- 1Department of Orthopedics, Zibo Central Hospital West Campus, Zibo, China
- 2Department of Hand and Foot Surgery, Zibo Central Hospital, Zibo, China
A Corrigendum on
A telomere-related signature for predicting prognosis and assessing immune microenvironment in osteosarcoma
by Li S, Zhang L and Zhang H (2025). Front. Pharmacol. 15:1532610. doi: 10.3389/fphar.2024.1532610
In the published article, there was an error in Figure 9 as published. The micrograph scale was omitted in Figure 9E, meaning that both the scale bar and duplication needed correction. As such, we magnified the ruler to make the Transwell analysis images clearer. The corrected Figure 9 and its caption appear below.
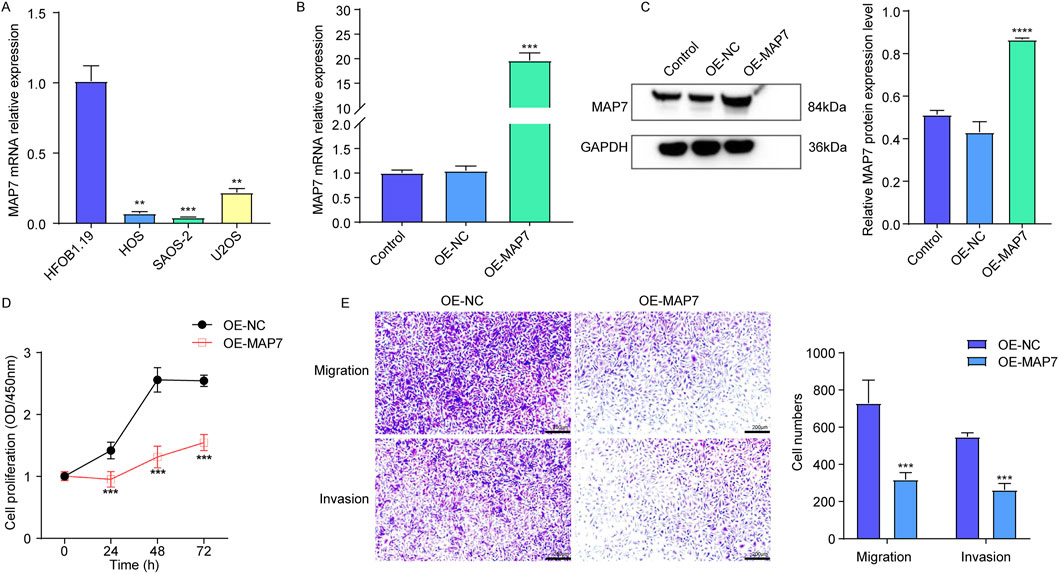
Figure 9. Overexpression of MAP7 might inhibit tumor proliferation, migration, and the invasion of osteosarcoma cells. (A) Expression of MAP7 in hFOB1.19, HOS, U-2OS, and SAOS-2 cells. Overexpression efficiency of MAP7 was evaluated by qRT-PCR (B) and Western blotting (C). (D) Cell proliferation was detected using CCK-8 assays. (E) The ability of cell migration and invasion was detected using the Transwell assay. * means P < 0.05, **P < 0.01, *** means P < 0.001, and **** means P < 0.0001.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: osteosarcoma, telomere-related signature, prognosis, immune microenvironment, drug sensitivity, MAP7
Citation: Li S, Zhang L and Zhang H (2025) Corrigendum: A telomere-related signature for predicting prognosis and assessing immune microenvironment in osteosarcoma. Front. Pharmacol. 16:1600217. doi: 10.3389/fphar.2025.1600217
Received: 26 March 2025; Accepted: 29 May 2025;
Published: 12 June 2025.
Edited and reviewed by:
John M. Seubert, University of Alberta, CanadaCopyright © 2025 Li, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiyang Zhang, emhhbmdvY2VhbjY2NkBzaW5hLmNvbQ==
 Shihao Li1
Shihao Li1 Lina Zhang
Lina Zhang Haiyang Zhang
Haiyang Zhang