- 1Department of Clinical Pharmacy, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
- 2DNA Extraction and Oligo Synthesis Unit, King Faisal Specialist Hospital and Research Center, Riyadh, Saudi Arabia
- 3Medicine-Hematology, King Khalid University Hospital, King Saud University, Riyadh, Saudi Arabia
- 4Translational and Clinical Research Institute, Newcastle University, Newcastle upon Tyne, United Kingdom
Background: Limited data are available on factors that affect warfarin dose requirement in Saudi patients. Saudis are among the underrepresented ethnic groups in warfarin pharmacogenetics research. The present study investigated the frequency of CYP2C9*2 and*3, CYP4F2 (G1347A) and VKORC1 –1639G>A genotypes and their impact on warfarin dose requirement in a cohort of Saudi patients requiring anticoagulation therapy.
Methods: 193 patients on chronic warfarin therapy and with stable anticoagulation took part in the study. Genotyping for VKORC1 1639G>A, CYP4F2 G1347A, CYP2C9*2 430C>T and CYP2C9*3 1075A>C were performed using TaqMan genotyping assays. Analysis of variance was carried out to determine the association between CYP2C9, CYP4F2, and VKORC1 genotype and warfarin dose requirement in two groups based on target INR range. Backward linear regression analysis identified genetic and clinical factors influencing doe requirements.
Results: Patients with CYP2C9 and VKORC1 polymorphisms required significantly lower warfarin doses compared to wild-type patients. Carriers of two mutant alleles required lower doses than those with one mutant allele. In contrast, CYP4F2 polymorphisms did not influence warfarin dose. Age and genetic variants in CYP2C9 and VKORC1 were negatively correlated with dose requirements, while body surface area (BSA) was positively correlated.
Conclusion: Saudi patients with polymorphisms in CYP2C9 and VKORC1 required lower warfarin doses than those with the wild-type allele. CYP4F2 polymorphism had no effect on warfarin dose requirement. Integrating patient clinical factors, including age and BSA, and genetic polymorphisms in CYP2C9 and VKORC1 provides the best estimation of factors contributing to warfarin dose in the Saudi patient population.
1 Introduction
For many years, vitamin K antagonists (VKAs), such as warfarin, have proven effective in preventing and treating arterial and venous thrombotic events. Warfarin exerts its anticoagulant effect via the inhibition of the vitamin K epoxide reductase (VKOR) enzyme, thus preventing reduction of vitamin K and reducing the synthesis of functional vitamin K–dependent clotting factors. Polymorphism in the VKORC1 gene is a major contributor to warfarin dose requirement. This gene is located on chromosome 16 and encodes the VKOR enzyme. VKORC1-1639A variant allele is associated with decreased warfarin dose requirement and increased risk of over anticoagulation (Bourgeois et al., 2016; Sconce et al., 2005; Scott et al., 2010).
Warfarin is a 1:1 racemic mixture of the R and S stereoisomers. The S enantiomer is approximately five times more potent than the R warfarin (Moyer et al., 2009). CYP2C9 is the main enzyme involved in the metabolism of (S)-warfarin (Kalman et al., 2016; Westervelt et al., 2014). More than 57 allelic variants of CYP2C9 have been identified (Niinuma et al., 2014). CYP2C9*2 and CYP2C9*3 are the most common SNPs found to affect warfarin metabolism. Individuals carrying the CYP2C9*2 or CYP2C9*3 alleles usually have decreased enzymatic activity and therefore slower warfarin metabolism, decreased warfarin clearance, increase warfarin sensitivity and bleeding risk (Gulseth et al., 2009; Kamali and Wynne, 2010; Lane et al., 2012; Moyer et al., 2009; Sconce et al., 2005).
Monitoring therapeutic response to VKAs is necessary because of the risk of thrombosis and bleeding associated with under- and over-anticoagulation, respectively (Wigle et al., 2013). The International Normalized Ratio (INR) is the most common method for monitoring VKAs therapy. Deviations from target INR range increases the risk of bleeding or stroke particularly during the first weeks of therapy (Homme et al., 2008).
Achieving and maintaining optimal anticoagulation is challenging due to the wide inter- and intra-individual variation in warfarin response and dose requirements. Although over 60 years have passed since warfarin was first used as an anticoagulant, the factors that contribute to the variability in dose requirement (up to 20-fold in daily dose requirement) have not yet been fully identified (Wen and Lee, 2013). Demographic, clinical, and genetic factors are shown to contribute to warfarin dose requirement (Kamali and Wynne, 2010). CYP2C9 and VKORC1 are reported to be the most important genetic determinants of warfarin dose requirements in different patient populations (Borgiani et al., 2009; Cen et al., 2010; Tatarunas et al., 2014). Other genetic variations, including that in the CYP4F2 gene, have been associated with warfarin dose but to a lesser extent. Genetic polymorphisms in VKORC1 -1639G>A, CYP2C9*2, and CYP2C9*3, together with clinical factors such as age and sex, explain up to 57% of the variability in warfarin dose requirement (Cavallari and Perera, 2012; McDonald et al., 2009; Zhang et al., 2017).
Significant differences exist in warfarin dose requirements between different ethnic groups, whereby Asians and African Americans require lower and higher warfarin doses, respectively, compared with Caucasians (Cavallari and Perera, 2012; Kamali and Wynne, 2010). Saudis are among the underrepresented ethnic groups in warfarin pharmacogenetics research. Only a few studies in Saudi patients have thus far reported the prevalence of CYP2C9 and VKORC1 polymorphisms and their impact on warfarin dose requirement and patient response (Al Ammari et al., 2020; Alzahrani et al., 2013; Mirghani et al., 2011; Mizzi et al., 2016; Saour et al., 2011; Al-Saikhan et al., 2018). To date, no studies have explored the effect of CYP4F2 genetic polymorphisms on warfarin dose requirements in Saudi patients. The present study was designed to a) investigate the frequency of CYP2C9*2 and *3, CYP4F2 (G1347A) and VKORC1 –1639G>A genotypes in the Saudi population, b) to evaluate the impact of polymorphisms in CYP2C9, CYP4F2 and VKORC1 genes on warfarin dose requirement, and c) to explore the association between CYP2C9*2,*3, CYP4F2 (G1347A) and VKORC1 –1639G>A genotypes along with other non-genetic factors and warfarin dose.
2 Materials and methods
2.1 Study design
This cross-sectional study was conducted at the King Khaled University Hospital (KKUH) anticoagulation clinic. Ethical approval was obtained from the Institutional Review Board in November 2017. All patients signed informed consent before participation. DNA extraction and genotyping were performed according to established methods at King Faisal Specialist Hospital and Research Centre (KFSHRC).
2.2 Participants
Between March 2018 and October 2019, 193 patients on warfarin therapy were recruited from the anticoagulation clinic at KKUH. The patients included were 18 years old or older, on at least 12 months of warfarin therapy, and had stable anticoagulation. A warfarin dose requirement that has been constant for at least three prior clinic visits spanning a minimum of 3 months and an INR within the target range are considered signs of stable anticoagulation. Patients were excluded if they had abnormal hepatic or renal function or were receiving medications known to interact with warfarin.
2.3 Data collection
Patient data were obtained using a self-report questionnaire and hospital records. The questionnaire comprised demographic information and warfarin-related information (indication, duration of use, recommended INR range). Demographic information comprised gender, age, weight, height, marital status, education, and place of residence. Data were also obtained from participants’ medical records, including comorbidities, concomitant medical therapy, previous stroke, and bleeding events.
2.4 DNA extraction and genotyping
DNA extraction was performed at KFSHRC according to the manufacturer’s protocol using the PureGene DNA extraction kit (Qiagen Sciences, Germantown, Maryland, United States). A Nanodrop ND-1000 spectrophotometer (Wilmington, DE, United States) was used to determine DNA concentration and purity. Genotyping for VKORC1 1639G>A (rs9923231; C_30403261_20), CYP4F2 G1347A (rs2108622; C_16179493_40), CYP2C9*2 430C>T (rs1799853; C_25625805_10) and CYP2C9*3 1075A>C (rs1057910; C_27104892_10) was performed using TaqMan genotyping assays according to the standard protocol provided by Applied Biosystems. The TaqMan real-time polymerase chain reaction (PCR) method results of 96 DNA samples were validated by Sanger sequencing. Primers for PCR were designed at the Oligonucleotide Synthesis Unit of the Genetics department at KFSHRC. Primer details are described in Table 1.
2.5 Data analysis
Data were analyzed using SPSS version 27 (IBM Corp- Chicago- IL- United States). Continuous variables are described as medians, means, and standard deviations. Categorical variables were summarized with numbers and percentages. Hardy-Weinberg equilibrium was assessed using the following equation: p2+2pq + q2 = 1, where p2 = frequency of homozygous wild genotype; 2pq = frequency of heterozygous genotype; q2 = frequency of homozygous mutant genotype.
The chi-squared test was used to compare observed genotype frequencies with expected frequencies. Analysis of variance (ANOVA) was carried out to determine the association between CYP2C9, CYP4F2, and VKORC1 genotype and warfarin dose requirement in two groups based on target INR range. Logarithmic transformation was performed to normalize warfarin doses. Data for each target INR group (2.0–3.0 and 2.5–3.5) were analyzed separately. Backward linear regression analysis was performed to identify genetic and clinical factors contributing to warfarin dose requirements. Log-transformed warfarin doses were used as a dependent variable to ensure that the values were normally distributed. Patient age, gender, BSA, smoking status, hypertension (HTN), diabetes mellitus (DM), myocardial infarction (MI), heart failure (HF), hypothyroidism, renal impairment, beta-blockers, antiplatelet, CYP2C9, VKORC1 and CYP4F2 were treated as independent variables.
3 Results
3.1 Baseline characteristics
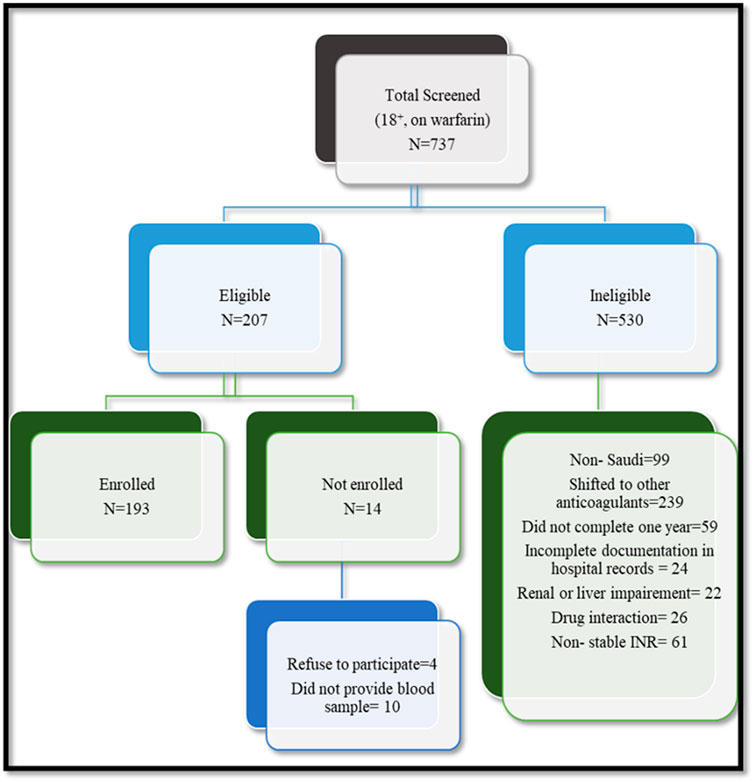
Out of 207 patients on long-term warfarin therapy identified for possible study participation, 193 (93.2%) met the inclusion criteria, signed informed consent, and answered the questionnaire, and each provided a blood (5 mL) sample. Figure 1 shows the flow chart for patient recruitment.
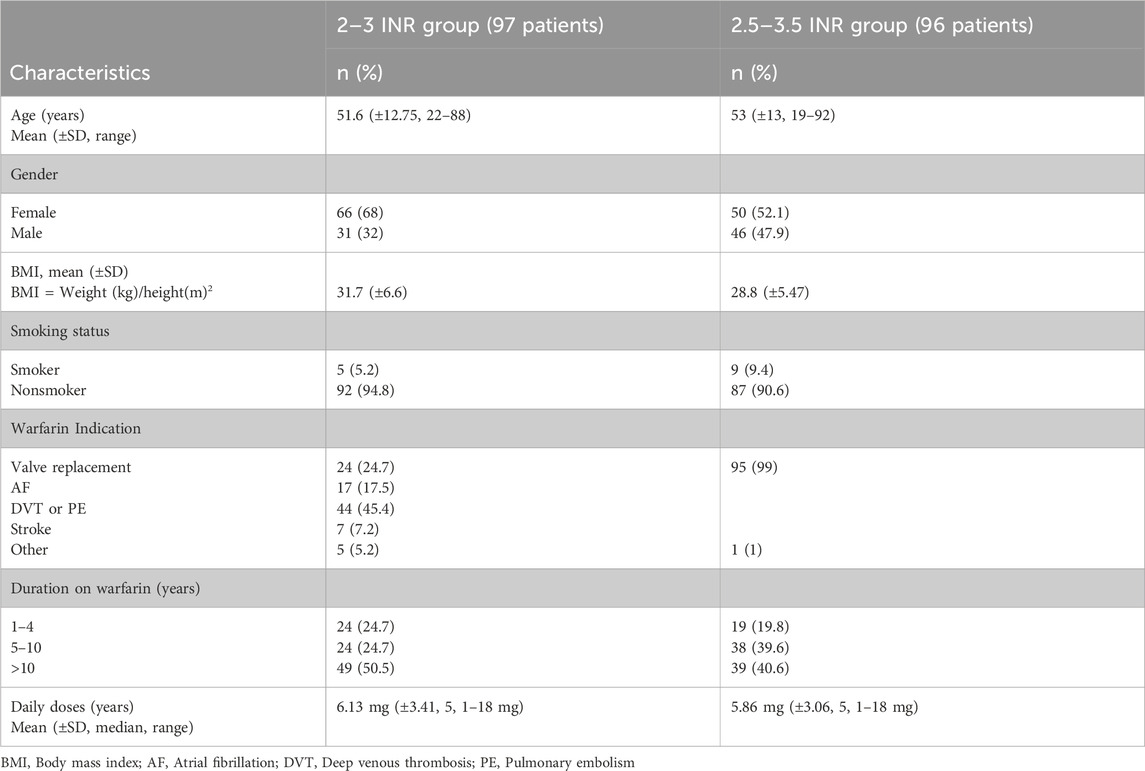
Table 2 represents the demographic and clinical characteristics of the patients which were analyzed separately based on the target INR range. Ninety-six patients had mechanical prosthetic heart valves with target INR 2.5–3.5. Their mean (±SD) age was 53 (±13) years (range: 19–92 years; median = 54 years). 52.1% of the participants in this group were female and 40.6% were on warfarin therapy for over 10 years with a mean (±SD, median, range) daily doses of warfarin of 5.86 mg (±3.06, 5, 1–18 mg). The remaining 97 patients in the cohort had a target INR 2.0–3.0. Their mean (±SD) age was 51.6 (±12.75) years (range: 22–88 years; median = 52 years). Approximately 68% of the participants in this group were female. Deep vein thrombosis/pulmonary embolism was the most common cause of warfarin use (45.4%). Other indications for anticoagulation were valvular replacement, atrial fibrillation, stroke and other indications in 24.7%, 17.5%, 7.2 and 5.2 of patients, respectively. About 50% of the participants have received warfarin therapy for more than 10 years. The mean (±SD, median, range) daily doses of warfarin in the group was 6.13 mg (±3.41, 5, 1–18 mg).
3.2 CYP2C9, VKORC1, and CYP4F2 genotype frequencies
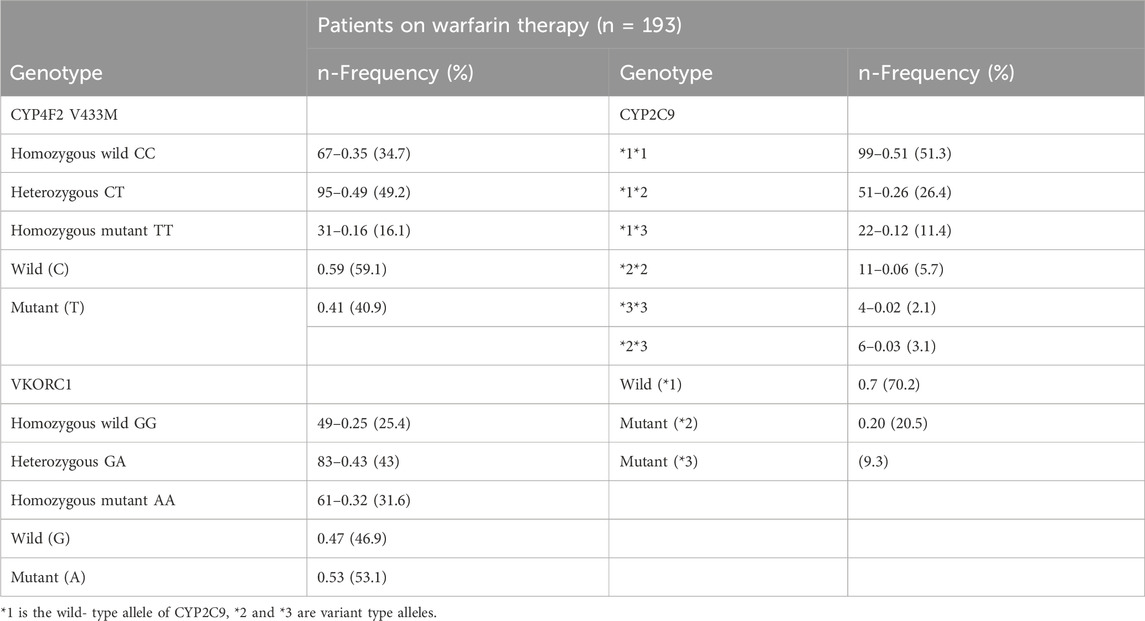
Table 3 summarizes the Saudi participants’ genotype distribution for VKORC1, CYP2C9*2 and *3 and CYP4F2. The VKORC1, CYP2C9*2 and *3 and CYP4F2 genotype were in Hardy-Weinberg equilibrium. The results of PCR were in 100% concordance with those obtained by the Sanger sequencing method.
The results showed that the CYP2C9*1*1 genotype was observed in 51.3% of participants, making it the most prevalent genotype. CYP2C9 *2 (20.5) was more common than CYP2C9*3 (9.3). Approximately 43% of the study patients were heterozygous (GA) for the VKORC1 genotype, 31% homozygous mutant alleles, and 25% homozygous wild-type genotypes. With regard to CYP4F2 V433M, 49.2% of the patients were of the heterozygous (CT) genotype, and ∼34% were homozygous wild-type (CC) alleles, while 16% were homozygous mutant (TT) alleles.
3.3 Impact of CYP2C9, VKORC1 and CYP4F2 on warfarin dose requirement
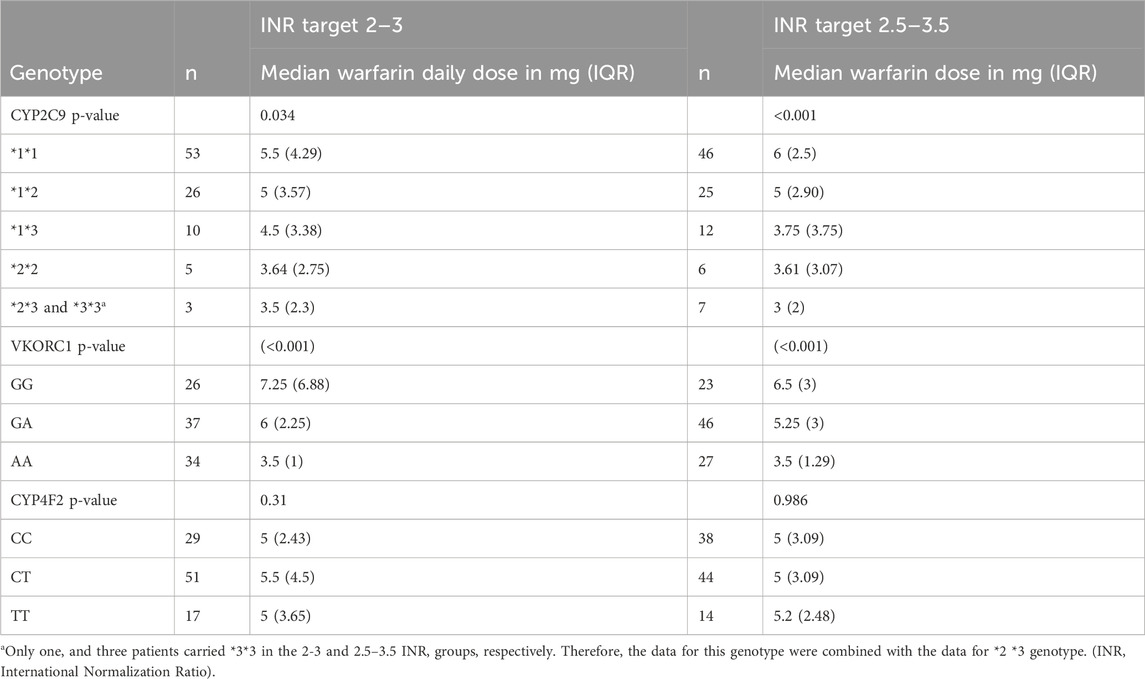
Warfarin dose requirements were significantly lower in patients with polymorphisms in CYP2C9 (Figure 2) and VKORC1 (Figure 3) than in wild-type patients (one way-ANOVA). Additionally, carriers of two mutant alleles had significantly lower dosage requirements than those of one mutant allele. In contrast, polymorphism in CYP4F2 did not affect warfarin dose requirements (Table 4).
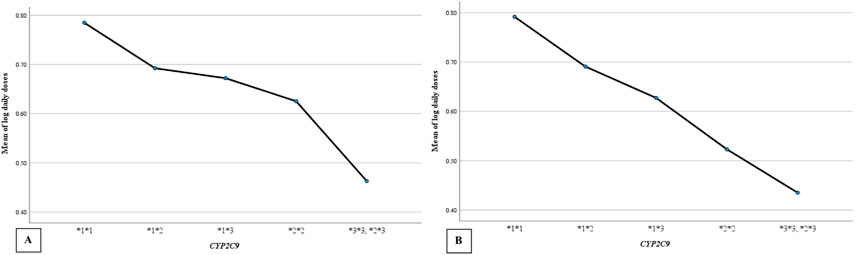
Figure 2. The mean daily warfarin dose requirement between CYP2C9 wild-type and variant genotypes. (A) target INR 2–3, (B) target INR 2.5–3.5.
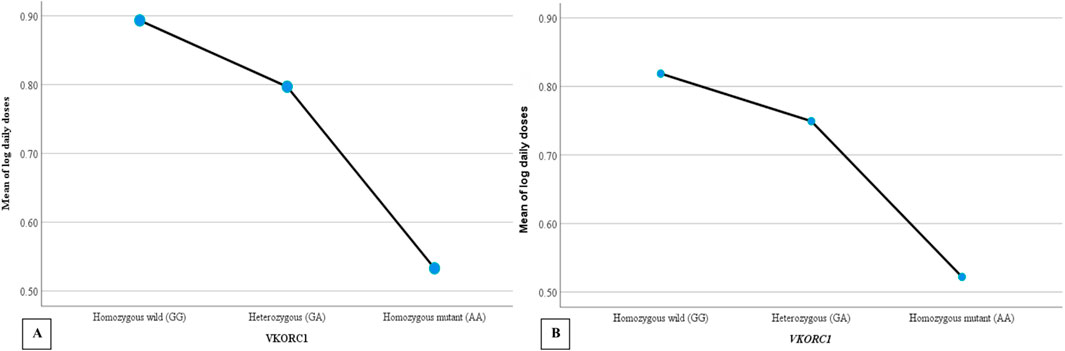
Figure 3. The mean daily warfarin dose requirement between VKORC1 wild-type and variant genotypes. (A) Target INR 2–3, (B) Target INR 2.5–3.5.
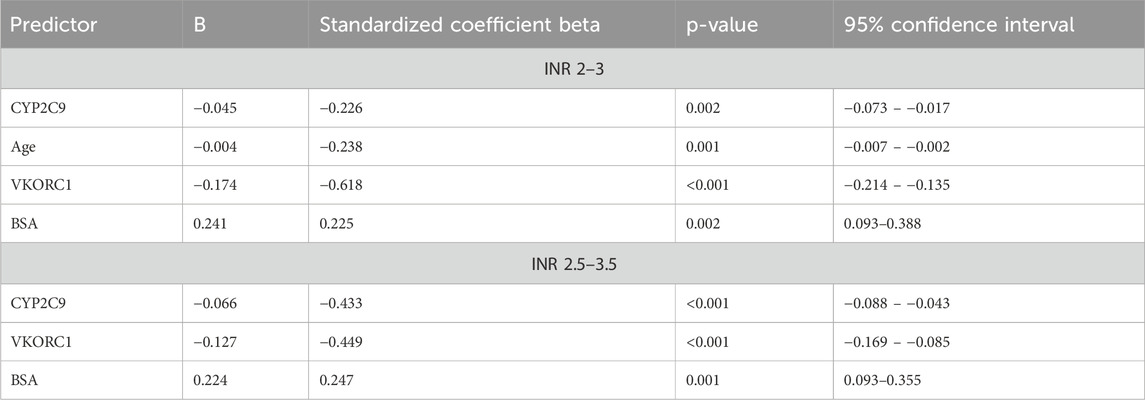
Backward linear regression analysis was used to identify genetic and clinical factors contributing to warfarin dose requirements using log daily dose as a dependent variable and patients’ age, gender, smoking status, hypertension, diabetes, myocardial infarction, heart failure, hypothyroidism, renal impairment, beta-blockers, antiplatelet, CYP2C9, VKORC1, and CYP4F2 as independent variables (Table 5). Age, BSA, CYP2C9, and VKORC1 accounted for 54.6% of the interindividual variability in warfarin dose requirement in patients with a target INR of 2.0–3.0. In patients with a target INR of 2.5–3.5, BSA, CYP2C9, and VKORC1 explained 51.2% of warfarin dose requirements. Age and polymorphisms in CYP2C9 and VKORC1 were negatively correlated with warfarin dose, while BSA was positively correlated with dose requirement.
4 Discussion
Warfarin use is complicated by the risk of bleeding and thrombosis owing to its narrow therapeutic window and variable dose response. Several demographics, clinical and genetic factors are known to significantly affect response to warfarin therapy. These include mainly patient’s age, gender, body size, comorbidities, concurrent medications, genetics, smoking as well as drug and food interactions (Apostolakis et al., 2013). Clinical factors together with genetic polymorphism in VKORC1 -1639G>A, CYP2C9*2, CYP2C9*3 and CYP4F2 V433M are responsible for 55%–57% of the variability in anticoagulation response to warfarin among Caucasians and 25% in African Americans (Cavallari and Perera, 2012; McDonald et al., 2009; Zhang et al., 2017).
Polymorphisms in the CYP2C9, VKORC1, and CYP4F2 genes have been found to contribute to the variability in warfarin dosage requirements in several ethnic groups. It is regarded that patients with polymorphisms in the VKORC1 gene have reduced expression of the VKOR enzyme. This type of mutation puts patients at higher risk of bleeding associated with VKAs therapy, necessitating lower warfarin doses. CYP2C9 is the major enzyme responsible for warfarin metabolism. Two major CYP2C9 variant alleles (CYP2C9 *2 and *3) affecting warfarin metabolism have been identified thus far in the literature. Carriers of the CYP2C9*2 and CYP2C9*3 variant alleles require lower warfarin doses than carriers of the wild-type CYP2C9 allele (Borgiani et al., 2009; Cen et al., 2010; Tatarunas et al., 2014). The CYP2C9*3 variant allele leads to the expression of CYP2C9 with reduced activity to a greater extent than the CYP2C9*2 variant allele. On the other hand, carriers of the CYP4F2 allele have been reported to require higher warfarin doses (Caldwell et al., 2008). However, CYP4F2 polymorphism has less impact on warfarin dose than those of the CYP2C9 and VKORC1 genes.
People from Saudi Arabia are among the ethnic groups that have not been adequately studied in warfarin pharmacogenetics research. To our knowledge, this is the first study investigating the prevalence of the CYP4F2 V433M polymorphism and its impact on the warfarin response among Saudi patients. Moreover, very few studies to date have investigated the prevalence of CYP2C9 and VKORC1 and their effect on warfarin dose requirements and anticoagulation response (Alzahrani et al., 2013; Al Ammari et al., 2020; Mirghani et al., 2011; Saour et al., 2011; Mizzi et al., 2016). Therefore, we aimed to investigate the frequency of the VKORC1 –1639G>A, CYP2C9*2 and *3 alleles, and CYP4F2 (G1347A) polymorphism and the association between VKORC1, CYP2C9 and CYP4F2 genotypes and warfarin dose requirement in this population.
The frequency of CYP2C9, VKORC1, and CYP4F2 genotypes varies among different ethnic populations. Americans and Europeans have higher CY2C9*2 and CY2C9*3 allele frequencies than African-American and Asian populations. The frequency of CYP2C9*1, *2, and * 3 are present in approximately 80%, 13%, and 7% of Caucasian individuals, respectively. Southeast Asians have a somewhat higher percentage of CYP2C9*3 (2%–10%), while CYP2C9*2 is almost absent. CYP2C9*2 (3%–5%) and *3 (1%–2%) polymorphisms are rare in African Americans (Gulseth et al., 2009; Westervelt et al., 2014). CYP2C9*1*1 was the most common genotype, while *3*3 was the least common in our population of Saudi patients. Additionally, the frequencies of the heterozygous alleles *1*2 and *1*3 were higher than those of the homozygous alleles *2*2 and *3*3. CYP2C9 *1*1 was present in 51.3% of patients on warfarin treatment. This was slightly lower than that reported in other studies of Saudi patients of 68.7% (Alzahrani et al., 2013), 64.1% (Mirghani et al., 2011), and 63.4% (Saour et al., 2011). However, the genotypic frequency of 26.4% for CYP2C9 *1*2 in anticoagulated patients was similar to the 26.7% reported by Alzahrani et al. (2013) did not report any patients on warfarin in their study with homozygous mutant allele genotypes (*2*2 and *3*3). The VKORC1-1639A variant allele is prevalent in approximately 67%, 40%, and 11% of Asians, African Americans and Caucasians, respectively. In the current study, approximately 43% of participants carried the heterozygous VKORC1-GA genotype, 31% carried the homozygous mutant (AA) allele, and 25% carried the homozygous wild-type allele (GG). This finding is similar to the findings of Al Ammari et al. (2020), who reported the following frequencies: GA (45.3%), AA (34%), and GG (20.7%). Regarding the CYP4F2 V433M genotype, 49.2% of the anticoagulated patients carry the heterozygous (CT) allele. Almost 34% of the participants carry the homozygous wild-type (CC) allele, while 16% carry the homozygous mutant (TT) allele. The frequency of the mutant CYP4F2 T allele in this study was 40.9%, while it was approximately 30% in white and Asian people and 7% in black individuals (Caldwell et al., 2008).
The association between CYP2C9*2 and *3, CYP4F2 (G1347A), and VKORC1 –1639G>A genotypes and warfarin dose requirements varies among populations. Compared to Caucasians, warfarin doses are higher among African Americans and lower among Asians. The main reason is the different gene frequencies reported in different racial groups. For instance, VKORC1(-1639A), CYP2C9*2, and *3 alleles have been found to be significantly associated with lower warfarin dose requirements in several populations. The frequency of VKORC1 (-1639A), CYP2C9*2, and *3 is lower in African–Americans than in Asians and Caucasians (Cavallari and Perera, 2012). CYP4F2 has also been found to affect warfarin dosage requirements in white and Asian population but not in Indians, Egyptians, Brazilians, and Black ethnic groups (Danese et al., 2019). This study found that patients carrying the CYP2C9*2, *3, and VKORC1 A allele required lower doses than those carrying the wild-type CYP2C9 and VKORC1 G allele. This study is the first to evaluate the impact of CYP4F2 polymorphism on warfarin dose requirements and warfarin response in a Saudi patient population. In contrast to CYP2C9 and VKORC1, CYP4F2 polymorphism was not found to be related to warfarin dose requirement.
There is a lack of research on the impact of the combined effect of clinical and genetic factors on warfarin dosing in the Saudi population. The association between genetic polymorphisms and stable maintenance doses was not properly assessed in previous studies of Saudi patients. Al Ammari et al. (2021) assessed the relationship between polymorphisms in VKORC1 and warfarin doses during the initiation phase (first 10 days of therapy). Al-Saikhan and colleagues published two separate pieces of research to assess the effects of CYP2C9 and VKORC1 1173C>T SNP on warfarin dose variability in 112 and 164 Saudi patients, respectively. However, the combined effect of genotypes and clinical factors was not addressed (Al-Saikhan and Abd-Elaziz, 2018). Saour et al. (2011) evaluated the effects of CYP2C9 polymorphism on the requirements of warfarin doses in Saudi individuals who received 2 mg or less of warfarin daily. Associations with doses greater than 2 mg were not assessed.
The present study demonstrated that age, BSA, CYP2C9*2 and *3, and VKORC1 (–1639G>A) variants contribute significantly to warfarin dose requirements. Age and variants of CYP2C9 and VKORC1 alleles were negatively correlated with warfarin doses, while BSA positively correlated with dosage requirements. It has been reported that CYP2C9 and VKORC1 variant alleles cause significant differences in warfarin doses in Caucasians, Asians, and African Americans; however, VKORC1 is known to be less common in African–Americans (Cavallari and Perera, 2012). The results of this study are similar to those of other previous studies in Caucasian populations. In a study by Kamali et al. (2004), age and CYP2C9*2 and *3 alleles significantly contributed to warfarin dose requirements. Sconce et al. (2005) reported that warfarin doses are negatively correlated with age, body weight, height, body surface area, VKORC1 (–1639G>A), CYP2C9*2 and *3 alleles. Genetic polymorphisms in CYP2C9 and VKORC1, together with age and height, were responsible for 54% of the variation in warfarin dose. Caldwell et al. (2008) suggested that clinical factors and polymorphisms in CYP2C9 and VKORC1 explained 54% of the interindividual variability in warfarin response. Bourgeois et al. (2016) performed a prospective cohort study to identify clinical and genetic factors contributing to warfarin response in 711 British patients. The multifactorial analysis of genetic (VKORC1 and CYP2C9) and clinical factors explained 57.89% of the variation in the mean weekly warfarin dose and 56.97% in the stable mean weekly dose. Analyzing 117 studies involving patients of various ethnic origins in a meta-analysis by Jorgensen et al. (2012) revealed that CYP2C9 and VKORC1 variant alleles significantly contribute to warfarin dose requirements in most ethnicities. Loebstein et al. (2001) established that older patients on warfarin treatment required significantly lower doses than younger patients. Gage et al. (2008) identified predictors of warfarin dose as VKORC1 −1639/3673 G>A (−28%), BSA (+11% per 0.25 m2), CYP2C9*3 (−33%), CYP2C9*2 (−19%), age (−7% for each decade), target INR (+11% for every 0.5-unit increase), amiodarone (−22%), smoking status (+10%), race (−9%) and current thrombosis (+7%). Similar to some previous studies (Rieder et al., 2005; Wadelius et al., 2005; Bourgeois et al., 2016), we found that the contribution of VKORC1 to warfarin dosage requirements was greater than that of CYP2C9.
The strengths of this study lie in the rigorous criteria that were used for participant selection. Eligibility criteria included stable INR control based upon a minimum period of 3 months. In addition, patients with factors that might affect anticoagulation control were excluded, all of which are important in determining the appropriate warfarin dose for each patient. Furthermore, sequencing was performed on random samples to validate the results of the real-time PCR with 100% concordance. Both real-time PCR genotyping and sequencing provide accurate genotyping analysis
The main limitation in our study is that sample size calculation was not performed, so the study may lack the power to detect the possible significant impact of CYP4F2 on warfarin dose requirements. Further studies are required to examine the effect of CYP4F2 polymorphism on warfarin dose requirements in a larger sample of Saudi patients.
5 Conclusion
This work contributes to the existing knowledge of how genetics affects the response to warfarin by providing new data regarding an underrepresented ethnic group, Saudi Arabia. This study showed that Saudi patients with polymorphisms in CYP2C9 and VKORC1 required lower warfarin doses than those with the wild-type allele. Integrating patient clinical factors, including age and BSA, and genetic polymorphisms in CYP2C9 and VKORC1 provides the best estimation of factors contributing to warfarin dose in this population.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by King Khalid University Hospital Institutional Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
SJ: Conceptualization, Formal Analysis, Investigation, Methodology, Project administration, Visualization, Writing – original draft, Writing – review and editing. MA: Conceptualization, Methodology, Project administration, Supervision, Validation, Writing – original draft, Writing – review and editing. DB: Investigation, Resources, Writing – review and editing. AA: Conceptualization, Resources, Writing – review and editing. NA: Investigation, Writing – review and editing. FK: Conceptualization, Methodology, Project administration, Supervision, Validation, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
We acknowledge Dr. Peter Avery, Senior Lecturer in Statistics (School of Mathematics and Statistics, Newcastle University) for his assistance with the statistical analyses of the data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Al Ammari, M., AlBalwi, M., Sultana, K., Alabdulkareem, I. B., Almuzzaini, B., Almakhlafi, N. S., et al. (2020). The effect of the VKORC1 promoter variant on warfarin responsiveness in the Saudi WArfarin Pharmacogenetic (SWAP) cohort. Sci. Rep. 10 (1), 11613. doi:10.1038/s41598-020-68519-9
Al Ammari, M., AlThiab, K., AlJohani, M., Sultana, K., Maklhafi, N., AlOnazi, H., et al. (2021). Tele-pharmacy Anticoagulation clinic during COVID-19 pandemic: patient outcomes. Front. Pharmacol. 12p, 652482. doi:10.3389/fphar.2021.652482
Al-Saikhan, F., Abd-Elaziz, M., and Ashour, R. (2018). Influence of vitamin K epoxide reductase complex 1 polymorphism on warfarin therapy in a cohort study of Saudi patients. Int. J. Pharmacol. 18, 414–420. doi:10.3923/ijp.2018.415.420
Al-Saikhan, F., Abd-Elaziz, M. A. R., Hamdy Asho, R., and Langaee, T. (2018). Impact of cytochrome P450 2C9 polymorphism on warfarin therapy in Saudi population. Int. J. Pharmacol. 14, 566–571. doi:10.3923/ijp.2018.566.571
Alzahrani, A. M., Ragia, G., Hanieh, H., and Manolopoulos, V. G. (2013). Genotyping of CYP2C9 and VKORC1 in the Arabic population of Al-ahsa, Saudi Arabia. BioMed Res. Int. 2013, 315980. doi:10.1155/2013/315980
Apostolakis, S., Sullivan, R. M., Olshansky, B., and Lip, G. Y. H. (2013). Factors affecting quality of anticoagulation control among patients with atrial fibrillation on warfarin: the SAMe-TT₂R₂ score. Chest 144 (5), 1555–1563. doi:10.1378/chest.13-0054
Borgiani, P., Ciccacci, C., Forte, V., Sirianni, E., Novelli, L., Bramanti, P., et al. (2009). CYP4F2 genetic variant (rs2108622) significantly contributes to warfarin dosing variability in the Italian population. Pharmacogenomics 10 (2), 261–266. doi:10.2217/14622416.10.2.261
Bourgeois, S., Jorgensen, A., Zhang, E. J., Hanson, A., Gillman, M. S., Bumpstead, S., et al. (2016). A multi-factorial analysis of response to warfarin in a UK prospective cohort. Genome Med. 8 (1), 2. doi:10.1186/s13073-015-0255-y
Caldwell, M. D., Awad, T., Johnson, J. A., Gage, B. F., Falkowski, M., Gardina, P., et al. (2008). CYP4F2 genetic variant alters required warfarin dose. Blood 111 (8), 4106–4112. doi:10.1182/blood-2007-11-122010
Cavallari, L. H., and Perera, M. A. (2012). The future of warfarin pharmacogenetics in under-represented minority groups. Future Cardiol. 8 (4), 563–576. doi:10.2217/fca.12.31
Cen, H.-J., Zeng, W.-T., Leng, X.-Y., Huang, M., Chen, X., Li, J.-L., et al. (2010). CYP4F2 rs2108622: a minor significant genetic factor of warfarin dose in Han Chinese patients with mechanical heart valve replacement. Br. J. Clin. Pharmacol. 70 (2), 234–240. doi:10.1111/j.1365-2125.2010.03698.x
Danese, E., Raimondi, S., Montagnana, M., Tagetti, A., Langaee, T., Borgiani, P., et al. (2019). Effect of CYP4F2, VKORC1, and CYP2C9 in influencing coumarin dose: a single-patient data meta-analysis in more than 15,000 individuals. Clin. Pharmacol. Ther. 105 (6), 1477–1491. doi:10.1002/cpt.1323
Gage, B. F., Eby, C., Johnson, J. A., Deych, E., Rieder, M. J., Ridker, P. M., et al. (2008). Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin. Pharmacol. Ther. 84 (3), 326–331. doi:10.1038/clpt.2008.10
Gulseth, M. P., Grice, G. R., and Dager, W. E. (2009). Pharmacogenomics of warfarin: uncovering a piece of the warfarin mystery. Am. J. health-system Pharm. 66 (2), 123–133. doi:10.2146/ajhp080127
Homme, M. B., Reynolds, K. K., Valdes, R., and Linder, M. W. (2008). Dynamic pharmacogenetic models in anticoagulation therapy. Clin. Laboratory Med. 28 (4), 539–552. doi:10.1016/j.cll.2008.10.002
Jorgensen, A. L., FitzGerald, R. J., Oyee, J., Pirmohamed, M., and Williamson, P. R. (2012). Influence of CYP2C9 and VKORC1 on patient response to warfarin: a systematic review and meta-analysis. PloS one 7 (8), e44064. doi:10.1371/journal.pone.0044064
Kalman, L. V., Agundez, J., Appell, M. L., Black, J. L., Bell, G. C., Boukouvala, S., et al. (2016). Pharmacogenetic allele nomenclature: International workgroup recommendations for test result reporting. Clin Pharmacol Ther, 99, 172–185. doi:10.1002/cpt.280
Kamali, F., Khan, T. I., King, B. P., Frearson, R., Kesteven, P., Wood, P., et al. (2004). Contribution of age, body size, and CYP2C9 genotype to anticoagulant response to warfarin. Clin. Pharmacol. Ther. 75 (3), 204–212. doi:10.1016/j.clpt.2003.10.001
Kamali, F., and Wynne, H. (2010). Pharmacogenetics of warfarin. Annu. Rev. Med. 61pp, 63–75. doi:10.1146/annurev.med.070808.170037
Lane, S., Al-Zubiedi, S., Hatch, E., Matthews, I., Jorgensen, A. L., Deloukas, P., et al. (2012). The population pharmacokinetics of R- and S- warfarin: effect of genetic and clinical factors. Br J Clin Pharmacol, 73, 66–76. doi:10.1111/j.1365–2125.2011.04051.x
Loebstein, R., Yonath, H., Peleg, D., Almog, S., Rotenberg, M., Lubetsky, A., et al. (2001). Interindividual variability in sensitivity to warfarin--Nature or nurture? Clin. Pharmacol. Ther. 70 (2), 159–164. doi:10.1067/mcp.2001.117444
McDonald, M. G., Rieder, M. J., Nakano, M., Hsia, C. K., and Rettie, A. E. (2009). CYP4F2 is a vitamin K1 oxidase: an explanation for altered warfarin dose in carriers of the V433M variant. Mol. Pharmacol. 75 (6), 1337–1346. doi:10.1124/mol.109.054833
Mirghani, R. A., Chowdhary, G., and Elghazali, G. (2011). Distribution of the major cytochrome P450 (CYP) 2C9 genetic variants in a Saudi population. Basic and Clin. Pharmacol. and Toxicol. 109 (2), 111–114. doi:10.1111/j.1742-7843.2011.00692.x
Mizzi, C., Dalabira, E., Kumuthini, J., Dzimiri, N., Balogh, I., Başak, N., et al. (2016). A European spectrum of pharmacogenomic biomarkers: implications for clinical pharmacogenomics. PloS one 11 (9), e0162866. doi:10.1371/journal.pone.0162866
Moyer, T. P. P., O'Kane, D. J. P., Baudhuin, L. M. P., Wiley, C. L. P., Fortini, A. M. D., Fisher, P. K. M. A., et al. (2009). Warfarin Sensitivity Genotyping: A Review of the Literature and Summary of Patient Experience. Mayo Clin Proc, 84, 1079–1094. doi:10.4065/mcp.2009.0278
Niinuma, Y., Saito, T., Takahashi, M., Tsukada, C., Ito, M., Hirasawa, N., et al. (2014). Functional characterization of 32 CYP2C9 allelic variants. Pharmacogenomics J. 14 (2), 107–114. doi:10.1038/tpj.2013.22
Rieder, M. J., Reiner, A. P., Gage, B. F., Nickerson, D. A., Eby, C. S., Mcleod, H. L., et al. (2005). Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med, 352, 2285–2293. doi:10.1056/NEJMoa044503
Saour, J. N., Shereen, A. W., Saour, B. J., and Mammo, L. A. (2011). CYP2C9 polymorphism studies in the Saudi population. Saudi Med. J. 32 (4), 347–352.
Sconce, E. A., Khan, T. I., Wynne, H. A., Avery, P., Monkhouse, L., King, B. P., et al. (2005). The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood 106 (7), 2329–2333. doi:10.1182/blood-2005-03-1108
Scott, S. A., Khasawneh, R., Peter, I., Kornreich, R., and Desnick, R. J. (2010). Combined CYP2C9, VKORC1 and CYP4F2 frequencies among racial and ethnic groups. Pharmacogenomics 11 (6), 781–791. doi:10.2217/pgs.10.49
Tatarunas, V., Lesauskaite, V., Veikutiene, A., Grybauskas, P., Jakuska, P., Jankauskiene, L., et al. (2014). The effect of CYP2C9, VKORC1 and CYP4F2 polymorphism and of clinical factors on warfarin dosage during initiation and long-term treatment after heart valve surgery. J. thrombosis thrombolysis 37 (2), 177–185. doi:10.1007/s11239-013-0940-x
Wadelius, M., Chen, L. Y., Downes, K., Ghori, J., Hunt, S., Eriksson, N., et al. (2005). Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 5, 262–270. doi:10.1038/sj.tpj.6500313
Wen, M.-S., and Lee, M. T. M. (2013). Warfarin pharmacogenetics: new life for an old drug. Acta Cardiol. Sin. 29 (3), 235–242.
Westervelt, P., Cho, K., Bright, D. R., and Kisor, D. F. (2014). Drug-gene interactions: inherent variability in drug maintenance dose requirements. P and T a peer-reviewed J. formulary Manag. 39 (9), 630–637.
Wigle, P., Hein, B., Bloomfield, H. E., Tubb, M., and Doherty, M. (2013). Updated guidelines on outpatient anticoagulation. Am. Fam. physician 87 (8), 556–566.
Keywords: warfarin, CYP2C9, VKORC1, CYP4F2, polymorphisms
Citation: Jokhab S, AlRasheed MM, Bakheet D, AlMomen A, AlAboud N and Kamali F (2025) The impact of CYP2C9, VKORC1, and CYP4F2 polymorphisms on warfarin dose requirement in Saudi patients. Front. Pharmacol. 16:1547142. doi: 10.3389/fphar.2025.1547142
Received: 17 December 2024; Accepted: 18 April 2025;
Published: 30 April 2025.
Edited by:
Alessio Squassina, University of Cagliari, ItalyReviewed by:
MyeongJin Yi, National Institute of Environmental Health Sciences (NIH), United StatesValeria Conti, University of Salerno, Italy
Copyright © 2025 Jokhab, AlRasheed, Bakheet, AlMomen, AlAboud and Kamali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Salha Jokhab, c2pva2hhYkBrc3UuZWR1LnNh
†ORCID: Salha Jokhab, orcid.org/0000-0002-7771-1500
 Salha Jokhab
Salha Jokhab Maha M. AlRasheed
Maha M. AlRasheed Dana Bakheet
Dana Bakheet Abdulkareem AlMomen3
Abdulkareem AlMomen3 Nouf AlAboud
Nouf AlAboud