Abstract
The development of new antimicrobial therapeutic strategies requires urgent attention to prevent the tens of millions of deaths predicted to occur by 2050 as a result of multidrug-resistant (MDR) bacterial infections. This study aimed to discover new tamoxifen derivatives with antimicrobial potential, particularly targeting methicillin-resistant Staphylococcus aureus (MRSA). The minimum inhibitory concentration (MIC) of 22 tamoxifen derivatives was determined against S. aureus reference and MRSA strains using microdilution assays. The antibacterial effects of selected tamoxifen derivatives against MRSA (USA7 strain) were assessed through bacterial growth assays. Additionally, bacterial membrane permeability and molecular dynamics (MD) simulation assays were performed. The MIC of the tamoxifen derivatives against reference S. aureus and MRSA strains ranged from to 16 to >64 μg/mL. Bacterial growth assays demonstrated that tamoxifen derivatives 2, 5, and 6, the only compounds bearing the electron-donating hydroxyl group in the para position on both phenyl rings of the tamoxifen skeleton, dose-dependently reduced the growth of the USA7 strain. Moreover, treatment of MRSA with derivatives 2 and 5 resulted in a slight increase of membrane permeabilization. Extensive MD simulations on the interaction between 5 and 6 and the S. aureus membrane model suggest that the compounds do not act by destabilizing the membrane integrity. These findings suggest that tamoxifen derivatives exhibit antibacterial activity against MRSA, potentially broadening the spectrum of available drug treatments for combating antimicrobial-resistant S. aureus.
1 Introduction
In the last decades, Gram-positive bacteria have demonstrated an increasing antimicrobial resistance, driven by various genomic, transcriptomic, and proteomic adaptations (Yelin and Kishony, 2018). This alarming trend aligns with a concerning decline in the development of new antibiotics, a phenomenon often referred to as the “Post-Antibiotic Era” (Bagley and Outterson, 2017). This situation underscores the urgent need for effective solutions, a concern highlighted by numerous institutions. Consequently, there is a growing demand for innovative antimicrobial therapeutic approaches, including the exploration of non-antibiotic compounds and drug repurposing, both as monotherapy and in combination with the limited clinically relevant antibiotics currently available (Canturri and Smani, 2022).
Among the various strategies employed in the discovery of new antibacterial agents, drug repurposing is one of the most exploited (Canturri and Smani, 2022; Schadich et al., 2022; Ćavar Zeljković et al., 2022; Schadich et al., 2025). A very representative example of such approach is tamoxifen, a well-known chemotherapic drug, widely used for decades as the gold standard for the treatment of estrogen receptor positive breast cancers and related metastatic forms (Green and Carroll, 2007). Recently, tamoxifen has exhibited relevant antibacterial properties against a range of pathogenic microorganisms, including Gram-positive Staphylococcus epidermidis, and Enterococcus faecalis and Gram-negative Escherichia coli and Acinetobacter baumannii (Miró-Canturri et al., 2021a; Miró-Canturri et al., 2021b). This antimicrobial effect may be ascribed to the cytochrome P450-mediated metabolism of tamoxifen, resulting in the generation of three major metabolites: N-desmethyltamoxifen, 4-hydroxytamoxifen and endoxifen (Miró-Canturri et al., 2021a; Klein et al., 2013).
Only a limited number of studies have reported the activity of these metabolites against various infectious agents (Miró-Canturri et al., 2021a; Miró-Canturri et al., 2021b; Selyunin et al., 2019; Montoya and Krysan, 2018; Weinstock et al., 2019; Buts et al., 2014; Chen et al., 2014). Among these, 4-hydroxytamoxifen has garnered attention for its chemical behavior as a weak base, since it has been observed that such property is responsible for the protection of cells and mice against lethal Shiga toxin 1 (STx1) or Shiga toxin 2 (STx2) toxicosis (Selyunin et al., 2019). It has also demonstrated efficacy against Plasmodium falciparum and Cryptococcus neoformans (Weinstock et al., 2019; Buts et al., 2014). Furthermore, when used in monotherapy, 4-hydroxytamoxifen has displayed activity against Mycobacterium tuberculosis (with a MIC50 of approximately 2.5–5 μg/mL) (Chen et al., 2014). Endoxifen’s activity was studied against C. neoformans, revealing a MIC of 4 μg/mL (Buts et al., 2014). The combination of N-desmethyltamoxifen, 4-hydroxytamoxifen and endoxifen exhibited MIC50 values of 8 and 16 μg/mL against clinical isolates of A. baumannii and E. coli, and MIC50 values of 1 and 2 μg/mL against clinical isolates of S. epidermidis and E. faecalis (Miró-Canturri et al., 2021a; Miró-Canturri et al., 2021b).
To identify new chemical compounds with antimicrobial potential, a collection of 22 tamoxifen derivatives, bearing different electron-withdrawing or electron-donating substituents on the aromatic rings A and B (Table 1, supplementary data) was tested against methicillin-resistant S. aureus (MRSA).
TABLE 1

|
|||||||
|---|---|---|---|---|---|---|---|
| Compound | Structure | R1 | R2 | R3 | R4 | S. aureus ATCC 1556 | |
| MIC (μg/mL) | MIC (µM) | ||||||
| 1 | A | H | H | H | H | >64 | >214 |
| 2 | A | OH | CON(CH2CH3)2 | H | OH | 32 | 74.3 |
| 3 | A | OBn | COCH2CH3 | H | OBn | >64 | >113 |
| 4 | B | OBn | COCH2CH3 | H | OBn | >64 | >113 |
| 5 | A | OH | CON(CH3)2 | H | OH | 32 | 79.5 |
| 6 | B | OH | CON(CH3)2 | H | OH | 16 | 39.8 |
| 7 | A | OCH3 | COiPropyl | H | H | >64 | >160 |
| 8 | B | OCH3 | COiPropyl | H | H | >64 | >160 |
| 9 | A | H | COiPropyl | H | H | >64 | >173 |
| 10 | B | H | COiPropyl | H | H | >64 | >173 |
| 11 | B | H | COiPropyl | H | OBn | >64 | >134 |
| 12 | A | H | COiPropyl | H | OBn | >64 | >134 |
| 13 | A | OH | COiPropyl | Cl | Cl | >64 | >141 |
| 14 | B | OH | COiPropyl | Cl | Cl | >64 | >141 |
| 15 | A | OCH3 | COiPropyl | H | OBn | >64 | >126 |
| 16 | B | OCH3 | COiPropyl | H | OBn | >64 | >126 |
| 17 | B | OCH3 | COiPropyl | H | OH | >64 | >154 |
| 18 | A | OCH3 | COiPropyl | H | OH | >64 | >154 |
| 19 | A | OH | COiPropyl | H | H | >64 | >166 |
| 20 | B | OH | COiPropyl | H | H | >64 | >166 |
| 21 | A | OBn | COiPropyl | H | OBn | >64 | >110 |
| 22 | B | OBn | COiPropyl | H | OBn | >64 | >110 |
MIC values of tamoxifen derivatives 1–22 for standard reference S. aureus strain.
2 Materials and methods
2.1 Bacterial strains
A total of 7 MRSA clinical isolates and 1 S. aureus reference ATCC 1556 strain were used in this study (Docobo Pérez, 2009).
2.2 Antimicrobial agent and tamoxifen derivatives
The small library of tamoxifen derivatives was prepared as described in literature, starting from appropriately substituted benzophenones which underwent McMurry olefination (Christodoulou et al., 2013; Christodoulou et al., 2016). The structures of the tested compounds are presented in Table 1.
2.3 In vitro susceptibility testing
The MICs of tamoxifen derivatives (from 16 to 64 μg/mL and from 39.8 to 214 μM) were determined against reference and MRSA strains in two independent experiments using the broth microdilution method, in accordance with the standard guidelines of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (European Committee on Antimicrobial Susceptibility Testing, 2022). A 5 × 105 cfu/mL inoculum of each strain was cultured in Luria Bertani (LB) and added to U bottom microtiter plates (Deltlab, Spain) containing the studied tamoxifen derivatives. The plates were incubated for 18 h at 37°C. Pseudomonas aeruginosa ATCC 27853 was used as the positive control strain.
2.4 Bacterial growth curves
To determine the antibacterial and synergistic effects, duplicate bacterial growth curves were performed for MRSA USA7 strain. A 1/200 dilution of an overnight bacterial cultures grown in LB at 37°C with continuous agitation at 180 rpm was performed in LB in 96-well plate in the presence of 1×, 2× and 4× MIC of compounds 2, 5 and 6. A drug-free broth was evaluated in parallel as control. Absorbance measurements at 600 nm every 20 min for 24 h were conducted using a Tecan spectrophotometer (model XYZ-2000, Austria).
2.5 Membrane permeability assays
The bacterial cells were grown in LB broth and incubated in the absence or presence of 1) 1x MIC of compounds 2, 5 and 6 for 3 h. The pellet was harvested by ultracentrifugation at 4600 g for 15 min. The bacterial cells were washed with PBS 1X, and after centrifugation in the same condition described before, the pellet was resuspended in 100 µL of PBS 1X containing 10 μL of Ethidium Homodimer-1 (ThermoFisher, United States). After 10 min of incubation, 100 μL was placed into a 96-well plate to measure fluorescence for 300 min using a Typhoon FLA 9000 laser scanner (GE Healthcare Life Sciences, United States) and quantified using ImageQuant TL software (GE Healthcare Life Sciences, United States) (Miró-Canturri et al., 2020).
2.6 Molecular dynamics simulations assays
The symmetric lipid bilayer membrane of S. aureus was built using the CHARMM-GUI Membrane Builder Tool (Li et al., 2021; Jo et al., 2008). This system was designed to mimic the phospholipid composition of the S. aureus membrane, which consists of 56.8% phosphatidylglycerol (PG), 37.9% Lys-PG, and 5.3% diphosphatidylglycerol (DPG), also referred as Cardiolipin (CL), in agreement with previous molecular dynamics (MD) studies (Kim et al., 2018; Princiotto et al., 2024; Witzke et al., 2016). Each lipid bilayer contained a total of 95 lipid molecules, positioned with their centers at z = 0., surrounded by a water layer with a thickness of 50 Å. The system was neutralized using Na+ and Cl-ions at a concentration of 0.145 M, as suggested by CHARMM-GUI.
Derivatives 5 and 6 were drawn using the Sketchpad powered by Marvin JS included in the ligand reader and modeler module of CHARMM-GUI, and were parameterized using the standard CHARMM force field (FF) (Kim et al., 2017). The Multicomponent Assembler tool was used to randomly distribute the small molecules under investigation within the solvent area. The membrane systems containing these small molecules were then parameterized with a membrane thickness of 50 Å and a box XY length of 79.50 Å (Huang et al., 2016; Lee et al., 2020). Following system construction with CHARMM-GUI, the topology and coordinate files were generated for AMBER (Lee et al., 2016; Brooks et al., 2009).
To investigate the interaction between 5 and 6 with the S. aureus membrane model, MD simulations were conducted using AMBER22 (Salomon-Ferrer et al., 2013; Case et al., 2005). The initial system was energy minimized for a total of 40000 steps., with the first 1500 steps employing the steepest descent algorithm, followed by the conjugate gradient algorithm for the remaining steps. A non-bonded cut-off of 10Å was used. After energy minimization, each system was gradually heated to 300 K over 900 ps at constant volume using the Langevin thermostat with a collision frequency of 2 ps-1, and then left at 300 K at constant volume for 200 ps. Box density was equilibrated at constant pressure and constant temperature (300 K) over 1 ns using the Berendsen barostat.
Following density equilibration, a preliminary 50 ns MD simulation was conducted at constant pressure. Subsequently, MD trajectories were generated for 500 ns; two independent replicates for each compound were run In all MD simulations, no positional restraints were applied.
Analysis of MD trajectories was performed using the CPPTRAJ (Roe and Cheatham, 2013) program from the AmberTools package. This analysis included calculation of the mass densities of derivatives within the system along the z-axis. Small molecules interactions with the membrane were visually inspected with PyMol (Delano, 2002).
2.7 Statistical analysis
Group data are presented as means ± standard errors of the means (SEM). The one-way ANOVA test was used to determine differences between means using the GraphPad Prism 9. A p-value <0.05 was considered significant.
3 Results
3.1 Antimicrobial activity of tamoxifen derivatives
Twenty-two tamoxifen derivatives (Table 1) were subjected to evaluation to determine their MIC against S. aureus ATCC 1556 reference strain. Only the tamoxifen derivatives 2, 5 and 6 showed MIC for S. aureus ATCC 1556 strain ranging from 16 to 32 μg/mL (39.8–79.5 μM), while the rest of the compounds did not show MIC values < 64 μg/mL (110–214 μM) (Table 1).
A comparison between the biological results and the chemical structures of the tamoxifen derivatives allowed some interesting structure-activity relationships outcomes. In particular, the most active compounds 2, 5 and 6 share the presence of the hydroxyl group in para position on both phenyl rings, A and B. All the derivatives bearing the hydroxyl group on only one of the phenyl rings resulted inactive (compounds 13, 14, 17, 18, 19 and 20). In the case of no substituents on the phenyl rings (compounds 1, 9 and 10) or in presence of electron-donating substituents diverse from the hydroxyl group on one or both phenyl rings A and B, the derivatives resulted inactive as well (compounds 3, 4, 7, 8, 11, 12, 15, 16, 21 and 22).
The most promising derivatives were selected on the base of a MIC ≤64 μg/mL and were evaluated against 7 clinical isolates of MRSA to extend their biological evaluation to the antimicrobial effect on resistant strains. As shown in Table 2, the MICs of tamoxifen derivatives 2, 5 and 6 for MRSA strains ranged from 16 to 64 μg/mL.
TABLE 2
| Strain | 2 | 5 | 6 | |||
|---|---|---|---|---|---|---|
| MIC (μg/mL) | MIC (µM) | MIC (μg/mL) | MIC (µM) | MIC (μg/mL) | MIC (µM) | |
| USA1 | 32 | 74.3 | 32 | 79.5 | 16 | 39.8 |
| USA2 | 32 | 74.3 | 64 | 159 | 16 | 39.8 |
| USA3 | 32 | 74.3 | 64 | 159 | 16 | 39.8 |
| USA4 | 32 | 74.3 | 64 | 159 | 16 | 39.8 |
| USA5 | 64 | 148 | 64 | 159 | 16 | 39.8 |
| USA6 | 64 | 148 | 64 | 159 | 16 | 39.8 |
| USA7 | 32 | 74.3 | 64 | 159 | 16 | 39.8 |
MIC values of tamoxifen derivatives 2, 5, and 6 against methicillin-resistant S. aureus clinical isolates.
3.2 Time dependent antibacterial effects
Using bacterial growth, we examined the antibacterial activity of the selected tamoxifen derivatives against MRSA USA7 strain. Compounds 2, 5 and 6 at concentrations of 1×, 2× and 4× MIC, reduced the growth of this strain in a concentration-dependent manner during 24 h. The inhibitory effects of tamoxifen derivatives 2 and 5 at 1×, 2× and 4× MIC on bacterial were more pronounced and began earlier than with tamoxifen derivative 6 (Figure 1).
FIGURE 1

Antibacterial activity of tamoxifen derivatives against clinical methicillin-resistant S. aureus. Bacterial growth curves of MRSA USA7 strain in the presence of 1×, 2× and 4× MIC of tamoxifen derivative 2, 5 or 6 for 24 h. Data are represented as mean from two independent. Replicates and experiments. COL: colistin.
3.3 Effect of tamoxifen derivatives on the bacterial cell membrane
In order to determine the mode of action of the selected tamoxifen derivatives, their effect on the membrane permeability of USA7 strain was evaluated by incubation with ethidium homodimer-1, a fluorescent marker known to enter bacterial cells when the membrane integrity is compromised.
Fluorescence monitoring using a Typhoon FLA scanner for 3 h showed a slight increase in the cellular fluorescence of the USA7 strain when treated with sub-MIC of the tamoxifen derivatives 2 and 5, but not for 6. (Figure 2).
FIGURE 2
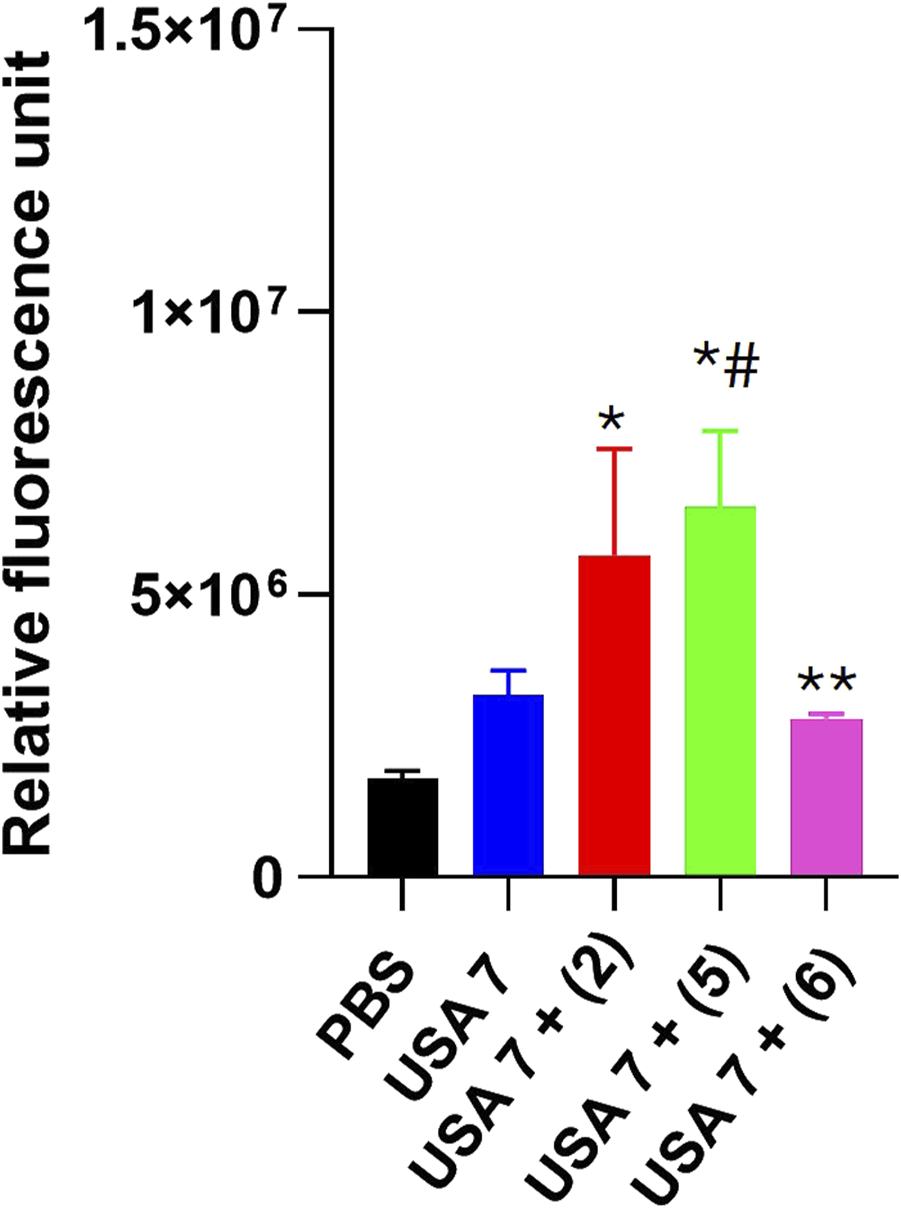
Effect of tamoxifen derivatives on the bacterial permeability against clinical methicillin-resistant S. aureus. The membrane permeabilization of MRSA USA7 strain in the presence of 1xMIC of tamoxifen derivative 2, 5 or 6, incubated for 10 min, was quantified by Typhon Scanner. Data are represented as mean ± SEM from three independent replicates and experiments. *P < 0.05 vs. PBS, #P < 0.05 vs. USA7, *P < 0.05 vs. USA7 + (5).
3.4 Tamoxifen derivatives interaction with bacterial membrane
To investigate the interaction between tamoxifen derivates 5 and 6 and the bacterial membrane model, and to assess how subtle structural modifications (i.e., cis/trans conformation) might affect their ability to embed within the target, unbiased MD simulations were carried out starting from the small molecules being placed in the solvent area (Li et al., 2021; Jo et al., 2008; Piggot et al., 2011; Kim et al., 2018; Princiotto et al., 2024; Witzke et al., 2016; Joodaki et al., 2022).
Different from previous findings with different scaffolds (Princiotto et al., 2024), for derivatives 5 and 6 the density plots clearly evidence that the molecules are unable to penetrate deeply into the membrane model (Figures 3A,B). In fact, density peaks of the two derivatives slightly exceed the density peak of the phosphates that represent the membrane outermost layer. These results suggest that the S. aureus membrane might not be the molecular target of 5 and 6 antibacterial efficacy.
FIGURE 3
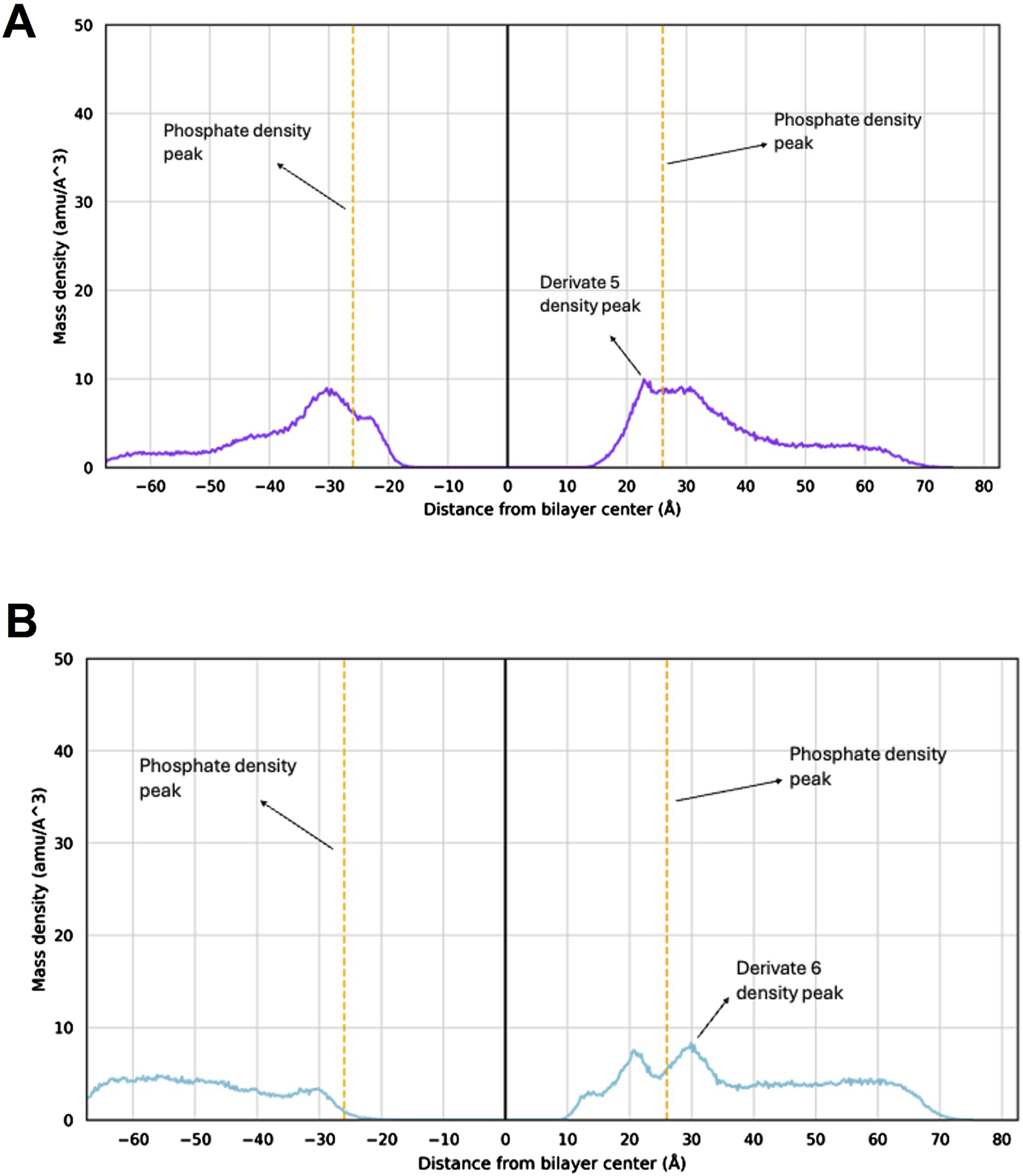
Mass density profile of 5 and 6 within the S. aureus membrane model. Mass density profile of 5 (A) and 6 (B) with the corresponding density peaks highlighted. The density peaks of the lipid phosphate groups are shown as orange dashed lines, with maximum density at −26 Å and +26 Å. The mass center of the membrane model is at point 0. The molecules’ mass density peaks indicate the position of each compound within the bilayer system as extracted from the MD simulations.
To further provide atomistic details of this outcome, the distance between each molecule and the lipid bilayer was monitored along MD trajectories. Then, the MD frame corresponding to the smallest intermolecular distance was extracted and visually inspected. A labile H-bond interaction between the phenol moiety of the molecules and the protonated ammino group (i.e., R-NH3+) of a phosphatidylglycerol residues at the membrane interface with the solvent was observed (Figures 4A,B). Unlike 5, derivative 6 also establishes an additional interaction with the -OH group of a phosphatidylglycerol residue (Figure 4B). However, this interaction is not stable in time and failed to promote molecules embedding into the S. aureus membrane model within the simulation time, which further reinforce the hypothesis that 5 and 6 exploit their antibacterial potentials through the interaction with target that differ from the bacterial membrane.
FIGURE 4
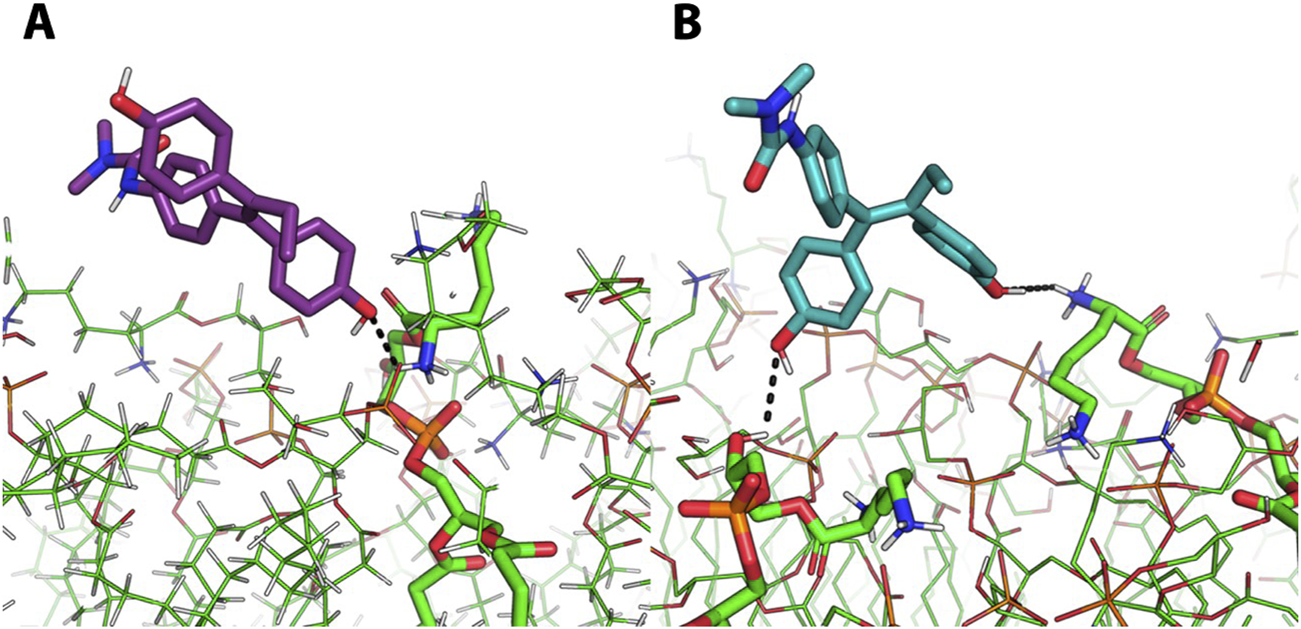
Representative of the most representative frame of MD simulations of molecules 5 and 6. Representative MD frames corresponding to the minimum distances between derivate 5 (A, purple sticks) or derivate 6 (B, light blue sticks) and the S. aureus membrane model. The interactions between these two derivates and the NH3+ moiety (A,B) and -OH moiety (B) of phosphatidylglycerol residues is represented by black dashed lines.
4 Discussion
The rise of multidrug resistant S. aureus has led to a wide use of the last options for the treatment of severe infections caused by this microorganism, and to the consequent acquired antimicrobial resistance worldwide in the last decades (Abebe and Birhanu, 2023). In previous studies, tamoxifen and its metabolites garnered attention as a potential repurposed drug for treatment of infectious diseases (Montoya and Krysan, 2018).
Due to the antibacterial effect of tamoxifen and its metabolites N-desmethyltamoxifen, 4-hydroxytamoxifen and endoxifen against S. epidermidis and A. baumannii (Miró-Canturri et al., 2021a; Miró-Canturri et al., 2021b), we hypothesized that tamoxifen derivatives may enhance this antibacterial activity against MRSA. For this purpose, twenty-two tamoxifen derivatives were tested against S. aureus reference strain.
Three tamoxifen derivatives 2, 5 and 6 in monotherapy showed antibacterial activity against MRSA strains being the MIC between 16 and 64 μg/mL, which overall fell within the range of other known antibiotics such as amikacin, teicoplanin, sulfonamides nitrofurantoin (European Committee on Antimicrobial Susceptibility Testing, 2024; CLSI, 2024). However, none of the rest of tested tamoxifen derivatives exhibited activity against MRSA. Reasons for this difference could be related to the chemical structure of these derivatives, being 2, 5 and 6 the only compounds bearing the electron-donating hydroxyl group in para position on both phenyl rings A and B.
The antibacterial activity of tamoxifen derivatives 2 and 5 at 1x, 2x and 4x MIC against MRSA USA7 strain was more pronounced and began earlier than with tamoxifen derivative 6. This result may be related to the trans stereochemistry (structure A) of compounds 2 and 5 and their ability to act on the plasma membrane permeability of S. aureus, as observed with tamoxifen metabolites against Gram-negative bacteria (Miró-Canturri et al., 2021a). Membrane permeability assays indicate that tamoxifen derivatives 2 and 5, but not 6, produced a slight increase in membrane permeability of MRSA. In support of this hypothesis, tamoxifen has been shown to increase the membrane permeability of Streptococcus pneumoniae by perturbing the phospholipid bilayer (Ortiz-Miravalles et al., 2023). Notably, MD simulations were carried out to simulate atomistic details of the interaction between 5 and 6 with the S. aureus membrane model, showing that, although an interaction does occur, the membrane integrity is not perturbed by the compounds.
As such, the S. aureus membrane might not be the molecular target of these derivatives. Alternative mechanisms of action, rather than the perturbation of the plasma membrane, cannot be ruled out. Of note, in other microorganisms such as fungi, the mechanism of action of the tamoxifen is well-documented and involves its binding to calmodulin (Dolan et al., 2009; Butts et al., 2014). Moreover, the tamoxifen metabolite, 4-hydroxytamoxifen, can potentially inhibit bacterial phospholipase D in P. aeruginosa (Scott et al., 2015). Additional research focusing on understanding the mechanism of action of the tested tamoxifen derivatives against MRSA would be of significant interest since different chemical structures resulted active on the aforementioned strain, thus providing a better therapeutic efficacy.
The antimicrobial activity of the selected derivatives identified in this work hints at a promising potential that deserves to be further explored in vivo after determining their pharmacokinetic parameters. However, in vitro bacterial growth showed a progressive regrowth of the MRSA USA7 strain after treatment with these tamoxifen derivatives, suggesting that acquired resistance could take place. Nevertheless, it should be noted that the MICs of tamoxifen derivatives 2, 5, and 6 against the USA7 strain in this bacterial growth condition are 32, 32, and 16 μg/mL, respectively. These values are lower than the concentrations originally tested, which were set at 2x and 4 MIC for those derivatives. In other words, the growth of the USA7 strain is inhibited at lower concentrations than expected based on the initial MIC, suggesting that tamoxifen derivatives 2, 5, and 6 are more effective under these specific growth conditions. Further investigations, including the determination of tamoxifen derivatives concentration during the bacterial growth assay, are necessary to better understand the regrowth of this strain in their presence. In addition, elucidating the mechanism of action of these derivatives and determining their optimal dosage could ensure therapeutic effectiveness in treating severe infections caused by multidrug-resistant S. aureus.
Finally, the prospect of repurposing tamoxifen derivatives as antimicrobial agents introduces a significant challenge: the need to dissociate their antimicrobial properties from tamoxifen’s original anti-estrogen activity. To achieve this, any tamoxifen derivative intended for use as an antimicrobial must be designed or modified to lose its capacity to interact with estrogen receptors while preserving its antimicrobial potency.
5 Conclusion
The findings of this study provide new insights into the use of a diverse group of tamoxifen derivatives against MRSA, a pathogen for which treatment options are severely restricted. We demonstrated that tamoxifen derivatives exhibit in vitro antibacterial activity against both reference strain and clinical isolates of S. aureus, without targeting the bacterial membrane. Moreover, their activity correlates with the presence of a hydroxyl group in the para position on both derivative phenyl rings.
Statements
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://zenodo.org/records/14965780.
Author contributions
IM: Formal Analysis, Writing – original draft, Methodology. JF: Formal Analysis, Methodology, Writing – review and editing. AH: Formal Analysis, Methodology, Writing – review and editing. SP: Formal Analysis, Writing – review and editing, Methodology. LC: Methodology, Writing – original draft. MM: Writing – review and editing, Formal Analysis. SD: Writing – review and editing. MC: Formal Analysis, Writing – review and editing, Funding acquisition, Supervision, Writing – original draft. YS: Conceptualization, Formal Analysis, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by the Ministerio de Ciencia e Innovación, Agencia Estatal de Investigación, Fondo Europeo de Desarrollo Regional, MCIN/AEI/10.13039/501100011033/FEDER, and (Grant CEX2020-001088-M-20-5) UE (grant PID2022-136357OB-I00), the Consejería de Universidad, Investigación e Innovación de la Junta de Andalucía (grant ProyExcel_00116), and the Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad (grant PI19/01009), cofinanced by the European Development Regional Fund (A way to achieve Europe, Operative Program Intelligent Growth, 2014–2020). This study has also received funding from the EU’s NextGeneration EU-MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (project no. PE00000007, INF-ACT) (MM, LC).
Acknowledgments
We thank José María Marimon Ortiz de Zárate for sharing the MRSA clinical isolates, Cayetana Martín and the Proteomic facility of the Andalusian Center of Developmental Biology for their technical help and the COST Action CA21145 – European Network for diagnosis and treatment of antibiotic-resistant bacterial infections (EURESTOP).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2025.1549288/full#supplementary-material
References
1
Abebe A. A. Birhanu A. G. (2023). Methicillin resistant Staphylococcus aureus: molecular mechanisms underlying drug resistance development and novel strategies to combat. Infect. Drug Resist.16, 7641–7662. 10.2147/IDR.S428103
2
Bagley N. Outterson K. (2017). We will miss antibiotics when they’re Gone. Available online at: www.nytimes.com/2017/01/18/opinion/how-to-avoid-a-post-antibiotic-world.html (Accessed April 15, 2021).
3
Brooks B. R. Brooks C. L. Mackerell A. D. Nilsson L. Petrella R. J. Roux B. et al (2009). CHARMM: the biomolecular simulation program. J. Comput. Chem.30 (10), 1545–1614. 10.1002/jcc.21287
4
Butts A. Koselny K. Chabrier-Roselló Y. Semighini C. P. Brown J. C. Wang X. et al (2014). Estrogen receptor antagonists are anti-cryptococcal agents that directly bind EF hand proteins and synergize with fluconazole in vivo. mBio5, 007655–e813. 10.1128/mBio.00765-13
5
Canturri A. M. Smani Y. (2022). Anthelmintic drugs for repurposing against Gram-negative bacilli infections. Curr. Med. Chem.30, 59–71. 10.2174/0929867329666220714092916
6
Case D. A. Cheatham T. E. Darden T. Gohlke H. Luo R. Merz K. M. et al (2005). The Amber biomolecular simulation programs. J. Comput. Chem.26 (16), 1668–1688. 10.1002/jcc.20290
7
Ćavar Zeljković S. Schadich E. Džubák P. Hajdúch M. Tarkowski P. (2022). Antiviral activity of selected lamiaceae essential oils and their monoterpenes against SARS-Cov-2. Front. Pharmacol.13, 893634. 10.3389/fphar.2022.893634
8
Chen F. C. Liao Y. C. Huang J. M. Lin C. H. Chen Y. Y. Dou H. Y. et al (2014). Pros and cons of the tuberculosis drugome approach-an empirical analysis. PLoS One9, e100829. 10.1371/journal.pone.0100829
9
Christodoulou M. S. Fokialakis N. Passarella D. García-Argáez A. N. Gia O. M. Pongratz I. et al (2013). Synthesis and biological evaluation of novel tamoxifen analogues. Bioorg. Med. Chem.21, 4120–4131. 10.1016/j.bmc.2013.05.012
10
Christodoulou M. S. Zarate M. Ricci F. Damia G. Pieraccini S. Dapiaggi F. et al (2016). 4-(1,2-diarylbut-1-en-1-yl)isobutyranilide derivatives as inhibitors of topoisomerase II. Eur. J. Med. Chem.118, 79–89. 10.1016/j.ejmech.2016.03.090
11
Clinical and Laboratory Standards Institute (CLSI) (2024). Performance standards for antimicrobial susceptibility testing. 34th ed. CLSI supplement M100.
12
Delano W. L. (2002). The PyMOL molecular graphics system. CCP4 Newsl. protein Crystallogr. DeLano Sci. San Carlos, CA, USA. Available online at: http://www.pymol.org.
13
Docobo Pérez F. (2009). Tratamiento de la neumonía experimental por Staphylococus aureus. Estudios de eficacia terapéutica de cotrimoxazol, cloxacilina, linezolid y vancomicina frente a cepas S. aureus sensible y resistente a meticilin. Ph.D. Thesis. Seville, Spain: Seville University.
14
Dolan K. Montgomery S. Buchheit B. Didone L. Wellington M. Krysan D. J. et al (2009). Antifungal activity of tamoxifen: in vitro and in vivo activities and 363 mechanistic characterization. Antimicrob Agents Chemother. 53, 3337–3346.
15
European Committee on Antimicrobial Susceptibility Testing (2022). European antimicrobial breakpoints. Basel: EUCAST.
16
European Committee on Antimicrobial Susceptibility Testing (2024). European antimicrobial breakpoints. Basel: EUCAST. Available online at: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_14.0_Breakpoint_Tables.pdf.
17
Green K. A. Carroll J. S. (2007). Oestrogen-receptor-mediated transcription and the influence of co-factors and chromatin state. Nat. Rev. Cancer7, 713–722. 10.1038/nrc2211
18
Huang J. Rauscher S. Nawrocki G. Ran T. Feig M. de Groot B. L. et al (2016). CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods14 (1), 71–73. 10.1038/nmeth.4067
19
Jo S. Kim T. Iyer V. G. Im W. (2008). CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem.29 (11), 1859–1865. 10.1002/jcc.20945
20
Joodaki F. Martin L. M. Greenfield M. L. (2022). Generation and computational characterization of a complex Staphylococcus aureus lipid bilayer. Langmuir38 (31), 9481–9499. 10.1021/acs.langmuir.2c00483
21
Kim S. Lee J. Jo S. Brooks C. L. Lee H. S. Im W. (2017). CHARMM-GUI ligand reader and modeler for CHARMM force field generation of small molecules. J. Comput. Chem.38 (21), 1879–1886. 10.1002/jcc.24829
22
Kim W. Zhu W. Hendricks G. L. Van Tyne D. Steele A. D. Keohane C. E. et al (2018). A new class of synthetic retinoid antibiotics effective against bacterial persisters. Nature556 (7699), 103–107. 10.1038/nature26157
23
Klein D. J. Thorn C. F. Desta Z. Flockhart D. A. Altman R. B. Klein T. E. (2013). PharmGKB summary: tamoxifen pathway, pharmacokinetics. Pharmacogenet. Genomics23, 643–647. 10.1097/FPC.0b013e3283656bc1
24
Lee J. Cheng X. Swails J. M. Yeom M. S. Eastman P. K. Lemkul J. A. et al (2016). CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations using the CHARMM36 additive force field. J. Chem. Theory Comput.12 (1), 405–413. 10.1021/acs.jctc.5b00935
25
Lee J. Hitzenberger M. Rieger M. Kern N. R. Zacharias M. Im W. (2020). CHARMM-GUI supports the Amber force fields. J. Chem. Phys.153 (3), 035103. 10.1063/5.0012280
26
Li Y. Liu J. Gumbart J. C. (2021). Preparing membrane proteins for simulation using CHARMM-GUI. In: Methods Mol. Biol. Vol 2302, 237, 251. 10.1007/978-1-0716-1394-8_13
27
Miró-Canturri A. Ayerbe-Algaba R. Vila-Domínguez A. Jiménez-Mejías M. E. Pachón J. Smani Y. (2021b). Repurposing of the tamoxifen metabolites to combat infections by multidrug-resistant gram-negative bacilli. Antibiotics10, 336. 10.3390/antibiotics10030336
28
Miró-Canturri A. Ayerbe-Algaba R. Villodres Á. R. Pachón J. Smani Y. (2020). Repositioning rafoxanide to treat Gram-negative bacilli infections. J. Antimicrob. Chemother.75, 1895–1905. 10.1093/jac/dkaa103
29
Miró-Canturri A. Vila-Domínguez A. Caretero-Ledesma M. Ayerber-Algaba R. Pachón J. Jiménez-Mejías M. E. et al (2021a). Repurposing of the tamoxifen metabolites to treat methicillin-resistant Staphylococcus epidermidis and vancomycin-resistant Enterococcus faecalis infections. Microbiol. Spectr.9, e0040321. 10.1128/Spectrum.00403-21
30
Montoya M. C. Krysan D. J. (2018). Repurposing estrogen receptor antagonists for the treatment of infectious disease. mBio9. 10.1128/mBio.02272-18
31
Ortiz-Miravalles L. Sánchez-Angulo M. Sanz J. M. Maestro B. (2023). Drug repositioning as a therapeutic strategy against Streptococcus pneumoniae: cell membrane as potential target. Int. J. Mol. Sci.24 (6), 5831. 10.3390/ijms24065831
32
Piggot T. J. Holdbrook D. A. Khalid S. (2011). Electroporation of the E. coli and S. aureus membranes: molecular dynamics simulations of complex bacterial membranes. J. Physic. Chemi. B115 (45), 13381–13388. 10.1021/jp207013v
33
Princiotto S. Casciaro B. G. Temprano A. Musso L. Sacchi F. Loffredo M. R. et al (2024). The antimicrobial potential of adarotene derivatives against Staphylococcus aureus strains. Bioorg. Chem.145, 107227. 10.1016/j.bioorg.2024.107227
34
Roe D. R. Cheatham T. E. (2013). PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput.9 (7), 3084–3095. 10.1021/ct400341p
35
Salomon-Ferrer R. Case D. A. Walker R. C. (2013). An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci.3 (2), 198–210. 10.1002/wcms.1121
36
Schadich E. Kaczorová D. Béres T. Džubák P. Hajdúch M. Tarkowski P. et al (2025). Secondary metabolite profiles and anti-SARS-CoV-2 activity of ethanolic extracts from nine genotypes of Cannabis sativa L. Arch. Pharm. Weinh.358 (1), e2400607. 10.1002/ardp.202400607
37
Schadich E. Nylén S. Gurská S. Kotulová J. Andronati S. Pavlovsky V. et al (2022). Activity of 1-aryl-4-(naphthalimidoalkyl) piperazine derivatives against Leishmania major and Leishmania mexicana. Parasitol. Int.91, 102647. 10.1016/j.parint.2022.102647
38
Scott S. A. Spencer C. T. O'Reilly M. C. Brown K. A. Lavieri R. R. Cho C. H. Jung D. I. Larock R. C. Brown H. A. Lindsley C. W. (2015). Discovery of desketoraloxifene analogues as inhibitors of mammalian, Pseudomonas aeruginosa, and nape phospholipase D enzymes. ACS Chem. Biol. 10, 421–432.
39
Selyunin A. S. Hutchens S. McHardy S. F. Mukhopadhyay S. (2019). Tamoxifen blocks retrograde trafficking of Shiga toxin 1 and 2 and protects against lethal toxicosis. Life Sci. Alliance2, e201900439. 10.26508/lsa.201900439
40
Weinstock A. Gallego-Delgado J. Gomes C. Sherman J. Nikain C. Gonzalez S. et al (2019). Tamoxifen activity against Plasmodium in vitro and in mice. Malar. J.18, 378. 10.1186/s12936-019-3012-7
41
Witzke S. Petersen M. Carpenter T. S. Khalid S. (2016). Molecular dynamics simulations reveal the conformational flexibility of lipid II and its loose association with the defensin plectin in the Staphylococcus aureus membrane. Biochemistry55 (23), 3303–3314. 10.1021/acs.biochem.5b01315
42
Yelin I. Kishony R. (2018). Antibiotic resistance. Cell.172, 1136–1136.e1. 10.1016/j.cell.2018.02.018
Summary
Keywords
Staphylococcus aureus , tamoxifen derivatives, resistance, infection, treatment
Citation
Molina Panadero I, Falcón Torres J, Hmadcha A, Princiotto S, Cutarella L, Mori M, Dallavalle S, Christodoulou MS and Smani Y (2025) Antibacterial activity of tamoxifen derivatives against methicillin-resistant Staphylococcus aureus. Front. Pharmacol. 16:1549288. doi: 10.3389/fphar.2025.1549288
Received
20 December 2024
Accepted
11 April 2025
Published
30 April 2025
Volume
16 - 2025
Edited by
Thaigarajan Parumasivam, University of Science Malaysia (USM), Malaysia
Reviewed by
Ermin Schadich, Palacký University, Czechia
Hendrik W. Van Veen, University of Cambridge, United Kingdom
Updates
Copyright
© 2025 Molina Panadero, Falcón Torres, Hmadcha, Princiotto, Cutarella, Mori, Dallavalle, Christodoulou and Smani.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Younes Smani, ysma@upo.es; Michael S. Christodoulou, michail.christodoulou@unimi.it
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.