- 1School of Medicine, Tel-Aviv University, Tel Aviv, Israel
- 2Sagol School of Neuroscience, Tel-Aviv University, Tel Aviv, Israel
- 3The Adelson School of Medicine, Ariel University, Ariel, Israel
- 4Neonatology Department, Laniado Hospital, Netanya, Israel
Background: G-proteins areindispensable regulators of cellular signaling, with G-protein-gated inwardly rectifying potassium channels (GIRK) as key effectors. GNB1 encephalopathy (GNB1E) is a congenital neurological syndrome resulting from mutations in the GNB1 gene, encoding the Gβ1 subunit of G-proteins trimer (Gαβγ). GNB1E manifests as a global developmental delay, accompanied by tonus disturbances, ataxia, and epilepsy.
Methods: We utilized the Xenopus laevis oocyte heterologous expression system to investigate the impact of the L95P mutation in Gβ1 (Gβ1-L95P) on the activation of neuronal GIRK channels GIRK2 and GIRK1/2. Mutant and wild-type (WT) Gβ1 RNAs were co-injected with RNAs encoding the Gγ2 and GIRK channel subunits. The expression levels of both Gβ1 and the channel proteins, as well as the channel activity, were systematically monitored. Additionally, rigid-body docking was used to model the GIRK1/2–Gβγ complex, evaluating L95P’s effect on channel–Gβγ interaction, Gβγ stability, and Gβγ–effector affinity.
Results: . Gβ1-L95P exhibited reduced protein expression compared to WT. Even after RNA adjustments to restore comparable membrane localization, the mutant failed to effectively activate GIRK2 and GIRK1/2. Structural analysis revealed that L95 was not consistent in the Gβγ–effector interface. Thermodynamic calculations suggested that the mutation primarily destabilized Gβ1 and Gβ1–effector complex.
Conclusion: Gβ1-L95P leads to both reduced protein expression and impaired function in the GIRK–Gβγ interaction system. The later effect can be attributed to the changes associated with protein misfolding.
Introduction
Heterotrimeric G-proteins (composed of Gα, Gβ, and Gγ subunits) and G-protein-coupled receptors (GPCRs) constitute integral components of GPCR signaling (Pierce et al., 2002). G-proteins interact with the effectors, including ion channels, enzymes, and various other regulatory proteins, and they play a significant role in diverse physiological processes such as neurotransmission, cardiovascular regulation, endocrine system, metabolism, immune response, cell growth and differentiation, and other intricate intracellular communication pathways (Oldham and Hamm, 2008). Gβγ subunit dimers, or Gβγ subunits for simplicity, in particular, are involved in the activation of GIRK channels (G-protein-activated inwardly rectifying potassium channel), among other functions (Logothetis et al., 1987). Upon activation, these channels induce hyperpolarization of the plasma membrane in cardiac cells and neurons. This activation is crucial for mediating the inhibitory effects of various neurotransmitters, including GABA (via GABA-B receptors), acetylcholine (via M2 muscarinic receptors), opioids (via opioid receptors), and others. The modulation of GIRK channels by Gβγ subunits represents a key mechanism in the regulation of neuronal excitability and the intricate balance of neurotransmission in the central nervous system (Campos-Rios et al., 2022; Cui et al., 2021; Luo et al., 2022).
Over the past decade, a cohort of patients harboring mutations in the Gβ1 subunit was described (Petrovski et al., 2016; Bassan et al., 2018). This group, identified with early-onset epilepsy, has been termed GNB1 encephalopathy (GNB1-E). Notable characteristics of these patients encompass developmental delay (DD) or intellectual disability (ID), frequently observed infantile hypotonia progressing to hypertonia, and structural brain abnormalities, accompanied by a spectrum of seizure types such as tonic, absence, myoclonic, generalized tonic-clonic, and focal seizures, along with epileptic spasms. Importantly, GNB1 has emerged as a potential candidate gene associated with West syndrome, and several individuals with GNB1-E have had West syndrome or infantile spasms (Da Silva et al., 2021; Endo et al., 2020; Hemati et al., 2018a; Jones et al., 2019; Schultz-Rogers et al., 2020; Revah-Politi et al., 2020).
GNB1-E is an autosomal dominant rare genetic disorder, with approximately 70 documented cases worldwide. The reported mutations associated with GNB1-E consist of 36 variants affecting 25 residues (Revah-Politi et al., 2020; Lansdon and Saunders, 2021). This suggests the existence of mutational hotspots, where certain amino acid positions are particularly susceptible to genetic alterations associated with GNB1-E. The most prevalent mutation, I80T, has been identified in at least 17 patients (Endo et al., 2020; Tsuji et al., 2023; Hemati et al., 2018b). Our prior investigations extensively examined the I80T, I80N, and K78R mutations using the Xenopus laevis oocyte heterologous expression system. We found that the K78R mutation exhibits a gain of expression alongside a partial loss of function when interacting with GIRK1/2, whereas I80T and I80N mutations are characterized by a partial loss of expression with a concurrent loss of function in GIRK2 activation (Reddy et al., 2021).
Moreover, we extended our studies to a mouse model harboring the K78R mutation, demonstrating a recapitulation of developmental delay and epileptic features observed in human cases. Notably, ethosuximide, a potent GIRK inhibitor, was found to suppress spike-wave discharges (SWDs) in vivo and restore normal network behavior in vitro, highlighting potential therapeutic avenues for managing GNB1-E-associated symptoms (Colombo et al., 2023; Shalomov et al., 2025).
In light of our previous findings, we continued to study the impact of GNB1 mutants on GIRK channels’ activity and expressions in Xenopus laevis oocytes. In the current work, we characterized the GIRK1/2 and GIRK2 structural–functional relationship with L95P mutation of Gβ1, Gβ1-L95P (c.284T > C in exon 7). As of now, L95P is the second most prevalent mutation (seven documented cases: five female, one male, and one unknown) among the GNB1-E patients after I80T (Endo et al., 2020; Lansdon and Saunders, 2021; Tsuji et al., 2023; Hemati et al., 2018b). L95P is strongly associated with cleft palate compared to other GNB1 mutations. L95P is also associated with developmental delay and seizures (Petrovski et al., 2016; Endo et al., 2020; Hemati et al., 2018a). Here, we report that L95P mutation is associated with combined loss of expression and loss of function of GNB1.
Materials and methods
Xenopus laevis frog maintenance and oocyte collection
The experiments were authorized by and conducted in accordance with the guidelines provided by the Tel Aviv University Institutional Animal Care and Use Committee under permits #01-16-104 and 01-20-083. Female Xenopus laevis frogs were cared for and subjected to procedures such as oocyte defolliculation, incubation, and RNA injection, following established protocols (Dascal and Lotan, 1992). The frogs were housed in tanks with dechlorinated water and maintained under a 10-h light/14-h dark cycle at 19°C ± 2°C. Ovary portions were extracted through a small incision in the abdomen under anesthesia (0.2%–0.25% solution of tricaine methanesulphonate). Following suturing, frogs were placed in a separate tank to recover from anesthesia before being transferred to a postoperative animals’ tank. The frogs exhibited no signs of postoperative distress and were allowed a recovery period of at least 3 months. After three to four surgeries, anaesthetized frogs were humanely euthanized through decapitation and double pithing.
Oocytes underwent defolliculation using collagenase (type 1A, Sigma) in a Ca-free ND96 solution (see below). Following a 2–4 h shaking incubation at room temperature, the oocytes were washed and transferred to a Petri dish with fresh ND96 solution and placed in an incubator at 20°C overnight. The subsequent day, visually healthy oocytes were sorted into a fresh dish and maintained in the incubator using NDE solution (ND96 supplemented with 2.5 mM pyruvate and 50 μg/mL gentamicin) at 20°C until RNA injection or further utilization, with daily solution changes. RNA injection followed established procedures (Rubinstein et al., 2009), involving the injection of 50 nL of RNA into healthy oocytes, which were then incubated for 2–4 days in NDE solution. The standard ND96 solution, with a pH of 7.6–7.8, comprised 96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 5 mM HEPES, with NaOH titration. CaCl2 was excluded in Ca2+-free ND96.
DNA constructs and RNA
The DNA constructs utilized for experiments in Xenopus oocytes were inserted into high-expression oocyte vectors pGEM-HE or pGEM-HJ, following previously established methods (Berlin et al., 2011; Rishal et al., 2005). Most DNA constructs were reported earlier, including bovine Gβ1, bovine Gγ2, rat GIRK1, mouse GIRK2, YFP-GIRK1 (rat), and mouse GIRK2-YFP (Yakubovich et al., 2015; Tabak et al., 2019). The Gβ1L95P point mutation was introduced to bovine Gβ1 through PCR-site directed mutagenesis using standard procedures with the PWO master PCR kit (Roche # 03789403001). DNA preparation employed the Wizard® Plus SV Miniprep kit (Promega # A1460). RNA synthesis followed established protocols (Rishal et al., 2003), with the amounts of injected RNA specified in the text and figure legends.
Giant membrane patches (GMPs)
Giant membrane patches from oocyte membranes were prepared and visualized following established procedures (Singer-Lahat et al., 2000; Kahanovitch et al., 2014). Oocytes were manually devitellinized using fine forceps in a hypertonic solution (containing in mM: NaCl 56, KCl 150, MgCl2 4, and HEPES 10; pH 7.6). The devitellinized oocytes were placed on a Thermanox™ coverslip (Nunc, Roskilde, Denmark) submerged in a Ca2+-free ND96 solution, with their animal pole facing the coverslip, for 10–20 min. Suctioning with a Pasteur pipette was then performed, resulting in a giant plasma membrane patch attached to the coverslip, with the cytosolic face oriented toward the external medium. The coverslip was thoroughly rinsed with fresh ND96 solution and fixed using 4% formaldehyde for 30 min. Fixed giant plasma membrane patches underwent immunostaining in 5% milk in phosphate buffer solution (PBS). Nonspecific binding was blocked with donkey IgG at a 1:200 ratio (Jackson ImmunoResearch, West Grove, PA, United States). Rabbit anti-Gβ1 antibody (Abcam, ab 137635) was applied at a 1:300 dilution for 45 min at 37°C. Subsequently, a DyLight 650-labeled secondary antibody (Goat Anti-rabbit IgG, 1:200; Abcam, ab 96886) was applied for 30 min at 37°C, followed by PBS washing and mounting on a slide for visualization. Immunostained slides were stored at 4°C for no longer than a week.
Confocal imaging
Confocal imaging was conducted using Zeiss LSM 710 or Leica TCS SP8 confocal microscopes equipped with a ×20 objective. For whole oocytes, the image was focused on the animal (dark) hemisphere at the equator. YFP was excited with the 514 nm line of the argon laser, and emission was collected at 535–546 nm. Fiji, an ImageJ-based software (https://imagej.net/ij/), was employed for image analysis. Fluorescent signals were averaged from three regions of interest (ROIs) at the plasma membrane (PM) and three similar ROIs from the coverslip outside the oocyte’s image. The average background signal was subtracted from the average PM signal in each oocyte, followed by subtracting the average net signal from the membrane of uninjected (naïve) oocytes.
For confocal imaging of proteins in giant plasma membrane patches (GMPs), DyLight 650 was excited using a 633 nm laser, and emission was collected at 663–673 nm. Images were centered on the edges of the membrane patches to observe and subtract background fluorescence from the coverslip. Fiji software (https://imagej.net/software/fiji/) was used for image analysis, selecting two ROIs: one covering most of the membrane patch within the field of view and another comprising background fluorescence, which was subtracted from the signal obtained from the patch. The average signal from giant plasma membrane patches of native oocytes’ membranes in the same experiment, immunostained using the same protocol, was subtracted from all groups.
Two electrode voltage clamp (TEVC)
All experiments were conducted within a temperature range of 20°C–22°C, following procedures outlined in a prior work (Rubinstein et al., 2009). Currents were recorded at −80 mV, filtered at 20 Hz, and sampled at rates of 5 or 10 kHz. Whole-cell GIRK currents in oocytes were assessed using two-electrode voltage clamp (TEVC) with GeneClamp 500 (Molecular Devices, Sunnyvale, CA, United States) utilizing agarose cushion electrodes filled with 3M KCl, featuring resistances of 0.1–0.8 MΩ for the current electrode and 0.2–1.5 MΩ for the voltage electrode.
For measuring GIRK currents through direct activation by Gβγ, oocytes were injected with RNAs of GIRK1 and GIRK2 (0.05 ng) or GIRK2 (2 ng) or YFP-GIRK1 and GIRK2 (0.5 ng) or GIRK2-YFP (2 ng), along with the specified amounts of Gβ1 RNA. The Gγ2 RNA amount was maintained at 1/5 of Gβ to achieve roughly equal molar ratios of Gβ and Gγ RNAs. GIRK channel currents were measured in ND96 (2 mM K or LK) solution or high-K+ solution (HK24), featuring the following composition in mM: 24 KCl, 72 NaCl, 1 CaCl2, 1 MgCl2, and 5 HEPES. The pH of all solutions ranged from 7.4 to 7.6. The recording protocol is shown in Supplementary Figure S1.
Quantification and statistical analysis
Statistical analyses were carried out using GraphPad Prism version 10 for Windows (GraphPad Software, La Jolla, California United States). Two-group comparisons employed the t-test when the data passed both the Shapiro–Wilk normality test and the equal variance test; otherwise, the Mann–Whitney rank sum test was utilized. Multiple group comparisons were conducted using one-way ANOVA (ANOVA on ranks when data did not follow a normal distribution). Tukey’s test was applied for normally distributed data, whereas the Kruskal–Wallis test was used otherwise.
The data in the graphs are presented as mean ± SEM. Statistical differences are indicated as follows: asterisks (∗) denote comparisons between channels with WT Gβ and mutant groups; the octothorpe sign (#) indicates comparisons with the channel alone (no Gβγ) group and Gβγ groups. Significance levels are represented as follows: ∗ or #, p < 0.05; ∗∗ or ##, p < 0.01; ∗∗∗ or ###, p < 0.001; ∗∗∗∗ or ####, p < 0.0001.
Structural analysis of Gβγ–effector interaction
Interface and thermodynamic analysis
PDB files were analyzed in PRODIGY (Xue et al., 2016), and lists of amino acids located in the interface between two protein molecules (i.e., Gβγ and channel or other effector) were created. Interface amino acids data are defined by the PRODIGY server as residues in two protein molecules at 5.5 Å and below distance from each other. In order to predict a possible role of L95P mutation in the stability of Gβ subunit and its influence on protein–protein interactions in Gβγ–effector complexes, structural models were submitted to the MCSM server (Pires et al., 2014). The change in protein stability or in the affinity of the protein–protein interaction was defined as significant in case ΔΔG was larger than 1 kcal/mol, which corresponds to ∼5 fold change in dissociation constant at 25°C (Berg et al., 2012) (∼0.6 kcal/mol is considered the noise threshold (Kessel and Ben-Tal, 2018)).
Gβγ docking procedure
For docking, we utilized the Gβγ structure from the crystal structure of GIRK2/Gβγ complex [4KFM (Whorton and MacKinnon, 2013)], separated from the channel, and a tentative structure of GIRK1/2 heterotetramer described by Gazgalis et al. generated based on homology modeling (Gazgalis et al., 2022). For Gβγ docking, we utilized the rigid body docking algorithm implemented at the ClusPro 2.0 server (Kozakov et al., 2017). No space limitations were imposed during the docking procedure. As a result of the docking procedure, 120 clusters of possible GIRK1/2–Gβγ models were generated and segregated according to balanced, hydrophobic favorite, electrostatic favorite, and van der Waals favorite energy scores. The corresponding centroid models of all clusters were further analyzed utilizing a method similar to that described by Mahajan et al. (2013). In particular, we generated a GIRK1/2 membrane-embedded structure utilizing OPMM (Lomize et al., 2012) and aligned each of the docking models with this structure. Consequently, all models in which any Gβγ atom was located above the lower membrane leaflet and all models in which the C-terminal α-carbon of Gγ was more than 30Å from the lower membrane leaflet were excluded from further analysis. These criteria assured that Gβγ structures implemented for further analysis would be sufficiently close to the plasma membrane, as expected from geranylgeranyl modification of Gγ C-terminus (Hibino et al., 2010).
Results
L95P mutation leads to a partial loss of expression of Gβ1 protein
In this study, groups of oocytes were injected with varying amounts of either WT Gβ1 or Gβ1-L95P mRNA, together with Gγ and channel mRNA (GIRK2 or GIRK1/2). The levels of Gβ expression in the plasma membrane were quantified using the giant membrane patch methodology, which allows to visualize and monitor plasma membrane-attached Gβγ with a Gβ antibody. Concurrently, oocytes from the same RNA injection groups were employed for electrophysiological experiments. Representative pictures obtained from confocal imaging for Gβ1-L95P and WT Gβ expressed with GIRK2 and GIRK1/2 channels, respectively, are shown in Figures 1A, 2A. Notably, regardless of which channel was co-expressed with the Gβγ under investigation, the mutant Gβγ consistently exhibited lower expression levels, as illustrated in Figures 1B, 2B. Using higher amounts of injected Gβ1-L95P RNA, we could attain levels of expression comparable to the levels of WT Gβ obtained with lower doses of RNA, both in GIRK2 and GIRK1/2 channel-expressing oocytes (Figures 1B, 2B). This result suggests a partial loss of Gβ1 protein expression, indicating either reduced synthesis of Gβ1-L95P or an increased rate of its degradation, or a combination of both.
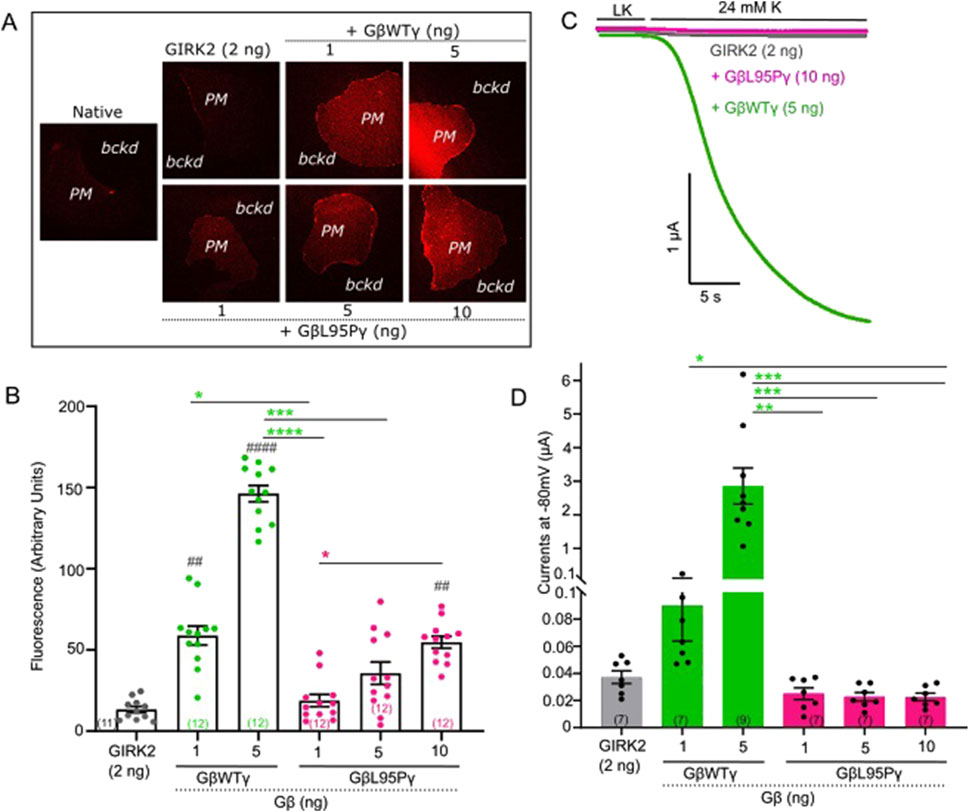
Figure 1. (A) Representative images of membrane patches at Gβ RNA doses 1 and 5 ng for GβWT and 1, 5, and 10 ng for GβL95P in the presence of GIRK2 channels. Plasma membrane patches were stained with an antibody against Gβ1. Membranes are seen as brighter-colored areas, and the background is black. (PM, plasma membrane; bckd, background). Number of frog donors (N) = 1. (B) Surface expression of GβWT and GβL95P mutant in the presence of GIRK2, measured in GMP. Data shown are the mean fluorescence intensity produced by the expressed Gβ, after subtraction of the average signal observed in channel-alone oocytes. (GMP, giant membrane patches). Statistical analysis: one-way ANOVA ####p < 0.0001, ###p < 0.001, ##p < 0.01, and #p < 0.05 relative to channel-only group and ****p < 0.0001, ***p < 0.001, **p < 0.01, and *p < 0.05 relative to GβWT. (C) Representative current traces of GIRK2 channels. (D) Summary of GIRK2 activation by GβWT and GβL95P. Statistical analysis: one-way ANOVA ####p < 0.0001, ###p < 0.001, ##p < 0.01, and #p < 0.05 relative to channel-only group and ****p < 0.0001, ***p < 0.001, **p < 0.01, and *p < 0.05 relative to GβWT.
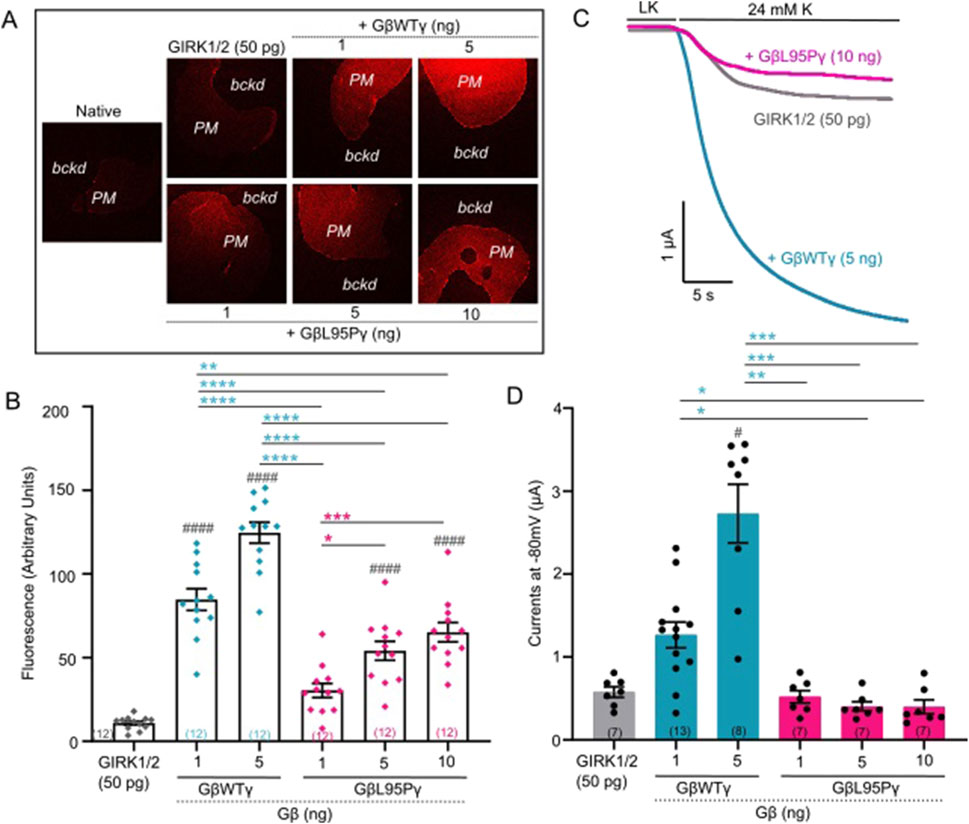
Figure 2. (A) Representative images of membrane patches at Gβ RNA doses 1 and 5 ng for GβWT and 1, 5, and 10 ng for GβL95P in the presence of GIRK1/2 channels. Plasma membrane patches were stained with an antibody against Gβ1. Membranes are seen as brighter-colored areas, and the background is black. (PM, plasma membrane; bckd, background). Number of frog donors (N) = 1. (B) Surface expression of GβWT and GβL95P mutant in the presence of GIRK1/2, measured in GMP. Data shown are the mean fluorescence intensity produced by the expressed Gβ, after subtraction of the average signal observed in channel alone oocytes (GMP, giant membrane patches). Statistical analysis: one-way ANOVA ####p < 0.0001, ###p < 0.001, ##p < 0.01, and #p < 0.05 relative to channel-only group and ****p < 0.0001, ***p < 0.001, **p < 0.01, and *p < 0.05 relative to GβWT. (C) Representative current traces of GIRK1/2 channels. (D) Summary of GIRK1/2 activation by GβWT and GβL95P. Statistical analysis: one-way ANOVA ####p < 0.0001, ###p < 0.001, ##p < 0.01, and #p < 0.05 relative to channel-only group and ****p < 0.0001, ***p < 0.001, **p < 0.01, and *p < 0.05 relative to GβWT.
Gβ1-L95P fails to activate GIRK channels even when the protein levels are similar to WT Gβ1
Having established that expression levels comparable to WT Gβ1 (1 ng RNA) can be achieved for Gβ1-L95P with a higher dose (10 ng RNA), we proceeded to investigate the ability of Gβ1-L95P (coexpressed with WT Gγ) to activate GIRK2 (Figures 1C,D) and GIRK1/2 (Figures 2C,D) channels. Despite achieving sufficient expression, Gβ1-L95P failed to activate both GIRK2 and GIRK1/2 channels.
GNB1 L95P does not reduce the expression of GIRK channels in the membrane
When levels of expression of GβL95P and GβWT were close to each other, GβL95P failed to activate the channels. As decreased channel expression could yield a similar outcome, our subsequent focus was on investigating the effect of L95P on channel expression in the plasma membrane. We expressed YFP-labeled GIRK channels together with either WT Gβ1 or Gβ1-L95P. The measurement of fluorescence intensity served as a reporter of channel density.
Our findings revealed a ∼2-fold increase in GIRK2-YFP channel expression in oocytes expressing either mutant or wild-type Gβγ (Figures 3A,B). On the other hand, there was no significant change in YFP-GIRK1/2 expression when compared to channel density without Gβγ co-expression, regardless of whether Gβγ was wild type or L95P (Figures 4A,B). Importantly, the selected RNA doses ensured comparable expression levels for both WT Gβ1 and Gβ1-L95P.
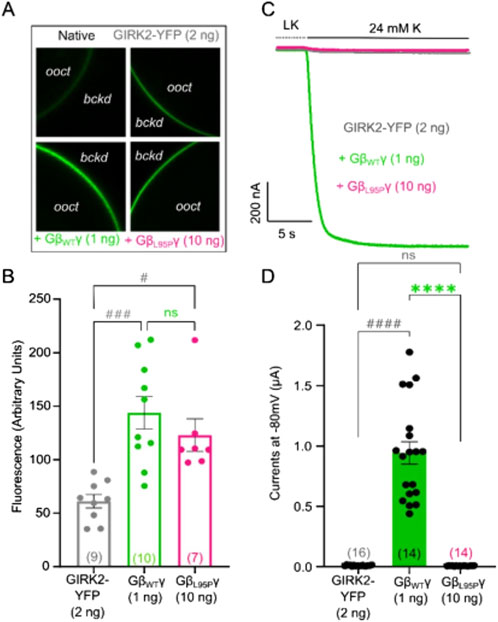
Figure 3. (A) Representative confocal images of oocytes expressing GIRK2-YFP channels. (B) Surface expression of GIRK2-YFP channels in the presence of GβWT (1 ng) and GβL95P (10 ng) mutant. Data shown are the mean fluorescence intensity produced by the expressed GIRK2, after subtraction of the average signal observed in native oocytes (ooct, oocyte; bckd, background). (C) Representative current traces of GIRK2-YFP channels. (D) Summary of GIRK2 activation by GβWT (1 ng) and GβL95P (10 ng). Statistical analysis: one-way ordinary ANOVA (Tukey’s test) or Kruskal–Wallis test ####p < 0.0001, ###p < 0.001, ##p < 0.01, and #p < 0.05 relative to channel-only group and ****p < 0.0001, ***p < 0.001, **p < 0.01, and *p < 0.05 relative to GβWT; ns, not significant. Number of frog donors (N) = 1.
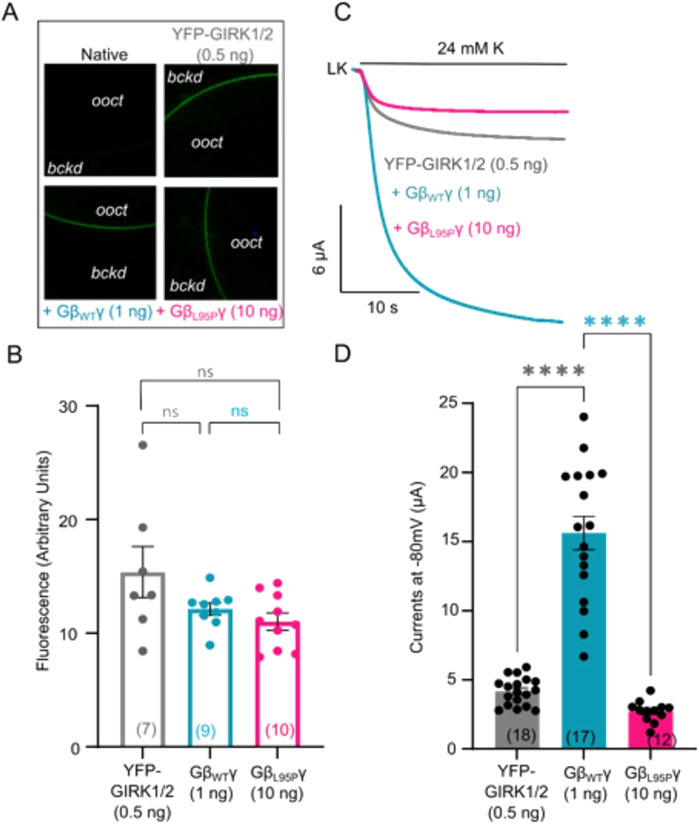
Figure 4. (A) Representative confocal images of oocytes expressing YFP-GIRK1/2 channels. (B) Surface expression of YFP-GIRK1/2 channels in the presence of GβWT (1 ng) and GβL95P (10 ng) mutant. Data shown are the mean fluorescence intensity produced by the expressed GIRK1/2 (ooct, oocyte; bckd, background). (C) Representative current traces of YFP-GIRK1/2 channels. (D) Summary of GIRK1/2 activation by GβWT (1 ng) and GβL95P (10 ng). Statistical analysis: one-way ordinary ANOVA (Tukey’s test) ****p < 0.0001, ***p < 0.001, **p < 0.01, and *p < 0.05; ns, not significant. Number of frog donors (N) = 1.
Simultaneous electrophysiological experiments revealed a lack of channel activation by Gβ1-L95P, as demonstrated in both GIRK2 (Figures 3C,D) and GIRK1/2 (Figures 4C,D), when the channel expression in the membrane was unaltered or even increased compared to WT Gβ1.
Structural analysis of L95P mutation
Initially, we analyzed the existing crystal structure of GIRK2–Gβγ complex (protein data bank accession number 4kfm). Based on coordinates from 4kfm, we generated the list of amino acids that are expected to be in the interface between GIRK2 and Gβγ utilizing the PRODIGY server (in Figure 5A, the amino acids in the interface between GIRK2 and Gβγ are shown as surfaces and L95 is shown as spheres). Notably, in the case of 4kfm, L95 is in the GIRK2–Gβγ interface (Figure 5B). Subsequently, we utilized the same criterion as in the PRODIGY algorithm (distance equal or less than 5.5 Å) in order to generate a contact list of possible contacts of L95, utilizing the INTAA server (Figure 5C). This assumption seems to be reasonable in light of the length of a non-covalent interaction (Kessel and Ben-Tal, 2018). Based on this list, we estimated the possible network of amino acids and non-covalent interactions of L95 utilizing the RINMAKER server (Figure 5D). A summary of amino acids network interacting with L95 is provided in Supplementary Table S1. The analysis procedure mentioned above was repeated for L95P mutation-containing structure that was generated on the base of 4kfm coordinates utilizing protein MutationExplorer server implementing the fixbb function of Rosetta for energy minimization. A cartoon representation of the expected structure of L95P mutant is shown in Figure 5E. Similar to the above, amino acid network and interface analysis were also conducted for the mutant structure (Figures 5F–H and Supplementary Table S1). It can be seen that there is a considerable decrease in amino acids expected to interact with proline in position 95 than with wild-type leucine. Moreover, we conducted limited analysis of the energetic impact of L95P mutation utilizing the MCSM server. It can be seen from Figure 8C that L95P mutation has an energetically significant impact on 4kfm stability. However, the effect of this mutation on GIRK2–Gβγ interaction affinity is expected to be rather modest and is near the thermal noise threshold.
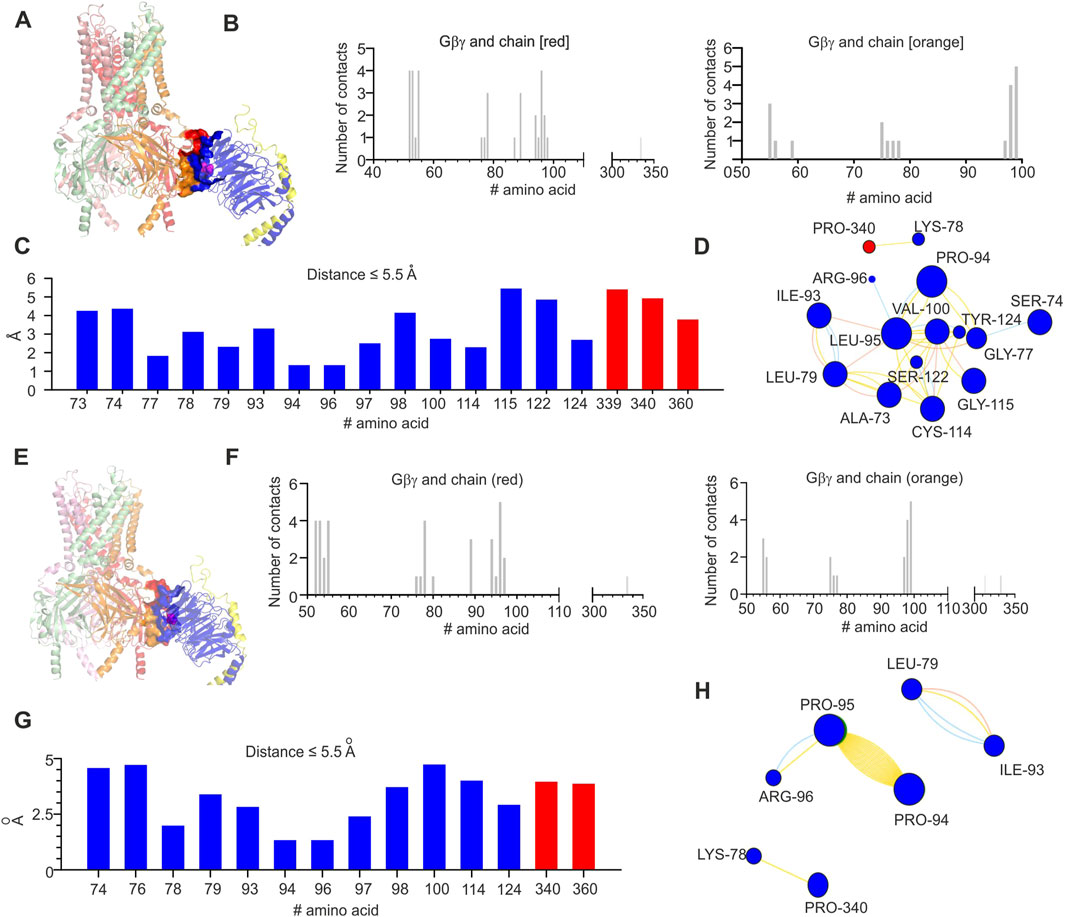
Figure 5. Structural analysis of the impact of L95P mutation on GIRK2-Gβγ complex. (A) Crystal structure of GIRK2–Gβγ complex (obtained from 4KFM, three Gβγ molecules were removed); L95 is shown as spheres and colored magenta. (B) Interface of GIRK–Gβγ complex, with data generated by the PRODIGY server; selection criterion: all amino acids ≤5.5 Å. The left plot corresponds to the chain colored red in A, and the right plot corresponds to the chain marked orange in (A). Note that L95 is in contact with some amino acids from the chain marked red. (C) Amino acids in contact with L95, same criterion as in B, with data obtained from analysis of GIRK–Gβγ wild-type utilizing the INTAA server. Amino acids that belong to Gβ are colored blue, and amino acids that belong to GIRK2 are marked red. (D) Analysis of non-covalent interactions of amino acids that are expected to be in contact with L95 (data shown in C); network is generated by the RINMAKER server. Specific interactions of L95 are summarized in Supplementary Table S1. (E) Predicted coordinates of GIRK2–GβL95Pγ complex, with data generated by the MutationExplorer server utilizing fast Rosetta fixbb function; P95 is shown as spheres and colored magenta. (F) Interface of GIRK–GβL95Pγ complex, with data generated by the PRODIGY server; selection criterion: all amino acids ≤5.5 Å. The left plot corresponds to the chain colored red in E, and the right plot corresponds to the chain colored orange in (E). Note that P95 is in contact with the chain marked red. (G) Amino acids in contact with P95, same criterion as in F, with data obtained from analysis of GIRK–GβL95Pγ utilizing the INTAA server. Amino acids that belong to Gβ are colored blue, and amino acids that belong to GIRK2 are colored red. (H) Analysis of non-covalent interactions of amino acids that are expected to be in contact with P95 (data shown in G); network is generated by the RINMAKER server. Specific interactions of P95 are summarized in Supplementary Table S1.
Analysis of docked GIRK1/2–Gβγ structures
In order to investigate energetic and structural consequences of the L95P mutation for a heterotetrameric GIRK channel, we analyzed a predicted model of GIRK1/2–Gβγ complex that was generated utilizing homology modeling and docking methods. We utilized the model of GIRK1/2 published by Gazgalis et al. (2022) and generated a rigid body docking model of GIRK1/2–Gβγ (see Materials and Methods). We implemented selection criteria similar to those described by Mahajan et al. (2013) in order to assure acceptable location of Gβγ relative to the plasma membrane. This procedure rendered 19 model clusters (see Materials and Methods), of which two models, which correspond to centroids of the largest cluster and the cluster with the best energy score, were further analyzed (Figures 6, 7).
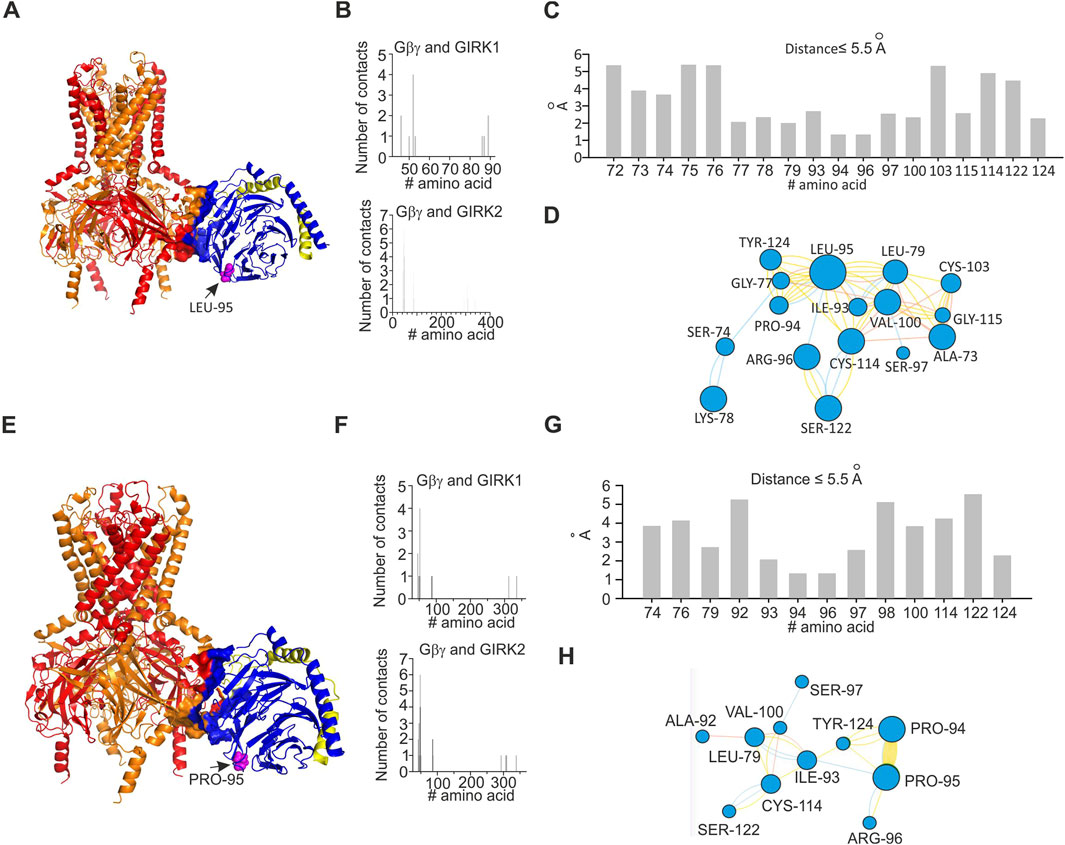
Figure 6. Structural analysis of the impact of L95P mutation on GIRK1/2–Gβγ complex. Analysis was conducted utilizing coordinates of the best scoring GIRK1/2–Gβγ complex. (A) Predicted structure of wild-type GIRK1/2–Gβγ complex (single Gβγ molecule is docked); L95 is shown as spheres and colored magenta, GIRK1 subunits are colored red, and GIRK2 subunits are colored orange. (B) Interface of GIRK–Gβγ complex, with data generated by the PRODIGY server; selection criterion: all amino acids are at ≤5.5 Å distance. The upper plot corresponds to GIRK1–Gβγ interface. The lower plot corresponds to GIRK2–Gβγ interface. Note that L95 is neither in GIRK1 nor in GIRK2 interface. (C) Amino acids in contact with L95, same criterion as in (B), with data obtained from analysis of GIRK1/2–Gβγ wild-type utilizing the INTAA server. As L95 is not part of the channel–Gβγ interface, only amino acids that belong to Gβγ are shown. (D) Analysis of non-covalent interactions of amino acids expected to be in contact with L95 (data shown in C); network is generated by the RINMAKER server. Specific interactions of L95 are summarized in Supplementary Table S1. (E) Predicted structure of GIRK1/2–GβL95Pγ complex, with data generated by the MutationExplorer server utilizing fast Rosetta fixbb function; P95 is shown as spheres and colored magenta. (F) Interface of GIRK–GβL95Pγ complex, with data generated by the PRODIGY server; selection criterion: all amino acids are at ≤5.5 Å distance. The upper plot corresponds to GIRK1–GβL95Pγ interface, and the lower plot corresponds to GIRK2–GβL95Pγ interface. Note that P95 is neither in GIRK1 nor in GIRK2 interface. (G) Amino acids in contact with P95, same criterion as in (F), with data obtained from analysis of GIRK–GβL95Pγ utilizing the INTAA server. Only amino acids that belong to Gβγ are shown. (H) Analysis of non-covalent interactions of amino acids that are expected to be in contact with P95 (data shown in (G)); network is generated by the RINMAKER server. Specific interactions of P95 are summarized in Supplementary Table S1.
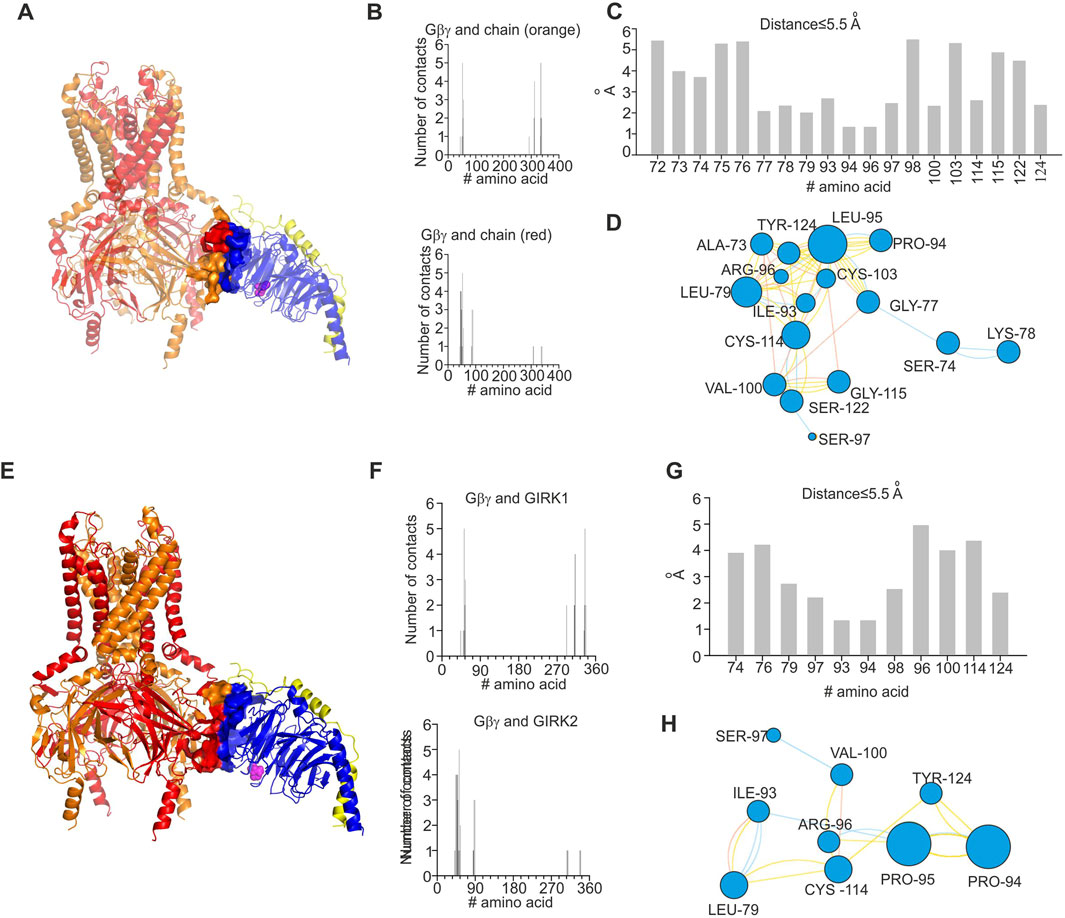
Figure 7. Structural analysis of impact of L95P mutation on GIRK1/2–Gβγ complex. Analysis was conducted utilizing coordinates of the largest cluster of GIRK1/2–Gβγ complex. (A) Structure of wild-type GIRK1/2–Gβγ complex (single Gβγ molecule is docked); L95 is shown as spheres and colored magenta, GIRK1 subunits are colored red, and GIRK2 subunits are colored orange. (B) Interface of GIRK–Gβγ complex, with data generated by the PRODIGY server; selection criterion: all amino acids ≤5.5 Å. The upper plot corresponds to GIRK1–Gβγ interface. The lower plot corresponds to GIRK2–Gβγ interface. Note that L95 is neither in GIRK1 nor in GIRK2 interface. (C) Amino acids in contact with L95, same criterion as in B, with data obtained from analysis of GIRK1/2–Gβγ wild-type utilizing the INTAA server. Only amino acids that belong to Gβγ are shown. (D) Analysis of non-covalent interactions of amino acids that are expected to be in contact with L95 (data shown in (C)); network is generated by the RINMAKER server. Specific interactions of L95 are summarized in Supplementary Table S1. (E) Predicted coordinates of GIRK1/2–GβL95Pγ complex, with data generated by the MutationExplorer server utilizing fast Rosetta fixbb function; P95 is shown as spheres and colored magenta. (F) Interface of GIRK–GβL95Pγ complex, with data generated by the PRODIGY server; selection criterion: all amino acids ≤5.5 Å. The upper plot corresponds GIRK1–GβL95Pγ interface, and the lower plot corresponds to GIRK2–GβL95Pγ interface. Note that P95 is neither in GIRK1 nor in GIRK2 interface. (G) Amino acids in contact with P95, same criterion as in F, with data obtained from analysis of GIRK–GβL95Pγ utilizing the INTAA server. Only amino acids that belong to Gβγ are shown. (H) Analysis of non-covalent interactions of amino acids that are expected to be in contact with P95 (data shown in (G)); network is generated by the RINMAKER server. Specific interactions of P95 are summarized in Supplementary Table S1.
Analysis of the two selected GIRK1/2–Gβγ models, based on the centroid of the largest cluster (lc) and the best energy score (bs), revealed that in both cases, L95 is not part of the GIRK–Gβγ interface (Figures 6, 7). This was consistent across both models, suggesting that the interaction between GIRK1/2 and Gβγ differs from that observed in the GIRK2–Gβγ complex. Additionally, the impact of L95P mutation on amino acids network interacting with that in position 95 in both models is quite similar to that observed for the 4kfm crystal structure. From Supplementary Table S1, it can be seen that at least seven amino acids interact with the wild-type leucine in position 95, whereas 2–4 amino acids remain interacting with proline in the same position in the mutant Gβ. In addition, from the energetic point of view, L95P is expected to be highly destabilizing for both GIRK1/2–Gβγ predicted models, whereas the effect of this mutation on Gβγ–GIRK1/2 interaction affinity is near the thermal noise threshold (Figure 8C).
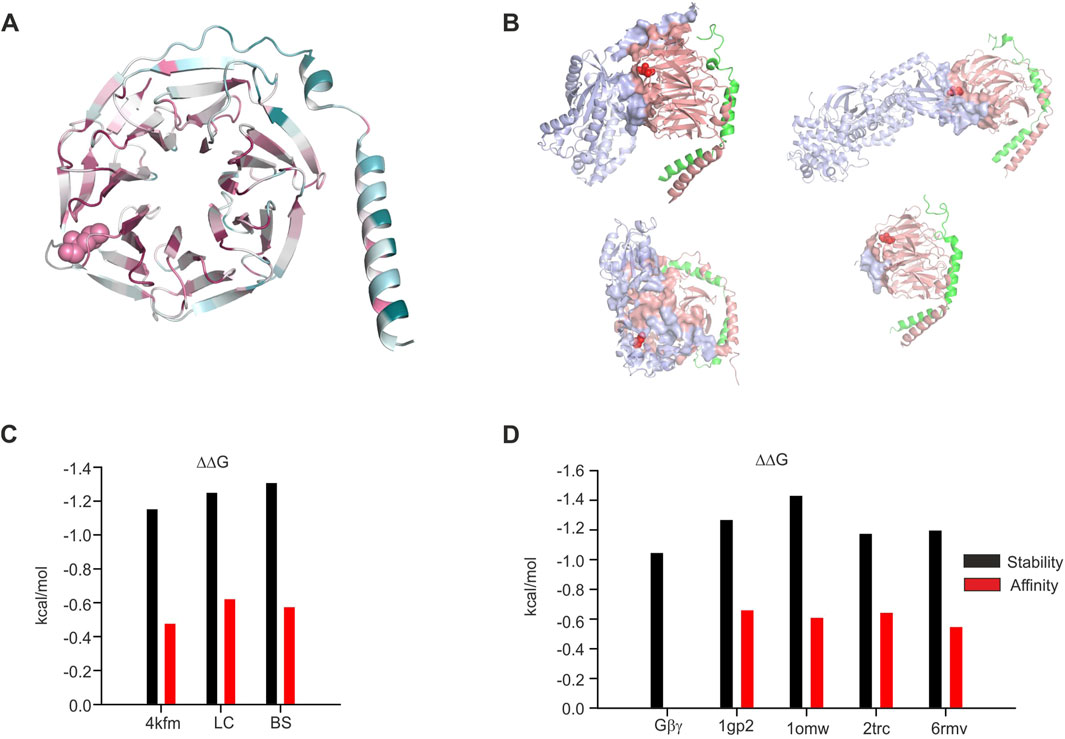
Figure 8. Structural analysis of the impact of L95P mutation on Gβγ−effector complexes. (A) Colored scale representation of Gβ1 generated in the ConSurf server (Ashkenazy et al., 2016). The scale is 1–9 with more evolutionary conserved amino acids colored by a more darker color. L95 is shown as spheres. (B) Gβγ−effector interface analysis. All interfaces are generated by the PRODIGY server. L95 is shown as red spheres; upper left—Gα1GDPGβγ (1gp2), upper right—β-adrenergic kinase–Gβγ complex (1omw), lower left—phosducin–Gβγ complex (2trc), and lower right—TRPM3–Gβγ (6rmv). (C) Effect of L95P mutation on protein stability and Gβγ affinity, GIRK crystal structure, and models. (D) Effect of L95P mutation on protein stability and Gβγ affinity and other effectors. All data in (C,D) were calculated utilizing the MCSM server.
Analysis of L95P mutation in other structures
In order to further understand the possible role of L95P mutation in Gβ1, we conducted structural and energetic analysis of this mutation in isolated Gβ1 and also in other crystal structures that contain this or homologous proteins. In particular, we analyzed the crystal structures of β-adrenergic kinase2 (β-ARK)-Gβγ (1omw) (Thal et al., 2011; Tesmer et al., 2010; Lodowski et al., 2003), Gαi1-Gβγ (1gp2) (Wall et al., 1995), and TRPM3- Gβγ (6rmv) (Behrendt et al., 2020) complexes and also of the phosducin-Gβγ complex (2trc) (Gaudet et al., 1996).
Initially, we analyzed the evolutional conservation of amino acids in Gβ1 utilizing the ConSurf server (Figure 8A), yielding L95 to be a highly conserved residue (color scale 8 with normalized conservation score of – 0.897). Thereafter, we studied the role of L95 in Gβγ interactions with other effectors mentioned above. We utilized the PRODIGY webserver for the detection of amino acids present in the protein–protein interface. The protein–protein interfaces for these structures are shown in Figure 8B. Out of four examined structures, L95 was found in the protein–protein interface only in one (β-ARK-Gβγ, 1omw).
Consequently, we estimated the possible impact of L95P mutation on protein stability and protein–protein affinity (Figure 8D). It can be seen that in all structures, the predicted energetic cost of the mutation is higher than the RT value at 25 Co (0.6 kcal/mol), and for the β-ARK-Gβγ complex, it is higher than 1.4 kcal/mol. On the other hand, the predicted impact of L95P mutation on affinity change is rather low, ∼0.6 kcal/mol (very close to the RT value, which is considered to be close to thermal noise effect).
Discussion
In order to study the effect of L95P mutation, we utilized heterologous expression (X. laevis oocytes), which enables strict control on both protein level expression and comparison of various possible ligand–effector pairs (Dascal and Lotan, 1992). We demonstrated that the L95P mutation exerts a dual effect by impairing both the expression and function of Gβ1 as a GIRK channel activating molecule. Our heterologous expression experiments revealed a clear reduction in expression levels of the Gβ1 protein, requiring significantly higher amounts of L95P mRNA than wild-type Gβγ RNA to achieve similar expression in the plasma membrane (Figures 1B, 3B). In addition to the loss of expression, we have shown that L95P mutation also impairs GIRK channel function. Such a combined effect of mutation both on protein expression and protein function has been described for other Gβ1 mutations by Reddy et al. (2021). Electrophysiological experiments with GIRK2 and GIRK1/2 channels showed that even when L95P expression levels matched those of wild-type Gβγ, the mutant protein failed to produce equivalent current levels.
The loss of expression effect is also supported by structural analysis as we demonstrated a destabilizing impact of L95P mutation observed in both the 4kfm crystal structure of GIRK2–Gβγ complex and in the predicted models of GIRK1/2–Gβγ. A similar effect was also shown for other Gβγ–effector complexes. This phenomenon, that is, protein misfolding that results from point mutation and subsequent rapid degradation, is described in many known pathologies such as phenylketonuria, cystic fibrosis, short chain acyl-CoA dehydrogenase deficiency disease (Gregersen et al., 2000), and others.
The loss of function effect of L95P mutation is more difficult to explain. In general, ion channel activity (formulated as open probability, for example) is assumed to be a function of maximal open probability and some kind of either hyperbolic or, in more complicated cases, sigmoid function which consists of an efficacy parameter and the concentration of activating ligand (Hille, 2001). In the case of GIRK channels, such a relationship includes channel affinity to Gβγ, maximal open probability, and allosteric coefficients relevant to channel activity changes due to binding of several Gβγ subunits (Touhara et al., 2016; Wang et al., 2016). According to the analysis of various Gβγ effectors, including GIRK2 (from existing crystal structure) and GIRK1/2 (from predicted models), there is no substantial change in Gβγ affinity to the channel induced by L95P mutation.
The aforementioned findings may have several interpretations, and among them is an impact of L95P on the maximal open probability of GIRK1/2 and GIRK2 in a situation similar to partial agonist scheme (Kenakin, 2009). However, this scenario does not agree well with all experimental findings. In a situation of similar affinity of L95P mutant to GIRK channel and lower maximal open probability, the expected effect of L95P expression would be a decrease of channel activity below the level observed before mutant co-expression. It must be taken into consideration that at least GIRK1/2 basal activity is highly (∼90%) Gβγ-depedent, as it was shown by Rishal et al. (2005). Correspondingly, the expression of L95P mutant with equal affinity to wild-type Gβγ and a reduced maximal open probability would lead to the appearance of two channel populations: one corresponding to the maximal open probability of wild-type Gβγ and the other to that of a mutant. Altogether, the above assumption of decreased maximal open probability and no change in Gβγ affinity to GIRK is expected to reduce the basal GIRK1/2 current, which was not observed experimentally.
At this point, we hypothesize that the loss of function effect of L95P mutation may also be associated with the protein misfolding phenomenon, which was described above as a reason for the loss of expression effect of L95P. Misfolded protein molecules frequently fail to interact with their receptors due to aberrant intracellular localization, aggregation, and removal by proofreading systems such as chaperons (Oikonomou and Hendershot, 2020). It must be emphasized that previously described mutations on Gβ (in particular I80T and I80N) were shown to also be loss of expression mutations (Reddy et al., 2021; Colombo et al., 2023), with possible similarity of L95P and I80T/N impact on protein stability.
In summary, the L95P mutation exerts a dual effect on the GIRK–Gβγ system, affecting both protein density and function. Despite L95 not being directly involved in GIRK–Gβγ structural interactions, the study highlights the importance of expression systems such as X. laevis oocytes in investigating disease-causing missense mutations. These systems allow precise control and measurement of protein expression and enable electrophysiological studies to assess the functional consequences of such mutations (Hatcher-Solis et al., 2014).
Detailed analysis of point mutations effect on the G-protein subunit–effector interaction conducted in this study and in other studies such as those for Gβγ (Reddy et al., 2021; Colombo et al., 2023) and those for Gα (Knight et al., 2023) can provide important information for further development of treatment strategies of complex medical conditions arising from those mutations (Shalomov et al., 2025).
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by the Tel Aviv University Institutional Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
HR: conceptualization, data curation, formal analysis, investigation, methodology, project administration, software, supervision, validation, visualization, writing – original draft, and writing – review and editing. TK-R: data curation, investigation, writing – original draft, and writing – review and editing. GT: data curation, investigation, writing – original draft, and writing – review and editing. ND: conceptualization, data curation, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, writing – original draft, and writing – review and editing. DY: conceptualization, data curation, formal analysis, investigation, methodology, software, supervision, validation, visualization, writing – original draft, and writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Israel Science Foundation (ISF), grant # 581/22 (ND).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2025.1592012/full#supplementary-material
References
Ashkenazy, H., Abadi, S., Martz, E., Chay, O., Mayrose, I., Pupko, T., et al. (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350. doi:10.1093/nar/gkw408
Bassan, H., Petrovski, S., Bilancia, C. G., Ramsey, K., Griffin, N. G., Bier, L., et al. (2018). Refining the phenotype associated with GNB1 mutations: Clinical data on 18 newly identified patients and review of the literature. Am. J. Med. Genet. A 176, 2259–2275.
Behrendt, M., Gruss, F., Enzeroth, R., Dembla, S., Zhao, S., Crassous, P. A., et al. (2020). The structural basis for an on-off switch controlling Gβγ-mediated inhibition of TRPM3 channels. Proc. Natl. Acad. Sci. U. S. A. 117, 29090–29100. doi:10.1073/pnas.2001177117
Berg, J. M., Tymoczko, J. L., and Stryer, L. (2012). Biochemistry/Jeremy M. Berg, John L. Tymoczko, Lubert Stryer; with Gregory J. Gatto, Jr. New York: W. H. Freeman.
Berlin, S., Tsemakhovich, V. A., Castel, R., Ivanina, T., Dessauer, C. W., Keren-Raifman, T., et al. (2011). Two distinct aspects of coupling between Gα(i) protein and G protein-activated K+ channel (GIRK) revealed by fluorescently labeled Gα(i3) protein subunits. J. Biol. Chem. 286, 33223–33235. doi:10.1074/jbc.M111.271056
Campos-Rios, A., Rueda-Ruzafa, L., and Lamas, J. A. (2022). The relevance of GIRK channels in heart function. Membranes 12, 1119. doi:10.3390/membranes12111119
Colombo, S., Reddy, H. P., Petri, S., Williams, D. J., Shalomov, B., Dhindsa, R. S., et al. (2023). Epilepsy in a mouse model of GNB1 encephalopathy arises from altered potassium (GIRK) channel signaling and is alleviated by a GIRK inhibitor. Front. Cell Neurosci. 17, 1175895. doi:10.3389/fncel.2023.1175895
Cui, M., Cantwell, L., Zorn, A., and Logothetis, D. E. (2021). Kir Channel molecular physiology, pharmacology, and therapeutic implications. pharmacology 267, 277–356. doi:10.1007/164_2021_501
Dascal, N., and Lotan, E. (1992). Expression of exogenous ion channels and neurotransmitter receptors in RNA-injected xenopus oocytes. Springer protocols. Series: Methods In Molecular Biology. Protocols in Molecular Neurobiology. NJ: Humana Totowa
Da Silva, J. D., Costa, M. D., Almeida, B., Lopes, F., Maciel, P., and Teixeira-Castro, A. (2021). Case report: a novel GNB1 mutation causes global developmental delay with intellectual disability and behavioral disorders. Front. Neurol. 12, 735549. doi:10.3389/fneur.2021.735549
Endo, W., Ikemoto, S., Togashi, N., Miyabayashi, T., Nakajima, E., Hamano, S. I., et al. (2020). Phenotype-genotype correlations in patients with GNB1 gene variants, including the first three reported Japanese patients to exhibit spastic diplegia, dyskinetic quadriplegia, and infantile spasms. Brain Dev. 42, 199–204. doi:10.1016/j.braindev.2019.10.006
Gaudet, R., Bohm, A., and Sigler, P. B. (1996). Crystal structure at 2.4 angstroms resolution of the complex of transducin betagamma and its regulator, phosducin. Phosducin Cell 87, 577–588. doi:10.1016/s0092-8674(00)81376-8
Gazgalis, D., Cantwell, L., Xu, Y., Thakur, G. A., Cui, M., Guarnieri, F., et al. (2022). Use of a molecular switch probe to activate or inhibit GIRK1 heteromers in silico reveals a novel gating mechanism. Int. J. Mol. Sci. 23, 10820. doi:10.3390/ijms231810820
Gregersen, N., Bross, P., Jorgensen, M. M., Corydon, T. J., and Andresen, B. S. (2000). Defective folding and rapid degradation of mutant proteins is a common disease mechanism in genetic disorders. J. Inherit. Metab. Dis. 23, 441–447. doi:10.1023/a:1005663728291
Hatcher-Solis, C., Fribourg, M., Spyridaki, K., Younkin, J., Ellaithy, A., Xiang, G., et al. (2014). G protein-coupled receptor signaling to Kir channels in Xenopus oocytes. Curr. Pharm. Biotechnol. 15, 987–995. doi:10.2174/1389201015666141031111916
Hemati, P., Revah-Politi, A., Bassan, H., Petrovski, S., Bilancia, C. G., Ramsey, K., et al. (2018a). Refining the phenotype associated with GNB1 mutations: clinical data on 18 newly identified patients and review of the literature. Am. J. Med. Genet. A 176, 2259–2275. doi:10.1002/ajmg.a.40472
Hemati, P., Revah-Politi, A., Bassan, H., Petrovski, S., Bilancia, C. G., Ramsey, K., et al. (2018b). Refining the phenotype associated with GNB1 mutations: clinical data on 18 newly identified patients and review of the literature. Am. J. Med. Genet. A 176, 2259–2275. doi:10.1002/ajmg.a.40472
Hibino, H., Inanobe, A., Furutani, K., Murakami, S., Findlay, I., and Kurachi, Y. (2010). Inwardly rectifying potassium channels: their structure, function, and physiological roles. Physiol. Rev. 90, 291–366. doi:10.1152/physrev.00021.2009
Jones, H. F., Morales-Briceño, H., Barwick, K., Lewis, J., Sanchis-Juan, A., Raymond, F. L., et al. (2019). Myoclonus-dystonia caused by GNB1 mutation responsive to deep brain stimulation American. J. Med. Genet. Part A 34, 1079–1080. doi:10.1002/ajmg.a.40472
Kahanovitch, U., Tsemakhovich, V., Berlin, S., Rubinstein, M., Styr, B., Castel, R., et al. (2014). Recruitment of Gβγ controls the basal activity of G-protein coupled inwardly rectifying potassium (GIRK) channels: crucial role of distal C terminus of GIRK1. J. Physiol. 592, 5373–5390. doi:10.1113/jphysiol.2014.283218
Kenakin, T. P. (2009). A pharmacology primer: theory, applications, and methods. Cambridge, MA: Academic Press/Elsevier.
Kessel, A., and Ben-Tal, N. (2018). Introduction to proteins: structure, function, and motion. Boca Raton, FL: Chapman and Hall/CRC.
Knight, K. M., Obarow, E. G., Wei, W., Mani, S., Esteller, M. I., Cui, M., et al. (2023). Molecular annotation of G protein variants in a neurological disorder, Cell Rep. 42, 113462 doi:10.1016/j.celrep.2023.113462
Kozakov, D., Hall, D. R., Xia, B., Porter, K. A., Padhorny, D., Yueh, C., et al. (2017). The ClusPro web server for protein-protein docking. Nat. Protoc. 12, 255–278. doi:10.1038/nprot.2016.169
Lansdon, L. A., and Saunders, C. J. (2021). Genotype-phenotype correlation in GNB1-related neurodevelopmental disorder: potential association of p.Leu95Pro with cleft palate. Am. J. Med. Genet. A 185, 1341–1343. doi:10.1002/ajmg.a.62080
Lodowski, D. T., Pitcher, J. A., Capel, W. D., Lefkowitz, R. J., and Tesmer, J. J. (2003). Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gbetagamma. Science 300, 1256–1262. doi:10.1126/science.1082348
Logothetis, D. E., Kurachi, Y., Galper, J., Neer, E. J., and Clapham, D. E. (1987). The beta gamma subunits of GTP-binding proteins activate the muscarinic K+ channel in heart. Nature 325, 321–326. doi:10.1038/325321a0
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I., and Lomize, A. L. (2012). OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic acids Res. 40, D370–D376. doi:10.1093/nar/gkr703
Luo, H., Marron Fernandez de Velasco, E., and Wickman, K. (2022). Neuronal G protein-gated K(+) channels. Am. J. Physiol. Cell Physiol. 323, C439–C460. doi:10.1152/ajpcell.00102.2022
Mahajan, R., Ha, J., Zhang, M., Kawano, T., Kozasa, T., and Logothetis, D. E. (2013). A computational model predicts that Gβγ acts at a cleft between channel subunits to activate GIRK1 channels. Sci. Signal 6, ra69. doi:10.1126/scisignal.2004075
Oikonomou, C., and Hendershot, L. M. (2020). Disposing of misfolded ER proteins: a troubled substrate's way out of the ER. Mol. Cell Endocrinol. 500, 110630. doi:10.1016/j.mce.2019.110630
Oldham, W. M., and Hamm, H. E. (2008). Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 9, 60–71. doi:10.1038/nrm2299
Petrovski, S., Küry, S., Myers, C. T., Anyane-Yeboa, K., Cogné, B., Bialer, M., et al. (2016). Germline de novo mutations in GNB1 cause severe neurodevelopmental disability, hypotonia, and seizures. Am. J. Hum. Genet. 98, 1001–1010. doi:10.1016/j.ajhg.2016.03.011
Pierce, K. L., Premont, R. T., and Lefkowitz, R. J. (2002). Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 3, 639–650. doi:10.1038/nrm908
Pires, D. E., Ascher, D. B., and Blundell, T. L. (2014). mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 30, 335–342. doi:10.1093/bioinformatics/btt691
Reddy, H. P., Yakubovich, D., Keren-Raifman, T., Tabak, G., Tsemakhovich, V. A., Pedersen, M. H., et al. (2021). Encephalopathy-causing mutations in Gβ1 (GNB1) alter regulation of neuronal GIRK channels. iScience 24, 103018. doi:10.1016/j.isci.2021.103018
Revah-Politi, A., Sands, T. T., Colombo, S., Goldstein, D. B., and Anyane-Yeboa, K. (2020). GNB1 Encephalopathy. GeneReviews®. Editors M. P. Adam, J. Feldman, G. M. Mirzaa, R. A. Pagon, S. E. Wallace, and A. Amemiya Seattle, WA: University of Washington, Seattle. 1993–2025.
Rishal, I., Keren-Raifman, T., Yakubovich, D., Ivanina, T., Dessauer, C. W., Slepak, V. Z., et al. (2003). Na+ promotes the dissociation between Galpha GDP and Gbeta gamma, activating G protein-gated K+ channels. J. Biol. Chem. 278, 3840–3845. doi:10.1074/jbc.C200605200
Rishal, I., Porozov, Y., Yakubovich, D., Varon, D., and Dascal, N. (2005). Gbetagamma-dependent and Gbetagamma-independent basal activity of G protein-activated K+ channels. J. Biol. Chem. 280, 16685–16694. doi:10.1074/jbc.M412196200
Rubinstein, M., Peleg, S., Berlin, S., Brass, D., Keren-Raifman, T., Dessauer, C. W., et al. (2009). Divergent regulation of GIRK1 and GIRK2 subunits of the neuronal G protein gated K+ channel by GalphaiGDP and Gbetagamma. J. Physiol. 587, 3473–3491. doi:10.1113/jphysiol.2009.173229
Schultz-Rogers, L., Masuho, I., Pinto, E. V. F., Schmitz, C. T., Schwab, T. L., Clark, K. J., et al. (2020). Haploinsufficiency as a disease mechanism in GNB1-associated neurodevelopmental disorder 8, e1477 doi:10.1002/mgg3.1477
Shalomov, B., Friesacher, T., Yakubovich, D., Combista, J. C., Reddy, H. P., Dabbah, S., et al. (2025). Ethosuximide: subunit- and Gβγ-dependent blocker and reporter of allosteric changes in GIRK channels. Br. J. Pharmacol. 182, 1704–1718.
Singer-Lahat, D., Dascal, N., Mittelman, L., Peleg, S., and Lotan, I. (2000). Imaging plasma membrane proteins in large membrane patches of Xenopus oocytes. Pflugers Arch. 440, 627–633. doi:10.1007/s004240000341
Tabak, G., Keren-Raifman, T., Kahanovitch, U., and Dascal, N. (2019). Mutual action by Gγ and Gβ for optimal activation of GIRK channels in a channel subunit-specific manner. Sci. Rep. 9, 508. doi:10.1038/s41598-018-36833-y
Tesmer, J. J., Tesmer, V. M., Lodowski, D. T., Steinhagen, H., and Huber, J. (2010). Structure of human G protein-coupled receptor kinase 2 in complex with the kinase inhibitor balanol. J. Med. Chem. 53, 1867–1870. doi:10.1021/jm9017515
Thal, D. M., Yeow, R. Y., Schoenau, C., Huber, J., and Tesmer, J. J. G. (2011). Molecular mechanism of selectivity among G protein-coupled receptor kinase 2 inhibitors. Mol. Pharmacol. 80, 294–303. doi:10.1124/mol.111.071522
Touhara, K. K., Wang, W., and MacKinnon, R. (2016). The GIRK1 subunit potentiates G protein activation of cardiac GIRK1/4 hetero-tetramers. Elife 5, e15750. doi:10.7554/eLife.15750
Tsuji, M., Ikeda, A., Tsuyusaki, Y., Iai, M., Kurosawa, K., Kosaki, K., et al. (2023). Atypical clinical course in two patients with GNB1 variants who developed acute encephalopathy. Brain Dev. 45, 462–466. doi:10.1016/j.braindev.2023.06.005
Wall, M. A., Coleman, D. E., Lee, E., Iñiguez-Lluhi, J. A., Posner, B. A., Gilman, A. G., et al. (1995). The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell 83, 1047–1058. doi:10.1016/0092-8674(95)90220-1
Wang, W., Touhara, K. K., Weir, K., Bean, B. P., and MacKinnon, R. (2016). Cooperative regulation by G proteins and Na(+) of neuronal GIRK2 K(+) channels. Elife 5, e15751. doi:10.7554/eLife.15751
Whorton, M. R., and MacKinnon, R. (2013). X-ray structure of the mammalian GIRK2-βγ G-protein complex. Nature 498, 190–197. doi:10.1038/nature12241
Xue, L. C., Rodrigues, J. P., Kastritis, P. L., Bonvin, A. M., and Vangone, A. (2016). PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 32, 3676–3678. doi:10.1093/bioinformatics/btw514
Yakubovich, D., Berlin, S., Kahanovitch, U., Rubinstein, M., Farhy-Tselnicker, I., Styr, B., et al. (2015). A quantitative model of the GIRK1/2 channel reveals that its basal and evoked activities are controlled by unequal stoichiometry of Gα and Gβγ. PLoS Comput. Biol. 11, e1004598. doi:10.1371/journal.pcbi.1004598
Keywords: GIRK channels, GNB1, mutation, modeling, docking
Citation: Reddy HP, Keren-Raifman T, Tabak G, Dascal N and Yakubovich D (2025) Loss of expression and function of Gβγ by GNB1 encephalopathy-associated L95P mutation of the Gβ1 subunit. Front. Pharmacol. 16:1592012. doi: 10.3389/fphar.2025.1592012
Received: 11 March 2025; Accepted: 11 April 2025;
Published: 09 May 2025.
Edited by:
Petra Scholze, Medical University of Vienna, AustriaReviewed by:
Jacy Wagnon, The Ohio State University, United StatesArmando Alberola-Die, University of Alicante, Spain
Copyright © 2025 Reddy, Keren-Raifman, Tabak, Dascal and Yakubovich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel Yakubovich, ZGFuaWVseWFAYXJpZWwuYWMuaWw=; Haritha P. Reddy, aGFyaXRoYTg5cmVkZHlAZ21haWwuY29t
†Present address: Haritha P. Reddy, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark
 Haritha P. Reddy
Haritha P. Reddy Tal Keren-Raifman1
Tal Keren-Raifman1 Nathan Dascal
Nathan Dascal Daniel Yakubovich
Daniel Yakubovich