Abstract
The Sec61 complex, which is located on the membrane of the mammalian endoplasmic reticulum (ER), serves as a pivotal component of protein transport channels. It plays a central role in the transport of nascent peptides and precursor peptides to the ER. This process includes the directed movement of precursor peptides to the ER membrane and the opening of the Sec61 transduction channel for translocation. The Sec61 channel not only plays a key role in transporting peptides into cells but also acts as a passive ER Ca2+ leak channel. In addition, the mutation, amplification and overexpression of Sec genes are closely related to the development of various genetic diseases and cancers. Over the past few decades, studies have elucidated the function of the Sec61 protein in the pathogenesis of diseases such as cancer, and Sec61 inhibitors have been developed for their treatment. This review describes the structure of Sec61 and its function in transporting ER transmembrane proteins and further summarizes the role of this gene in disease and recent advancements in Sec61 inhibitors. This study provides novel insights into the involvement of Sec61 in disease etiology and lays the groundwork for future treatment modalities targeting this pivotal protein complex.
1 Introduction
In eukaryotic cells, the folding and assembly of most secreted proteins and transmembrane proteins are completed in the endoplasmic reticulum (ER). The entry of nascent peptides into and across the ER membrane is a highly conserved process that requires the participation of various translocation proteins, including the ER channel protein Sec61 (Lang et al., 2019). Depending on the translocation of precursor proteins during or after ribosome synthesis, their transport across the endoplasmic reticulum membrane can be classified into two modes: cotranslational transport (Rapoport et al., 1996; Matlack et al., 1998) and posttranslational transport (Schlenstedt and Zimmermann, 1987; Kutay et al., 1995) (Figure 1A). During cotranslational transport, the signal peptide (SP) emerging from the ribosome is recognized by the cytosolic signal recognition particle (SRP), which causes translation to slow down. Then, SRP interacts with the SRP receptor (SR) located in the ER membrane to form a ribosome - nascent chain complex, which is guided to the Sec61 complex on the ER membrane (Rapoport et al., 2017). Translation resumes, and the nascent polypeptide chain enters the ER lumen through the Sec61 channel. The SP is cleaved by a signal peptidase located in the ER lumen, and the polypeptide chain undergoes folding and modification within the ER. Meanwhile, guanosine triphosphate (GTP) hydrolysis enables SRP and SR to return to their original state, preparing for the next round of cotranslational translocation (Haßdenteufel et al., 2019). In the posttranslational transport pathway, secretory proteins with fewer hydrophobic signal sequences bypass the SRP and complete translation on cytoplasmic free ribosomes (Schorr et al., 2020; Schlenstedt et al., 1990). Subsequently, signal proteins are guided into the ER by the Sec61 channel and the Sec62/Sec63 complex, after which they undergo folding and covalent modification (Deshaies et al., 1991; Kreil, 1981). In this process, as an endoplasmic reticulum lumen chaperone protein, the immunoglobulin heavy chain binding protein BiP/GRP78 acts as a “one-way valve” to ensure the unidirectional transport of signal proteins to ER tubes through the Sec61 channel (Dudek et al., 2009; Alder et al., 2005). This paper reviews the function of the Sec61 protein in cotranslational transport and further focuses on recent studies that provided the first insights into the functional role and therapeutic relevance of Sec61 in human diseases.
FIGURE 1
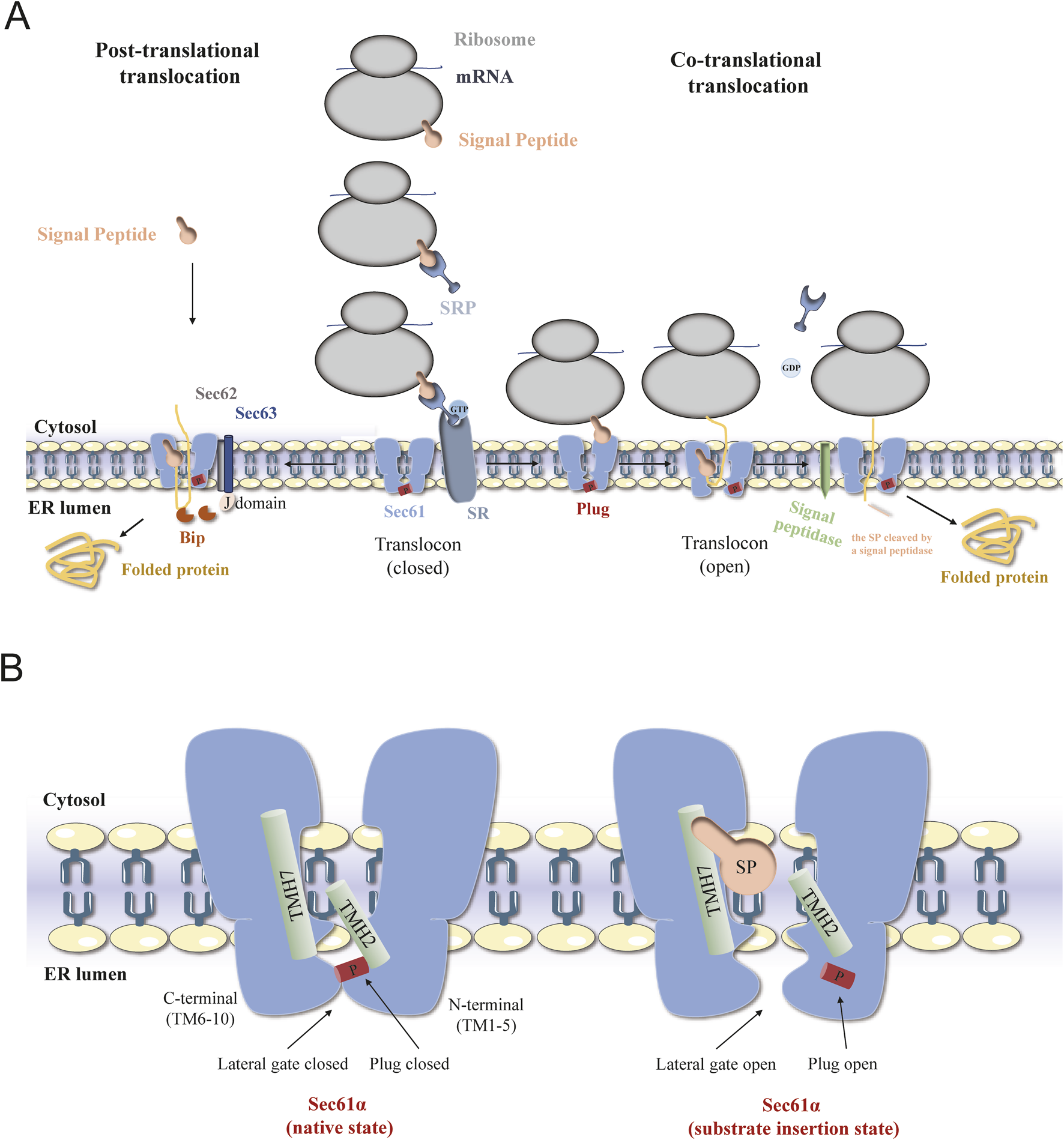
The schematic diagram of Sec61 translocation process and translocation conformations. (A) Schematic diagrams of the mechanisms of posttranslational transport (left) and cotranslational transport (right) of proteins mediated by the Sec61 complex in eukaryotic cells. (B) Overview of different structural states of SEC61α translocon. Conformation of Sec61α in the Sec61 complex in the resting state (left) and during translocation (right).
2 Structure and function of Sec61
The eukaryotic Sec61 complex is composed of three subunits, Sec61α, Sec61β and Sec61γ (referred to as Sec61p, Sbh1 and Sss1, respectively, in Saccharomyces cerevisiae) (Van den Berg et al., 2004). The Sec61α and Sec61γ subunit sequences are highly conserved and are critical for protein transport and cell viability, while the Sec61β subunit has low sequence homology and a limited role in the function of the channel (Tsukazaki et al., 2008). Sec61α, the largest subunit, contains ten transmembrane domains (TMDs). Transmembrane helices (TMs) 1-5 and TMs 6–10 form the central pore plug - like domains. These TM domains collectively create a clam - shaped pore with a central ring, facilitating protein translocation. On the luminal side, a short helix (TM2a) forms a displaceable plug domain that can seal the pore (Heinrich et al., 2000). Initial photocrosslinking experiments showed that the Sec61α subunit was surrounded by the peptide chain as it passed through the channel (Mothes et al., 1994). Sec61β and Sec61γ are single transmembrane proteins that belong to the tail-anchored protein family. The Sec61 channel exhibits a characteristic clamshell-like topology, with the central pore opening into the lipid phase through a lateral gate formed between TM2 and TM7 (Li et al., 2016). The channel in the Sec61 complex has a larger opening at the side gate to facilitate the passage of signal sequences in the form of α-helixes. Currently, the cryo-electron microscopic structure of the Sec61 complex has been used to determine several functional conformations (Bai et al., 2020). A comparison of the open, shifted, and idle conformations of Sec61α suggested that changes in the lateral gate conformation may be due to an unstable hydrogen bond network of pore ring residues located on TM2, TM5, TM7, and TM10. According to the two cryo-EM images of the mammalian ribosome-bound Sec61 complex (cotranslational mode), a hydrophobic signal peptide was observed to occupy the space between TM2 and TM7 and was eventually inserted into the lipid bilayer with a helical structure (Weng et al., 2021). To analyze the conformational changes in the Sec61 channel, Sun et al. conducted molecular dynamics (MD) simulations of the mammalian Sec61 channel and revealed that after the signal peptide chain enters the lipid bilayer, the side gate can quickly return to the partially closed state, and the conformational dynamics of the side gate, the pore loop and the plug domain are interrelated (Sun et al., 2019). Molecular docking and experimental results indicated that the hydrophobic core of the SRP substrate-dependent signal anchor was more inclined to occupy the space between the C-terminus of TM2 and the N-terminus of TM7 than was the hydrophobic core of the SRP substrate-independent signal peptide (Salmaso and Moro, 2018; Bhadra and Helms, 2021). These results indicate that the translocation process is dependent on the interaction of the targeting sequence with the side gate (Figure 1B).
The core Sec61 complex is highly dynamic. It can interact with a variety of molecular machines and enzymes to form multiple unique subcomplexes, each with distinct client proteins. These subcomplexes include key players in canonical translocation like the translocon - associated protein complex (TRAP), translocating - chain - associating membrane protein (TRAM), oligosaccharyltransferase (OST), and ribosome (Itskanov and Park, 2023; Pfeffer et al., 2014). There are also recently identified ones such as the EMC (ER membrane protein complex), GEL (Guided entry of tail-anchored proteins and EMC-Like) complex, PAT (protein associated with translocon), and BOS (Back of Sec61) complexes (Page et al., 2024). Studies show that in the biogenesis of tail - anchored (TA) proteins via either the GET or EMC pathway, the hydrophobicity of the transmembrane domain (TMD) is the key determinant, which is especially important for the insertion of multi - pass membrane proteins, including GPCRs. Notably, the EMC complex can substitute for Sec61 in inserting type III membrane proteins (Page et al., 2024). By associating with these complexes, the Sec61 complex is activated, a driving force for translocation is generated, and polypeptide translocation is coupled with other processes such as translation, post - translational modifications, and protein folding. Thus, the Sec61 complex has evolved to be regulated in a substrate - and partner complex - dependent manner to ensure the efficiency and accuracy of protein transport and insertion.
The Sec61 complex on the ER membrane serves as the primary entry channel for nascent polypeptides and possesses three protein transport functions: (1) by forming a protein transport channel; (2) by recognizing a functional signal sequence; and (3) by serving as the main ribosome receptor. During the ER targeting and transport processes, Sec61 enters the open state and interacts with a variety of other protein complexes on the cytoplasmic surface and on the ER membrane. This process is promoted not only by its substrate, amino terminus or transmembrane helical region of the signal peptide (SP) but also by translation transport ribosomes, TRAP and Sec62/Sec63 complex (Kalies et al., 2008; Almagro Armenteros et al., 2019; Voorhees and Hegde, 2016; Pfeffer et al., 2017; Itskanov et al., 2021). If proteins are not correctly folded in the ER, they trigger the unfolded protein response (UPR), or they are transported back to the proteasome for ER-associated protein degradation (ERAD) (Pilla et al., 2017; Moon et al., 2018; Fregno and Molinari, 2019). In addition, the Sec61 channel is considered a passive Ca2+ leak channel on the ER membrane that allows Ca2+ efflux from the ER in all nucleated cells (Van Coppenolle et al., 2004). In the case of severe and prolonged protein misfolding and aggregation, BiP isolates misfolded and aggregated peptides, leading to sustained Ca2+ leakage through the open Sec61 channel. Research indicates that various human genetic diseases and tumor diseases are caused by Sec61 point mutations and are associated with Sec61 channel gating dysfunction, which will be reviewed in detail later (Lang et al., 2017; Sicking et al., 2021; Linxweiler et al., 2017).
3 Sec61 proteins and diseases
In recent years, mutations and overexpression of Sec61 have been linked to numerous human diseases.
3.1 SEC61 mutation and genetic diseases
Diabetes mellitus (DM) is a metabolic disorder characterized by elevated blood glucose levels due to either an inadequate response to insulin or insufficient insulin production. According to a recent study on diabetes from 1999 to 2022, the global number of adults with diabetes reached 828 million in 2022, over four times the 1990 number. In terms of countries, in 2022, of the 828 million adults with diabetes globally, 212 million were in India (over a quarter), 148 million in China, 42 million in the US, 36 million in Pakistan, 25 million in Indonesia, and 22 million in Brazil (NCD Risk Factor Collaboration NCD-RisC, 2024). Proinsulin enters the ER through the Sec61-mediated cotranslational transport pathway (Liu et al., 2014). A mutation at the Y344H site of the SEC61A1 gene (where histidine at position 344 is replaced by tyrosine) can cause ER stress in the pancreatic islets of C57BL/6 mice. This leads to the apoptosis of pancreatic islet β-cells, results in insufficient insulin secretion, and eventually causes diabetes (Lloyd et al., 2010). However, when screening substrates, researchers have shown that compared with normal mice, heterozygous SEC61α+/Y344 and homozygous SEC61αY344H/Y344H mice exhibit reduced expression levels of the ERj3 protein in pancreatic and hepatic tissues (Schorr et al., 2020). In HeLa cells, when wild-type SEC61α is replaced with the corresponding mutant SEC61αY344H, ER calcium leakage increases and is no longer affected by the BiP concentration (Schäuble et al., 2012). This finding suggested that BiP normally mediates the closure of the Sec61 channel to limit Ca2+ leakage from the ER. Therefore, in different tissues of adult mice, ERj3 was confirmed to enter the mammalian ER by interacting with Sec61α TM7 in a BiP-dependent manner. Notably, many studies have confirmed that mutations and deletions of other resident ER proteins can also affect the biosynthesis of proinsulin and insulin and contribute to the development of DM, such as the deletion of the Hsp40-type accessory chaperones ERj4 and ERj5 of BiP or mutations of the BiP-interacting proteins proline-rich receptor-like protein kinase (PERK) and TRAP (Fritz et al., 2014; Dong et al., 2008; Harding et al., 2001; Huang et al., 2021).
Autosomal-dominant tubulo-interstitial kidney disease (ADTKD) is a monogenic disease characterized by renal tubular damage and interstitial fibrosis without glomerular damage and can lead to chronic progressive loss of renal function, which inevitably leads to end-stage renal disease (Devuyst et al., 2019). A SEC61A1 heterozygous mutation was detected in an ADTKD family, as were missense mutations at V67G (located in the plug domain) and T185A (located near the TM 5-hole loop). Both of these mutations affect important functional and conserved residues in Sec61, thereby causing renal tubule atrophy. These data were confirmed in a study of zebrafish embryos; replacement of either of these two variants affects the development of the anterior kidney and results in a coiling defect in the anterior tubules, which is consistent with the renal tubular atrophy observed in patients (Bolar et al., 2016). Moreover, in HEK293 cells, these two mutants caused the Sec61 protein to aggregate into clumps in the ER and appear in the Golgi apparatus. This abnormal protein may be mislocated to the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) and subjected to ER-related protein degradation. Therefore, Sec61α is essential for the development and maintenance of tubular tissues in the nephron.
Common variable immune deficiency (CVID) is a group of diseases of different origins that are usually characterized by impaired B-cell differentiation and function, resulting in low levels of immunoglobulin production and leading to respiratory infections in patients with repeated severe infections of multiple systems (Schubert et al., 2018). SEC61A1 deficiency is one underlying cause of CVID and is attributed to the missense mutation Sec61α V85D (in TM2) and the premature stop nonsense mutation E381* (in TM8). In addition, the overexpression of Sec61α V85D in HeLa cells not only affected the cotranslational transport of proteins but also greatly increased Ca2+ leakage from the ER, precipitating ER stress and irreversible UPR. Despite normal peripheral B and T-cell subsets in the two mutations, the production of SEC61α V85D selectively impaired the survival of cells in the plasma cell lineage because SEC61A1 is a target gene of XBP1s during plasma cell differentiation. B-cell lines transformed with EBV harboring the SEC61A1 mutation secreted less immunoglobulin. Currently, patients are responding well to immunoglobulin replacement therapy.
Severe congenital neutropenia (SCN) includes a group of genetically heterogeneous congenital immune deficiencies that are characterized by the arrest of granulocyte production and differentiation at the promyelocytic stage. Starting in early childhood, the absolute number of circulating mature neutrophils is low, predisposing SCN patients to life-threatening and recurrent infections (Skokowa et al., 2017). In patients with autosomal dominant severe congenital neutropenia (ADSCN), the identified heterozygous SEC61A1 mutations included two missense mutations, which led to amino acid substitutions, namely, V67G (in the plug domain) and Q92R (in TM2) (Van Nieuwenhove et al., 2020). Interestingly, neutropenia was also observed in patients with ADTKD harboring the SEC61α V67G mutation. Similarly, patients with the Q92R mutation not only had features of the SCN but also had B-cell maturation defects. In contrast, the kidney morphology of patients with the Q92R mutation was normal, and renal function also remained normal. These two mutations were both observed to cause reduced cellular Sec61 levels due to protein instability and dysregulation of calcium homeostasis. In addition, in myeloid leukemia HL-60 cells, after replacement of wild-type Sec61α with the Q92R mutant, calcium leakage from the ER increased, and differentiation into CD11b+ and CD16+ cells decreased, suggesting that the UPR was dysregulated. This finding was confirmed by ex vivo single-cell analysis (Van Nieuwenhove et al., 2020) (Table 1). In addition to mutations at the SEC61A1 locus causing SCN, the SRP also plays a critical role in neutrophil development. It has been found that human genetic defects in SRP19, SRPRA, and SRP54 cause severe congenital neutropenia (Linder et al., 2023; Bellanné-Chantelot et al., 2018).
TABLE 1
| Sec61A mutation site | Mutation position | Primary disease | Pathogenic mechanism | References |
|---|---|---|---|---|
| Y344H | TM7 | DM | Pancreatic islets and islet β-cell apoptosis, insufficient insulin secretion | Schorr et al. (2020), Lloyd et al. (2010), Schäuble et al. (2012) |
| V67G | Located in the plug-like domain between TM1 and TM2 | ADTKD, SCN | Renal tubular atrophy, neutropenia | Bolar et al. (2016) |
| T185A | TM5 | ADTKD | Renal tubular atrophy | Bolar et al. (2016) |
| V85D | In TM2 | CVID | Normal peripheral B and T-cell subsets, reduced plasma cells and reduced immunoglobulin | Schubert et al. (2018) |
| E381* | In TM8 | CVID | Reduced immunoglobulin | Schubert et al. (2018) |
| Q92R | In TM2 | SCN, CVID | Other leukopenia, B-cell maturation defect, and the morphology and function of kidney were normal | Van Nieuwenhove et al. (2020) |
SEC61 mutation and genetic diseases.
3.2 Sec61 and cancer
There are many reports on the upregulation of SEC61 gene expression in malignant tumors. Sec61α is highly expressed in esophageal cancer, but this expression is not related to patient prognosis (Bachmann et al., 2019). Fan and his colleagues reported that the expression of Sec61β was significantly increased in colorectal cancer (CRC) patients. The Sec61β autoantibody level in the plasma of patients was also significantly greater than that in the plasma of healthy controls, suggesting that the Sec61β autoantibody can be used as a new serum marker of CRC, especially in the early stage (Fan et al., 2011). In addition, according to whole-exome sequencing of polycystic liver disease (PCLD), in addition to the two most common genes, PRKCHS and SEC63, deletion of the Sec61β gene results in severely reduced expression of polycystin-1, which is encoded by the PKD1 gene. It is also a pathogenic inducer of polycystic liver disease (Besse et al., 2017). Lu et al. used quantitative polymerase chain reaction (PCR) to measure gene-level changes in 43 human glioblastoma patients and reported that 77% of patients had increased expression of SEC61G, while the expression of the genes encoding SEC61A1 and SEC61B did not differ from that of healthy individuals (Lu et al., 2009). On the basis of the statistical analysis of sequencing data from the Cancer Genome Atlas (TCGA) and the Chinese Glioma Genome Atlas (CGGA) cohorts, researchers have shown that high SEC61G expression is significantly associated with poor prognosis in glioblastoma patients. SEC61G may be used as a new prognostic marker for predicting the survival and treatment response of patients with glioblastoma (Liu et al., 2019). In addition, SEC61γ is a prognostic marker for hepatocellular carcinoma (HCC). SEC61G expression is significantly upregulated in HCC and is associated with patient survival (Gao et al., 2020). By analyzing the COSMIC database, six SEC61γ mutations were found in highly conserved residues in eukaryotes. The R24I mutation was found in a patient with colorectal cancer, the K27E and I64T mutations were found in endometrial cancer patients, the A39V mutation was found in pancreatic cancer patients, and the L56F and H58R mutations were found in lung cancer patients (Witham et al., 2021). The cancer-associated SEC61γ mutants are proposed to alter ion transport across the channel, such as GSH, Mn2+ and Ca2+, yet they do not impair the ability of this protein complex to transfer secreted proteins to the ER (Witham et al., 2021) (Figure 2).
FIGURE 2
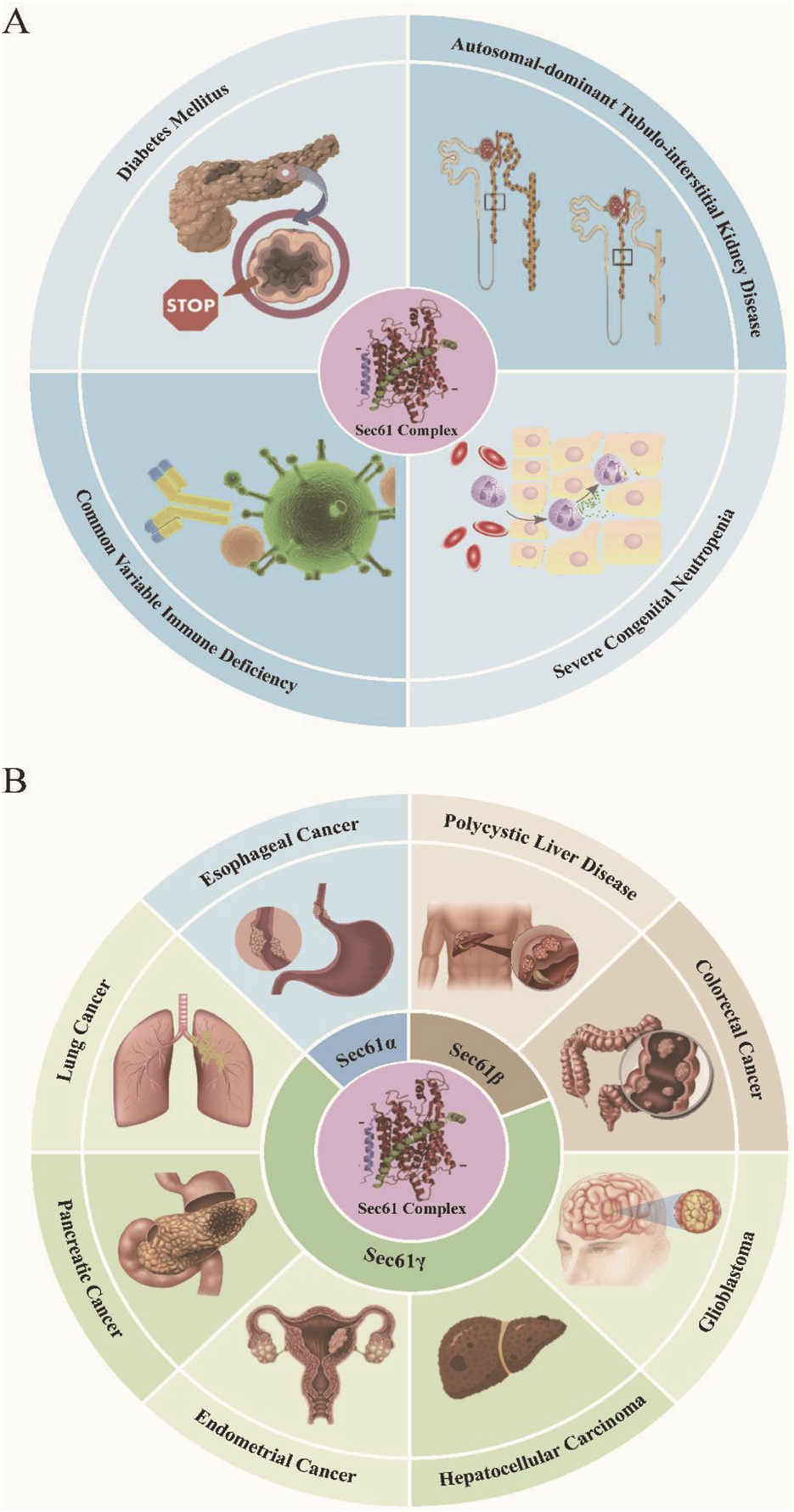
Mutations and overexpression of SEC61 gene in numerous human genetic disease (A) and cancer (B).
4 Small molecule inhibitors of Sec61
Studies have shown that selective inhibitors of secreted proteins can prevent cotranslational translocations by directly targeting Sec61 (Garrison et al., 2005). The correct translocation of proteins is critical for the normal functioning of cells. Since many inhibitors share the binding region of the Sec61α subunit (Paatero et al., 2016; Zong et al., 2019; Baron et al., 2016), Sec61α shows great potential as a molecular target for the treatment of various conditions, such as cancer, immune disorders, and viral infections (Pauwels et al., 2021a). These Sec61-dependent inhibitors are classified into substrate-selective inhibitors, such as HUN-7293, CAM741, Cotransin, and CADA, and broad-spectrum inhibitors, including Mycolactone, Exotoxin A, Apratoxin A, Coibamide A, Ipomoeassin F, Decatransin and Eeyarestatins (Table 2). These inhibitors have been proven to block the translocation of signal proteins and inhibit the leakage of Ca2+ through the Sec61 channel (Linxweiler et al., 2017; Haßdenteufel et al., 2018). In fact, SEC61a mutants that are resistant to one inhibitor usually also develop resistance to other inhibitors. Nevertheless, different inhibitors exhibit different specificities for transporters in different species and block the translocation of different types of substrates, which provides the possibility of developing therapeutically effective selective transporter modulators.
TABLE 2
| Inhibitor | Molecular formula | PubChem CID | Substrate selectivity | Chemical structures |
|---|---|---|---|---|
| HUN-7293 | C52H82N8O8 | 10931051 | VCAM-1,ICAM-1,E-selectin |

|
| CAM741 | C56H91N7O11 | 102353267 | VCAM-1,VEGF |

|
| Cotransin | C42H68N6O8 | 25068231 | Angiotensinogen,VCAM-1,p-selectin,β-lactamase,CRF1,ETBR,AQP2,TNFα,HER3 |

|
| CADA | C31H39N3O4S2 | 466371 | huCD4, SORT, DNAJC3, PTK7, ERLEC1, 4-1BB |

|
| Mycolactone | C44H70O9 | 5282079 | Broad-spectrum |

|
| Exotoxin A | C25H25N9O2 | 135345207 | Broad-spectrum |

|
| Apratoxin A | C45H69N5O8S | 6326668 | Broad-spectrum |

|
| Coibamide A | C65H110N10O16 | 24881184 | Broad-spectrum |

|
| Ipomoeassin F | C44H62O15 | 25258999 | Broad-spectrum |

|
| Decatransin | C63H109N9O12 | 166642447 | Broad-spectrum |

|
| Eeyaresatin | C27H25Cl2N7O7 | 5003929 | Broad-spectrum |

|
Chemical information for the Sec61 inhibitors.
4.1 SEC61 substrate-selective inhibitors
4.1.1 HUN-7293, CAM741 and cotransin
Cell adhesion molecules play an important role in the immune response by regulating the migration of leukocytes at sites of inflammation and interactions between cells. The first substrate-selective inhibitor discovered was HUN-7293, which inhibits the expression of three endothelial cell adhesion molecules: intercellular adhesion molecule 1 (ICAM-1), vascular cell adhesion molecule (VCAM-1), and E-selectin (Foster et al., 1994). CAM741 is an analog of Hun-7293 that is thought to interfere with the correct positioning of the VCAM - one signal peptide at the lateral gate upon its insertion into the translocase after being released from the SRP. Another substrate of CAM741 is the vascular endothelial growth factor (VEGF) (Harant et al., 2007). It can also prevent signal proteins from entering the lumen of the ER, indicating that this compound inhibits the signal peptide-dependent gating of the Sec61 channel (Westendorf et al., 2011). Another HUN-7293 analog named cotransin, inhibits the co-translational translocation of specific substrate proteins across the ER membrane. Numerous substrates have been identified for cotransin, including angiotensinogen, VCAM-1, p-selectin, β-lactamase, corticotropin-releasing factor 1 (CRF1), endothelin B receptor (ETBR), aquaporin 2 (AQP2), TNFα, and others (Garrison et al., 2005; Westendorf et al., 2011; Mackinnon et al., 2014; Klein et al., 2015; Maifeld et al., 2011). Considering the pivotal roles of VCAM-1, ICAM-1, and TNFα in cellular immune responses, HUN7293 and its related molecules CAM741 and cotransin are potentially promising as immunosuppressive agents (Maifeld et al., 2011). Additionally, cotransin has been found to target the oncoprotein human epidermal growth factor receptor 3 (HER3), which implies that cotransin may also possess anticancer activity (Ruiz-Saenz et al., 2015). Researchers have demonstrated the importance of Sec61 in supporting the replication of influenza A virus (IAV), human immunodeficiency virus (HIV), and dengue virus by using Cotransin to block the Sec61 channel, suggesting that inhibiting protein translocation across the ER is a potential antiviral strategy (Shah et al., 2018; Heaton et al., 2016).
4.1.2 Cyclotriazadisulfonamide
Cyclotriazadisulfonamide (CADA) is a synthetic small-molecule translocation inhibitor and the first such inhibitor found to directly bind to signal peptides. To date, six substrates of CADA have been identified (Claeys et al., 2021; Pauwels et al., 2021b): huCD4, SORT, DNAJC3, PTK7, ERLEC1, and 4-1BB (CD137), highlighting its substrate selectivity. By acting in a signal peptide-dependent manner, CADA inhibits the co-translational translocation of the type I integral membrane protein human CD4 (huCD4) across the endoplasmic reticulum (Vermeire et al., 2002). This mechanism allows CADA to specifically target proteins with signal peptides, thereby affecting their intracellular transport and localization. In addition, CADA can also significantly downregulate the expression of huCD4 on the surface of various cells, such as monocytes, T cells, and other lymphocytes (Pauwels et al., 2021b; Vermeire et al., 2002; Vermeire and Schols, 2003). As huCD4 is the primary receptor for HIV entry into host cells, CADA’s presence makes it difficult for HIV to bind to huCD4, thus inhibiting viral infection and replication and demonstrating significant antiviral effects (Vermeire et al., 2004). CADA also suppresses the expression of other membrane proteins related to viral entry, such as SORT, further enhancing its antiviral activity (Pauwels et al., 2021b). Moreover, CADA exerts immunosuppressive effects by inhibiting the secretion of multiple cytokines and reducing the levels of CD25, phosphorylated STAT5, and CTPS-1 (Claeys et al., 2021). Its impact is particularly pronounced in CD8+ T cell subsets, where it inhibits cell-mediated lympholysis. Notably, this effect is associated with CADA-induced upregulation of CD137, as CADA upregulates CD137 to suppress immune cell activation and function, thereby achieving an immunosuppressive effect (Claeys et al., 2021). Furthermore, CADA is associated with a reduction in progranulin - induced breast cancer stem cell proliferation, indicating its potential as an anticancer agent (Berger et al., 2021).
4.2 Sec61 broad-spectrum inhibitors
4.2.1 Mycolactone
Mycolactone inhibits the co-translational translocation of proteins into the ER through a mechanism that does not compromise the ER’s structural integrity (Hall et al., 2014). Secreted by the pathogen Mycobacterium ulcerans, this diffusible, lipid-like exotoxin forms a stable complex with Sec61a (Baron et al., 2016; Yotsu et al., 2018; Demangel and High, 2018). At nanomolar concentrations, it can block the co-translational transport of secreted proteins, such as various inflammatory mediators and cytokines, and inhibit the co-translational translocation of Sec61-dependent secreted proteins (McKenna et al., 2016). Mycolactone inhibits the translocation stage following ribosome contact with the translocon, affecting signal peptide interactions. This potent and stable inhibitor provides an opportunity to observe transporters in an inhibited state. Mycolactone can serve as a therapeutic drug with minimal side effects for multiple myeloma (MM), and it can effectively reduce the resistance of MM to proteasome inhibitors and immunomodulatory drugs. It induces ER stress in vitro, leading to the death of MM cell lines. In immunodeficient mice transplanted with MM cells, both primary and relapsed MM tumors were killed, and the growth of MM xenografts was delayed (Domenger et al., 2022).
4.2.2 Exotoxin A
Exotoxin A of Pseudomonas aeruginosa is a cytotoxic ADP-ribosyltransferase. It enters the cytoplasm of eukaryotic cells via endocytosis and retrograde transport. Moreover, it inhibits the retrograde export of immunogenic peptides from the endoplasmic reticulum to the cytoplasm (Zehner et al., 2015). Exotoxin A also competes with the cytoplasmic protein calmodulin (CaM) for binding to the N-terminus of Sec61α, thereby closing the Sec61 channel, preventing Ca2+ leakage, and terminating the cotranslational and posttranslational transport of signal proteins (Schäuble et al., 2014). Currently, the effectiveness of exotoxin A in antitumor applications has been validated in clinical studies. However, it can also damage the immune system of infected patients and may cause pneumonia or sepsis. At present, recombinant toxins with improved immunogenicity and reduced toxicity can be constructed through genetic engineering techniques to enhance their efficacy and reduce adverse effects (Burkhardt et al., 2019).
4.2.3 Apratoxin A and Coibamide A
Apratoxin A and coibamide A are natural secondary metabolites isolated from marine cyanobacteria (Tranter et al., 2020). They are produced by a nonribosomal peptide synthetase. Apratoxin A was found to be a cytotoxic antitumor drug capable of inhibiting the growth of various cancer cells, such as osteosarcoma and breast cancer cells, by inducing G1 cell cycle arrest and apoptosis (Liu et al., 2009). Coibamide A can reduce the drug resistance of tumors and prevent autophagic flux through the inhibition of autophagosome-lysosome fusion, thereby leading to caspase-independent death in tumor cells (Shi et al., 2021). It can also inhibit the migration, invasion, and cell cycle progression of glioblastoma and breast cancer cells (Hau et al., 2013). In addition, coibamide A possesses broad - spectrum activity, with a substrate overlap with apratoxin A, as exemplified by their shared targeting of HER/ErbB family proteins (Kazemi et al., 2021).
4.2.4 Ipomoeassin F
Ipomoeassin F (Ipo-F) is a natural plant-derived resin glycoside cytotoxin that directly binds to Sec61α and exhibits potent anticancer activity in human breast cancer cells (MCF7) and lymphoma cells (U937) (Roboti et al., 2021; Zong et al., 2015). In vitro translocation assays showed that Ipo-F blocks all Sec61 substrates but does not inhibit insertion/translocation of tail-anchor, type III membrane proteins or short secretory proteins which can translocate independently of Sec61 (Zong et al., 2019). In addition to its anticancer efficacy, Ipo-F has been reported to exhibit antiviral activity by inhibiting the cotranslation of the SARS-CoV-2 spike protein and the host cell membrane receptor ACE2 (O'Keefe et al., 2021).
4.2.5 Decatransin
Decatransin, a fungal-derived highly N-methylated cycloundecalactone peptide, exerts non-selective and broad-spectrum inhibitory effects on the translocation of polypeptides into the ER (Ohsawa et al., 2022). Studies have demonstrated that decatransin effectively suppresses the proliferation of cells by blocking Sec61-dependent protein translocation into the ER. This inhibitory mechanism operates independently of SRP-mediated recognition and SR-directed targeting processes, and is applicable to both co-translational and post-translational translocation pathways (Junne et al., 2015). Genetic studies have identified multiple decatransin-resistant mutations in Sec61α1, with the majority localized within the plug domain. Notably, the Q129L mutation in yeast Sec61α (orthologous to Q127L in human Sec61α) confers strong to moderate resistance against decatransin, suggesting that this residue plays a critical role in decatransin binding and function (Itskanov et al., 2023).
4.2.6 Eeyarestatin
Eeyarestatin (ES), including eeyarestatin I (ESI) and eeyarestatin II (ESII), is an ERAD inhibitor (Cross et al., 2009). As the Sec61 translocon is closely linked to ERAD, inhibiting ER protein transfer may also block the retrotranslocation of misfolded proteins (Wang et al., 2010). In Alzheimer’s, Parkinson’s, prion, and Huntington’s diseases, protein degradation is impaired (Smith, 2018). ES inhibits ERAD, causing misfolded protein accumulation in the ER and inducing ER stress, offering a potential intervention strategy (Fiebiger et al., 2004). ES’s anticancer potential is also promising. ESI, like bortezomib, kills tumor cells by disrupting ER homeostasis and inducing ER stress. In NSCLC xenograft models, inhibiting valosin-containing proteins reduces tumor growth (Valle et al., 2011). Moreover, ESI combined with proteasome inhibitors like bortezomib shows enhanced antitumor effects. In antimicrobial research, ES24, an ES analog, inhibits SecYEG-dependent protein translocation and membrane insertion in E. coli (Steenhuis et al., 2021). Notably, ESI at 0.2–5 μM for 4 h eliminates the infectivity of Zika and Usutu viruses in a dose- and time-dependent manner, highlighting ES’s potential in antiviral therapy (Rodrigo et al., 2022). Overall, ES shows broad application potential in disease treatment.
5 Challenges and prospects
In this review, we have summarized the structure and function of the Sec61 protein and the role of SEC61 mutations in genetic diseases and cancer. As research on Sec61 continues to grow, Sec61 has become increasingly recognized as a therapeutic target for genetic diseases and cancers. However, the role of the Sec61 protein and its related proteins in the disease process is still unclear, and little is known about the cotranslational and posttranslational substrate proteins involved. With the development of high-throughput omics technology, we can screen substrates of the Sec61 complex and their modification methods via various methods. Currently, Sec61 inhibitors are still in the clinical trial stage as targeted therapies for cancer. Additional research is needed to determine whether the role of Sec61 in ER protein import and/or Ca2+ homeostasis is related to the observed clinical course of human cancers. Therefore, exploring the specific roles of the Sec61 complex in different tissues will play a crucial role in the therapeutic application of Sec61 inhibitors.
Statements
Author contributions
JX: Conceptualization, Methodology, Writing – original draft, Writing – review and editing. KY: Software, Validation, Writing – review and editing. SL: Methodology, Validation, Writing – review and editing. PG: Supervision, Visualization, Writing – review and editing. SS: Funding acquisition, Project administration, Resources, Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by the National Natural Science Foundation of China (grant No. 82370868 and 82070818) and Taishan Scholar Project special funding (grant No. tsqn202211330).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Alder N. N. Shen Y. Brodsky J. L. Hendershot L. M. Johnson A. E. (2005). The molecular mechanisms underlying BiP-mediated gating of the Sec61 translocon of the endoplasmic reticulum. J. Cell Biol.168 (3), 389–399. 10.1083/jcb.200409174
2
Almagro Armenteros J. J. Salvatore M. Emanuelsson O. Winther O. von Heijne G. Elofsson A. et al (2019). Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance2 (5), e201900429. 10.26508/lsa.201900429
3
Bachmann K. Bockhorn M. Mann O. Gebauer F. Blessmann M. Izbicki J. R. et al (2019). Aberrant expression of Sec61α in esophageal cancers. J. Cancer Res. Clin. Oncol.145 (8), 2039–2044. 10.1007/s00432-019-02955-7
4
Bai L. You Q. Feng X. Kovach A. Li H. (2020). Structure of the ER membrane complex, a transmembrane-domain insertase. Nature584 (7821), 475–478. 10.1038/s41586-020-2389-3
5
Baron L. Paatero A. O. Morel J. D. Impens F. Guenin-Macé L. Saint-Auret S. et al (2016). Mycolactone subverts immunity by selectively blocking the Sec61 translocon. J. Exp. Med.213 (13), 2885–2896. 10.1084/jem.20160662
6
Bellanné-Chantelot C. Schmaltz-Panneau B. Marty C. Fenneteau O. Callebaut I. Clauin S. et al (2018). Mutations in the SRP54 gene cause severe congenital neutropenia as well as Shwachman-Diamond-like syndrome. Blood132 (12), 1318–1331. 10.1182/blood-2017-12-820308
7
Berger K. Pauwels E. Parkinson G. Landberg G. Le T. Demillo V. G. et al (2021). Reduction of progranulin-induced breast cancer stem cell propagation by sortilin-targeting cyclotriazadisulfonamide (CADA) compounds. J. Med. Chem.64 (17), 12865–12876. 10.1021/acs.jmedchem.1c00943
8
Besse W. Dong K. Choi J. Punia S. Fedeles S. V. Choi M. et al (2017). Isolated polycystic liver disease genes define effectors of polycystin-1 function. J. Clin. Invest127 (5), 1772–1785. 10.1172/JCI90129
9
Bhadra P. Helms V. (2021). Molecular modeling of signal peptide recognition by eukaryotic Sec complexes. Int. J. Mol. Sci.22 (19), 10705. 10.3390/ijms221910705
10
Bolar N. A. Golzio C. Živná M. Hayot G. Van Hemelrijk C. Schepers D. et al (2016). Heterozygous loss-of-function SEC61A1 mutations cause autosomal-dominant tubulo-interstitial and glomerulocystic kidney disease with anemia. Am. J. Hum. Genet.99 (1), 174–187. 10.1016/j.ajhg.2016.05.028
11
Burkhardt M. Reiter K. Nguyen V. Suzuki M. Herrera R. Duffy P. E. et al (2019). Assessment of the impact of manufacturing changes on the physicochemical properties of the recombinant vaccine carrier ExoProtein A. Vaccine37 (38), 5762–5769. 10.1016/j.vaccine.2018.09.037
12
Claeys E. Pauwels E. Humblet-Baron S. Provinciael B. Schols D. Waer M. et al (2021). Small molecule cyclotriazadisulfonamide abrogates the upregulation of the human receptors CD4 and 4-1BB and suppresses in vitro activation and proliferation of T lymphocytes. Front. Immunol.12, 650731. 10.3389/fimmu.2021.650731
13
Cross B. C. McKibbin C. Callan A. C. Roboti P. Piacenti M. Rabu C. et al (2009). Eeyarestatin I inhibits Sec61-mediated protein translocation at the endoplasmic reticulum. J. Cell Sci.122 (Pt 23), 4393–4400. 10.1242/jcs.054494
14
Demangel C. High S. (2018). Sec61 blockade by mycolactone: a central mechanism in Buruli ulcer disease. Biol. Cell110 (11), 237–248. 10.1111/boc.201800030
15
Deshaies R. J. Sanders S. L. Feldheim D. A. Schekman R. (1991). Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature349 (6312), 806–808. 10.1038/349806a0
16
Devuyst O. Olinger E. Weber S. Eckardt K. U. Kmoch S. Rampoldi L. et al (2019). Autosomal dominant tubulointerstitial kidney disease. Nat. Rev. Dis. Prim.5 (1), 60. 10.1038/s41572-019-0109-9
17
Domenger A. Choisy C. Baron L. Mayau V. Perthame E. Deriano L. et al (2022). The Sec61 translocon is a therapeutic vulnerability in multiple myeloma. EMBO Mol. Med.14 (3), e14740. 10.15252/emmm.202114740
18
Dong M. Bridges J. P. Apsley K. Xu Y. Weaver T. E. (2008). ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol. Biol. Cell19 (6), 2620–2630. 10.1091/mbc.e07-07-0674
19
Dudek J. Benedix J. Cappel S. Greiner M. Jalal C. Müller L. et al (2009). Functions and pathologies of BiP and its interaction partners. Cell Mol. Life Sci.66 (9), 1556–1569. 10.1007/s00018-009-8745-y
20
Fan C. W. Chan C. C. Chen K. T. Twu J. Huang Y. S. Han C. L. et al (2011). Identification of SEC61β and its autoantibody as biomarkers for colorectal cancer. Clin. Chim. Acta412 (11-12), 887–893. 10.1016/j.cca.2011.01.012
21
Fiebiger E. Hirsch C. Vyas J. M. Gordon E. Ploegh H. L. Tortorella D. (2004). Dissection of the dislocation pathway for type I membrane proteins with a new small molecule inhibitor, eeyarestatin. Mol. Biol. Cell15 (4), 1635–1646. 10.1091/mbc.e03-07-0506
22
Foster C. A. Dreyfuss M. Mandak B. Meingassner J. G. Naegeli H. U. Nussbaumer A. et al (1994). Pharmacological modulation of endothelial cell-associated adhesion molecule expression: implications for future treatment of dermatological diseases. J. Dermatol21 (11), 847–854. 10.1111/j.1346-8138.1994.tb03300.x
23
Fregno I. Molinari M. (2019). Proteasomal and lysosomal clearance of faulty secretory proteins: ER-associated degradation (ERAD) and ER-to-lysosome-associated degradation (ERLAD) pathways. Crit. Rev. Biochem. Mol. Biol.54 (2), 153–163. 10.1080/10409238.2019.1610351
24
Fritz J. M. Dong M. Apsley K. S. Martin E. P. Na C. L. Sitaraman S. et al (2014). Deficiency of the BiP cochaperone ERdj4 causes constitutive endoplasmic reticulum stress and metabolic defects. Mol. Biol. Cell25 (4), 431–440. 10.1091/mbc.E13-06-0319
25
Gao H. Niu W. He Z. Gao C. Peng C. Niu J. (2020). SEC61G plays an oncogenic role in hepatocellular carcinoma cells. Cell Cycle19 (23), 3348–3361. 10.1080/15384101.2020.1843816
26
Garrison J. L. Kunkel E. J. Hegde R. S. Taunton J. (2005). A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature436 (7048), 285–289. 10.1038/nature03821
27
Hall B. S. Hill K. McKenna M. Ogbechi J. High S. Willis A. E. et al (2014). The pathogenic mechanism of the Mycobacterium ulcerans virulence factor, mycolactone, depends on blockade of protein translocation into the ER. PLoS Pathog.10 (4), e1004061. 10.1371/journal.ppat.1004061
28
Harant H. Wolff B. Schreiner E. P. Oberhauser B. Hofer L. Lettner N. et al (2007). Inhibition of vascular endothelial growth factor cotranslational translocation by the cyclopeptolide CAM741. Mol. Pharmacol.71 (6), 1657–1665. 10.1124/mol.107.034249
29
Harding H. P. Zeng H. Zhang Y. Jungries R. Chung P. Plesken H. et al (2001). Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol. Cell7 (6), 1153–1163. 10.1016/s1097-2765(01)00264-7
30
Hau A. M. Greenwood J. A. Löhr C. V. Serrill J. D. Proteau P. J. Ganley I. G. et al (2013). Coibamide A induces mTOR-independent autophagy and cell death in human glioblastoma cells. PLoS One8 (6), e65250. 10.1371/journal.pone.0065250
31
Haßdenteufel S. Johnson N. Paton A. W. Paton J. C. High S. Zimmermann R. (2018). Chaperone-mediated Sec61 channel gating during ER import of small precursor proteins overcomes Sec61 inhibitor-reinforced energy barrier. Cell Rep.23 (5), 1373–1386. 10.1016/j.celrep.2018.03.122
32
Haßdenteufel S. Nguyen D. Helms V. Lang S. Zimmermann R. (2019). ER import of small human presecretory proteins: components and mechanisms. FEBS Lett.593 (18), 2506–2524. 10.1002/1873-3468.13542
33
Heaton N. S. Moshkina N. Fenouil R. Gardner T. J. Aguirre S. Shah P. S. et al (2016). Targeting viral proteostasis limits influenza virus, HIV, and dengue virus infection. Immunity44 (1), 46–58. 10.1016/j.immuni.2015.12.017
34
Heinrich S. U. Mothes W. Brunner J. Rapoport T. A. (2000). The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell102 (2), 233–244. 10.1016/s0092-8674(00)00028-3
35
Huang Y. Xu X. Arvan P. Liu M. (2021). Deficient endoplasmic reticulum translocon-associated protein complex limits the biosynthesis of proinsulin and insulin. Faseb J.35 (5), e21515. 10.1096/fj.202002774R
36
Itskanov S. Kuo K. M. Gumbart J. C. Park E. (2021). Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62. Nat. Struct. Mol. Biol.28 (2), 162–172. 10.1038/s41594-020-00541-x
37
Itskanov S. Park E. (2023). Mechanism of protein translocation by the Sec61 translocon complex. Cold Spring Harb. Perspect. Biol.15 (1), a041250. 10.1101/cshperspect.a041250
38
Itskanov S. Wang L. Junne T. Sherriff R. Xiao L. Blanchard N. et al (2023). A common mechanism of Sec61 translocon inhibition by small molecules. Nat. Chem. Biol.19 (9), 1063–1071. 10.1038/s41589-023-01337-y
39
Junne T. Wong J. Studer C. Aust T. Bauer B. W. Beibel M. et al (2015). Decatransin, a new natural product inhibiting protein translocation at the Sec61/SecYEG translocon. J. Cell Sci.128 (6), 1217–1229. 10.1242/jcs.165746
40
Kalies K. U. Stokes V. Hartmann E. (2008). A single Sec61-complex functions as a protein-conducting channel. Biochim. Biophys. Acta1783 (12), 2375–2383. 10.1016/j.bbamcr.2008.08.005
41
Kazemi S. Kawaguchi S. Badr C. E. Mattos D. R. Ruiz-Saenz A. Serrill J. D. et al (2021). Targeting of HER/ErbB family proteins using broad spectrum Sec61 inhibitors coibamide A and apratoxin A. Biochem. Pharmacol.183, 114317. 10.1016/j.bcp.2020.114317
42
Klein W. Westendorf C. Schmidt A. Conill-Cortés M. Rutz C. Blohs M. et al (2015). Defining a conformational consensus motif in cotransin-sensitive signal sequences: a proteomic and site-directed mutagenesis study. PLoS One10 (3), e0120886. 10.1371/journal.pone.0120886
43
Kreil G. (1981). Transfer of proteins across membranes. Annu. Rev. Biochem.50, 317–348. 10.1146/annurev.bi.50.070181.001533
44
Kutay U. Ahnert-Hilger G. Hartmann E. Wiedenmann B. Rapoport T. A. (1995). Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. Embo J.14 (2), 217–223. 10.1002/j.1460-2075.1995.tb06994.x
45
Lang S. Nguyen D. Pfeffer S. Förster F. Helms V. Zimmermann R. (2019). Functions and mechanisms of the human ribosome-translocon complex. Subcell. Biochem.93, 83–141. 10.1007/978-3-030-28151-9_4
46
Lang S. Pfeffer S. Lee P. H. Cavalié A. Helms V. Förster F. et al (2017). An update on Sec61 channel functions, mechanisms, and related diseases. Front. Physiol.8, 887. 10.3389/fphys.2017.00887
47
Li L. Park E. Ling J. Ingram J. Ploegh H. Rapoport T. A. (2016). Crystal structure of a substrate-engaged SecY protein-translocation channel. Nature531 (7594), 395–399. 10.1038/nature17163
48
Linder M. I. Mizoguchi Y. Hesse S. Csaba G. Tatematsu M. Łyszkiewicz M. et al (2023). Human genetic defects in SRP19 and SRPRA cause severe congenital neutropenia with distinctive proteome changes. Blood141 (6), 645–658. 10.1182/blood.2022016783
49
Linxweiler M. Schick B. Zimmermann R. (2017). Let's talk about Secs: Sec61, Sec62 and Sec63 in signal transduction, oncology and personalized medicine. Signal Transduct. Target Ther.2, 17002. 10.1038/sigtrans.2017.2
50
Liu B. Liu J. Liao Y. Jin C. Zhang Z. Zhao J. et al (2019). Identification of SEC61G as a novel prognostic marker for predicting survival and response to therapies in patients with glioblastoma. Med. Sci. Monit.25, 3624–3635. 10.12659/MSM.916648
51
Liu M. Wright J. Guo H. Xiong Y. Arvan P. (2014). Proinsulin entry and transit through the endoplasmic reticulum in pancreatic beta cells. Vitam. Horm.95, 35–62. 10.1016/B978-0-12-800174-5.00002-8
52
Liu Y. Law B. K. Luesch H. (2009). Apratoxin a reversibly inhibits the secretory pathway by preventing cotranslational translocation. Mol. Pharmacol.76 (1), 91–104. 10.1124/mol.109.056085
53
Lloyd D. J. Wheeler M. C. Gekakis N. (2010). A point mutation in Sec61alpha1 leads to diabetes and hepatosteatosis in mice. Diabetes59 (2), 460–470. 10.2337/db08-1362
54
Lu Z. Zhou L. Killela P. Rasheed A. B. Di C. Poe W. E. et al (2009). Glioblastoma proto-oncogene SEC61gamma is required for tumor cell survival and response to endoplasmic reticulum stress. Cancer Res.69 (23), 9105–9111. 10.1158/0008-5472.CAN-09-2775
55
Mackinnon A. L. Paavilainen V. O. Sharma A. Hegde R. S. Taunton J. (2014). An allosteric Sec61 inhibitor traps nascent transmembrane helices at the lateral gate. Elife3, e01483. 10.7554/eLife.01483
56
Maifeld S. V. MacKinnon A. L. Garrison J. L. Sharma A. Kunkel E. J. Hegde R. S. et al (2011). Secretory protein profiling reveals TNF-α inactivation by selective and promiscuous Sec61 modulators. Chem. Biol.18 (9), 1082–1088. 10.1016/j.chembiol.2011.06.015
57
Matlack K. E. Mothes W. Rapoport T. A. (1998). Protein translocation: tunnel vision. Cell92 (3), 381–390. 10.1016/s0092-8674(00)80930-7
58
McKenna M. Simmonds R. E. High S. (2016). Mechanistic insights into the inhibition of Sec61-dependent co- and post-translational translocation by mycolactone. J. Cell Sci.129 (7), 1404–1415. 10.1242/jcs.182352
59
Moon H. W. Han H. G. Jeon Y. J. (2018). Protein quality control in the endoplasmic reticulum and cancer. Int. J. Mol. Sci.19 (10), 3020. 10.3390/ijms19103020
60
Mothes W. Prehn S. Rapoport T. A. (1994). Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. Embo J.13 (17), 3973–3982. 10.1002/j.1460-2075.1994.tb06713.x
61
NCD Risk Factor Collaboration NCD-RisC (2024). Worldwide trends in diabetes prevalence and treatment from 1990 to 2022: a pooled analysis of 1108 population-representative studies with 141 million participants. Lancet404 (10467), 2077–2093. 10.1016/S0140-6736(24)02317-1
62
Ohsawa K. Fukaya S. Doi T. (2022). Total synthesis and structural determination of cyclodepsipeptide decatransin. Org. Lett.24 (30), 5552–5556. 10.1021/acs.orglett.2c02085
63
O'Keefe S. Roboti P. Duah K. B. Zong G. Schneider H. Shi W. Q. et al (2021). Ipomoeassin-F inhibits the in vitro biogenesis of the SARS-CoV-2 spike protein and its host cell membrane receptor. J. Cell Sci.134 (4), jcs257758. 10.1242/jcs.257758
64
Paatero A. O. Kellosalo J. Dunyak B. M. Almaliti J. Gestwicki J. E. Gerwick W. H. et al (2016). Apratoxin kills cells by direct blockade of the Sec61 protein translocation channel. Cell Chem. Biol.23 (5), 561–566. 10.1016/j.chembiol.2016.04.008
65
Page K. R. Nguyen V. N. Pleiner T. Tomaleri G. P. Wang M. L. Guna A. et al (2024). Role of a holo-insertase complex in the biogenesis of biophysically diverse ER membrane proteins. Mol. Cell84 (17), 3302–3319.e11. 10.1016/j.molcel.2024.08.005
66
Pauwels E. Rutz C. Provinciael B. Stroobants J. Schols D. Hartmann E. et al (2021b). A proteomic study on the membrane protein fraction of T cells confirms high substrate selectivity for the ER translocation inhibitor cyclotriazadisulfonamide. Mol. Cell Proteomics20, 100144. 10.1016/j.mcpro.2021.100144
67
Pauwels E. Schülein R. Vermeire K. (2021a). Inhibitors of the Sec61 complex and novel high throughput screening strategies to target the protein translocation pathway. Int. J. Mol. Sci.22 (21), 12007. 10.3390/ijms222112007
68
Pfeffer S. Dudek J. Gogala M. Schorr S. Linxweiler J. Lang S. et al (2014). Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon. Nat. Commun.5, 3072. 10.1038/ncomms4072
69
Pfeffer S. Dudek J. Schaffer M. Ng B. G. Albert S. Plitzko J. M. et al (2017). Dissecting the molecular organization of the translocon-associated protein complex. Nat. Commun.8, 14516. 10.1038/ncomms14516
70
Pilla E. Schneider K. Bertolotti A. (2017). Coping with protein quality control failure. Annu. Rev. Cell Dev. Biol.33, 439–465. 10.1146/annurev-cellbio-111315-125334
71
Rapoport T. A. Li L. Park E. (2017). Structural and mechanistic insights into protein translocation. Annu. Rev. Cell Dev. Biol.33, 369–390. 10.1146/annurev-cellbio-100616-060439
72
Rapoport T. A. Rolls M. M. Jungnickel B. (1996). Approaching the mechanism of protein transport across the ER membrane. Curr. Opin. Cell Biol.8 (4), 499–504. 10.1016/s0955-0674(96)80027-5
73
Roboti P. O'Keefe S. Duah K. B. Shi W. Q. High S. (2021). Ipomoeassin-F disrupts multiple aspects of secretory protein biogenesis. Sci. Rep.11 (1), 11562. 10.1038/s41598-021-91107-4
74
Rodrigo I. Ballesta C. Nunes E. B. Pérez P. García-Arriaza J. Arias A. (2022). Eeyarestatin I an inhibitor of the valosin-containing protein, exhibits potent virucidal activity against the flaviviruses. Antivir. Res.207, 105416. 10.1016/j.antiviral.2022.105416
75
Ruiz-Saenz A. Sandhu M. Carrasco Y. Maglathlin R. L. Taunton J. Moasser M. M. (2015). Targeting HER3 by interfering with its Sec61-mediated cotranslational insertion into the endoplasmic reticulum. Oncogene34 (41), 5288–5294. 10.1038/onc.2014.455
76
Salmaso V. Moro S. (2018). Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: an Overview. Front. Pharmacol.9, 923. 10.3389/fphar.2018.00923
77
Schäuble N. Cavalié A. Zimmermann R. Jung M. (2014). Interaction of Pseudomonas aeruginosa Exotoxin A with the human Sec61 complex suppresses passive calcium efflux from the endoplasmic reticulum. Channels (Austin)8 (1), 76–83. 10.4161/chan.26526
78
Schäuble N. Lang S. Jung M. Cappel S. Schorr S. Ulucan Ö. et al (2012). BiP-mediated closing of the Sec61 channel limits Ca2+ leakage from the ER. Embo J.31 (15), 3282–3296. 10.1038/emboj.2012.189
79
Schlenstedt G. Gudmundsson G. H. Boman H. G. Zimmermann R. (1990). A large presecretory protein translocates both cotranslationally, using signal recognition particle and ribosome, and post-translationally, without these ribonucleoparticles, when synthesized in the presence of mammalian microsomes. J. Biol. Chem.265 (23), 13960–13968. 10.1016/s0021-9258(18)77442-5
80
Schlenstedt G. Zimmermann R. (1987). Import of frog prepropeptide GLa into microsomes requires ATP but does not involve docking protein or ribosomes. Embo J.6 (3), 699–703. 10.1002/j.1460-2075.1987.tb04810.x
81
Schorr S. Nguyen D. Haßdenteufel S. Nagaraj N. Cavalié A. Greiner M. et al (2020). Identification of signal peptide features for substrate specificity in human Sec62/Sec63-dependent ER protein import. Febs J.287 (21), 4612–4640. 10.1111/febs.15274
82
Schubert D. Klein M. C. Hassdenteufel S. Caballero-Oteyza A. Yang L. Proietti M. et al (2018). Plasma cell deficiency in human subjects with heterozygous mutations in Sec61 translocon alpha 1 subunit (SEC61A1). J. Allergy Clin. Immunol.141 (4), 1427–1438. 10.1016/j.jaci.2017.06.042
83
Shah P. S. Link N. Jang G. M. Sharp P. P. Zhu T. Swaney D. L. et al (2018). Comparative flavivirus-host protein interaction mapping reveals mechanisms of dengue and Zika virus pathogenesis. Cell175 (7), 1931–1945. 10.1016/j.cell.2018.11.028
84
Shi W. Lu D. Wu C. Li M. Ding Z. Li Y. et al (2021). Coibamide A kills cancer cells through inhibiting autophagy. Biochem. Biophys. Res. Commun.547, 52–58. 10.1016/j.bbrc.2021.01.112
85
Sicking M. Lang S. Bochen F. Roos A. Drenth J. P. H. Zakaria M. et al (2021). Complexity and specificity of Sec61-channelopathies: human diseases affecting gating of the Sec61 complex. Cells10 (5), 1036. 10.3390/cells10051036
86
Skokowa J. Dale D. C. Touw I. P. Zeidler C. Welte K. (2017). Severe congenital neutropenias. Nat. Rev. Dis. Prim.3, 17032. 10.1038/nrdp.2017.32
87
Smith D. M. (2018). Could a common mechanism of protein degradation impairment underlie many neurodegenerative diseases?J. Exp. Neurosci.12, 1179069518794675. 10.1177/1179069518794675
88
Steenhuis M. Koningstein G. M. Oswald J. Pick T. O'Keefe S. Koch H. G. et al (2021). Eeyarestatin 24 impairs SecYEG-dependent protein trafficking and inhibits growth of clinically relevant pathogens. Mol. Microbiol.115 (1), 28–40. 10.1111/mmi.14589
89
Sun S. Wang S. Tong Z. Yao X. Gao J. (2019). A molecular dynamics study on the resilience of Sec61 channel from open to closed state. RSC Adv.9 (26), 14876–14883. 10.1039/c9ra01684h
90
Tranter D. Paatero A. O. Kawaguchi S. Kazemi S. Serrill J. D. Kellosalo J. et al (2020). Coibamide A targets Sec61 to prevent biogenesis of secretory and membrane proteins. ACS Chem. Biol.15 (8), 2125–2136. 10.1021/acschembio.0c00325
91
Tsukazaki T. Mori H. Fukai S. Ishitani R. Mori T. Dohmae N. et al (2008). Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature455 (7215), 988–991. 10.1038/nature07421
92
Valle C. W. Min T. Bodas M. Mazur S. Begum S. Tang D. et al (2011). Critical role of VCP/p97 in the pathogenesis and progression of non-small cell lung carcinoma. PLoS One6 (12), e29073. 10.1371/journal.pone.0029073
93
Van Coppenolle F. Vanden Abeele F. Slomianny C. Flourakis M. Hesketh J. Dewailly E. et al (2004). Ribosome-translocon complex mediates calcium leakage from endoplasmic reticulum stores. J. Cell Sci.117 (Pt 18), 4135–4142. 10.1242/jcs.01274
94
Van den Berg B. Clemons W. M. Collinson I. Modis Y. Hartmann E. Harrison S. C. et al (2004). X-ray structure of a protein-conducting channel. Nature427 (6969), 36–44. 10.1038/nature02218
95
Van Nieuwenhove E. Barber J. S. Neumann J. Smeets E. Willemsen M. Pasciuto E. et al (2020). Defective Sec61α1 underlies a novel cause of autosomal dominant severe congenital neutropenia. J. Allergy Clin. Immunol.146 (5), 1180–1193. 10.1016/j.jaci.2020.03.034
96
Vermeire K. Princen K. Hatse S. De Clercq E. Dey K. Bell T. W. et al (2004). CADA, a novel CD4-targeted HIV inhibitor, is synergistic with various anti-HIV drugs in vitro. Aids18 (16), 2115–2125. 10.1097/00002030-200411050-00003
97
Vermeire K. Schols D. (2003). Specific CD4 down-modulating compounds with potent anti-HIV activity. J. Leukoc. Biol.74 (5), 667–675. 10.1189/jlb.0403177
98
Vermeire K. Zhang Y. Princen K. Hatse S. Samala M. F. Dey K. et al (2002). CADA inhibits human immunodeficiency virus and human herpesvirus 7 replication by down-modulation of the cellular CD4 receptor. Virology302 (2), 342–353. 10.1006/viro.2002.1624
99
Voorhees R. M. Hegde R. S. (2016). Structure of the Sec61 channel opened by a signal sequence. Science351 (6268), 88–91. 10.1126/science.aad4992
100
Wang Q. Shinkre B. A. Lee J. G. Weniger M. A. Liu Y. Chen W. et al (2010). The ERAD inhibitor Eeyarestatin I is a bifunctional compound with a membrane-binding domain and a p97/VCP inhibitory group. PLoS One5 (11), e15479. 10.1371/journal.pone.0015479
101
Weng T. H. Steinchen W. Beatrix B. Berninghausen O. Becker T. Bange G. et al (2021). Architecture of the active post-translational Sec translocon. Embo J.40 (3), e105643. 10.15252/embj.2020105643
102
Westendorf C. Schmidt A. Coin I. Furkert J. Ridelis I. Zampatis D. et al (2011). Inhibition of biosynthesis of human endothelin B receptor by the cyclodepsipeptide cotransin. J. Biol. Chem.286 (41), 35588–35600. 10.1074/jbc.M111.239244
103
Witham C. M. Paxman A. L. Baklous L. Steuart R. F. L. Schulz B. L. Mousley C. J. (2021). Cancer associated mutations in Sec61γ alter the permeability of the ER translocase. PLoS Genet.17 (8), e1009780. 10.1371/journal.pgen.1009780
104
Yotsu R. R. Suzuki K. Simmonds R. E. Bedimo R. Ablordey A. Yeboah-Manu D. et al (2018). Buruli ulcer: a review of the current knowledge. Curr. Trop. Med. Rep.5 (4), 247–256. 10.1007/s40475-018-0166-2
105
Zehner M. Marschall A. L. Bos E. Schloetel J. G. Kreer C. Fehrenschild D. et al (2015). The translocon protein Sec61 mediates antigen transport from endosomes in the cytosol for cross-presentation to CD8(+) T cells. Immunity42 (5), 850–863. 10.1016/j.immuni.2015.04.008
106
Zong G. Barber E. Aljewari H. Zhou J. Hu Z. Du Y. et al (2015). Total synthesis and biological evaluation of Ipomoeassin F and its unnatural 11r-epimer. J. Org. Chem.80 (18), 9279–9291. 10.1021/acs.joc.5b01765
107
Zong G. Hu Z. O'Keefe S. Tranter D. Iannotti M. J. Baron L. et al (2019). Ipomoeassin F binds Sec61α to inhibit protein translocation. J. Am. Chem. Soc.141 (21), 8450–8461. 10.1021/jacs.8b13506
Summary
Keywords
Sec61, endoplasmic reticulum, genetic disease, tumor, inhibitor
Citation
Xin J, Yin K, Li S, Gu P and Shao S (2025) Exploring the ER channel protein Sec61: recent advances in pathophysiological significance and novel pharmacological inhibitors. Front. Pharmacol. 16:1580086. doi: 10.3389/fphar.2025.1580086
Received
20 February 2025
Accepted
22 May 2025
Published
04 June 2025
Volume
16 - 2025
Edited by
Jose Antonio Poveda Larrosa, Miguel Hernández University of Elche, Spain
Reviewed by
Manigandan Venkatesan, The University of Texas Health Science Center at San Antonio, United States
Martin Pool, The University of Manchester, United Kingdom
Updates
Copyright
© 2025 Xin, Yin, Li, Gu and Shao.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shanshan Shao, shaoshanshan11@126.com
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.