-analysis summarized the current research on using machine learning to predict ADEs, focusing on oncology-related treatments, including chemotherapy and tumor-targeting drugs. Among the ADEs analyzed, drug-related liver and kidney dysfunction was the most predicted due to its high incidence and potential to cause treatment interruptions. The study found that ML methods, particularly boosting algorithms such as XGBoost, GBM, and GBDT, effectively predict ADEs. Given the variability between studies, there is a need for more standardized research on different ADEs to ensure the accuracy and robustness of these predictive tools. Future research should prioritize multicenter studies incorporating diverse data types and evaluate the impact of artificial intelligence predictive models in real-world clinical settings.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
QH: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. YC: Methodology, Writing–original draft. DZ: Data curation, Writing–original draft. ZH: Methodology, Validation, Writing–review and editing. TX: Conceptualization, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Sichuan Science and Technology Support Program (grant number: 2023NSFSC1696), and the Science and Technology project of Chengdu Health Commission (grant number: 2022020). This research was supported by National Key Clinical Specialties Construction Program.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1497397/full#supplementary-material
References
Adel, N. (2017). Overview of chemotherapy-induced nausea and vomiting and evidence-based therapies. Am. J. Manag. Care 23, S259–S265.
Ahn, J. M., Kim, J., and Kim, K. (2023). Ensemble machine learning of gradient boosting (XGBoost, LightGBM, CatBoost) and attention-based CNN-LSTM for harmful algal blooms forecasting. Toxins (Basel) 15, 608. doi:10.3390/toxins15100608
AHQR (2024). “Methods guide for effectiveness and comparative effectiveness reviews,” in Methods guide for effectiveness and comparative effectiveness reviews. Available at: https://effectivehealthcare.ahrq.gov/sites/default/files/pdf/cer-methods-guide_overview.pdf.
Asai, Y., Ooi, H., and Sato, Y. (2023). Risk evaluation of carbapenem-induced liver injury based on machine learning analysis. J. Infect. Chemother. 29, 660–666. doi:10.1016/j.jiac.2023.03.007
Bae, J.-H., Baek, Y.-H., Lee, J.-E., Song, I., Lee, J.-H., and Shin, J.-Y. (2020). Machine learning for detection of safety signals from spontaneous reporting system data: example of nivolumab and docetaxel. Front. Pharmacol. 11, 602365. doi:10.3389/fphar.2020.602365
Bates, D. W., Boyle, D. L., Vander Vliet, M. B., Schneider, J., and Leape, L. (1995). Relationship between medication errors and adverse drug events. J. Gen. Intern Med. 10, 199–205. doi:10.1007/BF02600255
Blagus, R., and Lusa, L. (2013). SMOTE for high-dimensional class-imbalanced data. BMC Bioinforma. 14, 106. doi:10.1186/1471-2105-14-106
Cabitza, F., and Campagner, A. (2021). The need to separate the wheat from the chaff in medical informatics: introducing a comprehensive checklist for the (self)-assessment of medical AI studies. Int. J. Med. Inf. 153, 104510. doi:10.1016/j.ijmedinf.2021.104510
Chambers, P., Watson, M., Bridgewater, J., Forster, M. D., Roylance, R., Burgoyne, R., et al. (2023). Personalising monitoring for chemotherapy patients through predicting deterioration in renal and hepatic function. Cancer Med. 12, 17856–17865. doi:10.1002/cam4.6418
Chang, W.-T., Liu, C.-F., Feng, Y.-H., Liao, C.-T., Wang, J.-J., Chen, Z.-C., et al. (2022). An artificial intelligence approach for predicting cardiotoxicity in breast cancer patients receiving anthracycline. Arch. Toxicol. 96, 2731–2737. doi:10.1007/s00204-022-03341-y
Chen, C., Yin, C., Wang, Y., Zeng, J., Wang, S., Bao, Y., et al. (2023). XGBoost-based machine learning test improves the accuracy of hemorrhage prediction among geriatric patients with long-term administration of rivaroxaban. BMC Geriatr. 23, 418. doi:10.1186/s12877-023-04049-z
Cheng, M., Lin, R., Bai, N., Zhang, Y., Wang, H., Guo, M., et al. (2023). Deep learning for predicting the risk of immune checkpoint inhibitor-related pneumonitis in lung cancer. Clin. Radiol. 78, e377–e385. doi:10.1016/j.crad.2022.12.013
Chiu, L.-W., Ku, Y.-E., Chan, F.-Y., Lie, W.-N., Chao, H.-J., Wang, S.-Y., et al. (2024). Machine learning algorithms to predict colistin-induced nephrotoxicity from electronic health records in patients with multidrug-resistant Gram-negative infection. Int. J. Antimicrob. Agents 64, 107175. doi:10.1016/j.ijantimicag.2024.107175
Choi, H., Choi, B., Han, S., Lee, M., Shin, G.-T., Kim, H., et al. (2024). Applicable machine learning model for predicting contrast-induced nephropathy based on pre-catheterization variables. Intern Med. 63, 773–780. doi:10.2169/internalmedicine.1459-22
Deo, R. C. (2015). Machine learning in medicine. Circulation 132, 1920–1930. doi:10.1161/CIRCULATIONAHA.115.001593
Dmitriev, A., Filimonov, D., Lagunin, A., Karasev, D., Pogodin, P., Rudik, A., et al. (2019). Prediction of severity of drug-drug interactions caused by enzyme inhibition and activation. Molecules 24, 3955. doi:10.3390/molecules24213955
Dong, X., Rashidian, S., Wang, Y., Hajagos, J., Zhao, X., Rosenthal, R. N., et al. (2019). Machine learning based opioid overdose prediction using electronic health records. AMIA Annu. Symp. Proc. 2019, 389–398.
Food and Drug Administration (FDA) (2024). Preventable adverse drug reactions: a focus on drug interactions. Available at: https://www.fda.gov/drugs/drug-interactions-labeling/preventable-adverse-drug-reactions-focus-drug-interactions.
Gong, L., Gong, J., Sun, X., Yu, L., Liao, B., Chen, X., et al. (2023). Identification and prediction of immune checkpoint inhibitors-related pneumonitis by machine learning. Front. Immunol. 14, 1138489. doi:10.3389/fimmu.2023.1138489
González-Recio, O., Jiménez-Montero, J. A., and Alenda, R. (2013). The gradient boosting algorithm and random boosting for genome-assisted evaluation in large data sets. J. Dairy Sci. 96, 614–624. doi:10.3168/jds.2012-5630
Goyal, J., Ng, D. Q., Zhang, K., Chan, A., Lee, J., Zheng, K., et al. (2023). Using machine learning to develop a clinical prediction model for SSRI-associated bleeding: a feasibility study. BMC Med. Inf. Decis. Mak. 23, 105. doi:10.1186/s12911-023-02206-3
Gumusay, O., Callan, J., and Rugo, H. S. (2022). Immunotherapy toxicity: identification and management. Breast Cancer Res. Treat. 192, 1–17. doi:10.1007/s10549-021-06480-5
Güven, A. T., Özdede, M., Şener, Y. Z., Yıldırım, A. O., Altıntop, S. E., Yeşilyurt, B., et al. (2023). Evaluation of machine learning algorithms for renin-angiotensin-aldosterone system inhibitors associated renal adverse event prediction. Eur. J. Intern Med. 114, 74–83. doi:10.1016/j.ejim.2023.05.021
Hartigan-Go, K., and Wong, J. (2000). “Inclusion of therapeutic failures as adverse drug reactions,” in Side effects of drugs annual.
Heilbroner, S. P., Few, R., Mueller, J., Chalwa, J., Charest, F., Suryadevara, S., et al. (2021). Predicting cardiac adverse events in patients receiving immune checkpoint inhibitors: a machine learning approach. J. Immunother. Cancer 9, e002545. doi:10.1136/jitc-2021-002545
Henriksen, P. A. (2018). Anthracycline cardiotoxicity: an update on mechanisms, monitoring and prevention. Heart 104, 971–977. doi:10.1136/heartjnl-2017-312103
Heo, S., Yu, J. Y., Kang, E. A., Shin, H., Ryu, K., Kim, C., et al. (2023). Development and verification of time-series deep learning for drug-induced liver injury detection in patients taking angiotensin II receptor blockers: a multicenter distributed research network approach. Healthc. Inf. Res. 29, 246–255. doi:10.4258/hir.2023.29.3.246
Herrin, J., Abraham, N. S., Yao, X., Noseworthy, P. A., Inselman, J., Shah, N. D., et al. (2021). Comparative effectiveness of machine learning approaches for predicting gastrointestinal bleeds in patients receiving antithrombotic treatment. JAMA Netw. Open 4, e2110703. doi:10.1001/jamanetworkopen.2021.10703
Horrigan, J., Bhumi, S., Miller, D., Jafri, M., and Tadros, M. (2023). Management of opioid-induced constipation in older adults. J. Clin. Gastroenterol. 57, 39–47. doi:10.1097/MCG.0000000000001801
Hu, J., and Szymczak, S. (2023). A review on longitudinal data analysis with random forest. Brief. Bioinform 24, bbad002. doi:10.1093/bib/bbad002
Hu, Q., Wang, H., and Xu, T. (2023). Predicting hepatotoxicity associated with low-dose methotrexate using machine learning. J. Clin. Med. 12, 1599. doi:10.3390/jcm12041599
Huang, R., Cai, Y., He, Y., Yu, Z., Zhao, L., Wang, T., et al. (2023). Predictive model of oxaliplatin-induced liver injury based on artificial neural network and logistic regression. J. Clin. Transl. Hepatol. 11, 1455–1464. doi:10.14218/JCTH.2023.00399
Huang, S.-H., Chu, C.-Y., Hsu, Y.-C., Wang, S.-Y., Kuo, L.-N., Bai, K.-J., et al. (2022). How platinum-induced nephrotoxicity occurs? Machine learning prediction in non-small cell lung cancer patients. Comput. Methods Programs Biomed. 221, 106839. doi:10.1016/j.cmpb.2022.106839
Imai, S., Takekuma, Y., Kashiwagi, H., Miyai, T., Kobayashi, M., Iseki, K., et al. (2020). Validation of the usefulness of artificial neural networks for risk prediction of adverse drug reactions used for individual patients in clinical practice. Plos One 15, e0236789. doi:10.1371/journal.pone.0236789
Imai, S., Yamada, T., Kasashi, K., Ishiguro, N., Kobayashi, M., and Iseki, K. (2019). Construction of a flow chart-like risk prediction model of ganciclovir-induced neutropaenia including severity grade: a data mining approach using decision tree. J. Clin. Pharm. Ther. 44, 726–734. doi:10.1111/jcpt.12852
Jeong, J., Han, H., Ro, D. H., Han, H.-S., and Won, S. (2023). Development of prediction model using machine-learning algorithms for nonsteroidal anti-inflammatory drug-induced gastric ulcer in osteoarthritis patients: retrospective cohort study of a nationwide south Korean cohort. Clin. Orthop. Surg. 15, 678–689. doi:10.4055/cios22240
Ji, H.-H., Song, L., Xiao, J.-W., Guo, Y.-X., Wei, P., Tang, T.-T., et al. (2018). Adverse drug events in Chinese pediatric inpatients and associated risk factors: a retrospective review using the Global Trigger Tool. Sci. Rep. 8, 2573. doi:10.1038/s41598-018-20868-2
Jiang, D., Song, Z., Liu, P., Wang, Z., and Zhao, R. (2023). A prediction model for severe hematological toxicity of BTK inhibitors. Ann. Hematol. 102, 2765–2777. doi:10.1007/s00277-023-05371-7
Khanna, A. K., Bergese, S. D., Jungquist, C. R., Morimatsu, H., Uezono, S., Lee, S., et al. (2020). Prediction of opioid-induced respiratory depression on inpatient wards using continuous capnography and oximetry: an international prospective, observational trial. Anesth. Analg. 131, 1012–1024. doi:10.1213/ANE.0000000000004788
Kim, J. S., Han, J.-M., Cho, Y.-S., Choi, K.-H., and Gwak, H.-S. (2021). Machine learning approaches to predict hepatotoxicity risk in patients receiving nilotinib. Molecules 26, 3300. doi:10.3390/molecules26113300
Kim, W., Cho, Y.-A., Kim, D.-C., Jo, A.-R., Min, K.-H., and Lee, K.-E. (2021). Factors associated with thyroid-related adverse events in patients receiving PD-1 or PD-L1 inhibitors using machine learning models. Cancers (Basel) 13, 5465. doi:10.3390/cancers13215465
Lai, N.-H., Shen, W.-C., Lee, C.-N., Chang, J.-C., Hsu, M.-C., Kuo, L.-N., et al. (2020). Comparison of the predictive outcomes for anti-tuberculosis drug-induced hepatotoxicity by different machine learning techniques. Comput. Methods Programs Biomed. 188, 105307. doi:10.1016/j.cmpb.2019.105307
Lee, S., Heo, K.-N., Lee, M. Y., Ah, Y.-M., Shin, J., and Lee, J.-Y. (2023). Derivation and validation of a risk prediction score for nonsteroidal anti-inflammatory drug-related serious gastrointestinal complications in the elderly. Br. J. Clin. Pharmacol. 89, 2216–2223. doi:10.1111/bcp.15696
Lewinson, R. T., Meyers, D. E., Vallerand, I. A., Suo, A., Dean, M. L., Cheng, T., et al. (2021). Machine learning for prediction of cutaneous adverse events in patients receiving anti-PD-1 immunotherapy. J. Am. Acad. Dermatol 84, 183–185. doi:10.1016/j.jaad.2020.04.069
Li, C., Chen, L., Chou, C., Ngorsuraches, S., and Qian, J. (2022). Using machine learning approaches to predict short-term risk of cardiotoxicity among patients with colorectal cancer after starting fluoropyrimidine-based chemotherapy. Cardiovasc Toxicol. 22, 130–140. doi:10.1007/s12012-021-09708-4
Liao, K.-M., Liu, C.-F., Chen, C.-J., Feng, J.-Y., Shu, C.-C., and Ma, Y.-S. (2023). Using an artificial intelligence approach to predict the adverse effects and prognosis of tuberculosis. Diagn. (Basel) 13, 1075. doi:10.3390/diagnostics13061075
Lippenszky, L., Mittendorf, K. F., Kiss, Z., LeNoue-Newton, M. L., Napan-Molina, P., Rahman, P., et al. (2024). Prediction of effectiveness and toxicities of immune checkpoint inhibitors using real-world patient data. JCO Clin. Cancer Inf. 8, e2300207. doi:10.1200/CCI.23.00207
Liu, L., Yu, Y., Fei, Z., Li, M., Wu, F.-X., Li, H.-D., et al. (2018). An interpretable boosting model to predict side effects of analgesics for osteoarthritis. BMC Syst. Biol. 12, 105. doi:10.1186/s12918-018-0624-4
Livshits, Z., Rao, R. B., and Smith, S. W. (2014). An approach to chemotherapy-associated toxicity. Emerg. Med. Clin. North Am. 32, 167–203. doi:10.1016/j.emc.2013.09.002
Lu, Y.-T., Chao, H.-J., Chiang, Y.-C., and Chen, H.-Y. (2023). Explainable machine learning techniques to predict amiodarone-induced thyroid dysfunction risk: multicenter, retrospective study with external validation. J. Med. Internet Res. 25, e43734. doi:10.2196/43734
Ma, J., Wang, Y., Ma, S., and Li, J. (2023). The investigation and prediction of voriconazole-associated hepatotoxicity under therapeutic drug Monitoring. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2023, 1–4. doi:10.1109/EMBC40787.2023.10340343
Ma, X., Mo, C., Li, Y., Chen, X., and Gui, C. (2023). Prediction of the development of contrast-induced nephropathy following percutaneous coronary artery intervention by machine learning. Acta Cardiol. 78, 912–921. doi:10.1080/00015385.2023.2198937
Madias, N. E., and Harrington, J. T. (1978). Platinum nephrotoxicity. Am. J. Med. 65, 307–314. doi:10.1016/0002-9343(78)90825-2
Mantripragada, A. S., Teja, S. P., Katasani, R. R., Joshi, P., V, M., and Ramesh, R. (2021). Prediction of adverse drug reactions using drug convolutional neural networks. J. Bioinform Comput. Biol. 19, 2050046. doi:10.1142/S0219720020500468
Mao, J., Chao, K., Jiang, F.-L., Ye, X.-P., Yang, T., Li, P., et al. (2023). Comparison and development of machine learning for thalidomide-induced peripheral neuropathy prediction of refractory Crohn’s disease in Chinese population. World J. Gastroenterol. 29, 3855–3870. doi:10.3748/wjg.v29.i24.3855
Maray, I., Álvarez-Asteinza, C., Fernández-Laguna, C. L., Macía-Rivas, L., Carbajales-Álvarez, M., and Lozano-Blazquez, A. (2023). Dose reduction and toxicity of lenalidomide-dexamethasone in multiple myeloma: a machine-learning prediction model. J. Oncol. Pharm. Pract. 30, 1051–1056. doi:10.1177/10781552231200795
Martins, F., Sofiya, L., Sykiotis, G. P., Lamine, F., Maillard, M., Fraga, M., et al. (2019). Adverse effects of immune-checkpoint inhibitors: epidemiology, management and surveillance. Nat. Rev. Clin. Oncol. 16, 563–580. doi:10.1038/s41571-019-0218-0
Matsumoto, N., Mizuno, T., Ando, Y., Kato, K., Nakanishi, M., Nakai, T., et al. (2024). Prediction model for severe thrombocytopenia induced by gemcitabine plus cisplatin combination therapy in patients with urothelial cancer. Clin. Drug Investig. 44, 357–366. doi:10.1007/s40261-024-01361-3
Mehran, R., Aymong, E. D., Nikolsky, E., Lasic, Z., Iakovou, I., Fahy, M., et al. (2004). A simple risk score for prediction of contrast-induced nephropathy after percutaneous coronary intervention: development and initial validation. J. Am. Coll. Cardiol. 44, 1393–1399. doi:10.1016/j.jacc.2004.06.068
Moher, D., Liberati, A., Tetzlaff, J., and Altman, D. G.PRISMA Group (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6, e1000097. doi:10.1371/journal.pmed.1000097
Mora, D., Mateo, J., Nieto, J. A., Bikdeli, B., Yamashita, Y., Barco, S., et al. (2023). Machine learning to predict major bleeding during anticoagulation for venous thromboembolism: possibilities and limitations. Br. J. Haematol. 201, 971–981. doi:10.1111/bjh.18737
Mu, F., Cui, C., Tang, M., Guo, G., Zhang, H., Ge, J., et al. (2022). Analysis of a machine learning-based risk stratification scheme for acute kidney injury in vancomycin. Front. Pharmacol. 13, 1027230. doi:10.3389/fphar.2022.1027230
Nebeker, J. R., Barach, P., and Samore, M. H. (2004). Clarifying adverse drug events: a clinician’s guide to terminology, documentation, and reporting. Ann. Intern Med. 140, 795–801. doi:10.7326/0003-4819-140-10-200405180-00009
Nguyen, Q. T. N., Phan, P. T., Lin, S.-J., Hsu, M.-H., Li, Y.-C. J., Hsu, J. C., et al. (2024). Machine-learning based risk assessment for cancer therapy-related cardiac adverse events among breast cancer patients. Stud. Health Technol. Inf. 310, 1006–1010. doi:10.3233/SHTI231116
Noda, T., Mizuno, S., Mogushi, K., Hase, T., Iida, Y., Takeuchi, K., et al. (2024). Development of a predictive model for nephrotoxicity during tacrolimus treatment using machine learning methods. Br. J. Clin. Pharmacol. 90, 675–683. doi:10.1111/bcp.15953
Okawa, T., Mizuno, T., Hanabusa, S., Ikeda, T., Mizokami, F., Koseki, T., et al. (2022). Prediction model of acute kidney injury induced by cisplatin in older adults using a machine learning algorithm. PLoS One 17, e0262021. doi:10.1371/journal.pone.0262021
On, J., Park, H.-A., and Yoo, S. (2022). Development of a prediction models for chemotherapy-induced adverse drug reactions: a retrospective observational study using electronic health records. Eur. J. Oncol. Nurs. 56, 102066. doi:10.1016/j.ejon.2021.102066
Patel, A., Doernberg, S. B., Zack, T., Butte, A. J., and Radtke, K. K. (2024). Predictive modeling of drug-related adverse events with real-world data: a case study of linezolid hematologic outcomes. Clin. Pharmacol. Ther. 115, 847–859. doi:10.1002/cpt.3201
Pérez-Herrero, E., and Fernández-Medarde, A. (2015). Advanced targeted therapies in cancer: drug nanocarriers, the future of chemotherapy. Eur. J. Pharm. Biopharm. 93, 52–79. doi:10.1016/j.ejpb.2015.03.018
Ruiz Sarrias, O., Gónzalez Deza, C., Rodríguez Rodríguez, J., Arrizibita Iriarte, O., Vizcay Atienza, A., Zumárraga Lizundia, T., et al. (2023). Predicting severe haematological toxicity in gastrointestinal cancer patients undergoing 5-FU-based chemotherapy: a bayesian network approach. Cancers (Basel) 15, 4206. doi:10.3390/cancers15174206
Sacerdote, P. (2008). Opioid-induced immunosuppression. Curr. Opin. Support Palliat. Care 2, 14–18. doi:10.1097/SPC.0b013e3282f5272e
Sahni, V., Choudhury, D., and Ahmed, Z. (2009). Chemotherapy-associated renal dysfunction. Nat. Rev. Nephrol. 5, 450–462. doi:10.1038/nrneph.2009.97
Sarkar, U., López, A., Maselli, J. H., and Gonzales, R. (2011). Adverse drug events in U.S. adult ambulatory medical care. Health Serv. Res. 46, 1517–1533. doi:10.1111/j.1475-6773.2011.01269.x
Satheeshkumar, P. S., El-Dallal, M., and Mohan, M. P. (2021). Feature selection and predicting chemotherapy-induced ulcerative mucositis using machine learning methods. Int. J. Med. Inf. 154, 104563. doi:10.1016/j.ijmedinf.2021.104563
Schirrmacher, V. (2019). From chemotherapy to biological therapy: a review of novel concepts to reduce the side effects of systemic cancer treatment (Review). Int. J. Oncol. 54, 407–419. doi:10.3892/ijo.2018.4661
Simon, S. T., Mandair, D., Tiwari, P., and Rosenberg, M. A. (2021). Prediction of drug-induced long QT syndrome using machine learning applied to harmonized electronic health record data. J. Cardiovasc Pharmacol. Ther. 26, 335–340. doi:10.1177/1074248421995348
Subramanian, S., Tumlin, J., Bapat, B., and Zyczynski, T. (2007). Economic burden of contrast-induced nephropathy: implications for prevention strategies. J. Med. Econ. 10, 119–134. doi:10.3111/200710119134
Sun, L., Zhu, W., Chen, X., Jiang, J., Ji, Y., Liu, N., et al. (2020). Machine learning to predict contrast-induced acute kidney injury in patients with acute myocardial infarction. Front. Med. (Lausanne) 7, 592007. doi:10.3389/fmed.2020.592007
Surendran, S., B, M. C., Gilvaz, V., Manyam, P. K., Panicker, K., and Pradeep, M. (2024). Prediction of liver enzyme elevation using supervised machine learning in patients with rheumatoid arthritis on treatment with methotrexate. Cureus 16, e52110. doi:10.7759/cureus.52110
Tamma, P. D., Avdic, E., Li, D. X., Dzintars, K., and Cosgrove, S. E. (2017). Association of adverse events with antibiotic use in hospitalized patients. Jama Intern Med. 177, 1308–1315. doi:10.1001/jamainternmed.2017.1938
Tostmann, A., Boeree, M. J., Aarnoutse, R. E., de Lange, W. C. M., van der Ven, A. J. A. M., and Dekhuijzen, R. (2008). Antituberculosis drug-induced hepatotoxicity: concise up-to-date review. J. Gastroenterol. Hepatol. 23, 192–202. doi:10.1111/j.1440-1746.2007.05207.x
Venäläinen, M. S., Heervä, E., Hirvonen, O., Saraei, S., Suomi, T., Mikkola, T., et al. (2022). Improved risk prediction of chemotherapy-induced neutropenia-model development and validation with real-world data. Cancer Med. 11, 654–663. doi:10.1002/cam4.4465
Wu, X.-W., Zhang, J.-Y., Chang, H., Song, X.-W., Wen, Y.-L., Long, E.-W., et al. (2022). Develop an ADR prediction system of Chinese herbal injections containing Panax notoginseng saponin: a nested case-control study using machine learning. BMJ Open 12, e061457. doi:10.1136/bmjopen-2022-061457
Xiao, Y., Chen, Y., Huang, R., Jiang, F., Zhou, J., and Yang, T. (2024). Interpretable machine learning in predicting drug-induced liver injury among tuberculosis patients: model development and validation study. BMC Med. Res. Methodol. 24, 92. doi:10.1186/s12874-024-02214-5
Xie, Q., Wang, X., Pei, J., Wu, Y., Guo, Q., Su, Y., et al. (2022). Machine learning-based prediction models for delirium: a systematic review and meta-analysis. J. Am. Med. Dir. Assoc. 23, 1655–1668.e6. doi:10.1016/j.jamda.2022.06.020
Xu, Y., Li, X., Wu, D., Zhang, Z., and Jiang, A. (2022). Machine learning-based model for prediction of hemorrhage transformation in acute ischemic stroke after alteplase. Front. Neurol. 13, 897903. doi:10.3389/fneur.2022.897903
Yagi, R., Goto, S., Himeno, Y., Katsumata, Y., Hashimoto, M., MacRae, C. A., et al. (2024). Artificial intelligence-enabled prediction of chemotherapy-induced cardiotoxicity from baseline electrocardiograms. Nat. Commun. 15, 2536. doi:10.1038/s41467-024-45733-x
Yan, P., Duan, S.-B., Luo, X.-Q., Zhang, N.-Y., and Deng, Y.-H. (2023). Development and validation of a deep neural network-based model to predict acute kidney injury following intravenous administration of iodinated contrast media in hospitalized patients with chronic kidney disease: a multicohort analysis. Nephrol. Dial. Transpl. 38, 352–361. doi:10.1093/ndt/gfac049
Yoo, J. H., Sharma, V., Kim, J.-W., McMakin, D. L., Hong, S.-B., Zalesky, A., et al. (2020). Prediction of sleep side effects following methylphenidate treatment in ADHD youth. Neuroimage Clin. 26, 102030. doi:10.1016/j.nicl.2019.102030
Zhang, J., Cui, X., Yang, C., Zhong, D., Sun, Y., Yue, X., et al. (2023). A deep learning-based interpretable decision tool for predicting high risk of chemotherapy-induced nausea and vomiting in cancer patients prescribed highly emetogenic chemotherapy. Cancer Med. 12, 18306–18316. doi:10.1002/cam4.6428
Zhao, X., Peng, Q., Hu, D., Li, W., Ji, Q., Dong, Q., et al. (2024). Prediction of risk factors for linezolid-induced thrombocytopenia based on neural network model. Front. Pharmacol. 15, 1292828. doi:10.3389/fphar.2024.1292828
Zhou, F., Lu, Y., Xu, Y., Li, J., Zhang, S., Lin, Y., et al. (2023). Correlation between neutrophil-to-lymphocyte ratio and contrast-induced acute kidney injury and the establishment of machine-learning-based predictive models. Ren. Fail 45, 2258983. doi:10.1080/0886022X.2023.2258983
Keywords: systematic review, adverse drug event, prediction model, machine learning algorithm, electronic medical record data
Citation: Hu Q, Chen Y, Zou D, He Z and Xu T (2024) Predicting adverse drug event using machine learning based on electronic health records: a systematic review and meta-analysis. Front. Pharmacol. 15:1497397. doi: 10.3389/fphar.2024.1497397
Received: 17 September 2024; Accepted: 21 October 2024;
Published: 13 November 2024.
Edited by:
Shusen Sun, Western New England University, United StatesReviewed by:
Sheraz Ali, University of Tasmania, AustraliaZiran Li, University of California, San Francisco, United States
Copyright © 2024 Hu, Chen, Zou, He and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyao He, emhpeWFvaGVAc2N1LmVkdS5jbg==; Ting Xu, dGluZ3gyMDA5QDE2My5jb20=
 Qiaozhi Hu
Qiaozhi Hu Yuxian Chen1
Yuxian Chen1 Dan Zou
Dan Zou Zhiyao He
Zhiyao He Ting Xu
Ting Xu