- 1Department of Biosciences and Bioengineering, Indian Institute of Technology Jodhpur, Jodhpur, Rajasthan, India
- 2Department of Biochemistry, University of Delhi South Campus, New Delhi, India
- 3Department of Pharmacy, Birla Institute of Technology and Science Pilani, Pilani, Rajasthan, India
- 4Department of Neurochemistry, National Institute of Mental Health and Neuro Sciences, Institute of National Importance, Bangalore, Karnataka, India
- 5Institute of Nano Science and Technology, Mohali, Punjab, India
Autophagy is a degradative process that makes rapid turnover of old and impaired proteins and organelles possible. It is highly instigated by stress signals, like starvation, and contributes to the cell’s homeostasis. Autophagy performs a crucial function in keeping cell genomic integrity stable. Impaired autophagic flux is implicated in neurodegenerative diseases, abnormal ageing, and cancerous diseases. In diseases like cancer, autophagy performs a dualistic function; it can have both a tumor-suppressive and supportive role. Autophagy in the initial phases of tumorigenesis maintains the integrity of the genome and, if it fails, leads to cell death, thus having a tumor-suppressive role. Meanwhile, autophagy also imparts the function of the pro-survival mechanism in the latter stages of tumorigenesis and supports the cancerous cells in surviving conditions like hypoxia and increased oxidative stress. Autophagy also helps cancerous cells develop drug resistance in some cases. Thus, modulation of the autophagic mechanism is a possible therapeutic strategy in cancer therapy as its inhibition can sensitise cancer cells to anti-cancerous drugs. The promotion of autophagy, in some cases, can also safeguard cells from toxic protein aggregation and enhanced oxidative stress. Excessive autophagy can result in autophagic cell death. Autophagy also regulates several cellular processes and cell death pathways, like apoptosis. Therefore, an in-depth knowledge of the autophagy process and its regulating molecules is critically important. Pharmaceutical small molecules or cellular target modulation can help modulate the cellular autophagy process in the context of specific disease conditions.
1 Introduction
The term “autophagy” translates to “eating of oneself”, Christian de Duve in 1963 proposed the term, describing the presence of membraned (single or double) vesicles containing cytoplasmic contents and organelles in different stages of degradation (De Duve and Wattiaux, 1966). Some evidence suggests that the term autophagy was well used before 1963. A French physiologist named M. Anselmier used the term in the 1860s to refer to a phenomenon of a body’s self-nourishment system, which offers a person nourishment in extreme starvation by feeding on oneself (Ktistakis, 2017). Autophagy is a conserved and regulated process in cells of eukaryotes implicated in the lysosome-dependent recycling and degradation of cellular components impaired or senescent (Klionsky and Emr, 2000; Levine and Klionsky, 2004). Studies of autophagy in earlier days were based on morphological observations; therefore, the observations made were of later stages of autophagy before or after the lysosomal fusion with autophagosome. Using radioactive probes helped illustrate the earlier and intermediatory phases of autophagy (Gordon et al., 1988). Autophagy is a bulk degradative mechanism where constituents from cytoplasm are sequestered into an autophagosome (double-membraned) and transferred to lysosomal lumen for clearance (Yoshimori et al., 2004). As research on autophagy advanced, the selective forms of autophagy were also reported, where specific organelles (ER and mitochondria) or proteins are targeted for clearance (Bolender and Weibel, 1973; Lemasters et al., 1998; Beaulaton and Lockshin, 1977).
Besides cytoplasmic components, extracellular material and portions of the plasma membrane are also delivered to lysosomes through endocytosis (Tooze et al., 2014). The large and hydrophilic molecular cargoes need to be transported into the lumen of the lysosome via membrane barriers. Different forms of autophagy were observed that deliver these cargoes into the lysosomal lumen via membrane barriers. The macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA) are major autophagy forms that have been well studied (Yang and Klionsky, 2010). In macroautophagy and microautophagy, the inner autophagosomal layer and intra-luminal-vesicle layer, respectively, cause the cargo to be exposed to lysosomal luminal hydrolysing enzymes, but in CMA, the substrate is directly translocated into the lumen through putative pores (Mizushima et al., 2008). The studies in the field of autophagy up to the 1990s were focused on biochemical, morphological, and physiological aspects of autophagy; the advancement in autophagy research then elucidated molecular elements of autophagy. Autophagy-related genes (ATGs) that are implicated in modulation and proper functioning of autophagy were identified (Tsukada and Ohsumi, 1993; Klionsky et al., 2003). More than 40 ATG genes are reported so far, of which 16 are core ATG genes. These genes are conserved and crucial for macroautophagy and other forms of autophagy (Furukawa et al., 2024; Levine and Kroemer, 2019).
The modulation of autophagy is controlled by various signalling pathways and molecular regulators (He and Klionsky, 2009). It is well established that this includes one such negative regulator of autophagy, especially in circumstances of nutrient abundance, referred to as mTOR pathway, having been shown to phosphorylate, thus inactivating the ULK1 complex. ULK1 is the core complex crucial for autophagosome formation. Either nutrient starvation or stress decreases the activity of mTOR, initiating autophagy (Saxton and Sabatini, 2017). The physiological pathway of activation of autophagy through AMPK signalling necessitates instigation of ULK1 complex and a reduced state of mTOR activity, which are generated by reduced cellular energy levels (Alers et al., 2012). This complex, class III phosphatidylinositol 3-kinase (PI3-K), comprises proteins Beclin-1, ATG14L, and VPS34; it participates in the production of autophagosomes that are further mediated by other proteins, including Bcl-2. Bcl-2 can interact with Beclin-1 and functions as a negative modulator for autophagy (McKnight and Yue, 2013; Kang et al., 2011; Funderburk et al., 2010). In addition, transcriptional mechanisms that involve transcription factors such as TFEB, FOXO, and p53 affect levels of many autophagy-associated genes in response to various stress stimuli (Di Malta et al., 2019).
Furthermore, under intracellular nutrient starvation conditions, autophagy is a significant source of intracellular amino acids, lipids, and other key molecules that result from breakdown of cellular constituents. This is a key process for upkeep of the cell’s energy supply and for the continuation of cellular function (He, 2022). It is absolutely crucial for clearing impaired organelles, misfolded proteins, and potentially harmful cellular debris (Gómez-Virgilio et al., 2022). Autophagy holds a substantial part in processes of embryogenesis and differentiation and in elimination of surplus cellular components to reorganize cellular structures (Tettamanti et al., 2019). Moreover, autophagy interacts with immune system by involving in the clearance of intracellular pathogens, antigen presentation to immune cells, and also in inflammatory responses (Deretic et al., 2013). Apart from its degradative functions, autophagy and its associated molecules are engaged in a whole array of other physiological activities (Wang and Qin, 2013). The proper physiological function of autophagy is necessary to upkeep cell’s homeostasis and to respond to stressors (Gómez-Virgilio et al., 2022). Many physiological and pathological disease states are related to autophagy, including dysfunctional ageing, neurodegenerative diseases, infections, and cancer (Ortega et al., 2024).
The impaired autophagy mechanism is reported in numerous disorders and pathological conditions (Khandia et al., 2019). There are reports present that suggest a positive role of autophagy in cellular proliferation (Wang et al., 2020). Improper or excessive cellular proliferation is a hallmark of cancer (Holland, 2010). There are reports present suggesting that autophagy downregulation in cancer may inhibit proliferation (Wei et al., 2011; Sheng et al., 2018). The mechanistic details of autophagy in regulating cellular proliferation are not fully elucidated, although there is some evidence present suggesting some cell cycle proteins can modulate autophagy and vice versa (Zheng et al., 2019). This complex autophagy role is also observed based on the different types and stages of tumors; it may exhibit tumor suppressor characteristics or promote survival of tumors (Bhutia et al., 2013). Therefore, though autophagy eliminates the damaged organelles and proteins that may promote tumorigenesis, tumor cells can use autophagy to sustain stressors like nutrient starvation (Mathew et al., 2007). Impaired autophagy flux is reported in various neuronal degenerative disorders like Alzheimer’s, Huntington’s, and Parkinson’s, where failure of autophagy causes inadequate clearance of misfolded polypeptides and impaired organelles in neurons and leads to neuronal dysfunction and degeneration (Guo et al., 2018). Metabolic diseases are also linked with autophagy impairment, such as obesity, diabetes, and liver diseases, as autophagy coordinates lipid metabolism, insulin sensitivity, and mitochondrial functions (Jakubek et al., 2024).
2 Overview of the autophagy process
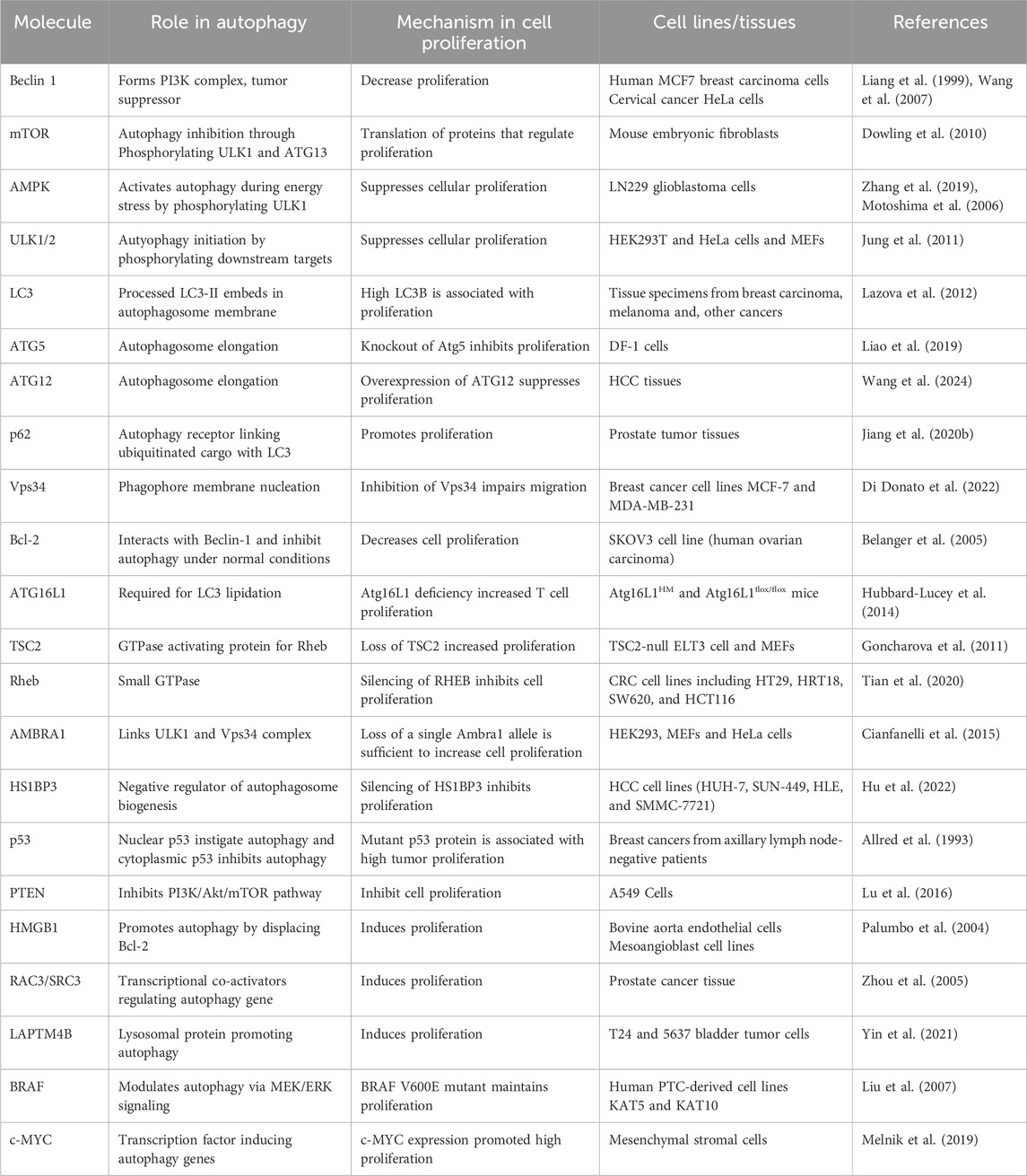
As discussed above, autophagy is a very well-modulated mechanism strictly conserved from yeast to humans (Delorme-Axford and Klionsky, 2018). It has received much attention due to its roles in several physiological processes and its implications in many disorders, like cancer (Ryter et al., 2013). Well-characterised form of autophagy is macroautophagy, which utilises vesicles called autophagosomes (double-membrane) to sequester cytoplasmic contents like impaired organelles, misfolded proteins and other cellular debris (Levine and Kroemer, 2008). Macroautophagy is regulated mainly through some complexes and systems. The PI3K complex, the ULK1 complex, and the ATG8 system are among them. Specific sensor molecules like mTOR and AMPK modulate the processes of autophagy. The PI3K complex is essential in autophagy initiation as it is involved in forming the autophagosome. The ULK1 complex (serine/threonine kinase) is instigated upon stress like starvation and works downstream of the mTOR pathway, further facilitating onset of autophagy by phosphorylation of proteins related to autophagy. After the phagophore is formed, elongation and closing of autophagosome membrane are facilitated by the ATG8 system, which consists of LC3 and GABARAPs. Overall, these complexes and molecules form the core machinery, which is crucial for coordinating the autophagy process to ensure the preservation of homeostasis and proper stress response (Feng et al., 2014). After the formation of the autophagosome, it sequesters the cytoplasmic materials and matures; after that, autophagosome is fused with lysosome, initiating the clearance of contents in presence of hydrolysing enzymes. This is vital for degrading damaged organelles, proteins, and cellular health Figure 1 (Wang et al., 2003).
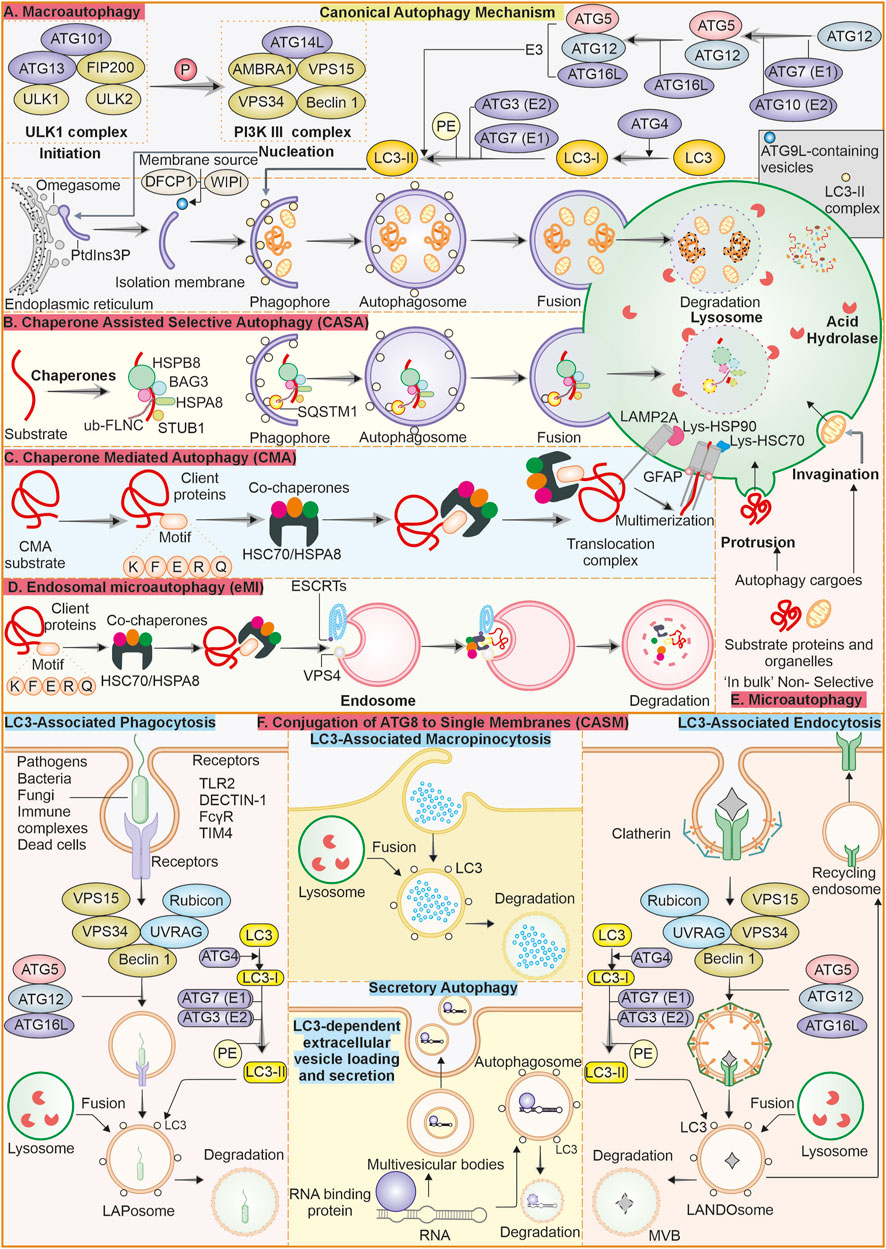
Figure 1. Overview of the autophagy process and its various types: (A) Macroautophagy: Macroautophagy is a well-described canonical form of the bulk degradation process. It initiates with forming of a phagophore, that closes and forms an autophagosome, which further fuses with the lysosome to clear its contents through degradation. (B) Chaperone-assisted selective autophagy (CASA): Selective autophagy where chaperones like HSPB8, BAG3 and HSPA8 identify the cargo and sequester into autophagosome and later fuse with lysosome for degradation. (C) Chaperone-mediated autophagy (CMA): This form of autophagy, with HSPA8 chaperone and co-chaperones selectively targets the substrate having a KFERQ motif, and this system transfers the substrate to the lysosome lumen for degradation through LAMP2A. (D) Endosomal microautophagy (eMI): It is similar to CMA and depends on chaperones and co-chaperones to selectively identify the cargo; after that, it enters the late endosome for degradation instead of the autophagosome. (E) Microautophagy: This form of autophagy does not require significant machinery. Here, the lysosomal membrane either invaginates or protrudes to engulf the cargo and transfer it to the lumen, where hydrolyzing enzymes are present. (F) Conjugation of ATG8 to single membranes (CASM): ATG8 family proteins, when conjugated to single membrane vesicles and internalize the cargo, is collectively called CASM. There are different forms of these non-canonical autophagy mechanisms like LC3-associated phagocytosis, LC3-associated macropinocytosis and LC3-associated endocytosis. There is one secretory form of autophagy called LC3-dependent extracellular vesicle loading and secretion, which is involved in the packaging and export of RNA and proteins.
In contrast, microautophagy is defined by the protrusion or invagination of lysosomes membrane and engulfing of substarte inside lumen (Mijaljica et al., 2012; Wang et al., 2023). Lysosomes’ direct uptake of cellular contents represents a central factor ensuring cell homeostasis at basal conditions. Endosomal microautophagy (eMI) is a unique autophagy type characterized by late endosomes direct internalisation of cytoplasmic components by membrane’s invaginations, thus eliminating the use of autophagosomes (Olsvik et al., 2019). This mechanism is vital for cellular homeostasis because it selectively degrades damaged proteins, organelles, and other cytoplasmic components. It, therefore, regulates many cellular processes, such as immune response and cell signalling. eMI ensures efficient recycling of cellular components and maintains cellular health and functionality. eMI can be bulk or selective as it relies on interaction of hsc70 with KFERQ-like motif present in substrate to translocate them to the LE/MVB membrane by interaction of phosphatidylserine. The machinery for ESCRT consists of Vps4 and Tsg101, which aids in the internalization of substrates into ILV. The substrates may get degraded within the LE/MVB or after the LE/MVB fuses with the lysosome (Morozova et al., 2016; Sahu et al., 2011).
The selective type of autophagy is CMA, where specific proteins are targeted for degradation (Dice, 2007). This process includes the chaperone Hsc70’s identification of motif KFERQ-like sequence in targeted protein. Targeted protein is translocated across lysosome’s membrane by receptor lysosome-associated membrane protein 2A (LAMP-2A), where it is degraded (Majeski et al., 2004). There is another autophagy type called Chaperone-assisted selective autophagy (CASA), which is a process that focuses on selective degradation of damaged and misfolded proteins. The core CASA complex consists of three central proteins: HSPA8, HSPB8, and BAG3, all of which are crucial for maintaining proteostasis in neurons and muscle cells. Mutations in such genes lead to diseases ranging from (cardio) myopathies to neurodegenerative diseases (Tedesco et al., 2023).
3 Non-canonical autophagy
In macroautophagy, the double-membraned autophagosome formation and maturation depend on conjugation of ATG8 proteins like GABARAPs and LC3s to phosphatidylethanolamine (PE) (Ichimura et al., 2000; Johansen and Lamark, 2020). Various studies have also reported that ATG8 has autophagy-independent functions (Galluzzi and Green, 2019; Nieto-Torres et al., 2021). It is also noted that ATG8 can be recruited by phosphatidylserine (PS) into single membrane vesicles, termed Conjugation of ATG8 to single membranes (CASM) (Durgan et al., 2021). The studies have suggested that parallel functions of ATG8 proteins leading to either cargo degradation or secretion are called non-canonical autophagy (Nieto-Torres et al., 2021; Codogno et al., 2012). LC3-associated phagocytosis (LAP) combine phagocytosis with LC3 protein recruitment, thus facilitating the LAPosomes fusion with lysosomes to improve the clearance and removal of pathogens within LAPosomes. It differs from xenophagy, in which intracellular pathogens, as well as other foreign material, are targeted for degradation (Herb et al., 2020). Both LAP and xenophagy utilize the same LC3 conjugation machinery, including ATG3, ATG7, and an ATG12-ATG16L1-ATG5 complex. However, the mechanism of induction is different; generation of PI(3)P on membranes, involvement of ROS, and conjugation of LC3 to membranes (single) are all different features of LAP. Some components of the PI3K complex are shared between LAP and autophagy, but recruitment of LC3 requires Rubicon and UVRAG proteins (Heckmann and Green, 2019).
LC3-associated endocytosis, or LANDO, is initiated by the identification of substrate through TREM2 and TLR (cell surface receptors), consequently internalizing clathrin-mediated endosomes. Process of LC3 recruitment to LANDOsome is parallel to LAP. Unlike LAP, LANDO is characterized by multiple endpoints that include fusion of the LANDOsomes with the lysosome, clearing of the ligand, and the cell surface recptor is recycled back to membrane (Heckmann et al., 2019). LC3-dependent extracellular vesicle loading and secretion (LDELS) is a type of secretory autophagy. It is reported as LC3 conjugation machinery mediating incorporation of cargo into the secretory vesicles that are exported outside the cell. Reports have shown that RNA-binding proteins are major cargos for this process, affecting non-coding RNAs (ncRNAs) and snoRNA secretion. This pathway requires ATG proteins and Rab27a to prevent the accumulation of receptors for autophagic cargo (Leidal et al., 2020; Eng et al., 2021). Cells react to damage of the plasma membrane by instigating macropinocytosis-related proteins at the injury site. Following sealing, they produce large macropinosomes at the repair site, which are marked with LC3B protein. The process is reliant on Rubicon, ATG7, and p62. Shrinking of the internalized macropinosome by osmotic draining and fusion with lysosomes. This is reported as a form of macropinocytosis, LC3-associated macropinocytosis (LAM), which helps remove damaged materials and restore membrane integrity (Sønder et al., 2021).
4 Autophagy implicated in diseases associated with cellular proliferation
The process of cellular proliferation is crucial for cell growth and division, as well as tissue repair and development. Recently, there have been several reports suggesting roles of autophagy in tissue repair and regeneration (Moreno-Blas et al., 2025). Excessive or improper cellular proliferation may become a risk factor for several diseases, ranging from cancer to inflammatory diseases. Mutations in oncogenes and tumor suppressor genes create an imbalance between cell death and cell division, resulting in excessive cellular proliferation (Dakal et al., 2024). RAS and MYC are oncogenes, and certain muations in these genes result in promotion of prolonged proliferation in absence of any external stimuli. In RAS-driven pancreatic, colorectal and lung cancers, autophagy helps in maintaining tumor growth by metabolic reprogramming and maintaining mitochondrial integrity, as can be observed in autophagy-deficient RAS mutant cells accumulating impaired mitochondria and impaired energy production. ATG7 deletion in BRAF V600E-driven lung tumors, mitochondrial impairment, along with metabolic dysfunction is observed (Ariosa et al., 2021; Mathiassen et al., 2017). In murine models with ATG16L1 (keratinocyte-specific), deletion exacerbated the skin inflammation and tumorigenesis. The deficiency of autophagy in these keratinocytes sensitizes them towards TNF-dependent apoptosis, altering hair follicle stem cell activation and wound healing (Van Hove et al., 2023). In liver fibrosis, the activation of hepatic stellate cells (HSC) is necessary. Autophagy in HSC instigates their transdifferentiation into myofibroblasts (collagen secreting) by degrading lipid droplets. Autophagy inhibition reduced the deposition of collagen in mouse models (Chen et al., 2025; Lucantoni et al., 2021). In chronic kidney diseases role of autophagy is observed where in renal fibroblasts, autophagy drives extracellular matrix production through TGF-β/SMAD4 signalling (Ruby et al., 2023 Lucantoni et al., 2021). The autophagy deficiency also impairs the clearance of bacteria, which exacerbates inflammation, whereas TNF-α inhibition may restore autophagy, resulting in reduced keratinocyte proliferation. CD4+ T cells in rheumatoid arthritis show increased autophagy, which supports their survival and inflammatory activities. In collagen-induced models, ATG5 knockout reduced the T cell proliferation and alleviated arthritis (Lin et al., 2024; Zhao et al., 2024).
Autophagy is implicated in various other cellular processes, and thus, its role in other disease models also varies. Some of the disease models that are extensively researched are cardiovascular diseases, neurodegenerative disorders, metabolic diseases, infectious diseases, cancer and congenital disorders. In models of cardiovascular diseases, ATG5/ATG7 knockout mice pressure overload (TAC) was utilised, showing dual role of autophagy where at basal level, autophagy maintains homeostasis and dysregulation causes exacerbated heart failure (Schiattarella et al., 2016; Nishida et al., 2009). Autophagy is involved with maintaining the cardiomyocyte health during the stress conditions but becomes detrimental when it is dysregulated. During myocardial ischemia, autophagy is reported to clear the damaged organelles and mitigate ROS production and apoptosis (Jiang et al., 2023). Pressure overload model studies showed that autophagy clears the polyubiquitinated proteins in cardiomyocytes, which results in prevention of accumulation of toxic aggregates. Mutant α-B-crystallin in desmin-associated cardiomyopathy disrupts the proteostasis, autophagic clearance of aggregates delays heart failure (Hill, 2011). Models of neuronal degenerative disorders studies in SENDA patients and a Pink1/Prkn Knockout mouse model of Parkinson’s autophagy were observed to prevent protein aggregation, and the impairment of autophagy caused neuronal death (Kingwell, 2013; Klionsky et al., 2021). Autophagy is also indicated to be disturbed in metabolic disorders like type 2 diabetes, as it has a protective role on β-cell functioning and insulin sensitivity during stress conditions. Studies on ATG7 haploinsufficient mice model reported an exacerbated intolerance towards glucose and hepatic steatosis under high-fat diet. This observation associates deficiency of autophagy to the progression of diabetes (Park et al., 2022). In congenital disorders, mutation in autophagy-related genes may cause multi-system disorders. For instance, in Vici syndrome, the EPG5 mutation disrupts autophagosome and lysosome fusion, resulting in cardiomyopathy and neurological disorders (Dafsari et al., 2025). WDR45 defects may impair the autophagy in neuronal cells causing SENDA to have Parkinsonian features (Kingwell, 2013). In infectious diseases, like HIV infection, autophagy is reported to clear the viral components; furthermore, it is also subverted by virus-promoting persistence. The HIV-1 Tat protein reduces the maturation of autophagosome while the Nef also impairs autophagy, which further enables viral replication (Lamsira et al., 2025). A list of autophagy related molecules and their roles in cellular proliferation is presented in Table 1.
5 Autophagy functioning in tumor suppression
Autophagy plays several roles in the mechanisms involved in both suppression of cancer initiation and progression. Among its more significant functions in the context of preventing cancer, the ability to upkeep cell’s homeostasis in stressful conditions is essential (Vitto et al., 2022). The degradation and recycling of impaired organelles and polypeptides help in avoiding build-up of damages that precede instability of genome as a potential causative agent in cancer (Miller and Thorburn, 2021; Vessoni et al., 2013). Autophagy promotes cell survival in adverse conditions, including nutrient starvation, hypoxia, and oxidative stress, which contribute to uncontrolled tumorigenicity (Mizushima et al., 2008; Rakesh et al., 2022; Lee et al., 2012). Autophagy clears potentially oncogenic material and maintains cellular homeostasis; autophagy in the initial stages of tumorigenesis guards against cancer initiation (Patergnani et al., 2021). Autophagy is also said to regulate cell death pathways by ensuring that apoptosis is controlled and prevents the uncontrolled proliferation of cells, a characteristic of cancer (Verma et al., 2021; Liu et al., 2023). Enhanced excessive autophagy can cause autophagic cell death when the cells get extensively damaged or under stress. Cell death via autophagy removes damaged or potentially malignant cells that should not contribute to accumulating additional mutations leading to cancer cells (Kanzawa et al., 2005). Functional autophagy in cellular quality control is such that only healthy cells are maintained to restrain the possibility of carcinogenesis Figure 2 (Yang et al., 2011; Linder and Kögel, 2019).
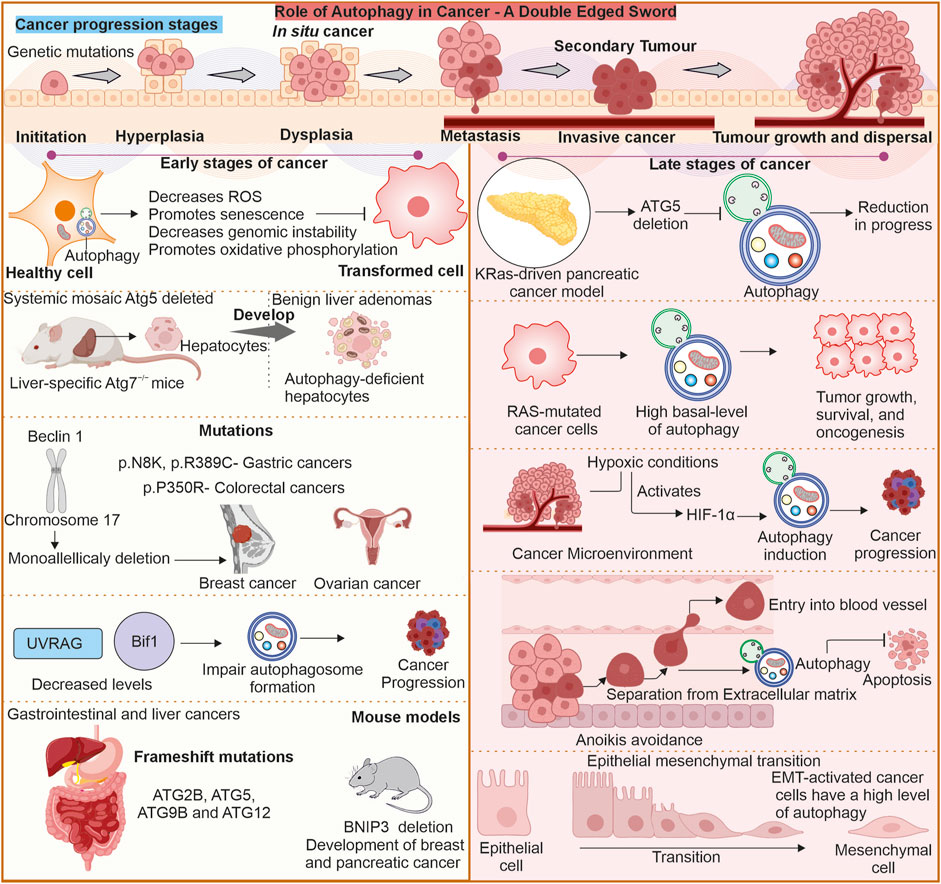
Figure 2. Role of autophagy in tumor proliferation a double-edged sword: In cancer autophagy can play dualistic role. It can have anti-proliferative role in early stages of cancer progression. The deletion or mutation of various ATG genes can support spontaneous tumor generation suggesting their anti-proliferative role. Similarly, once tumor is established autophagy supports the tumor cells by providing protection against stress conditions like hypoxia.
The inhibition or downregulation of autophagy may promote tumorigenesis. Some observations have been reported where this association between autophagy and tumorigenesis is established (Takahashi et al., 2007). The ATG knockout models show role of ATGs and Beclin-1, as well as autophagy inhibition, in promoting a higher incidence of tumorigenesis. Tumor cells, under stressful conditions, can escape primary tumor through metastasis and invasion. In primary tumor autophagy is associated with tumor cell necrosis and inflammation inhibition (Kenific et al., 2010). Autophagy is also involved in suppression of epithelial-to-mesenchymal transition (EMT) by degrading p62 and TWIST1, an EMT transcription factor (Qiang et al., 2023). Autophagy has a dualistic role in cancer, and thus it can either enhance or reduce the invasion and metastasis of tumor cells (Lisanti et al., 2010). Initially, autophagy enhances cancer cell survival by reducing necrosis and also reduces the infiltration of macrophages (Mowers et al., 2018; DeNardo et al., 2009; Bingle et al., 2002; Xia et al., 2020). Autophagy can also enhance cancer cell invasion and migration (Sharifi et al., 2016). Chaperone-mediated autophagy is also associated with tumor cell metastasis, as CMA inhibition results in reduced metastasis via the downregulation of migration and anoikis resistance (Kon et al., 2011). In breast cancer, there is an association between CMA and metastasis (Han et al., 2017). CMA is reported to clear HSD17B4 protein implicated in cellular invasion and migration (Zhang et al., 2017). Downregulated CMA by LAMP2A modulation may reduce or inhibit the tumor growth (Han et al., 2017).
There are evidence present that suggest that in many cases of cancer, the autophagy process is compromised. The earlier evidence came from studies on hemizygous mouse models for Beclin-1, which are prone to tumor development. Human BECN1 gene encodes for Beclin-1 protein, which is needed for autophagy, is located on chromosome 17q21, with monoallelic deletions reported in breast malignancies and ovarian and prostate cancers (Aita et al., 1999). The experiment on breast cancer cells also elucidated a frequent loss in Beclin-1 (Aita et al., 1999; Liang et al., 1999; Yue et al., 2003). Apart from this, the deletion of ATG7 can trigger tumorigenesis in the liver (Takamura et al., 2011). Autophagy loss in the liver may promote the generation of hepatocyte-derived progenitor cells involved in tumor initiation (Barthet et al., 2021). This evidence suggests that impairment of autophagy may help establish the tumor cells. Nevertheless, it is still unknown whether the suppression of autophagy is responsible for tumorigenesis or provides the microenvironment for tumorigenesis. Tumor suppressor transcription factors like p53 also interact with autophagy pathways. The autophagy modulation by p53 can be either negative or positive. p53, which induces cell cycle arrest and apoptosis after detecting damage to DNA, can increase autophagy (Rahman et al., 2022). At the basal level, p53, when in cytoplasm, suppresses autophagy (Tasdemir et al., 2008).
The point mutations in Beclin-1 are rare, and some mutations like p. N8K, p. R389C in gastric and p. P350R colorectal cancer are observed (Lee et al., 2007). Bif-1, or Endophilin B1, interlates with Beclin 1 through UVRAG as well as is a positive modulator of PI3KC3. In conditions like starvation, Bif-1 localises to autophagosomes in response, colocalising with ATG5 and LC3. Moreover, the absence of Bif-1 inhibits autophagosome production. It is reported that the knockout models of Bif-1 can increase the probability of spontaneous tumor generation (Takahashi et al., 2007). Similarly, UVRAG also imparts an essential function in suppressing tumorigenesis. A frameshift mutation in UVRAG is reported in cancer conditions expressing UVRAGFS. The Mice model with UVRAGFS was reported to have spontaneous tumorigenesis (Quach et al., 2019). Autophagy-related gene 4 (ATG4) protein, a crucial protein for autophagy process which, is directly implicated in the forming of autophagosomes. The ATG4 protein imparts a very important role in processing and lipidating LC3, which is necessary for the proper maturation of autophagosomes (Satoo et al., 2009). Recently, an important connection between ATG4B and the progression of cancer has been observed. More and more evidence indicates the overexpression of ATG4B in various cancer types, which makes it a prospective candidate for anticancer therapeutic strategies (Fu et al., 2019). Some reports also suggest that frameshift mutations in ATG genes cause cancer of colorectal and gastrointestinal (Kang et al., 2009). Other studies suggest that deletion of BNIP3 can cause breast cancer (Chourasia et al., 2015).
6 Role of autophagy in tumor promotion
The autophagy process is also reported to promote the established tumor’s progression and survival. The reported increased LC3 puncta levels and autophagosome accumulation in some tumor tissue suggest the increased autophagy supporting tumorigenesis (Fujii et al., 2008). Increased autophagy in cancer cells may support adaptation to harsh microenvironment of an established tumor characterised by nutrient scarcity, hypoxia, and oxidative stress (Yang et al., 2015). Autophagy delivers intracellular building blocks from degrading and recycling the components inside the cell, forming a principal source requisite for cell survival and proliferation. Thus, it helps sustain cancer cells’ metabolic needs in stress conditions (Karantza-Wadsworth et al., 2007; Degenhardt et al., 2006). In the hypoxic regions of tumors, autophagy helps remove damaged mitochondria via mitophagy, decreasing ROS quantity and preventing cell death (Zaarour et al., 2021). Consequently, this survival mechanism allows cancerous cells to sustain in microenvironment and promote tumor development (Poillet-Perez et al., 2015).
The autophagy involvement in cancer cell proliferation has been highlighted by multiple studies. In RAS-mutant cancer cases, autophagy is essential for providing glutamine by degrading various cellular components. This glutamine is converted to α-ketoglutarate through glutaminase (Vitto et al., 2022; Chavez-Dominguez et al., 2020). Mitophagy prevents the accumulation of ROS, thus maintaining the mitochondrial health required for proliferation in pancreatic ductal adenocarcinoma (Vitto et al., 2022; Mowers et al., 2018). Autophagy is also involved in anoikis resistance. During the metastasis phase, detached cancer cells evade anoikis, which is detachment-induced apoptosis caused by the degradation of pro-apoptotic signals by autophagy (Tonkin-Reeves et al., 2023; Sui et al., 2013). Apart from supporting cancer cell survival and metastatic spread, autophagy also increases their motility. Autophagy clears proteins like paxillin through LC3 interaction. Paxillin is a focal adhesion protein, enabling disassembly of cell matrix adhesion which further facilitates migration in breast cancer and melanoma (Mowers et al., 2018). Similarly, in TGF-β-induced autophagy instigates EMT in non-small cell lung carcinoma, further enhancing invasive potential (Hwang et al., 2022). Autophagy also helps in evading immune surveillance as it reduces MHC-I expression on tumor cells, which results in impaired CD8+ T cells recognition (Bustos et al., 2020).
In addition, autophagy is a significant aspect that enhances the survival of cancer stem cells (CSCs). CSCs have capabilities of differentiation and self-renewal, forming a subpopulation of cells. These cells are associated with recurrence, metastasis of tumors, and resistance of cancers to primary therapies. Autophagy supports CSC metabolic modulation by providing them with all the required nutrients and energy for survival and proliferation. Autophagy through the maintenance of stemness and survival of CSC contributes to tumor heterogeneity and the aggressive nature of the cancers (Rahman et al., 2024; Wang et al., 2022). Studies on mice have given proof of complex functioning of autophagy in cancer development. Reports show that mice with an ATG5 gene mosaic deletion develop non-cancerous liver adenomas, which do not form malignant tumors (Takamura et al., 2011). The observations proposed that autophagy probably has a key part in modulating formation of tumors at early stages, probably as a process that limits malignant transformation. Reports also imply that mice genetically modified to express oncogenic KRas within lung tissue, in combination with the deletion of ATG5 or ATG7 genes, demonstrate an elevated incidence of benign tumors. Significantly, these tumors do not result to malignant forms, further highlighting complex regulatory role of autophagy in advancing tumor (Guo et al., 2013; Rao et al., 2014).
Studies have identified a crucial observation that cells with Ras mutations are highly reliant on autophagic process for growth and survival. Specifically, numerous human cancer cell lines that carry activating Ras mutations display constitutive high levels of autophagy in vitro, suggesting metabolic adaptation (Guo et al., 2011). Autophagy dependence would be a promising therapeutic concept that arises from these studies. Autophagy and mitochondrial metabolism would then be targeted to develop novel therapeutic approaches against such aggressively behaving cancer types (Guo et al., 2011). Some reports suggest that in fibroblast cells, the activated HIF1α promotes autophagy, which further can help survive the fibroblast, and increased tumor mass and volume are also observed. Activated HIF1α also instigated expression of BNIP3L and BNIP3. Interestingly, the activated HIF1α shows a reduction of tumor mass and volume in breast cancer cells. Thus, activation of HIF1α may promote or suppress tumorigenesis based on cancer type (Chiavarina et al., 2010). A significant characteristic of metastatic cancer cells is their resistance to anoikis, a kind of apoptosis linked to inadequate adhesion to the extracellular matrix (ECM) (Taddei et al., 2012). PI3K/Akt and Ras/MAPK, growth factor pathways, are instigated in some cancer cells to evade anoikis. While autophagy-mediated cell death was first linked to anoikis, studies also corroborate the protective role of autophagy in this process (Kenific et al., 2010).
A process that is somewhat parallel to developmental process of embryo, where alteration in cellular morphologies characterized as EMT is found to impart a function in tumorigenic process (Brabletz et al., 2018). EMT is important in tumor invasion and metastasis because it allows epithelial cells to become mesenchymal cells, which increases motility as well as the ability to spread to other organs (Ribatti et al., 2020). It is also associated with the development of chemoresistance towards anticancer drugs (Zheng et al., 2015). A complicated relation exists between EMT and autophagy, as autophagy can play a dualistic role (Rojas-Sanchez et al., 2019). Reports suggest that autophagy can promote EMT in certain conditions; like in breast cancer cells, autophagy can promote EMT by instigating the TGF-β/Smad signalling pathway (Hwang et al., 2022; Hao et al., 2019). Thus, it is observed that alongside the EMT transition, elevated autophagy flux is also reported (Singla et al., 2017). Therefore, targeting autophagy inhibition can result in the blocking of EMT (Colella et al., 2019).
Given that autophagy is observed to impart a dualistic function in cancer, therapeutic targeting should be carefully designed and context-specific (Kumar et al., 2022). Enhancing autophagy at early stages of tumor development may prevent tumor initiation and progression by promoting cellular quality control and immune surveillance. In contrast, suppression of autophagy in established tumors may enhance cancer cell sensitivity to treatment and minimise the capability of cancerous cells to thrive in stressful conditions. Context and stage of cancer play an important role and must be well considered in developing autophagy-targeted therapies (Li et al., 2020).
7 Role of autophagy in tumor cell dormancy and immune modulation
Tumor cell dormancy can be described as wherein cells remain in quiescent state for a prolonged period of time. This dormancy is responsible for cancer recurrence and poses a hindrance in developing an effective, viable therapeutic strategy (Tufail et al., 2025). The mechanism of induction of dormancy in cancer cells is not well understood. There are many reports present that suggest the role of MAPK signalling in induction of cancer cell dormancy. The autophagy role in cellular dormancy has been shown in C. elegans; the developmentally quiescent larval state dauer diapause, is adopted in response to unfavourable conditions (Meléndez et al., 2003). Knockout of several autophagy genes in C. elegans resulted in death during dauer phase, indicating an important role of autophagy. Some reports suggest a diapause-like state in tumor cells when exposed to drugs (Rehman et al., 2021; Dhimolea et al., 2021). The study suggests diapause-like state (DLS) is autophagy-mediated. Moreover, in DLS tumor cells, MAPK pathway was observed to be activated. Mitophagy involvement is reported in cell renewal, migration, dormancy, and invasion (Smith and Macleod, 2019; Nazio et al., 2019; Jahangiri et al., 2021). A tumor suppressor, TSSC4, in glioblastoma and breast cancer inhibits formation of tumorsphere by downregulating autophagy (Chen et al., 2022).
Several signaling pathways regulate dormant and quiescent CSCs, including p53, retinoblastoma and some CDK inhibitors like p57, p27, p21 (Tiwari et al., 2024; Zeuner, 2015). Autophagy is needed to promote disseminated tumor cells (Vera-Ramirez et al., 2018; Sosa et al., 2013). In ovarian cancer, ARHI (tumor suppressor) instigates autophagy by PI3K-AKT signaling pathway inhibition, inducing tumor cell dormancy (He et al., 2015). Similarly, in colorectal cancer, downregulation of PLK4 is linked with autophagy induction and results in a dormant state via MAPK pathway (Tian et al., 2023). In breast cancer models, it is reported that ATG7 gene is essential for autophagy instigation in dormant tumor cells (Vera-Ramirez et al., 2018). Studies conducted on breast cancer have shown dual autophagy role in cancer dormancy. Autophagy induced by chemotherapy may facilitate tumor relapse, whereas autophagy intrinsic to cells delays tumor relapse (Aqbi et al., 2018). Autophagy inhibition can induce tumor dormancy emergence (La Belle Flynn et al., 2019). In some conditions, autophagy inhibition may prolong tumor dormancy (Aqbi et al., 2018).
Reports suggest that elevated autophagy in tumor cells can provide a pro-tumor function in tumor microenvironment (Yang et al., 2018; Katheder et al., 2017). The deficiency of autophagy in immunocompetent mouse produces an antitumor effect, but not in immune deficient mouse (Yamamoto et al., 2020; Eng et al., 2016). Autophagy is reported to increase the antigen cross-presentation by dendritic cells, a step in instigating cytotoxic T lymphocytes. Autophagosome derived from tumors sequesters ubiquitinated antigens like OVA and gp100 are engulfed by dendritic cells are then processed for MHC-I presentation priming CD8+ T cells (Mowers et al., 2018; Yi et al., 2012). The autophagy is crucial for this process; if autophagy is inhibited in dendritic cells, it results in downregulated MHC-I facilitated antigen presentation and thus impairing T cell response (Yang et al., 2023; De Souza et al., 2020). Autophagy promotes clearance of MHC-I through NBR1, a cargo receptor that assists tumor cells to evade immune detection. The inhibition of autophagy by chloroquine can restore the surface expression of MHC-I, resulting in increased T cell recognition (Yamamoto et al., 2020). Specific chemokines facilitate the trafficking of subsets of immune cell into the tumor microenvironment (Nagarsheth et al., 2017). The autophagy is reported to modulate the chemokines expression in tumor cell and thus can also alter the immune response by regulating migration of immune cells to tumor. In tumor associated macrophages the autophagy is induced by CCL2 and IL-6 which promotes the M2 polarization and TGF- β1 secretion resulting in metastasis (Kuo et al., 2022). For T cell survival, basal level of autophagy is vital whereas enhanced autophagy in conditions like chronic infection promotes T cell exhaustion through increased level of PD-1 (De Souza et al., 2020). Similarly, the CD8+ T cells, which are ATG5 deficient, shows increased glycolysis and histone methylation enhancing the production of IFN and TNF (DeVorkin et al., 2019).
8 Multidimensional approach to target autophagy
Autophagy is the primary process maintaining cellular homeostasis and stress adaptation. Under nutrient-deprivation conditions, autophagy delivers amino acids, lipids, and other crucial molecules through degrading and recycling cellular components. Autophagy’s involvement apparently provides promise in therapeutic applications to counter various complex disorders such as cancer and neuronal degenerative diseases. Both activation and inhibition of autophagy are possible. Cellular stress, starvation, and mTOR suppression stimulate autophagy (He et al., 2018), whereas several targets upstream (VPS34, Beclin 1, and ULK1 inhibitors) and downstream of autophagosome-lysosomal fusion location suppress cellular autophagy (Pavlinov et al., 2020). Some reports show that inhibition of autophagic process can cause cancer cells to be more sensitive to anticancer drugs, as it increases chemosensitivity (Yang et al., 2011). Effective targeting of modulation of autophagy can only be achieved by gaining deeper knowledge of the process involved, as represented in Figure 3.
Figure 3. Multimodal approach to target autophagy in cancer: (A) Genetic manipulation: The autophagy is regulated by several autophagy-related proteins, and loss of these proteins (ATG5, ATG7) can cause suppressed autophagy, and enhanced expression of proteins like Beclin-1 can also elevate cellular autophagy. Targeting these gene targets with miRNA or gene replacement can help modulate autophagy in cells. (B) Modulation of autophagy through upstream or downstream targets: The Autophagy pathway can be modulated by other upstream or downstream targets of autophagy, like mTOR and AMPK. Likewise, other factors like caloric restriction chaperones and transcription factors, as well as some organelles like mitochondria, can be targeted as they can modulate autophagy levels in cells. (C) Nanotechnologies: Advanced nanotechnologies can help deliver specific small molecules directly to the cells, helping in modulating autophagy. (D) Systems biology and (E) Artificial intelligence: Can help identify novel targets for efficiently targeting autophagy modulation. (F) Small pharmacological molecules: The small molecules can directly interact with the autophagy-associated proteins or pathways and produce autophagy-modulating results. Small molecules: Metformin (Gao et al., 2020), Perifosine (Fu et al., 2009), Rapamycin (Ballou and Lin, 2008), Spautin-1 (Shao et al., 2014), 3-Methyladenine (Kosic et al., 2021), Verteporfin (Donohue et al., 2011), Thapsigargin (Nelson et al., 1994), Flusipirlene (Xia et al., 2010), Verapamil (Kania et al., 2017), Lithium (Sarkar et al., 2005), poly-D-glutamic acid (Hart et al., 1983), Bafilomycin A1 (Mauvezin and Neufeld, 2015), Taxol (Sonee et al., 1998), Monensin (Stenseth and Thyberg, 1989), E64d and pepstatin (Yang et al., 2013).
The identification and creation of new autophagy modulators would require to explore specific molecular targets. Particular elements that are implicated in autophagic process or interact with autophagy-related proteins can serve as biomarkers for several diseases. To achieve these goals, a multimodal approach is needed where different fields of science can either combine to establish and analyse large-scale data from genomics or proteomics to develop novel therapeutic targets to modulate autophagy or develop a better delivery system for small pharmaceutical compounds for specific targeting. Another area through which autophagy may be targeted includes genetic engineering. Studies can be conducted using gene editing in the form of CRISPR-Cas9 to manipulate the genes related to autophagy to identify their functions and specific regulators (O’Prey et al., 2017). Caloric restriction as well as intermittent fasting are reported to induce autophagy and, thus, cellular wellbeing (Beaulaton and Lockshin, 1977). Such dietary regimens are proposed to work as treatments that may induce longevity and prevent age-related diseases. In summary, autophagy represents a crucial cellular mechanism indispensable for upkeeping homeostasis, adaptation to stress, and physiological functions in cells.
8.1 Artificial intelligence and systems biology
The artificial intelligence methodology holds the potential for accelerating drug development by predicting potential autophagy modulators and therapeutic targets-a phenomenon noted in many fields of drug discovery (Rashid, 2021). Systems biology represents a data-integrative approach that aggregates information from all tiers of biological organization-genomics, proteomics and metabolomics in an effort to build models that explain cellular processes (Wu et al., 2024). Therefore, systems biology may make it easier to outline complex networks involved in the autophagy process, making it simpler to identify regulatory nodes and potential targets for therapy. This may allow us to realize process of autophagy regulation and integration with other cellular pathways and how the alteration of autophagy might promote oncogenesis (Jegga et al., 2011; Shi et al., 2013). Machine learning, in particular, helps facilitate the analysis of such huge datasets that the studies of system biology produce. AI algorithms can predict impacts on cancer cells due to the modulated autophagy, challenging old drug paradigms and improving treatment plans. For instance, it could examine mRNA profiles of the cancers to determine which autophagy-related genes are over or under-expressed in those cancers and help tailor drugs to these specific pathways. The machine learning programs can discriminate between cancer cells by analysing immunohistochemistry images (He et al., 2020).
8.2 miRNA in autophagy regulation
These microRNAs (miRNA) are small non-coding RNA that modulate the levels of genes by binding to their 3′untranslated region in target mRNA, which subsequently leads to mRNA clearance or even inhibition of translation (O'Brien et al., 2018). In autophagy, miRNAs are involved, targeting key genes linked with autophagy mechanism. For instance, miR-30a targets the mRNA of Beclin 1 (BECN1), a core element of autophagy initiation complex, thus suppressing autophagy. Similarly, miR-101 targets the mRNA of RAB5A and ATG4D, which is known to induce autophagy, thereby suppressing autophagy (Zhu et al., 2009; Wang et al., 2018; Frankel et al., 2011). Such examples demonstrate the intricate network through which miRNAs regulate autophagy. In cancer, miRNAs may play either oncogenic or tumor-suppressive roles by regulating autophagy. For instance, while miR-21 is commonly overexpressed in cancers and promotes autophagy through targeting several autophagy-related genes to enhance the survival of cancer cells, miR-122 is often underexpressed in some cancers. It may regulate autophagy to inhibit tumor growth (Yan et al., 2008; Zhang et al., 2024; Saadh et al., 2024; Wang et al., 2019).
8.3 Autophagy modulation through nanotechnologies
Nanotechnology has now introduced novel and innovative methods of autophagy targeting, mainly with references to cancer treatment. The nanoparticle designs lead toward the targeted delivery of autophagy-modulating drugs, thus providing increased efficacy and reduced side effects for the treatments. Targeted delivery ensures that a therapeutic agent reaches a point of action while enhancing its impact at the same time and maintaining minimum levels of injury to tissues (Paskeh et al., 2022). Several studies have reported phototherapy and autophagy relations. Phototherapy can cause the instigation of autophagy and suppress cancerous cell apoptosis. Studies have employed autophagy inhibitors to sensitize tumor cells to phototherapy. One method reported is to disrupt lysosomes with a nanogel (SiPT) containing organosilica nanodots (OSiNDs), photosensitizer TPPS, and PEG-PLE. SiPT accumulates in lysosomes, and TPPS produces ROS under 532 nm laser irradiation, promoting tumor cell apoptosis and inhibiting autophagy through lysosome destruction. This combined approach blocks autophagy, hence making phototherapy more effective. Despite the enhanced localization of tumors, SiPT still retains distribution in other parts, which explains the need for targeting strategies (Zhang X. et al., 2020). Similarly, nanoparticles loaded with PD-L1 inhibitors are also a therapeutic strategy (Tran and Phuong Tran, 2022). A study reported that the formulation of folic acid, graphene oxide (GO), SNX-2112, is a newly designed GFS. The effect of this is that when under 808 nm laser light (NIR), GFS releases SNX-2112 to inhibit HSP90, thereby causing death from autophagy. Flow cytometry reports indicated that a combination of NIR and 3-MA-based autophagy inhibition further enhanced cell death (Deng et al., 2020).
9 Pharmacological and other interventions in targeting autophagy in cancer
Pharmacological agents targeting to modulate autophagy are one of the potential options in cancer treatments. Small molecules influence autophagy by targeting autophagy proteins directly or indirectly. Autophagy has a dual nature, which was realized during several studies of cancer biology, and its role depends on either stage or type of cancer (Yun et al., 2020). One of the approaches used when targeting autophagy in chemotherapy is the use of inhibitory drugs for autophagy. Hydroxychloroquine and chloroquine are well-known inhibitors of autophagy. These inhibitors prevent fusion of the autophagosome and lysosome, effectively preventing clearance of contents of autophagic vesicle. Inhibition of autophagy sensitises cancerous cells to chemotherapy and radiation stress and damage (Mauthe et al., 2018a). The rationale behind the therapy is that blocking the protective process of autophagy reduces the cancer cell’s capability to respond to the stressful environment resulting from the treatment, and it will increase cell death, thereby improving the outcomes. It is also to be noted that when insulin and amino acids are present, autophagy process is inhibited (Liu et al., 2009). Another important class of drugs in this regard is mTOR inhibitors. The mTOR is a crucial negative autophagy modulator. Drugs such as rapamycin and its derivatives, known as rapalogs, inhibit mTOR, thereby activating autophagy (Ballou and Lin, 2008). Molecules like everolimus, Torin 1 and Temsirolimus are also mTOR inhibitors (Witzig et al., 2015; Thoreen et al., 2009; Malizzia and Hsu, 2008). In some cases, excessive autophagy stimulation can result in autophagy-dependent cell death, which could be beneficial in some cases. This would create a metabolic stress environment that is detrimental to the survival of the cancerous cells (Fulda and Kögel, 2015).
Moreover, in some conditions, autophagy activators act as therapeutic agents. Activation of AMPK by small molecules like metformin drugs can induce autophagy (Goel et al., 2022). AMPK functions as an energy sensor to degrade cellular components and make the cells generate more energy at a low cellular level (Hardie et al., 2012). Therefore, autophagy activators can contribute towards the inhibition of growth by enhancing clearing and recycling of damaged cellular constituents. This strategy may particularly be significant in cancers in the initial stages as enhanced autophagy can help limit inflammation and genetic instability, induce cell death in transformed cells and uphold homeostasis (Yang et al., 2011).
Combination therapies have become a useful tool for making targeted therapy against cancer effective. The addition of autophagic process inhibitors process combined with other anticancer treatments overcomes the mechanisms of drug resistance and thus leads to improving patient outcomes (Mele et al., 2020). Thus, chloroquine combined with chemotherapy or targeted therapy is highly effective in making the protective autophagy pathway of cancer cells cytotoxic effects. In other words, this synergistic approach utilises the strengths of more than one therapy to kill cancerous cells more efficiently whilst limiting the chances of treatment resistance (Varisli et al., 2020; Agalakova, 2024). Another therapeutic approach is to use novel small-molecule compounds that target proteins associated with autophagy. These compounds could modulate autophagy with much greater precision and much fewer side effects than broad-range drugs. For instance, SBI-0206965, a ULK1 inhibitor, affects autophagy by targeting specific components of the machinery of autophagy; it is also reported to inhibit AMPK, which offers a potential for more effective and targeted use of cancer therapies (Knudsen et al., 2020). Similarly, other compounds like MRT67307 and MRT68921 are also ULK1 and ULK2 inhibitors (Petherick et al., 2015). Several other small molecules and drugs can target various autophagy-related proteins; they may induce or suppress autophagy flux by acting at different stages and steps of autophagy, as shown in Figure 4.
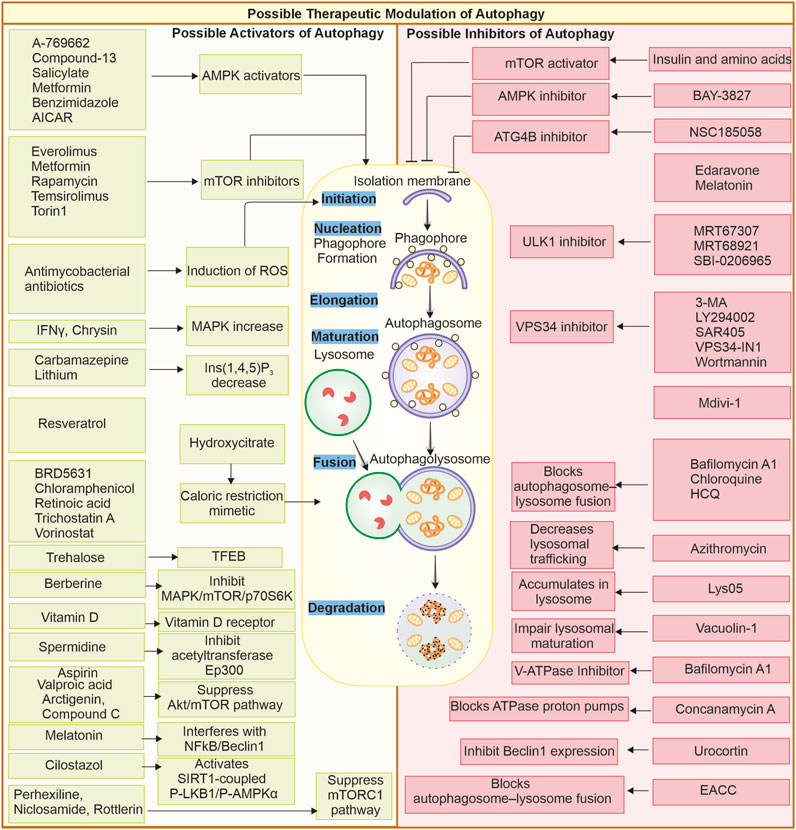
Figure 4. Possible therapeutic modulation of autophagy through small molecules: Small molecules can modulate autophagy; they can either be inhibitors or promoters. Some of the activators and inhibitors are represented. Autophagy activators: A-769662 (Walter et al., 2016), AICAR, benzimidazole, Salicylate, Compound-13 (Kim et al., 2016), Everolimus (Haas et al., 2019), Metformin, Rapamycin (Kim and Guan, 2015), Temsirolimus, Torin1 (Chandrika et al., 2017), Melatonin (Das et al., 2024), Antimycobacterial antibiotics (Kim et al., 2012), Chrysin (Jung et al., 2022), IFN-γ (Matsuzawa et al., 2012), Carbamazepine (Adikesavalu et al., 2021), Lithium (Sarkar et al., 2005), Hydroxycitrate (Piffoux et al., 2021), Resveratrol, Spermidine (Morselli et al., 2011), BRD5631 (Kuo et al., 2015), Chloramphenicol (Hsu et al., 2019), Retinoic acid (Rajawat et al., 2010), Trehalose (Rusmini et al., 2019), Trichostatin A (Liu et al., 2021), Vorinostat (Patra et al., 2022) Arctigenin (Jiang Y.a. et al., 2020), Aspirin (Peng et al., 2023), Compound C (Vucicevic et al., 2011), Berberine (Zhang Q. et al., 2020), Vitamin D (Wu and Sun, 2011), Valproic acid (Xia et al., 2016), Cilostazol (Park et al., 2016), Perhexiline, Niclosamide, Rottlerin (Balgi et al., 2009). Autophagy inhibitors: Amino acids and insulin (Shen et al., 2021), BAY-3827 (Hawley et al., 2023), NSC185058 (Ryan et al., 2024), Edaravone (Yin et al., 2019), Melatonin (Luo et al., 2020; Boga et al., 2019), MRT67307, MRT68921 (Petherick et al., 2015), SBI-0206965 (Knudsen et al., 2020; Egan et al., 2015), 3-MA (Heckmann et al., 2013; Wu et al., 2010) LY294002 (Wang et al., 2016; Blommaart et al., 1997), SAR405 (Pasquier, 2015), VPS34-IN1 (Meunier et al., 2020), Wortmannin (Yang et al., 2013; Takatsuka et al., 2004), Mdivi-1 (Aishwarya et al., 2020), Chloroquine, HCQ (Maycotte et al., 2012; Mukhopadhyay et al., 2011; Mauthe et al., 2018b), Lys05 (McAfee et al., 2012), Azithromycin (Takano et al., 2023), Concanamycin A, Bafilomycin A1 (Mauvezin and Neufeld, 2015) (Yano et al., 2015), Vacuolin-1 (Lu et al., 2014), Urocortin (Valentim et al., 2006), EACC (Vats and Manjithaya, 2019).
Some specific autophagy inhibitors are chloroquine and hydroxychloroquine, these compounds work by blocking fusion between autophagosome and lysosome through alkalinizing the lysosomes, which further reduces autophagy flux. The advantage of such compounds is that they are FDA-approved for use in cases of malaria and autoimmune disease; thus, repurposing such drugs can accelerate the clinical approval process (Mao et al., 2018; Verbaanderd et al., 2017). Another advantage of such drugs is that they have broad anticancer activity. CQ and HCQ are reported to be effective in various cancers like pancreatic, glioblastoma, esophageal, and breast, causing cell death (Cai et al., 2018). These drugs are also reported to be utilised in combination therapy and increase the efficiency of chemotherapy by overcoming the hypoxia induced resistance (Verbaanderd et al., 2017). There is also some evidence suggesting these drugs can restore MHC-1 expression in pancreatic cancer (Yamamoto et al. 2020; Mauthe et al., 2018a). There are few limitations of these drugs as they may have non desired effects like disruption of Golgi and at higher doses, can cause retinal damage (Yusuf et al., 2023; Mauthe et al., 2018a). Lys05 is a dimeric derivative of chloroquine that can accumulate in the lysosome and inhibit autophagy. This compound has some advantages, like higher potency than CQ in inhibiting autophagy and decreasing tumor growth. Under hypoxic conditions can target acute myeloid leukemia by suppressing mitophagy and impairing mitochondria. It can be used in a combination therapy with cytarabine and azacytidine in AML, clearing CD34+CD38 leukemic stem cells. There are some limitations of this compound; its high dose causes intestinal toxicity (Amaravadi and Winkler, 2012; Wang et al., 2024). Specific autophagy inhibitors target autophagy molecules; ULK-101, an autophagy inhibitor that inhibits specifically the ULK1 kinase and interrupts the autophagy initiation. Some advantages of such inhibitors are that they suppress the formation of autophagosomes, avoiding any compensatory mechanism observed with lysosomal inhibitors. ULK-101 shows reduced off-target effects and can be used in combination with nutrient deprivation in some KRAS mutant lung cancer. There is limited clinical data present when compared to CQ (Martin et al., 2018).
These developments emphasise the requirement for disentangling autophagy molecular mechanisms of toward novel and accurate therapeutic modalities. Gene therapy has emerged as another promising avenue for directly modulating the autophagy pathway in cancer by introducing genetic material that is known to either enhance or impair autophagy. There are several autophagy genes reported to be either mutated in diseases or implicated in complex diseases (Levine and Kroemer, 2019; Koveitypour et al., 2019; Amaravadi et al., 2019). This strategy may be applied to treat cancers with particular genetic alterations in autophagy-related genes. For example, restored mutated or downregulated autophagy-related genes in cancer cells can serve to enhance autophagic cell death and inhibit cancer development. This approach may enhance the efficacy of gene manipulation for targeted and highly efficient cancer therapy.
Nano technology’s drug delivery system has innovative targeting schemes to enhance the drug’s ability to target autophagy pathways to cancer therapy. Nanoparticles can be engineered to target the cells directly by delivering autophagy modulators. This means that the treatments will have a higher efficacy and lower side effects. For example, nanoparticles might be engineered to deliver autophagy inhibitors, which would then be controlled at the tumor site. Such a targeted delivery system would enable therapeutic agents to be delivered to the intended site of therapeutic action, hence increasing their effectiveness while reducing the adverse impact on other normal tissues. This is an approach that combines nanotechnology with the therapeutic value of modulating autophagy to offer advanced options for the treatment of cancer (Paskeh et al., 2022). Some dietary supplements and natural compounds have also been proven to modulate autophagy. For instance, resveratrol is a polyphenol contained in grapes and red wine; it activates autophagy through the AMPK pathway (Pasinetti et al., 2015). Spermidine, the naturally occurring product in diverse food sources, seems to favour the autophagy process with links to increased lifespan from model organism studies (Satarker et al., 2024; Hofer et al., 2024). Consumption of these compounds as supplements could improve levels of activity for autophagic machinery to promote cell health. Adding these natural compounds to cancer therapy will enable the investigation of new and complementary ways of modulating autophagy, thus leading to better treatment outcomes. Other precise ways of targeting autophagy-related genes are through the use of antisense oligonucleotides (ASOs). ASOs are small, nucleic acids strands (synthetic) that will bind to the target gene’s mRNA and halt protein synthesis, thus modulating autophagy activity (Dhuri et al., 2020; Hung et al., 2024). This approach allows a tight control over expression of autophagy-associated genes and hence may be used for promoting or inhibiting the processes of autophagy within the cancer cells. By focusing on specific genes in the autophagy pathway, ASOs offer the power to modulate and thus develop targeted cancer therapies against such diseases.
10 Conclusion and future outlook
Autophagy is a cellular degradative process that clears out worn-out proteins and organelles. It maintains cellular homeostasis as well as genomic stability by clearing out cellular toxic waste and regulating ROS levels. Autophagy is also implicated in coordinating with the immune system. Autophagy is also implicated in metabolism modulation. It is a critical mechanism in cells activated during stress response and is mainly a pro-survival process. Autophagy is also required to replenish the amino acid pools during starvation by recycling the cellular organelles and proteins. Impaired autophagy in several diseases, like neurodegenerative diseases and in different cancers, has been reported. In diseases like cancer, autophagy can perform a complex dualistic function. In initial stage of tumor formation, autophagy acts as a cellular protector, clears out cell debris, maintains genomic stability, and may clear out initial transforming cells, thus acting as a tumor suppressor. Its evidence is present when autophagy-associated proteins like ATG5, ATG7, and Beclin-1 are impaired, and then spontaneous tumors are reported, suggesting their tumor suppressive function. Meanwhile, in many cancerous cells, heightened levels of autophagy and autophagy-related proteins are reported to help cancer cells survive stresses like hypoxia and oxidative stress. Autophagy also provides cancer cells chemoresistance against anticancer drugs, thus functioning as tumor-supportive.
This makes autophagy a lucrative target for developing therapeutics against cancer. Autophagy is a multistep process that requires several molecules for regulation; thus, all these regulatory molecules and steps can be used to modulate autophagy. There are several reported pharmacological drugs that inhibit autophagy and present multiple approaches to cancer therapy. Autophagy can be modulated through autophagic inducers or inhibitors and novel small-molecule anti-cancer agents designed to instigate apoptosis in cancerous cells as well as increase the therapeutic efficacy of the main treatment strategies. Genetic therapies, nanoparticle delivery systems, dietary supplements, antisense oligonucleotides, and high-throughput screening methods have also been utilized to suppress autophagy in cancer. Personalized medicine approaches allow for the refinement of targeted therapies directed at autophagy regulation, tailored to the patient-specific characteristics. The personalized approaches, in conjunction with continuous clinical research, have the potential to improve treatment approaches and further enhance the patients’ outcomes. The progress in our understanding of autophagy biology will be the foundation of innovative and effective therapies in the field of oncology. Various pharmacological interventions are likely to play a very important role.
High-throughput cell-based assays are essential tools for screening of large libraries of compounds to identify potential autophagy modulators. It can be used to evaluate the impact of various compounds on the autophagic process by real-time measurements of autophagy flux. The rise in drug development and the introduction of new cancer treatments are increased by identifying new drugs that either promote or inhibit the autophagy process. The strategy will thus utilise advanced screening technologies coupled with deep autophagy biology insights to select some promising candidates. In this approach, personalised medicine allows the stratification of patients according to genetic and molecular profiling, selecting only those with the potential for benefit. For instance, tumors with a high basal autophagy rate or carrying particular genetic alterations in autophagy-related genes are probably more susceptible to inhibition of autophagy. Personalised medicine through tailored treatment could thus provide a better therapeutic response and reduce the risk of drug resistance. Thus, multidimensional approaches can be developed to advance research in this field in the future.
Author contributions
PK: Data curation, Formal Analysis, Investigation, Resources, Validation, Writing – original draft. AC: Data curation, Formal Analysis, Investigation, Methodology, Writing – review and editing. SK: Data curation, Formal Analysis, Investigation, Methodology, Validation, Writing – review and editing. YAJ: Data curation, Formal Analysis, Investigation, Methodology, Validation, Writing – review and editing. VKP: Investigation, Methodology, Resources, Writing – review and editing. DC: Formal Analysis, Investigation, Methodology, Resources, Writing – review and editing. SC: Formal Analysis, Investigation, Methodology, Resources, Writing – review and editing. RKV: Formal Analysis, Investigation, Methodology, Resources, Writing – review and editing. AM: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The work was supported by BRNS grant to (AM) 54/14/16/2020-BRNS-Government of India.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adikesavalu, H., Gopalaswamy, R., Kumar, A., Ranganathan, U. D., and Shanmugam, S. (2021). Autophagy induction as a host-directed therapeutic strategy against Mycobacterium tuberculosis infection. Medicina (Kaunas) 57 (6), 522. doi:10.3390/medicina57060522
Agalakova, N. I. (2024). Chloroquine and chemotherapeutic compounds in experimental cancer treatment. Int. J. Mol. Sci. 25 (2), 945. doi:10.3390/ijms25020945
Aishwarya, R., Alam, S., Abdullah, C. S., Morshed, M., Nitu, S. S., Panchatcharam, M., et al. (2020). Pleiotropic effects of mdivi-1 in altering mitochondrial dynamics, respiration, and autophagy in cardiomyocytes. Redox Biol. 36, 101660. doi:10.1016/j.redox.2020.101660
Aita, V. M., Liang, X. H., Murty, V. V., Pincus, D. L., Yu, W., Cayanis, E., et al. (1999). Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 59 (1), 59–65. doi:10.1006/geno.1999.5851
Alers, S., Löffler, A. S., Wesselborg, S., and Stork, B. (2012). Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol. Cell. Biol. 32 (1), 2–11. doi:10.1128/MCB.06159-11
Allred, D. C., Clark, G. M., Elledge, R., Fuqua, S. A., Brown, R. W., Chamness, G. C., et al. (1993). Association of p53 protein expression with tumor cell proliferation rate and clinical outcome in node-negative breast cancer. J. Natl. Cancer Inst. 85 (3), 200–206. doi:10.1093/jnci/85.3.200
Amaravadi, R. K., Kimmelman, A. C., and Debnath, J. J. (2019). Targeting autophagy in cancer: recent advances and future directions. Cancer Discov. 9 (9), 1167–1181. doi:10.1158/2159-8290.CD-19-0292
Amaravadi, R. K., and Winkler, J. D. J. A. (2012). Lys05: a new lysosomal autophagy inhibitor. Autophagy 8 (9), 1383–1384. doi:10.4161/auto.20958
Aqbi, H. F., Tyutyunyk-Massey, L., Keim, R. C., Butler, S. E., Thekkudan, T., Joshi, S., et al. (2018). Autophagy-deficient breast cancer shows early tumor recurrence and escape from dormancy. Oncotarget 9 (31), 22113–22122. doi:10.18632/oncotarget.25197
Ariosa, A. R., Lahiri, V., Lei, Y., Yang, Y., Yin, Z., Zhang, Z., et al. (2021). A perspective on the role of autophagy in cancer. Biochim. Biophys. Acta. Mol. Basis Dis. 1867 (12), 166262. doi:10.1016/j.bbadis.2021.166262
Balgi, A. D., Fonseca, B. D., Donohue, E., Tsang, T. C. F., Lajoie, P., Proud, C. G., et al. (2009). Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One 4 (9), e7124. doi:10.1371/journal.pone.0007124
Ballou, L. M., and Lin, R. Z. J. J. o.c.b. (2008). Rapamycin and mTOR kinase inhibitors. J. Chem. Biol. 1 (1), 27–36. doi:10.1007/s12154-008-0003-5
Barthet, V. J., Brucoli, M., Ladds, M. J. G. W., Nössing, C., Kiourtis, C., Baudot, A. D., et al. (2021). Autophagy suppresses the formation of hepatocyte-derived cancer-initiating ductular progenitor cells in the liver. Sci. Adv. 7 (23), eabf9141. doi:10.1126/sciadv.abf9141
Beaulaton, J., and Lockshin, R. A. (1977). Ultrastructural study of the normal degeneration of the intersegmental muscles of Anthereae polyphemus and Manduca sexta (Insecta, Lepidoptera) with particular reference of cellular autophagy. J. Morphol. 154 (1), 39–57. doi:10.1002/jmor.1051540104
Belanger, S., Côté, M., Lane, D., L’Espérance, S., Rancourt, C., and Piché, A. (2005). Bcl-2 decreases cell proliferation and promotes accumulation of cells in S phase without affecting the rate of apoptosis in human ovarian carcinoma cells. Gynecol. Oncol. 97 (3), 796–806. doi:10.1016/j.ygyno.2005.02.018
Bhutia, S. K., Mukhopadhyay, S., Sinha, N., Das, D. N., Panda, P. K., Patra, S. K., et al. (2013). Autophagy: cancer’s friend or foe? Adv. Cancer Res. 118, 61–95. doi:10.1016/B978-0-12-407173-5.00003-0
Bingle, L., Brown, N. J., and Lewis, C. E. (2002). The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J. Pathol. 196 (3), 254–265. doi:10.1002/path.1027
Blommaart, E. F., Krause, U., Schellens, J. P., Vreeling-Sindelárová, H., and Meijer, A. J. (1997). The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 243 (1-2), 240–246. doi:10.1111/j.1432-1033.1997.0240a.x
Boga, J. A., Caballero, B., Potes, Y., Perez-Martinez, Z., Reiter, R. J., Vega-Naredo, I., et al. (2019). Therapeutic potential of melatonin related to its role as an autophagy regulator: a review. J. Pineal Res. 66 (1), e12534. doi:10.1111/jpi.12534
Bolender, R. P., and Weibel, E. R. J. T. J. o.c.b. (1973). A morphometric study of the removal of phenobarbital-induced membranes from hepatocytes after cessation of threatment. J. Cell Biol. 56 (3), 746–761. doi:10.1083/jcb.56.3.746
Brabletz, T., Kalluri, R., Nieto, M. A., and Weinberg, R. A. (2018). EMT in cancer. Nat. Rev. Cancer 18 (2), 128–134. doi:10.1038/nrc.2017.118
Bustos, S. O., Antunes, F., Rangel, M. C., and Chammas, R., (2020). Emerging autophagy functions shape the tumor microenvironment and play a role in cancer progression - implications for cancer therapy, Front. Oncol. 10, 606436. doi:10.3389/fonc.2020.606436
Cai, Y., Cai, J., Ma, Q., Xu, Y., Zou, J., Xu, L., et al. (2018). Chloroquine affects autophagy to achieve an anticancer effect in EC109 esophageal carcinoma cells in vitro. Oncol. Lett. 15 (1), 1143–1148. doi:10.3892/ol.2017.7415
Chandrika, G., Natesh, K., Ranade, D., Chugh, A., and Shastry, P. (2017). Mammalian target of rapamycin inhibitors, temsirolimus and torin 1, attenuate stemness-associated properties and expression of mesenchymal markers promoted by phorbol-myristate-acetate and oncostatin-M in glioblastoma cells. Tumour Biol. 39 (3), 1010428317695921. doi:10.1177/1010428317695921
Chavez-Dominguez, R., Perez-Medina, M., Lopez-Gonzalez, J. S., Galicia-Velasco, M., and Aguilar-Cazares, D., (2020). The double-edge sword of autophagy in cancer: from tumor suppression to pro-tumor activity, Front. Oncol. 10, 578418. doi:10.3389/fonc.2020.578418
Chen, Y., Wang, Z., Ma, Q., and Sun, C. (2025). The role of autophagy in fibrosis: mechanisms, progression and therapeutic potential. Int. J. Mol. Med. 55 (4), 1–16. doi:10.3892/ijmm.2025.5502
Chen, Y., Zhang, Z., Henson, E. S., Cuddihy, A., Haigh, K., Wang, R., et al. (2022). Autophagy inhibition by TSSC4 (tumor suppressing subtransferable candidate 4) contributes to sustainable cancer cell growth. Autophagy 18 (6), 1274–1296. doi:10.1080/15548627.2021.1973338
Chiavarina, B., Whitaker-Menezes, D., Migneco, G., Martinez-Outschoorn, U. E., Pavlides, S., Howell, A., et al. (2010). HIF1-alpha functions as a tumor promoter in cancer-associated fibroblasts, and as a tumor suppressor in breast cancer cells: autophagy drives compartment-specific oncogenesis. Cell Cycle 9 (17), 3534–3551. doi:10.4161/cc.9.17.12908
Chourasia, A. H., Tracy, K., Frankenberger, C., Boland, M. L., Sharifi, M. N., Drake, L. E., et al. (2015). Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 16 (9), 1145–1163. doi:10.15252/embr.201540759
Cianfanelli, V., Fuoco, C., Lorente, M., Salazar, M., Quondamatteo, F., Gherardini, P. F., et al. (2015). AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat. Cell Biol. 17 (1), 706–730. doi:10.1038/ncb3171
Codogno, P., Mehrpour, M., and Proikas-Cezanne, T. J. (2012). Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat. Rev. Mol. Cell Biol. 13 (1), 7–12. doi:10.1038/nrm3249
Colella, B., Faienza, F., and Di Bartolomeo, S. J. C. (2019). EMT regulation by autophagy: a new perspective in glioblastoma biology. Cancers (Basel). 11 (3), 312. doi:10.3390/cancers11030312
Dafsari, H. S., Martinelli, D., Saffari, A., Ebrahimi-Fakhari, D., Fanto, M., Dionisi-Vici, C., et al. (2025). An update on autophagy disorders. J. Inherit. Metab. Dis. 48 (1), e12798. doi:10.1002/jimd.12798
Dakal, T. C., Dhabhai, B., Pant, A., Moar, K., Chaudhary, K., Yadav, V., et al. (2024). Oncogenes and tumor suppressor genes: functions and roles in cancers. MedComm (2020). 5 (6), e582. doi:10.1002/mco2.582
Das, N., Mukherjee, S., Das, A., Gupta, P., Bandyopadhyay, A., and Chattopadhyay, S. (2024). Intra-tumor ROS amplification by melatonin interferes in the apoptosis-autophagy-inflammation-EMT collusion in the breast tumor microenvironment. Heliyon 10 (1), e23870. doi:10.1016/j.heliyon.2023.e23870
De Duve, C., and Wattiaux, R. J. (1966). Functions of lysosomes. Annu. Rev. Physiol. 28 (1), 435–492. doi:10.1146/annurev.ph.28.030166.002251
Degenhardt, K., Mathew, R., Beaudoin, B., Bray, K., Anderson, D., Chen, G., et al. (2006). Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10 (1), 51–64. doi:10.1016/j.ccr.2006.06.001
Delorme-Axford, E., and Klionsky, D. J. (2018). Transcriptional and post-transcriptional regulation of autophagy in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 293 (15), 5396–5403. doi:10.1074/jbc.R117.804641
DeNardo, D. G., Barreto, J. B., Andreu, P., Vasquez, L., Tawfik, D., Kolhatkar, N., et al. (2009). CD4+ T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 16 (2), 91–102. doi:10.1016/j.ccr.2009.06.018
Deng, X., Guan, W., Qing, X., Yang, W., Que, Y., Tan, L., et al. (2020). Ultrafast low-temperature photothermal therapy activates autophagy and recovers immunity for efficient antitumor treatment. ACS Appl. Mater. Interfaces 12 (4), 4265–4275. doi:10.1021/acsami.9b19148
Deretic, V., Saitoh, T., and Akira, S. J. (2013). Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 13 (10), 722–737. doi:10.1038/nri3532
De Souza, A. S. C., Gonçalves, L. B., Lepique, A. P., and de Araujo-Souza, P. S., (2020). The role of autophagy in tumor immunology-complex mechanisms that may be explored therapeutically, Front. Oncol., 10, 603661. doi:10.3389/fonc.2020.603661
DeVorkin, L., Pavey, N., Carleton, G., Comber, A., Ho, C., Lim, J., et al. (2019). Autophagy regulation of metabolism is required for CD8+ T cell anti-tumor immunity. Cell Rep. 27 (2), 502–513. e5. doi:10.1016/j.celrep.2019.03.037
Dhimolea, E., de Matos Simoes, R., Kansara, D., Al’Khafaji, A., Bouyssou, J., Weng, X., et al. (2021). An embryonic diapause-like adaptation with suppressed Myc activity enables tumor treatment persistence. Cancer Cell 39 (2), 240–256. e11. doi:10.1016/j.ccell.2020.12.002
Dhuri, K., Bechtold, C., Quijano, E., Pham, H., Gupta, A., Vikram, A., et al. (2020). Antisense oligonucleotides: an emerging area in drug discovery and development. J. Clin. Med. 9 (6), 2004. doi:10.3390/jcm9062004
Dice, J. F. J. A. (2007). Chaperone-mediated autophagy. Autophagy 3 (4), 295–299. doi:10.4161/auto.4144
Di Donato, M., Giovannelli, P., Migliaccio, A., and Bilancio, A. (2022). Inhibition of Vps34 and p110δ PI3K impairs migration, invasion and three-dimensional spheroid growth in breast cancer cells. Int. J. Mol. Sci. 23 (16), 9008. doi:10.3390/ijms23169008
Di Malta, C., Cinque, L., and Settembre, C. (2019). Transcriptional regulation of autophagy: mechanisms and diseases, Front. Cell Dev. Biol. 7, 114. doi:10.3389/fcell.2019.00114
Donohue, E., Tovey, A., Vogl, A. W., Arns, S., Sternberg, E., Young, R. N., et al. (2011). Inhibition of autophagosome formation by the benzoporphyrin derivative verteporfin. J. Biol. Chem. 286 (9), 7290–7300. doi:10.1074/jbc.M110.139915
Dowling, R. J., Topisirovic, I., Alain, T., Bidinosti, M., Fonseca, B. D., Petroulakis, E., et al. (2010). mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science (1979). 328 (5982), 1172–1176. doi:10.1126/science.1187532
Durgan, J., Lystad, A. H., Sloan, K., Carlsson, S. R., Wilson, M. I., Marcassa, E., et al. (2021). Non-canonical autophagy drives alternative ATG8 conjugation to phosphatidylserine. Mol. Cell 81 (9), 2031–2040. e8. doi:10.1016/j.molcel.2021.03.020
Egan, D. F., Chun, M. G. H., Vamos, M., Zou, H., Rong, J., Miller, C. J., et al. (2015). Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell 59 (2), 285–297. doi:10.1016/j.molcel.2015.05.031
Eng, C. H., Wang, Z., Tkach, D., Toral-Barza, L., Ugwonali, S., Liu, S., et al. (2016). Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc. Natl. Acad. Sci. U. S. A. 113 (1), 182–187. doi:10.1073/pnas.1515617113
Eng, G. W. L., Zheng, Y., Yap, D. W. T., Teo, A. Y. T., and Cheong, J. K. (2021). Autophagy and ncRNAs: dangerous liaisons in the crosstalk between the tumor and its microenvironment. Cancers (Basel). 14 (1), 20. doi:10.3390/cancers14010020
Feng, Y., He, D., Yao, Z., and Klionsky, D. J. (2014). The machinery of macroautophagy. Cell Res. 24 (1), 24–41. doi:10.1038/cr.2013.168
Frankel, L. B., Wen, J., Lees, M., Høyer-Hansen, M., Farkas, T., Krogh, A., et al. (2011). microRNA-101 is a potent inhibitor of autophagy. EMBO J. 30 (22), 4628–4641. doi:10.1038/emboj.2011.331
Fu, L., Kim, Y. A., Wang, X., Wu, X., Yue, P., Lonial, S., et al. (2009). Perifosine inhibits mammalian target of rapamycin signaling through facilitating degradation of major components in the mTOR axis and induces autophagy. Cancer Res. 69 (23), 8967–8976. doi:10.1158/0008-5472.CAN-09-2190
Fu, Y., Huang, Z., Hong, L., Lu, J. H., Feng, D., Yin, X. M., et al. (2019). Targeting ATG4 in cancer therapy. Cancers (Basel). 11 (5), 649. doi:10.3390/cancers11050649
Fujii, S., Mitsunaga, S., Yamazaki, M., Hasebe, T., Ishii, G., Kojima, M., et al. (2008). Autophagy is activated in pancreatic cancer cells and correlates with poor patient outcome. Cancer Sci. 99 (9), 1813–1819. doi:10.1111/j.1349-7006.2008.00893.x
Fulda, S., and Kögel, D. J. O. (2015). Cell death by autophagy: emerging molecular mechanisms and implications for cancer therapy. Oncogene 34 (40), 5105–5113. doi:10.1038/onc.2014.458
Funderburk, S. F., Wang, Q. J., and Yue, Z. J. T. i.c.b. (2010). The Beclin 1–VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol. 20 (6), 355–362. doi:10.1016/j.tcb.2010.03.002
Furukawa, K., Ginevskaia, T., and Kanki, T. J. (2024). Re-exploration of all ATG genes. Autophagy Rep. 3 (1), 2386194. doi:10.1080/27694127.2024.2386194
Galluzzi, L., and Green, D. R. J. C. (2019). Autophagy-independent functions of the autophagy machinery. Cell 177 (7), 1682–1699. doi:10.1016/j.cell.2019.05.026
Gao, C., Fang, L., Zhang, H., Zhang, W.-S., Li, X.-O., and Du, S.-Y. (2020). Metformin induces autophagy via the AMPK-mTOR signaling pathway in human hepatocellular carcinoma cells. Cancer Manag. Res. 12, 5803–5811. doi:10.1101/2024.11.24.625107
Goel, S., Singh, R., Singh, V., Singh, H., Kumari, P., Chopra, H., et al. (2022). Metformin: activation of 5' AMP-activated protein kinase and its emerging potential beyond anti-hyperglycemic action, Front. Genet. 13, 1022739. doi:10.3389/fgene.2022.1022739
Gómez-Virgilio, L., Silva-Lucero, M. d. C., Flores-Morelos, D. S., Gallardo-Nieto, J., Lopez-Toledo, G., Abarca-Fernandez, A. M., et al. (2022). Autophagy: a key regulator of homeostasis and disease: an overview of molecular mechanisms and modulators. Cells 11 (15), 2262. doi:10.3390/cells11152262
Goncharova, E. A., Goncharov, D. A., Li, H., Pimtong, W., Lu, S., Khavin, I., et al. (2011). mTORC2 is required for proliferation and survival of TSC2-null cells. Mol. Cell Biol. 31 (12), 2484–2498. doi:10.1128/MCB.01061-10
Gordon, P. B., and Seglen, P. O. (1988). Prelysosomal convergence of autophagic and endocytic pathways. Biochem. Biophys. Res. Commun. 151 (1), 40–47.
Guo, F., Liu, X., Cai, H., and Le, W. (2018). Autophagy in neurodegenerative diseases: pathogenesis and therapy. Brain Pathol. 28 (1), 3–13. doi:10.1111/bpa.12545
Guo, J. Y., Chen, H. Y., Mathew, R., Fan, J., Strohecker, A. M., Karsli-Uzunbas, G., et al. (2011). Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 25 (5), 460–470. doi:10.1101/gad.2016311
Guo, J. Y., Karsli-Uzunbas, G., Mathew, R., Aisner, S. C., Kamphorst, J. J., Strohecker, A. M., et al. (2013). Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 27 (13), 1447–1461. doi:10.1101/gad.219642.113
Haas, N. B., Appleman, L. J., Stein, M., Redlinger, M., Wilks, M., Xu, X., et al. (2019). Autophagy inhibition to augment mTOR inhibition: a phase I/II trial of everolimus and hydroxychloroquine in patients with previously treated renal cell carcinoma. Clin. Cancer Res. 25 (7), 2080–2087. doi:10.1158/1078-0432.CCR-18-2204
Han, Q., Deng, Y., Chen, S., Chen, R., Yang, M., Zhang, Z., et al. (2017). Downregulation of ATG5-dependent macroautophagy by chaperone-mediated autophagy promotes breast cancer cell metastasis. Sci. Rep. 7 (1), 4759. doi:10.1038/s41598-017-04994-x
Hao, Y., Baker, D., and Ten Dijke, P. J. I. j.o.m.s. (2019). TGF-β-mediated epithelial-mesenchymal transition and cancer metastasis. Int. J. Mol. Sci. 20 (11), 2767. doi:10.3390/ijms20112767
Hardie, D. G., Ross, F. A., and Hawley, S. A. (2012). AMP-activated protein kinase: a target for drugs both ancient and modern. Chem. Biol. 19 (10), 1222–1236. doi:10.1016/j.chembiol.2012.08.019
Hart, P. D. a., Young, M. R., Jordan, M. M., Perkins, W. J., and Geisow, M. J. (1983). Chemical inhibitors of phagosome-lysosome fusion in cultured macrophages also inhibit saltatory lysosomal movements. A combined microscopic and computer study. J. Exp. Med. 158 (2), 477–492. doi:10.1084/jem.158.2.477
Hawley, S. A., Russell, F. M., Ross, F. A., and Hardie, D. G. (2023). BAY-3827 and SBI-0206965: potent AMPK inhibitors that paradoxically increase Thr172 phosphorylation. Int. J. Mol. Sci. 25 (1), 453. doi:10.3390/ijms25010453
He, C., Dong, X., Zhai, B., Jiang, X., Dong, D., Li, B., et al. (2015). MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget 6 (30), 28867–28881. doi:10.18632/oncotarget.4814
He, C., and Klionsky, D. J. J. A. r.o.g. (2009). Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43 (1), 67–93. doi:10.1146/annurev-genet-102808-114910
He, C. J. C. B. (2022). Balancing nutrient and energy demand and supply via autophagy. Curr. Biol. 32 (12), R684–R696. doi:10.1016/j.cub.2022.04.071
He, L., Zhang, J., Zhao, J., Ma, N., Kim, S. W., Qiao, S., et al. (2018). Autophagy: the last defense against cellular nutritional stress. Adv. Nutr. 9 (4), 493–504. doi:10.1093/advances/nmy011
He, Z., Liu, H., Moch, H., and Simon, H. U. (2020). Machine learning with autophagy-related proteins for discriminating renal cell carcinoma subtypes. Sci. Rep. 10 (1), 720. doi:10.1038/s41598-020-57670-y
Heckmann, B. L., and Green, D. R. (2019). LC3-associated phagocytosis at a glance. J. Cell Sci. 132 (5), jcs222984. doi:10.1242/jcs.222984
Heckmann, B. L., Teubner, B. J. W., Tummers, B., Boada-Romero, E., Harris, L., Yang, M., et al. (2019). LC3-associated endocytosis facilitates β-amyloid clearance and mitigates neurodegeneration in murine Alzheimer’s disease. Cell 178 (3), 536–551. e14. doi:10.1016/j.cell.2019.05.056
Heckmann, B. L., Yang, X., Zhang, X., and Liu, J. (2013). The autophagic inhibitor 3-methyladenine potently stimulates PKA-dependent lipolysis in adipocytes. Br. J. Pharmacol. 168 (1), 163–171. doi:10.1111/j.1476-5381.2012.02110.x
Herb, M., Gluschko, A., and Schramm, M. (2020). “LC3-associated phagocytosis-The highway to hell for phagocytosed microbes,” in Seminars in cell and developmental biology (Amsterdam, Netherlands: Elsevier).
Hofer, S. J., Daskalaki, I., Bergmann, M., Friščić, J., Zimmermann, A., Mueller, M. I., et al. (2024). Spermidine is essential for fasting-mediated autophagy and longevity. Nat. Cell Biol. 26 (9), 1571–1584. doi:10.1038/s41556-024-01468-x
Hsu, H.-L., Liao, P. L., Cheng, Y. W., Huang, S. H., Wu, C. H., Li, C. H., et al. (2019). Chloramphenicol induces autophagy and inhibits the hypoxia inducible factor-1 alpha pathway in non-small cell lung cancer cells. Int. J. Mol. Sci. 20 (1), 157. doi:10.3390/ijms20010157
Hu, X., Pan, H., Zhou, S., Pang, Q., Wang, Y., Zhu, C., et al. (2022). HS1BP3, transcriptionally regulated by ESR1, promotes hepatocellular carcinoma progression. Biochem. Biophys. Res. Commun. 623, 111–119. doi:10.1016/j.bbrc.2022.07.047
Hubbard-Lucey, V. M., Shono, Y., Maurer, K., West, M. L., Singer, N. V., Ziegler, C. G. K., et al. (2014). Autophagy gene Atg16L1 prevents lethal T cell alloreactivity mediated by dendritic cells. Immunity 41 (4), 579–591. doi:10.1016/j.immuni.2014.09.011
Hung, C., Fertan, E., Livesey, F. J., Klenerman, D., and Patani, R. (2022). APP antisense oligonucleotides reduce amyloid-β aggregation and rescue endolysosomal dysfunction in Alzheimer’s. Brain 147 (17), 2325–2333. doi:10.1093/brain/awae092
Hwang, J. S., Lai, T. H., Ahmed, M., Pham, T. M., Elashkar, O., Bahar, E., et al. (2022). Regulation of TGF-β 1-induced EMT by autophagy-dependent energy metabolism in cancer cells. Cancers (Basel). 14 (19), 4845. doi:10.3390/cancers14194845
Ichimura, Y., Kirisako, T., Takao, T., Satomi, Y., Shimonishi, Y., Ishihara, N., et al. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492. doi:10.1038/35044114
Jahangiri, L., Ishola, T., Pucci, P., Trigg, R. M., Pereira, J., Williams, J. A., et al. (2021). The role of autophagy and lncRNAs in the maintenance of cancer stem cells. Cancers (Basel) 13 (6), 1239. doi:10.3390/cancers13061239
Jakubek, P., Pakula, B., Rossmeisl, M., Pinton, P., Rimessi, A., and Wieckowski, M. R. (2024). Autophagy alterations in obesity, type 2 diabetes, and metabolic dysfunction-associated steatotic liver disease: the evidence from human studies. Intern. Emerg. Med. 19 (5), 1473–1491. doi:10.1007/s11739-024-03700-w
Jegga, A. G., Schneider, L., Ouyang, X., and Zhang, J. (2011). Systems biology of the autophagy-lysosomal pathway. Autophagy 7 (5), 477–489. doi:10.4161/auto.7.5.14811
Jiang, B., Zhou, X., Yang, T., Wang, L., Feng, L., Wang, Z., et al. (2023). The role of autophagy in cardiovascular disease: cross-interference of signaling pathways and underlying therapeutic targets. Front. Cardiovasc. Med. 10, 1088575. doi:10.3389/fcvm.2023.1088575
Jiang, G., Liang, X., Huang, Y., Lan, Z., Zhang, Z., Su, Z., et al. (2020b). p62 promotes proliferation, apoptosis-resistance and invasion of prostate cancer cells through the Keap1/Nrf2/ARE axis. Oncol. Rep. 43 (5), 1547–1557. doi:10.3892/or.2020.7527
Jiang, Y. a., Liu, J., Hong, W., Fei, X., and Liu, R. (2020a). Arctigenin inhibits glioblastoma proliferation through the AKT/mTOR pathway and induces autophagy. Biomed. Res. Int. 2020 (1), 3542613. doi:10.1155/2020/3542613
Johansen, T., and Lamark, T. J. (2020). Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol. 432 (1), 80–103. doi:10.1016/j.jmb.2019.07.016
Jung, C. H., Seo, M., Otto, N. M., and Kim, D. H. (2011). ULK1 inhibits the kinase activity of mTORC1 and cell proliferation. Autophagy 7 (10), 1212–1221. doi:10.4161/auto.7.10.16660
Jung, G.-H., Lee, J. H., Han, S. H., Woo, J. S., Choi, E. Y., Jeon, S. J., et al. (2022). Chrysin induces apoptosis via the MAPK pathway and regulates ERK/mTOR-mediated autophagy in MC-3 cells. Int. J. Mol. Sci. 23 (24), 15747. doi:10.3390/ijms232415747
Kang, M. R., Kim, M. S., Oh, J. E., Kim, Y. R., Song, S. Y., Kim, S. S., et al. (2009). Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J. Pathol. 217 (5), 702–706. doi:10.1002/path.2509
Kang, R., Zeh, H. J., Lotze, M. T., and Tang, D. (2011). The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 18 (4), 571–580. doi:10.1038/cdd.2010.191
Kania, E., Pająk, B., O’Prey, J., Sierra Gonzalez, P., Litwiniuk, A., Urbańska, K., et al. (2017). Verapamil treatment induces cytoprotective autophagy by modulating cellular metabolism. 284 (9), 1370–1387. doi:10.1111/febs.14064
Kanzawa, T., Zhang, L., Xiao, L., Germano, I. M., Kondo, Y., and Kondo, S. (2005). Arsenic trioxide induces autophagic cell death in malignant glioma cells by upregulation of mitochondrial cell death protein BNIP3. Oncogene 24 (6), 980–991. doi:10.1038/sj.onc.1208095
Karantza-Wadsworth, V., Patel, S., Kravchuk, O., Chen, G., Mathew, R., Jin, S., et al. (2007). Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 21 (13), 1621–1635. doi:10.1101/gad.1565707
Katheder, N. S., Khezri, R., O’Farrell, F., Schultz, S. W., Jain, A., Rahman, M. M., et al. (2017). Microenvironmental autophagy promotes tumour growth. Nature 541, 417–420. doi:10.1038/nature20815
Kenific, C. M., Thorburn, A., and Debnath, J. J. (2010). Autophagy and metastasis: another double-edged sword. Curr. Opin. Cell Biol. 22 (2), 241–245. doi:10.1016/j.ceb.2009.10.008
Khandia, R., Dadar, M., Munjal, A., Dhama, K., Karthik, K., Tiwari, R., et al. (2019). A comprehensive review of autophagy and its various roles in infectious, non-infectious, and lifestyle diseases: current knowledge and prospects for disease prevention, novel drug design, and therapy. Cells 8 (7), 674. doi:10.3390/cells8070674
Kim, J., Yang, G., Kim, Y., and Ha, J. (2016). AMPK activators: mechanisms of action and physiological activities. Exp. Mol. Med. 48 (4), e224. doi:10.1038/emm.2016.16
Kim, J.-J., Lee, H. M., Shin, D. M., Kim, W., Yuk, J. M., Jin, H. S., et al. (2012). Host cell autophagy activated by antibiotics is required for their effective antimycobacterial drug action. Cell Host Microbe 11 (5), 457–468. doi:10.1016/j.chom.2012.03.008
Kim, Y. C., and Guan, K.-L. J. T. (2015). mTOR: a pharmacologic target for autophagy regulation. J. Clin. Invest. 125 (1), 25–32. doi:10.1172/JCI73939
Kingwell, K. J. N. (2013). Mutations in autophagy gene cause a rare and severe neurodegenerative disease. Nat. Rev. Neurol. 9 (4), 182. doi:10.1038/nrneurol.2013.42
Klionsky, D. J., Cregg, J. M., Dunn, W. A., Emr, S. D., Sakai, Y., Sandoval, I. V., et al. (2003). A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5 (4), 539–545. doi:10.1016/s1534-5807(03)00296-x
Klionsky, D. J., and Emr, S. D. J. S. (2000). Autophagy as a regulated pathway of cellular degradation. Science (1979). 290 (5497), 1717–1721. doi:10.1126/science.290.5497.1717
Klionsky, D. J., Petroni, G., Amaravadi, R. K., Baehrecke, E. H., Ballabio, A., Boya, P., et al. (2021). Autophagy in major human diseases. 40 (19), e108863. doi:10.15252/embj.2021108863
Knudsen, J. R., Madsen, A. B., Persson, K. W., Henríquez-Olguín, C., Li, Z., and Jensen, T. E. (2020). The ULK1/2 and AMPK inhibitor SBI-0206965 blocks AICAR and insulin-stimulated glucose transport. Int. J. Mol. Sci. 21 (7), 2344. doi:10.3390/ijms21072344
Kon, M., Kiffin, R., Koga, H., Chapochnick, J., Macian, F., Varticovski, L., et al. (2011). Chaperone-mediated autophagy is required for tumor growth. Sci. Transl. Med. 3 (109), 109ra117. doi:10.1126/scitranslmed.3003182
Kosic, M., Paunovic, V., Ristic, B., Mircic, A., Bosnjak, M., Stevanovic, D., et al. (2021). 3-Methyladenine prevents energy stress-induced necrotic death of melanoma cells through autophagy-independent mechanisms. J. Pharmacol. Sci. 147 (1), 156–167. doi:10.1016/j.jphs.2021.06.003
Koveitypour, Z., Panahi, F., Vakilian, M., Peymani, M., Seyed Forootan, F., Nasr Esfahani, M. H., et al. (2019). Signaling pathways involved in colorectal cancer progression. Cell Biosci. 9 (1), 97. doi:10.1186/s13578-019-0361-4
Ktistakis, N. T. (2017). In praise of M. Anselmier who first used the term “autophagie” in 1859. Autophagy, (Milton Park, Abingdon-on-Thames, UK: Taylor & Francis, 2015–2017. doi:10.1080/15548627.2017.1367473
Kumar, P., Jagtap, Y. A., Patwa, S. M., Kinger, S., Dubey, A. R., Prajapati, V. K., et al. (2022). Autophagy based cellular physiological strategies target oncogenic progression. J. Cell. Physiol. 237 (1), 258–277. doi:10.1002/jcp.30567
Kuo, S.-Y., Castoreno, A. B., Aldrich, L. N., Lassen, K. G., Goel, G., Dančík, V., et al. (2015). Small-molecule enhancers of autophagy modulate cellular disease phenotypes suggested by human genetics. Proc. Natl. Acad. Sci. U. S. A. 112 (31), E4281–E4287. doi:10.1073/pnas.1512289112
Kuo, W. T., Chang, J. M., Chen, C. C., Tsao, N., and Chang, C. P. (2022). Autophagy drives plasticity and functional polarization of tumor-associated macrophages. IUBMB Life 74 (2), 157–169. doi:10.1002/iub.2543
La Belle Flynn, A., Calhoun, B. C., Sharma, A., Chang, J. C., Almasan, A., and Schiemann, W. P. (2019). Autophagy inhibition elicits emergence from metastatic dormancy by inducing and stabilizing Pfkfb3 expression. Nat. Commun. 10 (1), 3668. doi:10.1038/s41467-019-11640-9
Lamsira, H. K., Sabatini, A., Ciolfi, S., Ciccosanti, F., Sacchi, A., Piacentini, M., et al. (2025). Autophagy and programmed cell death modalities interplay in HIV pathogenesis. Cells 14 (5), 351. doi:10.3390/cells14050351
Lazova, R., Camp, R. L., Klump, V., Siddiqui, S. F., Amaravadi, R. K., and Pawelek, J. M. (2012). Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin. Cancer Res. 18 (2), 370–379. doi:10.1158/1078-0432.CCR-11-1282
Lee, J., Giordano, S., and Zhang, J. J. B. J. (2012). Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem. J. 441 (2), 523–540. doi:10.1042/BJ20111451
Lee, J. W., Jeong, E. G., Lee, S. H., Yoo, N. J., and Lee, S. H. (2007). Somatic mutations of BECN1, an autophagy-related gene, in human cancers. APMIS 115 (6), 750–756. doi:10.1111/j.1600-0463.2007.apm_640.x
Leidal, A. M., Huang, H. H., Marsh, T., Solvik, T., Zhang, D., Ye, J., et al. (2020). The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 22 (2), 187–199. doi:10.1038/s41556-019-0450-y
Lemasters, J. J., Nieminen, A. L., Qian, T., Trost, L. C., Elmore, S. P., Nishimura, Y., et al. (1998). The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim. Biophys. Acta 1366 (1-2), 177–196. doi:10.1016/s0005-2728(98)00112-1
Levine, B., and Klionsky, D. J. J. (2004). Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6 (4), 463–477. doi:10.1016/s1534-5807(04)00099-1
Levine, B., and Kroemer, G. J. C. (2008). SnapShot: macroautophagy. Cell 132 (1), e162. e1–e162.e3. doi:10.1016/j.cell.2007.12.026
Levine, B., and Kroemer, G. J. C. (2019). Biological functions of autophagy genes: a disease perspective. Cell 176 (1), 11–42. doi:10.1016/j.cell.2018.09.048
Li, X., He, S., and Ma, B. J. M. c. (2020). Autophagy and autophagy-related proteins in cancer. Mol. Cancer 19 (1), 12. doi:10.1186/s12943-020-1138-4
Liang, X. H., Jackson, S., Seaman, M., Brown, K., Kempkes, B., Hibshoosh, H., et al. (1999). Induction of autophagy and inhibition of tumorigenesis by Beclin 1. Nature 402 (6762), 672–676. doi:10.1038/45257
Liao, Z., Dai, Z., Cai, C., Zhang, X., Li, A., Zhang, H., et al. (2019). Knockout of Atg5 inhibits proliferation and promotes apoptosis of DF-1 cells. In Vitro Cell. Dev. Biol. Anim. 55 (5), 341–348. doi:10.1007/s11626-019-00342-7
Lin, Y., Wu, X., Yang, Y., Wu, Y., Xiang, L., and Zhang, C., (2024). The multifaceted role of autophagy in skin autoimmune disorders: a guardian or culprit? Front. Immunol. 15, 1343987, doi:10.3389/fimmu.2024.1343987
Linder, B., and Kögel, D. J. B. (2019). Autophagy in cancer cell death. Biol. (Basel). 8 (4), 82. doi:10.3390/biology8040082
Lisanti, M. P., Martinez-Outschoorn, U. E., Chiavarina, B., Pavlides, S., Whitaker-Menezes, D., Tsirigos, A., et al. (2010). Understanding the “lethal” drivers of tumor-stroma co-evolution: emerging role (s) for hypoxia, oxidative stress and autophagy/mitophagy in the tumor microenvironment. Cancer Biol. Ther. 10 (6), 537–542. doi:10.4161/cbt.10.6.13370
Liu, D., Liu, Z., Condouris, S., and Xing, M. (2007). BRAF V600E maintains proliferation, transformation, and tumorigenicity of BRAF-mutant papillary thyroid cancer cells. J. Clin. Endocrinol. Metab. 92 (6), 2264–2271. doi:10.1210/jc.2006-1613
Liu, H.-Y., Han, J., Cao, S. Y., Hong, T., Zhuo, D., Shi, J., et al. (2009). Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J. Biol. Chem. 284 (45), 31484–31492. doi:10.1074/jbc.M109.033936
Liu, J.-H., Cao, Y. M., Rong, Z. P., Ding, J., and Pan, X. (2021). Trichostatin A induces autophagy in cervical cancer cells by regulating the PRMT5-STC1-TRPV6-JNK pathway. Pharmacology 106 (1-2), 60–69. doi:10.1159/000507937
Liu, S., Yao, S., Yang, H., and Wang, Y. (2023). Autophagy: regulator of cell death. Cell Death Dis. 14 (10), 648. doi:10.1038/s41419-023-06154-8
Lu, X. X., Cao, L. Y., Chen, X., Xiao, J., Zou, Y., and Chen, Q. (2016). PTEN inhibits cell proliferation, promotes cell apoptosis, and induces cell cycle arrest via downregulating the PI3K/AKT/hTERT pathway in lung adenocarcinoma A549 cells. Biomed. Res. Int. 2016, 2476842. doi:10.1155/2016/2476842
Lu, Y., Dong, S., Hao, B., Li, C., Zhu, K., Guo, W., et al. (2014). Vacuolin-1 potently and reversibly inhibits autophagosome-lysosome fusion by activating RAB5A. Autophagy 10 (11), 1895–1905. doi:10.4161/auto.32200
Lucantoni, F., Martínez-Cerezuela, A., Gruevska, A., Moragrega, Á. B., Víctor, V. M., Esplugues, J. V., et al. (2021). Understanding the implication of autophagy in the activation of hepatic stellate cells in liver fibrosis: are we there yet? J. Pathol. 254 (3), 216–228. doi:10.1002/path.5678
Luo, F., Sandhu, A. F., Rungratanawanich, W., Williams, G. E., Akbar, M., Zhou, S., et al. (2020). Melatonin and autophagy in aging-related neurodegenerative diseases. Int. J. Mol. Sci. 21 (19), 7174. doi:10.3390/ijms21197174
Majeski, A. E., and Majeski, J. (2004). Mechanisms of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 36 (12), 2435–2444.
Malizzia, L. J., and Hsu, A. (2008). Temsirolimus, an mTOR inhibitor for treatment of patients with advanced renal cell carcinoma. Clin. J. Oncol. Nurs. 12 (4), 639–646. doi:10.1188/08.cjon.639-646
Mao, I.-C., Lin, C. Y., Wu, C. L., Kor, C. T., and Chang, C. C. (2018). Hydroxychloroquine and risk of development of cancers: a nationwide population-based cohort study. Ther. Clin. Risk Manag. 14, 1435–1443. doi:10.2147/TCRM.S175581
Martin, K. R., Celano, S. L., Solitro, A. R., Gunaydin, H., Scott, M., O’Hagan, R. C., et al. (2018). A potent and selective ULK1 inhibitor suppresses autophagy and sensitizes cancer cells to nutrient stress, iScience 8, 74–84. doi:10.1016/j.isci.2018.09.012
Mathew, R., Karantza-Wadsworth, V., and White, E. J. N. R. C. (2007). Role of autophagy in cancer. Nat. Rev. Cancer 7 (12), 961–967. doi:10.1038/nrc2254
Mathiassen, S. G., De Zio, D., and Cecconi, F. J. F. i.o., (2017). Autophagy and the cell cycle: a complex landscape, Front. Oncol. 7, 51. doi:10.3389/fonc.2017.00051
Matsuzawa, T., Kim, B. H., Shenoy, A. R., Kamitani, S., Miyake, M., and Macmicking, J. D. (2012). IFN-γ elicits macrophage autophagy via the p38 MAPK signaling pathway. J. Immunol. 189 (2), 813–818. doi:10.4049/jimmunol.1102041
Mauthe, M., Orhon, I., Rocchi, C., Zhou, X., Luhr, M., Hijlkema, K. J., et al. (2018a). Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14 (8), 1435–1455. doi:10.1080/15548627.2018.1474314
Mauthe, M., Orhon, I., Rocchi, C., Zhou, X., Luhr, M., Hijlkema, K. J., et al. (2018b). Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14 (8), 1435–1455. doi:10.1080/15548627.2018.1474314
Mauvezin, C., and Neufeld, T. P. J. A. (2015). Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy 11 (8), 1437–1438. doi:10.1080/15548627.2015.1066957
Maycotte, P., Aryal, S., Cummings, C. T., Thorburn, J., Morgan, M. J., and Thorburn, A. (2012). Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy 8 (2), 200–212. doi:10.4161/auto.8.2.18554
McAfee, Q., Zhang, Z., Samanta, A., Levi, S. M., Ma, X. H., Piao, S., et al. (2012). Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc. Natl. Acad. Sci. U. S. A. 109 (21), 8253–8258. doi:10.1073/pnas.1118193109
McKnight, N. C., and Yue, Z. (2013). Beclin 1, an essential component and master regulator of PI3K-III in health and disease. Curr. Pathobiol. Rep. 1, 231–238.
Mele, L., Del Vecchio, V., Liccardo, D., Prisco, C., Schwerdtfeger, M., Robinson, N., et al. (2020). The role of autophagy in resistance to targeted therapies, Cancer Treat. Rev. 88, 102043. doi:10.1016/j.ctrv.2020.102043
Meléndez, A., Tallóczy, Z., Seaman, M., Eskelinen, E. L., Hall, D. H., and Levine, B. (2003). Autophagy genes are essential for dauer development and life-span extension in C. elegans. Sci. (1979). 301 (5638), 1387–1391. doi:10.1126/science.1087782
Melnik, S., Werth, N., Boeuf, S., Hahn, E. M., Gotterbarm, T., Anton, M., et al. (2019). Impact of c-MYC expression on proliferation, differentiation, and risk of neoplastic transformation of human mesenchymal stromal cells. Stem Cell Res. Ther. 10 (1), 73. doi:10.1186/s13287-019-1187-z
Meunier, G., Birsen, R., Cazelles, C., Belhadj, M., Cantero-Aguilar, L., Kosmider, O., et al. (2020). Antileukemic activity of the VPS34-IN1 inhibitor in acute myeloid leukemia. Oncogenesis 9 (10), 94. doi:10.1038/s41389-020-00278-8
Mijaljica, D., Prescott, M., and Devenish, R. J. J. (2012). The intriguing life of autophagosomes. Int. J. Mol. Sci. 13 (3), 3618–3635. doi:10.3390/ijms13033618
Miller, D. R., and Thorburn, A. J. (2021). Autophagy and organelle homeostasis in cancer. Dev. Cell 56 (7), 906–918. doi:10.1016/j.devcel.2021.02.010
Mizushima, N., Levine, B., Cuervo, A. M., and Klionsky, D. J. (2008). Autophagy fights disease through cellular self-digestion. Nature 451 (7182), 1069–1075. doi:10.1038/nature06639
Moreno-Blas, D., Adell, T., and González-Estévez, C. J. C. (2025). Autophagy in tissue repair and regeneration. Cells 14 (4), 282.
Morozova, K., Clement, C. C., Kaushik, S., Stiller, B., Arias, E., Ahmad, A., et al. (2016). Structural and biological interaction of hsc-70 protein with phosphatidylserine in endosomal microautophagy. J. Biol. Chem. 291 (35), 18096–18106. doi:10.1074/jbc.M116.736744
Morselli, E., Mariño, G., Bennetzen, M. V., Eisenberg, T., Megalou, E., Schroeder, S., et al. (2011). Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 192 (4), 615–629. doi:10.1083/jcb.201008167
Motoshima, H., Goldstein, B. J., Igata, M., and Araki, E. (2006). AMPK and cell proliferation–AMPK as a therapeutic target for atherosclerosis and cancer. J. Physiol. 574 (1), 63–71. doi:10.1113/jphysiol.2006.108324
Mowers, E. E., Sharifi, M. N., and Macleod, K. F. (2018). Functions of autophagy in the tumor microenvironment and cancer metastasis. FEBS J. 285 (10), 1751–1766. doi:10.1111/febs.14388
Mukhopadhyay, A., Helgason, G. V., Karvela, M., and Holyoake, T. L. (2011). Hydroxychloroquine for chronic myeloid leukemia: complete cure on the horizon? Expert Rev. Hematol. 4 (4), 369–371. doi:10.1586/ehm.11.34
Nagarsheth, N., Wicha, M. S., and Zou, W. J. (2017). Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 17 (9), 559–572. doi:10.1038/nri.2017.49
Nazio, F., Bordi, M., Cianfanelli, V., Locatelli, F., and Cecconi, F. (2019). Autophagy and cancer stem cells: molecular mechanisms and therapeutic applications. Cell Death Differ. 26 (4), 690–702. doi:10.1038/s41418-019-0292-y
Nelson, E., Li, C. C., Bangalore, R., Benson, T., Kass, R. S., and Hinkle, P. M. (1994). Inhibition of L-type calcium-channel activity by thapsigargin and 2, 5-t-butylhydroquinone, but not by cyclopiazonic acid. Biochem. J. 302 (1), 147–154. doi:10.1042/bj3020147
Nieto-Torres, J. L., Leidal, A. M., Debnath, J., and Hansen, M. (2021). Beyond autophagy: the expanding roles of ATG8 proteins. Trends Biochem. Sci. 46 (8), 673–686. doi:10.1016/j.tibs.2021.01.004
Nishida, K., Kyoi, S., Yamaguchi, O., Sadoshima, J., and Otsu, K. (2009). The role of autophagy in the heart. Cell Death Differ. 16 (1), 31–38. doi:10.1038/cdd.2008.163
O’Brien, J., Hayder, H., Zayed, Y., and Pneg, C. (2018). Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne). 9, 402. doi:10.3389/fendo.2018.00402
Olsvik, H. L., Svenning, S., Abudu, Y. P., Brech, A., Stenmark, H., Johansen, T., et al. (2019). Endosomal microautophagy is an integrated part of the autophagic response to amino acid starvation. Autophagy 15 (1), 182–183. doi:10.1080/15548627.2018.1532265
O’Prey, J., Sakamaki, J., Baudot, A. D., New, M., Van Acker, T., Tooze, S. A., et al. (2017). Application of CRISPR/Cas9 to Autophagy Research. Methods Enzymol. 588, 79–108. doi:10.1016/bs.mie.2016.09.076
Ortega, M. A., Fraile-Martinez, O., de Leon-Oliva, D., Boaru, D. L., Lopez-Gonzalez, L., García-Montero, C., et al. (2024). Autophagy in its (proper) context: molecular basis, biological relevance, pharmacological modulation, and lifestyle medicine. Int. J. Biol. Sci. 20 (7), 2532–2554. doi:10.7150/ijbs.95122
Palumbo, R., Sampaolesi, M., De Marchis, F., Tonlorenzi, R., Colombetti, S., Mondino, A., et al. (2004). Extracellular HMGB1, a signal of tissue damage, induces mesoangioblast migration and proliferation. J. Cell Biol. 164 (3), 441–449. doi:10.1083/jcb.200304135
Park, K., and Lee, M.-S. (2022). Current status of autophagy enhancers in metabolic disorders and other diseases. Front Cell Dev. Biol. 10, 811701.
Park, S. Y., Lee, H. R., Lee, W. S., Shin, H. K., Kim, H. Y., Hong, K. W., et al. (2016). Cilostazol modulates autophagic degradation of β-amyloid peptide via SIRT1-coupled LKB1/AMPKα signaling in neuronal cells. PLoS One 11 (8), e0160620. doi:10.1371/journal.pone.0160620
Pasinetti, G. M., Wang, J., Ho, L., Zhao, W., and Dubner, L. (2015). Roles of resveratrol and other grape-derived polyphenols in Alzheimer’s disease prevention and treatment. Biochim. Biophys. Acta 1852 (6), 1202–1208. doi:10.1016/j.bbadis.2014.10.006
Paskeh, M. D. A., Entezari, M., Clark, C., Zabolian, A., Ranjbar, E., Farahani, M. V., et al. (2022). Targeted regulation of autophagy using nanoparticles: new insight into cancer therapy. Biochim. Biophys. Acta. Mol. Basis Dis. 1868 (3), 166326. doi:10.1016/j.bbadis.2021.166326
Pasquier, B. J. A. (2015). SAR405, a PIK3C3/Vps34 inhibitor that prevents autophagy and synergizes with MTOR inhibition in tumor cells. Autophagy 11 (4), 725–726. doi:10.1080/15548627.2015.1033601
Patergnani, S., Missiroli, S., Morciano, G., Perrone, M., Mantovani, C. M., Anania, G., et al. (2021). Understanding the role of autophagy in cancer formation and progression is a real opportunity to treat and cure human cancers. Cancers (Basel). 13 (22), 5622. doi:10.3390/cancers13225622
Patra, S., Praharaj, P. P., Klionsky, D. J., and Bhutia, S. K. (2022). Vorinostat in autophagic cell death: a critical insight into autophagy-mediated,-associated and-dependent cell death for cancer prevention. Drug Discov. Today 27 (1), 269–279. doi:10.1016/j.drudis.2021.08.004
Pavlinov, I., Salkovski, M., and Aldrich, L. N. (2020). Beclin 1–ATG14L protein–protein interaction inhibitor selectively inhibits autophagy through disruption of VPS34 complex I. J. Am. Chem. Soc. 142 (18), 8174–8182. doi:10.1021/jacs.9b12705
Peng, J., Xiao, X., Li, S., Lyu, X., Gong, H., Tan, S., et al. (2023). Aspirin alleviates pulmonary fibrosis through PI3K/AKT/mTOR-mediated autophagy pathway, Exp. Gerontol. 172, 112085. doi:10.1016/j.exger.2023.112085
Petherick, K. J., Conway, O. J. L., Mpamhanga, C., Osborne, S. A., Kamal, A., Saxty, B., et al. (2015). Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J. Biol. Chem. 290 (18), 28726–11383. doi:10.1074/jbc.A114.627778
Piffoux, M., Eriau, E., and Cassier, P. A. J. (2021). Autophagy as a therapeutic target in pancreatic cancer. Br. J. Cancer 124 (2), 333–344. doi:10.1038/s41416-020-01039-5
Poillet-Perez, L., Despouy, G., Delage-Mourroux, R., and Boyer-Guittaut, M. (2015). Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy, Redox Biol. 4, 184–192. doi:10.1016/j.redox.2014.12.003
Qiang, L., Zhao, B., Ming, M., Wang, N., He, T. C., Hwang, S., et al. (2023). Autophagy regulates tumor growth and metastasis. bioRxiv. doi:10.1101/2023.10.31.564991
Quach, C., Song, Y., Guo, H., Li, S., Maazi, H., Fung, M., et al. (2019). A truncating mutation in the autophagy gene UVRAG drives inflammation and tumorigenesis in mice. Nat. Commun. 10 (1), 5681. doi:10.1038/s41467-019-13475-w
Rahman, M. A., Apu, E. H., Rakib-Uz-Zaman, S. M., Chakraborti, S., Bhajan, S. K., Taleb, S. A., et al. (2024). Exploring importance and regulation of autophagy in cancer stem cells and stem cell-based therapies. Cells 13 (11), 958. doi:10.3390/cells13110958
Rahman, M. A., Park, M. N., Rahman, M. H., Rashid, M. M., Islam, R., Uddin, M. J., et al. (2022). p53 modulation of autophagy signaling in cancer therapies: perspectives mechanism and therapeutic targets, Front. Cell Dev. Biol. 10, 761080. doi:10.3389/fcell.2022.761080
Rajawat, Y., Hilioti, Z., and Bossis, I. J. A. (2010). Autophagy: a target for retinoic acids. Autophagy 6 (8), 1224–1226. doi:10.4161/auto.6.8.13793
Rakesh, R., PriyaDharshini, L. C., Sakthivel, K. M., and Rasmi, R. R. (2022). Role and regulation of autophagy in cancer. Biochim. Biophys. Acta. Mol. Basis Dis. 1868 (7), 166400. doi:10.1016/j.bbadis.2022.166400
Rao, S., Tortola, L., Perlot, T., Wirnsberger, G., Novatchkova, M., Nitsch, R., et al. (2014). A dual role for autophagy in a murine model of lung cancer. Nat. Commun. 5 (1), 3056. doi:10.1038/ncomms4056
Rashid, M. B. (2021). Artificial intelligence effecting a paradigm shift in drug development. SLAS Technol. 26 (1), 3–15. doi:10.1177/2472630320956931
Rehman, S. K., Haynes, J., Collignon, E., Brown, K. R., Wang, Y., Nixon, A. M. L., et al. (2021). Colorectal cancer cells enter a diapause-like DTP state to survive chemotherapy. Cell 184 (1), 226–242. e21. doi:10.1016/j.cell.2020.11.018
Ribatti, D., Tamma, R., and Annese, T. J. (2020). Epithelial-mesenchymal transition in cancer: a historical overview. Transl. Oncol. 13 (6), 100773. doi:10.1016/j.tranon.2020.100773
Rojas-Sanchez, G., Cotzomi-Ortega, I., Pazos-Salazar, N. G., Reyes-Leyva, J., and Maycotte, P. (2019). Autophagy and its relationship to epithelial to mesenchymal transition: when autophagy inhibition for cancer therapy turns counterproductive. Biol. (Basel). 8 (4), 71. doi:10.3390/biology8040071
Ruby, M., Gifford, C. C., Pandey, R., Raj, V. S., Sabbisetti, V. S., Ajay, A. K., et al. (2023). Autophagy as a Therapeutic Target for Chronic Kidney Disease and the Roles of TGF-β1 in Autophagy and Kidney Fibrosis. Cells. 12 (3), 412. doi:10.3390/cells12030412
Rusmini, P., Cortese, K., Crippa, V., Cristofani, R., Cicardi, M. E., Ferrari, V., et al. (2019). Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration. Autophagy 15 (4), 631–651. doi:10.1080/15548627.2018.1535292
Ryan, P. J., Uranga, S., Stanelle, S. T., Lewis, M. H., O’Reilly, C. L., Cardin, J. M., et al. (2024). The autophagy inhibitor NSC185058 suppresses mTORC1-mediated protein anabolism in cultured skeletal muscle. Sci. Rep. 14 (1), 8094. doi:10.1038/s41598-024-58716-1
Ryter, S. W., Cloonan, S. M., and Choi, A. M. K. (2013). Autophagy: a critical regulator of cellular metabolism and homeostasis. Mol. Cells 36 (1), 7–16. doi:10.1007/s10059-013-0140-8
Saadh, M. J., Mahdi, M. S., Allela, O. Q. B., Alazzawi, T. S., Ubaid, M., Rakhimov, N. M., et al. (2024). Critical role of miR-21/exosomal miR-21 in autophagy pathway. Pathol Res Pract. 257, 155275. doi:10.1016/j.prp.2024.155275
Sahu, R., Kaushik, S., Clement, C. C., Cannizzo, E. S., Scharf, B., Follenzi, A., et al. (2011). Microautophagy of cytosolic proteins by late endosomes. Dev. Cell 20 (1), 131–139. doi:10.1016/j.devcel.2010.12.003
Sarkar, S., Floto, R. A., Berger, Z., Imarisio, S., Cordenier, A., Pasco, M., et al. (2005). Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 170 (7), 1101–1111. doi:10.1083/jcb.200504035
Satarker, S., Wilson, J., Kolathur, K. K., Mudgal, J., Lewis, S. A., Arora, D., et al. (2024). Spermidine as an epigenetic regulator of autophagy in neurodegenerative disorders. Eur J Pharmacol. 979, 176823. doi:10.1016/j.ejphar.2024.176823
Satoo, K., Noda, N. N., Kumeta, H., Fujioka, Y., Mizushima, N., Ohsumi, Y., et al. (2009). The structure of Atg4B–LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. 28 (9), 1341–1350. doi:10.1038/emboj.2009.80
Saxton, R. A., and Sabatini, D. M. J. C. (2017). mTOR signaling in growth, metabolism, and disease. Cell 168 (6), 960–976. doi:10.1016/j.cell.2017.02.004
Schiattarella, G. G., and Hill, J. A. (2016). Therapeutic targeting of autophagy in cardiovascular disease. J. Mol. Cell. Cardiol. 95, 86–93.
Shao, S., Li, S., Qin, Y., Wang, X., Yang, Y., Bai, H., et al. (2014). Spautin-1, a novel autophagy inhibitor, enhances imatinib-induced apoptosis in chronic myeloid leukemia. Int. J. Oncol. 44 (5), 1661–1668. doi:10.3892/ijo.2014.2313
Sharifi, M. N., Mowers, E. E., Drake, L. E., Collier, C., Chen, H., Zamora, M., et al. (2016). Autophagy promotes focal adhesion disassembly and cell motility of metastatic tumor cells through the direct interaction of paxillin with LC3. Cell Rep. 15 (8), 1660–1672. doi:10.1016/j.celrep.2016.04.065
Shen, J. Z., Wu, G., and Guo, S. (2021). Amino acids in autophagy: regulation and function. Adv Exp Med Biol. 1332, 51–66. doi:10.1007/978-3-030-74180-8_4
Sheng, X., Zhu, P., Qin, J., and Li, Q. (2018). The biological role of autophagy in regulating and controlling the proliferation of liver cancer cells induced by bufalin. Oncol. Rep. 39 (6), 2931–2941. doi:10.3892/or.2018.6365
Shi, Z., Li, C. Y., Zhao, S., Yu, Y., An, N., Liu, Y. X., et al. (2013). A systems biology analysis of autophagy in cancer therapy. Cancer Lett. 337 (2), 149–160. doi:10.1016/j.canlet.2013.06.004
Singla, M., and Bhattacharyya, S. (2017). Autophagy as a potential therapeutic target during epithelial to mesenchymal transition in renal cell carcinoma: an in vitro study. Biomed. Pharmacother. 94, 332–340. doi:10.1016/j.biopha.2017.07.070
Smith, A. G., and Macleod, K. F. (2019). Autophagy, cancer stem cells and drug resistance. J. Pathol. 247 (5), 708–718. doi:10.1002/path.5222
Sønder, S. L., Häger, S. C., Heitmann, A. S. B., Frankel, L. B., Dias, C., Simonsen, A. C., et al. (2021). Restructuring of the plasma membrane upon damage by LC3-associated macropinocytosis. Sci. Adv. 7 (27), eabg1969. doi:10.1126/sciadv.abg1969
Sonee, M., Barrón, E., Yarber, F. A., and Hamm-Alvarez, S. F. (1998). Taxol inhibits endosomal-lysosomal membrane trafficking at two distinct steps in CV-1 cells. Am. J. Physiol. 275 (6), C1630–C1639. doi:10.1152/ajpcell.1998.275.6.C1630
Sosa, M. S., Bragado, P., Debnath, J., Aguirre-Ghiso, J. A., et al. (2013). Regulation of tumor cell dormancy by tissue microenvironments and autophagy. Adv Exp Med Biol. 73–89. doi:10.1007/978-1-4614-1445-2_5
Stenseth, K., and Thyberg, J. J. (1989). Monensin and chloroquine inhibit transfer to lysosomes of endocytosed macromolecules in cultured mouse peritoneal macrophages. Eur. J. Cell Biol. 49 (2), 326–333. Available online at: https://europepmc.org/article/med/2776777
Sui, X., Chen, R., Wang, Z., Huang, Z., Kong, N., Zhang, M., et al. (2013). Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 4 (10), e838. doi:10.1038/cddis.2013.350
Taddei, M. L., Giannoni, E., Fiaschi, T., and Chiarugi, P. (2012). Anoikis: an emerging hallmark in health and diseases. J. Pathol. 226 (2), 380–393. doi:10.1002/path.3000
Takahashi, Y., Coppola, D., Matsushita, N., Cualing, H. D., Sun, M., Sato, Y., et al. (2007). Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 9 (10), 1142–1151. doi:10.1038/ncb1634
Takamura, A., Komatsu, M., Hara, T., Sakamoto, A., Kishi, C., Waguri, S., et al. (2011). Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 25 (8), 795–800. doi:10.1101/gad.2016211
Takano, N., Hiramoto, M., Yamada, Y., Kokuba, H., Tokuhisa, M., Hino, H., et al. (2023). Azithromycin, a potent autophagy inhibitor for cancer therapy, perturbs cytoskeletal protein dynamics. Br. J. Cancer 128 (10), 1838–1849. doi:10.1038/s41416-023-02210-4
Takatsuka, C., Inoue, Y., Matsuoka, K., and Moriyasu, Y. (2004). 3-methyladenine inhibits autophagy in tobacco culture cells under sucrose starvation conditions. Plant Cell Physiol. 45 (3), 265–274. doi:10.1093/pcp/pch031
Tasdemir, E., Maiuri, M. C., Galluzzi, L., Vitale, I., Djavaheri-Mergny, M., D’Amelio, M., et al. (2008). Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 10 (6), 676–687. doi:10.1038/ncb1730
Tedesco, B., Vendredy, L., Timmerman, V., and Poletti, A. (2023). The chaperone-assisted selective autophagy complex dynamics and dysfunctions. Autophagy 19 (6), 1619–1641. doi:10.1080/15548627.2022.2160564
Tettamanti, G., Carata, E., Montali, A., Dini, L., and Fimia, G. M. (2019). Autophagy in development and regeneration: role in tissue remodelling and cell survival. Eur. Zool. J. 86 (1), 113–131. doi:10.1080/24750263.2019.1601271
Thoreen, C. C., Kang, S. A., Chang, J. W., Liu, Q., Zhang, J., Gao, Y., et al. (2009). An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284 (12), 8023–8032. doi:10.1074/jbc.M900301200
Tian, X., He, Y., Qi, L., Liu, D., Zhou, D., Liu, Y., et al. (2023). Autophagy inhibition contributes to apoptosis of PLK4 downregulation-induced dormant cells in colorectal cancer. Int. J. Biol. Sci. 19 (9), 2817–2834. doi:10.7150/ijbs.79949
Tian, Y., Shen, L., Yang, J., Wan, X., and Ouyang, M. (2020). Silencing of RHEB inhibits cell proliferation and promotes apoptosis in colorectal cancer cells via inhibition of the mTOR signaling pathway. J. Cell Physiol. 235 (1), 442–453. doi:10.1002/jcp.28984
Tiwari, M., Srivastava, P., Abbas, S., Jegatheesan, J., Ranjan, A., Sharma, S., et al. (2024). Emerging role of autophagy in governing cellular dormancy, metabolic functions, and therapeutic responses of cancer stem cells. Cells 13 (5), 447. doi:10.3390/cells13050447
Tonkin-Reeves, A., Giuliani, C. M., and Price, J. T., (2023). Inhibition of autophagy; an opportunity for the treatment of cancer resistance, Front. Cell Dev. Biol. 11, 1177440. doi:10.3389/fcell.2023.1177440
Tooze, S. A., Abada, A., and Elazar, Z. J. (2014). Endocytosis and autophagy: exploitation or cooperation? Cold Spring Harb. Perspect. Biol. 6 (5), a018358. doi:10.1101/cshperspect.a018358
Tran, T. H., and Phuong Tran, T. T. (2022). Targeting the PD-1/PD-L1 axis for cancer treatment: a review on nanotechnology. R. Soc. Open Sci. 9 (4), 211991. doi:10.1098/rsos.211991
Tsukada, M., and Ohsumi, Y. J. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333 (1-2), 169–174. doi:10.1016/0014-5793(93)80398-e
Tufail, M., Jiang, C.-H., and Li, N. J. (2025). Tumor dormancy and relapse: understanding the molecular mechanisms of cancer recurrence. Mil. Med. Res. 12 (1), 7. doi:10.1186/s40779-025-00595-2
Valentim, L., Laurence, K. M., Townsend, P. A., Carroll, C. J., Soond, S., Scarabelli, T. M., et al. (2006). Urocortin inhibits Beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J. Mol. Cell Cardiol. 40 (6), 846–852. doi:10.1016/j.yjmcc.2006.03.428
Van Hove, L., Toniolo, A., Ghiasloo, M., Lecomte, K., Boone, F., Ciers, M., et al. (2023). Autophagy critically controls skin inflammation and apoptosis-induced stem cell activation. Autophagy 19 (11), 2958–2971. doi:10.1080/15548627.2023.2247742
Varisli, L., Cen, O., and Vlahopoulos, S. J. I. (2020). Dissecting pharmacological effects of chloroquine in cancer treatment: interference with inflammatory signaling pathways. Immunology 159 (3), 257–278. doi:10.1111/imm.13160
Vats, S., and Manjithaya, R. J. (2019). A reversible autophagy inhibitor blocks autophagosome–lysosome fusion by preventing Stx17 loading onto autophagosomes. Mol. Biol. Cell 30 (17), 2283–2295. doi:10.1091/mbc.E18-08-0482
Vera-Ramirez, L., Vodnala, S. K., Nini, R., Hunter, K. W., and Green, J. E. (2018). Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat. Commun. 9 (1), 1944. doi:10.1038/s41467-018-04070-6
Verbaanderd, C., Maes, H., Schaaf, M. B., Sukhatme, V. P., Pantziarka, P., Sukhatme, V., et al. (2017). Repurposing drugs in oncology (ReDO)-chloroquine and hydroxychloroquine as anti-cancer agents, Ecancermedicalscience 11, 781. doi:10.3332/ecancer.2017.781
Verma, A. K., Bharti, P. S., Rafat, S., Bhatt, D., Goyal, Y., Pandey, K. K., et al. (2021). Autophagy paradox of cancer: role, regulation, and duality. Oxid. Med. Cell. Longev. 2021 (1), 8832541. doi:10.1155/2021/8832541
Vessoni, A., Filippi-Chiela, E. C., Menck, C. F., and Lenz, G. (2013). Autophagy and genomic integrity. Cell Death Differ. 20 (11), 1444–1454. doi:10.1038/cdd.2013.103
Vitto, V. A. M., Bianchin, S., Zolondick, A. A., Pellielo, G., Rimessi, A., Chianese, D., et al. (2022). Molecular mechanisms of autophagy in cancer development, progression, and therapy. Biomedicines 10 (7), 1596. doi:10.3390/biomedicines10071596
Vucicevic, L., Misirkic, M., Janjetovic, K., Vilimanovich, U., Sudar, E., Isenovic, E., et al. (2011). Compound C induces protective autophagy in cancer cells through AMPK inhibition-independent blockade of Akt/mTOR pathway. Autophagy 7 (1), 40–50. doi:10.4161/auto.7.1.13883
Walter, C., Clemens, L. E., Müller, A. J., Fallier-Becker, P., Proikas-Cezanne, T., Riess, O., et al. (2016). Activation of AMPK-induced autophagy ameliorates Huntington disease pathology in vitro, Neuropharmacology 108, 24–38. doi:10.1016/j.neuropharm.2016.04.041
Wang, E. S., Fay, H. R. S., Dykstra, K. M., Phelps, M. N., Johnson, M., Harrigan, A., et al. (2024). Autophagy inhibition induces AML cell death and enhances the efficacy of chemotherapy under hypoxia. doi:10.1101/2024.11.24.625107
Wang, C.-W., and Klionsky, D. J. J. M. m., (2003). The molecular mechanism of autophagy, Mol. Med. 9, 65–76. doi:10.1007/bf03402040
Wang, C.-Z., Deng, F., Li, H., Wang, D. D., Zhang, W., Ding, L., et al. (2018). MiR-101: A potential therapeutic target of cancers. Am. J. Transl. Res. 10 (11), 3310–3321. Available online at: www.ajtr.org
Wang, H., Huang, C., Cai, H., and Ling, Q. (2024). The role of autophagy related 12 (ATG12) in the progression of hepatocellular carcinoma and its prognostic value. Discov. Oncol. 15 (1), 842–916. doi:10.1007/s12672-024-01731-5
Wang, J., Kaplan, N., Wang, S., Yang, W., Wang, L., He, C., et al. (2020). Autophagy plays a positive role in induction of epidermal proliferation. FASEB J. 34 (8), 10657–10667. doi:10.1096/fj.202000770RR
Wang, L., Klionsky, D. J., and Shen, H.-M. (2023). The emerging mechanisms and functions of microautophagy. Nat. Rev. Mol. Cell Biol. 24 (3), 186–203. doi:10.1038/s41580-022-00529-z
Wang, L., Wu, W., Chen, J., Li, Y., Xu, M., and Cai, Y. (2019). miR-122 and miR-199 synergistically promote autophagy in oral lichen planus by targeting the Akt/mTOR pathway. Int. J. Mol. Med. 43 (3), 1373–1381. doi:10.3892/ijmm.2019.4068
Wang, X., Lee, J., and Xie, C. J. C. (2022). Autophagy regulation on cancer stem cell maintenance, metastasis, and therapy resistance. Cancers (Basel). 14 (2), 381. doi:10.3390/cancers14020381
Wang, Y., Kuramitsu, Y., Baron, B., Kitagawa, T., Tokuda, K., Akada, J., et al. (2016). PI3K inhibitor LY294002, as opposed to wortmannin, enhances AKT phosphorylation in gemcitabine-resistant pancreatic cancer cells. Int. J. Oncol. 50 (2), 606–612. doi:10.3892/ijo.2016.3804
Wang, Y., and Qin, Z.-h.J. A. P. S. (2013). Coordination of autophagy with other cellular activities. Acta Pharmacol. Sin. 34 (5), 585–594. doi:10.1038/aps.2012.194
Wang, Z.-h., Xu, L., Duan, Z. l., Zeng, L. q., Yan, N. h., and Peng, Z. l. (2007). Beclin 1-mediated macroautophagy involves regulation of caspase-9 expression in cervical cancer HeLa cells. Gynecol. Oncol. 107 (1), 107–113. doi:10.1016/j.ygyno.2007.05.034
Wei, H., Wei, S., Gan, B., Peng, X., Zou, W., and Guan, J. L. (2011). Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 25 (14), 1510–1527. doi:10.1101/gad.2051011
Witzig, T. E., Reeder, C., Han, J. J., LaPlant, B., Stenson, M., Tun, H. W., et al. (2015). The mTORC1 inhibitor everolimus has antitumor activity in vitro and produces tumor responses in patients with relapsed T-cell lymphoma. Blood 126 (3), 328–335. doi:10.1182/blood-2015-02-629543
Wu, L., Jin, W., Yu, H., and Liu, B., (2024). Modulating autophagy to treat diseases: a revisited review on in silico methods, J. Adv. Res. 58, 175–191. doi:10.1016/j.jare.2023.05.002
Wu, S., and Sun, J. (2011). Vitamin D, vitamin D receptor, and macroautophagy in inflammation and infection. Discov. Med. 11 (59), 325–335.
Wu, Y. T., Tan, H. L., Shui, G., Bauvy, C., Huang, Q., Wenk, M. R., et al. (2010). Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J. Biol. Chem. 285 (14), 10850–10861. doi:10.1074/jbc.M109.080796
Xia, H., Li, S., Li, X., Wang, W., Bian, Y., Wei, S., et al. (2020). Autophagic adaptation to oxidative stress alters peritoneal residential macrophage survival and ovarian cancer metastasis. JCI Insight 5 (18), e141115. doi:10.1172/jci.insight.141115
Xia, H.-g., Zhang, L., Chen, G., Zhang, T., Liu, J., Jin, M., et al. (2010). Control of basal autophagy by calpain1 mediated cleavage of ATG5. Autophagy 6 (1), 61–66. doi:10.4161/auto.6.1.10326
Xia, Q., Zheng, Y., Jiang, W., Huang, Z., Wang, M., Rodriguez, R., et al. (2016). Valproic acid induces autophagy by suppressing the Akt/mTOR pathway in human prostate cancer cells. Oncol. Lett. 12 (3), 1826–1832. doi:10.3892/ol.2016.4880
Yamamoto, K., Venida, A., Yano, J., Biancur, D. E., Kakiuchi, M., Gupta, S., et al. (2020). Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 581 (7806), 100–105. doi:10.1038/s41586-020-2229-5
Yan, L.-X., Huang, X. F., Shao, Q., Huang, M. Y., Deng, L., Wu, Q. L., et al. (2008). MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 14 (11), 2348–2360. doi:10.1261/rna.1034808
Yang, A., Herter-Sprie, G., Zhang, H., Lin, E. Y., Biancur, D., Wang, X., et al. (2018). Autophagy sustains pancreatic cancer growth through both cell-autonomous and nonautonomous mechanisms. Cancer Discov. 8 (3), 276–287. doi:10.1158/2159-8290.CD-17-0952
Yang, T., Zhang, Y., Chen, J., and Sun, L., (2023). Crosstalk between autophagy and immune cell infiltration in the tumor microenvironment, Front. Med. 10, 1125692. doi:10.3389/fmed.2023.1125692
Yang, X., Yu, D. D., Yan, F., Jing, Y. Y., Han, Z. P., Sun, K., et al. (2015). The role of autophagy induced by tumor microenvironment in different cells and stages of cancer. Cell Biosci. 5, 14–11. doi:10.1186/s13578-015-0005-2
Yang, Y.-p., Hu, L. f., Zheng, H. f., Mao, C. j., Hu, W. d., Xiong, K. p., et al. (2013). Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacol. Sin. 34 (5), 625–635. doi:10.1038/aps.2013.5
Yang, Z., and Klionsky, D. J. J. C. o.i.c.b. (2010). Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22 (2), 124–131. doi:10.1016/j.ceb.2009.11.014
Yang, Z. J., Chee, C. E., Huang, S., and Sinicrope, F. A. (2011). The role of autophagy in cancer: therapeutic implications. Mol. Cancer Ther. 10 (9), 1533–1541. doi:10.1158/1535-7163.MCT-11-0047
Yano, K., Yanagisawa, T., Mukae, K., Niwa, Y., Inoue, Y., and Moriyasu, Y. (2015). Dissection of autophagy in tobacco BY-2 cells under sucrose starvation conditions using the vacuolar H+-ATPase inhibitor concanamycin A and the autophagy-related protein Atg8. Plant Signal. Behav. 10 (11), e1082699. doi:10.1080/15592324.2015.1082699
Yi, Y., Zhou, Z., Shu, S., Fang, Y., Twitty, C., Hilton, T. L., et al. (2012). Autophagy-assisted antigen cross-presentation: autophagosome as the argo of shared tumor-specific antigens and DAMPs. Oncoimmunology 1 (6), 976–978. doi:10.4161/onci.20059
Yin, J., Zhou, Z., Chen, J., Wang, Q., Tang, P., Ding, Q., et al. (2019). Edaravone inhibits autophagy after neuronal oxygen-glucose deprivation/recovery injury. Int. J. Neurosci. 129 (5), 501–510. doi:10.1080/00207454.2018.1550399
Yin, Y., Fan, Y., Yu, G., and Du, Y. (2021). LAPTM4B promotes the progression of bladder cancer by stimulating cell proliferation and invasion. Oncol. Lett. 22 (5), 765. doi:10.3892/ol.2021.13026
Yoshimori, T. (2004). Autophagy: a regulated bulk degradation process inside cells. Biochem. Biophys. Res. Commun. 313 (2), 453–458.
Yue, Z., Jin, S., Yang, C., Levine, A. J., and Heintz, N. (2003). Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. U. S. A. 100 (25), 15077–15082. doi:10.1073/pnas.2436255100
Yun, C. W., Jeon, J., Go, G., Lee, J. H., and Lee, S. H. (2020). The dual role of autophagy in cancer development and a therapeutic strategy for cancer by targeting autophagy. Int. J. Mol. Sci. 22 (1), 179. doi:10.3390/ijms22010179
Yusuf, I. H., Charbel Issa, P., and Ahn, S. J. (2023). Hydroxychloroquine-induced Retinal Toxicity. Front Pharmacol. 14, 1196783. doi:10.3389/fphar.2023.1196783
Zaarour, R. F., Azakir, B., Hajam, E. Y., Nawafleh, H., Zeinelabdin, N. A., Engelsen, A. S. T., et al. (2021). Role of hypoxia-mediated autophagy in tumor cell death and survival. Cancers (Basel). 13 (3), 533. doi:10.3390/cancers13030533
Zeuner, A. (2015). The secret life of quiescent cancer stem cells. Mol. Cell Oncol. 2 (1), e968067. doi:10.4161/23723548.2014.968067
Zhang, J., Wu, L., Ding, R., Deng, X., and Chen, Z. (2024). Role of miRNA-122 in cancer. Int. J. Oncol. 65 (3), 83. doi:10.3892/ijo.2024.5671
Zhang, Q., Wang, X., Cao, S., Sun, Y., He, X., Jiang, B., et al. (2020b). Berberine represses human gastric cancer cell growth in vitro and in vivo by inducing cytostatic autophagy via inhibition of MAPK/mTOR/p70S6K and Akt signaling pathways. p70S6K Akt Signal. Pathw. 128, 110245. doi:10.1016/j.biopha.2020.110245
Zhang, S., Sheng, H., Zhang, X., Qi, Q., Chan, C. B., Li, L., et al. (2019). Cellular energy stress induces AMPK-mediated regulation of glioblastoma cell proliferation by PIKE-A phosphorylation. Cell Death Dis. 10 (3), 222. doi:10.1038/s41419-019-1452-1
Zhang, X., Chen, X., Guo, Y., Jia, H.-r., Jiang, Y. W., and Wu, F. G. (2020a). Endosome/lysosome-detained supramolecular nanogels as an efflux retarder and autophagy inhibitor for repeated photodynamic therapy of multidrug-resistant cancer. Nanoscale Horiz. 5 (3), 481–487.
Zhang, Y., Xu, Y. Y., Yao, C. B., Li, J. T., Zhao, X. N., Yang, H. B., et al. (2017). Acetylation targets HSD17B4 for degradation via the CMA pathway in response to estrone. Autophagy 13 (3), 538–553. doi:10.1080/15548627.2016.1268302
Zhao, X., Ma, D., Yang, B., Wang, Y., and Zhang, L., (2024). Research progress of T cell autophagy in autoimmune diseases, Front. Immunol. 15, 1425443. doi:10.3389/fimmu.2024.1425443
Zheng, K., Kitazato, K., and Wang, Y. (2019). Selective autophagy regulates cell cycle in cancer therapy. Theranostics 9 (1), 104–125. doi:10.7150/thno.30308
Zheng, X., Carstens, J. L., Kim, J., Scheible, M., Kaye, J., Sugimoto, H., et al. (2015). Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527 (7579), 525–530. doi:10.1038/nature16064
Zhou, H. J., Yan, J., Luo, W., Ayala, G., Lin, S. H., Erdem, H., et al. (2005). SRC-3 is required for prostate cancer cell proliferation and survival. Cancer Res. 65 (17), 7976–7983. doi:10.1158/0008-5472.CAN-04-4076
Zhu, H., Wu, H., Liu, X., Lin, B., Chen, Y., Ren, X., et al. (2009). Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy 5 (6), 816–823. doi:10.4161/auto.9064
Glossary
3-MA 3-methyladenine
AICAR 5-Aminoimidazole-4-carboxamide ribonucleotide
AMPK AMP-activated protein kinase
ATG autophagy related
BAG Bcl2-associated athanogene
Bcl2 B-cell leukemia/lymphoma 2
BIF1 Bax-interacting factor 1
CDK Cyclin-dependent protein kinase
CMA Chaperone-mediated autophagy
CQ chloroquine
CSCs Cancer stem cells
eMI endosomal microautophagy
EMT epithelial to mesenchymal transition
ER endoplasmic reticulum
ESCRT endosomal sorting complex required for transport
FIP200 Focal adhesion kinase family interacting protein of 200 kD
HCQ hydroxychloroquine
HIF1 Hypoxia inducible factor 1
Hip Hsc70-interacting protein
Hop Hsc70-Hsp90 organizing protein
HSP heat shock protein
ILV intralumenal vesicles
KFERQ Lysine, phenylalanine, Glutamic acid, Arginine, Glutamine
LAMP-2A Lysosome-associated membrane protein type 2a
LC3 Microtubule-associated protein light chain 3
LE/MVB late endosomes/multivesicular bodies
mTOR mechanistic target of rapamycin
PE phosphatidylethanolamine
PEG-PLE methoxy-poly (ethylene glycol)-poly (L-glutamic acid sodium salt) (PEG-PLE)
PI3K Phosphoinositide 3-kinase
PI3P Phosphoinositide-3-phosphate
SENDA static encephalopathy of childhood with neurodegeneration in adulthood
SQSTM1 sequestosome
TCA tricarboxylic acid cycle
TME tumor microenvironment
TPPS tetraphenylporphinesulfonate
ULK1 unc-51-like autophagy activating kinase 1
UVRAG Ultraviolet radiation resistance-associated gene.
Keywords: autophagy, autophagy inhibitors, autophagy activators, autophagy modulation cancer, cancer therapeutics
Citation: Kumar P, Choudhary A, Kinger S, Jagtap YA, Prajapati VK, Chitkara D, Chinnathambi S, Verma RK and Mishra A (2025) Autophagy as a potential therapeutic target in regulating improper cellular proliferation. Front. Pharmacol. 16:1579183. doi: 10.3389/fphar.2025.1579183
Received: 18 February 2025; Accepted: 24 April 2025;
Published: 15 May 2025.
Edited by:
Valerie R. Wiersma, University Medical Center Groningen, NetherlandsReviewed by:
Santanu Maji, Virginia Commonwealth University, United StatesZhiwei Xu, Jiangsu University, China
Copyright © 2025 Kumar, Choudhary, Kinger, Jagtap, Prajapati, Chitkara, Chinnathambi, Verma and Mishra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amit Mishra, YW1pdEBpaXRqLmFjLmlu
 Prashant Kumar1
Prashant Kumar1 Vijay Kumar Prajapati
Vijay Kumar Prajapati Rahul Kumar Verma
Rahul Kumar Verma Amit Mishra
Amit Mishra