- 1Department of Medical College, Chengdu University of Traditional Chinese Medicine, Chengdu, Sichuan, China
- 2Department of Medical Cosmetology, Chengdu University of Traditional Chinese Medicine, Chengdu, Sichuan, China
Introduction: Hypertrophic scars (HSs) are characterized by complex mechanisms and impose substantial economic and psychological burdens on patients with wounds. Recent studies have reported that various extracts from traditional Chinese medicines can help prevent and treat HSs. Scar healing ointment (SHO), a modified traditional Chinese prescription applied externally, has demonstrated potential in the clinical treatment of HSs, though its underlying mechanisms remain unexplored.
Methods: In this study, we systematically identified the active ingredients of the SHO formula and their potential targets using multiple databases (TCMSP, HERB, UniProt, GeneCards, DisGeNet, OMIM, PharmGKB, TTD) and explored the possible underlying mechanisms by which SHO treats HSs using bioinformatic analyses, including protein-protein interaction (PPI) network analysis, GO and KEGG enrichment analyses, and molecular docking.
Results: Our results indicated that the primary active ingredients in the SHO formula include quercetin, beta-sitosterol, kaempferol, stigmasterol, luteolin, alloimperatorin, acacetin, and (E)-2,3-bis(7-methoxy-2-oxochromen-8-yl)prop-2-enal. Protein-protein interaction network analysis revealed that the hub target proteins of the SHO formula are AKT1, MAPK1, CCND1, TP53, GSK3B, BCL2, CDKN1A, ESR1, and MYC. GO and KEGG enrichment analyses showed that these hub target genes are involved in processes and pathways related to apoptosis and responses to oxidants. Molecular docking analysis demonstrated that the MAPK1-stigmasterol and ESR1-alloimperatorin complexes exhibited strong binding affinities (–5.31 and –6.09) and formed multiple hydrogen bonds (3 and 2, respectively).
Discussion: These findings suggest that SHO may exert its effects by modulating MAPK1 and ESR1 proteins, thereby contributing to the prevention and treatment of HSs. This study offers new drugs and target candidates for the prevention and treatment of HSs and provides theoretical support for further research and application of the SHO formula. Nevertheless, additional in vivo and in vitro studies are necessary to validate these mechanisms.
Introduction
The skin is the largest organ in the human body and is highly susceptible to physical injury such as surgeries, traumas, and burns (Coentro et al., 2019). To maintain life, the skin must heal quickly, involving a complex physiologic process where the body attempts to replace injured skin with newly generated tissue and restore its barrier functions (Mony et al., 2023). This process involves four stages: hemostasis, inflammation, proliferation, and remodeling, as well as communication between many different types of cells (Gurtner et al., 2008). Under normal conditions, these stages occur in a highly sequential and finite manner, and the myofibroblasts undergo apoptosis during the remodeling stage, leaving a relatively acellular scar at the healed sites (Bharadia et al., 2023). However, this process sometimes does not occur sequentially and finitely, resulting in aberrant wound-healing processes and subsequent fibroproliferative disorders and hypertrophic scars (HSs) (Gurtner et al., 2008; Limandjaja et al., 2021). HSs are characterized by excessive extracellular matrix (ECM), especially collagen, deposition during the wound healing process, leading to raised, often red, thickened scars that remain within the boundaries of the original wound (Diegelmann and Evans, 2004; Lingzh et al., 2020). These scars can be painful and itchy, symptoms that may worsen with elevated ambient temperature, emotional arousal, or consumption of spicy foods (Bharadia et al., 2023; Jeschke et al., 2023), affecting 30%–90% of patients with wounds (Bharadia et al., 2023; Limandjaja et al., 2021) and posing serious social, functional, and economic burdens due to their appearance, contractures, and the costs associated with scar treatment (Thombs et al., 2008; Leblebici et al., 2006; Mcphail et al., 2022). Considering the significant portion of the population suffering from HSs, effectively preventing the formation of HSs during wound healing or removing already formed HSs is of great importance.
Currently, there are two mature clinical measures for preventing and treating the formation of HSs. The first is known as first-line therapy, represented by silicone gel sheeting (Bharadia et al., 2023). For example, silicone gel has been reported to reduce the expression of transforming growth factor (TGF)-β1, platelet-derived growth factor, and basic fibroblast growth factor (b-FGF) 4 months after surgery for surgical scars (Choi et al., 2015). However, other studies have reported that silicone gel can also upregulate the expression of b-FGF in the dermis of silicone gel sheet-treated scars (O’brien and Jones, 2013). The second measure is known as second-line therapy, which includes laser therapy as well as surgical excision in combination with intralesional corticosteroid injections postoperatively (Bharadia et al., 2023; Atiyeh, 2020). For instance, it has been reported that long-pulsed Nd:YAG laser and pulsed dye can lighten the color, reduce the thickness and tension of HSs, and relieve symptoms such as pain and pruritus (Amini-Nik et al., 2018).
In recent years, traditional Chinese medicine (TCM), including various extracts, has been proven effective as a potential therapy for treating HSs due to its roles in promoting skin regeneration and resisting skin aging. For example, many traditional plant-based products, such as quercetin, onion extract, resveratrol, epigallocatechin gallate, oleanolic acid, and curcumin, have been used clinically, providing relief to patients to varying extents (Bharadia et al., 2023). Scar healing ointment (SHO) is a TCM prescription for the external treatment of scarring. It includes ingredients like Wubeizi (Galla Chinensis), Wumei (Mume Fructus), Baizhi (Angelicae Dahuricae), Shechuangzi (Fructus Cnidii), Qianliguang (Senecio Scandens), and Digupi (Cortex Lycii), and has been clinically effective in preventing the formation of HSs and removing already formed HSs. However, the underlying mechanisms of its effects on HS prevention and treatment remain unclear.
Network pharmacology is a promising approach to reveal the pharmacological mechanisms of Chinese medicine formulas (Zhao et al., 2023) and predict their potential targets for specific diseases (Zhang et al., 2017; Kou and Ma, 2024). Molecular docking is a significant method in structural molecular biology and computer-aided drug design (Saikia and Bordoloi, 2019). Therefore, in the present study, we aim to analyze the active ingredients and molecular targets of SHO using network pharmacology and molecular docking methods to explore its potential underlying mechanisms for the prevention and treatment of HSs. Our study will provide new drug and target options for the prevention and treatment of HSs, and offer theoretical support for further research and promotion of SHO.
Materials and methods
Identification and screening of active ingredients and their protein targets
The Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, https://tcmsp.91medicine.cn/) (Ru et al., 2014) was used to identify all ingredients in the six herbs comprising the SHO formula, namely, Wubeizi, Wumei, Baizhi, Shechuangzi, Digupi, and Qianliguang. The names of these herbs were used as keywords to retrieve all components. As a result, the ingredients of Wubeizi, Wumei, Baizhi, Shechuangzi, and Digupi were obtained from the TCMSP database. However, the keyword “Qianliguang” was not available in TCMSP, so its ingredients were retrieved from the HERB database (http://herb.ac.cn/) (Fang et al., 2021). To ensure consistency, we cross-referenced the chemical abstracts service (CAS) registry number of the ingredients with a list in both databases, and only ingredients present in the TCMSP database were included as active ingredients of Qianliguang. Since the TCMSP database does not provide dermal permeability parameters for the ingredients, we screened these components using the pharmacokinetic parameters oral bioavailability (OB) ≥ 30% and drug-likeness (DL) ≥ 0.18, in accordance with previous studies (Fan et al., 2024; Han et al., 2022), to facilitate subsequent analysis. Afterward, the protein targets associated with the screened active ingredients were retrieved from the TCMSP database, and the gene names of these targets were further extracted using the UniProt database (https://www.uniprot.org/) (UniProt, 2021). To reflect the complex relationships between active ingredients and their potential target genes, we employed the Cytoscape software (v3.9.1; http://www.cytoscape.org/) (Shannon et al., 2003) to establish a visual herb-ingredient-target gene network.
Predicting the targets of HSs and protein-protein interactions (PPI) analysis
We gathered information on HSs-associated gene targets from the GeneCards database (https://www.genecards.org/), Disease-Gene Network database (DisGeNet; https://www.disgenet.com/), Online Mendelian Inheritance in Man database (OMIM; https://www.omim.org/), Pharmacogenomics Knowledgebase (PharmGKB; https://www.pharmgkb.org/), and Therapeutic Target Database (TTD, https://db.idrblab.org/ttd/) using “hypertrophic scars” as the keyword. The shared gene targets between the HSs-associated gene targets and SHO-associated gene targets were identified as the potential targets of SHO for HSs. These potential gene targets were put into STRING (v12.0; https://string-db.org/cgi/input.pl) to construct the PPI network interaction (interaction score ≥0.900) referencing previous research (Szklarczyk et al., 2019; Islam et al., 2018), which was visualized using Cytoscape software. The classical degree value was used as the standard to measure the importance of nodes in the PPI network, and the CytoNCA plug-in (Tang et al., 2015) in the Cytoscape software was used to calculate a series of degree values. The median degree value calculated from the CytoNCA plug-in, rounded to the nearest integer, was used as the threshold for screening hub genes, referencing previous studies (Fan et al., 2024; Han et al., 2022).
GO and KEGG enrichment analyses
The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.kegg.jp/) enrichment analyses of these SHO-related potential targets were performed using the “ClusterProfiler” package (v 4.10.1) in the RStudio software (v 2023.12.1). GO terms and KEGG pathways with adjusted p-value <0.05 were considered to be significantly enriched.
Ingredient-target molecular docking
The three-dimensional (3D) structures of the core target proteins were downloaded from the Research Collaboratory for Structural Bioinformatics Protein Data Bank database (PDB; https://www.rcsb.org/). AutoDock Tools software (v1.5.7; https://autodocksuite.scripps.edu/adt/) was then used to process these structures by removing water molecules, isolating proteins, adding nonpolar hydrogen atoms, and calculating Gasteiger charges. The processed structures were saved as PDBQT files to serve as receptors. The 2-dimensional (2D) structures of selected ingredients were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). These 2D structures were then converted into PDB format and saved as docking ligands using the Chem 3D software (v22.0.0.22). The active site for molecular docking was determined based on the ligand coordinates in the target protein complex. The ligand was set to be flexible, while the receptor remained rigid. AutoDock Vina (v1.1.2) was used to dock these ligands with the receptors. For the docking results, multiple conformations were generated, and the conformation with the best affinity was selected as the final docking conformation. Conformations with affinity <−5 were then visualized using Pymol (v 2.3) software.
Results
Active ingredients and potential target genes of SHO
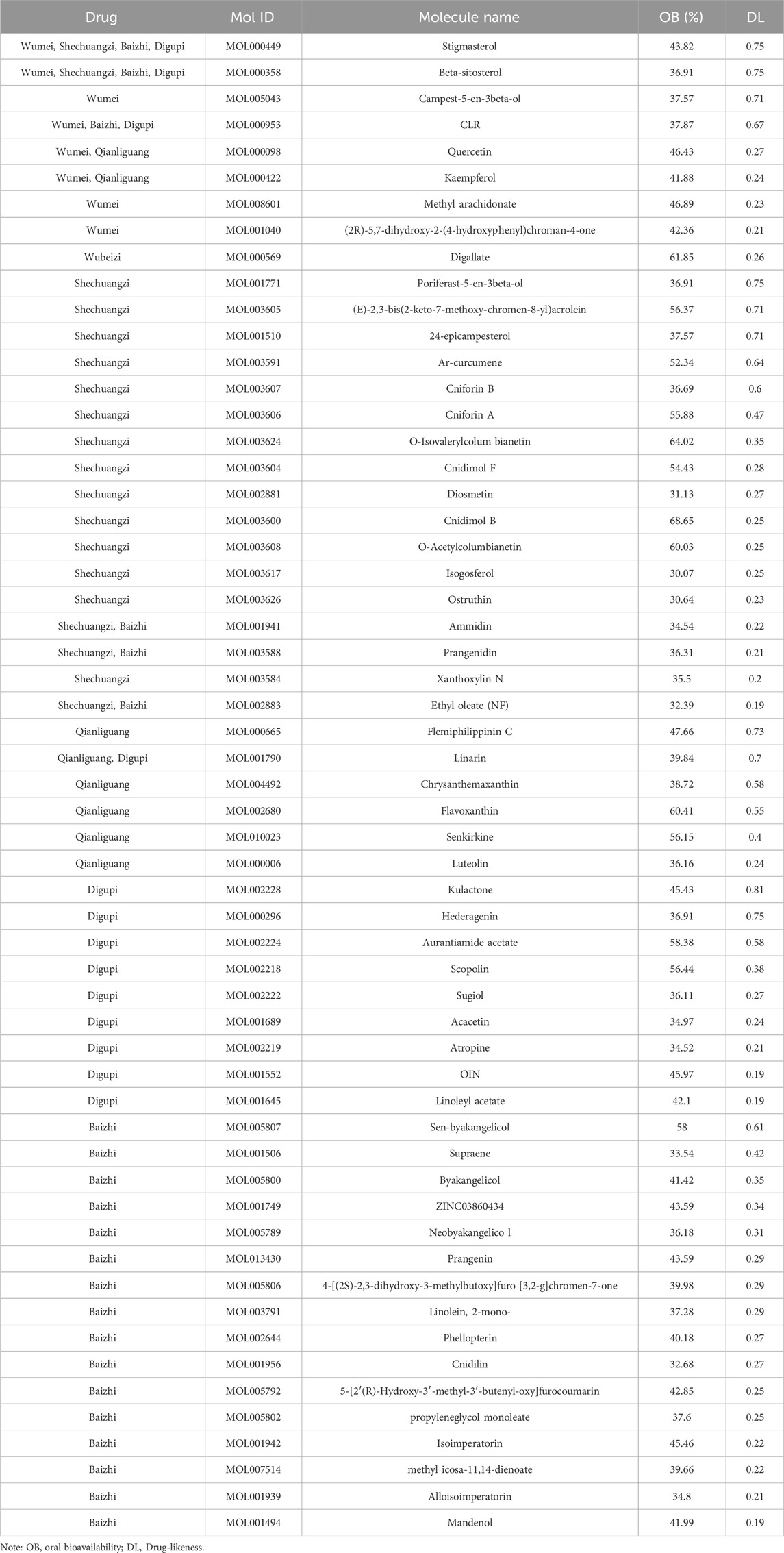
From the six herbs, a total of 386 active ingredients were retrieved from the TCMSP database: three in Wubeizi, 43 in Wumei, 114 in Shechuangzi, 222 in Baizhi, 20 in Qianliguang, and 38 in Digupi. After screening for OB ≥ 30% and DL ≥ 0.18, a total of 57 active ingredients were obtained: one in Wubeizi, eight in Wumei, 19 in Shechuangzi, eight in Qianliguang, 13 in Digupi, and 22 in Baizhi (Table 1). Detailed information about the retrieved active ingredients in the six herbs is shown in Supplementary Table S1. Then, a total of 145 target genes associated with these active ingredients were further retrieved from the TCMSP database (Supplementary Table S2).
HSs associated genes and potential target genes of SHO for HSs
We retrieved a total of 705 genes associated with HSs from five databases: 30 from the DisGeNet database, 197 from the OMIM database, 135 from the PharmGKB database, four from the TTD database, and 416 from the GeneCards database (Figure 1A). Detailed information about these HSs-associated genes is provided in Supplementary Table S3. Subsequently, it was identified that 42 core genes were shared between the 145 target genes associated with the 57 active ingredients and the 705 genes associated with HSs (Figure 1B). Supplementary Table S4 provides detailed information about the shared and unique genes between these active ingredients-associated genes and HSs-associated genes.
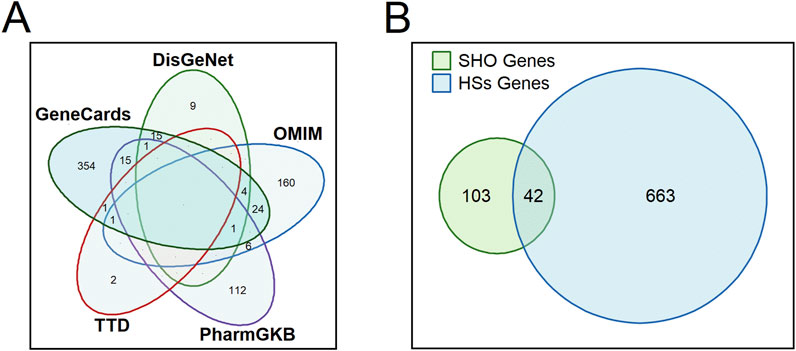
Figure 1. Venn diagrams showing the number of retrieved hypertrophic scars-associated genes from each database (A) and shared genes between the 145 target genes associated with the 57 active ingredients and the 705 genes associated with HSs. SHO, scar healing ointment; HSs, hypertrophic scars. (B) displays a Venn diagram of SHO genes and HSs genes, with overlapping regions labeled to indicate the number of shared genes (n = 42), where SHO refers to scar healing ointment and HSs refers to hypertrophic scars.
Herb-ingredient-target gene network
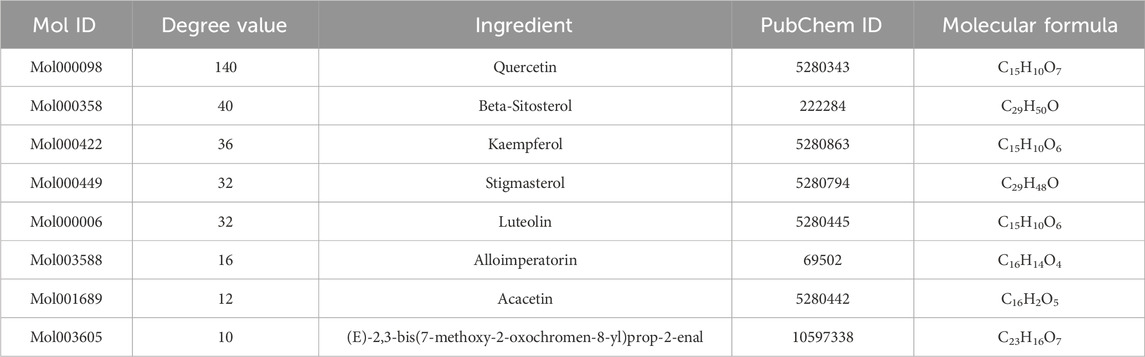
After removing 14 active ingredients without target genes, an herb-ingredient-target gene network was constructed. This network contains 91 nodes, including six herbs, 43 active ingredients, and 42 target genes, as well as 442 edges (Figure 2). Based on their degree values (≥10), we considered Mol000098, Mol000358, Mol000422, Mol000449, Mol000006, Mol003588, Mol001689, and Mol003605 as the core active ingredients in SHO. More detailed information about these active ingredients is provided in Table 2.
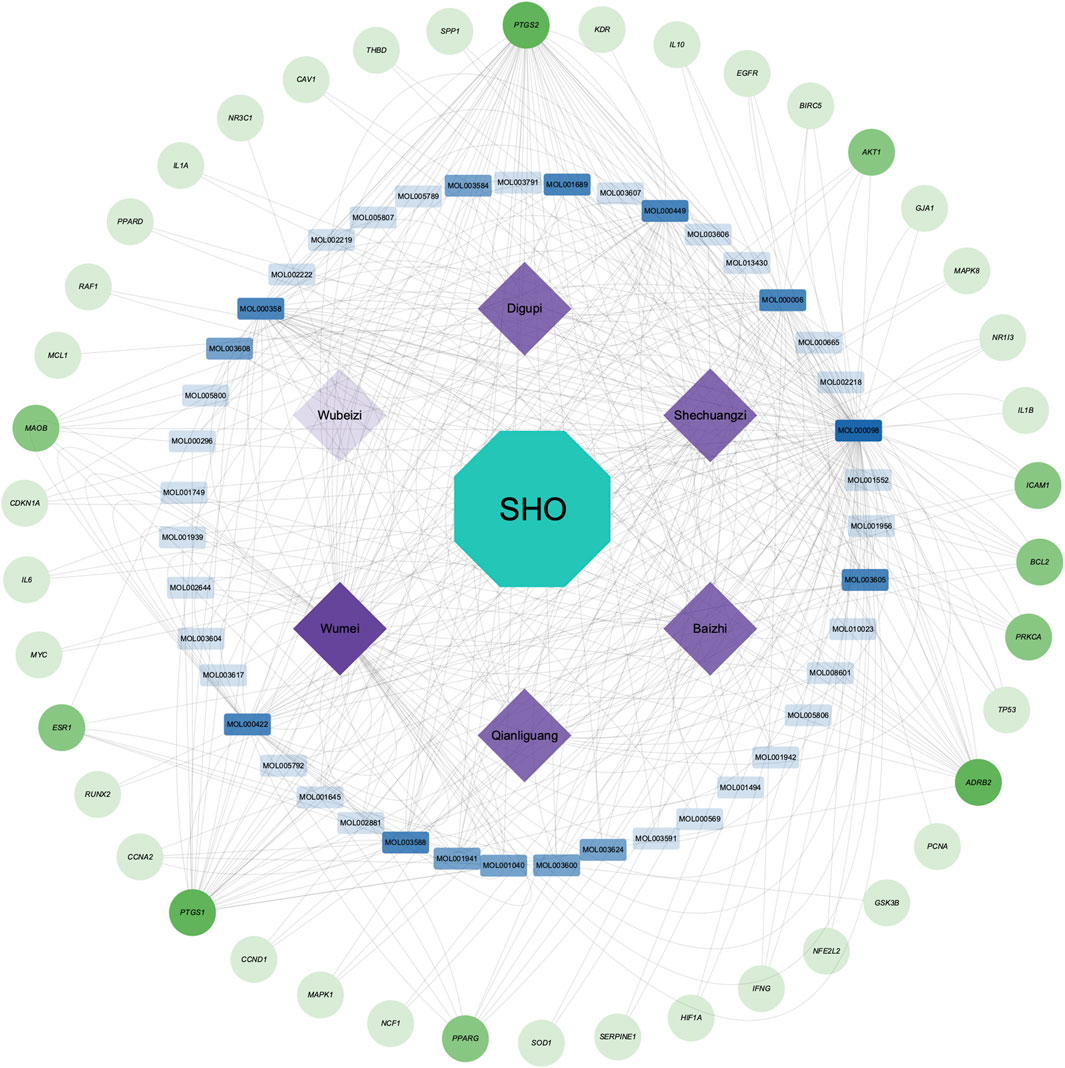
Figure 2. Herb-ingredient-target gene network of scar healing ointment (SHO). The green circle nodes in the outmost circle represent the 42 target genes, the blue rectangular nodes in the inner circle represent the 43 active ingredients with target genes, and the rhombic purple nodes in the innermost circle represent the six herbal medicines in the SHO. The transparency of the nodes reflects their Degree values: the lower transparency indicates higher Degree values.
PPI network and hub genes
Based on the 42 core genes, we constructed a PPI network containing 36 nodes and 93 edges (Figure 3). After one round of screening, nine hub genes were identified, namely, AKT1, MAPK1, CCND1, TP53, GSK3B, BCL2, CDKN1A, ESR1, and MYC. Supplementary Table S5 provides detailed scoring data used for hub gene screening.
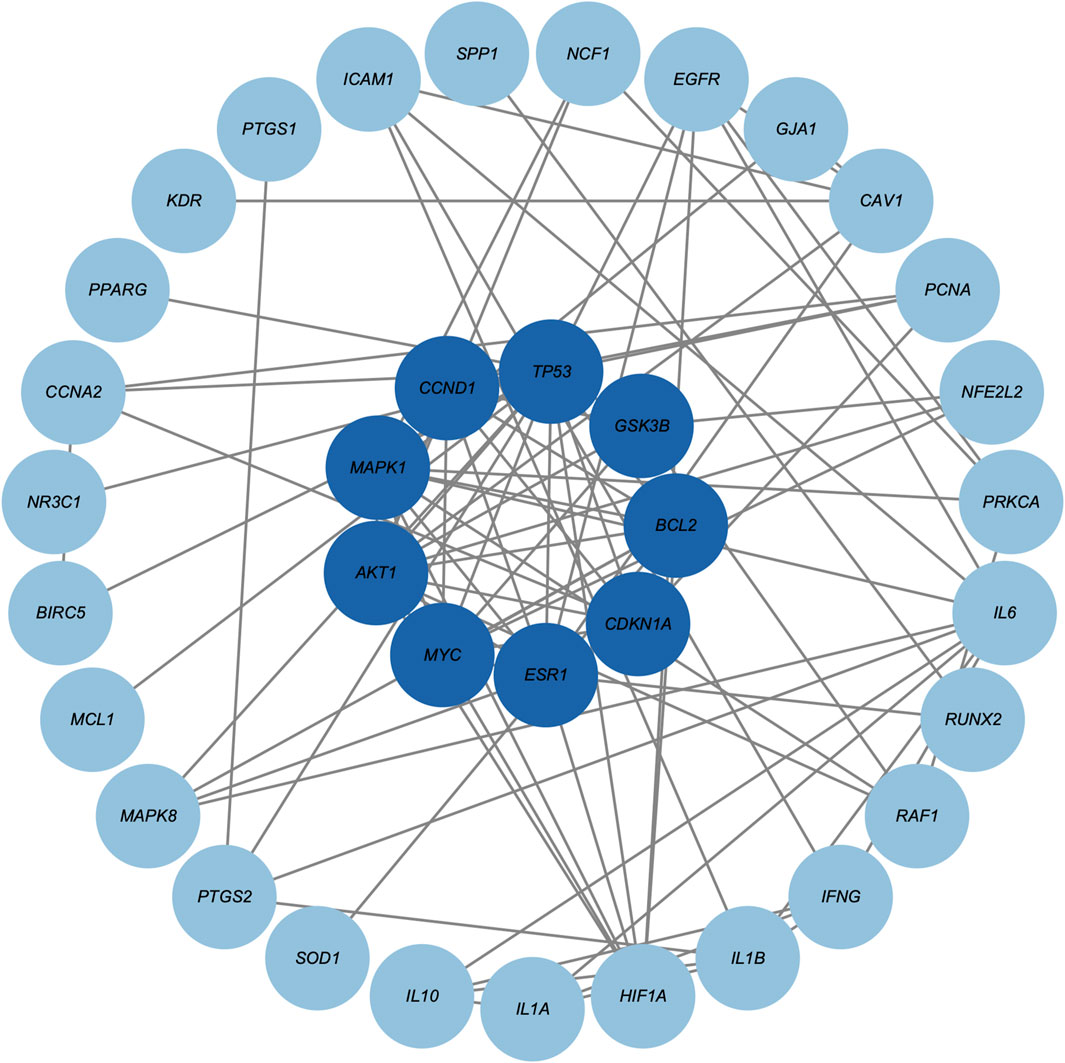
Figure 3. Protein-protein interaction network of core genes. Darker blue circle nodes in the inner circle indicate the nine screened hub genes.
GO and KEGG enrichment analyses
The 42 core target genes were significantly enriched (adjusted p < 0.05) into 1597 GO terms: 1842 biological process (BP) terms, 22 cellular component (CC) terms, and 93 molecular function (MF) terms. Figure 4A shows the top 10 most enriched BP, CC, and MF terms, including regulation of apoptotic signaling pathway, negative regulation of apoptotic signaling pathway, response to oxidative stress, response to reactive oxygen species, cellular response to peptide, cellular response to oxygen levels, cellular response to chemical stress, miRNA transcription, regulation of miRNA transcription, regulation of intrinsic apoptotic signaling pathway, et al. Supplementary Table S6 provides detailed information about these significantly enriched GO terms. KEGG enrichment analysis revealed 219 significantly enriched (adjusted p < 0.05) KEGG pathways: 20 in Cellular Processes, 29 in Environmental Information Processing, six in Genetic Information Processing, 90 in Human Diseases, eight in Metabolism, and 65 in Organismal Systems. Figure 4B shows the 30 most enriched KEGG pathways, including Colorectal cancer, AGE-RAGE signaling pathway in diabetic complications, Fluid shear stress and atherosclerosis, Hepatitis B, Kaposi sarcoma-associated herpesvirus infection, Proteoglycans in cancer, Endometrial cancer, et al. Supplementary Table S7 provides detailed information about these significantly enriched KEGG pathways.
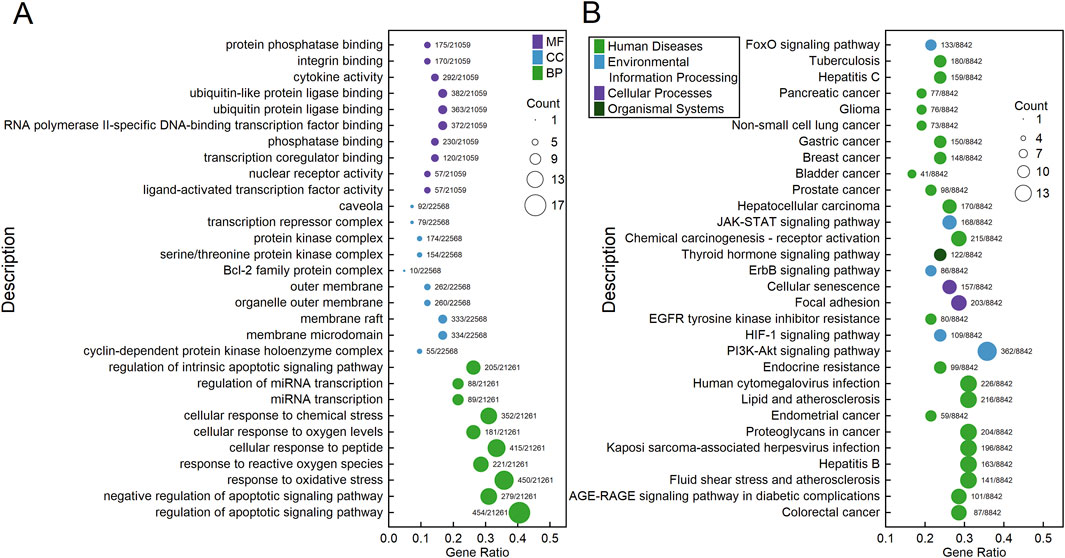
Figure 4. Bubble charts showing the results of GO (A) and KEGG (B) enrichment analyses of the 42 core genes. Each bubble represents a GO term (A) or a KEGG pathway (B). The fill color of the bubbles indicates their categories, and the size of the bubbles indicates the count of genes enriched into that GO term (A) or KEGG pathway (B). The “Gene Ratio” represents the ratio of the count of enriched genes to the total number of genes. The label alongside each bubble indicates the counts of genes in that GO term (A) or KEGG pathway (B) relative to the total number of genes in the category.
Molecular docking
We docked proteins coded by these nine hub genes with the eight core active ingredients, generating a total of 72 conformation complexes (Figure 5A). The results revealed that three out of these 72 conformation complexes exhibited affinities lower than −5. Specifically, the Cyclin D1 (CCND1)-stigmasterol complex had an affinity of −5.34 and formed three hydrogen bonds at the ASP-100, ALA-92, and LYS-99 residues (Figure 5B). The mitogen-activated protein kinase 1 (MAPK1)-stigmasterol complex had an affinity of −5.31 but did not form hydrogen bonds (Figure 5C). Lastly, the estrogen receptor 1 (ESR1)-alloimperatorin complex had an affinity of −6.09 and formed two hydrogen bonds at the LEU-346 and LEU-387 residues (Figure 5D).
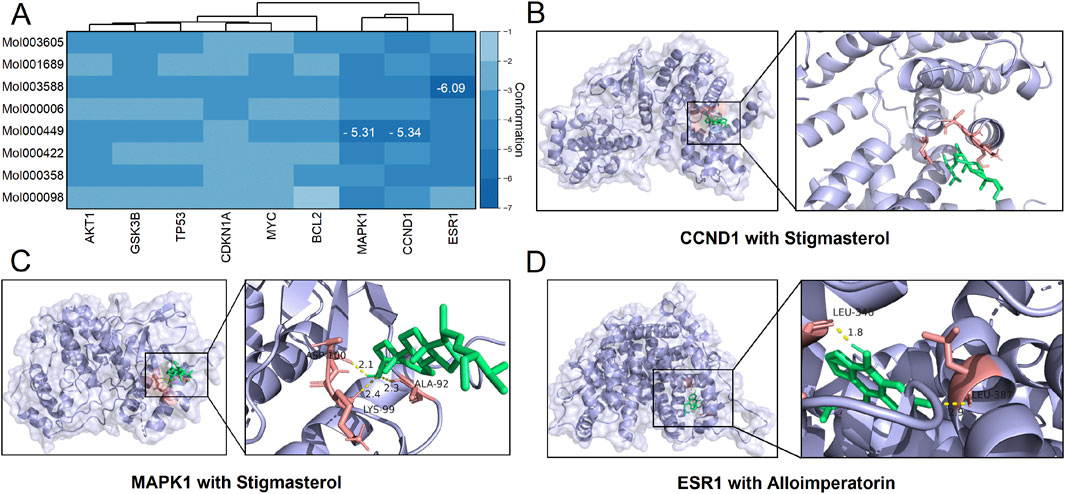
Figure 5. Results of molecular docking. (A) A clustered heatmap showing the affinities between each pair of docking conformations. (B–D) Molecular docking diagrams showing the conformation complexes of stigmasterol with Cyclin D1 (CCND1) and mitogen-activated protein kinase 1 (MAPK1), as well as the complex of alloimperatorin with estrogen receptor 1 (ESR1).
Discussion
HSs are a condition with complex mechanisms that impose significant economic and psychological burdens on patients with wounds (Mony et al., 2023; Bharadia et al., 2023; Mcphail et al., 2022). Recently, various extracts from multiple TCM have been reported to show effects in preventing and treating HSs (Ning et al., 2021; Chen et al., 2022). SHO is a TCM prescription used for the external treatment of scarring, showing potential effects in treating HSs. However, the underlying mechanisms of SHO’s effects on HSs remain unexplored. In this study, using bioinformatic methods, we systematically explored the potential mechanisms of SHO in the treatment of HSs.
The SHO formula, developed from decades of clinical experience, has been used for external treatment of scaring since the 1960s, successfully treating thousands of patients with HSs. The SHO formula includes Galla Chinensis, Fructus Mume, Radix Angelicae Dahuricae, Fructus Cnidii, Herba Senecionis Scandentis, and Cortex Lycii. Based on the TCM theory of “qi and blood stasis, and toxic accumulation”, it follows the principle of “softening hardness, dissipating nodules, and detoxifying”. Galla Chinensis, the principal herb, has astringent, toxic-expelling, and hardness-softening properties, helping to reduce swelling and scar formation. Modern pharmacology shows it has powerful antioxidant properties, protecting biological macromolecules from damage (Ren et al., 2021). Fructus Mume, sour and astringent, complements Galla Chinensis by reducing scars (Zhang et al., 2023). Radix Angelicae Dahuricae promotes blood circulation, dissipates blood stasis, reduces swelling, and generates new tissue. Fructus Cnidii clears heat, dries dampness, dispels wind, and relieves itching. Herba Senecionis Scandentis clears heat, detoxifies, kills parasites, and relieves itching. Together, these herbs have antibacterial and anti-inflammatory effects, theoretically capable of controlling local inflammation, reducing pain and itching, and accelerating wound healing, thereby shortening the treatment duration.
We first retrieved the active ingredients in the SHO formula from the TCMSP database, and HSs-related target genes from five databases. Since there was no information about Senecio Scandens in the TCMSP database, we obtained its active ingredients from the HERB database. To ensure the rigor of our analyses, we cross-validated these active ingredients with the TCMSP database and used only those certified by the TCMSP database in our subsequent analyses. After a series of analyses, we identified 57 active ingredients and predicted 42 potential targets in the six herbs pf SHO (Table 1; Figure 1). Notably, since SHO is an external ointment, dermal permeability parameters should ideally be used as screening criteria. However, due to the limited number of network pharmacology studies focused on external drug applications, most databases do not provide dermal permeability parameters for active ingredients, and such data are generally unavailable. Consequently, this study employed pharmacokinetic parameters commonly used in other network pharmacology studies (Fan et al., 2024; Han et al., 2022) as the screening criteria. This limitation may affect the reliability of our findings and highlights the importance of incorporating dermal permeability parameters in future research. Network analyses indicated that SHO might affect HSs by regulating proteins such as AKT1, MAPK1, CCND1, TP53, GSK3B, BCL2, CDKN1A, ESR1, and MYC (Figure 3). GO and KEFF enrichment analyses showed that these targets are involved in multiple categories and pathways, such as the cytokine activity, regulation of apoptotic signaling pathway, response to oxidative stress, response to a steroid hormone, et al. (Supplementary Table S6; Supplementary Table S7), all of which are considered mechanisms of HSs (Mony et al., 2023; Gurtner et al., 2008; Diegelmann and Evans, 2004). Considering that SHO is used for external treatment, it is likely to act by influencing the response of myofibroblasts to regulatory factors in the microenvironment, such as cytokines and reactive oxygen species.
To further investigate the underlying molecular mechanisms, we performed molecular docking analyses of these hub target proteins and the core active ingredients in the SHO formula (Figure 2; Table 2). Although we identified three complexes with high affinities (Figure 5A), one of them did not form hydrogen bonds (Figure 5B), indicating that this conformation is unstable. The remaining two conformations, the MAPK1-stigmasterol complex, and the ESR1-alloimperatorin complex, showed high affinities and formed hydrogen bonds (Figures 5C,D), suggesting they are the most likely underlying molecular mechanisms of SHO’s effects on HSs. It should be noted that our docking results only offer preliminary indications of potential interactions. To more accurately assess binding stability, further validation through molecular dynamics simulations and binding free energy calculations is required.
Indeed, inhibition of the TGF-β1-ERK/JNK pathway has previously been found to be involved in scar formation. For example, basic fibroblast growth factor stimulates the proliferation of human dermal fibroblasts via the ERK1/2 and JNK pathways (Makino et al., 2010), and loureirin B has been found to inhibit the formation of HSs by inhibiting the TGF-β1-ERK/JNK pathway (He et al., 2015). Additionally, ESR1 is also identified to play important roles in skin aging and wound healing process (Saville and Hardman, 2014; Smith, 2017). These findings suggest that the mechanisms by which SHOW treats HSs may involve the regulation of ERK2 and ESR1 proteins. However, it is important to note that molecular docking results alone do not provide definitive evidence of molecular interactions. To address this limitation, binding assays should be conducted in wet-lab experiments to confirm these interactions, and further validation of the underlying mechanisms is warranted through both in vivo and in vitro studies.
Conclusion
In summary, this study analyzed the effective targets and action pathways of the SHO formula and preliminarily explored its molecular mechanisms in treating HSs using network pharmacology and molecular docking methods. Our findings revealed that the main active ingredients of SHO include quercetin, beta-sitosterol, kaempferol, stigmasterol, luteolin, alloimperatorin, acacetin, and (E)-2,3-bis(7-methoxy-2-oxochromen-8-yl)prop-2-enal. The hub target proteins identified were AKT1, MAPK1, CCND1, TP53, GSK3B, BCL2, CDKN1A, ESR1, and MYC. GO and KEGG enrichment analyses indicated that these hub target genes are involved in processes and pathways related to apoptosis and responses to oxidants. Molecular docking analysis identified the MAPK1-stigmasterol and ESR1-alloimperatorin complexes as having high affinities and forming hydrogen bonds, suggesting that the SHO formula may affect the MAPK1 and ESR1 proteins, thereby preventing and treating HSs. Our study will provide new drug and target options for the prevention and treatment of HSs, and offer theoretical support for further research and promotion of SHO. However, further in vivo and in vitro studies are warranted to validate these mechanisms.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
LJ: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review and editing. XZ: Investigation, Methodology, Writing – original draft, Writing – review and editing. HL: Investigation, Methodology, Writing – original draft, Writing – review and editing. JW: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was financially supported by a “study on the optimal formulation and evaluation of clinical efficacy of keloid cream in the treatment of proliferative scarring” set by Chengdu University of Chinese Medicine (21SX06).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2025.1511570/full#supplementary-material
References
Amini-Nik, S., Yousuf, Y., and Jeschke, M. G. (2018). Scar management in burn injuries using drug delivery and molecular signaling: current treatments and future directions. Adv. Drug Deliv. Rev. 123, 135–154. doi:10.1016/j.addr.2017.07.017
Atiyeh, B. S. (2020). Nonsurgical management of hypertrophic scars: evidence-based therapies, standard practices, and emerging methods. Aesthetic Plast. Surg. 44 (4), 1320–1344. doi:10.1007/s00266-020-01820-0
Bharadia, S. K., Burnett, L., and Gabriel, V. (2023). Hypertrophic scar. Phys. Med. Rehabilitation Clin. N. Am. 34 (4), 783–798. doi:10.1016/j.pmr.2023.05.002
Chen, D., Li, Q., Zhang, H., Kou, F., Li, Q., Lyu, C., et al. (2022). Traditional Chinese medicine for hypertrophic scars-A review of the therapeutic methods and potential effects. Front. Pharmacol. 13, 1025602. doi:10.3389/fphar.2022.1025602
Choi, J., Lee, E. H., Park, S. W., and Chang, H. (2015). Regulation of transforming growth factor β1, platelet-derived growth factor, and basic fibroblast growth factor by silicone gel sheeting in early-stage scarring. Arch. Plast. Surg. 42 (1), 20–27. doi:10.5999/aps.2015.42.1.20
Coentro, J. Q., Pugliese, E., Hanley, G., Raghunath, M., and Zeugolis, D. I. (2019). Current and upcoming therapies to modulate skin scarring and fibrosis. Adv. Drug Deliv. Rev. 146, 37–59. doi:10.1016/j.addr.2018.08.009
Diegelmann, R. F., and Evans, M. C. (2004). Wound healing: an overview of acute, fibrotic and delayed healing. Front. Biosci. 9, 283–289. doi:10.2741/1184
Fan, Q.-Q., Zhai, B.-T., Zhang, D., Zhang, X. F., Cheng, J. X., Guo, D. Y., et al. (2024). Study on the underlying mechanism of yinhua gout granules in the treatment of gouty arthritis by integrating transcriptomics and network pharmacology. Drug Des. Dev. Ther. 18, 3089–3112. doi:10.2147/DDDT.S475442
Fang, S., Dong, L., Liu, L., Guo, J., Zhao, L., Zhang, J., et al. (2021). HERB: a high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Res. 49 (D1), D1197–D1206. doi:10.1093/nar/gkaa1063
Gurtner, G. C., Werner, S., Barrandon, Y., and Longaker, M. T. (2008). Wound repair and regeneration. Nature 453 (7193), 314–321. doi:10.1038/nature07039
Han, X. X., Tian, Y. G., Liu, X. F., Zhao, D., Du, X. H., Dong, H. R., et al. (2022). Network pharmacology combined with pharmacodynamics revealed the anti-inflammatory mechanism of Tanreqing capsule against acute-exacerbation chronic obstructive pulmonary disease. Sci. Rep. 12 (1), 13967. doi:10.1038/s41598-022-18326-1
He, T., Bai, X., Yang, L., Fan, L., Li, Y., Su, L., et al. (2015). Loureirin B inhibits hypertrophic scar formation via inhibition of the TGF-β1-ERK/JNK pathway. Cell. Physiology Biochem. 37 (2), 666–676. doi:10.1159/000430385
Islam, M. A., Khandker, S. S., Alam, F., Kamal, M. A., and Gan, S. H. (2018). Genetic risk factors in thrombotic primary antiphospholipid syndrome: a systematic review with bioinformatic analyses. Autoimmun. Rev. 17 (3), 226–243. doi:10.1016/j.autrev.2017.10.014
Jeschke, M. G., Wood, F. M., Middelkoop, E., Bayat, A., Teot, L., Ogawa, R., et al. (2023). Scars. Nat. Rev. 9 (1), 64. doi:10.1038/s41572-023-00474-x
Kou, H., and Ma, J. (2024). Network pharmacology prediction and molecular docking-based strategy to explore the potential mechanism of Duhuo-Jisheng pair against osteoarthritis. J. Pharm. Pharmacol. 76 (7), 743–752. doi:10.1093/jpp/rgad117
Leblebici, B., Adam, M., Bağiş, S., Tarim, A. M., Noyan, T., Akman, M. N., et al. (2006). Quality of life after burn injury: the impact of joint contracture. J. Burn Care and Res. 27 (6), 864–868. doi:10.1097/01.BCR.0000245652.26648.36
Limandjaja, G. C., Niessen, F. B., Scheper, R. J., and Gibbs, S. (2021). Hypertrophic scars and keloids: overview of the evidence and practical guide for differentiating between these abnormal scars. Exp. Dermatol 30 (1), 146–161. doi:10.1111/exd.14121
Lingzhi, Z., Meirong, L., and Xiaobing, F. (2020). Biological approaches for hypertrophic scars. Int. Wound J. 17 (2), 405–418. doi:10.1111/iwj.13286
Makino, T., Jinnin, M., Muchemwa, F., Fukushima, S., Kogushi-Nishi, H., Moriya, C., et al. (2010). Basic fibroblast growth factor stimulates the proliferation of human dermal fibroblasts via the ERK1/2 and JNK pathways. Br. J. Dermatology 162 (4), 717–723. doi:10.1111/j.1365-2133.2009.09581.x
Mcphail, S. M., Wiseman, J., Simons, M., Kimble, R., and Tyack, Z. (2022). Cost-effectiveness of scar management post-burn: a trial-based economic evaluation of three intervention models. Sci. Rep. 12 (1), 18601. doi:10.1038/s41598-022-22488-3
Mony, M. P., Harmon, K. A., Hess, R., Dorafshar, A. H., and Shafikhani, S. H. (2023). An updated review of hypertrophic scarring. Cells 12 (5), 678. doi:10.3390/cells12050678
Ning, X., Wiraja, C., Chew, W. T. S., Fan, C., and Xu, C. (2021). Transdermal delivery of Chinese herbal medicine extract using dissolvable microneedles for hypertrophic scar treatment. Acta Pharm. Sin. B 11 (9), 2937–2944. doi:10.1016/j.apsb.2021.03.016
O'brien, L., and Jones, D. J. (2013). Silicone gel sheeting for preventing and treating hypertrophic and keloid scars. Cochrane Database Syst. Rev. 2013 (9), Cd003826. doi:10.1002/14651858.CD003826.pub3
Ren, Y. Y., Zhang, X. R., Li, T. N., Zeng, Y. J., Wang, J., and Huang, Q. W. (2021). Galla Chinensis, a Traditional Chinese Medicine: comprehensive review of botany, traditional uses, chemical composition, pharmacology and toxicology. J. Ethnopharmacol. 278, 114247. doi:10.1016/j.jep.2021.114247
Ru, J., Li, P., Wang, J., Zhou, W., Li, B., Huang, C., et al. (2014). TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform 6, 13. doi:10.1186/1758-2946-6-13
Saikia, S., and Bordoloi, M. (2019). Molecular docking: challenges, advances and its use in drug discovery perspective. Curr. Drug Targets 20 (5), 501–521. doi:10.2174/1389450119666181022153016
Saville, C. R., and Hardman, M. J. (2014). “The role of estrogen deficiency in skin aging and wound healing,” in Skin, mucosa and menopause: management of clinical issues (Springer), 71–88.
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Smith, M. (2017). Exploring the effects of estrogen receptor beta polymorphisms on wound repair. United Kingdom: The University of Manchester.
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic acids Res. 47 (D1), D607-D613–D13. doi:10.1093/nar/gky1131
Tang, Y., Li, M., Wang, J., Pan, Y., and Wu, F. X. (2015). CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 127, 67–72. doi:10.1016/j.biosystems.2014.11.005
Thombs, B. D., Notes, L. D., Lawrence, J. W., Magyar-Russell, G., Bresnick, M. G., and Fauerbach, J. A. (2008). From survival to socialization: a longitudinal study of body image in survivors of severe burn injury. J. Psychosomatic Res. 64 (2), 205–212. doi:10.1016/j.jpsychores.2007.09.003
UniProt (2021). The universal protein knowledgebase in 2021. Nucleic Acids Res., 49(D1): D480-d9. doi:10.1093/nar/gkaa1100
Zhang, J., Li, Z., Zhang, Y., Guo, Y. L., Zhu, Y. R., Xia, W. X., et al. (2023). Mume Fructus (Prunus mume Sieb. et Zucc.) extract accelerates colonic mucosal healing of mice with colitis induced by dextran sulfate sodium through potentiation of cPLA2-mediated lysophosphatidylcholine synthesis. Phytomedicine 119, 154985. doi:10.1016/j.phymed.2023.154985
Zhang, Q., Yu, H., Qi, J., Tang, D., Chen, X., Wan, J. B., et al. (2017). Natural formulas and the nature of formulas: exploring potential therapeutic targets based on traditional Chinese herbal formulas. PLoS One 12 (2), e0171628. doi:10.1371/journal.pone.0171628
Keywords: hypertrophic scars, traditional Chinese medicine, scar healing ointment, network pharmacology, molecular docking
Citation: Jiang L, Zeng X, Li H and Wu J (2025) Network pharmacology and molecular docking analysis on molecular targets and mechanisms of scar healing ointment in the treatment of hypertrophic scars. Front. Pharmacol. 16:1511570. doi: 10.3389/fphar.2025.1511570
Received: 15 October 2024; Accepted: 22 July 2025;
Published: 06 August 2025.
Edited by:
Masaoki Kawasumi, University of Washington, United StatesCopyright © 2025 Jiang, Zeng, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingping Wu, amluZ3Bpbmd3dTIwMjJAMTYzLmNvbQ==
 Lijuan Jiang1
Lijuan Jiang1 Jingping Wu
Jingping Wu