Abstract
Seven novel clerodane diterpenoids, designated tinotanoids I–O (1−7), were isolated from the tuberous roots of Paratinospora sagittata (Oliv.) Wei Wang [syn.: Tinospora sagittata var. yunnanensis (S.Y.Hu) H.S.Lo; Menispermaceae]. The structural elucidation of these new metabolites, including their absolute configurations, was achieved through advanced spectroscopic methods such as mass spectrometry (MS), nuclear magnetic resonance (NMR), electronic circular dichroism (ECD), and X-ray crystallography. Evaluation of their anti-inflammatory activity revealed that metabolites 3 and 4 exhibited potent inhibition of nitric oxide (NO) production in lipopolysaccharide (LPS)-activated RAW264.7 macrophages, with IC50 values of 12.5 ± 0.5 and 16.4 ± 0.7 μM, respectively, which are better than those of the positive controls, quercetin and dexamethasone. Additionally, metabolites 3 and 4 effectively inhibited the pro-inflammatory cytokines TNF-α and IL-6 in a dose-dependent manner.
1 Introduction
The Tinospora genus is abundant in clerodane diterpenoids (Zhang J.-S. et al., 2021; Zhang et al., 2022; Zhu et al., 2022; Zhu et al., 2023; Nishidono and Tanaka, 2024; Song J.-Q. et al., 2024), alkaloids (Huang et al., 2023; Liang et al., 2024), and lignans (Huang et al., 2012; Zhang J. S. et al., 2021), showcasing a wide array of biological activities, including anti-inflammatory (Zhu et al., 2023; Song Y.-Z. et al., 2024), antitumor (Mantaj et al., 2015; Zhang et al., 2022), antibacterial (Royani et al., 2023; Song J.-Q. et al., 2024), immunomodulation (Ao et al., 2024), and anti-diabetic (Lam et al., 2023; Mishra et al., 2023; Kabbashi et al., 2024) activities. Tinospora sagittata, one of the six Chinese native species of Tinospora, is widespread across southern China. Jin Guo Lan, a revered traditional Chinese medicine documented in each year’s edition of the Chinese Pharmacopoeia, is carefully crafted from the tuberous roots of T. sagittata and T. capillipes. These roots are highly valued for their efficacy in clearing heat and detoxifying the body, a property that has also been harnessed in the traditional ethnomedicine practiced by the “Dai” people (Huang et al., 2010; Zhang et al., 2016). In this study, we examined the metabolites of T. sagittata, sourced from Xishuangbanna, Yunnan Province. Our findings include the successful isolation and detailed characterization of seven clerodane diterpenoids from the plant (Figure 1), together with an assessment of their potential anti-inflammatory activity.
FIGURE 1
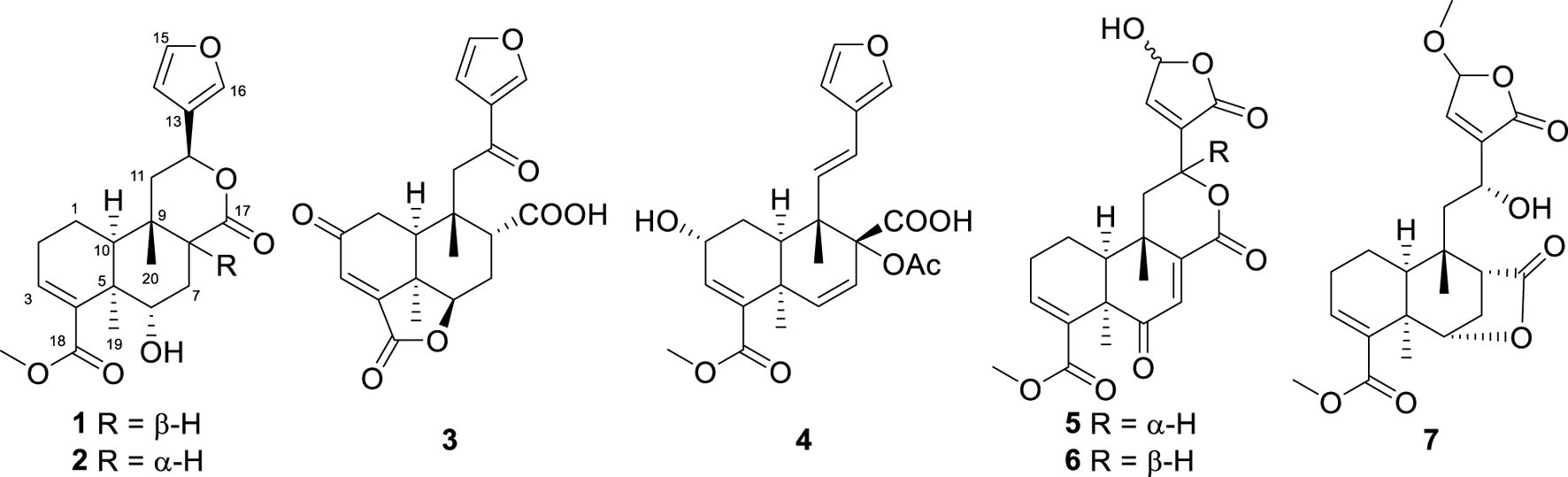
Structures of metabolites 1−7.
2 Materials and methods
2.1 General experimental procedures
X-ray diffraction data were acquired using an Agilent Xcalibur Nova X-ray diffractometer. Melting points were determined using an X-4 apparatus and reported without correction. Optical rotations were measured using a Perkin–Elmer 341 Polarimeter. NMR spectra were acquired using a Bruker AM-400 Spectrometer at 25°C. Electrospray ionization mass spectrometry (ESIMS) and high-resolution electrospray ionization mass spectrometry (HRESIMS) were performed using a Finnigan LCQ Deca system. Absorbance measurements were taken at 490 and 540 nm using a microplate reader (Molecular Devices, United States) and analyzed using SoftMax Pro 5 software (Molecular Devices, United States). For high-performance liquid chromatography (HPLC), a Shimadzu LC-20AT System coupled with an SPD-M20A PDA detector was used, and a YMC-pack ODS-A column (250 × 10 mm, S-5 μm, 12 nm) was used for semi-preparative HPLC. Column chromatography (CC) was conducted using silica gel (300–400 mesh, Qingdao Haiyang Chemical Co. Ltd.), reversed-phase C18 (RP-C18) silica gel (12 nm, S-50 μm, YMC Co. Ltd.), and MCI gel (CHP20P, 75–150 μm, Mitsubishi Chemical Industries Ltd.). All solvents were of analytical grade, sourced from Guangzhou Chemical Reagents Company, Ltd.
2.2 Plant materials
The tuberous roots of Paratinospora sagittata (Oliv.) Wei Wang [syn.: Tinospora sagittata var. yunnanensis (S.Y.Hu) H.S.Lo, Menispermaceae] were collected in August 2022 from Xishuangbanna, Yunnan province, China. The plant materials were authenticated by Jing Liu. A voucher specimen has been deposited at the Shandong Laboratory of Yantai Drug Discovery (accession number: JGL).
2.3 Extraction and isolation
The air-dried powder of the tuberous roots of T. sagittata (10 kg) was subjected to extraction by soaking in 95% ethanol at ambient temperature, with the process repeated three times at weekly intervals. Following this, the ethanol extract was concentrated to yield 900 g of dried material. This residue was then reconstituted by stirring in 5.0 L of water and subsequently subjected to liquid–liquid extraction with an equal volume of ethyl acetate in a separating funnel. The ethyl acetate layer was further fractionated using D101 macroporous resin column chromatography with a gradient of ethanol–water mixtures (30%, 60%, and 90%) to yield three distinct fractions. The 60% EtOH elution (140 g) was fractionated using silica gel CC and eluted with petroleum ether–EtOAc (5:1 to 0:1) to produce seven subfractions (A–G). Fraction E (10.0 g) was separated using silica gel CC (CH2Cl2–MeOH, 40:1 to 1:1) to produce eight subfractions (E1–E8), and fraction E5 was fractionated using RP-18 CC (MeOH–H2O, 30%–60%) to yield another eight subfractions (E4A–E4H). Fraction E4D was purified via Sephadex LH-20 column chromatography with methanol as the eluent, resulting in the isolation of metabolite 7 (40 mg). Fraction E4F was further separated using silica gel column chromatography with a gradient of petroleum ether and acetone (1:1 to 1:2) to yield four subfractions (E4F1–E4F4). Subsequently, subfractions E4F1 and E4F2 were purified using HPLC with the same solvent system (33% MeCN–H2O), leading to the isolation of metabolite 3 (2.2 mg; tR = 19.5 min) and 4 (8.5 mg; tR = 14.5 min), respectively. Fraction E6 was fractionated using silica gel CC (petroleum ether–acetone, 1:1 to 1:2) to yield three subfractions (E6A–E6C). Fraction E6B was purified using HPLC (36% MeCN–H2O) to produce 1 (4.0 mg; tR = 14.5 min) and 2 (2.0 mg; tR = 16 min), while E6C was also processed using HPLC (32% MeCN–H2O) to yield 5 (1.5 mg; tR = 18.5 min) and 6 (2 mg; tR = 19.0 min).
2.4 Spectroscopic data
Tinotanoid I (1): Colorless crystal, CCDC 2411568; Flack parameter, 0.10 (5); other crystal data are provided in Supplementary Table S1 in SI; m.p. 136−138°C; [α]25D −180 (c 0.05, MeOH); UV (MeOH) λmax (log ε), 209 nm (3.46) ; ECD (c 5.4 × 10−4 M, MeOH) λmax (Δε), 239 nm (−11.49); 1H and 13C NMR data are provided in Tables 1, 2; HRESIMS m/z 375.1805 [M + H]+ (calcd for C21H27O6+, 375.1802).
TABLE 1
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| 1 | 2.07, m | 2.08, m | 2.93, dd (18.6, 6.9) | 2.29, m | 2.17, dt (15.6, 8.7) | 2.12, td (15.6, 8.9) | 1.94, m (2H) |
| 1.77, m | 1.78, m | 2.69, d (18.6) | 1.85, m | 1.89, m | 1.95, m | ||
| 2 | 2.37, m | 2.41, m | 4.43, m | 2.44, m (2H) | 2.44, m (2H) | 2.39, m | |
| 2.32, m | 2.34, m | 2.31, m | |||||
| 3 | 6.77, t (3.6) | 6.61, t (3.9) | 6.66, s | 6.70, brs | 6.78, t (4.0) | 6.76, t (4.0) | 7.02, brt (3.5) |
| 6 | 4.62, brd (4.4) | 4.83, brs | 5.08, dd (10.6, 7.3) | 6.49, d (10.3) | 5.51, d (6.1) | ||
| 7 | 2.51, dt (14.4, 4.4) | 2.04, m | 2.53, m | 6.32, d (10.3) | 6.65, s | 6.98, s | 2.17, ddd (12.0, 6.1, 5.5) |
| 1.89, dd (14.4, 5.6) | 1.66, ddd (14.2, 12.3, 1.7) | 1.54, m | 1.98, m | ||||
| 8 | 2.21, dd (5.6,4.4) | 3.32, dd (12.1, 2.7) | 3.22, t (3.5) | 2.71, t (5.5) | |||
| 10 | 2.28, brd (6.4) | 2.08, m | 2.65, d (6.9) | 2.54, brd (5.6) | 2.30, dd (6.7, 1.7) | 2.01, dd (6.6, 1.0) | 1.51, m |
| 11 | 2.26, dd (14.3, 1.8) | 2.08, dd (13.9, 6.1) | 3.19, d (15.1) | 6.14, d (16.3) | 2.49, dd (14.5, 3.1) | 2.63, dd (13.9, 3.0) | 2.23, dd (15.1, 7.0) |
| 1.64, dd (14.3, 12.6) | 1.95, dd (13.9, 11.4) | 2.57, d (15.1) | 1.82, dd (14.5, 12.0) | 1.56, m | 1.76, d (15.1) | ||
| 12 | 5.55, dd (12.6, 1.8) | 5.39, dd (11.4, 6.1) | 6.22, d (16.3) | 4.93, ddd (12.0, 3.1, 1.0) | 5.42, ddd (12.1, 3.0, 1.0) | 4.89, d (7.0) | |
| 14 | 6.44, brs | 6.42, d (1.0) | 6.75, d (1.3) | 6.56, brs | 7.31, dd (1.4, 1.0) | 7.28, brs | 7.03, s |
| 15 | 7.41, brs | 7.42, t (1.6) | 7.48, t (1.3) | 7.35, brs | 6.22, brs | 6.21, brs | 5.28, s |
| 16 | 7.49, brs | 7.46, brs | 8.10, brs | 7.40, brs | |||
| 19 | 1.38, s | 1.37, s | 1.56, s | 1.54, s | 1.44, s | 1.41, s | 1.34, s |
| 20 | 1.14, s | 1.05, s | 0.97, s | 0.99, s | 1.26, s | 1.37, s | 1.22, s |
| 18-OMe | 3.74, s | 3.71, s | 3.74, s | 3.70, s | 3.71, s | 3.72, s | |
| 15-OMe | 3.56, s | ||||||
| OAc | 2.07, s |
1H NMR data of metabolites 1−7 in CDCl3 (recorded at 600 MHz; δ in ppm; J in Hz).
TABLE 2
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| 1 | 16.9 | 16.8 | 35.5 | 29.6 | 17.8 | 17.3 | 16.4 |
| 2 | 23.9 | 23.4 | 197.3 | 64.3 | 23.0 | 22.8 | 24.1 |
| 3 | 140.6 | 140.1 | 128.6 | 140.1 | 139.0 | 138.5 | 142.6 |
| 4 | 136.8 | 136.4 | 153.0 | 136.9 | 134.2 | 134.6 | 134.1 |
| 5 | 39.4 | 40.0 | 40.6 | 37.1 | 47.2 | 46.5 | 39.2 |
| 6 | 69.3 | 68.9 | 82.5 | 136.1 | 200.2 | 200.5 | 83.3 |
| 7 | 28.8 | 27.1 | 29.0 | 124.4 | 131.0 | 133.2 | 29.5 |
| 8 | 45.7 | 40.7 | 47.0 | 82.5 | 148.2 | 145.3 | 47.6 |
| 9 | 35.7 | 37.7 | 39.1 | 44.6 | 38.1 | 37.4 | 39.5 |
| 10 | 36.8 | 45.7 | 46.4 | 44.1 | 48.5 | 48.8 | 45.1 |
| 11 | 42.8 | 45.8 | 46.5 | 132.2 | 42.5 | 40.0 | 44.8 |
| 12 | 69.7 | 70.3 | 195.2 | 120.0 | 71.1 | 71.3 | 64.8 |
| 13 | 125.1 | 124.5 | 129.0 | 124.4 | 135.0 | 136.5 | 140.0 |
| 14 | 108.6 | 108.6 | 108.5 | 107.8 | 145.9 | 145.5 | 142.2 |
| 15 | 143.6 | 143.6 | 148.6 | 143.3 | 97.6 | 97.6 | 103.0 |
| 16 | 139.7 | 139.5 | 145.1 | 139.6 | 168.5 | 168.5 | 170.8 |
| 17 | 174.6 | 175.3 | 175.4 | 166.9 | 166.5 | 163.3 | 179.1 |
| 18 | 168.0 | 167.7 | 168.1 | 172.5 | 167.0 | 167.1 | 166.7 |
| 19 | 27.0 | 28.1 | 28.1 | 30.8 | 26.1 | 26.5 | 27.1 |
| 20 | 27.6 | 21.8 | 21.1 | 16.9 | 26.2 | 19.3 | 21.8 |
| 15-OMe | 57.2 | ||||||
| 18-OMe | 51.7 | 51.7 | 51.8 | 52.1 | 52.1 | 51.7 | |
| OAc | 169.7 | ||||||
| 21.0 |
13C NMR data of metabolites 1−7 in CDCl3 (151 MHz; δ in ppm).
Tinotanoid J (2): White solid; [α]25D −33.1 (c 0.10, MeOH); UV (MeOH) λmax (log ε), 208 nm (3.52); ECD (c 6.7 × 10−4 M, MeOH) λmax (Δε), 241 nm (−4.13) and 214 nm (+2.95); 1H and 13C NMR data are listed in Tables 1, 2, respectively; HRESIMS m/z 375.1804 [M + H]+ (calcd for C21H27O6+, 375.1802).
Tinotanoid K (3): Colorless oil; [α]25D −48.7 (c 0.30, MeOH); UV (MeOH) λmax (log ε), 209 nm (3.61) and 240 nm (3.25); ECD (c 2.7 × 10−4 M, MeOH) λmax (Δε), 281 nm (+6.78) and 232 nm (−7.79); 1H and 13C NMR data are listed in Tables 1, 2, respectively; HRESIMS m/z 373.1286 [M + H]+ (calcd for C20H21O7+, 373.1282).
Tinotanoid L (4): Colorless oil; [α]25D −89.1 (c 0.20, MeOH); UV (MeOH) λmax (log ε), 211 nm (3.81); ECD (c 2.3 × 10−4 M, MeOH) λmax (Δε), 253 nm (−1.52) and 218 nm (+14.22); 1H and 13C NMR data are listed in Tables 1, 2, respectively; HRESIMS m/z 429.1551 [M − H]− (calcd for C23H25O8−, 429.1555).
Tinotanoid M (5): Colorless oil; [α]25D +11.5 (c 0.20, MeOH); UV (MeOH) λmax (log ε), 206 nm (3.44); ECD (c 5.0 × 10−4 M, MeOH) λmax (Δε), 268 nm (−2.14), 236 nm (+10.61), and 215 nm (−4.70); 1H and 13C NMR data are listed in Tables 1, 2, respectively; HRESIMS m/z 401.1244 [M − H]− (calcd for C21H21O8−, 401.1242).
Tinotanoid N (6): Colorless oil; [α]25D +8.7 (c 0.20, MeOH); UV (MeOH) λmax (log ε), 207 nm (3.59); ECD (c 5.0 × 10−4 M, MeOH) λmax (Δε), 268 nm (−2.12), 238 nm (+10.48), and 216 nm (−4.25); 1H and 13C NMR data are listed in Tables 1, 2, respectively; HRESIMS m/z 401.1243 [M − H]− (calcd for C21H21O8−, 401.1242).
Tinotanoid O (7): Colorless oil; [α]25D −30.6 (c 0.20, MeOH); UV (MeOH) λmax (log ε), 211 nm (3.65); ECD (c 4.8 × 10−4 M, MeOH) λmax (Δε), 249 nm (−1.85) and 215 nm (+2.64); 1H and 13C NMR data are listed in Tables 1, 2, respectively; HRESIMS m/z 443.1676 [M + Na]+ (calcd for C22H28O8Na+, 443.1676).
2.5 ECD and NMR calculation procedure
The absolute configurations of metabolites 2−4 were assigned by theoretical ECD calculation using the time-dependent density functional theory (TDDFT) approach. The initial conformations of the target molecules were generated using the MM2 force field within the ChemDraw Pro 14.1 software suite. Subsequent conformational searches were conducted using a hybrid torsional/low-mode sampling approach, employing MMFFs within an energy window of 2.58 kcal/mol. This was achieved through the conformational search module integrated into Maestro 10.2 software. The re-optimization of these conformations, followed by TD-DFT calculations on the refined structures, was executed using Gaussian 09 at the B3LYP/6-311G (d,p) level of theory in a vacuum. To ensure that the re-optimized conformers represented energy minima, a frequency analysis was also performed. Finally, SpecDis 1.64 software was used to generate the Boltzmann-weighted average ECD spectra.
NMR calculations for metabolite 4 were performed using the gauge-including atomic orbitals (GIAO) approach at the B3LYP/6-31G(d) level of theory, with the polarizable continuum model (PCM) used to simulate chloroform as the solvent. The NMR data for the lowest energy conformers of each metabolite were averaged based on the principles of Boltzmann distribution. The correlation between experimental and calculated NMR data was assessed using the enhanced probability method DP4+, and the results are provided in the accompanying supplementary data file.
2.6 Analysis of NO production
RAW264.7 macrophages were plated into 96-well plates at a density of 8 × 104 cells/well for 24 h and then pre-incubated with varying concentrations of test metabolites for 1 h prior to stimulation with lipopolysaccharide (LPS) at a concentration of 0.1 μg/mL for 24 h. The concentration of nitric oxide (NO) in the culture supernatant was then quantified using a Griess reagent kit. Absorbance (A) at 540 nm was measured using a multifunction microplate reader. The inhibition of cell growth was calculated according to the following formula: Inhibition (%) = [1 − (ALPS + sample − Auntreated) / (ALPS − Auntreated)] × 100. The experiments were performed in triplicates, and the data were presented as the mean ± SD. Quercetin was used as a positive control.
2.7 Cytotoxicity assay
The cytotoxicity of the isolated metabolites toward RAW264.7 cells was evaluated using an MTT assay. RAW264.7 cells were planted in 96-well plates (8 × 103/well) for 24 h. Subsequently, the cells were exposed to the test metabolites, which were initially dissolved in DMSO and then diluted in DMEM to a final volume of 100 μL per well, achieving a concentration of 50 μM for the metabolites and maintaining a 1% DMSO concentration. Wells without cells containing only 100 μL of DMEM served as the blank control. After 24 h, 20 μL of the MTT solution was added to each well. After incubation for 4 h, the medium was removed, and 100 μL of DMSO was added to each well; then, absorbance (A) was detected at 490 nm using a microplate reader. The inhibition of cell growth was calculated according to the following formula: % Inhibition = [1− (Asample − Ablank) / (Asolvent − Ablank)] × 100.
2.8 Analysis of TNF-α and IL-6 production
RAW264.7 macrophages were pretreated with 3 and 4 for 1 h prior to the stimulation with LPS (0.1 μg/mL) for 6 h. The concentrations of TNF-α and IL-6 in the culture medium were determined using commercial ELISA kits, according to the instructions of each manufacturer.
3 Results and discussion
Metabolite 1, a colorless crystal, had the molecular formula C21H26O6, as established by HRESIMS, which showed an ion at m/z 375.1805 [M + H]+ (calcd 375.1802), corresponding to nine degrees of unsaturation. The 1H NMR data (Table 1) displayed signals for three methyl singlets [δH 1.14, 1.38, and 3.74 (each 3H, s)], two oxymethine protons [δH 4.62 (1H, d, J = 5.1 Hz) and 5.55 (1H, dd, J = 12.6, 1.8 Hz)], a β-furyl ring [δH 7.49 (1H, brs), 7.41 (1H, brs), and 6.44 (1H, brs)], an olefinic proton [δH 6.77 (1H, t, J = 3.6 Hz)], and a series of aliphatic multiplets. The 13C NMR spectrum, associated with DEPT experiments, resolved 21 carbon resonances attributable to two carbonyl groups (δC 174.6 and 168.0), a β-furyl ring (δC 143.6, 139.7, 125.1, and 108.6), a trisubstituted double bond (δC 140.6 and 136.8), three methyls (one oxygenated), four sp3 methylenes, four sp3 methines (two oxygenated), and two sp3 quaternary carbons (Table 2). The aforementioned information was characteristic of a clerodane diterpenoid, which showed high similarity to tinopanoid R, a known clerodane recently reported from plants of the same genus (Zhu et al., 2023). Compared with tinopanoid R, the major differences in metabolite 1 were the appearance of an additional methylene (δC 28.8) and an additional methine (δC 45.7), instead of a trisubstituted double bond (δC 141.0 and 135.0) in tinopanoid R. This was supported by the key 1H–1H COSY correlations from H-6 (δH 4.62) to H-7 (δH 2.51 and 1.89) and H-7 to H-8 (δH 2.21) (Figure 2). The relative configuration of metabolite 1 was assigned mainly by the NOESY correlations (Figure 3). NOESY cross peaks between H-10/H3-19 and H-12/H3-19 and the correlation from H3-19 to H-7a (δH at 2.51) provided clear evidence for the co-facial orientation of these protons. These protons were arbitrarily assigned to α-orientation. Additionally, NOESY correlations from the other proton of H2-7 (H-7b: δH 1.89) to H3-20 and H-8 were ascertained to be in the opposite orientation. These correlations indicated that H3-20 and H-8 are spatially oriented in the β-conformation in the molecule. Finally, the absolute configuration of metabolite 1 was determined to be 5R, 6S, 8R, 9S, 10S, 12S by single-crystal X-ray diffraction analysis (Figure 4). Metabolite 1 was thus established as depicted and given the trivial name tinotanoid I, following tinotanoids A–H, which were reported from the same species.
FIGURE 2
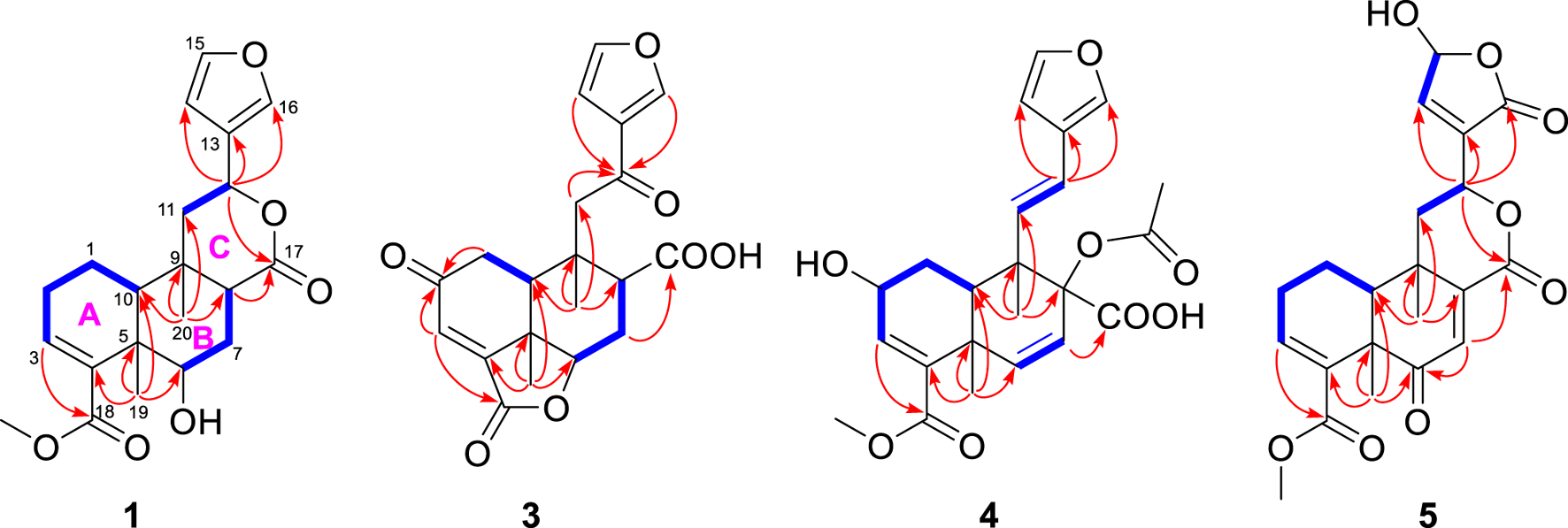
Selected 1H−1H COSY (‐) and HMBC (→) correlations of 1, 3, 4, and 5.
FIGURE 3
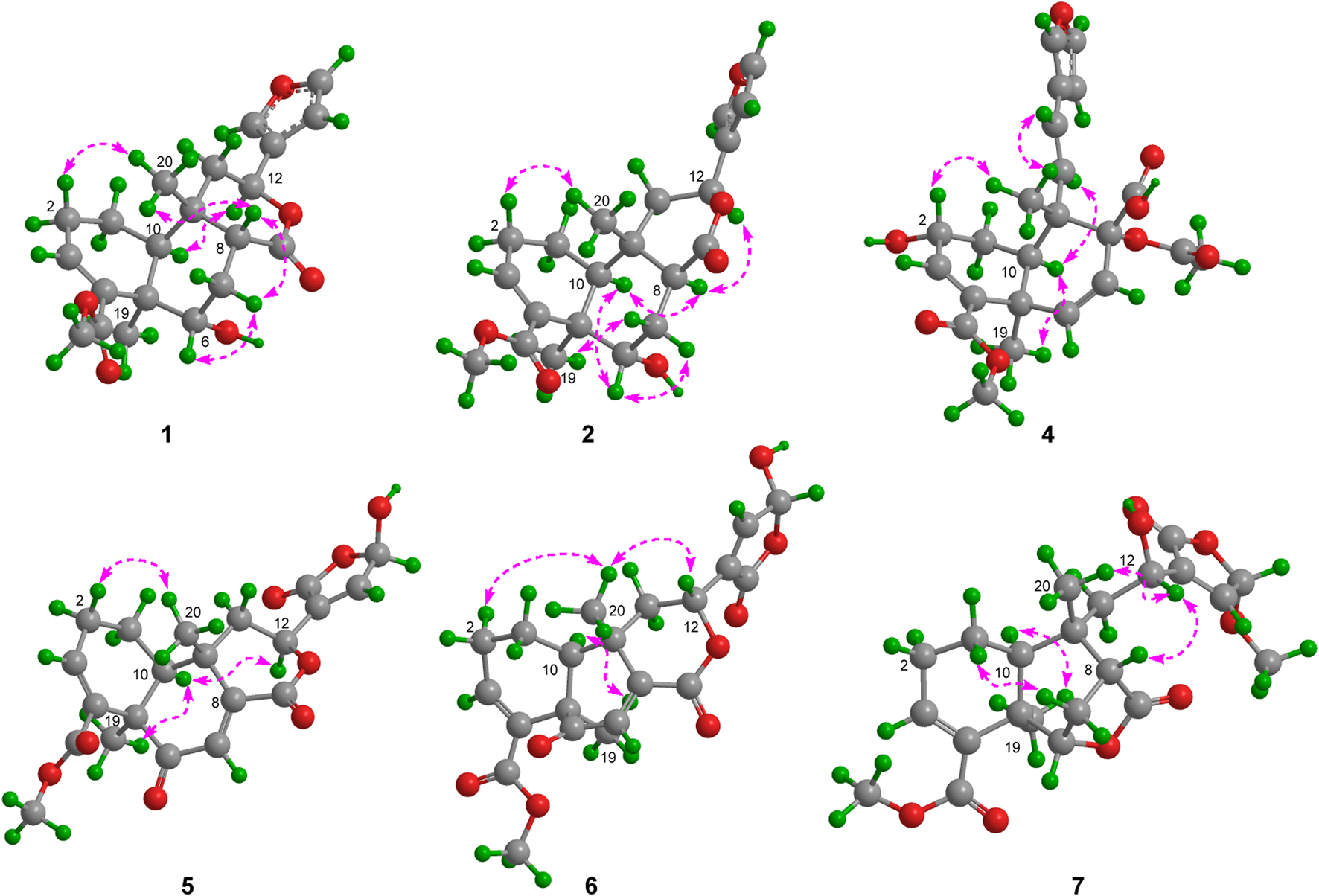
Selected NOESY correlations of metabolites 1, 2, and 4−7.
FIGURE 4
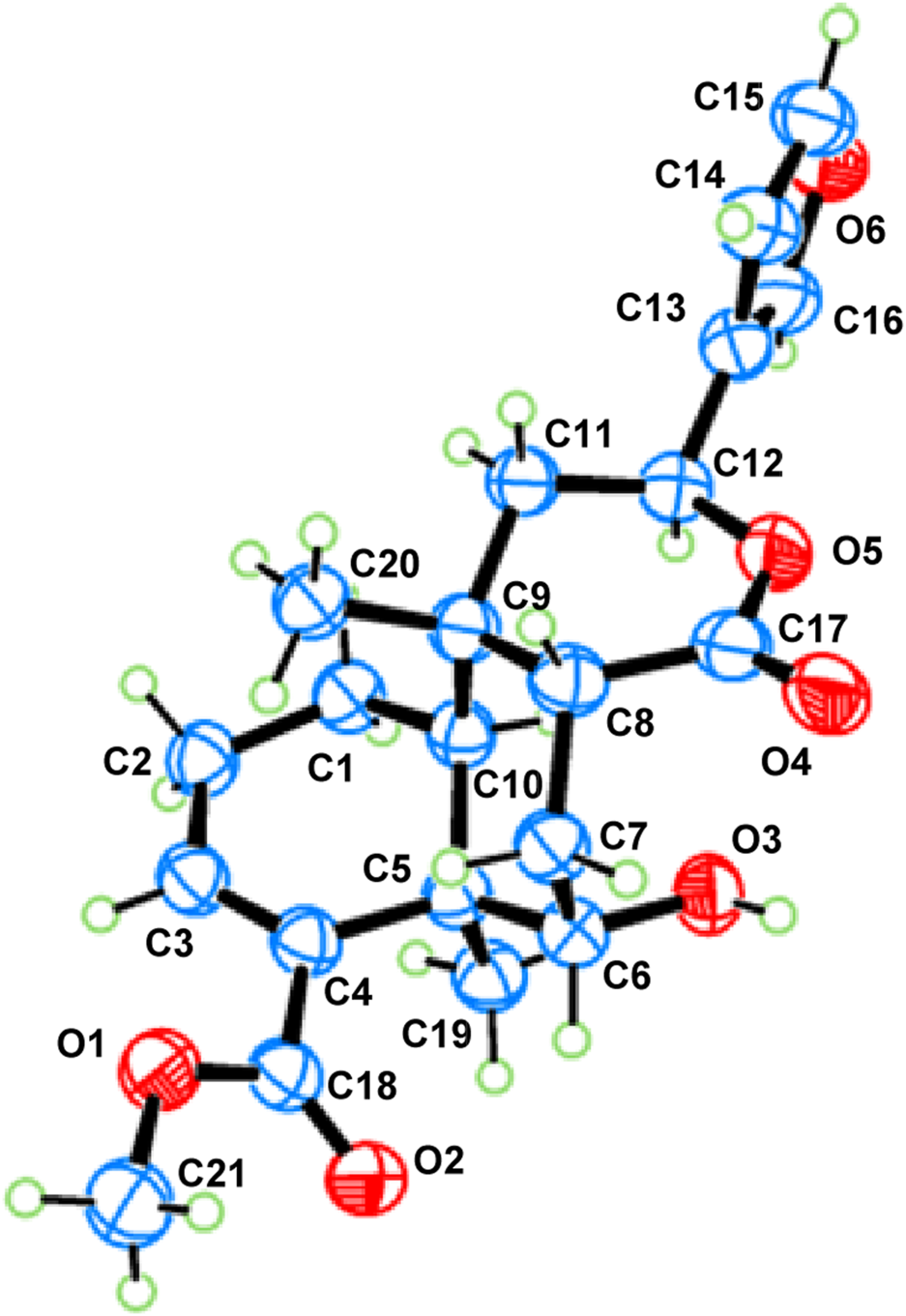
ORTEP drawing of metabolite 1.
Metabolite 2, a colorless gum, had the molecular formula C21H26O6, as determined by the HR-ESIMS and 13C NMR data (Table 2). The 1D NMR spectra of metabolite 2 exhibited most of the structural features found in metabolite 1, with the major difference occurring in the signals around C-8. A detailed interpretation of 2D NMR data revealed that metabolite 2 shared the same planar structure as metabolite 1. NOESY correlations from H-10 to H-8 and H3-19 revealed that H-8 was in the same orientation as H-10 and H3-19 and was in the α-orientation. Thus, metabolite 2 was supposed to be the 8-epimer of 1, which was also supported by the different coupling constants of H-8 (J = 5.6, 4.4 Hz in 1; J = 12.1, 2.7 Hz in 2). The absolute configuration of metabolite 2 was assigned as 5R, 6S, 8S, 9S, 10S, 12S by ECD calculation (Figure 5). Metabolite 2 was given the trivial name tinotanoid J.
FIGURE 5
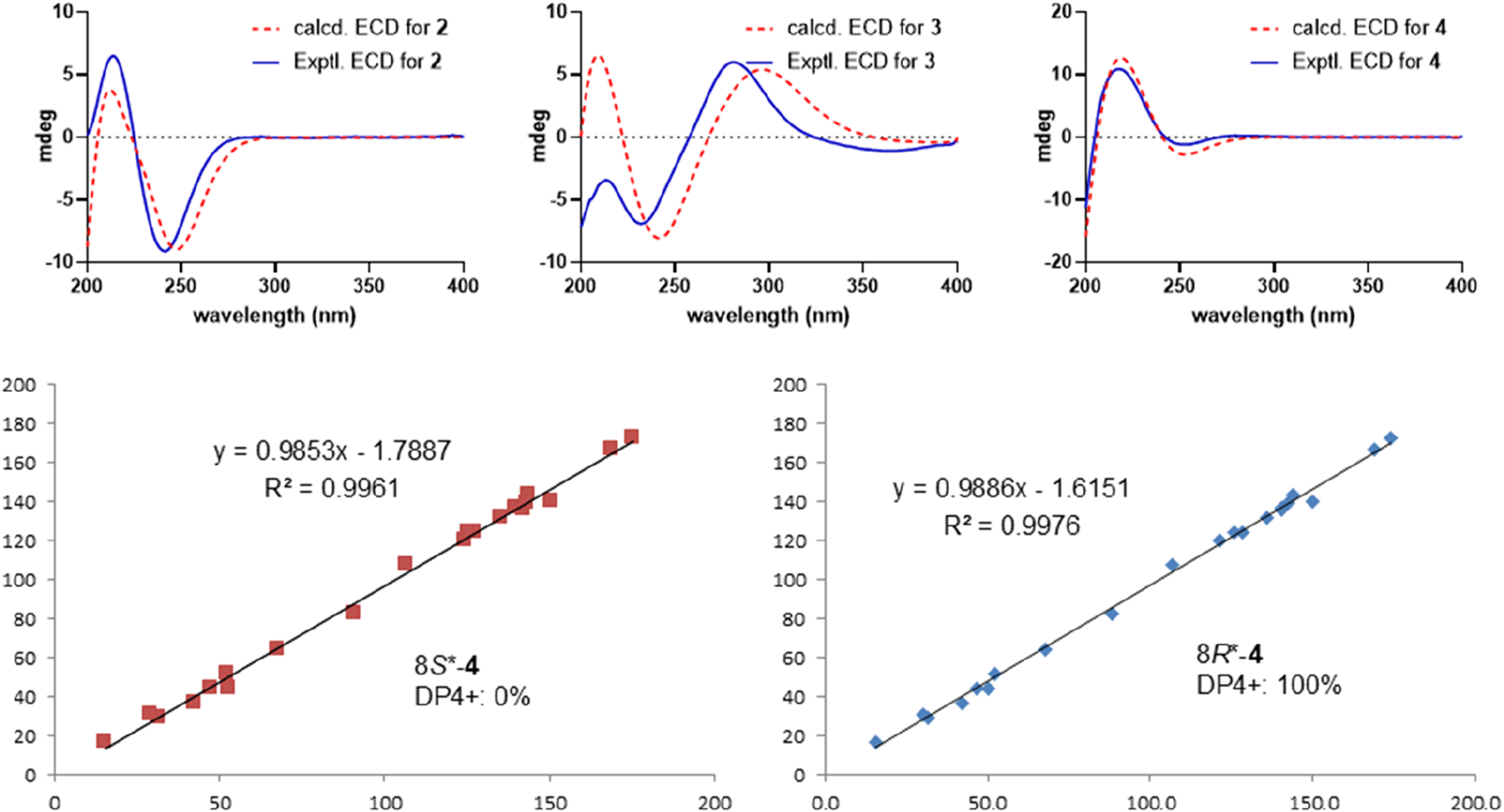
Experimental and calculated ECD spectra for metabolites 2−4 (above); NMR calculations with DP4+ probability analysis for metabolite 4 (below).
According to the HRESIMS ion peak at m/z 373.1286 [M + H]+ (calcd for C20H21O7+, 373.1282) and 13C NMR data, the molecular formula of tinotanoid K (3) was determined as C20H20O7 with 11 indices of hydrogen deficiency. The 1D NMR data (Tables 1, 2) suggested that metabolite 3 is a clerodane diterpenoid, exhibiting a high degree of similarity to the NMR characteristics of tinocapill B (Song Y.-Z. et al., 2024). Compared with the known tinocapill B, it was observed that metabolite 3 features a ketone group in place of the methylene unit present in the known metabolite. Key HMBC correlations from H-1 (δH 2.93 and 2.69), H-3 (δH 6.66), and H-10 (δH 2.65) to the additional ketone group (δC 197.3) revealed that the ketone group was at the C-2 position. The planar structure of metabolite 3 was further secured by a detailed interpretation of its 2D NMR data. The relative configuration of metabolite 3 was assigned to be the same as that of tinocapill B by comparing their 1D NMR data and analyzing its NOESY data. The absolute configuration of metabolite 3 was assigned as 5R, 6R, 8R, 9S, 10S by ECD calculation (Figure 5).
Metabolite 4, a colorless gum, had the molecular formula C23H26O8, as established by HRESIMS, which showed an ion at m/z 429.1551 [M − H]− (calcd 429.1555), corresponding to 11 degrees of unsaturation. The 1D NMR data (Tables 1, 2) and HSQC spectrum showed signals for a β-furyl ring group [δH 7.40 (1H, brs), 7.35 (1H, brs), and 6.56 (1H, brs); δC 143.3, 139.6, 124.4, and 107.8], two disubstituted double bonds [δH 6.49 (1H, d, J = 10.3 Hz), 6.32 (1H, d, J = 10.3 Hz), 6.22 (1H, d, J = 16.3 Hz), and 6.14 (1H, d, J = 16.3 Hz); δC 136.1, 132.2, 124.4, and 120.0], a trisubstituted double bond [δH 6.70 (1H, brs); δC 140.1 and 136.9], two carbonyl groups [δC 172.5 and 166.9], an acetoxy group [δH 2.07 (3H, s); δC 169.7 and 21.0], a methoxy group [δH 3.74 (3H, s); δC 51.8], and two methyl groups [δH 1.54 and 0.99 (each 3H, s); δC 30.8 and 16.9]. The aforementioned data of metabolite 4 exhibited most of the structural features of a clerodane diterpenoid and were quite similar to those of crispene F (Al et al., 2018). Compared with crispene F, the major differences in crispene metabolite 4 were due to the appearance of an additional trans-disubstituted double bond [δH 6.22 (1H, d, J = 16.3 Hz) and 6.14 (1H, d, J = 16.3 Hz)] instead of one methylene and an oxymethine in crispene F. NMR comparison of the two metabolites revealed that the most signals in rings A and B were quite similar. Key HMBC correlations from the proton in the additional trans-disubstituted double bond [δH 6.22 and 6.14] to C-10 and C-13 suggested that the double bond was at the C-11/C-12 position. Moreover, the chemical shift at C-8 (δC 82.5) was downfield-shifted compared to that (δC 78.4) in crispene F; the absence of HMBC correlations from the oxymethine to the acetoxy group indicated that 8-OH in crispene F was acetylated. A detailed 2D NMR correlations further confirmed the planar structure of metabolite 4. The relative configuration of metabolite 4 was assigned the same as crispene F by 1D NMR and NOESY data. In particular, the stereochemistry in C-8 was not determined; the authors later established this configuration via NMR calculations and analyzed it using the DP4+ probability method. As shown in Figure 5, the final DP4+ score of 8R* (100.0%) allowed the assignment of the relative configurations of C-8. Finally, the absolute configuration of metabolite 4 was assigned by TD-DFT ECD calculation. The experimental ECD curve exhibited comparable cotton effects to the calculated ECD spectrum of (2R, 5R, 8R, 9S, 10R)-isomer of metabolite 4. Thus, the structure of metabolite 4 was confidently assigned as indicated and named tinotanoid L.
Metabolite 5, a colorless oil, had the molecular formula C21H22O8, as determined by 13C NMR and HRESIMS data. The 1H and 13C NMR data (Tables 1, 2) of metabolite 5 showed a ketone group (δC 200.2), two carbonyl groups [δC 167.0 and 166.5], a β-butenolide moiety [δH 7.31 (1H, brs) and 6.22 (1H, brs); δC 168.5, 145.9, 135.0, and 97.6], two trisubstituted double bonds [δH 6.78 (1H, t, J = 4.0 Hz) and 6.65 (1H, s); δC 148.2, 139.0, 134.2, and 131.0], a methoxy group [δH 3.70 (3H, s); δC 52.1], and two methyl groups [δH 1.44 and 1.26 (each 3H, s); δC 26.2 and 26.1]. The abovementioned data were quite similar to those of tinocordifoliol A (Hoang et al., 2024), with notable differences being the absence of the β-furyl ring in tinocordifoliol A and the presence of a β-butenolide moiety in metabolite 5. HMBC correlations from H-12 (δH 4.93) to the carbons of the β-butenolide moiety (δC 168.5, 145.9, and 135.0) supported the above elucidation. 2D NMR analysis established the molecular structure of metabolite 5. The relative configuration of metabolite 5 was determined by 1D NMR data, compared to the known metabolite, along with the NOESY spectrum (Figure 3). Key NOESY correlations from H3-19 to H-10 assigned these protons as having α-orientation, similar to those of tinocordifoliol A. Thus, NOESY correlations from H-12 to H-10 revealed that H-12 and H-10 were co-facial and in the α-orientation. Thus, metabolite 5 was named tinotanoid M.
HRESIMS analysis revealed that the molecular ion at m/z 401.1243 of tinotanoid N (6) was consistent with the molecular formula C21H22O8 (calcd 401.1242), indicating that it was an isomer of metabolite 5. Comprehensive analyses of the NMR spectrum revealed that metabolite 6 shared the same planar structure as metabolite 5. The configuration of H-10/H3-19 in the α-orientation, along with H3-20 in the β-orientation, was assigned as the same as in metabolite 5, according to the biosynthesis consideration. A comprehensive analysis of NOESY correlations revealed the signals of H-12/H3-20, indicating that H-12 was in the β-orientation. Thus, metabolite 6 was assigned as 12-isomer of metabolite 5.
Metabolites 5 and 6 are both expected to be racemates at the C-15 position as a result of the hemiacetal functionality, leading to dynamic interconversion at this site. Nonetheless, spectral data indicate a high degree of purity for both metabolites, potentially due to hydrogen bonding interactions. The exact mechanism behind this phenomenon is a subject for further investigation. The absolute configurations of 5 and 6 were determined by ECD calculation (see Supplementary Figure S50).
Metabolite 7 had the molecular formula C22H24O8. The NMR spectra of metabolite 7 showed the signals of a β-butenolide moiety [δH 7.03 (1H, s) and 5.78 (1H, s); δC 170.7, 142.2, 140.0, and 103.0], two carbonyl groups [δC 179.1 and 166.7], a trisubstituted double bond [δH 7.02 (1H, brt, J = 3.5 Hz); δC 142.6 and 134.1], two methoxy groups [δH 3.72 and 3.56 (each 3H, s); δC 57.2 and 51.7], and two methyl groups [δH 1.34 and 1.22 (each 3H, s); δC 27.1 and 21.8] (Tables 1, 2). The abovementioned data were quite similar to those of tinocapill C, except for the absence of the ethyl unit, indicating that metabolite 7 was the deethylated product of tinocapill C (Song J.-Q. et al., 2024). A detailed 2D NMR analysis revealed the planar structure of metabolite 7. The relative configuration of metabolite 7 was assigned the same as that of tinocapill C by their highly matched 1D NMR data and NOESY correlations. In particular, NOESY correlations from H-12 to H-8 and H3-20, along with the similar 1D NMR data of tinocapill C and metabolite 7 [δH 4.92 and δC 64.4 in tinocapill C; δH 4.89 and δC 64.8 in metabolite 7], indicated that H-12 was β-oriented. Thus, metabolite 7 was named tinotanoid O.
Metabolites 1−7 were assessed for their ability to inhibit NO production in LPS-stimulated RAW264.7 macrophages. Quercetin, a recognized natural inhibitor of NO, and dexamethasone, a broad-spectrum anti-inflammatory drug, were employed as positive controls with IC50 values of 21.0 ± 0.6 and 24.5 ± 1.9 μM, respectively. The results presented in Table 3 indicate that metabolites 3 and 4 demonstrated significant inhibitory effects, with IC50 values of 12.5 ± 0.5 and 16.4 ± 0.7 μM, respectively. In contrast, the remaining metabolites, with IC50 values exceeding 50 μM, were deemed inactive. To rule out the possibility that the inhibitory effects were due to cytotoxicity, the viability of RAW264.7 cells treated with the active metabolites was evaluated using the MTT assay. No appreciable cytotoxicity was noted, with cell survival rates exceeding 90% at all the concentrations below 50 μM, suggesting that the inhibitory activities observed were not a result of cellular toxicity.
TABLE 3
| No. | IC50 (μM) | No. | IC50 (μM) |
|---|---|---|---|
| 1 | >50 | 2 | >50 |
| 3 | 12.5 ± 0.5 | 4 | 16.4 ± 0.7 |
| 5 | >50 | 6 | >50 |
| 7 | >50 | Quercetina | 21.0 ± 0.6 |
| Dexamethasonea | 24.5 ± 1.9 | a positive control | |
IC50 values of the active metabolites on LPS-induced NO production in RAW264.7 cells.
An overabundance of pro-inflammatory mediators, including TNF-α, IL-1, IL-6, NO, and PGE2, secreted by macrophages, can precipitate pathological conditions and is associated with various chronic inflammatory diseases (Chan et al., 2015). Notably, TNF-α and IL-6 are considered key biomarkers for evaluating the inflammatory state. To delve into the anti-inflammatory capabilities of metabolites 3 and 4, their ability to inhibit LPS-induced expression of TNF-α and IL-6 in RAW 264.7 cells was examined. Figure 6 reveals that the elevated levels of TNF-α and IL-6 following LPS stimulation were significantly reduced by pretreatment with metabolites 3 and 4 with a dose-dependent inhibition. These findings indicate that metabolites 3 and 4 have the potential to mitigate the inflammatory response in RAW264.7 cells.
FIGURE 6
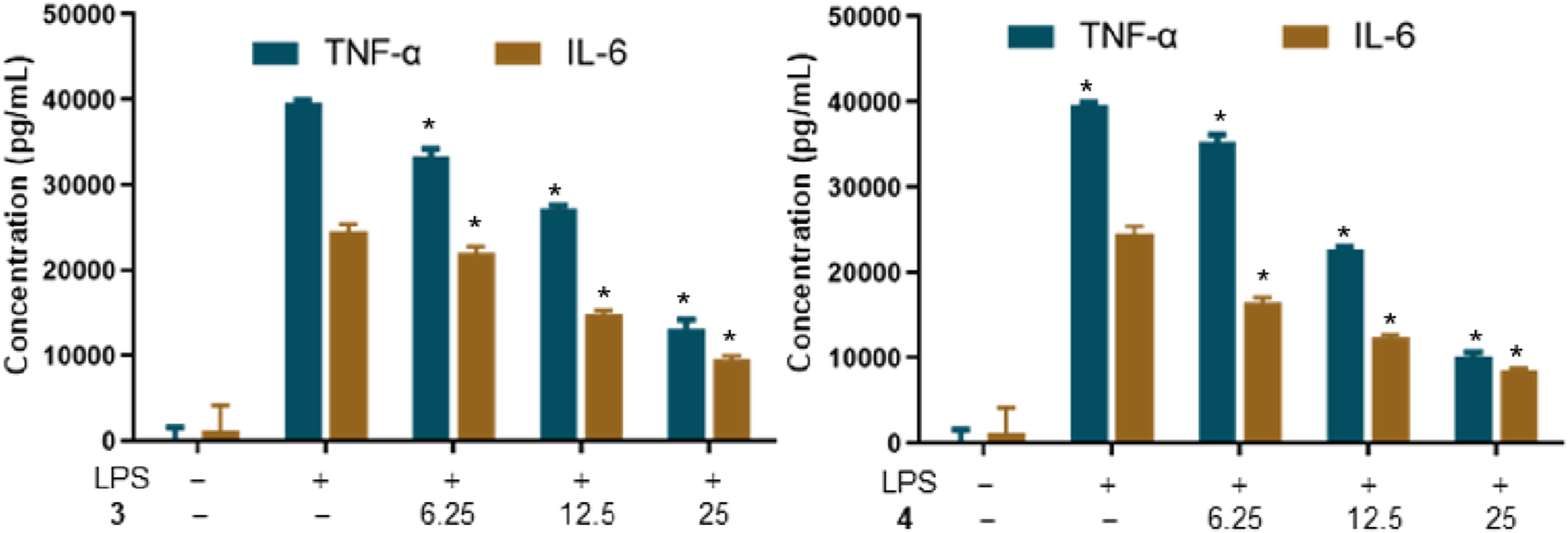
Effects of metabolites 3 and 4 on the production of TNF-α in LPS-induced RAW264.7 macrophages (*p < 0.05 versus the group treated with LPS only).
4 Conclusion
Tinospora sagittata, a revered traditional Chinese medicinal botanical drug, is renowned for its potent anti-inflammatory properties. Previous studies have demonstrated that clerodane diterpenoids extracted from T. sagittata exhibit a range of anti-inflammatory effects (Li et al., 2012; Song Y.-Z. et al., 2024). In the current study, seven previously unreported clerodane diterpenoids were successfully isolated from the tuberous roots of T. sagittata var. yunnanensis, two of which showed potent anti-inflammatory activity. These findings suggest that the anti-inflammatory properties of T. sagittata may be attributed to clerodane diterpenoids. Additionally, the specific clerodane diterpenoids identified in this study could serve as promising candidates for further pharmacological investigation and may contribute to the advancement of botanical drugs in the treatment of inflammatory diseases. Furthermore, exploring the mechanisms by which these clerodane diterpenoids modulate key inflammatory pathways, such as NF-κB and MAPK, could be a promising strategy to enhance their pharmacological significance.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author contributions
HL: writing–original draft, data curation, formal analysis, investigation, and software. JL: data curation, investigation, validation, and writing–original draft. CF: conceptualization, project administration, resources, validation, visualization, and writing–review and editing. SN: conceptualization, funding acquisition, project administration, resources, supervision, validation, and writing–review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the Natural Science Foundation of Shandong, China (ZR2023MH196).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2025.1561954/full#supplementary-material
References
1
Al N. M. A. Hossain T. Ahsan M. Jamshidi S. Hasan C. M. Rahman K. M. (2018). Crispenes F and G, cis-clerodane furanoditerpenoids from Tinospora crispa, inhibit STAT3 dimerization. J. Nat. Prod.81 (2), 236–242. 10.1021/acs.jnatprod.7b00377
2
Ao R. Li M.-Y. Yang F.-F. Bao J. Zhang J.-S. Zhang H. (2024). Targeted discovery of clerodane diterpenoids from Tinospora sinensis as immunomodulatory agents. Fitoterapia178, 106174. 10.1016/j.fitote.2024.106174
3
Chan P.-M. Tan Y.-S. Chua K.-H. Sabaratnam V. Kuppusamy U. R. (2015). Attenuation of inflammatory mediators (TNF-α and nitric oxide) and up-regulation of IL-10 by wild and domesticated basidiocarps of Amauroderma rugosum (blume and T. Nees) torrend in LPS-stimulated RAW264.7 cells. Plos One10 (10), e0139593. 10.1371/journal.pone.0139593
4
Hoang N. N. Hoshino S. Kodama T. Nakashima Y. Do K. M. Thao H. X. et al (2024). Three neo-clerodane diterpenoids from Tinospora cordifolia stems and their arginase inhibitory activities. Chem. Pharm. Bull.72 (6), 540–546. 10.1248/cpb.c24-00240
5
Huang F. M. Gongpan P. Ji K. L. Zhou L. Song Q. S. Fan Q. F. (2023). One novel alkaloid from the stems of Tinospora crispa. Nat. Prod. Res., 1–6. 10.1080/14786419.2023.2272023
6
Huang X. Z. Cheng C. M. Dai Y. Fu G. M. Guo J. M. Liang H. et al (2012). A novel lignan glycoside with antioxidant activity from Tinospora sagittata var. yunnanensis. Nat. Prod. Res.26 (20), 1876–1880. 10.1080/14786419.2011.619190
7
Huang X.-Z. Cheng C.-M. Dai Y. Fu G.-M. Guo J.-M. Yin Y. et al (2010). A novel 18-norclerodane diterpenoid from the roots of Tinospora sagittata var. yunnanensis. Molecules15 (11), 8360–8365. 10.3390/molecules15118360
8
Kabbashi A. S. Sattar M. A. Aamer M. Siddiqui N. N. Kamran M. Fayaz A. et al (2024). Clerodane furanoditerpenoids from Tinospora bakis (A.rich.) miers (Menispermaceae). Molecules29 (1), 154. 10.3390/molecules29010154
9
Lam S.-H. Liu H.-K. Chung S.-Y. Chang J.-L. Hong M.-X. Kuo S.-C. et al (2023). Diterpenoids and their glycosides from the stems of Tinospora crispa with beta-cell protective activity. J. Nat. Prod.86 (6), 1437–1448. 10.1021/acs.jnatprod.3c00114
10
Li W. Huang C. Li S. Ma F. Li Q. Asada Y. et al (2012). Clerodane diterpenoids from Tinospora sagittata (oliv.) gagnep. Planta Med.78 (1), 82–85. 10.1055/s-0031-1280163
11
Liang Y. Guo S. B. Guo Y. H. Xu C. Wang G. Y. Zhang W. et al (2024). A pair of new enantiomeric alkaloids from Tinospora sinensis. Phytochem. Lett.60, 179–183. 10.1016/j.phytol.2024.02.009
12
Mantaj J. Rahman S. M. A. Bokshi B. Hasan C. M. Jackson P. J. M. Parsons R. B. et al (2015). Crispene E, a cis-clerodane diterpene inhibits STAT3 dimerization in breast cancer cells. Org. Biomol. Chem.13 (13), 3882–3886. 10.1039/c5ob00052a
13
Mishra A. Sharma K. Pandey J. Dev K. Kadan S. Sahai M. et al (2023). Tinosporaside from Tinospora cordifolia encourages skeletal muscle glucose transport through both PI-3-Kinase- and AMPK-dependent mechanisms. Molecules28 (2), 483. 10.3390/molecules28020483
14
Nishidono Y. Tanaka K. (2024). Structural revision of tinotufolins from Tinospora crispa leaves guided by empirical rules and DFT calculations. J. Nat. Prod.87 (4), 774–782. 10.1021/acs.jnatprod.3c00902
15
Royani A. Hanafi M. Lotulung P. D. N. Prastya M. E. Verma C. Manaf A. et al (2023). Isolation and identification of bioactive compounds from Tinospora cordifolia stem extracts as antibacterial materials in seawater environments. Arab. J. Chem.16 (9), 105014. 10.1016/j.arabjc.2023.105014
16
Song J.-Q. Deng L. Zhu Y.-L. Zhou H. Kong X.-Q. Zhang L.-J. et al (2024a). Clerodane diterpenoids with in-vitro anti-neuroinflammatory activity from the tuberous root of Tinospora sagittata (Menispermaceae). Phytochemistry218, 113932. 10.1016/j.phytochem.2023.113932
17
Song Y.-Z. Yuan C. Li W.-J. Song Q.-J. Zhu W.-H. Zhang J. (2024b). New clerodane diterpenoids from Tinospora capillipes with antibacterial and anti-inflammatory properties. Chem. Biodivers.21, e202401033. 10.1002/cbdv.202401033
18
Zhang G. Ma H. Hu S. Xu H. Yang B. Yang Q. et al (2016). Clerodane-type diterpenoids from tuberous roots of Tinospora sagittata (Oliv.) Gagnep. Fitoterapia110, 59–65. 10.1016/j.fitote.2016.02.012
19
Zhang J.-S. Hu Y. Song K.-S. Wu F. Zhu K. Xu D.-F. et al (2021a). Diterpenoid glucosides with cystathionine γ-lyase inhibitory activity from Tinospora sinensis. Bioorg. Chem.116, 105400. 10.1016/j.bioorg.2021.105400
20
Zhang J. S. Xu D. F. Cao X. X. Wang Y. Y. Zhang H. (2021b). Lignans with NO inhibitory activity from Tinospora sinensis. Chin. J. Nat. Med.19 (7), 500–504. 10.1016/S1875-5364(21)60049-3
21
Zhang J.-S. Xu D.-F. Wang Y.-Y. Ma R.-F. Zhang H. (2022). Clerodane furanoditerpenoids from the stems of Tinospora sinensis. Arch. Pharm. Res.45 (5), 328–339. 10.1007/s12272-022-01383-5
22
Zhu Y.-L. Deng L. Dai X.-Y. Song J.-Q. Zhu Y. Liu T. et al (2023). Tinopanoids K-T, clerodane diterpenoids with anti-inflammatory activity from Tinospora crispa. Bioorg. Chem.140, 106812. 10.1016/j.bioorg.2023.106812
23
Zhu Y.-L. Deng L. Song J.-Q. Zhu Y. Yuan R.-W. Fan X.-Z. et al (2022). Clerodane diterpenoids with anti-inflammatory and synergistic antibacterial activities from Tinospora crispa. Org. Chem. Front.9 (24), 6945–6957. 10.1039/d2qo01437h
Summary
Keywords
clerodane, diterpenoid, Paratinospora sagittata , Tinospora sagittata , anti-inflammatory activity
Citation
Li H, Liu J, Fang C and Ning S (2025) Clerodane diterpenoids with anti-inflammatory activity from the tuberous root of Paratinospora sagittata (Oliv.) Wei Wang. Front. Pharmacol. 16:1561954. doi: 10.3389/fphar.2025.1561954
Received
17 January 2025
Accepted
10 March 2025
Published
20 March 2025
Volume
16 - 2025
Edited by
Yan-Hui Fu, Hainan Normal University, China
Reviewed by
Xu-Dong Zhou, Hunan University of Chinese Medicine, China
Zhixin Wang, China Academy of Chinese Medical Sciences, China
Chuan Li, Hainan University, China
Updates
Copyright
© 2025 Li, Liu, Fang and Ning.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shanglei Ning, kingning2010@hotmail.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.